Abstract
Protein synthesis is a complex, multi-step process that involves large conformational changes of the ribosome and protein factors of translation. Over the last decade, Förster resonance energy transfer (FRET) has become instrumental for studying structural rearrangements of the translational apparatus. Here, we discuss the design of ensemble and single-molecule (sm) FRET assays of translation. We describe a number of experimental strategies that can be used to introduce fluorophores into the ribosome, tRNA, mRNA and protein factors of translation. Alternative approaches to tethering of translation components to the microscope slide in smFRET experiments are also reviewed. Finally, we discuss possible challenges in the interpretation of FRET data and ways to address these challenges.
Keywords: ensemble FRET, single-molecule FRET, ribosome, fluorescence, labeling
1. Introduction
Protein synthesis is a complex, multi-step process catalyzed by the ribosome. Understanding the molecular mechanism of protein synthesis remains one of the most challenging problems in molecular biology because of the size and structural complexity of the ribosome. The main phases of protein synthesis, namely initiation, elongation, termination and recycling, are mediated by the interaction of the ribosome with a number of protein factors of translation. The ribosome, tRNA, mRNA, and translational factors undergo coordinated large-scale conformational changes. For example, during translation elongation, tRNA and mRNA are moved through the ribosome along a path of >100 Å in a process that involves the formation of multiple intermediates [1]. tRNA/mRNA translocation is accompanied by dramatic structural rearrangements of the ribosome including forward and reverse rotations between the large and small ribosomal subunits, “swiveling” motions of the “head” domain of the small ribosomal subunit, and the inward/outward fluctuations of the L1 stalk of the large ribosomal subunit [1].
Over the last two decades, X-ray crystallography and cryo-electron microscopy (cryo-EM) have provided high-resolution snapshots of different functional states of the ribosome and ribosomal ligands, which have revolutionized our mechanistic understanding of protein synthesis. However, X-ray crystallography and cryo-EM (with the exception of recently emerged time-resolved cryo-EM [2, 3]) reveal neither the sequence of structural rearrangements nor the rates of these rearrangements. In addition, a number of factors, such as crystal lattice in X-ray crystallography, interactions of the ribosome with the water-air interface and the supporting film in cryo-EM microscopy, may alter the structure of ribosomal functional complexes. Hence, methods that reveal the structural dynamics of the translational apparatus in solution and in real time are indispensable for investigating protein synthesis.
Förster resonance energy transfer (FRET) is a popular and powerful tool for studying conformational dynamics within macromolecules in solution. FRET is the non-radiative transfer of energy from a donor fluorophore to an acceptor molecule over distances of 10–100 Å. FRET efficiency (E) is inversely proportional to the sixth power of the distance between donor and acceptor (R): E = R06/(R06+R6). (R0, the Förster radius is the distance at which the efficiency of energy transfer is 0.5; R0 depends on photophysical properties of the specific donor-acceptor pair). Thus, FRET can be used to follow conformational changes within a macromolecule labeled with a donor and an acceptor. If two interacting macromolecules are labeled with a donor and an acceptor, respectively, then FRET can also be used to detect binding/dissociation events.
Different applications of FRET have been extensively used to study protein synthesis over the last fifteen years. The average FRET efficiency over the entire ensemble of ribosomes (or factors of translation) is typically measured in the bulk of the sample using conventional fluorescence spectroscopy. Combining fluorescence detection with fast-mixing devices (e.g. stopped-flow apparatus) allows for measuring the rates of conformational changes and binding/dissociation under pre-equilibrium conditions [4–8]. Finally, the behavior of individual molecules that is undetectable in bulk measurements because of ensemble averaging is revealed by single-molecule FRET (smFRET) microscopy (Fig. 1). Single-molecule fluorescence is typically measured using total internal reflection fluorescence (TIRF) microscopy, confocal microscopy or zero-mode waveguides [9], which use different ways to minimize excitation volume and, thus, excite fluorescence in individual molecules while leaving bulk of the sample non-fluorescent. Because macromolecules quickly diffuse, they need to be tethered to the microscope slide, in order to measure FRET on milliseconds-to-minutes time scale. Similar to the ensemble experiments, smFRET can be measured under both equilibrium and pre-equilibrium conditions. smFRET measurements are especially important in studies of protein synthesis because the population of translating ribosomes is highly dynamic and asynchronous.
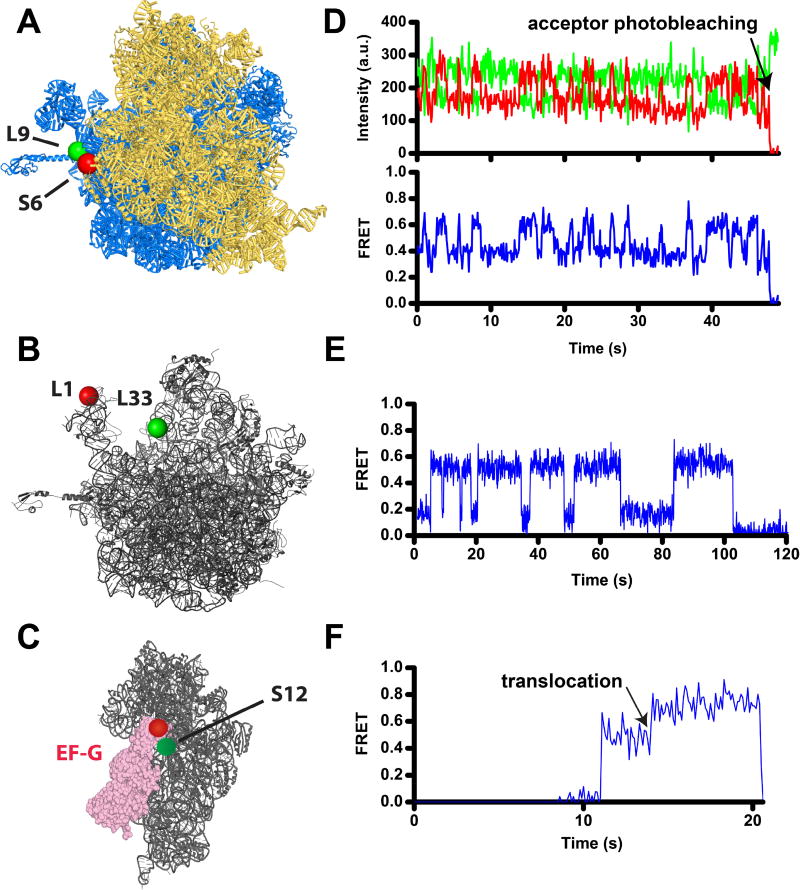
Figure 1. Following translocation and ribosome dynamics using smFRET.
A) Intersubinit rotation is followed by FRET between a donor (Cy3, green) and an acceptor (Cy5, red) attached to ribosomal proteins bL9 and bS6 on the large (blue) and small (gold) ribosomal subunits, respectively [80, 106, 122]. View from the solvent side of the small subunit of the 70S ribosome from E. coli in the nonrotated conformation (PDBID 4V9D [123]). In this view, the formation of the rotated conformation of the ribosome results in counterclockwise rotation of the small subunit. B) The movement of the 50S L1 stalk of the large subunit (grey) is followed by FRET between Cy5 (red) and Cy3 (green) attached to proteins uL1 and bL33, respectively [84, 122] (PDBID 4V51 [124]). C) Docking of domain IV of EF-G (pink) to the A site of the small subunit (grey) during translocation is followed by FRET between Cy3 (red) and Cy5 (red) attached to EF-G and protein uS12, respectively [28]. The small subunit of the posttranslocated ribosome is viewed from the subunit interface (PDBID 4V5F [125]). D–F) smFRET traces show fluorescence intensities for Cy3 (green) and Cy5 (red) (D), and FRET efficiency (blue, D–F). D) S6/L9 smFRET trace shows spontaneous fluctuations of a ribosome, which contains deacylated tRNAfMet in the P site, between the rotated and nonrotated conformations that correspond to 0.4 and 0.6 FRET states. E) L1/L33 smFRET trace shows fluctuations of the L1 stalk in a ribosome, which contains deacylated tRNAfMet in the P site, between the open and closed conformations that correspond to 0.2 and 0.5 FRET states. F) S12-EF-G smFRET trace shows a transition from 0.55 to 0.8 FRET that corresponds to the movement of domain IV toward the A site of the small subunit during translocation of tRNA/mRNA.
In order to follow translation using FRET in an in vitro reconstituted translation system, small, bright and photostable organic fluorescent dyes are typically introduced into the ribosome, factors of translation, tRNA or mRNA. In this article, we review general considerations for designing FRET assays of translation, different approaches to fluorescent labeling of the components of the translational apparatus, immobilization of translating ribosomes on the microscope slide for smFRET measurements, and possible challenges in the interpretation of FRET data.
2. Design of FRET assays
In order to design an effective FRET assay, a number of considerations need to be assessed including choice of fluorophores, labeling strategy, the distance between sites of labeling and preservation of biological activity of the translational machinery.
Cyanine fluorophores, Cy3 and Cy5, are the most common donor and acceptor dyes, respectively, used in ensemble and smFRET measurements due to their high photostability and brightness [10]. Another advantage of the Cy3–Cy5 pair is a significant spectral separation between donor and acceptor fluorescence. Amine-, thiol and aldehyde-/ketone- reactive derivatives of Cy3 and Cy5 are sold by a number of suppliers, including GE Healthcare and Click Chemistry Tools. In addition, other dyes with spectroscopic properties that are similar to Cy3 and Cy5 are sold by Invitrogen (Alexa Fluor 555 and Alexa Fluor 647), Thermo Scientific (DyLight 550 and DyLight 650) and Sigma Aldrich (Atto 550 and Atto 647). Derivatives of cyanine dyes with enhanced photostability (LD550 and LD650) are offered by Lumidyne technologies (http://lumidynetechnologies.com/) [11].
Analysis of X-ray crystal and cryo-EM structures of the ribosome captured at different steps of translation reveals structural rearrangements that can be followed with FRET. Furthermore, cryo-EM and X-ray crystal structures of the ribosome and translation factors guide the selection of labeling sites. Amino acid residues in proteins or nucleotides in RNA that are chosen for labeling should be solvent exposed to ensure efficient conjugation with fluorophores. RNA nucleotides or protein residues that are located in the vicinity of active sites and directly interact with other components of the translational machinery should not be used for labeling in order to protect translational activity. In addition to analysis of structural information, alignment of multiple RNA or protein sequences from a variety of species can be used to identify nucleotides or amino acids that are not evolutionary conserved and, thus, unlikely to be functionally important [12]. A number of biochemical assays can be used to examine whether fluorescently-labeled components of the translational machinery maintain their activity [12].
Another important consideration in designing a new FRET assay is the distances between labeling sites. The distance between the donor and acceptor fluorophores should preferably be near R0 for the given donor-acceptor pair (~56–60Å for Cy3/Cy5 pair[13, 14]), where the FRET efficiency is the most sensitive to changes in distance between fluorophores. It is not practical to use FRET to measure changes in distance outside of the range of 0.6R0 to 1.5R0 [10] because respective changes in fluorescence intensities of donor and acceptor will be too small for accurate measurement in relevant time scales.
Another challenge in designing FRET assays is that FRET provides only a one-dimensional measure of multi-dimensional structural rearrangements [15]. Similar FRET changes might result from distinct and potentially uncoupled conformational changes. For example, energy transfer between fluorophores, which are attached to protein uS13 at the head domain of the small (30S) ribosomal subunit and protein uL5 at the core of the large (50S) subunit, are sensitive to the ratchet-like rotation between ribosomal subunits parallel to the plane of the intersubunit interface [16]. (Here and elsewhere, letters u, b and e indicate: a ribosomal protein that is universally conserved (u), or if a protein occurs only in bacteria (b) or eukaryotes (e)) [17]. However, the same FRET pair is also sensitive to the forward- and back-swiveling motions of the 30S head domain relative to the rest of the 30S subunit around the axis that is orthogonal to the axis of intersubunit rotation [16]. Therefore, multiple FRET pairs might be needed to accurately describe complex structural rearrangements of the ribosome and ribosomal factors. Computational energy landscape analyses of ribosomal structures can guide the design of FRET pairs, which are sensitive to specific conformational transitions in the ribosome [15].
3. Labeling components of translational apparatus
Fluorescent labeling of tRNA, factors of translation and the ribosome has been used to develop a number of FRET assays, which revealed tRNA dynamics, conformational rearrangements of the ribosome and ribosomal factors during initiation, elongation, and termination phases of protein synthesis in bacteria (Table 1) [18–20]. Below we compare advantages and limitations of various methods for site-specific labeling of translational machinery.
Table 1.
smFRET assays used to study bacterial translation.
| Sites of labeling with donor and acceptor fluorophores |
Structural rearrangements detected by each FRET pair |
References |
|---|---|---|
| Elbows of the A- and P-site tRNAs | Stepwise tRNA translocation through the ribosome | [105] |
| P-site tRNA elbow and protein L1 | Stepwise tRNA translocation through the ribosome / inward-outward movement of the 50S L1 stalk | [79] |
| Proteins S6 and L9 | Intersubunit rotation | [106] |
| h44 of 16S and H101 of 23S rRNA | Intersubunit rotation | [120] |
| Proteins L1 and L33 | Inward-outward movement of the 50S L1 stalk | [84] |
| Proteins L1 and L9 | Inward-outward movement of the 50S L1 stalk | [83] |
| Protein L11 and tRNA elbow | Stepwise tRNA translation through the ribosome / movement of the 50S L11 stalk | [81] |
| Protein L27 and tRNA elbow | Stepwise tRNA translation through the ribosome | [82] |
| Proteins S13 and L1 | Intersubunit rotation / inward-outward movement of the 50S L1 stalk | [121] |
| Protein S13 and tRNA elbow | tRNA translocation / movement of the head domain of the 30S | [16] |
| Proteins S13 and L5 | Movement of the head domain of the 30S / intersubunit rotation | [16] |
| Protein S12 and tRNA elbow | Stepwise tRNA translocation through the ribosome | [89] |
| Protein S12 and domain IV of EF-G | EF-G rearrangements during tRNA translocation | [28] |
| tRNA elbow and C-terminus of EF-G | tRNA translocation / EF-G rearrangements during tRNA translocation | [26] |
| Domains II and IV of EF-G | Interdomain rearrangements in EF-G | [30] |
| N- and C-terminal domains of IF3 | Interdomain rearrangements of IF3 | [27] |
| IF2 and protein L11 | Subunit joining / IF2 rearrangements during translation initiation | [29] |
| IF2 and P-site tRNA | IF2 rearrangements during translation initiation | [31] |
3.1. tRNA labeling with fluorophores
tRNAs are translocated from the A to P to E sites of the ribosome in a stepwise process that involves the formation of multiple intermediates [1]. Fluorescently labeled tRNAs were used to detect the intermediates of tRNA accommodation to the A site and tRNA translocation in a number of ensemble and smFRET assays [18–21].
Most commonly, fluorophores are introduced using naturally occurring post-transcriptional modifications of nucleotides in native tRNAs. For example, 3-(3-amino-3-carboxypropyl) uridine at position 47 (Acp3U47) in E. coli tRNAPhe and tRNALys can be specifically modified with N-hydroxysuccinimide (NHS) ester of Cy3 and Cy5 [12, 22]. 4-thiouridine at position 8 (S4U8), which is common to many E. coli tRNAs, has been previously labeled in E. coli tRNAfMet and tRNAVal using Cy3- or Cy5-maleimide [12, 23]. Dihydrouridine (D residue) is another tRNA base that occurs in almost all tRNAs and can be labeled by NaBH4 treatment followed by the reaction with hydrazide derivatives of Cy3 or Cy5 [24].
Fluorescent labeling of post-transcriptionally modified tRNA bases has a number of limitations including a relatively low yield of conjugated product. The labeling efficiency of 4-thiouridine and dihydrouridine with cyanine dyes typically does not exceed 50% [12, 24]. Labeling of S4U8 was also reported to reduce EF-Tu-dependent aa-tRNA binding to the ribosome [24]. Currently, only a few native tRNAs from E. coli and S. cerevisiae are commercially available: yeast tRNAPhe (from Sigma and Chemical Block), and E coli tRNAfMet, tRNAMet, tRNAPhe, tRNALys, tRNAVal, tRNATyr, tRNAGlu, and tRNAArg (from MP Biomedicals and Chemical Block). In addition, many tRNAs contain two or more 4-thiouridines and dihydrouridines, thus, precluding labeling of a unique position in tRNA. A comprehensive list of post-transcriptional modifications in native tRNAs from multiple species can be found at http://modomics.genesilico.pl/ [25]. Nevertheless, to our knowledge, no other potent and cost-effective approaches to tRNA labeling with cyanine dyes have been used in FRET experiments published to date.
3.2. Fluorescent labeling of protein factors of translation
Translation initiation, elongation, and termination factors (IFs, EFs, and RFs, respectively) mediate the corresponding phases of protein synthesis. During protein synthesis, translation factors likely undergo large conformational changes, which can be studied by FRET. Indeed, FRET experiments revealed significant structural rearrangements of elongation factor G (EF-G) (Fig. 1C, F), and initiation factors 2 and 3 (IF2 and IF3) [26–31]. Site-specific labeling of translation factors for FRET experiments can be done through several strategies: 1) alkylation of cysteine residues with maleimide derivatives of fluorophores, 2) modification of genetically encoded unnatural amino acids, such as para-acetyl-L-phenylalanine, and 3) enzymatic labeling of signal peptides fused to translation factors.
The amino group of lysine and sulfhydryl group of cysteine can be modified with NHS-ester and maleimide derivatives of fluorophores, respectively [32]. Due to the rare occurrence of cysteine in most proteins, cysteine modification by maleimide is commonly used in site-specific labeling [12, 32]. A cysteine residue can be introduced by mutagenesis at a desired position. Likewise, naturally occurring cysteines can be replaced with other amino acids. Sometimes sulfhydryl groups of naturally occurring cysteines are partially buried, react poorly with maleimide and, thus, do not have to be substituted. Reactivity of naturally occurring cysteines can be tested using Ellman's reagent (5,5'-dithiobis-(2-nitrobenzoic acid), DTNB) [33, 34]. To introduce both a donor and an acceptor fluorophore into the same protein, two cysteine resides can be reacted with a mixture of equimolar amounts of donor and acceptor dyes. This procedure produces doubly-labeled species with mixed donor-acceptor orientations between the two positions as well as species that contain two donor or two acceptor fluorophores. If such heterogeneity of the sample is not desirable then a cysteine and an unnatural amino acid (UAA) can be used instead of two cysteines to prepare doubly-labeled protein. UAAs can also be employed if naturally occurring cysteines cannot be substituted in the protein without alteration of its biological activity.
To date, over 200 different UAAs have been incorporated into proteins produced in bacterial, yeast and mammalian systems [35, 36]. UAAs are typically introduced by the suppressor tRNA, which recognizes the amber stop codon (UAG) inserted into protein Open Reading Frame (ORF) at a desired position. The suppressor tRNA is charged with a UAA by a specific genetically-engineered aminoacyl-tRNA synthetase, which is co-expressed with the suppressor tRNA. For example, para-acetyl-L-phenylalanine (p-AcPhe) can be modified by hydroxylamine derivatives of fluorophores with great selectivity and high yield of fluorescently-labeled product [37, 38]. In smFRET studies of ribosomal translocation, p-AcPhe was used to label domain IV of EF-G [38].
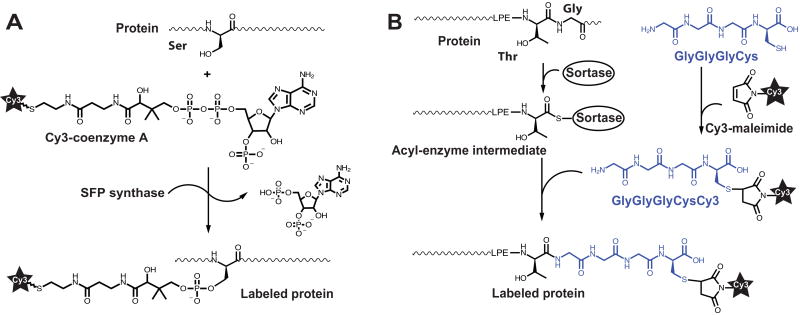
In addition to UAA-based methods, fluorescent labeling can be achieved using enzymatic labeling of short polypeptide tags genetically introduced into the target protein [39–42]. For example, bacterial phosphopantetheinyl transferases (Sfp) catalyze the transfer of a phosphopantetheinyl (Ppant) group from coenzyme A (CoA) to a serine residue within 11-residue peptide (ybbR tag, DSLEFIASKLA) derived from natural substrates of Sfp, the peptidyl and acyl carrier proteins (Fig. 2A) [39, 43]. To obtain fluorescent substrate for Sfp-catalyzed reaction, sulfhydril group of CoA is modified with maleimide derivatives of Cy3 or Cy5 (Fig. 2A). Sfp-specific tags can be genetically introduced at the N-terminus, C-terminus or in an unstructured internal loop of proteins. For example, Sfp-catalyzed labeling of the C-terminus of EF-G was used in smFRET studies of ribosomal translocation [26].
Figure 2. Protein labeling via (A) phosphopantetheinyl transferase Sfp (A) or sortase A (SrtA) reactions (B).
(A) The serine residue in the ybbR tag (DSLEFIASKLA) is labeled with fluorophores conjugated with coenzyme A (Cy3-CoA). (B) The five-amino acid (LPETG) of SrtA recognition site is modified with the GlyGlyGlyCys peptide conjugated to maleimide-Cy3.
In addition to Sfp, transpeptidase sortases from Gram-positive bacteria, like a transpeptidase sortase A (SrtA) from Staphylococcus aureus, can also be repurposed for enzymatic fluorescent labeling. SrtA specifically recognizes the pentapeptide sequence LPETG and mediates the cleavage of the amide bond between threonine and glycine (Fig. 2B)[40, 44]. Resulting thioacyl intermediate reacts with an amino group of an oligoglycine (Gly) probe conjugated to Cy3/Cy5. Sortase-compatible fluorescent probe can be obtained by solid-phase synthesis of NH2-Gly-Gly-Gly-Cys peptide, followed by Cy3/5-maleimide coupling to the Cys residue (Fig. 2B) [45, 46].
Enzymatic fluorophore-protein coupling is a versatile method that may be indispensable for the labeling of large proteins, which have multiple-naturally occurring cysteine residues.
3.3. Fluorescent labeling of mRNA
Structured elements within mRNA play fundamental roles in translational control of gene expression [47]. FRET can provide insights into the dynamics of structured elements of mRNA that regulate translation, such as riboswitches, frameshift-inducing RNA hairpins/pseudoknots and internal ribosome entry sites (IRESs) [48–51]. Donor and acceptor fluorophores can be incorporated into short (60–120 nt) RNA molecules through solid phase chemical synthesis provided by various companies, such as IDT and Dharmacon. Fluorophores can be introduced into synthetic RNAs through modification of the phosphate backbone or nucleotide bases at the 5’end, the 3’ end or internal sites [52]. The preferred method for the internal labeling of synthetic RNA is introducing an aminoallyl-modified uridine, which can be coupled with N-hydroxysuccinimide (NHS)-ester derivatives of fluorescent dyes [52]. Unlike cyanine dyes incorporated into the phosphate backbone of RNA, fluorophores conjugated to aminoallyl-uridine do not destabilize basepairing interactions within mRNA [10].
The length of chemically synthesized and fluorescently-labeled RNA is limited to ~60 nt (~120 nt in the case of 5’-labeled RNAs) because of prohibitively high cost and low yield for the synthesis of longer RNA molecules. Longer (>60 nt) RNA molecules are synthesized via in vitro transcription catalyzed by bacteriophage SP6, T3 or T7 RNA polymerase [52]. The most commonly used T7 RNA polymerase has a strong preference for guanosines at the first and second transcribed positions [52]. Furthermore, T7 polymerase typically adds one, two or three extra non-templated nucleotides to the RNA 3’end in a fraction of the transcript [52]. If homogeneity at the transcript 3’ end is essential for a given study, the sequence encoding cis-cleaving ribozymes can be inserted downstream of the 3’ end of transcription template[53, 54]. Alternatively, DNAzyme [55] or RNaseH-mediated cleavage [56] can be used to generate homogeneous 3’ ends of T7 polymerase transcripts [52]. Below we review different strategies for fluorescent labeling of in vitro transcribed RNAs.
3.3.1. 5’ end labeling of RNA
The 5’ and 3’ ends of in vitro transcribed RNA have unique 5’ tri-phosphate and the 3’ ribose cis diols groups, which can be modified with fluorophores. A sulfhydryl-modified guanosine monophosphates (5'-deoxy-5'-thioguanosine-5'-monophosphorothioate; GSMP) can be co-transcriptionally incorporated at the 5' end of RNA [57]. With a ratio of GSMP: GTP: ATP: CTP: UTP = 50:1:1:1:1, GSMP used as the first transcript nucleotide with 100% efficiency without incorporation into internal positions [57]. Sulfhydryl group of GSMP is subsequently conjugated to maleimide derivatives of fluorophores. However, to our knowledge, GSMP is not commercially available that limits the practical utility of this method.
A maleimide-reactive thiol group can also be enzymatically introduced at the 5’ end of the RNA using T4 polynucleotide kinase (T4 PNK) [58, 59]. The in vitro transcribed RNA is initially dephosphorylated by calf intestinal alkaline phosphatase. Then T4 PNK introduces a sulfhydryl group at the 5’ transcript end by catalyzing the transfer of the γ-phosphorothioate group from 5’-O-(3-thio) adenosine triphosphate (ATPγS) to the 5’-hydroxyl of RNA [60]. The labeling efficiency for this method typically does not exceed 50–80% [58, 59].
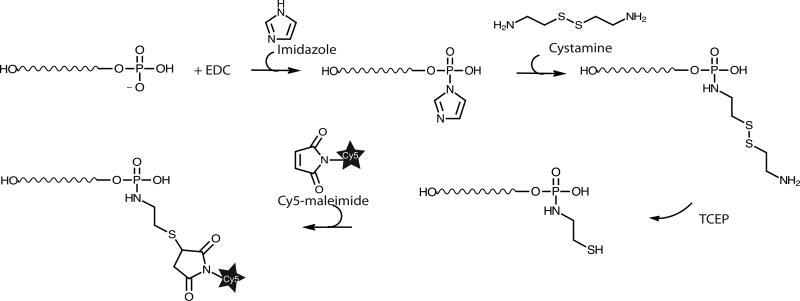
The higher labeling yield can be achieved by modifying the 5’ phosphate group of the in vitro transcribed RNA using carbodiimide, imidazole, cystamine and maleimide derivative of fluorophore (Fig. 3) [32, 61]. The 5’ γ phosphate group of RNA is first chemically modified and conjugated to amine-containing molecules using crosslinker EDC (1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide) and imidazole. Then, the phosphorimidazolide intermediate at the 5’ end of the RNA reacts with cystamine (H2N-CH2-CH2-S-S-CH2-CH2-NH2). Finally, the disulfide bond is reduced with tris(2-carboxyethyl)phosphine (TCEP) or other reducing reagents, yielding a reactive 5’-sulhydryl-RNA. To avoid the loss of fluorophores due to spontaneous hydrolysis of γ and β phosphate groups at the 5’ end, the γ and β phosphates can be removed by the bacterial 5´ RNA Pyrophosphohydrolase (RppH, New England Biolabs) before modifying the 5’ α phosphate with EDC and imidazole. We can reproducibly achieve 100% yield of labeling of either the 5’ γ or α phosphates in the in vitro transcribed mRNA using the following protocol:
-
1
Dissolve cystamine in 0.1 M imidazole (Sigma, ca no I5513), pH 6.0 to the final concentration of 0.25 M.
-
2
Mix 1–2 mg of EDC (Fisher Scientific, ca no PI22980) dissolved in 20 µl of 0.1M imidazole, pH 6.0, 20 µl of 50 µM RNA (1 nmole) and 5µl of 0.25 M cystamine (Sigma, ca no 30050). Incubate at room temperature for 2 hours in the dark.
-
2
Remove EDC and cystamine using 1 ml G-25 spin column equilibrated with 10 mM sodium phosphate, pH 7.2, 0.15 M NaCl, 10 mM EDTA.
-
6
Add 1 M TCEP hydrochloride (Fisher Scientific, ca no T2556) to the final concentration of 0.1 M to release sulfhydryl groups. TCEP treatment results in RNA precipitation.
-
7
Resuspend the RNA pellet in 50 µl of 10 mM sodium phosphate, pH 7.2, 0.15 M NaCl, 10 mM EDTA; add 30 mM maleimide-Cy3/Cy5 dissolved in dimethylformamide to the final concentration of 8 mM; and incubate for 2 hours at room temperature in the dark. The labeled RNA is then purified by denaturing PAGE.
Figure 3. Chemical modification and labeling of the 5’ phosphate group of an in vitro transcribed RNA using carbodiimide (EDC), imidazole, cystamine and maleimide-Cy5.
Please see details in the text.
3.3.2. 3’ end labeling
The 3’ end of in vitro transcribed RNAs can be labeled with high (90–100%) efficiency via periodate oxidation [62, 63]. Sodium periodate oxidizes the 3’ ribose cis diols and creates highly-reactive aldehydes groups, which can be conjugated to amine- or hydrazide-derivatives of fluorophores. The resulting labile hydrazone linkage between fluorophore and RNA can be stabilized by the reaction with cyanoborohydride [32]. The downside of this approach is unwanted side-reactions that may involve periodate or the highly reactive aldehyde groups created during the 3’ ribose oxidation [64].
Alternatively, the 3’ end of in vitro transcribed RNAs can be conjugated to Cy3/Cy5-cytidine-3’,5’-bisphosphate (Cy3/5-pCp; Jena Bioscience) via the one-step reaction catalyzed by T4 RNA ligase [65, 66]. We find that the efficiency of Cy3/5-pCp labeling of transcripts, which are > 200 nt long, often does not exceed 15–30%. Altering the RNA/ Cy3/5- ratio and adding DMSO to destabilize RNA structure, which interferes with the reaction, improve labeling efficiency. Although the effectiveness of labeling with pCp and T4 RNA ligase varies from RNA to RNA, this method is still attractive because of its simplicity and high stability of produced dye-RNA linkage.
3.3.3. mRNA labeling at internal positions
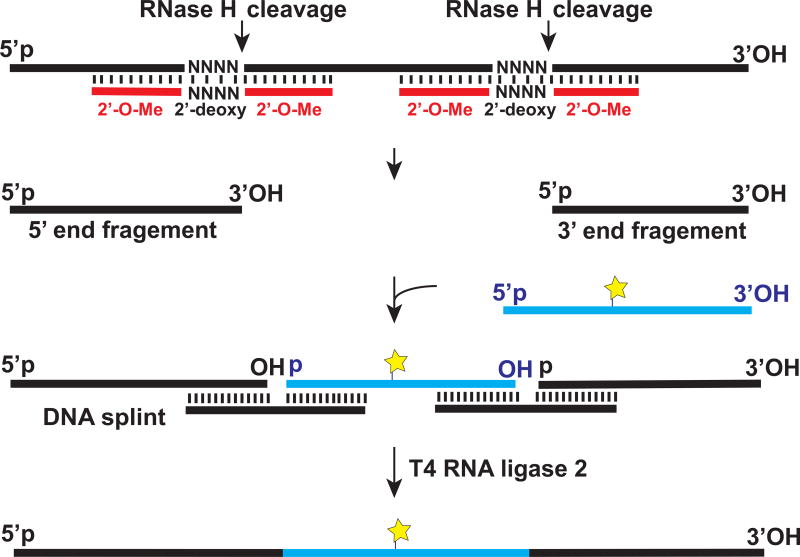
Introducing one or two fluorophores at specific internal positions of in vitro transcribed RNA that may be desired in certain FRET experiments is not straightforward but it can be achieved through the splinted ligation approach [56] (Fig. 4). As an example, the 60 nt-long internal segment of an in vitro transcribed RNA can be replaced by a chemically synthesized RNA fragment that contains an aminoallyl-uridine, which is introduced at a desired position and conjugated to a fluorophore. To that end, the 60 nt-long internal segment is first excised from the in vitro transcribed RNA by RNaseH cleavage directed by two chimeric 2’-O-me RNA/DNA hybrid targeting oligonucleotides (Fig. 4). Resulting 5’ and 3’ end fragments of the in vitro transcribed RNA are then spliced together with the chemically synthesized, fluorescently-labeled RNA fragment. This ligation reaction is facilitated by hybridization with complementary bridging DNA oligonucleotides (DNA splints) and catalyzed by T4 DNA ligase or T4 RNA ligase 2. Details of fluorescent labeling via splinted ligation can be found elsewhere [56].
Figure 4. Introducing a fluorophore into an in vitro transcribed RNA by splinted ligation.
Please see details in the text.
In addition to splinted ligation, labeling at specific internal positions of in vitro transcribed RNA can be mediated by a complementary DNA oligonucleotide containing a reactive group at either the 5’ or 3’ ends [67, 68]. However, these oligonucleotide-guided mRNA labeling strategies have not yet been widely used in FRET studies.
3.4. Strategies for ribosome labeling
Conformational rearrangements of bacterial ribosome that accompany protein synthesis have been extensively investigated via FRET (Fig. 1, Table 1). In these studies, a number of experimental approaches for fluorescent labeling of ribosomal proteins and ribosomal (r)RNA have been developed.
The Hardesty’s group, which pioneered the use of FRET for probing of the ribosomal structure, labeled the 3’ end of 16S rRNA through periodate oxidation and then reconstituted the small (30S) subunit from fluorescently-labeled 16S rRNA and a mixture of 30S proteins (TP30) [69, 70]. More recently, a different strategy for labeling rRNA was developed: fluorescently-labeled oligonucleotides were hybridized with 22–23 nucleotide-long extensions of 16S and 23 rRNA, which were introduced via mutagenesis to replace the loops in surface-exposed rRNA helices [71]. This approach, which was originally developed for labeling E. coli ribosome, in principle, can be employed to incorporate fluorophores into ribosomes from any species. Indeed, this method was successfully used to label yeast ribosomes [72]. However, the RNA insertions engineered into ribosomal rRNA may potentially affect ribosome structure, structural dynamics and translational activity. Another potential caveated of this labeling strategy is that hybridization of fluorescent oligonucleotide with rRNA is reversible. This could be particularly problematic in smFRET experiments done using TIRF microscopy, in which fluorescently-labeled molecules are used at concentrations below 10 nM to prevent overcrowding on the microscope slide and limit background fluorescence. Finally, high intrinsic dynamics of terminal loops of a number rRNA helices, such as h44 of 16S rRNA [73], can interfere with FRET detection of large-scale conformational changes of the ribosome, e.g. intersubunit rotation.
Instead of rRNA labeling, fluorophores can be attached to ribosomal proteins. Most commonly, a ribosomal protein conjugated to a fluorophore is introduced into ribosomes, which lack the same protein due to either chemical or genetic manipulations. Proteins bS21 and bL12 can be selectively depleted from E. coli ribosomes by NH4Cl and NH4Cl/ethanol treatments, respectively [69, 74]. In addition, there are a number of ribosomal proteins that are not essential for cell viability and can therefore be deleted from the bacterial genome one or two at a time. 13 knockout E. coli strains lacking a single ribosomal protein can be purchased from the Keio strain collection (https://cgsc2.biology.yale.edu/index.php) [75]: ΔbS6, ΔuS15, ΔbS20, Δb21, ΔuL1, ΔbL9, ΔuL11, ΔbL25, ΔbL31, ΔbL32, ΔbL33, ΔbL35 and ΔbL36. In addition, uS9, uS13, uS17, uL15, bL21, uL24, bL27, uL29, uL30 and bL34 were also shown to be nonessential in E. coli [76, 77].
As an example, a single-cysteine recombinant variant of the deleted ribosomal protein can be expressed, purified, conjugated to a maleimide derivative of a donor or acceptor fluorophore, and then incorporated into ribosomes isolated form the knockout strain through in vitro reconstitution. This approach has been used to introduce fluorescent dyes into the E. coli ribosome at proteins bS6 [78], uL1 [79], bL9 [80], uL11 [81] and bL27 [82]. Double-knockout strains ΔuL1/ΔbL9 [83] and ΔuL1/ΔbL33 [84] were also successfully used for site-specific fluorescent labeling of E. coli ribosomes. However, especially in eukaryotes, there are a limited number of non-essential ribosomal proteins, which restrict the choice of sites for fluorescent labeling using aforementioned strategy. For example, in buddying yeast, there are only eight non-essential ribosomal proteins (eS25, eL22, eL24, uL24, eL29, eL38, eL39 and eL41), deletion of which results in moderate or no growth defect [85].
By contrast, the complete reconstitution of the 30S subunit from 16S rRNA and a mixture of individually purified ribosomal proteins, one or two of which are labeled with fluorophores, offers unparalleled flexibility in the choice of labeling sites on the small subunit of the bacterial ribosome [86, 87]. This approach was successfully employed to create FRET constructs for studies of ratchet-like intersubunit rotation of the ribosome, rearrangements within the 30S subunit, and the movement of elongation EF-G relative to the 30S A site [28, 80, 87, 88]. Nevertheless, labeling the 30S subunit through the complete reconstitution can be challenging. Overexpression of several 30S proteins results in the formation of inclusion bodies, which are poorly soluble in such potent denaturants as 8 M urea and 6 M guanidine hydrochloride [86, 87]. The yield of fluorescently-labeled ribosomes, which are also active in tRNA and mRNA binding, is variable and depends on the identity of fluorescently-labeled protein and purity of other ribosomal proteins used in the reconstitution. Furthermore, this approach is only applicable to the 30S subunit labeling since no protocol for reconstitution from a mixture of individually purified ribosomal proteins has been developed for either 50S subunit of the bacterial ribosome or subunits of eukaryotic ribosomes.
Limitations of partial and complete reconstitution procedures can be overcome by enzymatic labeling. As described in section 3.2., an 11 amino acid-long (ybbR) peptide tag for in vitro enzymatic labeling by Sfp phosphopantethenyl transferase can be genetically introduced at the N-terminus, C-terminus or in an unstructured internal loop of a ribosomal protein [43]. Using this approach, it is possible to label essential ribosomal proteins, such as protein uS12 [89] and uL5 [16]. A potential downside of this method is that the insertion of ybbR peptide tag may alter the function of the ribosomal protein chosen for labeling and its incorporation into the ribosome during ribosome biogenesis. Encountering this problem is quite probable since the N- and C-termini of many ribosomal proteins make extensive interactions with rRNA [90]. Nevertheless, enzymatic labeling promises to be the key technology to fluorescent labeling of eukaryotic ribosome in future studies.
Another promising approach to fluorescent labeling of the ribosome is genetically fusing the SNAP tag to the N- or C-terminus of a ribosomal protein. The SNAP tag is a 20 kDa mutant variant of the DNA repair protein O6-alkylguanine-DNA alkyltransferase that reacts specifically and rapidly with benzylguanine derivatives of fluorescent dyes, leading to irreversible covalent labeling of the SNAP-tag with a fluorophore [91]. SNAP-tag reactive analogues of Cy3 and Cy5 (SNAP-549 and SNAP-649) are sold by New England Biolabs. Similar to SNAP tag, DHFR and Halo protein tags bind small organic fluorophores and can also be used for fluorescent labeling [92–94].
A unique advantage of SNAP-tagging technology is that the fluorophore can be introduced into the ribosome in cells or cell extracts. Thus, SNAP-tagged ribosomes can potentially be studied by single-molecule microscopy in cell extracts eliminating the need for reconstituting the translation system from purified components that is quite challenging in the case of the eukaryotic translation system [95, 96]. Because of the relatively large size of the SNAP protein, SNAP-tagging may not be suitable for FRET studies of conformational changes of the ribosome. Nevertheless, the SNAP-tag can be used for smFRET detection of ribosome interactions with other components of the translational machinery. For example, smFRET between a fluorophore attached to mRNA and SNAP-tag fused to ribosomal protein eS25 were recently used for measuring kinetics of the recruitment of human small (40S) ribosomal subunit to hepatitis C IRES [97]. Likewise, SNAP-tag fused to ribosomal protein uL5 was employed in smFRET kinetic measurements of cricket paralysis virus IRES binding to yeast ribosomes [98].
4. Tethering in single-molecule experiments
To study translation via smFRET measurements on a milliseconds-to-minutes time scale, translating ribosomes have to be tethered to a microscope slide. Biotin/neutravidin/biotin complex is usually employed to bridge ribosomes and slide surface [10]. Tetrameric proteins neutravidin [99] and streptavidin can simultaneously bind up to four biotin moieties with extremely high affinity (Kd for biotin/neutravidin interaction is 10−15 M [100]). To prepare biotinylated slides, amino silane-modified slide surface is typically coated by a mixture of NHS ester of polyethylene glycol (PEG), which reduces non-specific binding of RNA and proteins, and NHS-ester of biotin-PEG [10]. Alternatively, slides can be coated with dimethyldichlorosilane (DSS)/Tween-20 and biotinylated bovine serum albumin (BSA) [101].
Ribosomes can be biotinylated by periodate oxidation of the 3’ end of 23S rRNA followed by conjugation with biotin-hydrazide [102]. Ribosomes were also tethered by genetically fusing the C terminus of ribosomal protein uS15 to a 15 amino-acid peptide derived from the biotin carboxyl carrier protein (BCCP) subunit of acetyl-CoA carboxylase [103], which can be biotinylated both in vitro and in vivo by biotin ligase (BirA) [104]. However, direct immobilization of biotinylated ribosomes may produce artifacts arising from the interaction between the ribosome and the slide surface. For this reason, tethering translating ribosomes to the slide via immobilization of ribosome-bound mRNA is more widely used in smFRET experiments. To that end, ~20 nt-long biotinylated DNA oligo (DNA handles) are typically annealed to mRNA (Fig. 5A) [105, 106]. We prefer to design optimal DNA handles for mRNA immobilization using the OligoWalk program from the RNAstructure software package (http://rna.urmc.rochester.edu/servers/oligowalk2/help.html) [107]. This program helps to avoid DNA handles and mRNA sequences with stable intramolecular secondary structure. Ribosome immobilization via mRNA tethering was used in smFRET experiments to examine structural dynamics of the ribosome, tRNA and ribosome-bound translation factors.
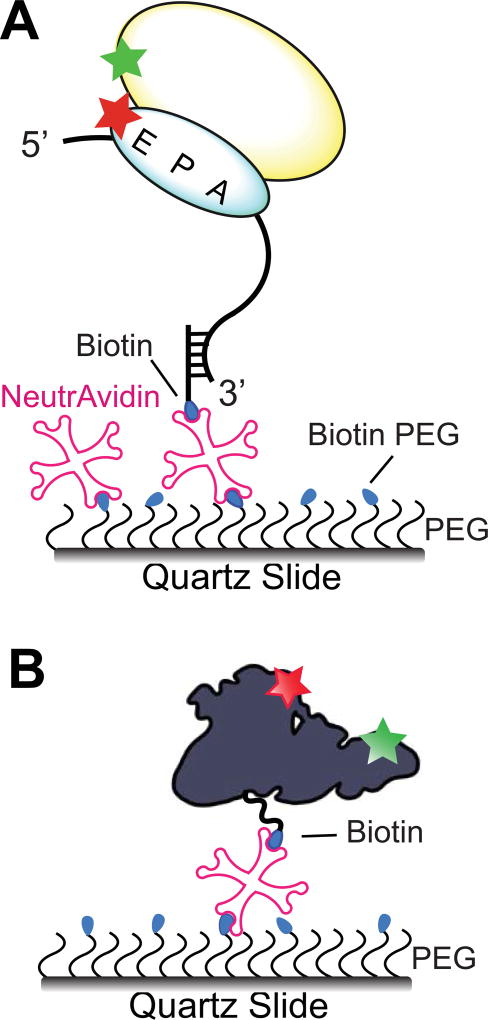
Figure 5. Surface immobilization strategies for smFRET experiments.
Mixture of biotin-PEG-NHS ester and PEG-NHS ester is covalently attached to amino-silanized slide surface. (A) S6Cy5/L9Cy3-labeled 70S ribosome is tethered to the quartz slide by hybridization of mRNA with a biotinylated DNA oligo bound to neutravidin [106]. (B) EF-G, domains II and IV of which are labeled with Cy3 and Cy5, is immobilized via C-terminal biotinylated BCCP tag [30].
smFRET probing of ribosome-free conformations of doubly-labeled translation factors requires their direct immobilization, which can be achieved by genetically fusing the BCCP biotinylation tag to either N- or C-termini of a translation factor (Fig. 5B) [30]. Alternatively, a translation factor can be immobilized using commercially-available biotinylated antibodies specific to the hexa-histidine (6-his) tag or other affinity purification polypeptide tags. In order to immobilize 6-his tagged proteins, specific biotinylated antibodies can be substituted with biotinyl-NTA resin (biotin-X NTA, Biotium) [10, 108, 109].
5. Interpretation of FRET data
5.1. Determination of FRET efficiency
In the ensemble FRET experiments, FRET efficiency is typically determined from measurements of (i) quantum yields of donor in the presence and absence of acceptor; (ii) fluorescence lifetimes of donor; or (iii) enhanced acceptor fluorescence [110]. We prefer to deduce FRET efficiency from the value of ratioA, which is the ratio of FRET signal from the acceptor and the signal from directly excited acceptor [110]. RatioA value is measured by exciting fluorescence in a doubly-labeled sample at wavelengths near the excitation maxima of the donor and acceptor fluorophores, respectively (Fig. 6). Unlike FRET values determined from quantum yields or fluorescence lifetimes of donor measured in the presence and absence of acceptor, ratioA value is measured in a single doubly-labeled sample. Therefore, ratioA values are concentration-independent and are not affected by pipetting errors. Because of the high experimental reproducibility of ratioA values (a typical average standard error is 0.001) the ratioA method is well suited for measuring small changes in FRET efficiency [111]. FRET efficiency (E) can be determined from the ratioA according to the equation:
where γ is the fraction of molecules labeled with both donor and acceptor dye; β is fraction of molecules labeled with acceptor only, ε(λ) is the extinction coefficient at wavelength λ, and the subscripts D and A denote donor or acceptor. λ1 is near maximum of donor excitation, λ2 is near maximum of acceptor excitation [110]. In the case of random labeling (i.e. when labeling with donor dye is independent of labeling with acceptor dye), the fraction of acceptor-labeled species that is also labeled with donor ((γ+ β)/γ) simplifies to 1/d+ where d+ is the fraction of molecules labeled with donor. The efficiency of labeling for each preparation can be determined from absorbance spectra [110]. The ratio εA(λ1)/εA(λ2) accounts for the contribution of direct excitation of acceptor at λ1 (i.e. in the absence of FRET between donor and acceptor, ratioA=εA(λ1)/εA(λ2)).
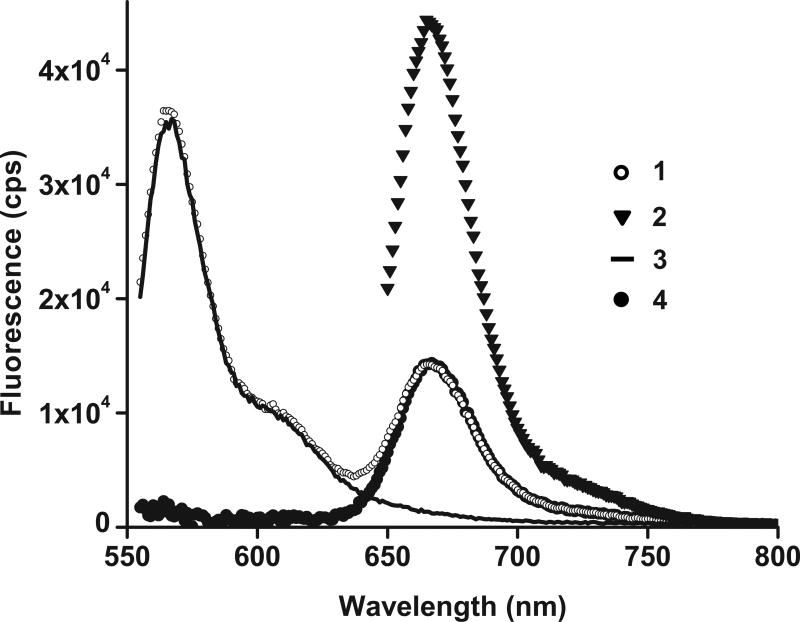
Figure 6. Measuring ensemble FRET by RatioA method.
Spectra used for calculation of the ratioA in doubly-labeled EF-G•GTP (EF-G_301–541_Cy3/Cy5) [30]: (1) the doubly labeled sample emission excited at 540 nm (open circles); (2) the doubly labeled sample emission excited at 635 nm (triangles); (3) normalized emission of donor-only labeled sample excited at 540 nm (solid line); (4) the extracted acceptor fluorescence (black circles) obtained by subtracting (3) from (1). The ratioA is determined from (4) divided by (2).
In smFRET measurements, apparent FRET efficiency (Eapp) is determined from the ratio of intensities of donor (ID) and acceptor (IA) fluorescence: Eapp=IA/(ID+IA) (Fig. 1D). Actual FRET efficiency (E) is determined using expression E=IA/(ID+γ•IA), where the parameter γ accounts for the differences in quantum yield and detection efficiency between the donor and the acceptor [10]. Parameter γ is determined from the change of acceptor fluorescence intensity (ΔIA) normalized by the change in donor fluorescence intensity (ΔID) upon acceptor photobleaching (γ= ΔIA/ΔID) (Fig. 1D) [10]. For Cy3/Cy5 pair γ is typically ~1 and, thus, apparent and actual FRET efficiency are equal [10]. However, if quantum yield of acceptor fluorophore is significantly affected by the local micro-environment at the labeling site, then γ will deviate from 1 and needs to be accurately determined to calculate actual FRET efficiency [112].
5.2. Uncertainties in FRET distance measurements
The distance between fluorophores (R) can be determined from FRET efficiency using expression . The Förster radius R0 is:
where θD is the quantum yield of the donor dye, κ2 describes the relative orientation between the fluorophores, J(λ) is the integral of the spectral overlap of the donor emission spectrum with the acceptor absorption spectrum, and n is the refractive index of the medium [10].
The R0 for Cy3/Cy5 pair is usually assumed to be ~56–60 Å [13, 14]. However, interactions of donor and acceptor fluorophores with the labeling sites may lead to a significant deviation in R0 from the reference value [10]. This occurs due to changes in the orientation factor κ2 and the quantum yield of donor (θD) induced by the unique micro-environment at the labeling site. The orientation factor κ2 is usually assumed to be equal to 2/3 because donor and acceptor orientations are anticipated to be randomized by rotational diffusion prior to energy transfer. When experimentally measured fluorescence anisotropy of both fluorophores is below 0.2, motions of fluorophores are fairly unrestricted and 2/3 is an accurate approximation of κ2 [10, 113, 114]. However, fluorescence anisotropy of Cy3 and Cy5 attached to proteins and nucleic acids is usually > 0.2 [10] and, thus, the actual κ2 value, which can theoretically range from 0 to 4, is less certain.
Because of an uncertainty in the orientation factor κ2, estimating distances from FRET data is prone to significant error. Nevertheless, FRET usually remains a monotonic function of distance even when motions of fluorophores are somewhat restricted [10, 115, 116]. The local micro-environment of both donor and acceptor is rarely altered by the transition between different conformations of the macromolecule [111]. In other words, while actual R0 might deviate from its reference value, it is unlikely to be affected by conformational changes of the macromolecule. Accordingly, FRET is more suitable for detecting changes in the distance between fluorophores than for estimating the distance itself [111].
If observed FRET changes are large and consistent with known biochemical and structural data, then these FRET changes likely correspond to actual conformational rearrangements of macromolecule and do not arise from perturbations of the fluorescent properties of the fluorophores. Nevertheless, a number of control experiments can be performed to test this assumption. For example, in the absence of site-specific perturbations of the fluorescent properties of the fluorophores, the reversal of dye pair orientation (i.e. switching sites to which the donor and acceptor are attached) should not affect observed FRET values. If anisotropy of both donor and acceptor remain unchanged between different functional states of the fluorescently-labeled macromolecule, then significant changes in κ2 value during conformational changes of the macromolecule are unlikely.
Protein binding in the vicinity (< 40 Å) of Cy3 and other cyanine fluorophores (DyLight 547, DyLight 647, Cy5 and Alexa Fluor 647) can increase fluorescence by slowing down isomerization of the fluorophore from the photo-active trans- to the photo-inactive cis-state [117, 118]. Such protein-induced fluorescence enhancement (PIFE) increases the donor quantum yield and R0. PIFE may affect FRET studies of translation: for example, PIFE might be induced by the binding of translation factor in the vicinity of Cy3-labeled ribosomal protein in Cy3–Cy5 labeled ribosomes. Highest reported levels of protein-induced Cy3 enhancement (2.5–3.5 -fold increase in quantum yield of Cy3 [118]) result in ~1.2-fold increase in R0 [119]. Therefore, in the worst case scenario, protein binding may produce an increase in observed FRET in the absence of conformational changes. The magnitude of protein-induced fluorescence enhancement (θD,enhanced/ θD,non-enhanced) can be determined in the experiments performed with donor-only labeled ribosomes. Then corrected R0 value can easily be calculated as R0, corrected ≈ R0, reference•(θD,enhanced/ ΔD,non-enhanced)1/6 [119]. Alternatively, Cy3B or Atto550, which do not undergo PIFE, can be used instead of Cy3 [119].
6. Conclusions
Over the last fifteen years, dramatic progress has been made in FRET studies of structural dynamics of the bacterial ribosome. However, many important mechanistic details of bacterial translation await further investigation. For example, the sequence of structural rearrangements of tRNA, mRNA, EF-G and the ribosome during ribosomal translocation has not been fully elucidated. Likewise, FRET measurements may provide additional insights into the molecular mechanism of translation regulation in bacteria via programmed ribosome frameshifting and mRNA riboswitches. Finally, some of the strategies developed for fluorescent labeling and FRET measurements for the bacterial system can now be extended to investigation of protein synthesis in eukaryotes, which is more complex and less well understood than bacterial translation. In particular, FRET studies will likely bring crucial advances in the understanding of translation initiation in eukaryotes, many mechanistic aspects of which are poorly elucidated.
Highlights.
FRET is a powerful tool for studies of molecular mechanisms of translation.
Fluorophores can be introduced into the translational machinery through a number of methods.
Important considerations for the appropriate interpretation of FRET data are discussed.
Acknowledgments
We apologize to all authors whose works have not been cited due to space limitation. We thank Paul Whitford for his comments on the manuscript. The work of the D.N.E. lab was supported by grant from the US National Institute of Health no. GM-099719. We thank Enea Salsi, Zoe Netter, Elie Farah and Clarence Ling for their contributions to smFRET and fluorescent labeling experiments done in the D.N.E. lab.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ling C, Ermolenko DN. Structural insights into ribosome translocation. Wiley Interdiscip Rev RNA. 2016;7(5):620–36. doi: 10.1002/wrna.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Z, Shaikh TR, Barnard D, Meng X, Mohamed H, Yassin A, Mannella CA, Agrawal RK, Lu TM, Wagenknecht T. Monolithic microfluidic mixing-spraying devices for time-resolved cryo-electron microscopy. J Struct Biol. 2009;168(3):388–95. doi: 10.1016/j.jsb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen B, Kaledhonkar S, Sun M, Shen B, Lu Z, Barnard D, Lu TM, Gonzalez RL, Jr, Frank J. Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure. 2015;23(6):1097–105. doi: 10.1016/j.str.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milon P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. Real-time assembly landscape of bacterial 30S translation initiation complex. Nat Struct Mol Biol. 2012;19(6):609–15. doi: 10.1038/nsmb.2285. [DOI] [PubMed] [Google Scholar]
- 5.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385(6611):37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 6.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18(4):457–62. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holtkamp W, Cunha CE, Peske F, Konevega AL, Wintermeyer W, Rodnina MV. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014;33(9):1073–85. doi: 10.1002/embj.201387465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25(4):519–29. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tinoco I, Jr, Gonzalez RL., Jr Biological mechanisms, one molecule at a time. Genes Dev. 2011;25(12):1205–31. doi: 10.1101/gad.2050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–16. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altman RB, Terry DS, Zhou Z, Zheng Q, Geggier P, Kolster RA, Zhao Y, Javitch JA, Warren JD, Blanchard SC. Cyanine fluorophore derivatives with enhanced photostability. Nat Methods. 2011;9(1):68–71. doi: 10.1038/nmeth.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fei J, Wang J, Sternberg SH, MacDougall DD, Elvekrog MM, Pulukkunat DK, Englander MT, Gonzalez RL., Jr A highly purified, fluorescently labeled in vitro translation system for single-molecule studies of protein synthesis. Methods Enzymol. 2010;472:221–59. doi: 10.1016/S0076-6879(10)72008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietrich A, Buschmann V, Muller C, Sauer M. Fluorescence resonance energy transfer (FRET) and competing processes in donor-acceptor substituted DNA strands: a comparative study of ensemble and single-molecule data. J Biotechnol. 2002;82(3):211–31. doi: 10.1016/s1389-0352(01)00039-3. [DOI] [PubMed] [Google Scholar]
- 14.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87(2):1328–37. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen K, Whitford PC. Challenges in describing ribosome dynamics. Phys Biol. 2017;14(2):023001. doi: 10.1088/1478-3975/aa626b. [DOI] [PubMed] [Google Scholar]
- 16.Wasserman MR, Alejo JL, Altman RB, Blanchard SC. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol. 2016;23(4):333–41. doi: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, McAlear MA, Moore PB, Noller HF, Ortega J, Panse VG, Ramakrishnan V, Spahn CM, Steitz TA, Tchorzewski M, Tollervey D, Warren AJ, Williamson JR, Wilson D, Yonath A, Yusupov M. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–9. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchard SC. Single-molecule observations of ribosome function. Curr Opin Struct Biol. 2009;19(1):103–9. doi: 10.1016/j.sbi.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caban K, Gonzalez RL., Jr The emerging role of rectified thermal fluctuations in initiator aa-tRNA- and start codon selection during translation initiation. Biochimie. 2015;114:30–8. doi: 10.1016/j.biochi.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prabhakar A, Choi J, Wang J, Petrov A, Puglisi JD. Dynamic basis of fidelity and speed in translation: Coordinated multistep mechanisms of elongation and termination. Protein Sci. 2017;26(7):1352–1362. doi: 10.1002/pro.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HK, Liu F, Fei J, Bustamante C, Gonzalez RL, Jr, Tinoco I., Jr A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc Natl Acad Sci U S A. 2014;111(15):5538–43. doi: 10.1073/pnas.1403457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Petrov A, Johansson M, Tsai A, O'Leary SE, Puglisi JD. Dynamic pathways of-1 translational frameshifting. Nature. 2014;512(7514):328–32. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaur J, Raj M, Cooperman BS. Fluorescent labeling of tRNA dihydrouridine residues: Mechanism and distribution. RNA. 2011;17(7):1393–400. doi: 10.1261/rna.2670811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010;17(12):1470–7. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elvekrog MM, Gonzalez RL., Jr Conformational selection of translation initiation factor 3 signals proper substrate selection. Nat Struct Mol Biol. 2013;20(5):628–33. doi: 10.1038/nsmb.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salsi E, Farah E, Dann J, Ermolenko DN. Following movement of domain IV of elongation factor G during ribosomal translocation. Proc Natl Acad Sci U S A. 2014;111(42):15060–5. doi: 10.1073/pnas.1410873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDougall DD, Gonzalez RL., Jr Translation initiation factor 3 regulates switching between different modes of ribosomal subunit joining. J Mol Biol. 2015;427(9):1801–18. doi: 10.1016/j.jmb.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salsi E, Farah E, Netter Z, Dann J, Ermolenko DN. Movement of elongation factor G between compact and extended conformations. J Mol Biol. 2015;427(2):454–67. doi: 10.1016/j.jmb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Caban K, Gonzalez RL., Jr Ribosomal initiation complex-driven changes in the stability and dynamics of initiation factor 2 regulate the fidelity of translation initiation. J Mol Biol. 2015;427(9):1819–34. doi: 10.1016/j.jmb.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermanson GT. Bioconjugate techniques. (2) 2008 [Google Scholar]
- 33.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82(1):70–7. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 34.Simpson RJ. Estimation of Free Thiols and Disulfide Bonds Using Ellman's Reagent. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot4699. pdb prot4699. [DOI] [PubMed] [Google Scholar]
- 35.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 36.Xiao H, Schultz PG. At the Interface of Chemical and Biological Synthesis: An Expanded Genetic Code. Cold Spring Harb Perspect Biol. 2016;8(9) doi: 10.1101/cshperspect.a023945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301(5635):964–7. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 38.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29(4):770–81. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat Protoc. 2006;1(1):280–5. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 40.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3(11):707–8. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 41.Taki M, Shiota M, Taira K. Transglutaminase-mediated N- and C-terminal fluorescein labeling of a protein can support the native activity of the modified protein. Protein Eng Des Sel. 2004;17(2):119–26. doi: 10.1093/protein/gzh015. [DOI] [PubMed] [Google Scholar]
- 42.Rashidian M, Dozier JK, Distefano MD. Enzymatic labeling of proteins: techniques and approaches. Bioconjug Chem. 2013;24(8):1277–94. doi: 10.1021/bc400102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, Kelleher NL, Kolter R, Walsh CT. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc Natl Acad Sci U S A. 2005;102(44):15815–20. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popp MW, Ploegh HL. Making and breaking peptide bonds: protein engineering using sortase. Angew Chem Int Ed Engl. 2011;50(22):5024–32. doi: 10.1002/anie.201008267. [DOI] [PubMed] [Google Scholar]
- 45.Kurosaki T, Li W, Hoque M, Popp MW, Ermolenko DN, Tian B, Maquat LE. A posttranslational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014;28(17):1900–16. doi: 10.1101/gad.245506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popp MW. Site-specific labeling of proteins via sortase: protocols for the molecular biologist. Methods Mol Biol. 2015;1266:185–98. doi: 10.1007/978-1-4939-2272-7_13. [DOI] [PubMed] [Google Scholar]
- 47.Mauger DM, Siegfried NA, Weeks KM. The genetic code as expressed through relationships between mRNA structure and protein function. FEBS Lett. 2013;587(8):1180–8. doi: 10.1016/j.febslet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13(8):857–68. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 49.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7(6):393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 50.Suddala KC, Walter NG. Riboswitch structure and dynamics by smFRET microscopy. Methods Enzymol. 2014;549:343–73. doi: 10.1016/B978-0-12-801122-5.00015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boerneke MA, Hermann T. Conformational flexibility of viral RNA switches studied by FRET. Methods. 2015;91:35–9. doi: 10.1016/j.ymeth.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solomatin S, Herschlag D. Methods of site-specific labeling of RNA with fluorescent dyes. Methods Enzymol. 2009;469:47–68. doi: 10.1016/S0076-6879(09)69003-0. [DOI] [PubMed] [Google Scholar]
- 53.Price SR, Ito N, Oubridge C, Avis JM, Nagai K. Crystallization of RNA-protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J Mol Biol. 1995;249(2):398–408. doi: 10.1006/jmbi.1995.0305. [DOI] [PubMed] [Google Scholar]
- 54.Ferre-D'Amare AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5' and 3' heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24(5):977–8. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci U S A. 1997;94(9):4262–6. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akiyama BM, Stone MD. Assembly of complex RNAs by splinted ligation. Methods Enzymol. 2009;469:27–46. doi: 10.1016/S0076-6879(09)69002-9. [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Sun L, Cui Z, Gottlieb RL, Zhang B. 5'-sulfhydryl-modified RNA: initiator synthesis, in vitro transcription, and enzymatic incorporation. Bioconjug Chem. 2001;12(6):939–48. doi: 10.1021/bc015504g. [DOI] [PubMed] [Google Scholar]
- 58.Czworkowski J, Odom OW, Hardesty B. Fluorescence study of the topology of messenger RNA bound to the 30S ribosomal subunit of Escherichia coli. Biochemistry. 1991;30(19):4821–30. doi: 10.1021/bi00233a026. [DOI] [PubMed] [Google Scholar]
- 59.Zearfoss NR, Ryder SP. End-labeling oligonucleotides with chemical tags after synthesis. Methods Mol Biol. 2012;941:181–93. doi: 10.1007/978-1-62703-113-4_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang LK, Lima CD, Shuman S. Structure and mechanism of T4 polynucleotide kinase: an RNA repair enzyme. EMBO J. 2002;21(14):3873–80. doi: 10.1093/emboj/cdf397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghosh SS, Kao PM, McCue AW, Chappelle HL. Use of maleimide-thiol coupling chemistry for efficient syntheses of oligonucleotide-enzyme conjugate hybridization probes. Bioconjug Chem. 1990;1(1):71–6. doi: 10.1021/bc00001a009. [DOI] [PubMed] [Google Scholar]
- 62.Whitfeld PR. Application of the periodate method for the analysis of nucleotide sequence to tobacco mosaic virus RNA. Biochim Biophys Acta. 1965;108(2):202–10. doi: 10.1016/0005-2787(65)90004-3. [DOI] [PubMed] [Google Scholar]
- 63.Qiu C, Liu WY, Xu YZ. Fluorescence labeling of short RNA by oxidation at the 3'-end. Methods Mol Biol. 2015;1297:113–20. doi: 10.1007/978-1-4939-2562-9_8. [DOI] [PubMed] [Google Scholar]
- 64.LoPachin RM, Gavin T. Molecular mechanisms of aldehyde toxicity: a chemical perspective. Chem Res Toxicol. 2014;27(7):1081–91. doi: 10.1021/tx5001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.England TE, Gumport RI, Uhlenbeck OC. Dinucleoside pyrophosphate are substrates for T4-induced RNA ligase. Proc Natl Acad Sci U S A. 1977;74(11):4839–42. doi: 10.1073/pnas.74.11.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richardson RW, Gumport RI. Biotin and fluorescent labeling of RNA using T4 RNA ligase. Nucleic Acids Res. 1983;11(18):6167–84. doi: 10.1093/nar/11.18.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Babaylova ES, Malygin AA, Lomzov AA, Pyshnyi DV, Yulikov M, Jeschke G, Krumkacheva OA, Fedin MV, Karpova GG, Bagryanskaya EG. Complementary-addressed site-directed spin labeling of long natural RNAs. Nucleic Acids Res. 2016;44(16):7935–43. doi: 10.1093/nar/gkw516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao M, Steffen FD, Borner R, Schaffer MF, Sigel RKO, Freisinger E. Site-specific dual-color labeling of long RNAs for single-molecule spectroscopy. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Odom OW, Dabbs ER, Dionne C, Muller M, Hardesty B. The distance between S1, S21, and the 3' end of 16S RNA in 30S ribosomal subunits. The effect of poly(uridylic acid) and 50S subunits on these distances. Eur J Biochem. 1984;142(2):261–7. doi: 10.1111/j.1432-1033.1984.tb08280.x. [DOI] [PubMed] [Google Scholar]
- 70.Odom OW, Stoffler G, Hardesty B. Movement of the 3'-end of 16 S RNA towards S21 during activation of 30 S ribosomal subunits. FEBS Lett. 1984;173(1):155–8. doi: 10.1016/0014-5793(84)81037-6. [DOI] [PubMed] [Google Scholar]
- 71.Dorywalska M, Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33(1):182–9. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petrov A, Puglisi JD. Site-specific labeling of Saccharomyces cerevisiae ribosomes for single-molecule manipulations. Nucleic Acids Res. 2010;38(13):e143. doi: 10.1093/nar/gkq390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Korostelev A, Noller HF. Analysis of structural dynamics in the ribosome by TLS crystallographic refinement. J Mol Biol. 2007;373(4):1058–70. doi: 10.1016/j.jmb.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24(24):4316–23. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG. Systematic chromosomal deletion of bacterial ribosomal protein genes. J Mol Biol. 2011;413(4):751–61. doi: 10.1016/j.jmb.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bubunenko M, Baker T, Court DL. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J Bacteriol. 2007;189(7):2844–53. doi: 10.1128/JB.01713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Svidritskiy E, Ling C, Ermolenko DN, Korostelev AA. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proc Natl Acad Sci U S A. 2013;110(30):12283–8. doi: 10.1073/pnas.1304922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30(3):348–59. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 80.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of Intersubunit Movement of the Ribosome in Solution Using FRET. J Mol Biol. 2007;370(3):530–40. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 81.Chen C, Stevens B, Kaur J, Cabral D, Liu H, Wang Y, Zhang H, Rosenblum G, Smilansky Z, Goldman YE, Cooperman BS. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol Cell. 2011;42(3):367–77. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ly CT, Altuntop ME, Wang Y. Single-molecule study of viomycin's inhibition mechanism on ribosome translocation. Biochemistry. 2010;49(45):9732–8. doi: 10.1021/bi101029g. [DOI] [PubMed] [Google Scholar]
- 83.Fei J, Bronson JE, Hofman JM, Srinivas RL, Wiggins CH, Gonzalez RL., Jr Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci U S A. 2009;106(37):15702–7. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A. 2009;106(8):2571–6. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Steffen KK, McCormick MA, Pham KM, MacKay VL, Delaney JR, Murakami CJ, Kaeberlein M, Kennedy BK. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics. 2012;191(1):107–18. doi: 10.1534/genetics.111.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Culver GM, Noller HF. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA. 1999;5(6):832–43. doi: 10.1017/s1355838299990714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hickerson R, Majumdar ZK, Baucom A, Clegg RM, Noller HF. Measurement of internal movements within the 30 S ribosomal subunit using Forster resonance energy transfer. J Mol Biol. 2005;354(2):459–72. doi: 10.1016/j.jmb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 88.Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A. 2012;109(50):20391–4. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Juette MF, Terry DS, Wasserman MR, Altman RB, Zhou Z, Zhao H, Blanchard SC. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat Methods. 2016;13(4):341–4. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316(3):725–68. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 91.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21(1):86–9. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 92.Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331(6022):1289–95. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holguin A, Lospitao E, Lopez M, de Arellano ER, Pena MJ, del Romero J, Martin C, Soriano V. Genetic characterization of complex inter-recombinant HIV-1 strains circulating in Spain and reliability of distinct rapid subtyping tools. J Med Virol. 2008;80(3):383–91. doi: 10.1002/jmv.21105. [DOI] [PubMed] [Google Scholar]
- 94.Miller LW, Cai Y, Sheetz MP, Cornish VW. In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat Methods. 2005;2(4):255–7. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 95.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125(6):1125–36. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 96.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–45. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 97.Fuchs G, Petrov AN, Marceau CD, Popov LM, Chen J, O'Leary SE, Wang R, Carette JE, Sarnow P, Puglisi JD. Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proc Natl Acad Sci U S A. 2015;112(2):319–25. doi: 10.1073/pnas.1421328111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petrov A, Grosely R, Chen J, O'Leary SE, Puglisi JD. Multiple Parallel Pathways of Translation Initiation on the CrPV IRES. Mol Cell. 2016;62(1):92–103. doi: 10.1016/j.molcel.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marttila AT, Laitinen OH, Airenne KJ, Kulik T, Bayer EA, Wilchek M, Kulomaa MS. Recombinant NeutraLite avidin: a non-glycosylated, acidic mutant of chicken avidin that exhibits high affinity for biotin and low non-specific binding properties. FEBS Lett. 2000;467(1):31–6. doi: 10.1016/s0014-5793(00)01119-4. [DOI] [PubMed] [Google Scholar]
- 100.Green NM. Avidin. 1. The Use of (14-C)Biotin for Kinetic Studies and for Assay. Biochem J. 1963;89:585–91. doi: 10.1042/bj0890585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hua B, Han KY, Zhou R, Kim H, Shi X, Abeysirigunawardena SC, Jain A, Singh D, Aggarwal V, Woodson SA, Ha T. An improved surface passivation method for single-molecule studies. Nat Methods. 2014;11(12):1233–6. doi: 10.1038/nmeth.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stapulionis R, Wang Y, Dempsey GT, Khudaravalli R, Nielsen KM, Cooperman BS, Goldman YE, Knudsen CR. Fast in vitro translation system immobilized on a surface via specific biotinylation of the ribosome. Biol Chem. 2008;389(9):1239–49. doi: 10.1515/BC.2008.141. [DOI] [PubMed] [Google Scholar]
- 103.Liu T, Kaplan A, Alexander L, Yan S, Wen JD, Lancaster L, Wickersham CE, Fredrick K, Noller H, Tinoco I, Bustamante CJ. Direct measurement of the mechanical work during translocation by the ribosome. Elife. 2014;3:e03406. doi: 10.7554/eLife.03406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8(4):921–9. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101(35):12893–8. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30(5):578–88. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lu ZJ, Mathews DH. OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008;36(Web Server issue):W104–8. doi: 10.1093/nar/gkn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao Y, Terry D, Shi L, Weinstein H, Blanchard SC, Javitch JA. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature. 2010;465(7295):188–93. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Agrawal AA, Salsi E, Chatrikhi R, Henderson S, Jenkins JL, Green MR, Ermolenko DN, Kielkopf CL. An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal. Nat Commun. 2016;7:10950. doi: 10.1038/ncomms10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–88. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 111.Majumdar ZK, Hickerson R, Noller HF, Clegg RM. Measurements of internal distance changes of the 30S ribosome using FRET with multiple donor-acceptor pairs: quantitative spectroscopic methods. J Mol Biol. 2005;351(5):1123–45. doi: 10.1016/j.jmb.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 112.Chung HS, Louis JM, Eaton WA. Distinguishing between protein dynamics and dye photophysics in single-molecule FRET experiments. Biophys J. 2010;98(4):696–706. doi: 10.1016/j.bpj.2009.12.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haas E, Katchalski-Katzir E, Steinberg IZ. Effect of the orientation of donor and acceptor on the probability of energy transfer involving electronic transitions of mixed polarization. Biochemistry. 1978;17(23):5064–70. doi: 10.1021/bi00616a032. [DOI] [PubMed] [Google Scholar]
- 114.Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem. 1978;47:819–46. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 115.Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DM. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci U S A. 2008;105(32):11176–81. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA. Polyproline and the "spectroscopic ruler" revisited with single-molecule fluorescence. Proc Natl Acad Sci U S A. 2005;102(8):2754–9. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stennett EM, Ciuba MA, Lin S, Levitus M. Demystifying PIFE: The Photophysics Behind the Protein-Induced Fluorescence Enhancement Phenomenon in Cy3. J Phys Chem Lett. 2015;6(10):1819–23. doi: 10.1021/acs.jpclett.5b00613. [DOI] [PubMed] [Google Scholar]
- 118.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A. 2011;108(18):7414–8. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ploetz E, Lerner E, Husada F, Roelfs M, Chung S, Hohlbein J, Weiss S, Cordes T. Forster resonance energy transfer and protein-induced fluorescence enhancement as synergetic multi-scale molecular rulers. Sci Rep. 2016;6:33257. doi: 10.1038/srep33257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105(40):15364–9. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang L, Pulk A, Wasserman MR, Feldman MB, Altman RB, Cate JH, Blanchard SC. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol. 2012;19(9):957–63. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ling C, Ermolenko DN. Initiation factor 2 stabilizes the ribosome in a semirotated conformation. Proc Natl Acad Sci U S A. 2015;112(52):15874–9. doi: 10.1073/pnas.1520337112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–4. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 125.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326(5953):694–9. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]