Abstract
Microorganisms associated with plants are highly diverse and can produce a large number of secondary metabolites, with antimicrobial, anti-parasitic and cytotoxic activities. We are particularly interested in exploring endophytes from medicinal plants found in the Pantanal, a unique and widely unexplored wetland in Brazil. In a bio-prospecting study, strains LGMF1213 and LGMF1215 were isolated as endophytes from Vochysia divergens, and by morphological and molecular phylogenetic analyses were characterized as Phaeophleospora vochysiae sp. nov. The chemical assessment of this species reveals three major compounds with high biological activity, cercoscosporin (1), isocercosporin (2) and the new compound 3-(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone (3). Besides the isolation of P. vochysiae as endophyte, the production of cercosporin compounds suggest that under specific conditions this species causes leaf spots, and may turn into a pathogen, since leaf spots are commonly caused by species of Cercospora that produce related compounds. In addition, the new compound 3-(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone showed considerable antimicrobial activity and low cytotoxicity, which needs further exploration.
Introduction
Endophytes are recognized as microorganisms that for all or part of their lifetime colonize internal plant tissues1, and using recent definitions, we refer to the term endophytes only in context of the microorganism habitat, with no definition of function (Hardoim et al. 2015). Endophytes associated with medicinal plants have been shown to possess a high potential to produce new bioactive metabolites, which often are valuable for plant growth promotion and/or biological control2,3.
Cataloguing microorganisms from unexplored biomes is an effective strategy to discover new species, often also producers of new secondary metabolites. Our groups are interested to explore the biodiversity of endophytes from medicinal plants found in the Pantanal, a unique and little explored wetland region in Brazil2,4,5, with seasonal inundation enduring more than 200 days a year6. Only few plants are able to tolerate such long periods of flooding, among them Vochysia divergens Pohl, a medicinal plant used in folk medicine to treat respiratory and gastric diseases7, and producer of antimicrobial compounds8,9.
In our search for novel bioactive compounds, we isolated two endophytic strains from V. divergens, with characteristic of the genus Phaeophleospora. The genus Phaeophleospora Rangel10 is an anamorph of the Mycosphaerellaceae and it is based on P. eugeniae, which was isolated in Brazil from leaf spots of Eugenia uniflora. To date, most species of this genus have been associated with leaf spot diseases in various plants, e.g., Myrtaceae, Polypodiaceae, Elaeocarpaceae, Plantaginaceae, Proteaceae and Apocynaceae11–14. So far, only two Phaeophleospora species, P. stramenti and P. eucalypticola, were found as endophytes from Eucalyptus sp11 and Eugenia uniflora15, respectively. Despite the fact that the genus Phaeophleospora was described as early as 1916, there were no reports of any bioactive secondary metabolites, and its metabolic potential remained unknown.
Based on morphological and phylogenetic analyses we describe here Phaeophleospora vochysiae as a new species within the genus Phaeophleospora, studied its secondary metabolites, and linked these to biological activities.
Results
Strains Isolation and Identification
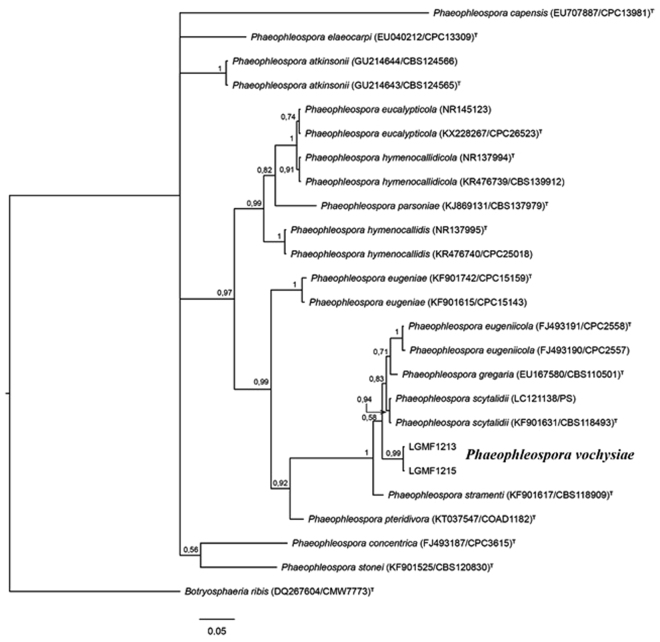
Based on endophyte isolation protocol, strains LMGF1213 and LGMF1215 were isolated in June of 2010, from two different V. divergens asymptomatic plants, located in Nhecolândia, Pantanal, Brazil. The alignment of internal transcribed spacer region ITS1-5.8S-ITS2 comprised of 539 characters, 259 (48%) of these characters were constant, 122 (22.6%) were parsimony-uninformative and 158 (29.4%) characters were parsimony informative (Genbank access number: KY754582 and KY754582). The Bayesian phylogenetic analysis of ITS region showed strains LGMF1213 and LGMF1215 are in the same clade as P. eugeniicola, P. gregaria, P. scytalidii and P. stramenti, with a strong probability support (1.00), sharing 99% of ITS1–5.8S–ITS2 rDNA sequence similarity with species P. scytalidii (GenBank LC121138, Identities = 463/467). However, isolates LGMF1213 and LGMF1215 are located in a single branch (supported by 0.99 of probability), different from the Phaeophleospora species previously described (Fig. 1), and were characterized as a new species, Phaeophleospora vochysiae (Fig. 1). We also analyzed the large subunit ribosomal RNA (LSU) (Genbank access number: MG214701 and MG214702), elongation factor 1-alpha (TEF) (Genbank access number: MG190362 and MG190363) and acting (ACT) (Genbank access number: MG214703 and MG214704) genes (Fig. S38–40). As expected, LSU phylogenic analysis showed similar topology than ITS analysis, however with lower resolutions (Fig. S40). For the TEF gene, sequences of P. eugeniicola, P. gregaria, P. scytalidii and P. stramenti were not available, so our isolates clustered with the type species of the genus Phaeophleospora (P. eugeniae) (Fig. S39). ACT analysis that revealed strains LGMF1213 and LGMF1215 to be in the same clade as P. eucalypti, P. destructans, P. gregaria and P. scytalidii, however, in a separated branch (Fig. S38), supporting the proposition of LGMF1213 and LGMF1215 as a new species within the genus Phaeophleospora. A multilocus sequence analysis was not performed, since the sequence of other genes than ITS were not available for all the type strains.
Figure 1.
Bayesian phylogenetic tree based on ITS sequence of rRNA gene of LGMF1215 and LGMF1213 and 15 described species of Phaeophleospora genus. Values on the node indicate Bayesian posterior probabilities. The species Botryosphaeria ribis was used as outgroup. Scale bar indicates the number of substitutions per site.
Taxonomic description
Phaeophleospora vochysiae
Savi & Glienke, sp. nov. Mycobank: MB821497. Fig. 2.
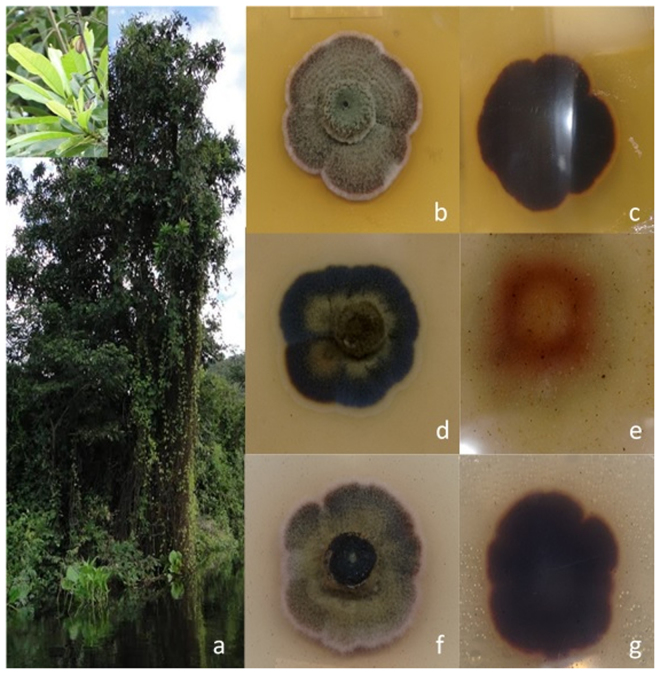
Figure 2.
Host and morphology of Phaeophleospora vochysiae LGMF1215 on Malt Extract Agar (MEA), Oatmeal Agar (OA) and Potato Dextrose Agar (PDA) at 28 °C after 14 days. (a) Vochysia divergens (b and c) Colony on MEA surface and reverse; (d and e) colony on OA surface and reverse; (f and g) colony on PDA surface and reverse.
Etymology. Name after the host genus from which it was isolated, Vochysia.
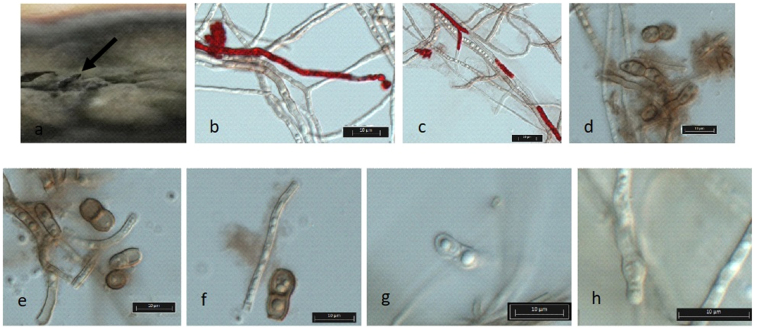
Mycelium consisting of septate, branched, verruculose hyphae, 2–3 μm wide, in some points with red pigment inside, due the presence of cercosporin (1) and isocercosporin (2) (Fig. 3). Codiomata pycnidial, black, globose to subglobose. Conidiophores inside of pycnidium are aggregated arising from the upper cells of an irregular brown stroma up to 30 μm. Conidiogenous cells terminal, unbranched, medium brown, smooth to verruculose, 9.3 (7.4–11.2) × 5.0 (3.9–6.0) μm. Conidia solitary, or in simple chains, brown, verruculose, subcylindrical to oval, apex obtuse, base subtruncate, 1–2 septate, frequently constricted at the septa, 8.8 (7.0–13.7) × 4.7 (3.8–6.6) μm.
Figure 3.
Macro- and micro-morphology of Phaeophleospora vochysiae LGMF1215. (a) Colony with pycnidium (arrow). (b and c) Mycelium consisting of septate, branched, verruculose hyphae in some points with red pigment inside. (d) Conidiophore and conidium. (e–g) Conidia. (h) Conidiophore.
Culture characteristics: Colonies spreading, erumpent, lobed margins and moderate aerial mycelium, reaching 27 mm after 14 days at 28 °C. On Potato Dextrose Agar (PDA) medium, the surface is olivaceus-grey with vinaceus margin, the reverse side red due to a diffuse pigment. On oatmeal agar (OA) surface olivaceus-grey with yellow center, and on malt extract agar (MEA) green-grey and reverse iron-grey (Fig. 2).
Specimen examined: Brazil, Nhecolandia, Pantanal, Mato Grosso do Sul, S18°10.07′, W57°23.03′, on Vochysia divergens leaf, 9 Feb. 2010, D.C. Savi (Holotype: UPCB90660 (Herbarium of the Department of Botany code, University Federal of Paraná), ex-type culture LGMF1215 (Laboratory of Genetics of Microorganism Culture Collection code, University Federal of Paraná).
Notes: Strain LGMF1215 was isolated from healthy leaf tissue of Vochysia divergens, ITS sequence GenBank KY754582.
Secondary metabolites
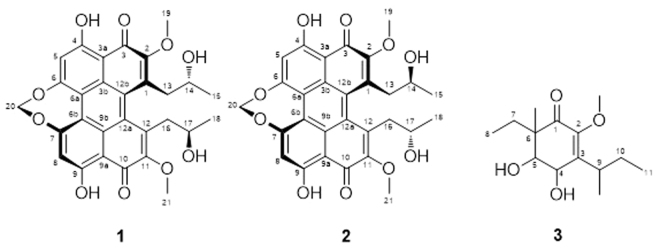
A large-scale fermentation of the strain LGMF1215 in Potato Dextrose-medium produced 6.1 g of mycelium crude extract, and 3.7 g of culture filtrate extract. The purification of these extracts using various chromatographic techniques resulted two known compounds, (+)-cercosporin (1)16,17 and (+)-isocercosporin (2)17, plus one new compound, 3-(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone (3) (Fig. 4 and S1). The chemical structure of compounds 1-3 were determined by cumulative 1D and 2D NMR spectroscopy, high resolution mass spectrometry (HRMS) (Table 1 and 2 and Fig. S2-S37), and by comparison with literature data16,17.
Figure 4.
Chemical structures of compounds 1–3.
Table 1.
Physicochemical properties of compounds 1–3 isolated from cultivation of strain LGMF1215.
| (+)-Cercosporin (1) | (+)-Isocercosporin (2) | Compound 3 | |
|---|---|---|---|
| Molecular Formula | C29H26O10 | C29H26O10 | C14H24O4 |
| Appearance | Red solid, UV absorbing (254 nm) | Red solid, UV absorbing (254 nm) | Colorless oil, UV absorbing (254 nm) |
| 2n NaOH | Dark-green | Dark-green | |
| HPLC-Rt a) | 21.45 (min) | 21.85 (min) | 18.19 (min) |
| (+)-APCI-MS: m/z | 535 [M + H]+ | 535 [M + H]+ | 257 [M + H]+ |
| (+)-HRESI-MS: m/z | |||
| Found | 535.1598 [M + H]+ | 535.1599 [M + H]+ | 257.1748 [M + H]+ |
| Calcd. | 535.1599 for C29H27O10 [M + H]+ | 535.1599 for C29H27O10 [M + H]+ | 257.1747 for C14H25O4 [M + H]+ |
| (−)-HRESI-MS: m/z | |||
| Found | 533.1441 [M‒H]− | 533.1444 [M‒H]− | 255.1596 [M‒H]−, 291.1361 [M + Cl]− |
| Calcd. | 533.1453 for C29H25O10 [M‒H]− | 533.1453 for C29H25O10 [M‒H]− | 255.1601 for C14H23O4 [M‒H]− 291.1368 for C14H24ClO4 [M + Cl]− |
| UV/Vis λmax | 220, 270, 450, 470 (sh), 570 (sh) nm | 220, 270, 450, 470 (sh), 570 (sh) nm | 250 nm |
Table 2.
13C (100 MHz) and 1H (400 MHz) NMR Spectroscopic Data of Compounds 1–3 in CDCl3 (δ in ppm) isolated from cultivation of strain LGMF1215, compared with the literature data.
| Position | (+)-Cercosporin (1) | (+)-Isocercosporin (2) | Data reported in literature for compound 127 | Data reported in literature for compound 228 | Position | Compound 3 | |||
|---|---|---|---|---|---|---|---|---|---|
| δC, type | δH (mult.) | δC, type | δH (mult.) | δH (mult.) | δH (mult.) | δC, type | δH (mult.) | ||
| 1,12 | 135.4, C | 136.8, C | 1 | 198.5, C | |||||
| 2, 11 | 153.0, C | 153.4, C | 2 | 148.1, C | |||||
| 3, 10 | 182.0, C | 181.9, C | 2-OCH3 | 59.7, CH3 | 3.62 (s) | ||||
| 3a, 9a | 108.4, C | 108.6, C | 3 | 144.7, C | |||||
| 3b, 9b | 128.1, C | 127.6, C | 4 | 68.8, CH | 4.56 (m) | ||||
| 4, 9 | 167.7, C | 167.8, C | 5 | 74.4, C | 3.87 (brd, 3.6) | ||||
| 4,9-OH | 14.81 (s) | 14.89 (s) | 14.86 (s) | 14.91 (s) | 6 | 50.4, C | |||
| 5, 8 | 109.5, CH | 7.06 (s) | 109.4, CH | 7.03 (s) | 7.07 (s) | 7.02 (s) | 6-CH3 | 18.9, CH3 | 1.08 (s) |
| 6, 7 | 163.6, C | 163.6, C | 7 | 25.4, CH2 | 1.81 (m), 1.73 (m) | ||||
| 6a, 6b | 113.1, C | 113.2, C | 8 | 7.7, CH3 | 0.86 (t, 7.6) | ||||
| 12a, 12b | 130.7, C | 131.8, C | 9 | 35.9, CH | 2.80 (m) | ||||
| 13, 16 | 42.4, CH2 | 3.57 (dd, 13.0, 7.1) 2.88 (dd, 12.9, 5.9) |
42.5, CH2 | 3.49 (dd, 14.0, 3.2) 2.85 (dd, 13.5, 8.4) |
3.59 (dd, 13.1, 7.0) 2.90 (dd, 13.1, 6.0) |
3.51 (dd, 13.3, 3.4) 2.87 (dd, 13.3, 8.2) |
9-CH3 | 18.1, CH3 | 1.19 (d, 6.9) |
| 14, 17 | 68.3, CH | 3.36 (m) | 69.8, CH | 3.68 (m) | 3.39 (m) | 3.70 (m) | 10 | 29.1, CH2 | 1.68 (m), 1.58 (m) |
| 15, 18 | 23.6, CH3 | 0.62 (d, 6.0) | 24.0, CH3 | 0.95 (d, 6.0) | 0.65 (d, 6.1) | 0.97 (d, 6.2) | 11 | 13.1, CH3 | 0.91 (t, 7.5) |
| 19, 21 | 61.4, CH3 | 4.19 (s) | 61.2, CH3 | 4.21 (s) | 4.21 (s) | 4.23 (s) | |||
| 20 | 92.8, CH2 | 5.72 (s) | 92.8, CH2 | 5.71 (s) | 5.75 (s) | 5.73 (s) | |||
δH (mult., J in [Hz]); Assignments supported by 2D HSQC and HMBC experiments.
The physicochemical properties of compounds 1–3 are summarized in Table 1. Compound 3 was obtained as colorless oil using various chromatographic techniques (see Supporting Information, Fig. S1). The molecular formula of 3 was deduced as C14H24O4 on the basis of (+) and (−)-HRESI-MS [m/z 257.1748 [M + H]+ (calcd. for C14H25O4, 257.1747) and 255.1596 [M‒H]− (calcd. for C14H23O4, 255.1601)] and NMR data, revealing three double bond equivalents (DBE). The proton NMR spectrum of 3 in CDCl3 (Table 2) was rich in aliphatic proton signals, including two oxymethines (δ 4.56, m, 1 H; δ 3.87, brd, J = 3.6, 1 H), one methoxy (δ 3.62, s, 3 H), one methine (δ 2.80, m), two methylene (δ 1.85-1.50, 4 H), one singlet methyl (δ 3.62, s, 3 H) and four methyl signals of which two were observed as triplets at δ 0.91 (J = 7.5, 3 H) and 0.86 (J = 7.6, 3 H), suggesting the presence of two ethyl residues in 3. The 13C NMR/HSQC spectra of 3 (Table 2) displayed 14 carbon signals corresponding to five methyl, two methylene, three methine and four quaternary carbon signals. In addition, the presence of three downfield quaternary carbon signals (δ 198.5, 148.1 and 144.7) indicate the presence of an α-β-unsaturated double bond with a carbonyl group (δ 198.5) in compound 3. The attachment of the methoxy group at 2-position was confirmed through the 3J HMBC correlation observed from 2-OCH3 (δH 3.62) to C-2 (δC 148.1). The attachment of the ethyl/sec-butyl residues at 6-/3-positions, respectively, were established through the cumulative analyses of 1H1,H-COSY, TOCSY and HMBC spectra (Figures S2 and S3). The HMBC correlations observed from 6-CH3 (δH 1.08) to C-1 (δC 198.5), CH-5 (δC 74.4), C-6 (δC 50.4) and CH2-7 (δC 25.4) confirm the attachment of the methyl group at 6-position. All of the remaining HMBC correlations (Figure S2) and NMR data (Table 2) are in full agreement with structure 3. Finally, the relative configuration at the stereocenters were indirectly established through the analyses of NOESY correlations (Figure S3), and by comparison to those of the reported cyclopentenone ring-contracted analogues, such as similin A (4) (produced by the fungus Sporormiella similis)18, phomapentenone A (5) (produced by the fungus Phoma sp. NRRL 25697)19 and boydone B (6) (produced by the plant associated Pseudallescheria boydii NTOU2362)20 (Figure S41). Thus, thorough cumulative analyses of 1D (1H13,C) and 2D (HSQC1,H1,H-COSY, TOCSY, HMBC and NOESY) NMR spectra established the structure of 3 as -(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone (Fig. 4).
Antibacterial and Antifungal Activities
The antimicrobial activity of the three crude extracts produced in the PD, Czapeck and MEA culture media were evaluated in order to select the best culture conditions to produce bioactive secondary metabolites. Extracts from PD showed the best results, with potent activity against the phytopathogen Phyllosticta citricarpa, moderate activity against methicillin sensitive and resistant Staphylococcus aureus, and low activity against the phytopathogen Colletotrichum abscissum (Table 3). The isolated compounds 1–3 (from cultures in PD medium), also displayed both, antibacterial activity against sensitive and resistant S. aureus and antifungal activity against the citrus phytopathogen Phyllosticta citricarpa (Table 3).
Table 3.
Inhibition zones (in millimeters) of LGMF1215 crude extracts of different culture media and compounds 1~3 tested antibacterial and antifungal assays at 100 μg/disc.
| Extracts/compounds | Staphylococcus aureus | MRSA | Escherichia coli | Phyllosticta citricarpa | Colletotrichum abscissum |
|---|---|---|---|---|---|
| Extract produced using PD | 13 | 12 | 11 | 32 | 50 |
| Extract produced using Czapeck | — | — | — | 2 | 4 |
| Extract produced using MEA | 8 | — | 6 | 10 | 15 |
| Cercosporin 1 | 46 | 45 | 28 | 30 | Ne |
| Isocercosporin 2 | 35 | 27 | 20 | 28 | Ne |
| 3-(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone 3 | 14 | 14 | — | 30 | Ne |
| Control | 30 | 0 | 35 | 37 | 82 |
–denotes no measurable halo, antibacterial control: Ampicillin (1 mg/disc), Antifungal control: Derosal (1 mg/disc), ne: not evaluated.
Cytotoxicity activity
The cytotoxic activities of compounds 1–3 were determined, using PC3 (prostate) and A549 (non-small cell lung) human cancer cell lines (Fig. 5). Cell viability assays showed that cercosporin (1) and isocercosporin (2), were highly cytotoxic, IC50 = 2.82 μM (A549) and 0.42 μM (PC3), and IC50 = 19.21 μM (A549) and 2.10 μM (PC3), respectively. In contrast, compound 3-(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone (3) showed no cytotoxicity up to 60 μM concentration against these cell lines.
Figure 5.
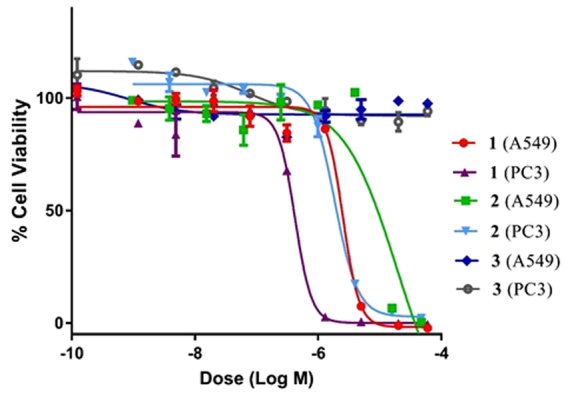
Dose-response viability assay of compounds 1–3 isolated from strain LGMF1215 against A549 (lung) and PC3 (prostate) human cancer cell lines for 72 hrs treatments. Note: each line represents a different treatment with a specific cell line and compound.
Discussion
Endophytic fungi are an important resource of secondary metabolites5,21. To better access this important source of bioactive molecules it is essential to explore the diversity of endophytes and to catalogue their species occurrence in different ecosystems5. Strains LGMF1215 and LGMF1213 were isolated from healthy leaves of a medicinal plant, Vochysia divergens, commonly found in flooding areas of the Pantanal (Brazil), and were characterized as a new species of Phaeophleospora genus, namely P. vochysiae.
Characterization of Phaeophleospora species traditionally was based on morphology15 in comparison with the type species P. eugeniae10. In the past, the asexual morphs of Phaeophleospora were characterized as pycnidial22, however, the recently described P. pteridivora has a sporodochial asexual morph23. Guatimosim et al.23 suggested that the genus Phaeophleospora should be defined based on phylogeny analysis instead, due to its high morphological diversity. So far, 19 species of Phaeophleospora were described, however, only 15 were accepted and validated based on ITS phylogenetic analysis (Table S1), but sufficient information to distinguish species within the genus Phaeophleospora was revealed11,12,23,24. We suggest a multilocus analysis as valuable tool to better understand evolutionary factors and intra- and inter-specific relationships of the Phaeophleospora species. We also suggest TEF1 and ACT as candidate genes for this analysis, in view of the number of informative sites compared to ITS and LSU sequences (Fig. 1, S-38-40). In the phylogenetic analysis, P. vochysiae is in the same clade (1) as P. gregaria, P. scytaliddi, P. stramenti and P. eugeniicola, but in a single branch, supported by high probability value (Fig. 1). Besides the phylogenetic support, P. vochysiae also showed difference in conidia size and growth rates from the P. scytaliddi (Table S2), the closest related species (99% of similarity in ITS sequence). P. scytaliddi has an elliptic conidium25, longer and narrowed compared to P. vochysiae and, different from the others species in this clade, only P. vochysiae produced a red pigment associated with the hypha (Fig. 3). P. gregaria also produced a red pigment in culture media, however it is not hypha associated26. The other species in the clade were associated with leaf spot diseases (Table S1), and, except for P. gregaria, which was described in Australia, they were all isolated in Brazil, from plants belonging to the Myrtaceae family11,26. This is the first isolation of a Phaeophleospora species from plants belonging to the family Vochyseaceae (Vochysia divergens). In contrast to those other species, the two strains of P. vochysiae were isolated from asymptomatic plants.
Phaeophleospora is an anamorph of the Mycosphaerellaceae26, and although other genera from the family of Mycosphaerella were recognized as producers of various secondary metabolites with antibacterial, antifungal and anti-parasitic activity27–29, there has been no report of secondary metabolites isolated from the species Phaeophleospora. Therefore, as a new species belonging to a genus without metabolite studies, and isolated from a region exposed to high hydric stress, we hoped for an interesting spectrum of secondary metabolites from P. vochysiae, including new metabolites. Two known compounds, cercosporin and isocercosporin, and a new compound, 3-(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone, were produced in large amounts by the strain LGMF1215 (Fig. 3, S1). These compounds showed antibacterial activity against MSSA and MRSA, and high antifungal activity. Since MRSA strains have acquired resistance to many antibiotics and are associated with higher human death rate than those caused by HIV and influenza combined30, the search for new compounds with activity against this pathogen has great importance. In addition to the activity against MRSA, the new compound 3-(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone (3) showed complete inhibition of mycelial growth of the phytopathogen P. citricarpa. On the other hand, compound 3 showed no cytotoxic activity against the human tumor cells evaluated, which may suggest a more selective, and less toxic antimicrobial activity (Table 3, Fig. 4). Therefore, compound 3 may be used for the control of the phytopathogen P. citricarpa, the causative agent of citrus black spot (CBS), a disease subjected to phytosanitary regulations in the European Union31.
The phytotoxic compounds cercosporin and isocercosporin are perylenequinones isolated for the first time in 1981 and 1991, from Cercospora beticola and Scolecotrichum graminis, respectively16,17, while 3-(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone (3) was never found before, only similar cyclopentenone derivatives, i.e. ring-contracted analogues compared to 3, such as similin A (4, produced by the fungus Sporormiella similis)18, phomapentenone A (5, produced by the fungus Phoma sp. NRRL 25697)19 and boydone B (6, produced by the plant associated Pseudallescheria boydii NTOU2362)20 (Figure S41). The cercosporins are photosensitizing compounds that interact with molecular oxygen to produce highly toxic singlet oxygen32. Nearly all bacteria and fungi are susceptible to cercosporin cell damage, however, the species of Cercospora16,33 and Phaeophleospora vochysiae are resistant. The resistance against cercosporins by P. vochysiae and the host Vochysia divergens, can be associated with different factors, the most common are the production of enzymes or compounds that quench or block the formation of oxygen species, such as carotenoids34 or antioxidants35. In addition, some studies suggest that cercosporin associated with the Cercospora hyphae is present in the reduced form, which makes the compound nontoxic, or not photoactived36. The association of this compound with the hyphae was also observed here in P. vochysiae (Fig. 3), and can be one explanation for the isolation of this species from healthy tissues from V. divergens. However, we cannot exclude the possibility of P. vochysiae to cause disease in certain unfavorable conditions.
In conclusion, the species P. vochysiae, isolated from healthy tissues of the medicinal plant Vochysia divergens, was described by morphological and phylogenetic analyses, and was characterized as a source of highly active secondary metabolites, including the new compound 3-(sec-butyl)-6-ethyl-4,5-dihydroxy-2-methoxy-6-methylcyclohex-2-enone (3). This new compound showed activity against MRSA and P. citricarpa, but no cytotoxicity. P. vochysiae also produced the phytotoxic compounds cercosporin (1) and isocercosporin (2) in culture conditions, suggesting the potential of this species to cause diseases, under specific conditions or in different host plants. More studies are necessary to find out whether the cercosporins are associated with the pathological mechanisms involved in the leaf spot diseases caused by other Phaeophleospora species, e.g., in Eucalyptus spp.
Material and Methods
Taxonomy
Strains LGMF1213 and LGMF1215 were isolated from V. divergens leaves collected in the Pantanal, in the region of Nhecolândia (S18°10.07′, W57°23.03′) in Brazil. For the isolation of endophytes, leaves with no marks, scratches or wounds were collected. To eliminate epiphytic microorganisms, a purification protocol of six steps was used37. The leaves were fragmented and inoculated in Petri dishes with medium PDA (Potato Dextrose Agar). The plates were incubated at 28 °C for 30 days, and the growth was verified daily. The cultures were deposited in the Laboratory of Genetics of Microorganisms (LabGeM) culture collection, Federal University of Paraná, Curitiba, Paraná, Brazil (http://www.labgem.ufpr.br/).
For the macro- and microscopic analysis, strain LGMF1215 was grown in plates in Potato Dextrose Agar (IBI Scientific), Oatmeal Agar (IBI Scientific) and Malt Extract Agar (Bacto Difco) culture medium at 23 °C and 28 °C, respectively, for 40 days, as described by Crous et al.12. Culture characteristics were studied from cultures 21 days after inoculation. Microscopic preparations were performed in distilled water, with 50 measurements per structure in microscope Axio Imager Z2 (Carl Zeiss, Jena, DE), equipped with Metafer 4/VSlide software (Metasystems, Altlussheim, DE), using differential phase interference contrast (DIC) illumination with software support, ImageJ.
Genomic DNA extraction was carried out using the UltraClean™ Microbial DNA Kit (MO Bio, Carlsbad, CA, USA). The internal transcribed spacer region (ITS) 1, 5.8 S, ITS2, LSU, elongation factor (TEF-1) and actin were amplified using the primers ITS4 and V9G38, LR0R and LR523, EF-728F and EF-986R23, ACT-512F and ACT-783 R18, respectively. The PCR product was purified using the enzymes EXO1 and FastAP (ThermoFisher, Waltham, MA USA). The PCR product was sequenced using BigDye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems, Foster City, CA, USA), and sequences were analyzed on an ABI3500 DNA Sequencer (Applied Biosystems, Foster City, CA, USA). The sequence was compared with type specimens sequences available in the Genbank database of NCBI (http://www.ncbi.nlm.nih.gov/) and Mycobank (http://www.mycobank.org/), and aligned using CLUSTAL_X v.1.8139. Bayesian inference of the phylogeny was performed in MrBayes version 3.2.140, with permutations allowed until a frequency of division ≤0.01 was reached. The general time-reversible (GTR) substitution model was used. FigTree version 1.4.241 was used to edit the phylogenetic trees that were constructed. Sequences obtained in this study were deposited in GenBank with the accession numbers listed in Table S1.
Fermentation, Extraction and Isolation
Phaeophleospora vochysiae LGMF1215 was cultivated on PDA-agar plates at 28 °C for 7 days. Fragments of agar (5/flask) were used to inoculate two Erlenmeyer flasks (500 mL) containing 250 mL of PD (IBI Scientific), MEA (Bacto Difco) and Czapeck media (Bacto Difco), and cultured for 21 days, at 27 °C and 250 rpm. Cultures were extracted with EtOAc (3 × 500 mL) and then the recovered organics were evaporated in vacuo at 40 °C. The antibacterial activity of crude extracts was determined and the best conditions were selected for a large-scale culture.
Large-scale fermentation (10 L) was performed using Potato Dextrose broth (IBI Scientific). The obtained red/black culture broth was separated by filtration using Whatman paper n 4. The biomass (mycelium) was extracted with MeOH/EtOAc (5 × 1000 mL) and then the recovered organics were evaporated in vacuo at 40 °C to yield 6.1 g of crude extract. The aqueous fraction was extracted with EtOAc (5 × 500 mL) and the recovered organics were evaporated in vacuo at 40 °C to yield 3.7 g of broth (culture filtrate) extract. The mycelium crude extract was subjected to Sephadex LH-20 (MeOH; 1 × 20 cm), and further purified by HPLC to afford compound 1 (3 mg) (Fig. S2). The EtOAc fraction was also subjected to Sephadex LH-20 (MeOH; 1 × 20 cm) and purified by HPLC to yield compounds 1 (2.6 mg), 2 (1.3 mg) and 3 (7.0 mg) in pure forms (Fig. S1).
Antimicrobial and Antifungal Activity
The Gram-positive bacteria Staphylococcus aureus (ATCC 25923), methicillin-resistant Staphylococcus aureus (MRSA) (BACHC-MRSA) were maintained in lysogeny broth (LB) liquid media and Mueller-Hinton agar (Bacto Difco, USA). A loopful of each organism was inoculated into a 7 mL culture of LB broth and incubated in a 37 °C orbital shaker at 200 rpm for 10 hours. Each test organism was streaked on a sterile Mueller-Hinton agar plate with a sterile cotton swab. Compounds 1–3 were dissolved in methanol and aliquoted in 100 μg amounts per each 6 mm sterile filter disc and were allowed to dry in a laminar flow hood. The discs were placed on the plates, which were then incubated for 24 hours at 37 °C. The resulting inhibition zones were measured in millimeters.
The fungal strains Phyllosticta citricarpa LGMF06 and Colletotrichum abscissum LGMF1268 were used in disc diffusion assays. Solutions of amphotericin B (positive control) and tested compounds were dissolved in MeOH. Each sterile paper disc was loaded with 10 µL solution and was allowed to dry in the biosafety cabinet for 4 h. The dried discs were then placed on the PDA plate following the homogeneous distribution of fungus. MeOH was used as a negative control. The plates were incubated at 24 °C for 7 and 21 days, when the inhibition zones were measured.
Cytotoxic activity
To assess the viability of human lung non-small cell carcinoma A549 and prostate adenocarcinoma PC3 cell against compounds 1–3 the conversion of resazurin (7-hydroxy-10-oxido-phenoxazin-10-ium-3-one) to its fluorescent product resorufin was monitored. DMEM/F-12 Kaighn’s modification media (Life Technologies, NY, USA) with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL, streptomycin, 2 mM L-glutamine was used to grow A549 and PC3 cells (ATCC, Manassas, VA, USA). Cells were seeded at a density of 5 × 103 cells per well in 96-well clear bottom culture plates (Corning, NY, USA), incubated 24 hours at 37 °C in a humidified atmosphere containing 5% CO2 and subsequently exposed to known toxins (1.5 mM hydrogen peroxide or 10 μg/mL actinomycin D, positive control) and test compounds for 72 hours. To assess cell viability, 150 μM of resazurin (Sigma, St. Louis, MO, USA) was added to each well, plates were shaken briefly for 10 seconds and incubated for another 3 hours at 37 °C to allow viable cells to convert resazurin into resorufin. The fluorescence intensity for resorufin was detected on a scanning microplate spectrofluorometer FLUOstar Omega (BMG Labtech, Cary, NC, USA) using an excitation wavelength of 560 nm and an emission wavelength of 590 nm.
Electronic supplementary material
Acknowledgements
This work was supported by NIH grants CA 91091, GM 105977 and an Endowed University Professorship in Pharmacy to J.R. This work was also supported by the University of Kentucky Markey Cancer Center, the National Center for Advancing Translational Sciences (UL1TR001998) and NIH grants R01 GM115261 to J. S. T. Additional support came from Fundação Araucária de Apoio e Desenvolvimento Científico e Tecnológico do Paraná – Brazil, grant 441/2012–23510, CNPq-Brazil grant 486016/2011–0 to C.G., and CAPES-Brazil – a grant to D.C.S. We also thank Lisandra S. F. Maba and the Laboratório de Microscopia de Fluorescência Convencional e Confocal, UFPR, for image acquisitions.
Author Contributions
C.G. and J.R. coordinated the project and edited the manuscript. D.C.S. isolated the fungus, extracted genomic DNA and phylogenetic analyses. D.C.S. and K.A.S contributed to the large-scale cultivation, compounds isolation and manuscript preparation. J.S.T. contributed to the compound isolation/identification and consultation. L.V.P. performed the cytotoxicity evaluation. C.G. and F.M.W.R.G. contributed to the morphological analysis.
Competing Interests
The authors declare no competing interests.
Footnotes
Daiani C. Savi and Khaled A. Shaaban contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21400-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chirlei Glienke, Email: ch.glienke@gmail.com.
Jürgen Rohr, jrohr2@email.uky.edu.
References
- 1.Hardoim PR, et al. The hidden world within plants: ecological and evolutionary considerations for defining function of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015;79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savi DC, et al. Microbispora sp. LGMF259 endophytic actinomycete isolated from Vochysia divergens (Pantanal, Brazil) producing β-carbolines and indoles with biological activity. Curr. Microbiol. 2015;70:345–354. doi: 10.1007/s00284-014-0724-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopes R, et al. Endophytic bacteria isolated from common bean (Phaseolus vulgaris) exhibiting high variability showed antimicrobial activity and quorum sensing inhibition. Curr. Microbiol. 2015;71:509–516. doi: 10.1007/s00284-015-0879-6. [DOI] [PubMed] [Google Scholar]
- 4.Savi DC, Aluizio R, Galli-Terasawa L, Kava V, Glienke C. 16S-gyrB-rpoB multilocus sequence analysis for species identification in the genus Microbispora. Antonie Van Leeuwenhoek. 2016;109:801–815. doi: 10.1007/s10482-016-0680-y. [DOI] [PubMed] [Google Scholar]
- 5.Hokama, Y. M. et al. Endophytic fungi isolated from Vochysia divergens in the Pantanal, Mato Grosso do Sul: diversity, phylogeny, and biocontrol of Phyllosticta citricarpa. In: Endophytic fungi: diversity, characterization and biocontrol. (Ed Hughes E) chap3 (Hauppauge: Nova Publishers, 2016).
- 6.Junk WJ, et al. The comparative biodiversity of seven globally important wetlands: a synthesis. Aquat. Sci. 2006;68:400–414. doi: 10.1007/s00027-006-0856-z. [DOI] [Google Scholar]
- 7.Pott, A. & Pott, V. J. Plantas do Pantanal. (Corumbá, Embrapa, 1994).
- 8.Honda NK, Cruz AB, Messana I. Antibacterial activity and phytochemical analysis of Vochysia divergens (Vochysiaceae) J. Ethnopharmacol. 1995;47:97–100. doi: 10.1016/0378-8741(95)01260-K. [DOI] [PubMed] [Google Scholar]
- 9.Hess SC, Monache FD. Divergioic acid, a triterpene from Vochysia divergens. J. Braz. Chem. Soc. 1999;10:104–106. doi: 10.1590/S0103-50531999000200005. [DOI] [Google Scholar]
- 10.Crous PW, Wingfield MJ. Colletogloeopsis, a new coelomycete genus to accommodate anamorphs of two species of Mycosphaerella on Eucalyptus. Can. J. Bot. 1997;75:667–674. doi: 10.1139/b97-074. [DOI] [Google Scholar]
- 11.Quaedvlieg W, et al. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia. 2014;33:1–40. doi: 10.3767/003158514X681981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crous PW, et al. Fungal planet description sheets: 320-370. Persoonia. 2015;34:167–266. doi: 10.3767/003158515X688433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennycook, S. R. & McKenzie, E. H. C. Scoleciasis atkinsonii, an earlier name for Phaeophleospora hebes; and a note on G. H. Cunningahams epithets hebe and pseudopanax. Myctaxon82, 145–146.
- 14.Taylor JE, Cannon PF, David JC, Crous PW. Two new Phaeophleospora species associated with leaf spots of Proteaceae. South Afr. J. Bot. 2001;67:39–43. doi: 10.1016/S0254-6299(15)31088-7. [DOI] [Google Scholar]
- 15.Taylor JE, Crous PW. Phaeophleospora faureae comb. nov. associated with leaf spots on Faurea saligna (Proteaceae), with a key to species of Phaeophleospora. Fungal Diver. 1999;3:153–158. [Google Scholar]
- 16.Steinkamp MP, Martin SS, Hoefert LL, Ruppel EG. Ultrastructure of lesions produced in leaves of Beta vulgaris by cercosporin, a toxin from Cercospora beticola. Phytopathol. 1981;71:1272–1281. [Google Scholar]
- 17.Tabuchi FH, Ajimi AT, Ichihara A. (+)-Isocercosporin, a phytotoxic compound isolated from Scolecotrichum graminis. Agric. Biol. Chem. 1991;55:2675–2676. [Google Scholar]
- 18.Weber HA, Swenson DC, Gloer JB. Similins A and B: New antifungal metabolites from the coprophilous. Sporomeilla similis. Tetrahedron Lett. 1992;33:1157–1160. doi: 10.1016/S0040-4039(00)91884-7. [DOI] [Google Scholar]
- 19.Che Y, Gloer JB, Wicklow DT. Phomadecalins A-D and Phomapentenone A: New bioactive metabolites from Phoma sp. NRRL 25697, a fungal colonist of Hypoxylon stromata. J. Nat. Prod. 2002;65:399–402. doi: 10.1021/np010519o. [DOI] [PubMed] [Google Scholar]
- 20.Chang YC, et al. Polyketides from the littoral plant associated fungus. Pseudollescheria boydii. J. Nat. Prod. 2013;76:1796–1800. doi: 10.1021/np400192q. [DOI] [PubMed] [Google Scholar]
- 21.Tonial, F. et al. Biological activity of Diaporthe terebinthifolii extracts against Phyllosticta citricarpa. FEMS Microbiol. Lett. 10.1093/femsle/fnx026, (2017). [DOI] [PubMed]
- 22.Walker, J. Notes on plant parasitic fungi. I. Proc. Linn. Soc. 87, 162–176 (1962).
- 23.Guatimosim E, Schwartsburd PB, Barreto RW, Crous PW. Novel fungi from an old niche: Cercosporoid and related sexual morphs on ferns. Persoonia. 2016;37:106–141. doi: 10.3767/003158516X690934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crous PW, et al. Fungal planet description sheets: 214–280. Persoonia. 2014;32:184–306. doi: 10.3767/003158514X682395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crous PW, Wingfield MJ, Mansilla JP, Alfenas AC, Groenewald JZ. Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. II. Syst. Micol. 2006;55:99–131. doi: 10.3114/sim.55.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Groenewald JZ. Novel species of Mycosphaerellaceae and Teratosphaeriaceae. Persoonia. 2009;23:119–146. doi: 10.3767/003158509X479531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Assante G, Nasini G, Zhang S, Bradshaw RE. A novel secondary metabolite from the Eucalyptus pathogen Mycosphaerella cryptica. For. Path. 2009;39:289–292. doi: 10.1111/j.1439-0329.2008.00582.x. [DOI] [Google Scholar]
- 28.Moreno E, et al. Chemical constituents of the new endophytic fungus Mycosphaerella sp. nov. and their anti-parasitic activity. Nat. Prod. Commun. 2011;6:835–840. [PMC free article] [PubMed] [Google Scholar]
- 29.Pusztahelyi T, Holb IJ, Pocsi I. Secondary metabolites in fungus-plant interaction. Front. Plant. Sci. 2015;6:573. doi: 10.3389/fpls.2015.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magill SS, et al. Emerging infections program healthcare-associated infections and antimicrobial use prevalence survey team. N. Engl. J. Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos PJC, et al. Diaporthe endophytica and D. terebinthifolii from medicinal plants for biological control of Phyllosticta citricarpa. Microbiol. Res. 2016;186:153–160. doi: 10.1016/j.micres.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Daub ME, Herrero S, Chung KR. Reactive oxygen species in plant pathogenesis: the role of Perylenequinone photosensitizers. Antiox. Redox. Sing. 2013;19:970–989. doi: 10.1089/ars.2012.5080. [DOI] [PubMed] [Google Scholar]
- 33.Beseli A, Noar R, Daub ME. Characterization of Cercospora nicotianae hypothetical proteins in Cercosporin resistance. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0140676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tripathy BC, Oelmuller R. Reactive oxygen species generation and signaling in plants. Plant. Signal. Behav. 2012;72:1621–1633. doi: 10.4161/psb.22455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song R, et al. Phytoalexin phenalenone derivate inactivate mosquito larvae and root-knot nematode as Type-II photosensitizer. Sci. Rep. 2017;7:42–58. doi: 10.1038/s41598-017-00093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson JM, Poland JA, Benson BM, Stromberg EL, Nelson RJ. Resistance to gray leaf spot of maize: genetic architecture and mechanisms elucidated through nested association mapping and near-isogenic line analysis. PLoS Gent. 2015;11:e1005045. doi: 10.1371/journal.pgen.1005045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrini O. Taxonomy of endophytic fungi of aerial plant tissues. Microbiol. 1986;2:175–187. [Google Scholar]
- 38.White JF, Jr., Morrow AC. Endophyte-host associations in forage grasses. XII. A fungal endophyte of Trichachne insularis belonging to Pseudocercosporella. Mycol. 1990;82:218–226. [Google Scholar]
- 39.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2011;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rambaut, A. Tree Figure Drawing Tool Version 1.4.0. Institute of Evolutionary Biology, University of Edinburgh, [http://tree.bio.ed.ac.uk/software/figtree/)] (2006–2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.