Abstract
Prognostic and predictive factors are indispensable tools in the treatment of patients with neoplastic disease. For the most part, such factors rely on a few specific cell surface, histological, or gross pathologic features. Gene expression assays have the potential to supplement what were previously a few distinct features with many thousands of features. We have developed Bayesian regression models that provide predictive capability based on gene expression data derived from DNA microarray analysis of a series of primary breast cancer samples. These patterns have the capacity to discriminate breast tumors on the basis of estrogen receptor status and also on the categorized lymph node status. Importantly, we assess the utility and validity of such models in predicting the status of tumors in crossvalidation determinations. The practical value of such approaches relies on the ability not only to assess relative probabilities of clinical outcomes for future samples but also to provide an honest assessment of the uncertainties associated with such predictive classifications on the basis of the selection of gene subsets for each validation analysis. This latter point is of critical importance in the ability to apply these methodologies to clinical assessment of tumor phenotype.
Recent studies demonstrate that gene expression information generated by DNA microarray analysis of human tumors can provide molecular phenotyping that identifies distinct tumor classifications not evident by traditional histopathological methods (1–7). The promise of such information lies in the potential to inform and so improve clinical decisions and strategies used to treat patients with neoplastic disease. Traditional methods of phenotypic characterization are often limited and do not have the ability to discern subtle differences that may be of importance for developing a better understanding of the tumor and advancing therapeutic strategies for the treatment of disease. We have taken a two-pronged approach in creating a statistical method that provides robust probabilistic prediction and classification of tumors based on gene expression data and also permits formal assessment of the uncertainties inherent in any predictive model. Such an approach is critical in an arena where clinicians must gauge their certainty of a tumor's phenotypic properties against the potential morbidities of specific interventions.
We have applied this approach to breast cancer, a disease where further molecular characterization is needed to improve diagnostic and therapeutic strategies. Numerous studies have correlated genetic alterations with clinical outcome including a strong correlation between the amplification of the erbB-2 receptor gene (Her-2) and poor clinical outcome (8, 9). In addition, overexpression of erbB-2 is a strong predictor of response to adriamycin-based therapy (10). Nevertheless, such correlations are few and often do not adequately define tumor subtypes. The inability to define a subclass of tumor type that may be refractory to standard therapies restricts the development of new, more efficacious therapeutic strategies.
The analysis of gene expression represents an indirect measure of the genetic alterations in tumors because, in most instances, these alterations affect gene regulatory pathways. Given the tremendous complexity that can be scored by measuring gene expression with DNA microarrays, together with the absence of bias in assumptions as to what type of pathway might be affected in a particular tumor, the analysis of gene expression profiles offers the potential to impact clinical decision-making based on more precise determinations of tumor cell phenotypes. It is critical that such analyses characterize the inherent variability and the resulting uncertainty about the predicted clinical status of tumors with out-of-sample predictions to properly assess the potential utility of such information in therapeutic decision making.
Experimental Procedures
Breast Tumor Samples.
Primary breast tumors from the Duke Breast Cancer SPORE frozen tissue bank were selected for this study on the basis of several criteria. Tumors were either positive for both the estrogen and progesterone receptors or negative for both receptors. Each tumor was diagnosed as invasive ductal carcinoma and was between 1.5 and 5 cm in maximal dimension. In each case, a diagnostic axillary lymph node dissection was performed. Each potential tumor was examined by hematoxylin/eosin staining and only those that were >60% tumor (on a per-cell basis), with few infiltrating lymphocytes or necrotic tissue, were carried on for RNA extraction. The final collection of tumors consisted of 13 estrogen receptor (ER)+ lymph node (LN)+ tumors, 12 ER− LN+ tumors, 12 ER+ LN− tumors, and 12 ER− LN− tumors (details can be found in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org).
RNA Preparation.
Approximately 30 mg of frozen breast tumor tissue was added to a chilled BioPulverizer H tube (Bio101) (Q-Biogene, La Jolla, CA). Lysis buffer from the Qiagen (Chatsworth, CA) RNeasy Mini kit was added, and the tissue was homogenized for 20 sec in a MiniBeadbeater (Biospec Products, Bartlesville, OK). Tubes were spun briefly to pellet the garnet mixture and reduce foam. The lysate was transferred to a new 1.5-ml tube by using a syringe and 21-gauge needle, followed by passage through the needle 10 times to shear genomic DNA. Total RNA was extracted by using the Qiagen RNeasy Mini kit. Two extractions were performed for each tumor, and total RNA was pooled at the end of the RNeasy protocol, followed by a precipitation step to reduce volume. Quality of the RNA was checked by visualization of the 28S:18S ribosomal RNA ratio on a 1% agarose gel.
Affymetrix genechip Analysis.
The targets for Affymetrix DNA microarray analysis were prepared according to the manufacturer's instructions. All assays used the human HuGeneFL genechip microarray. Arrays were hybridized with the targets at 45°C for 16 h and then washed and stained by using the genechip Fluidics. DNA chips were scanned with the genechip scanner, and signals obtained by the scanning were processed by genechip Expression Analysis algorithm (version 3.2) (Affymetrix, Santa Clara, CA).
Statistical Methods.
Analysis uses binary regression models combined with singular value decompositions (SVDs) and with stochastic regularization by using Bayesian analysis (M.W., unpublished work), as discussed and referenced in Experimental Procedures, which are published as supporting information on the PNAS web site. The classification probability for each of the two possible outcomes for each sample is structured as a probit regression model in which the expression levels of genes are scored by regression parameters in a regression vector b. Analysis estimates this regression vector and the resulting classification probabilities for both training and validation samples. The estimated regression vector itself is important not only in defining the predictive classification but also in scoring genes as to their contribution to the classification.
Results
Tumor samples are derived from the Duke Breast Cancer SPORE tissue resource of frozen tumors together with all pertinent clinical and pathologic information. The collection of samples includes mostly Stage II cancers and above (see Table 2). All cancer samples have the same histology (invasive ductal carcinoma), and each is between 1.5 and 5 cm in their largest dimension. The tumor samples were chosen to include roughly an equal representation of hormone receptor-positive versus hormone receptor-negative cancers. All tissues were screened for tumor content, and cases that contained less than 60% tumor cells were excluded. We analyzed bulk tumor samples without attempt to separate tumor cells from other contributing cell types. We have made use of Affymetrix Human Gene FL genechip DNA arrays. RNA from each of the samples was converted to target following established procedures (described in Experimental Procedures) and then used to hybridize to the genechip arrays. The hybridized chips were then processed and analyzed as described in Experimental Procedures.
Classification of Tumor Samples on the Basis of ER Status.
The initial 49 tumors were classified as ER+ or ER− via immunohistochemistry (IHC) at time of diagnosis and then later via protein immunoblotting assay for ER to check the IHC results. In five cases, the IHC and blot test conflicted. These five cases and an additional four of the tumors selected randomly were separated from the rest to be treated as validation samples to be predicted on the basis of analysis of the remaining training cases. Of the latter, two were rejected due to failed array hybridization, leaving 18 ER+ and 20 ER−, as determined by both IHC and immunoblotting. The five cases of conflicting biological test results raise concerns about sample heterogeneity and the status of these tumors, hence it makes sense to treat them as of uncertain status and explore the expression-based predictions of status by using the statistical model.
By using the ER outcomes of only the 38 training arrays, we first implemented a simple screen to identify the 100 genes maximally correlated with outcome. This screening strategy aims to reduce noise contributed by irrelevant or unexpressed genes by an initial selection process, and the choice of the number 100 was determined by repeat experimentation. This screen computed sample correlation coefficients between genes and ER+/ER− binary outcomes and selected those genes giving the 100 largest absolute values of this correlation. Alternative methods, such as selecting genes according to maximum differences between mean ranks in the two outcome groups, give similar results. Some form of gene selection for reducing noise is required. Analysis of the full set of genes implies that all aspects of variation are incorporated in the SVD analysis, and the computed singular factors are influenced by the noise affecting each and every gene. Use of the full gene set generally means that the discriminatory ability of resulting factors is clouded by such noise. In the ER analysis using all genes, results are broadly similar to those reported here but for the fact that all predictive probabilities have much higher associated uncertainties, and one or two tumors are much less well classified. More ambiguous classification is a result of the much higher level of noise influencing the analysis. Screening to a smaller, relevant, discriminatory subset of genes is guaranteed to reduce such unwanted noise, with cleaner and more accurate results. We note that, in some applied contexts, the levels of extraneous noise may be lower than in the complex and challenging case of breast cancer; we have experienced this, for example, in our analysis (not reported here) of the Massachusetts Institute of Technology leukemia data set (4), which is in this respect a less challenging problem of predictive discrimination than is breast cancer. Further, current statistical research involves development of refined models that aim to address this question by automatically selecting the most discriminatory genes within the analysis of all genes, rather than via an initial screening process; such a formal modeling approach offers the potential for more incisive noise reduction and hence improved prediction, but, until such approaches are available, some form of prescreen is needed to address noise reduction.
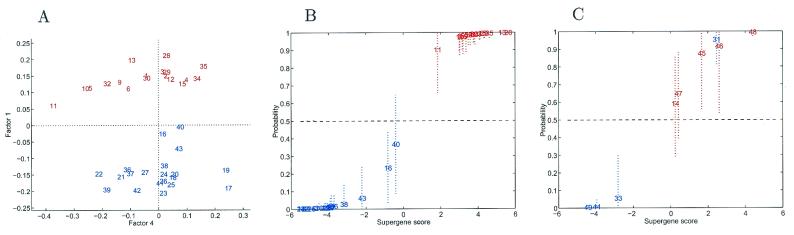
The binary regression model was then fitted to the set of 100 selected genes by using the resulting SVD factors on the basis of these 100 genes. Fig. 1A shows that the first of the resulting “supergene” factors provides a good discrimination between the ER+ and ER− cases. That this discrimination is related to many genes among the 100 is clear from inferences on the gene regression vector b indicating many significant values (not illustrated here). Fig. 1B depicts the estimates of classification probabilities for the training cases together with 90% probability intervals illustrating the degree of uncertainty. This figure must be interpreted carefully; it shows fitted classification probabilities for each of the 38 training cases, thereby illustrating the in-sample discrimination rather than prediction, and provides a useful visual assessment of how clearly the samples are discriminated.
Figure 1.
Factor analysis for ER+/ER− comparison. (A) Pairwise factor analysis. Breast tumors depicted in a scatter plot on two dominant factors underlying 100 genes selected in pure discrimination of the training cases. Each tumor is indicated by a simple index number (see Table 2) and is color coded, with red indicating ER+ cases and blue indicating ER− cases. Only the tumors in the training set are plotted. Factor 1 is clearly discriminatory (Factor 4 is chosen purely for display purposes). (B) Fitted classification probabilities for training cases from the factor regression analysis. The values on the horizontal axis are estimates of the overall factor score in the regression. The corresponding values on the vertical axis are fitted/estimated classification probabilities, with corresponding 90% probability intervals marked as dashed lines to indicate uncertainty about these estimated values. Color coding is as described in A. (C) Predictive probabilities for ER status of each tumor in the validation sample. The analysis was based on the selected subset of 100 genes in the full training sample analysis. Color coding is as described in A.
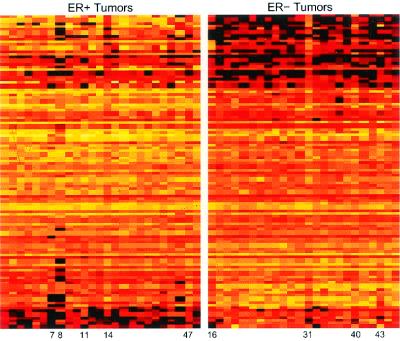
Genes can be ordered by the absolute values of the estimated regression vector b vector to provide an assessment of their relevance in the discrimination. The 100 genes, along with estimated regression parameters, are published as Table 3 in the supporting information on the PNAS web site. Fig. 2 depicts expression levels of the genes, with each row representing an individual gene, ordered from top to bottom according to the absolute values of the estimated regression coefficients. The group of genes includes some that function in the ER pathway, including the ER gene itself as well as a number of known targets for ER (Table 1). Several others contribute to the discrimination inversely with ER+ status (negative coefficients); some of these encode proteins known to have inverse relationships with ER function, such as maspin and glutathione S-transferase-Pi. Also included are genes that are not regulated by ER but that are known to function in concert with ER, such as those encoding HNF3α and androgen receptor; although the model is not designed to discover regulatory mechanisms, these factor models may generate clues about relationships among genes that do indeed relate to underlying functional pathways.
Figure 2.
Expression levels of top 100 genes providing pure discrimination of ER status. Expression levels are depicted by color coding, with black representing the lowest level, followed by red, orange, yellow, and then white as the highest level of expression. Each column in the figure represents all 100 genes from an individual tumor sample, which are grouped according to determined ER status. Each row represents an individual gene, ordered from top to bottom according to regression coefficients (see Table 3).
Table 1.
Genes that contribute to discrimination of ER status
| Rank | Weight | Unigene cluster | Estrogen relation | Ref. |
|---|---|---|---|---|
| 1 | 0.08 | Trefoil factor 1 (pS2) | Estrogen induced | 15, 16 |
| 2 | 0.079 | ER | ER | |
| 3 | 0.067 | Cytochrome P450, subfamily IIB | ||
| 4 | 0.064 | Trefoil factor 3 | Estrogen induced | 17 |
| 5 | 0.061 | (Insulin-like growth factor) | Estrogen induced | 18, 19 |
| 6 | 0.057 | Human clone 23948 mRNA sequence | ||
| 7 | 0.056 | Microtubule-associated protein τ | Estrogen induced | 20 |
| 8 | 0.055 | Hepsin | ||
| 9 | 0.048 | GATA-binding protein 3 | Co-expressed with ER | 21–23 |
| 10 | 0.047 | v-myb avian myeloblastosis viral oncogene homolog | Estrogen induced | 24, 25 |
| 11 | −0.043 | Serine proteinase inhibitor, clade B, member 5 (Maspin) | Induced by tamoxifen; inverse with ER | 26, 27 |
| 12 | 0.041 | N-acetyltransferase 1 | ||
| 13 | −0.041 | S100 calcium-binding protein A9 | ||
| 14 | −0.041 | Retinoic acid receptor responder 1 | ||
| 15 | −0.039 | Small inducible cytokine subfamily D, member 1 | ||
| 16 | 0.039 | Hepatocyte nuclear factor 3 α | Synergistic with ER | 28 |
| 17 | 0.038 | 37-kDa leucine-rich repeat protein | ||
| 18 | 0.038 | (Androgen receptor) | Physical interaction with ER | 29 |
| 19 | −0.038 | Cathepsin C | ||
| 20 | 0.037 | Inositol polyphosphate-4-phosphatase, type II, 105 kD | ||
| 21 | 0.036 | Purinergic receptor P2X, ligand-gated ion channel, 4 | Estrogen biosynthesis | 30, 31 |
| 22 | −0.036 | KIAA0125 gene product | ||
| 23 | 0.036 | (Neuropeptide Y receptor Y1) | ||
| 24 | 0.035 | Meis (mouse) homolog 3 | ||
| 25 | 0.035 | LIV-1 protein | Estrogen induced | 32 |
| 26 | 0.034 | (CCAAT displacement protein) | ||
| 27 | 0.032 | Postmeiotic segregation increased 2-like 3 | ||
| 28 | −0.031 | Secretory leukocyte protease inhibitor | ||
| 29 | 0.029 | Carboxypeptidase B1 | ||
| 30 | 0.027 | KIAA0430 gene product | ||
| 31 | −0.027 | Glutathione S-transferase π | Inverse relation with ER | 33 |
| 32 | 0.025 | GATA-binding protein 3 | Estrogen induced | 21–23 |
| 33 | 0.023 | X-Box-binding protein 1 | ||
| 34 | −0.023 | Lactate dehydrogenase B | ||
| 35 | 0.022 | 5T4 oncofetal trophoblast glycoprotein | ||
| 36 | 0.022 | (Fructose-1,6-biphosphatase) | ||
| 37 | 0.021 | Androgen receptor | Physical interaction with ER | 29 |
| 38 | 0.021 | Cysteine-rich protein 1 | ||
| 39 | 0.021 | Cytochrome C oxidase subunit vic | ||
| 40 | −0.02 | Singed (Drosophila)-like |
Genes are listed according to the discriminatory ranking, with gene 1 having the greatest weight in the discrimination. Negative values indicate an inverse correlation with ER+ status (and thus a positive correlation with ER− status).
Fig. 1C illustrates the formal and “honest” predictions for the nine validation tumors. Some are quite surely predicted as of either ER+ or ER− status, but those in the central region are of uncertain status, and the probability intervals reflect this uncertainty. Tumor samples 45 and 46 were determined as ER− by IHC at time of diagnosis but as ER+ by the later immunoblotting. This change in ER status could reflect an initial borderline reading at the time of diagnosis that was more clearly positive by immunoblot assay, or it could reflect tumor heterogeneity that influenced the assay based on sampling differences. On the basis of the statistical analysis, the expression profiles are strongly consistent with the immunoblotting results. Tumor samples 14, 31, and 33 were initially determined to be ER+ by IHC but ER− by the later immunoblotting. Again, this difference could reflect tumor heterogeneity. In these cases, the statistical analysis indicates an expression profile consistent with the initial determination of a positive ER status for tumor 31 and the subsequent immunoblotting result of negative ER status for tumor 33; for tumor 14, the expression profile yields an uncertain prediction. This analysis highlights the use of model predictions as a third test to compare with the IHC and immunoblotting results. For the cases when the two biological tests conflict, the analysis sometimes agrees with IHC and sometimes with immunoblotting, and in some it indicates a high degree of uncertainty about ER status. This kind of information is then available for review by oncologists; in some cases, ER status is simply difficult to determine, because of either within-tumor heterogeneity or changes over time in protein levels. The model analysis is appropriately reflecting the ambiguities in such borderline cases.
Crossvalidation Analysis of ER Status and Honest Prediction.
A major practical interest and potential clinical value of such statistical analyses lies in the ability to predict the status of new cases on the basis of a gene expression profile, that is, to provide a rational theoretically well-founded estimate of the probability of ER status for any new case, accompanied by a realistic assessment of uncertainty. Such uncertainties may be high due to limited information and population heterogeneity, and it is critical that this uncertainty be reported and communicated to clinical researchers and clinicians along with point estimates of outcome probabilities.
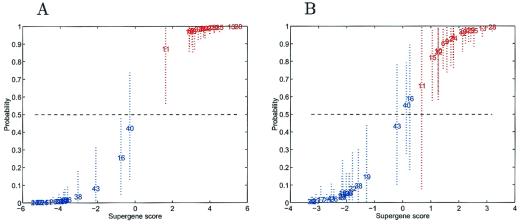
By using the set of 100 genes selected from the full training sample study, the regression model was repeatedly refitted to the training data, each time removing the ER status of one of the tumors and then estimating the classification probability for that tumor. This is a standard “one-at-a-time” crossvalidation analysis; the status of each tumor in the training sample is predicted on the basis of the remaining cases. Fig. 3A displays the predictions in a format similar to Fig. 1B, with similar results. There is more uncertainty about tumors in the mid-range, because these are now predictions rather than fitted values. However, it is important to note that the results in Fig. 3A do not provide a reliable guide to the true predictive value, because they are based on the prescreened subset of 100 genes that utilize the known ER status of all cases and so bias toward a potentially over-optimistic discrimination.
Figure 3.
Out-of-sample crossvalidation predictions of ER status. (A) One-at-a-time crossvalidation predictions of classification probabilities for training cases from the factor regression analysis. The values on the horizontal axis are estimates of the overall factor score in the regression. The corresponding values on the vertical axis are estimated classification probabilities with corresponding 90% probability intervals marked as dashed lines to indicate uncertainty about these estimated values. The analysis and predictions for each tumor are based on the screened subset of 100 most discriminatory genes to parallel current practice in expression studies by other groups. (B) One-at-a-time crossvalidation predictions of classification probabilities for training cases in the ER study, in a format similar to that of A. In this instance, each case is predicted only on the basis of the ER status of the remaining training tumors, with the subset of 100 genes reselected in each case. The figure presents the resulting honest uncertainties about the extent of true predictive accuracy in a practical setting, reflecting inherent variability due to heterogeneity of expression profiles.
For a true predictive assessment, gene screening and selection may be performed separately in each “hold-one-out” analysis, so mirroring the real-life circumstances that will be faced in using such models and methods to predict future outcomes. In each of the 38 analyses, the “hold-one-out” analysis leads to a different subset of 100 screened genes. These subsets are highly overlapping but also show up additional genes case by case, reflecting sample variability and inherent heterogeneity in expression profiles. Fig. 3B illustrates the results; uncertainty intervals tend to be fairly wide for tumors whose predicted probabilities are in the central region, nearer 0.5 than 0 or 1, reflecting the ambiguity discovered in the expression profiles of these cases relative to the 100 genes found to be most discriminatory among the other 37 cases. These “uncertain” cases are of obvious special interest for further study. Case 16 clearly has an expression profile more in accord with those of the ER+ cases than with those sharing its designated ER− status. This case has a low level of expression of the ER gene, consistent with its ER− determination, but with relatively elevated levels of other genes in the top group, such as a marginally elevated level of pS2. Cases 40 and 43 share similar expression characteristics with tumor 16, exhibiting elevated levels of several known estrogen-regulated genes. In some cases, the discrepancy in clinical classification versus molecular classification is evident from the expression data. The ER− cases 16, 40, and 43, which are most borderline, exhibit patterns that lie somewhere between the ER+ and ER−, as does the ER+ case of tumor 11. Tumor 31, whose laboratory ER status determinations were conflicting, strongly exhibits a pattern consistent with an ER+ state.
With these exceptions, the predictive accuracy of the analysis is very high. In particular, 34 of 38 are predicted accurately with a high degree of confidence. Thus, not only do these expression patterns, derived from regression analysis, have the capacity to classify on the basis of ER status, but they also have an ability to honestly predict the ER status of unknown samples, demonstrating the validity of the link between expression and clinical phenotype. Note again the clear differences between this display and that of Fig. 1B and the extent to which the clean classification in Fig. 3A is shown to be less reliable than is suggested when compared with the more relevant and appropriate results in Fig. 3B. In particular, the latter highlights the increased uncertainties about cases 16, 40, and 43 in the middle ground.
Classification of Breast Cancer Based on Lymph Node Status.
The analysis of ER status demonstrates the power to predict the status of samples with associated assessments of predictive uncertainties. A second analysis concerns the clinically important issue of metastatic spread of the tumor. The determination of the extent of lymph node involvement in primary breast cancer is the single most important risk factor in disease outcome (11), and here the analysis compares primary cancers that have not spread beyond the breast to ones that have metastasized to the axillary lymph nodes at the time of diagnosis. The potential power in making this determination from the primary cancer is significant in those instances where a positive lymph node might be missed or where a tumor is poised to metastasize to the lymph node but has not yet done so.
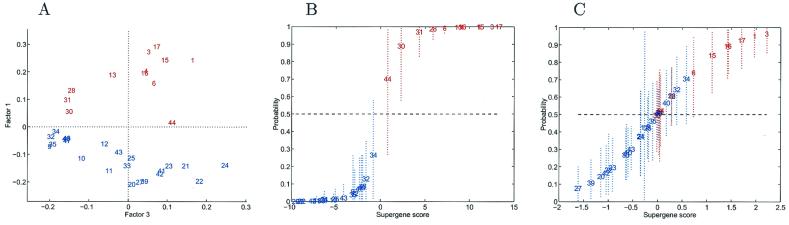
We identified tumors as “reported negative” when no positive lymph nodes were discovered and “reported positive” for tumors with at least three identifiably positive nodes, resulting in 12 reported positives (1) and 22 reported negatives (0). After screening to select the “top 100” most correlated genes, the first factor of the SVD provides discrimination according to nodal status (Fig. 4A). The crossvalidation probabilities from the binary regression model analysis, together with estimated uncertainties, are shown in Fig. 4B. As in the ER study, this first analysis uses the overall screened subset of 100 genes; it is of interest to demonstrate the very clear discriminatory ability of this subset of genes and hence underscore the potential for underlying biological interpretation. This analysis again provides a good classification based on lymph node status, quite comparable to that for the ER discrimination.
Figure 4.
Analysis for nodal comparisons. (A) Pairwise factor analysis. Breast tumors depicted in a scatter plot on two dominant factors underlying 100 genes selected in pure discrimination according to nodal status. Each tumor is indicated by a simple index number (see Table 2) and is color coded, with red indicating node positive cases with at least three identified positive nodes and blue indicating lymph node negative cases. Factor 1 is clearly discriminatory (Factor 3 is chosen purely for display purposes). (B) One-at-a-time crossvalidation predictions of classification probabilities in nodal analysis. The values on the horizontal axis are estimates of the overall factor score in the regression. The corresponding values on the vertical axis are estimated classification probabilities, with corresponding 90% probability intervals marked as dashed lines to indicate uncertainty about these estimated values. The analysis and predictions for each tumor are based on the screened subset of 100 most discriminatory genes. (C) One-at-a-time crossvalidation predictions in the nodal study, in a format similar to that of A. Each case is predicted only on the basis of the nodal status of the remaining training tumors, with the subset of 100 genes reselected in each case. As such, the analysis exhibits the resulting uncertainties about the extent of true predictive accuracy in a practical setting, reflecting inherent variability due to heterogeneity of expression profiles.
Fig. 4C illustrates the practically relevant crossvalidation analysis that adopts a screen to select potentially different genes for each hold-out case. The differences relative to the situation in Fig. 4B are clear—several tumors whose predictions are now moved into the mid-range of ≈0.5 probability and increased uncertainties about predicted probabilities. The screened subsets of 100 most discriminatory genes vary more widely than that seen in the ER analysis as we move across tumors, reflecting higher levels of natural variation in gene expression patterns with respect to nodal status. All of the reportedly positive cases have estimated probabilities appropriately above 0.5, although some are close to that boundary with moderate uncertainty. Perhaps most interesting are the few reportedly negative cases whose predicted probabilities slightly exceed 0.5. Cases like this are of paramount interest, because identifying genomic predictors of the progression from node negative to positive is a major goal from the viewpoint of potential therapeutic implications. These cases could, in principle, represent tumors that have metastasized but were missed in the nodal determination; or, these could be cases that have not yet metastasized but are poised to do so. This analysis of nodal status provides a clear illustration of the importance of honest crossvalidatory studies of predictions in gauging the validity of the classification. Whereas Fig. 4 A and B show clean separation on the basis of nodal status, honest crossvalidation predictions reveal realistic levels of uncertainty, likely due to heterogeneity in the profiles and the clinical phenotypes, and stress the importance of the validation studies to verify the significance of the classification. Nevertheless, it remains true that the analysis does identify gene expression patterns that have predictive capability. Clearly, it is the analysis of those tumors in the uncertain region that must be the focus of further studies.
Discussion
Recent studies of breast cancer (7, 12), leukemia (4), and lymphoma (5) have shown that the analysis of patterns of gene expression has the capacity to classify tumors as well as to define tumor subtypes. The analyses presented here further demonstrate that clinically relevant phenotypes can be determined for primary breast tumor samples through the analysis of gene expression. We take gene expression phenotyping further in developing predictive analyses that brings gene expression analysis to real-world clinical applicability, facilitating the use of complex gene expression patterns as discrete prognostic or predictive factors. Similar studies have used gene expression profiles in out-of-sample crossvalidation studies, and most approaches use some form of initial gene screening to select discriminatory subsets; we have stressed and illustrated the practical importance of repeating such gene screening exercises within each crossvalidation to adequately assess realistic uncertainties about predictions and avoid misleading confidence in predictive accuracy and validity.
Classifications of leukemias and lymphomas that have been achieved in recent analyses of gene expression patterns represent a significant step in the development of methodologies to phenotype tumors (4, 5). The analysis of breast cancer phenotypes likely represents a context of considerably more biological heterogeneity, reflecting subtle aspects of tumor phenotype. As such, that the crossvalidation predictions reveal tumors with an uncertain classification, particularly for the lymph node analysis, is not unexpected. Indeed, it would be surprising to find that such an analysis would yield two cleanly separated groups. In this context, it is critical to develop methods, as we report here, that not only validate classifications with out-of-sample crossvalidation methods, but that also provide appropriate and adequate assessments of the inherent uncertainties found with such predictions. The predictive or prognostic capacity demonstrated here is particularly relevant because clinical decision making depends on a rational, theoretically well-founded model for assessing clinical data from new patients. Because such prognostic and predictive factors are couched in probabilistic language, clinicians can make judgments on the basis of unbiased assessments of the uncertainties in a classification.
The assay of ER status by immunohistochemistry is far from perfect and can produce erroneous results, as highlighted by our study. In addition, such assays would not score alterations that disable the ER pathway, as opposed to the receptor itself. Thus, if the clinically significant determination is the status of the pathway, not just the status of ER itself, then measurements of gene expression profiles that reflect the activity of the pathway could provide an important advance in understanding the behavior of breast cancers. The finding that the group of genes that contribute most weight to the discrimination includes not only ER and ER pathway genes but also genes that encode proteins that synergize with ER, such as HNF3α and androgen receptor, points to the potential power of the analysis in identifying functionally significant relationships.
An additional important benefit of these analyses is the potential for identifying gene pathways underlying an observed phenotype. A key point is the capacity to identify not just highly expressed genes but genes whose expression highly correlates with the phenotype, regardless of level of expression. Perhaps most important is the fact that these analyses identify not only genes expected to be involved in the phenotype (ER-regulated genes), thus validating the process, but also genes for which a connection is not immediately clear. It is the identification of this latter group of genes that represents a major focus of these studies—the use of expression analysis to identify genes that highly correlate with the observed phenotype, thus providing additional insight into the underlying biological pathways.
Finally, we note that the presence of metastatic breast cancer in axillary lymph nodes is the most significant factor in overall survival (11). Although the determination of lymph node status is relatively routine, the surgical procedure is highly invasive, and selectivity in the process of identifying nodes for examination induces biases that suggest some reported negatives may indeed be truly positive (13, 14). Further, the ability to accurately predict axillary lymph node status on the basis of an expression profile of the primary tumor may obviate the need for axillary lymph node dissection and the significant morbidity associated with this procedure. Perhaps of more significance is the patient with truly negative lymph nodes but with a primary tumor that is poised to metastasize. Much more data are needed to determine the precision of the predictive capability for lymph node status, but it is clearly possible that a gene expression profile could predict metastatic potential even in the absence of reportedly positive nodes.
Supplementary Material
Acknowledgments
We are grateful to the editor and two reviewers for their comments on an earlier version of the paper. This work was supported by the Duke SPORE in Breast Cancer (CA 68438), the Early Detection Research Network (CA 84955), and pilot project funds from the Duke Comprehensive Cancer Center. J.R.N. is an Investigator of the Howard Hughes Medical Institute. R.S. and H.Z. were partially supported as postdoctoral fellows of the National Institute of Statistical Sciences.
Abbreviations
- ER
estrogen receptor
- IHC
immunohistochemistry
- SVD
singular value decomposition
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Alon U, Barkai N, Notterman D A, Gish K, Ybarra S, Mack D, Levine A J. Proc Natl Acad Sci USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Nature (London) 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 3.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 4.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 5.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 6.Khan J, Simon R, Bittner M, Chen Y, Leighton S B, Pohida T, Smith P D, Jiang Y, Gooden G C, Trent J M, et al. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 7.Perou C M, Jeffrey S S, van de Rijn M, Rees C A, Eisen M B, Ross D T, Pergamenschikov A, Williams C F, Zhu S X, Lee J C F, et al. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciocca D R, Fujimura F K, Tandon A K, Clark G M, Mark C, Lee Chen G J, Pounds G J, Vendely P, Owens M A, Pandian M R. J Natl Cancer Inst. 1992;84:1279–1282. doi: 10.1093/jnci/84.16.1279. [DOI] [PubMed] [Google Scholar]
- 9.Tandon A K, Clark G M, Chamness G C, Ullrich A, McGuire W L. J Clin Oncol. 1989;7:1120–1128. doi: 10.1200/JCO.1989.7.8.1120. [DOI] [PubMed] [Google Scholar]
- 10.Muss H B, Thor A D, Berry D A, Kute T, Liu E T, Lerner F, Cirrincione C T, Budman D R, Wood W C, Barcos M. N Engl J Med. 1994;331:211. doi: 10.1056/NEJM199405053301802. [DOI] [PubMed] [Google Scholar]
- 11.Shek L L, Godolphin W. Cancer Res. 1988;48:5565–5569. [PubMed] [Google Scholar]
- 12.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi O-P, et al. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 13.Kjaergaard J, Blichert-Toft M, Andersen J A, Rank F, Pedersen B V. Br J Surg. 1985;72:365–367. doi: 10.1002/bjs.1800720511. [DOI] [PubMed] [Google Scholar]
- 14.Hill A D, Tran K N, Akhurst T, Yeung H, Yeh S D, Rosen P P, Borgen P I, Cody H S. Ann Surg. 1999;231:148–149. doi: 10.1097/00000658-199904000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeltsch J M, Roberts M, Schatz C, Garnier J M, Brown A M, Chambon P. Nucleic Acids Res. 1987;15:1401–1414. doi: 10.1093/nar/15.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry M, Nunez A M, Chambon P. Proc Natl Acad Sci USA. 1989;86:1218–1222. doi: 10.1073/pnas.86.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May F E, Westley B R. J Pathol. 1997;182:404–413. doi: 10.1002/(SICI)1096-9896(199708)182:4<404::AID-PATH875>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Richmond R S, Carlson C S, Register T C, Shanker G, Loeser R F. Arthritis Rheum. 2000;43:2081–2090. doi: 10.1002/1529-0131(200009)43:9<2081::AID-ANR20>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 19.Cardona-Gomez G P, Chowen J A, Garcia-Segura L M. J Neurobiol. 2000;43:269–281. doi: 10.1002/(sici)1097-4695(20000605)43:3<269::aid-neu5>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 20.Matsuno A, Takekoshi S, Sanno N, Utsunomiya H, Ohsugi Y, Saito N, Kanemitsu H, Tamura A, Nagashima T, Osamura R Y, et al. J Histochem Cytochem. 1997;45:805–813. doi: 10.1177/002215549704500605. [DOI] [PubMed] [Google Scholar]
- 21.Hoch R V, Thompson D A, Baker R J, Weigel R J. Int J Cancer. 1999;84:122–128. doi: 10.1002/(sici)1097-0215(19990420)84:2<122::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 22.Yang G P, Ross D T, Kuang W W, Brown P O, Weigel R J. Nucleic Acids Res. 1999;27:1517–1523. doi: 10.1093/nar/27.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertucci F, Houlgatte R, Benziane A, Granjeaud S, Adelaide J, Tagett R, Loriod B, Jacquemier J, Viens P, Jordan B, et al. Hum Mol Genet. 2000;9:2981–2991. doi: 10.1093/hmg/9.20.2981. [DOI] [PubMed] [Google Scholar]
- 24.Jeng M H, Shupnik M A, Bender T P, Westin E H, Bandyopadhyay D, Kumar R, Masamura S, Santen R J. Endocrinology. 1998;139:4164–4174. doi: 10.1210/endo.139.10.6229. [DOI] [PubMed] [Google Scholar]
- 25.Gudas J M, Klein R C, Oka M, Cowan K H. Clin Cancer Res. 1995;1:235–243. [PubMed] [Google Scholar]
- 26.Shao Z M, Radziszewski W J, Barsky S H. Cancer Lett. 2000;157:133–144. doi: 10.1016/s0304-3835(00)00466-3. [DOI] [PubMed] [Google Scholar]
- 27.Martin K J, Kritzman B M, Price L M, Koh B, Kwan C P, Zhang X, Mackay A, O'Hare M J, Kaelin C M, Mutter G L, et al. Cancer Res. 2000;60:2232–2238. [PubMed] [Google Scholar]
- 28.Robyr D, Gegonne A, Wolffe A P, Wahli W. J Biol Chem. 2000;275:28291–28300. doi: 10.1074/jbc.M002726200. [DOI] [PubMed] [Google Scholar]
- 29.Panet-Raymond V, Gottlieb B, Beitel L K, Pinsky L, Trifiro M A. Mol Cell Endocrinol. 2000;167:139–150. doi: 10.1016/s0303-7207(00)00279-3. [DOI] [PubMed] [Google Scholar]
- 30.Gillman T A, Pennefather J N. Clin Exp Pharmacol Physiol. 1998;25:592–599. doi: 10.1111/j.1440-1681.1998.tb02257.x. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt M, Loffler G. Eur J Immunol. 1998;252:147–154. doi: 10.1046/j.1432-1327.1998.2520147.x. [DOI] [PubMed] [Google Scholar]
- 32.el-Tanani M K, Green C D. Mol Cell Endocrinol. 1996;121:29–35. doi: 10.1016/0303-7207(96)03844-0. [DOI] [PubMed] [Google Scholar]
- 33.Gilbert L, Elwood L J, Merino M, Masood S, Barnes R, Steinberg S M, Lazarous D F, Pierce L, d'Angelo T, Moscow J A, et al. J Clin Oncol. 1993;11:49–58. doi: 10.1200/JCO.1993.11.1.49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.