Abstract
A growing number of single-nucleotide polymorphisms (SNPs) have been associated with body mass index (BMI) and obesity, but whether the effects of these obesity-susceptibility loci are uniform across the BMI distribution remains unclear. We studied the effects of 37 BMI-associated SNPs in 75,230 adults of European ancestry across BMI percentiles by using conditional quantile regression (CQR) and meta-regression (MR) models. The effects of nine SNPs (24%)—rs1421085 (FTO; p = 8.69 × 10−15), rs6235 (PCSK1; p = 7.11 × 10−6), rs7903146 (TCF7L2; p = 9.60 × 10−6), rs11873305 (MC4R; p = 5.08 × 10−5), rs12617233 (FANCL; p = 5.30 × 10−5), rs11672660 (GIPR; p = 1.64 × 10−4), rs997295 (MAP2K5; p = 3.25 × 10−4), rs6499653 (FTO; p = 6.23 × 10−4), and rs3824755 (NT5C2; p = 7.90 × 10−4)—increased significantly across the sample BMI distribution. We showed that such increases stemmed from unadjusted gene interactions that enhanced the effects of SNPs in persons with a high BMI. When 125 height-associated SNPs were analyzed for comparison, only one (<1%), rs6219 (IGF1, p = 1.80 × 10−4), showed effects that varied significantly across height percentiles. Cumulative gene scores of these SNPs (GS-BMI and GS-height) showed that only GS-BMI had effects that increased significantly across the sample distribution (BMI: p = 7.03 × 10−37; height: p = 0.499). Overall, these findings underscore the importance of gene-gene and gene-environment interactions in shaping the genetic architecture of BMI and advance a method for detecting such interactions by using only the sample outcome distribution.
Keywords: body mass index, height, polygenic inheritance, gene score, quantitative trait distribution, variable allele penetrance, conditional quantile regression, epistasis, gene-environment interactions, missing heritability
Introduction
Obesity is a prominent risk factor for osteoarthritis, hypertension, type 2 diabetes (T2D), cardiovascular disease, and certain psychological disorders and cancers.1, 2 The rise in obesity has coincided with “obesogenic” societal and environmental changes that include increased consumption of high-calorie foods, an increasingly sedentary lifestyle, and urbanization.2, 3, 4 Genetic factors are also known to play an important role in obesity, given that 50%–80% of body mass index (BMI) variation can be ascribed to genetics (heritability).5, 6 Moreover, genome-wide association studies (GWASs) have identified ∼140 polygenic loci that are directly associated with BMI or obesity.7
The role of individual and compound gene-environment (GXE) and gene-gene (GXG) interactions in determining BMI has not been fully elucidated. The study of BMI-associated GXG interactions has been impeded by statistical and computational limitations, although promising new approaches have recently been proposed.8, 9, 10 On the other hand, several lines of evidence suggest that GXE interactions could play an important role in shaping BMI. First, estimates of the heritability of BMI are influenced by environmental exposures.11 One study reported that the heritability of BMI is increased in persons born after the obesogenic transition, whereas another reported that the heritability of BMI is correlated with the population prevalence of obesity.12, 13 More recently, the cumulative gene score from 29 BMI-associated single-nucleotide polymorphisms (SNPs) showed a positive interaction effect with birth year.14 Interactions between the genetic determinants of BMI and obesogenic environmental factors readily explain why both estimates of BMI heritability and cumulative SNP effects are enhanced in permissive environments. Second, specific interactions between BMI-associated SNPs and environmental factors have been documented.11 Physical activity and energy intake have been reported to modify the effects of SNPs within the fat-mass- and obesity-associated gene FTO (MIM: 610966).15, 16, 17, 18, 19 Importantly, FTO (rs1421085) has been shown to jointly interact with diet, physical activity, salt and alcohol consumption, and sleep duration.20 Thus, a subset of genetic variants could affect BMI through a mixture of direct effects and compound interactions. As such, investigating individual environmental factors might not capture the full range of environmental modification for a given SNP.21, 22
In this report, we advanced a statistical framework to assess the effects of single and mixed GXE and GXG interactions on the association between SNPs and BMI. Specifically, we applied conditional quantile regression (CQR) to investigate the effects of 37 BMI-associated SNPs at multiple percentiles of the sample BMI distribution in 75,230 adults of European ancestry (EA).23, 24 Variability in SNP effects across these BMI percentiles was demonstrated to result from unadjusted interactions and was modeled by meta-regression (MR).25, 26 In this way, we used CQR and MR to collect evidence of unadjusted interactions directly from the sample distribution of BMI without measures of specific environmental factors. A secondary analysis of 125 established height-associated SNPs is also included for comparison.
Subjects and Methods
Participants and Phenotypes
The sample population included participants from the following studies: Atherosclerosis Risk in Communities (ARIC; phs000280.v3.p1), Coronary Artery Risk Development in Young Adults (CARDIA; phs000285.v3.p2), Cardiovascular Health Study (CHS; phs000287.v6.p1), EpiDREAM, the Framingham Cohort (phs000007.v29.p10), Multi-Ethnic Study of Atherosclerosis (MESA; phs000209.v13.p3), Genetic Epidemiology of COPD (COPDGene; phs000179.v5.p2), Electronic Medical Records and Genomics (eMERGE) II (phs000888.v1.p1), and the Women's Health Initiative (WHI; phs000200.v10.p3). Measurements collected from participants below the age of 18 years or above the age of 92 years were excluded (<1% collectively). For studies with repeated measures across multiple time points or visits, the median height and the median weight were extracted along with the corresponding age at these median values. We calculated BMI by dividing the median weight (in kg) by the square of the average measures of height (m). Diabetic status was indicated by one of the following criteria: (1) physician report or self-report of physician diagnosis, (2) report of taking diabetes medication, (3) fasting plasma glucose ≥ 126 mg/dL (7 mM), or (4) 2 hr glucose ≥ 200 mg/dL (11 mM) during an oral glucose-tolerance test.27 Obesity categories including normal weight (NW) and overweight (OW), as well as obesity classes I, II, and III (Ob-I, Ob-II, and Ob-III, respectively), were specified according to World Health Organization guidelines.28 Analyses were restricted to participants of self-reported EA with a combined sample size of n = 75,230. Summary statistics are presented in Table S1. This project was approved by a local ethics committee (Hamilton Integrated Research Ethics Board), and participant-level data access was granted through the Database of Genotypes and Phenotypes (dbGaP) after approval was provided by study-specific data-access committees. All analyses are consistent with study-specific data-use certifications.
Sample Quality Control
Detailed genotyping procedures for EpiDREAM and studies from the Candidate Gene Association Resource (CARe) project, including ARIC (phs000557.v2.p1), CARDIA (phs000613.v1.p2), CHS (phs000377.v4.p1), the Framingham Cohort (phs000282.v17.p10), and MESA (phs000283.v7.p3), are presented elsewhere.29, 30 Genotyping was performed with the gene-centric HumanCVD Genotyping BeadChip with 49,320 markers concentrated in ∼2,100 loci related to metabolism and cardiovascular disease.31 This limited scope of analysis was motivated by the availability of a greater sample size, as well as the high computational cost of fitting CQR models. Samples with sex discordance, an array-wide call rate below 95%–98%, and/or an average heterozygosity beyond 3 standard deviations of the mean heterozygosity were removed.32, 33 Family members were defined by identity by descent (IBD, ) above 0.5, and those with a lower call rate were removed so that only one member of each family group was retained for analysis (Table S2). Samples from COPDGene (phs000765.v1.p2) were genotyped with the Illumina HumanHap550 (v3) genotyping BeadChip (Illumina) with 561,466 markers, and QC procedures were performed as above except that cryptic relatedness was defined by IBD > 0.1875.34, 35 Genotypes from the WHI study (phs000746.v1.p3) and eMERGE II (phs000888.v1.p1) were composed of an imputed dataset, and samples from related or duplicate participants were removed. Analyses of the WHI dataset were conducted on each sub-study (WHI Memory Study [WHIMS], WHI Genomics and Randomized Trials Network [GARNET], HIPFX [Hip Fracture GWAS], MOPMAP, and Genetics and Epidemiology of Colorectal Cancer Consortium [GECCO]). A summary of sample quality control (QC), along with a complete list of datasets (and accession numbers) and additional details on these studies, is provided in Table S2.
SNP Selection and Marker QC
We identified SNPs that had previously been associated with BMI, obesity, and height by searching the GWAS Catalog and GIANT Consortium data files and screening the literature.36, 37, 38, 39 A.A. and D.M. conducted literature screening independently to maximize SNP attainment. For GWAS SNPs, only associations with p < 5 × 10−8 were considered. These SNPs were sorted into correlated linkage disequilibrium (LD, R2 > 0.1) blocks on the basis of genomic sequences from EA populations (1000 Genomes Project phase 3), and the strongest association SNP on the HumanCVD Genotyping BeadChip was selected.31, 40 Proxy SNPs (R2 > 0.9) were identified for SNPs not represented on the array. Thus, 39 BMI- and 129 height-associated SNPs were identified. For studies that used different genotyping platforms, the original association SNPs (39 BMI and 129 height) were screened and proxied as described above on each genotyping platform. For SNPs that mapped to the same gene, we screened them jointly with conditional regression analysis to test for independent associations with quantitative traits (BMI or height), and only SNPs that maintained associations were retained.41 However, SNPs in FTO (rs1421085 and rs6499653) and PCSK1 (MIM: 162150; rs6232 and rs6235) were exempted from exclusion as a result of prior evidence in the literature of independent associations with BMI.42, 43, 44 In total, 37 BMI- and 125 height-associated independent SNPs were identified and selected for further analysis. SNP call rate, minor allele frequency (MAF), and exact tests of Hardy-Weinberg equilibrium (HWE) in EA populations are presented in Tables S3 and S4. Within each study, SNPs with a call rate < 90% or HWE p value < 1 × 10−6 were excluded from analysis. In addition, only SNPs imputed with high quality were retained for analysis (R2 > 0.7 for WHI and info score > 0.7 for eMERGE II).45 SNP genotypes were encoded per the effect alleles and modeled additively for individual analyses.
Gene Scores
The cumulative gene score (GS) was calculated for all BMI- and height-associated SNPs (GS-BMI and GS-height, respectively). An un-weighted GS was utilized because weights can be biased and context dependent.46, 47 No GS was calculated for participants with more than 10% missing genotypes; otherwise, missing SNP genotypes were imputed with the arithmetic average genotype at each missing SNP. In addition to being associated with BMI, GIPR (MIM: 137241; rs10423928, LD R2 = 1 with rs11672660 in EA), TCF7L2 (MIM: 602228; rs7903146), TOMM40 (MIM: 608061) and APOE (MIM: 107741) (both rs2075650), HMGCR (MIM: 142910; rs4604177, LD R2 = 0.63 with rs6453133 in EA), PCSK1 (rs6235), CDKAL1 (MIM: 611259; rs9356744), and KCNQ1 (MIM: 607542; rs2283228) have also been associated with several co-morbidities of obesity, including glucose homeostasis, T2D, increased lipid levels, and heightened C-reactive protein (CRP) levels.48, 49, 50, 51, 52, 53, 54, 55 To mitigate potential biases stemming from these comorbidities at higher BMI percentiles, we also calculated a GS excluding these seven SNPs: GS-BMI (stringent). Finally, GSs for both BMI and height were calculated without imputation of missing genotypes: GS-BMI (no imputation) and GS-height (no imputation). GS-BMI (stringent), GS-BMI (no imputation), and GS-height (no imputation) were tested by sensitivity analysis.
Statistical Analysis
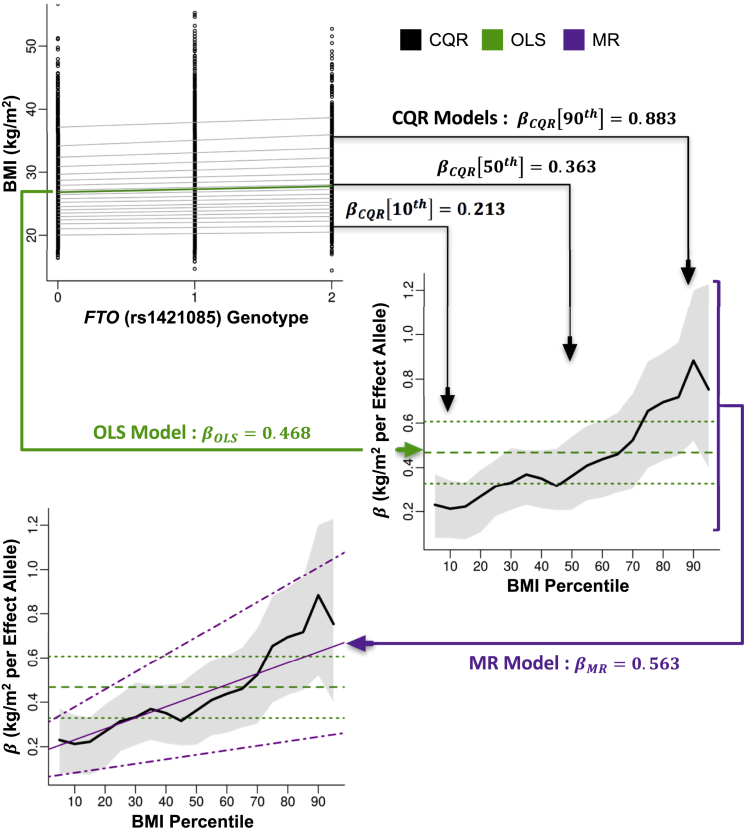
A statistical framework combining CQR and MR was used to model variation in the effects of SNPs under single and mixed GXE and GXG interactions (see Supplemental Note).24, 26 Like ordinary least-squares (OLS) models, CQR models can assume a linear relationship and provide intercept and slope estimates for a series of pre-specified percentiles.23, 24 Therefore, CQR can be applied to produce a comprehensive evaluation of the effects of a SNP across the sample distribution of a quantitative trait (e.g., BMI or height). A piecewise linear plot for the series of CQR estimates at different percentiles provides a useful visual summary of their variation along the sample distribution.23, 24 Figure 1 shows a working example of CQR and MR in comparison with OLS for FTO (rs1421085) in the ARIC CARe study.
Figure 1.
Working Example of Conditional Quantile Regression
BMI (kg/m2) was plotted against the number of effect alleles of FTO (rs1421085) in the ARIC CARe study (top left). An ordinary least-squares (OLS) model of the mean effect of this SNP on BMI was plotted (solid green line). Conditional quantile regression (CQR) models, fitted at every fifth percentile of BMI, show the effects of this SNP at these BMI percentiles (solid gray lines). The slopes (βOLS, horizontal dashed green line; βCQR, thick black line; kg/m2 per effect allele) from these models were then plotted against the BMI percentile at which they were fitted (middle right). 95% confidence intervals for these estimates were also plotted (OLS, horizontal dotted green line; CQR, shaded gray region). The change in CQR estimates across BMI percentiles was modeled with meta-regression (MR). The MR slope (βMR, kg/m2 per effect allele per BMI percentile, thin magenta line) and the 95% confidence intervals (dotted magenta lines) were plotted (bottom left).
Under conditions where true single and mixed GXE and GXG interactions are unadjusted, SNPs will shift both the location and scale (variance) of the sample outcome distribution (see Supplemental Note).56 These shifts in scale result in detectable variations of CQR estimates collected from percentiles across the sample outcome distribution. It follows that CQR estimates for a SNP are constant (i.e., equal) across percentiles if all unadjusted interaction effects are zero. Thus, the association between SNPs and an outcome under unadjusted interactions essentially reduces to modeling variability in CQR estimates. This can be effectively achieved with MR.25, 26 In this context, MR is basically a regression model where the CQR estimates from across the sample outcome distribution represent the dependent variable, and the percentiles at which these CQR estimates were calculated represent the independent variable (Figure 1). Additional details on CQR and MR, as well as simulations and an analytic description of this statistical framework, are presented in the Supplemental Note and Figures S1 and S2.
OLS models were used to verify the associations of SNPs and GSs with BMI and height in the sample populations included in this study. CQR models were fitted at every fifth percentile of the distribution of BMI and height for each SNP. We used a total of 10,000 Markov-chain-marginal-bootstrap replicates to compute confidence intervals (CIs) and the cross-percentile variance-covariance matrix for CQR estimates.57, 58, 59 The proportion of the trait variance explained by GS-BMI and GS-height in CQR models was also calculated.60 We computed hypothesis test statistics in MR (by assuming normality) to estimate the effects of percentiles on changes in mean CQR estimates for each SNP. The set of percentiles (5th–95th) was re-centered at the 50th percentile so that the intercept of the MR models corresponded to the main effect of the SNP at the median. Lastly, the effects of each SNP and the GS on the risk of specific BMI categories (NW versus OW, NW versus Ob-I, NW versus Ob-II, and NW versus Ob-III) were estimated with logistic regression.
All regression models were performed by one-step individual-participant-data meta-analysis (also known as “joint-analysis” or “mega-analysis”).61, 62 This method was chosen on the basis of access to individual participant data and the fact that CQR analyses refer to the conditional sample distribution.63 This means that analyses on separate studies correspond to their conditional distributions, and it would not be appropriate to combine them by using meta-analysis of their summary statistics. All models were adjusted for age (years), sex (female = 0, male = 1), and study (factor). For BMI analysis, age was modeled quadratically (age and age squared) as in previous reports.14, 20 Analyses of the associations of SNPs and GSs with BMI (37 SNPs + GS = 38) and height (125 SNPs + GS = 126) were subject to multiple-testing correction using Bonferroni-adjusted p value thresholds of p < 0.05/38 = 1.32 × 10−3 and p < 0.05/126 = 3.97 × 10−4, respectively.64 QC and statistical analyses were conducted with PLINK v1.90b3.42 and R v3.3.2.32, 33, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75 CQR models were fitted with quantreg, and MR models were fitted with metafor.76, 77 Additional packages used in the analysis include pracma, doParallel, foreach, and data.table.78, 79, 80, 81 An extended version of this work appears online.82
Results
Figure 1 depicts a step-by-step analysis of FTO (rs1421085) in the ARIC CARe study. In the top left panel, we fitted an OLS model (green) to determine the mean effects of the FTO genotype on BMI (βOLS, kg/m2 per effect allele) and fitted CQR models (gray) evenly across the sample BMI distribution (every fifth percentile) to determine the effects of the FTO genotype at each BMI percentile (βCQR, kg/m2 per effect allele). In the middle right panel, the estimates (βOLS and βCQR) and 95% CIs from these models are collected and plotted against the BMI percentile at which they were fitted. In the bottom left panel, MR analysis (magenta) models variation in the CQR estimates across the sample BMI distribution, and MR estimates (βMR, kg/m2 per effect allele per BMI percentile) are plotted along with 95% CIs. Presenting the results of OLS, CQR, and MR in this way is useful for summarizing the purpose of each analysis and contrasting possible differences between them.
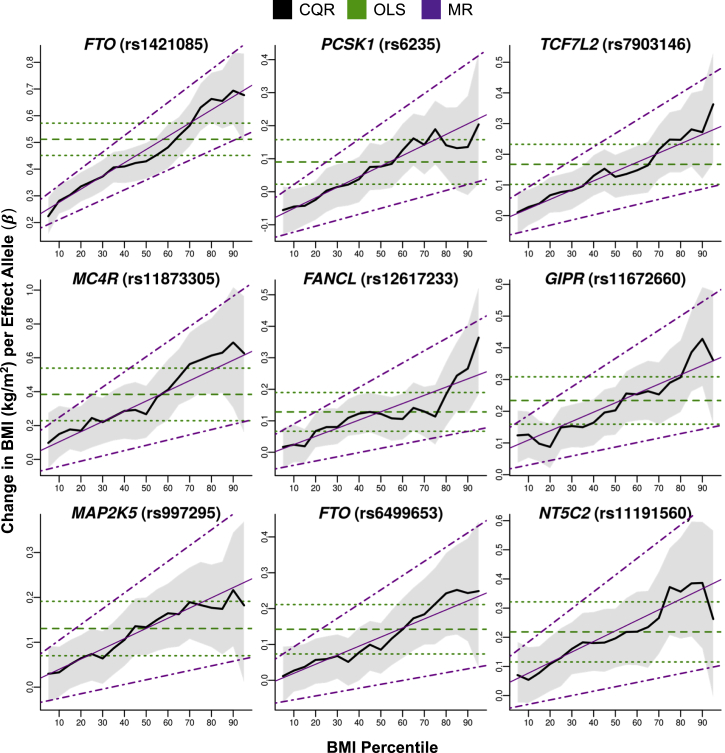
Initially, OLS models were fitted for each of 37 BMI-associated SNPs, and all but one were verified to increase BMI in this study sample (Table 1). We then fitted CQR models at regular intervals of the BMI distribution to explore whether the effects of SNPs on BMI varied across the sample distribution (Table S5). We plotted CQR estimates for each SNP against the BMI percentiles at which they were produced to provide a visual summary of the CQR results (Figure 2 and Figure S3). Several SNPs—including rs1421085 (FTO), rs6235 (PCSK1), rs7903146 (TCF7L2), rs11873305 (MC4R [MIM: 155541]), rs12617233 (FANCL [MIM: 608111]), rs11672660 (GIPR), rs997295 (MAP2K5 [MIM: 602520]), rs6499653 (FTO), and rs3824755 (NT5C2 [MIM: 600417])—had effects that appeared to increase across the distribution of BMI.
Table 1.
BMI-Associated SNP Information and Results from OLS Models
| SNP | Gene (OMIM) | Chromosome Position | E/O | PMID | βOLS[95% CI] | p Value |
|---|---|---|---|---|---|---|
| rs1421085 | FTO (610966) | chr16: 53,800,954 | C/T | 17658951 | 0.512 [0.451, 0.572] | 5.88 × 10−62 |
| rs10767664 | BDNF (113505) | chr11: 27,725,986 | A/T | 20935630 | 0.246 [0.172, 0.319] | 5.89 × 10−11 |
| rs11672660 | GIPR (137241) | chr19: 46,180,184 | C/T | 25673413 | 0.234 [0.159, 0.309] | 8.16 × 10−10 |
| rs4788099 | SH2B1 (608937) | chr16: 28,855,727 | G/A | 23001569 | 0.180 [0.113, 0.246] | 1.13 × 10−7 |
| rs7903146 | TCF7L2 (602228) | chr10: 114,758,349 | C/T | 25673413 | 0.167 [0.102, 0.232] | 5.36 × 10−7 |
| rs2075650 | TOMM40 (608061) | chr19: 45,395,619 | A/G | 23001569 | 0.218 [0.131, 0.305] | 9.75 × 10−7 |
| rs11873305 | MC4R (155541) | chr18: 58,049,192 | A/C | 25673413 | 0.384 [0.229, 0.539] | 1.23 × 10−6 |
| rs997295 | MAP2K5 (602520) | chr15: 68,016,343 | T/G | 23001569 | 0.131 [0.070, 0.191] | 2.40 × 10−5 |
| rs3824755 | NT5C2 (600417) | chr10: 104,595,849 | C/G | 25673413 | 0.218 [0.115, 0.321] | 3.32 × 10−5 |
| rs12617233 | FANCL (608111) | chr2: 59,039,998 | C/T | 23001569 | 0.128 [0.067, 0.190] | 4.34 × 10−5 |
| rs6499653 | FTO (610966) | chr16: 53,877,592 | T/C | 25673413 | 0.142 [0.073, 0.211] | 5.19 × 10−5 |
| rs1788826 | NPC1 (607623) | chr18: 21,154,024 | G/A | 25673413 | 0.124 [0.061, 0.186] | 1.08 × 10−4 |
| rs17066846 | MC4R (155541) | chr18: 58,044,818 | G/T | 25673413 | 0.144 [0.068, 0.220] | 2.09 × 10−4 |
| rs6453133 | HMGCR (142910) | chr5: 74,692,776 | A/G | 25673413 | 0.124 [0.058, 0.189] | 2.18 × 10−4 |
| rs739564 | IQCK | chr16: 19,740,237 | A/G | 25673413 | 0.147 [0.067, 0.227] | 2.97 × 10−4 |
| rs2272903 | TFAP2B (601601) | chr6: 50,786,571 | G/A | 23001569 | 0.173 [0.076, 0.270] | 4.77 × 10−4 |
| rs7553158 | TNNI3K (613932) | chr1: 75,005,238 | G/A | 25673413 | 0.102 [0.042, 0.162] | 8.40 × 10−4 |
| rs11570094 | SPI1 (165170) | chr11: 47,359,706 | A/C | 25673413 | 0.107 [0.041, 0.172] | 1.37 × 10−3 |
| rs4946932 | FOXO3 (602681) | chr6: 108,974,746 | C/A | 25673413 | 0.107 [0.041, 0.174] | 1.57 × 10−3 |
| rs2819347 | LMOD1 (602715) | chr1: 201,884,288 | G/C | 25673413 | 0.101 [0.037, 0.165] | 1.89 × 10−3 |
| rs2836754 | ETS2 (164740) | chr21: 40,291,740 | C/T | 25673413 | 0.099 [0.033, 0.164] | 3.20 × 10−3 |
| rs2984618 | TAL1 (187040) | chr1: 47,690,438 | T/G | 25673413 | 0.087 [0.026, 0.148] | 5.17 × 10−3 |
| rs11208662 | LEPR (601007) | chr1: 65,987,164 | C/G | 23563609 | 0.139 [0.037, 0.242] | 7.66 × 10−3 |
| rs6235 | PCSK1 (162150) | chr5: 95,728,898 | G/C | 18604207 | 0.090 [0.023, 0.158] | 8.82 × 10−3 |
| rs9356744 | CDKAL1 (611259) | chr6: 20,685,486 | T/C | 22344219 | 0.071 [0.005, 0.137] | 0.035 |
| rs7988412 | MTIF3 | chr13: 28,000,282 | T/C | 25673413 | 0.090 [0.005, 0.175] | 0.037 |
| rs1780050 | NEXN (613121) | chr1: 78,400,540 | A/C | 25673413 | 0.063 [0.002, 0.124] | 0.042 |
| rs526134 | USP37 | chr2: 219,402,371 | G/A | 25673413 | 0.066 [0.000, 0.132] | 0.049 |
| rs980828 | NOS1AP (605551) | chr1: 162,306,415 | G/T | 25133637 | 0.050 [−0.010, 0.110] | 0.100 |
| rs17001561 | SCARB2 | chr4: 77,096,118 | A/G | 25673413 | 0.070 [−0.017, 0.157] | 0.113 |
| rs6232 | PCSK1 (162150) | chr5: 95,751,785 | C/T | 18604207 | 0.095 [−0.041, 0.232] | 0.172 |
| rs749767 | KAT8 (609912) | chr16: 31,124,407 | A/G | 25673413 | 0.042 [−0.022, 0.105] | 0.199 |
| rs1211166 | NTRK2 (600456) | chr9: 87,285,992 | A/G | 23001569 | 0.041 [−0.034, 0.116] | 0.289 |
| rs2535633 | ITIH4 (600564) | chr3: 52,859,630 | G/C | 24861553 | 0.024 [−0.037, 0.085] | 0.437 |
| rs10144353 | PRKCH (605437) | chr14: 61,911,157 | T/C | 23563609 | 0.044 [−0.067, 0.155] | 0.441 |
| rs1561288 | ADCY3 (600291) | chr2: 25,369,002 | C/T | 23669352 | 0.024 [−0.047, 0.095] | 0.507 |
| rs2283228 | KCNQ1 (607542) | chr11: 2,849,530 | C/A | 24861553 | −0.037 [−0.159, 0.085] | 0.550 |
37 BMI-predisposing SNPs were selected for analysis. The effect and other (E and O, respectively) alleles were based on original discovery studies (PMID), and SNPs were coded by BMI-increasing or obesity-predisposing alleles. The indicated positions are based on GRCh37, and all alleles are on the positive strand. The association between these SNPs and BMI was assessed by OLS models that were adjusted for age, age squared, sex, and study. βOLS is the effect size (kg/m2 per effect allele), and 95% CIs are the 95% confidence intervals.
Figure 2.
The Effects of BMI-Associated SNPs across the Sample BMI Distribution
CQR models of BMI-associated SNPs were fitted every fifth percentile of BMI and adjusted for age, age squared, sex, and study. Estimates of the change in BMI (kg/m2) per effect allele (βCQR) from these models were plotted against the BMI percentile (thick black line) along with the 95% confidence intervals (shaded gray region). The results from OLS models (βOLS, kg/m2 per effect allele, horizontal dashed green line) and the 95% confidence intervals (horizontal dotted green lines) were also plotted for comparison. The change in CQR estimates across BMI percentiles was modeled with MR, and estimates from MR (βMR, kg/m2 per effect allele per BMI percentile, thin magenta line) and the 95% confidence intervals (dotted magenta lines) were plotted. MR analysis detected significant (p < 1.32 × 10−3) increases in the effects of these SNPs across the sample BMI distribution.
Single or mixed SNP interactions that are not adjusted in regression models will produce variability in CQR estimates along the distribution of the outcome (see Supplemental Note). This variability can be detected and quantified with MR.25, 26 Simulations showed that the power to detect such interactions by using CQR and MR was not affected by the MAF or the main effects of the SNPs, but it increased with the number of interactions as well as the main effects of the interacting covariate (see Supplemental Note and Figure S1). Yaghootkar et al. recently showed that differences in the prevalence of disease outcomes (e.g., the outcome of T2D) between sample and general populations can bias regression estimates of the main effects of SNPs on risk factors (e.g., BMI).83 However, the variability of CQR estimates across the sample distribution is not affected by biased main effects when CQR models are adjusted for disease status (see Supplemental Note). This was supported by simulations showing that the prevalence of disease outcomes in sample populations had negligible effects on the power and type I error rate for detecting unadjusted interactions when CQR models were adjusted for disease status (see Supplemental Note and Figure S2).
We fitted MR models to assess the variability in the CQR estimates of BMI-associated SNPs along the sample distribution of BMI (Table 2, Figure 2, and Figure S3). Significant positive associations (p < 1.32 × 10−3) between BMI percentile and CQR estimates were detected for 9 of 37 SNPs (24%): rs1421085 (FTO; βMR [95% CI] = 0.49 [0.37, 0.62], p = 8.69 × 10−15), rs6235 (PCSK1; 0.32 [0.18, 0.46], 7.11 × 10−6), rs7903146 (TCF7L2; 0.30 [0.17, 0.44], 9.60 × 10−6), rs11873305 (MC4R; 0.60 [0.31, 0.89], 5.08 × 10−5), rs12617233 (FANCL; 0.26 [0.13, 0.39], 5.30 × 10−5), rs11672660 (GIPR; 0.29 [0.14, 0.45], 1.64 × 10−4), rs997295 (MAP2K5; 0.23 [0.10, 0.35], 3.25 × 10−4), rs6499653 (FTO; 0.25 [0.11, 0.40], 6.23 × 10−4), and rs3824755 (NT5C2; 0.36 [0.15, 0.57], 7.90 × 10−4). The estimates from MR (βMR) quantify changes in the impact of each SNP on BMI across the sample distribution. For these 37 SNPs, the median βMR value [Q1, Q3] was 0.135 [0.094, 0.217] kg/m2 per effect allele per BMI percentile. In this statistical framework, βMR is equal to zero if all SNP interaction effects are also equal to zero (see Supplemental Note). Positive βMR estimates indicate that the effects of SNPs vary systemically by BMI percentile because unadjusted interactions are inflating the effects of SNPs in participants with a high BMI.
Table 2.
Quantifying the Effect of BMI Percentile on CQR Estimates by Using MR
| SNP | Gene (MIM) | RI50 | βMR[95% CI] | p Value |
|---|---|---|---|---|
| rs1421085 | FTO (610966) | 0.473 | 0.495 [0.370, 0.620] | 8.69 × 10−15∗ |
| rs6235 | PCSK1 (162150) | 0.078 | 0.320 [0.180, 0.459] | 7.11 × 10−6∗ |
| rs7903146 | TCF7L2 (602228) | 0.144 | 0.303 [0.169, 0.437] | 9.60 × 10−6∗ |
| rs11873305 | MC4R (155541) | 0.344 | 0.603 [0.311, 0.895] | 5.08 × 10−5∗ |
| rs12617233 | FANCL (608111) | 0.129 | 0.261 [0.134, 0.387] | 5.30 × 10−5∗ |
| rs11672660 | GIPR (137241) | 0.227 | 0.294 [0.141, 0.447] | 1.64 × 10−4∗ |
| rs997295 | MAP2K5 (602520) | 0.131 | 0.228 [0.103, 0.352] | 3.25 × 10−4∗ |
| rs6499653 | FTO (610966) | 0.121 | 0.253 [0.108, 0.398] | 6.23 × 10−4∗ |
| rs3824755 | NT5C2 (600417) | 0.222 | 0.362 [0.151, 0.574] | 7.90 × 10−4∗ |
| rs7553158 | TNNI3K (613932) | 0.099 | 0.196 [0.071, 0.322] | 2.12 × 10−3 |
| rs10767664 | BDNF (113505) | 0.247 | 0.217 [0.064, 0.370] | 5.50 × 10−3 |
| rs4788099 | SH2B1 (608937) | 0.151 | 0.194 [0.057, 0.332] | 5.59 × 10−3 |
| rs17066846 | MC4R (155541) | 0.124 | 0.215 [0.063, 0.367] | 5.61 × 10−3 |
| rs9356744 | CDKAL1 (611259) | 0.063 | 0.186 [0.050, 0.322] | 7.35 × 10−3 |
| rs6453133 | HMGCR (142910) | 0.130 | 0.177 [0.040, 0.314] | 0.011 |
| rs2819347 | LMOD1 (602715) | 0.111 | 0.137 [0.004, 0.269] | 0.044 |
| rs2075650 | TOMM40 (608061) | 0.283 | 0.161 [−0.019, 0.341] | 0.079 |
| rs4946932 | FOXO3 (602681) | 0.106 | 0.120 [−0.016, 0.256] | 0.084 |
| rs2984618 | TAL1 (187040) | 0.069 | 0.108 [−0.019, 0.235] | 0.095 |
| rs980828 | NOS1AP (605551) | 0.024 | 0.095 [−0.030, 0.220] | 0.135 |
| rs1788826 | NPC1 (607623) | 0.109 | 0.094 [−0.036, 0.224] | 0.156 |
| rs11570094 | SPI1 (165170) | 0.103 | 0.096 [−0.039, 0.231] | 0.163 |
| rs7988412 | MTIF3 | 0.088 | 0.109 [−0.062, 0.280] | 0.212 |
| rs2283228 | KCNQ1 (607542) | 0.003 | 0.147 [−0.094, 0.388] | 0.232 |
| rs739564 | IQCK | 0.122 | 0.100 [−0.065, 0.265] | 0.234 |
| rs526134 | USP37 | 0.062 | 0.079 [−0.055, 0.212] | 0.247 |
| rs2272903 | TFAP2B (601601) | 0.145 | 0.113 [−0.084, 0.310] | 0.261 |
| rs2836754 | ETS2 (164740) | 0.086 | 0.073 [−0.060, 0.206] | 0.280 |
| rs2535633 | ITIH4 (600564) | 0.016 | 0.068 [−0.059, 0.194] | 0.296 |
| rs11208662 | LEPR (601007) | 0.142 | 0.111 [−0.105, 0.327] | 0.314 |
| rs6232 | PCSK1 (162150) | 0.075 | 0.133 [−0.137, 0.404] | 0.334 |
| rs749767 | KAT8 (609912) | 0.048 | 0.058 [−0.075, 0.191] | 0.390 |
| rs1561288 | ADCY3 (600291) | 0.027 | −0.037 [−0.185, 0.112] | 0.627 |
| rs10144353 | PRKCH (605437) | 0.043 | 0.049 [−0.171, 0.269] | 0.662 |
| rs1211166 | NTRK2 (600456) | 0.029 | −0.027 [−0.179, 0.126] | 0.731 |
| rs17001561 | SCARB2 | 0.068 | −0.020 [−0.194, 0.154] | 0.824 |
| rs1780050 | NEXN (613121) | 0.045 | 0.010 [−0.117, 0.136] | 0.883 |
MR was used to model variability in the CQR estimates across BMI percentiles. Note that the percentiles were re-centered around the 50th percentile so that the intercept from MR models would correspond to the main effect of the SNP at the median. Asterisks (∗) denote statistical significance at the Bonferroni-adjusted threshold of p < 1.32 × 10−3, RI50 is the re-centered intercept of the MR models, βMR is the effect of BMI percentile on CQR estimates (kg/m2 per effect allele per BMI percentile), and 95% CIs are the 95% confidence intervals.
Given that height is known to be highly heritable, analyses were extended to height for comparison with the BMI results.22, 84, 85 OLS models were fitted for each of 125 height-associated SNPs, and all but two were verified to increase height (Table S6). CQR and MR were used to estimate variation in the effects of these SNPs on height as described previously (Figure S4 and Table S7). Only one height-associated SNP, rs6219 (IGF1 [MIM: 147440], βMR [95% CI] = 0.48 [0.23, 0.73], p = 1.80 × 10−4), showed significantly (p < 3.97 × 10−4) increased effects along the sample height distribution (Table S8). For height-associated SNPs, the median βMR value [Q1, Q3] was 0.002 [−0.056, 0.085] cm per effect allele per height percentile. Thus, CQR estimates for height-associated SNPs were predominantly consistent across height percentiles, and <1% showed evidence of unadjusted interactions, whereas 24% of BMI-associated SNPs did.
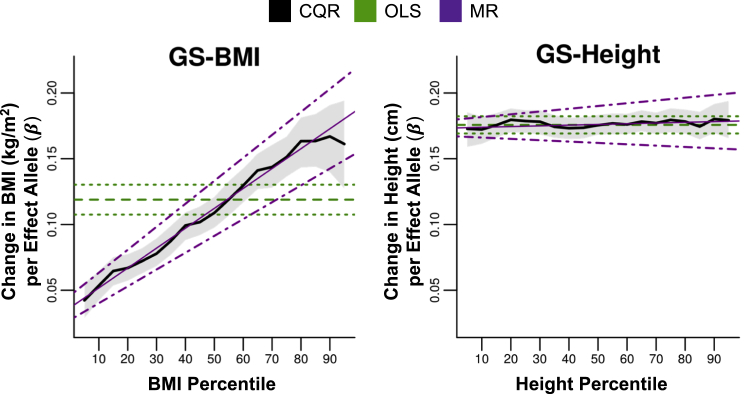
We combined BMI- and height-associated SNPs into GSs (GS-BMI and GS-height, respectively) to examine the overall association of these SNPs across the sample distribution. OLS models were used to verify the positive association between GS-BMI and GS-height and their respective traits (Table 3). CQR models for GS-BMI showed steadily increasing effects with increasing percentiles, whereas CQR models for GS-height did not vary across percentiles (Figure 3). MR analysis indicated that percentiles were significantly and positively associated with CQR estimates for GS-BMI (βMR [95% CI] = 0.15 [0.13, 0.17], 7.03 × 10−37) but not GS-height (0.01 [−0.01, 0.02], 0.499) (Table 3). At the 10th and 90th BMI percentiles, each additional effect allele of GS-BMI increased BMI by 0.054 and 0.167 kg/m2 (3.1-fold increase), respectively, whereas each additional allele of GS-height increased height by 0.172 and 0.180 kg/m2, respectively (Tables S5 and S7). Thus, in 1.73-m-tall persons at the tenth BMI percentile, carrying ten additional BMI-increasing alleles was associated with 1.6 kg of extra weight, whereas at the 90th BMI percentile, this was associated with 5.0 kg of extra weight. Furthermore, at the 10th and 90th BMI percentiles, the proportion of trait variance explained by GS-BMI increased (2.7-fold from 0.130% to 0.357%), whereas that of GS-height was stable (1.825% to 1.822%) (Tables S5 and S7). These results support the conclusion that the impact of BMI-associated SNPs was larger for individuals with high BMI, whereas the impact of height-associated SNPs varied little by height.
Table 3.
Analysis of GS-BMI and GS-Height
| SNP |
OLS Models |
MR Models |
|||
|---|---|---|---|---|---|
| βOLS[95% CI] | p Value | RI50 | βMR[95% CI] | p Value | |
| GS-BMI | 0.119 [0.108, 0.130] | 3.48 × 10−93 | 0.112 | 0.151 [0.128, 0.175] | 7.03 × 10−37∗ |
| GS-height | 0.176 [0.169, 0.182] | <2.2 × 10−308 | 0.176 | 0.005 [−0.010, 0.021] | 0.499 |
BMI- and height-associated SNPs were combined into gene scores (GS-BMI and GS-height, respectively). As in Table 1, the results from OLS models are presented. Furthermore, as in Table 2, MR analysis was applied to quantify the effects of trait (BMI and height) percentile on the CQR estimates for GS-BMI and GS-height, respectively. The asterisk (∗) denotes statistical significance at the Bonferroni-adjusted threshold of p < 1.32 × 10−3 for GS-BMI and p < 3.97 × 10−4 for GS-height. βOLS is the effect size (GS-BMI, kg/m2 per effect allele; GS-height, cm per effect allele) from OLS models, RI50 is the re-centered intercept of the MR models (same unit as for βOLS), βMR is the effect size (GS-BMI, kg/m2 per effect allele per BMI percentile; GS-height, cm per effect allele per height percentile) from MR models, and 95% CIs are the 95% confidence intervals.
Figure 3.
The Effects of GS-BMI and GS-Height across the Sample Distribution of BMI and Height, Respectively
As in Figure 2, CQR models of GS-BMI and GS-height were plotted against the BMI percentile and height percentile, respectively. The thick black line is the estimated change in each trait per effect allele (GS-BMI, βCQR, kg/m2 per effect allele; GS-height, βCQR, cm per effect allele), and the shaded gray region represents the 95% confidence intervals. Also plotted are the OLS regression estimates (GS-BMI, βOLS in kg/m2 per effect allele; GS-height, βOLS, cm per effect allele, horizontal dashed green line) and 95% confidence intervals (horizontal dotted green lines). The change in CQR estimates across outcome percentiles was modeled with MR. Estimates from MR (GS-BMI, βMR, kg/m2 per effect allele per BMI percentile; GS-height, βMR, cm per effect allele per height percentile; thin magenta line) and the 95% confidence intervals (dotted magenta lines) were also plotted.
Excluding seven SNPs that have also been associated with comorbidities of obesity from the gene score GS-BMI (stringent) did not alter the pattern of increasing effects across the sample BMI distribution (Figure S5).48, 49, 50, 51, 52, 53, 54, 55 Moreover, MR analysis indicated that BMI percentile was significantly and positively associated with the CQR estimates for GS-BMI (stringent) (βMR [95% CI] = 0.14 [0.11, 0.16], p = 2.18 × 10−23). In addition, CQR models were refitted with adjustment for diabetic status because this had been shown to mitigate the effects of possible stratification within the sample population (see Supplemental Note and Figure S2). Of the nine SNPs whose effects showed significant increases across the sample BMI distribution (Table 2 and Figure 2), three have also been associated with glucose homeostasis and T2D, namely, GIPR (rs11672660), TCF7L2 (rs7903146), and PCSK1 (rs6235).48, 50, 53 Refitting CQR models with adjustment for diabetic status had little impact on the results from MR analysis of these SNPs or GS-BMI (Table S9). Additional sensitivity analysis that included linearly modeling the effects of age or testing fewer percentiles (i.e., every 10th percentile from the 5th to 95th BMI percentiles) also showed no substantial changes to MR results (Table S9). Furthermore, calculating the GS for each trait without imputing missing genotypes did not affect results for GS-BMI or GS-height (Figure S5). Finally, the results from CQR were compared with those obtained from conventional subgroup analysis. To this end, the effect of genotype on the risk of OW, Ob-I, Ob-II, and Ob-III was evaluated separately with logistic regression (Table S10). The odds ratios of each SNP for each category were plotted against the BMI percentiles of the corresponding category, and CQR estimates were then overlaid on these bar plots. The patterns from logistic regression models across BMI categories were qualitatively consistent with the patterns from CQR models at comparable BMI percentiles (Figure S6).
Discussion
The aim of this study was to investigate variations in the effect of 37 BMI-associated SNPs across the distribution of BMI. We introduced a method that applies CQR to model the effects of SNPs at different percentiles of the sample BMI distribution and estimates variability in these effects by using MR. CQR estimates at different percentiles were shown to be uniform if all unadjusted SNP interactions were zero (see Supplemental Note). It follows that SNPs whose CQR estimates vary significantly across the sample BMI distribution are regulated by such interactions.
CQR analysis revealed distinct profiles of associations of BMI SNPs across the sample BMI distribution. Several of these SNPs had effects that increased steadily at higher BMI percentiles, whereas others had uniform effects that varied little across BMI percentiles (Figure 2 and Figure S3). One other study has used CQR to investigate the association between BMI and FTO (rs1558902) and a GS in a modest sample of adults.86 The patterns reported by that study are consistent with the results reported here.86 Two other studies used CQR to investigate the effects of SNPs on BMI in European children, and their results are also comparable with those here.87, 88 Overall, the high degree of correspondence between previously reported CQR results from European children and those from adults presented here emphasizes the robustness of these findings. Furthermore, the patterns observed with CQR analysis were compared with those from conventional logistic regression (subgroup analysis), given that Berndt et al. have demonstrated that the genetic architecture of BMI strongly overlaps BMI categories (Table S10).89 Across BMI categories, the patterns from logistic regression were largely consistent with those from CQR (Figure S6). CQR overcomes several of the limitations of subgroup analysis by utilizing all sample data to estimate regression parameters on the same scale as the continuous outcome, and comparing CQR estimates from different quantiles is relatively intuitive and easy.23, 89
MR was applied in order to model changes in the effects of BMI SNPs across the sample BMI distribution.25, 26 Results from MR showed that BMI percentile was positively and significantly associated with CQR estimates for 9 of 37 SNPs (24%). In addition, nominal associations were also observed for several other SNPs, and the median βMR [Q1, Q3] was 0.135 [0.094, 0.217] kg/m2 per effect allele per BMI percentile (Table 2 and Figure S3). This is supported by the GS-BMI analysis, which also showed significantly increasing effects across the sample BMI distribution (Figure 3 and Table 3). These findings indicate that unadjusted interactions enhanced the effects of BMI-associated SNPs at higher BMI levels. Modeling the effects of age linearly or considering fewer BMI percentiles (i.e., every tenth rather than every fifth percentile) had minimal effects on these results (Table S9).
There is evidence that differences in disease prevalence (e.g., in T2D) between sample and general populations can result in the stratification of secondary traits (e.g., BMI) that are risk factors for disease.83 This stratification can compromise regression estimates of the main effects of SNPs on secondary traits, and naively adjusting regression models for disease status might not adequately address this.83 Although the main effects of SNPs from disease-adjusted regression models are susceptible to stratification bias, the variation of SNP effects across the sample distribution is not (see Supplemental Note). This was evident in simulations showing that stratification had little effect on the power and type I error rate of MR analysis when CQR models were adjusted for disease status (Figure S2). Because GIPR (rs11672660), TCF7L2 (rs7903146), and PCSK1 (rs6235) have been associated with glucose homeostasis and T2D, CQR models were refitted with adjustment for diabetic status and analyzed by MR.48, 50, 53 These SNPs and the GS continued to show significantly increasing effects across the sample BMI distribution with this adjustment, demonstrating that the results were not an artifact of possible sample stratification (Table S9). Although estimating the variability of disease-adjusted CQR estimates across the sample distribution by using MR is robust to stratification bias, future studies aimed at estimating the main effects of SNPs by using CQR should implement methods to address this potential source of bias.90 A total of 7 of the 37 obesity-predisposing loci that were selected for analysis have also been associated with comorbidities of obesity, including glucose homeostasis, T2D, increased lipid levels, and heightened CRP levels.48, 49, 50, 51, 52, 53, 54, 55 Excluding these SNPs from the GS did not alter the pattern observed across the sample BMI distribution or affect the results from MR analysis, suggesting that these findings do not stem from the influence of comorbidities at high BMI levels (Figure S5).
Although BMI was the primary focus of this report, these analyses were also applied to height. This was important because analysis of height could shed light on the nature of the unadjusted interactions that were detected. BMI is a composite of both height and weight—height is one of the most heritable complex human traits, and weight is strongly influenced by environmental exposures and behavior.11, 91 If unadjusted interactions in the effects of BMI-associated SNPs are predominantly due to GXG interactions, then it is reasonable to suppose that these unadjusted interactions would be detected at a similar frequency in other quantitative traits such as height. On the other hand, if GXE interactions predominate, then these unadjusted interactions might be less frequently detected in quantitative traits with a smaller environmental component (i.e., height). CQR models for 125 height-associated SNPs were mostly uniform and exhibited little variability across height percentiles (Figure S4). Only one significant association between height percentiles and CQR estimates for height SNPs was detected by MR, and the median βMR [Q1, Q3] was 0.002 [−0.056, 0.085] cm per effect allele per height percentile (Table S8). Moreover, the effects of GS-height did not vary along the sample height distribution, which suggests that unadjusted interactions do not affect the genetic architecture of height to the same extent that they do for BMI (Table 3 and Figure 3). The simplest explanation for the discrepancy between the results for GS-BMI and GS-height is that the unadjusted interactions detected from GS-BMI were predominantly GXE interactions.
GXE interactions for SNPs in FTO have been reported for physical activity, food intake, dietary salt, alcohol consumption, and sleep duration.92, 93, 94, 95 In addition, the association between TCF7L2 (rs12255372) and BMI was modulated by fat intake in a weight-loss trial.96 Our analyses also pointed to significant interactions for FTO (rs1421085) and TCF7L2 (rs7903146) but suggested that such interactions might extend to additional BMI-associated SNPs—including rs6235 (PCSK1), rs11873305 (MC4R), rs12617233 (FANCL), rs11672660 (GIPR), rs997295 (MAP2K5), rs6499653 (FTO), and rs3824755 (NT5C2)—and GS-BMI. This is entirely consistent with a report showing that the effects of GS-BMI (29 SNPs) were enhanced by increased greater exposure to obesogenic environments and another demonstrating interactions between GS-BMI (69 SNPs) and several obesogenic drivers, including socio-economic status, TV watching, “Westernized” diets, and physical activity.14, 97 These reports also support the argument that the unadjusted interactions detected for BMI SNPs are predominately GXE interactions. Environmental modification of the effects of genetic variants raises the possibility that preventive measures, sustained lifestyle modifications, and therapeutic interventions could attenuate some of the genetic predisposition to unhealthy BMI. Indeed, the overall effect of BMI SNPs is minimal at low BMI levels (Figures 2 and 3). If weight gain leads to a genetically driven “vicious circle,” then weight loss can lead to a genetically driven “virtuous circle.” Investigating additional BMI-associated SNPs by using CQR and MR to uncover the full extent of unadjusted interactions in the architecture of BMI will be the focus of future studies.
This study is the largest yet to apply CQR to examine how the effects of SNPs vary with BMI, and it establishes quantitative support for hitherto qualitative descriptions of CQR. The combined utility of CQR and MR presents a contemporary statistical framework to cue hypotheses on gene interactions, better define clinical risks associated with genetic profiles, and prioritize clinical targets. Future studies aimed at distinguishing variants whose effects are modified by unadjusted interactions from those with fixed effects could advance the field of precision medicine. With the combined application of CQR and MR, this can now be achieved solely with information contained within the sample outcome distribution.
Acknowledgments
We thank Aihua Li for her assistance in database management and work using SPSS. Sebastien Robiou-du-Pont was supported by the Heart and Stroke Foundation of Ontario. Sonia S. Anand holds the Heart and Stroke Foundation of Ontario Michael G. DeGroote Chair in Population Health and a Canada Research Chair in Ethnicity and Cardiovascular Disease, Hertzel C. Gerstein holds the Aventis PHRI Chair in Diabetes, Salim Yusuf holds the Heart and Stroke Endowed Chair in Cardiovascular Research, and David Meyre holds a Canada Research Chair in Genetics of Obesity. Further acknowledgments are presented in the Supplemental Data.
Published: December 7, 2017
Footnotes
Supplemental Data include one Supplemental Note, six figures, and ten tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.10.007.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
GWAS Catalog, https://www.ebi.ac.uk/gwas/
OMIM, http://omim.org/
R statistical software, http://www.r-project.org/
Supplemental Data
References
- 1.Wang Y.C., McPherson K., Marsh T., Gortmaker S.L., Brown M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet. 2011;378:815–825. doi: 10.1016/S0140-6736(11)60814-3. [DOI] [PubMed] [Google Scholar]
- 2.Must A., Spadano J., Coakley E.H., Field A.E., Colditz G., Dietz W.H. The disease burden associated with overweight and obesity. JAMA. 1999;282:1523–1529. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- 3.Hill J.O., Peters J.C. Environmental contributions to the obesity epidemic. Science. 1998;280:1371–1374. doi: 10.1126/science.280.5368.1371. [DOI] [PubMed] [Google Scholar]
- 4.Misra A., Khurana L. Obesity and the metabolic syndrome in developing countries. J. Clin. Endocrinol. Metab. 2008;93(11, Suppl 1):S9–S30. doi: 10.1210/jc.2008-1595. [DOI] [PubMed] [Google Scholar]
- 5.Stunkard A.J., Harris J.R., Pedersen N.L., McClearn G.E. The body-mass index of twins who have been reared apart. N. Engl. J. Med. 1990;322:1483–1487. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- 6.Wardle J., Carnell S., Haworth C.M., Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am. J. Clin. Nutr. 2008;87:398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- 7.Pigeyre M., Yazdi F.T., Kaur Y., Meyre D. Recent progress in genetics, epigenetics and metagenomics unveils the pathophysiology of human obesity. Clin. Sci. 2016;130:943–986. doi: 10.1042/CS20160136. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert-Diamond D., Moore J.H. Analysis of gene-gene interactions. Curr. Protoc. Hum. Genet. 2011;Chapter 1:14. doi: 10.1002/0471142905.hg0114s70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordell H.J. Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De R., Verma S.S., Drenos F., Holzinger E.R., Holmes M.V., Hall M.A., Crosslin D.R., Carrell D.S., Hakonarson H., Jarvik G. Identifying gene-gene interactions that are highly associated with Body Mass Index using Quantitative Multifactor Dimensionality Reduction (QMDR) BioData Min. 2015;8:41. doi: 10.1186/s13040-015-0074-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddon H., Guéant J.-L., Meyre D. The importance of gene-environment interactions in human obesity. Clin. Sci. 2016;130:1571–1597. doi: 10.1042/CS20160221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rokholm B., Silventoinen K., Ängquist L., Skytthe A., Kyvik K.O., Sørensen T.I.A. Increased genetic variance of BMI with a higher prevalence of obesity. PLoS ONE. 2011;6:e20816. doi: 10.1371/journal.pone.0020816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rokholm B., Silventoinen K., Tynelius P., Gamborg M., Sørensen T.I.A., Rasmussen F. Increasing genetic variance of body mass index during the Swedish obesity epidemic. PLoS ONE. 2011;6:e27135. doi: 10.1371/journal.pone.0027135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter S., Mejía-Guevara I., Estrada K., Liu S.Y., Glymour M.M. Association of a Genetic Risk Score With Body Mass Index Across Different Birth Cohorts. JAMA. 2016;316:63–69. doi: 10.1001/jama.2016.8729. [DOI] [PubMed] [Google Scholar]
- 15.Kilpeläinen T.O., Qi L., Brage S., Sharp S.J., Sonestedt E., Demerath E., Ahmad T., Mora S., Kaakinen M., Sandholt C.H. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8:e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad T., Lee I.M., Paré G., Chasman D.I., Rose L., Ridker P.M., Mora S. Lifestyle interaction with fat mass and obesity-associated (FTO) genotype and risk of obesity in apparently healthy U.S. women. Diabetes Care. 2011;34:675–680. doi: 10.2337/dc10-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad S., Rukh G., Varga T.V., Ali A., Kurbasic A., Shungin D., Ericson U., Koivula R.W., Chu A.Y., Rose L.M., InterAct Consortium. DIRECT Consortium Gene × physical activity interactions in obesity: combined analysis of 111,421 individuals of European ancestry. PLoS Genet. 2013;9:e1003607. doi: 10.1371/journal.pgen.1003607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreasen C.H., Stender-Petersen K.L., Mogensen M.S., Torekov S.S., Wegner L., Andersen G., Nielsen A.L., Albrechtsen A., Borch-Johnsen K., Rasmussen S.S. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57:95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- 19.Xi B., Wang C., Wu L., Zhang M., Shen Y., Zhao X., Wang X., Mi J. Influence of physical inactivity on associations between single nucleotide polymorphisms and genetic predisposition to childhood obesity. Am. J. Epidemiol. 2011;173:1256–1262. doi: 10.1093/aje/kwr008. [DOI] [PubMed] [Google Scholar]
- 20.Young A.I., Wauthier F., Donnelly P. Multiple novel gene-by-environment interactions modify the effect of FTO variants on body mass index. Nat. Commun. 2016;7:12724. doi: 10.1038/ncomms12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demerath E.W., Choh A.C., Johnson W., Curran J.E., Lee M., Bellis C., Dyer T.D., Czerwinski S.A., Blangero J., Towne B. The positive association of obesity variants with adulthood adiposity strengthens over an 80-year period: a gene-by-birth year interaction. Hum. Hered. 2013;75:175–185. doi: 10.1159/000351742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson M.R., Hemani G., Medina-Gomez C., Mezzavilla M., Esko T., Shakhbazov K., Powell J.E., Vinkhuyzen A., Berndt S.I., Gustafsson S. Population genetic differentiation of height and body mass index across Europe. Nat. Genet. 2015;47:1357–1362. doi: 10.1038/ng.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koenker R., Hallock K. Quantile regression. J. Econ. Perspect. 2001;15:143–156. [Google Scholar]
- 24.Koenker R. Cambridge University Press; 2005. Quantile Regression. [Google Scholar]
- 25.Thompson S.G., Higgins J.P.T. How should meta-regression analyses be undertaken and interpreted? Stat. Med. 2002;21:1559–1573. doi: 10.1002/sim.1187. [DOI] [PubMed] [Google Scholar]
- 26.Borenstein M., Hedges L.V., Higgins J.P.T., Rothstein H.R. Introduction to Meta-Analysis. John Wiley & Sons, Ltd; 2009. Meta-Regression. [Google Scholar]
- 27.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33(Suppl 1):S62–S69. doi: 10.2337/dc10-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization (2000). Obesity: preventing and managing the global epidemic. A report of a WHO consultation. http://www.who.int/nutrition/publications/obesity/WHO_TRS_894/en/. [PubMed]
- 29.Anand S.S., Dagenais G.R., Mohan V., Diaz R., Probstfield J., Freeman R., Shaw J., Lanas F., Avezum A., Budaj A., EpiDREAM Investigators Glucose levels are associated with cardiovascular disease and death in an international cohort of normal glycaemic and dysglycaemic men and women: the EpiDREAM cohort study. Eur. J. Prev. Cardiol. 2012;19:755–764. doi: 10.1177/1741826711409327. [DOI] [PubMed] [Google Scholar]
- 30.Musunuru K., Lettre G., Young T., Farlow D.N., Pirruccello J.P., Ejebe K.G., Keating B.J., Yang Q., Chen M.-H., Lapchyk N., NHLBI Candidate Gene Association Resource Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3:267–275. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keating B.J., Tischfield S., Murray S.S., Bhangale T., Price T.S., Glessner J.T., Galver L., Barrett J.C., Grant S.F.A., Farlow D.N. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weale M.E. Quality control for genome-wide association studies. Methods Mol. Biol. 2010;628:341–372. doi: 10.1007/978-1-60327-367-1_19. [DOI] [PubMed] [Google Scholar]
- 33.Anderson C.A., Pettersson F.H., Clarke G.M., Cardon L.R., Morris A.P., Zondervan K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pillai S.G., Ge D., Zhu G., Kong X., Shianna K.V., Need A.C., Feng S., Hersh C.P., Bakke P., Gulsvik A., ICGN Investigators A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu G., Warren L., Aponte J., Gulsvik A., Bakke P., Anderson W.H., Lomas D.A., Silverman E.K., Pillai S.G., International COPD Genetics Network (ICGN) Investigators The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am. J. Respir. Crit. Care Med. 2007;176:167–173. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 36.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Mägi R., MAGIC. Procardis Consortium Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J., Genetic Investigation of ANthropometric Traits (GIANT) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benzinou M., Creemers J.W.M., Choquet H., Lobbens S., Dina C., Durand E., Guerardel A., Boutin P., Jouret B., Heude B. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat. Genet. 2008;40:943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 43.Thorleifsson G., Walters G.B., Gudbjartsson D.F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 44.Rivera M., Cohen-Woods S., Kapur K., Breen G., Ng M.Y., Butler A.W., Craddock N., Gill M., Korszun A., Maier W. Depressive disorder moderates the effect of the FTO gene on body mass index. Mol. Psychiatry. 2012;17:604–611. doi: 10.1038/mp.2011.45. [DOI] [PubMed] [Google Scholar]
- 45.Verma S.S., de Andrade M., Tromp G., Kuivaniemi H., Pugh E., Namjou-Khales B., Mukherjee S., Jarvik G.P., Kottyan L.C., Burt A. Imputation and quality control steps for combining multiple genome-wide datasets. Front. Genet. 2014;5:370. doi: 10.3389/fgene.2014.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janssens A.C.J.W., Moonesinghe R., Yang Q., Steyerberg E.W., van Duijn C.M., Khoury M.J. The impact of genotype frequencies on the clinical validity of genomic profiling for predicting common chronic diseases. Genet. Med. 2007;9:528–535. doi: 10.1097/gim.0b013e31812eece0. [DOI] [PubMed] [Google Scholar]
- 47.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 49.Saxena R., Elbers C.C., Guo Y., Peter I., Gaunt T.R., Mega J.L., Lanktree M.B., Tare A., Castillo B.A., Li Y.R., Look AHEAD Research Group. DIAGRAM consortium Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am. J. Hum. Genet. 2012;90:410–425. doi: 10.1016/j.ajhg.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 51.Aulchenko Y.S., Ripatti S., Lindqvist I., Boomsma D., Heid I.M., Pramstaller P.P., Penninx B.W.J.H., Janssens A.C.J.W., Wilson J.F., Spector T., ENGAGE Consortium Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spracklen C.N., Chen P., Kim Y.J., Wang X., Cai H., Li S., Long J., Wu Y., Wang Y.X., Takeuchi F. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum. Mol. Genet. 2017;26:1770–1784. doi: 10.1093/hmg/ddx062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strawbridge R.J., Dupuis J., Prokopenko I., Barker A., Ahlqvist E., Rybin D., Petrie J.R., Travers M.E., Bouatia-Naji N., Dimas A.S., DIAGRAM Consortium. GIANT Consortium. MuTHER Consortium. CARDIoGRAM Consortium. C4D Consortium Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hwang J.-Y., Sim X., Wu Y., Liang J., Tabara Y., Hu C., Hara K., Tam C.H.T., Cai Q., Zhao Q. Genome-wide association meta-analysis identifies novel variants associated with fasting plasma glucose in East Asians. Diabetes. 2015;64:291–298. doi: 10.2337/db14-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ng M.C.Y., Shriner D., Chen B.H., Li J., Chen W.-M., Guo X., Liu J., Bielinski S.J., Yanek L.R., Nalls M.A., FIND Consortium. eMERGE Consortium. DIAGRAM Consortium. MuTHER Consortium. MEta-analysis of type 2 DIabetes in African Americans Consortium Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 2014;10:e1004517. doi: 10.1371/journal.pgen.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paré G., Cook N.R., Ridker P.M., Chasman D.I. On the use of variance per genotype as a tool to identify quantitative trait interaction effects: a report from the Women’s Genome Health Study. PLoS Genet. 2010;6:e1000981. doi: 10.1371/journal.pgen.1000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He X., Hu F. Markov chain marginal bootstrap. J. Am. Stat. Assoc. 2002;97:783–795. [Google Scholar]
- 58.Kocherginsky M., He X., Mu Y. Practical Confidence Intervals for Regression Quantiles. J. Comput. Graph. Stat. 2005;14:41–55. [Google Scholar]
- 59.Koenker R., Bassett G., Jr. Robust tests for heteroscedasticity based on regression quantiles. Econometrica. 1982;50:43–61. [Google Scholar]
- 60.Koenker R., Machado J. Goodness of fit and related inference processes for quantile regression. JASA. 1999;94:1296–1310. [Google Scholar]
- 61.Sung Y.J., Schwander K., Arnett D.K., Kardia S.L.R., Rankinen T., Bouchard C., Boerwinkle E., Hunt S.C., Rao D.C. An empirical comparison of meta-analysis and mega-analysis of individual participant data for identifying gene-environment interactions. Genet. Epidemiol. 2014;38:369–378. doi: 10.1002/gepi.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riley R.D., Lambert P.C., Abo-Zaid G. Meta-analysis of individual participant data: rationale, conduct, and reporting. BMJ. 2010;340:c221. doi: 10.1136/bmj.c221. [DOI] [PubMed] [Google Scholar]
- 63.Borah B.J., Basu A. Highlighting differences between conditional and unconditional quantile regression approaches through an application to assess medication adherence. Health Econ. 2013;22:1052–1070. doi: 10.1002/hec.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feise R.J. Do multiple outcome measures require p-value adjustment? BMC Med. Res. Methodol. 2002;2:8. doi: 10.1186/1471-2288-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lanktree M.B., Guo Y., Murtaza M., Glessner J.T., Bailey S.D., Onland-Moret N.C., Lettre G., Ongen H., Rajagopalan R., Johnson T., Hugh Watkins on behalf of PROCARDIS. Meena Kumari on behalf of the Whitehall II Study and the WHII 50K Group Meta-analysis of Dense Genecentric Association Studies Reveals Common and Uncommon Variants Associated with Height. Am. J. Hum. Genet. 2011;88:6–18. doi: 10.1016/j.ajhg.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo Y., Lanktree M.B., Taylor K.C., Hakonarson H., Lange L.A., Keating B.J., IBC 50K SNP array BMI Consortium Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum. Mol. Genet. 2013;22:184–201. doi: 10.1093/hmg/dds396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R Development Core Team (2014). R: A language and environment for statistical computing (R Foundation for Statistical Computing).
- 68.Purcell, S., and Chang, C. (2016). PLINK v1.90b3.42 64-bit. http://www.cog-genomics.org/plink/1.9/.
- 69.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wigginton J.E., Cutler D.J., Abecasis G.R. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taliun D., Gamper J., Pattaro C. Efficient haplotype block recognition of very long and dense genetic sequences. BMC Bioinformatics. 2014;15:10. doi: 10.1186/1471-2105-15-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graffelman J., Moreno V. The mid p-value in exact tests for Hardy-Weinberg equilibrium. Stat. Appl. Genet. Mol. Biol. 2013;12:433–448. doi: 10.1515/sagmb-2012-0039. [DOI] [PubMed] [Google Scholar]
- 75.Gaunt T.R., Rodríguez S., Day I.N. Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool ‘CubeX’. BMC Bioinformatics. 2007;8:428. doi: 10.1186/1471-2105-8-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koenker, R. (2013). quantreg: Quantile Regression (R Foundation for Statistical Computing).
- 77.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010;36:1–48. [Google Scholar]
- 78.Borchers, H.W. (2015). pracma: Practical Numerical Math Functions. R package version 1.8.3 (R Foundation for Statistical Computing).
- 79.Analytics, R., and Weston, S. (2014). doParallel: Foreach Parallel Adaptor for the Parallel Package. R package version 1.0.8 (R Foundation for Statistical Computing).
- 80.Dowle, M., Short, T., and Lianoglou, S. (2014). data.table: Extension of Data. R package version 1.9.4 (R Foundation for Statistical Computing).
- 81.Warnes, G.R., Bolker, B., Gorjanc, G., Grothendieck, G., Korosec, A., Lumley, T., MacQueen, D., Magnusson, A., Rogers, J., et al. (2014). gdata: Various R Programming Tools for Data Manipulation (R Foundation for Statistical Computing).
- 82.Abadi A., Alyass A., Robiou du Pont S., Ben Bolker, Singh P., Mohan V., Diaz R., Engert J.C., Gerstein H.C., Anand S.S., Meyre D. Penetrance of polygenic obesity susceptibility loci across the body mass index distribution: an update on scaling effects. BioRxiv. 2017 doi: 10.1016/j.ajhg.2017.10.007. https://doi.org/10.1101/225128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yaghootkar H., Bancks M.P., Jones S.E., McDaid A., Beaumont R., Donnelly L., Wood A.R., Campbell A., Tyrrell J., Hocking L.J. Quantifying the extent to which index event biases influence large genetic association studies. Hum. Mol. Genet. 2017;26:1018–1030. doi: 10.1093/hmg/ddw433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Silventoinen K., Sammalisto S., Perola M., Boomsma D.I., Cornes B.K., Davis C., Dunkel L., De Lange M., Harris J.R., Hjelmborg J.V.B. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Res. 2003;6:399–408. doi: 10.1375/136905203770326402. [DOI] [PubMed] [Google Scholar]
- 85.Hemani G., Yang J., Vinkhuyzen A., Powell J.E., Willemsen G., Hottenga J.-J., Abdellaoui A., Mangino M., Valdes A.M., Medland S.E. Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs. Am. J. Hum. Genet. 2013;93:865–875. doi: 10.1016/j.ajhg.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams P.T. Quantile-specific penetrance of genes affecting lipoproteins, adiposity and height. PLoS ONE. 2012;7:e28764. doi: 10.1371/journal.pone.0028764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beyerlein A., von Kries R., Ness A.R., Ong K.K. Genetic markers of obesity risk: stronger associations with body composition in overweight compared to normal-weight children. PLoS ONE. 2011;6:e19057. doi: 10.1371/journal.pone.0019057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mitchell J.A., Hakonarson H., Rebbeck T.R., Grant S.F.A. Obesity-susceptibility loci and the tails of the pediatric BMI distribution. Obesity (Silver Spring) 2013;21:1256–1260. doi: 10.1002/oby.20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berndt S.I., Gustafsson S., Mägi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L., Croteau-Chonka D.C., Day F.R. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei Y., Song X., Liu M., Ionita-Laza I. Quantile Regression in the Secondary Analysis of Case–Control Data. J. Am. Stat. Assoc. 2016;111:344–354. doi: 10.1080/01621459.2015.1008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A.A.E., Lee S.H., Robinson M.R., Perry J.R.B., Nolte I.M., van Vliet-Ostaptchouk J.V., LifeLines Cohort Study Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reddon H., Gerstein H.C., Engert J.C., Mohan V., Bosch J., Desai D., Bailey S.D., Diaz R., Yusuf S., Anand S.S., Meyre D. Physical activity and genetic predisposition to obesity in a multiethnic longitudinal study. Sci. Rep. 2016;6:18672. doi: 10.1038/srep18672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Corella D., Arnett D.K., Tucker K.L., Kabagambe E.K., Tsai M., Parnell L.D., Lai C.-Q., Lee Y.-C., Warodomwichit D., Hopkins P.N., Ordovas J.M. A high intake of saturated fatty acids strengthens the association between the fat mass and obesity-associated gene and BMI. J. Nutr. 2011;141:2219–2225. doi: 10.3945/jn.111.143826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lappalainen T., Lindström J., Paananen J., Eriksson J.G., Karhunen L., Tuomilehto J., Uusitupa M. Association of the fat mass and obesity-associated (FTO) gene variant (rs9939609) with dietary intake in the Finnish Diabetes Prevention Study. Br. J. Nutr. 2012;108:1859–1865. doi: 10.1017/S0007114511007410. [DOI] [PubMed] [Google Scholar]
- 95.Qi Q., Kilpeläinen T.O., Downer M.K., Tanaka T., Smith C.E., Sluijs I., Sonestedt E., Chu A.Y., Renström F., Lin X. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Hum. Mol. Genet. 2014;23:6961–6972. doi: 10.1093/hmg/ddu411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mattei J., Qi Q., Hu F.B., Sacks F.M., Qi L. TCF7L2 genetic variants modulate the effect of dietary fat intake on changes in body composition during a weight-loss intervention. Am. J. Clin. Nutr. 2012;96:1129–1136. doi: 10.3945/ajcn.112.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tyrrell J., Wood A.R., Ames R.M., Yaghootkar H., Beaumont R.N., Jones S.E., Tuke M.A., Ruth K.S., Freathy R.M., Davey Smith G. Gene-obesogenic environment interactions in the UK Biobank study. Int. J. Epidemiol. 2017;46:559–575. doi: 10.1093/ije/dyw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.