Abstract
Background
Anaplasma phagocytophilum is a zoonotic tick-borne intracellular alpha-proteobacterium causing tick-borne fever, which leads to significant economic losses in domestic ruminants in Europe. Its epidemiological cycles are complex and reservoir host species of bovine strains have not yet been identified. Given that little genetic information is available on strains circulating within a defined bovine environment, our objective was to assess the genetic diversity of A. phagocytophilum obtained from the same farms over time.
Methods
Blood samplings were performed several times in two European herds. In the French herd, 169 EDTA-blood samples were obtained from 115 cows (32 were sampled two to four times). In the German herd, 20 cows were sampled six times (120 EDTA-blood samples). The presence of A. phagocytophilum DNA was assessed using a qPCR targeting msp2. The positive DNA samples underwent MLST at nine genetic markers (typA, ctrA, msp4, pleD, recG, polA, groEL, gyrA, and ankA). For each locus, sequences were aligned with available bacterial sequences derived from cattle, horse, dog, and roe deer hosts, and concatenated neighbor joining trees were constructed using three to six loci.
Results
Around 20% (57/289) of samples were positive. Forty positive samples from 23 French and six German cows (11 of them being positive at two time points) were sequenced. Six loci (typA, ctrA, msp4, pleD, recG, and polA) allowed to build concatenated phylogenetic trees, which led to two distinct groups of bovine variants in the French herd (hereafter called A and B), whereas only group A was detected in the German herd. In 42% of French samples, double chromatogram peaks were encountered in up to four loci. Eleven cows were found infected three weeks to 17 months after first sampling and harboured a new variant belonging to one or the other group.
Conclusions
Our results demonstrate the occurrence of two major bovine strain groups and the simultaneous infection of single cows by more than one A. phagocytophilum strain. This challenges the role of cattle as reservoirs for A. phagocytophilum. This role may be facilitated via long-term bacterial persistence in individual cows and active circulation at the herd scale.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2661-7) contains supplementary material, which is available to authorized users.
Keywords: Anaplasma phagocytophilum, Tick-borne disease, Bovine, France, Germany, MLST, Reservoir
Background
Anaplasma phagocytophilum is a zoonotic intracellular alpha-proteobacterium transmitted by ticks belonging to the Ixodes persulcatus complex. In Europe, the exophilic hard tick Ixodes ricinus is the main vector of A. phagocytophilum [1]. This bacterium infects a wide range of vertebrate hosts, including wild and domestic ruminants, dogs, horses, rodents and humans [2]. For decades it has been known that A. phagocytophilum causes tick-borne fever (TBF) in domestic ruminants in Europe [3, 4].
TBF outbreaks usually occur following the first time cattle graze in an endemic area [5]. The main clinical signs are hyperthermia, anorexia, mild to moderate depression, a sudden drop in milk yield, and abortions in pregnant females. Respiratory signs such as coughing, polypnea, nasal discharge, and abnormal lung sounds have also been described [6]. For European farms, this infection leads to significant economic losses [7], whereas TBF has never been described in the USA to date [8].
Anaplasma phagocytophilum epidemiological cycles are complex and differ greatly between the USA and Europe [8]. Several genetic lineages are described and some are thought to have specific host tropism [2]. Transovarial transmission has not yet been described in the vector tick under natural conditions [2]. Therefore, exploiting mammalian hosts as reservoirs is essential for A. phagocytophilum survival [8]. Several wild ruminant and rodent species are suspected to be involved [8].
In Europe, wild ruminants, especially red deer, are currently thought to carry strains infecting cattle [8–13]. Moreover, experimental infections in sheep - which are among the most frequently infected domestic species in Europe [14] - demonstrated long-term bacterial persistence in sheep peripheral blood [15], suggesting that they could also be reservoirs of sheep-infecting strains. However, proof that cattle play a role as reservoirs for cow-infecting strains has not yet been obtained.
The development of many novel molecular typing methods in recent years offers an unparalleled opportunity to generate new information about the complex epidemiology of A. phagocytophilum and its genetic diversity [8]. Among these techniques, multi-locus sequence typing (MLST), based on PCR amplification and sequencing of several housekeeping genes, is widely recognized as a reliable phylogenetic method for bacterial molecular characterization, specifically A. phagocytophilum, for which few other appropriate multi-loci typing methods are currently available [8, 9].
The objectives of this study were to explore the molecular epidemiology of A. phagocytophilum strains from cattle by longitudinally assessing their circulation, persistence, and genetic diversity in individual cattle and within herds from both France and Germany.
Methods
Animal sampling
French samples
In 2008, early clinical signs of disease, including fever, paresis, weight loss and abortions, and leading some animals to death, were observed in cattle (Bos taurus) in a farm near Gien, Sologne, France. Initial testing for intoxication returned negative results. When tick-borne fever was suggested as a possible cause, animals were treated with oxytetracycline without any further diagnostic investigation, and they recovered.
Since then, TBF has been continually clinically suspected on the farm, but clinical consequences have been less severe as animals were quickly treated. In October 2014, in March, May and October 2016, and in May 2017, blood sampling was preferentially performed on young cattle, and on cattle that were positive during previous samplings. In total, 169 EDTA-blood samples were obtained from 115 cows (Table 1). Thirty-two animals were sampled several times (between two to four times) during the study.
Table 1.
Detection of A. phagocytophilum DNA by real-time PCR in cattle from Sologne, France
| Date of sampling (year/month/day) | No. of cows sampled | No. of positive individuals (%) |
|---|---|---|
| 2014/10/09 | 32 | 7 (21.9) |
| 2016/03/03 | 34 | 6 (17.6) |
| 2016/05/17 | 40 | 9 (22.5) |
| 2016/10/26 | 34 | 3 (8.8) |
| 2017/05/16 | 29 | 6 (20.7) |
| Total no. of samples | 169 | 31 (18.3) |
German samples
In 2009 and 2010, several cows from a farm in Meschede, in North-Rhine Westphalia, presented with high fever, decreased milk yield, lower limb edema, and anorexia, one to two weeks after having been brought to pasture. After initially suspecting a pasture infection, A. phagocytophilum was finally detected in the herd by PCR and blood smears [16]. In 2011, cows and heifers of the herd were sampled six times, for a period of up to five months, from the point at which they first developed fever [17]. In total, 120 EDTA-blood samples were used for this study (Table 2) [17].
Table 2.
Detection of A. phagocytophilum DNA by real-time PCR in cattle from Meschede, Germany. The same 20 animals (including 19 heifers) were sampled over time
| Date of sampling (year/month/day) | No. of cows sampled | No. of positive individuals (%) |
|---|---|---|
| 2011/05/17–22 | 20 | 6 (30.0) |
| 2011/05/30–2011/06/04 | 20 | 7 (35.0) |
| 2011/06/14–22 | 20 | 7 (35.0) |
| 2011/07/09 | 20 | 2 (10.0) |
| 2011/09/07 | 20 | 3 (15.0) |
| 2011/10/03 | 20 | 1 (5.0) |
| Total no. of samples | 120 | 26 (21.7) |
DNA extraction and qPCR
DNA from EDTA-blood samples from French cattle was extracted using the NucleoSpin Blood QuickPure kit (Macherey-Nagel, Hoerdt, France), and DNA from German EDTA-blood samples with the Qiagen DNA Minikit (Qiagen, Hilden, Germany), both according to the manufacturer’s instructions. DNA extracts were stored at -20 °C prior to testing.
The presence of specific A. phagocytophilum DNA was assessed with a qPCR targeting a 77 bp fragment of the major surface protein 2 (msp2) gene, according to the protocol of Courtney et al. [18]. Positive (DNA from an A. phagocytophilum (Webster strain)-infected tick cell line IRE-CTVM20) and negative (molecular biology grade water) controls were included in each PCR run.
Genotyping
In order to type the A. phagocytophilum-positive samples, the nine loci selected in a recently published A. phagocytophilum MLST study were used: typA, ctrA (APH 1099–1100), msp4, pleD, recG, polA, groEL, gyrA, and ankA [9]. PCRs for the nine loci were performed according to Chastagner et al. [9]. A nested PCR was then performed for all loci except pleD, using 5 μl of the initial PCR product. Amplicons were separated by electrophoresis on 1.5% agarose gels and stained with ethidium bromide for imaging. If the initial amplification yielded sufficient DNA of the correct size, this PCR product was used for sequencing. Otherwise, the shorter fragment obtained with internal primers was used. The bands were excised from the gel and purified using NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel), following the manufacturer’s instructions. Nucleotide sequences were obtained from PCR products by Sanger sequencing in both directions (Eurofins Genomics, Ivry-sur-Seine, France). Sequencing results were manually edited in Bioedit software version 7.2.5 (Ibis Biosciences, Carlsbad, USA). IUPAC codes were used to indicate ambiguous loci.
Construction of phylogenetic trees
Nucleotide sequences were aligned using the MEGA7 software (Molecular Evolutionary Genetics Analysis version 7.0.18 [19]), with the ClustalW program. In order to build phylogenetic trees, the sequences of each locus obtained in this study were aligned with A. phagocytophilum sequences from cow, horse, dog, and roe deer hosts from Chastagner et al. [9] (GenBank: KJ832158–KJ833031) (Additional file 1: Table S1). Moreover, sequences of American human, dog, rodent, and tick A. phagocytophilum strains (HZ, Webster, HGE1, Dog2, JM and CRT), and a Norwegian sheep (Ovis aries) strain (Norway_variant2), were added to the alignments [9].
Phylogenetic trees were built after concatenation of three to six loci. Concatenated sequences were imported and neighbor joining trees (NJ trees) were constructed from the alignment using BioNumerics version 7.6.1 software (Applied Maths, Sint-Martens-Latem, Belgium). A profile is defined here as A. phagocytophilum sequences obtained from chromatograms for three to six loci. A phylogenetic tree based on the concatenation of six markers was built for samples with complete profiles, i.e. complete sequences without ambiguities of all six genetic markers, with the addition of concatenated sequences from 40 cows and two horses obtained by Chastagner et al. [9] and HZ, Webster, Dog2, JM, and Norway_variant2 strains.
Due to missing data or ambiguous sites for some loci, other trees based on the concatenation of five loci (typA, ctrA, pleD, recG and polA), four loci (typA, ctrA, msp4 and pleD; typA, ctrA, msp4 and polA; typA, ctrA, msp4 and recG), and three loci (typA, ctrA and msp4) were built. Other host species samples (six more horses, three dogs, five roe deer, HGE1, and CRT strains) from Chastagner et al. [9], which had generated sequences for only three or four loci, were added to the corresponding concatenated trees.
Results
Detection of A. phagocytophilum DNA
French samples
Altogether, 31/169 (18.3%) blood samples from 23 cattle (20/23 were heifers) were positive for specific A. phagocytophilum DNA with msp2-real-time PCR. The percentage of infected samples varied from 8.8% (October 2016) to 22.5% (May 2016) (Table 1). Of the 32 cows sampled at least twice during the experiment, eight yielded positive results at two successive sampling dates: four of them were positive two months apart between March and May 2016, three were positive after seven months between October 2016 and May 2017, and a bull was still found to be infected after 17 months (Table 3).
Table 3.
French cattle found to be infected twice during the sampling period
| Bovine ID | Sampling date (year/month/day) | Time span between the two positive samples (months) | ||||
|---|---|---|---|---|---|---|
| 2014/10/09 | 2016/03/03 | 2016/05/17 | 2016/10/26 | 2017/05/16 | ||
| BV2802 | Pos | Pos | Neg | na | na | 17 |
| BV0001 | na | Pos | Pos | Neg | Neg | 2.5 |
| BV0006 | na | Pos | Pos | Neg | Neg | 2.5 |
| BV0016 | na | Pos | Pos | Neg | Neg | 2.5 |
| BV9949 | na | Pos | Pos | Neg | Neg | 2.5 |
| BV0012 | na | na | na | Pos | Pos | 7 |
| BV0047 | na | na | na | Pos | Pos | 7 |
| BV0048 | na | na | na | Pos | Pos | 7 |
Abbreviations: Pos blood sample positive with msp2 qPCR; Neg blood sample negative with msp2 qPCR; na not applicable (no sampling at this date)
German samples
The number of positive cattle at each sampling date varied from one (5%) to seven (35%) (Table 2). Altogether, 26/120 (21.7%) samples were found positive for specific A. phagocytophilum DNA with msp2-real-time PCR. One positive sample from the study performed in 2010 [16], and eight samples from five cows sampled in 2011 [17] were available for genotyping (Table 4).
Table 4.
German samples used for molecular typing and dates of sampling
| Bovine ID | Dates of positive samples used for typing | Time span between the two positive results | |
|---|---|---|---|
| First sampling date (year/month/day) | Second sampling date (year/month/day) | ||
| BV34 | 2010/10/17 | na | na |
| BV46 | 2011/05/22 | 2011/08/21 | 3 months |
| BV49 | 2011/05/19 | na | na |
| BV57 | 2011/06/04 | na | na |
| BV58 | 2011/05/17 | 2011/07/09 | 1.5 months |
| BV61 | 2011/06/22 | 2011/07/09 | 3 weeks |
Abbreviation: na not applicable
Genotyping and polymorphism analysis
Amplicons obtained with external or internal primers were sent for sequencing. For six loci (typA, ctrA, msp4, pleD, recG, and polA), the number of available sequences varied from 35 to 40 (all samples generated results for the typA locus). Sequence lengths after alignment ranged from 309 (recG) to 597 bp (pleD) (Table 5). Regarding groEL, gyrA, and ankA, only few samples gave sequencing results. Consequently, consecutive analyses were made using the loci typA, ctrA, msp4, pleD, recG, and polA.
Table 5.
Genetic diversity observed at each locus. Amplicon lengths with external or internal primers were obtained with Primer-BLAST (NCBI), apart from the PCR products with recG external primers, and polA external and internal primers, which were estimated on agarose gels
| typA | ctrA | msp4 | pleD | recG | polA | |
|---|---|---|---|---|---|---|
| No. of complete sequences | 40 | 39 | 36 | 36 | 35 | 37 |
| No. of sequences without ambiguities | 31 | 39 | 28 | 33 | 33 | 31 |
| No. of sequences with ambiguities (%) | 9 (22.5) | 0 (0) | 8 (22.2) | 3 (8.3) | 2 (5.7) | 6 (16.2) |
| Amplicon length with external primers (bp) | 1455 | 1453 | 558 | 1102 | 515 | ~490 |
| Amplicon length with internal primers (bp) | 550 | 574 | 471 | na | ~480 | ~460 |
| Alignment length (bp) | 385 | 317 | 337 | 597 | 309 | 331 |
| No. of polymorphic sites | 5 | 6 | 11 | 20 | 6 | 7 |
| Proportion of polymorphic sites | 1.30 | 1.89 | 3.26 | 3.35 | 1.94 | 2.11 |
| No. of alleles at each locus | 7 | 6 | 7 | 3 | 8 | 8 |
Abbreviation: na not applicable
Loci with the highest percentage of polymorphic sites were pleD (3.35%) and msp4 (3.26%). The number of alleles for each locus varied from three (pleD) to eight (polA and recG). In 42% of French samples (13/31), double chromatograms peaks were encountered in up to four loci (typA, msp4, pleD, recG, and/or polA), which were confirmed by repeat sequencing on new amplicons. Ambiguities were only identified at particular polymorphic sites. In those samples with double peaks, the majority occurred at either typA or msp4 loci, and five samples had ambiguous sites at both loci (Tables 5, 6). We excluded all samples with sequences presenting at least one ambiguous site from subsequent analyses, to avoid incongruence between loci.
Table 6.
Complete sequencing results for the six loci and ambiguity occurrences. Those cattle for whom all six loci sequences were obtained are in bold. The strain clustering within the two major groups, based on the concatenation of five (all but msp4) or six loci, is indicated in the last column
| Bovine ID | Sampling date (year/month/day) | Date of birth | typA | ctrA | msp4 | pleD | recG | polA | Group in NJ tree |
|---|---|---|---|---|---|---|---|---|---|
| Twice-positive French samples | |||||||||
| BV2802 (Bull) | 2014/10/09 | 2011/11/27 | OK | OK | OK | OK | OK | OK | A |
| BV2802 (Bull) | 2016/03/04 | OK | OK | OK | OK | OK | OK | B | |
| BV9949 | 2016/03/04 | 2014/03/12 | OK | OK | OK | OK | OK | OK | A |
| BV9949 | 2016/05/17 | 1 AS | OK | OK | OK | OK | OK | ||
| BV0001 | 2016/03/04 | 2015/04/05 | 1 AS | OK | 6 AS | OK | OK | 2 AS | |
| BV0001 | 2016/05/17 | 3 AS | OK | 9 AS | 19 AS | OK | 1 AS | ||
| BV0006 | 2016/03/04 | 2015/04/27 | 1 AS | OK | OK | OK | OK | OK | |
| BV0006 | 2016/05/17 | OK | OK | 9 AS | OK | OK | 2 AS | ||
| BV0016 | 2016/03/04 | 2015/11/14 | OK | na | 4 AS | OK | OK | na | |
| BV0016 | 2016/05/17 | OK | OK | OK | OK | na | 2 AS | ||
| BV0012 | 2016/10/25 | 2015/07/09 | OK | OK | OK | OK | OK | OK | B |
| BV0012 | 2017/05/16 | OK | OK | OK | OK | OK | OK | A | |
| BV0047 | 2016/10/25 | 2016/08/29 | OK | OK | OK | OK | OK | OK | A |
| BV0047 | 2017/05/16 | OK | OK | OK | OK | OK | OK | A | |
| BV0048 | 2016/10/25 | 2016/08/31 | OK | OK | OK | OK | OK | OK | A |
| BV0048 | 2017/05/16 | OK | OK | OK | OK | OK | OK | B | |
| Other French samples | |||||||||
| BV1551 | 2014/10/09 | 2003/01/02 | OK | OK | OK | OK | OK | OK | B |
| BV9932 | 2014/10/09 | 2013/05/03 | OK | OK | OK | OK | OK | OK | A |
| BV9940 | 2014/10/09 | 2013/05/16 (dead 2015/02/23) | 2 AS | OK | 3 AS | 19 AS | na | OK | |
| BV9942 | 2014/10/09 | 2013/05/27 | OK | OK | OK | OK | na | OK | |
| BV9952 | 2014/10/09 | 2014/04/13 | 3 AS | OK | 5 AS | OK | na | OK | |
| BV9873 | 2014/10/09 | 2012/03/11 (dead 2016/06/09) | OK | OK | 2 AS | na | na | na | |
| BV9959 | 2016/03/04 | 2014/05/05 | OK | na | OK | na | OK | OK | |
| BV2193 | 2016/05/17 | na | OK | OK | OK | OK | OK | OK | A |
| BV2302 | 2016/05/17 | na | OK | OK | OK | OK | OK | OK | B |
| BV3559 | 2016/05/17 | na | OK | OK | OK | OK | OK | OK | A |
| BV0013 | 2016/05/17 | 2015/07/09 | 1 AS | OK | 10 AS | OK | 2 AS | 1 AS | |
| BV9955 | 2016/05/17 | 2014/04/15 | OK | OK | OK | OK | OK | OK | B |
| BV0027 | 2017/05/16 | 2016/04/13 | 5 AS | OK | OK | OK | OK | OK | |
| BV0049 | 2017/05/16 | 2016/10/05 | 5 AS | OK | OK | 19 AS | 1 AS | 2 AS | |
| BV0056 | 2017/05/16 | 2016/11/23 | OK | na | OK | OK | OK | OK | |
| Twice-positive German samples | |||||||||
| BV46 | 2011/05/22 | 2008/08/27 | OK | OK | na | OK | OK | OK | A |
| BV46 | 2011/08/21 | OK | OK | OK | OK | OK | OK | A | |
| BV58 | 2011/05/17 | 2008/08/18 | OK | OK | na | OK | OK | OK | A |
| BV58 | 2011/07/09 | OK | OK | OK | OK | OK | na | A | |
| BV61 | 2011/06/22 | 2008/10/15 | OK | OK | na | OK | OK | OK | A |
| BV61 | 2011/07/09 | OK | OK | na | OK | OK | OK | A | |
| Other German samples | |||||||||
| BV49 | 2011/05/19 | 2008/03/12 | OK | OK | OK | OK | OK | OK | A |
| BV34 | 2010/10/17 | 2007/10/14 | OK | OK | OK | OK | OK | OK | A |
| BV57 | 2011/06/04 | 2007/11/27 | OK | OK | OK | OK | OK | OK | A |
Abbreviations: OK complete sequence for the locus, without double peaks, AS ambiguous sites, na not available
Nineteen samples (four German samples from four cows, and 15 French samples from 11 cows) generated complete profiles for the six markers, without any ambiguities (Table 6).
The sequences obtained in this study are available in the GenBank database under the accession numbers MF580591–MF580636.
Phylogenetic analyses
Nineteen samples from our study and sequences from Chastagner et al. [7] were included in the phylogenetic tree based on the concatenation of six loci (Fig. 1). Building this tree allowed identifying groups of isolates and variants within groups. Variants are defined here as bacteria that evolved from the same bacterial cell. In the case of intra-cellular bacteria such as A. phagocytophilum, mutations (and possibly recombinations) occur at a high rate in single strains within an infected host. A group therefore gathers together variants that have highly similar profiles according to phylogenetic analyses, and that probably share the same ancestor. Two groups of bovine variants (A and B) were obtained, which were clearly distinct from the American strains, from the French horse strains, and from the Norwegian sheep strain (Table 6). Both groups were detected within the French herd, whereas only group A was present in the German herd. Interestingly, four French cows with positive results at two different time points enabled the comparison of two entire profiles from the same animal. Three of these cows (BV0012, BV0048 and BV2802), had a different profile at each time point which clustered into separate groups. Conversely, the two profiles from the fourth cow (BV0047) both clustered into group A.
Fig. 1.
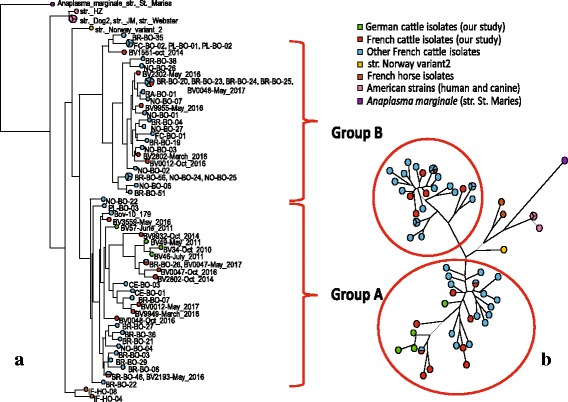
Neighbor-joining trees obtained with six loci concatenation. Each circle represents a unique sequence type. Logarithmic scale is used. To increase the readability of the second tree (b), names of samples have been removed, but can be found in the first tree (a). German cattle samples are in green, whereas the French are in red (our study), or in blue (Chastagner et al. study [9]). The two groups of cattle strains are identified and are clearly distinct from horse strains and American strains
Four German samples with indecipherable msp4 sequences were added to the above-mentioned samples in order to build a phylogenetic tree based on concatenation of five loci (Additional file 2: Figure S1). Clustering reconfirmed the presence of two cattle groups, and the additional German samples clustered into group A. Interestingly, the profiles from BV46 and BV61 which were positive at two time points, differed between the two sampling dates. The two heifers were first infected by a variant from the same group, and subsequently (three weeks for BV61 and three months for BV46) they were both infected by another variant, which again shared the same profile.
The tree based on typA, ctrA, msp4, and pleD concatenation (Additional file 3: Figure S2) was consistent with the two other trees, whereas bovine groups became mixed when using the other loci concatenations (Additional file 4 Figure S3, Additional file 5: Figure S4, Additional file 6: Figure S5). Sequences from roe deer on one hand, and from dogs and horses on the other, also clustered into two distinct groups.
Discussion
Our results reveal the co-existence of two major groups of bovine A. phagocytophilum strains among all cattle samples analyzed in this study. These groups did not associate with geographical origin, as cattle from the study by Chastagner et al. [9] originated from several French regions and clustered into either group. Moreover, these two groups remained stable over time according to MLST profiles, as identical sequences were obtained 31 months apart in the French herd. Although the bovine strains of these two groups are clearly distinct in NJ trees, the variant diversity within them remains high, as has already been described [20]. The variability of A. phagocytophilum strains within defined sheep flocks was already mentioned [10, 21–23]. Even so, there is no molecular typing data from any longitudinal study of A. phagocytophilum infection in cattle herds.
The six loci used in this study were sufficient to discriminate isolates belonging to the two groups, variants within the same group and successive isolates from the same animal. Indeed, the two strains from BV61, which were obtained only three weeks apart, were separated by using the concatenation of only five markers (typA, ctrA, pleD, recG, and polA). Given the irregularity in the collection of samples over time and given the possible interference of antibiotic treatments, this study was not intended to determine the intra-herd infection rate but instead to collect data on the molecular characteristics of circulating A. phagocytophilum strains within one given herd. Indeed, two groups, with variants within them, were clearly observed. Nevertheless, the use of additional markers could help to discriminate isolates obtained from the same animal at very close successive dates. Thus, another longitudinal study with more frequent and regular sampling would be very interesting in order to study in depth the succession of strains and/or the evolution of strains with time within one given animal.
The two groups of A. phagocytophilum strains co-circulated within the French farm, whereas only one was observed in the German herd. The presence of variants from only one group in the German herd could be related to the fact that they are dairy cows (red and black Holsteins, German Simmental and hybrids), whereas the French herd are beef cattle (Limousines). Dairy cows are more confined, have less extensive grazing pastures, and return every day to the stable, so may be less likely to contract infections by different A. phagocytophilum strains through tick bites.
In addition, in 42% of French samples (13/31), double chromatogram peaks were encountered. The presence of these ambiguities suggests the simultaneous occurrence of two A. phagocytophilum profiles resulting from two different alleles at some positions. In some cases, these double peaks could be the consequence of punctual mutations occurring during a prolonged infection by a single strain that evolved over time within the host, leading to a final co-existence of two variants. This is likely the case when only one or two ambiguities at a single locus are observed in a sample. Nevertheless, other cases were suggestive of simultaneous infection by two or more distinct strains, as several samples contained sequences with many successive ambiguities (5/5 for typA, 10/11 for msp4, and 19/20 for pleD). This could result from the simultaneous transmission of several strains by the same tick, or from several ticks biting the same cow, leading to several independent inoculations of different strains. Interestingly, four of the eight French cattle that were positive at two different time points presented ambiguities in up to four loci, and for three cows, this occurred on both sampling dates. In future studies, performing PCR products cloning could lead to the identification of strains that are associated in multiple infections and could help to investigate whether specific strains are more likely to be present in simultaneous infections.
The presence of multiple infections has already been described [9, 12, 21, 23, 24] and is considered to be a major drawback in typing methods [12]. The MLST study of Huhn et al. [12] demonstrated that double infections are more frequently found in wild ruminants, and another study indicated that 60.7% of infected French roe deer harbored two or three genetic variants [24]. Chastagner et al. [9] reported that approximately 30% of roe deer samples presented multiple infections, compared to very few cattle samples. In addition, in a longitudinal study involving lambs, only 4 out of 85 PCR-positive samples (4.7%) were infected with multiple A. phagocytophilum strains [21]. In our study, more than 40% of A. phagocytophilum sequences obtained from cattle in France presented double peaks, a higher frequency than that which was previously reported [9, 12]. Assuming that ticks can harbor several A. phagocytophilum strains, animals with frequent tick exposure could be at risk of multiple infections. In Sologne, ticks were frequently found on cattle, so it was assumed that infection pressure with multiple strains could be high. Conversely, none of the A. phagocytophilum sequences obtained from cattle in Germany presented double peaks. This could be due to the fact that all variants circulating within the farm belonged to the same group. Nevertheless, such typing methods are only able to detect simultaneous infections by multiple strains if one variant does not dominate the pool, so the number of multiple infections could be underestimated.
This work also raises the question whether cattle can act as reservoirs for A. phagocytophilum. Eight French cattle and three German cows remained positive after several weeks or months. Three French cattle (BV0012, BV0048, and BV2802) were found to be successively infected by strains from each group, but the order of infection was not always the same (group A strain then group B strain, or vice versa) (Table 6). These results demonstrate that neither of the two groups appears to be predominant, and instead may favor reinfection by a different variant, which strongly suggests the existence of complex interactions between cattle and A. phagocytophilum, as well as a lack of cross protection against other A. phagocytophilum strains. Conversely, for BV0047 (French heifer), BV46 and BV61 (German heifers), both the initial and subsequent infective strains belonged to the same group and were genetically similar. These results favor a long-term infection by the same variant, which could have evolved with time, perhaps to counteract the host immune response. It is noteworthy to mention here that one of the two German samples (BV61) also displayed another msp2 sequence although samples were taken only three weeks apart [13]. Future work should aim to sample animals daily or weekly in order to determine if long-term infections result from the evolution and divergence of the initial strain and/or by reinfection with another variant.
Moreover, as several animals were infected at each sampling time point, it could be hypothesized that a reservoir could exist at the herd scale. Such a reservoir would facilitate tick reinfection following tick inactivity periods, subsequent transmission to other cattle and the circulation of various strains within the herd. An experimental study in sheep demonstrated that A. phagocytophilum can persist in the tissues of this theoretical reservoir, even though blood PCR analyses failed to detect infection [25]. It has also been shown that sheep are efficient reservoirs of A. phagocytophilum even during the post-acute phase of infection [26], and thus these bacteria could periodically circulate in the blood and infect feeding ticks. Further studies are required to confirm the hypothesis that cattle could be considered as reservoirs, whether via long-term A. phagocytophilum persistence at the animal scale, and/or via active A. phagocytophilum circulation at herd scale, taking into account strain evolution in infected animals. This hypothesis is already reinforced by both our own results and by several previous studies.
First, in our study, none of the French and German bovine strains clustered with A. phagocytophilum strains from other host species, which clustered into two distinct phylogenetic groups, separating horse and dog sequences from roe deer sequences. This was not unexpected as only a few cattle sequences had been associated with these two groups in a previous study [9]. Secondly, recent multiple-locus variable number tandem repeat analysis showed that cattle strains could be divided into two groups [10]. In the aforementioned study, the first group clustered only with red deer (Cervus elaphus) strains, whereas the second clustered with strains isolated from various host species (roe deer, horses, dogs, humans, and sheep). Cattle may represent accidental hosts for variants belonging to the second group, which also included sheep, another domestic ruminant species already recognized as an A. phagocytophilum reservoir species for its own strains [8]. These data are very interesting as cattle and sheep do not usually share grazing pastures in France and Germany. It is therefore unlikely that sheep are potential reservoirs of cattle strains in these countries. Conversely, both red deer (suspected reservoir hosts of cattle variants [8–13]) and cattle herds themselves (current results) could be implicated according to ecological contexts, in the long term persistence of cattle-infecting strains. This role does not contradict the development of protective immunity at the scale of the individual animal [27], as indicated by the young mean age of positive cattle in our study. In recently infected animals, there could be a window of time where A. phagocytophilum may escape the immune response via antigenic variations and other mechanisms. Moreover, according to our results, it seems likely that a new strain is able to co-exist with another strain and/or with the immune response previously developed by a cow against another strain.
Conclusions
For the first time, our study demonstrates the co-circulation of two major groups of A. phagocytophilum strains within a cattle herd in France and the presence of multiple variants within the group(s) circulating in France and in Germany. Eleven cows were found infected several weeks to several months after previous sampling, with a variant belonging or not to the same group. Moreover, 42% of French cattle presented ambiguous sequence reads at one or several loci, suggestive of simultaneous infections with multiple strains. Interestingly, at each sampling time point, A. phagocytophilum DNA was detected in blood samples from several cattle without clinical signs. This suggests that cattle could play the role of reservoir for strains with bovine host tropism, at least at the herd level.
Additional files
DNA samples from Chastagner et al. [9] used for the alignments. The available sequences for each locus are indicated by an “X”. (XLSX 12 kb)
NJ tree obtained using the concatenation of typA, ctrA, pleD, recG, and polA. Legends as in Fig. 1. (PDF 1383 kb)
NJ tree obtained using the concatenation of typA, ctrA, msp4, and pleD. Legends as in Fig. 1. (PDF 1399 kb)
NJ tree obtained using the concatenation of typA, ctrA, msp4, and recG. Legends as in Fig. 1. (PDF 1395 kb)
NJ tree obtained using the concatenation of typA, ctrA, msp4, and polA. Legends as in Fig. 1. (PDF 1434 kb)
NJ tree obtained using the concatenation of typA, ctrA, and msp4. Legends as in Fig. 1. (PDF 1438 kb)
Acknowledgements
The authors thank Juliette Bavouzet, Perrine Lequitte-Charransol, Christelle Gandouin, and Corinne Bouillin for their technical support. We also thank Rémi Nantois and Brigitte Macé, the French cattle owners and the family of the owner of the German cattle herd for their great cooperation and for allowing sampling of their cattle. The work of MP and CS was performed under the umbrella of the EurNegVec COST Action TD1301.
Funding
This work was supported by the French Ministry of Agriculture, the French Agency for Food, Environmental and Occupational Health and Safety (Anses) and the French National Institute for Agricultural Research (INRA).
Availability of data and materials
All data supporting the conclusions of this article are included within the article and its additional files. The sequences were submitted to the GenBank database under the accession numbers MF580591–MF580636.
Abbreviations
- MLST
Multi-locus sequence typing
- msp2
Major surface protein 2
- NJ
Neighbor joining
- TBF
Tick-borne fever
Authors’ contributions
ACL contributed to experimental design, to French cattle sample collection, to genotyping data analysis, was responsible for laboratory work and drafted the manuscript. CR and MK participated in French cattle sample collection, in laboratory work and genotyping data analysis, as well as critical revision of the manuscript. TD participated in laboratory work and critical revision of the manuscript. GG helped perform phylogenetic analysis and participated in critical revision of manuscript. BD helped with genotyping analysis and critical revision of the manuscript. MP, CS, and MN contributed to German cattle sample collection and were involved in critical revision of the manuscript. HJB and NH were responsible for the conception and contributed to experimental design and for drafting the manuscript. All authors read and approved the final manuscript.
Ethics approval
The domestic animals used in this study met the definition of “farm animals”. They are not covered by French regulations to date (Décret 2013-118 dated the 1st February 2013, from the French Ministry of Agriculture). The owners of the animals provided permission for cattle sampling during our study. In order that breeding farms remained anonymous, samples were de-identified in this publication. The sampling of German cattle was performed for diagnostic purposes as 2011 was the first season the previously unknown Schmallenberg virus emerged and sickened cattle in Germany and in other European countries. The town of Schmallenberg where the virus was first detected and described is only 25 km away from our studied farm.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2661-7) contains supplementary material, which is available to authorized users.
Contributor Information
Anne-Claire Lagrée, Email: anne-claire.lagree@vet-alfort.fr, Email: aclairelecorre@yahoo.fr.
Clotilde Rouxel, Email: clotilde.rouxel@vet-alfort.fr.
Maëllys Kevin, Email: maellys.kevin.ext@anses.fr.
Thibaud Dugat, Email: thibaud.dugat.pro@gmail.com.
Guillaume Girault, Email: guillaume.girault@anses.fr.
Benoît Durand, Email: Benoit.DURAND@anses.fr.
Martin Pfeffer, Email: pfeffer@vetmed.uni-leipzig.de.
Cornelia Silaghi, Email: cornelia.silaghi@fli.de.
Marion Nieder, Email: marion_nieder@gmx.de.
Henri-Jean Boulouis, Email: henri-jean.boulouis@vet-alfort.fr.
Nadia Haddad, Email: nadia.haddad@vet-alfort.fr.
References
- 1.Rizzoli A, Silaghi C, Obiegala A, Rudolf I, Hubalek Z, Földvari G, et al. Ixodes ricinus and its transmitted pathogens in urban and peri-urban areas in Europe: new hazards and relevance for public health. Front Public Health. 2014;2:251. doi: 10.3389/fpubh.2014.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum - a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol. 2013;3:31. doi: 10.3389/fcimb.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon WS, Brownlee A, Wilson DR, MacLeod J. Tick-borne fever: a hitherto undescribed disease of sheep. J Comp Pathol Ther. 1932;45:301–307. doi: 10.1016/S0368-1742(32)80025-1. [DOI] [Google Scholar]
- 4.Foggie A. Studies on the infectious agent of tick-borne fever in sheep. J Pathol. 1951;63:1–15. doi: 10.1002/path.1700630103. [DOI] [PubMed] [Google Scholar]
- 5.Pusterla N, Pusterla JB, Braun U, Lutz H. Serological, hematologic, and PCR studies of cattle in an area of Switzerland in which tick-borne fever (caused by Ehrlichia phagocytophila) is endemic. Clin Diagn Lab Immunol. 1998;5:325–327. doi: 10.1128/cdli.5.3.325-327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pusterla N, Braun U. Clinical findings in cows after experimental infection with Ehrlichia phagocytophila. Transbound Emerg Dis. 1997;44:385–390. doi: 10.1111/j.1439-0442.1997.tb01123.x. [DOI] [PubMed] [Google Scholar]
- 7.Woldehiwet Z. The natural history of Anaplasma phagocytophilum. Vet Parasitol. 2010;167:108–122. doi: 10.1016/j.vetpar.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Dugat T, Lagrée A-C, Maillard R, Boulouis H-J, Haddad N. Opening the black box of Anaplasma phagocytophilum diversity: current situation and future perspectives. Front Cell Infect Microbiol. 2015;5:61. doi: 10.3389/fcimb.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chastagner A, Dugat T, Vourc’h G, Verheyden H, Legrand L, Bachy V, et al. Multilocus sequence analysis of Anaplasma phagocytophilum reveals three distinct lineages with different host ranges in clinically ill French cattle. Vet Res. 2014;45:114. doi: 10.1186/s13567-014-0114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dugat T, Chastagner A, Lagrée A-C, Petit E, Durand B, Thierry S, et al. A new multiple-locus variable-number tandem repeat analysis reveals different clusters for Anaplasma phagocytophilum circulating in domestic and wild ruminants. Parasit Vectors. 2014;7:439. doi: 10.1186/1756-3305-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dugat T, Zanella G, Véran L, Lesage C, Girault G, Durand B, et al. Multiple-locus variable-number tandem repeat analysis potentially reveals the existence of two groups of Anaplasma phagocytophilum circulating in cattle in France with different wild reservoirs. Parasit Vectors. 2016;9:596. doi: 10.1186/s13071-016-1888-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huhn C, Winter C, Wolfsperger T, Wüppenhorst N, Strašek Smrdel K, Skuballa J, et al. Analysis of the population structure of Anaplasma phagocytophilum using multilocus sequence typing. PLoS One. 2014;9:e93725. doi: 10.1371/journal.pone.0093725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scharf W, Schauer S, Freyburger F, Petrovec M, Schaarschmidt-Kiener D, Liebisch G, et al. Distinct host species correlate with Anaplasma phagocytophilum ankA gene clusters. J Clin Microbiol. 2011;49:790–796. doi: 10.1128/JCM.02051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woldehiwet Z. Anaplasma phagocytophilum in ruminants in Europe. Ann N Y Acad Sci. 2006;1078:446–460. doi: 10.1196/annals.1374.084. [DOI] [PubMed] [Google Scholar]
- 15.Thomas RJ, Birtles RJ, Radford AD, Woldehiwet Z. Recurrent bacteraemia in sheep infected persistently with Anaplasma phagocytophilum. J Comp Pathol. 2012;147:360–367. doi: 10.1016/j.jcpa.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Nieder M, Silaghi C, Hamel D, Pfister K, Schmäschke R, Pfeffer M, et al. Tick-borne fever caused by Anaplasma phagocytophilum in Germany. Tierärztl Prax Grosstiere. 2012;40:101–106. [PubMed] [Google Scholar]
- 17.Silaghi C, Nieder M, Sauter-Louis C, Knubben-Schweizer G, Pfister K, Pfeffer M. Epidemiology, genetic variants and clinical course of natural infections with Anaplasma phagocytophilum in a dairy cattle herd. Parasit Vectors. 2018;11:20. doi: 10.1186/s13071-017-2570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Courtney JW, Kostelnik LM, Zeidner NS, Massung RF. Multiplex real-time PCR for detection of Anaplasma phagocytophilum and Borrelia burgdorferi. J Clin Microbiol. 2004;42:3164–3168. doi: 10.1128/JCM.42.7.3164-3168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. [DOI] [PMC free article] [PubMed]
- 20.Bown KJ, Lambin X, Ogden NH, Petrovec M, Shaw SE, Woldehiwet Z, et al. High-resolution genetic fingerprinting of European strains of Anaplasma phagocytophilum by use of multilocus variable-number tandem-repeat analysis. J Clin Microbiol. 2007;45:1771–1776. doi: 10.1128/JCM.00365-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladbury GAF, Stuen S, Thomas R, Bown KJ, Woldehiwet Z, Granquist EG, et al. Dynamic transmission of numerous Anaplasma phagocytophilum genotypes among lambs in an infected sheep flock in an area of anaplasmosis endemicity. J Clin Microbiol. 2008;46:1686–1691. doi: 10.1128/JCM.02068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derdáková M, Štefančíková A, Špitalská E, Tarageľová V, Košťálová T, Hrkľová G, et al. Emergence and genetic variability of Anaplasma species in small ruminants and ticks from central Europe. Vet Microbiol. 2011;153:293–298. doi: 10.1016/j.vetmic.2011.05.044. [DOI] [PubMed] [Google Scholar]
- 23.Stuen S, Van De Pol I, Bergstrom K, Schouls LM. Identification of Anaplasma phagocytophila (formerly Ehrlichia phagocytophila) variants in blood from sheep in Norway. J Clin Microbiol. 2002;40:3192–3197. doi: 10.1128/JCM.40.9.3192-3197.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jouglin M, Chagneau S, Faille F, Verheyden H, Bastian S, Malandrin L. Detecting and characterizing mixed infections with genetic variants of Anaplasma phagocytophilum in roe deer (Capreolus capreolus) by developing an ankA cluster-specific nested PCR. Parasit Vectors. 2017;10:377. doi: 10.1186/s13071-017-2316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuen S, Casey ANJ, Woldehiwet Z, French NP, Ogden NH. Detection by the polymerase chain reaction of Anaplasma phagocytophilum in tissues of persistently infected sheep. J Comp Pathol 2006;134:101–104. [DOI] [PubMed]
- 26.Ogden NH, Casey ANJ, Woldehiwet Z, French NP. Transmission of Anaplasma phagocytophilum to Ixodes ricinus ticks from sheep in the acute and post-acute phases of infection. Infect Immun. 2003;71:2071–2078. doi: 10.1128/IAI.71.4.2071-2078.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown WC, Barbet AF. Persistent infections and immunity in ruminants to arthropod-borne bacteria in the family Anaplasmataceae. Ann Rev Anim Biosci. 2016;4:177–197. doi: 10.1146/annurev-animal-022513-114206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA samples from Chastagner et al. [9] used for the alignments. The available sequences for each locus are indicated by an “X”. (XLSX 12 kb)
NJ tree obtained using the concatenation of typA, ctrA, pleD, recG, and polA. Legends as in Fig. 1. (PDF 1383 kb)
NJ tree obtained using the concatenation of typA, ctrA, msp4, and pleD. Legends as in Fig. 1. (PDF 1399 kb)
NJ tree obtained using the concatenation of typA, ctrA, msp4, and recG. Legends as in Fig. 1. (PDF 1395 kb)
NJ tree obtained using the concatenation of typA, ctrA, msp4, and polA. Legends as in Fig. 1. (PDF 1434 kb)
NJ tree obtained using the concatenation of typA, ctrA, and msp4. Legends as in Fig. 1. (PDF 1438 kb)
Data Availability Statement
All data supporting the conclusions of this article are included within the article and its additional files. The sequences were submitted to the GenBank database under the accession numbers MF580591–MF580636.


