Abstract
Ixodes ricinus ticks are vectors of numerous human and animal pathogens. They are host generalists able to feed on more than 300 vertebrate species. The prevalence of tick-borne pathogens is influenced by host–vector–pathogen interactions that results in spatial distribution of infection risk. Broad-range polymerase chain reaction electrospray ionization mass spectrometry (PCR/ESI-MS) was used to analyze 435 I. ricinus nymphs from four localities in the south of the Czech Republic for the species identification of tick-borne pathogens. Borrelia burgdorferi sensu lato spirochetes were the most common pathogen detected in the ticks; 21% of ticks were positive for a single genospecies and 2% were co-infected with two genospecies. Other tick-borne pathogens detected included Rickettsia helvetica (3.9%), R. monacensis (0.2%), Anaplasma phagocytophilum (2.8%), Babesia venatorum (0.9%), and Ba. microti (0.5%). The vertebrate host of the ticks was determined using PCR followed by reverse line blot hybridization from the tick's blood-meal remnants. The host was identified for 61% of ticks. DNA of two hosts was detected in 16% of samples with successful host identification. The majority of ticks had fed on artiodactyls (50.7%) followed by rodents (28.6%) and birds (7.8%). Other host species were wild boar, deer, squirrels, field mice and voles.
Keywords: tick, Ixodes ricinus, PCR-ESI/MS, Borrelia, host, Lyme borreliosis, Babesia, Anaplasma, Rickettsia
Prevalence of multiple human and veterinary pathogens was estimated in host-seeking nymphal Ixodes ricinus ticks, and their previous vertebrate host was determined.
INTRODUCTION
Ticks harbor a wide variety of viruses, bacteria and protozoans, some of which can be transmitted to vertebrate hosts, including humans, and cause disease (Parola and Raoult 2001; Charrel et al.2004). Ixodes ricinus is the most widespread human biting tick in Europe. It is the vector of multiple pathogens including the Borrelia burgdorferi sensu lato complex, which cause Lyme borreliosis; the intracellular bacteria Anaplasma phagocytophilum, which cause granulocytic anaplasmosis (Woldehiwet 2006); several species of Rickettsia (Karbowiak et al.2016); and protozoan parasites of the genus Babesia (Hunfeld and Brade 2004; Yabsley and Shock 2013). In nature, ticks transmit these pathogens between species of vertebrate blood-meal hosts. Humans and some species of animals are the accidental, and frequently ‘dead end’, hosts. The species of the tick blood-meal hosts differ in their abundance, in the level of tick infestation, and in the efficiency of pathogen gain, amplification and transmission (Perez et al.2012; Geller et al.2013; Hofmeester et al.2016). Therefore, the knowledge of the impact of a particular host species on pathogen circulation is of substantial importance for the estimation of infection risk and preventive measures design.
Ixodes ricinus is a host generalist able to feed on more than 300 vertebrate species including rodents (Kozuch et al.1967; Matuschka, Richter and Spielman 1991; Estrada-Pena et al.2005; Perez et al.2012), birds (Estrada-Pena et al.2005; Geller et al.2013; Lommano et al.2014), insectivores (Kozuch et al.1967; Matuschka, Richter and Spielman 1991; Perez et al.2012), artiodactyls (Kiffner et al.2010; Kjelland et al.2011) and reptiles (Matuschka, Richter and Spielman 1991; Majlathova et al.2006). This behavior makes it difficult to decipher the complex system of zoonotic pathogen circulation among the populations of hosts and tick vectors. Host trapping methods frequently focus only on selected species and are influenced by differences in trapping effort and efficiency. Therefore, the contribution of the individual species to the overall pool of tick hosts is not easy to assess from this type of data. Molecular biology methods, which allow host identification in questing ticks, have potential to overcome these obstacles. These techniques are primarily based on polymerase chain reaction (PCR) amplification of host DNA from blood remnants in the questing tick using universal primers. Host species identification is performed by the means of reverse hybridization (reverse line blotting) (Kirstein and Gray 1996; Gray et al.1999; Pichon et al.2003; Humair et al.2007), restriction fragment polymorphism analyses (Kirstein and Gray 1996; Wodecka, Rymaszewska and Skotarczak 2013), or sequencing (Gariepy et al.2012). Currently available DNA-based methods differ in the target nucleotide sequence as well as in the spectrum of hosts they are able to detect and identify at the taxonomic level (Estrada-Pena et al.2005; Pichon et al.2005; Humair et al.2007; Kent 2009). DNA-based host identification methods are generally able to identify the host in approximately 30%–60% of the samples (Pichon et al.2005; 2006, Humair et al.2007; Allan et al.2010; Wodecka, Rymaszewska and Skotarczak 2013; Léger et al.2015). In this work, we identified host DNA from I. ricinus blood remnants by a reverse line blotting procedure described by Humair et al. (2007) using species-, genus-, and a group of species-specific probes described by Moran Cadenas et al. (2007).
To survey the prevalence of tick-borne pathogens in I. ricinus ticks, we used a broad-range PCR electrospray ionization mass spectrometry (PCR/ESI-MS) assay designed to detect a wide-range of tick-borne organisms. We have previously applied this technique for the detection of vector-borne pathogens such as Borrelia, Ehrlichia, Powassan virus, Babesia spp. and canine heartworm from ticks and/or clinical specimens (Crowder et al.2010b, 2012; Eshoo et al.2010, 2012, 2014). In addition, this PCR/ESI-MS technique was used to detect Borrelia DNA in ticks used for the xenodiagnoses of patients with Lyme disease (Marques et al.2014). Following a multilocus broad-range PCR, automated electrospray ionization mass spectrometry is used to determine the masses of the PCR amplicons. From these masses, basecount signatures (i.e. the number of A’s, G’s, C’s, and T’s) are determined for each primer pair, which are then matched to a database to identify the microorganism or microorganisms present in the sample (Ecker et al.2006a). This technique identifies pathogens, including those involved in co-infections, genotypes pathogens, and can identify new genetic variants. For example, we previously demonstrated our ability to distinguish B. burgdorferi genotypes, even when present in mixtures of genotypes (Crowder et al.2010a), and we also identified a novel and widespread Anaplasmataceae species in a survey of I. pacificus ticks in California using our PCR/ESI-MS method (Eshoo et al.2015). This same approach can be used to detect and identify a wide range of vector-borne pathogens in a single test (Eshoo et al.2014).
In the present work, we found that B. burgdorferi sensu lato group were the most common microorganisms detected in the I. ricinus nymphal ticks in the Southern Czech Republic. Of 435 specimens, 100 (23%) were positive for B. burgdorferi. In 91% of the positive detections, we observed a single genospecies, whereas 9% of these were simultaneously infected with two genospecies. Other microorganisms detected included A. phagocytophilum, Babesia spp. and Rickettsia spp. Identification of the previous host was successful in 61% of the I. ricinus nymphs with artiodactyls and rodents being the most prevalent host species.
MATERIALS AND METHODS
Tick collection and nucleic acid extractions
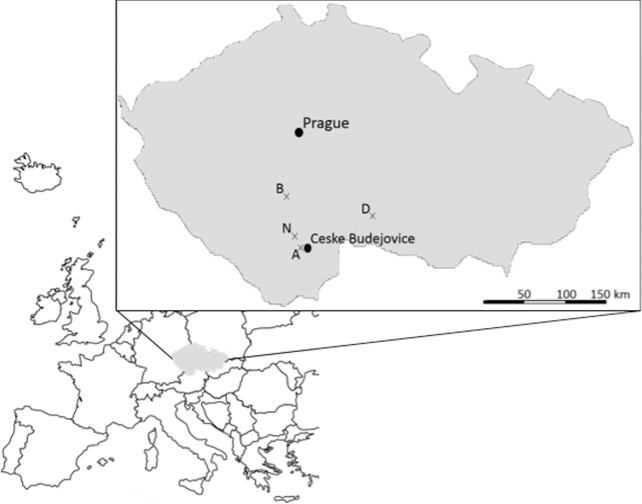
A total of 435 I. ricinus nymphs were collected by flagging in mixed and deciduous forests at four sites in South Bohemia region of the Czech Republic during the summer of 2010 (Fig. 1). A total of 153 ticks were collected from locality Zavadilka (48°58΄47.32″N, 14°25΄47.41″E; 415 m above the sea level) between 28 May 2010 and 8 June 2010. This sampling site is in a peri-urban mixed forest near recreational and residential housing areas of the city of Ceske Budejovice. In the tree stratum, Picea abies, Pinus sylvestris, Quercus spp. prevail, accompanied by Tilia cordata, Populus tremula, Corylus avellana. The density of undergrowth and height of litter layer are variable. On 8 August 2010, 89 ticks were collected from Netolice (49°2΄28.29″N, 14°10΄41.90″E; 470 m a. s. l.). The habitat is slightly sloped with orientation toward the south and is markedly drier compared to the other collection sites. Deciduous trees predominate (T. cordata, Quercus spp., B. pendula, Fraxinus excelsior, Fagus sylvatica), although formations of P. abies are also present. The undergrowth consists mainly of grass species (Calamagrostis epigejos, Dactylis glomerata, Poa annua) and herbs (Holcus mollis, Veronica officinalis). On 8 June 2010, 100 ticks were collected from Blatná (49°26΄42.54″N, 13°52΄56.57″E; 480 m a. s. l.). The terrain is flat and covered by variegated vegetation of an ecotonous character. Vegetation cover consists of deciduous trees (Acer platanoides, P. tremula, C. avellana, Salix caprea, Q. petraea, Carpinus betulus, Prunus avium). In the undergrowth, the grass species (Poa nemorosa, Festuca ovina, C. epigejos, D. glomerata) together with Dryopteris filix-mas, V. officinalis and Lamium spp. are observed. On 10 June 2010, 93 ticks were collected from Dačice (49°5΄56.90″N, 15°26΄24.06″E; 610 m a. s. l.). This sampling site is characterized by dense shrubby vegetation of deciduous tree species (Q. robur mainly) with minor representation of P. abies. The herbaceous stratum is also dense and rich in species (Stellaria holostea, Lathyrus vernus, Pulmonaria officinalis, Hypericum perforatum, Campanula trachelium, Rubus idaeus). Sample homogenization and total nucleic acids extraction was performed using a previously described Qiagen column-based protocol, with the substitution of the Qiagen DNeasy column (Qiagen, Valencia, CA, USA) for the Qiagen Virus Minelute columns (Crowder et al.2010b). Nucleic acids were extracted from the ticks at the University of South Bohemia, and the extracts shipped overnight at room temperature in screw capped tubes to Ibis Biosciences for analysis on the PCR/ESI-MS system using the broad-range vector-borne microorganism detection assay. The extracts were stabilized prior to shipping using either RNAstable (Biomatrica, San Diego, CA, USA) according to the manufacturer's instructions or with 1–2 μL/sample SUPERase-in (Invitrogen, Carlsbad, CA, USA).
Figure 1.
Tick sampling sites located in the southern part of the Czech Republic. A, Zavadilka; B, Blatná; D, Dačice; N, Netolice.
Identification and characterization of tick-borne pathogens by PCR/ESI-MS
Detection of tick-borne pathogens was performed using a PCR/ESI-MS assay using nine broad-range PCR primer pairs designed to amplify genomic material from bacterial and protozoan vector-borne organisms (Table 1). Primer pairs, BCT3517 and INV4855 were employed in a single multiplexed reaction. Primer pairs BCT2328, BCT3511, INV4443, BCT3514, BCT1083, BCT3570 and BCT3575 were employed in individual singleplex PCR reactions (Eshoo et al.2015). Internal positive controls made from cloned synthetic DNA (BlueHeron Biotechnology, Bothell, WA, USA) were included in each PCR reaction at 20 copies per reaction. The internal controls were designed to be identical to the expected amplicon for one of the primer pairs in the reaction with the exception of a 5-base pair deletion to enable the control to be distinguished from the target-derived amplicon.
Table 1.
PCR/MS-ESI and DNA sequencing primers and targets.
| Primer pair | Primer ID | Primer sequence | Target | Target clade/genus | Purpose | Reference |
|---|---|---|---|---|---|---|
| BCT2328 | BCT5602F | TGAGGGTTTTATGCTTAAAGTTGGTTTTATTGGTT | asd | F. tularensis | PCR/ESI-MS | (Crowder et al.2012) |
| BCT5603R | TGATTCGATCATACGAGACATTAAAACTGAG | |||||
| INV4855 | INV10812F | TGAGAGAAATCGTACACATTCAAGCGGG | β-tubulin | All Babesia spp. | PCR/ESI-MS | (Crowder et al.2012) |
| INV10813R | TCCATGTTCGTCGGAGATGACTTCCCA | |||||
| INV4443 | INV10034F | TGCGCAAATTACCCAATCCTGACAC | 18S rRNA | All Babesia spp. | PCR/ESI-MS | (Crowder et al.2012) |
| INV10035R | TCCAGACTTGCCCTCCAATTGGTA | |||||
| BCT3511 | BCT8229F | TGCATTTGAAAGCTTGGCATTGCC | gyrB | All Spirochetes | PCR/ESI-MS | (Crowder et al.2012) |
| BCT8230R | TCATTTTAGCACTTCCTCCAGCAGAATC | |||||
| BCT3514 | BCT8235F | TTTGGTACCACAAAGGAATGGGA | rpoC | All Spirochetes | PCR/ESI-MS | (Crowder et al.2012) |
| BCT8236R | TGCGAGCTCTATATGCCCCAT | |||||
| BCT3517 | BCT8241F | TGCTGAAGAGCTTGGAATGCA | flagellin | All Spirochetes | PCR/ESI-MS | (Crowder et al.2012) |
| BCT8242R | TACAGCAATTGCTTCATCTTGATTTGC | |||||
| BCT1083 | BCT2764F | TAAGAGCGCACCGGTAAGTTGG | RNaseP | All Rickettsia spp. | PCR/ESI-MS | (Crowder et al.2012) |
| BCT2763R | TCAAGCGATCTACCCGCATTACAA | |||||
| BCT3570 | BCT8336F | TGCATGCAGATCATGAACAGAATGC | gltA | Alphaproteobacteria | PCR/ESI-MS | (Crowder et al.2012) |
| BCT8337R | TCCACCATGAGCTGGTCCCCA | |||||
| BCT3575 | BCT8346F | TGCATCACTTGGTTGATGATAAGATACATGC | rpoB | Alphaproteobacteria | PCR/ESI-MS | (Crowder et al.2012) |
| BCT8347R | TCACCAAAACGCTGACCACCAAA | |||||
| Ap-msp4 | F | M13F-ICAGTMTGYGCYTGCTCCCT | msp4 | Anaplasma spp. | Sequencing | This study |
| R | M13R-CCTTAIYTGAAMISIAATCTTGCTCC | |||||
| Ap-groEL | F | M13F-GAIAIIACTGAYGGTATGCAGTTTG | GroEL | Anaplasma spp. | Sequencing | This study |
| R | M13R-CYAIMCIYTCYYTMAGYTTTTCCTT | |||||
| 18S rRNA-M13 | F | M13F-GACTAGGGATTGGAGGTC | 18S rRNA | Babesia spp. | Sequencing | (Blaschitz et al.2008) |
| R | M13R-GAATAATTCACCGGATCACTC | |||||
| ITS1-M13 | F | M13F-CGAGTGATCCGGTGAATTATTC | ITS1 | Babesia spp. | Sequencing | (Blaschitz et al.2008) |
| R | M13R-CCTTCATCGTTGTGTGAGCC | |||||
| ITS2-M13 | F | M13F-GGCTCACACAACGATGAAGG | ITS2 | Babesia spp. | Sequencing | (Blaschitz et al.2008) |
| R | M13R-CTCGCCGTTACTAAGGGAATC | |||||
| HSP70-M13 | F | M13F-GCTATTGGTATTGACTTGGG | hsp70 | Babesia spp. | Sequencing | (Blaschitz et al.2008) |
| R | M13R-CCTTCATCTTGATAAGGACC | |||||
| Bb-16S | F | M13F-CGCTGGCAGTGCGTCTTAAG | 16S rRNA | B. burgdorferi s. l. | Sequencing | This study |
| R | M13R-GCGTCAGTCTTGACCCAGAAGTTC | |||||
| Bb-ITS | F | M13F-CTGCGAGTTCGCGGGAGA | ITS | B. burgdorferi s. l. | Sequencing | This study |
| R | M13R-TCCTAGGCATTCACCATA | |||||
| rrs-rrlA IGS | F | GTATGTTTAGTGAGGGGGGTG | IGS Outer | B. miyamotoi | Sequencing | (Bunikis et al.2004) |
| R | GGATCATAGCTCAGGTGGTTAG | |||||
| rrs-rrlA IGS-M13 | F | M13F-AGGGGGGTGAAGTCGTAACAAG | IGS Inner | B. miyamotoi | Sequencing | (Bunikis et al.2004) |
| R | M13R-GTCTGATAAACCTGAGGTCGGA | |||||
| M13 | F | CCCAGTCACGACGTTGTAAAACG | M13 | Sequencing | (Eshoo et al.2007) | |
| R | AGCGGATAACAATTTCACACAGG |
PCR was performed in a 50 μL reaction volume with 10 μL nucleic acid extract in a reaction mix as previously described (Crowder et al.2010a). PCR cycling conditions were the same as those reported previously (Eshoo et al.2007). PCR amplicons were analyzed by ESI-MS, and base composition analyses were performed on a research-use-only ESI-MS instrument (Abbott Molecular, Des Plaines, IL, USA) as previously described (Ecker et al.2005, 2006b, 2010; Crowder et al.2010a, 2012; Eshoo et al.2010; Rounds et al.2012).
The Babesia venatorum PCR/ESI-MS signature was confirmed by Sanger sequencing of a 613-nt portion of the 18S rDNA, the ITS1 and ITS2 regions (∼550 and ∼400 nt, respectively), and a 333-nt segment of the HSP70 gene as previously described (Blaschitz et al.2008). All sequencing PCR reactions were performed using Platinum Taq High Fidelity (Invitrogen) in Platinum Taq buffer with 200 μM of each dNTP, 2 mM MgSO4 and 250 nM of each primer. Sequencing PCR reactions were cycled with the following conditions: 95°C for 2 min; 8 cycles of 95°C for 15 s, 50°C for 45 s (increasing 0.6°C per cycle) and 68°C for 90 s; 37 cycles of 95°C for 15 s, 60°C for 15 s, and 68°C for 60 s; followed by 4 min at 68°C. For samples requiring cloning, PCR products were ligated and cloned using the ZeroBlunt TOPO PCR Cloning kit (Invitrogen) according to the manufacturer's instructions. Individual clones were purified and sequenced by SeqWright (Houston, TX, USA) using SP6 and T7 promoter primers.
Borrelia genotyping
All Borrelia-positive specimens were genotyped by a previously described multilocus PCR/ESI-MS Borrelia genotyping assay, which targets eight loci in the Borrelia genome to enable genotype assignment (Crowder et al.2010a). Borrelia garinii identification requires amplification of only seven primers as it is frequently not detectable with primer pair BCT3514. For Borrelia species with fewer than 20 genomes/PCR reaction as quantified by the PCR/ESI-MS instrument, the nucleic acid extracts were treated with an isothermal amplification prior to genotyping as previously described (Eshoo et al.2012).
Tick host identification
Identification of the host DNA from blood remnants in the tick was carried out using reverse line blotting according to Humair et al. (2007). Briefly, a portion of 12S rDNA was amplified using a universal pair of primers with the reverse primer labeled on the 5΄ end with biotin. PCR was performed in 25-μL reactions using PPP Premix (Top-Bio, Prague, Czech Republic), 0.4 μM primer 12S-6F (CAAACTGGGATTAGATACC) and 0.4 μM primer B-12S-9R (biotin-AGAACAGGCTCCTCTAG) (Humair et al.2007), and 10 μL of tick extractions as source of template DNA brought to volume with PCR-grade water. The amplification was carried out according to the cycling profile previously reported (Humair et al.2007). Subsequently, the PCR products were hybridized to species-, genus- and group of species-specific probes described previously (Moran Cadenas et al.2007); Rattus rattus and Neomys anomalus probes were not used. The probes were bound to Biodyne C membranes and hybridized with the labeled PCR products. After washing and binding of streptavidin-linked horseradish peroxidase, the hybridized PCR products were visualized on a sensitive film using chemiluminescence (Humair et al.2007).
Reverse line blotting is a highly sensitive technique that requires multiple steps and hence tends to be sensitive to cross-contamination among the samples, external contamination from environment, and surface contamination of the ticks. The possibility of contamination can never be fully excluded, but measures were taken to minimize the risk and monitor possible false positive results. Pre-PCR and post-PCR processes were spatially separated and were performed at different times. All nucleic acid extractions performed in a biohazard box (class II), and all PCR mastermixes were prepared in a PCR box. All surfaces were treated before and after work with UV light (20 min), DNA remover solution (Minerva Biolabs, Berlin Germany), and were subsequently washed with dH2O. Filtered tips were used for pipetting. In each set of samples, a negative extraction control (a blank sample without a tick) was included that entered the analysis on the level of sample homogenization (i.e. prior to nucleic acid extraction). Thus, the negative controls were subjected to all the components (except the tick) and all steps of the process from sample homogenization to analysis.
Statistics
The data were analyzed using χ2 or Fisher´s exact test in case of low frequencies (GraphPad Software, Inc., San Diego, California, USA, version 5.04). Differences with P < 0.05 were considered significant.
RESULTS
Tick collections and broad-range detection PCR/ESI-MS
A total of 435 I. ricinus nymphs were collected during a 10-week period over the summer of 2010 from four sites in the South Bohemia region of the Czech Republic. Tick extracts were analyzed using a broad-range vector-borne assay designed to detect and identify to the species level a wide range of vector-borne pathogens and other microorganisms, notably spirochetes (including all Borrelia species), Anaplasma spp., Ehrlichia sp., Rickettsia spp., Franciscella sp., Babesia spp. and tick species-specific endosymbionts.
The B. burgdorferi sensu lato group was the most common group of organisms found in the ticks with 100 of the 435 (23%) ticks positive for this pathogen. Infections by a single genospecies were detected in 91 of these 100 (91%) ticks, whereas nine (9%) of the B. burgdorferi-positive ticks were simultaneously infected with two genospecies (Table 2). The most prevalent B. burgdorferi genospecies (multiple infections included) was B. afzelii (38/109 or 34.9% of all B. burgdorferi s.l. positive identifications), followed by B. burgdorferi s.s. (32/109 or 29.4%) and B. garinii (23/109 or 21.1%). Borrelia valaisiana was observed in 8 of 109 (7.3%) of identifications and was not found at study site Dačice. Borrelia lusitaniae was found at only one site, Netolice, and accounted for eight (7.3%) of identifications overall. The relapsing fever-like spirochete, B. miyamotoi, was detected in 9 of the 435 (2.1%) ticks (Table 2).
Table 2.
Identification of microorganisms in nymphal Ixodes ricinus ticks using PCR/ESI-MS.
| Site | ||||||
|---|---|---|---|---|---|---|
| Blatná | Dačice | Netolice | Zavadilka | Overall | ||
| Number of ticks | 100 | 93 | 89 | 153 | 435 | |
| Pathogen | ||||||
| Anaplasma phagocytophilum | 3 | 0 | 2 | 4 | 9 | |
| Babesia microti | 0 | 0 | 1 | 0 | 1 | |
| Babesia venatorum | 0 | 1 | 0 | 2 | 3 | |
| Babesia sp. unknown | 0 | 0 | 1 | 0 | 1 | |
| Borrelia afzelii | 4 | 4 | 5 | 19 | 32 | |
| Borrelia burgdorferi s.s. | 10 | 8 | 2 | 6 | 26 | |
| Borrelia garinii | 7 | 2 | 3 | 4 | 16 | |
| Borrelia lusitaniae | 0 | 0 | 7 | 0 | 7 | |
| Single infections | Borrelia valaisiana | 2 | 0 | 1 | 0 | 3 |
| Borrelia miyamotoi | 2 | 2 | 0 | 2 | 6 | |
| Rickettsia monacensis | 0 | 0 | 0 | 1 | 1 | |
| Rickettsia helvetica | 4 | 1 | 3 | 4 | 12 | |
| B. burgdorferi s.s. and B. afzelii | 0 | 3 | 0 | 0 | 3 | |
| B. garinii and B. valaisiana | 1 | 0 | 1 | 2 | 4 | |
| B. lusitaniae and B. valaisiana | 0 | 0 | 1 | 0 | 1 | |
| B. miyamotoi and B. garinii | 0 | 0 | 0 | 1 | 1 | |
| B. miyamotoi and A. phagocytophilum | 0 | 0 | 0 | 1 | 1 | |
| B. burgdorferi s.s. and R. helvetica | 0 | 0 | 0 | 1 | 1 | |
| B. afzelii and R. helvetica | 1 | 0 | 0 | 0 | 1 | |
| B. garinii and R. helvetica | 0 | 0 | 1 | 0 | 1 | |
| Co-infections | B. garinii and Babesia venatorum | 1 | 0 | 0 | 0 | 1 |
| B. afzelii and Babesia microti | 0 | 0 | 0 | 1 | 1 | |
| B. burgdorferi s.s. and B. afzelii and R. helvetica | 1 | 0 | 0 | 0 | 1 | |
| B. burg. s.s. and B. miyamotoi and A. phagocytophilum | 0 | 1 | 0 | 0 | 1 | |
| A. phagocytophilum and R. helvetica | 0 | 0 | 0 | 1 | 1 | |
| Total infected | 36 | 22 | 28 | 49 | 135 | |
| % infected | 36.0% | 23.7% | 31.5% | 32.0% | 31.0% | |
In addition to the Borrelia species mentioned above, several other organisms were found in the ticks. Anaplasma phagocytophilum was detected at all four tick collection sites and was observed in 12 of 435 (2.8%) ticks (Table 2). Two variants of A. phagocytophilum were detected; both shared the same basecount for primer pair BCT3575 (A22 G33 C20 T27), but differed by a single nucleotide polymorphism in the region amplified by primer pair BCT3570 (A23 G39 C27 T36 and A24 G38 C27 T36).
Babesia spp. were found in 7 of 435 (1.6%) I. ricinus nymphs. Babesia microti was detected once at Netolice and once at Zavadilka with primer pairs INV4855 and INV4443, producing the expected basecounts of A29 G22 C15 T20 and A45 G32 C25 T28, respectively. A different basecount signature was found in four ticks, one each from Blatná and Dačice and two from Zavadilka with these same primers, producing novel basecounts of A26 G24 C20 T16 and A44 G32 C26 T27, respectively. By sequencing of the 18S, ITS1, ITS2 and HSP70 loci from one of the representative positive specimens, this signature was determined to represent Ba. venatorum (formerly called Babesia sp. EU1). The 18S rDNA sequence exhibited identity of 611 or 613 nucleotides with Babesia sp. ‘venatorum’ isolate DP-1569 (accession number JX042320.1), and the ITS1 sequence was identical to that of Babesia sp. ‘venatorum’ (accession number HM113372.1). The 408-nt ITS2 and 33-nt HSP70 sequences from our Ba. venatorum were identical to accession numbers EU185802.1 and EU185813.1 from a previously described Babesia sp. GoA3. However, no ITS2 or HSP70 sequences exist in GenBank for Ba. venatorum, nor do any 18S rDNA or ITS1 sequences exist for Babesia sp. GoA3. Given the close match of the 18S rDNA and the ITS region to Ba. venatorum, our Babesia signature is considered to be that of the widely reported Ba. venatorum.
Rickettsia spp. were found in 18 of 435 (4.1%) of the tick samples. Two species of Rickettsia were detected: R. helvetica and R. monacensis. The former was observed at all four sites in 17 (3.9%) of ticks (Table 2); the basecounts for primer pairs BCT1083 and BCT3570 (A41 G33 C30 T31 and A29 G30 C29 T37, respectively) matched those found in German ticks (Eshoo et al.2014). We also detected R. monacensis (BCT1083: A42 G33 C24 T30; BCT3570: A28 G30 C32 T35) in one tick from Zavadilka (Table 2). None of the samples tested were positive for Franciscella tularensis. No significant differences were found in the prevalence rate of the individual pathogens among the sampling sites.
Borrelia genotyping
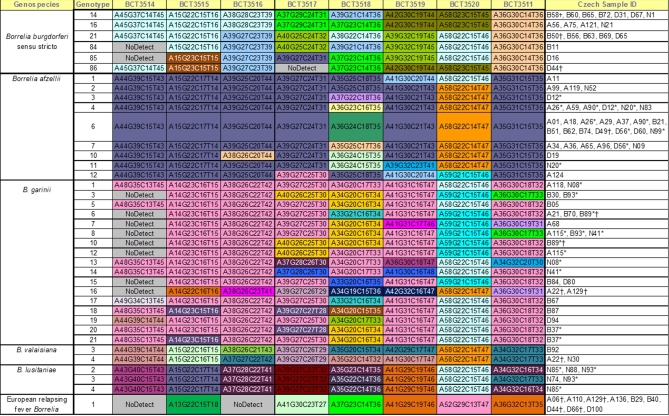
The B. burgdorferi s.l. positive specimens were further characterized using a previously described PCR/ESI-MS based Borrelia genotyping assay (Crowder et al.2010a). As the nymph extracts contained a limited amount of Borrelia DNA, most Borrelia positive specimens were subjected to a nested isothermal amplification to increase the amount of material available for genotyping (Eshoo et al.2012). Of the B. burgdorferi sensu stricto (s.s.)-positive ticks, there were two genotypes that differed from the majority of the detections (Fig. 2). The first variant, found in two ticks from Dačice, had a unique basecount at primer pair BCT3515 (A15 G23 C15 T15). The second variant, found in five ticks from Blatná and one from Dačice, had a unique basecount at primer pair BCT3517 (A40 G25 C24 T32).
Figure 2.
Borrelia burgdorferi s.l. genotypes detected among I. ricinus ticks from the Czech Republic. Each column represents basecounts of a specific PCR product (primer pairs marked with BCT and number). ‘*’ indicates more than one genotype of particular genospecies present; ‘†’ indicates more than one B. burgdorferi s.l. genospecies detected. Sample ID code: A, Zavadilka; B, Blatna, D, Dacice; N, Netolice.
Co-infections
In the 435 ticks tested, 18 (4.1%) contained more than one organism (including different B. burgdorferi s.l. genospecies) (Table 2). Of these, 16 (89% of co-infections) harbored two organisms and two (11%) harbored three. The most frequent co-infection was two B. burgdorferi s.l. genospecies (50.0%), followed by a B. burgdorferi s.l. genospecies and R. helvetica (22%).
Tick host identification
Aliquots of samples analyzed by PCR/ESI-MS were subjected to host identification by reverse line blotting. From the total number of 435 samples of questing I. ricinus nymphs, identification of the previous host was successful in 266 samples (61.1%). In 42 (15.8%) of cases, DNA of two different hosts was identified, resulting in a total of 308 host identifications. The differences in identification success rate among the localities (range 56.0% to 64.5%) were not statistically significant (Table 3). Artiodactyls (50.7%) and rodents (28.6%) were the most prevalent host species, while birds, small predatory mammals, and insectivores were less frequently a source of blood-meal for larval ticks (Table 3). Most of the hosts were identified only to the group level. Sus scrofa (105/156 or 67.3%) and Capreolus capreolus (2/156 or 1.3%) were detected among artiodactyl host species. Sciurus vulgaris (10/119 or 8.4%) was the most frequently identified rodent species followed by Apodemus spp. (5/119 or 4.2%), Microtus spp. (5/119 or 4.2%) and Myodes (Clethrionomys) glareolus (Carleton et al.2014) (1/119 or 0.8%). The only avian species-specific identification was achieved by a single probe specific for Turdus spp. and Parus spp. host DNA (3/24 or 12.5%). Mustella spp. (8/8) and Neomys sp. (1/1) were detected among predatory mammals and insectivores, respectively.
Table 3.
Tick hosts by sampling site.
| Zavadilka | Blatna | Dacice | Netolice | Total | |
|---|---|---|---|---|---|
| Identification success rate | 60.8% | 56% | 64.5% | 64% | 61.1% |
| (93/153) | (56/100) | (60/93) | (57/89) | (266/435) | |
| Rodents | 37.6% | 31.7% | 26.9% | **62.3% | 28.6% |
| (41/109) | (19/60) | (21/78) | (28/61) | (119/308) | |
| Artiodactyls | 54.1% | 60% | 53.9% | **31.15% | 50.7% |
| (59/109) | (36/60) | (42/78) | (19/61) | (156/308) | |
| Birds | 5.5%* | 5.0% | *15.4% | 4.9% | 7.8% |
| (6/109) | (3/60) | (12/78) | (3/61) | (24/308) | |
| Predatory mammals | 1.8% | 3.3% | 3.9% | 1.6% | 2.6% |
| (2/109) | (2/60) | (3/78) | (1/61) | (8/308) | |
| Insectivores | 0.9% | 0.0% | 0.0% | 0.0% | 0.3% |
| (1/109) | (0/60) | (0/78) | (0/61) | (1/308) |
Identification success rate is the number of positively identified samples out of the number of ticks tested; proportional representation of groups of host species is the number of identifications of a particular host out of all successful identifications including two hosts identified in one sample. Statistically significant results are indicated by *P < 0.05 and **P < 0.01.
The composition of the tick host fauna differed among the individual sampling sites. Locality Netolice presented with a statistically significant higher portion of rodent blood-fed ticks (Fisher exact test; P < 0.01) and a relatively lower frequency of artiodactyl host species (Fisher exact test; P < 0.01) than other sites. Ticks sampled in locality Dačice had significantly more frequently fed on birds than ticks from the Zavadilka study site (Fisher exact test; P < 0.01); differences from other localities were not statistically significant (Table 3).
Tick hosts and pathogens
From the 266 ticks with only one host species identified, 72 carried at least one pathogen (including four samples with two pathogens). The prevalence rate of B. burgdorferi s.l. was 23.9% in rodent-fed ticks, 26.1% in ticks fed on artiodactyl blood and 16.7% in bird-fed ticks (Table 4). The differences in pathogen prevalence rate among ticks fed on different groups of hosts were not statistically significant (Table 5). Ticks infected by B. afzelii had fed on rodents or artiodactyls but not on birds. Avian blood-fed ticks were infected either by B. garinii (1/2) or a combination of B. garinii and B. valaisiana (1/2). Borrelia garinii and/or B. valaisiana infected ticks were found also in rodent or artiodactyl-fed ticks, but with lower frequency. The difference for B. garinii but not for B. valaisiana was statistically significant (Fisher exact test; P < 0.05). Nymphs positive for DNA of B. lusitaniae had fed on rodents (2/21) or artiodactyls (1/30) as larvae. Rickettsia helvetica infected ticks contained DNA from all three groups of hosts. Babesia spp. were found in nymphs that fed on rodents (Ba. Microti, 1/88, Ba. Venatorum, 1/88) and artiodactyls (Ba. venatorum, 2/;119 Ba. microti, 1/;119 Babesia sp. unidentified, 1/119). In addition, A. phagocytophilum-positive ticks also obtained their blood-meal from rodents (2/88) or artiodactyls (2/119) (Table 4). No statistically significant differences were found in the frequency of occurrence of co-infections by multiple pathogens (including different genospecies of B. burgdorferi s.l.) in samples with only one host identified (10/72; 16%) compared to those positive for multiple hosts (2/42; 4.8%).
Table 4.
Pathogen prevalence in ticks fed on a specific group of hosts.
| Rodents | Artiodactyls | Birds | Unidentified | |
|---|---|---|---|---|
| Borrelia burgdorferi s.l. | 23.9% | 26.1% | 16.7% | 23.1% |
| (21/88) | (31/119) | (3/12) | (39/169) | |
| Borrelia miyamotoi | 1.1% | 0.0% | 0.0% | 4.1% |
| (1/88) | (0/119) | (0/12) | (7/169) | |
| Babesia sp. | 2.3% | 3.4% | 0% | 0.6% |
| (2/88) | (4/119) | (0/12) | (1/169) | |
| Anaplasma phagocytophilum | 2.3% | 1.7% | 0.0% | 4.1% |
| (2/88) | (2/119) | (0/12) | (7/169) | |
| Rickettsia sp. | 5.7% | 4.2% | 8.3% | 4.1% |
| (5/88) | (5/119) | (1/12) | (7/169) |
Only samples with a single detected host are included (number of positive samples/number of samples with particular host detected); samples with positive pathogen detection in which the host could not be identified are labeled as unidentified.
Table 5.
Representation of genospecies of Borrelia burgdorferi s.l. among different groups of Ixodes ricinus hosts.
| Rodents | Artiodactyls | Birds | |
|---|---|---|---|
| B. burgdorferi s.s. | 33.3% | 33.3% | 0.0% |
| (7/21) | (10/30) | (0/2) | |
| B. afzelii | 33.3% | 33.3% | 0.0% |
| (7/21) | (10/30) | (0/2) | |
| B. garinii | 19.1% | 10.0% | 50.0%* |
| (4/21) | (3/30) | (1/2) | |
| B. valaisiana | 0% | 6.7% | 0.0% |
| (0/21) | (2/30) | (0/2) | |
| B. lusitaniae | 9.5% | 3.3% | 0% |
| (2/21) | (1/30) | (0/2) | |
| B. garinii + B. valaisiana | 4.8% | 6.7% | 50% |
| (1/21) | (2/30) | (1/2) | |
| B. burgdorferi s.s. + B. afzeliiB. afzelii | 0% | 3.3% | 0% |
| (0/21) | (1/30) | (0/2) | |
| B. valaisana + B. lusitaniae | 0% | 3.3% | 0% |
| (0/21) | (1/30) | (0/2) |
Number of positive samples of a genospecies/total number B. burgdorferi s.l. positive ticks fed on particular host are reported. ‘*’ indicates that higher frequency of B. garinii occurrence among bird fed ticks compared to other hosts is statistically significant (P < 0.05).
DISCUSSION
In this study, 435 questing nymphal I. ricinus ticks were tested using a broad-range assay designed to detect a variety of tick-borne pathogens. The prevalence of B. burgdorferi s.l. infected ticks (23%) was higher than average prevalence rates previously reported in nymphal I. ricinus in Central Europe (16.7%) (Strnad et al.2017). The difference may be due to the employment in this study of more sensitive molecular biology based methods (PCR/ESI-MS) or to seasonal or geographical differences.
Most of the B. burgdorferi s.l. infected ticks were infected by a single genospecies (91%). Most of the multiple infections consisted of B. garinii and B. valaisiana or B. burgdorferi s.s. and B. afzelii. Since the questing nymphs likely obtained only a single blood-meal as larvae and since B. burgdorferi genospecies show a certain level of host specificity (Kurtenbach et al.1998b, 2002), these combinations of genospecies might have been obtained from a single co-infected host during a single feeding. Co-infections by genospecies associated with different species of hosts (B. afzelii and B. lusitaniae) were also detected, however. Such findings might be explained by interrupted larval feeding on one infected host followed by successful feeding on another host infected by a different genospecies of B. burgdorferi. Infected and seemingly unfed larvae of I. ricinus have been reported in multiple studies (Nazzi et al.2010; Kalmar et al.2013; Tappe et al.2014; van Duijvendijk et al.2016). Detection of DNA of multiple host species in up to 17% of questing nymphal ticks in this study as well as previous studies (Humair et al.2007; Moran Cadenas et al.2007; Collini et al.2016) supports the possibility of interrupted feeding.
The majority of the Borrelia-positive ticks were infected by one of the unequivocally pathogenic genospecies (B. afzelii, B. garinii, B. burgdorferi s.s.) and therefore represent a significant health risk for humans. Moreover, 2% of the ticks, similar to the percentage reported previously (Crowder et al.2014) were infected with B. miyamotoi. This bacterium causes mild febrile disease in immunocompetent humans (Platonov et al.2011; Chowdri et al.2013; Krause et al.2013) but serious infections of the central nervous system in immunocompromised patients (Gugliotta et al.2013; Hovius et al.2013). This spirochete was previously detected in European I. ricinus ticks with a prevalence ranging from 0% to 3.85% (Gern et al.2010; Richter and Matuschka 2012; Potkonjak et al.2016).
Apart from Borrelia species, other human pathogens were detected in the questing nymphs. Anaplasma phagocytophilum, a causative agent of human granulocytic anaplasmosis, was found at all the sampling sites with prevalence ranging from 1% to 3%. The prevalence rates in nymphal I. ricinus vary significantly among different studies (<0.8% to ∼9%), possibly due to differences in study sites and years of sampling (Stuen, Granquist and Silaghi 2013). Despite relatively high seroprevalence in human population, diagnosed human disease cases are still a rare event in Europe. It was suggested previously that this discrepancy is associated with the occurrence of specific strains of the bacterium, which are unable to cause disease in humans, under-diagnosis of the infection, or antibody cross-reactivity (Dumler et al.2005; Silaghi et al.2012).
The prevalence of Babesia spp. was 1.6%, which is in concordance with other findings in I. ricinus in Central Europe (0.3%–4.1%) (Rudolf et al.2005; Schorn et al.2011; Venclikova et al.2015; Hamsikova et al.2016). Babesia microti and Ba. venatorum were previously found in I. ricinus (Silaghi et al.2012; Venclikova et al.2015; Hamsikova et al.2016). Babesia venatorum was reported to cause disease in immunocompromised patients (Hildebrandt, Gray and Hunfeld 2013). In Europe, Ba. microti is mostly associated with imported human cases (Hildebrandt, Gray and Hunfeld 2013). Nevertheless, there are also confirmed autochthonous (Hildebrandt et al.2007) and probable autochthonous (Arsuaga et al.2016; Moniuszko-Malinowska et al.2016) infections by European lineages of Ba. microti. In one tick in our study a basecount matching Ba. gibsoni was detected by one primer pair, but we were not able to confirm the identity by sequencing. Therefore, we refer to this sample as Babesia sp. in the results. The presence of Ba. gibsoni is unlikely since Mediterranean Rhipicephalus sanguineus ticks are considered to be the main vectors.
The spotted fever group Rickettsia, R. monacensis was detected at a single locality in a single tick, whereas R. helvetica was omnipresent. Rickettsia helvetica belongs to commonly occurring species in I. ricinus ticks (Karbowiak et al.2016). Rickettsia monacensis is less prevalent, frequently reported in ticks sampled from birds (Elfving et al.2010; Biernat et al.2016; Mărcuţan et al.2016). Both these Rickettsia species are considered generally non-pathogenic and have rarely been identified in association with human disease cases with variable in severity (Fournier et al.2000; Jado et al.2007; Nilsson, Elfving and Påhlson 2010). None of the samples was positive for F. tularensis. Although several studies report presence of this bacterium, in I. ricinus ticks in Central and Western Europe, the prevalence is reaching only up to 0.2% even in tularemia active foci (Hubalek et al.1996; Gehringer et al.2013). South Bohemia is not a known tularemia endemic region.
Identification of the host in questing exophilic ticks presents a complex methodological challenge, since the blood-meal acquired in the previous developmental stage is already digested and the remnants, which allow host species detection, are usually present in a limited quantity and quality (Kirstein and Gray 1996). Therefore, sensitivity of the identification method is crucial. Multiple molecular biology methods of blood-meal analysis in questing ticks have been developed. Generally, these methods are based on amplification of a multicopy (usually mitochondrial) gene target such as 12S rDNA (Humair et al.2007; Wodecka, Rymaszewska and Skotarczak 2013), 18S rDNA (Pichon et al.2003; Allan et al.2010) or cytB (Kirstein and Gray 1996; Gray et al.1999). Techniques differ in the method of analysis (reverse line blotting, sequencing, restriction fragment length polymorphism, real-time PCR coupled with high-resolution melting analysis) and have variable levels of sensitivity and specificity.
In our study, the identification of the tick host was possible in 61% of the samples, which is in the range reported in previous studies (33%–65%) (Gray et al.1999; Pichon et al.2003, 2005; Estrada-Pena et al.2005; Pichon et al.2006; Wodecka, Rymaszewska and Skotarczak 2013; Collini et al.2016) but higher than in studies using the same method of analysis (49%, 43% and 39% as reported in Humair et al.2007; Moran Cadenas et al.2007; Burri et al.2011, respectively). In previous studies, the identification efficiency varied significantly according to the sampling locality (Moran Cadenas et al.2007) and sampling season (Pichon et al.2005), indicating the influence of temperature (Moran Cadenas et al.2007) and/or period post molting (Pichon et al.2005; Léger et al.2015). No statistically significant differences in the identification success rates were observed that correlated with localities or month of collection. The previously reported efficient DNA extraction procedure (Crowder et al.2010b) used here may have led to the higher success rate as previous studies with lower identification rates used an ammonium hydroxide extraction method (Pichon et al.2003, 2005; Estrada-Pena et al.2005; Humair et al.2007; Moran Cadenas et al.2007; Burri et al.2011). It was shown previously by direct comparison, that DNA extraction influences the host identification rate (Allan et al.2010; Collini et al.2016) with differences due to final concentration of the target DNA in the homogenate or eluate, extent of contamination with PCR inhibitors, or extent of damage to the integrity of the DNA during extraction.
As expected, the portion of samples with multiple hosts is higher in those studies with higher host identification rates. In our study, DNA of two host species was detected in 16% of the samples, even though only questing I. ricinus nymphs, which presumably fed only once as larvae, were tested. Similar results were reported for questing nymphal I. ricinus (Moran Cadenas et al.2007; Collini et al.2015, 2016) and questing nymphal Amblyomma americanum (Allan et al.2010). Interrupted feeding and subsequent infestation of a different host species might be an explanation for such results. It was shown previously that ixodid ticks are able to reattach and successfully complete feeding until engorgement after being prematurely detached from the host (Shih and Spielman 1993; Wang, Henbest and Nuttall 1999).
A high host identification rate as well as a high prevalence of multiple host detections could be due to contamination. A positive signal in negative extraction controls was rarely detected in our analysis and was never confirmed in repeated analysis, indicating that contamination of the DNA extract is unlikely. Therefore, we are convinced that the difference in identification success rate is not due to contamination. Further, the extraction method used here seems to produce a more efficient template for host identification (Allan et al.2010). Although we have used a relatively old method (Humair et al.2007) of host identification, a recently presented PCR—high-resolution melting analysis brings very similar results concerning identification success (65% and 55% using Qiagen silica-based extraction kit) and portion of samples with multiple host species detected (24% and 11%) (Collini et al.2015, 2016).
The hosts most frequently identified at all but one study site were artiodactyls (31%–60%) followed by rodents (27%–62%). Birds were less frequently identified as hosts, although variations were present among the study sites. The high proportion of artiodactyls is in agreement with findings of other studies employing blood-meal analysis (Pichon et al.2003; Humair et al.2007; Moran Cadenas et al.2007; Burri et al.2011; Wodecka, Rymaszewska and Skotarczak 2013). Large mammals, which are frequently infested by ticks of all stages, are able to support large numbers of ticks (Ruiz-Fons et al.2006; Carpi et al.2008; Kiffner et al.2010; Vor et al.2010). Thus, they may serve as a globally important source of blood-meal despite their relatively lower abundance compared to rodents (Hofmeester et al.2016). In different study sites, different patterns of proportional representation among rodent, artiodactyl and bird species were observed, presumably reflecting the overall representation of the species determined by natural conditions in the particular habitat. Similar results showing high proportion of bird-fed (Estrada-Pena et al.2005; Pichon et al.2005) as well as artiodactyl-fed ticks (Moran Cadenas et al.2007; Wodecka, Rymaszewska and Skotarczak 2013) were reported from different locations in Europe.
Hosts were rarely identified to the species level, although it is known that species of deer, birds, Apodemus spp., Microtus spp. and Myodes glareolus occur in the area of South Bohemia. Since the identification success rate on species level was similar in a study using the same set of probes (Moran Cadenas et al.2007), it seems that the current probes do not cover the real sequence variability of the target species. The probes should be further optimized or new probes added to enable species-level identification of tick hosts. This is especially needed for the artiodactyl probe, which was originally validated using only artiodactyl DNA but not on artiodactyls-fed ticks (Humair et al.2007).
Comparison of the prevalence of tick-borne pathogens among groups of ticks that fed on different hosts shows effectiveness of pathogen transmission from particular host species to feeding ticks. Exclusion of samples with multiple hosts reduced the sample size and statistically significant differences among different host groups were not detected. Surprisingly, a similar proportion of B. burgdorferi s.l.-positive samples was found among ticks fed on rodents and artiodactyls, particularly wild boar. Similar results were reported in other studies based on blood-meal analyses (Estrada-Pena et al.2005; Moran Cadenas et al.2007; Wodecka, Rymaszewska and Skotarczak 2013). In contrast, multiple laboratory and field studies indicate that ungulates are generally incompetent as B. burgdorferi reservoirs (Matuschka et al.1993; Talleklint and Jaenson 1994; Nelson et al.2000; Kjelland et al.2011). Borrelia burgdorferi spirochetes are present in the blood and skin of artiodactyls including wild boar (Pichon et al.2000; Faria et al.2015; Ebani et al.2016; VanBik et al.2017; Zhai et al.2017), and some of the ticks are infected (Kjelland et al.2011; Pacilly et al.2014; Silaghi, Pfister and Overzier 2014), but the prevalence rates in partially fed ticks are markedly lower compared to questing ticks. Since the probes specific for artiodactyla and wild boar seemed to have appropriate specificity (Humair et al.2007) and similar results were obtained in a study using different approach (Wodecka, Rymaszewska and Skotarczak 2013), it seems improbable that these results are due to false identification of the host. We speculate that part of the difference in the prevalence in questing and fed ticks may be caused by reduced number of spirochetes after feeding, which (together with PCR inhibitors present in the fed tick) could result in lower prevalence rate. After the blood digestion and molting, the spirochetes amplify and are readily detected in the questing ticks (Jacquet et al.2017). An analysis of B. burgdorferi s.l. prevalence in fed and unfed larval ticks sampled from multiple host species compared to analysis in questing nymphal ticks from the same locality would address this hypothesis.
The associations between B. burgdorferi genospecies and vertebrate hosts reported previously in multiple host trapping and laboratory transmission experiments (Kurtenbach et al.1998a,b; Perez et al.2012) were not completely recapitulated in our study. Nevertheless, the affinity of B. afzelii for rodents and of B. garinii and B. valaisiana for avian hosts was confirmed and is in concordance with conclusions of other blood-meal analysis based studies (Pichon et al.2005; Moran Cadenas et al.2007; Wodecka, Rymaszewska and Skotarczak 2013). In our study, B. bavariensis was not differentiated from B. garinii, since B. bavariensis was not yet established as a separate genospecies at the time of the sample analysis. Therefore, some B. garinii identifications in rodent-fed ticks might be in fact be the rodent-associated genospecies B. bavariensis. Based on currently available data, the prevalence of B. bavariensis is generally low (approximately 4.5% of B. burgdorferi infected ticks) among European I. ricinus tick populations (Lommano et al.2012; Herrmann, Gern and Voordouw 2013; Glatz et al.2014; Daniel et al.2016) and therefore should not substantially influence the results. In addition, trapping studies also report some exceptions from the general genospecies associations. Borrelia afzelii and B. bavariensis were identified in bird-fed ticks (including larvae) (Geller et al.2013; Lommano et al.2014), and B. garinii and B. burgdorferi s.s. were found in lizard-fed ticks (Majlathova et al.2006). Borrelia garinii (distinguished from B. bavariensis) was detected in rodent tissues (Khanakah et al.2006; Pisanu et al.2014; Hamsikova et al.2017). Borrelia lusitaniae was also found in artiodactyl-fed ticks, although lizards are considered the main reservoir host of this Borrelia genospecies (Majlathova et al.2006; Richter and Matuschka 2006).
Because of our relatively small sample size, we have limited data on host association of the less frequent tick-borne pathogens and conditional pathogens. In general, our data correspond with current knowledge. Borrelia miyamotoi was found in a tick that fed on rodents, which is the most probable reservoir host of this spirochete (Barbour et al.2009; Cosson et al.2014). Rickettsia helvetica was detected in ticks that fed on rodent, artiodactyl and avian host species. In concordance with this finding, R. helvetica has been detected in the blood of a variety of vertebrate animals (wild boar, deer and rodents) (Karbowiak et al.2016). No specific reservoir host has been identified for this Rickettsia species. Apparently, transovarial and transstadial transmission in ticks contributes to the natural circulation of the bacterium (Sprong et al.2009).
Anaplasma phagocytophilum in Europe occurs in four distinct ecotypes and has been detected in a broad range of hosts including rodents and artiodactyls (Silaghi et al.2012; Jahfari et al.2014), as confirmed in our study. We detected Ba. microti in a rodent-fed tick, its common host species (Silaghi et al.2012; Hamsikova et al.2016). Babesia venatorum was found in artiodactyl- and rodent-fed ticks in our study, although to date only roe deer have been reported as the host of this Babesia species (Duh et al.2005; Bonnet et al.2007; Overzier et al.2013; Andersson et al.2016). Nevertheless, transovarial transmission was reported for Babesia venatorum in I. ricinus ticks (Bonnet et al.2007), and thus the presence of the pathogen in the tick may not necessarily be the result of an interaction of the tick with the last host.
Molecular methods of blood-meal analysis are still the only available techniques that bring objective, overall assessment of the tick host species spectrum and the proportional representation of different host species. Some of our results are in disagreement with generally accepted opinions on I. ricinus ecology and pathogen transmission, in particular, the presence of DNA from multiple hosts in questing nymphs and the unexpected host associations of B. burgdorferi s.l. and its particular genospecies. Careful validation of the sensitivity and specificity of the host-identification methods is needed whenever applied in a new environment. A study employing blood-meal analysis of questing nymphs combined with on host tick sampling and pathogen detection should reveal the cause of the discrepancies as well as the true species composition of a particular tick population.
Acknowledgements
The authors would like to thank Prof. Lise Gern and Caroline Burri (Laboratory of Eco-epidemiology of Parasites, University of Neuchatel) for practical introduction into the tick blood-meal analysis. We would also like to thank Jacqueline R. Wyatt of J & L Scientific Editing for the editing of this manuscript.
FUNDING
This work was supported by National Institutes of Health [grant number 2R44AI077156], by Abbott, by the European Union 7th Framework program [project 278976 ANTIGONE], and by the Ministry of Education, Youth and Sports of the Czech Republic under the National Program of Sustainability I (NPU I) [project LO1218].
Conflict of interest. MWE, CDC, HEC, MAR, MRM, CM and DJE are all employees of Ibis Biosciences, an Abbott Company, which developed the PCR/ESI-MS assays and instrumentation used in these studies. Assays described are for research use only.
REFERENCES
- Allan BF, Goessling LS, Storch GA. et al. Blood meal analysis to identify reservoir hosts for Amblyomma americanum Ticks. Emerg Infect Dis 2010;16:433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MO, Bergvall UA, Chirico J. et al. Molecular detection of Babesia capreoli and Babesia venatorum in wild Swedish roe deer, Capreolus capreolus. Parasite Vector 2016;9:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsuaga M, Gonzalez LM, Lobo CA. et al. First report of Babesia microti-caused babesiosis in Spain. Vector-Borne Zoonot 2016;16:677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Bunikis J, Travinsky B. et al. Niche Partitioning of Borrelia burgdorferi and Borrelia miyamotoi in the same tick vector and mammalian reservoir species. Am J Trop Med Hyg 2009;81:1120–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat B, Stańczak J, Michalik J. et al. Rickettsia helvetica and R. monacensis infections in immature Ixodes ricinus ticks derived from sylvatic passerine birds in west-central Poland. Parasitol Res 2016;115:3469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschitz M, Narodoslavsky-Gföller M, Kanzler M. et al. Babesia species occurring in Austrian Ixodes ricinus ticks. Appl Environ Microb 2008;74:4841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet S, Jouglin M, L’Hostis M. et al. Babesia sp. EU1 from roe reer and transmission within Ixodes ricinus. Emerg Infect Dis 2007;13:1208–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis J, Garpmo U, Tsao J. et al. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiol Read Engl 2004;150:1741–55. [DOI] [PubMed] [Google Scholar]
- Burri C, Bastic V, Maeder G. et al. Microclimate and the zoonotic cycle of tick-borne encephalitis virus in Switzerland. J Med Entomol 2011;48:615–27. [DOI] [PubMed] [Google Scholar]
- Carleton MD, Gardner AL, Pavlinov IY. et al. The valid generic name for red-backed voles (Muroidea: Cricetidae: Arvicolinae): restatement of the case for Myodes Pallas, 1811. J Mammal 2014;95:943–59. [Google Scholar]
- Carpi G, Cagnacci F, Neteler M. et al. Tick infestation on roe deer in relation to geographic and remotely sensed climatic variables in a tick-borne encephalitis endemic area. Epidemiol Infect 2008;136:1416–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel RN, Attoui H, Butenko AM. et al. Tick-borne virus diseases of human interest in Europe. Clin Microbiol Infec 2004;10:1040–55. [DOI] [PubMed] [Google Scholar]
- Chowdri HR, Gugliotta JL, Berardi VP. et al. Borrelia miyamotoi infection presenting as human granulocytic anaplasmosis: a case report. Ann Intern Med 2013;159:21–7. [DOI] [PubMed] [Google Scholar]
- Collini M, Albonico F, Hauffe HC. et al. Identifying the last bloodmeal of questing sheep tick nymphs (Ixodes ricinus L.) using high resolution melting analysis. Vet Parasitol 2015;210:194–205. [DOI] [PubMed] [Google Scholar]
- Collini M, Albonico F, Rosà R. et al. Identification of Ixodes ricinus blood meals using an automated protocol with high resolution melting analysis (HRMA) reveals the importance of domestic dogs as larval tick hosts in Italian alpine forests. Parasite Vector 2016;9:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson J-F, Michelet L, Chotte J. et al. Genetic characterization of the human relapsing fever spirochete Borrelia miyamotoi in vectors and animal reservoirs of Lyme disease spirochetes in France. Parasite Vector 2014;7:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder CD, Carolan HE, Rounds MA. et al. Prevalence of Borrelia miyamotoi in Ixodes Ticks in Europe and the United States. Emerg Infect Dis 2014;20:1678–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder CD, Matthews HE, Rounds MA. et al. Detection of heartworm infection in dogs via PCR amplification and electrospray ionization mass spectrometry of nucleic acid extracts from whole blood samples. Am J Vet Res 2012;73:854–9. [DOI] [PubMed] [Google Scholar]
- Crowder CD, Matthews HE, Schutzer S. et al. Genotypic variation and mixtures of Lyme borrelia in Ixodes ticks from North America and Europe. PLoS One 2010a;5, DOI: 10.1371/journal.pone.0010650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder CD, Rounds MA, Phillipson CA. et al. Extraction of total nucleic acids from ticks for the detection of bacterial and viral pathogens. J Med Entomol 2010b;47:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel M, Rudenko N, Golovchenko M. et al. The occurrence of Ixodes ricinus ticks and important tick-borne pathogens in areas with high tick-borne encephalitis prevalence in different altitudinal levels of the Czech Republic Part II. Ixodes ricinus ticks and genospecies of Borrelia burgdorferi sensu lato complex. Epidemiol Mikrobi Im 2016;65:182–92. [PubMed] [Google Scholar]
- Duh D, Petrovec M, Bidovec A. et al. Cervids as Babesiae hosts, Slovenia. Emerg Infect Dis 2005;11:1121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumler JS, Choi K-S, Garcia-Garcia JC. et al. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg Infect Dis 2005;11:1828–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebani VV, Rocchigiani G, Bertelloni F. et al. Molecular survey on the presence of zoonotic arthropod-borne pathogens in wild red deer (Cervus elaphus). Comp Immunol Microb 2016;47:77–80. [DOI] [PubMed] [Google Scholar]
- Ecker DJ, Drader JJ, Gutierrez J. et al. The Ibis T5000 Universal Biosensor: an automated platform for pathogen identification and strain typing. JALA J Assoc Lab Aut 2006a;11:341–51. [Google Scholar]
- Ecker JA, Massire C, Hall TA. et al. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J Clin Microbiol 2006b;44:2921–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker DJ, Sampath R, Blyn LB. et al. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc Natl Acad Sci U S A 2005;102:8012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker DJ, Sampath R, Li H. et al. New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev Mol Diagn 2010;10:399–415. [DOI] [PubMed] [Google Scholar]
- Elfving K, Olsen B, Bergström S. et al. Dissemination of spotted fever Rickettsia agents in Europe by migrating birds. PLoS One 2010;5:e8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshoo MW, Carolan HE, Massire C. et al. Survey of Ixodes pacificus ticks in California reveals a diversity of microorganisms and a novel and widespread Anaplasmataceae species. PLoS One 2015;10:e0135828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshoo MW, Crowder CC, Rebman AW. et al. Direct molecular detection and genotyping of Borrelia burgdorferi from whole blood of patients with early Lyme disease. PLoS One 2012;7:e36825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshoo MW, Crowder CD, Carolan HE. et al. Broad-range survey of tick-borne pathogens in southern Germany reveals a high prevalence of Babesia microti and a diversity of other tick-borne pathogens. Vector-Borne Zoonot 2014;14:584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshoo MW, Crowder CD, Li H. et al. Detection and identification of Ehrlichia species in blood by use of PCR and electrospray ionization mass spectrometry. J Clin Microbiol 2010;48:472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshoo MW, Whitehouse CA, Zoll ST. et al. Direct broad-range detection of alphaviruses in mosquito extracts. Virology 2007;368:286–95. [DOI] [PubMed] [Google Scholar]
- Estrada-Pena A, Osacar JJ, Pichon B. et al. Hosts and pathogen detection for immature stages of Ixodes ricinus (Acari: Ixodidae) in north-central Spain. Exp Appl Acarol 2005;37:257–68. [DOI] [PubMed] [Google Scholar]
- Faria AS, Paiva-Cardoso M das N, Nunes M. et al. First detection of Borrelia burgdorferi sensu lato DNA in serum of the wild boar (Sus scrofa) in northern Portugal by nested-PCR. EcoHealth 2015;12:183–7. [DOI] [PubMed] [Google Scholar]
- Fournier PE, Grunnenberger F, Jaulhac B. et al. Evidence of Rickettsia helvetica infection in humans, eastern France. Emerg Infect Dis 2000;6:389–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariepy TD, Lindsay R, Ogden N. et al. Identifying the last supper: utility of the DNA barcode library for bloodmeal identification in ticks. Mol Ecol Resour 2012;12:646–52. [DOI] [PubMed] [Google Scholar]
- Gehringer H, Schacht E, Maylaender N. et al. Presence of an emerging subclone of Francisella tularensis holarctica in Ixodes ricinus ticks from south-western Germany. Ticks Tick-borne Dis 2013;4:93–100. [DOI] [PubMed] [Google Scholar]
- Geller J, Nazarova L, Katargina O. et al. Tick-borne pathogens in ticks feeding on migratory passerines in western part of Estonia. Vector-Borne Zoonot 2013;13:443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gern L, Douet V, López Z. et al. Diversity of Borrelia genospecies in Ixodes ricinus ticks in a Lyme borreliosis endemic area in Switzerland identified by using new probes for reverse line blotting. Ticks Tick-Borne Dis 2010;1:23–9. [DOI] [PubMed] [Google Scholar]
- Glatz M, Muellegger RR, Hizo-Teufel C. et al. Low prevalence of Borrelia bavariensis in Ixodes ricinus ticks in southeastern Austria. Ticks Tick-Borne Dis 2014;5:649–50. [DOI] [PubMed] [Google Scholar]
- Gray JS, Kirstein F, Robertson JN. et al. Borrelia burgdorferi sensu lato in Ixodes ricinus ticks and rodents in a recreational park in south-western Ireland. Exp Appl Acarol 1999;23:717–29. [DOI] [PubMed] [Google Scholar]
- Gugliotta JL, Goethert HK, Berardi VP. et al. Meningoencephalitis from Borrelia miyamotoi in an immunocompromised patient. New Engl J Med 2013;368:240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamsikova Z, Coipan C, Mahrikova L. et al. Borrelia miyamotoi and co-infection with Borrelia afzelii in Ixodes ricinus ticks and rodents from Slovakia. Microb Ecol 2017;73:1000–8. [DOI] [PubMed] [Google Scholar]
- Hamsikova Z, Kazimirova M, Harustiakova D. et al. Babesia spp. in ticks and wildlife in different habitat types of Slovakia. Parasite Vector 2016;9:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C, Gern L, Voordouw MJ. Species co-occurrence patterns among Lyme borreliosis pathogens in the tick vector Ixodes ricinus. Appl Environ Microb 2013;79:7273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt A, Gray JS, Hunfeld K-P. Human Babesiosis in Europe: what clinicians need to know. Infection 2013;41:1057–72. [DOI] [PubMed] [Google Scholar]
- Hildebrandt A, Hunfeld K-P, Baier M. et al. First confirmed autochthonous case of human Babesia microti infection in Europe. Eur J Clin Microbiol 2007;26, DOI: 10.1007/s10096-007-0333-1. [DOI] [PubMed] [Google Scholar]
- Hofmeester TR, Coipan EC, van Wieren SE. et al. Few vertebrate species dominate the Borrelia burgdorferi s. l. life cycle. Environ Res Lett 2016;11:043001. [Google Scholar]
- Hovius JWR, de Wever B, Sohne M. et al. A case of meningoencephalitis by the relapsing fever spirochaete Borrelia miyamotoi in Europe. Lancet 2013;382:658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubalek Z, Treml F, Halouzka J. et al. Frequent isolation of Francisella tularensis from Dermacentor reticulatus ticks in an enzootic focus of tularaemia. Med Vet Entomol 1996;10:241–6. [DOI] [PubMed] [Google Scholar]
- Humair P-F, Douet V, Morán Cadenas F. et al. Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J Med Entomol 2007;44:869–80. [DOI] [PubMed] [Google Scholar]
- Hunfeld KP, Brade V. Zoonotic Babesia: possibly emerging pathogens to be considered for tick-infested humans in Central Europe. Int J Med Microbiol IJMM 2004;293(Suppl 37):93–103. [DOI] [PubMed] [Google Scholar]
- Jacquet M, Genné D, Belli A. et al. The abundance of the Lyme disease pathogen Borrelia afzelii declines over time in the tick vector Ixodes ricinus. Parasite Vector 2017;10, DOI: 10.1186/s13071-017-2187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jado I, Oteo JA, Aldámiz M. et al. Rickettsia monacensis and Human Disease, Spain. Emerg Infect Dis 2007;13:1405–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahfari S, Coipan EC, Fonville M. et al. Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasite Vector 2014;7:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar Z, Mihalca AD, Dumitrache MO. et al. Geographical distribution and prevalence of Borrelia burgdorferi genospecies in questing Ixodes ricinus from Romania: a countrywide study. Ticks Tick-Borne Dis 2013;4:403–8. [DOI] [PubMed] [Google Scholar]
- Karbowiak G, Biernat B, Stańczak J. et al. The role of particular tick developmental stages in the circulation of tick-borne pathogens affecting humans in Central Europe. 3. Rickettsiae. Ann Parasitol 2016;62:89–100. [DOI] [PubMed] [Google Scholar]
- Kent RJ. Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol Ecol Resour 2009;9:4–18. [DOI] [PubMed] [Google Scholar]
- Khanakah G, Kocianová E, Vyrosteková V. et al. Seasonal variations in detecting Borrelia burgdorferi sensu lato in rodents from north eastern Austria. Wien Klin Wochenschr 2006;118:754–8. [DOI] [PubMed] [Google Scholar]
- Kiffner C, Lödige C, Alings M. et al. Abundance estimation of Ixodes ticks (Acari: Ixodidae) on roe deer (Capreolus capreolus). Exp Appl Acarol 2010;52:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Gray JS. A molecular marker for the identification of the zoonotic reservoirs of Lyme borreliosis by analysis of the blood meal in its European vector Ixodes ricinus 1996;62:4060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjelland V, Ytrehus B, Stuen S. et al. Prevalence of Borrelia burgdorferi in Ixodes ricinus ticks collected from moose (Alces alces) and roe deer (Capreolus capreolus) in southern Norway. Ticks Tick-Borne Dis 2011;2:99–103. [DOI] [PubMed] [Google Scholar]
- Kozuch O, Grešíková M, Nosek J. et al. The role of small rodents and hedgehogs in a natural focus of tick-borne encephalitis. Bull World Health Organ 1967;36:61–6. [PMC free article] [PubMed] [Google Scholar]
- Krause PJ, Narasimhan S, Wormser GP. et al. Human Borrelia miyamotoi infection in the United States. New Engl J Med 2013;368:291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtenbach K, Carey D, Hoodless AN. et al. Competence of pheasants as reservoirs for Lyme disease spirochetes. J Med Entomol 1998a;35:77–81. [DOI] [PubMed] [Google Scholar]
- Kurtenbach K, De Michelis S, Etti S. et al. Host association of Borrelia burgdorferi sensu lato–the key role of host complement. Trends Microbiol 2002;10:74–9. [DOI] [PubMed] [Google Scholar]
- Kurtenbach K, Sewell H-S, Ogden NH. et al. Serum complement sensitivity as a key factor in Lyme disease ecology. Infect Immun 1998b;66:1248–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léger E, Liu X, Masseglia S. et al. Reliability of molecular host-identification methods for ticks: an experimental in vitro study with Ixodes ricinus. Parasite Vector 2015;8, DOI: 10.1186/s13071-015-1043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommano E, Bertaiola L, Dupasquier C. et al. Infections and coinfections of questing Ixodes ricinus ticks by emerging zoonotic pathogens in western Switzerland. Appl Environ Microb 2012;78:4606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommano E, Dvorak C, Vallotton L. et al. Tick-borne pathogens in ticks collected from breeding and migratory birds in Switzerland. Ticks Tick-Borne Dis 2014;5:871–82. [DOI] [PubMed] [Google Scholar]
- Majlathova V, Majlath I, Derdakova M. et al. Borrelia lusitaniae and green lizards (Lacerta viridis), karst region, Slovakia. Emerg Infect Dis 2006;12:1895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mărcuţan I-D, Kalmár Z, Ionică AM. et al. Spotted fever group rickettsiae in ticks of migratory birds in Romania. Parasite Vector 2016;9, DOI: 10.1186/s13071-016-1565-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A, Telford SR, Turk S-P. et al. Xenodiagnosis to detect Borrelia burgdorferi infection: a first-in-human study. Clin Infect Dis 2014;58:937–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuschka FR, Heiler M, Eiffert H. et al. Diversionary role of hoofed game in the transmission of Lyme disease spirochetes. Am J Trop Med Hyg 1993;48:693–9. [DOI] [PubMed] [Google Scholar]
- Matuschka FR, Richter D, Spielman A. Differential detachment from resting hosts of replete larval and nymphal Ixodes ticks. J Parasitol 1991;77:341–5. [PubMed] [Google Scholar]
- Moniuszko-Malinowska A, Swiecicka I, Dunaj J. et al. Infection with Babesia microti in humans with non-specific symptoms in North East Poland. Infect Dis 2016;48:537–43. [DOI] [PubMed] [Google Scholar]
- Moran Cadenas F, Rais O, Humair P-F. et al. Identification of host bloodmeal source and Borrelia burgdorferi sensu lato in field-collected Ixodes ricinus ticks in Chaumont (Switzerland). J Med Entomol 2007;44:1109–17. [DOI] [PubMed] [Google Scholar]
- Nazzi F, Martinelli E, Del Fabbro S. et al. Ticks and Lyme borreliosis in an alpine area in northeast Italy. Med Vet Entomol 2010;24:220–6. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Rooney S, Miller NJ. et al. Complement-mediated killing of Borrelia burgdorferi by nonimmune sera from sika deer. J Parasitol 2000;86:1232–8. [DOI] [PubMed] [Google Scholar]
- Nilsson K, Elfving K, Påhlson C. Rickettsia helvetica in patient with meningitis, Sweden, 2006. Emerg Infect Dis 2010;16:490–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overzier E, Pfister K, Herb I. et al. Detection of tick-borne pathogens in roe deer (Capreolus capreolus), in questing ticks (Ixodes ricinus), and in ticks infesting roe deer in southern Germany. Ticks Tick-Borne Dis 2013;4:320–8. [DOI] [PubMed] [Google Scholar]
- Pacilly FCA, Benning ME, Jacobs F. et al. Blood feeding on large grazers affects the transmission of Borrelia burgdorferi sensu lato by Ixodes ricinus. Ticks Tick-Borne Dis 2014;5:810–7. [DOI] [PubMed] [Google Scholar]
- Parola P, Raoult D. Tick-borne bacterial diseases emerging in Europe. Clin Microbiol Infec 2001;7:80–3. [DOI] [PubMed] [Google Scholar]
- Perez D, Kneubühler Y, Rais O. et al. Seasonality of Ixodes ricinus ticks on vegetation and on rodents and Borrelia burgdorferi sensu lato genospecies diversity in two Lyme borreliosis–endemic areas in Switzerland. Vector-Borne Zoonot 2012;12:633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon B, Egan D, Rogers M. et al. Detection and identification of pathogens and host DNA in unfed host-seeking Ixodes ricinus L. (Acari: Ixodidae). J Med Entomol 2003;40:723–31. [DOI] [PubMed] [Google Scholar]
- Pichon B, Gilot B, Pérez-Eid C. et al. Detection of spirochaetes of Borrelia burgdorferi complexe in the skin of cervids by PCR and culture. Eur J Epidemiol 2000;16:869–73. [DOI] [PubMed] [Google Scholar]
- Pichon B, Kahl O, Hammer B. et al. Pathogens and host DNA in Ixodes ricinus nymphal ticks from a German forest. Vector-Borne Zoonot Larchmt N 2006;6:382–7. [DOI] [PubMed] [Google Scholar]
- Pichon B, Rogers M, Egan D. et al. Blood-meal analysis for the identification of reservoir hosts of tick-borne pathogens in Ireland. Vector-Borne Zoonot Larchmt N 2005;5:172–80. [DOI] [PubMed] [Google Scholar]
- Pisanu B, Chapuis J-L, Dozières A. et al. High prevalence of Borrelia burgdorferi s.l. in the European red squirrel Sciurus vulgaris in France. Ticks Tick-Borne Dis 2014;5:1–6. [DOI] [PubMed] [Google Scholar]
- Platonov AE, Karan LS, Kolyasnikova NM. et al. Humans infected with relapsing fever spirochete Borrelia miyamotoi, Russia. Emerg Infect Dis 2011;17:1816–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkonjak A, Kleinerman G, Gutiérrez R. et al. Occurrence of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks with first identification of Borrelia miyamotoi in Vojvodina, Serbia. Vector-Borne Zoonot Larchmt N 2016;16:631–5. [DOI] [PubMed] [Google Scholar]
- Richter D, Matuschka F-R. Perpetuation of the Lyme disease spirochete Borrelia lusitaniae by lizards. Appl Environ Microb 2006;72:4627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter D, Matuschka F-R. “Candidatus Neoehrlichia mikurensis,” Anaplasma phagocytophilum, and Lyme disease spirochetes in questing European vector ticks and in feeding ticks removed from people. J Clin Microbiol 2012;50:943–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounds MA, Crowder CD, Matthews HE et al. . Identification of endosymbionts in ticks by broad-range polymerase chain reaction and electrospray ionization mass spectrometry. J Med Entomol 2012;49:843–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf I, Golovchenko M, Sikutová S. et al. Babesia microti (Piroplasmida: Babesiidae) in nymphal Ixodes ricinus (Acari: Ixodidae) in the Czech Republic. Folia Parasitol (Praha) 2005;52:274–6. [DOI] [PubMed] [Google Scholar]
- Ruiz-Fons F, Fernandez-de-Mera IG, Acevedo P. et al. Ixodid ticks parasitizing Iberian red deer (Cervus elaphus hispanicus) and European wild boar (Sus scrofa) from Spain: geographical and temporal distribution. Vet Parasitol 2006;140:133–42. [DOI] [PubMed] [Google Scholar]
- Schorn S, Pfister K, Reulen H. et al. Occurrence of Babesia spp., Rickettsia spp. and Bartonella spp. in Ixodes ricinus in Bavarian public parks, Germany. Parasite Vector 2011;4:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CM, Spielman A. Accelerated transmission of Lyme disease spirochetes by partially fed vector ticks. J Clin Microbiol 1993;31:2878–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silaghi C, Pfister K, Overzier E. Molecular investigation for bacterial and protozoan tick-borne pathogens in wild boars (Sus scrofa) from southern Germany. Vector-Borne Zoonot 2014;14:371–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silaghi C, Woll D, Hamel D. et al. Babesia spp. and Anaplasma phagocytophilum in questing ticks, ticks parasitizing rodents and the parasitized rodents – Analyzing the host-pathogen-vector interface in a metropolitan area. Parasite Vector 2012;5:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprong H, Wielinga PR, Fonville M. et al. Ixodes ricinus ticks are reservoir hosts for Rickettsia helvetica and potentially carry flea-borne Rickettsia species. Parasite Vector 2009;2:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad M, Honig V, Ruzek D. et al. Europe-wide meta-analysis of Borrelia burgdorferi sensu lato prevalence in questing Ixodes ricinus ticks. Appl Environ Microb 2017;83, DOI: 10.1128/AEM.00609-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum—a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol 2013;3, DOI: 10.3389/fcimb.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talleklint L, Jaenson TG. Transmission of Borrelia burgdorferi s.l. from mammal reservoirs to the primary vector of Lyme borreliosis, Ixodes ricinus (Acari: Ixodidae), in Sweden. J Med Entomol 1994;31:880–6. [DOI] [PubMed] [Google Scholar]
- Tappe J, Jordan D, Janecek E. et al. Revisited: Borrelia burgdorferi sensu lato infections in hard ticks (Ixodes ricinus) in the city of Hanover (Germany). Parasite Vector 2014;7:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijvendijk G, Coipan C, Wagemakers A. et al. Larvae of Ixodes ricinus transmit Borrelia afzelii and B. miyamotoi to vertebrate hosts. Parasite Vector 2016;9:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBik D, Lee SH, Seo MG. et al. Borrelia species detected in ticks feeding on wild Korean water deer (Hydropotes inermis) using molecular and genotypic analyses. J Med Entomol 2017, DOI: 10.1093/jme/tjx106. [DOI] [PubMed] [Google Scholar]
- Venclikova K, Mendel J, Betasova L. et al. First evidence of Babesia venatorum and Babesia capreoli in questing Ixodes ricinus ticks in the Czech Republic. Ann Agric Environ Med 2015;22:212–4. [DOI] [PubMed] [Google Scholar]
- Vor T, Kiffner C, Hagedorn P. et al. Tick burden on European roe deer (Capreolus capreolus). Exp Appl Acarol 2010;51:405–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Henbest PJ, Nuttall PA. Successful interrupted feeding of adult Rhipicephalus appendiculatus (Ixodidae) is accompanied by re-programming of salivary gland protein expression. Parasitology 1999;119:143–9. [DOI] [PubMed] [Google Scholar]
- Wodecka B, Rymaszewska A, Skotarczak B. Host and pathogen DNA identification in blood meals of nymphal Ixodes ricinus ticks from forest parks and rural forests of Poland. Exp Appl Acarol 2013;62:543–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldehiwet Z. Anaplasma phagocytophilum in ruminants in Europe. Ann N Y Acad Sci 2006;1078:446–60. [DOI] [PubMed] [Google Scholar]
- Yabsley MJ, Shock BC. Natural history of zoonotic Babesia: role of wildlife reservoirs. Int J Parasitol Parasites Wildl 2013;2:18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Niu Q, Yang J. et al. Identification and molecular survey of Borrelia burgdorferi sensu lato in sika deer (Cervus nippon) from Jilin Province, north-eastern China. Acta Trop 2017;166:54–7. [DOI] [PubMed] [Google Scholar]