Summary
Tandem zinc finger (ZF) proteins are the largest and most rapidly diverging family of DNA-binding transcription regulators in mammals. ZFP568 represses a transcript of placental-specific insulin like growth factor 2 (Igf2-P0) in mice. ZFP568 binds a 24-base pair sequence-specific element upstream of Igf2-P0 via the eleven-ZF array. Both DNA and protein conformations deviate from the conventional one finger-three bases recognition, with individual ZFs contacting 2, 3, or 4 bases and recognizing thymine on the opposite strand. These interactions arise from a shortened minor groove caused by an AT-rich stretch, suggesting adaptability of ZF arrays to sequence variations. Despite conservation in mammals, mutations at Igf2 and ZFP568 reduce their binding affinity in Chimpanzee and humans. Our studies provide important insights into the evolutionary and structural dynamics of ZF-DNA interactions that play a key role in mammalian development and evolution.
eTOC Blurb
Evolutionary and structure-function dynamics of zinc finger-DNA interactions reveal unconventional recognition codes and co-evolution of ZFP568 and its target gene Igf2 in mammals.
Introduction
The Krüppel-associated box (KRAB) domain containing Cys2–His2 (C2H2) zinc finger (ZF) proteins are the largest family of transcription factors/regulators in vertebrates that expanded and diversified during vertebrate evolution (Huntley et al., 2006; Imbeault et al., 2017; Najafabadi et al., 2015). Many of these genes are preferentially expressed in brain and embryonic cells (Gregg et al., 2010; Quenneville et al., 2012), where epigenetic reprogramming takes place. KRAB-ZF proteins act mostly as transcriptional repressors (Meylan et al., 2011) via KRAB associated recruitment of the corepressor protein KAP1 (Friedman et al., 1996; Ozato et al., 2008). The KAP1 associated cofactors include histone deacetylases, histone H3 lysine 9 (H3K9) methyltransferase SETDB1, heterochromatin protein 1, and DNA methyltransferases (Ideraabdullah and Bartolomei, 2011; Iyengar and Farnham, 2011; Peng et al., 2000; Schultz et al., 2002). Examination of the hundreds of human or mouse KRAB-ZF proteins shows they contain tandem ZFs ranging from 3 to ~35 fingers, with an average array size of 11–13 fingers (Liu et al., 2013).
An evolutionary analysis revealed that KRAB-ZF genes first appeared in a common ancestor of ceolecanth, birds, and tetrapods, and continued to expand, diversify and turnover during evolution (Imbeault et al., 2017). ChIP-seq analyses in human embryonic kidney 293T cells have demonstrated that approximately two thirds of human KRAB-ZF proteins bind to transposable elements (Imbeault et al., 2017; Schmitges et al., 2016). These studies suggest that new KRAB-ZF proteins emerge in response to invading transposable elements, followed by eventual deterioration of the KRAB-ZF proteins as target elements decay by genetic drift, except in cases where the KRAB-ZF proteins may have been integrated into an important gene regulatory network. In support of this model, several dozen KRAB-ZF proteins that emerged in eutherian mammals have retained DNA binding fingerprints, at least one of which, Zfp57, has maintained its ancestral function in regulating imprint control regions in diverse mammals including humans (Li et al., 2008; Liu et al., 2012; Mackay et al., 2008; Quenneville et al., 2011). Despite the growing data on their binding sites, only a handful of KRAB-ZF proteins have been studied in detail, and these have been shown to carry out diverse functions including control of expression of transposable elements, maintenance of genomic imprints, and determination of meiotic recombination hotspots (Castro-Diaz et al., 2014; Davies et al., 2016; Garcia-Garcia et al., 2008; Jacobs et al., 2014; Krebs et al., 2012; Mihola et al., 2009; Thomas and Schneider, 2011; Wolf and Goff, 2009).
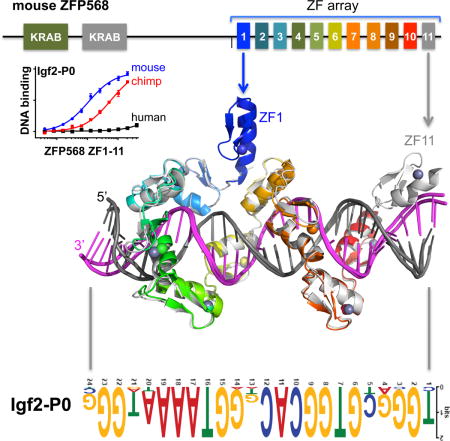
ZFP568 regulates convergent extension and extraembryonic tissue morphogenesis during gastrulation (Garcia-Garcia et al., 2008; Shibata et al., 2011; Shibata and Garcia-Garcia, 2011) and is a direct repressor of a placental specific transcript of insulin-like growth factor 2 (called Igf2-P0) in early mouse development (Yang et al., 2017). ZFP568 comprises three broad regions: two N-terminal KRAB domains, followed by an uncharacterized region, and a C-terminal tandem array of 11 fingers (Figure 1A), and functions via direct binding to a ~24 nucleotide motif found upstream of the Igf2-P0 promoter where it establishes H3K9 methylation-dependent silencing (Yang et al., 2017). Interestingly, ZNF568, the human ortholog of mouse Zfp568, was identified in a screen for rapidly evolving genes in humans relative to chimpanzees and has three allele variants within human population (Chien et al., 2012). All three alleles have accumulated substitutions that potentially disrupt the function of the KRAB and ZF domains. We thus sought to explore whether ZFP568-based silencing of Igf2-P0 was generally a conserved feature in mammals and whether this function might be lost in humans.
Figure 1. Conservation of ZFP568 and its Igf2-P0 binding site in eutheria.
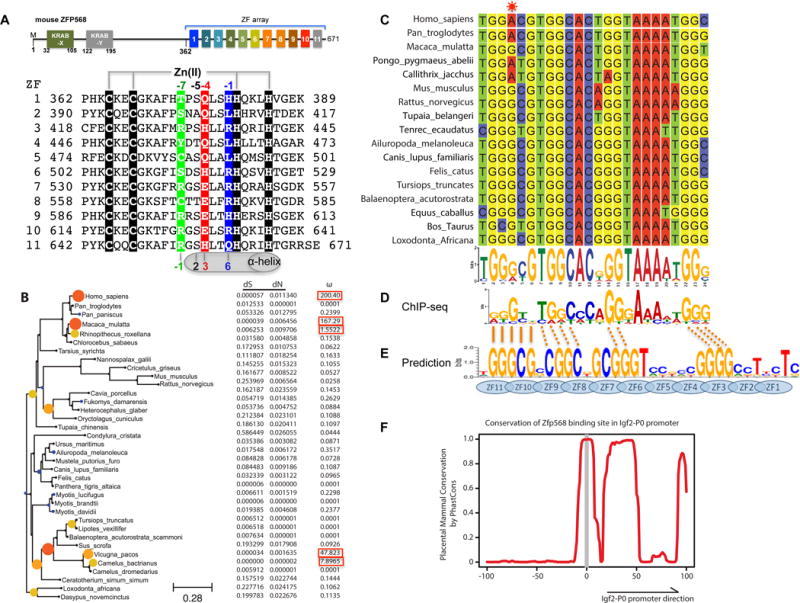
(A) Schematic representation of mouse ZFP568 and sequence alignment of its 11 zinc fingers. The four Zn-coordinating residues of each finger are highlighted with white letters against black. In conventional C2H2 ZF proteins, the amino acids occupying four key ‘canonical’ positions of the helix, namely −1, 2, 3 and 6, specify a DNA target sequence of three or four adjacent DNA base pairs (bottom). This structure-based numbering scheme refers to the position immediately prior to the helix (−1) and positions within the helix (2, 3 and 6). To reduce possible ambiguity, we use the first zinc-coordination His in each finger as reference position 0, with residues prior to this, at sequence positions −1 (blue), −4 (red), −5, and −7 (green), corresponding to the 6, 3, 2 and −1 of the structure-based numbering (compare top and bottom). The new numbering scheme (residues at positions −1, −4, and −7) corresponds to the 5′-middle-3′ of each DNA triplet element.
(B) Maximum likelihood phylogenetic tree and selection pressure for ZFP568 zinc finger domains in mammals. The ω (dN/dS) value for each branch is indicated by the color and size of the round disk on the node, with darker color and larger size for higher ω value. Human ZNF568 H allele was used for this analysis.
(C) Alignment of the Igf2-P0 locus from 17 representative placenta mammals.
(D) mZFP568 consensus binding motif as determined by ChIP-seq.
(E) Prediction made by polynomial SVM (support vector machines)-based algorithm.
(F) Plot of placental mammal conservation in a 100bp window flanking the Igf2-P0 site. See also Figure S1 and Table S1.
In conventional C2H2 ZF proteins, each finger comprises two β strands and a helix and interacts with three adjacent DNA base pairs (Choo and Klug, 1997; Pavletich and Pabo, 1991; Wolfe et al., 2000). When bound to DNA, the helix of each ZF lies in the DNA major groove, and side chains from specific amino acids within the N-terminal portion of each helix and the preceding loop make hydrogen-bond (H-bond) contacts with the bases of primarily one DNA strand. The identities of these amino acids are the principle determinants of the DNA sequence recognized in a well-characterized three-fingered system (Gupta et al., 2014; Persikov and Singh, 2014; Persikov et al., 2015) (Figure 1A). The one finger-three base rule is preserved for five to six finger tandem arrays either in a designed six-fingered protein or in PRDM9 and CTCF (Hashimoto et al., 2017; Patel et al., 2016b; Segal et al., 2006). Here we show that the 11-finger tandem array of mouse ZFP568 (mZFP568) deviates from this rule by interacting with 2, 3 or 4 bases per finger and by interacting with the opposite strand for a stretch of five thymine bases.
Results
ZFP568 orthologs are highly conserved in mammals and retain Igf2-P0 binding function
To identify ZFP568 orthologs we used reciprocal BLAST (Altschul et al., 1990) with the mZFP568 as a query and identified orthologs in all eutherian mammals examined but not platypus or marsupials (Table S1). Most orthologs had the same basic structure of one or two KRAB domains at the N-terminus with an 11-finger array at the C-terminus (Figures 1A and S1A). Sequence alignments showed considerable conservation of base-interacting residues relative to other amino acids within the zinc fingers with the exception of the first two fingers (Figure S1B). To determine whether ZFP568 orthologs were therefore evolving under selective pressure, we calculated the non-synonymous to synonymous substitution rate (dN/dS ratio). We found that ZFP568 orthologs were largely evolving under purifying selection with a dN/dS less than 1 (Figure 1B). One notable exception is three allele variants of human ZNF568, which evolve under positive selection with very high dN/dS (Figure 1B).
In mice, the primary function of mZFP568 is the repression of the placental specific transcript Igf2-P0 via the direct binding to an upstream 24 nucleotide sequence that matches the mZFP568 binding motif inferred from ChIP-seq data (Yang et al., 2017). We searched the genomes of mammals using BLAT (Kent, 2002) and found the binding site to be highly conserved, with 15 of the 24 nucleotides to be invariant amongst the seventeen species examined (Figure 1C). Importantly, these residues match the invariant nucleotides of the ChIP-seq motif (Figure 1D). The consensus sequence is generally rich in guanines, but contains a stretch of 4-5 adenines towards the 3′ end. We note that the consensus sequence does not match the predicted DNA-binding specificity of mZFP568 by a computational algorithm (Persikov and Singh, 2014), particularly for the poly-adenine sequence (Figure 1E). Using PhastCons (Siepel et al., 2005), we could determine that just upstream of the binding site there was very little conservation in mammals, whereas downstream conservation increased, consistent with the downstream region being part of the Igf2-P0 first exon as well as the final exon of the Igf2-antisense transcript (Figure 1F).
Ten out of eleven ZFs of mZFP568 contribute to the binding of the Igf2-P0 sequence
We generated a construct of mZfp568 that includes the entire 11-ZF DNA binding domain and analyzed its binding to three double-stranded oligonucleotides (oligos): the mouse and chimpanzee Igf2-P0 sequences and an arbitrary negative control that partially overlaps the consensus (Figure 2A). Fluorescence polarization was used to measure the dissociation constants (KD) of the three oligos (Patel et al., 2016a). ZF1-11 displayed approximately 4-fold greater binding affinity for the mouse Igf2 sequence than for the chimp sequence (Figure 2B), which deviates from the mouse sequence at only 3 locations within the 24-bp consensus including a Gua-to-Ade substitution at position 4 (Figure 2A). There is no measureable binding to the negative control (which shares 8/24 bp with the mouse Igf2-P0) (Figure 2A). These results confirm the presumed specificity of ZF1-11 of mZFP568 towards its own Igf2-P0 sequence.
Figure 2. mZFP568 binds Igf2-P0 sequence.
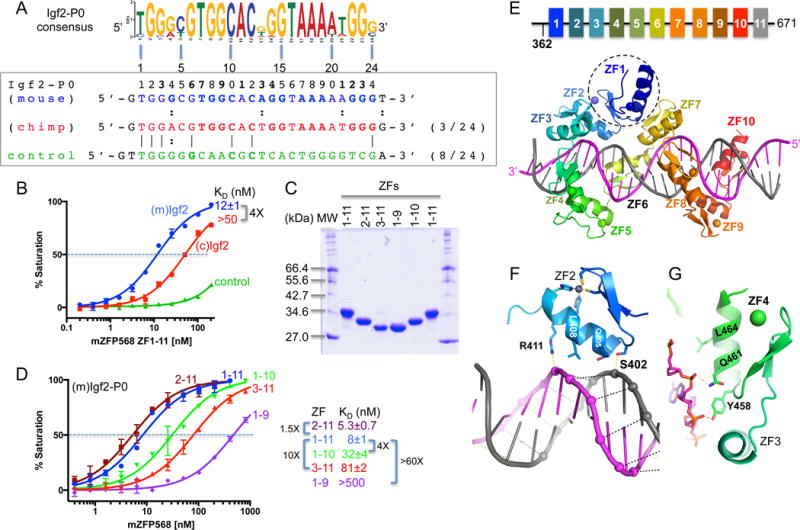
(A) The Igf2-P0 consensus sequence is aligned with three DNA sequences used for binding assays (mouse Igf2-P0, chimp Igf2-P0, and control).
(B) Binding affinities of mZFP568 against the three oligos.
(C) A 12% SDS-PAGE showing purified ZF proteins used in the study.
(D) The binding of five ZF fragments against mIgf2-P0 sequence.
(E) A model of ZF1-10 (colored) binding mIgf2-P0. ZF1 (circled) is involved in intra-molecular interactions between ZF2 and ZF7.
(F–G) ZF2 (F) and ZF4 (G) involvement in DNA phosphate backbone interactions at the AT-rich stretch.
As mentioned, in conventional C2H2 ZF proteins, each finger interacts mainly with three adjacent DNA bases (Choo and Klug, 1997; Hashimoto et al., 2017; Patel et al., 2016b; Pavletich and Pabo, 1991; Wolfe et al., 2000), we estimated that 8 fingers might be sufficient to interact with the 24-bp oligo and thus generated constructs that deleted one or two fingers from either N-or C-terminus (Figure 2C). Deleting one or two C-terminal fingers caused the affinity for Igf2-P0 to decrease by a factor of ~4 with ZF1-10 and more than 60-fold with ZF1-9 (Figure 2D), indicating both C-terminal fingers, particularly ZF10, interact favorably with the Igf2-P0 sequence. Similarly, deleting the N-terminal two fingers (ZF3-11) reduced affinity by a factor of ~10, whereas ZF2-11 had a slightly enhanced affinity (1.5X) than that of ZF1-11 (Figure 2D), indicating that ZF2, but not ZF1, is involved in DNA binding. Therefore, it appears that at least ten of 11 fingers (ZF2-11) are required for high affinity binding with Igf2-P0 sequence.
Structural Investigations
To investigate the molecular mechanism by which the mZFP568 tandem ZF array recognizes its target DNA sequence, we crystallized ZF1-11 bound with a 28-bp oligo containing the Igf2-P0 sequence in crystallographic space group C2221. The structure was solved to a resolution of 2.09 Å (Table S2). Although ZF1-11 was used for crystallization, the last finger, ZF11, could not be observed in the structure. We thus utilized ZF1-10, which crystallized in a different space group (P1) in complex with a 26-bp oligo, in a resolution of 2.06 Å comparable to that of ZF1-11 (Table S2). In addition, we crystallized the same 28-bp oligo in complex with ZF2-11, which included an ordered ZF11 in the co-crystal structure (Figure S2A). Thus, we observed structural information for the entire 11 fingers by superimposing the structures of ZF1-10 and ZF2-11 (Figure S2B). Pairwise comparison revealed that the three complex structures are highly similar, with a root-mean-square deviation (RMSD) of less than 1 Å when comparing nine to ten common ZFs (Figure S2, B and E). The fact that we crystallized the ZF array (with or without ZF1 or ZF11) in two space groups with very different crystal packing lattices (Figure S2, C and D), yet resulted in nearly identical structures (Figure S2E), suggests that crystal packing force does not influence overall protein-DNA conformation. Here, we will describe structural information for fingers 1-10 in the space group C2221 (Figure 2E).
As expected, ZF1 is not involved in DNA binding, but instead is involved in intra-molecular interactions bridging between ZF2 and ZF7 (Figure 2E). Deletion of ZF1, which increased DNA binding slightly (Figure 2D), might “relax” ZF2 and ZF7 to a more intimate DNA interacting conformation. Interestingly, ZF1 of ZFP568 is also the least conserved of the 11 fingers in mammals (Figure S1B). In addition, ZF2 and ZF4 are mainly involved in DNA backbone phosphate interactions (Figures 2, F-G), which leaves eight fingers (ZF3, ZF5-11) for base recognition of the 24-bp consensus motif. Among them, fingers near the ends (ZF3, ZF10 and ZF11) follow the one-finger-three base rule, whereas ZF8 and ZF9 recognize two bases each, ZF6 and ZF7 each span four-bases, and ZF5 together with ZF6 recognize thymines of the opposite strand for a stretch of five A:T bases pairs (Figure 3).
Figure 3. Schematic representation of ZF1-11 interactions with DNA.
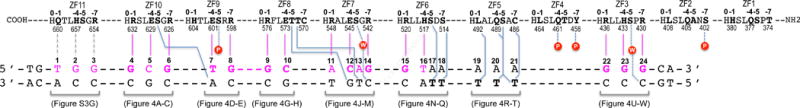
The top four lines indicate amino acids of each finger (ZF1 to ZF11) from N- to C-terminus (right to left). Relative positions within each finger and residue numbers are indicated respectively above and below the sequence. The second set of three lines indicates the sequence of the double-stranded oligo used for crystallization. The base pair matching the consensus sequence are numbered as 1–24.
Base specific interactions
The convention that we used for numbering nucleotides and amino acids is that the 24-bp oligo is numbered 1–24 from 5′ (left) to 3′ (right), with the recognition sequence as the “top” strand (colored magenta in Figure 3), whereas the protein sequence runs in the opposite direction, from carboxyl (COOH) to amino (NH2) termini. While ZF11 from the structure of ZF2-11 interacts with the 5′ sequence (T1G2G3) (Figure S3A), ZF10 from the structure of ZF1-10 interacts with the following 3-base triplet (G4C5G6), followed by other fingers, and ends with ZF3 interaction the 3′ sequence (G22G23G24) (Figure 3). Within each finger, we use the first zinc-coordinating His in each finger as a reference position 0, residues preceding this, at positions −1, −4, and −7, are potential base-interacting residues typically forming H-bonds with the 5′ base, central base, and 3′ base, respectively, of each DNA triplet (Figure 1A).
Conserved Arg-Gua interactions
The most dominant base-specific interaction observed is the Arg-Gua contacts. Eleven G:C base pairs in the Igf2-P0 sequence (11/24) are recognized by nine arginines and two histidine residues, e.g., R654 and H657 of ZF11 (Figure S3A), R632 and R626 of ZF10 (Figures 4, A and C), R598 of ZF9 (Figure 4E), R576 of ZF8 (Figure 4G), R542 of ZF7 (Figure 4M), R520 of ZF6 (Figure 4N); and R436, H433 and R430 of ZF3 (Figures 4, U-W). In accordance with apposition of Arg with Gua as the most common mechanism for Gua recognition (Luscombe et al., 2001; Patel et al., 2016b; Vanamee et al., 2005), the terminal Nη1 and Nη2 groups of arginine donate H-bonds to the O6 and N7 atoms of guanines, respectively (for example, see Figure 4A). Similarly, the Nε2 group of H443 of ZF3 donate one H-bond to guanine N7 and the adjacent ring Cε1 atom donates a C–H…O type of interaction to the O6 atom (Figure 4V) (Horowitz and Trievel, 2012), forming a bidentate H-bond interaction. When R548 of ZF7 encounters an A:T at base pair position 11, the side chain of R548 adopts an alternative conformation and interacts with the DNA phosphate group (Figure 4J). This observation suggests that the hydrogen bonds involving bases are adaptable in the sense that the participating amino acids (Arg in this case) can alter conformation to suit the substrate, a feature that likely contributes to the variability observed at the base pair positions 6 and 11 by the ChIP data (comparing Figure 1D to Figures 4, C and J).
Figure 4. mZFP568 ZF3-10 form base-specific contacts.
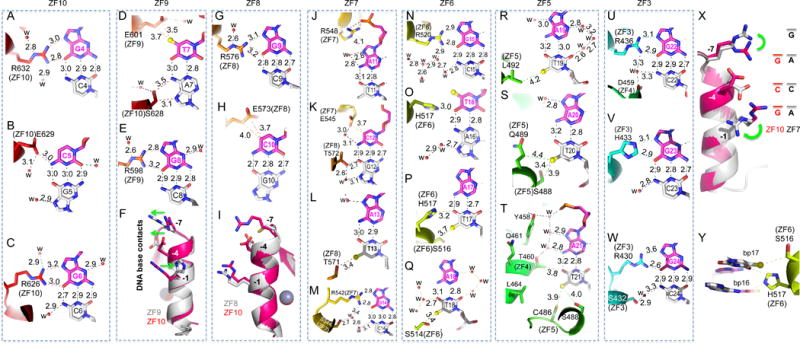
(A–C) ZF10 interacts with a 3-bp triplet (G4-C5-G6). R632 interacts with G4 (A). E629 interacts with C5 (B). R626 interacts with G6 (C).
(D–E) ZF9 interacts with two base pairs (T7-G8). In addition to E601 interaction with T7, S638 at position −5 of ZF10 interacts with A7 of the bottom strand (D). Despite conservation of Ser/Thr at position −5 in each ZF (Figure 1A), the way in which it interacts with DNA differs from finger to finger but most of them interact with the bottom strand.
(F) Superimposition of ZF10 (3-base binder in magenta/red color) and ZF9 (2-base binder in grey color). Note the opposite movement (indicating by green arrows) of base-interacting residues at position −1 and positions −4 and −7 of ZF9.
(G–H) ZF8 interacts with two base pairs (G9-C10). R576 interacts with G9 (G) and E573 interacts with C10 (H).
(I) Superimposition of ZF10 (in magenta/red color) and ZF8 (in grey color) indicates that the small side chain of C570 of ZF8 at position −1 is too far away for direct base contact.
(J–M) ZF7 spans four base pairs (A11-C12-A13-G14). R548 forms a salt bridge with a DNA backbone phosphate group (J). E545, together with T572 of ZF8, interact with the C12:G12 base pair (K). T571 at position −5 of ZF8 interacts with T13 of bottom strand (L). R542 interacts with G14 (M).
(N–Q) ZF6 spans four base pairs (G15-T16-A17-A18). R520 interacts with G15 (N). H517 bridges between T16 and T17 (O and P). S514 interacts with T18 of the bottom strand (Q).
(R–T) ZF5 interaction with three A:T base pairs (A19:T19-A21:T21). L492 interacts with T19 (R). S488 and Q489 interact with T20 (S). Besides C486 and S488 of ZF5, a layer of water molecules separates Y458 and T460 of ZF4 and the A21:T21 base pair (T).
(U–W) ZF3 interacts with three G:C base pairs (G22-G24). R436 interacts with G22 (U). H422 interacts with G23 (V). R430 interacts with G24 (W).
(X) Superimposition of ZF10 (in magenta/red color) and ZF7 (in grey color). Note the different conformations of two arginine residues at positions −1 and −7.
(Y) H517 bridges between two base pairs 15 and 16.
Conserved Glu-pyrimidine interactions
After the Arg-Gua pair, the second most used amino acid-base interaction by mZFP568 is glutamate with pyrimidines, e.g., the E629 of ZF10 interacting with C5 (Figure 4B), the E601 of ZF9 interaction with T7 (Figure 4D), the E573 of ZF8 interacting with C10 (Figure 4H), and the E545 of ZF7 interacting with C12 (Figure 4K). Although all four glutamate residues are located at position −4 of each respective finger, the side chain adopts different conformations to form an H-bond with the cytosine exocyclic amino group N4 (Figure 4B), a van der Waals contact with thymine methyl group (Figure 4D) or van der Waals contacts with the ring carbon C5 atom (Figures 4, H and K). It seems that ZF9 and ZF8, each recognizing two bases pairs, T7G8 and G9C10 respectively, maximize the interactions with DNA by optimizing the Arg-Gua and Glu-pyrimidine contacts. Superimposition of ZF10 (a 3-bp binder) and ZF9 (a 2-bp binder) indicates that H604 at position −1 of ZF9 rotates its side chain away from DNA (Figure 4F), allowing E601 at position −4 and R598 at position −7 to move along the opposite direction and interact with T7 and G8 respectively (Figure 4, D and E). Comparison between ZF10 and ZF8 shows the smaller side chain of C570 at position −7 of ZF8 is positioned too far away from the DNA base to form an interaction (Figure 4I). We note that cysteine (together with proline, phenylanine and tryptophan) is not typically used for base-interacting positions by survey of hundreds of two-finger modules from both natural and artificial sources (Gupta et al., 2014). However, unique to ZF8, T571, adjacent to C570, makes a van der Waals contact to the methyl group of T13 of the opposite strand (Figure 4L). In the ChIP-seq sequence motif and the Igf2-P0 consensus motif, base pair 13 is a variable position (Figure 1D). Substitution of A13-to-G13 in mouse Igf2-P0 shows slightly reduced DNA binding (Figure S4).
When Arg meets an Ade
ZF10 and ZF7 have identical residues (Arg, Glu, and Arg) at base-interacting positions (−1, −4 and −7). ZF10 uses the three residues to recognize a triple of G4C5G6 (Figures 4, A-C). When ZF7 is presented with the A11C12A13 sequence, the two arginine residues respond very differently (Figure 4X). R548 at position −1 of ZF7 rotates the side chain away from DNA base and forms a charge-charge interaction with the backbone phosphate group of A11 (Figure 4J). R542 at position −7 simply rotates its side-chain guanidine group pointing to the next base (Figure 4X), which happens to be a Gua (G14 in Figure 4M), extending interactions to a span of four base pairs (Figure 4X). In the ChIP-seq sequence motif, position 14 is an invariant guanine (Figure 1D). This property of ZFP568 can be traced to the ability of specific residues in each ZF unit to adopt alternative conformations, allowing it to establish versatile H-bonds with some bases but not with others (Hashimoto et al., 2017; Patel et al., 2016b). Examples of the versatile binding by C2H2 ZF include PRDM9, another KRAB-ZF protein, where side chain conformational switch allows identical ZF modules to recognize different sequences (Patel et al., 2017).
When His meets a Thy
Like ZF3 and ZF11, ZF6 has a His at position −4. Unlike the His-Gua interaction seen in ZF3 and ZF11 (Figures 4V and S3A), when H517 of ZF6 meets a T:A base pair at position 16, the imidazole ring of H517 rotates approximately 90° to bridge between T16 and T17 from opposite strands, effectively occupying two base pairs (Figure 4Y), or spanning four base pairs by the single unit of ZF6. Although the histidine side chain is relatively rigid, the ability to rotate along the side chain torsion angles allowed the residue to adopt at least three different conformations in response to sequence variation: the H433 of ZF3 interaction with G23 (Figure 4V), H517 of ZF6 bridging between two A:T base pairs (Figure 4Y), and H604 of ZF9 devoid of specific interaction (Figure 4F).
Methyl specific interaction with A:T rich sequence
Within the placental-specific promoter, Igf2-P0, there is a stretch of five A:T base pairs (positions 17-21), with A17 being an invariant adenine in the ChIP-seq motif (Figure 1D). Unexpectedly, instead of the adenines of the top strand, the five thymines of the bottom strand are contacted by ZF5-6 via van der Waals interactions with the thymine methyl groups (Figure 3). S516 of ZF6 interacts with the methyl group of T17; S514 of ZF6 interacts with T18; L792 of ZF5 interacts with T19; S488 and Q489 (via the Cβ atom) of ZF5 interact with T20, and C486 and S488 of ZF5 interacts with T21 (Figures 4, P-T). In addition, H517 of ZF6 provides the imidazole ring Cε1 atom for a C–H…O type of interaction to the O4 atom of T17 (Figure 4P). Such interactions with a stretch of five bases of the non-recognition strand have not been described in classical ZF-DNA complexes.
Following the A:T rich sequence are three G:C base pairs, which are recognized by arginine residues at positions −1 and −7 and a histidine at position −4 of ZF3, all involving specific bidentate Gua–Arg and Gua–His interactions (Figures 4, U-W). ZF4, which possesses hydrophobic (L464), glutamine (Q461) and aromatic (Y458) residues at the corresponding positions, none of which are known for specific interaction with Gua, is pushed out of register with Q461 and Y458 making two backbone phosphate contacts (Figures 2G and S5). A layer of water molecules mediates the adjoining contacts between ZF4 residues Y458 and T460 and DNA base pair A21:T21 (Figure 4T).
Narrower DNA minor groove at the A:T rich sequence
We compared the DNA base-interacting ZF3-10 of mZFP568-DNA co-crystal structure (Figure 5A) to that of human PRDM9 allele-A, a related KRAB-ZF protein, in complex with its target DNA (Patel et al., 2016b). In the structure of PRDM9A-DNA (PDB 5EGB), the classic 4-finger array follows the right-handed twist of the DNA, with each finger recognizing a 3-bp triplet, thus occupying the major groove for a total 12-bp. Superimposition of the two structures immediately revealed the major difference between the two structures lies in the DNA conformation with a narrower minor groove at the A:T rich sequence (~9Å), in comparison to ~13.5Å observed in other regions of the DNA or in standard B DNA structures (Figures 5, B and C; Table S3) (El Hassan and Calladine, 1998). Minor-groove narrowing is often associated with A-tracts within AT-rich sequences (Rohs et al., 2009). The narrower minor groove is accompanied by poorly aligned N-terminal 4-finger fragment ZF3-6 with PRDM9A (RMSD >5Å over 106 pairs of Cα atoms), whereas the last 4-finger fragment ZF7-10 superimposed well with PRDM9A resulting in a RMSD of ~1.9Å for 108 pairs of Cα atoms (Figures 5, D and E).
Figure 5. DNA and protein conformational changes.
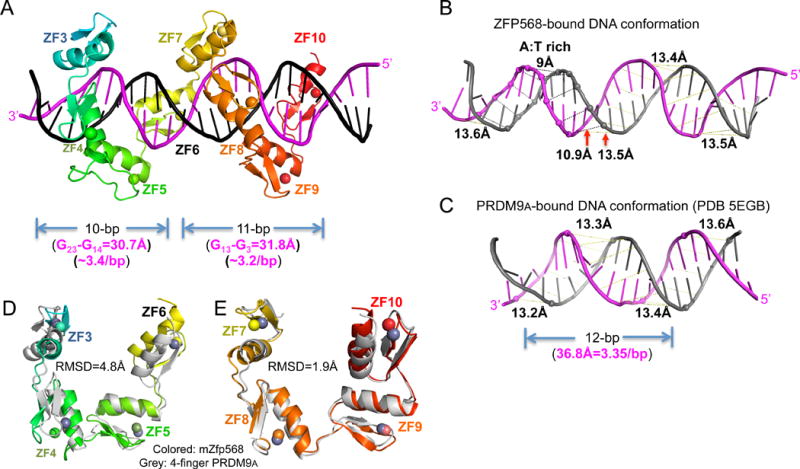
(A) After removing ZF1-2, ZF3-10 could be divided into two 4-finger fragments with each contacting 10 or 11 base pairs.
(B) mZFP568-bound DNA has a narrower minor groove spanning the A:T rich sequence.
(C) For comparison, PRDM9A-bound DNA has a consistent minor groove width (PDB 5EGB).
(D–E) Superimposition of 4-finger array of PRDM9A with that of ZF3-6 (D) and ZF7-10 (E). See also Figure S5 and Table S3.
Chimp ZFP568 ortholog has reduced binding affinity to its own Igf2-P0
Our evolutionary analysis indicated that mZFP568 dependent binding and repression of Igf2–P0 appear to be maintained in most mammals. We cloned mZFP568 orthologs from chimpanzee, pig, elephant, and the human H, C1, and C2 alleles (Figure 6A), and tested the ability of ZFP568 orthologs to repress a reporter luciferase plasmid that contains either the mouse Igf2-P0 binding site or a larger Igf2-P0 promoter upstream of an SV40 promoter. While mZFP568 strongly silenced both reporters, chimp, pig, and elephant ZFP568 could significantly repress them, but the human alleles could not (Figures 6B and S6B).
Figure 6. Chimp ZFP568 has reduced binding affinity to its own Igf2-P0.
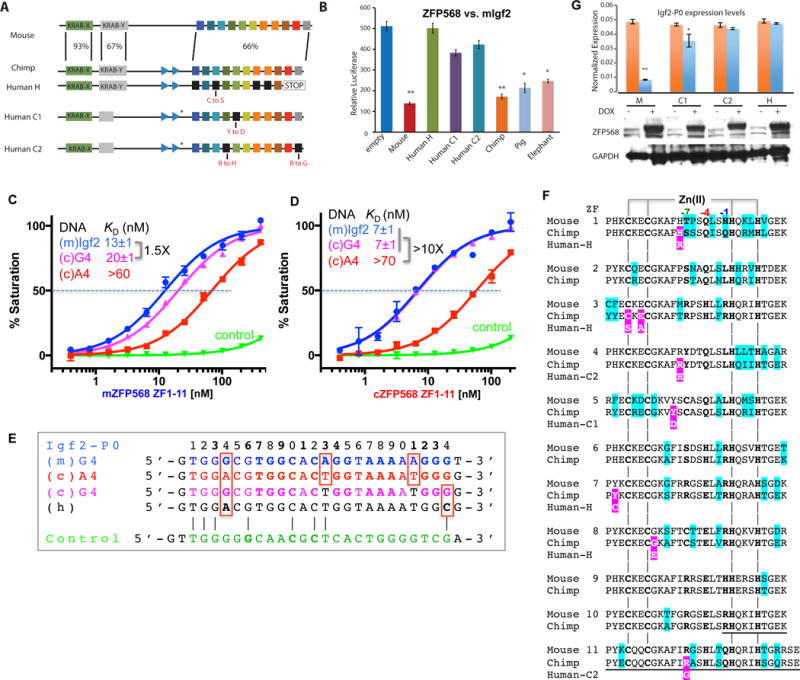
(A) Zfp568 ortholog in chimpanzee compared with mouse with percent conservation within each domain indicated, including the mutation N-terminal to ZF1 in the linker region (marked with a *).
(B) Luciferase assay to test mouse, human alleles H, C1, C2, chimp, pig and elephant ZFP568 repression activity against the mouse Igf2-P0 binding site cloned upstream of an SV40 promoter. Data in panels B and G are shown as mean±SD, t-test, *p<0.05, **p<0.01, n=3.
(C) The A4-to-G4 change at position 4 shows enhanced DNA binding by mZFP568.
(D) The A4-to-G4 change at position 4 recuperates the DNA binding by chimp ZFP568 to the same magnitude as that of mouse Igf2-P0 sequence.
(E) Chimp Igf2-P0 (A4 sequence) deviates from mouse Igf2-P0 at three locations including an Ade instead of Gua at position 4.
(F) Pairwise sequence alignment between mouse and chimp ZFP568 orthologs. The sequence variations are highlighted in cyan. The deviations of human alleles from the chimp sequence are highlighted with white letters against magenta. The underlined sequences in ZF10 and ZF11 are deleted in human allele H.
(G) Zfp568KO/KO ESCs were “rescued” by infection with DOX inducible lentiviral constructs expressing mouse ZFP568 (M) or human ZNF568 proteins encoded by the H, C1, or C2 alleles. ZFP568 Western blots (below) were performed to demonstrate similar expression levels of the rescue constructs. The relative levels of Igf2-P0 were determined by RT-qPCR.
As mentioned, mZFP568 ZF domain displays a 4-5 fold stronger binding affinity for the mouse Igf2-P0 than for the corresponding chimp sequence (Figures 2B and 5C). Unexpectedly, chimp ZFP568 ZF domain binds 10 times stronger for the mouse Igf2-P0 than for its own sequence (Figure 6D). The chimp Igf2-P0 deviates from the mouse sequence at three locations within the 24-bp consensus including G-to-A at position 4, and A-to-T at positions 13 and 21 (Figure 6E). Because positions 13 and 21 are variable in the ChIP data (Figure 1D), we replaced the A:T base pair at position 4 with a G:C base pair in the chimp Igf2-P0, in essence to mimic the mouse sequence. Chimp ZFP568 ZF domain binds the two oligos (mouse Igf2-P0 and A4-to-G4 substituted chimp sequence) indistinguishably (Figures 6, C and D). Pairwise sequence alignment between mouse and chimp ZFP568 proteins indicates that despite the variability between the two orthologs at every finger, there is no difference at the base-interacting residues of ZF2-11, at positions −7, −4, and −1 (Figure 6F). It is the single G:C to A:T change of DNA sequence at position 4 of chimp Igf2-P0 that is the determinant for the reduced binding by chimp ZFP568 to its own Igf2-P0 sequence.
Human ZNF568 alleles fail to bind and repress Igf2-p0
Humans have at least three allelic variants of ZNF568, with a truncation of the second KRAB domain in the C1 and C2 alleles, and a loss of the final two zinc fingers in the H allele. In addition, relative to chimpanzee, human variants have a number of substitutions within zinc fingers specific to each allele or in the linker region (Figure 6, A and F). Human embryonic stem (ES) cells lack H3K9me3 at the IGF2-P0 region, consistent with the lack of a ZNF568-based repression in humans (Figure S6A). Unlike orthologs for mouse and chimp, all three human alleles failed to repress reporters containing either the mouse Igf2-P0 binding site or a larger mouse specific Igf2-P0 promoter (Figures 6B and S6B). Human H and C2 alleles could not restore repression of Igf2-P0 when expressed in Zfp568 mutant mouse ES cells, whereas the C1 allele displayed very weak repression activity (Figure 6G). Likewise the human alleles H and C2 could not bind and the C1 allele bound very weakly to the IGF2-P0 upstream region when expressed in human 293T cells, in contrast to the mZFP568, which bound more strongly (Figure S6C). Consistent with these findings, the human H and C2 orthologs of ZFP568 failed to repress reporters containing the human IGF2-P0 sequence and the C1 allele only repressed weakly, whereas the mouse, chimp, and pig ZFP568 orthologs all repressed the reporter more significantly, which is similar to what observed for the mouse Igf2-P0 sequence (Figures 6B and S6D). The loss of function of allele H in repression of mouse Igf2-P0 is consistent with our in vitro binding data, where deletions of the two C-terminal fingers of mZFP568 caused more than 60-fold reduction in the affinity for Igf2-P0 (Figure 2D). In addition to the loss of two C-terminal fingers, allele H suffers further divergence from the chimp ortholog, including a substitution of a Zn-coordinating cysteine to serine in ZF3 (Figure S3, D-G), which would further destabilize the ZF array.
We generated a series of individual mutants to determine the contribution of individual substitutions to the human C alleles to the loss of Igf2-P0 repressive function using reporter gene studies. Introducing the tyrosine to aspartate (Y-to-D) substitution within ZF5 of the C1 allele to Chimp ZFP568 partially reduced repressive activity, whereas the mutation in the linker region had no effect (Figure 7A, lines 3 and 4). Splicing mutations in the C1 alleles that lead to truncation of the second human KRAB domain further reduced repressive activity (Figure 7A, lines 5 and 6), consistent with the ability of both KRAB domains to bind to KAP1 and repress reporter genes when fused to a heterologous zinc finger protein ZFP809 (Figures S6, E and F). When the Y-to-D substitution within ZF5 in human C1 allele was reverted, the repression activity was significantly restored (Figure 7A, lines 7 and 8).
Figure 7. Human ZNF568 alleles fail to repress Igf2-P0.
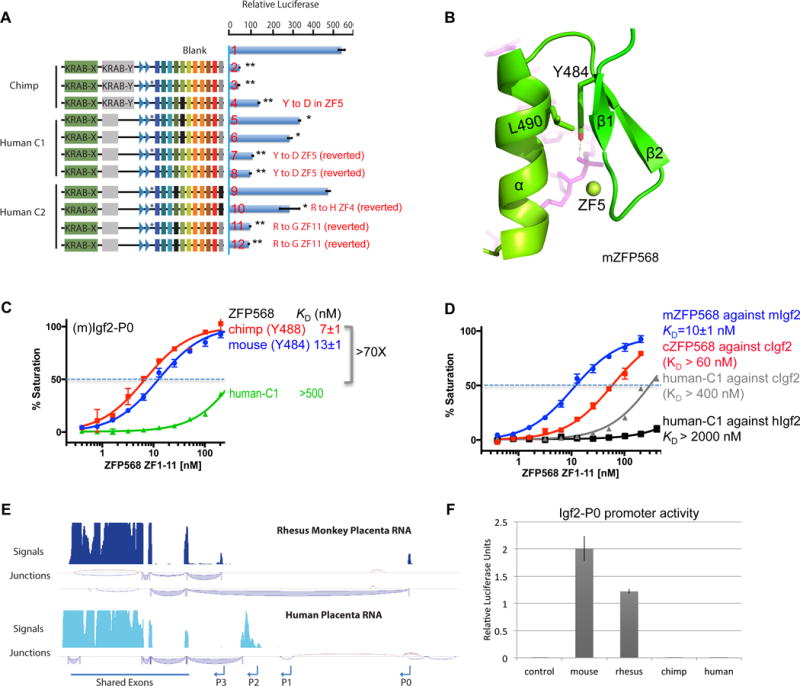
(A) The indicated ZFP568 wild type (lines 2, 5 and 9) and mutant proteins in reporter luciferase assays against the mouse Igf2-P0. The mutation N-terminal to ZF1 in the linker region in human alleles compared with chimp is marked with a *. Data in panels A and F are shown as mean±SD, t-test, *p<0.05, **p<0.01, n=3.
(B) Y484 and L490 of ZF4 in mZFP568 are packed against each other via a hydrophobic force.
(C) The ZF domain of human–C1 shows much reduced DNA binding.
(D) Comparison of binding affinities by mZFP568, cZFP568 and human-C1 against their respective Igf2-P0 sequences.
(E) Rhesus macaque and human placenta RNA-seq tracks at the Igf2 locus. The most abundant junctions are shown below the signal tracks.
(F) Relative luciferase activity of Igf2-P0 reporters cloned from indicated species transfected into 293T cells.
In the structure of mZFP568, the corresponding residue is Y484, which together with L490, forms a local hydrophobic core that is likely important for the stability of the finger (Figure 7B). The corresponding residues of Y484 and L490 in each ZF are highly conserved (F/Y and L/I) (Figure 6F). The extra hydroxyl group in Tyr, versus the typical phenylalanine, forms an additional interaction with the DNA phosphate group (Figure 7B). Substitution of this conserved tyrosine by aspartate, a negatively charged residue, would likely disrupt both the hydrophobic core and the DNA phosphate interaction, which results in at least 70-fold loss of affinity with the mouse Igf2-P0 (Figure 7C).
The C2 allele completely lost the repressor activity in the luciferase assay (Figure 7A, line 9). Reverting the histidine substitution in ZF4 back to arginine showed slight repressor activity (Figure 7A, line 10), yet reverting the glycine substitution in ZF11 back to arginine almost completely restored repressor activity (Figure 7A, lines 11 and 12), suggesting the R-to-G change in ZF11 is the main reason C2 allele lost the ability to repress Igf2-P0. The corresponding residues in the mZFP568 are R457 of ZF4 and R654 of ZF11. In the current structure, R457 of ZF4 interacts with the DNA phosphate group (Figure S3B). The R-to-H substitution probably would maintain the charge-charge interaction. On the other hand, R654 of ZF11 at position −7 is a base-interacting residue, recognizing guanine G3 (Figure S3A). The substitution of the arginine to glycine (the smallest and most flexible amid acid) would lose the base contact as well as introduce flexibility to the helix. These data indicate that a combination of deletions to the second KRAB domain and zinc fingers (ZF10-11), and substitutions of Zn-coordinating and base-interacting residues contributes to the loss of Igf2-P0 silencing activity of human ZNF568 alleles.
The Igf2-P0 transcript and promoter activity are lost in humans
Our finding that humans lack a functional ZFP568 allele with the ability to strongly bind and suppress Igf2-P0 is inconsistent with the notion that mammals require an Igf2-P0 repressor for embryonic survival through gastrulation, which we have observed in the mouse (Yang et al., 2017). In addition, we have shown that chimp ZFP568 ZF domain has diminished binding to its own Igf2-P0 (Figure 7D), mainly due to the G4-to-A4 change of the chimp Igf2-P0 sequence, suggesting chimps also have diminished ability to suppress Igf2-P0. Human Igf2-P0 sequence is identical to that of chimp, except the G24 to C24 change at the last base pair (Figures 1C and 6E). As shown in Figure 7D, the ZF domain of the human C1 allele (mutation Y484D) completely lost binding to human Igf2-P0. Since silencing Igf2-P0 is essential for gastrulation, we asked whether changes to human Igf2-P0 promoter may have rendered a ZFP568-like repressor unnecessary in humans. RNA-seq analysis confirmed that rhesus macaques also express high levels of Igf2-P0 in term placentas, but strikingly, humans do not (Figure 7E). We analyzed gene expression datasets from a number of human fetal and adult tissues and could find no convincing expression of the Igf2-P0 transcript (Figure S7A). Furthermore, the human and chimp IGF2-P0 promoters have significantly reduced transcriptional activity in human 293T cells relative to the mouse and rhesus Igf2-P0 promoters (Figure 7F), despite generally high conservation among human, chimp and rhesus (Figure S7B). Thus changes to Igf2-P0 activity in a common ancestor of chimps and humans could have reduced selective pressure to maintain a functional ZFP568 repressor/Igf2-P0 binding site in the human lineage.
DISCUSSION
DNA recognition by proteins is essential for specific expression or repression of genes in any living organisms. The principle of proteins recognizing DNA sequences by contacts in the major groove has been known for several decades (Seeman et al., 1976). In C2H2-type zinc finger proteins, since the determination of the first structure reported for a three-finger protein in complex with DNA more than 25 years ago (Pavletich and Pabo, 1991), the DNA recognition process is sufficiently understood to define a DNA recognition code (Choo and Klug, 1997; Wolfe et al., 2000), which led to designed zinc finger nucleases for genomic engineering (Chandrasegaran and Carroll, 2016). However, it is evident from our study that the prediction of DNA-binding specificity for ZF arrays containing large number of fingers (>10) is still in its infancy, particularly for AT-rich sequences (Figure 1E).
The presence of A-tract sequences is often associated with minor groove narrowing (Rohs et al., 2009). The readout of this sequence–dependent DNA deformation by mZFP568 involves altered alignment of the ZF array and exploits the strongly enhanced negative electrostatic potential of the DNA due to the proximity of negatively charged phosphate groups in narrower minor grooves. ZF2 spans the narrowed minor groove (Figure 2F), and together with ZF4, provide side chains as well as the amino end of the helix dipole moment for interactions with the DNA backbone (Figures 2, F-G). It is interesting to note that the phosphate-interacting residues of ZF2 and ZF4 are located at the same positions (−7, −4 and −1) of base-interacting residues in other ZFs. The similar prodigy had been observed in E. coli lac repressor DNA binding domain and bacteriophage T4 DNA methyltransferase, where the same set of protein residues can switch from an electrostatic interaction with the DNA backbone in a nonspecific complex to a specific binding mode with DNA base pairs in the cognate complex (Horton et al., 2005; Kalodimos et al., 2004). These findings indicate that the ability of ZFP568 to detect local variations in DNA shape and electrostatic potential.
Our structure also revealed that the fingers near the ends (ZF3, ZF10 and ZF11) follow the one-finger-three base rule, recognizing GC-rich sequence involving highly specific Arg-Gua and His-Gua interactions, whereas the fingers in the middle vary from 2-base to 4-base recognition patterns. It is possible that the specific contacts with the ZF array are formed sequentially, or even uni-directionally along the DNA, for example, initiate from the ZF3 binding to the 3′ end of DNA before spreading. We did observe that at least ten of 11 fingers (ZF2-11) are required for high affinity binding with Igf2-P0 sequence and the extreme C-terminal end ZF11 is more flexible than the rest (Figure S2A). A similar observation was made for Egr1/Zif268 that either the first or the last ZF dissociated from DNA during the search for specific sequence (Zandarashvili et al., 2015) and for PRDM9 allele A, where the C-terminal ZF12, essential for DNA binding, was not visible in the structure (Patel et al., 2016b). Alternatively, every finger could participate in concert for the formation of cognate complex. A solution study using a single molecule approach might afford snapshots to discern a possible mechanism (directional vs. random).
Our study provides insights into the co-evolution of ZFP568 and its target gene, Igf2, in mammals. We demonstrated that ZFP568 is an essential gene product that functions primarily by suppressing Igf2-P0 during gastrulation (Yang et al., 2017). In mice, the Igf2-P0 accounts for ~10% of placental Igf2 expression, and although it is not required for survival it plays an important role in maintaining maternal/fetal growth balance (Constancia et al., 2000; Constancia et al., 2002; Moore et al., 1997). Our finding that ZFP568 appeared in a common ancestor of eutherian mammals, ZFP568 orthologs are evolving under purifying selection and the Igf2-P0 binding site is highly conserved suggests their evolution was a key adaptation regulating viviparity. However ZFP568 based suppression of Igf2-P0 has not been strictly maintained as both chimpanzees and humans have accumulated substitutions and other mutations to both the Igf2-P0 binding site and ZFP568 itself, which reduced or completely prevented their association. Perhaps the expression/repression of placental-specific transcript of growth factors is important for polyzygotic species (mice and pigs), but less so for monozygotic species (chimp, human). Importantly, our structural studies rationalize how each “mutation” disrupts interactions. The fact that humans and chimpanzees survive without using ZFP568 to suppress Igf2-P0 at gastrulation could be explained by our finding that the larger Igf2-P0 promoter activity has been lost in chimps and humans. In humans, ZNF568 has alternate transcript isoforms that include a splice variant that uses the same upstream exons spliced to an entirely separate zinc finger array that is most similar to the mouse Zfp74. In mice, Zfp74 is directly adjacent to Zfp568. The DNA binding properties and function of this protein in mice and humans is unknown. Human ZNF568 might follow the path of many KRAB-ZF proteins that eventually decay by genetic drift once they are no longer required, the finding that the human alleles of ZNF568 are very rapidly evolving and are associated with human brain size at birth suggest that these proteins may be neofunctionalizing in humans (Chien et al., 2012).
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Xiaodong Cheng (xcheng5@mdanderson.org).
METHOD DETAILS
Evolutionary analysis
ZFP568 orthologs (Table S1) were obtained using BLAST. Orthologs were confirmed using reciprocal BLAST and synteny analysis where possible. Specifically we used BLAST to determine if the adjacent genes of mouse ZFP568 (Zfp27 and Zfp47) were syntenic in other species, and confirmed perfect synteny for all 11 species with good genome assemblies (Homo_sapiens, Pan_troglodytes, Chlorocebus_sabaeus, Canis_lupus_familiaris, Rattus_norvegicus, Mus_musculus, Oryctolagus_cuniculus, Felis_catus, Mustela_putorius_furo, Sus_scrofa and Macaca_mulatta). The protein sequences were scanned using ScanProsite (de Castro et al., 2006) to detect KRAB and C2H2 zinc finger motifs. To improve the sensitivity, both pattern and profile based scanning were performed, and then the results were merged together. When overlapping motifs were predicted by both approaches, only the one from pattern scanning were retained for visualization.
Global alignment of protein sequences were generated with Clustal W 2.1 (Larkin et al., 2007) with default settings. The conservation score for each position was calculated as the Jensen-Shannon divergence (Capra and Singh, 2007). Based on the protein sequence alignment, the maximum likelihood phylogenetic tree was constructed using MEGA7 (Kumar et al., 2016) with the Jones-Taylor-Thornton (JTT) model. Bootstrap values from 1,000 replicates were used to assess the robust of the tree topology. The tree was rooted with Loxodonta Africana and Dasypus Novemcinctus, which are two basal species for mammals.
We mapped the corresponding nucleotide sequences to the protein sequence alignment site by site to generate the codon alignment. Then, the codon alignment of zinc finger regions was extracted with custom PERL script according to the ScanProsite result. Based on the codon alignment for zinc finger regions, the dN/dS value for each branch was estimated with the free-ratio model (model = 1, NS Sites = 0) using codeml as implemented in PAML 4.8 (Xu and Yang, 2013). The phylogenetic tree along with the estimated dN/dS ratio was visualized using ETE 3 (Huerta-Cepas et al., 2016). The Igf2-P0 binding site from mouse was used to search the genomes of mammals using BLAT for alignment. Conservation scores flanking the Igf2-P0 binding site were determined using PhastCons (Siepel et al., 2005). Full Igf2-P0 sequences (that begin just upstream of ZFP568 binding sites and end near the transcription start site of Igf2-P0 transcript in mouse) for human chimp, mouse, and rhesus are shown in Table S4.
Cell culture and lentiviral production/infection
293T cells were cultured in DMEM containing 10% FBS. ZFP568KO/KO ESCs were generated as described (Yang et al., 2017) and cultured in ESM medium containing DMEM, 15% FBS, 10mM HEPES, NEAA, 100 μM β-mercaptoethanol, 2 mM L-glutamine, 10ng/mL LIF as described. GFP-tagged ZFP568 from mouse or human ZNF568 H, C1, and C2 alleles were sub-cloned into the pCW57.1 vector (Addgene #41393). pCW57.1 containing GFP-ZFP568 were then cotransfected into 293T cells with packaging plasmids psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) to produce lentiviral particles, which were collected from supernatant after 48 hours. Lentiviral particles were concentrated using Lenti-X (Clontech) according to manufacturers instructions. Lentiviruses were then added to ZFP568 KO/KO ESCs, which were treated with 1ug/ml puromycin after 48 hours. Resistant cells were expanded and treated with doxycyxline (1ug/ml) for 48 hours to induce protein expression before RNA analysis.
RNA extraction, qRT-PCR
RNA was prepared from cells using the RNeasy Plus Mini or Micro kit (Qiagen) with on-column DNase digestion. For qRT–PCR analysis, cDNA was generated using SuperScript III (Invitrogen) and random hexamer priming. qPCR was performed using SYBRGreen master mix (Applied Biosystems). All reactions were performed in triplicate. Standard curves were generated for each primer pair, and expression levels were performed relative to Gapdh. The Gapdh primers are TGT TCC TAC CCC CAA TGT GT (forward) and GGT CCT CAG TGT AGC CCA AG (reverse). The IGF2-P0 primers are TTT ATC CAC CGT CCG GGA AC (forward) and GCA GTC GTC GTA GTC GTT CT (reverse).
ChIP-qPCR
ChIP was performed from 293T cells infected with Lentiviral particles expressing GFP-ZFP568 plasmids as previously described (O’Geen et al., 2010). Crosslinked chromatin from ~50 million nuclei equivalents was sheared to 200-500 base pairs using a Bioruptor Sonicator (Diagenode), and immunoprecipitated with GFP antibodies (Thermofisher A-11121). Purified DNA was subject to qPCR using SYBR green and primers at the putative Igf2–P0 binding site (forward TGG CAG TTA CTT GAC ACC CTG, reverse CTG TAC CGG GAG CAC CTA AA) or at a negative control region at the Znf180 gene (forward TGA TGC ACA ATA AGT CGA GCA, reverse TGC AGT CAA TGT GGG AAG TC).
CoIP assays
HA-tagged KAP1 (Iyengar et al., 2011) was co-expressed with 3X Flag tagged ZFP568 expression constructs in HEK293T cells using Lipofectamine 2000 (Invitrogen). HEK293T cells were lysed in 1% NP40 lysis/IP buffer containing 150 mM NaCl, 10 mM Tris-HCl pH7.2, and protease inhibitors. Proteins were immunoprecipitated using anti-Flag M2 agarose beads (Sigma), washed extensively with IP buffer, and subject to western blot using anti Flag M2 or anti HA antibodies.
Luciferase assays
Reporter Luciferase constructs containing the full Igf2-P0 promoter from mouse and Igf2-P0 ZFP568 binding site upstream of an SV40 promoter were described previously (Yang et al., 2017). Luciferase reporter containing the PBSpro site (which is bound by ZFP809) was described previously (Wolf et al., 2015). Igf2-P0 promoter regions corresponding to chimp, rhesus macaque and human reference genomic DNA (~400 bp, shown in Table S4) were synthesized by Integrated DNA Technologies and cloned into the pGL3-basic vector by in-fusion cloning (Clontech). These reporter plasmids were cotransfected with different ZFP568 constructs (cloned into the pcDNA3 vector, Invitrogen) with either GFP or 3X Flag tags into 293T cells using Lipofectamine 2000 (Invitrogen) together with the Renilla luciferase-expressing pRL vector for internal normalization. Western blots were performed to ensure equivalent expression. Luciferase activity was measured two days after transfection using the dual-luciferase reporter assay system (Promega) and plotted as relative luciferase units.
Recombinant protein expression and purification
The genes encompassing the C-terminal array of ZF1-11 of ZFP568 from mouse (NP_001028527.2) and chimp orthologs (XP_009433702.1) were synthesized and sub-cloned into pGEX-6p1. Constructs for mouse ZFP568 ZF2-11, 3-11, 1-10 and 1-9 were generated by PCR from mouse ZFP568 ZF1-11 plasmid DNA and sub-cloned into pGEX-6p1 vector. The protein was expressed as Glutathione S-transferase (GST) tagged fusion proteins in Escherichia coli BL21 (DE3) Codon-plus RIL cells.
Cells were grown in LB media at 37°C until the OD600 reached to 0.5 when the temperature was lowered to 16°C. The culture was supplemented with 100 μM ZnCl2, and induced by 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight. Cells were harvested by centrifugation and re-suspended into lysis buffer containing 20 mM Tris (pH 7.5), 700 mM NaCl, 5% glycerol, 25 μM ZnCl2, 0.5 mM tris(2-carboxyethyl) phosphine (TCEP) and 0.1 mM phenylmethylsulphoyl fluoride (PMSF). Cells were lysed by sonication for 8 minutes total with 1 sec on and 2 sec off cycle. Lysate was treated with neutralized polyethylenimine (Sigma) to final concentration of 0.1% and clarified by centrifugation. Clear lysate was loaded onto Glutathione Sepharose 4B column (GE healthcare) and GST-tagged protein eluted from the column with buffer containing 100 mM Tris (pH 8.0), 500 mM NaCl, 5% glycerol, 25 μM ZnCl2, 0.5 mM TCEP and 20 mM reduced glutathione. GST tag was removed by treating the eluted protein with ~100 μg of Precission protease (purified in-house) at 4°C overnight. Cleaved protein was diluted to ~350 mM NaCl concentration and further purified to homogeneity by ion-exchange chromatography on tandem Hitrap Q-SP columns (GE Healthcare). Most of the protein flowed through the Q column onto the SP column from which it was eluted as a single peak at ~0.8 M NaCl using a linear gradient of NaCl from 0.35 M to 1 M.
Mutant Y488D of chimp ZFP568 ZF1-11 was generated using site directed mutagenesis protocol, confirmed by sequencing and purified using the same protocol.
Fluorescence-based DNA binding assay
DNA binding was performed using fluorescence polarization assay (Patel et al., 2016a). Various amounts of ZF protein was incubated with 5 nM of 6-carboxyfluorescein (FAM)-labeled double-stranded DNA probe in buffer containing 20 mM Tris (pH 7.5), 260 mM NaCl, 5% glycerol, 25 μM ZnCl2 and 0.5 mM TCEP, with a final volume of 50 μL at room temperature for 30 min. The fluorescence polarization was measured using a Synergy 4 Microplate Reader (BioTek). Curves were fit individually using the equation [mP] = [maximum mP] × [C]/(KD + [C]) + [baseline mP], where mP is millipolarization and [C] is protein concentration. Fitting the experimental data to the equation in Graph-pad prim software (version 6.0) derived KD values (from two experimental replicates). Final data was plotted as % saturation vs protein concentration. For those binding curves that did not reach saturation, under the conditions used, the lower limit of the binding affinity was estimated.
Crystallography
Purified mouse ZFP568 ZF1-11, 1-10, 2-11 were incubated with the double-stranded DNA at equimolar ratio to final concentration of 25 μM on ice in 20 mM Tris (pH 7.5), 500 mM NaCl, 5% glycerol, 25 μM ZnCl2 and 0.5 mM TCEP. The protein-DNA complex was formed by dialysis against the same buffer components with 250 mM NaCl but without ZnCl2. The complex was further concentrated up to ~0.6 mM prior to crystallization. An aliquot of protein-DNA complex (0.2 μl) was mixed with equal volume of mother liquor by a PHOENIX liquid handler robot (Art Robbins Instruments). Good diffracting crystals grew overnight at 16°C with different DNAs under different conditions. The best diffracting crystals obtained by the hanging drop vapor diffusion method were under the conditions:
| Crystallization conditions for mZFP568-DNA complexes | ||
|---|---|---|
| Oligos used for crystallization | mZFP568 | Crystallization conditions |
| 5′TGTGGGCGTGGCACAGGTAAAAAGGGCA3′ 3′ACACCCGCACCGTGTCCATTTTTCCCGT5′ |
ZF1-11 | 10% Isopropanol 0.1M Bicine (pH 8.5) 30% polyethylene glycol 1500 |
| 5′TGTGGGCGTGGCACAGGTAAAAAGGGCA3′ 3′ACACCCGCACCGTGTCCATTTTTCCCGT5′ |
ZF2-11 | 0.2 M calcium acetate 16% polyethylene glycol 3350 |
| 5′GTGGGCGTGGCACAGGTAAAAAGGGC3′ 3′CACCCGCACCGTGTCCATTTTTCCCG5′ |
ZF1-10 | 8% Tacsimate (pH 5.0) 20% polyethylene glycol 3350 |
The crystals were flash frozen with liquid nitrogen using 20% ethylene glycol as cryoprotactant. X-ray diffraction data were collected at SER-CAT 22-ID beamline of the Advanced Photon Source (Argonne National Laboratory, Argonne, IL) and were processed by HKL2000 (Otwinowski et al., 2003). The Autosolve module of PHENIX (Adams et al., 2010) was used for initial crystallographic phasing by single-wavelength anomalous dispersion of Zn signals. The initial electron density revealed clear visible DNA molecules, and a DNA model made by the “make-na server” (http://structure.usc.edu/make-na/index.html) was placed into the density and refined using the PHENIX REFINE module. This partial model was then used during searches for ZFs, which was placed into density by molecular replacement using the PHASER-MR algorithm. The structure was further refined using PHENIX and the model manually adjusted by COOT (Emsley and Cowtan, 2004). Structure quality was analyzed and validated by the PDB validation server. Molecular graphics were generated using PyMol (DeLano Scientific, LLC).
DATA AND SOFTWARE AVAILABLBILITY
The X-ray structures (coordinates and structure factor files) of mZFP568 ZFs with bound DNA have been submitted to PDB under accession number 5V3M (ZF1-11 in complex with 28-bp DNA), 5V3J (ZF1-10 in complex with 26-bp DNA), and 5WJQ (ZF2-11 in complex with 28-bp DNA).
Supplementary Material
Figure S1. Sequence alignment, Related to Figure 1.
(A) Global alignment of ZFP568 ortholog protein sequences with domain structures. The conservation score for each position (bottom) was calculated as the Jensen-Shannon divergence (Capra and Singh, 2007).
(B) Zinc fingerprint alignment for each zinc finger at potential base-interacting positions −1, −4, −5, −7 (see Figure 1A). Consensus sequences (bottom) were generated by MEME (Bailey et al., 2009).
Figure S2. Structures of ZF1-10 and ZF2-11, Related to Figure 2.
(A) Structure of ZF2-11 in complex with 28-bp oligonucleotide, displayed by crystallographic heatmap, from low to high thermal B-factors (blue, cyan, green, yellow, and red). Note that ZF11 has higher temperature-dependent atomic vibrations or static disorder in the crystal lattice.
(B) A complete model of mZFP568 ZF1-11 binding Igf2-P0 sequence, generated by superimposing the common nine fingers (ZF2-10) from structures of ZF1-10 and ZF2-11, resulting in a root-mean-square deviation (RMSD) of 0.7 Å overlapping 222 pairs of Cα atoms.
(C–D) Crystal packing contacts of two ZF protein-DNA complexes in the space groups C2221 (A) and P1 (B). The C2221 space group contains one protein-DNA complex per crystallographic asymmetric unit, whereas the P1 space group contains two such complexes. Note the two symmetry-related complexes in P1 have difference orientations (rainbow color scheme from ZF1-to-ZF10 with blue-to-red) from that of C2221.
(E) Superimposition of two structures of ZF1-10 in complexes in C2221 (colored) and P1 (grey) with 28-bp and 26-bp oligonucleotides, respectively, with an RMSD of less than 1 Å when comparing 231 pairs of Cα atoms.
Figure S3. Locations of human ZNF678 point mutations in the corresponding mZFP568 structure, Related to Figures 3, 4, 6 and 7.
(A) R654 and H547 of ZF11 from the structure of ZF2-11 recognizes G3 and G2, respectively. The side chain of Q660 forms H-bonds with T1.
(B) R457 of ZF4 interacts with a DNA backbone phosphate group.
(C) Mutations in human alleles (H, C1 and C2) in the corresponding mZFP568 are highlighted.
(D) H373 of ZF1 points to solvent. Like R457 of ZF4 (panel B), the corresponding position of H373 in each ZF is quite a variable and accommodates a variety of amino acids (see Figure 6F).
(E) C421 of ZF3 is one of the zinc ligands, and the C421S mutant would disturb the structural rigidity to the finger. The corresponding position of E423 of ZF3 is a conserved acidic or polar residue (E/D/Q) in each ZF (Figure 6F), which may provide additional electrostatic interaction with positively charged Zn(II) ion. The substitution of glutamate-to-alanine (E423A) would weaken its interaction the metal ion.
(F) Y531 of ZF, located in the beginning of strand β1, is involved in intra-molecule interactions with ZF2, forming a H-bond network with the counterpart tyrosine Y391 of ZF2, the main-chain carbonyl oxygen atoms of P401 of ZF2 and R541 of ZF7 (via a water molecule).
(G) G565 of ZF8 is located in the beginning of strand β2, providing flexibility for the loop between two strands. Although conserved in almost all ZFs, ZF5 of mZFP568 does contain a negatively charged aspartate in the same position (see Figure 6E).
Figure S4. Substitution of A13-to-G13 in mouse Igf2-P0, related to Figure 4.
It is interesting to note that ZF4 (and ZF5) possesses at least two amino acids, Gln at position −4 and Tyr at position −7 (Figure 1A), known to interact with adenine and thymine respectively. Juxtaposition of Gln (or Asn) with Ade is a common mechanism for recognition of this base (Luscombe et al., 2001), as seen in a few classic examples, including Gln44 of lambda repressor (PDB 3BDN), Gln28 of repressor of phage 434 (PDB 2OR1), Gln16 and Gln44 of a designed zinc finger protein (PDB 1MEY), and more recently Gln418 of human CTCF (Hashimoto et al., 2017). In addition, the aromatic ring of a tyrosine contacting a thymine methyl group has been used by the zinc finger protein ZNF217 (PDB 4F2J), although interactions of this type have not been often described in classical ZF-DNA complexes. We reasoned that ZF4 and ZF5 could potentially interact with A:T rich sequence in a classical fashion. The “irregularity” observed from the current structure could be the result of the expanded 4-bp recognition by ZF7, particularly R542 skipping an A:T at bp position 13 to interact with an invariant G:C at position 14 (Figure 4X). Because bp position 13 is a variable in the ChIP data and in the Igf2-P0 consensus sequence (Figure 2A), we substituted the G:C base pair for A:T and ask whether we can convert this “irregular” ZF7 (a 4-bp reader) to a canonical 3-bp reader. ZF7 contains exactly the same recognition residues, R-E-R, as ZF10, which recognizes the canonical 3 bases (shown in Figure 4X) and we anticipated that R542-Gua interaction would restore the 3-bp recognition of ZF7 and subsequently that of ZF6 to ZF4, which could potentially interact with the A:T rich sequence in a classic fashion resulting in a substantial increase in binding affinity (MODEL 1). However, the opposite was observed, in that only a slight reduction in affinity (1.5x) was observed for the mutant oligo. This suggested that the 4 base spacing for ZF7 was probably maintained in the binding of the mutant oligo, without gross rearrangement of the rest of the ZFs, even if the local rearrangement does occur at base pair position 13 (MODEL 2).
Figure S5. ZF4 is out of register, Related to Figure 5.
(A–B) We generated a classic three-finger fragment of PRDM9A (by deleting one of the terminal finger) and scanned it through ZF3-10 of mZFP568. The three-finger array aligns well with ZF7-9 or ZF8-10 (B), but it attempts to skip ZF4 and makes an arrangement with the three fingers of ZF3, 5, and 6 (A), suggesting that it is ZF4 that is out of register.
(C–D) ZF3 follows the conventional rule of one-finger-three bases, so we superimposed the single ZF3 to that of PRDM9A (RMSD=0.3Å), and asked what is the conformational difference for the following fingers. ZF4 rotates its helix approximately 20° perpendicular to the DNA axis with the amino end fixed and the carboxyl end of the helix moving approximately 5Å (C). This movement is sufficient to displace the base-interacting residues of ZF4 out of touch to the base (D) and allows a layer of water molecules to diffuse into the space (Figure 4T).
(E–F) The conformational change is extended to ZF5, which moves towards ZF4 by ~6Å (approximately one helical turn) and upwards ~3Å (F). This movement in two dimensions allows ZF5 to interact with the opposite strand of three A:T base pairs immediately after three G:C base pairs by ZF3 (Figure 3).
Figure S6. Systematic analysis of ZFP568 repression activity in human and other mammals, Related to Figure 6.
(A) ChIP-seq tracks of indicated histone marks at the IGF2 locus in human H1 ESCs (data from Broad/ENCODE). The IGF2-P0 promoter region is highlighted in gray.
(B) Luciferase assay to test orthologs of mouse, human H, C1, C2, chimp, pig and elephant ZFP568 repression activities against the mouse Igf2-P0 native promoter. Data in panels B, C, D and F are shown as mean ± s.d., t-test, *p<0.05, **p<0.01, n=3.
(C) ChIP-qPCR analysis of GFP, mouse ZFP568 (M), human ZNF568 H, C1, C2 alleles occupancy at Igf2 and a negative control promoter of another ZF gene (Znf180).
(D) Luciferase assay to test mouse, human alleles H, C1, C2, chimp, pig and elephant ZFP568 repression activity against the human IGF2-P0 binding site cloned upstream of an SV40 promoter.
(E) Co-immunoprecipitation assay of human 293T cells transfected with plasmids expressing Flag-tagged mouse ZFP568 or human ZNF568 KRAB domains and HA-tagged KAP1. Cell lysates were immunoprecipitated with anti-Flag antibodies. The cell lysates and immunoprecipitates were analyzed by immunoblotting with antibodies as indicated.
(F) The indicated KRAB-domains from a control ZF protein (ZFP809) or human ZNF568 alleles were cloned upstream of the zinc fingers of ZFP809, and tested in reporter luciferase assays against the PBS-pro sequence (which is bound by ZFP809). The relative luciferase activity and fold repression are plotted for each chimeric protein.
Figure S7. Loss of IGF2-P0 transcript in humans, Related to Figure 7.
(A) IGF2 gene expression track in different human tissues. No IGF2-P0 expression was detected in any tissues examined.
(B) Sequence alignment of Igf2-P0 promoter regions (used in Figure 7F). Position of the ZFP568 binding site is indicated and the sequence variations are colored in cyan. Pairwise comparison indicated three nucleotide changes between human and chimp (highlighted in the chimp sequence), 14 nucleotide changes between chimp and rhesus (highlighted in the rhesus sequence), and more extensive differences exist between rhesus and mouse throughout the region examined including deletions and insertions.
Table S1. Protein Sequences for mammalian ZFP568 orthologs, Related to Figure 1
Table S2. Summary of X-ray data, Related to Figures 3 and 4
Table S3. DNA Minor and major groove widths, Related to Figure 5
Table S4. Igf2 promoter sequences including binding motif and cloning sites, Related to 7
Highlights.
ZFP568 and its Igf2-P0 binding activity is conserved in eutheria
Mouse ZFP568 11-finger array makes numerous non-canonical ZF-DNA interactions
ZFP568 forms versatile contacts in response to sequence-specific deformation in DNA
Chimp and human ZFP568 have weakened or abolished binding to their Igf2-P0 sequence
Acknowledgments
We thank C.K-James Shen for providing human ZNF568 constructs and helpful discussion and B. Baker of New England Biolabs for synthesizing the oligonucleotides. The Department of Biochemistry of Emory University School of Medicine supported the use of SER-CAT beamlines. This work was supported by grants from the National Institutes of Health GM049245-24 (X.Z. and X.C.), 1ZIAHD008933 (T.S.M), and CPRIT-RR160029 (X.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.P. performed crystallographic and in vitro binding assays; P.Y. and M.T. performed reporter assays, ChIP-qPCR, Co-IP, and evolutionary analyses; with assistance from M.P. (cloning, purification and crystallization), Y.W. (rescue studies), D.H. and G.W. (luciferase assays), and J.R.H. (crystallography). M-A.S. performed computational and evolutionary analysis. T.S.M. initiated this collaborative work and together with X.Z. and X.C. organized and designed the scope of the study. All authors were involved in analyzing data and preparing the manuscript.
Declaration of Interests
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information includes seven figures and four tables and can be found with this article online at https://
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 2014;28:1397–1409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasegaran S, Carroll D. Origins of Programmable Nucleases for Genome Engineering. J Mol Biol. 2016;428:963–989. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien HC, Wang HY, Su YN, Lai KY, Lu LC, Chen PC, Tsai SF, Wu CI, Hsieh WS, Shen CK. Targeted disruption in mice of a neural stem cell-maintaining, KRAB-Zn finger-encoding gene that has rapidly evolved in the human lineage. PLoS ONE. 2012;7:e47481. doi: 10.1371/journal.pone.0047481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y, Klug A. Physical basis of a protein-DNA recognition code. Curr Opin Struct Biol. 1997;7:117–125. doi: 10.1016/s0959-440x(97)80015-2. [DOI] [PubMed] [Google Scholar]
- Constancia M, Dean W, Lopes S, Moore T, Kelsey G, Reik W. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat Genet. 2000;26:203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- Constancia M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, Stewart F, Kelsey G, Fowden A, Sibley C, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417:945–948. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- Davies B, Hatton E, Altemose N, Hussin JG, Pratto F, Zhang G, Hinch AG, Moralli D, Biggs D, Diaz R, et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature. 2016;530:171–176. doi: 10.1038/nature16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassan MA, Calladine CR. Two distinct modes of protein-induced bending in DNA. J Mol Biol. 1998;282:331–343. doi: 10.1006/jmbi.1998.1994. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia MJ, Shibata M, Anderson KV. Chato, a KRAB zinc-finger protein, regulates convergent extension in the mouse embryo. Development. 2008;135:3053–3062. doi: 10.1242/dev.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Christensen RG, Bell HA, Goodwin M, Patel RY, Pandey M, Enuameh MS, Rayla AL, Zhu C, Thibodeau-Beganny S, et al. An improved predictive recognition model for Cys2-His2 zinc finger proteins. Nucleic Acids Res. 2014;42:4800–4812. doi: 10.1093/nar/gku132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Wang D, Horton JR, Zhang X, Corces VG, Cheng X. Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA. Mol Cell. 2017;66:711–720. doi: 10.1016/j.molcel.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S, Trievel RC. Carbon-oxygen hydrogen bonding in biological structure and function. J Biol Chem. 2012;287:41576–41582. doi: 10.1074/jbc.R112.418574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng X. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of dam methyltransferase. Cell. 2005;121:349–361. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Molecular biology and evolution. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideraabdullah FY, Bartolomei MS. ZFP57: KAPturing DNA methylation at imprinted loci. Mol Cell. 2011;44:341–342. doi: 10.1016/j.molcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Imbeault M, Helleboid PY, Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017;543:550–554. doi: 10.1038/nature21683. [DOI] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Ivanov AV, Jin VX, Rauscher FJ, 3rd, Farnham PJ. Functional analysis of KAP1 genomic recruitment. Mol Cell Biol. 2011;31:1833–1847. doi: 10.1128/MCB.01331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs CJ, Schultz DC, Robins DM. The KRAB zinc finger protein RSL1 regulates sex- and tissue-specific promoter methylation and dynamic hormone-responsive chromatin configuration. Mol Cell Biol. 2012;32:3732–3742. doi: 10.1128/MCB.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular biology and evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Developmental cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Toh H, Sasaki H, Zhang X, Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang X, Blumenthal RM, Cheng X. A common mode of recognition for methylated CpG. Trends Biochem Sci. 2013;38:177–183. doi: 10.1016/j.tibs.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet. 2008;40:949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- Meylan S, Groner AC, Ambrosini G, Malani N, Quenneville S, Zangger N, Kapopoulou A, Kauzlaric A, Rougemont J, Ciuffi A, et al. A gene-rich, transcriptionally active environment and the pre-deposition of repressive marks are predictive of susceptibility to KRAB/KAP1-mediated silencing. BMC Genomics. 2011;12:378. doi: 10.1186/1471-2164-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci U S A. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- O’Geen H, Frietze S, Farnham PJ. Using ChIP-seq technology to identify targets of zinc finger transcription factors. Methods Mol Biol. 2010;649:437–455. doi: 10.1007/978-1-60761-753-2_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Hashimoto H, Zhang X, Cheng X. Characterization of How DNA Modifications Affect DNA Binding by C2H2 Zinc Finger Proteins. Methods Enzymol. 2016a;573:387–401. doi: 10.1016/bs.mie.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Horton JR, Wilson GG, Zhang X, Cheng X. Structural basis for human PRDM9 action at recombination hot spots. Genes Dev. 2016b;30:257–265. doi: 10.1101/gad.274928.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Zhang X, Blumenthal RM, Cheng X. Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence. J Biol Chem. 2017;292:15994–16002. doi: 10.1074/jbc.M117.805754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ., 3rd Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- Persikov AV, Singh M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014;42:97–108. doi: 10.1093/nar/gkt890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov AV, Wetzel JL, Rowland EF, Oakes BL, Xu DJ, Singh M, Noyes MB. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 2015;43:1965–1984. doi: 10.1093/nar/gku1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell reports. 2012;2:766–773. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Radovani E, Najafabadi HS, Barazandeh M, Campitelli LF, Yin Y, Jolma A, Zhong G, Guo H, Kanagalingam T, et al. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 2016;26:1742–1752. doi: 10.1101/gr.209643.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, Crotty JW, Bhakta MS, Barbas CF, 3rd, Horton NC. Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA. J Mol Biol. 2006;363:405–421. doi: 10.1016/j.jmb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Shibata M, Blauvelt KE, Liem KF, Jr, Garcia-Garcia MJ. TRIM28 is required by the mouse KRAB domain protein ZFP568 to control convergent extension and morphogenesis of extra-embryonic tissues. Development. 2011;138:5333–5343. doi: 10.1242/dev.072546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Garcia-Garcia MJ. The mouse KRAB zinc-finger protein CHATO is required in embryonic-derived tissues to control yolk sac and placenta morphogenesis. Developmental biology. 2011;349:331–341. doi: 10.1016/j.ydbio.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanamee ES, Viadiu H, Kucera R, Dorner L, Picone S, Schildkraut I, Aggarwal AK. A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA. Embo J. 2005;24:4198–4208. doi: 10.1038/sj.emboj.7600880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer EM, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015;29:538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Xu B, Yang Z. PAMLX: a graphical user interface for PAML. Molecular biology and evolution. 2013;30:2723–2724. doi: 10.1093/molbev/mst179. [DOI] [PubMed] [Google Scholar]
- Yang P, Wang Y, Hoang D, Tinkham M, Patel A, Sun MA, Wolf G, Baker M, Chien HC, Lai KN, et al. A placental growth factor is silenced in mouse embryos by the zinc finger protein ZFP568. Science. 2017;356:757–759. doi: 10.1126/science.aah6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandarashvili L, Esadze A, Vuzman D, Kemme CA, Levy Y, Iwahara J. Balancing between affinity and speed in target DNA search by zinc-finger proteins via modulation of dynamic conformational ensemble. Proc Natl Acad Sci U S A. 2015;112:E5142–5149. doi: 10.1073/pnas.1507726112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence alignment, Related to Figure 1.
(A) Global alignment of ZFP568 ortholog protein sequences with domain structures. The conservation score for each position (bottom) was calculated as the Jensen-Shannon divergence (Capra and Singh, 2007).
(B) Zinc fingerprint alignment for each zinc finger at potential base-interacting positions −1, −4, −5, −7 (see Figure 1A). Consensus sequences (bottom) were generated by MEME (Bailey et al., 2009).
Figure S2. Structures of ZF1-10 and ZF2-11, Related to Figure 2.
(A) Structure of ZF2-11 in complex with 28-bp oligonucleotide, displayed by crystallographic heatmap, from low to high thermal B-factors (blue, cyan, green, yellow, and red). Note that ZF11 has higher temperature-dependent atomic vibrations or static disorder in the crystal lattice.
(B) A complete model of mZFP568 ZF1-11 binding Igf2-P0 sequence, generated by superimposing the common nine fingers (ZF2-10) from structures of ZF1-10 and ZF2-11, resulting in a root-mean-square deviation (RMSD) of 0.7 Å overlapping 222 pairs of Cα atoms.
(C–D) Crystal packing contacts of two ZF protein-DNA complexes in the space groups C2221 (A) and P1 (B). The C2221 space group contains one protein-DNA complex per crystallographic asymmetric unit, whereas the P1 space group contains two such complexes. Note the two symmetry-related complexes in P1 have difference orientations (rainbow color scheme from ZF1-to-ZF10 with blue-to-red) from that of C2221.
(E) Superimposition of two structures of ZF1-10 in complexes in C2221 (colored) and P1 (grey) with 28-bp and 26-bp oligonucleotides, respectively, with an RMSD of less than 1 Å when comparing 231 pairs of Cα atoms.
Figure S3. Locations of human ZNF678 point mutations in the corresponding mZFP568 structure, Related to Figures 3, 4, 6 and 7.
(A) R654 and H547 of ZF11 from the structure of ZF2-11 recognizes G3 and G2, respectively. The side chain of Q660 forms H-bonds with T1.
(B) R457 of ZF4 interacts with a DNA backbone phosphate group.
(C) Mutations in human alleles (H, C1 and C2) in the corresponding mZFP568 are highlighted.
(D) H373 of ZF1 points to solvent. Like R457 of ZF4 (panel B), the corresponding position of H373 in each ZF is quite a variable and accommodates a variety of amino acids (see Figure 6F).
(E) C421 of ZF3 is one of the zinc ligands, and the C421S mutant would disturb the structural rigidity to the finger. The corresponding position of E423 of ZF3 is a conserved acidic or polar residue (E/D/Q) in each ZF (Figure 6F), which may provide additional electrostatic interaction with positively charged Zn(II) ion. The substitution of glutamate-to-alanine (E423A) would weaken its interaction the metal ion.
(F) Y531 of ZF, located in the beginning of strand β1, is involved in intra-molecule interactions with ZF2, forming a H-bond network with the counterpart tyrosine Y391 of ZF2, the main-chain carbonyl oxygen atoms of P401 of ZF2 and R541 of ZF7 (via a water molecule).
(G) G565 of ZF8 is located in the beginning of strand β2, providing flexibility for the loop between two strands. Although conserved in almost all ZFs, ZF5 of mZFP568 does contain a negatively charged aspartate in the same position (see Figure 6E).
Figure S4. Substitution of A13-to-G13 in mouse Igf2-P0, related to Figure 4.
It is interesting to note that ZF4 (and ZF5) possesses at least two amino acids, Gln at position −4 and Tyr at position −7 (Figure 1A), known to interact with adenine and thymine respectively. Juxtaposition of Gln (or Asn) with Ade is a common mechanism for recognition of this base (Luscombe et al., 2001), as seen in a few classic examples, including Gln44 of lambda repressor (PDB 3BDN), Gln28 of repressor of phage 434 (PDB 2OR1), Gln16 and Gln44 of a designed zinc finger protein (PDB 1MEY), and more recently Gln418 of human CTCF (Hashimoto et al., 2017). In addition, the aromatic ring of a tyrosine contacting a thymine methyl group has been used by the zinc finger protein ZNF217 (PDB 4F2J), although interactions of this type have not been often described in classical ZF-DNA complexes. We reasoned that ZF4 and ZF5 could potentially interact with A:T rich sequence in a classical fashion. The “irregularity” observed from the current structure could be the result of the expanded 4-bp recognition by ZF7, particularly R542 skipping an A:T at bp position 13 to interact with an invariant G:C at position 14 (Figure 4X). Because bp position 13 is a variable in the ChIP data and in the Igf2-P0 consensus sequence (Figure 2A), we substituted the G:C base pair for A:T and ask whether we can convert this “irregular” ZF7 (a 4-bp reader) to a canonical 3-bp reader. ZF7 contains exactly the same recognition residues, R-E-R, as ZF10, which recognizes the canonical 3 bases (shown in Figure 4X) and we anticipated that R542-Gua interaction would restore the 3-bp recognition of ZF7 and subsequently that of ZF6 to ZF4, which could potentially interact with the A:T rich sequence in a classic fashion resulting in a substantial increase in binding affinity (MODEL 1). However, the opposite was observed, in that only a slight reduction in affinity (1.5x) was observed for the mutant oligo. This suggested that the 4 base spacing for ZF7 was probably maintained in the binding of the mutant oligo, without gross rearrangement of the rest of the ZFs, even if the local rearrangement does occur at base pair position 13 (MODEL 2).
Figure S5. ZF4 is out of register, Related to Figure 5.
(A–B) We generated a classic three-finger fragment of PRDM9A (by deleting one of the terminal finger) and scanned it through ZF3-10 of mZFP568. The three-finger array aligns well with ZF7-9 or ZF8-10 (B), but it attempts to skip ZF4 and makes an arrangement with the three fingers of ZF3, 5, and 6 (A), suggesting that it is ZF4 that is out of register.
(C–D) ZF3 follows the conventional rule of one-finger-three bases, so we superimposed the single ZF3 to that of PRDM9A (RMSD=0.3Å), and asked what is the conformational difference for the following fingers. ZF4 rotates its helix approximately 20° perpendicular to the DNA axis with the amino end fixed and the carboxyl end of the helix moving approximately 5Å (C). This movement is sufficient to displace the base-interacting residues of ZF4 out of touch to the base (D) and allows a layer of water molecules to diffuse into the space (Figure 4T).
(E–F) The conformational change is extended to ZF5, which moves towards ZF4 by ~6Å (approximately one helical turn) and upwards ~3Å (F). This movement in two dimensions allows ZF5 to interact with the opposite strand of three A:T base pairs immediately after three G:C base pairs by ZF3 (Figure 3).
Figure S6. Systematic analysis of ZFP568 repression activity in human and other mammals, Related to Figure 6.
(A) ChIP-seq tracks of indicated histone marks at the IGF2 locus in human H1 ESCs (data from Broad/ENCODE). The IGF2-P0 promoter region is highlighted in gray.
(B) Luciferase assay to test orthologs of mouse, human H, C1, C2, chimp, pig and elephant ZFP568 repression activities against the mouse Igf2-P0 native promoter. Data in panels B, C, D and F are shown as mean ± s.d., t-test, *p<0.05, **p<0.01, n=3.
(C) ChIP-qPCR analysis of GFP, mouse ZFP568 (M), human ZNF568 H, C1, C2 alleles occupancy at Igf2 and a negative control promoter of another ZF gene (Znf180).
(D) Luciferase assay to test mouse, human alleles H, C1, C2, chimp, pig and elephant ZFP568 repression activity against the human IGF2-P0 binding site cloned upstream of an SV40 promoter.
(E) Co-immunoprecipitation assay of human 293T cells transfected with plasmids expressing Flag-tagged mouse ZFP568 or human ZNF568 KRAB domains and HA-tagged KAP1. Cell lysates were immunoprecipitated with anti-Flag antibodies. The cell lysates and immunoprecipitates were analyzed by immunoblotting with antibodies as indicated.
(F) The indicated KRAB-domains from a control ZF protein (ZFP809) or human ZNF568 alleles were cloned upstream of the zinc fingers of ZFP809, and tested in reporter luciferase assays against the PBS-pro sequence (which is bound by ZFP809). The relative luciferase activity and fold repression are plotted for each chimeric protein.
Figure S7. Loss of IGF2-P0 transcript in humans, Related to Figure 7.
(A) IGF2 gene expression track in different human tissues. No IGF2-P0 expression was detected in any tissues examined.
(B) Sequence alignment of Igf2-P0 promoter regions (used in Figure 7F). Position of the ZFP568 binding site is indicated and the sequence variations are colored in cyan. Pairwise comparison indicated three nucleotide changes between human and chimp (highlighted in the chimp sequence), 14 nucleotide changes between chimp and rhesus (highlighted in the rhesus sequence), and more extensive differences exist between rhesus and mouse throughout the region examined including deletions and insertions.
Table S1. Protein Sequences for mammalian ZFP568 orthologs, Related to Figure 1
Table S2. Summary of X-ray data, Related to Figures 3 and 4
Table S3. DNA Minor and major groove widths, Related to Figure 5
Table S4. Igf2 promoter sequences including binding motif and cloning sites, Related to 7


