Abstract
Meiosis is a highly regulated process, partly due to the need to break and then repair DNA as part of the meiotic program. Post-translational modifications are widely used during meiotic events to regulate steps such as protein complex formation, checkpoint activation, and protein attenuation. In this paper, we investigate how proteins that are obligatory components of the SUMO (small ubiquitin-like modifier) pathway, one such post-translational modification, affect the Caenorhabditis elegans germline. We show that UBC-9, the E2 conjugation enzyme, and the C. elegans homolog of SUMO, SMO-1, localize to germline nuclei throughout prophase I. Mutant analysis of smo-1 and ubc-9 revealed increased recombination intermediates throughout the germline, originating during the mitotic divisions. SUMOylation mutants also showed late meiotic defects including defects in the restructuring of oocyte bivalents and endomitotic oocytes. Increased rates of noninterfering crossovers were observed in ubc-9 heterozygotes, even though interfering crossovers were unaffected. We have also identified a physical interaction between UBC-9 and DNA repair protein MRE-11. ubc-9 and mre-11 null mutants exhibited similar phenotypes at germline mitotic nuclei and were synthetically sick. These phenotypes and genetic interactions were specific to MRE-11 null mutants as opposed to RAD-50 or resection-defective MRE-11. We propose that the SUMOylation pathway acts redundantly with MRE-11, and in this process MRE-11 likely plays a structural role.
Keywords: meiosis, SUMOylation, DNA damage repair, germline, ubc-9
THE Caenorhabditis elegans germline is a dynamic organ that contains both mitotically dividing nuclei and nuclei undergoing meiosis (Hubbard 2005). The distal tip of hermaphroditic gonads contains mitotic nuclei that transition into meiosis, the process by which eggs and sperm are formed. Meiosis involves two rounds of cellular division; the first is reductional and the second equational. Meiosis initiates when homologous chromosomes pair and the synaptonemal complex (SC), a proteinaceous zipper-like structure, forms to hold homologs together (Dernburg et al. 1998; Plug et al. 1998; Walker and Hawley 2000). At this early stage of meiosis, SPO-11 forms DNA double-stranded breaks (DSBs) as the first step in homologous recombination (HR) (Klapholz et al. 1985; Cao et al. 1990; Malone et al. 1991; Keeney et al. 1997; McKim and Hayashi-Hagihara 1998; Romanienko and Camerini-Otero 1999). These DSBs are initially processed by the MRE11, RAD50, and NBS1/Xrs2 (MRN/X) complex. RAD-50’s coiled-coil and hook domains hold the broken ends together, while MRE11 binds DNA, nicks the DNA upstream of SPO11, and resects the DNA leaving a short 3’ overhang (Usui et al. 1998; de Jager et al. 2001; Borde et al. 2004; Milman et al. 2009; Hohl et al. 2011). The latter activity of the MRN/X complex removes SPO11 bound to the 5’ end of the DNA and is required for long-range resection performed by other nucleases. The single-stranded binding protein RPA initially covers the single-stranded DNA (ssDNA), and then is replaced by the recombinase RAD51 and/or its meiosis-specific ortholog DMC1 (Bishop et al. 1992; Shinohara et al. 1992; Bishop 1994; Habu et al. 1996; Dresser et al. 1997; Yoshida et al. 1998). MSH4/5 then localizes to sites that will become interfering crossovers along with other crossover-promoting proteins, including COSA-1 in worms (Hollingsworth et al. 1995; Paquis-Flucklinger et al. 1997; Bocker et al. 1999; Novak et al. 2001; Snowden et al. 2004; Yokoo et al. 2012). Following crossover formation, chromosomes restructure, while the SC disassembles. Chiasmata (the physical manifestation of crossovers) hold the chromosomes together through the end of diakinesis (Rasmussen and Holm 1984; Lawrie et al. 1995; Moens and Spyropoulos 1995; Bascom-Slack et al. 1997).
Although the function of many meiotic proteins is understood, the complex regulation of HR is still under investigation. Many of the canonical HR proteins are modified post-translationally, by phosphorylation, ubiquitination, or SUMOylation (Falck et al. 2012; Lu et al. 2012; Bologna et al. 2015; Ismail et al. 2015; Parameswaran et al. 2015; Luo et al. 2016; Silva et al. 2016; Tomimatsu et al. 2017). SUMOylation is a modification that involves the transfer of a small polypeptide called small ubiquitin-like modifier (SUMO) to a target protein (Choudhury and Li 1997; Lapenta et al. 1997; Mahajan et al. 1997; Chen et al. 1998; Huang et al. 1998; Tanaka et al. 1999). Yeast and C. elegans have a single SUMO moiety, while mammals have three SUMO isoforms (Su and Li 2002). SUMOylation is used to modify protein function; it can be used to stabilize protein complexes, localize target proteins to specific cell organelles, or signal for degradation (similar to ubiquitination). The SUMOylation pathway consists of an E1 activating enzyme that binds SUMO (Haas et al. 1982). E1 then transfers SUMO to an E2 conjugating enzyme, which can transfer SUMO directly to a target protein (Bernier-Villamor et al. 2002). Alternatively, E2 can transfer SUMO to an E3 ligase that then SUMOylates the target protein (Yunus and Lima 2009). C. elegans uses the E1 enzymes UBA-2 and AOS-1, a single E2 enzyme UBC-9, and two canonical E3 ligases GEI-17 and ZK1248.11 (Holway et al. 2006; Pelisch and Hay 2016). A target protein can be monoSUMOylated, or polySUMOylated (Rojas-Fernandez et al. 2014; Horigome et al. 2016). The latter can be branched or straight depending on context. SUMO chains can help create larger structures that may hold macromolecules together in vivo (Tatham et al. 2001; Bylebyl et al. 2003; Windecker and Ulrich 2008; Skilton et al. 2009; Guzzo et al. 2012; Rojas-Fernandez et al. 2014; Chen et al. 2016). SUMOylation is a regulated process, and SUMO can be added to or removed from target proteins depending on cellular input. SUMO moieties that are conjugated to target proteins can then interact noncovalently with other proteins’ SUMO-interacting motifs (SIMs) to form functional complexes (Song et al. 2004).
Evidence that SUMOylation is required for proper meiosis has been found from studies in Saccharomyces cerevisiae and mice (Voelkel-Meiman et al. 2013; Chen et al. 2016). In yeast, immunofluorescence reveals that both UBC9 (E2) and SUMO localize to the SC during early prophase I, and that deletion of either protein prevents normal SC assembly (Hooker and Roeder 2006; Voelkel-Meiman et al. 2013). UBC9 interacts with proteins that act in meiotic recombination in mammals and yeast, in a step required for RAD-51 filament assembly. RNF212 is an E3 SUMO ligase that has been shown to regulate meiotic recombination in mice (Rao et al. 2017). RNF212 acts by stabilizing recombination sites through controlling HEI10-mediated proteasomal degradation of proteins promoting noncrossover pathways. RAD51 and UBC9 colocalize during spermatogenesis in mice (Kovalenko et al. 1996). In yeast, Rad52 and Rad59 are both SUMOylated; this is required for Rad52 stabilization and for Rad52/Rad59 function in loading Rad51 onto ssDNA (Sacher et al. 2006; Altmannova et al. 2010).
SUMOylation in mitosis has been examined in many organisms including yeast and mammals. In mammals, three SUMO paralogs with distinct but overlapping functions are expressed (Citro and Chiocca 2013). SUMO1 is important for all the major pathways of DSB repair, and the other two paralogs, SUMO2 and SUMO3, are required for classic nonhomologous end joining (Hu and Parvin 2014). In mammalian tissue culture, RNF4 forms SUMO/ubiquitin hybrid chains through its SIM domain and is required for localization of BRCA1 and RAP80 to DNA damage foci (Guzzo et al. 2012). RNF4 also recognizes polySUMOylated proteins including RPA, BRCA1, 53BP1, and MDC1 in vivo, and is responsible for targeting them for degradation via the ubiquitin pathway (Galanty et al. 2012; Yin et al. 2012; Vyas et al. 2013). RNF4 in mammals responds to replication stress, as it is localized at stalled replication forks when ATR is depleted (Ragland et al. 2013). Rad52/Rad59 are also targets for SUMOylation in mitosis, where they plays a role in repair pathway choice (Silva et al. 2016). In yeast mitosis, Rad52/Rad59 SUMOylation regulates Srs2, which itself is SUMOylated. All three SUMOylated proteins affect the crossover vss. gene conversion decision in HR as well as the choice between the single-strand annealing pathway and HR. This pathway choice could be mediated by the Rad52/59 complex role in Rad51 loading (Altmannova et al. 2010; Silva et al. 2016). Rad52/Rad59 is not the only target of SUMOylation in DNA repair. Studies in yeast have demonstrated that both Mre11 and Xrs2 of the MRX complex are SUMOylated upon DNA damage (Cremona et al. 2012; Sarangi et al. 2015). Mre11 in yeast uses SIMs to associate with SUMO on other proteins (Chen et al. 2016). SIMs may help hold the complex together in a structurally functional conformation.
SUMOylation has been most extensively studied in the C. elegans soma in the context of development. UBC-9 and/or SUMO play a role in vulva development, telomere localization, dosage compensation, pharynx development, ER stress response, adherens junction and cytoplasmic intermediate filament assembly, nuclear localization of FIGL-1, and in targeting sensory receptors in primary cilia (Broday et al. 2004; Leight et al. 2005; Roy Chowdhuri et al. 2006; Kaminsky et al. 2009; Li et al. 2012; Ferreira et al. 2013; Pferdehirt and Meyer 2013; Ward et al. 2013; Lim et al. 2014; Tsur et al. 2015). In the germline, the SUMO pathway was examined by the analysis of smo-1(ok359) null mutants and was reported not to be required for crossover formation, but led to SC disassembly defects (Bhalla et al. 2008). Depleting ubc-9 via RNAi (RNA interference) confers a radiation-sensitive phenotype in C. elegans (Boulton et al. 2004). UBC-9 has also been shown to physically interact with RAD-51, BRC-1, and BRD-1, a functional partner of BRC-1, through yeast two-hybrid and in vitro pull-down assays (Boulton et al. 2004). Despite these physical interactions with UBC-9, assays were unable to detect direct interactions between the SUMO moiety and RAD-51 or BRC-1 (Boulton et al. 2004).
In this paper, we demonstrate that SUMOylation is important for proper C. elegans germline function, with both premeiotic replication and meiosis negatively affected when SUMO-1 or UBC-9 function is abrogated. We find that DNA breaks are increased in mitotically dividing nuclei in the premeiotic tip (PMT) and fail to repair before transitioning into meiosis. Prophase I nuclei have meiotic progression defects and show an increase in recombination intermediates. Oocytes in SUMOylation mutants fail to properly form in both smo-1 and ubc-9 null mutants, resulting in sterility. Multiple germline defects in SUMO pathway mutants indicate potential roles for SUMO in the DNA damage repair (DDR) in response to replication fork collapse in the PMT and roles in meiotic HR.
Materials and Methods
Strains
All strains were cultured at 20° on plates containing OP50. N2 worms were used as the control strain for all experiments (Brenner 1974). All mutant alleles and transgenic lines used were outcrossed six times into the N2 background. Wild-type Hawaiian line CB4856 hermaphrodites were used for SNP recombination mapping and crossed to ubc-9(tm2610)/nT1 males (Swan et al. 2002). The nT1 balancer is a reciprocal translocation of chromosomes VI and V, and homozygous balancer progeny are lethal (Edgley et al. 2006). Mutant lines and transgenic lines used were: ubc-9(tm2610), spo-11(ok79), mre-11(iow1), mre-11(ok179), smo-1(ok359), fgpIs20 [(pAA64) pie-1p::mCherry::smo-1(GG) + unc-119(+), fgpIs21 [(pAA64) pie-1p::mCherry::smo-1(GA) + unc-119(+)], meIs8 [pie-1p::gfp::cosa-1 + unc-119(+)]II, ubc-9(iow31[3xflag::ubc-9])IV/nT1[qIs51] (IV;V), and mre-11(iow45[mre-11::gfp]).
Yeast two-hybrid
Two C. elegans mre-11 cDNA fragments were cloned into the pLexA gateway vector (plasmid #11345; Addgene) and pACT2.2. The C. elegans yeast two-hybrid library was generated by Guy Caldwell (University of Alabama) and obtained from Addgene (plasmid # 11523). The mre-11 C-terminal truncation contained the first 1263 bp of cDNA encoding the first 421 amino acids of MRE-11. The mre-11 N-terminal truncation contained 1021 bp of cDNA encoding the last 339 amino acids. Putative interactions were detected through screening for histidine reporter gene expression and further validated via X-gal (5-bromo-4-chloro-3-indolyl-beta-D-galacto-pyranoside) assays. Colonies with strong growth in the absence of histidine and blue color when incubated with X-gal were sequenced. For ubc-9/mre-11 interaction validation, a full-length mre-11 cDNA was cloned, and a targeted yeast two-hybrid assay was conducted between full-length and both N- and C-terminal truncations of MRE-11 coupled with full-length UBC-9 cloned into the pAct2.2 gateway vector (plasmid # 11346; Addgene). Interactions were detected through X-gal assay expression (Supplemental Material, Figure S1 in File S1).
Clustered regularly interspaced short palindromic repeats (CRISPR)/CAS9 generation of mre-11::gfp, 3Xflag::ubc-9 and rad-51::3Xflag lines
The gfp repair template was cloned into the pDD282 C-terminal gfp plasmid with a hygromycin cassette (Dickinson and Goldstein 2016). On either side of the break, > 500-bp homology arms were cloned via PCR. The single-guide RNA (sgRNA) sequence used for mre-11::gfp was 5′-GTTCTCGAAGTAGACTGTGG-AGG-3′. Silent mutations at the protospacer adjacent motif (PAM) site of the repair template were introduced to prevent cutting after gfp integration, per the standard protocol. The 3Xflag::ubc-9 repair template used was a 200-bp single-stranded oligodeoxynucleotides (ssODN) ordered from Integrated DNA Technologies. The ssODN sequence was: 5′-ccacttctcttttacaaatttgatatttttcagtgtaaccgaacaaaaATGgactacaaagaccatgacggtgattataaagatcatgaTatcgaTtacaaggatgacgatgacaagTCGGGAATTGCTGCAGGACGCCTCGCTGAGGAGAGAAAACACTGGCGAAAGgtgagaattttatcattacatggcaagtcggg-3′. Lowercase letters after the ATG indicate the 3Xflag sequence [uppercase letters indicate silent mutations made in the 3Xflag sequence to create EcoRV and ClaI sites, as referenced in Paix and Wang (2014)]. sgRNA was cloned into the pIK198 vector (Katic et al. 2015). pIK198 was generated by Iskra Katic (plasmid # 65629; Addgene). The sgRNA sequence used is 5′-CAGGACGCCTCGCGGAAGAA-AGG-3′. The PAM site sequences are provided after the dash for reference. 3Xflag insertion can be detected in a 2% agarose gel run at 105 V for 1.5 hr after PCR using primers F: 5′-CTGACAAGTGTCACGAACACG-3′ and R: 5′-CGGGAAATCGTCCTTGAAGAG-3′ at 55.3° for 40 sec. Wild-type size: 591- bp PCR product; flag insertion size: 655- bp PCR product. 3Xflag::ubc-9 worms are balanced with nT1 because of developmental problems resulting in larval lethality in homozygous offspring. 3Xflag::ubc-9 homozygotes or ubc-9(tm2610) /3Xflag::ubc-9 are viable and develop normal gonads (as determined by DAPI morphology, synapsis, bivalent formation, and RAD-51 kinetics), but 3Xflag::ubc-9 or ubc-9(tm2610) /3Xflag::ubc-9 produce no viable progeny, indicating that 3Xflag::ubc-9 creates a protein that is functional in the germline but not in early embryonic development. Since 3Xflag::ubc-9 still functions normally in the germline, anti-FLAG staining can be used to examine the UBC-9 pattern of localization. The rad-51::3Xflag line was created by using an ssODN repair template to insert the 3Xflag sequence just prior to the stop codon. The ssODN sequence is: 5′-CAATCACGAATCATGGTATTGAGGACGCACGCGAAGACgactacaaagaccatgacggtgattataaagatcatgaTatcgaTtacaaggatgacgatgacaagTAGccgttcgtttttctttttcttatcaaacttca-3′. ssODN was injected with a C-termina sgRNA, 5′-TGGTATTGAGGACGCACGCG-3′, and recombinant Cas9 protein (University of California, Berkeley), along with the dpy-10 co-injection marker. Injections were performed on ubc-9(tm2610)/nT1(GFP) hermaphrodites. GFP+ F1 offspring that were dumpy or rollers were singled and then insertions were screened via PCR after 3 days. Primers used for screening were F: 5′-GAAGCCGAAGCGACCTACTCA-3′ and R: 5′-CATGAGGGGCAGGCG-3′, used at 62° with an extension time of 20 sec. A 2% gel was used to separate bands, with wild-type = 225 bp and FLAG insertion = 300 bp. A heterozygote line was established and GFP− worms were screened to determine whether the FLAG tag was on the nonbalancer chromosome. Sequencing determined whether the tag was in frame.
Immunostaining and microscopy
Gonads were dissected and immunostained as described in Colaiacovo et al. (2003). Both whole worms and dissected gonads were prepared 20-hr post-L4. Antibodies used were as follows: rabbit anti-RAD-51 (1:10,000; ModEncode), preabsorbed mouse anti-SMO-1 (1:10 Developmental Studies Hybridoma Bank 6F2), mouse anti-FLAG (1:100; Sigma [Sigma Chemical], St. Louis, MO), rabbit anti-phospho histone 3 (PH3) (1:5000; Upstate Biotechnologies), and rabbit anti-SUN-1 (1:5000; Sdix). Secondary antibodies used were Alexa Fluor 488-anti-rabbit antibody (1:500) and Alexa Fluor 550-anti-mouse antibody (1:500). Whole worms were ethanol-fixed for COSA-1:GFP imaging and stained with DAPI [1:2000 dilution of a 5-mg/ml DAPI stock in PBS tween (PBST)] and mounted with Vectashield mounting medium. syp-3::gfp, mCherry::smo-1 lines and mre-11:gfp worms were dissected, frozen on dry ice blocks, fixed for 15 min in 4% paraformaldehyde in the dark, then washed with PBST for 5 min, stained with DAPI for 10 min, and washed in PBST for 10 min. Slides were mounted with Vectashield.
Images were taken with a DeltaVision wide-field fluorescence microscope system (Applied Precision) with an Olympus 100×/1.40-numerical aperture lenses. Optical sections were collected at 0.20-μm increments with a coolSNAPHQ camera (Photometrics) and softWoRx software (Applied Precision), and deconvolved using softWoRx 5.0.0 software. Images are projections through three-dimensional data stacks of whole nuclei (15–30 0.2-mm slices/stack).
Focus quantification
RAD-51 focus quantification was performed at four stages: PMT, the transition zone (TZ) that includes leptotene and zygotene, early pachytene (EP), and mid/late pachytene (MLP). Nuclear morphology (DAPI staining) and SYP-1 antibody staining were used to identify the four stages in the germline (Yin and Smolikove 2013). GFP::COSA-1 foci were counted in zone 7 in late pachytene, before diplotene.
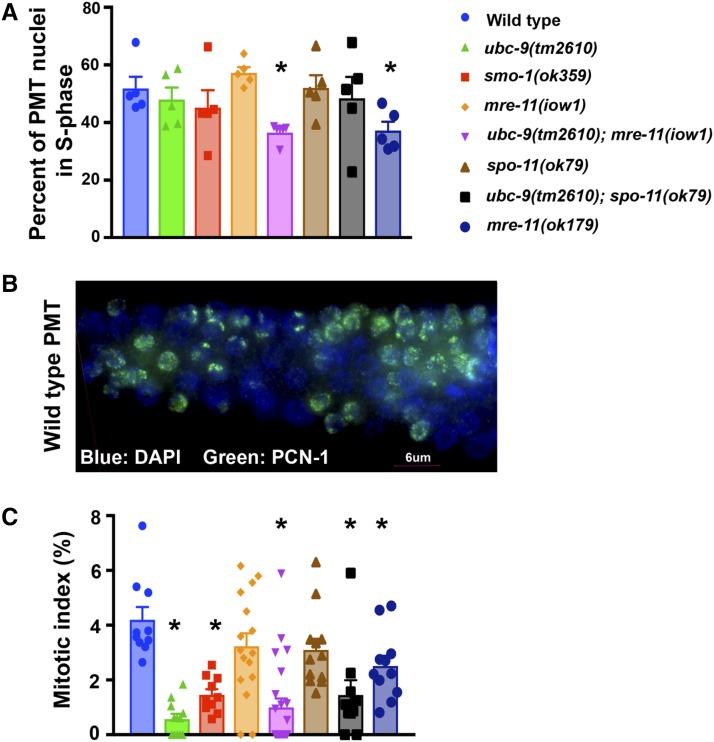
Mitotic index calculation
At least 10 gonads were analyzed per genotype to calculate the average mitotic index (MI). PH3 staining marks metaphase nuclei in the PMT. The ratio of stained nuclei/total PMT nuclei was calculated as the MI.
Length and width gonad measurements
Whole worms were ethanol-fixed and stained with DAPI, then mounted with Vectashield. Images were taken at 10× magnification. Gonad length was measured in ImageJ. Length measurement was defined as the distance from the beginning of the PMT to the beginning of diplotene. Width measurements were taken at two points: the 50% mark in the PMT and the 50% mark from the TZ to the end of pachytene. The genotypes measured were not significantly different in overall body size.
EMO phenotype measurements
Whole worms were fixed with ethanol and stained with DAPI. Worms were mounted with Vectashield. A minimum of 10 gonads was analyzed for each genotype. Endomitotic oocyte (EMO) was defined as prematurely replicating nuclei that did not display bivalents but instead produced a single decondensed mass of DNA (Iwasaki et al. 1996). The number of endomitoic (Cremona et al. 2012) diakinesis oocytes were counted and divided by the total number of diakinesis oocytes in the germline.
UBC-9/SUN-1 colocalization measurements
Fifteen nuclei were measured in total from three different gonads that were stained for FLAG:UBC-9 and SUN-1 and DAPI. Two intensity peaks were measured per nucleus for each protein for 15 nuclei measured. Images were analyzed in FIJI using the Plot profile function. Z-stacks were used to determine the approximate halfway point through the nucleus, and then a line was drawn through both walls of the nuclear envelope. The plot profile data was then downloaded into Excel (Microsoft, Redmond, WA) for all three channels, and intensity peaks were determined for SUN-1 and UBC-9. Positive or negative values were determined based on whether the UBC-9 peak was inside or outside of the SUN-1 peak relative to the center of the nucleus. If UBC-9’s peak signal was outside of the SUN-1 peak relative to the center of the nucleus, the measurement was negative. If UBC-9’s peak fell inside of the SUN-1 peak relative to the center of the nucleus, then the measurement was positive.
Camptothecin/hydroxyurea adult assay
aaSeeded OP50 worm plates were exposed to UV light for 1 hr (placed on a UV illuminator from Fisher) to kill the bacteria so to prevent the metabolism of the camptothecin (CPT) or hydroxyurea (HU). Then, a final concentration of either 5.25 mM HU diluted in M9 or 350 nM CPT (10 mM stock CPT diluted in DMSO, final concentration diluted in M9 pH6) was plated and allowed to soak in overnight in the dark. One day post-L4 worms of each genotype were placed on the treated plates for up to 8 hr in 2 hr increments in the dark. Control worms were dissected immediately without being exposed to any chemicals. Worms were promptly dissected according to normal protocols and stained with RAD-51 antibody (1:10,000). Three to nine gonads were counted for each genotype and treatment.
Larval lethality assay
A single heterozygote L4 worm of the nT1 balancer was added per plate with a small lawn of spread OP50 (1 cm2) that was grown overnight at room temperature. The worm was transferred to a new plate every 24 hr for 4 days. Eggs were counted after 24 hr, and larvae and adults were counted for a total of 6 days after egg lay to account for delayed development. Both GFP+ and GFP− worms were counted under a GFP dissection scope to collect data from heterozygote (GFP+) and homozygote (GFP−) mutant offspring.
Recombination frequency mapping
ubc-9(tm2610)/nT1 males in the N2 background were crossed to CB4856 hermaphrodites to create ubc-9(tm2610)/+ heterozygotes with N2/CB4856 SNP distribution. Twelve GFP− F1 L4 hermaphrodites were moved to individual plates and let lay for 3 days. Eight F2 offspring were lysed from each of the 12 F1’s for a total of 96 offspring per cross that were analyzed. The F1 worms were also lysed and analyzed for SNP heterozygosity. Two SNPs were analyzed on chromosome II at location −18 and 11 using Dra1 digestion. At location −18, N2: 263 + 112 bp PCR product; CB4856: 375 bp PCR product. At location 11, N2: 483 bp PCR product; CB4856: 352 + 132 bp PCR product. N2 and CB4856 lines were used as controls to ensure complete DraI (New England Biolabs, Beverly, MA) digestion. The formula used to calculate the recombination frequency is p = 1−(1−R)1/2. R = [(number of animals heterozygous for one marker and homozygous for the other) + 2 × (number of animals homozygous for recombinant chromosomes)]/total number of animals scored (Brenner 1974; Bazan and Hillers 2011).
Data availability
Strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
MRE-11 and UBC-9 physically interact in a yeast two-hybrid assay
To better understand how MRE-11 is regulated in meiosis, we aimed to identify proteins physically interacting with C. elegans MRE-11. We conducted a yeast two-hybrid screen using C-terminal and N-terminal fragments of MRE-11 as the bait. UBC-9 was identified as an interacting protein; the yeast two-hybrid interaction was also found with full-length MRE-11 (Figure S1 in File S1). ubc-9 encodes the SUMO E2 conjugating enzyme.
UBC-9 and SMO-1 localize to the nuclear periphery in the C. elegans germline
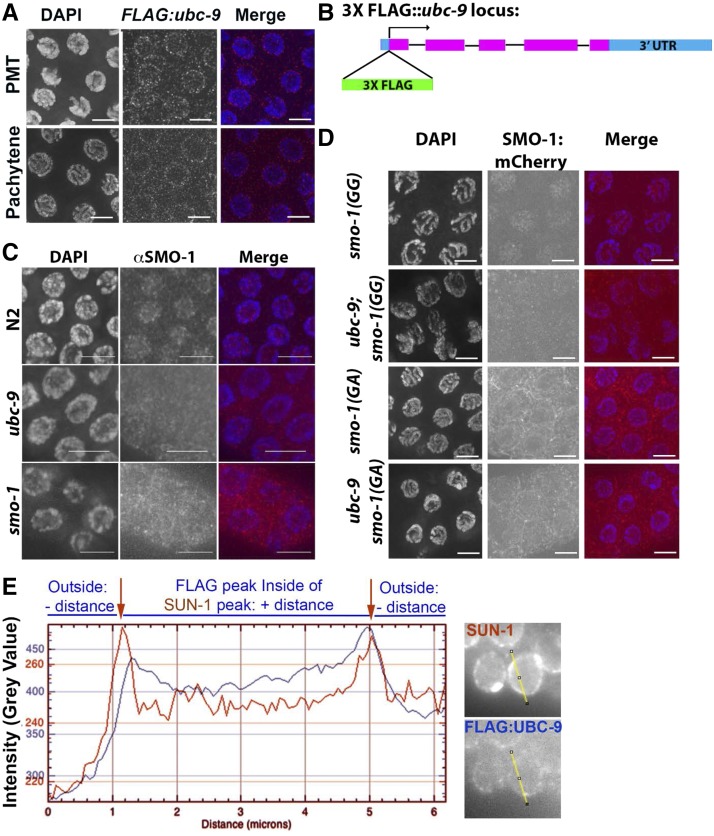
MRE-11 is a protein required for meiotic recombination in C. elegans, as found in other organisms (Chin and Villeneuve 2001; Goodyer et al. 2008). If UBC-9 interacts with MRE-11 in vivo, they should both be expressed in the same tissue. Previous studies have indicated that both SUMO-1 and UBC-9 are expressed in the germline, but it was not clear in which stage of meiotic prophase I they were expressed (Pelisch et al. 2014,,2017). We created a 3Xflag-tagged ubc-9 line via CRISPR/Cas9 homology recombination repair (Figure 1, A and B). The 3Xflag tag was inserted at the N-terminus of ubc-9 directly following the ATG start codon. flag::ubc-9 worms have normal gonad morphology and RAD-51 staining, but do not produce viable progeny, indicating that the FLAG::UBC-9 protein is dysfunctional only for processes required for development after oocyte formation (Materials and Methods and Figure S2A in File S1). Anti-FLAG immunostaining of 3Xflag::ubc-9 homozygotes shows that UBC-9 localizes to nuclei in the PMT through late pachytene in a punctate pattern with an enrichment near the nuclear membrane (Figure 1A). FLAG staining was not observed in wild-type worms that do not contain the flag tag (Figure S2B in File S1). UBC-9 was absent from diplotene and diakinesis nuclei (data not shown).
Figure 1.
UBC-9 and SMO-1 localize to the nucleus in the C. elegans germline. (A) 3XFLAG::UBC-9 localizes to the nuclear periphery throughout the germline in the wild-type (WT) strain. Premeiotic tip (PMT) and mid/late pachytene are shown. DAPI is blue, 3XFLAG::UBC-9 is red. (B) Schematic of 3Xflag insertion in the N-terminus of the endogenous ubc-9 locus. The tag is inserted directly after the start codon. (C) Endogenous SMO-1 localization in WT, ubc-9(tm2610), and smo-1(ok359) strains. Nuclear localization is present in WT, but in ubc-9 mutants staining is diffuse with no nuclear preference. smo-1 mutants have low levels of background staining with no nuclear enrichment. DAPI is blue, SMO-1 is red. (D) smo-1::mCherry lines with a WT sequence (GG), or a nonconjugatable point mutation (GA), were expressed in WT and ubc-9 backgrounds. Nuclear mCherry expression is visible in the WT smo-1(GG) strain, but not when ubc-9 is mutated. smo-1(GA) strains have diffuse cytosolic staining in both WT and ubc-9 mutants. All images are midpachytene stage. (E) Graph of signal intensity peaks. Representative lines used for analysis taken 50% through the nucleus for SUN-1 and 3XFLAG::UBC-9 are shown.
To observe where in the C. elegans gonad the SUMO protein, SMO-1, localizes, we used an antibody for SMO-1 and found a diffuse nuclear localization pattern (Figure 1C and Figure S2C in File S1). To test for specificity of staining we analyzed two mutants: ubc-9(tm2610), a 315- bp deletion mutant that encompasses the region encoding the SUMO-binding site in UBC-9, and smo-1(ok359), a complete deletion of the only C. elegans SUMO gene. The smo-1 gene is essential, but smo-1(ok359) homozygous progeny produced from balanced hermaphrodites are viable and reach adulthood likely due to maternally provided SMO-1 (Broday et al. 2004). SMO-1 nuclear staining was specific, as both smo-1(ok359) and ubc-9(tm2610) null mutants lacked nuclear staining. We also utilized two transgenic lines containing an mCherry::SUMO-1 fusion transgene with a wild-type endogenous smo-1 gene generated by bombardment (Pelisch et al. 2014). One transgenic line contained a wild-type smo-1 gene [SMO-1(GG)], whereas the other had a point mutation allele for the last amino acid, rendering the SMO-1 protein unconjugatable [SMO-1(GA)]. SMO-1(GG) showed nuclear localization using the anti-SUMO antibody, similar to what we observed with the antibody staining for wild-type strains (Figure 1D). The mutant form SMO-1(GA) showed cytoplasmic, but not nuclear, localization in a filament-like pattern. (Figure 1D). In a ubc-9(tm2610) mutant background, neither the wild-type [SMO-1(GG)] nor the unconjugatable form [SMO-1(GA)] localized to the nucleus (Figure 1D). Only if the SUMO E2 enzyme and conjugatable SUMO are present can SUMO be found in the nucleus. We therefore presume that SUMO and UBC-9 modify a germline nuclear target(s).
The localization pattern of 3XFLAG::UBC-9 is enriched at the nuclear envelope, but anti-FLAG staining alone did not identify whether UBC-9 was inside or outside of the nuclear envelope. We costained with a SUN-1 antibody (that marks the inner nuclear membrane) and a FLAG antibody to identify whether UBC-9 localizes close to the nuclear membrane (Figure 1D and Figure S2D in File S1) (Tzur et al. 2006; Penkner et al. 2007; Minn et al. 2009). Figure 1D and Figure S2D in File S1 indicate that the positions of UBC-9 and SUN-1 are not significantly different, confirming that UBC-9 is associated with the nuclear membrane. Our data suggest that UBC-9 may in fact be located on the inner membrane (Figure 1D).
Gonads in SUMOylation mutants are smaller than wild-type
Both smo-1(ok359) and ubc-9(tm2610) mutant worms are completely sterile and do not lay eggs. Their protruding vulva phenotype could potentially explain the lack of egg laying (Horvitz and Sternberg 1991; Greenwald 1997; Broday et al. 2004). However, defects in germline development could also contribute to the sterility observed. In the nematode C. elegans germline, prophase I occurs along the germline in spatiotemporal organization; the syncytial germline nuclei are generated at the distal tip and then flow toward the proximal end of the gonad as nuclei progress through meiosis. Examination of ubc-9(tm2610) and smo-1(ok359) mutant germlines via DAPI staining revealed that these mutants contain a germline with most stages of meiosis with several defects (discussed in detail below). Gonads of ubc-9(tm2610) and smo-1(ok359) homozygotes were 33.1 and 36.4% shorter in length and 33.4 and 59.6% smaller in width at midpachytene, respectively, compared to wild-type (Figure S3 in File S1 and Tables S1 and S2 in File S3).
DSBs are increased in SUMOylation pathway mutants and atypical RAD-51 foci are observed
The SC assembled without any observable defects in both ubc-9(tm2610) and smo-1(ok359) mutants. This is in agreement with a previous study describing no SC assembly defect in smo-1(ok359) mutants (Bhalla et al. 2008) and with the lack of SC-specific localization of UBC-9 and SMO-1 in C. elegans (this study). Wild-type nuclear SMO-1 localization, coupled with reduced germline size without SC phenotypes in SUMOylation mutants, suggests that the SUMO pathway in C. elegans may be involved in other aspects of germline function such as DNA repair in the PMT and/or meiotic recombination. A role in these pathways is suggested by the physical interaction found between UBC-9 and MRE-11 in the yeast two-hybrid assay.
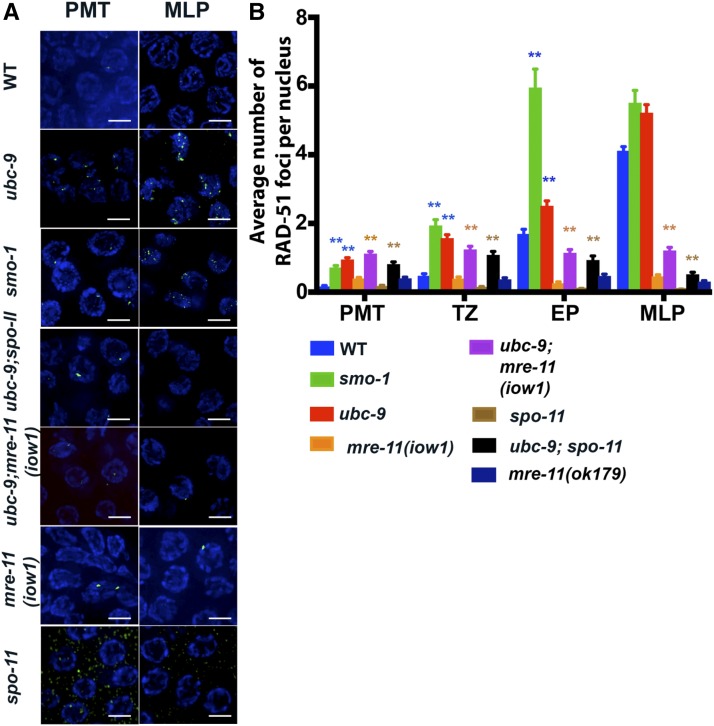
C. elegans lacks a DMC1 homolog; therefore RAD-51 is the only recombinase present in meiosis (Takanami et al. 1998). We dissected gonads to stain for RAD-51, which measures the number of ssDNA recombination intermediates to which RAD-51 can bind (Figure 2,Figure S4 in File S1, and Tables S3 and S4 in File S3). RAD-51 is essential for HR (Shinohara et al. 1992). The presence of wild-type SC in ubc-9(tm2610) and smo-1(ok359) mutants allows the precise staging of meiotic nuclei based on SC morphology (see below) (Yin et al. 2016). SYP-1 antibody was used to place germline nuclei into four stages: the PMT that lacks SYP-1 staining, the TZ that has SYP-1 puncta, EP that contain regions of linear SYP-1 that are not full-length, and MLP that contains fully linearized SYP-1 along the entire chromosome. RAD-51 foci were increased in the PMT in both ubc-9(tm2610) and smo-1(ok359) mutants (0.16 ± 0.02 foci/nucleus in wild-type to 0.95 ± 0.05 and 0.71 ± 0.06 foci on average, respectively, Figure 2). This indicates that SUMOylation plays a role in the PMT and that a lack of SUMO leads to increased ssDNA loaded with RAD-51. RAD-51 foci in the PMT could be generated by collapsed replication forks. A similar increase is also observed in TZ (wild-type 0.48 ± 0.06, ubc-9(tm2610) 1.57 ± 0.1, and smo-1(ok359) 1.94 ± 0.17 foci/nucleus) and EP nuclei (wild-type 1.7 ± 0.13, ubc-9(tm2610) 2.51 ± 0.14, and smo-1(ok359) 5.95 ± 0.54 foci/nucleus). Foci also are seen in both diplotene and diakinesis in the SUMOylation mutants, but are absent from these stages in wild-type worms.
Figure 2.
RAD-51 foci numbers are increased in SUMOylation mutants. (A) Representative images of RAD-51 foci from the premeiotic tip (PMT) and mid/late pachytene (MLP) of wild-type (WT) and mutants. RAD-51 is green, DAPI is blue. (B) RAD-51 foci data count each type of aberrant RAD-51 focus (singlet, doublet, triplet, quadruplet, and string) as one focus. The colors of significance stars are what each mutant is compared to [i.e., ubc-9(tm2610) has blue stars above it because it is being compared to WT (blue bar)]. Full pairwise comparison charts with significance are found in the supplements. Error bars signify the mean ± SEM. Three full gonads were counted per genotype except ubc-9(tm2610) single mutants where five full gonads were counted. n-values: PMT range from 236 to 667 nuclei counted per genotype, transition zone (TZ) range from 129 to 268 nuclei counted, early pachytene (EP) range from 69 to 271 nuclei counted, and MLP range from 199 to 660 nuclei counted.
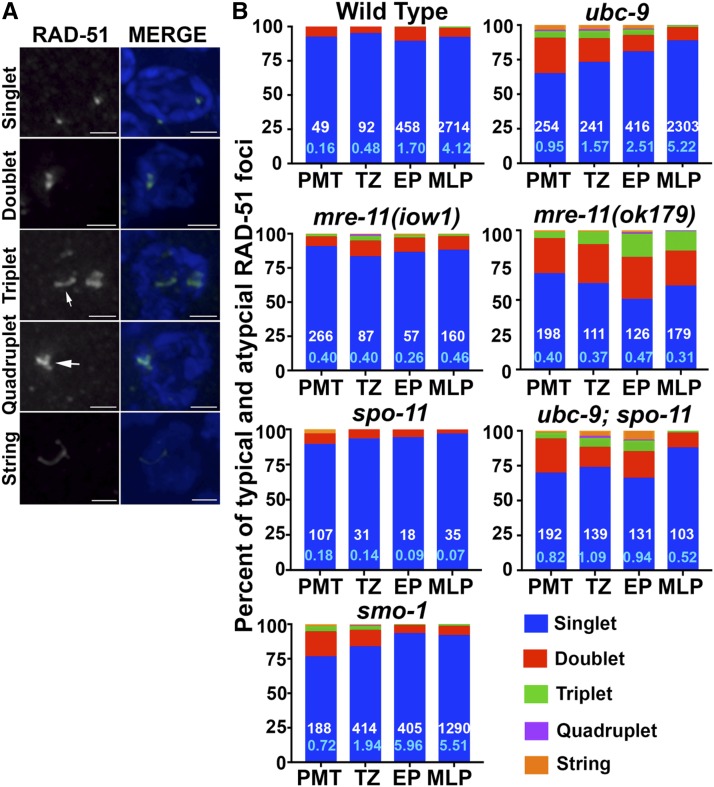
In addition to the increase in RAD-51 foci, atypical RAD-51 forms were observed in SUMOylation mutants. Wild-type RAD-51 foci primarily appear as single round dots, but we observed atypical RAD-51 foci that formed detectable doublets, triplets, quadruplets, and long string-like filaments in the two SUMOylation mutants (Figure 3 and Table S5 in File S3). Doublets, triplets, and quadruplets were categorized as two, three, or four foci, respectively that are clearly discernable but in contact with each other and have no space between them. We also observed strings: RAD-51 structures that are smooth, long structures that resemble filaments. Wild-type gonads had 8.1% doublets in the PMT and 0–10.2% atypical foci throughout meiotic nuclei (Figure 3B). Although both ubc-9(tm2610) and smo-1(ok359) mutants exhibit atypical RAD-51 structures in addition to normal foci, there are differences between the frequencies found in the two strains. In the PMT, 25.6% of foci are doublets in ubc-9(tm2610) mutants, whereas 18.1% of foci are doublets in smo-1(ok359) mutants. For triplets, quadruplets, and strings, both ubc-9(tm2610) and smo-1(ok359) mutants have roughly twice as many atypical forms as wild-type in almost all meiotic stages (Figure 3B and Figure S5 in File S1). The data in Figure 2 was generated for each form counted as 1 (i.e., doublet, triplet, or quadruplet foci, or strings, were all counted as one focus). This is the standard technique for analyzing RAD-51 foci in the C. elegans germline (Yin and Smolikove 2013). We wondered whether counting of atypical foci as several foci would affect the outcome. We performed an alternative count of the same data counting each discernible part of an aberrant focus as one (Figure S4 in File S1). The conclusions from this method of counting were consistent with the ones drawn from Figure 2; ubc-9 and smo-1 mutants have increased levels of RAD-51 foci (see Discussion).
Figure 3.
The fraction of alternate RAD-51 foci is increased in SUMOylation mutants and mre-11 null mutants. (A) Representative images for each type of RAD-51 focus. The singlet picture is from a wild-type germline. Doublets, triplets, and quadruplet images were taken from ubc-9(tm2610) germlines and string image was taken from a smo-1(ok359) germline. Arrows point to the focus of interest. All images were taken from the premeiotic tip (PMT). (B) All graphs are percentage of total foci for the five RAD-51 categories. PMT, TZ (transition zone), EP (early pachytene), and MLP (mid/late pachytene). White numbers at the bottom of the blue bars indicate the total number of foci scored (both typical and atypical) included in that stage, while the light blue number is the average number of foci per stage and genotype.
RAD-51 strings could be due to elevated DSBs in SUMOylation mutant backgrounds. To test this, we irradiated ubc-9(tm2610) and wild-type worms to create increased DSBs in the germline. There was no increase in the percentage of total nuclei with string-like structures, in either the wild-type or ubc-9(tm2610) mutant, compared to nonirradiated worms (Figure S5 in File S1 and Table S6 in File S3).
mre-11 mutants exhibit differences in aberrant RAD-51 focus patterns
Null mre-11(ok179) mutants have fewer RAD-51 foci in meiotic nuclei in all stages because they cannot make DSBs to initiate recombination. However, they do have a small increase in RAD-51 foci numbers in the PMT compared to wild-type (0.40 ± 0.03 compared to 0.17 ± 0.02 P < 0.0001); the latter is capable of forming DSBs but cannot process them, leading to low levels of RAD-51 foci throughout meiosis (Figure 2B). Furthermore, mre-11(ok179) mutants have a high percentage (25–30%) of RAD-51 doublets in all four germline stages, compared to doublets in both wild-type (5.4–10.3%) and mre-11(iow1) (7.1–11.5%) (Figure 3 and Table S5 in File S3).
In SUMOylation mutants, repair of recombination intermediates generated in mitosis is not completed before meiotic entry
Because both SUMOylation mutants exhibited an increase in RAD-51 foci in the PMT, we asked whether PMT foci were being carried over into meiosis. SPO-11 is the meiosis-specific protein required for the formation of meiotic DSBs. The numbers of RAD-51 foci in the PMT in ubc-9(tm2610);spo-11(ok79) double mutant was comparable to that of ubc-9(tm2610) mutants (Figure 2B). Similar numbers of RAD-51 were observed in meiotic zones (lower than found in ubc-9 mutants and higher than in spo-11(ok79) single mutants). This data indicated that the elevated levels of RAD-51 foci in ubc-9 mutants are caused by unrepaired DSBs carried over from mitosis to meiotic prophase.
Due to the increase in nuclear RAD-51 foci in SUMOylation mutants, we tested for a global increase in RAD-51 protein in ubc-9(tm2610) mutants compared to wild-type strains by performing a western blot for FLAG-tagged RAD-51 (Figure S6B in File S2). We generated a rad-51::flag line in the ubc-9(tm2610)/nT1 background. Anti-FLAG staining showed RAD-51::FLAG foci that colocalized with antibody for RAD-51 (Figure S6B in File S2). Balanced and homozygous ubc-9(tm2610) strains had no detectable differences in the levels of RAD-51::FLAG protein, indicating that a loss of SUMOylation does not affect RAD-51 protein levels.
A synthetic sick interaction between ubc-9 and mre-11 null mutations
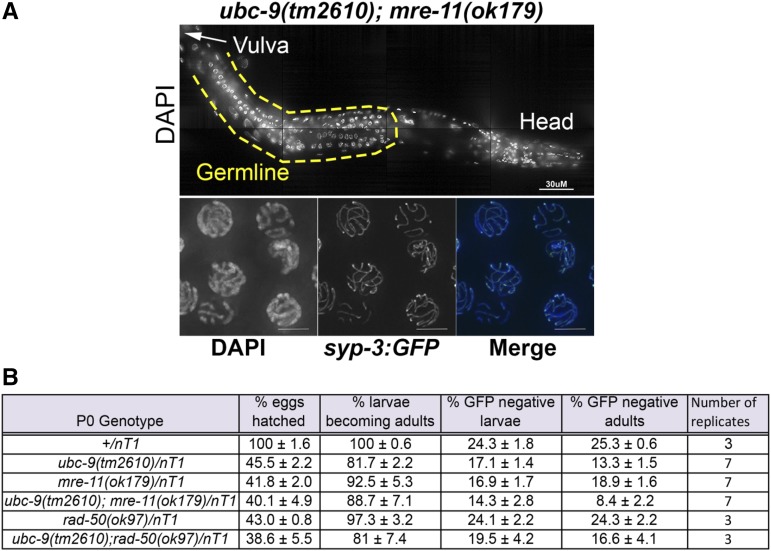
In C. elegans meiosis, MRE-11 is required both for DSB formation and for subsequent resection; both of these activities are absent in the null mre-11(ok179) mutant. Deleting mre-11 in the ubc-9(tm2610) background caused developmental and germline defects that were more severe than the ones observed in ubc-9(tm2610) single mutants (Figure 4 and Table S7 in File S3). nT1 is a balancer translocation chromosome that covers both ubc-9 and mre-11 on chromosomes IV and V, respectively (Edgley et al. 2006). nT1 is lethal when homozygous. A +/nT1 strain produced 24.3% homozygous wild-type larvae. A ubc-9/nT1 strain produced less (17.3%) homozygous ubc-9 progeny (P = 0.0012, Fisher’s exact test). Homozygous ubc-9(tm2610) progeny were also less likely to reach adulthood (14.4% in ubc-9(tm2610), compared to 25.3% in wild-type, P < 0.0001). Progeny survival in the mre-11(ok179)/nT1 strain was also reduced compared to wild-type (18.9% reach adulthood compared to 25.3% in wild-type, P = 0.0053). The double ubc-9(tm2610);mre-11(ok179) mutant progeny have further reduced larval and adult survival compared to wild-type (14.3% larval survival vs. 24.3% for wild-type; 8.4% adult survival vs. 25.3% in wild-type, P < 0.0001 for both). In addition, the double mutant had reduced survival to adulthood compared to each single mutant (8.4% vs. 14.4% in ubc-9 and 18.9% in mre-11(ok179), P = 0.02 and P < 0.0001, respectively) (Figure 4 and Table S6 in File S3).
Figure 4.
ubc-9(tm2610); mre-11(ok179) double mutants have underdeveloped germlines and have slowed larval development and survival. (A) DAPI-stained half worm of ubc-9(tm2610); mre-11(ok179) double mutant. The middle of the worm is oriented to the left (arrow labeled “vulva” points to its location). The germline is outlined in a yellow dashed line. syp-3::gfp was crossed into the double-mutant background and whole worms were fixed. SYP-3::GFP linearization was observed in nuclei toward the end of the germline indicating that pairing occurs in these mutants. (B) Larval lethality of broods laid from balanced hermaphrodites of various genotypes. All genotypes are balanced over nT1(gfp). Between three and seven replicates of each genotype were counted. Pairwise comparisons (Fisher’s exact t-test) are in the supplement. Percentages of GFP+/− larvae and GFP+/− adults were calculated to determine if there is increased lethality among the GFP− (homozygous) offspring. Mean ± SEM is given for all percentages.
We observed developmental effects in the ubc-9(tm2610); mre-11(ok179) double mutants that were not apparent in the single mutants. Double-mutant worms that develop into adults take longer to reach adulthood (5–6 days after egg lay compared to 4 days in either single mutant). We observed that 100% of the double mutants that survived to adulthood were uncoordinated in their movement; this compares with 0% of ubc-9(tm2610) and mre-11(ok179) single mutants.
The ubc-9(tm2610); mre-11(ok179) double-mutant gonads did not develop to normal wild-type volume and contained nuclei with abnormal DNA morphology (Figure 4A). Diakinesis was completely absent in this germline or there were up to two misshapen oocytes after late pachytene. We were not able to dissect ubc-9; mre-11(ok179) gonads due to their small size, precluding staining for RAD-51. To test if the double-mutant nuclei were still capable of entering meiosis, the assembly of the SC was used as a marker. We created a syp-3::gfp; ubc-9(tm2610); mre-11(ok179) strain to visualize synapsis in whole worms without dissection. Despite abnormal DNA morphology, SYP-3 localized to chromosomes in a linear manner in “pachytene-like” nuclei (Figure 4A). This indicates that the double-mutant germline contains meiotic nuclei. Taken together, these data indicate that progression through meiosis in ubc-9(tm2610); mre-11(ok179) gonads is severely affected, although meiotic entry does occur.
rad-50 or mre-11(iow1) mutations do not lead to synthetic sickness with ubc-9
In meiotic recombination, MRX/N has multiple roles, including structural roles such as tethering broken DNA and catalytic roles (i.e., making DSBs and processing them to remove SPO-11 and create ssDNA) (Johzuka and Ogawa 1995; Nairz and Klein 1997; Moreau et al. 1999; Paull and Gellert 1999; van der Linden et al. 2009; Deshpande et al. 2014, 2016; Rojowska et al. 2014). One possibility for the deleterious phenotype observed in the germline in ubc-9(tm2610); mre-11(ok179) double mutants is due to the complete loss of all MRE-11 roles. Alternatively, it could be due to a specific MRE-11 function. To test the role of meiotic DSB formation in the synthetic sick germline phenotype, we created a double mutant in which only the resection activity of MRE-11 is compromised in the ubc-9 null background. ubc-9(tm2610); mre-11(iow1) double mutants formed germlines comparable in size to ubc-9(tm2610) and exhibited a RAD-51 localization pattern similar to that observed in the ubc-9(tm2610);spo-11(ok79) strain (Figure 2 and Figure S4 in File S1). This data indicates that the synthetic lethal interaction between ubc-9 and mre-11 is dependent on MRE-11’s function outside of its catalytic activity.
To determine if the synthetic sick interaction was specific to ubc-9 and mre-11, or if any member of the MRX/N complex would produce the same result, we created a ubc-9(tm2610); rad-50(ok97) double mutant and assessed gonad morphology and larval lethality in this mutant as was done in Figure 4B. rad-50(ok97) is a null mutation. There was no difference in hatch or growth rate in ubc-9; rad-50 double mutants compared to ubc-9 mutants (Figure 4B and Figure S6 in File S2). Germline morphology of the ubc-9; rad-50 double mutants was indistinguishable from that of ubc-9 single mutants (Figure 4 and data not shown). These data indicate that the sickness found in ubc-9(tm2610); mre-11(ok179) double mutants is specifically due to the absence of MRE-11 rather than the complete MRX/N complex (see Discussion).
UBC-9 localization is MRE-11-independent
The MRX/N complex is known to localize to DNA since it acts directly at sites of DNA damage (Desai-Mehta et al. 2001; Robison et al. 2004). If MRE-11 and UBC-9 interact in vivo, the nuclear localization of one may be dependent on the other. We created a mre-11::gfp strain through CRISPR/Cas9 genome editing to visualize MRE-11 localization in a ubc-9(tm2610) mutant background. mre-11::gfp worms are a viable and fertile strain, indicating that insertion of gfp likely does not perturb any function of MRE-11. In wild-type strains, MRE-11::GFP localizes to meiotic nuclei in all stages of meiosis through the end of pachytene (Figure S7 in File S2), similarly to what is found in immunolocalization studies (Goodyer et al. 2008). We did not see a difference in the MRE-11 localization pattern between wild-type and ubc-9(tm2610) strains (Figure S7 in File S2). We then tested whether FLAG::UBC-9 localization was affected in mre-11(ok179) mutants. FLAG::UBC-9 was still able to localize in pachytene nuclei in the absence of mre-11 (Figure S8 in File S2). The intensity of FLAG::UBC-9 in PMT nuclei was measured and no significant change was detected. We conclude that UBC-9 and MRE-11 localize to meiotic nuclei independently of each other.
Mitotic proliferation is decreased in SUMOylation mutants
The increase in RAD-51 foci in the PMT in SUMOylation mutants suggests that the repair of DNA damage generated during replication (such as collapsed replication forks) is SUMOylation-dependent. DNA damage can trigger cell cycle arrest leading to reduced mitotic proliferation, which could explain the reduction in germline size (Gartner et al. 2000). To test this hypothesis, we examined the percentage of M- and S-phase nuclei in the PMT in our mutants.
To determine how S-phase is affected in SUMOylation mutants, we stained dissected gonads with an antibody to PCN-1, C. elegans PCNA (Kim and Michael 2008). PCNA is essential for replication and a good marker for S-phase nuclei (Celis et al. 1987; Prelich et al. 1987). We calculated the percentage of PCN-1-positive cells in the PMT in all genetic backgrounds examined in this paper (Figure 5A and Table S8 in File S3). Both the mre-11(ok179) strain and the ubc-9;mre-11(iow1) double-mutant strain had fewer S-phase nuclei compared to the wild-type strain (36.8 and 36.9% compared to 50.5%; P < 0.0001 for both comparisons, n = 5 PMTs/genotype). No difference in the percentage of S-phase nuclei was found for the other genotypes (Figure 5A and Table S8 in File S3).
Figure 5.
SUMOylation mutants have wild-type levels of PCN-1-positive nuclei but exhibit a decrease in M-phase nuclei. (A) Germlines of various genotypes were stained with an antibody to PCN-1 and the number of PCN-1+ nuclei counted; the number was divided by the total number of nuclei in the premeiotic tip (PMT). Five PMTs of each genotype were counted. (B) Example of PCN-1 staining in a wild-type PMT. The distal tip of the gonad is oriented to the left. PCN-1 is green, DAPI is blue. (C) Germlines were stained for phospho-histone 3 (PH3). The mitotic index was calculated as PH3+ PMT nuclei/total PMT nuclei. At least 10 PMTs were counted per genotype. Error bars signify the mean ± SEM. Pairwise comparisons can be found in the supplement (Fisher’s exact test).
PH3 in worms marks metaphase nuclei (Hendzel et al. 1997). The percentage of PH3-positive nuclei is defined as the MI (MI = PH3+ PMT nuclei/total PMT nuclei × 100), which correlates with the percentage of mitotically dividing germline nuclei (Crittenden and Kimble 2008). Wild-type worms had an MI of 4%, which was significantly higher than that of both SUMOylation mutants (Figure 5B and Table S9 in File S3; 1.66%, P = 0.0003 for ubc-9(tm2610), and 0.57%, P < 0.0001 for smo-1(ok359) mutants). The total number of nuclei present in the PMT of SUMOylation mutants was also reduced compared to wild-type (average: wild-type 205.8, ubc-9(tm2610) 144.5, and smo-1(ok359) 121).
mre-11 and ubc-9 mutants are delayed in RAD-51 loading in response to replication stress
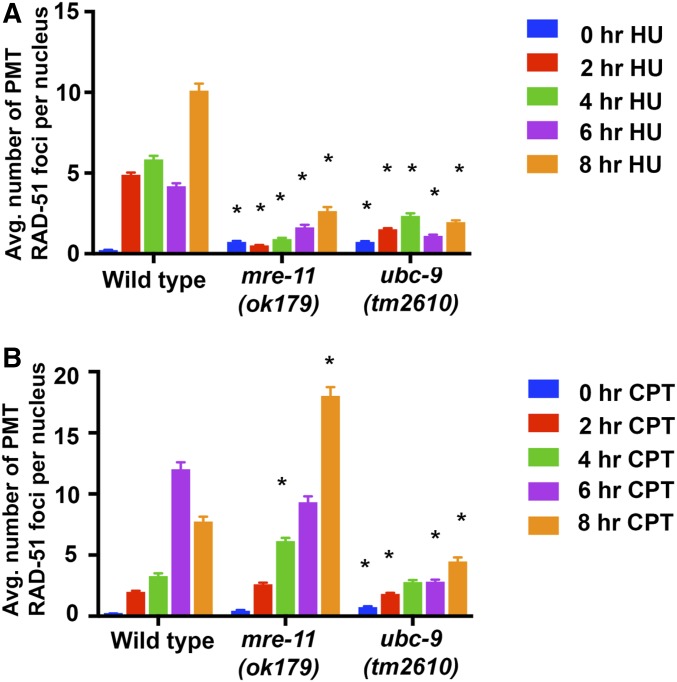
SUMOlyation mutants had reduced PMT nuclei in S-phase and an increased number of RAD-51 foci. Both of these results are consistent with the presence of increased damage in the PMT. We introduced replication stress in wild-type, ubc-9(tm2610), and mre-11(ok179) mutants by using the chemicals HU and CPT. HU depletes dNTP pools and causes polymerase to stall, while CPT inhibits topoisomerase I (TOP-1) (Fox 1985; Porter and Champoux 1989; Gedik and Collins 1990) (Figure 6). These chemicals test the reaction of ubc-9 and mre-11 null mutants to replication stresses (Kim and Colaiacovo 2014, 2015). Since neither ubc-9 nor mre-11 null mutants produce viable offspring, it is not possible to test the effects of these chemicals on progeny numbers. Instead, we analyzed the effect of HU and CTP on RAD-51 foci in the gonad following an 8-hr exposure. As expected, wild-type worms had increased RAD-51 foci in response to both drugs compared to controls. The mre-11(ok179) mutant exposed to HU did not show a significant increase in PMT RAD-51 foci compared to wild-type nuclei and only began to increase in RAD-51 foci at the 6-hr time-point (Figure 6A). CPT response was comparable to wild-type in the mre-11(ok179) background (Figure 6B). ubc-9(tm2610) mutants exposed to HU showed minimal increase over 8 hr, while exposure to CTP led to a smaller increase compared to nonexposed nuclei, but still significantly less than wild-type at 8 hr (Figure 6). This data indicates that both MRE-11 and UBC-9 are required for the formation of the ssDNA-RAD-51 filament following replication stress at the time intervals examined.
Figure 6.
ubc-9(tm2610) and mre-11(ok179) mutants show defects in average (Avg.) RAD-51 recruitment following HU (hydroxyurea) and CPT (camptothecin) exposure in premeiotic tip (PMT) nuclei. (A) Graph of RAD-51 foci counts after 5.25 mM HU exposure over an 8-hr period. Controls were not exposed to any chemical. At least three PMTs were counted per genotype (826–1626 total PMT nuclei counted per treatment). Stars indicate P < 0.05 compared to wild-type at the equivalent time-point (Mann–Whitney U test). Other pairwise comparisons can be found in supplemental tables. (B) Graph of RAD-51 foci counts after 350 nM CPT exposure over an 8-hr period. Three to eight PMTs were counted in each of the time-points and for each genotype (668–1719 total PMT nuclei counted per treatment). Error bars signify the mean ± SEM. Stars indicate P < 0.05 compared to wild-type at the equivalent time-point (Mann–Whitney U test).
Interfering crossovers are not affected in ubc-9 mutants, however noninterfering crossover pathways may be increased
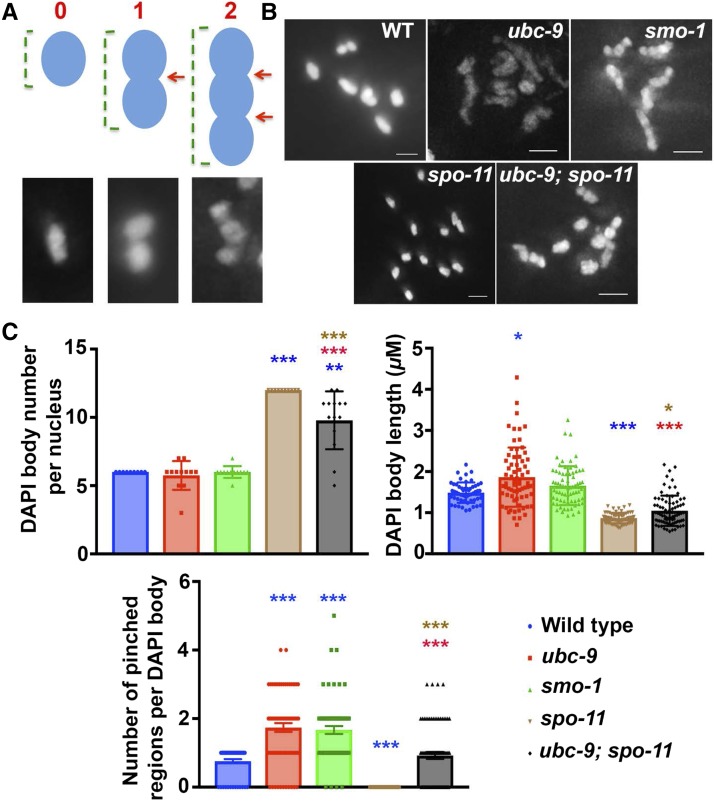
C. elegans is an organism in which crossover interference is complete; HR ultimately results in a single interfering crossover per chromosome pair, for a total of six crossovers per nucleus (Goldstein and Slaton 1982). DAPI bodies observed in ubc-9 mutants look aberrant, with elongated, multiple pinched regions along the chromosomes instead of the cruciform structure observed in wild-type (Figure 7, A and B). The abnormal ubc-9 morphology could be explained if there were more than one crossover per bivalent in SUMOylation mutants.
Figure 7.
spo-11 mutant univalent phenotype is partially rescued in ubc-9;spo-11 double-mutant diakinesis. (A) Schematic of how DAPI bodies were measured in this figure. Univalents do not have any pinched regions, normal bivalents (middle) have one pinched region (red arrow), and an example of an aberrant DAPI body with more than one pinched region is displayed on the right. Green brackets indicate how the length measurement was obtained in (C). (B) Representative images of diakinesis nuclei from all genotypes measured. Bar, 1.5 μM. (C) Graphs of all five genotypes measuring the number of DAPI bodies per nucleus, length of each DAPI body within a nucleus, and the number of pinched regions per DAPI body within a nucleus. Between 10 and 14 nuclei were analyzed, and for individual DAPI body measurements 54–79 DAPI bodies were measured per genotype. Mann–Whitney U test was performed on all pairwise groups and the stars above data indicate significance. The color corresponds to the genotype with which the data is significantly different [i.e., blue stars are being compared with wild-type (WT)]. *** P < 0.0001, ** P < 0.001, and *P < 0.05.
To test this hypothesis, we first tested if the pinched regions are a visual manifestation of crossovers in our mutants. Based on RAD-51 foci counts, it is likely that in ubc-9; spo-11 double mutants, DSBs are reduced but not eliminated (since DSBs are carried over from mitosis to meiosis). These mutants still maintain the aberrant chromosomal morphology of ubc-9(tm2610) mutants but are expected to have less crossovers. If pinched regions are synonymous to crossovers, then their frequency should decrease in ubc-9; spo-11 double mutants compared to ubc-9(tm2610) mutants. Indeed, DAPI body counts in ubc-9; spo-11 double mutants are intermediate between that found in each single mutant, indicating that crossovers are present but reduced in the double mutants (Figure 7C, left top). Accordingly, the number of pinched regions is reduced as well (Figure 7C, right top). There is a correlation between bivalent length and the number of pinched regions; bigger DAPI bodies that are more likely to be bivalents are more likely to have increased numbers of pinched regions (Figure 7C, bottom and Figure S10 in File S2).
To test if crossovers are increased in ubc-9(tm2610) mutants, we created a cosa-1::gfp; ubc-9(tm2610) strain. COSA-1 marks interfering crossover sites in C. elegans (Yokoo et al. 2012). Six COSA-1 foci are observed per nucleus in a wild-type background; six COSA-1 foci are also observed per nucleus in ubc-9(tm2610) mutants (Figure S9 in File S2 and Table S10 in File S3). Elevated levels of recombination could still occur, either as noninterfering crossovers or as events resolved as noncrossovers.
We next measured recombination frequency to assess both interfering and noninterfering crossovers. In wild-type C. elegans, all crossovers are interfering, but some mutants show increases in noninterfering crossovers without affecting interfering crossovers (Youds et al. 2010). It is not possible to measure recombination frequencies in null SUMOylation mutants because they are sterile. Therefore, we examined a heterozygous ubc-9(tm2610)/+ strain in the hope that it might be haploinsufficient. A heterozygous ubc-9(tm2610)/+ strain was generated from a cross between balanced heterozygotes, ubc-9(tm2610)/nT1 in the Bristol background and wild-type males from the Hawaiian C. elegans background. Recombination frequencies were compared to wild-type hermaphrodites generated by a cross between N2 (Bristol) and Hawaiian strains (Table S11 in File S3). We performed SNP analysis for two loci on chromosome II that have been published to be 29 map units (m.u.) apart (Wicks et al. 2001; Swan et al. 2002). F2 progeny were scored for recombination events. The wild-type worms had a recombination frequency of 33 m.u, which is not significantly different from the expected recombination frequency of 29 m.u. However, the ubc-9(tm2610) heterozygotes had a recombination frequency of 50.3 m.u., which is significantly higher than the wild-type background cross (P = 0.0016) and the published recombination frequency (p). Additional analysis was performed for chromosome I across a smaller genomic interval. Expected recombination frequency was 11 m.u., wild-type was 20.4 m.u., and ubc-9 heterozygotes had a recombination frequency of 37.8 m.u. The difference between the wild-type and the ubc-9 heterozygotes was significant (P = 0.014). These data are consistent with the view that recombination frequency is increased in ubc-9 heterozygotes. These data support the hypothesis that even though interfering crossovers are not affected by SUMOylation loss, total crossovers appear to be increased, suggesting that SUMOylation normally plays a role in downregulating crossover numbers in C. elegans.
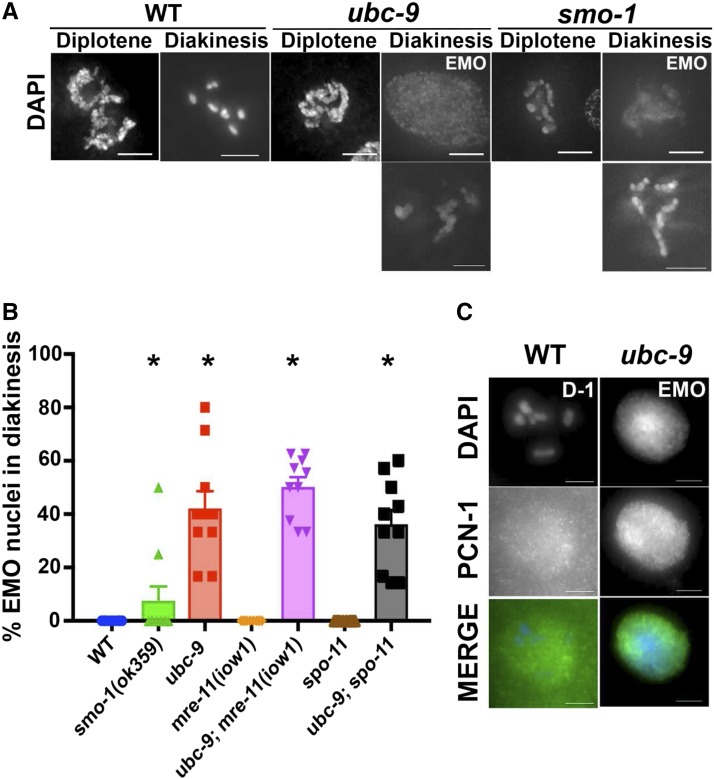
EMO nuclei are found in SUMOylation mutants
Diakinesis contains an average of 8.5 ± 0.27 nuclei in wild-type germlines. ubc-9(tm2610) mutants have an average of 5.4 ± 0.34 total diakinesis nuclei and smo-1(ok359) mutants have an average of 3.9 ± 0.36 total diakinesis nuclei. Mutant ubc-9 diakinesis nuclei also contained endomitotic nuclei (Cremona et al. 2012) (Figure 7). These are diakinesis oocytes that prematurely replicate their DNA prior to the end of meiosis I leading to a decondensed mass of DNA, and present as a bright ball of DNA stained with DAPI (Iwasaki et al. 1996). EMO nuclei were found in both single SUMOylation mutants, while they were never found in wild-type worms of the same age (Figure 7 and Table S12 in File S3). ubc-9(tm2610) mutants had at least one EMO nucleus in diakinesis, with an average of 40% of total nuclei in diakinesis being EMO. This phenotype was not as prevalent in smo-1(ok359) mutants, with only 10% of total diakinesis nuclei being EMO. All ubc-9(tm2610) EMO nuclei stained brightly for replication protein PCN-1 compared to the haze of PCN-1 not localized to chromatin in wild-type diakinesis nuclei (Figure 7C).
We wondered whether the EMO phenotype observed in SUMOlyation mutants might be caused by the increased level of recombination that we had observed (see above). To test this, we analyzed the double-mutant strains ubc-9(tm2610) mre-11(iow1) and ubc-9(tm2610) spo-11 for EMO nuclei in diakinesis. The spo11 mutation prevents all meiotic recombination, and the mre-11(iow1) mutation blocks recombination after DSB formation. EMO formation was not detectably altered compared to the single ubc-9 mutant background. These data indicate that SUMOylation is important to prevent premature replication independently of its role in controlling recombination and preventing DNA damage repair (Figure 8).
Figure 8.
SUMOylation mutants have aberrant chromosomal morphology and EMO nuclei at diakinesis. (A) Representative images of diplotene and diakinesis nuclei in wild-type (WT), ubc-9(tm2610), and smo-1(ok359) strains. Both EMO and non-EMO phenotypes are shown for mutants. Mutant non-EMO oocytes still had abnormal morphology compared to WT. Points on the graph indicate individual data points, with the bar indicating the mean of all data points. (B) Percentage of EMO nuclei counted from DAPI-stained whole worms. At least 10 worms from each genotype were counted. Error bars signify the mean ± SEM. (C) Representative EMO oocyte from a ubc-9(tm2610) gonad staining strongly for PCN-1. WT oocytes have low levels of background PCN-1 staining not associated with chromatin. Ten WT D-1 oocytes and 11 EMO oocytes were counted for PCN-1 staining. Points on the graph indicate individual data points, with the bar indicating the mean of all data points.
Discussion
The SUMO protein is a modification present in all eukaryotes that is highly conserved at the amino acid level. SUMO has a wider range of functions than its structural cousin, ubiquitin. Despite the important role SUMOylation plays in development, it is not essential for viability (M+Z−) in C. elegans. In this paper, we discovered a number of roles for SUMOylation in the C. elegans germline; SUMOylation is important for nuclear proliferation by preventing the accumulation of unrepaired DNA damage, affects the number of crossovers, and prevents replication in late meiotic prophase I nuclei. We also described a genetic interaction between ubc-9 and mre-11 in C. elegans that was previously unknown.
SUMOylation pathway proteins localize to germline nuclei and genetically interact with mre-11
SUMO and UBC-9 proteins are enriched in the nucleus and UBC-9 is present at the nuclear periphery in wild-type germlines [our study and Pelisch et al. (2017)]. Analysis of the ubc-9 mutation combined with the mre-11 null mutation implies that the double mutant displays a synergistic effect. The phenotypes conferred by the mre-11(iow1) and mre-11(ok179) alleles, when coupled with the ubc-9(tm2610) allele, suggest that resection activity is not responsible for the different phenotypes. Neither allele has resection activity. One explanation could be a structural role for MRE-11. This hypothesis is consistent with MRE-11 interacting with multiple proteins that are key players in various aspects of DSB repair (Williams et al. 2007, 2009). The ubc-9(tm2610);mre-11(ok179) synergistic interaction would be consistent with the MRE-11 and UBC-9 proteins affecting the same biological function, albeit with distinct mechanisms.
SUMOylation is required both for preventing DNA damage and for the proliferation of mitotically cycling cells in the hermaphrodite germline
Mice with mutations in SUMO1, SUMO2, or UBC9 arrest in early embryonic development and do not survive past birth (Nacerddine et al. 2005). Developmental defects are also seen in zebrafish where SUMOylation defects are associated with increased apoptosis, polyploidization, and defects in the G2/M-phase of the cell cycle; there is no effect on S-phase (Nowak and Hammerschmidt 2006). Staining for the PCNA homolog (PCN-1) in the PMT of the C. elegans germline showed no significant difference between SUMOylation mutants and a wild-type strain in the fraction of nuclei in S-phase, but we did observe that the M-phase fraction of nuclei was reduced in SUMOylation mutants. A reduction in M-phase nuclei could stem from reduced M-entry or accelerated M-exit. Because SUMOylation mutant germlines contain fewer overall nuclei, we favor the former explanation.
One explanation for the PMT defects observed in SUMOylation mutants is the activation of a G2 (G2/M) checkpoint that recognizes defects occurring in S-phase. The smo-1(ok359) and ubc-9(tm2610) mutants both show increased numbers of RAD-51 foci in the PMT indicating increased DNA damage. Since PMT cells are cycling (∼50% are in S-phase, Figure 5), we posit that the damage is caused by replication fork stalling and collapse (Cha and Kleckner 2002). Many replication and repair proteins (including RPA, PCNA, BLM, and TopII) are known to be SUMOylated in other organisms upon replication stalling (Pfander et al. 2005; Takahashi et al. 2008; Gali et al. 2012; Ouyang et al. 2013; Wu and Zou 2016). If this type of SUMOylation occurs in worms, the increase in DNA damage in the PMT may result from aberrant function of one or more of these proteins. So far, we have excluded RAD-51 as a SUMOylation target, while PCNA/PCN-1 was excluded by another group (Kim and Michael 2008). It is likely that ZTF-8, which acts in the 9-1-1 complex, is a target of SUMOylation (Kim and Colaiacovo 2015). However, the impairment of meiotic functions in ztf-8 mutants (in the absence of DNA damage) is less severe compared to what is found in ubc-9 mutants, indicating that ZTF-8 is not the only germline mitotic target of UBC-9 (Kim and Colaiacovo 2015). The complete array of S-phase SUMOylation targets in the C. elegans PMT awaits discovery.
Despite the presence of DNA damage in S-phase cells in SUMO mutants, the fraction of nuclei in S-phase is not affected by SUMOylation mutations. This implies that neither entry into, nor progression through, S-phase is affected by SUMOylation mutations; it further suggests that the mutations do not cause activation of an S-phase checkpoint. In mammalian cells, the E3 SUMO ligase PIAS3 is required to SUMOylate ATR so that it can be activated through autophosphorylation in response to DNA damage to establish the S-phase checkpoint. (Wu and Zou 2016). Because ATR in C. elegans is also required for the S-phase checkpoint, the mammalian mechanism may explain the worm S-phase result (Garcia-Muse and Boulton 2005). In worm SUMOylation mutants, S-phase checkpoints appeared disabled and permitted cell cycle progression in the presence of damage. Because M-phase nuclei are decreased, the C. elegans G2/M checkpoint appears to be functional, and therefore not dependent on SUMOylation to function. We have also shown that the response to HU, in terms of RAD-51 focus formation, was attenuated in ubc-9 mutants. This is consistent with the view that SUMOylation is needed for effective recruitment of RAD-51 following HU exposure.
To conclude, we propose that UBC-9 is required for the recruitment of RAD-51 after excess damage, to prevent replication fork collapse at the mitotic proliferating zone/PMT. In its absence, unrepaired DNA damage in the PMT triggers cell cycle arrest (likely at the G2/M transition) and thus reduces germline proliferation, leading to decreases in germline size. Since RAD-51 loading is impaired after excess damage but not prevented, the increase in DNA damage due to unrepaired DSBs ultimately causes and increase in the overall levels of RAD-51 foci.
Does SUMOylation play different roles in different types of cell visions in C. elegans?
In the germline, SMO-1 shows diffused nuclear localization [Pelisch and Hay (2016) and this study] but moves to the midbivalent regions upon nuclear envelope breakdown (Pelisch and Hay 2016]. In mitosis, SMO-1 shows a similar pattern, moving from diffused nuclear localization to specific localization on metaphase chromosomes (Pelisch et al. 2014). AIR-2 and KLP-19 proteins, which are present in the midbivalent region, are likely targets for SUMOylation, which is consistent with the localization pattern of SMO-1 (Pelisch and Hay 2016). Due to the early roles SUMOylation plays in the germline, studies of the meiotic and mitotic divisions of the embryo were performed by RNAi knockdown of ubc-9. These studies established the importance of SUMOylation in the congression of chromosomes at the metaphase plate, and therefore identified the key roles that they play in meiosis (Pelisch and Hay 2016). SUMOylation also controls the mitotic divisions in the embryo, targeting AIR-2, as found for mitosis (Pelisch et al. 2014). These studies reveal the importance of SUMOylation outside the germline and explain the embryonic lethality caused by the ubc-9::flag, which is likely to compromise the embryonic but not the germline function of SUMOylation.
We observed a reduction in M-phase nuclei in germline mitotic nuclei of both smo-1 and ubc-9 mutants, which is consistent with the reduction in the number of germline nuclei. This may indicate a conserved metaphase function of SUMOylation in the mitotic germline as found in other dividing cells. However, we did not observe SMO-1 localized specifically to metaphase PMT chromosomes, nor an obvious congression defect in mitotic germline nuclei of smo-1 and ubc-9 mutants (Pelisch et al. 2014; Pelisch and Hay 2016). Due to the evident increase in RAD-51 foci in both smo-1 and ubc-9 mutants, we suggest that the proliferation defects in these mutants are due to checkpoint activation that prohibits M-phase entry and not due to defects in congression.
RAD-51 focus formation is altered in SUMO mutants
RAD-51 foci are typically present as individually defined foci of symmetrical appearance. However, in the SUMO mutants, the fraction of such atypical RAD-51 foci increased. Aberrant foci have been observed in mammalian cells when Rad51 was overexpressed, as well as in maize mutants defective in chromosome pairing during meiosis (Franklin et al. 1999; Raderschall et al. 2002; Pawlowski et al. 2003). Most of the atypical RAD-51 foci in single SUMOylation mutants are found in the PMT and then decrease throughout meiosis. After irradiation, the increased number of RAD-51 foci limits scoring of most classes of atypical foci with the exception of strings. Because the number of RAD-51 strings does not increase after radiation, their appearance in the PMT appears to be due to specific lesions, perhaps collapsed replication forks, which happen more frequently in SUMOylation mutants.
We hypothesize that atypical and typical foci represent two states of the same molecular event: a collapsed replication fork. In these events, a typical focus will contain two regions of ssDNA loaded with RAD-51, likely on sister chromatids, held together in close proximity. These RAD-51 foci on two sister chromatids would not be held together properly in our SUMOylation mutants. Doublets and other atypical RAD-51 foci could therefore represent a defect in the tethering of broken DNA molecules initiating from the same event of a collapsed replication fork, eventually leading to the broken DNA dissociating from one another.
ubc-9(tm2610);mre-11(ok179) double-mutant synthetic sickness is due to a function of MRE-11 other than resection
Double mutants lacking both MRE-11 and UBC-9 were slow-growing and fewer worms reached adulthood; they also exhibited a smaller germline compared to each single mutant, presumably due to less proliferation in the PMT. These results together point to an additive effect of MRE-11 and the SUMOylation pathway in promoting proper nuclear division prior to prophase I. These defects were not found in ubc-9(tm2610);mre-11(iow1) or ubc-9(tm2610);rad-50(ok97) double mutants, indicating that the role of MRE-11 in PMT events is specific to functions or structures still present in the mre-11(iow1) point mutant. One possibility is that the key role of the MRE-11 protein is a structural role in recruiting other proteins essential for DSB repair after S-phase damage (Lee and Paull 2005; Zheng et al. 2009; Shim et al. 2010; Nimonkar et al. 2011). If this were true, the different phenotype conferred by a rad-50 null mutation suggests that the structural role is specific to MRE-11, not the MRX/N complex.
mre-11(iow1) single mutants have a small increase in atypical RAD-51 foci numbers compared to wild-type, but a larger effect is observed in mre-11(ok179) gonads. There is evidence in humans that MRE11 and the MRN/X complex are needed not only for resection during HR, but also during replication for tethering DNA, similar to the cohesin complex (de Jager et al. 2001; Seeber et al. 2016). In the PMT, the resection activity of MRE-11 is not absolutely essential due to the activity of other nucleases (Hayashi et al. 2007; Yin and Smolikove 2013) but, if MRE-11 protein is absent, the complex could not tether DNA molecules initiating from the same collapsed replication fork event, allowing the DNA ends to drift away from each other. The synthetic sickness of the ubc-9; mre-11(ok179) double mutants prevented us from analyzing RAD-51 localization in the PMT.
SUMOylation in yeast and mammalian cells is known to be involved in MRN/X complex function through either direct SUMOylation of the MRN/X complex proteins or through noncovalent SUMO interactions via Mre11’s SIM domain in yeast (Chen et al. 2016). If this is true in C. elegans, this may explain why we uncovered UBC-9 and MRE-11 as physically interacting proteins by the yeast two-hybrid assay. It is possible that the absence of SUMOylation may destabilize the MRX/N complex in C. elegans, leading to atypical RAD-51 foci and increased lethality, but other SUMOylation functions would be MRE-11-independent. The latter could explain the synthetic sickness of the ubc-9(tm2610);mre-11(ok179) double mutants. If so, MRE-11 noncovalent SUMO interaction would only be responsible for MRE-11’s mitotic functions. This will explain why ubc-9 and mre-11 null mutants share phenotypes in PMT nuclei and not in prophase I.
The fraction of S-phase nuclei in the PMT were significantly decreased in mre-11(ok179) mutants; this could be due to MRE-11’s role in replication initiation and/or to intra-S checkpoint activation (Olson et al. 2007). The delay in the loading of RAD-51 protein in mre-11 mutants subjected to DNA damage agents is consistent with its role in resection to form ssDNA.
SUMOylation may play a role in meiotic recombination
We have shown that SUMOylation plays a role in germline proliferation in the PMT; this explains why SUMO mutants have reduced germline size. However, SUMOylation clearly has a role in later meiotic events. Our studies suggest that these meiotic roles are reserved for late events in DSB repair. Analysis of RAD-51 foci numbers indicates that the increase in meiotic foci in SUMOylation mutants compared to wild-type is primarily due to the effect of these mutations on PMT nuclei rather than a direct effect on meiosis. SUMOylation may also affect later meiotic events, such as crossover regulation. The number of interfering crossovers (measured by COSA-1 foci) is unchanged in the ubc-9(tm2610) mutant, but noninterfering crossovers are increased. One interpretation is that some recombination events that normally would be resolved as noncrossovers are altered (Barber et al. 2008; Getz et al. 2008; Youds et al. 2010). The proposed increase in total crossovers is consistent with the appearance of diakinesis chromosomes that appear to have multiple chiasmata. These observations suggest that SUMOylation is involved in the mechanism regulating the outcome of recombination events. SUMOylation in mouse meiosis promotes crossovers at the expense of noncrossovers, likely by stabilizing the MSH-4/5 complex (Qiao et al. 2014; Rao et al. 2017). In C. elegans, the antirecombinase RTEL-1, along with DCC complex components, has been suggested to control the levels of noninterfering crossovers (Barber et al. 2008; Youds et al. 2010; Pferdehirt and Meyer 2013). It would be interesting if they were SUMOylation targets.
Some diakinesis nuclei in SUMOylation mutants appear to be undergoing replication based on PCNA localizing to the DNA. This observation suggests that SUMOylation could be required for repressing replication in the later stages of prophase I. There is no specific evidence that replication in late prophase I occurs in SUMOylation pathway mutants in other organisms, but in Xenopus the SUMO2/3 target is cyclin E, an early S-phase activator (Bonne-Andrea et al. 2013). When SUMO2/3 is depleted, increased origin firing occurs in the Xenopus cell-free system. The EMO phenotype that is observed in some diakinesis nuclei in SUMOylation mutants in late prophase I must contribute to their sterility. Since UBC-9 and SUMO were not detectable in diakinesis oocytes, their function in repressing rereplication must be established at earlier stages.
Conserved and divergent roles of SUMOylation in meiosis
C. elegans UBC-9 and SMO-1 are visible as peripheral or diffuse nuclear stains, respectively. These patterns differ from both yeast Ubc9 and mammalian Smct3 that localize to the SC (Kovalenko et al. 1996; Klug et al. 2013). SUMO chains that form along the SC in yeast stabilize the SC, promoting its proper assembly. The yeast SC protein Zip1 interacts directly with the SUMO-conjugated domain of SC protein Zip3 (Cheng et al. 2006). Unlike the role for SUMOylation in SC assembly in yeast, we and others (Bhalla et al. 2008) have not observed any defects in SC formation in SUMOylation mutants in C. elegans. In C. elegans, ZHP-3 was implicated in SUMO-mediated disassembly of the SC (Bhalla et al. 2008); although this may be a direct effect, aberrant SC localization in diakinesis can also stem from defects in bivalent restructuring in ubc-9 mutants.
Study of the meiotic roles of the SUMOylation is difficult in vertebrates because null mutants are unable to complete embryonic development even when maternally loaded proteins are present in the early embryo (Mukai et al. 2006). In mammalian meiosis, the SUMO ligase RNF212 is required for proper Msh4/5 localization and crossover designation during HR (Reynolds et al. 2013). RNF212 is essential for crossover formation and SUMO is hypothesized to selectively stabilize crossover proteins at sites of recombination. We have also proposed that SUMOylation regulates crossover formation in C. elegans; however, this role is different: SUMOylation suppresses noninterfering crossovers in C. elegans, as opposed to promoting interfering crossovers as found in mouse.
Do SMO-1 and UBC-9 have similar but not identical functions?
smo-1 and ubc-9 encode for the only SUMO and E2 in the SUMOylation pathway of C. elegans. Thus, ubc-9(tm2610) and smo-1(ok359) mutants are expected to exhibit the same phenotypes. Our analysis indicates that these two mutants indeed show very similar phenotypes. However, these phenotypes are not identical in their severity and most of the time ubc-9(tm2610) exhibits the more extreme phenotype. The smo-1(ok359) allele is a deletion of the whole reading frame of the gene, while the ubc-9(tm2610) allele removes just the sequences that encode for the catalytic domain of UBC-9. Therefore, the difference in the severity of the phenotypes cannot be explained by smo-1(ok359) mutants retaining SUMO activity. In other organisms, Ubc9 has catalysis-independent function (Chakrabarti et al. 1999; Poukka et al. 1999; Kurtzman and Schechter 2001). Therefore, loss-of-function mutations in the two genes may not lead to identical phenotypes. However, the fact that the overall phenotypes are very similar between the ubc-9(tm2610) and smo-1(ok359) mutants indicates that if there is a catalysis-independent function for Ubc9, it has a relatively minor role. It is also possible that the ubc-9(tm2610) allele is not a complete deletion and some gain-of-function of the residual truncated protein modifies the phenotypes caused by the removal of SUMOylation in the germline. Again, if this is so, the effects are relatively small. A third option is that SUMO/SMO-1 is not depleted as quickly as UBC-9 during development and some maternally contributed SUMO is still present in the germline that will explain the differences.
Conclusions
Our studies are consistent with a central role for SUMO in DNA metabolism in worms, consistent with its established roles in mitosis and meiosis in many organisms. From our observations, we hypothesize that the meiotic functions of SUMOylation may vary substantially among different organisms, and to fully understand the breadth of its involvement it will need to be examined in multiple biological systems.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.118.300787/-/DC1.
Acknowledgments
Some strains and clones were kindly provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD-010440), and the C. elegans Reverse Genetics Core Facility at the University of British Columbia, which is part of the International C. elegans Gene Knockout Consortium. We thank the National Bioresource Project for the Experimental Animal “Nematode C. elegans”, Japan for providing the tm2610 allele. We thank M. Michael for the PCN-1 antibody and M. Wheat for generation of the mre-11::gfp strain. We thank the Radiation and Free Radical Research Core Facility in the Carver College of Medicine for the irradiation service. This work was funded by NIH grant number 1R01 GM-112657 (to S.S.).
Footnotes
Communicating editor: J. Engebrecht
Literature Cited
- Altmannova V., Eckert-Boulet N., Arneric M., Kolesar P., Chaloupkova R., et al. , 2010. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 38: 4708–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber L. J., Youds J. L., Ward J. D., McIlwraith M. J., O’Neil N. J., et al. , 2008. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135: 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom-Slack C. A., Ross L. O., Dawson D. S., 1997. Chiasmata, crossovers, and meiotic chromosome segregation. Adv. Genet. 35: 253–284. [DOI] [PubMed] [Google Scholar]
- Bazan G. C., Hillers K. J., 2011. SNP-based mapping of crossover recombination in Caenorhabditis elegans. Methods Mol. Biol. 745: 207–222. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D., 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356. [DOI] [PubMed] [Google Scholar]
- Bhalla N., Wynne D. J., Jantsch V., Dernburg A. F., 2008. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 4: e1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., 1994. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N., 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456. [DOI] [PubMed] [Google Scholar]
- Bocker T., Barusevicius A., Snowden T., Rasio D., Guerrette S., et al. , 1999. hMSH5: a human MutS homologue that forms a novel heterodimer with hMSH4 and is expressed during spermatogenesis. Cancer Res. 59: 816–822. [PubMed] [Google Scholar]
- Bologna S., Altmannova V., Valtorta E., Koenig C., Liberali P., et al. , 2015. Sumoylation regulates EXO1 stability and processing of DNA damage. Nat. Cell Biol. 14: 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne-Andrea C., Kahli M., Mechali F., Lemaitre J. M., Bossis G., et al. , 2013. SUMO2/3 modification of cyclin E contributes to the control of replication origin firing. Nat. Commun. 4: 1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V., Lin W., Novikov E., Petrini J. H., Lichten M., et al. , 2004. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell 13: 389–401. [DOI] [PubMed] [Google Scholar]
- Boulton S. J., Martin J. S., Polanowska J., Hill D. E., Gartner A., et al. , 2004. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr. Biol. 14: 33–39. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L., Kolotuev I., Didier C., Bhoumik A., Gupta B. P., et al. , 2004. The small ubiquitin-like modifier (SUMO) is required for gonadal and uterine-vulval morphogenesis in Caenorhabditis elegans. Genes Dev. 18: 2380–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylebyl G. R., Belichenko I., Johnson E. S., 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278: 44113–44120. [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N., 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Celis J. E., Madsen P., Celis A., Nielsen H. V., Gesser B., 1987. Cyclin (PCNA, auxiliary protein of DNA polymerase delta) is a central component of the pathway(s) leading to DNA replication and cell division. FEBS Lett. 220: 1–7. [DOI] [PubMed] [Google Scholar]
- Cha R. S., Kleckner N., 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S. R., Sood R., Ganguly S., Bohlander S., Shen Z., et al. , 1999. Modulation of TEL transcription activity by interaction with the ubiquitin-conjugating enzyme UBC9. Proc. Natl. Acad. Sci. USA 96: 7467–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Mannen H., Li S. S., 1998. Characterization of mouse ubiquitin-like SMT3A and SMT3B cDNAs and gene/pseudogenes. Biochem. Mol. Biol. Int. 46: 1161–1174. [DOI] [PubMed] [Google Scholar]
- Chen Y. J., Chuang Y. C., Chuang C. N., Cheng Y. H., Chang C. R., et al. , 2016. S. cerevisiae Mre11 recruits conjugated SUMO moieties to facilitate the assembly and function of the Mre11-Rad50-Xrs2 complex. Nucleic Acids Res. 44: 2199–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. H., Lo Y. H., Liang S. S., Ti S. C., Lin F. M., et al. , 2006. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20: 2067–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin G. M., Villeneuve A. M., 2001. C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev. 15: 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury B. K., Li S. S., 1997. Identification and characterization of the SMT3 cDNA and gene from nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 234: 788–791. [DOI] [PubMed] [Google Scholar]
- Citro S., Chiocca S., 2013. Sumo paralogs: redundancy and divergencies. Front. Biosci. (Schol. Ed.) 5: 544–553. [DOI] [PubMed] [Google Scholar]
- Colaiacovo M. P., MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A., et al. , 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Cremona C. A., Sarangi P., Yang Y., Hang L. E., Rahman S., et al. , 2012. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell 45: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Kimble J., 2008. Analysis of the C. elegans germline stem cell region. Methods Mol. Biol. 450: 27–44. [DOI] [PubMed] [Google Scholar]
- de Jager M., Dronkert M. L., Modesti M., Beerens C. E., Kanaar R., et al. , 2001. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 29: 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., et al. , 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Desai-Mehta A., Cerosaletti K. M., Concannon P., 2001. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol. Cell. Biol. 21: 2184–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande R. A., Williams G. J., Limbo O., Williams R. S., Kuhnlein J., et al. , 2014. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 33: 482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande R. A., Williams G. J., Limbo O., Williams R. S., Kuhnlein J., et al. , 2016. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 35: 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Goldstein B., 2016. CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202: 885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser M. E., Ewing D. J., Conrad M. N., Dominguez A. M., Barstead R., et al. , 1997. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley M. L., Baillie D. L., Riddle D. L., Rose A. M., 2006. Genetic balancers (April 6, 2006). WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.89.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J., Forment J. V., Coates J., Mistrik M., Lukas J., et al. , 2012. CDK targeting of NBS1 promotes DNA-end resection, replication restart and homologous recombination. EMBO Rep. 13: 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira H. C., Towbin B. D., Jegou T., Gasser S. M., 2013. The shelterin protein POT-1 anchors Caenorhabditis elegans telomeres through SUN-1 at the nuclear periphery. J. Cell Biol. 203: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R. M., 1985. Changes in deoxynucleoside triphosphate pools induced by inhibitors and modulators of ribonucleotide reductase. Pharmacol. Ther. 30: 31–42. [DOI] [PubMed] [Google Scholar]
- Franklin A. E., McElver J., Sunjevaric I., Rothstein R., Bowen B., et al. , 1999. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11: 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Jackson S. P., 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26: 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gali H., Juhasz S., Morocz M., Hajdu I., Fatyol K., et al. , 2012. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 40: 6049–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T., Boulton S. J., 2005. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 24: 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M. O., 2000. A conserved checkpoint pathway mediates DNA damage--induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. [DOI] [PubMed] [Google Scholar]
- Gedik C. M., Collins A. R., 1990. Comparison of effects of fostriecin, novobiocin, and camptothecin, inhibitors of DNA topoisomerases, on DNA replication and repair in human cells. Nucleic Acids Res. 18: 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz T. J., Banse S. A., Young L. S., Banse A. V., Swanson J., et al. , 2008. Reduced mismatch repair of heteroduplexes reveals “non”-interfering crossing over in wild-type Saccharomyces cerevisiae. Genetics 178: 1251–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein P., Slaton D. E., 1982. The synaptonemal complexes of Caenorhabditis elegans: comparison of wild-type and mutant strains and pachytene karyotype analysis of wild-type. Chromosoma 84: 585–597. [DOI] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J. D., Boulton S. J., et al. , 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell 14: 263–274. [DOI] [PubMed] [Google Scholar]
- Greenwald I., 1997. Development of the vulva in C. elegans II, edited by Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Guzzo C. M., Berndsen C. E., Zhu J., Gupta V., Datta A., et al. , 2012. RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci. Signal. 5: ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. L., Warms J. V., Hershko A., Rose I. A., 1982. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 257: 2543–2548. [PubMed] [Google Scholar]
- Habu T., Taki T., West A., Nishimune Y., Morita T., 1996. The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res. 24: 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Chin G. M., Villeneuve A. M., 2007. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 3: e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., et al. , 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106: 348–360. [DOI] [PubMed] [Google Scholar]
- Hohl M., Kwon Y., Galvan S. M., Xue X., Tous C., et al. , 2011. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat. Struct. Mol. Biol. 18: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Ponte L., Halsey C., 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9: 1728–1739. [DOI] [PubMed] [Google Scholar]
- Holway A. H., Kim S. H., La Volpe A., Michael W. M., 2006. Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. J. Cell Biol. 172: 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker G. W., Roeder G. S., 2006. A role for SUMO in meiotic chromosome synapsis. Curr. Biol. 16: 1238–1243. [DOI] [PubMed] [Google Scholar]
- Horigome C., Bustard D. E., Marcomini I., Delgoshaie N., Tsai-Pflugfelder M., et al. , 2016. PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev. 30: 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H. R., Sternberg P. W., 1991. Multiple intercellular signalling systems control the development of the Caenorhabditis elegans vulva. Nature 351: 535–541. [DOI] [PubMed] [Google Scholar]
- Hu Y., Parvin J. D., 2014. Small ubiquitin-like modifier (SUMO) isoforms and conjugation-independent function in DNA double-strand break repair pathways. J. Biol. Chem. 289: 21289–21295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. W., Tsoi S. C., Sun Y. H., Li S. S., 1998. Identification and characterization of the SMT3 cDNA and gene encoding ubiquitin-like protein from Drosophila melanogaster. Biochem. Mol. Biol. Int. 46: 775–785. [DOI] [PubMed] [Google Scholar]
- Hubbard, E. J. A., and G. David, 2005 Introduction to the germ line (September 1, 2005). WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.18.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Ismail I. H., Gagne J. P., Genois M. M., Strickfaden H., McDonald D., et al. , 2015. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat. Cell Biol. 17: 1446–1457. [DOI] [PubMed] [Google Scholar]
- Iwasaki K., McCarter J., Francis R., Schedl T., 1996. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J. Cell Biol. 134: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K., Ogawa H., 1995. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139: 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky R., Denison C., Bening-Abu-Shach U., Chisholm A. D., Gygi S. P., et al. , 2009. SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans. Dev. Cell 17: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Xu L., Ciosk R., 2015. CRISPR/Cas9 genome gediting in Caenorhabditis elegans: evaluation of templates for homology-mediated hmrepair and knock-kins by homology-hindependent DNA repair. G3 5: 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kim H. M., Colaiacovo M. P., 2014. ZTF-8 interacts with the 9-1-1 complex and is required for DNA damage response and double-strand break repair in the C. elegans germline. PLoS Genet. 10: e1004723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Colaiacovo M. P., 2015. DNA damage sensitivity dsassays in Caenorhabditis elegans. Bio Protoc. 5: e1487. [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Michael W. M., 2008. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol. Cell 32: 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S., Waddell C. S., Esposito R. E., 1985. The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110: 187–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug H., Xaver M., Chaugule V. K., Koidl S., Mittler G., et al. , 2013. Ubc9 sumoylation controls SUMO chain formation and meiotic synapsis in Saccharomyces cerevisiae. Mol. Cell 50: 625–636. [DOI] [PubMed] [Google Scholar]
- Kovalenko O. V., Plug A. W., Haaf T., Gonda D. K., Ashley T., et al. , 1996. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 93: 2958–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman A. L., Schechter N., 2001. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc. Natl. Acad. Sci. USA 98: 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapenta V., Chiurazzi P., van der Spek P., Pizzuti A., Hanaoka F., et al. , 1997. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics 40: 362–366. [DOI] [PubMed] [Google Scholar]
- Lawrie N. M., Tease C., Hulten M. A., 1995. Chiasma frequency, distribution and interference maps of mouse autosomes. Chromosoma 104: 308–314. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Paull T. T., 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554. [DOI] [PubMed] [Google Scholar]
- Leight E. R., Glossip D., Kornfeld K., 2005. Sumoylation of LIN-1 promotes transcriptional repression and inhibition of vulval cell fates. Development 132: 1047–1056. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Wei Q., Zhang Y., Ling K., et al. , 2012. SUMOylation of the small GTPase ARL-13 promotes ciliary targeting of sensory receptors. J. Cell Biol. 199: 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y., Lee D., Kalichamy K., Hong S. E., Michalak M., et al. , 2014. Sumoylation regulates ER stress response by modulating calreticulin gene expression in XBP-1-dependent mode in Caenorhabditis elegans. Int. J. Biochem. Cell Biol. 53: 399–408. [DOI] [PubMed] [Google Scholar]
- Lu C. S., Truong L. N., Aslanian A., Shi L. Z., Li Y., et al. , 2012. The RING finger protein RNF8 ubiquitinates Nbs1 to promote DNA double-strand break repair by homologous recombination. J. Biol. Chem. 287: 43984–43994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Li L., Li Y., Wu C., Yin Y., et al. , 2016. A phosphorylation-deubiquitination cascade regulates the BRCA2-RAD51 axis in homologous recombination. Genes Dev. 30: 2581–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R., Delphin C., Guan T., Gerace L., Melchior F., 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88: 97–107. [DOI] [PubMed] [Google Scholar]
- Malone R. E., Bullard S., Hermiston M., Rieger R., Cool M., et al. , 1991. Isolation of mutants defective in early steps of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 128: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Hayashi-Hagihara A., 1998. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12: 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N., Higuchi E., Smith G. R., 2009. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol. Cell. Biol. 29: 5998–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn I. L., Rolls M. M., Hanna-Rose W., Malone C. J., 2009. SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol. Biol. Cell 20: 4586–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P. B., Spyropoulos B., 1995. Immunocytology of chiasmata and chromosomal disjunction at mouse meiosis. Chromosoma 104: 175–182. [DOI] [PubMed] [Google Scholar]
- Moreau S., Ferguson J. R., Symington L. S., 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M., Kitadate Y., Arita K., Shigenobu S., Kobayashi S., 2006. Expression of meiotic genes in the germline progenitors of Drosophila embryos. Gene Expr. Patterns 6: 256–266. [DOI] [PubMed] [Google Scholar]
- Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., et al. , 2005. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9: 769–779. [DOI] [PubMed] [Google Scholar]
- Nairz K., Klein F., 1997. mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 11: 2272–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., et al. , 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak J. E., Ross-Macdonald P. B., Roeder G. S., 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158: 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M., Hammerschmidt M., 2006. Ubc9 regulates mitosis and cell survival during zebrafish development. Mol. Biol. Cell 17: 5324–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E., Nievera C. J., Liu E., Lee A. Y., Chen L., et al. , 2007. The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol. Cell. Biol. 27: 6053–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang K. J., Yagle M. K., Matunis M. J., Ellis N. A., 2013. BLM SUMOylation regulates ssDNA accumulation at stalled replication forks. Front. Genet. 4: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C. Y., Calidas D., et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics 198: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquis-Flucklinger V., Santucci-Darmanin S., Paul R., Saunieres A., Turc-Carel C., et al. , 1997. Cloning and expression analysis of a meiosis-specific MutS homolog: the human MSH4 gene. Genomics 44: 188–194. [DOI] [PubMed] [Google Scholar]
- Parameswaran B., Chiang H. C., Lu Y., Coates J., Deng C. X., et al. , 2015. Damage-induced BRCA1 phosphorylation by Chk2 contributes to the timing of end resection. Cell Cycle 14: 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T. T., Gellert M., 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13: 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski W. P., Golubovskaya I. N., Cande W. Z., 2003. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15: 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisch F., Hay R. T., 2016. Tools to study SUMO conjugation in Caenorhabditis elegans. Methods Mol. Biol. 1475: 233–256. [DOI] [PubMed] [Google Scholar]
- Pelisch F., Sonneville R., Pourkarimi E., Agostinho A., Blow J. J., et al. , 2014. Dynamic SUMO modification regulates mitotic chromosome assembly and cell cycle progression in Caenorhabditis elegans. Nat. Commun. 5: 5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisch F., Tammsalu T., Wang B., Jaffray E. G., Gartner A., et al. , 2017. A SUMO-dependent protein network regulates chromosome dpnrccongression during oocyte omeiosis. Mol. Cell 65: 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., et al. , 2007. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell 12: 873–885. [DOI] [PubMed] [Google Scholar]
- Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S., 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433. [DOI] [PubMed] [Google Scholar]
- Pferdehirt R. R., Meyer B. J., 2013. SUMOylation is essential for sex-specific assembly and function of the Caenorhabditis elegans dosage compensation complex on X chromosomes. Proc. Natl. Acad. Sci. USA 110: E3810–E3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plug A. W., Peters A. H., Keegan K. S., Hoekstra M. F., de Boer P., et al. , 1998. Changes in protein composition of meiotic nodules during mammalian meiosis. J. Cell Sci. 111: 413–423. [DOI] [PubMed] [Google Scholar]
- Porter S. E., Champoux J. J., 1989. The basis for camptothecin enhancement of DNA breakage by eukaryotic topoisomerase I. Nucleic Acids Res. 17: 8521–8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H., Aarnisalo P., Karvonen U., Palvimo J. J., Janne O. A., 1999. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J. Biol. Chem. 274: 19441–19446. [DOI] [PubMed] [Google Scholar]
- Prelich G., Kostura M., Marshak D. R., Mathews M. B., Stillman B., 1987. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature 326: 471–475. [DOI] [PubMed] [Google Scholar]
- Qiao H., Prasada Rao H. B., Yang Y., Fong J. H., Cloutier J. M., et al. , 2014. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46: 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderschall E., Bazarov A., Cao J., Lurz R., Smith A., et al. , 2002. Formation of higher-order nuclear Rad51 structures is functionally linked to p21 expression and protection from DNA damage-induced apoptosis. J. Cell Sci. 115: 153–164. [DOI] [PubMed] [Google Scholar]
- Ragland R. L., Patel S., Rivard R. S., Smith K., Peters A. A., et al. , 2013. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 27: 2259–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H. B., Qiao H., Bhatt S. K., Bailey L. R., Tran H. D., et al. , 2017. A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355: 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. W., Holm P. B., 1984. The synaptonemal complex, recombination nodules and chiasmata in human spermatocytes. Symp. Soc. Exp. Biol. 38: 271–292. [PubMed] [Google Scholar]
- Reynolds A., Qiao H., Yang Y., Chen J. K., Jackson N., et al. , 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison J. G., Elliott J., Dixon K., Oakley G. G., 2004. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 279: 34802–34810. [DOI] [PubMed] [Google Scholar]
- Rojas-Fernandez A., Plechanovova A., Hattersley N., Jaffray E., Tatham M. H., et al. , 2014. SUMO chain-induced dimerization activates RNF4. Mol. Cell 53: 880–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojowska A., Lammens K., Seifert F. U., Direnberger C., Feldmann H., et al. , 2014. Structure of the Rad50 DNA double-strand break repair protein in complex with DNA. EMBO J. 33: 2847–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko P. J., Camerini-Otero R. D., 1999. Cloning, characterization, and localization of mouse and human SPO11. Genomics 61: 156–169. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhuri S., Crum T., Woollard A., Aslam S., Okkema P. G., 2006. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev. Biol. 295: 664–677. [DOI] [PubMed] [Google Scholar]
- Sacher M., Pfander B., Hoege C., Jentsch S., 2006. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8: 1284–1290. [DOI] [PubMed] [Google Scholar]
- Sarangi P., Steinacher R., Altmannova V., Fu Q., Paull T. T., et al. , 2015. Sumoylation influences DNA break repair partly by increasing the solubility of a conserved end resection protein. PLoS Genet. 11: e1004899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A., Hegnauer A. M., Hustedt N., Deshpande I., Poli J., et al. , 2016. RPA mediates recruitment of MRX to forks and double-strand dbreaks to hold sister chromatids together. Mol. Cell 64: 951–966. [DOI] [PubMed] [Google Scholar]
- Shim E. Y., Chung W. H., Nicolette M. L., Zhang Y., Davis M., et al. , 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29: 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T., 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470. [DOI] [PubMed] [Google Scholar]
- Silva S., Altmannova V., Eckert-Boulet N., Kolesar P., Gallina I., et al. , 2016. SUMOylation of Rad52-Rad59 synergistically change the outcome of mitotic recombination. DNA Repair (Amst.) 42: 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skilton A., Ho J. C., Mercer B., Outwin E., Watts F. Z., 2009. SUMO chain formation is required for response to replication arrest in S. pombe. PLoS One 4: e6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T., Acharya S., Butz C., Berardini M., Fishel R., 2004. hMSH4-hMSH5 recognizes holliday hjunctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y., 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA 101: 14373–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H. L., Li S. S., 2002. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene 296: 65–73. [DOI] [PubMed] [Google Scholar]
- Swan K. A., Curtis D. E., McKusick K. B., Voinov A. V., Mapa F. A., et al. , 2002. High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 12: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Dulev S., Liu X., Hiller N. J., Zhao X., et al. , 2008. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet. 4: e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanami T., Sato S., Ishihara T., Katsura I., Takahashi H., et al. , 1998. Characterization of a Caenorhabditis elegans recA-like gene Ce-rdh-1 involved in meiotic recombination. DNA Res. 5: 373–377. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nishide J., Okazaki K., Kato H., Niwa O., et al. , 1999. Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol. Cell. Biol. 19: 8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., et al. , 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276: 35368–35374. [DOI] [PubMed] [Google Scholar]
- Tomimatsu N., Mukherjee B., Harris J. L., Boffo F. L., Hardebeck M. C., et al. , 2017. DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J. Biol. Chem. 292: 10779–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsur A., Bening Abu-Shach U., Broday L., 2015. ULP-2 SUMO protease regulates E-cadherin recruitment to adherens junctions. Dev. Cell 35: 63–77. [DOI] [PubMed] [Google Scholar]
- Tzur Y. B., Margalit A., Melamed-Book N., Gruenbaum Y., 2006. Matefin/SUN-1 is a nuclear envelope receptor for CED-4 during Caenorhabditis elegans apoptosis. Proc. Natl. Acad. Sci. USA 103: 13397–13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T., Ohta T., Oshiumi H., Tomizawa J., Ogawa H., et al. , 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95: 705–716. [DOI] [PubMed] [Google Scholar]
- van der Linden E., Sanchez H., Kinoshita E., Kanaar R., Wyman C., 2009. RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucleic Acids Res. 37: 1580–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Taylor L. F., Mukherjee P., Humphryes N., Tsubouchi H., et al. , 2013. SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLoS Genet. 9: e1003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas R., Kumar R., Clermont F., Helfricht A., Kalev P., et al. , 2013. RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ. 20: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. Y., Hawley R. S., 2000. Hanging on to your homolog: the roles of pairing, synapsis and recombination in the maintenance of homolog adhesion. Chromosoma 109: 3–9. [DOI] [PubMed] [Google Scholar]
- Ward J. D., Bojanala N., Bernal T., Ashrafi K., Asahina M., et al. , 2013. Sumoylated NHR-25/NR5A regulates cell fate during C. elegans vulval development. PLoS Genet. 9: e1003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H., 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Williams R. S., Williams J. S., Tainer J. A., 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85: 509–520. [DOI] [PubMed] [Google Scholar]
- Williams R. S., Dodson G. E., Limbo O., Yamada Y., Williams J. S., et al. , 2009. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windecker H., Ulrich H. D., 2008. Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae. J. Mol. Biol. 376: 221–231. [DOI] [PubMed] [Google Scholar]
- Wu C. S., Zou L., 2016. The SUMO (small subiquitin-like modifier) ligase PIAS3 primes ATR for checkpoint activation. J. Biol. Chem. 291: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Smolikove S., 2013. Impaired resection of meiotic double-strand breaks channels repair to nonhomologous end joining in Caenorhabditis elegans. Mol. Cell. Biol. 33: 2732–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Seifert A., Chua J. S., Maure J. F., Golebiowski F., et al. , 2012. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26: 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Donlevy S., Smolikove S., 2016. Coordination of recombination with meiotic progression in the Caenorhabditis elegans germline by KIN-18, a TAO kinase that regulates the timing of MPK-1 signaling. Genetics 202: 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K. A., Nabeshima K., Drake M., Arur S., et al. , 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Kondoh G., Matsuda Y., Habu T., Nishimune Y., et al. , 1998. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell 1: 707–718. [DOI] [PubMed] [Google Scholar]
- Youds J. L., Mets D. G., McIlwraith M. J., Martin J. S., Ward J. D., et al. , 2010. RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327: 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus A. A., Lima C. D., 2009. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Kanagaraj R., Mihaljevic B., Schwendener S., Sartori A. A., et al. , 2009. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 37: 2645–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.