Abstract
The impact of inherited and somatic mutations on messenger RNA (mRNA) structure remains poorly understood. Recent technological advances that leverage next-generation sequencing to obtain experimental structure data, such as SHAPE-MaP, can reveal structural effects of mutations, especially when these data are incorporated into structure modeling. Here, we analyze the ability of SHAPE-MaP to detect the relatively subtle structural changes caused by single-nucleotide mutations. We find that allele-specific sorting greatly improved our detection ability. Thus, we used SHAPE-MaP with a novel combination of clone-free robotic mutagenesis and allele-specific sorting to perform a rapid, comprehensive survey of noncoding somatic and inherited riboSNitches in two cancer-associated mRNAs, TPT1 and LCP1. Using rigorous thermodynamic modeling of the Boltzmann suboptimal ensemble, we identified a subset of mutations that change TPT1 and LCP1 RNA structure, with approximately 14% of all variants identified as riboSNitches. To confirm that these in vitro structures were biologically relevant, we tested how dependent TPT1 and LCP1 mRNA structures were on their environments. We performed SHAPE-MaP on TPT1 and LCP1 mRNAs in the presence or absence of cellular proteins and found that both mRNAs have similar overall folds in all conditions. RiboSNitches identified within these mRNAs in vitro likely exist under biological conditions. Overall, these data reveal a robust mRNA structural landscape where differences in environmental conditions and most sequence variants do not significantly alter RNA structural ensembles. Finally, predicting riboSNitches in mRNAs from sequence alone remains particularly challenging; these data will provide the community with benchmarks for further algorithmic development.
Keywords: RNA secondary structure, SHAPE-MaP, TPT1, LCP1, riboSNitch, somatic mutation
INTRODUCTION
RiboSnitches are single-nucleotide variants (SNVs) that cause changes in RNA secondary structure (Halvorsen et al. 2010; Wan et al. 2014; Gotea et al. 2015; Kutchko et al. 2015; Lu et al. 2015; Solem et al. 2015). These riboSNitches result in RNAs with different structures, leading to potentially different regulatory and functional abilities. Approximately 15% of inherited SNVs change RNA structure in family trio transcriptome-wide structural experiments (Wan et al. 2014). However, in general the function of structure in messenger RNAs (mRNAs) is unclear (Bartel 2009; Scharff et al. 2011; Dethoff et al. 2012; Li et al. 2014). Certain specific structures, such as the Iron Responsive Element and the Histone Stem–Loop, play central roles in post-transcriptional regulation of the mRNAs in which they occur (Gallie et al. 1996; Marzluff et al. 2008; Ma et al. 2012). For instance, the Iron Response Element within FTL mRNA normally forms a hairpin structure and is regulated by an iron response element binding protein (Burdon et al. 2007). Several inherited SNVs within FTL alter the structure of this element and are associated with hyperferritinemia cataract syndrome (Halvorsen et al. 2010; Martin et al. 2012).
The coupling of next-generation sequencing with chemical and enzymatic probing, such as in SHAPE (Selective 2′ Hydroxyl Acylation by Primer Extension) methodology (Siegfried et al. 2014; Lu and Chang 2016; Zubradt et al. 2017) enables structure analysis at unprecedented scale, including transcriptome-wide RNA secondary structure determination (Kertesz et al. 2010; Underwood et al. 2010; Zheng et al. 2010; Lucks et al. 2011; Ding et al. 2014; Incarnato et al. 2014; Rouskin et al. 2014; Wan et al. 2014; Del Campo et al. 2015). However, the final structures and even existence of RNA secondary structure under biological conditions remains controversial as secondary structure probing within cells yields significant variability depending on the approach (Ding et al. 2014; Rouskin et al. 2014; Spitale et al. 2015; Watters et al. 2016a; Lee et al. 2017). Although some well-studied small RNAs, such as the hairpin ribozyme and bacterial RNase-P, fold to near native conformations in the absence of cellular components (Donahue et al. 2000; Lindell et al. 2005), the specific effects of the cellular environment on RNA structure remain poorly understood (Schroeder et al. 2004; Mahen et al. 2010; Zemora and Waldsich 2010; Kubota et al. 2015; Smola et al. 2015a, 2016). We know that during translation ribosomes assisted by helicases must unfold both the mRNA coding region and, likely, structured elements in the untranslated regions (UTRs). However, it is a thermodynamic reality that, when not actively unfolded, single-stranded RNA will rapidly form intramolecular base pairs (Das et al. 2003). Thus, we expect that different subpopulations of mRNAs exist in the cellular environment, and, if we probe their structures in bulk, we will observe signals averaged over the ensemble of subpopulations, including structures specific to cellular and in vitro conditions.
We chose to look in-depth at two specific mRNAs, TPT1 and LCP1. The first mRNA, TPT1 (tumor protein, translationally controlled 1), was originally characterized as a sequestered mRNA that is translationally induced during growth conditions (Gross et al. 1989). We are particularly interested in TPT1 because it is thought to have extensive secondary structure, even to the extent of activating the double-strand RNA recognition protein PKR (Bommer et al. 2002; Nussbaum et al. 2002). Although not known to be as structured as TPT1, LCP1 (lymphocyte cytosolic protein 1) is also over-expressed in many different cancers and may be involved in cell mobility (Shinomiya 2012; Van Audenhove et al. 2016).
We develop here a combined experimental and computational method to confidently detect riboSNitches arising from inherited SNVs as well as from somatic mutations identified in cancers. We take advantage of recent chemical probing techniques that allow us to rapidly obtain high-resolution structural information on full-length transcripts (Siegfried et al. 2014; Smola et al. 2015b), while also facilitating a rapid, allele-specific sorting of reads for rapid and accurate riboSNitch detection. We compare the ability of traditional SHAPE-MaP to detect riboSNitches and find that using allele-specific sorting decreases the background noise and improves riboSNitch detection. Combined with a thermodynamically rigorous framework that enables us to use experimental SHAPE data as pseudo-free energies in nearest neighbor RNA folding free energy models, we are able to directly visualize the Boltzmann suboptimal ensemble and how these variants affect the ensemble (Deigan et al. 2009; Hajdin et al. 2013; Woods et al. 2017). Finally, to verify that in vitro determined riboSNitches are biologically relevant, we test how stable the structures are within TPT1 and LCP1 mRNAs by performing SHAPE-MaP in the presence and absence of cellular proteins. With these techniques, we determined that TPT1 and LCP1 mRNAs are structurally robust with high correlations between protein-bound and -unbound experiments. We also identified five riboSNitches, including two arising from somatic mutations within the coding sequencing of LCP1 and three within the UTRs of TPT1.
RESULTS
Variation and conservation within TPT1 and LCP1 mRNAs
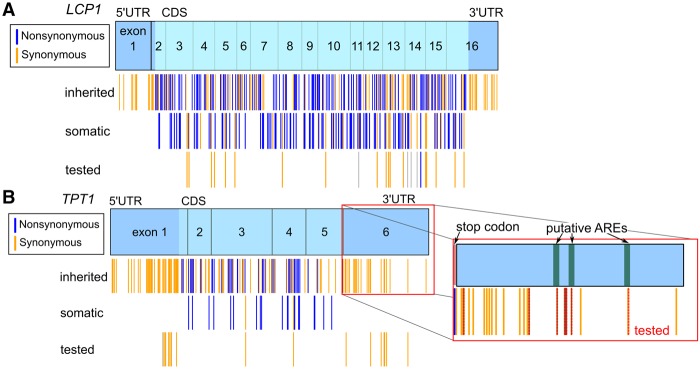
Like many other human genes, LCP1 and TPT1 harbor somatic mutations from various cancers and have extensive inherited variation (Fig. 1A,B; Sherry et al. 2001; Forbes et al. 2015). We are particularly interested in the potential effects of these mutations on RNA structure. We performed in vitro SHAPE-MaP experiments on a subset of somatic and inherited SNVs. Our goal was to screen a broad range of genetic and somatic mutations to assess potential consequences on RNA structure. We obtained somatic mutations from the Catalogue of Somatic Mutations in Cancer (COSMIC) database, which contains primarily exome sequencing from a variety of different cancers, and we obtained inherited polymorphisms from the National Center for Biotechnology Information (dbSNP) database. We focused our experimental analysis on synonymous somatic mutations in the coding sequence because these mutations will not affect the protein product and are therefore more likely to be functional riboSNitches (Shabalina et al. 2013; Hunt et al. 2014; Supek et al. 2014; Gotea et al. 2015). LCP1 has only one isoform and a long coding region with 21 synonymous somatic mutations (Fig. 1A). In the most commonly expressed isoform of TPT1 (NM_003295), there are only two synonymous mutations in the coding sequence (Fig. 1B). We therefore expanded our experimental investigation into a subset of inherited SNVs from the 5′ and 3′ UTRs, concentrating on putative functional regions such as predicted AU-rich elements (AREs) (Fig. 1B). Our selected subsets of somatic and inherited variants did not have different conservation scores from randomly selected nucleotides (nt) as a group in either LCP1 or TPT1.
FIGURE 1.
Variation within LCP1 and TPT1. (A) LCP1 mRNA is long (coding sequence of 1884 nt) and is composed of 16 exons. (B) TPT1 has three isoforms and multiple alternative polyadenylation sites, of which NM_003295 with an early polyadenylation site is the most prevalent (∼1000 nt in length). All identified inherited SNVs and somatic mutations within these genes are shown (blue, nonsynonymous; gold, synonymous). We tested all synonymous somatic mutations within the LCP1 and TPT1 CDS regions and selected inherited variants.
Allele-specific sorting greatly improves riboSNitch detection
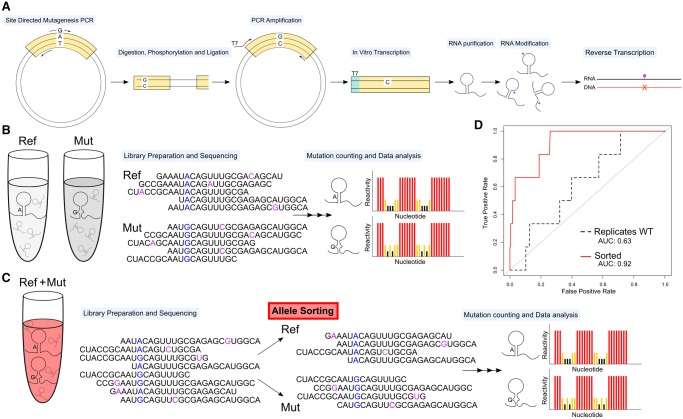
We were able to obtain SHAPE data on 37 selected variants within LCP1 and TPT1 using a novel, high-throughput SHAPE-MaP protocol (Siegfried et al. 2014; Smola et al. 2015b). Briefly, we used a clone-free, site-directed mutagenesis technique to create selected variants, transcribed the RNA variants, probed the RNA with 1M7 or the negative control, DMSO, and proceeded with error-prone reverse transcription to fix the adduct locations as mutations in the cDNA (Fig. 2A,C). Since the mutagenesis step (Fig. 2A) is not 100% efficient, a percentage of wild-type (WT) sequence remains in the amplification, and WT RNA is spiked into the reaction, resulting in both alleles being simultaneously probed. This is in contrast to a strategy in which both alleles are probed in separate tubes, effectively introducing replicate variability into their SHAPE signals (Fig. 2B). The novel experimental protocol used here takes advantage of SHAPE-MaP's read-through reverse transcriptase step and allows allele-specific sorting of reads (Fig. 2C). Therefore, our strategy for obtaining matched SHAPE data for both alleles uses an in vitro mutagenesis approach, enabling us to solve the structures of each allele by sorting reads prior to generating SHAPE data. This approach has the advantage of being internally consistent, i.e., each allele is probed under identical conditions, increasing the sensitivity of the assay.
FIGURE 2.
High-throughput strategy to identify riboSNitches is improved by clone-free allele-specific sorting. (A) We performed site-directed mutagenesis followed by SHAPE-MaP. (B) In a traditional experiment, reference and mutant RNAs are modified and prepared for sequencing in separate tubes. (C) In our novel protocol, reference and mutant simultaneously undergo modification. The sequencing reads are sorted based on the reference/mutant allele and SHAPE reactivities are calculated from the sorted reads. (D) By simulating the traditional experimental approach, we have poor differentiation (AUC 0.63, dotted line), while simulating our novel within sample approach results in a dramatic improvement (AUC 0.92, red line).
To evaluate the improvement resulting from modification of both alleles in the same tube we estimated the false discovery rate of riboSNitches in replicate WT SHAPE data. We compared each WT SHAPE data set to the total average WT SHAPE. We expect that replicates of the same RNA sequence should have no riboSNitches and that any differences in the data are due to replicate noise and are therefore false positives (only manually confirmed nonchanger samples were used to create the average total WT SHAPE in these comparisons). We used the classSNitch algorithm, which establishes if two SHAPE traces are different using a random forest classification algorithm (Woods and Laederach 2017). ClassSNitch classified 52% of these WT to WT comparisons as riboSNitches, suggesting a false discovery rate of a traditional nonallele-sorted experiment at 0.52.
We also generated within-sample data sets that were ratiometrically normalized to match our allele specific sorting to control for variations in sequencing depth, which are known to impact signal to noise in SHAPE-MaP data (Siegfried et al. 2014). In this case, we expect that within-sample data sets should have no riboSNitches and that any differences in the data are therefore false positives. We used the classSNitch algorithm (Woods and Laederach 2017) and found that within sample comparison dramatically lowered the false positive rate, with only 9% of samples falsely identified as a riboSNitch. We performed receiver operator curve (ROC) analysis to control for classSNitch sensitivity on the ratiometrically sorted (red line, Fig. 2D), and replicate WT (black dotted line, Fig. 2D) data sets. The result is a remarkable improvement in riboSNitch detection accuracy (area under the curve, AUC) increase from 0.63 to 0.92.
Adding allele-specific sorting to the SHAPE-MaP procedure greatly improves the detection of single-nucleotide mutations that change RNA structure. This technique is not without cost, however, as only reads that span the mutation site or are paired with a read spanning the mutation site can be accurately sorted. This restricts SHAPE data to within a few hundred nucleotides of the mutation. The vast majority of base pairs even in highly structured RNAs like the Ribosome tend to be short-range; thus it is reasonable to expect riboSNitches to be local and allele-specific sorting adequate for detection of the majority of structure changes (Doshi et al. 2004; Kladwang et al. 2011a,b; Cordero et al. 2012; Woods and Laederach 2017). In addition, improvements in sequencing technology readily increase read lengths, and as data are collected with longer reads, allele-specific sorting will allow the detection of larger structural changes.
Identification of five riboSNitches within TPT1 and LCP1 mRNAs
We analyzed 37 allele-sorted data sets using the novel experimental protocol illustrated in Figure 2. We identified five riboSNitches using classSNitch in this data set (Woods and Laederach 2017). Our data are consistent with previous transcriptome-wide secondary structure analysis on a family trio that estimated that 15% of SNPs are likely riboSNitches (Wan et al. 2014). We find that 2/19 somatic mutations and 3/18 SNPs are riboSNitches with an overall ratio of 14% (Supplemental Table S1). A recent analysis of mutations within structured RNAs revealed significant differences in their susceptibility to mutation with an upper bound of over 60% of mutations disrupting the Lariat Capping Ribozyme structure (Woods and Laederach 2017). Nonetheless, on average 19% of mutations disrupted structure within these structured RNAs, suggesting that TPT1 and LCP1 structure are, on average, similarly robust to mutation as other RNAs. Furthermore, our data suggests that somatic mutations and inherited variation do not differ in their propensity for structural change. However, we do not find the UTRs or coding sequences to have a significant difference in riboSNitch frequency, implying that the likelihood of a point mutation changing a structure is dependent on the individual RNA's susceptibility to change (Woods and Laederach 2017).
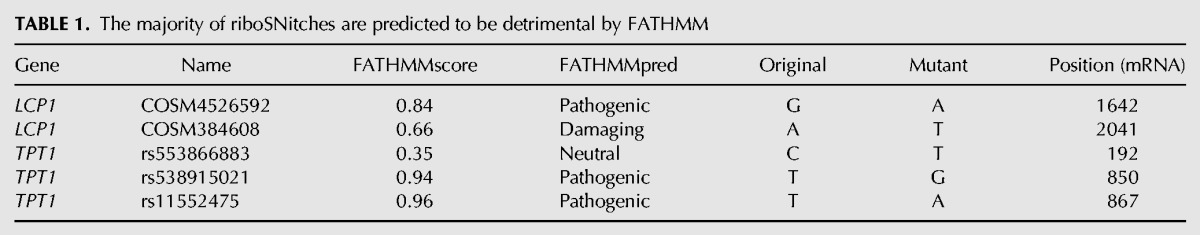
We used the functional variant detection algorithm FATHMM-MKL to predict the consequences of the five riboSNitches we identified. Identifying which SNPs and mutations are biologically relevant is exceptionally difficult, and FATHMM-MKL is one of the few predictive algorithms to include noncoding mutations. FATHMM-MKL uses conservation, histone markers, and other features of the genome to assess whether a variant is likely to have a pathogenic effect. Scores are reported as P-values on a scale of 0–1, with higher numbers meaning more likely to be detrimental and lower numbers equating to neutral (Shihab et al. 2013, 2015). Although these predictive scores require experimental validation, FATHMM-MKL predictions perform well when compared with several databases of pathogenic mutations such as ClinVar and HGMD (Shihab et al. 2015). Four of the riboSNitches were predicted to be detrimental (Table 1). Within LCP1, the somatic riboSNitches COSM4526592 and COSM384608 had scores of 0.84 and 0.66, respectively, and, within TPT1, the inherited riboSNitches rs538915021 and rs11552475 had scores of 0.94 and 0.96 (Table 1). Although rs553866883 within TPT1 had a neutral score (0.35), it is possible that its ability to change the structure of TPT1 will make it more likely to be pathogenic in a manner not currently measured by FATHMM-MKL. FATHMM-MKL also predicted that many of nonchangers are harmful, as expected based on our growing knowledge of how synonymous and noncoding variations can influence pathogenesis even without altering RNA secondary structure (Supplemental Table S1; Hunt et al. 2014; Supek et al. 2014; Khurana et al. 2016).
TABLE 1.
The majority of riboSNitches are predicted to be detrimental by FATHMM
We analyzed the structure change in all five riboSNitches by visualizing the ensemble of RNA structures. RNA is much more flexible than protein or DNA, usually existing as an ensemble of different structures instead of one stable conformation. We used the software ensemblerna to model and visualize these ensembles (Woods et al. 2017). Ensemblerna uses SHAPE data to guide a secondary structure folding algorithm. Then ensemblerna takes the set of structures, groups them, and counts the number of structures in each cluster before mapping the clusters onto a stable conformational space. To aid in understanding, we also show the medoid structure for the main clusters. Most riboSNitches cause a shift in the population of clusters and they sometimes cause formation of a new cluster. Due to sequencing restrictions we only obtained high depth information 100 nt around each mutation and may have missed larger structural effects from the five riboSNitches we detected. However, we expect the majority of structure changes to be local (Doshi et al. 2004; Kladwang et al. 2011a,b; Cordero et al. 2012; Woods and Laederach 2017).
Somatic riboSNitches in the coding region of LCP1 mRNA
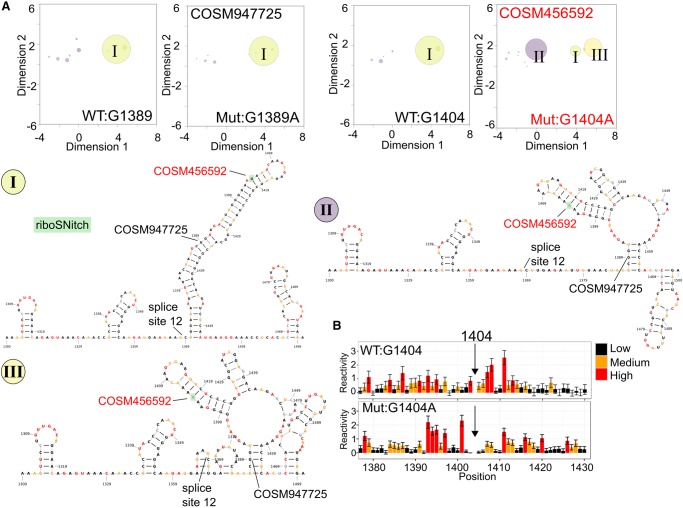
In the LCP1 coding region (around position 1400), the mRNA folds as one group of related structures, shown with a representative secondary structure (Fig. 3A, large yellow bubble I). This group predominates within reference sequences and in another nonchanger mutation in the same region, COSM947725, which is highly likely to be neutral (FATHMM-MKL score: 0.06). However, the riboSNitch G1404A (COSM4526592), which is predicted to be pathogenic (FATHMM-MKL score: 0.84), causes a decrease in the population of the normal cluster and drives an increase in population of two additional clusters of structures (Fig. 3A, purple and light yellow bubbles II and III). The loss of a large dsRNA region is shown as the ensemble moves from the main WT cluster to alternative clusters (Fig. 3A). The normalized SHAPE data around the mutation site show a relatively local change with an increase in reactivity before the mutation site and a decrease in reactivity after the mutation site (Fig. 3B). We also analyzed the riboSNitch COSM384608 within the coding region of LCP1, which is predicted to be detrimental (FATHMM-MKL score: 0.66). Normally, this region exists as many different structure clusters (Supplemental Fig. S1A–C), but the riboSNitch COSM384608 causes a collapse of the ensemble into one main group (Supplemental Fig. S1D).
FIGURE 3.
The riboSNitch COSM4526592 shifts the LCP1 mRNA ensemble from one predominant structure group into three groups. (A) The SHAPE-guided structural ensembles for WT (G1389), Mutant (G1389A, nonchanger) and WT (G1404) have one main structure cluster (green bubble, I). The riboSNitch G1404A (COSM4526592), labeled in red, shifts the ensemble into two new structure clusters (purple, II; yellow, III). Each cluster is labeled in the ensemble plots (A) and illustrated with the medoid structures from the cluster (I–III). These medoid structures are example secondary structures for each cluster. The major allele structure is colored by WT SHAPE reactivity (I), whereas the nucleotides in the alternative conformations are colored by mutant reactivities (II and III). (B) Normalized SHAPE data around the riboSNitch show an increase in reactivity before the mutation and a decrease afterward in the mutant profile (bottom).
Inherited rare variant riboSNitches in the 5′ and 3′ UTRs of TPT1 mRNA
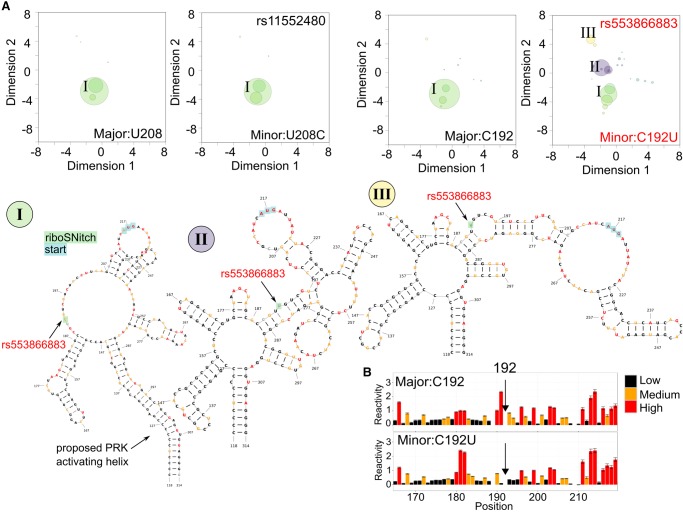
Within the 5′ UTR of TPT1, there is one main group of similar structures (Fig. 4A, green bubble I). This region includes both the start codon and the location where PKR is predicted to bind TPT1 and cause translational repression. We observed this grouping of similar structures in mRNAs with the nonchanger SNV rs11552489 and with the major allele sequence for both the nonchanger and the riboSNitch rs553866883. However, the riboSNitch C192U (rs553866883) induces an increase in two additional clusters that are poorly populated in the major allele (Fig. 4A, bubbles II and III). Exemplary secondary structures from these clusters (Fig. 4A, I,II) indicate that the large dsRNA region preceding the variant has decreased in the alternative conformations and the variant itself moves from a relatively unstructured region into a more structured area.
FIGURE 4.
The riboSNitch rs553866883 shifts the TPT1 mRNA ensemble from one predominant structure cluster into three. (A) The SHAPE-guided structural ensembles for major allele (U208), minor allele (U208C, rs11552489, nonchanger), major allele C192 all show one main structure cluster (green, I). The riboSNitch C192U (rs553866883), labeled in red, shifts the ensemble to form two new structure clusters that were previously not populated (purple, II; yellow, III). Each cluster is labeled in the ensemble plots and illustrated with the medoid structures from the group (I–III). The medoid secondary structure for the main cluster is colored by WT SHAPE reactivity while nucleotides in the alternative conformations (II and III) are colored by mutant reactivities. Points of references from the full-length structure are highlighted. (B) Comparison of normalized SHAPE data for the minor allele profile (bottom) compared to the major allele (top) around the riboSNitch.
The SHAPE data around the riboSNitch C192U indicate that this mutation causes a local change with a decrease in reactivity (increase in structure) before and after the SNP (Fig. 4B). The nonchanging SNV U208C (rs11552489) is predicted to be pathogenic (FATHMM-MKL score: 0.88), whereas the riboSNitch C192U (rs553866883) has a neutral FATHMM-MKL score (score: 0.35). In addition, two other nonchanger SNVs close by are also predicted to be pathogenic (rs776089085 and rs770667436, scores: 0.94 and 0.86) (Supplemental Table S1). We propose that the riboSNitch C192U (rs553866883) belongs to a group of SNVs that have functional importance within the 5′ UTR of TPT1, and they may not have been identified previously as pathogenic because of its cryptic effect on mRNA structure.
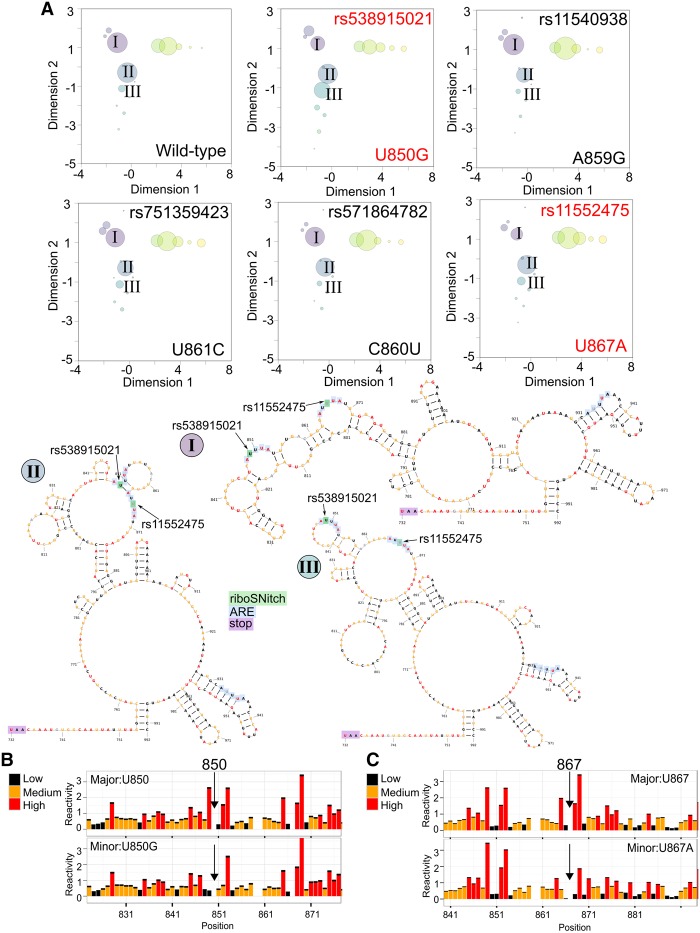
Two additional riboSNitches occur within the 3′ UTR of TPT1, which forms three main structure clusters that exist in relatively equal proportions (Fig. 5A, purple—I, blue—II and yellow bubbles) and one existing but less populated cluster (Fig. 5A, teal—III). The example medoid structures for the labeled structures illustrate the flexibility of this relatively open region (Fig. 5A). The riboSNitches U850G (rs538915021) and U867A (rs11552475) are both highly predicted to cause functional change to TPT1 mRNA (FATHMM-MKL scores >0.94). RiboSNitch U850G (rs538915021) induces an increase in a secondary cluster that is not well populated in the reference (Fig. 5A bubble III), whereas riboSNitch U867A (rs11552475) induces a reduction in the main clusters (Fig. 5A bubble I and III). The SHAPE data for both riboSNitches indicate local change with a decrease in reactivity before or after the SNV, respectively (Fig. 5B,C). Three other SNVs within the same region do not alter the structural ensemble (Fig. 5A), but they are all predicted to be pathogenic (FATHMM-MKL scores >0.91) (Supplemental Table S1). The two riboSNitches occur within proposed AREs, and the other nonchangers are in close proximity. We measured the stability of GFP-TPT1 3′ UTR mRNAs with these variants and ARE-disrupting mutations, but we did not detect any differences (Supplemental Fig. S2). These AREs may be tissue specific and not functional in HEK-293 cells.
FIGURE 5.
The riboSNitches rs538915021 and rs11552475 alter the proportions of the 3′ UTR TPT1 mRNA structure clusters. (A) The SHAPE-guided structural ensemble for the reference sequence (control) is shown next to two riboSNitches (U850G and U867A). The riboSNitch U850G (rs538915021) causes an increase in population of the III cluster. The riboSNitch U867A (rs11552475) causes a decrease in the population of the I cluster. Three nonchangers are shown below (rs11540938, rs571864782, and rs751359423) that all occur within the same region. (I–III) Three representative medoid secondary structures are shown. Each cluster is labeled in the ensemble plots. The nucleotides in all structures are colored by WT SHAPE reactivity. (B,C) Comparison of normalized SHAPE data for the major and minor allele of the riboSNitches U850G (rs538915021) and U867A (rs11552475).
The robust nature of highly structured TPT1 mRNA
We performed probing experiments on TPT1 to determine how the presence of proteins affects the TPT1 structural ensemble. We reacted cellular mRNAs with the 1M7 SHAPE chemical probe that reacts with accessible 2′ hydroxyl groups and compared reactivity rates to a background (solvent-only) control (Wilkinson et al. 2006; Siegfried et al. 2014). To mimic the cellular environment, but maximize the reactivity of the probe, we treated cellular lysates with 1M7 and compared the results with mRNAs treated after extraction and in the absence of normal RNA-binding proteins. We also in vitro transcribed TPT1 mRNA and compared the structural results of our protein-bound mRNA to artificially generated mRNA in the absence of any RNA-binding proteins. We used the SHAPE-MaP protocol in which SHAPE adducts are converted to mutations during library preparation, and the locations of adducts are identified as mutations through next-generation sequencing (Siegfried et al. 2014; Smola et al. 2015b). The SHAPE-MaP data sets used to create our secondary structure models for TPT1 are available in the supplement (Supplemental Figs. S1–S3; Supplemental Files S1–S4, http://bit.ly/2sXWL3K; Sansone et al. 2012).
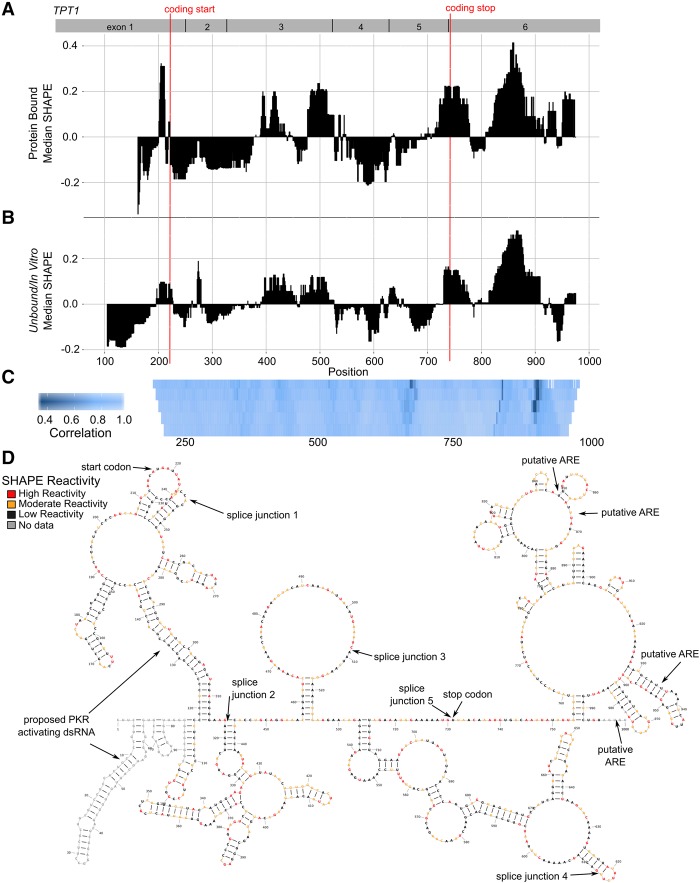
TPT1 was originally characterized as a sequestered mRNA that is translationally induced during growth conditions (Gross et al. 1989). TPT1 mRNA activates the double-strand RNA recognition protein PKR leading to repression of TPT1 protein translation, presumably through extensive double-stranded RNA structures within the 5′ UTR (Bommer et al. 2002; Nussbaum et al. 2002). TPT1 is over-expressed in cancers and, although the mechanism is not fully elucidated, its protein product is believed to function as an antiapoptotic factor controlled by mTORC1 (Amson et al. 2013; Chen et al. 2013; Acunzo et al. 2014; Bommer et al. 2015; Thébault et al. 2016). We mapped the median SHAPE reactivity across TPT1 to determine regions of high and low reactivity for the protein bound RNP (Fig. 6A) and naked RNA (Fig. 6B) conditions. This approach to visualizing the data enables us to identify regions of relatively high median SHAPE (less structure, more complex conformational ensemble) and relatively low median SHAPE (more structure, more likely a single conformation) (Smola et al. 2016). We treated the protein:mRNA complex with 1M7 (bound), extracted RNA from cells, removing all proteins (unbound), or we transcribed RNA with T7 RNA polymerase and then treated with 1M7 (in vitro) (Supplemental Figs. S3, S4). The unbound and in vitro samples had high agreement, as did all replicates (Supplemental Fig. S3D,O), allowing us to develop a robust structural model by merging and averaging the data to form a comprehensive in vitro SHAPE over nearly the entire transcript (Fig. 6D). We summarize further specifics of our correlation analysis in Supplemental Figures S3 and S5 and Supplemental Table S1. In general, the median reactivities for protein-bound, unbound and in vitro transcribed mRNA followed the same profile across TPT1, as corroborated by high levels of correlation between the samples, with an average correlation around 0.85 (Fig. 6C). For TPT1 mRNA, we conclude that the overall fold of the mRNA is similar in all conditions examined, but specific smaller regions adopt different conformations based on conditions.
FIGURE 6.
TPT1 mRNA secondary structures are similar in the presence and absence of protein. (A) Median reactivity data from RNA in cellular lysate over a moving window of 40 nt indicating structured (low median SHAPE) and unstructured regions (high median SHAPE) within corresponds closely with the pattern of (B) median reactivity data from naked RNA. A 5′ to 3′ decrease in median SHAPE is observed, consistent with a highly structured 5′ UTR and less structured 3′ UTR. (C) Supportive of the overall similar median reactivities, pattern correlation of SHAPE reactivity is high (>0.85 overall). We performed correlations over multiple windows, from 10 to 50, bottom to top, with lighter blue indicating higher correlation. Regions of difference are noticeable as darker bands in the CDS and 3′ UTR. (D) TPT1 secondary structure model informed with SHAPE reactivies from unbound RNA modification. Regions of interest are labeled, including the start codon, stop codon, proposed PKR activating helices in the 5′ UTR, and putative AREs in the 3′ UTR.
Using a model secondary structure informed with extensive in vitro SHAPE data, we found that TPT1 is highly structured in the 5′ UTR, with more than 60% of the bases involved in pairing, and it is relatively open in the 3′ UTR, with less than 40% of the bases involved in base-pairing interactions. This observation correlates with the 5′ to 3′ increase in median SHAPE values plotted in Figure 6B. TPT1 mRNA activates the double-stranded RNA (dsRNA) recognition protein PKR (Bommer et al. 2002; Nussbaum et al. 2002). PKR primarily recognizes viral dsRNA and inhibits viral translation, but PKR also interacts with a variety of cellular dsRNA-containing mRNAs, including the TNF-α and IFN-γ (Osman et al. 1999; Cohen-Chalamish et al. 2009; Hull and Bevilacqua 2016). Activation of PKR depends on a dsRNA region of at least 30 bases to allow binding of two or more PKR molecules (Manche et al. 1992; Zheng and Bevilacqua 2004; Lemaire et al. 2008). The highly structured 5′ UTR and first exon region of TPT1 contain 95 base pairs (bp), primarily as two imperfect helices in close proximity, composed of 23 and 25 bp (Fig. 6D). This dsRNA region is likely to bind and activate PKR, resulting in translational inhibition of TPT1 mRNA.
Structured coding region of LCP1 mRNA
LCP1 is over-expressed in many different cancers and may be involved in cell mobility (Shinomiya 2012; Van Audenhove et al. 2016). Although little is known about the molecular mechanisms that regulate LCP1 transcription and translation, a variant in the promoter region of LCP1 causes a decreased risk for prostrate cancer (Chen et al. 2016) while other variants in LCP1 are proposed as biomarkers for colorectal cancer recurrence and eQTLs (Garge et al. 2010; Ning et al. 2014). We performed probing experiments on LCP1 mRNA to determine how the presence of proteins affects structural ensemble and how dependent LCP1 structure is upon cellular conditions. We treated the LCP1 mRNA in complex with its native RNA-binding proteins with the 1M7 SHAPE reagent and compared the results with 1M7 treatment of in vitro transcribed LCP1 mRNA (Fig. 7A,B). We used the SHAPE-MaP protocol where 1M7 adducts are converted to mutations during library preparation, and identified with next-generation sequencing (Siegfried et al. 2014; Smola et al. 2015b). An overview of the different conditions used and raw data histograms are summarized in Supplemental Figure S5, and raw data are available in Supplemental Files S3 and S4 (also available on http://bit.ly/2sXWL3K) (Sansone et al. 2012).
FIGURE 7.
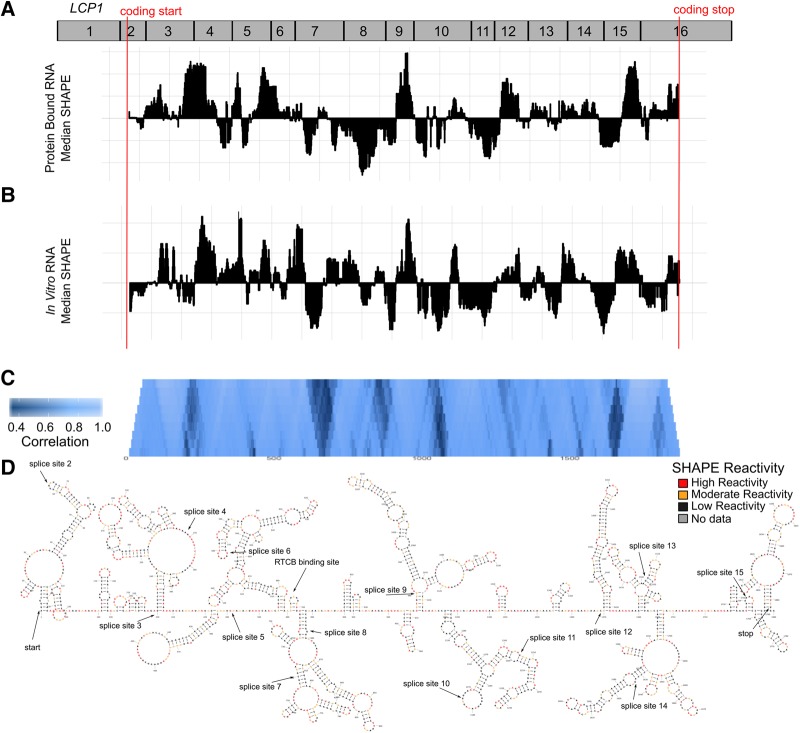
LCP1 mRNA secondary structures are similar in the presence and absence of protein. (A) LCP1 RNPs were probed and analyzed to generate median reactivity data. Low SHAPE regions indicate likely structured regions, while high SHAPE regions are likely unstructured. The median reactivity within the CDS of LCP1 corresponds closely with the pattern of (B) naked RNA reactivity data. (C) The pattern of SHAPE reactivity is highly correlated (>0.82 overall). Correlations are performed over multiple windows, from 10 to 100, and lighter blue indicates higher correlation. Various regions of difference can be seen throughout the CDS as darker bands. (D) LCP1 secondary structure model for the entire 1884 nt CDS using SHAPE data from unbound RNA to direct RNA structure prediction. Regions of interest are labeled, including splice junctions and potential RNA-binding protein sites.
LCP1 has similar overall structures under cellular-like conditions with protein complexes and under in vitro transcribed conditions (Fig. 7A–C). The median reactivity profiles for LCP1 mRNA in these environments were highly correlated with an average correlation coefficient of 0.82 (Fig. 7C). We derived a SHAPE-informed secondary structure model with the comprehensive in vitro SHAPE data set for LCP1 mRNA (Supplemental Fig. S7D). LCP1 is structured, but it does not have as many contiguous helices as TPT1. In PAR-Clip analyses, only the RNA-binding protein RCTB was found to interact with the LCP1 mRNA coding sequence, although AGO2, LIN28B, IGF2BP1, and MOV10 can interact with the UTRs (Yang et al. 2015). Due to its extensive structure, we expect that post-transcriptional regulation is important for LCP1 function.
Environmentally dependent mRNA structures
Differences between structures from the same RNA as measured on the bench versus within the cell have been attributed to RNA-binding proteins, N-6-methyl adenosine modification, and active cellular unwinding, such as that associated with translation (Rouskin et al. 2014; Smola et al. 2015b; Spitale et al. 2015). Thus, we expect differences between TPT1 and LCP1 mRNAs in a cellular-like environment versus mRNAs transcribed in vitro with only T7 polymerase. This expectation is demonstrated best in our TPT1 data sets with the lowest average correlation occurring between protein-bound TPT1 and in vitro transcribed TPT1 (Supplemental Table S2). We observed that the structure of TPT1 transcribed in the cell, but removed from the cellular environment before probing, was in between RNA structures derived from experiments with more cellular protein-bound TPT1 and in vitro transcribed TPT1 (Supplemental Table S2). Because LCP1 is not highly expressed, we do not have extensive coverage of this mRNA from transcriptome-wide experiments, however, within regions of adequate depth, LCP1 transcribed within the cell did correlate better with protein-bound LCP1 mRNA (Supplemental Fig. S5K, orange line), whereas the T7-transcribed in vitro LCP1 data were much less correlated (Supplemental Fig. S5K, maroon and red lines). The similarities between the natively transcribed mRNAs, with or without protein binding, imply that transient protein-induced structural changes are not the only factors involved in differences between cellular and in vitro RNA structure, and additional properties such as cotranscriptional folding or splicing-influenced structures may also be important (Jin et al. 2011; McManus and Graveley 2011; Lai et al. 2013; Watters et al. 2016b).
To better understand the importance of environmentally dependent structures, we looked at whether these regions fell into particular categories. We found no connection between environmentally dependent structures and regions of high or low structure, RNA-binding protein regions, or splice sites (Supplemental Table S3; Supplemental Fig. S6). We analyzed the conservation of LCP1 and TPT1 mRNA sequences using scored alignments of 100 vertebrate sequences from the UCSC Table Browser (Karolchik et al. 2004; Siepel et al. 2005). PhyloP scores detect both conserved (positive values) and accelerated evolution (negative values) at the level of individual bases (Pollard et al. 2010). Both LCP1 and TPT1 are conserved, as the majority of PhyloP scores are above zero (Supplemental Fig. S7A,B), although LCP1 has more nucleotides undergoing accelerated evolution compared with TPT1 (negative values, Supplemental Fig. S6B). When we compared regions with environmentally dependent structures with conservation phyloP scores we noticed that environmentally sensitive regions (pink) tended to be more conserved than insensitive regions (blue) in both TPT1 and LCP1 (Supplemental Fig. S7). This trend is also present when analyzed in reverse, i.e., nucleotides with high PhyloP and high conservation are, on average, more environmentally similar (Supplemental Table S3). Although not statistically significant, this correlation suggests that the most highly conserved regions appear to undergo the most significant conformational rearrangements. How cellular conditions impact RNA secondary structure is an active area of research (Ding et al. 2014; Rouskin et al. 2014; Spitale et al. 2015; Smola et al. 2016; Watters et al. 2016a), and conservation may be key to understanding just how the environment affects RNA structures.
DISCUSSION
Natural and somatic noncoding variations have the potential to yield significant insight into structure–function relationships in eukaryotic transcripts (Wan et al. 2014; Solem et al. 2015). In some cases, variants affecting UTR structure form a riboSNitch and cause human disease phenotypes (Halvorsen et al. 2010; Martin et al. 2012; Jafarifar et al. 2014; Rogler et al. 2014; Kutchko et al. 2015). However, it is likely that the vast majority of natural and somatic genetic variation is phenotypically benign. We still have a poor understanding of how natural noncoding genetic variation affects specific mRNA structures, and we are limited in our ability to predict these changes computationally (Ritz et al. 2012; Corley et al. 2015). Chemical probing experiments, especially high-accuracy approaches like SHAPE-MaP, detect structured regions in RNAs directly and enable us to identify the small subset of somatic and inherited variants that cause structural changes. Identification of these riboSNitches will further our understanding of the changes in RNA structure and lead to understanding which mutations and riboSNitches are pathogenic.
The median SHAPE values for TPT1 and LCP1 (Fig. 6,7) reveal that significant regions of mRNA have low median values with and without protein binding, consistent with a high degree of structure. These SHAPE data fit with our understanding of RNA folding thermodynamics that suggest that bases will pair if they are complementary (Mathews et al. 1999). Classic folding experiments with scrambled sequences demonstrate that even random RNAs will adopt stable secondary structures in complementary regions (Woodside et al. 2006). Thus, it is not surprising to find structured regions even within the coding sequence of mRNAs. Still, we do not understand how somatic or inherited variation will affect the fold of an mRNA in either highly structured or disordered regions. Our data suggest that the overall structural effects of somatic and inherited mutations do not depend on the local degree of structuredness (as measured by local median SHAPE, Supplemental Fig. S6) or functional context (UTR versus coding sequence, or even ARE). In fact, the majority of variants have no measurable effect on mRNA structure; consistently only around 15% of variants, either naturally occurring or man-made, resulted in even local structure change (Woods and Laederach 2017). Thus, the global folds of the TPT1 and LCP1 mRNAs appear relatively robust to variation. This insensitivity to variation contrasts with more highly structured RNAs such as elements of the ribosome and two ribozymes, for which recent analogous mutational analyses revealed significantly higher rates of structural disruption (over 60% for the Lariat Capping Ribozyme [Cheng et al. 2015; Woods and Laederach 2017]).
The LCP1 and TPT1 folds are also remarkably consistent between environmental conditions. Our data suggest that the fundamental structural features observed in vitro are preserved in the cell. The SHAPE-MaP approach improves the signal-to-noise by sequencing deeper and rigorously defines differences between protein-bound mRNA structures and in vitro mRNA folding. The extent of RNA structure change between cellular and in vitro conditions has been controversial and varies widely between studies and RNAs (Ding et al. 2014; Rouskin et al. 2014; Spitale et al. 2015; Watters et al. 2016a). We do identify multiple regions that are highly sensitive to environmental conditions (Figs. 6, 7; Supplemental Fig. S7) in both TPT1 and LCP1 mRNAs, and these regions tend to occur in regions with higher sequence conservation. The cellular environment causes differences between in vivo and in vitro SHAPE reactivity by rearranging the RNA structure, often through binding of RNA-binding proteins but also through many other mechanisms (Jin et al. 2011; McManus and Graveley 2011; Lai et al. 2013; Rouskin et al. 2014; Smola et al. 2015b; Spitale et al. 2015; Watters et al. 2016b). Our observation that these regions have higher sequence conservation suggests that interactions with the cellular environment may exert selective pressure on the RNA sequence. However, these differences do not occur with higher frequency in regions with low or high SHAPE reactivity (Supplemental Table S3), suggesting that interactions with the cellular environment are not limited to regions folding to single, well-defined conformations that have low median SHAPE (Smola et al. 2016).
We have shown that dissection of the molecular details of any mRNA requires high-resolution analysis using techniques such as allele-specific SHAPE-MaP that offer sufficient signal-to-noise (Fig. 2D) to infer specific structural ensemble models (Figs. 3–5). Furthermore, at present, SHAPE is the only structure probing technique for which there exists a validated, nucleotide resolution, thermodynamic correction for the nearest-neighbor rules (Deigan et al. 2009; Hajdin et al. 2013). We are beginning to unravel the intricacies of mRNA structural ensembles by performing allele-specific sorting and obtaining SHAPE ensemble models for a riboSNitch in single, clone-free experiments. The ensembles we visualize (Figs. 3–5) reveal the significant complexity of mRNA structures and the challenges faced when attempting to understand structure/function relationships in these regions. Furthermore, it is evident that specific variants and somatic mutations will alter the ensemble. The experimental and computational tools we propose and which we have used in this study provide a rigorous approach to dissect the complex interplay of ensemble thermodynamics, sequence selection, and RNA structure in driving noncoding function in the transcriptome.
MATERIALS AND METHODS
High-throughput multiplexed SHAPE treatment
Our modified high-throughput SHAPE-MaP protocol was performed on a Tecan Freedom Evo. We purchased mRNA clones of TPT1 and LCP1 (Origene—SC323772 and SC118739) and designed primers to introduce variants into select regions (Supplemental Table S6). Site-directed mutagenesis was performed with an NEB Q5 Site-Directed Mutagenesis Kit, but without customary transformation or cloning. Instead, we PCR-amplified the site-directed target after ligation using primers spanning the entire mRNA; the forward primer included a T7 promoter (NEB Q5 Hotstart). Ampure bead purification was performed to purify the DNA (Beckman Coulter—Ampure XP). Then we performed in vitro transcription with the T7 polymerase to synthesize RNA (NEB T7 Polymerase). To remove DNA, the sample was treated with TurboDNAse for 15 min at 37°C (ThermoFisher Scientific TurboDNAse Kit). Ampure bead purification was performed to purify the RNA (Beckman Coulter—RNAClean XP). To fold the RNA, samples were incubated at 37°C for 10 min in buffer containing 100 mM Na-HEPES, pH 8.0, 100 mM NaCl, and 10 mM MgCl2. The RNA was incubated for 5 min at 37°C with 10% dimethyl sulfoxide (DMSO) or with 10 mM 1-methyl-7-nitroisatoic anhydride (1M7) in DMSO. Ampure bead purification was performed to purify the modified RNA (Beckman Coulter—RNAclean XP).
Selection of variants and their functional significance
We obtained somatic mutations from the COSMIC database (v72) (Forbes et al. 2015) and designed primers to recreate all synonymous variants within TPT1 and LCP1 (Supplemental Table S6). Inherited SNVs within TPT1 were obtained from dbSNP (Sherry et al. 2001), and primers were designed in the same manner (Supplemental Table S6). We used the FATHMM-MKL webserver to extract predicted functional significance from all tested variants in TPT1 and LCP1 (Shihab et al. 2013, 2015). FATHMM-MKL estimates significance based on a variety of factors including conservation, histone modifications, transcription factor binding sites, and open chromatin. High FATHMM-MKL P-values equate to more likely deleterious substitutions, while lower P-values are more likely to be neutral.
Protein-bound mRNA SHAPE treatment
Protein-bound mRNA SHAPE MaP experiments were performed with lymphoblastoid cell lines (1000 Genomes cell lines—NA07037, NA12003, NA19098, NA19099) (1000 Genomes Project Consortium 2015) or Tet-Off HEK-293 cells (Clontech, Tet-off cell line). For mutant analysis of the 3′ UTR of TPT1, HEK-293 cells were transfected with WT or mutant GFP-TPT1 3′ UTR constructs (GenScript, Clontech pTRE-TIGHT, NEB Q5 Site-Directed Mutagenesis Kit). HEK-293 cells were washed in PBS, trypsinized, and resuspended in complete media. Lymphoblastoid cell lines were pelleted by centrifugation. For all cell types, approximately 50 million cells were resuspended in 1 mL of folding buffer (same buffer as in vitro SHAPE protocol) and supplemented with 400 U murine RNAse inhibitor (NEB). Cells briefly sonicated at 10% power for three 10-sec intervals (Fisher Scientific Sonic Dismembrator Model 500). The lysates were incubated at 37°C for 5 min and then immediately modified. SHAPE treatment was performed for a period of 5 min with an addition of DMSO or three separate additions of 1M7 with a final concentration of 30 mM 1M7, 10% DMSO. RNA was extracted from the lysates, DNAse digested and depleted of ribosomal RNA (ThermoFisher TRIzol, 5PRIME PhaseLock Heavy, Invitrogen Purelink RNA columns, ThermoFisher Purelink DNase Set, and ThermoFisher Scientific RiboMinus Eukaryote System v2 from Life Technologies).
Extracted mRNA SHAPE treatment
SHAPE-MaP experiments were performed with the same cell lines (above) on natively transcribed mRNAs. RNA was extracted by standard TRIzol purification and DNase digestion (ThermoFisher TRIzol, 5PRIME PhaseLock Heavy, Invitrogen Purelink RNA columns, ThermoFisher Purelink DNase Set) and incubated at 37°C for 10 min in folding buffer (above, in vitro SHAPE treatment). SHAPE treatment was performed for 5 min with an addition of DMSO or three separate additions of 1M7 with a final concentration of 30 mM 1M7, 10% DMSO. We performed buffer exchange (GE illustra MicroSpin G-50 Columns) before depletino of ribosomal RNA (RiboMinus Eukaryote System v2 from Life Technologies).
Reverse transcription and library preparation
We performed SHAPE-Map reverse transcription with SuperScript II, random nonamers and error-prone conditions for all samples (Siegfried et al. 2014; Smola et al. 2015b) (ThermoFisher Scientific SuperScript II, NEB random nonamers). The samples were purified with Ampure XP beads or G-50 columns to isolate the cDNA (Beckman-Coulter, GE illustra MicroSpin G-50 Columns). For transcriptome-wide structure probing, we performed second strand synthesis (NEBNext Second Strand Synthesis Module) and either Nextera or Nextera XT library preparation samples (Nextera DNA Sample Preparation Kit, Nextera XT DNA Sample Preparation Kit and Index Kits from Illumina). For gene specific structure probing, we designed primers specific to the 3′ region, coding sequence, and 5′ region of the mRNA and PCR-amplified these regions after reverse transcription (NEB Q5 HotStart). We performed secondary PCR to add TruSeq barcodes. Sequencing for the T7 transcribed samples was performed on HiSeq2500 as paired end, 2 × 50 read multiplex run. Sequencing for the natively transcribed samples was performed on HiSeq2500 as paired end, 2 × 100 read multiplex runs. TruSeq libraries were sequenced as necessary for their designed length, primarily as paired end 2 × 300 read multiplex runs on a MiSeq instrument.
SHAPE data analysis
For T7 transcribed samples and gene-specific TruSeq samples, we used bowtie2 (v2.2.9) to align SHAPE reads to either LCP1 or TPT1 mRNA (Langmead and Salzberg 2012). For transcriptome-wide experiments, we aligned reads to the entire genome (hg38). For riboSNitch analysis, we sorted the reads into WT or mutant based on the nucleotide at the mutation site, resulting in loss of SHAPE reactivity at the site of the mutation, but allowing us to separate the WT and mutant reads and determine overall reactivity for the entire region. The ShapeMapper pipeline to calculate mutation frequency has been previously described (Siegfried et al. 2014; Smola et al. 2015b). Briefly, we used the ShapeMapper algorithm to calculate the mutation frequency in the 1M7-treated sample, correcting for mutation frequency in the background (DMSO only) sample. We normalized the corrected reactivity by a multiplier based on the reactivity distribution of the full-length SERPINA1 transcript. To examine the broad, overall SHAPE reactivity, we averaged select data sets (defined in each section, available as Supplemental Files S1–S4 and on http://bit.ly/2sXWL3K) and calculated the median reactivity for 40–50 nt sliding windows (Pollom et al. 2013). We used this SHAPE reactivity to inform a minimum free energy structure using RNAstructure with a maximum pairing distance of 200/300 nt (Fig. 1; Supplemental Fig. S4). Incorporating SHAPE reactivities as a pseudo-free energy term in the nearest neighbor thermodynamic model of RNA folding improves the predictive capability of the model (Deigan et al. 2009; Hajdin et al. 2013; Sukosd et al. 2013).
Identification of riboSNitches
To identify riboSNitches, we analyzed SHAPE traces for differences between WT controls and mutants manually and with classSNitch. Sequencing data were collected as 2 × 50 reads, thus, we restricted our analysis to 50–150 nt around the mutation. ClassSNitch uses a random forest algorithm based on expert classification to identify riboSNitches based on a comparison control and variant using normalized reactivities for each (Woods and Laederach 2017). To mimic a traditional between replicate analysis we compared the full-length data set for all nonchangers with allele-specific nonchangers (false positives) or allele-specific changers (true positives). To obtain the improvement offered by allele-specific sorting, we ratiometrically split each sample into 75% and 25% reads before calculating SHAPE reactivity. This splitting simulates the ratio of mutant to WT RNA modified and sequenced in the experiment, where there are more reads assigned to the mutant than to the WT. We compared these split reads to each other (no differences expected, false positives) and to split reads from manually identified riboSNitches (true positives). We used a ROC curve to quantify the improvement conferred by allele-specific sorting. To obtain the final set of riboSNitches we used allele-specific sorting with ClassSNitch. For riboSNitches within the 3′ UTR of TPT1 we also performed a replicate-based experiment that agreed with the results of high quality allele-specific experiments. We identified five mutants as riboSNitches, all of which were verified by eye and the classSNitch algorithm (Table 1).
Ensemble-based analysis of riboSNitches
We analyzed how the ensemble of RNA structures changed between the WT and mutant based on SHAPE-directed secondary structure predictions. Ensemblerna creates a conformational map of structures based on RNA sequence and visualizes them with multidimensional scaling that incorporates normalized SHAPE data, comparing the WT and mutant within the same space (Woods et al. 2017). Conformational maps were restricted to 50–200 nt of allele-specific data within the mutation, based on the sequencing data. The medoid structures for each cluster were used as representative secondary structure in all instances. All the classSNitch-identified riboSNitches showed differences in their secondary structure ensembles in mutant samples when compared with WT.
RNA stability assay
We measured RNA stability with a doxycycline-inducible construct including GFP upstream of the 3′ UTR of TPT1 in a Tet-Off HEK-293 cell system (Genscript Gene Synthesis, Clontech pTRE-TIGHT) (Ysla et al. 2008). Tet-Off HEK293 cells were plated at a concentration of 150,000 cells/mL in Tet-free media supplemented with 100 μg/mL G418 followed by overnight incubation. Cells were transfected with WT of mutant 3′ UTR-GFP constructs using serum-free DMEM, 50 ng of construct, 350 ng of carrier DNA, and Transit (Mirus Transit). The cells were incubated for 24 h before the addition of 2 μg/mL of doxycycline. We sampled at 2, 4, and 6 h after addition of doxycycline and performed a standard TRIzol extraction to isolate RNA (Clontech Doxycycline, ThermoFisher TRIzol, 5PRIME PhaseLock Heavy, Invitrogen Purelink RNA columns). DNAse digestion and qRT-PCR were performed with the TaqMan Gene Expression Kit (Turbo DNAse, TaqMan Gene Expression kit) and TaqMan Gene Expression Probes from Applied BioSciences. We used probes that detected GAPDH and GFP (ThermoFisher Scientific, eGFP probe Mr04097229_mr and GAPDH probe Hs02758991_g1) on an ABI 7000 sequence detector system (Applied Biosystems). Data were analyzed by normalizing an average of triplicate samples with GAPDH, removing outliers, calculating expression, and comparing doxycycline-treated cells with untreated cells. Where ΔCT = GFP−GAPDH, the relative expression is (−ΔCTdox)2 ÷ (−ΔCTuntreated)2 .
Differences in RNA structure between environmental conditions
We downloaded both PhastCon and PhyloP-scored alignments of 100 vertebrates from the UCSC Table Browser (Karolchik et al. 2004; Siepel et al. 2005). We also downloaded PAR-Clip data for TPT1 and LCP1 from CLIP-db (Yang et al. 2015). Splice sites were defined as in the UCSC gtf files for each transcript (Karolchik et al. 2004). We defined highly similar ex vivo/in vitro regions as those with a correlation higher than 0.75 standard deviations from the mean and dissimilar regions as those with correlation lower than 0.75 standard deviations from the mean (40 nt window). We used bootstrapping to determine whether actual versus expected numbers of splice sites, RNA-binding protein sites, and conserved nucleotides occurred within similar or dissimilar regions of ex vivo/in vitro correlation. Other standard deviation groupings for highly similar or dissimilar nucleotides did not substantially change our correlations.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Lieb, Weeks, and Marzluff laboratories for instrument sharing, Silvia Ramos for advice on qRT-PCR, and Katrina Kutchko for assistance with R and scripting. We also thank Nick Shalosky for cell storage and growth at the Tissue Culture Facility of the UNC Lineberger Comprehensive Cancer Center. This work was supported by the US National Institutes of Health (grant numbers HL111527, GM101237, and HG008133); and the American Cancer Society Lee National Denim Day Postdoctoral Fellowship (PF-15-133-01-RMC) to L.L.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.064469.117.
Freely available online through the RNA Open Access option.
REFERENCES
- 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acunzo J, Baylot V, So A, Rocchi P. 2014. TCTP as therapeutic target in cancers. Cancer Treat Rev 40: 760–769. [DOI] [PubMed] [Google Scholar]
- Amson R, Karp JE, Telerman A. 2013. Lessons from tumor reversion for cancer treatment. Curr Opin Oncol 25: 59–65. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer U-A, Borovjagin AV, Greagg MA, Jeffrey IW, Russell P, Laing KG, Lee M, Clemens MJ. 2002. The mRNA of the translationally controlled tumor protein P23/TCTP is a highly structured RNA, which activates the dsRNA-dependent protein kinase PKR. RNA 8: 478–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer U-A, Iadevaia V, Chen J, Knoch B, Engel M, Proud CG. 2015. Growth-factor dependent expression of the translationally controlled tumour protein TCTP is regulated through the PI3-K/Akt/mTORC1 signalling pathway. Cell Signal 27: 1557–1568. [DOI] [PubMed] [Google Scholar]
- Burdon KP, Sharma S, Chen CS, Dimasi DP, Mackey DA, Craig JE. 2007. A novel deletion in the FTL gene causes hereditary hyperferritinemia cataract syndrome (HHCS) by alteration of the transcription start site. Hum Mutat 28: 742. [DOI] [PubMed] [Google Scholar]
- Chen W, Wang H, Tao S, Zheng Y, Wu W, Lian F, Jaramillo M, Fang D, Zhang DD. 2013. Tumor protein translationally controlled 1 is a p53 target gene that promotes cell survival. Cell Cycle 12: 2321–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Cai Q, He W, Li Z, Zhou F, Liu Z, Zhong G, Chen X, Zhao Y, Dong W, et al. 2016. An NKX3.1 binding site polymorphism in the l-plastin promoter leads to differential gene expression in human prostate cancer. Int J Cancer 138: 74–86. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Chou FC, Kladwang W, Tian S, Cordero P, Das R. 2015. Consistent global structures of complex RNA states through multidimensional chemical mapping. eLife 4: e07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Chalamish S, Hasson A, Weinberg D, Namer LS, Banai Y, Osman F, Kaempfer R. 2009. Dynamic refolding of IFN-γ mRNA enables it to function as PKR activator and translation template. Nat Chem Biol 5: 896–903. [DOI] [PubMed] [Google Scholar]
- Cordero P, Lucks JB, Das R. 2012. An RNA Mapping DataBase for curating RNA structure mapping experiments. Bioinformatics 28: 3006–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley M, Solem A, Qu K, Chang HY, Laederach A. 2015. Detecting riboSNitches with RNA folding algorithms: a genome-wide benchmark. Nucleic Acids Res 43: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, Maskel GS, Seifert S, Mochrie SG, Thiyagarajan P, et al. 2003. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J Mol Biol 332: 311–319. [DOI] [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci 106: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo C, Bartholomaus A, Fedyunin I, Ignatova Z. 2015. Secondary structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet 11: e1005613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. 2012. Functional complexity and regulation through RNA dynamics. Nature 482: 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. 2014. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505: 696–700. [DOI] [PubMed] [Google Scholar]
- Donahue CP, Yadava RS, Nesbitt SM, Fedor MJ. 2000. The kinetic mechanism of the hairpin ribozyme in vivo: influence of RNA helix stability on intracellular cleavage kinetics. J Mol Biol 295: 693–707. [DOI] [PubMed] [Google Scholar]
- Doshi KJ, Cannone JJ, Cobaugh CW, Gutell RR. 2004. Evaluation of the suitability of free-energy minimization using nearest-neighbor energy parameters for RNA secondary structure prediction. BMC Bioinformatics 5: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. 2015. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 43: D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Lewis NJ, Marzluff WF. 1996. The histone 3′-terminal stem–loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res 24: 1954–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garge N, Pan H, Rowland MD, Cargile BJ, Zhang X, Cooley PC, Page GP, Bunger MK. 2010. Identification of quantitative trait loci underlying proteome variation in human lymphoblastoid cells. Mol Cell Proteomics 9: 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V, Gartner JJ, Qutob N, Elnitski L, Samuels Y. 2015. The functional relevance of somatic synonymous mutations in melanoma and other cancers. Pigment Cell Melanoma Res 28: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B, Gaestel M, Böhm H, Bielka H. 1989. cDNA sequence coding for a translationally controlled human tumor protein. Nucleic Acids Res 17: 8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. 2013. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci 110: 5498–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A. 2010. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet 6: e1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Bevilacqua PC. 2016. Discriminating self and non-self by RNA: roles for RNA structure, misfolding, and modification in regulating the innate immune sensor PKR. Acc Chem Res 49: 1242–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt RC, Simhadri VL, Iandoli M, Sauna ZE, Kimchi-Sarfaty C. 2014. Exposing synonymous mutations. Trends Genet 30: 308–321. [DOI] [PubMed] [Google Scholar]
- Incarnato D, Neri F, Anselmi F, Oliviero S. 2014. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol 15: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarifar F, Dietrich RC, Hiznay JM, Padgett RA. 2014. Biochemical defects in minor spliceosome function in the developmental disorder MOPD I. RNA 20: 1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Yang Y, Zhang P. 2011. New insights into RNA secondary structure in the alternative splicing of pre-mRNAs. RNA Biol 8: 450–457. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature 467: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin MA, Gerstein M. 2016. Role of non-coding sequence variants in cancer. Nat Rev Genet 17: 93–108. [DOI] [PubMed] [Google Scholar]
- Kladwang W, Cordero P, Das R. 2011a. A mutate-and-map strategy accurately infers the base pairs of a 35-nucleotide model RNA. RNA 17: 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, VanLang CC, Cordero P, Das R. 2011b. A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat Chem 3: 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota M, Tran C, Spitale RC. 2015. Progress and challenges for chemical probing of RNA structure inside living cells. Nat Chem Biol 11: 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchko KM, Sanders W, Ziehr B, Phillips G, Solem A, Halvorsen M, Weeks KM, Moorman N, Laederach A. 2015. Multiple conformations are a conserved and regulatory feature of the RB1 5′ UTR. RNA 21: 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D, Proctor JR, Meyer IM. 2013. On the importance of cotranscriptional RNA structure formation. RNA 19: 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Flynn RA, Kadina A, Guo JK, Kool ET, Chang HY. 2017. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA 23: 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire PA, Anderson E, Lary J, Cole JL. 2008. Mechanism of PKR activation by dsRNA. J Mol Biol 381: 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kazan H, Lipshitz HD, Morris QD. 2014. Finding the target sites of RNA-binding proteins. Wiley Interdiscip Rev RNA 5: 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell M, Brannvall M, Wagner EG, Kirsebom LA. 2005. Lead(II) cleavage analysis of RNase P RNA in vivo. RNA 11: 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Chang HY. 2016. Decoding the RNA structurome. Curr Opin Struct Biol 36: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y-F, Mauger DM, Goldstein DB, Urban TJ, Weeks KM, Bradrick SS. 2015. IFNL3 mRNA structure is remodeled by a functional non-coding polymorphism associated with hepatitis C virus clearance. Sci Rep 5: 16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. 2011. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc Natl Acad Sci 108: 11063–11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Haldar S, Khan MA, Sharma SD, Merrick WC, Theil EC, Goss DJ. 2012. Fe2+ binds iron responsive element-RNA, selectively changing protein-binding affinities and regulating mRNA repression and activation. Proc Natl Acad Sci 109: 8417–8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahen EM, Watson PY, Cottrell JW, Fedor MJ. 2010. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol 8: e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche L, Green SR, Schmedt C, Mathews MB. 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol 12: 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JS, Halvorsen M, Davis-Neulander L, Ritz J, Gopinath C, Beauregard A, Laederach A. 2012. Structural effects of linkage disequilibrium on the transcriptome. RNA 18: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940. [DOI] [PubMed] [Google Scholar]
- McManus CJ, Graveley BR. 2011. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev 21: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Gerger A, Zhang W, Hanna DL, Yang D, Winder T, Wakatsuki T, Labonte MJ, Stintzing S, Volz N, et al. 2014. Plastin polymorphisms predict gender- and stage-specific colon cancer recurrence after adjuvant chemotherapy. Mol Cancer Ther 13: 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum JM, Gunnery S, Mathews MB. 2002. The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res 30: 1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Jarrous N, Ben-Asouli Y, Kaempfer R. 1999. A cis-acting element in the 3′-untranslated region of human TNF-α mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev 13: 3280–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollom E, Dang KK, Potter EL, Gorelick RJ, Burch CL, Weeks KM, Swanstrom R. 2013. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog 9: e1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz J, Martin JS, Laederach A. 2012. Evaluating our ability to predict the structural disruption of RNA by SNPs. BMC Genomics 4(13 Suppl): S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler LE, Kosmyna B, Moskowitz D, Bebawee R, Rahimzadeh J, Kutchko K, Laederach A, Notarangelo LD, Giliani S, Bouhassira E, et al. 2014. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage–hair hypoplasia. Hum Mol Genet 23: 368–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. 2014. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505: 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone SA, Rocca-Serra P, Field D, Maguire E, Taylor C, Hofmann O, Fang H, Neumann S, Tong W, Amaral-Zettler L, et al. 2012. Toward interoperable bioscience data. Nat Genet 44: 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Childs L, Walther D, Bock R. 2011. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet 7: e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, Barta A, Semrad K. 2004. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol 5: 908–919. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Spiridonov NA, Kashina A. 2013. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res 41: 2073–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR. 2013. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 34: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day INM, Gaunt TR, Campbell C. 2015. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 31: 1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya H. 2012. Plastin family of actin-bundling proteins: its functions in leukocytes, neurons, intestines, and cancer. Int J Cell Biol 2012: 213492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Calabrese JM, Weeks KM. 2015a. Detection of RNA–protein interactions in living cells with SHAPE. Biochemistry 54: 6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. 2015b. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc 10: 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. 2016. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci 113: 10322–10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem AC, Halvorsen M, Ramos SB, Laederach A. 2015. The potential of the riboSNitch in personalized medicine. Wiley Interdiscip Rev RNA 6: 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung J-W, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. 2015. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519: 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukosd Z, Swenson MS, Kjems J, Heitsch CE. 2013. Evaluating the accuracy of SHAPE-directed RNA secondary structure predictions. Nucleic Acids Res 41: 2807–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. 2014. Synonymous mutations frequently act as driver mutations in human cancers. Cell 156: 1324–1335. [DOI] [PubMed] [Google Scholar]
- Thébault S, Agez M, Chi X, Stojko J, Cura V, Telerman SB, Maillet L, Gautier F, Billas-Massobrio I, Birck C, et al. 2016. TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL. Sci Rep 6: 19725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. 2010. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods 7: 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Audenhove I, Denert M, Boucherie C, Pieters L, Cornelissen M, Gettemans J. 2016. Fascin rigidity and L-plastin flexibility cooperate in cancer cell invadopodia and filopodia. J Biol Chem 291: 9148–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. 2014. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505: 706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Abbott TR, Lucks JB. 2016a. Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Res 44: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Strobel EJ, Yu AM, Lis JT, Lucks JB. 2016b. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat Struct Mol Biol 23: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616. [DOI] [PubMed] [Google Scholar]
- Woods CT, Laederach A. 2017. Classification of RNA structure change by “gazing” at experimental data. Bioinformatics 33: 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CT, Lackey L, Williams B, Dokholyan NV, Gotz D, Laederach A. 2017. Comparative visualization of the RNA suboptimal conformational ensemble in vivo. Biophys J 113: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. 2006. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci 103: 6190–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-CT, Di C, Hu B, Zhou M, Liu Y, Song N, Li Y, Umetsu J, Lu ZJ. 2015. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genomics 16: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ysla RM, Wilson GM, Brewer G. 2008. Chapter 3. Assays of adenylate uridylate-rich element-mediated mRNA decay in cells. Methods Enzymol 449: 47–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemora G, Waldsich C. 2010. RNA folding in living cells. RNA Biol 7: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Bevilacqua PC. 2004. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA 10: 1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, Cao K, Wang L-S, Gregory BD. 2010. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet 6: e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. 2017. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods 14: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.