Abstract
Aggressive breast cancer is difficult to treat as it is unresponsive to many hormone-based therapies; therefore, it is imperative to identify novel, targetable regulators of progression. Long non-coding RNAs (lncRNAs) are important regulators in breast cancer and have great potential as therapeutic targets; however, little is known about how the majority of lncRNAs function within breast cancer. This study, characterizes a novel lncRNA, MANCR (mitotically-associated long non-coding RNA; LINC00704), which is upregulated in breast cancer patient specimens and cells. Depletion of MANCR in triple-negative breast cancer (TNBC) cells significantly decreases cell proliferation and viability, with concomitant increases in DNA damage. Transcriptome analysis, based on RNA sequencing (RNA-seq), following MANCR knockdown reveals significant differences in the expression of >2000 transcripts, and gene set enrichment analysis (GSEA) identifies changes in multiple categories related to cell cycle regulation. Furthermore, MANCR expression is highest in mitotic cells by both RT-qPCR and RNA in situ hybridization. Consistent with a role in cell cycle regulation, MANCR-depleted cells have a lower mitotic index and higher incidences of defective cytokinesis and cell death. Taken together, these data reveal a role for the novel lncRNA, MANCR, in genomic stability of aggressive breast cancer, and identify it as a potential therapeutic target.
Implications
The novel lncRNA, MANCR (LINC00704), is upregulated in breast cancer and is functionally linked with cell proliferation, viability, and genomic stability.
Keywords: MANCR, LINC00704, mitosis, DNA damage, cytokinesis
Introduction
Breast cancer is a major health concern worldwide; it is the most frequently diagnosed cancer and the leading cause of cancer-related death in women (1). There are several subtypes of breast cancer, defined by their expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (Her2). The most common subtypes, luminal A and B are both ER+/PR+ and have the best prognosis, as they are generally low-grade and responsive to hormone therapy (2). The Her2 over-expression subtype grows faster, but is responsive to targeted drug treatments such as Herceptin (3). A third subtype, basal or triple negative breast cancer (TNBC), is characterized as being ER-/PR-/Her2-. Although TNBC is diagnosed in only 15% of breast cancer patients, it is the most aggressive subtype, unresponsive to treatment, and usually has a poor prognosis (4). As such, it remains critical to identify and characterize novel, targetable regulators of breast cancer. One novel class of regulators that hold great clinical potential are the epigenetic regulators, long non-coding RNAs (lncRNA).
lncRNAs encompass a broad class of transcripts loosely defined as being greater than 200 nucleotides in length and lacking protein-coding potential. They are transcribed by RNA polymerase II, can have multiple splice variants and their expression is regulated by transcription factors. Expression of lncRNAs is more cell-type and tissue specific than their mRNA protein-coding counterparts (5). Once dismissed as transcriptional noise, lncRNAs are involved in a variety of biological processes, such as X-chromosome inactivation by Xist (6,7), genomic imprinting by H19 (8,9), and mammary epithelial differentiation by Zfas1 (10).
lncRNAs are often deregulated in cancer (11). HOTAIR is a well-studied lncRNA that is overexpressed in a variety of cancers (12). It promotes cancer invasiveness and metastasis by interacting with Polycomb Repressive Complex 2 (PRC2), altering histone H3K27me3 and silencing anti-metastatic genes (13). In addition to pan-cancer lncRNAs, several studies have identified signatures of lncRNAs associated with specific breast cancer stages and subtypes (14-17). While these studies have identified hundreds of cancer-associated lncRNAs, such as DSCAM-AS1, an ER-regulated lncRNA that has been implicated in luminal breast cancer and tamoxifen resistance (18,19), relatively few have been functionally studied.
Here, we characterize a novel lncRNA associated with aggressive breast cancer and significantly reduced patient survival, designated MANCR (Mitotically Associated Non-Coding RNA). MANCR was originally identified in our transcriptome profiling of a series of breast cancer cell lines (20). We demonstrate overexpression of MANCR in TNBC cell lines as well as in patient samples, compared to very low levels in normal mammary epithelial and early stage (ER+) positive breast cancer cells. The results of our functional analyses show a striking reduction in cell growth and an induction of cell death upon MANCR knockdown in MDA-MB-231 cells. Additionally, loss of MANCR leads to increased incidence of DNA damage and defective cytokinesis. These results reveal that MANCR may have a cytoprotective role in supporting and/or sustaining breast cancer growth.
Materials and Methods
Cell culture
MCF-10A cells were grown in DMEM/F12 (Corning: 10090CV) supplemented with 5% horse serum (Gibco: 16050), 10 ug/ml human insulin (Sigma Aldrich: I-1882), 20 ng/ml recombinant hEGF (Peprotech: AF-100-15), 100 ng/ml cholera toxin (Sigma: C-8052), 0.5 ug/ml hydrocortisone (Sigma: H-0888), 1% penicillin/streptomycin (Life Technologies: 15140-122), and 2 mM glutamine (Life Technologies: 25030-081). MDA-MB-231 cells were grown in DMEM/F12 supplemented with 10% fetal bovine serum (Atlanta Biologicals, S11550), 1% penicillin/streptomycin, and 2 mM glutamine.
Cell line authentication
MCF-10A cells were a kind gift from Jeffrey Nickerson in 2011. MDA-MB-231 cells were purchased from ATCC in 2006. MCF-10A and MDA-MB-231 cell lines were validated by short tandem repeat (STR) analysis in 2012 using the Promega GenePrint 10 System at the UVM Cancer Center DNA Analysis Facility. Cell lines tested negative for mycoplasma using MycoAlert (Lonza: LT07-28) in August 2016. All experiments were performed within 10 passages after thawing cells.
RNA extraction
Total RNA was isolated from cells using Trizol (Life Technologies) and purified using the Direct-zol RNA kit with DNaseI treatment (Zymo Research, Irvine, CA, USA: R2050) according to the manufacturer's instructions. RNA quality and quantity were assessed using the RNA 6000 Nano Kit with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA quantity was further assessed using a Nanodrop2000 (Thermo Scientific, Lafayette, CO) and Qubit HS RNA assay (Thermo Fisher Scientific).
Library and sequencing preparation
Total RNA was depleted of ribosomal RNA, reverse transcribed and strand-specific adapters added using the TruSeq Stranded Total RNA Library Prep kit with Ribo-Zero Gold (Illumina, San Diego, CA, USA) according to manufacturer's protocol except to reduce over-amplification, final cDNA libraries were amplified using the Real-time Library Amplification Kit (Kapa Biosystems, Wilmington, MA, USA). Generated cDNA libraries were assayed for quality using the High Sensitivity DNA Kit on the Agilent 2100 Bioanalyzer (Agilent Technologies) and sequenced as single-end 100 bp reads (IlluminaHiSeq1000, UVM Advanced Genome Technologies Core).
Bioinformatics analysis
Sequence files (fastq) were mapped to the most recent assemblies of the human genome (hg38) using STAR (21). Expression counts were determined by HTSeq (22) with Gencode v25 gene annotation (23). Differential expression was analyzed by DESeq2 (24). Correlation between replicates and differential gene expression between samples was assessed by principal component analysis (PCA). Differentially expressed mRNAs after MANCR knockdown in MDA-MB-231 cells and differentially expressed mRNAs between MCF-10A and MDA-MB-231 cells (GSE75168) (25) were analyzed for gene set enrichment using Gene Set Enrichment Analysis (GSEA) for the GO biological processes v6 Molecular Signatures Database (MSigDB) using default parameters with the exception that permutation type was set to gene set (26,27). To create the gene set enrichment map, GSEA results were input into the EnrichmentMap app (28) for Cytoscape (www.cytoscape.org). The following parameters were used: P-value ≤ 0.0001, FDR Q-value ≤ 0.05, Overlap Coefficient ≥ 0.8. RNA-Seq datasets have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE102155.
Gene expression analysis
Complementary DNA (cDNA) was synthesized from isolated RNA using the Superscript III First-Strand Synthesis System according to the manufacturer's instructions (Life Technologies, Carlsbad, CA). Quantitative RT-PCR (RT-qPCR) was performed using gene-specific primers and SYBR Green Master Mix (Bio Rad, Hercules, CA) in an Applied Biosystems Viia 7 system (Life Technologies, Carlsbad, CA). After normalization to the reference genes glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and actin, relative expression levels of each target gene were calculated using the comparative CT (ΔΔCT) method. See Supplemental Table 1 for oligonucleotide primer sequences used for RT-qPCR.
Transfections
MDA-MB-231 cells were transfected at 70-80% confluence with 25 nM negative control A GapmeR (Control ASO) or GapmeRs targeting MANCR ASO_1 (5′-CCGAAACTTGCCATTT-3′) and ASO_2 (5′-CGAGTGGTGAGTGGAT-3′) (Exiqon, Woburn, MA) using DharmaFECT 4 (Dharmacon) according to the manufacturer's protocol. Cells were harvested for RNA and protein analyses 24 or 48 hr after transfection.
Western blot
Adherent cells were directly lysed in RIPA buffer (50 mM Tris pH 8, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% Triton X-100) plus cOmplete, protease inhibitor cocktail (Roche). Proteins were separated by 12% SDS-PAGE and transferred to 0.45 μm PVDF membranes (Millipore, Billerica, MA). Blots were blocked using 5% milk in TBST and incubated with primary antibodies overnight at 4°C, then incubated with HRP-conjugated secondary antibodies for 1 hr. Proteins were detected with an ECL reagent and the Chemidoc XRS+ imaging system (Bio Rad, Hercules, CA). The following primary antibodies were used: anti-cleaved caspase 3 (rabbit polyclonal, 1:1000) (Cell Signaling, #9664) and anti-alpha-tubulin (mouse monoclonal, 1:500) (Sigma, #T9026). Secondary antibodies used were: HRP-goat anti-mouse and HRP-goat anti-rabbit (Santa Cruz Biotechnology, Dallas, TX).
Growth assay
Cells were plated in one 6-well plate per time point and transfected as described above. Cells were trypsinized and counted using a Countess Automated Cell Counter at each time point; 0 hr is at transfection, 24 hr after transfection, and 48 hr after transfection.
Cell synchronization by nocodazole block
Cells were plated in one 100 mm plate per time point and allowed to adhere. Cells were treated with 100 ng/ml nocodazole for 18 hr to synchronize cells in G2/M. After 18 hr, cells were released from the block: media was collected and spun down at 200g for 5 min, cells were washed twice with PBS, and were re-plated in fresh media. At each time point; 0 hr (at release), 6 hr, 12 hr, 18 hr and 24 hr, cells were harvested by media collection and trypsinization, spun down, and washed twice with PBS. Harvested cells were split into two batches, one for gene expression analysis and one for cell cycle analysis by flow cytometry.
Flow cytometry analysis
Cells were harvested by trypsinization and fixed in ice cold 75% ethanol for 30 min at 4°C. Then cells were permeabilized with permeabilization buffer (0.25% Triton X-100 in PBS) for 15 min at room temperature. For mitotic indexing, cells were incubated with AF647- conjugated antibody against H3S28p (BD Biosciences: 558609) diluted 1:50 in permeabilization buffer for 30 min at room temperature in the dark. For mitotic indexing and cell cycle analysis, cells were stained with propidium iodide (PI/RNase staining buffer, BD Biosciences: 550825) for 15 min at room temperature in the dark. Flow cytometry was performed using an LSRII instrument (BD Biosciences). Flowjo v10 (Ashland, OR, http://www.flowjo.com/) was used to determine the percent of H3S28P-positive cells and to display DNA histograms.
RNA in situ hybridization
RNA chromogenic in situ hybridization (RNA CISH) was performed using RNAscope reagents, a HybEz oven, and a probe targeting MANCR (Hs-LINC00704, cat# 411081) (Advanced Cell Diagnostics, Hayward, California, USA), according to the manufacturer's protocols. Positive control assays were performed using a Homo sapiens PPIB probe, and negative control assays were performed using an Escherichia coli dapB probe. Slides were imaged with a Zeiss Axioscope bright-field microscope, and images were captured using Zen2012 software (Zeiss Inc.)
RNA fluorescence in situ hybridization (RNA FISH) was performed using ViewRNA ISH reagents and a custom designed probe targeting MANCR (Affymetrix), according to the manufacturer's protocol. The nuclei were counterstained with DAPI. RNase A pretreatment was included to confirm probe hybridization to RNA. Images were obtained using a Zeiss LSM 510 META confocal microscope using a 63× oil immersion objective. Image analyses were performed using Volocity software (PerkinElmer).
Immunofluorescence
Cells grown on coverslips were fixed in 1% paraformaldehyde in methanol on ice for 10 minutes. Fixed cells were immunofluorescently labeled with the following primary and secondary antibodies:anti-53BP1 (rabbit polyclonal, 1:200) (Santa Cruz Biotechnology: sc-22760), anti-γH2AX-S139 (mouse monoclonal, 1:200) (EMD Millipore: 05-636), goat anti-mouse IgG (H+L) Alexa Fluor 594, and goat anti-rabbitIgG (H+L) Alexa Fluor 488. The nuclei were counterstained with DAPI. Cells were imaged on a Zeiss AxioImager. Z2 equipped with Hamamatsu CCD camera, and images were captured using Zen2012 software. Image analyses were performed using ImageJ (https://imagej.nih.gov/).
Live cell imaging
MDA-MB-231 cells were cultured in 4-chambered, glass bottom 35 mm dishes (Greiner Bio-One: cat# 627975). Cells were transfected with Control ASO (2 chambers) or MANCR ASO_2 (2 chambers) as described above, and 16 hr later were changed to CO2-independent media with 10% FBS (Life Technologies) for imaging. Multiple fields of cells (n ≥ 4/chamber) were imaged at 2-minute intervals by differential interference contrast microscopy for up to 16 hours on a temperature controlled Eclipse Ti microscope (Nikon) equipped with Clara CCD and iXon X3 EMCCD cameras (Andor), Plan APO 40× 0.95 NA objective, and NIS Elements software (Nikon).
Gene expression database mining
Level 3 data from The Cancer Genome Atlas (TCGA)-BRCA (29) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (30,31) was accessed using cBioPortal for cancer genomics (www.cbioportal.org) (32,33).
Statistical analyses
Statistical analyses were performed using GraphPad Prism v7.01.
Results
Identification of the TNBC-associated lncRNA MANCR
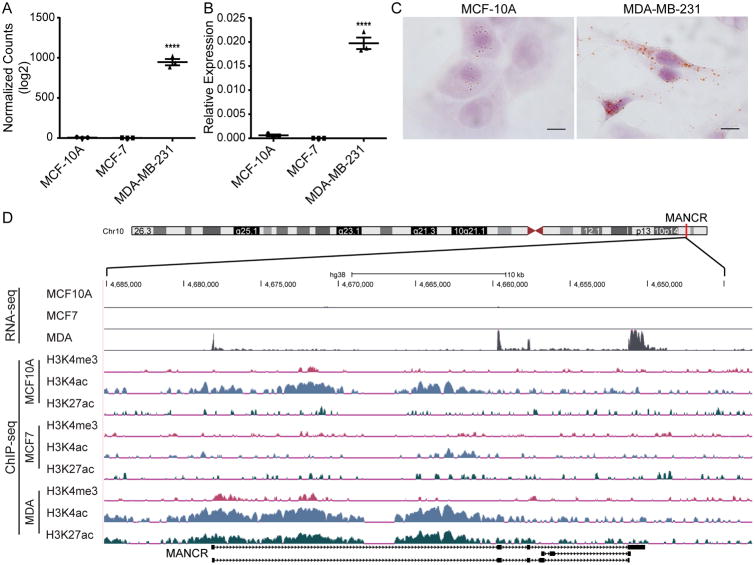
MANCR (also termed LINC00704) is an intergenic lncRNA encoded at chromosome 10p15.1 (Fig 1); the nearest protein-coding gene is more than 100kb away (AKR1E2, aldo-keto reductase family 1 member E2). MANCR was selected for further investigation based on its high expression by both RNA-seq (Fig 1A) and RT-qPCR (Fig 1B) in MDA-MB-231 cells compared to MCF-10A and MCF-7. To gain insight into MANCR function, we first examined the cellular localization of MANCR by performing RNA in situ hybridization (RNA-ISH) on MCF-10A and MDA-MB-231 cells. RNA-ISH revealed that MANCR is both nuclear and cytoplasmic, and provided further confirmation that expression is higher in MDA-MB-231 than MCF-10A cells (Fig 1C). To confirm transcriptional activation of MANCR in MDA-MB-231, we examined histone modifications that are associated with activation, H3K4 tri-methylation (H3K4me3), H3K4 acetylation (H3K4ac), and H3K27 acetylation (H3K27ac) that were previously profiled in breast cancer cells (25,34). In MDA-MB-231 cells, there are H3K4me3, H3K4ac, and H3K27ac marks across the gene, indicative of active transcription. The MCF-10A cells have only the H3K4ac mark, whereas the MCF-7 cells (ER+) lack the three histone marks, consistent with lower expression in these two cell lines (Fig 1D).
Figure 1. MANCR expression is increased in MDA-MB-231 cells.
A) Log2 values of DESeq2 normalized counts of MANCR from RNA-seq in MCF-10A, MCF-7, and MDA-MB-231 cells. Data are presented as mean ± S.E. Statistical significance was determined using one-way ANOVA with multiple comparisons. ****p<0.0001. B) MANCR expression by RT-qPCR in three breast cell lines, MCF-10A (normal-like), MCF-7 (luminal), and MDA-MB-231 (basal), n=3. MANCR expression is relative to GAPDH and ACTB. Data are presented as mean ± S.E. Statistical significance was determined using one-way ANOVA with multiple comparisons. ****p<0.0001. C) Representative images from RNA-CISH of MANCR in MCF-10A (left panel) and MDA-MB-231 cells (right panel). Brown spots indicate expressed RNA, cells are counterstained with hematoxylin. Scale bars represent 5 μm. D) UCSC genome browser tracks (genome build hg38) of RNA-seq and H3K4me3, H3K4ac, and H3K27ac ChIP-seq peaks across the MANCR gene locus in MCF-10A, MCF-7, and MDA-MB-231 cells. The scale for the RNA-seq tracks is 0-50 reads per million, purple bars indicate peaks that go beyond the scale. The scale for all ChIP-seq tracks is 0-2 log10 enrichment. MANCR is located at Chr10 p15.1 (indicated by the red bar on the chromosome ideogram) on the negative strand; the depicted tracks have been flipped to read left to right.
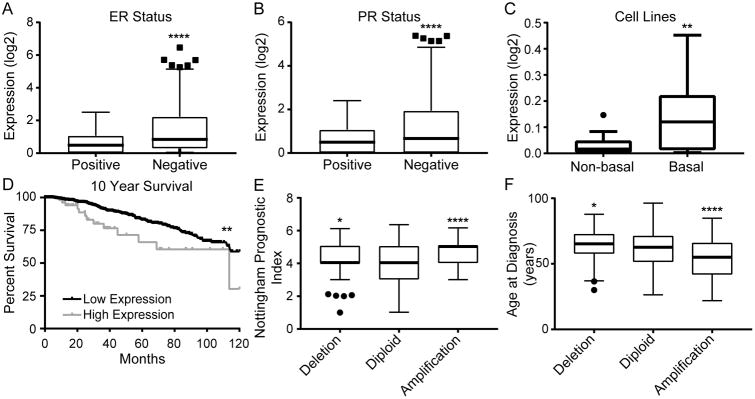
To determine the clinical relevance of MANCR, we queried breast cancer patient sample data from The Cancer Genome Atlas (TCGA) Research Network (29) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (30,31) using cBioPortal (32,33). Analysis of TCGA data revealed that MANCR expression is higher in ER- and PR- tumors (Fig 2A and B). We observed a similar trend in a panel of breast cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE); MANCR expression is higher in basal breast cancer cell lines compared to non-basal cell lines (Fig 2C). Furthermore, patients with high MANCR expression have a ten-year survival of 30% as compared to 59% for patients with low MANCR expression (Fig 2D). The five-year survival is also significantly decreased from 83% in patients with low expression to 66% in patients with high expression. Consistent with decreased survival, the copy number variant analysis from METABRIC revealed that patients with amplified MANCR have a higher Nottingham Prognostic Index, indicating a worse prognosis (Fig 2E). Finally, patients with amplification were also diagnosed at a younger age, a mean of 54 yr compared to 61.6 yr for patients with diploid tumors (Fig 2F). Taken together, these data suggest that MANCR contributes to a more aggressive disease phenotype in breast cancer patients.
Figure 2. High levels of MANCR are associated with a poor prognosis in breast cancer patients.
A,B) Log2 expression levels of MANCR by RNA-seq in TCGA breast cancer patient samples separated according to (A) estrogen receptor (ER) status; positive n=530, negative n=144; or (B) progesterone receptor (PR) status; positive n=463, negative n=214. Data are presented as median, interquartile range (IQR), and ± 1.5IQR. Statistical significance was determined using unpaired t-test. ****p<0.0001. C) Log2 expression levels by RNA-seq of MANCR in Cancer Cell Line Encyclopedia (CCLE) breast cancer cell lines that have been identified as either non-basal or basal. Data are presented as median, IQR, and ± 1.5IQR. Statistical significance was determined using unpaired t-test. **p<0.01. D) Kaplan-Meier curve of ten-year survival data for breast cancer patients stratified by MANCR expression; 56 patients with high expression and 904 patients with low expression. Statistical significance was determined using log-rank test. **p<0.01. E,F) Nottingham Prognostic Index (E) and age at diagnosis (F) in METABRIC breast cancer patient samples categorized by their MANCR copy number variation; deletion n=116, diploid n=1481, amplification n=91. Data are presented as median, IQR, and ± 1.5IQR. Statistical significance was determined using one-way ANOVA with multiple comparisons. *p<0.05, ****p<0.0001.
MANCR alters cell survival
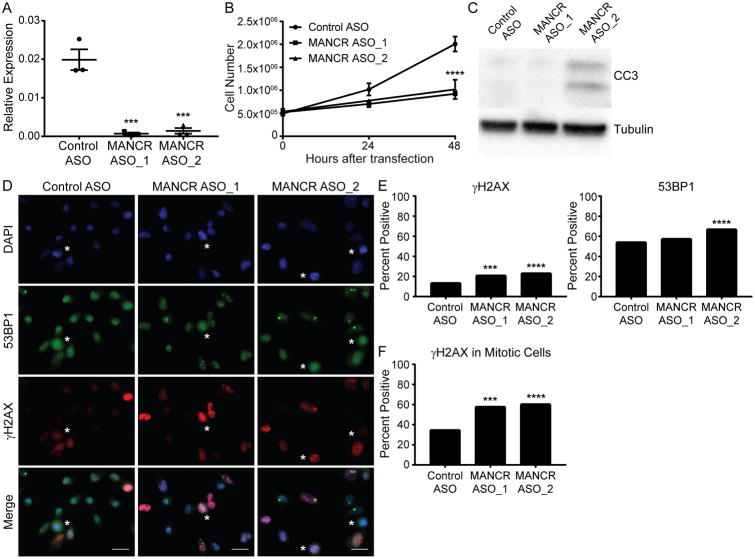
The high expression of MANCR in ER-/PR- patient samples and basal cell lines led us to investigate whether MANCR is related to the aggressive cancer cell phenotype of the triple negative MDA-MB-231 cells. We examined the function of MANCR in these cells by knockdown using two custom designed, locked nucleic acid (LNA) modified anti-sense oligonucleotides (ASO) targeting MANCR. The two ASOs consistently achieved greater than 90% knockdown of MANCR expression at 24 hr after transfection when compared to negative control (Control ASO) by both RT-qPCR (Fig 3A) and RNA-FISH (Supplemental Fig S1A). The knockdown was maintained at 48 hr after transfection (Supplemental Fig S1B). Depletion of MANCR caused a decrease in the growth rate of MDA-MB-231 cells as compared to controls by 24 hr after transfection, and a further decrease was observed at 48 hr (Fig 3B). We also saw an increase in cell death as confirmed by western blot for cleaved caspase 3 (a marker of apoptosis). Cells with knock down of MANCR by ASO_2 expressed higher protein levels of cleaved caspase 3 (CC3) than control cells at 24 hours (Fig 3C). Combined, these results suggest that MANCR affects growth and survival of MDA-MB-231 cells.
Figure 3. MANCR depletion affects cell survival.
A) MDA-MB-231 cells transfected with control ASO or an ASO directed against MANCR (MANCR ASO_1 or ASO_2) were analyzed for MANCR expression by RT-qPCR, 24 hr after transfection, n=3. MANCR expression is relative to GAPDH and ACTB. Data are presented as mean ± S.E. Statistical significance was determined using one-way ANOVA with multiple comparisons. ***p<0.001. B) Proliferation curves of MDA-MB-231 cells with control ASO or ASO-mediated knockdown of MANCR. Data are presented as mean ± S.E. Statistical significance was determined using two-way ANOVA with multiple comparisons. ****p<0.0001. C) Western blot analysis of cleaved caspase 3 (CC3) protein expression in cell lysates from MDA-MB-231 cells 24 hr after transfection with control ASO or ASOs targeting MANCR. D) Immunofluorescence microscopy of MDA-MB-231 cells 24 hr after transfection with control ASO (left panels) or MANCR ASO (middle and right panels). 53BP1 (green), γ-H2AX (red), nuclei were counterstained with DAPI (blue). Mitotic cells are indicated by a white asterisk. Scale bars represent 20 μm. E) Quantification of cells (n>400) positive for γ-H2AX and 53BP1 from 2 experiments. Statistical significance was determined using a chi-square test. ***p<0.001, ****p<0.0001. F) Quantification of mitotic cells with persistent γ-H2AX staining in control ASO (n= 120), MANCR ASO_1 (n= 120) and ASO_2 (n= 120) from 2 experiments. Statistical significance was determined using a chi-square test. ***p<0.001, ****p<0.0001.
A possible cause for increased cell death is DNA damage that cannot be resolved. Aggressive breast cancer typically has genomic instability with a basal level of DNA damage (35). We evaluated double strand breaks (DSBs) by immunostaining for γH2AX, the phosphorylated form of histone H2AX that marks DSBs, and 53BP1 which is required for the DNA damage response. By 24 hr, MANCR knockdown cells showed increases in both γH2AX and 53BP1 compared to control cells, indicative of increased DSBs (Fig 3D and E). γH2AX foci persisted in ∼60% of mitotic cells in the knockdown conditions compared to ∼35% of mitotic cells in the control (Fig 3F), suggestive of defective DNA repair and loss of the G2/M checkpoint.
Depletion of MANCR affects expression of key cell cycle regulators
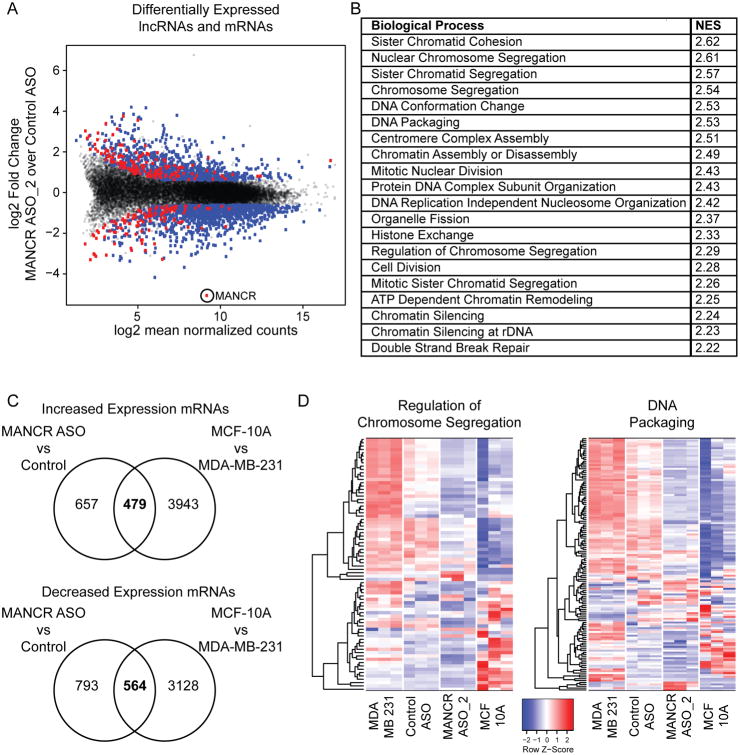
To investigate the global effect of MANCR on gene expression, we performed transcriptome profiling after MANCR knockdown with ASO_2 in MDA-MB-231 cells. We found 2493 genes with a ≥1.5-fold difference in expression in cells treated with MANCR ASO_2 versus Control ASO (Fig 4A). Of these, 980 mRNAs were upregulated and 1240 were downregulated (blue); 156 lncRNAs were upregulated and 117 were downregulated (red). As expected, MANCR was the most downregulated gene after knockdown. A panel of genes was validated for expression by RT-qPCR following knockdown with MANCR ASO_1 or ASO_2 (Supplemental Fig S2A-C). Gene Set Enrichment Analysis (GSEA) of protein coding genes revealed several biological processes that were affected by MANCR depletion. Using a stringent false discovery rate (FDR) cutoff of <5%, the upregulated genes were not significantly enriched for any gene sets; whereas the downregulated genes were enriched in 88 gene sets (Supplemental Table 2). In genes downregulated by MANCR depletion, the top 20 enriched biological processes were mainly related to mitosis (Fig 4B). These results are consistent with the decreased cell proliferation and survival that we observed after MANCR knockdown (Fig 3).
Figure 4. Depletion of MANCR affects gene expression.
A) MA plot displaying differentially expressed mRNAs (blue circles) and lncRNAs (red circles) in MDA-MB-231 with MANCR knockdown (MANCR ASO_2) compared to control MDA-MB-231 cells (Control ASO); gray dots represent genes that did not make the fold change (≥1.5) or p-value (<0.05) cutoffs. Data is from three biological replicates. B) Top 20 gene sets and their normalized enrichment scores (NES) from gene set enrichment analysis of differentially expressed mRNAs using the Molecular Signatures Database (MSigDB) Hallmark gene sets. C) The number of genes with either increased (upper panel) or decreased (lower panel) expression (fold change ≥1.5) in both MANCR ASO_2 vs Control and MCF-10A vs MDA-MB-231 cells. D) Heatmaps showing the expression of the 79 genes (rows) in the MSigDB “Biological Processes: Regulation of Chromosome Segregation” gene set (left panel) and the 136 genes in the “Biological Processes: DNA Packaging” gene set (right panel) for MDA-MB-231, Control ASO, MANCR ASO_2, and MCF-10A cells (columns). The color scale of the heatmaps represents the relative expression of each gene.
MANCR expression is increased in aggressive tumors compared to normal tissue or more benign tumors (Fig 2), and its depletion causes significant gene expression changes. Therefore, we examined whether MANCR supports an aggressive breast cancer phenotype by determining if depletion of MANCR returns MDA-MB-231 cells to a more normal-like profile. Gene expression changes after MANCR knockdown in MDA-MB-231 were compared to those between MDA-MB-231 and the normal-like MCF-10A cells (Fig 4C). Many of the genes that were more highly expressed in MCF-10A than in MDA-MB-231 cells were also upregulated by MANCR knockdown in MDA-MB-231 cells (42%). Likewise, 40% of the genes that were downregulated by MANCR knockdown had lower expression levels in MCF-10A than in MDA-MB-231 cells (Fig 4C).
Many more genes were differentially expressed between MCF-10A and MDA-MB-231 cells than between MANCR knockdown and control cells. However, when gene ontologies of the differentially expressed genes were compared between the two groups, there was remarkable overlap of the most significantly enriched gene sets from each group (Supplemental Fig S3 and Supplemental Table 2). Notably, nearly all of the overlapping gene sets were related to cell cycle, and similar gene expression changes occurred. For example, many of the genes in the “regulation of chromosome segregation” and “DNA packaging” gene expression sets had decreased expression levels in both MANCR ASO_2 and MCF-10A cells compared to Control ASO and MDA-MB-231 cells (Fig 4D). These data suggest that MANCR is involved in aberrant cell cycle regulation that drives tumor growth.
MANCR expression is periodic and its depletion causes cytokinesis defects
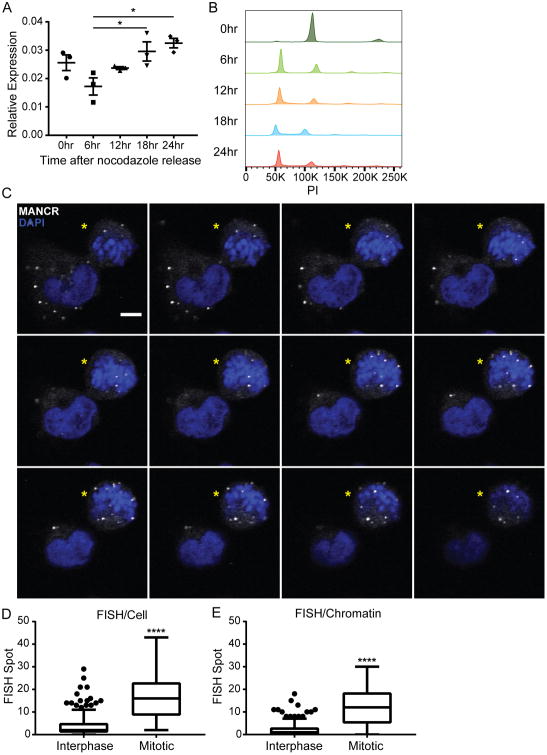
Deregulated cell cycle and growth are hallmarks of tumorigenesis (36). Several lncRNAs, such as MALAT1, H19, and lncRNA-RoR, have been associated with cell cycle regulation (37-39). Many genes, including lncRNAs, that are involved in cell cycle regulation have a periodic expression pattern (40,41). MDA-MB-231 cells were synchronized and RT-qPCR analysis confirmed that MANCR RNA levels oscillate during the cell cycle. We observed high levels at 0 hr after release from synchronization (prometaphase) that decreased by 6 hr after release (G1) and then steadily increased (Fig 5A) as the cells progressed through the cell cycle (Fig 5B). In addition, RNA-FISH of MDA-MB-231 cells revealed that MANCR RNA is variable in the cell population; interphase cells had the lowest expression while early mitotic cells had the highest expression (Fig 5C). The localization of MANCR was also cell cycle dependent. It was mostly cytoplasmic in interphase cells; but primarily localized to chromatin in mitotic cells (Fig 5D and E).
Figure 5. MANCR expression is highest in mitotic cells.
A) Synchronized MDA-MB-231 cells were analyzed for MANCR expression by RT-qPCR every 6 hr after release from an 18 hr nocodazole block. 0 hr is at release, n=3. Data are presented as mean ± S.E. Statistical significance was determined using one-way ANOVA with multiple comparisons. *p<0.05. B) Representative cell cycle distribution of synchronized MDA-MB-231 cells in A. C) Confocal microscopy slices (0.39 μm) from RNA-FISH for MANCR expression in interphase and mitotic (yellow stars) MDA-MB-231 cells. MANCR (white), DAPI (blue). Scale bars represent 5 μm. D,E) Quantification of FISH puncta per cell (D) and FISH puncta per chromatin (E) in interphase (n=283) and mitotic (n=21) cells pooled from 3 experiments. Data are presented as median, IQR, and ± 1.5IQR. Statistical significance was determined using unpaired t-test. ****p<0.0001.
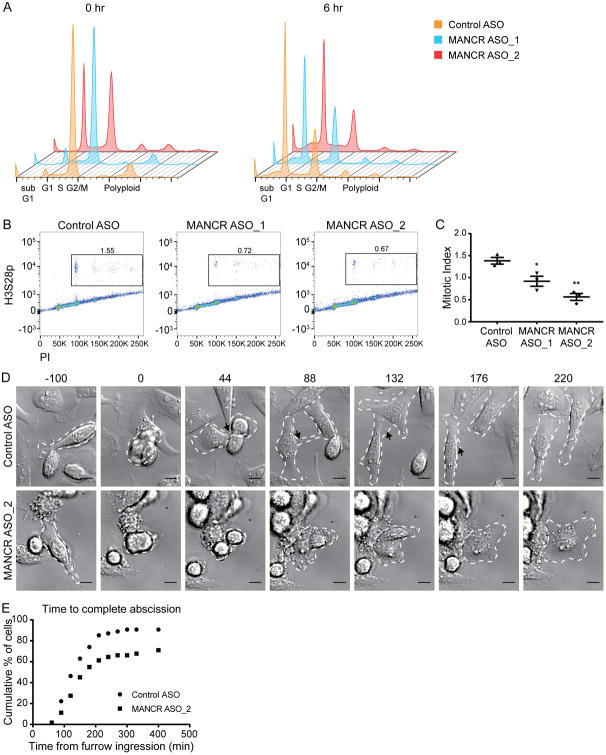
Our GSEA results implicating MANCR in cell cycle control (Fig 4) and its increased expression in mitotic cells (Fig 5) led us to investigate whether MANCR knockdown affects cell cycle. This was tested by flow cytometric analysis for DNA content of nocodazole synchronized cells after MANCR knockdown (Fig 6A). At 0 hr after release from nocodazole, the Control ASO and MANCR ASO_1 cells were arrested at G2/M. In contrast, ASO_2 cells were only partially blocked at 0 hr, with populations in both G1 and G2/M. This result indicates that a subset of cells escaped the nocodazole block, suggesting a defective spindle assembly checkpoint. By 6 hr after release, ASO_1 cells had an increased G2/M population compared to Control ASO cells indicating a delay in mitotic progression. In addition, MANCR-depleted (ASO_1 and ASO_2) cells had a smaller G1 population at 6 hr after release, again indicating a delay in progression. Knockdown with either ASO_1 or ASO_2 resulted in an increased sub-G1 population relative to control that continued to accumulate over 24 hr (Supplemental Fig S4A). This result is consistent with the observed decrease in cell survival (Fig 3).
Figure 6. MANCR depletion causes cytokinesis defects.
A) Representative cell cycle distribution of Control and MANCR-depleted cells that were synchronized 18 hr after transfection, then fixed and stained with PI every 6 hr after release from an 18 hr nocodazole block. 0 hr (left) is at release (36 hr after transfection). B) Flow cytometry of control and MANCR-depleted cells that were fixed 48 hr after transfection and stained for phosphohistone H3 (H3S28p) and propidium iodide (PI). Boxed area indicates the mitotic cell population. C) Quantification of mitotic indexing in B, n=3. Data are presented as mean ± S.E. Statistical significance was determined using one-way ANOVA with multiple comparisons. *p<0.05, **p<0.01. D) Control and MANCR depleted cells were imaged by differential interference contrast microscopy over a 16 hr period. Time 0 min is at furrow ingression. White dashed lines outline dividing cells. Black arrowheads highlight midbody. Scale bars represent 10 μm. E) Distribution of the time from furrow ingression to complete abscission in control (n=54) and MANCR-depleted (n=62) cells from 2 transfections. Statistical significance was determined using log-rank test. **p<0.01.
Since MANCR knockdown had an apparent effect on mitosis, we determined the mitotic index of MANCR-depleted cells by staining for PI and histone H3 serine 28 phosphorylation (H3pSer28), a mitotic marker. We observed a significant decrease in the percentage of M-phase cells after ASO-mediated knockdown of MANCR, from 1.38% in control cells, to 0.92% and 0.56% for ASO_1 and ASO_2, respectively (Fig 6B and C). Taken together, these results indicate that MANCR is necessary for proper cell cycle control.
The observed decrease in mitotic cells following MANCR knockdown prompted us to investigate what mitotic stage MANCR depletion was affecting. We performed live cell imaging in MDA-MB-231 cells following MANCR knockdown with a focus on mitotic cells (Supplemental Videos 1 and 2). While we did not observe a difference in the time from nuclear envelope break down (NEBD) to anaphase onset between control and MANCR knock down cells (Supplemental Fig S4B and C), there was a significant decrease in the percent of mitotic cells that completed cytokinesis upon MANCR knock down (Fig 6D and E). In figure 6D, video stills from live cell imaging demonstrate normal cytokinesis in control cells and defective cytokinesis in MANCR knockdown cells. Furrow ingression is marked as time 0. At 44 min, two daughter cells have formed in both the control and knockdown conditions. By 88 min, the daughter cells in the control condition have both spread onto the plate and the midbody is clearly visible; the control cells continue to separate and complete abscission at 176 min. In contrast, the daughter cells in the MANCR knockdown condition have fused by 88 min, resulting in a binucleate cell. This result indicates that MANCR depletion negatively affects cytokinesis in MDA-MB-231 cells.
Discussion
The association of lncRNAs with cancer progression has revealed novel mechanisms of dysregulation and consequences in cancer cells. Here, we describe a previously uncharacterized lncRNA, MANCR, that is upregulated in triple negative breast cancer cells and affects cell cycle and survival. The key findings of our characterization of MANCR are related to an aggressive breast cancer phenotype and significantly reduced patient survival. We show that MANCR is highly expressed in TNBC cells with an exceptionally intense staining by RNA-FISH in mitotic cells. Insights into MANCR's mechanisms were established by its depletion in MDA-MB-231 cells that resulted in inhibited growth, increased cell death and DNA damage, significant changes in expression of cell cycle regulators, and defective cytokinesis. We conclude from these findings that the abnormal expression of MANCR in aggressive breast cancer cells contributes to the dysregulated cancer phenotype.
The cellular localization of lncRNAs has been related to function (42). RNA-CISH and RNA-FISH revealed that MANCR localized to both the cytoplasm and chromatin; however, the ratio of localization depended on the cell cycle phase with a higher percentage of MANCR associating with chromatin during mitosis. Our observation of increased RNA levels in mitotic cells suggested a functional property of MANCR in cell division. This prediction was validated by time-lapse imaging of live MANCR-depleted cells that revealed defects in completing mitosis. These cells attempted to produce daughter cells, but defects in cell division resulted in multi-nucleated cells, cell death, or mitotic exit. Indeed our transcriptome analysis from knockdown of MANCR informed genes and pathways contributing to the observed defects in cell division. It is important to note that while the gene expression changes identified by RNA-seq reflected alterations to the cell cycle, it remains unclear whether the changes were a direct consequence of MANCR depletion or due to cell cycle arrest. Further studies are needed to investigate the potential role of MANCR in the regulation of genes related to cell cycle.
The GSEA of biological processes affected by MANCR depletion revealed several interesting results. There was more significant enrichment of gene sets that were downregulated by MANCR depletion than upregulated. This may indicate that MANCR functions primarily as an activator of gene expression. The enrichment of gene sets related to biological processes such as chromatin remodeling and spindle assembly suggests a possible role for MANCR during late interphase/ early mitosis. Consistent with a possible defect in the spindle assembly checkpoint (SAC), when treated with nocodazole, a smaller population of MANCR ASO_2-depleted cells arrested in G2/M compared to control (Fig 6). However, there was no effect on the time between nuclear envelope breakdown and anaphase onset in MANCR-depleted cells (Supplemental Fig S4), suggestive of incomplete disruption of the SAC. Recently, it has been reported that two pathways, KNL1-Bubs and RZZ, function in parallel to activate the SAC (43). MANCR depletion could affect one of these arms to partially disrupt the SAC.
Biological processes related to DNA repair were also significantly enriched in genes downregulated by MANCR-depletion. This is consistent with the increased DNA damage that we observed following MANCR knockdown (Fig 3) which may be related to the mutated status of p53 in MDA-MB-231 cells (44). Absence of functional p53 would allow MANCR-depleted cells with increased DNA damage to bypass the normal cell cycle checkpoints, leading to ever increasing genomic instability and eventual cell death.
The most significantly enriched gene sets were almost exclusively related to cell cycle. The top four enriched gene sets all associated with chromosome segregation, suggesting MANCR involvement in this biological process. Failures in chromosome segregation during anaphase lead to DNA damage, formation of chromatin bridges, and defective cytokinesis (45). Our identification of increased DNA damage markers (Fig 3) and cytokinesis defects (Fig 6) following MANCR depletion supports a possible role for MANCR in chromosome segregation.
In conclusion, our results indicate that the previously uncharacterized lncRNA, MANCR (LINC00704), affects genomic stability and cell cycle progression in aggressive breast cancer cells as demonstrated by increased DNA damage, gene expression changes, and cell division defects upon MANCR depletion. Importantly, our in silico analysis of breast cancer patient data revealed that MANCR is clinically relevant. This finding is consistent with a previous study that identified MANCR in a signature of 9 upregulated lncRNAs with prognostic potential for breast cancer relapse (46); however, Liu et al did not functionally characterize MANCR. As it appears to support cell survival, MANCR has the potential to be a novel therapeutic target. According to data from the Genotype-Tissue Expression (GTEx) Project, MANCR expression is very low in most normal tissues with the exception of the spleen. Thus, MANCR may be a promising target in aggressive breast cancer, as inhibition would likely have limited side effects.
Supplementary Material
Acknowledgments
Studies reported here were supported by NIH/NIAMS R01AR039588 (G.S. Stein), NIH/NCI P01CA082834 Project 1 (J.L. Stein), NIH/NCI P01CA082834 Project 3 (J.B. Lian), NIH/NCI U01CA196383 (J.L. Stein), NIH/NIDCR R37DE012528 (J.B. Lian), NIH/NIGMS R01GM121491 (J. Stumpff), Susan G. Komen CCR16377648 (J. Stumpff), Institutional Research Grant 126773-IRG-14-196-01-IRG, awarded to the University of Vermont Cancer Center from the American Cancer Society (C.E. Tye), Lake Champlain Cancer Research Organization/University of Vermont Cancer Center Program Juckett Postdoctoral Fellowship (K.M. Tracy), and The Charlotte Perelman Fund for Cancer Research (G.S. Stein). This research project was supported, in part, by the University of Vermont Cancer Center Shared Resources. The next-generation sequencing and STR analysis were performed in the University of Vermont Cancer Center Advanced Genome Technologies Core and was supported by the University of Vermont Cancer Center, Lake Champlain Cancer Research Organization, and the University of Vermont College of Medicine. Flow cytometry data were collected at the Harry Hood Bassett Flow Cytometry and Cell Sorting Facility, University of Vermont. The authors wish to thank Nicole Bishop and Nicole Bouffard for their assistance with imaging work that was performed at the Microscopy Imaging Center at the University of Vermont. The authors also thank Cindy Fonseca for her assistance with live cell imaging.
Footnotes
COI disclosure statement: The authors declare no potential conflicts of interest
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Metzger-Filho O, Sun Z, Viale G, Price KN, Crivellari D, Snyder RD, et al. Patterns of Recurrence and outcome according to breast cancer subtypes in lymph node-negative disease: results from international breast cancer study group trials VIII and IX. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:3083–90. doi: 10.1200/JCO.2012.46.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loibl S, Gianni L. HER2-positive breast cancer. Lancet (London, England) 2017;389:2415–29. doi: 10.1016/S0140-6736(16)32417-5. [DOI] [PubMed] [Google Scholar]
- 4.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–26. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 7.Jeon Y, Sarma K, Lee JT. New and Xisting regulatory mechanisms of X chromosome inactivation. Current opinion in genetics & development. 2012;22:62–71. doi: 10.1016/j.gde.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harbor perspectives in biology. 2014;6 doi: 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–5. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 10.Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA (New York, NY) 2011;17:878–91. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer research. 2017 doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer biology & medicine. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradford JR, Cox A, Bernard P, Camp NJ. Consensus Analysis of Whole Transcriptome Profiles from Two Breast Cancer Patient Cohorts Reveals Long Non-Coding RNAs Associated with Intrinsic Subtype and the Tumour Microenvironment. PloS one. 2016;11:e0163238. doi: 10.1371/journal.pone.0163238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo W, Wang Q, Zhan Y, Chen X, Yu Q, Zhang J, et al. Transcriptome sequencing uncovers a three-long noncoding RNA signature in predicting breast cancer survival. Scientific reports. 2016;6:27931. doi: 10.1038/srep27931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu YR, Jiang YZ, Xu XE, Hu X, Yu KD, Shao ZM. Comprehensive Transcriptome Profiling Reveals Multigene Signatures in Triple-Negative Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:1653–62. doi: 10.1158/1078-0432.CCR-15-1555. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M, Zhong L, Xu W, Sun Y, Zhang Z, Zhao H, et al. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Scientific reports. 2016;6:31038. doi: 10.1038/srep31038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miano V, Ferrero G, Reineri S, Caizzi L, Annaratone L, Ricci L, et al. Luminal long non-coding RNAs regulated by estrogen receptor alpha in a ligand-independent manner show functional roles in breast cancer. Oncotarget. 2016;7:3201–16. doi: 10.18632/oncotarget.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niknafs YS, Han S, Ma T, Speers C, Zhang C, Wilder-Romans K, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nature communications. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tracy KM, Tye CE, Page NA, Fritz AJ, Stein JL, Lian JB, et al. Selective expression of long non-coding RNAs in a breast cancer cell progression model. Journal of cellular physiology. 2017 doi: 10.1002/jcp.25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messier TL, Gordon JA, Boyd JR, Tye CE, Browne G, Stein JL, et al. Histone H3 lysine 4 acetylation and methylation dynamics define breast cancer subtypes. Oncotarget. 2016;7:5094–109. doi: 10.18632/oncotarget.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS one. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nature communications. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Messier TL, Boyd JR, Gordon JA, Stein JL, Lian JB, Stein GS. Oncofetal Epigenetic Bivalency in Breast Cancer Cells: H3K4 and H3K27 Tri-Methylation as a Biomarker for Phenotypic Plasticity. Journal of cellular physiology. 2016;231:2474–81. doi: 10.1002/jcp.25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR. Genomic instability in breast cancer: pathogenesis and clinical implications. Molecular oncology. 2010;4:255–66. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS genetics. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. The Journal of biological chemistry. 2005;280:29625–36. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell research. 2013;23:340–50. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature genetics. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breeden LL. Cyclin transcription: Timing is everything. Current biology : CB. 2000;10:R586–8. doi: 10.1016/s0960-9822(00)00634-5. [DOI] [PubMed] [Google Scholar]
- 42.Chen LL. Linking Long Noncoding RNA Localization and Function. Trends in biochemical sciences. 2016;41:761–72. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Silio V, McAinsh AD, Millar JB. KNL1-Bubs and RZZ Provide Two Separable Pathways for Checkpoint Activation at Human Kinetochores. Developmental cell. 2015;35:600–13. doi: 10.1016/j.devcel.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Bartek J, Iggo R, Gannon J, Lane DP. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene. 1990;5:893–9. [PubMed] [Google Scholar]
- 45.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science (New York, NY) 2011;333:1895–8. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Li J, Koirala P, Ding X, Chen B, Wang Y, et al. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget. 2016;7:20584–96. doi: 10.18632/oncotarget.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.