Summary
The evolutionary stability of synthetic genetic circuits is key to both the understanding and application of genetic control elements. One useful but challenging situation is a switch between life and death depending on environment. Here are presented “essentializer” and “cryodeath” circuits, which act as kill switches in Escherichia coli. The essentializer element induces cell death upon the loss of a bi-stable cI/Cro memory switch. Cryodeath makes use of a cold-inducible promoter to express a toxin. We employ rational design and a toxin/antitoxin titering approach to produce and screen a small library of potential constructs, in order to select for constructs that are evolutionarily stable. Both kill switches were shown to maintain functionality in vitro for at least 140 generations. Additionally, cryodeath was shown to control the growth environment of a population, with an escape frequency of less than 1 in 105 after ten days of growth in the mammalian gut.
Keywords: Containment, Library, Synthetic Biology, Lambda, CspA, Promoter, Toxin, Antitoxin, Cold Shock
Introduction
As synthetic biology makes advances in producing real world applications using genetically engineered micro-organisms, the issue of biological containment becomes increasingly important. Safeguards have previously been developed that require the addition of a survival factor to maintain viability in a bacterial population. Approaches include inducing an auxotrophy for a particular metabolite(Gallagher et al., 2015, Steidler et al., 2003), repressing the expression of an essential gene (Cai et al., 2015; Chan et al., 2015, Gallagher et al., 2015) or re-writing the genetic code to ensure dependency on a synthetic amino acid (Rovner et al., 2015). However, future applications of synthetic biology look beyond the confines of a laboratory. Strains have already been developed that can degrade inorganic polymers to reduce waste (Yoshida et al., 2016), provide sustenance or energy during space travel (Menezes et al., 2014, Montague et al., 2012, Way et al., 2011), or that colonize the mammalian gut to help diagnose and treat pathogenic infections (Kotula et al., 2014, Steidler, 2003). For these applications, a new form of containment is required for uncontrolled environments, one that does not require human monitoring or input.
Evolutionary instability is an inherent flaw in any biological control using a lethal effect to control environmental growth. Microorganisms rapidly evolve to remove any genetic element that reduces fitness (Knudsen & Karlstrom, 1991, Molin et al., 1993). “Kill switches” are defined as artificial systems that result in cell death under certain conditions. Several kill switches have been explored for containment of engineered microbes, but necessarily involve lethal genes that are induced in designated non-permissive conditions. Thus any kill switch with leaky, low level expression of a toxin in permissive conditions may be quickly disabled in rapidly growing microbes. Although a host of effective kill switches have been described, most evolve to lose functionality within days (Chan et al., 2015), or have no data supporting their longevity (Ahrenholtz et al., 1994, Caliando & Voigt, 2015, Callura et al., 2010, Djordjevic et al., 1997, Kong et al., 2008, Piraner et al., 2016). One exception is a multi-layered kill switch (Gallagher et al., 2015), which is stable for at least 110 generations but requires external supplementation of survival factors. Mutational loss of a microbial kill switch occurs in the context of an asexually reproducing population subject to “periodic selection”(Atwood, 1951, Maddamsetti et al., 2015 Novick & Szilard, 1950). In this regime, selection occurs at one locus at a time, and loss of a synthetic-biological device implies that it is the most deleterious element in a genome. To be evolutionarily stable, an engineered element can have a small fitness cost, provided that its selection coefficient is less than 1–10%, the typical fitness advantage associated with mutations that drive adaptive sweeps (Maddamsetti et al., 2015).
To create evolutionarily stable kill switches, we used an approach for varying the level of expression in a toxin/antitoxin system. Small rationally designed libraries were created with key bases modified in the promoter and RBS sites of both toxin and antitoxin. This system is broadly applicable in kill switch designs with diverse forms of regulation. We demonstrate this approach with two unrelated control systems: (1) the early termination of transgenics that lose function of another engineered module; and (2), the temperature-dependent termination of a transgenic microorganism that resides in the mammalian gut. In doing so, we explore a limited rationally designed space to achieve the optimized behavior of genetic circuits.
Results
General design strategy
An environmentally sensitive kill switch needs to be highly lethal in non-permissive conditions, but stable enough in permissive conditions to avoid conferring an evolutionary disadvantage. Achieving intended levels of protein repression can be more straightforward when a molecular ‘sponge’ is included to titrate low levels of an expressed toxin (Bläsi & Young, 1996; Nathan, Glenn, & Johan, 2016). In our system, this has been achieved through toxin/antitoxin pairings where the antitoxin sequesters toxin expressed during permissive states (Figure 1A), but does not prevent cell death after a non-permissive event. The toxin is placed under the control of an environmentally sensitive promoter, capable of strong repression in permissive conditions and high levels of expression in non-permissive conditions (Figure 1B). Even with highly stringent repression of a promoter, there will always be leaky expression of the intended gene product. For a toxin lethal enough to make an effective kill switch, any unintended expression would have a negative effect on cell survival and decrease evolutionary stability. For this study we utilized the type II toxin-antitoxin system CcdB/CcdA. CcdB is a lethal toxin for many Enterobacteriaceae (Wright, Stan, & Ellis, 2013). It targets the GyrA subunit of DNA gyrase, arresting it in the intermediate stage of action after a double-stranded break has been induced, resulting in cell death. (Madl et al., 2006).
Figure 1. Overview of kill switch design concept.
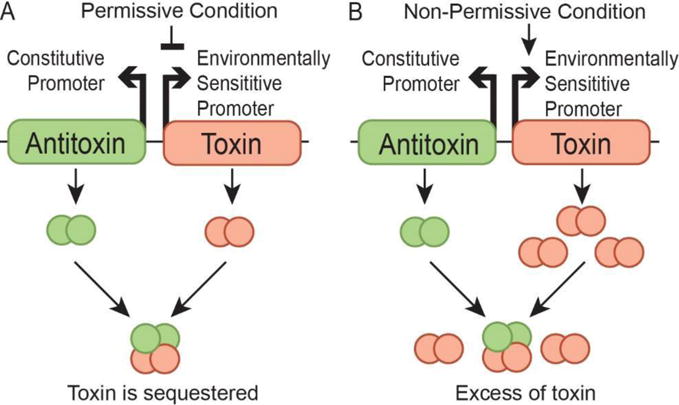
A) In permissive conditions the toxin is repressed. The antitoxin is expressed at a constitutive low level to accommodate for any leaky expression of the toxin. B) Upon a change of environment to non-permissive conditions the repression is lifted and toxin expression increases. The low level of antitoxin expression is no longer capable of preventing a lethal level of free toxic
We calculated the probability that expressed [toxin] > [antitoxin] for when the average number of transcription events for each gene varies from 1 to 30 (Figure S6A). To delineate between the mutating and non-mutating regions of the landscape (Figure S6B), we assume that in an asexually reproducing population, only the most strongly deleterious mutation will actually be selected against (Atwood, 1951; Koch, 1974). This results in conservation of sequences that may be mildly deleterious. Such mutations causing adaptive sweeps have selection coefficient of about 2-5% per generation (Koch, 1974; Maddamsetti et al., 2015; Novick & Szilard, 1950). We therefore defined the ‘non-selected’ region of the kill-switch landscape as when the element incorrectly enters the killing state less than 1% of the time.
An ‘Essentializer’ Element to Select for the Preservation of a Transgenic Cassette
In previous work, we have published a bistable genetic switch, the ‘memory element’, which is able to stably record and maintain an output from an exogenous signal (Kotula et al., 2014). The memory element is required to maintain its function for extensive periods of time, over which synthetic circuits are prone to mutation and deletion (Sleight et al., 2010). The memory element relies upon the bacteriophage lambda transcription factors cI and Cro. cI binds preferentially to the operator regions OR1 and OR2, whilst Cro binds preferentially to OR3. Binding of cI to OR1 and OR2 represses the expression of cro, and vice versa binding of Cro to OR3 represses the expression of cI.
The essentializer element kill switch was designed using the toxin-antitoxin system CcdB/CcdA, and would select for the presence of the memory element. For the essentializer element, the lambda phage operator sites were reordered and positioned over the lambda PR promoter such that binding of either cI or Cro would repress the expression of the toxin (Figure 2A and 2B), but in the absence of expression of both transcription factors the toxin would be expressed (Figure 2C). Furthermore, the binding sites OR1 and OR2 were modified to increase the binding affinity of cI (Sarai & Takeda, 1989, Takeda et al., 1989). A modified OR3 was placed downstream of the -10 region for the PR/toxin promoter in a position analogous to lacO within lacP; a protein binding to the major groove of these bases is expected to sterically prevent RNA polymerase binding (Murakami et al., 2002). The operators are separated by 6-7 bases, which should allow cooperative binding of cI to either OR1-OR2 or OR1-OR3 in this configuration (Ptashne et al., 1980).
Figure 2. Design and construction of the essentializer element kill switch.
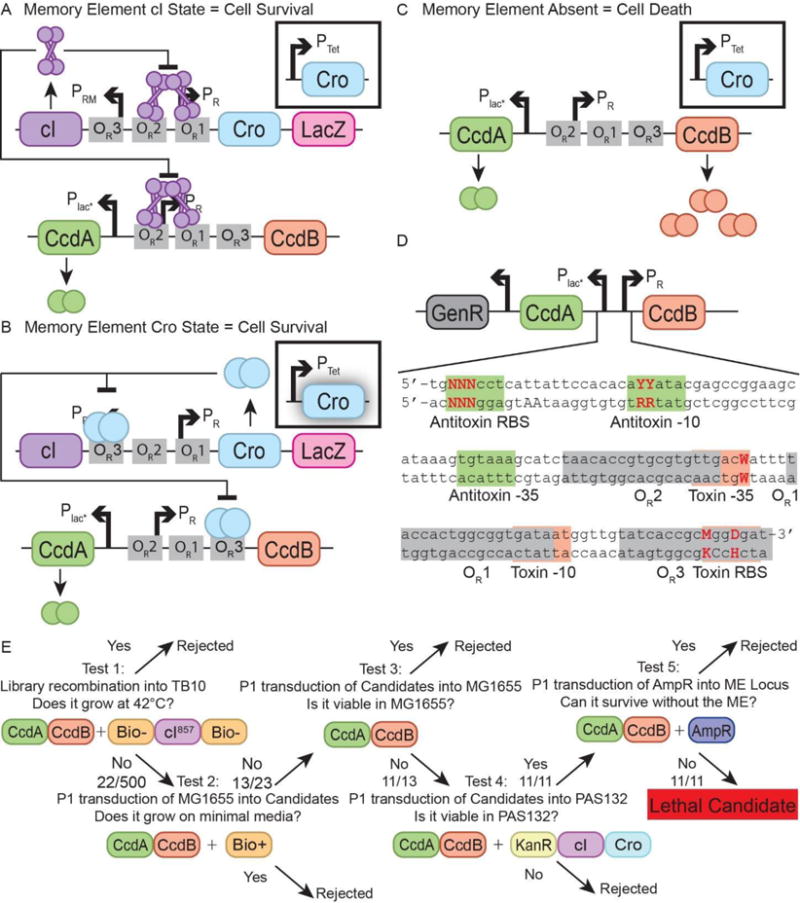
A) Expression of cI represses expression of cro and lacZ in the memory element (top cassette), whilst simultaneously repressing expression of ccdB in the essentializer element (bottom cassette). ccdA is expressed at a constitutive low level. B) Exposure to tetracycline leads to a pulse of expression of cro from the trigger element (boxed off). Expression of cro allows for the expression of lacZ, whilst simultaneously repressing cI and ccdB. C) Memory element is absent. Without repression from cI or cro, ccdB is expressed at lethal levels. D) The engineered region encompassing the regulatory DNA for ccdB and ccdA. Highlighted are loci that have a strong impact on expression level for both the toxin (red) and antitoxin (green), as well as operator binding sites for cI and Cro (grey). Key bases that have been varied are emphasized. N = A, C, T or G; Y = C or T, W = A or T; M = A or C; D = A, T or G. E) Overview of the screening process for identifying lethal essentializer element candidates. Displayed are the relevant genotype after each step has been completed and the fraction of candidates that passed a given screen. See also Figure S1.
A specific design goal was to quantitatively adjust the levels of toxin and antitoxin expression so that when cI and Cro proteins are absent, toxin expression is sufficient to kill the cell, but when either Cro or cI protein is present, the toxin is sufficiently repressed such that cell growth is not affected. This should be true even allowing for stochastic binding of repressor or Cro to the operators and expression of the antitoxin. To achieve this goal, a small rationally designed library of essentializer element candidates was constructed to introduce degeneracy at key locations in the regulatory region of the kill switch (Figure 2D). Three bases were varied in the RBS for the antitoxin, each with 4 different possible nucleotides; two bases were varied in the -10 region of the antitoxin promoter, each with 2 different possible nucleotides; one base was varied in the -35 region of the toxin promoter, with 2 different possible nucleotides; two bases were varied in the RBS for the toxin, one with 2 possible nucleotides and the other with 3. This resulted in a library size of 4ˆ3 × 3ˆ1 × 2ˆ4 = 3072 potential combinations. The promoter variations were designed based on (Mulligan et al., 1984), who estimated that, for example, the possible non- preferred bases in the 4th and 5th positions in the consensus −10 region (TATAAT) cause a mild and roughly equal decrement in promoter strength, so that incorporating only two variants would generate maximal functional variation while limiting the number of candidates to be manually screened. Variations in the toxin RBS were chosen to potentially preserve cI and Cro-binding at the overlapping OR3 element (Sarai & Takeda, 1989; Takeda et al., 1989). A construct was also made with a frame shift mutation in the toxin open reading frame to act as a non-lethal control (“EE toxin mutant”).
The essentializer element candidate pool yielded constructs that depended upon the memory element for survival. A series of tests was designed to select for candidates that survived in the permissive conditions (presence of memory element in the cI state) but failed to grow in non- permissive conditions (absence of memory element) (Figure 2E). In test 1, the essentializer element library was recombined into the lambda red expressing strain TB10. TB10 expresses cI857, a temperature sensitive mutant of cI (Caulcott & Rhodes, 1986) which is active at 30 °C but inactive at 42 °C, therefore repressing ccdB in a temperature dependent manner. Any recombinant that failed to grow at 42°C but survived at 30°C was considered potentially lethal. Test 2 used a P1 transduction of a wild type MG1655 lysate to remove cI857, allowing the strain to regain an operational biotin operon whilst removing the lambda red machinery. Successful growth on minimal media without biotin therefore implied a non-lethal essentializer element as cI857 would no longer be present to repress. Test 3 involved P1 transducing each essentializer element candidate into MG1655, relying on the distance between the loci of cI857 and the essentializer element to prevent them being co-transduced. With no source of cI or Cro in MG1655, if a candidate failed to produce any transductants, it was a candidate. Test 4 transduced the essentializer elements into PAS132, a strain containing the memory element, as well as an ATc responsive trigger element (Kotula et al., 2014) preset to the cI state. If a candidate produced transductants here, but had failed to produce transductants in test 3, it was a candidate. Candidates were sequenced across the antitoxin, toxin and regulatory region to determine the exact sequence (Figure S1 and Tables S1-S2), and termed candidates EE01-EE11. All other sequenced candidates were termed EE12- EE31.
The new candidate strains containing the essentializer element in a PAS132 background showed selection for maintaining the memory element. The fifth test inserted ampicillin resistance by P1 transduction into the candidate strains approximately 3000 bp distal to the memory element, likely replacing it. The more common transductant result of removing the memory element would result in only ampicillin resistance and the essentializer element remaining, but a rare transduction event could insert ampicillin resistance without removing the memory element and its associated kanamycin resistance cassette (Figure 3A). If a candidate contained a functional essentializer element, it would not be able to survive without the memory element. In this case only the rare transductant would be viable, whereas controls would have a majority of common transductants with a low proportion of rare transductants. In all 11 candidates, only the rare transductant result was recovered out of 52 transductants screened for growth on ampicillin and subsequently restreaked onto kanamycin (Figure 3B red bars). It should be noted that this assay only tests for the expression of either cI or Cro, and does not account for other potential failures of the memory element.
Figure 3. Analysis of essentializer element candidates.
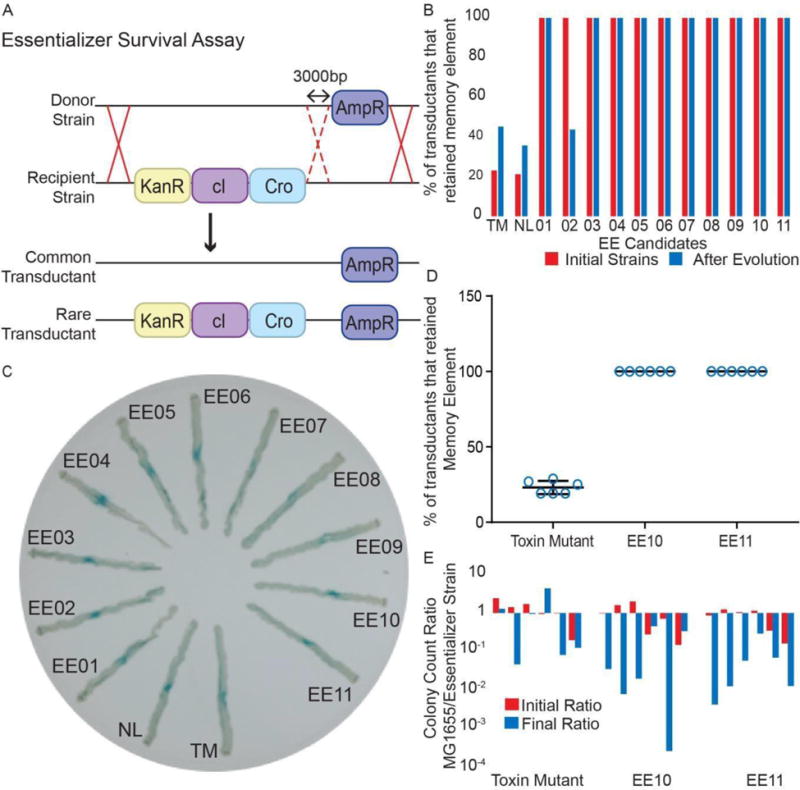
A) Transduction Assay. A donor strain with ampicillin resistance ~3000 bp from the memory element was used to remove the memory element. Due to the spacing between the loci of the cassettes a small subset of transductants would have both cassettes. B) Percentage of colonies that retained the memory element after the transduction assay (Figure 3A), both before (red) and after (blue) passaging for 140 generations. TM = toxin mutant and NL = EE non-lethal. C) Candidate essentializer strains were streaked on a plate spanning sub and super induction levels of ATc. Super induction of Cro near the center represses lacZ expression from the memory element. An intermediate expression level of Cro allows stable switching to the cro state and lacZ expression. Sub induction levels results in remaining in the cI state. D) Six biological repeats of the transduction assay (Figure 3A) for candidates EE10 and EE11. E) Six biological repeats of a competitive growth assay conducted over 70 generations to compare the fitness of parental (MG1655) and engineered bacterial strains. See also Tables S1-3.
The lethal activity of essentializer element candidates remained stable over extended growth periods in the cI state (Figure 3B). Candidates were passaged for approximately 140 generations and then the test outlined in Figure 3A was used to assay whether the memory element was still essential, by screening 52 transductants for each candidate. Only candidate EE02, when subcultured, failed to maintain its selection for preservation of the memory element (Figure 3B). Sequencing of isolated clones from EE01-EE10 subcultures revealed multiple mutations in the regulatory region of both the toxin and the antitoxin for candidates EE02 and EE06, but no such mutations in the other candidates. It appears that the mutations in the regulatory region of EE06 alters the expression rate of the toxin and antitoxin but does not prevent the lethal effect of the essentializer. This result might be expected if there is a selection for some level of toxin expression in the presence of the antitoxin. The nature of the essentializer limits the screening for potential escapees to the transduction-based assay (Figure 3A). However, it can be inferred that had an essentializer mutation arisen within the population of candidates EE1, EE3-5 and EE7-10, such mutations were not able to confer a growth advantage and become a significant proportion of the population within the 140 generation time frame. During 140 cell divisions, it is expected that at least one adaptive sweep may occur (Maddamsetti et al., 2015, Novick & Szilard, 1950).
Upon switching to the Cro state of the memory element, candidates maintained viability with the presence of Cro rather than cI. PAS132 contains a trigger element, allowing for the expression of cro under a tetracycline sensitive promoter Ptet (Kotula et al., 2014). If grown in the presence of anhydrotetracycline (ATc), the memory element switches from the cI state to the Cro state through expression of the Ptet cro trigger. All candidates were streaked on a gradient of ATc spanning sub- induction to super-induction levels. At high ATc concentrations, Cro binds to the OR1 and OR2 operator regions in addition to its preferred OR3, repressing itself in the memory element design. For this reason, switching to the Cro state can only occur in a narrow range of Cro concentrations. In Figure 3C we see narrow strips of blue for each candidate, indicating the strain has survived the memory element flipping from the cI state to the Cro state, and subsequently allowed expression of the reporter beta- galactosidase (LacZ).
The essentializer element candidates EE10 and EE11 were selected for more in depth assays of their evolutionary stability. Each candidate was passaged for approximately 140 generations, along with the toxin-defective control. The candidates were subsequently tested for transductional removal of the memory element (Figure 3A). For both EE10 and EE11, 52 of 52 colonies screened for each biological repeat maintained the memory element, compared with about 20% for the toxin mutant control (Figure 3D). In addition, co-cultures of roughly equal starting ratio of the essentializer candidates and the parental MG1655 strain were grown and passaged for approximately 70 generations. The ratio of essentializer strain to parental MG1655 was measured by comparing colony- forming units before and after growth across 6 biological repeats (Figure 3E). Strains EE10 and EE11 generally outgrew MG1655, implying that the essentializer element does not confer a significant growth disadvantage, and suggests that expression of certain ratios of toxin plus antitoxin could confer a selective advantage.
The sequences of the artificial regulatory regions were determined in essentializer elements EE01-EE11, as well as for several elements that failed at various points during the screening process. Overall, about 2/3 of the sequences had deletions of 1 or more bases in this region (Table S2). The remainder of ccdA and ccdB were not sequenced, and it is possible that additional mutations could have occurred in these genes or in the host genome. In five cases (the lethal candidates EE01 and EE07 as well as the 3 non-lethal candidates EE13, EE15 and EE27) the 5′-most operator (OR2) had a deletion and yet the strains are viable to different extents, indicating that loss of this operator is not required for tight repression and suggesting that the system is somewhat overdesigned. However, it is difficult to define a structure-activity relationship between the regulatory region sequences and the phenotypes observed, possibly because the widely varying ribosome binding sites may lead to changes in mRNA structure.
To test whether an antitoxin is valuable in these constructions, we designed versions of EE10 and EE11 with the antitoxin ORF removed. These constructs were genomically inserted into TB10 by recombineering. Eight and seven (respectively) recombinant colonies of the toxin-only EE10 and EE11 strains were picked and grown at 42°C to determine if they could survive without the repression from cI857. Two strains that did select for the presence of the memory element were passaged for approximately 140 generations, before being subjected to the transduction assay (Figure 3A). All biological repeats for both constructs retained their selection for the memory element after this period of growth (Figure S5).
Design and Construction of the Temperature Sensitive Kill Switch ‘Cryodeath’
To further test our approach to balancing toxin/ antitoxin expression, we designed a kill switch that would respond to temperature. This would allow confinement of an engineered bacterium to an environment with a defined temperature, such as the mammalian gut. The temperature sensitive regulatory region of cold shock protein A (PcspA) is modular and confers temperature-regulated expression of a protein of interest (Lee et al., 1994). PcspA contains a constitutive promoter that has a high rate of transcription at all temperatures (Yamanaka, 1999). This is followed by a long 5′ untranslated region (UTR) of 159 bp that adopts an unstable secondary structure at 37°C and is rapidly degraded by RNaseE (Mitta et al., 1997), but at lower temperatures forms a stable configuration that allows translation (Fang et al., 1997, Giuliodori et al., 2010). The cold-shock regulatory region also includes a downstream box (DB) element located within the first 13 amino acids of the open reading frame of cspA that enhances translation during cold shock by binding to the anti-DB sequence of 16S rRNA (Etchegaray & Inouye, 1999, Mitta et al., 1997) (Figure 4A).
Figure 4. Structure and modularity of PcspA.
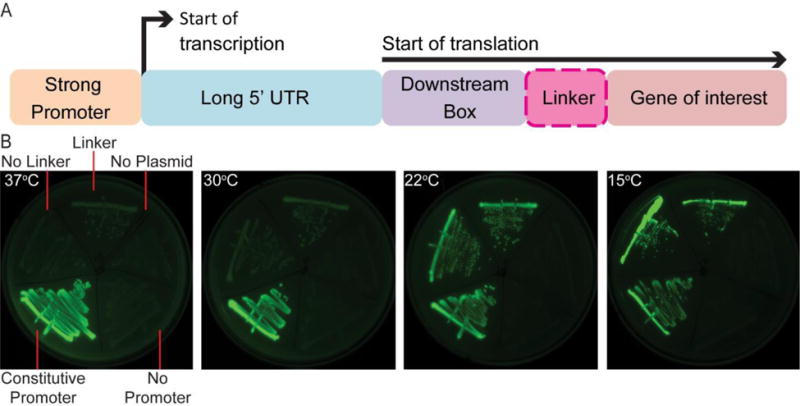
A) The regulatory region of cold shock protein A (CspA). B) Expression of GFP under PcspA at 37°C, 30°C, 22°C, and 15° C for 10 days. GFP expression level is shown with and without a linker, under PrpsL, with no promoter and with no plasmid present. Images are from the same picture. See also Figure S2.
We demonstrated the utility of this isolated regulatory region by inducing GFP in a temperature sensitive fashion. As it was unclear whether a particular gene of interest would be affected by an additional 13 amino acids on the amino end, we generated two versions of the regulatory region fused to GFP - one with a linker (N-GGGGS-C) between the truncated CspA and GFP designed to minimize interaction of the DB element, and one with no linker (Figure 4A). When assayed for expression at 37°C, no discernible expression was observed compared to negative controls. However, to a small extent at 30°C, and greater extent at room temperature (referred to as 22°C) and 15°C, the level of induction was visibly increased both with and without the linker (Figure 4B and S2), suggesting that the regulatory region functioned in a temperature sensitive manner.
Rational design was used to generate small libraries to introduce variation at key locations in the regulatory region of the toxin and antitoxin. The ccdB coding region was placed after the cspA regulatory region, both with and without a linker. In a similar design to the essentializer element, the ccdA coding region was placed after a modified, constitutive LacUV5 promoter (Plac*) (Malan & McClure, 1984) (Figures 5A and 5B). A Gentamycin resistance cassette was used for selection purposes. To achieve a range of expression levels for the toxin and antitoxin, ten bases were varied - three in the RBS of the antitoxin, two in the -10 promoter region of the antitoxin, one in the -35 promoter region of the toxin, two in the -10 promoter region of the toxin, and two in the RBS of the toxin (Figure 5C). Each varied position had the possibility of 2 different nucleotides, making a possible 2ˆ10 = 1024 different constructs. The bases chosen to be varied followed a similar logic to that of the essentializer element, generally with bases of less importance from within the -10 and -35 regions, according to previous published analysis of E. coli promoter and ribosome binding sites (Mulligan et al., 1984; Shultzaberger et al., 2001, Shultzaberger et al., 2007).
Figure 5. Design of the temperature sensitive kill switch cryodeath.
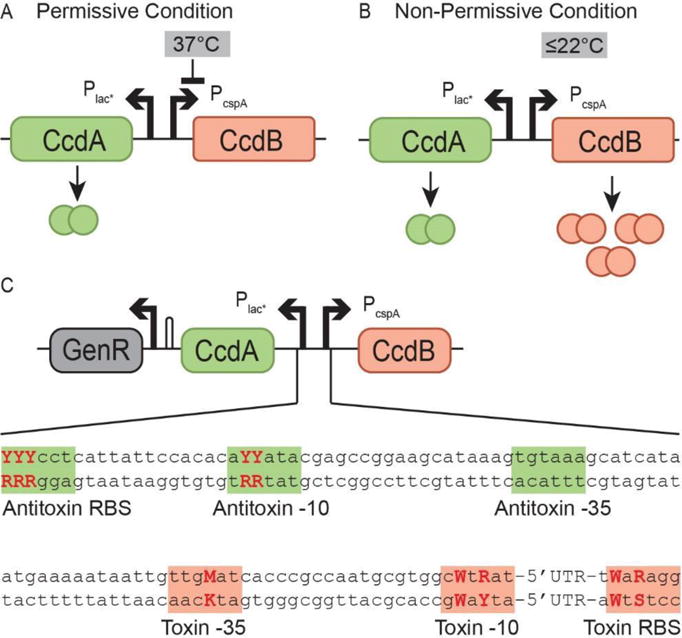
A) At 37 °C translation of CcdB is limited allowing cell survival. B) At colder temperatures, expression of CcdB increases, resulting in cell death C) The engineered region encompassing the regulatory DNA for CcdB and CcdA. Highlighted are segments that have a strong impact on expression level for both the toxin (red) and antitoxin (green). Varied bases are emphasized. Y = C or T; M = A or C; W = A or T; R = A or G. See also Figure S3.
To screen for cold-sensitive toxin induced death, E. coli strain Dh10β was transformed with the linear fragment libraries, using the lambda red genes expressed on plasmid pKD46 to enhance recombination. After an initial selection for gentamicin resistance, 26 unique candidates were identified; 2 with no linker and 24 with a linker. The 26 candidates were colony-purified on LB agar and tested on plates at the non-permissive temperatures of 30°C, 22° and 15°C as well as the permissive temperature, 37°C. Of the 26 candidates, ten failed to grow or grew poorly at <37°C, and were selected for further analysis (CD01-CD10). Non-lethal candidates were termed CD11-CD26. All except CD01 and CD11 contained a linker. As lethal candidates were identified both with and without a linker, the linker has no or little effect on the function of CcdB. CD12 was subsequently used as a negative control, referred to in this text as “CD non-lethal”. The candidates were sequenced across the antitoxin, toxin, and regulatory region, toxin and antitoxin open reading frames (Table S5). Although most candidates had retained the expected sequence across the regulatory region, only varying at the intended positions, candidate CD10 had a 9 bp deletion partially including and upstream of the toxin RBS (Figure S3). Modifying bases in this region has previously been noted to affect the stability of the CspA mRNA (Fang et al., 1997).
Evolutionarily stable circuits with the intended phenotype were identified. A survival assay comparing cfu at room temperature to that at 37°C was used to quantify the extent of population death, termed the survival ratio (Figure 6A). After an initial assay was made at room temperature, the candidates were passaged in permissive conditions for approximately 140 generations in LB before a second assay for cold-sensitivity was performed at room temperature. The pre- and post-survival ratios for each candidate were compared to determine which candidates induced the highest level of population death, and which candidates maintained their lethality after the opportunity for evolution (Figure 6B). CD01 and CD10 had low survival ratios of approximately 10−4 both before and after the period of growth, and were therefore chosen for further analysis. All candidates were sequenced across the antitoxin, toxin and regulatory region after the period of growth, with no mutations. For the candidates that have lost lethality, this implies that a mutation has occurred in another location in the genome to prevent the effect of CcdB.
Figure 6. Analysis of cryodeath cadidates.
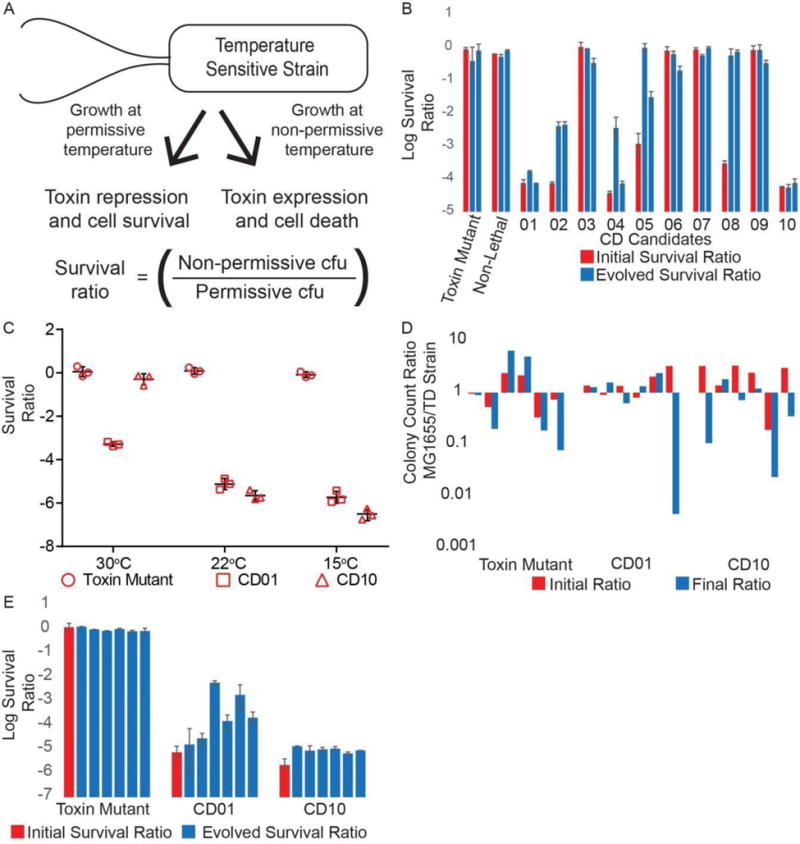
A) Survival assay used to test the extent of population termination at non-permissive temperatures. B) Survival ratio of the ten temperature sensitive candidates in DH10β (CD1-CD10) before and after 140 generations of growth at a permissive temperature. Data represents average of two technical repeats, with error bars showing range. Two biological repeats after a period of growth are shown. Non-Lethal = CD non-lethal. C) Survival ratio of candidates CD01 and CD10 in MG1655 at 30 °C, 22 °C, and 15 °C. Three technical repeats of each condition are shown. D) Six biological repeats of a competitive growth assay of 70 generations to compare the fitness of parental (MG1655) and engineered bacterial strains. E) Six biological replicates of survival ratio of candidates CD01 and CD10 in MG1655 after 140 generations, each biological replicate data point is the average of 3 technical replicates plotted with 3 technical repeats. Initial survival ratios are from the data collected for Figure 6C. See also Figure S4 and Table S5.
To identify what these modes of escape could be, we used whole genome sequencing to identify mutations that arose in the cryodeath strain CD08 after 140 generations. Two colonies were identified from separate passaging repeats, and were assayed to ensure they were not cold sensitive (Figure 6A). These were sequenced and compared to the genome of CD08 before the period of growth. Of the two strains, one had no noticeable alterations between the parent and evolved strain, and the second had a singlxe SNP in the 5′ UTR of the native cspA gene, as well as an approximately 500 bp deletion in the ORF of gtrS (Table S6). It is not immediately apparent what effect these mutations would have.
The cryodeath kill switch candidates were transferred to a different genetic background for use in the mammalian gut. After transfer by P1 transduction from DH10β into MG1655, a survival assay was used to measure lethality in both strains at 30 °C, 22 °C and 15 °C. In DH10β, both CD01 and CD10 induced no death compared to the toxin mutant control at 30 °C, a survival ratio ranging from 10−4 to 10−5 at room temperature and 10−5 at 15 °C (Figure S4). In MG1655, survival ratios were over an order of magnitude lower, 10−5 to 10−6 at 22 °C, and 10−6 for candidate CD10 at 15 °C (Figure 6C). Candidate CD01 also had a substantial drop in survival ratio at 30 °C, (10−3).
The cryodeath candidates showed no disadvantage in growth rate when compared to wild type MG1655. Co-cultures were grown in minimal media for approximately 70 generations. The population ratio of kill switch strain to MG1655 was measured by comparing cfu before and after growth across 6 biological repeats (Figure 6D). Across the repeats, it varied which strain gained an evolutionary advantage over the time period, implying that a spontaneous mutation independent of the kill switch was responsible for the advantage.
One cryodeath strain was stable after an extensive period of growth. An evolutionary stability experiment was repeated with 6 biological repeats of CD01, CD10 and the toxin mutant control. Again the candidates were grown for approximately 140 generations and then assayed at room temperature to determine their evolved survival ratio. CD10 maintained its survival ratio at around 10−5 after the period of growth across all 6 biological repeats. The survival ratio for CD01 decreased by at least an order of magnitude in four cultures (Figure 6E) suggesting that a certain proportion of the population had lost or modified the lethal phenotype. As the candidate displaying the most robust evolutionary stability, CD10 was chosen as the most desirable cryodeath construct. To test if the antitoxin was necessary to maintain stability for CD10, we attempted to construct a version with no antitoxin. However, any such attempt resulted in a frame shift appearing in the ccdB ORF and a non-cold sensitive strain (Figure S5 and Table S8).
To determine the escape modes of CD10, we isolated 5 different escapees from the survival assay (Figure 6A), conducted at 22 °C. Their genomes were sequenced and assayed for differences to the parent strain, CD10. Of the 5, one had a frame shift mutation in yedN, one had a large insertion in the 5′ UTR of ccdB, one had a 9 bp insertion between the -10 and -35 of the ccdB promoter and 2 had no discernable differences when compared to CD10 (Table S6). Except for the mutation in yedN, it is easily deduced how these mutations would have an effect on toxin expression at 22 °C. We have no definitive explanation for the strains seemingly lacking a genomic mutation (although an unstable tandem duplication (Roth, 1978) would be difficult to detect by whole-genome sequencing). However, we note that in this selection we isolated promoter mutants that may have partial activity, while when selecting in the absence of the antitoxin, we isolated mutations in the toxin coding sequence. These observations suggest that expression of the antitoxin in the absence of the toxin may be somewhat deleterious.
CD10 induced efficient and stable population death upon defecation from the mammalian gut. Streptomycin resistance was introduced to CD10 and the toxin mutant control in order to facilitate colonization of the mouse gut by P1 transducing a mutated rpsL from a spontaneously streptomycin resistant strain. CD10 and the toxin mutant negative control were each gavaged into 3 separate Balb C mice after 24 hours of streptomycin treatment, and fecal samples were collected before gavaging, 24 hours after gavaging and 10 days after gavaging. The survival assay (Figure 6A) was then carried out on cultures grown from these fecal samples. When compared to earlier in vitro experiments CD10 samples showed a similar level of lethality with a survival ratio of 10−5 across all mice and time points (Figure 7B). No bacteria were isolated on streptomycin plates from the sample collected before gavaging.
Figure 7. Testing cryodeath in vivo.
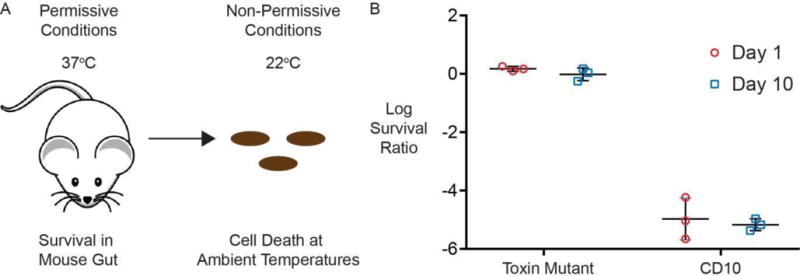
A) Containment of transgenic bacteria to mammalian gut by temperature sensitive induction of cryodeath. B) Survival assay of cultures grown from feces of 3 mice gavaged with toxin mutant and three mice gavaged with CD10 in MG1655, both 1 day and 10 days after gavaging. Each data point is a separate biological repeat, made from the average of three technical repeats.
Discussion
We have constructed two inducible toxin/antitoxin kill switch systems for controlling the environment or conditions in which a genetically engineered strain of E. coli can survive (Figure 1). The first is the “essentializer” element, a kill switch that links cell survival to the presence of the memory element, an engineered genetic circuit that is normally not essential (Figures 2 and 3). The second, “cryodeath,” responds to environmental temperature, allowing growth at 37°C but resulting in a survival ratio of less than 10−5 at 22°C and below (Figures 5, 6, and 7). Using these two systems, it is possible to limit the growth conditions of transgenic E. coli without the need for human monitoring or input.
The design criteria for these kill switches involve multiple phenotypes, including a lack of detrimental effect on viability and growth, which collectively do not lend themselves to high- throughput screening. Our approached therefore relied on rational design and construction of small libraries whose members could be extensively tested. Promoter and ribosome binding site expression levels were varied to create a pool of candidates with different levels of toxin and antitoxin production. Overlap extension PCR or Gibson assembly with degenerate primers were used to vary primarily those bases expected to modulate the quantitative level of gene expression but not affect regulation. This method was chosen over random mutagenesis technique such as error prone PCR, as the limited pool of potential candidates could be screened in its entirety if needed, and because an intense random mutagenesis would likely create primarily loss-of-function mutations that could mask mutations of interest. In addition, as many of the possible nucleotides have a well characterized effect on expression (Mulligan et al., 1984, Shultzaberger et al., 2001, Shultzaberger et al., 2007), we could limit the potential candidates to those that were more likely to perform within the desired range of expression. Another alternative approach, use of wild- type promoters with varying strengths, was not chosen because such promoters may be regulated in an unknown manner, and because the chosen approach was technically simpler, faster, and less expensive. The success of modifying two different regulatory regions indicates this technique could be applied to any regulatory region where the influence of each individual base can be identified with a reasonable degree of accuracy, i.e. any promoter in E. coli or other well characterized organisms. Due to the error prone nature of oligo synthesis, many of the constructs sequenced had mutations likely to affect regulatory function, including the most stable cryodeath candidate CD10.
Both kill switches maintain functionality after at least 140 generations (20 passages) in vitro (Figure 3 and 6). The cryodeath CD10 element maintained 100% of its functionality over this time (Figure 6). In addition, CD10 maintained its lethality when used in vivo to limit bacterial growth after expulsion from the mammalian digestive tract in a mouse model. Under these conditions, cryodeath remained functional for at least 10 days (Figure 7). Of the 10 cryodeath strains that originally showed a temperature sensitivity at 22 °C, only 1 proved to be stable after an opportunity to evolve as opposed to 10 out of 11 essentializer candidates. This can likely be attributed to two causes; a lower basal expression level from PR compared to PcspA, and the greater opportunity for the unstable essentializer candidates to lose their lethality throughout the more extensive screening process. These observations indicate that the kill switches designed here are likely to be genetically stable. In this context, it is important to note that the phenomenon of “periodic selection” has a conserving effect on evolution in asexual populations (Atwood, 1951; Novick & Szilard, 1950). Essentially, for any given environment, favorable mutations will arise in such a population and the mutants will overtake the parental population. Based on inspection of data from long-term cultures (Maddamsetti et al., 2015, Novick & Szilard, 1950), a typical selection coefficient for such an overtaking mutation is about 1-10%. If a kill switch is negatively selected with a coefficient of 0.5%, then in a long-term culture of bacteria containing a kill switch, there will arise unlinked mutations with a selection coefficient of at least 1% and mutations in the kill switch with a selection coefficient of 0.5%; bacteria with the unlinked mutation, which will in general have a non-mutant kill switch, will overtake the population and thus the kill switch will be preserved even though it is slightly deleterious.
Our working hypothesis is that the presence of the antitoxin increases the stability of the system by negating the deleterious evolutionary pressure of leaky toxin expression. To test this, we designed constructs identical to our best performing kill switches (EE10, EE11 and CD10) except with the antitoxin ORF removed. The CD10 homolog failed to yield a stable cold-sensitive strain (Figure S5 and Table S8) implying that basal level of toxin expression at permissive temperatures prevents E. coli survival without the antitoxin. For the essentializer homologs, while constructing a strain with the memory element and a toxin-only cassette over 25% of recombinants tested unable to select for the presence of a cI repressor (Table S4). However, once the toxin only cassettes were combined with the memory element, they were stable over an equal time length to that tested for EE10 and EE11. It is likely that cI857 has a lower strength of repression than its wild type counterpart (Angeles, 1976) and whilst the antitoxin does not appear necessary for EE10 and EE11 to maintain their functionality, it was likely beneficial during construction when repression is less than optimal.
The cryodeath system consistently reported a survival ratio of at least 10−5 at 22°C (Figure 6 and 7) and can reach as low a survival ratio as 10−6 at 15°C (Figure 6C). PcspA has been shown to induce a 16 fold increase in expression as high as 27°C, indicating the potential for a lethal level of induction at higher temperatures (Hoynes-O’Connor 2017). Previously published kill switches using the toxin CcdB as the sole source of cell death in E. coli have a survival ratio of approximately 10−3 ((Chan et al., 2015; Piraner et al., 2016). This disparity in escape frequency can be partially explained by the fact that the target of CcdB, the GyrA subunit of DNA gyrase, is one of the genes upregulated by the cold shock response (Jones 1992). In addition, the fact that our approach yielded a kill switch that induced a higher frequency of population death can be attributed to the screening of a rationally designed small library.
An important design requirement is that kill switches should not be deleterious to growth of the host organism in permissive conditions. This means that stochastic variation in expression of the toxin and antitoxin genes should not allow greater expression of toxin even in rare circumstances. Natural toxin-antitoxin systems avoid spontaneous deletion because the antitoxin is much less stable than the toxin, so a stochastic decrease in antitoxin expression will not be significantly averaged over time. The antitoxin promoter used here is based on the lac promoter, which initiates transcription about once per minute when induced (Bremer, 1975), corresponding to an average of 20 events/generation in rapidly growing cells. A cell will have a 2-fold reduction in antitoxin expression in about 1% of all such periods (P<=10) when the average is 20), so that if the toxin is regulated by a promoter with an induction ratio of >5, our mutation-and-screening approach should yield satisfactory kill-switches with a high probability (Figure S6).
A variety of promoters responding to different environmental conditions have been identified in E. coli, and our system could be modified to include an alternative mode of death induced by pH (Chou, 1995), oxygen level (Cotte, 1990), or nutrient abundance (Yansura & Henner, 1990). Combined with an independent toxin-antitoxin system (Yamaguchi & Inouye, 2011), the escape frequency of the system would be decreased whilst maintaining a high level of evolutionary stability.
Star Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Pamela Silver (pamela_silver@hms.harvard.edu)
Experimental model and subject details
E. coli K12 strain MG1655
Used as the bases for all strains, except for the initial screen of cryodeath constructs that was conducted in DH10β. Maintained using established protocols for E. coli. Strains that contained cryodeath candidates were maintained at a constant temperature of 37 °C unless being assayed, or kept as glycerol stocks at −80 °C.
E. coli K12 strain TB10
A derivative of MG1655, with a large section of the lambda prophage genome inserted into biotin operon. Used to recombine the essentializer library. cI has been mutated to the temperature sensitive variant, cI857, allowing for temperature sensitive induction of the lambda red genes. Maintained using established protocols for E. coli, except all growth was kept at 30 °C unless being assayed.
E. coli strain DH10β containing plasmid pKD46
E. coli strain used to recombine cryodeath library. Maintained using established protocols for E. coli. Before cryodeath integration, strain was maintained at 30 °C to allow for pKD46 propogation. After cryodeath integration, strains were maintained at 37 °C unless being assayed, or kept as glycerol stocks at −80 °C.
Mouse Strain BalbC
Approval for animal work came from the Harvard Medical School IACUC under protocol 04966. Seven week old female BalbC mice from Charles River Laboratories were given two weeks to acclimatize to the facility upon delivery. Three mice were used to test CD10 and three used for the negative control, kept in two respective cages. Maintained using the standard conditions of the facility.
Method Details
Media and growth conditions
Unless otherwise specified LB media with relevant antibiotic was use for all growth conditions. For the competitive growth assay, screening for lethality of the essentializer candidates and for the initial evolutionary screen of the essentializer candidates, M9 minimal media supplemented with 1 mM MgSO4, 1 μg/ml thiamine hydrochloride, 0.4% w/v glucose, 100 μMCaCl2 was used. For all evolution experiments and survival assays no antibiotic were used (except for streptomycin in the fecal sample survival assay). For mouse survival assays, MacConkey lactose with streptomycin was used. For plating the competitive growth assay, MacConkey rhamnose without antibiotic was used to differentiate between wild type MG1655 and strains with kill switches inserted into the rhamnose operon. Unless otherwise stated, in all instances of antibiotic use the following concentrations were employed: Amp 100 μg/ml, Kan 50 μg/ml, Gent 10 μg/ml, Strep 100ug/ml.
Cold-sensitive reporter plasmid design
The PcspA regulatory region was amplified using PCR from the K12 genome using primers TS3 and TS4 or TS6 to add homology to the plasmid pUA66, and incorporate the GGGGS linker in the case of TS6. pUA66 GFP was linearized by PCR using primers TS1 and TS2 and combined with PcspA promoters using a NEB Gibson assembly kit according to the manufacturers guidelines, to create plasmids with temperature sensitive expression of GFP.
Construction of kill switch libraries
All primer sequences are given in Table S7. To construct essentializer variants (Figure 2D) a cassette was ordered as a Gblock from Integrated DNA Technologies with the sequence “original EE sequence” (Table S1). The degenerate oligo FS1 as well as primers FS2, FS3 and FS4 were used to amplify the cassette in two parts, which were subsequently combined using overlap extension PCR and amplified with FS3 and FS4. For the cryodeath kill switch, the degenerate primers TS12 and TS13 or TS18 were used to amplify PcspA with and without a linker respectively. Primers TS14-TS17 were used to amplify “original EE sequence” in two parts, which were subsequently combined with the degenerate PcspA amplicons using Gibson assembly and amplified using TS14 and TS17. In initial experiments, we found that the stitched product appeared to be correct based on gel electrophoresis, but failed to produce transformants upon recombineering (see below). We hypothesized that during the stitching amplification, the amount of DNA product exceeded the dNTPs and/or primers, with the result that the final PCR cycles simply involved denaturation and reannealing of full-length single strands, which would in general contain numerous mismatches. Upon transformation and recombination into the genome, such mismatches would be expected to undergo mismatch repair in a strand-independent manner, resulting in double-stranded breaks as gap-repairing polymerases meet. To avoid this problem, the reaction was passed through a Zymo Clean and Concentrator™ kit before being added to fresh PCR reagents for an additional cycle with primers FS3 and FS4 or TS14 and TS17 for the essentializer and cryodeath libraries respectively. This ensured that the predominant PCR product was a matching double stranded helix. After this modification, the frequency of transformant isolation increased dramatically.
P1 transduction
P1 lysates conducted according to previously published methods (L. C. Thomason, Costantino, & Court, 2007) with lysate of greater than 109 pfu. Overnight cultures of donor strains were grown at relevant temperatures in LB, before being back diluted into 50 fold into 5 ml of LB supplemented with 0.2% w/v glucose, 20 mM MgCl2 and 5 mM CaCl2. After approximately 45 minutes of growth at the relevant temperature, between 10 and 100 ul of high titre (pfu > 109) lisate was added and the culture was grown until lysis had occurred. Lysates were then centrifuged at 4000 rpm for 5 minutes and the supernatant filtered using a 0.2 micron filter. Lysates were stored for up to 6 months at 4°C.
Recombineering
For both kill switches, electrocompetent cells were prepared for transformation using previously published methds (L. Thomason et al., 2007). The degenerate essentializer library was transformed into the recombinant strain TB10 that had been induced for 15 minutes in a 42°C water bath with manual shaking every two minutes. For the cryodeath degenerate library, the recombinant plasmid pKD46 (Datsenko & Wanner, 2000) was transformed into Thermo Scientific MAX Efficiency DH10β electrocompetent cells. Cells were induced for 2-3 hours in the presence of 10 mM L-arabinose. Approximately 100 ng of DNA was combined with 50 μl of cells in a 0.1 cm cuvette, and electroporated using EC1 setting on a Biorad Micropulser. Cells were recovered in SOC medium for one hour before being spread on the relevant antibiotic plate.
Temperature sensitive survival assay
All colonies from the recombination of the two temperature sensitive libraries (with and without a linker) were grown at 37 °C, 30 °C, 22 °C and 15 °C. 5ml overnight cultures were made at 37 °C of all samples from the 37 °C plate, and combined 1:1 with 50% glycerol at 37 °C before being immediately transferred to a −80°C freezer. Unless being assayed for growth at lower temperatures, all subsequent experiments involving temperature sensitive strains were prepared in a constant 37 °C environment. For the strains that showed variation of growth at different temperatures, the glycerol stocks were used to make overnight cultures at 37 °C in 5ml of LB. This was diluted by a factor of 10 into 4 ml of LB in a 15 ml culture tube, and grown at 37 °C until ~OD600 1.0 was reached. Serial dilutions of the cultures were made and plated onto a number of LB plates corresponding to the number of temperatures being assayed. Separate plates were then incubated for up to 15 days at various temperatures, and the cfu at each temperature compared to the cfu at 37 °C was termed the survival ratio.
Testing evolutionary stability of kill switch strains
For the essentializer candidates, glycerol stocks were made of the 11 candidates that proved lethal from the extensive screening outlined in Figure 2E. These glycerol stocks were used to inoculate 2 ml of M9 minimal media without antibiotic. After 24 hours of growth, 20 μl was used to inoculate 2 ml of M9 minimal media. This was repeated for 20 passages. A P1 transduction was then used to potentially replace the memory element with ampicillin resistance (Figure 3A). Essentially, an ampicillin-resistance cassette was placed in the mhpC ORF, such that in a typical transduction, the ampicillin resistance and kanamycin resistance markers are about 80% linked. For each candidate (EE01-EE10), 52 ampicillin-resistant transductants were streaked onto ampicillin/kanamycin plates to determine whether the memory element was still present. If 100% of transductants retained the memory element, it was deemed that the essentializer was still producing a selective pressure to keep the memory element.
Competitive Growth Assay
Overnight cultures of MG1655, Toxin Mutant, EE10, EE11, CD01 and CD10 were grown to stationary phase. 100ul of each culture was then used to inoculate M9 minimal media, and glycerol stocks were made of this initial inoculation. Cultures were passaged once every 24 hours diluting 1:1000 into M9 minimal media. After 8 days, glycerol stocks were made of the final culture. Glycerol stocks were then defrosted and plated directly onto MacConkey Rhamnose plates. As the two kill switches had been inserted into the Rhamnose operon, they could be distinguished as making white colonies on Rhamnose MacConkey compared to red colonies produced by MG1655. Cfu were used to determine the ratio of kill switch strain to MG1655.
Testing Temperature Sensitive Kill Switch in the Mammalian Gut
Approval for animal work came from the Harvard Medical School IACUC under protocol 04966. Seven week old female BalbC mice from Charles River Laboratories were given two weeks to acclimatize to the facility upon delivery. 500 ug/ml of streptomycin was added to their drinking water and maintained throughout the experiment. The Toxin mutant and strain CD10 were gavaged at a concentration of approximately 108 cfu/ml, each into 3 separate mice. Feces was collected between 2-6pm from day 0 (immediately before gavaging) to day 10, and immediately placed on dry ice before being transferred to a −80 °C freezer. Feces was resupended in PBS by shaking at 4 °C for 1 hr, then 100 μl was used to inoculate 5ml of MacConkey lactose broth with streptomycin and grown overnight. The following day this overnight culture was diluted by a factor of 10 into MacConkey lactose and grown to approximately OD 1.0. Cultures were then serial diluted and plated at 37 °C and room temperature to compare cfu at permissive and non-permissive temperatures.
Quantification and statistical analysis
For all graphs, error bars represent the range of data collected. For survival assays, data points were omitted if contamination was observed, or if the cfu from several different dilutions was equivalent, indicating a range of values for a particular cultures cfu spanning multiple orders of magnitude. In such cases a dilution error is suspected, and alternative technical repeats were used instead.
Supplementary Material
Acknowledgments
We thank Georg Gerber for help with building a theoretical model for kill switch design. We thank Sarah Boswell for guidance with preparing Miseq samples for whole genome sequencing. We thank Sean Wilson from the laboratory of Prof. Ethan Garner for help with image analysis.
Funding:
This work was supported by Defense Advanced Research Projects Agency Grant HR0011-15-C-0094 and funds from the Wyss Institute for Biologically Inspired Engineering. FS acknowledges funding from NIH training grant [5T32GM007598].
Footnotes
Contributions:
Conceptualization, F.S. and J.W.; Investigation, F.S., L.B., S.O., E.R., and J.O.; Writing – Original Draft, F.S.; Writing – Review & Editing, F.S., J.W., P.S., L.B., and J.O.; Funding Acquisition, P.S. and J.W.
References
- Ahrenholtz I, Lorenz MG, Wackernagel W. A conditional suicide system in Escherichia coli based on the intracellular degradation of DNA. Applied and Environmental Microbiology. 1994;60(10):3746–3751. doi: 10.1128/aem.60.10.3746-3751.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles L. Heat-sensitive DNA-binding Activity of the cI Product of Bacteriophage Lambda. 1976;302:299–302. doi: 10.1007/BF00701254. [DOI] [PubMed] [Google Scholar]
- Atwood BYKC. strain 15. 1951:146–155. [Google Scholar]
- Bläsi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Molecular Microbiology. 1996;21(4):675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- Bremer H. Analysis of Enzyme Induction in Bacteria. 1975;152:243–254. doi: 10.1042/bj1520243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Agmon N, Choi WJ, Ubide A, Stracquadanio G, Caravelli K, Boeke JD. Intrinsic biocontainment: multiplex genome safeguards combine transcriptional and recombinational control of essential yeast genes. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(6):1803–8. doi: 10.1073/pnas.1424704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliando BJ, Voigt CA. Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nature Communications. 2015;6:1–10. doi: 10.1038/ncomms7989. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ. Physiology Using Synthetic Riboregulators. Proceedings of the National Academy of Sciences. 2010;107(36):15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulcott CA, Rhodes M. Temperature-induced synthesis of recombinant proteins. Trends in Biotechnology 1986 [Google Scholar]
- Chan CTY, Lee JW, Cameron DE, Bashor CJ, Collins JJ. “Deadman” and “Passcode” microbial kill switches for bacterial containment. Nature Chemical Biology. 2015;12(2):82–86. doi: 10.1038/nchembio.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CH, Aristidou aa, Meng SY, Bennett GN, San KY. Characterization of a pH- inducible promoter system for high-level expression of recombinant proteins in Escherichia coli. Biotechnology and Bioengineering. 1995;47(2):186–92. doi: 10.1002/bit.260470210. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Chepuri V, Gennis RB, Gunsalus RP. Cytochrome o (cyoABCDE) and d (cydAB) Oxidase Gene Expression in Escherichia coli Is Regulated by Oxygen, pH, and the fnr. Gene Product. 1990;172(11):6333–6338. doi: 10.1128/jb.172.11.6333-6338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic GM, Sullivan DJO, Walker SA, Conkling MA, Carolina N. A Triggered-Suicide System Designed as a Defense against Bacteriophages. 1997;179(21):6741–6748. doi: 10.1128/jb.179.21.6741-6748.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Inouye M. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. Journal of Biological Chemistry. 1999;274(15):10079–10085. doi: 10.1074/jbc.274.15.10079. [DOI] [PubMed] [Google Scholar]
- Fang L, Jiang W, Bae W, Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37° C by mRNA stabilization. Molecular Microbiology. 1997;23(2):355–64. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- Gallagher RR, Patel JR, Interiano AL, Rovner AJ, Isaacs FJ. Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Research. 2015;43(3):1945–54. doi: 10.1093/nar/gku1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliodori AM, Di Pietro F, Marzi S, Masquida B, Wagner R, Romby P, Pon CL. The cspA mRNA Is a Thermosensor that Modulates Translation of the Cold-Shock Protein CspA. Molecular Cell. 2010;37(1):21–33. doi: 10.1016/j.molcel.2009.11.033. [DOI] [PubMed] [Google Scholar]
- Hoynes-O’Connor A, Shopera T, Hinman K, Creamer JP, Moon TS. Enabling complex genetic circuits to respond to extrinsic environmental signals. Biotechnology and Bioengineering. 2017;9999(xxx):1–6. doi: 10.1002/bit.26279. [DOI] [PubMed] [Google Scholar]
- Jones PG, Krah R, Tafuri SR, Wolffe AP. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J Bacteriol. 1992;174(18):5798–5802. doi: 10.1128/jb.174.18.5798-5802.1992. Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen SM, Karlstrom OH. Development of efficient suicide mechanisms for biological containment of bacteria. Applied and Environmental Microbiology. 1991;57(1):85–92. doi: 10.1128/aem.57.1.85-92.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL. The pertinence of the periodic selection phenomenon to prokaryote evolution. Genetics. 1974;77(1):127–142. doi: 10.1093/genetics/77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Wanda S, Zhang X, Bollen W, Tinge SA, Roland KL, Curtiss R. Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. 2008;105(27):9361–9366. doi: 10.1073/pnas.0803801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, Way JC, Silver PA. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(13):4838–43. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Xie A, Jiang W, Etchegaray JP, Jones PG, Inouye M. Family of the major cold- shock protein, CspA (CS7.4), of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Molecular Microbiology. 1994;11(5):833–839. doi: 10.1111/j.1365-2958.1994.tb00361.x. [DOI] [PubMed] [Google Scholar]
- Maddamsetti R, Lenski RE, Barrick JE. Adaptation, Clonal Interference, and Frequency-Dependent Interactions in a Long-Term Evolution Experiment with Escherichia coli. 2015;200:619–631. doi: 10.1534/genetics.115.176677. June. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl T, Van Melderen L, Mine N, Respondek M, Oberer M, Keller W, Zangger K. Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA. Journal of Molecular Biology. 2006;364(2):170–185. doi: 10.1016/j.jmb.2006.08.082. [DOI] [PubMed] [Google Scholar]
- Malan TP, McClure WR. Dual promoter control of the escherichia coli lactose operon. Cell. 1984;39(1):173–180. doi: 10.1016/0092-8674(84)90203-4. [DOI] [PubMed] [Google Scholar]
- Menezes AA, Cumbers J, Hogan JA, Arkin AP. Towards synthetic biological approaches to resource utilization on space missions. Journal of The Royal Society Interface. 2014;12(102):20140715–20140715. doi: 10.1098/rsif.2014.0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitta M, Fang L, Inouye M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Molecular Microbiology. 1997;26(2):321–335. doi: 10.1046/j.1365-2958.1997.5771943.x. [DOI] [PubMed] [Google Scholar]
- Molin S, Boe L, Jensen LB, Kristensen CS, Givskov M, Ramos JL, Bej AK. Suicidal genetic elements and their use in biological containment of bacteria. Annu Rev Microbiol. 1993;47:139–166. doi: 10.1146/annurev.mi.47.100193.001035. [DOI] [PubMed] [Google Scholar]
- Montague M, McArthur GHth, Cockell CS, Held J, Marshall W, Sherman LA, Cumbers J. The role of synthetic biology for in situ resource utilization (ISRU) Astrobiology. 2012;12(12):1135–1142. doi: 10.1089/ast.2012.0829. [DOI] [PubMed] [Google Scholar]
- Mulligan ME, Hawley DK, Entriken R, McClure WR. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Research. 1984;12(1):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell Ea, Muzzin O, Darst Sa. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science (New York, NY) 2002;296(5571):1285–1290. doi: 10.1126/science.1069595. https://doi.org/10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Nathan LP, Glenn DL, Johan V. Synchronous long-term oscillations in a synthetic gene circuit. Nature. 2016;538(7626):1–4. doi: 10.1038/nature19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOVICK A, SZILARD L. Experiments with the Chemostat on spontaneous mutations of bacteria. Proceedings of the National Academy of Sciences of the United States of America. 1950;36(12):708–19. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piraner DI, Abedi MH, Moser BA, Lee-Gosselin A, Shapiro MG. Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat Chem Biol. 2016:1–8. doi: 10.1038/nchembio.2233. advance on(November) [DOI] [PubMed] [Google Scholar]
- Ptashne M, Jeffrey A, Johnson AD, Maurer R, Meyer BJ, Pabo CO, Sauer RT. How the lambda repressor and cro work. Cell. 1980;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Roth JR. Tandem Genetic Duplications in Salmonella typhimurium : Amplification of the Histidine Operon. 1978:53–71. doi: 10.1016/0022-2836(78)90279-6. [DOI] [PubMed] [Google Scholar]
- Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Isaacs FJ. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518(7537):89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarai A, Takeda Y. Lambda repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(17):6513–7. doi: 10.1073/pnas.86.17.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultzaberger RK, Bucheimer RE, Rudd KE, Schneider TD. Anatomy of Escherichia coli ribosome binding sites. Journal of Molecular Biology. 2001;313:215–228. doi: 10.1006/jmbi.2001.5040. [DOI] [PubMed] [Google Scholar]
- Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli ??70 promoters. Nucleic Acids Research. 2007;35(3):771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleight SC, Bartley Ba, Lieviant Ja, Sauro HM. Designing and engineering evolutionary robust genetic circuits. Journal of Biological Engineering. 2010;4(1):12. doi: 10.1186/1754-1611-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidler L. Genetically engineered probiotics. Bailliere’s Best Practice and Research in Clinical Gastroenterology. 2003;17(5):861–876. doi: 10.1016/s1521-6918(03)00072-6. [DOI] [PubMed] [Google Scholar]
- Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B, Remaut E. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol. 2003;21(7):785–789. doi: 10.1038/nbt840. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Sarai a, Rivera VM. Analysis of the sequence-specific interactions between Cro repressor and operator DNA by systematic base substitution experiments. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:439–443. doi: 10.1073/pnas.86.2.439. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JC, Silver Pa, Howard RJ. Sun-driven microbial synthesis of chemicals in space. International Journal of Astrobiology. 2011;10(4):359–364. [Google Scholar]
- Wright O, Delmans M, Stan GB, Ellis T. GeneGuard: A modular plasmid system designed for biosafety. ACS Synthetic Biology. 2015;4(3):307–316. doi: 10.1021/sb500234s. [DOI] [PubMed] [Google Scholar]
- Wright O, Stan GB, Ellis T. Building-in biosafety for synthetic biology. Microbiology (United Kingdom) 2013;159(PART7):1221–1235. doi: 10.1099/mic.0.066308-0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin- antitoxin systems. Nat Rev Microbiol. 2011;9(11):779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- Yamanaka K. Cold Shock Response in Escherichia coli JMMB Symposium. J Mol Microbiol Biotechnol. 1999;1(2):193–202. [PubMed] [Google Scholar]
- Yansura DG, Henner DJ. Use of Escherichia coli trp promoter for direct expression of proteins. Methods in Enzymology. 1990;185(1981):54–60. doi: 10.1016/0076-6879(90)85007-b. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Oda K. A bacterium that degrades and assimilates poly(ethylene terephthalate) Science. 2016;351(6278):1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


