Abstract
Analysis of chromatin accessibility can reveal transcriptional regulatory sequences, but heterogeneity of primary tissues poses a significant challenge in mapping the precise chromatin landscape in specific cell types. Here we report single-nucleus ATAC-seq, a combinatorial barcoding-assisted single-cell assay for transposase-accessible chromatin that is optimized for use on flash-frozen primary tissue samples. We apply this technique to the mouse forebrain through eight developmental stages. Through analysis of more than 15,000 nuclei, we identify 20 distinct cell populations corresponding to major neuronal and non-neuronal cell types. We further define cell-type-specific transcriptional regulatory sequences, infer potential master transcriptional regulators and delineate developmental changes in forebrain cellular composition. Our results provide insight into the molecular and cellular dynamics that underlie forebrain development in the mouse and establish technical and analytical frameworks that are broadly applicable to other heterogeneous tissues.
Transcriptional regulatory elements in the genome (cis-regulatory elements) play fundamental roles in development and disease1,2. Analysis of chromatin accessibility in primary tissues using assays such as DNase-seq3,4 and ATAC-seq5,6 has identified millions of candidate cis-regulatory elements in the human and mouse genomes2,7. However, we still lack precise information about the cis-regulatory elements in specific cell types, because previous experiments performed on heterogeneous tissue samples yielded an ensemble average signal from multiple constituent cell types. In some cases, specific cell types can be isolated from heterogeneous tissues using protein markers6,8–10, but a more general strategy is needed to enable the study of cell-type-specific gene regulation on a larger scale.
In theory, single-cell-based chromatin accessibility studies can be used for unbiased identification of subpopulations in a heterogeneous biological sample and for identification of the regulatory elements active in each subpopulation. Indeed, proof of principle has been reported using cultured mammalian cells and cryopreserved blood cell types11–13. However, to make these approaches more widely applicable, it is necessary to optimize them for primary tissues. One major difficulty in working with primary tissues is that they are typically preserved by flash freezing, which is not amenable to the isolation of intact single cells. Here we show that it is possible to isolate single nuclei from frozen tissues and assay chromatin accessibility in these nuclei in a massively parallel manner.
Results
Method optimization and computational analysis framework
We adopted a combinatorial barcoding assisted single cell ATAC-seq strategy12 and optimized it for frozen tissue samples (see Methods). Compared to previous reports12, key modifications were made to maximally preserve nuclei integrity during sample processing and optimize transposase-mediated fragmentation of chromatin in individual nuclei (Supplementary Figs. 1–3). We applied this modified protocol, hereafter referred to as snATAC-seq (single-nucleus ATAC-seq), to forebrain tissue from 8-week-old adult mice (postnatal day (P) 56) and from mouse embryos at seven developmental stages from embryonic day (E) 11.5 to birth (P0; Fig. 1a,b). DNA libraries were sequenced to near-saturation as indicated by a read duplication rate of 36–73% per sample (Supplementary Table 1). The barcode collision rate, which assesses the probability of two nuclei sharing the same barcode combination, was ~16%, slightly higher than expected and reported before (Supplementary Fig. 3c)12. We filtered out low-quality datasets using three stringent quality-control criteria including read depth (Supplementary Fig. 3d), recovery rate of constitutively accessible promoters in each nucleus (Supplementary Fig. 3e) and signal-over-noise ratio estimated by fraction of reads in peak regions (Supplementary Fig. 3f and Methods). In total, 15,767 high-quality snATAC-seq datasets were obtained. The median read depth per nucleus ranged from 9,275 to 18,397, with the median promoter coverage at 11.6% and the median fraction of reads in peak regions at 22% (Supplementary Tables 2 and 3). Our protocol maintains the extraordinary scalability of combinatorial indexing, while featuring a ~6-fold increase in read depth per nucleus compared to previous reports (Supplementary Table 3). The high quality of the single-nucleus chromatin accessibility maps was supported by strong concordance between the aggregate snATAC-seq data and bulk ATAC-seq data (R > 0.9) and excellent reproducibility between independent snATAC-seq experiments (R > 0.91; Fig. 1c and Supplementary Fig. 4).
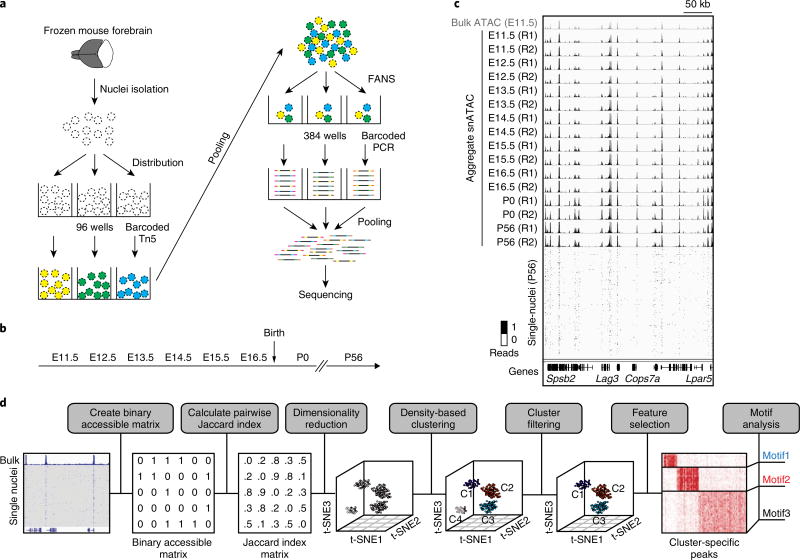
Fig 1. Overview of the experimental and computational procedures of snATAC-seq.
a, Following nuclei isolation from frozen forebrain tissue biopsies, tagmentation of 4,500 permeabilized nuclei was carried out using barcoded Tn5 in 96-well plates. After pooling, 25 nuclei were sorted into each well of a 384-well plate, and PCR was carried out to introduce the second set of barcodes. FANS, fluorescence-assisted nuclei sorting. b, Overview of the developmental timepoints examined in the current study. E, embryonic; P, postnatal. c, Chromatin accessibility profiles of aggregate snATAC-seq (black tracks) agree with bulk ATAC-seq (gray, top track) and are consistent between independent experiments. R1, replicate 1; R2, replicate 2. d, Framework of computational analysis of snATAC-seq data.
The snATAC-seq profiles from each forebrain tissue arise from a mixture of distinct cell types. Enhancer regions are well known to display cell-type-dependent chromatin accessibility14, and are more effective at classifying cell types than promoters or transcriptomic data11 (Supplementary Fig. 5a,b). Thus, we focused on transcriptional start sites (TSS)-distal accessible chromatin regions (defined as all genomic elements outside a 2-kb window upstream from the TSS), corresponding to putative enhancers, to group individual nucleus profiles into distinct cell types. We developed a computational framework to uncover distinct cell types from the snATAC-seq datasets without requiring prior knowledge (Methods). First, we determined the open chromatin regions from the bulk ATAC-seq profiles of mouse forebrain tissue at seven fetal development timepoints and at maturity, resulting in a total of 140,103 TSS-distal elements (Fig. 1d, Methods and Supplementary Table 4). Next, we constructed a binary accessibility matrix of open chromatin regions, using 0 or 1 to indicate the absence or presence of a read at each open chromatin region in each nucleus (Fig. 1d). We then calculated the pairwise similarity between cells using a Jaccard index and applied a nonlinear dimensionality reduction method, t-SNE15, to project the Jaccard index matrix to a low-dimension space (Fig. 1d)16. The final t-SNE plot depicts cell types as distinct clusters in a three-dimensional space (Fig. 1d).
Identification of forebrain cell types from snATAC-seq profiles
We applied this computational framework first to 3,033 high-quality snATAC-seq profiles obtained from the adult forebrain (Fig. 2a and Supplementary Table 2). As a negative control, we included 200 ‘shuffled’ nuclear profiles (Supplementary Fig. 5c,d and Online Methods). This analysis revealed 10 total clusters. As expected, the shuffled nuclei formed a distinct cluster with low intracluster similarity. In addition, one other cluster showing low intracluster similarity likely represented low-quality nuclei or accessibility profiles resulting from barcode collision events (Supplementary Fig. 3c). After eliminating these nuclei, we determined eight distinct cell-type clusters from the adult forebrain (Fig. 2a and Supplementary Fig. 5c,d). Notably, the clustering results were highly reproducible for two independent experiments (Supplementary Fig. 5e,f).
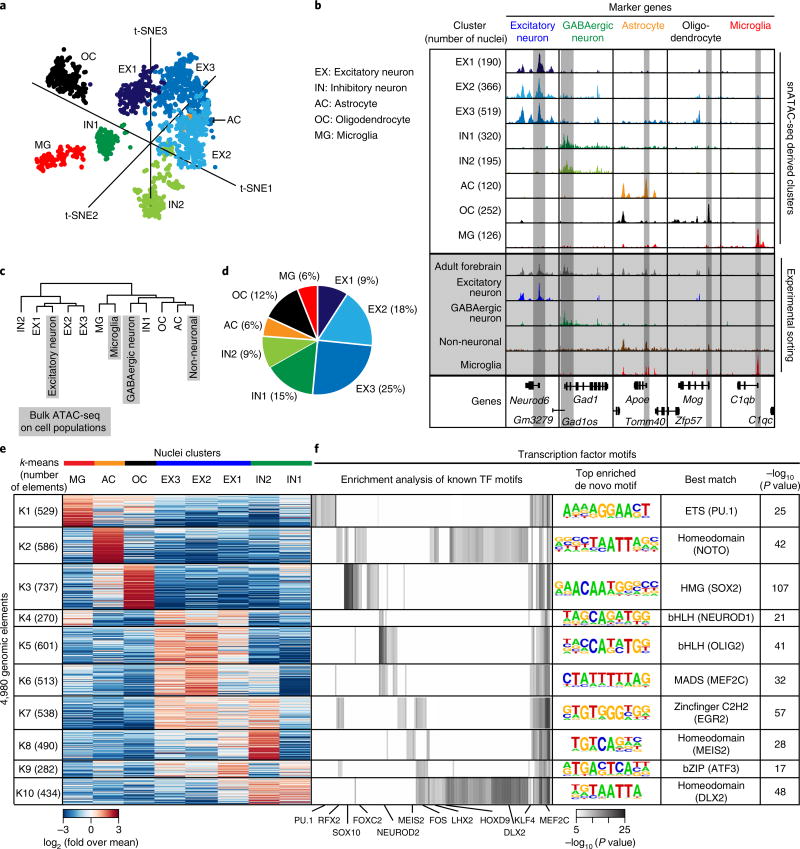
Fig 2. Delineation of cell types in the P56 mouse forebrain and identification of potential master regulators of each cell type.
a, Clustering of single nuclei from both experiments revealed eight different cell groups in adult forebrain. b, Aggregate chromatin accessibility profiles for each cell cluster and the bulk ATAC-seq for the sorted cell populations or the whole forebrain at several marker gene loci (bulk data are shaded in gray). c, Hierarchical clustering of aggregate snATAC-seq data and bulk ATAC-seq datasets. d, Cellular composition of adult forebrain derived from snATAC-seq data. e, k-means clustering of 4,980 genomic elements based on chromatin accessibility. f, Enrichment analysis for transcription factor motifs in each cell group. For enrichment of known motifs, one-tailed Fisher’s exact test was used to calculate significance49. Displayed P values are Bonferroni corrected for multiple testing. For de novo motif enrichment testing, a hypergeometric test was used50. Displayed P values are not corrected for multiple testing.
To categorize each cluster, we generated aggregate chromatin accessibility maps for each cluster and examined the patterns of chromatin accessibility at known cell type marker genes. We found three clusters with chromatin accessibility at Neurod6 and other excitatory neuron-specific genes17 (clusters EX1–3; Fig. 2b and Supplementary Fig. 6a); two clusters with accessibility at the gene locus of Gad1 likely representing inhibitory neurons (clusters IN1 and 2; Fig. 2b and Supplementary Fig. 6a)18; one cluster with accessibility at the Apoe locus and other known astroglia markers19 (cluster AC; Fig. 2b); one cluster with accessibility at the Mog gene locus and other oligodendrocyte marker genes20 (cluster OC; Fig. 2b); and one microglia cluster with accessibility at genes encoding complement factors including the gene C1qb (cluster MG; Fig. 2b and Supplementary Fig. 6c–e)21. We also compared the aggregate chromatin accessibility maps for each cluster to previously published maps from sorted excitatory neurons6, GABAergic neurons8, microglia21 and NeuN-negative nuclei (which mostly comprise non-neuronal cells including astrocytes and oligodendrocytes22; Fig. 2b and Supplementary Fig. 7a–c). Consistent with the accessibility patterns at marker gene loci, we observed that clusters EX1–3 were highly similar to sorted excitatory neurons. To further characterize the distinct excitatory neuron clusters, we compared EX1–3 with published bulk ATAC-seq data from different cortical layers and from dentate gyrus8,23. Notably, we found that EX1 and EX3 were more similar to upper and lower cortical layers, respectively, whereas EX2 showed properties of dentate gyrus neurons (Supplementary Fig. 8a). Cluster IN1 was highly similar to sorted cortical GABAergic neurons. Unexpectedly, IN2 was more similar to sorted excitatory neurons than cortical GABAergic neurons. Distinctions between the inhibitory neuron clusters (IN1 and IN2) were not clear at this stage, but came into focus later when we analyzed transcription factor (TF) motifs enriched in the accessible chromatin regions (described below). Clusters OC and AC resembled sorted NeuN-negative cells, and cluster MG is similar to sorted microglia (Fig. 2b,c).
According to our snATAC-seq data, the adult mouse forebrain consists of 52% excitatory neurons, 24% inhibitory neurons, 12% oligodendrocytes and 6% astrocytes and microglia, respectively (Fig. 2d). Since cell type proportions vary between different forebrain regions, for example, cortex and hippocampus17, the percentages derived from snATAC-seq represent an average of all forebrain regions (Fig. 2e and Supplementary Fig. 7d,e). The predominance of neuronal nuclei derived from adult forebrain tissue was confirmed by flow cytometry analysis stained against the postmitotic neuron marker NeuN22 (Fig. 2e and Supplementary Figs. 6b and 7b,e).
Delineation of the cis-regulatory landscape of specific cell types in the adult forebrain
The power of snATAC-seq is not simply to delineate cell types but also to reveal the cis-regulatory landscape within each cell type. To this end, we calculated the cell-type-specificity of each putative cis-regulatory element (i.e., chromatin accessibility region) using a Shannon entropy index (Supplementary Fig. 9). As expected, proximal promoter elements were accessible in more cell types, while the distal enhancer elements showed significantly higher cell-type-specificity (median value of 4.2% for proximal elements vs. 0.4% for distal elements; Supplementary Fig. 9a,b,d). We next developed a feature selection method (see Methods) to identify the subset of elements that could best distinguish the eight cell type clusters from each other. This approach identified 4,980 elements showing clear cell-type-dependent accessibility (Fig. 2e). To gain insight into the key transcriptional regulators and pathways active in each cell type, we performed k-means clustering followed by motif enrichment analysis for these genomic elements (Fig. 2e,f, Supplementary Fig. 9d and Supplementary Table 4). For each cell type, we observed an enrichment of binding motifs corresponding to key TFs (Fig. 2f). For example, the binding motif for ETS-factor PU.1 was enriched in MG elements24, motifs for SOX proteins were enriched in OC elements25, bHLH motifs were enriched in EX1–3 elements and DLX homeodomain factor motifs were enriched in IN elements (Fig. 2f)26. Moreover, this analysis revealed an important difference between the inhibitory neuron clusters IN1 and IN2. We found that a binding motif for MEIS factors was enriched in a subset of elements specific to IN2. Previous reports showed that MEIS2 plays a major role in generation of medium spiny neurons, the main GABAergic neurons in the striatum27. Accordingly, we identified gene loci of Ppp1r1b and Drd1, which encode markers of medium spiny neurons, as highly accessible in IN2 but not IN1 (Supplementary Fig. 10)27. These data suggest that IN2 may represent medium spiny neurons, while IN1 could represent a distinct class of GABAergic neurons. We also identified motifs that were differentially enriched between EX1, EX2 and EX3. Notably, regions specific for EX1 and 3 were enriched for motifs from the Forkhead family, and EX2 was enriched for motifs recognized by MEF2C (Supplementary Fig. 8c and Supplementary Table 4), which has been shown to play an important role in hippocampus mediated memory28. A comparison with data from cell-type-specific differentially methylated regions identified by single cell DNA-methylation analysis of neurons showed that both methods were able to identify inhibitory and excitatory neuron specific elements (Supplementary Fig. 11)29.
Profiling embryonic forebrain development using snATAC-seq
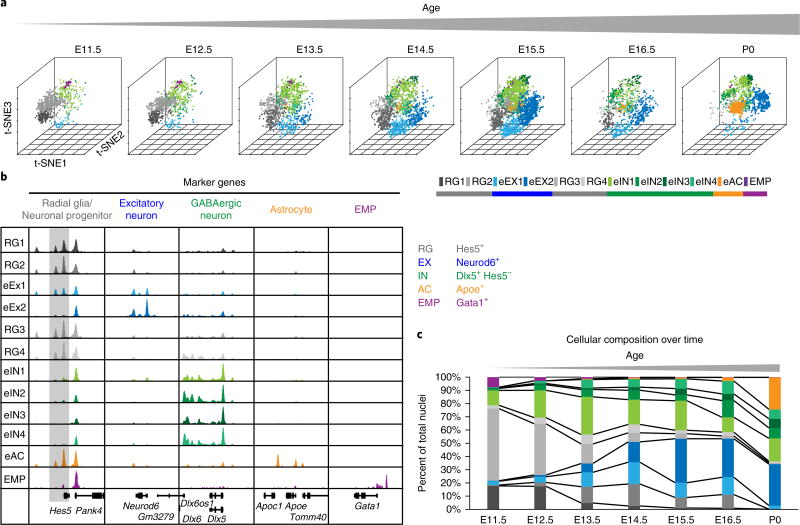
We next extended our framework by analyzing the snATAC-seq profiles derived from fetal mouse forebrains at seven developmental stages (Fig. 1b), seeking to reveal developmental dynamics of transcriptional regulation at the cellular level. The developmental stages examined cover key events from the onset of neurogenesis to gliogenesis30. From 12,733 high-quality snATAC-seq profiles, we identified 12 distinct subpopulations (Fig. 3a) that exhibited changes in abundance through development (Fig. 3a–c). This broad cell-type classification allowed us to profile the dynamic cis-regulatory landscape of forebrain development. Based on chromatin accessibility profiles at gene loci of known marker genes, we assigned these cell populations to radial glia, excitatory neurons, inhibitory neurons, astrocytes and erythromyeloid progenitors (EMP; Fig. 3b)24,31. Notably, the EMP cluster was restricted to E11.5, whereas the astrocyte cluster was present after E16.5 and expanded dramatically around birth (Fig. 3b,c)24,30, highlighting two developmental processes: invasion of myeloid cells into the brain before neurogenesis and gliogenesis succeeding neurogenesis after E16.530. Mature excitatory neurons (eEX2) were indicated by increased accessibility at Neurod6, which encodes a postmitotic neuron marker, and absence of signal at the Hes5 gene, which encodes a Notch effector and a marker gene for neuronal progenitors (Fig. 3b,c)30,31. This cell type expanded in abundance between E12.5 and E13.5 and followed the emergence of early differentiating neurons (eEX1; Fig. 3b,c). Notably, inhibitory-neuron-like cells were already present at E11.5 (Fig. 3b).
Fig 3. snATAC-seq analysis reveals the timing of neurogenesis and gliogenesis during embryonic forebrain development.
a, Clustering of single nuclei from both independent experiments revealed 12 different cell groups with changing relative abundance. b, Aggregate chromatin accessibility profiles for cell clusters and at marker gene loci used to assign cell types. For better visualization, the Hes5 gene locus is shaded gray. c, Quantification of cellular composition during forebrain development.
Identification of lineage-specific transcriptional regulators during embryonic forebrain development
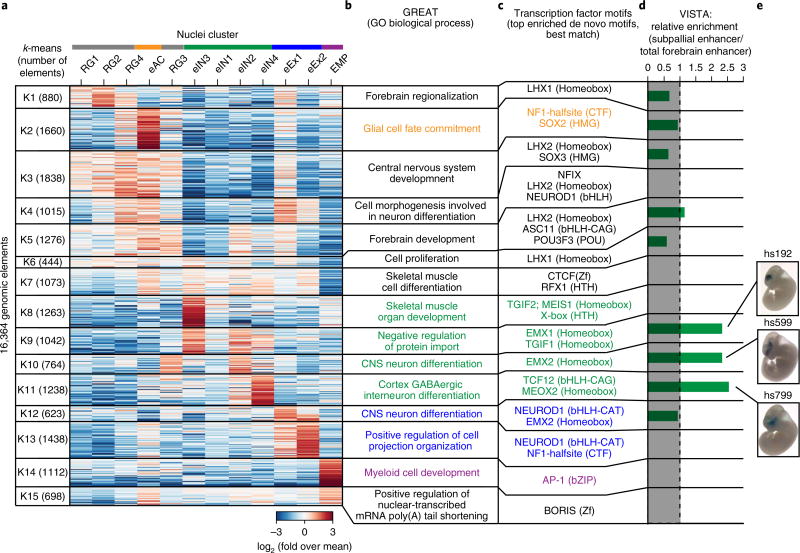
To identify the transcriptional regulatory sequences in each subpopulation, we identified 16,364 genomic elements that showed cell-population-specific chromatin accessibility and best separated the cell subpopulations (Fig. 4a and Supplementary Table 4). To further characterize these elements, we performed motif enrichment analysis and gene ontology analysis of each cluster using GREAT32. Our analysis showed that genomic elements that were mostly associated with radial glia like cell groups (RG1–4; Fig. 4a) fell into regulatory regions of genes involved in early forebrain developmental processes, including forebrain regionalization (Fig. 4b; K1), central nervous system development (Fig. 4b; K3) or forebrain development (Fig. 4b; K5). These elements were enriched for homeobox motifs corresponding to LHX-transcription factors including LHX2 (Fig. 4c; K1, K3 and K5), which is critical for generating correct neuron numbers by regulating proliferation of neural progenitors33 and for temporally promoting neurogenesis over astrogliagenesis34. Notably, one of these clusters was also enriched for both the proneural bHLH transcription factor ASCL1 (Mash1) and its co-regulator POU3F3 (Brn1; Fig. 4c; K5)35. ASCL1 is required for normal proliferation of neural progenitor cells36 and implicated in a DLX1/2-associated network that promotes GABAergic neurogenesis37. In line with this, associated genomic elements were also accessible in one inhibitory neuron cluster (eIN2; Fig. 4c; K5).
Fig 4. snATAC-seq analysis uncovers cis-regulatory elements and transcriptional regulators of lineage specification in the developing forebrain.
a, A heat map shows the results of k-means clustering of 16,364 candidate cis-regulatory elements based on chromatin accessibility in different cell types. b, Gene ontology analysis of each cell type using GREAT32. c, Transcription factor motifs enriched in each group50. d, Enrichment of enhancers that were functionally validated as part of the VISTA database45. e, Representative images of transgenic mouse embryos showing LacZ reporter gene expression under control of the indicated subpallial enhancers. Pictures were downloaded from the VISTA database45.
We also identified transcriptional regulators that were specifically associated either with neurogenesis or gliogenesis during forebrain development. For example, the early astrocyte (eAC)-specific elements were located in open chromatin regions near genes involved in glia cell fate commitment, and the top enriched transcription factor motif was NF1-halfsite (Fig. 4a–c; K2). Previous studies show that NF1 transcription factor NF1A alone is capable of specifying glia cells to the astrocyte lineage25. NFIX is another NF1 family member with proneural function38. This motif was enriched together with the bHLH transcription factor NEUROD1, binding sites mainly in open chromatin regions found in the excitatory neuron cell population (Fig. 4c; K4, K12, K13)31. Based on chromatin accessibility profiles at marker gene loci, we had previously assigned two cell clusters to the excitatory neuron lineage (eEX1 and eEX2; Fig. 3b). Compared to cluster eEX2, eEX1 showed increased accessibility at both radial glia associated open chromatin (Figs. 3b and 4a; K4) and chromatin regions associated with CNS neuron differentiation (Fig. 4a; K12). In addition, eEX1 nuclei preceded the emergence of eEX2 nuclei during development (Fig. 3c). These findings indicated that eEX1 might represent a transitional state during excitatory neuron differentiation.
The bHLH transcription factor family consists of several subfamilies that recognize different DNA motifs39. NEUROD1 belongs to a subfamily of transcription factors that bind to a central CAT motif, whereas other transcription factors, such as TCF12, preferentially bind to a CAG motif39. Our snATAC-seq profiles revealed an enrichment of the TCF12-binding motif in regions associated with cortex GABAergic interneuron differentiation, in contrast to the excitatory neuron associated enrichment for NEUROD1 (Fig. 4a–c; K4, K11–K13)26,31,40. Analysis of specific genomic elements of the inhibitory neuron cluster eIN3 showed a notable bias in proximity to genes associated with ‘skeletal muscle and organ development’ (Fig. 4a,b; K8). More detailed analysis revealed that the underlying genes Mef2c/d and Foxp1/2, as well as Drd2/3, encode transcription factors and dopamine receptors, indicating differentiating striatal medium spiny neurons41,42. This finding was consistent with the enrichment for MEIS-homeodomain factors in these regions (Fig. 4c; K8) comparable to the medium spiny neuron cluster in adult forebrain (Fig. 2e,f (K8) and Supplementary Fig. 10). Further, genomic elements specific to the EMP cluster were associated with genes involved in myeloid cell development (Fig. 4a–c; K14) and enriched for motifs of the ubiquitous AP-1 transcription factor complexes that have been described as playing a role in shaping the enhancer landscape of macrophages43.
Finally, we attempted to identify developmental dynamics of elements within each cell cluster (Supplementary Fig. 11). Our analysis revealed between 41 and 2,114 dynamic genomic elements for each cell type (Supplementary Fig. 12c–g). Regions that are more accessible after birth (P0) compared to early timepoints were enriched for the RFX1 motif in GABAergic neurons, including the cluster eIN1 as well as the excitatory neuron cluster eEX2 (Supplementary Fig. 12d,e), indicating a general role of the evolutionarily conserved RFX factors in perinatal adaptation of brain cells. Several family members, including RFX1, are expressed in the brain and have been implicated in regulating cilia, for example, in sensory neurons44.
Functional and anatomical annotation of identified candidate cis-regulatory elements
While assessment of open chromatin plays an important role in predicting regulatory elements in the genome2,7, it does not provide direct information of functional activity. To address this point, we asked whether cluster-specific transposase-accessible chromatin in the embryonic forebrain overlaps with genomic elements tested in reporter assays to validate enhancer activity in mouse embryonic forebrain in vivo45. First, we focused our analysis on all genomic elements with validated functional activity in the forebrain and on a subset shown to be active only in the subpallium46,47. The subpallium is a brain region that gives rise to GABAergic and cholinergic neurons46. In total, 63.1% (275 of 436) of all forebrain enhancers and 64.8% (59 of 91) of subpallial enhancers were represented in our subset of genomic elements, indicating a high degree of sensitivity. Next, we calculated the relative enrichment of subpallial enhancers over total forebrain enhancers for each cluster. Notably, subpallial enhancers were only enriched in clusters K9–11, which were assigned to the GABAergic neuron lineage (Fig. 4d,e and Supplementary Fig. 13). Next, we found that elements mainly accessible in radial glia cells were active in pallial regions (K1, 3 and 4; Fig. 4a and Supplementary Fig. 13). Unexpectedly, elements of cluster K5 were active in dorsal and lateral pallial regions, as well as in the lateral ganglionic eminence, indicating conserved roles for these genomic elements in a wide variety of regions in the developing forebrain (Fig. 4a and Supplementary Fig. 13). Integration of genomic elements identified by snATAC-seq in specific cell clusters with transgenic enhancer assays confirmed the high specificity and sensitivity of snATAC-seq in identifying cell populations and their underlying regulatory elements.
Discussion
Tissue heterogeneity has been a significant hurdle in the dissection of gene regulatory programs driving mammalian development. While single-cell-based analysis of chromatin accessibility has been reported, a major challenge lies in the published methods’ requirement for fresh cell populations, whereas most biological samples in tissue banks are either frozen or in formalin fixed paraffin embedded blocks. We report here a general approach (snATAC-seq) and a computational framework that can be used to dissect cellular heterogeneity and delineate cell-type-specific gene regulatory sequences in snap-frozen primary tissues. We applied snATAC-seq to heterogeneous forebrain tissue from adult and embryonic mice and resolved specific cell types in these samples. Similarly to other approaches, such as single-cell RNA-seq17,48 and single-cell DNA-methylation analysis29, snATAC-seq can be used to identify cell types de novo in heterogeneous tissue, facilitating generation of cell atlases in the brain and other tissues. In addition, snATAC-seq catalogs the candidate enhancers for each cell type, enabling the dissection of gene regulatory programs without the need to purify specific cell types. As such, this method is particularly suitable for studying cell populations in complex tissues where cellular surface markers are not available. The current framework allows analysis of major cell types with a relative abundance of at least 5%, as shown for microglia in the adult forebrain. It is expected that increasing the number of cells profiled per experiment will linearly increase the sensitivity of cell-type detection. Indeed, the presented combinatorial barcoding protocol can be scaled up to > 5,000 high-quality nuclei per experiment simply by working in 384-well plates rather than 96-well plates. Increasing the number of barcodes during tagmentation will also help to lower the final barcode collision rate without limiting the throughput12.
Through integrative analysis of single-nuclei chromatin accessibility profiles, we tracked changes in the relative proportions of these cell types during development, identified putative regulatory elements active within each cell type and used those regulatory elements to reveal key TFs in specific forebrain cell types. Therefore, our results provide a unique view of the cell-type-specific cis-regulatory landscape in the forebrain. We expect that with larger cell numbers in the future it will be possible to uncover previously unknown regulatory elements in rare cell types. Moreover, applying snATAC-seq to human tissue samples and integration with genomic variants may reveal relative contributions of distinct cell types to diseases like schizophrenia or Alzheimer’s. We anticipate that our snATAC-seq approach will be a valuable tool for analysis of other brain regions and non-neuronal tissues and will help to pave the way to a better understanding of mammalian developmental programs.
Methods
Mouse tissues
All animal experiments were approved by the Lawrence Berkeley National Laboratory Animal Welfare and Research Committee or the University of California, San Diego, Institutional Animal Care and Use Committee. Forebrains from embryonic mice (E11.5–E16.5) and early postnatal mice (P0) were dissected from one pregnant female or one litter at a time and combined. For breeding, animals were purchased from Charles River Laboratories (C57BL/6NCrl strain) or Taconic Biosciences (C57BL/6NTac strain) for E14.5 and P0. Breeding animals for other timepoints were received from Charles River Laboratories (C57BL/6NCrl). Dissected tissues were flash frozen in a dry ice ethanol bath. For the adult time point (P56), forebrains from 8-week old male C57BL/6NCrl mice (Charles River Laboratories) were dissected and flash frozen in liquid nitrogen separately. Tissues were pulverized in liquid nitrogen using pestle and mortar. For each timepoint, two replicates were processed (n = 2 per timepoint).
Transposome generation
To generate A/B transposomes, A and B oligos were annealed to common pMENTs oligos (95 °C for 2 min, cooled to 14 °C at a cooling rate of 0.1 °C/s) separately (Supplementary Table 5). Next, barcoded transposons were mixed in a 1:1 molar ratio with unloaded transposase Tn5, which was generated at Illumina. Mixture was incubated for 30 min at room temperature (20–25 °C, with an average of 23 °C). Finally, A and B transposomes were mixed. For combinatorial barcoding, we used eight different A transposons and 12 distinct B transposons, which eventually resulted in 96 barcode combinations (Supplementary Table 5)51.
Combinatorial barcoding assisted single-nuclei ATAC-seq
Combinatorial ATAC-seq was performed as described previously with modifications12. We transferred 5–20 mg frozen tissue to a 1.5-mL Lobind tube (Eppendorf) in 1 mL NPB (5% BSA (Sigma), 0.2% IGEPAL-CA630 (Sigma), cOmplete (Roche), 1 mM DTT in PBS) and incubated for 15 min at 4 °C. Nuclei suspension was filtered over a 30-µ m Cell-Tric (Sysmex) and centrifuged for 5 min at 500 g. The nuclei pellet was resuspended in 500 µ L of 1.1× DMF buffer (36.3 mM Tris-acetate (pH = 7.8), 72.6 mM potassium-acetate, 11 mM Mg-acetate, 17.6% DMF) and nuclei were counted using a hemocytometer. Concentration was adjusted to 500/µ L, and 4,500 nuclei were dispensed into each well of a 96-well plate. For tagmentation, 1 µ L barcoded Tn5 transposome (0.25 µ M, Supplementary Table 5)51 was added to each well, mixed 5 times and incubated for 60 min at 37 °C with shaking (500 rpm). To quench the reaction, 10 µ L of 40-mM EDTA were added to each well and the plate was incubated at 37 °C for 15 min with shaking (500 rpm). We then added 20 µ L sort buffer (2% BSA, 2 mM EDTA in PBS) to each well, and all wells were combined afterwards. The nuclei suspension was filtered using a 30-µ m CellTric (Sysmex) into a FACS tube, and 3 µ M Draq7 (Cell Signaling) was added. Using a SH800 sorter (Sony), 25 nuclei were sorted per well into four 96-well plates (total of 384 wells) containing 18.5 µ L EB (50 pM Primer i7 (Supplementary Table 5), 200 ng BSA (Sigma)). Sort plates were shortly spun down. After addition of 2 µ L 0.2% SDS, samples were incubated at 55 °C for 7 min with shaking (500 rpm). We added 2.5 µ L 10% Triton-X to each well to quench the SDS. Finally, 2 µ L 25 µ M Primer i5 (Supplementary Table 5) and 25 µ L NEBNext High-Fidelity 2× PCR Master Mix (NEB) were added and samples were PCR-amplified for 11 cycles (72 °C 5 min, 98 °C 30 s, (98 °C 10 s, 63 °C 30 s, 72 °C 60 s) × 11, held at 72 °C). Following PCR, all wells were combined (around 15.5 mL) and mixed with 80 mL PB including pH-indicator (1:2,500, Qiagen) and 4 mL sodium-acetate (3 M, pH = 5.2). Purification was carried out on four columns following the MinElute PCR Purification Kit manual (Qiagen). DNA was eluted with 15 µ L EB, and eluate from all four columns was combined in a LoBind Tube (Eppendorf). For Ampure XP Bead (Beckmann Coulter) cleanup 170 µ L EB buffer and 110 µ L Ampure XP Beads (0.55× ) were added to 30 µ L eluate. After incubation at room temperature for 5 min and magnetic separation, supernatant was transferred to a new tube and another 190 µ L Ampure XP Beads (1.5× ) were added. After incubation, beads were washed twice on the magnet using 500 µ L 80% EtOH. After drying the beads for 7 min at room temperature, library was eluted with 20 µ L EB (Qiagen). Libraries were quantified using a Qubit fluorimeter (Life technologies) and the nucleosomal pattern was verified using Tapestation (High Sensitivity D1000, Agilent). We loaded 25 pM library per lane of a HiSeq2500 sequencer (Illumina) using custom sequencing primers (Supplementary Table 5)51 and following read lengths: 50 + 43 + 37 + 50 (Read1 + Index1 + Index2 + Read2). The first 8 bp of Index1 correspond to the p7 barcode and the last 8 bp to the i7 barcode. The first 8 bp of Index2 correspond to the i5 barcode and the last 8 bp to the p5 barcode. Since Index1 and 2 each contain 2 barcodes separated by a common linker sequence, we generated a spike-in library using different transposon and PCR primer sequences to balance the bases within each detection cycle (Supplementary Table 5). For the human–mouse mixture experiment, E15.5 forebrain and GM12878 nuclei were mixed in a 1:1 ratio before tagmentation. Samples were processed as above with the exceptions that only 96 wells were used after nuclei sorting and PCR amplification was performed for 13 cycles. The final library was loaded at 15 pM and sequenced using a MiSeq (Illumina) with the following read lengths: PE 44 + 43 + 37 + 44 (Read1 + Index1 + Index2 + Read2).
Cell culture
GM12878 (Coriell Institute for Medical Research) cells were cultured in RPMI1640 medium (Thermo Fisher Scientific) containing 2 mM l-glutamine (Thermo Fisher Scientific), 15% fetal bovine serum (Gemini Bioproducts) and 1% penicillin–streptomycin (Thermo Fisher Scientific) in T25 flasks (Corning) at 37 °C under 5% carbon dioxide. For the snATAC-seq mixture experiment, cells were harvested by centrifugation, washed with PBS (Thermo Fisher Scientific) and resuspended in NPB (5% BSA (Sigma), 0.2% IGEPAL-CA630 (Sigma), cOmplete (Roche), 1 mM DTT in PBS). Samples were incubated 5 min at 4 °C, and finally nuclei were pelleted by centrifugation (500 g, 5 min, 4 °C). The nuclei pellet was resuspended in 500 µ l of 1.1× DMF buffer (36.3 mM Tris-acetate (pH = 7.8), 72.6 mM potassium-acetate, 11 mM Mg-acetate, 17.6% DMF), and nuclei were counted using a hemocytometer.
NeuN-negative sorting
We resuspended 10 mg adult forebrain tissue (P56) in 500 µ L lysis buffer (0.5% BSA, 0.1% Triton-X, cOmplete (Roche), 1 mM DTT in PBS) and incubated it for 10 min at 4 °C. After spinning down (5 min, 500 g), the sample was resuspended in 500 µ L staining buffer (0.5% BSA in PBS). The nuclei suspension was incubated with anti-NeuN antibody (1:5,000, MAB377, Lot 2806074, EMD Millipore) for 30 min at 4 °C. After centrifugation, nuclei were resuspended in 500 µ L staining buffer (0.5% BSA in PBS) containing antimouse Alexa Fluor-488 antibody (1:1,000, A11001, Lot 1696425, Thermo Fisher Scientific). After incubating for 30 min at 4 °C, nuclei were pelleted (5 min, 500 g) and resuspended in 700 uL sort buffer (1% BSA, 1 mM EDTA in PBS). After filtration into a FACS tube, 5 uL DRAQ7 (Cell Signaling Technologies) were added and NeuN-negative nuclei were sorted using a SH800 sorter (Sony) into 5% BSA (Sigma) in PBS.
ATAC-seq
ATAC-seq was performed on 20,000 sorted nuclei, as described previously, with minor modifications52. After adding IGEPAL-CA630 (Sigma) in a final concentration of 0.1%, nuclei were pelleted for 15 min at 1,000 g. The pellet was resuspended in 19 µ L 1.1× DMF buffer (36.3 mM Tris-acetate (pH = 7.8), 72.6 mM potassium-acetate, 11 mM Mg-acetate, 17.6% DMF). After addition of 1 µ L Tn5 transposomes (0.5 µ M), tagmentation was performed at 37 °C for 60 min with shaking (500 rpm). Next, samples were purified using MinElute columns (Qiagen), PCR-amplified for 8–10 cycles with NEBNext High-Fidelity 2× PCR Master Mix (NEB, 72 °C 5 min, 98 °C 30 s, (98 °C 10 s, 63 °C 30 s, 72 °C 60 s) per cycle, held at 72 °C). Amplified libraries were purified using MinElute columns (Qiagen) and Ampure XP Bead (Beckmann Coulter). Sequencing was carried out on a HiSeq2500 or 4000 (50 bp PE, Illumina).
Data analysis
Single-nuclei ATAC-seq data processing pipeline:
Step 1. Alignment. Paired-end sequencing reads were aligned to mm10 reference genome using Bowtie253 in paired-end mode with the following parameters: bowtie2 -p 5 -t -X2000–no-mixed–no-discordant.
Step 2. Alignment filtering. Nonuniquely mapped (MAPQ < 30) and improperly paired (flag = 1,804) alignments were filtered.
Step 3. Barcode error correction. Each barcode consists of four 8-bp long indexes (i5, i7, p5 and p7). Reads with barcode combinations containing more than one mismatch (or one edit distance) for any index were removed. Any index with less than one mismatch was changed to its closest index.
Step 4. Reads separation. Reads were separated into individual cells based on the barcode combination (Supplementary Tables 1 and 5).
Step 5. Mark and remove PCR duplicates. For individual cells, we sorted reads based on the genomic coordinates using ‘samtools sort’54, then marked and removed PCR duplicates using Picard tools (MarkDuplicates).
Step 6. Mitochondrial reads removal. Reads mapped to the mitochondrial genome were filtered.
Step 7. Adjusting position of Tn5 insertion. All reads aligning to the positive strand were offset by + 4 bp, and all reads aligning to the negative strand were offset –5 bp
Step 8. Quality assessment of each single cell. Coverage of constitutively accessible promoters (promoters that are accessible across all tissues/cell line from ENCODE DHS) was calculated, and the number of reads and signal-over-noise ratio estimated by a ‘reads in peaks’ ratio for each cell.
Step 9. Cell selection. We only kept cells that passed our threshold: (i) coverage of constitutively accessible promoter > 10%; (ii) number of reads > 1,000; (iii) reads in peak ratio greater than estimation from corresponding bulk ATAC-seq level (https://www.encodeproject.org/search/?type=Experiment&lab.title=Bing+Ren%2C+UCSD&assay_title=ATAC-seq&organ_slims=brain).
Step 10. Replicates separation. Selected cells were separated into two replicates based on the predefined barcode combination.
Single-nuclei ATAC-seq cluster analysis
Cluster analysis partitions cells into groups such that cells from the same group have higher similarity than cells from different groups. Here, we developed a pipeline to obtain cell clusters (https://github.com/r3fang/snATAC). We first generated a catalog of accessible chromatin regions using bulk ATAC-seq data and created a binary accessible matrix. Chromatin sites were assigned a value of 1 for a given cell if there was a read detected within the peak region. Next, we calculated the pairwise Jaccard index between every two cells on the basis of overlapping open chromatin regions. Next, we applied a nonlinear dimensionality reduction method (t-SNE) to map the high-dimensional structure to a 3-D space15. This transforms high-dimensional structures to dense data clouds in a low-dimensional space, allowing partitioning of cells using a density-based clustering method16. We then identified the optimal number of cell clusters using the Dunn index55. Finally, we compared our cluster results to those of the shuffled set to further verify that our cluster result was not driven by library complexity or other confounding factors.
Step 1. Determining accessible chromatin sites in single cells. To catalog accessible chromatin sites in individual cells, we first created a reference map of open chromatin sites determined by bulk ATAC-seq. The chromatin accessibility maps from different timepoints (from E11.5 to P56) were merged into a single reference file using BEDtools56. For clustering of single cells, we tested clustering performance using accessible promoters (2 kb upstream of TSS) and distal elements, respectively, and found that clusters by distal elements outperformed promoters with lower Kullback-Leibler divergence (Supplementary Fig. 5). Therefore, we decided to only focus on distal genomic elements as features to perform clustering. Reads in individual cells overlapping with accessible sites were identified. We generated an accessible matrix of the read counts overlapping each individual accessible sites (columns) in each cell (row).
Step 2. Binary accessible matrix. We next converted the chromatin accessibility matrix to a binary matrix MN×D, in which Mij is 1 if any read in cell i is mapped to region j.
Step 3. Jaccard index matrix. Jaccard index matrixes JN×N were calculated between every two cells in which Jij measures the commonly shared open chromatin regions between cell Ci and Cj as follows:
Diagonal elements of JN×N are set to 0 as required by t-SNE analysis.
Step 4. Dimensionality reduction using t-SNE. Using Jaccard index matrix JN×N as input, we next applied t-SNE to map the N-dimensional data to a 3-D space15. Since t-SNE has a nonconvex objective function, it is possible for different runs to yield different solutions15. Thus, we ran t-SNE several times with different initiations and used the result with the lowest Kullback-Leibler divergence and best visualization. In a previous study, sequencing depth was a confounding factor and highly correlated with the first principle component of PCA analysis (Pearson correlation > 0.95)12. However, we did not observe correlation between sequencing depth and any of the t-SNE dimensions. We expected that the coherent structure of the open chromatin landscape of cells with high similarity would rely on a continuous and smooth 3-D structure and cells for different groups would locate to distinct parts of the plot. We used t-SNE to transform the high-dimensional structures to dense data clouds in the 3-D space15. Finally, we applied a density-based clustering method to identify different cell populations within the embedded 3-D space16.
Step 5. Density-based clustering. We applied a density-based clustering method to partition cells into groups in the embedded 3-D space16. The method identifies cluster centers that are characterized by two properties: (i) high local density ρi and (ii) large distance δi from points of higher density, which are centers of the clusters16. Any cells that showed values above defined thresholds (ρ0, δ0) were considered as centers of cluster. Next, the rest of cells were assigned to the center as described here16. Clearly, different thresholds (ρ0, δ0) will generate different number of clusters. To find the optimal number of clusters, we adopted the method developed by Habib et al. to evaluate the quality of different cluster results55.
Step 6. Number of clusters. In detail, Habib’s method applied the Dunn index to quantify the quality of cluster result as following55:
in which Δ (Ci, Cj) represents the intercluster distance between cluster Ci and Cj, Δ (Ck) represents the intracluster distance of cluster Ck. We used the ‘MaxStep’ distance developed by Habib et al. to calculate the distance for the Dunn index55. Finally, we iterated all possible (ρ0, δ0) combinations that yield different clusters and calculated their Dunn index. The clustering result with the highest Dunn index was chosen as final cluster (Algorithm 3).
Algorithm 1.
Cluster assignment
| Input: local density (ρ) and local distance (δ) for every cell; pairwise Euclidean distance in embedded 3-D space (D). |
| Output: Cluster assignment (C) |
| Let n be the total number of cells |
| Let Cbest be an empty array of length n |
| Let DIbest = −INF |
| for ρ0 from 0 to max(ρ) do |
| for δ0 from 0 to max(δ) do |
| choose cells whose ρ(i) and δ(i) are greater than ρ0, δ0 as Centers |
| C = cluster_assignment(D,Centers)* |
| if Dunn(D,C) > DIbest |
| DIbest = Dunn(D,C) |
| Cbest = C |
| end |
| end |
| end |
| Return Cbest |
cluster_assignment(D, Centers) and Dunn(D,C) were described in ref. 55.
Step 7. Shuffled cells. Due to the limited genome coverage of each single cell, cells may cluster according to their sequencing depth rather than ‘true’ covariation12. To verify that our cluster results were not driven by such artifacts, we compared our results to a simulated dataset. For this dataset, binary accessible sites within each cell were randomly shuffled across all accessible sites. In other words, we shuffled the data and removed the biological significance, but maintained the distribution of sequencing depth across cells. Shuffled cells were uniformly distributed as a ‘ball’ in the embedded 3-D space without clear partition of cells. However, we did observe a small portion of cells that tend to form a cluster but did not pass the cutoff (ρ0, δ0) used for the P56 forebrain dataset12.
Identification of cluster-specific features
We next developed a computational method that combines stability selection with LASSO57 to identify genomic elements (features) that potentially distinguish cells belonging to different clusters. LASSO regression enables sparse feature selections through the use of an L1 penalty. However, LASSO regression often does not result in a robust set of selected features and is sensitive to data perturbation. This is especially true when features are correlated. To overcome these limitations, we adopted a stable lasso to robustly identify features that distinguish every two cell clusters (Algorithm 2)57. Finally, we combined all identified features that distinguish different cell types to identify genomic elements (features) that potentially distinguish cells belonging to different clusters.
Algorithm 2.
Identification of cluster specific elements
| Input: X∈R(n,p) (binary matrix), Y∈{0, 1}n(cluster label), α (subsampling rate), β (perturbation rate), T (iteration) | |
| Output: importance score for each feature | |
| for t = 1 to T do: | |
| Randomly perturb the data: | |
| Draw a subset (Xt, Yt) of α of (X, Y) | |
| Draw a vector w~U ([β, 1]p) | |
| Reweight the features: | |
| Compute the LASSO path of length α · n | |
| Keep the selection matrix St where | |
|
| |
| end for loop; |
Compute the feature importance
Bulk ATAC-seq
Paired-end sequencing reads were aligned to the mm10 reference genome using Bowtie2 in paired-end mode, with following parameters: bowtie2 -p 5 -t -X2000–no-mixed–no-discordant (ref. 53), and PCR duplicates were removed using samtools54. Next, mitochondrial reads were removed and the position of alignments adjusted58. For visualization the ‘bamCoverage’ utility from deepTools2 was used59.
Hierarchical clustering of ATAC-seq profiles in adult forebrain
DeepTools2 was used for correlation analysis and hierarchical clustering of ATAC-seq profiles from cell clusters and sorted cell types in the adult forebrain59. First, we computed read coverage for each dataset against the merged list of genomic elements that separate two cell clusters in the adult forebrain using the ‘multiBamSummary’ utility. Next we used ‘plotCorrelation’ to generate hierarchical clustering using Spearman correlation coefficient between two clusters59.
Accessibility analysis and clustering of genomic elements
To cluster genomic elements based on their accessibility profile, we used promoter distal elements that were capable of distinguishing two cell clusters. For each feature, we extended the summits identified by MACS260 in both directions by 250 bp and generated a union set of elements using the ‘mergeBED’ function of BEDTools v2.17.056. Next, we intersected cluster-specific bam files with the peak list using the ‘coverageBED’ function of BEDTools56. We discarded elements that had fewer than five reads on average. After adding a pseudocount of one, we calculated cluster-specific RPM (reads per million sequenced reads) values for each genomic element. We divided the RPM value for a given cluster by the average value of all clusters (fold over mean) and finally log3-transformed the data. The generated matrix was used for k-means clustering of the elements using Ward’s method. We performed this analysis for all adult clusters, as well as for the excitatory neuron clusters and the 12 developmental cell clusters. A list of elements for each analysis can be found in Supplementary Table 4. To compare clusters of genomic elements in the adult forebrain with previously described single-cell DNA-methylation data29, we calculated the fraction of cell-type-specific differentially methylated regions (DMR) with each cluster using the ‘intersectBED’ function of BEDTools56 and normalized it by the total number of elements. Since Luo et al.29 focused on frontal cortex and specifically purified neurons, we centered the comparison on clusters associated with excitatory and inhibitory neurons.
Motif enrichment analysis
To identify potential regulators of chromatin accessibility, we performed motif analysis using the AME utility of the MEME suite49. For enrichment of known motifs, one-tailed Fisher’s exact test was used to calculate significance. P values were corrected by the Bonferroni method for multiple testing. A P value cutoff of < 10−5 was chosen for known motifs from the JASPAR database (JASPAR_CORE_2016_vertebrates.meme)61. For identification of de novo motifs, the HOMER tool was used with its default settings50.
Annotation of genomic elements
The GREAT algorithm was used to annotate distal genomic elements using the following settings to define the regulatory region of a gene: basal + extension (constitutive, 1 kb upstream and 0.1 kb downstream, up to 500-kb max extension)32. Gene ontology categories ‘molecular function’ and ‘biological processes’ were used.
Analysis of dynamic chromatin accessibility within a cell cluster
First, the ATAC-seq reads were counted in all peaks for each stage, cell type and replicate. For each cell cluster, only stages with more than 250,000 reads overlapping ATACseq peaks and more than 50 nuclei were used for dynamic analysis. Peaks with greater than 1 read per million reads (RPM) in at least two samples were kept. We used edgeR62 to assess the significance of difference between adjacent stages for cell clusters with at least 4 of 7 stages passing filtering criteria. P values were corrected using the Bonferroni method. Peaks with a Bonferroni P value < 0.05 were called dynamic peaks. The total numbers of dynamic peaks in each cell type are listed in Supplementary Fig. 11c. For each cell type, the read counts in each peak were normalized to a unit vector (i.e., values were divided by the square root of the sum of the squares of the values). k-means clustering was used for cell clusters with more than 200 dynamic elements (k = 3). Motif enrichment analysis was performed for each peak cluster using HOMER50.
VISTA analysis
Genomic locations of 484 VISTA validated elements45 were downloaded from https://enhancer.lbl.gov using the search term ‘forebrain’. Genomic locations were converted from mm9 to mm10 using the ‘liftOver’ tool (minimum rematch ratio of 0.95)63. Of these, 91 showed specific activity in the subpallium46. To identify developmental clusters that were enriched for subpallial enhancers, we first calculated the ratio of elements per k-means cluster overlapping with the total forebrain enhancer list and the subpallial subset separately. Finally, we calculated the relative enrichment using the ratio of subpallial over the complete forebrain regions. For anatomical annotation of distinct clusters, we intersected these regions with enhancers that are active in specific areas in the developing mouse forebrain47. After filtering clusters with fewer than five overlapping regions, we performed a binomial test to identify anatomical regions enriched for each cluster. The enrichment score is defined as –log10(binomial P value).
External datasets
Published ATAC-seq data of sorted excitatory neurons (GSM1541964, GSM1541965)6, GABAergic neurons (GSM2333635, GSM2333636)8, microglia (GSM2104286)21, neurons of the dentate gyrus (GSM2179990, GSM2179991)23 and distinct cortical layers (Layer2/3: GSM2333632, GSM2333633; Layer 4: GSM2333644, GSM2333645; Layer 5: GSM2333641, GSM2333642; Layer 6, GSM2333638, GSM2333639)8 were reprocessed. In addition, bulk ATAC-seq data for embryonic time points generated by the ENCODE consortium were analyzed for comparison (https://www.encodeproject.org/search/?searchTerm=atac+forebrain).
Statistics
No statistical methods were used to predetermine sample sizes, and we have not formally tested the distribution of the data. There was no randomization of the samples, and investigators were not blinded toward the developmental time point investigated. However, clustering of single nuclei based on chromatin accessibility was performed in an unbiased manner. Cell types were assigned afterwards. Low-quality nuclei were excluded from downstream analysis as outlined above.
Distal genomic elements to separate two cell clusters were identified using a stable LASSO approach57. A negative binomial test was used to identify promoters enriched in a specific cell clusters to enable annotation. To identify differentially accessible sites within a given cell type between developmental stages, a negative binomial test was used and the resulting P value was corrected using the Bonferroni method62. Motif enrichment for known transcription factor motifs in different sets of genomic elements was performed using a one-tailed Fisher’s exact test in combination with Bonferroni correction for multiple testing49. For significance testing of enrichment of de novo motifs, a hypergeometric test was used without correction for multiple testing50.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Accession codes
Raw and processed data have been deposited to NCBI Gene Expression Omnibus with the accession number GSE1000333. Data analysis pipeline can be downloaded at https://github.com/r3fang/snATAC.
Data availability
Raw and processed data to support the findings of this study have been deposited to NCBI Gene Expression Omnibus with the accession number GSE1000333.
Code availability
The scripts and pipeline for the analysis can be found at https://github.com/r3fang/snATAC.
Supplementary Material
Acknowledgments
We thank B. Li for bioinformatic support. We thank M. He and T. Osothprarop for providing the Tn5 enzyme. We thank D. Gao for sequencing on the MiSeq. This study was funded in part by the National Human Genome Research Institute (U54HG006997 to B.R.), National Institute Mental Health (1U19MH114831 to B.R., U01MH098977 to K.Z.), NIH (2P50 GM085764 to B.R.), and the Ludwig Institute for Cancer Research (to B.R.). S.P. was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (DFG, PR 1668/1-1). R.R. was supported by a Ruth L. Kirschstein National Research Service Award NIH/NCI T32 CA009523. Research conducted at the E.O. Lawrence Berkeley National Laboratory was performed under US Department of Energy Contract DE-AC02-05CH11231, University of California.
Footnotes
Author contributions
The study was conceived and designed by B.R., S.P., R.F. and K.Z.; it was overseen by B.R. and K.Z. Experiments were performed by S.P., B.C.S. and H.H.; tissue collection by V.A., D.E.D., A.V., L.A.P. and S.P.; sequencing performed by S.K.; computational strategy developed by R.F. and S.P., data analysis performed by S.P., R.F., Yu. Z., R.R., Ya. Z. and D.U.G. The manuscript was written by S.P., R.F. and B.R. A.V., L.A.P. and K.Z. provided input and edited the manuscript. All authors discussed results and commented on the manuscript.
Competing interests
The authors declare no competing financial interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41593-018-0079-3.
References
- 1.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium, E. P., ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurano MT, et al. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 2015;47:1393–1401. doi: 10.1038/ng.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 6.Mo A, et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray LT, et al. Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex. eLife. 2017;6:e21883. doi: 10.7554/eLife.21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilsbach R, et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 2014;5:5288. doi: 10.1038/ncomms6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cusanovich DA, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vierstra J, et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Maaten L, Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 16.Rodriguez A, Laio A. Machine learning. Clustering by fast search and find of density peaks. Science. 2014;344:1492–1496. doi: 10.1126/science.1242072. [DOI] [PubMed] [Google Scholar]
- 17.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 18.La Manno G, et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell. 2016;167:566–580.e19. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rousseau A, et al. Expression of oligodendroglial and astrocytic lineage markers in diffuse gliomas: use of YKL-40, ApoE, ASCL1, and NKX2-2. J. Neuropathol. Exp. Neurol. 2006;65:1149–1156. doi: 10.1097/01.jnen.0000248543.90304.2b. [DOI] [PubMed] [Google Scholar]
- 20.Pernet V, Joly S, Christ F, Dimou L, Schwab ME. Nogo-A and myelin-associated glycoprotein differently regulate oligodendrocyte maturation and myelin formation. J. Neurosci. 2008;28:7435–7444. doi: 10.1523/JNEUROSCI.0727-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matcovitch-Natan O, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 22.Huttner HB, et al. The age and genomic integrity of neurons after cortical stroke in humans. Nat. Neurosci. 2014;17:801–803. doi: 10.1038/nn.3706. [DOI] [PubMed] [Google Scholar]
- 23.Su Y, et al. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat. Neurosci. 2017;20:476–483. doi: 10.1038/nn.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 25.Glasgow SM, et al. Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat. Neurosci. 2014;17:1322–1329. doi: 10.1038/nn.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nord AS, Pattabiraman K, Visel A, Rubenstein JL. Genomic perspectives of transcriptional regulation in forebrain development. Neuron. 2015;85:27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan F, et al. Efficient generation of region-specific forebrain neurons from human pluripotent stem cells under highly defined condition. Sci. Rep. 2015;5:18550. doi: 10.1038/srep18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbosa AC, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc. Natl Acad. Sci. USA. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo C, et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357:600–604. doi: 10.1126/science.aan3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martynoga B, Drechsel D, Guillemot F. Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harb. Perspect. Biol. 2012;4:a008359. doi: 10.1101/cshperspect.a008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollen AA, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian L, et al. Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc. Natl Acad. Sci. USA. 2011;108:E265–E274. doi: 10.1073/pnas.1101109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu LC, et al. Lhx2 regulates the timing of β -catenin-dependent cortical neurogenesis. Proc. Natl. Acad. Sci. USA. 2015;112:12199–12204. doi: 10.1073/pnas.1507145112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castro DS, et al. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Castro DS, et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long JE, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb. Cortex. 2009;19:i96–i106. doi: 10.1093/cercor/bhp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heng YH, et al. NFIX regulates neural progenitor cell differentiation during hippocampal morphogenesis. Cereb. Cortex. 2014;24:261–279. doi: 10.1093/cercor/bhs307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Hori K, et al. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 2008;22:166–178. doi: 10.1101/gad.1628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian X, Kai L, Hockberger PE, Wokosin DL, Surmeier DJ. MEF-2 regulates activity-dependent spine loss in striatopallidal medium spiny neurons. Mol. Cell. Neurosci. 2010;44:94–108. doi: 10.1016/j.mcn.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onorati M, et al. Molecular and functional definition of the developing human striatum. Nat. Neurosci. 2014;17:1804–1815. doi: 10.1038/nn.3860. [DOI] [PubMed] [Google Scholar]
- 43.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Choksi SP, Lauter G, Swoboda P, Roy S. Switching on cilia: transcriptional networks regulating ciliogenesis. Development. 2014;141:1427–1441. doi: 10.1242/dev.074666. [DOI] [PubMed] [Google Scholar]
- 45.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silberberg SN, et al. Subpallial enhancer transgenic lines: a data and tool resource to study transcriptional regulation of GABAergic cell fate. Neuron. 2016;92:59–74. doi: 10.1016/j.neuron.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visel A, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–73. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amini S, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Genet. 2014;46:1343–1349. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Habib N, et al. Div-seq: single-nucleus RNA-seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Series B Stat. Methodol. 1996;58:267–288. [Google Scholar]
- 58.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mathelier A, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tyner C, et al. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res. 2017;45:D626–D634. doi: 10.1093/nar/gkw1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data to support the findings of this study have been deposited to NCBI Gene Expression Omnibus with the accession number GSE1000333.



 URI:
URI: