NMDA receptor agonists have been used for many years to generate animal models of polymicrogyria, a malformation of cortical development. Fry et al. identify de novo GRIN1 mutations in eleven patients with severe bilateral polymicrogyria. Polymicrogyria-associated GRIN1 mutations cluster in specific protein domains and significantly alter NMDA receptor function.
Keywords: polymicrogyria, GRIN1, GluN1, NR1, N-methyl-d-aspartate receptor
Abstract
Polymicrogyria is a malformation of cortical development. The aetiology of polymicrogyria remains poorly understood. Using whole-exome sequencing we found de novo heterozygous missense GRIN1 mutations in 2 of 57 parent-offspring trios with polymicrogyria. We found nine further de novo missense GRIN1 mutations in additional cortical malformation patients. Shared features in the patients were extensive bilateral polymicrogyria associated with severe developmental delay, postnatal microcephaly, cortical visual impairment and intractable epilepsy. GRIN1 encodes GluN1, the essential subunit of the N-methyl-d-aspartate receptor. The polymicrogyria-associated GRIN1 mutations tended to cluster in the S2 region (part of the ligand-binding domain of GluN1) or the adjacent M3 helix. These regions are rarely mutated in the normal population or in GRIN1 patients without polymicrogyria. Using two-electrode and whole-cell voltage-clamp analysis, we showed that the polymicrogyria-associated GRIN1 mutations significantly alter the in vitro activity of the receptor. Three of the mutations increased agonist potency while one reduced proton inhibition of the receptor. These results are striking because previous GRIN1 mutations have generally caused loss of function, and because N-methyl-d-aspartate receptor agonists have been used for many years to generate animal models of polymicrogyria. Overall, our results expand the phenotypic spectrum associated with GRIN1 mutations and highlight the important role of N-methyl-d-aspartate receptor signalling in the pathogenesis of polymicrogyria.
Introduction
Malformations of cortical development (MCDs) are a spectrum of brain abnormalities that occur due to disruption of the intricate developmental processes that form the cerebral cortex. Although rare, MCDs have a major impact on the lives of patients and their families. Polymicrogyria is a subtype of MCD characterized, macroscopically, by an excessive number of small cortical folds (gyri). At a microscopic level, polymicrogyria is associated with abnormal cortical architecture including thinning or loss of cortical layers (Squier and Jansen, 2014; Jansen et al., 2016). The clinical effects of polymicrogyria depend on the extent and regions of the brain affected. Common consequences include intellectual disability, epilepsy, spasticity and cortical visual impairment (Stutterd and Leventer, 2014). Polymicrogyria can result from non-genetic events such as hypoxic-ischaemic insults or congenital infections. In addition, a range of chromosomal and single-gene disorders have been identified in polymicrogyria patients (Jansen and Andermann, 2005; Stutterd and Leventer, 2014). Despite these discoveries, the underlying cause of the malformation remains unknown in the majority of patients.
Polymicrogyria is usually a sporadic disorder, with most patients having no family history. Polymicrogyria also causes a significant loss of reproductive fitness. This suggests a role for de novo mutations in the aetiology of the disorder. Recent candidate gene and exome sequencing studies have shown that de novo mutations cause polymicrogyria in some patients (Jaglin et al., 2009; Riviere et al., 2012; Mirzaa et al., 2014). Based on these observations we took the approach of exome sequencing in a cohort of 57 parent-offspring trios. This strategy led us to identify two likely causal variants in GRIN1 in patients with extensive bilateral polymicrogyria. GRIN1 encodes GluN1, the obligatory subunit of the N-methyl-d-aspartate (NMDA) receptor, an ionotropic glutamate receptor that is highly expressed in the foetal brain (Law et al., 2003). Mutations in GRIN2B, a gene that encodes a different NMDA receptor subunit, have recently been reported in MCD patients (Platzer et al., 2017). Therefore, having observed two de novo GRIN1 mutations in patients with polymicrogyria, we searched for additional GRIN1 mutations in MCD patients. Furthermore, we examined the functional impact of polymicrogyria-associated GRIN1 mutations by computer-based protein structure modelling and in vitro electrophysiological analysis.
Materials and methods
Patients
Patients 1 and 2 were part of a cohort of 57 unrelated proband-parent trios recruited from Clinical Genetic and Paediatric Neurology clinics around the UK. All probands had polymicrogyria demonstrated by MRI and confirmed by review of the neuroradiology. The probands had no known cause for their polymicrogyria and normal array comparative genome hybridization. Parents were judged to be unaffected based on history and a brief physical examination. Most parents had not undergone brain imaging. For more details about the UK cohort see Supplementary Table 1. The study was approved by the Research Ethics Committee for Wales (09/MRE09/51). Informed consent was obtained from all participants (or their parents/legal guardians) prior to testing. Patients 3, 4, and 6–11 were ascertained through a request to collaborators and members of the European Network on Brain malformations looking for similar patients. Patients 3 and 4 underwent trio-based whole exome sequencing as part of an ongoing research program in France. Patients 6–8 had trio-based exome sequencing performed during their clinical diagnostic workup. Patients 9–11 were part of a cohort of 211 polymicrogyria patients who underwent targeted sequencing of the GRIN1 gene as part of a US-based research program. Patient 5 had trio-based whole exome sequencing performed on a clinical basis. He was ascertained due to appearing in a poster at the European Paediatric Neurology Society Congress 2015.
Exome sequencing and in silico prediction
Standard approaches to exome sequencing and variant filtering were used. Detailed descriptions are given in the Supplementary material. Predictions of the functional impact of the de novo mutations were made by a range of analysis programs. These included PhyloP (Pollard et al., 2010), SIFT (Kumar et al., 2009), PolyPhen-2 (Adzhubei et al., 2010), MutationTaster (Schwarz et al., 2014), CADD v1.3 (Kircher et al., 2014) and M-CAP (Jagadeesh et al., 2016). We searched the ExAC database for each variant (release 0.3, 14 January 2016) (Lek et al., 2016). Genomic coordinates are based on genome build hg19/GRCh37 (February 2009). Coding and protein positions of the GRIN1 mutations are based on GenBank accession codes NM_007327.3 (ENST00000371561.3) and NP_015566.1 respectively.
Homology modelling
Structural modelling of wild-type and mutant GluN1, and wild-type GluN2A proteins was carried out using a previously-described homology-based modelling pipeline (Mullins, 2012). This approach uses the solved structure of a homologous template to model the native folds of a target sequence. The target sequences selected were wild-type GluN1 (based on Q05586, Uniprot Isoform 3 FASTA file) and wild-type GluN2A (based on Q12879, Uniprot Isoform 1 FASTA file). The template for both Q05568 and Q12879 was the crystal structure of the GluN1a/GluN2B NMDA receptor (4PE5 chain A) from Rattus norvegicus (Karakas and Furukawa, 2014). The 4PE5 chain A contains the amino terminal domain, agonist binding domain, and transmembrane domain of the wild-type channel tetramer. We constructed our models using two GluN1 and two GluN2A subunits arranged as 1-2-1-2. Target and template underwent structural and consensus alignment using T-Coffee (Notredame et al., 2000). Homology modelling was performed by MODELLER (Webb and Sali, 2016). The putative structure was refined to improve the accuracy of non-conserved regions, optimize bond geometries and remove unfavourable contacts. Structural models were viewed and analysed using the UCSF Chimera software (https://www.cgl.ucsf.edu/chimera/) (Pettersen et al., 2004). Each mutant receptor was superimposed onto the wild-type model. The effects on the transmembrane helices were assessed by measuring the displacement, between the two models, of residues at the ends of each helix (from alpha carbon atoms). The residues were 559, 581, 615, 606, 655, 630, 810 and 828. The superimposed models were also used to calculate root-mean-square deviation (RMSD) values for nine domains (using all backbone atoms in the specified residues): amino terminal (residues 23–394), first and second ligand binding domains (S1 and S2; residues 395–544 and 658–808, respectively), transmembrane domains one to four (M1–M4; residues 560–580, 606–615, 637–657, and 813–833), DRPEER motif (658–663) and SYTANLAAF motif (646–654). UCSF Chimera FindHBond function (using the default relaxation) was used to predict hydrogen bonding between the glycine ligand and the glycine-binding residues of GluN1. UniProt lists these as 516–518, 523, 688 and 732.
Expression plasmids and mutagenesis
For two-electrode voltage clamp recordings, the cDNA for human wild-type NMDA subunits GluN1, GluN2A, and GluN2B (GenBank accession codes: NP_015566, NP_000824 and NP_000825, respectively) were subcloned into pCI-neo (Hedegaard et al., 2012). Mutant GluN1-Y647C, GluN1-R659T, GluN1-N674I, GluN1-D789N and GluN1-R794Q were generated by site-directed mutagenesis using the QuikChange™ protocol with Pfu DNA polymerase (Agilent Technologies). The parental strand was replicated with the desired mismatch incorporated into the primer (Yuan et al., 2005). Methylated parental DNA template was digested with Dpn I. The nicked double-stranded mutant DNA was transformed into TOP10 Competent Cells (Life Technologies). The mutations were verified by sequencing through the region of the mutations. For whole-cell voltage clamp recordings the cDNA for human wild-type GRIN1 and GRIN2B (Myc-DDK-tagged, based on GenBank accession codes NM_007327 and NM_000834, respectively) were subcloned into pCMV6-Entry (OriGene Technologies, catalogue numbers RC216458 and RC223623). Mutant GluN1-N674I was generated in pCMV6-GluN1 by site-directed mutagenesis using the QuikChange™ mutagenesis kit as described above and verified by Sanger sequencing.
Two-electrode voltage clamp recordings
Two-electrode voltage clamp recordings were performed as previously described (Hansen et al., 2013; Yuan et al., 2014; Chen et al., 2017b). Briefly, coding RNA for wild-type and mutant GluN1 was synthesized in vitro from linearized template cDNA and injected into Xenopus laevis oocytes (Ecocyte). Following injection, the oocytes were stored at 15–19°C in Barth’s solution containing (in mM) 88 NaCl, 2.4 NaHCO3, 1 KCl, 0.33 Ca(NO3)2, 0.41 CaCl2, 0.82 MgSO4 and 5 Tris/HCl (pH 7.4 with NaOH). Voltage-clamp recordings were performed 2–4 days post-injection at room temperature (23°C). The recording solution contained (in mM) 90 NaCl, 1 KCl, 10 HEPES, 0.5 BaCl2 and 0.01 EDTA (pH 7.4 with NaOH). Voltage and current electrodes were filled with 0.3 and 3.0 M KCl, respectively, and current responses were recorded at a holding potential of −40 mV (unless otherwise stated). Data acquisition and voltage control were accomplished with a two-electrode voltage-clamp amplifier (OC725, Warner Instrument). NMDA receptor agonists (glutamate or glycine) and antagonists (Mg2+ or H+) were applied to the oocyte using a computer-controlled eight-modular valve positioner (Digital MVP Valve, Hamilton). Glutamate (100 μM) and glycine (100 μM) were used in all oocyte experiments unless otherwise stated. The agonist concentration-response curves were fitted with:
| (1) |
where EC50 is the agonist concentration that elicited a half-maximal response and nH is the Hill slope. IC50 values for Mg2+ were obtained by fitting the concentration-response data with:
| (2) |
where IC50 is the concentration that produces a half-maximal effect, and minimum is the degree of residual inhibition at a saturating concentration of Mg2+. Data were expressed as mean ± standard error of the mean (SEM).
Preparation and transfection of HEK 293 cells
HEK 293 cells (ATCC) were plated onto glass coverslips coated in 100 µg/ml poly-d-lysine and incubated at 37°C (5% CO2 in DMEM/F12 1:1 media) supplemented with 10% foetal bovine serum, 2 mM glutamine, 50 U/ml penicillin and 50 µg/ml streptomycin. The cells were co-transfected with plasmid cDNAs encoding green fluorescent protein (GFP), GluN2B, and GluN1 or GluN1–N674I. For transfection, media was changed to S-MEM media with 1% foetal bovine serum, 0.5 mM glutamine and 0.5 mM CaCl2. Mixed pCMV plasmid DNA (0.25 μg each of GluN2B, GluN1 or GluN1-N674I, and 0.125 μg nucGFP) with 50 μl Opti-MEM™ and 1 μl Lipofectamine™ 2000 (Life Technologies) was added to each well and the mixture incubated at 37°C for 4 h. Media was then changed to transfection media plus 100 μM D-AP5 (R&D systems), a competitive NMDA receptor antagonist. The cells were inspected by fluorescence microscope 16–24 h post-transfection. Cells expressing GFP were used for whole-cell voltage-clamp recordings.
Whole-cell voltage clamp recordings
Whole-cell voltage clamp recordings were performed as previously described (Jiang et al., 2005; Thompson et al., 2012). Membrane potential of transfected cells were held at −60 mV (at room temperature, 23°C) using a patch clamp amplifier (EPC10, HEKA). Borosilicate glass microelectrodes, 5–5.5 MΩ were filled with intracellular solution containing (in mM) 117 KCl, 10 NaCl, 11 HEPES, 11 EGTA, 2 MgCl2, 1 CaCl2, 2 Na2ATP (pH 7.2 with KOH). The extracellular solution contained (in mM) 135 NaCl, 5 KCl, 5 HEPES, 10 glucose, 1.2 MgCl2 and 1.25 CaCl2 (pH 7.4 with NaOH). Solution exchange was achieved with a rapid solution changer (BioLogic) and data collected using PatchMaster software (HEKA). Peak currents were defined as the maximal amplitude of response during the agonist application; responses were plotted as current density (pA/pF). Concentration-response curves were constructed and fitted by Equation 1 using Prism 7.
Results
GRIN1 mutations in patients with polymicrogyria
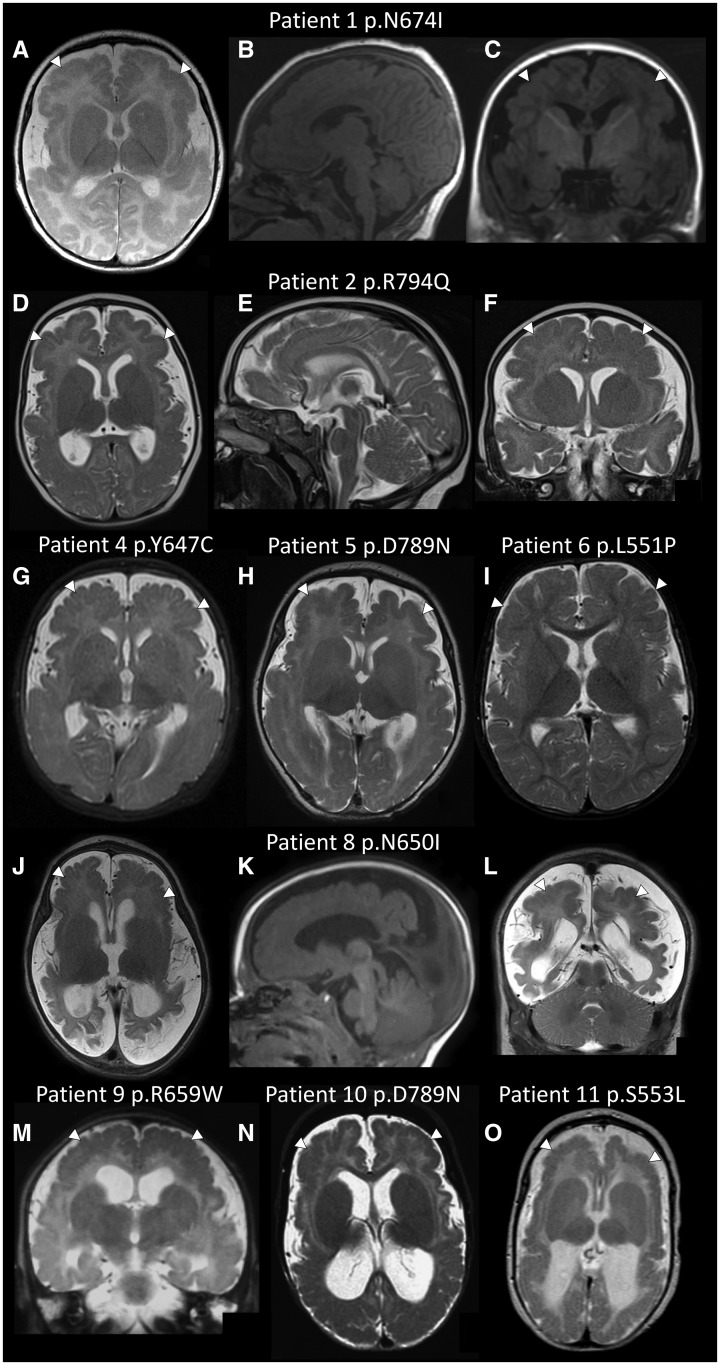
We performed whole-exome sequencing in 57 unrelated individuals with polymicrogyria and their unaffected parents. Two of the polymicrogyria patients (Patients 1 and 2) had de novo missense mutations in GRIN1 [c.2021A>T, p.(Asn674Ile) and c.2381G>A, p.(Arg794Gln)]. Apart from known polymicrogyria genes, GRIN1 was the only gene with de novo mutations in more than one patient. (A manuscript describing all de novo mutations in the cohort is in preparation). Given the predicted rate of de novo missense mutations in GRIN1 (from ExAC: 4.4 × 10−5) the observation of more than one de novo GRIN1 missense mutation in 57 subjects has a P-value of 3.1 × 10−6 (binomial test). This is close to a strict Bonferroni-corrected experiment-wide P-value threshold of 2.5 × 10−6 per gene (Kiezun et al., 2012). We found nine further GRIN1 missense mutations in additional MCD patients (Table 1). The 11 missense mutations were all de novo, affected highly-conserved residues (Supplementary Fig. 1) and in silico predictions suggested they were all deleterious (Supplementary Table 2). Two of the mutations [c.1975C>T, p.(Arg659Trp) and c.2365G>A, p.(Asp789Asn)] were recurrent. Shared clinical features in the live-born patients were severe or profound developmental delay, postnatal microcephaly, cortical visual impairment and treatment-resistant epilepsy. A summary of the clinical features is given in Table 1 (detailed case reports are provided in the Supplementary material). Some previous GRIN1 patients have been noted to have abnormal eye movements resembling oculogyric crises and stereotypic hand movements. At least four of our series had similar features (abnormal movements in Patient 4, stereotypic movements in Patient 6, and episodes of gaze deviation in Patients 8 and 10). Abnormal movements may have been present in the other patients but either unreported or misinterpreted (e.g. as seizures or roving eye movements). Magnetic resonance images from all subjects (apart from Patients 3 and 7) were available for review (Fig. 1). The patients demonstrated an extensive bilateral cortical malformation similar in appearance to tubulinopathy- or GRIN2B-associated dysgyria (Platzer et al., 2017). None of the brains have been examined histologically but the magnetic resonance appearance was most consistent with polymicrogyria. The distribution of the polymicrogyria was typically diffuse (frontal, perisylvian, parietal and temporal) and bilateral but with some occipital sparing. Patients 8 and 9 had polymicrogyria in the frontal and parietal regions but the extent of perisylvian involvement was unclear from the available images. Patient 6 had predominantly perisylvian polymicrogyria with some frontal, parietal and temporal involvement (grade 2 bilateral perisylvian polymicrogyria). This milder cortical malformation correlated with the patient’s milder phenotype (sitting and walking, but still severely delayed). Additional magnetic resonance findings in the patients were increased extra-axial spaces (particularly anteriorly, 9/11), enlarged lateral ventricles (8/11), reduced white matter volume (9/11), thinning of the corpus callosum (4/11) and abnormal hippocampi (3/11).
Table 1.
Clinical features of patients with GRIN1 mutations and polymicrogyria
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Male | Female | Female | Male | Male | Female | Male | Female | Male | Male | Female |
| Age at last review | 9 y 2 m | 2 y 5 m | 4 y 7 m | 19 m | 3 y 6 m | 9 y | 22 w gestation | 20 m | Died at 14 y | 17 y | Died at 8 y |
| Mutation | c.2021A>T | c.2381G>A | c.1975C>T | c.1940A>G | c.2365G>A | c.1652T>C | c.1958C>G | c.1949A>T | c.1975C>T | c.2365G>A | c.1658C>T |
| p.(Asn674Ile) | p.(Arg794Gln) | p.(Arg659Trp) | p.(Tyr647Cys) | p.(Asp789Asn) | p.(Leu551Pro) | p.(Ala653Gly) | p.(Asn650Ile) | p.(Arg659Trp) | p.(Asp789Asn) | p.(Ser553Leu) | |
| Birth OFC | n/a | n/a | +0.0 SD | −0.9 SD | −2.5 SD | −0.8 SD | n/a | +0.59 SD | n/a | −4.9 SD | n/a |
| Last OFC | −3.6 SD | −5.2 SD | −7.1 SD | −1.5 SD | −6.5 SD | n/a | −1.6 SD | −5.7 SD | n/a | −6.7 SD at 16 m | −7.1 SD at 3 y |
| Development | Profound delay. Not sitting or walking. Vocalizing. | Severe delay. Good head control. No babbling. | Profound delay. Not sitting or walking. No babbling. | Severe delay. Not sitting. Abnormal movements. | Profound delay. No motor or speech development. | Severe delay. Sitting. Walks with frame. Stereotypies. | n/a | Severe delay. Not sitting or walking. No babbling. | Severe delay. Not sitting or walking. | Severe delay. Not sitting or walking. Vocalizing. | Severe delay. Not sitting or walking. |
| CVI | Yes | Yes | Yes | Yes | Yes | Yes | n/a | Yes | n/a | Yes | Yes |
| Seizure onset | 6 w | 9 m | 2 m | 3 m | 1 w | <1 y | n/a | <1 m | 5 w | ? <1 m | 2 w |
| Initial seizure type | Myoclonic | Generalized tonic-clonic | Spasms | Tonic | Grimacing | Spasms | n/a | Tonic, gaze deviation | Tonic | Gaze deviation from 3 y | Tonic |
| Neurology | Spastic tetraplegia, axial hypotonia | Spastic tetraplegia, axial hypotonia | Spastic tetraplegia, axial hypotonia | Pseudobulbar palsy, hypotonia | Spastic tetraplegia, axial hypotonia | Spastic tetraplegia, axial hypotonia | n/a | Spastic tetraplegia, axial hypotonia | Mild scoliosis | Spastic tetraplegia, axial hypotonia | Spastic tetraplegia, axial hypotonia |
| Cortex | Extensive bilateral PMG with occipital sparing | Extensive bilateral PMG with occipital sparing | Diffuse bilateral PMG. | Extensive bilateral PMG with occipital sparing | Extensive bilateral PMG with occipital sparing | Bilateral perisylvian PMG, frontal, parietal and temporal spread | Abnormal thinning and sulcation of the cerebral cortex | Fronto-parietal PMG | Extensive bilateral PMG with occipital sparing | Diffuse bilateral PMG | Diffuse bilateral PMG |
| Corpus callosum | Normal | Normal | Thin | Normal | Normal | Normal | Hypoplastic | Thin | Normal | Normal | Thin |
| Lateral ventricles | Normal | Mildly enlarged | n/a | Mildly enlarged | Mildly enlarged | Normal | Large | Large | Moderately enlarged | Enlarged | Enlarged |
| Hippocampi | Normal | Normal | n/a | Normal | Abnormal | Normal | Normal | Abnormal | Normal | Normal | Thin leaves |
Ages: weeks (w); months (m) and years (y); CVI = cortical visual impairment; n/a = not available/applicable; OFC = occipital frontal circumference; PMG = polymicrogyria.
Figure 1.
Polymicrogyria in patients with GRIN1 mutations. Axial, midline sagittal and coronal brain magnetic resonance images for Patient 1 at age 2 months (A–C) and Patient 2 at age 5 months (D–F); axial magnetic resonance images for Patient 4 at age 3 months (G), Patient 5 at age 6 weeks (H) and Patient 6 at age 8 months (I); axial, sagittal and coronal images for Patient 8 at age 3 months (J–L); a coronal image for Patient 9 at age 4 months (M); axial images from Patient 10 at age 8 months (N) and Patient 11 at age 2 months (O). Images B, C and K are T1-weighted. All other images are T2-weighted. The images demonstrate bilateral extensive polymicrogyria (white arrows) more severe anteriorly. Note the increased extra-axial spaces and enlarged lateral ventricles (in most images apart from I) suggesting cerebral volume loss.
Lemke et al. (2016) reviewed the MRI findings of 19 previous GRIN1 patients. None were noted to have polymicrogyria (delayed formation of sulci was observed in one patient with homozygous GRIN1 nonsense mutations) (Lemke et al., 2016). To ensure our findings were not simply due to differences in the interpretation of radiology we reviewed MRI brain images from four previous non-polymicrogyria patients with GRIN1 mutations (p.Asp552Glu, p.Met641Ile, p.Gly815Arg and p.Gly827Arg) (Ohba et al., 2015; Lemke et al., 2016). This confirmed the absence of polymicrogyria.
Clustering of polymicrogyria-associated GRIN1 mutations
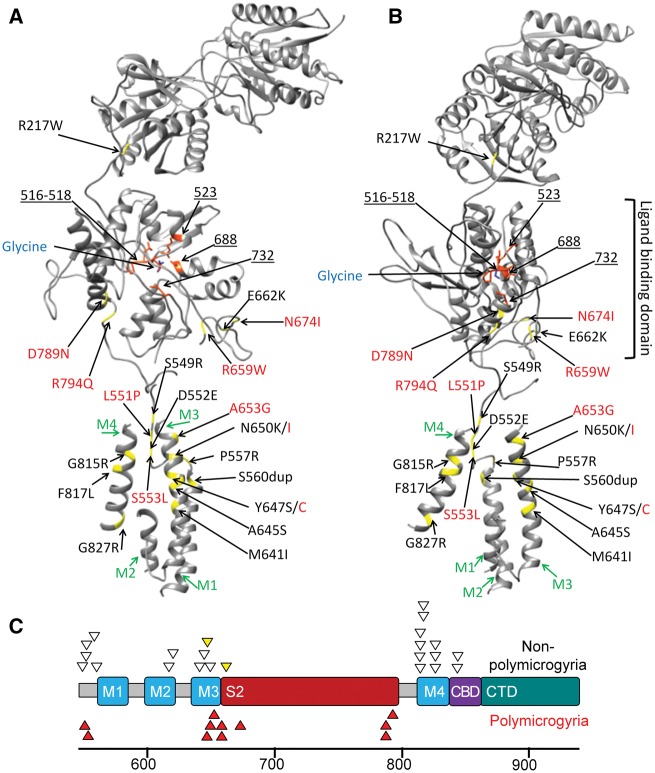
We compared the positions of the 11 polymicrogyria-associated GRIN1 mutations with 16 different heterozygous proven (or likely) de novo GRIN1 mutations previously reported in 23 patients with non-syndromic intellectual disability and epileptic encephalopathy (Fig. 2) (Lemke et al., 2016). We observed that most polymicrogyria-associated mutations occurred in the S2 domain of GluN1 or the adjacent M3 helix. The proportion of mutation in these two domains (9/11) was greater than expected based on the distribution of previous GRIN1 mutations (5/23; P = 0.002, Fisher’s exact test). Furthermore, no polymicrogyria-associated mutations were observed in M4, a domain where nearly half of all previous mutations were located.
Figure 2.
Position of the GRIN1 mutations. (A) This ribbon diagram of the GluN1 subunit of the NMDA receptor demonstrates the location of the binding site of glycine in the ligand binding domain, transmembrane helices (M1-M4 in green) and glycine binding residues of GluN1 (orange residues and blue underlined numbers). Mutated residues are in yellow. Polymicrogyria-associated mutations are in red text and previous GRIN1 mutations are in black text. (B) The same ribbon model rotated 90 degrees in the axial plane. (C) A model of the C-terminal end of GRIN1. In addition to showing M1-M4, the model shows the position of the second ligand binding domain (S2), calmodulin biding domain (CBD) and the C-terminal domain (CTD). Codon position is listed below the model. Heterozygous GRIN1 variants reported or reviewed by Lemke et al. (2016) (non-polymicrogyria, white triangles) are shown above the model. The two yellow triangles represent subjects with CT brain imaging only. Polymicrogyria-associated GRIN1 mutations (red triangles) cluster in the S2 domain (6/11) or in the adjacent Lurcher motif of M3 (3/11).
Six polymicrogyria-associated GRIN1 mutations [p.Arg659Trp (two mutations), p.Asn674Ile, p.Asp789Asn (two mutations) and p.Arg794Gln] were located in the S2 domain. The S2 domain forms part of the ligand-binding domain of GluN1 and has been shown to be highly intolerant to variation (Ogden et al., 2017). The recurrent p.Arg659Trp mutation is in the DRPEER motif of S2 (residues 658–663). This motif is close to the extracellular end of M3, near the channel pore entrance. Only two previous GRIN1 mutations have been located in S2 (p.Glu662Lys and p.Ser688Tyr) (Hamdan et al., 2011; Zehavi et al., 2017). Neither patient was reported to have polymicrogyria although the p.Glu662Lys patient only had a CT brain scan (MRI was not done) (Hamdan et al., 2011). CT generally lacks the resolution to detect polymicrogyria.
Three polymicrogyria-associated GRIN1 mutations (p.Tyr647Cys, Asn650Ile and p.Ala653Gly) were in close proximity to the S2 domain in a region of highly-conserved residues at the extracellular end of M3. These residues (646–654) are known as the Lurcher motif (SYTANLAAF). This motif is thought to act as the major permeation barrier at the intersection of four M3 helices (Murthy et al., 2012). Different mutations affecting residues 647 and 650 have previously been reported in patients with epileptic encephalopathy (Epi4K Consortium et al., 2013; Ohba et al., 2015). Neither was reported to have polymicrogyria although the p.Tyr647Ser patient only had a CT brain scan. The final two polymicrogyria-associated mutations (p.Leu551Pro and p.Ser553Leu) were located in the S1-M1 linker region. The tertiary structure of GluN1 means the residues are located close to the extracellular end of the M3 helix (Fig. 2). It has recently been proposed that the pre-M1 region is close enough to M3 to interact as a key gating element (Sobolevsky et al., 2009; Chen et al., 2017b; Ogden et al., 2017).
Modelling the structural effects of GRIN1 mutations
We hypothesized that polymicrogyria-associated and previous GRIN1 mutations might have different effects on the structure of GluN1. To study this we developed a 3D model of the GluN1/GluN2A NMDA receptor based on the rat GluN1/GluN2B tetramer (Fig. 2). We simulated models for the nine polymicrogyria-associated GRIN1 mutations and 16 previous GRIN1 mutations. We measured each mutant model in three ways: (i) the displacement, between mutant and wild-type, of the extracellular and intracellular ends of each transmembrane helix; (ii) the displacement, between the mutant and wild-type, of nine key domains. A RMSD value (a measure of average distance between the two superimposed structures) was calculated for each domain; and (iii) the number of hydrogen bonds between the glycine ligand and the glycine-binding residues of GluN1.
Analysis of transmembrane helix positions (Supplementary Table 3) and domain-specific RMSD values (Supplementary Table 4) revealed no consistent differences between previous and polymicrogyria-associated GRIN1 mutations. The regions of GluN1 most affected by GRIN1 mutations in both groups were M2, M3 (particularly the extracellular end) and S2. All GRIN1 mutants had RMSD values >2 Å for the S2 domain and DRPEER motif. The extracellular end of the M3 helix (the part of the helix involved in the channel pore entrance) was displaced >1 Å for all GRIN1 mutants apart from polymicrogyria-associated p.Asp789Asn (0.67 Å). The p.Asp789Asn mutation stood out from other mutations in causing the least displacement of transmembrane helices (all ≤1.01 Å) and the lowest RMSD value for the M3 helix (1.17 Å). The p.Asp789Asn mutation still had its greatest effects on the S2 domain (RMSD 3.06 Å) where it was located. The polymicrogyria-associated p.Leu551Pro and p.Ser553Leu mutations were located in the S1-M1 linker region but had their greatest effects on the RMSD values of the M2, M3 and S2 domains. On average polymicrogyria-associated mutations generated slightly more hydrogen bonds with glycine (6.8) compared with previous mutations (4.8; P = 0.02, Mann-Whitney U-test) and wild-type (4) (Table 2). However, the effects varied between mutations. All polymicrogyria-associated mutations increased the number of hydrogen bonds with glycine apart from p.Asp789Asn, which formed one less than wild-type.
Table 2.
Effect of GRIN1 mutations on hydrogen bonds between the glycine ligand and the glycine-binding residues of GluN1
| GluN1 residue | Pro516 | Thr518 | Arg523 | Ser687 | Ser688 | Asp732 | Bond total |
|---|---|---|---|---|---|---|---|
| Wild-type | − | ×× | × | − | × | − | 4 |
| Previous GRIN1 mutations | |||||||
| S549R | − | − | ×× | × | × | × | 5 |
| D552E | − | − | ×× | − | × | × | 4 |
| P557R | − | × | − | − | × | × | 3 |
| S560dup | − | × | × | − | ×(689) | ×(733) | 4 |
| G618R | − | − | − | − | ×× | × | 3 |
| G620R | − | ××× | − | − | × | × | 5 |
| M641I | − | ×× | ×× | × | × | ×× | 8 |
| A645S | − | × | ×× | − | × | − | 4 |
| Y647S | − | − | − | × | ×× | ×× | 5 |
| N650K | − | − | − | × | × | × | 3 |
| E662K | − | × | × | − | × | × | 4 |
| G815R | − | × | × | − | ×× | × | 5 |
| G815V | − | ××× | ××× | − | × | × | 8 |
| F817L | − | − | − | − | ×× | × | 3 |
| G827R | − | ×× | ×× | − | × | × | 6 |
| R844C | − | ×× | ×× | − | × | × | 6 |
| Polymicrogyria-associated GRIN1 mutations | |||||||
| L551P | × | ××× | × | − | ×× | ×× | 9 |
| S553L | − | ×× | ×× | ×× | − | − | 6 |
| Y647C | − | ×× | × | × | × | × | 6 |
| N650I | − | ×× | × | × | ×× | × | 7 |
| A653G | − | ××× | ××× | − | × | ×× | 9 |
| R659W | − | ×× | × | − | ×× | × | 6 |
| N674I | − | ××× | ××× | − | ×× | ×× | 10 |
| D789N | − | − | − | − | ×× | × | 3 |
| R794Q | − | − | − | × | ×× | ×× | 5 |
Number of hydrogen bonds between glycine and specified residues: − (none), ×(one), ×× (two), ××× (three). All predicted hydrogen bonds were <3 Å in length.
Impact of GRIN1 mutations on NMDA receptor function
To investigate whether polymicrogyria-associated GRIN1 mutations influence NMDA receptor function in vitro we undertook site-directed mutagenesis to introduce five of the GRIN1 mutations (p.Tyr647Cys, p.Arg659Trp, p.Asn674Ile, p.Asp789Asn and p.Arg794Gln) into cDNA encoding human GluN1. We then expressed wild-type and mutant GluN1 with either human wild-type GluN2A or GluN2B in Xenopus oocytes and evaluated the effects of these mutants on pharmacological properties of NMDA receptors by using two-electrode voltage clamp recordings, including agonist potency (glutamate and glycine), as well as magnesium and proton sensitivity. The concentration that produced a half-maximal current response (EC50) was determined by measuring the current response to a range of glutamate (in the presence of 100 µM glycine) and glycine (in the presence of 100 µM glutamate) concentrations at a holding potential of −40 mV. The magnesium sensitivity (IC50) was determined by measuring the effect of different magnesium concentrations on agonist-evoked currents (by 100 µM glutamate and 100 µM glycine) at a holding potential of −60 mV. The proton sensitivity was evaluated as the percentage of receptor activity at pH 6.8 compared with receptor activity at pH 7.6 (holding potential of −40 mV).
The p.Arg659Trp and p.Arg794Gln mutations had similar effects; both increased the potency (reduced EC50 values) of agonists, and in particular, glutamate. The EC50 of mutant receptors was reduced to 10–20% of wild-type levels (Table 3). The potency of glycine was also increased: EC50 was 71% of wild-type for GluN1-R794Q/GluN2A, 61% for GluN1-R794Q/GluN2B, 36% for GluN1-R659W/GluN2A and 15% for GluN1-R659W/GluN2B. There were no detectable differences in Mg2+ blockade. Proton inhibition was not statistically significant apart from a reduced block in GluN1-R659W/GluN2A (70% mutant versus 52% wild-type).
Table 3.
Summary of two-electrode voltage clamp data
| Constructs | Glu EC50, µM (n) | Mutant/ WT, % | Gly EC50, µM (n) | Mutant/ WT, % | Mg2+ IC50, µM (n) | Mutant/ WT, % | %, pH 6.8/ pH 7.6 | %, Mutant/WT |
|---|---|---|---|---|---|---|---|---|
| WT GluN1/GluN2A | 3.3 ± 0.04 (6) | 1.3 ± 0.05 (6) | 24 ± 5.9 (5) | 51 ± 1.7 (6) | ||||
| GluN1-R794Q/GluN2A | 0.68 ± 0.06 (6)* | 21 | 0.92 ± 0.09 (6)* | 71 | 31 ± 5.4 (6) | 129 | 47 ± 0.9 (6) | 92 |
| WT GluN1/GluN2B | 1.7 ± 0.12 (7) | 0.33 ± 0.05 (6) | 18 ± 0.53 (6) | 16 ± 0.6 (5) | ||||
| GluN1-R794Q//GluN2B | 0.21 ± 0.03 (14)* | 12 | 0.20 ± 0.02 (12)* | 61 | 17 ± 1.7 (7) | 94 | 17 ± 1.0 (7) | 106 |
| WT GluN1/GluN2A | 3.2 ± 0.04 (6) | 1.3 ± 0.01 (7) | 24 ± 5.9 (5) | 48 ± 6.4 (5) | ||||
| GluN1-N674I/GluN2A | 2.2 ± 0.14 (6) | 69 | 1.6 ± 0.08 (6) | 123 | 34 ± 5.3 (6) | 142 | 72 ± 3.2 (6)* | 150 |
| WT GluN1/GluN2B | 1.5 ± 0.17 (10) | 0.38 ± 0.06 (6) | 26 ± 3.7 (8) | 16 ± 0.9 (6) | ||||
| GluN1-N674I/GluN2B | 1.9 ± 0.15 (14) | 126 | 0.86 ± 0.05 (11)* | 226 | 24 ± 2.6 (6) | 92 | 50 ± 1.3 (6)* | 313 |
| WT GluN1/GluN2A | 3.9 ± 0.28 (10) | 1.0 ± 0.11 (11) | 29 ± 4.1 (10) | 52 ± 1.1 (12) | ||||
| GluN1-R659W/GluN2A | 0.43 ± 0.03 (18)* | 11 | 0.36 ± 0.09 (10)* | 36 | 37 ± 3.5 (7) | 128 | 70 ± 1.6 (11)* | 135 |
| WT GluN1/GluN2B | 1.3 ± 0.09 (9) | 0.33 ± 0.03 (12) | 18 ± 0.5 (6) | 16 ± 0.8 (10) | ||||
| GluN1-R659W/GluN2B | 0.25 ± 0.04 (11)* | 19 | 0.05 ± 0.01 (14)* | 15 | 20 ± 3.4 (6) | 111 | 15 ± 1.7 (10) | 94 |
| WT GluN1/GluN2A | 3.8 ± 0.51 (6) | 1.3 ± 0.06 (9) | 24 ± 3.1 (6) | 41 ± 2.0 (6) | ||||
| GluN1-Y647C/GluN2A | 0.06 ± 0.03 (5)* | 1.6 | 0.06 ± 0.01 (6)* | 4.6 | 9.0 ± 1.0 (5)* | 38 | 35 ± 2.1 (6)* | 85 |
| WT GluN1/GluN2B | 1.7 ± 0.08 (7) | 0.41 ± 0.05 (6) | 21 ± 3.1 (5) | 15 ± 2.5 (6) | ||||
| GluN1-Y647C/GluN2B | 0.04 ± 0.01 (6)* | 2.4 | 0.046 ± 0.015 (9)* | 11 | 17 ± 4.3 (6) | 81 | 30 ± 3.5 (6)* | 200 |
Data were from two-electrode voltage-clamp recordings on Xenopus oocytes at −40 mV holding potential (except for Mg2+ at −60 mV) and expressed as mean ± SEM (n); WT = wild-type; *P < 0.05, unpaired t-test, compared to the corresponding data from wild-type receptors recorded on the same day.
The p.Tyr647Cys mutation demonstrated a profound increase in the potency of glutamate with an EC50 of just 1.6% of wild-type for GluN1-Y647C/GluN2A and 2.4% for GluN1-Y647C/GluN2B. There was a similarly increased sensitivity to glycine (EC50 4.6% for GluN1-Y647C/GluN2A; 11.2% for GluN1-Y647C/GluN2B). There was a statistically significant increase in Mg2+ blockade for GluN1-Y647C/GluN2A (IC50 38% of wild-type) but not GluN1-Y647C/GluN2B (IC50 81% of wild-type) (Table 3). There were also significant but variable effects on proton inhibition: slightly increased in GluN1-Y647C/GluN2A (35% mutant versus 41% wild-type) but reduced in GluN1-Y647C/GluN2B (30% mutant versus 15% wild-type).
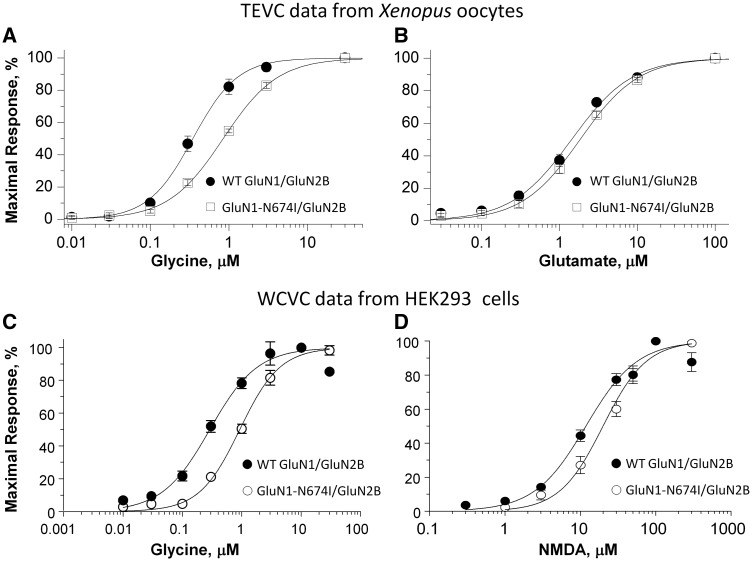
The p.Asn674Ile mutant had a different profile of effects. The EC50 for glycine was unchanged for GluN1-N674I/GluN2A but increased to 226% for GluN1-N674I/GluN2B (Table 3 and Fig. 3A). This indicated a reduced potency for glycine. The EC50 of glutamate was not significantly altered for GluN1-N674I/GluN2A or GluN1-N674I/GluN2B (Table 3 and Fig. 3B). However, p.Asn674Ile did demonstrate significantly reduced proton inhibition (72% GluN1-N674I/GluN2A versus 48% wild-type GluN1/GluN2A; 50% GluN1-N674I/GluN2B versus 16% wild-type GluN1/GluN2B). There was no significant difference in Mg2+ blockade for p.Asn674Ile receptors. To confirm that p.Asn674Ile reduced glycine potency we assessed the mutation in a second, separate in vitro heterologous expression setting. We co-expressed wild-type and mutant GluN1 with human GluN2B in HEK 293 cells and evaluated the concentration-effect curves for agonist (NMDA) and co-agonist (glycine) using whole-cell voltage clamp. The whole-cell voltage clamp results (Fig. 3C and D) were consistent with the two-electrode voltage clamp findings and demonstrated a pronounced loss of sensitivity to glycine (EC50 310% of wild-type; GluN1-N674I/Glu2B: 0.93 µM, n = 8; versus wild-type GluN1/Glu2B: 0.30 µM, n = 8; P < 0.0001; unpaired t-test) and a moderate reduction in the potency of NMDA (EC50 164% of wild-type; GluN1-N674I/Glu2B: 19.71 µM, n = 6; versus wild-type GluN1/Glu2B: 12.05 µM, n = 8; P = 0.01; unpaired t-test).
Figure 3.
The p.Asn674Ile mutation changes the response of the NMDA receptor to agonists. The top graphs display concentration-response curves for (A) glycine (in the presence of 100 µM glutamate) and (B) glutamate (in the presence of 100 µM glycine) determined by two-electrode voltage-clamp (TEVC) recordings from Xenopus oocytes expressing either wild-type (WT)-GluN1/GluN2B or GluN1-N674I /GluN2B. The bottom graphs display concentration-response curves for (C) glycine (in the presence of 100 µM NMDA) and (D) NMDA (in the presence of 100 µM glycine) determined by whole-cell voltage-clamp (WCVC) recordings from transfected HEK 293 cells expressing either WT-GluN1/GluN2B or GluN1-N674I /GluN2B. Error bars represent SEM.
Multiple attempts (mutagenesis, RNA syntheses, RNA injection, and recordings) were made to express p.Asp789Asn mutant receptors (as both GluN1/GluN2A and GluN1/GluN2B) in Xenopus oocytes. The current amplitudes at saturating agonist concentrations (up to 1 mM glutamate and 3 mM glycine at holding potential of −40 mV) were too small to characterize the effects on NMDA receptor pharmacological properties (GluN1-D789N/Glu2A: 13 ± 2.8 nA, n = 14; versus wild-type GluN1/Glu2A: 855 ± 199 nA, n = 14; GluN1-D789N/Glu2B: 11 ± 3.1 nA, n = 14; versus wild-type GluN1/Glu2B: 585 ± 156 nA, n = 14). We therefore evaluated the expression levels of GluN1-D789N using a β-lactamase activity assay in transiently-transfected HEK cells (Supplementary material) (Swanger et al., 2016). The ratio of surface-to-total protein levels for the mutant GluN1-D789N was reduced to 71 ± 4.2 % (n = 4; P = 0.04; unpaired t-test) of wild-type when co-expressed with wild-type GluN2A, and had no significant change (115 ± 46 %; n = 8; P = 0.46; unpaired t-test) when co-expressed with wild-type GluN2B (Supplementary Fig. 2), suggesting that the complete loss of current responses for GluN1-D789N is not due to a trafficking defect or lack of surface expression but rather a functional change in the membrane-bound receptor.
Discussion
By using exome data from parent-offspring trios we identified 2 of 57 polymicrogyria patients with de novo missense mutations in GRIN1. Another nine de novo missense GRIN1 mutations were identified in additional MCD patients. Shared features in the patients were extensive bilateral polymicrogyria, severe or profound developmental delay, postnatal microcephaly, cortical visual impairment and treatment-resistant epilepsy. De novo missense mutations in GRIN1 have previously been reported in patients with non-syndromic intellectual disability (Hamdan et al., 2011; Redin et al., 2014; Zhu et al., 2015; Lemke et al., 2016; Rossi et al., 2017), movement disorders (Ohba et al., 2015; Chen et al., 2017a; Zehavi et al., 2017), epileptic encephalopathy (Epi4K Consortium et al., 2013), and cerebral visual impairment (Bosch et al., 2016). The MRI features seen in previous GRIN1 patients were non-specific volume loss and generalized cerebral atrophy but not polymicrogyria (Lemke et al., 2016). We note that at least two previous GRIN1 patients only had CT brain scans, which cannot reliably detect polymicrogyria. This raises the possibility that some previous GRIN1 patients may have had unrecognized polymicrogyria. It is also possible that apparently non-polymicrogyria GRIN1 patients (by MRI) may have subtle structural brain abnormalities that are below the resolution of current scanning technology.
The NMDA receptor is a tetrameric heteromultimer that comprises two GluN1 subunits (encoded by GRIN1) and two variable GluN2 subunits, which are encoded by GRIN2A, GRIN2B, GRIN2C or GRIN2D. Like GluN1, GluN2B is extensively expressed in the cerebral cortex during foetal development (Liu et al., 2004). GRIN2B mutations have been found in patients with autism (O’Roak et al., 2011; Tarabeux et al., 2011), cerebral visual impairment (Bosch et al., 2016), West syndrome (Lemke et al., 2014) and intellectual disability (Endele et al., 2010; Adams et al., 2014; Lemke et al., 2014; Hu et al., 2016; Swanger et al., 2016). GRIN2B mutations have also recently been observed in MCD patients (Platzer et al., 2017). In contrast to the foetus, the main GluN2 subunit of the adult brain is GluN2A. This change occurs during early postnatal development and is thought to be a key developmental switch (Liu et al., 2004). GRIN2A mutations have been found in patients with epilepsy-aphasia spectrum disorders (Carvill et al., 2013; Lemke et al., 2013; Lesca et al., 2013; Gao et al., 2017), early-onset epileptic encephalopathy (Endele et al., 2010; Pierson et al., 2014; Yuan et al., 2014; Swanger et al., 2016) and schizophrenia (Tarabeux et al., 2011). However, consistent with its mainly postnatal expression, GRIN2A mutations have not been reported to cause MCDs.
The reason why some GRIN1 patients get polymicrogyria is uncertain. This may require additional factors in the patient’s genetic background or environmental conditions during gestation. Only two of our series were known to have had testing for cytomegalovirus. Patient 5 had negative postnatal serology. Patient 6 had positive testing (urine PCR and serum IgM) at 4 months of age. However, his mother had negative serology at 36 weeks, which suggests the infection occurred postnatally. Other patients were not tested because they lacked typical features of cytomegalovirus infection (e.g. rash, retinitis, brain calcification or deafness). It has not been possible to retrospectively screen the other patients for cytomegalovirus. We observed that polymicrogyria-associated GRIN1 mutations clustered in the S2 or M3 domains, regions that are significantly depleted of variation in control populations (Swanger et al., 2016; Ogden et al., 2017). In addition, the GluN1 S2 domain has rarely been mutated in non-polymicrogyria GRIN1 patients. The GluN1 S2 domain is critical to the binding of glycine, the co-agonist of the NMDA receptor. Therefore, the observation of this putative genotype–phenotype correlation suggests that polymicrogyria and non-polymicrogyria mutations may have different effects on co-agonist binding or the activation state of the receptor. Inspecting the 3D model of GluN1 suggests the polymicrogyria-associated mutations in S2 reside close to the extracellular end of the M3 transmembrane helix that forms a bundle crossing that occludes ion permeation. These variants may therefore alter gating. The polymicrogyria-associated p.Arg659Trp mutation was located in the DRPEER motif of S2. This motif is a highly charged motif thought to influence the relative permeability of Ca2+ ions through the channel (Watanabe et al., 2002). Similarly, the three M3 mutations (p.Tyr647Cys, p.Asn650Ile and p.Ala653Gly) had milder effects on the wider structure of the receptor but were located in the highly-conserved Lurcher motif, which controls NMDA receptor gating.
Our structural modelling found polymicrogyria-associated mutations were associated with an increase in the average number of hydrogen bonds between the glycine ligand and the glycine-binding residues of GluN1. The formation of additional hydrogen bonds may alter the kinetics of co-agonist binding (e.g. increased affinity). No other stark differences between polymicrogyria-associated and previous GRIN1 mutations were observed. The measurements used (transmembrane helix position and domain-specific RMSD) may not have been sensitive to key differences between the two groups. In addition, the structural modelling suggests there is heterogeneity in how polymicrogyria-associated mutations affect GluN1 structure. This may have confounded comparison between the two groups.
Two-electrode voltage clamp analysis showed that three of the polymicrogyria-associated mutations (p.Tyr647Cys, p.Arg659Trp and p.Arg794Gln) significantly increased the potency of both glutamate and glycine. This increased potency may mean that mutant receptors can be activated at lower concentrations of agonist than wild-type receptor. Several lines of evidence link excess NMDA receptor signalling to polymicrogyria. The intracerebral injection of ibotenate has been used for decades to generate in vivo models of epilepsy and cortical malformations including polymicrogyria (Marret et al., 1996; Takano et al., 2004). Ibotenate is an agonist of both NMDA and glutamatergic metabotropic receptors. Intracerebral ibotenate injection in newborn mice (Marret et al., 1995), hamsters (Marret et al., 1996; Takano et al., 2004) and rats (Takano and Matsui, 2015) causes a range of grey and white matter changes, including polymicrogyria-like lesions, similar to those seen following perinatal hypoxic/ischaemic insults. Over-stimulation of NMDA receptors is thought to lead to excitotoxicity due to excess calcium influx through the receptor channel (Choi et al., 1988; Zhou et al., 2013). Profound gain-of-function NMDA receptor subunit mutations are excitotoxic when expressed in vitro (Li et al., 2016; Ogden et al., 2017). NMDA receptor-related excitotoxicity has been implicated in hypoxic/ischaemic events (Simon et al., 1984; Rothman and Olney, 1986). Hypoxic/ischaemic events during foetal brain development are a well-recognized cause of polymicrogyria in humans. Calcium influx through the NMDA receptor can lead to activation of a range of cellular pathways including the pro-survival PI3K-AKT pathway (Lafon-Cazal et al., 2002; Wang et al., 2012). Activating mutations in components of the PI3K-AKT pathway have been found in polymicrogyria patients (Riviere et al., 2012; Mirzaa et al., 2014). MCDs including polymicrogyria are a feature of Zellweger syndrome, a rare metabolic disorder caused by peroxisomal dysfunction. Analysis of a mouse model of Zellweger syndrome showed that the neuronal migration abnormalities seen in the mice were due to NMDA receptor-mediated calcium mobilization (Gressens et al., 2000). Another metabolic disorder associated with polymicrogyria is glycine encephalopathy (Dobyns, 1989). The hyperglycinaemia in this condition may enhance the excitotoxic activity of glutamate acting through the NMDA receptor (Subramanian et al., 2015).
In contrast to the three potential gain-of-function polymicrogyria-associated GRIN1 mutations, most previous GRIN1 mutations caused dominant-negative effects resulting in a significant loss of receptor function (Lemke et al., 2016). Animal models suggest that NMDA receptor hypofunction is less likely to disturb gross cortical structure. Mice homozygous for GluN1 null alleles die soon after birth from respiratory problems but do not have severe abnormalities of neuronal migration (Messersmith et al., 1997). Similarly, mice with a GluN1 deletion limited to excitatory cortical neurons have only subtle disturbance of cortical structure (Iwasato et al., 2000). However, a simple model of gain-of-function GRIN1 mutations causing polymicrogyria is challenged by the results for p.Asn674Ile. Both two-electrode and whole-cell voltage clamp analyses were consistent in showing this mutation decreased the potency of glycine and possibly (to a lesser extent) glutamate. How can we explain this apparently paradoxical finding? There are a number of possible mechanisms that might explain why p.Asn674Ile causes polymicrogyria.
First, the p.Asn674Ile mutation caused a significant loss of proton inhibition. GluN2B receptors are inhibited with an IC50 at physiological pH. Therefore loss of proton inhibition will potentiate responses even at resting pH. This may mean neurons are prone to excess calcium influx, a problem that will be exacerbated in low pH environments (e.g. during oxidative stress). Recent work has shown the importance of the balance of signalling through synaptic and extra-synaptic NMDA receptors (Zhou et al., 2015). Activation of synaptic NMDA receptors activates pro-survival pathways. In contrast, massive and prolonged co-activation of both synaptic and extrasynaptic receptors leads to cell death. If p.Asn674Ile blunts the response of synaptic receptors (e.g. through reduced agonist potency) while promoting signalling at extrasynaptic receptors (e.g. through reduced proton inhibition) this may tip the balance in some neurons towards apoptosis. Alternatively, polymicrogyria may be a consequence of disturbed NMDA receptor signalling regardless of whether there is gain- or loss-of-function. There is evidence from in vitro studies that a low-level background of NMDA activation is needed to support neuronal survival (Zhou et al., 2015), guide radial migration (Behar et al., 1999; Hirai et al., 1999) and promote neuronal differentiation (Yoneyama et al., 2008). Transient delivery of an NMDA antagonist to a focal area of the cortex of newborn rats disturbs cortical lamination and generates heterotopic cell clusters (Reiprich et al., 2005). A range of NMDA antagonists, including ethanol, have been shown to induce apoptosis in the brains of developing rats (Olney et al., 2002). This is of relevance as polymicrogyria has occasionally been reported in patients with foetal alcohol syndrome (Reinhardt et al., 2010). Finally, it remains possible that p.Asn674Ile (and other polymicrogyria-associated GRIN1 mutations) may have additional effects that have not been captured by our electrophysiological analysis.
If polymicrogyria-associated GRIN1 mutations mainly cause gain of function while non-polymicrogyria mutations cause loss of function it raises the question why the two types cause such similar phenotypes (severe developmental delay, spasticity, early onset seizures, postnatal microcephaly, cerebral visual impairment and stereotypic movements). A potential explanation is that gain-of-function mutations may cause cell death (due to excitotoxicity) soon after neurons begin expressing NMDA receptors. Early cell loss would thin the foetal cortex (leading to polymicrogyria) and result in a postnatal cortex depleted of cells expressing NMDA receptors. In contrast, loss-of-function mutations may cause cell death more gradually (e.g. due to loss of NMDA-mediated pro-survival signalling) missing the key window when polymicrogyria occurs (<22 weeks). Having cells in the postnatal cortex which are insensitive to glutamate (due to loss-of-function mutations) may be functionally equivalent to the cells being absent (due to gain-of-function mutations). The evidence of cerebral atrophy observed in several patients was present in infancy and progressed on subsequent scans (Patient 11). The atrophy is likely due to the direct effects of mutations (excitotoxicity or loss of pro-survival signalling) as well as damage from frequent seizures.
In conclusion, we have found de novo GRIN1 missense mutations in patients with extensive bilateral polymicrogyria. Our results provide evidence for a genotype–phenotype correlation with most polymicrogyria-associated GRIN1 mutations clustering in the S2 or M3 domains, regions of the protein rarely mutated in non-polymicrogyria patients or the normal population. In addition, we showed that polymicrogyria-associated GRIN1 mutations significantly alter in vitro NMDA receptor function. Our results confirm the importance of de novo mutations in the aetiology of MCDs and polymicrogyria; expand the phenotypic spectrum associated with GRIN1 mutations; demonstrate similarities between human polymicrogyria and animal models of the disorder; and highlight the important role of NMDA signalling in the pathogenesis of polymicrogyria.
Supplementary Material
Acknowledgements
The authors would like to thank the patients, families, clinicians and scientists who contributed to this work. We would particularly like to thank the GRIN1 family group for providing access to the MRI scans of their members.
Funding
This work was funded by the Newlife Foundation for Disabled Children (Grant Reference: 11-12/04). This publication is a result of the European Network on Brain Malformations (COST Action CA16118), a network funded by COST (European Cooperation in Science and Technology). The project was also supported by the Wales Epilepsy Research Network and the Wales Gene Park. H.Y. was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development under Award Number R01HD082373; S.F.T. was supported by NIH-NINDS R01NS036654, R01NS065371, and R24NS092989. G.M.M. was supported by NIH-NINDS K08NS092898. We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z and Medical Research Council Hub grant G0900747 91070) for the generation of the Sequencing data. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviations
- EC50
concentration of agonist required for half-maximal effect
- IC50
concentration of antagonist required for half-maximal inhibition
- MCD
malformation of cortical development
- NMDA
N-methyl-d-aspartate
- RMSD
root-mean-square deviation
References
- Adams DR, Yuan H, Holyoak T, Arajs KH, Hakimi P, Markello TC, et al. Three rare diseases in one Sib pair: RAI1, PCK1, GRIN2B mutations associated with Smith-Magenis Syndrome, cytosolic PEPCK deficiency and NMDA receptor glutamate insensitivity. Mol Genet Metab 2014; 113: 161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar TN, Scott CA, Greene CL, Wen X, Smith SV, Maric D, et al. Glutamate acting at NMDA receptors stimulates embryonic cortical neuronal migration. J Neurosci 1999; 19: 4449–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch DG, Boonstra FN, de Leeuw N, Pfundt R, Nillesen WM, de Ligt J, et al. Novel genetic causes for cerebral visual impairment. Eur J Hum Genet 2016; 24: 660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvill GL, Regan BM, Yendle SC, O’Roak BJ, Lozovaya N, Bruneau N, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet 2013; 45: 1073–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Shieh C, Swanger SA, Tankovic A, Au M, McGuire M, et al. GRIN1 mutation associated with intellectual disability alters NMDA receptor trafficking and function. J Hum Genet 2017a; 62: 589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Tankovic A, Burger PB, Kusumoto H, Traynelis SF, Yuan H. Functional evaluation of a de novo GRIN2A mutation identified in a patient with profound global developmental delay and refractory epilepsy. Mol Pharmacol 2017b; 91: 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Koh JY, Peters S. Pharmacology of glutamate neurotoxicity in cortical cell culture: attenuation by NMDA antagonists. J Neurosci 1988; 8: 185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobyns WB. Agenesis of the corpus callosum and gyral malformations are frequent manifestations of nonketotic hyperglycinemia. Neurology 1989; 39: 817–20. [DOI] [PubMed] [Google Scholar]
- Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet 2010; 42: 1021–6. [DOI] [PubMed] [Google Scholar]
- Epi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS, Berkovic SF, Cossette P, Delanty N, et al. De novo mutations in epileptic encephalopathies. Nature 2013; 501: 217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Tankovic A, Zhang Y, Kusumoto H, Zhang J, Chen W, et al. A de novo loss-of-function GRIN2A mutation associated with childhood focal epilepsy and acquired epileptic aphasia. PLoS One 2017; 12: e0170818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressens P, Baes M, Leroux P, Lombet A, Van Veldhoven P, Janssen A, et al. Neuronal migration disorder in Zellweger mice is secondary to glutamate receptor dysfunction. Ann Neurol 2000; 48: 336–43. [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 2011; 88: 306–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jørgensen L, Vance KM, et al. Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol Pharmacol 2013; 84: 114–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard M, Hansen KB, Andersen KT, Brauner-Osborne H, Traynelis SF. Molecular pharmacology of human NMDA receptors. Neurochem Int 2012; 61: 601–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K, Yoshioka H, Kihara M, Hasegawa K, Sakamoto T, Sawada T, et al. Inhibiting neuronal migration by blocking NMDA receptors in the embryonic rat cerebral cortex: a tissue culture study. Brain Res Dev Brain Res 1999; 114: 63–7. [DOI] [PubMed] [Google Scholar]
- Hu C, Chen W, Myers SJ, Yuan H, Traynelis SF. Human GRIN2B variants in neurodevelopmental disorders. J Pharmacol Sci 2016; 132: 115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasato T, Datwani A, Wolf AM, Nishiyama H, Taguchi Y, Tonegawa S, et al. Cortex-restricted disruption of NMDAR1 impairs neuronal patterns in the barrel cortex. Nature 2000; 406: 726–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesh KA, Wenger AM, Berger MJ, Guturu H, Stenson PD, Cooper DN, et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet 2016; 48: 1581–6. [DOI] [PubMed] [Google Scholar]
- Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet 2009; 41: 746–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen A, Andermann E. Genetics of the polymicrogyria syndromes. J Med Genet 2005; 42: 369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AC, Robitaille Y, Honavar M, Mullatti N, Leventer RJ, Andermann E, et al. The histopathology of polymicrogyria: a series of 71 brain autopsy studies. Dev Med Child Neurol 2016; 58: 39–48. [DOI] [PubMed] [Google Scholar]
- Jiang LH, Rassendren F, Mackenzie A, Zhang YH, Surprenant A, North RA. N-methyl-D-glucamine and propidium dyes utilize different permeation pathways at rat P2X(7) receptors. Am J Physiol Cell Physiol 2005; 289: C1295–302. [DOI] [PubMed] [Google Scholar]
- Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 2014; 344: 992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet 2012; 44: 623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–81. [DOI] [PubMed] [Google Scholar]
- Lafon-Cazal M, Perez V, Bockaert J, Marin P. Akt mediates the anti-apoptotic effect of NMDA but not that induced by potassium depolarization in cultured cerebellar granule cells. Eur J Neurosci 2002; 16: 575–83. [DOI] [PubMed] [Google Scholar]
- Law AJ, Weickert CS, Webster MJ, Herman MM, Kleinman JE, Harrison PJ. Expression of NMDA receptor NR1, NR2A and NR2B subunit mRNAs during development of the human hippocampal formation. Eur J Neurosci 2003; 18: 1197–205. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JR, Geider K, Helbig KL, Heyne HO, Schutz H, Hentschel J, et al. Delineating the GRIN1 phenotypic spectrum: a distinct genetic NMDA receptor encephalopathy. Neurology 2016; 86: 2171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JR, Hendrickx R, Geider K, Laube B, Schwake M, Harvey RJ, et al. GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann Neurol 2014; 75: 147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JR, Lal D, Reinthaler EM, Steiner I, Nothnagel M, Alber M, et al. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat Genet 2013; 45: 1067–72. [DOI] [PubMed] [Google Scholar]
- Lesca G, Rudolf G, Bruneau N, Lozovaya N, Labalme A, Boutry-Kryza N, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet 2013; 45: 1061–6. [DOI] [PubMed] [Google Scholar]
- Li D, Yuan H, Ortiz-Gonzalez XR, Marsh ED, Tian L, McCormick EM, et al. GRIN2D recurrent de novo dominant mutation causes a severe epileptic encephalopathy treatable with NMDA receptor channel blockers. Am J Hum Genet 2016; 99: 802–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XB, Murray KD, Jones EG. Switching of NMDA receptor 2A and 2B subunits at thalamic and cortical synapses during early postnatal development. J Neurosci 2004; 24: 8885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marret S, Gressens P, Evrard P. Arrest of neuronal migration by excitatory amino acids in hamster developing brain. Proc Natl Acad Sci USA 1996; 93: 15463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marret S, Mukendi R, Gadisseux JF, Gressens P, Evrard P. Effect of ibotenate on brain development: an excitotoxic mouse model of microgyria and posthypoxic-like lesions. J Neuropathol Exp Neurol 1995; 54: 358–70. [DOI] [PubMed] [Google Scholar]
- Messersmith EK, Feller MB, Zhang H, Shatz CJ. Migration of neocortical neurons in the absence of functional NMDA receptors. Mol Cell Neurosci 1997; 9: 347–57. [DOI] [PubMed] [Google Scholar]
- Mirzaa GM, Parry DA, Fry AE, Giamanco KA, Schwartzentruber J, Vanstone M, et al. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat Genet 2014; 46: 510–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JG. Structural modelling pipelines in next generation sequencing projects. Adv Protein Chem Struct Biol 2012; 89: 117–67. [DOI] [PubMed] [Google Scholar]
- Murthy SE, Shogan T, Page JC, Kasperek EM, Popescu GK. Probing the activation sequence of NMDA receptors with lurcher mutations. J Gen Physiol 2012; 140: 267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 2000; 302: 205–17. [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet 2011; 43: 585–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden KK, Chen W, Swanger SA, McDaniel MJ, Fan LZ, Hu C, et al. Molecular mechanism of disease-associated mutations in the pre-M1 helix of NMDA receptors and potential rescue pharmacology. PLoS Genet 2017; 13: e1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba C, Shiina M, Tohyama J, Haginoya K, Lerman-Sagie T, Okamoto N, et al. GRIN1 mutations cause encephalopathy with infantile-onset epilepsy, and hyperkinetic and stereotyped movement disorders. Epilepsia 2015; 56: 841–8. [DOI] [PubMed] [Google Scholar]
- Olney JW, Wozniak DF, Jevtovic-Todorovic V, Farber NB, Bittigau P, Ikonomidou C. Drug-induced apoptotic neurodegeneration in the developing brain. Brain Pathol 2002; 12: 488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 2004; 25: 1605–12. [DOI] [PubMed] [Google Scholar]
- Pierson TM, Yuan H, Marsh ED, Fuentes-Fajardo K, Adams DR, Markello T, et al. GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Ann Clin Transl Neurol 2014; 1: 190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer K, Yuan H, Schütz H, Winschel A, Chen W, Hu C, et al. GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J Med Genet 2017; 54: 460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 2010; 20: 110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redin C, Gérard B, Lauer J, Herenger Y, Muller J, Quartier A, et al. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet 2014; 51: 724–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt K, Mohr A, Gartner J, Spohr HL, Brockmann K. Polymicrogyria in fetal alcohol syndrome. Birth Defects Res A Clin Mol Teratol 2010; 88: 128–31. [DOI] [PubMed] [Google Scholar]
- Reiprich P, Kilb W, Luhmann HJ. Neonatal NMDA receptor blockade disturbs neuronal migration in rat somatosensory cortex in vivo. Cereb Cortex 2005; 15: 349–58. [DOI] [PubMed] [Google Scholar]
- Riviere JB, Mirzaa GM, O’Roak BJ, Beddaoui M, Alcantara D, Conway RL, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet 2012; 44: 934–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Chatron N, Labalme A, Ville D, Carneiro M, Edery P, et al. Novel homozygous missense variant of GRIN1 in two sibs with intellectual disability and autistic features without epilepsy. Eur J Hum Genet 2017; 25: 376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman SM, Olney JW. Glutamate and the pathophysiology of hypoxic—ischemic brain damage. Ann Neurol 1986; 19: 105–11. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11: 361–2. [DOI] [PubMed] [Google Scholar]
- Simon RP, Swan JH, Griffiths T, Meldrum BS. Blockade of N-methyl-D-aspartate receptors may protect against ischemic damage in the brain. Science 1984; 226: 850–2. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 2009; 462: 745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squier W, Jansen A. Polymicrogyria: pathology, fetal origins and mechanisms. Acta Neuropathol Commun 2014; 2: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutterd CA, Leventer RJ. Polymicrogyria: a common and heterogeneous malformation of cortical development. Am J Med Genet C Semin Med Genet 2014; 166C: 227–39. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Kadiyala P, Hariharan P, Neeraj E. A rare case of glycine encephalopathy unveiled by valproate therapy. J Pediatr Neurosci 2015; 10: 143–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanger SA, Chen W, Wells G, Burger PB, Tankovic A, Bhattacharya S, et al. Mechanistic insight into NMDA receptor dysregulation by rare variants in the GluN2A and GluN2B agonist binding domains. Am J Hum Genet 2016; 99: 1261–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Matsui K. Increased expression of GAP43 in interneurons in a rat model of experimental polymicrogyria. J Child Neurol 2015; 30: 716–28. [DOI] [PubMed] [Google Scholar]
- Takano T, Sawai C, Takeuchi Y. Radial and tangential neuronal migration disorder in ibotenate-induced cortical lesions in hamsters: immunohistochemical study of reelin, vimentin, and calretinin. J Child Neurol 2004; 19: 107–15. [DOI] [PubMed] [Google Scholar]
- Tarabeux J, Kebir O, Gauthier J, Hamdan FF, Xiong L, Piton A, et al. Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry 2011; 1: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BA, Storm MP, Hewinson J, Hogg S, Welham MJ, MacKenzie AB. A novel role for P2X7 receptor signalling in the survival of mouse embryonic stem cells. Cell Signal 2012; 24: 770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YB, Wang JJ, Wang SH, Liu SS, Cao JY, Li XM, et al. Adaptor protein APPL1 couples synaptic NMDA receptor with neuronal prosurvival phosphatidylinositol 3-kinase/Akt pathway. J Neurosci 2012; 32: 11919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe J, Beck C, Kuner T, Premkumar LS, Wollmuth LP. DRPEER: a motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J Neurosci 2002; 22: 10209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A. Comparative protein structure modeling using modeller. Curr Protoc Bioinformatics 2016; 54: 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Nakamichi N, Fukui M, Kitayama T, Georgiev DD, Makanga JO, et al. Promotion of neuronal differentiation through activation of N-methyl-D-aspartate receptors transiently expressed by undifferentiated neural progenitor cells in fetal rat neocortex. J Neurosci Res 2008; 86: 2392–402. [DOI] [PubMed] [Google Scholar]
- Yuan H, Erreger K, Dravid SM, Traynelis SF. Conserved structural and functional control of N-methyl-D-aspartate receptor gating by transmembrane domain M3. J Biol Chem 2005; 280: 29708–16. [DOI] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Zhang J, Pierson TM, Markello TC, Fajardo KV, et al. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat Commun 2014; 5: 3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehavi Y, Mandel H, Zehavi A, Rashid MA, Straussberg R, Jabur B, et al. De novo GRIN1 mutations: an emerging cause of severe early infantile encephalopathy. Eur J Med Genet 2017; 60: 317–320. [DOI] [PubMed] [Google Scholar]
- Zhou X, Chen Z, Yun W, Ren J, Li C, Wang H. Extrasynaptic NMDA receptor in excitotoxicity: function revisited. Neuroscientist 2015; 21: 337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hollern D, Liao J, Andrechek E, Wang H. NMDA receptor-mediated excitotoxicity depends on the coactivation of synaptic and extrasynaptic receptors. Cell Death Dis 2013; 4: e560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med 2015; 17: 774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.