Abstract
Salinity stress is one of the major abiotic stresses limiting crop production in arid and semi-arid regions. Interest is increasing in the application of PGPRs (plant growth promoting rhizobacteria) to ameliorate stresses such as salinity stress in crop production. The identification of salt-tolerant, or halophilic, PGPRs has the potential to promote saline soil-based agriculture. Halophytes are a useful reservoir of halotolerant bacteria with plant growth-promoting capabilities. Here, we review recent studies on the use of halophilic PGPRs to stimulate plant growth and increase the tolerance of non-halophytic crops to salinity. These studies illustrate that halophilic PGPRs from the rhizosphere of halophytic species can be effective bio-inoculants for promoting the production of non-halophytic species in saline soils. These studies support the viability of bioinoculation with halophilic PGPRs as a strategy for the sustainable enhancement of non-halophytic crop growth. The potential of this strategy is discussed within the context of ensuring sustainable food production for a world with an increasing population and continuing climate change. We also explore future research needs for using halotolerant PGPRs under salinity stress.
Keywords: salinity, salinity-sensitive crop, halophytes, salt-tolerant, halophilic PGPRs, saline soil-based agriculture
Introduction
Food security is a fundamental need of all societies. The global population is projected to increase to around 10 billion people within the next 50 years (Godfray et al., 2010). To meet the additional food demand, an estimated 50% increase in yields of the major food crops will be required (Godfray et al., 2010). Whereas, the world's population is increasing, agricultural soils are decreasing about 1–2% every year in global arid and semi-arid zones due to soil salinity (Kafi and Khan, 2008). The low rainfall and high temperature characteristic of these zones promote high salinity (Shrivastava and Kumar, 2015), and this salinity has become an important factor limiting the growth of salt-sensitive plants and even some halophytes (Hasegawa et al., 2000; Sobhanian et al., 2011). Salinity stress has resulted in up to a 70% decrease in yield of important crops like wheat, maize, rice, and barley (Acquaah, 2007). Moreover, salinity stress is predicted to increase further in many regions due to global climate change. The costs associated with this stress are potentially enormous, estimated at US$12 billion per annum globally, and rising (Qadir et al., 2008; Dodd and Pérez-Alfocea, 2012).
A decrease in the availability of fertile land and the consequent extensive reuse of irrigated lands have driven the rapid development of saline soil-based agriculture in recent years (Zhu et al., 2011). Whereas, plants that are salt-resistant can produce significant yields in saline soils, many agricultural crops, and trees exhibit a low tolerance to salt (Glenn et al., 1991). Future agricultural production in these salt-affected agricultural environments thus requires the development of salt-tolerant food and fiber crops (Rozema and Flowers, 2008; Joshi et al., 2015). Traditional breeding and genetic engineering approaches have had only limited successes in developing salinity-resistant plants, despite significant efforts (Munns and Tester, 2008; Schubert et al., 2009; Dodd and Pérez-Alfocea, 2012; Joshi et al., 2015; Krishna et al., 2015). These efforts are complicated by the fact that salinity affects several facets of plant physiology (Dodd and Pérez-Alfocea, 2012; Kumari et al., 2015).
An alternative strategy to crop improvement to enhance salt tolerance may be to introduce salt-tolerant microbes that augment crop growth (Dodd and Pérez-Alfocea, 2012). Soil salinity-tolerant microorganisms have been found to increase the growth of many crops grown in salt-affected soils, which suggests that this approach may succeed where developing salt-tolerant germplasm has not (Dodd and Pérez-Alfocea, 2012). Identifying and using salinity-tolerant microorganisms could not only enhance the salt tolerance of crops but also reduce pressure on arable lands. Among the microorganisms associated with plants, plant growth-promoting rhizobacteria (PGPRs) have been effective at improving plant stress tolerance (Etesami and Beattie, 2017; Etesami, 2018). Yang et al. (2009) coined the term “Induced Systemic Tolerance” to describe the tolerance to abiotic stresses that is elicited by PGPRs in plants. Previous reports have reviewed the effects of PGPRs in relieving abiotic stress in various crop plants (Dutta and Khurana, 2015; Etesami and Beattie, 2017). The ability of PGPRs to transform nutrients and increase plant tolerance to abiotic stress is influenced by environmental conditions, including the climate, weather, and soil characteristics (e.g., high salinity), and by interactions with other microbial flora in the soil (Giongo et al., 2008). For example, the performance of phosphorus-solubilizing microorganisms (PSMs) is strongly affected by environmental factors, especially stress factors (Yoon et al., 2001; Sánchez-Porro et al., 2009). Upadhyay et al. (2009) found that PGPRs lose plant growth-promoting (PGP) traits with increasing salinity in vitro. Thus, the use of halotolerant PGPRs that are selected based on both high salt tolerance and efficiency in expressing PGP traits could significantly advance our ability to grow crops in environments with natural or induced salinity (Zhu et al., 2011). Rhizobacteria isolated from saline habitats have been shown to be more efficient at enhancing plant tolerance to salt than PGPRs isolated from non-saline habitats (Paul and Nair, 2008; Egamberdieva and Kucharova, 2009; Khan et al., 2016). There is now clear evidence that PGPRs associated with plants growing in harsh environmental conditions help those plants tolerate abiotic stresses (Lucero et al., 2008, 2011; Rodriguez et al., 2008; Lau and Lennon, 2012; Marasco et al., 2012; Kaplan et al., 2013). Moreover, recent advances in plant–bacterial interactions indicate that plants can shape the microbiome in the rhizosphere and endosphere (i.e., the zone within the roots; Berendsen et al., 2012). Under stress conditions, plants can require the presence of associated bacteria to tolerate stress and therefore grow and become established in an ecosystem (Hardoim et al., 2008). Symbiotic bacteria exist in all plants, and this relationship may be a key factor involved in plant stress tolerance. In fact, local adaptation of plants to their environment is driven by the genetic differentiation among closely associated PGPRs (Rodriguez and Redman, 2008). Transplanting various plant species in the absence of bacteria is notoriously difficult (Leifert et al., 1989), and this difficulty supports the importance of bacteria to plant growth, including under stressful conditions.
Halophytes are extremely salt tolerant plants—they usually grow and survive in environments with salinity concentrations as high as 5 g l−1 (Joshi et al., 2015). Halophytes play an important role in protecting ecosystems due to their remediation abilities. Halophytic plants have evolved various strategies to live in saline environments. These strategies include the production of compatible solutes to increase the osmotic pressure in the cytoplasm, the accumulation of Na+ in the vacuole, and the exclusion of Na+ from cells (Flowers and Colmer, 2008). They also have evolved an ability to exploit the benefits provided by endophytes and rhizosphere microorganisms (Sgroy et al., 2009; Ruppel et al., 2013).
The rhizosphere of halophytic plants serves as a reservoir for various groups of salt-tolerant rhizobacteria that could enhance the growth of crops under salinity stress (Jha et al., 2012, 2015; Shukla et al., 2012; Bharti et al., 2013; Ramadoss et al., 2013; Goswami et al., 2014; Sharma et al., 2016; Yuan et al., 2016). Like halophytic plants, salt-tolerant rhizobacteria have evolved various strategies to live in high saline environments. An important strategy is the ability to accumulate compatible osmolytes to maintain intracellular osmotic balance (Nabti et al., 2015; Sharma et al., 2016). These bacteria exhibit multiple stress-related traits that may contribute to their plant protective capabilities under growth inhibiting levels of salt (Rohban et al., 2009; Siddikee et al., 2010; Bharti et al., 2013; Sharma et al., 2016). In this review, we present the attempts thus far to isolate halotolerant PGPRs that bestow salt tolerance to agricultural crops. We offer a view of the ability of PGPRs to increase plant tolerance to salt and facilitate plant growth, as well as their potential to be isolated from the rhizosphere of halophytes. Lastly, we highlight the future application of these PGPRs as bio-inoculants in saline soil-based agriculture. A key concept in this review is that the range of PGPRs with multiple PGP traits that exist in the rhizosphere of halophytic plants is a valuable resource for improving crop tolerance to salinity and promoting saline soil-based agriculture in the future.
Halophytes
Plants can grow at high levels of soil salinity although the extent of growth inhibition varies among plant species. Plants are classified into glycophytes (salt-sensitive plants) and halophytes (salt-loving plants) based on their tolerance to salinity. Halophytes are plants which naturally survive in salt-contaminated environments and can tolerate salinity concentrations as high as 1 M NaCl (Flowers and Colmer, 2008; Kumari et al., 2015). About 1% of the total flora of the world (both dicots and monocots) are halophytic plants. These are distributed primarily in arid, semi-arid inlands, and high salinity wetlands along the tropical and sub-tropical coasts (Kumari et al., 2015). Halophytes have salt-responsive genes and proteins to counteract the adverse effects of salinity, while glycophytes cannot tolerate high salinity (Askari et al., 2006; Yu et al., 2011). Depending on their resistance and demand for sodium salts (NaCl), halophyte plants can be known as obligate or facultative halophytes (Kumari et al., 2015). Facultative halophytes can grow under freshwater conditions, whereas obligate halophytes need some salt to survive and grow (Kumari et al., 2015). Hydro-halophytes and xero-halophytes are another division for halophytes. Hydro-halophytes can grow in aquatic conditions or on wet soil, and xero-halphytes can grow in habitats where the soil is always saline and dry (Kumari et al., 2015). Most herbal varieties in desert areas are xero-halophytes and many of them are succulent (Kumari et al., 2015). Because halophytes flourish in high salinity conditions, they are considered to be extremophiles (Kosová et al., 2013).
Halophytes employ several mechanisms to adjust to soil salinity (Shabala, 2013; Zhang and Shi, 2013; Flowers and Colmer, 2015; Joshi et al., 2015; Kumari et al., 2015). These mechanisms include complex molecular, biochemical, physiological, and morphological changes (Wang et al., 2001) such as (i) modulating plant hormones (Parida and Das, 2005; Gupta and Huang, 2014) like IAA, jasmonic acid (JA), gibberellin (GA), ethylene (ET), and abscisic acid (ABA), and inducing enzymes related to their biosynthesis; (ii) synthesizing compatible solutes and osmoprotectants (Sanchez et al., 2008; Flowers and Colmer, 2015; Slama et al., 2015); (iii) controlling ion absorption, especially potassium (K) ions, by roots and ion transfer to leaves. Owing to their role in maintaining an osmotic balance, K+ ions play an important role in closing and opening stomata and as co-factors for many enzymes; (iv) selective accumulation or removal of ions (Mahajan and Tuteja, 2005); (v) producing nitric oxide (NO) (Del Río, 2015); (vi) activating antioxidant enzymes and producing antioxidant compounds (Ozgur et al., 2013; Wang et al., 2013); (vii) producing polyamines (Takahashi and Kakehi, 2009); (viii) altering photosynthetic pathways (Stepien and Johnson, 2009; Uzilday et al., 2014); (ix) compartmentalizing ions at the cellular and whole-plant levels (Pang et al., 2010; Shabala and Mackay, 2011); and (x) regulating the expression of genes involved in plant salinity tolerance. In terms of gene regulation, halophytic plants respond to salt stress by up-regulating a large number of genes and transcription factors (Kawasaki et al., 2001; Lim et al., 2010; Gupta and Huang, 2014; Kumari et al., 2015), and these can be grouped into the following functional categories: (i) senescence-associated genes (e.g., SAG); (ii) ion transport or homeostasis genes (e.g., SOS genes, AtNHX1, and H+-ATPase); (iii) molecular chaperones (e.g., HSP genes); and (iv) dehydration-related transcription factors (e.g., DREB) (Gupta and Huang, 2014).
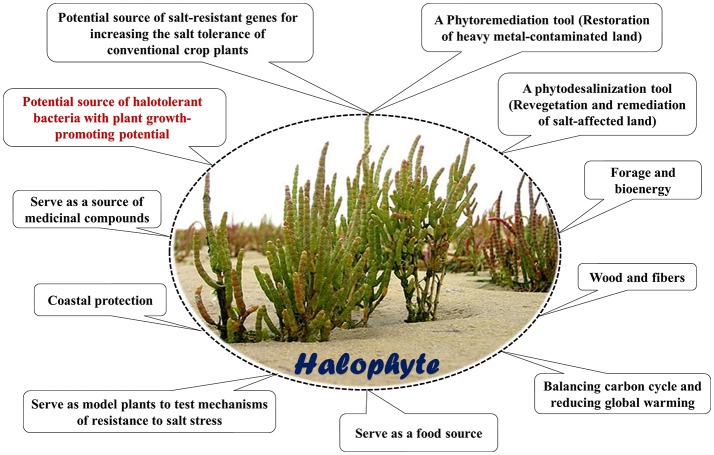
Interest in salinity tolerant and halophytic plants is because of a trend toward increasing salinity in agricultural soils in the arid and semi-arid regions of the world. The potential use of halophytes and other salt-tolerant species would allow the production of crops in these areas. Halophytes have many potential uses (Figure 1; Gago et al., 2011; Manousaki and Kalogerakis, 2011; Ksouri et al., 2012; Rozema and Schat, 2013; Hasanuzzaman et al., 2014; Song and Wang, 2014; Cheeseman, 2015; Jesus et al., 2015; Akinshina et al., 2016; Himabindu et al., 2016), including their use as a reservoir for isolating halotolerant PGPRs.
Figure 1.
Some potential use of halophytes.
Halotolerant PGPRs
Eukaryotic and prokaryotic micro-organisms, including fungi, bacteria, and archaea, are able to adapt to a range of changes in external osmolarity (Ruppel et al., 2013). Halotolerant bacteria are able to grow in environments with a wide range of salinities, from 1 to 33% NaCl, as well as in the absence of NaCl (Larsen, 1986; Khan et al., 2016). They are therefore well-suited to grow in the rhizosphere of halophytes where there are often low water potentials due to salt stress in dry climates (Upadhyay et al., 2009; Ruppel et al., 2013). Interestingly, PGPRs isolated from environmental extremes maintain their PGP traits even in the presence of high salt concentrations. For example, Zhu et al. (2011) isolated a high phosphorus-solubilizing halotolerant PGPR, Kushneria sp. YCWA18, from the sediment of Daqiao saltern on the eastern coast of China that was able to grow on a solid medium containing 20% (w/v) of sodium chloride. Tiwari et al. (2011) also isolated PGPRs that were halotolerant based on their ability to tolerate 2–25% NaCl; these included Bacillus pumilus, Pseudomonas mendocina, Arthrobacter sp., Halomonas sp., and Nitrinicola lacisaponensis with plant growth-promoting traits like phosphorus (P) solubilization and the ability to produce IAA, siderophores, and 1-aminocyclopropane-1-carboxylate (ACC) deaminase. These are considered PGP traits due to their ability to provide P to the plant under P-limiting conditions, promote plant growth by functioning as a phytohormone (IAA), provide Fe to the plant via chelation and uptake (siderophores), and deplete a precursor to the plant stress hormone ethylene (ACC deaminase). Distinct genera of halotolerant bacteria have been isolated from distinct halophytic plants such as Rosa rugosa (Bibi et al., 2011), Salicornia bigelovii (Rueda-Puente et al., 2010), Salicornia brachiate (Jha et al., 2012), Halocnemum strobilaceum (Al-Mailem et al., 2010), Acacia spp. (Boukhatem et al., 2012), Sesuvium portulacastrum (Bian et al., 2011; Anburaj et al., 2012), and Avicennia marina (El-Tarabily and Youssef, 2010), and from a wide range of habitats such as extreme alkali-saline soils, desert soils, and saline soils (Antón et al., 2002; Ventosa et al., 2008; Abou-Elela et al., 2010; Shi et al., 2012; Zhou et al., 2012; Ruppel et al., 2013). Many of these halotolerant bacteria exhibited an ability to promote plant growth (Table 1).
Table 1.
Potential application of PGPRs, with multiple plant growth promoting (PGP) traits, associated to halophytes to promote growth and enhance salinity tolerance of non-halophyte and halophyte plants.
| Halotolerant bacteria | Host halophyte | PGP activity | Inoculated plant | Plant response | References |
|---|---|---|---|---|---|
| Bacillus alcalophilus, B. thuringiensis, and Gracilibacillus saliphilus | Arthrocnemum macrostachyum | IAA production, siderophore production, and phosphate solubilization | A. macrostachyum | Mitigated the effects of high salinity on plant growth and physiological performance. | Navarro-Torre et al., 2017 |
| Micrococcus yunnanensis, Planococcus rifietoensis, and Variovorax paradoxus | Seven species of halophytes | N2 fixation, IAA production, siderophore production, phosphate solubilization, and ACC deaminase activity | Sugar beet (Beta vulgaris L.) | An increase in salt stress tolerance, seed germination (%), and plant biomass, and photosynthetic capacity, and a decrease in stress-induced ethylene production at different NaCl concentrations (50–125 mM). | Zhou et al., 2017 |
| Dietzia natronolimnaea STR1 | Not reported | – | Wheat (Triticum aestivum L.) | Increased wheat tolerance to salt stress by improved wheat growth in terms of plant dry weight and plant height (higher biomass, shoot, and root elongation), increased photosynthetic pigments, enhanced content of enzymes catalase and ascorbate peroxidase, and increased the gene expression of the antioxidants compared to un-inoculated plants. | Bharti et al., 2016 |
| Bacillus, Pantoea, Marinobacterium, Acinetobacter, Enterobacter, Pseudomonas, Rhizobium, and Sinorhizobium | Psoralea corylifolia L. | IAA production and siderophore production | Wheat (Triticum aestivum L.) | Enhanced seed germination and root length of wheat | Sorty et al., 2016 |
| Klebsiella, Pseudomonas, Agrobacterium, and Ochrobactrum | Arthrocnemum indicum | IAA production, N2 fixation, phosphate solubilization, ACC deaminase activity, and HCN production | Peanut | A significant increase in total N content (up to 76%), maintained ion homeostasis, accumulated less ROS, and enhanced plant growth compared to non-inoculated seedlings. | Sharma et al., 2016 |
| Serratia marcescens and B. cereus | Aster tripolium L. | IAA production, N2 fixation, siderophore production, and ACC deaminase activity | – | – | Szymańska, et al., 2016 |
| Pseudomonas sp. | Suaeda salsa | – | Cucumber and rice | Increase in plant growth and salt tolerance of plant. | Yuan et al., 2016 |
| Bacillus endophyticus, B. tequilensis, Planococcus rifietoensis, Variovorax paradoxus, and Arthrobacter agilis | Salicornia europaea | IAA production, phosphate solubilization, and ACC deaminase activity | S. europaea | Increase in germination percentage by 7–11%, in shoot length by 13–22%, in plant root length by 44–57%, and in fresh weight by 21–54%. | Zhao et al., 2016 |
| Arthrobacter pascens | Atriplex leucoclada | Phosphate solubilization and siderophore production | Maize | Increase in shoot and root length, in shoot and root fresh and dry weight, in osmolytes (e.g., sugar and proline), and in antioxidant enzymes activity (e.g., superoxide dismutase, peroxidase, catalase and ascorbate peroxidase) of maize plant. | Ullah and Bano, 2015 |
| Suaeda fruticosa | |||||
| Bacillus, Pseudomonas, Klebsiella, Serratia, Arthrobacter, Streptomyces, Isoptericola, and Microbacterium | Limonium sinense | N2 fixation, IAA production, phosphate solubilization, and ACC deaminase activity | L. sinense | Significant increase in plant root length, shoot length, leaf number, and leaf area as compared to the non-inoculated control. | Qin et al., 2014 |
| Chromohalobacter, Marinococcus, Halobacillus, Nesterenkonia, Halomonas, Oceanobacillus, and Virgibacillus | Salicornia strobilacea | IAA, N2 fixation, phosphate solubilization, and ACC deaminase activity | – | – | Mapelli et al., 2013 |
| Rhodococcus fascians | Salicornia sp. | N2 fixation | – | – | Jafari et al., 2012 |
| Brachybacterium saurashtrense sp., Zhihengliuella sp., Brevibacterium casei, Haererehalobacter sp., Halomonas sp., Vibrio sp., Cronobacter sakazakii, Pseudomonas spp., Rhizobium radiobacter, Mesorhizobium sp., and Bacillus sp. | Salicornia brachiata | N2 fixation, IAA production, phosphate solubilization, and ACC deaminase activity | S. brachiata | Increase in percent germination at 0–0.5 mol l−1 NaCl concentrations and significant increases in root length, shoot length, vigor index and the fresh weight of S. brachiate. | Jha et al., 2012 |
| Agrobacterium tumefaciens, Zhinguelliuella, Brachybacterium saurashtrense, Brevibacterium casei, Haererohalobacter, and Vibrio | Salicornia brachiata | IAA production, N2 fixation, phosphate solubilization, siderophore production, and ACC deaminase activity | Arachis hypogaea | Increase in plant length, shoot length, root length, shoot dry weight, root dry weight, and total biomass compared to un-inoculated plants, increase in the percentage of water content in the shoots and roots in inoculated plants compared to un-inoculated plants, and increase in amino acids, K+/Na+ ratio, and content of Ca2+, P, N, and IAA of the inoculated plants. | Shukla et al., 2012 |
| Brachybacterium saurashtrense sp. | Salicornia brachiata | N2 fixation, IAA production, siderophore production, and ACC deaminase activity | – | – | Gontia et al., 2011 |
| Rhizobium spp. and Bacillus spp. | Salicornia bigelovii | N2 fixation | – | – | Rueda-Puente et al., 2010 |
| Brevibacterium epidermidis, B. iodinum, Arthrobacter nicotianae, Zhihengliuella alba, Micrococcus yunnanensis, Oceanimonas smirnovii, Bacillus licheniformis, B. stratosphericus, B. aryabhattai, and Corynebacterium variabile | Not reported | N2 fixation, IAA production, and ACC deaminase activity | Canola | Increase in root length between 5.2 and 47.8%, and in root dry weight between 16.2 and 43%, in comparison with the un-inoculated canola plant. | Siddikee et al., 2010 |
| Pseudomonas oryzihabitans, Pseudomonas sp., Pantoea agglomerans and Pseudomonas putida | Suaeda salsa | IAA production, gibberellic acid production, abscisic acid production, phosphate solubilization, ACC deaminase activity, siderophore production, and antifungal activity | – | – | Teng et al., 2010 |
| Lysinibacillus fusiformis, B. subtilis, Brevibacterium halotolerans, B. licheniformis, B. pumilus, Achromobacter xylosoxidans, and P. putida | Prosopis strombulifera | N2 fixation, IAA production, siderophore production, ACC deaminase activity, gibberellin production, antifungal activity, and protease activity | – | – | Sgroy et al., 2009 |
| Pseudomonas pseudoalcaligenes | Salicornia europea | N2 fixation | S. europea | Increase in the chlorophyll content and N content of S. europea. | Ozawa et al., 2007 |
| Halomonas maura | Salicornia sp. | N2 fixation | – | – | Argandona et al., 2005 |
| Klebsiella pneumoniae | Salicornia bigelovii | N2 fixation | S. bigelovii | Increase in germination, early seedling growth, fresh and dry weights and the length of roots of S. bigelovii. | Rueda-Puente et al., 2003 |
| Azospirillum halopraeferens sp. Nov. | Kallar grass | N2 fixation | – | – | Reinhold et al., 1987 |
Halotolerant bacteria employ a range of strategies to grow and survive in saline habitats (Etesami and Beattie, 2017). These strategies include (i) minimizing the uptake of salt due to compositional properties of the cell membrane or cell wall; (ii) regulating intracellular ion concentrations by pumping ions out of the cell through electrogenic Na+/H+ antiporters and K+/Na+ ion transporters for osmotic adjustment; (iii) accumulating compatible solutes such as sucrose, trehalose, glycosyl glycerol, and glycine betaine by endogenous biosynthesis; (iv) producing proteins and enzymes that are adapted to high concentrations of solute ions; (v) increasing the energetic capacity; and (vi) producing exopolysaccharides (EPS) that help the development of hydrating biofilms (Sandhya et al., 2010; Ruppel et al., 2013; Qin et al., 2016). In addition to these strategies, fundamental cellular properties of halophytes may enhance their halotolerance, including their high GC content and a high proportion of proteins that exhibit a low hydrophobicity, a low tendency to form helices, and a high tendency to form stabilizing coil structures (Jacob, 2012; Szymańska, et al., 2016).
Several reports have shown that halotolerant PGPRs effectively improve growth of various agricultural crops under salinity stress conditions (Figure 2; Mayak et al., 2004a; Nabti et al., 2010; Shukla et al., 2012; Goswami et al., 2014; Ji et al., 2014; Kim et al., 2014; Kaushal and Wani, 2016; Orhan, 2016; Qin et al., 2016; Singh and Jha, 2016; Etesami, 2018). Mechanisms by which they improve growth have been predicted or shown to include (i) activating plant antioxidant defense machinery by upregulating the activity of key enzymes such as superoxide dismutase (SOD), peroxidase, and catalase (CAT) that scavenge excess reactive oxygen species (ROS), and protect the plants from salt toxicity (Jha and Subramanian, 2014; Islam et al., 2016; Qin et al., 2016); (ii) improving plant nutrition by fixing atmospheric nitrogen (N2), solubilizing P or K, producing siderophores for Fe uptake (Dodd and Pérez-Alfocea, 2012; Etesami and Beattie, 2017; Etesami, 2018); (iii) increasing the efficiency of inoculated plants to take up select ions for maintaining a high K+/Na+ ratio; this can directly reduce the accumulation of toxic ions such as Na+ and Cl− and improve the nutritional status of both macronutrients and micronutrients by regulating ion transporter expression and/or activity (Giri et al., 2007; Zuccarini and Okurowska, 2008; Shukla et al., 2012; Islam et al., 2016; Etesami, 2018); (iv) decreasing plant Na+ accumulation by excreting EPS to bind cations (especially Na+) in roots and prevent their translocation to leaves; this helps promote a physical barrier called a rhizosheath around the roots (Ashraf et al., 2004; Dodd and Pérez-Alfocea, 2012; Qin et al., 2016; Etesami and Beattie, 2017). EPS-producing-halotolerant PGPRs enhance the soil structure by promoting soil aggregation, which results in water retention and increased provision of nutrients to plants. EPS can also alleviate plant salt stress by binding Na+; this binding is due to the hydroxyl, sulfhydryl, carboxyl and phosphoryl functional groups characteristic of bacterial EPS (Watanabe et al., 2003; Nunkaew et al., 2015). Aeromonas hydrophila/caviae, Bacillus sp., Planococcus rifietoensis, Halomonas variabilis, Burkholderia, Enterobacter, Microbacterium, and Paenibacillus are some of the halotolerant PGPRs that produce EPS and facilitate biofilm formation (Upadhyay et al., 2011; Qurashi and Sabri, 2012; Ruppel et al., 2013; Khan et al., 2016); (v) synthesizing the enzyme ACC deaminase, which converts the plant ethylene precursor ACC to ammonia and α-ketobutyrate (Etesami and Beattie, 2017), thus reducing the accumulation of ethylene in the plant and avoiding ethylene-mediated growth inhibition in response to abiotic stresses such as salinity (Etesami et al., 2014; Glick, 2014; Singh et al., 2015); (vi) changing root architecture and morphology, hydraulic conductance, and hormone status (Arora et al., 2006, 2012). These root changes, which may result from increased IAA, can facilitate the uptake of more nutrients and provide access to a more extensive network of soil water (Vacheron et al., 2013; Goswami et al., 2014); (vii) emitting stress-related volatile compounds that enhance plant biomass and survival under severe drought stress (Timmusk et al., 2014); (viii) accumulating osmolytes such as amino acids and their derivatives (e.g., glutamate, proline, peptides, and N-acetylated amino acids), quaternary amines (e.g., glycine betaine and carnitine), and sugars (e.g., sucrose and trehalose) (Creus et al., 2004); (ix) preserving higher stomatal conductance and photosynthetic activities (del Amor and Cuadra-Crespo, 2012), which can reduce the accumulation of toxic ions (Na+ and Cl−) and improve the ratio of K+: Na+ in the leaf (Pérez-Alfocea et al., 2010); (x) inducing the expression of stress-responsive genes. In particular, halotolerant PGPRs cause up-regulation of stress tolerance genes (Kaushal and Wani, 2016; Etesami and Beattie, 2017) such as RAB18 (LEA), the RD29A and RD29B regulons of ABA-responsive elements (ABRE), and dehydration responsive elements (DRE), as well as the transcription factor DREB2b DRE binding protein. They also can induce genes that encode proteins related to energy metabolism and cell division, particularly amino acid metabolism and the tricarboxylic acid cycle (Banaei-Asl et al., 2015; Qin et al., 2016). The halotolerant PGPRs Azospirillum brasilense, Pantoea agglomerans, and Bacillus megaterium can help plants decrease their cellular water potential by increasing the expression of genes PIP2, ZmPIP1-1, and HvPIP2-1, which are involved in producing aquaporins. Aquaporins are water channel proteins in the plasma membranes of plant cells that contribute to the transfer of water into the plant (Marulanda et al., 2010; Zawoznik et al., 2011; Gond et al., 2015; Moshelion et al., 2015). PGPRs induction of aquaporins may encourage plants to continue to take up water from salt-affected soils (Qin et al., 2016). Furthermore, the PGPR B. subtilis can also decrease the absorption of excessive amounts of Na+ by the roots of plants by down-regulating expression of the high-affinity K+ transporter (HKT1) in the roots of salinity-affected plants (Zhang et al., 2008; Qin et al., 2016). In addition, these halotolerant PGPRs facilitate shoot-to-root Na+ recirculation by triggering the induction of HKT1 in shoots (Zhang et al., 2008); and (xi) protecting plants from phytopathogens, such as by producing extracellular enzymes to hydrolyze fungal cell walls, synthesizing antimicrobial compounds, producing Fe-chelating siderophores to starve phytopathogens for Fe, excluding pathogens via competition for nutrients and sites on root, and inducing systemic resistance (Glick and Bashan, 1997; Bhattacharyya and Jha, 2012; Etesami, 2018).
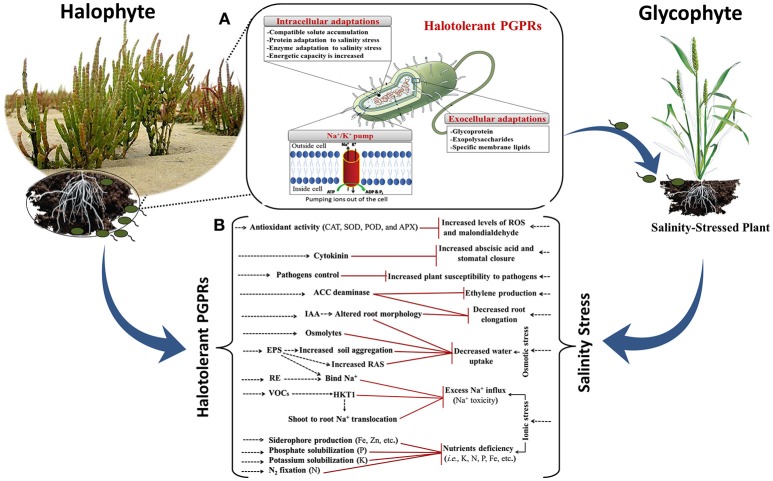
Figure 2.
(A) Schematic overview of the mechanisms developed by halotolerant plant growth promoting rhizobacteria (PGPRs) to live and survive in highly salinity conditions. For more details, see this reference (Ruppel et al., 2013). (B), Beneficial attributes of halotolerant PGPRs toward salinity stress tolerance in non-halophyte crops grown in saline soils. Red arrows indicate rhizobacterial components negating salinity stress effects. Halotolerant PGPRs increase the K+/Na+ ratio by selectively enhancing K+ uptake and avoiding translocation of toxic Na+ under saline conditions. These bacteria are capable of increasing the antioxidative systems in plants for reactive oxygen species (ROS) scavenging such as enzymatic components of superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), peroxidase (POD), and glutathione reductase (GR) and non-enzymatic components of cysteine, glutathione and ascorbicacid. 1-aminocyclopropane-1-carboxylate (ACC)-deaminase producing PGPRs decrease the excessive ethylene production in plants caused by salinity stress and thereby eliminate the negative effect of ethylene on roots. Production of phytohormones increases the overall growth and also alters root characteristics (i.e., alteration of root proliferation, metabolism and respiration rate) to facilitate uptake of water and nutrients. Phytohormone indole-3-acetic acid (IAA) also increases the size of aerial parts of the plants. Production of osmoprotectants (i.e., proline, polyamines, glutamate, total free amino acids, etc.) by PGPR also contributes to salinity stress tolerance in PGPRs-inoculated plants. Exopolysaccharides (EPS) bind the toxic Na+ and restrict Na+ influx into roots. Soil aggregation due to production of EPS or alteration of root exudates (RE) hydrates the rhizosphere and helps in enhancing uptake of water and nutrients. EPS also increase root adhering-soil (RAS). Volatile organic compounds (VOCs) can trigger induction of high affinity K+ transporter (HKT1) in shoots and reduction of HKT1 in roots, limiting Na+ entry into roots and facilitating shoot-to-root Na+ recirculation. For more details, see these references (Dutta and Khurana, 2015; Kaushal and Wani, 2016; Qin et al., 2016; Sáenz-Mata et al., 2016).
Inoculating crops with halotolerant PGPRs isolated from halophytes has been successful at improving crop growth and tolerance under salt stress conditions (Shukla et al., 2012; Khan et al., 2016). Halotolerant PGPRs can provide many benefits to plants, including helping halophytes and glycophytes overcome salt stress (Table 1). For example, salt- tolerant PGPRs isolated from rhizospheric soil of the halophytes Haloxylon salicornicum, Lespedeza bicolor, Atriplex leucoclada, Suaeda fruticosa, and Salicornica virginica also enhanced the growth of salinity-stressed maize (Ullah and Bano, 2015). These plants exhibited an accumulation of osmolytes (e.g., sugar and proline) and increase in antioxidant enzyme activity (e.g., SOD, peroxidase, CAT, and ascorbate peroxidase) as compared to un-inoculated plants. Similarly, a study by Siddikee et al. (2010) showed that, following the inoculation of canola seedlings with halotolerant bacterial isolates isolated from halophytic plants under salt stress in gnotobiotic conditions, the plants exhibited significantly increased growth, as shown by a 35–43% increase in dry weight and 29–47% increase in root length. The studies shown in Table 1 illustrate that PGPRs isolated from the rhizosphere of halophytic species can be used as effective bio-inoculants for non-halophytic crops grown under salt stress.
Halophytes and ACC deaminase-producing PGPRs
Ethylene is a plant growth regulator and stress hormone (Mayak et al., 2004b; Pierik et al., 2007) that is produced by almost all plant species. This gaseous growth hormone has a key role in causing physiological changes in plants at the molecular level. The production of ethylene is significantly enhanced in response to environmental stresses such as drought and salinity. Excessive ethylene inhibits root growth and, as a consequence, limits further growth of the plant. High ethylene levels in nodules is also associated with decreased N2 fixation (Ma et al., 2002). Although ethylene production near roots is constantly modulated during plant growth and development (Mayak et al., 2004a; Mahajan and Tuteja, 2005; Gamalero and Glick, 2015), reducing stress-induced ethylene levels alleviates some effects of stress on plants (Glick, 2004; Etesami and Beattie, 2017).
As described earlier, PGPRs that secrete the enzyme ACC deaminase can reduce ethylene levels by metabolizing ACC, a precursor of plant-produced ethylene, into α-ketobutyrate and ammonia (Etesami and Beattie, 2017). Plants inoculated with ACC deaminase-producing PGPRs often exhibit extended root growth, attributed to reductions in ethylene, and enhanced resistance to salinity stress (Mayak et al., 2004a,b; Cheng et al., 2007; Glick et al., 2007; Zahir et al., 2009; Nadeem et al., 2010; Barnawal et al., 2012; Jha et al., 2012; Etesami and Beattie, 2017). These PGPRs can also influence plant ethylene homeostasis by altering the expression of genes encoding the ethylene synthesis enzymes ACC synthase and ACC oxidase (Tsukanova et al., 2017).
Although salinity has been associated with the loss in ACC deaminase production by some PGPRs (Upadhyay et al., 2009), at least some salt-tolerant PGPRs isolated from saline environments appear to maintain ACC deaminase production based on documentation of their beneficial properties in helping plants overcome salinity stress by reducing ethylene levels (Mayak et al., 2004a). For example, 25 out of 140 halotolerant bacterial isolates from coastal soils of the South Korean Yellow Sea showed ACC deaminase activity (Siddikee et al., 2010); these bacterial isolates belonged to the genera of Arthrobacter, Bacillus, Brevibacterium, Corynebacterium, Exiguobacterium, Halomonas, Micrococcus, Oceanimonas, Planococcus, and Zhihengliuella. ACC deaminase-producing PGPRs isolated from saline environments alleviated salinity stress in a variety of plants. For example, the ACC deaminase-producing PGPR strains P. fluorescens N3 and P. putida Q7 promoted the growth of maize roots by 3.3-fold, and maize shoots by 2.3-fold, respectively, under salinity stress as compared to un-inoculated controls (Kausar and Shahzad, 2006; Khan et al., 2016). Similarly, inoculation of legume plants with ACC deaminase-producing rhizobia isolated from saline soils promoted nodule formation (Shaharoona et al., 2006), and inoculation of wheat plants with the PGPR strain A. brasilense FP2 from saline soils resulted in a decrease in the expression of the plant ACC oxidase (Camilios-Neto et al., 2014).
In addition to halotolerant bacteria isolated from saline environments, halotolerant bacteria isolated from various halophytic species exhibit ACC deaminase production (Table 1; Siddikee et al., 2010; Jha et al., 2012; Zhou et al., 2017). ACC deaminase-producing PGPRs isolated from halophytes have been found to alleviate salinity stress and increase plant growth for both halophytes and salinity-sensitive crop plants (Table 1). For example, novel diazotrophic halotolerant bacteria isolated from the roots of Salicornia brachiata featured ACC deaminase activity and these isolates included Brachybacterium saurashtrense, Brevibacteriumcasei, Cronobacter sakazakii, Haererehalobacter, Halomonas, Mesorhizobium, Pseudomonas, Rhizobium radiobacter, Vibrio, and Zhihengliuella (Jha et al., 2012). Moreover, growth parameters of S. brachiate increased significantly under salt stress after re-inoculation with B. saurashtrense and Pseudomonas (Jha et al., 2012). In another study (El-Tarabily and Youssef, 2010), one out of 62 bacterial isolates from the A. marina rhizosphere exhibited a high level of ACC deaminase activity. Following inoculation of this isolate, identified as P. maricaloris, plant seedlings exhibited a decrease in the endogenous levels of ACC and improved growth undersalinity stress. Following the inoculation of red pepper plants with the ACC deaminase-producing halotolerant PGPRs Brevibacterium iodinum, Zhihengliuela alba, and Bacillus licheniformis isolated from halophytes, ethylene levels in the plants decreased by 44, 53, and 57%, respectively. Furthermore, their salt tolerance, as assessed using a salt tolerance index, increased significantly compared to non-inoculated plants (Siddikee et al., 2011). These studies illustrate that habitat-adapted ACC deaminase-producing PGPRs associated with halophytes can mitigate the effects of salinity stress on crops and reduce ethylene to below growth-inhibitory levels (Jha et al., 2012).
Considerable attention has been given to the isolation of ACC deaminase-producing salt-tolerant PGPRs for their use in promoting plant growth in saline environments (Hardoim et al., 2008; Nadeem et al., 2010; Ali et al., 2014). Methods of isolating such PGPRs are well-established (Penrose and Glick, 2003). A rapid and efficient approach to their isolation is using polymerase chain reaction (PCR)-based screening for the ACC deaminase-encoding gene acdS coupled to a colorimetric ninhydrin assay to measure ACC (Nikolic et al., 2011; Jasim et al., 2015; Li et al., 2015b; Qin et al., 2016). Interestingly, recent results suggest that endophytic bacteria are more able to produce the enzyme ACC deaminase than PGPRs isolated from other habitats, including the surfaces of leaves and roots and from non-rhizosphere soil (Bruto et al., 2014; Qin et al., 2016). Future research that compares the bio-activity of ACC deaminase-producing bacteria isolated from various habitats, including distinct tissues of halophytic plants, would be useful.
Halophytes and phytohormone-producing PGPRs
Phytohormones regulate the protective response of plants to biotic and abiotic stresses (Raghavan et al., 2006), and also the development and tolerance to diverse environmental stresses including salinity stress (Ryu and Cho, 2015). Plant responses to salt stress include an array of changes at the molecular, biochemical, and physiological levels (Manchanda and Garg, 2008; Ahmad et al., 2013; Kumari et al., 2015), and depend upon environmental conditions, soil properties, and plant growth stage (Zhu et al., 1992). Previous studies (Dodd and Pérez-Alfocea, 2012; Khan et al., 2016) indicate that salinity can either diminish (300 mM NaCl) (Dunlap and Binzel, 1996) or increase (100 mM NaCl) (Albacete et al., 2008) endogenous IAA levels in roots. Plants can also respond to exogenous phytohormones, and these can relieve the adverse effects of salinity (Singh and Jain, 1982; Zahir et al., 2010). Thus, exogenous application of phytohormones and their precursors provides an attractive approach to counter salt stress conditions by changing the balance of endogenous levels of hormones (Ilangumaran and Smith, 2017). This was illustrated in a study showing that treating wheat seeds with IAA reduced the detrimental effects of salinity stress on wheat growth (Datta et al., 1997). In addition to stimulating root proliferation, which can enhance growth and salt tolerance (Dodd and Pérez-Alfocea, 2012), IAA can help maintain leaf growth, which helps prevent salinity-induced limitations in plant productivity (Munns, 2002; Albacete et al., 2008). IAA has also been reported to enhance the protection of bacterial cells against abiotic stresses such as high salt concentrations (Bianco et al., 2006).
PGPRs may enhance plant growth, in part, by modulating the plant hormonal balance (Ilangumaran and Smith, 2017; Tsukanova et al., 2017). IAA production is a relatively common trait of most salt-tolerant PGPRs (Dodd et al., 2010), and IAA-producing PGPRs can increase the fitness of plants grown in salt-affected soils (Tiwari et al., 2011). PGPRs may improve crop salt tolerance by altering hormonal root–shoot signaling (Yang et al., 2009). The ability to modify plant stress levels by providing IAA, which influences the development of lateral roots, has previously been reported for halotolerant-bacteria isolated from coastal soils (Siddikee et al., 2010), halophyte roots in Argentina (Sgroy et al., 2009), highly saline habitats (Tiwari et al., 2011), the halophyte Prosopis strombulifera (Piccoli et al., 2011), the rhizosphere of halophytic weeds from the Pakistani Khewra salt range (Naz et al., 2009), halotolerant plants from a Chinese coastal sandbank (Bian et al., 2011), and the rhizosphere of C. annum growing in desert areas (Marasco et al., 2012). Some IAA-producing salt-tolerant PGPRs isolated from halophytes are shown in Table 1, as is their potential as a tool for promoting the salt tolerance of halophytes and glycophytes. For example, Tiwari et al. (2011) demonstrated that inoculation of wheat with IAA-producing salt-tolerant Halomonas sp. resulted in a higher IAA content in the rhizosphere of treated plants than control plants and increased plant growth. In another study, the IAA-overproducing strain Sinorhizobium meliloti ameliorated the reduced growth of Medicago truncatula in saline soils (Bianco and Defez, 2009). This work was further supported by Egamberdieva (2009). These studies clearly show that managing IAA production in halophytic and non-halophytic plants by endophytic and rhizosphere bacteria may be an important tool in conferring salt tolerance.
Cytokinins (CKs) are also involved in the development of plant resistance to biotic and abiotic stresses (Großkinsky et al., 2011; O'Brien and Benková, 2013). CK production is a relatively common trait of PGPRs (Dodd et al., 2010). PGPRs can influence plant CK concentration by synthesizing CK or altering CK homeostasis in the plant (Arshad and Frankenberger, 1991; de Garcia Salamone et al., 2005; Glick, 2012; Pallai et al., 2012; Kapoor and Kaur, 2016). The Platycladus orientalis plants inoculated with a CK-producing PGPR strain B. subtilis had increased CK levels in the shoots and were more resistant to drought (Liu et al., 2013). Increased growth of drought-stressed lettuce plants inoculated with a CK-producing B. subtilis strain suggested modulation of root-to-shoot CK signaling (Arkhipova et al., 2007). The ability of PGPRs to synthesize CK or alter plant CK homeostasis highlights the importance of understanding how PGPRs stimulate growth and increase plant resistance to salinity.
Gibberellic acid (GA) positively regulates cell division and elongation, hypocotyl and stem growth, and leaf and root meristem size (Guo et al., 2015; Wang et al., 2015; Martínez et al., 2016). GA signaling is a key factor in the inhibition of plant growth under stress (Magome and Kamiya, 2016; Martínez et al., 2016). PGPRs can influence the endogenous GA levels in plants (Bottini et al., 2004; Kang et al., 2014a; Shahzad et al., 2016). Some PGPR strains, such as B. amyloliquefaciens RWL-1, Promicromonospora sp. SE188, Leifsonia soli SE134, and Enterococcus faecium LKE12, can synthesize GA (Bottini et al., 2004; Kang et al., 2012, 2014a; Lee et al., 2015; Shahzad et al., 2016). After inoculation of plants with the GA-producing PGPR strains, B. cereus MJ-1 (Joo et al., 2005) and Promicromonospora sp. SE188, the amount of endogenous GA in the shoots increased (Kang et al., 2014a). Some bacterial isolates from the halophyte P. strombulifera (Piccoli et al., 2011) and the rhizosphere of halophytic weeds from the Pakistani Khewra salt range showed the ability to produce GA (Naz et al., 2009), as did the PGPR strains B. licheniformis, Lysinibacillus fusiformis, Achromobacter xylosoxidans, and Brevibacterium halotolerans isolated from the halophyte P. strombulifera (Sgroy et al., 2009).
Abscisic acid (ABA) is an important plant stress hormone that is synthesized in response to abiotic stresses and activates the genes responsible for stress resistance (Sah et al., 2016). This hormone plays an important role in alleviating salinity stress by mediating stomatal, and thereby photosynthetic, responses to high salinity (Dodd and Pérez-Alfocea, 2012). It also plays a crucial role in plant-PGPR interactions (Dodd, 2003). Many PGPRs produce ABA in vitro (Dodd et al., 2010); these include A. brasilense, B. licheniformis, Novosphingobium sp., P. fluorescens, Rhodococcus sp. P1Y, and Variovorax paradoxus (Sgroy et al., 2009; Jiang et al., 2012; Belimov et al., 2014; Salomon et al., 2014; Cohen et al., 2015). PGPRs can also produce ABA under salinity stress conditions and increase growth of salinized plants (Naz et al., 2009). For example, in a study, following inoculation of plants with ABA-producing strains such as B. licheniformis Rt4M10, P. fluorescens Rt6M10, A. brasilense Sp 245, the internal ABA content increased and inoculated plants become more resistant to drought compared to un-inoculated plants (Salomon et al., 2014; Cohen et al., 2015). In addition, inoculation with ABA-producing PGPRs often decreased the accumulation and concentration of ABA in roots and significantly altered the long-distance signaling of shoot-to-root ABA transport in the phloem and the root-to-shoot ABA transport in the xylem (Dodd and Pérez-Alfocea, 2012; Jiang et al., 2012; Belimov et al., 2014; Qin et al., 2016); the resulting changes in ABA levels may mitigate the plant's sensitivity to water scarcity. Recently, the two rhizospheric bacteria Rhodococcus sp. and Novosphingobium sp. were found to metabolize ABA in vitro (Belimov et al., 2014; Qin et al., 2016), suggesting a mechanism for decreasing plant ABA concentrations. Interestingly, disrupting plant ABA homeostasis can influence the activity of halotolerant PGPRs, as shown by wild-type tomato plants that exhibited enhanced growth, and ABA-deficient mutant plants that exhibited reduced growth, in response to B. megaterium inoculation (Porcel et al., 2014; Qin et al., 2016). Collectively, these results suggest that ABA-producing halotolerant PGPRs, ABA-metabolizing halotolerant PGPRs, and general halotolerant PGPRs will act differently in adjusting plant ABA status and thus may result in variable plant responses to salinity stress. ABA production has also been reported in bacterial isolates from halophytes, including from the rhizosphere of halophytic weeds from the salt range of Pakistani Khewra (Naz et al., 2009) and the halophyte P. strombulifera (Piccoli et al., 2011). L. fusiformis, B. subtilis, B. halotolerans, B. licheniformis, B. pumilus, A. xylosoxidans, and Pseudomonas putida are some ABA-producing bacteria isolated from the halophyte P. strombulifera (Sgroy et al., 2009). Relatively little is known of the role of ABA in plant-bacterial interactions. The ability of PGPRs to alter ABA levels in plants suggest opportunities to use these bacteria to influence plant growth and abiotic stress resistance, and highlights a need for more research to understand how PGPRs influence plant ABA signal transduction components.
Jasmonic acid (JA) is also involved in abiotic stress resistance (Ahmad et al., 2016). Several endophytic PGPRs synthesize JA and salicylic acid (SA) (Forchetti et al., 2007; Chen et al., 2014). Inoculating plants with the PGPR strains P. fluorescens Pf4, P. aeruginosa Pag (Singh et al., 2003), and B. amyloliquefaciens LJ02 (Li et al., 2015a) resulted in a rise in the endogenous levels of SA in various plant tissues. Inoculation of Vitis vinifera with the PGPR strain Burkholderia phytofirmans PsJN also led to SA accumulation (Bordiec et al., 2010), as did inoculation with the GA-producing PGPR strains Promicromonospora sp. SE188 (Kang et al., 2012) and B. amyloliquefaciens RWL-1 (Shahzad et al., 2016).
Although there is some evidence that PGPRs improved plant salt tolerance by altering the endogenous hormone status (Kang et al., 2014b; Sahoo et al., 2014; Qin et al., 2016; Ilangumaran and Smith, 2017), little is known about how PGPRs influence this process. We have a similar knowledge deficit regarding the potential for halotolerant PGPRs to synthesize many of these phytohormones and to produce them in vitro or in planta. Bacterial isolates from halophytes have thus far been screened primarily for IAA synthesis, among the hormones discussed. However, the roles of GA, ABA, CK, SA, and JA in the physiology of plant halotolerance indicates that future research on how bacterial isolates from halophytes influence phytohormone homeostasis in plants may be fruitful.
Halophytes and phosphate-solubilizing PGPRs
Phosphorus is one of the major essential macronutrients for plants. Although organic and inorganic P are abundant in soils, P availability is limited due to its presence in insoluble forms. Whereas, P comprises about 0.05% (w/w) of soils, often only 0.1% of the total P is available to plants because of poor solubility and its fixation in soil (Goldstein, 1986). In both saline soil-based and fertile soil-based agriculture, intensive cultivation strongly depletes soil nutrients. The use of inorganic NPK fertilizers increases soil salinity, particularly when coupled with saline irrigation. Phosphate-solubilizing halotolerant PGPRs provide an opportunity to enhance P availability to plants without exacerbating soil salinity levels. Phosphate-solubilizing PGPRs can solubilize insoluble phosphates via various mechanisms like chelation, ion exchange, and acidification by secreting low molecular weight organic acids (Sharma et al., 2013; Etesami, 2018). In salt-affected soils, inoculation with phosphate-solubilizing halotolerant PGPRs improved plant growth and suppressed the adverse effects of salt (Giri et al., 2004). Following the inoculation of Solanum lycopersicum plants with Achromobacter piechaudii, plant P content and water use efficiency increased under salinity stress (Mayak et al., 2004a). Similarly, inoculation of wheat with B. aquimaris increased plant P content under salinity stress in the field (Upadhyay and Singh, 2015). Both studies suggest that phosphate-solubilizing PGPRs solubilize insoluble P in saline soils. Halotolerant bacteria isolated from halophytes also exhibit P solubilization activity (Table 1). A screen of the mangrove A. marina rhizosphere identified 129 bacterial strains with the ability to solubilize rock phosphate, with Oceanobacillus picturae able to mobilize 97% of this mineral (El-Tarabily and Youssef, 2010). Bacteria isolated from halophytes, including Arthrobacter, Bacillus, Azospirillum, Vibrio, Phyllobacterium, and O. picturae, were shown to solubilize Ca3(PO4)2, AlPO4, and FePO4 (Bashan et al., 2000; Banerjee et al., 2010; El-Tarabily and Youssef, 2010; Yasmin and Bano, 2011) and increase the P content in both halophytes and glycophytes under salinity stress (Table 1). When the halophytes S. bigelovii and S. bigelovii were inoculated with various halotolerant PGPRs, including Azospirillum, Vibrio, Bacillus, and Phyllobacterium, the P content of the foliage increased (Bashan et al., 2000). This increased P content in plant tissues may help ameliorate the growth-restraining effects of salinity.
Halophytes and siderophore-producing PGPRs
Iron is a micronutrient that is a component of many enzymes involved in biochemical processes, including respiration, photosynthesis, and N2 fixation (Kobayashi and Nishizawa, 2012; Abbas et al., 2015). Iron availability is very low in calcareous and saline sodic soils throughout the world (Rabhi et al., 2007; Abbas et al., 2015). These soils suppress the availability of most micronutrients, including iron, and suppress plant growth by concurrent salinity and iron deficiency stresses (Yousfi et al., 2007; Abbas et al., 2015). PGPRs often secrete siderophores, which are small, high-affinity Fe(III)-chelating compounds that scavenge iron, and the iron–siderophore complexes can be easily accessed by plants (Kloepper et al., 1980). Siderophore production by halotolerant PGPRs isolated from halophytes has been reported (Table 1); however, the ability of these strains to increase the availability of iron and other micro-elements, such as Zn, Mn, and Cu, to plants is not yet known.
Halophytes and N2-fixing PGPRs
Most agricultural systems depend on the application of exogenous nitrogen, as it is often the nutrient that most limits productivity (Vitousek and Howarth, 1991). The productivity of halophytic crop species can also be limited by a lack of available N in saline soils. For legumes, nitrogen fixation is more sensitive than plant growth to soil salinity (Djekoun and Planchon, 1991), and all stages in nodule formation and nodule function are negatively affected by salinity (de la Peña and Pueyo, 2012; Bruning and Rozema, 2013). Salinity can interfere with plant N nutrition and thus decrease the N content of plant tissues (Naidoo, 1987), as illustrated by salinity-mediated repression of ammonium and nitrate uptake and assimilation (Ullrich, 2002). Typically, farmers use chemical fertilizers to compensate for a lack of soil N; however, the excessive use of inorganic fertilizers may increase salinity, severely degrade the soil structure, and change the composition of the soil microflora (Akhavan-Kharazian et al., 1991; Rueda-Puente et al., 2003). Salinity also results in low soil microbial activity due to osmotic stress and ion toxicity. Increases in soil salinity in many parts of the world are therefore limiting plant productivity and the benefits accrued from biological N2 fixation (Jha et al., 2012). Salt-tolerant N2-fixing PGPRs can tolerate osmotic stress by producing osmolytes that allow them to maintain their cell turgor and metabolism (Yan et al., 2015). N2 fixation by salt-tolerant bacteria associated with the roots of halophytes is an important source of available N in saline soils. Furthermore, these roots are a source of halotolerant N2-fixing bacteria with plant growth-promoting potential (Table 1; Rueda-Puente et al., 2003; Jha et al., 2012; Sharma et al., 2016), some of which have been found to increase the growth of halophytes as well as non-halophytic crops in saline soils (Table 1). The potential benefits of biological N2-fixers to halophytes and salt-sensitive crops (Rueda-Puente et al., 2003; Jha et al., 2012) highlight the interest in exploring N2-fixing halotolerant PGPRs as potential bio-fertilizer resources for saline soil-based agriculture.
Halophytes and PGPRs that control phytopathogens
In addition to disrupting plant physiology and morphology, soil salinity increases plant susceptibility to pathogens (Besri, 1993). Plant diseases are a major constraint to crop yields but can potentially be controlled biologically by using PGPRs. Biological control using PGPRs offers a more eco-friendly approach to disease management than agricultural chemicals (Compant et al., 2010; Etesami and Alikhani, 2018). Some mechanisms that PGPRs use to counter the deleterious effects of phytopathogens include (Olanrewaju et al., 2017): (i) the synthesis of one or more antimicrobial metabolites (Couillerot et al., 2009), many of which have been reported in PGPRs of the genera Bacillus and Pseudomonas. These metabolites may serve as cytotoxic, antifungal, antibacterial, phytotoxic, antihelminthic, antiviral, antioxidant, and/or antitumor agents (Olanrewaju et al., 2017); (ii) the production of fungal cell wall-degrading enzymes (Chernin et al., 1995) such as lipase, which can degrade some fungal cell wall-associated lipids, β-1,3-glucanase, which can degrade cell wall carbohydrates, chitinase, which can degrade the integral fungal cell wall component chitin (Husson et al., 2017), and protease, which can degrade cell wall proteins (Vaddepalli et al., 2017); (iii) competition either for nutrients or for binding sites on plant roots (Barahona et al., 2011); such competition can limit phytopathogen growth or binding to the plant thereby making it difficult for the pathogen to proliferate (Olanrewaju et al., 2017); (iv) the synthesis of hydrogen cyanide, which when produced by bio-control PGPRs such as Rhizobium, Pseudomonas, Alcaligenes, Bacillus, and Aeromonas, inhibits cytochrome C oxidase as well as other important metalloenzymes (Nandi et al., 2017); (v) activation of induced systemic resistance, which is a resistance mechanism in plants (Van Loon et al., 1998; Halfeld-Vieira et al., 2006) in which exposure of plants to specific microbes, such as some biocontrol PGPRs, primes the plant to react faster and more strongly to a subsequent pathogen attack (Olanrewaju et al., 2017). Induction of systemic resistance provides strong protection coordinated by phytohormone signaling pathways (Pieterse et al., 2012, 2014; Walters et al., 2013); (vi) quorum quenching, which is the disruption of signaling among pathogens. This may occur via the production of signal-degrading enzymes such as lactonase, and the subsequent loss of disruption of signaling may minimize pathogen virulence (Olanrewaju et al., 2017); and (vii) synthesis of siderophores (Olanrewaju et al., 2017), which can prevent or reduce pathogen proliferation by reducing the iron available to pathogens (Shen et al., 2013). The siderophores from PGPRs have been found, at least in some cases, to have a higher affinity for Fe3+ than the siderophores from fungal pathogens (Kloepper et al., 1980), thus giving the PGPRs a competitive advantage for iron.
Halophilic PGPRs may also provide biological control of phytopathogens. Many can produce antibiotics and antifungal metabolites, as shown in the halophilic bacteria B. subtilis, B. cereus, B. pumilus, B. licheniformis, Halomonas elongate, and Halobacillus halophilus, which antagonize phytopathogenic fungi such as Fusarium sambucinum, F. roseum var. sambucinum, F. oxysporum, F. moniliforme, F. graminearum, Penicillium citrinum, Aspergillus flavus, and Botrytis cinerea; these organisms have been shown to produce antibiotics, proteases, chitinases, and β−1,3-glucanases (Niehaus et al., 1999; Sadfi et al., 2001, 2002; Sadfi-Zouaoui et al., 2008; Essghaier et al., 2009; Siddikee et al., 2010; Berrada et al., 2012; Ruppel et al., 2013; Goswami et al., 2014; Singh and Jha, 2016). For example, the strains B. halotolerans Ps9 and B. pumilus Ps19, which were isolated from the halophyte P. strombulifera, exhibited protease activity and inhibited the growth of the phytopathogenic fungus Alternaria sp. by more than 50%, at least on plates (Sgroy et al., 2009). Similarly, a halotolerant PGPR Pseudomonas sp. strain isolated from the halophyte Suaeda salsa suppressed the growth of the phytopathogenic fungi Fusarium oxysporum f. sp. cucumerinum and F. oxysporum f. sp. conglutinans (Teng et al., 2010). The biological control potential of halophilic bacteria may be correlated with their production of membrane-bound or extracellular hydrolytic enzymes (Sadfi-Zouaoui et al., 2008). Although antagonistic halotolerant PGPRs may provide an ecologically friendly alternative to synthetic fungicides, research is needed to evaluate that antagonistic potential of halotolerant PGPRs against phytopathogens, and the severity of the disease pressure by these pathogens, in saline environments (Sadfi-Zouaoui et al., 2008).
Conclusions and future prospects
This review has highlighted the potential for halophytes to be used as an isolation source for halotolerant PGPRs, including PGPRs that exhibit PGP traits such as IAA production, phosphate solubilization, siderophore production, N2 fixation, ACC deaminase activity, and control of phytopathogens. Halotolerant PGPRs isolated from the endosphere or rhizosphere of halophytes can be used to enhance the growth, and possibly the yield, of halophytic and non-halophytic crops (Sáenz-Mata et al., 2016). Crop inoculation with halotolerant PGPRs is therefore a viable strategy for sustainable crop production in salinity-based agriculture, which includes crop production in arid and semiarid environments (Khan et al., 2016). Several avenues of research would move us closer to adopting this strategy for salinity-based agriculture:
Although some beneficial effects of halotolerant PGPRs on salinity-affected plants are known, many of the underlying physiological and molecular mechanisms contributing to enhanced plant growth and halotolerance are not. Knowledge of these mechanisms, and the portfolio of traits optimal for inoculum performance, would contribute to designing agronomic applications of these bacteria for saline-based agriculture (Dodd and Pérez-Alfocea, 2012).
Knowledge of how the endogenous bacterial and fungal microbiomes of halophytes contribute to halophyte resistance to extreme salinity would provide insights into optimal applications of introduced halotolerant PGPRs.
Increasing global food production requires improved crop production not only in saline soils, but also in areas where the irrigation water is contaminated with salt (Ruppel et al., 2013). This is an increasing problem in coastal zones and thus will be increasingly important in many parts of the world. Halophytes should be explored as a reservoir for halotolerant PGPRs for uses under these conditions as well as in saline soils.
Since the diversity of halotolerant PGPRs in salt-affected soils and in the microbiome of halophytic plants depends on soil parameters and plant species (Qin et al., 2016; Szymańska, et al., 2016), further studies on the diversity of the microbial communities in the rhizosphere and endosphere of various halophytic plant species are needed to clarify and describe these ecological associations in saline soil-based agriculture.
Knowledge of the signaling mechanisms and factors influencing the interactions between halotolerant PGPRs and halophytes and glycophytes in the field will provide a better understanding of the ecology of these bacteria and how they have promoted halophyte adaptation to high salinity environments (Egamberdiyeva and Islam, 2008; Khan et al., 2016).
Knowledge of the biochemical and physiological characteristics of PGPRs associated with halophytes could facilitate strategies for plant protection and remediation of saline soils (Ruppel et al., 2013; Egamberdieva and Lugtenberg, 2014; Khan et al., 2016).
Agricultural inoculants, including those for bio-stimulation, often vary in efficacy due, in part, to their strong dependence on environmental context for activity. Although the isolation of halotolerant PGPRs from halophytes in saline soils should increase the probability that the strains are active in saline soils (Khan et al., 2009), knowledge of the key environmental traits that influence their activity could help reduce variation in efficacy. Moreover, isolating PGPRs from roots under conditions of high alkalinity, acidity or salinity, drought, high and low temperatures, and flooded conditions could provide strains or traits that are efficacious in plant protection or growth promotion under diverse agricultural conditions (Khan et al., 2016).
Knowledge of the molecular mechanisms by which salt-tolerant PGPRs increase plant resistance to salinity may suggest genetic approaches to engineer bacteria with enhanced abilities to stimulate plant growth and salinity tolerance, as well as plants that are improved in their ability to interact with halotolerant PGPRs (Khan et al., 2016).
Knowledge of the endophytic and rhizospheric fungi associated with halophytes and their impacts on halophyte growth and survival may contribute to additional strategies for protecting halophyte and non-halophyte plants in saline soils (Sharma et al., 2016).
To increase our fundamental knowledge of microbial interactions with halophytes, investigations are needed that address the specificity of halophyte-microbe interactions, the effect of root exudates on these interactions, and the effect of root exudates on gene expression related to plant growth promotion and biological control.
Lastly, the development of halotolerant PGPRs that can sustainably improve plant growth under diverse high salinity crop production conditions requires that the performance of these strains be examined over long periods (at least 2 years) on a scale that is relevant to crop production and under field conditions that provide a diversity of soil conditions and environmental stresses. Sustainable improvements in crop productivity may benefit from strategies that combine PGPRs with stress-tolerant beneficial fungi, and that involve co-inoculating multiple PGPRs that alleviate distinct stresses. The latter is particularly appealing given the co-occurrence of many stresses, such as drought, salinity, and heavy metal contamination, in field soils. Importantly, halotolerant PGPRs that are used effectively in agriculture may also contribute to applications for phytoremediation, phytodesalinization, bio-fertilization, and biological control.
Author contributions
HE gathered literature and prepared the manuscript. GB revised and approved the final version to be published.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank University of Tehran and Iowa State University for providing the necessary facilities for this study.
References
- Abbas G., Saqib M., Akhtar J. (2015). Interactive effects of salinity and iron deficiency on different rice genotypes. J. Plant Nutr. Soil Sci. 178, 306–311. 10.1002/jpln.201400358 [DOI] [Google Scholar]
- Abou-Elela S. I., Kamel M. M., Fawzy M. E. (2010). Biological treatment of saline wastewater using a salt-tolerant microorganism. Desalination 250, 1–5. 10.1016/j.desal.2009.03.022 [DOI] [Google Scholar]
- Acquaah G. (2007). Principles of Plant Genetics and Breeding. Oxford: Blackwell. [Google Scholar]
- Ahmad P., Azooz M. M., Prasad M. N. V. (eds.). (2013). Salt Stress in Plants. New York, NY: Springer, 1–495. 10.1007/978-1-4614-6108-1 [DOI] [Google Scholar]
- Ahmad P., Rasool S., Gul A., Sheikh S. A., Akram N. A., Ashraf M., et al. (2016). Jasmonates: multifunctional roles in stress tolerance. Front. Plant Sci. 7:813. 10.3389/fpls.2016.00813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhavan-Kharazian M., Campbell W. F., Jurinak J. J., Dudley L. M. (1991). Effects of CaSO4, CaCl2, and NaCl on leaf nitrogen, nodule weight, and acetylene reduction activity in Phaseolus vulgaris L. Arid Land Res. Manage. 5, 97–103. [Google Scholar]
- Akinshina N., Azizov A., Karasyova T., Klose E. (2016). On the issue of halophytes as energy plants in saline environment. Biomass Bioener. 91, 306–311. 10.1016/j.biombioe.2016.05.034 [DOI] [Google Scholar]
- Albacete A., Ghanem M. E., Martínez-Andújar C., Acosta M., Sánchez-Bravo J., Martínez V., et al. (2008). Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 59, 4119–4131. 10.1093/jxb/ern251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Charles T. C., Glick B. R. (2014). Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 80, 160–167. 10.1016/j.plaphy.2014.04.003 [DOI] [PubMed] [Google Scholar]
- Al-Mailem D. M., Sorkhoh N. A., Marafie M., Al-Awadhi H., Eliyas M., Radwan S. S. (2010). Oil phytoremediation potential of hypersaline coasts of the Arabian Gulf using rhizosphere technology. Bioresour. Technol. 101, 5786–5792. 10.1016/j.biortech.2010.02.082 [DOI] [PubMed] [Google Scholar]
- Anburaj R., Nabeel M. A., Sivakumar T., Kathiresan K. (2012). The role of rhizobacteria in salinity effects on biochemical constituents of the halophyte Sesuvium portulacastrum. Russ. J. Plant Physiol. 59, 115–119. 10.1134/S1021443712010025 [DOI] [Google Scholar]
- Antón J., Oren A., Benlloch S., Rodríguez-Valera F., Amann R., Rosselló-Mora R. (2002). Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Microbiol. 52, 485–491. 10.1099/00207713-52-2-485 [DOI] [PubMed] [Google Scholar]
- Argandona M., Fernandez-Carazo R., Llamas I., Martinez-Checa F., Caba J. M., Quesada E., et al. (2005). The moderately halophilic bacterium Halomonas maura is a free-living diazotroph. FEMS Microbiol. Lett. 244, 69–74. 10.1016/j.femsle.2005.01.019 [DOI] [PubMed] [Google Scholar]
- Arkhipova T. N., Prinsen E., Veselov S. U., Martinenko E. V., Melentiev A. I., Kudoyarova G. R. (2007). Cytokinin producing bacteria enhance plant growth in drying soil. Plant Soil 292, 305–315. 10.1007/s11104-007-9233-5 [DOI] [Google Scholar]
- Arora N. K., Singhal V., Maheshwari D. K. (2006). Salinity-induced accumulation of poly-β-hydroxybutyrate in rhizobia indicating its role in cell protection. World J. Microbiol. Biotechnol. 22, 603–606. 10.1007/s11274-005-9077-1 [DOI] [Google Scholar]
- Arora N. K., Tewari S., Singh S., Lal N., Maheshwari D. K. (2012). PGPR for protection of plant health under saline conditions, in Bacteria in Agrobiology: Stress Management, ed Maheshwari D. K. (Berlin; Heidelberg: Springer-Verlag; ), 239–258. [Google Scholar]
- Arshad M., Frankenberger W. T., Jr. (1991). Microbial production of plant hormones, in The Rhizosphere and Plant Growth, eds Keister D. L., Cregan P. B. (Dordrecht: Springer; ), 327–334. [Google Scholar]
- Ashraf M., Hasnain S., Berge O., Mahmood T. (2004). Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol. Fertil. Soils 40, 157–162. 10.1007/s00374-004-0766-y [DOI] [Google Scholar]
- Askari H., Edqvist J., Hajheidari M., Kafi M., Salekdeh G. H. (2006). Effects of salinity levels on proteome of Suaeda aegyptiaca leaves. Proteomics 6, 2542–2554. 10.1002/pmic.200500328 [DOI] [PubMed] [Google Scholar]
- Banaei-Asl F., Bandehagh A., Uliaei E. D., Farajzadeh D., Sakata K., Mustafa G., et al. (2015). Proteomic analysis of canola root inoculated with bacteria under salt stress. J. Proteomics 124, 88–111. 10.1016/j.jprot.2015.04.009 [DOI] [PubMed] [Google Scholar]
- Banerjee S., Palit R., Sengupta C., Standing D. (2010). Stress induced phosphate solubilization By ‘arthrobacter’ sp. and ‘Bacillus’ sp. isolated from tomato rhizosphere. Aust. J. Crop Sci. 4, 378. [Google Scholar]
- Barahona E., Navazo A., Martínez-Granero F., Zea-Bonilla T., Pérez-Jiménez R. M., Martín M., et al. (2011). Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl. Environ. Microbiol. 77, 5412–5419. 10.1128/AEM.00320-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnawal D., Bharti N., Maji D., Chanotiya C. S., Kalra A. (2012). 1-Aminocyclopropane-1-carboxylic acid (ACC) deaminase-containing rhizobacteria protect Ocimum sanctum plants during waterlogging stress via reduced ethylene generation. Plant Physiol. Biochem. 58, 227–235. 10.1016/j.plaphy.2012.07.008 [DOI] [PubMed] [Google Scholar]
- Bashan Y., Moreno M., Troyo E. (2000). Growth promotion of the seawater-irrigated oilseed halophyte Salicornia bigelovii inoculated with mangrove rhizosphere bacteria and halotolerant Azospirillum spp. Biol. Fertil. Soils 32, 265–272. 10.1007/s003740000246 [DOI] [Google Scholar]
- Belimov A. A., Dodd I. C., Safronova V. I., Dumova V. A., Shaposhnikov A. I., Ladatko A. G., et al. (2014). Abscisic acid metabolizing rhizobacteria decrease ABA concentrations in planta and alter plant growth. Plant Physiol. Biochem. 74, 84–91. 10.1016/j.plaphy.2013.10.032 [DOI] [PubMed] [Google Scholar]
- Berendsen R. L., Pieterse C. M. J., Bakker P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Berrada I., Benkhemmar O., Swings J., Bendaou N., Amar M. (2012). Selection of halophilic bacteria for biological control of tomato gray mould caused by Botrytis cinerea. Phytopathol. Mediterr. 51, 625–630. 10.14601/Phytopathol_Mediterr-10627 [DOI] [Google Scholar]
- Besri M. (1993). Effects of salinity on plant diseases development, in Towards the Rational Use of High Salinity Tolerant Plants, eds Lieth H., Al Masoom A. A. (Springer Netherlands; ), 67–74. [Google Scholar]
- Bharti N., Pandey S. S., Barnawal D., Patel V. K., Kalra A. (2016). Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 6:34768. 10.1038/srep34768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti N., Yadav D., Barnawal D., Maji D., Kalra A. (2013). Exiguobacterium oxidotolerans, a halotolerant plant growth promoting rhizobacteria, improves yield and content of secondary metabolites in Bacopa monnieri (L.) Pennell under primary and secondary salt stress. World J. Microbiol. Biotechnol. 29, 379–387. 10.1007/s11274-012-1192-1 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya P. N., Jha D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. 10.1007/s11274-011-0979-9 [DOI] [PubMed] [Google Scholar]
- Bian G., Zhang Y., Qin S., Xing K., Xie H., Jiang J. (2011). Isolation and biodiversity of heavy metal tolerant endophytic bacteria from halotolerant plant species located in coastal shoal of Nantong. Wei Sheng Wu Xue Bao 51, 1538–1547. [PubMed] [Google Scholar]
- Bianco C., Defez R. (2009). Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 60, 3097–3107. 10.1093/jxb/erp140 [DOI] [PubMed] [Google Scholar]
- Bianco C., Imperlini E., Calogero R., Senatore B., Amoresano A., Carpentieri A., et al. (2006). Indole-3-acetic acid improves Escherichia coli's defences to stress. Arch. Microbiol. 185, 373–382. 10.1007/s00203-006-0103-y [DOI] [PubMed] [Google Scholar]
- Bibi F., Chung E. J., Yoon H. S., Song G. C., Jeon C. O., Chung Y. R. (2011). Haloferula luteola sp. nov., an endophytic bacterium isolated from the root of a halophyte, Rosa rugosa, and emended description of the genus Haloferula. Int. J. Syst. Evol. Microbiol. 61, 1837–1841. 10.1099/ijs.0.022772-0 [DOI] [PubMed] [Google Scholar]
- Bordiec S., Paquis S., Lacroix H., Dhondt S., Ait Barka E., Kauffmann S., et al. (2010). Comparative analysis of defence responses induced by the endophytic plant growth-promoting rhizobacterium Burkholderia phytofirmans strain PsJN and the non-host bacterium Pseudomonas syringae pv. pisi in grapevine cell suspensions. J. Exp. Bot. 62, 595–603. 10.1093/jxb/erq291 [DOI] [PubMed] [Google Scholar]
- Bottini R., Cassán F., Piccoli P. (2004). Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 65, 497–503. 10.1007/s00253-004-1696-1 [DOI] [PubMed] [Google Scholar]
- Boukhatem Z. F., Domergue O., Bekki A., Merabet C., Sekkour S., Bouazza F., et al. (2012). Symbiotic characterization and diversity of rhizobia associated with native and introduced acacias in arid and semi-arid regions in Algeria. FEMS Microbiol. Ecol. 80, 534–547. 10.1111/j.1574-6941.2012.01315.x [DOI] [PubMed] [Google Scholar]
- Bruning B., Rozema J. (2013). Symbiotic nitrogen fixation in legumes: perspectives for saline agriculture. Environ. Exp. Bot. 92, 134–143. 10.1016/j.envexpbot.2012.09.001 [DOI] [Google Scholar]
- Bruto M., Prigent-Combaret C., Muller D., Moënne-Loccoz Y. (2014). Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 4:6261. 10.1038/srep06261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilios-Neto D., Bonato P., Wassem R., Tadra-Sfeir M. Z., Brusamarello-Santos L. C. C., Valdameri G., et al. (2014). Dual RNA-seq transcriptional analysis of wheat roots colonized by Azospirillum brasilense reveals up-regulation of nutrient acquisition and cell cycle genes. BMC Genomics 15:378. 10.1186/1471-2164-15-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman J. M. (2015). The evolution of halophytes, glycophytes and crops, and its implications for food security under saline conditions. New Phytol. 206, 557–570. 10.1111/nph.13217 [DOI] [PubMed] [Google Scholar]
- Chen Y., Fan J.-B., Du L., Xu H., Zhang Q.-H., He Y.-Q. (2014). The application of phosphate solubilizing endophyte Pantoea dispersa triggers the microbial community in red acidic soil. Appl. Soil Ecol. 84, 235–244. 10.1016/j.apsoil.2014.05.014 [DOI] [Google Scholar]
- Cheng Z., Park E., Glick B. R. (2007). 1-Aminocyclopropane-1-carboxylate deaminase from Pseudomonas putida UW4 facilitates the growth of canola in the presence of salt. Can. J. Microbiol. 53, 912–918. 10.1139/W07-050 [DOI] [PubMed] [Google Scholar]
- Chernin L., Ismailov Z., Haran S., Chet I. (1995). Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl. Environ. Microbiol. 61, 1720–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A. C., Bottini R., Pontin M., Berli F. J., Moreno D., Boccanlandro H., et al. (2015). Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant. 153, 79–90. 10.1111/ppl.12221 [DOI] [PubMed] [Google Scholar]
- Compant S., Clément C., Sessitsch A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. 10.1016/j.soilbio.2009.11.024 [DOI] [Google Scholar]
- Couillerot O., Prigent-Combaret C., Caballero-Mellado J., Moënne-Loccoz Y. (2009). Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett. Appl. Microbiol. 48, 505–512. 10.1111/j.1472-765X.2009.02566.x [DOI] [PubMed] [Google Scholar]
- Creus C. M., Sueldo R. J., Barassi C. A. (2004). Water relations and yield in Azospirillum-inoculated wheat exposed to drought in the field. Can. J. Bot. 82, 273–281. 10.1139/b03-119 [DOI] [Google Scholar]
- Datta K. S., Varma S. K., Angrish R., Kumar B., Kumari P. (1997). Alleviation of salt stress by plant growth regulators in Triticum aestivum L. Biol. Plant. 40, 269–275. 10.1023/A:1001076805595 [DOI] [Google Scholar]
- de Garcia Salamone I. E., Hynes R. K., Nelson L. M. (2005). Role of cytokinins in plant growth promotion by rhizosphere bacteria, in PGPR: Biocontrol and Biofertilization, ed Siddiqui Z. A. (Springer Netherlands; ), 173–195. [Google Scholar]
- del Amor F. M., Cuadra-Crespo P. (2012). Plant growth-promoting bacteria as a tool to improve salinity tolerance in sweet pepper. Funct. Plant Biol. 39, 82–90. 10.1071/FP11173 [DOI] [PubMed] [Google Scholar]
- de la Peña T. C., Pueyo J. J. (2012). Legumes in the reclamation of marginal soils, from cultivar and inoculant selection to transgenic approaches. Agron. Sustain. Dev. 32, 65–91. 10.1007/s13593-011-0024-2 [DOI] [Google Scholar]
- Del Río L. A. (2015). ROS and RNS in plant physiology: an overview. J. Exp. Bot. 66, 2827–2837. 10.1093/jxb/erv099 [DOI] [PubMed] [Google Scholar]
- Djekoun A., Planchon C. (1991). Water status effect on dinitrogen fixation and photosynthesis in soybean. Agron. J. 83, 316–322. 10.2134/agronj1991.00021962008300020011x [DOI] [Google Scholar]
- Dodd I. C. (2003). Hormonal interactions and stomatal responses. J. Plant Growth Regul. 22, 32–46. 10.1007/s00344-003-0023-x [DOI] [Google Scholar]
- Dodd I. C., Pérez-Alfocea F. (2012). Microbial amelioration of crop salinity stress. J. Exp. Bot. 63, 3415–3428. 10.1093/jxb/ers033 [DOI] [PubMed] [Google Scholar]
- Dodd I. C., Zinovkina N. Y., Safronova V. I., Belimov A. A. (2010). Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 157, 361–379. 10.1111/j.1744-7348.2010.00439.x [DOI] [Google Scholar]
- Dunlap J. R., Binzel M. L. (1996). NaCI reduces indole-3-acetic acid levels in the roots of tomato plants independent of stress-induced abscisic acid. Plant Physiol. 112, 379–384. 10.1104/pp.112.1.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S., Khurana S. M. P. (2015). Plant growth-promoting rhizobacteria for alleviating abiotic stresses in medicinal plants, in Plant-Growth-Promoting Rhizobacteria (PGPR) and Medicinal Plants, eds Egamberdieva D., Shrivastava S., Varma A. (Springer; ), 167–200. [Google Scholar]
- Egamberdieva D. (2009). Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Physiol. Plant. 31, 861–864. 10.1007/s11738-009-0297-0 [DOI] [Google Scholar]
- Egamberdieva D., Kucharova Z. (2009). Selection for root colonising bacteria stimulating wheat growth in saline soils. Biol. Fertil. Soils 45, 563–571. 10.1007/s00374-009-0366-y [DOI] [Google Scholar]
- Egamberdieva D., Lugtenberg B. (2014). Use of plant growth-promoting rhizobacteria to alleviate salinity stress in plants, in Use of Microbes for the Alleviation of Soil Stresses, Vol. 1, ed Miransari M. (New York, NY: Springer; ), 73–96. [Google Scholar]
- Egamberdiyeva D., Islam K. R. (2008). Salt-tolerant rhizobacteria: plant growth promoting traits and physiological characterization within ecologically stressed environments, in Plant-Bacteria Interactions: Strategies and Techniques to Promote Plant Growth, eds Ahmad I., Pichtel J., Hayat S. (Weinheim: Wiley-VCH Verlag GmbH & Co., KGaA; ), 257–281. 10.1002/9783527621989.ch14 [DOI] [Google Scholar]
- El-Tarabily K. A., Youssef T. (2010). Enhancement of morphological, anatomical and physiological characteristics of seedlings of the mangrove Avicennia marina inoculated with a native phosphate-solubilizing isolate of Oceanobacillus picturae under greenhouse conditions. Plant Soil 332, 147–162. 10.1007/s11104-010-0280-y [DOI] [Google Scholar]
- Essghaier B., Fardeau M.-L., Cayol J.-L., Hajlaoui M. R., Boudabous A., Jijakli H., et al. (2009). Biological control of grey mould in strawberry fruits by halophilic bacteria. J. Appl. Microbiol. 106, 833–846. 10.1111/j.1365-2672.2008.04053.x [DOI] [PubMed] [Google Scholar]
- Etesami H. (2018). Can interaction between silicon and plant growth promoting rhizobacteria benefit in alleviating abiotic and biotic stresses in crop plants? Agric. Ecosyst. Environ. 253, 98–112. 10.1016/j.agee.2017.11.007 [DOI] [Google Scholar]
- Etesami H., Alikhani H. A. (2018). Bacillus species as the most promising bacterial biocontrol agents in rhizosphere and endorhiza of plants grown in rotation with each other. Eur. J. Plant Pathol. 150, 497–506. 10.1007/s10658-017-1276-8 [DOI] [Google Scholar]
- Etesami H., Beattie G. A. (2017). Plant-microbe interactions in adaptation of agricultural crops to abiotic stress conditions, in Probiotics and Plant Health, eds Kumar V., Kumar M., Sharma S., Prasad R. (Singapore: Springer; ), 163–200. [Google Scholar]
- Etesami H., Hosseini H. M., Alikhani H. A. (2014). Bacterial biosynthesis of 1-aminocyclopropane-1-caboxylate (ACC) deaminase, a useful trait to elongation and endophytic colonization of the roots of rice under constant flooded conditions. Physiol. Mol. Biol. Plants 20, 425–434. 10.1007/s12298-014-0251-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers T. J., Colmer T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179, 945–963. 10.1111/j.1469-8137.2008.02531.x [DOI] [PubMed] [Google Scholar]
- Flowers T. J., Colmer T. D. (2015). Plant salt tolerance: adaptations in halophytes. Ann. Bot. 115, 327–331. 10.1093/aob/mcu267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchetti G., Masciarelli O., Alemano S., Alvarez D., Abdala G. (2007). Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 76, 1145–1152. 10.1007/s00253-007-1077-7 [DOI] [PubMed] [Google Scholar]
- Gago C., Sousa A. R., Juliao M., Miguel G., Antunes D. C., Panagopoulos T. (2011). Sustainable use of energy in the storage of halophytes used for food. Int. J. Ener. Environ. 5, 592–599. [Google Scholar]
- Gamalero E., Glick B. R. (2015). Bacterial modulation of plant ethylene levels. Plant Physiol. 169, 13–22. 10.1104/pp.15.00284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giongo A., Ambrosini A., Vargas L. K., Freire J. R. J., Bodanese-Zanettini M. H., Passaglia L. M. P. (2008). Evaluation of genetic diversity of bradyrhizobia strains nodulating soybean [Glycine max (L.) Merrill] isolated from South Brazilian fields. Appl. Soil Ecol. 38, 261–269. 10.1016/j.apsoil.2007.10.016 [DOI] [Google Scholar]
- Giri B., Kapoor R., Agarwal L., Mukerji K. G. (2004). Preinoculation with arbuscular mycorrhizae helps Acacia auriculiformis grow in degraded Indian wasteland soil. Commun. Soil Sci. Plant Anal. 35, 193–204. 10.1081/CSS-120027643 [DOI] [Google Scholar]
- Giri B., Kapoor R., Mukerji K. G. (2007). Improved tolerance of Acacia nilotica to salt stress by arbuscular mycorrhiza, Glomus fasciculatum may be partly related to elevated K/Na ratios in root and shoot tissues. Microb. Ecol. 54, 753–760. 10.1007/s00248-007-9239-9 [DOI] [PubMed] [Google Scholar]
- Glenn E. P., O'Leary J. W., Watson M. C., Thompson T. L., Kuehl R. O. (1991). Salicornia bigelovii Torr.: an oilseed halophyte for seawater irrigation. Science 251, 1065–1068. [DOI] [PubMed] [Google Scholar]
- Glick B. R. (2004). Bacterial ACC deaminase and the alleviation of plant stress. Adv. Appl. Microbiol. 56, 291–312. 10.1016/S0065-2164(04)56009-4 [DOI] [PubMed] [Google Scholar]
- Glick B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012:963401. 10.6064/2012/963401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. 10.1016/j.micres.2013.09.009 [DOI] [PubMed] [Google Scholar]
- Glick B. R., Bashan Y. (1997). Genetic manipulation of plant growth-promoting bacteria to enhance biocontrol of phytopathogens. Biotechnol. Adv. 15, 353–378. 10.1016/S0734-9750(97)00004-9 [DOI] [PubMed] [Google Scholar]
- Glick B. R., Cheng Z., Czarny J., Duan J. (2007). Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur. J. Plant Pathol. 119, 329–339. 10.1007/s10658-007-9162-4 [DOI] [Google Scholar]
- Godfray H. C. J., Beddington J. R., Crute I. R., Haddad L., Lawrence D., Muir J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science 327, 812–818. 10.1126/science.1185383 [DOI] [PubMed] [Google Scholar]
- Goldstein A. H. (1986). Bacterial solubilization of mineral phosphates: historical perspective and future prospects. Am. J. Altern. Agric. 1, 51–57. 10.1017/S0889189300000886 [DOI] [Google Scholar]
- Gond S. K., Torres M. S., Bergen M. S., Helsel Z., White J. F. (2015). Induction of salt tolerance and up-regulation of aquaporin genes in tropical corn by rhizobacterium Pantoea agglomerans. Lett. Appl. Microbiol. 60, 392–399. 10.1111/lam.12385 [DOI] [PubMed] [Google Scholar]
- Gontia I., Kavita K., Schmid M., Hartmann A., Jha B. (2011). Brachybacterium saurashtrense sp. nov., a halotolerant root-associated bacterium with plant growth-promoting potential. Int. J. Syst. Evol. Microbiol. 61, 2799–2804. 10.1099/ijs.0.023176-0 [DOI] [PubMed] [Google Scholar]
- Goswami D., Dhandhukia P., Patel P., Thakker J. N. (2014). Screening of PGPR from saline desert of Kutch: growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol. Res. 169, 66–75. 10.1016/j.micres.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Großkinsky D. K., Naseem M., Abdelmohsen U. R., Plickert N., Engelke T., Griebel T., et al. (2011). Cytokinins mediate resistance against Pseudomonas syringae in tobacco through increased antimicrobial phytoalexin synthesis independent of salicylic acid signaling. Plant Physiol. 157, 815–830. 10.1104/pp.111.182931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Wang Y., Liu H., Hu P., Jia Y., Zhang C., et al. (2015). Exogenous GA3 application enhances xylem development and induces the expression of secondary wall biosynthesis related genes in Betula platyphylla. Int. J. Mol. Sci. 16, 22960–22975. 10.3390/ijms160922960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta B., Huang B. (2014). Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J. Genomics 2014:701596. 10.1155/2014/701596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfeld-Vieira B. d. A., Vieira Júnior J. R., Romeiro R. d. S., Silva H. S. A., Baracat-Pereira M. C. (2006). Induction of systemic resistance in tomato by the autochthonous phylloplane resident Bacillus cereus. Pesqui. Agropecu. Bras. 41, 1247–1252. 10.1590/S0100-204X2006000800006 [DOI] [Google Scholar]
- Hardoim P. R., van Overbeek L. S., Elsas J. D. V. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. 10.1016/j.tim.2008.07.008 [DOI] [PubMed] [Google Scholar]
- Hasanuzzaman M., Nahar K., Alam M. M., Bhowmik P. C., Hossain M. A., Rahman M. M., et al. (2014). Potential use of halophytes to remediate saline soils. Biomed. Res. Int. 2014:589341. 10.1155/2014/589341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa P. M., Bressan R. A., Zhu J.-K., Bohnert H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Biol. 51, 463–499. 10.1146/annurev.arplant.51.1.463 [DOI] [PubMed] [Google Scholar]
- Himabindu Y., Chakradhar T., Reddy M. C., Kanygin A., Redding K. E., Chandrasekhar T. (2016). Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 124, 39–63. 10.1016/j.envexpbot.2015.11.010 [DOI] [Google Scholar]
- Husson E., Hadad C., Huet G., Laclef S., Lesur D., Lambertyn V., et al. (2017). The effect of room temperature ionic liquids on the selective biocatalytic hydrolysis of chitin via sequential or simultaneous strategies. Green Chem. 19, 4122–4131. 10.1039/C7GC01471F [DOI] [Google Scholar]
- Ilangumaran G., Smith D. L. (2017). Plant growth promoting rhizobacteria in amelioration of salinity stress: a systems biology perspective. Front. Plant Sci. 8:1768. 10.3389/fpls.2017.01768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam F., Yasmeen T., Arif M. S., Ali S., Ali B., Hameed S., et al. (2016). Plant growth promoting bacteria confer salt tolerance in Vigna radiata by up-regulating antioxidant defense and biological soil fertility. Plant Growth Regul. 80, 23–36. 10.1007/s10725-015-0142-y [DOI] [Google Scholar]
- Jacob J. H. (2012). Classification of halophilic heterotrophic bacteria thriving in the jordanian dead sea littoral zone. J. Biol. Sci. 12, 246–252. 10.3923/jbs.2012.246.252 [DOI] [Google Scholar]
- Jafari B., Hanifezadeh M., Parvin M. S. J. (2012). Molecular study of bacteria associated with Salicornia symbiotic bacteria as a candidate for Hormozgan salty zone culturing by Persian Gulf water irrigation. Afr. J. Microbiol. Res. 6, 4687–4695. 10.5897/AJMR11.1132 [DOI] [Google Scholar]
- Jasim B., Anish M. C., Shimil V., Jyothis M., Radhakrishnan E. K. (2015). Studies on plant growth promoting properties of fruit-associated bacteria from Elettaria cardamomum and molecular analysis of ACC deaminase gene. Appl. Biochem. Biotechnol. 177, 175–189. 10.1007/s12010-015-1736-6 [DOI] [PubMed] [Google Scholar]
- Jesus J. M., Danko A. S., Fiúza A., Borges M.-T. (2015). Phytoremediation of salt-affected soils: a review of processes, applicability, and the impact of climate change. Environ. Sci. Pollut. Res. 22, 6511–6525. 10.1007/s11356-015-4205-4 [DOI] [PubMed] [Google Scholar]
- Jha B., Gontia I., Hartmann A. (2012). The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant Soil 356, 265–277. 10.1007/s11104-011-0877-9 [DOI] [Google Scholar]
- Jha B., Singh V. K., Weiss A., Hartmann A., Schmid M. (2015). Zhihengliuella somnathii sp. nov., a halotolerant actinobacterium from the rhizosphere of a halophyte Salicornia brachiata. Int. J. Syst. Evol. Microbiol. 65, 3137–3142. 10.1099/ijsem.0.000391 [DOI] [PubMed] [Google Scholar]
- Jha Y., Subramanian R. B. (2014). PGPR regulate caspase-like activity, programmed cell death, and antioxidant enzyme activity in paddy under salinity. Physiol. Mol. Biol. Plants 20, 201–207. 10.1007/s12298-014-0224-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S. H., Gururani M. A., Chun S.-C. (2014). Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol. Res. 169, 83–98. 10.1016/j.micres.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Jiang F., Chen L., Belimov A. A., Shaposhnikov A. I., Gong F., Meng X., et al. (2012). Multiple impacts of the plant growth-promoting rhizobacterium Variovorax paradoxus 5C-2 on nutrient and ABA relations of Pisum sativum. J. Exp. Bot. 63, 6421–6430. 10.1093/jxb/ers301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo G.-J., Kim Y.-M., Kim J.-T., Rhee I.-K., Kim J.-H., Lee I.-J. (2005). Gibberellins-producing rhizobacteria increase endogenous gibberellins content and promote growth of red peppers. J. Microbiol. 43, 510–515. [PubMed] [Google Scholar]
- Joshi R., Mangu V. R., Bedre R., Sanchez L., Pilcher W., Zandkarimi H., et al. (2015). Salt adaptation mechanisms of halophytes: improvement of salt tolerance in crop plants, in Elucidation of Abiotic Stress Signaling in Plants, ed Pandey G. K. (New York, NY: Springer; ), 243–279. [Google Scholar]
- Kafi M., Khan M. A. (2008). Crop and Forage Production using Saline Waters. Daya Books. [Google Scholar]
- Kang S.-M., Khan A. L., Hamayun M., Hussain J., Joo G.-J., You Y.-H., et al. (2012). Gibberellin-producing Promicromonospora sp. SE188 improves Solanum lycopersicum plant growth and influences endogenous plant hormones. J. Microbiol. 50, 902. 10.1007/s12275-012-2273-4 [DOI] [PubMed] [Google Scholar]
- Kang S.-M., Khan A. L., You Y.-H., Kim J.-G., Kamran M., Lee I.-J. (2014a). Gibberellin production by newly isolated strain Leifsonia soli SE134 and its potential to promote plant growth. J. Microbiol. Biotechnol. 24, 106–112. 10.4014/jmb.1304.04015 [DOI] [PubMed] [Google Scholar]
- Kang S.-M., Radhakrishnan R., Khan A. L., Kim M.-J., Park J.-M., Kim B.-R., et al. (2014b). Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal and stress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol. Biochem. 84, 115–124. 10.1016/j.plaphy.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Kaplan D., Maymon M., Agapakis C. M., Lee A., Wang A., Prigge B. A., et al. (2013). A survey of the microbial community in the rhizosphere of two dominant shrubs of the Negev Desert highlands, Zygophyllum dumosum (Zygophyllaceae) and Atriplex halimus (Amaranthaceae), using cultivation-dependent and cultivation-independent methods. Am. J. Bot. 100, 1713–1725. 10.3732/ajb.1200615 [DOI] [PubMed] [Google Scholar]
- Kapoor R., Kaur M. (2016). Cytokinins production by fluorescent Pseudomonas isolated from rhizospheric soils of Malus and Pyrus. Afr. J. Microbiol. Res. 10, 1274–1279. 10.5897/AJMR2016.8211 [DOI] [Google Scholar]
- Kausar R., Shahzad S. M. (2006). Effect of ACC-deaminase containing rhizobacteria on growth promotion of maize under salinity stress. J Agric. Soc. Sci. 2, 216–218. [Google Scholar]
- Kaushal M., Wani S. P. (2016). Rhizobacterial-plant interactions: strategies ensuring plant growth promotion under drought and salinity stress. Agric. Ecosyst. Environ. 231, 68–78. 10.1016/j.agee.2016.06.031 [DOI] [Google Scholar]
- Kawasaki S., Borchert C., Deyholos M., Wang H., Brazille S., Kawai K., et al. (2001). Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13, 889–905. 10.1105/tpc.13.4.889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. A., Boër B., Özturk M., Clüsener-Godt M., Gul B., Breckle S.-W. (2016). Sabkha Ecosystems: Vol. V: The Americas. Springer. [Google Scholar]
- Khan M. S., Zaidi A., Wani P. A., Ahemad M., Oves M. (2009). Functional diversity among plant growth-promoting rhizobacteria: current status, in Microbial Strategies for Crop Improvement, eds Khan M. S., Zaidi A., Musarrat J. (Berlin; Heidelberg: Springer; ), 105–132. [Google Scholar]
- Kim K., Jang Y.-J., Lee S.-M., Oh B.-T., Chae J.-C., Lee K.-J. (2014). Alleviation of salt stress by Enterobacter sp. EJ01 in tomato and Arabidopsis is accompanied by up-regulation of conserved salinity responsive factors in plants. Mol. Cells 37, 109–117. 10.14348/molcells.2014.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper J. W., Leong J., Teintze M., Schroth M. N. (1980). Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286, 885–886. 10.1038/286885a0 [DOI] [Google Scholar]
- Kobayashi T., Nishizawa N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. 10.1146/annurev-arplant-042811-105522 [DOI] [PubMed] [Google Scholar]
- Kosová K., Vítámvás P., Urban M. O., Prášil I. T. (2013). Plant proteome responses to salinity stress–comparison of glycophytes and halophytes. Funct. Plant Biol. 40, 775–786. 10.1071/FP12375 [DOI] [PubMed] [Google Scholar]
- Krishna G., Singh B. K., Kim E. K., Morya V. K., Ramteke P. W. (2015). Progress in genetic engineering of peanut (Arachis hypogaea L.)—A review. Plant Biotechnol. J. 13, 147–162. 10.1111/pbi.12339 [DOI] [PubMed] [Google Scholar]
- Ksouri R., Ksouri W. M., Jallali I., Debez A., Magné C., Hiroko I., et al. (2012). Medicinal halophytes: potent source of health promoting biomolecules with medical, nutraceutical and food applications. Crit. Rev. Biotechnol. 32, 289–326. 10.3109/07388551.2011.630647 [DOI] [PubMed] [Google Scholar]
- Kumari A., Das P., Parida A. K., Agarwal P. K. (2015). Proteomics, metabolomics, and ionomics perspectives of salinity tolerance in halophytes. Front. Plant Sci. 6:537. 10.3389/fpls.2015.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen H. (1986). Halophilic and halotolerant microorganisms-an overview and historical perspective. FEMS Microbiol. Lett. 39, 3–7. 10.1111/j.1574-6968.1986.tb01835.x [DOI] [Google Scholar]
- Lau J. A., Lennon J. T. (2012). Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. 109, 14058–14062. 10.1073/pnas.1202319109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-E., Radhakrishnan R., Kang S.-M., You Y.-H., Joo G.-J., Lee I.-J., et al. (2015). Enterococcus faecium LKE12 cell-free extract accelerates host plant growth via gibberellin and indole-3-acetic acid secretion. J. Microbiol. Biotechnol. 25, 1467–1475. 10.4014/jmb.1502.02011 [DOI] [PubMed] [Google Scholar]
- Leifert C., Waites W. M., Nicholas J. R. (1989). Bacterial contaminants of micropropagated plant cultures. J. Appl. Bacteriol. 67, 353–361. 10.1111/j.1365-2672.1989.tb02505.x [DOI] [Google Scholar]
- Li Y., Gu Y., Li J., Xu M., Wei Q., Wang Y. (2015a). Biocontrol agent Bacillus amyloliquefaciens LJ02 induces systemic resistance against cucurbits powdery mildew. Front. Microbiol. 6:883. 10.3389/fmicb.2015.00883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chang S., Ye S., Chen M., Lin L., Li Y., et al. (2015b). Differentiation of 1-aminocyclopropane-1-carboxylate (ACC) deaminase from its homologs is the key for identifying bacteria containing ACC deaminase. FEMS Microbiol. Ecol. 91:fiv112. 10.1093/femsec/fiv112 [DOI] [PubMed] [Google Scholar]
- Lim G. H., Zhang X., Chung M. S., Lee D. J., Woo Y. M., Cheong H. S., et al. (2010). A putative novel transcription factor, AtSKIP, is involved in abscisic acid signalling and confers salt and osmotic tolerance in Arabidopsis. New Phytol. 185, 103–113. 10.1111/j.1469-8137.2009.03032.x [DOI] [PubMed] [Google Scholar]
- Liu F., Xing S., Ma H., Du Z., Ma B. (2013). Cytokinin-producing, plant growth-promoting rhizobacteria that confer resistance to drought stress in Platycladus orientalis container seedlings. Appl. Microbiol. Biotechnol. 97, 9155–9164. 10.1007/s00253-013-5193-2 [DOI] [PubMed] [Google Scholar]
- Lucero M. E., Barrow J. R., Osuna P., Reyes I., Duke S. E. (2008). Enhancing native grass productivity by cocultivating with endophyte-laden calli. Rangeland Ecol. Manage. 61, 124–130. 10.2111/06-144R3.1 [DOI] [Google Scholar]
- Lucero M. E., Unc A., Cooke P., Dowd S., Sun S. (2011). Endophyte microbiome diversity in micropropagated Atriplex canescens and Atriplex torreyi var griffithsii. PLoS ONE 6:e17693. 10.1371/journal.pone.0017693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Penrose D. M., Glick B. R. (2002). Strategies used by rhizobia to lower plant ethylene levels and increase nodulation. Can. J. Microbiol. 48, 947–954. 10.1139/w02-100 [DOI] [PubMed] [Google Scholar]
- Magome H., Kamiya Y. (2016). Inactivation processes: Gibberellins, in Annual Plant Reviews, Vol. 49, eds Hedden P., Thomas S. G. (Chichester: John Wiley & Sons; ) 73–94. 10.1002/9781119210436.ch3 [DOI] [Google Scholar]
- Mahajan S., Tuteja N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. 10.1016/j.abb.2005.10.018 [DOI] [PubMed] [Google Scholar]
- Manchanda G., Garg N. (2008). Salinity and its effects on the functional biology of legumes. Acta Physiol. Plant. 30, 595–618. 10.1007/s11738-008-0173-3 [DOI] [Google Scholar]
- Manousaki E., Kalogerakis N. (2011). Halophytes present new opportunities in phytoremediation of heavy metals and saline soils. Ind. Eng. Chem. Res. 50, 656–660. 10.1021/ie100270x [DOI] [Google Scholar]
- Mapelli F., Marasco R., Rolli E., Barbato M., Cherif H., Guesmi A., et al. (2013). Potential for plant growth promotion of rhizobacteria associated with Salicornia growing in Tunisian hypersaline soils. Biomed Res. Int. 2013:248078. 10.1155/2013/248078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco R., Rolli E., Ettoumi B., Vigani G., Mapelli F., Borin S., et al. (2012). A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS ONE 7:e48479. 10.1371/journal.pone.0048479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C., Espinosa-Ruiz A., Prat S. (2016). Gibberellins and plant vegetative growth. Annu. Plant Rev. 49, 285–322. 10.1002/9781119210436.ch10 [DOI] [Google Scholar]
- Marulanda A., Azcón R., Chaumont F., Ruiz-Lozano J. M., Aroca R. (2010). Regulation of plasma membrane aquaporins by inoculation with a Bacillus megaterium strain in maize (Zea mays L.) plants under unstressed and salt-stressed conditions. Planta 232, 533–543. 10.1007/s00425-010-1196-8 [DOI] [PubMed] [Google Scholar]
- Mayak S., Tirosh T., Glick B. R. (2004a). Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 42, 565–572. 10.1016/j.plaphy.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Mayak S., Tirosh T., Glick B. R. (2004b). Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci. 166, 525–530. 10.1016/j.plantsci.2003.10.025 [DOI] [Google Scholar]
- Moshelion M., Halperin O., Wallach R., Oren R. A. M., Way D. A. (2015). Role of aquaporins in determining transpiration and photosynthesis in water-stressed plants: crop water-use efficiency, growth and yield. Plant Cell Environ. 38, 1785–1793. 10.1111/pce.12410 [DOI] [PubMed] [Google Scholar]
- Munns R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. 10.1046/j.0016-8025.2001.00808.x [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Nabti E., Sahnoune M., Ghoul M., Fischer D., Hofmann A., Rothballer M., et al. (2010). Restoration of growth of durum wheat (Triticum durum var. waha) under saline conditions due to inoculation with the rhizosphere bacterium Azospirillum brasilense NH and extracts of the marine alga Ulva lactuca. J. Plant Growth Regul. 29, 6–22. 10.1007/s00344-009-9107-6 [DOI] [Google Scholar]
- Nabti E., Schmid M., Hartmann A. (2015). Application of halotolerant bacteria to restore plant growth under salt stress, in Halophiles, eds Maheshwari D. K., Saraf M. (Cham: Springer; ), 235–259. [Google Scholar]
- Nadeem S. M., Zahir Z. A., Naveed M., Ashraf M. (2010). Microbial ACC-deaminase: prospects and applications for inducing salt tolerance in plants. CRC Crit. Rev. Plant Sci. 29, 360–393. 10.1080/07352689.2010.524518 [DOI] [Google Scholar]
- Naidoo G. (1987). Effects of salinity and nitrogen on growth and water relations in the mangrove, Avicennia marina (Forsk.) Vierh. New Phytol. 107, 317–325. 10.1111/j.1469-8137.1987.tb00183.x [DOI] [PubMed] [Google Scholar]
- Nandi M., Selin C., Brawerman G., Fernando W. G. D., de Kievit T. (2017). Hydrogen cyanide, which contributes to Pseudomonas chlororaphis strain PA23 biocontrol, is upregulated in the presence of glycine. Biol. Control 108, 47–54. 10.1016/j.biocontrol.2017.02.008 [DOI] [Google Scholar]
- Navarro-Torre S., Barcia-Piedras J. M., Mateos-Naranjo E., Redondo-Gómez S., Camacho M., Caviedes M. A., et al. (2017). Assessing the role of endophytic bacteria in the halophyte Arthrocnemum macrostachyum salt tolerance. Plant Biol. 19, 249–256. 10.1111/plb.12521 [DOI] [PubMed] [Google Scholar]
- Naz I., Bano A., Ul-Hassan T. (2009). Isolation of phytohormones producing plant growth promoting rhizobacteria from weeds growing in Khewra salt range, Pakistan and their implication in providing salt tolerance to Glycine max L. Afr. J. Biotechnol. 8, 5762–5768. 10.5897/AJB09.1176 [DOI] [Google Scholar]
- Niehaus F., Bertoldo C., Kähler M., Antranikian G. (1999). Extremophiles as a source of novel enzymes for industrial application. Appl. Microbiol. Biotechnol. 51, 711–729. 10.1007/s002530051456 [DOI] [PubMed] [Google Scholar]
- Nikolic B., Schwab H., Sessitsch A. (2011). Metagenomic analysis of the 1-aminocyclopropane-1-carboxylate deaminase gene (acdS) operon of an uncultured bacterial endophyte colonizing Solanum tuberosum L. Arch. Microbiol. 193, 665–676. 10.1007/s00203-011-0703-z [DOI] [PubMed] [Google Scholar]
- Nunkaew T., Kantachote D., Nitoda T., Kanzaki H., Ritchie R. J. (2015). Characterization of exopolymeric substances from selected Rhodopseudomonas palustris strains and their ability to adsorb sodium ions. Carbohydr. Polym. 115, 334–341. 10.1016/j.carbpol.2014.08.099 [DOI] [PubMed] [Google Scholar]
- O'Brien J. A., Benková E. (2013). Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 4:451. 10.3389/fpls.2013.00451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanrewaju O. S., Glick B. R., Babalola O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33:197. 10.1007/s11274-017-2364-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhan F. (2016). Alleviation of salt stress by halotolerant and halophilic plant growth-promoting bacteria in wheat (Triticum aestivum). Braz. J. Microbiol. 47, 621–627. 10.1016/j.bjm.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T., Wu J., Fujii S. (2007). Effect of inoculation with a strain of Pseudomonas pseudoalcaligenes isolated from the endorhizosphere of Salicornia europea on salt tolerance of the glasswort. Soil Sci. Plant Nutr. 53, 12–16. 10.1111/j.1747-0765.2007.00098.x [DOI] [Google Scholar]
- Ozgur R., Uzilday B., Sekmen A. H., Turkan I. (2013). Reactive oxygen species regulation and antioxidant defence in halophytes. Funct. Plant Biol. 40, 832–847. 10.1071/FP12389 [DOI] [PubMed] [Google Scholar]
- Pallai R., Hynes R. K., Verma B., Nelson L. M. (2012). Phytohormone production and colonization of canola (Brassica napus L.) roots by Pseudomonas fluorescens 6-8 under gnotobiotic conditions. Can. J. Microbiol. 58, 170–178. 10.1139/w11-120 [DOI] [PubMed] [Google Scholar]
- Pang Q., Chen S., Dai S., Chen Y., Wang Y., Yan X. (2010). Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599. 10.1021/pr100034f [DOI] [PubMed] [Google Scholar]
- Parida A. K., Das A. B. (2005). Salt tolerance and salinity effects on plants: a review. Ecotoxicol. Environ. Saf. 60, 324–349. 10.1016/j.ecoenv.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Paul D., Nair S. (2008). Stress adaptations in a plant growth promoting rhizobacterium (PGPR) with increasing salinity in the coastal agricultural soils. J. Basic Microbiol. 48, 378–384. 10.1002/jobm.200700365 [DOI] [PubMed] [Google Scholar]
- Penrose D. M., Glick B. R. (2003). Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 118, 10–15. 10.1034/j.1399-3054.2003.00086.x [DOI] [PubMed] [Google Scholar]
- Pérez-Alfocea F., Albacete A., Ghanem M. E., Dodd I. C. (2010). Hormonal regulation of source–sink relations to maintain crop productivity under salinity: a case study of root-to-shoot signalling in tomato. Funct. Plant Biol. 37, 592–603. 10.1071/FP10012 [DOI] [Google Scholar]
- Piccoli P., Travaglia C., Cohen A., Sosa L., Cornejo P., Masuelli R., et al. (2011). An endophytic bacterium isolated from roots of the halophyte Prosopis strombulifera produces ABA, IAA, gibberellins A1 and A3 and jasmonic acid in chemically-defined culture medium. Plant Growth Regul. 64, 207–210. 10.1007/s10725-010-9536-z [DOI] [Google Scholar]
- Pierik R., Sasidharan R., Voesenek L. A. C. J. (2007). Growth control by ethylene: adjusting phenotypes to the environment. J. Plant Growth Regul. 26, 188–200. 10.1007/s00344-006-0124-4 [DOI] [Google Scholar]
- Pieterse C. M. J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Zamioudis C., Berendsen R. L., Weller D. M., Van Wees S. C. M., Bakker P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. 10.1146/annurev-phyto-082712-102340 [DOI] [PubMed] [Google Scholar]
- Porcel R., Zamarreño Á. M., García-Mina J. M., Aroca R. (2014). Involvement of plant endogenous ABA in Bacillus megaterium PGPR activity in tomato plants. BMC Plant Biol. 14:36. 10.1186/1471-2229-14-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadir M., Tubeileh A., Akhtar J., Larbi A., Minhas P. S., Khan M. A. (2008). Productivity enhancement of salt-affected environments through crop diversification. Land Degrad. Dev. 19, 429–453. 10.1002/ldr.853 [DOI] [Google Scholar]
- Qin S., Zhang Y.-J., Yuan B., Xu P.-Y., Xing K., Wang J., et al. (2014). Isolation of ACC deaminase-producing habitat-adapted symbiotic bacteria associated with halophyte Limonium sinense (Girard) Kuntze and evaluating their plant growth-promoting activity under salt stress. Plant Soil 374, 753–766. 10.1007/s11104-013-1918-3 [DOI] [Google Scholar]
- Qin Y., Druzhinina I. S., Pan X., Yuan Z. (2016). Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 34, 1245–1259. 10.1016/j.biotechadv.2016.08.005 [DOI] [PubMed] [Google Scholar]
- Qurashi A. W., Sabri A. N. (2012). Bacterial exopolysaccharide and biofilm formation stimulate chickpea growth and soil aggregation under salt stress. Braz. J. Microbiol. 43, 1183–1191. 10.1590/S1517-83822012000300046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi M., Barhoumi Z., Ksouri R., Abdelly C., Gharsalli M. (2007). Interactive effects of salinity and iron deficiency in Medicago ciliaris. C. R. Biol. 330, 779–788. 10.1016/j.crvi.2007.08.007 [DOI] [PubMed] [Google Scholar]
- Raghavan C., Ong E. K., Dalling M. J., Stevenson T. W. (2006). Regulation of genes associated with auxin, ethylene and ABA pathways by 2, 4-dichlorophenoxyacetic acid in Arabidopsis. Funct. Integr. Genomics 6, 60–70. 10.1007/s10142-005-0012-1 [DOI] [PubMed] [Google Scholar]
- Ramadoss D., Lakkineni V. K., Bose P., Ali S., Annapurna K. (2013). Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. SpringerPlus 2:6. 10.1186/2193-1801-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold B., Hurek T., Fendrik I., Pot B., Gillis M., Kersters K., et al. (1987). Azospirillum halopraeferens sp. nov., a nitrogen-fixing organism associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth). Int. J. Syst. Evol. Microbiol. 37, 43–51. [Google Scholar]
- Rodriguez R. J., Henson J., Van Volkenburgh E., Hoy M., Wright L., Beckwith F., et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2:404. 10.1038/ismej.2007.106 [DOI] [PubMed] [Google Scholar]
- Rodriguez R., Redman R. (2008). More than 400 million years of evolution and some plants still can't make it on their own: plant stress tolerance via fungal symbiosis. J. Exp. Bot. 59, 1109–1114. 10.1093/jxb/erm342 [DOI] [PubMed] [Google Scholar]
- Rohban R., Amoozegar M. A., Ventosa A. (2009). Screening and isolation of halophilic bacteria producing extracellular hydrolyses from Howz Soltan Lake, Iran. J. Ind. Microbiol. Biotechnol. 36, 333–340. 10.1007/s10295-008-0500-0 [DOI] [PubMed] [Google Scholar]
- Rozema J., Flowers T. (2008). Crops for a salinized world. Science 322, 1478–1480. 10.1126/science.1168572 [DOI] [PubMed] [Google Scholar]
- Rozema J., Schat H. (2013). Salt tolerance of halophytes, research questions reviewed in the perspective of saline agriculture. Environ. Exp. Bot. 92, 83–95. 10.1016/j.envexpbot.2012.08.004 [DOI] [Google Scholar]
- Rueda-Puente E., Castellanos T., Troyo-Diéguez E., Díaz de León-Alvarez J. L., Murillo-Amador B. (2003). Effects of a nitrogen-fixing indigenous bacterium (Klebsiella pneumoniae) on the growth and development of the halophyte Salicornia bigelovii as a new crop for saline environments. J. Agron. Crop Sci. 189, 323–332. 10.1046/j.1439-037X.2003.00051.x [DOI] [Google Scholar]
- Rueda-Puente E. O., Castellanos-Cervantes T., Díaz de León-Álvarez J. L., Preciado-Rangel P., Almaguer-Vargas G. (2010). Bacterial community of rhizosphere associated to the annual halophyte Salicornia bigelovii (Torr.). Terra Latinoamericana 28, 345–353. [Google Scholar]
- Ruppel S., Franken P., Witzel K. (2013). Properties of the halophyte microbiome and their implications for plant salt tolerance. Funct. Plant Biol. 40, 940–951. 10.1071/FP12355 [DOI] [PubMed] [Google Scholar]
- Ryu H., Cho Y.-G. (2015). Plant hormones in salt stress tolerance. J. Plant Biol. 58, 147–155. 10.1007/s12374-015-0103-z [DOI] [Google Scholar]
- Sadfi N., Chérif M., Fliss I., Boudabbous A., Antoun H. (2001). Evaluation of bacterial isolates from salty soils and Bacillus thuringiensis strains for the biocontrol of Fusarium dry rot of potato tubers. J. Plant Pathol. 83, 101–117. 10.4454/jpp.v83i2.1118 [DOI] [Google Scholar]
- Sadfi N., Cherif M., Hajlaoui M. R., Boudabbous A. (2002). Biological control of the potato tubers dry rot caused by Fusarium roseum var. sambucinum under greenhouse, field and storage conditions using Bacillus spp. isolates. J. Phytopathol. 150, 640–648. 10.1046/j.1439-0434.2002.00811.x [DOI] [Google Scholar]
- Sadfi-Zouaoui N., Essghaier B., Hajlaoui M. R., Fardeau M.-L., Cayaol J. L., Ollivier B., et al. (2008). Ability of moderately halophilic bacteria to control grey mould disease on tomato fruits. J. Phytopathol. 156, 42–52. 10.1111/j.1439-0434.2007.01329.x [DOI] [Google Scholar]
- Sáenz-Mata J., Palacio-Rodríguez R., Sánchez-Galván H., Balagurusamy N. (2016). Plant growth promoting rhizobacteria associated to halophytes: potential applications in agriculture, in Sabkha Ecosystems Volume V: The Americas, eds Khan M. A., Boër B., Ȫzturk M., Clüsener-Godt M., Gul B., Breckle S.-W. (Cham: Springer; ), 411–425. [Google Scholar]
- Sah S. K., Reddy K. R., Li J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. 10.3389/fpls.2016.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo R. K., Ansari M. W., Pradhan M., Dangar T. K., Mohanty S., Tuteja N. (2014). A novel Azotobacter vinellandii (SRI Az 3) functions in salinity stress tolerance in rice. Plant Signal. Behav. 9, 511–523. 10.4161/psb.29377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M. V., Bottini R., de Souza Filho G. A., Cohen A. C., Moreno D., Gil M., et al. (2014). Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant. 151, 359–374. 10.1111/ppl.12117 [DOI] [PubMed] [Google Scholar]
- Sanchez D. H., Siahpoosh M. R., Roessner U., Udvardi M., Kopka J. (2008). Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol. Plant. 132, 209–219. 10.1111/j.1399-3054.2007.00993.x [DOI] [PubMed] [Google Scholar]
- Sánchez-Porro C., Rafael R., Soto-Ramírez N., Márquez M. C., Montalvo-Rodríguez R., Ventosa A. (2009). Description of Kushneria aurantia gen. nov., sp. nov., a novel member of the family Halomonadaceae, and a proposal for reclassification of Halomonas marisflavi as Kushneria marisflavi comb. nov., of Halomonas indalinina as Kushneria indalinina comb. nov. and of Halomonas avicenniae as Kushneria avicenniae comb. nov. Int. J. Syst. Evol. Microbiol. 59, 397–405. 10.1099/ijs.0.001461-0 [DOI] [PubMed] [Google Scholar]
- Sandhya V., Ali S. Z., Venkateswarlu B., Reddy G., Grover M. (2010). Effect of osmotic stress on plant growth promoting Pseudomonas spp. Arch. Microbiol. 192, 867–876. 10.1007/s00203-010-0613-5 [DOI] [PubMed] [Google Scholar]
- Schubert S., Neubert A., Schierholt A., Sümer A., Zörb C. (2009). Development of salt-resistant maize hybrids: the combination of physiological strategies using conventional breeding methods. Plant Sci. 177, 196–202. 10.1016/j.plantsci.2009.05.011 [DOI] [Google Scholar]
- Sgroy V., Cassán F., Masciarelli O., Del Papa M. F., Lagares A., Luna V. (2009). Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl. Microbiol. Biotechnol. 85, 371–381. 10.1007/s00253-009-2116-3 [DOI] [PubMed] [Google Scholar]
- Shabala S. (2013). Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 112, 1209–1221. 10.1093/aob/mct205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S. N., Mackay A. S. (2011). Ion transport in halophytes. Adv. Bot. Res. 57, 151–187. 10.1016/B978-0-12-387692-8.00005-9 [DOI] [Google Scholar]
- Shaharoona B., Arshad M., Zahir Z. A. (2006). Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Lett. Appl. Microbiol. 42, 155–159. 10.1111/j.1472-765X.2005.01827.x [DOI] [PubMed] [Google Scholar]
- Shahzad R., Waqas M., Khan A. L., Asaf S., Khan M. A., Kang S.-M., et al. (2016). Seed-borne endophytic Bacillus amyloliquefaciens RWL-1 produces gibberellins and regulates endogenous phytohormones of Oryza sativa. Plant Physiol. Biochem. 106, 236–243. 10.1016/j.plaphy.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Sharma S. B., Sayyed R. Z., Trivedi M. H., Gobi T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2:587. 10.1186/2193-1801-2-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Kulkarni J., Jha B. (2016). Halotolerant rhizobacteria promote growth and enhance salinity tolerance in peanut. Front. Microbiol. 7:1600. 10.3389/fmicb.2016.01600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Hu H., Peng H., Wang W., Zhang X. (2013). Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genomics 14:271. 10.1186/1471-2164-14-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Takano T., Liu S. (2012). Isolation and characterization of novel bacterial taxa from extreme alkali-saline soil. World J. Microbiol. Biotechnol. 28, 2147–2157. 10.1007/s11274-012-1020-7 [DOI] [PubMed] [Google Scholar]
- Shrivastava P., Kumar R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. 10.1016/j.sjbs.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P. S., Agarwal P. K., Jha B. (2012). Improved salinity tolerance of Arachishypogaea (L.) by the interaction of halotolerant plant-growth-promoting rhizobacteria. J. Plant Growth Regul. 31, 195–206. 10.1007/s00344-011-9231-y [DOI] [Google Scholar]
- Siddikee M. A., Glick B. R., Chauhan P. S., Jong Yim W., Sa T. (2011). Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing 1-aminocyclopropane-1-carboxylic acid deaminase activity. Plant Physiol. Biochem. 49, 427–434. 10.1016/j.plaphy.2011.01.015 [DOI] [PubMed] [Google Scholar]
- Siddikee M. A., Chauhan P. S., Anandham R., Han G.-H., Sa T. (2010). Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J. Microbiol. Biotechnol. 20, 1577–1584. 10.4014/jmb.1007.07011 [DOI] [PubMed] [Google Scholar]
- Singh G., Jain S. (1982). Effect of some growth regulators on certain biochemical parameters during seed development in chickpea under salinity. Indian J. Plant Physiol. 25, 167–179. [Google Scholar]
- Singh R. P., Jha P. N. (2016). A halotolerant bacterium Bacillus licheniformis HSW-16 augments induced systemic tolerance to salt stress in wheat plant (Triticum aestivum). Front. Plant Sci. 7:1890. 10.3389/fpls.2016.01890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. P., Shelke G. M., Kumar A., Jha P. N. (2015). Biochemistry and genetics of ACC deaminase: a weapon to “stress ethylene” produced in plants. Front. Microbiol. 6:937 10.3389/fmicb.2015.00937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U. P., Sarma B. K., Singh D. P. (2003). Effect of plant growth-promoting rhizobacteria and culture filtrate of Sclerotium rolfsii on phenolic and salicylic acid contents in chickpea (Cicer arietinum). Curr. Microbiol. 46, 0131–0140. 10.1007/s00284-002-3834-2 [DOI] [PubMed] [Google Scholar]
- Slama I., Abdelly C., Bouchereau A., Flowers T., Savouré A. (2015). Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 115, 433–447. 10.1093/aob/mcu239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhanian H., Aghaei K., Komatsu S. (2011). Changes in the plant proteome resulting from salt stress: toward the creation of salt-tolerant crops? J. Proteomics 74, 1323–1337. 10.1016/j.jprot.2011.03.018 [DOI] [PubMed] [Google Scholar]
- Song J., Wang B. (2014). Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 115, 541–553. 10.1093/aob/mcu194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorty A. M., Meena K. K., Choudhary K., Bitla U. M., Minhas P. S., Krishnani K. K. (2016). Effect of plant growth promoting bacteria associated with halophytic weed (Psoralea corylifolia L) on germination and seedling growth of wheat under saline conditions. Appl. Biochem. Biotechnol. 180, 872–882. 10.1007/s12010-016-2139-z [DOI] [PubMed] [Google Scholar]
- Stepien P., Johnson G. N. (2009). Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte Thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiol. 149, 1154–1165. 10.1104/pp.108.132407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymańska S., Płociniczak T., Piotrowska-Seget Z., Złoch M., Ruppel S., Hrynkiewicz K. (2016). Metabolic potential and community structure of endophytic and rhizosphere bacteria associated with the roots of the halophyte Aster tripolium L. Microbiol. Res. 182, 68–79. 10.1016/j.micres.2015.09.007 [DOI] [PubMed] [Google Scholar]
- Takahashi T., Kakehi J.-I. (2009). Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 105, 1–6. 10.1093/aob/mcp259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Liu Y., Zhao L. (2010). Isolation, identification and characterization of ACC deaminase-containing endophytic bacteria from halophyte Suaeda salsa. Wei Sheng Wu Xue Bao 50, 1503–1509. [PubMed] [Google Scholar]
- Timmusk S., El-Daim I. A. A., Copolovici L., Tanilas T., Kännaste A., Behers L., et al. (2014). Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: enhanced biomass production and reduced emissions of stress volatiles. PLoS ONE 9:e96086. 10.1371/journal.pone.0096086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Singh P., Tiwari R., Meena K. K., Yandigeri M., Singh D. P., et al. (2011). Salt-tolerant rhizobacteria-mediated induced tolerance in wheat (Triticum aestivum) and chemical diversity in rhizosphere enhance plant growth. Biol. Fertil. Soils 47:907 10.1007/s00374-011-0598-5 [DOI] [Google Scholar]
- Tsukanova K. A., Chebotar V., Meyer J. J. M., Bibikova T. N. (2017). Effect of plant growth-promoting Rhizobacteria on plant hormone homeostasis. South Afr. J. Bot. 113, 91–102. 10.1016/j.sajb.2017.07.007 [DOI] [Google Scholar]
- Ullah S., Bano A. (2015). Isolation of plant-growth-promoting rhizobacteria from rhizospheric soil of halophytes and their impact on maize (Zea mays L.) under induced soil salinity. Can. J. Microbiol. 61, 307–313. 10.1139/cjm-2014-0668 [DOI] [PubMed] [Google Scholar]
- Ullrich W. R. (2002). Salinity and nitrogen nutrition, in Salinity: Environment - Plants - Molecules, eds Läuchli A., Lüttge U. (Dordrecht: Springer; ), 229–248. [Google Scholar]
- Upadhyay S. K., Singh D. P. (2015). Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 17, 288–293. 10.1111/plb.12173 [DOI] [PubMed] [Google Scholar]
- Upadhyay S. K., Singh D. P., Saikia R. (2009). Genetic diversity of plant growth promoting rhizobacteria isolated from rhizospheric soil of wheat under saline condition. Curr. Microbiol. 59, 489–496. 10.1007/s00284-009-9464-1 [DOI] [PubMed] [Google Scholar]
- Upadhyay S. K., Singh J. S., Singh D. P. (2011). Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 21, 214–222. 10.1016/S1002-0160(11)60120-3 [DOI] [Google Scholar]
- Uzilday B., Ozgur R., Sekmen A. H., Yildiztugay E., Turkan I. (2014). Changes in the alternative electron sinks and antioxidant defence in chloroplasts of the extreme halophyte Eutrema parvulum (Thellungiella parvula) under salinity. Ann. Bot. 115, 449–463. 10.1093/aob/mcu184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron J., Desbrosses G., Bouffaud M.-L., Touraine B., Moënne-Loccoz Y., Muller D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4:356. 10.3389/fpls.2013.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaddepalli P., Fulton L., Wieland J., Wassmer K., Schaeffer M., Ranf S., et al. (2017). The cell wall-localized atypical β-1, 3 glucanase ZERZAUST controls tissue morphogenesis in Arabidopsis thaliana. Development 144, 2259–2269. 10.1242/dev.152231 [DOI] [PubMed] [Google Scholar]
- Van Loon L. C., Bakker P., Pieterse C. M. J. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. 10.1146/annurev.phyto.36.1.453 [DOI] [PubMed] [Google Scholar]
- Ventosa A., Mellado E., Sanchez-Porro C., Marquez M. C. (2008). Halophilic and halotolerant micro-organisms from soils, in Microbiology of Extreme Soils, eds Dion P., Nautiyal C. S. (Berlin; Heidelberg: Springer; ), 87–115. [Google Scholar]
- Vitousek P. M., Howarth R. W. (1991). Nitrogen limitation on land and in the sea: how can it occur? Biogeochemistry 13, 87–115. [Google Scholar]
- Walters D. R., Ratsep J., Havis N. D. (2013). Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot. 64, 1263–1280. 10.1093/jxb/ert026 [DOI] [PubMed] [Google Scholar]
- Wang G.-L., Que F., Xu Z.-S., Wang F., Xiong A.-S. (2015). Exogenous gibberellin altered morphology, anatomic and transcriptional regulatory networks of hormones in carrot root and shoot. BMC Plant Biol. 15:290. 10.1186/s12870-015-0679-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chang L., Wang B., Wang D., Li P., Wang L., et al. (2013). Comparative proteomics of Thellungiella halophila leaves from plants subjected to salinity reveals the importance of chloroplastic starch and soluble sugars in halophyte salt tolerance. Mol. Cell. Proteomics 12, 2174–2195. 10.1074/mcp.M112.022475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Mopper S., Hasenstein K. H. (2001). Effects of salinity on endogenous ABA, IAA, JA, and SA in Iris hexagona. J. Chem. Ecol. 27, 327–342. 10.1023/A:1005632506230 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Kawahara K., Sasaki K., Noparatnaraporn N. (2003). Biosorption of cadmium ions using a photosynthetic bacterium, Rhodobacter sphaeroides S and a marine photosynthetic bacterium, Rhodovulum sp. and their biosorption kinetics. J. Biosci. Bioeng. 95, 374–378. 10.1016/S1389-1723(03)80070-1 [DOI] [PubMed] [Google Scholar]
- Yan N., Marschner P., Cao W., Zuo C., Qin W. (2015). Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 3, 316–323. 10.1016/j.iswcr.2015.11.003 [DOI] [Google Scholar]
- Yang J., Kloepper J. W., Ryu C.-M. (2009). Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14, 1–4. 10.1016/j.tplants.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Yasmin H., Bano A. (2011). Isolation and characterization of phosphate solubilizing bacteria from rhizosphere soil of weeds of khewra salt range and attock. Pak. J. Bot 43, 1663–1668. [Google Scholar]
- Yoon J.-H., Choi S. H., Lee K.-C., Kho Y. H., Kang K. H., Park Y.-H. (2001). Halomonas marisflavae sp. nov., a halophilic bacterium isolated from the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 51, 1171–1177. 10.1099/00207713-51-3-1171 [DOI] [PubMed] [Google Scholar]
- Yousfi S., Mahmoudi H., Abdelly C., Gharsalli M. (2007). Effect of salt on physiological responses of barley to iron deficiency. Plant Physiol. Biochem. 45, 309–314. 10.1016/j.plaphy.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Yu J., Chen S., Zhao Q., Wang T., Yang C., Diaz C., et al. (2011). Physiological and proteomic analysis of salinity tolerance in Puccinellia tenuiflora. J. Proteome Res. 10, 3852–3870. 10.1021/pr101102p [DOI] [PubMed] [Google Scholar]
- Yuan Z., Druzhinina I. S., Labbé J., Redman R., Qin Y., Rodriguez R., et al. (2016). Specialized microbiome of a halophyte and its role in helping non-host plants to withstand salinity. Sci. Rep. 6:32467. 10.1038/srep32467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir Z. A., Ghani U., Naveed M., Nadeem S. M., Asghar H. N. (2009). Comparative effectiveness of Pseudomonas and Serratia sp. containing ACC-deaminase for improving growth and yield of wheat (Triticum aestivum L.) under salt-stressed conditions. Arch. Microbiol. 191, 415–424. 10.1007/s00203-009-0466-y [DOI] [PubMed] [Google Scholar]
- Zahir Z. A., Shah M. K., Naveed M., Akhter M. J. (2010). Substrate-dependent auxin production by Rhizobium phaseoli improves the growth and yield of Vigna radiata L. under salt stress conditions. J. Microbiol. Biotechnol. 20, 1288–1294. 10.4014/jmb.1002.02010 [DOI] [PubMed] [Google Scholar]
- Zawoznik M. S., Ameneiros M., Benavides M. P., Vázquez S., Groppa M. D. (2011). Response to saline stress and aquaporin expression in Azospirillum-inoculated barley seedlings. Appl. Microbiol. Biotechnol. 90, 1389–1397. 10.1007/s00253-011-3162-1 [DOI] [PubMed] [Google Scholar]
- Zhang H., Kim M.-S., Sun Y., Dowd S. E., Shi H., Paré P. W. (2008). Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol. Plant-Microbe Interact. 21, 737–744. 10.1094/MPMI-21-6-0737 [DOI] [PubMed] [Google Scholar]
- Zhang J.-L., Shi H. (2013). Physiological and molecular mechanisms of plant salt tolerance. Photosyn. Res. 115, 1–22. 10.1007/s11120-013-9813-6 [DOI] [PubMed] [Google Scholar]
- Zhao S., Zhou N., Zhao Z.-Y., Zhang K., Wu G.-H., Tian C.-Y. (2016). Isolation of endophytic plant growth-promoting bacteria associated with the halophyte Salicornia europaea and evaluation of their promoting activity under salt stress. Curr. Microbiol. 73, 574–581. 10.1007/s00284-016-1096-7 [DOI] [PubMed] [Google Scholar]
- Zhou M., Chen W., Chen H., Wei G. (2012). Draft genome sequence of Mesorhizobium alhagi CCNWXJ12-2T, a novel salt-resistant species isolated from the Desert of Northwestern China. J. Bacteriol. 194, 1261–1262. 10.1128/JB.06635-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Zhao S., Tian C. Y. (2017). Effect of halotolerant rhizobacteria isolated from halophytes on the growth of sugar beet (Beta vulgaris L.) under salt stress. FEMS Microbiol. Lett. 364:fnx091. 10.1093/femsle/fnx091 [DOI] [PubMed] [Google Scholar]
- Zhu F., Qu L., Hong X., Sun X. (2011). Isolation and characterization of a phosphate-solubilizing halophilic bacterium Kushneria sp. YCWA18 from Daqiao Saltern on the coast of Yellow Sea of China. Evid. Based Complement. Altern. Med. 2011:615032. 10.1155/2011/615032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Shearer G., Kohl D. H. (1992). Proline fed to intact soybean plants influences acetylene reducing activity and content and metabolism of proline in bacteroids. Plant Physiol. 98, 1020–1028. 10.1104/pp.98.3.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarini P., Okurowska P. (2008). Effects of mycorrhizal colonization and fertilization on growth and photosynthesis of sweet basil under salt stress. J. Plant Nutr. 31, 497–513. 10.1080/01904160801895027 [DOI] [Google Scholar]