Abstract
Background
Chlamydomonas reinhardtii is an unicellular green alga used for functional genomics studies and heterologous protein expression. A major hindrance in these studies is the low level and instability of expression of nuclear transgenes, due to their rearrangement and/or silencing over time.
Results
We constructed dedicated vectors for Agrobacterium-mediated transformation carrying, within the T-DNA borders, the Paromomycin (Paro) selectable marker and an expression cassette containing the Luciferase (Luc) reporter gene. These vectors and newly developed co-cultivation methods were used to compare the efficiency, stability and insertion sites of Agrobacterium- versus electroporation-mediated transformation. The influence of different transformation methods, of the cell wall, of the virulence of different Agrobacterium strains, and of transgene orientation with respect to T-DNA borders were assessed. False positive transformants were more frequent in Agrobacterium-mediated transformation compared to electroporation, compensating for the slightly lower proportion of silenced transformants observed in Agrobacterium-mediated transformation than in electroporation. The proportion of silenced transformants remained stable after 20 cycles of subculture in selective medium. Next generation sequencing confirmed the nuclear insertion points, which occurred in exons or untraslated regions (UTRs) for 10 out of 10 Agrobacterium-mediated and 9 out of 13 of electroporation-mediated insertions. Electroporation also resulted in higher numbers of insertions at multiple loci.
Conclusions
Due to its labor-intensive nature, Agrobacterium transformation of Chlamydomonas does not present significant advantages over electroporation, with the possible exception of its use in insertional mutagenesis, due to the higher proportion of within-gene, single-locus insertions. Our data indirectly support the hypothesis that rearrangement of transforming DNA occurs in the Chlamydomonas cell, rather than in the extracellular space as previously proposed.
Electronic supplementary material
The online version of this article (10.1186/s12896-018-0416-3) contains supplementary material, which is available to authorized users.
Keywords: Chlamydomonas, Agrobacterium, Gene expression, Luciferase, Silencing
Background
Chlamydomonas reinhardtii is widely used for functional genomics studies as well as for heterologous protein production. Significant progress has been made since the first transformation reports using complementation of the nitrate reductase (nit1) and argininosuccinate lyase (arg7) mutations [1, 2]. A large number of selectable markers and promoters have been used for nuclear transformation, and delivery of foreign DNA has been obtained through a variety of methods, such as particle bombardment [1], agitation with glass beads or silicon carbide whiskers [3, 4], electroporation [5], or Agrobacterium tumefaciens [6–8]. Despite recent advances in genetic transformation, the expression of foreign genes in the nuclear genome of wild-type Chlamydomonas remains challenging due to transgene rearrangement and silencing [8–10].
In this scenario, the possibility to transform Chlamydomonas by Agrobacterium tumefaciens is appealing, given the lower levels of transgene rearrangement and silencing reported in plants. Agrobacterium tumefaciens is a pathogenic soil bacterium that has evolved the capacity to transfer a segment of DNA (the T-DNA) from the tumor-inducing (Ti) plasmid into the nucleus of a plant cell. An advantage in using Agrobacterium for plant transformation is the reduction in transgene copy number, DNA rearrangements and transgene silencing [11, 12].
Nuclear transformation of Chlamydomonas reinhardtii by Agrobacterium tumefaciens has been reported, using vectors containing the Cauliflower Mosaic Virus 35S promoter and non-codon-optimized reporter genes [6]. The reported transformation efficiency was 50-fold higher than that obtained with the glass beads method. Further improvements of the transformation efficiency were described in a second report [7] using modified transformation vectors, culture media and Agrobacterium strains. No data on the stability of expression over time of the reporter gene were reported in either case.
In the present work, we describe the construction of transformation vectors carrying a selectable marker (Paro gene [13]) and a reporter gene (Renilla reniformis luciferase (Luc) [14]) which are codon optimized for Chlamydomonas nuclear expression and placed under the control of strong Chlamydomonas promoters: 70 kDa heat shock protein/ribulose bisphosphate carboxylase/oxygenase small subunit (HSP70/RBCS2) [15] and Photosystem I reaction center subunit II (PSAD) [16]. The effects of different transformation protocols, Agrobacterium strains, and orientation of the transgenes with respect to the T-DNA right border were studied, and compared with results obtained via electroporation.
Results
Development of vectors and protocols for Agrobacterium-mediated transformation of C. reinhardtii
First, we constructed the pAgroR vector, optimized for Agrobacterium-mediated Chlamydomonas transformation. This vector harbors the Paro gene, conferring paromomycin resistance [13] and an empty expression cassette consisting of the PSAD promoter and terminator [16] flanking a multiple cloning site (Fig. 1a). The vector was introduced in two Agrobacterium strains with low and high virulence, respectively, LBA4404 and C58C1 [17] and used for transformation of the cell wall-proficient CC125(+) and the cell wall-deficient cw15 Chlamydomonas strains.
Fig. 1.
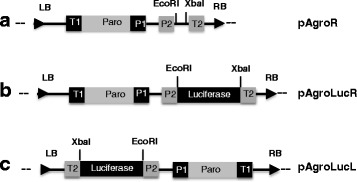
Schematic map of the vectors used for Agrobacterium-mediated transformation. Only the portion between the LB and RB (T-DNA) is shown. All vectors contain the Paro gene, conferring resistance to paromomycin under the control of the strong HSP70/RBCS2 hybrid promoter (P1) and RBCS2 terminator (T1). pAgroR (a) contains an expression cassette containing the PSAD promoter (P2) and terminator (T2) sequences. In pAgroLucR (b) the Luc coding sequence is cloned in the expression cassette. In pAgroLucL (c) the Paro and Luc cassettes have an inverted orientation with respect to pAgroLucR. For further details see Material and Methods
In the first round of experiments we followed published co-cultivation protocols [6, 7], but we were unable to obtain paromomycin-resistant colonies. Since Agrobacterium virulence (vir) genes are known to be induced by acidic pH and nutrient-poor growth media [18], we developed a different transformation method, based on the recommendations by Gelvin [19]. This method consists in growing Agrobacterium cells for the first 24 h in AB medium and then overnight in AB induction medium [19]. Co-cultivation of Agrobacterium with Chlamydomonas is carried out on solid AB induction medium for 48 h (for further details see Materials and Methods).
Using this method, cw15 cells were successfully transformed using both the LBA4404 and C58C1 Agrobacterium strains, while CC125(+) cells could be transformed only with the more virulent C58C1 strain (Table 1). In general, higher transformation efficiencies were obtained using cell wall-deficient Chlamydomonas cells and highly virulent Agrobacterium (Table 1). Transformation was confirmed by PCR analysis using oligonucleotides directed to the Paro gene (See Additional file 1: Figure S1 for an example). Sixty-seven to 92 % (67–92%) of paromomycin-resistant colonies were PCR-positive for the Paro gene (Table 1).
Table 1.
Efficiency of different Agrobacterium-mediated transformation methods. Two different Agrobacterium strains (LBA4404 and C58C1), containing the pAgroR vector and two Chlamydomonas strains (CC125 and cw15) were used. Transformation efficiency is expressed as the number of colonies resistant to paromomycin/108 cells transformed. The presence of the Paro transgene was tested by PCR and is expressed as the percentage of Paro-resistant colonies testing positive in the PCR assay. Kanamycin (Kan)-positive transformants, indicative of bacterial contamination, were excluded from the analysis. Results of two independent experiments for each protocol/strain combination are reported. At least 50 independent transformants for each experiment were analyzed by PCR
| Chlamydomonas strain | Agrobacterium strain | Transformation efficiency (colonies/108 cells) | Positivity to Paro (PCR) |
|---|---|---|---|
| cw15 | LBA4404 | 31 | 75% |
| 16 | 92% | ||
| cw15 | C58C1 | 25 | 75% |
| 33 | 67% | ||
| CC125 | LBA4404 | 1 | 0% |
| 0 | / | ||
| CC125 | C58C1 | 6 | 86% |
| 14 | 92% |
We conducted a second round of experiments using the pAgroLucR vector, carrying a codon-optimized luciferase (Luc) gene [14] in the PSAD expression cassette (Fig. 1b). This vector was used for either Agrobacterium-mediated transformation or electroporation of the cw15 strain (Table 2). Even with the optimized protocol, Agrobacterium-mediated transformation efficiencies were 2.5- to 60-fold lower with respect to electroporation using the same plasmid (Table 2). Also, while 100% of electroporated Paro-resistant colonies tested are positive for the presence of the Paro gene in the PCR assay, only 65% to 97% of colonies obtained after co-cultivation with Agrobacterium did so, indicating a high percentage of false positive transformants in the latter protocol (Table 2).
Table 2.
Comparison of electroporation and Agrobacterium-mediated transformation. Transformation efficiencies are expressed as the number of paromomycin resistant colonies/108 cells transformed. Presence of Paro and Luc transgenes was tested by PCR. Transformants containing the Luc transgene were further analyzed for Luc enzymatic activity (last column). Results of two independent experiments are reported. At least 96 independent transformants were analyzed for each experiment
| Transformation method | Plasmid | Transformants/108 cells | Paro-positive transformants (PCR) | Luc-positive transformants (PCR) | Transformants exhibiting Luc activitya |
|---|---|---|---|---|---|
| Electroporation | pAgroLucR | 120 | 100% | 26% | 15% |
| 250 | 100% | 33% | 20% | ||
| Agrobacterium (C58C1) | pAgroLucR | 4 | 97% | 16% | 13% |
| 52 | 65% | 13% | 11% | ||
| Agrobacterium (C58C1) | pAgroLucL | 31 | 93% | 23% | 20% |
| 12 | 49% | 13% | 10% |
aLuc activity > 3-fold that of the untransformed control
Transgene rearrangements
Next, we tested the maintenance of the Luc transgene in the Paro-positive transformants obtained either through electroporation or through co-cultivation with Agrobacterium. The results (Additional file 2: Figure S2 shows the analysis of a subset of transformants) show that, although most transformants tested are PCR-positive for the Paro gene, only few carry the Luc gene. The frequency of Luc-positive colonies was tested in two independent experiments and compared to that obtained in electroporation experiments using the same vector. The results (Table 2) indicate that only 13%–16% of Paro-resistant colonies obtained through Agrobacterium transformation carry a complete Luc gene, compared to 26%–33% obtained through electroporation with the same vector. Thus, in contrast with the results obtained in higher plants [12], Agrobacterium-mediated transformation of Chlamydomonas does not alleviate, compared to electroporation, the rearrangement of transgenes cloned within the T-DNA borders.
PCR reactions on a set of nine independent transformants (Fig. 2, Additional file 3: Figure S3 and Additional file 4: Figure S4) with nested pairs of oligonucleotides spanning the whole T-DNA region (Fig. 2a) indicated that, both in Agrobacterium-mediated transformation and in electroporation, deletion frequency is highest close to the right border (RB), i.e. away from the Paro selectable marker (Fig. 2b).
Fig. 2.
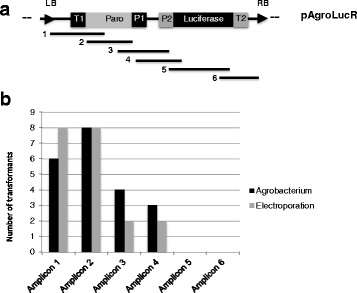
Deletions along the T-DNA in transformants obtained with electroporation or Agrobacterium-mediated transformation. Chlamydomonas cw15 cells were transformed with the pAgroLucR plasmid, using Agrobacterium or electroporation. Integrity of the inserted T-DNA was tested by PCR amplification with nested pairs of oligonucleotides spanning the T-DNA (Panel a) in a set of nine independent transformants. The histogram (Panel b) shows that there is a gradient of deletion from the LB (Amplicon1) to the RB (Amplicon 6) independently of the transformation method used (Agrobacterium, black; electroporation, gray). Oligonucleotide sequences are reported in Additional file 7: Table S2
Influence of orientation with respect to the T-DNA borders on transgene rearrangements
T-DNA integration occurs in a directional way, resulting in asymmetric deletion frequencies in regions close to the right and left borders (RB and LB) [20]. In order to investigate if the position of the Luc gene with respect to LB and RB influences the frequency of deletions at this locus, we constructed a second vector, pAgroLucL, in which the orientation of the Paro and Luc genes is inverted with respect to pAgroLucR (Fig. 1c). The results (Table 2) indicate a slight increase in the number of the transformants containing an intact Luc transgene when the pAgroLucL construct was used. Deletions coming from the RB and LB as well as more internal deletions were tested via PCR (Fig. 3, Additional file 5: Figure S5 and Additional file 6: Figure S6). The results show that deletions of the Luc transgene are more frequent (82% vs 62%) when it flanks the RB compared to the LB.
Fig. 3.
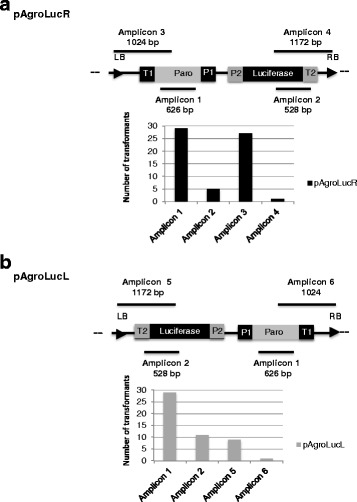
T-DNA deletion pattern in pAgroLucR and pAgroLucL transformants. The pAgroLucR and pAgroLucL plasmids were used for Agrobacterium mediated transformation and integrity of the T-DNA in the resulting transformants was tested via PCR with appropriate oligonucleotide pairs. Panels a (pAgroLucR) and b (pAgroLucL) show the position of the amplicons (top in each box) and the frequency of transformants showing amplification for each amplicon (bottom in each box). A set of 29 independent transformants were tested for each construct. All transformants contain amplicon 1, corresponding to the Paro gene. When Luc is cloned next to the LB is retained at higher frequency (pAgroLucL, amplicons 2 and 5 in Panel b) with respect to the RB (pAgroLucR, amplicons 2 and 4). See also Additional file 5: Figure S5 and Additional file 6: Figure S6. Oligonucleotide sequences are reported in Additional file 7: Table S3
Transgene expression and its stability over time under selective pressure
Next, we asked whether transgene rearrangement or transgene silencing were the major causes resulting in loss of transgene expression. In order to answer this question, cells containing an intact Luc gene as judged by PCR were subjected to 3 subsequent subcultures on selective medium in order to get rid of the Agrobacterium cells and then assayed for Luc activity. The results (Table 2 and Additional file 7: Table S4) indicate that a large fraction of transformants containing an intact Luc transgene also showed some level of Luc activity (at least three times higher than the background) and that gene silencing may be slightly more active during electroporation than during Agrobacterium-mediated transformation.
In order to follow the stability of Luc expression in transformants grown under selective pressure, cells were subcultured on solid medium containing paromomycin at 1-week intervals, and Luc activity was measured at the 3rd and 20th subculture cycle (Additional file 7: Table S4). In all three cases (electroporation and co-cultivation with Agrobacterium carrying pAgroLucR and pAgroLucL vectors), the percentage of Luc-positive transformants showing luciferase activity does not drop significantly from the 3rd to the 20th subculture cycle. Quantitative Luc activity data on the twenty highest expressors (Fig. 4) show that enzymatic activity is quite stable over time, indicating that neither the method of transformation (electroporation or Agrobacterium) nor the vector (pAgroLucL or pAgroLucR) influences Luc expression levels and their stability over subsequent generations on selective medium.
Fig. 4.
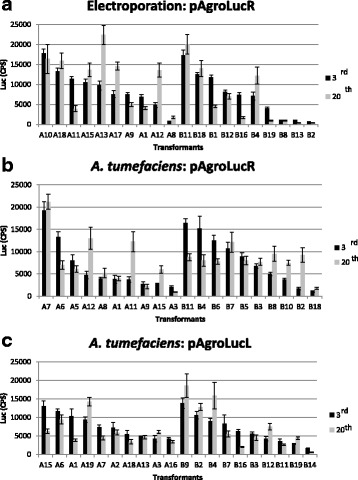
Temporal stability of Luc activity in Chlamydomonas transformants obtained through electroporation or Agrobacterium-mediated transformation. Twenty Chlamydomonas transformants showing the highest level of luciferase expression were collected from two experiments (experiments A and B: ten transformants chosen for each experiment, see Additional file 7: Table S4) and analyzed for luciferase expression at the 3rd (black bars) and the 20th (gray bars) cycles of subculture after the initial transformation event. a Electroporation with pAgroLucR b Agrobacterium-mediated trasformation with pAgroLucR c Agrobacterium-mediated trasformation with pAgroLucL. Luminescence is expressed as CPS normalized for 105 cells
Whole genome sequencing of electroporation- vs Agrobacterium-generated transformants
In order to identify the insertion points of transformants obtained with the different methods, we performed whole genomic sequencing of twenty transformants in the cw15 background, ten obtained by Agrobacterium and 10 by electroporation using the pAgroLucR plasmid. We selected 5 high and 5 low Luc expressors for each transformation method (Agrobacterium or electroporation) in order to assess any influence of the genomic context of the insertion on transgene expression. Paired-end Illumina reads (2 × 100 nucleotides long) were generated to give an average coverage of about 20×. The reads were aligned on the Chlamydomonas genome and the pAgroLucR vector sequences and the insertion points where determined using paired ends mapping to both the algal genome and the vector. The results are summarized in Table 3. One of the Agrobacterium transformants did not carry a detectable insertion, presumably because the insertion occurred in a genomic region not present in the available genome assembly. The remainder of the insertions occurred, for the most part, in genes (5′ and 3′ UTR and exons) with only a minority occurring in intergenic regions. One Agrobacterium and three electroporation transformants carried insertions in multiple chromosomal regions. Examples of DNA integration are given in Fig. 5 for one low and one high Luc expressor obtained by both Agrobacterium-mediated transformation and electroporation.
Table 3.
Information on the insertion sites and Luc expression of the different transformants. Five low and five high Luc expressors were collected and analyzed for the characterization of the insertion site both for Agrobacterium transformation and electroporation. The first column reports the name of the transformant selected (see Additional file 7: Table S4). Luc activity of the transformants at the 3rd and 20th subculture in selective medium is expressed as average values of CPS (counts per second) normalized for 105 cells ± standard deviation of three biological replicates. Chr #: chromosome number in which DNA integration occurred. Insertion points were determined from the Chlamydomonas reinhardtii structural annotation v5.5 [32]. Gene annotation was taken from the Chlamydomonas reinhardtii functional annotation v 5.5 [32]. Gene expression refers to RNA-Seq experiments conducted on wild type cells grown in TAP medium [32, 36] and are reported in FPKM (fragments per kilobase per million mapped fragments)
| Transformant | Luc activity 3rd subculture | Luc activity 20th subculture | Chr # | Insertion point | Gene name | Gene annotation | Gene expression (FPKM) |
|---|---|---|---|---|---|---|---|
| CPS ± sd | CPS ± sd | ||||||
| Transformation method: Agrobacterium | |||||||
| B 20 | 9 ± 2 | 8 ± 1 | 8 | Gene (3′ UTR) | Cre08.g381050 | Senescence-associated gene 12 | 4.65 |
| A 14 | 38 ± 11 | 18 ± 2 | 1 | Gene (exon) | Cre01.g051900 | Ubiquinol-cytochrome C reductase iron-sulfur subunit | 166.65 |
| B 9 | 5 ± 1 | 64 ± 12 | 12 | Gene (5′UTR) | Cre12.g486000 | n.a. | 1.49 |
| B 12 | 7 ± 1 | 69 ± 13 | 17 | Gene (exon) | Cre17.g736700 | n.a. | 0 |
| A 18 | 48 ± 10 | 16 ± 2 | 1 | Gene (exon) | Cre01.g015250 | DNA binding; DNA-directed DNA polymerases | 4.13 |
| A 6 | 13,306 ± 1218 | 6979 ± 926 | 3 | Gene (exon) | Cre03.g204200 | n.a. | 0 |
| A 7 | 19,240 ± 1945 | 21,210 ± 1694 | n.d. | n.d. | n.d. | n.d. | n.d. |
| B 4 | 15,202 ± 2795 | 8006 ± 1259 | 9 | Gene (5′UTR) | Cre09.g398067 | Rotamase FKBP 1 | 9.45 |
| B 6 | 12,460 ± 1239 | 7751 ± 643 | 17 | Gene (exon) | Cre17.g699600 Cre08.g377150 | Sedoheptulose-bisphosphatase | 14.62 |
| 8 | Gene (exon) | n.a. | 18.19 | ||||
| B 11 | 16,458 ± 892 | 8762 ± 831 | 8 | Gene (3′ UTR) | Cre08.g379400 | n.a. | 2.95 |
| Transformation method: Electroporation | |||||||
| A 2 | 7 ± 3 | 49 ± 6 | 12 | Gene (3′ UTR) | Cre12.g560450 | n.a. | 3.08 |
| A 3 | 2 ± 0 | 68 ± 9 | 1 | Gene (exon) | Cre01.g014150 | MATE efflux family protein | 5.20 |
| 4 | Gene (exon) | Cre04.g223550 | n.a. | 0.42 | |||
| A 4 | 4 ± 1 | 24 ± 5 | 13 | Gene: 3′ UTR | Cre13.g569850 | Ammonium transporter 1;2 | 0.89 |
| A 5 | 37 ± 11 | 61 ± 7 | 12 | Gene (exon) | Cre12.g559450 | Calcium-dependent lipid-binding family protein | 37.98 |
| A 7 | 1 ± 1 | 21 ± 5 | 11 | Gene (exon) | Cre11.g467556 | n.a. | 2.73 |
| 5 | Gene (5′UTR) | Cre05.g232100 | n.a. | 6.87 | |||
| A 10 | 17,849 ± 1022 | 16,393 ± 3650 | 3 | Gene (3′ UTR) | Cre03.g186050 | n.a. | 1.26 |
| A 11 | 11,347 ± 645 | 3932 ± 771 | 2 | 2 Genes, both 3′UTR | Cre02.g098700; | ABC-2 type transporter family protein | 0.63 |
| Intergenic region | Cre02.g098750 | STELAR K+ outward rectifier | 2.77 | ||||
| 8 | / | / | |||||
| B 11 | 17,253 ± 1316 | 20,099 ± 2464 | 3 | Gene (intron) | Cre03.g165700 | Pyruvate decarboxylase-2 | 87.52 |
| A 18 | 13,348 ± 753 | 15,958 ± 1918 | 3 | Intergenic region | / | / | / |
| B 18 | 12,482 ± 499 | 13,977 ± 1959 | 16 | Intergenic region | / | / | / |
n.d. not determined, n.a. not available
Fig. 5.
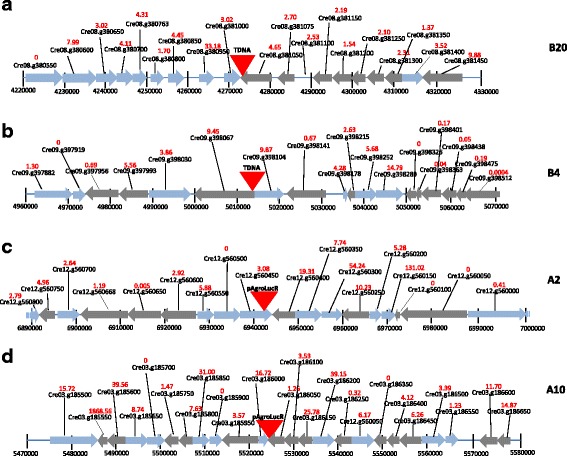
Examples of DNA insertion in transgenic C. reinhardtii clones. a and b Low and high Luc expressor transformants obtained by Agrobacterium-mediated transformation. c and d Low and high Luc expressor transformants obtained by electroporation. Insertions are shown as red triangles. Transformant names are shown on the right. Genes are indicated by arrows oriented according to their transcription direction and are labeled with gene IDs [32]. Gene expression (FPKM) in C. reinhardtii cells grown in TAP medium [32, 36] is reported in red above each gene ID
Discussion
Although Agrobacterium mediated transformation in Chlamydomonas was first reported in 2004 [6], up to now it has not been widely adopted. Previous publications [6–8], demonstrate the successful expression of exogenous genes and, in one case, stable integration in the nucleus. It has been proposed that, similar to plants, transformation of Chlamydomonas by Agrobacterium could provide stable integration and lower copy numbers, potentially leading to fewer problems with transgene co-suppression and instability. However, no data on the rearrangements/insertion points and copy number are available for Agrobacterium-mediated transformants and, most importantly, no comparative data are available with widely used methods, such as electroporation.
The experiments described here indicate that the cw15 strain is more susceptible to Agrobacterium transformation than the CC125 strain. This may be due to the fact that cw15 is a cell wall deficient mutant and may be more susceptible to Agrobacterium infection. Analysis of a large set of transformants indicated that, regardless of the method of transformation used, just a small percentage (10% to 20%) of the transformants show significant Luc expression (Table 2). In the case of electroporation, all the transformants analyzed contain the Paro selectable gene, while about 1/3 also contain a non-rearranged Luc gene. Transformation with Agrobacterium results in a higher number of false positive colonies (Table 2), lacking both genes. This may be due to low level expression of the Paro gene in Agrobacterium, leading to inactivation of paromomycin during the co-cultivation step, or to the long incubation of plates from Agrobacterium-mediated transformation, leading to light-mediated degradation of paromomycin, or to interference by the antibiotics used to remove Agrobacterium cells.
In cells transformed with pAgroLucR a gradient of deletions is observed, with very high deletion frequencies near the RB, i.e. away from the selectable marker gene (Fig. 2, Additional file 3: Figure S3 and Additional file 4: Figure S4). A slight effect of the LB and RB on deletion frequencies was observed, with the RB being more prone to deletion than LB, as observed in plants [20]. Luminometric assays performed at the 3th and 20th cycles of subculture in selective media after transformation revealed that the percentage of colonies expressing luciferase gene is relatively stable over time (Additional file 7: Table S4 and Fig. 4). This suggests that the major factors limiting Luc gene expression are the reporter gene rearrangements during the transformation step. We can not, however, exclude completely the occurrence of silencing phenomena in the first three subculture steps, since we analyzed Luc activity only after the third subculture in selective medium, to remove residual bacterial cells.
Sequencing of five high Luc and five low Luc expressors for each transformation method did not suggest any clear difference in insertion patterns or any correlation between Luc expression and the genomic region in which the insertion occurred (Table 3). A higher number of Agrobacterium insertions (10 out of 10) occurred in transcribed regions (exons and UTRs), compared to electroporation (9 out of 13). This is much higher than what reported in large scale insertional mutagenesis projects, in which only 50% of insertions were in exons and UTRs [21]. Also, Agrobacterium displayed a lower number of multiple insertions (1 out of 10 transformants) compared to electroporation (3 out of 10 transformants). This indicates that the optimized Agrobacterium-mediated transformation protocol reported here may be useful for insertional mutagenesis projects in Chlamydomonas.
Zhang et al. have proposed a model to explain the large rearrangements to which exogenous DNA is subjected during Chlamydomonas transformation. According to this model, the exogenous DNA is cleaved extracellularly by an endonuclease and then ligated to Chlamydomonas DNA present in the medium due to cell lysis, before it is taken up by the cell and inserted in the nuclear genome [21]. Our data only partially support this model. It is known that T-DNA is transferred from Agrobacterium to the recipient cell nucleus by means of a bacterial pilus, as a single-stranded complex with the virulence proteins VirD2 and VirE2, the former forming a covalent bond with the 5′ end of the T-DNA molecule [17]. During this process, it is unlikely that the T-DNA is exposed to extracellular endonucleases, and thus, if the cleavage was extracellular, we should observe a much lower frequency of DNA rearrangements in Agrobacterium-mediated transformants. Also if, as has been proposed, the mechanisms mediating transgene rearrangement in Chlamydomonas have an evolutionary significance as a defense against pathogenic bacteria and viruses, it makes more evolutionary sense that these mechanisms act within the Chlamydomonas cell, where the bacterial or viral DNA is naked, rather than in the extracellular space, where it is contained within a bacterial cell or a viral capsid. We thus propose that the introduction of exogenous DNA in the Chlamydomonas cell triggers an endonucleolytic mechanism aimed at its degradation, and that this mechanism preferably acts within the nucleus, when the nucleoprotein T-DNA complex is disassembled and the T-DNA is naked, before insertion into the nuclear DNA.
Conclusions
Chlamydomonas has been proposed as a system for heterologous protein expression [22]. However, low levels of expression and rearrangements of the transforming DNA prevent it from being a competitive production platform with respect to bacterial, fungal or animal cells. In the present work we compared Agrobacterium-mediated transformation with electroporation of Chlamydomonas in order to verify if the former can alleviate these problems. Regardless of the method used to transform Chlamydomonas cells, extensive rearrangements occur at the Luc gene, which is not under selective pressure. This drawback was only partially compensated by the reduced Luc silencing in Agrobacterium transformants. Thus, differently from what observed in plants, Agrobacterium-mediated transformation does not present significant advantages in terms of higher or more stable expression. In order to increase transgene stability and expression, mutants affected in transgene silencing [23] or in exogenous DNA cleavage can be of great help. The knowledge gained from Agrobacterium-mediated transformation may be instrumental in the latter endeavour.
Methods
Chlamydomonas strains and culture conditions
The Chlamydomonas reinhardtii cell wall-deficient cw15 and cell wall-proficient CC125 (+) strains [24], were used for all experiments. Cells were grown photomixotrophically in Tris Acetate Phosphate (TAP) medium [24] at 25 °C under continuous irradiation with fluorescent white light (60 μE m− 2 s− 1). In the case of cw15 strain the growth medium was supplemented with 1% (w/v) D-sorbitol.
For luciferase assay, the transformants were grown in 1.5 ml of TAP medium at 160 rpm at 80 μE m− 2 s−1on a rotary shaker until they reach the stationary phase (about 48 hs) in 24-well blocks (Qiagen, cat. n. 19,583). The cultures were then diluted 1:20 in 4 ml of TAP medium and grown for 48 h. The plates were covered with Breathe-Easy membrane (Sigma-Aldrich, cat. n. Z763624-100EA) to prevent evaporation without limiting gaseous and light exchange. Frozen cell pellets relative to 200 μl of each culture were re-suspended in 40 μl of lysis buffer (Renilla Luciferase Assay System, Promega, cat. E2820), lysed at room temperature for 15 min on a rotary shaker (750 rpm) and then incubated on ice until assayed activity. Data were normalized using optical absorbance of at least two biological replicates for each transformant.
Plasmid construction
In order to obtain the pAgroR (Fig. 1, panel a) and pAgroL plasmids (map not shown), pSL18 [25] was digested with NotI and KpnI. The digestion produces two fragments, 3255 and 2867 bp long, that were blunt ended using the Klenow Fragment (Roche, cat. n. 11,008,404,001). The 3255 bp fragment containing the paromomycin expression cassette and the PSAD promoter and terminator regulatory elements was gel purified and used for the next cloning step. The plasmid pKCRTI [26] (derived from pCAMBIA 1390) was digested with BglII and XhoI releasing four fragments of 13, 865, 3391 and 6906 bp long. The digestion was subjected to filling in by Klenow and the 6906 bp fragment containing all the genetic elements required for Agrobacterium propagation and transfer into the host nucleus (including the LB and RB) was gel purified. The two fragments of 3255 bp, originating from pSL18, and 6906 bp, originating from pKCRTI, were blunt cloned and PCR was performed using specific oligonucleotides to discriminate the two different orientations of the fragments. The suffix R and L in pAgro vectors refers to the position of the empty PSAD expression cassette respect to the borders (R: the PSAD cassette is close to the RB; L: the PSAD cassette is close to the LB). To build pAgroLucR and pAgroLucL plasmids, Renilla reniformis luciferase coding sequence was isolated by PCR from plasmid PSAD:cRLuc [27] with the following oligos: crLuc-EcoRI for CAGCGAATTCATGGCCAGCAAGGTGTACGAC and cRLuc-XbaI rev CAGTCTAGATTACGTATCGTTCTTCAGC (EcoRI and XbaI restriction sites are reported in bold, gene specific sequence in italics is underlined). The PCR product was then cloned EcoRI-XbaI into pAgroR and pAgroL generating respectively pAgroLucR and pAgroLucL (Fig. 1, panel b and c). The bacterial strain used for cloning was XL1Blue from Statagene. Bacterial transformation was carried out by electroporation.
DNA extraction from Chlamydomonas cells and PCR analysis
Single colonies of Chlamydomonas cells were picked and resuspended in 200 μl of 5% (w/v) chelex solution (Sigma-Aldrich, cat. n. C7901-25G) in a 96-well plate. The plate was incubated 10 min at 100 °C, 10 min on ice and centrifuged at 6000 g for 3 min. One microliter of the supernatant was used to perform PCR analysis. The sequences of oligonucleotides used to screen Chlamydomonas transformants are reported in Additional file 7: Table S1. β-tubulin was used as positive control for DNA extraction, paromomycin oligonucleotides were used to screen the positivity to the antibiotic, kanamycin oligonucleotides were used to evaluate residual Agrobacterium contamination while cRLuc for 710 and PSAD Ter 276 were used to check the presence of luciferase gene.
Chlamydomonas colonies were analysed by PCR after the three transfers in 1,5% TAP agar plate containing 10 μg/μl paramomycin. In the case of Chlamydomonas transformants obtained through co-cultivation method 500 μg/μl cefotaxime and 500 μg/μl carbenicillin were added in order to remove residual Agrobacterium cells. Ninety-six colonies were analyzed by PCR for each transformation experiment.
Chlamydomonas electroporation
Chlamydomonas cells were cultured in TAP medium to 3–6 × 106 cells/ml and then centrifuged at 3000 g for 5 min at 4 °C. The pellet was washed once with cold TAP containing 60 mM D-sorbitol and resuspended at final concentration of 2 × 108 cells/ml. 250 μl (5 × 107 cells) of this culture was mixed with 200 ng of linearized vector in a 0,4 cm cuvette (BIORAD, cat. n. 165–2081) and electroporated at 25 μF, 2000 V/cm. The time constant ranged between 13 and 16 ms. The electroporated cells were recovered overnight in TAP medium at low light (30 μE m− 2 s− 1) and then plated on 1% TAP agar plate containing 10 μg/μl paramomycin in the case of vectors described in Fig. 1. For electroporation, the pAgroLucR vector (Fig. 1b) was linearized with AgeI. Two independent electroporation experiments were performed, for each electroporation ten transformations were carried out. Ninety-six transformants for each transformation were analysed by PCR.
Agrobacterium-mediated transformation with published methods
In the first set of experiments, Chlamydomonas cells were transformed according to [28]. Briefly, LBA4404 (Takara cat. n° 9115) and C58C1 Agrobacterium cells, kindly donated by Prof. Edgardo Filippone, were transformed through electroporation with the pAgroR vector (Fig. 1, Panel a) and plated on LB containing 100 μg/ml kanamycin and 100 μg/ml rifampicin (LBA4404 strain) or 200 μg/ml kanamycin and 100 μg/ml rifampicin (C58C1 strain) and grown for 48 h at 28 °C. Five single colonies from each transformation plate were inoculated in 1.5 ml of LB medium containing the appropriate antibiotics and analysed by PCR in order to check the presence of the right and left borders with the following pairs of oligonucleotides: PSAD Ter for GATTTCGCTGATTGATACGG and R border rev TAAACGCTCTTTTCTCTTAGG, producing an amplicon of 894 bp, L border for TGGCAGGATATATTGTGGTG and Paro box rev CTGGACTGGGAGCGGTGT, producing an amplicon of 1039 bp. All the five picked colonies showed the right amplicons and one of them was inoculated in LB medium plus antibiotics to prepare glycerol stocks. For each transformation experiment 100 μl of glycerol stock were inoculated in 15 ml liquid YEB medium - containing 30 μg/ml rifampicin and 100 μg/ml kanamycin - and grown for 24 h at 28 °C. The cells where then centrifuged at 2700 g for 30 min and resuspended in liquid TAP medium containing 100 μM acetosyringone to an A595 of 0.5. 107 Chlamydomonas cells were plated on a 90 mm petri dish and grown for two days at 80 μE m− 2 s− 1 in continuous light to allow a lawn of cells to be formed. 200 μl of the bacterial suspension obtained as described above was spread on the lawn of Chlamydomonas cells for co-cultivation. After co-cultivation for 48 h at 30 μE m− 2 s− 1 cells were harvested at 1500 g for 5 min washed twice with liquid TAP medium containing 500 μg/μl cefotaxime by centrifugation at 1500 g for 5 min. Finally the cells were resuspended in TAP at 5 × 107 cells/ml concentration and 1 ml was then plated onto 1% agar TAP plates containing 10 μg/μl paramomycin, 500 μg/μl cefotaxime and 500 μg/μl carbenicillin. Paromomycin is the selective antibiotic for transformation while cefotaxime and carbenicillin were used to kill Agrobacterium cells. Single colonies appeared after 7 days of growth at 30 μE m− 2 s− 1. These colonies were picked and transferred on new TAP agar plates containing all the three antibiotics. Even if we could observe several colonies on the primary transformation plate, after two or three transfers on selective medium they died, probably because they were false positive or because the vector did not integrate stably into the nucleus. In a second round of experiments we followed the protocol described in [7], but in this case we could not observe any colony on the primary transformation plates.
Development of a modified protocol for Agrobacterium-mediated transformation
In order to set up a protocol for Chlamydomonas transformation through Agrobacterium in our laboratory, we followed the indications by [19]. Glycerol stocks of the Agrobacterium strains LBA4404 and C58C1 transformed with pAgroR vector obtained as described in the previous paragraph were inoculated in 15 ml of AB medium (pH 7) [29] containing 0.5% (w/v) glucose, 30 μg/ml rifampicin and 100 μg/ml kanamycin. The flasks were grown at 28 °C for 24 h, then centrifuged at 2700 g for 30 min and finally resuspended in 40 ml of AB induction medium (AB medium containing 0.5% glucose, 20 μM MES pH 5,6 and 100 μM acetosyringone) and grown overnight at 28 °C. Before co-cultivation the cell density was adjusted to A595 = 0,5 with AB induction medium. Fifty ml of Chlamydomonas culture grown for 2 days in TAP medium at approximately 107 cells/ml were centrifuged at 1500 g for 5 min and resuspended in AB medium at a final cell density of 108 cells/ml. For co-cultivation, 100 μl of microalgae corresponding to 107 cells and 40 μl of Agrobacterium were spotted on 1% agar AB induction medium plates. In total 14 spots, corresponding to 108 Chlamydomonas cells, were plated on each 1% agar AB induction medium plate and the plates were grown at 30 μE m− 2 s− 1 for 48 h. The cells were then harvested from each plate, washed twice with liquid TAP medium containing 500 μg/μl cefotaxime, centrifuged at 1500 g for 5 min, resuspended in liquid TAP medium at a density of 5 × 107 cells/ml and plated on two TAP agar plates containing 10 μg/μl paramomycin, 500 μg/μl cefotaxime and 500 μg/μl carbenicillin. After 1 week of growth, colonies started to appear. Transformation efficiency is expressed as an average of the number of paromomycin resistant colonies obtained per 108 of Chlamydomonas cells.
Paromomycin resistant colonies were transferred for three subculture rounds on 1.5% TAP agar plates containing the selective antibiotics and then analyzed by PCR. Two independent transformation experiments for each vector were performed and 96 colonies for each experiment were analysed by PCR.
Luciferase assay
Luciferase assay was performed using the Renilla Luciferase Assay System (Promega, cat. n. E2820) as described in [27]. The luminescence of the wild type (background luminescence) was subtracted from the luminescence of the transformants. All the transformants showing a level of Luc activity > 3 fold than the background were considered Luc-positive. The luminescence values reported are relative to 105 cells. At least two biological and two technical replicates were analyzed for each transformant.
Whole genome sequencing and bioinformatic analyses
Total DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Cat No. 69104) from pellets obtained from 8 ml of mid-log phase cultures (concentration of 5 × 106 cells/ml), grown in TAP medium as described above.
DNA quality and concentration was checked by running samples on 0.5% agarose gel and by fluorometry (Qubit, ThermoFisher Scientific), respectively. For each sample, one microgram of DNA was used to generate paired-end Illumina libraries, which were then sequenced on a HiSeq 2500 Illumina sequencer generating about 8 millions paired-end reads on average. Reads were cleaned of adapters and quality trimmed with Cutadapt v. 1.8.1 [30] and Trimmomatic v. 0.33 [31]. A composite reference was prepared by combining Chlamydomonas reinhardtii genome v. 5.5 [32] and the pAgroLucR vector sequence. All libraries were aligned to the reference using Bowtie2 v. 2.2.7 [33] as single ends, to avoid any bias due to pairing in the alignment results (Additional file 7: Table S5). In order to isolate the insertion signals, only pairs with a hit both on the alga genome and the vector were retained. Finally, for each transformed sample, all signals were compared to the wild type and insertion points were called with MACS v. 2.1.1 [34].
Additional files
Figure S1. PCR confirmation of the presence of the Paro selectable marker. Fourteen Paro resistant colonies (1–14) obtained by co-cultivation of the cw15 strain with Agrobacterium C58C1 cells harbouring the pAgroR plasmid were analyzed by PCR for the presence of the endogenous β-tub gene (used as positive control for DNA extraction), Paro gene (selectable marker) and Kan gene (diagnostic of residual Agrobacterium contamination). Transformants 1–12 and 14 have the Paro gene, while the results on transformant 13 are inconclusive since the colony is still contaminated by Agrobacterium. Wt: cw15 strain; P: pAgroR plasmid; C+: positive cw15 transformant; C-: no DNA. M: 1 Kb Plus ladder (Life Technologies). Oligonucleotide sequences are shown in Additional file 7: Table S1. (PPTX 380 kb)
Figure S2. Retention of the Luc transgene in a set of Chlamydomonas colonies transformed with the pAgroLucR plasmid. cw15 cells were co-cultivated with the Agrobacterium C58C1 strain harboring the pAgroLucR plasmid. The DNA extracted was analyzed for the presence of the genes shown in Additional file 1: Figure S1 plus the Luc gene. The vast majority of the transformants, although positive for the presence of the Paro gene, do not contain an intact Luc gene. A and B are two independent control transformants, positive for the presence of the Paro and Luc genes. P: pAgroLucR plasmid. C-: negative control. M: 1 Kb Plus DNA Ladder (Life Technologies). Oligonucleotide sequences are shown in Additional file 7: Table S1. (PPTX 1875 kb)
Figure S3. PCR analysis on a set of nine independent transformants obtained through Agrobacterium to study the deletion pattern long the T-DNA. Chlamydomonas cw15 cells were co-cultivated with Agrobacterium C58C1 strain carrying the pAgroLucR plasmid. Six PCR reactions were performed on extracted DNA with nested pairs of oligonucleotides annealing in the T-DNA from the LB to the RB (Panel A). The results (Panel B) show that there is a gradient of deletions from the LB to the RB. β-tubulin was used as positive control for DNA extraction. M: 1 Kb Plus DNA Ladder (Life Technologies); wt: cw15 strain; P: pAgroLucR; −: negative control. Oligonucleotide sequences are reported in Additional file 7: Table S2. (PPTX 204 kb)
Figure S4. PCR analysis on a set of nine independent transformants obtained through electroporation to study the deletion pattern along the T-DNA. Chlamydomonas cw15 cells were electroporated with the pAgroLucR plasmid. Six PCR reactions were performed on extracted DNA with nested pairs of oligonucleotides annealing in the T-DNA from the LB to the RB (Panel A). The results (Panel B) show that there is a gradient of deletions from the LB to the RB. β-tubulin was used as positive control for DNA extraction. M: 1 Kb Plus DNA Ladder (Life Technologies); wt: cw15 strain; P: pAgroLucR; −: negative control. Oligonucleotide sequences are reported in Additional file 7: Table S2. (PPTX 128 kb)
Figure S5. Deletion pattern on the T-DNA in the pAgroLucR transformants obtained though co-cultivation of Chlamydomonas with Agrobacterium cells transformed with the pAgroLucR plasmid. The figure shows respectively a PCR analysis of a set of 29 independent cw15 transformants obtained with C58C1 Agrobacterium cells carrying the pAgroLucR vector. Wt: cw15, P: pAgroLucR plasmid; C-: negative control. Oligonucleotide sequences are reported in Additional file 7: Table S3. (PPTX 1196 kb)
Figure S6. Deletion pattern on the T-DNA in the pAgroLucL transformats obtained though co-cultivation of Chlamydomonas with Agrobacterium cells transformed with the pAgroLucL plasmid. The Figure shows a PCR analysis of a set of 29 independent transformants obtained co-cultivating cw15 cells with C58C1 Agrobacterium cells carrying the pAgroLucL vector. Wt: cw15, P: pAgroLucL plasmid; C-: negative control. Oligonucleotide sequences are reported in Additional file 7: Table S3. (PPTX 2320 kb)
Table S1. Oligonucleotides used to screen Chlamydomonas transformants. Table S2. Oligonucleotides used to study the T-DNA deletion pattern. Table S3. Oligonucleotides used to study the influence of R and L borders on T-DNA rearrangements. Table S4. Luciferase activity data at the 3rd and 20th subcultures. Table S5. NGS library mapping statistics. (DOCX 75 kb)
Acknowledgements
Part of the computing resources and the related technical support used for this work have been kindly provided by CRESCO/ENEAGRID High Performance Computing infrastructure and its staff [35]. This work was supported by the Italian Ministry of Agriculture, Hydrobio project.
Funding
This work was financially supported by the Italian Ministry of Agriculture, Hydrobio project. The funding body had no role in study design, in the collection, analysis and interpretation of data, in the writing the Ms. and in the decision to submit the article for publication.
Availability of data and materials
The raw data relative to Tables 1 and 2 are available upon request. The Illumina sequence data have been submitted as Bioproject [PRJNA395035] to NCBI sequence read archive under accession number [SRP113153].
Abbreviations
- Chr
Chromosome
- CPS
Counts per second
- FPKM
Fragments per kilobase per million mapped fragments
- HSP70/RBCS2
70 kDa heat shock protein/ribulose bisphosphate carboxylase/oxygenase small subunit
- LB
Left border
- Luc
Luciferase
- nt
Nucleotides
- Paro
Paromomycin
- PSAD
Photosystem I reaction center subunit II
- RB
Right border
- TAP
Tris Acetate Phosphate
- UTR
Untraslated region
- β-tub
β-tubulin
Authors’ contributions
PM, OCD, SV, PP, GA and PF produced data. PM, OCD, GA, PF and GG analyzed data. PF and GG coordinated the study and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12896-018-0416-3) contains supplementary material, which is available to authorized users.
Contributor Information
Paola Mini, Email: paola.mini@enea.it.
Olivia Costantina Demurtas, Email: olivia.demurtas@enea.it.
Silvia Valentini, Email: valentini.bros@libero.it.
Patrizia Pallara, Email: patrizia.pallara@enea.it.
Giuseppe Aprea, Email: giuseppe.aprea@enea.it.
Paola Ferrante, Email: paola.ferrante@enea.it.
Giovanni Giuliano, Email: giovanni.giuliano@enea.it.
References
- 1.Kindle KL, Schnell RA, Fernandez E, Lefebvre PA. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989;109(6 Pt 1):2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debuchy R, Purton S, Rochaix JD. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 1989;8(10):2803–2809. doi: 10.1002/j.1460-2075.1989.tb08426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1990;87(3):1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunahay TG. Transformation of Chlamydomonas reinhardtii with silicon carbide whiskers. BioTechniques. 1993;15(3):452-455, 457-458, 460. [PubMed] [Google Scholar]
- 5.Shimogawara K, Fujiwara S, Grossman A, Usuda H. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics. 1998;148(4):1821–1828. doi: 10.1093/genetics/148.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar SV, Misquitta RW, Reddy VS, Rao BJ, Rajam MV. Genetic transformation of the green alga—Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 2004;166(3):731–738. doi: 10.1016/j.plantsci.2003.11.012. [DOI] [Google Scholar]
- 7.Pratheesh PT, Vineetha M, Kurup GM. An efficient protocol for the Agrobacterium-mediated genetic transformation of microalga Chlamydomonas reinhardtii. Mol Biotechnol. 2014;56(6):507–15. doi: 10.1007/s12033-013-9720-2. [DOI] [PubMed] [Google Scholar]
- 8.Barahimipour R, Neupert J, Bock R. Efficient expression of nuclear transgenes in the green alga Chlamydomonas: synthesis of an HIV antigen and development of a new selectable marker. Plant Mol Biol. 2016;90(4–5):403–418. doi: 10.1007/s11103-015-0425-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Specht E, Miyake-Stoner S, Mayfield S. Micro-algae come of age as a platform for recombinant protein production. Biotechnol Lett. 2010;32(10):1373–1383. doi: 10.1007/s10529-010-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neupert J, Shao N, Lu Y, Bock R. Genetic transformation of the model green alga Chlamydomonas reinhardtii. Methods Mol Biol. 2012;847:35–47. doi: 10.1007/978-1-61779-558-9_4. [DOI] [PubMed] [Google Scholar]
- 11.Valvekens D, Montagu MV, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci U S A. 1988;85(15):5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai S, Zheng P, Marmey P, Zhang S, Tian W, Chen S, Beachy RN, Fauquet C. Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Mol Breed. 2001;7(1):25–33. doi: 10.1023/A:1009687511633. [DOI] [Google Scholar]
- 13.Sizova I, Fuhrmann M, Hegemann P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 2001;277(1–2):221–229. doi: 10.1016/S0378-1119(01)00616-3. [DOI] [PubMed] [Google Scholar]
- 14.Fuhrmann M, Hausherr A, Ferbitz L, Schodl T, Heitzer M, Hegemann P. Monitoring dynamic expression of nuclear genes in Chlamydomonas reinhardtii by using a synthetic luciferase reporter gene. Plant Mol Biol. 2004;55(6):869–881. doi: 10.1007/s11103-005-2150-1. [DOI] [PubMed] [Google Scholar]
- 15.Schroda M, Blocker D, Beck CF. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21(2):121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- 16.Fischer N, Rochaix JD. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Gen Genomics. 2001;265(5):888–894. doi: 10.1007/s004380100485. [DOI] [PubMed] [Google Scholar]
- 17.Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67(1):16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vernade D, Herrera-Estrella A, Wang K, Van Montagu M. Glycine Betaine allows enhanced induction of the Agrobacterium tumefaciens vir genes by Acetosyringone at low pH. J Bacteriol. 1988;170(12):5822–5829. doi: 10.1128/jb.170.12.5822-5829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelvin SB. Agrobacterium virulence gene induction. In: Wang K, editor. Agrobacterium protocols. Totowa: Humana Press; 2006. [Google Scholar]
- 20.Tzfira T, Li J, Lacroix B, Citovsky V. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 2004;20(8):375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Zhang R, Patena W, Armbruster U, Gang SS, Blum SR, Jonikas MC. High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and Endonucleolytic cleavage of transforming DNA. Plant Cell. 2014;26(4):1398–1409. doi: 10.1105/tpc.114.124099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasala BA, Muto M, Lee PA, Jager M, Cardoso RM, Behnke CA, Kirk P, Hokanson CA, Crea R, Mendez M, et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol J. 2010;8(6):719–733. doi: 10.1111/j.1467-7652.2010.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neupert J, Karcher D, Bock R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009;57(6):1140–1150. doi: 10.1111/j.1365-313X.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- 24.Harris EH. The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use. 1989. [DOI] [PubMed] [Google Scholar]
- 25.Depège N, Bellafiore S, Rochaix JD. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299(5612):1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- 26.Diretto G, Al-Babili S, Tavazza R, Papacchioli V, Beyer P, Giuliano G. Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS One. 2007;2:e350. doi: 10.1371/journal.pone.0000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrante P, Catalanotti C, Bonente G, Giuliano G. An optimized, chemically regulated gene expression system for Chlamydomonas. PLoS One. 2008;3(9):e3200. doi: 10.1371/journal.pone.0003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajam MV, Kumar SV. Green Alga (Chlamydomonas reinhardtii) Methods Mol Biol. 2006;344:421–433. doi: 10.1385/1-59745-131-2:421. [DOI] [PubMed] [Google Scholar]
- 29.Miller JH. Experiments in molecular genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 30.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 31.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ponti G, Palombi F, Abate D, Ambrosino F, Aprea G, Bastianelli T, Beone F, Bertini R, Bracco G, Caporicci M. High Performance Computing & Simulation (HPCS), 2014 International Conference on. 2014. The role of medium size facilities in the HPC ecosystem: the case of the new CRESCO4 cluster integrated in the ENEAGRID infrastructure; pp. 1030–1033. [Google Scholar]
- 36.Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, et al. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell. 2011;23(4):1273–1292. doi: 10.1105/tpc.111.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. PCR confirmation of the presence of the Paro selectable marker. Fourteen Paro resistant colonies (1–14) obtained by co-cultivation of the cw15 strain with Agrobacterium C58C1 cells harbouring the pAgroR plasmid were analyzed by PCR for the presence of the endogenous β-tub gene (used as positive control for DNA extraction), Paro gene (selectable marker) and Kan gene (diagnostic of residual Agrobacterium contamination). Transformants 1–12 and 14 have the Paro gene, while the results on transformant 13 are inconclusive since the colony is still contaminated by Agrobacterium. Wt: cw15 strain; P: pAgroR plasmid; C+: positive cw15 transformant; C-: no DNA. M: 1 Kb Plus ladder (Life Technologies). Oligonucleotide sequences are shown in Additional file 7: Table S1. (PPTX 380 kb)
Figure S2. Retention of the Luc transgene in a set of Chlamydomonas colonies transformed with the pAgroLucR plasmid. cw15 cells were co-cultivated with the Agrobacterium C58C1 strain harboring the pAgroLucR plasmid. The DNA extracted was analyzed for the presence of the genes shown in Additional file 1: Figure S1 plus the Luc gene. The vast majority of the transformants, although positive for the presence of the Paro gene, do not contain an intact Luc gene. A and B are two independent control transformants, positive for the presence of the Paro and Luc genes. P: pAgroLucR plasmid. C-: negative control. M: 1 Kb Plus DNA Ladder (Life Technologies). Oligonucleotide sequences are shown in Additional file 7: Table S1. (PPTX 1875 kb)
Figure S3. PCR analysis on a set of nine independent transformants obtained through Agrobacterium to study the deletion pattern long the T-DNA. Chlamydomonas cw15 cells were co-cultivated with Agrobacterium C58C1 strain carrying the pAgroLucR plasmid. Six PCR reactions were performed on extracted DNA with nested pairs of oligonucleotides annealing in the T-DNA from the LB to the RB (Panel A). The results (Panel B) show that there is a gradient of deletions from the LB to the RB. β-tubulin was used as positive control for DNA extraction. M: 1 Kb Plus DNA Ladder (Life Technologies); wt: cw15 strain; P: pAgroLucR; −: negative control. Oligonucleotide sequences are reported in Additional file 7: Table S2. (PPTX 204 kb)
Figure S4. PCR analysis on a set of nine independent transformants obtained through electroporation to study the deletion pattern along the T-DNA. Chlamydomonas cw15 cells were electroporated with the pAgroLucR plasmid. Six PCR reactions were performed on extracted DNA with nested pairs of oligonucleotides annealing in the T-DNA from the LB to the RB (Panel A). The results (Panel B) show that there is a gradient of deletions from the LB to the RB. β-tubulin was used as positive control for DNA extraction. M: 1 Kb Plus DNA Ladder (Life Technologies); wt: cw15 strain; P: pAgroLucR; −: negative control. Oligonucleotide sequences are reported in Additional file 7: Table S2. (PPTX 128 kb)
Figure S5. Deletion pattern on the T-DNA in the pAgroLucR transformants obtained though co-cultivation of Chlamydomonas with Agrobacterium cells transformed with the pAgroLucR plasmid. The figure shows respectively a PCR analysis of a set of 29 independent cw15 transformants obtained with C58C1 Agrobacterium cells carrying the pAgroLucR vector. Wt: cw15, P: pAgroLucR plasmid; C-: negative control. Oligonucleotide sequences are reported in Additional file 7: Table S3. (PPTX 1196 kb)
Figure S6. Deletion pattern on the T-DNA in the pAgroLucL transformats obtained though co-cultivation of Chlamydomonas with Agrobacterium cells transformed with the pAgroLucL plasmid. The Figure shows a PCR analysis of a set of 29 independent transformants obtained co-cultivating cw15 cells with C58C1 Agrobacterium cells carrying the pAgroLucL vector. Wt: cw15, P: pAgroLucL plasmid; C-: negative control. Oligonucleotide sequences are reported in Additional file 7: Table S3. (PPTX 2320 kb)
Table S1. Oligonucleotides used to screen Chlamydomonas transformants. Table S2. Oligonucleotides used to study the T-DNA deletion pattern. Table S3. Oligonucleotides used to study the influence of R and L borders on T-DNA rearrangements. Table S4. Luciferase activity data at the 3rd and 20th subcultures. Table S5. NGS library mapping statistics. (DOCX 75 kb)
Data Availability Statement
The raw data relative to Tables 1 and 2 are available upon request. The Illumina sequence data have been submitted as Bioproject [PRJNA395035] to NCBI sequence read archive under accession number [SRP113153].


