Abstract
Pancreatic ductal adenocarcinoma (PDAC) is usually detected late in the disease process. Clinical work-up through imaging and tissue biopsies is often complex and expensive due to a paucity of reliable biomarkers. Here, we used an advanced multiplexed plasmonic assay to analyze circulating, tumor-derived extracellular vesicles (tEV) in over 100 clinical populations. Using EV based protein marker profiling, we identified a signature of 5 markers (PDACEV signature) for PDAC detection. In our prospective cohort, the accuracy for the PDACEV signature was 84% (95% confidence interval, CI: 69–93%), but only 63–72% for single marker screening. GPC1 alone had a sensitivity of 82% (CI: 60–95%) and a specificity of 52% (CI: 30–74%) while the PDACEV signature showed a sensitivity of 86% (CI: 65–97%) and a specificity of 81% (CI: 58–95%). We show that the PDACEV signature of tEV offered higher sensitivity, specificity, and accuracy than existing serum (CA 19-9) or single tEV marker analyses. This approach should enhance the diagnosis of pancreatic cancer.
Keywords: pancreatic cancer, extracellular vesicles, diagnosis, chip sensor
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death in the United States, with a 5-year survival rate below 10%. Most newly diagnosed patients’ (>80%) tumors are considered unresectable(1). Earlier detection could increase survival by an estimated 30–40%(2), while more reliable and real-time assessment of treatment effects could prolong survival and/or improve quality-of-life. Detecting serum levels of CA 19-9 is currently the best established blood test for PDAC and has a pooled sensitivity of 75.4% (95% confidence interval, CI: 73.4–77.4%) and a specificity of 77.6% (CI: 75.4–79.7%)(3). Although often used to follow treatment response, CA 19-9 is a poor biomarker for early detection, commonly rises late in the disease, and may be elevated in non-malignant conditions such as biliary obstruction and pancreatitis(4). Recent modeling studies of future assay performance cite a minimum sensitivity of 88% and specificity of 85% to prolong patient survival and demonstrate cost effectiveness(2). Various approaches to achieve this are being explored(5; 6), including the use of CA242(3), circulating tumor cells(7), circulating tumor-derived extracellular vesicles including exosomes(8), metabolites(9), proteomic analyses(10; 11) and circulating DNA(12). Beyond the inherent technical challenges of these advanced analyses, a key question is whether early reported findings can be validated independently in larger sets of patients.
Tumor-derived extracellular vesicles (tEVs) offer an attractive approach to monitor cancers using “liquid biopsies”. tEVs are relatively more abundant than other circulating biomarkers, structurally more stable, and contain protein and mRNA profiles that highly reflect parental cancer cells(13–15). Experimental studies have shed light on the composition and functional roles of tEVs(16; 17) for: 1) diagnosis(13; 18; 19); 2) long range communication(16); and 3) host cell interaction(20). Rapid and accurate analysis of tEVs in clinical samples is often hampered by three practical challenges: lengthy isolation procedures requiring ultracentrifugation to isolate tEVs, general unavailability of ultra-sensitive assay systems to analyze large clinical cohorts for multiple markers, and specific cancer markers that separate tEVs from host cell EVs. Here, we developed an advanced plasmonic sensing system for higher throughput analysis of clinical samples to directly address the shortcomings often found in translational clinical analyses. The basic operating principle relies on measuring spectral shifts of resonant light transmission through periodically arranged gold nanopores to which tEVs are captured by immunoaffinity. In proof-of principle cancer experiments, we showed that tEVs can indeed be detected by plasmonic sensors(14). However, the original prototype was manually operated and had limited throughput and chip production rates thus preventing widespread clinical use. As a result, we devised a multi-parametric system incorporating significantly larger numbers of sensing arrays (>100 sensing spots) and automatic operation to enable routine clinical sample analyses. Intrigued by recent reports(8; 21), we set out to determine key protein profiles of tEVs in 135 patients undergoing surgery for pancreatic pathologies. Dictated by clinical needs, we were particularly interested in defining practical and reliable tEV marker sets for PDAC diagnosis.
RESULTS
Nanoplasmonic sensors for high-throughput, sensitive analyses of EVs
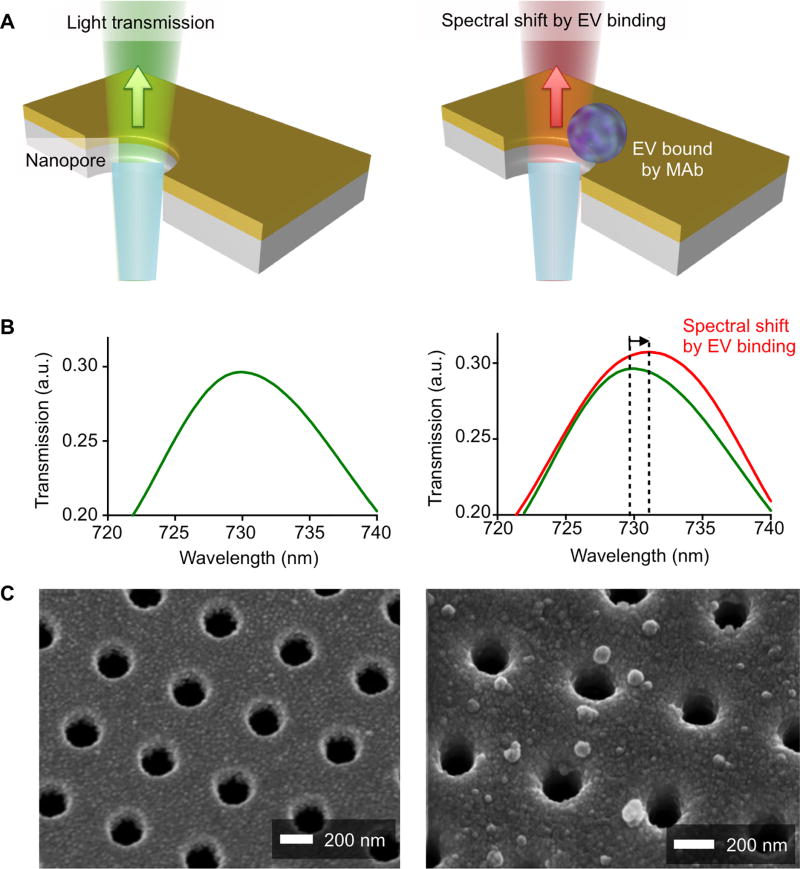
Fig. 1 summarizes the working principle of the nanoplasmonic sensor (NPS) assay, specifically designed for clinical workflows, small clinical sample amounts, and high throughput detection. The sensor chip contains periodically arranged nanopores (200 nm in diameter and 500 nm in periodicity) patterned in a 100 nm-thick gold film. The function of the pores is to transmit light shown onto the gold surface (Fig. 1A). When EVs are bound to the vicinity of these pores via specific antibodies, the wavelength of the transited light shifts to the red. It is this red-shift that is detected by sensors and reflects the amount of EV bound (Figs. 1B and C).
Fig. 1. Working principle of the plasmon sensor chip (NPS) for tumor derived extracellular vesicles.
(A) EV binding to the nanopore surface via monoclonal antibody immobilized on the gold surface causes a spectral shift of light transmitted through the nanopores. (B) The spectral shift of resonance peak in light transmission is measured to quantify the amount of EVs captured on the nanopore surface. a.u., arbitrary unit. (C) Scanning electron micrographs (SEMs) show the periodically arranged nanopore array and EVs captured on the surface. Each nanohole has a diameter of 200 nm and a periodicity of 500 nm.
The sensor chip is easily scalable to larger arrays of over 1,000 sensing sites; multiple sensor chips can be made through batch fabrication processes (see the Method for details). We prepared a customized chip with 100 detection sites to yield data for 25 antibodies/markers in quadruplicate. Antibody and EV solutions were printed in as small as 100 nL spots on the sensor chip through a molecular printing method. A piezoelectrical microscope stage was incorporated into the system for scanning sensing arrays and collecting transmission spectra. Both printing and measurements are operated automatically to improve assay throughput and reduce variation among users. Overall, the smaller chip size, higher spot density and smaller measurement volumes represent a 25× increase in sensitivity compared to a previous prototype (120 µL for 12 markers vs 10 µL for 25 markers). Fig. S1 shows the NPS chip with 100 sensing arrays and the integrated set-up optimized for processing clinical samples.
tEV composition in PDAC models mirrors that of parental cells
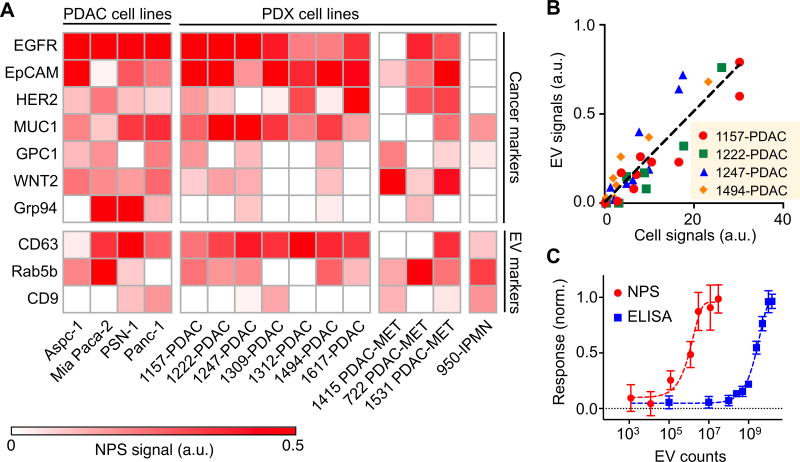
A number of putative cell-associated PDAC cancer markers have been described for individual patients, but using single markers in entire cohorts generally has insufficient sensitivity or specificity. Proteome analyses have identified soluble markers(11; 22–24) while profiling studies have identified cell surface(25–27) or exosomal markers(8; 28; 29). In order to calibrate and validate the new plasmonic sensing system, we investigated 15 putative cancer and EV markers (Table S1) by performing flow cytometry on whole pancreatic cancer cells (Fig. S2). Based on the cell data, we eliminated some of the non-cancer specific markers and performed NPS measurements on tEVs (Fig. 2A). Beyond the commonly used PDAC cell lines, we also investigated 11 patient-derived tumor xenograft (PDX) models of PDAC, metastatic PDAC and intraductal papillary mucinous neoplasm (IPMN) (Fig. 2A). Our data show good correlation between expression patterns seen in whole cells and tEV (Spearman correlation coefficient r = 0.86 for 1157-PDAC, 1222-PDAC, 1247-PDAC and 1494-PDAC; Fig. 2B). The tEV assays by the NPS chip are on the order of 102 more sensitive than the gold standard ELISA assay for this analysis (Fig. 2C).
Fig. 2. In vitro profiling of tEV markers on cell line-derived EVs.
(A) The molecular expression of cancer markers (EGFR, EpCAM, HER2, MUC1, GPC1, WNT2. Grp94) and EV markers (CD63, Rab5b, CD9) were characterized on EVs derived from 4 cancer cell lines and 11 patient-derived xenograft (PDX) cell lines including PDAC, metastatic PDAC (PDAC-MET) and IPMN. (B) Correlation of protein levels measured between EVs and their parental cell lines (1157-PDAC, 1222-PDAC, 1247-PDAC and 1494-PDAC). a.u., arbitrary unit. (C) Sensitivity comparison between NPS and the gold standard ELISA. The responses were normalized against the values of highest concentrations.
Establishing a PDAC tEV panel
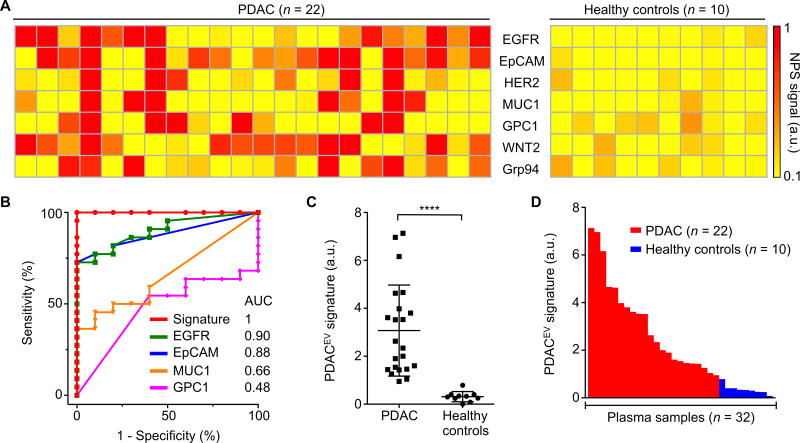
We next collected plasma from 32 patients enrolled into a training cohort involving 22 cases of PDAC and 10 healthy controls. Fig. 3A summarizes each patient for the chosen tEV markers including pan-cancer markers (EGFR, EpCAM, HER2, MUC1) and putative PDAC markers (GPC1, WNT2, Grp94(30)). Using receiver operating characteristic (ROC) analyses, we determined sensitivity, specificity and accuracy for each marker individually and also in combination (Fig. 3B, Table 2). We observed that no single marker achieved sufficiently high sensitivity and specificity. Therefore we reasoned that a combination of multiple markers would be necessary. Indeed, a previously identified generic quad marker cancer signature(31) (EGFR, EpCAM, HER2, MUC1) had high sensitivity (91%), specificity (100%), and accuracy (94%). When we replaced HER2 with putative PDAC markers (GPC1 and WNT2), we could further improve the sensitivity and specificity (Table 2). This new PDACEV signature, representing an unweighted sum of EGFR, EpCAM, MUC1, GPC1 and WNT2 signals, showed the accuracy of 100% in this training cohort (Figs. 3C and 3D). Because of the limited sample size (n=32) we also tested all four and five marker combinations in the prospective cohort.
Fig. 3. Molecular profiling of plasma EV for a training cohort.
(A) Putative cancer markers (EGFR, EpCAM, HER2, Muc1) and PDAC markers (GPC1, WNT2, Grp94 and B7-H3) were profiled on EVs collected from 22 PDAC patients and 10 healthy controls. (B) ROC curves were calculated for single protein markers as well as for the PDACEV signature combination to determine optimum EV threshold values. AUC, area under the curve. (C) A combined marker panel (EGFR, EpCAM, MUC1, GPC1, WNT2) was established as a PDACEV signature that showed 100% accuracy for the training cohort in distinguishing PDAC from healthy controls. P value was determined by Mann-Whitney test. ****P < 0.0001. (D) A waterfall plot shows the PDACEV signature signals sorted from high (left) to low (right). Each column represents a different patient sample (red, malignant; blue, benign). a.u., arbitrary unit.
Table 2. Statistical analyses of EV markers for training and prospective cohorts.
95% confidence intervals are indicated in parentheses.
| Training cohort (n=32) | Prospective cohort (n=43) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Biomarker(s) | n | Cutoff | AUC | Sens (%) | Spec (%) | Acc (%) | Sens (%) | Spec (%) | Acc (%) |
| EGFR | 1 | 0.15 (0.01–0.24) | 0.90 (0.79–1) | 73 | 100 | 81 | 59 (36–79) | 76 (53–92) | 67 (51–81) |
| EpCAM | 1 | 0.28 (0.01–0.34) | 0.88 (0.77–0.99) | 73 | 100 | 81 | 45 (24–68) | 95 (76–100) | 70 (54–83) |
| HER2 | 1 | 0.13 (0.03–0.32) | 0.72 (0.55–0.89) | 59 | 90 | 69 | 59 (36–79) | 85 (64–97) | 72 (56–85) |
| MUC1 | 1 | 0.34 (0.02–0.53) | 0.66 (0.48–0.84) | 36 | 100 | 56 | 36 (17–59) | 90 (70–99) | 63 (47–77) |
| GPC1 | 1 | 0.04 (0.04–0.68) | 0.48 (0.28–0.67) | 55 | 60 | 56 | 82 (60–95) | 52 (30–74) | 67 (51–81) |
| WNT2 | 1 | 0.18 (0.09–0.48) | 0.84 (0.71–0.96) | 77 | 90 | 81 | 64 (41–83) | 76 (53–92) | 70 (54–83) |
| Grp94 | 1 | 0.10 (0.02. 0.46) | 0.73 (0.55–0.90) | 73 | 70 | 72 | 55 (32–76) | 71 (48–89) | 63 (47–77) |
| B7-H3 | 1 | 0.19 (0.02. 0.23) | 0.75 (0.58–0.93) | 50 | 100 | 59 | NA | NA | NA |
|
| |||||||||
| EGFR + EpCAM + HER2 + MUC1 | 4 | 0.67 (0.29–0.68) | 0.99 (0.97–1) | 91 | 100 | 94 | 86 (65–97) | 86 (64–97) | 86 (72–95) |
| EGFR + EpCAM + GPC1 + WNT2 | 4 | 0.74 (0.65–0.84) | 1.0 | 100 | 100 | 100 | 82 (60–95) | 90 (70–99) | 86 (72–95) |
| EGFR + EpCAM + MUC1 + GPC1 + WNT2 | 5 | 0.87 (0.68–1.00) | 1.0 | 100 | 100 | 100 | 86 (65–97) | 81 (58–95) | 84 (69–93) |
| EGFR + EpCAM + HER2 + MUC1 + GPC1 + WNT2 | 6 | 0.89 (0.73–1.00) | 1.0 | 100 | 100 | 100 | 95 (77–100) | 81 (58–95) | 88 (75–96) |
A number of observations were of particular interest. Some of the chosen markers highly expressed in EVs did not provide diagnostic information (Rab5b, CD9, CD63, Fig. S3) and were thus eliminated from the ensuing prospective study. Second, GPC1 was not specific for PDAC in our cohort and had a lower accuracy as a single marker than marker combinations, similar to other markers tested. These findings did not change by using alternative commercially available GPC1 antibodies, all of which were validated prior to their use (Table S2). Third, other putative PDAC markers such as WNT2 showed better accuracy than GPC1.
Validation cohort
We next analyzed a prospective cohort of 43 patients undergoing surgery for pancreatic (n=35) or other abdominal indications (n=8, age-matched control group). In all 38 patients operated for pancreatic indications, tissue was available for clinical pathology interpretation (n=22 for PDAC, n=8 for pancreatitis, n=5, benign cystic tumor). We obtained 2–10 mL of plasma from each patient on the day of or immediately preceding surgery and NPS measurements were performed using identical markers from our training cohorts (see Materials and Methods for details).
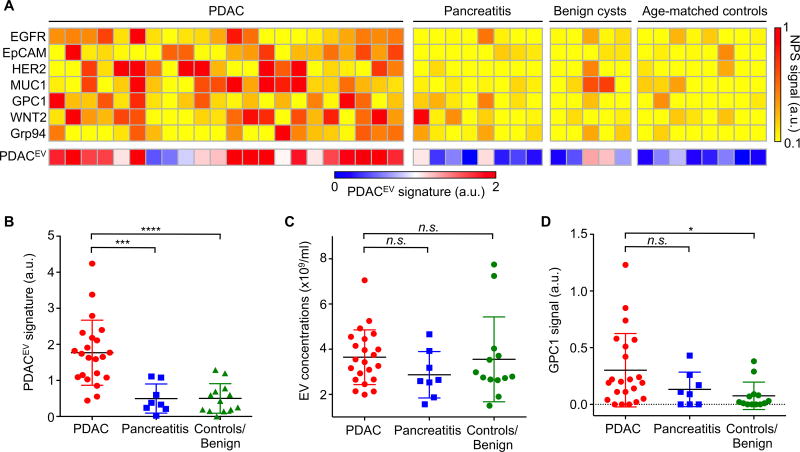
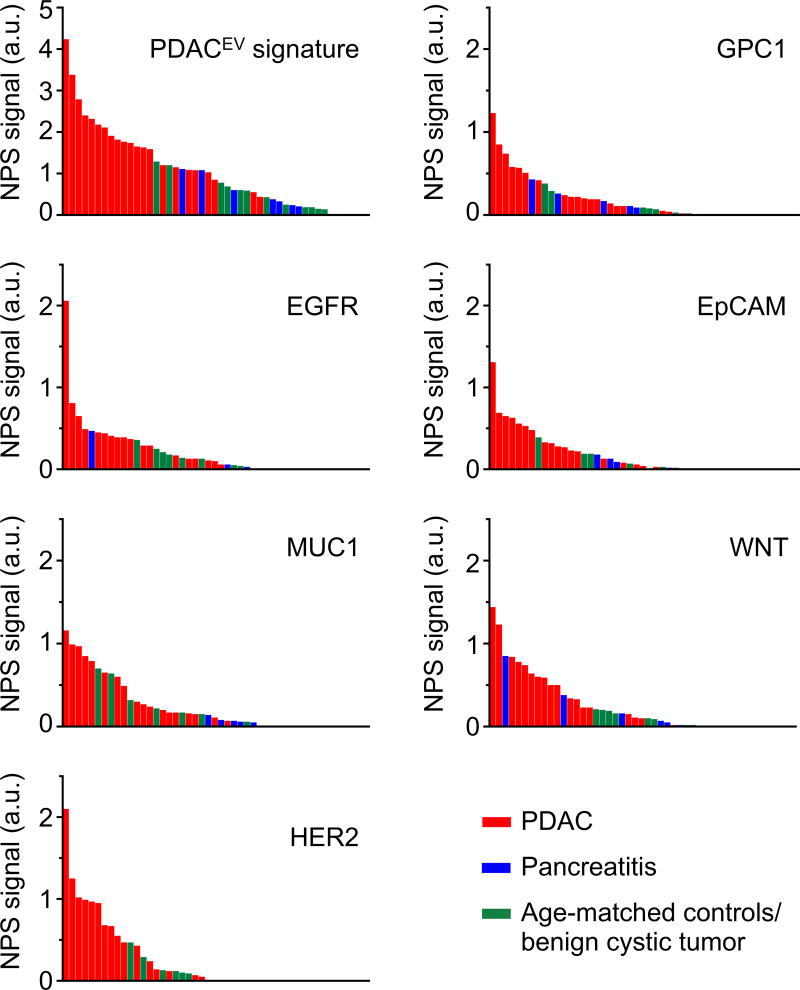
Fig. 4A summarizes the performance of the PDAC markers in differentiating PDAC from pancreatitis, benign and control patient groups. Analyzing the heatmap of EV markers, once again demonstrated that no single patient had similar markers elevated. Rather, it was the combination of the five markers comprising the PDACEV signature that resulted in an overall accuracy of 84%. In this prospective cohort, the PDACEV signature (EGFR, EpCAM, MUC1, WNT2, and GPC1) identified in the training cohort showed a sensitivity of 86% (CI: 65–97%) and a specificity of 81% (CI: 58–95%, Fig. 4B, Table 2), while total EV concentrations were not significantly different between the groups (Dunn’s multiple comparisons test, p=0.16 for PDAC and pancreatitis, p=0.78 for PDAC and control, Fig. 4C). Furthermore, GPC1 was not statistically significant in differentiating PDAC from pancreatitis (p=0.31) but was slightly higher in PDAC when compared to the control group (median values of 0.20 for PDAC and 0.02 for the control group; p=0.018, Fig. 4D). Fig. 5 displays the experimental data of single and marker combinations as a waterfall plot. Table 2 summarizes the diagnostic accuracies of all markers and combinations in this prospective cohort.
Fig. 4. The PDACEV signature differentiation of PDAC patients from pancreatitis and control patient groups.
(A) Heatmap analysis of EV markers. The PDACEV signature is defined as a combined marker panel of EGFR, EpCAM, MUC1, GPC1 and WNT2. (B–D) The established PDACEV signature signals (B), EV concentrations (C) and single GPC1 signal (D) as measured for plasma EVs collected from 22 PDAC patients, 8 pancreatitis, 5 benign cystic tumor, and 8 age matched controls. Pairwise comparison p values were determined by the Dunn’s multiple comparisons test. *P < 0.05, ***P < 0.001, ****P < 0.0001. n.s., not significant. a.u., arbitrary unit.
Fig. 5. Distribution of EV protein marker signals.
Waterfall plots show EV protein levels of each of the different biomarkers sorted from high (left) to low (right). Each column represents a different patient sample (red, PDAC, n=22; blue, pancreatitis, n=8; green, age-matched controls and benign cystic tumors, n=13). a.u., arbitrary unit.
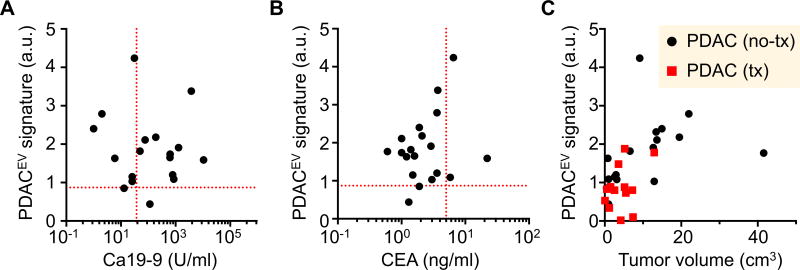
We next correlated EV analyses to clinical gold standard serum biomarkers (CA19-9 and CEA) in patients with PDAC. The PDACEV signature was not correlated with either CA19-9 (Spearman correlation coefficient r = −0.28; p = 0.26) or CEA (r < 0.001, p > 0.99) (Fig. 6A). In our cohort, only 61% of PDAC patients (11 out of 18) showed an elevated level of CA19-9 (> 37U/ml, threshold value used in clinic) while 89% (16 out of 18) had high PDACEV values (> 0.87 NPS signal, Table 2). For CEA, only 17% of PDAC patients (3 out of 18) were positive (>5 ng/ml, Fig. 6B). Finally, we compared the PDACEV signature against tumor size, showing a modest correlation for the signature (Spearman correlation coefficient r = 0.58; p = 0.018) and little correlation (r = −0.09; p = 0.62) between EV counts and tumor size (Figs. 6C and S4).
Fig. 6. Comparison of EV analyses with conventional clinical metrics.
The PDACEV signature values are correlated to serum biomarkers (CA 19-9, A and CEA, B) on patients with PDAC and the tumor diameter (C). The dashed red lines indicate the threshold values to be positive (CA 19-9, 37 U/ml; CEA, 5 ng/ml; PDACEV signature, 0.87). a.u., arbitrary unit.
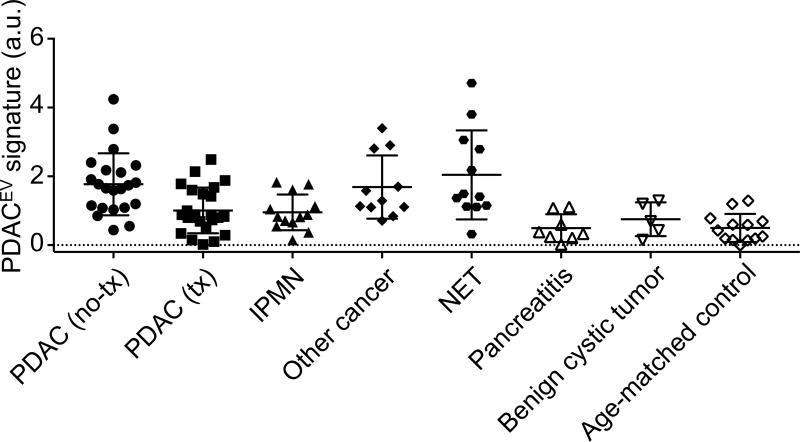
To further expand the clinical utility of tEV analyses we also studied several additional patient cohorts (n = 69, Table 1, Figs. 7, S5 and S6): i) PDAC treated with neoadjuvant regimen (n = 24), ii) IPMN (n = 14), iii) other GI cancers mimicking pancreatic duodenal cancers (n = 11) iv) neuroendocrine tumors (NET, n = 12) and v) benign cystic tumors (n = 5). Patients with PDAC treated with neoadjuvant regimen had lower EV signatures (median values, 1.70 vs 0.86; Mann Whitney test, p = 0.015, Fig. S7A) as a group compared to the untreated PDAC group, likely reflecting the smaller tumor mass and/or favorable treatment response.
Table 1. Summary of patient cohorts.
| Characteristic | Training cohort | Prospective Cohort | Total | ||
|---|---|---|---|---|---|
| Malignant | Benign | Malignant | Benign | ||
| Total cases | 22 | 10 | 82 | 21 | 135 |
|
| |||||
| Subtypes | |||||
|
| |||||
| PDAC untreated | 13 | - | 22 | - | 35 |
|
| |||||
| PDAC neoadjuvant tx | 9 | - | 24 | - | 33 |
|
| |||||
| IPMN inter/high grade | - | - | 13 | - | 13 |
|
| |||||
| NET | - | - | 12 | - | 12 |
|
| |||||
| Other cancer | - | - | 11 | - | 11 |
|
| |||||
| Benign cystic tumor | - | - | - | 5 | 5 |
|
| |||||
| Pancreatitis | - | - | - | 8 | 8 |
|
| |||||
| Controls | - | 10 | - | 8 | 18 |
|
| |||||
| Age | |||||
|
| |||||
| Median | 68 | 48 | 65 | 57 | 63 |
|
| |||||
| Range | 47–88 | 23–82 | 17–84 | 19–91 | 17–91 |
|
| |||||
| Sex | |||||
|
| |||||
| Male | 8 | 4 | 47 | 14 | 73 |
|
| |||||
| Female | 14 | 1 | 40 | 14 | 69 |
|
| |||||
| CA19-9 | |||||
|
| |||||
| PDAC untreated | 1148 (1–6684) | - | 1,006 (1–10,625) | - | 1064 (1–10,625) |
|
| |||||
| PDAC neoadjuvant tx | 2258 (19–17,101) | - | 657 (4–7,730) | - | 1175 (4–17,101) |
|
| |||||
| IPMN | - | - | 10.6 (1–26) | - | 10.6 (1–26) |
|
| |||||
| CEA | |||||
|
| |||||
| PDAC untreated | 7.0 (2–12) | - | 3.4 (0.6–22.1) | - | 5.0 (0.6–22.1) |
|
| |||||
| PDAC neoadjuvant tx | 11.0 (1–53) | - | 56.4 (0.7–1003) | - | 40 (0.7–1003) |
|
| |||||
| IPMN | - | - | 2.1 (0.7–3.3) | - | 2.1 (0.7–3.3) |
|
| |||||
| Stage | |||||
|
| |||||
| I | 0 | - | 10 | - | 10 |
|
| |||||
| II | 1 | - | 42 | - | 43 |
|
| |||||
| III | 5 | - | 5 | - | 10 |
|
| |||||
| IV | 16 | - | 10 | - | 26 |
|
| |||||
| Co-therapies PDAC | |||||
|
| |||||
| Folforinox/XRT | 4 | - | 21 | - | 25 |
|
| |||||
| Gemcitabine | 5 | - | 1 | - | 6 |
|
| |||||
| Other | - | - | 2 | - | 2 |
Fig. 7. EV analyses for patients with different types of pancreatic-related diseases.
The PDACEV signature values are measured for patient cohorts (n = 103 including i) PDAC without treatment (n = 22), ii) PDAC treated with neoadjuvant regimen (n = 24), ii) IPMN (n = 13), iv) other GI cancers mimicking pancreatic duodenal cancers (n = 11), v) neuroendocrine tumors (NET, n = 12), vii) pancreatitis (n = 8), viii) benign cystic tumors and (n = 5) and ix) age-matched control (n = 18). a.u., arbitrary unit.
We also studied a number of cases of intraductal papillary mucinous neoplasms (IPMN), which grows within the pancreatic ducts and is characterized by the production of thick mucinous fluid. IPMN are important because some progress to invasive cancer and therefore represent windows of opportunity to treat before aggressive and difficult to manage cancer develops. Our cohort contained 11 cases of intermediate and high grade IPMN and 2 cases of low grade IPMN (Fig. 7, Table 2). As shown in Fig. S7B, IPMN had an elevated PDACEV signature compared to age-matched controls (Dunn’s multiple comparisons test, p < 0.0001), but were lower compared to PDAC (p = 0.022).
The validation cohort included a limited number of other pancreatic cancers or cancers that can mimic pancreatic symptomatology. These included neuroendocrine tumors (NET) and gastroduodenal cancers. Again, while the numbers are limited, most of the malignancies tested positive for some of the extravesicular markers (e.g. among 23 patients, 18 positive for EGFR and/or EpCAM). These findings are in line with other observations. For example, EpCAM has been evaluated as a CTC detection marker in NET populations(32; 33). Of particular interest was the fact that 9 out of 12 NET tested positive for SSTR2 expression on EVs while all PDAC patients (n = 22) and age-matched healthy controls (n = 8) were negative for SSTR2 with a threshold value of 0.15. (Dunn’s multiple comparisons test, p < 0.0001 between PDAC and NET; p = 0.0018 between NET and control, Fig. S7C). Finally, we investigated a limited number of patients with benign mucinous tumors but no detectable malignancy. In these cases, we observed PDACEV signatures similar to the age-matched control group (Mann Whitney test, p = 0.35, Fig. S7D).
DISCUSSION
Extracellular vesicles are attractive as circulating biomarkers given their abundance, relative stability and similar molecular make-up to parental cells(13–15). Despite these apparent advantages, it has been difficult to define single tumor specific EV markers (mRNA, DNA or protein)(8; 28; 34), validate purported malignancy biomarkers in larger patient cohorts(8; 13; 34), implement lengthy purification procedures (ultracentrifugation) into the clinical workflow(13), and commercialize cost-effective technologies. Here, we show in a sizable pancreatic data set that single EV protein biomarkers are unlikely to be sufficiently accurate to improve patient management. No individual putative protein tEV marker (EGFR, EpCAM, MUC1, GPC1, WNT2) yielded sensitivities above 86% (CI: 65–97%) and specificities above 81% (CI: 58–95%) to be considered cost effective(2). Indeed, many had much lower sensitivities/specificities including GPC1 despite prior studies(5; 8); unfortunately the previously used GPC1 antibody is no longer commercially available. Therefore, there is a possibility that the discrepancy could be attributed to the antibody used in the study.
Based on the hypothesis that tumoral heterogeneity will require multiplexed biomarkers for clinical use, we set out to define protein signatures representative of epithelial and pancreatic cancers(25). We initially surveyed about 50 proteins of potential interest and discarded all but 10 following feasibility studies in PDAC cells lines and PDX models. These 10 EV markers included 7 tEV markers (EGFR, EpCAM, MUC1, HER2, GPC1, WNT2, Grp94) and 3 pan EV markers (CD63, CD9, Rab5b). The pan EV markers were excluded from the tumor diagnostic marker panel and were solely used to confirm the presence of EV in a given sample. From an initial training dataset, we further refined the markers to 5 essential ones, which then informed our PDACEV signature (EGFR, EpCAM, Muc1, GPC1, WNT2). The signature was defined as the unweighted sum of each marker level, with a score of > 0.85 suggesting PDAC. It is conceivable to further improve on this panel by identifying additional molecular tumor markers present on the EV surface. A number of proteomic approaches have been used to identify putative markers, but validation work remains to be done. It would also be attractive to expand the panel to intravesicular markers such as mutant KRAS protein, but this would require EV lysis and further NPS assay optimization.
Applying the above PDACEV signature to 43 patients, we show an overall sensitivity of 86% for detecting PDAC and a specificity of 81% for differentiating PDAC from other pancreatic diseases (Table 2). The accuracy was 84% (CI: 69–93%). The relatively high accuracy is most likely attributed to the selection of protein EV markers, the surgical patient cohorts enrolled and ease of measurements, i.e. reduced analytical failures. The last point of particular interest is that existing EV analyses are cumbersome and often require large sample amounts. In contradistinction, we set out to develop a miniaturized sensing technology with automated microarray spotter and scanning stage to perform measurements at scales that are clinically feasible and affordable. The NPS measurements performed here requires ~ 10 minutes of measurement/analysis time and currently costs $60 (Chip cost $42, antibody cost $18) per patient sample. Since the majority of current costs are driven by manual manufacture of chips and antibodies, it is expected that real costs will scale downward significantly with bulk fabrication. The current limits of the NPS technology as developed in this study are i) the need for EV purification and concentration prior to measurement, ii) lower sensitivity for intravesicular markers, iii) the need for high quality antibodies which are necessary for capture and iv) the composition of the markers panel. With further optimization and commercialization, all these points could be addressed and further improved.
A number of previous studies have investigated tEV as a diagnostic cancer marker both by protein and nucleic acid analysis(8; 13–15). Remaining questions include: i) whether these results hold up in larger patient cohorts and ii) how cost effective and practical are newer analytical techniques. For example, Melo et al. explored the use of GPC1 as a single-marker for detection of PDAC from EVs(8). Similarly, several studies have explored the serum proteome of PDAC(22–24) with the goal of providing more advanced diagnostic tools to guide clinicians. While studies are ongoing, measurement of tEV appears to be a promising venue for pancreatic diagnoses.
The current study was designed as a feasibility study to focus on some of the pressing questions in surgical oncology. In future studies we plan to expand tEV analysis to assess treatment efficacy. While the current study was not designed to investigate this, such work has been done for other cancers(14; 15; 35). For example, we have shown in prior research that longitudinal tEV profiling is feasible and can be informative in treatment assessment(14; 15; 35). Our long term plan also includes serially analyzing high-risk subjects longitudinally for PDAC development, an endeavor that will require larger data sets and multi-year follow-ups.
Materials and Methods
Cell lines
AsPC-1, MIA PaCa-2, PSN-1, and PANC-1 cell lines were purchased from ATCC. AsPC-1 and PSN-1 cells were maintained in RPMI-1640 medium. MIA PaCa-2 and PANC-1 cells were maintained in Dulbecco’s Modified Eagle’s medium. All cell line medium was supplemented with 10% fetal bovine serum, 100 I.U. penicillin, and 100 µg/ml streptomycin. PDAC PDX cell lines were a kind gift from Dr. Carlos Fernandez del Castillo and were all maintained in a 50/50 mix of Dulbecco’s Modified Eagle’s and Ham’s F-12 medium supplemented as above.
Selection of markers
Several PDAC proteomic studies have been described in the literature(10; 11; 22–24; 29) or are available online (http://wlab.ethz.ch/cspa/; https://www.proteomicsdb.org/#projects/4256; pancreaticcancerdatabase.org/publications.php). Both of these literature sources were analyzed to define EV marker candidates. To derive the marker set we surveyed these available databases, three vesicle databases (vesiclepedia, EVpedia and ExoCarta), and the literature for published markers. The putative “hits” were then screened using hundreds of commercially available antibodies (see Table S1). We eliminated targets that were not specific for cancer cells, yielded only duplicative information or were primarily intravesicular proteins which we were not able to capture efficiently. From the initial 50 candidate markers, we selected 10 after feasibility studies with PDAC cells lines and PDX models (Fig. S2). We decided to take forward 7 tumor markers, all of them vesicle surface markers that can be used for chip capture. In addition, we assayed for three ubiquitous EV markers, CD9, CD63 and Rab5b.
Antibody and biotinylation
All antibodies used in these studies are listed in Supplemental Table 1. For biotinylation, all antibodies (50 µg in 100 µl PBS) were first passed through 0.5ml 7MWCO Zeba Spin Desalting columns (ThermoFisher, 89882) to remove sodium azide. EZ-Link Sulfo-NHS-LC-Biotin (ThermoFisher, 21327) was used for antibody biotinylation according to the manufacturers’ instructions. Briefly, antibodies were mixed with a 20-fold molar excess of 10 mM Biotin for 30 min at room temperature. Excess Biotin was then removed using a second Zeba Desalting column. Antibody concentration was checked using a NanoDrop spectrophotometer (ThermoFisher).
Flow cytometry
Antibodies were tested and compared to EV signals from NPS using flow cytometry. On the day of EV collection from cell lines, a portion of the remaining cells were trypsinized for flow cytometry. 500,000 to 1,000,000 cells per condition were fixed in 4% paraformaldehyde in PBS (Electron Microscopy Sciences, 15710-S) for 10 min at room temperature. Cells were washed twice with PBS plus 0.5% BSA. Antibodies were diluted to 10 µg/ml in 100 µl PBS plus 0.5% BSA and incubated with cells for 1 hr at room temperature. Cells were washed twice with PBS plus 0.5% BSA and then incubated with appropriate AlexaFluor 488 secondary antibody diluted 1:1000 in PBS plus 0.5% BSA for 30 min at room temperature. Cells were washed twice with PBS plus 0.5% BSA. Fluorescent signal was measured using a FACSCalibur flow cytometer (BD Biosciences) and compared to appropriate isotype controls and secondary antibody only signal using the following: (signal primary antibody - signal isotype control)/signal secondary antibody.
EV isolation from cell culture
Cells were grown for 48 hrs in normal growth media supplemented with 5% EV-depleted FBS (ThermoFisher, A2720801). Conditioned media was collected in 50 ml tubes and centrifuged at 300 × g for 10 min. Media was filtered through a 0.22 µm cellulose acetate vacuum filter (Corning, 430767) and then aliquoted to ultracentrifuge tubes (Beckman, 344058). Media was centrifuged at 100,000 × g for 70 min to pellet EVs. The pellet was washed with PBS and re-pelleted by centrifugation at 100,000 × g for 70 min. EVs were resuspended in an appropriate volume of PBS and stored at −80C until NPS measurement.
Sample collection
There current study was designed to prospectively obtain fresh samples and then closely correlate them to pathological and clinical information. All clinical data was entered into a unified database and used for blinded analyses by the biobank coordinator at MGH. The biospecimen collection was optimized for EV analysis and included the following steps: (1) collect whole blood into one 10 mL purple-top EDTA tube, 2) mix blood by inverting the tube 10 times, 3) store vacutainer tubes upright at 4°C and process within 1 hour of blood collection, 4) centrifuge blood samples for 10 min at 400 × g at 4°C, 5) collect the plasma layer in a 15ml conical tube with a pipette without disturbing the buffy coat, 6) centrifuge the plasma layer for 10 min at 1100 × g at 4°C, 7) pipette the plasma into a 15 mL labeled conical tube, 8) store at −80°C until processing.
EV isolation from plasma
Plasma was thawed, aliquoted to ultracentrifuge tubes, and diluted to 30–35ml total volume in PBS. Plasma was initially centrifuged at 14,000 × g for 20 min to pellet cell debris. Cleared supernatant was passed through a 0.22 µm PVDF (Millipore) syringe filter into an ultracentrifuge tube. EVs were then pelleted by ultracentrifugation at 100,000 × g for 70 min. The pellet was resuspended in PBS and centrifuged again at 100,000 × g for 70 min. The final EV pellets were resuspended in 300 µl PBS and stored at −80°C until NPS measurement.
EV size measurements
Nanoparticle tracking analysis (NTA, Nanosight, Malvern) was used to assess EV size and concentration. Measurements were done as reported in the literature(36). Briefly, samples were diluted in PBS (generally a 1:100 dilution). Five, 30 sec videos were recording using the following settings for all measurements: threshold 1,482, gain 680. Videos were processed and the highest and lowest EV concentrations were excluded.
NPS fabrication
We used interference lithography to prepare NPS devices (Fig. S1). First, periodic nanohole patterns were made on a double-polished 4-inch Si wafer coated with a 125 nm silicon nitride (SiN) layer. The patterned wafer was dry-etched using reactive ion etching (RIE) to create nanoholes in the SiN layer. In this step, only a partial layer was etched to protect the front Si surface from the subsequent silicon etching with potassium hydroxide (KOH). The opposite Si backside was lithographically patterned to define sensing sites and wet-etched with KOH at 80°C. Patterned wafers were diced into individual NPS chips with each chip containing 100 (10 × 10) measurement sites. After removing the remaining SiN layer, a 100 nm Au film with a 2 nm Ti adhesion layer was directly deposited on the patterned SiN side. After the EV assays, the metal films were removed by Au etchant and hydrogen fluoride (HF) solutions to regenerate chips. After cleaning the patterned Si templates, fresh metal films were deposited on the regenerated Si templates.
NPS measurement
The fabricated Au chip was first incubated with a 1:3 mixture of linear polyethylene glycol (PEG) (final 10 mM in PBS) for overnight at room temperature: one was end-functionalized with thiol and biotin (1 kDa, Nanocs), and the other was methyl- and sulfhydryl-terminated (0.2 kDa, Fisher scientific). After washing in PBS, the chip was secondary incubated with neutravidin (Thermo scientific, 50 µg/mL in PBS with 0.2% BSA) for 40 min at room temperature. Finally, after washing, 0.5 µL of biotinylated antibodies (10 µg/ml in PBS with 0.2% BSA) were added to individual nanopore arrays by using a microarray spotter (DigiLab, Inc) and incubated for 40 min at room temperature with humidity. The autibody-conjugated chip was washed in PBS, then measured by spectrometer (USB4000-UV-VIS-ES, Ocean Optics, Inc.) as a baseline spectrum.
For EV detection, EV samples (0.5 µL, in 1% BSA) were spotted onto individual sensor arrays using the micoarray spotter and incubated in a humidity chamber for 50 min at room temperature. The chip was washed with PBS to remove unbound EVs and light transmission of each nanopore array was measured. A custom-built software program (MATLAB R2015a, MathWorks Inc.) was used to analyze spectral shifts after EV binding. A set of control arrays with isotype control antibodies was used to measure signals due to non-specific binding; these background signals were subtracted from the positive arrays.
Patients
Between 2015 and 2016, 135 patients underwent surgical resection of pancreatic neoplasm or other abdominal abnormalities. Through an Institutional Review Board approved protocol at the Massachusetts General Hospital (PI: CFC), blood samples were acquired. All samples were anonymized and only age, gender, medical history and final pathological diagnosis were recorded. All samples were processed in blinded fashion by operators blinded to the sample type.
Statistics
The Spearman correlation coefficient was used to quantify the correlations between different variables. Group differences were tested using the nonparametric Mann-Whitney test for two groups and the Kruskal-Wallis test for more than two groups; p-values for pairwise comparisons were obtained using the Dunn’s multiple comparison test. Receiver operating curves (ROC) were constructed for individual markers and selected marker combinations to describe the accuracy of detecting cancer. The cutoff points were selected using Youden’s index, which maximizes the sum of sensitivity and specificity. We used data from the training cohort (n=32) to select the optimal cutoff points associated with individual markers and marker combinations and then evaluated the sensitivity, specificity and accuracy of predicting tumor status associated with the optimal cutoff points using data from the prospective cohort (n=43). Selection of marker combinations were informed by literature, biological information and data-driven statistical procedures. One set of markers were selected through fitting the least absolute shrinkage and selection operator (lasso) paths for regularized logistic regression(37) to the training cohort where the tuning parameter was selected through a 10-fold cross-validation(38). For marker combinations, the sums of selected markers were used to predict tumor status. Of note, although the lasso procedure suggested a linear combination of markers with the weights being the estimated coefficients, the associated uncertainty with these estimated coefficients was large. We therefore used the unweighted sums for all marker combinations for ease of implementation in practice. Confidence intervals for AUC were calculated using the Delong Method (39) and for the cut points, the stratified Bootstrap percentile method. Exact confidence intervals for sensitivity, specificity and accuracy were obtained based on binomial distributions. All tests were two-sided and a P value < 0.05 was considered statistically significant. Analyses were formed using R versions 3.3.2 and GraphPad Prism 7.
Supplementary Material
Acknowledgments
We would like to thank our clinical colleagues involved in the clinical care of the patients reported here. Our thanks also go to Allison Roberts for help with screening antibodies prior to this clinical study.
Funding: Part of this study was funded by a grant from the Lustgarten Foundation (R.W.), NIH R01CA204019 (R.W.), P01CA069246 (R.W.), K99CA201248 (H.I.), R01HL113156 (H.L.), R21CA205322 (H.L.), and a pilot grant from the Andrew L. Warshaw, M.D. Institute for Pancreatic Cancer Research at MGH (K.Y., H.I.). C.P. was supported by the CaNCURE program, Northeastern University, NIH (R25CA17174650).
Footnotes
Fig. S1: NPS set-up.
Fig. S2: Comparison of PDAC and EV markers on cells and EVs.
Fig. S3: Extracellular vesicle markers in a training cohort.
Fig. S4: Comparison of EV counts with conventional clinical metrics.
Fig. S5: NPS signal from validation cohort tEVs.
Fig. S6: EV counts for patients with different types of pancreatic-related diseases.
Fig. S7: Comparison of the PDAVEV signature in different patient cohorts.
Table S1: Antibodies used in flow cytometry and NPS.
Author contributions: K.S.Y., H.I., and S.H. designed and performed all experiments; I.P., C.C. and C.F.D.C. collected patient samples; A.F.D.C., S.C., C.H., and C.P. assisted with data collection; K.Y., H.I., S.H., R. Wang, R.Y., and H.L. analyzed the data; S.F. provided new reagents; R.W., K.Y., H.I., C.P., H.L. provided funding; K.Y., H.I., H.L., C.C., and R.W. wrote the paper.
Competing interests: The authors declare no competing financial interests.
References
- 1.Rahib L, Fleshman JM, Matrisian LM, Berlin JD. Evaluation of Pancreatic Cancer Clinical Trials and Benchmarks for Clinically Meaningful Future Trials: A Systematic Review. JAMA Oncol. 2016;2:1209–1216. doi: 10.1001/jamaoncol.2016.0585. [DOI] [PubMed] [Google Scholar]
- 2.Ghatnekar O, Andersson R, Svensson M, Persson U, Ringdahl U, Zeilon P, Borrebaeck CA. Modelling the benefits of early diagnosis of pancreatic cancer using a biomarker signature. Int J Cancer. 2013;133:2392–2397. doi: 10.1002/ijc.28256. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Yang J, Li H, Wu Y, Zhang H, Chen W. Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: a meta-analysis. Int J Clin Exp Med. 2015;8:11683–11691. [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy MJ, Sturgeon C, Lamerz R, Haglund C, Holubec VL, Klapdor R, Nicolini A, Topolcan O, Heinemann V. Tumor markers in pancreatic cancer: a European Group on Tumor Markers (EGTM) status report. Ann Oncol. 2010;21:441–447. doi: 10.1093/annonc/mdp332. [DOI] [PubMed] [Google Scholar]
- 5.Seufferlein T, Mayerle J. Pancreatic cancer in 2015: Precision medicine in pancreatic cancer--fact or fiction. Nat Rev Gastroenterol Hepatol. 2016;13:74–75. doi: 10.1038/nrgastro.2015.215. [DOI] [PubMed] [Google Scholar]
- 6.Kelly KA, Hollingsworth MA, Brand RE, Liu CH, Singh VK, Srivastava S, Wasan AD, Yadav D, Andersen DK. Advances in Biomedical Imaging, Bioengineering, and Related Technologies for the Development of Biomarkers of Pancreatic Disease: Summary of a National Institute of Diabetes and Digestive and Kidney Diseases and National Institute of Biomedical Imaging and Bioengineering Workshop. Pancreas. 2015;44:1185–1194. doi: 10.1097/MPA.0000000000000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagrath S, Jack RM, Sahai V, Simeone DM. Opportunities and Challenges for Circulating Pancreatic Tumor Cells. Gastroenterology. 2016;151:412–426. doi: 10.1053/j.gastro.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 8.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan C, Clish CB, Wu C, Mayers JR, Kraft P, Townsend MK, Zhang M, Tworoger SS, Bao Y, Qian ZR, Rubinson DA, Ng K, Giovannucci EL, Ogino S, Stampfer MJ, Gaziano JM, Ma J, Sesso HD, Anderson GL, Cochrane BB, Manson JE, Torrence ME, Kimmelman AC, Amundadottir LT, Vander Heiden MG, Fuchs CS, Wolpin BM. Circulating Metabolites and Survival Among Patients With Pancreatic Cancer. J Natl Cancer Inst. 2016;108:djv409. doi: 10.1093/jnci/djv409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, Yang JY, Broom BM, Verhaak RG, Kane DW, Wakefield C, Weinstein JN, Mills GB, Liang H. TCPA: a resource for cancer functional proteomics data. Nat Methods. 2013;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capello M, Bantis LE, Scelo G, Zhao Y, Li P, Dhillon DS, Patel NJ, Kundnani DL, Wang H, Abbruzzese JL, Maitra A, Tempero MA, Brand R, Brennan L, Feng E, Taguchi I, Janout V, Firpo MA, Mulvihill SJ, Katz MH, Hanash SM. Sequential Validation of Blood-Based Protein Biomarker Candidates for Early-Stage Pancreatic Cancer. J Natl Cancer Inst. 2017;109:djw266. doi: 10.1093/jnci/djw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih LM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Théry C. Cancer: Diagnosis by extracellular vesicles. Nature. 2015;523:161–162. doi: 10.1038/nature14626. [DOI] [PubMed] [Google Scholar]
- 14.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32:490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, García-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin SJ, Smith JA, Rezniczek GA, Pan S, Chen R, Brentnall TA, Wiche G, Kelly KA. Unexpected gain of function for the scaffolding protein plectin due to mislocalization in pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:19414–19419. doi: 10.1073/pnas.1309720110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alderton GK. Diagnosis: Fishing for exosomes. Nat Rev Cancer. 2015;15:453. doi: 10.1038/nrc3990. [DOI] [PubMed] [Google Scholar]
- 19.Rahbari M, Rahbari N, Reissfelder C, Weitz J, Kahlert C. Exosomes: novel implications in diagnosis and treatment of gastrointestinal cancer. Langenbecks Arch Surg. 2016;401:1097–1110. doi: 10.1007/s00423-016-1468-2. [DOI] [PubMed] [Google Scholar]
- 20.Pucci F, Garris C, Lai CP, Newton A, Pfirschke C, Engblom C, Alvarez D, Sprachman M, Evavold C, Magnuson A, von Andrian UH, Glatz K, Breakefield XO, Mempel TR, Weissleder R, Pittet MJ. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science. 2016;352:242–246. doi: 10.1126/science.aaf1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herreros-Villanueva M, Bujanda L. Glypican-1 in exosomes as biomarker for early detection of pancreatic cancer. Ann Transl Med. 2016;4:64. doi: 10.3978/j.issn.2305-5839.2015.10.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faca VM, Song KS, Wang H, Zhang Q, Krasnoselsky AL, Newcomb LF, Plentz RR, Gurumurthy S, Redston MS, Pitteri SJ, Pereira-Faca SR, Ireton RC, Katayama H, Glukhova V, Phanstiel D, Brenner DE, Anderson MA, Misek D, Scholler N, Urban ND, Barnett MJ, Edelstein C, Goodman GE, Thornquist MD, McIntosh MW, DePinho RA, Bardeesy N, Hanash SM. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koopmann J, Zhang Z, White N, Rosenzweig J, Fedarko N, Jagannath S, Canto MI, Yeo CJ, Chan DW, Goggins M. Serum diagnosis of pancreatic adenocarcinoma using surface-enhanced laser desorption and ionization mass spectrometry. Clin Cancer Res. 2004;10:860–868. doi: 10.1158/1078-0432.ccr-1167-3. [DOI] [PubMed] [Google Scholar]
- 24.Yanagisawa K, Tomida S, Matsuo K, Arima C, Kusumegi M, Yokoyama Y, Ko SB, Mizuno N, Kawahara T, Kuroyanagi Y, Takeuchi T, Goto H, Yamao K, Nagino M, Tajima K, Takahashi T. Seven-signal proteomic signature for detection of operable pancreatic ductal adenocarcinoma and their discrimination from autoimmune pancreatitis. Int J Proteomics. 2012;2012:510397. doi: 10.1155/2012/510397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang JJ, Kimchi ET, Staveley-O’Carroll KF, Tan D. Diagnostic and prognostic biomarkers in pancreatic carcinoma. Int J Clin Exp Pathol. 2009;2:1–10. [PMC free article] [PubMed] [Google Scholar]
- 26.Costello E, Greenhalf W, Neoptolemos JP. New biomarkers and targets in pancreatic cancer and their application to treatment. Nat Rev Gastroenterol Hepatol. 2012;9:435–444. doi: 10.1038/nrgastro.2012.119. [DOI] [PubMed] [Google Scholar]
- 27.Kim MS, Kuppireddy SV, Sakamuri S, Singal M, Getnet D, Harsha HC, Goel R, Balakrishnan L, Jacob HK, Kashyap MK, Tankala SG, Maitra A, Iacobuzio-Donahue CA, Jaffee E, Goggins MG, Velculscu VE, Hruban RH, Pandey A. Rapid characterization of candidate biomarkers for pancreatic cancer using cell microarrays (CMAs) J Proteome Res. 2012;11:5556–5563. doi: 10.1021/pr300483r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, Giese NA, Kalthoff H, Becker T, Büchler MW, Zöller M. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136:2616–2627. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 29.Klein-Scory S, Tehrani MM, Eilert-Micus C, Adamczyk KA, Wojtalewicz N, Schnölzer M, Hahn SA, Schmiegel W, Schwarte-Waldhoff I. New insights in the composition of extracellular vesicles from pancreatic cancer cells: implications for biomarkers and functions. Proteome Sci. 2014;12:50. doi: 10.1186/s12953-014-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Wang X, Ferrone CR, Schwab JH, Ferrone S. Intracellular antigens as targets for antibody based immunotherapy of malignant diseases. Mol Oncol. 2015;9:1982–1993. doi: 10.1016/j.molonc.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haun JB, Castro CM, Wang R, Peterson VM, Marinelli BS, Lee H, Weissleder R. Micro-NMR for rapid molecular analysis of human tumor samples. Sci Transl Med. 2011;3:71ra16. doi: 10.1126/scitranslmed.3002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan MS, Kirkwood A, Tsigani T, Garcia-Hernandez J, Hartley JA, Caplin ME, Meyer T. Circulating tumor cells as prognostic markers in neuroendocrine tumors. J Clin Oncol. 2013;31:365–372. doi: 10.1200/JCO.2012.44.2905. [DOI] [PubMed] [Google Scholar]
- 33.Oberg K, Modlin IM, De Herder W, Pavel M, Klimstra D, Frilling A, Metz DC, Heaney A, Kwekkeboom D, Strosberg J, Meyer T, Moss SF, Washington K, Wolin E, Liu E, Goldenring J. Consensus on biomarkers for neuroendocrine tumour disease. Lancet Oncol. 2015;16:e435–436. doi: 10.1016/S1470-2045(15)00186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diamandis EP, Plebani M. Glypican-1 as a highly sensitive and specific pancreatic cancer biomarker. Clin Chem Lab Med. 2016;54:e1–2. doi: 10.1515/cclm-2015-0773. [DOI] [PubMed] [Google Scholar]
- 35.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardiner C, Ferreira YJ, Dragovic RA, Redman CW, Sargent IL. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Statist Soc B. 1996;58:267–288. [Google Scholar]
- 38.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 39.DeLong ER, Delong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 40.Fauci JM, Sabbatino F, Wang YY, Londoño-Joshi AI, Straughn JM, Jr, Landen CN, Ferrone S, Buchsbaum DJ. Monoclonal antibody-based immunotherapy of ovarian cancer: Targeting ovarian cancer cells with the B7-H3-specific mAb 376.96. Gynecol Oncol. 2014;132:203–210. doi: 10.1016/j.ygyno.2013.10.038. [DOI] [PubMed] [Google Scholar]
- 41.Imai K, Wilson BS, Bigotti A, Natali PG, Ferrone S. A 94,000-dalton glycoprotein expressed by human melanoma and carcinoma cells. J Natl Cancer Inst. 1982;68:761–769. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.