Abstract
Target-specific treatment modalities are currently not available for triple-negative breast cancer (TNBC), and acquired chemotherapy resistance is a primary obstacle for the treatment of these tumors. Here we employed derivatives of BT-549 and MDA-MB-468 TNBC cell lines that were adapted to grow in the presence of either 5-Fluorouracil, Doxorubicin or Docetaxel in an aim to identify molecular pathways involved in the adaptation to drug-induced cell killing. All six drug-adapted BT-549 and MDA-MB-468 cell lines displayed cross resistance to chemotherapy and decreased apoptosis sensitivity. Expression of the anti-apoptotic co-chaperone BAG3 was notably enhanced in two thirds (4/6) of the six resistant lines simultaneously with higher expression of HSP70 in comparison to parental controls. Doxorubicin-resistant BT-549 (BT-549rDOX20) and 5-Fluorouracil-resistant MDA-MB-468 (MDA-MB-468r5-FU2000) cells were chosen for further analysis with the autophagy inhibitor Bafilomycin A1 and lentiviral depletion of ATG5, indicating that enhanced cytoprotective autophagy partially contributes to increased drug resistance and cell survival. Stable lentiviral BAG3 depletion was associated with a robust down-regulation of Mcl-1, Bcl-2 and Bcl-xL, restoration of drug-induced apoptosis and reduced cell adhesion in these cells, and these death-sensitizing effects could be mimicked with the BAG3/Hsp70 interaction inhibitor YM-1 and by KRIBB11, a selective transcriptional inhibitor of HSF-1. Furthermore, BAG3 depletion was able to revert the EMT-like transcriptional changes observed in BT-549rDOX20 and MDA-MB-468r5-FU2000 cells. In summary, genetic and pharmacological interference with BAG3 is capable to resensitize TNBC cells to treatment, underscoring its relevance for cell death resistance and as a target to overcome therapy resistance of breast cancer.
Abbreviations: TNBC, Triple negative breast cancer; DOX, Doxorubicin; DOC, Docetaxel; 5-FU, 5-Fluorouracil; Baf A1, Bafilomycin A1; STS, Staurosporine; BAG3, Bcl-2-associated athanogene 3; HSP70, Heat shock protein 70; HSF1, Heat shock factor 1; Par, Parental; Ctrl, Control; KD, Knockdown
Introduction
Despite advances in screening techniques leading to early detection of breast cancer, resistance to tumor therapy is still a major challenge in treatment of this disease, and recurrence rates are very high [1], [2]. Drug resistance is broadly classified into two types; 1) de novo (intrinsic) drug resistance in patients that do not respond to conventional therapies, and 2) acquired resistance in patients developed during treatment [3]. Intrinsic and acquired therapy resistances are major challenges for the successful treatment of patients, in particular those with triple-negative breast cancer (TNBC) [4]. TNBC is a subtype of epithelial breast cancer that doesn’t express estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) [5]. Only 15-20% of the total population of breast cancers is triple negative, but these are highly aggressive and metastatic. Due to the absence of specific therapeutic targets, treatment strategies against this tumor subtype are severely limited. As a consequence, current treatment of these tumors is restricted to chemotherapy, frequently leading to development of therapy resistance and recurrent disease [6]. Acquired drug resistance of tumor cells can be driven by a plethora of different mechanisms, like increased drug efflux, tumor cell heterogeneity, inactivation of apoptosis, increased DNA repair, angiogenesis, altered metabolism and stress-induced genetic or epigenetic alterations after drug exposure [3], [7], [8], [9], [10], [11]. Among these mechanisms, the adaptation of cancer cells to different cellular stress conditions (as induced by anti-cancer drugs) play a particularly important role for therapy resistance. A better understanding of the underlying resistance mechanisms are urgently required to identify new targets for treatment in an aim to improve clinical outcomes of TNBC.
Resistance to cell death caused by defects in apoptotic pathways and overexpression of anti-apoptotic proteins is a general hallmark of cancer [12], [13], [14]. Pro- and anti-apoptotic members of the Bcl-2 family are key regulators of apoptotic cell death. The Bcl-2 family proteins can be classified into three subfamilies: (i) the pro-apoptotic BH3-only proteins which have only one domain in common, the alpha helical BH3 domain; (ii) the pro-apoptotic Bax-like proteins which contain three such domains (BH1,2,3) and (iii) the anti-apoptotic Bcl-2-like proteins that contain 4 homology domains (BH1-4) and are regularly overexpressed in cancer. Bax and Bak trigger mitochondrial outer membrane permeabilization (MOMP) that is required for the release of pro-apoptotic factors from the mitochondria into the cytosol. This intrinsic apoptosis pathway is kept in check by the pro-survival Bcl-2 family members (Bcl-2, Bcl-xL, Mcl-1, Bcl-w and Bfl-1) [15], [16], [17].
The Hsp70 co-chaperone and anti-apoptotic protein BAG3 (also called Bis) is a member of the Bcl-2-associated anthanogene (BAG) protein family. This highly conserved family of co-chaperone interacts with the ATPase domain of heat shock protein 70 (HSP70) through a specific structural domain – the BAG domain [18]. BAG3 regulates several key hallmarks of cancer, including cell survival, cell adhesion, metastasis, angiogenesis and regulation of proteostasis [19], [20]. A key mechanism promoting its anti-apoptotic function is represented by BAG3-dependent stabilization of the pro-survival Bcl-2 family members including Bcl-2, Bcl-xL and Mcl-1, thereby supporting the anti-apoptotic function of these proteins [21], [22]. BAG3 expression has been reported to be elevated in various tumors including breast cancer, and we could previously show that estrogen receptor α (ERα) regulates a non-canonical type of autophagy that involves the function of BAG3 and provides stress resistance in ERα-expressing breast cancer cells [23].
Here we investigated the cellular mechanisms promoting enhanced chemotherapy resistance in TNBC cells adapted to growth in the presence of the clinically relevant chemotherapeutic agents 5-Flourouracil (5-FU), Doxorubicin (DOX) and Docetaxel (DOC). We demonstrate that increased apoptosis resistance is associated with enhanced cytoprotective autophagy, elevated expression of the oncogenic co-chaperone BAG3, BAG3-dependent stabilization of pro-survival Bcl-2 proteins and induction of EMT-like changes in gene expression. We also show that genetic and pharmacological interference with BAG3 function is capable to resensitize cells to apoptosis, underscoring the relevance of BAG3 as a target to overcome therapy resistance in TNBC.
Materials and Methods
Materials
Staurosporine (STS) was obtained from Alexis Biochemicals (San Diego, CA, USA). KRIBB11 ((N(2)-(1H-indazole-5-yl)-N(6)-methyl-3-nitropyridine-2,6-diamine)) was acquired from Calbiochem (Darmstadt, Germany). Bafilomycin A1 (Baf A1), YM-1 (2-((Z)-((E)-3-Ethyl-5-(3-methylbenzo[d]thiazol-2(3H)-ylidene)-4-oxothiazolidin-2-ylidene)methyl)-1-methylpyridin-1-ium chloride), p-HEMA (Poly (2-hydroxyethyl methacrylate)), Doxorubicin hydrochloride (DOX), Docetaxel (DOC), 5-Fluorouracil (5-FU), 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium (MTT), ABT-737 and all other chemicals or biochemicals were purchased from Sigma-Aldrich (Munich, Germany).
Cell lines and Culture
The triple negative parental human breast cancer cell lines BT-549 and MDA-MB-468 were obtained from ATCC/LGC Promochem GmbH (Wesel, Germany). The chemoresistant cell lines were established by continuous exposure to increasing concentrations of the respective drugs to the parental cell lines as previously described [24], [25] and derived from the Resistant Cancer Cell Line (RCCL) collection (www.kent.ac.uk/stms/cmp/RCCL/RCCLabout.html). 5-Fluorouracil (5-FU) resistant BT-549 and MDA-MB-468 sublines were cultured under continuous presence of 2000 ng/ml of 5-FU and named as BT-549r5-FU2000 and MDA-MB-468r5-FU2000 respectively, BT-549 and MDA-MB-468 cell lines were adapted to 40 ng/ml and 20 ng/ml of Docetaxel (DOC) and named as BT-549rDOC40 and MDA-MB-468rDOC20respectively, whereas Doxorubicin (DOX) resistant BT-549 and MDA-MB-468 cell lines were cultivated under the presence of 20 ng/ml and 200 ng/ml of DOX and named as BT-549rDOX20 and MDA-MB-468rDOX200 respectively. BT-549 parental and chemoresistant cells were cultured in Iscove modified Dulbecco’s medium (IMDM) supplemented with 10% Fetal Calf Serum (FCS), 4 mM L-glutamine, 10*2 IU/ml penicillin and 100 μg/ml streptomycin, whereas MDA-MB-468 parental and chemoresistant cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with F-12 Nutrient Mixture (Ham) supplemented with 10% FCS, 4 mM L-glutamine, 20 mM HEPES, 10*2 IU/ml penicillin and 100 μg/ml streptomycin (all: Gibco/Invitrogen, Karlsruhe, Germany) and cultures were maintained in a humidified 37°C and 5% CO2 incubator.
Lentiviral Transduction
Lentiviral vector stocks specific for BAG3 (SHCLNV-NM_ 004281; TRCN0000007294; Sigma-Aldrich) and ATG5 (SHCLNG-NM_004849; TRCN0000151474; Sigma-Aldrich) were used for transduction of both the parental and resistant cell lines of BT-549 and MDA-MB-468 breast cancer cells. Five different small hairpins sequences were included to set the target. (SHC002; Sigma-Aldrich), the pLKO.1-puro Non-Mammalian shRNA control plasmid DNA was used as a negative control. Transduction was executed as previously reported [26].
Cell Viability (MTT) Assay
3,000 cells suspended in 100 μl of medium were seeded per well in 96-well-plates and cultured for 24 h before onset of treatment. After completion of the treatment period, 20 μl of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) from 5 mg/ml of stock solution were added into 100 μl of medium in each well, followed by 3 h of incubation in a humidified 37°C and 5% CO2 incubator. Following 3 h of incubation, the medium containing MTT reagent was discarded, the formazan crystals formed after MTT treatment were solubilized by adding 100 μl of the mixture of isopropanol/1M HCl (24:1) and gently shaken for 30 min in dark condition. Then absorbance was measured at 560 nm using microplate reader (TECAN GENios, Crailsheim, Germany).
Flow Cytometry
For quantitative estimation of cell death, flow cytometry was performed as previously described [26].
SDS-PAGE and Western Blotting
SDS-PAGE and Western blotting were performed as recently reported [27]. After blocking in 5% BSA for 1 h at room temperature, the nitrocellulose membranes were incubated overnight at 4°C with primary antibodies directed against BAG3 (rabbit, 1:5000, Abnova, Heidelberg, Germany), LC3 (rabbit, 1:1000, Thermo Fisher Scientific, Rockford, USA), p62 (mouse, 1:1000, BD Biosciences, USA), GAPDH (mouse, 1:10,000, Calbiochem) and rest were against Bcl-2, Mcl-1, Bcl-xL, HSP70, HSF-1, Bak, Bax, ATG5, Beclin-1, pFAK (Tyr397), FAK, HSP60, E-cadherin and N-cadherin which were purchased from Cell Signaling Technology (Danvers, USA) raised in rabbit and used in the dilution of 1:1000. Following incubation, primary antibodies were detected by using respective secondary antibodies coupled with infrared dyes in green (800 CW) or red (680 RD) (IRDye goat anti-rabbit or anti-mouse from LICOR Biosciences, Bad Homburg, Germany diluted in 1:10,000 in 3% BSA for 1 h at room temperature and the signals were detected using the LI-COR Odyssey Infrared Imager (LI-COR Biosciences, Bad Homburg, Germany).
Confocal Microscopy
Cells seeded on chamber slide were fixed with paraformaldehyde (4% PFA) after completion of the respective treatments and permeabilized with 0.1% Triton X-100. For assessment of Cellular morphology, cells were stained with Texas Red-X phalloidin (ThermoFisher Scientific, Darmstadt, Germany). DAPI (Applichem, Darmstadt, Germany) was used for nuclear staining in all cases. After mounting on microscope slides, cells were finally analyzed using Nikon C1i confocal microscope.
Quantitative Real-Time PCR
Total RNA isolation and quantitative real-time PCR (qPCR) was executed as previously described [28]. The StepOnePlus™ Real-Time PCR System (Life Technologies, Darmstadt, Germany) was used for the detection of fluorescence signal above the threshold (Ct) value followed by normalization of fluorescence intensity of the samples were performed to the amplification value of the control gene TATA box binding protein (TBP). All the primers for qPCR were purchased from Applied Biosystems (Darmstadt, Germany). Following primers were used for qPCR: TaqMan® Gene Expression Assay primer for BAG3(Hs00188713_m1), CDH1 (Hs01023895_m1), CDH2 (Hs00169953_m1), SNAI1 (Hs00195591_m1), SNAI2(Hs00161904_m1), TWIST1(Hs01675818_S1), TWIST2 (Hs02379973_s1).
poly-HEMA Coating for Suspension Cultures
poly-HEMA (p-HEMA) solution was prepared by suspending 1mg of p-HEMA powder in 1 ml of 95% of ethanol followed by thorough mixing at 50°C and 750 rpm in Thermomixer comfort (Eppendorf) for overnight. Then the homogenized solution was filtered and pipetted into 24 well plates and allowed to dry inside the safety hood for two days with opened lids. Before the cells were seeded, the wells were properly washed with sterile PBS and finally the cells were allowed to remain in suspension condition throughout the experiment.
Invasion (Boyden-Chamber) Assay
Before seeding the cells into the chamber, the matrigel (Corning transwell insert with 8 μm pore, Corning, Tewksbury, MA) was rehydrated with 500 μl of FCS-free medium for 2 hours inside the humidified 37°C and 5% CO2 incubator. Then approximately 750 μl of medium containing 10% FCS was added into the wells of the 24-well cell culture insert companion plate. After discarding the rehydration medium, the transwell inserts were put into the wells of the cell cultured insert companion plate carefully by avoiding air bubble. Then approximately 20,000 cells suspended in 500 μl medium containing 2% FCS were seeded into the matrigel coated insert. After 20 hours of incubation, the cells on the top of the inserts were removed by using Q-tips and the cells attached to the bottom of the inserts were fixed with methanol for 2 minutes followed by staining for another 2 minutes with 0.1% crystal violate. The chambers were washed three four times in purified water and allowed to air dry. Then 6 pictures of each matrigel were captured with a Nikon TS100 inverted microscope equipped with a charge-coupled device (CCD) camera followed by counting of stained cells by using ImageJ software. Three matrigel chambers were used for each condition and each experiment was repeated 3 times.
Migration (Scratch) Assay
400,000 cells were seeded per well in 6 well cultured plates. After 24 hours of incubation, 10 μg/ml of Mitomycin C (Sigma-Aldrich) was added for 2 hours to prevent cell proliferation. Then a scratch was made using 200 μl pipette tip followed by washing with PBS. Pictures of the scratch at 0 hour time point were taken and the same positions were captured after 20 hours of incubation. The number of cells migrated into the scratch area were counted using ImageJ software. Each experiment was repeated for 3 times.
Subcellular Fractionation
The digitonin-based subcellular fractionation technique was employed for the separation of cytosolic and mitochondrial fractions as previously described [29].
Global Proteomics Analysis
For global proteomic analysis, the cells were grown until ~70% confluency. For each cell line, three independent samples were taken. Samples for LC-MS/MS analysis were prepared according to Kulak et al. with minor modifications [30]. In brief, cell lysis and protein denaturation were performed by boiling the samples in 6 M GnHCl, 100 mM Tris pH 8.5, 10 mM TCEP and 40 mM ChlAA. Proteins were digested with Lys-C for 3 hours, followed by tryptic digestion overnight. Tryptic peptides were desalted and concentrated using STAGE-Tips (Empore C18, 3M).
Peptides were separated on an easy nLC 1200 (ThermoFisher Scientific) and a 15 cm long, 75μm ID fused-silica column, which has been packed in house with 1.9 μm C18 particles (Dr. Maisch), and kept at 45°C using an integrated column oven (Sonation). Peptides were eluted by a non-linear gradient from 4% to 48% acetonitrile over 135 minutes and directly sprayed into a QExactive HF mass-spectrometer equipped with a nanoFlex ion source (ThermoFisher Scientific). Precursor ions were analyzed with a resolution of 60,000 and the 15 most abundant ions were subjected to HCD fragmentation and resulting fragments were analyzed with a resolution of 15,000. Single charged ions and ions with unassigned charge states were not taken into account for fragmentation and dynamic exclusion was set to 20 s.
Data analysis was done with MaxQuant and essentially default settings [31]. Fragment spectra were searched against the Uniprot human reference proteome (version “December 2017”), with a false discovery rate of 1% on PSM and protein level and at least one unique peptide. Fold changes were determined by LFQ quantification with the “match between runs”-option being activated [32]. Statistical significant changes between parental and resistant cell-lines were determined with Perseus using a Two-sample t-test with a permutation based FDR of 5% and a s0 of 0.6 on log2 transformed LFQ values [33].
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD008522 [34]. ProteomeXchange provides globally co-ordinate proteomics data submission and dissemination [34].
Statistics
Data are represented as means ± SEM. For statistical analysis, t-test (two-tailed) was applied by using (GraphPad Prism, GraphPad Software, Inc., La Jolla, CA, USA). p<0.05 was considered to be statistically significant and was denoted with asterisks or hashtags.
Results
BAG3 is Overexpressed in Chemoresistant Breast Cancer Cells
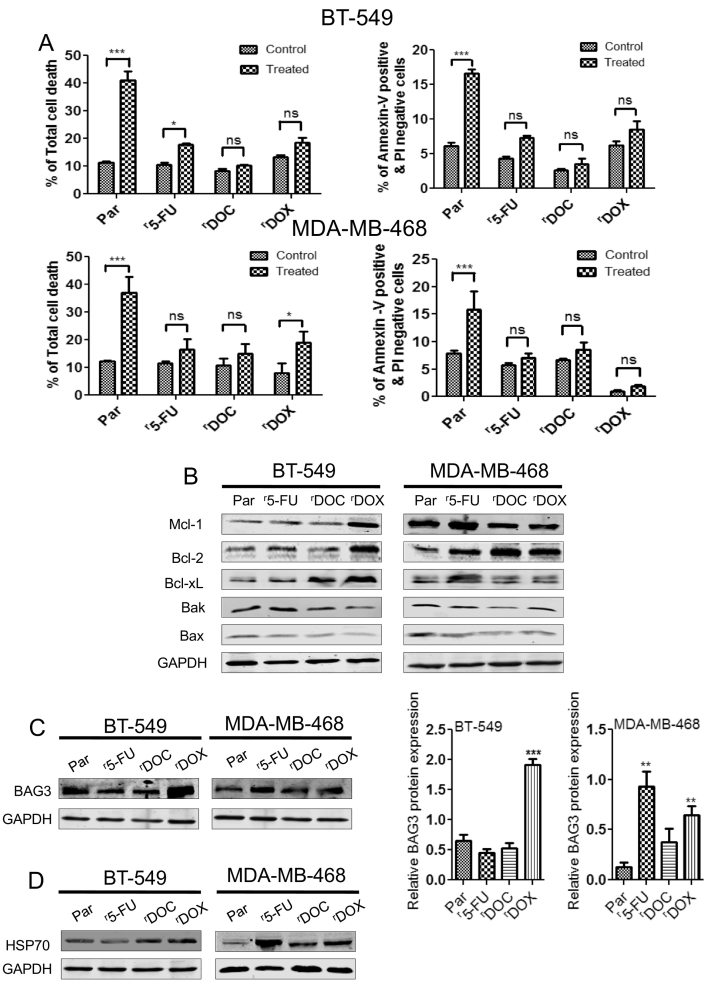
In our study, the triple negative breast cancer cell line BT-549 was adapted to growth in medium containing either 2000 ng/ml of 5-FU, 40 ng/ml of DOC or 20 ng/ml of DOX; the MDA-MB-468 cell line was similarly adapted to growth in the presence of 2000 ng/ml of 5-FU, 20 ng/ml of DOC or 200 ng/ml of DOX. The 50% inhibitory concentration (IC50) value of each drug was obtained by MTT assays. Table 1 depicts the IC50 value of each drug and the level of cross resistance of the BT-549Par (parental control cells), BT-549r5-FU2000 (adapted to 5-fluorouracil), BT-549rDOC40 (adapted to Docetaxel), BT-549rDOX20 (adapted to Doxorubicin), MDA-MB-468Par (parental control cells), MDA-MB-468r5-FU2000 (adapted to 5-fluorouracil), MDA-MB-468rDOC20 (adapted to Docetaxel), and MDA-MB-468rDOX200 (adapted to Doxorubicin) cell lines to the chemotherapeutic drugs 5-FU, DOC and DOX. All drug-adapted cell lines displayed increased resistance to the other two chemotherapeutic agents at a varying degree. Further we used staurosporine (STS), a well-recognized apoptotic cell death inducer to investigate the possible changes in the general sensitivity to apoptosis in chemoresistant cell lines and analyzed early apoptosis and total cell death by FACS-based Annexin V/PI-staining. FACS data (Figure 1A) revealed that all drug-adapted BT-549 and MDA-MB-468 cell lines are significantly more resistant to STS compared to their parental counterparts, indicating decreased apoptosis sensitivity. Expression analysis of anti-apoptotic and pro-apoptotic Bcl-2 family members by western blot unveiled a prominent increase of Bcl-2, Bcl-xL and Mcl-1 proteins in BT-549rDOX20 and MDA-MB-468r5-FU2000 cells whereas Bak and Bax expression is decreased in almost all the resistant cells compared to their parental controls (Figure 1B). Since we found an increased expression of anti-apoptotic Bcl-2 family proteins in chemoresistant BT-549rDOX20 and MDA-MB-468r5-FU2000 cells, we also treated these cells with the selective BH3-mimetic ABT-737 to determine whether the drug resistant cells are sensitive to inhibition of Bcl-2 and Bcl-xL. Interestingly, our cell viability assay indicates that both the resistant lines are sensitive to ABT-737, with an increased sensitivity of MDA-MB-468r5-FU2000 cells in comparison to control cells (Figure S1). Further, we investigated the expression levels of HSP70 and BAG3, a HSP70 co-chaperone in all the parental and chemoresistant cells. Western blot analysis revealed a pronounced increase of BAG3 and HSP70 in most of the chemoresistant cell lines, with BT-549rDOX20 (Figure 1C) and MDA-MB-468r5-FU2000 (Figure 1D) cells showing the highest expression compared to their parental control cells, respectively.
Table 1.
Establishment of Breast Cancer Cellular Resistant Models
| IC50 ± SEM | BT-549 |
|||
|---|---|---|---|---|
| Par | r5-FU | rDOC | rDOX | |
| 5-Fluorouracil (μg/ml) | 0.63 ± 0.21 | 5.89 ± 0.47 | 4.24 ± 1.39 | 3.37 ± 0.13 |
| Docetaxel (ng/ml) | 8.12 ± 0.84 | 12.82 ± 1.67 | 212.56 ± 1.20 | 37.09 ± 2.18 |
| Doxorubicin (ng/ml) | 9.46 ± 1.62 | 15.97 ± 1.26 | 180.6 ± 2.09 | 84.38 ± 1.92 |
| IC50 ± SEM | MDA-MB-468 |
|||
|---|---|---|---|---|
| Par | r5-FU | rDOC | rDOX | |
| 5-Fluorouracil (μg/ml) | 1.16 ± 0.14 | 18.97 ± 0.29 | 5.97 ± 0.07 | 4.28 ± 0.83 |
| Docetaxel (ng/ml) | 9.74 ± 1.02 | 12. 62 ± 0.86 | 63.07 ± 1.93 | 13.47 ± 0.32 |
| Doxorubicin (ng/ml) | 116 ± 1.69 | 129.20 ± 1.95 | 338.47 ± 1.40 | 536.47 ± 1.79 |
Breast cancer cell line BT-549 was adapted to grow isolatedly with 2000 ng/ml of 5-FU, 40 ng/ml of DOC and 20 ng/ml of DOX; MDA-MB-468 cell line was grown similarly with 2000 ng/ml of 5-FU, 20 ng/ml of DOC and 200 ng/ml of DOX. Then 50% inhibitory concentration (IC50) value of each drug was determined by MTT assay. The values are means of three independent experiments performed in triplicate ± SEM. p values<0.05 were considered to be statistically significant.
Figure 1.
Overexpression of BAG3, HSP70 and other anti-apoptotic proteins and down-regulation of pro-apoptotic proteins in chemoresistant breast cancer cells.
(A) BT-549 and MDA-MB-468 parental and chemoresistant cell lines were treated with 3 μM of apoptotic cell death inducer staurosporine (STS) and DMSO (0.1%) as control for 6 hours followed by % of total cell death and % of Annexin-V positive & PI negative cells were estimated by flow cytometry. Data are representative of three independent experiments performed in triplicate. Columns represent means ± SEM. Statistical significance; *p<0.05, ** p<0.01, *** p<0.001 and ns not significant compared to controls (DMSO). (B) Western blot analysis shows anti-apoptotic proteins (Bcl-2, Bcl-xL, and Mcl-1) are over expressed and pro-apoptotic proteins (Bax and Bak) are down regulated in chemoresistant cell lines compared to the parental counterparts. (C) BAG3 and (D) HSP70 are highly expressed in various chemoresistant cell lines. GAPDH was used as loading control. Densitometric analysis of relative BAG3 protein expression was performed in BT-549 and MDA-MB-468 parental and chemoresistant cell lines. Columns represent means ± SEM. Statistical significance; ** p<0.01, *** p<0.001 compared to parental control cells.
Attenuation of Autophagy Increases the Sensitivity of Chemoresistant Breast Cancer Cells
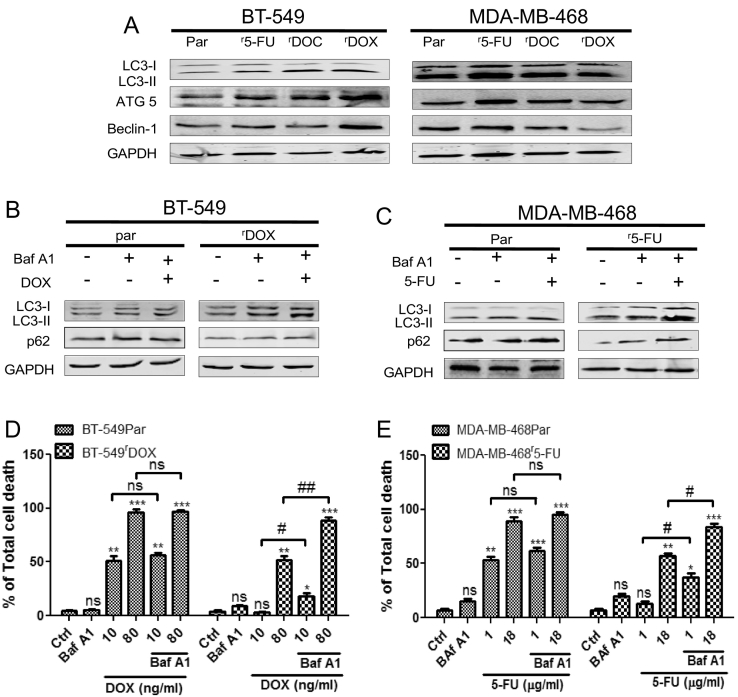
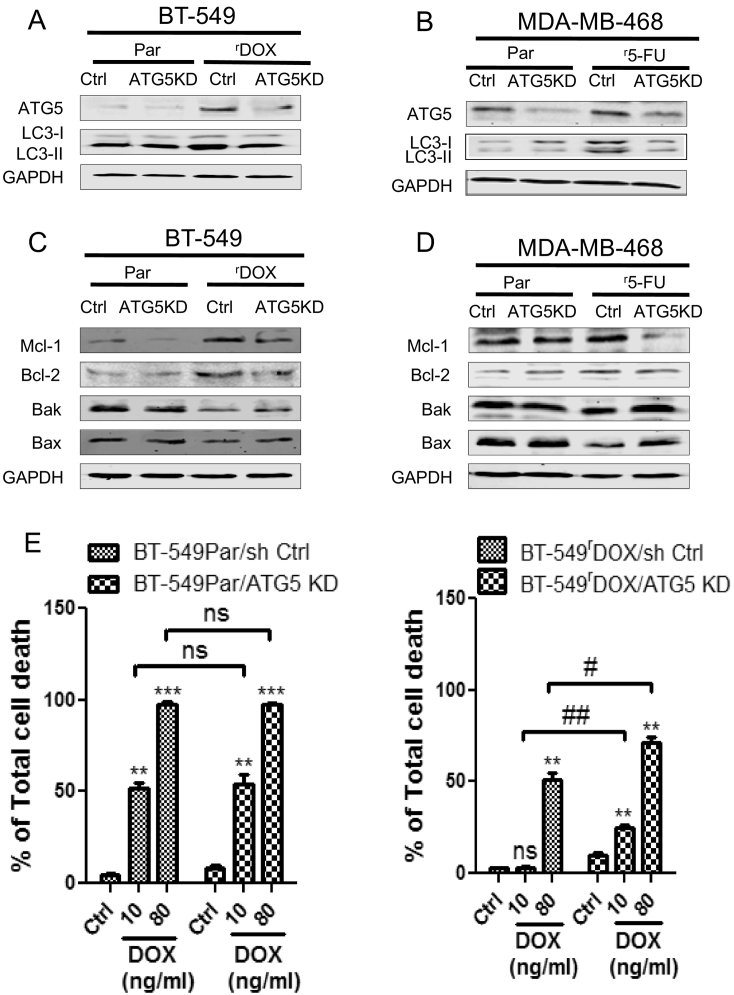
One of the potential mechanisms contributing to cell survival in chemoresistance is increased autophagy [35]. To further investigate this possibility in our cell models, we explored the basal autophagy status in all the parental and chemoresistant cells as a starting point. Most of the chemoresistant cells exhibited slightly enhanced levels of autophagy marker proteins like LC3-II, ATG5 and Beclin-1 compared to their parental controls, with the most pronounced changes observed in BT-549rDOX20 and MDA-MB-468r5-FU2000 cells (Figure 2A) that were chosen for a subsequent, more detailed analysis. Western blot analysis revealed a shift from LC3-I to LC3-II and accumulation of p62 after combined treatment with Bafilomycin A1 and the respective drugs, i.e. DOX in BT-549Par and BT-549rDOX20 cells (Figure 2B), and 5-FU in MDA-MB-468Par and MDA-MB-468r5-FU2000 cells (Figure 2C). The LC3 shift and accumulation of p62 appeared to be more prevalent in the chemoresistant cells, suggesting that the autophagic flux may be increased in these cells compared to parental controls. The effect of autophagy is highly context-dependent and it can promote either cell survival or cell death [36]. In order to evaluate the death-modulating role of autophagy in our chemoresistant cells, a combined treatment of Bafilomycin A1 and the chemotherapeutic drugs was performed. BT-549 cells were treated with 10 ng/ml of DOX (approximate IC50 of BT-549Par) and 80 ng/ml of DOX (approximate IC50 of BT-549rDOX20), MDA-MB-468 cells were treated with 1 μg/ml of 5-FU (approximate IC50 of MDA-MB-468Par), and 18 μg/ml of 5-FU (approximate IC50 of MDA-MB-468r5-FU2000) (see Table 1). Then total cell death was assessed by FACS analysis. FACS data revealed a significant increase of total cell death in BT-549rDOX20 (Figure 2D) and MDA-MB-468r5-FU2000 cells (Figure 2E) after combined treatment with chemotherapeutics and Bafilomycin A1, whereas the amount of cell death in parental cells remained largely unaltered. We also genetically inhibited autophagy by a stable lentiviral ATG5 knockdown (ATG5 KD). ATG5 is a key molecule in the early stage of autophagosome formation [37] and depletion of ATG5 is an efficient way to interfere with induction of macroautophagy. Stable ATG5 knockdowns were established both in the BT-549par and BT-549rDOX20 cells (Figure 3A), as well as in MDA-MB-468Par and MDA-MB-468r5-FU2000 cells (Figure 3B). Knockdown of ATG5 decreased the LC3-II level in BT-549rDOX20 (Figure 3A) and MDA-MB-468r5-FU2000 (Figure 3B) cells. Further, we found a decrease in the expression of Mcl-1 and Bcl-2 proteins following knockdown of ATG5 in BT-549rDOX20cells (Figure 3C) and also in MDA-MB-468r5-FU2000 (Figure 3D) cells. For assessment of cell death after ATG5 KD, we performed a dose escalation experiment with DOX for 72 h in the BT-549Par/sh Ctrl, BT-549Par/ATG5 KD, BT-549rDOX20/sh Ctrl and BT-549rDOX20/ATG5 KD cells followed by FACS analysis of total cell death. FACS data revealed a minor, but significant increase of total cell death in the BT-549rDOX20/ATG5 KD cells compared to BT-549rDOX20/sh Ctrl, whereas the amount of cell death in BT-549par/ATG5 KD compared to BT-549par/sh Ctrl remained unaltered (Figure 3E). These observations confirm our previous results obtained with pharmacological inhibition of autophagy by Bafilomycin A1. Collectively, we conclude that increased autophagy in chemoresistant cells acts in a cytoprotective manner and may partially contribute to enhanced therapy resistance.
Figure 2.
Induction of prosurvival autophagy in chemoresistant cells and pharmacological inhibition of autophagy augments the sensitivity of chemoresistant breast cancer cells
(A) Basal level of autophagy was increased in chemoresistant cells as LC3-II, ATG5 and Beclin-1 proteins expression were increased in chemoresistant cells compared to their respective parental cells in western blot analysis. (B) Autophagic flux was determined by the accumulation of LC3-II and p62 after combined treatment of DOX (10 ng/ml) for 72 h and autophagic flux inhibitor Baf A1 (25 nM) for 4 h in both the BT-549Par and BT-549rDOX20 cells and (C) similarly in MDA-MB-468Par and MDA-MB-468r5-FU2000 cell lines after combined treatment of 5-FU (1 μg/ml) for 72 h and Baf A1 (25 nM) for 4 h. GAPDH was used as loading control. (D) Breast cancer cell lines BT-549Par and BT-549rDOX20 were treated with 10 ng/ml and 80 ng/ml of DOX for 72 h with or without Baf A1 (25 nM for 4 h), (E) similarly MDA-MB-468Par and MDA-MB-468r5-FU2000 cell lines were treated with 1 μg/ml and 18 μg/ml of 5-FU for 72 h with or without Baf A1 (25 nM for 4 h). Total cell death was quantified by Annexin V/PI double staining followed by flow cytometry. Columns represent means of three independent experiments performed in triplicate ± SEM. Statistical significance: * p<0.05, ** p<0.01, *** p<0.001 and ns not significant compared to respective controls (Ctrls); # p<0.05, ## p<0.01 and ns not significant with Baf A1 treatment compared to without Baf A1 treatment.
Figure 3.
Knockdown of ATG5 sensitizes chemoresistant breast cancer cells
(A) Stable lentiviral knockdowns of ATG5 were established in both the BT-549Par and BT-549rDOX20 cells and (B) also in the MDA-MB-468Par and MDA-MB-468r5-FU2000 cells. Stable lentiviral transduction of control vector was used as control (Ctrl). Knockdowns were confirmed by western blot. Knockdown of ATG5 reduced LC3-II protein expression in BT-549rDOX20 and MDA-MB-468r5-FU2000 cells. GAPDH was used as loading control in western blot. (C) Down regulation of ATG5 reduces the protein expression of Mcl-1 and other anti-apoptotic proteins, whereas augments the expression of pro-apoptotic proteins likes Bak and Bax in BT-549rDOX20and (D) MDA-MB-468r5-FU2000 cells respectively. GAPDH served as loading control in western blot. (E) BT-549rDOX20/ATG5 KD cell exhibited significantly higher levels of total cell death compared to the BT-549Par cells after 72 hours of DOX treatment in a dose dependent manner. 0.1% DMSO for 72 h was used as control. Cell death was determined by Annexin V/PI double staining followed by flow cytometry. Data are means ± SEM of three different experiments each performed in triplicate. * p<0.05 ** p<0.01, *** p<0.001 and ns not significant compared to respective controls (Ctrls); # p<0.05, ## p<0.01 and ns not significant of ATG5 KD compared to respective sh Ctrls.
Lentiviral Depletion of BAG3 Resensitizes Chemoresistant Breast Cancer Cells to Therapy
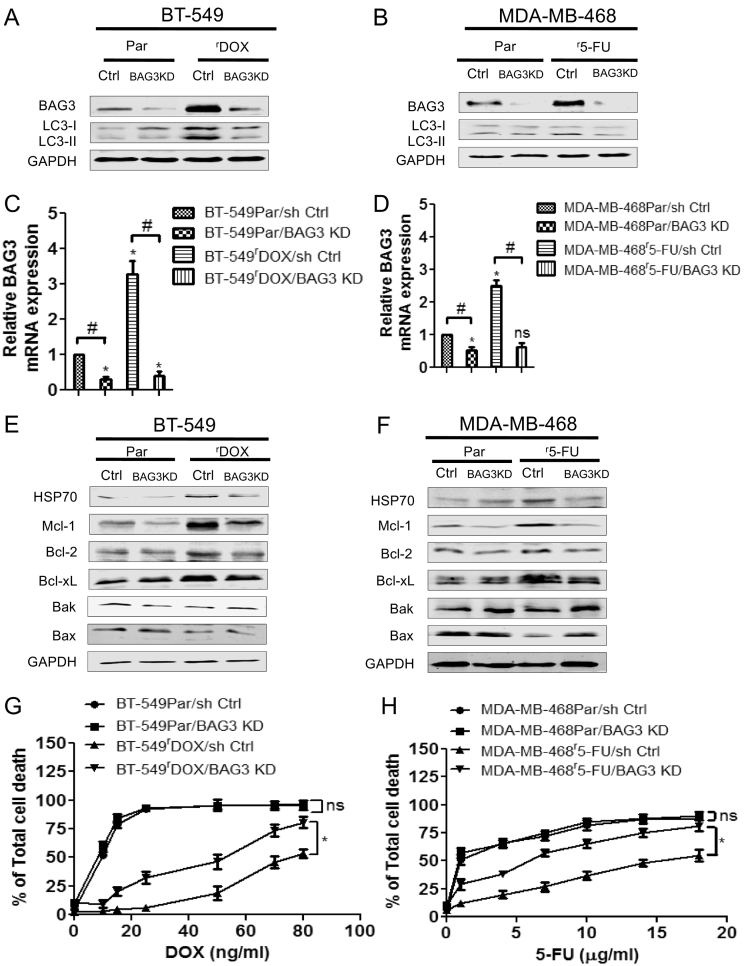
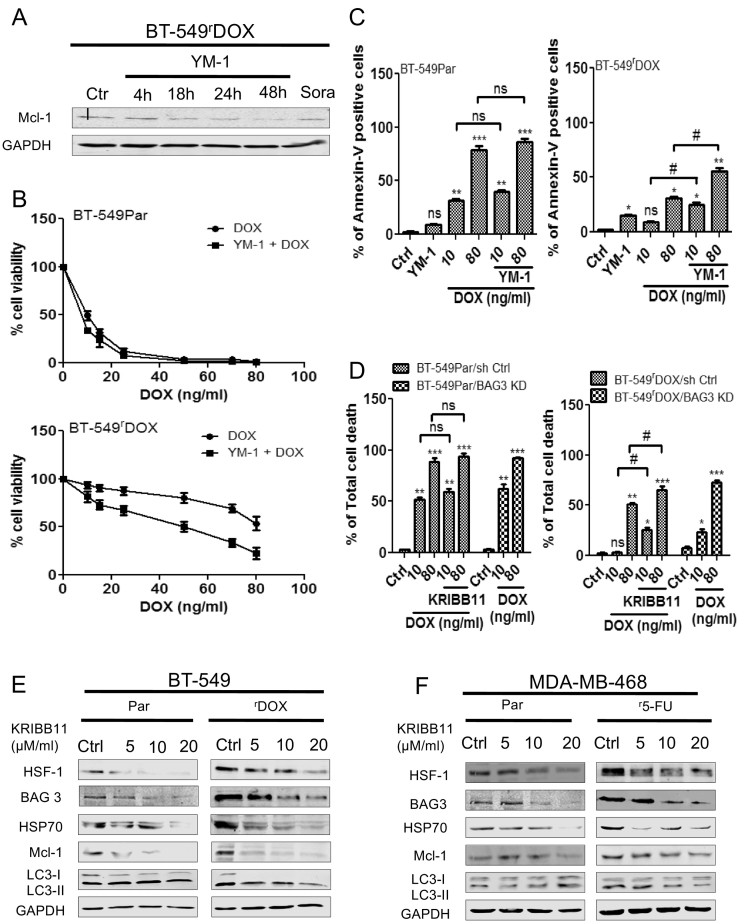
So far we had observed that the higher expression of BAG3 protein is associated with simultaneously increased basal levels of autophagy and enhanced apoptosis resistance in BT-549rDOX20 and MDA-MB-468r5-FU2000 cells compared to their parental counterparts. BAG3 has key functions in regulation of both autophagy and apoptosis. It maintains cellular homeostasis by selective degradation of damaged proteins after recruiting the macroautophagic pathway [38]. Importantly, it also antagonizes activation of the intrinsic apoptosis pathway through stabilization of Bcl-2 proteins [22]. To further delineate the role of BAG3 as a resistance factor in chemoresistant cells, we established stable lentiviral BAG3 knockdowns in the BT-549Par and BT-549rDOX20 cells (Figure 4A), and in MDA-MB-468Par and MDA-MB-468r5-FU2000 cells (Figure 4B), confirmed by western blot and qPCR in BT-549Par and BT-549rDOX20 (Figure 4C), as well as MDA-MB-468Par and MDA-MB-468r5-FU2000 (Figure 4D) cells. The level of LC3-II was significantly decreased in the BT-549rDOX20/BAG3 KD cells (Figure 4A) and a similar result was also obtained in MDA-MB-468r5-FU2000/BAG3 KD cells (Figure 4B). Our western blot data indicates that HSP70and the anti-apoptotic proteins Mcl-1, Bcl-2 and Bcl-xL are decreased in BT-549rDOX20/BAG3 KD cells (Figure 4E) and MDA-MB-468r5-FU2000/BAG3 KD cells (Figure 4F) to levels comparable to those observed in parental control cells. These data suggest that interference with BAG3 is able to effectively counteract the chemoresistance-associated increase of Mcl-1, Bcl-2 and Bcl-xL expression. It was previously proposed that BAG3 may also sequester pro-apoptotic BAX in the cytoplasm and serve to inhibit its mitochondrial translocation in glioblastoma cells [21]. Consistent with this hypothesis, we found that mitochondrial BAX was increased in BAG3-depleted MDA-MB-468r5-FU2000cells (Figure S2). To analyze whether knockdown of BAG3 KD also reactivates the sensitivity of the chemoresistant cells to the respective drugs, we performed cell death assays following DOX treatment in BT-549 cell lines and 5-FU treatment in MDA-MB-468 cell lines, respectively. Indeed, our FACS data reveal that both BT-549rDOX20/BAG3 KD (Figure 4G) and MDA-MB-468r5-FU2000/BAG3 KD (Figure 4H) cells are significantly resensitized to the respective drugs, which was not observed in parental BAG3 KD cells compared to their controls.
Figure 4.
Depletion of BAG3 sensitizes the chemoresistant breast cancer cells by attenuating protective autophagy
(A) Stable lentiviral knockdowns of BAG3 were established in both the BT-549-Par and BT-549rDOX20 cells and (B) also in the MDA-MB-468Par and MDA-MB-468r5-FU2000 cells. Stable lentiviral transduction of empty vector was used as control (Ctrl). Knockdowns were confirmed by western blot. Knockdown of BAG3 reduced LC3-II protein expression in BT-549rDOX20 and MDA-MB-468r5-FU2000 cells. GAPDH served as loading control in western blot. (C) Relative BAG3 mRNA expression was also determined by quantitative PCR (qPCR) after stable lentiviral knockdowns of BAG3 in BT-549Par and BT-549rDOX20 cell lines and (D) similarly in MDA-MB-468 cells respectively. qPCR data represent means of three independent experiments± SEM (n = 3). Significant BAG3 mRNA expression compared to parental sh Ctrls are marked by asterisks: * p<0.05, ** p<0.01 and ns not significant. Significant differences between BAG3 KD and respective sh Ctrls are denoted by hashtags: # p<0.05, ##p<0.01 and ns not significant. (E) Knockdown of BAG3 reduced the protein expression of HSP70, Mcl-1 and other anti-apoptotic proteins whereas augments the expression of pro-apoptotic proteins like Bak and Bax in BT-549rDOX20 and (F) MDA-MB-468r5-FU2000 cells respectively. GAPDH served as loading control. (G) BT-549rDOX20/BAG3 KD and (H) MDA-MB-468r5-FU2000/BAG3 KD cells exhibited significantly higher levels of total cell death compared to their parental counterparts after 72 hours of DOX and 5-FU treatment in a dose dependent manner respectively. 0.1% DMSO for 72 h was used as control. Cell death was determined by Annexin V/PI double staining followed by flow cytometry. Data represent means of three independent experiments ± SEM (n = 3). *p<0.05 and ns not significant of BAG3 KD compared to respective sh Ctrls.
Drug Resensitization by Pharmacological Interference with the HSF1/HSP70/BAG3 Pathway
It was recently reported that BAG3 acts as a critical molecule in HSP70-modulated cancer signaling. Thus, targeting the interaction of these two molecules may represent a promising therapeutic strategy [39]. For selective disruption of the HSP70/BAG3 complex, we used YM-1 according to Colvin et al. [39]. YM-1 treatment of BT-549rDOX20 cells dramatically reduced Mcl-1 levels in a time-dependent manner (Figure 5A). Combined treatment with YM-1 and DOX applied at different concentrations synergized in limiting cell viability in BT-549rDOX20 cells, but not in the parental BT-549 cells, as confirmed by MTT assays (Figure 5B). Furthermore, combined treatment of YM-1 and DOX significantly increased cell death in BT-549rDOX20 cells, as detected by Annexin-V staining followed by FACS analysis (Figure 5C). Please note that in this experiment, PI staining could not be performed due to the autofluorescence properties of YM-1. The stress induced transcription factor heat shock factor 1 (HSF-1) acts upstream of HSP70 and its co-chaperone BAG3. Since BAG3/HSP70 interaction stabilizes Mcl-1, inhibition of HSF-1 should lead to rapid degradation of Mcl-1 [22]. To further decipher the importance of the HSF-1/HSP70/BAG3 pathway for cell death resistance, the HSF-1 inhibitor KRIBB11 was used in comparison to the experiments previously executed with knockdown of BAG3. The combined treatment of KRIBB11 and DOX significantly increased apoptosis in BT-549rDOX20/sh Ctrl cells; with overall cell death reaching a very similar extent in comparison to BT-549rDOX20/BAG3 KD cells treated with DOX (Figure 5D). The expression of BAG3, HSP70, and Mcl-1 and LC3-II proteins gradually decreased in BT-549rDOX20 (Figure 5E) and MDA-MB-468r5-FU2000 cells (Figure 5F) in a dose-dependent manner after 48 h treatment of KRIBB11.
Figure 5.
HSP70/BAG3 interaction inhibitor YM-1 and HSF-1 inhibitor KRIBB11 sensitize the chemoresistant breast cancer cells
(A) Dissociation of the HSP70/BAG3 complex by YM-1 (5 μM) decreased Mcl-1 expression in BT-549rDOX20 cells after 4 h, 18 h, 24 h, 48 h treatment in western blot analysis. 0.1% DMSO for 48 h was used as Control (Ctrl) for the solvent, whereas Sorafenib (Sora) 5 μM for 48 h was used as positive control. GAPDH was used as loading control in western blot. (B) % of cell viability was analyzed by MTT assay in BT-549Par and BT-549rDOX20 cells after 2 h pre-treatment of 5 μM of YM-1 for 48 h followed by DOX for 72 h. DMSO 0.1 % for 48 h was used as control (Ctrl) for the solvent. (C) Breast cancer cell lines BT-549Par and BT-549rDOX20 were treated with 10 ng/ml and 80 ng/ml of DOX for 72 h with or without YM-1 (5 μM, 48 h). 0.1 % DMSO for 48 h was used as control (Ctrl) for the solvent. Then cell death was analyzed by Annexin-V positive staining in flow cytometry. Columns represent means of three independent experiments ± SEM (n = 3). Statistical significance: * p<0.05, ** p<0.01, *** p<0.001 and ns not significant compared to ctrls (0.1 % DMSO); # p<0.05, ## p<0.01 and ns not significant with combined treatment of YM-1 and DOX compared to DOX treatment alone. (D) Combined treatment of KRIBB11 (20 μM, 48 h) and DOX significantly augment the total cell death in BT-549rDOX20/sh Ctrl cells compared to DOX treatment alone. 0.1 % DMSO for 48 h was used as control (Ctrl) for the solvent. Cell death was determined by Annexin V/PI double staining followed by flow cytometry. Columns represent means of three independent experiments ± SEM (n = 3). Statistical significance: * p<0.05, ** p<0.01, *** p<0.001 and ns not significant compared to ctrls (0.1 % DMSO); # p<0.05, ## p<0.01 and ns not significant with combined treatment of KRIBB11 and DOX compared to DOX treatment alone. (E) KRIBB11 treatment decreased BAG3, HSP70, Mcl-1 and LC3 II in a dose dependent manner in BT-549rDOX20 and (F) MDA-MB-468r5-FU2000 cells respectively. Cells were treated with 5 μM, 10 μM and 20 μM of KRIBB11 for 48 h. DMSO (0.1 %, 48 h) was used as control (Ctrl). GAPDH served as loading control for western blot.
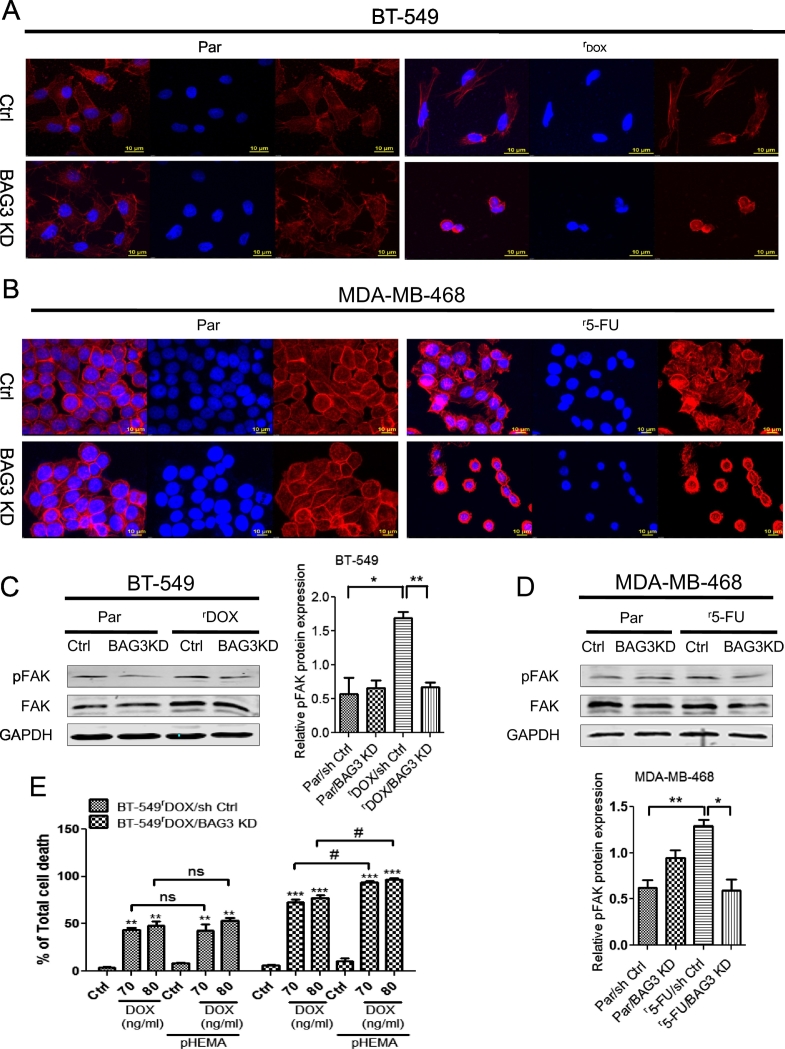
BAG3 Depletion Alters Cell Matrix Adhesion in Chemoresistant Breast Cancer Cells
It was previously reported that BAG3 significantly contributed to maintenance of the cell adhesion properties [40]. To explore the effect of BAG3 knockdown on cell adhesion properties of our drug-resistant cells, we performed F-actin staining by using Texas Red-X phalloidin followed by confocal microscopy. Confocal imaging revealed strong alterations in cell morphology in BT-549rDOX20/BAG3 KD cells (Figure 6A), while no obvious morphological changes were found in the BT-549Par/BAG3 KD cells. BT-549rDOX20/BAG3 depleted cells were more rounded and loosely attached, whereas the BAG3-proficient cells were more flattened, bigger in size and tightly attached. Similar results were also obtained in MDA-MB-468r5-FU2000/BAG3 KD cells (Figure 6B). Western blot analysis revealed that the increase in FAK phosphorylation at Tyr397 residue in BT-549rDOX20cells (Figure 6C) and MDA-MB-468r5-FU2000 cells (Figure 6D) was reduced after BAG3 knockdown. To further assess the sensitivity of the BT-549rDOX20/BAG3 depleted cells to DOX after matrix detachment, cell adhesion was hindered by coating the cell culture plates with p-HEMA, so that all cells were cultured in suspension. Cell death was enhanced in suspension cultures of BAG3-depleted BT-549rDOX20 cells after 72 h DOX treatment whereas no significant changes of cell death were noticed in the BAG3 proficient BT-549rDOX20 cells in comparison to controls (Figure 6E).
Figure 6.
BAG3 depletion alters cell matrix adhesion in chemoresistant breast cancer cells
(A) Knockdown of BAG3 altered cellular morphology and actin cytoskeletal distribution in BT-549rDOX20 and (B) MDA-MB-468r5-FU2000 cells compared to their respective parental counterparts in confocal imaging. For staining of F-actin, Texas Red-X phalloidin was used whereas nuclei were stained with DAPI. Scale bar 10 μm. (C) FAK phosphorylation was reduced in BT-549rDOX20/BAG3 KD and (D) MDA-MB-468r5-FU2000/BAG3 KD cells respectively. GAPDH served as loading control in the western blot. Densitometric analysis of relative pFAK protein expression was performed in control and BAG3 KD of both BT-549 and MDA-MB-468 parental and chemoresistant cell lines. Columns represent means ± SEM. Statistical significance; * p<0.05, ** p<0.01, *** p<0.001 compared to controls (E) BT-549rDOX20/BAG3 KD cells cultured in suspension exhibited more sensitivity to DOX treatment in a dose dependent manner. Cell cultured dishes were coated with pHEMA to prevent cell adhesion. Water (0.1%, 72 h) was used as control (Ctrl) for the solvent. Cell death was determined by Annexin V/PI double staining followed by flow cytometry. Columns represent means of three independent experiments performed in triplicate ± SEM. Statistical significance: * p<0.05, ** p<0.01, *** p<0.001 and ns not significant compared to ctrls (0.1 % water); # p<0.05 and ns not significant with combined treatment of DOX and pHEMA compared to DOX treatment alone.
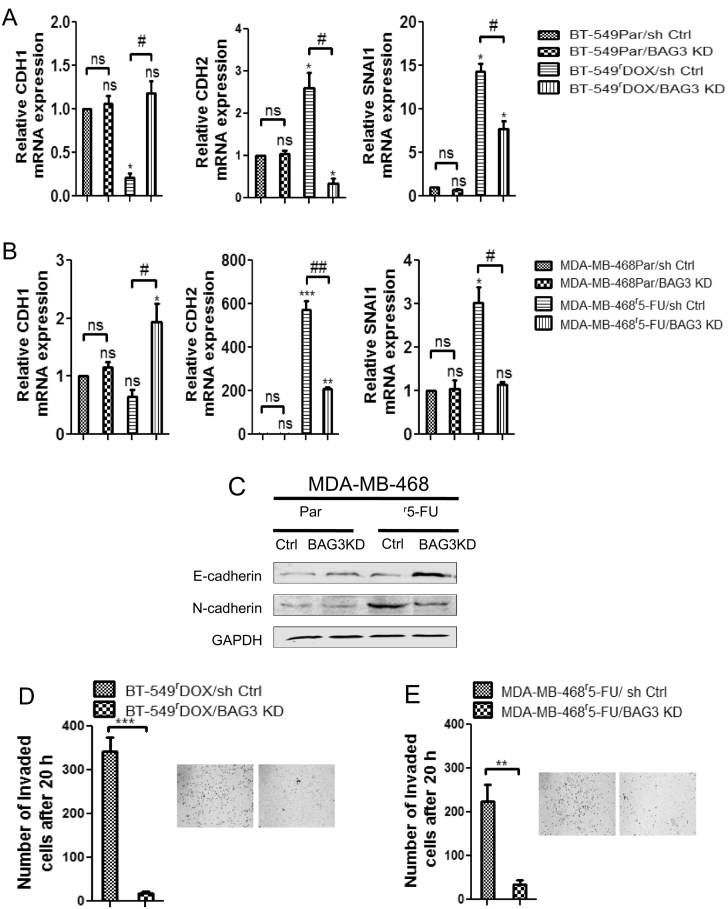
Depletion of BAG3 Reverses EMT-Associated Expression Changes in Chemoresistant Breast Cancer Cells
Recently the role of BAG3in controlling epithelial-mesenchymal transition (EMT) has come to attention of the scientific community [19]. To further investigate this phenomenon in chemoresistant cells, we performed qPCR to analyze the relative mRNA expression levels of several key genes involved in EMT. qPCR data revealed that CDH1 (encoding E-cadherin) mRNA expression was strongly increased whereas CDH2 (encoding N-cadherin) and SNAI1 (encoding Snail) expression was significantly decreased in BAG3-depleted BT-549rDOX20 cells (Figure 7A) and MDA-MB-468r5-FU2000 cells (Figure 7B), consistent with the hypothesis that BAG3 promotes EMT. We also found reduced mRNA expression of genes encoding the transcription factors SNAI2 (Snail2), TWIST1 and TWIST2 in BAG3-depleted BT-549rDOX20 (Figure S3A) and similarly in MDA-MB-468r5-FU2000cells (Figure S3B). We did not find any significant changes in the mRNA expression of the above genes in the parental BAG3-depleted cells compared to their controls, except the increase of TWIST1 mRNA expression in BAG3-depleted parental MDA-MB-468 cells (Figure S3B). To further validate our qPCR data, we performed western blot analysis revealing a strong up-regulation of E-cadherin and down-regulation of N-cadherin in BAG3-depleted MDA-MB-468r5-FU2000 cells (Figure 7C) which corroborated our qPCR data. To investigate whether depletion of BAG3 has any impact on cell invasion and migration properties of the chemoresistant cells, we performed Boyden-chamber assays and scratch assays. BAG3-depleted BT-549rDOX20 cells exhibited significantly reduced invasiveness compared to the BAG3-proficient BT-549rDOX20 cells after 20 h of incubation (Figure 7D), and similar results were also obtained in MDA-MB-468r5-FU2000/BAG3 KD cells (Figure 7E). Our scratch assays also revealed a significantly reduced motility of BAG3-depleted BT-549rDOX20 cells (Figure S4A) and MDA-MB-468r5-FU2000 cells (Figure S4B) after 20 h incubation compared to their controls.
Figure 7.
Knockdown of BAG3 reverses the EMT phenomena and simultaneously suppresses invasion in chemoresistant breast cancer cells
(A) Knockdown of BAG3 reduced the relative CDH2, SNAI1 mRNA expression, simultaneously increased CDH1 mRNA expression in BT-549rDOX20/BAG3 KD and (B) MDA-MB-468r5-FU2000/BAG3 KD cells in qPCR respectively. qPCR data represent means of three independent experiments ± SEM (n = 3). Significant mRNA expression compared to parental sh Ctrls are marked by asterisks: * p<0.05, ** p<0.01 and ns not significant. Significant differences between BAG3 KD and respective sh Ctrls are denoted by hashtags: # p<0.05, ## p<0.01 and ns not significant. (C) Expression of E-cadherin protein was increased whereas N-cadherin expression was decreased in MDA-MB-468r5-FU2000/BAG3 KD cells in western blot. GAPDH served as loading control. (D) Number of invaded cells was decreased in BT-549rDOX20/BAG3 KD and (E) MDA-MB-468r5-FU2000/BAG3 KD cells. Invasion assay was performed for 20 h followed by stained with 1% crystal violet, bright field image was taken and invaded cells were counted by using ImageJ software. Columns represent means of three independent experiments ± SEM (n = 3). Statistical significance of invasion: ** p<0.01, *** p<0.001 and ns not significant with BAG3 KD compared to sh Ctrls.
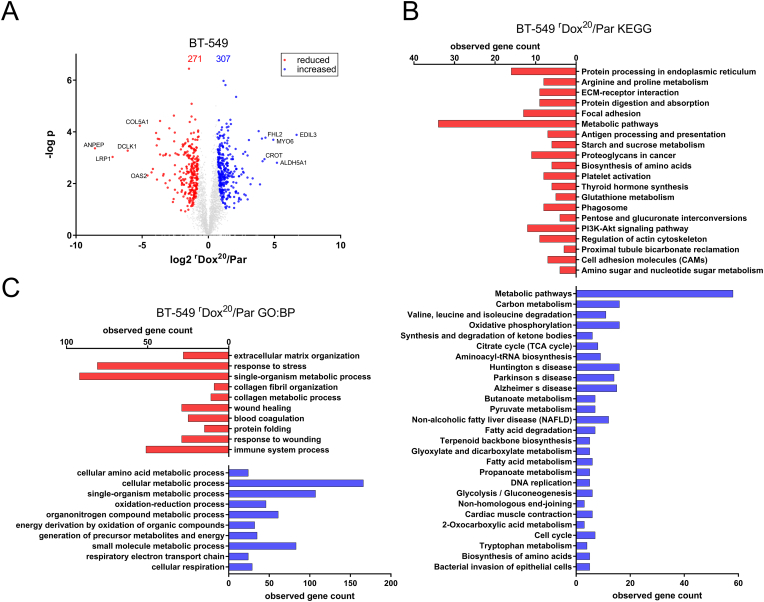
Proteomic Analysis Reveals a Gene Signature Associated with Tumor Aggressiveness
The development of drug resistance is a very complex mechanism including alterations of many different processes and proteins. Thus, we performed global proteomic analysis of BT-549rDOX20 compared to BT-549Par using label-free quantification. This analysis showed that out of the 4906 reproducibly quantified proteins groups, 271 and 307 were significantly under- and overrepresented in BT-549rDOX20 compared to BT-549Par cells (Figure 8A), respectively. The 5 most increased and decreased proteins are also depicted in Figure 8A, the complete list of significantly changed proteins is provided as a supplemental file (Table S1). Bioinformatics analysis using the freely available STRING-platform (string-db.org); [41] was carried to analyze Gene Ontology (GO) terms that were significantly enriched among the reduced or increased proteins (Figure 8A). In total 20 and 27 KEGG pathways were significantly overrepresented among reduced and increased protein groups respectively (Figure 8B). Reduced pathways include focal adhesion and cell adhesion molecules (CAMs). Increased pathways include citrate cycle (TCA) and oxidative phosphorylation, two metabolic pathways that are correlated with tumor aggressiveness [42], but also DNA replication and cell cycle, indicating increased proliferation. The analyses of biological processes (GO:BP; Figure 8C) revealed that among the top 10 processes which were overrepresented among the reduced proteins, extracellular matrix organization (GO.0030198) and response to stress (GO.0006950) are the highest ranking processes. Notably, other processes whose associated proteins were significantly enriched are related to regulation of apoptosis (e.g.: regulation of extrinsic apoptotic signaling pathway (GO.2001236) and regulation of apoptotic signaling pathway (GO.2001233)) and cell adhesion (e.g.: positive regulation of cell adhesion (GO.0045785) and cell-cell adhesion mediated by integrin (GO.0033631)). On the other hand many increased processes are related to an enhanced metabolism, including cellular amino acid metabolic process (GO.0006520), cellular metabolic process (GO.0044237) and also cellular respiration (GO.0045333). Other processes are related to an increased glucose and ATP metabolism (e.g.: gluconeogenesis (GO.0006094), ATP metabolic process (GO.0046034)), replication (DNA strand elongation involved in DNA replication (GO.0006271)). Basically, each of the 10 most increased and decreased single proteins found have previously been related to tumorigenesis or tumor progression (Table 2), thus exemplifying that treatment resistance is accompanied by a more aggressive phenotype.
Figure 8.
Proteomic analysis reveals a gene signature associated with tumor aggressiveness in BT-549rDOX20 cells
(A) Volcano-Plot showing the protein ratios (in log2) as a function of the –log p-value of label-free quantification proteomic-data from BT-549rDOX20 cells compared to BT-459Par. A total of 4906 proteins were quantified, of those 271 (red dots) and 307 (blue dots) were significantly reduced and increased, respectively. (B and C) Bioinformatic cluster analysis using String (string-db.org; [41]) was performed. (B) Shows all KEGG pathways that are overrepresented among reduced (red) or increased protein groups (blue). (C) Shows the 10 highest ranking Gene Ontology (GO) biological processes (GO:BP) that are overrepresented among proteins with significantly reduced (red) or increased amount (blue).
Table 2.
The 10 Highest Ranking Proteins are Listed Along with the NCBI Gene ID, the Log2 Ratio of BT-549rDOX20 Over BT-549Par and the –Log P Value, as Well as a Brief Summary of the Known Function of These Proteins in Cancer
| Official gene name | Gene-ID | log2 ratio | –log p value | Function |
|---|---|---|---|---|
| ANPEP | 290 | -8.567 | 3.364 | Silenced in prostate cancer [53] |
| LRP1 | 4035 | -7.233 | 3.030 | Cancer cell survival and metastasis [54] |
| DCLK1 | 9201 | -6.092 | 3.274 | Associated with favorable prognosis in breast cancer [55] |
| COL5A1 | 1289 | -5.178 | 4.235 | Marker gene for TNBC classification [56] |
| OAS2 | 4939 | -4.594 | 2.313 | Higher expression in relapsed tumors [57] |
| STEAP3 | 55240 | -4.294 | 2.433 | Part of a metastasis gene signature [58] |
| PLA2G4A | 5321 | -4.190 | 2.565 | Associated with adverse prognosis [59] |
| ITPR1 | 3708 | -3.948 | 3.958 | Breast cancer susceptibility locus [60],[61] |
| SERPINB5 | 5268 | -3.900 | 3.477 | Tumor suppressor in breast cancer [62] |
| DTX3L | 151636 | -3.881 | 2.156 | Associated with overall survival and autophagy signature [63] |
| Official gene name | Gene-ID | log2 ratio | –log p value | Known relevance for breast cancer |
|---|---|---|---|---|
| EDIL3 | 10085 | 6.668 | 3.885 | Cell invasion and metastasis [64] |
| ALDH5A1 | 7915 | 5.188 | 2.805 | Stem-like phenotype [65]; detoxifaction [66] |
| MYO6 | 4646 | 4.908 | 3.694 | Oncogenic in breast cancer [67] |
| FHL2 | 2274 | 4.317 | 3.774 | Oncogenesis and tumor progression [68], [69] |
| CROT | 54677 | 4.242 | 2.945 | Correlates with drug resistance [70] |
| LMCD1 | 29995 | 4.101 | 2.864 | Involved in tumor recurrence [71] |
| GNG11 | 2791 | 4.049 | 3.740 | Cell migration and metastasis [72] |
| KYNU | 8942 | 3.908 | 1.968 | Metastasis [73] |
| CUX1 | 1523 | 3.804 | 4.027 | Migration, invasion and apoptosis-resistance [74], [75] |
| TAGLN | 6876 | 3.349 | 2.201 | Epithelial mesenchymal transition [76] |
Discussion
Here we demonstrate that overexpression of BAG3 is associated with chemotherapy resistance of triple negative BT-549 and MDA-MB-468 breast cancer cells that were adapted to growth in the presence of the clinically applied chemotherapeutic agents 5-FU, DOX and DOC. Furthermore, we provide evidence that the increased apoptosis resistance of these cells is associated with BAG3-dependent stabilization of the pro-survival Bcl-2 family proteins Bcl-2, Bcl-xL and Mcl-1, induction of EMT-like transcriptional changes and enhanced cytoprotective autophagy that partially contributes to increased cell survival. In line with our findings, overexpression of BAG3 has been reported in several types of human cancers, such as glioblastoma [21], lung carcinoma [43], pancreatic carcinoma [44], leukemia [45], and thyroid carcinoma [46], compared with very low basal levels of BAG3 in non-malignant cells [43]. Consistent with the hypothesis that BAG3 may represent a key resistance factor in breast cancer; a very recent study demonstrated that enhanced BAG3 expression correlates with poor patient survival in this tumor entity [47].
All drug-adapted BT-549 and MDA-MB-468 cell lines displayed cross resistance to chemotherapy and the apoptosis inducer STS, indicating an increase in general apoptosis resistance. BAG3 expression was visibly elevated in all three resistant MDA-MB-468 lines and one of the BT-549 lines. To further address the role of BAG3 in cell death resistance, we performed stable lentiviral BAG3 depletion in the BT-549rDOX20 and MDA-MB-468r5-FU2000 cell lines. Interestingly, BAG3 depletion led to a robust down-regulation of Mcl-1, Bcl-2 and Bcl-xL and apoptosis was efficiently restored exemplifying its crucial role in apoptosis resistance.
The levels of the autophagy markers LC3-II, ATG5 and Beclin-1 were also found to be elevated in BT-549rDOX20 and MDA-MB-468r5-FU2000 cells. In our chemoresistant cell models, expression of both Bcl-2 and Beclin-1 was elevated simultaneously, suggesting that interaction of Bcl-2 and Beclin-1 may have no significant effect on the regulation of autophagy in these cells. This notion is consistent with previously reported findings pointing to an indirect effect of Bcl-2 on autophagy that is mediated by inhibiting Bak and Bax [48]. Since pro-survival autophagy is a key mechanism underlying drug resistance of cancer cells in many paradigms [35], we investigated the potential contribution of cytoprotective autophagy in chemotherapy resistance. To this end, we applied the pharmacological autophagy inhibitor Baf A1 and established stable lentiviral knockdowns of ATG5 to block the non-selective macroautophagy pathway. In our experiments, treatment with Baf A1 and knockdown of ATG5 significantly increased apoptotic cell death after treatment with DOX and 5-FU in BT-549rDOX20 and MDA-MB-468r5-FU2000 resistant cells respectively, but not in parental cells, indicating that enhanced autophagy may indeed partially contribute to drug resistance.
In a similar fashion, lentiviral knockdown of BAG3 significantly resensitized both resistant cell lines to drug treatment, indicating that BAG3 overexpression is involved in acquired drug resistance. Interestingly, BAG3 depletion was associated with decreased levels of both LC3-I and LC3-II, indicating that BAG3 overexpression may contribute to enhanced autophagy in the resistant setting. Of note, BAG3 was previously shown to regulate total LC3 levels at the translational level, possibly leading to enhanced basal autophagy [49]. We also found that depletion of BAG3 in chemoresistant cells facilitates the translocation of BAX to mitochondria suggesting that BAG3-dependent sequestration of BAX in the cytoplasmic compartment [21] may contribute to the anti-apoptotic effects of BAG3 in breast cancer chemoresistant cells.
In order to overcome the proposed BAG3-driven resistance, we also pharmacologically targeted the BAG3/HSP70/Mcl-1 signaling axis. The BAG3 gene is a transcriptional target of the stress-induced transcription factor HSF1. KRIBB11 was found to selectively inhibit the transcriptional activity of HSF1 [50], which is required for expression of both BAG3 and its interactor HSP70. Indeed, we could observe significant synergistic effects on cell death in the combination therapy with KRIBB11 and DOX in the BT-549rDOX20 cell line. To further scrutinize these pro-apoptotic effects of BAG3 inhibition, we employed the specific HSP70/BAG3 small molecule inhibitor YM-1 that prevents formation and function of the HSP70-BAG3 module [39]. Inhibition of BAG3 by YM-1 significantly decreased the protein levels of Mcl-1 and was able to mimic the sensitizing effect of BAG3 depletion on apoptosis after combined treatment with DOX in BT-549rDOX20 cells. These data underscore the pivotal role of the HSP70 interaction in promoting the anti-apoptotic function of BAG3 and highlight the relevance of this complex as a therapeutic target in breast cancer.
Global proteomic analysis of BT-549rDOX20 revealed major changes in several pathways/biological processes implicated in tumorigenesis and tumor progression, including cell adhesion. Besides its anti-apoptotic function, BAG3 was also suggested to support cell adhesion and motility [40], and to mediate resistance to anoikis, a special form of apoptosis induced by matrix detachment [27]. The ability of BAG3 to regulate cell adhesion was proposed to rely on multiple interactors (e.g. PDZGEF2 and CCN1) of BAG3 through different structural domains in this context [51]. Moreover, BAG3 was implicated in cytoskeleton organization by regulating actin folding via interaction with CCT, a cytosolic chaperonin [52]. The important role of BAG3 in cytoskeleton organization was also observed in this study, as demonstrated by gross re-arrangements of the cytoskeletal structure in BAG3-depleted cells that was visualized by actin staining followed by confocal microscopy. In particular, BAG3 depletion was accompanied with a more rounded and loosely attached cellular morphology. A role of BAG3 in controlling epithelial-mesenchymal transition (EMT) was recently proposed [19]. Indeed, we observed an EMT-like expressional shift in chemotherapy-resistant cells compared to parental controls in our cell models. This shift was associated with increased expression of CDH2 (N-cadherin), SNAI1 (Snail1), SNAI2 (Snail2), TWIST1 and TWIST2 and decreased expression of CDH1 (E-cadherin). Consistent with the proposed function of BAG3 in EMT modulation, BAG3 depletion induced a back shift in the expression pattern of these genes to a more epithelial like state and also reduced the invasiveness and motility in the resistant cell models.
Our findings obtained in established cell lines and their drug-resistant derivatives represent a first step in elucidating specific mechanisms of acquired therapy resistance. These data open the avenue for follow-up investigations in patient-derived cultures and in vivo models. Collectively, we demonstrate that BAG3 plays a major role for the cell death resistance of breast cancer cells and their response to chemotherapy, as well as their aggressive growth characteristics. Based on the findings of our study, we propose that by stabilizing anti-apoptotic Bcl-2 family members and promoting EMT-like changes, the HSF1/HSP70/BAG3 pathway may play a pivotal role for therapy resistance of breast cancer. Pharmacological intervention with BAG3 and HSP70 function is an interesting approach for combined therapies aimed at synergistically inducing apoptosis in advanced breast cancer and may aid the design of new strategies aimed at overcoming the resistance to current breast cancer therapy.
The following are the supplementary data related to this article.
Complete list of significantly changed proteins in global proteomics analysis
Chemoresistant breast cancer cells are more sensitive to the selective BH3-mimetic ABT-737. Cell viability was analyzed by MTT assay in (A) BT-549Par and BT-549rDOX20 cells and (B) MDA-MB-468Par and MDA-MB-468r5-FU2000 cells after 48 h treatment with ABT-737. DMSO (0.1 %, for 48 h) was used as control (Ctrl) for the solvent.
Depletion of BAG3 increases translocation of BAX to mitochondria in chemoresistant breast cancer cells. Cells were harvested and the mitochondrial and cytosolic fractions were separated by digitonin-based subcellular fractionation in control and BAG3 KD MDA-MB-468r5-FU2000 cells followed by western blot for detection of BAX. GAPDH and HSP60 served as loading controls for the cytosolic and mitochondrial fractions, respectively.
Knockdown of BAG3 reduces the mRNA expression of SNAI2, TWIST1, TWIST2 in BT-549rDOX20/BAG3 KD and MDA-MB-468r5-FU2000/BAG3 KD cells. (A) Knockdown of BAG3 reduced the relative SNAI1, TWIST1, TWIST2 mRNA expression in BT-549rDOX20/BAG3 KD and (B) MDA-MB-468r5-FU2000/BAG3 KD cells in qPCR respectively. qPCR data represent means of three independent experiments± SEM (n = 3). Significant mRNA expression compared to parental sh Ctrls are marked by asterisks: *P < .05 and ns not significant. Significant differences between BAG3 KD and respective sh Ctrls are denoted by hashtags: #P < .05 and ns not significant.
Depletion of BAG3 reduces the migration of breast cancer chemoresistant cells. (A) Number of migrated cells was decreased in BT-549rDOX20/BAG3 KD and (B) MDA-MB-468r5-FU2000/BAG3 KD cells. Migration assay was performed for 20 h followed by bright field image was taken in x40, scale bar 200 μm and migrated cells were counted by using ImageJ software. Columns represent means of three independent experiments ± SEM (n = 3). Statistical significance of migration: *P < .05, ***P < .001 and ns not significant with BAG3 KD compared to sh Ctrls.
Acknowledgments
Acknowledgements
We would like to thank Gabriele Köpf and Hildegard König for outstanding technical support.
Funding Sources
We would like to thank German Academic Exchange Service (DAAD), Germany and Department of Science and Technology (DST-INSPIRE- IF130677), Govt. of India for providing scholarships. This study was supported by the Deutsche Forschungsgemeinschaft (SFB 1177 on selective autophagy).
Conflict of Interest
The authors declare no conflict of interest
References
- 1.Martin HL, Smith L, Tomlinson DC. Multidrug-resistant breast cancer: current perspectives. Breast Cancer (Dove Med Press) 2014;6:1–13. doi: 10.2147/BCTT.S37638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yardley DA. Drug resistance and the role of combination chemotherapy in improving patient outcomes. Int J Breast Cancer. 2013;2013:137414. doi: 10.1155/2013/137414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 4.O'Reilly EA, Gubbins L, Sharma S, Tully R, Guang MH, Weiner-Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell M. The fate of chemoresistance in triple negative breast cancer (TNBC) BBA Clin. 2015;3:257–275. doi: 10.1016/j.bbacli.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, Sledge GW, Carey LA. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8018. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- 6.Guestini F, McNamara KM, Ishida T, Sasano H. Triple negative breast cancer chemosensitivity and chemoresistance: current advances in biomarkers indentification. Expert Opin Ther Targets. 2016;20:705–720. doi: 10.1517/14728222.2016.1125469. [DOI] [PubMed] [Google Scholar]
- 7.Coley HM. Mechanisms and strategies to overcome chemotherapy resistance in metastatic breast cancer. Cancer Treat Rev. 2008;34:378–390. doi: 10.1016/j.ctrv.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Luqmani YA. Mechanisms of drug resistance in cancer chemotherapy. Med Princ Pract. 2005;14(Suppl. 1):35–48. doi: 10.1159/000086183. [DOI] [PubMed] [Google Scholar]
- 9.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 10.Gillet JP, Gottesman MM. Mechanisms of multidrug resistance in cancer. Methods Mol Biol. 2010;596:47–76. doi: 10.1007/978-1-60761-416-6_4. [DOI] [PubMed] [Google Scholar]
- 11.Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, Sarkar S. Drug resistance in cancer: an overview. Cancers. 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 15.Delbridge AR, Strasser A. The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ. 2015;22:1071–1080. doi: 10.1038/cdd.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick JM, Soane L. Multiple functions of BCL-2 family proteins. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng JH, Viacava Follis A, Kriwacki RW, Moldoveanu T. Discoveries and controversies in BCL-2 protein-mediated apoptosis. FEBS J. 2016;283:2690–2700. doi: 10.1111/febs.13527. [DOI] [PubMed] [Google Scholar]
- 18.Rosati A, Graziano V, De Laurenzi V, Pascale M, Turco MC. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2:e141. doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao H, Cheng S, Tong R, Lv Z, Ding C, Du C, Xie H, Zhou L, Wu J, Zheng S. BAG3 regulates epithelial-mesenchymal transition and angiogenesis in human hepatocellular carcinoma. Lab Investig. 2014;94:252–261. doi: 10.1038/labinvest.2013.151. [DOI] [PubMed] [Google Scholar]
- 20.Sturner E, Behl C. The role of the multifunctional BAG3 protein in cellular protein quality control and in disease. Front Mol Neurosci. 2017;10:177–194. doi: 10.3389/fnmol.2017.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Festa M, Del Valle L, Khalili K, Franco R, Scognamiglio G, Graziano V, De Laurenzi V, Turco MC, Rosati A. BAG3 protein is overexpressed in human glioblastoma and is a potential target for therapy. Am J Pathol. 2011;178:2504–2512. doi: 10.1016/j.ajpath.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boiani M, Daniel C, Liu X, Hogarty MD, Marnett LJ. The stress protein BAG3 stabilizes Mcl-1 protein and promotes survival of cancer cells and resistance to antagonist ABT-737. J Biol Chem. 2013;288:6980–6990. doi: 10.1074/jbc.M112.414177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felzen V, Hiebel C, Koziollek-Drechsler I, Reissig S, Wolfrum U, Kogel D, Brandts C, Behl C, Morawe T. Estrogen receptor alpha regulates non-canonical autophagy that provides stress resistance to neuroblastoma and breast cancer cells and involves BAG3 function. Cell Death Dis. 2015;6:e1812. doi: 10.1038/cddis.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaelis M, Agha B, Rothweiler F, Loschmann N, Voges Y, Mittelbronn M, Starzetz T, Harter PN, Abhari BA, Fulda S. Identification of flubendazole as potential anti-neuroblastoma compound in a large cell line screen. Sci Rep. 2015;5:8202–8210. doi: 10.1038/srep08202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaelis M, Rothweiler F, Barth S, Cinatl J, van Rikxoort M, Loschmann N, Voges Y, Breitling R, von Deimling A, Rodel F. Adaptation of cancer cells from different entities to the MDM2 inhibitor nutlin-3 results in the emergence of p53-mutated multi-drug-resistant cancer cells. Cell Death Dis. 2011;2:e243. doi: 10.1038/cddis.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonietti P, Gessler F, Dussmann H, Reimertz C, Mittelbronn M, Prehn JH, Kogel D. AT-101 simultaneously triggers apoptosis and a cytoprotective type of autophagy irrespective of expression levels and the subcellular localization of Bcl-xL and Bcl-2 in MCF7 cells. Biochim Biophys Acta. 2016;1863:499–509. doi: 10.1016/j.bbamcr.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Antonietti P, Linder B, Hehlgans S, Mildenberger IC, Burger MC, Fulda S, Steinbach JP, Gessler F, Rodel F, Mittelbronn M. Interference with the HSF1/HSP70/BAG3 Pathway Primes Glioma Cells to Matrix Detachment and BH3 Mimetic-Induced Apoptosis. Mol Cancer Ther. 2017;16:156–168. doi: 10.1158/1535-7163.MCT-16-0262. [DOI] [PubMed] [Google Scholar]
- 28.Mohrenz IV, Antonietti P, Pusch S, Capper D, Balss J, Voigt S, Weissert S, Mukrowsky A, Frank J, Senft C. Isocitrate dehydrogenase 1 mutant R132H sensitizes glioma cells to BCNU-induced oxidative stress and cell death. Apoptosis. 2013;18:1416–1425. doi: 10.1007/s10495-013-0877-8. [DOI] [PubMed] [Google Scholar]
- 29.Adrain C, Creagh EM, Martin SJ. Apoptosis-associated release of Smac/DIABLO from mitochondria requires active caspases and is blocked by Bcl-2. EMBO J. 2001;20:6627–6636. doi: 10.1093/emboj/20.23.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 31.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 32.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 34.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fulda S, Kogel D. Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene. 2015;34:5105–5113. doi: 10.1038/onc.2014.458. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colvin TA, Gabai VL, Gong J, Calderwood SK, Li H, Gummuluru S, Matchuk ON, Smirnova SG, Orlova NV, Zamulaeva IA. Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 2014;74:4731–4740. doi: 10.1158/0008-5472.CAN-14-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwasaki M, Homma S, Hishiya A, Dolezal SJ, Reed JC, Takayama S. BAG3 regulates motility and adhesion of epithelial cancer cells. Cancer Res. 2007;67:10252–10259. doi: 10.1158/0008-5472.CAN-07-0618. [DOI] [PubMed] [Google Scholar]
- 41.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solaini G, Sgarbi G, Baracca A. Oxidative phosphorylation in cancer cells. Biochim Biophys Acta. 2011;1807:534–542. doi: 10.1016/j.bbabio.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Chiappetta G, Basile A, Barbieri A, Falco A, Rosati A, Festa M, Pasquinelli R, Califano D, Palma G, Costanzo R. The anti-apoptotic BAG3 protein is expressed in lung carcinomas and regulates small cell lung carcinoma (SCLC) tumor growth. Oncotarget. 2014;5:6846–6853. doi: 10.18632/oncotarget.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Q, Ozawa F, Friess H, Zimmermann A, Takayama S, Reed JC, Kleeff J, Buchler MW. The anti-apoptotic protein BAG-3 is overexpressed in pancreatic cancer and induced by heat stress in pancreatic cancer cell lines. FEBS Lett. 2001;503:151–157. doi: 10.1016/s0014-5793(01)02728-4. [DOI] [PubMed] [Google Scholar]
- 45.Zhu H, Wu W, Fu Y, Shen W, Miao K, Hong M, Xu W, Young KH, Liu P, Li J. Overexpressed BAG3 is a potential therapeutic target in chronic lymphocytic leukemia. Ann Hematol. 2014;93:425–435. doi: 10.1007/s00277-013-1883-1. [DOI] [PubMed] [Google Scholar]
- 46.Chiappetta G, Ammirante M, Basile A, Rosati A, Festa M, Monaco M, Vuttariello E, Pasquinelli R, Arra C, Zerilli M. The antiapoptotic protein BAG3 is expressed in thyroid carcinomas and modulates apoptosis mediated by tumor necrosis factor-related apoptosis-inducing ligand. J Clin Endocrinol Metab. 2007;92:1159–1163. doi: 10.1210/jc.2006-1712. [DOI] [PubMed] [Google Scholar]
- 47.Liu BQ, Zhang S, Li S, An MX, Li C, Yan J, Wang JM, Wang HQ. BAG3 promotes stem cell-like phenotype in breast cancer by upregulation of CXCR4 via interaction with its transcript. Cell Death Dis. 2017;8:e2933. doi: 10.1038/cddis.2017.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindqvist LM, Heinlein M, Huang DC, Vaux DL. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc Natl Acad Sci U S A. 2014;111:8512–8517. doi: 10.1073/pnas.1406425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez AE, Lopez-Crisosto C, Pena-Oyarzun D, Salas D, Parra V, Quiroga C, Morawe T, Chiong M, Behl C, Lavandero S. BAG3 regulates total MAP1LC3B protein levels through a translational but not transcriptional mechanism. Autophagy. 2016;12:287–296. doi: 10.1080/15548627.2015.1124225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon YJ, Kim JA, Shin KD, Shin DS, Han YM, Lee YJ, Lee JS, Kwon BM, Han DC. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J Biol Chem. 2011;286:1737–1747. doi: 10.1074/jbc.M110.179440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwasaki M, Tanaka R, Hishiya A, Homma S, Reed JC, Takayama S. BAG3 directly associates with guanine nucleotide exchange factor of Rap1, PDZGEF2, and regulates cell adhesion. Biochem Biophys Res Commun. 2010;400:413–418. doi: 10.1016/j.bbrc.2010.08.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fontanella B, Birolo L, Infusini G, Cirulli C, Marzullo L, Pucci P, Turco MC, Tosco A. The co-chaperone BAG3 interacts with the cytosolic chaperonin CCT: new hints for actin folding. Int J Biochem Cell Biol. 2010;42:641–650. doi: 10.1016/j.biocel.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Sorensen KD, Abildgaard MO, Haldrup C, Ulhoi BP, Kristensen H, Strand S, Parker C, Hoyer S, Borre M, Orntoft TF. Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br J Cancer. 2013;108:420–428. doi: 10.1038/bjc.2012.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montel V, Gaultier A, Lester RD, Campana WM, Gonias SL. The low-density lipoprotein receptor-related protein regulates cancer cell survival and metastasis development. Cancer Res. 2007;67:9817–9824. doi: 10.1158/0008-5472.CAN-07-0683. [DOI] [PubMed] [Google Scholar]
- 55.Liu YH, Tsang JY, Ni YB, Hlaing T, Chan SK, Chan KF, Ko CW, Mujtaba SS, Tse GM. Doublecortin-like kinase 1 expression associates with breast cancer with neuroendocrine differentiation. Oncotarget. 2016;7:1464–1476. doi: 10.18632/oncotarget.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, Gaetje R, Solbach C, Ahr A, Metzler D. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Callari M, Musella V, Di Buduo E, Sensi M, Miodini P, Dugo M, Orlandi R, Agresti R, Paolini B, Carcangiu ML. Subtype-dependent prognostic relevance of an interferon-induced pathway metagene in node-negative breast cancer. Mol Oncol. 2014;8:1278–1289. doi: 10.1016/j.molonc.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Savci-Heijink CD, Halfwerk H, Koster J, van de Vijver MJ. A novel gene expression signature for bone metastasis in breast carcinomas. Breast Cancer Res Treat. 2016;156:249–259. doi: 10.1007/s10549-016-3741-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caiazza F, McCarthy NS, Young L, Hill AD, Harvey BJ, Thomas W. Cytosolic phospholipase A2-alpha expression in breast cancer is associated with EGFR expression and correlates with an adverse prognosis in luminal tumours. Br J Cancer. 2011;104:338–344. doi: 10.1038/sj.bjc.6606025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(353-361):361.e351–361.e352. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Purrington KS, Slettedahl S, Bolla MK, Michailidou K, Czene K, Nevanlinna H, Bojesen SE, Andrulis IL, Cox A, Hall P. Genetic variation in mitotic regulatory pathway genes is associated with breast tumor grade. Hum Mol Genet. 2014;23:6034–6046. doi: 10.1093/hmg/ddu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maass N, Nagasaki K, Ziebart M, Mundhenke C, Jonat W. Expression and regulation of tumor suppressor gene maspin in breast cancer. Clin Breast Cancer. 2002;3:281–287. doi: 10.3816/CBC.2002.n.032. [DOI] [PubMed] [Google Scholar]
- 63.Gu Y, Li P, Peng F, Zhang M, Zhang Y, Liang H, Zhao W, Qi L, Wang H, Wang C. Autophagy-related prognostic signature for breast cancer. Mol Carcinog. 2016;55:292–299. doi: 10.1002/mc.22278. [DOI] [PubMed] [Google Scholar]
- 64.Lee JE, Moon PG, Cho YE, Kim YB, Kim IS, Park H, Baek MC. Identification of EDIL3 on extracellular vesicles involved in breast cancer cell invasion. J Proteome. 2016;131:17–28. doi: 10.1016/j.jprot.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 65.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 66.Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. 2016;7:11018–11032. doi: 10.18632/oncotarget.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H, Wang B, Zhu W, Yang Z. Lentivirus-mediated knockdown of myosin vi inhibits cell proliferation of breast cancer cell. Cancer Biother Radiopharm. 2015;30:330–335. doi: 10.1089/cbr.2014.1759. [DOI] [PubMed] [Google Scholar]
- 68.Gabriel B, Fischer DC, Orlowska-Volk M, zur Hausen A, Schule R, Muller JM, Hasenburg A. Expression of the transcriptional coregulator FHL2 in human breast cancer: a clinicopathologic study. J Soc Gynecol Investig. 2006;13:69–75. doi: 10.1016/j.jsgi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 69.Cao CY, Mok SW, Cheng VW, Tsui SK. The FHL2 regulation in the transcriptional circuitry of human cancers. Gene. 2015;572:1–7. doi: 10.1016/j.gene.2015.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hansen SN, Ehlers NS, Zhu S, Thomsen MB, Nielsen RL, Liu D, Wang G, Hou Y, Zhang X, Xu X. The stepwise evolution of the exome during acquisition of docetaxel resistance in breast cancer cells. BMC Genomics. 2016;17:442–456. doi: 10.1186/s12864-016-2749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang CY, Lin SC, Su WH, Ho CM, Jou YS. Somatic LMCD1 mutations promoted cell migration and tumor metastasis in hepatocellular carcinoma. Oncogene. 2012;31:2640–2652. doi: 10.1038/onc.2011.440. [DOI] [PubMed] [Google Scholar]
- 72.Chou HL, Yao CT, Su SL, Lee CY, Hu KY, Terng HJ, Shih YW, Chang YT, Lu YF, Chang CW. Gene expression profiling of breast cancer survivability by pooled cDNA microarray analysis using logistic regression, artificial neural networks and decision trees. BMC Bioinformatics. 2013;14:100–110. doi: 10.1186/1471-2105-14-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou L, Gao HF, Liu DS, Feng JY, Gao DD, Xia W. Gene expression profiling of brain metastatic cell from triple negative breast cancer: Understanding the molecular events. Gene. 2018;640:21–27. doi: 10.1016/j.gene.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 74.Vadnais C, Shooshtarizadeh P, Rajadurai CV, Lesurf R, Hulea L, Davoudi S, Cadieux C, Hallett M, Park M, Nepveu A. Autocrine activation of the Wnt/beta-catenin pathway by CUX1 and GLIS1 in breast cancers. Biol Open. 2014;3:937–946. doi: 10.1242/bio.20148193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ripka S, Neesse A, Riedel J, Bug E, Aigner A, Poulsom R, Fulda S, Neoptolemos J, Greenhalf W, Barth P. CUX1: target of Akt signalling and mediator of resistance to apoptosis in pancreatic cancer. Gut. 2010;59:1101–1110. doi: 10.1136/gut.2009.189720. [DOI] [PubMed] [Google Scholar]
- 76.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete list of significantly changed proteins in global proteomics analysis
Chemoresistant breast cancer cells are more sensitive to the selective BH3-mimetic ABT-737. Cell viability was analyzed by MTT assay in (A) BT-549Par and BT-549rDOX20 cells and (B) MDA-MB-468Par and MDA-MB-468r5-FU2000 cells after 48 h treatment with ABT-737. DMSO (0.1 %, for 48 h) was used as control (Ctrl) for the solvent.
Depletion of BAG3 increases translocation of BAX to mitochondria in chemoresistant breast cancer cells. Cells were harvested and the mitochondrial and cytosolic fractions were separated by digitonin-based subcellular fractionation in control and BAG3 KD MDA-MB-468r5-FU2000 cells followed by western blot for detection of BAX. GAPDH and HSP60 served as loading controls for the cytosolic and mitochondrial fractions, respectively.
Knockdown of BAG3 reduces the mRNA expression of SNAI2, TWIST1, TWIST2 in BT-549rDOX20/BAG3 KD and MDA-MB-468r5-FU2000/BAG3 KD cells. (A) Knockdown of BAG3 reduced the relative SNAI1, TWIST1, TWIST2 mRNA expression in BT-549rDOX20/BAG3 KD and (B) MDA-MB-468r5-FU2000/BAG3 KD cells in qPCR respectively. qPCR data represent means of three independent experiments± SEM (n = 3). Significant mRNA expression compared to parental sh Ctrls are marked by asterisks: *P < .05 and ns not significant. Significant differences between BAG3 KD and respective sh Ctrls are denoted by hashtags: #P < .05 and ns not significant.
Depletion of BAG3 reduces the migration of breast cancer chemoresistant cells. (A) Number of migrated cells was decreased in BT-549rDOX20/BAG3 KD and (B) MDA-MB-468r5-FU2000/BAG3 KD cells. Migration assay was performed for 20 h followed by bright field image was taken in x40, scale bar 200 μm and migrated cells were counted by using ImageJ software. Columns represent means of three independent experiments ± SEM (n = 3). Statistical significance of migration: *P < .05, ***P < .001 and ns not significant with BAG3 KD compared to sh Ctrls.