Abstract
Objective
Pancreatic ductal adenocarcinoma (PDAC) is a therapy recalcitrant disease with the worst survival rate of common solid tumours. Preclinical models that accurately reflect the genetic and biological diversity of PDAC will be important for delineating features of tumour biology and therapeutic vulnerabilities.
Design
27 primary PDAC tumours were employed for genetic analysis and development of tumour models. Tumour tissue was used for derivation of xenografts and cell lines. Exome sequencing was performed on the originating tumour and developed models. RNA sequencing, histological and functional analyses were employed to determine the relationship of the patient-derived models to clinical presentation of PDAC.
Results
The cohort employed captured the genetic diversity of PDAC. From most cases, both cell lines and xenograft models were developed. Exome sequencing confirmed preservation of the primary tumour mutations in developed cell lines, which remained stable with extended passaging. The level of genetic conservation in the cell lines was comparable to that observed with patient-derived xenograft (PDX) models. Unlike historically established PDAC cancer cell lines, patient-derived models recapitulated the histological architecture of the primary tumour and exhibited metastatic spread similar to that observed clinically. Detailed genetic analyses of tumours and derived models revealed features of ex vivo evolution and the clonal architecture of PDAC. Functional analysis was used to elucidate therapeutic vulnerabilities of relevance to treatment of PDAC.
Conclusions
These data illustrate that with the appropriate methods it is possible to develop cell lines that maintain genetic features of PDAC. Such models serve as important substrates for analysing the significance of genetic variants and create a unique biorepository of annotated cell lines and xenografts that were established simultaneously from same primary tumour. These models can be used to infer genetic and empirically determined therapeutic sensitivities that would be germane to the patient.
Keywords: PANCREATIC CANCER, CANCER GENETICS, ONCOGENES
Significance of this study.
What is already known on this subject?
Pancreatic cancer is genetically diverse.
Patient-derived xenografts are considered the most reliable avatar of individual patient tumour.
Pancreatic cancer cell lines are believed to poorly represent the tumour of origin.
Establishment of cell lines is associated with significant genetic drift.
What are the new findings?
Newly developed cancer cell lines harbour strong genetic conservation with pancreatic tumour of origin.
These cell lines maintain unique features relative to tumour biology.
Suites of models representative of genetic diversity of disease can be developed.
The models provide insights into processes of genetic evolution.
Models can be used to dissect genetic and patient-selective therapeutic vulnerabilities.
How might it impact on clinical practice in the foreseeable future?
The cell lines developed in this manuscript accurately recapitulate the genetic features of pancreatic cancer and can be used to functionally study genes as a basis for rational therapy development. Since these cell lines are amenable to high-throughput drug screening, they also provide a means to identify new therapeutic vulnerabilities at the individual patient level and provide a distinct model of precision therapy.
Background
Pancreatic ductal adenocarcinoma (PDAC) has the worst prognosis of any common solid tumours with a 5-year survival of approximately 6% and is expected to become the second leading cause of cancer deaths by 2030.1–3 Many patients present with advanced disease and even with complete resection long-term survival is poor, as tumours recur in the majority of patients.4 These features of PDAC have driven the need for effective systemic treatments to control disseminated disease. The approved systemic therapies have a relatively modest impact on survival and, in spite of extensive preclinical studies and numerous clinical trials, PDAC remains a therapy-recalcitrant disease.1 2 5 The treatment of PDAC is largely dependent on chemotherapy. In part, this is because the PDAC genetic landscape is dominated by currently non-actionable KRAS mutations and tumour suppressor losses.6–11 Therefore, there is a significant need for a better understanding of PDAC biology and clinically relevant treatment interventions for patients.1 6 12
The aetiology of PDAC is complex and associated with a myriad of factors including genetic predisposition, environmental exposure and chronic inflammation.13 14 These events shape the genetic landscape of the tumour from mutation burden and spectra to chromosomal instability.8 11 15 Transcriptional profiling has suggested that PDAC cases cluster into several specific subtypes.16–19 Sequencing analyses revealed a complex genetic architecture, where most PDAC cases have multiple alterations in a distinct spectrum of cancer-associated pathways including cell cycle, chromatin remodelling, WNT signalling, MYC, NOTCH signalling, Hedgehog signalling and DNA damage repair.8 10 11 This complexity has led to consideration of each tumour case as a unique disease entity.
A limited repertoire of existing PDAC models has been cited as one of the main barriers in developing effective treatments, and has driven significant interest in generating new models of disease. Of the approximately 40 established PDAC cell lines that are available, only 15 are commonly used to model PDAC.20 21 For the majority of established cell lines, there are no means to determine how reflective these cells are of the genetic features of the tumour from which they were derived, and the extent to which they have diverged after years in culture. Additionally, select PDAC molecular subtypes (eg, exocrine) are not represented among established cell lines.17 In recognition of these shortcomings, genetically engineered mouse models provide a clean canvas on which to evaluate the impact of specific genetic events on the aetiology and progression of PDAC.20 22 23 Notably, mutant KRAS mouse models are widely employed to model the aetiology, progression and treatment of pancreatic cancer.23–25 While likely representative of a subset of PDAC cases, these models likely do not reflect the genetic complexity and diversity of clinical PDAC cases with their myriad genetic alterations beyond KRAS. Patient-derived xenografts (PDX) and tumour organoids represent other models of PDAC.26–28 A number of these models have been recently developed, however, their stability over time and relationship to the primary tumour in many cases is marginally characterised.28–30
Here, a suite of patient-derived models was established and extensively characterised in relation to primary tumour genetics. These efforts demonstrated that low passage cell lines genetically recapitulate the primary tumour and maintain genetic features of PDAC in a fashion comparable to that observed in PDX models. They can also enable to evaluate tumour heterogeneity and evolution and afford the opportunity to explore the functional significance of specific genetic events or gene expression subtypes as observed in the primary tumour.
Results
Model derivation platform
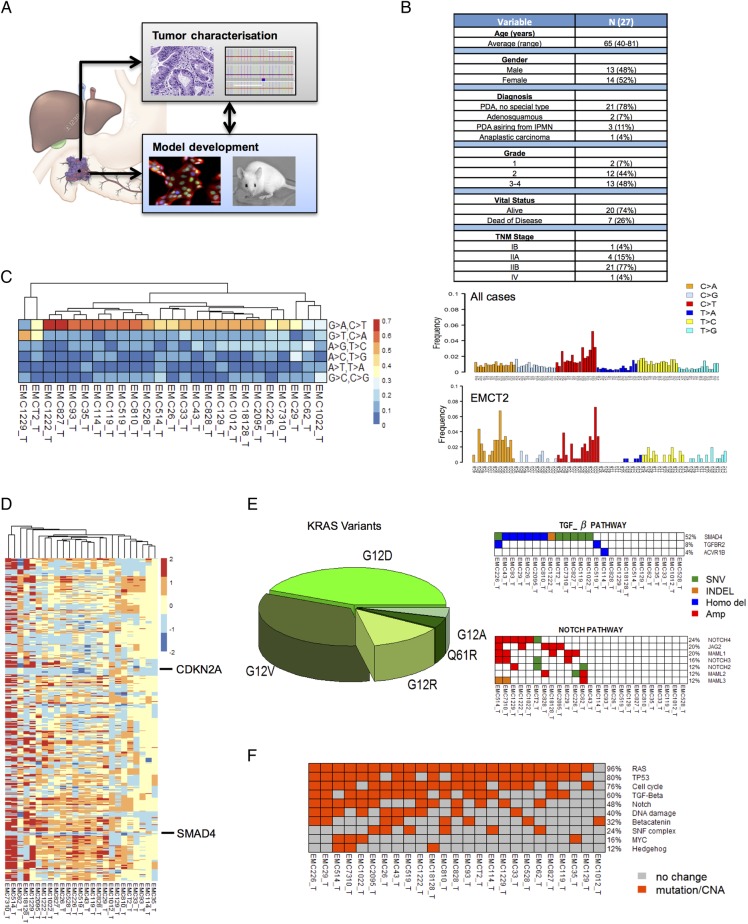
A pipeline was developed where patients with a diagnosis of PDAC were consented for the collection of tumour tissue, genetic studies and models development (figure 1A). Tumour tissue was triaged for genetic analysis by whole exome sequencing, as well as the development of cell lines and PDXs. Resultant models were characterised by exome and RNA sequencing. This pipeline was applied to a collection of 27 unselected PDAC cases (see figure 1B and online supplementary data). The majority of cases in the study represented PDAC of no special type; there were also two adenosquamous carcinomas, one anaplastic carcinoma and three invasive carcinomas arising from intraductal papillary mucinous neoplasm. A total of 54 PDX tumours and 56 cell lines derived from these cases were subjected to exome sequencing, with the majority of cell lines sequenced in both early and late passages.
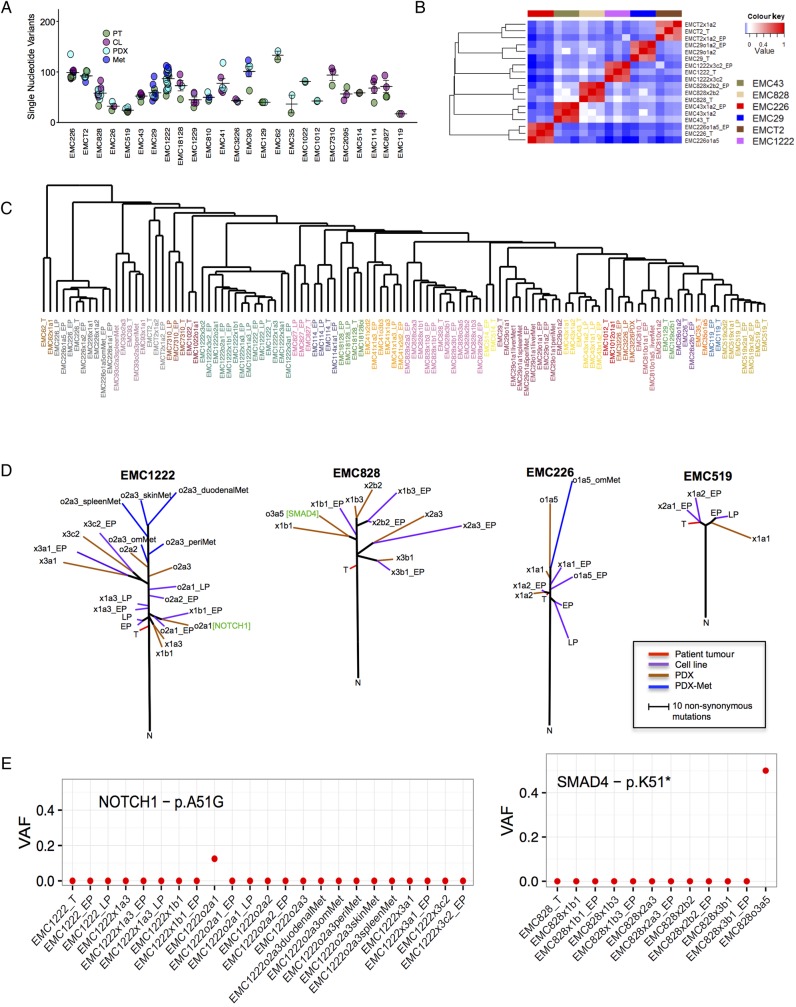
Figure 1.
Genetic analysis of the pancreatic ductal adenocarcinoma (PDAC) tumour specimens. (A) Schematic of the overall pipeline employed. Tumour tissue not required for diagnosis was used for characterisation of the patient tumour. Parallel tissue was employed for development of patient-derived xenograft (PDX) and cell line models. Models were used to evaluate drug sensitivities that could then inform genetic or empirically defined sensitivities. (B) Summary of the clinical and pathological features of the patient cohort. TNM, tumour, node, metastases. (C) Mutational spectra in the cohort of sequenced cases, overall trinucleotide alterations through the entire cohort. The EMCT2 tumour was from a smoker with the tumour exhibiting increased incidence of C>A transversions. (D) Summary of copy number alterations as determined by genomic identification of significant targets in cancer (GISTIC) analysis, the position of CDKN2A and SMAD4 deletions are indicated. (E) KRAS mutant allele distribution through the cohort. Summary of events targeting cell cycle, tumour growth factor-β (TGF-β) and NOTCH signalling pathways. Blue denotes homozygous deletion, red denotes amplification, orange denotes tumour-specific insertion deletion (INDEL) and green indicates tumour-specific single nucleotide variants (SNV). (F) Overall distribution of genetic events targeting oncogenic pathways in PDAC cases sequenced.
gutjnl-2016-313133supp001.pdf (49.3MB, pdf)
Genetic diversity of PDAC tumours
The PDAC tumours exhibited diverse mutational spectra consistent with previously published studies11 15 (figure 1C). The majority of cases were dominated by C>T transitions associated with age-related mutational events.15 PDAC cases arising in smokers exhibited enrichment of C>A transversions as indicated by the EMCT2 and EMC1229 cases (figure 1C). The tumours also captured the distinct chromosomal instability profiles observed across PDAC cases,8 9 11 with frequent deletion of CDKN2A and SMAD4 (see figure 1D and online supplementary figure S1). The cohort harboured multiple different KRAS mutations and aberrations in multiple signalling pathways that are representative of PDAC (see figure 1E, online supplementary figure S1 and online supplementary data). In this context, PDAC cases employed for models development recapitulate the genetic diversity of PDAC that has been described8 10 11 (see figure 1F, online supplementary figure S1 and online supplementary data).
Generation and characterisation of patient-derived models
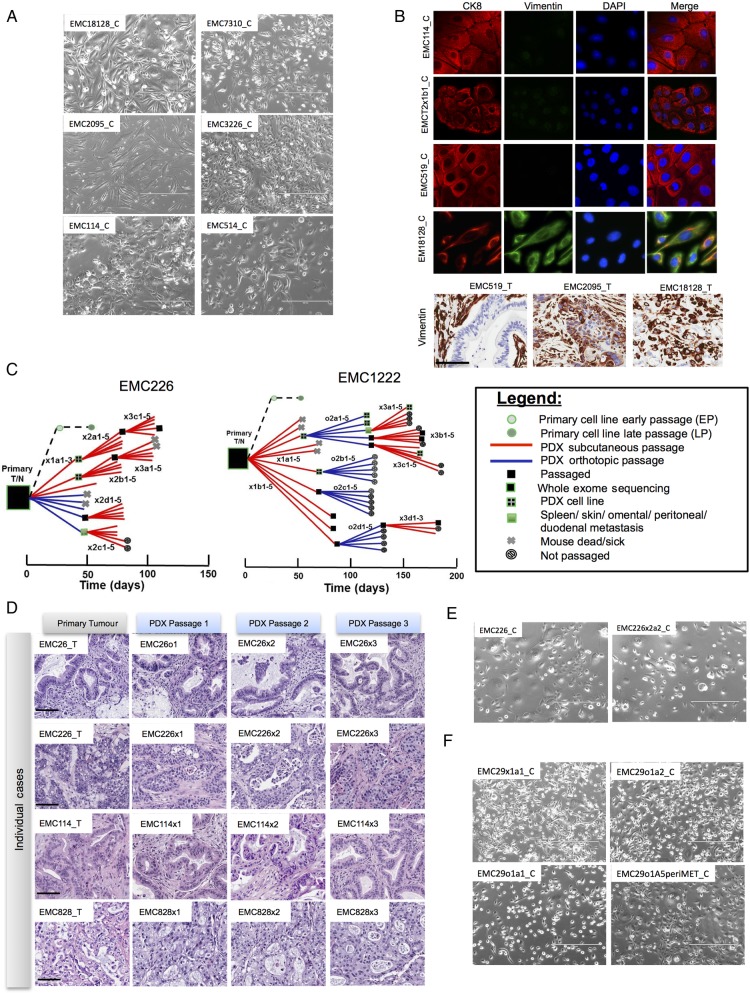
Primary cell lines were established from PDAC (denoted by case identifier with extension ‘_C’). Optimised culture conditions were developed so that pure tumour cell populations could be isolated in 8–12 weeks on collagen matrix (extended methods). Primary cell lines had distinct morphological features ranging from typical epithelial (eg, EMC114_C) to mesenchymal (eg, EMC2095_C) patterns (see figure 2A and online supplementary figure S2). The majority of the cell lines exhibited epithelial characteristics, as indicated by the presence of CK8 and low levels of vimentin expression (see figure 2B and online supplementary figure S2). However, several cell lines (eg, EMC18128_C) exhibited features of epithelial-mesenchymal transition as indicated by co-expression of CK8 and vimentin (see figure 2B and online supplementary figure S2). These phenotypes including the expression of CK8 (see online supplementary figure S2) and overall cell morphology (not shown) remained stable with extended passage (ie, up to passage 40). Features of mesenchymal phenotype as observed in the cell lines could be detected in the primary tumour (see figure 2B and online supplementary figure S2).
Figure 2.
Generation of cell lines from pancreatic ductal adenocarcinoma (PDAC) tumours. (A) Representative phase contrast images of cell lines derived from independent cases. Scale bar is 400 µm. (B) Representative immunofluorescence staining of CK8 and vimentin from developed cell lines. Vimentin staining in the clinical tumours employed to generate cell lines. Scale bar is 100 µm. (C) Representative pedigree charts of patient-derived xenograft (PDX) development. The derivation of cell lines and exome sequencing of the models are indicated by the legend. (D) Representative H&E stained sections of the primary tumour and PDX tissue from three passages. Scale bar is 100 µm. (E) Representative phase contrast images of cell lines derived from independent PDAC cases and associated PDX. Scale bar is 400 µm. (F) Representative phase contrast images of cell lines derived from multiple PDX models and associated metastases. Scale bar is 400 µm.
In parallel to the development of the cell lines, tumour tissue from patients was dissected and implanted either subcutaneously (denoted by ‘x’ following case identifier) or orthotopically (denoted by ‘o’ following case identifier) in NSG mice. The time to tumour engraftment varied across cases as summarised in pedigree charts showing detailed transplant history for each model (see figure 2C and online supplementary figure S2). The PDX models exhibited a striking conservation of the primary tumour histoarchitecture, including desmoplastic stroma, and remained stable across at least three passages (see figure 2D and online supplementary figure S2). This feature of tumour biology is important, as stromal compartment is believed to be critical for disease progression, metastatic spread and therapeutic sensitivity.25 31 Cell lines were developed as xenografts were passaged. In select cases, tumour cell lines were developed from both the primary tumour and related PDX, allowing for direct comparison of primary tumour-derived and PDX-derived lines. In this setting, there was a marked conservation in overall cell morphology (see figure 2E, F and online supplementary figure S2).
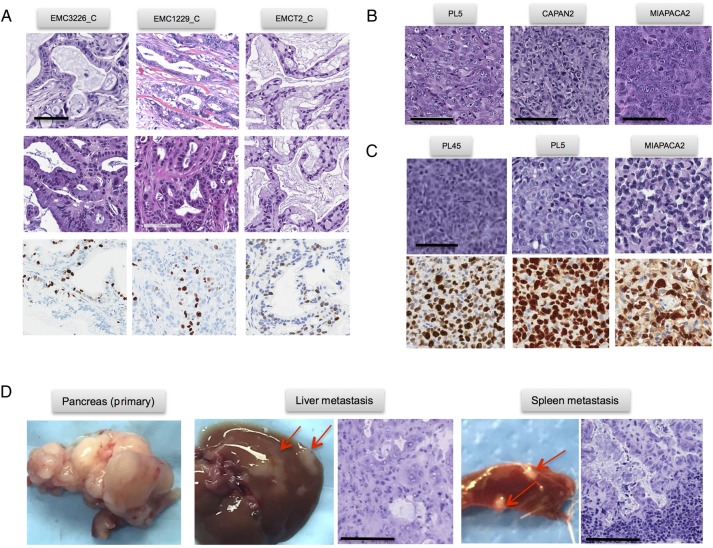
The patient-derived cell lines were tumourigenic and gave rise to tumours when orthotopically injected into NSG mice in four of five models tested. Interestingly, xenograft tumours derived from injected patient-derived cell lines recapitulated the histoarchitecture of the primary tumour including gland formation and reconstituted desmoplastic stroma (figure 3A). These data suggest that patient-derived tumour cells have the capacity to programme microenvironment and re-establish stromal compartment through mobilisation of murine tissue elements. The xenografts arising from these cell lines demonstrated a level of Ki67 labelling that is consistent with that observed in clinical cases. In contrast, established commonly used pancreatic cancer cell lines (eg, PL5, MiaPaca2 and PL45) did not induce stromal reaction and generated tumour masses composed of sheets of tumour cells when implanted orthotopically. These models also exhibited an exceedingly high Ki67 index (figure 3B, C). Thus, the patient-derived cell line models developed in this study retain several key features that are not apparent in established models. Additionally, the orthotopic PDXs gave rise to metastatic spread to visceral organs consistent with the biology of PDAC, thereby providing a model to study disease progression (figure 3D).
Figure 3.
Phenotypic features of patient-derived models. (A) Representative images of orthotopically injected patient-derived cell lines. H&E and Ki67 staining are shown, scale bar is 100 µm. (B and C) Representative images of orthotopically injected established cell lines. H&E and Ki67 staining are shown, scale bar is 100 µm. (C) Orthotopic patient-derived xenograft (PDX) model showing the metastatic spread to liver and spleen. Scale bar is 100 µm.
Genetic characterisation of patient-derived models
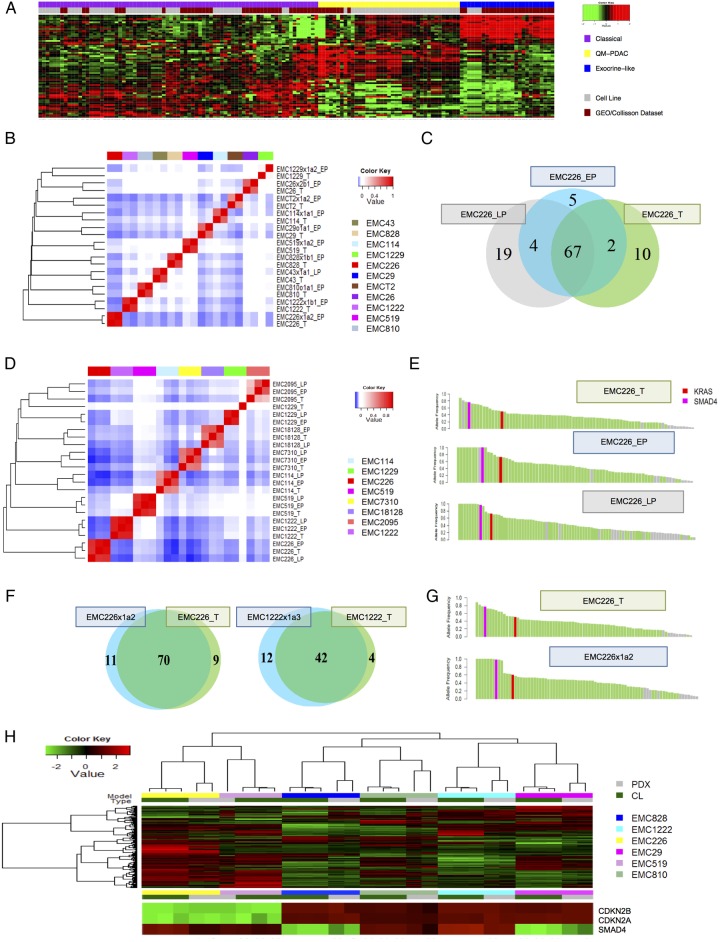
To determine the relationship of the gene expression patterns observed in patient-derived cell lines and those observed in clinical PDAC specimens, RNA sequencing was performed on the cell lines and their association with PDAC subtypes was evaluated.17 19 PDAC cell lines exhibited classical, quasi-mesenchymal and exocrine-like gene expression profiles observed in clinical PDAC samples, indicating that they represent the phenotypic diversity of PDAC (see figure 4A and online supplementary figure S3). Importantly, the exocrine-like subtype is not represented among established PDAC cell lines.17
Figure 4.
Molecular and genetic conservation of patient-derived cell lines. (A) Clustering analysis of cell lines (grey colour bar) versus pancreatic ductal adenocarcinoma (PDAC) cases that exhibit classical (purple), exocrine-like (yellow) and quasi-mesenchymal (blue) gene expression programmes. Cases and genes were clustered based on Euclidian distance. (B) Spearman's correlation analysis of single nucleotide variants (SNVs) from early passage cell lines and associated patient tumour or patient-derived xenograft (PDX). (C) Representative Venn diagram of the patient tumour and early and late passage cell lines. (D) Spearman's correlation analysis of SNVs from early and late passage cell lines and associated patient tumour or PDX. (E) Allele frequency plot illustrating the conservation of high frequency genetic events between the primary tumour and cell lines at early and late passage. Green bars denote genetic events in common while grey bars denote unique events, fuchsia and red denote SMAD4 and KRAS mutations, respectively. (F) Venn diagrams summarising the overlap in SNVs between representative primary tumours and PDX model. (G) Allele frequency plot illustrating the conservation of high frequency genetic events between the primary tumour and PDX. Green bars denote genes in common while grey bars denote unique events, fuchsia and red denote SMAD4 and KRAS mutations, respectively. (H) Unsupervised clustering of gene expression was performed using genes of high variance between the cell line and PDX models from the same clinical cases. The inset shows the expression of CDKN2A/CDKN2B and SMAD4 in relation to the cell lines and PDX models.
It is a common perception that cell culture selects for substantial genetic alterations that compromise the ability of cell lines to inform the biology of the tumour. To characterise genetic features of the cell lines and PDX models, whole exome sequencing was performed at an average of 124x coverage depth (see online supplementary data). To eliminate the presence of contaminating mouse genetic material from PDXs and the general errors associated with paralogs, we developed an algorithm, mouse and paralog exterminator (MAPEX). This algorithm uses basic local alignment search tool (BLAST) to selectively eliminate mouse genes and paralogous miscalls in the analysis of variants (manuscript in preparation). The analysis of the cell lines developed herein indicated that they were genetically highly consistent with the tumours from which they were derived. This was determined by correlation analysis of the somatic mutation events detected in the primary tumour and the cell line (see figure 4B and online supplementary figure S3). Analysis of the shared genes indicated that common PDAC oncogenic driver mutations (eg, KRAS, TP53 and SMAD4) and high allele frequency mutations were preserved in the cell lines and remained stable for up to 40 passages (see figure 4C–E and online supplementary figure S3). The non-conserved mutations generally had a low allele frequency and did not commonly occur in characterised cancer-related genes (see figure 4E and online supplementary figure S3). For example, there was no evidence for selective TP53 mutation during cell line derivation.
Several recent large studies advocated using PDXs as models that more faithfully recapitulate genetic features of the primary tumours.30 32 However, direct and adequately powered comparison of PDXs and primary cell lines with the parental tumour has not been performed. To address this knowledge gap, we first analysed the preservation of genetic architecture between PDXs and the tumours from which they originated (figure 4F). Across all models, >75% of non-synonymous mutational events (single nucleotide variants (SNVs)) were conserved between the primary tumours and the PDX models, and as with the cell lines, genetic events with a higher allele frequency were conserved (see figure 4F, G, online supplementary figure supplementary figure S3). These analyses suggested that the conservation level determined using statistically based variant calling methods (eg, MuTect), as performed here, was likely an underestimate. Additionally, in rare cases (ie, EMC1229) due to the histoarchitecture of the tumour it was very difficult to obtain high tumour purity and that limited effective variant calling and was likely responsible for the limited overlap of that case with the derived models.
The PDX remained closely related over at least three passages, and the majority of SNVs were preserved in PDXs originating from different areas of primary tumour (see online supplementary figure S3). Metastatic deposits from selected mice were also evaluated by exome sequencing. In these cases, the majority of mutations occurred in the primary tumour before metastasis emerged, and were therefore present in all metastases that were evaluated (see online supplementary figure S3). Importantly, the genetic conservation of cell lines with PDX was also evidenced by overall maintenance of gene expression features (figure 4H). Six cell lines that were RNA sequenced were clustered based on gene expression variance, and then PDX models derived from the same primary tumour were evaluated. As shown, the PDX clustered with the cell lines that were derived from the same tumour. Not surprisingly, the expression of genetically lost genes (eg, CDKN2A and SMAD4) was highly correlated in PDX and cell line (figure 4H).
Using patient-derived models to gain insights into tumour evolution
In total, over 100 individual PDX and cell line models derived from 27 clinical cases were sequenced (figure 5A). Direct comparison of PDX and cell lines with the parental tumour demonstrated that a large number of mutations were omnipresent, including alterations in known PDAC driver genes (ie, KRAS, TP53 and SMAD4) and multiple other genes that have been associated with pancreatic cancer (eg, ARID1A and RBM10) (see online supplementary figure S4 and supplementary data). Correspondingly, Spearman's correlation analysis showed that each primary tumour and derived PDX and cell line clustered together (figure 5B). In order to evaluate multiple cases together, gene level SNV data were used for unsupervised clustering based on Euclidean distance. As shown, clinical cases and their derivative models clustered into discrete clades that included cell lines, xenografts and even metastasis derived from the same primary tumour (figure 5C). These data indicate that each series of models is most closely related to the tumour of origin. To investigate tumour evolution across specific models, phylogenetic analysis was employed. As could be expected, earlier passage models (both cell lines and PDXs) are more closely related to the clinical case (figure 5D). In this context, the variance between tumours and models within the same passage is similar to the variance observed between distinct geographical locations reported in PDAC, and was generally consistent with the concept that resected PDAC harbours mutations indicative of metastatic potential.33 34 With passaging in mice and with metastasis development there was evolutionary divergence and acquisition of select cancer-associated events, suggesting that these events may provide increased fitness. Analysis of the unique genes that appeared in the models indicated enrichment for genes associated with differentiation and transcription (see online supplementary figure S5). Detailed analysis of raw sequence reads for NOTCH1 and SMAD4 mutations indicated that the mutations did not exist in the primary tumour and therefore most likely arose ex vivo (figure 5E). Interestingly, events that may not necessarily have functional significance exhibited a limited allele frequency (eg, NOTCH1), while mutation of SMAD4 dominated the PDX in which the mutation arose (figure 5E). These data suggest that there is selection for genetic events that are associated with PDAC progression or poor prognosis.11 35 The data also underscore the necessity of evaluating the genetics of tumour models, and suggest that evolutionary features observed ex vivo could inform the functionality of these events in patient tumours.
Figure 5.
Composite genetic analysis of models and evolution. (A) Summary of exome sequencing data from over 100 primary tumours and associated models. Similarity in mutation burden is summarised in the scatter plot. (B) Spearman's correlation of single nucleotide variants from the patient tumour, patient-derived xenograft (PDX) and cell line from the same case. (C) Unsupervised clustering of cases based on mutated genes and Euclidian distance. Samples are colour-coded for the same case. (D) Phylogenetic analysis of the indicated models, with multiple cell lines and PDX sequenced. The trees are rooted in the normal tissue, the primary tumour and associated models are indicated by colour bar. (E) The allele frequency was determined for each of the noted cases and models. VAF, variant allele frequencies.
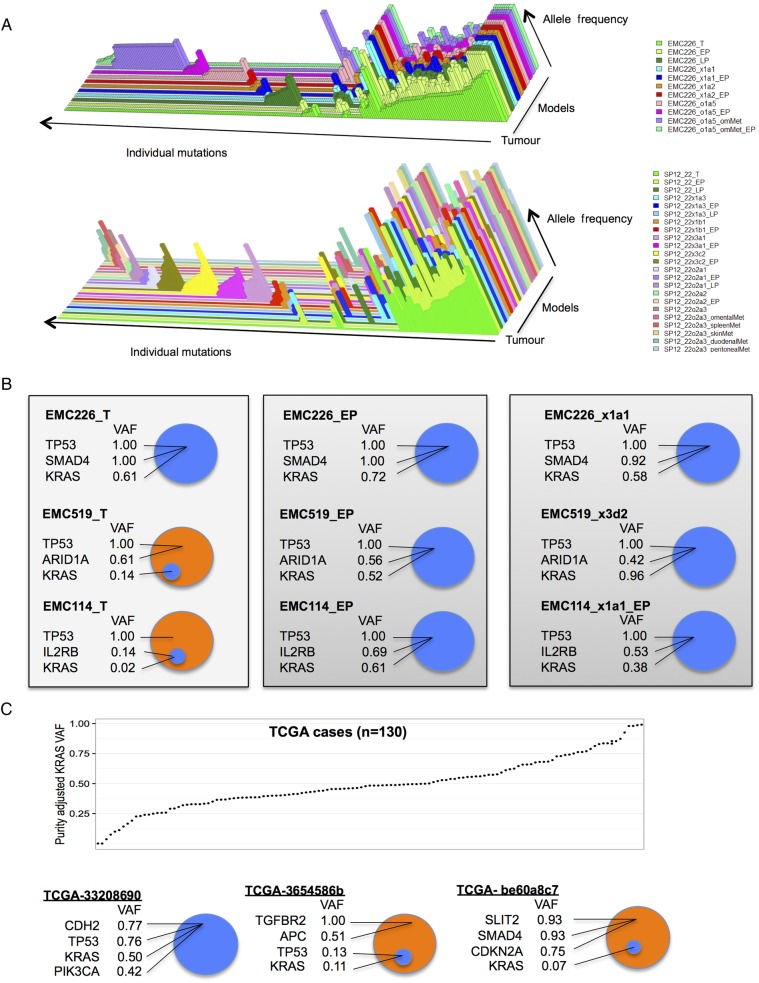
Inferring tumour heterogeneity and dominant clones
Sequencing analysis reveals features of tumour heterogeneity based on mutant allele frequencies within a given tumour. As shown in the three-dimensional plots from select models, the core genetic features of the primary tumour with high allele frequency are conserved through multiple models including cell lines, PDXs and metastatic nodules of the PDX (figure 6A). In these cases, KRAS, TP53 and/or SMAD4 mutations harbour relatively high allelic frequencies. This point was apparent through the entirety of the cohort, where the average common mutant allele frequency was 0.54, while the tumour-specific or model-specific allele frequency was 0.13 and 0.25, respectively. Although there was a marked conservation of genetic features, the models exhibited subclonal unique events that had relatively low allele frequencies. Discordant events could represent expansion of rare populations present in primary tumour (eg, below level of detection) or acquisition of new alterations during model establishment and propagation. As discussed above, in a few cases mutations, present in later passage and not called by MuTect in the primary tumour, could be detected on visual inspection of sequence reads. Preservation of somatic variants between tumour and PDX or cell line was further evaluated in pairwise comparisons of the primary tumours with all models originating from the same primary tumour (see online supplementary figure S4). We observed many cases, exemplified by EMC226, where KRAS, SMAD4 and TP53 mutant allele frequencies in the tumour are comparable for oncogene and tumour suppressors, respectively and these ratios are preserved in the derived models (see figure 6B and online supplementary figure S4). However, there were cases that exhibited high mutant allele frequencies for specific cancer genes, but very low mutant allele frequency for KRAS (figure 6B). These data indicate that in a subset of PDAC cases, KRAS represented a subclonal event. Interestingly, in each case with a subclonal KRAS mutation in the primary tumour, we observed expansion of the KRAS mutation to fully clonal in both cell line and PDX models (figure 6B). These data suggest that KRAS mutations provide a significant fitness advantage during model derivation. To determine if this finding was particular to our cohort, the TCGA pancreatic cancer data were interrogated for KRAS mutant allele frequencies. As expected, the majority of cases have an approximately 0.5 allele frequency suggesting that KRAS mutation is preserved through most of the tumour cells. However, there were several cases with KRAS allele frequencies <0.2 (figure 6C). Importantly, high-frequency tumour-associated mutations occurred in cases with low-frequency KRAS mutant alleles. The totality of the data indicates that in most cases, pancreatic cancer harboured a dominant KRAS-driven clone. However, in a subset of cases KRAS was not present in the dominant clone in the parental PDAC tumour.
Figure 6.
Allelic frequency and clonal features of KRAS mutation in pancreatic ductal adenocarcinoma (PDAC). (A) Three-dimensional plots from primary tumour (front) and multiple models (behind). Each mutational event is summarised as a bar, and the raw allele frequency is represented by the height of the bar. (B) Normalised variant allele frequencies (VAF) of the indicated genes are shown for the tumour case and derived models. (C) Tumour purity adjusted KRAS VAF shown across the TCGA cohort of 130 cases. Select TCGA cases, where KRAS mutation is a common event throughout the tumour, and two cases where KRAS mutation is subclonal relative to other cancer-related mutations are shown.
Using patient-derived cell lines to interrogate functional features
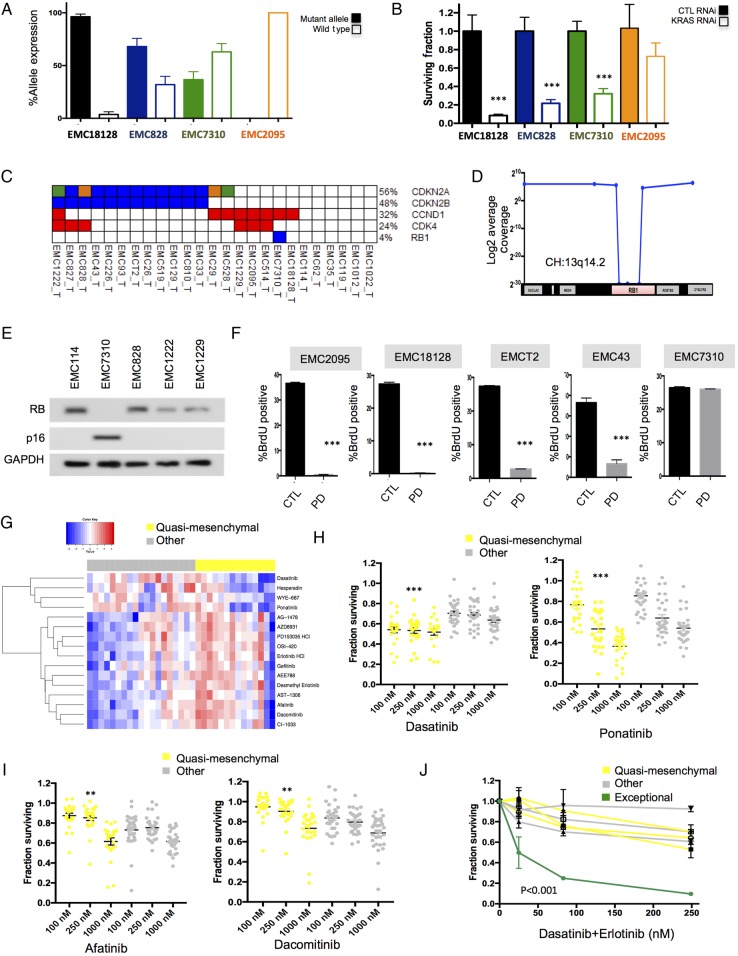
Since the models were molecularly annotated, we could employ them to interrogate the functional significance of specific genetic events. Oncogenic KRAS mutations are known to be a key driver in PDAC and it has been postulated that loss of expression of the wild-type allele of KRAS is important for aggressive growth and disease progression. In the analysis of cell lines there was a diverse expression of the wild-type versus mutant allele (figure 7A). Thus, there was not a requirement for losing the wild-type allele. Interestingly, in three tested cell lines with varying levels of wild-type KRAS expression, viability was still dependent on KRAS as determined by knockdown analysis (figure 7B). In contrast, a model that was wild-type for KRAS (EMC2095_C) was largely resistant to the effects of KRAS knockdown. Another genetic hallmark of PDAC is the loss of the CDK4/6 inhibitor p16ink4a that is encoded by the CDKN2A gene. This event occurred frequently in tumours via homozygous deletions or mutations (figure 7C). However, one of the PDAC cases harboured an intragenic deletion in the RB1 tumour suppressor (figure 7D). In the cell line derived from that tumour, the loss of retinoblastoma tumor suppressor protein (RB) and compensatory elevation of p16ink4a expression were observed (figure 7E). Although most of primary PDAC cell lines are sensitive to CDK4/6 inhibition, the model with the RB1 deletion, as expected, was resistant (figure 7F). These data indicate that the genetically annotated models can be used to interrogate specific genetic features of pancreatic cancer that drive biological features of disease or represent potential therapeutic targets.
Figure 7.
Defining tumour selective sensitivity profiles. (A) The expression of the mutant and wild-type KRAS allele was determined by analysis of RNA sequencing data. These data revealed three principle forms of expression. Those solely expressing mutant KRAS (black bars), those biased towards mutant KRAS (blue bars) and those with approximately equal expression of both wild-type and mutant KRAS (green bars). All data are from triplicate RNA sequencing, the mean and SD are shown. (B) The knockdown of KRAS was performed in models exhibiting various KRAS expression patterns. The surviving fraction was determined by cell-titre glow (n=5). Data show the average and SD. Data were statistically significant as determined by unpaired t-test, ***p<0.001. (C) Alterations of genes in RB pathway. (D) Schematic of 13q14.2 locus and the intragenic deletion of RB1 in the case EMC7310. (E) Immunoblotting for RB, p16ink4a and GAPDH in the indicated cell lines. (F) Flow cytometry analysis of the response to PD-0332991 in the indicated cell lines. Mean and SD of BrdU-positive cells is shown (n=3). Data were statistically significant as determined by unpaired t-test, ***p<0.001. (G) Heatmap of the area under the curve of drug sensitivity to the indicated agents in quasi-mesenchymal subtype versus other patient-derived cell lines. (H and I) Scatter plots of drug sensitivity of quasi-mesenchymal subtype versus other patient-derived cell lines for the indicated agents. **p<0.01, ***p<0.001 determined by two-way analysis of variance. (J) Dose-response for the indicated cell lines with the dasatinib+erlotinib at the indicated concentrations.
As an alternative to directed interrogation of genetic events, the models can be used to determine if specific PDAC subtypes are associated with therapeutic sensitivity (see figure 7G and online supplementary figure S6). For this analysis, we interrogated specific therapeutic sensitivities associated with the quasi-mesenchymal subtype of PDAC. A collection of 305 cancer drugs was screened across all primary cell lines as well as a panel of established cell lines, as we have recently published.36 By unbiased correlation analysis we identified agents that were selective to established models, or selective to quasi-mesenchymal cell lines (see figure 7G and online supplementary figure S6). Established cell lines were sensitive chemotherapy (eg, paclitaxel), yet markedly resistant to other targeted therapies (eg, ABT-737) (see online supplementary figure S6). The quasi-mesenchymal cells were generally resistant to epidermal growth factor receptor (EGFR) inhibitors (figure 7I). This finding is consistent with previously published studies.17 In contrast, the multityrosine kinase inhibitors (eg, ponatinib and dasatinib) were selectively active in the quasi-mesenchymal group (figure 7H). In spite of these general subtype-specific responses, there remained substantial variation of sensitivity between individual patient-derived models within a given subtype. Notably, one of the quasi-mesenchymal cell lines was also sensitive to EGFR inhibitors (figure 7G). Consonantly, when compared across all models analysed, this specific cell line model (denoted in green) was exceptionally sensitive to the combination of dasatinib and erlotinib (figure 7J). These data suggest that while there are ‘subtype-related sensitivities’, each tumour appears to have its own distinct vulnerability.
Discussion
PDAC represents a major clinical challenge that is denoted by the terrible prognosis and failure of multiple clinical trials. The shortcomings of cell line models have been cited as a reason for high failure rate of oncology drugs. Here, we show that patient-derived models (cell lines and PDX) can be developed from PDAC with diverse histological and genetic features expanding the compendium of existing models. This included cell lines with recently described PDAC mutations (eg, ARID1A, GNAS, RBM10 and KDM6A) and those derived from well-differentiated tumours. This is also the first study that comprehensively compares genetics of cell lines and PDX models derived from the same originating tumour and directly addresses question of genetic fidelity of cell lines.
PDAC is genetically diverse and individual tumour genomes are highly complex.8 11 This is apparent in veritably all cases, where KRAS mutations combine with a plethora of other genetic events that are cancer relevant. As such, PDAC represents a complex genetic compendium, which would be almost impossible to emulate with engineered models. In most cases, multiple different aberrations in core signalling pathways (eg, tumour growth factor-β, cell cycle, WNT, chromatin remodelling) are observed in addition to KRAS. Presumably, this genetic milieu drives the biology and therapeutic recalcitrance of PDAC. Developing models reflective of these genetic features will be important for fully understanding the biology of PDAC.
While all commonly deployed PDAC cell lines are by definition ‘patient-derived’, the similarity to the tumour of derivation is largely unknown. As a result of the absence of a normal control, a comprehensive genetic analysis is limited. Correspondingly, in the absence of a primary tumour, it is impossible to know the extent to which they recapitulate the primary tumour characteristics. There is also a concern that long-term growth of cancer cell lines in a non-physiological environment results in genetic changes that are not reflective of originating tumour genetics. To mitigate effects of culture, all cell models herein were isolated on collagen matrix and there was no evidence for phenotypic drift associated with establishment of the cell line. Models arose as epithelial or mesenchymal like cells consistent with known expression of vimentin in a subset of clinical PDAC cases.37 These cell lines were amenable to downstream analyses including RNA interference transfection and drug sensitivity assays.
The modest relationship of cell lines to the cancers from which they had been derived has been cited as a reason for the high failure rate of clinical trials. Here, we extensively compared the genetics of pancreatic cancer cell lines and the parental tumours to evaluate the extent of genetic drift that occurs during cell line establishment/propagation. These data showed that cell lines generally maintained the oncogenic drivers and high-allele frequency genetic events of the primary tumour. The level of genetic conservation was reproducibly identical to that observed in PDX models, and remained stable for at least 40 passages. In phylogenetic analysis, cell lines surprisingly yielded less genetic evolution with passage than PDX models. Presumably, this indicates that using the methods employed here, the stress of culture does not significantly select for mutations; while in the tumour environment there is more selective pressure. This selective pressure interestingly yielded mutations in known PDAC tumour suppressors, and suggests that such ‘ex vivo’ progression could represent a means to model PDAC tumour progression. For example, we identified the occurrence of model selective SMAD4 loss that was acquired with tumour passaging in PDX. These data also underscore the importance of maintaining genetic annotation of both PDX and cell lines.
Studying features of tumour progression and heterogeneity in solid tumours can be challenging. Most studies have achieved this by either distinct geographical sampling of resected tumour tissue, the analysis of autopsy samples or multiple biopsies. In PDAC, the analysis of metastatic deposits from autopsy cases demonstrated significant genetic conservation between the primary and metastatic disease.38 These findings are largely consistent with the supposition that even in a setting of resectable PDAC metastasis has occurred in majority of cases. Therefore, rapidly developed models from resected disease could serve as important avatars of the recurrent disease that almost invariably emerges in PDAC. Importantly, the timing of the development and characterisation of cell lines is such that the information could ostensibly be employed to inform treatment of recurrent disease.
While PDAC genomes are being sequenced and there is growing understanding of the significant genetic events,8 11 16 the model for PDAC progression from, pancreatic intraepithelial neoplasia (PanIN), has remained largely unchanged. Studies applying sequencing to analyse PanINs suggest a gradual expansion of the KRAS mutated clone during progression, with acquisition of additional genetic or epigenetic changes.39 40 41 From existing sequencing data, it is clear that the vast majority of PDAC cases harbour high KRAS mutant allele frequency as would be expected for the initiating driver mutation. However, approximately 5%–10% of cases carry KRAS mutant allele frequencies that are clearly subclonal, present in a small fraction of tumour cells. Tumour purity impacts mutation detection and it is known that PDAC is characterised by abundant non-neoplastic cells in tumour microenvironment. To define tumour purity in this study, we applied a bench-marking strategy relative to other well-known cancer-associated genes (eg, TP53 mutation). This method had advantages relative to qpure algorithm previously used in PDAC, which operated under the assumption that KRAS is most common genetic event in all PDAC cases. In this study, and in TCGA data between 5% and 10% of PDAC cases will harbour only a small fraction of mutant KRAS alleles (eg, allelic ratio of mutant to wild-type allele as low as 0.02). By subsequently analysing models derived from PDAC containing small percentage of KRAS mutated cells, we could determine the fate of the minor clone containing KRAS mutations. In all models interrogated, the presence of KRAS dominated the models. This finding suggests that there could be two distinct routes to PDAC development, one which is driven by KRAS and another mechanism that ultimately selects for KRAS mutations latter in disease progression.
Many of the genetic features of PDAC remain an enigma. For example, how mutations impacting splicing or chromatin remodelling contribute to disease is still scantly understood,8 but appear to have prognostic significance.11 Models with the specific tumour-associated mutations provide an opportunity to address functionality by gene editing and other approaches that address the impact of endogenous genes. In reference to therapeutic intervention, they can clearly inform dependence on KRAS or requirement of CDK4/6 activity for cell cycle progression. In this context, cell lines offer many advantages that facilitate high-throughput approaches, and can be used to interrogate the mechanistic impact of tumour-associated variants whose functional significance remains unclear. Here, we showed that while molecular subtypes of PDAC had differential sensitivities to therapeutic agents, there was considerable interindividual difference in drug sensitivity that became particularly relevant when considering combination therapies. The cell line-identified vulnerabilities can then be interrogated within the PDX. Our group and others have recently published on this general approach.19 36 42 Thus, integrated banks of patient-derived models that are genetically annotated will serve as an important resource to further the interrogation of tumour biology and will lend much-needed insights to the treatment of disease.
Methods
Detailed description of all methods is provided in the supplemental information. The sequencing data are available through dbGaP (PRJNA289986).
Acknowledgments
The authors thank all members of the Knudsen and Witkiewicz laboratory for thought-provoking discussion, technical assistance and help with manuscript preparation.
Footnotes
Contributors: AKW and ESK are co-corresponding authors and jointly directed the project. CE, UB, PV, BM and CM contributed to data generation. UB, PV and BM provided informatics support and analysis of sequencing data. JM provided logistical support and clinical assistance. CE, CM, ESK and AKW developed the cell and xenograft models in the study. AKW and ESK wrote much of the study with all authors contributing.
Funding: The work is supported by grants to ESK and AKW from the National Institutes of Health. Grants from NCI: CA211878, CA023074-S2, and CA142543-05S2.
Competing interests: None declared.
Ethics approval: IRB at University of Texas Southwestern.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: This is present in the Methods section of the manuscript for the acquisition of data or models presented.
References
- 1.Paulson AS, Tran Cao HS, Tempero MA, et al. . Therapeutic advances in pancreatic cancer. Gastroenterology 2013;144:1316–26. 10.1053/j.gastro.2013.01.078 [DOI] [PubMed] [Google Scholar]
- 2.Kleger A, Perkhofer L, Seufferlein T. Smarter drugs emerging in pancreatic cancer therapy. Ann Oncol 2014;25:1260–70. 10.1093/annonc/mdu013 [DOI] [PubMed] [Google Scholar]
- 3.Almhanna K, Philip PA. Defining new paradigms for the treatment of pancreatic cancer. Curr Treat Options Oncol 2011;12:111–25. 10.1007/s11864-011-0150-8 [DOI] [PubMed] [Google Scholar]
- 4.Yeo CJ, Cameron JL, Lillemoe KD, et al. . Pancreaticoduodenectomy for cancer of the head of the pancreas. 201 patients. Ann Surg 1995;221:721–31; discussion 31–3 10.1097/00000658-199506000-00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philip PA, Mooney M, Jaffe D, et al. . Consensus report of The National Cancer Institute clinical trials planning meeting on pancreas cancer treatment. J Clin Oncol 2009;27:5660–9. 10.1200/JCO.2009.21.9022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knudsen ES, O'Reilly EM, Brody JR, et al. . Genetic diversity of pancreatic ductal adenocarcinoma and opportunities for precision medicine. Gastroenterology 2016;150:48–63. 10.1053/j.gastro.2015.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saif MW. Advancements in the management of pancreatic cancer: 2013. JOP 2013;14:112–18. [DOI] [PubMed] [Google Scholar]
- 8.Waddell N, Pajic M, Patch AM, et al. . Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495–501. 10.1038/nature14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biankin AV, Waddell N, Kassahn KS, et al. . Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399–405. 10.1038/nature11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones S, Zhang X, Parsons DW, et al. . Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801–6. 10.1126/science.1164368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witkiewicz AK, McMillan EA, Balaji U, et al. . Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744 10.1038/ncomms7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol 2015;12:319–34. 10.1038/nrclinonc.2015.53 [DOI] [PubMed] [Google Scholar]
- 13.Vincent A, Herman J, Schulick R, et al. . Pancreatic cancer. Lancet 2011;378:607–20. 10.1016/S0140-6736(10)62307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol 2008;3:157–88. 10.1146/annurev.pathmechdis.3.121806.154305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. . Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey P, Chang DK, Nones K, et al. . Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. 10.1038/nature16965 [DOI] [PubMed] [Google Scholar]
- 17.Collisson EA, Sadanandam A, Olson P, et al. . Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17: 500–3. 10.1038/nm.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moffitt RA, Marayati R, Flate EL, et al. . Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168–78. 10.1038/ng.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noll EM, Eisen C, Stenzinger A, et al. . CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med 2016;22:278–87. 10.1038/nm.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang CI, Boj SF, Clevers H, et al. . Preclinical models of pancreatic ductal adenocarcinoma. J Pathol 2016;238:197–204. 10.1002/path.4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barretina J, Caponigro G, Stransky N, et al. . The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7. 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herreros-Villanueva M, Hijona E, Cosme A, et al. . Mouse models of pancreatic cancer. World J Gastroenterol 2012;18:1286–94. 10.3748/wjg.v18.i12.1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuveson DA, Hingorani SR. Ductal pancreatic cancer in humans and mice. Cold Spring Harb Symp Quant Biol 2005;70:65–72. 10.1101/sqb.2005.70.040 [DOI] [PubMed] [Google Scholar]
- 24.Day CP, Merlino G, Van Dyke T. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell 2015;163:39–53. 10.1016/j.cell.2015.08.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olive KP, Jacobetz MA, Davidson CJ, et al. . Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009;324:1457–61. 10.1126/science.1171362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer 2015;15:311–16. 10.1038/nrc3944 [DOI] [PubMed] [Google Scholar]
- 27.Boj SF, Hwang CI, Baker LA, et al. . Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324–38. 10.1016/j.cell.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang L, Holtzinger A, Jagan I, et al. . Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med 2015;21:1364–71. 10.1038/nm.3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hidalgo M, Bruckheimer E, Rajeshkumar NV, et al. . A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol Cancer Ther 2011;10:1311–16. 10.1158/1535-7163.MCT-11-0233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao H, Korn JM, Ferretti S, et al. . High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 2015;21:1318–25. 10.1038/nm.3954 [DOI] [PubMed] [Google Scholar]
- 31.Neesse A, Michl P, Frese KK, et al. . Stromal biology and therapy in pancreatic cancer. Gut 2011;60:861–8. 10.1136/gut.2010.226092 [DOI] [PubMed] [Google Scholar]
- 32.Townsend EC, Murakami MA, Christodoulou A, et al. . The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell 2016;29:574–86. 10.1016/j.ccell.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yates LR, Gerstung M, Knappskog S, et al. . Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 2015;21:751–9. 10.1038/nm.3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerlinger M, Rowan AJ, Horswell S, et al. . Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcea G, Neal CP, Pattenden CJ, et al. . Molecular prognostic markers in pancreatic cancer: a systematic review. Eur J Cancer 2005;41:2213–36. 10.1016/j.ejca.2005.04.044 [DOI] [PubMed] [Google Scholar]
- 36.Witkiewicz AK, Balaji U, Eslinger C, et al. . Integrated patient-derived models delineate individualized therapeutic vulnerabilities of pancreatic cancer. Cell Rep 2016;16:2017–31. 10.1016/j.celrep.2016.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Gu F, Ma N, et al. . CIP2A expression is associated with altered expression of epithelial-mesenchymal transition markers and predictive of poor prognosis in pancreatic ductal adenocarcinoma. Tumour Biol 2013;34:2309–13. 10.1007/s13277-013-0775-2 [DOI] [PubMed] [Google Scholar]
- 38.Yachida S, Jones S, Bozic I, et al. . Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010;467:1114–17. 10.1038/nature09515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hruban RH, Wilentz RE, Kern SE. Genetic progression in the pancreatic ducts. Am J Pathol 2000;156:1821–5. 10.1016/S0002-9440(10)65054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hruban RH, Goggins M, Parsons J, et al. . Progression model for pancreatic cancer. Clin Cancer Res 2000;6:2969–72. [PubMed] [Google Scholar]
- 41.Brosens LA, Hackeng WM, Offerhaus GJ, et al. . Pancreatic adenocarcinoma pathology: changing “landscape”. J Gastrointest Oncol 2015;6:358–74. 10.3978/j.issn.2078-6891.2015.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruna A, Rueda OM, Greenwood W, et al. . A Biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 2016;167:260–74.e22. 10.1016/j.cell.2016.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2016-313133supp001.pdf (49.3MB, pdf)