Summary
Viruses that are typically benign sometimes invade the brainstem in otherwise healthy children. We report bi-allelic DBR1 mutations in unrelated patients from different ethnicities, each of whom had brainstem infection due to herpes simplex virus 1 (HSV1), influenza virus, or norovirus. DBR1 encodes the only known RNA lariat debranching enzyme. We show that DBR1 expression is ubiquitous, but strongest in the spinal cord and brainstem. We also show that all DBR1 mutant alleles are severely hypomorphic, in terms of expression and function. The fibroblasts of DBR1-mutated patients contain higher RNA lariat levels than control cells, this difference becoming even more marked during HSV1 infection. Finally, we show that the patients’ fibroblasts are highly susceptible to HSV1. RNA lariat accumulation and viral susceptibility are rescued by wild-type DBR1. Autosomal recessive, partial DBR1 deficiency underlies viral infection of the brainstem in humans through the disruption of tissue-specific and cell-intrinsic immunity to viruses.
Introduction
The incidence of viral infections of the central nervous system (CNS) is approximately 1–2 cases per 100,000 individuals each year (Jmor et al., 2008), corresponding to an estimated cumulative lifetime incidence of ~1/1,000 births. At least 20 viruses, including common viruses, such as herpes simplex viruses (HSV), influenza viruses (IV), and noroviruses (NV), can cause encephalitis (Stahl et al., 2011). Viral infections of the CNS often strike children, resulting in high mortality and severe sequelae. Intriguingly, most of the causal viruses are typically innocuous in human populations, rarely causing encephalitis. The pathogenesis of viral encephalitis in otherwise healthy individuals remained elusive until our discovery that isolated HSV1 encephalitis (HSE) can result from single-gene inborn errors of TLR3-dependent, interferon (IFN)-α/β-mediated immunity (Casrouge et al., 2006; Guo et al., 2011; Zhang and Casanova 2015). Consistent with the lack of HSV1 dissemination in children with HSE, the TLR3 pathway is redundant for innate and adaptive hematopoietic immunity, including responses to dsRNA and viruses in most leukocyte subsets (Casrouge et al., 2006; Guo et al., 2011). By contrast, TLR3 is essential for human fibroblast and induced pluripotent stem cell (iPSC)-derived cortical neuron and oligodendrocyte responses to dsRNA and HSV1 (Lafaille et al., 2012). Interestingly, all children with HSE due to inborn errors of TLR3 immunity only display lesions of the frontal and temporal lobes. This suggests that HSE in these patients results from a disruption of cell-intrinsic immunity in the forebrain.
It remains unclear whether viral infections of other regions of the brain, including those caused by HSV1, are due to other defects impairing intrinsic immunity in these specific regions. Cells from other regions of the CNS may not rely on TLR3 to control HSV1 infection. In this context, we were intrigued by brainstem HSV1 infections, which occur in about 5% of patients with HSE (Jubelt et al., 2011). In about half of these brainstem HSV1 infections, the brainstem is the only region affected (Jubelt et al., 2011). None of the patients with TLR3 pathway deficiencies in our study had brainstem lesions. Moreover, other common viruses, including IV, human herpesvirus 6, Japanese encephalitis virus, and enteroviruses (especially enterovirus 71), can cause brainstem encephalitis (Jubelt et al., 2011). In most patients with these devastating forms of viral encephalitis, the brainstem is the only region of the CNS affected. We thus tested the hypothesis that brainstem viral infections caused by a broad range of viruses may result from inborn errors of brainstem-intrinsic immunity.
Results
Biallelic DBR1 coding sequence mutations in patients with brainstem viral encephalitis
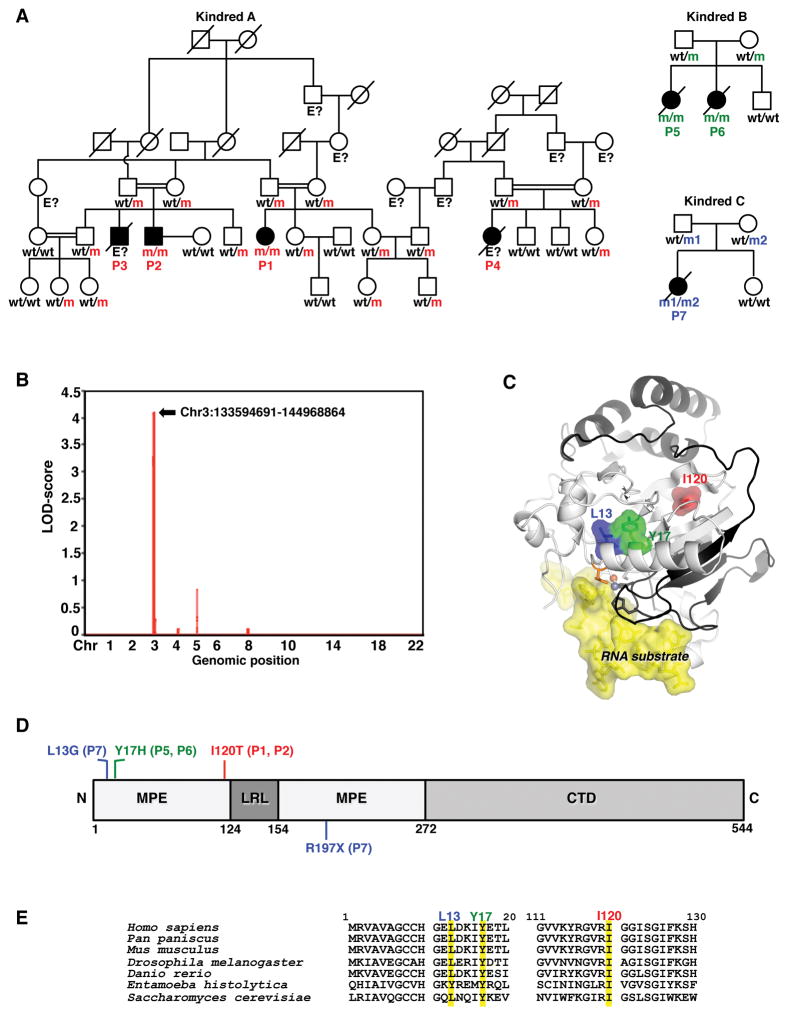
We evaluated seven patients from three unrelated families. Five had proven and two had suspected brainstem viral encephalitis, caused by HSV1 (a dsDNA virus of the Herpesviridae family), influenza B virus (IBV, a negative-stranded ssRNA virus of the Orthomyxoviridae family), or NV (a positive-stranded ssRNA virus of the Caliciviridae family). We first evaluated a multiplex Arab kindred living in Israel (kindred A, Figure 1A), and a multiplex family originating from and living in Portugal (kindred B, Figure 1A). In kindred A, two first cousins (P1 and P2) suffered from brainstem HSV1 encephalitis at the ages of 12 and 5 years, respectively. One of P2’s brothers (P3) and another relative of P1 and P2 (P4) also died of suspected herpes encephalitis at the ages of seven years and 10 months, respectively (Figure 1A, Data S1). The parents of P1 were consanguineous, as were those of P2 and P3, and those of P4. In kindred B, which was not known to be consanguineous, two siblings (P5, P6) died from brainstem encephalitis due to proven and suspected IBV, respectively, at the ages of two years and one year, respectively (Figure 1A, Data S1, Figure S1A–C). We hypothesized that brainstem viral encephalitis in these two families was due to an autosomal recessive single-gene defect with complete clinical penetrance. Genome-wide linkage (GWL) analysis revealed a single linked region with a LOD score of 4.11, spanning 10.58 Mb on chromosome 3 and common to all four patients (P1, P2, P5, P6) for whom genomic DNA was available (Figure 1B, Table S1). We also performed whole-exome sequencing (WES) for these four patients. We searched for genes harboring homozygous non-synonymous or splice-site variations, with a minor allele frequency (MAF) below 0.01 in the 1000 Genome and ExAC databases, and our own exome database (4,440 exomes to date) containing data for patients with other types of infectious diseases. We identified DBR1 as the only gene carrying such variants in both kindreds: c.359T>C (p.I120T) in P1 and P2, and c.49T>C (p.Y17H) in P5 and P6 (Table S2). This gene is located within the linked interval on chromosome 3. Sanger sequencing confirmed that the familial segregation of both mutations was consistent with an autosomal recessive trait with complete penetrance (Figure 1A, Figure S1D). We then studied a third, non-consanguineous, kindred from Japan with sporadic disease, in which one child (P7, a girl) developed brainstem encephalitis due to NV infection at the age of six months (kindred C in Figure 1A, Figure S1C). P7 had additional clinical manifestations, including intrauterine growth retardation, mental retardation, curly hair, and congenital neutropenia (Figure S1E, Data S1). WES for P7 revealed missense (c.37–38CT>GG; p.L13G) and nonsense (c.589C>T; p.R197X) mutations of DBR1, which were shown by Sanger sequencing to be inherited from her mother and father, respectively (Figure 1A, Figure S1F). The four mutations are in three DBR1 exons, L13G and Y17H in exon 1, I120T in exon 3, and R197X in exon 5. The four mutations are 11bp, 3,070bp, and 4,471bp apart from each other. Moreover, no transcripts other than the DBR1 mRNA are known for the DBR1 locus. Overall, rare biallelic missense and nonsense mutations of DBR1 were found in five patients from three kindreds of different ethnicities with brainstem encephalitis caused by three different viruses, but not in the 29 healthy relatives of these patients tested, consistent with an autosomal recessive trait with complete penetrance.
Figure 1. Bi-allelic DBR1 mutations in patients with brainstem viral encephalitis from three kindreds.
A) Family pedigrees with allele segregation. The patients, in black, are homozygous for the I120T (‘m’ in red) or the Y17H (‘m’ in green) mutation, or compound heterozygous for the R197X and L13G mutations (‘m1’ and ‘m2’, respectively, in blue). The other family members were either heterozygous for one mutant allele or wild-type (wt). B) Whole-genome linkage analysis for kindreds A and B. C) Predicted 3-dimensional structure of human DBR1 protein. The residues affected by the three missense mutations found in viral encephalitis patients are highlighted in red (I120), green (Y17H) or blue (L13). This human DBR1 homology model was built from PDB 5K78. A 16-mer RNA substrate is shown as a yellow area. Residues 1–197 are shown in gray, and residues 198–363 are shown in black. H10, shown in orange, is a key active site residue that binds the Zn cofactor and stacks with the branch point adenosine of intron lariats. The Fe and Zn cofactors are shown in orange and gray, respectively. D) Schematic representation of the structure of the human DBR1 protein, and location of the mutations. MPE: metallophosphoesterase core domain. LRL: lariat recognition loop. (LRL). CTD: C-terminal domain. E) Human DBR1 gene sequence, for the regions containing the three patient-specific missense mutations and the corresponding regions for the other six species studied. The nucleotides affected by the mutations are shaded in yellow.
See also Figure S1–2, Data S1, and Table S1–3.
The DBR1 mutations are predicted to be pathogenic
DBR1 encodes debranching enzyme 1 (DBR1), which was first discovered in yeast (Chapman and Boeke, 1991). DBR1 is the only known RNA lariat-debranching enzyme in humans. It hydrolyzes 2′5′-phosphodiester linkages at the branch points of intron lariat RNAs, facilitating their rapid turnover (Chapman and Boeke, 1991; Jacquier and Rosbash, 1986; Nam et al., 1994; Nam et al., 1997). The L13G, Y17H, and I120T mutations were apparently private to kindreds C, B, and A, respectively, as they were not found in the 1000 Genomes Project, GnomAD, or in-house databases. The R197X mutation from kindred C was heterozygous in only six individuals from the GnomAD database (MAF=0.0000217). Fifty-three of the 138,632 individuals in GnomAD were homozygous for a rare (MAF<0.01) non-synonymous DBR1 mutation, but only one of these individuals was homozygous for a mutation predicted to be deleterious by all three computational tools used: combined annotation–dependent depletion (CADD), PolyPhen-2, and sorting intolerant from tolerant (SIFT) (Adzhubei, 2010; Kircher et al., 2014; Kumar et al., 2009) (Figure S1G, Table S3). Human DBR1 is a highly conserved gene, with a gene damage index (GDI) of 2.47, suggesting that damaging mutations of this gene are likely to be pathogenic (Itan et al., 2015). Based on inferences from the three-dimensional structure of the DBR1 protein in Entamoeba histolytica (Montemayor et al., 2014), human DBR1 is predicted to contain a metallophosphoesterase (MPE) core domain, into which a lariat recognition loop (LRL) is inserted, and a C-terminal domain (CTD) (Figure 1C, D). The MPE domain, which contains the four residues affected by the mutations in patients, is less tolerant of sequence variation than the whole protein in the general population (Gussow et al., 2016) (Figure S1H). It has also been strongly conserved throughout evolution (Montemayor et al., 2014). The L13G, Y17H, and I120T missense mutations affect highly conserved residues (Figure 1E). The L13G and Y17H substitutions are predicted to destabilize the hydrophobic core in an area adjacent to the key active site residues C8 and H10 (Clark et al., 2016; Montemayor et al., 2014). The I120T substitution is predicted to destabilize the hydrophobic core by introducing a polar residue into a non-polar environment. The introduction of a stop codon in place of R197 would result in a C-terminally truncated protein lacking key active site residues and secondary structure elements (Figure 1C). All four mutations are predicted to be highly damaging (Figure S1G) (Itan et al., 2016). The human DBR1 mRNA is found throughout the body, but its levels are highest in the PNS and CNS (http://biogps.org/). We found that DBR1 protein levels were highest in human spinal cord and brainstem (Figure S2A–E). The brainstem contains the nuclei of secondary nerves projecting onto diverse organs of the human body. The brainstem lesions in all five patients with DBR1 mutations are thus consistent with both the neurotropic nature of HSV1, IBV, and NV infections, and the expression profile of DBR1 in humans. Collectively, these findings suggest that the bi-allelic DBR1 mutations were responsible for the patients’ susceptibility to viral infections of the brainstem.
Impaired expression and function of the mutant DBR1 alleles in Escherichia coli
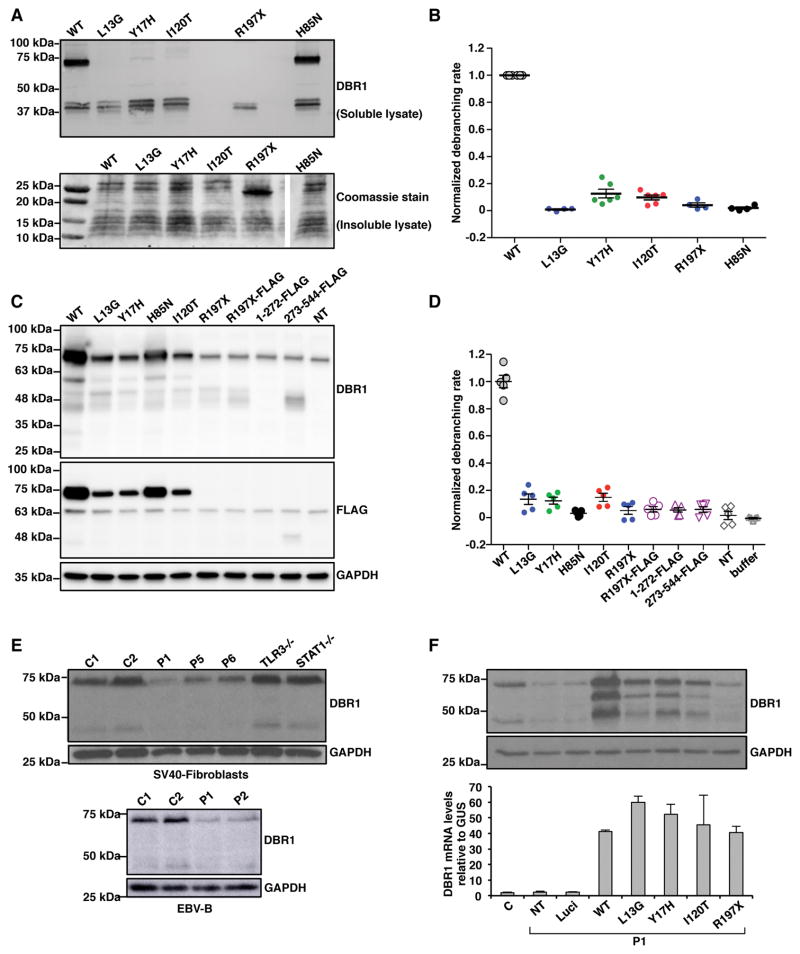
We assessed the impact of the L13G, Y17H, I120T, and R197X mutations on the production and function of human DBR1 protein. We first produced His-tagged mutant and wild-type (WT) DBR1 proteins in Escherichia coli (E. coli). WT DBR1 was detected in the soluble fraction of E. coli lysates as two species of different molecular weights (MW) on western blots with a DBR1-specific polyclonal antibody (pAb) (Figure 2A). A band at ~70 kDa corresponded to the full-length DBR1, and a band at ~40 kDa corresponded to a cleaved form of DBR1. The predicted MW of full-length DBR1 (544 aa) is ~60 kDa, and that of the predicted MPE domain (from residues 1 to ~272) is ~30 kDa. When the L13G, Y17H, and I120T DBR1 alleles were expressed in E. coli, very little full-length DBR1 was detected (only 5–10% the amount observed for the WT). R197X allele yielded no detectable full-length protein. The 40 kDa form was produced in normal amounts from the three missense alleles and in small amounts from the R197X allele (Figure 2A). A band of ~20 kDa from the insoluble fraction of R197X-expressing E. coli lysate was clearly visible on Coomassie-stained gels (Figure 2A). The H85N variant is a synthetic allele encoding a protein that lacks a key catalytic histidine residue but remains stable, as demonstrated by the similar amounts of the 70 kDa protein for the WT and H85N alleles. We assessed the enzymatic activity of the soluble lysates by measuring the debranching of a synthetic RNA lariat mimic (Katolik et al., 2017). The function of the L13G mutant was highly impaired, with <1% the activity of the WT, whereas the Y17H, I120T, and R197X mutants had activity levels 13%, 9%, and 4% those of the WT, respectively (Figure 2B). The H85N variant had 2% the WT level of activity. Overall, enzymatic activity appeared to be correlated with the presence of the 70 kDa form on western blots. This suggests that the loss of enzymatic activity of the L13G, Y17H, and I120T mutant proteins results primarily from an impairment of protein folding and/or lower thermodynamic stability of the full-length protein than of the WT enzyme. The R197X allele resulted in a loss of expression of the full-length protein and was profoundly hypomorphic in this system.
Figure 2. Impaired production and function of the patient-specific DBR1 mutant proteins.
A) Wild-type (WT) and mutant DBR1 protein levels in E. coli. Levels of WT, L13G, Y17H, I120T or R197X DBR1 in the soluble fraction of lysates were assessed by western blotting (upper panel), with an anti-DBR1 polyclonal antibody (pAb). The lower panel shows a Coomassie-stained gel of the insoluble fraction of the lysate. B) Debranching activity of the WT and mutant DBR1 proteins in the soluble fraction of E. coli lysates. The results of 4–6 independent biological replicates are plotted. C) HEK293T cells were transfected with plasmids containing C-terminal FLAG-tagged WT, L13G, Y17H, I120T or R197X cDNA, R197X cDNA with FLAG-coding sequences inserted in-between amino acid positions 196 and 197 (R197X-FLAG), or plasmids encoding C-terminal FLAG-tagged a.a. 1-272 or aa. 273-544 DBR1. The cell lysates were analyzed for western blotting with an anti-DBR1 pAb and an anti-FLAG monoclonal antibody. D) Debranching activity of HEK293T cell lysates with and without transfection with various DBR1 constructs. E) DBR1 protein levels, as assessed by western blotting, in SV40-fibroblasts (upper panel) from healthy controls (C) and patients with biallelic DBR1 mutations (I120T/I120T for P1, Y17H/Y17H for P5 and P6). DBR1 protein levels in EBV-B cells (lower panel) from healthy controls, P1 and P2 with homozygous I120T DBR1 mutations. F) DBR1 protein and mRNA levels, as assessed by western blotting (upper panel) and RT-qPCR (lower panel), respectively, in SV40-fibroblasts from a healthy control, from P1 with and without stable transfection with a pTRIP-RFP vector without DBR1 cDNA, or with WT or mutant DBR1 cDNA. NT: non-transfected, Luci: luciferase plasmid. In C), E) and F), the internal expression control was GAPDH. Each experiment shown in E–F is representative of at least three independent experiments.
See also Figure S3.
Lower levels of mutant DBR1 proteins in eukaryotic cells
We expressed C-terminal FLAG-tagged WT, L13G, Y17H, I120T, and R197X mutant forms of DBR1, and N-terminal (aa.1-272) or C-terminal (aa.273-544) fragments of DBR1, in HEK293T cells, to rule out the possibility that the mutant proteins were unstable in E. coli but functional in human cells, and to resolve the ambiguity as to which protein product (70 kDa or 40 kDa) was active. The catalytically inactive H85N mutant was used as a control. No human or mouse DBR1-knockout cell lines are available, consistent with the growth defect observed following CRISPR/Cas9-mediated DBR1 knockout in human cells (Findlay et al., 2014). HEK293T cells have low levels of endogenous DBR1 protein (Figure 2C). By contrast to the results obtained for E. coli, in which both the 70 and 40 kDa species were observed, the 70 kDa band strongly predominated in HEK293T cells, consistent with probable stabilization of the full-length DBR1 protein in mammalian cells. The overexpression of WT DBR1 led to the detection of a clear 70 kDa band on western blots probed with a FLAG-specific monoclonal antibody (mAb), and the DBR1-specific pAb (Figure 2C). The same 70 kDa band was observed for the L13G, Y17H, I120T, and H85N mutants, whereas no protein product was detected for R197X and aa.1-272 DBR1. The L13G, Y17H and I120T mutants produced only small amounts of full-length protein (anti-FLAG band intensities were ~25% those for the WT) (Figure 2C). The 40 kDa band was very weak for WT DBR1, and barely detectable for the three missense alleles and the nonsense alleles (Figure 2C). For aa.273-544, a ~45 kDa band was detected with antibodies against FLAG or DBR1 (Figure 2C). We checked transgene expression levels, by quantifying DBR1 mRNA levels by RT-qPCR. These mRNA levels were found to be similar in WT- and mutant DBR1-transfected cells (Figure S3A). These results suggest that the full-length DBR1 protein is the predominant form in human cells. The four DBR1 mutations result in a full (R197X), or partial loss of the full-length protein (L13G, Y17H, I120T), probably due to lower stability.
Impaired function of the mutant DBR1 proteins in eukaryotic cells
Robust debranching enzymatic activity was detected in the lysates of HEK293T cells expressing WT DBR1, whereas non-transfected cells had ~2% the activity of cells expressing WT DBR1. After the subtraction of background DBR1 activity, H85N-transfected cells had 1% WT levels of activity, whereas R197X, 1-272, and 273-544 each had ~5% WT levels of activity. Cells expressing the L13G, Y17H, and I120T mutants displayed ~11–14% WT levels of activity (Figure 2D). Overall, there was a clear relationship between the amount of full-length 70 kDa protein produced and debranching activity (Figure 2C,D). A semi-quantitative correction of activity according to protein levels suggested that, if the mutant and WT proteins were produced in equal amounts, the mutants would have ~50% WT levels of activity. Thus, the overall defect of the mutant DBR1 in human cells results from both an enzymatic defect and lower protein levels (Figure 2C,D; S3B). Similar results were obtained when the Y17H, I120T, and WT DBR1 alleles were expressed in Sf9 insect cells (Figure S3C,D). Moreover, transformation of the dbr1-1-null yeast with the yeast I12IT dbr1 allele (corresponding to human I120T) only partially attenuated the intron lariat accumulation phenotype (Figure S3E), whereas WT dbr1 fully rescued this phenotype (Chapman and Boeke, 1991). Collectively, these data indicate that the patients with DBR1 mutations from the three kindreds were homozygous for a hypomorphic allele (I120T, Y17H) or compound heterozygous for two even more severely hypomorphic alleles (L13G and R197X). The more severe and complex clinical phenotype of P7 (kindred C) is consistent with the patient’s more severe DBR1 genotype.
Low levels of DBR1 proteins in fibroblasts from human patients
We then assessed DBR1 protein and mRNA levels in SV40-transformed dermal fibroblast cells (SV40-fibroblasts) and EBV-transformed B (EBV-B) cells from P1 and P2 (I120T/I120T, kindred A), P5 and P6 (Y17H/Y17H, kindred B), and healthy controls. No cells were available for P7. Consistent with the data obtained for overexpression in E. coli, Sf9, and HEK293T cells, endogenous DBR1 was detected as two species of different MW by western blotting with a DBR1-specific pAb (Figure 2E): a ~70 kDa and a ~40 kDa form. As observed in HEK293T cells, the ~70 kDa species predominated in human fibroblasts. DBR1 protein levels were low in SV40-fibroblasts from P1, P5, and P6, as shown by western blotting (Figure 2E). Low DBR1 protein levels were also observed in EBV-B cells from P1 and P2 (Figure 2E). However, DBR1 mRNA levels in DBR1-mutated SV40-fibroblasts or EBV-B cells were similar to those in the corresponding healthy controls (Figure S3F). Moreover, the stable transfection of P1 SV40-fibroblasts with L13G, Y17H, I120T, or R197X cDNA did not restore DBR1 protein levels to those detected after transfection with WT cDNA, despite the production of similar amounts of DBR1 mRNA (Figure 2F). Collectively, these data for patient cells confirm the results obtained for the expression of individual alleles. Patients with biallelic DBR1 mutations had very low levels of DBR1 protein in the two cell types tested.
Impaired function of DBR1 in fibroblasts from patients
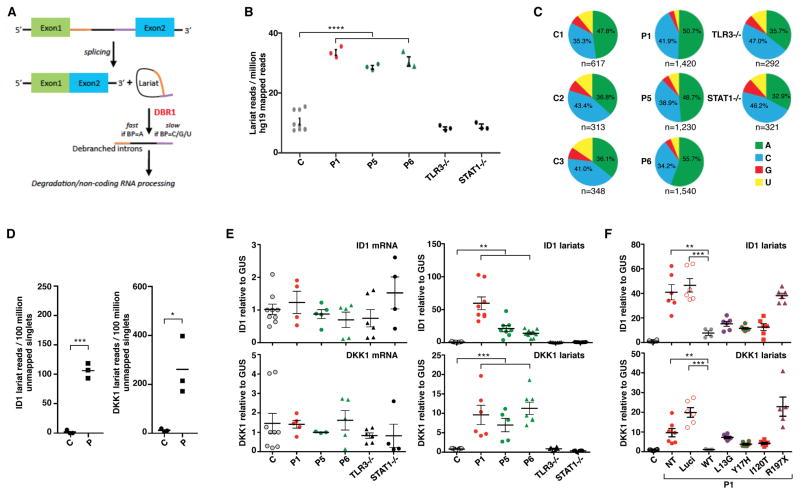
As DBR1 is the only known RNA lariat-debranching enzyme, we hypothesized that biallelic hypomorphic DBR1 mutations would lead to an increase in the abundance of intronic RNA lariats in human cells (Figure 3A). We performed RNA-Seq on primary fibroblasts from a DBR1 I120T/I120T patient (P1), two DBR1 Y17H/Y17H patients (P5 and P6), three healthy controls, and two patients with AR complete TLR3 or STAT1 deficiency (TLR3−/−, STAT1−/−) (Chapgier et al., 2006; Guo et al., 2011). We used a lariat read-count analysis strategy (Taggart et al., 2017) to identify and quantify intronic lariats. The abundance of intronic RNA lariats was high in samples from patients, reaching a level about three times that in healthy controls, and in TLR3−/−, and STAT1−/− fibroblasts (Figure 3B). DBR1 has a strong substrate specificity for ‘adenine (A)’ branchpoints (Nam et al., 1994). We therefore also evaluated branchpoint nucleotide composition in the identified lariat sequences. Samples from patients contained significantly more ‘A’ branchpoints than samples from controls (Figure 3C), providing further evidence for lower levels of DBR1 activity in the cells of patients. We also measured lariat abundance by the ‘LaSSO’ method (Bitton et al., 2014), which, as applied, identifies ‘A’ branchpoint lariat sequences in RNA-Seq reads. This approach also showed that there were more lariats in patient fibroblasts than in control cells (Figure S3G). Lariat sequences derived from a diverse collection of genes were detected in patient samples, but we also observed a large number of unique lariat events derived from DKK1, and, to a lesser extent, ID1 transcripts (Figure 3D, Figure S3H). We then validated our RNA-Seq findings by RT-qPCR for these two apparently abundant intronic RNA lariats (derived from DKK1 and ID1 transcripts) in total RNA from SV40-fibroblasts. Both lariats were present at higher levels in SV40-fibroblasts from P1, P5 and P6 than in cells from healthy controls and TLR3−/− or STAT1−/− individuals, despite the presence of similar amounts of the corresponding mature mRNA transcripts (Figure 3E). The stable expression of exogenous WT DBR1 rescued this cellular phenotype in SV40-fibroblasts from P1 (Figure 3F). The overexpression of Y17H, I120T, or L13G cDNA decreased DKK1 and ID1 lariat levels to intermediate levels, whereas the overexpression of R197X cDNA did not (Figure 3F). Overall, our data demonstrate that homozygosity for the hypomorphic I120T or Y17H DBR1 mutations, and, by inference, compound heterozygosity for the L13G and R197X mutations, decrease but do not abolish DBR1 protein levels and intron lariat debranching activity. These bi-allelic mutations therefore underlie a partial form of AR DBR1 deficiency, in terms of both expression and function, in patients from the three kindreds.
Figure 3. AR partial DBR1 deficiency leads to intronic RNA lariat accumulation in patient fibroblasts.
A) Schematic representation of DBR1 activity in intronic lariat RNA debranching. B) Lariat branchpoint-traversing read counts, obtained from total RNA-Seq data with the lariat read-count method and normalized against hg19 mapped read pairs, for three healthy controls, P1, P5, P6, and TLR3−/− and STAT1−/− patients. Each dot represents one technical replicate for one sequenced sample. C) An analysis of DBR1 specificity for branchpoint nucleotide composition. Pie charts depicting the ratio of ‘A’, ‘C’, ‘G’, and ‘U’ branchpoints in recovered branchpoint-traversing reads. Chi-square test was performed to compare the proportion of ‘A’ branchpoints to non-‘A’ branchpoints in aggregated control versus patient samples (p<0.0001). D) ID1 and DKK1 RNA lariat read counts, obtained from total RNA-Seq data by the LaSSO method and normalized against unmapped singlets, for three healthy controls, and three patients. E) mRNA (left panels) and intronic lariat levels (right panels) for ID1 (upper panels) and DKK1 (lower panels), measured by RT-qPCR, in SV40-fibroblasts from healthy controls, P1, P5, P6, and TLR3−/− and STAT1−/− patients. F) Intronic lariat levels for ID1 (upper panel) and DKK1 (lower panel), measured by RT-qPCR, in SV40-fibroblasts from healthy controls, fibroblasts from P1 with and without stable transfection with the pTRIP-RFP vector without the insertion of DBR1 cDNA, or expressing WT or mutant DBR1. NT: non-transfected, Luci: luciferase plasmid. In E and F, each dot represents one technical replicate, from three independent experiments. In B–F), * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
See also Figure S3.
Intact TLR3- and IFN-responsive pathways in the patients’ fibroblasts
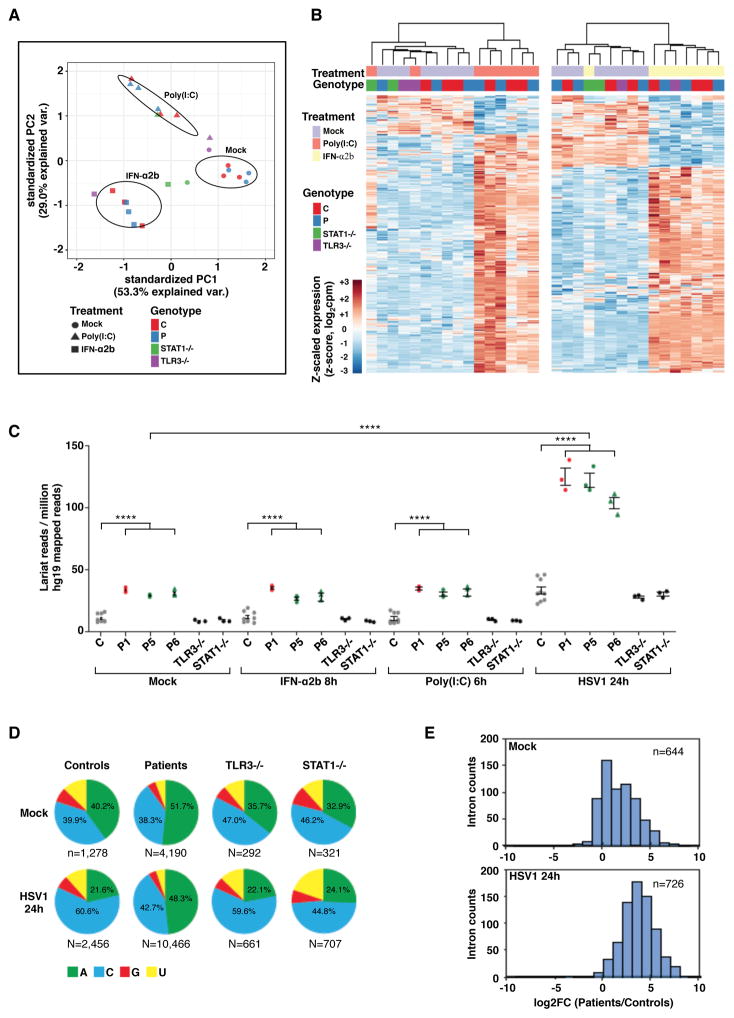
The pathogenesis of brainstem viral encephalitis in patients with bi-allelic DBR1 mutations probably involves an impairment of cell- and brainstem-intrinsic antiviral immunity, rather than a defect of the innate or adaptive immunity mediated by leukocytes. We investigated the possibility that DBR1 deficiency might underlie viral encephalitis by a novel mechanism, by comparing antiviral responses in SV40-fibroblasts derived from patients with DBR1 mutations, and patients with AR complete TLR3 or STAT1 deficiency, genetic etiologies of childhood cortical HSE, either alone (TLR3) or in combination with mycobacterial disease (STAT1) (Chapgier et al., 2006; Dupuis et al., 2003; Guo et al., 2011). Unlike TLR3−/− cells, fibroblasts from P1, P5, and P6 produced normal amounts of IFN-λ1 and IL-6 following stimulation with various concentrations of polyinosinic:polycytidylic acid (poly(I:C)), a synthetic TLR3 agonist that mimics dsRNA (Figure S4A). DBR1-deficient fibroblast responses to intracellularly delivered poly(I:C) (activator of cytosolic dsRNA sensors RIG-I and MDA5), or T7-GFP (agonist of RIG-I (Pichlmair et al., 2006), were also similar to those of healthy control cells (Figure S4A,B). We performed a more comprehensive assessment of TLR3 and IFN responses in DBR1-deficient fibroblasts, by using RNA-Seq to measure gene expression in primary fibroblasts from P1, P5, P6, and corresponding controls, with and without stimulation with poly(I:C) or IFN-α2b. The stimulation of P1, P5 and P6 fibroblasts with IFN-α2b or poly(I:C) induced changes in gene expression similar to those observed in healthy control cells (Figure 4A,B; S5C). Furthermore, gene set (Molecular Signatures Database, hallmark sets) enrichment testing revealed no obvious deficiencies in common signaling pathway and/or transcription network induction (Figure S4D). TLR3−/− and STAT1−/− cells served as negative controls for poly(I:C) and IFN-α2b responses, respectively (Figure 4A,B). These data suggest that the TLR3- and IFN-α-mediated signaling pathways remain intact in fibroblasts with AR partial DBR1 deficiency. Therefore, the mechanism underlying the pathogenesis of brainstem viral encephalitis in patients with DBR1 mutations is different from that in patients with mutations affecting the TLR3 and STAT1 pathways, consistent with the two different anatomical sites of infection within the CNS.
Figure 4. Intact TLR3- and IFN-responsive pathways, and very high RNA lariat levels in DBR1-deficient fibroblasts following HSV1 infection.
A) Principal component analysis (PCA) of RNA-Seq-quantified gene expression in primary fibroblasts from three healthy controls, three DBR1-mutated patients (P1, P5 and P6), one TLR3−/− and one STAT1−/− patient. Cells were mock-treated, stimulated with 25 μg/ml poly(I:C) for 6 hours, or stimulated with 100 IU/ml IFN-α2b for 8 hours. B) Heatmaps of RNA-Seq-quantified gene expression (z-score scaled log2 read counts per million, cpm) in primary fibroblasts from healthy controls, DBR1-mutated patients, TLR3−/− and STAT1−/− patients, with stimulations as described in A). Each heatmap includes genes differentially expressed (FDR 0.01, > 2-fold difference) in response to the indicated stimulus relative to mock-treated samples in the healthy control or DBR1-mutated groups. Hierarchical clustering (complete method) on Euclidean distance values. C) Lariat branchpoint-traversing read counts, obtained with the lariat read-count method and normalized against hg19 mapped read pairs, for primary fibroblasts with or without stimulation with poly(I:C) or IFN-α2b as described in A), or 24 hours of infection with HSV1, for three healthy controls (C), three DBR1-mutated patients, and TLR3−/− and STAT1−/− patients. We performed t-tests to compare intronic RNA lariat levels in samples from patients or healthy controls after infection with HSV1 with the corresponding uninfected samples (p<0.0001 in each case). **** p<0.0001. D) An analysis of DBR1 specificity in ‘A’, ‘C’, ‘G’, and ‘U’ branchpoint nucleotide composition, for primary fibroblasts with or without 24 hours of infection with HSV1, for controls (C) and patients. Chi-square test was used to compare the proportion of ‘A’ to non-‘A’ branchpoints in aggregated control versus patient samples after infection with HSV1 (p<0.0001). E) Distribution of intronic read density enrichment for three healthy controls versus three DBR1-mutated patients, in the absence of infection (top) and after HSV1 infection (bottom).
Very high RNA lariat levels in patients’ fibroblasts following HSV1 infection
As a first approach to deciphering the mechanism of brainstem viral encephalitis in DBR1-deficient patients, we assessed host RNA lariat levels upon HSV1 infection at a multiplicity of infection (MOI) of 1, or following stimulation with IFN-α2b or poly(I:C), in primary fibroblasts from healthy controls or DBR1-deficient patients. Treatment with IFN-α2b or poly(I:C) did not affect the levels of intronic RNA lariats in any of the control or DBR1-deficient patient samples assayed (Figure 4C). As expected, HSV1 infection resulted in a similar redistribution of RNA-Seq reads from host mRNA to HSV1 mRNA in fibroblasts from all healthy controls and patients (8 hours or 24 hours of HSV1 infection, Figure S5A). After 24 hours of HSV-1 infection, the detected lariat levels had increased in both control and patient samples (3.2- to 3.8-fold increase relative to the corresponding non-infected samples, Figure 4C). It has previously been reported that HSV1 interferes with the human pre-mRNA splicing machinery, resulting in splicing inhibition or alternative splicing of some genes (Roizman et al., 2013; Tang et al., 2016). Strikingly, these high lariat levels were particularly marked in fibroblasts from DBR1-deficient patients. In addition, after 24 hours of HSV1 infection, the enrichment in ‘A’ branchpoint lariats was found to be more marked in DBR1-deficient fibroblasts than in healthy control, TLR3−/− or STAT1−/− fibroblasts (Figure 4D). Similar results were obtained by the ‘LaSSO’ method (Figure S5B,C). Interestingly, intronic lariat reads derived from HSV1 transcripts were also found to be about twice as abundant in fibroblasts from DBR1-deficient patients than in healthy control, TLR3−/− or STAT1−/− fibroblasts, after 24 hours of HSV1 infection (Figure S5D). We used the Shapeshifter tool to perform 644 individual comparisons of intron abundance in samples from controls and from DBR1-deficient patients. These comparisons included comparisons for DKK1 and ID1 transcripts, for which the lariat was more abundant than the message post-infection (Figure S5E and manuscript under review). Intron counts were higher in samples from patients than in those from controls, probably at least partly due to the accumulation of lariats, and this difference was particularly marked after 24 hours of HSV1 infection (Figure 4E). For DKK1- and ID1-derived lariats, an analysis for individual introns (Figure S5F) reflected the trends observed in the overall data (Figure 4C,E). These results suggest that DBR1 may control cell- and brainstem-intrinsic defenses by regulating cellular and/or viral RNA lariat metabolism during viral infection.
Impaired control of viruses by the patients’ fibroblasts
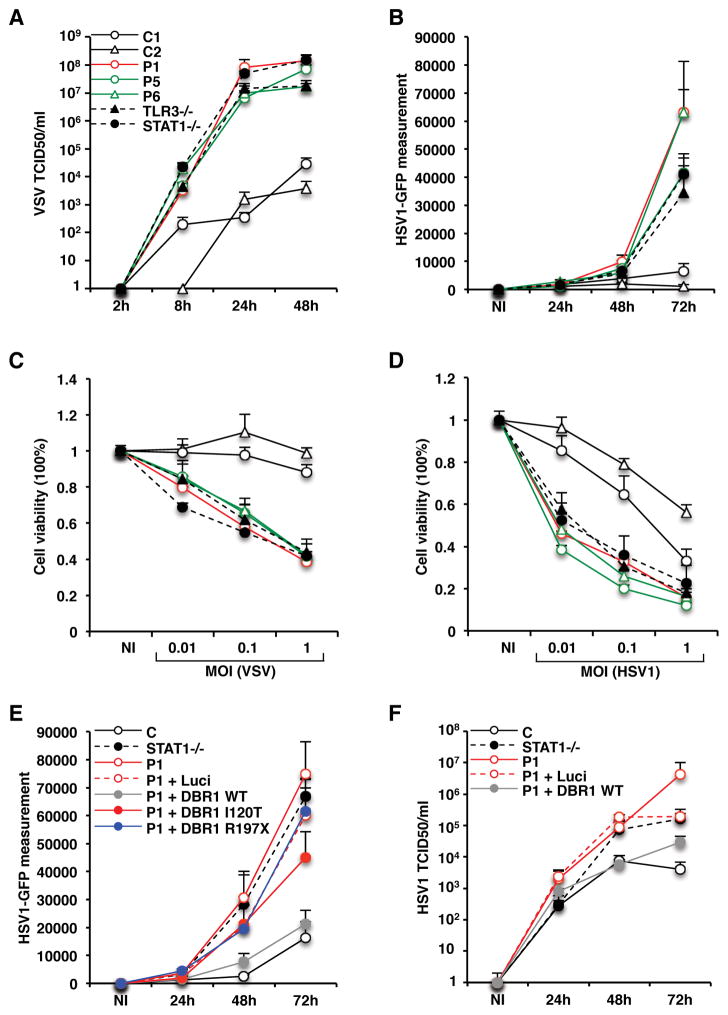
We then studied susceptibility to viral infection in DBR1-mutated SV40-fibroblasts. In addition to HSV1, as in our previous studies of HSE, we also included VSV, as a neurotropic virus capable of infecting and replicating in human fibroblasts, and exerting cytopathic effects (Casrouge et al., 2006; Guo et al., 2011). Similarly high levels of replication were observed for VSV (MOI of 1) and HSV1 (MOI of 0.01) in DBR1-mutated SV40-fibroblasts from P1, P5, and P6, as high as in TLR3−/− or STAT1−/− SV40-fibroblasts, and much higher than those in healthy control cells (Figure 5A, B, Figure S6A). Higher levels of virus-induced cell death following VSV or HSV1 infection with various MOI were also observed in SV40-fibroblasts from all patients tested (Figure 5C,D). The stable transfection of DBR1 I120T/I120T fibroblasts from P1 with WT DBR1 rescued both DBR1 protein levels (Figure 2F) and HSV1 replication phenotype, as demonstrated by the levels of GFP expression following infection with a GFP-expressing HSV1, HSV1 genome copy number upon infection with unmodified HSV1, and the titration of infectious HSV1 by the TCID50 method, on Vero cells (Figure 5E,F; S7B). No such effect was observed with the R197X DBR1. Transfection with the I120T allele partially rescued the HSV1 replication phenotype (Figure 5E; Figure S6B). Together with the high intronic RNA lariat levels of DBR1-mutated fibroblasts, which increased still further during HSV1 infection, these data demonstrate that DBR1-mutated fibroblasts are intrinsically susceptible to viruses. These data provide support for a causal relationship between partial DBR1 deficiency and susceptibility to brainstem viral encephalitis through the impairment of a DBR1-dependent mechanism operating in the cells of the brainstem or its afferent peripheral nerves. Our findings therefore identify inborn errors of DBR1-dependent cell-intrinsic immunity as the first genetic etiology of brainstem viral encephalitis, as opposed to inborn errors of TLR3-dependent immunity, which underlie forebrain viral encephalitis.
Figure 5. Enhanced viral susceptibility in DBR1-deficient fibroblasts.
A,B) Levels of VSV and HSV1 replication, at various time points, following infection with VSV at a MOI of 1, or a MOI of 0.01 for HSV1-GFP, as measured in the TCID50 assay and GFP intensity assay, respectively, in SV40-fibroblasts from healthy controls (C1, C2), P1, P5 and P6, and TLR3−/− and STAT1−/− patients. NI: not infected. C,D) Viability of SV40-fibroblasts following 24 hours of infection with VSV or 72 hours of infection with HSV1, at various MOI, in cells from healthy controls, P1, P5 and P6, and TLR3−/− or STAT1−/− patients. T-tests were performed to compare the virus levels or cell viability in samples from patients with that from healthy controls for the latest point during the time course (A,B) or the highest MOI tested (C,D) (p<0.05 in D, p<0.01 in each other case). E) HSV1-GFP replication levels, as assessed on the basis of GFP intensity, in SV40-fibroblasts from healthy controls, from P1 with and without stable transfection with the pTRIP-RFP vector without the insertion of DBR1 cDNA, or expressing WT or mutant DBR1, following 24, 48 and 72 hours of infection with HSV1-GFP at a MOI of 0.01. F) HSV1 replication levels, as assessed in the TCID50 assay, in SV40-fibroblasts from healthy controls, from P1 with and without stable transfection with the pTRIP-RFP vector without the insertion of DBR1 cDNA, or expressing WT DBR1, following 24, 48 and 72 hours of infection with HSV1 at a MOI of 0.01. Mean values from at least three independent experiments are shown in each panel of A–F.
See also Figure S6.
Discussion
This study provides experimental evidence of a causal relationship between a human DBR1 genotype and a recessive phenotype of brainstem viral encephalitis. A form of AR DBR1 deficiency that is severe but partial, is present in three unrelated kindreds with brainstem encephalitis. This deficiency results both in accumulation of lariats and greater susceptibility of the patients’ cells to viral infection. In yeasts, deletion of the dbr1 gene results in the accumulation of large amounts of intronic RNA lariats in both Schizosaccharomyces pombe and Saccharomyces cerevisiae. However, a growth defect was observed with S. pombe but not S. cerevisiae, presumably because S. pombe has more than 10 times more intron-containing genes (40% of all genes) than S. cerevisiae (2.5%) (Chapman and Boeke, 1991; Kim et al., 2000; Nam et al., 1997). Complete Dbr1 deficiency in mice is probably embryonic lethal, as no homozygous Dbr1 null mouse has ever been reported (Zheng et al., 2015; IMPC project: https://www.mousephenotype.org/). The four patients from kindreds A and B, who are respectively homozygous for the I120T and Y17H hypomorphic alleles, developed brainstem encephalitis but were otherwise healthy. By contrast, the patient from kindred C had a broader phenotype, as she carried the two most deleterious mutant alleles (L13G and R197X). Residual DBR1 activity in patients from these three kindreds was probably sufficient for normal pre- and postnatal development. Alternatively, there may be DBR1-independent RNA lariat debranching and degradation pathways, compensating, to some extent, for the defect of DBR1 function in these patients (Danin-Kreiselman et al., 2003; Garrey et al., 2014; Tseng and Cheng, 2013). Either way, human DBR1 deficiency in these patients was severe but partial and manifested as brainstem viral encephalitis in childhood, indicating a requirement for full DBR1 activity for protective antiviral immunity in the brainstem.
A remarkable feature of partial DBR1 deficiency is that it disrupts immunity in a small, specific anatomical territory. The discovery of DBR1 deficiency in children with brainstem viral encephalitis adds weight to the emerging paradigm that single-gene inborn errors of immunity can underlie severe infections in otherwise healthy individuals, especially in children (Casanova, 2015a, b). It also supports the emerging notion that cell-intrinsic mechanisms in non-hematopoietic cells can be critical for the control of viral illness. Examples include forebrain HSV1 encephalitis, due to TLR3 pathway mutations that disrupt CNS neuron- and oligodendrocyte-intrinsic immunity (Casrouge et al., 2006; Zhang and Casanova, 2015), and severe influenza pneumonia, due to mutations in IRF7 that disrupt cell-intrinsic immunity to influenza A virus in both fibroblasts and iPSC-derived pulmonary epithelial cells (Ciancanelli et al., 2015). HSV1 infection of the frontal or temporal lobes in the forebrain, and infections of the brainstem with various viruses, including HSV1, may thus be due to single-gene inborn errors of antiviral immunity. TLR3 governs a pathway controlling CNS- and cortical neuron-intrinsic immunity to HSV1, whereas DBR1 probably defines another pathway controlling brainstem-intrinsic immunity to viruses as diverse as HSV1, IBV, and NV. Host defense against other viruses capable of infecting the brainstem, such as Japanese encephalitis virus and enterovirus 71 (Jubelt et al., 2011), might also normally require unimpaired DBR1 activity. A recent study proposed DBR1 as a potential therapeutic target in amyotrophic lateral sclerosis (ALS), as the sequestration of TDP-43 by intron lariats accumulating in the cytoplasm of neurons may abolish its ALS-causing toxicity (Armakola et al., 2012). Our study suggests that caution is required when considering this approach to ALS treatment.
Our findings reveal an unexpected connection between a ubiquitous, host cell pre-mRNA processing mechanism and an antiviral, cell-intrinsic defense function in the brainstem. Partial DBR1 deficiency did not disrupt the TLR3-IFN-α/β and IFN-α/βR-STAT1 pathways, which have been shown to be involved in the molecular basis of forebrain HSE pathogenesis (Zhang and Casanova, 2015). Further studies are required to dissect the mechanisms by which DBR1 governs host antiviral defense in the brainstem. At the cellular level, DBR1 may be required in the brainstem, in TG cells, or both, and in neurons or other cell types. At the molecular level, the accumulation of intron lariats due to DBR1 deficiency may impair virus recognition by host cells, thereby damaging cell-intrinsic defenses against viral invasion. DBR1 may also regulate the processing of some protein-coding RNAs or non-coding RNAs (Han et al., 2017; Murray et al., 2014; Ooi et al., 1998; Petfalski et al., 1998; Sedger, 2013) essential for the control of viral infection in the brainstem. Alternatively, it may interfere with the processing of viral RNA lariats (Galvis et al., 2017; Perng and Jones; Plotch and Krug, 1986), thereby controlling the switch between lytic and latent infection in the viral life cycle. As HSV1 and IV interfere with the host RNA processing machinery, facilitating viral gene expression and replication (Rivas et al., 2016; Roizman et al., 2013; Tang et al., 2016), DBR1 may protect host cells against these viral processes. Finally, hitherto unknown functions of DBR1 might be involved. Elucidating the human genetic basis of the various forms of viral encephalitis, as illustrated by the deficiency of the dsRNA-sensing TLR3 pathway in forebrain HSE and deficiency of the housekeeping protein DBR1 in brainstem infections with multiple viruses, has both clinical and biological implications.
STAR Methods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-human DBR1 antibody | ProteinTech | Cat# 16019-1-AP |
| Rabbit polyclonal anti-human DBR1 antibody | Santa-Cruz, Custom-produced | N/A |
| Mouse monoclonal anti-human GAPDH antibody | Santa-Cruz | Cat# sc-365062 |
| Mouse monoclonal anti-FLAG antibody | SIGMA | Cat# F1804 |
| Goat anti-human polyclonal anti-IFN-λ1 antibody | R&D | Cat# AF1598 |
| Human IFN-λ1 biotinylated antibody | R&D | Cat# BAF1598 |
| Bacterial and Virus Strains | ||
| HSV1 (KOS strain) | ATCC | Cat# VR1493 |
| HSV1-GFP (based on KOS strain) | Dr Desai laboratory, Desai et al., 1998 | N/A |
| VSV (Indiana strain) | This laboratory | N/A |
| Escherichia coli (strain BL21) | Life Technologies | Cat# C600003 |
| Biological Samples | ||
| Human Heart Protein Medley | ClonTech | Cat# 635302 |
| Human Smooth Muscle Protein Medley | ClonTech | Cat# 635333 |
| Human Stomach Protein Medley | ClonTech | Cat# 635313 |
| Human Lymph Node Protein Medley | ClonTech | Cat# 635316 |
| Human Spleen Protein Medley | ClonTech | Cat# 635312 |
| Human Thymus Protein Medley | ClonTech | Cat# 635350 |
| Human Fetal Liver Protein Medley | ClonTech | Cat# 635342 |
| Human Kidney Protein Medley | ClonTech | Cat# 635303 |
| Human Brain Cerebral Cortex Protein Medley | ClonTech | Cat# 635323 |
| Human Brain Hippocampus Protein Medley | ClonTech | Cat# 635319 |
| Human Brain Frontal Lobe Protein Medley | ClonTech | Cat# 635318 |
| Human Brain Temporal Lobe Protein Medley | ClonTech | Cat# 635321 |
| Human Brain Hypothalamus Protein Medley | ClonTech | Cat# 635320 |
| Human Brain Cerebellum Protein Medley | ClonTech | Cat# 635326 |
| Human Brain medulla oblongata Protein Medley | ClonTech | Cat# 635346 |
| Human Brainstem Protein Medley | ClonTech | Cat# 635325 |
| Human Spinal Cord Protein Medley | ClonTech | Cat# 635324 |
| Human Skin Protein Medley | ClonTech | Cat# 635355 |
| Human Brain medulla oblongata lysate | ProSci-Inc | Cat# XBL-10111 |
| Human Spinal Cord lysate | ProSci-Inc | Cat# 1377 |
| C57/BalbC mouse tissue lysates | This manuscript | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant wild-type and mutant human DBR1 proteins | This manuscript | N/A |
| Recombinant IFN-α2b | Schering-Plough | Intron® A |
| Polyinosine-polycytidylic acid (Poly(I:C)) | GE Healthcare | Cat# 27-4729-01 |
| Random hexamers | Invitrogen | Cat# N8080127 |
| Multiscribe reverse transcriptase | Applied Biosystems | Cat# 4308226 |
| TaqMan universal PCR master mix | Applied Biosystems | Cat# 4304437 |
| Extremgene 9 reagent | Sigma-Aldrich | Cat# XTG9-RO |
| Lipofectamine-3000 | Life Technologies | Cat# L3000001 |
| Critical Commercial Assays | ||
| miRNeasy Mini Kit | QIAGEN | Cat# 217004 |
| QuantiTect SYBR Green PCR kit | QIAGEN | Cat# 204141 |
| Complete Lysis-M EDTA-free | Sigma-Aldrich | Cat# 4719964001 |
| RiboZero TruSeq Stranded Total RNA Library Prep Kit | Illumina | Cat# RS-122-2201 |
| TruSeq DNA Sample Prep Kit | Illumina | Cat# FC-121-2001 |
| Illumina HiSeq 2000 | Illumina | Cat# HiSeq 2000 |
| Illumina NextSeq 500 | Illumina | Cat# NextSeq 500 |
| Genome-wide SNP 6.0 array | Affymetrix | Cat# 901182 |
| SuperScript III Cells-Direct kit | Life Technologies | Cat# 18080200 |
| Cell lysis buffer (10X) | Cell Signaling Technologies | Cat# 9803 |
| Bac-to-Bac system | Life Technologies | Cat# A11099 |
| Resazurin oxidoreduction (TOX-8) | Sigma-Aldrich | Cat# TOX8 Sigma |
| IL-6 ELISA kit | R&D | Cat# DY206-05 |
| Deposited Data | ||
| Genome Reference Consortium Human Build hg19 | N/A | |
| The Genome Aggregation Database (gnomAD) | Large scale human genome and exome sequencing database | http://gnomad.broadinstitute.org/about |
| The Exome Aggregation Consortium (ExAC) | Large scale human exome sequencing database | http://exac.broadinstitute.org/ |
| 1000 Genome Project | International genome sample resource | http://www.internationalgenome.org/ |
| Raw and analyzed whole-exome sequencing and RNA-Seq data | This manuscript | SRA: SRP130621 |
| Experimental Models: Cell Lines | ||
| DBR1 I120T/I120T human fibroblasts | This manuscript | N/A |
| DBR1 Y17H/Y17H human fibroblasts | This manuscript | N/A |
| DBR1 I120T/I120T human EBV-B cell lines | This manuscript | N/A |
| HEK293T cells | ATCC | Cat# CRL11268 |
| Sf9 cells | Expression Systems | Cat# 94-006S |
| Dbr1-1 yeast | Dr Boeke laboratory, Chapman and Boeke, 1991 | N/A |
| Experimental Models: Organisms/Strains | ||
| N/A | ||
| Oligonucleotides | ||
| Primer for human ID1 intron lariats RT-qPCR (forward): CCACTTCCGTCCCATCCTT | This manuscript | N/A |
| Primer for human ID1 intron lariats RT-qPCR (reverse, branch point): GGTCGGATCTGGATCTCACTTGG | This manuscript | N/A |
| Primer for human DKK1 intron lariats RT-qPCR (forward): GAGGGAGTAGAACGTGCTGA | This manuscript | N/A |
| Primer for human DKK1 intron lariats RT-qPCR (reverse, branch point): GCCCGACCCCTCTCACTGAG | This manuscript | N/A |
| Primer for human GUS mRNA RT-qPCR (forward): CACCAGGATCCACCTCTGAT | This manuscript | N/A |
| Primer for human GUS mRNA RT-qPCR (reverse): TCCAAATGAGCTCTCCAACC | This manuscript | N/A |
| Primer for HSV1 UL30 qPCR (forward): CATCACCGACCCGGAGAGGGAC | This manuscript | N/A |
| Primer for HSV1 UL30 qPCR (reverse): GGGCCAGGCGCTTGTTGGTGTA | This manuscript | N/A |
| Probe for HSV1 UL30 qPCR: 5′-6FAM-CCGCCGAACTGAGCAGACACCCGCGC-ZEN/lowa Black FQ | This manuscript | N/A |
| Primers for yeast Actin intron lariat Northern-Blot probe generation | Dr Boeke laboratory, Chapman and Boeke, 1991 | N/A |
| Recombinant DNA | ||
| pTRIP-DBR1-RFP plasmids containing wild-type or mutant human DBR1 cDNA | This manuscript | N/A |
| pcDNA3-DBR1-FLAG plasmids containing wild-type or mutant human DBR1 cDNA | This manuscript | N/A |
| pET expression plasmids containing wild-type or mutant human DBR1 cDAN | This manuscript | N/A |
| pRS315 expression plasmids containing wild-type or mutant yeast Dbr1 cDNA | This manuscript | N/A |
| Software and Algorithms | ||
| Graph Pad Prism 5 | GraphPad software | https://www.graphpad.com/scientific-software/prism/ |
| GeneTools image analysis software | Syngene | http://www.syngene.com/genetools-software-download/ |
| Genome Analysis Toolkit (GATK) | Genome variant call software | https://software.broadinstitute.org/gatk/ |
| Affymetrix Power Tools | Genotype calling software | www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx |
| Merlin | Parametric multipoint linkage analysis software. Abecassis et al., 2002 | http://csg.sph.umich.edu/abecasis/merlin/reference.html |
| Iterative inverted alignment method for branchpoint-traversing lariat reads identification | Taggart et al., 2017 | http://fairbrother.biomed.brown.edu/data/Lariat2016/ |
| STAR RNA-Seq aligner | Dobin et al., 2013 | http://code.google.com/p/rna-star/ |
| HTseq-count (v0.6.1, Ensembl v75 transcript annotations) | Anders et al., 2015 | http://www-huber.embl.de/HTSeq |
| Gene-set enrichment testing for Hallmark (H) gene sets (Molecular Signatures Database v6.1) | Liberzon et al., 2015 | http://software.broadinstitute.org/gsea/msigdb |
| DESeq2 package (v1.14.1) | Love et al., 2014 | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| voom/limma package (v 3.22.7) | Law et al., 2014 | https://bioconductor.riken.jp/packages/3.0/bioc/html/limma.html |
| Other | ||
| N/A | ||
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shen-Ying Zhang (shzh289@rockefeller.edu). The human subject fibroblast and EBV-B cell lines generated from this study may require execution of a simple material transfer agreement (MTA) with INSERM. The human DBR1 cDNA containing plasmids generated from this study may require execution of a MTA with the Rockefeller University.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
All human patients were living in and followed up in their countries of origin (Israel for P1–P4, Portugal for P5 and P6, Japan for P7). P1, P4–7 are females. P2 and P3 are males. 29 other family members from their families, including 15 females and 14 males, also participated in the genetic study (see Figure 1A). Informed consent was obtained in the home country of each patient, in accordance with local regulations and a human subjects research protocol approved by the institutional review board (IRB) of INSERM. Experiments using samples from human subjects were conducted in the United States and France, in accordance with local regulations and with the approval of the IRB of The Rockefeller University, and the IRB of INSERM and AFSSAPS, respectively.
Animals
C57/BalbC mice were obtained from the Jackson Laboratory. All experiments were performed under approval of the UTHSCSA Institutional Animal Care and Use Committee, in adherence with the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, and the Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Cell lines
Fibroblast cell lines and EBV-B cell lines from controls and patients
Primary human fibroblasts were obtained from skin biopsy specimens from patients (P1, P5, P6, all females), and were cultured in DMEM medium (Gibco BRL, Invitrogen) supplemented with 10% fetal calf serum (FCS) (Gibco BRL, Invitrogen). To create immortalized SV40-transformed fibroblast cell lines (SV40-fibroblasts), 4 μg of a plasmid containing T antigen DNA was used to transfect about 5 millions of cells by electroporation. The cells were then placed into two fresh 75 cm2 flasks containing 12 ml of DMEM medium (Gibco BRL, Invitrogen) supplemented with 10% FCS (Gibco BRL, Invitrogen). SV40-fibroblasts clones appeared after about 15 days. They were then grown and passaged for experimental usage. To create Epstein-Barr virus (EBV)-transformed B-cell lines (EBV-B), fresh blood samples were obtained from patients (P1, a female; P2, a male), peripheral blood mononuclear cells (PBMC) were isolated on Ficoll-Paque PLUS (GE Healthcare, USA). PBMCs were then infected with EBV, and kept in culture in RPMI medium (Gibco BRL, Invitrogen) supplemented with 10% FCS (Gibco BRL, Invitrogen). EBV-B clones will appear in about 15 days. They could then be grown and passaged for experimental usage. SV40-fibroblasts from three healthy controls (2 females and 1 male) and EBV-B cell lines from other two healthy controls (1 female and 1 male) have been generated using the same methods through previous studies (Casrouge et al., 2006; Chapgier et al., 2006; Dupuis et al., 2013; Guo et al. 2011) of the laboratory and also used in this study.
Other cell lines
HEK293T cells (ATCC) were maintained in DMEM supplemented with 10% FCS.
METHOD DETAILS
Genome-wide linkage analysis
Ten members of the Israeli family (P1, P2, their four parents, their three healthy siblings including two sisters and one brother, a grandmother of P1) and five members of the Portuguese family (P5, P6, their two parents, their healthy brother) were genotyped with the Affymetrix genome-wide SNP 6.0 array. Genotype calling was achieved with Affymetrix Power Tools (www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx) for all the genotyped family members. We discarded monomorphic SNPs, SNPs with a call rate lower than 100% and SNPs presenting more than one Mendelian inconsistency in the family. SNPs were further filtered with population-based filters. We then used the high-quality SNP markers to carry out linkage analysis, assuming autosomal recessive inheritance with complete penetrance (homozygosity mapping). Parametric multipoint linkage analysis was carried out with Merlin (Abecasis et al., 2002).
Whole-exome sequencing
DNA (3 μg) was extracted from cells and sheared with a Covaris S2 Ultrasonicator (Covaris). An adapter-ligated library was prepared with the TruSeq DNA Sample Prep Kit (Illumina). Exome capture was performed with the SureSelect Human All Exon 37 Mb kit (Agilent Technologies) for P1 and P2, the 50 Mb kit for P5 and the 71 Mb kit for P6. Paired-end sequencing was performed on an Illumina HiSeq 2000 (Illumina), generating 100-base reads. The sequences were aligned with the human genome reference sequence (hg19 build), with BWA aligner. Downstream processing was carried out with the Genome Analysis Toolkit (GATK), SAM tools and Picard Tools (http://picard.sourceforge.net). Variant calls were made with GATK UnifiedGenotyper. All calls with a Phred-scaled SNP quality ≤20 were filtered out.
Molecular genetics
DNA was isolated by phenol-chloroform (Sigma-Aldrich, USA) extraction, according to the kit manufacturer’s instructions. Total RNA was extracted from primary fibroblast cells or Simian virus 40-immortalized fibroblast cell lines (SV40-fibroblasts), with Trizol (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA was reverse-transcribed directly, with oligo-dT (Invitrogen). Polymerase chain reaction (PCR) was carried out with Taq polymerase (Invitrogen) and the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, California, USA). The exons of DBR1 were amplified by PCR. The PCR products were purified by ultracentrifugation through Sephadex G-50 Superfine resin (Amersham-Pharmacia-Biotech) and sequenced with the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). Sequencing products were purified by centrifugation through Sephadex G-50 Superfine resin and sequences were analyzed with an ABI Prism 3700 apparatus (Applied Biosystems). The mutation was confirmed by the analysis of genomic DNA extracted from leukocytes and primary fibroblasts, and of cDNA from SV40-transformed fibroblasts.
Measurement of DBR1 transcript levels by RT-qPCR
Gene expression was assessed by measuring transcript levels. Total RNA was extracted from SV40-fibroblasts, EBV-B cells and HEK293T cells. RNA was reverse-transcribed directly, with random hexamers (Invitrogen)(for RNA from SV40-fibroblasts or EBV-B cells) or with the SuperScript III Cells-Direct kit (Life Technologies, for HEK293T cells), to determine mRNA levels for DBR1. Reverse transcription-quantitative PCR (RT-qPCR) was performed with Applied Biosystems 2 x universal Taqman reaction mixture and Assays-on-Demand™ probe/primer combinations, in an ABI PRISM® 7700 Sequence Detection System. The GUS gene was used for normalization. Quantitative PCR efficiency was estimated at ~87%, 94%, and 102% for each of the probe/primer combinations, for DBR1 (Hs01113902_m1 and Hs01113907_m1) and GUS Hs00939627_m1, respectively. Results are expressed according to the ΔCt method, as described by the manufacturer. Three independent experiments were performed, for measurement of DBR1 transcript levels in SV40-fibroblasts or EBV-B cells, using SV40-fibroblasts from three patients (P1, P5, P6) and two healthy controls, and EBV-B cells from two patients (P1, P2) and two healthy controls, technical duplicates were used in each experiment. For measurement of DBR1 transcript levels in HEK293T cells, one experiment was performed using biological triplicates for each condition.
Heterologous expression of DBR1 and debranching activity measurements
E. coli experiments
The cDNA sequences encoding the WT, L13G, Y17H, H85N, I120T, and R197X DBR1 variants were inserted into a modified pET expression plasmid encoding an 8x-histidine tag downstream from DBR1, resulting in the production of a fusion protein with a C-terminal His-tag. For R197X, the His-tag was at the N-terminus. The expression plasmids were checked by sequencing and used to transform Escherichia coli (E. coli) strain BL21 (Life Technologies). Strains were cultured in 10 mL of Terrific Broth, to an OD600 of ~1, at which point the temperature was reduced to 17°C, and IPTG was added to a final concentration of 1 mM. After 18 hours of induction, the cells were harvested by centrifugation and lysed with BugBuster reagent (EMD) supplemented with a cocktail of protease inhibitors (Roche, USA). Soluble and insoluble fractions were analyzed by SDS-PAGE followed by western blotting with a custom-produced rabbit polyclonal antibody against DBR1. Debranching activity was measured by adding 2 μL of soluble lysate to 18 μL of 2 μM AK88 substrate (dissolved in 50 mM HEPES pH 7, 100 mM NaCl, 1 mM TCEP) (Katolik et al., 2017). Fluorescence intensity (with excitation at 488 nm, and detection above 520 nm) was measured with a plate reader (BMG). The rates, in RFU/s were normalized relative to the WT samples. The entire experiment was performed four to six times.
Human embryonic kidney 293T (HEK293T) cell experiments
The cDNA sequences encoding WT, L13G, Y17H, H85N, I120T, and R197X, aa.1-272, and aa.273-544 DBR1 were subcloned into a pcDNA3 plasmid modified to generate C-terminal FLAG-tagged fusion proteins. HEK293T cells were purchased from ATCC, and cultured in OptiMem media (Gibco, 51985034) supplemented with 5% FCS. Cells were transfected with lipid-based transfection reagents according to the manufacturer’s instructions (Lipofectamine-3000, Life Technologies), using 1.25 μg plasmid DNA/well (6-well plates). They were harvested 48-hours post-transfection in either the Roche Complete Lysis-M EDTA-free kit (Sigma-Aldrich) (for activity assays), or 2x Laemmli sample buffer (for electrophoresis). Lysates were diluted 10-fold in assay buffer, (50 mM HEPES pH 7, 100 mM NaCl, 1 mM TCEP), and assayed as described above. The experiment was performed twice, on different days, and a total of 5 biological replicates (individual wells of 6-well plates) are plotted in Figure 2D. Gene expression levels were quantified by synthesizing cDNAs from transfected cells (SuperScript III Cells-Direct kit, Life Technologies), and quantifying DBR1 expression relative to GUS, as described above.
Insect cell experiments
For the insect cell culture experiments, recombinant baculoviruses were created with the Bac-to-Bac system (Life Technologies), according to the manufacturer’s instructions. Sf9 cells were infected with high-titer viruses, harvested 48 hours after infection and assayed as described for the E. coli cultures. The entire experiment was performed two times.
I121T phenotype in yeast
A dbr1-null (dbr1-1) yeast strain, KM196, was transformed with three plasmids: the empty vector pRS315, a wild-type dbr1 expression plasmid, pKC49, and a dbr1-I121T mutant (analogous to the I120T mutation of DBR1 in humans, derived from pKC49) plasmid, produced by site-directed mutagenesis. Total RNA was prepared by the acid phenol method, with extraction in CHCl3 and electrophoresis in a 1.2% agarose/formaldehyde-RNA gel. A northern blot was performed with the indicated 32P-labeled probe recognizing the intron and flanking exon sequences of the yeast actin gene (Chapman and Boeke, 1991). The probe was generated by PCR with the following primers: forward primer: 5′-cctctctcttgtattcttccttcc-3′, reverse primer: 5′-gacgtgagtaacaccatcacc-3′. The experiment was performed once with biological triplicates for each condition.
Generation of the custom-made DBR1 antibody
Residues 1-502 of human DBR1 were produced in E. coli and purified to homogeneity. The purified protein was used as an antigen for the immunization of rabbits (Rockland Immunochemicals), and the resulting immune serum was purified against full-length recombinant human DBR1 covalently coupled to agarose beads (Pierce AminoLink). This antibody was validated by blotting cell lysates overexpressing DBR1 along with control cells, through the use of siRNAs targeting endogenous DBR1 in cultured cells, by cross-validation with commercial Dbr1 antibodies (Santa Cruz and ProteinTech), and through antigen-blocking experiments (Figure S2C). The specificity of the custom-produced DBR1 antibody was confirmed by probing identical blots of control samples with purified antibody, or purified antibody+soluble antigen (30-fold excess of purified recombinant full-length human DBR1, Figure S2C). No visible bands were present after preincubation of the antigen with the custom-made DBR1 antibody, demonstrating the DBR1-specificity of the epitopes identified in Figure S2A.
Western blotting of human tissues for DBR1
Human tissue lysates were purchased from ClonTech (Figure S2A,B) or ProSci-Inc (Figure S2C and data not shown), and separated by electrophoresis in a 4–20% gradient gel, which was then probed with the custom-produced anti-DBR1 antibody (see above). The immunoblots were then stripped, and reprobed with a GAPDH antibody. We used 15μg of ProSci-Inc protein lysate and 2 μg of ClonTech lysate (Figure S2A, C). The hDBR1 signal was very strong in the medulla oblongata and spinal cord. We therefore optimized the western blots to show these samples clearly, but this masked the ubiquitous expression of Dbr1 detected at higher protein loads (i.e. 15 μg), as the dynamic range of hDBR1 expression was too broad for capture on a single western blot. Duplicate gels were stained with Coomassie blue to confirm equal protein loading and normalization (Figure S2B). The western blotting was performed twice with the lysates from ClonTech (Figure S2A) and twice with the lysates from ProSci-Inc (Figure S2C and data not shown).
Western blotting of mouse tissues for DBR1
A six-month-old male C57/BalbC mouse was killed and its tissues were flash-frozen on dry ice. Tissues were homogenized in PBS with protease inhibitors (Roche), a Bullet Blender and NAVY bead kits (Next-Advance). After homogenization, a 1/10 volume of 10 x cell lysis buffer (Cell Signaling Technologies) was added, and the homogenates were sonicated and centrifuged at 12,000 x g for 15 min at 4°C. The protein concentration of the resulting lysate was measured in a Bradford assay (Pierce 23236), and adjusted to 2 μg/ul. We loaded 15 μg of protein per lane. Proteins were separated by SDS-PAGE and transferred onto PVDF membranes. Duplicate gels were stained with Coomassie blue for normalization (Figure S2E). Similar results were obtained when the experiment was repeated with a three-month-old male C57Bl/6 mouse and commercially available lysates from AMS-bio (data not shown).
Western blotting for DBR1 in human cell lines
Total cell extracts were prepared from SV40-fibroblasts or EBV-B cells from patients or healthy controls, or from HEK293T cells (ATCC) with and without transfection of pCDNA3 plasmids containing WT or mutant human DBR1 cDNA. Equal amounts of protein from each sample were separated by SDS-PAGE and blotted onto PVDF membranes (Bio-Rad). Immunoblots were then probed with an anti-human DBR1 antibody (ProteinTech), the custom DBR1 antibody, or anti-FLAG M2 mAb (SIGMA). Membranes were stripped and reprobed with GAPDH antibody (Santa-Cruz), to control for protein loading. Antibody binding was detected by enhanced chemiluminescence (ECL; Amersham-Pharmacia-Biotech). The experiments were independently performed three times, in SV40-fibroblasts and EBV-B cells, using SV40-fibroblasts from three patients (P1, P5, P6) and two healthy controls; in EBV-B cells from two patients (P1, P2) and two healthy controls; and twice in HEK293T cells.
Quantification of western blots
The GeneTools program (Syngene) was used to quantify the western blots and Coomassie-stained gels shown in Figures 2 and Figure S2. We boxed whole lanes for Coomassie gels, and an area around the ~70 kDa hDbr1 band on the anti-FLAG blot. The hDBR1 antibody identifies multiple hDBR1 species (reflecting processing/breakdown species that appear in mouse and human tissues), so we quantified total immunoreactive material by boxing the ~80-30 kDa region for quantification. In all cases, quantification was standardized by setting the value of an empty lane to 0, and that of the darkest lane to 100 (arbitrary value). As the human tissues samples were too numerous to be run on a single gel, the protein marker lane was used to scale the intensities of the two gels. Before the plotting of graphs, values were normalized to 1 by dividing all quantified values by the highest value in the series. We normalized blots for total protein loading, by dividing the normalized quantities of immunoreactive material by the amount of Coomassie blue-stained protein to obtain a semi-quantitative immunoreactive material/total protein ratio.
Total RNA sequencing and analysis
Total RNA was extracted from primary fibroblasts from three DBR1-mutated patients (P1, P5, P6, all females), one TLR3-deficient patient (a male, Guo et al., 2011) and one STAT1-deficient patient (a male, Chapgier et al., 2006), and three healthy controls (two females and one male), with the miRNeasy Mini Kit (QIAGEN), and treated with DNAse (Ambion) to remove residual genomic DNA. RNA-Seq libraries were prepared with the Illumina RiboZero TruSeq Stranded Total RNA Library Prep Kit (Illumina) and sequenced on the Illumina NextSeq platform in the 150 nt, paired-end configuration. Each library was sequenced three times. For the identification of lariat reads and the analysis of branchpoint nucleotides, we first identified branchpoint-traversing lariat reads with an iterative inverted alignment method described elsewhere (http://fairbrother.biomed.brown.edu/data/Lariat2016/) (Taggart et al., 2017). Lariat reads were identified in three sequencing runs for each of the control, P1, P5, P6, TLR3−/−, and STAT1−/− samples, with and without 24 hours of HSV1 infection. Forward alignment was conducted with the STAR RNA-Seq aligner (Dobin et al., 2013). Lariat read recovery rate was plotted as the number of recovered lariat reads per million hg19-mapped reads in a given sample. We used t-tests to assess the significance of differences in lariat read recovery rate. The branchpoint nucleotide was identified by correcting for RT skipping and removing reads derived from template switches and fully circularized introns by the alignment of reads to U2-premRNA interaction models, as previously described (Taggart et al., 2017). Chi-square tests were used to assess the significance of differences in branchpoint nucleotides.
We determined the abundance of individual introns by the ShapeShifter method (Taggart et al, submitted, 2017). Briefly, an unsupervised machine learning approach was used for stabilized lariat intron read density shape. A k-means clustering approach over the 5′ end of all human introns was applied to publicly available ENCODE RNAseq samples (total RNA, whole-cell fraction). The 5′ end window of the intron consists of positions 5–50 of each intron, with a minimum of 10 reads traversing this window. The learning curve was used as a heuristic for the calling of stabilized introns in future samples. This pipeline was applied to three sequencing runs of control, P1, P5, P6, TLR3−/−, and STAT1−/− samples with and without 24 hours of HSV1 infection. If a given intron was called as having a stabilized intron shape in two of the three replicates for a given sample, it was considered to be a stable intron. Intron levels were calculated as the number of intronic reads per million mapped reads per kilobase of intron. The change in a particular intron abundance was calculated as the log2 fold-change in the mean of P1, P5, P6 replicates divided by the mean of the control replicates. We performed t-tests to assess the significance of differences in intronic coverage.
For intron lariat assessment, we also used a slightly modified version of the LaSSO approach (Bitton et al., 2014). Briefly, we first created a sequence database containing all possible lariats with an adenine (A) breakpoint located within 50 nucleotides of the end of an intron (human reference genome hg19, Ensembl v75 transcript annotations). Following standard RNA-Seq read mapping by TopHat with stringent alignment parameters, all unmapped reads (single reads not mapping to the reference genome or mapping with an incorrect orientation of paired-end mates) were mapped to the LaSSO lariat database. Lariat mapping results were then filtered such that only those alignments with a paired-end read mate that could be aligned (BLAT) within the same transcript (intron or exon) and with distinct start-end coordinates were selected as unique lariat events.
For RNA-Seq gene expression analyses, gene-level read counts were obtained with htseq-count (v0.6.1, Ensembl v75 transcript annotations) (Anders et al., 2015) and analyzed with the voom/limma package (v 3.22.7) (Law et al., 2014). Differential gene expression analysis incorporated a linear model with a factor for genotype group (healthy control, DBR1-mutant, TLR3−/−, STAT1−/−) and treatment conditions (mock, poly(I:C), IFN-α2b). Patient blocking was applied with the duplicateCorrelation function. Genes were considered to be differentially expressed in response to a given stimulus if they had an adjusted (Benjamini-Hochberg) p value < 0.01 and log2 fold-change >1 or <-1 in any genotype group (F test). Gene-set enrichment testing for Hallmark (H) gene sets (Molecular Signatures Database v6.1, http://software.broadinstitute.org/gsea/msigdb), (Liberzon et al., 2015) was performed with the MROAST tool (Wu et al., 2010), incorporating the same linear model factors described above. Gene sets were considered to display significant enrichment if the adjusted (Benjamini-Hochberg) p value was < 0.01 in pairwise comparisons. Gene set activity scores (Figure S4D) were generated as the proportion of the genes from the set contributing to enrichment, as reported in MROAST output.
For principal component analysis (PCA, Figure 4A), read counts were normalized and variance was stabilized by regularized log transformation with the rlog function from the DESeq2 package (v1.14.1, (Love et al., 2014)). Patient-specific effects were corrected with the removeBatchEffect function in limma. PCA was performed on the 2000 expressed genes displaying the greatest variance across all samples.
For gene expression heatmaps (Figure 4B), regularized log-transformed read counts (as above, no correction for patient effects) for genes differentially expressed in response to the indicated stimulus (significant in any healthy control or DBR1-mutated group, as described above) were z-scaled and hierarchical clustering was performed on Euclidean distance values, according to the complete method.
For stimulus response fold-change scatter plots (Figure S4C), moderated fold-change values were extracted from limma TopTable output for the corresponding contrast.
For quantification of the relative levels of human and HSV1 transcripts (Figure S5A), transcript levels (TPM) were quantified with the Salmon pseudo-alignment tool (v0.8.2, (Patro et al., 2017)) with a combined reference transcriptome annotation incorporating human and HSV1 RNAs (Ensembl v75 and JQ673480.1, respectively).
RT-qPCR on ID1 and DKK1 RNA transcripts and intron lariats
For the quantification of ID1 and DKK1 intron lariats, total RNA was isolated from the patient’s SV40 fibroblasts with the miRNeasy mini kit (Qiagene). We reverse-transcribed 100–500 ng RNA with 0.5 μM branch point-specific reverse primer for ID1 and DKK1 or reverse primers for β-glucuronidase (GUS) for standardization. The branch point-specific reverse primers were complementary to the 5′ sequence of the first intron of ID1 or DKK1, with the addition of four nucleotides at the 3′ end of the primer, complementary to the branch-point sequence, for specific reverse transcription of the lariat form. The cDNA samples were used for real-time PCR (SYBR Green PCR kit, Qiagen). PCR amplification was performed with the StepOnePlus real-time PCR system (Applied Biosystems). Primers for the ID1 lariat: CCACTTCCGTCCCATCCTT (forward) and GGTCGGATCTGGATCTCACTTGG (reverse, branch point). Primers for the DKK1 lariat: GAGGGAGTAGAACGTGCTGA (forward) and GCCCGACCCCTCTCACTGAG (reverse, branch point). Primers for GUS mRNA: CACCAGGATCCACCTCTGAT (forward) and TCCAAATGAGCTCTCCAACC (reverse). Primers for the ID1 mRNA are: AAACGTGCTGCTCTACGACA (forward) and CTCCAACTGAAGGTCCCTGA (reverse). Primers for the DKK1 mRNA: CCTTGGATGGGTATTCCAGA (forward) and CCTGAGGCACAGTCTGATGA (reverse). The qPCR products obtained with the ID1 or DKK1 lariats primers were separated by electrophoresis in a 2% agarose gel and stained with GreenGlo™ Safe DNA Dye, to confirm that the products were of the expected size. The qPCR products were further sequenced to validate the specificity of lariat form amplification. The efficiency of qPCR for ID1 and DKK1 lariats was about 107% and 118%, respectively, and that for ID1, DKK1 and GUS mRNA was about 97%, 95% and 106%, respectively. The level of ID1 and DKK1 lariats and transcripts was normalized relative to that of GUS transcripts, calculated according to the ΔCt method. The data shown in Figure 3E–F was obtained from three to four independent experiments, with biological duplicates for each condition.
Plasmid transfection of human fibroblasts
The WT DBR1 cDNA was inserted into the pTRIP-RFP expression vector by the Gateway method (Invitrogen). Mutagenesis was performed to generate the patient-specific I120T, Y17H, L13G and R197X mutants, with the pDNOR221 vector, and the mutant DBR1 cDNA was then inserted into the pTRIP-DBR1-RFP vector by the Gateway method (Invitrogen). All plasmids were Sanger-sequenced to confirm the WT or mutant full-length DBR1 cDNA sequences. SV40-fibroblast cells from P1 were transfected with pTRIP-RFP or pTRIP-DBR1-RFP vectors containing the wild-type or mutant DBR1 cDNA, in the presence of Extremgene 9 reagent (Sigma-Aldrich). For stable transfection, cells were cultured under puromycin selection pressure (Invitrogen, 0.4 μg/mL) for two weeks. RT-qPCR and western blotting were performed to confirm the expression of the DBR1 gene used for transfection.
Stimulation and ELISA
We used a synthetic analog of dsRNA, polyinosine-polycytidylic acid (poly(I:C)) (GE Healthcare), as a nonspecific agonist of TLR3 and MDA5/RIG-I. We used T7-GFP RNA as an agonist of RIG-I (Pichlmair et al., 2006). SV40-fibroblast cells were activated in 24-well plates, at a density of 105 cells/well, for 24 hours, with various doses of poly(I:C) (1, 5 or 25 μg/mL). Cells were stimulated with 25 μg/mL poly(I:C) in the presence of Lipofectamine 2000 to activate MDA5/RIG-I signaling. The production of IFN-λ1 and IL-6 was assessed by ELISA. The IFN-λ1 ELISA was developed in the laboratory. Briefly, plates were coated by incubation overnight at 4°C with 1 μg/mL anti-human IFN-λ1 pAb (R&D Systems, Minneapolis, MN), and the concentration of IFN-λ1 in the supernatant was determined by incubation with a secondary human IFN-λ1 biotinylated antibody (R&D Systems) at a concentration of 400 μg/mL. The production of IL-6 was assessed with the R&D IL-6 ELISA kit (R&D). The data shown in Figure S4A were generated from four independent experiments, and that shown in Figure S4B were obtained from two independent experiments.
Quantification of viral replication
For VSV infection, 105 SV40-fibroblasts per well were added to 24-well plates and infected with VSV (Indiana strain), at an MOI of 1, in DMEM supplemented with 2% FCS. After incubation for 30 minutes, the cells were washed and incubated in 500 μL of medium. Supernatants were obtained at the 2 h, 8 h, 24 h and 48 h time points and frozen. VSV titers were determined by calculating the 50% end point (TCID50), as described by Reed and Muench, after the inoculation of Vero cell cultures in 96-well plates. For HSV-1 (KOS strain, ATCC) infection, 105 SV40-fibroblasts per well were added to 24-well plates and infected with HSV-1, at an MOI of 0.01, in DMEM supplemented with 2% FCS. After 2 hours, the cells were washed and incubated in 500 μL of medium. Cells were collected at 2 h, 24 h, 48 h and 72 h and frozen. In addition, supernatants were obtained at the 2 h, 8h, 24 h and 48 h time points and frozen. HSV-1 titers were determined by calculating the TCID50, as described by Reed and Muench, after the inoculation of Vero cell cultures in 96-well plates. HSV1 UL30 gene copy numbers for HSV1-infected cells were also determined with a standard-curve based qPCR method developed in the laboratory. The primers for HSV1 UL30 gene amplification were CATCACCGACCCGGAGAGGGAC (forward) and GGGCCAGGCGCTTGTTGGTGTA (reverse). A TaqMan probe labeled with 6FAM at the 5′ end and with ZEN/lowa Black FQ at the 3′ end was used, the probe sequence is CCGCCGAACTGAGCAGACACCCGCGC. The RNase P (4316844, Applied Biosystems) copy number for each sample was used for normalization. QPCR was performed with Applied Biosystems 2 x universal Taqman reaction mixture, in an ABI PRISM® 7700 Sequence Detection System. For HSV1-GFP infection, 2×104 SV40-transformed fibroblasts per well were added to 96-well plates and infected with HSV1-GFP, at various MOI, in DMEM supplemented with 2% FCS. HSV1-GFP was a gift from Dr. Desai. In this virus, GFP is fused to the HSV1 UL35 open reading frame encoding a capsid protein designated VP26, resulting in a so-called K26GFP HSV1 (KOS7), in which the VP26-GFP fusion protein is incorporated into intranuclear capsids and mature virions in infected cells. This virus can infect and grow in cell cultures as efficiently as the parental virus, and the infected cells display fluorescence, which can be used as a marker for the quantification of viral replication (Desai and Person, 1998). Cells were incubated for two hours, then washed and incubated in 200 μl of culture medium. The GFP fluorescence of the samples was quantified at the 24 h, 48 h, and 72 h time points. All experiments were independently repeated at least three times.
Cell viability assay
The viability of SV40-fibroblasts was assessed by resazurin oxidoreduction (TOX-8) (Sigma-Aldrich). Cells were plated, in triplicate, in DMEM supplemented with 10% FCS, in 96-well flat-bottomed plates (2 × 104 cells/well); 18 hours later, the cells were infected by incubation for 24 hours with VSV or for 72 hours with HSV-1 at various MOI. Resazurin dye solution was then added, at a volume one tenth that of the culture medium, and the samples were incubated for an additional two hours at 37°C. Fluorescence was then measured at a wavelength of 590 nm, using an excitation wavelength of 560 nm. Background fluorescence, calculated for dye and complete medium alone (in the absence of cells) was then subtracted from the values for all the other samples; 100% viability corresponds to the fluorescence of uninfected cells. All experiments were independently repeated at least three times.
QUANTIFICATION AND STATISTICAL ANALYSIS
For RT-qPCR, relative levels of gene mRNA expression or RNA lariats were determined using the ΔCt method relative to that of a housekeeping gene (GUS). For RNA lariat level assessment in primary fibroblasts total RNA-Seq data, an iterative inverted alignment method (http://fairbrother.biomed.brown.edu/data/Lariat2016/) (Taggart et al., 2017) and a LaSSO approach (Bitton et al., 2014) were used. More details are provided in the ‘Total RNA sequencing and analysis’ section in METHOD DETAILS. For virus replication level quantification, VSV or HSV1 titers were determined by calculating the 50% end point (TCID50), as described by Reed and Muench. QPCR method was also used, by quantifying HSV-1 UL30 gene copy numbers for HSV1-infected cells, as well as a GFP measurement method by using a HSV1-GFP which provides GFP expression levels as a marker for the quantification of viral replication (Desai and Person, 1998). More details are provided in the ‘Quantification of viral replication’ section in METHOD DETAILS. Results are presented as means±standard deviation, unless stated otherwise. Mean values were compared between control cells and cells from the patients, by two-tailed Student’s t-tests, unless stated otherwise. Chi-square tests were used to compare the proportion of ‘A’ branchpoints in RNA-Seq data for control and patient fibroblasts. Information about number of cell lines tested and number of experimental replication can be found in the legends to Figure 3B–F, Figure 4C–D, Figure 5A–D, Figure S3G and Figrue S5B. Statistical analysis was performed in Graph Pad PRISM 5, including normality tests of the data when appropriate. More details about statistical analysis and adjustment, that have been used in fibroblast total RNA-sequencing data analysis, are provided in ‘Total RNA sequencing and analysis’ section in METHOD DETAILS. Statistical significance was denoted with * (p<0.05), ** (p<0.01), *** (p<0.001), and **** (p<0.0001) in the figures and figures legends.
DATA AND SOFTWARE AVAILABILITY
The NCBI SRA repository accession number for the whole-exome sequencing and RNA-Seq data reported in this paper is SRA: SRP130621.
Supplementary Material
Supplementary clinical information for the patients studied, Related to Figure 1.
A) Photomicrograph of a brain sample from P5, showing symmetric bilateral thalamic and brainstem lesions (parenchymal softening and brown discoloration). B) Brain histology, showing perivascular plasma exudation and neuronal and glial necrotic changes in the thalamus in P5, and petechial hemorrhage in the pons in P6. C) CT-scan images of the brains of P5 and P7, showing brain lesions affecting the brainstem. D) Histogram representation of the DBR1 mutations, confirmed by Sanger sequencing on genomic DNA from leukocytes (P1, P2) or primary fibroblasts (P5, P6). The mutations in P1, P5 and P6 were also confirmed by the Sanger sequencing of cDNA from SV40-fibroblasts (data not shown). E) Photograph of P7, showing her curly hair. F) Histogram representation of the DBR1 mutations, confirmed by Sanger sequencing on genomic DNA from leukocytes, for P7 and her parents. G) Representation of all the nonsynonymous DBR1 variants reported in the GenomAD database, and the three DBR1 missense variants found in the patients with viral encephalitis studied here. Each of these three variants is private to one of the three kindreds. The minor allele frequency and CADD PHRED score of each variant are shown. CADD MSC of DBR1: the 95% confidence interval mutational significance cutoff CADD score of DBR1 (http://pec630.rockefeller.edu:8080/MSC/). H) Exon subRVIS (subregion residual variation intolerance score) scores for DBR1, as predicted by the subRVIS website (http://www.subrvis.org/). The subRVIS percentile corresponds to the RVIS score percentiles of the eight DBR1 gene exons across the genome. The subRVIS percentiles of exons 1, 2, 3, 4, 5, 7 of the DBR1 gene are below 35%, the general threshold below which an exon is likely harbor disease-causing mutations. The locations of the four DBR1 mutations in patients with brainstem viral encephalitis are indicated with red (kindred A), green (kindred B) or blue (kindred C) arrows.
A) Assessment of DBR1 protein levels in diverse human tissues, by western blotting with a polyclonal antibody (pAb) against human DBR1 (upper panel). GAPDH blots show tissue integrity (middle panel), but, as GAPDH levels vary across tissues, we opted to use duplicate Coomassie blue-stained gels (lower panel) for quantification. B) Quantification of blots in A), normalized according to total protein loading based on Coomassie blue staining. C) For confirmation of the specificity of the custom DBR1 antibody, we performed an antigen-blocking experiment on key samples. When soluble antigen (full-length recombinant DBR1 protein) was incubated with the primary antibody beforehand, no bands were observed on the blot (lower panel), demonstrating that the fragments identified (upper panel) contained DBR1-specific epitopes. D) Assessment of DBR1 protein levels in diverse mouse tissues, by western blotting with a pAb against DBR1 (upper panel), GAPDH blots show tissue integrity (middle panel); the Coomassie blue-stained gel (lower panel) was used for quantification. E) Quantification of the blot in D), normalized according to total protein loading based on Coomassie blue staining.
A) DBR1 mRNA levels in HEK293T cells transfected with WT or mutant DBR1 cDNA-containing plasmids, assessed by RT-qPCR with one set of probe/primer combination spanning exons 2–3 (upper panel) and another set of probe/primer combination spanning exons 7–8 (lower panel) of DBR1. B) The debranching activity of WT or mutant DBR1, as presented in Figure 2D, was normalized against the intensity of the signal for full-length DBR1 as detected with the FLAG antibody. C) Levels of WT or mutant DBR1 protein in Sf9 cells, as assessed by western blotting. D) RNA lariat debranching activity of WT and mutant DBR1 proteins in Sf9 cells, as assessed by the rate of debranching for a synthetic Y-shaped RNA lariat mimic, in the presence of WT or mutant DBR1 protein. The lariat debranching activity of the mutant DBR1 proteins was normalized against that of the WT protein. The data presented here are representative of two independent experiments. E) Analysis of the Saccharomyces cerevisiae dbr1-I121T mutation, analogous to the I120T mutation of DBR1 in humans. Northern blotting with an actin intron plus exon probe was performed, to identify the accumulating intron. Strong accumulation of the 0.3 kb excised introns was observed in the yeast dbr1-1 loss-of-function mutant transformed with an empty vector. This intron accumulation phenotype was rescued by a plasmid containing the WT dbr1 gene. For the yeast mutant harboring the dbr1-I121T mutant plasmid, mild intron lariat accumulation was observed, indicating that this mutation was hypomorphic. For each group, we analyzed three independent clones. F) DBR1 mRNA levels, as assessed by RT-qPCR, in SV40-fibroblasts or EBV-B cells from healthy controls (C), and patients with DBR1 mutations (P1, P5 and P6 for SV40-fibroblasts, P1 and P2 for EBV-B). G) Unique intronic RNA lariat counts (LaSSO workflow), obtained from primary fibroblast total RNA-Seq data and normalized against unmapped read pairs, for three healthy controls, P1 (I120T/I120T), P5 (Y17H/Y17H), P6 (Y17H/Y17H), and TLR3−/− and STAT1−/− patients. We performed t-tests to compare RNA lariat levels in patients with those in healthy controls. ** p<0.01. H) Gene diversity of the unique intronic lariats detected in each primary fibroblast RNA-Seq sample from controls, P1, P5, P6, and TLR3−/− and STAT1−/− patients. The number of unique intronic lariat events detected in each sample is indicated in the center of each donut plot. Each “wedge” represents a different gene in which a lariat was detected.
A) IFN-λ1 (upper panel) and IL-6 (lower panel) production, as measured by ELISA, in SV40-fibroblasts from P1, P5 and P6 with DBR1 mutations, a TLR3−/− patient, and four healthy controls, with and without stimulation with various doses of poly(I:C) stimulation (1, 5, 25 μg/mL), or with 25 μg/mL poly(I:C) in the presence of Lipofectamine. NS: not stimulated. B) IFN-λ1 (upper panel) and IL-6 (lower panel) production, as measured by ELISA, in SV40-fibroblasts from P1 and P5 with DBR1 mutations, a TLR3−/− patient, a NEMO IP patient, and two healthy controls, with and without stimulation with various doses of T7-GFP (1, 10, 100 ng/mL), in the presence of Lipofectamine. C) Scatter plots of fold-changes in gene expression (RNA-Seq) following stimulation with 25 μg/ml poly(I:C) for 6 hours (left panel) or 100 IU/ml IFN-α2b for 8 hours (right panel), in primary fibroblasts from DBR1-deficient patients (n=3) and healthy controls (n=3). Moderated fold-changes are expressed relative to the corresponding mock-treated samples. Each point represents a single gene, and each plot includes genes identified as differentially expressed (FDR 0.01, > 2-fold difference) in response to the indicated stimulus relative to mock-treated samples in the healthy control or DBR1-mutated groups. D) Gene set enrichment testing for DBR1-deficient (n =3) and healthy control (n =3) primary fibroblasts simulated with poly(I:C) for 6 hours or IFN-α2b for 8 hours, relative to the corresponding mock-treated samples. Hallmark gene sets from the Molecular Signature Database were analyzed for enrichment in each genotype/stimulus condition, and are included here if significant enrichment (FDR 0.01) was detected for any particular comparison. For each genotype/stimulus condition, circle size is proportional to enrichment significance (−log10 p value) and circle color represents gene set “activity score” (red indicates the set of genes are upregulated in response to the stimulus, blue indicates the set of genes downregulated in response to the stimulus; the intensity of the color indicates the proportion of genes in the set contributing to set enrichment).
A) Transcriptome composition analysis based on RNA-Seq quantification, in primary fibroblasts from healthy controls, DBR1-deficient patients (P1, P5 and P6) and TLR3−/− and STAT1−/− patients, mock-treated or 8 or 24 hours after infection with HSV1. B) Unique intronic RNA lariat counts (LaSSO workflow), from total primary fibroblast RNA-Seq data, normalized against unmapped read singlets, for healthy controls (n=3), DBR1-deficient patients (n=3) and TLR3−/− (n=1) and STAT1−/− (n=1) patients. Cells were mock-treated, stimulated with poly(I:C) for 6 hours, stimulated with IFN-α2b for 8 hours, or infected with HSV1 for 8 or 24 hours. We performed t-tests to compare the RNA lariat levels in patients with those in healthy controls under each set of stimulation conditions. * p<0.05, ** p< 0.01, *** p< 0.001. C) The gene diversity of the unique intronic lariats (LaSSO workflow) detected in each primary fibroblast RNA-Seq sample, under each set of stimulation conditions, for controls, P1, P5, P6, and TLR3−/− and STAT1−/− patients. The number of unique intronic lariat events detected in each sample is indicated in the center of each donut plot. Each “wedge” represents a different gene in which a lariat was detected. D) HSV1 RNA lariat counts (LaSSO workflow), from total primary fibroblast RNA-Seq data normalized against unmapped read singlets, for healthy controls (n= 3), DBR1-deficient patients (n= 3) and TLR3−/− (n=1) and STAT1−/− (n=1) patients, 24 hours after infection with HSV1. E) Intronic read density plots over two typical introns, ID1 (upper panels) and DKK1 (lower panels), for healthy control 1 (C1) and P1 (I120T/I120T), from primary fibroblast RNA-Seq data in the absence (left panels) and presence (right panels) of HSV1 infection for 24 hours. F) RNA-Seq lariat profiling for introns ID1 (upper panels) and DKK1 (lower panels), for three healthy controls (C), three DBR1-mutated patients (P1, P5 and P6), and TLR3−/− and STAT1−/− patients. Lariat abundance was profiled with branchpoint-traversing reads normalized against hg19-mapped reads (left panels, p<0.0001 for all comparisons of patients with controls, t-test). Intronic read coverage was determined as the number of intronic reads per million mapped hg19 reads per kb of intron (right panels, p<0.0002 for all comparisons of patients with controls, t-test).
A) HSV1 replication levels were assessed by qPCR, to determine HSV1 UL30 gene copy numbers, with normalization against the copy number of the RNase P gene, at various time points after infection with HSV1 at a MOI of 0.1, in SV40-fibroblasts from healthy controls (C), P1, P5 and P6, and TLR3−/− and STAT1−/− patients. The data shown are mean values from two independent experiments, with technical duplicates in each experiment. B) HSV1 replication levels were assessed by qPCR, to determine HSV1 UL30 gene copy number, with normalization against RNase P gene copy number, at various time points after infection with HSV-1 at a MOI of 0.1, in healthy controls, and in SV40-fibroblasts from P1 with and without stable transfection with the pTRIP-RFP vector without the insertion of DBR1 cDNA, or with the expression of the WT, I120T or R197X DBR1. The experiment shown is representative of three independent experiments carried out.
Table S1. List of genes located in the homozygous interval on chromosome 3 identified by WGL, Related to Figure 1.
Table S2. List of homozygous rare nonsynonymous variations in the two pairs of patients from kindred A and B, Related to Figure 1.
Table S3. List of nonsynonymous DBR1 variants for which homozygous carriers have been reported in the GnomAD database, Related to Figure 1.
Highlights.
Bi-allelic DBR1 mutations are found in patients with viral infection of the brainstem
The DBR1 mutant alleles are severely hypomorphic in terms of expression and function
The levels of RNA lariats levels are increased in DBR1-mutated patients’ fibroblasts
Cell-intrinsic immunity to viruses is impaired in DBR1-mutated patients’ fibroblasts
Acknowledgments
We thank the patients and their families; the members of the laboratory for helpful discussions; Tatiana Kochetkov for technical assistance; Lei Shang, Benedetta Bigio, Vimel Ratinna, Bertrand Boisson, and Jason Banfelder for computational assistance; Dominick Papandrea, Cécile Patissier, and Yelena Nemirovskaya for administrative assistance. This work was funded in part by the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), Clinical and Translational Science Award program grant number 8UL1TR000043, NIH grants 5R21NS084255, 5R01AI088364 and 5R01NS072381, the ANR grants HSEIEIER and ANR-10-LABX-62-IBEID, an award from D.I.M. Pathologies infectieuses humaines (DIM150012), a grant from Starr Foundation Tri-Institutional Stem Cell Initiative (2014-005), a Grant from the Japan Society for the Promotion of Science (16H05355), a grant from the Japanese Agency for Medical Research and Development, AMED, the Rockefeller University, INSERM, Paris Descartes University, and the St Giles Foundation. NEC was supported by NIH T-32 AG021890. KD and LDN are supported by the NIH. BRR was supported in part by the NIH and then John C. Whitehead Presidential Fellowship.
Footnotes
Author Contributions
SYZ and JLC designed and supervised the study. SYZ, NEC, CAF, EP, MJC, SO, AT, AB, FGL, MT, AC, JL, SV, BZ, SS, EJGR, KD, MH, KM, SPH, ND, SAH, LL, RH, CEV, AK, SA, WF performed the experiments and analyzed the data. SO, HM, PG, AD, OO, MH, EF, RF, MK, LD and AE saw and enrolled the patients. GAS, VG, MJD, NEC, LA, BRR, and JH helped to design the experiments and analyzed the data. SYZ and JLC wrote the manuscript, with the assistance of all coauthors.
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Adzhubei I, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen LL, Mork N, Reinert LS, Kofod-Olsen E, Narita R, Jorgensen SE, Skipper KA, Honing K, Gad HH, Ostergaard L, et al. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J Exp Med. 2015;212:1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z, Fang X, Shorter J, Krogan NJ, Finkbeiner S, et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet. 2012;44:1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton DA, Rallis C, Jeffares DC, Smith GC, Chen YY, Codlin S, Marguerat S, Bahler J. LaSSO, a strategy for genome-wide mapping of intronic lariats and branch points using RNA-seq. Genome Res. 2014;24:1169–1179. doi: 10.1101/gr.166819.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JL. Human genetic basis of interindividual variability in the course of infection. Proc Natl Acad Sci U S A. 2015a;112:E7118–7127. doi: 10.1073/pnas.1521644112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci U S A. 2015b;112:E7128–7137. doi: 10.1073/pnas.1521651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, Alcais A, Picard C, Mahfoufi N, Nicolas N, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- Chapgier A, Wynn RF, Jouanguy E, Filipe-Santos O, Zhang S, Feinberg J, Hawkins K, Casanova JL, Arkwright PD. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol. 2006;176:5078–5083. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- Chapman KB, Boeke JD. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65:483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- Ciancanelli MJ, Huang SX, Luthra P, Garner H, Itan Y, Volpi S, Lafaille FG, Trouillet C, Schmolke M, Albrecht RA, et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NE, Katolik A, Roberts KM, Taylor AB, Holloway SP, Schuermann JP, Montemayor EJ, Stevens SW, Fitzpatrick PF, Damha MJ, et al. Metal dependence and branched RNA cocrystal structures of the RNA lariat debranching enzyme Dbr1. Proc Natl Acad Sci U S A. 2016;113:14727–14732. doi: 10.1073/pnas.1612729114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvis AE, Fisher HE, Fan H, Camerini D. Conformational Changes in the 5′ End of the HIV-1 Genome Dependent on the Debranching Enzyme DBR1 During Early Stages of Infection. J Virol. 2017 doi: 10.1128/JVI.01377-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrey SM, Katolik A, Prekeris M, Li X, York K, Bernards S, Fields S, Zhao R, Damha MJ, Hesselberth JR. A homolog of lariat-debranching enzyme modulates turnover of branched RNA. RNA. 2014;20:1337–1348. doi: 10.1261/rna.044602.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Audry M, Ciancanelli M, Alsina L, Azevedo J, Herman M, Anguiano E, Sancho-Shimizu V, Lorenzo L, Pauwels E, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208:2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussow AB, Petrovski S, Wang Q, Allen AS, Goldstein DB. The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol. 2016;17:9. doi: 10.1186/s13059-016-0869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Park HK, Ching T, Panneerselvam J, Wang H, Shen Y, Zhang J, Li L, Che R, Garmire L, et al. Human DBR1 modulates the recycling of snRNPs to affect alternative RNA splicing and contributes to the suppression of cancer development. Oncogene. 2017 doi: 10.1038/onc.2017.150. [DOI] [PMC free article] [PubMed]
- Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez-Barricarte R, Scott E, Shah I, Stenson PD, Gleeson J, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods. 2016;13:109–110. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y, Shang L, Boisson B, Patin E, Bolze A, Moncada-Velez M, Scott E, Ciancanelli MJ, Lafaille FG, Markle JG, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci U S A. 2015;112:13615–13620. doi: 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A, Rosbash M. RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc Natl Acad Sci U S A. 1986;83:5835–5839. doi: 10.1073/pnas.83.16.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jmor F, Emsley HC, Fischer M, Solomon T, Lewthwaite P. The incidence of acute encephalitis syndrome in Western industrialised and tropical countries. Virology journal. 2008;5:134. doi: 10.1186/1743-422X-5-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelt B, Mihai C, Li TM, Veerapaneni P. Rhombencephalitis/brainstem encephalitis. Current neurology and neuroscience reports. 2011;11:543–552. doi: 10.1007/s11910-011-0228-5. [DOI] [PubMed] [Google Scholar]
- Katolik A, Clark NE, Tago N, Montemayor EJ, Hart PJ, Damha MJ. Fluorescent Branched RNAs for High-Throughput Analysis of Dbr1 Enzyme Kinetics and Inhibition. ACS chemical biology. 2017a;12:622–627. doi: 10.1021/acschembio.6b00971. [DOI] [PubMed] [Google Scholar]
- Kim JW, Kim HC, Kim GM, Yang JM, Boeke JD, Nam K. Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. Nucleic Acids Res. 2000;28:3666–3673. doi: 10.1093/nar/28.18.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lafaille FG, Pessach IM, Zhang SY, Ciancanelli MJ, Herman M, Abhyankar A, Ying SW, Keros S, Goldstein PA, Mostoslavsky G, et al. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature. 2012;491:769–773. doi: 10.1038/nature11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montemayor EJ, Katolik A, Clark NE, Taylor AB, Schuermann JP, Combs DJ, Johnsson R, Holloway SP, Stevens SW, Damha MJ, et al. Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1. Nucleic Acids Res. 2014;42:10845–10855. doi: 10.1093/nar/gku725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JL, Sheng J, Rubin DH. A role for H/ACA and C/D small nucleolar RNAs in viral replication. Molecular biotechnology. 2014;56:429–437. doi: 10.1007/s12033-013-9730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K, Hudson RH, Chapman KB, Ganeshan K, Damha MJ, Boeke JD. Yeast lariat debranching enzyme. Substrate and sequence specificity. J Biol Chem. 1994;269:20613–20621. [PubMed] [Google Scholar]
- Nam K, Lee G, Trambley J, Devine SE, Boeke JD. Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol Cell Biol. 1997;17:809–818. doi: 10.1128/mcb.17.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Samarsky DA, Fournier MJ, Boeke JD. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA. 1998;4:1096–1110. doi: 10.1017/s1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Jones C. Towards an understanding of the herpes simplex virus type 1 latency-reactivation cycle. Interdiscip Perspect Infect Dis. 2010:262415. doi: 10.1155/2010/262415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Plotch SJ, Krug RM. In vitro splicing of influenza viral NS1 mRNA and NS1-beta-globin chimeras: possible mechanisms for the control of viral mRNA splicing. Proc Natl Acad Sci U S A. 1986;83:5444–5448. doi: 10.1073/pnas.83.15.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas HG, Schmaling SK, Gaglia MM. Shutoff of Host Gene Expression in Influenza A Virus and Herpesviruses: Similar Mechanisms and Common Themes. Viruses. 2016;8:102. doi: 10.3390/v8040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Knipe DM, Whitley RJ. Herpes simplex viruses. Fields Virology. 2013;2:1823–1879. [Google Scholar]
- Sedger LM. microRNA control of interferons and interferon induced anti-viral activity. Mol Immunol. 2013;56:781–793. doi: 10.1016/j.molimm.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Stahl JP, Mailles A, Dacheux L, Morand P. Epidemiology of viral encephalitis in 2011. Medecine et maladies infectieuses. 2011;41:453–464. doi: 10.1016/j.medmal.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Taggart AJ, Lin CL, Shrestha B, Heintzelman C, Kim S, Fairbrother WG. Large-scale analysis of branchpoint usage across species and cell lines. Genome Res. 2017;27:639–649. doi: 10.1101/gr.202820.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Patel A, Krause PR. Herpes simplex virus ICP27 regulates alternative pre-mRNA polyadenylation and splicing in a sequence-dependent manner. Proc Natl Acad Sci U S A. 2016;113:12256–12261. doi: 10.1073/pnas.1609695113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CK, Cheng SC. The spliceosome catalyzes debranching in competition with reverse of the first chemical reaction. RNA. 2013;19:971–981. doi: 10.1261/rna.038638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SY, Casanova JL. Inborn errors underlying herpes simplex encephalitis: From TLR3 to IRF3. J Exp Med. 2015;212:1342–1343. doi: 10.1084/jem.2129insight4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Vuong BQ, Vaidyanathan B, Lin JY, Huang FT, Chaudhuri J. Non-coding RNA Generated following Lariat Debranching Mediates Targeting of AID to DNA. Cell. 2015;161:762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary clinical information for the patients studied, Related to Figure 1.
A) Photomicrograph of a brain sample from P5, showing symmetric bilateral thalamic and brainstem lesions (parenchymal softening and brown discoloration). B) Brain histology, showing perivascular plasma exudation and neuronal and glial necrotic changes in the thalamus in P5, and petechial hemorrhage in the pons in P6. C) CT-scan images of the brains of P5 and P7, showing brain lesions affecting the brainstem. D) Histogram representation of the DBR1 mutations, confirmed by Sanger sequencing on genomic DNA from leukocytes (P1, P2) or primary fibroblasts (P5, P6). The mutations in P1, P5 and P6 were also confirmed by the Sanger sequencing of cDNA from SV40-fibroblasts (data not shown). E) Photograph of P7, showing her curly hair. F) Histogram representation of the DBR1 mutations, confirmed by Sanger sequencing on genomic DNA from leukocytes, for P7 and her parents. G) Representation of all the nonsynonymous DBR1 variants reported in the GenomAD database, and the three DBR1 missense variants found in the patients with viral encephalitis studied here. Each of these three variants is private to one of the three kindreds. The minor allele frequency and CADD PHRED score of each variant are shown. CADD MSC of DBR1: the 95% confidence interval mutational significance cutoff CADD score of DBR1 (http://pec630.rockefeller.edu:8080/MSC/). H) Exon subRVIS (subregion residual variation intolerance score) scores for DBR1, as predicted by the subRVIS website (http://www.subrvis.org/). The subRVIS percentile corresponds to the RVIS score percentiles of the eight DBR1 gene exons across the genome. The subRVIS percentiles of exons 1, 2, 3, 4, 5, 7 of the DBR1 gene are below 35%, the general threshold below which an exon is likely harbor disease-causing mutations. The locations of the four DBR1 mutations in patients with brainstem viral encephalitis are indicated with red (kindred A), green (kindred B) or blue (kindred C) arrows.
A) Assessment of DBR1 protein levels in diverse human tissues, by western blotting with a polyclonal antibody (pAb) against human DBR1 (upper panel). GAPDH blots show tissue integrity (middle panel), but, as GAPDH levels vary across tissues, we opted to use duplicate Coomassie blue-stained gels (lower panel) for quantification. B) Quantification of blots in A), normalized according to total protein loading based on Coomassie blue staining. C) For confirmation of the specificity of the custom DBR1 antibody, we performed an antigen-blocking experiment on key samples. When soluble antigen (full-length recombinant DBR1 protein) was incubated with the primary antibody beforehand, no bands were observed on the blot (lower panel), demonstrating that the fragments identified (upper panel) contained DBR1-specific epitopes. D) Assessment of DBR1 protein levels in diverse mouse tissues, by western blotting with a pAb against DBR1 (upper panel), GAPDH blots show tissue integrity (middle panel); the Coomassie blue-stained gel (lower panel) was used for quantification. E) Quantification of the blot in D), normalized according to total protein loading based on Coomassie blue staining.
A) DBR1 mRNA levels in HEK293T cells transfected with WT or mutant DBR1 cDNA-containing plasmids, assessed by RT-qPCR with one set of probe/primer combination spanning exons 2–3 (upper panel) and another set of probe/primer combination spanning exons 7–8 (lower panel) of DBR1. B) The debranching activity of WT or mutant DBR1, as presented in Figure 2D, was normalized against the intensity of the signal for full-length DBR1 as detected with the FLAG antibody. C) Levels of WT or mutant DBR1 protein in Sf9 cells, as assessed by western blotting. D) RNA lariat debranching activity of WT and mutant DBR1 proteins in Sf9 cells, as assessed by the rate of debranching for a synthetic Y-shaped RNA lariat mimic, in the presence of WT or mutant DBR1 protein. The lariat debranching activity of the mutant DBR1 proteins was normalized against that of the WT protein. The data presented here are representative of two independent experiments. E) Analysis of the Saccharomyces cerevisiae dbr1-I121T mutation, analogous to the I120T mutation of DBR1 in humans. Northern blotting with an actin intron plus exon probe was performed, to identify the accumulating intron. Strong accumulation of the 0.3 kb excised introns was observed in the yeast dbr1-1 loss-of-function mutant transformed with an empty vector. This intron accumulation phenotype was rescued by a plasmid containing the WT dbr1 gene. For the yeast mutant harboring the dbr1-I121T mutant plasmid, mild intron lariat accumulation was observed, indicating that this mutation was hypomorphic. For each group, we analyzed three independent clones. F) DBR1 mRNA levels, as assessed by RT-qPCR, in SV40-fibroblasts or EBV-B cells from healthy controls (C), and patients with DBR1 mutations (P1, P5 and P6 for SV40-fibroblasts, P1 and P2 for EBV-B). G) Unique intronic RNA lariat counts (LaSSO workflow), obtained from primary fibroblast total RNA-Seq data and normalized against unmapped read pairs, for three healthy controls, P1 (I120T/I120T), P5 (Y17H/Y17H), P6 (Y17H/Y17H), and TLR3−/− and STAT1−/− patients. We performed t-tests to compare RNA lariat levels in patients with those in healthy controls. ** p<0.01. H) Gene diversity of the unique intronic lariats detected in each primary fibroblast RNA-Seq sample from controls, P1, P5, P6, and TLR3−/− and STAT1−/− patients. The number of unique intronic lariat events detected in each sample is indicated in the center of each donut plot. Each “wedge” represents a different gene in which a lariat was detected.
A) IFN-λ1 (upper panel) and IL-6 (lower panel) production, as measured by ELISA, in SV40-fibroblasts from P1, P5 and P6 with DBR1 mutations, a TLR3−/− patient, and four healthy controls, with and without stimulation with various doses of poly(I:C) stimulation (1, 5, 25 μg/mL), or with 25 μg/mL poly(I:C) in the presence of Lipofectamine. NS: not stimulated. B) IFN-λ1 (upper panel) and IL-6 (lower panel) production, as measured by ELISA, in SV40-fibroblasts from P1 and P5 with DBR1 mutations, a TLR3−/− patient, a NEMO IP patient, and two healthy controls, with and without stimulation with various doses of T7-GFP (1, 10, 100 ng/mL), in the presence of Lipofectamine. C) Scatter plots of fold-changes in gene expression (RNA-Seq) following stimulation with 25 μg/ml poly(I:C) for 6 hours (left panel) or 100 IU/ml IFN-α2b for 8 hours (right panel), in primary fibroblasts from DBR1-deficient patients (n=3) and healthy controls (n=3). Moderated fold-changes are expressed relative to the corresponding mock-treated samples. Each point represents a single gene, and each plot includes genes identified as differentially expressed (FDR 0.01, > 2-fold difference) in response to the indicated stimulus relative to mock-treated samples in the healthy control or DBR1-mutated groups. D) Gene set enrichment testing for DBR1-deficient (n =3) and healthy control (n =3) primary fibroblasts simulated with poly(I:C) for 6 hours or IFN-α2b for 8 hours, relative to the corresponding mock-treated samples. Hallmark gene sets from the Molecular Signature Database were analyzed for enrichment in each genotype/stimulus condition, and are included here if significant enrichment (FDR 0.01) was detected for any particular comparison. For each genotype/stimulus condition, circle size is proportional to enrichment significance (−log10 p value) and circle color represents gene set “activity score” (red indicates the set of genes are upregulated in response to the stimulus, blue indicates the set of genes downregulated in response to the stimulus; the intensity of the color indicates the proportion of genes in the set contributing to set enrichment).
A) Transcriptome composition analysis based on RNA-Seq quantification, in primary fibroblasts from healthy controls, DBR1-deficient patients (P1, P5 and P6) and TLR3−/− and STAT1−/− patients, mock-treated or 8 or 24 hours after infection with HSV1. B) Unique intronic RNA lariat counts (LaSSO workflow), from total primary fibroblast RNA-Seq data, normalized against unmapped read singlets, for healthy controls (n=3), DBR1-deficient patients (n=3) and TLR3−/− (n=1) and STAT1−/− (n=1) patients. Cells were mock-treated, stimulated with poly(I:C) for 6 hours, stimulated with IFN-α2b for 8 hours, or infected with HSV1 for 8 or 24 hours. We performed t-tests to compare the RNA lariat levels in patients with those in healthy controls under each set of stimulation conditions. * p<0.05, ** p< 0.01, *** p< 0.001. C) The gene diversity of the unique intronic lariats (LaSSO workflow) detected in each primary fibroblast RNA-Seq sample, under each set of stimulation conditions, for controls, P1, P5, P6, and TLR3−/− and STAT1−/− patients. The number of unique intronic lariat events detected in each sample is indicated in the center of each donut plot. Each “wedge” represents a different gene in which a lariat was detected. D) HSV1 RNA lariat counts (LaSSO workflow), from total primary fibroblast RNA-Seq data normalized against unmapped read singlets, for healthy controls (n= 3), DBR1-deficient patients (n= 3) and TLR3−/− (n=1) and STAT1−/− (n=1) patients, 24 hours after infection with HSV1. E) Intronic read density plots over two typical introns, ID1 (upper panels) and DKK1 (lower panels), for healthy control 1 (C1) and P1 (I120T/I120T), from primary fibroblast RNA-Seq data in the absence (left panels) and presence (right panels) of HSV1 infection for 24 hours. F) RNA-Seq lariat profiling for introns ID1 (upper panels) and DKK1 (lower panels), for three healthy controls (C), three DBR1-mutated patients (P1, P5 and P6), and TLR3−/− and STAT1−/− patients. Lariat abundance was profiled with branchpoint-traversing reads normalized against hg19-mapped reads (left panels, p<0.0001 for all comparisons of patients with controls, t-test). Intronic read coverage was determined as the number of intronic reads per million mapped hg19 reads per kb of intron (right panels, p<0.0002 for all comparisons of patients with controls, t-test).
A) HSV1 replication levels were assessed by qPCR, to determine HSV1 UL30 gene copy numbers, with normalization against the copy number of the RNase P gene, at various time points after infection with HSV1 at a MOI of 0.1, in SV40-fibroblasts from healthy controls (C), P1, P5 and P6, and TLR3−/− and STAT1−/− patients. The data shown are mean values from two independent experiments, with technical duplicates in each experiment. B) HSV1 replication levels were assessed by qPCR, to determine HSV1 UL30 gene copy number, with normalization against RNase P gene copy number, at various time points after infection with HSV-1 at a MOI of 0.1, in healthy controls, and in SV40-fibroblasts from P1 with and without stable transfection with the pTRIP-RFP vector without the insertion of DBR1 cDNA, or with the expression of the WT, I120T or R197X DBR1. The experiment shown is representative of three independent experiments carried out.
Table S1. List of genes located in the homozygous interval on chromosome 3 identified by WGL, Related to Figure 1.
Table S2. List of homozygous rare nonsynonymous variations in the two pairs of patients from kindred A and B, Related to Figure 1.
Table S3. List of nonsynonymous DBR1 variants for which homozygous carriers have been reported in the GnomAD database, Related to Figure 1.