Abstract
Chromosome dosage plays a significant role in reproductive isolation and speciation in both plants and animals, but underlying mechanisms are largely obscure1. Transposable elements can promote hybridity through maternal small RNA2, and have been postulated to regulate dosage response via neighboring imprinted genes3,4. Here, we show that a highly conserved microRNA in plants, miR845, targets the tRNAMet primer-binding site (PBS) of LTR-retrotransposons in Arabidopsis pollen, and triggers the accumulation of 21 to 22-nucleotide small RNA in a dose dependent fashion via RNA polymerase IV. We show that these epigenetically activated small-interfering RNAs (easiRNAs) mediate hybridization barriers between diploid seed parents and tetraploid pollen parents (“the triploid block”), and that natural variation for miR845 may account for “endosperm balance” allowing formation of triploid seeds. Targeting the PBS with small RNA is a common mechanism for transposon control in mammals and plants, and provides a uniquely sensitive means to monitor chromosome dosage and imprinting in the developing seed.
Epigenetic silencing of transposable elements (TEs) in flowering plants is regulated by cytosine methylation (mC), guided in part by RNA-directed DNA methylation (RdDM) in three sequence contexts (mCG, mCHG and mCHH, where H is A, C or T)5. RdDM undergoes reprogramming in the male germline6 but is restored through double fertilization of the egg cell and the central cell (from the embryo sac), by two sperm cells (from pollen grains), producing a diploid embryo and a triploid endosperm, respectively7. In pollen8–10 and endosperm11,12 companion cells, namely the haploid vegetative nucleus (VN) and diploid central cell (CC), de-methylation of TEs flanking imprinted genes promotes fertility and seed viability10,13,. Reprogramming is accompanied by mobilization of easiRNA from TE transcripts into neighboring germ cells8–10,14, but the biological significance of these small RNAs has remained unclear. easiRNA are triggered by microRNAs (miRNAs), most notably miR845a (21-nt) and miR845b (22-nt) that target multiple transposons expressed in Arabidopsis pollen9,15 (Fig. 1a). miR845a and miR845b target most Gypsy and Copia retrotransposons at the 18nt primer-binding site (PBS) (Fig. 1b and Supplementary Table 1), where tRNAs initiate reverse transcription. MIR845 is highly conserved in plants16, but is not present in many Brassicaceae, including the perennial Arabis alpina where these Gypsy transposons have contributed to a massive genome expansion17. MIR845 orthologs in drought-stressed rice leaves18 and diploid strawberry19, may have been derived from tRNAiMet 19, but MIR845 in Arabidopsis appears to have been derived by truncation and inversion of a 5′ LTR plus PBS (Fig. 1b). Intriguingly, in mammalian cells, abundant 18 to 22-nt 3′CCA tRNA fragments also match endogenous retroviruses at the PBS, and strongly suppress retrotransposition, suggesting an ancient mechanism for transposon control20.
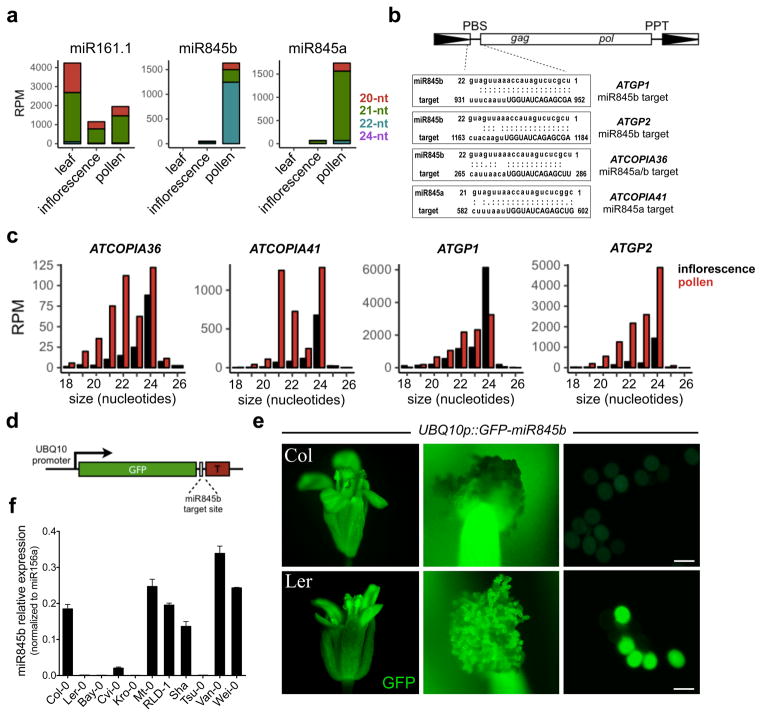
Figure 1. miR845 family is expressed in pollen and targets retrotransposons.
(a) miR845a and miR845b are preferentially expressed in mature pollen, consistent with complete absence in leaves and low levels in inflorescence tissue. (b) miR845 targets the PBS (uppercase) of retrotransposons where tRNAs bind to initiate reverse transcription. Predicted targets include Gypsy and Copia elements. The full list of targets is presented in Supplementary Table 1. (c) miR845 targets accumulate high levels of secondary 21- and 22-nt easiRNA in pollen. RPM, reads per million. (d) GFP sensor construct includes 3′UTR with a miR845b target site and driven by the UBIQUITIN10 (UBQ10) promoter. (e) Strong GFP fluorescence was detected in floral organs of 8 independent transgenic lines, but not in wild type Col-0 pollen. The same reporter is not silenced in Ler-0 pollen, where miR845b is not expressed. Scale bars represent 30 μm. (f) The MIR845 haplotype in Ler-0 is also found in other Arabidopsis accessions such as Bay-0, Kro-0 and Tsu-0 that produce low levels of miR845b, while Col-like accessions express high levels of miR845b in pollen. Bars represent mean Ct values of miR845b levels as measured by quantitative RT-PCR (qRT-PCR) and normalized to the levels of miR156a (n=2 technical replicates). Error bars represent range.
Gypsy and Copia retrotransposons generate easiRNA in pollen (Fig. 1c), as does a ubiquitously expressed GFP sensor incorporating a 3′UTR miR845b target site (Fig. 1d). Strikingly, this sensor is silenced in pollen (Fig. 1e) and sperm cells (Supplementary Fig. 1b) of the Columbia accession (Col-0), but not in Landsberg (Ler-0). In Ler-0, MIR845a has been deleted completely, while MIR845b has a single nucleotide polymorphism (SNP) in the complementary miRNA* sequence that potentially impairs miRNA processing (Supplementary Fig. 2a–f). The Ler-0 MIR845 haplotype is conserved in Kro-0, Bay-0 and Tsu-0, and levels of miR845b are also depleted in these accessions (Fig. 1f). To address the potential effect of miR845 on TE silencing, we compared Col-0 and Ler-0 pollen transcriptomes and found that TE transcripts are overall more abundant in Ler-0 pollen (Supplementary Fig. 3a), including miR845 targets such as ATGP2 and ATCOPIA36 that have escaped silencing in this ecotype (Supplementary Fig. 3c). Further, miR845-targeted TEs such as ATGP2 and ATCOPIA41 produce easiRNA in Col-0 pollen, while other TE families such as ATCOPIA63 are expressed and produce easiRNA specifically in Ler-0 (Supplementary Fig. 3b). Other differences between Col-0 and Ler-0 pollen may reflect TE copy number variation, as well as indirect effects on easiRNA production from TEs without miR845 target sites (such as AGO1 stability, or integration of DNA TEs near an existing LTR)21.
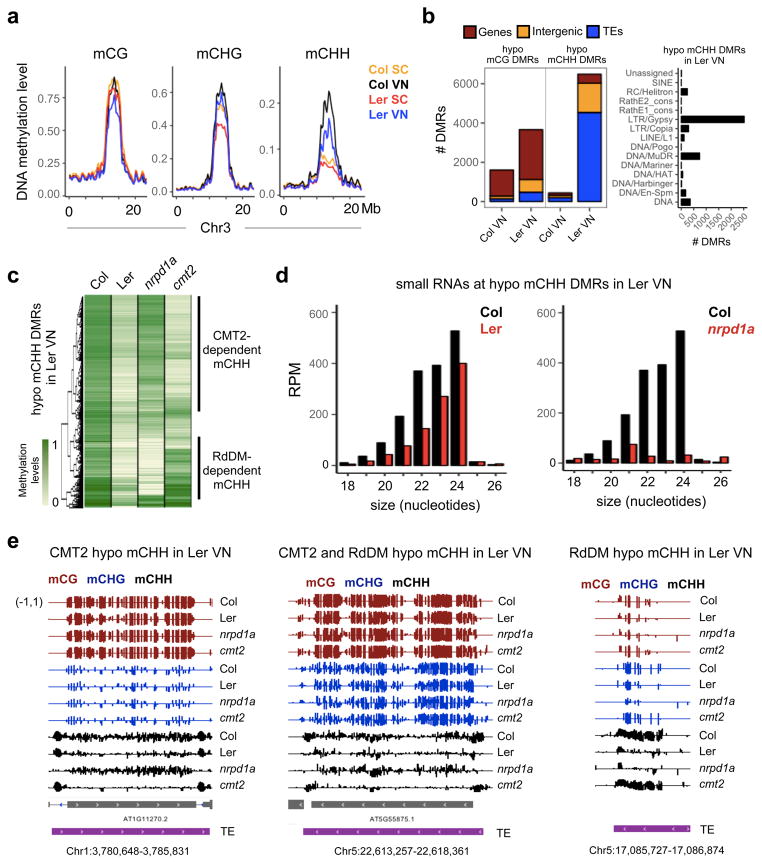
easiRNAs have been associated with RdDM activity15,22, and bisulfite sequencing of FACS-sorted Ler-0 pollen nuclei revealed lower levels of mCHH than in Col-0 VN (Fig. 2a), resembling seed methylomes in this respect23. This difference is particularly striking in pericentromeric heterochromatin, where the chromomethylases CMT2 and CMT3 maintain mCHH and mCHG, respectively (Fig. 2a). In contrast, the levels of mCHH in Col-0 and Ler-0 SC nuclei were found to be identical and very low (Fig. 2a)8,10,24. In the VN, we found approximately 6000 differentially methylated regions (DMRs) for mCHH between Col-0 and Ler-0, which overlapped primarily with TEs (including predicted miR845 targets) (Fig. 2b and Supplementary Table 2). The majority of VN DMRs were hypermethylated in Col-0 (Fig. 2b), but not in the RdDM mutant nrpd1a (largest subunit of RNA Polymerase IV) or cmt2, indicating targeting by both CMT2 and Pol IV-dependent siRNAs (Fig. 2c–e).
Figure 2. Natural variation in DNA methylation levels in Col-0 and Ler-0 pollen nuclei.
(a) Bisulfite sequencing of FACS-sorted Col-0 and Ler-0 pollen nuclei revealed decreased mCHH levels in Ler-0 VN, compared to Col-0 VN. (b) Differentially methylated regions (DMRs) between Col-0 and Ler-0 VN were detected for CG and CHH methylation. mCHH hypomethylated DMR in Ler-0 VN overlapped primarily with TE features, particularly the superfamilies LTR/Gypsy and DNA/MuDR. (c) Hypomethylated mCHH DMRs in Ler-0 VN overlapped with hypomethylated loci in nrpd1a (Pol IV) and cmt2 mutant VN (both in Col-0 background). (d) Small RNA in Col-0 pollen matching hypomethylated mCHH DMRs in Ler-0 VN are depleted in wild type Ler-0 pollen, and dependent on Pol IV (nrpd1a) (21/22 and 24-nt). (e) Genome browser tracks of CG, CHG and CHH methylation levels in the VN of Col-0, Ler-0, nrpd1a and cmt2 pollen, illustrating CMT2 and RdDM-targeted TEs where CHH methylation was lost in Ler-0 VN.
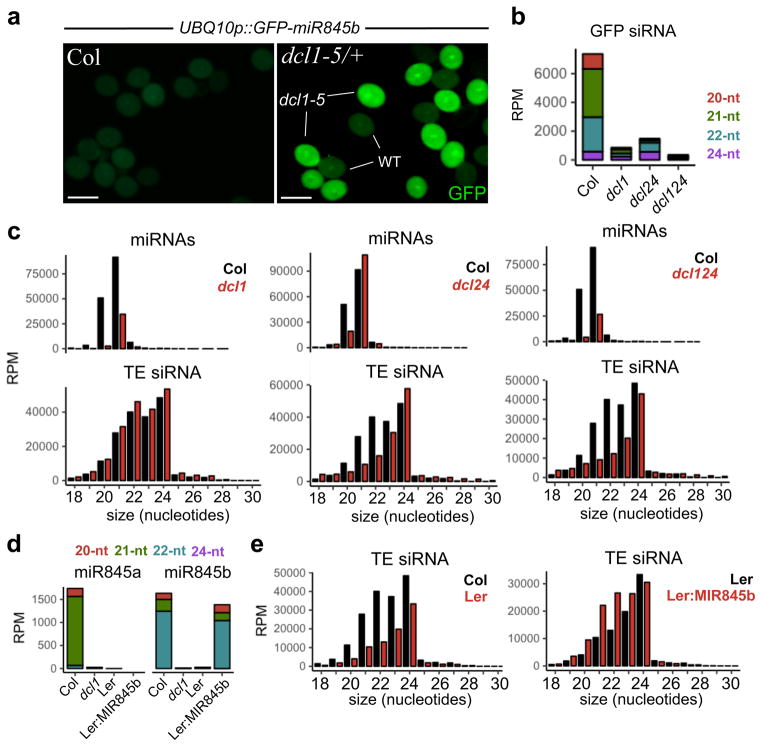
In Arabidopsis, most 20 to 22-nt miRNAs are processed by DICER-LIKE1 (DCL1) and loaded into ARGONAUTE1 (AGO1), while secondary siRNAs require a “double hit” with two 21-nt miRNAs or a single hit with a 22-nt miRNA to trigger production from target transcripts, via the RNA-dependent RNA polymerase RDR6, DCL2 and DCL425. As expected, in plants heterozygous for the null dcl1-5 mutant allele or the strong hypomorphic ago1-9 allele, GFP sensor expression was restored in mutant pollen (Fig. 3a and Supplementary Fig. 1c). We therefore used dcl1-5/+ and dcl1/+,dcl2/dcl4 mutant backgrounds to purify wild-type, dcl1, dcl2/4 and dcl1/2/4 pollen by fluorescence-activated cell sorting (FACS) (Supplementary Fig. 1d). Small RNA sequencing confirmed that miRNAs were depleted in dcl1 mutant pollen, including miR845a and miR845b (Fig. 3c,d and Supplementary Fig. 1e), while secondary siRNAs derived from the GFP transcript were depleted in dcl1, dcl24 and dcl124 pollen (Fig. 3b). However, loss of easiRNAs was only observed in dcl2/4 pollen, but not in dcl1 mutants (Fig. 3c), including 21/22-nt small RNAs produced from 5′ LTRs upstream of the PBS (miR845b target site) (Supplementary Fig. 4a), thus suggesting that miR845b triggers the production of GFP siRNAs and easiRNAs through different pathways. In somatic tissues, small RNAs matching LTRs are 24-nt in length and produced by Pol IV, RDR2, and DCL325, raising the possibility that 21/22-nt easiRNA in pollen are dependent on Pol IV. Indeed, small RNA from nrpd1a mutant pollen lost siRNAs for the majority of TEs in all size classes (Supplementary Fig. 4b), indicating that easiRNA biogenesis in pollen at miR845 targets depends on Pol IV, DCL2 and DCL4. Similar pathways have been described under certain types of genotoxic stress26,27, and in wild-type Arabidopsis siliques, where 21-nt “disiRNA” depend on a nuclear isoform of DCL4 that uses a hypomethylated promoter28, also found in both Col-0 and Ler-0 VN. disiRNA depend on RDR2 for biosynthesis28, resembling some miRNA-dependent secondary siRNAs29, though a role for miRNA in disiRNA biogenesis has not been proposed28.
Figure 3. miR845b-dependent easiRNA biogenesis from transgenes and transposons.
(a) GFP sensor including 3′UTR with a miR845b target site and driven by the UBIQUITIN10 (UBQ10) promoter in wild type and dcl1-5/+ heterozygous background. GFP expression was restored in dcl1 pollen, allowing FACS-purification of wild type, dcl1, dcl2/4 and dcl1/2/4 pollen grains. Scale bars represent 30 μm. (b) Loss of GFP siRNA was detected in dcl1, dcl2/4 and dcl1/2/4 pollen grains, indicating that miR845b triggers DCL2/4-dependent secondary siRNA from the GFP transgene. (c) Small RNA sequencing from wild type and mutant FACS-sorted pollen revealed that 21- and 22-nt TE siRNA were lost in the dcl2/4 mutants, while miRNAs were depleted in dcl1. (d) miR845a and miR845b were depleted in dcl1 mutant and Ler-0 pollen, but miR845b was restored in transgenic Ler-0 plants expressing Col-MIR845b (Ler:MIR845b). (e) 21- and 22-nt TE-derived siRNA levels were also depleted in wild-type Ler-0 pollen, but restored in transgenic Ler:MIR845b. RPM, reads per million.
Thus, retention of easiRNA in mature dcl1 pollen (Fig. 3c) where both miR845a and miR845b are down-regulated (Supplementary Fig. 1e), suggests that miR845 triggers Pol IV-easiRNA biogenesis either during meiosis or early at the onset of gametogenesis, and not in the VN9,15. In order to confirm that miR845 is able to trigger Pol IV-easiRNA biogenesis, we transformed Ler-0 wild-type plants with the Col-MIR845b locus and observed specific up-regulation of 21- and 22-nt TE-derived siRNAs, resembling Col-0 pollen (Fig. 3e). Interestingly, restored easiRNA biogenesis in Ler:MIR845b pollen did not result in significant changes in CHH methylation (Supplementary Fig. 5b), supporting the idea that easiRNAs accumulate in the sperm cells9,14 where RdDM and CMT2 pathways are not active8,24.
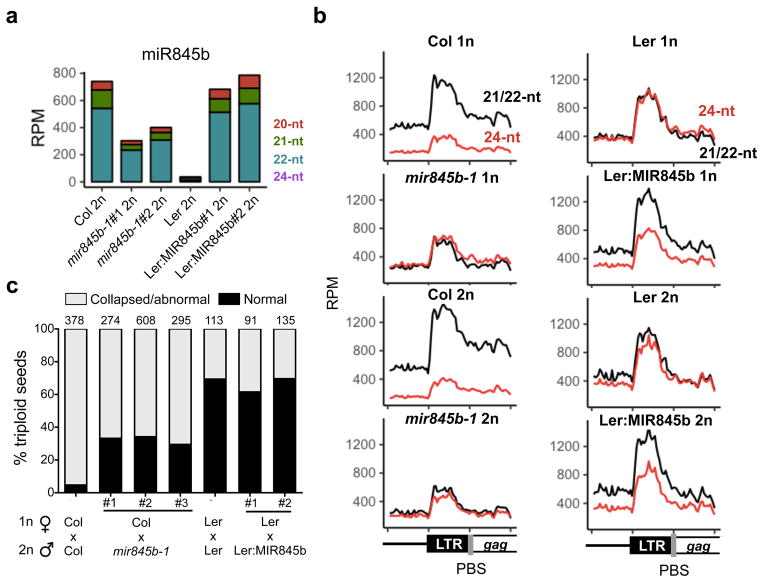
Parental small RNA differences can build strong barriers to hybridization2, and we have previously speculated that they might play a role in interspecific and interploid hybridization barriers3. Spontaneous chromosome doubling (polyploidization) is common in plants and is a major pathway towards reproductive isolation and speciation4,30. This is because hybrid seeds collapse as a result of unbalanced expression of imprinted genes in the endosperm, a phenomenon known as the “triploid block”. The “endosperm balance number” hypothesis further postulates the existence of multiple loci responsible for seed collapse in different strains4. The triploid block can be conveniently demonstrated in Arabidopsis using the omission of second division (osd1) mutant that forms unreduced diploid male and female gametes that are self-fertile31,32. When diploid osd1 pollen are crossed to wild-type seed parents, triploid seeds with tetraploid endosperm abort at high frequency depending on genetic background31,32, mimicking seed from tetraploid pollen parents33,34. In order to test whether miR845b-directed easiRNA biogenesis is involved in the triploid block response, we used a line carrying a T-DNA insertion at the MIR845b locus (mir845b-1) in Col-0 in which miR845b was down-regulated by roughly one half (Fig. 4a), and 21/22-nt easiRNAs were much lower than in wild-type Col-0 (Fig. 4b), resembling dcl2/4 mutants and wild type Ler-0 in this respect. Thus miR845b-directed easiRNA biogenesis is dose-sensitive, which is consistent with a role in endosperm balance. Pollinations of wild-type Col-0 seed parents with osd1-1 homozygous pollen gives rise to approximately 5% of viable seeds, while pollinations with osd1-1/mir845b-1 pollen had significantly increased seed viability at 35% (Fig. 4c). Interestingly, previous studies have shown that the triploid block response is much weaker in Ler-033,34, where miR845-dependent easiRNAs are lost. Therefore, we crossed pollen from osd1-2 (Ler-0 background) and osd1-2 expressing Col-miR845b (osd1-2:MIR845b) to Ler-0 wild-type female parents. However, easiRNA up-regulation in Ler-0 diploid pollen (Fig. 4a,b) was not sufficient to restore the triploid block, since the levels of viable triploid seeds remained comparable (Fig. 4c). We conclude that miR845b-dependent easiRNAs are required but not sufficient for the triploid block, suggesting additional regulatory mechanisms balance parental gene dosage in Ler-0 endosperm, as previously reported32,34.
Figure 4. miR845b-dependent easiRNA is required for the triploid block.
(a) miR845b was down-regulated 2-fold in two biologically independent replicates of diploid pollen (2n) from a T-DNA insertion mutant at the MIR845b locus (mir845b-1, Col-0 background), and abolished entirely in Ler-0. Expression of Col-MIR845b in Ler-0 2n pollen (osd1-2 mutant background, Ler:MIR845b) restored miR845b levels in the pollen of two biologically independent transgenic lines. (b) TE-derived siRNAs were mapped to aligned 5′ regions (metaplots) of LTR Gypsy elements. 21/22-nt easiRNA were depleted in mir845b-1 pollen and wild-type Ler-0 pollen compared to Col-0, but restored in Ler:MIR845b. These effects were even more pronounced in osd1-2 diploid pollen (2n). (c) Wild type Col-0 seed parents were pollinated with osd1-1 diploid (2n) pollen leading to the production of triploid seeds that abort at high frequency. When osd1-1/mir845b-1 double mutant (2n) pollen was used, seed viability increased to 35%. When osd1-2 Ler-0 (2n) pollen was used, the triploid block was also partially suppressed with 30% abnormal or collapsed seeds. However, expression of Col-MIR845b in osd1-2 Ler-0 2n pollen (Ler:MIR845b) was not sufficient to restore the triploid block. The numbers on top of each bar shows the number of seeds counted for each cross.
Thus paternally expressed miR845b stimulates dose-dependent biogenesis of 21/22-nt secondary siRNAs via Pol IV transcription, to mediate the triploid block response. Interestingly, the ratio of 21/22-nt vs 24-nt siRNA is much higher in interploid hybrid seeds with paternal excess33–35 (Supplementary Fig. 6c), suggesting that 21/22-nt easiRNAs might stabilize unbalanced genomic imprinting in interploid seeds. In previous studies, reduced levels of maternal 24-nt siRNAs resulted in up-regulation of maternally expressed genes (MEGs) but did not suppress triploid seed abortion35, while loss-of-function mutations in several paternally expressed genes (PEGs) are strong suppressors of the triploid block response32,36. PEGs and TEs are also expressed in crosses between Arabidopsis species37,38, implicating them in seed abortion upon both interspecific and interploidy hybridizations. One possibility is that paternal easiRNAs are involved in silencing MEGs, which include important components of the Polycomb Repressive Complex (PRC2) such as MEDEA (MEA) and FERTILIZATION INDEPENDENT SEED 2 (FIS2). PEGs are silenced by PRC2-mediated H3K27me3 in the endosperm39, and so would be upregulated in triploid seeds (Supplementary Fig. 7). This idea is supported by the fact that MEGs accumulate 21/22-nt easiRNAs in interploid seeds with paternal excess, thus suggesting they are paternal in origin35 (Supplementary Fig. 6a–c), while maternal expression of MEA and FIS2 is specifically repressed in Col, but not Ler triploid seed40. Another possibility is that easiRNA promotes the expression of PEGs, by antagonizing RdDM activity at imprinted loci. In an accompanying paper, Martinez et al. show that paternal Pol IV mutations suppress the triploid block41, which is consistent with these mechanisms15.
Dose-dependent miRNA loci might contribute to “endosperm balance number”, which has been classically implicated in the triploid block and measures maternal as well as paternal chromosome dose4. In maize, maternal doubling induced before or even after fertilization (by doubling the fertilized central cell) causes seed failure, suggesting that the relative dosage of cytoplasmic components in the gametophyte to genomic targets in the endosperm is also important42,43. In Arabidopsis, maternal excess causes reduced triploid seed size, and accumulation of Pol IV-dependent small RNA35, including disiRNA28. It will be interesting to determine whether small RNAs inherited from the maternal gametophyte also mediate dosage response3. Thus plant germline easiRNAs promote hybrid failure in interploids, targeting the same sequences as tRNA fragments that regulate endogenous retroviruses in early mammalian development20. Similarly, Drosophila germline piRNA promote hybrid viability2, while protist scnRNA mediate interactions between micro- and macronuclear genomes in the germline44, suggesting transposon recognition by small RNA is an ancient mechanism for chromosomal interaction.
Online Methods
Plant material and growth conditions
Plants were grown under long day conditions at 22 °C. Seeds were always surface sterilized with sodium hypochlorite, sowed on Murashige and Skoog (MS) medium and stratified for 3 days at 4°C. Seedlings were transplanted to soil two weeks after germination and grown under long day conditions at 22 °C. We used the following Arabidopsis mutant alleles: dcl1-5 (CS16069, Col-0 background), dcl2-1 and dcl4-2 (CS16391, Col-0), ago1-9 45 (originally in Ler, but introgressed in Col for 6 generations of backcrossing), mir845b-1 (SAIL_172_A08, Col-0), osd1-2 (GT21481, Ler-0), nrpd1a-3 (Salk_128428, Col-0) and osd1-3 (Koncz collection, Col-0)31. The osd1-1 mutant was kindly provided by Raphael Mercier. This mutant allele was originally identified in the Nossen background, and introgressed into Col by repeated backcrosses over five generations. The mir845b-1 tetraploid plant was obtained by colchicine treatment as previously described46. Additionally to Col-0 and Ler-0, we used the following ecotypes: Tsu-0 (CS1564), Kro-0 (CS1301), Bay-0 (CS954), Cvi-0 (CS902), RLD-1 (CS28687), Wei-0 (CS76301), Van-0 (CS28796), Sha (CS28737), Mt-0 (CS28502).
Transgene cloning
The UBQ10 promoter and miR845b target site promoter were cloned into a GFP reporter vector obtained from the VIB Department of Plant Systems Biology, UGent, by using Gateway technology (Life Technologies). Ectopic expression of MIR845a and MIR845b in Col-0 was performed by gateway cloning both MIR genes into pB2GW7. Expression of Col-MIR845b in Ler-0 was performed by PCR amplification of 1kb fragment of genomic DNA flanking MIR845b in Col-0, which was cloned into pMDC123 by Gateway cloning system. Primers used in this study are listed in Supplementary Table 3.
Pollen collection and FACS
Pollen was collected in 1.5mL eppendorf tubes by vortexing open flowers in pollen extraction buffer (PEB, 10 mM CaCl2, 2 mM MES, 1 mM KCl, 1% H3BO3, 10%) for 3 min 47, followed by filtration through a 30um mesh (Partec/Sysmex) and centrifugation at 5,000g for 1 min. Pollen was suspended in 50ul of PEB, immediately frozen in liquid nitrogen and stored at -80°C until RNA extraction. Purification of pollen expressing the GFP-miR845b sensor in dcl1-5/+ and dcl1/+,dcl2/dcl4 mutants was performed by vortexing open flowers in 1 mL of PEB and filtering through a 30um mesh before FACS. Pollen population was identified in SSC/FSC scatter plots, and GFP fluorescence was analyzed by excitation with a 488nm laser and detected with a 530/30 bandpass filter (Supplementary Fig. 1d).
RNA purification, qPCR analysis and sequencing
Pollen total RNA was extracted using Trizol LS reagent (Life Technologies) by vortexing with glass beads for 5 minutes, and concentrated with Direct-zol columns (Zymo Research). miRNA-qPCR was performed using the Quantimir kit (System Biosciences) following manufacturer instructions. Libraries for RNA sequencing from Col-0 and Ler-0 pollen were prepared using rRNA-depleted total RNA samples and the ScriptSeq-v2 RNA-Seq Library Prep kit (Epicenter/Illumina), following manufacturer instructions. cDNA libraries were sequenced on a HiSeq2500 instrument. FastQ files were processed and mapped to TAIR10 genome using SubRead 48, normalized by calculating the number of paired-end reads per kilobase of transcript per million of mapped reads (FPKM), and analyzed using R scripts. Small RNAs were purified by running total RNA on acrylamide gels (15% polyacrylamide, 7M urea) and performing size selection of approximately 18 to 30-nt region using a small RNA ladder (Zymo Research). The small RNA fraction was isolated from acrylamide gel plugs by grinding with a plastic pestle in Trizol LS (Life Technologies), and concentrated using Direct-zol columns (Zymo Research). Libraries were prepared using the TruSeq Small RNA Sample Preparation Kit (Illumina) following manufacturer instructions, barcoded and sequenced in Illumina HiSeq 2500, NextSeq500 or MiSeq platforms depending on sample pooling strategies and desired sequencing depth. After de-multiplexing, 36-nt reads were pre-processed by trimming the 3′ adapter and filtering collapsed reads according to length and quality. Filtered reads were mapped to the Arabidopsis TAIR10 genome annotation (or GFP transgene) with bowtie reporting all multi-mappers. Only perfect match reads were used for down-stream analysis, and reads mapped to multiple genomic locations where normalized by dividing non-redundant read counts by the number of genomic hits, and subsequently calculating the number of reads per million of filtered (18–30nt) and perfectly mapped reads. Additional downstream analyses were performed using R scripts. A summary of all small RNA sequencing data generated in this study is presented in Supplementary Table 4. Inflorescence small RNA data was previously reported22. Total RNA and small RNA sequencing data discussed in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the accession numbers GSE106113, GSE106114 and GSE106115.
Bisulfite sequencing and DNA methylation analysis
Pollen nuclei were isolated as previously described47,49. Approximately 50.000 nuclei were purified by FACS and used to construct sequencing libraries of bisulfite-treated DNA using the Pico Methyl-Seq kit (Zymo Research), according to manufacturer instructions. Non-directional libraries were sequenced on HiSeq and NextSeq platforms. Single end 100 and/or 76 reads were preprocessed using Trimmomatic50 to remove adapters, trim the first 10 nucleotides and split the read in half. Preprocessed C/T- and G/A-converted reads were mapped to C/T- and G/A-converted TAIR10 genome allowing two mismatches. Perl and Python scripts were used to recover the sequence of each mapped read and calculate methylation at each individual cytosine covered by 3 or more reads. Differentially methylated regions (DMRs) were defined as 100bp bins containing at least 4, 6 or 8 differentially methylated mCGs, mCHGs or mCHHs, respectively, and absolute methylation difference of 0.35. DMRs localizing 200bp of each other were merged. A summary of all genome-wide bisulfite sequencing data generated in this study is presented in table S4, and is accessible through the NCBI’s Gene Expression Omnibus Series GSE106112. Bisulfite sequencing data of cmt2 mutant VN was previously reported24.
Supplementary Material
Acknowledgments
We thank all members of the Martienssen laboratory for discussions. Research in the Martienssen laboratory is supported by the US National Institutes of Health (NIH) grant R01 GM067014, The National Science Foundation Plant Genome Research Program, and by the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation. The authors acknowledge assistance from the Cold Spring Harbor Laboratory Shared Resources, which are funded in part by the Cancer Center Support Grant (5PP30CA045508). Research in the Köhler laboratory was supported by a European Research Council Starting Independent Researcher grant, a grant from the Swedish Science Foundation and a grant from the Knut and Alice Wallenberg Foundation (to C.K.). G.M. was supported by a Marie Curie IOF Postdoctoral Fellowship (PIOF-GA-2012-330069). Sequencing for the Köhler laboratory was performed by the SNP&SEQ Technology Platform, Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council (VRRFI) and the Knut and Alice Wallenberg Foundation.
Footnotes
Author contributions
F.B., C.K. and R.A.M designed the study. F.B., J.S.P., F.V.E., P.W. and G.M performed experiments, and F.B. analyzed the data. All authors contributed with ideas and discussion. F.B. and R.A.M prepared the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Data Availability Statement
All sequencing datasets presented in this study are accessible through the NCBI’s Gene Expression Omnibus Series GSE106117
References
- 1.Birchler JA, Veitia RA. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109:14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senti KA, Brennecke J. The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet. 2010;26:499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martienssen RA. Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytol. 2010;186:46–53. doi: 10.1111/j.1469-8137.2010.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köhler C, Mittelsten Scheid O, Erilova A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 2010;26:142–148. doi: 10.1016/j.tig.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heard E, Martienssen RA. Transgenerational Epigenetic Inheritance: Myths and Mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dresselhaus T, Sprunck S, Wessel GM. Fertilization Mechanisms in Flowering Plants. Curr Biol. 2016;26:R125–39. doi: 10.1016/j.cub.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehring M, Bubb KL, Henikoff S. Extensive Demethylation of Repetitive Elements During Seed Development Underlies Gene Imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh TF, et al. Genome-Wide Demethylation of Arabidopsis Endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoft VK, et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci USA. 2011;108:8042–8047. doi: 10.1073/pnas.1105117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez G, Panda K, Köhler C, Slotkin RK. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nature Plants. 2016;2:16030. doi: 10.1038/nplants.2014.23. Published online: 2 February 2015. [DOI] [PubMed] [Google Scholar]
- 15.Creasey KM, et al. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature. 2014;508:411–415. doi: 10.1038/nature13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathore P, Geeta R, Das S. Microsynteny and phylogenetic analysis of tandemly organised miRNA families across five members of Brassicaceae reveals complex retention and loss history. Plant Sci. 2016;247:35–48. doi: 10.1016/j.plantsci.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Willing EM, et al. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nature Plants. 2015;1:14023. doi: 10.1038/nplants.2014.23. Published online: 2 February 2015. [DOI] [PubMed] [Google Scholar]
- 18.Zhou L, et al. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot. 2010;61:4157–4168. doi: 10.1093/jxb/erq237. [DOI] [PubMed] [Google Scholar]
- 19.Šurbanovski N, Brilli M, Moser M, Si-Ammour A. A highly specific microRNA-mediated mechanism silences LTR retrotransposons of strawberry. Plant J. 2016;85:70–82. doi: 10.1111/tpj.13090. [DOI] [PubMed] [Google Scholar]
- 20.Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen RA. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell. 2017;170:61–71.e11. doi: 10.1016/j.cell.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quadrana L, et al. The Arabidopsis thaliana mobilome and its impact at the species level. Elife. 2016;5:e15716. doi: 10.7554/eLife.15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nuthikattu S, et al. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 2013;162:116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pignatta D, et al. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. Elife. 2014;3:e03198. doi: 10.7554/eLife.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh PH, et al. Arabidopsis male sexual lineage exhibits more robust maintenance of CG methylation than somatic tissues. Proc Natl Acad Sci USA. 2016;113:15132–15137. doi: 10.1073/pnas.1619074114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nature Rev Mol Cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei W, et al. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Schalk C, et al. Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc Natl Acad Sci USA. 2017;114:E2965–E2974. doi: 10.1073/pnas.1618834114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pumplin N, et al. DNA Methylation Influences the Expression of DICER-LIKE4 Isoforms, Which Encode Proteins of Alternative Localization and Function. Plant Cell. 2016;28:2786–2804. doi: 10.1105/tpc.16.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ronemus M, Vaughn MW, Martienssen RA. MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell. 2006;18:1559–1574. doi: 10.1105/tpc.106.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmickl R, Koch MA. Arabidopsis hybrid speciation processes. Proc Natl Acad Sci USA. 2011;108:14192–14197. doi: 10.1073/pnas.1104212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.d’Erfurth I, et al. Turning meiosis into mitosis. PloS Biol. 2009;7:e1000124. doi: 10.1371/journal.pbio.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kradolfer D, Wolff P, Jiang H, Siretskiy A, Köhler C. An Imprinted Gene Underlies Postzygotic Reproductive Isolation in Arabidopsis thaliana. Dev Cell. 2013;26:525–535. doi: 10.1016/j.devcel.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Scott RJ, Spielman M, Bailey J, Dickinson HG. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development. 1998;125:3329–3341. doi: 10.1242/dev.125.17.3329. [DOI] [PubMed] [Google Scholar]
- 34.Dilkes BP, et al. The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PloS Biol. 2008;6:2707–2720. doi: 10.1371/journal.pbio.0060308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J, Zhang C, Baulcombe DC, Chen ZJ. Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc Natl Acad Sci USA. 2012;109:5529–5534. doi: 10.1073/pnas.1203094109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolff P, Jiang H, Wang G, Santos-González J, Köhler C. Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. Elife. 2015;4 doi: 10.7554/eLife.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Josefsson C, Dilkes B, Comai L. Parent-Dependent Loss of Gene Silencing during Interspecies Hybridization. Curr Biol. 2006;16:1322–1328. doi: 10.1016/j.cub.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 38.Kirkbride RC, et al. An Epigenetic Role for Disrupted Paternal Gene Expression in Postzygotic Seed Abortion in Arabidopsis Interspecific Hybrids. Mol Plant. 2015;8:1766–1775. doi: 10.1016/j.molp.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Mozgová I, Köhler C, Hennig L. Keeping the gate closed: functions of the polycomb repressive complex PRC2 in development. Plant J. 2015;83:121–132. doi: 10.1111/tpj.12828. [DOI] [PubMed] [Google Scholar]
- 40.Jullien PE, Berger F. Parental genome dosage imbalance deregulates imprinting in Arabidopsis. PLoS Genet. 2010;6:e1000885. doi: 10.1371/journal.pgen.1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez G, et al. Paternal easiRNAs establish the triploid block in Arabidopsis. 2017 [Google Scholar]
- 42.Kato A, Birchler JA. Induction of tetraploid derivatives of maize inbred lines by nitrous oxide gas treatment. J Hered. 2006;97:39–44. doi: 10.1093/jhered/esj007. [DOI] [PubMed] [Google Scholar]
- 43.Birchler JA. Interploidy hybridization barrier of endosperm as a dosage interaction. Front Plant Sci. 2014;5:281. doi: 10.3389/fpls.2014.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duharcourt S, Lepère G, Meyer E. Developmental genome rearrangements in ciliates: a natural genomic subtraction mediated by non-coding transcripts. Trends Genet. 2009;25:344–350. doi: 10.1016/j.tig.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Kidner CA, Martienssen RA. The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Dev Biol. 2005;280:504–517. doi: 10.1016/j.ydbio.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 46.Schatlowski N, et al. Hypomethylated pollen bypasses the interploidy hybridization barrier in Arabidopsis. Plant Cell. 2014;26:3556–3568. doi: 10.1105/tpc.114.130120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borges F, et al. FACS-based purification of Arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods. 2012;8:44. doi: 10.1186/1746-4811-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoft VK, et al. SYBR Green-activated sorting of Arabidopsis pollen nuclei based on different DNA/RNA content. Plant Reprod. 2015;28:61–72. doi: 10.1007/s00497-015-0258-2. [DOI] [PubMed] [Google Scholar]
- 50.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.