Abstract
Plastidic acetyl-coenzyme A (CoA) carboxylase (ACCase) catalyzes the first committed reaction of de novo fatty acid biosynthesis. This heteromeric enzyme is composed of one plastid-coded subunit (β-carboxyltransferase) and three nuclear-coded subunits (biotin carboxy-carrier, biotin carboxylase, and α-carboxyltransferase). We report the primary structure of the Arabidopsis α-carboxyltransferase and β-carboxyltransferase subunits deduced from nucleotide sequences of the respective genes and/or cDNA. Co-immunoprecipitation experiments confirm that the α-carboxyltransferase and β-carboxyltransferase subunits are physically associated. The plant α-carboxyltransferases have gained a C-terminal domain relative to eubacteria, possibly via the evolutionary acquisition of a single exon. This C-terminal domain is divergent among plants and may have a structural function rather than being essential for catalysis. The four ACCase subunit mRNAs accumulate to the highest levels in tissues and cells that are actively synthesizing fatty acids, which are used either for membrane biogenesis in rapidly growing tissues or for oil accumulation in developing embryos. Development coordinately affects changes in the accumulation of the ACCase subunit mRNAs so that these four mRNAs maintain a constant molar stoichiometric ratio. These data indicate that the long-term, developmentally regulated expression of the heteromeric ACCase is in part controlled by a mechanism(s) that coordinately affects the steady-state concentrations of each subunit mRNA.
De novo fatty acid synthesis is a fundamental process required for the biogenesis of membrane and storage lipids. Using acetyl-coenzyme A (CoA) as the initial primer, this process occurs by the sequential condensation of two-carbon units, which are derived from malonyl-CoA. The generation of malonyl-CoA from acetyl-CoA is the first committed step in fatty acid biosynthesis. This reaction is catalyzed by the biotin-containing enzyme acetyl-CoA carboxylase (ACCase).
ACCase catalyzes a two-step reaction that requires a non-catalytic biotin-containing component and two catalytic functions, all three of which are recognizable as conserved structural domains. These domains are: biotin carboxyl-carrier (BCC), biotin carboxylase (BCase), and carboxyltransferase (CTase). The biotin prosthetic group, which is covalently bound to the BCC subunit, is absolutely essential to the enzymatic function of ACCase as the intermediate carrier of the carboxyl group that carboxylates acetyl-CoA (Lane et al., 1974). The two half-reactions that result in the carboxylation of acetyl-CoA are: (1) carboxylation of the biotin prosthetic group, which is catalyzed by the BCase domain, and (2) transfer of the carboxyl group from carboxy-biotin to acetyl-CoA to form malonyl-CoA, which is catalyzed by the CTase domain.
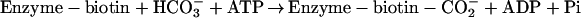 |
1 |
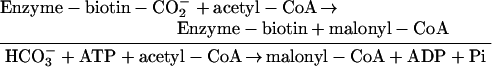 |
2 |
Two physically distinct ACCases have been identified in plants: the heteromeric and homomeric forms (Li and Cronan, 1992d; Gornicki et al., 1993; Sasaki et al., 1993, 1995; Alban et al., 1994; Konishi and Sasaki, 1994; Roesler et al., 1994; Schulte et al., 1994; Shorrosh et al., 1994, 1995, 1996; Choi et al., 1995; Yanai et al., 1995). These two enzymes differ in the quaternary organization of their structural domains. In the heteromeric ACCase, the three domains occur on separate dissociable proteins, whereas in the homomeric ACCase these domains are sequentially ordered on a single polypeptide. Heteromeric ACCases also occur in most eubacteria (Guchhait et al., 1974; Hunaiti and Kolattukudy, 1982; Kondo et al., 1991; Li and Cronan, 1992b, 1992c), whereas homomeric ACCases occur in the cytosol of fungi (Walid et al., 1992; Hablacher et al., 1993) and animals (Lopez-Casillas et al., 1988; Takai et al., 1988).
In contrast to other organisms, plants contain ACCases in two separate subcellular compartments, the plastids and the cytosol. In most plants (excluding Graminae), the heteromeric ACCase is plastidic, whereas the homomeric ACCase is cytosolic (Kannangara and Stumpf, 1972; Li and Cronan, 1992a; Sasaki et al., 1993; Konishi and Sasaki, 1994; Alban et al., 1994; Choi et al., 1995; Shorrosh et al., 1995, 1996; Konishi et al., 1996). Graminae do not contain the heteromeric ACCase, but contain homomeric isoforms in plastids and cytosol (Egli et al., 1993; Konishi et al., 1996; Gornicki et al., 1997). Because membranes are impermeable to acyl-CoAs (Jacobson and Stumpf, 1972; Benjamin et al., 1983; Banhegyi et al., 1996), these two ACCases generate physically isolated pools of malonyl-CoA. The plastidic malonyl-CoA pool is the precursor of de novo fatty acid biosynthesis, which produces 16 and 18 carbon fatty acids. The cytosolic malonyl-CoA pool is used for the elongation of these fatty acids to 20 carbons and longer. (In aerial portions of terrestrial plants these very-long-chain fatty acids are used for the biosynthesis of epicuticular waxes, and in seeds of some plants they are deposited in the seed oil.) In addition, cytosolic malonyl-CoA is utilized for the biosynthesis of flavonoids, stilbenoids, malonic acid, and malonyl derivatives (of d-amino acids, 1-aminocyclopropane carboxylic acid, etc.) (Conn, 1981; Nikolau et al., 1984). Therefore, to accommodate the multiplicity of malonyl-CoA-requiring processes and to regulate the supply of malonyl-CoA for these biosynthetic processes, plants have compartmentalized the generation of this intermediate at the cellular and subcellular level.
The plastidic heteromeric ACCase consists of four subunits: BCC, BCase, α-CT, and β-CT. To date, genes for two of these subunits have been characterized from Arabidopsis, the CAC1 gene, which codes for BCC (Choi et al., 1995; Ke et al., 1997), and the CAC2 gene, which codes for BCase (Bao et al., 1997; Sun et al., 1997). In this report, we describe the isolation and characterization of the CAC3 gene (GenBank accession no. AF056970), which codes for α-CT, and the plastidic accD gene (GenBank accession no. AF056971), which codes for β-CT. To approach the regulation of ACCase gene expression, we determined the spatial and temporal pattern of CAC1, CAC2, CAC3, and accD mRNA accumulation during Arabidopsis silique development. These studies indicate that the ACCase subunit mRNAs accumulate to highest levels in cells and tissues undergoing rapid growth and/or seed oil biogenesis. Furthermore, these studies establish that the subunit mRNAs accumulate at a constant molar stoichiometric ratio, which implies that the three nuclear and one plastidic ACCase gene must communicate in order to establish coordinate expression.
MATERIALS AND METHODS
Plant Materials
Arabidopsis Heynh (ecotype Columbia) seeds were germinated in sterile soil, and plants were grown under continuous light as described previously (Ke et al., 1997). The first three leaves of 17-d-old seedlings, flower buds, and open flowers were harvested and frozen in liquid N2 for RNA isolation or processed for in situ hybridization. Developing siliques were staged relative to the day of flowering (DAF). After the first two flowers had opened, subsequent flowers were tagged with colored threads at the time of flowering. Siliques were collected each day between 1 and 15 DAF, and were either frozen in liquid N2 for RNA isolation or processed for in situ hybridization.
Isolation of cDNA and Genomic Clones
The following materials were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus): (a) two Arabidopsis genomic libraries, one from the ecotype Landsberg erecta (Voytas et al., 1990) cloned in the vector λFIX, and the other from the ecotype Wassilewskija cloned in the cosmid vector pOCA18 (Olszewski et al., 1988); (b) a 3- to 6-kb size-selected cDNA library in the vector λZAP II prepared from polyadenylated RNA isolated from 3-d-old Arabidopsis (ecotype Columbia) seedling hypocotyls (Kieber et al., 1993); and (c) the expressed sequence tag cDNA clone GBGe16 (R. Mache, F. Quigley, F. Thomas, and D. Yu, unpublished data).
Hybridization screening of bacteriophage and cosmid libraries was performed according to standard methods (Sambrook et al., 1989). Clones that hybridized to 32P-labeled probes were plaque or colony purified. DNA restriction fragments that hybridized to the probes were isolated and subcloned into pBluescript SK for sequencing.
DNA sequencing was performed at the Iowa State University Nucleic Acids Facility with automated sequencing equipment from Applied Biosystems (Foster City, CA). Both strands of all DNA fragments were sequenced twice. DNA primers used for sequencing were synthesized at the Iowa State University Nucleic Acids Facility. Computer-based analysis of gene promoter sequences used the PLACE database (Higo et al., 1998).
Expression of Recombinant Proteins in Escherichia coli
Recombinant CAC3- and accD-coded proteins (α-CT and β-CT, respectively) were produced in E. coli using the pET30 expression vectors. This resulted in the production of chimeric recombinant polypeptides that contained a short N-terminal “tag,” which consists of a polyhistidyl sequence (Van Dyke et al., 1992), the S·Tag-peptide (Kim and Raines, 1993), and two protease cleavage sites (His·Tag-thrombin cleavage site-S·Tag-enterokinase cleavage site).
For CAC3, the NcoI-NotI fragment encompassing nucleotides 443 to 2,853 of the CAC3 cDNA (GenBank accession no. AF056969), which codes for the predicted mature polypeptide (from the 67th residue) was isolated. The NcoI overhang was filled-in with Klenow, and the DNA fragment was cloned in-frame with the N-terminal “tags” of the pET-30c expression vector at the unique EcoRV/NotI sites; the resulting plasmid was called pET-CAC3.
To express accD, the clone was subjected to PCR-based, site-directed mutagenesis to create a unique NcoI site at the translational start site. This was achieved with the primer 5′-GGCCAGAAGCTCCATGGAAAAA-3′, which is complementary to the sequence around the translation start codon, ATG, but contains an NcoI site (underlined). From the resulting mutagenized plasmid, the 1.67-kb NcoI-SacI accD-containing fragment was cloned in the appropriate sites of pET30a (the SacI site is vector-derived). The resulting plasmid was called pET-accD. The pET-derived recombinant plasmids were introduced into the E. coli strain BL21(DE3), and protein production was induced with isopropylthio-β-galactoside. Proteins were extracted from the cell pellets, fractionated into soluble and insoluble fractions, and the expressed proteins were purified by affinity chromatography with a His·Bind column, as described by the manufacturer (Novagen, Madison, WI). Recombinant expressed proteins were detected by SDS-PAGE and western analysis using S-protein alkaline phosphatase conjugate to identify the S·Tag.
Immunological Methods
Antibodies were generated in New Zealand White female rabbits by injecting with purified recombinant α-CT and β-CT proteins. Approximately 1 mg of purified protein was emulsified with Freund's complete adjuvant and injected intradermally at multiple sites on the backs of the rabbits. A month after the initial injection and at 2-week intervals thereafter, the rabbits were challenged each time with muscular injections of 0.5 mg of protein emulsified in Freund's incomplete adjuvant. One week after each of these latter injections, 2 to 3 mL of blood was withdrawn from the ear of each rabbit. The blood was allowed to coagulate, and the serum was collected after centrifugation at 4,000g for 10 min. The serum was stored in small aliquots at −20°C.
Immunological detection of proteins was carried out following SDS-PAGE (Laemmli, 1970). Proteins were electrophoretically transferred to a nitrocellulose membrane (Kyhse-Andersen, 1984) and detected using antisera and 125I-protein A.
For immunoinhibition studies, Arabidopsis leaf extracts were incubated on ice for 1 h with increasing amounts of either the anti-serum or preimmune serum. Equal aliquots of a suspension of Protein A-agarose were added to each reaction tube and the resulting mixtures were further incubated on ice for 1 h. Following centrifugation at 10,000g for 1 min, a sample of each supernatant was assayed for residual ACCase activity (Choi et al., 1995).
RNA Analysis
RNA was extracted from Arabidopsis leaves, buds, flowers, and siliques as described previously (Weaver et al., 1995). Non-radioactive sense RNA concentration standards of CAC1, CAC2, CAC3, and accD RNAs were obtained by in vitro transcription from the respective pBSK full-length cDNA clones. The RNA concentrations were determined from A280 and A260 and by comparison against standards of known concentration following ethidium bromide staining of gels.
Ten micrograms of RNA isolated from each Arabidopsis tissue sample, and a range (0.01–10 pg) of CAC1, CAC2, CAC3, and accD RNA standards were fractionated by electrophoresis in formaldehyde-containing agarose gels (Ke et al., 1997). After transfer of the RNA to nylon membranes (Magna Lift, MSI, Westbourough, MA), hybridizations were conducted in a buffer containing 50% (v/v) formamide at 65°C for 12 to 16 h using 32P-labeled probes. 32P-Labeled antisense RNA probes were transcribed from vectors containing the following inserts: the 730 nucleotides at the 3′ end of the CAC1 cDNA (Choi et al., 1995; Ke et al., 1997); the 195 nucleotides at the 5′ end of CAC2 cDNA (Sun et al., 1997); the CAC3 (GBGe16) cDNA; and the 1 kb at the 3′ end of the accD genomic clone. Hybridized membranes were rinsed twice with 2× SSC, 2% (w/v) SDS for 10 min each time at room temperature, and then washed twice with 0.1× SSC, 0.1% (w/v) SDS for 20 min each time at 65°C. The membranes were exposed to a phosphor screen (Molecular Dynamics, Sunnyvale, CA) for 4 h, and the radioactivity in each band was quantified with a phosphor imager (Storm 840, Molecular Dynamics). The amount of each mRNA in each tissue sample was quantified by comparing the intensities of the mRNA bands with those of the RNA concentration standards.
In Situ Hybridization
Arabidopsis siliques were harvested daily between 1 and 15 DAF. Siliques were cut into 3- to 4-mm-long pieces, fixed, and sectioned as previously described (Wang et al., 1995a; Ke et al., 1997). 35S-Labeled antisense and sense RNA probes were transcribed from vectors containing the following inserts: the 730 nucleotides at the 3′ end of the CAC1 cDNA (Choi et al., 1995; Ke et al., 1997); the 195 nucleotides at the 5′ end of CAC2 cDNA (Sun et al., 1997); the CAC3 (GBGe16) cDNA; and the 1 kb at the 3′ end of the accD genomic clone. After hybridization and washing, the tissue sections were coated with Kodak NTB2 emulsion (Eastman-Kodak, Rochester, NY), exposed for 2 to 4 d, and developed. Slides were stained with toluidine blue to detect cellular structure. Photographs were taken with a microscope (Orthopha, Leitz, Wetzlar, Germany) using bright-field optics. In situ hybridizations were repeated three times (using 20–30 silique sections each time). Two sets of plant materials that had been independently processed were used, all with similar results. Control slides containing sections of the same siliques were routinely hybridized with the four sense RNA probes simultaneously with the antisense RNA hybridizations. Virtually no signal was detected in the control slides. Although in situ hybridizations were carried out using material collected every DAF through d 15, for space considerations, data presented in figures are from alternate days only.
RESULTS
The α-CT Subunits of Plant Heteromeric ACCase Contain a Divergent C-Terminal Extension Relative to the Eubacterial α-CT Subunits
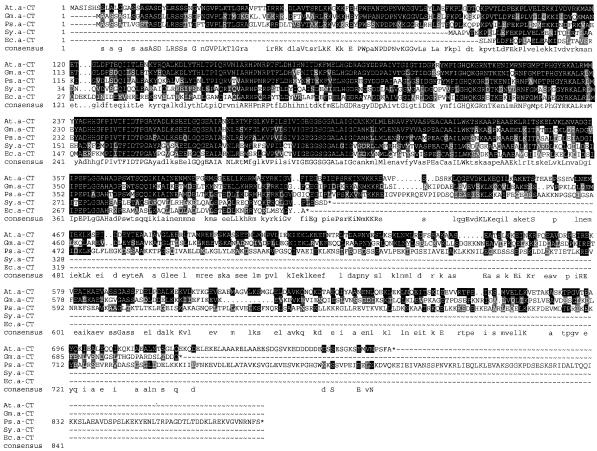
The 300-nucleotide sequence at the 5′ end of the Arabidopsis expressed sequence tag cDNA clone GBGe16 shows significant sequence similarity to the E. coli accA gene that codes for the α-CT subunit. Because GBGe16 is a partial cDNA, it was used as a probe to screen an Arabidopsis cDNA library (Kieber et al., 1993). This resulted in the isolation of 10 hybridizing clones. The largest of these, pCAC3, is 2,852 nucleotides in length and is a full-length copy of the CAC3 mRNA. Beginning at nucleotide 248 of the pCAC3 sequence is a 2,310-nucleotide open reading frame (ORF) that codes for a polypeptide of 769 amino acids with a calculated molecular mass of 85,305 D. This ORF is followed by a 295-nucleotide 3′-untranslated region. Comparison of the deduced amino acid sequence of the CAC3 ORF with previously published sequences indicated that the CAC3 protein shares highest sequence similarity with the α-CT subunit of the heteromeric ACCase of E. coli (Li and Cronan, 1992a), pea (Shorrosh et al., 1996), cyanobacterium (Gornicki et al., 1993), and soybean (Reverdatto et al., 1999) (Fig. 1).
Figure 1.
Primary structure of α-CT. The amino acid sequence of the Arabidopsis α-CT (At.α-CT) is compared with the corresponding sequences from soybean (Gm.α-CT), pea (Ps.α-CT), Synechocystis (Sy.α-CT), and E. coli (Ec.α-CT). Residues that are identical to those of the Arabidopsis sequence are shaded in black, whereas conservative substitutions are shaded in gray. The CONSENSUS sequence identifies residues that are either conserved in the Arabidopsis and any other presented sequence (lowercase) or are conserved in all presented sequences (uppercase).
Sequence similarities indicate that pCAC3 codes for the α-CT subunit of the heteromeric ACCase of Arabidopsis. However, because several biotin enzymes are present in plants (Wurtele and Nikolau, 1990; Guan et al., 1999), and the sequences of their carboxyltransferase subunits are unknown, the enzymatic function of the CAC3-encoded protein had to be determined. Therefore, we conducted immunoprecipitation assays to confirm that CAC3 codes for the α-CT subunit of the heteromeric ACCase. Antiserum directed against the E. coli-expressed CAC3 protein reacts with a single 85-kD polypeptide in Arabidopsis leaf extracts (Fig. 2A). The Mr of this polypeptide is very close to that predicted from the CAC3 sequence. Arabidopsis leaf extracts were incubated with increasing amounts of either the anti-CAC3 serum or preimmune serum, and then with protein A-agarose. Following centrifugation to remove antigen-antibody complexes, samples of the supernatants were assayed for residual ACCase activity. While up to 5 μL of preimmune serum had no effect on ACCase activity, 2 μL of anti-CAC3 serum was sufficient to remove 50% of the activity, and 5 μL of anti-CAC3 serum removed 65% of the ACCase activity (Fig. 2B). The remaining ACCase activity was resistant to immunoprecipitation, presumably due to the activity of the cytosolic ACCase isozyme, which is immunologically distinct from the heteromeric ACCase. These data demonstrate that CAC3 encodes the α-CT subunit of the heteromeric ACCase of Arabidopsis.
Figure 2.
CAC3 codes for the α-CT subunit of the heteromeric ACCase. A, Western-blot analysis of an Arabidopsis leaf extract probed with antiserum directed against bacterially produced recombinant protein coded by the CAC3 cDNA. A single, 85-kD polypeptide was detected. B, Increasing amounts of preimmune (○) or anti-CAC3 (●) serum were incubated with Arabidopsis leaf extracts. Following the addition of protein A-agarose, antigen-antibody complexes were removed by centrifugation. Supernatants were assayed for residual ACCase activity.
The α-CT protein contains an N-terminal extension of 87 amino acids, relative to the E. coli and Synechocystis accA sequences (Fig. 1). As expected, this extension has characteristics of a plastidic stromal-targeting transit peptide: it is rich in hydroxylated residues (Ser and Thr) and in small hydrophobic residues (Ala), but lacks acidic residues and thus has an overall positive charge (Keegstra et al., 1989). The PSORT algorithm (Nakai and Horton, 1999) predicts that α-CT is a chloroplast stromal protein.
Comparison of the Arabidopsis, pea, soybean, Synechocystis, and E. coli α-CT sequences indicates that the mature eukaryotic proteins can be divided into two domains, which are distinguished by their degree of sequence conservation. The N-terminal domain (residues 94–426 of the Arabidopsis α-CT) is highly conserved and matches the sequence of the eubacterial α-CT proteins.
In contrast, the C-terminal domain (residues 426–769 of the CAC3-coded α-CT) is less conserved among the plant proteins and is absent from the eubacterial α-CT proteins.
The first 250 residues of this C-terminal domain (residues 426–680 in Arabidopsis) have very similar hydrophobicity profiles despite substantial sequence divergence. The remainder of these proteins (from residue 681 of Arabidopsis) are extremely divergent with regard to both length (Arabidopsis = 89 residues; pea = 160 residues; soybean = 40 residues) and sequence. A common theme throughout these C-terminal domains is the occurrence of nested repetitive sequences, although the specific sequences and relative positions of these repetitions are not conserved among the plant α-CTs. The largest of these repetitive sequences occurs in the pea α-CT, and the duplication partially overlaps itself (residues 453–489 are duplicated at positions 481–517, with 83% conservation between the duplications). This repeat is flanked by a second duplication (residues 438–458 are duplicated at positions 521–541, with 81% conservation). A shorter repeat motif that occurs in the C-terminal domain of all three plant α-CTs has the sequence [hyph][E/D]-[K/R]- [hyph]- [K/R], where “hyph” stands for any hydrophobic residue. This sequence occurs four, three, and six times in the Arabidopsis, soybean, and pea sequences, respectively. Imperfectly repeated motifs rich in E, L, and K residues are present seven, six, and 13 times in the Arabidopsis, soybean, and pea sequences, respectively.
The relative lack of sequence conservation among the plant-specific C-terminal domains, and its absence from the eubacterial α-CTs, indicates that this domain may not be required for catalysis, but, rather, may have some as-yet-unknown structural function. Consistent with this hypothesis is the finding that despite the reduced sequence identity between the plant C-terminal domains, they are predicted to share common secondary structural features, i.e. similar hydrophobicity profiles, an overabundance of charged residues (and thus high hydrophilicity), and nested repetitive sequences. An additional feature consistent with these domains having a structural role is that they are predicted by the PROTEINPREDICT algorithm (Rost and Sander, 1993) to be rich in α-helices and not be in a globular configuration. Finally, this plant-specific C-terminal domain shares sequence similarity with myosin and other structural proteins.
α-CT and β-CT Subunits Are Physically Associated
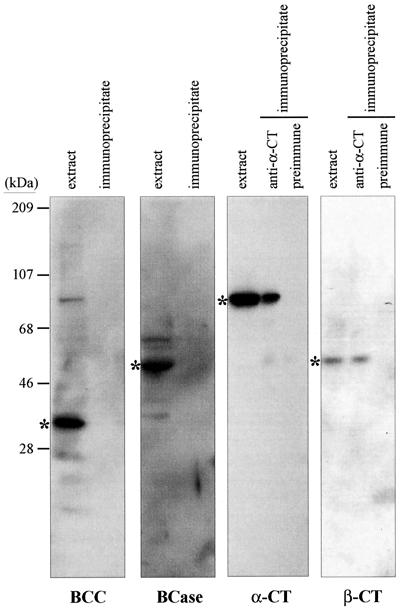
The heteromeric ACCase of E. coli readily dissociates into three components: BCC, BCase, and CTase. The CTase component is a heteromeric tetramer with an α4β4 quaternary structure (Guchhait et al., 1974; Li and Cronan, 1992a). The quaternary organization of the plant heteromeric ACCase is still unresolved; however, the pea enzyme appears to dissociate into at least two components, one containing the BCC and BCase subunits and the second containing the α-CT and β-CT subunits (Sasaki et al., 1993; Alban et al., 1994; Choi et al., 1995; Shorrosh et al., 1995, 1996). To ascertain whether the α-CT subunit is associated with other subunits of the heteromeric ACCase of Arabidopsis, crude extracts and immunoprecipitates (obtained as described for Fig. 2B) were analyzed by western blotting using antisera against each of the heteromeric ACCase polypeptides (Fig. 3). The α-CT and β-CT subunits, but not the BCC or BCase subunits, were co-immunoprecipitated by the anti-α-CT serum. These data indicate that the α-CT and β-CT subunits are in a more tightly associated complex with each other than with the BCC and BCase subunits, which is consistent with the previous characterizations of the pea and E. coli heteromeric ACCase.
Figure 3.
Physical association between the α-CT and β-CT subunits of the heteromeric ACCase. Preimmune or anti-CAC3 serum was incubated with Arabidopsis leaf extracts. Antibody-antigen complexes were bound to protein A-agarose and collected by centrifugation. The resulting pellets were boiled in the presence of 2% (w/v) SDS and, following centrifugation, aliquots of the supernatants were subjected to western-blot analyses. These blots were sequentially probed with anti-BCC, anti-BCase, anti-α-CT, and anti-β-CT sera. The positions of the BCC, BCase, α-CT, and β-CT polypeptides are indicated by asterisks (*).
α-CT Subunit Is a Stromal Protein Whose Localization Is Unaffected by Illumination
The α-CT subunit of pea heteromeric ACCase was originally identified as a chloroplast inner envelope protein of unknown function (Hirsch and Soll, 1995). Consistent with this, fractionation of pea leaf extracts indicated that the α-CT subunit is primarily (about 90%) associated with cellular membranes (Shorrosh et al., 1996). It is not clear how to reconcile findings that α-CT subunit is located in the inner plastid membrane (Shorrosh et al., 1996), whereas ACCase activity, BCase, and BCC are soluble (Sasaki et al., 1993; Alban et al., 1994; Shorrosh et al., 1995; Ke et al., 1997). Because the α-CT and β-CT subunits are in a complex (see Fig. 3), it is possible that this CTase complex is membrane associated under some conditions in the plant, while the BCC and BCase subunits remain soluble.
We determined directly whether the α-CT subunit of Arabidopsis is a membrane-bound polypeptide. In addition, because of indications that light affects the activity of ACCase (Sauer and Heise, 1984; Sasaki et al., 1997; Hunter and Ohlrogge, 1998; Kozaki and Sasaki, 1999), we surmised that light might effect a possible association of the α-CT subunit with membranes. Therefore, we examined the location of the α-CT subunit in illuminated and in dark-treated plants. Extracts were prepared from leaves of Arabidopsis plants that had either been grown under continuous illumination for 11 d or under continuous illumination for 10 d and then transferred to darkness for 24 h. These extracts were fractionated by centrifugation at 100,000g for 1 h, and the supernatant and pellet (50 μg of protein each) were subjected to SDS-PAGE and western-blot analysis with anti-α-CT serum. Virtually all of the α-CT subunit protein was recovered in the soluble fraction, regardless of the illumination status of the plants from which the extract was prepared (Fig. 4). These data indicate that the α-CT subunit of Arabidopsis is a soluble, stromal protein and its solubility is unaffected by illumination.
Figure 4.
α-CT subunit of the heteromeric ACCase is not membrane associated. Arabidopsis plants were grown either under continuous illumination for 11 d (L) or under continuous illumination for 10 d and subsequently moved to a dark chamber for 1 d (D). Proteins extracted from these two sets of plants were separated into a soluble (S) and membrane-bound (M) fraction by centrifugation. Aliquots of these two fractions containing equal amounts of protein (50 μg) were subjected to SDS-PAGE and western analysis with anti-α-CT serum.
Characterization of the CAC3 Gene
Approximately 40,000 recombinant bacteriophage from an Arabidopsis genomic library (Voytas et al., 1990) were screened by hybridization with the pGBGe16 cDNA. Five of 11 hybridizing clones were plaque purified and further analyzed. Restriction digests and Southern-blot hybridization analyses using 5′- and 3′-end-specific probes from the CAC3 cDNA revealed that these five clones contained overlapping segments of the Arabidopsis genome, and that two of these clones contained both ends of the CAC3 gene; inserts from these phage were subcloned into pBluescript SK and sequenced.
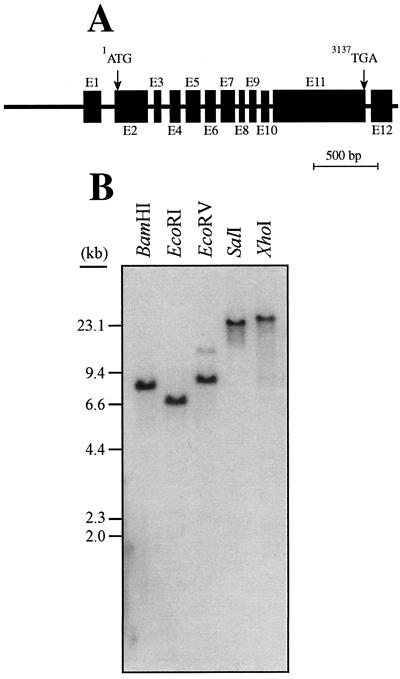
In total, more than 6-kb of a contiguous stretch of Arabidopsis genome was sequenced. Comparison of this sequence to the CAC3 cDNA sequence identified the structure of the CAC3 gene (Fig. 5A). The CAC3 gene is interrupted by 11 introns that range from 73 to 203 nucleotides in length. The nucleotide sequences at the intron-exon-intron junctions follow characteristic patterns observed in other plant genes (Brown, 1989; Ghislain et al., 1994). The exception to this rule is the 3′-end of intron 3, which has the sequence 5′-TCC↓G.
Figure 5.
The CAC3 gene of Arabidopsis, which codes for the α-CT subunit of the heteromeric ACCase. A, Schematic representation of a 6,280-bp Arabidopsis genomic fragment that contains the CAC3 gene. This fragment contains the potential promoter of the CAC3 gene (1,056-bp sequence 5′ of the CAC3 coding sequence). Positions of the translational start (1ATG) and stop (3137TGA) codons are indicated. Nucleotides are numbered relative to the translational start codon. Exons are represented by shaded boxes and their positions are: E1 (−450 to −269; E2 (−65–339); E3 (441–515); E4 (649–756); E5 (849–1,009); E6 (1,103–1,217); E7 (1,299–1,453); E8 (1,539–1,590); E9 (1,672–1,740); E10 (1,829–1,906); E11 (1,980–3,159); and E12 (3,244–3,494). Non-exonic sequences are represented by solid lines. B, Arabidopsis DNA was digested with the indicated restriction endonucleases and subjected to Southern-blot analysis. The blot was probed with 32P-labeled CAC3 cDNA.
Interestingly, the first and last introns of the CAC3 gene are positioned within the 5′- and 3′-untranslated regions of the gene; this is a rather rare occurrence and its significance is still unknown. The portions of the Arabidopsis α-CT protein that are missing from the eubacterial α-CT proteins are coded by exon 2 and exon 11 of the CAC3 gene. Exon 2 codes for the plastidic transit peptide, and would be expected to have been acquired after the event that led to endosymbiotic acquisition and refinement of the plastid. Also intriguing is the indication that the entire C-terminal domain of the plant α-CT subunit, which is absent from eubacterial α-CT proteins, appears to have been due to a post-endosymbiotic acquisition of a single exon (exon 11).
Southern-blot analysis of Arabidopsis genomic DNA shows single hybridizing bands of the size predicted from the CAC3 sequence for BamHI, EcoRI, SalI, and XhoI digests (Fig. 5B). Furthermore, EcoRV digestion generates two hybridizing bands of different intensities, which is consistent with the CAC3 gene sequence, which contains a unique EcoRV site near the 3′-end of the gene. These data indicate that the CAC3 gene occurs only once in the Arabidopsis genome.
Isolation of the accD Gene Coding for the β-CT Subunit of the Arabidopsis Heteromeric ACCase
To isolate the Arabidopsis plastid-encoded accD gene, a cosmid Arabidopsis library (Olszewski et al., 1988) was screened by hybridization with an accD-containing DNA fragment from tobacco (the 780-bp XmnI-BamHI DNA fragment from position 59, 876–60, 655 of the tobacco chloroplast genome). This resulted in the isolation of a single hybridizing clone, from which a 2.95-kb HindIII-KpnI accD-hybridizing fragment was sequenced. This fragment contains an incomplete 98-bp ORF at its HindIII-end, identical to the C-terminal 32 residues of rbcL (Ohyama et al., 1986; Shinozaki et al., 1986), followed by an ORF of 1,464 bp. The amino acid sequence of this 1,464-bp ORF shares 37% identity with E. coli accD polypeptide, and 48%, 67%, and 91% identity with the pea, tobacco, and rape accD polypeptides, respectively (Wen, 1997). These results indicate that this 2.95-kb fragment is from the Arabidopsis plastid genome and includes the accD gene. Antiserum made to the expressed protein coded by accD reacts with a single 55-kD polypeptide in a leaf extract from Arabidopsis, and this antiserum specifically inhibits ACCase activity, directly confirming that the gene encodes the β-CT subunit of the heteromeric ACCase (Wen, 1997).
Coordinate Accumulation of the CAC1, CAC2, CAC3, and accD mRNAs
As a first step in elucidating the mechanisms that regulate the heteromeric ACCase in Arabidopsis, we investigated the temporal changes in the accumulation of the four ACCase subunit mRNAs (CAC1, CAC2, CAC3, and accD) during silique development and compared this pattern with the accumulation of these mRNAs in young, expanding leaves. For each mRNA, we compared the intensity of each northern-blot hybridization band with hybridization intensities obtained from serial dilutions of in vitro-generated RNA standards of known concentrations. By including such standards, we were able to absolutely quantify the accumulation of each ACCase subunit mRNA.
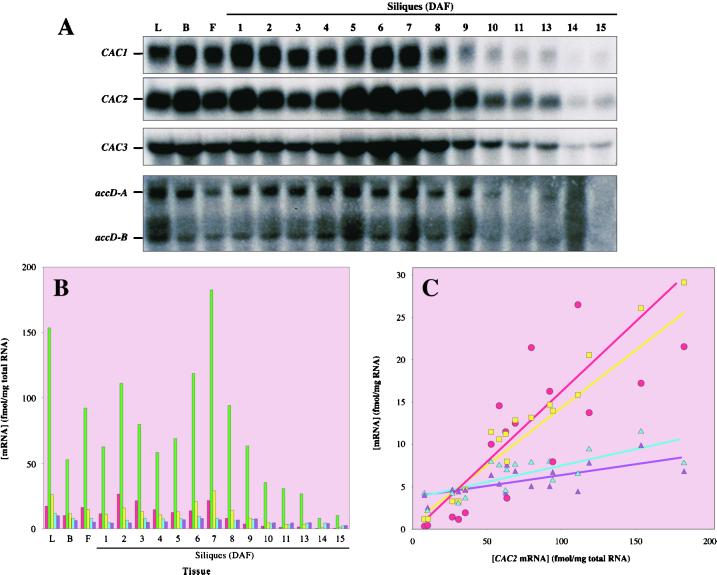
Hybridization of northern blots with the CAC1, CAC2, and CAC3 antisense RNAs revealed the accumulation of each of the corresponding mRNAs (Fig. 6A). The sizes of these mRNAs (1.1, 2.0, and 2.8 kb, respectively) are consistent with previous characterizations (Choi et al., 1995; Bao et al., 1997; Sun et al., 1997) and the size of the full-length CAC3 cDNA. Hybridizations with the accD probe revealed two mRNAs, accD-A (2.3 kb) and accD-B (1.5 kb). This was not an unexpected finding, as multiple accD mRNAs had previously been detected in pea and Arabidopsis leaves (Woodbury et al., 1988; Meurer et al., 1996) and rape seed embryos (Elborough et al., 1996). Both the accD-A andaccD-B mRNAs are sufficiently large to encode the β-CT subunit. The significance of multiple accD transcripts in the regulation of ACCase expression is not clear, but both the accD-A and accD-B mRNAs accumulate with a similar pattern.
Figure 6.
Coordinate temporal changes in accumulation of mRNAs coding the heteromeric ACCase subunits. A, RNA blots were hybridized with CAC1-, CAC2-, CAC3-, and accD-specific 32P-labeled antisense RNA probes. RNA was isolated from young expanding leaves (L), flower buds (B), flowers (F), and developing siliques at the indicated DAF. B, The concentration of the CAC1, CAC2, CAC3, and accD mRNAs was determined by comparing the intensity of hybridization of each mRNA to the hybridization intensity obtained with RNA concentration standards, as described in “Materials and Methods.” The average values from three independent determinations are shown. The sd for each determination ranged between 10% and 15% of the indicated values. CAC1 (red), CAC2 (green), CAC3 (yellow), accD-A (blue), or accD-B (purple) mRNAs. C, For each of the tissues analyzed in A and B, the concentrations of the CAC1 (red), CAC3 (yellow), accD-A (blue), or accD-B (purple) mRNAs are plotted against the concentration of the CAC2 mRNA. Linear regression analyses indicate that there is a linear relationship between the concentrations of each of these mRNAs with correlation coefficients of greater than 0.85.
During silique development, there are two maxima in the accumulation of the ACCase subunit mRNAs (Fig. 6, A and B). The first occurs at 2 DAF, when siliques are most rapidly expanding, and the second at 7 DAF, a time of near-maximal oil accumulation in the embryos. Subsequently, as siliques mature (9–12 DAF), the accumulation of these mRNAs declines to about 5% of peak levels. These modulations in steady-state levels appear to occur coordinately for all five ACCase mRNAs.
This coordination is even more evident from analysis of the data, as shown in Figure 6C. For each sample point shown in Figure 6B, the concentrations of the CAC1, CAC3, accD-A, and accD-B mRNAs are plotted against the concentration of the CAC2 mRNA (Fig. 6C). These plots reveal the linear relationship between the concentration of the CAC1, CAC3, accD-A, or accD-B mRNAs, and the concentration of the CAC2 mRNA, indicating that the ACCase subunit mRNAs accumulate at a constant molar ratio. The molar ratio between these mRNAs was calculated to be CAC1:CAC2:CAC3:accD-A:accD-B = 0.14:1.0:0.17:0.04:0.02. This ratio is also maintained in organs other than developing siliques, namely, expanding leaves, flower buds, and flowers.
In addition, we used the quantitative data in Figure 6 to calculate the absolute abundance of each of the ACCase mRNAs. These range from 0.5 to 170 fmol/mg total RNA. Presuming that an average RNA is 2,000 bases, 10 fmol/mg of total RNA is equivalent to 0.00066 mol % of total RNA. (If 1% of the total RNA is mRNA, this figure translates to 0.066 mol % of cellular mRNA. At its highest levels, the CAC2 mRNA accumulates at up to 1 mol % of mRNA.)
Silique Development Induces Coordinate Changes in the Spatial and Temporal Patterns of CAC1, CAC2, CAC3, and accD mRNA Accumulation
To obtain more detailed insights into the regulation of ACCase expression, we investigated the spatial distribution of the CAC1, CAC2, CAC3, and accD mRNAs in developing siliques by in situ hybridization (Fig. 7). These analyses indicated that silique development induced both quantitative and qualitative changes in the accumulation of the ACCase subunit mRNAs. Most dramatically, development coordinately affected changes in the spatial distribution of all of these ACCase mRNAs.
Figure 7.
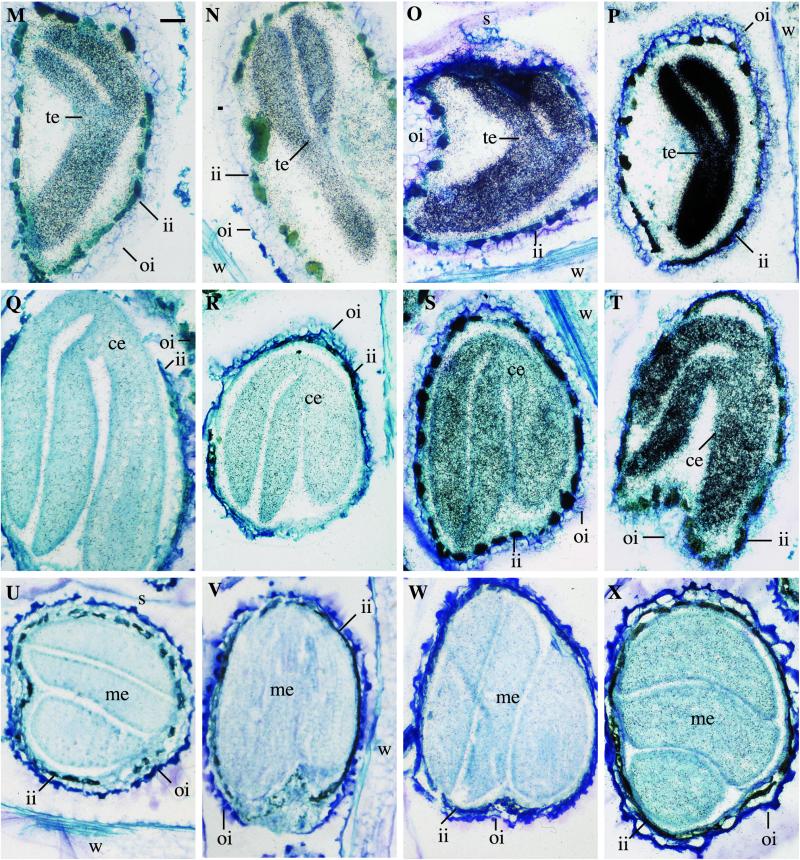
Cellular distribution of mRNAs coding the heteromeric ACCase subunits in developing siliques of Arabidopsis. Silique sections containing ovules and enclosed embryos were hybridized with 35S-labeled antisense RNA probes as described in “Materials and Methods.” Hybridizations depict the accumulation of: CAC1 mRNA (A, E, I, M, Q, and U); CAC2 mRNA (B, F, J, N, R, and V); CAC3 mRNA (C, G, K, O, S, and W); and accD mRNA (D, H, L, P, T, and X). Siliques are sampled at 1 DAF (A–D); 3 DAF (E–H); 5 DAF (I–L); 7 DAF (M–P); 9 DAF (Q–T); and 12 DAF (U–X). Sections are stained with toluidine blue O. Hybridization signal is observed as black spots. w, Silique wall; ii, inner integument of ovule; oi, outer integument of ovule; o, ovule; ge, globular embryo; he, heart embryo; te, torpedo embryo;ce, curled embryo; me, mature embryo; s, central septum. Bar = 65 μm.
In very young siliques (1 DAF; Fig. 7, A–D), which are rapidly expanding, all four of the ACCase mRNAs are evenly distributed among the silique tissues, including the cells of the silique walls, central septum, and developing ovules (integuments and endosperm). In 3 DAF siliques (Fig. 7, E–H), which are almost fully expanded, the accumulation of ACCase mRNAs is reduced in the silique walls, integuments of the ovules, and the central septum compared with 1 DAF siliques. In these 3 DAF siliques, the embryo within the ovule has grown to the globular stage of development and is of sufficient size that it is readily apparent; the accumulation of the ACCase mRNAs is concentrated within these embryos. By 5 DAF, the embryos have developed to the heart stage and are beginning to accumulate oil; the ACCase mRNAs are highly concentrated within these embryos (Fig. 7, I–L). In the other silique tissues (silique walls, integuments of the ovules, and the central septum), the accumulation of the ACCase mRNAs is barely above background. From 7 DAF and later, ACCase mRNAs are not detectable in the non-embryo tissues of the siliques, and the entire hybridization signal for each mRNA is concentrated solely within the embryo (Fig. 7, M–P). Maximal accumulation of the ACCase mRNAs within the developing embryo occurs at the torpedo stage of development (i.e. in siliques at 7 DAF). At this stage, the rate of oil accumulation in the embryos is maximal. Subsequently, as the siliques mature, the embryos within develop to the walking stick stage by 9 DAF (Fig. 7, Q–T) and to maturity by 12 DAF (Fig. 7, U–Y), just prior to the initiation of desiccation. During these later stages of development, the accumulation of all of the ACCase mRNAs declines in the embryo.
DISCUSSION
In plants, de novo fatty acid biosynthesis from acetyl-CoA occurs solely in plastids. Although many studies have attempted to ascertain whether ACCase has a major role in controlling plant fatty acid biosynthesis, it is not yet clear how this process is regulated. This is in part due to the fact that plants contain two ACCases that are differentially compartmentalized at the subcellular level. Furthermore, at least in non-Graminae species, these two ACCase isozymes are structurally distinct and the plastidic isozyme is heteromeric, unstable, and dissociates during isolation (e.g. Sasaki et al., 1993; Alban et al., 1995; Shorrosh et al., 1996). These complicating characteristics, which have become apparent only in the last 6 years, have precluded the direct demonstration that ACCase regulates fatty acid biosynthesis in plants.
To address the questions of whether and how ACCase may regulate de novo fatty acid biosynthesis, we have focused on the characterization of the heteromeric ACCase of Arabidopsis. We report the isolation of the CAC3 and accD genes and the CAC3 cDNA. In combination with previous characterizations of the CAC1 (Choi et al., 1995; Ke et al., 1997) and CAC2 (Bao et al., 1997; Sun et al., 1997) genes, this work completes the isolation of all four genes that code for the heteromeric ACCase of Arabidopsis. This body of work shows that in Arabidopsis, each of the four ACCase subunits is probably encoded by single-copy genes, three of which are nuclear and one of which is plastidic. This simplified genetic organization compared with other plant species (Shorrosh et al., 1995; Elborough et al., 1996; Reverdatto et al., 1999) should facilitate the molecular dissection of the mechanism by which ACCase is regulated.
Investigations to date indicate that ACCase regulation is complex (Post-Beittenmiller et al., 1991, 1992; Shintani and Ohlrogge, 1995; Ke et al., 1997; Roesler et al., 1997; Roughan, 1997; Sun et al., 1997; Caffrey et al., 1998) and may encompass many different levels of control. Potential post-translational regulatory mechanisms that affect ACCase activity on a short time frame include the biotinylation of the BCC subunit (as is the case for the biotin-containing subunit of 3-methylcrotonyl-CoA carboxylase; Wang et al., 1995b); the targeting, import, and assembly of the three nuclear encoded subunits (BCC, BCase, and α-CT) with the plastid encoded subunit (β-CT); phosphorylation (Savage and Ohlrogge, 1999); and the biochemical modulation of enzymatic activity (Eastwell and Stumpf, 1983; Sasaki et al., 1997; Hunter and Ohlrogge, 1998; Kozaki and Sasaki, 1999).
In contrast, developmentally induced modulations of ACCase occur in a longer time frame and are likely to be the consequence of the regulation of pre-translational processes. During seed maturation, fatty acids are rapidly biosynthesized and deposited as seed oils. To gain insight into how ACCase expression is regulated during this development, we examined the temporal and spatial patterns of ACCase mRNA accumulation in developing siliques.
These data indicate that the CAC1, CAC2, CAC3, and accD mRNAs accumulate to maximal levels in cells just prior to or at the stage when they would be expected to be actively synthesizing fatty acids. Furthermore, the accumulation of these mRNAs declines when fatty acid biosynthesis would be expected to be decreasing. Specifically, siliques undergo rapid growth and expansion early in their development, between 1 and 2 DAF (Bowman, 1994). During this time, the cells of the silique walls, central septum, and integuments would be expected to be synthesizing fatty acids, which would be used for the deposition of membranes to support the growth of the silique. Thus, the finding that the maximal accumulation of the ACCase mRNAs in the non-embryonic cells of the silique occurs at 1 DAF is consistent with the expected demand of malonyl-CoA generation needed for membrane lipid biogenesis by these cells.
By 3 DAF, when the growth of the silique and associated membrane deposition decrease, the accumulation of the ACCase mRNAs declines in the non-embryonic cells of the siliques. In contrast, between 3 and 9 DAF, the embryos within the developing seeds undergo transition from globular embryos of several hundred cells to mature embryos containing thousands of cells. Juxtaposed on this embryonic growth is the accumulation of oil, which occurs at maximal rates at between 5 and 8 DAF (Mansfield and Briarty, 1991, 1992; Bowman, 1994). By maturity, about 30% of the fresh weight of the seed is oil (H.R. Qian, E. Wurtele, and B. Nikolau, unpublished data). Consistent with the expected demand for malonyl-CoA generation in the embryos, the accumulation of the ACCase mRNAs in the embryos is substantial between 3 and 5 DAF and peaks by 7 DAF. Subsequently, as the rate of oil deposition declines, the accumulation of these mRNAs decreases. Thus, the highest accumulation of the ACCase mRNAs occurs in cells undergoing rapid cell division and growth and/or accumulating large quantities of oil.
The spatial and temporal pattern of accumulation of the ACCase mRNAs contrasts with patterns of mRNAs coding for enzymes not related to fatty acid biosynthesis. These include mRNAs coding for seed storage proteins (Raynal et al., 1999), and the metabolic enzymes methylcrotonyl-CoA carboxylase (J. Ke, B. Nikolau, and E. Wurtele, unpublished data), acetyl-CoA synthetase, homomeric acetyl-CoA carboxylase, ATP-citrate lyase, and biotin synthase (Ke, 1997). However, the accumulation pattern of ACCase mRNAs is nearly identical to that of plastidic pyruvate dehydrogenase, which is consistent with a role of this enzyme in generating acetyl-CoA for fatty acid biosynthesis in seeds (Ke, 1997; Wurtele et al., 1997).
These analyses indicate that during silique development, the temporal and spatial patterns of CAC1, CAC2, CAC3, and accD mRNA accumulation are coordinately altered. This finding contrasts with earlier work finding that the accumulation of BCase, BCC, and α-CT subunit proteins does not maintain a strict stoichiometric ratio when the expression of the BCase is up-regulated or down-regulated in leaves of transgenic tobacco (Shintani et al., 1997). The difference in the conclusions of these two studies may be due to differences in experimental systems and/or could reflect more complex layered regulatory mechanisms.
The quantitative analyses presented herein indicate that CAC2 mRNA accumulates to levels about 7-fold higher than the CAC1 and CAC3 mRNAs, and 15-fold higher than the accD mRNAs (accD-A plus accD-B). If the stoichiometric ratio of these mRNAs reflects even in part the levels of the respective subunit proteins, the BCase subunit would be present in excess. Our data substantiate the suggestion of Shintani et al. (1997) that the BCase subunit accumulates in excess, and explains why a substantial decrease in the accumulation of the BCase subunit is required to bring about even a conditional phenotype in transgenic tobacco.
Our data indicate that during silique development, ACCase expression is regulated by mechanism(s) that coordinately affect the steady-state concentrations of each of the ACCase mRNAs. Reporter transgene analyses of the CAC2 (Bao et al. 1997) and CAC1 (Choi et al., 1997; J.-K. Choi and B. Nikolau, unpublished data) promoters implicate transcriptional regulation as an important mechanism that establishes the changing patterns in the accumulation of these mRNAs. Maintaining transcriptional coordination would require communication between the three nuclear ACCase subunit genes (CAC1, CAC2, and CAC3) and the plastidic subunit gene (accD). This communication, at least between the three nuclear genes, could be mediated by common trans-acting factor(s) that interact directly or indirectly with each of the three nuclear ACCase genes and coordinately regulate their transcription.
If there is a direct interaction between this hypothesized coordinating factor(s) and each nuclear ACCase gene, a binding site for the coordinating factor(s) would occur on all three nuclear ACCase genes and may be recognizable as a conserved nucleotide sequence motif. Indeed, the promoters of the CAC1 (Ke et al., 1997), CAC2 (Bao et al., 1997; Sun et al., 1997), and CAC3 genes have several potential transcriptional regulatory motifs in common. These include: multiple E-boxes, which are important for light- and seed-specific expression (Stalberg et al., 1996); Gt1 consensus sequences, which are involved in light regulation (Zhou, 1999); and a motif that is common to promoters of napin genes of B. napus and binds nuclear protein(s), but has no established function (Ericson et al., 1991). Detailed molecular studies are required to functionally identify whether one or more of these motifs provide binding sites for coordinating regulatory factor(s). That each ACCase subunit in Arabidopsis is probably coded by a single-copy gene provides a major advantage in using this species for investigating coordinating mechanisms of ACCase expression.
Mechanisms to explain the coordination between the nuclear (CAC1, CAC2, and CAC3) and the plastidic (accD) ACCase subunit genes involve additional complexities associated with two distinct subcellular genomes and two distinct subcellular compartments. The interplay between nuclear- and plastid-coded genetic information generates the wide variety of plastids that occur in different organs and tissues (e.g. chloroplasts, amyloplasts, etioplasts, chromoplasts, and oilseed plastids). Parallel to these developmentally induced changes in plastid morphology and function are concomitant changes in macromolecular composition. Most studies of plastid development have focused on chloroplasts, in particular using photosynthetic genes as molecular tools (Leon et al., 1998; Pyke, 1999). Because the heteromeric ACCase accumulates in many plastid types, studies of its biogenesis can provide the means for elucidating the mechanisms of nuclear-plastid interactions that control the development of a variety of plastids. Specifically, ACCase can be useful in understanding the development of seed plastids and the mechanisms that influence the partitioning of carbon into starch and oil in seeds.
Footnotes
This work was supported by the Hatch Act and State of Iowa funds and by a U.S. Department of Agriculture-National Research Initiative competitive grant (no. 97–01912 to B.J.N. and E.S.W.), by a grant from the Iowa Soybean Promotion Board (to E.S.W. and B.J.N.), and by a research award to J.K. from the Iowa State University Molecular, Cellular, and Developmental Biology graduate program. This is journal paper no. J–18656 of the Iowa Agriculture and Home Economics Experiment Station, Ames, project nos. 2,997 and 2,913.
LITERATURE CITED
- Alban C, Baldet P, Douce R. Localization and characterization of two structurally different forms of acetyl-CoA carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxyproprionate herbicides. Biochem J. 1994;300:557–565. doi: 10.1042/bj3000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alban C, Julien J, Job D, Douce R. Isolation and characterization of biotin carboxylase from pea chloroplasts. Plant Physiol. 1995;109:927–935. doi: 10.1104/pp.109.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banhegyi G, Csala M, Mandl J, Burchell A, Burchell B, Marcolongo P, Fulceri R, Benedetti A. Fatty acyl-CoA esters and the permeability of rat liver microsomal vesicles. Biochem J. 1996;320:343–344. doi: 10.1042/bj3200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Shorrosh BS, Ohlrogge JB. Isolation and characterization of an Arabidopsis biotin carboxylase gene and its promoter. Plant Mol Biol. 1997;35:539–550. doi: 10.1023/a:1005881006620. [DOI] [PubMed] [Google Scholar]
- Benjamin AM, Murthy CR, Quastel JH. Calcium-dependent release of acetyl-coenzyme A from liver mitochondria. Can J Physiol Pharmacol. 1983;61:154–158. doi: 10.1139/y83-023. [DOI] [PubMed] [Google Scholar]
- Bowman J. Arabidopsis. An Atlas of Morphology and Development. New York: Springer-Verlag; 1994. [Google Scholar]
- Brown JW. A catalogue of splice junction and putative branch point sequences from plant introns. Nucleic Acids Res. 1989;14:9549–9559. doi: 10.1093/nar/14.24.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey JJ, Choi J-K, Wurtele ES, Nikolau BJ. Tissue distribution of acetyl-CoA carboxylases in leaves of leek (Allium porrum L.) J Plant Physiol. 1998;153:265–269. [Google Scholar]
- Choi J-K, Ke J, McKean AL, Weaver LM, Wen T-N, Sun J, Diez T, Yu F, Guan X, Wurtele ES, Nikolau BJ. Molecular biology of biotin-containing enzymes required in lipid metabolism. In: Williams JP, Khan MU, Lem NW, editors. Physiology, Biochemistry and Molecular Biology of Plant Lipids. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 363–367. [Google Scholar]
- Choi J-K, Yu F, Wurtele ES, Nikolau BJ. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of the chloroplastic acetyl-coenzyme A carboxylase. Plant Physiol. 1995;109:619–625. doi: 10.1104/pp.109.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn EE. The Biochemistry of Plants: A Comprehensive Treatise. 7, Secondary Plant Metabolism. New York: Academic Press; 1981. [Google Scholar]
- Eastwell K, Stumpf PK. Regulation of plant acetyl-CoA carboxylase by adenylate nucleotides. Plant Physiol. 1983;72:50–55. doi: 10.1104/pp.72.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli MA, Gengenbach BG, Gronwald JW, Somers DA, Wyse DL. Characterization of maize acetyl-coenzyme A carboxylase. Plant Physiol. 1993;101:499–506. doi: 10.1104/pp.101.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elborough KM, Winz R, Deka RK, Markham JE, White AJ, Rawsthone SR, Slabas AR. Biotin carboxyl carrier protein and carboxyltransferase subunits of the multi-subunit form of acetyl-CoA carboxylase from Brassica napus: cloning and analysis of expression during oilseed rape embryogenesis. Biochem J. 1996;315:103–112. doi: 10.1042/bj3150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson ML, Muren E, Gustavsson H-O, Josefsson L-G, Rask L. Analysis of the promoter region of napin genes from Brassica napus demonstrates binding of nuclear protein in vitro to a conserved sequence motif. Eur J Biochem. 1991;197:741–746. doi: 10.1111/j.1432-1033.1991.tb15966.x. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Frankard V, Vandenbossche D, Matthews BF, Jacobs M. Molecular analysis of the aspartate kinase-homoserine dehydrogenase gene from Arabidopsis thaliana. Plant Mol Biol. 1994;24:835–851. doi: 10.1007/BF00014439. [DOI] [PubMed] [Google Scholar]
- Gornicki P, Faris J, King I, Podkowinski J, Gill B, Haselkorn R. Plastid-localized acetyl-CoA carboxylase of bread wheat is encoded by a single gene on each of the three ancestral chromosome sets. Proc Natl Acad Sci USA. 1997;94:14179–14184. doi: 10.1073/pnas.94.25.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornicki P, Scappino LA, Haselkorn R. Genes for two subunits of acetyl coenzyme A carboxylase of Anabaena sp. strain PCC7120: biotin carboxylase and biotin carboxyl carrier protein. J Bacteriol. 1993;175:5268–5272. doi: 10.1128/jb.175.16.5268-5272.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Diez T, Prasad KT, Nikolau BJ, Wurtele ES. Discovery and characterization of geranoyl-CoA carboxylase in plants. Archives Biochem Biophys. 1999;362:12–21. doi: 10.1006/abbi.1998.1014. [DOI] [PubMed] [Google Scholar]
- Guchhait RB, Polakis SE, Dimaroth P, Stoll E, Moss J, Lane MD. Acetyl coenzyme A carboxylase system of Escherichia coli. J Biol Chem. 1974;249:6633–6645. [PubMed] [Google Scholar]
- Hablacher M, Ivessa AS, Paltauf F, Kohlwein SD. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Higo H. PLACE: a database of plant cis-regulatory DNA elements. Nucleic Acids Res. 1998;26:358–359. doi: 10.1093/nar/26.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Soll J. Import of a new chloroplast inner envelope protein is greatly stimulated by potassium phosphate. Plant Mol Biol. 1995;27:1173–1181. doi: 10.1007/BF00020890. [DOI] [PubMed] [Google Scholar]
- Hunaiti AR, Kolattukudy PE. Isolation and characterization of an acyl-coenzyme A carboxylase from an erythromycin-producing Streptomyces erythreus. Arch Biochem Biophys. 1982;216:362–371. doi: 10.1016/0003-9861(82)90222-3. [DOI] [PubMed] [Google Scholar]
- Hunter SC, Ohlrogge JB. Regulation of spinach chloroplast acetyl-CoA carboxylase. Arch Biochem Biophys. 1998;359:170–178. doi: 10.1006/abbi.1998.0900. [DOI] [PubMed] [Google Scholar]
- Jacobson BS, Stumpf PK. Fat metabolism in higher plants: LV. Acetate uptake and accumulation by class I and class II chloroplasts from Spinacia oleracea. Arch Biochem Biophys. 1972;153:656–663. doi: 10.1016/0003-9861(72)90384-0. [DOI] [PubMed] [Google Scholar]
- Kannangara CG, Stumpf PK. Fat metabolism in higher plants: LIV. Prokaryotic type acetyl-CoA carboxylase in spinach chloroplasts. Arch Biochem Biophys. 1972;152:83–91. doi: 10.1016/0003-9861(72)90196-8. [DOI] [PubMed] [Google Scholar]
- Ke J. Expression of genes encoding acetyl-coA carboxylase, biotin synthase, and acetyl-CoA generating enzymes in Arabidopsis thaliana. PhD thesis. Ames: Iowa State University; 1997. [Google Scholar]
- Ke J, Choi J-K, Smith M, Horner HT, Nikolau BJ, Wurtele ES. Structure of the CAC1 gene and in situ characterization of its expression: the Arabidopsis thaliana gene coding for the biotin-containing subunit of the plastidic acetyl-coenzyme A carboxylase. Plant Physiol. 1997;113:357–365. doi: 10.1104/pp.113.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Olsen LJ, Theg SM. Chloroplastic precursors and their transport across the envelope membranes. Plant Mol Biol. 1989;40:471–501. [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim J-S, Raines RT. Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci. 1993;2:348–356. doi: 10.1002/pro.5560020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Shiratsuchi K, Yoshimoto T, Masuda T, Kitazono A, Tsuru D, Anai M, Sekiguchi M, Tanabe T. Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc Natl Acad Sci USA. 1991;88:9730–9733. doi: 10.1073/pnas.88.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Sasaki Y. Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc Natl Acad Sci USA. 1994;91:3598–3601. doi: 10.1073/pnas.91.9.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Shinohara K, Yamada K, Sasaki Y. Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and eukaryotic forms of this enzyme. Plant Cell Physiol. 1996;37:117–122. doi: 10.1093/oxfordjournals.pcp.a028920. [DOI] [PubMed] [Google Scholar]
- Kozaki A, Sasaki Y. Light-dependent changes in redox status of the plastidic acetyl-CoA carboxylase and its regulatory component. Biochem J. 1999;339:541–546. [PMC free article] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lane MC, Moss J, Polakis SE. Acetyl coenzyme A carboxylase. Curr Top Cell Regul. 1974;8:139–195. [PubMed] [Google Scholar]
- Leon P, Arroyo A, Mackenzie S. Nuclear control of plastid and mitochondrial development. Annu Rev Plant Physiol Mol Biol. 1998;49:453–480. doi: 10.1146/annurev.arplant.49.1.453. [DOI] [PubMed] [Google Scholar]
- Li S-J, Cronan JE. The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992a;267:16841–16847. [PubMed] [Google Scholar]
- Li S-J, Cronan JE., Jr The gene encoding the biotin carboxylase subunit of E. coli acetyl-CoA carboxylase. J Biol Chem. 1992b;267:855–863. [PubMed] [Google Scholar]
- Li S-J, Cronan JE., Jr The genes encoding the two carboxyltransferase subunits of E. coli acetyl-CoA carboxylase. J Biol Chem. 1992c;267:16841–16847. [PubMed] [Google Scholar]
- Li S-J, Cronan JE., Jr Putative zinc finger protein encoded by a conserved chloroplast gene is very likely a subunit of a biotin-dependent carboxylase. Plant Mol Biol. 1992d;20:759–761. doi: 10.1007/BF00027147. [DOI] [PubMed] [Google Scholar]
- Lopez-Casillas F, Bai D-H, Luo X, Kong I-S, Hermodson MA, Kim K-H. Structure of the coding sequence and primary amino acid sequence of acetyl-coenzyme A carboxylase. Proc Natl Acad Sci USA. 1988;85:5784–5788. doi: 10.1073/pnas.85.16.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. Early embryogenesis in Arabidopsis thaliana II. The developing embryo. Can J Bot. 1991;69:461–476. [Google Scholar]
- Mansfield SG, Briarty LG. Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can J Bot. 1992;70:151–164. [Google Scholar]
- Meurer J, Berger A, Westhoff P. A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell. 1996;8:1193–1207. doi: 10.1105/tpc.8.7.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nikolau BJ, Wurtele ES, Stumpf PK. Tissue distribution of acetyl-coenzyme A carboxylase in leaves. Plant Physiol. 1984;75:895–901. doi: 10.1104/pp.75.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, Aota S-I, Inokuchi H, Ozeki H. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. [Google Scholar]
- Olszewski NE, Martin FB, Ausubel FM. Specialized binary vector for plant transformation: expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res. 1988;16:10765–10782. doi: 10.1093/nar/16.22.10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller D, Jaworski JG, Ohlrogge JB. In vivo pools of free and acylated acyl carrier proteins in spinach. J Biol Chem. 1991;266:1858–1863. [PubMed] [Google Scholar]
- Post-Beittenmiller D, Roughan G, Ohlrogge JB. Regulation of plant fatty acid biosynthesis. Plant Physiol. 1992;100:923–930. doi: 10.1104/pp.100.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA. Plastid division and development. Plant Cell. 1999;11:549–556. doi: 10.1105/tpc.11.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal M, Guilleminot J, Gueguen C, Cooke R, Delseny M, Gruber V. Structure, organization and expression of two closely related novel Lea (late-embryogenesis-abundant) genes in Arabidopsis thaliana. Plant Mol Biol. 1999;40:153–165. doi: 10.1023/a:1026403215270. [DOI] [PubMed] [Google Scholar]
- Reverdatto S, Beilinson V, Nielsen NC. A multisubunit acetyl coenzyme A carboxylase from soybean. Plant Physiol. 1999;119:961–978. doi: 10.1104/pp.119.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler K, Shintani D, Savage L, Boddupalli S, Ohlrogge J. Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseed. Plant Physiol. 1997;113:75–81. doi: 10.1104/pp.113.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler KR, Shorrosh BS, Ohlrogge JB. Structure and expression of an Arabidopsis acetyl-coenzyme A carboxylase gene. Plant Physiol. 1994;105:611–617. doi: 10.1104/pp.105.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Roughan PG. Stromal concentrations of coenzyme A and its esters are insufficient to account for rates of chloroplast fatty acid synthesis: evidence for substrate channelling within the chloroplast fatty acid synthase. Biochem J. 1997;327:267–273. doi: 10.1042/bj3270267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sasaki Y, Hakamada K, Suama Y, Nagano Y, Furusawa I, Matsuno R. Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J Biol Chem. 1993;268:25118–25123. [PubMed] [Google Scholar]
- Sasaki Y, Konishi T, Nagano Y. The compartmentation of acetyl-coenzyme A carboxylase in plants. Plant Physiol. 1995;108:445–449. doi: 10.1104/pp.108.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M. Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1997;94:11096–11101. doi: 10.1073/pnas.94.20.11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer A, Heise K-P. Regulation of acetyl-coenzyme A carboxylase and acetyl-coenzyme A synthetase in spinach chloroplasts. Z Naturforsch. 1984;39c:268–275. [Google Scholar]
- Savage LJ, Ohlrogge JB. Phosphorylation of pea chloroplast acetyl-CoA carboxylase. Plant J. 1999;18:521–527. doi: 10.1046/j.1365-313x.1999.00478.x. [DOI] [PubMed] [Google Scholar]
- Schulte W, Schell J, Topfer R. A gene encoding acetyl-coenzyme A carboxylase from Brassica napus. Plant Physiol. 1994;106:793–794. doi: 10.1104/pp.106.2.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chungwongse J, Obakata J, Yamaguch-Shinozaki K, Ohto C, Torazawa K, Meng B-Y, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura J. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani D, Roesler K, Shorrosh B, Savage L, Ohlrogge J. Antisense expression and overexpression of biotin carboxylase in tobacco leaves. Plant Physiol. 1997;114:881–886. doi: 10.1104/pp.114.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani DK, Ohlrogge JB. Feedback inhibition of fatty acid synthesis in tobacco suspension cells. Plant J. 1995;7:577–587. [Google Scholar]
- Shorrosh BS, Dixon RA, Ohlrogge JB. Molecular cloning, characterization, and elicitation of acetyl-CoA carboxylase from alfalfa. Proc Natl Acad Sci USA. 1994;91:4323–4327. doi: 10.1073/pnas.91.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrosh BS, Roesler KR, Shintani D, van de Loo FL, Ohlrogge JB. Structural analysis, plastid localization, and expression of the biotin carboxylase subunit of acetyl-coenzyme A carboxylase from tobacco. Plant Physiol. 1995;108:805–812. doi: 10.1104/pp.108.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrosh BS, Savage LJ, Soll J, Ohlrogge JB. The pea chloroplast membrane-associated protein, IEP96, is a subunit of acetyl-CoA carboxylase. Plant J. 1996;10:261–268. doi: 10.1046/j.1365-313x.1996.10020261.x. [DOI] [PubMed] [Google Scholar]
- Stalberg K, Allerstom M, Ezcurra I, Ablov S, Rask L. Disruption of an overlapping E-box/Abre motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta. 1996;199:515–519. doi: 10.1007/BF00195181. [DOI] [PubMed] [Google Scholar]
- Sun J, Ke J, Johnson JL, Nikolau BJ, Wurtele ES. Biochemical and molecular biological characterization of CAC2: the Arabidopsis thaliana gene coding for the biotin carboxylase subunit of plastidic acetyl-coenzyme A carboxylase. Plant Physiol. 1997;115:1371–1383. doi: 10.1104/pp.115.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T, Yokoyama C, Wada K, Tanabe T. Primary structure of chicken liver acetyl-coenzyme A carboxylase deduced from cDNA sequence. J Biol Chem. 1988;263:2651–2657. [PubMed] [Google Scholar]
- Van Dyke MW, Sirito M, Sawadogo M. Single-step purification of bacterially expressed polypeptides containing an oligo-histidine domain. Gene. 1992;111:99–104. doi: 10.1016/0378-1119(92)90608-r. [DOI] [PubMed] [Google Scholar]
- Voytas DF, Konieczny A, Cummings MP, Ausubel FM. The structure, distribution and evolution of the Ta1 retrotransposable element family of Arabidopsis thaliana. Genetics. 1990;126:713–721. doi: 10.1093/genetics/126.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walid A-F, Chirala SS, Wakil SJ. Cloning of the yeast FAS3 gene and primary structure of yeast acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1992;89:4534–4538. doi: 10.1073/pnas.89.10.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-Q, Colbert JT, Wurtele ES. A molecular marker of the root cortical ground meristem is present in both root and coleorhiza of the maize embryo. Am J Bot. 1995a;82:1083–1088. [Google Scholar]
- Wang X, Wurtele ES, Nikolau BJ. Regulation of β-methylcrotonyl-coenzyme A carboxylase activity by biotinylation of the apoenzyme. Plant Physiol. 1995b;108:1133–1139. doi: 10.1104/pp.108.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Lebrun L, Franklin A, Huang L, Hoffman N, Wurtele ES, Nikolau BJ. 3-Methylcrotonyl-CoA carboxylase of Arabidopsis thaliana: isolation and characterization of cDNA coding for the biotinylated subunit. Plant Physiol. 1995;107:1013–1014. doi: 10.1104/pp.107.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury NW, Roberts LL, Palmer JD, Thompson WF. A transcription map of the pea chloroplast genome. Curr Genet. 1988;14:75–89. [Google Scholar]
- Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]
- Wurtele ES, Behal RH, Cui X, Ke J, Johnson JL, Lui F, Nikolau BJ, Oliver BJ, Schnable PS. In: Molecular Biology Of Acetyl-CoA Generation. Advances in Plant Lipids. Sanchex J, Cerdalmedo E, Martinez-Force E, editors. Seville, Spain: University of Sevilla Press; 1997. pp. 54–56. [Google Scholar]
- Wurtele ES, Nikolau BJ. Plants contain multiple biotin enzymes: discovery of 3-methylcrotonyl-CoA carboxylase, propionyl-CoA carboxylase and pyruvate carboxylase in the plant kingdom. Arch Biochem Biophys. 1990;278:179–186. doi: 10.1016/0003-9861(90)90246-u. [DOI] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N. Genomic organization of 251 kDa acetyl-CoA carboxylase genes in Arabidopsis: tandem gene organization has made two differentially expressed isozymes. Plant Cell Physiol. 1995;36:779–787. doi: 10.1093/oxfordjournals.pcp.a078822. [DOI] [PubMed] [Google Scholar]
- Zhou D-X. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 1999;4:1360–1385. doi: 10.1016/s1360-1385(99)01418-1. [DOI] [PubMed] [Google Scholar]