Abstract
Clostridium difficile is the primary cause of nosocomial diarrhea and pseudomembranous colitis. It produces dormant spores, which serve as an infectious vehicle responsible for transmission of the disease and persistence of the organism in the environment. In Bacillus subtilis, the sin locus coding SinR (113 aa) and SinI (57 aa) is responsible for sporulation inhibition. In B. subtilis, SinR mainly acts as a repressor of its target genes to control sporulation, biofilm formation, and autolysis. SinI is an inhibitor of SinR, so their interaction determines whether SinR can inhibit its target gene expression. The C. difficile genome carries two sinR homologs in the operon that we named sinR and sinR’, coding for SinR (112 aa) and SinR’ (105 aa), respectively. In this study, we constructed and characterized sin locus mutants in two different C. difficile strains R20291 and JIR8094, to decipher the locus’s role in C. difficile physiology. Transcriptome analysis of the sinRR’ mutants revealed their pleiotropic roles in controlling several pathways including sporulation, toxin production, and motility in C. difficile. Through various genetic and biochemical experiments, we have shown that SinR can regulate transcription of key regulators in these pathways, which includes sigD, spo0A, and codY. We have found that SinR’ acts as an antagonist to SinR by blocking its repressor activity. Using a hamster model, we have also demonstrated that the sin locus is needed for successful C. difficile infection. This study reveals the sin locus as a central link that connects the gene regulatory networks of sporulation, toxin production, and motility; three key pathways that are important for C. difficile pathogenesis.
Author summary
In Bacillus subtilis, sporulation, competence and biofilm formation are regulated by a pleiotropic regulator called SinR. Two sinR homologs are present in C. difficile genome as an operon and henceforth labeled as sinR and sinR’. Our detailed investigation revealed that in C. difficile, the SinR and SinR’ are key master regulators needed for the regulation of several pathways including sporulation, toxin production, and motility.
Introduction
Clostridium difficile, a major nosocomial pathogen, is the causative agent of antibiotic-associated diarrhea and pseudomembranous colitis [1, 2]. Every year, nearly half a million cases of C. difficile infections (CDI) occur in the United States and result in approximately 14,000 deaths [3]. C. difficile toxins damage the colonic epithelium, which results in moderate to severe diarrhea [4]. Recent studies have shown that these toxins are essential for C. difficile pathogenesis [4–7]. Due to the strictly anaerobic nature of the vegetative cell, C. difficile survives outside the host in the form of dormant spores, which are highly resilient and resistant to most disinfectants. Thus, C. difficile spores are critical for its host to host transmission and persistence in the hospital environment [8].
C. difficile Toxins A and B are encoded by the tcdA and tcdB genes respectively, and their expression is dependent on TcdR, an alternative RNA polymerase sigma factor [9–11]. Environmental stresses, such as alteration of the redox potential, high temperature, or limitation of nutrients like glucose, and biotin, modulate toxin production by influencing the expression of tcdR [9–12]. Similar to toxin production, the sporulation pathway in C. difficile is also known to be influenced by nutrient availability and uptake [13, 14]. The regulators involved in controlling toxin synthesis in response to nutrients are the global regulatory proteins CcpA and CodY [14–18]. Among them, CcpA mediates glucose-dependent toxin gene repression [15, 16], and CodY blocks the transcription of toxin genes during the exponential growth phase of the bacterial culture [17, 18]. Other than affecting toxin production, mutations in codY and ccpA were also found to affect sporulation [13, 16]. Other genes that are known to influence both toxin production and sporulation include spo0A, sigH, and rstA [19–22]. New evidence suggests that the toxin, motility, and sporulation regulatory networks are linked together in C. difficile [19, 23, 24]. The sigma factor SigD needed for transcription of the flagellar operon was identified to regulate tcdR transcription to influence toxin production [25, 26] positively. Mutations in spo0A, rstA, and sigH also influenced motility along with toxin production and sporulation [19–22]. This study identified that mutation of the sin locus in C. difficile could affect toxin production and sporulation along with motility and thus reports a new regulatory element of this network.
In Bacillus subtilis, the sin (sporulation inhibitor) locus codes for two proteins SinR and SinI and regulates several genes involved in sporulation, motility, competency, proteolysis, and biofilm formation [27–31]. In this study, we have created C. difficile sin locus mutants in two different strains. Using RNA-Seq analysis, we compared the transcriptome of the mutants with respective parent strains to identify and assess the transcriptional regulation of sin locus coded regulators. Follow up phenotypic analyses and complementation experiments showed that the Sin regulators in C. difficile are also pleiotropic as in B. subtilis. Here, their regulatory roles in toxin production, sporulation, and motility were further investigated and discussed.
Results
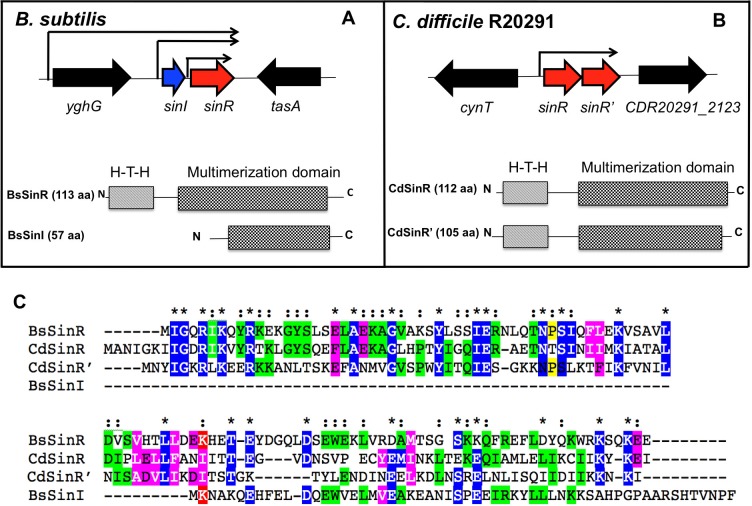
Comparison of C. difficile and B. subtilis sin loci
In B. subtilis, the sin locus carries two small ORFs, sinI and sinR [32, 33] (Fig 1A). B. subtilis SinR (BsSinR) is a DNA-binding protein that binds to a conserved DNA sequence upstream of the translational start site of target genes to negatively control their transcription. SinI, encoded by a gene adjacent to sinR, has an antagonistic relationship with SinR and binds directly to the SinR protein to inhibit its activity. This causes the pathways that were repressed by SinR to switch on. In B. subtilis, SinR contains 113 aa, and the DNA binding domain is located at the N-terminus part, which spans from residues 5–61 [32, 33] (Fig 1A). The C-terminal part of SinR forms alpha-helices and is responsible for multimerization and SinI interaction. The SinI protein, on the other hand, resembles a truncated SinR without the DNA binding region and carries only the alpha-helical structure to drive the hetero-dimerization of SinR-SinI complex [32–34]. In C. difficile the sin locus contains two ORFs CDR20291_2121 and CDR20291 _2122 (in C. difficile R20291 reference genome), which codes for proteins that are 43% and 35% identical to B. subtilis SinR, respectively (Fig 1B). Both these proteins are predicted to be DNA-binding since they carry HTH (Helix-Turn-Helix) domains in their N-terminal regions. Hence we named CDR20291_2121 as sinR and CDR20291_ CD2122 as sinR’. The C. difficile SinR (CdSinR) contains 112 amino acids, and its predicted HTH domain spans residues 11 to 66. The SinR’ (CdSinR’) protein carries 105 aa, and its predicted HTH domain spans from residues 7 to 62 (Fig 1B). Both CdSinR and CdSinR’ shows the highest homology to BsSinR in this DNA-binding domain, where within the 50 residues of HTH domain, 13 of them are identical and 19 of them represent conservative substitutions (Fig 1C). CdSinR and CdSinR’ shows similarity with each other (33% identity) only in their N terminal DNA binding domain. The C terminus multimerization domains of these proteins show variations, and there is less similarity of CdSinR and CdSinR’ to BsSinR and each other in this region.
Fig 1.
Genetic organization of genes in the sin locus in Bacillus subtilis (A) and C. difficile R20291 strain (B). The different domains within Sin proteins are presented below. (C) Sequence alignment of the C. difficile SinR (CdSinR) and SinR’ (CdSinR’) with Bacillus subtilis SinR (BsSinR) and SinI (BsSinI) using ClustalW.
In various Bacillus sp. SinR homologs are known to control the expression of the genes adjacent to the sin loci. Thus, identifying genes adjacent to the sin loci were helpful in predicting at least a few functions of the Sin regulators in these bacterial species. For example, in B. subtilis, the sin locus is adjacent to the tapA-sipW-tasA operon, and SinR represses the expression of this operon whose products are involved in the production of the biofilm matrix [31]. In Bacillus anthracis, the sin locus is next to calY that codes for camelysin, a cell surface associated protease, and SinR in this species is known to repress the calY expression [35]. In C. difficile, the sin locus is located in between cynT (codes for carbonic anhydrase) and CDR20291_2123 (unknown function) (Fig 1B) and is not close to any other genes that are known to be essential for virulence in this pathogen. Thus, the location of the sin locus in C. difficile chromosome did not provide us any clues about its possible functions. To get more information about the locus and its role in C. difficile physiology we decided to construct and characterize mutants in sin locus.
Construction and verification of sinRR’ mutants in C. difficile strains JIR8094 and R20291
An erythromycin resistant marker was introduced in the sinR at nucleotide 141 using Clostron, a TargeTron-based group II intron in C. difficile JIR8094 [36] and R20291 strains [37]. The presence of the retargeted intron in the correct gene in both mutant strains was confirmed by PCR (S1 Fig). In B. subtilis, three different promoters drive the transcription of the sin genes [33]. In B. subtilis, the polycistronic sinIR transcript is produced from two different promoters, and the sinR transcript is driven from an independent promoter immediately downstream of sinI (Fig 1A) [33]. In C. difficile, the operon upstream of sin locus transcribes in the opposite direction, and no read-through transcription of sin locus is possible from its promoter (Fig 1B). Using cDNA prepared from the JIR8094 and the mutant strain, we performed RT-PCR analysis and checked for the presence of sinR, sinR’ and sinRR’ transcripts (S2 Fig). We could detect sinR, sinR’ and also the read through sinRR’ transcripts, which confirmed that the sinR and sinR’ are transcribed as a single transcript (S2 Fig). When the same analysis was performed using the mutant strain cDNA both the sinR, sinR’ and sinRR’ transcripts were absent (S2 Fig). The QRT-PCR analysis of the sinR mutant showed significant reduction of both sinR and sinR’ transcript levels (S2 Fig). It also revealed that similar to B. subtilis, the C. difficile sin locus is expressed between late-exponential and early-stationary growth phase (10 to 12 h) (S2 Fig). Similar results were obtained in RT-PCR analyses of cDNA from the R20291 strain (S2 Fig). When we performed the western blot analysis using the SinR and SinR’ specific antibodies (see M&M), both SinR and SinR’ were found to be absent in the mutant (S3 Fig). Our western blot and the RT-PCR results together suggest that sinR and sinR’ are part of an operon. However, there is a possibility that sinR’ could have an independent promoter coded within the sinR coding region, which was not expressed in the growth conditions tested. Since the insertion of the intron in sinR (first gene in the operon) disrupted both sinR, sinR’ transcripts, and SinR, SinR’ production in the growth conditions tested, we named the mutant strains with the disrupted sinR gene as JIR8094::sinRR’ and R20291::sinRR’.
Impact of sinRR’ inactivation in C. difficile
We first analyzed the impact of sin locus inactivation on the growth of C. difficile in TY medium. During the exponential phase of the growth, both parents and mutants grew at a similar rate. However, when they entered the stationary phase, we observed a decrease in the turbidity of the mutant cultures as measured as OD@600 nm (S3 Fig). We performed the Triton X-100 autolysis assay to check the influence of SinRR’ on global autolysis of C. difficile [25]. We used the 16h old stationary phase culture to perform this assay, where the R20291::sinRR’ lysed at a faster rate compared to the parent (S4 Fig). These results suggested that inactivation of sinRR’ induced autolysis in C. difficile. In B. subtilis, SinR along with another regulatory protein SlrR represses the expression of lytA-lytB-lytC and lytF autolysins [38]. Our initial observation of lysis phenotype in the sinRR’ mutants suggested that like B. subtilis SinR, C. difficile SinR might also be controlling the autolysin genes. In B. subtilis the SinR is a pleiotropic regulator and controls various pathways including autolysis [29–31, 33, 38, 39]. We suspected that SinR and SinR’ in C. difficile might also regulate several targets to control multiple functions. Hence, to identify the sinRR’ regulated pathways in C. difficile, we performed the transcriptome analysis of the sinRR’ mutants in comparison with their respective parents.
Assessment of the sinRR’ regulon in C. difficile
Based on the growth pattern of the sinRR’ mutants (S3 Fig) and the expression kinetics of sinRR’ in the parent strains (S2 Fig), we decided to compare the transcriptomes of mutant strains with their respective parent strains during the early stationary phase (i.e., 12 h of growth) in TY medium. We used three biological replicates and genes were considered differentially expressed if the fold change was ≥ log2 1.5 and their adjusted p-value was ≤0.05.
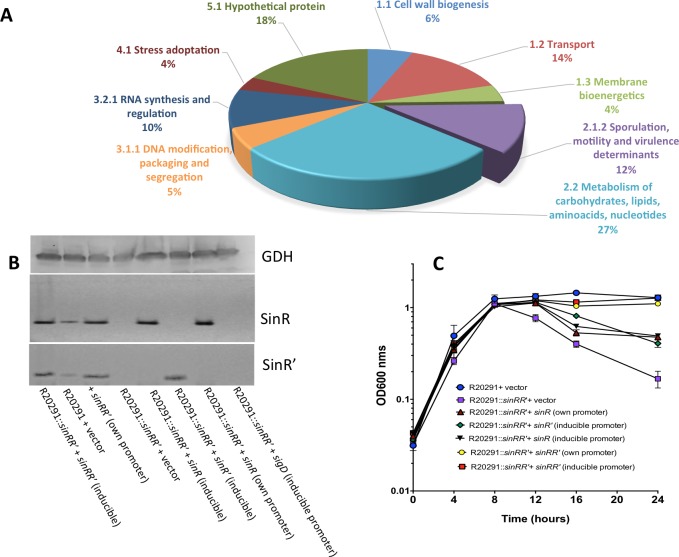
In the RNA seq analysis, it was observed that 437 and 425 genes were over-expressed in R20291::sinRR’ and in JIR8094::sinRR’ mutant strains, respectively, while 668 and 208 genes were under-expressed in R20291::sinRR’ and JIR8094::sinRR’ mutant strains, respectively. Results from the transcriptome analysis confirm that as in B. subtilis, SinRR’ in C. difficile also regulates a wide range of genes involved in several pathways including sporulation, motility, metabolism, membrane transport, stress response and toxin synthesis (Fig 2A). A list of genes identified to be differentially regulated in mutants R20291::sinRR’ and JIR8094::sinRR’ compared to their parent strains are listed in S4, S5, S6 and S7 Tables respectively. To test and validate the transcriptome profiles, we performed relevant phenotypic assays and functional analysis with parent and mutant strains for major pathways (sporulation, motility, toxin production and autolysis) that were suggested to be regulated by SinR and SinR’.
Fig 2. Characterization of sin locus (sinRR’) mutant in C. difficile.
(A) Functional categorization of genes affected by sin locus mutation in R20291 strains based on RNA seq data. (B) Western blot analysis with SinR and SinR’ specific antibodies demonstrating the absence of both SinR and SinR’ in the sinRR’ mutants and their presence after the complementation. GDH detection using anti-GDH antibodies was used as loading control. (C) Growth curve of the parent (R20291), sinRR’ mutant and the sinRR’ mutant complemented strains in TY medium. The data shown are means ± standard errors of three replicates.
We have included following strains in the phenotypic analysis: parent strain, sinRR’ mutant, sinRR’ mutant with pRGL311 (plasmid with sinRR’ under its native promoter), and sinRR’ mutant with pRG334 (plasmid with sinRR’ under the inducible promoter). To determine the independent role of SinR and SinR’ in the phenotypes, the sinRR’ mutant with plasmids: pRG300 (sinR gene alone with its promoter region); pRG310 (sinR under the inducible promoter); and pRG306 (sinR’ alone under the inducible promoter) were used. Western blot analysis with SinR and SinR’ specific antibodies were performed to confirm their expressions from the constructs, and the sinRR’ mutant with vector alone was used as negative controls (Fig 2B). Growth curve analysis showed when sinRR’ was expressed from its promoter or the inducible promoter in the sinRR’ mutant, no autolysis was observed, and they grew similar as the wild type (Fig 2C and S4 Fig). In the Triton X-100 autolysis assay, a partial recovery from autolysis was observed when either SinR or SinR’ alone was expressed in the mutant (S4 Fig).
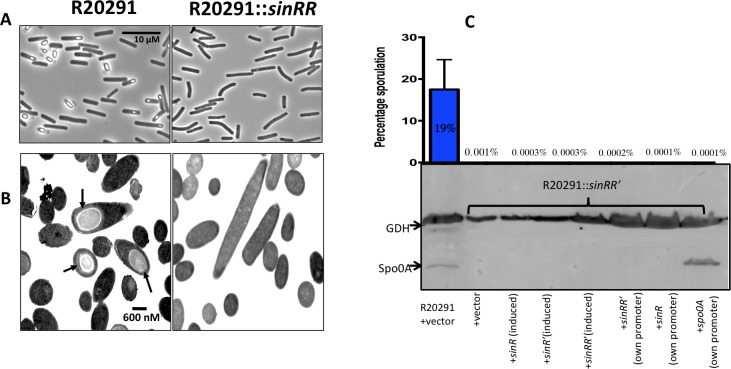
C. difficile sinRR’ mutants are asporogenic
To determine the role of sinRR’ on sporulation, we grew the test strains on 70:30 sporulation agar for 30h. Initial analysis through phase contrast microscopy detected no spores in R20291::sinRR’ (Fig 3A). Transmission electron microscopy (TEM) further confirmed this observation (Fig 3B). Fully mature spores could be detected in R20291, whereas the sinRR’ mutant cells were devoid of any spores. Similar results were obtained for JIR8094::sinRR’ mutant as well (S5 Fig).
Fig 3. Sporulation in sinRR’ mutant.
(A) Phase contrast microscopy of paraformaldehyde-fixed R20291::sinRR’ strains revealed no spores. (B) R20291::sinRR’ was asporogenic as shown in representative TEM images in comparison with the parent strain. Black arrows indicate mature spores in parent strains. C. Asporulation phenotype of sinRR’ mutant could not be complemented. Sporulation frequency (CFU/ml of ethanol resistant spores) of R20291, sinRR’ mutant and mutant complemented with different constructs were determined. The sinRR’ mutant strain expressing spo0A from its own promoter was also included in this analysis. Below the sporulation frequency graph is the multiplex-western blot analysis of sinRR’ mutant complemented strain proteins using Spo0A and GDH specific antibodies.
We performed ethanol treatment based sporulation efficiency assay where the ability of the bacteria to produce viable spores were analyzed by counting the total number of CFU (Colony Forming Units) following ethanol treatment. The mean sporulation efficiency of the parental strain R20291 was 18.7% (Fig 3C). The sinRR’ mutant strain did not produce any spores, and the percentage of sporulation was near zero. We were surprised by the observation that expression of either sinRR’ or sinR/sinR’ alone also did not revive the sporulation in the sinRR’ mutants (Fig 3C).
Sporulation in C. difficile is initiated with the activation of Spo0A, which in turn triggers early sporulation gene transcription [22, 40]. Transcripts of spo0A were 3.5-fold and 2.9-fold under-expressed in JIR8094::sinRR’ and in R20291::sinRR’ strains respectively, when compared to parent strains. We performed western blot analysis with the Spo0A specific antibodies [41]. We detected GDH (glutamate dehydrogenase) for loading control since its production was found to be unaffected in the sinRR’ mutants. Western blot analysis showed that in R20291::sinRR’ the Spo0A was absent or below the detectable level (Fig 3C, S5 Fig). Lower production of Spo0A can result in down-regulation of all sporulation genes under its control. Our transcriptomic data indeed found many sporulation-associated genes to be affected (Tables 1, S4 and S6) in the sinRR’ mutant. The QRT-PCR analysis performed on selected sporulation genes confirmed their down-regulation in the sinRR’ mutants (S8 Table).
Table 1. Under-expressed sporulation genes in R20291::sinRR’.
| Locus Tag | Gene | Protein Name | Fold-Change: R20291/sinRR’ mutant |
log2 ratio | Known/predicted sigma factor needed for expression | Adjusted p value |
|---|---|---|---|---|---|---|
| CDR20291_0104 | cwlD | Germination N-acetylmuramoyl-L-alanine amidase, Autolysin | 67.7 | 6.1 | SigE | 1.01E-09 |
| CDR20291_0125 | spoIIID | Stage III sporulation protein D | 68.0 | 6.1 | SigE | 6.24E-05 |
| CDR20291_0128 | putative sporulation protein yyac, DUF1256 family | 50.2 | 5.6 | SigE | 0.005 |
|
| CDR20291_0213 | hypothetical protein | 3041.9 | 11.6 | SigE | 6.05E-14 |
|
| CDR20291_0316 | spore coat assembly asparagine rich protein | 81.8 | 6.4 | SigE | 0.00057 |
|
| CDR20291_0713 | Putative sporulation protein YunB | 63.0 | 6.0 | SigE | 0.00666 |
|
| CDR20291_1005 | Putative membrane protein, BDBH YlbJ involved in spore cortex formation | 5.7 | 2.5 | SigE | 5.93E-06 |
|
| CDR20291_1030 | spoIIIAA | Stage III sporulation protein AA | 18464.8 | 14.2 | SigE | 0.00005 |
| CDR20291_1031 | spoIIIAB | Stage III sporulation protein AB | 90.8 | 6.5 | SigE | 8.33E-06 |
| CDR20291_1033 | spoIIIAD | Stage III sporulation protein AD | 3571.7 | 11.8 | SigE | 1.32E-08 |
| CDR20291_1034 | spoIIIAE | Stage III sporulation protein AE | 13.9 | 3.8 | SigE | 1.48E-09 |
| CDR20291_1035 | spoiIIIAF | Stage III sporulation protein AF | 119.5 | 6.9 | SigE | 0.00825 |
| CDR20291_1036 | spoIIIAG | Stage III sporulation protein AG | 794.0 | 9.6 | SigE | 2.08E-09 |
| CDR20291_1051 | spoIVB | Stage IV sporualtion protein AB | 6.2 | 2.6 | SigE, SigG | 6.61E-06 |
| CDR20291_1073 | Putative phage protein, skin element | 98.9 | 6.6 | SigE | 0.00821 |
|
| CDR20291_1282 | cotE | Spore coat protein CotE peroxiredoxin/chitinase | 18.7 | 4.2 | SigE | 0.00029 |
| CDR20291_1360 | cotB | Spore outer coat layer protein CotB | 332.0 | 8.4 | SigE | 1.03E-07 |
| CDR20291_2146 | cspC | Subtilisin-like serine germination related protease- CspC | 16.3 | 4.0 | SigE | 0.00921 |
| CDR20291_2147 | cspBA | Subtilisin like serine germination related protease, CspB | 19.5 | 4.3 | SigE | 7.88E-07 |
| CDR20291_2289 | cotJA | Putative spore coat protein | 103 | 7.0 | SigE | 0.00074 |
| CDR20291_2291 | cotD | Spore coat protein CotD manganese catalase | 139.6 | 7.1 | SigE | 0.056 |
| CDR20291_2334 | spoIV | Stage IV sporulation protein | 4.1 | 2.0 | SigE | 9.981E-06 |
| CDR20291_2335 | putative sporulation protein yyac | 1593.4 | 10.6 | SigE | 0.00097 | |
| CDR20291_2513 | spoIVA | Stage IV sporulation protein AA | 309.1 | 8.3 | SigE | 2.35E-06 |
| CDR20291_2573 | spoIIE | Stage II sporulation protein E | 36.13938 | 5.17 | SigE | 7.04E-05 |
| CDR20291_3331 | Putative spore protein | 3.1 | 1.6 | SigE | 6.75E-09 |
|
| CDR20291_3376 | spmB | Spore maturation protein B | 41.4 | 5.4 | SigE | 0.00081 |
| CDR20291_3377 | spmA | Spore maturation protein A | 21.5 | 4.4 | SigE | 2.80E-12 |
| CDR20291_3404 | sipL | SpoIVA interacting protein | 18.3 | 4.2 | SigE | 8.71E-05 |
| CDR20291_0124 | spoIIQ | Stage II sporulation protein Q | 56.9 | 5.8 | SigF | 0.05 |
| CDR20291_0213 | hypothetical protein | 3041.9 | 11.6 | SigE, SigF | 6.05E-14 |
|
| CDR20291_0316 | spore coat assembly asparagine-rich protein | 81.8 | 6.4 | SigE, SigF | 0.0005 |
|
| CDR20291_2362 | spoIIP | Stage II sporulation protein P | 15.1 | 3.9 | SigF | 2.67E-11 |
| CDR20291_2363 | gpr | Spore endopeptidase | 28.8 | 4.8 | SigF | 1.83E-07 |
| CDR20291_2576 | sspA | small acid-soluble spore protein A | 196.8 | 7.6 | SigG, SigF | 5.77E-07 |
| CDR20291_3080 | small acid-soluble spore protein | 9.0 | 3.2 | SigG, SigF | 6.98E-06 |
|
| CDR20291_3107 | sspB | small acid-soluble spore protein B | 305.5 | 8.3 | SigG, SigE, SigF | 5.95E-08 |
| CDR20291_3400 | sleB | Putative spore cortex-lytic enzyme | 14.0 | 3.8 | SigF | 5.79E-09 |
| CDR20291_2530 | sigG | RNA polymease sigma-G factor | 44.0 | 5.5 | SigG | 2.10E-11 |
| CDR20291_0702 | spoVAC | Stage V sporulation protein VAC | 86.6 | 6.4 | SigG | 0.000105 |
| CDR20291_0703 | spoVAD | Stage V sporualtion protein VAD | 84.5 | 6.4 | SigG | 0.000161 |
| CDR20291_3080 | small acid-soluble spore protein | 9.0 | 3.2 | SigG | 6.98E-06 |
|
| CDR20291_3336 | spoVT | Stage V sporulation protein T | 309.1 | 8.3 | SigG | 9.59E-10 |
| CDR20291_0476 | sleC | SleC- spore peptidoglycan hydrolase/ germinant receptor complex | 642.4 | 9.3 | SigE, SigK | 0.00506 |
| CDR20291_0926 | cdeC | Cysteine rich exosporium protein | 101.6 | 6.7 | SigE, SigF, SigK | 1.05E-06 |
| CDR20291_1282 | cotE | Spore coat protein CotE peroxiredoxin/chitinase | 18.7 | 4.2 | SigE, SigK | 0.00029 |
| CDR20291_2289 | cotJA | Putative spore coat protein | 103 | 7.0 | SigE, SigK | 0.00074 |
| CDR20291_2291 | cotD | Spore coat protein CotD manganese catalase | 139.6 | 7.1 | SigE, SigK | 0.056 |
| CDR20291_3090 | bclA2 | exosprium glycoprotein | 8.4 | 3.1 | SigE, SigF, SigK | 0.00288 |
| CDR20291_3193 | bclA3 | Putative exosporium glycoprotein | 13.1 | 3.7 | SigE, SigF, SigK | 0.00543 |
Since our transcriptome analysis and western blot analysis revealed a lower Spo0A in R20291::sinRR’, we decided to test whether the asporogenic phenotype of the sinRR’ mutants is due to the lower production of Spo0A. We expressed spo0A from its native promoter (pRGL312) in the R20291::sinRR’ and production of Spo0A in sinRR’ mutants was verified through the western blot analysis using Spo0A specific antibodies (Fig 3C) [41]. To our surprise, production of Spo0A in the sinRR’ mutants did not induce the sporulation in the R20291::sinRR’ strain (Fig 3C). For sporulation to proceed normally, the Spo0A protein should get activated by phosphorylation [42]. Spo0A~P then acts as a transcriptional activator for many downstream genes in the sporulation pathway that includes sigma factors, the forespore specific sigF, and the mother cell-specific sigE [22, 40, 42]. We performed QRT-PCR to detect the transcripts of Spo0A~P activated sigF and sigE genes. We did not observe increases in sigF and sigE transcript levels in the spo0A expressing sinRR’ mutant when compared to the sinRR’ mutant with vector alone control. This result suggests that activation of Spo0A to Spo0A~P is affected in the sinRR’ mutant.
In Bacillus sp., the pathway that controls Spo0A phosphorylation is well characterized [43–47]. In Clostridia, the components of this phosphorelay are absent, and it has been hypothesized that sporulation-associated sensor kinases may directly phosphorylate the Spo0A for its activation. In C. difficile, four orphan kinases (CD630_01352, CD630_2492, CD630_01579, and CD630_1949) are present, among which, the CD630_1579 kinase was shown to phosphorylate Spo0A in vitro, and the CD630_2492 mutant was found to be less efficient in sporulation [48]. In the transcriptome data, the CD630_1579 and the CD630_ 2492 kinases were to be under-expressed ~1.5-fold and ~3-fold, respectively, in the JIR8094::sinRR’ mutant. However, their homologs CDR20291_1476 and CDR20291_2385 in the R20291::sinRR’ were not affected suggesting that these kinases might not be the main reason for Spo0A inactivation in the sinRR’ mutants. Since the regulatory network of Spo0A activation is largely unknown, there is a possibility that unknown kinases could have been affected in sinRR’ mutants.
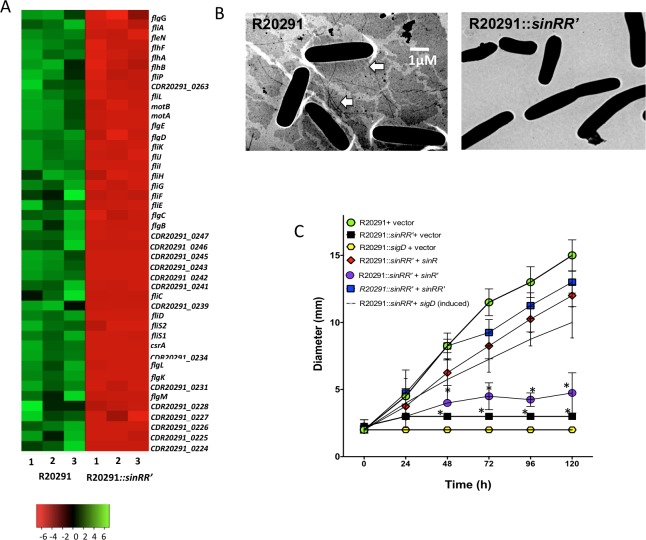
C. difficile sinRR’ mutants are non-motile
The JIR8094 strain was intrinsically non-motile due to mutations within the flagellar operon [49]. Hence, we choose only R20291 and R20291::sinRR’ to perform motility-related experiments. The R20291::sigD mutant and the R20291::sinRR’ strains with vector alone (pRPF185) were used as the controls. Exponentially growing bacterial cultures were spotted on BHI with 0.3% agar and was incubated at 37°C for 36h to monitor motility. The bacterial cultures expressing sinRR’, or sinR or sinR’ from the tet-inducible promoters were spotted on BHI with 50 ng/ml of ATc and 0.3% agar. In the motility assays, the R20291::sinRR’ strain was defective in motility (Fig 4C and S6 Fig). The transcriptome analysis supported our observation, where sigD, the sigma factor needed for the transcription of the flagellar operons, was found to be 14-fold under-expressed in the R20291::sinRR’ (Fig 4A, S4 Table) along with other motility-related genes. Electron microscopic analysis followed by negative staining failed to detect flagellar structures in the R20291::sinRR’ (Fig 4B). A dot blot analysis with FliC (the flagellar structural protein) specific antibodies also confirmed the absence of flagella in the R20291::sinRR’ strain (S6 Fig). Expression of sinRR’ from its promoter or the inducible promoter revived the motility (Fig 4C). Interestingly, expression of SinR alone was sufficient to revive the motility in the R20291::sinRR’ strain, whereas the SinR’ expression alone did not have any effect (Fig 4C).
Fig 4. Mutation in the sin locus affects C. difficile flagellar synthesis.
(A) Heat map showing the lower expression of flagellar and motility-related genes in the R2091::sinRR’ mutant compared to the parent. Color intensity in each cell represents corresponding Log2 expression values in the color scale bar. (B) Transmission electron micrographs of negatively stained C. difficile cells. White arrows point to flagella. (C) Motility of R20291, sinRR’ mutant and the sinRR’ mutant complemented strains in BHIS with 0.3% agar. The sigD mutant and the sinRR’ mutant expressing sigD from an inducible promoter were included in this analysis. The swim diameters (mm) was measured every 24 h for a total of 120 h is shown and the data shown are means ± standard errors of three biological replicates. The experiments were repeated at least three times independently (*, p≤0.05 by a two-tailed Student's t-test).
SigD is needed for the transcription of the flagellar operon in C. difficile [25, 26]. To determine whether the non-motile phenotype of sinRR’ mutant is due to the reduced levels of sigD in the sinRR’ mutants, we expressed sigD from the tetracycline-inducible promoter by introducing the construct pRGL291 into the R20291::sinRR’ strain (S1). We observed motility was partially restored in the R20291::sinRR’ when the sigD expression was induced (Fig 4C), suggesting that sinRR’ controls motility by controlling the expression of sigD in C. difficile.
C. difficile sinRR’ mutants produce less toxins than their parent strains
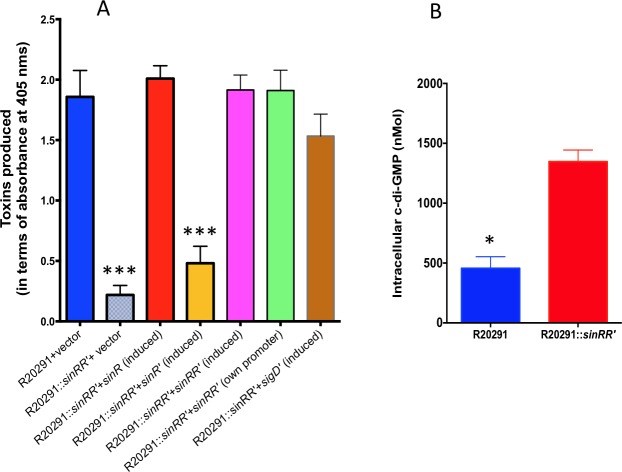
The transcriptome analysis and the follow-up QRT-PCR (Fig 5A, Table 2, S4, S6 and S8 Tables) result suggested sin locus’s role in toxin gene regulation. Toxin ELISA was performed with the cytosolic protein extracts of sinRR’ mutants and their respective parent strains.
Fig 5. SinRR’ positively influences the expression of PaLoC genes.
(A) Quantification of toxins in parent R20291 and the sinRR’ mutant complemented strains using toxins specific ELISA. The data shown are means ± standard errors of three replicates. Statistical significance was tested using one-way ANOVA, followed by Dunnett’s multiple comparisons test comparing values to the average of the parent with vector control (*** <0.0005 p-value). (B) Increased intracellular levels of c-di-GMP in the sinRR’ mutant. Statistical analysis was performed using two-tailed t-test (* <0.05 p-value).
Table 2. Expression levels of PaLoc genes and their regulators in R20291::sinRR’.
| Gene | Known or predicted function | RNA-Seq Analysis Fold-change WT/mutant |
Q-RT-PCR Analysis Fold-change WT/mutant |
Expression in sinRR’ mutant | ||||
|---|---|---|---|---|---|---|---|---|
| Actual | Log2 ratio | Adj.p value | Actual | Log2 ratio | Adj.p value | |||
| tcdR | Sigma factor for toxin genes | 32.9 | 5.0 | 1.11E-16 | 6.09 | 2.06 | 5.35E-03 | Under-expressed |
| tcdB | Toxin B | 88.4 | 6.5 | 5.80E-12 | 8.02 | 3.00 | 1.94E-11 | Under-expressed |
| tcdE | Holin like protein | 44.2 | 5.5 | 6.57E-05 | 6.29 | 2.65 | 3.32E-04 | Under-expressed |
| tcdA | Toxin A | 13.1 | 3.7 | 3.04E-04 | 7.89 | 2.98 | 0.00452 | Under-expressed |
| sigD | Sigma factor for flagellar operon | 14.4 | 3.8 | 4.13E-08 | 24.76 | 4.63 | 6.39E-03 | Under-expressed |
| dccA | Diguanylate cyclase | 0.10 | -3.3 | 2.05E-24 | 0.008 | -6.93 | 9.67E-04 | Over-expressed |
| codY | GTP sensing transcriptional regulator | 0.35 | -1.5 | 7.94E-18 | 0.12 | -3.03 | 2.23E-05 | Over-expressed |
| ccpA | transcriptional regulator | 1.29 | 0.4 | 3.45E-03 | 1.95 | -0.97 | 8.45E-08 | No significant change |
Bacterial cultures expressing either sinRR’ or sinR/sinR’ alone from the tetracycline-inducible promoter were grown for 6h in TY medium and were induced with 50ng/ml of ATc for 5 hours. Cytosolic proteins harvested from these induced cultures were used for toxin ELISA. We observed a six-fold reduction in toxin production (Fig 5A) in the R20291::sinRR’ when compared to the R20291 strain. In JIR8094::sinRR’ however, a moderate two-fold reduction in toxin level was recorded when compared to the parent strain (S7 Fig). Expression of sinRR’ in the mutants brought the toxin production back to the level comparable to the parent strains. As we observed in the motility assay, expression of sinR alone was sufficient to bring back the toxin production in the sinRR’ mutant, while expression of sinR’ did not show any effect. In C. difficile, SigD positively regulates tcdR, the sigma factor needed for toxin gene transcription [25, 26]. Interestingly, the expression of sigD from an inducible promoter revived the toxin production in sinRR’ mutants, suggesting that sinRR’ controls both toxin production and motility by regulating sigD in C. difficile.
Elevated c-di-GMP levels are present in sinRR’ mutant
We observed that SigD expression in the sinRR’ mutants partially recovered both the motility and the toxin production in that strain (Fig 4C and Fig 5A). The main question that arises from this observation is how SinR controls sigD expression. The sigD gene is part of the flagellar operon, whose transcription is directly controlled by the intracellular cyclic di-GMP (c-di-GMP) concentration [26, 50]. Within the cells, the c-di-GMP is synthesized from two molecules of GTP by diguanylate cyclases (DGCs) and is hydrolyzed by phosphodiesterases (PDEs) [50, 51]. The functionality of several of these C. difficile DGCs and PDEs has been confirmed by expressing them heterologously in Vibrio cholerae, where they resulted in phenotypes (biofilm formation and motility) that correspond to elevated or lowered levels of intracellular c-di-GMP [51]. In C. difficile when CD630_1420 (dccA) was expressed from an inducible promoter, it resulted in elevated levels of intracellular c-di-GMP and reduced bacterial motility [50]. In R20291::sinRR’, ten-fold more (-3.3 Log2 fold) dccA (CDR2029_1267) transcript was observed (S5 Table) compared to parent. We measured the intracellular concentration of c-di-GMP (S8 Fig) and observed a nearly three-fold increase in the c-di-GMP concentration in the sinRR’ mutant compared to the parent R20291 strain (Fig 5B). This elevated intracellular level of c-di-GMP in sinRR’ mutants can block the sigD expression, which in turn will result in reduced motility and toxin production (Figs 4C and 5B). Hence, when sigD was expressed from the tetracycline-inducible promoter (which is not affected by c-di-GMP concentration), motility and toxin production in the sinRR’ mutant could be revived. These two findings corroborate our conclusion that elevated levels of c-di-GMP in sinRR’ mutant plays a major role in controlling its toxin production and motility. We are currently performing experiments to test whether SinR can directly regulate dccA in C. difficile.
Inactivation of SinR' results in hyper-sporulation, higher toxin production, and motility than the parent strain
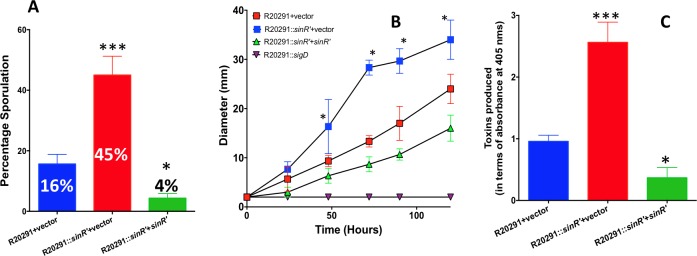
Results from the sinR and sinR’ complementation experiments showed that expression of SinR alone could revive the toxin production and the motility in the R20291::sinRR’ strain, whereas SinR’ expression alone did not have any effect on the toxin production or the motility (Figs 4C and 5A). These results suggested that among SinR and SinR’, only SinR can directly influence the toxin production and the motility, which raised the question on the role of SinR’ in these pathways. To find the answer, we created a sinR’ mutant which expressed SinR in the absence of SinR’ (S9 Fig). Our repeated attempts to create a sinR’ mutant using the similar technique in the JIR8094 background failed for unknown reasons. Mutation in sinR’ was confirmed by PCR (S9 Fig) and western blot analysis using SinR’ specific antibodies. As expected the SinR’ mutant produced SinR protein, but not the SinR’ (S9 Fig). The R20291::sinR’ grew almost similar to the parent strain and did not show any profound autolysis phenotype as the R20291::sinRR’ (S6 Fig). We performed the assays to measure sporulation, motility and toxin production in the R20291::sinR’. In the sporulation assay, it was found that R20291::sinR’ produced nearly three-fold more spores than the parent R20291 strain (Fig 6A). The R20291::sinR’ was more motile than the R20291 strain (Fig 6B). Similarly, a 2.5-fold increase in the toxin production was observed in the R20291::sinR’ when compared to the parent strain (Fig 6C). These initial results revealed that SinR’ can negatively influence sporulation, toxin production, and motility. In our complementation of R20291::sinRR’ we showed that presence of SinR’ alone in the C. difficile cells in the absence of SinR could not influence either toxin production or the motility (Fig 4C and Fig 5A). Hence, SinR’ must be influencing these pathways through its action on SinR. For example, if SinR’ is an inhibitor of SinR then the absence of SinR’ in the R20291::sinR’ would result in increased SinR activity, which in turn may result in increased sporulation, toxin production and motility in this strain. To test this hypothesis, we performed two experiments. First, tested the effect of over-expressed SinR in the wild-type strain; Second, we checked for physical interaction of SinR with SinR’ proteins by performing pull-down experiments.
Fig 6. Characterization of C. difficile R20291::sinR’.
(A) C. difficile cultures were grown in 70:30 medium for 30 h under anaerobic conditions and Sporulation frequency (CFU/ml of ethanol resistant spores) of R20291, sinR’ mutant was determined. The data shown are means ± standard errors of three biological replicates. (B) Motility assays of the C. difficile R20291, sinR’ mutant and complemented sinR’ mutant. The experiments were repeated at least three times independently (*, P≤0.05 by a two-tailed Student's t-test). (C) Toxin production measured by ELISA. Statistical analysis was performed using one way-ANOVA with Dunnett’s multiple comparisons test comparing values to the average of the parent with vector control (***<0.0005, *< 0.05 p-value).
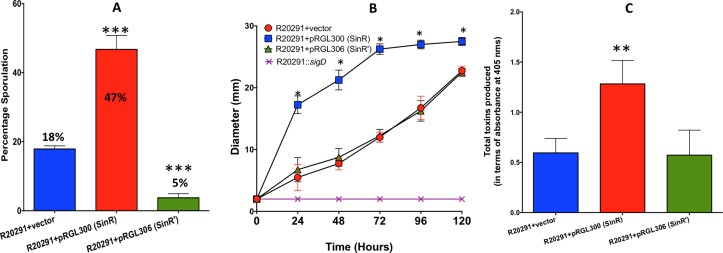
Overexpression of SinR in the wildtype strain R20291 results in hyper- sporulation and increased the toxin production and motility
The plasmid construct with either sinR (pRG300) or sinR’ (pRG306) under tetracycline-inducible promoter were introduced into R20291 parent strain and were tested for their toxin production, sporulation, and motility upon induction with ATc. The R20291 strain with the vector alone was used as the control in these assays. To perform the sporulation assay, we used bacterial cultures grown in 70:30 medium supplemented with 50 ng/ml of ATc for 36 hours. Sporulation efficiency was enumerated as described in the method section. Overexpression of sinR in R20291 strain increased its sporulation efficiency 2.5-fold (45%) when compared to the control strain, where the average sporulation efficiency was 18%. Overproduction of SinR’ in R20291, however, reduced the sporulation efficiency to 5% (Fig 7A). Overproduction of SinR in R20291 resulted in increased motility as well (Fig 7B). In C. difficile, toxin production is minimal during exponential phase (~4 to 8h) of the bacterial culture and reaches its maximum during the stationary phase (12h -16h) [9]. To detect any positive influence of both SinR and SinR’ on toxin production in the parent strain, we chose to use the 8h time point. The bacterial cultures were grown for 6h in TY medium and were induced with 50 ng/ml of ATc for two hours before harvesting their cytosolic protein for Toxin ELISA. Results from these experiments showed that overexpression of sinR resulted in a nearly 2.5-fold increase in the toxin production in the R20291 strain when compared to the R20291 with vector alone control (Fig 7C). No significant effect on toxin production was observed when sinR’ was overexpressed in R20291 (Fig 7C). This could be because sin locus is expressed only during the early stationary phase (10-12h) in C. difficile (S2 Fig). We performed toxin ELISA at 8h time-point when SinR is predicted to be lower in the bacterial cells. If SinR’ acts on toxin production primarily by repressing SinR, then overexpression of SinR’ at this time-point will not have any effect on toxin production. Nevertheless, results from this overexpression studies demonstrated that increased SinR content in C. difficile could result in increased toxin production, motility, and sporulation.
Fig 7. Effect of sinR or sinR’ overexpression in the R20291 strain.
The sinR or the sinR’ gene was cloned under tetracycline-inducible promoter and the resulting plasmid constructs were introduced into wildtype (WT) R20291 strain for overexpression. (A) Toxin ELISA, (B) Motility assay (C) Sporulation frequency. The data shown are means ± standard errors of three biological replicates. Statistical analysis was performed using one way-ANOVA with Dunnett’s multiple comparisons test comparing values to the average of the parent with vector control (***<0.0005, *< 0.05 p-value).
SinR’ interacts with SinR
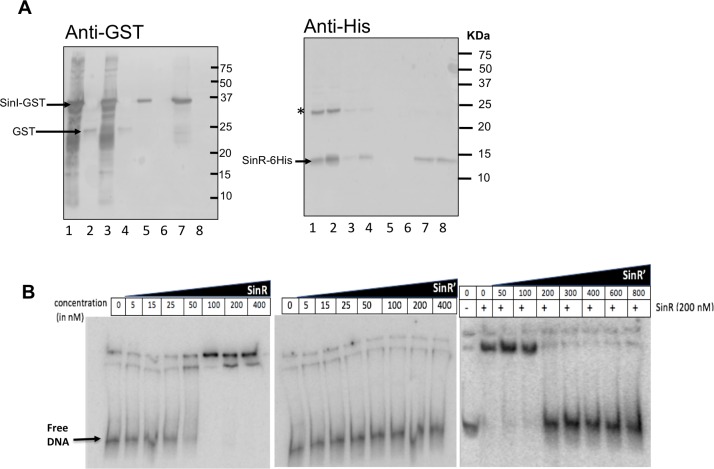
In B. subtilis, SinR monomers bind with each other to form a homotetramer, which would then bind to upstream sequences of the target genes to repress their expression [34, 52]. SinI in B. subtilis binds with SinR and prevents the SinR homotetramer formation and thus blocks its activity [52]. To test the protein-protein interaction of C. difficile SinR with SinR’, we performed GST pull-down experiments using SinR-6His and SinR’-GST. Purified SinR-6His protein was mixed with crude lysates from E. coli expressing SinR’-GST. When we passed this mixture through the Ni++ affinity chromatography column, we pulled out SinR-6His along with SinR’-GST, suggesting the tight association of SinR with SinR’ (Fig 8A, lanes 5, 7). In control, the GST alone did not interact with the SinR-6His (Fig 8A, lanes 6, 8), confirming protein specific interaction between SinR with SinR’. These results provided compelling evidence that SinR’ affects toxin production and sporulation indirectly by binding with SinR to inhibit its activity on its target genes.
Fig 8. SinR’ interacts with SinR.
(A) In vitro, protein-protein interactions indicate that SinR’ binds tightly to SinR. GST-tagged SinR’ protein was incubated with SinR-6His proteins and purified using Ni++ agarose affinity columns. The elutes were probed with anti-GST and with anti-His antibodies. Lanes details are as follows: Input 1: Mixture of SinR’-GST expressing E.coli lysate with purified SinR-6His. 1.Input 2: Mixture of GST expressing E. coli lysate with purified SinR-6His. 2. Unbound from input 1 after passing through Ni++ column. 3. Unbound from input 2 after passing through Ni++ column. 4. Elute with 50 mm imidazole (SinR’-GST + SinR-6His). 5. Elute with 50 mM imidazole (GST+SinR-6His) 6. Elute with 200 mM imidazole (SinR’-GST + SinR-6His). 7. Elute with 200 mM imidazole (GST+SinR-6His). * indicates SinR-His dimer. (B) Interactions of SinR with codY promoter region. EMSA analysis of SinR-6His, SinR’-6His, a mixture of SinR’-6His and SinR-His binding to codY probe.
SinR binds to codY promoter region
Transcriptome analysis of the R20291::sinRR’ showed up-regulation of codY, an important global regulator by ~3 to 30 fold compared to parent strains (S5 Table, S8 Table). CodY is highly conserved in many Gram-positive bacteria [53–55]. In B. subtilis it regulates several metabolic genes and controls competence, sporulation, and motility [56–58]. In C. difficile, the codY mutant produced more toxins and spores than the parent strains and thus it is a repressor of these pathways [14, 17, 18]. We hypothesized that many phenotypes and transcriptional changes we observe in the sinRR’ mutant could be related to the up-regulation of codY in these mutant strains. To investigate whether SinR and SinR’ or both controls codY expression by binding to the promoter region of codY, we carried out electrophoretic mobility shift assays (EMSAs). We used radiolabeled DNA probe that contained the putative promoter region of the codY gene and performed binding reactions using purified SinR-6His or SinR’-6His proteins. First, we tested SinR alone at increasing concentrations and found that it can shift the probe when used above 100 nM concentration (Fig 7B). When SinR’ was used similarly, it was unable to cause the mobility shift of the probe, even at the highest concentration (Fig 7B). We then tested whether SinR’ would prevent SinR from binding to the codY promoter region. To do this, we used increasing amounts of SinR’, in the presence of a fixed amount of SinR (Fig 7B). The results show that the presence of SinR’ in the reaction mix could prevent SinR from binding to the DNA. As a negative control, we used a DNA probe that contained the promoter region of gluD, which codes for glutamate dehydrogenase (GDH). Neither SinR nor SinR’ was able to shift the control DNA even at the highest concentrations tested (S10 Fig). Based on these results, we conclude that SinR binds specifically to codY promoter region to control its transcription. This result also provided evidence that the SinR’ interaction with SinR prevents its regulatory activity on its target gene.
CodY regulates sin locus expression
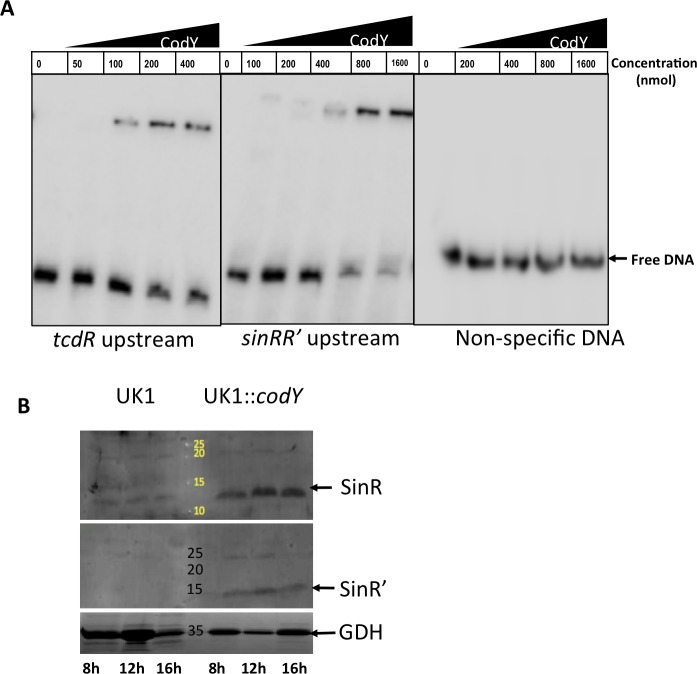
In a recent study, CodY was found to negatively regulate sinRR’ expression in the C. difficile 630Δerm strain [14]. A CodY putative binding site was identified in the sin locus upstream sequence, and reporter fusions with the sin locus promoter revealed the CodY could negatively regulate sin locus expression in this strain. However, in the UK1 strain (belongs to the ribotype 027 as R20291), the promoter fusion revealed a positive regulation of sin locus by CodY. Because of these contradictory observations, one could not conclude whether CodY regulates sin locus. To examine the role of CodY on sin locus expression, we performed EMSA with purified CodY-6His and the putative CodY binding region upstream of sin locus. An oligonucleotide with putative CodY binding sequence upstream of sinR was synthesized (ORG 721) (S2 Table) and was radioactively labeled with [γ- 32 P] dATP. A double-stranded DNA probe was generated after annealing with the complementary oligonucleotide (ORG722). It is worth noting no sequence difference was found within this putative sin promoter regions of the UK1, R20291, JIR8094 and 630Δerm genomes. We also generated probes with a known CodY binding sequence upstream of the tcdR gene (using ORG719 and ORG720) and with non-specific sequence (ORG702 and ORG723) as positive and negative controls respectively. EMSA was performed by incubating the radioactively labeled probes with varying concentrations of purified CodY-6His. We found that CodY could bind to the sequence upstream of sin locus at the concentration of 400 nM (Fig 9A). As expected the shift was observed with the positive control probe, while no shift could be observed with the non-specific DNA probe even with high protein concentrations (Fig 9A). Binding of CodY to its targets most of the time results in repression of their transcription [17, 18, 58]. However, there are few targets where CodY was found to promote transcription [58]. To check whether CodY has any positive influence on sin locus expression in UK1 strain as reported [14], we performed western blot analysis and looked for SinR and SinR’ in UK1 strain and its codY mutant (UK1::codY). Results showed that SinR and SinR’ protein content in the UK1::codY mutant was higher than in the UK1 parent strain (Fig 9B). Our data demonstrate that CodY has a negative impact on SinR and SinR’ production in this strain. Since our repeated attempts to create codY mutants in R20291 and JIR8094 strains failed, we could not include them in this analysis. Nonetheless, our results from the EMSA and the western blot analyses corroborate the negative regulation of the sin locus by CodY.
Fig 9. CodY controls the sin locus expression.
(A) CodY-6His binding to sin locus promoter region. The tcdR upstream and a non-specific DNA probe was as positive and negative controls respectively. (B) Western blot analysis of UK1 and UK1::codY mutants to detect SinR and SinR’ proteins.
R20291::sinRR’ is less virulent in hamster
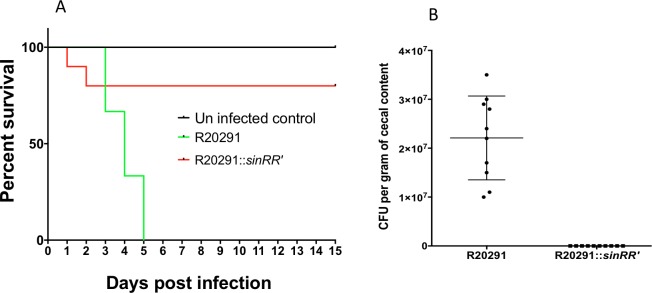
Since the C. difficile sin locus was found to be important for the regulation of many important pathways under in vitro growth conditions, we wanted to determine its significance in C. difficile pathogenesis. We used the hamster model in which C. difficile infection is known to cause severe disease signs [59, 60]. Syrian hamsters were gavaged with 2,000 vegetative cells of C. difficile strain R20291 or with R20291::sinRR’ and monitored for C. difficile infection. Fecal pellets were collected daily until animals developed diarrheal symptoms. All ten animals infected with parental strain R20291 succumbed to the disease within five days after bacterial challenge. Two of the ten animals infected with R20291::sinRR’ exhibited disease symptoms within two days after challenge (Fig 10A). Diseased hamsters were sacrificed (see M&M), and their cecal contents were collected for toxin ELISA and CFU count. All surviving sinRR’ mutant infected hamsters (8 in total) and uninfected control hamsters were also sacrificed fifteen days post-infection, and their cecal contents were also tested for toxins and C. difficile cells. Toxins could be detected (S11 Fig) in the cecal contents of all the diseased hamsters (10 from R20291 group and two from R20291::sinRR’ group), which confirmed the occurrence of CDI in them. However, toxins could not be detected in the eight hamsters that survived the R20291::sinRR’ challenge. The cecal contents of R20291 infected hamsters contained nearly 107 colony-forming units per gram. No C. difficile could be recovered from the cecal contents of any of the R20291::sinRR’ challenged animals, including of the two hamsters that came down with CDI in this group (Fig 10B). If the sinRR’ mutant lyses in vivo, as we observed in in vitro growth conditions, it could explain why we could not recover any C. difficile cells but could detect toxins in the cecal contents of that two hamsters that came down with the disease after sinRR’ mutant challenge. Since nearly 80% of the animals survived the R20291::sinRR’ challenge, we conclude that members of the SinRR’ regulon are needed for C. difficile successful pathogenesis.
Fig 10. Disrupting sinRR’ decreases morbidity in hamster models of C. difficile infection.
(A) Kaplan-Meier survival curve of clindamycin-treated Syrian golden hamsters inoculated with 2,000 vegetative cells of C. difficile R20291 (n = 10) or sinRR’ mutant (n = 10). Six animals were used as an uninfected control. Animals were monitored every four hours for the symptoms of lethargy, poor fur coat, wet tail or hunched posture. Moribund animals were euthanized and log-rank statistical analysis was performed; p<0.001. (B) Total number of C. difficile colony forming units (CFU) /gm of cecal contents recovered postmortem.
Discussion
This study aims to decipher the role of the SinRR’ regulators in C. difficile physiology. In C. difficile, there has been no data explaining their function, except for a few expression analyses, where mutations in sigH, tcdR, codY, spo0A, opp, app were found to affect the expression of the sin locus [13, 14, 21, 22, 60]. Initial clues about the role of SinR and SinR’ in sporulation came from the work performed by Saujet et al. where they showed increased expression of sinR in the asporogenous sigH mutant, suggesting it to be a negative regulator of sporulation as in the case of B. subtilis [22]. However, sinR was found to be up-regulated in the hyper-sporulating oligopeptide transporter opp-app mutant and was down-regulated in the hypo-sporulation tcdR mutant [13, 60]. These later studies suggested the positive influence of SinR on sporulation. In this work, we mutated the sin locus in two different C. difficile strains and conclusively showed that unlike B. subtilis SinR, which inhibits sporulation, C. difficile SinR has a positive effect on sporulation.
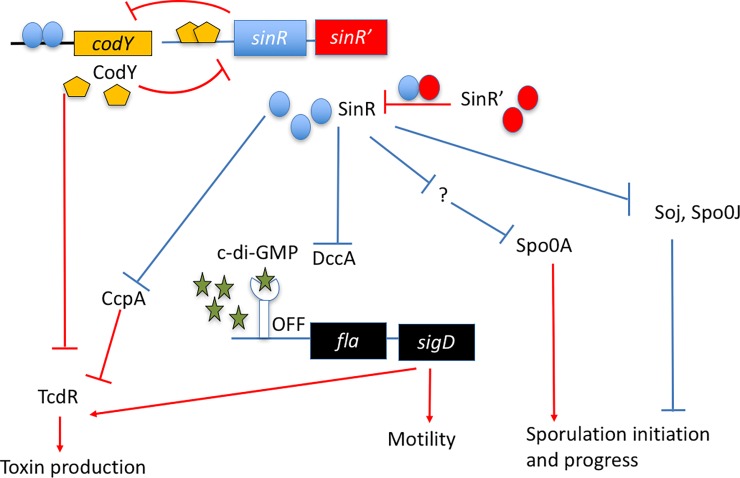
Transcriptome analysis of sinRR’ mutants revealed that in addition to sporulation, genes involved in motility, transport, stress response, cell wall biogenesis, and various metabolic pathways were also affected. It is worth noting that cynT, the gene adjacent to sin locus (Fig 1B), is one among the many metabolic genes that were found to be down-regulated in the sinRR’ mutants (S4 and S6 Tables). The analysis also revealed that the sin locus mutations could affect the transcription of many important regulators, including codY, sigD, spo0A, and tcdR. This observation compelled us to hypothesize that SinRR’ might be indirectly influencing transcription of many of these genes by controlling their regulators. For example, changing in the transcription of codY, a global regulator can affect the gene regulatory circuits of various pathways.
CodY is known to be a sensor of the metabolic state of the cell. During the exponential growth phase, when the nutrients are abundant, CodY binds to branched-chain amino acids (BCAAs), and GTP and acts primarily as a repressor of various alternative metabolic pathways [17, 18]. When nutrients become limited in the cell, CodY is no longer bound by the cofactors and the transcriptional repression by CodY is alleviated on its targets. In C. difficile, CodY controls toxin production and sporulation in addition to metabolic pathways [14, 17, 18]. The transcription of codY was found to be up-regulated in the R20291::sinRR’ (S5 Table), (Fig 11). This observation of increased codY transcription in the asporogenic sinRR’ C. difficile mutant is consistent with the recent findings that a C. difficile codY mutant hyper- sporulates [14]. To test whether increased CodY activity in the mutant is the reason for its lower toxin production and sporulation, we tried to isolate a sinRR’-codY double mutant and were unsuccessful even after several attempts. However, our EMSA experiments with purified SinR and codY upstream DNA showed that the SinR could specifically bind to this region, possibly to repress its transcription. We have also shown that purified CodY, in turn, can bind with the upstream region of sin locus upstream region to control its expression. Since the sin locus codes for both sinR and its antagonist sinR’, SinR repression on codY would be moderate when compared to CodY’s repression on the sin locus. Also, when the cells enter the stationary phase, CodY repression on the sin locus may be alleviated in the absence of its co-substrates and will result in the sin locus expression, which we found to be essential for sporulation initiation. We performed dot blot analysis with cytosolic proteins of R20291 and R20291::sinRR’ and determined that CodY in R20291::sinRR’ was only moderately higher than R20291 (S12 Fig). This could be due to the cell to cell variation in gene expression within the test population. For example, only 18% of the R20291 population enters sporulation in the growth conditions we tested. In C. difficile, only cells with low or inactive CodY enter sporulation. If we consider sporulation as an indirect measure for inactive CodY in a bacterial cell, we can say that the CodY production or activity was affected only in a fraction of cells in the parent population. To overcome this issue, we compared the CodY content in R20291::sinR’ cells (which produce more SinR) with R20291::sinRR’. It is worth to note that nearly 50% of R20291::sinR’ culture enters sporulation. Nearly two-fold more CodY could be detected in R20291::sinRR’ cells when compared to R20291::sinR’ cells. Other than modulating CodY content in C. difficile, SinR could also affect the CodY activity indirectly by affecting the concentrations of CodY substrates (BCAA and GTP). The transcriptome analysis indeed showed numerous metabolic genes to be affected in the sinRR’ mutant.
Fig 11. Schematic diagram showing sin locus regulation of genes involved in toxin production, sporulation, and motility in C. difficile.
Known genetic interactions are marked in red and the predicted interactions are marked in blue.
In the JIR8094::sinRR’ mutant, codY was not among the differentially regulated genes. However, in this strain ccpA was up-regulated nearly 13.5-fold (S7 Table). Similar to CodY, CcpA also represses toxin gene expression in C. difficile [15, 16]. Thus lower toxin production in JIR8094::sinRR’ could be due to the higher CcpA activity in this mutant (Fig 11). We are currently testing whether ccpA is directly regulated by SinRR’. We are also setting up experiments to check whether increased CcpA has any role in controlling codY expression in the JIR8094::sinRR’ strain.
SigD is one other regulator whose expression was found to be affected in the sinRR’ mutants. In C. difficile, the sigD expression is repressed by elevated levels of c-di-GMP [50]. The enzyme, diguanylate cyclase coded by dccA synthesize c-di-GMP from GTP. In this study, we have shown the expression of dccA is up-regulated in sinRR’ mutant (S5 Table and Table 2) and the observation of three-fold higher intracellular concentration of c-di-GMP in the sinRR’ mutant, corroborated the transcriptome data. These results suggest that SinR and SinR’ regulates motility and toxin production indirectly by regulating the c-di-GMP production. Another scenario that can result in higher intracellular c-di-GMP concentration is when c-di-GMP degrading phosphodiesterases are reduced within the cell. In C. difficile, pdcA codes for a c-di-GMP phosphodiesterases, and it was recently identified to be repressed by CodY [61]. RNA-Seq analysis did not identify pdcA as one among the differentially regulated genes in R20291::sinRR’ strain. However, it was under-expressed nearly 4-fold in JIR8094::sinRR’ (S6 Table) mutant. Increased CodY activity in the sinRR’ mutant could indirectly result in increased c-di-GMP concentration, which in turn can suppress toxin production and motility.
In B. subtilis, the SinR’s repressor’s activity on its target genes is inhibited by SinI, which is coded in the same operon (Fig 1A). In B. subtilis the polycistronic sinRI transcripts are produced from two upstream promoters. The monocistronic sinR transcripts are driven from a promoter located within the coding region of sinI. Regulating the transcription rate of sinRI and sinR helps B. subtilis to control its SinR and SinI content. Our RT-PCR and QRT-PCR analysis detected sinRR’ transcripts in C. difficile. We have also shown that disrupting sinR by insertion mutagenesis affects both sinR and sinR’ transcription. These results suggest that sinRR’ is transcribe as a bicistronic message. However, there is a possibility that sinR’ may have an independent promoter within sinR coding sequence as in B. subtilis. Our QRT-PCR analysis repeatedly detected lower levels of sinR’, sinRR’ transcripts than the sinR transcripts. Western blot analysis also revealed lower levels of SinR’ than the SinR in growth conditions tested (Fig 2B-lane 2and S9C Fig). There is a possibility that mRNA degradation from the 3’ end can result in lower levels of sinR’ transcripts, which in turn can result in lower levels of SinR’ than SinR. We did not detect any secondary structures upstream of sinR’ that can influence its translation rate or translation initiation. We, however, noted that the RBS of sinR’ are just two nucleotides away from the sinR stop codon. Ribosome complex occupying the sinR stop codon can prevent the assembly of new ribosome complex at the sinR’ RBS to initiate translation. Since SinR’ has a DNA binding domain, it is also possible that SinR’ may work as direct regulator independently from SinR and may have its own targets for regulation. In such case, SinR’ may not always be available to inhibit SinR function. A transcriptome analysis of sinR’ mutant and its comparison with sinRR’ transcriptome may help us to identify direct targets of SinR’.
In B. subtilis, other than SinI, SinR also interacts with SlrR and SlrA to regulate genes involved in matrix formation (the eps and tap-sipW-tas operon), autolysis (lytABC) and motility (hag, encoding flagellin) [29, 31]. In B. subtilis, the SlrR is a DNA binding protein, and it is homologous to SinR. Conversely, SlrA is a small protein devoid of any DNA binding domains and is homologous to SinI [38, 39]. While SlrR can form heterodimers with SinR to repress lytABC and hag expression, it can also inhibit SinR’s repression activity on eps and tap-sipW-tas operons [38, 39, 62] which are needed for biofilm formation. The C. difficile sin locus codes for two DNA binding proteins SinR and SinR’ and their interactions resembles the interaction between B. subtilis SinR-SlrR. Similar to B. subtilis SlrR, C. difficile SinR’ carries a DNA binding domain and it will be interesting to analyze whether SinR-SinR’ complexes together are needed for the repression of any genes. It is important to note that the autolysis phenotype of sinRR’ mutant was complemented only when both SinR and SinR’ were expressed (Fig 2C). This suggests that like B. subtilis SinR-SlrR complex, SinR-SinR’ complex together repress autolysis in C. difficile. No lytABC homologs could be identified in C. difficile genome, and the precise reason for autolysis in sinRR’ mutant is not clear yet. However, the RNA-Seq data revealed that nearly 6% of the differentially expressed genes in sinRR’ mutant plays a role in cell wall synthesis or assembly. This highlights that SinR and SinR’ play an important regulatory role in this pathway.
Among the phenotypes tested, asporogenesis of the sinRR’ mutant was the only one we could not complement. Even the expression of spo0A failed to initiate sporulation in this mutant. Transcripts of Spo0A~P activated sigE and sigF did not show any increase when spo0A was expressed in the sinRR’ mutant, suggesting the Spo0A remain unphosphorylated and inactive. We are currently performing additional experiments to test this hypothesis.
Another regulatory checkpoint for sporulation initiation is chromosomal DNA replication and segregation. This is achieved through the action of Soj and Spo0J in B. subtilis, where they repress sporulation until chromosomal segregation has occurred. They block the spo0A dependent transcription in B. subtilis [63]. The spo0J and soj homologs in C. difficile are CD630_3671 and CD630_3672, respectively in an operon, which also carries CD630_3673, an additional Spo0J-like orthologue. In both JIR8094::sinRR’ and R20291::sinRR’, all three genes were up-regulated ~3 fold. Hence, the inactivation of Spo0A could result partly because of the up-regulation of the soj operon in the sinRR’ mutants (Fig 11). But the function of soj and spo0J in C. difficile should be determined before we can speculate their roles in asporogenesis of sinRR’ mutants.
BLAST search revealed that SinRR’ to be unique to C. difficile and its close relative Clostridium sordellii. The sin locus is absent in other Clostridia. Even though sporulation-specific sigma factors appear to be conserved among Clostridia, recent studies have suggested that sporulation initiation and regulation of C. difficile to be distinct [64, 65]. Since the sin locus appears to play a significant role in sporulation initiation and regulation, it is reasonable to speculate its presence could be one of the reasons why the regulation of sporulation initiation is distinct in C. difficile.
In summary, our study supports earlier reports that in C. difficile, virulence, sporulation, metabolism and motility pathways are inter-connected [13–24]. While many regulators in this network are yet to be identified, here we present the evidence that SinRR’ play a central role in this regulatory network. SinR regulates multiple pathways by controlling other global regulators. Finding genes that are directly under SinR regulation may lead to the identification of new regulatory genes and gene products that are important for C. difficile pathogenesis.
Materials and methods
Ethics statement
All animal procedures were performed with prior approval from the KSU Institutional Animal Care and Use Committee (protocol #3657). Animals showing signs of disease were euthanized by CO2 asphyxia followed by thoracotomy as a secondary means of death, in accordance with Panel on Euthanasia of the American Veterinary Medical Association. Kansas State University is accredited by AAALAC International (Unit #000667) and files an Assurance Statement with the NIH Office of Laboratory Animal Welfare (OLAW). KSU Animal Welfare Assurance Number is D16-00369 (A3609-01), and USDA Certificate Number is 48-R-0001. Kansas State University utilizes the United States Government Principles for the utilization and care of vertebrate animals used in testing, research and training guidelines for appropriate animal use in a research and teaching setting.
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in S1 Table and cloning strategies used are listed in S1 Text. Clostridium difficile strains were grown anaerobically (10% H2, 10% C02 and 80% N2) in TY (Tryptose and Yeast extract) agar or broth as described previously [60, 66]. Erythromycin (Erm; 2.5 μg ml-1), Lincomycin (Linc 20ug/ml), Cefoxitin (Cef; 25 μg/ml), thiamphenicol (Thio; 15 μg ml-1) were added to culture medium whenever necessary. Sporulation was induced in respective C. difficile strains by growing them in 70:30 sporulation medium (63 g Bacto-Peptone, 3.5 g Protease-Peptone, 11.1 g BHI, 1.5 g Yeast-Extract, 1.06 g Tris base, 0.7 g NH4SO4, 15 g agar per liter) [67]. Escherichia coli strain S17-1 [68] was used for conjugation and cultured aerobically in Luria-Bertani (LB) broth and supplemented with chloramphenicol (25μg ml-1) or ampicillin (100μg ml-1) as indicated.
Construction of C. difficile mutant strains
ClosTron gene knockout system [69] was used to construct sinRR’ and sinR’ mutants. For sinRR’ disruption, the group II intron insertion site between nucleotides 141 and 142 in sinR gene in the antisense orientation was selected using the Perutka algorithm, a Web-based design tool available at http://www.Clostron.com. For sinR’ mutant construction, the group II intron insertion site between nucleotides 129 and 130 in the sense direction was selected. The designed retargeted intron was cloned into pMTL007-CE5 as described previously [59, 70]. The resulting plasmids pMTL007-CE5::Cdi-sinR-141s or pMTL007-CE5::Cdi-sinR’-129s was transferred into C. difficile cells by conjugation as described earlier [59, 70]. The potential Ll.ltrB insertions within the target genes in the C. difficile chromosome was conferred by the selection of erythromycin or lincomycin resistant transconjugants in 5 μg ml-1erythromycin or 20 μg ml-1 lincomycin plates. PCR using gene-specific primers (S2 Table) in combination with the EBS-U universal and ERM primers was performed to identify putative C. difficile mutants.
General DNA techniques
DNeasy Blood and Tissue Kit (Qiagen) was used to extract chromosomal DNA from the C. difficile cultures. Primers used throughout the study are listed in S2 Table and S3 Table. Geneclean Kit (mpbio) was used to gel extract the PCR products, and QIAprep Spin Miniprep Kit (Qiagen) was used to extract plasmid DNA. Standard procedures were used to perform routine cloning.
Sporulation efficiency assays
Sporulation assays were performed in 70:30 sporulation medium as described previously [60]. C. difficile strains were grown on 70:30 sporulation agar. After 30 h of growth, cells were scraped from the plates and suspended in 70:30 sporulation liquid medium to an OD600 of 1.0. Cells were immediately serially diluted and plated onto TY agar with 0.1% taurocholate to enumerate viable vegetative cells and spores. To determine the number of spores present, 500μl of the samples from each culture were mixed 1:1 with 95% ethanol and incubated for 1hour to kill all the vegetative cells. The ethanol-treated samples were then serially diluted, plated on TY agar with 0.1% taurocholate and incubated at 37°C for 24 to 48 hours to enumerate the number of spores. Dividing the number of spores by the total number of CFU and multiplying the value by 100 determined the percentage of ethanol-resistant spores. The results were based on a minimum of three biological replicates.
Phase-contrast microscopy
C. difficile strains were grown in 70:30 medium as described above. At indicated time points, 1 ml of culture was removed from the anaerobic chamber, centrifuged at 17,000g for 1min and suspended in 30μl of sterile PBS. A thin layer of 0.7% agarose was applied to the surface of slide and 2μl of concentrated culture was placed on it. Phase contrast microscopy was performed using 100x oil immersion objective on OLYMPUS BX41 microscope. The PixeLINK camera was used to acquire the view of at least three fields for each strain.
Transmission electron microscopy
All steps in sample preparation were performed at room temperature and solutions were prepared in 1X PBS (phosphate-buffered saline) unless indicated otherwise. For transmission electron microscopy, cells (1010) were fixed overnight in a solution of 2% glutaraldehyde and 2% paraformaldehyde. The cells were thoroughly rinsed with 1X PBS (5 minutes each) and post-fixed with 1% osmium tetroxide with constant rotation for 1–2 hours. The samples were then washed thrice with 1X PBS (5 minutes each), enblock stained with 2% Uranyl acetate in water for 1hr with light protection, and finally washed three times (5 min each) with distilled water. The cells were further dehydrated in a graded 50% -100% acetone series (vol/vol) for 5 minutes and infiltrated in graded EMBED 812/Araldite resin (Electron Microscopy Sciences) at RT with constant rotation. Thin sections of polymerized resin were placed on copper grids and stained with 2% alcoholic uranyl acetate and Reynolds' lead citrate respectively. Sections were examined with a transmission electron microscope (Philips CM100) and regions containing the cross-section of the cells were photographed at 80 kV for image analysis.
To visualize the flagella, whole bacterial cells harvested from overnight cultures were processed as above and were negatively stained with 2% uranyl acetate before transmission electron microscopy analysis.
RNA-Seq analysis
We isolated total RNA from three biological replicates of each strain belonging to early-stationary phase (12 hours after inoculation) and quality was checked using Agilent 2100 Bioanalyzer. The RNA-Seq was performed as previously described [60]. Briefly, we depleted the rRNA content in the selected samples using Epicenter Bacterial Ribo-Zero kit. Strand-specific single end cDNA libraries were prepared using Truseq Small Stranded Total RNA sample prep kit Illumina as per the manufacturers’ instructions. Illumina HiSeq2000 sequencer (multiplexing three samples per lane) was used to sequence libraries. Sequences were cleaned with AlienTrimmer [71] of adapter sequences. Only high-quality sequences with a minimum of 30 nucleotides in length were considered for further analysis. Cleaned genes were aligned to reference genomes (FN545816.1 and AM180355.1) using Bowtie (version 1.0.1) [25, 60, 72]. DESeq2 version 1.8.3 was used to perform normalization and differential analysis. Genes were considered differentially expressed if the fold change was ≥ log2 1.5 and their adjusted p-value was ≤0.05.
Cloning, expression, and purification of SinR-6His, SinR’-6His, and CodY-6His proteins in E. coli
SinR, SinR’ and CodY proteins were overexpressed in Rosetta E. coli DE3 cells using pET16B expression system. The ORFs for cloning were PCR amplified from JIR8094 chromosome using gene-specific primers (listed in S2 Table), and the amplified gene fragments were then digested with Xho1 and BamH1 to clone into pET16B digested with the same enzymes. The resulting plasmids were then transformed into E.coli Rosetta DE3 (Novagen) competent cells to obtain recombinant strains. To overexpress SinR-6His, and SinR’-6His, the E. coli recombinant strains were grown at 37°C in LB medium containing chloramphenicol (25μg ml-1) and ampicillin (100ug ml-1). Protein expression was achieved by inducing with 1mM IPTG at 17°C overnight. Cells were harvested by centrifugation, and the 6His-tagged proteins were purified by affinity chromatography on Ni++ agarose (Sigma-Aldrich) beads following the manufacturer’s recommendations.
Antibody production
The anti-SinR used in this study was raised against SinR-His6 in rabbits by Lampire Biologicals (Everett, PA). The anti-SinR’ was raised against SinR’-His6 in mice by Lampire Biologicals (Everett, PA).
Western blot analysis
C. difficile cells for western blot analysis were harvested and washed in 1x PBS solution before suspending in sample buffer (Tris 80mM; SDS 2%; and Glycerol 10%) for sonication. Whole cell extracts were then heated at 100°C for 7 min and centrifuged at 17,000 g for 1 min, and the proteins were separated by SDS-PAGE and electro-blotted onto PVDF membrane. Immobilized proteins in the membranes were then probed with specific antibodies at a dilution of 1:10,000. The blot was subsequently probed with HRP-conjugated secondary antibodies at a dilution of 1:10000. Immuno-detection of proteins was performed with ECL Kit (Thermo Scientific) following the manufacturer’s recommendations and were developed using Typhoon 9100 scanner.
Toxin ELISA
Cytosolic toxins from 16h old C. difficile cultures grown in TY medium were measured as described previously [70, 73]. In brief, one ml of C. difficile cultures were harvested and suspended in 200 μl of sterile PBS, sonicated and centrifuged to harvest the cytosolic protein. One hundred μg of cytosolic proteins was used to measure the relative toxin levels using C. difficile premier Toxin A &B ELISA kit from Meridian Diagnostics Inc. (Cincinnati, OH).
Motility assay
C. difficile cultures were grown until mid-exponential phase at 37°C. After adjusting their OD@600 to 0.5, 3μl of each strain was inoculated by stabbing or spotting into BHI medium with 0.3% w/v agar in tubes and plates respectively. After incubation at 37°C, the motility was quantified by measuring the radius of the cultures at different time points. Motility assay was performed in 4 replicates and independently repeated at least three times.
SinR-6His; SinI-GST pull-down experiment
To express SinR’-GST protein we cloned the sinR’ gene in the pGST-parallel2 expression system [74]. First, the sinR’ gene was PCR amplified using primers ORG619 and ORG620 (S2 Table) and R20291 chromosomal DNA as a template. The PCR fragments were then cloned in between NcoI and SalI sites of the pGST-parallel2 vector. The resulting plasmid was then transformed into E.coli Rosetta DE3 competent cells to obtain recombinant strain. To overexpress SinR’-GST, E. coli recombinant strains were grown at 37°C in LB medium containing chloramphenicol (25μg ml-1). Protein expression was achieved by inducing with 1mM IPTG at 17°C overnight with mild agitation. To perform the pull-down experiment, 200 μgs of whole cell lysate proteins from the E. coli cells expressing SinR’-GST was mixed with ~20 μgs of purified SinR-6His protein and incubated at 4°C for 1hr. The mixture was then passed through the Ni++ affinity column (Sigma-Aldrich) to trap and elute SinR-6His protein. Whole lysates from E. coli cells expressing GST alone was also mixed with purified SinR-6His protein, and this control mixture was processed in the same way as the test sample. The elutes from Ni++ columns were then separated by SDS-PAGE and were electro blotted onto PVDF membrane. Membranes with immobilized proteins were then probed with either Anti-6His antibodies at 1:10,000 dilution or with anti-GST antibodies at the dilution of 1:5000. Immunodetection of proteins was performed with Pierce ECL 2 Western blotting Substrate Kit (Thermo Scientific) and the Typhoon 9100 scanner.
Electrophoretic mobility shift assay (EMSAs)
SinR and SinR’ binding was performed with radioactively labeled DNA probes. The codY upstream and the gluD upstream regions were amplified using primer pairs ORG629- ORG630 and ORG72-ORG73, respectively and the products were cloned into a pGEMT cloning vector. The region was then excised from the plasmid construct using EcoRI and was radiolabeled using Klenow fragment of DNA polymerase I (NEB. labs) and [α- 32 P]dATP-6000 Ci/mmol (PerkinElmer Life Sciences). Binding experiments with radioactively labelled codY upstream DNA with SinR-6His or SinR’-6His was performed using reaction buffer containing 10 mM Tris–HCl (pH 8.0), 0.1 mM DTT, 150 mM KCl, 0.5mM EDTA, 0.1% Triton X-100 and 12.5% glycerol. For binding experiments containing both SinR and SinR’, proteins were mixed in the reaction buffer at a specified concentration and were incubated at room temperature for 30 minutes before adding the DNA probe. Reactions were loaded onto a 6% native polyacrylamide gel in 1XTBE (Tris/Borate/EDTA) and subjected to electrophoresis at 100 V for 45 minutes. Gels were then dried, and the autoradiography was performed with Molecular Dynamics Phosphor-Imager technology.
For the CodY binding experiments, the upstream region of the sin locus with the predicted CodY binding sequence (shown as underlined) 5’ TAGAAA ATTTTTTTAATTTTCAAAATATATTCTACATATCTAA was synthesized and was labeled with [γ- 32 P]dATP-6000 Ci/mmol (PerkinElmer Life Sciences) using T4 polynucleotide kinase. It was then annealed with the complementary oligo to generate double-stranded DNA probe. Known CodY binding sequence upstream of the tcdR gene was similarly synthesized (S2 Table) and used as a positive control. A non-specific double-stranded DNA was used as negative control (S2 Table). The DNA-protein binding reactions were carried out at room temperature for 30 min in 10μl volume containing 1x binding buffer [10mM Tris pH 7.5, 50mM KCl, 50μg BSA, 0.05% NP40, 10% Glycerol, 10 mM GTP and 2mM ILV (Isoleucine, Leucine and Valine), 100 μg/ml poly dI-dC and 800nM of DNA probe with varying concentration of purified CodY protein. DNA probe in reaction buffer was incubated for 10 min at RT before adding purified CodY-6His protein. The reaction was stopped by adding 5ul of gel loading buffer and electrophoresed at 100V for 1.5 h using 6% 1XTBE gel in 0.5X TBE buffer containing 10 mM ILV. Gels were then dried, and the autoradiography was performed with Molecular Dynamics Phosphor-Imager technology.
Hamster model for C. difficile pathogenesis
Syrian golden hamsters (100–120 g) were used for C. difficile infection. Upon their arrival, fecal pellets were collected from all hamsters, homogenized in 1 ml saline, and examined for C. difficile by plating on CCFA-TA (Cycloserine Cefoxitin Fructose Agar- 0.1% Taurocholate) to ensure that the animals did not harbor indigenous C. difficile. After this initial screen, they were housed individually in sterile cages with ad libitum access to food and water for the duration of the study. Hamsters were first gavaged with 30 mg/kg clindamycin [59, 75]. C. difficile infection was initiated five days after clindamycin administration by gavage with vegetative cells. We used vegetative C. difficile cells because of the test strain R20291::sinRR’ is asporogenic and do not produce any spores. Bacterial inoculums were standardized and prepared immediately before challenge as described in our earlier study [59]. They were transported in independent 1.5 ml Eppendorf tubes to the vivarium using the Remel AnaeroPack system (one box for each strain) to maintain viability. Immediately before and after infecting the animal, a 10 μL sample of the inoculum was plated onto TY agar with cefoxitin to confirm the bacterial count and viability. There were five groups of animals, including the uninfected control group. Ten animals per group were used for the infection. Approximately, 2000 C. difficile vegetative cells of R20291 strain and R20291::sinRR’ were used for the animal challenge. In the uninfected control (group 5) only five animals were used, and they received only antibiotics and sterile PBS. Animals were monitored for signs of disease (lethargy, poor fur coat, sunken eyes, hunched posture, and wet tail) every four hours (six times per day) throughout the study period. Hamsters were scored from 1 to 5 for the signs mentioned above (1-normal and 5-severe). Fresh fecal pellets were collected daily from every animal to monitor C. difficile colonization until they began developing diarrheal symptoms. Hamsters showing signs of severe disease (a cumulative score of 12 or above) were euthanized by CO2 asphyxiation. Surviving hamsters were euthanized 15 days after C. difficile infection. Thoracotomy was performed as a secondary mean of death. The cecal contents from these hamsters were collected in 15ml Nalgene tubes, secured air tight and were transported to the lab using Remel AnaeroPack system. They were then immediately subjected to CFU enumeration. For CFU enumeration, the daily fecal samples or the cecal contents collected post-mortem were resuspended in 1X PBS, serially diluted and plated onto CCFA agar with 0.1% Taurocholate (CCFA-TA). The CFU were counted after 48 h of incubation. The survival data of the challenged animals were graphed as Kaplan-Meier survival analyses and compared for statistical significance using the log-rank test using GraphPad Prism 6 software (GraphPad Software, San Diego, CA).
Supporting information
(A) Schematic representation of ClostTron (group II intron)- mediated disruption of the sinR gene in C. difficile. (B) PCR verification of the intron insertion, conducted with intron-specific primer EBS universal [EBS(U)] with sinR—specific primers ORG-549 and ORG-550.
(TIF)
(A) RT-PCR results of sinRR’, sinR and sinR’ using cDNA, RNA and genomic DNA prepared from C. difficile JIR8094 and JIR8094::sinRR’. (B) Schematic representation of gene structure in sin locus and the location of primer design site for each gene products respectively. (C) RT-PCR results of sin locus transcripts in JIR8094 and JIR8094::sinRR’ strains collected at different time points. The representative results from three independent experiments are shown.
(TIF)
(A) Western blot analysis of parent and mutants using SinR and SinR’ specific antibodies. (B) Growth curve of parent and the mutant strains.
(TIF)
Triton X-100 induced autolysis of R20291::sinRR’ at the stationary phase showing rapid lysis compared to the parent strain. Expression of sinRR’ prevented autolysis in sinRR’ mutant. The autolysis is expressed as percent initial absorbance at an optical density of 600nm. Error bars indicate ± standard deviation. The experiments were repeated at least three times independently (*, p≤0.05 by a two-tailed Student's t-test).
(TIF)
(A) Phase contrast microscopy of JIR8094 and JIR8094::sinRR’ cells. (B) JIR8904::sinRR’ mutant was asporogenic as shown in the representative TEM images in comparison with the parent strain. Black arrows indicate mature spores in the parent strain. (C) Sporulation frequency of JIR8094 and JIR8094::sinRR’ strains. The data shown are mean ± standard errors of three replicates. *** p< 0.0005 (by two-tailed student’s t-test). (D) Western blot analysis demonstrating lower Spo0A expression in the sinRR’ mutant.
(TIF)
(A) Dot blot analysis of R20291, R20291::sinRR’ proteins using FliC and GDH (internal control) specific antibody. (B) Swimming motility of the R20291 and R20291::sinRR’ strain showing the non-motile phenotype of sinRR’ mutant in BHIS with 0.3% agar.
(TIF)
Toxin ELISA performed with cytosolic proteins harvested from JIR8094 and JIR8094::sinRR’ mutant. The data shown are mean ± standard errors of three replicates. ** p< 0.005 (by two-tailed student’s t-test).
(TIF)
(A) The c-di-GMP peak in HPLC. (B) The standard curve was constructed by analyzing samples containing a predetermined amount of c-di-GMP and their respective peak area. (C) Analysis of intracellular nucleotide pools prepared from R20291 and R20291::sinRR’ cells. Arrows indicate the peak corresponding to c-di-GMP.
(TIF)
(A) PCR verification of the intron insertion verified with intron-specific primer EBS universal [EBS(U)] with gene-specific primers ORG-553 and ORG-554. (B) Schematic representation of ClostTron (group II intron)- mediated disruption of the sinR’ gene in C. difficile R20291. (C) Western blot analysis of R20291 and R20291::sinR’ proteins using SinR and SinR’ specific antibodies. (D) Growth curve of parent R20291 and sinR’ mutant in TY medium showing no autolysis of sinR’ mutant.
(TIF)
(TIF)
Cecal contents harvested upon post-mortem were analyzed using C. difficile premier Toxin A &B ELISA kit from Meridian Diagnostics Inc. (Cincinnati, OH), following manufacturer’s instruction. Negative control from the ELISA kit used along with the test samples. Each bar represents one animal.
(TIF)
UK::codY mutant was used as a control.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank following investigators for sharing their lab resources: David Ho, Rockefeller University, for FliC antibody; Wiep Klass, Leids University and Aimee Shen, Tufts University for Spo0A antibody; Linc Sonenshein, Tufts University for CodY antibody and UK::codY mutant; Nigel Minton, University of Nottingham, for the plasmid pMTL007C-E5; Robert Fagan for the vector pRPF185. We thank Jose E. Lopez for technical assistance throughout the study and Yusuf Ceftci, Varun Govind for editing the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
RG is supported by 1R15AI122173 from NIAID. Funds from the Johnson Cancer Center-KSU and a pilot project to RG from CBID-KU (1P20GM113117-01) also supported this work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Curry SR. Clostridium difficile. Clin Lab Med. 2017;37(2):341–69. doi: 10.1016/j.cll.2017.01.007 . [DOI] [PubMed] [Google Scholar]
- 2.Napolitano LM, Edmiston CE Jr. Clostridium difficile disease: Diagnosis, pathogenesis, and treatment update. Surgery. 2017. doi: 10.1016/j.surg.2017.01.018 . [DOI] [PubMed] [Google Scholar]
- 3.CDC. Antibiotic resistance threats in the United States, 2013. 2013; Available from:. http://wwwcdcgov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508pdf.
- 4.Carter GP, Rood JI, Lyras D. The role of toxin A and toxin B in Clostridium difficile-associated disease: Past and present perspectives. Gut Microbes. 2010;1(1):58–64. doi: 10.4161/gmic.1.1.10768 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467(7316):711–3. doi: 10.1038/nature09397 . [DOI] [PubMed] [Google Scholar]
- 6.Kuehne SA, Collery MM, Kelly ML, Cartman ST, Cockayne A, Minton NP. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209(1):83–6. doi: 10.1093/infdis/jit426 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458(7242):1176–9. doi: 10.1038/nature07822 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun. 2012;80(8):2704–11. doi: 10.1128/IAI.00147-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupuy B, Sonenshein AL. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol. 1998;27(1):107–20. . [DOI] [PubMed] [Google Scholar]
- 10.Mani N, Dupuy B. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci U S A. 2001;98(10):5844–9. doi: 10.1073/pnas.101126598 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, et al. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J Bacteriol. 2002;184(21):5971–8. doi: 10.1128/JB.184.21.5971-5978.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlsson S, Dupuy B, Mukherjee K, Norin E, Burman LG, Akerlund T. Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect Immun. 2003;71(4):1784–93. doi: 10.1128/IAI.71.4.1784-1793.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards AN, Nawrocki KL, McBride SM. Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect Immun. 2014;82(10):4276–91. doi: 10.1128/IAI.02323-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nawrocki KL, Edwards AN, Daou N, Bouillaut L, McBride SM. CodY-Dependent Regulation of Sporulation in Clostridium difficile. J Bacteriol. 2016;198(15):2113–30. doi: 10.1128/JB.00220-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, et al. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 2012;40(21):10701–18. doi: 10.1093/nar/gks864 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antunes A, Martin-Verstraete I, Dupuy B. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol. 2011;79(4):882–99. doi: 10.1111/j.1365-2958.2010.07495.x . [DOI] [PubMed] [Google Scholar]
- 17.Dineen SS, McBride SM, Sonenshein AL. Integration of metabolism and virulence by Clostridium difficile CodY. J Bacteriol. 2010;192(20):5350–62. doi: 10.1128/JB.00341-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol. 2007;66(1):206–19. doi: 10.1111/j.1365-2958.2007.05906.x . [DOI] [PubMed] [Google Scholar]
- 19.Edwards AN, Tamayo R, McBride SM. A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol Microbiol. 2016;100(6):954–71. doi: 10.1111/mmi.13361 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D. Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile. PLoS One. 2013;8(11):e79666 doi: 10.1371/journal.pone.0079666 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, et al. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics. 2014;15:160 doi: 10.1186/1471-2164-15-160 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol. 2011;193(13):3186–96. doi: 10.1128/JB.00272-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anjuwon-Foster BR, Tamayo R. A genetic switch controls the production of flagella and toxins in Clostridium difficile. PLoS Genet. 2017;13(3):e1006701 doi: 10.1371/journal.pgen.1006701 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aubry A, Hussack G, Chen W, KuoLee R, Twine SM, Fulton KM, et al. Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infect Immun. 2012;80(10):3521–32. doi: 10.1128/IAI.00224-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Meouche I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, et al. Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One. 2013;8(12):e83748 doi: 10.1371/journal.pone.0083748 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J Bacteriol. 2013;195(22):5174–85. doi: 10.1128/JB.00501-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barilla D, Caramori T, Galizzi A. Coupling of flagellin gene transcription to flagellar assembly in Bacillus subtilis. J Bacteriol. 1994;176(15):4558–64. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda A, Sekiguchi J. High-level transcription of the major Bacillus subtilis autolysin operon depends on expression of the sigma D gene and is affected by a sin (flaD) mutation. J Bacteriol. 1993;175(3):795–801. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu F, Kearns DB, McLoon A, Chai Y, Kolter R, Losick R. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;68(5):1117–27. doi: 10.1111/j.1365-2958.2008.06201.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodgire P, Dixit M, Rao KK. ScoC and SinR negatively regulate epr by corepression in Bacillus subtilis. J Bacteriol. 2006;188(17):6425–8. doi: 10.1128/JB.00427-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59(4):1216–28. doi: 10.1111/j.1365-2958.2005.05019.x . [DOI] [PubMed] [Google Scholar]
- 32.Gaur NK, Cabane K, Smith I. Structure and expression of the Bacillus subtilis sin operon. J Bacteriol. 1988;170(3):1046–53. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voigt CA, Wolf DM, Arkin AP. The Bacillus subtilis sin operon: an evolvable network motif. Genetics. 2005;169(3):1187–202. doi: 10.1534/genetics.104.031955 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Draughn GL, Bobay BG, Stowe SD, Olson AL, Feldmann EA, Thompson RJ, et al. Solution structures of biofilm-controlling proteins SinI and SinR from Bacillus subtilis reveal details of DNA-binding and regulatory mechanism. The FASEB Journal. 2017;31(1 Supplement):907.2–.2. [Google Scholar]
- 35.Pflughoeft KJ, Sumby P, Koehler TM. Bacillus anthracis sin locus and regulation of secreted proteases. J Bacteriol. 2011;193(3):631–9. doi: 10.1128/JB.01083-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connor JR, Lyras D, Farrow KA, Adams V, Powell DR, Hinds J, et al. Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol. 2006;61(5):1335–51. doi: 10.1111/j.1365-2958.2006.05315.x . [DOI] [PubMed] [Google Scholar]
- 37.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10(9):R102 doi: 10.1186/gb-2009-10-9-r102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chai Y, Norman T, Kolter R, Losick R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 2010;24(8):754–65. doi: 10.1101/gad.1915010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chai Y, Norman T, Kolter R, Losick R. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 2011;30(7):1402–13. doi: 10.1038/emboj.2011.36 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, et al. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet. 2013;9(8):e1003660 doi: 10.1371/journal.pgen.1003660 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK. C. difficile 630Deltaerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS One. 2012;7(10):e48608 doi: 10.1371/journal.pone.0048608 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olmedo G, Ninfa EG, Stock J, Youngman P. Novel mutations that alter the regulation of sporulation in Bacillus subtilis. Evidence that phosphorylation of regulatory protein SpoOA controls the initiation of sporulation. J Mol Biol. 1990;215(3):359–72. . [DOI] [PubMed] [Google Scholar]
- 43.Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64(3):545–52. . [DOI] [PubMed] [Google Scholar]
- 44.Jiang M, Grau R, Perego M. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J Bacteriol. 2000;182(2):303–10. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.LeDeaux JR, Grossman AD. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177(1):166–75. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perego M, Cole SP, Burbulys D, Trach K, Hoch JA. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989;171(11):6187–96. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trach K, Burbulys D, Strauch M, Wu JJ, Dhillon N, Jonas R, et al. Control of the initiation of sporulation in Bacillus subtilis by a phosphorelay. Res Microbiol. 1991;142(7–8):815–23. . [DOI] [PubMed] [Google Scholar]
- 48.Underwood S, Guan S, Vijayasubhash V, Baines SD, Graham L, Lewis RJ, et al. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J Bacteriol. 2009;191(23):7296–305. doi: 10.1128/JB.00882-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collery MM, Kuehne SA, McBride SM, Kelly ML, Monot M, Cockayne A, et al. What's a SNP between friends: The influence of single nucleotide polymorphisms on virulence and phenotypes of Clostridium difficile strain 630 and derivatives. Virulence. 2016:1–15. doi: 10.1080/21505594.2016.1237333 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J Bacteriol. 2012;194(13):3307–16. doi: 10.1128/JB.00100-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bordeleau E, Fortier LC, Malouin F, Burrus V. c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 2011;7(3):e1002039 doi: 10.1371/journal.pgen.1002039 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newman JA, Rodrigues C, Lewis RJ. Molecular basis of the activity of SinR protein, the master regulator of biofilm formation in Bacillus subtilis. J Biol Chem. 2013;288(15):10766–78. doi: 10.1074/jbc.M113.455592 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, et al. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol Microbiol. 2007;63(5):1453–67. doi: 10.1111/j.1365-2958.2007.05597.x . [DOI] [PubMed] [Google Scholar]
- 54.Chateau A, van Schaik W, Joseph P, Handke LD, McBride SM, Smeets FM, et al. Identification of CodY targets in Bacillus anthracis by genome-wide in vitro binding analysis. J Bacteriol. 2013;195(6):1204–13. doi: 10.1128/JB.02041-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, et al. Direct targets of CodY in Staphylococcus aureus. J Bacteriol. 2010;192(11):2861–77. doi: 10.1128/JB.00220-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Handke LD, Shivers RP, Sonenshein AL. Interaction of Bacillus subtilis CodY with GTP. J Bacteriol. 2008;190(3):798–806. doi: 10.1128/JB.01115-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 2001;15(9):1093–103. doi: 10.1101/gad.874201 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol. 2005;8(2):203–7. doi: 10.1016/j.mib.2005.01.001 . [DOI] [PubMed] [Google Scholar]
- 59.Girinathan BP, Braun S, Sirigireddy AR, Lopez JE, Govind R. Importance of Glutamate Dehydrogenase (GDH) in Clostridium difficile Colonization In Vivo. PLoS One. 2016;11(7):e0160107 doi: 10.1371/journal.pone.0160107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Girinathan BP, Monot M, Boyle D, McAllister KN, Sorg JA, Dupuy B, et al. Effect of tcdR Mutation on Sporulation in the Epidemic Clostridium difficile Strain R20291. mSphere. 2017;2(1). doi: 10.1128/mSphere.00383-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Purcell EB, McKee RW, Courson DS, Garrett EM, McBride SM, Cheney RE, et al. A Nutrient-Regulated Cyclic Diguanylate Phosphodiesterase Controls Clostridium difficile Biofilm and Toxin Production during Stationary Phase. Infect Immun. 2017;85(9). doi: 10.1128/IAI.00347-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11(3):157–68. doi: 10.1038/nrmicro2960 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ireton K, Gunther NWt, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176(17):5320–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pereira FC, Saujet L, Tome AR, Serrano M, Monot M, Couture-Tosi E, et al. The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet. 2013;9(10):e1003782 doi: 10.1371/journal.pgen.1003782 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, et al. Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet. 2013;9(10):e1003756 doi: 10.1371/journal.pgen.1003756 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Girinathan BP, Braun SE, Govind R. Clostridium difficile glutamate dehydrogenase is a secreted enzyme that confers resistance to H2O2. Microbiology. 2014;160(Pt 1):47–55. doi: 10.1099/mic.0.071365-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Putnam EE, Nock AM, Lawley TD, Shen A. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J Bacteriol. 2013;195(6):1214–25. doi: 10.1128/JB.02181-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teng F, Murray BE, Weinstock GM. Conjugal transfer of plasmid DNA from Escherichia coli to enterococci: a method to make insertion mutations. Plasmid. 1998;39(3):182–6. doi: 10.1006/plas.1998.1336 . [DOI] [PubMed] [Google Scholar]
- 69.Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, et al. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J Microbiol Methods. 2010;80(1):49–55. doi: 10.1016/j.mimet.2009.10.018 . [DOI] [PubMed] [Google Scholar]
- 70.Govind R, Dupuy B. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog. 2012;8(6):e1002727 doi: 10.1371/journal.ppat.1002727 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Criscuolo A, Brisse S. AlienTrimmer: a tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics. 2013;102(5–6):500–6. doi: 10.1016/j.ygeno.2013.07.011 . [DOI] [PubMed] [Google Scholar]
- 72.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013;9(5):e1003493 doi: 10.1371/journal.pgen.1003493 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Govind R, Fitzwater L, Nichols R. Observations on the Role of TcdE Isoforms in Clostridium difficile Toxin Secretion. J Bacteriol. 2015;197(15):2600–9. doi: 10.1128/JB.00224-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of "parallel" expression vectors. Protein Expr Purif. 1999;15(1):34–9. doi: 10.1006/prep.1998.1003 . [DOI] [PubMed] [Google Scholar]
- 75.Sambol SP, Tang JK, Merrigan MM, Johnson S, Gerding DN. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J Infect Dis. 2001;183(12):1760–6. doi: 10.1086/320736 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic representation of ClostTron (group II intron)- mediated disruption of the sinR gene in C. difficile. (B) PCR verification of the intron insertion, conducted with intron-specific primer EBS universal [EBS(U)] with sinR—specific primers ORG-549 and ORG-550.
(TIF)
(A) RT-PCR results of sinRR’, sinR and sinR’ using cDNA, RNA and genomic DNA prepared from C. difficile JIR8094 and JIR8094::sinRR’. (B) Schematic representation of gene structure in sin locus and the location of primer design site for each gene products respectively. (C) RT-PCR results of sin locus transcripts in JIR8094 and JIR8094::sinRR’ strains collected at different time points. The representative results from three independent experiments are shown.
(TIF)
(A) Western blot analysis of parent and mutants using SinR and SinR’ specific antibodies. (B) Growth curve of parent and the mutant strains.
(TIF)
Triton X-100 induced autolysis of R20291::sinRR’ at the stationary phase showing rapid lysis compared to the parent strain. Expression of sinRR’ prevented autolysis in sinRR’ mutant. The autolysis is expressed as percent initial absorbance at an optical density of 600nm. Error bars indicate ± standard deviation. The experiments were repeated at least three times independently (*, p≤0.05 by a two-tailed Student's t-test).
(TIF)
(A) Phase contrast microscopy of JIR8094 and JIR8094::sinRR’ cells. (B) JIR8904::sinRR’ mutant was asporogenic as shown in the representative TEM images in comparison with the parent strain. Black arrows indicate mature spores in the parent strain. (C) Sporulation frequency of JIR8094 and JIR8094::sinRR’ strains. The data shown are mean ± standard errors of three replicates. *** p< 0.0005 (by two-tailed student’s t-test). (D) Western blot analysis demonstrating lower Spo0A expression in the sinRR’ mutant.
(TIF)
(A) Dot blot analysis of R20291, R20291::sinRR’ proteins using FliC and GDH (internal control) specific antibody. (B) Swimming motility of the R20291 and R20291::sinRR’ strain showing the non-motile phenotype of sinRR’ mutant in BHIS with 0.3% agar.
(TIF)
Toxin ELISA performed with cytosolic proteins harvested from JIR8094 and JIR8094::sinRR’ mutant. The data shown are mean ± standard errors of three replicates. ** p< 0.005 (by two-tailed student’s t-test).
(TIF)
(A) The c-di-GMP peak in HPLC. (B) The standard curve was constructed by analyzing samples containing a predetermined amount of c-di-GMP and their respective peak area. (C) Analysis of intracellular nucleotide pools prepared from R20291 and R20291::sinRR’ cells. Arrows indicate the peak corresponding to c-di-GMP.
(TIF)
(A) PCR verification of the intron insertion verified with intron-specific primer EBS universal [EBS(U)] with gene-specific primers ORG-553 and ORG-554. (B) Schematic representation of ClostTron (group II intron)- mediated disruption of the sinR’ gene in C. difficile R20291. (C) Western blot analysis of R20291 and R20291::sinR’ proteins using SinR and SinR’ specific antibodies. (D) Growth curve of parent R20291 and sinR’ mutant in TY medium showing no autolysis of sinR’ mutant.
(TIF)
(TIF)
Cecal contents harvested upon post-mortem were analyzed using C. difficile premier Toxin A &B ELISA kit from Meridian Diagnostics Inc. (Cincinnati, OH), following manufacturer’s instruction. Negative control from the ELISA kit used along with the test samples. Each bar represents one animal.
(TIF)
UK::codY mutant was used as a control.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.