ABSTRACT
Botrytis cinerea is a plant-pathogenic fungus producing apothecia as sexual fruiting bodies. To study the function of mating type (MAT) genes, single-gene deletion mutants were generated in both genes of the MAT1-1 locus and both genes of the MAT1-2 locus. Deletion mutants in two MAT genes were entirely sterile, while mutants in the other two MAT genes were able to develop stipes but never formed an apothecial disk. Little was known about the reprogramming of gene expression during apothecium development. We analyzed transcriptomes of sclerotia, three stages of apothecium development (primordia, stipes, and apothecial disks), and ascospores by RNA sequencing. Ten secondary metabolite gene clusters were upregulated at the onset of sexual development and downregulated in ascospores released from apothecia. Notably, more than 3,900 genes were differentially expressed in ascospores compared to mature apothecial disks. Among the genes that were upregulated in ascospores were numerous genes encoding virulence factors, which reveals that ascospores are transcriptionally primed for infection prior to their arrival on a host plant. Strikingly, the massive transcriptional changes at the initiation and completion of the sexual cycle often affected clusters of genes, rather than randomly dispersed genes. Thirty-five clusters of genes were jointly upregulated during the onset of sexual reproduction, while 99 clusters of genes (comprising >900 genes) were jointly downregulated in ascospores. These transcriptional changes coincided with changes in expression of genes encoding enzymes participating in chromatin organization, hinting at the occurrence of massive epigenetic regulation of gene expression during sexual reproduction.
KEYWORDS: ascospore, epigenetic regulation, plant disease, sexual reproduction, transcriptome
IMPORTANCE
Fungal fruiting bodies are formed by sexual reproduction. We studied the development of fruiting bodies (“apothecia”) of the ubiquitous plant-pathogenic ascomycete Botrytis cinerea. The role of mating type genes in apothecium development was investigated by targeted mutation. Two genes are essential for the initiation of sexual development; mutants in these genes are sterile. Two other genes were not essential for development of stipes; however, they were essential for stipes to develop a disk and produce sexual ascospores. We examined gene expression profiles during apothecium development, as well as in ascospores sampled from apothecia. We provide the first study ever of the transcriptome of pure ascospores in a filamentous fungus. The expression of numerous genes involved in plant infection was induced in the ascospores, implying that ascospores are developmentally primed for infection before their release from apothecia.
INTRODUCTION
Sexual reproduction permits organisms to generate new combinations of alleles and thereby offers the possibility of increasing the fitness of (part of) their offspring. Sexual spores of fungi can survive under adverse conditions (1, 2), but their main biological function is in dispersal. Sexual spores of plant-pathogenic fungi can be dispersed by rain or wind, sometimes over long distances, and can subsequently serve as inoculum for new infections (3–6). In ascomycete fungi, sexual compatibility is determined by two opposite mating type (MAT) loci, designated MAT1-1 and MAT1-2 (7–9). Ascomycetes can have either heterothallic or homothallic sexual reproduction systems. In a heterothallic species, mating occurs between two isolates with opposite mating types. The opposite MAT loci of ascomycetes are generally present at the same chromosomal location and therefore genetically allelic; however, they lack sequence similarity and are often referred to as “idiomorphs” rather than alleles (10). Each idiomorph contains at least one gene encoding a transcription factor. By convention, the MAT1-1 idiomorph encodes a protein with an α domain, while MAT1-2 encodes a protein with a high-mobility-group box (HMG box) (9, 11). Homothallic species are capable of self-fertilization, as they carry both core MAT genes (MAT1-1-1 and MAT1-2-1) in one genome (12–15), usually next to each other, although the model fungus Aspergillus nidulans serves as an exception since its core MAT genes are located on separate chromosomes (16).
Botrytis cinerea is a heterothallic ascomycete fungus in the class Leotiomycetes, order Helotiales, family Sclerotiniaceae. A gapless community-annotated genome sequence is available (17). As a member of the Helotiales, B. cinerea develops a fruiting body called an apothecium (18, 19), consisting of an exposed hymenium on top of a stipe. Apothecia are carpogenic from a sclerotium, a melanized survival structure that contains a reservoir of nutrients to support apothecium development (20) and usually resides in plant residues in the topsoil layer. By their emergence from sclerotia and their phototropic growth (21), apothecia of Sclerotiniaceae function as an elevated launch platform that facilitates the discharge of ascospores (22), which act as primary inoculum for plant infection, especially in early spring when the asexual conidia are scarce.
The B. cinerea MAT1-1 locus contains two genes: MAT1-1-1, encoding the α domain protein, and MAT1-1-5, of unknown function. The MAT1-2 locus also contains two genes: MAT-1-2-1, encoding the HMG box protein, and a second gene of unknown function. This B. cinerea gene was initially named MAT1-2-4 (15) and has an ortholog in Sclerotinia sclerotiorum; however, an ongoing revision of ascomycete MAT genes resulted in their being renamed MAT1-2-10 (9). For this reason, the new name will be adopted for these B. cinerea and S. sclerotiorum genes from here onward. Both MAT loci in B. cinerea are flanked by the APN2 gene (ortholog of the Saccharomyces cerevisae gene encoding a DNA lyase) and the SLA2 gene (ortholog of the S. cerevisiae gene encoding a cytoskeletal protein), which are convergently transcribed toward the MAT loci (15, 23). Except for several Dothideomycetes, most filamentous ascomycetes with heterothallic reproductive lifestyles possess MAT loci that are also surrounded by APN2 and SLA2 genes, suggesting that this represents the ancestral configuration in ascomycete fungi (7, 9). S. sclerotiorum, a close relative of B. cinerea, has a homothallic reproductive mode and carries four linked MAT genes (MAT1-1-1, MAT1-1-5, MAT1-2-1, and MAT1-2-10), which are equivalent to the heterothallic MAT1-1 and MAT1-2 loci in B. cinerea (15). Similarly, the homothallic species Sordaria macrospora carries four linked MAT genes (MAT1-1-1, MAT1-1-2, MAT1-1-3, and MAT1-2-1), equivalent to the genes in both idiomorphs of its heterothallic relative Neurospora crassa (24, 25).
Several studies have focused on unraveling the functions of mating type genes in ascomycetes by targeted mutagenesis, primarily in model fungi (26–31). Doughan and Rollins (32) recently described a functional analysis of genes in the MAT locus of S. sclerotiorum and reported that mutants in the MAT1-1-1, MAT1-1-5, and MAT1-2-1 genes were entirely sterile, while mutants in the MAT1-2-10 gene were delayed in carpogenic germination and formed apothecia with aberrant morphology. So far, the functions of the four genes in the B. cinerea MAT locus in apothecium development have not been studied. The genome sequence and gene annotations of B. cinerea (15, 17) enabled the molecular dissection of apothecium development in B. cinerea, at both the transcriptional and the functional level.
This study aimed to identify the functions of B. cinerea MAT genes in apothecium development by targeted deletion and to perform a genome-wide transcriptome analysis during sexual reproduction using RNA sequencing (RNA-seq), which currently is the preferred method because of its sensitivity and quantitative accuracy (33–35) as well as its affordability. Expression profiling by RNA-seq was reported in numerous filamentous fungi (e.g., references 36 to 40); however, RNA-seq analyses in the context of sexual development are less abundant (41–45). Expression profiles observed during B. cinerea fruiting body development revealed concerted upregulation in the sexual ascospores of numerous genes, many of which are involved in interactions with plants, suggesting developmental priming for host invasion during sexual reproduction. Hints that epigenetic changes play a role in this regulation were obtained.
RESULTS AND DISCUSSION
Development of Botrytis cinerea apothecia.
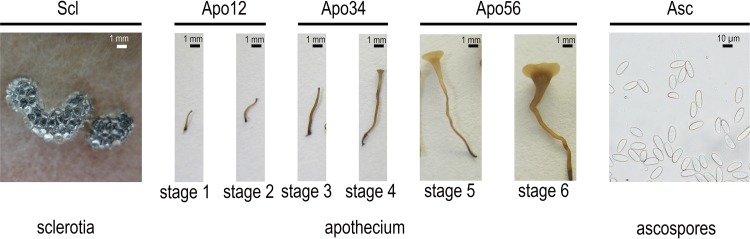
Fertilization of vernalized sclerotia of B. cinerea isolate SAS405 (MAT1-2) with microconidia of B. cinerea isolate SAS56 (MAT1-1), followed by incubation under appropriate conditions (18, 19), resulted in the formation of apothecia with asci and ascospores. Crosses were effective reciprocally, i.e., regardless of which isolate was used as the maternal parent (sclerotia) or the paternal parent (microconidia). Sexual structures began to emerge from sclerotia at 20 to 30 days postfertilization (dpf) and reached maturity at 30 to 60 days postemergence. Apothecial development was divided into six stages (Fig. 1): primordia emerging from sclerotium (stage 1, 20 to 30 dpf); primordia extending into stipes (stage 2, 24 to 35 dpf); fully extended stipes before onset of tip swelling (stage 3, 30 to 45 dpf); stipes with swollen tips before apothecial disk expansion (stage 4, 35 to 60 dpf); immature apothecial disk with a diameter of <3 mm and a pale color (stage 5, 40 to 70 dpf); and mature disk with a diameter of >5 mm and a light brown color and filled with asci containing eight ascospores (stage 6, 50 to 90 dpf).
FIG 1 .
Different stages of sexual reproduction in Botrytis cinerea. Following fertilization of asexual resting structures (sclerotia), apothecium development is divided into six stages: primordia emerging (stage 1), primordia extending (stage 2), extended stipes before tip swelling (stage 3), stipes with swollen tips (stage 4), immature apothecium (stage 5), and mature apothecium with asci and ascospores (stage 6). Pure ascospores sampled from mature apothecial disks are shown in the right-hand image. The five samples used for transcriptome analyses are indicated at the top.
Functional analysis of the MAT genes by targeted deletion.
In order to study the function of mating type genes MAT1-1-1, MAT1-1-5, MAT1-2-1, and MAT1-2-10 in apothecium development, deletion mutants were generated by replacing the coding region of each gene with a hygromycin resistance cassette in wild-type strains SAS56 and SAS405 (see Fig. S1 in the supplemental material). The MAT1-1-5 gene was also deleted in the genetic background of strain B05.10. Between three and six independent deletion mutants were obtained for each of the four MAT genes. All phenotypic analyses were carried out on three independent deletion mutants for each gene, and results were identical for independent mutants. To analyze whether the mutants had additional ectopic integrations, the copy number of the hygromycin resistance cassette was determined by quantitative PCR (qPCR) on genomic DNA and normalized to the single-copy housekeeping gene Bcrpl5. The deletion mutants had a single copy of the hph gene with the exception of one MAT1-2-10 mutant (out of three tested) which contained ~10 additional copies in an unknown location(s). Growth rates and morphology of asexual structures (mycelium, sclerotia, and conidia) of all ΔMAT1-1-1, ΔMAT1-1-5, ΔMAT1-2-1, and ΔMAT1-2-10 deletion mutants (including the ΔMAT1-2-10 mutant with additional copies of the hph gene) were indistinguishable from those of the corresponding wild type. Reciprocal crosses were set up using two wild-type strains, SAS56 and SAS405, and four single-gene mutant strains (ΔMAT1-1-1, ΔMAT1-1-5, ΔMAT1-2-1, or ΔMAT1-2-10) in all relevant combinations (Table 1). In the control cross of wild-type sclerotia (acting as maternal parent) fertilized with wild-type microconidia (as paternal parent), apothecia developed as described above. In contrast, apothecia never developed and there was no sign of outgrowth of primordia when crosses were performed between wild-type strain SAS405 and the ΔMAT1-1-1 mutant or between wild-type strain SAS56 and the ΔMAT1-2-1 mutant (Fig. 2). Failure to develop sexual structures was also observed in reciprocal crosses, indicating that the MAT1-1-1 and MAT1-2-1 genes are essential for the initiation of sexual development, in both maternal and paternal tissues. Loss of fertility was also reported for deletion mutants in the MAT1-1-1 or the MAT1-2-1 gene of S. sclerotiorum (a close relative of B. cinerea), though ascogonia were normally formed in these mutants (32). We did not investigate the presence of ascogonia in these B. cinerea mutants, since it is unknown when and where the fertilization occurs in the sclerotia during the 4-week incubation period following fertilization.
TABLE 1 .
Crosses performed between B. cinerea deletion mutants in MAT genes
| Type of cross | Sclerotia (maternal parent) |
Microconidia (paternal parent) |
Primordia + stipes |
Apothecial disks |
Asci and ascospores |
|---|---|---|---|---|---|
| WTd × WT | SAS56 | SAS405 | Yes | Yes | Yes |
| WT × WT | SAS405 | SAS56 | Yes | Yes | Yes |
| WT × mutant | SAS56 | ΔMAT1-2-1a | No | No | No |
| WT × mutant | SAS56 | ΔMAT1-2-10a | Yes | No | No |
| WT × mutant | SAS405 | ΔMAT1-1-1b | No | No | No |
| WT × mutant | SAS405 | ΔMAT1-1-5b | Yes | No | No |
| Mutant × WT | ΔMAT1-2-1a | SAS56 | No | No | No |
| Mutant × WT | ΔMAT1-2-10a | SAS56 | Yes | No | No |
| Mutant × WT | ΔMAT1-1-1b | SAS405 | No | No | No |
| Mutant × WT | ΔMAT1-1-5b | SAS405 | Yes | No | No |
| Mutant × mutant | ΔMAT1-2-1a | ΔMAT1-1-1b | No | No | No |
| Mutant × mutant | ΔMAT1-2-1a | ΔMAT1-1-5b | No | No | No |
| Mutant × mutant | ΔMAT1-2-10a | ΔMAT1-1-1b | No | No | No |
| Mutant × mutant | ΔMAT1-2-10a | ΔMAT1-1-5b | Yes | No | No |
| Mutant × mutant | ΔMAT1-1-1b | ΔMAT1-2-1a | No | No | No |
| Mutant × mutant | ΔMAT1-1-1b | ΔMAT1-2-10a | No | No | No |
| Mutant × mutant | ΔMAT1-1-5b | ΔMAT1-2-1a | No | No | No |
| Mutant × mutant | ΔMAT1-1-5b | ΔMAT1-2-10a | Yes | No | No |
| Control | ΔMAT1-2-1a | SMWc | No | No | No |
| Control | ΔMAT1-2-10a | SMW | No | No | No |
| Control | ΔMAT1-1-1b | SMW | No | No | No |
| Control | ΔMAT1-1-5b | SMW | No | No | No |
| Control | SAS56 | SMW | No | No | No |
| Control | SAS405 | SMW | No | No | No |
Mutant created in genetic background of SAS405.
Mutant created in genetic background of SAS56.
SMW, sterile Milli-Q water.
WT, wild type.
FIG 2 .
Results of crosses with B. cinerea mutants in MAT genes. The maternal parent (sclerotia) is mentioned first, and the paternal parent (microconidia) is second. The six images below the dashed line show the results for reciprocal crosses of the six images above. For each mutated target gene, results of one mutant are shown; identical results were obtained with two additional, independent deletion mutants.
Schematic representation of strategy for targeted deletion of MAT genes. Knockout of BcMAT1-1-1, BcMAT1-1-5, BcMAT1-2-1, and BcMAT1-2-10 by targeted gene replacement. Organization of BcMAT1-1-1, BcMAT1-1-5, BcMAT1-2-1, and BcMAT1-2-10 locus before and after homologous recombination. Orientations of the target gene and HPH are indicated by white and gray arrows, respectively. Upstream and downstream flanks of target genes are shown with open boxes. PCR analysis of wild-type strains SAS56 and SAS405 and knockout mutant strains. The genomic DNA of each strain was used to verify 5′ and 3′ homologous recombination and the absence of targeted genes in corresponding deletion mutants, respectively. Download FIG S1, PDF file, 0.3 MB (354.9KB, pdf) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
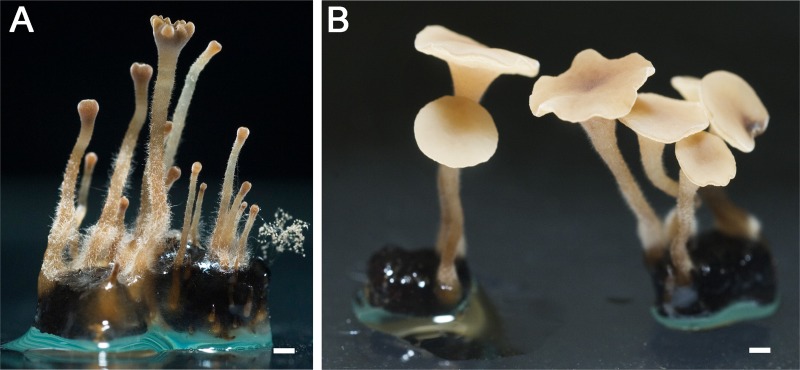
Sexual behavior of the ΔMAT1-1-5 and ΔMAT1-2-10 deletion mutants was distinct from that of ΔMAT1-1-1 and ΔMAT1-2-1 mutants (Fig. 2). Aberrant development of apothecia was observed in crosses between wild-type strain SAS405 and the ΔMAT1-1-5 mutant, as well as in crosses between wild-type strain SAS56 and the ΔMAT1-2-10 mutant. The emergence of primordia and development of stipes occurred similarly as in crosses between two wild-type strains; however, the stipes failed to develop into disks. Stipes did swell at the tip similarly to the wild type but failed to expand laterally. After several weeks of extended incubation, the stipes developed lobed, indented structures at the top (Fig. 3A), clearly distinct from wild-type apothecial disks (Fig. 3B). In these defective structures, obtained in crosses with ΔMAT1-1-5 and ΔMAT1-2-10 mutants, the development of croziers, asci, and ascospores was never observed. The failure of mutant stipes to make the developmental switch to apothecial disks was observed for three independent knockout mutants for either gene, and it occurred likewise in reciprocal crosses. Crosses between a ΔMAT1-1-5 mutant and a ΔMAT1-2-10 mutant yielded an identical defective phenotype as crosses between either single mutant and their corresponding wild-type mating partner (Fig. 2), suggesting that these two genes jointly control the transition from stipe to disk development.
FIG 3 .
Closeup of defective stipes formed in a cross between wild-type sclerotia of SAS405 and a ΔMAT1-1-5 mutant (A) and of a fully developed wild-type apothecium (B). The mutant stipe is blocked in transition to the apothecial disk and forms only a lobed, indented structure at the tip of the stipe. Crosses between wild-type sclerotia of SAS56 and MAT1-2-10 deletion mutants, as well as between a MAT1-1-5 deletion mutant and a MAT1-2-10 deletion mutant, result in very similar phenotypes. White bar, 1 mm.
The phenotype of these B. cinerea mutants differed from that of the corresponding mutants in S. sclerotiorum, in which the ΔMAT1-1-5 mutant was entirely sterile and the ΔMAT1-2-10 mutant showed severely delayed carpogenic germination and aberrant apothecium morphology (32). The transition from stipe to apothecial disk is typified by the formation of crozier cells in which karyogamy occurs (46, 47), and the resulting diploid nuclei subsequently enter meiosis to eventually form eight ascospores in an ascus. We propose that in B. cinerea, MAT1-1-5 and MAT1-2-10 act as (possibly dimeric) regulators that directly or indirectly control the formation of crozier cells and that the absence of either of these proteins results in failure to proceed to karyogamy, which blocks further downstream processes such as apothecial disk expansion. It remains to be demonstrated whether the MAT1-1-5 and MAT1-2-10 proteins indeed physically interact and how they impact on transcription and sexual development.
Attempts were made to complement the phenotype of MAT gene mutants. With the exception of the MAT1-1-5 gene, complementation constructs in which wild-type genes, including flanking regulatory sequences, were cloned into vector pNR4 containing a nourseothricin resistance cassette (48) appeared unstable in Escherichia coli. Complementation could thus not be performed for the ΔMAT1-1-1, ΔMAT1-2-1, and ΔMAT1-2-10 mutants. For the MAT1-1-5 gene, the complementation construct was transformed into a ΔMAT1-1-5 mutant. Three independent transformants with an ectopic insertion of the intact MAT1-1-5 gene (including flanking sequences) in the ΔMAT1-1-5 mutant background were tested in crosses, and all failed to show recovery of normal mating behavior. Ectopic integration of the intact MAT1-1-5 gene may have resulted in an inadequate level or temporal pattern of transcription, either because of the wrong chromatin context or because of the occurrence of meiotic silencing by unpaired DNA (MSUD) in an ectopic location, as was reported for Neurospora crassa crosses using a mutant strain that contains the matA gene in an ectopic location (49). The MAT1-1-5-complemented transformants were not analyzed further.
Sampling for analysis of gene expression during sexual development.
B. cinerea apothecia are an attractive resource for transcriptome analysis compared to other ascocarps such as cleistothecia, perithecia, and pseudothecia. Apothecia develop from sclerotia in an aqueous environment (18, 19), and the distinct developmental stages are in the millimeter-to-centimeter size range (Fig. 1). Hence, fairly pure tissue samples could easily be obtained. The fruiting body tissue samples were devoid of vegetative mycelium and sclerotial tissue. The use of laser microdissection to obtain ascocarp tissues, as performed in Sordaria macrospora (50), was thus not required for B. cinerea. Samples representing five different stages of sexual development were used for RNA extraction (Fig. 1): (i) 4-week-old sclerotia prior to vernalization, (ii) primordia of stages 1 and 2 that were dissected from sclerotia, (iii) stipes of stages 3 and 4, (iv) apothecial disks of stages 5 and 6, and (v) pure ascospores obtained from mature apothecia. The samples were designated Scl, Apo12, Apo34, Apo56, and Asc, respectively. In addition, we sampled for RNA extraction the elongated stipes with a swelling (stage Apo34) from a cross between wild-type strain SAS405 and the ΔMAT1-1-5 deletion mutant and from a cross between wild-type strain B05.10 and the ΔMAT1-2-10 deletion mutant, which are defective in apothecial disk development (Fig. 2). Two biological replicates of each RNA sample were used for cDNA synthesis, quantitative real-time PCR (qRT-PCR), and RNA sequencing.
Expression of mating type genes during development of apothecia.
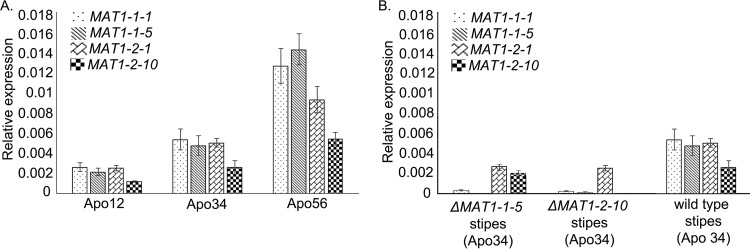
First, qRT-PCR analysis was performed to determine the transcript levels of the MAT1-1-1, MAT1-1-5, MAT1-2-1, and MAT1-2-10 genes over three stages of apothecium development (Apo12, Apo34, and Apo56) in a wild-type cross between SAS56 and SAS405. Transcript levels were normalized to the constitutively expressed β-tubulin gene BctubA. Figure 4A shows that the transcript levels of all four MAT genes were lowest in primordia (Apo12), slightly increased in stipes (Apo34), and peaked in apothecial disks (Apo56). Transcript levels of all MAT genes were also quantified in stipes defective in apothecial disk development, obtained from crosses between a wild-type strain and a ΔMAT1-1-5 or ΔMAT1-2-10 deletion mutant (Fig. 4B). In ΔMAT1-1-5 stipes, the transcript of the MAT1-1-5 gene was undetectable, as expected, but also transcript levels of the MAT1-1-1 and MAT1-2-1 genes were lower than in stipes from the cross between two wild-type isolates. In ΔMAT1-2-10 stipes, the transcript of the MAT1-2-10 gene was undetectable, but also the transcript levels of the other three MAT genes were lower than in stipes from the cross between two wild-type isolates. As discussed above, it is assumed that the mutant stipes failed to undergo karyogamy and therefore remained blocked in the dikaryotic stage. The expression profiles of MAT genes in the mutant stipes indicate that the absence of one MAT gene in a nucleus derived from one of the mating partners affects transcript levels of MAT genes in the nucleus derived from the opposite partner. How transcriptional cross talk between opposite MAT alleles in separate nuclei is accomplished remains to be studied.
FIG 4 .
qRT-PCR analysis of B. cinerea MAT genes, relative to the internal standard BctubA. (A) Expression in three stages of apothecium development in a wild-type cross: primordia (left), stipes (middle), and disks (right). (B) Expression levels in mutant stipes (ΔMAT1-1-5, left; ΔMAT1-2-10, middle) compared to wild-type stipes in an equivalent stage (Apo34, right). Expression data from Apo34 wild-type stipes are the same between panels A and B.
Whole-genome transcriptome analysis during development of apothecia.
RNA sequencing was performed on the five samples, each in two biological replicates. The library size and mapping efficiency of reads on the B. cinerea genome are listed in Table S1. Over 94% of the 11,700 gene models in the B. cinerea genome (17) were represented by at least one read in each individual sample; in every sample, at least 79% of all genes had counts per million (CPM) of ≥1. The transcriptome of sample Apo56 was the most diverse, with >90% of the gene models displaying CPM of ≥1. This sample also contained the greatest diversity of cell types, as material taken from apothecial disks was composed of cell layers of excipulum and hymenium, of dikaryotic cells, and of diploid cells undergoing meiosis as well as asci and ascospores in different stages of development and maturity. Sclerotia and ascospore samples, on the other hand, represent more homogeneous tissue types; it thus is logical that their expression profile was less diverse.
Mapping efficiency of RNA-seq reads on the Botrytis cinerea (strain B05.10) genome. Five samples represent sclerotia (Scl), apothecial primordia (Apo12), apothecial stipes (Apo34), apothecial disks (Apo56), and ascospores (Asc); biological replicates are labeled A and B. The number and proportion of genes with CPM values of ≥1 are given. Download TABLE S1, XLSX file, 0.01 MB (10.5KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of B. cinerea orthologs to genes involved in sexual development of model fungi.
Transcript levels of B. cinerea homologs to genes in Aspergillus nidulans and Sordaria macrospora that were previously listed as activators or repressors of sexual development in these fungi were examined (51, 52). Blast searches identified 88 B. cinerea homologs for 70 of these A. nidulans and S. macrospora genes (Table S2). Expression of these B. cinerea genes was detectable in all samples tested, with CPM values ranging from 0.1 to 6,282. Table S2 presents the expression profiles of the B. cinerea genes, grouped on the basis of their proposed role (in A. nidulans) in perception of environmental signals, mating processes and signal transduction, transcription factors, endogenous physiological processes, and ascospore production and maturation, according to the work of Dyer and O’Gorman (51). Expression profiles of a subset of these genes are discussed below in more detail. The Bcwcl2 gene and the Bcvel1 gene are putative positive regulators of sexual reproduction, and their transcript levels increased 2- to 3-fold in B. cinerea primordia and mature stipes, compared to sclerotia and ascospores, while transcripts of Bcwcl1 and Bcvel2 genes were less abundant and barely changed during sexual development. Genes encoding components of the B. cinerea heterotrimeric G protein complex (Gα1, Gβ, and Gγ but not Gα2 and Gα3) showed slight increases in transcript levels in apothecial tissues compared to sclerotia and ascospores. Furthermore, the G protein signaling regulator and a phosducin-like chaperone showed ~8-fold increases in transcript levels at the transition from sclerotia to apothecial primordia. The transcript levels of transcription factor genes BcnsdD, BcfhpA, and BcnrdA/msnA (positive regulators of sexual reproduction in A. nidulans) increased strongly in all stages of apothecial development and decreased in ascospores, while expression patterns of BcflbC and BcstuA (negative regulators of sexual reproduction in A. nidulans) showed an almost inverse pattern. The expression profiles of B. cinerea genes orthologous to S. macrospora genes required for sexual development (52) were also examined (Table S2). The transcript level of the gene encoding ATP citrate lyase subunit ACL1 increased 3-fold in the transition from sclerotia to primordia, remained constant in apothecia, and further increased in ascospores. The ortholog of S. macrospora pro4/leu1 (β-isopropylmalate dehydrogenase) was expressed abundantly in primordia and stipes but at low levels in sclerotia, apothecial disks, and ascospores; BcnoxD, the ortholog of S. macrospora pro41, was highly expressed during all stages of apothecial development but at low levels in sclerotia and ascospores.
Transcript levels of Botrytis cinerea orthologs to genes in Aspergillus nidulans and Sordaria macrospora previously listed as activators or repressors of sexual development (51, 52). The A. nidulans and S. macrospora gene symbols, locus identifier, and their proposed functions and domains are given in columns A to C; whether those genes are considered activators or repressors of sexual development is indicated in column D, with the relevant reference provided in column E. Gene identifiers (IDs) for the B. cinerea orthologs are given in column F, with their expression values in the five tissue samples (in counts per million mapped reads [CPM]) presented in columns G to K as averages from two biological replicates. Download TABLE S2, XLSX file, 0.03 MB (28.5KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In summary, a number of B. cinerea genes which, based on functions described in A. nidulans, likely act as positive regulators of sexual development had high transcript levels in early stages of apothecium development, which dropped in mature apothecial disks and ascospores. Conversely, several B. cinerea genes that, based on functions described in A. nidulans, likely act as negative regulators of sexual development had high transcript levels in sclerotia and primordia, which dropped in later stages of apothecium development and ascospores. Although such patterns would be in agreement with proposed positive or negative regulatory roles of these genes in sexual development, validation of their role in sexual reproduction would require functional analysis. Such expression patterns were, however, not observed for all B. cinerea genes that (based on functions in A. nidulans) might act as positive or negative regulators in sexual development. In several cases, the expression level was constant or showed a distinct pattern. It should, in this context, be considered that the role of a particular gene as a regulator of sexual development does not necessarily imply that such a gene is differentially transcribed during sexual development.
Genes differentially expressed between consecutive stages of sexual development.
Changes in gene expression during developmental transitions were analyzed by comparing expression levels between subsequent stages of apothecial development (Scl and Apo12, Apo12 and Apo34, Apo34 and Apo56, and Apo56 and Asc). Differentially expressed genes [adjusted P value of <0.05 and log2(fold change [FC]) of >2 or <−2) that were identified are recorded in Table 2 and listed in Tables S3 to S6. Pronounced changes in transcript levels were detected in the transition from sclerotia to primordia (>2,500 differentially expressed genes) and even more so between apothecial disks and ascospores, in which >3,900 genes were differentially expressed (i.e., one-third of the 11,700 genes in the B. cinerea genome). In contrast, only 75 genes were differentially expressed between primordia and stipes, whereas between stipes and apothecial disks, the number of differentially expressed genes was 594.
TABLE 2 .
Differentially expressed genes during developmental transitions
| Comparison | No. of genes |
|
|---|---|---|
| Upregulated [log2(FC) > 2] | Downregulated [log2(FC) <− 2] | |
| Scl vs Apo12 | 1,570 | 1,011 |
| Apo12 vs Apo34 | 37 | 38 |
| Apo34 vs Apo56 | 318 | 276 |
| Apo56 vs Asc | 1,424 | 2,485 |
Differentially expressed genes in the transition from sclerotia to apothecial primordia. Panels S3a and S3b list the upregulated and downregulated genes, respectively; their expression values in both biological replicates of both samples (in counts per million mapped reads [CPM]); the log2 of the fold change (logFC); average log2 of the CPM values between the four RNA samples (logCPM); test statistic of the likelihood-ratio test (LR); the uncorrected and Benjamini-Hochberg-corrected P value of the likelihood test; the B. cinerea gene name; and a description resulting from manual annotation. Panels S3c and S3d present GO enrichment analyses for the upregulated and downregulated genes, respectively. Download TABLE S3, XLSX file, 0.5 MB (487.2KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
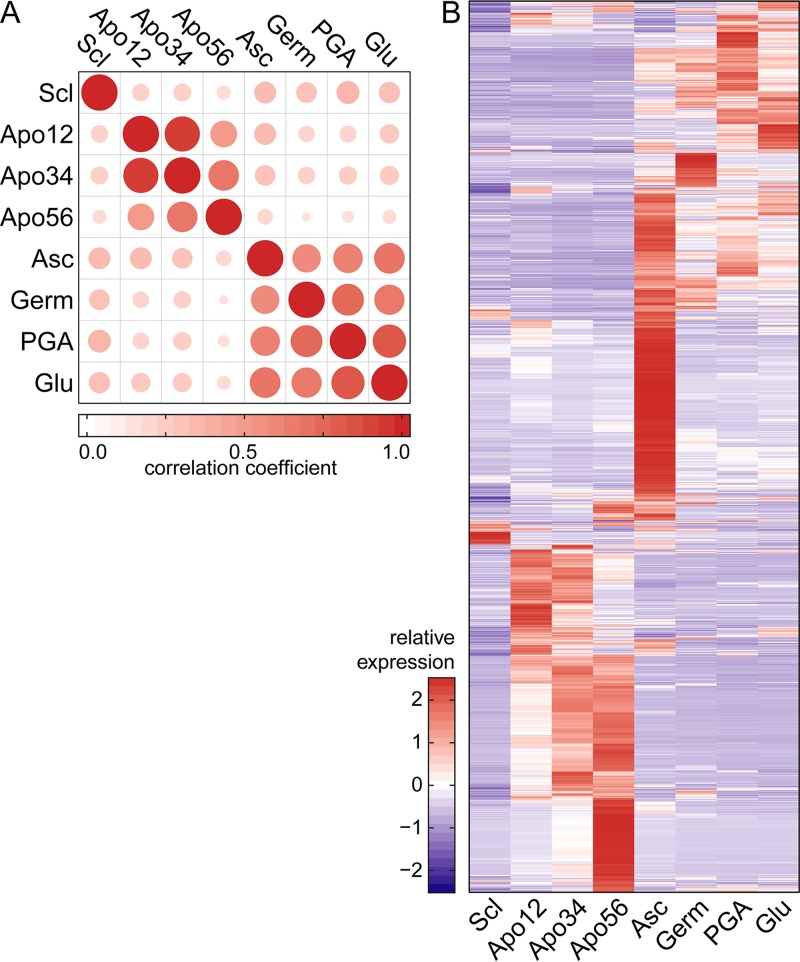
In order to examine whether the expression patterns during sexual development were distinct from asexual development, we determined pairwise correlations between CPM values in the five stages of sexual development reported here and three published data sets of asexual tissues, representing RNA from young germlings (conidia inoculated in liquid medium and grown for 12 h) (53) or from mycelium grown in polygalacturonate or glucose (54) (Fig. 5A). The correlations between samples were generally low (<0.5) with some exceptions. The transcriptomes in stages Apo12 and Apo34 were very similar to one another, and these tissues are morphologically alike. All other pairwise correlations between sexual and asexual stages were <0.75, with sclerotia displaying the most dissimilar expression profile.
FIG 5 .
Comparison of transcriptomes of five stages of sexual development with three asexual tissues (Germ, germinating conidia [53]; PGA, mycelium on polygalacturonate-containing medium [54]; Glu, mycelium on glucose-containing medium [54]). For each gene in these comparisons, the mean CPM was calculated over available replicates. (A) Pairwise correlations between CPM values of B. cinerea total transcriptome samples. Dot size and color represent the Pearson correlation coefficient. (B) Relative expression (Z-scores) of subset of 3,084 genes that were differentially expressed in at least one of the sexual development stages (Scl, Apo12, Apo34, Apo56, and Asc) compared to asexual tissues.
The expression levels of all 3,084 genes that are differentially expressed during at least one of the five stages of sexual development were plotted in a heat map and compared to levels in the asexual tissues. The heat map (Fig. 5B) shows that the expression patterns of this gene set differed markedly between each of the sexual stages and the three nonsexual stages. Features of the sets of differentially expressed genes are discussed below. Gene ontology (GO) enrichment analysis was carried out on the four sets of differentially expressed genes (adjusted P value of <0.05, any fold change).
Differentially expressed genes in the transition from sclerotia to primordia.
The transition from sclerotia to primordia was associated with significantly increased transcript levels of 1,570 genes and reduced transcript levels of 1,011 genes (Table S3). A major change in secondary metabolism was observed. Among the upregulated genes (Table S3a) were 10 distinct secondary metabolite (SM) gene clusters, involved in production of polyketides (6 clusters), sesquiterpenes (2 clusters), and nonribosomal peptides (2 clusters), for all of which the SMs that they produce are unknown. Concomitantly, there was downregulation (Table S3b) of 5 SM clusters involved in the production of polyketides (2 clusters) and nonribosomal peptides (3 clusters). Among the upregulated genes (Table S3a) were also several regulatory genes, such as the MAT1-1-1 and MAT1-1-5 genes, two homeobox genes (Bchox4 and Bchox8 [55]), the Velvet complex subunit Bcvel4, the Vivid-like putative light sensor, the Medusa-like transcriptional regulator, and the light-responsive GATA-type transcription factor Bcltf1 (56); several genes involved in histone modification and DNA methylation (see below); hydrophobin genes Bhp1, Bhp2, and Bhl1 (57); five laccase genes; and 19 transporter genes of various classes. Among the downregulated genes (Table S3b) were, besides genes in five SM clusters mentioned above, regulatory genes such as HMG-box transcription factor-encoding gene Bcste11 (58) and the mitogen-activated protein (MAP) kinase gene Bmp3 (59), several genes involved in histone modification (see below), four glutathione S-transferases, the hydrophobin gene Bhp3, the melanin biosynthetic tetrahydroxynaphthalene reductase gene Bcbrn1 (60, 61), the sclerotium-specific lectin gene Bcssp1, and 28 transporters of various classes.
GO term enrichment analysis on the upregulated genes showed overrepresentation of (among others) the terms ribosome biogenesis, DNA replication, microtubule-based movement, fatty acid biosynthetic process, lipid biosynthetic process, oxidation-reduction process, tricarboxylic acid cycle, and aerobic respiration (Table S3c). GO term enrichment analysis on the downregulated genes showed an overrepresentation of (among others) the terms oxidation-reduction process, hexose metabolic process, and transmembrane transport (Table S3d).
Differentially expressed genes in the transition from primordia to stipes.
The transition from primordia (Apo12) to stipes (Apo34) was associated with significantly different transcript levels of 75 genes (Tables 2 and S4a and b). Among the 38 upregulated genes were 13 genes (34%) of unknown function that are specific to B. cinerea or the family Sclerotiniaceae. Among the 37 downregulated genes were a gene (Bcin04g02570) encoding a small secreted cysteine-rich protein (86-amino-acid mature protein with 13 Cys residues of which four occur in two Cys–Cys pairs) that does resemble, but not precisely match, the pattern for hydrophobin-like proteins; two genes encoding glycosyltransferases (CAZY GT2 and GT4 family proteins); and two genes in an SM gene cluster comprising polyketide synthase gene Bcpks15, of which the product is unknown. GO term enrichment analysis detected overrepresentation of the term response to stress in the set of upregulated genes (Table S4c). GO term enrichment analysis on the set of downregulated genes (Table S4d) showed an overrepresentation of (among others) the terms ribosome biogenesis, rRNA processing, and oxidation-reduction process.
Differentially expressed genes in the transition from apothecial primordia to stipes. Panels S4a and S4b list the upregulated and downregulated genes, respectively; their expression values in both biological replicates of both samples (in counts per million mapped reads [CPM]); the log2 of the fold change (logFC); average log2 of the CPM values between the four RNA samples (logCPM); test statistic of the likelihood-ratio test (LR); the uncorrected and Benjamini-Hochberg-corrected P value of the likelihood test; the B. cinerea gene name; and a description resulting from manual annotation. Panels S4c and S4d present GO enrichment analyses for the upregulated and downregulated genes, respectively. Download TABLE S4, XLSX file, 0.4 MB (384.4KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed genes in the transition from stipes to apothecial disks.
The transition from stipes (Apo34) to apothecial disks (Apo56) was associated with significantly increased transcript levels of 318 genes and significantly reduced transcript levels of 276 genes (Tables 2 and S5). Among the upregulated genes (Table S5a) were the gene encoding the activator of meiotic anaphase-promoting complex Bcama1; the hydrophobin gene Bhp3 (57); several transporter-encoding genes, including the aquaporin genes Bcaqp2 and Bcaqp5 (62), the ABC transporter gene Bmr3, and hexose transporter genes Bchxt2, Bchxt5, and Bchxt9 (63); and a bZIP transcription factor gene. Among the downregulated genes (Table S5b) were the endopolygalacturonase gene Bcpg6, two nonribosomal peptide synthase genes (Bcnrps5 and Bcnrps8), the oxaloacetate hydrolase gene Bcoah1 (64), the gene encoding the transcriptional regulator of sexual development nosA (ortholog of S. macrospora pro1 [52]), the aspartyl proteinase gene Bcap9 (65), the laccase gene Bclcc10, the DNA repair protein RAD1-encoding gene, the ceratoplatanin gene Bcspl1 (66), and the sclerotium-specific lectin gene Bcssp1.
Differentially expressed genes in the transition from apothecial stipes to disks. Panels S5a and S5b list the upregulated and downregulated genes, respectively; their expression values in both biological replicates of both samples (in counts per million mapped reads [CPM]); the log2 of the fold change (logFC); average log2 of the CPM values between the four RNA samples (logCPM); test statistic of the likelihood-ratio test (LR); the uncorrected and Benjamini-Hochberg-corrected P value of the likelihood test; the B. cinerea gene name; and a description resulting from manual annotation. Panels S5c and S5d present GO enrichment analyses for the upregulated and downregulated genes, respectively. Download TABLE S5, XLSX file, 0.1 MB (127.8KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GO term enrichment analysis on the set of upregulated genes showed an overrepresentation of (among others) the terms cellular chemical homeostasis, amino acid transmembrane transport, heme metabolic process, oxidation-reduction process, cellular biogenic amine metabolic process, dephosphorylation, carbohydrate metabolic process, autophagy, organic anion transport, phosphatidylinositol metabolic process, and aspartate family amino acid biosynthetic process (Table S5c). GO term enrichment analysis on the set of downregulated genes showed an overrepresentation of (among others) the terms oxidation-reduction process, carbohydrate metabolic process, potassium ion transport, sterol metabolic process, and response to oxidative stress (Table S5d).
Gene expression in mutant stipes blocked in transition to the apothecial disk.
The transcriptomes of mutant stipes, obtained in crosses with ΔMAT1-1-5 and ΔMAT1-2-10 deletion mutants, were determined by RNA-seq and compared to one another and to the transcriptome of wild-type stipes at the equivalent stage (Apo34). A total of 1,310 genes were differentially expressed between mutant stipes from the SAS405 × ΔMAT1-1-5 mutant cross and stipes from a wild-type cross (Tables 3 and S6). Among this total set, 854 genes were downregulated and 456 genes were upregulated in the mutant stipes (blocked in transition to the apothecial disk), compared to stipes from the cross between two wild-type strains (Table S6a and b). In the cross between wild-type strain B05.10 and the ΔMAT1-2-10 mutant, 985 genes were downregulated and 152 genes were upregulated in mutant stipes, compared to stipes in a cross between wild-type strains (Table S6c and d).
TABLE 3 .
Differentially expressed genes in stipes of MAT gene mutants compared to wild-type stipes
| Comparison | No. of genes |
|
|---|---|---|
| Downregulated [log2(FC) > 2] | Upregulated [log2(FC) <− 2] | |
| ΔMAT1-1-5 vs wild type | 854 | 456 |
| ΔMAT1-2-10 vs wild type | 985 | 152 |
Differentially expressed genes in stipes obtained with MAT gene mutants. Panels S6a and S6b list the downregulated and upregulated genes, respectively, in stipes obtained in an SAS405 × ΔMAT1-1-5 cross, compared to stipes from a wild-type cross, SAS405 × SAS56. Panels S6c and S6d list the downregulated and upregulated genes, respectively, in stipes obtained in an SAS56 × ΔMAT1-2-10 cross, compared to stipes from a wild-type cross, SAS405 × SAS56. Panel S6e lists the overlap between downregulated genes in panels S6a and S6c, while panel S6f lists the overlap between upregulated genes in panels S6b and S6d. Download TABLE S6, XLSX file, 0.3 MB (345.4KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
There was some overlap between the differentially expressed genes in both mutant stipes: compared to wild-type stipes, 87 genes were downregulated in both the SAS405 × ΔMAT1-1-5 cross and the B05.10 × ΔMAT1-2-10 cross, while 26 genes were upregulated in both mutant stipes (Table S6e and f). This list of 87 genes was further compared with the genes that are, in crosses between two wild-type isolates, upregulated in the transition from stipes (stage Apo34) to mature apothecial disks (stage Apo56). Among the genes that were not upregulated in both mutant stipes while they were upregulated in wild-type apothecia were one Zn2Cys6 transcription factor-encoding gene, four major facilitator superfamily (MFS) transporter genes, the MAT1-1-1 and MAT1-2-1 genes, a HIT/MYND domain containing gene, and a G-protein-coupled receptor (BcGPCR2), as well as 24 genes of unknown function that are specific either to Botrytis or to the family Sclerotiniaceae.
Differentially expressed genes in the transition from apothecial disks to ascospores.
Between the apothecial disks (Apo56) and the ascospore sample, as many as 3,909 genes were differentially expressed, of which 37% were upregulated and 63% were downregulated (Tables 2 and S7a and b). Among the upregulated genes were 57 genes encoding ribosomal proteins and 24 other genes involved in ribosome biogenesis, as well as 10 translation initiation and elongation factor-encoding genes and 9 mitochondrial protein-encoding genes. The gene set also contains the Bcnop1 gene, encoding a histone glutamine methyltransferase; the heterotrimeric G protein β subunit gene Bcgbl1; and five calcineurin-dependent genes (67) as well as the calcineurin regulator calcipressin gene (68). GO term enrichment analysis on the set of upregulated genes showed an overrepresentation of >200 terms (Table S7c), while GO term enrichment analysis on the downregulated genes showed an overrepresentation of 42 terms (Table S7d). A substantial number of genes upregulated in ascospores were of special interest, despite not being reflected in the GO enrichment analysis by lack of annotation in the “biological process” domain. These genes are discussed in more detail below.
Differentially expressed genes in the transition from apothecial disks to ascospores. Panels S7a and S7b list the upregulated and downregulated genes, respectively; their expression values in both biological replicates of both samples (in counts per million mapped reads [CPM]); the log2 of the fold change (logFC); average log2 of the CPM values between the four RNA samples (logCPM); test statistic of the likelihood-ratio test (LR); the uncorrected and Benjamini-Hochberg-corrected P value of the likelihood test; the B. cinerea gene name; and a description resulting from manual annotation. Panels S7c and S7d present GO enrichment analyses for the upregulated and downregulated genes, respectively. Download TABLE S7, XLSX file, 1 MB (1.1MB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ascospores are transcriptionally primed for host plant invasion.
Inspection of the list of genes upregulated in ascospores revealed numerous genes that encode enzymes potentially contributing to plant invasion, such as components of the fungal pectinolytic machinery: endopolygalacturonase genes Bcpg2 and Bcpg6 (69); the pectin methylesterase gene Bcpme2 (70); the galacturonate reductase gene Bcgar2 (48); aspartyl proteinase genes Bcap3, Bcap4, and Bcap14 (65); laccase genes Bclcc2 and Bclcc8, the former encoding an enzyme that can oxidize the grapevine phytoalexin resveratrol (71); five ABC multidrug efflux transporter-encoding genes, BcatrA (72), Bmr1 (73), BcatrO (74), BcatrD (75), and BcatrB, involved in the tolerance of B. cinerea to fungicides and plant defense compounds (72, 76), as well as the Zn2Cys6 transcription factor Bcmrr1, which regulates BcatrB expression (77); and genes encoding proteins involved in signal transduction pathways—the heterotrimeric G protein subunit Gβ1, a phosphoinositide biosynthetic gene, the regulator of calcineurin (calcipressin) (68), the two-component histidine kinase Bos1 (78, 79), and the G-protein-coupled receptor Bcgpr1. The list also comprises five hexose transporter-encoding genes (63), the oxalic acid biosynthetic gene Bcoah1 (64), the superoxide dismutase gene Bcsod3, and six CND (calcineurin-dependent) genes (67). Finally, several genes encoding chromatin-modifying enzymes were also upregulated, such as the histone glutamine methyltransferase gene Bcnop1, the histone deacetylase gene Bchst2, and the histone chaperone gene Bcnap1 (see below).
Among the genes that were downregulated between mature apothecial disks and ascospore samples were several genes encoding histone-modifying enzymes and C-5 cytosine-specific DNA methylases (see below), as well as the Dicer-like gene Bcdcl1 (80); as many as 15 SM clusters (six nonribosomal peptide synthase [NRPS] clusters, six polyketide synthase [PKS] clusters, one sesquiterpene synthase [STC] cluster, the PHS1 carotenoid cluster, and the YGH1 cluster involved in melanin biosynthesis); the homeobox gene Bchox6 (53); regulators of sexual development such as the MAT1-1-1 gene, the NADPH oxidase gene BcnoxD (ortholog of S. macrospora pro41 [81]), the transcriptional regulator nosA (ortholog of S. macrospora pro1), and the Bcspo11 gene, encoding a protein that initiates meiotic recombination; the opsin gene Bop1; six laccase genes; and three aquaporin genes, Bcaqp5, Bcaqp8, and Bcaqp9 (62).
Spatially clustered coregulation of transcription during sexual development.
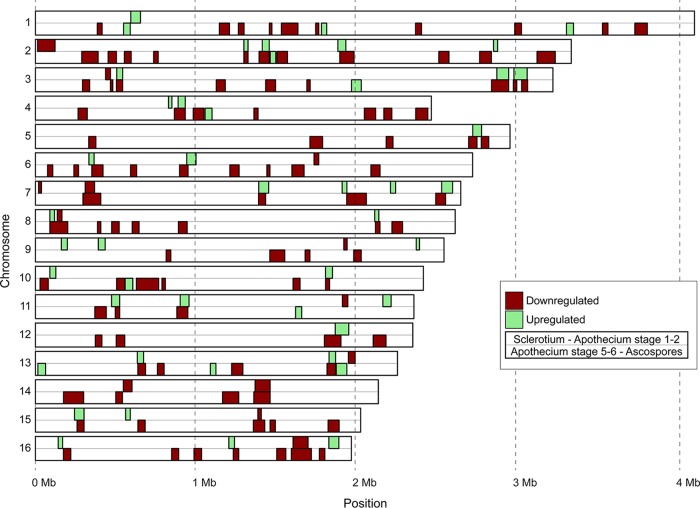
Using sliding windows of ≥12 genes, more than 150 cases were observed in which at least 50% of genes were significantly coregulated (either jointly up- or jointly downregulated) during the same developmental transition (Table 4). At the onset of sexual development, during the transition from sclerotia to primordia, 35 coregulated clusters were identified with an average length of 44.7 kb and containing on average 8 upregulated genes. Even more notably, there were 99 clusters of downregulated genes at the completion of sexual reproduction (the transition from apothecial disks to ascospores), with an average length of 56.5 kb and containing on average 9.3 downregulated genes (Table 4; Fig. 6). The largest of these clusters was 135 kb in length and contained as many as 24 genes. Altogether, these 99 clusters contained 923 genes (Table S8), representing 37% of the 2,486 genes that were downregulated during the transition from apothecial disks to ascospores. Of the 35 clusters that were upregulated in the transition from sclerotia to primordia, 23 (66%) overlapped at least partially with the 99 clusters that were downregulated in ascospores, which was especially notable in chromosomes 2, 3, and 13 (Fig. 6).
TABLE 4 .
Clustering of coregulated genes during transitions in fruiting body developmenta
| Comparison | No. of genes |
|
|---|---|---|
| Upregulated | Downregulated | |
| Scl vs Apo12 | 35 | 13 |
| Apo12 vs Apo34 | 0 | 0 |
| Apo34 vs Apo56 | 0 | 2 |
| Apo56 vs Asc | 11 | 99 |
Clusters are defined as blocks of at least 12 consecutive genes, of which at least 50% show simultaneous up- or downregulation.
FIG 6 .
CROC analysis for identification of clusters of coregulated genes. Core chromosomes of B. cinerea strain B05.10 are displayed vertically with Chr1 at the top and Chr16 at the bottom. Size markers are provided at the bottom. Clusters of upregulated genes are displayed in green, while clusters of downregulated genes are displayed in red. For each chromosome, two rows are provided: the top row displays clusters of genes coregulated during the transition from sclerotia to primordia, while the bottom row displays clusters of genes coregulated during the transition from apothecial disks to ascospores. Minichromosomes 17 and 18 (17) did not contain coregulated clusters and were omitted for simplicity.
CROC analysis for identification of clusters of coregulated genes. Clusters were defined as sliding windows of ≥12 genes, of which at least 50% are jointly upregulated or downregulated. Each table provides information about the chromosome number, the start and end coordinates of the cluster, the adjusted P value for clustering (threshold was set at a P value of <0.05), and the gene IDs of the coexpressed genes in the cluster. CROC output is provided for upregulated genes in the transition from sclerotia to primordia (panel S8a), downregulated genes in the transition from sclerotia to primordia (panel S8b), downregulated genes in the transition from primordia to stipes (panel S8c), upregulated genes in the transition from apothecial disks to ascsopores (panel S8d), and downregulated genes in the transition from apothecial disks to ascsopores (panel S8e). Download TABLE S8, XLSX file, 0.03 MB (36.8KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
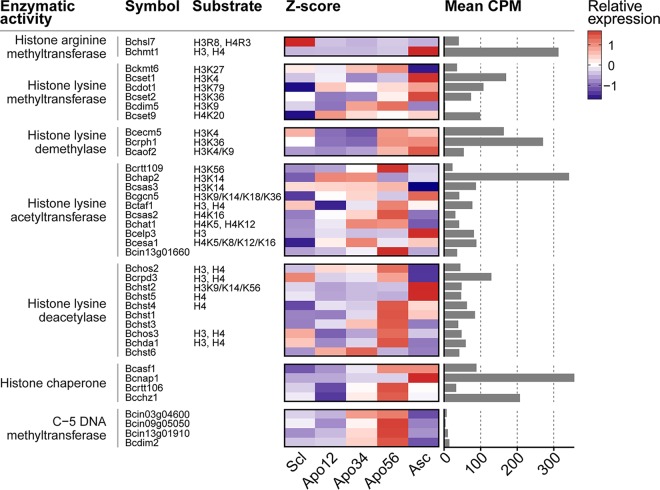
The spatial pattern in transcriptional changes, especially during the onset and the end of sexual development, suggested the occurrence of changes in chromatin organization during the developmental transitions in apothecium development. The importance of histone acetylation and chromatin remodeling in meiotic recombination was reported in the fission yeast, Schizosaccharomyces pombe (82). Furthermore, the histone chaperone ASF1 in Sordaria macrospora was shown to be essential for the formation of mature perithecia (83). We examined the transcriptional profiles of 39 genes encoding histone-modifying enzyzmes or DNA methylases in the five stages of sexual development (Fig. 7). Half of these genes showed the highest expression level in the mature apothecia, in which meiosis was taking place. Notably, all four genes encoding C-5 cytosine-specific DNA methyltransferase and six of the 10 histone lysine deacetylase genes showed a clear peak of expression in this sample. In contrast, genes encoding two histone lysine methyltransferases, a histone lysine acetyltransferase, and two other histone lysine deacetylases, as well as a histone chaperone, displayed a strong peak of expression in the ascospores. Finally, one histone arginine methyltransferase gene was predominantly expressed in sclerotia. Chromatin immunoprecipitation sequencing (ChIP-seq) and bisulfite sequencing experiments in the different stages of B. cinerea apothecium development will be required to explore the changes in chromatin architecture during these developmental transitions. The roles of histone-modifying enzymes and DNA methylases in sexual reproduction can be validated by targeted mutagenesis (if such mutants are viable).
FIG 7 .
Transcript abundance of enzyme-encoding genes involved in histone modification and DNA methylation, at five stages in apothecium development. Columns from left to right indicate the respective annotated enzymatic activity, assigned gene symbol, the modified residue on the histone, the Z-score-transformed expression profiles over the five determined life stages, and the mean CPM values over the five stages to relate the relative changes to absolute, respectively. Functional annotations are taken from reference 95.
Conclusion.
This study presents the first transcriptional analysis of sexual development in the Helotiales and a unique view on the transcriptome of pure ascospores of a filamentous ascomycete. The comparison of expression profiles of consecutive stages of sexual development revealed massive changes in transcript levels in the transition from sclerotia to primordia (the onset of sexual development) and even more so in the transition from mature disks to ascospores (completion of sexual reproduction). The number and nature of genes that were upregulated in the ascospores indicate that these spores are transcriptionally primed for the production of virulence factors prior to their first encounter with a host plant. B. cinerea ascospores thus do not land on plant tissues totally naive but possess a reservoir of virulence factors at their arrival, which facilitates a fast and effective invasion. Such an observation does not preclude that virulence genes might be further upregulated during germination of the ascospores on host tissue. It will be interesting to study whether the induction of virulence genes also occurs during development of asexual conidia, which are as infectious as ascospores and form important propagules for dispersal of B. cinerea to neighboring host plants. The present study also provided evidence for a clear spatial clustering in transcriptional changes during the onset and termination of sexual development, which merits further studies on epigenetic changes during B. cinerea apothecium development.
MATERIALS AND METHODS
Strains.
Wild-type B. cinerea strains SAS56 (reference MAT1-1 strain) and SAS405 (reference MAT1-2 strain) were used (18, 84), as well as strain B05.10 (MAT1-1), for which a gapless reference genome sequence was determined (17). Strains were plated on malt extract agar (MEA; Oxoid, Basingstoke, United Kingdom). For obtaining sclerotia, cultures were grown at 15°C in darkness. For obtaining conidia, cultures were grown at 20°C for 3 to 4 days and then exposed for 1 day to near-UV light and incubated for another week before conidiospores were collected.
Deletion of mating type genes.
The MAT1-1-1, MAT1-1-5, MAT1-2-1, and MAT1-2-10 genes were individually targeted for mutagenesis by homologous recombination (see Fig. S1). MAT1-1-1 and MAT1-1-5 genes were deleted in SAS56, whereas the MAT1-2-1 and MAT1-2-10 genes were deleted in SAS405. In addition, the MAT1-1-5 gene was deleted in B05.10. The strategies for generating deletion constructs, B. cinerea protoplast transformation, and PCR-based screening of transformants were as described previously (70). Table S9 contains primers that were used for amplification of gene deletion fragments. The hygromycin resistance (HPH) cassette, amplified from vector pLOB7 (48) with primer pair Cassette-5/Cassette-3, was used as a selection marker. Genomic DNA of transformants was analyzed for the presence/absence of the wild-type target gene by amplifying the target genes MAT1-1-1, MAT1-1-5, MAT1-2-1, and MAT1-2-10 using primer combinations MAT111-5.1/MAT111-3.1, MAT115-5.1/MAT115-3.1, MAT121-5.1/MAT121-3.1, and MAT124-5.1/MAT124-3.1, respectively.
Primers used in this study. Download TABLE S9, XLSX file, 0.01 MB (13.3KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DNA extraction.
Freeze-dried mycelium was used for DNA extraction, using a Gentra Puregene DNA purification kit (Qiagen, Venlo, The Netherlands), according to manufacturer’s instructions.
Sexual crosses.
Sexual crosses were performed using wild-type strains (SAS56, SAS405, and B05.10) and four mutants: the ΔMAT1-1-1, ΔMAT1-1-5, ΔMAT1-2-1, and ΔMAT1-2-10 strains. Crosses were set up using previously described protocols (18, 19). Sclerotia (incubated at 0°C for 1 month) were sampled from MEA plates, and their surfaces were cleaned with water using a soft toothbrush. Three to five sclerotia of ~1 cm in size were placed in single wells in a six-well microtiter plate. Suspensions of microconidia and hyphal fragments were prepared by pouring sterile water onto the plate from which sclerotia were removed, followed by gentle rubbing with a plastic spatula. Sclerotia in the microtiter plate were fertilized with this suspension (3 ml per well). The plate was sealed with Parafilm and incubated at 12°C in candescent light with a 12-h photoperiod. Apothecia developed over a period of 60 to 90 days. Ascospores were sampled from mature apothecia by gently crushing apothecial disks in water and filtering the suspension over glass wool to separate hymenium tissue and large debris from the ascospores (in the flowthrough). The suspension of ascospores was centrifuged at 250 × g for 10 min, and the pellet was resuspended in water for rinsing and centrifuged again.
RNA extraction and RNA sequencing.
Samples of three different developmental stages of apothecia, as well as of ascospores, were freeze-dried, and total RNA was isolated using the Nucleospin RNA plant kit (Macherey-Nagel, Düren, Germany), according to the manufacturer’s instructions. For isolation of RNA from sclerotia, freeze-dried sclerotia were frozen in liquid nitrogen and ground with a mortar and pestle. Total RNA was isolated with TRIzol reagent (Life Technologies, Grand Island, NY, USA) in combination with the Nucleospin RNA plant kit, according to manufacturer’s instructions. Integrity of RNA was monitored by gel electrophoresis, and the concentration determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). All samples were collected in two biological replicates. Twenty micrograms of RNA from each sample was used by the Beijing Genome Institute (Hong Kong, China) for library construction (300- to 700-nucleotide [nt] insert sizes) and sequenced using an Illumina HiSeq 2000 platform. Paired-end RNA-seq reads of 90 nt were assessed for quality using FastQC v0.10.1 (Babraham Institute; http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were trimmed to 80 nt using FASTX-Toolkit v0.13.2 (http://hannonlab.cshl.edu/fastx_toolkit).
cDNA synthesis and qRT-PCR.
First-strand cDNA was synthesized from 1 µg total RNA with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Leiden, The Netherlands) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed using an ABI7300 PCR machine (Applied Biosystems, Foster City, CA, USA) in combination with the qPCR SensiMix kit (Bioline, London, United Kingdom) using primers listed in Table S9. qRT-PCR conditions were as follows: an initial 95°C denaturation step for 10 min, followed by denaturation for 15 s at 95°C and annealing/extension for 1 min at 60°C, for 40 cycles. The data were analyzed on the 7300 System SDS software (Applied Biosystems, Foster City, CA, USA). The gene expression values were normalized to the B. cinerea tubulin gene, BctubA (85).
Read mapping and quantification of expression.
RNA-seq reads of the five stages of sexual development and three asexual tissues (53, 54) were trimmed and mapped onto the Botrytis cinerea B05.10 genome (17) using HISAT v2.0.3-beta (86). The mapping efficiency ranged from 90.5 to 95.9% (Table S1). Expression was quantified using Rsubread v1.16.1 (87), by considering reads that were fully aligned to exons, based on the manually curated genome annotations (http://fungi.ensembl.org/Botrytis_cinerea). Reads overlapping multiple exons were also counted multiple times. Consecutively, genes expressed at low levels (i.e., read counts of <10 in all samples) were filtered. The read counts were normalized by using the trimmed mean of M-values method from the EdgeR package v3.8.6 deriving counts per million (CPM) read values (88, 89). Subsequently, differential gene expression between pairs of consecutive RNA-seq samples was assessed by fitting generalized linear models (GLMs) to the expression data under a negative binomial distribution, weighting count values for replicate effects and sample dispersion. Resulting P values were corrected for multiple testing using the Benjamini-Hochberg method (90). The functional annotations of genes with a log2(FC) of >2 (or <−2) between two samples, at an adjusted P value of <0.05, were manually inspected. For visual inspection and comparison of expression profiles, the CPM expression values were scaled using a Z-score transformation: Z = (X − μ)/σ. Here, X is the CPM value of the respective gene at a particular stage, and μ and σ represent the mean and standard deviation for this gene over the five stages, respectively.
Enrichment tests.
To test for overrepresented gene ontology (GO) terms in each set of differentially expressed genes, GO enrichment analysis was performed. The total set of proteins from B. cinerea was annotated with GO terms using InterProScan v5.18.57 (91). Using the R package TopGO v2.18.0 (92), Fisher’s exact tests were performed on the GO terms associated with the significantly up- and downregulated genes (adjusted P value of <0.05) between every consecutive pair of conditions. The resulting lists of significantly overrepresented GO terms were trimmed using REVIGO (93).
Differentially expressed gene clusters.
To identify coexpressed gene clusters on the genome, a sliding window analysis was performed using CROC software (94). For the sliding window, a step size of one gene and a window size of 12 genes were applied, of which at least 6 genes must be either up- or downregulated for these to be considered for further analysis. Consecutively, on these windows a hypergeometric test was performed to test for a nonrandom distribution of the up- or downregulated genes, which were considered coexpressed gene clusters. The resulting P values were corrected using a Benjamini-Hochberg multiple testing correction method (88).
Data availability.
RNA-seq reads from this study are deposited in the NCBI SRA archive under PRJNA351919 (SRP093589). Accession numbers for data sets are as follows: sclerotia of strain SAS405, SRS1810933; primordia Apo12, SRS1810937; stipes Apo34, SRS1810938; apothecial disks Apo56, SRS1810941; ascospores, SRS1812737.
ACKNOWLEDGMENTS
We are grateful to José Abrantes Teixeira Duarte for his contribution to deletion of the MAT1-1-5 gene, Michael Seidl for advice in analyses and for feedback on the manuscript, Luigi Faino for stimulating discussion, and Harold Meijer for feedback on the manuscript.
R.B.T. was supported by the Malaysian Ministry of Education and the Universiti Putra Malaysia; research by J.H.M.S. was funded by Syngenta Crop Protection AG, Switzerland; research by J.V. was funded by the Xunta de Galicia, Spain.
Footnotes
Citation Rodenburg SYA, Terhem RB, Veloso J, Stassen JHM, van Kan JAL. 2018. Functional analysis of mating type genes and transcriptome analysis during fruiting body development of Botrytis cinerea. mBio 9:e01939-17. https://doi.org/10.1128/mBio.01939-17.
REFERENCES
- 1.Clarkson JP, Staveley J, Phelps K, Young CS, Whipps JM. 2003. Ascospore release and survival in Sclerotinia sclerotiorum. Mycol Res 107:213–222. doi: 10.1017/S0953756203007159. [DOI] [PubMed] [Google Scholar]
- 2.Pearson RC, Siegfried W, Bodmer M, Schüepp H. 1991. Ascospore discharge and survival in Pseudopezicula tracheiphila, causal agent of Rotbrenner of grape. J Phytopathol 132:177–185. doi: 10.1111/j.1439-0434.1991.tb00110.x. [DOI] [Google Scholar]
- 3.Burt PJA, Rosenberg LJ, Rutter J, Ramirez F, Gonzales H. 1999. Forecasting the airborne spread of Mycosphaerella fijiensis, a cause of black Sigatoka disease on banana: estimations of numbers of perithecia and ascospores. Ann Appl Biol 135:369–377. doi: 10.1111/j.1744-7348.1999.tb00863.x. [DOI] [Google Scholar]
- 4.Guerin L, Froidefond G, Xu X-M. 2001. Seasonal patterns of dispersal of ascospores of Cryphonectria parasitica (chestnut blight). Plant Pathol 50:717–724. doi: 10.1046/j.1365-3059.2001.00600.x. [DOI] [Google Scholar]
- 5.Hunter T, Coker RR, Royle DJ. 1999. The teleomorph stage, Mycosphaerella graminicola, in epidemics of Septoria tritici blotch on winter wheat in the UK. Plant Pathol 48:51–57. doi: 10.1046/j.1365-3059.1999.00310.x. [DOI] [Google Scholar]
- 6.Timmermann V, Børja I, Hietala AM, Kirisits T, Solheim H. 2011. Ash dieback: pathogen spread and diurnal patterns of ascospore dispersal, with special emphasis on Norway. EPPO Bull 41:14–20. doi: 10.1111/j.1365-2338.2010.02429.x. [DOI] [Google Scholar]
- 7.Butler G. 2007. The evolution of MAT: the Ascomycetes, p 3–18. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in fungi: molecular determination and evolutionary implications. ASM Press, Washington, DC. [Google Scholar]
- 8.Debuchy R, Berteaux-Lecellier V, Silar P. 2010. Mating systems and sexual morphogenesis in ascomycetes, p 501–535. In Borkovich KA (ed), Cellular and molecular biology of filamentous fungi. ASM Press, Washington, DC. [Google Scholar]
- 9.Wilken PM, Steenkamp ET, Wingfield MJ, De Beer ZW, Wingfield BD. 2017. Which MAT gene? Pezizomycotina (Ascomycota) mating-type gene nomenclature reconsidered. Fungal Biol Rev 31:199–211. doi: 10.1016/j.fbr.2017.05.003. [DOI] [Google Scholar]
- 10.Metzenberg RL, Glass NL. 1990. Mating type and mating strategies in Neurospora. Bioessays 12:53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- 11.Martin SH, Wingfield BD, Wingfield MJ, Steenkamp ET. 2011. Structure and evolution of the Fusarium mating type locus: new insights from the Gibberella fujikuroi complex. Fungal Genet Biol 48:731–740. doi: 10.1016/j.fgb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Glass NL, Smith ML. 1994. Structure and function of a mating-type gene from the homothallic species Neurospora africana. Mol Gen Genet 244:401–409. doi: 10.1007/BF00286692. [DOI] [PubMed] [Google Scholar]
- 13.Yun SH, Berbee ML, Yoder OC, Turgeon BG. 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci U S A 96:5592–5597. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yun SH, Arie T, Kaneko I, Yoder OC, Turgeon BG. 2000. Molecular organization of mating type loci in heterothallic, homothallic, and asexual Gibberella/Fusarium species. Fungal Genet Biol 31:7–20. doi: 10.1006/fgbi.2000.1226. [DOI] [PubMed] [Google Scholar]
- 15.Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier JM, Quévillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun M-H, Dickman M. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paoletti M, Seymour FA, Alcocer MJC, Kaur N, Calvo AM, Archer DB, Dyer PS. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol 17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 17.van Kan JAL, Stassen JHM, Mosbach A, van der Lee TAJ, Faino L, Farmer AD, Papasotiriou DG, Zhou S, Seidl MF, Cottam E, Edel D, Hahn M, Schwartz DC, Dietrich RA, Widdison S, Scalliet G. 2017. A gapless genome sequence of the fungus Botrytis cinerea. Mol Plant Pathol 18:75–89. doi: 10.1111/mpp.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faretra F, Antonacci E, Pollastro S. 1988. Sexual behaviour and mating system of Botryotinia fuckeliana, teleomorph of Botrytis cinerea. Microbiology 134:2543–2550. doi: 10.1099/00221287-134-9-2543. [DOI] [Google Scholar]
- 19.van der Vlugt-Bergmans CJB, Brandwagt BF, Vant’t Klooster JW, Wagemakers CAM, van Kan JAL. 1993. Genetic variation and segregation of DNA polymorphisms in Botrytis cinerea. Mycol Res 97:1193–1200. doi: 10.1016/S0953-7562(09)81284-7. [DOI] [Google Scholar]
- 20.Li M, Liang X, Rollins JA. 2012. Sclerotinia sclerotiorum γ-glutamyl transpeptidase (Ss-Ggt1) is required for regulating glutathione accumulation and development of sclerotia and compound appressoria. Mol Plant Microbe Interact 25:412–420. doi: 10.1094/MPMI-06-11-0159. [DOI] [PubMed] [Google Scholar]
- 21.Sun P, Yang XB. 2000. Light, temperature, and moisture effects on apothecium production of Sclerotinia sclerotiorum. Plant Dis 84:1287–1293. doi: 10.1094/PDIS.2000.84.12.1287. [DOI] [PubMed] [Google Scholar]
- 22.Pöggeler S, Nowrousian M, Kück U. 2006. Fruiting-body development in ascomycetes, p 325–355. In Kües U, Fischer R (ed), The Mycota I: growth, differentiation and sexuality. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 23.De Miccolis Angelini RM, Rotolo C, Pollastro S, Faretra F. 2016. Molecular analysis of the mating type (MAT1) locus in strains of the heterothallic ascomycete Botrytis cinerea. Plant Pathol 65:1321–1332. doi: 10.1111/ppa.12509. [DOI] [Google Scholar]
- 24.Pöggeler S, Kück U. 2000. Comparative analysis of mating-type loci from Neurospora crassa and Sordaria macrospora: identification of novel transcribed ORFs. Mol Gen Genet 263:292–301. doi: 10.1007/s004380051171. [DOI] [PubMed] [Google Scholar]
- 25.Pöggeler S, Risch S, Kück U, Osiewacz HD. 1997. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic Ascomycete. Genetics 147:567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira AV-b, An Z, Metzenberg RL, Glass NL. 1998. Characterization of mat A-2, mat A-3 and ΔmatA mating-type mutants of Neurospora crassa. Genetics 148:1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnaise S, Zickler D, Le Bilcot S, Poisier C, Debuchy R. 2001. Mutations in mating-type genes of the heterothallic fungus Podospora anserina lead to self-fertility. Genetics 159:545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klix V, Nowrousian M, Ringelberg C, Loros JJ, Dunlap JC, Pöggeler S. 2010. Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot Cell 9:894–905. doi: 10.1128/EC.00019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desjardins AE, Brown DW, Yun SH, Proctor RH, Lee T, Plattner RD, Lu SW, Turgeon BG. 2004. Deletion and complementation of the mating type (MAT) locus of the wheat head blight pathogen Gibberella zeae. Appl Environ Microbiol 70:2437–2444. doi: 10.1128/AEM.70.4.2437-2444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HK, Cho EJ, Lee S, Lee YS, Yun SH. 2012. Functional analyses of individual mating-type transcripts at MAT loci in Fusarium graminearum and Fusarium asiaticum. FEMS Microbiol Lett 337:89–96. doi: 10.1111/1574-6968.12012. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Q, Hou R, Juanyu, Zhang, Ma J, Wu Z, Wang GH, Wang CF, Xu JR. 2013. The MAT locus genes play different roles in sexual reproduction and pathogenesis in Fusarium graminearum. PLoS One 8:e66980. doi: 10.1371/journal.pone.0066980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doughan B, Rollins JA. 2016. Characterization of MAT gene functions in the life cycle of Sclerotinia sclerotiorum reveals a lineage-specific MAT gene functioning in apothecium morphogenesis. Fungal Biol 120:1105–1117. doi: 10.1016/j.funbio.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, Menzel C, Chen W, Li Y, Zeng R, Khaitovich P. 2009. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics 10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bidard F, Aït Benkhali J, Coppin E, Imbeaud S, Grognet P, Delacroix H, Debuchy R. 2011. Genome-wide gene expression profiling of fertilization competent mycelium in opposite mating types in the heterothallic fungus Podospora anserina. PLoS One 6:e21476. doi: 10.1371/journal.pone.0021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orshinsky AM, Hu J, Opiyo SO, Reddyvari-Channarayappa V, Mitchell TK, Boehm MJ. 2012. RNA-Seq analysis of the Sclerotinia homoeocarpa-creeping bentgrass pathosystem. PLoS One 7:e41150. doi: 10.1371/journal.pone.0041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steindorff AS, Ramada MH, Coelho AS, Miller RN, Pappas GJ, Ulhoa CJ, Noronha EF. 2014. Identification of mycoparasitism-related genes against the phytopathogen Sclerotinia sclerotiorum through transcriptome and expression profile analysis in Trichoderma harzianum. BMC Genomics 15:204. doi: 10.1186/1471-2164-15-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang ZX, Zhou XZ, Meng HM, Liu YJ, Zhou Q, Huang B. 2014. Comparative transcriptomic analysis of the heat stress response in the filamentous fungus Metarhizium anisopliae using RNA-Seq. Appl Microbiol Biotechnol 98:5589–5597. doi: 10.1007/s00253-014-5763-y. [DOI] [PubMed] [Google Scholar]
- 40.Soanes DM, Chakrabarti A, Paszkiewicz KH, Dawe AL, Talbot NJ. 2012. Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae. PLoS Pathog 8:e1002514. doi: 10.1371/journal.ppat.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sikhakolli UR, López-Giráldez F, Li N, Common R, Townsend JP, Trail F. 2012. Transcriptome analyses during fruiting body formation in Fusarium graminearum and Fusarium verticillioides reflect species life history and ecology. Fungal Genet Biol 49:663–673. doi: 10.1016/j.fgb.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Traeger S, Altegoer F, Freitag M, Gabaldon T, Kempken F, Kumar A, Marcet-Houben M, Pöggeler S, Stajich JE, Nowrousian M. 2013. The genome and development-dependent transcriptomes of Pyronema confluens: a window into fungal evolution. PLoS Genet 9:e1003820. doi: 10.1371/journal.pgen.1003820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becker K, Beer C, Freitag M, Kück U. 2015. Genome-wide identification of target genes of a mating-type alpha-domain transcription factor reveals functions beyond sexual development. Mol Microbiol 96:1002–1022. doi: 10.1111/mmi.12987. [DOI] [PubMed] [Google Scholar]
- 44.Kim HK, Jo SM, Kim GY, Kim DW, Kim YK, Yun SH. 2015. A large-scale functional analysis of putative target genes of mating-type loci provides insight into the regulation of sexual development of the cereal pathogen Fusarium graminearum. PLoS Genet 11:e1005486. doi: 10.1371/journal.pgen.1005486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ait Benkhali J, Coppin E, Brun S, Peraza-Reyes L, Martin T, Dixelius C, Lazar N, van Tilbeurgh H, Debuchy R. 2013. A network of HMG-box transcription factors regulates sexual cycle in the fungus Podospora anserina. PLoS Genet 9:e1003642. doi: 10.1371/journal.pgen.1003642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malagnac F, Wendel B, Goyon C, Faugeron G, Zickler D, Rossignol JL, Noyer-Weidner M, Vollmayr P, Trautner TA, Walter J. 1997. A gene essential for de novo methylation and development in Ascobolus reveals a new type of eukaryotic DNA methyltransferase structure. Cell 91:281–290. doi: 10.1016/S0092-8674(00)80410-9. [DOI] [PubMed] [Google Scholar]
- 47.Coppin E, Berteaux-Lecellier V, Bidard F, Brun S, Ruprich-Robert G, Espagne E, Aït-Benkhali J, Goarin A, Nesseir A, Planamente S, Debuchy R, Silar P. 2012. Systematic deletion of homeobox genes in Podospora anserina uncovers their roles in shaping the fruiting body. PLoS One 7:e37488. doi: 10.1371/journal.pone.0037488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Thiewes H, van Kan JAL. 2011. The D-galacturonic acid catabolic pathway in Botrytis cinerea. Fungal Genet Biol 48:990–997. doi: 10.1016/j.fgb.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Shiu PKT, Raju NB, Zickler D, Metzenberg RL. 2001. Meiotic silencing by unpaired DNA. Cell 107:905–916. doi: 10.1016/S0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 50.Teichert I, Wolff G, Kück U, Nowrousian M. 2012. Combining laser microdissection and RNA-seq to chart the transcriptional landscape of fungal development. BMC Genomics 13:511. doi: 10.1186/1471-2164-13-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dyer PS, O’Gorman CM. 2012. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev 36:165–192. doi: 10.1111/j.1574-6976.2011.00308.x. [DOI] [PubMed] [Google Scholar]
- 52.Teichert I, Nowrousian M, Pöggeler S, Kück U. 2014. The filamentous fungus Sordaria macrospora as a genetic model to study fruiting body development. Adv Genet 87:199–244. doi: 10.1016/B978-0-12-800149-3.00004-4. [DOI] [PubMed] [Google Scholar]
- 53.Haile ZM, Pilati S, Sonego P, Malacarne G, Vrhovsek U, Engelen K, Tudzynski P, Zottini M, Baraldi E, Moser C. 2017. Molecular analysis of the early interaction between the grapevine flower and Botrytis cinerea reveals that prompt activation of specific host pathways leads to fungus quiescence. Plant Cell Environ 40:1409–1428. doi: 10.1111/pce.12937. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Hua C, Stassen JHM, Chatterjee S, Cornelissen M, van Kan JAL. 2014. Genome-wide analysis of pectate-induced gene expression in Botrytis cinerea: identification and functional analysis of putative D-galacturonate transporters. Fungal Genet Biol 72:182–191. doi: 10.1016/j.fgb.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Antal Z, Rascle C, Cimerman A, Viaud M, Billon-Grand G, Choquer M, Bruel C. 2012. The homeobox BcHOX8 gene in Botrytis cinerea regulates vegetative growth and morphology. PLoS One 7:e48134. doi: 10.1371/journal.pone.0048134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schumacher J, Simon A, Cohrs KC, Viaud M, Tudzynski P. 2014. The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genet 10:e1004040. doi: 10.1371/journal.pgen.1004040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosbach A, Leroch M, Mendgen KW, Hahn M. 2011. Lack of evidence for a role of hydrophobins in conferring surface hydrophobicity to conidia and hyphae of Botrytis cinerea. BMC Microbiol 11:10. doi: 10.1186/1471-2180-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schamber A, Leroch M, Diwo J, Mendgen K, Hahn M. 2010. The role of mitogen-activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea. Mol Plant Pathol 11:105–119. doi: 10.1111/j.1364-3703.2009.00579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rui O, Hahn M. 2007. The Slt2-type MAP kinase Bmp3 of Botrytis cinerea is required for normal saprotrophic growth, conidiation, plant surface sensing and host tissue colonization. Mol Plant Pathol 8:173–184. doi: 10.1111/j.1364-3703.2007.00383.x. [DOI] [PubMed] [Google Scholar]
- 60.Schumacher J. 2016. DHN melanin biosynthesis in the plant pathogenic fungus Botrytis cinerea is based on two developmentally regulated key enzyme (PKS)-encoding genes. Mol Microbiol 99:729–748. doi: 10.1111/mmi.13262. [DOI] [PubMed] [Google Scholar]
- 61.Zhang C, He Y, Zhu P, Chen L, Wang Y, Ni B, Xu L. 2015. Loss of Bcbrn1 and Bcpks13 in Botrytis cinerea not only blocks melanization but also increases vegetative growth and virulence. Mol Plant Microbe Interact 28:1091–1101. doi: 10.1094/MPMI-04-15-0085-R. [DOI] [PubMed] [Google Scholar]
- 62.An B, Li B, Li H, Zhang Z, Qin G, Tian S. 2016. Aquaporin8 regulates cellular development and reactive oxygen species production, a critical component of virulence in Botrytis cinerea. New Phytol 209:1668–1680. doi: 10.1111/nph.13721. [DOI] [PubMed] [Google Scholar]
- 63.Dulermo T, Rascle C, Chinnici G, Gout E, Bligny R, Cotton P. 2009. Dynamic carbon transfer during pathogenesis of sunflower by the necrotrophic fungus Botrytis cinerea: from plant hexoses to mannitol. New Phytol 183:1149–1162. doi: 10.1111/j.1469-8137.2009.02890.x. [DOI] [PubMed] [Google Scholar]
- 64.Han Y, Joosten HJ, Niu W, Zhao Z, Mariano PS, McCalman M, Van Kan J, Schaap PJ, Dunaway-Mariano D. 2007. Oxaloacetate hydrolase, the C-C bond lyase of oxalate secreting fungi. J Biol Chem 282:9581–9590. doi: 10.1074/jbc.M608961200. [DOI] [PubMed] [Google Scholar]
- 65.ten Have A, Espino JJ, Dekkers E, Van Sluyter SC, Brito N, Kay J, González C, van Kan JAL. 2010. The Botrytis cinerea aspartic proteinase family. Fungal Genet Biol 47:53–65. doi: 10.1016/j.fgb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 66.Frías M, González C, Brito N. 2011. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol 192:483–495. doi: 10.1111/j.1469-8137.2011.03802.x. [DOI] [PubMed] [Google Scholar]
- 67.Viaud M, Brunet-Simon A, Brygoo Y, Pradier JM, Levis C. 2003. Cyclophilin A and calcineurin functions investigated by gene inactivation, cyclosporin A inhibition and cDNA arrays approaches in the phytopathogenic fungus Botrytis cinerea. Mol Microbiol 50:1451–1465. doi: 10.1046/j.1365-2958.2003.03798.x. [DOI] [PubMed] [Google Scholar]
- 68.Harren K, Schumacher J, Tudzynski B. 2012. The Ca2+/calcineurin-dependent signaling pathway in the gray mold Botrytis cinerea: the role of calcipressin in modulating calcineurin activity. PLoS One 7:e41761. doi: 10.1371/journal.pone.0041761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kars I, Krooshof GH, Wagemakers L, Joosten R, Benen JAE, van Kan JAL. 2005. Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J 43:213–225. doi: 10.1111/j.1365-313X.2005.02436.x. [DOI] [PubMed] [Google Scholar]
- 70.Kars I, McCalman M, Wagemakers L, van Kan JAL. 2005. Functional analysis of Botrytis cinerea pectin methylesterase genes by PCR-based targeted mutagenesis: Bcpme1 and Bcpme2 are dispensable for virulence of strain B05.10. Mol Plant Pathol 6:641–652. doi: 10.1111/j.1364-3703.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- 71.Schouten A, Wagemakers L, Stefanato FL, van der Kaaij RM, van Kan JAL. 2002. Resveratrol acts as a natural profungicide and induces self-intoxication by a specific laccase. Mol Microbiol 43:883–894. doi: 10.1046/j.1365-2958.2002.02801.x. [DOI] [PubMed] [Google Scholar]
- 72.Del Sorbo G, Ruocco M, Schoonbeek HJ, Scala F, Pane C, Vinale F, de Waard MA. 2008. Cloning and functional characterization of BcatrA, a gene encoding an ABC transporter of the plant pathogenic fungus Botryotinia fuckeliana (Botrytis cinerea). Mycol Res 112:737–746. doi: 10.1016/j.mycres.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 73.Makizumi Y, Takeda S, Matsuzaki Y, Nakaune R, Hamamoto H, Akutsu K, Hibi T. 2002. Cloning and selective toxicant-induced expression of BMR1 and BMR3, novel ABC transporter genes in Botrytis cinerea. J Gen Plant Pathol 68:338–341. doi: 10.1007/PL00013100. [DOI] [Google Scholar]
- 74.Pane C, Rekab D, Firrao G, Ruocco M, Scala F. 2008. A novel gene coding for an ABC transporter in Botrytis cinerea (Botryotinia fuckeliana) is involved in resistance to H2O2. J Plant Pathol 90:453–462. [Google Scholar]
- 75.Hayashi K, Schoonbeek HJ, De Waard MA. 2002. Expression of the ABC transporter BcatrD from Botrytis cinerea reduces sensitivity to sterol demethylation inhibitor fungicides. Pestic Biochem Physiol 73:110–121. doi: 10.1016/S0048-3575(02)00015-9. [DOI] [Google Scholar]
- 76.Schoonbeek H, Del Sorbo G, De Waard MA. 2001. The ABC transporter BcatrB affects the sensitivity of Botrytis cinerea to the phytoalexin resveratrol and the fungicide fenpiclonil. Mol Plant Microbe Interact 14:562–571. doi: 10.1094/MPMI.2001.14.4.562. [DOI] [PubMed] [Google Scholar]
- 77.Kretschmer M, Leroch M, Mosbach A, Walker AS, Fillinger S, Mernke D, Schoonbeek HJ, Pradier JM, Leroux P, de Waard MA, Hahn M. 2009. Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog 5:e1000696. doi: 10.1371/journal.ppat.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Viaud M, Fillinger S, Liu W, Polepalli JS, Le Pêcheur P, Kunduru AR, Leroux P, Legendre L. 2006. A class III histidine kinase acts as a novel virulence factor in Botrytis cinerea. Mol Plant Microbe Interact 19:1042–1050. doi: 10.1094/MPMI-19-1042. [DOI] [PubMed] [Google Scholar]
- 79.Cui W, Beever RE, Parkes SL, Weeds PL, Templeton MD. 2002. An osmosensing histidine kinase mediates dicarboximide fungicide resistance in Botryotinia fuckeliana (Botrytis cinerea). Fungal Genet Biol 36:187–198. doi: 10.1016/S1087-1845(02)00009-9. [DOI] [PubMed] [Google Scholar]
- 80.Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin HL. 2013. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Siegmund U, Marschall R, Tudzynski P. 2015. BcNoxD, a putative ER protein, is a new component of the NADPH oxidase complex in Botrytis cinerea. Mol Microbiol 95:988–1005. doi: 10.1111/mmi.12869. [DOI] [PubMed] [Google Scholar]
- 82.Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K. 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J 23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gesing S, Schindler D, Fränzel B, Wolters D, Nowrousian M. 2012. The histone chaperone ASF1 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol Microbiol 84:748–765. doi: 10.1111/j.1365-2958.2012.08058.x. [DOI] [PubMed] [Google Scholar]
- 84.Faretra F, Antonacci E. 1987. Production of apothecia of Botryotinia fuckeliana (de Bary) Whetz under controlled environmental conditions. Phytopathol Mediterr 26:29–35. [Google Scholar]
- 85.Benito EP, ten Have A, van ’t Klooster JW, van Kan JAL. 1998. Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea. Eur J Plant Pathol 104:207–220. doi: 10.1023/A:1008698116106. [DOI] [Google Scholar]
- 86.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao Y, Smyth GK, Shi W. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Stat Methodol 57:289–300. [Google Scholar]
- 91.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alexa A, Rahnenfuhrer J. 2010. TopGO: enrichment analysis for gene ontology. http://bioconductor.org/packages/release/bioc/html/topGO.html.
- 93.Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pignatelli M, Serras F, Moya A, Guigó R, Corominas M. 2009. CROC: finding chromosomal clusters in eukaryotic genomes. Bioinformatics 25:1552–1553. doi: 10.1093/bioinformatics/btp248. [DOI] [PubMed] [Google Scholar]
- 95.Porquier A. 2016. Etude des mécanismes de régulation du métabolisme secondaire chez Botrytis cinerea. PhD thesis Universités Paris-Saclay and Paris-Sud, Paris, France. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of strategy for targeted deletion of MAT genes. Knockout of BcMAT1-1-1, BcMAT1-1-5, BcMAT1-2-1, and BcMAT1-2-10 by targeted gene replacement. Organization of BcMAT1-1-1, BcMAT1-1-5, BcMAT1-2-1, and BcMAT1-2-10 locus before and after homologous recombination. Orientations of the target gene and HPH are indicated by white and gray arrows, respectively. Upstream and downstream flanks of target genes are shown with open boxes. PCR analysis of wild-type strains SAS56 and SAS405 and knockout mutant strains. The genomic DNA of each strain was used to verify 5′ and 3′ homologous recombination and the absence of targeted genes in corresponding deletion mutants, respectively. Download FIG S1, PDF file, 0.3 MB (354.9KB, pdf) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mapping efficiency of RNA-seq reads on the Botrytis cinerea (strain B05.10) genome. Five samples represent sclerotia (Scl), apothecial primordia (Apo12), apothecial stipes (Apo34), apothecial disks (Apo56), and ascospores (Asc); biological replicates are labeled A and B. The number and proportion of genes with CPM values of ≥1 are given. Download TABLE S1, XLSX file, 0.01 MB (10.5KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcript levels of Botrytis cinerea orthologs to genes in Aspergillus nidulans and Sordaria macrospora previously listed as activators or repressors of sexual development (51, 52). The A. nidulans and S. macrospora gene symbols, locus identifier, and their proposed functions and domains are given in columns A to C; whether those genes are considered activators or repressors of sexual development is indicated in column D, with the relevant reference provided in column E. Gene identifiers (IDs) for the B. cinerea orthologs are given in column F, with their expression values in the five tissue samples (in counts per million mapped reads [CPM]) presented in columns G to K as averages from two biological replicates. Download TABLE S2, XLSX file, 0.03 MB (28.5KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed genes in the transition from sclerotia to apothecial primordia. Panels S3a and S3b list the upregulated and downregulated genes, respectively; their expression values in both biological replicates of both samples (in counts per million mapped reads [CPM]); the log2 of the fold change (logFC); average log2 of the CPM values between the four RNA samples (logCPM); test statistic of the likelihood-ratio test (LR); the uncorrected and Benjamini-Hochberg-corrected P value of the likelihood test; the B. cinerea gene name; and a description resulting from manual annotation. Panels S3c and S3d present GO enrichment analyses for the upregulated and downregulated genes, respectively. Download TABLE S3, XLSX file, 0.5 MB (487.2KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed genes in the transition from apothecial primordia to stipes. Panels S4a and S4b list the upregulated and downregulated genes, respectively; their expression values in both biological replicates of both samples (in counts per million mapped reads [CPM]); the log2 of the fold change (logFC); average log2 of the CPM values between the four RNA samples (logCPM); test statistic of the likelihood-ratio test (LR); the uncorrected and Benjamini-Hochberg-corrected P value of the likelihood test; the B. cinerea gene name; and a description resulting from manual annotation. Panels S4c and S4d present GO enrichment analyses for the upregulated and downregulated genes, respectively. Download TABLE S4, XLSX file, 0.4 MB (384.4KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed genes in the transition from apothecial stipes to disks. Panels S5a and S5b list the upregulated and downregulated genes, respectively; their expression values in both biological replicates of both samples (in counts per million mapped reads [CPM]); the log2 of the fold change (logFC); average log2 of the CPM values between the four RNA samples (logCPM); test statistic of the likelihood-ratio test (LR); the uncorrected and Benjamini-Hochberg-corrected P value of the likelihood test; the B. cinerea gene name; and a description resulting from manual annotation. Panels S5c and S5d present GO enrichment analyses for the upregulated and downregulated genes, respectively. Download TABLE S5, XLSX file, 0.1 MB (127.8KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed genes in stipes obtained with MAT gene mutants. Panels S6a and S6b list the downregulated and upregulated genes, respectively, in stipes obtained in an SAS405 × ΔMAT1-1-5 cross, compared to stipes from a wild-type cross, SAS405 × SAS56. Panels S6c and S6d list the downregulated and upregulated genes, respectively, in stipes obtained in an SAS56 × ΔMAT1-2-10 cross, compared to stipes from a wild-type cross, SAS405 × SAS56. Panel S6e lists the overlap between downregulated genes in panels S6a and S6c, while panel S6f lists the overlap between upregulated genes in panels S6b and S6d. Download TABLE S6, XLSX file, 0.3 MB (345.4KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed genes in the transition from apothecial disks to ascospores. Panels S7a and S7b list the upregulated and downregulated genes, respectively; their expression values in both biological replicates of both samples (in counts per million mapped reads [CPM]); the log2 of the fold change (logFC); average log2 of the CPM values between the four RNA samples (logCPM); test statistic of the likelihood-ratio test (LR); the uncorrected and Benjamini-Hochberg-corrected P value of the likelihood test; the B. cinerea gene name; and a description resulting from manual annotation. Panels S7c and S7d present GO enrichment analyses for the upregulated and downregulated genes, respectively. Download TABLE S7, XLSX file, 1 MB (1.1MB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CROC analysis for identification of clusters of coregulated genes. Clusters were defined as sliding windows of ≥12 genes, of which at least 50% are jointly upregulated or downregulated. Each table provides information about the chromosome number, the start and end coordinates of the cluster, the adjusted P value for clustering (threshold was set at a P value of <0.05), and the gene IDs of the coexpressed genes in the cluster. CROC output is provided for upregulated genes in the transition from sclerotia to primordia (panel S8a), downregulated genes in the transition from sclerotia to primordia (panel S8b), downregulated genes in the transition from primordia to stipes (panel S8c), upregulated genes in the transition from apothecial disks to ascsopores (panel S8d), and downregulated genes in the transition from apothecial disks to ascsopores (panel S8e). Download TABLE S8, XLSX file, 0.03 MB (36.8KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S9, XLSX file, 0.01 MB (13.3KB, xlsx) .
Copyright © 2018 Rodenburg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
RNA-seq reads from this study are deposited in the NCBI SRA archive under PRJNA351919 (SRP093589). Accession numbers for data sets are as follows: sclerotia of strain SAS405, SRS1810933; primordia Apo12, SRS1810937; stipes Apo34, SRS1810938; apothecial disks Apo56, SRS1810941; ascospores, SRS1812737.