Abstract
Spatial organization is a hallmark of all living systems. Even bacteria, the smallest forms of cellular life, display defined shapes and complex internal organization showcasing a highly structured genome, cytoskeletal filaments, localized scaffolding structures, dynamic spatial patterns, active transport, and occasionally intracellular organelles. Spatial order is required for faithful and efficient cellular replication, and offers a powerful means for the development of unique biological properties. Here, we discuss organizational features of bacterial cells and highlight how bacteria have evolved diverse spatial mechanisms to overcome challenges cells face as self-replicating entities.
INTRODUCTION
The second law of thermodynamics dictates that isolated systems evolve toward greater entropy. Organisms, however, are not isolated systems. They exchange energy and matter with the environment, driving them far from thermodynamic equilibrium. As a result, living systems can achieve a stable organized state. Spatial organization is, in fact, evident at all levels of biological complexity.
Bacterial cells are no exception. Spatial order is readily apparent in the highly reproducible cell geometries observed in the bacterial world, ranging from spherical and rod shapes to helical, branched and complex spiny morphologies. Geometrical order in bacterial shapes has been known for a long time, first unveiled by Antonie van Leeuwenhoek at the dawn of microscopy over three centuries ago. However, an appreciation for the internal organization of bacterial cells only emerged in the last 15-20 years, spurred by advances in imaging techniques. In fact, as we will illustrate, bacteria exhibit many of the cell biological complexities once thought to be unique to eukaryotic cells. Granted, apart from notable exceptions, bacteria lack the membrane-enclosed organelles that help organize the interior of eukaryotic cells. Instead, bacteria have adopted an open plan architectural design of their cytoplasmic space where spatial organization arises in the absence of membrane boundaries.
The first glimpses of intracellular organization in bacteria came with the discoveries that bacterial cells can cluster chemoreceptors at specific locations (Alley et al., 1992; Maddock and Shapiro, 1993), possess cytoskeletal structures (Bi and Lutkenhaus, 1991; Jones et al., 2001; van den Ent et al., 2001; Williamson, 1974), feature protein oscillations (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999b), and spatially organize and actively segregate chromosomal regions (Glaser et al., 1997; Gordon et al., 1997; Teleman et al., 1998; Webb et al., 1997). These pioneering works were followed by a wealth of studies showcasing the diversity of cell biology among bacterial species and demonstrating the pervasive role of spatial order in many aspects of bacterial life. Cellular organization has emerged as an integral element of the cellular processes that must take place for bacteria to successfully self-replicate in their environment. Bacteria are famous (or infamous) for their proliferative potential. They are also experts in cellular specialization, which has allowed them to colonize almost every corner of the Earth. While much of this colonization success stems from diversification of metabolic functions, diversification of cellular organization is also an important contributing factor.
Because of space constraints, we are unable to describe the vast array of exciting cell biological observations that have been reported in bacteria. This review is not meant to be exhaustive. Our goal is to describe, with illustrative examples, organizational features and self-organizing properties under two different contexts. First, we will discuss the bacterial architecture in the context of the universal tasks that cells must perform to achieve efficient and faithful self-replication. Second, we will discuss spatial organization in the context of specialized biological functions that bacteria have evolved to survive and thrive in their ecological niches.
SHAPING THE CELL
Bacterial cells generally adopt well-defined physical shapes. Cell geometry impacts various aspects of bacterial physiology, such as nutrient uptake, motility, colonization, and pathogenesis (Kysela et al., 2016; Yang et al., 2016). Another critical, but often unappreciated, aspect of cell shape is that it provides geometric features that bacteria use to establish the intracellular organization necessary for various cellular processes, as we will discuss throughout the review. Thus, reproducing a specific cell geometry is a crucial task that bacteria must accomplish at every division cycle.
Cell shape is typically endowed by the peptidoglycan (PG), a major component of the cell wall, that consists of glycan strands crosslinked by peptides. This meshwork, also referred to as the sacculus, encases the cytoplasmic membrane and bears stress generated from cytoplasmic turgor pressure. A loss of PG integrity (e.g., following exposure to β-lactam antibiotics) results in loss of cell shape and eventually lysis. Isolated PG sacculi generally preserve the morphological features of the cells from which they were purified, indicating that the morphology of a cell is dictated by the morphogenesis of the PG sacculus.
Cell division: duplicating round shapes or generating new cell poles
How do PG sacculi obtain their forms? This spatial problem is solved through cellular organization, usually using cytoskeletal elements and scaffolding structures that spatially regulate PG synthesis and remodeling. For instance, part of cell geometry is generated during the cell division process, which involves septal (inward) PG synthesis. In spherical and ovoid bacteria, septal PG growth plays an important role in reproducing the round shape of the cell at every generation, though the cell cycle pattern of PG growth can vary among species (Figure 1Ai) (Monteiro et al., 2015; Turner et al., 2010; Wheeler et al., 2011; Zhou et al., 2015; Zhou et al., 2016).
Figure 1. Shaping the cell.
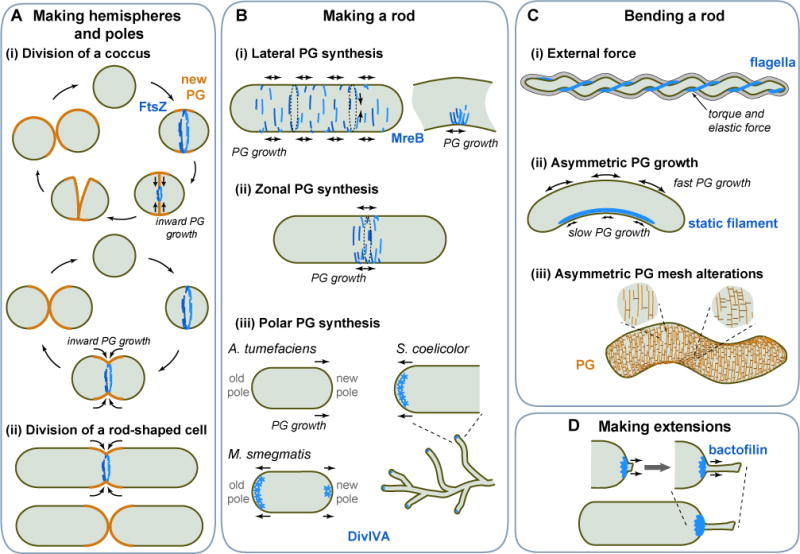
A. (i) FtsZ-dependent inward growth of PG during division produces new cell halves in spherical bacteria. Two types of cell cycle are shown. The top panel showcases septal PG synthesis followed by an abrupt separation of daughter cells, as in Staphylococcus aureus. The bottom panel illustrates the hypothetical case of septal PG synthesis occurring concurrently with cell constriction. A good candidate for this cell cycle pattern is Moraxella catarrhalis (Zhou et al, 2016). (ii) Septal PG growth gives rise to cell poles in rod-shaped bacteria. B. Rod shapes can be created by MreB-dependent (i) lateral or (ii) zonal PG incorporation. Preferential localization of MreB to the inner curve of a bent cell cylinder has been proposed to help restore straight shape. (iii) Rod shape can also derive from polar growth, which, in some bacteria, is DivIVA-dependent. C. Curved cell shapes can be achieved (i) by a filamentous structure such as periplasmic flagella applying an external force on the PG, (ii) through asymmetric PG growth between lateral walls caused by a static protein filament, (iii) via local modifications of the PG mesh, affecting its material properties. D. Stalk formation is achieved through localized PG growth with the assistance of a bactofilin structure.
Apart from a few known exceptions (Jeske et al., 2015; Liechti et al., 2016; Ouellette et al., 2012; Pilhofer et al., 2013), the orchestration of PG synthesis during division is entrusted to the highly conserved FtsZ protein. FtsZ is a GTPase and a structural tubulin homolog that polymerizes upon GTP binding (Bramhill and Thompson, 1994; de Boer et al., 1992; Erickson et al., 1996; Lowe and Amos, 1998; Mukherjee et al., 1993; Mukherjee and Lutkenhaus, 1994; RayChaudhuri and Park, 1992). FtsZ polymers attach to the cytoplasmic membrane through auxiliary proteins, such as FtsA (Pichoff and Lutkenhaus, 2005), and accumulate at the site of division, where they form a ring-like structure (Z-ring) (Figure 1A) (Bi and Lutkenhaus, 1991; Levin and Losick, 1996; Ma et al., 1996).
The structure of the Z-ring (continuous vs. discontinuous) and the exact arrangement of FtsZ polymers within the ring (length, number, orientation, etc.) are the subject of intense debate and may vary between species (Fu et al., 2010; Holden et al., 2014; Jennings et al., 2011; Li et al., 2007; Rowlett and Margolin, 2014; Si et al., 2013; Strauss et al., 2012; Szwedziak et al., 2014; Yao et al., 2017). It is, however, well accepted that the Z-ring provides a scaffold for the recruitment of a number of other division proteins, including components of the PG synthesis machinery that build septal PG during Z-ring constriction (den Blaauwen et al., 2017; Typas et al., 2011).
The Z-ring scaffold is a dynamic structure (Strauss et al., 2012; Stricker et al., 2002). FtsZ polymers move circumferentially around the septal plane (Bisson-Filho et al., 2017; Yang et al., 2017). This motion is achieved by treadmilling, a GTP-hydrolysis-dependent process in which FtsZ monomers are added at one end of the polymer while being removed from the other end. FtsZ treadmilling, which is also observed in vitro (Loose and Mitchison, 2014), is thought to position and distribute PG synthases around the septal plane (Bisson-Filho et al., 2017; Yang et al., 2017).
How the Z-ring constricts and coordinates PG synthesis inward remains an open question. In vitro reconstitution experiments have demonstrated that FtsZ filaments alone can pinch the membrane of tubular liposomes (Osawa et al., 2008; Osawa and Erickson, 2013; Szwedziak et al., 2014). Thus, an attractive possibility is that FtsZ filaments locally pinch the cytoplasmic membrane, perhaps through filament motion, sliding, or bending (Erickson and Osawa, 2017; Lan et al., 2009; Li et al., 2007; Szwedziak et al., 2014). Membrane invagination would help redirect PG synthesis inward, and reiteration of this process would produce concentric PG rings of decreasing size.
Cell elongation: reproducing rod shapes
Rod-shaped bacteria must add a phase of PG growth during which the cylindrical portion of the cell elongates. This can be achieved by inserting new PG material while maintaining the cell width constant. In many rod-shaped bacteria, this process is regulated by the ATPase MreB, a structural homolog of actin that polymerizes upon ATP binding (Jones et al., 2001; van den Ent et al., 2001).
How MreB polymers spatially control PG morphogenesis is not fully understood. Inside cells, MreB filaments bind to the cytoplasmic face of the membrane (Dempwolff et al., 2011; Salje et al., 2011) and connect to the PG synthesis machinery through transmembrane proteins, such as RodZ and MreC/D (Typas et al., 2011). MreB filaments, which are largely oriented orthogonal to the long axis of the cell (Olshausen et al., 2013; Ouzounov et al., 2016), move along the cell circumference together with the PG machinery (Figure 1Bi) (Dominguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011). MreB motion is unlikely to result from treadmilling because MreB forms antiparallel double-stranded filaments in vitro (van den Ent et al., 2014), indicating that both ends are equivalent. Instead, circumferential motion depends on PG synthesis (Dominguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011), suggesting that the processivity of PG synthetic enzymes sustains MreB motion. Presumably, the orientation of the MreB filaments helps direct the addition of circular (likely discontinuous) bands of PG oriented perpendicular to the cell length. What sets the width of these PG hoops remains unclear. It does not require a preexisting shape template, as rod shape can arise from spherical PG-deficient cells following restoration of PG synthesis (Kawai et al., 2014; Lederberg, 1956; Miller et al., 1968).
At the cellular scale, the spatial distribution of MreB dictates where new PG will be added. Several patterns have been reported for rod-shaped bacteria. For example, in Bacillus subtilis and Escherichia coli, MreB filaments are distributed throughout the cylindrical portion of the cell. As a result, PG material is inserted along the entire cell cylinder, in a growth mode referred to as “dispersed” or “lateral” PG synthesis (Figure 1Bi). What prevents PG growth at the poles? The answer is likely related to cell geometry. For instance, the conical shape of anionic phospholipids (e.g., cardiolipin) favors their accumulation in the curved membrane of cell poles (Mukhopadhyay et al., 2008; Renner and Weibel, 2011), which presumably repels MreB filaments, as they tend to avoid anionic phospholipids (Kawazura et al., 2017). On a finer scale, MreB filaments accumulate at the inner curvature of cell bends (Ursell et al., 2014), potentially due to their circumferential motion (Wong et al., 2017). The resulting local accumulation of PG material (Ursell et al., 2014) or PG-strain-dependent growth (Wong et al., 2017) is thought to help straighten the cell cylinder from accidental bends (Figure 1Bi).
While the lateral mode of PG synthesis is common among rod-shaped species, cell elongation can also derive from PG growth in more confined cellular regions, as in Borrelia species (Jutras et al., 2016). In Caulobacter crescentus, MreB localization and addition of PG material shift during the cell cycle from a dispersed distribution along the cell body to a narrow central zone in an FtsZ-dependent manner (Figure 1Bii) (Aaron et al., 2007; Figge et al., 2004; Gitai et al., 2004).
Given the function of MreB in PG morphogenesis, it is not surprising that most spherical bacteria do not possess an MreB homolog (Pinho et al., 2013). However, some rod-shaped bacteria also lack MreB. Examples include members of Mycobacteriaceae and Rhizobiales. These bacteria display a “polar” or “subpolar” mode of growth. For example, Agrobacterium tumefaciens elongates from the “new” pole, which was formed by the last division (Figure 1Biii) (Brown et al., 2012). In Mycobacterium smegmatis, PG incorporation occurs at both polar regions, although possibly at a different rate (Figure 1Biii) (Aldridge et al., 2012; Joyce et al., 2012; Kang et al., 2008; Meniche et al., 2014; Santi et al., 2013; Thanky et al., 2007). Polar growth also occurs in the mycelium-forming bacterium Streptomyces coelicolor (Flardh, 2003). This bacterium possesses MreB, yet it uses an MreB-independent mechanism to direct polar cell wall growth (Mazza et al., 2006). This mechanism relies on DivIVA, another type of polymerizing protein that self-assembles into a matrix at the incipient growing tips (Figure 1Biii) (Flardh, 2003) with the help of the intermediate filament-like proteins, FilP and Scy (Fuchino et al., 2013; Holmes et al., 2013).
Curvature generation: producing crescent, sigmoid, and helical shapes
Various bacteria form curved rods or spirals. One way to curve a rod is by physically bending it, a strategy that the Lyme disease-causing spirochete Borrelia burgdorferi has adopted. This pathogen forms long cells with a flat-wave morphology. The curved morphology is mechanically imparted by the flagella located between the cytoplasmic and outer membranes where flagella wrap around the PG (Charon et al., 2009; Dombrowski et al., 2009; Goldstein et al., 1996; Goldstein et al., 1994; Motaleb et al., 2000). The stiffness of the helical flagella relative to that of the cell cylinder causes the cell to deform into a flat-wave shape (Figure 1Ci). Flagellar rotation propagates waves of cell deformations along the cell length, propelling the cell forward. A flagellar mutant loses the ability to swim and becomes rod-shaped, showcasing the moonlighting skeletal function of the flagella.
Bending the PG to shape the cell is an unusual strategy. More commonly, bacterial curved morphologies are imprinted into the PG fabric itself. Cell curvature can be achieved during growth by adding more PG material to one lateral wall relative to the other (Figure 1Cii). Studies in curved bacteria suggest that anisotropy in PG growth between lateral walls can be achieved with the help of self-assembling proteins, such as crescentin in C. crescentus and CrvA in Vibrio cholerae (Ausmees et al., 2003; Bartlett et al., 2017). These proteins spontaneously polymerize into filaments in vitro (i.e., without the need of nucleotides) and form static structures that are asymmetrically localized inside the cell. Evidence suggests that these structures generate curvature by mechanically reducing PG synthesis on the side of the cell where the protein structure attaches (Figure 1Cii) (Bartlett et al., 2017; Cabeen et al., 2009), though the precise mechanism is not fully understood. Perhaps the elastic property of the filamentous structure locally opposes the stretching of the PG fabric exerted by the turgor pressure (Cabeen et al., 2009). Reduced mechanical stress on the PG on one side of the cell relative to the other presumably causes an anisotropy in PG growth.
The commonalities between crescentin and CrvA illustrate an interesting case of convergent evolution, as these proteins have important distinguishing features. Like traditional cytoskeletal proteins, crescentin is cytoplasmic. Furthermore, crescentin is a coiled-coil rich protein that shares the domain organization, the biochemical properties and the in vivo dynamics of eukaryotic intermediate filament proteins (Ausmees et al., 2003; Cabeen et al., 2011; Charbon et al., 2009). CrvA, on the other hand, is periplasmic, and shares little similarity to intermediate filament proteins (apart from two relatively short coiled-coil regions) or to other known cytoskeletal proteins (Bartlett et al., 2017).
Some bacteria combine cytoskeletal and PG-modifying functions to curve their PG fabric. For example, Helicobacter pylori uses cytoplasmic intermediate filament-like proteins (coiled-coil-rich proteins Ccrp59, Ccrp1143, Ccrp58 and Ccrp1142) to curve its cells (Specht et al., 2011; Waidner et al., 2009), presumably through anisotropic PG growth. In addition, this bacterium modulates its helical twist through the activity of PG crosslink hydrolases (Sycuro et al., 2010). The PG-relaxing activity of these enzymes affects the material properties of the load-bearing PG mesh. Modeling shows that if these alterations are spatially ordered (e.g., along a helical path), they will lead to curved morphologies under turgor pressure.
Expansion of shape complexity
Spatial regulation of PG morphogenesis can yield a wide diversity of cell shapes (Kysela et al., 2016), though the underlying mechanisms are poorly understood. In some cases, it involves cytoskeletal proteins unique to the bacterial world, such as bactofilins (Lin and Thanbichler, 2013). This is illustrated in C. crescentus, which adopts an exotic shape by forming a long, thin cell extension (stalk) at a cell pole (Figure 1D). Full extension of the polar stalk partially depends on bactofilins BacA and BacB (Kuhn et al., 2010). These two proteins self-assemble into a sheet-like structure at the cell pole where a PG synthase is recruited (Figure 1D). Studies of stalk morphogenesis have also provided insight into how morphological transitions may arise during evolution. For instance, changes in the regulation and protein sequence of a morphogen (SpmX) are thought to have driven the evolution of stalk positioning from polar (C. crescentus) to subpolar (Asticcacaulis excentricus) and bilateral (Asticcacaulis biprosthecum) (Jiang et al.,2014).
2. PACKING AND DECODING THE GENOME
Cell geometry defines the intracellular space. An important challenge that all cells face is to pack their oversized genome into this limited space without compromising DNA processes. The contour length of a bacterial chromosome is ~1,000 times larger than the micrometer-sized cell (Figure 2Ai; see also Box 1). To overcome this packing problem, bacteria condense their chromosome(s) into a spatially ordered, yet pliable, three-dimensional (3D) structure composed of domains within domains (Badrinarayanan et al., 2015; Dame and Tark-Dame, 2016; Wang and Rudner, 2014).
Figure 2. Folding and decoding the genome.
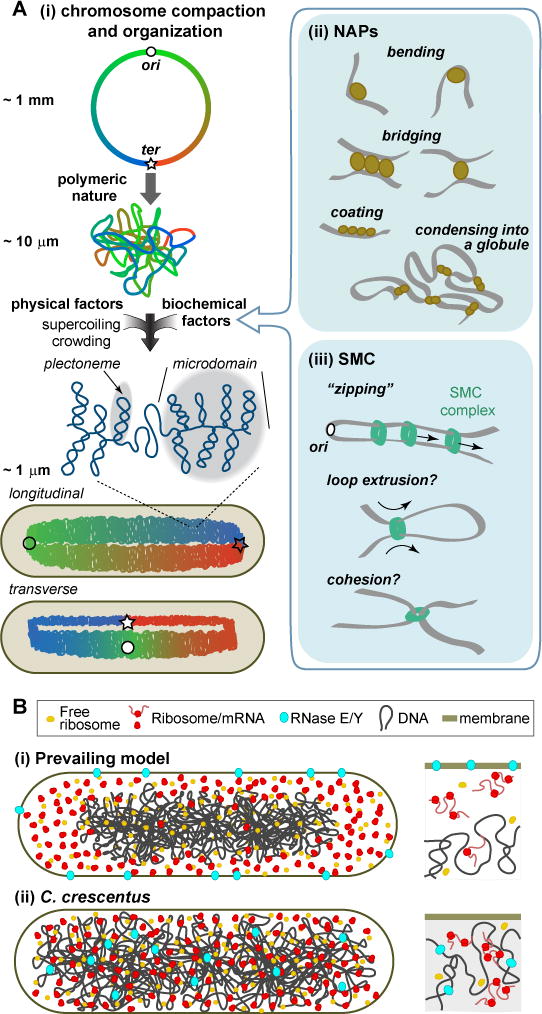
A. (i) Bacterial chromosomes can fold into two major configurations: longitudinal or transverse. Folding is driven by physical factors and DNA-binding proteins such as (ii) NAPs and (iii) SMC complexes. SMC complexes may travel in pairs (Badrinarayanan et al., 2012; Wang et al., 2017), but just a single complex is shown for simplicity. B. (i) Prevailing view of nucleoid (transcription), polysome (translation) and RNase E or Y (mRNA degradation) arrangement in bacterial cells. (ii) Unconventional organization of the chromosomal DNA, ribosomes and RNase E in C. crescentus.
BOX 1. PHYSICAL CONSIDERATIONS.
Here, we briefly describe some physical aspects that contribute to intracellular organization.
Polymeric nature of the DNA
Most bacteria have a single circular chromosome. Given that the distance between adjacent nucleotides is 1/3 nm, the contour length of a typical bacterial chromosome (4.5×106 base pairs, bp) is 1.5 mm. At the cellular scale, the chromosome can be modeled as a very long flexible polymer, as the persistence (rigidity) length (Lp) of double-stranded DNA is small (~150 bp or ~50 nm) relative to the length of the chromosome (1.5 mm). This polymeric nature emphasizes the role of entropic effects, as the chromosome has an intrinsic tendency to adopt a conformation that maximizes available degrees of freedom for its segments. These entropic effects would drive spontaneous condensation of the chromosome into a soft globule of size L~Lp(N)1/2 ≈ 10 μm, considering the chromosome as an ideal polymer of length N in persistence-length units.
Crowding and excluded volume effects on chromosome organization
Inside cells, the DNA is not in an ideal solvent. The high macromolecular crowding of the cytoplasm causes the collapse of large polymer loops through excluded volume effects. This effect is entropy-driven; a decrease in the conformational freedom of the polymer is overcompensated by the gain in accessible volume by the large number of crowding particles.
DNA supercoiling
Another crucial aspect of DNA physics is that the DNA is an oriented helix with a natural pitch of about 10.5 bp per twist. Addition or removal of twists imposes a torsional (mechanical) strain. In the case of a circular DNA, the strain is partially released by the folding of the DNA into superhelices. A classic (albeit outdated) analogy for this supercoiling effect is the coiling of a telephone cord generated by repetitive rotations of the phone’s handset.
Elastic properties of the chromosome
From a statistical physics perspective, the probability to find a particle (a chromosomal locus, in our case) at position x is given by the Boltzmann distribution: , where E(x) is the potential energy of the particle, k is the Boltzmann constant, and T is the absolute temperature. From the experimental measurement of such a distribution, one can infer the energy potential E(x) in which the particle moves. For chromosomal loci, experiments show that the probability of finding a chromosomal locus away (at position x) from its mean position x0 follows a Gaussian distribution: , with the deviation (σ) from the mean position being ~ 100 nm (Lim et al., 2014; Surovtsev et al., 2016). This implies that a chromosomal locus moves in a “harmonic” potential E(x) = a(x−x0)2 with a = kT/2σ2 and with the particle experiencing a force F = 2a(x−x0), i.e., an elastic force with a spring constant ksp = 2a = kT/σ2. In other words, the chromosomal locus moves like a particle on a spring experiencing a characteristic force: F = ksρσ ~ 0.04 pN.
ParA-dependent translocating force on plasmid cargos
A force F applied to a particle in a viscous liquid is related to the particle velocity v as: , where D is the diffusion coefficient of the particle. The average velocity of a single plasmid cargo can be estimated from experiments as v = 2l/t = 0.007 μm/s, where l is the length of nucleoid (~ 2 μm) and t is the period of plasmid oscillation (~10 min) (Ah-Seng et al., 2013; Ringgaard et al., 2009). Given the experimentally measured diffusion coefficient of plasmids D ~ 0.001 μm2/s (Ietswaart et al., 2014; Surovtsev et al., 2016), the estimated translocating force on plasmid cargos is F~ 0.03 pN.
Random distribution and inheritance of cellular components
If a cellular component in n copies are distributed randomly in the cell, the probability to find all copies in the same half of the cell is .
Chromosome organization
How do the compaction and domain organization of the bacterial chromosome come about? Physics offers a partial explanation. First, polymer dynamics predicts that even without a cell envelope or other physical confinement, the chromosome should spontaneously shape into a globule (Figure 2Ai), accounting for an ~100-fold compaction (see Box 1). Second, the high macromolecular crowding of the cytoplasm results in additional compacting forces on the DNA polymer through excluded volume effects (see Box 1) (Cunha et al., 2001; Murphy and Zimmerman, 1997; Odijk, 1998). Third, torsional stress leads to supercoiling, i.e., DNA folding into superhelices (plectonemes) (Figure 2Ai, see Box 1). Various DNA-related activities mitigate or enhance these physical effects. For example, topoisomerases and RNA polymerases modulate torsional stress, thus affecting DNA supercoiling.
Biochemical factors also play important roles in compacting and spatially organizing the chromosome. While bacteria lack histones, they produce various nucleoid-associated proteins (NAPs), which are DNA-binding proteins that generate local and long-distance changes in chromosome organization (Figure 2Aii). The presence and abundance of specific NAPs depend on the species and the growth phase of the culture (e.g., exponential vs. stationary phase) (Dorman, 2014). Some NAPs (e.g., FIS in E. coli and IHF in B. subtilis) change DNA geometry locally by generating kinks in the DNA polymer (Figure 2Aii) (e.g., Thompson and Landy, 1988). Other NAPs (e.g., H-NS in E. coli) bridge different DNA segments and stabilize domains by simultaneously binding to multiple sites (Figure 2Aii) (e.g., Dame et al., 2006). NAPs can have multiple functions. For example, H-NS and HU have also been reported to make a “polymeric coat” on stretches of DNA (Figure 2Aii), locally altering the bending rigidity of the DNA (e.g., Amit et al., 2003; Sagi et al., 2004; van Noort et al., 2004). Other, less abundant DNA-binding proteins organize specific DNA regions on a smaller scale. For example, MatP organizes the terminus (ter) region of the E. coli chromosome (Mercier et al., 2008). In vitro observations suggest that MatP creates a cross-connected DNA globule around ter through bivalent bridging of multiple high-affinity binding sites (Figure 2Aii) (Dupaigne et al., 2012).
The chromosome is also actively shaped by the Structural Maintenance of Chromosome (SMC) complex, which forms a structure that encircles DNA (Figure 2Aiii) (Gruber, 2014; Kleine Borgmann and Graumann, 2014; Nolivos and Sherratt, 2014). Typically, the SMC complex consists of the ATPase SMC and the auxiliary proteins ScpA and ScpB (Mascarenhas et al., 2002; Soppa et al., 2002). E. coli lacks these proteins, but forms an analogous and distantly related complex made of MukB, MukE and MukF (Yamazoe et al., 1999). In B. subtilis, Streptococcus pneumoniae and C. crescentus, the SMC complex preferentially loads onto the chromosome at the parS sequence near the origin of replication (ori) (Gruber and Errington, 2009; Minnen et al., 2011; Sullivan et al., 2009; Tran et al., 2017), in a process that requires ATP binding and probably ATP hydrolysis (Wilhelm et al., 2015). The loaded SMC complex then translocates to distal DNA in an ATP hydrolysis-dependent manner (Figure 2Aiii) (Minnen et al., 2016). The current model in B. subtilis suggests that continuous loading of SMC complexes, followed by their active translocation from ori to ter, “zips” and thereby aligns the chromosome arms (Figure 2Aiii) (Marbouty et al., 2015; Wang et al., 2017; Wang et al., 2015). SMC complexes are also thought to mediate intra-arm contact (e.g., Marbouty et al., 2015), which, with active translocation, would result in DNA loop extrusion (Figure 2Aiii).
Together, physical and biochemical factors shape the chromosome into a domain-within-a-domain structure (Figure 2Ai), which was recently mapped at high resolution in chromosome conformation capture experiments (Le et al., 2013; Marbouty et al., 2014; Marbouty et al., 2015; Val et al., 2016; Wang et al., 2015). At the cellular scale, the chromosome maintains a general linear arrangement inside the cell, such that the cytoplasmic position of each gene locus can be predicted based on its chromosomal coordinate (Figure 2Ai) (David et al., 2014; Vallet-Gely and Boccard, 2013; Viollier et al., 2004; Wiggins et al., 2010). The orientation of the chromosome during the non-replicating phase can, however, vary. In the “longitudinal” arrangement, ori and ter are located at opposite cell ends, with the left and right chromosomal arms side-by-side (Figure 2Ai) (Deghelt et al., 2014; Fogel and Waldor, 2005; Harms et al., 2013; Jensen and Shapiro, 1999; Srivastava et al., 2006; Teleman et al., 1998; Vallet-Gely and Boccard, 2013). In this arrangement, the left and right arms are thought to wrap around each other (Umbarger et al., 2011). In the “transverse” arrangement, ori and ter are in the middle of the cell, with the left and right arms at opposite ends (Figure 2Ai) (Nielsen et al., 2006; Niki et al., 2000; Wang et al., 2006). Some bacteria can switch between longitudinal and transverse arrangements depending on the cell cycle stage and the growth conditions (Bates and Kleckner, 2005; Cass et al., 2016; Niki et al., 2000; Wang et al., 2014a).
Chromosomal organization and compaction result in a discrete, albeit dynamic, coiled structure whose shape depends on the geometry of the cell (Fisher et al., 2013; Hadizadeh Yazdi et al., 2012). This compacted DNA structure is called the nucleoid.
Spatial organization of gene expression
The nucleoid and cell geometry are the most salient features of bacterial cell architecture. These two cellular features serve as foundational elements from which other aspects of cellular organization can develop.
To start with, the nucleoid and cell geometry help spatially organize the transfer of genetic information from DNA to proteins. This is best studied in rod-shaped bacteria, where the nucleoid is centrally located in the cytoplasm leaving the cell poles largely free of DNA (Figure 2Bi). The open plan of the bacterial cytoplasm means that ribosomes have direct access to the DNA. Free ribosomes diffuse through the nucleoid (Sanamrad et al., 2014) and load onto nascent mRNAs, such that translation initiates during transcription. However, as translation proceeds, polysomes (mRNAs loaded with multiple ribosomes) relocate outside of the nucleoid, resulting in ribosome accumulation at the poles (Figure 2Bi), as shown in E. coli, B. subtilis and Lactococcus lactis (Bakshi et al., 2012; Lewis et al., 2000; van Gijtenbeek et al., 2016). The partial physical separation between the nucleoid and ribosomes suggests that transcription and translation are partially separated in space, despite the lack of a nuclear envelope.
mRNA degradation is also under spatial control. The major component of the degradation machinery (RNase E in E. coli and RNase Y in B. subtilis) is enriched at the cytoplasmic membrane (Figure 2Bi) (Lehnik-Habrink et al., 2011; Liou et al., 2001). This membrane localization indicates that even though mRNA degradation can begin on nascent mRNAs during transcription (Morikawa and Imamoto, 1969; Morse et al., 1969), most of mRNA degradation likely occurs near the membrane, away from the nucleoid. In E. coli, mRNAs encoding inner membrane proteins are found in greater abundance near the membrane; accordingly, they have shorter lifetimes relative to mRNAs located elsewhere (Moffitt et al., 2016). Some mRNAs have also been shown to localize to specific regions where their protein products are needed (dos Santos et al., 2012; Nevo-Dinur et al., 2011). Thus, in spite of their open plan layout, bacteria spatially regulate processes involved in the transfer of genetic information (transcription, translation and mRNA degradation), drawing a parallel to eukaryotic biology.
What accounts for the physical separation between polysomes and the condensed chromosome meshwork? In the prevailing model, this separation has been, at least in part, attributed to excluded volume effects (Castellana et al., 2016; Mondal et al., 2011). This explanation makes physical sense: the DNA polymer avoids the cell boundary to maximize conformational entropy, and the polysomes occupy the available space outside the DNA to maximize translational entropy. But apparent contradictions are found in bacteria such as C. crescentus, which exhibits a fundamentally different spatial organization of gene expression. In this bacterium, DNA and ribosomes are spread throughout the cytoplasm, and RNase E appears to associate (directly or indirectly) with the DNA instead of the membrane (Figure 2Bii) (Briegel et al., 2006; Montero Llopis et al., 2010). The same physical laws, including excluded volume effects, should apply to C. crescentus. Yet, we do not see the DNA of C. crescentus at the center of the cell with polysomes enriched at the cell poles, as in the prevailing model. Solving this mystery will help us understand the evolutionary constraints and physiological consequences of spatially organizing gene expression.
PARTITIONING THE CELLULAR CONTENT
Decoding the genome creates biomass, and biomass must be properly partitioned between daughter cells at every generation. In this respect, the small size of bacterial cells presents a big advantage, as diffusion—a passive process driven by thermal fluctuations—is an effective means of molecular transport over short distances. For freely-diffusing components, diffusion will randomize position, and for any component in high-copy number, random distribution will ensure that each daughter cell gets about half of the cellular components at division. However, diffusion alone is not a reliable means of partitioning low-copy-number cellular components, as the probability that a daughter cell receives zero copies increases exponentially with decreasing copy number (see Box 1). The solution, here again, is intracellular organization: bacteria have evolved means of spatially ordering low-copy-number cellular components inside cells to ensure their proper partitioning between daughter cells. Development of this spatial order generally depends on either the nucleoid or cell geometry, as exemplified below.
Bipolar distribution of low-copy-number components
In rod-shaped bacteria, one way to ensure inheritance of a low-copy-number factor upon division is by segregating copies to opposite ends of the cell. This strategy is used by a subset of low-copy-number plasmids whose segregation mechanisms are based on active transport and dynamic cytoskeletal filaments made of distant actin or tubulin homologs (Fink and Aylett, 2017; Gayathri and Harne, 2017). This is illustrated by the ParM/R system encoded by the low-copy-number E. coli plasmid R1 (Moller-Jensen et al., 2003). ParM is an actin homolog that polymerizes into a bipolar spindle comprised of two or more antiparallel filaments (Bharat et al., 2015). In an interesting evolutionary twist, the ParM structure behaves more like microtubules than actin filaments by displaying dynamic instability, switching from steady elongation to rapid shrinking (Figure 3Ai) (Garner et al., 2004). This dynamic property provides a “search-and-capture” mechanism, as shrinking stops when one end of the ParM filaments becomes bound by the plasmid via the ParR protein (Garner et al., 2007; Gayathri et al., 2012). Disassembly from the other filament end is prevented through antiparallel pairing with another ParM filament (Gayathri et al., 2012). With both ends unable to disassemble, the bipolar spindle continues growing, pushing sister plasmids apart (Figure 3Ai). The rod-shaped geometry of the cell plays a critical role in this mechanism. Through elongation, the spindle naturally orients itself along the long axis of the cell, leading to plasmid translocation toward opposite cell poles.
Figure 3. Partitioning cellular components.
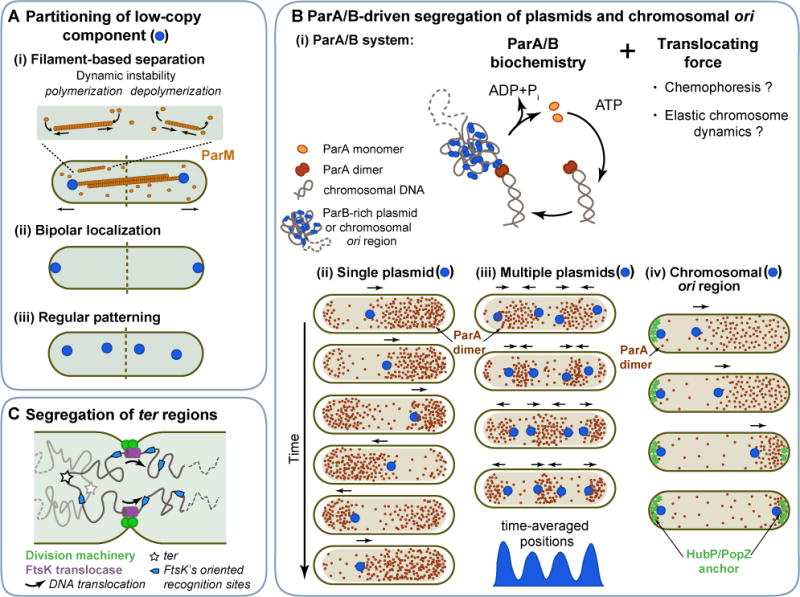
A. Different approaches ensure faithful partitioning of low-copy-number cellular components between daughter cells: (i) Cytoskeleton-based active transport of component copies to opposite poles, as illustrated with the plasmid ParM/R system. (ii) Localization of copies at opposite poles. (iii) Regular distribution of copies along the long cell axis. B. (i) Current models of the ParA/B DNA partitioning system propose different sources of the translocating force, which, together with the ParA/B biochemistry, drive (ii) oscillations of a single plasmid, (iii) regular patterning of multiple plasmid copies, and (iv) segregation of chromosomal ori regions in some bacteria. C. Segregation of chromosomal ter regions across the septal plane is driven by the hexameric FtsK translocase, which interacts with the division machinery. FstK recognizes DNA sequences on the chromosome that have an ori-to-ter orientation, which determines the directionality of DNA translocation across the septal plane.
Other segregation mechanisms that depend on actin- or tubulin-like proteins have been described (Fink and Lowe, 2015; Ni et al., 2010; Polka et al., 2014), which differ in their mechanism of generating polymer instability. For example, in the case of the Bacillus thuringiensis pBtoxis plasmid system, filaments of the tubulin-like protein TubZ do not display dynamic instability. Instead, they treadmill (Fink and Lowe, 2015; Larsen et al., 2007). In the case of the B. subtilis plasmid pLS32 system (Becker et al., 2006), the actin-like protein AlfA polymerizes into stable bundles of filaments (Polka et al., 2009; Popp et al., 2010), and their destabilization is driven by the accessory protein AlfB (Polka et al., 2014).
It is also worth noting that some cellular components (e.g., chemoreceptor arrays) preferentially localize at the cell pole membranes (Figure 3Aii). Any mechanism underlying such bipolar localization could work as a potential partitioning mechanism because each daughter cell inherits a cell pole from the mother cell. Such mechanisms do not necessarily involve protein filaments. We will present several examples in a later section (see “Functionalization of the cell poles”).
Equidistant distribution of low-copy-number components
Segregating low-copy-number components to opposite ends of the cell is not the only way to achieve proper partitioning. Equidistant distribution of component copies along the long axis of the cell will also ensure equal inheritance between daughter cells (Figure 3Aiii). Such spatial patterning has been reported for low-copy-number plasmids (Adachi et al., 2006; Ebersbach et al., 2006; Lioy et al., 2015; Sengupta et al., 2010), storage granules in C. crescentus, Pseudomonas aeruginosa and Pseudomonas putida (Galan et al., 2011; Henry and Crosson, 2013; Racki et al., 2017), cytoplasmic chemotaxis protein clusters in Rhodobacter sphaeroides (Thompson et al., 2006), and carboxysomes (protein organelles for carbon fixation) in Synechococcus elongatus (Savage et al., 2010).
The best studied system driving such a striking spatial pattern is the ParA/B partitioning system, which is expressed by a large number of low-copy-number plasmids (Gerdes et al., 2000). Note that ParA/B systems are also expressed by many bacterial chromosomes (Livny et al., 2007), as discussed later. How do ParA/B systems regularly space plasmids? Once again, the mechanism involves active transport of plasmids. However, here, the active transport does not appear to involve cytoskeletal filaments (Hwang et al., 2013; Lim et al., 2014; Vecchiarelli et al., 2013). Furthermore, the plasmids travel within the nucleoid volume (Le Gall et al., 2016). A large body of work has revealed that ParA/B-dependent transport relies on the following core biochemical features of the DNA-binding protein ParB and the P-loop ATPase ParA (Baxter and Funnell, 2014). ParB associates with the plasmid cargo through the recognition of a specific DNA sequence, whereas ParA binds to the nucleoid non-specifically in an ATP-bound dimeric form. The ParB-decorated cargo diffuses until it interacts with one or more nucleoid-bound ParA dimers, resulting in a complex. Each ParA-ParB interaction is transient because ParB stimulates ParA ATPase activity, releasing freely diffusible ParA monomers. Following nucleotide exchange and redimerization, ParA rebinds to the nucleoid, restarting the cycle (Figure 3Bi). Iteration of this cycle leads to the ParB-decorated cargo being “pulled” by a retracting ParA concentration gradient (Figure 3Bii) (Fogel and Waldor, 2006; Hatano et al., 2007; Ringgaard et al., 2009). These correlated dynamics lead to oscillatory behaviors of the plasmid and ParA waves across the nucleoid when a single plasmid cargo is present (Figure 3Bii). When multiple plasmids are present, the correlated dynamics between ParA and the ParB-decorated plasmids create depletion zones of ParA between approaching plasmids (Surovtsev et al., 2016). This depletion presumably causes plasmids to change direction each time they come close to each other, resulting in localized motion and equidistant distribution of plasmids on time average (Figure 3Biii).
A key question remains: how does ParA “pull” the ParB-decorated plasmids? Computational simulations suggest that ParA/B biochemistry and plasmid diffusion alone cannot account for the observed active transport and patterning—a translocating force is required (Figure 3Bi) (Ietswaart et al., 2014; Surovtsev et al., 2016). Indeed, all recent mathematical models that reproduce the active motion and spatial patterning observed in vivo include a non-Brownian, ParA-dependent force (Hu et al., 2017; Ietswaart et al., 2014; Jindal and Emberly, 2015; Sugawara and Kaneko, 2011; Surovtsev et al., 2016; Walter et al., 2017). But what is the source of the force? In some models, motion is proposed to be driven by “chemophoresis” (or “proteophoresis”) in which the force originates from ParB and ParA binding in a chemical (ParA) gradient (Sugawara and Kaneko, 2011; Walter et al., 2017). Such a force is predicted by theoretical thermodynamic considerations (Sugawara and Kaneko, 2011), though the scale of this force inside cells remains to be determined. In other models, motion is driven by an elastic force originating from chromosomal fluctuations (Hu et al., 2017; Surovtsev et al., 2016). The compacted chromosome behaves as an elastic body, with chromosomal loci (and therefore anything bound to them) experiencing an elastic force on the scale of 0.04 pN (see Box 1) (Surovtsev et al., 2016; Wiggins et al., 2010). Importantly, this chromosomal force is on the same scale as the translocating force plasmids experience inside cells (see Box 1). The chromosomal elastic force would act on the plasmid cargo whenever it interacts with the nucleoid via ParA/ParB interaction.
Chromosome segregation
A low-copy-number component that all cells must partition without fail is the chromosome. Unlike in eukaryotes, bacterial chromosome segregation happens simultaneously with DNA replication, and can be divided into three major steps: a) separation and translocation of the freshly duplicated ori regions, b) segregation of the bulk of the chromosome, and c) separation of the ter regions (Badrinarayanan et al., 2015; Bouet et al., 2014; Wang and Rudner, 2014). Here again, intracellular organization facilitates proper chromosome segregation. This is particularly apparent in the first and last steps of chromosome segregation, which will therefore be the focus of our discussion.
Segregation of the ori regions dictates the directionality of chromosome segregation and sets up the final configuration of the chromosome. In some bacteria, ori translocation is driven or assisted by a chromosomally-encoded ParA/B system that is closely related to the ParA/B plasmid system described above. For example, in V. cholerae (for the large chromosome) and C. crescentus, one of the replicated ParB-bound ori regions stays at one pole while the other moves to the opposite pole in the wake of a retracting ParA wave over the nucleoid (Figure 3Biv) (Fogel and Waldor, 2006; Ptacin et al., 2010; Schofield et al., 2010; Shebelut et al., 2009). The translocated ParB-bound ori is then anchored to the pole through a direct interaction between ParB and a pole-organizing element, PopZ in C. crescentus (Bowman et al., 2008; Ebersbach et al., 2008) and HubP in V. cholerae (Yamaichi et al., 2012). In Actinobacteria such as Corynebacterium glutaminicum, the polar anchor for the ParB-bound ori likely is DivIVA (Donovan et al., 2012).
The correlated localization dynamics between the ParB-bound ori region and the ParA waves are visually similar to one leg of the single-plasmid oscillation described above (Figure 3Bii). The underlying mechanism is likely similar: the ParB-bound ori region harnesses the chemophoretic force and/or the chromosomal elastic force through iterative interactions with nucleoid-bound ParA to propel itself along the ParA gradient (Lim et al., 2014; Sugawara and Kaneko, 2011). Remarkably, the ParA and ParB localization patterns can differ among species. For instance, instead of being anchored at the cell poles, ParB-bound ori displays a near-midcell position in M. smegmatis or a subpolar position in M. xanthus during the non-replicating phase (Ginda et al., 2017; Harms et al., 2013; Iniesta, 2014; Trojanowski et al., 2015). Furthermore, instead of localizing to the nucleoid, ParA is largely confined to the large DNA-free regions between the cell poles and the nucleoid in M. xanthus (Harms et al., 2013; Iniesta, 2014). These differences suggest a diversification in ParA/B-dependent mechanisms, highlighting the importance of studying different bacterial models.
Other bacteria, such as E. coli, lack parA and parB genes (Livny et al., 2007). In these cases, other non-exclusive physical phenomena play a role in the translocation of the chromosomal ori regions. Polymer dynamics in confined space predicts that duplicated ori regions will naturally separate from each other by entropic forces (Youngren et al., 2014), especially if these ori regions are condensed into compact domains (e.g., via NAP and SMC activity) (Marko, 2009). In B. subtilis, self-condensation and disentanglement of ori-proximal loops may also be actively driven by SMC complexes that load close to ori via the ParB homolog (Spo0J) and then thread left and right chromosomal segments presumably by a loop-extrusion mechanism (Gruber and Errington, 2009; Marbouty et al., 2015; Sullivan et al., 2009; Wang et al., 2017). Inactivation of SMC leads to interconnected sisters ori (Gruber et al., 2014; Wang et al., 2014b). A “snap-release” mechanism has been proposed for ori segregation in E. coli, based on the observation that sister ori regions remain in close proximity for an extended amount of time before they abruptly separate (Bates and Kleckner, 2005; Joshi et al., 2011). Release of accumulated mechanical stress, perhaps due to the entropic repulsion of polymer segments, is thought to drive the sister ori regions apart (Fisher et al., 2013). Similar physical phenomena are thought to also underlie bulk chromosome segregation (Fisher et al., 2013; Hadizadeh Yazdi et al., 2012; Hong and McAdams, 2011; Hong et al., 2013; Jun and Mulder, 2006; Junier et al., 2014; Lampo et al., 2015; Marko, 2009; Youngren et al., 2014).
To ensure that chromosomes are fully segregated before division completes, DNA “pumps” made of proteins such as FtsK (SpoIIIE during B. subtilis sporulation) actively translocate DNA from one daughter cell compartment to the other (Figure 3C) (Aussel et al., 2002; Lesterlin et al., 2008; Sharp and Pogliano, 2002). FtsK associates with the division machinery at the growing septum (Wang and Lutkenhaus, 1998; Yu et al., 1998) and recognizes specific sequences on the chromosome (Bigot et al., 2006; Bigot et al., 2005; Levy et al., 2005; Lowe et al., 2008; Pease et al., 2005). The ori-to-ter orientation of these recognition sites sets the directionality of DNA translocation relative to the septum (Figure 3C). Effectively, FtsK transforms the orientation of the replicating chromosome and the geometry of the dividing cell into a directed translocation of chromosomal DNA across the division plane.
DIVIDING AT THE RIGHT PLACE
Speaking of the division plane, another critical event that occurs at every generation is the selection of the division site. While some bacteria divide asymmetrically, most divide in the middle of the cell. But how is the midcell point identified? This inherently geometrical problem is, not surprisingly, solved through intracellular organization. Selection of the division site occurs through positive and/or negative spatial regulation of Z-ring assembly (Monahan et al., 2014; Rowlett and Margolin, 2015). Positive regulation involves protein complexes or cell-wall markers that recruit FtsZ to the middle of the cell. In contrast, negative regulation generally involves protein patterns that restrict FtsZ ring formation everywhere except the cell midpoint. Bacteria often have multiple redundant mechanisms of Z-ring positioning (Bailey et al., 2014; Rodrigues and Harry, 2012), and the mechanisms themselves vary among species. However, a recurring theme is, again, the reliance on the nucleoid or cell geometry to provide spatial cues, as illustrated by three examples below.
Negative regulation by nucleoid occlusion
The Z-ring preferentially assembles in DNA-free-regions through a phenomenon known as “nucleoid occlusion”. Unrelated DNA-binding proteins, SlmA in E. coli and Noc in B. subtilis, contribute to nucleoid occlusion (Bernhardt and de Boer, 2005; Wu and Errington, 2004). SlmA depolymerizes FtsZ polymers (Cabre et al., 2015; Cho et al., 2011; Du and Lutkenhaus, 2014) or prevents FtsZ polymers from coalescing into a Z-ring (Tonthat et al., 2011; Tonthat et al., 2013). Noc is thought to interact with both the nucleoid and the cytoplasmic membrane to form large membrane/DNA complexes that physically occlude Z-ring assembly and cell constriction (Adams et al., 2015). These inhibitory activities are spatially regulated by the intracellular arrangement of the chromosome. While Noc and SlmA bind across the chromosome, their binding sites are specifically depleted in the ter domain (Figure 4A) (Cho et al., 2011; Tonthat et al., 2011; Wu et al., 2009). This uneven distribution of binding sites helps prevent assembly of the functional Z-ring in the middle region of the cell until segregation of replicated ori regions leaves the ter region at midcell (Figure 4A).
Figure 4. Selecting the division site.
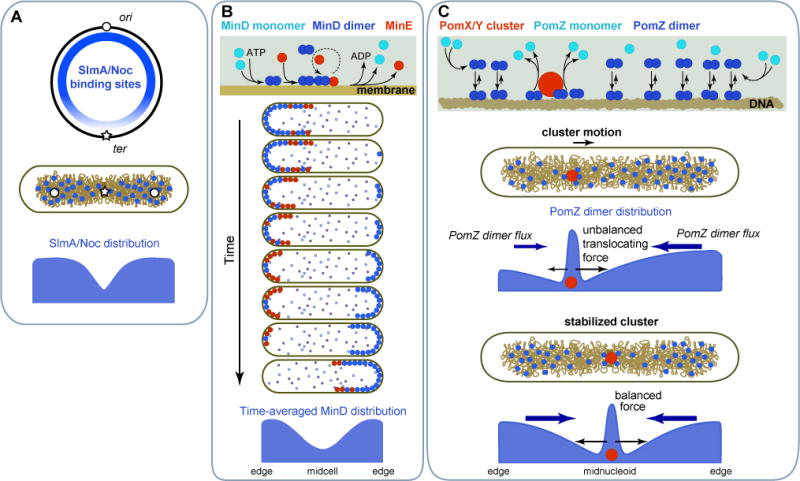
A. Nucleoid occlusion of Z-ring formation is facilitated by the asymmetry in DNA-binding sites of FtsZ inhibitors SlmA (E. coli) or Noc (B. subtilis) along the chromosome. B. The E. coli Min system prevents Z-ring formation in the polar regions through protein oscillations driven by a reaction-diffusion mechanism. These oscillations result in time-averaged accumulation of an FtsZ-assembly inhibitor at the polar regions. C. The Pom system in M. xanthus identifies midcell by positioning an FtsZ assembly activating complex, the PomX/Y cluster, at mid-nucleoid through a localized-sink/distributed-source mechanism coupled to a translocating force of chemophoretic or chromosomal origin.
Negative regulation by the E. coli Min system
E. coli uses cell geometry and a reaction-diffusion mechanism to establish a pole-to-pole oscillating wave of the FtsZ polymerization inhibitor MinC (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a). Oscillations of MinC from one end of the cell to other results in a minimum of MinC concentration and inhibitory activity at the middle of the cell on time average, thereby confining Z-ring assembly to that location. MinC oscillations are driven by the antagonistic relationship between two other Min proteins, the P-loop ATPase MinD and its regulator MinE (Kretschmer and Schwille, 2016; Rowlett and Margolin, 2015). While details vary among current models (Bonny et al., 2013; Park et al., 2011; Vecchiarelli et al., 2016; Wu et al., 2016), the core features of the oscillatory mechanism are as follows (Figure 4B). MinD associates with the cytoplasmic membrane upon ATP binding and dimerization. Membrane-associated MinD recruits MinE. This is followed by enhanced binding of MinD to the membrane. MinE antagonizes MinD’s membrane binding by stimulating its ATPase activity, causing the release of MinD monomers into the cytoplasm. Following nucleotide exchange and diffusion, MinD redimerizes and rebinds to the membrane. The antagonism between MinD and MinE, coupled with the positive feedback of MinD binding to the membrane, drives oscillations of MinD and MinE between the most distant locations of the cell, the cell poles (Figure 4B). The FtsZ inhibitor MinC rides on the oscillatory waves simply by interacting with MinD.
Positive regulation by the Myxococcus xanthus Pom system
M. xanthus uses another P-loop ATPase, PomZ, but, in this case, for positive regulation of Z-ring formation (Schumacher et al., 2017; Treuner-Lange et al., 2013). PomZ is a ParA-like protein. Thus, instead of binding to the membrane like MinD, PomZ binds to the nucleoid upon ATP binding and dimerization (Figure 4C). PomZ dimers also associate with a protein cluster of PomX and PomY, which synergistically stimulates PomZ monomerization. After fast cytoplasmic diffusion and nucleotide exchange, PomZ reforms dimers and rebinds to the nucleoid. As a result, the PomX/Y cluster acts as a localized sink for PomZ dimers while the nucleoid serves as a broadly distributed source, resulting in a diffusive flux of PomZ dimers into the PomX/Y cluster (Figure 4C). When the cluster is away from midcell, the fluxes on each side of the cluster are unequal, resulting in a higher accumulation of PomZ dimers on one side of the cluster relative to the other. This asymmetry results in active translocation of the cluster toward the higher concentration of PomZ dimers. The translocating force likely emanates from chemophoresis or the elastic properties of the chromosome, as discussed above for the ParA/B system involved in DNA segregation. The asymmetry and the force vanish when the PomX/Y cluster reaches the middle of the nucleoid (i.e., midcell) because the fluxes of PomZ dimers from each side of the cluster become equivalent (Figure 4C) (Schumacher et al., 2017). Stabilization of the cluster at midcell stimulates Z-ring assembly through an unknown mechanism.
SPECIALIZING THE CELL
As illustrated in the sections above, spatial organization of the bacterial cell is an integral element of processes required for cellular replication. Often, different bacteria have evolved different spatial mechanisms to complete the same task. Over the course of evolution, diversification of cellular organization has also offered ways for bacteria to expand their metabolic capability and evolve new phenotypic traits. In some cases, cell specialization has come from the functionalization of specific cellular regions, in particular, the cell poles. In others, the development of new biological properties has been associated with cellular compartmentalization, generally through the formation of endomembranes and intracellular organelles.
Functionalization of the cell poles
While the vast majority of bacterial proteins have dispersed distribution in the cell, some proteins accumulate at one or both cell poles in diverse non-spherical bacteria. These pole-localized proteins are involved in a variety of functions including chemotaxis, motility, pathogenesis, cell differentiation, cell-cycle regulation, energy production, and secretion. Polar localization can offer various benefits. For example, in the aquatic bacterium C. crescentus, polar protein localization establishes cellular asymmetry and creates regulatory hubs that coordinate developmental programs with cell cycle progression (Kirkpatrick and Viollier, 2012; Lasker et al., 2016). During each cell cycle, C. crescentus produces two distinct daughter cells: a motile cell with pili and a flagellum for exploring new territories, and a sessile cell with an appendage (stalk) and an adhesin (holdfast) for colonizing the local environment. In this organism, temporally orchestrated localization of regulatory and signaling proteins at a specific pole couples the synthesis or activation of polar organelles (flagellum, pili, holdfast, and stalk) with DNA replication, chromosome segregation or cell division (Figure 5Ai). Functionalization of each cell pole with distinct regulatory pathways is necessary to accommodate the polar morphogenesis and the dimorphic lifestyle of this bacterium.
Figure 5. Specializing the cell.
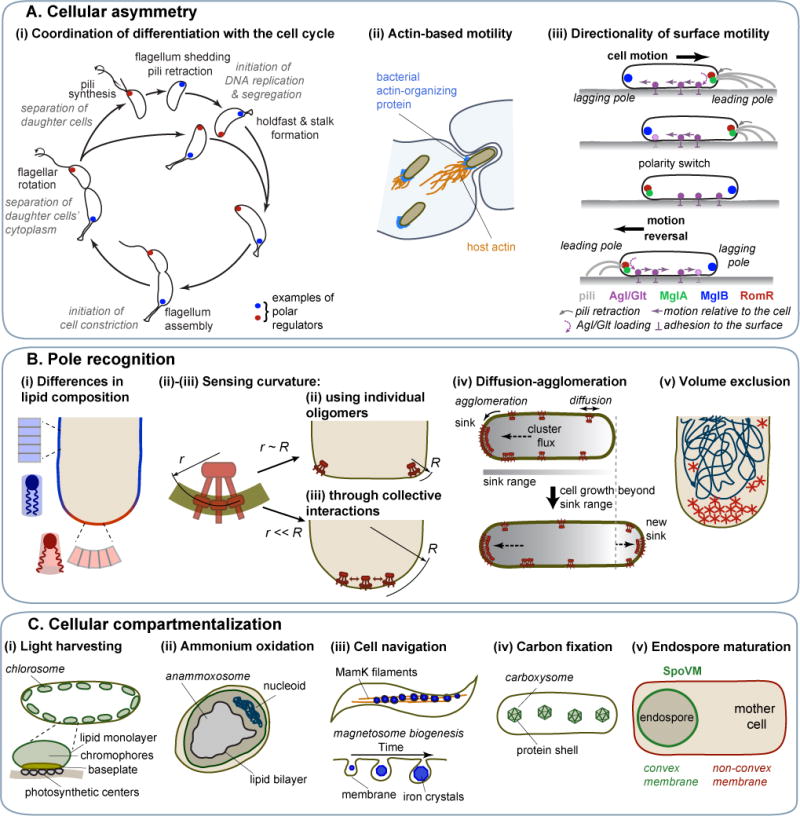
A. Protein localization at a specific cell pole results in cellular asymmetry that underlies (i) the coordination between developmental and cell cycle events in C. crescentus (the polar localization of only two histidine kinases, DivJ and PleC, are shown for clarity (Wheeler and Shapiro, 1999); in reality, there are at least a dozen regulators that display polar accumulation during the cell cycle), (ii) the actin-based motility of some intracellular pathogens and their cell-to-cell spread in eukaryotic host tissues, (iii) the directional motility in M. xanthus. B. Pole recognition can be based on (i) differences in lipid composition between membrane regions of different curvatures, (ii) sensing of high curvature by individual protein oligomers, (iii) sensing of moderate curvature through collective oligomer interactions, (iv) interplay between protein cluster diffusion and protein cluster agglomeration, (v) volume exclusion that promotes formation of large protein structures outside the nucleoid. C. Examples of bacterial microcompartments: (i) chlorosomes bound by a single lipid layer and a protein baseplate, (ii) an anammoxosome enclosed by a lipid bilayer, (iii) iron-containing magnetosomes, (iv) carboxysomes, which confine reactions involved in carbon fixation in a protein shell, (v) an endospore maturing into a spore inside the mother cell. B. subtilis SpoVM localizes to the outer surface of the endospore to facilitate spore coat formation.
Another striking example of cell specialization through pole functionalization is the actin-based motility that propels diverse intracellular pathogens in the cytosol of eukaryotic host cells, allowing them to spread to neighboring cells (Figure 5Aii). In these pathogens, polymerization of the host actin occurs at one cell pole and is usually promoted by an asymmetrically distributed actin-organizing protein, such as ActA in Listeria monocytogenes, IcsA/VirG in Shigella flexneri, BimA in Burkholderia pseudomallei and Sca2 in Rickettsia parkeri (Goldberg et al., 1993; Haglund et al., 2010; Kocks et al., 1993; Stevens et al., 2005).
How is polar localization achieved? Some bacteria first reduce the problem by relying on pole-organizing proteins. For example, B. subtilis DivIVA, C. crescentus PopZ, and V. cholerae HubP recruit multiple proteins at the poles either by interacting directly with them or by self-assembling into a multivalent scaffolding matrix (Bowman et al., 2008; Ebersbach et al., 2008; Holmes et al., 2016; Lenarcic et al., 2009; Ramamurthi and Losick, 2009; Yamaichi et al., 2012). But how do pole-organizing proteins (or other polar proteins that do not use their help) accumulate at the pole in the first place? Various physical mechanisms relying on cell geometry (cell pole shape) or the nucleoid have been proposed.
Such a mechanism is based on protein interaction with lipids, like cardiolipin, that preferentially accumulate at curved polar membranes due to their conical shape (Figure 5Bi) (Lin and Weibel, 2016; Mukhopadhyay et al., 2008), as mentioned earlier. Another geometry-dependent mechanism is based on proteins directly sensing membrane curvature. For example, a transmembrane protein oligomer can gauge the curvature of the membrane when the curvature radius of the membrane is on the same scale as the intrinsic curvature of the protein oligomer, as shown for cone-shaped trimers-of-dimers of the B. subtilis chemoreceptor TlpA (Figure 5Bii) (Strahl et al., 2015). Similar membrane curvature recognition by bent oligomers may underlie DivIVA localization (Lenarcic et al., 2009; Oliva et al., 2010; Ramamurthi and Losick, 2009). Since the cell poles of E. coli do not display the sharp corners of B. subtilis cell poles, they lack membranes with a curvature radius small enough to recruit individual oligomers. One model proposes that curvature sensing and polar localization of E. coli chemoreceptors are achieved through the clustering of trimers-of-dimers (Figure 5Biii) (Draper and Liphardt, 2017), potentially with the help of the Tol-Pal complex (Santos et al., 2014). The requirement for clustering is an example of spatial pattern emerging from collective interactions, a concept usually referred to as collective phenomena in physics. Another model for the polar accumulation of E. coli chemoreceptors does not involve membrane curvature; instead, it is based on the idea that large chemoreceptor clusters act as a sink for diffusing smaller clusters through agglomeration (Figure 5Biv) (Thiem and Sourjik, 2008; Wang et al., 2008). Small clusters can develop into a new large one only when the cell grows sufficiently for the cluster to be out of range of the sink (the old large cluster). Since one of the cell poles is always the most distant location from any arbitrary position in the cell, a large cluster will develop there.
For the polar matrix-assembling PopZ protein in C. crescentus, theoretical and experimental evidence suggests that nucleoid-dependent size exclusion effects are at play (Ebersbach et al., 2008; Laloux and Jacobs-Wagner, 2013; Saberi and Emberly, 2010). In C. crescentus, the nucleoid spreads throughout most of the cell (Figure 2Bii), limiting the volume available for large structures, such as the PopZ matrix, to the DNA-free cell tips (Figure 5Bv). When there is a pre-existing matrix (from the previous division cycle), PopZ self-affinity dictates continuous growth of this matrix. To break this pattern and nucleate PopZ assembly at the opposite pole, C. crescentus is thought to exploit the local accumulation of the ParA protein during chromosomal ori segregation (Laloux and Jacobs-Wagner, 2013). ParA would recruit PopZ oligomers through direct interaction, seeding the assembly of a new pole-organizing matrix right on time to attach the segregating ori region. Other molecular factors may be involved in regulating PopZ localization (Berge et al., 2016; Ptacin et al., 2014).
In diverse rod-shaped bacteria, the two poles are not functionally equivalent. This is readily obvious in any bacterium that exhibits unipolar features or properties (e.g., Figure 5Ai-ii). Pole identity is generally defined by the age of the pole: the “new” pole results from the latest division whereas the “old” pole originates from an earlier division. Little is known about how pole identity is achieved and reset at every division. In C. crescentus, TipN, a “birthmark” protein, regulates flagellar biosynthesis and marks the new pole until the end of the cell cycle, when it relocates to the division site to mark the new pole of the daughter cells at birth (Huitema et al., 2006; Lam et al., 2006). TipN relocation to the developing poles is dependent on the Tol-Pal complex, which bridges the cell envelope layers during division (Yeh et al., 2010). In Agrobacterium tumefaciens, PopZ plays a similar birthmark function to mark the location of polar growth at the new pole (Grangeon et al., 2015).
The functional identity of the pole is not always associated with its age. Powerful examples are the motility systems of M. xanthus, in which the identity of the pole (leading vs. lagging) is associated with the direction of cell motion on surfaces (Schumacher and Sogaard-Andersen, 2017). This soil bacterium has two motility systems that are important for individual and social cell behaviors such as chemotaxis, group predation, and multicellular development. The so-called S-motility system relies on type IV pili, which are protein filaments that extrude from the leading pole. Cycles of pili extension, surface attachment, and retraction result in cell motion (Figure 5Aiii). The A-motility machinery, which consists of the cell envelope Agl-Glt complex, also assembles at the leading pole (Figure 5Aiii). Its active translocation toward the lagging pole, combined with its adhesion to the underlying surface at focal adhesion sites, results in propulsion of the cell in the opposite direction (Faure et al., 2016; Mignot et al., 2007). When the A-motility machinery reaches the lagging pole, it disassembles. Upon sensing environmental cues through the Frz chemosensory system, both the Agl-Glt complex and the type IV pili switch their polarity axis (Keilberg et al., 2012; Zhang et al., 2012). The leading pole becomes the lagging one, and vice versa, resulting in a reversal of motility direction (Figure 5Aiii). The leading-lagging polarity control system consists of three proteins, MglA, MglB and RomR, which simultaneously switch pole location to direct cell motion reversal (Figure 5Aiii) (Keilberg et al., 2012; Leonardy et al., 2010; Mauriello et al., 2010; Miertzschke et al., 2011; Treuner-Lange et al., 2015; Zhang et al., 2010; Zhang et al., 2012). MglA is a Ras-like GTPase that recruits the motility engines when bound to GTP. MglA-GTP is excluded from the lagging pole by the GTPase-activating protein (GAP) MglB and is recruited to the leading pole by the response regulator RomR. The involvement of a small GTPase and a GAP in the polarity of a bacterial motility system draws a remarkable parallel to the cell polarity role of small GTPases and GAPs in eukaryotic cell migration (Artemenko et al., 2014).
Compartmentalization of the cytoplasm
A number of diverse species deviate from the conventional open plan of the bacterial cytoplasm by forming endomembranes and specialized intracellular compartments that confer unique biological properties. For example, green sulfur bacteria (phylum Chlorobi) and filamentous anoxygenic phototrophs (phylum Chloroflexi) build intracellular organelles known as chlorosomes comprised of a lipid monolayer and a proteinaceous baseplate (Figure 5Ci) (Nielsen et al., 2016). These chlorosomes serve as ultra-efficient light-harvesting antennae that allow the organism to perform photosynthesis in near complete darkness (Beatty et al., 2005; Manske et al., 2005). But perhaps the bacterial organelle most similar to a eukaryotic membrane-enclosed organelle is the anammoxosome found inside anaerobic ammonium oxidizing planctomyces (Figure 5Cii) (Neumann et al., 2014). The purpose of this compartment is to catalyze anaerobic ammonium oxidation and to prevent the extremely toxic and unstable intermediate hydrazine (a rocket fuel propellant!) from reacting with the cytoplasmic milieu. Hydrazine readily diffuses across most biological membranes. However, the anammoxosome membrane is rich in ladderane lipids (lipids containing fused cyclobutane rings), which creates an exceptionally dense membrane that limits hydrazine diffusion (Sinninghe Damste et al., 2002).
Another remarkable example of specialization through organelle formation is found in magnetotactic bacteria. These organisms form magnetosomes, which are made of magnetic iron crystals surrounded by a membrane derived from cytoplasmic membrane invaginations (Figure 5Ciii) (Uebe and Schuler, 2016). In some Magnetospirillum species, filaments of the actin homolog MamK help align the magnetosomes along the cell body through an interaction with MamJ (Figure 5Ciii) (Komeili et al., 2006; Pradel et al., 2006; Scheffel et al., 2006). This cellular-scale order effectively endows the cell with a compass. Such a built-in navigation system allows the cell to orient itself along the Earth’s magnetic field and swim toward a position in the water column where growth is optimal.
Many bacteria can also form intracellular organelles surrounded by a protein shell, offering unique properties compared to membrane-bound organelles. For example, different types of aquatic bacteria (e.g., cyanobacteria and anoxygenic phototrophic bacteria) carry hollow gas-filled proteinaceous vesicles in their cytoplasm (Pfeifer, 2012). The rigid protein wall, which primarily consists a highly hydrophobic protein (GvpA), is impermeable to liquid water, but permeable to gases (Sivertsen et al., 2010; Walsby, 1969). As a result, the vesicles fill with air and cells become buoyant, preventing them from sinking and adjusting them to the right water depth.
Most protein-based intracellular organelles in bacteria are, however, not filled with air. Instead, they encapsulate enzymes (Bobik et al., 2015). Genomic analyses suggest that about 20% bacteria across phyla can compartmentalize metabolic reactions within a protein shell (Abdul-Rahman et al., 2013). The best characterized bacterial microcompartments (BMCs) are the cyanobacterial carboxysomes (Figure 5Civ), which encapsulate CO2-fixing enzymes, and the enteric metabolosomes that carry out ethanolamine and propanediol degradation (Bobik et al., 1999; Kofoid et al., 1999; Shively et al., 1973). The selectively permeable protein shell of BMCs serves not only to colocalize enzymes of a specific pathway, but also to confine toxic intermediates and volatile compounds(Bobik et al., 2015). For instance, carboxysomes help reduce the loss of CO2 (Dou et al., 2008), a small, nonpolar molecule that readily crosses lipid membranes. The microcompartments for ethanolamine and propanediol utilization are thought to sequester a volatile aldehyde intermediate that is toxic to the cell (Havemann et al., 2002; Penrod and Roth, 2006; Sampson and Bobik, 2008). Structural studies of carboxysomes have shown that the icosahedral protein shell shares assembly principles with viral capsids and consists of facets of tightly packed hexamers and vertices of pentamers (Kerfeld et al., 2005; Tanaka et al., 2008; Tsai et al., 2007). Pores within the protein shell are thought to control what goes in and out of the microcompartment (Cai et al., 2013; Klein et al., 2009; Sagermann et al., 2009). BMCs typically range from 80 to 500 nm in size. Similar, but smaller (20-40 nm) proteinaceous compartments called encapsulins are also widespread in bacteria (Giessen and Silver, 2017; Sutter et al., 2008). Protein targeting to the BMC or encapsulin interior only requires a short peptide sequence, allowing encapsulation of foreign proteins through genetic engineering (Cassidy-Amstutz et al., 2016; Choudhary et al., 2012; Fan and Bobik, 2011; Rurup et al., 2014; Tamura et al., 2015). This property, together with their stability (e.g., to temperature, pH or denaturants), makes BMCs and encapsulins promising platforms for biotechnology and biomedical applications (Giessen, 2016; Giessen and Silver, 2016).
An extreme case of cytoplasmic compartmentalization in bacteria is endospore formation, a developmental process that members of the Bacillus and Clostridium genera undergo in response to starvation. The endospore is a cell maturing into a resilient spore within the cytoplasm of a nurturing “mother” cell (Figure 5Cv). This organization presents a new cell geometrical cue—a convex membrane inside the cell—that can be exploited for protein localization. This is elegantly illustrated by SpoVM, a small protein that specifically accumulates to the endospore’s outer surface by sensing, through a cooperative interaction, small differences in lipid packing between convex and non-convex membrane curvatures (Figure 5Cv) (Gill et al., 2015; Ramamurthi et al., 2009). This geometry-driven localization of SpoVM provides positional information for the deposition of a protective proteinaceous coat on the developing spore.
IN CLOSING
The examples presented above demonstrate how ingrained cellular organization is in many aspects of bacterial replication and lifestyle. Order at the cellular scale emerges from interactions at the molecular scale through protein polymerization, reaction-diffusion mechanisms, excluded volume effects, and collective phenomena. The examples also highlight the crucial role that physical factors play in organizing the cell.
A recurring theme in bacterial cellular organization is the reliance of self-organizing mechanisms on ATPases or GTPases. These proteins render the system dynamic by cycling between different states. At the same time, the activity of these proteins pushes the system out of thermodynamic equilibrium by continuously consuming ATP or GTP. This less appreciated aspect of their activity allows cells to maintain spatial order by “fighting back” against the natural increase in entropy.
Another theme is the role of the nucleoid and cell geometry in providing spatial cues for the development of finer organization. This dependence emphasizes the importance of studying how geometrical cell features (e.g., cell shape, cell size, curved membranes) and aspects of nucleoid organization (nucleoid shape, DNA compaction, chromosome orientation, and spatial gene positioning) are established and maintained. Most of our current knowledge comes from studies of rod-shaped, monoploid bacteria, and much remains to be learned from them. In recent years, bacteria with different shapes or ploidy have gained momentum as experimental systems. Further expanding the repertoire of bacterial models will be important to develop a complete understanding.
In the context of cell biology, bacterial systems have emerged from virtual irrelevance into excellent experimental models for investigating how spatial order can spontaneously arise from disorder. In the field of residential and office architecture, an open layout provides flexibility and efficient use of the space. Perhaps, by analogy, the open plan of the bacterial cytoplasm offers a flexible platform for the evolution of different intracellular organizations. The study of evolutionary divergent species has, indeed, taught us that bacteria have often found different solutions to the same biological problem. In this context, the breadth of bacterial diversity gifts us with a unique opportunity to explore the space of design principles and to study the evolution of cellular organization. Understanding the self-emergent and evolvable organization of the cell will bring us closer to understanding life itself.
Acknowledgments
We apologize to our colleagues whose work was not discussed because of space constraints. We thank Drs. Ethan Garner, Peter Graumann, Stephan Gruber, and Jie Xiao, for helpful discussions. We also thank Drs. Brandon Jutras and Irnov Irnov for their help on early drafts. In addition, we thank Dr. Jean-Yves Matroule for providing a drawing of the C. crescentus cell cycle. We are also grateful to the reviewers and all the members of the Jacobs-Wagner lab for critical reading of this manuscript. The research in the Jacobs-Wagner lab is funded in part by the National Institutes of Health (R01 GM065835). C.J.-W. is a Howard Hughes Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–52. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Abdul-Rahman F, Petit E, Blanchard JL. Distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. J Phylogen Evolution Biol. 2013;1:118. [Google Scholar]
- Adachi S, Hori K, Hiraga S. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J Mol Biol. 2006;356:850–63. doi: 10.1016/j.jmb.2005.11.088. [DOI] [PubMed] [Google Scholar]
- Adams DW, Wu LJ, Errington J. Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J. 2015;34:491–501. doi: 10.15252/embj.201490177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah-Seng Y, Rech J, Lane D, Bouet JY. Defining the role of ATP hydrolysis in mitotic segregation of bacterial plasmids. PLoS Genet. 2013;9:e1003956. doi: 10.1371/journal.pgen.1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335:100–4. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley MR, Maddock JR, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–36. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J. 2003;84:2467–73. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemenko Y, Lampert TJ, Devreotes PN. Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell Mol Life Sci. 2014;71:3711–47. doi: 10.1007/s00018-014-1638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–13. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- Badrinarayanan A, Le TB, Laub MT. Bacterial chromosome organization and segregation. Annu Rev Cell Dev Biol. 2015;31:171–99. doi: 10.1146/annurev-cellbio-100814-125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science. 2012;338:528–31. doi: 10.1126/science.1227126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Mannik J. Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet. 2014;10:e1004504. doi: 10.1371/journal.pgen.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol Microbiol. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett TM, Bratton BP, Duvshani A, Miguel A, Sheng Y, Martin NR, Nguyen JP, Persat A, Desmarais SM, VanNieuwenhze MS, et al. A Periplasmic polymer curves Vibrio cholerae and promotes pathogenesis. Cell. 2017;168:172–85 e15. doi: 10.1016/j.cell.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter JC, Funnell BE. Plasmid partition mechanisms. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.PLAS-0023-2014. PLAS-0023-2014. [DOI] [PubMed] [Google Scholar]
- Beatty JT, Overmann J, Lince MT, Manske AK, Lang AS, Blankenship RE, Van Dover CL, Martinson TA, Plumley FG. An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. Proc Natl Acad Sci USA. 2005;102:9306–10. doi: 10.1073/pnas.0503674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker E, Herrera NC, Gunderson FQ, Derman AI, Dance AL, Sims J, Larsen RA, Pogliano J. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 2006;25:5919–31. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge M, Campagne S, Mignolet J, Holden S, Theraulaz L, Manley S, Allain FH, Viollier PH. Modularity and determinants of a (bi-)polarization control system from free-living and obligate intracellular bacteria. Elife. 2016;5:e20640. doi: 10.7554/eLife.20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell. 2005;18:555–64. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Murshudov GN, Sachse C, Lowe J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 2015;523:106–10. doi: 10.1038/nature14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–4. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bigot S, Saleh OA, Cornet F, Allemand JF, Barre FX. Oriented loading of FtsK on KOPS. Nat Struct Mol Biol. 2006;13:1026–8. doi: 10.1038/nsmb1159. [DOI] [PubMed] [Google Scholar]
- Bigot S, Saleh OA, Lesterlin C, Pages C, El Karoui M, Dennis C, Grigoriev M, Allemand JF, Barre FX, Cornet F. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 2005;24:3770–80. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355:739–43. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J Bacteriol. 1999;181:5967–75. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA, Lehman BP, Yeates TO. Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol. 2015;98:193–207. doi: 10.1111/mmi.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonny M, Fischer-Friedrich E, Loose M, Schwille P, Kruse K. Membrane binding of MinE allows for a comprehensive description of Min-protein pattern formation. PLoS Comput Biol. 2013;9:e1003347. doi: 10.1371/journal.pcbi.1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet JY, Stouf M, Lebailly E, Cornet F. Mechanisms for chromosome segregation. Curr Opin Microbiol. 2014;22:60–5. doi: 10.1016/j.mib.2014.09.013. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–55. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D, Thompson CM. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc Natl Acad Sci USA. 1994;91:5813–7. doi: 10.1073/pnas.91.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol. 2006;62:5–14. doi: 10.1111/j.1365-2958.2006.05355.x. [DOI] [PubMed] [Google Scholar]
- Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA. 2012;109:1697–701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009;28:1208–19. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Herrmann H, Jacobs-Wagner C. The domain organization of the bacterial intermediate filament-like protein crescentin is important for assembly and function. Cytoskeleton (Hoboken) 2011;68:205–19. doi: 10.1002/cm.20505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabre EJ, Monterroso B, Alfonso C, Sanchez-Gorostiaga A, Reija B, Jimenez M, Vicente M, Zorrilla S, Rivas G. The nucleoid occlusion SlmA protein accelerates the disassembly of the FtsZ Protein polymers without affecting their GTPase activity. PLoS One. 2015;10:e0126434. doi: 10.1371/journal.pone.0126434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment. J Biol Chem. 2013;288:16055–63. doi: 10.1074/jbc.M113.456897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass JA, Kuwada NJ, Traxler B, Wiggins PA. Escherichia coli chromosomal loci segregate from midcell with universal dynamics. Biophys J. 2016;110:2597–609. doi: 10.1016/j.bpj.2016.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Amstutz C, Oltrogge L, Going CC, Lee A, Teng P, Quintanilla D, East-Seletsky A, Williams ER, Savage DF. Identification of a minimal peptide tag for in vivo and in vitro loading of encapsulin. Biochemistry. 2016;55:3461–8. doi: 10.1021/acs.biochem.6b00294. [DOI] [PubMed] [Google Scholar]
- Castellana M, Hsin-Jung Li S, Wingreen NS. Spatial organization of bacterial transcription and translation. Proc Natl Acad Sci USA. 2016;113:9286–91. doi: 10.1073/pnas.1604995113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbon G, Cabeen MT, Jacobs-Wagner C. Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 2009;23:1131–44. doi: 10.1101/gad.1795509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, Wolgemuth CW, Limberger RJ, Rowe N. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol. 2009;191:600–7. doi: 10.1128/JB.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, McManus HR, Dove SL, Bernhardt TG. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci USA. 2011;108:3773–8. doi: 10.1073/pnas.1018674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. Engineered protein nano-compartments for targeted enzyme localization. PLoS One. 2012;7:e33342. doi: 10.1371/journal.pone.0033342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha S, Woldringh CL, Odijk T. Polymer-mediated compaction and internal dynamics of isolated Escherichia coli nucleoids. J Struct Biol. 2001;136:53–66. doi: 10.1006/jsbi.2001.4420. [DOI] [PubMed] [Google Scholar]
- Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–90. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- Dame RT, Tark-Dame M. Bacterial chromatin: converging views at different scales. Curr Opin Cell Biol. 2016;40:60–5. doi: 10.1016/j.ceb.2016.02.015. [DOI] [PubMed] [Google Scholar]
- David A, Demarre G, Muresan L, Paly E, Barre FX, Possoz C. The two Cisacting sites, parS1 and oriC1, contribute to the longitudinal organisation of Vibrio cholerae chromosome I. PLoS Genet. 2014;10:e1004448. doi: 10.1371/journal.pgen.1004448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell division protein FtsZ is a GTPase. Nature. 1992;359:254–56. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- Deghelt M, Mullier C, Sternon JF, Francis N, Laloux G, Dotreppe D, Van der Henst C, Jacobs-Wagner C, Letesson JJ, De Bolle X. G1-arrested newborn cells are the predominant infectious form of the pathogen Brucella abortus. Nat Commun. 2014;5:4366. doi: 10.1038/ncomms5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempwolff F, Reimold C, Reth M, Graumann PL. Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system. PLoS One. 2011;6:e27035. doi: 10.1371/journal.pone.0027035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T, Hamoen LW, Levin PA. The divisome at 25: the road ahead. Curr Opin Microbiol. 2017;36:85–94. doi: 10.1016/j.mib.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski C, Kan W, Motaleb MA, Charon NW, Goldstein RE, Wolgemuth CW. The elastic basis for the shape of Borrelia burgdorferi. Biophys J. 2009;96:4409–17. doi: 10.1016/j.bpj.2009.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–8. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- Donovan C, Sieger B, Kramer R, Bramkamp M. A synthetic Escherichia coli system identifies a conserved origin tethering factor in Actinobacteria. Mol Microbiol. 2012;84:105–16. doi: 10.1111/j.1365-2958.2012.08011.x. [DOI] [PubMed] [Google Scholar]
- Dorman CJ. Function of nucleoid-associated proteins in chromosome structuring and transcriptional regulation. J Mol Microbiol Biotechnol. 2014;24:316–31. doi: 10.1159/000368850. [DOI] [PubMed] [Google Scholar]
- dos Santos VT, Bisson-Filho AW, Gueiros-Filho FJ. DivIVA-mediated polar localization of ComN, a posttranscriptional regulator of Bacillus subtilis. J Bacteriol. 2012;194:3661–9. doi: 10.1128/JB.05879-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J Biol Chem. 2008;283:10377–84. doi: 10.1074/jbc.M709285200. [DOI] [PubMed] [Google Scholar]
- Draper W, Liphardt J. Origins of chemoreceptor curvature sorting in Escherichia coli. Nat Commun. 2017;8:14838. doi: 10.1038/ncomms14838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Lutkenhaus J. SlmA antagonism of FtsZ assembly employs a two-pronged mechanism like MinCD. PLoS Genet. 2014;10:e1004460. doi: 10.1371/journal.pgen.1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupaigne P, Tonthat NK, Espeli O, Whitfill T, Boccard F, Schumacher MA. Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome. Mol Cell. 2012;48:560–71. doi: 10.1016/j.molcel.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:956–68. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Ringgaard S, Moller-Jensen J, Wang Q, Sherratt DJ, Gerdes K. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol Microbiol. 2006;61:1428–42. doi: 10.1111/j.1365-2958.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Osawa M. FtsZ Constriction Force - Curved Protofilaments Bending Membranes. Subcell Biochem. 2017;84:139–60. doi: 10.1007/978-3-319-53047-5_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–23. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Bobik TA. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol. 2011;193:5623–8. doi: 10.1128/JB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure LM, Fiche JB, Espinosa L, Ducret A, Anantharaman V, Luciano J, Lhospice S, Islam ST, Treguier J, Sotes M, et al. The mechanism of force transmission at bacterial focal adhesion complexes. Nature. 2016;539:530–35. doi: 10.1038/nature20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–32. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- Fink G, Aylett CHS. Tubulin-like proteins in prokaryotic DNA positioning. Subcell Biochem. 2017;84:323–56. doi: 10.1007/978-3-319-53047-5_11. [DOI] [PubMed] [Google Scholar]
- Fink G, Lowe J. Reconstitution of a prokaryotic minus end-tracking system using TubRC centromeric complexes and tubulin-like protein TubZ filaments. Proc Natl Acad Sci USA. 2015;112:E1845–50. doi: 10.1073/pnas.1423746112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JK, Bourniquel A, Witz G, Weiner B, Prentiss M, Kleckner N. Fourdimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell. 2013;153:882–95. doi: 10.1016/j.cell.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flardh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Mol Microbiol. 2003;49:1523–36. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol Microbiol. 2005;55:125–36. doi: 10.1111/j.1365-2958.2004.04379.x. [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–82. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchino K, Bagchi S, Cantlay S, Sandblad L, Wu D, Bergman J, Kamali-Moghaddam M, Flardh K, Ausmees N. Dynamic gradients of an intermediate filament-like cytoskeleton are recruited by a polarity landmark during apical growth. Proc Natl Acad Sci USA. 2013;110:E1889–97. doi: 10.1073/pnas.1305358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan B, Dinjaski N, Maestro B, de Eugenio LI, Escapa IF, Sanz JM, Garcia JL, Prieto MA. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol Microbiol. 2011;79:402–18. doi: 10.1111/j.1365-2958.2010.07450.x. [DOI] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–5. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Mullins RD. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 2004;306:1021–5. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–4. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayathri P, Fujii T, Moller-Jensen J, van den Ent F, Namba K, Lowe J. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science. 2012;338:1334–7. doi: 10.1126/science.1229091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayathri P, Harne S. Structure and dynamics of actin-like cytomotive filaments in plasmid segregation. Subcell Biochem. 2017;84:299–321. doi: 10.1007/978-3-319-53047-5_10. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–66. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- Giessen TW. Encapsulins: microbial nanocompartments with applications in biomedicine, nanobiotechnology and materials science. Curr Opin Chem Biol. 2016;34:1–10. doi: 10.1016/j.cbpa.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Giessen TW, Silver PA. Encapsulation as a strategy for the design of biological compartmentalization. Journal of Molecular Biology. 2016;428:916–27. doi: 10.1016/j.jmb.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Giessen TW, Silver PA. Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat Microbiol. 2017;2:17029. doi: 10.1038/nmicrobiol.2017.29. [DOI] [PubMed] [Google Scholar]
- Gill RL, Jr, Castaing JP, Hsin J, Tan IS, Wang X, Huang KC, Tian F, Ramamurthi KS. Structural basis for the geometry-driven localization of a small protein. Proc Natl Acad Sci USA. 2015;112:E1908–15. doi: 10.1073/pnas.1423868112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginda K, Santi I, Bousbaine D, Zakrzewska-Czerwinska J, Jakimowicz D, McKinney J. The studies of ParA and ParB dynamics reveal asymmetry of chromosome segregation in mycobacteria. Mol Microbiol. 2017;105:453–68. doi: 10.1111/mmi.13712. [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA. 2004;101:8643–8. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–8. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- Goldberg MB, Barzu O, Parsot C, Sansonetti PJ. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J Bacteriol. 1993;175:2189–96. doi: 10.1128/jb.175.8.2189-2196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein SF, Buttle KF, Charon NW. Structural analysis of the Leptospiraceae and Borrelia burgdorferi by high-voltage electron microscopy. J Bacteriol. 1996;178:6539–45. doi: 10.1128/jb.178.22.6539-6545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein SF, Charon NW, Kreiling JA. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc Natl Acad Sci USA. 1994;91:3433–7. doi: 10.1073/pnas.91.8.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight A, Losick R, Murray AW, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–21. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- Grangeon R, Zupan JR, Anderson-Furgeson J, Zambryski PC. PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2015;112:11666–71. doi: 10.1073/pnas.1515544112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S. Multilayer chromosome organization through DNA bending, bridging and extrusion. Curr Opin Microbiol. 2014;22:102–10. doi: 10.1016/j.mib.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–96. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Gruber S, Veening JW, Bach J, Blettinger M, Bramkamp M, Errington J. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Curr Biol. 2014;24:293–8. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadizadeh Yazdi N, Guet CC, Johnson RC, Marko JF. Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol Microbiol. 2012;86:1318–33. doi: 10.1111/mmi.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12:1057–63. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A, Treuner-Lange A, Schumacher D, Sogaard-Andersen L. Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLoS Genet. 2013;9:e1003802. doi: 10.1371/journal.pgen.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T, Yamaichi Y, Niki H. Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol Microbiol. 2007;64:1198–213. doi: 10.1111/j.1365-2958.2007.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havemann GD, Sampson EM, Bobik TA. PduA is a shell protein of polyhedral organelles involved in coenzyme B(12)-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2. J Bacteriol. 2002;184:1253–61. doi: 10.1128/JB.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JT, Crosson S. Chromosome replication and segregation govern the biogenesis and inheritance of inorganic polyphosphate granules. Mol Biol Cell. 2013;24:3177–86. doi: 10.1091/mbc.E13-04-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden SJ, Pengo T, Meibom KL, Fernandez Fernandez C, Collier J, Manley S. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci USA. 2014;111:4566–71. doi: 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes JA, Follett SE, Wang H, Meadows CP, Varga K, Bowman GR. Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc Natl Acad Sci USA. 2016;113:12490–95. doi: 10.1073/pnas.1602380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes NA, Walshaw J, Leggett RM, Thibessard A, Dalton KA, Gillespie MD, Hemmings AM, Gust B, Kelemen GH. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc Natl Acad Sci USA. 2013;110:E397–406. doi: 10.1073/pnas.1210657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, McAdams HH. Compaction and transport properties of newly replicated Caulobacter crescentus DNA. Mol Microbiol. 2011;82:1349–58. doi: 10.1111/j.1365-2958.2011.07899.x. [DOI] [PubMed] [Google Scholar]
- Hong SH, Toro E, Mortensen KI, de la Rosa MA, Doniach S, Shapiro L, Spakowitz AJ, McAdams HH. Caulobacter chromosome in vivo configuration matches model predictions for a supercoiled polymer in a cell-like confinement. Proc Natl Acad Sci USA. 2013;110:1674–9. doi: 10.1073/pnas.1220824110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Vecchiarelli AG, Mizuuchi K, Neuman KC, Liu J. Brownian ratchet mechanism for faithful segregation of low-copy-number plasmids. Biophys J. 2017;112:1489–502. doi: 10.1016/j.bpj.2017.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–37. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Hwang LC, Vecchiarelli AG, Han YW, Mizuuchi M, Harada Y, Funnell BE, Mizuuchi K. ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J. 2013;32:1238–49. doi: 10.1038/emboj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ietswaart R, Szardenings F, Gerdes K, Howard M. Competing ParA structures space bacterial plasmids equally over the nucleoid. PLoS Comput Biol. 2014;10:e1004009. doi: 10.1371/journal.pcbi.1004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA. ParABS system in chromosome partitioning in the bacterium Myxococcus xanthus. PLoS One. 2014;9:e86897. doi: 10.1371/journal.pone.0086897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings PC, Cox GC, Monahan LG, Harry EJ. Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron. 2011;42:336–41. doi: 10.1016/j.micron.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci USA. 1999;96:10661–6. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske O, Schuler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H, et al. Planctomycetes do possess a peptidoglycan cell wall. Nat Commun. 2015;6 doi: 10.1038/ncomms8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Brown PJ, Ducret A, Brun YV. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature. 2014;506:489–93. doi: 10.1038/nature12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal L, Emberly E. Operational principles for the dynamics of the in vitro ParA-ParB system. PLoS Comput Biol. 2015;11:e1004651. doi: 10.1371/journal.pcbi.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–22. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Joshi MC, Bourniquel A, Fisher J, Ho BT, Magnan D, Kleckner N, Bates D. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc Natl Acad Sci USA. 2011;108:2765–70. doi: 10.1073/pnas.1019593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce G, Williams KJ, Robb M, Noens E, Tizzano B, Shahrezaei V, Robertson BD. Cell division site placement and asymmetric growth in mycobacteria. PLoS One. 2012;7:e44582. doi: 10.1371/journal.pone.0044582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–93. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier I, Boccard F, Espeli O. Polymer modeling of the E. coli genome reveals the involvement of locus positioning and macrodomain structuring for the control of chromosome conformation and segregation. Nucleic Acids Res. 2014;42:1461–73. doi: 10.1093/nar/gkt1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Scott M, Parry B, Biboy J, Gray J, Vollmer W, Jacobs-Wagner C. Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. Proc Natl Acad Sci USA. 2016;113:9162–70. doi: 10.1073/pnas.1610805113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154:725–35. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Mercier R, Errington J. Bacterial cell morphogenesis does not require a preexisting template structure. Curr Biol. 2014;24:863–7. doi: 10.1016/j.cub.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazura T, Matsumoto K, Kojima K, Kato F, Kanai T, Niki H, Shiomi D. Exclusion of assembled MreB by anionic phospholipids at cell poles confers cell polarity for bidirectional growth. Mol Microbiol. 2017;104:472–86. doi: 10.1111/mmi.13639. [DOI] [PubMed] [Google Scholar]
- Keilberg D, Wuichet K, Drescher F, Sogaard-Andersen L. A response regulator interfaces between the Frz chemosensory system and the MglA/MglB GTPase/GAP module to regulate polarity in Myxococcus xanthus. PLoS Genet. 2012;8:e1002951. doi: 10.1371/journal.pgen.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–8. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CL, Viollier PH. Decoding Caulobacter development. FEMS Microbiol Rev. 2012;36:193–205. doi: 10.1111/j.1574-6976.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392:319–33. doi: 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- Kleine Borgmann LA, Graumann PL. Structural maintenance of chromosome complex in bacteria. J Mol Microbiol Biotechnol. 2014;24:384–95. doi: 10.1159/000368931. [DOI] [PubMed] [Google Scholar]
- Kocks C, Hellio R, Gounon P, Ohayon H, Cossart P. Polarized distribution of Listeria monocytogenes surface protein ActA at the site of directional actin assembly. J Cell Sci. 1993;105(Pt 3):699–710. doi: 10.1242/jcs.105.3.699. [DOI] [PubMed] [Google Scholar]
- Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–29. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–5. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Kretschmer S, Schwille P. Pattern formation on membranes and its role in bacterial cell division. Curr Opin Cell Biol. 2016;38:52–9. doi: 10.1016/j.ceb.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Kuhn J, Briegel A, Morschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010;29:327–39. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kysela DT, Randich AM, Caccamo PD, Brun YV. Diversity Takes Shape: Understanding the mechanistic and adaptive basis of bacterial morphology. PLoS Biol. 2016;14:e1002565. doi: 10.1371/journal.pbio.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloux G, Jacobs-Wagner C. Spatiotemporal control of PopZ localization through cell cycle-coupled multimerization. J Cell Biol. 2013;201:827–41. doi: 10.1083/jcb.201303036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–23. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Lampo TJ, Kuwada NJ, Wiggins PA, Spakowitz AJ. Physical modeling of chromosome segregation in escherichia coli reveals impact of force and DNA relaxation. Biophys J. 2015;108:146–53. doi: 10.1016/j.bpj.2014.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. Condensation of FtsZ filaments can drive bacterial cell division. Proc Natl Acad Sci USA. 2009;106:121–6. doi: 10.1073/pnas.0807963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–52. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Mann TH, Shapiro L. An intracellular compass spatially coordinates cell cycle modules in Caulobacter crescentus. Curr Opin Microbiol. 2016;33:131–39. doi: 10.1016/j.mib.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall A, Cattoni DI, Guilhas B, Mathieu-Demaziere C, Oudjedi L, Fiche JB, Rech J, Abrahamsson S, Murray H, Bouet JY, et al. Bacterial partition complexes segregate within the volume of the nucleoid. Nat Commun. 2016;7:12107. doi: 10.1038/ncomms12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TB, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–4. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J. Bacterial protoplasts induced by penicillin. Proc Natl Acad Sci USA. 1956;42:574–7. doi: 10.1073/pnas.42.9.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, Commichau FM, Lewis RJ, Stulke J. RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J Bacteriol. 2011;193:5431–41. doi: 10.1128/JB.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–82. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, Sogaard-Andersen L. Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J. 2010;29:2276–89. doi: 10.1038/emboj.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterlin C, Pages C, Dubarry N, Dasgupta S, Cornet F. Asymmetry of chromosome Replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet. 2008;4:e1000288. doi: 10.1371/journal.pgen.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–88. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- Levy O, Ptacin JL, Pease PJ, Gore J, Eisen MB, Bustamante C, Cozzarelli NR. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc Natl Acad Sci USA. 2005;102:17618–23. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ, Thaker SD, Errington J. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 2000;19:710–8. doi: 10.1093/emboj/19.4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti G, Kuru E, Packiam M, Hsu YP, Tekkam S, Hall E, Rittichier JT, VanNieuwenhze M, Brun YV, Maurelli AT. Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog. 2016;12:e1005590. doi: 10.1371/journal.ppat.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HC, Surovtsev IV, Beltran BG, Huang F, Bewersdorf J, Jacobs-Wagner C. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. Elife. 2014;3:e02758. doi: 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Thanbichler M. Nucleotide-independent cytoskeletal scaffolds in bacteria. Cytoskeleton (Hoboken) 2013;70:409–23. doi: 10.1002/cm.21126. [DOI] [PubMed] [Google Scholar]
- Lin TY, Weibel DB. Organization and function of anionic phospholipids in bacteria. Appl Microbiol Biotechnol. 2016;100:4255–67. doi: 10.1007/s00253-016-7468-x. [DOI] [PubMed] [Google Scholar]
- Liou GG, Jane WN, Cohen SN, Lin NS, Lin-Chao S. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc Natl Acad Sci USA. 2001;98:63–8. doi: 10.1073/pnas.011535498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy VS, Volante A, Soberon NE, Lurz R, Ayora S, Alonso JC. ParAB Partition Dynamics in Firmicutes: Nucleoid-bound ParA captures and tethers ParB-plasmid complexes. PLoS One. 2015;10:e0131943. doi: 10.1371/journal.pone.0131943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol. 2007;189:8693–703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–6. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–3003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–23. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Manske AK, Glaeser J, Kuypers MM, Overmann J. Physiology and phylogeny of green sulfur bacteria forming a monospecific phototrophic assemblage at a depth of 100 meters in the Black Sea. Appl Environ Microbiol. 2005;71:8049–60. doi: 10.1128/AEM.71.12.8049-8060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty M, Cournac A, Flot JF, Marie-Nelly H, Mozziconacci J, Koszul R. Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. Elife. 2014;3:e03318. doi: 10.7554/eLife.03318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty M, Le Gall A, Cattoni DI, Cournac A, Koh A, Fiche JB, Mozziconacci J, Murray H, Koszul R, Nollmann M. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol Cell. 2015;59:588–602. doi: 10.1016/j.molcel.2015.07.020. [DOI] [PubMed] [Google Scholar]
- Marko JF. Linking topology of tethered polymer rings with applications to chromosome segregation and estimation of the knotting length. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;79:051905. doi: 10.1103/PhysRevE.79.051905. [DOI] [PubMed] [Google Scholar]
- Mascarenhas J, Soppa J, Strunnikov AV, Graumann PL. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 2002;21:3108–18. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriello EM, Mouhamar F, Nan B, Ducret A, Dai D, Zusman DR, Mignot T. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 2010;29:315–26. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza P, Noens EE, Schirner K, Grantcharova N, Mommaas AM, Koerten HK, Muth G, Flardh K, van Wezel GP, Wohlleben W. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol Microbiol. 2006;60:838–52. doi: 10.1111/j.1365-2958.2006.05134.x. [DOI] [PubMed] [Google Scholar]
- Meniche X, Otten R, Siegrist MS, Baer CE, Murphy KC, Bertozzi CR, Sassetti CM. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA. 2014;111:E3243–51. doi: 10.1073/pnas.1402158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, Espeli O. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135:475–85. doi: 10.1016/j.cell.2008.08.031. [DOI] [PubMed] [Google Scholar]
- Miertzschke M, Koerner C, Vetter IR, Keilberg D, Hot E, Leonardy S, Sogaard-Andersen L, Wittinghofer A. Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity. EMBO J. 2011;30:4185–97. doi: 10.1038/emboj.2011.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–6. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IL, Wiebe W, Landman OE. Gelatin-induced reversion of protoplasts of Bacillus subtilis to the bacillary form: photomicrographic study. J Bacteriol. 1968;96:2171–4. doi: 10.1128/jb.96.6.2171-2174.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol. 2011;81:676–88. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- Minnen A, Burmann F, Wilhelm L, Anchimiuk A, Diebold-Durand ML, Gruber S. Control of Smc coiled coil architecture by the ATPase heads facilitates targeting to chromosomal ParB/parS and release onto flanking DNA. Cell Rep. 2016;14:2003–16. doi: 10.1016/j.celrep.2016.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Pandey S, Boettiger AN, Wang S, Zhuang X. Spatial organization shapes the turnover of a bacterial transcriptome. Elife. 2016;5:e13065. doi: 10.7554/eLife.13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol Cell. 2003;12:1477–87. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- Monahan LG, Liew AT, Bottomley AL, Harry EJ. Division site positioning in bacteria: one size does not fit all. Front Microbiol. 2014;5:19. doi: 10.3389/fmicb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal J, Bratton BP, Li Y, Yethiraj A, Weisshaar JC. Entropy-based mechanism of ribosome-nucleoid segregation in E. coli cells. Biophys J. 2011;100:2605–13. doi: 10.1016/j.bpj.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro JM, Fernandes PB, Vaz F, Pereira AR, Tavares AC, Ferreira MT, Pereira PM, Veiga H, Kuru E, VanNieuwenhze MS, et al. Cell shape dynamics during the staphylococcal cell cycle. Nat Commun. 2015;6:8055. doi: 10.1038/ncomms9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa N, Imamoto F. Degradation of tryptophan messenger. On the degradation of messenger RNA for the tryptophan operon in Escherichia coli. Nature. 1969;223:37–40. doi: 10.1038/223037a0. [DOI] [PubMed] [Google Scholar]
- Morse DE, Mosteller R, Baker RF, Yanofsky C. Direction of in vivo degradation of tryptophan messenger RNA–a correction. Nature. 1969;223:40–3. doi: 10.1038/223040a0. [DOI] [PubMed] [Google Scholar]
- Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci USA. 2000;97:10899–904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division Protein FtsZ is a guanine nucleotide-binding protein. Proc Natl Acad Sci U S A. 1993;90:1053–57. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–8. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Huang KC, Wingreen NS. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J. 2008;95:1034–49. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LD, Zimmerman SB. Stabilization of compact spermidine nucleoids from Escherichia coli under crowded conditions: implications for in vivo nucleoid structure. J Struct Biol. 1997;119:336–46. doi: 10.1006/jsbi.1997.3884. [DOI] [PubMed] [Google Scholar]
- Neumann S, Wessels HJ, Rijpstra WI, Sinninghe Damste JS, Kartal B, Jetten MS, van Niftrik L. Isolation and characterization of a prokaryotic cell organelle from the anammox bacterium Kuenenia stuttgartiensis. Mol Microbiol. 2014;94:794–802. doi: 10.1111/mmi.12816. [DOI] [PubMed] [Google Scholar]
- Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science. 2011;331:1081–4. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- Ni L, Xu W, Kumaraswami M, Schumacher MA. Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc Natl Acad Sci USA. 2010;107:11763–8. doi: 10.1073/pnas.1003817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Molecular Microbiology. 2006;62:331–38. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- Nielsen JT, Kulminskaya NV, Bjerring M, Linnanto JM, Ratsep M, Pedersen MO, Lambrev PH, Dorogi M, Garab G, Thomsen K, et al. In situ high-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum. Nat Commun. 2016;7:12454. doi: 10.1038/ncomms12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–23. [PMC free article] [PubMed] [Google Scholar]
- Nolivos S, Sherratt D. The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol Rev. 2014;38:380–92. doi: 10.1111/1574-6976.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odijk T. Osmotic compaction of supercoiled DNA into a bacterial nucleoid. Biophys Chem. 1998;73:23–9. doi: 10.1016/s0301-4622(98)00115-x. [DOI] [PubMed] [Google Scholar]
- Oliva MA, Halbedel S, Freund SM, Dutow P, Leonard TA, Veprintsev DB, Hamoen LW, Lowe J. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 2010;29:1988–2001. doi: 10.1038/emboj.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshausen PV, Defeu Soufo HJ, Wicker K, Heintzmann R, Graumann PL, Rohrbach A. Superresolution imaging of dynamic MreB filaments in B. subtilis—a multiple-motor-driven transport? Biophys J. 2013;105:1171–81. doi: 10.1016/j.bpj.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–4. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USA. 2013;110:11000–4. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette SP, Karimova G, Subtil A, Ladant D. Chlamydia co-opts the rod shapedetermining proteins MreB and Pbp2 for cell division. Mol Microbiol. 2012;85:164–78. doi: 10.1111/j.1365-2958.2012.08100.x. [DOI] [PubMed] [Google Scholar]
- Ouzounov N, Nguyen JP, Bratton BP, Jacobowitz D, Gitai Z, Shaevitz JW. MreB orientation correlates with cell diameter in Escherichia coli. Biophys J. 2016;111:1035–43. doi: 10.1016/j.bpj.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KT, Wu W, Battaile KP, Lovell S, Holyoak T, Lutkenhaus J. The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell. 2011;146:396–407. doi: 10.1016/j.cell.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, Bustamante C, Cozzarelli NR. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–90. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–74. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F. Distribution, formation and regulation of gas vesicles. Nat Rev Microbiol. 2012;10:705–15. doi: 10.1038/nrmicro2834. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Molecular Microbiology. 2005;55:1722–34. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, Brun YV, VanNieuwenhze MS, Vollmer W, Horn M, et al. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun. 2013;4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho MG, Kjos M, Veening JW. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol. 2013;11:601–14. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- Polka JK, Kollman JM, Agard DA, Mullins RD. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J Bacteriol. 2009;191:6219–30. doi: 10.1128/JB.00676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polka JK, Kollman JM, Mullins RD. Accessory factors promote AlfA-dependent plasmid segregation by regulating filament nucleation, disassembly, and bundling. Proc Natl Acad Sci USA. 2014;111:2176–81. doi: 10.1073/pnas.1304127111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp D, Narita A, Ghoshdastider U, Maeda K, Maeda Y, Oda T, Fujisawa T, Onishi H, Ito K, Robinson RC. Polymeric structures and dynamic properties of the bacterial actin AlfA. J Mol Biol. 2010;397:1031–41. doi: 10.1016/j.jmb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Pradel N, Santini CL, Bernadac A, Fukumori Y, Wu LF. Biogenesis of actin-like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc Natl Acad Sci USA. 2006;103:17485–9. doi: 10.1073/pnas.0603760103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Gahlmann A, Bowman GR, Perez AM, von Diezmann AR, Eckart MR, Moerner WE, Shapiro L. Bacterial scaffold directs pole-specific centromere segregation. Proc Natl Acad Sci USA. 2014;111:E2046–55. doi: 10.1073/pnas.1405188111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–8. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki LR, Tocheva EI, Dieterle MG, Sullivan MC, Jensen GJ, Newman DK. Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2017;114:E2440–E49. doi: 10.1073/pnas.1615575114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–7. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci USA. 2009;106:13541–5. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999a;181:6419–24. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999b;96:4971–6. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D, Park JT. Escherichia coli cell division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–4. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Renner LD, Weibel DB. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc Natl Acad Sci USA. 2011;108:6264–9. doi: 10.1073/pnas.1015757108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA. 2009;106:19369–74. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues CD, Harry EJ. The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet. 2012;8:e1002561. doi: 10.1371/journal.pgen.1002561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett VW, Margolin W. 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys J. 2014;107:L17–20. doi: 10.1016/j.bpj.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett VW, Margolin W. The Min system and other nucleoid-independent regulators of Z ring positioning. Front Microbiol. 2015;6:478. doi: 10.3389/fmicb.2015.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rurup WF, Snijder J, Koay MS, Heck AJ, Cornelissen JJ. Self-sorting of foreign proteins in a bacterial nanocompartment. J Am Chem Soc. 2014;136:3828–32. doi: 10.1021/ja410891c. [DOI] [PubMed] [Google Scholar]
- Saberi S, Emberly E. Chromosome driven spatial patterning of proteins in bacteria. PLoS Comput Biol. 2010;6:e1000986. doi: 10.1371/journal.pcbi.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagermann M, Ohtaki A, Nikolakakis K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci U S A. 2009;106:8883–7. doi: 10.1073/pnas.0902324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi D, Friedman N, Vorgias C, Oppenheim AB, Stavans J. Modulation of DNA conformations through the formation of alternative high-order HU-DNA complexes. J Mol Biol. 2004;341:419–28. doi: 10.1016/j.jmb.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Salje J, van den Ent F, de Boer P, Lowe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–87. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–71. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanamrad A, Persson F, Lundius EG, Fange D, Gynna AH, Elf J. Single-particle tracking reveals that free ribosomal subunits are not excluded from the Escherichia coli nucleoid. Proc Natl Acad Sci USA. 2014;111:11413–8. doi: 10.1073/pnas.1411558111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat Commun. 2013;4:2470. doi: 10.1038/ncomms3470. [DOI] [PubMed] [Google Scholar]
- Santos TM, Lin TY, Rajendran M, Anderson SM, Weibel DB. Polar localization of Escherichia coli chemoreceptors requires an intact Tol-Pal complex. Mol Microbiol. 2014;92:985–1004. doi: 10.1111/mmi.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–61. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–4. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29:3068–81. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher D, Bergeler S, Harms A, Vonck J, Huneke-Vogt S, Frey E, Sogaard-Andersen L. The PomXYZ Proteins self-organize on the bacterial nucleoid to stimulate cell division. Dev Cell. 2017;41:299–314 e13. doi: 10.1016/j.devcel.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Schumacher D, Sogaard-Andersen L. Regulation of cell polarity in motility and cell division in Myxococcus xanthus. Annu Rev Microbiol. 2017;71:61–78. doi: 10.1146/annurev-micro-102215-095415. [DOI] [PubMed] [Google Scholar]
- Sengupta M, Nielsen HJ, Youngren B, Austin S. P1 plasmid segregation: accurate redistribution by dynamic plasmid pairing and separation. J Bacteriol. 2010;192:1175–83. doi: 10.1128/JB.01245-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science. 2002;295:137–9. doi: 10.1126/science.1066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shebelut CW, Jensen RB, Gitai Z. Growth conditions regulate the requirements for Caulobacter chromosome segregation. J Bacteriol. 2009;191:1097–100. doi: 10.1128/JB.00862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–6. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- Si F, Busiek K, Margolin W, Sun SX. Organization of FtsZ filaments in the bacterial division ring measured from polarized fluorescence microscopy. Biophys J. 2013;105:1976–86. doi: 10.1016/j.bpj.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinninghe Damste JS, Strous M, Rijpstra WI, Hopmans EC, Geenevasen JA, van Duin AC, van Niftrik LA, Jetten MS. Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nature. 2002;419:708–12. doi: 10.1038/nature01128. [DOI] [PubMed] [Google Scholar]
- Sivertsen AC, Bayro MJ, Belenky M, Griffin RG, Herzfeld J. Solid-state NMR characterization of gas vesicle structure. Biophys J. 2010;99:1932–9. doi: 10.1016/j.bpj.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J, Kobayashi K, Noirot-Gros MF, Oesterhelt D, Ehrlich SD, Dervyn E, Ogasawara N, Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- Specht M, Schatzle S, Graumann PL, Waidner B. Helicobacter pylori possesses four coiled-coil-rich proteins that form extended filamentous structures and control cell shape and motility. J Bacteriol. 2011;193:4523–30. doi: 10.1128/JB.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P, Fekete RA, Chattoraj DK. Segregation of the replication terminus of the two Vibrio cholerae chromosomes. J Bacteriol. 2006;188:1060–70. doi: 10.1128/JB.188.3.1060-1070.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MP, Stevens JM, Jeng RL, Taylor LA, Wood MW, Hawes P, Monaghan P, Welch MD, Galyov EE. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol Microbiol. 2005;56:40–53. doi: 10.1111/j.1365-2958.2004.04528.x. [DOI] [PubMed] [Google Scholar]
- Strahl H, Ronneau S, Gonzalez BS, Klutsch D, Schaffner-Barbero C, Hamoen LW. Transmembrane protein sorting driven by membrane curvature. Nat Commun. 2015;6:8728. doi: 10.1038/ncomms9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci USA. 2002;99:3171–5. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara T, Kaneko K. Chemophoresis as a driving force for intracellular organization: Theory and application to plasmid partitioning. Biophysics (Nagoya-shi) 2011;7:77–88. doi: 10.2142/biophysics.7.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtsev IV, Campos M, Jacobs-Wagner C. DNA-relay mechanism is sufficient to explain ParA-dependent intracellular transport and patterning of single and multiple cargos. Proc Natl Acad Sci USA. 2016;113:E7268–E76. doi: 10.1073/pnas.1616118113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15:939–47. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–33. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Bharat TA, Tsim M, Lowe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura A, Fukutani Y, Takami T, Fujii M, Nakaguchi Y, Murakami Y, Noguchi K, Yohda M, Odaka M. Packaging guest proteins into the encapsulin nanocompartment from Rhodococcus erythropolis N771. Biotechnol Bioeng. 2015;112:13–20. doi: 10.1002/bit.25322. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–6. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Graumann PL, Lin DC, Grossman AD, Losick R. Chromosome arrangement within a bacterium. Curr Biol. 1998;8:1102–9. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- Thanky NR, Young DB, Robertson BD. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis (Edinb) 2007;87:231–6. doi: 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Thiem S, Sourjik V. Stochastic assembly of chemoreceptor clusters in Escherichia coli. Mol Microbiol. 2008;68:1228–36. doi: 10.1111/j.1365-2958.2008.06227.x. [DOI] [PubMed] [Google Scholar]
- Thompson JF, Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988;16:9687–705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR, Wadhams GH, Armitage JP. The positioning of cytoplasmic protein clusters in bacteria. Proc Natl Acad Sci USA. 2006;103:8209–14. doi: 10.1073/pnas.0600919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011;30:154–64. doi: 10.1038/emboj.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonthat NK, Milam SL, Chinnam N, Whitfill T, Margolin W, Schumacher MA. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc Natl Acad Sci USA. 2013;110:10586–91. doi: 10.1073/pnas.1221036110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NT, Laub MT, Le TBK. SMC progressively aligns chromosomal arms in Caulobacter crescentus but is antagonized by convergent transcription. Cell Rep. 2017;20:2057–71. doi: 10.1016/j.celrep.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuner-Lange A, Aguiluz K, van der Does C, Gomez-Santos N, Harms A, Schumacher D, Lenz P, Hoppert M, Kahnt J, Munoz-Dorado J, et al. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol Microbiol. 2013;87:235–53. doi: 10.1111/mmi.12094. [DOI] [PubMed] [Google Scholar]
- Treuner-Lange A, Macia E, Guzzo M, Hot E, Faure LM, Jakobczak B, Espinosa L, Alcor D, Ducret A, Keilberg D, et al. The small G-protein MglA connects to the MreB actin cytoskeleton at bacterial focal adhesions. J Cell Biol. 2015;210:243–56. doi: 10.1083/jcb.201412047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski D, Ginda K, Pioro M, Holowka J, Skut P, Jakimowicz D, Zakrzewska-Czerwinska J. Choreography of the Mycobacterium replication machinery during the cell cycle. MBio. 2015;6:e02125–14. doi: 10.1128/mBio.02125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RD, Ratcliffe EC, Wheeler R, Golestanian R, Hobbs JK, Foster SJ. Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nat Commun. 2010;1:26. doi: 10.1038/ncomms1025. [DOI] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–36. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebe R, Schuler D. Magnetosome biogenesis in magnetotactic bacteria. Nat Rev Microbiol. 2016;14:621–37. doi: 10.1038/nrmicro.2016.99. [DOI] [PubMed] [Google Scholar]
- Umbarger MA, Toro E, Wright MA, Porreca GJ, Bau D, Hong SH, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44:252–64. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, Gitai Z, Shaevitz JW, Huang KC. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci USA. 2014;111:E1025–34. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val ME, Marbouty M, de Lemos Martins F, Kennedy SP, Kemble H, Bland MJ, Possoz C, Koszul R, Skovgaard O, Mazel D. A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae. Sci Adv. 2016;2:e1501914. doi: 10.1126/sciadv.1501914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I, Boccard F. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 2013;9:e1003492. doi: 10.1371/journal.pgen.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Izore T, Bharat TA, Johnson CM, Lowe J. Bacterial actin MreB forms antiparallel double filaments. Elife. 2014;3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gijtenbeek LA, Robinson A, van Oijen AM, Poolman B, Kok J. On the spatial organization of mRNA, plasmids, and ribosomes in a bacterial host overexpressing membrane proteins. PLoS Genet. 2016;12:e1006523. doi: 10.1371/journal.pgen.1006523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc Natl Acad Sci USA. 2004;101:6969–74. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA. 2011;108:15822–7. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Hwang LC, Mizuuchi K. Cell-free study of F plasmid partition provides evidence for cargo transport by a diffusion-ratchet mechanism. Proc Natl Acad Sci USA. 2013;110:E1390–7. doi: 10.1073/pnas.1302745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Li M, Mizuuchi M, Hwang LC, Seol Y, Neuman KC, Mizuuchi K. Membrane-bound MinDE complex acts as a toggle switch that drives Min oscillation coupled to cytoplasmic depletion of MinD. Proc Natl Acad Sci USA. 2016;113:E1479–88. doi: 10.1073/pnas.1600644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–62. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidner B, Specht M, Dempwolff F, Haeberer K, Schaetzle S, Speth V, Kist M, Graumann PL. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS Pathog. 2009;5:e1000669. doi: 10.1371/journal.ppat.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsby AE. Permeability of Blue-Green Algal Gas-Vacuole Membranes to Gas. Proceedings of the Royal Society Series B-Biological Sciences. 1969;173:235–+. [Google Scholar]
- Walter JC, Dorignac J, Lorman V, Rech J, Bouet JY, Nollmann M, Palmeri J, Parmeggiani A, Geniet F. Surfing on protein waves: Proteophoresis as a mechanism for bacterial genome partitioning. Phys Rev Lett. 2017;119:028101. doi: 10.1103/PhysRevLett.119.028101. [DOI] [PubMed] [Google Scholar]
- Wang H, Wingreen NS, Mukhopadhyay R. Self-organized periodicity of protein clusters in growing bacteria. Phys Rev Lett. 2008;101:218101. doi: 10.1103/PhysRevLett.101.218101. [DOI] [PubMed] [Google Scholar]
- Wang LL, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–40. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Brandao HB, Le TB, Laub MT, Rudner DZ. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science. 2017;355:524–27. doi: 10.1126/science.aai8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Le TB, Lajoie BR, Dekker J, Laub MT, Rudner DZ. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 2015;29:1661–75. doi: 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu X, Possoz C, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20:1727–31. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Montero Llopis P, Rudner DZ. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci USA. 2014a;111:12877–82. doi: 10.1073/pnas.1407461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Rudner DZ. Spatial organization of bacterial chromosomes. Curr Opin Microbiol. 2014;22:66–72. doi: 10.1016/j.mib.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang OW, Riley EP, Rudner DZ. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol. 2014b;24:287–92. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–74. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol Microbiol. 2011;82:1096–109. doi: 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell. 1999;4:683–94. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J. Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci USA. 2010;107:4991–5. doi: 10.1073/pnas.0912062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm L, Burmann F, Minnen A, Shin HC, Toseland CP, Oh BH, Gruber S. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. Elife. 2015;4:e06659. doi: 10.7554/eLife.06659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson DL. Unusual fibrils from the spirochete-like sex ratio organism. J Bacteriol. 1974;117:904–6. doi: 10.1128/jb.117.2.904-906.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F, Renner LD, Ozbaykal G, Paulose J, Weibel DB, van Teeffelen S, Amir A. Mechanical strain sensing implicated in cell shape recovery in Escherichia coli. Nat Microbiol. 2017;2:17115. doi: 10.1038/nmicrobiol.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Halatek J, Reiter M, Kingma E, Frey E, Dekker C. Multistability and dynamic transitions of intracellular Min protein patterns. Mol Syst Biol. 2016;12:873. doi: 10.15252/msb.20156724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–25. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–52. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Bruckner R, Ringgaard S, Moll A, Cameron DE, Briegel A, Jensen GJ, Davis BM, Waldor MK. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 2012;26:2348–60. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe M, Onogi T, Sunako Y, Niki H, Yamanaka K, Ichimura T, Hiraga S. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 1999;18:5873–84. doi: 10.1093/emboj/18.21.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Blair KM, Salama NR. Staying in shape: The impact of cell shape on bacterial survival in diverse environments. Microbiol Mol Biol Rev. 2016;80:187–203. doi: 10.1128/MMBR.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science. 2017;355:744–47. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Jewett AI, Chang YW, Oikonomou CM, Beeby M, Iancu CV, Briegel A, Ghosal D, Jensen GJ. Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis. EMBO J. 2017;36:1577–89. doi: 10.15252/embj.201696235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol. 2010;192:4847–58. doi: 10.1128/JB.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren B, Nielsen HJ, Jun S, Austin S. The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev. 2014;28:71–84. doi: 10.1101/gad.231050.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Tran AH, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Franco M, Ducret A, Mignot T. A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol. 2010;8:e1000430. doi: 10.1371/journal.pbio.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guzzo M, Ducret A, Li YZ, Mignot T. A dynamic response regulator protein modulates G-protein-dependent polarity in the bacterium Myxococcus xanthus. PLoS Genet. 2012;8:e1002872. doi: 10.1371/journal.pgen.1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Halladin DK, Rojas ER, Koslover EF, Lee TK, Huang KC, Theriot JA. Bacterial division. Mechanical crack propagation drives millisecond daughter cell separation in Staphylococcus aureus. Science. 2015;348:574–8. doi: 10.1126/science.aaa1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Halladin DK, Theriot JA. Fast mechanically driven daughter cell separation is widespread in Actinobacteria. MBio. 2016;7:e00952–16. doi: 10.1128/mBio.00952-16. [DOI] [PMC free article] [PubMed] [Google Scholar]


