Significance
FISH enables researchers to visualize the subcellular distribution of RNA and DNA molecules in individual cells. The recent development of FISH methods employing probes composed of synthetic DNA oligonucleotides (oligos) allows researchers to tightly control aspects of probe design such as binding energy and genomic specificity. Although oligo FISH probes are central to many recently developed massively multiplexed and superresolution imaging methods, no dedicated computational utility exists to facilitate the design of such probes on the genome-wide scale. Here, we introduce a streamlined pipeline for the rapid, genome-scale design of oligo FISH probes and validate our approach by using conventional and superresolution imaging. Our method provides a framework with which to design oligo-based hybridization experiments.
Keywords: oligonucleotide, FISH, in situ, superresolution, oligo
Abstract
Oligonucleotide (oligo)-based FISH has emerged as an important tool for the study of chromosome organization and gene expression and has been empowered by the commercial availability of highly complex pools of oligos. However, a dedicated bioinformatic design utility has yet to be created specifically for the purpose of identifying optimal oligo FISH probe sequences on the genome-wide scale. Here, we introduce OligoMiner, a rapid and robust computational pipeline for the genome-scale design of oligo FISH probes that affords the scientist exact control over the parameters of each probe. Our streamlined method uses standard bioinformatic file formats, allowing users to seamlessly integrate new and existing utilities into the pipeline as desired, and introduces a method for evaluating the specificity of each probe molecule that connects simulated hybridization energetics to rapidly generated sequence alignments using supervised machine learning. We demonstrate the scalability of our approach by performing genome-scale probe discovery in numerous model organism genomes and showcase the performance of the resulting probes with diffraction-limited and single-molecule superresolution imaging of chromosomal and RNA targets. We anticipate that this pipeline will make the FISH probe design process much more accessible and will more broadly facilitate the design of pools of hybridization probes for a variety of applications.
FISH is a powerful single-cell technique that harnesses the specificity afforded by Watson–Crick base pairing to reveal the abundance and positioning of cellular RNA and DNA molecules in fixed samples. Originally introduced as a radioactive in situ hybridization method in the late 1960s (1–3), FISH has undergone a series of optimizations that have improved its detection efficiency and sensitivity (4–7). Many of these refinements have centered on the preparation and labeling of the probe material, which traditionally has been derived from cellular DNA or RNA, and include the introduction of the nick translation method that increases the specific activity of labeling (8, 9) and the development of suppressive hybridization techniques that limit background originating from repetitive sequences contained in many probes (10).
More recently, advances in DNA synthesis technology have afforded researchers the opportunity to construct FISH probes entirely from synthetic oligonucleotides (oligos). Oligo probes offer many potential advantages, as they can be selected to have specific thermodynamic properties, engineered to avoid repetitive sequences, designed against any sequenced genome, and endowed with many different types and densities of labels. Whereas the use of oligo probes was initially restricted to the interrogation of multicopy targets such as repetitive DNA (11–13) and mRNA (14–16) with the use of one to a few dozen oligo probes, the recent development of oligo libraries produced by massively parallel array synthesis (17) has empowered a new generation of FISH technologies able to target single-copy chromosomal regions with highly complex libraries of hundreds to many thousands of oligo probes (18–20).
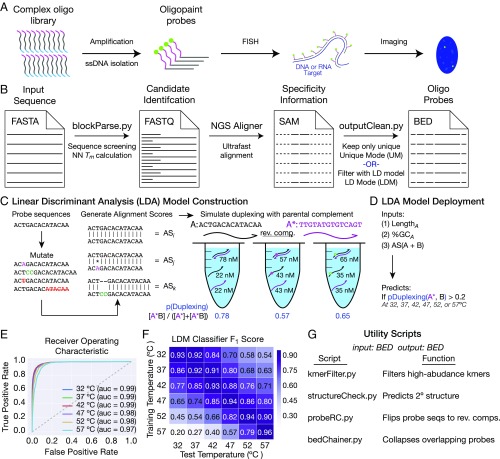
We have previously introduced Oligopaints, a method for the generation of highly efficient probes for RNA FISH and DNA FISH from libraries composed of dozens to many thousands of unique oligo species (20). In the Oligopaints approach, these libraries are encoded such that each molecule contains a short region of homology (∼30–50 bases) to the RNA or DNA target flanked by PCR primers (Fig. 1A). Following PCR amplification, ssDNA probes can be generated by a number of approaches, including nicking endonuclease treatment followed by gel extraction (20, 21), in vitro transcription followed by reverse transcription (22–24), and digestion by λ-exonuclease (25). Ultimately, these molecular biological approaches produce pools of ssDNA probes that can be labeled directly through the use of a fluorophore-conjugated primer during the PCR or reverse-transcription steps (Fig. 1A) or indirectly through the inclusion of a binding site for a fluorescently labeled “secondary” oligo that can be added during or after the FISH experiment (25).
Fig. 1.
Implementation of OligoMiner. (A) Schematic overview of the Oligopaints. (B) Schematic overview of the OligoMiner pipeline. (C and D) Schematic overviews of LDA model (C) creation and (D) implementation. (E) Receiver operating characteristic curves for each temperature-specific LDA model. auc, area under the curve. (F) Heat map showing the support-weighted F1 score for each temperature-specific LDA model when tested against validation data simulated at each of the six indicated temperatures. (G) Description of utility scripts also developed as part of OligoMiner.
A key feature of Oligopaints is their programmability, wherein the genomic and nongenomic sequences that compose each probe oligo can be specified precisely. This fine level of control has enabled several important technical advances in FISH imaging, including the single-molecule superresolution imaging of chromosome structure at nonrepetitive targets (25–27), the development of probes that can distinguish genomically unique regions of homologous chromosomes (25), and the introduction of a method able to label dozens of chromosomal loci (28). The general programmability of oligo FISH probes has also enabled the creation of related methods that use aspects of the Oligopaints approach to enable the highly multiplexed visualization of dozens to >1,000 distinct mRNA species in the same sample (22, 29) and >10,000 mRNA species in vitro (30).
Despite the rapid maturation of new FISH technologies reliant on oligo probes, comparatively little progress has been made in the development of computational tools to facilitate the design of these oligos. Such programs seek to identify optimal probe sequences within a block of input sequence based on thermodynamic properties such as melting temperature (Tm) while attempting to identify and exclude probes that are likely to hybridize at sites other than the intended target. Although computational utilities exist to create small numbers of oligo probes against targets such as bacterial rRNA (31, 32) and to design large pools of oligo pairs such as PCR primers (33–36) or padlock probes (37, 38), to our knowledge, no bioinformatic utility has been created for the explicit purpose of designing oligo hybridization probes at the genome-wide scale. Consequently, older utilities such as the microarray design program OligoArray (39) have been repurposed to facilitate probe design. Although OligoArray has produced effective oligo FISH probes (20, 25), it can only provide limited throughput, with large genomes such as those of human and mouse taking 1–2 mo of continuous cluster computing to mine with a single set of parameters (20) and smaller regions still requiring hours of cluster computing to complete. Additionally, OligoArray offers the user a limited amount of control over the probe discovery process, as users interact only with a compiled executable Java Archive file and cannot change the nature or order of steps taken or the values of many of parameters used for thermodynamic calculations and specificity checking.
Here, we introduce OligoMiner, a rapid and flexible genome-scale design environment for oligo hybridization probes. The modular, open-source OligoMiner pipeline is written in Python using Biopython (40) and uses standard bioinformatic file formats at each step in the probe mining process, greatly simplifying probe discovery. Additionally, OligoMiner introduces a method of assessing probe specificity that employs supervised machine learning to predict thermodynamic behavior from genome-scale sequence alignment information. The OligoMiner pipeline can readily be deployed on any sequenced genome and can mine the entirety of the human genome in minutes to hours and smaller <10-Mb regions in mere minutes on a standard desktop or laptop computer, greatly reducing the time and computational resource cost of probe discovery. We also demonstrate the effectiveness of probes produced by our approach with conventional and single-molecule superresolution microscopy.
Results
Identification of Candidate Probes.
OligoMiner is a collection of Python scripts that facilitates the design of oligo-based hybridization probes. These scripts are designed to be run in a command-line environment, where they can be integrated into existing workflows with other bioinformatic utilities. OligoMiner can be used on any modern Windows, Macintosh, or Linux system and requires no direct knowledge of programming. To execute the probe-design process, users simply need to prepare a suitable input file and call the relevant scripts in the pipeline. The scripts, a “readme” document describing this workflow and providing installation and utilization instructions, and example input and output files can be found at oligominer.net. The contents of the readme document are also provided in Dataset S1. Importantly, OligoMiner allows the user to specify a broad range of parameters, including the alignment method used to check specificity, providing substantially more flexibility than OligoArray (Fig. S1).
The OligoMiner workflow begins with a FASTA-formatted input file (41) containing the genomic sequence to be searched for probes, which can be masked by a program such as RepeatMasker (42) to exclude regions containing repetitive elements. This input file is first passed to the blockParse script (Fig. 1B and Fig. S2), which screens for prohibited sequences such as homopolymeric runs and “N” bases and allows users to specify allowable ranges of probe length, percent G + C content (GC%), and adjusted Tm calculated by using nearest-neighbor thermodynamics (43). Candidate probe sequences passing all checks are outputted in FASTQ format (44) to facilitate input into next-generation sequencing (NGS) alignment programs such as Bowtie/Bowtie2 (45, 46) and BWA (47), which can be used to assess off-target potential. Importantly, these NGS alignment programs are optimized for the extremely rapid alignment of millions of short sequences to a reference genome in parallel, thereby allowing the specificity-check step of the pipeline to proceed much more quickly than approaches like OligoArray that use BLAST (48) in serial.
Predicting Probe Specificity.
Ultrafast alignment programs can provide a wealth of information about the relatedness of a given input sequence to other sequences present in a genome assembly. OligoMiner allows users to evaluate probe specificity by using two distinct approaches, in either case using the script outputClean to process the Sequence Alignment/Map (SAM) file (49) produced by the alignment program and outputting Browser Extendable Data (BED) format (50) files; BED files are designed for visualizing sequence features in genome browsers and are fully compatible with our previously described tools that facilitate the design and ordering of Oligopaint probe libraries (20) (genetics.med.harvard.edu/oligopaints) and utilities such as BEDTools (51) (Fig. 1B and Fig. S3). The first approach, termed Unique Mode (UM), uses the number of reported alignments to differentiate between candidate probes predicted to only have one genomic target from those with multiple potential binding sites; candidates with more than one reported alignment or that fail to align are filtered, whereas candidate probes that align uniquely are passed to the output. Unique Mode thereby enables users to experiment with different groups of alignment parameters to find an optimal combination for a given application.
Ideally, the thermodynamics of hybridization between a candidate probe and potential off-target sites would be modeled in silico and employed as a means of identifying probe oligos likely to only bind their intended targets in a given set of reaction conditions. Although powerful utilities such as NUPACK (52–54) are capable of performing such simulations, the limited throughput of these programs renders a direct thermodynamic approach impractical for genome-scale probe design. However, we hypothesized that features in rapidly calculated data such as alignment scores may be predictive of thermodynamic behavior and could therefore serve as a proxy for the information that would be produced by thermodynamic simulations. Inspired by this idea, we first selected 800 “probe” sequences identified by blockParse in the human hg38 assembly that represented three commonly used probe length ranges (26–32, 35–41, 40–46 nt; Methods). To simulate the types of binding sites that these “probes” might encounter in situ during a FISH experiment in a complex genome, we next generated 406,014 variant versions of the “probe” sequences in silico that each contained one or more point mutation, insertion, deletion, or large truncation, creating, in combination with the 800 “probe” sequences, a pool of 406,814 “target sites” (Methods and Fig. 1C). We then aligned each “probe” to its corresponding “target sites” in pairwise alignments by using Bowtie2 with ultrasensitive settings (Methods), generating a set of 406,814 alignment scores (Fig. 1C). In parallel, we also computed the probability of a duplex forming between each “probe” and each of its corresponding “target sites” in FISH conditions (2× SSC, 50% formamide at 32, 37, 42, 47, 52, or 57 °C) in pairwise test tube simulations by using NUPACK (Methods and Fig. 1C).
To connect our alignment scores and duplexing probabilities, we next performed supervised machine learning by using linear discriminant analysis (LDA) on 60% of the combined datasets with scikit-learn (55). Specifically, we built six temperature-specific LDA models that predict whether the duplexing probability of a “probe”–“target site” pair will be above a threshold level of 0.2 (i.e., less than fivefold weaker than a fully paired duplex) given the length and GC% of the “probe” sequence and the score of the alignment of the two sequences (Methods and Fig. 1D). We tested these LDA models on the remaining 40% of the data and found that all six performed exceptionally well, with each producing areas under receiver operating characteristic curves of ≥0.97 (Fig. 1E) and support-weighted F1 scores ≥0.92 (Fig. 1F and Fig. S4). Notably, all six models also performed strongly when tested against data simulated at hybridization temperatures 5 °C higher or lower than the training temperature (support-weighted F1 score range, 0.79–0.92; mean, 0.86; Fig. 1F), indicating that the models are all capable of predicting duplexing behavior over a relatively broad range of reaction conditions. Collectively, our data argue that the LDA model identifies potentially problematic “probe”–“target site” interactions (i.e., those with a probability of duplexing >0.2) effectively as well as the much slower thermodynamic simulations. We have integrated the six LDA models into outputClean to create the second specificity evaluating approach, “LDA Mode” (LDM): candidate probes are first aligned to the reference genome of interest by using the same Bowtie2 scoring settings used to construct the LDA models (Methods), and the resulting SAM file is processed by a selected temperature-specific LDA model such that candidate probes predicted to have more than one thermodynamically relevant target site (i.e., probability of duplexing >0.2) are filtered (Fig. 1B and Fig. S3).
Postprocessing Functionalities.
We have written a series of utility scripts to augment the core OligoMiner pipeline (Fig. 1G). These utility scripts accept and return BED files, making them compatible with output files created by outputClean (Fig. 1B) and files created by the previous Oligopaint probe discovery method (20) and adding additional functionalities. For instance, kmerFilter enables the user to perform another layer of specificity checking by calling Jellyfish (56) to screen probe sequences for the presence of high-abundance k-mers (e.g., 16mers or 18mers) that may be missed by alignment programs because of their short lengths and could lead to off-target binding (57, 58). Users can also identify and filter probe sequences predicted to adopt secondary structures in a given set of experimental conditions by using structureCheck, which depends on NUPACK. Several additional tools facilitate the processing of probe files for specific applications, including the conversion of probe sequences to their reverse complements by probeRC for strand-specific DNA or RNA FISH and the collapsing of overlapping probes by bedChainer for the design of high-density probe sets. Finally, we have created additional modularity with pair of scripts, “fastqToBed” and “bedToFastq,” that allow users to convert between the BED and FASTQ format files.
Rapid Genome-Scale Probe Discovery.
To assess the scalability of OligoMiner, we performed genome-wide probe discovery in the human hg38 genome assembly. We first developed three sets of input parameters spanning a range of commonly used probe lengths and experimental conditions: a “coverage” set designed to maximize the number of probes discovered (26–32 nt length, 37 °C hybridization), a “stringent” set designed to maximize probe-binding affinity and thereby permit stringent hybridization and washing conditions (40–46 nt, 47 °C hybridization), and a “balance” set that seeks to compromise between coverage and binding affinity (35–41 nt, 42 °C hybridization; Fig. 2A). We next deployed OligoMiner by using these parameter settings in UM and LDM, in both cases using Bowtie2 for the alignment step and also including the optional kmerFilter specificity check (Methods). Excitingly, both approaches were able to mine the entire hg38 assembly very rapidly by using all three parameter sets, with UM averaging a rate of 1.70 Mb/min and a total time of 97 min per chromosome across all three parameter settings (Fig. 2 B and C) and LDM averaging a similar rate of 1.48 Mb/min and a total time of 104 min per chromosome (Fig. 2 C and D). These rates support mining the entire human genome in as little as 24–48 h if each chromosome was run in serial on a laptop or desktop computer and tens of minutes if parallel computing (e.g., ∼100–400 simultaneous jobs) was instead employed, in either case achieving a dramatic increase in speed from the 1–2 mo of parallel computing needed in our previous approach (20). Indeed, a direct comparison of probe discovery rates between OligoMiner and OligoArray revealed that OligoMiner provides a ∼50–100-fold increase over 10–100-kb intervals and a ∼800-fold increase over megabase-scale intervals (Fig. S5).
Fig. 2.
Genome-scale probe discovery with OligoMiner. (A) Description of three parameter sets used for genome-scale mining runs. (B–E) Box plots displaying overall mining times and rates for UM (B and C) and LDM (D and E). Each chromosome was run separately and reported, resulting in 24 data points per parameter setting and a total of 72 data points per plot. The mean rate or time for all 72 data points is displayed beneath each box plot. (F and G) Swarm plots displaying changes in probe density (i.e., probes per kilobase) that occurred over the course of the pipeline in UM (F) and LDM (G). bP, blockParse; kF, kmerFilter; oC, outputClean. (H) Swarm plot displaying probe densities in the C. elegans (ce11), D. melanogaster (dm6), zebrafish (danRer10), human (hg38), mouse (mm10), and A. thaliana (tair10) genome assemblies after whole-genome mining using LDM and kmerFilter.
The modularity of OligoMiner allows users to monitor how the parameters chosen at each step in the probe discovery process affect the final number of output probes. We have used this capability to examine changes in probe density (e.g., probes per kilobase) that occurred during the genome-wide probe discovery runs in hg38. As expected, blockParse discovered the highest density of candidate probes by using the “coverage” (c) settings, followed by “balance” (b) and “stringent” (s): c, 8.5; b, 5.7; s, 3.4 probes per kilobase; Fig. 2 F and G). However, we observed striking differences following outputClean depending on the mode used, with UM preserving the same order (c, 7.3; b, 4.7; s, 2.7 probes per kilobase) but the density of the “coverage” oligos plummeting in LDM (c, 2.6; b, 5.0; s, 3.0 probes per kilobase; Fig. 2 F and G and Dataset S2). We also observed large relative decreases in the density of “coverage” oligos following the application of kmerFilter, but only a modest reduction with the other sets (UM, c, 3.1; b, 4.6; s, 2.6 probes per kilobase; LDM, c, 1.6; b, 4.8; s, 2.9 probes per kilobase; Fig. 2 F and G); this effect is likely a result of the use of 16mer dictionary with “coverage” sets but an 18mer dictionary with the “balance” and “stringent” sets (Fig. 2A), a choice informed by differences in k-mer binding affinities at the different simulated hybridization temperatures (Fig. S6 and Dataset S2).
Collectively, our genome-scale hg38 probe sets are similar in probe density to previous sets designed with OligoArray (20, 25), and a direct comparison using a set of 3-Mb intervals revealed a high degree of concordance between the probes discovered by the two methods (80–96% of probes shared, R2 values of probes per kilobase of 0.86–0.97, n = 3; Fig. S5). Additionally, our results suggest that, when taking the thermodynamics of hybridization into account, longer oligo probes that can support higher hybridization temperatures can effectively provide higher probe densities, as observed with the UM and LDM “balance” sets (Fig. 2 F and G). Intriguingly, this phenomenon appears to depend on genome size and complexity; the same ordering of the three parameter sets was also observed in whole-genome probe discovery performed using LDM and kmerFilter in the mouse mm10 and zebrafish danRer10 assemblies, but the “coverage” set provided the highest densities in the smaller Drosophila melanogaster dm6, Caenorhabditis elegans ce11, and Arabidopsis thaliana tair10 assemblies (Fig. 2H and Dataset S2). The resulting probes discovered by these genome-scale probe discovery runs and additional LDM + kmerFilter whole-genome runs in the ce6, dm3, hg19, and mm9 assemblies are available on the Oligopaints Web site (genetics.med.harvard.edu/oligopaints).
OligoMiner Enables Conventional and Superresolution Imaging.
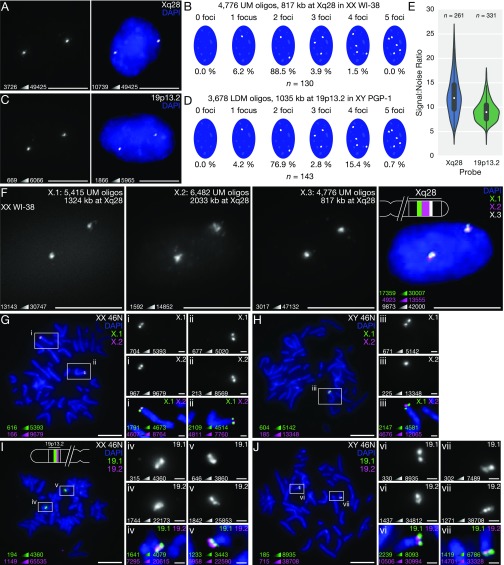
To test the efficacy of oligo probes designed with OligoMiner in situ, we first performed 3D FISH (59, 60) in XX 2N WI-38 human fetal lung fibroblasts with a set of 4,776 40–45mer Oligopaint probes designed using UM without kmerFilter targeting 817 kb at Xq28 (Table S1). In line with previous Oligopaint experiments using probes designed by OligoArray (20, 25), we observed highly efficient staining, with 100% of nuclei displaying at least one FISH signal and 88.5% of nuclei displaying two signals (n = 130; Fig. 3 A and B). We observed similarly efficient staining after performing 3D FISH in XY 2N PGP-1 fibroblasts with a set of 3,678 35–41mer Oligopaint probes designed using LDM with kmerFilter targeting 1,035 kb at 19p13.2 (100% nuclei with ≥1 signal, 76.9% with two signals, n = 143; Fig. 3 C and D and Table S1), illustrating the high labeling efficiency of probes produced by the UM and LDM approaches. Additionally, automated image analysis (61) revealed excellent signal:noise ratios (SNRs) for both probes (mean SNR, 12.3, n = 261 signals for Xq28; mean SNR, 9.2, n = 331 signals for 19p13.2; Fig. 3E), demonstrating the robustness of probes discovered with both modes. We also validated our ability to design custom hybridization patterns by performing 3D FISH with two additional sets of 40–45mer Oligopaint probes designed using UM without kmerFilter targeting Xq28 in WI-38 cells, which led to the expected three-color colocalization pattern in situ (Fig. 3F). Finally, we highlighted the specificity of our probes by performing two-color FISH on female and male metaphase spread chromosomes using two sets of Oligopaint probes targeting adjacent regions at Xq28 or 19p13.2 (Table S1), in all cases observing the expected number and distribution of signals (Fig. 3 G–J).
Fig. 3.
OligoMiner enables highly efficient FISH. (A and B) Representative single-channel minimum-maximum (min-max) contrasted image (Left) and two-color image with manual contrast adjustment (Right) (A) and signal number quantification (B) of 3D FISH experiment performed with a probe set consisting of 4,776 UM oligos targeting 817 kb at Xq28 in human XX 2N WI-38 fibroblasts. (C and D) Representative single-channel min-max contrasted image (C, Left) and two-color contrast-adjusted (C, Right) and signal number quantification (D) of 3D FISH experiment performed with a probe set consisting of 3,678 LDM oligos targeting 1,035 kb at 19p13.2 in human XY 2N PGP-1 fibroblasts. (E) Quantification of background-subtracted SNR for the Xq28 and 19p13.2 probes. (F) Three-color 3D FISH experiment performed using ATTO 488-labeled “X.1” (green), ATTO 565-labeled “X.2” (magenta), and Alexa Fluor 647-labeled “X.3” UM probe sets targeting adjacent regions on Xq28 in WI-38 fibroblasts. (G and H) Two-color metaphase FISH experiment performed using ATTO 488-labeled “X.1” (green) and Alexa Fluor 647-labeled “X.2” (magenta) UM probe sets targeting adjacent regions on Xq28 on XX 46N (G) and XY 46N (H) chromosome spreads. (I and J) Two-color metaphase FISH experiment performed using Alexa Fluor 647-labeled “19.1” (green) and Cy3B-labeled “19.2” (magenta) LDM probe sets targeting adjacent regions on 19p13.2 on XX 46N (I) and XY 46N (J) chromosome spreads. All images in are maximum-intensity projections in Z. DNA is stained with DAPI (blue) in multichannel images. In G–J, the multicolor images of the full spread and single-channel images (Inset) are min-max contrasted and the multichannel images (Inset) have manual contrast adjustments. (Scale bars: 10 µm; G–J, Inset, 1 µm.) For each image, the minimum and maximum pixel intensity value used to set the display scale is indicated in the lower left.
To further showcase the performance of oligos designed using OligoMiner in situ, we visualized 3D FISH by using stochastic optical reconstruction microscopy (STORM) (62) and DNA-based points accumulation in nanoscale topography (DNA-PAINT) (63): these single-molecule superresolution imaging techniques spatiotemporally isolate the fluorescent emissions of individual molecules and are capable of achieving <20-nm lateral and <50-nm axial resolution, which represent an order of magnitude or more below the diffraction limit (64). Specifically, we performed STORM imaging of Oligopaints (OligoSTORM) (25) of human 19p13.2 with two sets of 35–41mer oligos designed by using LDM with kmerFilter targeting a 1,035-kb region with 3,768 oligos (Fig. 4 A and B and Table S1) or a 20-kb region with 104 oligos (Fig. 4 C and D and Table S1) and, in both cases, were readily able to resolve the nanoscale morphologies of these foci, including features <40 nm (Fig. 4D), values comparable to those obtained by using probes designed by OligoArray (25). We also performed DNA-PAINT imaging of Oligopaints (OligoDNA-PAINT) (25) to visualize our 817-kb Xq28 probe set (Fig. 4 E and F and Table S1) and a set of 167 35–41mer oligos designed by using LDM with kmerFilter targeting the Xist RNA (65) (Fig. 4 G and H and Table S1), which also enabled us to reveal <40-nm structural features in the superresolved images (Fig. 4 F and H). Taken together, these superresolution experiments demonstrate the OligoMiner oligos can readily enable the single-molecule superresolution imaging of a broad range of target types and sizes.
Fig. 4.
Single-molecule superresolution imaging of OligoMiner oligos. (A and B) Diffraction-limited (A) and superresolved STORM (B) images of a probe set consisting of 3,678 LDM oligos targeting 1,035 kb at 19p13.2 in human XY 2N PGP-1 fibroblasts. (C and D) Diffraction-limited (C) and superresolved STORM (D) images of a probe set consisting of 104 LDM oligos targeting 20 kb at 19p13.2 in PGP-1 fibroblasts. (E and F) Diffraction-limited (E) and superresolved DNA-PAINT (F) images of a probe set consisting of 4,776 UM oligos targeting 817 kb at Xq28 in human XY 2N MRC-5 fibroblasts. (G and H) Diffraction-limited (G) and superresolved DNA-PAINT (H) images of a probe set consisting of 176 LDM oligos targeting 11 kb of the Xist RNA in human XX 2N WI-38 fibroblasts. (i–viii) Normalized single-molecule counts along the indicated 1D line traces (blue bars) and one- or two-component Gaussian fits to the underlying data (black lines). Superresolution data are presented using a “hot” color map in which single-molecule localization density scales from black (lowest) to red to yellow to white (highest). (Scale bars: 500 nm.) The minimum and maximum values of detected photons per square nanometer used to set the display scale is shown to right of each superresolution image, and the SD of the Gaussian blur used in the construction of each superresolution image is denoted in the top right corner.
Discussion
OligoMiner provides a framework for the rapid design of oligo hybridization probes on the genome-wide scale. We have demonstrated the ease and scalability of our pipeline by mining the human hg38 genome assembly with three distinct parameter sets and in two specificity-checking modes, a feat that would have otherwise required many months of cluster computing, and further highlighted the effectiveness of our approach with conventional and single-molecule superresolution imaging. Created by using open-source Python and Biopython and freely available via GitHub (oligominer.net), OligoMiner can readily be run on any standard laptop or desktop computer and exclusively uses standard bioinformatic file formats, providing users the opportunity to integrate OligoMiner scripts into existing pipelines and readily allowing additional and updated programs to be seamlessly integrated into the workflow. Critically, OligoMiner is capable of discovering the thousands to tens of thousands of oligo probes commonly ordered as pools from commercial suppliers in mere minutes, freeing the researcher to tailor the design of each probe set to the experimental question at hand instead of relying on preexisting collections of probe sequences obtained from previous probe mining runs or online databases (20). We expect the dramatic increase in speed and flexibility provided by OligoMiner will enable a much broader collection of research groups to use oligo FISH probes, including those working on developing new imaging technologies and in model organism systems not currently supported by existing probe collections. Moreover, we anticipate that OligoMiner could be employed more broadly to design hybridization probes for a wide range of experimental assays beyond in situ hybridization.
Methods
Genome Sequences.
The hg19, hg38, mm9, mm10, ce6, ce11, danRer10, dm3, and dm6 genome assemblies were downloaded with and without repeat masking from genome.ucsc.edu. The tair10 assembly was downloaded from www.arabidopsis.org/. To generate a repeat-masked version of tair10, transposable element locations identified by TASR (66) were converted to BED format and used as a guide for masking by pyfaidx (67).
Pipeline Construction and Implementation.
OligoMiner is written for Python 2.7 and depends on Biopython (40) and scikit-learn 0.17+ (55). Additional optional dependencies include Jellyfish 2.0+ (56) for k-mer screening and NUPACK 3.0 (52–54) for secondary structure analysis. To generate data for this study, scripts were executed locally in an OS X Anaconda Python 2.7 environment (Continuum Analytics) created with the command “conda create --name probeMining biopython scikit-learn” or in a CentOS Linux environment on the Orchestra High Performance Compute Cluster at Harvard Medical School.
LDA Model Construction.
Two sets of “probe” and “target site” sequences were used for the LDA model construction. For the first, all possible k-mers ≥8 were generated from 500 40–46mer sequences from hg38 chrX that were identified as candidate probes by blockParse, resulting in a total pool of 337,514 truncated and full-length sequences. In the second, 100 26–32mer, 100 35–41mer, and 100 40–46mer sequences from hg38 chr7 identified as candidate probes by blockParse were used as a starting pool of sequences. A Python script was then used to generate variant sequences containing 1–10 point mutations, 1–3 insertions of 1–6 bases each, or 1–3 deletions of 1–6 bases each, resulting in a total pool of 69,300 parental and variant sequences. These two pools were then combined to create a final pool of 406,814 sequences. To generate Bowtie2 alignment scores for each “probe”–“target-site” pairing, the “probe” sequence flanked by 3 “T” bases on both the 5′ and 3′ ends was used to create a Bowtie2 alignment index against which the “target-site” sequence was aligned by using the following settings: “--local -D 20 -R 3 -N 1 -L 10 -i S,1,0.5 --score-min G,1,1 -k 1.” To generate NUPACK duplexing probabilities for each pairing at a given temperature, the “complexes” executable was first called and given an input of the reverse complement of the “probe” sequence flanked by 3 “T” bases on both the 5′ and 3′ ends and the “target-site” sequence in a two-strand simulation with a maximum complex size of two strands. To account for FISH conditions, the Na+ concentration was set to 390 mM and the input temperature was increased by 31 °C (0.62 × 50) to account for the presence of 50% formamide. The resulting partition function outputted by “complexes” was then passed to the “concentrations” executable, with each strand being assigned an initial concentration of 1 µM. The percentage of the “probe” oligo contained in the “probe–target” complex was then stored as the duplexing probability. If the probability of duplexing was <0.2, the pairing was assigned to the “not likely to bind stably”/(−1) class; If the probability of duplexing was ≥0.2, the pairing was assigned to the “likely to bind stably”/(1) class. LDA model building, testing, and validation was performed by using scikit-learn 0.17 (55).
Whole-Genome Probe Discovery.
Genome assemblies in FASTA format without repeat masking were used to build Bowtie2 alignment indices and Jellyfish files. Repeat-masked input files were used for probe discovery. The blockParse script was run with the settings indicated in Fig. 2A and all other values set to their defaults (Fig. S2). Bowtie2 was run with “--very-sensitive-local –k 2 -t” in UM and “--local -D 20 -R 3 -N 1 -L 20 -i C,4 --score-min G,1,4 -k 2 -t” in LDM. The outputClean script was run with default values (Fig. S3) in LDM or UM. The kmerFilter script was used with the k-mer lengths indicated in Fig. 2A and “-k/–kmerThreshold” set to 5. To minimize file sizes and maximize speed, Jellyfish files were created such that k-mers occurring 0 or 1 time were not recorded and all kmers occurring >255 times were reported as “255”, i.e., the counts were recorded with 1 bit. Jellyfish hash size was set to the approximate size of the genome assembly, e.g., the command “jellyfish count -s 3300M -m 18 -o hg38_18.jf --out-counter-len 1 -L 2 hg38.fa” was used to create the 18mer dictionary for hg38. Bowtie 2.2.4 and Jellyfish 2.2.4 were used. The resulting probe files for all whole-genome runs described in Fig. 2, as well as whole-genome runs with the “c,” “b,” and “s” parameter sets in the hg19, mm9, dm3, and ce6 assemblies using LDM and kmerFilter, are available at oligominer.net.
Mining Speed Calculations.
Genome-scale hg38 mining runs were conducted on the Orchestra Compute Cluster, with each chromosome being run as its own individual job (i.e., without further parallelization) for each step in the probe design process (blockParse, Bowtie2, outputClean, kmerFilter). Wall clock times for the three OligoMiner Python scripts were reported via the Python “timeit” module and written to meta files by flagging the “-M/–Meta” option present in the three scripts. Bowtie2 wall clock time was reported by flagging the “-t” option and read from the printed output. Graphs presenting probe mining speed and probe densities were created in Python by using seaborn (68).
OligoArray vs. OligoMiner Mining Rate Comparison.
Eight 10-Mb intervals from the hg38 chromosome 1 scaffold (40,000,001–50,000,000, 50,000,001–60,000,000… 110,000,001–120,000,000) were selected. For each interval, the first 10 kb, 100 kb, 1 Mb, or the full 10 Mb were inputted into each pipeline. OligoArray 2.1 was run by using the settings “-l 36 -L 36 -g 36 -t 80 -T 85 -p 20 -P 80 -s 80 -x 80 -m “GGGGG;CCCCC;TTTTT;AAAAA”.” For OligoMiner, blockParse was run by using the settings “-l 36 -L 36 -S 0 -t 42 -T 47 -g 20 -G 80 -X “GGGGG;CCCCC;TTTTT;AAAAA”.” outputClean was run in LDM with “-T 42.” kmerFilter was then run with “-m 18 -k 5.” For OligoArray and OligoMiner, probe discovery was run on the Orchestra High Performance Compute Cluster and CPU run time was reported by the LSF job handling system.
OligoArray vs. OligoMiner Coverage Comparison.
Three intervals in hg38 were chose for analysis: (i) chr1 40,000,001–43,000,000, (ii) chr19 11,000,001–14,000,000, and (iii) chrX 149,000,001–152,000,000. Each pipeline was run with matched settings in an exhaustive search of the target region (i.e., overlapping probes were allowed). OligoArray was run by using the settings “-l 36 -L 36 -g 1 -t 80 -T 85 -p 20 -P 80 -s 80 -x 80 -m “GGGGG;CCCCC;TTTTT;AAAAA” -n 1005 -D 1000”. For OligoMiner, blockParse was run using the settings “-l 36 -L 36 -t 39.5 -T 44.5 -g 20 -G 80 -X “GGGGG;CCCCC;TTTTT;AAAAA” -O”. outputClean was run in LDM with “-T 42.” kmerFilter was then run with “-m 18 -k 5.” Linear regressions were calculated by using Python.
Oligopaint Probe Synthesis.
OligoMiner settings used to design each Oligopaint FISH probe set are provided in Table S1. Probe sets were synthesized by using the previously described gel extraction (20) (Xq28 probes) or T7 methods (22) (19p13.2 probes) and generated from complex oligo libraries ordered from Custom Array. A stepwise synthesis protocol is described in ref. 21. The Xist RNA FISH probe set was ordered as a set of individually column synthesized oligos from Integrated DNA Technologies. A list of primer sequences used is provided in Table S2.
Cell Culture.
Human WI-38 [CCL-75; American Type Culture Collection (ATCC)], MRC-5 (CCL-171; ATCC), and PGP-1 fibroblasts (GM23248; Coriell Institute) were grown at 37 °C in the presence of 5% CO2 in Dulbecco’s modified Eagle medium (no. 10564; Gibco) supplemented with 10% (vol/vol) serum (no. 10437; Gibco), 50 U/mL penicillin, and 50 μg/mL streptomycin (no. 15070; Gibco). The PGP-1 fibroblasts were also supplemented with MEM nonessential amino acids solution (no. 11140050; Gibco).
Three-Dimensional DNA FISH.
Three-dimensional DNA FISH (59, 60) was essentially performed as described previously (20, 21, 25, 26). WI-38, IMR-90, or PGP-1 fibroblasts were seeded at ∼20% confluence into the wells of Labtek-II Coverglass Chambers or ididi coverglass chambers or onto no.-1.5 coverglass and allowed to grow to ∼70–90% confluence in a mammalian tissue culture incubator. Samples were then rinsed with 1× PBS solution and fixed for 10 min in 1× PBS solution + 4% (wt/vol) paraformaldehyde and then rinsed again with 1× PBS solution. Samples were next permeabilized by a rinse in 1× PBS solution + 0.1% (vol/vol) Tween-20 followed by a 10-min incubation in 1× PBS solution + 0.5% (vol/vol) Triton X-100 and a 5-min incubation in 0.1 N HCl. Samples were then transferred to 2× SSC + 0.1% (vol/vol) Tween-20 (SSCT) and then to 2× SSCT + 50% (vol/vol) formamide. Samples were then incubated in 2× SSCT + 50% formamide at 60 °C for 20–60 min, after which a hybridization solution consisting of 2× SSCT, 50% formamide, 10% (wt/vol) dextran sulfate, 40 ng/µL RNase A (EN0531; Thermo Fisher), and Oligopaint FISH probe sets at 1.6 or 2.5 µM was added. Samples were denatured at 78 °C for 3 min on a water-immersed heat block or flat-block thermocycler (Mastercycler Nexus; Eppendorf) and then allowed to hybridize for more than 24 h at 47–52 °C in a humidified chamber placed in an air incubator or on a flat-block thermocycler. After hybridization, samples were washed in 2× SSCT at 60 °C for 5 min four times and in 2× SSCT at room temperature two times, and then transferred to 1× PBS solution. Unlabeled secondary oligos (25) and tertiary oligos bearing Alexa Fluor 405 and 647 dyes (26) (Table S2) at 0.5–1 µM were subsequently hybridized to the 19p13.2 samples for 1 h in 2× SSC + 30% formamide + 10% dextran sulfate at room temperature and washed three times for 5 min each in 2× SSC + 30% formamide. SlowFade Gold + DAPI (S36938; Thermo Fisher) was added to samples prepared for diffraction-limited imaging. Samples for superresolution imaging were stained in a 1-µg/mL DAPI solution in 1× PBS solution or 2× SSCT for 5 min at 37 °C, followed by a brief rinse in 1× PBS solution or 2× SSCT at room temperature.
RNA FISH.
RNA FISH was performed exactly as described for 3D DNA FISH, except that the 3-min denaturation at 78 °C was replaced with a 5-min incubation at 60 °C, RNase A was omitted from the hybridization buffer, and hybridization was carried out at 42 °C for 16 h.
Metaphase FISH.
Dry microscope slides containing human XX 46N or XY 46N metaphase spreads (AppliedGenetics Laboratories) were immediately immersed in 2× SSCT + 70% (vol/vol) formamide at 70 °C and incubated for 90 s. Slides were immediately transferred to ice-cold 70% (vol/vol) ethanol and incubated for 5 min, transferred to ice-cold 90% (vol/vol) ethanol and incubated for 5 min, then transferred to ice-cold 100% ethanol and incubated for 5 min. Samples were then removed from the 100% ethanol and allowed to air-dry. A hybridization solution (25 µL) consisting of 2× SSCT, 50% formamide, 10% (wt/vol) dextran sulfate, 40 ng/µL RNase A (EN0531; Thermo Fisher), and Oligopaint FISH probe sets at 1.6–3 µM was then added to each slide and sealed beneath a 22 × 22-mm coverslip by using rubber cement. Samples were allowed to hybridize overnight at 45 °C overnight in a humidified chamber. Samples were then washed for 15 min in 2× SSCT at 60 °C and then twice for 5 min each in 2× SSCT at room temperature. At this point, samples receiving the 19p13.2 probes were additionally hybridized with secondary and tertiary oligos at 1.4 µM each (Table S2) in 2× SSCT + 30% (vol/vol) formamide for 1 h at room temperature, washed for 10 min in 2× SSCT + 40% (vol/vol) formamide at room temperature, and then washed twice for 2 min each in 2× SSCT + 40% formamide at room temperature. Samples were mounted with SlowFade Gold + DAPI and sealed beneath a 22 × 30-mm coverslip by using nail polish.
Diffraction-Limited Imaging.
Diffraction-limited imaging of 3D DNA FISH samples was conducted on an inverted Zeiss Axio Observer Z1 using a 63× Plan-Apochromat Oil differential interference contrast (N.A. 1.40) objective. Samples were illuminated by using Colibri light source using a 365-nm, 470-nm, 555-nm, or 625-nm LED. DAPI was visualized by using a filter set composed of a 365-nm clean-up filter (Zeiss G 365), a 395-nm long-pass dichroic mirror (Zeiss FT 395), and a 445/50 nm band-pass emission filter (Zeiss BP 445/50). ATTO 488 was visualized by using a filter set composed of a 470/40-nm excitation filter (Zeiss BP 470/40), a 495-nm long-pass dichroic mirror (Zeiss FT 495), and a 525/50-nm band-pass emission filter (Zeiss BP 525/50). ATTO 565 was visualized by using a filter set composed of a 545/25-nm excitation filter (Zeiss BP 545/25), a 570-nm long-pass dichroic mirror (Zeiss FT 570), and a 605/70-nm band-pass emission filter (Zeiss BP 605/70). Alexa Fluor 647 was visualized by using a filter set composed of a 640/30-nm excitation filter (Zeiss BP 640/30), a 660-nm long-pass dichroic mirror (Zeiss FT 660), and a 690/50 nm band-pass emission filter (Zeiss BP 690/50). Images were acquired by using a Hamamatsu Orca-Flash 4.0 sCMOS camera with 6.5-µm pixels, resulting in an effective magnified pixel size of 103 nm. Z-stacks were acquired by using an interval of 240 nm. Images were processed by using Zeiss Zen software and Fiji/ImageJ (69). Metaphase FISH images were captured on a Nikon Eclipse Ti-E microscope by using a CFI PlanApo 100× Oil (N.A. 1.45) objective. Samples were illuminated by using a Spectra X LED system (Lumencor) using a 395/25-nm, 295-mW LED (DAPI); 470/24-nm, 196-mW LED (ATTO 488); 550/15-nm, 260-mW LED (Cy3B); or a 640/30-nm, 231-mW LED (Alexa Fluor 647). Illumination light was spectrally filtered and directed to the objective, and emission light was spectrally filtered and directed to the camera by one of four filter cubes from Semrock: BFP-A-Basic-NTE, DAPI; FITC-2024B-NTE-ZERO, ATTO 488; TRITC-B-NTE-0, Cy3B; or Cy5-4040C-NTE-ZERO, Alexa Fluor 647. Images were acquired by using an Andor Zyla 4.2+ sCMOS camera with 6.5-µm pixels, resulting in an effective magnified pixel size of 65 nm. Z-stacks were acquired by using an interval of 200 nm. Images were processed by using Nikon Elements software and Fiji/ImageJ.
Automated Quantification of FISH Signals.
Raw, multichannel .czi Z-stacks were inputted into Fiji/ImageJ, in which a macro was used to create maximum-intensity projection in Z .png images for the DAPI and FISH signal channels. These .png images were inputted into CellProfiler 3.0 (61), in which an automated image-analysis pipeline was constructed to identify nuclei FISH signals, the pixels in the FISH image overlapping with the nucleus but not part of the FISH foci (i.e., the nuclear background of the FISH signal), and the baseline background of pixels in the FISH image not overlapping with the FISH foci, nuclei, cell bodies, or other objects of increased intensity such as debris. A parent–child relationship was also established between nuclei and FISH signals. From these data, a background-subtracted SNR was calculated as follows: (mean FISH signal pixel intensity − mean baseline background pixel intensity)/(mean nuclear background pixel intensity − mean baseline background pixel intensity). The parent–child relationship was used to determine the number of FISH signals in each nucleus. The complete CellProfiler pipeline used, as well as example images for Xq28 and 19p13.2, are available at https://github.com/brianbeliveau/OligoMiner/tree/master/ImageQuantification.
STORM Imaging.
STORM imaging was performed on a commercial Nikon N-STORM 3.0 microscope featuring a Perfect Focus System and a motorized total internal reflection fluorescence (TIRF) illuminator at the Nikon Imaging Center located at Harvard Medical School. STORM was performed by using highly inclined and laminated optical sheet illumination (HILO) (70) and with pulsed activation of the 405-nm laser, followed by 647 nm, and then 561 nm. Light was focused through a CFI Apo TIRF 100× oil (N.A. 1.49) objective. The 561-nm laser was used at 2% (out of 50 mW) to image 200-nm orange FluoSpheres (F8809; Thermo Fisher), which were used as fiducial makers to facilitate drift correction. The 405-nm laser was used to enhance the blinking rate at 0–5% (out of 20 mW), and the 647-nm laser was used at 100% power (out of 125 mW measured at fiber optic). Emission light was spectrally filtered (Chroma ET600/50m for 561 nm; Chroma ET700/75m for 647 nm) and imaged on an EMCCD camera (Andor iXon ×3 DU-897) with 16-µm pixels using a CCD readout bandwidth of 10 MHz at 16 bit, 1 bit preamp gain, and no electron-multiplying gain on the center 256 × 256 or 186 × 190 pixels, resulting in an effective pixel size of 160 nm. A total of 6,250 or 12,500 10-ms frames were acquired. Single-molecule localization events were identified by using in-house MATLAB software (71) that calls a 2D fitting algorithm (72). Individual localization events were blurred with 2D Gaussian functions whose “sigma” parameter was set according to the global drift-independent localization precision as determined by nearest neighbor-based analysis (NeNA) (73). NeNA values were as follows: 19p13.2 1035 kb– 12.6 nm sigma, 29.6 nm supported resolution; 19p13.2 20 kb– 11.8 nm sigma, 27.6 nm supported resolution. One- and two-component Gaussian fits of the line traces presented in Fig. 4 A–D were calculated by using the “Gaussian Mixture Model” module in scikit-learn (55).
DNA-PAINT Imaging.
DNA-PAINT imaging was performed on a commercial Nikon N-STORM 3.0 microscope featuring a Perfect Focus System and a motorized TIRF illuminator. DNA-PAINT was performed by using HILO with 15–30% of a 200-mW, 561-nm laser (Coherent Sapphire) using a CFI Apo TIRF 100× oil (N.A. 1.49) objective at an effective power density of ∼0.5–1 kW/cm2. The 561-nm laser excitation light was passed through a clean-up filter (Chroma ZET561/10) and directed to the objective by using a multiband beam splitter (Chroma ZT405/488/561/647rpc). Emission light was spectrally filtered (Chroma ET600/50m) and imaged on an EMCCD camera (Andor iXon ×3 DU-897) with 16-µm pixels by using a CCD readout bandwidth of 3 MHz at 14 bit, 5.1 preamp gain, and no electron-multiplying gain on the center 256 × 256 pixels, resulting in an effective pixel size of 160 nm. A total of 15,000 100-ms frames were acquired for each image by using 1–3 nM of Cy3B-labeled 10mer oligo in 1× PBS solution + 125–500 nM NaCl. Gold nanoparticles (40 nm; no. 753637; Sigma-Aldrich) were used as fiducial markers to facilitate drift correction. Single-molecule localization events were identified by using in-house MATLAB software (71) that calls a 2D fitting algorithm (72). Individual localization events were blurred with 2D Gaussian functions whose “sigma” parameter was set according to the global drift-independent localization precision as determined by NeNA (73). NeNA values were as follows: Xq28– 5.6 nm sigma, 13.2 nm supported resolution; Xist RNA– 5.1 nm sigma, 12.0 nm supported resolution. One- and two-component Gaussian fits of the line traces presented in Fig. 4 E–H were calculated by using the “Gaussian Mixture Model” module in scikit-learn (55).
Supplementary Material
Acknowledgments
We thank Ninning Liu, Mingjie Dai, Thomas C. Ferrante, Josh Rosenberg, Nikhil Gopalkrishnan, Florian Schüder, Ralf Jungmann, Jesse Silverberg, Sungwook Woo, and members of the laboratories of P.Y. and C.-t.W. for helpful discussions; Jin Billy Li for the idea to use k-mer filtering as a means of specificity checking; and Geoffrey Fudenberg for assistance with the 19p13.2 probe design. This work was supported by National Institutes of Health Awards 1R01EB018659-01 (to P.Y.), 1-U01-MH106011-01 (to P.Y.), DP1GM106412 (to C.-t.W.), and R01HD091797 (to C.-t.W.); Office of Naval Research Awards N00014-13-1-0593 (to P.Y.), N00014-14-1-0610 (to P.Y.), N00014-16-1-2182 (to P.Y.), and N00014-16-1-2410 (to P.Y.); National Science Foundation (NSF) Awards CCF-1054898 and CCF-1317291 (to P.Y.); a Damon Runyon Cancer Research Foundation Fellowship (to B.J.B.); a Uehara Memorial Foundation Research Fellowship (to H.M.S.); postdoctoral fellowships from the European Molecular Biology Organization (to S.K.S.) and the Human Frontier Science Program (to S.K.S.); and NSF Graduate Research Fellowships (to J.Y.K. and S.C.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genome-scale probe mining sets are available at oligominer.net.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714530115/-/DCSupplemental.
References
- 1.Pardue ML, Gall JG. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci USA. 1969;64:600–604. doi: 10.1073/pnas.64.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.John HA, Birnstiel ML, Jones KW. RNA-DNA hybrids at the cytological level. Nature. 1969;223:582–587. doi: 10.1038/223582a0. [DOI] [PubMed] [Google Scholar]
- 3.Buongiorno-Nardelli M, Amaldi F. Autoradiographic detection of molecular hybrids between RNA and DNA in tissue sections. Nature. 1970;225:946–948. doi: 10.1038/225946a0. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence JB, Singer RH. Quantitative analysis of in situ hybridization methods for the detection of actin gene expression. Nucleic Acids Res. 1985;13:1777–1799. doi: 10.1093/nar/13.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Ploeg M. Cytochemical nucleic acid research during the twentieth century. Eur J Histochem. 2000;44:7–42. [PubMed] [Google Scholar]
- 6.Levsky JM, Singer RH. Fluorescence in situ hybridization: Past, present and future. J Cell Sci. 2003;116:2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 7.Riegel M. Human molecular cytogenetics: From cells to nucleotides. Genet Mol Biol. 2014;37(suppl):194–209. doi: 10.1590/s1415-47572014000200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rigby PWJ, Dieckmann M, Rhodes C, Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977;113:237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- 9.Langer PR, Waldrop AA, Ward DC. Enzymatic synthesis of biotin-labeled polynucleotides: Novel nucleic acid affinity probes. Proc Natl Acad Sci USA. 1981;78:6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landegent JE, Jansen in de Wal N, Dirks RW, Baao F, van der Ploeg M. Use of whole cosmid cloned genomic sequences for chromosomal localization by non-radioactive in situ hybridization. Hum Genet. 1987;77:366–370. doi: 10.1007/BF00291428. [DOI] [PubMed] [Google Scholar]
- 11.Moyzis RK, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matera AG, Ward DC. Oligonucleotide probes for the analysis of specific repetitive DNA sequences by fluorescence in situ hybridization. Hum Mol Genet. 1992;1:535–539. doi: 10.1093/hmg/1.7.535. [DOI] [PubMed] [Google Scholar]
- 13.Dernburg AF, et al. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 14.Dirks RW, et al. Simultaneous detection of different mRNA sequences coding for neuropeptide hormones by double in situ hybridization using FITC- and biotin-labeled oligonucleotides. J Histochem Cytochem. 1990;38:467–473. doi: 10.1177/38.4.2108203. [DOI] [PubMed] [Google Scholar]
- 15.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 16.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosuri S, Church GM. Large-scale de novo DNA synthesis: Technologies and applications. Nat Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada NA, et al. Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet Genome Res. 2011;132:248–254. doi: 10.1159/000322717. [DOI] [PubMed] [Google Scholar]
- 19.Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 2011;19:901–909. doi: 10.1007/s10577-011-9245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beliveau BJ, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci USA. 2012;109:21301–21306. doi: 10.1073/pnas.1213818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beliveau BJ, Apostolopoulos N, Wu CT. Visualizing genomes with Oligopaint, FISH probes. Curr Protoc Mol Biol. 2014;105:Unit 14.23.1. doi: 10.1002/0471142727.mb1423s105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murgha Y, et al. Combined in vitro transcription and reverse transcription to amplify and label complex synthetic oligonucleotide probe libraries. Biotechniques. 2015;58:301–307. doi: 10.2144/000114298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beliveau BJ, et al. In situ super-resolution imaging of genomic DNA with oligoSTORM and oligoDNA-PAINT. Methods Mol Biol. 2017;1663:231–252. doi: 10.1007/978-1-4939-7265-4_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beliveau BJ, et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun. 2015;6:7147. doi: 10.1038/ncomms8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boettiger AN, et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kundu S, et al. Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol Cell. 2017;65:432–446.e5. doi: 10.1016/j.molcel.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353:598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah S, Lubeck E, Zhou W, Cai L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron. 2016;92:342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eng CL, Shah S, Thomassie J, Cai L. Profiling the transcriptome with RNA SPOTs. Nat Methods. 2017;14:1153–1155. doi: 10.1038/nmeth.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pernthaler J, Glöckner FO, Schönhuber W, Amann R. Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes. Methods Microbiol. 2001;30:207–226. [Google Scholar]
- 32.Yilmaz LS, Parnerkar S, Noguera DR. mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl Environ Microbiol. 2011;77:1118–1122. doi: 10.1128/AEM.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogan PK, Cazcarro PM, Knoll JHM. Sequence-based design of single-copy genomic DNA probes for fluorescence in situ hybridization. Genome Res. 2001;11:1086–1094. doi: 10.1101/gr.171701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navin N, et al. PROBER: Oligonucleotide FISH probe design software. Bioinformatics. 2006;22:2437–2438. doi: 10.1093/bioinformatics/btl273. [DOI] [PubMed] [Google Scholar]
- 35.Nedbal J, Hobson PS, Fear DJ, Heintzmann R, Gould HJ. Comprehensive FISH probe design tool applied to imaging human immunoglobulin class switch recombination. PLoS One. 2012;7:e51675. doi: 10.1371/journal.pone.0051675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bienko M, et al. A versatile genome-scale PCR-based pipeline for high-definition DNA FISH. Nat Methods. 2013;10:122–124. doi: 10.1038/nmeth.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banér J, et al. Parallel gene analysis with allele-specific padlock probes and tag microarrays. Nucleic Acids Res. 2003;31:e103. doi: 10.1093/nar/gng104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenberg J, Nilsson M, Landegren U. ProbeMaker: An extensible framework for design of sets of oligonucleotide probes. BMC Bioinformatics. 2005;6:229. doi: 10.1186/1471-2105-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: Design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cock PJA, et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 42.Smit A, Hubley R, Green P. 2013 RepeatMasker Open-4.0. 2013–2015. Available at repeatmasker.org. Accessed September 23, 2015.
- 43.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg SL. Bowtie: An ultrafast memory-efficient short read aligner. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dirks RM, Pierce NA. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J Comput Chem. 2003;24:1664–1677. doi: 10.1002/jcc.10296. [DOI] [PubMed] [Google Scholar]
- 53.Dirks RM, Pierce NA. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J Comput Chem. 2004;25:1295–1304. doi: 10.1002/jcc.20057. [DOI] [PubMed] [Google Scholar]
- 54.Dirks RM, Bois JS, Schaeffer JM, Winfree E, Pierce NA. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 2007;49:65–88. [Google Scholar]
- 55.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. doi: 10.1007/s13398-014-0173-7.2. [DOI] [Google Scholar]
- 56.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arvey A, et al. Minimizing off-target signals in RNA fluorescent in situ hybridization. Nucleic Acids Res. 2010;38:e115. doi: 10.1093/nar/gkq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moffitt JR, et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci USA. 2016;113:11046–11051. doi: 10.1073/pnas.1612826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solovei I, et al. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH) Exp Cell Res. 2002;276:10–23. doi: 10.1006/excr.2002.5513. [DOI] [PubMed] [Google Scholar]
- 60.Solovei I, Cremer M. 3D-FISH on cultured cells combined with immunostaining. Methods Mol Biol. 2010;659:117–126. doi: 10.1007/978-1-60761-789-1_8. [DOI] [PubMed] [Google Scholar]
- 61.Carpenter AE, et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jungmann R, et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 64.Godin AG, Lounis B, Cognet L. Super-resolution microscopy approaches for live cell imaging. Biophys J. 2014;107:1777–1784. doi: 10.1016/j.bpj.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown CJ, et al. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 66.El Baidouri M, et al. A new approach for annotation of transposable elements using small RNA mapping. Nucleic Acids Res. 2015;43:e84. doi: 10.1093/nar/gkv257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shirley M, Ma Z, Pedersen B, Wheelan S. 2015. Efficient “pythonic” access to FASTA file using pyfaidx. PeerJ Prepr, 1–4.
- 68.Waskom M, et al. 2014. seaborn: v0.5.0 (November 2014). 10.5281/zenodo.12710.
- 69.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 71.Dai M, Jungmann R, Yin P. Optical imaging of individual biomolecules in densely packed clusters. Nat Nanotechnol. 2016;11:798–807. doi: 10.1038/nnano.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith CS, Joseph N, Rieger B, Lidke KA. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat Methods. 2010;7:373–375. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Endesfelder U, Malkusch S, Fricke F, Heilemann M. A simple method to estimate the average localization precision of a single-molecule localization microscopy experiment. Histochem Cell Biol. 2014;141:629–638. doi: 10.1007/s00418-014-1192-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.