Abstract
Background
Sugarcane bagasse has been proposed as a lignocellulosic residue for second-generation ethanol (2G) produced by breaking down biomass into fermentable sugars. The enzymatic cocktails for biomass degradation are mostly produced by fungi, but low cost and high efficiency can consolidate 2G technologies. A. fumigatus plays an important role in plant biomass degradation capabilities and recycling. To gain more insight into the divergence in gene expression during steam-exploded bagasse (SEB) breakdown, this study profiled the transcriptome of A. fumigatus by RNA sequencing to compare transcriptional profiles of A. fumigatus grown on media containing SEB or fructose as the sole carbon source. Secretome analysis was also performed using SDS-PAGE and LC-MS/MS.
Results
The maximum activities of cellulases (0.032 U mL-1), endo-1,4-β--xylanase (10.82 U mL-1) and endo-1,3-β glucanases (0.77 U mL-1) showed that functional CAZymes (carbohydrate-active enzymes) were secreted in the SEB culture conditions. Correlations between transcriptome and secretome data identified several CAZymes in A. fumigatus. Particular attention was given to CAZymes related to lignocellulose degradation and sugar transporters. Genes encoding glycoside hydrolase classes commonly expressed during the breakdown of cellulose, such as GH-5, 6, 7, 43, 45, and hemicellulose, such as GH-2, 10, 11, 30, 43, were found to be highly expressed in SEB conditions. Lytic polysaccharide monooxygenases (LPMO) classified as auxiliary activity families AA9 (GH61), CE (1, 4, 8, 15, 16), PL (1, 3, 4, 20) and GT (1, 2, 4, 8, 20, 35, 48) were also differentially expressed in this condition. Similarly, the most important enzymes related to biomass degradation, including endoxylanases, xyloglucanases, β-xylosidases, LPMOs, α-arabinofuranosidases, cellobiohydrolases, endoglucanases and β-glucosidases, were also identified in the secretome.
Conclusions
This is the first report of a transcriptome and secretome experiment of Aspergillus fumigatus in the degradation of pretreated sugarcane bagasse. The results suggest that this strain employs important strategies for this complex degradation process. It was possible to identify a set of genes and proteins that might be applied in several biotechnology fields. This knowledge can be exploited for the improvement of 2G ethanol production by the rational design of enzymatic cocktails.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4627-8) contains supplementary material, which is available to authorized users.
Keywords: Aspergillus fumigatus, Sugarcane bagasse, CAZymes, Lignocellulose breakdown, RNA-Seq, Secretome
Background
The demand for energy has increased continuously worldwide, which has raised concerns about sustainability and has prompted the search and development of advanced renewable and sustainable sources of energy [1]. Bioethanol has been noted as an alternative fuel to tackle these issues [1–3]. In Brazil, ethanol production relies on the fermentation of sucrose from sugarcane to yield the so-called first-generation (1G) bioethanol [4, 5]. The current Brazilian production is estimated at 30 billion liters per year, but the growing appeal of this fuel has called for investments in the development of new technologies to produce ethanol [6]. Large amounts of sugarcane straw and bagasse are generated every year in Brazil, so this biomass could be used as a substrate to produce 2G bioethanol, which in a few years will compete with 1G ethanol costs [6–11].
Lignocellulose is the most abundant material in nature. It consists of three major polymers: cellulose, hemicellulose and lignin. Cellulose, the main polymeric component of plant biomass, usually contains regions that are highly crystalline. It is a linear polymeric chain of over 10,000 D-glucose residues linked by β-1,4-glycosidic bonds [12–14]. The degradation of lignocellulose into fermentable sugars require many types of enzymes, e.g., β-glucosidases, cellobiohydrolases, endoglucanases, β-xylosidases, endo-β-1,4-xylanases, and numerous other auxiliary enzymes [5, 12, 15]. Due its recalcitrant characteristic, lignocellulose is difficult to degrade, even when enzymes work synergistically [12, 16].
Filamentous fungi such as Trichoderma reesei and Aspergillus niger play an important role in the secretion of enzymes known as CAZymes (carbohydrate-active enzymes), which can act synergistically and are the main source of enzymatic cocktails. Several studies have been conducted to optimize the current enzymatic cocktails and to reduce costs involved in 2G ethanol production [4, 12, 17–19]. The Aspergillus genus comprises over 250 species and has received much attention due numerous species secreting hydrolytic enzymes of interest to lignocellulosic biorefineries. A. fumigatus is an opportunistic and pathogenic fungus, and depending on immunological status of host, can lead to a variety of allergic reactions. However is an important producer of lignocellulolytic enzymes that act synergistically to increase the efficiency of the secreted enzymes. In addition, this fungus secretes thermostable glycosyl hydrolases, such as β-glucosidases (EC 3.2.1.21), endoglucanases (EC 3.2.1.4), cellobiohydrolases (EC 3.2.1.9), xylosidases (EC 3.2.1.37) and endoxylanases (EC 3.2.1.32), which can withstand elevated temperatures [20–23].
Previously, specific cellulose-, hemicellulose-, pectin-, and lignin-degrading enzymes were identified as secreted by A. fumigatus in the presence of different carbon sources (Avicel, cellulose, rice straw, starch, xylan, corn and soybean) that can be used in the lignocellulosic bioenergy industry [22, 24–26]. To gain more insight into how efficiently A. fumigatus AF293 can depolymerize the sugarcane bagasse, a complex biomass important for Brazilian 2G ethanol production, and to identify genes and proteins responsible for these lignocellulosic breakdown reactions, we examined the transcriptional response by RNA-Seq and proteomic profile by mass spectrometry (LC-MS/MS) of A. fumigatus that was cultivated in the presence of steam-exploded sugarcane bagasse (SEB).
Methods
Strains, media, and growth conditions
A. fumigatus AF293, gently donated by the Prof. Dr. Sérgio Akira Uyemura (University of São Paulo, BR), was grown on YAG medium (2% (w/v) dextrose, 0.5% (w/v) yeast extract, 0.1% (v/v) trace elements and 1.8% (w/v) agar) at 37 °C for two days. Spores were harvested and inoculated to a final concentration of 1 × 108 per 50 mL of YNB culture with 1% (w/v) fructose as the carbon source at 37 °C for 16 h (h) in a rotary shaker with agitation at 200 rpm. Afterward, the mycelia were transferred to 1% (w/v) SEB (47.5% cellulose; 9.0% hemicellulose and 34.3% lignin) or 1% (w/v) fructose as the carbon source for 3, 6, 12, 18, 24, 48 and 72 h. Fructose was used as a control in all experimental conditions [26]. Mycelia were harvested by filtration through Whatman grade 1 filters (GE Healthcare, Grandview Blvd. Waukesha, WI, USA), washed once with sterile cool water and kept at − 80 °C until RNA extraction. Supernatants were collected to measure enzymatic activity and determine the secretome. All the experiments described below were performed in three biological replicates.
Enzymatic activity assays
Specific xylanase (endo-1,4-β-xylanase) and cellulose (endo-1,4-β-glucanase) activities were performed with Azo-Xylan (Birchwood) and Azo-CM-Cellulose (both from Megazyme International, Bray, Ireland) as substrates, respectively, according to the manufacturer’s protocols. The enzymatic activities are represented as U mL− 1. All the reactions were performed in triplicate. The software Mega-Calc™ (Megazyme International) was used to determine the enzymatic activities.
Enzymatic activities were also measured by the dinitrosalicylic acid (DNS) assay [27]. Cellulase activities were measured with β-glucan and low-viscosity carboxymethylcellulose (CMC) as substrates, and xylanase activities were measured with the xyloglucan. Briefly, 20 μL of the supernatant from the samples grown in presence of 1% SEB for 24, 48 and 72 h were mixed with 30 μL of sodium acetate buffer 100 mM (pH 5.5) and 50 μL of substrate at 0.5% (w/v) final concentration to achieve a final volume of 100 μL. The reactions were incubated at 40 °C for 5 min for β-glucan and for 10 min for xyloglucan and CMC substrates. The reaction was stopped by adding 100 μL of DNS. All the reactions were performed in triplicate. The calculation of enzyme activities was based on a corresponding standard containing glucose. One unit (U) of enzymatic activity was defined as the amount of enzyme needed to liberate 1 μmol of reducing sugars per minute.
RNA isolation and cDNA synthesis
Fungal biomass was harvested at different times from SEB or fructose culture conditions, and mycelia were ground in liquid nitrogen using a mortar and pestle. Total RNA was purified by using the “Direct-zol™ RNA MiniPrep” kit according to the manufacturer’s instructions (Zymo Research, Irvine, CA, EUA) using the on-column DNAse treatment. RNA integrity was confirmed with a bioanalyzer by using the “Agilent RNA 6000 Nano” kit (Agilent Technologies, Santa Clara, CA, EUA) and the “Plant Total RNA Nano” protocol. RNA was quantified on a Qubit® 2.0 fluorimeter (Thermo Fisher Scientific, Waltham, MS, EUA) with the Qubit® RNA BR Assay kit (Thermo Fisher Scientific, Waltham, MS, EUA). cDNA was synthesized from 1 μg of mRNA using SuperScript® II Reverse Transcriptase (Invitrogen, Carlsbad, CA, EUA).
Library preparation and RNA sequencing
RNA sequencing libraries were prepared using the “TruSeq Stranded mRNA HT Sample Prep” kit (Illumina, San Diego, CA, EUA), mRNA enrichment was performed using magnetic beads coupled with oligo (dT). Sequencing was carried out in the HiSeq 2500 system (Illumina, San Diego, CA, EUA) at the NGS facility located at the Brazilian Bioethanol Science and Technology Laboratory (CTBE), Campinas, SP, Brazil.
Bioinformatic analysis of RNA-Seq data
FastQC [28] was used to check the quality of the sequencing reads visually. Removal of the remaining adapter sequences and quality trimming with a sliding window of size 4, minimum quality of 20, and length filtering (to keep reads with a length of at least 60 bp) were performed with Trimmomatic v0.32, [29]. Clean reads were screened against a database of ribosomal RNA with the aid of SortMeRNA [30]. High-quality reads were mapped in a strand-specific manner by using TopHat2 [31] against the genome sequence of A. fumigatus Af293 obtained from ASPGD [32, 33]. The number of exon-exon junctions at different levels of read subsampling was employed to confirm sequencing saturation with RSeQC [34]. Mapping of the reads to the features of the exons were summarized at the gene level by using the function featureCounts from the Rsubread v1.12.6 package [35] in R v3.0.2 [36] and the annotation file in GFF3 format from ASPGD. Differential gene expression was analyzed with edgeR [37] in R [36]. Briefly, genes with at least one CPM (counts per million) in at least three samples were kept for analysis, which was equivalent to removing genes with low and noisy expression. The expression values were normalized by the trimmed mean of M-values (TMM) method to account for differences in the composition of RNA [38]. After the dispersion was estimated and the biological coefficient of variation was computed, the differentially expressed genes were called by fitting a negative binomial model with generalized linear models (GLS) that included factors for the TMM and the dispersion estimates [37]. A likelihood ratio test was performed to provide a p-value for differential expression. The p-values were adjusted for multiple testing by the method of Benjamini-Hochberg, to control the false discovery rate (FDR) [39]. The full R script used for the analysis and the raw count matrix are available in Additional file 1: Figure S1 and Additional file 2: Table S1, respectively. Genes with FDR values lower than 0.05 and log2-fold changes greater than 1.0 or lower than − 1.0, i.e., a difference of twice the expression level in either direction, were considered differentially expressed.
Supernatant analysis by SDS-PAGE
Supernatants (50 mL) from SEB or fructose culture conditions were lyophilized until completely dry and re-suspended in 2 mL of buffer (Tris-HCl 50 mM, pH 6.8; 1 mM DTT; and 1 mM protease inhibitor), and 15 μL was separated by 10% SDS-PAGE (110 V, 90 min). The proteins were visualized by staining with 0.1% Coomassie Brilliant Blue R250 (w/v), which was followed by destaining with 45% methanol and 10% acetic acid solution (v/v). The protein concentration was determined by Bradford’s Assay (Bio-Rad Protein Assay Hercules, CA, EUA) [40]. Prior to mass spectrometry, all the bands from the SDS-PAGE gels were manually excised, reduced, alkylated, digested with trypsin, and purified (Promega, Madison, WI, EUA - V5111) according to a previously described method [41].
Identification of proteins by coupled system of the LC-MS/MS type
Peptides were sequenced on a Synapt G2 HDMS (Waters, Milfords, MS, EUA) mass spectrometer coupled to a UPLC NanoAcquity system with 1D technology (Waters, Milfords, MS, EUA) and captured by a C18 Symmetry column (5 μm, 180 μm × 20 mm) (Waters, Milfords, MS, EUA). The peptides were separated by using a 2–90% acetonitrile gradient in 0.1% formic acid and an HSS T3 analytical column (1.8 μm, 75 μm × 100 mm) (Waters, Milfords, MS, EUA) with a flow of 300 μL min− 1 for 120 min. The data were acquired on a Waters Synapt G2S Q-TOF mass spectrometer equipped with a NanoLockSpray (Waters, Milfords, MS, EUA). The experiments were performed in the HDMSE mode (data-independent analysis). The mass spectra were processed with the ProteinLynxGlobalServer (PLGS) software version 3.1. The proteins were identified by comparison to the Aspergillus UNIPROT database (207,966 proteins) [42]. The defined parameters were automatic tolerance for precursors and ion products, minimum of three corresponding ion fragments per peptide, minimum of seven corresponding ion fragments per protein, trypsin missed cleavage, carbamidomethylation as a fixed modification, oxidation of methionine as a variable modification, and 4% FDR peptide.
Protein analysis
Protein sequences were analyzed with the BLAST (basic local alignment search tool) software (http:ncbi.nlm.nih.gov/Blast.cgi). The subcellular localization of proteins was predicted by YLoc (interpretable subcellular localization prediction) (abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi) [43], and the presence of signals due to peptides of the secreted proteins was predicted by SignalP v.4.0 (http://www.cbs.dtu.dk/services?SignalP/) [44]. Additionally, Secretome Pv2.0 (http://www.cbs.dtu.dk/services/SecretomeP/) was used to define the proteins that were secreted by the non-classic pathway [45].
For CAZy enzyme identification, the proteins in the secretome were screened with a library of hidden Markov models by using HMMER3 [46] of carbohydrate-active enzymes obtained from dbCAN [47]. Hits were considered positive on the basis of the dbCAN recommendations.
qRT-PCR analysis
After RNA-Seq analysis, 4 DEGs (Differentially Expressed Genes) were selected, including sugar transporters and CAZymes, for qRT-PCR analysis. RNA was extracted and purified as previously described. cDNA was synthesized from 5 μg of RNA using SuperScript® II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). Quantitative PCR (qPCR) analyses were performed according to Semighini et al. [48]. The abundance of the respective mRNAs was normalized using β-tubulin probes. The primers for the investigated genes are listed in Additional file 3: Table S2.
Functional enrichment
Genes identified as differentially expressed were analyzed by FunCat functional enrichment [49]. The CAZy proteins from the secretome were classified according to the GO-Slim classifications from the AspGD based on the ontology “Molecular function” (GO Categorization Slim Mapper) [50].
Venn diagrams
The area-proportional Venn diagrams were drawn based on images generated with free online software [51].
Results
Enzymatic analysis
To evaluate the activity of enzymes produced by A. fumigatus in the presence of sugarcane bagasse, we performed enzymatic assays using xyloglucan, β-glucan, and CMC as substrates. Specific endo-1,4-β-xylanase and endo-1,4- β-glucanase activities, were also investigated using Azo-Xylan (Birchwood) and Azo-CM-Cellulose (both from Megazyme International, Bray, Ireland), respectively. The enzymes from supernatants derived from A. fumigatus cultures were capable of hydrolyzing cellulose (CMC) and hemicelluloses (β-glucan and xylan), but no activities were detected in xyloglucans (Fig. 1a-b).
Fig. 1.
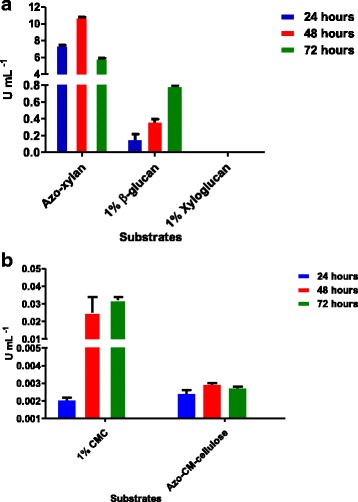
Enzyme activity. Enzymatic activities (U mL−1) of supernatants from A. fumigatus culture against different substrates after 24, 48 and 72 h growth on sugarcane bagasse. Each bar represents the mean and the standard deviation of values from three independent experiments. a Azo-xylan, β-glucan and xyloglucan; b 1% CMC and Azo-CMC
We observed that the activities depended on duration of growth, with maximum activities detected within three days, except for endo-1,4-β-D-xylanase, with peak activity after two days culture and no activities detected prior to 24 h of incubation. Maximum activities of cellulases (0.032 U mL− 1), endo-1,4-β--xylanase (10.82 U mL− 1) and endo-1,3-β glucanases (0.77 U mL− 1) were detected for A. fumigatus while in A. niger (0.002 U mL− 1; 2.3 U mL− 1 and 0.4 U mL− 1, respectively) and T. reesei RUT-C30 (0.0039 U mL− 1, 0.4 U mL− 1 and 0.1 U mL− 1, respectively) are described on the same biomass [4]. These results indicate that A. fumigatus is an excellent producer of an arsenal of hydrolytic enzymes, with activities superior to the hypercellulolytic strain T reesei RUT30-C.
To gain more insight into the hydrolytic enzymes of A. fumigatus specific for sugarcane bagasse breakdown, we selected the cultivation of 24 h (when we detected enzyme activity) because we were interested in the initial process of SEB breakdown. In this time we determinate the transcriptome and the secretome responses of this strain.
Analysis of the transcriptome of A. fumigatus under the influence of sugarcane bagasse as the substrate
To identify potential new genes involved in SEB breakdown, we analyzed the transcriptome by RNA-Seq after 24 h of cultivation. After RNA sequencing, each sample generated approximately 14 to 16 million paired-end reads. RNAseq data were analysed by comparing the mycelium grown on SEB and that grown on fructose. We observed 2227 genes differentially expressed (FDR < 0.05, |log2FC| > 1) in SEB where 1181 were upregulated, while 1045 were upregulated in fructose conditions (downregulated in SEB) (Additional file 4: Table S3). Gene ontology (GO) and the functional catalogue (FunCat) classified the differentially expressed genes functionally in 18 different enriched categories [40]. Two significant categories among upregulated genes were Metabolic Processes (GO:0008152) and Protein Synthesis (GO:0006412) and in downregulated genes Metabolic Processes (GO:0008152) and Energy (GO:0006112) (Additional file 5: Table S4). Genes related to the regulation of C-compound and carbohydrate metabolic processes represent the two GO terms commonly enriched for both up- and downregulated genes in the SEB condition, including genes encoding carbohydrate-active enzymes (CAZymes) and transporters. Given the importance of CAZymes for the degradation of biomass, we directed our efforts toward a better understanding of the transcriptional profile of these enzymes and sugar transporters.
We found 197 differentially expressed CAZyme genes classified based on the CAZy database (http://www.cazy.org) [52]. Concerning the 566 CAZyme genes predicted in the A. fumigatus AF293 genome categorized into the different classes (247 GHs, 105 GTs, 96 CEs, 59 AAs, 15 PLs, and 44 CBMs) (Additional file 6: Table S5), we concluded that 35% of CAZyme genes were differentially expressed in our data, which highlights the potential that a wide spectrum of hydrolytic enzymes were produced. However, glycosyl transferases appeared only in a very small percentage (~ 1%), suggesting a secondary role in polysaccharide degradation (Fig. 2a).
Fig. 2.
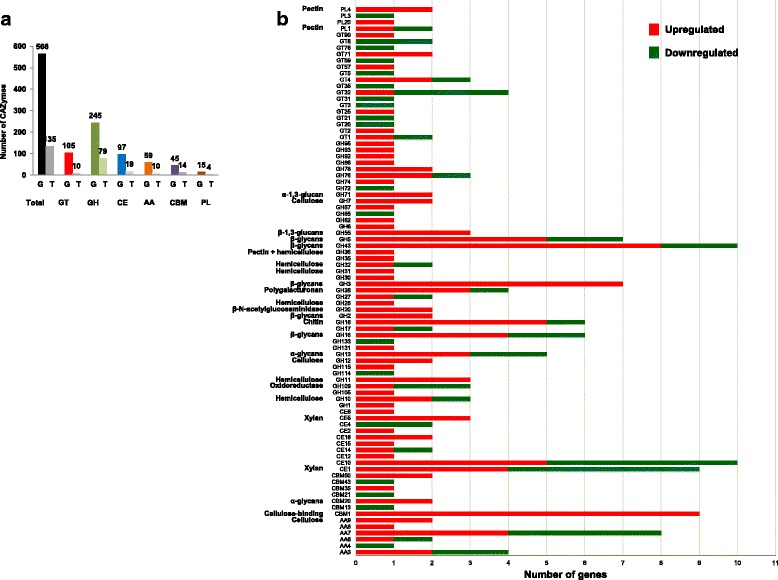
Differentially expressed CAZymes of A. fumigatus identified in RNA-Seq data. The total number of CAZymes and their respective families found in the genome (G) and upregulated in the transcriptome (T) (a). The classification of CAZymes families of up- and downregulated genes (b). AA, auxiliary activities; CBM, carbohydrate binding module; CE, carbohydrate esterase; GT, glycosyltransferases; PL, polysaccharide lyase; GH, glycoside hydrolases. Known substrates or activities of some CAZyme families are given
Among the 197 CAZymes differentially expressed, 135 genes were upregulated in SEB and 62 were downregulated in SEB (upregulated in fructose), classified into 67 and 41 families, respectively (Additional file 4: Table S3). The classes of upregulated genes were 40% glycoside hydrolases (GH), 10% carbohydrate esterases (CE), 7% carbohydrate-binding modules (CBM), 5% glycosyltransferases (GT), 5% auxiliary activities (AA) and 2% polysaccharide lyases (PL), while downregulated genes represented 11% GHs, 6% CEs, 1% CBMs, 8% GTs, 4% AAs, and 1% PLs.
For plant biomass degradation, many enzymes working synergistically are required for efficiency hydrolysis. For cellulose degradation, endoglucanases (EGs) catalyze the hydrolysis at random positions in less crystalline regions; cellobiohydrolases (CBHs) act on the reducing and non-reducing ends of the chains, releasing cellobiose, which is cleaved into glucose by β-glucosidases [53]. We observed a synergistic upregulation of endoglucanases (GH5, GH12, GH16 and CBM1), cellobiohydrolases (GH6 and GH7) and β-glycosidases (GH1 and GH3). In addition to the cellulose degradation enzymes, 32 genes involved in xylan hydrolysis were also upregulated, e.g., endoxylanases (GH10, GH11 and CBM1), xylosidases (GH3 and GH43) and acetylxylan esterases (CBM1, CE2, CE16) (Fig. 2b).
In addition, numerous other plant cell wall polysaccharide-degrading enzymes were also upregulated as described in Additional file 4: Table S3. Among the DEGs, GH11 endo-1,4-beta xylanase (log2FC = 10.39) appeared highly expressed, as well as CBM1 endoglucanase (log2FC = 9.57), extracellular glycosyl hydrolase/cellulose CBM 1 (log2FC = 9.35) and AA9 endo-1,4-beta-glucanase (log2FC = 9.21 and 8.75) (Table 1). Genes encoding delignification enzymes, such as laccase (Afu2g17530), cellobiose dehydrogenase (Afu2g17620), catalase (Afu2g18030), putative FAD-dependent oxygenase (Afu6g12070), and oxidoreductase enzymes, were also upregulated. Similarly, pectate lyases – PL1 (Afu2g00760), amylases – GH13 (Afu2g03230), and carboxypeptidases (Afu3g07040 and Afu5g01200) were detected in this study (Additional file 4: Table S3).
Taken together, endoglucanases, cellobiohydrolases and beta-glucosidases were significantly upregulated, suggesting the cellulose degradation potential of this strain, and the abundance of hemicellulases highlights, once again, the great potential of A. fumigatus in complex biomass deconstruction.
Table 1.
Main CAZymes related to biomass deconstruction upregulated in A. fumigatus AF293 transcriptome
| Gene ID | Gene Description | CAZy family | log2FC | Peptide Signal | Predicted substrate |
|---|---|---|---|---|---|
| Afu3g03870 | endo-1,4-beta-glucanase | AA9 | 9.21 | Y | cellulose |
| Afu4g07850 | endoglucanase | AA9 | 8.75 | Y | cellulose |
| Afu2g00920 | extracellular glycosyl hydrolase/cellulase | CBM1 | 9.35 | Y | arabinoxylan |
| Afu3g00420 | acetyl xylan esterase (Axe1) | CBM1 | 4.13 | Y | xylan |
| Afu6g01800 | endoglucanase | CBM1 | 9.57 | Y | cellulose |
| Afu6g03280 | swollenin | CBM1 | 7.51 | Y | cellulose |
| Afu6g11600 | endoglucanase | CBM1 | 8.11 | Y | cellulose |
| Afu6g13610 | endo-1,4-beta-xylanase | CBM1 | 9.20 | Y | xylan |
| Afu7g06740 | endoglucanase | CBM1 | 8.34 | N | cellulose |
| Afu8g06570 | acetyl xylan esterase | CBM1 | 7.57 | Y | xylan |
| Afu8g06830 | endoglucanase | CBM1 | 4.40 | Y | cellulose, β-1,4-glucan |
| Afu2g00690 | glucan 1,4-alpha-glucosidase | CBM20 | 2.94 | Y | starch |
| Afu4g10140 | glucoamylase | CBM20 | 1.14 | N | starch |
| Afu8g02510 | glycosyl hydrolase family 43 protein | CBM35 | 1.61 | Y | xylan, pectin |
| Afu2g14530 | esterase | CE1 | 2.16 | Y | xylan |
| Afu7g02380 | ferulic acid esterase (FaeA) | CE1 | 1.72 | Y | xylan |
| Afu2g00510 | cellulose-binding GDSL lipase/acylhydrolase | CE16 | 7.28 | Y | xylan, mannan |
| Afu2g00630 | cellulose-binding GDSL lipase/acylhydrolase | CE16 | 3.97 | Y | xylan, mannan |
| Afu2g09380 | cutinase | CE5 | 7.10 | Y | cutin |
| Afu2g14420 | cutinase | CE5 | 3.71 | Y | cutin |
| Afu4g03210 | cutinase | CE5 | 6.74 | Y | cutin |
| Afu1g14710 | beta-glucosidase | GH1 | 3.42 | N | cellulose |
| Afu3g15210 | endo-1,4-beta-xylanase | GH10 | 8.59 | Y | xylan |
| Afu4g09480 | extracellular endo-1,4-beta-xylanase | GH10 | 8.82 | Y | xylan |
| Afu3g00320 | endo-1,4-beta-xylanase (XlnA) | GH11 | 10.39 | Y | xylan |
| Afu3g00470 | endo-1,4-beta-xylanase | GH11 | 8.64 | Y | xylan |
| Afu6g12210 | endo-1,4-beta-xylanase (XynG1) | GH11 | 6.91 | Y | xylan |
| Afu7g06150 | endoglucanase | GH12 | 8.58 | Y | cellulose |
| Afu3g02090 | beta-xylosidase | GH3 | 4.17 | Y | xylan |
| Afu4g13770 | glycosyl hydrolase | GH3 | 1.55 | Y | cellulose |
| Afu5g07080 | beta-glucosidase | GH3 | 2.03 | Y | cellulose |
| Afu5g07190 | beta-glucosidase | GH3 | 2.24 | N | cellulose |
| Afu6g14490 | beta-glucosidase | GH3 | 2.58 | N | cellulose |
| Afu7g06140 | beta-D-glucoside glucohydrolase | GH3 | 3.03 | Y | cellulose |
| Afu8g02100 | beta-glucosidase | GH3 | 2.13 | Y | cellulose |
| Afu1g17320 | endo-arabinanase | GH43 | 4.50 | Y | pectin |
| Afu2g00930 | xylosidase | GH43 | 7.97 | N | xylan |
| Afu2g13190 | xylosidase: arabinofuranosidase | GH43 | 1.99 | N | xylan |
| Afu2g14750 | endo-arabinase | GH43 | 1.92 | Y | pectin |
| Afu3g01660 | glycosyl hydrolase, family 43 | GH43 | 2.24 | Y | xylan, pectin |
| Afu6g00770 | extracellular arabinanase | GH43 | 1.80 | Y | xylan, pectin |
| Afu6g14550 | xylosidase/arabinosidase | GH43 | 6.47 | N | xylan, pectin |
| Afu8g04710 | xylosidase | GH43 | 4.04 | N | xylan |
| Afu5g01830 | extracellular endoglucanase | GH5 | 2.00 | Y | cellulose |
| Afu6g07480 | endoglucanase | GH5 | 2.90 | Y | cellulose |
| Afu7g01070 | endo-1,4-beta-mannosidase | GH5 | 2.48 | N | mannan |
| Afu7g05610 | glucanase | GH5 | 5.07 | N | β-1,6-glucan |
| Afu8g07030 | endo-1,4-beta-mannosidase | GH5 | 1.82 | Y | mannan, galactomannan, glucomannan |
| Afu3g01910 | cellobiohydrolase | GH6 | 9.02 | Y | cellulose |
| Afu2g12770 | alpha-L-arabinofuranosidase | GH62 | 8.54 | Y | arabinoxylan, arabinogalactan |
| Afu5g14380 | Alpha-glucuronidase | GH67 | 2.58 | Y | xylan |
| Afu6g07070 | cellobiohydrolase celD | GH7 | 6.78 | Y | cellulose |
| Afu6g11610 | 1,4-beta-D-glucan-cellobiohydrolyase | GH7 | 9.83 | Y | cellulose |
| Afu8g01490 | endoglucanase | GH74 | 7.89 | Y | xyloglucan |
| Afu2g12830 | UDP-glucosyl transferase family protein | GT1 | 1.81 | N | UDP-glucosyl + acceptor |
| Afu8g02020 | glycosyltransferase | GT2 | 3.09 | N | – |
| Afu8g00650 | LPS glycosyltransferase | GT25 | 2.11 | N | UDP-glucose + lypopolysaccharide |
| Afu8g00640 | glycosyl transferase | GT32 | 3.46 | N | – |
| Afu1g06890 | alpha-1,2-mannosyltransferase (Alg11) | GT4 | 1.01 | N | GDP-mannose + Man3GlcNAc2-PP-dolichol or Man4GlcNAc2-PP-dolichol |
| Afu1g17030 | glycosyl transferase | GT4 | 2.05 | N | – |
| Afu3g07700 | glucosyltransferase | GT57 | 1.31 | N | dolichol-P-glucose + acceptor |
| Afu6g04450 | alpha-1,2-mannosyltransferase (Mnn2) | GT71 | 1.46 | N | GDP-mannose + heteroglycan |
| Afu6g14480 | alpha-1,3-mannosyltransferase | GT71 | 1.22 | N | GDP-mannose + heteroglycan |
| Afu2g00760 | pectate lyase A | PL1 | 1.10 | Y | pectin |
| Afu4g03780 | rhamnogalacturonase B | PL4 | 1.76 | Y | pectin |
| Afu8g00820 | rhamnogalacturonase | PL4 | 3.09 | Y | pectin |
Modulated: all genes with log2FC > 1 and < −1 in presence of SEB
Up: genes with Log2FC > 1 in presence of SEB
Down: genes with Log2FC < − 1 in presence of SEB
To validate RNAseq data and get additional information about the expression over time, we have performed qRT-PCR for 4 DEGs that encode enzymes essential to biomass degradation, Afu4g07850 (LPMO), Afu1g14710 (β-glucosidase), Afu6g11610 (1,4-β-D-glucan-cellobiohydrolase) and Afu3g02090 (β-xylosidase), during 3, 6, 12, 18 and 24 h of cultivation in SEB and fructose. The expression profiles of these genes behaved in different ways: Afu1g14710 and Afu3g02090 genes were strongly induced at the beginning (6 h) of the growth in SEB, and their expression decreased after 6 h, while Afu4g07850 had an increasing gene expression during the time course, and Afu6g11610 increased at 24 h (Fig. 3).
Fig. 3.
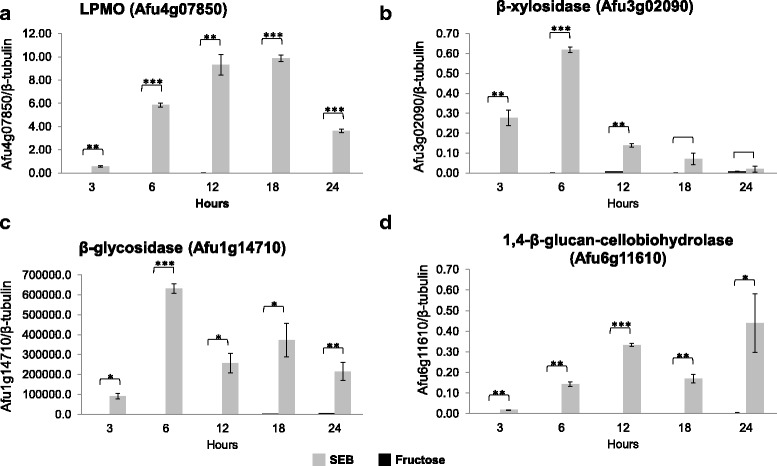
qRT-PCR. The expression levels of the Afu4g07850 (a), Afu3g02090 (b), Afu1g14710 (c), and Afu6g11610 (d) genes were determined after 3, 6, 12, 18, and 24 h of A. fumigatus growth in the presence of SEB 1% or fructose 1%. Each value represents the expression ratio relative to the expression of the β-tubulin gene. The data are the average of three replicates, and the bar indicates the standard deviation. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) (Student’s t-test)
Sugar transporters identified during RNA sequencing
Approximately 106 genes encoding sugar transporters have been reported in the Aspergillus genome, and only 88 genes were described as encoding sugar transporters in A. fumigatus strain Z5, which are distributed among the SP, FHS, SHS, and GPH families (the SP family includes 79 genes) [48]. Additionally, the genomes of filamentous fungi also encode large numbers of major facilitator superfamily (MFS) transporters. Among them, 25 transporters were differentially expressed on SEB, classified as encoding MFS hexose transporter, MFS and sugar transporter, UDP-Glc/Gal endoplasmic reticulum nucleotide sugar transporter, nucleotide sugar transporter, hexose transporter protein, high affinity glucose/hexose transporter, MFS glucose transporter, MFS lactose transporter, MFS maltose transporter, and xylose transporter (Fig. 4a).
Fig. 4.
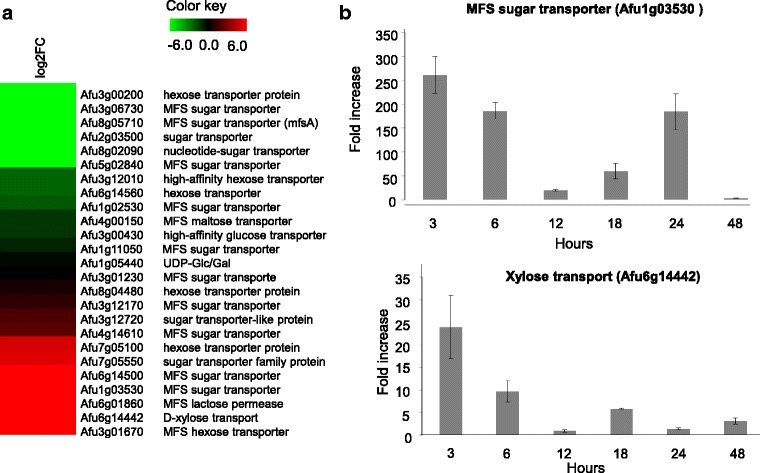
Sugar transporters differentially expressed in RNAseq. Heat map of up- and downregulated genes encoding sugar transporters (a). The color bar represents the log2FC values for each gene. Red color: upregulated genes; black color: unchanged genes; green color: downregulated genes. Expression profiles (qRT-PCR) of genes encoding sugar transporters (b) from A. fumigatus grown in the presence of 1% xylose. Each value represents a fold increase in the expression ratios compared to fungal growth in the presence of 1% fructose. Data are the average of three replicates, and the bar indicates the standard deviation
Orthologous genes in Aspergillus and non-Aspergillus species were identified by sequence analysis according to Aspergillus Genome Database (AspGD; http://www.aspgd.org) [32]. Three orthologous genes encoding possible putative xylose transporters (Afu1g03530, Afu4g14610, and Afu6g14442) and five related to cellobiose transporters (Afu3g01670, Afu6g14500, Afu6g14560, Afu7g05100, and Afu8g04480) have been identified in N. crassa, A. oryzae, A. niger, and A. nidulans (Additional file 7: Table S6).
To analyze the potential xylose transporters, we selected Afu1g03530 (log2FC = 3.43) and Afu6g14442 (log2FC = 4.6), which are orthologous to the xtrD xylose transporter of A. nidulans (An0250) [54] and show high similarity to transporters in other fungi, with conserved regions among different species [54–69]. To characterize the expression profile, A. fumigatus was grown in 1% xylose and 1% fructose as a carbon source for the time course (3, 6, 12, 18, 24, and 48 h). Afu6g14442 gene expression was highly induced after 3 and 6 h of cultivation in 1% xylose with an increase in up to 25-fold. On the other hand, the expression of Afu1g03530 increased to 250-, 180-, 25-, 60-, 200-, and 5-fold after 3, 6, 12, 18, 24, and 48 h, respectively (Fig. 4b). These results lead us to speculate that both genes could encode potential xylose transporters, which can be further better characterized.
Characterization of the secretome of A. fumigatus in the presence of sugarcane bagasse
Once the transcriptome was characterized, we analyzed the secreted protein profiles of A. fumigatus cultivated in the same condition by SDS-PAGE and LC-MS/MS. The total protein secreted by the fungi was approximately 300 μg mL− 1 in SEB versus 112 μg mL− 1 in the fungi grown on fructose (Additional file 8: Figure S2). In the SEB supernatant, we detected 128 secreted proteins, and only 44 were detected in the fructose supernatant (Additional file 9: Table S7), 27 of which are the same for both conditions (Fig. 5a).
Fig. 5.
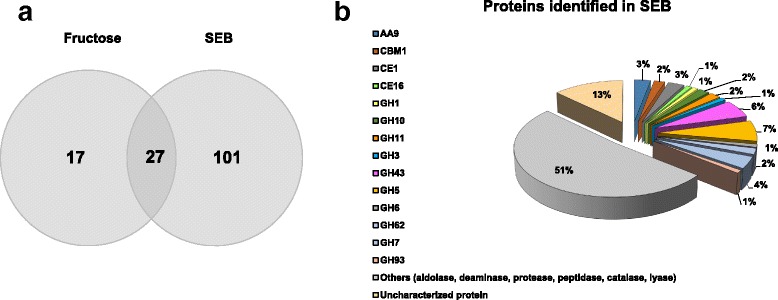
Proteins identified in the A. fumigatus secretome. Venn diagram of proteins found in the secretome of A. fumigatus grown on SEB or fructose (a). Percentage of CAZymes identified in the secretome classified according to their molecular function (b). CAZyme classification was from AspGD based on the facet of “Molecular function” [42]
As previously described, in the A. fumigatus genome, 566 CAZyme genes (461 proteins, excluding GTs) are predicted [25]; 271 of them are predicted to be secreted (Additional file 6: Table S5). The presence of a signal peptide in these proteins was inferred using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) [44]. Approximately 78 proteins identified in SEB correspond to CAZymes in the classes GH (46), CE (8), AA (8), PL (2), and CBM (15), which is 18.61% of the total CAZymes encoded by the A. fumigatus genome. The remaining proteins are classified as proteins of unknown functions (17%), hydrolases/peptidases/proteases/binding proteins (14%), oxidoreductases (6%), transferases (2%), and lyases (2%) (Fig. 5b). We have also identified intracellular proteins, suggesting cell lysis or an unknown mechanism.
The most commonly identified cellulases and hemicellulases were GH5 and GH43, respectively. When compared to previous data concerning the secretome of A. fumigatus, in different types of biomass [22–24], we detected CAZymes found exclusively in the SEB supernatant, such as GH10 (endo-1,4-β-xylanase), GH11 (Endo-1,4-β-xylanase (xyn11A), GH43 (β-glucosidase), GH43 (arabinase), GH47 (mannosidase), GH5 (endo-1,4-β-mannosidase) GH28 (xylogalacturonan), GH27 (α-galactosidase) GH62 (α-arabinofuranosidase) PL3 (pectate lyase) and CE1 (acetyl xylan esterase) (Table 2). Table S7 lists all these enzymes identified in both SEB and fructose conditions.
Table 2.
CAZymes detected in A. fumigatus SEB secretome related to biomass breakdown
| Uniprot Acession | Gene ID | Protein Name | CAZy Family | Score | SeqCover (%) | Peptide Signal | Substrate |
|---|---|---|---|---|---|---|---|
| Q6MYM8 | Afu1g12560 | endoglucanase | AA9 | 1158.51 | 13.67 | Y | cellulose |
| Q4WP32 | Afu4g07850 | endoglucanasea,c | AA9 | 11,713.03 | 47.2 | Y | cellulose |
| Q4X071 | Afu2g14490 | endoglucanase | AA9 | 1475.28 | 15.22 | Y | cellulose |
| D4AHU7 | Afu6g03280 | swollenin | CBM1 | 7631.56 | 32.14 | Y | cellulose |
| Q4WBW4 | Afu8g06570 | acetyl xylan esterase | CBM1 | 5408.98 | 9.16 | Y | xylan |
| B0Y7U1 | Afu6g09040 | feruloyl esterase | CE1 | 250.94 | 11.22 | Y | arabinoxylan, pectin |
| A4D9B6 | faeC | feruloyl esterase C | CE1 | 3223.58 | 37.87 | Y | xylan |
| Q4WIS4 | Afu2g00820 | extracellular GDSL-like lipase/acylhydrolasea | CE16 | 3089.24 | 22.45 | Y | xylan, mannan |
| Q4WRY0 | Afu1g14710 | beta-glucosidasea | GH1 | 4556.9 | 26.5 | N | cellulose |
| Q4WCM9 | Afu6g01800 | endoglucanasea | GH7/CBM1 | 6457.03 | 16.74 | Y | cellulose |
| Q0H904 | Afu4g09480 | endo-1,4-beta-xylanase C (xlnC)a | GH10 | 34,440.44 | 85.23 | Y | xylan |
| Q4WLG5 | Afu6g13610 | endo-1,4-beta-xylanasea,c | GH10/CBM1 | 7068.88 | 58.19 | Y | xylan |
| V5R355 | Afu3g00320 | endo-1,4-beta-xylanase (XlnA)a,c | GH11 | 10,011.55 | 37.04 | Y | xylan |
| B0Y8Q8 | Afu6g12210 | endo-1,4-beta-xylanase (XynG1) | GH11 | 1009.69 | 15.38 | Y | xylan |
| Q4WQR8 | Afu4g13770 | glycosyl hydrolase | GH3 | 327.91 | 7.06 | Y | cellulose |
| B0YDT3 | Afu6g00770 | extracellular arabinanase | GH43 | 4307.26 | 28.04 | Y | xylan, pectin |
| Q4WIR3 | Afu2g00930 | xylosidase/glycosyl hydrolasea,c | GH43 | 2555.85 | 13.28 | N | xylan |
| Q4WIU1 | Afu2g00650 | arabinosidasea,c | GH43 | 8662.27 | 27.8 | N | pectin |
| Q4X046 | Afu2g14750 | endo-arabinaseb | GH43 | 2990.38 | 34.88 | Y | pectin |
| Q4WCE5 | Afu8g04710 | xylosidasea | GH43 | 2270.88 | 24.77 | N | xylan |
| Q4WBJ5 | Afu8g02510 | glycosyl hydrolase family 43 proteinc | GH43/CBM35 | 583.34 | 22.2 | Y | xylan, pectin |
| Q4WD15 | Afu6g03150 | Uncharacterized protein, hydrolase activitya | GH5 | 2751.27 | 17.67 | Y | unknown |
| Q4WW63 | Afu5g14560 | Cellulase family proteinb | GH5 | 2371.35 | 20.15 | Y | cellulose |
| Q4WGN1 | Afu7g05610 | glucanasea | GH5 | 1540.22 | 16.6 | N | β-1,6-glucan |
| Q4WN62 | Afu6g07480 | endoglucanase | GH5 | 2409.74 | 28.1 | Y | cellulose |
| B0Y9E7 | Afu8g07030 | endo-1,4-beta-mannosidase | GH5/CBM1 | 649.86 | 16.21 | Y | mannan, galactomannan, glucomannan |
| F1DGF4 | Afu6g11600 | endoglucanasea,c | GH5/CBM1 | 8172.11 | 36.52 | Y | cellulose |
| Q4WE56 | Afu5g01830 | extracellular endoglucanasea | GH5/CBMX2 | 6764.9 | 19.83 | Y | cellulose |
| B0XWL3 | Afu3g01910 | cellobiohydrolasea,c | GH6/CBM1 | 6069.09 | 23.13 | Y | cellulose |
| Q4X0P5 | Afu2g12770 | alpha-L-arabinofuranosidasea | GH62 | 9479.28 | 40.66 | Y | arabinoxylan, arabinogalactan |
| Q4WIR4 | Afu2g00920 | extracellular glycosyl hydrolase/cellulasea,c | GH62/CBM1 | 3622.83 | 28.28 | Y | arabinoxylan |
| B0Y793 | Afu6g07070 | cellobiohydrolase celDa,c | GH7 | 21,820.01 | 54.65 | Y | cellulose |
| Q4WM08 | Afu6g11610 | 1,4-beta-D-glucan-cellobiohydrolyasea,b | GH7/CBM1 | 24,706.69 | 35.53 | Y | cellulose |
| T1YVP0 | N/A | Glucanasea | GH7/CBM1 | 6572.88 | 24.78 | Y | cellulose |
| Q4WLW1 | Afu6g12120 | BNR/Asp-box repeat domain protein | GH93 | 778.23 | 16.32 | Y | pectin |
We have also identified lignin-depolymerizing enzymes such as catalase-peroxidase, cellobiose dehydrogenase, catalase B, FAD-dependent oxidase, laccase, and Cu-Zn superoxide dismutase. Although the sugarcane bagasse employed here was treated by steam explosion, traces of lignin might have remained in the substrate, which justifies the secretion of these enzymes by the fungus.
We can conclude that the most important CAZymes (GH3, GH5, GH6, GH7, GH10, GH11, GH43, GH62, GH93, CE1, CE16 and AA9 (LPMO)) were secreted and play important roles in biomass degradation. For the first time, new proteins such as GH16 (endo-1,4-beta-glucanase), GH5 (endoglucanase), LPMO (AA9), swollenin and GH3 (β-glucosidase) were identified in the Aspergillus fumigatus secretome, probably because we used sugarcane bagasse as the source of carbon, and these enzymes can be specific to this complex biomass.
Integration of secretomics and transcriptomics
We observed weak correlations between transcriptome and secretome datasets, mainly because we chose the same time (24 h) to isolate mRNA and proteins. Considering that Aspergillus needs at least a few hours to translate mRNA to protein and to secrete it, these data provide an idea about which proteins are transcribed earlier or produced constitutively. Among the 1181 upregulated genes in transcriptome, 63 encoded proteins were detected in the secretome. As the same way, 16 of secreted proteins were identified as downregulated in RNAseq data (Fig. 6). In addition, the weak correlation observed could be the result of the influence of some factors that alter transcription and translation mechanisms [22, 23, 54].
Fig. 6.
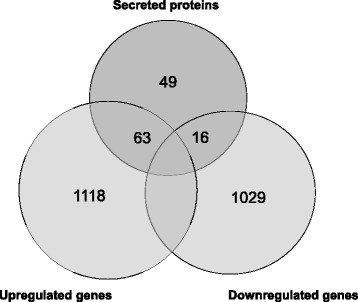
Intersection of differentially expressed genes and secreted proteins. Venn diagram of transcriptome (upregulated genes and downregulated genes) and proteins secreted by A. fumigatus when grown on SEB
The both data revealed a significant upregulation of secreted CAZymes, which is important for the observation of specific alterations triggered by different conditions. Among the 135 upregulated CAZyme genes, 48 encoded proteins were detected in the secretome, mainly cellulolytic and hemicellulolytic enzymes (Table 3). Four enzymes, β-1,3-endoglucanase EglC (Afu3g00270), FAD-dependent oxygenase (Afu3g00840), FAD/FMN-containing isoamyl alcohol oxidase MreA (Afu6g03620) and oligopeptidase family protein (Afu8g04730), were downregulated when the strain was cultivated in SEB but were detected in the secretome.
Table 3.
CAZymes identified in both transcriptome and secretome of A. fumigatus grown on SEB
| Gene ID | Gene Description | CAZy family | log2FC |
|---|---|---|---|
| Afu3g00320 | endo-1,4-beta-xylanase (XlnA) | GH11 | 10.39 |
| Afu6g11610 | 1,4-beta-D-glucan-cellobiohydrolyase | GH7 | 9.83 |
| Afu6g01800 | endoglucanase | CBM1 | 9.58 |
| Afu2g00920 | extracellular glycosyl hydrolase/cellulase | CBM1 | 9.35 |
| Afu6g13610 | endo-1,4-beta-xylanase | CBM1 | 9.21 |
| Afu6g12120 | BNR/Asp-box repeat domain protein | GH93 | 9.07 |
| Afu3g01910 | Putative cellobiohydrolase | GH6 | 9.02 |
| Afu4g09480 | extracellular endo-1,4-beta-xylanase | GH10 | 8.82 |
| Afu4g07850 | endoglucanase | AA9 | 8.75 |
| Afu2g12770 | alpha-L-arabinofuranosidase | GH62 | 8.54 |
| Afu6g11600 | endoglucanase | CBM1 | 8.11 |
| Afu2g00930 | xylosidase/glycosyl hydrolase | GH43 | 7.97 |
| Afu3g03080 | Endo-1,3(4)-beta-glucanase, putative | GH16 | 7.70 |
| Afu8g06570 | acetyl xylan esterase | CBM1 | 7.57 |
| Afu6g03280 | swollenin | CBM1 | 7.51 |
| Afu8g06890 | Probable endo-xylogalacturonan hydrolase A (xghA) | GH28 | 6.99 |
| Afu6g12210 | endo-1,4-beta-xylanase (XynG1) | GH11 | 6.91 |
| Afu6g07070 | cellobiohydrolase celD | GH7 | 6.78 |
| Afu5g01190 | Uncharacterized protein, alpha-L-fucosidase activity | GH65 | 6.49 |
| Afu2g17620 | Cellobiose dehydrogenase | AA8 | 6.45 |
| Afu2g15420 | Uncharacterized protein | GH131 | 6.41 |
| Afu6g14540 | Endo-1,3(4)-beta-glucanase, putative | GH16 | 6.37 |
| Afu8g01410 | Endochitinase B1 (chiB1) | GH18 | 5.79 |
| Afu2g03980 | Alpha-1,3-glucanase/mutanase, putative | GH71 | 5.38 |
| Afu3g14510 | Rhamnogalacturonan acetylesterase (RgaE) | CE12 | 5.19 |
| Afu7g05610 | glucanase | GH5 | 5.07 |
| Afu7g05140 | Class III chitinase, putative | GH18/CBM19 | 4.86 |
| Afu3g07160 | Class V chitinase, putative | GH18 | 4.25 |
| Afu3g00420 | acetyl xylan esterase (Axe1) | CBM1 | 4.13 |
| Afu8g04710 | xylosidase | GH43 | 4.04 |
| Afu6g12070 | FAD binding domain protein | AA7 | 3.48 |
| Afu1g14710 | beta-glucosidase | GH1 | 3.42 |
| Afu6g10130 | N, O-diacetyl muramidase, putative | GH25 | 3.23 |
| Afu1g14450 | Exo-1,3-beta-D-glucanase, putative (exgO) | GH55 | 3.19 |
| Afu2g00690 | glucan 1,4-alpha-glucosidase | CBM20 | 2.94 |
| Afu5g10930 | Uncharacterized protein | PL20 | 2.91 |
| Afu6g07480 | endoglucanase | GH5 | 2.9 |
| Afu5g10520 | Alpha-1,2-mannosidase family protein | GH92 | 2.86 |
| Afu8g07120 | Endo-1,6-beta-D-glucanase neg1 | GH30 | 2.76 |
| Afu2g14520 | Hydrolase, putative | GH2 | 2.67 |
| Afu1g05290 | Endo-1,3(4)-beta-glucanase, putative | GH16 | 2.38 |
| Afu2g14530 | esterase D | CE1 | 2.16 |
| Afu5g01830 | extracellular endoglucanase | GH5 | 2 |
| Afu2g14750 | endo-arabinase | GH43 | 1.92 |
| Afu8g07030 | endo-1,4-beta-mannosidase | GH5 | 1.82 |
| Afu8g02510 | glycosyl hydrolase family 43 protein | CBM35 | 1.61 |
| Afu4g13770 | glycosyl hydrolase | GH3 | 1.55 |
| Afu6g02560 | Alpha-galactosidase | GH27 | 1.36 |
The variation in expression during fungal growth in the presence of SEB was observed for some genes in the qRT-PCR data (Fig. 3). We observed that the gene expression depends on the duration of incubation, which explains the low correlation between the data of the secretome and transcriptome and supports the hypothesis that the approach used in this study is able to provide information about the modulation of gene expression of A. fumigatus in the presence of sugarcane bagasse. Genes encoding enzymes that are upregulated in the transcriptome (the highest log2FC) and are present in the proteome, as well, are described in Table 3. The main enzymes produced by the fungus are endo-β1,4-xylanase, cellobiohydrolase B, endoglucanase and arabinofuranosidase, which leads us to speculate that A. fumigatus spent its energy on the transcription and production of cellulolytic and xylanolytic compounds, which were probably being secreted for biomass degradation and the release of small sugars.
Discussion
In Brazil, sugarcane bagasse is an important agro-industrial residue. It is composed of cellulose (25–45%), hemicellulose (28–32%), lignin (15–25%) and a small percentage of other compounds [70, 71]. Despite this recalcitrant composition, efficient hydrolysis mechanisms allow the release of fermentable sugars from sugarcane bagasse and their later use to produce cellulosic ethanol [18, 72–74]. Transcriptomic and/or proteomic studies on filamentous fungi have been employed to understand and to improve enzymatic cocktails to deconstruct plant biomass. Such studies have revealed a huge repertoire of cellulases and hemicellulases [4, 18, 19, 75–77]. N. crassa, A. niger, T. reesei and A. nidulans are excellent enzyme producers for industrial applications, and several studies have focused on these fungi [78–82]. Understanding the molecular mechanisms through which fungi degrade plant biomass can improve the SEB saccharification step, which is important for Brazilian 2G ethanol production [4, 18, 19, 83, 84].
To gain more insight into new enzymes and to identify new genes specific to sugarcane biomass hydrolysis, we have chosen to investigate A. fumigatus because it is widely distributed in the environment, it can degrade plant biomass, and it is an excellent enzyme producer [21]. To the best of our knowledge, this is the first study on transcriptional response and secretome of A. fumigatus grown on sugarcane steam-exploded bagasse.
There are few omic studies on biomass hydrolysis by A. fumigatus [22–25], and only one study has analyzed the transcriptome of A. fumigatus when grown on polysaccharide substrates. Miao et al. [25] conducted a transcriptional study on the induction of CAZymes by this fungus grown on cellulose, oat spelt xylan, rice straw and sucrose. The authors showed that important genes are differentially expressed in each carbon source.
Here, we found few discrepancies in the number of induced genes when we used SEB as carbon source (Table 4). The main CAZyme families (GH1, GH3, GH5, GH6, GH7, GH10, GH11, GH12, GH43, GH62, GH67, GH74, AA9, CE3, CE5 and CE16 [85]) were also induced. However, important genes including GH45, GH51, GH54, GH93, GH115, and CE1 were downregulated in SEB. Furthermore, two important genes were exclusively induced in SEB: PL4 (Afu4g03780 and Afu8g00820), which plays a role in pectin breakdown, and CBM35 (Afu8g02510), which is known to bind primarily to xylan and mannans [86]. The distinct gene expression was most likely due to substrate composition and to cultivation time, which was 24 h for our analysis. Shorter cultivation times could point out new pattern to gene induction. In qRT-PCR data, we observed that 6 h is the beginning of gene expression, which might represent a standard mechanism in which A. fumigatus acts in contact with complex biomass sources and should contain more highly induced enzymes. These results also explain the percentage (35%) of differentially expressed CAZymes identified in the RNA-Seq data at 24 h.
Table 4.
Aspergillus fumigatus transcriptome studies using different carbon sources
| Strain | Technology | Carbon source | Time | Total number of Cazymes genes upregulated | Reference |
|---|---|---|---|---|---|
| A. fumigatus Z5 | Illumina | cellulose, xylan, rice straw, oat spelt | 16 h | 47, xylan; 143, rice straw; 157, cellulose | [25] |
| A. fumigatus AF293 | Illumina | sugarcane exploded bagasse | 24 h | 135, SEB | This work |
Because glycosyltransferases (GTs) contribute to fungal cell remodeling, the percentage of upregulated genes was low (~ 1%). Likewise, Miao et al. [25] described that genes encoding GTs are downregulated in Z5 strain, which supports the idea that GTs do not directly participate in the hydrolysis of complex biomass.
In this sense, the biomass itself has to be investigated in order to understand sugarcane biomass hydrolysis as well as possible. A similar work performed by Borin et al., 2017 [18] described a transcriptional response of A. niger and T. reesei grown in SEB for different periods. They found 190 upregulated CAZymes from 62 different families in A. niger, and 105 genes of 51 CAZyme families in T. reesei, whereas we detected 135 upregulated CAZymes in the A. fumigatus transcriptome herein. The number of genes induced by each microorganism was different and depended on time. A higher number of DEGs in A. niger and T. reesei was observed in 24 h of culture, and so we then compared our data at this duration, and again small differences in upregulated CAZymes were observed.
After biomass hydrolysis, which breaks down cellulose and hemicellulose into mono- or disaccharides, the released sugars need to be transported into the cells through a large number of sugar transporters, most of which have not been characterized yet [20, 21, 87]. One of the main challenges concerning biofuel production from lignocellulosic biomass is the inability of organisms to grow on, to transport, and to ferment sugars other than glucose (e.g., xylose and cellobiose). Gaining a better insight into potential xylose and/or cellobiose transporters seems to be a good approach to overcome this challenge [87]. These transporters represent an important industrial tool that can be applied to different industrial processes [88–90]. Additionally, the genomes of filamentous fungi also encode large numbers of major facilitator superfamily (MFS) transporters. For example, the T. reesei and A. nidulans genomes have been predicted to encode 164 and 357 proteins belonging to MFS, respectively, although the exact number of proteins involved in sugar transport remains unknown [91–93].
We were also interested in new sugar transporters, such as the xylose transporter. Until now, no sugar transporter for A. fumigatus related to biomass breakdown has been described. We verified that the 25 DEG homolog transporters had particular expression profiles, upregulated or downregulated, suggesting that SEB hydrolysis released enough glucose, xylose or cellobiose to regulate sugar transporter gene expression. Another interesting finding was that two sugar transporters (Afu6g14442 and Afu1g03530) were highly induced in SEB (25.5 times and 10 times, respectively) and in the presence of 1% xylose, which revealed that these sugar transporters could be specific xylose transporters in Aspergillus fumigatus. Overexpression of xylose transporters in S. cerevisiae is a fast way to use xylose and may improve ethanol productivity [94].
In addition to the transcriptome, we evaluated the proteins secreted by A. fumigatus by SDS-PAGE and LC-MS/MS. Similarly, three studies compared the secretome of A. fumigatus on complex substrates (Table 2) [22–24]. Liu et al., [24] identified 152 proteins on rice straw and 125 different proteins on Avicel. Adav et al., [23] quantified 73 proteins belonging to cellulases, glycoside hydrolases and amylases. We detected some secreted proteins that were also identified when A. fumigatus was grown on corn, wheat, soybean, Avicel, rice straw, xylan, and starch as carbon source. However, for the first time, we verified important secreted CAZymes like swollenin (CBM1), two putative endoglucanases (LPMO) (AA9), acetyl xylan esterase (CBM1), two feruloyl esterases (CE1), endo-1,4-beta-xylanase (GH11), endo-1,3(4)-beta-glucanase (GH16), glycosyl hydrolase (GH3), endoglucanase (GH5), arabinanase (GH43), endo-1,4-beta-mannosidase (GH5/CBM1), and arabinogalactan endo-1,4-beta-galactosidase (GH53). In this way, we can conclude that we detected similar amounts of proteins with those previously described, which may be specific for SEB biomass.
These data showed that, although the transcriptome data did not reveal potential new enzyme targets for the deconstruction of sugarcane biomass deconstruction, the secretome analyses indicated key enzymes that may be essencial for this hydrolysis and which act synergistically for efficient deconstruction. We clearly observed the need for accessory enzymes secreted as LPMO and swollenin, which have never been described in other A. fumigatus secretome analyses [95], which allowed us to conclude that the other secreted CAZymes together with AA9 identified in A. fumigatus form a potential arsenal of hydrolytic enzymes.
LPMOs have recently been implicated in lignocellulosic biomass degradation. Although these enzymes were first classified into the GH61 and CBM33 families, they are currently classified into the AA9 and AA10 families. These enzymes cleave the lignocellulosic biomass glycosidic bonds through an oxidative mechanism that provides new ends for the recognition of cellulases and for action on cellulose [95, 96]. In addition, AA9s have been identified in A. niger, M. thermophile, T. asperellum, and T. reesei secretome growth in sugarcane bagasse, which allows us to conclude that they play an important role in lignocellulosic biomass breakdown [4, 95–99].
Many studies have focused on LPMO enzymes, and some works have even characterized them, but no investigations into LPMO enzymes from A. fumigatus are available so far [96]. Hence, the role of most of these enzymes remains unclear, and AA expression during A. fumigatus growth on bagasse suggests that they play an important part in biomass degradation.
Together, the transcriptome and secretome have shown several enzymes that A. fumigatus uses to hydrolyze SEB and which most likely act synergistically to depolymerize cellulose and hemicellulose. In most Aspergillus species, distinct genes encode the same class of enzymes (isoenzymes) [77], as observed by the data regarding the A. fumigatus secretome and transcriptome. Our results suggest that complete hydrolysis of this lignocellulose biomass to simple sugars, such as glucose, xylose, and arabinose, requires the combined actions of several enzymes that have different substrate specificities and act synergistically. The great potential of this species is evident, and its enzymes can contribute to optimization of enzymatic cocktails for use in 2G bioethanol production.
Conclusion
Through these findings, it is suggested that different biomasses require a set of enzymes due its complexity and A. fumigatus Af293 is an excellent CAZymes producer for sugarcane biomass breakdown. The analysis of proteome and transcriptome revealed a set of CAZymes highly expressed and secreted, such as cellulases, hemicellulases, delignification and auxiliary enzymes necessary to SEB breakdown. In addition, from CAZymes proteins, LPMOs, which could contribute to better degradation of cellulose, were also detected in A. fumigatus secretome. Cellobiohydrolases, endoglucanases and LPMOs can act synergistically in cellulose depolymerization and LPMOs can be included in the most advanced enzymatic cocktails. Altogether, the data show that despite the pathogenicity of A. fumigatus, it can produce a wide variety of enzymes, which can be expressed in a nonpathogenic microorganism and may contribute to the optimization of currently marketed enzymatic cocktails for the viable production of 2G bioethanol.
Additional files
Figure S1. R script of RNA sequencing analysis. (PDF 2406 kb)
Table S1. Raw Count matrix of genes found in RNA-seq. (XLSX 585 kb)
Table S2. Sequence of primers from genes upregulated in RNA-seqdata selected for qRT-PCR. (XLSX 11 kb)
Table S3. Up and downregulated genes in SEB. (XLSX 314 kb)
Table S4. Functional enrichment analysis with FunCat of up- and downregulated genes in Aspergillus fumigatus grown in SEB. (XLSX 12 kb)
Table S5. CAZymes families in A. fumigatus genome and predicted peptide signal. (XLSX 32 kb)
Table S6. A. fumigatus sugar transporters modulated in RNA-seq data. (XLSX 13 kb)
Figure S2. Proteins from A. fumigatus secretome separated by SDS-PAGE. (PPTX 283 kb)
Table S7. Proteins detected in the secretome of Aspergillus fumigatus grown in SEB by coupled system type LC-MS/MS. (XLSX 37 kb)
Acknowledgments
The authors would like to thank Laboratório de Alto Desempenho em Sequenciamento e Robótica do Laboratório Nacional de Ciência e Tecnologia do Bioetanol (CTBE), Campinas, SP, Brazil and its technician Dr. Douglas Paixão for technical support with the Illumina HiSeq 2500.
Funding
This study was supported by Fundação de Amparo À Pesquisa do Estado de São Paulo (FAPESP, 2014/10466–0). AVB and LEG were funded by scholarships granted by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). PFG was funded by a scholarship granted by Coordenação de Aperfeiçoamento de Pessoal de Ensino Superior (CAPES).
Availability of data and materials
The datasets generated and/or analysed during this study are included in this published article and its Additional files 1, 2, 3, 4, 5, 6, 7, 8 and 9. The raw data from A. fumigatus RNAseq are available at the NCBI’s Gene Expression Omnibus under the accession code GSE95798 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE95798), the underlying strand-specific raw short reads were deposited at the NCBI’s Short Read Archive and are associated to the BioProject PRJNA376829 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA376829).
Abbreviations
- ASPGD
Aspergillus genome database
- bp
Base pair
- CAZymes
Carbohydrate-active enzymes
- cDNA
Complementary DNA
- CMC
Carboxymetilcellulose
- CPM
Counts per million
- DEGs
Differentially expressed genes
- DNA
Deoxyribonucleic acid
- DNS
Dinitrosalicylic acid
- DTT
Dithiothreitol;
- FDR
False rate discovery
- FPKM
Fragments per kilobase of exon per million mapped reads
- HCl
Hydrocloric acid
- kb
Kilobases
- log2FC
Log of fold-change in base 2
- LPMO
Lytic polysaccharides mono-oxygenase
- M
Molar (mol L− 1)
- m/z
Mass/charge relation
- min
Minute
- mM
Millimolar
- mRNA
Messenger RNA
- NGS
New generation sequencing
- PCR
Polymerase chain reaction
- pH
Hydrogenionic potential
- qRT-PCR
Real-time quantitative PCR
- RNA
Ribonucleic acid
- RNASeq
RNA sequencing
- rpm
Rotations per minute
- SDS
Sodium dodecyl sulphate
- SDS-PAGE
SDS-polyacrilamide gel electrophoresis
- SEB
Steam-exploded bagasse
- Tris
Tris- (hydroxymethyl) aminoethane
- U
Units
- V
Volts
- v/v
Volume/volume ratio
- w/v
Weight/volume ratio
- μg
Micrograms
- μM
Micromolar
Authors’ contributions
PFG and AVB contributed substantially to the experimental work, data analyses and were involved in writing the manuscript and preparation of figures/tables, and offered intellectual contributions. LEG carried out secretome experiments and intellectual contributions. ESS carried out RNAseq experiments, data analyses and offered intellectual contributions. DMRP performed the analysis of RNAseq data, offered intellectual contributions and wrote the manuscript. SAU offered intellectual contributions at various stages of this work and provided critical reviews of the manuscript. TMD participated in the design and coordination of the study, wrote the manuscript, and conducted the final analysis of the data. All the authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4627-8) contains supplementary material, which is available to authorized users.
Contributor Information
Paula Fagundes de Gouvêa, Email: paulafgouvea@yahoo.com.br.
Aline Vianna Bernardi, Email: alinevbernardi@gmail.com.
Luis Eduardo Gerolamo, Email: gerolamo00@hotmail.com.
Emerson de Souza Santos, Email: emerson_usp@yahoo.com.br.
Diego Mauricio Riaño-Pachón, Email: diriano@gmail.com.
Sergio Akira Uyemura, Email: suyemura@ffclrp.usp.br.
Taisa Magnani Dinamarco, Email: tdinamarco@ffclrp.usp.br.
References
- 1.Souza GM, Victoria RL, Joly CA, Verdade LM. Bioenergy & Sustainability: bridging the gaps. 2015. [Google Scholar]
- 2.Cheng JJ, Timilsina GR. Status and barriers of advanced biofuel technologies: a review. Renew En. 2011;36:3541–3549. doi: 10.1016/j.renene.2011.04.031. [DOI] [Google Scholar]
- 3.Moysés DN, Reis VC, de Almeida JR, de Moraes LM, Torres FA. Xylose fermentation by Saccharomyces cerevisiae: challenges and prospects. J Mol Sci. 2016;17:207. doi: 10.3390/ijms17030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borin GP, Sanchez CC, de Souza AP, de Santana ES, de Souza AT, Leme AFP, et al. Comparative secretome analysis of Trichoderma reesei and Aspergillus niger during growth on sugarcane biomass. PLoS One. 2015;10(6):e0129275. doi: 10.1371/journal.pone.0129275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novacana. http://www.novacana.com (2016) Accessed 20 Nov 2016.
- 6.Conab. http://www.conab.gov.br (2017) Accessed 10 Nov 2017.
- 7.Milanez AY, Nyko D, Valente MS, de Sousa LC, Bonomi A, de Jesus CDF, et al. De promessa a realidade: como o etanol celulósico pode revolucionar a indústria da cana-de-açúcar: uma avaliação do potencial competitivo e sugestões de política pública. BNDES Setorial, Rio de Janeiro. 2015;41:237–294. [Google Scholar]
- 8.Batalha LAR, Han Q, Jameel H, Chang H, Colodette JL, Gomes FJB. Production of fermentable sugars from sugarcane bagasse by enzymatic hydrolysis after autohydrolysis and mechanical refining. Bioresour Technol. 2015;180:97–105. doi: 10.1016/j.biortech.2014.12.060. [DOI] [PubMed] [Google Scholar]
- 9.Pereira SC, Maehara L, Machado CM, Farinas CS. 2G ethanol from the whole sugarcane lignocellulosic biomass. Biotechnol Biofuels. 2015;8:44–60. [DOI] [PMC free article] [PubMed]
- 10.Sindhu R, Binod P, Pandey A. Biological pretreatment of lignocellulosic biomass - an overview. Bioresour Technol. 2016;199:76–82. doi: 10.1016/j.biortech.2015.08.030. [DOI] [PubMed] [Google Scholar]
- 11.Sambusiti C, Licari A, Solhy A, Aboulkas A, Cacciaguerra T, Barakat A. One-pot dry chemo-mechanical deconstruction for bioethanol production from sugarcane bagasse. Bioresour Technol. 2015;181:200–206. doi: 10.1016/j.biortech.2015.01.058. [DOI] [PubMed] [Google Scholar]
- 12.Glass NL, Schmoll M, Cate JHD, Coradetti S. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Fan C, Hu H, Li Y, Sun D, Wang Y, Peng L. Genetic modification of plant cell walls to enhance biomass yield and biofuel production in bioenergy crops. Biotechnol Adv. 2016;34:997–1017. doi: 10.1016/j.biotechadv.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Sun Y, Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol. 2002;83:1–11. doi: 10.1016/S0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 15.Behera S, Arora R, Nandhagopal N, Kumar S. Importance of chemical pretreatment for bioconversion of lignocellulosic biomass. Renew Sustain En Rev. 2014;36:91–106. doi: 10.1016/j.rser.2014.04.047. [DOI] [Google Scholar]
- 16.Cragg SM, Beckham GT, Bruce NC, Bugg TD, Distel DL, Dupree P, et al. Lignocellulose degradation mechanisms across the tree of life. Curr Opin Chem Biol. 2015;29:108–119. doi: 10.1016/j.cbpa.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma RK, Arora DS. Fungal degradation of lignocellulosic residues: an aspect of improved nutritive quality. Crit Rev Microbiol. 2015;41(1):52–60. doi: 10.3109/1040841X.2013.791247. [DOI] [PubMed] [Google Scholar]
- 18.Borin GP, Sanchez CC, Santana ES, Zanini GK, dos Santos RAC, Pontes AO, et al. Comparative transcriptome analysis reveals different strategies for degradation of steam-exploded sugarcane bagasse by Aspergillus niger and Trichoderma reesei. BMC Genomics. 2017;18(1):501. doi: 10.1186/s12864-017-3857-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damasio AR, Rubio MV, Gonçalves TA, Persinoti GF, Segato F, Prade RA, et al. Xyloglucan breakdown by endo-xyloglucanase family 74 from Aspergillus fumigatus. Appl Microbiol Biotechnol. 2017;101:2893–2903. doi: 10.1007/s00253-016-8014-6. [DOI] [PubMed] [Google Scholar]
- 20.Culleton H, McKie V, de Vries RP. Physiological and molecular aspects of degradation of plant polysaccharides by fungi: what have we learned from Aspergillus? Biotechnol J. 2013;8:884–894. doi: 10.1002/biot.201200382. [DOI] [PubMed] [Google Scholar]
- 21.van den Brink J, de Vries RP. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 2011;91(6):1477–1492. doi: 10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma GP, Ouyang H, Wang Q, Luo Y, Shi B, Yang J, et al. Insight into enzymatic degradation of corn, wheat, and soybean cell wall cellulose using quantitative secretome analysis of Aspergillus fumigatus. J Proteome Res. 2016;15(12):4387–4402. doi: 10.1021/acs.jproteome.6b00465. [DOI] [PubMed] [Google Scholar]
- 23.Adav SS, Ravindran A, Sze SK. Quantitative proteomic study of aspergillus fumigatus secretome revealed deamidation of secretory enzymes. J Proteome. 2015;119:154–168. doi: 10.1016/j.jprot.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Li J, Zhao S, Zhang R, Wang M, Miao Y, et al. Secretome diversity and quantitative analysis of cellulolytic Aspergillus fumigatus Z5 in the presence of different carbon sources. Biotechnol Biofuels. 2013;6(1):149. doi: 10.1186/1754-6834-6-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao Y, Liu D, Li G, Li P, Xu Y, Shen Q, Zhang R. Genome-wide transcriptomic analysis of a superior biomass-degrading strain of a. Fumigatus revealed active lignocellulose-degrading genes. BMC Genomics. 2015;16:459. doi: 10.1186/s12864-015-1658-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amore A, Giacobbe S, Faraco V. Regulation of cellulase and hemicellulase gene expression in fungi. Curr Gen. 2013;14:230–249. doi: 10.2174/1389202911314040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 28.Andrew S. FastQC: a quality control tool for high throughput sequence data. 2010. [Google Scholar]
- 29.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28(24):3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 31.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, et al. The aspergillus genome database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014;42(1):D705–D710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 35.Liao Y, Smyth GK, Shi W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Development Core Team . R: a language and environment for statistical computing. 2015. [Google Scholar]
- 37.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glueck DH, Mandel J, Karimpour-Fard A, Hunter L, Keith E. Exact calculations of average power for the Benjamini-Hochberg procedure. Int J Biostat. 2008;4(1):11. doi: 10.2202/1557-4679.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammond JBW, Kruger NJ. The Bradford method for protein quantitation. In: Walker JM, editor. New protein techniques. Methods in molecular biology™, vol 3: Humana Press; 1988. [DOI] [PubMed]
- 41.Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, et al. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pundir S, Martin MJ, O’Donovan C, UniProt Consortium. UniProt tools. Curr Protoc Bioinformatics. 2016;53:1.29–1-15. www.uniprot.org. Accessed 10 Dec 2015. [DOI] [PMC free article] [PubMed]
- 43.Briesemeister S, Rahnenführer J, Kohlbacher O. Going from where to why--interpretable prediction of protein subcellular localization. Bioinformatics. 2010;26(9):1232–1238. doi: 10.1093/bioinformatics/btq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat methods. 2011;8(10):785–6. [DOI] [PubMed]
- 45.Bendtsen JD, Jensen LJ, Blom N, von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17(4):349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 46.Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, et al. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43(W1):W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin Y, Mao X, Yang JC, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semighini CP, Marins M, Goldman MHS, Goldman GH. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl Environ Microbiol. 2002;68(3):1351–1357. doi: 10.1128/AEM.68.3.1351-1357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, Wymore F, Wortman JR, Sherlock G. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.BioInforx. http://bioinforx.com/free/bxarrays/venndiagram.php Accessed 10 July 2016.
- 52.Carbohydrate Active Enzymes Database. Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 2014;42:D490–D495. http://www.cazy.org Accessed 26 Mar 2016. [DOI] [PMC free article] [PubMed]
- 53.Percival YHZ, Himmel ME, Mielenz JR. Outlook for cellulase improvement: screening and selection strategies. Biotechnol Adv. 2006;24:452–481. doi: 10.1016/j.biotechadv.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 54.Colabardini AC, Ries LN, Brown NA, Dos Reis TF, Savoldi M, Goldman MH, et al. Functional characterization of a xylose transporter in Aspergillus nidulans. Biotechnol Biofuels. 2014;7(1):46. doi: 10.1186/1754-6834-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Souza WR, de Gouvea PF, Savoldi M, Malavazi I, Bernardes LAS, Goldman MH, et al. Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotechnol Biofuels. 2011;4:40. doi: 10.1186/1754-6834-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer V, Arentshorst M, Flitter SJ, Nitsche BM, Kwon MJ, Reynaga-Peña CG, et al. Reconstruction of signaling networks regulating fungal morphogenesis by transcriptomics. Eukaryot Cell. 2009;8(11):1677–1691. doi: 10.1128/EC.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis DA, Bisson LF. The HXT1 gene product of Saccharomyces cerevisiae is a new member of the family of hexose transporters. Mol Cell Biol. 1991;11(7):3804–3813. doi: 10.1128/MCB.11.7.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan J, Chaturvedi V, Shen SH. Identification and phylogenetic analysis of a glucose transporter gene family from the human pathogenic yeast Candida albicans. J Mol Evol. 2002;55(3):336–346. doi: 10.1007/s00239-002-2330-4. [DOI] [PubMed] [Google Scholar]
- 59.Pengli C, Ruimeng G, Bang W, Jingen L, Li W, Chaoguang T, Yanhe M. Evidence of a critical role for cellodextrin transporter 2 (CDT-2) in both cellulose and hemicellulose degradation and utilization in Neurospora crassa. PLoS One. 2014;9(2):e89330. doi: 10.1371/journal.pone.0089330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamano K, Sano M, Yamane N, Terabayashi Y, Toda T, Sunagawa M, et al. Transcriptional regulation of genes on the non-syntenic blocks of Aspergillus oryzae and its functional relationship to solid-state cultivation. Fungal Genet Biol. 2008;45(2):139–151. doi: 10.1016/j.fgb.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 61.Wei H, Vienken K, Weber R, Bunting S, Requena N, Fischer R. A putative high affinity hexose transporter, hxtA, of Aspergillus nidulans is induced in vegetative hyphae upon starvation and in ascogenous hyphae during cleistothecium formation. Fungal Genet Biol. 2004;41(2):148–156. doi: 10.1016/j.fgb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 62.Kwon MJ, Jørgensen TR, Nitsche BM, Arentshorst M, Park J, Ram AF, Meyer V. The transcriptomic fingerprint of glucoamylase over-expression in Aspergillus niger. BMC Genomics. 2012;13:701. doi: 10.1186/1471-2164-13-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jørgensen TR, Goosen T, Hondel CA, Ram AF, Iversen JJ. Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway. BMC Genomics. 2009;10:44. doi: 10.1186/1471-2164-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guillemette T, van Peij N, Goosen T, Lanthaler K, Robson GD, van den Hondel CA, et al. Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics. 2007;8:158. doi: 10.1186/1471-2164-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noguchi Y, Sano M, Kanamaru K, Ko T, Takeuchi M, Kato M, Kobayashi T. Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl Microbiol Biotechnol. 2009;85(1):141–154. doi: 10.1007/s00253-009-2236-9. [DOI] [PubMed] [Google Scholar]
- 66.Andersen MR, Nielsen J. Current status of systems biology in aspergilli. Fungal Genet Biol. 2009; 10.1016/j.fgb.2008.07.006. [DOI] [PubMed]
- 67.Sá-Pessoa J, Amillis S, Casal M, Diallinas G. Expression and specificity profile of the major acetate transporter AcpA in Aspergillus nidulans. Fungal Genet Biol. 2015;76:93–103. doi: 10.1016/j.fgb.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 68.Salazar M, Vongsangnak W, Panagiotou G, Andersen MR, Nielsen J. Uncovering transcriptional regulation of glycerol metabolism in Aspergilli through genome-wide gene expression data analysis. Mol Gen Genomics. 2009;282(6):571–586. doi: 10.1007/s00438-009-0486-y. [DOI] [PubMed] [Google Scholar]
- 69.Fekete E, Karaffa L, Seiboth B, Fekete E, Kubicek CP, Flipphi M. Identification of a permease gene involved in lactose utilisation in Aspergillus nidulans. Fungal Genet Biol. 2012;49(6):415–425. doi: 10.1016/j.fgb.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 70.Lima MS, Damasio AR, Crnkovic PM, Pinto MR, da Silva AM, da Silva JC, et al. Co-cultivation of Aspergillus nidulans recombinant strains produces an enzymatic cocktail as alternative to alkaline sugarcane bagasse pretreatment. Front Microbiol. 2016;7:583. doi: 10.3389/fmicb.2016.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buckeridge MS. Seed cell wall storage polysaccharides: models to understand cell wall biosynthesis and degradation. Plant Physiol. 2010;154:1017–1023. doi: 10.1104/pp.110.158642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farzad S, Mandegari MA, Guo M, Haigh KF, Shah N, Görgens JF. Multi-product biorefineries from lignocelluloses: a pathway to revitalisation of the sugar industry? Biotechnol Biofuels. 2017;10:87. doi: 10.1186/s13068-017-0761-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Calderan-Rodrigues MJ, Jamet E, Douché T, Bonassi MB, Cataldi TR, Fonseca JG, et al. Cell wall proteome of sugarcane stems: comparison of a destructive and a non-destructive extraction method showed differences in glycoside hydrolases and peroxidases. BMC Plant Biol. 2016;16:14. doi: 10.1186/s12870-015-0677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buckeridge MS, de Souza AP. Breaking the “Glycomic code” of cell wall polysaccharides may improve second-generation bioenergy production from biomass. Bioenergy Res. 2014;7(4):1065–1073. doi: 10.1007/s12155-014-9460-6. [DOI] [Google Scholar]
- 75.Ribeiro DA, Cota J, Alvarez TM, Bruchli F, Bragato J, Pereira BM, et al. The Penicillium echinulatum secretome on sugar cane bagasse. PLoS One. 2012;7:e50571. doi: 10.1371/journal.pone.0050571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saykhedkar S, Ray A, Ayoubi-Canaan P, Hartson SD, Prade RA, Mort AJ. A time course analysis of the extracellular proteome of Aspergillus nidulans growing on sorghum Stover. Biotechnol Biofuels. 2012;5(1):52. doi: 10.1186/1754-6834-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benoit I, Culleton H, Zhou M, DiFalco M, Aguilar-Osorio G, Battaglia E, et al. Closely related fungi employ diverse enzymatic strategies to degrade plant biomass. Biotechnol Biofuels. 2015;8:107. doi: 10.1186/s13068-015-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 79.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 80.Pel HJ, De Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 81.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26(5):553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 82.Pontercorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AWJ. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 83.Vicentini R, Bottcher A, Brito MS, Dos Santos AB, Creste S, Landell G, et al. Large-scale transcriptome analysis of two sugarcane genotypes contrasting for lignin content. PLoS One. 2015;10(8):e0134909. doi: 10.1371/journal.pone.0134909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horta MA, Vicentini R, Delabona PS, Laborda P, Crucello A, Freitas S, et al. Transcriptome profile of Trichoderma harzianum IOC-3844 induced by sugarcane bagasse. PLoS One. 2014;9(2):e88689. doi: 10.1371/journal.pone.0088689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Segato F, Damasio AR, de Lucas RC, Squina FM, Prade RA. Genomics review of holocellulose deconstruction by Aspergilli. Microbiol Mol Biol Rev. 2014;78:588–613. doi: 10.1128/MMBR.00019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Montanier C, van Bueren AL, Dumon C, Flint JE, Correia MA, Prates JA, et al. Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc Natl Acad Sci U S A. 2009;106(9):3065–3070. doi: 10.1073/pnas.0808972106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dos Reis TF, de Lima PB, Parachin NS, Mingossi FB, Oliveira JVC, Ries LN, Goldman GH. Identification and characterization of putative xylose and cellobiose transporters in Aspergillus nidulans. Biotechnol Biofuels. 2016;9:204. doi: 10.1186/s13068-016-0611-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Turner TL, Kim H, Kong II, Liu JJ, Zhang GC, Jin YS. Engineering and evolution of Saccharomyces cerevisiae to produce biofuels and chemicals. Adv Biochem Eng Biotechnol. 2016; 10.1007/10_2016_22. [DOI] [PubMed]
- 89.Zhang J, Zhang B, Wang D, Gao X, Hong J. Improving xylitol production at elevated temperature with engineered Kluyveromyces marxianus through over-expressing transporters. Bioresour Technol. 2015;175:642–645. doi: 10.1016/j.biortech.2014.10.150. [DOI] [PubMed] [Google Scholar]
- 90.Zhang W, Yi ZL, Huang JF, Li FC, Hao B, Li M, et al. Three lignocellulose features that distinctively affect biomass enzymatic digestibility under NaOH and H2SO4 pretreatments in Miscanthus. Bioresour Technol. 2013;130:30–37. doi: 10.1016/j.biortech.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 91.Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, et al. Fungal cellulases. Chem Rev. 2015;115:1308–1448. doi: 10.1021/cr500351c. [DOI] [PubMed] [Google Scholar]
- 92.Jordan DB, Bowman MJ, Braker JD, Dien BS, Hector RE, Lee CC, et al. Plant cell walls to ethanol. Biochem J. 2012;442:241–252. doi: 10.1042/BJ20111922. [DOI] [PubMed] [Google Scholar]
- 93.Ferreira ME, Colombo AL, Paulsen I, Ren Q, Wortman J, Huang J, et al. The ergosterol biosynthesis pathway, transporter genes, and azole resistance in Aspergillus fumigatus. Med Mycol. 2005;43(Suppl 1):S313–S319. doi: 10.1080/13693780400029114. [DOI] [PubMed] [Google Scholar]
- 94.Hou J, Qiu C, Shen Y, Li H, Bao X. Engineering of Saccharomyces cerevisiae for the efficient co-utilization of glucose and xylose. FEMS Yeast Res. 2017;17:fox034. [DOI] [PubMed]
- 95.Berrin JG, Rosso MN, Abou HM. Fungal secretomics to probe the biological functions of lytic polysaccharide monooxygenases. Carbohydr Res. 2017;448:155–160. doi: 10.1016/j.carres.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 96.Monclaro AV, Filho EXF. Fungal lytic polysaccharide monooxygenases from family AA9: recent developments and application in lignocelullose breakdown. Int J Biol Macromol. 2017;102:771–778. doi: 10.1016/j.ijbiomac.2017.04.077. [DOI] [PubMed] [Google Scholar]
- 97.Dos Santos HB, Bezerra TMS, Pradella JGC, Delabona P, Lima D, Gomes E, et al. Myceliophthora thermophila M77 utilizes hydrolytic and oxidative mechanisms to deconstruct biomass. AMB Express. 2016;6(1):103. doi: 10.1186/s13568-016-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marx IJ, van Wyk N, Smit S, Jacobson D, Viljoen-Bloom M, Volschenk H. Comparative secretome analysis of Trichoderma asperellum S4F8 and Trichoderma reesei rut C30 during solid-state fermentation on sugarcane bagasse. Biotech Biofuels. 2013;6(1):172. doi: 10.1186/1754-6834-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Herpoël-Gimbert I, Margeot A, Dolla A, Jan G, Mollé D, Lignon S, et al. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotech Biofuels. 2008;1:18. doi: 10.1186/1754-6834-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. R script of RNA sequencing analysis. (PDF 2406 kb)
Table S1. Raw Count matrix of genes found in RNA-seq. (XLSX 585 kb)
Table S2. Sequence of primers from genes upregulated in RNA-seqdata selected for qRT-PCR. (XLSX 11 kb)
Table S3. Up and downregulated genes in SEB. (XLSX 314 kb)
Table S4. Functional enrichment analysis with FunCat of up- and downregulated genes in Aspergillus fumigatus grown in SEB. (XLSX 12 kb)
Table S5. CAZymes families in A. fumigatus genome and predicted peptide signal. (XLSX 32 kb)
Table S6. A. fumigatus sugar transporters modulated in RNA-seq data. (XLSX 13 kb)
Figure S2. Proteins from A. fumigatus secretome separated by SDS-PAGE. (PPTX 283 kb)
Table S7. Proteins detected in the secretome of Aspergillus fumigatus grown in SEB by coupled system type LC-MS/MS. (XLSX 37 kb)
Data Availability Statement
The datasets generated and/or analysed during this study are included in this published article and its Additional files 1, 2, 3, 4, 5, 6, 7, 8 and 9. The raw data from A. fumigatus RNAseq are available at the NCBI’s Gene Expression Omnibus under the accession code GSE95798 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE95798), the underlying strand-specific raw short reads were deposited at the NCBI’s Short Read Archive and are associated to the BioProject PRJNA376829 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA376829).


