Abstract
In E. coli chemosensory arrays, transmembrane receptors, a histidine autokinase CheA, and a scaffolding protein CheW interact to form an extended hexagonal lattice of signaling complexes. One interaction, previously assigned a crucial signaling role, occurs between chemoreceptors and the CheW-binding P5 domain of CheA. Structural studies showed a receptor helix fitting into a hydrophobic cleft at the boundary between P5 subdomains. Our work aimed to elucidate the in vivo roles of the receptor-P5 interface, employing as a model the interaction between E. coli CheA and Tsr, the serine chemoreceptor. Crosslinking assays confirmed P5 and Tsr contacts in vivo and their strict dependence on CheW. Moreover, the P5 domain only mediated CheA recruitment to polar receptor clusters if CheW was also present. Amino acid replacements at CheA.P5 cleft residues reduced CheA kinase activity, lowered serine response cooperativity and partially impaired chemotaxis. Pseudoreversion studies identified suppressors of P5 cleft defects at other P5 groove residues or at surface-exposed residues in P5 subdomain 1, which interacts with CheW in signaling complexes. Our results indicate that a high-affinity P5-receptor binding interaction is not essential for core complex function. Rather, P5 groove residues are probably required for proper cleft structure and/or dynamic behavior, which likely impact conformational communication between P5 subdomains and the strong binding interaction with CheW that is necessary for kinase activation. We propose a model for signal transmission in chemotaxis signaling complexes in which the CheW-receptor interface plays the key role in conveying signaling-related conformational changes from receptors to the CheA kinase.
Keywords: chemotaxis, chemoreceptor, autokinase, signal transduction
Graphical abstract

Introduction
Transmembrane chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs) enable motile bacteria and archaea to detect and follow chemical gradients in their environments [1]. MCPs control the activity of CheA, a cytoplasmic histidine autokinase, to communicate stimulus signals to the locomotor apparatus. Our current understanding of chemoreceptor signaling comes mainly from work with Escherichia coli and Salmonella typhimurium, whose chemotaxis machinery has been intensively studied for decades. In these systems, unliganded receptors activate CheA, which in turn donates phosphoryl groups to the CheY response regulator. Phospho-CheY (P-CheY) promotes clockwise rotation of the flagellar motors, causing random changes in swimming direction. Attractant-bound chemoreceptors down-regulate CheA, reducing intracellular P-CheY levels to promote forward swimming. P-CheY is actively dephosphorylated by a dedicated phosphatase, CheZ, ensuring rapid motor responses during travel in chemoeffector gradients. A sensory adaptation system adjusts the detection sensitivity of MCP chemoreceptors, enabling the cells to sense chemoeffector changes over a wide concentration range [2]. Adaptation occurs through reversible covalent modifications, catalyzed by the CheR methyltransferase and the CheB methylesterase, at specific residues in the receptor cytoplasmic kinase control domain.
The receptor core complex, the smallest signaling unit, comprises six homodimeric MCP molecules, organized as trimers of dimers, two CheW molecules and a multi-domain CheA dimer (Fig. 1a) [3]. In the core unit, the cytoplasmic tip of one receptor in each trimer binds a CheW molecule and another receptor in each trimer contacts the P5 domain in one CheA subunit (Fig. 1b). The CheA dimerization domains (P3/P3′) lie between the receptor trimers, whereas the remaining domains, P1 (phosphorylation site), P2 (CheY/CheB binding) and P4 (ATP binding), extend past the hairpin tips of the receptors. A strong binding interaction between CheA.P5 and CheW (interface 1) bridges the two receptor trimers in the core complex. A second but weaker CheA.P5-CheW interaction (interface 2) networks core complexes into hexagonal arrays (Fig. 1c) that typically localize in large clusters at the cell poles [4,5,6].
Fig. 1. Structural organization of receptor signaling complexes.
(a) The core signaling unit. The periplasmic sensing domains of the receptor molecules are at the top of the figure, external to the membrane (gray rectangle) and the intracellular compartment. CheA functions as a homodimer with five domains in each subunit: P1 (phosphorylation site); P2 (CheB and CheY binding); P3 (dimerization); P4 (ATP binding); P5 (CheW and receptor interaction). Note that CheA autophosphorylation is a trans reaction, involving interaction of a P1 domain in one subunit with a P4 domain in the other. Vertical white lines in the receptors and in the P3/P3' domains of CheA indicate the dimerization interface between the two protomers of the homodimers. One CheA.P5 domain and one CheW molecule lie behind the tips of the receptor trimers and are largely obscured in this view. The black circle at the P5-CheW junction indicates the interface 1 binding interaction between P5 subdomain 1 and CheW subdomain 2 that plays an essential role in assembly and function of the core complex.
(b) Top-down cross-section view of the core complex. Most of the trimer-stabilizing interactions between receptors involve residues in one subunit of each receptor dimer. CheW and the CheA.P5 domain are structural homologs; each comprises two similar subdomains. Black circles indicate the interface 1 binding interaction between P5 subdomain 1 and CheW subdomain 2. Black rectangles indicate the CheW-receptor interface. Black stars indicate the receptor-P5 contact surface studied in this work. (c) Three core complexes networked through interface 2 connections between P5 and CheW (white circles).
The CheA.P5 regulatory domain is essential for interaction of CheA with CheW and for receptor control of kinase activity [7,8,9,10]. P5 and CheW have similar protein folds, comprising two intertwined β-barrels (subdomains 1 and 2), sandwiching a central cleft or groove that interacts with the receptor tip [4,11,12,13] (Fig. 2a). In vitro crosslinking experiments in reconstituted arrays and mutational analyses of CheA.P5 cleft residues led to a proposal that the P5-receptor interface plays an important role in receptor-mediated activation and conformational control of CheA through a binding interaction [14,15].
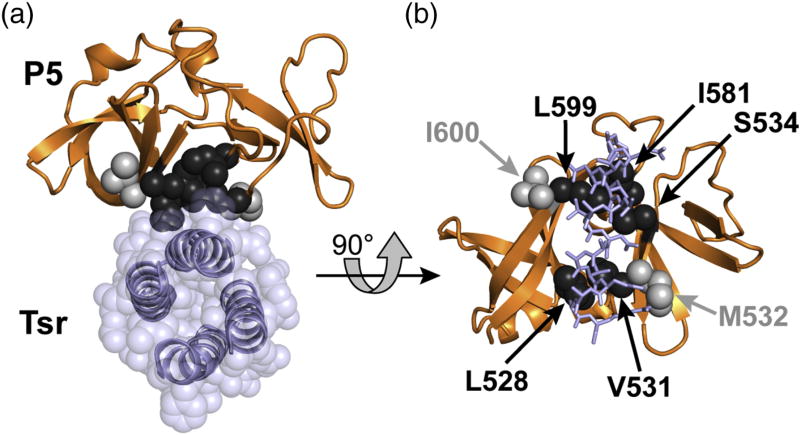
Fig. 2. Structural model of the CheA.P5-receptor interface.
Atomic coordinates for an E. coli P5-Tsr complex were modeled from a T. maritima ternary complex structure (PDB code 4JPB) [13] (see Materials and Methods).
(a) Top view of the P5-Tsr contact surface. The cytoplasmic tip of a receptor molecule, in this case Tsr (blue helix backbones, transparent blue atoms), inserts the N-helix of one subunit into a groove or cleft in P5 (orange backbone). The side-chain atoms of P5 cleft residues implicated in the receptor interaction are shown as black spheres (L528, V531, S534, I581, L599). Sidechain atoms of nearby interface control residues that project away from the cleft are shown as light gray spheres (M532, I600).
(b) Face-on view of the P5 cleft. Sidechains of receptor N-helix residues that contact P5 cleft residues are shown as blue sticks. The shading conventions for P5 residues follow those in panel (a).
The present work aimed to elucidate the in vivo roles of CheA kinase-receptor contacts by exploring the interaction between the E. coli CheA.P5 domain and Tsr, the serine chemoreceptor. An extensive mutational analysis of the CheA.P5 domain demonstrated that P5 cleft residues are indeed important for CheA kinase activation. However, our results indicate that kinase activation is not dependent on a high-affinity direct P5-receptor binding interaction. Accordingly, we propose a different role for P5 cleft residues in signaling and present a refined model for conformational signal transmission in core complexes and the chemosensory array.
Results
In vivo interactions between Tsr and CheA.P5 cleft residues
The P5 domain of CheA and the structurally homologous CheW protein each have two intertwined β-barrel subdomains [11,12]. Previous genetic, biochemical, and structural studies identified residues lining the cleft between the two subdomains in each protein as sites of interaction with a helix of the receptor hairpin tip [5,9,10,14,15,16,17,18]. When mapped onto the E. coli CheA.P5-Tsr interface modeled on the structure of a T. maritima signaling complex [13], P5 residues L528, V531, S534, I581, and L599 flank a groove between P5 subdomains 1 and 2 and their sidechains are in close proximity to receptor residues (Fig. 2).
We devised cysteine-directed crosslinking assays to probe for these P5-receptor contacts in vivo (Fig. 3). Single cysteine replacements were introduced at P5 groove residues L528, V531, S534, I581, and L599 in a functional, hemagglutinin antigen (HA)-tagged CheA variant that lacks its three native cysteines [19]. We also constructed cysteine reporters at two CheA control positions M532 and I600, whose sidechains project away from the P5-receptor interface (Fig. 2). In parallel, we constructed a set of Tsr reporters bearing single-cysteine replacements in the hairpin tip: at N-helix residues F373, N376, L380, A383, V384, A387 and G390, which project into or near the P5 cleft (Fig. 2), and at more distant C-helix residues, G395, A397, V398 or G401. We treated cells expressing all reporter pair combinations with Cu2+-phenanthroline to promote disulfide bond formation and detected crosslinking products by gel electrophoresis and immunoblotting. Our results were in close agreement with the P5-Tsr interface model: CheA-L528C, V531C, S534C and L599C readily formed disulfide bridges almost exclusively with nearby receptor N-helix cysteine residues (Fig 3). The CheA control position reporters yielded much reduced (M532C) or almost no (I600C) CheA-Tsr crosslinking. Unexpectedly, I581C, which the structural model predicted to be in close proximity to Tsr N-helix cysteine residues, did not form any crosslinks to Tsr. In all cases, no CheA-Tsr crosslinked products were detected in cells lacking CheW, indicating that in vivo the P5-Tsr interaction is strictly dependent on CheW (Fig. 3).
Fig. 3. In vivo P5-Tsr crosslinking assays.
HA-tagged CheA-Cys reporters were expressed alone from pGP34 (white columns) or co-expressed with CheW from pGP32 (gray columns), whereas Tsr-Cys reporters were expressed from plasmid pPA114. Cells of strain UU2806 [Δ(cheA-cheZ) Δ(tar, tap, tsr, trg, aer)] co-transformed with derivatives of pPA114 and pGP34 or pGP32 were grown and treated as detailed in Materials and Methods, and lysate proteins were separated by SDS/PAGE and probed with anti-HA antibody to detect CheA and CheA~Tsr crosslinking products. Histogram bars indicate the fraction of CheA subunits crosslinked to Tsr; error bars indicate the standard error of three measurements. Circles indicate the distances between the β-carbons of each pair of reporter sidechains, obtained from the atomic coordinates of the model shown in Fig. 2. Black circles indicate reporter site pairs closer than 10 Å.
Cellular clustering properties of P5-containing proteins
Previous studies of YFP-P5 fusion proteins suggested that the CheA.P5 domain could interact with chemoreceptors and mediate recruitment to receptor clusters in CheW-independent fashion [14,20]. Our P5-Tsr crosslinking experiments challenge that finding (Fig. 3). To clarify the issue, we carried out fluorescence microscopy localization studies in several cellular backgrounds, employing GFP-P5 or P5-GFP protein fusions (Fig 4). When expressed from a high copy-number, strong-promoter plasmid, like the one used in previous studies [14,20], GFP-P5 formed clusters even in a receptor-less strain (Fig 4, panel A), which implies that these expression conditions promoted protein aggregation. However, GFP-P5 and P5-GFP fusion proteins expressed from a low-copy, weak-promoter plasmid did not form aggregates in the absence of receptors (Fig. 4, panels B and C). They clustered at polar receptor patches only when CheW was present (Fig. 4, panels N and O), demonstrating that, at native stoichiometries, P5 cannot by itself establish stable cellular interactions with receptors. Similarly, GFP-CheA, but not GFP-CheAΔP5, exhibited polar clustering only in a strain containing both CheW and receptors (Fig. 4P and 4Q). We conclude that in vivo formation of the P5-receptor interface and the concomitant incorporation of CheA into signaling complexes are strictly dependent on CheW, which can bind directly to chemoreceptors (Fig. 4, panel L) [9,10] and to the CheA.P5 domain through interface 1 interactions [8,9,10].
Fig. 4. Cellular clustering assays of fluorescent CheA, P5 and CheW fusion proteins.
Plasmids expressing the fusion proteins indicated along the top of the figure were introduced into three ΔcheA host strains, listed on the left side of the figure, for clustering assays. pGP3 is a high copy-number plasmid with a strong lac promoter and expresses the GFP-P5 fusion protein at high cellular levels (double arrows). Plasmids pGP7, pGP8, pGP12, and pGP68 have low copy numbers and a tightly regulated salicylate-inducible promoter. In these experiments, their fusion protein products were expressed at approximately the native cellular level. pPA802 is also a low copy-number plasmid, with a tightly controlled IPTG-inducible promoter. Its product, YFP-CheW, was expressed at approximately native levels in these experiments. Fluorescence images were inverted and converted to grayscale to facilitate visualization of clusters (black spots).
Mutational analysis of receptor interface residues in the CheA-P5 domain
To generate all possible amino acid replacements at each targeted CheA.P5 residue, mutations were created by all-codon mutagenesis in plasmid pPM25, which co-expresses the cheA and cheW genes under control of a salicylate-inducible promoter [21]. Strain UU1607 [Δ(cheAW)] carrying the various mutant plasmids was tested for chemotaxis on tryptone soft agar plates. The replacement mutants (hereafter designated P5*) defined three functional groups: >60% of the wild-type colony size (50/95 mutants); 40–60% of wild-type size (20/95 mutants); or <40% of wild-type size (25/95 mutants) (Fig. 5a). Amino acids with charged (D, E, K, R) or bulky (F, Y, W) sidechains were the most deleterious (Fig. 5a and 5b). By constrast, tryptophan and glycine replacements at the M532 and I600 control positions had no deleterious effect on chemotaxis performance (Fig. S1a).
Fig. 5. Mutational survey of CheA.P5 residues at the receptor contact cleft.
(a) Functional phenotypes of CheA.P5 mutants. Mutant pPM25 plasmids were tested in strain UU1607 (ΔcheAW) for ability to support chemotaxis on tryptone soft agar plates (see Materials and Methods). The left-hand column lists the one-letter designations for amino acid replacements at each of the targeted residues. The size and shading of the circles indicate the chemotactic performance of each mutant: >60% of wild-type colony size with a ring of chemotactic cells at the colony edge (small, white); 40–60% of wild-type colony size with a chemotactic ring (medium, gray); <40% of wild-type colony size with no obvious ring of chemotactic cells (large, black).
(b) Examples of mutant CheA phenotypes on tryptone soft agar plates.
CheA.P5* proteins that failed to support good chemotaxis in soft agar tests nevertheless had wild-type steady-state cellular levels (Tables S1–S5), indicating that their functional defects were not due to poor expression or intracellular instability. We conclude that the mutant CheA proteins fold normally and have near-native in vivo structures. The P5* proteins also assembled core complexes and receptor clusters with wild-type efficiency (Tables S1–S5), indicating that their defects impair signaling functions rather than disrupt assembly or alter structure of the chemoreceptor array.
In vivo signaling properties of CheA.P5* mutants
We characterized the P5* mutants with in vivo CheA kinase assays based on Förster resonance energy transfer (FRET) between CFP-tagged CheZ (the FRET donor) and YFP-tagged CheY (the FRET acceptor) [22,23]. The CheZ phosphatase has low binding affinity for CheY, but high affinity for P-CheY, so at steady-state, their FRET interaction reflects CheA activity, the rate-limiting step in P-CheY production. The dose-response relationship of kinase activity to applied attractant stimuli yields three signaling parameters: K1/2, the attractant concentration that inhibits 50% of the CheA activity; the Hill coefficient, a measure of response cooperativity; and the overall level of receptor-stimulated kinase activity.
Altogether, we surveyed the FRET responses of 73 plasmid-borne P5* mutants in the Δ(cheAW) strain UU2784, which expresses wild-type Tsr as its only receptor and also lacks the CheR and CheB sensory adaptation enzymes: 17 L528 mutants (Table S1); 18 V531 mutants (Table S2); 11 S534 mutants (Table S3); 10 I581 mutants (Table S4); and 17 L599 mutants (Table S5). In UU2784, the Tsr subunits retain a [QEQEE] residue pattern at their five adaptation sites, with Q residues mimicking the kinase-on signaling properties of methylated E residues. In the adaptation-deficient host, the parental pPM25 plasmid produced a K1/2 response to serine of 17.4 ± 0.8 µM, with high cooperativity (Hill coefficient = 18 ± 3). (These averages and standard errors, derived from 6 independent experiments, typify the variation in parameter values from our FRET experiments [21,24,25,26,27,28]). Consistent with their wild-type chemotactic behavior (Fig. S1a), the CheA-M532G, -M532W, -I600G, and -I600W control mutants displayed response sensitivities and cooperativities closely comparable to those of wild-type CheA in strain UU2784 (Fig. S1b).
The signaling properties of P5* mutants in UU2784 were variable: Their K1/2 values ranged from 0.4× (L599Y) to 3.6× (V531D) of the wild type value; Hill coefficients ranged from 0.2× (S534W) to 1.3× (L528K) of the wild type value; and kinase activities ranged from 0.1× (S534R) to 1.2× (V531T) of the wild type value (Tables S1–S5). However, we detected no systematic differences between the UU2784 response parameters of P5* proteins that supported good chemotaxis in soft agar tests and those that supported poor performance (Fig. S2).
Conceivably, the high kinase activity of Tsr [QEQEE] core complexes in strain UU2784 might mask intrinsic signaling deficiencies in some P5* mutants. Accordingly, we also measured FRET response parameters for s subset of P5* mutants in the Δ(cheAW) strain UU2794, which expresses wild-type Tsr as its only receptor and contains the CheR and CheB sensory adaptation enzymes. This adaptation-proficient host, in which Tsr molecules average about one Q or methylated E site per subunit, closely resembles the Δ(cheAW) (cheRB+) UU1607 strain in which we assessed the chemotaxis performance of P5* mutants. The kinase activity of wild-type CheA signaling complexes in strain UU2794 was one-third of its UU2784 level (Tables S1–S5), resulting in lower response cooperativity (Hill coefficient = 2.6 ± 0.2), but much higher response sensitivity (K1/2 = 0.58 ± 0.04 µM; 8 independent experiments) than in the adaptation-deficient UU2784 host.
We chose 40 P5* mutants for FRET assays in UU2794 (CheRB+). Their signaling properties were also variable in this host, with K1/2 values ranging from 0.3× (I581E) to 2.5× (S534K) of wild type, Hill coefficients ranging from 0.3× (V531S) to 1.4× (L528Q) of wild type, and kinase activities ranging from 0.3× (L528G) to 1.2× (L599H) of wild type (Tables S1–S5). We noted no correlation between mutant response sensitivity in UU2794 and chemotaxis performance (Fig. S2), but both response cooperativity and kinase activity in UU2794 were generally lower in poorly chemotactic P5* mutants than in those with better chemotactic ability (Fig. S2). Thus, reduced kinase activity and/or lower response cooperativity may be responsible for the chemotaxis deficiencies of P5* mutants, although changes in sensory adaptation ability, not examined in the present study, could also be a contributing factor.
Sidechain tolerances of P5 cleft residues
Amino acid replacements at P5 residues whose sidechains contribute directly to a receptor binding interaction - for example, through hydrophobic or polar effects - should reduce interaction affinity. Moreover, bulky sidechain replacements at such P5 residues could also weaken the receptor binding interaction through steric clashes. Based on mutant functionality in an adaptation-proficient host, none of the surveyed P5 residue sidechains (L528, V531, S534, I581, L599) was individually essential for chemotaxis (Fig. 5a). The P5 residue most sensitive to sidechain character was S534. In an X-ray structure of the T. maritima P5-receptor complex, the residue at the corresponding S534 position makes a mainchain hydrogen bond to a receptor residue (corresponding to N381 in Tsr) [13]. This P5 position tolerated A, D, N, T, C, and G replacements (Fig. 5a), whereas sidechains with larger volume than the native serine (V, I, L, M, F, Y, W, H, R, K, E, Q) or restricted backbone motion (P) impaired chemotactic ability.
To better evaluate the sidechain contributions of the P5 cleft residues to core complex function, we compared the signaling properties of alanine and tryptophan replacement mutants at each position in UU2784, the adaptation-deficient FRET tester strain (Fig. 6). Tryptophan replacements at three positions (L528, S534, L599) shifted signal output toward the kinase-off state, reduced overall kinase activity, and drastically lowered response cooperativity. Alanine replacements at the same residues had little effect on these signaling parameters (Fig. 6; Tables S1, S3, S5). Thus, L528 and L599, like S534, lie at the contact interface with the receptor, but each individually is not critical to that interaction. Both A and W replacements at V531 lowered kinase activity, but had relatively modest effects on response cooperativity and sensitivity (Fig. 6; Table S2). Thus, the V531 sidechain seems to be primarily responsible for attaining a native level of kinase activity, at least with a homogeneous population of [QEQEE] receptor molecules. Whether the reduced kinase activity of the V531 mutants is due to an impaired receptor interaction is not yet clear. Finally, the A and W replacements at I581 had little effect on kinase activity or response cooperativity, with relatively modest changes in response sensitivity (Fig. 6; Table S4). In conclusion, the signaling defects of single-replacement P5* mutants might arise from impaired receptor interactions, but if so, the P5 cleft residues are not individually critical to that interaction.
Fig. 6. Serine dose-response behaviors of CheA.P5 mutants in adaptation-deficient cells.
Mutant pPM25 derivatives expressing alanine and tryptophan (A and W) replacements at P5 cleft residues were tested in the FRET reporter strain UU2784, which lacks the CheR and CheB sensory adaptation enzymes and expresses wild-type Tsr in the [QEQEE] modification state as its sole chemoreceptor. Shown are fits of the FRET dose-response data to a multi-site Hill equation, extrapolated to the X-axis limits for each plot. The dashed line shows the response curve for wild-type CheA with data points omitted to facilitate comparison to the mutant responses. The maximum kinase activity for each mutant, defined by a saturating serine stimulus (see Materials and Methods) was normalized to that for the wild-type control. Mean K1/2, Hill and relative kinase activity values ± standard errors in these experiments are listed in Tables S1–S5.
The four principal P5 cleft residues (L528, V531, I581, L599), all hydrophobic in character, nevertheless tolerate replacement with serine (Fig. 5a). If each native sidechain contributes to the overall receptor binding affinity, multiple serine replacements might produce a synergistic decline in chemotaxis performance. Accordingly, we constructed plasmid pPM25 derivatives encoding all possible combinations of double and triple serine replacements at these four residues. Serine was chosen for mutant replacements because single-serine mutants had almost wild-type chemotactic ability (Fig. 5 and Tables S1, S2, S4 and S5) and signaling properties (Fig. 7 and Tables S1, S2, S4 and S5). Multiple serine mutants were screened for chemotaxis in soft-agar plates in strain UU1607 (ΔcheAW). The double and triple serine mutants had near wild-type behavior, except for the V531S/L599S double mutant and triple mutants containing the V531S/L599S combination, which could not promote chemotaxis (Fig. S3 and Table S6). Although the V531S/L599S double mutant protein expressed at wild type levels, it probably contains a misfolded P5 domain because it failed to make ternary signaling complexes (Table S6). As expected, the chemotaxis-proficient triple serine-mutants L528S/I581S/L599S and L528S/V531S/I581S responded to serine in FRET experiments with ΔcheRB host cells (Fig. 7). However, their responses were compromised, especially that of the L528S/V531S/I581S mutant. Because CheA.P5 mutants with multiple serine replacements at cleft residues can still promote stimulus responses, we conclude that a stereo-specific P5-receptor binding interaction is not essential for kinase activation or control.
Fig. 7. Dose-response behaviors of P5* serine-replacement mutants.
Plasmid pPM25 derivatives were tested in the FRET reporter strain UU2784 (see Materials and Methods and the legend of Fig. 4). Dose-response data were fitted to a multi-site Hill equation, extrapolated to the X-axis limits for each plot. The dashed line shows the response curve for wild-type CheA with data points omitted to facilitate comparison to the mutant responses. The maximum kinase activity for each mutant, defined by a saturating serine stimulus (see Materials and Methods) was normalized to that for the wild-type control. Mean K1/2, Hill and relative kinase activity values ± standard errors in these experiments are listed in Tables S1 (L528S), S2 (V531S), S4 (I581S), S5 (L599S) and S6 (multiple S replacements).
upper panel: single replacement mutants;
lower panel: two triple replacement mutants that retain chemotactic ability (see Fig. S3).
Suppression analysis of P5 cleft mutants
Given that the CheA.P5 domain does not seem to promote a strong, CheW-independent binding interaction with the receptor, what is the functional basis for the impaired signaling properties of some P5 cleft mutants? To address this question, we first asked whether a structural change in the receptor could alleviate a P5* chemotaxis defect. The hunt for P5* suppressors in Tsr was unsuccessful (see Materials and Methods), suggesting that single-residue alterations of the receptor’s P5 contact surface cannot restore proper signaling behavior to CheA.P5* mutants.
Subsequently, we searched for second-site mutations in cheW or elsewhere in the cheA gene that could improve the impaired chemotactic performance of P5* mutants L528W, I581W, L599W, and S534R. We searched for suppressor mutations in plasmid pPM25 P5* derivatives by two different methods, one involved all-codon mutagenesis of targeted P5 cleft residue codons, the other involved random mutagenesis of the entire plasmid. In both cases, the mutagenized pPM25-P5* plasmids were transformed into strain UU1607 (ΔcheAW) and pseudorevertants with improved chemotactic ability were selected from pooled transformants on tryptone soft agar.
The all-codon approach yielded one or more suppressors for each starting P5* mutant (Fig. 8). Because the mutagenized plasmid pools contained all possible codon changes at the targeted sites, we conclude that P5 groove lesions are compensated only by a specific amino acid replacement at another cleft position (Fig 9). The P5-S534R mutant yielded a variety of suppressors at L599 and I581, all but one of them (I581G) introducing a polar sidechain replacement. We conclude that functional suppression of the S534R lesion may require dislodging the arginine sidechain from the P5 cleft through a hydrogen-bonding interaction with a polar sidechain at residue 581 or 599. Consistent with this view, the random mutagenesis approach yielded no S534R suppressors elsewhere in the P5 domain. In contrast, random mutagenesis of the L528W, I581W and L599W P5* mutants yielded suppressors at surface-exposed residues lying at P5-CheW interface 1 (Fig. 8). No suppressors were found in CheW or elsewhere in CheA, implying that P5 cleft lesions mainly perturb P5 interactions with CheW that take place at interface 1.
Fig. 8. Second-site suppressors of CheA.P5* lesions at the receptor interface cleft.
Second-site revertants were isolated from four P5* cleft mutants (labels inside ovals). The gray shade (L528W, I581W, L599) or blue color (S534R) of the parental mutants matches the single-letter labels for the amino acid replacements in their suppressors. The wild-type residues at the suppressor sites in P5 subdomain 1 are shown in space-fill mode in gray shades keyed to the parental mutant from which they arose. The two P5 subdomains and their corresponding cleft residues are shown in different colors.
Fig. 9. Dose-response behaviors of CheA.P5* pseudorevertants.
Mutant derivatives of plasmid pPM25 were tested in the FRET reporter strain UU2794, which contains the CheR and CheB sensory adaptation enzymes and expresses wild-type Tsr as its sole chemoreceptor. Dose-response data were fitted to a multi-site Hill equation, extrapolated to the X-axis limits for each plot. Data points for the wild-type response curves (dashed lines) were omitted to facilitate comparisons. The maximum kinase activity for each mutant, defined by a saturating serine stimulus (see Materials and Methods) was normalized to that for the wild-type control. Mean K1/2, Hill and relative kinase activity values ± standard errors in these experiments are listed in Table S7.
upper panel: the doubly mutant revertant (I581W/I600T) and its two component mutants;
lower panel: the doubly mutant revertant (I581W/V605A) and its two component mutants.
Suppressor mutants alone and in combination with their parental P5 defect were analyzed by in vivo FRET assays in strain UU2794 (cheRB+). P5 cleft mutants L528W, I581W and L599W showed low kinase activity values, consistent with the trend noted previously (Fig. S2). However, the suppressors invariably improved or fully restored kinase activity to the P5* mutants (Table S7). Two suppression patterns were evident: In one, the suppressor alone had normal kinase activity (e.g., I600T in Fig. 9); in the other, the suppressor alone exhibited impaired kinase activity, but improved kinase activity in combination with a P5* cleft lesion (e.g., V605A in Fig. 9).
Discussion
Role of P5 cleft residues in chemotaxis and signaling
Our results indicate that residues lying at the groove formed by CheA.P5 subdomains 1 and 2 influence the signaling properties of receptor arrays and chemotaxis performance in soft agar. All-codon mutational analyses of P5 groove residues revealed that a variety of amino acid replacements at L528, V531, S534, I581 and L599 reduced both kinase activity and chemotactic ability. In cells containing receptors plus CheW, CheA.P5 cleft mutants formed polar clusters, consistent with a previous in vitro study showing that P5 cleft lesions reduced kinase activity, but did not impair incorporation of the mutant CheA protein into receptor arrays [14]. Thus, P5 cleft residues play a role in kinase activity and/or signal transmission, but not in core complex assembly.
Several lines of evidence indicate that the CheA.P5 domain does not bind directly to receptors with an affinity comparable to the binding interaction between receptors and CheW, a P5 paralog: In vivo crosslinking between CheA and Tsr was strictly dependent on the presence of CheW, as was recruitment of CheA or the CheA.P5 domain to cellular receptor clusters. In contrast, a variety of experimental approaches have demonstrated that CheW binds to receptors in the absence of CheA [10,16,18,29,30,31]. Amino acid replacements at CheW residue positions that correspond to some of the P5 groove residues discussed in this report disrupt the CheW-receptor interaction [10,18,30].
The substantial differences in the receptor interaction properties of CheW and CheA.P5 imply that the signaling defects of P5 groove mutants do not arise from disruption of a direct, CheW-independent, P5-receptor binding interaction. Many P5 cleft mutants tolerated single amino acid replacements with only modest declines in chemotactic ability, indicating that none of the P5 residues that contact the receptor is a critical functional determinant. Moreover, CheA proteins with multiple P5 cleft alterations (e.g., three serine replacements) also supported chemotaxis. Because the P5-receptor contacts seen in ternary core complexes can tolerate a variety of P5 sidechains with little detriment to chemotaxis performance, we conclude that the P5 and receptor contact surfaces do not require a highly stereospecific match between interacting residues.
Tryptophan replacements at the P5 cleft residues produced the most severe functional defects, while still allowing normal core complex assembly. Conceivably, a very bulky sidechain could impair function by distorting the P5-receptor contact surface, but receptor-dependent steric clashes cannot account for the pseudoreversion patterns of such P5* mutants. We found that specific amino acid replacements at other P5 groove or subdomain 1 residues restored chemotaxis and kinase activity to defective P5 cleft tryptophan-replacement mutants. Our suppression study suggests that P5 lesions affect primarily P5 cleft structure and/or dynamic motions that impact the P5-CheW interface 1 interaction. A complete disruption of interface 1 affects both CheA kinase activity and array binding [8], whereas P5 cleft mutants form arrays normally. We conclude that their low kinase activity arises mainly from altered conformational interactions between CheW and the CheA.P5 domain at interface 1 and not directly from their structural changes at the P5-receptor interaction surface.
A new model for core complex signaling
The CheA.P5 domain and CheW both share the same overall protein fold and receptor contact surface, suggesting that kinase activation and control could be mediated by both P5-receptor and CheW-receptor contacts. Here, we demonstrated that stereospecific P5-receptor interactions are not critical for kinase activation and control. We propose, therefore, that the receptor-CheW interface is the principal route for transmission of signaling-related conformational changes between receptors and the CheA kinase (Fig. 10). This view is consistent with the previously documented essential role of CheW in receptor-mediated control of CheA [7,29,32,33,34] and a recent study indicating that control may occur through only one receptor dimer in each trimer of dimers [35]. Within the core complex, CheW signals probably modulate kinase activity through the interface 1 connection to the CheA.P5 domain [8] (Fig. 10). Conformational changes in subdomain 1 of P5 might in turn influence CheA activity through the linker connecting the P5 domain to the P4 ATP-binding domain [36]. Those signals might, for example, promote CheA dynamic motions needed for productive interaction between its substrate (P1) and catalytic (P4) domains [37]. Additionally, receptor conformational signals might travel through the P3/P3' dimerization domain of CheA [35], which also adjoins P4 through a critical linker segment [38].
Fig. 10. Proposed transmission routes of signaling-related conformational changes in chemoreceptor core complexes.
A top-down cross-section view of a core signaling complex is shown (see Fig. 1). The lower half of the cartoon shows P5-CheW interface 1 (black circle) and the interfaces between receptor dimers and CheW (black rectangle) and the CheA.P5 domain (gray rectangle). The upper half of the cartoon indicates a possible route for receptor conformational signals (large arrowhead) that control CheA activity. P5 conformational changes probably influence CheA activity through the P4–P5 linker (small dark gray arrow). In addition, receptor signals may reach the P4 domain through the P3–P4 linker (small light gray arrow). Our study shows that receptors do not modulate CheA activity through their direct contacts with the P5 domain. See text for further explanation.
CheW conformational signals propagate to neighboring core complexes through interface 2 connections [21]. The CheW-P5 interface 1 connection might also transmit conformational changes from P5 subdomain 1 through groove residues to subdomain 2, thereby influencing P5 interface 2 and array cooperativity (Fig. 10). Indeed, we noted that the majority of P5 groove mutants have reduced response cooperativity, implying fewer interface 2 connections (Fig. S2 and Tables S1–S5).
In summary, P5 cleft residues are important for CheA kinase activation, but not through direct binding contacts with the receptor, as previously proposed [14,15]. Instead, P5 groove residues ensure correct cleft structure and dynamics necessary for proper interface 1 interactions with CheW that lead to P5 conformational changes important for kinase activation and for transmission of stimulus signals to other core complexes through array interface 2.
Materials and Methods
Bacterial strains
All strains used in the study were derivatives of E. coli K-12 strain RP437 [39]. Their relevant genotypes were: UU1607 [Δ(cheA-cheW)2167] [21]; UU2682 [Δ(cheA-cheW-tar-tap) 4530 tsrΔ5547 aerΔ1 trgΔ4543]; UU2784 [Δ(cheA-cheW-tar-tap-cheR-cheB-cheY-cheZ)1214 aerΔ1 trgΔ4543] [21]; UU2794 [Δ(cheA-tar-tap)4530 Δ(cheY-cheZ)1214 aerΔ1 trgΔ4543]; UU1935 [mutD5 Δ(flhD-flhA)4 tsrΔ7028 trgΔ100] [40]; UU2806 [Δ(cheA-cheW-tar-tap-cheR-cheB-cheY-cheZ)1214 tsrΔ5547 aerΔ1 trgΔ4543] [21]; RP9535 [cheAΔ1643] [7].
Plasmids
Plasmids were: pKG116, a derivative of pACYC184 [41] that confers chloramphenicol resistance and gene expression inducible by sodium salicylate [42]; pPM25, a pKG116 derivative that co-expresses CheA and CheW proteins [21]; pGP7, a pKG116 derivative that expresses a GFP-P5 fusion; pGP8, a pKG116 derivative that expresses a P5-GFP fusion; pGP12, a pKG116 derivative expressing a GFP-CheA sandwich fusion in which GFP replaces the P2 domain; pGP68; a pGP12 derivative in which the CheA.P5 domain is deleted; pPA114, a pKG116 derivative that expresses Tsr; pRR48, a derivative of pBR322 [43] that confers ampicillin resistance and drives protein expression from an IPTG-inducible tac promoter with an ideal (perfectly palindromic) lacI operator that is under tight control of a plasmid-encoded LacI repressor [44]; pPA789, a pRR48 derivative that expresses a CFP-CheZ protein fusion [45]; pPA802, a pRR48 derivative that expresses a YFP-CheW protein fusion; pRR53, a pRR48 derivative that expresses Tsr [46]; pGP32, a pRR48 derivative that co-expresses a cysteine-less HA-tagged CheA and CheW; pGP34, a pRR48 derivative that expresses a cysteine-less HA-tagged CheA; pGP3, a derivative of pGFPuv (Clontech) expressing a GFP-P5 fusion; and pVS88, a derivative of pTrc99A that expresses CheY-YFP and CheZ-CFP under IPTG-inducible control [22].
Site-directed mutagenesis
Mutations in the cheA and tsr genes were generated by subjecting the corresponding plasmids to Quickchange PCR mutagenesis (Agilent). The presence of the desired mutation was confirmed by sequencing the entire targeted coding region.
Chemotaxis assays
Strain UU1607 transformed with pPM25 and its mutant derivatives was assessed for chemotactic ability on tryptone semi-solid agar plates (10 g/L tryptone, 5 g/L NaCl, 2,5 g/L agar) supplemented with 12.5 µg/ml chloramphenicol and 0.6 µM sodium salicylate as previously described [47]. Plates were incubated at 30°C for 7 h before assessing colony size and morphology.
Expression levels of mutant CheA proteins
Strain UU1607 harboring pPM25 and its mutant derivatives was diluted 100-fold from an overnight culture in fresh tryptone broth (10 g/L tryptone, 5 g/L NaCl) supplemented with 12.5 µg/ml chloramphenicol and 0.6 µM sodium salicylate and grown at 30°C for 6 hours. A culture aliquot of 1 ml was withdrawn, centrifuged and cells were resuspended and lysed in 100 µl of 1× Laemmli sample buffer [48]. CheA protein expression levels were determined by western blotting with a polyclonal rabbit CheA antiserum and normalized to a wild-type control.
Crosslinking Assays
Strain UU2806 co-transformed with cysteine-reporter derivatives of pPA114 and pGP32 (or pGP34) was grown at 30°C with aeration in tryptone broth supplemented with 12.5 µg/ml chloramphenicol, 50 µg/ml ampicillin, 0.6 µM sodium salicylate and 75 µM IPTG. At OD600nm ~0.5, a 1.5-ml aliquot of the culture was withdrawn, washed twice with 1 ml of PBS, finally resuspended in 1 ml of PBS and treated with 300 µM Cu2+-phenanthroline for 10 min at 35°C to induce disulfide formation, as previously described [21]. Whole cell lysates were separated by SDS/PAGE and CheA-containing species were detected by immunoblotting with a polyclonal anti-HA antibody (Pierce) and a Cy5-labeled secondary antibody (Invitrogen). Western blots were imaged under fluorescence mode in a Typhoon 9500 scanner (GE Healthcare), and bands were quantified with ImageQuant software (GE Healthcare).
Clustering assays
Localization studies using GFP-P5, P5-GFP, GFP-CheA, GFP-CheAΔP5, and YFP-CheW protein fusions were done in strains UU2806, UU1607 and RP9535, transformed with the corresponding plasmids. Cells bearing pKG116 derivatives pGP7 (GFP-P5), pGP8 (P5-GFP), pGP12 (GFP-CheA) or pGP68 (GFP-CheAΔP5) were grown in tryptone broth supplemented with 12.5 µg/ml chloramphenicol, 0.6 µM sodium salicylate and incubated at 30°C for 6 hours before imaging. Cells transformed with pGP3 or pPA802 were cultured in the same way but the medium was supplemented with 50 µg/ml ampicillin and 10 µM (for pGP3) or 150 µM IPTG (for pPA802),
Ternary complex formation by CheA-P5 mutants was assessed by studying the cellular localization of a CFP-CheZ reporter, which binds CheAS, an alternate cheA translation product [49], as described [50]. Strain UU2784 transformed with pPM25 (or pPM25-P5* derivatives) and pPA789 was diluted 100-fold from an overnight culture in fresh tryptone broth supplemented with 12.5 µg/ml chloramphenicol, 50 µg/ml ampicillin, 200 µM IPTG and 0.6 µM sodium salicylate and incubated at 30°C for 6 hours. In all cases, cells were analyzed by fluorescence microscopy as previously described [44].
In vivo FRET-based CheA kinase assays
The in vivo FRET-based kinase assay measures CheA activity-dependent interactions between CFP-tagged CheZ and YFP-tagged CheY, which are enhanced by CheA-dependent CheY phosphorylation. Cell preparation, flow cell assembly, stimulus protocol, FRET instrumentation and data analysis followed the method described by Sourjik, et al. [23] with minor modifications in our laboratory [25]. Strain UU2784 or UU2794 expressing the FRET protein pair (CheY-YFP and CheZ-CFP) from plasmid pVS88 and CheA variants from plasmid pPM25 were grown overnight at 37°C in tryptone broth containing 100 µg/ml ampicillin and 25 µg/ml chloramphenicol. Starter cultures were diluted 100-fold into fresh tryptone broth supplemented with 12.5 µg/ml chloramphenicol, 50 µg/ml ampicillin, 50 µM IPTG and 0.6 µM sodium salicylate and incubated at 30°C for 6 hours to mid-exponential phase (OD600nm = 0.45–0.55). Cells were washed with motility buffer (10 mM potassium phosphate, 10 mM sodium lactate, 0.1 mM potassium EDTA, 100 µM methionine, pH 7.0), attached to a round polylysine-coated coverslip, mounted in a flow cell and subjected to sequential addition and removal of serine diluted in motility buffer. The attached cells and all solutions were kept at 30°C throughout the experiment. The cell sample was excited at CFP wavelength and epifluorescent light emission from CFP (FRET donor) and yellow fluorescent protein (YFP; FRET acceptor) was measured by photon-counting photomultipliers. The ratio of YFP to CFP photon counts reports on CheA kinase activity, and changes in response to serine stimuli. A dose-response curve obtained by plotting the fractional changes in kinase activity versus applied serine concentrations was fitted to a multisite Hill equation (1 − [Ser]H/([Ser]H + K1/2H), where K1/2 is the attractant concentration that inhibits 50% of the kinase activity, and H, the Hill coefficient, reflects the extent of cooperativity of the response. Maximal relative kinase activity was calculated from the YFP/CFP ratio drop after exposing cells to a saturating dose of serine (100 µM) [23].
Isolation of P5* pseudorevertants
Three strategies were followed to identify second-site mutations that suppressed the chemotaxis defects of P5* mutants L528W, S534R, I581W and L599W. In the first, pPM25 derivatives encoding these CheA variants were mutagenized by passage through UU1935, a proofreading-deficient polymerase mutant strain [43]. Plasmids were purified from this strain and electroporated into UU1607 cells. Transformed cells were seeded in the middle of a tryptone soft-agar plate supplemented with 12.5 µg/ml chloramphenicol and 0.6 µM sodium salicylate. Plates were incubated overnight at 30°C to allow the appearance of chemotactic revertants. pPM25 mutant plasmids were purified from revertant colonies, sequenced and used to transform UU1607 cells for re-testing their suppressor properties in tryptone soft-agar plates. In the second strategy, pPM25 derivatives were subjected to targeted all-codon mutagenesis: for the L528W mutant the targeted residues were V531, I581 and L599; for I581W, the targeted residues were L528, V531 and L599; for L599W, the targeted residues were L528, V531 and I581; and for S534R the targeted residues were I581 and L599. The isolation of revertants and identification of suppressor mutations were done as described above.
To search for suppressor changes in Tsr, plasmid pRR53 was passed through UU1935 and introduced by electroporation into strain UU2682 bearing pPM25-L528W, -S534R, -I581W or -L599W. Pooled transformants were placed on tryptone soft-agar plates containing 150 µM IPTG, 50 µg/ml ampicillin, 12.5 µg/ml chloramphenicol and 0.6 µM sodium salicylate. The plates were incubated overnight at 30°C.
Protein modeling and structural display
The P5-Tsr complex was modeled based on the structure (PDB code 4JPB) for the Thermotoga maritima ternary complex between receptor fragment, CheA (P3-P4-P5 domains) and CheW [13]. The model was built according to the following steps: (i) homology modeling by the Phyre2 server [51] of the E. coli CheA.P5 domain, using as template the coordinates of the Thermotoga CheA.P5 domain extracted from PBD structure1B3Q; and (ii) superimposition of the crystal structure of E. coli serine receptor dimer (1QU7) and the modeled P5 domain onto Thermotoga structure (4JPB). Structure images were prepared with MacPymol.
Supplementary Material
Highlights.
-
▪
The P5 domain of kinase CheA plays essential roles in chemoreceptor signaling
-
▪
In signaling complexes receptor residues touch a cleft between CheA.P5 subdomains
-
▪
Amino acid replacements at key P5 cleft residues do not destroy chemotactic ability
-
▪
Receptors do not control CheA activity directly through contacts to the P5 domain
-
▪
Receptors control CheA by manipulating CheW and its interface 1 connection to P5
Acknowledgments
We thank three anonymous reviewers for constructive suggestions that improved the clarity of the manuscript. This work was supported by U.S. Public Health Service research grant GM19559 from the National Institute of General Medical Sciences. The Protein-DNA Core Facility at the University of Utah receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute.
Abbreviations
- IPTG
isopropyl-β-D-thiogalactopyranoside
- FRET
Förster resonance energy transfer
- HA
hemagglutinin antigen
- HAMP
histidine kinases, adenylate cyclases, methyl-accepting proteins, and phosphatases
- MCP
methyl-accepting chemotaxis protein
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- PBS
phosphate buffered saline
- PDB
protein data bank
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends. Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkinson JS, Hazelbauer GL, Falke JJ. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015;23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc. Natl. Acad. Sci. USA. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. USA. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc. Natl. Acad. Sci. USA. 2012;109:E1481–1488. doi: 10.1073/pnas.1200781109. http://dx.doi.org/10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassidy CK, Himes BA, Alvarez FJ, Ma J, Zhao G, Perilla JR, Schulten K, Zhang P. CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling. eLife. 2015;4 doi: 10.7554/eLife.08419. http://dx.doi.org/10.7554/eLife.08419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Parkinson JS. Mutational analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J. Bacteriol. 2006;188:3299–3307. doi: 10.1128/JB.188.9.3299-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natale AM, Duplantis JL, Piasta KN, Falke JJ. Structure, function, and on-off switching of a core unit contact between CheA kinase and CheW adaptor protein in the bacterial chemosensory array: A disulfide mapping and mutagenesis study. Biochemistry. 2013;52:7753–7765. doi: 10.1021/bi401159k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boukhvalova M, VanBruggen R, Stewart RC. CheA kinase and chemoreceptor interaction surfaces on CheW. J. Biol. Chem. 2002;277:23596–23603. doi: 10.1074/jbc.M202288200. [DOI] [PubMed] [Google Scholar]
- 10.Boukhvalova MS, Dahlquist FW, Stewart RC. CheW binding interactions with CheA and Tar: Importance for chemotaxis signaling in Escherichia coli. J. Biol. Chem. 2002;277:22251–22259. doi: 10.1074/jbc.M110908200. [DOI] [PubMed] [Google Scholar]
- 11.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 12.Griswold IJ, Zhou H, Matison M, Swanson RV, McIntosh LP, Simon MI, Dahlquist FW. The solution structure and interactions of CheW from Thermotoga maritima. Nat Struct Biol. 2002;9:121–125. doi: 10.1038/nsb753. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Fleetwood AD, Bayas C, Bilwes AM, Ortega DR, Falke JJ, Zhulin IB, Crane BR. The 3.2 A resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays. Biochemistry. 2013;52:3852–3865. doi: 10.1021/bi400383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piasta KN, Ulliman CJ, Slivka PF, Crane BR, Falke JJ. Defining a key receptor-CheA kinase contact and elucidating its function in the membrane-bound bacterial chemosensory array: a disulfide mapping and TAM-IDS Study. Biochemistry. 2013;52:3866–3880. doi: 10.1021/bi400385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Vu A, Lee K, Dahlquist FW. CheA-receptor interaction sites in bacterial chemotaxis. J. Mol. Biol. 2012;422:282–290. doi: 10.1016/j.jmb.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vu A, Wang X, Zhou H, Dahlquist FW. The receptor-CheW binding interface in bacterial chemotaxis. J. Mol. Biol. 2012;415:759–767. doi: 10.1016/j.jmb.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller AS, Kohout SC, Gilman KA, Falke JJ. CheA kinase of bacterial chemotaxis: chemical mapping of four essential docking sites. Biochemistry. 2006;45:8699–8711. doi: 10.1021/bi060580y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu JD, Parkinson JS. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J. Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, Parkinson JS. Cysteine-scanning analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J. Bacteriol. 2006;188:4321–4330. doi: 10.1128/JB.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kentner D, Thiem S, Hildenbeutel M, Sourjik V. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol. Microbiol. 2006;61:407–417. doi: 10.1111/j.1365-2958.2006.05250.x. [DOI] [PubMed] [Google Scholar]
- 21.Pinas GE, Frank V, Vaknin A, Parkinson JS. The source of high signal cooperativity in bacterial chemosensory arrays. Proc Natl Acad Sci USA. 2016;113:3335–3340. doi: 10.1073/pnas.1600216113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sourjik V, Vaknin A, Shimizu TS, Berg HC. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol. 2007;423:363–391. doi: 10.1016/S0076-6879(07)23017-4. [DOI] [PubMed] [Google Scholar]
- 24.Han XS, Parkinson JS. An unorthodox sensory adaptation site in the Escherichia coli serine chemoreceptor. J. Bacteriol. 2014;196:641–649. doi: 10.1128/JB.01164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai RZ, Parkinson JS. Functional suppression of HAMP domain signaling defects in the E. coli serine chemoreceptor. J. Mol. Biol. 2014;426:3642–3655. doi: 10.1016/j.jmb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitanovic S, Ames P, Parkinson JS. A Trigger Residue for Transmembrane Signaling in the Escherichia coli Serine Chemoreceptor. J. Bacteriol. 2015;197:2568–2579. doi: 10.1128/JB.00274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ames P, Hunter S, Parkinson JS. Evidence for a Helix-Clutch Mechanism of Transmembrane Signaling in a Bacterial Chemoreceptor. J. Mol. Biol. 2016;428:3776–3788. doi: 10.1016/j.jmb.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai RZ, Gosink KK, Parkinson JS. Signaling Consequences of Structural Lesions that Alter the Stability of Chemoreceptor Trimers of Dimers. J. Mol. Biol. 2017;429:823–835. doi: 10.1016/j.jmb.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gegner JA, Graham DR, Roth AF, Dahlquist FW. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 30.Cardozo MJ, Massazza DA, Parkinson JS, Studdert CA. Disruption of chemoreceptor signalling arrays by high levels of CheW, the receptor-kinase coupling protein. Mol. Microbiol. 2010;75:1171–1181. doi: 10.1111/j.1365-2958.2009.07032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedetta A, Parkinson JS, Studdert CA. Signalling-dependent interactions between the kinase-coupling protein CheW and chemoreceptors in living cells. Mol. Microbiol. 2014;93:1144–1155. doi: 10.1111/mmi.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu JD, Parkinson JS. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Levit M, Lurz R, Surette MG, Stock JB. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortega DR, Mo G, Lee K, Zhou H, Baudry J, Dahlquist FW, Zhulin IB. Conformational coupling between receptor and kinase binding sites through a conserved salt bridge in a signaling complex scaffold protein. PLoS Comput Biol. 2013;9:e1003337. doi: 10.1371/journal.pcbi.1003337. http://dx.doi.org/10.1371/journal.pcbi.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Hazelbauer GL. Selective allosteric coupling in core chemotaxis signaling complexes. Proc. Natl. Acad. Sci. USA. 2014;111:15940–15945. doi: 10.1073/pnas.1415184111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Wu C, Vu A, Shea JE, Dahlquist FW. Computational and experimental analyses reveal the essential roles of interdomain linkers in the biological function of chemotaxis histidine kinase CheA. J. Am. Chem. Soc. 2012;134:16107–16110. doi: 10.1021/ja3056694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briegel A, Ames P, Gumbart JC, Oikonomou CM, Parkinson JS, Jensen GJ. The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signalling state. Mol. Microbiol. 2013;89:831–841. doi: 10.1111/mmi.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Vallurupalli P, Vu A, Lee K, Sun S, Bai WJ, Wu C, Zhou H, Shea JE, Kay LE, Dahlquist FW. The linker between the dimerization and catalytic domains of the CheA histidine kinase propagates changes in structure and dynamics that are important for enzymatic activity. Biochemistry. 2014;53:855–861. doi: 10.1021/bi4012379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massazza DA, Parkinson JS, Studdert CA. Cross-linking evidence for motional constraints within chemoreceptor trimers of dimers. Biochemistry. 2011;50:820–827. doi: 10.1021/bi101483r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang ACY, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gosink KK, Buron-Barral M, Parkinson JS. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J. Bacteriol. 2006;188:3487–3493. doi: 10.1128/JB.188.10.3487-3493.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolivar F, Rodriguez R, Greene PJ, Betlach MC, Heyneker HL, Boyer HW. Construction and characterization of new cloning vehicles. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 44.Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ames P, Parkinson JS. Conformational suppression of inter-receptor signaling defects. Proc. Natl. Acad. Sci. USA. 2006;103:9292–9297. doi: 10.1073/pnas.0602135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkinson JS, Ames P, Studdert CA. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 2005;8:116–121. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Parkinson JS. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 1976;126:758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 49.Smith RA, Parkinson JS. Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1980;77:5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cantwell BJ, Draheim RR, Weart RB, Nguyen C, Stewart RC, Manson MD. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 2003;185:2354–2361. doi: 10.1128/JB.185.7.2354-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.