Abstract
Photosynthesis converts solar energy into chemical energy to sustain all life on earth by providing oxygen and food, and controlling the atmospheric carbon dioxide. During this process, the water-splitting and oxygen-evolving reaction is catalyzed by photosystem II (PSII), while photosystem I (PSI) generates the reducing power for the reduction of NADP+ to NADPH. Together with their peripheral light-harvesting complexes (LHCs), photosystems function as multisubunit supercomplexes located in the thylakoid membranes of cyanobacteria, algae, and plants. Recent advances in single-particle cryo-electron microscopy (cryoEM), X-ray free electron laser (XFEL) and other techniques have revealed unprecedented structural and catalytic details concerning the two supercomplexes. Several high-resolution structures of the complexes from plants were solved, and serial time-resolved crystallography and “radiation-damage-free” femtosecond XFEL also provided important insights into the mechanism of water oxidation. Here, we review these exciting advances in the studies of the photosystem supercomplexes with an emphasis on PSII-LHCII, propose presently unresolved problems in this field, and suggest potential tendencies for future studies.
Keywords: chloroplast, photosynthesis, structure, photosystem II protein complex, protein complex
Introduction
Photosynthesis carries out a series of biophysical and biochemical processes, finally converting solar energy into chemical energy. Oxygenic photosynthesis split water molecules to oxygen, which is indispensable for maintaining aerobic life on earth (Dismukes et al., 2001). It is believed that photosynthesis has evolved only once during the evolution history in cyanobacteria. For algae and higher plants, they acquired photosynthesis capacity via cyanobacteria endosymbionts which evolved to chloroplasts in plants (Tomioka and Sugiura, 1983). Plant leaves are the major organs of photosynthesis with about 100 chloroplasts in each mesophyll cell (Woodson, 2016). Due to their essential roles in light harvesting and energy production, chloroplasts are vital organelles of photosynthetic cells in algae and higher plants, acting as the suppliers of carbon sources and energy. Chloroplast is a large organelle with a complex structure harboring two outer membranes called the chloroplast envelope and a third extensively folded internal membrane system called thylakoid (Arvidsson and Sundby, 1999; Kirchhoff, 2013). The thylakoid membrane is composed of two morphologically distinct domains, the grana domain which is characterized by ∼5–20 layers of cylindrical stacks of thylakoid membrane disks (Mullineaux, 2005; Mustárdy et al., 2008), and the stroma lamellae domain which are stroma-exposed membrane pairs connecting the grana stacks (Dekker and Boekema, 2005).
There are two types of photosystems in cyanobacteria, algae and higher plants, called photosystem I (PSI, plastocyanin-ferredoxin oxidoreductase) and photosystem II (PSII, water-plastoquinone oxidoreductase), both of which are multisubunit membrane complexes. The PSI is located in the stroma lamella of thylakoid while the PSII is in the stacked grana domain (Albertsson, 2001; Dekker and Boekema, 2005). Each photosystem is composed of a core complex and a peripheral antenna system, light harvesting complex I (LHCI) for PSI and light harvesting complex II (LHCII) for PSII, respectively. Recently, new atom-resolution structures of the photosystems and detailed insights into the water-splitting process have been reported, with the development of single-particle cryo-electron microscopy, serial time-resolved crystallography and other techniques. Here, we will review the recent progress in the studies into the structures and functions of photosystems of different origins, with an emphasis on PSII-LHCII.
Biochemistry of Photosystems
The complete photosynthetic reactions in cyanobacteria, algae and plants are executed by four major protein supercomplexes including PSI, PSII, cytochrome b6f (plastoquinone-plastocyanin oxidoreductase) and F-ATPase (proton-motive force-driven ATP synthase) (Nelson and Benshem, 2004; Nelson and Yocum, 2006). Both PSI and PSII supercomplexes bind chlorophyll molecules to sense different spectrums and intensities of light (Remelli et al., 1999; Nelson and Yocum, 2006; Croce and van Amerongen, 2013; van Amerongen and Croce, 2013; Caffarri et al., 2014; Ruban, 2015). Light harvested by the chlorophylls and other pigments in PSI and PSII is transferred to the photosynthetic reaction center (RC), further inducing the excitation of chlorophylls known as P680 for PSII and P700 for PSI to initiate the proton translocation across the membrane (Nelson and Junge, 2015). In PSII, P680 undergoes charge separation and the generated electrons are transferred to the quinone acceptor pheophytin and plastoquinone sequentially (Grabolle and Dau, 2005; Johnson, 2016). Meanwhile, water molecule, the authentic electron donor, is oxidized to molecular oxygen and P680 is eventually reduced. After the reaction, the electrons are ultimately transferred to the thylakoid-embedded cytochrome b6f, which oxidizes plastoquinols to plastoquinones and reduces plastocyanins (Cramer et al., 1996). And then, the plastocyanin is oxidized by PSI, during which the reduced electron carrier protein ferredoxin, is used to reduce NADP+ to NADPH by ferredoxin–NADP+ reductase (FNR) enzyme (Brettel and Leibl, 2001; Sétif, 2001; Golbeck, 2006). Together, PSII generates the most positive redox potential, while PSI generates the powerful naturally occurring reductant NADPH (Hope, 2000; Holzwarth et al., 2006; Nelson, 2011). The photocatalytic activity of PSII and PSI is linked by the cytochrome b6f complex, and the proton-motive force generated during the process are utilized by the F-ATPase to generate ATP, which together with NADPH are supplied as energy compounds for sugar synthesis from carbon dioxide by the dark reaction (Pfannschmidt, 2003).
The first PSI structure from the thermophilic cyanobacterium Synechococcus elongatus (S. elongatus) displayed a complex with 12 protein subunits and 127 cofactors (96 chlorophylls, 22 carotenoids, 2 phylloquinones, 3 Fe4S4 clusters, and 4 lipids), providing the very first detailed molecular architecture of PSI (Fromme et al., 2001; Jordan et al., 2001). S. elongatus PSI is a trimer with a diameter of 210 Å and a maximum height of 90 Å (Jordan et al., 2001), while the plant PSI supercomplex is a monomer (Ben-Shem et al., 2003). The core complex is largely conserved from cyanobacteria to plants with nine membrane-embedded subunits, whereas the LHCI complexes are variable in subunit composition, binding pigments and sizes due to the different habitats of cyanobacteria, algae, and plants (Ben-Shem et al., 2003; Qin et al., 2015). However, although PSI and PSII evolved from the same ancestor, belonging to the same superfamily, their structures are largely different.
Structure of the Cyanobacterial PSII Supercomplex
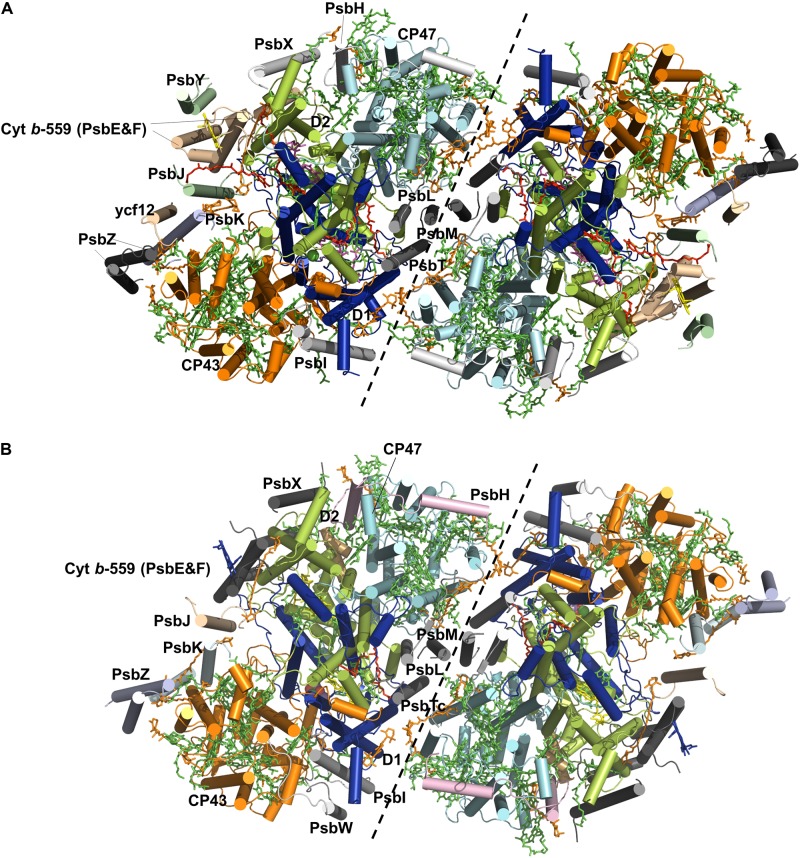
The PS II homodimer from Thermosynechococcus elongatus (T. elongatus) has dimensions of 105 Å in depth (45 Å in membrane), 205 Å in length, and 110 Å in width (Ferreira et al., 2004). The PSII supercomplex in cyanobacteria comprises the reaction center (RC) proteins D1 and D2, the antenna subunits CP47 and CP43, 13 membrane-intrinsic small subunits (PsbE, PsbF, PsbH-M, PsbN, PsbX, PsbY, PsbZ, and PsbYcf12) and 3 extrinsic subunits (PsbO, PsbU, and PsbV). The structures of D1 and D2 are similar to each other, both containing five helices all tilted against the membrane planes (Zouni et al., 2001; Kamiya and Shen, 2003), and they form the center of the PSII complex. CP43 and CP47 surround the D1-D2 core with similar structures of six helices, respectively. Afterward, Loll et al. (2005) provided the first complete structure of cyanobacterial photosystem II, providing a full glimpse of the PSII cofactors. They displayed the positions of 20 protein subunits and their interactions with 77 cofactors (Figure 1A). The overall structures of the supercomplex and protein subunits are similar to those previously reported (Zouni et al., 2001; Kamiya and Shen, 2003; Biesiadka et al., 2004). Lipids have long been thought to play a role in the assembly and function of PSII, and for the first time the authors showed the lipid integrally bound to PSII. Eleven lipids surrounding the RC form a belt to separate it from the antenna and small protein subunits, while the remaining lipids are mostly located at the monomer-monomer interface. The lipid-rich property renders PSII both structural flexibility for local mobility and convenience in subunit-subunit recognition (Guskov et al., 2009). Eleven carotenoid molecules were modeled as β-carotenes in all-trans configurations in their study and an additional Car15 was identified in the study by Guskov et al. (2009). In the study by Guskov et al. (2009), they also successfully assigned the small protein subunits Psbycf12, PsbY, and PsbX to the previously unassigned positions (Loll et al., 2005; Guskov et al., 2009). A summary of the subunit composition information, including the subunit-cofactor interactions in PSII from T. elongatus, is presented in Table 1 (Guskov et al., 2009). In 2009, Broser et al. presented the first structure of a monomeric form of PSII core complex (PSIIcc) with high oxygen-evolution capacity from T. elongatus (Broser et al., 2010). The assembly of the protein subunits, tetrapyrrole cofactors and the non-heme iron in the monomeric PSIIcc are all identical to those in the dimer structure.
FIGURE 1.
Overall structures of the PSII core complex from Tynechococcus elongatus (A) and spinach (B). The monomer-monomer interface is indicated by a black dashed line. Only the four large subunits and the intrinsic subunits of PSIIs are shown. The subunits D1 (blue), D2 (lime), CP43 (yellow), and CP47 (cyan) and the other small subunits are labeled in the monomer on the left. Cofactors are shown in sticks, including Chl (green), Car (orange), heme (light magenta) and lipids (red). The Mn cluster is shown in spheres.
Table 1.
Transmembrane (TM) helices, chlorophylls, and other cofactors of PSII RCs from cyanobacteria and plants1.
| Subunits | TM helices | Chlorophylls | Other cofactors in plant PSII RC | |||||
|---|---|---|---|---|---|---|---|---|
| Cyano | Plant | Cyano | Plant | Carotenoid | Heme | Lipid | Others | |
| A (D1) | 5 | 5 | 4 | 62 | 1 | 3 | 1 Mn4CaO5 | |
| B (CP47) | 6 | 6 | 16 | 16 | 3 | 2 | ||
| C (CP43) | 6 | 6 | 13 | 13 | 3 | 4 | ||
| D (D2) | 5 | 5 | 4 | 2 | 1 | 4 | 1 plastoquinone | |
| E | 1 | 1 | 13 | |||||
| F | 1 | 1 | ||||||
| H | 1 | 1 | 1 | 1 | ||||
| I | 1 | 1 | ||||||
| J | 1 | 1 | ||||||
| K | 1 | 1 | 1 | |||||
| L | 1 | 1 | 1 | |||||
| M | 1 | 1 | ||||||
| O | 0 | 0 | ||||||
| P | 0 | |||||||
| Q | 0 | |||||||
| T | 1 | |||||||
| U | 0 | |||||||
| V | 0 | |||||||
| W | 1 | |||||||
| Tc | 1 | |||||||
| Tn | 0 | |||||||
| X | 1 | 1 | ||||||
| Y | 1 | |||||||
| Z | 1 | 2 | 1 | |||||
| Ycf12 | 1 | |||||||
1The subunit composition and the number of TM helices are based on the structures of PSIIs from Tynechococcus elongatus and spinach (Guskov et al., 2009; Wei et al., 2016). 2These include 4 Chl a and 2 pheophytin. 3This heme is bound by both PsbE and PsbF.
During photosynthesis, water oxidation happens in the oxygen-evolving complex (OEC), which comprises the Mn4CaO5 cluster as the catalytic center. Water splitting is a process fulfilled in five consecutive stages named S0 to S4. It has been a model system for synthesizing catalysts for inorganic water oxidation and dioxygen evolution (Kanady and Agapie, 2011; Mukherjee et al., 2012). In the work of Ferreira et al. (2004), they reported that the OEC harbors a “cubane-like” Mn3CaO4 cluster linked to a fourth Mn by a mono-μ-oxo bridge which had not been specifically suggested before. However, neither water nor hydroxide could be observed to find the water oxidation site accurately in their study. In the first complete PSII structure, the Mn4Ca cluster was proposed as a “Y-shaped hook,” considerably differed from the “cubane-like” model (Loll et al., 2005). Then a 1.9-Å resolution X-ray structure of PSII from T. vulcanus revealed a clear picture of the Mn4CaO5 cluster, in which the electron densities for each metal ion and the oxo-bridged oxygen atoms were totally separated, thus allowing the clear assignment of each of the atoms. They found that the OEC Mn4CaO5 cluster displayed a “distorted chair” conformation with three Mn, one Ca and four oxygen atoms forming an asymmetric cubane-like seat base and the fourth Mn (Mn4) together with the fifth oxygen atom (O4) forming the chair back (Umena et al., 2011). Subsequently, a simultaneous femtosecond X-ray spectroscopy and diffraction of the PSII system showed that the electron density maps of the dark and illuminated states are similar with an overall correlation coefficient (CC) of 0.77 (a CC of 0 means no correlation; a CC of 1 indicates full correlation), suggesting no significant conformational changes between the S1 and S2 states (Kern et al., 2013). However, with a serial time-resolved crystallography, the authors acquired PSII structures in the dark S1 and putative S3 states, in which they found that the distance between the Mn3OxCa cubane and the distant protruding Mn (dangler Mn) increased in the putative S3 state, allowing the binding of the second water molecule during the S2 to S3 state transition (Kupitz et al., 2014). Moreover, with a “radiation-damage-free” femtosecond X-ray free electron laser (XFEL), Suga et al. (2015, 2017) found that the Mn-Mn and Mn-O distances showed marked differences in the OEC from XFEL: all the distances are about 0.1–0.2 Å shorter than those from the X-ray diffraction (XRD) structures. In addition, the position of O5 is also unusual. The results showed that it functions more as a hydroxide ion instead of a normal oxygen dianion to serve as one of the substrate oxygen atoms (Suga et al., 2015). Their recent work described the light-induced structural changes in PSII by two-flash illumination, modeled a sixth oxygen atom (O6) close to O5, and provided important implications for the O=O bond formation mechanism (Suga et al., 2017). Furthermore, it was reported that the chloride ion is essential for oxygen evolution, and there are two anion binding sites positioned on the two sides of the MnCa cluster with the same distance from the cluster to stabilize its structure (Kawakami et al., 2009).
Structure of the Plant PSII Supercomplex
Plant PSII has a similar overall structure of a dimeric supercomplex as cyanobacterial PSII. The first structure of plant PSII-LHCII complex from spinach was obtained 17 years ago at 17 Å (Nield et al., 2000), and recently a 3.2 Å spinach C2S2-type (C: PSII core complex; S: strongly associated LHCII trimer) supercomplex structure was reported with the development of single-particle cryo-electron microscopy techniques (Wei et al., 2016) (Figure 1B). More recently, structure of the dominant type of supercomplex in plants, the C2S2M2-type (M: moderately bound LHCIIs) was also solved at 2.7 and 3.2 Å for the stacked and unstacked forms from Pisum sativum (pea), and at 5.3 Å from Arabidopsis thaliana, respectively (Su et al., 2017; van Bezouwen et al., 2017). Compared to the structure of the spinach C2S2-type supercomplex, the structure reported by Su et al. (2017) was determined under more physiological conditions, containing three light-harvesting complex (LHC) monomers (CP29, CP26, and CP24) and two trimers (S-LHCII and M-LHCII) per core. In contrast, CP24 and M-LHCII are missing in the previous structure of Wei et al. (2016). The plant PSII indeed exhibits the same composition and organizations of the subunits and cofactors as their cyanobacterial counterparts (Table 1). The catalytic center within the core complex is composed of four largest membrane intrinsic subunits PsbA (D1), PsbB (CP47), PsbC (CP43), and PsbD (D2). Specifically, D1 and D2 form the photochemical RC, which is responsible for the charge separation and electron transfer, and CP47 and CP43 act as internal antenna proteins involved in light harvesting and energy transportation from peripheral antenna to the RC. In the core complex, there are also 12 low molecular-mass (MM) membrane-spanning subunits surrounding the reaction center, forming a belt-like structure. In the spinach PSII-LHCII complex, these subunits are PsbE, PsbF, PsbH-M, PsbTc, PsbW, PsbX, and PsbZ. Most of these subunits are structurally conserved with a single transmembrane helix except PsbZ with two helices. These subunits are essential for both the dimerization and stabilization of the core complex and the association between the core complex and the peripheral antenna complex. In addition, they bind cytochrome b-559 to protect the PSII complex from photo-damage. Three extrinsic subunits PsbO, PsbP, and PsbQ constitute the OEC, which also encompasses the luminal domain of CP43 and the C-terminal domain of D1, shielding the water splitting machinery. Among them, PsbO stabilizes the Mn complex while PsbP and PsbQ are involved in optimizing the oxygen evolution at physical concentration of calcium and chloride ions. Structure comparison also revealed that the flexible regions of these subunits experience significant conformational changes when they bind to the core complex (Wei et al., 2016). Outside the core complex is the LHCII, the structure of which in the PSII supercomplex is almost the same as the pea LHCII complex (Standfuss et al., 2005).
Plant LHCII occupies about 30% of total proteins in the chloroplast membrane, therefore representing the most abundant membrane protein on earth (Peter and Thornber, 1991; Standfuss et al., 2005). LHCII acts as a heterotrimer constituted by Lhcb1, Lhcb2, and Lhcb3. Each polypeptide spans the thylakoid membrane three times with its C terminus positioned on the luminal side (Kuhlbrandt et al., 1994). The LHCII complex is vital for both photosynthesis and chloroplast grana formation. For the first function, the LHCII heterotrimers are linked to the photosystem core complex by the minor antenna subunits Lhcb4 (CP29), Lhcb5 (CP26), and Lhcb6 (CP24). In spinach PSII-LHCII supercomplex, two LHCII heterotrimer together with two CP26 proteins flank the core dimer complex from both sides (Wei et al., 2016). Whereas in the C2S2M2 supercomplex from Arabidopsis and pea, there are four LHCII trimers, out of which the two strongly bound LHCII trimers (S2) along with CP26 and CP24, and the moderately bound trimers (M2) with CP29, together encompass the core complex for electron transportation (Su et al., 2017; van Bezouwen et al., 2017). For the second function, the stromal surface of the LHCII trimer is negatively charged whereas its N-terminal first 15 residues contain 4 positively charged residues. This striking charge pattern resembles a “Velcro-like” mode, guaranteeing non-specific interactions of LHCII trimers in the adjacent thylakoid membranes (Standfuss et al., 2005), which seems essential for the chloroplast grana formation. It has been reported that constitutively expression of Lhcb1 robustly increased grana stacks in the transgenic tobacco plants (Labate et al., 2004), while knock-down of Lhcb1 and Lhcb2 impede formation of grana stacks (Andersson et al., 2003; Garab, 2014, 2016). In a recent work, in order to figure out how PSII-LHCII supercomplexes interact with each other in the chloroplast thylakoid, the authors isolated the PSII-LHCII supercomplexes in an ionic concentration that resembles the chloroplast native environment, and found that most of the supercomplexes are existed in a paired C2S2M form (Albanese et al., 2017). This study provided new insights into how adjacent thylakoids might be linked to mediate the stacking of grana membranes by interactions between pairs of PSII-LHCII supercomplexes.
Cofactors within the PSII-LHCII supercomplex are indispensable for their appropriate functions. Similar to those in cyanobacteria, these cofactors mainly include chlorophylls, carotenoids, lipids etc. (Table 1). In spinach C2S2 PSII-LHCII supercomplex, there are in total 105 chlorophyll molecules, 28 β-carotenes and xanthophylls, one heme, one Mn4CaO5 cluster, one plastoquinone and numerous lipids. Interestingly, the LHCII monomer shows both amino acid sequence and structure similarities to those of CP29, however, the type, quantity and location of the chlorophylls they bind are significantly different (Liu et al., 2004; Standfuss et al., 2005; Pan et al., 2011). As the largest chromophore-bound antenna subunits, the LHCII harbors Chls which absorb solar radiation of different wavelengths of 660 ± 20 nm.
Conclusions and Perspectives
Photosynthesis plays very important roles in molecular oxygen production, atmospheric carbon dioxide control and global food supply. Structural information of the photosystems is invaluable for our understanding of photosynthesis, probably the most important process on earth. The information will also help design artificial photosynthetic system for the improvement of bioenergy production and the enhancement of agricultural productivity. Most recently, the structure of the largest light-harvesting complex, the phycobilisome (PBS) from Griffithsia pacifica was also reported (Zhang et al., 2017). As the main light-harvesting antenna in cyanobacteria and red algae, it exhibits a very fast energy transfer rate with a high quantum yield (Glazer, 1989). The structural information of the PBS will provide a firm basis for understanding its energy transfer pathways and further applications in the designs of artificial light-harvesting machineries.
Recent advances in single-particle cryo-EM have provided unprecedented structural information about these huge membrane complexes. However, there are also several open questions to be answered. First, the exact reaction mechanism underlying water oxidation and possible structural rearrangements during the S-state transitions still await the structures of PSII in more intermediate S states. Second, it is still not well understood why in PSII only one electron transfer chain is functional (as in bacterial RC), whereas in PSI both are functional (Santabarbara et al., 2010). Since static structures solved thus far has provided no conclusive clues in this respect, new studies investigating the dynamic nature of PSII might shed more light on this, which is very relevant to make PSII not only a proton pump but also the site of O2 evolution. Third, more structural information is needed to figure out the localizations and functions of PsbR and PsbS, PSII subunits that are essential for oxygen-evolving activity (Allahverdiyeva et al., 2007) and photoprotection of plants (Fan et al., 2015), respectively. Last, new high-resolution structures of the photosystems from cyanobacteria, algae, and plants will provide more insights into the evolution of oxygenic photosynthesis, based on which better artificial photosynthetic machineries could be developed.
Author Contributions
JG wrote the manuscript with the help of HW and QY. YF reviewed and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Grant Nos. 31400635 and 31670766).
References
- Albanese P., Melero R., Engel B. D., Grinzato A., Berto P., Manfredi M., et al. (2017). Pea PSII-LHCII supercomplexes form pairs by making connections across the stromal gap. Sci. Rep. 7:10067. 10.1038/s41598-017-10700-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsson P. (2001). A quantitative model of the domain structure of the photosynthetic membrane. Trends Plant Sci. 6 349–354. 10.1016/S1360-1385(01)02021-0 [DOI] [PubMed] [Google Scholar]
- Allahverdiyeva Y., Mamedov F., Suorsa M., Styring S., Vass I., Aro E. M. (2007). Insights into the function of PsbR protein in Arabidopsis thaliana. Biochim. Biophys. Acta 1767 677–685. 10.1016/j.bbabio.2007.01.011 [DOI] [PubMed] [Google Scholar]
- Andersson J., Wentworth M., Walters R. G., Howard C. A., Ruban A. V., Horton P., et al. (2003). Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II-effects on photosynthesis, grana stacking and fitness. Plant J. Cell Mol. Biol. 35 350–361. 10.1046/j.1365-313X.2003.01811.x [DOI] [PubMed] [Google Scholar]
- Arvidsson P. O., Sundby C. (1999). A model for the topology of the chloroplast thylakoid membrane. Funct. Plant Biol. 26 687–694. 10.1071/PP99072 [DOI] [Google Scholar]
- Ben-Shem A., Frolow F., Nelson N. (2003). Crystal structure of plant photosystem I. Nature 426 630–635. 10.1038/nature02200 [DOI] [PubMed] [Google Scholar]
- Biesiadka J., Loll B., Kern J., Irrgang K. D., Zouni A. (2004). Crystal structure of cyanobacterial photosystem II at 3.2 Å resolution: a closer look at the Mn-cluster. Phys. Chem. Chem. Phys. 6 4733–4736. 10.1039/B406989G [DOI] [Google Scholar]
- Brettel K., Leibl W. (2001). Electron transfer in photosystem I. Biochim. Biophys. Acta 1507 100–114. 10.1016/S0005-2728(01)00202-X [DOI] [PubMed] [Google Scholar]
- Broser M., Gabdulkhakov A., Kern J., Guskov A., Müh F., Saenger W., et al. (2010). Crystal structure of monomeric photosystem II from Thermosynechococcus elongatus at 3.6-Å resolution. J. Biol. Chem. 285 26255. 10.1074/jbc.M110.127589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarri S., Tibiletti T., Jennings R. C., Santabarbara S. (2014). A comparison between plant photosystem I and photosystem II architecture and functioning. Curr. Prot. Peptide Sci. 15 296–331. 10.2174/1389203715666140327102218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer W. A., Soriano G. M., Ponomarev M., Huang D., Zhang H., Martinez S. E., et al. (1996). Some new structural aspects and old controversies concerning the cytochrome b6f complex of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 477–508. 10.1146/annurev.arplant.47.1.477 [DOI] [PubMed] [Google Scholar]
- Croce R., van Amerongen H. (2013). Light-harvesting in photosystem I. Photosynth. Res. 116 153–166. 10.1007/s11120-013-9838-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. P., Boekema E. J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706 12–39. 10.1016/j.bbabio.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Dismukes G. C., Klimov V. V., Baranov S. V., Kozlov Y. N., DasGupta J., Tyryshkin A. (2001). The origin of atmospheric oxygen on Earth: the innovation of oxygenic photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 98 2170–2175. 10.1073/pnas.061514798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M., Li M., Liu Z., Cao P., Pan X., Zhang H., et al. (2015). Crystal structures of the PsbS protein essential for photoprotection in plants. Nat. Struct. Mol. Biol. 22 729–735. 10.1038/nsmb.3068 [DOI] [PubMed] [Google Scholar]
- Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303 1831–1838. 10.1126/science.1093087 [DOI] [PubMed] [Google Scholar]
- Fromme P., Jordan P., Krauss N. (2001). Structure of photosystem I. Biochim. Biophys. Acta 1507 5–31. 10.1016/S0005-2728(01)00195-5 [DOI] [PubMed] [Google Scholar]
- Garab G. (2014). Hierarchical organization and structural flexibility of thylakoid membranes. Biochim. Biophys. Acta 1837 481–494. 10.1016/j.bbabio.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Garab G. (2016). Self-assembly and structural-functional flexibility of oxygenic photosynthetic machineries: personal perspectives. Photosynth. Res. 127 131–150. 10.1007/s11120-015-0192-z [DOI] [PubMed] [Google Scholar]
- Glazer A. N. (1989). Directional energy transfer in a photosynthetic antenna. J. Biol. Chem. 264 1–4. [PubMed] [Google Scholar]
- Golbeck J. H. (2006). Photosystem I: The Light-Driven Plastocyanin: Ferredoxin Oxidoreductase. Dordrecht: Springer. [Google Scholar]
- Grabolle M., Dau H. (2005). Energetics of primary and secondary electron transfer in Photosystem II membrane particles of spinach revisited on basis of recombination-fluorescence measurements. Biochim. Biophys. Acta 1708 209–218. 10.1016/j.bbabio.2005.03.007 [DOI] [PubMed] [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16 334–342. 10.1038/nsmb.1559 [DOI] [PubMed] [Google Scholar]
- Holzwarth A. R., Müller M. G., Reus M., Nowaczyk M., Sander J., Rögner M. (2006). Kinetics and mechanism of electron transfer in intact photosystem II and in the isolated reaction center: pheophytin is the primary electron acceptor. Proc. Natl. Acad. Sci. U.S.A. 103 6895–6900. 10.1073/pnas.0505371103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope A. B. (2000). Electron transfers amongst cytochrome f, plastocyanin and photosystem I: kinetics and mechanisms. Biochim. Biophys. Acta 1456 5–26. 10.1016/S0005-2728(99)00101-2 [DOI] [PubMed] [Google Scholar]
- Johnson M. P. (2016). Photosynthesis. Essays Biochem. 60 255–273. 10.1042/EBC20160016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P., Fromme P., Witt H. T., Klukas O., Saenger W., Krauss N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411 909–917. 10.1038/35082000 [DOI] [PubMed] [Google Scholar]
- Kamiya N., Shen J. R. (2003). Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 100 98–103. 10.1073/pnas.0135651100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanady J. S., Agapie T. (2011). A synthetic model of the Mn3Ca subsite of the oxygen-evolving complex in photosystem II. Science 333 733–736. 10.1126/science.1206036 [DOI] [PubMed] [Google Scholar]
- Kawakami K., Umena Y., Kamiya N., Shen J. R. (2009). Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 106 8567. 10.1073/pnas.0812797106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern J., Alonso-Mori R., Tran R., Hattne J., Gildea R. J., Echols N., et al. (2013). Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature. Science 340 491–495. 10.1126/science.1234273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H. (2013). Architectural switches in plant thylakoid membranes. Photosynth. Res. 116 481–487. 10.1007/s11120-013-9843-0 [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W., Wang D. N., Fujiyoshi Y. (1994). Atomic model of plant light-harvesting complex by electron crystallography. Nature 367 614–621. 10.1038/367614a0 [DOI] [PubMed] [Google Scholar]
- Kupitz C., Basu S., Grotjohann I., Fromme R., Zatsepin N. A., Rendek K. N., et al. (2014). Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature 513 261–265. 10.1038/nature13453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labate M. T., Ko K., Ko Z. W., Pinto L. S., Real M. J., Romano M. R., et al. (2004). Constitutive expression of pea Lhcb 1-2 in tobacco affects plant development, morphology and photosynthetic capacity. Plant Mol. Biol. 55 701–714. 10.1007/s11103-004-1963-7 [DOI] [PubMed] [Google Scholar]
- Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., et al. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428 287–292. 10.1038/nature02373 [DOI] [PubMed] [Google Scholar]
- Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005). Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438 1040–1044. 10.1038/nature04224 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Stull J. A., Yano J., Stamatatos T. C., Pringouri K., Stich T. A., et al. (2012). Synthetic model of the asymmetric [Mn3CaO4] cubane core of the oxygen-evolving complex of photosystem II. Proc. Natl. Acad. Sci. U.S.A. 109 2257–2262. 10.1073/pnas.1115290109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux C. W. (2005). Function and evolution of grana. Trends Plant Sci. 10 521–525. 10.1016/j.tplants.2005.09.001 [DOI] [PubMed] [Google Scholar]
- Mustárdy L., Buttle K., Steinbach G., Garab G. (2008). The three-dimensional network of the thylakoid membranes in plants: quasihelical model of the granum-stroma assembly. Plant Cell 20 2552–2557. 10.1105/tpc.108.059147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N. (2011). Photosystems and global effects of oxygenic photosynthesis. Biochim. Biophys. Acta 1807 856–863. 10.1016/j.bbabio.2010.10.011 [DOI] [PubMed] [Google Scholar]
- Nelson N., Benshem A. (2004). The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5 971–982. 10.1038/nrm1525 [DOI] [PubMed] [Google Scholar]
- Nelson N., Junge W. (2015). Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 84 659–683. 10.1146/annurev-biochem-092914-041942 [DOI] [PubMed] [Google Scholar]
- Nelson N., Yocum C. F. (2006). Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 57 521–565. 10.1146/annurev.arplant.57.032905.105350 [DOI] [PubMed] [Google Scholar]
- Nield J., Orlova E. V., Morris E. P., Gowen B., van Heel M., Barber J. (2000). 3D map of the plant photosystem II supercomplex obtained by cryoelectronmicroscopy and single particle analysis. Nat. Struct. Biol. 7 44–47. 10.1038/71242 [DOI] [PubMed] [Google Scholar]
- Pan X., Li M., Wan T., Wang L., Jia C., Hou Z., et al. (2011). Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nat. Struct. Mol. Biol. 18 309–315. 10.1038/nsmb.2008 [DOI] [PubMed] [Google Scholar]
- Peter G. F., Thornber J. P. (1991). Biochemical composition and organization of higher plant photosystem II light-harvesting pigment-proteins. J. Biol. Chem. 266 16745–16754. [PubMed] [Google Scholar]
- Pfannschmidt T. (2003). Chloroplast redox signals: how photosynthesis controls its own genes. Trends Plant Sci. 8 33–41. 10.1016/S1360-1385(02)00005-5 [DOI] [PubMed] [Google Scholar]
- Qin X., Suga M., Kuang T., Shen J. R. (2015). Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348 989–995. 10.1126/science.aab0214 [DOI] [PubMed] [Google Scholar]
- Remelli R., Varotto C., Sandonà D., Croce R., Bassi R. (1999). Chlorophyll binding to monomeric light-harvesting complex. A mutation analysis of chromophore-binding residues. J. Biol. Chem. 274 33510–33521. 10.1074/jbc.274.47.33510 [DOI] [PubMed] [Google Scholar]
- Ruban A. V. (2015). Evolution under the sun: optimizing light harvesting in photosynthesis. J. Exp. Bot. 66 7–23. 10.1093/jxb/eru400 [DOI] [PubMed] [Google Scholar]
- Santabarbara S., Galuppini L., Casazza A. P. (2010). Bidirectional electron transfer in the reaction centre of photosystem I. J. Integr. Plant Biol. 52 735–749. 10.1111/j.1744-7909.2010.00977.x [DOI] [PubMed] [Google Scholar]
- Sétif P. (2001). Ferredoxin and flavodoxin reduction by photosystem I. Biochim. Biophys. Acta 1507 161–179. 10.1016/S0005-2728(01)00205-5 [DOI] [PubMed] [Google Scholar]
- Standfuss J., Ac T. V. S., Lamborghini M., Kühlbrandt W. (2005). Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Å resolution. EMBO J. 24 919–928. 10.1038/sj.emboj.7600585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Ma J., Wei X., Cao P., Zhu D., Chang W., et al. (2017). Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 357 815–820. 10.1126/science.aan0327 [DOI] [PubMed] [Google Scholar]
- Suga M., Akita F., Hirata K., Ueno G., Murakami H., Nakajima Y., et al. (2015). Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 517 99–103. 10.1038/nature13991 [DOI] [PubMed] [Google Scholar]
- Suga M., Akita F., Sugahara M., Kubo M., Nakajima Y., Nakane T., et al. (2017). Light-induced structural changes and the site of O = O bond formation in PSII caught by XFEL. Nature 543 131–135. 10.1038/nature21400 [DOI] [PubMed] [Google Scholar]
- Tomioka N., Sugiura M. (1983). The complete nucleotide sequence of a 16S ribosomal RNA gene from a blue-green alga, Anacystis nidulans. Mol. Gen. Genet. 191 46–50. 10.1007/BF00330888 [DOI] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J. R., Kamiya N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473 55–60. 10.1038/nature09913 [DOI] [PubMed] [Google Scholar]
- van Bezouwen L. S., Caffarri S., Kale R. S., Kouřil R., Thunnissen A. W. H., Oostergetel G. T., et al. (2017). Subunit and chlorophyll organization of the plant photosystem II supercomplex. Nat. Plants 3:17080. 10.1038/nplants.2017.80 [DOI] [PubMed] [Google Scholar]
- van Amerongen H., Croce R. (2013). Light harvesting in photosystem II. Photosynth. Res. 116 251–263. 10.1007/s11120-013-9824-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Su X., Cao P., Liu X., Chang W., Li M., et al. (2016). Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534 69–74. 10.1038/nature18020 [DOI] [PubMed] [Google Scholar]
- Woodson J. D. (2016). Chloroplast quality control – balancing energy production and stress. New Phytol. 212 36–41. 10.1111/nph.14134 [DOI] [PubMed] [Google Scholar]
- Zhang J., Ma J., Liu D., Qin S., Sun S., Zhao J., et al. (2017). Structure of phycobilisome from the red alga Griffithsia pacifica. Nature 551 57–63. 10.1038/nature24278 [DOI] [PubMed] [Google Scholar]
- Zouni A., Witt H. T., Kern J., Fromme P., Krauss N., Saenger W., et al. (2001). Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 409 739–743. 10.1038/35055589 [DOI] [PubMed] [Google Scholar]