Abstract
We present genome-wide microarray expression analysis of 11,000 genes in an aging potentially mitotic tissue, the liver. This organ has a major impact on health and homeostasis during aging. The effects of life- and health-span-extending caloric restriction (CR) on gene expression among young and old mice and between long-term CR (LT-CR) and short-term CR (ST-CR) were examined. This experimental design allowed us to accurately distinguish the effects of aging from those of CR on gene expression. Aging was accompanied by changes in gene expression associated with increased inflammation, cellular stress, and fibrosis, and reduced capacity for apoptosis, xenobiotic metabolism, normal cell-cycling, and DNA replication. LT-CR and just 4 weeks of ST-CR reversed the majority of these changes. LT-CR produced in young mice a pattern of gene expression that is a subset of the changes found in old LT-CR mice. It is possible that the early changes in gene expression, which extend into old age, are key to the life- and health-span-extending effects of CR. Further, ST-CR substantially shifted the “normo-aging” genomic profile of old control mice toward the “slow-aging” profile associated with LT-CR. Therefore, many of the genomic effects of CR are established rapidly. Thus, expression profiling should prove useful in quickly identifying CR- mimetic drugs and treatments.
Published microarray studies of mammalian aging have focused on the postmitotic tissues, muscle and brain (1–3). These studies found that aging was associated with changes in gene expression linked to the development of the characteristic age-related pathologies of these tissues. In contrast to muscle and brain, the liver is a potentially mitotic tissue that is thought to age well from a clinical perspective (4). The liver is the central organ for the regulation of glucose homeostasis, xenobiotic metabolism and detoxification, and steroid hormone biosynthesis and degradation. This organ also has a major impact on health and homeostasis through its control of serum protein composition. While differentiated hepatic functions are generally well maintained with age, changes do occur. Serum and biliary cholesterol rise, liver regeneration declines, hepatic drug clearance decreases, and liver volume and blood flow decrease (4). The resilience of the liver to aging, and its central role in the maintenance of whole body health and homeostasis make it an intriguing target for genome-wide expression analysis of aging.
Caloric restriction (CR) is the only intervention shown to extend lifespan in mammals (5). It is also the most effective means known of reducing cancer incidence and increasing the mean age of onset of age-related diseases and tumors (6). Our studies made use of an experimental design that allowed us to clearly distinguish the effects of diet from those of age on genome-wide expression patterns. Another distinctive aspect of the study allowed us to resolve changes in gene expression induced directly by CR from those that arise over time as a consequence of the interaction between CR and aging.
Materials and Methods
Study Design.
Female mice of the long-lived F1 hybrid strain C3B10RF1 were fed and maintained as described (7). Briefly, mice were weaned at 28 days, individually housed, given free access to water, and randomly assigned to study groups. Comparisons between five groups of mice were used to determine the effects of aging and CR on gene expression. Control young (7-month-old; n = 3) and old (27-month-old; n = 3) mice were fed 95 kcal of a semipurified control diet (Harlan Teklad, Madison, WI; no. TD94145) per week after weaning. Long-term CR (LT-CR) young (7-month-old; n = 3) and old (27-month-old; n = 3) mice were fed 53 kcal of a semipurified CR diet (Harlan Teklad; no. TD94146) per week after weaning. Short-term CR (ST-CR) mice were 34-month-old control mice that were switched to 80 kcal of CR diet for 2 weeks, followed by 53 kcal for 2 weeks (n = 3). The effects of age on gene expression in control mice were determined by comparison between results from the young control and the old control groups. The effects of LT-CR on gene expression were determined by comparison between results from the young control and the young LT-CR groups, and from the old control and the old LT-CR groups. The effects of ST-CR were determined by comparison between results from the old control and the ST-CR groups. Mice were fasted for 48 h before killing. Mice were killed by cervical dislocation, and the livers were rapidly excised and flash frozen in liquid nitrogen. No signs of pathology were detected in any of the animals used. All animal use protocols were approved by the institutional animal use committee of the University of California, Riverside.
Measurement of Specific mRNA Levels.
Total liver RNA was isolated from frozen tissue as described previously (7). Specific mRNA levels were determined by using Affymetrix Mu11KsubA and Mu11KsubB high-density oligonucleotide arrays containing targets for more than 11,000 mouse genes and expressed sequence tags (ESTs) according to the standard Affymetrix protocol (Affymetrix, Santa Clara, CA). More detailed information can be found in Supporting Materials and Methods, which is published on the PNAS web site, www.pnas.org.
Data Analysis.
Analysis was performed as previously described for Affymetrix microarray data (8). We performed pairwise comparisons between data derived from individual mice in experimental groups (n = 3), generating nine pairwise comparisons by using the Affymetrix GeneChip analysis suite v3.2. An average fold change, derived from all nine possible pairwise comparisons, of 1.7 or greater was used as the cut-off for significant differences in gene expression. This criterion was chosen because fold changes of this magnitude were routinely validated by Northern blotting. Thresholds between 1.6 and 2.0 are commonly used in similar studies (9, 10). The specific intragroup correlation coefficients were between 0.94 and 0.98 for all experimental groups. Gene names were obtained from the Jackson Laboratory Mouse Genome Informatics or TIGR database (August 1, 2001). Complete tables of the data will be posted on our web site upon publication of the manuscript. More details regarding Affymetrix microarray technology can be found in Supporting Materials and Methods on the PNAS web site, www.pnas.org.
Validation by Northern Blotting.
Changes in the expression level of eight genes from our array studies were confirmed by Northern blotting. Six of these eight genes did not change expression with age, and this result also was confirmed. More detail can be found in Supporting Materials and Methods, Fig. 4 and its legend, and Supporting Results and Discussion, all of which are published on the PNAS web site, www.pnas.org.
Results and Discussion
Microarray determination of the relative levels of mRNA (levels of expression) of more than 11,000 genes and ESTs in young (7 months) and old (27 months) control mice revealed that 46 known genes (0.9% of the known genes on the chip) changed expression during aging in the liver. Of these genes, the expression of 20 (43%) increased with age (Fig. 1), and the expression of 26 (57%) decreased with age (Fig. 2). Age-related changes in gene expression are often assumed to be deleterious, because they represent a departure from what is assumed to be a positive, young pattern of expression (e.g., refs. 1–3).
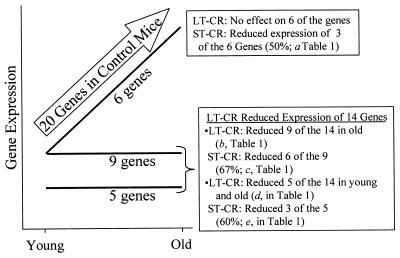
Figure 1.
LT- and ST-CR effects on the expression profile of 20 liver genes whose expression increases with age (arrow). LT-CR opposed the age-associated increase in the expression of 14 genes, in the manner shown by the lines labeled “9 genes” and “5 genes.” For the 9 genes, LT-CR returned expression to youthful levels in old mice. For the 5 genes, LT-CR reduced expression in both young and old mice. LT-CR had no effect on the expression of 6 of the 20 genes (line labeled “6 genes”). These genes are identified by the notation “NE” in Table 1. The effects of ST-CR are described in the text boxes.
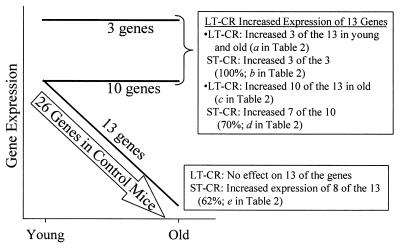
Figure 2.
Effects of LT- and ST-CR on the expression profile of 26 liver genes whose expression decreases with age (arrow). LT-CR opposed the age-associated decrease in the expression of 13 genes, in the manner shown by the lines labeled “3 genes” and “10 genes.” For the 3 genes, LT-CR increased expression in both young and old mice. For the 10 genes, LT-CR increased expression to youthful levels in old mice. LT-CR had no effect on the expression of 13 of the 26 genes (line labeled “13 genes”). These genes are identified by the notation “NE” in Table 2. The effects of ST-CR are described in the text boxes.
Genes Whose Expression Increased with Age.
Of the genes that increased expression with age, 40% were associated with inflammation (Table 1). These results suggest that inflammation may be a component of the aging process common to liver, neocortex and cerebellum (1). Lysozyme (Lyzs) is a myeloid cell-specific marker. Induction of this gene is normally associated with macrophage activation (11). The β-chain of complement component C1q (C1qb), a macrophage-expressed protein, is a part of the recognition set of the complement C system, the primary humoral mediator of antigen–antibody reactions (12). Macrophages, among other inflammatory cells, are involved in a large number of liver diseases, including cirrhosis, hepatitis, and sepsis- and endotoxin-induced liver injury (13).
Table 1.
Effects of LT- and ST-CR on expression of genes increased during aging in control mice
| GenBank no. | Age effect* | Gene | LT-CR†‡ | ST-CR‡§ |
|---|---|---|---|---|
| Inflammatory response | ||||
| L20276 | 1.9 | Bgn | −1.8b | −2.0c |
| U59807 | 1.8 | Cstb | −2.5b | −2.6c |
| U70139 | 5.3 | Ccr4 | −2.0b | −2.1c |
| K01925 | 1.9 | H2-Aa | −2.6b | −2.6c |
| M21050 | 1.7 | Lyzs | −2.5b | NE |
| M22531 | 1.9 | C1qb | −2.4b | NE |
| X16899 | 2.4 | Sap | NE | NE |
| U88327 | 3.0 | Cish2 | NE | NE |
| Stress response/chaperones | ||||
| J04633 | 1.7 | Hsp86-1 | (−2.8, −2.0)d | −2.9e |
| D78645 | 1.7 | Hspa5 | (−2.0, −2.3)d | −2.1e |
| M73329 | 1.9 | Grp58 | −1.9b | −2.1c |
| L07577 | 1.9 | Hsp25 | NE | −2.3a |
| U27830 | 2.3 | Stip1 | NE | −2.4a |
| Apoptosis regulators | ||||
| AF018268 | 1.9 | Api6 | (−2.1, −5.0)d | −1.9e |
| U44088 | 2.7 | Tdag | (−2.8, −7.4)d | NE |
| U35623 | 1.9 | Mcl1 | NE | NE |
| Miscellaneous | ||||
| M81445 | 1.8 | Gjb2 | (−1.8, −2.6)d | NE |
| Z48670 | 1.9 | Abcd2 | −2.0b | −2.1c |
| J00361 | 3.5 | Amy2 | −3.4b | NE |
| AF006492 | 4.4 | Zfpm1 | NE | −2.1a |
The numbers in this column represent the average fold of the age-related increase in specific mRNA derived from all nine possible pairwise comparisons among individual mice from 7- and 27-month-old control groups (n = 3).
The numbers in this column represent the average fold change in specific mRNA associated with LT-CR. When two values are reported, the first represents the decrease in specific mRNA associated with LT-CR in 7-month-old mice and the second value represents the difference found in 27-month-old mice. When one value is reported, it represents the difference in 27-month-old mice.
The superscript letter indicates the gene group described in Fig. 1. NE indicates CR had no effect on gene expression.
The average fold change in specific mRNA associated with ST-CR.
In the neocortex and cerebellum, complement component C1q C polypeptide (C1qc) is up-regulated with age as is the C1qb in the liver. The α polypeptide is regulated in the same manner in the neocortex. Lyzs also was up-regulated with age in neocortex and cerebellum, but neither gene was affected in muscle (1, 3). Changes in the expression of these genes are among the few that were common to liver, muscle, and brain.
Normal liver aging was associated with other gene expression changes consistent with pathogenesis. Old mice overexpressed the mRNA for biglycan (Bgn), a proteoglycan of the hepatic extracellular matrix, serum amyloid P-component (Sap), a glycoprotein present in all amyloid deposits, and cystatin B (Cstb), an inhibitor of cysteine proteinases (Table 1). In areas of inflammation, fibrogenic myofibroblasts express Bgn and other proteoglycans, leading to hepatic fibrosis (14). Sap is one of the major acute-phase reactants induced by inflammation in hepatocytes (15). Cystatins and their target enzymes play a role in many pathological events, including inflammatory disease (16). In the liver, an imbalance between cystatins and their targets can disregulate matrix degradation and accumulation, leading to hepatic fibrosis (17).
Twenty-five percent of the genes that increased expression with age were stress response proteins/chaperones (Table 1). We have previously reported an age-related increase in hepatic chaperone expression in mice (18, 19). Diverse physiological stresses, including oxidative injury, may be causally involved in aging (20). In mammals, the oxidative processes centered in the liver are major sources of free radicals. The induction of chaperone gene expression in the livers of aged mice may be a physiological adaptation to increased oxidative or possibly other stress during aging. Chaperones provide cytoprotective functions, including prevention of protein denaturation and aggregation, the repair of damaged structural and functional proteins (21), and promotion of the ubiquitination and proteasomal degradation of potentially toxic, damaged proteins produced by oxidative or glycoxidative processes (22, 23).
However, another function of chaperones may mitigate some of these benefits. Elevated chaperone levels prevent apoptosis (24), a function implicated in tumorigenesis (25). Our studies also found that aging increased the expression of other anti-apoptotic genes, myeloid cell leukemia sequence 1 (Mcl1), and apoptosis inhibitory protein 6 (Api6). These phenomena may explain why hepatocellular neoplasms are the most common lesions in older mice (26, 27).
Genes Whose Expression Decreased with Age.
Of the 26 genes that decreased expression with age in control mice, 23% are involved in DNA replication and the cell cycle (Table 2). Most of these have a negative effect on cell growth and division. Among these, the product of phosphatase and tensin homolog (Pten) gene is a tumor suppressor that induces cell-cycle arrest through inhibition of the phosphoinositide 3-kinase pathway (28). B cell translocation gene 2 (Btg2) is a tumor suppressor that increases expression in response to DNA damage (29). The murine gene product of the amino-terminal enhancer of split (Aes) is a potent corepressor of gene expression and cellular proliferation (30). Calcium-binding protein A11 (S100a10) binds to and regulates the activity of annexin II, which is involved in the transduction of calcium-related mitogenic signals (31). Insulin-like growth factor (IGF) binding protein 1 (Igfbp1) plays an important role in the negative regulation of the IGF-1 system, a stimulator of mitogenesis (32).
Table 2.
Effects of LT- and ST-CR on expression of genes decreased during aging of control mice
| GenBank no. | Age effect* | Gene | LT-CR†‡ | ST-CR‡§ |
|---|---|---|---|---|
| Cell cycle/DNA replication | ||||
| M64292 | −1.7 | Btg2 | 2.6c | 2.8d |
| X73359 | −4.6 | Aes | 7.4c | NE |
| U92437 | −1.7 | Pten | NE | 2.0e |
| M16465 | −1.9 | S100a10 | NE | 1.9e |
| X67493 | −2.4 | Igfbp1 | NE | 2.7e |
| L31783 | −2.5 | Umpk | NE | 3.3e |
| Xenobiotic metabolism | ||||
| U80819 | −4.5 | Gsto1 | (1.8, 4.9)a | 5.1b |
| AF026073 | −2.0 | Sultn | (2.0, 2.3)a | 3.6b |
| M77497 | −4.9 | Cyp2f2 | 2.2c | 6.9d |
| X00479 | −1.8 | Cyp1a2 | NE | NE |
| M60358 | −3.5 | Cyp2b13 | NE | NE |
| Major urinary proteins | ||||
| X03208 | −5.2 | Mup1 | 1.9c | 3.2d |
| M16359 | −3.8 | Mup3 | 2.4c | 3.3d |
| M16358 | −3.0 | Mup4 | 1.9c | 2.4d |
| M16360 | −5.0 | Mup5 | 2.4c | 4.6d |
| Miscellaneous | ||||
| L07645 | −2.5 | Hal | (3.5, 5.3)a | 5.5b |
| M58691 | −2.1 | Zfp36 | 1.9c | 2.1d |
| X61940 | −3.8 | Ptpn16 | 2.8c | NE |
| AF033565 | −2.0 | Clk3 | 3.2c | NE |
| D50032 | −2.3 | Ttgn2 | NE | 2.5e |
| U12473 | −2.1 | Chuk | NE | 2.3e |
| D00466 | −1.7 | Apoe | NE | 2.1e |
| M27796 | −3.0 | Car3 | NE | 4.5e |
| U02554 | −1.9 | Saa4 | NE | NE |
| ET61200 | −2.0 | Cry | NE | NE |
| U75530 | −2.7 | Eif4ebp2 | NE | NE |
The numbers in this column represent the average fold of the age-related change in specific mRNA derived from all nine possible pairwise comparisons among individual mice from 7- and 27-month-old control groups (n = 3).
The numbers in this column represent the average fold increase in specific mRNA associated with LT-CR. When two values are reported, the first represents the increase in specific mRNA associated with LT-CR in 7-month-old mice and the second value represents the difference found in 27-month-old mice. When one value is reported, it represents the difference in 27-month-old mice.
The superscript letter indicates the gene group described in Fig. 2. NE indicates CR had no effect on gene expression.
The average fold increase in specific mRNA associated with ST-CR.
Reduced expression of genes discussed above indicates that there is a general loss of negative cell growth control with age. Seventy-eight percent of the mice of this strain and sex fed the control diet used here die of some form of neoplasia, and the death rate from neoplasia accelerates dramatically with age (33). Approximately 21% of these mice die of hepatoma, mostly late in life. Decreased expression of the negative growth regulators and overexpression of the chaperone genes with age also are consistent with this higher incidence of hepatoma in aged mice.
Aging decreased expression of a second group of genes with antineoplastic potential, xenobiotic metabolism genes (Table 2). The genes for the phase I enzymes amine N-sulfotransferase (Sultn) and three cytochrome P450 isozymes, as well as the gene for the phase II enzyme glutathione S-transferase omega 1 (Gsto1) gene were negatively regulated by age. We and others have reported decreased expression of phase I enzyme genes in the liver of aged rodents (34–36). Decreased expression of such genes is likely responsible in part for the age-related decline in the xenobiotic-metabolizing capacity of the liver. This decline is a recognized source of adverse drug reactions in aged mammals (37). It may contribute to the increase in neoplasms with age in mice.
Aging was associated with decreased expression of other genes responsible for differentiated liver functions. Apolipoprotein E (Apoe) decreased expression with age (Table 2). This protein is mainly secreted by the liver, and it is required for clearance of lipoproteins from the blood. Disruption of Apoe is associated with severe atherosclerosis in mice (38). Thus, decreased Apoe expression with age may be related to an increase in atherosclerotic lesions in mice (39). There is a similar association between impaired or absent Apoe expression and hyperlipidemia in humans (38), where the expression of a specific Apoe subtype is associated with extreme longevity (40).
LT-CR Opposed Age-Associated Changes in Gene Expression.
LT-CR opposed the age-related increase in expression of 14 of the 20 genes that increased expression in control mice (70%; Fig. 1). Five of these 14 genes were affected by CR in both young and old mice. LT-CR suppressed the increase in 75% (6 of 8 genes) of the inflammatory response genes (Table 1). Consistent with decreased inflammatory response gene expression, CR delays the onset and diminishes the severity of autoimmune and inflammatory diseases in mice (41).
LT-CR opposed the age-related increase in the expression of 3 of the 5 stress response proteins (Table 1). We have previously shown that expression of at least 7 chaperones is negatively regulated in response to LT-CR (19). These data suggest that LT-CR reduces physiological stress on the liver. Further, as discussed above, reduced chaperone expression is proapoptotic and anti-neoplastic. Consistent with these results, LT-CR decreased expression of Api6 (Table 1). Together, these effects may explain the delayed onset of hepatoma in LT-CR mice (33).
LT-CR opposed the age-associated decrease in the expression of 13 of the 26 genes that decreased expression in control mice (50%; Fig. 2). Many of these genes are responsible for key differentiated functions of the liver. Reversal of the age-related decrease in the Btg2, Aes, Gsto1, Sultn, and cytochrome P450 2f2 (Cyp2f2) mRNAs also are consistent with the delayed onset of hepatoma in LT-CR mice (Table 2). Partial restoration of the hepatic drug-metabolizing and detoxifying functions of the liver may be a source of the anti-aging and anticancer effects of LT-CR.
ST-CR Reproduced the Majority of the Effects of LT-CR on Age-Responsive Gene Expression.
To determine the effects of ST-CR on gene expression, old control mice were shifted to CR for 4 weeks, and their microarray profiles were compared with those of old LT-CR and control mice. This approach directly tested the often-expressed hypothesis that LT-CR acts to slow aging by preventing age-related changes in gene expression (1, 3). Overall, ST-CR reproduced nearly 70% of the effects of LT-CR on genes that changed expression with age (Figs. 1 and 2; Tables 1 and 2). The effects of ST- and LT-CR were highly homologous in both direction and fold-change for these genes. Thus, CR rapidly reversed, rather than prevented, many age-related changes in gene expression. ST-CR induced a more youthful gene expression profile associated with longer life- and health-span. Expression profiling of these genes should prove useful in rapidly identifying CR-mimetic drugs and treatments.
ST-CR reproduced 100% of the LT-CR effects on xenobiotic metabolism, stress response/chaperone, and major urinary protein gene expression. It also reproduced 67% (4 of 6) of the effects of LT-CR on inflammatory response gene expression. These results suggest that CR may rapidly ameliorate inflammation and other stresses, even in very old animals. ST-CR also reproduced the effects of LT-CR on the expression of 50% of the cell-cycle/DNA replication and apoptosis genes. The combination of these effects on gene expression suggests that ST-CR may be capable of rapidly reproducing the antineoplastic effects of LT-CR in very old animals. This conclusion is consistent with studies showing that short-term fasting increased apoptosis of preneoplastic cells and preneoplastic lesions, and reduced rates of chemical carcinogenesis (42, 43). Thus, CR mimetics might be useful anticancer therapies. The effects of ST-CR on the expression of genes associated with xenobiotic metabolism suggest that ST-CR may rapidly restore some differentiated functions in tissues of older animals.
CR-Specific Reprogramming of Gene Expression.
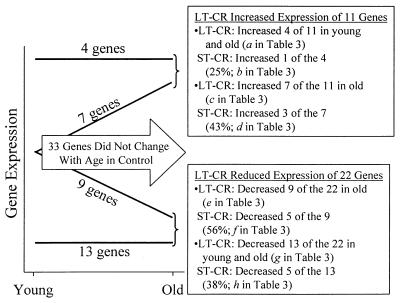
LT-CR reprogrammed the expression of 33 genes that were not differentially expressed with age in control mice (Table 3 and Fig. 3). Here we term these effects CR-specific changes to distinguish them from the effects of CR discussed above (Tables 1 and 2). LT-CR increased the expression of 11 of these genes (Fig. 3). Of the 11 genes, 7 increased expression only in old CR mice, and 4 increased expression in both young and old CR mice. LT-CR reduced the expression of 22 genes (Fig. 3). Of these genes, 9 were reduced only in the old, and 13 were reduced in both young and old mice.
Table 3.
LT- and ST-CR effects on gene expression unaffected by age in control mice
| GenBank no. | Gene | LT-CR*† | ST-CR†‡ |
|---|---|---|---|
| Energy metabolism/biosynthesis | |||
| J02623 | Got1 | (4.5, 2.3)a | NE |
| L07037 | Mgat1 | 4.6c | NE |
| M13966 | Apoa4 | (−1.9, −3.2)g | −4.8h |
| X13752 | Alad | (−3.1, −2.8)g | −4.5h |
| U49861 | Dio1 | (−2.1, −5.1)g | −5.5h |
| M65034 | Fabp2 | (−3.1, −5.4)g | −4.5h |
| X13135 | Fasn | (−2.7, −2.0)g | NE |
| AB008895 | Gpi1h | −2.2e | −2.0f |
| U67611 | Taldo1 | −2.0e | NE |
| AF010499 | Gamt | −2.3e | NE |
| M76727 | Pdha1 | −2.0e | NE |
| Apoptosis/cell growth and survival | |||
| AF011644 | Cdkap1 | 2.3c | 2.6d |
| U30840 | Vdac1 | 3.6c | 3.3d |
| Y13231 | Bak1 | 2.7c | NE |
| L75822 | Igfbp7 | 2.2c | NE |
| U04268 | Ly6e | (−2.5, −6.6)g | NE |
| AF027963 | Xbp1 | (−1.8, −2.2)g | NE |
| U28423 | Prkri | −1.9e | −2.0f |
| Xenobiotic metabolism | |||
| U20086 | Ubp1 | (2.7, 1.8)a | 1.9b |
| X63023 | Cyp3a13 | 2.1c | 4.7d |
| Y11638 | Cyp4a14 | 2.2c | NE |
| U38652 | Slc22a1 | (−1.8, −2.9)g | −5.0h |
| L11333 | Es31 | −2.8e | NE |
| Intracellular signaling | |||
| M33324 | Ghr | (−1.8, −2.9)g | NE |
| X61432 | Calm | −2.7e | −2.7f |
| X70533 | Cbg | −2.7e | −9.3f |
| ET61316 | Rgn | −2.3e | −2.2f |
| Stress response/chaperones | |||
| J05186 | Cai | (−1.8, −2.2)g | NE |
| X70303 | Psma2 | (−2.2, −2.5)g | NE |
| Miscellaneous | |||
| U14666 | Nr6a1 | (1.9, 2.1)a | NE |
| D64160 | Raet1a | (2.1, 2.7)a | NE |
| M17122 | C4bp | (−1.8, −2.4)g | NE |
| Z47088 | Tceb1l | (−1.9, −2.2)g | NE |
The numbers in this column represent the average fold increase or decrease in specific mRNA associated with LT-CR. When two values are reported, the first represents the change in specific mRNA associated with LT-CR in 7-month-old mice and the second value represents the difference found in 27-month-old mice. When one value is reported, it represents the difference in 27-month-old mice.
The superscript letter indicates the gene group described in Fig. 3. NE indicates CR had no effect on gene expression.
The average fold decrease or increase in specific mRNA associated with ST-CR.
Figure 3.
Effects of LT- and ST-CR on the expression profile of 33 liver genes whose expression remains unchanged during aging of control mice (arrow). Expression of 11 of the 33 genes increased in the LT-CR group, in the manner shown by the lines labeled “4 genes” and “7 genes.” For the 4 genes, LT-CR increased expression in both young and old mice. For the 7 genes, LT-CR increased expression only in old mice. Expression of 22 of the 33 genes decreased in the LT-CR group, in the manner shown by the lines labeled “9 genes” and “13 genes.” For the 9 genes, LT-CR decreased expression only in old mice. For the 13 genes, LT-CR decreased expression in both young and old mice. The effects of ST-CR are described in the text boxes.
One-third of the CR-specific genes code for key metabolic enzymes (Table 3). The LT-CR-related increase in glutamate–oxaloacetate transaminase 1 (Got1) and decrease in pyruvate dehydrogenase E1α-subunit (Pdha1) expression are consistent with our previous studies showing that LT-CR increases the enzymatic capacity of the liver for gluconeogenesis and the disposal of the byproducts of extrahepatic protein catabolism for energy production, while reducing the enzymatic capacity for glycolysis (44). Recently, we have shown that these effects persist in the hours after feeding (45). Therefore, these CR effects are consistent with theories of aging, such as the oxidative stress theory, which postulate that the accumulation of damaged proteins contributes to the rate of aging (44).
Reduced expression of Pdha1, fatty acid synthase (Fasn), fatty acid binding protein 2 (Fabp2), and transaldolase 1 (Taldo) genes suggests that CR mice have a reduced enzymatic capacity for fatty acid biosynthesis. Reduced apolipoprotein A-IV (Apoa4) expression may be a source of some of the health benefits of CR, because overexpression of this protein can be associated with increased vascular disease (46). Rapid reduction of Apoa4 expression by ST-CR suggests that these health benefits may start soon after shifting to a CR diet.
We found further evidence that CR modified energy utilization. LT- and ST-CR decreased the expression of iodothyronine deiodinase type I (Dio1). Down-regulation of this enzyme may be responsible for the reduced levels of circulating triiodothyronine (T3) found in CR rodents (47). Short-term treatment with low-calorie diets rapidly reduces circulating T3 levels in morbidly obese men, apparently by reducing type I deiodinase activity (48). Thus, similar negative regulation of Dio1 may occur in ST-CR humans. T3 influences a diverse set of energy-related processes in mammalian tissues, including oxidative phosphorylation and oxygen consumption. LT-CR also decreased the expression of guanidinoacetate methyltransferase (Gamt), which catalyzes the last step of creatine biosynthesis. The creatine synthesized by liver is thought to supply the creatine energy reservoir in tissues such as skeletal, cardiac, and smooth muscle, and brain (49). Thus, mice may adapt to LT-CR by reducing tissue energy stores throughout the body.
Genes associated with apoptosis, cell growth, and survival constituted 21% of the CR-specific genes (Table 3). LT- and ST-CR induced the expression of cyclin-dependent kinase 2-associated protein 1 (Cdkap1), a putative tumor-suppressor gene (50). Overexpression of this gene suggests that LT- and ST-CR enhance antiproliferative growth control. Consistent with this idea, IGF-binding protein 7 (Igfbp7) gene expression was induced by LT-CR. The product of this gene functions both as an IGF-binding protein and independently of IGF as a growth-suppressing factor (51). The expression of Igfbp1, which has antigrowth activity through its inhibition of IGF-1 signaling, was reduced by age and restored by ST-CR (Table 2). Thus, LT- and ST-CR may produce additional antiproliferative effects on preneoplastic cells of the liver through their effects on the expression of these IGF-binding protein family members.
LT-CR induced the expression of the Bcl2 homologous antagonist/killer (Bak1) and voltage-dependent anion channel 1 (Vdac1) (porin) genes. Bak1 is a pro-apoptotic member of the Bcl2 family of apoptosis regulators. It directly interacts with porin to release the pro-apoptotic factor cytochrome c from mitochondria, initiating apoptosis (52). Overexpression of porin in ST-CR mice is consistent with the increase in apoptosis and reduction in chemical carcinogenesis found in fasting rodents (42, 43). LT-CR decreased the expression of the anti-apoptotic genes: IFN-inducible double-stranded RNA-dependent inhibitor (Prkri), X-box binding protein (Xbp1), and lymphocyte antigen 6 complex, locus E (Ly6e) (53–55) (Table 3). Consistent with these effects, CR also up-regulated several cytochrome P450 genes, and the upstream binding protein 1 (Ubp1) gene (Table 3). The latter gene governs the rate of transcription of many P450 genes (Table 3). Overexpression of these genes and overexpression of three phase I and II genes in old LT- and ST-CR mice (Table 1) likely contribute to the positive health effects of CR.
Several significant intracellular signaling genes were down-regulated by CR (Table 3). LT- and ST-CR reduced calmodulin (calm) and regucalcin (Rgn) mRNA. Calmodulin is a central component of Ca2+-mediated signal transduction. It acts both directly and by means of protein kinases and phosphatases to regulate metabolism, cytoskeletal dynamics, and cellular proliferation. The decline in its expression may lead to a reduction in the calcium, second messenger-mediated, proliferative potential of CR liver. LT- and ST-CR also reduced expression of corticosteroid-binding globulin (Cbg), which transports and modulates the bioavailability of glucocorticoids. This effect of CR has been postulated to mediate the life- and health-span effects of CR by producing transient daily periods of mild hyperadrenocorticism (56).
LT-CR decreased the level of growth hormone receptor (Ghr) mRNA in young and old mice (Table 3). Declining plasma IGF-1 levels are often hypothesized to contribute to aging in animals and humans. The LT-CR-associated down-regulation of Ghr mRNA found here may be responsible for the decrease in its binding activity reported in the livers of LT-CR mice (57).
CR-Specific Reprogramming of Gene Expression in Young Mice.
In young mice, LT-CR established a CR-specific pattern of gene expression that was a subset of that found in old LT-CR mice (Table 4). Most of the effects of LT-CR on stress response, energy, and xenobiotic metabolism genes started in young mice. The inflammatory response genes were not overexpressed in young mice, and appropriately, no effect on expression of these genes was found until old age. Thus, CR seems to produce a gene expression profile early in life, which becomes embellished with age. These early changes may be keys to extended longevity. Rats that were CR only during their first year outlived rats that were CR only after their first year of life (58). Thus, it seems possible that the early effects of CR on gene expression may be more important to life- and health-span extension than the late effects.
Table 4.
Categories of CR-regulated genes in young and old mice
| Gene category | Young CR/old CR
|
|
|---|---|---|
| %* | Ratio† | |
| Stress response/chaperones | 80 | 4/5 |
| Energy metabolism/biosynthesis | 55 | 6/11 |
| Xenobiotic metabolism | 50 | 4/8 |
| Apoptosis/cell growth and survival | 44 | 4/9 |
| Intracellular signaling | 25 | 1/4 |
| Inflammatory response | 0 | 0/6 |
| Major urinary proteins | 0 | 0/4 |
| Cell cycle/DNA replication | 0 | 0/2 |
| Miscellaneous | 55 | 6/11 |
| Total | 42 | 25/60 |
The percentage of genes in each category that changed expression in young with respect to old CR mice.
The ratio between the number of genes that change expression in young and old CR mice.
CR Rapidly Induced a “Slow-Aging” Expression Profile.
ST-CR reproduced 64% (9 of 14) of the effects of LT-CR on genes that were induced during aging in control mice (Table 1). It reproduced 77% (10 of 13) of the effects of LT-CR on the expression of genes that were down-regulated during aging of control mice (Table 2). Forty-two percent (14 of 33) of the CR-specific changes in gene expression induced in old LT-CR mice were reproduced by ST-CR (Table 3). Overall, 55% of all of the effects of LT-CR on gene expression in old mice were reproduced by ST-CR. Thus, ST-CR reproduced half or more of the effects of LT-CR in 6 of the 8 categories of genes altered in expression by LT-CR (Table 5). These results raise the possibility that relatively brief treatments with drugs or nutraceuticals and other procedures can be used to search for CR mimetics.
Table 5.
Categories of ST- and LT-CR-regulated genes
| Gene category | ST-CR/LT-CR
|
|
|---|---|---|
| %* | Ratio† | |
| Major urinary proteins | 100 | 4/4 |
| Intracellular signaling | 75 | 3/4 |
| Xenobiotic metabolism | 75 | 6/8 |
| Inflammatory response | 67 | 4/6 |
| Stress response/chaperones | 60 | 3/5 |
| Cell cycle/DNA replication | 50 | 1/2 |
| Apoptosis/cell growth and survival | 44 | 4/9 |
| Energy metabolism/biosynthesis | 45 | 5/11 |
| Miscellaneous | 27 | 3/11 |
| Total | 55 | 33/60 |
The genes that changed expression with ST-CR as a percentage of those that changed with LT-CR.
The ratio between number of genes that change expression with ST- and LT-CR.
Tissue Specificity of Expression Profiles.
We compared the gene expression profiles found here with those reported for muscle and brain (1–3). Aging induced the expression of a number of stress response genes in liver, skeletal muscle, neocortex, and cerebellum. However, in the cortex and hypothalamus, aging reduced the expression of other stress response genes. Likewise, aging induced expression of inflammatory genes in the liver, neocortex, and cerebellum, but not in skeletal muscle, cortex, or hypothalamus. These results clearly show that aging is tissue-specific in its effects. They also suggest that tissues are subjected to different stresses during aging.
CR opposed the age-related induction of stress response genes in muscle, and stress response and inflammatory genes in liver, neocortex, and cerebellum. Thus, the amelioration of physiological stress appears to be a common anti-aging effect of CR. Aging and CR also altered the expression of genes involved in cell growth in liver and brain. However, aging and CR changed the expression of apoptosis-related genes only in the liver, which is consistent with the mitotic potential of this tissue. CR altered expression of genes involved with energy metabolism and biosynthesis in liver and brain, but not in muscle. Again, these results highlight the tissue specificity of both aging and CR. As more tissues in more species are studied, characteristic tissue-specific patterns of gene regulation by aging and CR may emerge.
Supplementary Material
Acknowledgments
We thank William Wachsman, Matt Hazel, the Veterans Affairs San Diego Health Care Systems GeneChip Core, and the University of California San Diego Cancer Center Microarray Shared Resource for performing the GeneChip assays. We also thank William Wachsman for helpful discussions.
Abbreviations
- CR
caloric restriction
- LT-CR
long-term CR
- ST-CR
short-term CR
- ESTs
expressed sequence tags
- IGF
insulin-like growth factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lee C K, Weindruch R, Prolla T A. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 2.Jiang C H, Tsien J Z, Schultz P G, Hu Y. Proc Natl Acad Sci USA. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C K, Klopp R G, Weindruch R, Prolla T A. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 4.Schmucker D L. J Gerontol A Biol Sci Med Sci. 1998;53:B315–B320. doi: 10.1093/gerona/53a.5.b315. [DOI] [PubMed] [Google Scholar]
- 5.McCay C M, Crowell M F, Maynard L A. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- 6.Weindruch R, Walford R L. The Retardation of Aging and Disease by Dietary Restriction. Springfield, IL: Thomas; 1988. [Google Scholar]
- 7.Dhahbi J M, Tillman J B, Cao S, Mote P L, Walford R L, Spindler S R. J Gerontol A Biol Sci Med Sci. 1998;53:B180–B185. doi: 10.1093/gerona/53a.3.b180. [DOI] [PubMed] [Google Scholar]
- 8.Lipshutz R J, Fodor S P, Gingeras T R, Lockhart D J. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 9.Rampon C, Jiang C H, Dong H, Tang Y P, Lockhart D J, Schultz P G, Tsien J Z, Hu Y. Proc Natl Acad Sci USA. 2000;97:12880–12884. doi: 10.1073/pnas.97.23.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaminski N, Allard J D, Pittet J F, Zuo F, Griffiths M J, Morris D, Huang X, Sheppard D, Heller R A. Proc Natl Acad Sci USA. 2000;97:1778–1783. doi: 10.1073/pnas.97.4.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keshav S, Chung P, Milon G, Gordon S. J Exp Med. 1991;174:1049–1058. doi: 10.1084/jem.174.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petry F, Reid K B, Loos M. FEBS Lett. 1989;258:89–93. doi: 10.1016/0014-5793(89)81622-9. [DOI] [PubMed] [Google Scholar]
- 13.Jaeschke H. Am J Physiol. 1997;273:G602–G611. doi: 10.1152/ajpgi.1997.273.3.G602. [DOI] [PubMed] [Google Scholar]
- 14.Friedman S L. N Engl J Med. 1993;328:1828–1835. doi: 10.1056/NEJM199306243282508. [DOI] [PubMed] [Google Scholar]
- 15.Pepys M B, Baltz M, Gomer K, Davies A J, Doenhoff M. Nature (London) 1979;278:259–261. doi: 10.1038/278259a0. [DOI] [PubMed] [Google Scholar]
- 16.Buttle D J, Abrahamson M, Burnett D, Mort J S, Barrett A J, Dando P M, Hill S L. Biochem J. 1991;276:325–331. doi: 10.1042/bj2760325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kos J, Lah T T. Oncol Rep. 1998;5:1349–1361. doi: 10.3892/or.5.6.1349. [DOI] [PubMed] [Google Scholar]
- 18.Spindler S R, Crew M D, Mote P L, Grizzle J M, Walford R L. J Nutr. 1990;120:1412–1417. doi: 10.1093/jn/120.11.1412. [DOI] [PubMed] [Google Scholar]
- 19.Dhahbi J M, Mote P L, Tillman J B, Walford R L, Spindler S R. J Nutr. 1997;127:1758–1764. doi: 10.1093/jn/127.9.1758. [DOI] [PubMed] [Google Scholar]
- 20.Harman D. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 21.Hartl F U. Nature (London) 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 22.Medina R, Wing S S, Goldberg A L. Biochem J. 1995;307:631–637. doi: 10.1042/bj3070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman M Y, Goldberg A L. EXS. 1996;77:57–78. doi: 10.1007/978-3-0348-9088-5_5. [DOI] [PubMed] [Google Scholar]
- 24.McMillan D R, Xiao X, Shao L, Graves K, Benjamin I J. J Biol Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- 25.Ciocca D R, Fuqua S A, Lock-Lim S, Toft D O, Welch W J, McGuire W L. Cancer Res. 1992;52:3648–3654. [PubMed] [Google Scholar]
- 26.Muskhelishvili L, Hart R W, Turturro A, James S J. Am J Pathol. 1995;147:20–24. [PMC free article] [PubMed] [Google Scholar]
- 27.Weindruch R, Walford R L. Science. 1982;215:1415–1418. doi: 10.1126/science.7063854. [DOI] [PubMed] [Google Scholar]
- 28.Cantley L C, Neel B G. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortes U, Moyret-Lalle C, Falette N, Duriez C, Ghissassi F E, Barnas C, Morel A P, Hainaut P, Magaud J P, Puisieux A. Mol Carcinog. 2000;27:57–64. [PubMed] [Google Scholar]
- 30.Pinto M, Lobe C G. J Biol Chem. 1996;271:33026–33031. doi: 10.1074/jbc.271.51.33026. [DOI] [PubMed] [Google Scholar]
- 31.Puisieux A, Ji J, Ozturk M. Biochem J. 1996;313:51–55. doi: 10.1042/bj3130051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frystyk J, Delhanty P J, Skjaerbaek C, Baxter R C. Am J Physiol. 1999;277:E245–E252. doi: 10.1152/ajpendo.1999.277.2.E245. [DOI] [PubMed] [Google Scholar]
- 33.Weindruch R, Walford R L, Fligiel S, Guthrie D. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- 34.Ali B, Walford R L, Imamura T. Life Sci. 1985;37:1387–1393. doi: 10.1016/0024-3205(85)90077-3. [DOI] [PubMed] [Google Scholar]
- 35.Mote P L, Grizzle J M, Walford R L, Spindler S R. Mech Ageing Dev. 1990;53:101–110. doi: 10.1016/0047-6374(90)90038-h. [DOI] [PubMed] [Google Scholar]
- 36.Mote P L, Grizzle J M, Walford R L, Spindler S R. J Gerontol. 1991;46:B95–B100. doi: 10.1093/geronj/46.3.b95. [DOI] [PubMed] [Google Scholar]
- 37.Schmucker D L. Pharmacol Rev. 1985;37:133–148. [PubMed] [Google Scholar]
- 38.Davignon J, Cohn J S, Mabile L, Bernier L. Clin Chim Acta. 1999;286:115–143. doi: 10.1016/s0009-8981(99)00097-2. [DOI] [PubMed] [Google Scholar]
- 39.Merat S, Fruebis J, Sutphin M, Silvestre M, Reaven P D. J Gerontol A Biol Sci Med Sci. 2000;55:B85–B94. doi: 10.1093/gerona/55.2.b85. [DOI] [PubMed] [Google Scholar]
- 40.Schachter F, Faure-Delanef L, Guenot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D. Nat Genet. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- 41.Cherry, Engelman R W, Wang B Y, Kinjoh K, El Badri N S, Good R A. Proc Soc Exp Biol Med. 1998;218:218–222. doi: 10.3181/00379727-218-44289. [DOI] [PubMed] [Google Scholar]
- 42.Pitot H C, Hikita H, Dragan Y, Sargent L, Haas M. Aliment Pharmacol Ther. 2000;14, Suppl. 1:153–160. doi: 10.1046/j.1365-2036.2000.014s1153.x. [DOI] [PubMed] [Google Scholar]
- 43.Hikita H, Vaughan J, Babcock K, Pitot H C. Toxicol Sci. 1999;52:17–23. doi: 10.1093/toxsci/52.2.17. [DOI] [PubMed] [Google Scholar]
- 44.Dhahbi J M, Mote P L, Wingo J, Tillman J B, Walford R L, Spindler S R. Am J Physiol. 1999;277:E352–E360. doi: 10.1152/ajpendo.1999.277.2.E352. [DOI] [PubMed] [Google Scholar]
- 45.Dhahbi J M, Mote P L, Wingo J, Rowley B C, Cao S X, Walford R, Spindler S R. Mech Ageing Dev. 2001;122:35–50. doi: 10.1016/s0047-6374(01)00230-5. [DOI] [PubMed] [Google Scholar]
- 46.Verges B L, Lagrost L, Vaillant G, Petit J M, Cohen M, Gambert P, Brun J M. Diabetes. 1997;46:125–132. doi: 10.2337/diab.46.1.125. [DOI] [PubMed] [Google Scholar]
- 47.Herlihy J T, Stacy C, Bertrand H A. Mech Ageing Dev. 1990;53:9–16. doi: 10.1016/0047-6374(90)90030-j. [DOI] [PubMed] [Google Scholar]
- 48.Katzeff H L, Yang M-U, Presta E, Leibel R L, Hirsch J, Van Itallie T B. Am J Clin Nutr. 1990;52:263–266. doi: 10.1093/ajcn/52.2.263. [DOI] [PubMed] [Google Scholar]
- 49.Gerber G B, Gerber G, Koszalka T R, Miller L L. J Biol Chem. 1962;237:2246–2250. [PubMed] [Google Scholar]
- 50.Todd R, McBride J, Tsuji T, Donoff R B, Nagai M, Chou M Y, Chiang T, Wong D T. FASEB J. 1995;9:1362–1370. doi: 10.1096/fasebj.9.13.7557027. [DOI] [PubMed] [Google Scholar]
- 51.Oh Y, Nagalla S R, Yamanaka Y, Kim H S, Wilson E, Rosenfeld R G. J Biol Chem. 1996;271:30322–30325. doi: 10.1074/jbc.271.48.30322. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu S, Tsujimoto Y. Proc Natl Acad Sci USA. 2000;97:577–582. doi: 10.1073/pnas.97.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang N M, Korth M J, Gale M, Wambach M, Der S D, Bandyopadhyay S K, Williams B R, Katze M G. Mol Cell Biol. 1999;19:4757–4765. doi: 10.1128/mcb.19.7.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reimold A M, Etkin A, Clauss I, Perkins A, Friend D S, Zhang J, Horton H F, Scott A, Orkin S H, Byrne M C, et al. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 55.Treister A, Sagi-Assif O, Meer M, Smorodinsky N I, Anavi R, Golan I, Meshel T, Kahana O, Eshel R, Katz B Z, et al. Int J Cancer. 1998;77:306–313. doi: 10.1002/(sici)1097-0215(19980717)77:2<306::aid-ijc22>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 56.Sabatino F, Masoro E J, McMahan C A, Kuhn R W. J Gerontol. 1991;46:B171–B179. doi: 10.1093/geronj/46.5.b171. [DOI] [PubMed] [Google Scholar]
- 57.Xu X, Sonntag W E. J Gerontol A Biol Sci Med Sci. 1996;51:B167–B174. doi: 10.1093/gerona/51a.2.b167. [DOI] [PubMed] [Google Scholar]
- 58.Stuchlikova E, Juricova-Horakova M, Deyl Z. Exp Gerontol. 1975;10:141–144. doi: 10.1016/0531-5565(75)90043-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.