Abstract
Plants have many, highly variable resistance (R) gene loci, which provide resistance to a variety of pathogens. The first R gene to be cloned, maize (Zea mays) Hm1, was published over 25 years ago, and since then, many different R genes have been identified and isolated. The encoded proteins have provided clues to the diverse molecular mechanisms underlying immunity. Here, we present a meta-analysis of 314 cloned R genes. The majority of R genes encode cell surface or intracellular receptors, and we distinguish nine molecular mechanisms by which R proteins can elevate or trigger disease resistance: direct (1) or indirect (2) perception of pathogen-derived molecules on the cell surface by receptor-like proteins and receptor-like kinases; direct (3) or indirect (4) intracellular detection of pathogen-derived molecules by nucleotide binding, leucine-rich repeat receptors, or detection through integrated domains (5); perception of transcription activator-like effectors through activation of executor genes (6); and active (7), passive (8), or host reprogramming-mediated (9) loss of susceptibility. Although the molecular mechanisms underlying the functions of R genes are only understood for a small proportion of known R genes, a clearer understanding of mechanisms is emerging and will be crucial for rational engineering and deployment of novel R genes.
INTRODUCTION
Plants can trigger an effective immune response to a wide variety of fungal, bacterial, and viral pathogens. The robustness of the plant immune system is remarkable, given that plants are sessile and unremittingly exposed to potential pathogens while their immunity does not rely on mobile immune cells and adaptive somatic variation, both of which are so effective in vertebrates.
In plants, resistance (R) genes play a key role in their remarkable immune responses. R genes are usually dominant (but sometimes recessive) genes that provide full or partial resistance to one or more pathogens. We include receptors of pathogen-associated molecular patterns (PAMPs) as R genes because they provide partial and sometimes even full resistance (Lacombe et al., 2010). R genes exist in natural plant populations and have been used by humankind since early crop domestication. Selection during domestication favored dominant R genes providing full resistance, but recessive R genes and R genes that provide partial resistance may provide more durable resistance. Most identified R genes are polymorphic in plant populations, which led to their initial characterization and use in plant breeding programs. However, individual plants have up to a few hundred R gene analogs that make no identified contribution to resistance. Many of these R gene analogs are also fixed in plant species and are thought to contribute to non-host resistance (Schulze-Lefert and Panstruga, 2011).
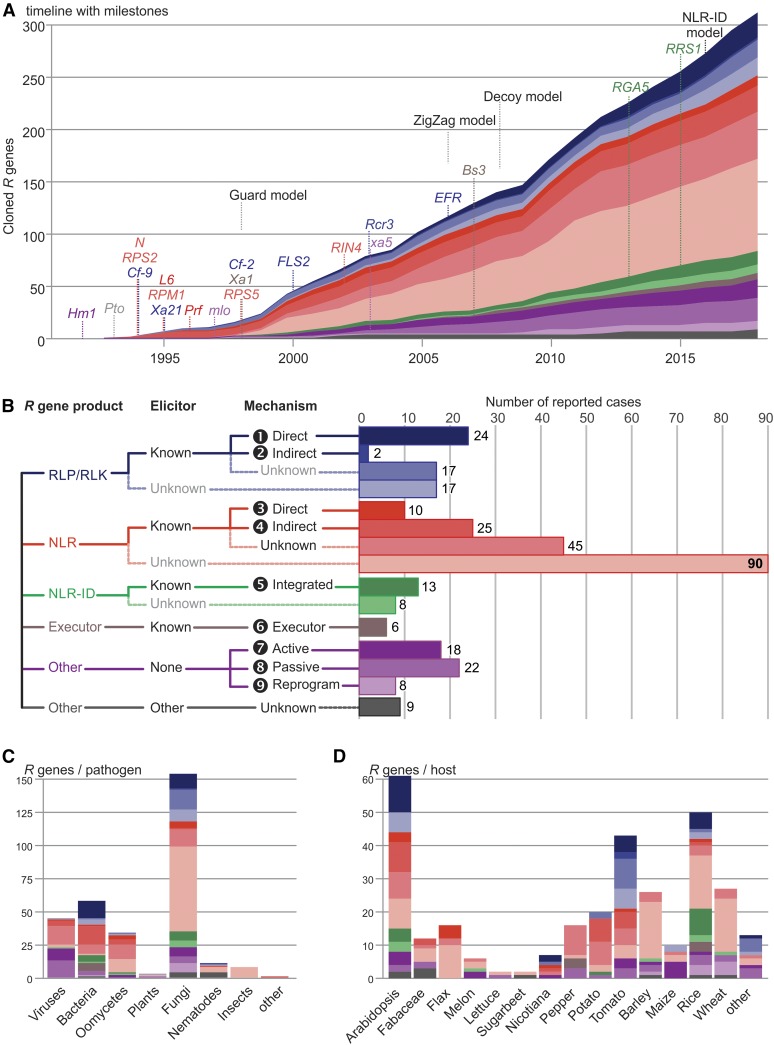
The cloning of the first R gene was published in 1992 and the number of cloned R genes has steadily increased over the subsequent 25 years (Figure 1A). The first cloned R gene, Hm1 from maize (Zea mays), encodes an enzyme that detoxifies Helminthosporium carbonum (HC) toxin from the fungal pathogen Cochliobolus carbonum (Johal and Briggs, 1992). The cloning of Hm1 was followed by the cloning of Pto from tomato (Solanum lycopersicum) in 1993 (Martin et al., 1993), and then, in 1994, Cf-9 from tomato (Jones et al., 1994), N from tobacco (Nicotiana tabacum; Whitham et al., 1994), and RPS2 from Arabidopsis thaliana (Bent et al., 1994; Mindrinos et al., 1994). Since then, hundreds of examples of cloned R genes have been published.
Figure 1.
Analysis of Molecular Mechanisms Underpinning R Genes.
A near-comprehensive literature search resulted in a database of R genes conferring immunity to a wide range of plant pathogens (Supplemental Data Set 1). This list was filtered to remove duplicates: R genes that code for receptors that perceive pathogen components were counted only once if this component is conserved between pathogens. By contrast, if an R gene codes for a receptor that perceives structurally or sequence unrelated pathogen-derived components, these were counted multiple times. Furthermore, alleles of R genes with slightly different recognition spectra were counted as multiple R genes, as were paralogs within species and orthologs in different species which are shown to be involved in immunity. Loss-of-susceptibility R genes that can act against several pathogens through an identical mechanism were counted only once. In addition, R genes that were shown to induce enhanced disease resistance when taken outside of their native context were removed, as were engineered R genes. Finally, some genes that are involved in autoimmunity resemble R genes, but these were not included unless these were proven to be involved in immunity against pathogens.
(A) A timeline summarizing the increased knowledge regarding R genes. The identified mechanisms were plotted cumulatively over time, with some of the milestones added. The date of the first cloned R gene was taken in case multiple host components are involved or the underlying molecular mechanism was elucidated later. These R genes were grouped according to the proposed mechanism of function as we understand it today.
(B) All identified R genes were grouped according to their proposed mechanism.
(C) The identified R genes were grouped by pathogen which they act against and colored by the molecular mechanism by which they function.
(D) The identified R genes were grouped by host species carrying them and colored by the molecular mechanism by which they function. Colors are explained in (B).
Now, after 25 years of R gene cloning, we look back and determine mechanistic trends in the function of R gene products. Through a comprehensive review, we identified 314 cloned functional R genes (Supplemental Table 1). A mechanism has been suggested for 128 of the 314 identified R gene products (41%; Table 1); among these, we distinguish nine distinct mechanisms (Table 1, Figure 1A), explained further in this review, and supported by examples.
Table 1. R Genes Classified by Nine Mechanisms.
| Mechanism | Description | R Genes (Plant Species) |
|---|---|---|
| 1: RLP/RLK, direct | Recognition triggered by direct interaction of a pathogen-derived effector and a cell surface RLK/RLP receptor. | EFR, FLS2, LORE, LYK3, LYK4, LYK5, LYM1/LYM3, LYM2, RBGP1, RLP23 (Arabidopsis), VvFLS2 (grapevine), NbCORE, NbFLS2 (N. benthamiana), CEBiP, LYP4/LYP6, OsFLS2 (rice), CORE, FLS3, LeEIX2, SlFLS2 (tomato) |
| 2: RLP/RLK, indirect | Recognition triggered either by effector binding to a host component or by effector-mediated modification of a host component, perceived by a cell surface RLK/RLP receptor. | Cf-2 (tomato) |
| Guardee/decoy: Rcr3 | ||
| RLP/RLK, unknown mechanism | Ve1 (tomato), StoVe1 (eggplant), HLVe1-2A (hop), NgVe1 (Nicotiana glutinosa), LepR3, RLM2 (oilseed rape), StuVe1, ELR (potato), XA21 (rice), 9DC1, 9DC2, 9DC3, Cf-4, Cf-5, Cf-9, Hcr9-4E, I, I-3 (tomato) | |
| 3: NLR, direct | Recognition triggered by direct interaction of a pathogen-derived component and an NLR. | RPP1-{EstA/Nda/ZdrA} (Arabidopsis), L5/L6/L7, M (flax), Roq1 (N. benthamiana), Pi-ta (rice), Sw-5b (tomato) |
| 4: NLR, indirect | Recognition triggered either by effector binding to a host component or by effector-mediated modification of a host component, perceived by an NLR. | HRT1, RPM1, RPS2, RPS5, SUMM2, ZAR1 (Arabidopsis), Gpa2, R2, R2-like, Rpi-abpt, Rpi-blb3, Rx1, Rx2 (potato), Rpg1-b, Rpg1r (soybean), N (tobacco), Prf (tomato) |
| Guardees/decoys: TIP, RIN4, PBS1, CRCK3, ZED1, ZRK3, RKS1, PBL2 (Arabidopsis), RanGAP2, BSL1 (soybean), GmRIN4 (tobacco), LescPth5, Fen, Pto (tomato) | ||
| NLR, unknown mechanism | RBA1, RCY1, RPP13-Nd-1, RPP13-UKID37, RPP13-UKID5, RPP5, RPS6, TAO1 (Arabidopsis), Mla1, Mla10, Mla13 (barley), Bs2 (black pepper), P, P2 (flax), Rxo1 (maize), Fom-2 (melon), L1, L1a, L1c, L2, L2b, L3, L4, Pvr4, Tsw (pepper), R3a, R3b, R8, Rpi-blb2, Rpi-vnt1.1, Rpi-vnt1.2, Rpi-vnt1.3 (potato), Pi9, Pib, Piz-t (rice), 3gG2 (soybean), N' (tobacco), Bs4, I2, Tm-2, Tm22 (tomato), Pm2, Pm3a, Pm3f (wheat) | |
| 5: NLR-ID | Recognition is triggered either by effector binding to a domain or by effector-mediated modification of a domain that is integrated in a host NLR. | RRS1B, RRS1-R, RRS1-S (Arabidopsis), R1 (potato), Pii-2, Pik-{1/h/p1/s}, RGA5-A, Xa1 (rice) |
| 6: Executor | Recognition triggered by transcriptional activation of the executor gene by a pathogen TAL effector. | Bs3, Bs3-E, Bs4C-R (pepper), Xa10, Xa23, Xa7 (rice) |
| 7: Other, active | Loss of susceptibility by directly disarming the pathogen by actively interrupting a key pathogenicity process. | JAX1, RTM1, RTM2, RTM3 (Arabidopsis), HvHm1 (barley), Hm1, Hm2, qMdr9.02, qRfg1, ZmTrxh (maize), At1, At2 (melon), IVR (N. benthamiana), PaLAR3 (Norway spruce), STV11-R (rice), Tm-1, Ty-1, Ty-3 (tomato) |
| 8: Other, passive | Loss of susceptibility by mutation in a host component, leading to the inability to manipulate the host. | rwm1, lov1 (Arabidopsis), Rym-4, Rym-5 (barley), retr01 (cabbage), bc-3 (French bean), Mo-1 (lettuce), sbm1 (pea), Pvr21, Pvr22, pvr6 (pepper), Eva1 (potato), xa13, xa25, xa5 (rice), LGS1, Pc (sorghum), Asc-1, Ty-5, Pot-1 (tomato), Tsn1, Snn1 (wheat) |
| 9: Reprogramming | Loss of susceptibility by a deregulated host. | mlo (barley), GH3-2, GH3-8, pi21 (rice), Lr34, Lr67, YrL693, Yr36 (wheat) |
Several R genes have multiple synonyms. This table summarizes only one name per cloned R gene. Only the name of the sensor NLR is mentioned in case of NLR-IDs. R genes that can perceive multiple sequence-unrelated effectors have been counted multiple times. Bold indicates key examples discussed in this work.
Many R genes confer recognition of pathogen-derived effectors and initiate effector-triggered immunity (ETI), which often involves the hypersensitive response (HR), a type of programmed cell death (Jones and Dangl, 2006). ETI is considered distinct from the other layer of induced immunity, PAMP-triggered immunity (PTI) (Jones and Dangl, 2006). In contrast to ETI, PTI involves recognition of conserved PAMPs and does not involve HR. In this review, we do not distinguish between PTI and ETI, as the PTI and ETI responses are very similar and the division has become hazy with the description of conserved effectors and polymorphic PAMPs (Thomma et al., 2011). We do, however, distinguish between extracellular and intracellular pathogen recognition mechanisms.
OVERVIEW AND OCCURRENCE OF THE NINE MECHANISMS
Although many cloned R genes have no proposed mechanism in the literature, they most likely fit within these nine mechanisms (Figure 1B). These mechanisms can broadly be divided into two clades: perception (I) and loss of susceptibility (II). Perception-based mechanisms can be further subdivided into three subclades: perception involving receptor-like proteins/kinases (RLPs/RLKs) (Ia); Nod-like receptors (NLRs) (Ib); and Executor genes (Ic). RLPs/RLKs in the apoplast can perceive PAMPs and effectors directly (subclade Ia, mechanism 1) or indirectly (subclade Ia, mechanism 2). Likewise, NLRs can perceive effectors directly (subclade Ib, mechanism 3) or indirectly (subclade Ib, mechanism 4), or through integrated domains (IDs) (subclade Ib, mechanism 5). Perception of transcription activator-like (TAL) effectors occurs by triggering expression of executor genes (subclade Ic, mechanism 6).
We identify three loss-of-susceptibility mechanisms (clade II): active loss of susceptibility by the expression of immune-related components or specific enzymes involved in detoxification/degradation of pathogen and pathogen-associated components (clade II, mechanism 7); passive loss of susceptibility through loss of interaction with key pathogen targets (clade II, mechanism 8); and loss of susceptibility by host-reprogramming leading to reduced pathogen growth (clade II, mechanism 9).
Of the cloned R genes, 61% encode NLRs (191/313), but for most NLRs the perception mechanism is either unclear (45/191) or the perceived component is unknown (98/191) (Figure 1B). The other main class of cloned R genes (19%, 60/314), encode RLPs/RLKs, which can recognize pathogen-derived components directly (24/60) or indirectly (2/60). Finally, several cloned R genes are executor genes (6/314) or lead to loss of susceptibility (48/314) (Figure 1B). Interestingly, with the exception of mechanism 6 (executor genes), the mechanisms are not specific for immunity against certain types of pathogens (Figure 1C) or only used by particular plant species or taxonomic groups (Figure 1D). Instead, these mechanisms are deployed against different kinds of pathogens and appear to be universally used in the plant kingdom. However, the distribution of these mechanisms may not reflect the natural frequencies of mechanisms because of a strong research bias toward model systems.
Subclade Ia: Extracellular Perception
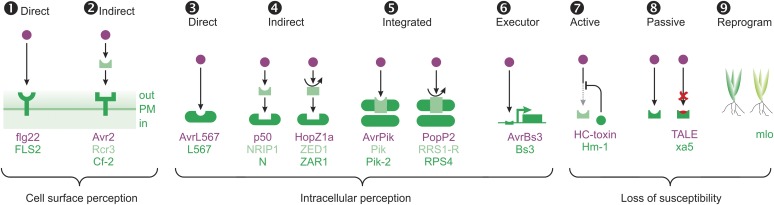
Plants perceive various PAMPs and effectors by R genes encoding for cell surface-localized RLPs or RLKs (Figure 2). This perception can be direct (mechanism 1) or indirect (mechanism 2).
Figure 2.
Nine Molecular Mechanisms Underpinning R Gene Functions.
Illustration of direct (1) and indirect (2) recognition at the cell surface; four different intracellular perception mechanisms (3–6); and three loss-of-susceptibility mechanisms (7–9). PAMPs and effectors are colored in purple, indirect receptors in light green, and direct receptors in dark green.
Mechanism 1: Direct Perception at the Cell Surface
Many PAMPs are perceived directly at the cell surface by RLPs and RLKs. The best studied PAMP in plants is bacterial flagellin (Felix et al., 1999). Flagellin fragment flg22 is perceived by the RLK FLAGELLIN-SENSITIVE2 (FLS2) in Arabidopsis (Gómez-Gómez and Boller, 2000). The flg22 peptide binds directly to FLS2 triggering the recruitment of BRI1-ASSOCIATED RECEPTOR KINASE (BAK1), a RLK (Chinchilla et al., 2007). The crystal structure of FLS2 in complex with BAK1 and flg22 has been elucidated, revealing that flg22 acts as a “molecular glue” inducing complex formation of FLS2 with BAK1 (Sun et al., 2013). FLS2 orthologs from different plant species perceive different parts of bacterial flagellin. For instance, tomato SlFLS2 recognizes a 15-amino acid epitope that overlaps with flg22 (Robatzek et al., 2007), whereas different flagellin epitopes (flgII-28 and CD2-1) are recognized by the RLK FLS3 in tomato and an uncharacterized receptor in rice (Oryza sativa), respectively (Hind et al., 2016; Katsuragi et al., 2015). Furthermore, FLS2 receptors from different plant species display different affinities for the conserved part of flagellin from different pathogens, possibly reflecting coevolution with specific bacterial pathogens (Trdá et al., 2014).
Many other microbial PAMPs are recognized directly similar to flagellin (reviewed in Zipfel, 2014). These include EF-Tu, chitin, lipopolysaccharide, peptidoglycan, and other components of the bacterial cell wall (Zipfel, 2014), as well as bacterial RNA (Lee et al., 2016), possibly by direct interaction with RLP/RLKs. Although often not considered as being R genes, PAMP-recognizing RLKs provide partial resistance and can even confer full resistance when transferred to other plant species (Lacombe et al., 2010).
Perception of extracellular effector proteins may also occur via direct interactions with RLPs or RLKs. For example, nlp20, a fragment of NECROSIS AND ETHYLENE-INDUCING PEPTIDE1 (NEP1), has been coimmunoprecipitated with RLP23 (Albert et al., 2015). NEP-LIKE PROTEINs (NLPs) are secreted by bacterial, oomycete, and fungal plant pathogens (Böhm et al., 2014; Oome et al., 2014). Many NLPs function in the necrotrophic phase of these pathogens and have a cytotoxic role. Noncytotoxic NLPs, on the other hand, are expressed early during infection and serve an unknown role during the initial stages of host colonization (Oome et al., 2014). Both cytotoxic (Böhm et al., 2014) and noncytotoxic NLPs (Oome et al., 2014) trigger immune responses via RLP23 through the conserved nlp20 fragment (Albert et al., 2015). RLP23 requires the RLKs SUPPRESSOR OF BIR1 1 and BAK1 for signaling, triggering a weak immune response (Albert et al., 2015).
A direct interaction has been demonstrated for the perception of the fungal ethylene-inducing xylanase (EIX) effector by the tomato RLP LeEIX2 (Ron and Avni, 2004; Rotblat et al., 2002). Expression of LeEIX2 in mammalian cells enables binding of EIX (Ron and Avni, 2004). Furthermore, several fungal endopolygalacturonases are perceived by the Arabidopsis RLP RBPG1 (Zhang et al., 2014), and these enzymes can be coimmunoprecipitated with RBPG1 (Zhang et al., 2014). In addition, perception of EIX and fungal endopolygalacturonases by LeEIX2 and RBPG1, respectively, is independent of their enzymatic functions (Rotblat et al., 2002; Zhang et al., 2014). A direct interaction is thought to also occur for the RaxX effector from Xanthomonas oryzae, which is perceived by the RLK encoded by the rice R gene Xa21 (Pruitt et al., 2015). Likewise, the Verticillium-derived effector AVE1 is perceived by the tomato R gene Ve1, which encodes an RLP (de Jonge et al., 2012). However, no direct interaction has been shown for these effectors and their cognate Xa21 and Ve1 receptors.
Taken together, these examples indicate that direct perception of PAMPs and effectors at the cell surface is a common strategy underlying the partial and full disease resistance provided by R genes.
Mechanism 2: Indirect Extracellular Effector Perception
Pathogen perception at the cell surface can also occur indirectly, by recognition of modified host factors. The key example of this mechanism is the recognition of the fungal pathogen Cladosporium fulvum (syn. Passalora fulva) by the tomato R gene product Cf-2, a RLP which has been introgressed into cultivated tomato from Solanum pimpinellifolium (Dixon et al., 1996). Cf-2 confers recognition of the fungal effector Avr2 from C. fulvum (Luderer et al., 2002), as well as the unrelated nematode effector GrVap1 from Globodera rostochiensis (Lozano-Torres et al., 2012). Avr2/GrVap1 perception requires Rcr3, which encodes a secreted papain-like Cys protease (Dixon et al., 2000). Both Avr2 and GrVAP1 function as protease inhibitors and directly interact with Rcr3 (Lozano-Torres et al., 2012; Rooney et al., 2005). Avr2 and GrVap1 act as virulence factors (van Esse et al., 2008; Lozano-Torres et al., 2014), likely by inhibition of various host proteases (Kaschani et al., 2010; Lozano-Torres et al., 2012, 2014; Shabab et al., 2008), including the Rcr3 paralog Pip1 (Shabab et al., 2008). These data suggest that Rcr3 might act as a decoy for the perception of Avr2 and GrVap1 (van der Hoorn and Kamoun, 2008). However, tomato lines lacking a functional Rcr3 allele are more susceptible to Phytophthora infestans, but do not display an altered susceptibility toward C. fulvum or Pseudomonas syringae (Ilyas et al., 2015). This indicates a role for Rcr3 beyond effector recognition. A second argument against Rcr3 being a decoy is that close Rcr3 orthologs are present in pepper (Capsicum annuum), potato (Solanum tuberosum), and tomato (Ilyas et al., 2015), whereas Cf-2 is probably an evolutionarily much younger gene present only in tomato (Dixon et al., 1996; Seear and Dixon, 2003).
So far, Avr2/GrVap1 perception by Cf-2 and Rcr3 is the only example of indirect perception of effectors in the apoplast. However, perception of other effectors in the apoplast might also be indirect. For example, a direct interaction of tomato RLP Cf-9 with the C. fulvum effector Avr9 could not be detected (Luderer et al., 2001), and Avr9 perception seems to involve an additional Solanaceae-specific factor known as the high-affinity binding site for Avr9 (Kooman-Gersmann et al., 1996, 1998). Likewise, perception of the Fusarium oxysporum Avr3 effector by the tomato RLK I-3 may require an additional host component present in tomato, but absent in tobacco and Nicotiana benthamiana (Catanzariti et al., 2015).
Subclade Ib: Intracellular Perception
In addition to extracellular perception of pathogens, plants are able to recognize effectors inside the cell (Figure 2). Most cloned R genes encode NLRs, which can perceive effectors either directly (mechanism 3) or indirectly (mechanism 4). However, some R genes encode NLRs containing additional IDs (NLR-IDs) required for effector recognition (mechanism 5) (Kroj et al., 2016; Sarris et al., 2016). Finally, some effectors activate executor genes (mechanism 6).
Mechanism 3: Direct Intracellular Effector Recognition—Trigger-Happy NLRs
Direct perception of effectors is not restricted to the cell surface, as several effectors have been convincingly demonstrated to directly interact with NLRs to trigger immune responses. ARABIDOPSIS THALIANA RECOGNIZED1 (ATR1), for example, is an effector from the oomycete pathogen Hyaloperonospora arabidopsidis, and ATR1 directly interacts with the Arabidopsis NLR RECOGNITION OF PERONOSPORA PARASITICA1 (RPP1), leading to its recognition (Krasileva et al., 2010). Various RPP1 alleles have distinct recognition specificities for different ATR1 alleles, and this specificity is mediated by the LRRs of RPP1 (Steinbrenner et al., 2015). The AvrL567 effector from the fungal pathogen Melampsora lini is another effector that directly interacts with its cognate receptor, the flax L5, L6, and L7 NLRs, which are encoded by allelic genes (Dodds et al., 2004, 2006). Different variants of AvrL567 are perceived differentially by these receptors (Dodds et al., 2006; Wang et al., 2007), and the specificity for these AvrL567 variants resides in the LRR domain of the NLR (Ravensdale et al., 2012; Wang et al., 2007).
Interestingly, effector perception is also affected by cooperative polymorphisms in other domains of the NLR besides the LRR (Ravensdale et al., 2012). This suggests a mechanism of perception where effector binding competes with intramolecular interactions (Ravensdale et al., 2012). The L5, L6, and L7 receptors exist in an equilibrium between “on” and “off” states, and effector binding stabilizes the “on” form, thereby initiating immune signaling (Bernoux et al., 2016). This equilibrium model may be more widely applicable to NLR signaling. Indeed, the NLR Sw-5b directly recognizes a small 21-amino acid PAMP-like peptide region from the viral movement protein NSm (NSm21), and it has been argued that binding of NSm21 destabilizes the interaction between the LRR and NB-ARC domain triggering recognition (Zhu et al., 2017).
In the case of RPP1 and L5/L6/L7, allelic NLR-encoding genes perceive allelic effector variants. However, it is not always the case that homologous and/or allelic NLRs perceive homologous effector proteins. For example, rice and barley (Hordeum vulgare) have homologous NLRs residing in the same R gene cluster; these NLRs perceive sequence-unrelated effectors from Magnaporthe oryzae and Blumeria graminis, respectively (Lu et al., 2016; Wu et al., 2015). In addition, potato orthologs of two different tomato NLR-encoding genes, tomato Sw-5b, which perceives a viral effector, or tomato I2, which perceives a fungal effector, can perceive oomycete effectors (Giannakopoulou et al., 2015; Vossen et al., 2016). The exact molecular mechanism by which sequence-unrelated effectors from different pathogens can be perceived directly by highly similar NLRs is unknown. In many cases, the specificity-determining region in the NLR is in the C-terminal LRR region (Shen et al., 2003), while in other cases mutations in regions other than the NLR are sufficient to alter specificity (Giannakopoulou et al., 2015). Therefore, it seems that small changes in several parts of the NLR can sensitize the NLR to destabilization by sequence-unrelated effectors, triggering recognition. In addition, evolving a different specificity to sequence-unrelated effectors may be facilitated by the fact that these effectors might adopt a similar fold. Indeed, several sequence-unrelated effectors from M. oryzae have been shown to adopt a similar fold (de Guillen et al., 2015; Maqbool et al., 2015). A structural understanding of effector binding by these NLRs would provide clues to the exact molecular mechanism of direct perception, but these experiments are notoriously challenging.
Mechanism 4: Indirect Intracellular Recognition—Decoys and Guardees
Many effectors are perceived indirectly by NLRs, through their interaction with—or enzymatic modification of—additional host components. These additional host proteins have also been called guardees or decoys to suggest that they are effector targets or mimics of effector targets, respectively.
An example of indirect interaction is the recognition of the 50-kD helicase (p50) domain of Tobacco mosaic virus by the tobacco NLR N, which requires NRIP1, a chloroplast-localized rhodanese sulfurtransferase (Caplan et al., 2008). NRIP1 is recruited to the cytoplasm by the p50 effector, and NRIP1 only interacts with N in the presence of the p50 effector. It is thought that N activation requires a prerecognition complex of NRIP1 and the p50 effector (Caplan et al., 2008).
An example of indirect detection via enzymatic host modification is the perception of the P. syringae Type-III effectors HopZ1a (Lewis et al., 2010) and HopF2a (Seto et al., 2017), and the Xanthomonas campestris Type-III effector AvrAC (syn. XopAc) (Wang et al., 2015b; Xu et al., 2008) by the functionally conserved NLR ZAR1 (Baudin et al., 2017; Lewis et al., 2010; Peele et al., 2014). HopZ1a is perceived through its acetyltransferase activity on the PM-localized pseudokinase HOPZ-ETI-DEFICIENT1 (ZED1), a receptor-like cytoplasmic kinase (RLCK) XII family member (Lewis et al., 2013). AvrAC is perceived through its activity on the RLCK VII family member PBL2 (Wang et al., 2015b). When PBL2 is uridylylated by AvrAC, it interacts with the ZED1-like RKS1, which in turn forms a stable complex with HOPZ-ACTIVATED RESISTANCE1 (ZAR1), leading to a weak HR (Wang et al., 2015b). HopF2a is an ADP-ribosyltransferase and its recognition requires ZAR1 and the ZED1-like ZRK3, which may act as an adaptor for effector recognition similar to RKS1 (Seto et al., 2017). ZED1 and PBL2 are thought to act as decoys for the perception of HopZ1a or AvrAC, respectively. Indeed, ZED1 does not contribute to immunity or susceptibility to P. syringae in the absence of ZAR1, indicating that ZED1 acts as a decoy (Lewis et al., 2013). By contrast, PBL2 is not a strict decoy, as it appears to be required for perception of certain PAMPs, including flg22 and elf18 (Zhang et al., 2010). It is yet undetermined whether PBL2 contributes to resistance or susceptibility to X. campestris in the absence of ZAR1. The exact molecular mechanism by which ZAR1 perceives modulation of various RLCKs is unknown. ZAR1 is a classic example of how one R gene product can perceive the action of multiple effectors from multiple pathogens through interactions with several host components, all of which may be able to recruit additional components.
Another functionally conserved NLR that indirectly perceives multiple effectors is tomato Prf (Bombarely et al., 2012; Salmeron et al., 1996; reviewed in Ntoukakis et al., 2014). Prf interacts with several RLCK VII family members, including Pto and Fen (Salmeron et al., 1996). Interaction with Pto leads to perception of the P. syringae Type-III effectors AvrPto and AvrPtoB, which bind Pto (Kim et al., 2002). AvrPto acts as a kinase inhibitor (Xing et al., 2007), while AvrPtoB acts as a weak kinase inhibitor and has an additional E3 ligase domain to target its interactors for destruction by the host proteasome (Cheng et al., 2011; Rosebrock et al., 2007). Prf also interacts with the Pto-homolog Fen, leading to immune signaling in response to the insecticide Fenthion (Martin et al., 1994). It is thought that Prf oligomerizes with several molecules of Pto and/or Pto homologs and that effector binding to one of these kinases in this complex would induce a series of transphosphorylation events resulting in an immune response (Ntoukakis et al., 2014). The oligomerization with different Pto homologs within one receptor complex could expand the recognition spectrum of Prf (Ntoukakis et al., 2014).
The NLRs ZAR1 and Prf each interact with multiple host proteins; by contrast, RPM1 INTERACTING PROTEIN4 (RIN4) is a host protein that interacts with multiple NLRs. RIN4 forms complexes with the NLRs RPS2 and RPM1 (Axtell and Staskawicz, 2003; Mackey et al., 2002). The P. syringae Type-III effectors AvrB and AvrRpm1 induce phosphorylation of RIN4 by recruiting the Arabidopsis RIPK protein kinase (Liu et al., 2011a; Mackey et al., 2002). The phosphorylation of RIN4 by RIPK at a conserved threonine residue (T166) reduces RIN4 interaction with the prolyl-peptidyl isomerase ROTAMASE CYP1 (ROC1) leading to an altered conformation of RIN4, which triggers RPM1-mediated immune signaling (Li et al., 2014). In the absence of RPM1, the phosphorylation of RIN4 enhances its role as a negative regulator of immunity (Lee et al., 2015; Liu et al., 2009; Wilton et al., 2010), indicating RIN4 is not a decoy. Cleavage of RIN4 by the P. syringae Type-III effector AvrRpt2 inhibits RPM1-mediated immune signaling (Kim et al., 2005). This cleavage requires AvrRpt2 maturation by eukaryotic prolyl-peptidyl isomerases such as ROC1 (Coaker et al., 2005), which is required for RPM1-mediated immune signaling. In response, the NLR RPS2 perceives RIN4 cleavage, triggering immune signaling (Axtell and Staskawicz, 2003). RIN4 binding to RPS2 keeps RPS2 in an “off” state, and cleavage releases the inhibition of RPS2 triggering immune signaling (Day et al., 2005). In addition, RPS2 weakly perceives AvrRpm1 (Kim et al., 2009). In the absence of RPM1, AvrRpt2-dependent cleavage of RIN4 does not contribute to virulence (Lim and Kunkel, 2004), indicating that in the absence of AvrB or AvrRpm1, RIN4 acts as a decoy for AvrRpt2. Soybean (Glycine max) has independently converged on a similar perception mechanism, where the soybean NLRs RPG1-b and RPG1r perceive the manipulation of the soybean RIN4 ortholog by AvrB and AvrRpm1, respectively (Ashfield et al., 2004; Kessens et al., 2014; Selote and Kachroo, 2010).
Other NLRs also perceive effectors indirectly using additional host proteins. The NLR RPS5, for example, perceives the P. syringae Type-III effector AvrPphB (Warren et al., 1998), a family C58 cysteine protease (Zhu et al., 2004). AvrPphB contributes to virulence by cleaving several RLCKs (Zhang et al., 2010), including RIPK, thereby inhibiting RPM1-mediated recognition of AvrB (Russell et al., 2015). The perception of AvrPphB by RPS5 requires PBS1, which encodes a RLCK VII family member (Ade et al., 2007; Shao et al., 2003; Swiderski and Innes, 2001). PBS1 binds to RPS5 to form a prerecognition complex (DeYoung et al., 2012), and subsequent cleavage of PBS1 by AvrPphB triggers a subtle conformational change that allows an exposed loop in PBS1 to trigger activation of RPS5 (DeYoung et al., 2012; Qi et al., 2014). It remains to be demonstrated whether PBS1 cleavage by AvrPphB contributes to the virulence of P. syringae in the absence of RPS5 and therefore whether PBS1 is a decoy. However, PBS1 may not be a decoy, as PBS1 contributes to PAMP-triggered immune responses (Zhang et al., 2010) and is under strong purifying selection, even though not all Arabidopsis accessions carry RPS5 (Caldwell and Michelmore, 2009), indicating a physiological function for PBS1 in the absence of RPS5.
Finally, sometimes NLRs monitor cellular homeostasis. The Arabidopsis NLR SUMM2, for example, perceives the phosphorylation status of the calmodulin binding RLCK CRCK3 (Zhang et al., 2017). CRCK3 is phosphorylated by a MAP kinase signaling cascade involving the MAP kinases MEKK1, MKK1, and MKK2, as well as MPK4 (Zhang et al., 2017). The CRCK3 phosphorylation status in the absence of MPK4, MKK1/MKK2, or MEKK1 kinase activity triggers an immune response that is dependent on SUMM2 (Zhang et al., 2012). The P. syringae Type-III effector HopAI1 inactivates MPK4, thereby triggering an immune response that is dependent on SUPPRESSOR OF MKK1 MKK2 2 (SUMM2; Zhang et al., 2012). SUMM2 is therefore an example of an NLR monitoring cellular homeostasis. Perception of an alteration of homeostasis, rather than the actual effector target itself, may allow for the perception of many effectors whose effects ultimately converge on the same host components. CRCK3 has no known function beyond its signaling role through SUMM2, suggesting it might act as a decoy (Zhang et al., 2012).
In conclusion, NLRs often trigger immune signaling by monitoring either (1) the interaction of effectors with host proteins, (2) the enzymatic modification of host proteins, or (3) cellular homeostasis. More bacterial effectors have been shown to be perceived indirectly by NLRs than effectors of other classes of pathogens (Figure 1C). This likely reflects a better understanding of bacterial effectors as well as the fact that bacteria are easier to manipulate, rather than a biological difference in how these different types of pathogens are perceived by plants. Although there are several clear candidates acting as guardees (RIN4 and PBL2) or decoys (ZED1 and Pto), it is unclear in other cases (PBS1 and CRCK3). Furthermore, several of these proposed decoys seem more conserved than the corresponding NLR.
Mechanism 5: NLR-IDs—Sensors Embedded within Receptors
Some NLRs carry integrated domains (NLR-IDs) (Kroj et al., 2016; Sarris et al., 2016) thought to be required for effector recognition. As these additional domains seem to have integrated repeatedly and recently, it is not unlikely they still possess their original activity in the absence of the recognized effector, unlike a decoy (Sarris et al., 2016). Furthermore, it remains to be demonstrated on a case-by-case basis whether the ID is required for effector perception. Three NLR-IDs have been studied in detail: Arabidopsis RRS1 and rice RGA5 and Pik. Remarkably, these NLR-IDs are coexpressed from the same promoter in opposing orientation with a NLR lacking the ID, and heterodimerization of these paired, coexpressed NLR proteins is required for signal transduction.
Arabidopsis RRS1 is a NLR containing an integrated C-terminal WRKY transcription factor domain and cooperates with the NLR RPS4 to trigger effector recognition. The WRKY domain of RRS1 interacts with the P. syringae Type-III effector AvrRps4, triggering RPS4-dependent immune responses (Sarris et al., 2015). PopP2 is a Ralstonia solanacearum Type-III effector of the YopJ family that acts as an acetyltransferase, acetylating key lysines in the RRS1 WRKY domain. In the Col-0 allele of RRS1 (RRS1-S), this acetylation blocks AvrRps4 recognition (Sarris et al., 2015), while the Nd-1 and Ws-2 alleles of RRS1 (RRS1-R) signal in response to this acetylation, in addition to recognizing AvrRps4 (Le Roux et al., 2015; Sarris et al., 2015). Furthermore, the RRS1-R allele recognizes a yet unidentified effector from the fungus Colletotrichum higginsianum (Sarris et al., 2015), and the RPS4-RRS1 locus also underlies a quantitative trait locus for resistance against X. campestris (Debieu et al., 2016). Therefore, RRS1 can recognize effectors from various bacterial and fungal pathogens. Furthermore, RRS1-R can perceive the direct interaction of AvrRps4 with its integrated WRKY domain, as well as perceive the enzymatic modification of this domain by PopP2. The biological significance of the RRS1 WRKY domain binding to W-box DNA sequences (Sarris et al., 2015) outside of the context of perceiving pathogen effectors is not known. Remarkably, AvrRps4 is also perceived in a similar way in Arabidopsis by a homologous pair of NLRs, designated RRS1B (containing a WRKY ID), and RPS4B (Saucet et al., 2015).
The rice RGA5 and Pik NLRs both contain an integrated RATX1 or HMA domain, which is related to the Saccharomyces cerevisiae copper binding protein ATX1 (Cesari et al., 2013). The HMA domain is located at the C terminus of RGA5 and between the CC and NB-ARC domain in Pik, representing independent integration events. RGA5 interacts with the M. oryzae effectors Avr-Pia and Avr-CO39 using its HMA-domain, and this triggers an RGA4-dependent immune response (Cesari et al., 2013). Alleles of the M. oryzae effector Avr-Pik bind to the HMA domain of Pik alleles (Maqbool et al., 2015), which together with Pik-2 triggers immune signaling (Ashikawa et al., 2008). Substitutions in the HMA-domain of different Pik alleles revealed a correlation between the recognition specificity of these different Pik alleles and the different Avr-Pik variants (Maqbool et al., 2015).
In conclusion, perception by NLR-IDs can be the result of direct binding of the effector or due to enzymatic activity of the effector on the integrated domain. In addition, studying NLR-IDs can provide valuable insight into the molecular targets of effectors, as these integrated domains likely represent domains targeted by these effectors (Kroj et al., 2016). All studied NLR-IDs require a genetically linked NLR to trigger signaling. However, genetically unpaired NLRs can also cooperate. In Arabidopsis, tobacco, and tomato, unlinked “helper” NLRs have been shown to act downstream of several canonical “sensor” NLRs (Bonardi et al., 2011; Peart et al., 2005; Wu et al., 2017). In addition, helper-NLRs are required for effector perception by RLPs (Fradin et al., 2009; Gabriëls et al., 2007), thereby linking effector perception in the apoplast to intracellular signaling by NLRs.
Mechanism 6: Executor Genes—Promoter Traps for TAL Effectors
Executor genes are R genes that are transcriptionally activated by transcription activator-like effectors (TALEs) produced by Xanthomonas species and confer immunity to the Xanthomonas strains carrying these TALEs. TALEs function by binding to specific DNA sequences, altering host transcription of key susceptibility factors. Executor genes function as a promoter trap for these TALEs, forcing them to promote transcription of genes involved in immunity. Promoters of executor genes therefore act as decoys, mimicking the promoter regions of these susceptibility factors, leading to the initiation of immune responses (van der Hoorn and Kamoun, 2008). So far, six executor genes have been cloned: rice Xa27 (Gu et al., 2005), Xa10 (Tian et al., 2014), and Xa23 (Wang et al., 2015a), and pepper Bs3/Bs3-E (Römer et al., 2007), and Bs4C-R (Strauss et al., 2012). Identified executor genes either encode for proteins with a catalytic function (Bs3 and Bs3-E encode for a putative flavin monooxygenase; Römer et al., 2007) or for proteins with multiple putative transmembrane domains (Xa27, Xa10, Xa23, and Bs4C-R) (Zhang et al., 2015). Importantly, increased understanding of the specificity of DNA binding by TALEs has enabled the development of synthetic executor genes mediating immunity against multiple Xanthomonas strains (Hummel et al., 2012; Römer et al., 2009; Zeng et al., 2015). This strategy could be used to engineer resistance against the pathogen R. solanacearum, which also expresses TALEs, known as RipTALs (de Lange et al., 2013).
Clade II: Loss of Susceptibility
Not all R genes encode proteins involved in mechanisms relying on perception. Several R genes rely on loss of susceptibility, sometimes also called susceptibility (S) genes (van Schie and Takken, 2014). Loss of susceptibility can occur in three ways (Figure 2): active (mechanism 7), passive (loss of interaction; mechanism 8), or by host reprogramming (by mutations in components of cellular pathways; mechanism 9). Many loss-of-susceptibility mechanisms of resistance are very durable, but can carry a cost for the host and therefore can lead to a yield penalty. When active and passive loss-of-susceptibility mechanisms become fixed in populations, they lead to Type-I non-host resistance, typically characterized by the absence of a strong immune response (Mysore and Ryu, 2004).
Mechanism 7: Active Loss of Susceptibility—Blocking Key Pathogen Strategies
R genes underlying active loss-of-susceptibility mechanisms encode host proteins that are directly involved in disarming the pathogen by actively interrupting a key pathogen process. Active loss-of-susceptibility mechanisms are diverse and can act broadly against many pathogens, or be specific to some. These active mechanisms are often constitutively expressed, but some can be upregulated upon immune signaling. In addition, active loss-of-susceptibility mechanism may also lead to the production of PAMPs and danger-associated molecular patterns (DAMPs), which are then perceived and amplify the original response. For example, Hm1, the first R gene cloned (Johal and Briggs, 1992), encodes a NADPH-dependent reductase that is specifically involved in detoxifying HC toxin (Johal and Briggs, 1992). HC toxin is produced by C. carbonum race 1 (CCR1), the cause of leaf blight and ear mold in maize. Orthologs of Hm1 are present in the grass family, and in barley such orthologs contribute to non-host resistance to CCR1 (Sindhu et al., 2008). Many different active loss-of-susceptibility mechanisms of resistance are employed against viruses. For example, the tomato Tm-1 gene product, which confers resistance to Tomato mosaic virus inhibits the replication of Tomato mosaic virus RNA by binding to replication proteins (Ishibashi et al., 2014). In addition, the allelic tomato Ty-1/Ty-3 resistance genes encode for γ-clade RNA-directed RNA polymerases, which induce RNA-directed DNA methylation to provide immunity against single strand DNA geminiviruses (Butterbach et al., 2014; Verlaan et al., 2013).
Mechanism 8: Passive Loss of Susceptibility—Losing Interaction with Key Host Targets
Loss of interaction of a key host susceptibility factor with a pathogen effector is a common mechanism underpinning recessive R genes. Indeed, half of the known R genes against viruses involve such a loss of interaction (Truniger and Aranda, 2009). The majority of the so far identified recessive R genes act against potyviruses (Kang et al., 2005) and encode mutant translation initiation factors of the 4E or 4G family, which are unable to interact with the cap structure on viral transcripts, thereby conferring resistance to potyviruses (Truniger and Aranda, 2009). In addition, one-third of the R genes against X. oryzae pv oryzae, the causal agent of bacterial blight, are inherited recessively (Liu et al., 2011b), and the underlying mechanisms seems often to involve loss of interaction with key host targets. A recessive mutation in the promoter of the rice xa13 allele does no longer facilitate its manipulation by the TALE AvrXa13, leading to loss of susceptibility (Chu et al., 2006). Likewise, a single amino acid mutation in the rice gamma subunit of transcription factor IIa (OsTFIIAγ5), encoded by the recessive xa5 resistance gene, confers resistance to various X. oryzae strains in adult plants (Iyer and McCouch, 2004) by restricting bacterial movement (Iyer-Pascuzzi et al., 2008). OsTFIIAγ5 directly interacts with TALEs and is required for their activity (Yuan et al., 2016). The xa5 mutation likely affects interaction with TALEs to varying degrees, thereby conferring varying degrees of resistance to different X. oryzae pathovars (Gu et al., 2009; Huang et al., 2016; Yuan et al., 2016).
Mechanism 9: Passive Loss of Susceptibility by Host Reprogramming
Host reprogramming by mutations in components of cellular pathways is a common strategy leading to durable resistance against a broad range of pathogens. This form of loss of susceptibility is often a recessive trait, but may also involve dominant-negative alleles. The genes involved in this category are often called Adult Plant Resistance (APR) genes, as the resistance they confer is often only effective in later stages of plant life (Ellis et al., 2014). Furthermore, APR genes often confer partial resistance against a broad range of pathogens (Ellis et al., 2014). It is important to note that the mechanisms underlying this type of resistance do not necessarily act only in the adult plant; therefore, the term APR genes does not fully cover this category. Furthermore, perception-based mechanisms as well as loss-of-interaction mechanisms may only function in the adult plant.
The prime example of a senescence-associated loss-of-susceptibility mechanism is mediated by recessive loss-of-function mlo (Mildew Locus O) alleles in both monocots and dicots (Hückelhoven and Panstruga, 2011). MLO encodes a plant-specific integral membrane protein with an unknown molecular function that acts as a negative regulator of cell death in response to both biotic and abiotic stress (Piffanelli et al., 2002). MLO loss-of-function alleles are associated with spontaneous cell death (Büschges et al., 1997). In both barley and Arabidopsis, MLO is coexpressed with PEN1, PEN2, and PEN3 (or its barley orthologs), which are required for an active response against powdery mildews (Humphry et al., 2010). MLO acts as a negative regulator of the PEN1/PEN2/PEN3 pathways (Humphry et al., 2010), and in both barley and Arabidopsis, these genes are required for mlo-dependent immunity (Humphry et al., 2006). Therefore, loss of a general suppressor of cell death can confer specific resistance to powdery mildews in both monocots and dicots by deregulation of the PEN1 and PEN2/PEN3 pathways. This principle has been used to engineer a transgene-free tomato mlo mutant resistant to powdery mildews (Nekrasov et al., 2017).
Analogous to the mlo mechanism, the recessive resistance toward rice blast is conferred by a loss-of-function mutation in Pi21, a gene encoding a HMA domain protein that acts to suppress the plant defense response (Fukuoka et al., 2009). It is interesting to note that HMA domain proteins are crucial virulence targets for different pathogenic fungi and that several sequence-unrelated effectors adopt a similar fold that converge on HMA domain proteins (de Guillen et al., 2015; Maqbool et al., 2015). Therefore, engineering loss-of-function alleles of HMA domain-encoding genes analogous to pi21 may confer broad resistance to various pathogenic fungi in unrelated plant species. Other genes conferring disease resistance through senescence-associated mechanisms include wheat (Triticum aestivum) Lr34, which encodes an ABC transporter and confers dominant adult resistance to leaf rust, stripe rust, and powdery mildew (Krattinger et al., 2009). In addition, wheat WKS1 (syn. Yr36) encodes a kinase and START domain-containing protein that confers dominant partial resistance to a variety of stripe rusts (Fu et al., 2009). WKS1 functions by reducing the ability of chloroplast thylakoid ascorbate peroxidases (tAPXs) to detoxify peroxides, which contributes to cell death (Gou et al., 2015). Finally, wheat Lr67 is a dominant R gene that confers partial adult resistance against all races of leaf rust, stripe rust, stem rust, and powdery mildew and is caused by a mutation in a predicted hexose transporter that differs by two amino acids from the susceptible allele (Moore et al., 2015). Lr67 displays a dominant-negative effect through heterodimerization with the susceptible allele product, reducing glucose uptake (Moore et al., 2015), ultimately leading to leaf tip necrosis (Herrera-Foessel et al., 2014). The resistance these genes confer is seemingly dependent on a deregulated initial immune response, resulting in a quicker and stronger immune response that is able to partially suppress the pathogen. However, it is possible that some of these genes in fact act through a different molecular mechanism.
DISCUSSION
Since the cloning and molecular characterization of the first R genes in the early 1990s, a steady amount of R genes and interactors has been identified (Figure 1A). Even still, while Phytozome lists ∼14 000 NLR-encoding genes, only 191 have been shown to act as R genes (Goodstein et al., 2012). With the advance of novel techniques such as SMRT RenSeq and other next-generation sequencing-based techniques (Steuernagel et al., 2016; Witek et al., 2016), the number of annotated R genes is expected to increase at a faster pace than before. Understanding the molecular mechanisms underlying these R genes, however, will still require careful case-by-case examination. Indeed, of the examined 314 R genes, only 128 have a proposed mechanism, based on an even lower number of detailed studies into molecular mechanisms. The underlying mechanisms of the remaining 186 R genes is yet unknown. Understanding these molecular mechanisms in addition to discovering more R genes is vital, as it allows us to transfer these traits to other species as well as rationally engineer disease resistance, thereby extending the recognition spectra of R genes to outside of what is found in nature. For instance, engineering of the AvrPphB cleavage site in PBS1 to that of other pathogen-derived proteases leads to their recognition in an RPS5-dependent manner (Kim et al., 2016). Likewise, Executor R genes have been engineered to increase their recognition spectra (Hummel et al., 2012; Römer et al., 2009; Zeng et al., 2015). In addition to engineering novel R genes, high-throughput screens with effectors on non-host plants can identify R genes conferring non-host resistance, which may be very durable when deployed in susceptible hosts.
Critically, the distinction between direct and indirect interactions between the pathogen-derived elicitor and the R gene product is not as easily defined as it seems, both experimentally and conceptually. Direct interactions are not necessarily proven by bimolecular fluorescence complementation, fluorescence resonance energy transfer, or coimmunoprecipitations as the detected complex may contain additional host components required for the interaction. Likewise, even interactions in yeast two-hybrid assays do not exclude conserved eukaryotic factors facilitating the interaction. Direct interactions are best proven with purified proteins and by including noninteracting mutant proteins as controls. A direct interaction is confirmed in vivo when these mutations also disrupt the interaction in vivo. These assays are challenging and have often not been performed. Second, the requirement of an additional host protein for the interaction does not exclude that there is a physical interaction between the R gene product and the pathogen-derived effector, for instance, when a host protein acts as a molecular glue to enhance the direct interaction. Therefore, caution must be taken in the interpretation of results.
We have frequently discussed the compliance with guard and decoy models in mechanisms 3, 5, and 6. The molecular mechanistic interpretation of the difference between the guard and decoy model boils down to the question of whether a putative decoy has a function in the absence of the R gene. However, even when a guard/decoy is shown to have a role in the absence of the R gene, this may result from a decoy function, e.g., when several R gene products converge on guarding the same decoy. Similarly, a decoy may still have a function in a pathogen interaction in the absence of a perceived effector if multiple independent effectors from that pathogen converge on the same decoy triggering recognition. Therefore, mechanistically it may be better to refer to a guarded protein acting as a decoy when there is no clear virulence role in targeting the guard/decoy in the absence of an R gene in a given pathogen interaction. In other words, decoys only function in specific interactions where they act as coreceptors by mimicking the elicitor target.
It is also interesting to point out genes that have not been found among cloned R genes. First, signaling components of defense pathways, such as EDS1, NPR1, etc., are not among cloned R genes. It might be that these genes are not polymorphic in nature as they are central hubs in interconnected signaling networks, and alterations in these signaling components could disrupt multiple pathways. R genes therefore seem to be at the edges of signaling pathways. Second, no pathogenesis-related (PR) genes have been found to underlie R genes. This could be because most PR genes more likely function in immunity in an additive and quantitative manner. Finally, no R gene encoding for DAMPs or their receptors have been identified. DAMPs are endogenous “danger” signals that are produced or released upon cellular stress, cellular damage, or nonphysiological cell death and can be involved in either triggering or reinforcing immune signaling (Yamada et al., 2016). Because pathogens can cause DAMP release, DAMPs can theoretically facilitate indirect perception of pathogens. It is possible that transfer of DAMPs and their recognition machinery to other species may confer enhanced resistance to specific pathogens, although this remains to be demonstrated.
In conclusion, the past 25 years has revealed the molecular identity of many R genes and uncovered the underlying molecular mechanisms governing their functioning. However, much remains to be discovered. Understanding the molecular and structural mechanisms involved in R gene mediated disease resistance will allow us to engineer resistance for the crops of the future.
Supplemental Data
Supplemental Data Set 1. List of R genes.
Acknowledgments
This work was supported by the European Research Council (616449), by the Clarendon Fund, and by the University of Oxford.
AUTHOR CONTRIBUTIONS
Both authors contributed to writing the article.
Footnotes
Articles can be viewed without a subscription.
References
- Ade J., DeYoung B.J., Golstein C., Innes R.W. (2007). Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 104: 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I., et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1: 15140. [DOI] [PubMed] [Google Scholar]
- Ashfield T., Ong L.E., Nobuta K., Schneider C.M., Innes R.W. (2004). Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikawa I., Hayashi N., Yamane H., Kanamori H., Wu J., Matsumoto T., Ono K., Yano M. (2008). Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180: 2267–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Staskawicz B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377. [DOI] [PubMed] [Google Scholar]
- Baudin M., Hassan J.A., Schreiber K.J., Lewis J.D. (2017). Analysis of the ZAR1 immune complex reveals determinants for immunity and molecular interactions. Plant Physiol. 174: 2038–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Kunkel B.N., Dahlbeck D., Brown K.L., Schmidt R., Giraudat J., Leung J., Staskawicz B.J. (1994). RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bernoux M., Burdett H., Williams S.J., Zhang X., Chen C., Newell K., Lawrence G.J., Kobe B., Ellis J.G., Anderson P.A., Dodds P.N. (2016). Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 28: 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H., Albert I., Oome S., Raaymakers T.M., Van den Ackerveken G., Nürnberger T. (2014). A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 10: e1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A., Rosli H.G., Vrebalov J., Moffett P., Mueller L.A., Martin G.B. (2012). A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 25: 1523–1530. [DOI] [PubMed] [Google Scholar]
- Bonardi V., Tang S., Stallmann A., Roberts M., Cherkis K., Dangl J.L. (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA 108: 16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büschges R., et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705. [DOI] [PubMed] [Google Scholar]
- Butterbach P., Verlaan M.G., Dullemans A., Lohuis D., Visser R.G.F., Bai Y., Kormelink R. (2014). Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proc. Natl. Acad. Sci. USA 111: 12942–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell K.S., Michelmore R.W. (2009). Arabidopsis thaliana genes encoding defense signaling and recognition proteins exhibit contrasting evolutionary dynamics. Genetics 181: 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J.L., Mamillapalli P., Burch-Smith T.M., Czymmek K., Dinesh-Kumar S.P. (2008). Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti A.-M., Lim G.T.T., Jones D.A. (2015). The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207: 106–118. [DOI] [PubMed] [Google Scholar]
- Cesari S., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25: 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W., Munkvold K.R., Gao H., Mathieu J., Schwizer S., Wang S., Yan Y.B., Wang J., Martin G.B., Chai J. (2011). Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III Effector. Cell Host Microbe 10: 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Chu Z., Yuan M., Yao J., Ge X., Yuan B., Xu C., Li X., Fu B., Li Z., Bennetzen J.L., Zhang Q., Wang S. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20: 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coaker G., Falick A., Staskawicz B. (2005). Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308: 548–550. [DOI] [PubMed] [Google Scholar]
- Day B., Dahlbeck D., Huang J., Chisholm S.T., Li D., Staskawicz B.J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17: 1292–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debieu M., Huard-Chauveau C., Genissel A., Roux F., Roby D. (2016). Quantitative disease resistance to the bacterial pathogen Xanthomonas campestris involves an Arabidopsis immune receptor pair and a gene of unknown function. Mol. Plant Pathol. 17: 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guillen K., Ortiz-Vallejo D., Gracy J., Fournier E., Kroj T., Padilla A. (2015). Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathog. 11: e1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R., van Esse H.P., Maruthachalam K., Bolton M.D., Santhanam P., Saber M.K., Zhang Z., Usami T., Lievens B., Subbarao K.V., Thomma B.P.H.J. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA 109: 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange O., Schreiber T., Schandry N., Radeck J., Braun K.H., Koszinowski J., Heuer H., Strauß A., Lahaye T. (2013). Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 199: 773–786. [DOI] [PubMed] [Google Scholar]
- DeYoung B.J., Qi D., Kim S.-H., Burke T.P., Innes R.W. (2012). Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell. Microbiol. 14: 1071–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M.S., Jones D.A., Keddie J.S., Thomas C.M., Harrison K., Jones J.D.G. (1996). The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84: 451–459. [DOI] [PubMed] [Google Scholar]
- Dixon M.S., Golstein C., Thomas C.M., van Der Biezen E.A., Jones J.D.G. (2000). Genetic complexity of pathogen perception by plants: the example of Rcr3, a tomato gene required specifically by Cf-2. Proc. Natl. Acad. Sci. USA 97: 8807–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.-M., Ayliffe M.A., Ellis J.G. (2004). The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16: 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.-M., Teh T., Wang C.-I.A., Ayliffe M.A., Kobe B., Ellis J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103: 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.G., Lagudah E.S., Spielmeyer W., Dodds P.N. (2014). The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 5: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276. [DOI] [PubMed] [Google Scholar]
- Fradin E.F., Zhang Z., Juarez Ayala J.C., Castroverde C.D.M., Nazar R.N., Robb J., Liu C.-M., Thomma B.P.H.J. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D., Uauy C., Distelfeld A., Blechl A., Epstein L., Chen X., Sela H., Fahima T., Dubcovsky J. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323: 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka S., Saka N., Koga H., Ono K., Shimizu T., Ebana K., Hayashi N., Takahashi A., Hirochika H., Okuno K., Yano M. (2009). Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325: 998–1001. [DOI] [PubMed] [Google Scholar]
- Gabriëls S.H.E.J., Vossen J.H., Ekengren S.K., van Ooijen G., Abd-El-Haliem A.M., van den Berg G.C.M., Rainey D.Y., Martin G.B., Takken F.L.W., de Wit P.J.G.M., Joosten M.H.A.J. (2007). An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J. 50: 14–28. [DOI] [PubMed] [Google Scholar]
- Giannakopoulou A., Steele J.F.C., Segretin M.E., Bozkurt T.O., Zhou J., Robatzek S., Banfield M.J., Pais M., Kamoun S. (2015). Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 28: 1316–1329. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J.-Y., et al. (2015). Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell 27: 1755–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu K., Yang B., Tian D., Wu L., Wang D., Sreekala C., Yang F., Chu Z., Wang G.-L., White F.F., Yin Z. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435: 1122–1125. [DOI] [PubMed] [Google Scholar]
- Gu K., Tian D., Qiu C., Yin Z. (2009). Transcription activator-like type III effector AvrXa27 depends on OsTFIIAgamma5 for the activation of Xa27 transcription in rice that triggers disease resistance to Xanthomonas oryzae pv. oryzae. Mol. Plant Pathol. 10: 829–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Foessel S.A., Singh R.P., Lillemo M., Huerta-Espino J., Bhavani S., Singh S., Lan C., Calvo-Salazar V., Lagudah E.S. (2014). Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 127: 781–789. [DOI] [PubMed] [Google Scholar]
- Hind S.R., et al. (2016). Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2: 16128. [DOI] [PubMed] [Google Scholar]
- Huang S., Antony G., Li T., Liu B., Obasa K., Yang B., White F.F. (2016). The broadly effective recessive resistance gene xa5 of rice is a virulence effector-dependent quantitative trait for bacterial blight. Plant J. 86: 186–194. [DOI] [PubMed] [Google Scholar]
- Hückelhoven R., Panstruga R. (2011). Cell biology of the plant-powdery mildew interaction. Curr. Opin. Plant Biol. 14: 738–746. [DOI] [PubMed] [Google Scholar]
- Hummel A.W., Doyle E.L., Bogdanove A.J. (2012). Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 195: 883–893. [DOI] [PubMed] [Google Scholar]
- Humphry M., Consonni C., Panstruga R. (2006). mlo-based powdery mildew immunity: silver bullet or simply non-host resistance? Mol. Plant Pathol. 7: 605–610. [DOI] [PubMed] [Google Scholar]
- Humphry M., Bednarek P., Kemmerling B., Koh S., Stein M., Göbel U., Stüber K., Piślewska-Bednarek M., Loraine A., Schulze-Lefert P., Somerville S., Panstruga R. (2010). A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc. Natl. Acad. Sci. USA 107: 21896–21901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas M., Hörger A.C., Bozkurt T.O., van den Burg H.A., Kaschani F., Kaiser M., Belhaj K., Smoker M., Joosten M.H.A.J., Kamoun S., van der Hoorn R.A.L. (2015). Functional divergence of two secreted immune proteases of tomato. Curr. Biol. 25: 2300–2306. [DOI] [PubMed] [Google Scholar]
- Ishibashi K., Kezuka Y., Kobayashi C., Kato M., Inoue T., Nonaka T., Ishikawa M., Matsumura H., Katoh E. (2014). Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1. Proc. Natl. Acad. Sci. USA 111: E3486–E3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., McCouch S.R. (2004). The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 17: 1348–1354. [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A.S., Jiang H., Huang L., McCouch S.R. (2008). Genetic and functional characterization of the rice bacterial blight disease resistance gene xa5. Phytopathology 98: 289–295. [DOI] [PubMed] [Google Scholar]
- Johal G.S., Briggs S.P. (1992). Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258: 985–987. [DOI] [PubMed] [Google Scholar]
- Jones D.A., Thomas C.M., Hammond-Kosack K.E., Balint-Kurti P.J., Jones J.D. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kang B.-C., Yeam I., Jahn M.M. (2005). Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43: 581–621. [DOI] [PubMed] [Google Scholar]
- Kaschani F., Shabab M., Bozkurt T., Shindo T., Schornack S., Gu C., Ilyas M., Win J., Kamoun S., van der Hoorn R.A.L. (2010). An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154: 1794–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y., Takai R., Furukawa T., Hirai H., Morimoto T., Katayama T., Murakami T., Che F.-S. (2015). CD2–1, the C-terminal region of flagellin, modulates the induction of immune responses in rice. Mol. Plant Microbe Interact. 28: 648–658. [DOI] [PubMed] [Google Scholar]
- Kessens R., Ashfield T., Kim S.H., Innes R.W. (2014). Determining the GmRIN4 requirements of the soybean disease resistance proteins Rpg1b and Rpg1r using a Nicotiana glutinosa-based agroinfiltration system. PLoS One 9: e108159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-S., Desveaux D., Singer A.U., Patel P., Sondek J., Dangl J.L. (2005). The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. USA 102: 6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.G., Geng X., Lee S.Y., Mackey D. (2009). The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the Arabidopsis nucleotide-binding leucine-rich repeat protein RPS2. Plant J. 57: 645–653. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Qi D., Ashfield T., Helm M., Innes R.W. (2016). Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351: 684–687. [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Lin N.-C., Martin G.B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598. [DOI] [PubMed] [Google Scholar]
- Kooman-Gersmann M., Honee G., Bonnema G., De Wit P. (1996). A high-affinity binding site for the AVR9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other solanaceous plants. Plant Cell 8: 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooman-Gersmann M., Vogelsang R., Vossen P., van den Hooven H.W., Mahé E., Honée G., de Wit P.J.G.M. (1998). Correlation between binding affinity and necrosis-inducing activity of mutant AVR9 peptide elicitors. Plant Physiol. 117: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva K.V., Dahlbeck D., Staskawicz B.J. (2010). Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger S.G., Lagudah E.S., Spielmeyer W., Singh R.P., Huerta-Espino J., McFadden H., Bossolini E., Selter L.L., Keller B. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323: 1360–1363. [DOI] [PubMed] [Google Scholar]
- Kroj T., Chanclud E., Michel-Romiti C., Grand X., Morel J.-B. (2016). Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210: 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S., Rougon-Cardoso A., Sherwood E., Peeters N., Dahlbeck D., van Esse H.P., Smoker M., Rallapalli G., Thomma B.P.H.J., Staskawicz B., Jones J.D.G., Zipfel C. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28: 365–369. [DOI] [PubMed] [Google Scholar]
- Lee B., Park Y.-S., Lee S., Song G.C., Ryu C.-M. (2016). Bacterial RNAs activate innate immunity in Arabidopsis. New Phytol. 209: 785–797. [DOI] [PubMed] [Google Scholar]
- Lee D., Bourdais G., Yu G., Robatzek S., Coaker G. (2015). Phosphorylation of the plant immune regulator RPM1-INTERACTING PROTEIN4 enhances plant plasma membrane H+-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell 27: 2042–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161: 1074–1088. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Wu R., Guttman D.S., Desveaux D. (2010). Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 6: e1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D., Lee A.H.-Y., Hassan J.A., Wan J., Hurley B., Jhingree J.R., Wang P.W., Lo T., Youn J.-Y., Guttman D.S., Desveaux D. (2013). The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc. Natl. Acad. Sci. USA 110: 18722–18727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., et al. (2014). Proline isomerization of the immune receptor-interacting protein RIN4 by a cyclophilin inhibits effector-triggered immunity in Arabidopsis. Cell Host Microbe 16: 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M.T.S., Kunkel B.N. (2004). The Pseudomonas syringae type III effector AvrRpt2 promotes virulence independently of RIN4, a predicted virulence target in Arabidopsis thaliana. Plant J. 40: 790–798. [DOI] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Fuglsang A.T., Palmgren M.G., Staskawicz B.J., Coaker G. (2009). RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Lin Z.-J.D., Coaker G. (2011a). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yuan M., Zhou Y., Li X., Xiao J., Wang S. (2011b). A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34: 1958–1969. [DOI] [PubMed] [Google Scholar]
- Lozano-Torres J.L., et al. (2012). Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. USA 109: 10119–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Torres J.L., Wilbers R.H.P., Warmerdam S., Finkers-Tomczak A., Diaz-Granados A., van Schaik C.C., Helder J., Bakker J., Goverse A., Schots A., Smant G. (2014). Apoplastic venom allergen-like proteins of cyst nematodes modulate the activation of basal plant innate immunity by cell surface receptors. PLoS Pathog. 10: e1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Kracher B., Saur I.M.L., Bauer S., Ellwood S.R., Wise R., Yaeno T., Maekawa T., Schulze-Lefert P. (2016). Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl. Acad. Sci. USA 113: E6486–E6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderer R., et al. (2001). No evidence for binding between resistance gene product Cf-9 of tomato and avirulence gene product AVR9 of Cladosporium fulvum. Mol. Plant Microbe Interact. 14: 867–876. [DOI] [PubMed] [Google Scholar]
- Luderer R., Takken F.L.W., de Wit P.J.G.M., Joosten M.H.A.J. (2002). Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45: 875–884. [DOI] [PubMed] [Google Scholar]
- Mackey D., Holt B.F. III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- Maqbool A., Saitoh H., Franceschetti M., Stevenson C.E.M., Uemura A., Kanzaki H., Kamoun S., Terauchi R., Banfield M.J. (2015). Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4: e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.B., Brommonschenkel S.H., Chunwongse J., Frary A., Ganal M.W., Spivey R., Wu T., Earle E.D., Tanksley S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262: 1432–1436. [DOI] [PubMed] [Google Scholar]
- Martin G.B., Frary A., Wu T., Brommonschenkel S., Chunwongse J., Earle E.D., Tanksley S.D. (1994). A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell 6: 1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M., Katagiri F., Yu G.-L., Ausubel F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099. [DOI] [PubMed] [Google Scholar]
- Moore J.W., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47: 1494–1498. [DOI] [PubMed] [Google Scholar]
- Mysore K.S., Ryu C.M. (2004). Nonhost resistance: how much do we know? Trends Plant Sci. 9: 97–104. [DOI] [PubMed] [Google Scholar]
- Nekrasov V., Wang C., Win J., Lanz C., Weigel D., Kamoun S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis V., Saur I.M., Conlan B., Rathjen J.P. (2014). The changing of the guard: the Pto/Prf receptor complex of tomato and pathogen recognition. Curr. Opin. Plant Biol. 20: 69–74. [DOI] [PubMed] [Google Scholar]
- Oome S., Raaymakers T.M., Cabral A., Samwel S., Böhm H., Albert I., Nürnberger T., Van den Ackerveken G. (2014). Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 16955–16960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart J.R., Mestre P., Lu R., Malcuit I., Baulcombe D.C. (2005). NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 15: 968–973. [DOI] [PubMed] [Google Scholar]
- Peele H.M., Guan N., Fogelqvist J., Dixelius C. (2014). Loss and retention of resistance genes in five species of the Brassicaceae family. BMC Plant Biol. 14: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P., Zhou F., Casais C., Orme J., Jarosch B., Schaffrath U., Collins N.C., Panstruga R., Schulze-Lefert P. (2002). The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129: 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt R.N., et al. (2015). The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 1: e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D., Dubiella U., Kim S.H., Sloss D.I., Dowen R.H., Dixon J.E., Innes R.W. (2014). Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on s-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol. 164: 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravensdale M., Bernoux M., Ve T., Kobe B., Thrall P.H., Ellis J.G., Dodds P.N. (2012). Intramolecular interaction influences binding of the Flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Pathog. 8: e1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Bittel P., Chinchilla D., Köchner P., Felix G., Shiu S.-H., Boller T. (2007). Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 64: 539–547. [DOI] [PubMed] [Google Scholar]
- Römer P., Hahn S., Jordan T., Strauss T., Bonas U., Lahaye T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318: 645–648. [DOI] [PubMed] [Google Scholar]
- Römer P., Recht S., Lahaye T. (2009). A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl. Acad. Sci. USA 106: 20526–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney H.C.E., Van’t Klooster J.W., van der Hoorn R.A.L., Joosten M.H.A.J., Jones J.D.G., de Wit P.J.G.M. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308: 1783–1786. [DOI] [PubMed] [Google Scholar]
- Rosebrock T.R., Zeng L., Brady J.J., Abramovitch R.B., Xiao F., Martin G.B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotblat B., Enshell-Seijffers D., Gershoni J.M., Schuster S., Avni A. (2002). Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J. 32: 1049–1055. [DOI] [PubMed] [Google Scholar]
- Russell A.R., Ashfield T., Innes R.W. (2015). Pseudomonas syringae effector AvrPphB suppresses AvrB-induced activation of RPM1 but not AvrRpm1-induced activation. Mol. Plant Microbe Interact. 28: 727–735. [DOI] [PubMed] [Google Scholar]
- Salmeron J.M., Oldroyd G.E.D., Rommens C.M.T., Scofield S.R., Kim H.-S., Lavelle D.T., Dahlbeck D., Staskawicz B.J. (1996). Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86: 123–133. [DOI] [PubMed] [Google Scholar]
- Sarris P.F., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161: 1089–1100. [DOI] [PubMed] [Google Scholar]
- Sarris P.F., Cevik V., Dagdas G., Jones J.D.G., Krasileva K.V. (2016). Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucet S.B., Ma Y., Sarris P.F., Furzer O.J., Sohn K.H., Jones J.D.G. (2015). Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 6: 6338. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P., Panstruga R. (2011). A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16: 117–125. [DOI] [PubMed] [Google Scholar]
- Seear P.J., Dixon M.S. (2003). Variable leucine-rich repeats of tomato disease resistance genes Cf-2 and Cf-5 determine specificity. Mol. Plant Pathol. 4: 199–202. [DOI] [PubMed] [Google Scholar]
- Selote D., Kachroo A. (2010). RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol. 153: 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto D., Koulena N., Lo T., Menna A., Guttman D.S., Desveaux D. (2017). Expanded type III effector recognition by the ZAR1 NLR protein using ZED1-related kinases. Nat. Plants 3: 17027. [DOI] [PubMed] [Google Scholar]
- Shabab M., Shindo T., Gu C., Kaschani F., Pansuriya T., Chintha R., Harzen A., Colby T., Kamoun S., van der Hoorn R.A.L. (2008). Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell 20: 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F., Golstein C., Ade J., Stoutemyer M., Dixon J.E., Innes R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shen Q.-H., Zhou F., Bieri S., Haizel T., Shirasu K., Schulze-Lefert P. (2003). Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu A., Chintamanani S., Brandt A.S., Zanis M., Scofield S.R., Johal G.S. (2008). A guardian of grasses: specific origin and conservation of a unique disease-resistance gene in the grass lineage. Proc. Natl. Acad. Sci. USA 105: 1762–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrenner A.D., Goritschnig S., Staskawicz B.J. (2015). Recognition and activation domains contribute to allele-specific responses of an Arabidopsis NLR receptor to an oomycete effector protein. PLoS Pathog. 11: e1004665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuernagel B., Periyannan S.K., Hernández-Pinzón I., Witek K., Rouse M.N., Yu G., Hatta A., Ayliffe M., Bariana H., Jones J.D.G., Lagudah E.S., Wulff B.B. (2016). Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34: 652–655. [DOI] [PubMed] [Google Scholar]
- Strauss T., et al. (2012). RNA-seq pinpoints a Xanthomonas TAL-effector activated resistance gene in a large-crop genome. Proc. Natl. Acad. Sci. USA 109: 19480–19485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A.P., Han Z., Hu Z., Zipfel C., Zhou J.-M., Chai J. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628. [DOI] [PubMed] [Google Scholar]
- Swiderski M.R., Innes R.W. (2001). The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26: 101–112. [DOI] [PubMed] [Google Scholar]
- Thomma B.P.H.J., Nürnberger T., Joosten M.H.A.J. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Wang J., Zeng X., Gu K., Qiu C., Yang X., Zhou Z., Goh M., Luo Y., Murata-Hori M., White F.F., Yin Z. (2014). The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 26: 497–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trdá L., Fernandez O., Boutrot F., Héloir M.-C., Kelloniemi J., Daire X., Adrian M., Clément C., Zipfel C., Dorey S., Poinssot B. (2014). The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol. 201: 1371–1384. [DOI] [PubMed] [Google Scholar]
- Truniger V., Aranda M.A. (2009). Recessive resistance to plant viruses. In Advances in Virus Research: Natural and Engineered Resistance to Plant Viruses, Part I, Vol. 75, G. Loebenstein and J.P. Carr, eds (Academic Press), pp. 119–231. [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A.L., Kamoun S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse H.P., Van’t Klooster J.W., Bolton M.D., Yadeta K.A., van Baarlen P., Boeren S., Vervoort J., de Wit P.J.G.M., Thomma B.P.H.J. (2008). The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20: 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie C.C., Takken F.L. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52: 551–581. [DOI] [PubMed] [Google Scholar]
- Verlaan M.G., Hutton S.F., Ibrahem R.M., Kormelink R., Visser R.G.F., Scott J.W., Edwards J.D., Bai Y. (2013). The Tomato Yellow Leaf Curl Virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 9: e1003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossen J.H., van Arkel G., Bergervoet M., Jo K.-R., Jacobsen E., Visser R.G.F. (2016). The Solanum demissum R8 late blight resistance gene is an Sw-5 homologue that has been deployed worldwide in late blight resistant varieties. Theor. Appl. Genet. 129: 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., et al. (2015a). XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 8: 290–302. [DOI] [PubMed] [Google Scholar]
- Wang C.-I.A., et al. (2007). Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell 19: 2898–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., et al. (2015b). The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18: 285–295. [DOI] [PubMed] [Google Scholar]
- Warren R.F., Henk A., Mowery P., Holub E., Innes R.W. (1998). A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10: 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S., Dinesh-Kumar S.P., Choi D., Hehl R., Corr C., Baker B. (1994). The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115. [DOI] [PubMed] [Google Scholar]
- Wilton M., Subramaniam R., Elmore J., Felsensteiner C., Coaker G., Desveaux D. (2010). The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. USA 107: 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witek K., Jupe F., Witek A.I., Baker D., Clark M.D., Jones J.D.G. (2016). Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol. 34: 656–660. [DOI] [PubMed] [Google Scholar]
- Wu C.-H., Abd-El-Haliem A., Bozkurt T.O., Belhaj K., Terauchi R., Vossen J.H., Kamoun S. (2017). NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 114: 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., et al. (2015). Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol. 206: 1463–1475. [DOI] [PubMed] [Google Scholar]
- Xing W., et al. (2007). The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449: 243–247. [DOI] [PubMed] [Google Scholar]
- Xu R.-Q., et al. (2008). AvrAC(Xcc8004), a type III effector with a leucine-rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col-0. J. Bacteriol. 190: 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Yamashita-Yamada M., Hirase T., Fujiwara T., Tsuda K., Hiruma K., Saijo Y. (2016). Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J. 35: 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Ke Y., Huang R., Ma L., Yang Z., Chu Z., Xiao J., Li X., Wang S. (2016). A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. eLife 5: e19605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Tian D., Gu K., Zhou Z., Yang X., Luo Y., White F.F., Yin Z. (2015). Genetic engineering of the Xa10 promoter for broad-spectrum and durable resistance to Xanthomonas oryzae pv. oryzae. Plant Biotechnol. J. 13: 993–1001. [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]
- Zhang J., Yin Z., White F. (2015). TAL effectors and the executor R genes. Front. Plant Sci. 6: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kars I., Essenstam B., Liebrand T.W.H., Wagemakers L., Elberse J., Tagkalaki P., Tjoitang D., van den Ackerveken G., van Kan J.A.L. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Liu Y., Huang H., Gao M., Wu D., Kong Q., Zhang Y. (2017). The NLR protein SUMM2 senses the disruption of an immune signaling MAP kinase cascade via CRCK3. EMBO Rep. 18: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Shao F., Innes R.W., Dixon J.E., Xu Z. (2004). The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site. Proc. Natl. Acad. Sci. USA 101: 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., et al. (2017). The intracellular immune receptor Sw-5b confers broad-spectrum resistance to tospoviruses through recognition of a conserved 21-amino-acid viral effector epitope. Plant Cell 29: 2214–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35: 345–351. [DOI] [PubMed] [Google Scholar]