Abstract
Complex proteoforms contain various primary structural alterations resulting from variations in genes, RNA, and proteins. Top-down mass spectrometry is commonly used for analyzing complex proteoforms because it provides whole sequence information of the proteoforms. Proteoform identification by top-down mass spectral database search is a challenging computational problem because the types and/or locations of some alterations in target proteoforms are in general unknown. Although spectral alignment and mass graph alignment algorithms have been proposed for identifying proteoforms with unknown alterations, they are extremely slow to align millions of spectra against tens of thousands of protein sequences in high throughput proteome level analyses. Many software tools in this area combine efficient protein sequence filtering algorithms and spectral alignment algorithms to speed up database search. As a result, the performance of these tools heavily relies on the sensitivity and efficiency of their filtering algorithms. Here we propose two efficient approximate spectrum-based filtering algorithms for proteoform identification. We evaluated the performances of the proposed algorithms and 4 existing ones on simulated and real top-down mass spectrometry data sets. Experimental results showed that the proposed algorithms outperformed the existing ones for complex proteoform identification. In addition, combining the proposed filtering algorithms and mass graph alignment algorithms identified many proteoforms missed by ProSightPC in proteome-level proteoform analyses.
Keywords: Top-down mass spectrometry, spectral identification, filtering algorithms
1 Introduction
Because of variations in genes, gene expression, and other biological processes, a gene may have various proteoforms [1], many of which contain multiple primary structural alterations (PSAs), such as amino acid substitutions, post-translational modifications (PTMs), and terminal truncations. PSAs play an important role in many diseases such as heart failure [2] and age-dependent memory impairment [3]. Proteoform identification and PSA localization are essential to understanding proteoform functions in cellular processes. For example, the combinatorial patterns of PSAs in histone proteins determine their gene regulatory functions [4, 5].
Top-down mass spectrometry (MS) is more efficient than bottom-up MS in identifying modified proteoforms and combinatorial patterns of PSAs because it analyzes intact proteoforms instead of short protein fragments [6]. Database search is the dominant method for top-down MS-based proteoform identification, in which top-down tandem mass (MS/MS) spectra are searched against a protein sequence database or an annotated database for spectral identification [7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21]. A list of software tools for proteoform identification by top-down MS can be found in Table S1 in the supplementary material.
Most protein databases contain only a reference sequence for a gene or transcript even though the gene or transcript may have many proteoforms. The target proteoform may have various alterations compared with its corresponding database sequence [18]. We study four types of alterations: terminal truncations, fixed PTMs, variable PTMs, and unexpected alterations. A terminal truncation removes a prefix or suffix of a protein sequence. A fixed PTM specifies its mass shift and modified amino acids in a protein sequence. A variable PTM specifies its mass shift and a set of amino acids that may be modified. The mass shift and location of an unexpected alteration are unknown.
ProSightPC [7] is a commonly used software tool for top-down MS-based proteoform identification. By constructing a “shotgun annotated” proteoform database containing known modified proteoforms, it efficiently identifies proteoforms in the database. In the Delta-M mode of ProSightPC, the delta is the difference between the precursor mass of the query spectrum and the theoretical precursor mass of the target database sequence. When the allowed delta is large and a proteoform with unexpected alterations has a long unmodified N-terminal or C-terminal fragment, mass spectra generated from the proteoform can be matched to its corresponding database sequence. In the sequence tag mode of ProSightPC, sequence tags are extracted from the query mass spectrum and searched against the proteoform database for proteoform identification.
Many software tools use spectral alignment to identify proteoforms with unexpected alterations [8, 12, 15, 16, 17, 18]. Let S be a query mass spectrum generated from a proteoform with unexpected alterations and P the unmodified database protein sequence of the proteoform. The spectral alignment algorithm finds an optimal alignment between S and P by inserting into P mass shifts corresponding to the unexpected alterations in the blind mode. When the spectrum S contains enough fragment masses, the alignment algorithm is capable of identifying and characterizing the proteoform. MS-Align+ [12] and TopPIC [16] are commonly used tools for identifying proteoforms with unexpected alterations using top-down MS. In these tools, variable PTMs are treated as unexpected alterations, making them inefficient in identifying ultramodified proteoforms with many variable PTMs. To address this problem, several spectral alignment algorithms, such as MS-Align-E [15], MSPathFinder [21], pTop [17], TopMG [18] have been proposed to identify proteoforms with many variable PTMs.
There are two main steps in spectral alignment-based software tools for identifying proteoforms with variable PTMs and/or unexpected alterations by database search. First, a filtering algorithm is used to filter out most candidate protein sequences in the database for the query mass spectrum. Second, a spectral alignment algorithm is employed to align the mass spectrum against each remaining candidate protein sequence to find the best scoring proteoform spectrum-match (PrSM) [8]. It is extremely slow to align mass spectra against tens of thousands of database protein sequences [12]. Therefore, the filtering step is indispensable in proteome-level analyses. A filtering algorithm is efficient if it keeps the correct target protein sequence as a candidate for spectral alignment.
Most proteoform identification methods allow fixed PTMs and terminal truncations in the target proteoform. There are several scenarios for the other two types of alterations: (1) neither variable PTMs nor unexpected alterations are allowed in the target proteoform; (2) only variable PTMs are allowed; (3) only unexpected alterations are allowed; and (4) both variable PTMs and unexpected alterations are allowed. In the first scenario, a candidate protein sequence (may be truncated) is filtered out if its molecular mass does not match the precursor mass of the query spectrum. In the last three scenarios, the precursor mass of the query spectrum may be different from the molecular mass of its corresponding database sequence. For the second scenario, one filtering method is to check if the difference between the precursor mass and the molecular mass can be explained by a combination of variable PTMs. In this paper, we focus on filtering methods for the last three scenarios.
There are three main approaches for protein sequence filtering. In the first approach, a large error tolerance is allowed between the precursor mass of the query spectrum and the molecular mass of the candidate sequence [22]. In top-down MS, the method is employed in the Delta-M mode in ProSightPC [7]. However, when the error tolerance is very large, the filtering method reports many candidates, significantly increasing the running time of database search.
The second approach is based on sequence tags, which were proposed by Mann et al. in a pioneer work in 1994 [23]. In this approach, sequence tags are generated from the query spectrum and searched against the database to find hits, based on which top candidates are selected. Sequence tags and gapped sequence tags have been widely and successfully used for bottom-up spectral interpretation [24, 25, 26, 27, 28, 29, 30]. In top-down MS, tag-based methods have been used in USTag [31], pTop [17], MSPathFinder [21], and the sequence tag mode in ProSightPC [7]. The accuracy of tag-based methods depends on whether the query spectrum contains consecutive fragment ions.
The third approach uses unmodified protein fragments (UPFs) and their matched fragment masses in the query spectrum to filter proteins [12, 16]. The idea is to find a mass shift for the fragment masses in the query spectrum such that many shifted fragment masses are explained by the unmodified target protein sequence. This method is computationally intensive. Fortunately, index-based algorithms [32, 33, 34] have been proposed to partially solve the problem. In top-down MS, UPF-based methods have been used in MS-Align+ [12] and TopPIC [16] and achieved satisfactory performance in identifying unexpected alterations. The three filtering approaches can be combined to improve filtering efficiency. For example, proteins can be filtered by combining a large error tolerance for the precursor mass and sequence tags extracted from the query spectrum.
The three filtering approaches are designed to identify proteoforms with a limited number (1 or 2 in most cases) of unexpected alterations. These methods may fail to keep the target database protein sequence in filtration when the target proteoform contains more than 2 variable PTMs and/or unexpected alterations.
In this paper, we propose two Approximate Spectrum-based Filtering (ASF) algorithms for identifying complex proteoforms with variable PTMs and those with both variable PTMs and unexpected alterations. Let F be the target proteoform and F′ a proteoform obtained from F by removing h variable PTMs. In the ASF algorithms, the query spectrum is transformed into an approximate spectrum of F′, which is searched against database sequences to find candidate proteins. After the transformation, the number of variable PTMs in the target proteoform is reduced by h (Fig. 1), significantly increasing filtering efficiency.
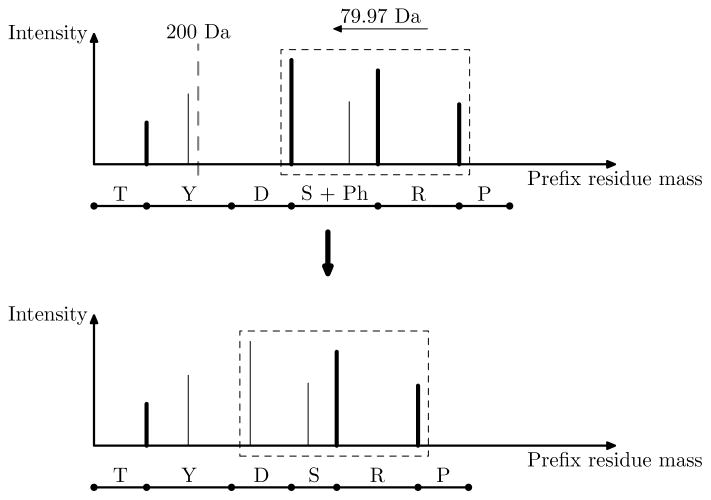
Figure 1.
A prefix residue mass spectrum (top) of the proteoform TYDS[Ph]RP with a phosphorylation site on the serine residue is transformed into an approximate prefix residue mass spectrum (bottom) of the unmodified protein TYDSPR. In the top spectrum, each peak represents a possible prefix residue mass extracted from the experimental spectrum, and bold peaks are those mapped to theoretical prefix residue masses of the proteoform TYDS[Ph]RP. The prefix residue mass 200 Da is a guessed prefix residue mass for the modification site. All peaks (in the box) with a mass larger than 200 Da are shifted to the left by 79.97 Da, which is the mass shift of a phosphorylation site. In the bottom spectrum, the two shifted bold peaks in the box are matched to prefix residue masses of TYDSRP, and the left most peak in the box is not matched to any prefix residue mass of TYDSRP because of the error in the estimated 200 Da for the modification site.
We evaluated the ASF algorithms and 4 existing ones for protein sequence filtration in top-down MS database search. Experiments on simulated data showed that the ASF algorithms outperformed the existing ones for complex proteoform identification. By combining the ASF and mass graph alignment algorithms [18], we identified many phosphorylated proteoforms missed by ProSightPC from a top-down MS data set of breast cancer xenograft samples.
2 Methods
A top-down MS/MS spectrum contains a list of peaks, each of which is represented as (m/z, intensity), where m/z and intensity are the mass-to-charge ratio and abundance of its corresponding fragment ion, respectively. The precursor mass of the MS/MS spectrum measures the molecular mass of the target proteoform. The first step in top-down spectral interpretation is usually spectral deconvolution [21, 35, 36, 37, 38, 39, 40, 41], which converts fragment ion peaks of various charge states and isotopomers into neutral monoisotopic fragment masses. A list of software tools for top-down spectral deconvolution can be found in Table S2 in the supplementary material. MS-Deconv [39] was used in the experiments for spectral deconvolution. In MS-Deconv, candidate isotopomer envelopes, each of which contains peaks from the same fragment ion with the same charge state, are first obtained by using the theoretical intensity distributions of these peaks, and are then selected by a dynamic programming algorithm. Finally, a neutral monoisotopic mass is computed for each selected isotopomer envelope. MS-Deconv often significantly simplifies top-down MS/MS spectra and converts a complex spectrum with thousands of peaks into a deconvoluted one with dozens or hundreds of fragment masses. We assume that the query spectrum is a deconvoluted top-down MS/MS spectrum in database search.
In the ASF algorithms, approximate spectra are first generated from the query spectrum and then searched against the protein database using the methods proposed in UPF-based filtering algorithms. We first review UPF-based filtering algorithms and then describe the ASF algorithms.
2.1 UPF-based filtering algorithms
We introduce some notations for describing UPF-based filtering algorithms. Let mass(a) be the residue mass of an amino acid a. The residue mass of a protein sequence P = a1a2 … an is the sum of the residue masses of its amino acids, that is, . The residue mass of the length-i prefix a1a2 … ai is a prefix residue mass of P, denoted by pi. The residue mass of the length-i suffix an−i+1 … an is a suffix residue mass of P, denoted by si. Specifically, the residue masses of the empty prefix and the empty suffix are 0, that is, p0 = 0 and s0 = 0. We denote the set of all prefix residue masses of P as Ppre = {p0, p1, …, pn} and the set of all suffix residue masses of P as Psuf = {s0, s1, …, sn}.
Let S be a deconvoluted top-down MS/MS spectrum with a precursor mass M. The set of deconvoluted neutral fragment masses of S are converted into a set of possible prefix (suffix) residue masses corresponding to the masses of proteoform prefixes (suffixes). When S is a collision-induced dissociation (CID) spectrum, both the prefix residue mass set and the suffix residue mass set contain the following two masses: 0 and M – mass(H2O), where mass(H2O) is the mass of a water molecule. In addition, for each fragment mass x, two masses x and M – x are added to the prefix residue mass set, and two masses x – mass(H2O) and M – x – mass(H2O) are added to the suffix residue mass set. The mass of a water molecule is deducted from x for suffix residue masses because the mass difference between a neutral y-ion fragment mass and its corresponding suffix residue mass is mass(H2O). The sets of fragment masses, prefix residue masses, and suffix residue masses of spectrum S are denoted as Sfra, Spre, and Ssuf, respectively. For example, when S is a CID spectrum with a precursor mass 302.17 Da and two neutral fragment masses 71.04 Da and 174.11 Da, the mass 0 and M – mass(H2O) = 284.16 are added into Spre and Ssuf. Spre = {0, 71.04, 128.06, 174.11, 231.13, 284.16} after the masses x and M – x for fragment masses x are added; Ssuf = {0, 53.03, 110.05, 156.10, 213.12, 284.16} after the masses x – mass(H2O) and M – x – mass(H2O) for x are added. Similarly, we use the most commonly observed fragment ion types to convert other types of deconvoluted spectra into prefix (suffix) residue masses. For example, we choose c, z-dot, and z-prime ions as the most commonly observed ones in electron-transfer dissociation (ETD) spectra, and each fragment mass in the deconvoluted spectrum is converted to three possible prefix residue masses based on the mass differences between the neutral prefix residue mass and its corresponding c, z-dot and z-prime fragment masses.
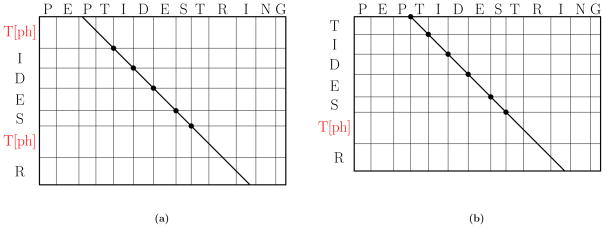
Two UPF-based filtering methods are implemented in TopPIC [16]. The first method is based on diagonal scores defined below. Let A, B be two set of masses. The mass counting score of A and B is the number of masses in A that match masses in B (within an error tolerance), denoted by C(A, B). Let shift(A, d) be the set of masses generated by adding a shift d to each mass in A. The diagonal score of A and B is the maximum mass counting score of A and B among all shift values (Fig. 2(a)), denoted by D(A, B) = maxd C(shift(A, d), B). Let P be an unmodified protein sequence and F a modified form of P with truncations and PTMs. A high diagonal score between Ppre and Fpre means that F contains a long unmodified fragment. For example, the proteoform T[Ph]IDEST[Ph]R in Fig. 2(a) contains an unmodified fragment IDES. When a CID spectrum of T[Ph]IDEST[Ph]R contains peaks of the b-ions b1, b2, …, b5, the diagonal score between the prefix residue masses of PEPTIDESTRING and those of the spectrum is at least 5. In the first method, the similarity score between a database protein sequence P and a deconvoluted spectrum S is defined as D(Ppre, Spre ).
Figure 2.
Diagonal scores and restricted diagonal scores. (a) The diagonal score between the prefix residue masses of PEPTIDESTRING and T[Ph]IDEST[Ph]R is 5, corresponding to the 5 dots in the diagonal. The score is obtained by shifting the prefix residue masses of PEPTIDESTRING by – 243.18 Da, which equals – mass(PEPT)+mass(T[Ph]). (b) The restricted diagonal score between the prefix residue mass of PEPTIDESTRING and TIDEST[Ph]R is 6. The score is obtained by shifting the prefix residue masses of PEPTIDESTRING by – 323.15 Da = – mass(PEP).
The second method is based on restricted diagonal scores. The restricted diagonal score of A and B is the maximum mass counting score among all non-positive shifts whose absolute values equal a mass in A (Fig. 2(b)), denoted by R(A, B) = maxd ∈A C(shift(A, – d), B). For example, when A is the set of prefix residue masses {0, 97.05, 226.09} of the peptide PE, R(A, B) = max{C(shift(A, 0), B), C(shift(A, – 97.05), B), C(shift(A, – 226.09), B)}. A high restricted diagonal score between Ppre and Fpre means that F contains a long unmodified prefix that is a substring of P. For example, the proteoform TIDEST[Ph]R in Fig. 2(b) contains an unmodified prefix TIDES that is a substring of PEPTIDESTRING. In contrast, the restricted diagonal score between the prefix residue masses of T[Ph]IDEST[Ph]R and those of PEPTIDESTRING is 1 because T[Ph]IDEST[Ph]R does not have a long unmodified prefix. Similarly, a high restricted diagonal score between Psuf and Fsuf means that F contains a long unmodified suffix that is a substring of P. In the second method, the similarity score between a protein sequence P and a deconvoluted spectrum S is defined as R(Ppre, Spre)+R(Psuf, Ssuf ), which is determined by the unmodified prefix and suffix of the target proteoform. Different from the computation of a diagonal score, only a small number of mass shifts are considered to compute a restricted diagonal score. As a result, the chance that a random spectrum protein pair has a high restricted diagonal score is significantly reduced compared with a high diagonal score. However, when the target proteoform has two modifications: one at the N-terminus and the other at the C-terminus, using the restricted diagonal score may fail to retain the target database protein sequence in filtration. The second method is efficient for identifying proteoforms with a long unmodified prefix or suffix.
In the two filtering methods, the two similarity scores are used to rank proteins in the database, and the top t proteins are reported as filtering results. The scores are computed using index-based algorithms [32]. The two methods are called UPF-DIAGONAL (the diagonal score) and UPF-RESTRICT (the restricted diagonal score), respectively.
2.2 ASF algorithms
In bottom-up MS, variable PTMs are often incorporated into database peptides to identify modified peptides. However, this approach is inefficient for top-down MS (see Section Discussion). In the proposed ASF algorithms, we incorporate variable PTMs into the query spectrum to improve the efficiency and sensitivity of protein filtration.
We use phosphorylation as an example to explain how to generate an approximate spectrum. Let δ be the mass shift of phosphorylation. Let P = a1 … ai … an be an unmodified protein sequence (may be truncated) and F a modified form of P with one phosphorylation site on the amino acid ai. The theoretical prefix residue mass spectrum Ppre = {p0, p1, …, pi, pi+1 …, pn} contains all prefix residue masses of P and the theoretical spectrum Fpre contains all prefix residue masses of F, that is, Fpre = {p0, p1, …, pi + δ, pi+1 + δ, …, pn + δ}. We can convert Fpre into Ppre by deducting δ from the prefix residue masses pi + δ, pi+1 + δ, …, pn + δ.
Let Spre be a prefix residue mass spectrum generated from an experimental spectrum of F. The precursor mass of the experimental spectrum is M. The spectrum Spre is similar to Fpre, but has missing and noise peaks. To simplify the analysis, we assume that Spre is a perfect spectrum, that is, Spre = Fpre = {p0, p1, …, pi + δ, pi+1 + δ, …, pn + δ}. In the ASF method, we try to convert Spre into an approximate spectrum of Ppre with limited information (Fig. 1): it is known that the target proteoform contains a phosphorylation, but the target protein sequence and the location of the phosphorylation site are unknown.
Because the modification site is unknown, we give k guesses for the prefix residue mass pi, the smallest prefix residue mass with the modification, and hope that one of the guesses is similar to pi. The mass pn+δ in Spre is the residue mass of the target proteoform, which equals M – mass(H2O). We divide the mass pn + δ into k intervals (0, l], (l, 2l], …, ((k – 1)l, kl] each with the same length . The k centers of the intervals are the guessed values for pi. For example, when pn + δ = 5000 Da and k = 2, the two intervals are (0, 2500] and (2500, 5000], and the two centers are 1250 and 3750.
For each guessed prefix residue mass q, we convert Spre into a spectrum conv(Spre, q) by deducting δ from all masses in Spre that are no less than q. In Fig. 1, the guessed prefix residue mass is 200 Da and all masses no less than 200 Da are shifted to the left by 79.97 Da. When q < pi, all masses in the mass intervals (0, q) and [pi, pn + δ] are correctly converted into their corresponding masses in Ppre, and all masses in the mass interval [q, pi) are not correctly converted. In Fig. 1, peaks in the mass intervals (0, 200) and [546.14, 799.29] are correctly converted into peaks of TYPDSRP, but the left most peak in the box is not correctly converted. The ratio between the length of the interval [q, pi) and pn + δ is called the conversion error ratio of conv(Spre, q). When q > pi, all masses in the mass intervals (0, pi) and [q, pn + δ] are correctly converted into their corresponding masses in Ppre, and all masses in the mass interval [pi, q) are not correctly converted. The conversion error ratio of conv(Spre, q) is the ratio between the length of the interval [pi, q) and pn + δ. The distance between pi and the best guessed value q* is no larger than , and the conversion error ratio of conv(Spre, q*) is no larger than . When k is large, conv(Spre, q*) is almost the same as Ppre and is called an approximate prefix residue mass spectrum of P. In practice, although Spre has missing and noise peaks, it is converted into an approximate prefix residue mass spectrum of P using the same method. The above method is used to generated approximate suffix residue mass spectra as well.
Next we extend the method to generate approximate spectra for proteoforms with g > 1 variable PTM sites. When the target proteoform F is ultramodified and the number g is large, it is impractical to enumerate all approximate spectra with g PTM sites. Let F′ be a proteoform that is obtained from F by removing h variable PTM sites. By using h (h < g) variable PTM sites in spectral conversion, we generate an approximate spectrum of F′ from Spre. Although the resulting spectrum is not an approximate spectrum of the protein sequence P, it is more similar to the theoretical spectrum of P compared with Spre. We treat the remaining g – h PTM sites in F′ as unexpected PTMs. Note that h is a user-specified parameter and not related to the number of PTM sites in the target proteoform.
To generate approximate spectra, we first choose h interval centers (each of the k centers can be chosen multiple times) as the guessed values of the prefix residue masses corresponding to the h PTM sites, then enumerate all possible combinations of the types of variable PTMs on the sites. For each configuration of h guessed prefix residue masses and guessed PTM types, we convert the spectrum Spre into an approximation spectrum. The total number of configurations is proportional to (kf)h, where f is the number of variable PTM types in database search. The UPF-RESTRICT and UPF-DIAGONAL methods are employed to search these approximate spectra against the protein database to find candidate proteins. The ASF method coupled with UPF-RESTRICT is called the ASF-RESTRICT algorithm (Fig. S1). Detailed steps for Step 4 in the algorithm is given in Fig. S2 in the supplementary material. To couple the ASF method with UPF-DIAGONAL, we replace the UPF-RESTRICT algorithm with the UPF-DIAGONAL algorithm in Step 5 of the ASF-RESTRICT algorithm. The ASF method with the UPF-DIAGONAL algorithm is referred to as the ASF-DIAGONAL algorithm.
To guarantee the efficiency of the method, the values of k, f and h need to be small. In the experiments, k = 3 was chosen based on the evaluation of speed and sensitivity of the ASF algorithms with various settings of k (see Section Parameter settings), and h was set as 1 or 2. The number f of variable PTM types is a parameter specified by the user.
3 Results
3.1 Data sets
Four top-down MS data sets were used in this study: the first was generated from Escherichia coli (EC) K-12 MG1655, the second from purified human histone H3 protein, the third from purified human histone H4 protein, and the fourth from breast tumor xenograft samples.
The EC data set was obtained using a liquid chromatography system coupled with an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, Waltham, MA). MS and MS/MS spectra were collected at a 60 000 resolution. The top 4 ions in each MS spectrum were selected for MS/MS analysis and the alternating fragmentation mode was used. In total, 2 027 CID and 2 027 ETD top-down MS/MS spectra were collected [16].
The histone H3 data set [42] was obtained using an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, Waltham, MA). Core histones were separated in the first dimension using a Jupiter C5 column and further separated in the second dimension by a weak cation exchange hydrophilic interaction LC (WCX-HILIC) using a PolyCAT A column. All acquisitions were performed with a 60 000 resolution. In total, 3 462 CID and 3 462 ETD top-down MS/MS spectra were collected.
The histone H4 data set [15] was generated using an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, Waltham, MA). Core histones were separated by a 2-dimensional reversed-phase and hydrophilic interaction liquid chromatography (RP-HILIC) system where the histone H4 protein was isolated in the first dimension. With a resolution of 60 000, a total of top-down MS/MS 1 626 CID and 1 626 ETD spectra were acquired.
The breast tumor xenograft data set [43] was generated using an Orbitrap Elite mass spectrometer (Thermo Scientific, Waltham, MA). Cryopulverization of the tumor xenografts was performed using the standard CPTAC protocols [44]. A basal-like (WHIM2) breast cancer sample and a luminal B (WHIM16) breast cancer sample [45, 46] were used for the experiments. Protein separation was achieved using a commercial GELFREE 8100 fractionation system (Expedeon, Cambridge, UK). With a resolution of 60 000, a total of 51 474 and 50 372 higher-energy collisional dissociation (HCD) top-down MS/MS spectra were collected from the WHIM2 and WHIM16 samples respectively.
3.2 Simulated data set
To evaluate the accuracy and speed of the filtering algorithms, a test data set of PrSMs with mutations (treated as PTMs) was generated from the EC data set. The proteome database of Escherichia coli K-12 MG1655 was downloaded from the UniProt database [47] (version Sept 12, 2016, 4 306 entries) and concatenated with a shuffled decoy database of the same size. The 4 054 top-down MS/MS spectra were deconvoluted by MS-Deconv [39] and then searched against the target-decoy concatenated EC proteome database using TopPIC [16]. Parameter settings of TopPIC are given in Table S3 in the supplementary material. A total of 874 PrSMs without PTMs (529 from CID and 345 from ETD) were identified with a 1% spectrum-level false discovery rate (FDR). The 874 PrSMs can be found in Table S4 in the supplementary material, and the histogram of the lengths of the identified proteoforms is given in Fig. S3 in the supplementary material.
For each identified PrSM between a spectrum S and a protein sequence P with a score x, we used the generating function method [48, 49] to compute the conditional spectral probability that the similarity score between the spectrum S and a random protein sequence is no less than x on the condition that the molecular mass of the random protein matches the precursor mass of S. In the generating function method, a dynamic programming algorithm is employed to efficiently and accurately compute the distribution of the similarity scores between the spectrum S and random proteins as well as the conditional spectral probability. The histogram of the conditional spectral probabilities of the identified PrSMs is given in Fig. S4 in the supplementary material.
The 874 PrSMs without PTMs were used to generate test PrSMs with random mutations. Let (P, S) be a PrSM between a spectrum S and a protein sequence P without PTMs. We randomly select an amino acid in P, then replace it with a random amino acid, resulting in a protein sequence P′ with a mutation. The mass difference between the original amino acid and the new one is required to be larger than 5 Da. In addition, a random sequence with no more than 20 amino acids is appended to the N-terminus of P′ and another random sequence with no more than 20 amino acids to the C-terminus of P′. The PrSM between the resulting sequence and S contains a PTM (mutation), an N-terminal truncation, and a C-terminal truncation. Using this method, a total of 13 110 test PrSMs (15 test PrSMs for each of the 874 PrSMs: 5 without terminal truncation, 5 with only an N- or C-terminal truncation, and 5 with both N- and C-terminal truncations) were generated. In addition, PrSMs with 2, 3, 4, 5 mutations were generated using a similar method. When two or more PTMs (mutations) were added to a protein sequence, the random mutations were chosen independently and were different in most cases. A total of 65 550 PrSMs (13 110 for each setting of the mutation numbers 1, 2, 3, 4, 5) were generated. All the experiments on the simulated data set were performed on a desktop with an Intel Core i7-3770 Quad-Core 3.4 GHz CPU and 16 GB memory.
3.3 Parameter settings
We tested the ASF-RESTRICT and ASF-DIAGONAL algorithms with various settings of the parameters k and h on the simulated PrSMs with 5 PTMs. The error tolerance for computing diagonal scores and restricted diagonal scores was 15 ppm. For each test PrSM with a mutated protein sequence P′ and a spectrum S, we replaced the unmodified protein sequence of P′ in the EC proteome database with P′, then used the ASF algorithms to search S against the proteome database, and finally reported t = 20 candidate proteins. If the 20 candidate proteins contain protein P′, we say the filtration is efficient. The efficiency rate of the filtering algorithm is the ratio between the number of PrSMs with efficient filtration and the total number of test PrSMs.
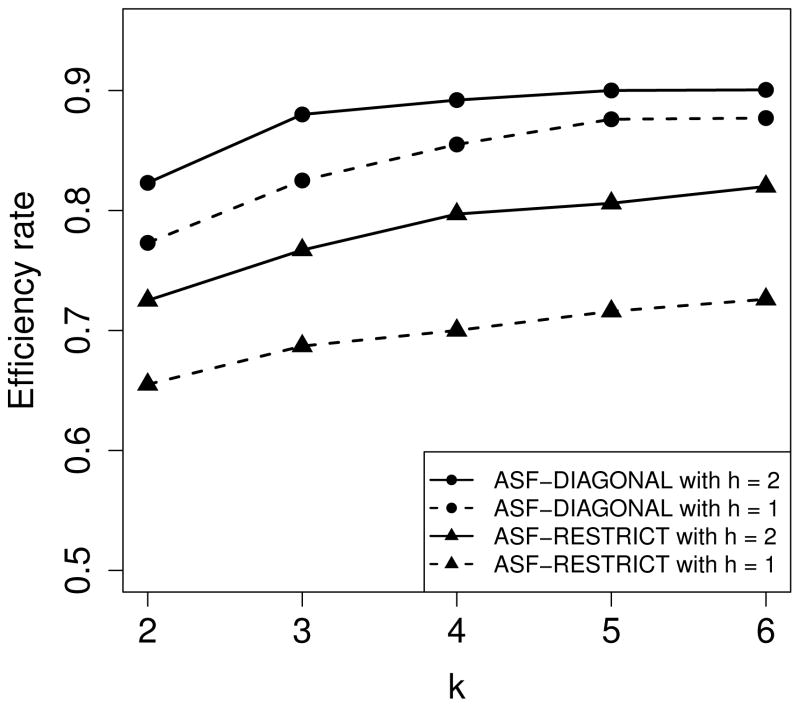
The efficiency rates and average running times (per spectrum) of the ASF algorithms with various settings for k = 2, 3, 4, 5,6 and h = 1, 2 are shown in Fig. 3 and Fig. S5 in the supplementary material. Removing two modification sites from the query spectrum (h = 2) achieved marginal improvement in the efficiency rate compared with removing one modification site (h = 1). However, the average running time of ASF-RESTRICT and ASF-DIAGONAL with h = 2 was more than 10 times slower than those with h = 1. When k increases, the efficiency rate increases, but the increase rate becomes less significant. In the ASF-based methods, each approximate spectrum is searched against the database sequentially, and the memory usage of the algorithms remains the same when the parameter settings of h and k increase and the number of generated approximate spectra increases. The memory usage of ASF-RESTRICT and ASF-DIAGONAL was less than 4 GB.
Figure 3.
The efficiency rates of the ASF algorithms with various settings k = 2, 3, 4, 5,6 and h = 1, 2 on the simulated PrSMs with 5 PTMs.
3.4 Evaluation on filtration efficiency
Two sequence tag-based filtering methods were compared with the UPF and ASF-based methods on the simulated PrSMs. The first method, which was employed in MS-Align+Tag [50], uses the long tag strategy. Long tags are first extracted from the query spectrum, then all length l (l = 4 in the experiments) substrings of the long tags are reported for protein sequence filtration. The second method, which is a part of MSPathFinder [21], extracts from the query spectrum all sequence tags with a length l between the minimum length lmin and the maximum length lmax, that is, lmin ≤ l ≤ lmax. In the experiment, lmin = 5 and lmax = 8. The two methods are called TAG-LONG (with the long tag strategy) and TAG-VAR (with tags of various lengths), respectively. Detailed description of the two tag-based methods can be found in the supplementary material.
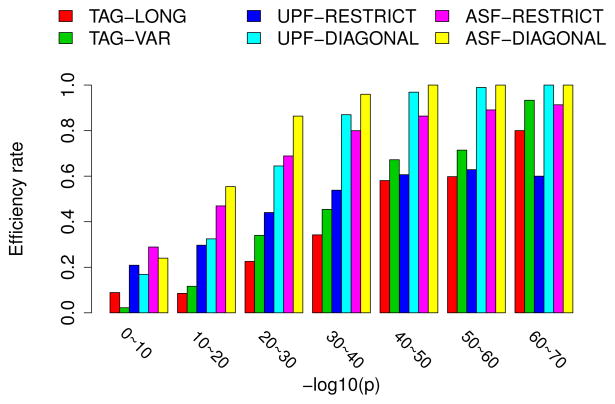
We tested the TAG-LONG, TAG-VAR, UPF-RESTRICT, UPF-DIAGONAL, ASF-RESTRICT and ASF-DIAGONAL algorithms on the simulated PrSMs with 5 PTMs. Parameter settings of the algorithms are given in Table S5 in the supplementary material. The ASF-DIAGONAL method achieved the best filtration efficiency rate 82.4%, while the filtration efficiency rates of the tag-based methods were below 40% and those of the UPF-based method were below 70% (Fig. S7 in the supplementary material). The ASF-DIAGONAL algorithm missed 528, 253, and 794 PrSMs efficiently filtered by UPF-RESTRICT, UPF-DIAGONAL, and ASF-RESTRICT, respectively (Fig. S8 in the supplementary material).
The efficiency rates of the filtering algorithms are related to the conditional spectral probabilities of test PrSMs (Fig. 4). Most PrSMs with a conditional spectral probability ≥ 10−30 have less than 30 matched masses, and protein sequence filtering for these PrSMs is more challenging than those with many matches masses. For PrSMs with a conditional spectral probability between 10−20 and 10−30, the efficiency rate of ASF-DIAGONAL was higher than 85%. For PrSMs with a conditional spectral probability between 10−10 and 10−20, the efficiency rate of the ASF-DIAGONAL algorithm was still higher than 50%. In addition, the filtration efficiency rates of ASF-based algorithms were similar on CID and ETD spectra (Fig. S9 in the supplementary material).
Figure 4.
Comparison of the filtration efficiency rates of the TAG-LONG, TAG-VAR, UPF-RESTRICT, UPF-DIAGONAL, ASF-RESTRICT and ASF-DIAGONAL algorithms on the simulated test PrSMs with 5 PTMs. The PrSMs are divided into 7 groups based on their conditional spectral probabilities p, and the efficiency rates for each group are compared.
The filtration efficiency rates of the algorithms for the simulated test PrSMs with 1, 2, 3, and 4 PTMs are shown in Fig. S10–S13 in the supplementary material. Because ASF-RESTRICT and ASF-DIAGONAL are designed for identifying proteoforms with multiple PTMs, they were not tested on the PrSMs with 1 PTM. ASF-RESTRICT outperformed the other algorithms on the test PrSMs with 2 or 3 PTMs, and ASF-DIAGONAL obtained the best performance on the test PrSMs with 4 or 5 PTMs. The main reason is that ASF-RESTRICT and ASF-DIAGONAL have complementary strengths in protein sequence filtration. When the proteoform that corresponds to the approximate spectrum contains only a small number of PTMs, it is highly possible that the proteoform has a long unmodified N-terminal or C-terminal fragment. Compared with ASF-DIAGONAL, ASF-RESTRICT is more efficient for identifying this type of proteoforms. ASF-DIAGONAL is more powerful than ASF-RESTRICT when the proteoform contains a long unmodified internal fragment. The experimental results show that combining the two methods can improve filtration efficiency.
The average running time of ASF-DIAGONAL (10.9 seconds) for one test PrSM was about 8 times of TAG-LONG (1.34 seconds) and TAG-VAR (1.35 seconds) and 13 times of UPF-DIAGONAL (0.85 seconds). Although ASF-DIAGONAL is slower than other filtering methods, its running time is still acceptable because the running time is similar to that of spectral alignment algorithms. The running time for aligning a mass spectrum with 20 candidate protein sequences is usually more than 20 seconds.
To test the filtering algorithms on large protein databases, we concatenated the EC proteoform database with the human proteome database downloaded from the UniProt database [47] (version Jul 9, 2016, 20 191 entries). The concatenated database contained 24 497 proteins. The filtration efficiency rates of ASF-RESTRICT and ASF-DIAGONAL were 61.6% and 70.6%, respectively, while those of the other four algorithms were below 55% (Fig. S14 in the supplementary material).
3.5 Evaluation on the histone data sets
The two human histone protein data sets were used to evaluate the filtering methods for identifying proteoforms with multiple PTMs. All the experiments on the histone data sets were performed on the same desktop used for the simulated data analyses. All the spectra of the histone H3 and H4 data sets were deconvoluted using MS-Deconv [39]. TopMG [18] was employed to align the histone H3 and H4 spectra against their corresponding histone H3 and H4 protein sequences. Five PTMs: acetylation, methylation, dimethylation, trimethylation, phosphorylation (Table S6 in the supplementary material) were used as variable PTMs in proteoform identification. Other parameter settings used in TopMG are given in Table S7 in the supplementary material. TopMG identified 3 205 and 1 087 PrSMs with at least 10 matched fragment ions from the histone H3 and H4 data sets, respectively (Table S8 and S9 in the supplementary material).
The tag-based, UPF-based, and ASF algorithms were tested on these identified PrSMs. For each identified PrSM of protein P and spectrum S, the filtering algorithm used the spectrum S to filter the UniProt human proteome database (version Jul 9, 2016, 20 191 entries) and reported 20 top candidate protein sequences. If the 20 protein sequences contain the target protein P (histone H3 or H4), the filtration is efficient. The five PTMs used in proteoform identification were treated as variable PTMs in the ASF algorithms, and parameter settings of the algorithms are provided in Table S5 in the supplementary material.
The filtration efficiency rates of the 6 filtering methods for the histone H3 and H4 PrSMs are summarized in Table 1. The filtration efficiency rates of the two tag-based methods were not as high as the UPF and ASF based methods. The main reason is that many spectra in the test PrSMs do not contain long consecutive fragment ions. The filtration efficiency rates of UPF-RESTRICT and ASF-RESTRICT were the highest among the 6 methods. Most of the histone H3 and H4 proteoforms have no more than 4 PTMs (Fig. S15(b) and Fig. S16(b) in the supplementary material), and most PTM sites on the histone H3 and H4 proteins lie in a short region near the N-terminus and can be treated as one large unexpected mass shift in protein filtering. UPF-RESTRICT and ASF-RESTRICT are efficient in filtering proteins for this type of spectra. As a result, ASF-RESTRICT outperformed ASF-DIAGONAL on the histone data sets. Compared with UPF-RESTRICT, ASF-RESTRICT improved the efficiency rate by about 9.7% for the histone H3 PrSMs and 2.6% for the histone H4 PrSMs. ASF-RESTRICT efficiently filtered 334 histone H3 PrSMs missed by UPF-RESTRICT and 1 094 histone H3 PrSMs missed by ASF-DIAGONAL (Fig. S17(a)). Similarly, ASF-RESTRICT outperformed ASF-DIAGONAL and UPF-RESTRICT on the histone H4 PrSMs (Fig. S17(b)). The Venn diagrams for the comparison of ASF-RESTRICT, ASF-DIAGONAL, TAG-LONG, and TAG-VAR can be found in Fig. S18 in the supplementary material. Compared with UPF-RESTRICT, ASF-RESTRICT achieved a better improvement on the histone H3 data set than the histone H4 data set. The main reason is that the quality of the histone H3 PrSMs is not as good as that of the histone H4 PrSMs. While 86.0% of the histone H3 PrSMs contain ≤ 25 matched fragment ions, only 29.7% of the histone H4 PrSMs contain ≤ 25 matched fragment ions (Fig. S15(a) and S16(a) in the supplementary material). Most of the PrSMs with ≤ 25 matched fragment ions have a relatively large conditional spectral probability. Compared with the UPF-based methods, the ASF algorithms achieve a better improvement in the filtration efficiency for PrSMs with large conditional spectral probabilities than those with very small ones (Fig. 4).
Table 1.
Comparison of the 6 filtering algorithms in the filtration efficiency rate using the 3 205 histone H3 PrSMs and the 1 087 histone H4 PrSMs
| H3 | H4 | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| # efficiently filtered PrSMs | Efficiency rate | Time (minutes) | # efficiently filtered PrSMs | Efficiency rate | Time (minutes) | |
| TAG-LONG | 210 | 6.6% | 91.8 | 563 | 51.8% | 73.9 |
| TAG-VAR | 415 | 13.0% | 92.1 | 583 | 53.6% | 73.9 |
| UPF-RESTRICT | 2019 | 63.0% | 35.7 | 1052 | 96.8% | 11.2 |
| UPF-DIAGONAL | 940 | 29.3% | 507.4 | 1014 | 93.3% | 87.7 |
| ASF-RESTRICT | 2313 | 72.2% | 400.7 | 1080 | 99.3% | 150.8 |
| ASF-DIAGONAL | 1235 | 38.5% | 4642.0 | 1036 | 95.3% | 1307.4 |
A total of 892 histone H3 PrSMs and 7 histone H4 PrSMs were missed by ASF-RESTRICT. The main reasons for inefficient filtration of these PrSMs are: (1) some PrSMs are of low quality and (2) some contain many PTM sites. Of the 899 histone PrSMs (892 histone H3 and 7 histone H4 PrSMs), 576 (64.1%) contain no more than 15 matched fragment ions. Of the other 323 PrSMs, 294 (91.0%) contain at least 4 variable PTM sites. Of the 29 remaining PrSMs, 28 have less than 22 matched fragment ions but more than 220 deconvoluted peaks and 1 has 125 deconvoluted peaks with 17 matched fragment ions, showing the low quality of the PrSMs.
The speed of the ASF algorithms is much slower than the other filtering methods. For the histone H3 data set, the running time of ASF-RESTRICT was about 11 times of UPF-RESTRICT, and the running time of ASF-DIAGONAL was about 11 times of ASF-RESTRICT and 130 times of UPF-RESTRICT. In practice, the ASF-based algorithms can be combined with other methods to speed up protein sequence filtration: fast filtering methods are used in the first round of spectral identification, and the ASF-based algorithms are employed to identify spectra that are elusive for the fast methods.
3.6 Phosphorylated proteoforms identified from the xenograft data set
The ASF algorithms were combined with TopMG [18] for proteome-wide complex proteoform identification. In the combined method, ASF-RESTRICT and ASF-DIAGONAL were employed to report top 20 candidate proteins separately for each query spectrum. The resulting proteins were aligned with the query spectrum using TopMG to find the best PrSM. We compared the performances of ProSightPC [7] and TopMG coupled with the ASF algorithms for identifying phosphorylated proteoforms on the breast cancer xenograft data set.
All the mass spectra from the WHIM2 and WHIM16 samples were deconvoluted by MS-Deconv [39]. Because the xenograft samples contain both mouse and human proteins, a multi-step database search approach was used for proteoform identification. While TopMG coupled with the ASF methods was used to identify phosphorylated proteoforms, TopPIC [16] was used to identify proteoforms without variable PTMs. The experiments were performed on a node with two 12-core Intel Xeon E5-2680 v3 CPUs and 256 GB memory on Carbonate, a parallel computing system at Indiana University. A total of 12 threads were used in the analysis. The running time for analyzing all the spectra was about 63 hours (3 hours for TopPIC and 60 hours for TopMG), of which 30 hours were used by the ASF algorithms. When multiple threads are used, the memory usage of the ASF algorithms is proportional to the number of threads. The maximum memory usage for analyzing the xenograft data set was 48 GB (4 GB for each thread).
Proteoforms identified by ProSightPC were obtained from a previous study [43], in which a customized version of cRAWler was used for spectral deconvolution and a five step database search was performed for proteoform identification. The third and fourth steps were to identify proteoforms with sample specific mutations and splicing events; the fifth step was to identify proteoforms with unexpected alterations. Because the last three steps were not designed to identify proteoforms with variable PTMs, we focused on only proteoforms identified in the first two steps.
Mouse proteoforms
In the first step of the ProSightPC analysis, the absolute mass mode was used to search all the deconvoluted spectra against a mouse proteoform database including proteoforms with PTMs, which was built based on the UniProt mouse proteome database (version May 2014) and its annotations. The error tolerances for precursor and fragment masses were set as 2.2 Da and 10 ppm, respectively. With a p-value cutoff 10−10, this step reported 648 proteoforms from 54 proteins, including 41 proteoforms without PTMs (N-terminal acetylation is allowed) and 24 phosphorylated proteoforms from 14 proteins. Some reported phosphorylated proteoforms are of the same protein and their precursor masses are the same (within an error tolerance). The only difference of these proteoforms is the locations of phosphorylation sites. The 24 phosphorylated proteoforms correspond to 15 distinct precursor masses.
In the first step of the analysis of TopPIC and TopMG, the mouse proteome database was downloaded from the UniProt database (version Nov 13, 2016, 16 840 entries) and concatenated with a shuffled decoy database of the same size. We first used TopPIC to search all the deconvoluted spectra against the target-decoy mouse database to identify proteoforms without variable PTMs and unexpected alterations (terminal truncations and N-terminal acetylation are allowed), then used TopMG to search the spectra unidentified by TopPIC against the database to identify phosphorylated proteoforms. In TopPIC, the error tolerances for precursor and fragment masses were set as 10 ppm. In the ASF algorithms, the parameter h was set as 1 and the error tolerance for computing filtering scores was set as 10 ppm. In TopMG, the error tolerances for precursor and fragment masses were set as 10 ppm and 0.1 Da respectively, and phosphorylation was used as the variable PTM. Other parameter settings of TopPIC and TopMG are given in Tables S10 and S11 in the supplementary material. With a 5% proteoform-level FDR, TopPIC identified 122 proteoforms from 105 proteins, and TopMG identified 45 proteoforms, including 41 phosphorylated proteoforms from 27 proteins and 4 proteoforms without phosphorylation sites (Tables S12–S14 in the supplementary material). The reason that the 4 unmodified proteoforms were missed by TopPIC is that TopPIC used a more stringent error tolerance for fragment masses compared with TopMG. Most of the identified phosphorylated proteoforms contain ≤ 3 phosphorylation sites (Fig. S19(a) in the supplementary material).
A total of 21 proteoforms without variable PTMs (some may contain terminal truncations and N-terminal acetylation) were identified by both ProSightPC and TopPIC. In addition, TopPIC identified 101 proteoforms missed by ProSightPC (Fig. S20(a) in the supplementary material). Because the spectral scan numbers of the proteoforms reported by ProSightPC were not available, we matched the molecular masses of the proteoforms to the precursor masses of the spectra reported by MS-Deconv with an error tolerance 2.2 Da to find candidate PrSMs. Of the 20 proteoforms missed by TopPIC, MS-Deconv failed to report corresponding deconvoluted spectra for 4 proteoforms. The molecular masses of the other 16 proteoforms were matched to the precursor masses of 242 deconvoluted spectra, but their corresponding PrSMs were not reported by TopPIC because their E-values were not highly significant. One main reason that ProSightPC missed many proteoforms identified by TopPIC is that truncations were not allowed in the first step of the ProSightPC analysis.
ProSightPC reported several proteoforms with the same molecular mass, but different PTM sites. Because it is a challenging problem to confidently localize PTM sites in top-down spectral identification, we decided not to directly compare proteoforms reported by the two tools. If a proteoform reported by ProSightPC and a proteoform reported by TopMG are of the same protein and have the same precursor mass (within an error tolerance), we say the two proteoforms match. We compared the numbers of distinct precursor masses corresponding to the proteoforms, not the numbers of proteoforms, reported by ProSightPC and TopMG. A total of 38 and 15 distinct precursor masses were reported by TopMG and ProSightPC, respectively. Only one phosphorylated proteoform (corresponding to one precursor mass) was reported by both TopMG and ProSightPC (Fig. S21(a)). Of the remaining 23 phosphorylated proteoforms (14 precursor masses) reported by ProSightPC, 4 did not have matched deconvoluted spectra reported by MS-Deconv, and 19 were matched to deconvoluted spectra, but their corresponding PrSMs were not reported by TopMG. ProSightPC missed many proteoforms reported by TopMG because the proteoform database (data warehouse) used in ProSightPC was incomplete. The proteoforms identified by TopMG include 37 highly confident ones with an E-value smaller than 10−10 (Fig. S19(b) in the supplementary material). Four proteoforms with significant E-values are provided in Fig. S22–S25 in the supplementary material. These identified proteoforms show that TopMG is efficient in identifying novel phosphorylated proteoforms.
Human proteoforms
In the second step of the ProSightPC analysis, the absolute mass and biomarker modes were used to search the spectra unidentified in the first step against a human proteoform database, which was built based on the human RefSeq database and protein annotations. The error tolerance for precursor masses was set as 2.2 Da in the absolute mass mode and 10 ppm in the biomarker mode; the error tolerance for fragment masses was set as 10 ppm in the two search modes. With a p-value cutoff 10−10, ProSightPC identified 685 proteoforms from 150 proteins1, including 147 proteoforms without PTMs (N-terminal acetylation is allowed) and 98 phosphorylated proteoforms from 26 proteins. The 98 phosphorylated proteoforms are matched to 35 distinct precursor masses.
In the second step of the analysis of TopPIC and TopMG, the human proteome database (version Jul 9, 2016, 20 191 entries) was downloaded from UniProt and concatenated with a shuffled decoy database with the same size. Using the same parameters in the first step, the spectra unidentified in the first step were searched against the human target-decoy database using TopPIC and TopMG. TopPIC identified 265 proteoforms from 190 proteins without variable PTMs, and TopMG identified 91 proteoforms from 64 proteins, including 82 phosphorylated proteoforms from 59 proteins (Tables S15–S17 in the supplementary material). Similar to the first step, most of the identified phosphorylated proteoforms contain ≤ 3 phosphorylation sites (Fig. S26(a) in the supplementary material).
The human database search of TopPIC identified 85 of the 147 human proteoforms without PTMs (except for terminal truncations and N-terminal acetylation) reported by ProSightPC (Fig. S20(b) in the supplementary material). Of the 62 proteoforms missed by TopPIC, 13 were identified by TopPIC in the mouse database search because they are the same as their homologous mouse proteins. Similar to mouse proteoforms, the main reasons for the remaining 49 proteoforms missed by TopPIC are the missing of matched deconvoluted spectra and large E-values of PrSMs. TopPIC also identified 180 proteoforms missed by ProSightPC.
A total of 80 and 35 distinct precursor masses were reported by TopMG and ProSightPC, including 14 ones reported by both the two tools (Fig. S21(b)). The proteoforms identified by TopMG include 47 proteoforms with an E-value smaller than 10−10 (Fig. S26(b) in the supplementary material). Four proteoforms with significant E-values are provided in Fig. S27–S30 in the supplementary material. Similar to the comparison on mouse phosphorylated proteoforms, TopMG identified many phosphorylated human proteoforms missed by the absolute mass and biomarker modes of ProSightPC. All the annotated PrSMs reported by TopPIC and TopMG can be found in the supplementary material.
4 Discussion and conclusions
In this paper, we proposed two ASF algorithms for protein filtration in proteoform identification by top-down MS and evaluated the performances of the ASF algorithms as well as two tag-based and two UPF-based filtering algorithms on simulated and real top-down MS data sets. The experimental results showed that the UPF-based filtering algorithms outperformed the tag-based algorithms and that the ASF algorithms achieved the best performance among the 6 evaluated algorithms in filtration efficiency. The ASF algorithms are efficient when the target proteoform contains truncations as well as many variable PTMs and/or unknown alterations. Specifically, the filtration efficiency of ASF-DIAGONAL is much higher than other methods for spectra with low sequence coverage. Although the ASF algorithms are the slowest, their speed is still acceptable in proteoform identification.
Both ASF-RESTRICT and ASF-DIAGONAL use approximate spectra in protein filtration, but they are designed for different scenarios. ASF-RESTRICT has a smaller search space than ASF-DIAGONAL. While the filtration efficiency of ASF-RESTRICT depends on if the corresponding proteoform of the approximate spectrum contains a long unmodified prefix or suffix, the filtration efficiency of ASF-DIAGONAL depends on if the corresponding proteoform of the approximate spectrum contains a long unmodified fragment (a prefix, a suffix, or an internal one). In practice, we suggest combining the two algorithms to achieve good filtration efficiency.
The parameters h, f, and k determine the search space, running time, and filtration efficiency of the ASF algorithms. When h, f, and k increases, the search space and running time increase. The experimental results demonstrate that using one variable PTM site in approximate spectrum generation (h = 1) significantly improves filtration efficiency for complex proteoforms with multiple variable PTMs compared with UPF-based methods. While using h = 2 achieves marginal improvement in filtration efficiency compared with h = 1, it significantly increases the running time. We suggest using h = 1 in most cases. When only one or two types of variable PTMs are used (f = 1 or 2) and many proteoforms are highly modified, h = 2 can be used to further improve filtration efficiency. To guarantee that the ASF algorithms are fast in protein filtration, we suggest that the settings of k and f should be no more than 5.
The ASF algorithms are proposed for proteoform identification in proteome-level proteomics studies in which all proteoforms in the sample are analyzed in an MS experiment. The types of PTMs of interest are known in many proteome-level proteomics studies. For example, phosphorylation is the PTM of interest and chosen as the variable PTM in the studies of phosphoproteins. In the discovery mode analysis, the types of PTMs of interest are unknown and it is a challenging problem to anticipate the types of PTMs that will be identified in proteoforms. To solve the problem, we first use spectral alignment algorithms, such as TopPIC, to identify proteoforms with mass shifts corresponding to unexpected alterations. If the number of occurrences of a specific mass shift, e.g. 80 Da, in identified proteoforms is large and the mass shift is explained by a PTM (80 Da is explained by phosphorylation), then we use the PTM as a variable one in the second round of database search to find proteoforms with the PTM.
The number of variable PTM types needs to be small to guarantee the fast speed of the ASF algorithms. A proteome level MS analysis may identify more than 10 types of PTMs, but each proteoform often contains only one or two types of PTMs. To identify these proteoforms, we can perform multiple rounds of database searches, and a small number of variable PTM types are selected in each round.
A proteoform may contain various alterations including terminal truncations, sequence mutations, fixed PTMs, variable PTMs, and unexpected alterations. The ASF algorithms are capable of filtering spectra of proteoforms with truncations, fixed PTMs, variable PTMs, and unexpected alterations. When sample specific protein databases are not available, sequence mutations are treated as unexpected alterations in protein filtration. When RNA-Seq data of the sample are available, sequence mutations obtained from RNA-Seq data can be incorporated into sample specific protein databases to improve filtration efficiency. When the target proteoform contains many variable PTM sites, most of them are treated as unexpected alterations in filtration because approximate spectra usually remove only one or two variable PTM sites (h = 1 or 2) in the proteoform.
Unexpected alterations and the alterations that are treated as unexpected ones in filtration are called filtration blind alterations. The number and locations of filtration blind alterations affect the filtration efficiency of the ASF algorithms. In general, the filtration efficiency decreases when the number of filtration blind alterations increases. ASF-DIAGONAL filters proteins using a long unmodified protein fragment. When a proteoform with many filtration bind alterations has a long fragment free of filtration bind alterations, it is highly possible that ASF-DIAGONAL is efficient for the proteoform. Similarly, when a proteoform with many filtration blind alterations contains a long prefix or suffix free of filtration blind alterations, it is highly possible that ASF-RESTRICT is efficient for the proteoform.
In proteome-level proteomics studies, proteoforms can be divided into three groups: (1) proteoforms with only variable PTMs, (2) proteoforms with only filtration blind alterations, and (3) proteoforms with both variable PTMs and filtration blind alterations. The ASF algorithms are designed to improve the sensitivity in proteoform identification in groups (1) and (3), but not in group (2). That is, the ASF algorithms work well for proteoforms with only variable PTMs, and those with both variable PTMs and unexpected alterations, not for proteoforms with only unexpected alterations.
In the ASF algorithms, the query spectrum is transformed into an approximate spectrum to reduce the number of variable PTMs in the match between the target database sequence and the spectrum. An alternative method is to incorporate variable PTMs into database sequences to generate a proteoform database. This approach has been widely used in PTM identification in bottom-up MS, but it is inefficient in top-down MS. Proteoforms analyzed in top-down MS are generally longer than peptides in bottom-up MS. Because long proteins often contain many possible modification sites, the size of a proteoform database may be extremely large. For example, when phosphorylation is the only variable PTM and one or two PTM sites (h = 2) are incorporated into each proteoform, the size of the proteoform database increases by more than 100 times compared with the original one.
The proposed ASF algorithms have some limitations. The first limitation is that the running time of the algorithms is an exponential function of the parameter h. In practice, a small number h (h = 1 or 2) is used to reduce the running time of the algorithms, limiting its ability to identify complex proteoforms with many variable PTM sites. The second limitation is that the ASF algorithms are inefficient for proteoforms with many PTM types. Using a large number (> 5) of variable PTM types significantly increases the running time of the algorithms. The third limitation is that peak intensities are ignored in computing diagonal scores and restricted diagonal scores. Incorporating peak intensities into similarity scores can further improve the performance of the filtering algorithms.
Availability
The proposed ASF algorithms have been integrated into TopMG, which is available at http://proteomics.informatics.iupui.edu/software/topmg/. The source code is available at https://github.com/toppic-suite/toppic-suite.
Supplementary Material
Significance of the study.
Identifying proteoforms with primary structural alterations is essential to understanding protein functions and related biological processes. In this study, we present new protein sequence filtering algorithms that outperform existing ones for top-down mass spectrometry-based proteoform identification. Combining the filtering algorithms and existing spectral alignment algorithms will significantly improve the sensitivity in proteoform identification and facilitate the studies of proteoforms with alterations.
Acknowledgments
The research was supported by the National Institute of General Medical Sciences, National Institutes of Health (NIH) through Grant R01GM118470. The authors declare no competing financial interest.
Footnotes
The supplementary Table S1 in Ref. [43] shows that the second step identified a mouse protein RS30, which may be an error in the table.
References
- 1.Smith LM, Kelleher NL Consortium for Top Down Proteomics. Proteoform: a single term describing protein complexity. Nature Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong X, Sumandea CA, Chen YC, Garcia-Cazarin ML, Zhang J, Balke CW, Sumandea MP, Ge Y. Augmented phosphorylation of cardiac troponin I in hypertensive heart failure. Journal of Biological Chemistry. 2012;287:848–857. doi: 10.1074/jbc.M111.293258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 4.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nature methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 5.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Molecular & Cellular Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catherman AD, Skinner OS, Kelleher NL. Top down proteomics: facts and perspectives. Biochemical and Biophysical Research Communications. 2014;445:683–93. doi: 10.1016/j.bbrc.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, Viswanathan V, Spaulding IT, Early BP, Bluhm EJ, Babai S, Kelleher NL. ProSight PTM 2.0: improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Research. 2007;35(Web Server issue):W701–W706. doi: 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank AM, Pesavento JJ, Mizzen CA, Kelleher NL, Pevzner PA. Interpreting top-down mass spectra using spectral alignment. Analytical Chemistry. 2008;80:2499–2505. doi: 10.1021/ac702324u. [DOI] [PubMed] [Google Scholar]
- 9.Tsai YS, Scherl A, Shaw JL, MacKay CL, Shaffer SA, Langridge-Smith PRR, Goodlett DR. Precursor ion independent algorithm for top-down shotgun proteomics. Journal of the American Society for Mass Spectrometry. 2009;20:2154–2166. doi: 10.1016/j.jasms.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Karabacak NM, Li L, Tiwari A, Hayward LJ, Hong P, Easterling ML, Agar JN. Sensitive and specific identification of wild type and variant proteins from 8 to 669 kDa using top-down mass spectrometry. Molecular & Cellular Proteomics. 2009;8:846–856. doi: 10.1074/mcp.M800099-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong W, Théberge R, Infusini G, Perlman DH, Costello CE, McComb ME. BUPID-top-down: database search and assignment of top-down MS/MS data. Proceedings of the 57th American Society Conference on Mass Spectrometry and Allied Topics; Philadelphia, PA. 2009. [Google Scholar]
- 12.Liu X, Sirotkin Y, Shen Y, Anderson G, Tsai YS, Ting YS, Goodlett DR, Smith RD, Bafna V, Pevzner PA. Protein identification using top-down spectra. Molecular & Cellular Proteomics. 2012;11:M111.008524. doi: 10.1074/mcp.M111.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Current Protocols in Bioinformatics. 2012;Chapter 13(Unit 13):20. doi: 10.1002/0471250953.bi1320s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Tian Z. Interpreting raw biological mass spectra using isotopic mass-to-charge ratio and envelope fingerprinting. Rapid Communications in Mass Spectrometry. 2013;27:1267– 1277. doi: 10.1002/rcm.6565. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Hengel S, Wu S, Tolić N, Paša-Tolić L, Pevzner PA. Identification of ultramodified proteins using top-down tandem mass spectra. Journal of Proteome Research. 2013;12:5830–5838. doi: 10.1021/pr400849y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kou Q, Xun L, Liu X. TopPIC: a software tool for top-down mass spectrometry-based proteoform identification and characterization. Bioinformatics. 2016;32:3495–3497. doi: 10.1093/bioinformatics/btw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun RX, Luo L, Wu L, Wang RM, Zeng WF, Chi H, Liu C, He SM. pTop 1.0: A high-accuracy and high-efficiency search engine for intact protein identification. Analytical Chemistry. 2016;88:3082–90. doi: 10.1021/acs.analchem.5b03963. [DOI] [PubMed] [Google Scholar]
- 18.Kou Q, Wu S, Tolić N, Paša-Tolić L, Liu Y, Liu X. A mass graph-based approach for the identification of modified proteoforms using top-down tandem mass spectra. Bioinformatics. 2016;33:1309–1316. doi: 10.1093/bioinformatics/btw806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai W, Guner H, Gregorich ZR, Chen AJ, Ayaz-Guner S, Peng Y, Valeja SG, Liu X, Ge Y. MASH Suite Pro: A comprehensive software tool for top-down proteomics. Molecular & Cellular Proteomics. 2016;15:703–714. doi: 10.1074/mcp.O115.054387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shortreed MR, Frey BL, Scalf M, Knoener RA, Cesnik AJ, Smith LM. Elucidating proteoform families from proteoform intact-mass and lysine-count measurements. Journal of Proteome Research. 2016;15:1213–1221. doi: 10.1021/acs.jproteome.5b01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J, Piehowski PD, Wilkins C, Zhou M, Mendoza J, Fujimoto GM, Gibbons BC, Shaw JB, Shen Y, Shukla AK, Moore RJ, Liu T, Petyuk VA, Tolić N, Paša-Tolić L, Smith RD, Payne SH, Kim S. Informed-Proteomics: open-source software package for top-down proteomics. Nature Methods. 2017;14:909–914. doi: 10.1038/nmeth.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chick JM, Kolippakkam D, Nusinow DP, Zhai B, Rad R, Huttlin EL, Gygi SP. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nature Biotechnology. 2015;33:743–749. doi: 10.1038/nbt.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Analytical chemistry. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 24.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Analytical Chemistry. 2005;77:4626–39. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 25.Frank A, Tanner S, Bafna V, Pevzner P. Peptide sequence tags for fast database search in mass-spectrometry. Journal of Proteome Research. 2005;4:1287–1295. doi: 10.1021/pr050011x. [DOI] [PubMed] [Google Scholar]
- 26.Cao X, Nesvizhskii AI. Improved sequence tag generation method for peptide identification in tandem mass spectrometry. Journal of Proteome Research. 2008;7:4422–4434. doi: 10.1021/pr800400q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabb DL, Ma Z-Q, Martin DB, Ham A-JL, Chambers MC. DirecTag: accurate sequence tags from peptide MS/MS through statistical scoring. Journal of Proteome Research. 2008 Sep;7(9):3838–3846. doi: 10.1021/pr800154p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Gupta N, Bandeira N, Pevzner PA. Spectral dictionaries integrating de novo peptide sequencing with database search of tandem mass spectra. Molecular & Cellular Proteomics. 2009;8:53–69. doi: 10.1074/mcp.M800103-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong K, Kim S, Bandeira N, Pevzner PA. Gapped spectral dictionaries and their applications for database searches of tandem mass spectra. Molecular & Cellular Proteomics. 2011;10:M110–002220. doi: 10.1074/mcp.M110.002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng F, Wang L, Liu X. An efficient algorithm for the blocked pattern matching problem. Bioinformatics. 2014;31:532–538. doi: 10.1093/bioinformatics/btu678. [DOI] [PubMed] [Google Scholar]
- 31.Shen Y, Tolić N, Hixson KK, Purvine SO, Anderson GA, Smith RD. De novo sequencing of unique sequence tags for discovery of post-translational modifications of proteins. Analytical Chemistry. 2008;80:7742–7754. doi: 10.1021/ac801123p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Mammana A, Bafna V. Speeding up tandem mass spectral identification using indexes. Bioinformatics. 2012;28:1692–1697. doi: 10.1093/bioinformatics/bts244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi H, He K, Yang B, Chen Z, Sun RX, Fan SB, Zhang K, Liu C, Yuan ZF, Wang QH, Liu SQ, Dong MQ, He S-M. pFind-Alioth: A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. Journal of Proteomics. 2015;125:89–97. doi: 10.1016/j.jprot.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nature Methods. 2017;14:513–520. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. Journal of the American Society for Mass Spectrometry. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 36.Zabrouskov V, Senko MW, Du Y, Leduc RD, Kelleher NL. New and automated MSn approaches for top-down identification of modified proteins. Journal of the American Society for Mass Spectrometry. 2005;16:2027–2038. doi: 10.1016/j.jasms.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics. 2008;24:1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carvalho PC, Xu T, Han X, Cociorva D, Barbosa VC, Yates JR., III YADA: a tool for taking the most out of high-resolution spectra. Bioinformatics. 2009;25:2734–2736. doi: 10.1093/bioinformatics/btp489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Inbar Y, Dorrestein PC, Wynne C, Edwards N, Souda P, Whitelegge JP, Bafna V, Pevzner PA. Deconvolution and database search of complex tandem mass spectra of intact proteins: a combinatorial approach. Molecular & Cellular Proteomics. 2010;9:2772–2782. doi: 10.1074/mcp.M110.002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slysz GW, Baker ES, Shah AR, Jaitly N, Anderson GA, Smith RD. The DeconTools framework: an application programming interface enabling flexibility in accurate mass and time tag workflows for proteomics and metabolomics. Proceedings of the 58th American Society Conference on Mass Spectrometry and Allied Topics; 2010. [Google Scholar]
- 41.Kou Q, Wu S, Liu X. A new scoring function for top-down spectral deconvolution. BMC Genomics. 2014;15:1140. doi: 10.1186/1471-2164-15-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian Z, Tolić N, Zhao R, Moore RJ, Hengel SM, Robinson EW, Stenoien DL, Wu S, Smith RD, Paša-Tolić L. Enhanced top-down characterization of histone post-translational modifications. Genome Biology. 2012;13:R86. doi: 10.1186/gb-2012-13-10-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ntai I, LeDuc RD, Fellers RT, Erdmann-Gilmore P, Davies SR, Rumsey J, Early BP, Thomas PM, Li S, Compton PD, Ellis MJC, Ruggles KV, Fenyö D, Boja ES, Rodriguez H, Townsend RR, Kelleher NL. Integrated bottom-up and top-down proteomics of patient-derived breast tumor xenografts. Molecular & Cellular Proteomics. 2016;15:45–56. doi: 10.1074/mcp.M114.047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, Clauser KR, Qiao JW, Gritsenko MA, Moore RJ, Levine DA, Townsend R, Erdmann-Gilmore P, Snider JE, Davies SR, Ruggles KV, Fenyo D, Kitchens RT, Li S, Olvera N, Dao F, Rodriguez H, Chan DW, Liebler D, White F, Rodland KD, Mills GB, Smith RD, Paulovich AG, Ellis M, Carr SA. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Molecular & Cellular Proteomics. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, DeSchryver K, Davies S, Guintoli T, Lin L, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Jr, Ivanovich DMT, Goodfellow JL, Perou PJ, Weinstock CM, Aft GM, Watson R, Ley M, Wilson TJ, Mardis RK, ER Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, Ding L, Griffith OL, Miller C, Larson D, Fulton RS, Harrison M, Mooney T, McMichael JF, Luo J, Tao Y, Goncalves R, Schlosberg C, Hiken JF, Saied L, Sanchez C, Giuntoli T, Bumb C, Cooper C, Kitchens RT, Lin A, Phommaly C, Davies SR, Zhang J, Kavuri MS, McEachern D, Dong YY, Ma C, Pluard T, Naughton M, Bose R, Suresh R, McDowell R, Michel L, Aft R, Gillanders W, DeSchryver K, Wilson RK, Wang S, Mills GB, Gonzalez-Angulo A, Edwards JR, Maher C, Perou CM, Mardis ER, Ellis MJ. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Reports. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Research. 2015;43(D1):D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. Journal of Proteome Research. 2008;7(8):3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Segar MW, Li SC, Kim S. Spectral probabilities of top-down tandem mass spectra. BMC genomics. 2014;15:S9. doi: 10.1186/1471-2164-15-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MS-Align+Tag. 2012 http://bioinf.spbau.ru/proteomics/ms-align-plus-tag.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.