Abstract
Magnesium-protoporphyrin IX chelatase (Mg-chelatase) is located at the branchpoint of tetrapyrrole biosynthesis, at which point protoporphyrin IX is distributed for the synthesis of chlorophyll and heme. We investigated the regulatory contribution of Mg-chelatase to the flow of metabolites. In plants, the enzyme complex consists of three subunits, designated CHL D, CHL I, and CHL H. Transgenic tobacco (Nicotiana tabacum) plants expressing antisense RNA for the Mg-chelatase subunit CHL H were analyzed to elucidate further the role of Mg-chelatase in the distribution of protoporphyrin IX into the branched tetrapyrrolic pathway. The transgenic plants displayed a reduced growth rate and chlorophyll deficiency. Both phenotypical properties were correlated with lower Mg-chelatase activity. Unexpectedly, less protoporphyrin IX and heme accumulated, and a decrease in 5-aminolevulinate (ALA)-synthesizing capacity and ALA dehydratase activity paralleled the progressive reduction in Mg-chelatase activity in the transformants compared with control plants. The reduced activities of the early enzymatic steps corresponded with lower levels of transcripts encoding glutamyl-tRNA reductase and ALA-dehydratase. The decreased expression and activities of early enzymes in the pathway could be explained by a feedback-controlled mechanism in response to lower Mg-chelatase activity. We discuss intercompartmental signaling that synchronizes the activities of the first steps in tetrapyrrolic metabolism with the late steps for the synthesis of end products.
Photosynthetic organisms synthesize (bacterio-) chlorophyll, heme, and phycobilines, the major tetrapyrroles in nature (von Wettstein et al., 1995; Grimm, 1998). The stepwise formation of protoporphyrin IX from 5-aminolevulinic acid is in principle identical in bacteria and plants. Two chelating enzymes compete subsequently for protoporphyrin IX. Ferrochelatase (Fe-chelatase) directs the substrate toward heme, which serves as a cofactor in different cellular processes. In cyanobacteria, protoheme is mainly metabolized to phycobiline, and in plants it is metabolized (to a lower extent) to the light-perceptive phytochromobiline. The first step unique in (bacterio-) chlorophyll formation is the insertion of Mg2+ into protoporphyrin IX. This step is catalyzed by magnesium-protoporphyrin IX-chelatase (Mg-chelatase). This enzyme consists of three subunits (in parentheses are the molecular masses of the proteins from Rhodobacter capsulatus and tobacco [Nicotiana tabacum], respectively) BchI/CHL I (40 and 42 kD), CHL D (60 and 83 kD), and CHL H (140 and 154 kD) (Zsebo and Hearst, 1984; Kruse et al., 1997; Papenbrock et al., 1997). The same subunits are structurally related in prokaryotes and eukaryotes. The first identified plant-coding sequence involved in Mg chelation was the T-DNA-tagged cs-sequence, which was equivalent to Chl I, although a function in chlorophyll synthesis had not been assigned at that time (Koncz et al., 1990). The first plant Chl H sequence was found by transposon-tagged gene inactivation (Hudson et al., 1993). The third subunit, CHL D, showed significant similarity to the cyanobacterial counterpart and was proven to be essential for the formation of an active enzyme complex of recombinant tobacco subunits (Papenbrock et al., 1997).
The catalytic insertion of Mg2+ into protoporphyrin IX is apparently a complex mechanism that differs from the insertion of ferrous iron into protoporphyrin IX by Fe-chelatase (Castelfranco et al., 1994). Mg chelation is catalyzed in an ATP-dependent, two-step reaction, and is still not entirely understood (Walker and Willows, 1997; Jensen et al., 1998; Gräfe et al., 1999). An initial activation step includes CHL I and CHL D (Gibson et al., 1995; Willows et al., 1996). An ATPase function was assigned to this protein complex (Hansson and Kannangara, 1997). The protoporphyrin IX-binding CHL H subunit subsequently interacts with the activation complex for the Mg-insertion into protoporphyrin IX, and then transfers Mg-protoporphyrin IX to Mg-protoporphyrin IX-methyltransferase (Gorchein, 1972; Hinchigeri et al., 1997).
In plants, tetrapyrrole metabolism is located in the chloroplasts. All enzymes involved are nuclear encoded. It is expected that a regulatory mechanism controls the genetic information of the nuclear-encoded enzymes to coordinate them with the required activities of the pathway in the plastids. The rate-limiting step of tetrapyrrole metabolism is ALA synthesis at the beginning of the pathway. After the conversion of eight ALA molecules into a cyclic porphyrin, protoporphyrin IX is directed into the chlorophyll- and heme-synthesizing branch. In angiosperms, chlorophyll is exclusively formed in the light. We address the question of how the metabolic flow and the distribution of protoporphyrin IX, which is light and developmentally dependent, adjusted to the requirements of varying amounts of chlorophyll and heme. A spatial separation of the heme- and chlorophyll-synthesizing pathway in plastids could play a role (Matringe et al., 1994), as could varying affinities of the two enzymes to the substrate and a difference in ATP requirement for the two chelation reactions (Guo et al., 1998; Jensen et al., 1998). Moreover, the antagonistic rhythmicity of Mg-chelatase and Fe-chelatase activities and ALA synthesis could contribute in light-/dark-grown tobacco plants to the distribution of protoporphyrin IX (Papenbrock et al., 1999). We describe the role of Mg-chelatase in the control of metabolic flow. Activities of Mg-chelatase and the early enzymes of the pathway are apparently matched to guarantee a balanced flow of tetrapyrrole precursors in the pathway and to avoid accumulation of harmful photosensitizing porphyrin intermediates. We obtained evidence for the intracellular interdependence of gene expression for enzymes of the early steps of the pathway and the metabolic activities in the Mg-porphyrin branch.
MATERIALS AND METHODS
Growth of Plants
Tobacco (Nicotiana tabacum) plants were grown in the greenhouse for 5 weeks before harvest. Unless stated otherwise, supplemental illumination was provided by 400-W high-pressure sodium vapor lamps to ensure a light intensity of 300 μmol m−2 s−1 over a 16-h photoperiod. All experiments were done with primary transformants.
Construction of the Transgene
A partial cDNA fragment encoding the tobacco CHL H subunit of Mg-chelatase (Kruse et al., 1997) was ligated into the SmaI-digested plant binary vector BinAR (Höfgen and Willmitzer, 1992). The Agrobacterium tumefaciens strain GV 2260 was transformed with the plasmid harboring the Chl H coding sequence in inverted orientation behind the cauliflower mosaic virus 35S promoter. The Chl H antisense gene construct was introduced into the tobacco genome (N. tabacum cv Samsun NN) by leaf disc transformation (Horsch et al., 1985).
RNA and DNA Analysis
Total RNA was extracted essentially as described previously (Chomczinsky and Sacchi, 1987). The methods for DNA extraction and for Southern- and northern-blot hybridization were carried out according to standard procedures (Sambrook et al., 1989). Aliquots of 15 μg of RNA and 20 μg of DNA were analyzed. After blotting, filters were probed with different cDNA fragments and labeled by random priming using [32P]dCTP (Gibco-BRL, Eggenstein, Germany).
Enzyme Assays
Activity of 5-aminolevulinic acid dehydratase was assayed as described previously (Smith, 1988). For assaying Mg-chelatase 12 g of freshly harvested plant material was homogenized in 75 mL of homogenization buffer consisting of 0.5 m sorbitol, 0.1 m Tris/HCl, pH 7.5, 0.1% (w/v) bovine serum albumin (BSA), and 1 mm dithiothreitol (DTT), filtered through two layers of cheesecloth and two layers of gauze, and centrifuged at 5,000g for 10 min. The pellet of chloroplasts was resuspended in 2.5 mL of homogenization buffer and used for enzyme activity measurements. Mg-chelatase was assayed as in Lee et al. (1992), with some modifications. The assay mix contained 40 mm MgCl2 and 20 mm ATP. Reactions were started by adding dimethyl sulfoxide (DMSO)-solved protoporphyrin IX to a final concentration of 100 μm, and stopped after 0, 15, 30, 45, and 60 min at 28°C by freezing tubes in liquid nitrogen. Assay samples were sequentially extracted with methanol, 50 mm potassium phosphate buffer (pH 7.8), and acetone:methanol:0.1 m NH4OH (10:9:1; v/v). Combined extracts were subjected to HPLC analysis using authentic standards for quantification of assay products (see below).
Determination of Porphyrins
Porphyrin extraction and HPLC analysis were done as described by Mock and Grimm (1997). Liquid ground leaf material (100 mg) was sequentially extracted with 50 mm potassium phosphate buffer (pH 7.8), methanol:0.1 m NH4OH (9:1; v/v), and acetone:0.1 m NH4OH (9:1; v/v). Extracts were diluted with an equal volume of methanol prior to HPLC analysis. Porphyrins were eluted with a linear gradient of solvent B (90% [w/v] methanol and 0.1 m ammonium acetate, pH 5.2) in solvent A (10% [w/v] methanol and 0.1 m ammonium acetate, pH 5.2) as follows: 0% to 100% over 12 min and 100% (w/v) solvent B for 13 min. Column eluent was monitored by fluorescence detection (λex 405/λem 625 and λex 420 nm/λem 595), and porphyrins were identified and quantified using authentic standards (Fluka, Milwaukee, WI; Porphyrin Products, Logan, UT). Mg-protoporphyrin IX monomethylester was a generous gift of Prof. W. Rüdiger (Botanical Institute of the Ludwig-Maximillian University, Munich).
Determination of Chlorophyll and Carotenoid Contents
Chlorophyll estimation was according to the method of Porra et al. (1989) in alkaline acetone extracts. Carotenoids were analyzed essentially as described previously (Kruse et al., 1995), except that an HPLC system equipped with photo-diode array detection (996 PDA, Waters, Milford, MA) was used. Authentic standards were used for quantification.
Determination of ALA-Synthesizing Capacity
Three leaf discs per sample were harvested from the fourth leaf of the plant, incubated in 40 mm levulinic acid in 20 mm phosphate buffer, pH 7.2, in the light for 6 h, and then frozen in liquid nitrogen. Samples were homogenized, given 1 mL of 20 mm K2HPO4/KH2PO4, pH 7.2, and centrifuged. One-hundred microliters of ethylacetoacetate was added to a 500-μL aliquot of the supernatant, which was subsequently boiled for 10 min and cooled on ice for 5 min. An equal volume of modified Ehrlichs reagent was added and color development was measured at 553 nm using the spectrophotometer (Mauzerall and Granick, 1956). Standard curves were used for calculating amounts of accumulated ALA.
Miscellaneous
Protein concentrations were determined according to the method of Bradford (1976). Heme was extracted as described by Hagège et al. (1992) and quantified using a molar absorption coefficient given by Castelfranco and Jones (1975).
RESULTS
Phenotypical Changes in Transgenic Plants Transformed with the Mg-Chelatase Subunit CHL H in Antisense Orientation
A 3.5-kb cDNA sequence encoding the CHL H subunit of Mg-chelatase from tobacco (Kruse et al., 1997) was inserted in reverse orientation behind a modified cauliflower mosaic virus 35S promoter and introduced into the tobacco genome by A. tumefaciens. Transformants showed a broad variety of phenotypes with reduced green pigmentation. Some completely bleached transformants could only be preserved in sugar-containing tissue culture. Approximately 70 transgenic lines of Chl H antisense transformants were cultivated in soil in the greenhouse. The transgenic seeds of some lines germinated slowly and the seedlings died within a few days after being transferred to soil. Four representative lines, Pa 1/14, Pa 1/54, Pa 1/56, and Pa 1/60, were selected for detailed analysis (Fig. 1, top). Their developmental growth and flowering were delayed, and fewer inflorescences and seeds were produced. The older leaves were also more bleached compared with the younger leaves. Individual leaves of each line displayed a characteristic pattern of lower pigmentation and showed a descending gradient of green pigments from the bottom to the leaf tip (Fig. 1, bottom).
Figure 1.
Primary tobacco transformants expressing Chl H antisense RNA and a wild-type plant (SNN) grown in the greenhouse at 300 μmol m−2 s−1 for 5 weeks. Top panel, Transformants with a progressive loss of chlorophyll. Left to right, Wild-type plant SNN, Pa 1/14, Pa 1/60, Pa 1/54, and Pa 1/56. Bottom panel, Sixth leaves of the same transformants (except of wild-type-like Pa 1/60) shown for comparison. Transgenic plants remain slightly green along the vascular bundles.
Antisense Inhibition of Chl H Expression Causes a Reduction in Mg-Chelatase Activity
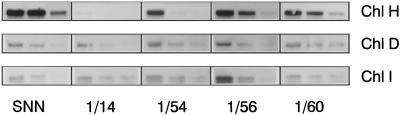
Total RNA was extracted from leaves 4, 6, and 8 (counting from the top of the plant) of wild-type plants and the selected lines Pa 1/14, Pa 1/54, Pa 1/56, and Pa 1/60, and analyzed by northern-blot hybridization. The Chl H transcript levels of transgenic and wild-type plants were compared. Effects of the Chl H antisense inhibition on the contents of Chl I and Chl D RNA were investigated in parallel (Fig. 2). The transcripts encoding all three Mg-chelatase subunits maximally accumulated in the youngest leaf of control plants and decreased subsequently toward older leaves. The Chl H mRNA contents were reduced in Chl H antisense plants compared with wild-type plants. The lower content of Chl H RNA was correlated with increasing chlorosis of the transgenic lines. The Chl H RNA content was close to the detection limit in the very pale yellow-green transformant Pa 1/14, whereas the abundance of Chl H mRNA of the wild-type-like transformant Pa 1/60 was only slightly diminished. Chl I and Chl D transcript levels were not altered in response to reduced Chl H mRNA in the transgenic lines compared with control plants.
Figure 2.
Northern-blot analysis of the expression of Mg-chelatase subunits. Plants were grown for 5 weeks in the greenhouse at 300 μmol m−2 s−1. For the determination of steady-state RNA levels of the three Mg-chelatase subunits in selected Chl H antisense plants, total RNA was extracted from leaves 4, 6, and 8 of wild-type plants and transformants. Fifteen micrograms of isolated RNA was subjected to northern-blot hybridization and subsequently hybridized with the Chl H, Chl D, and Chl I cDNA. The Chl H cDNA probe recognized both the endogenous transcript and the antisense RNA.
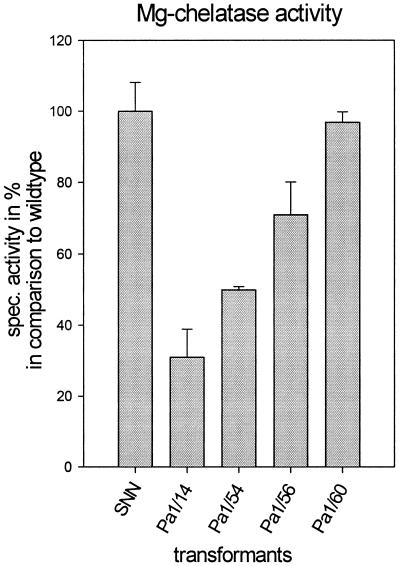
In tobacco plants, Mg-chelatase activity has its maximal activity in immature leaves. Mg-chelatase activity was determined from plastids of leaves 1 to 6 of 5-week-old wild-type and transformant plants. The Mg-chelatase activity of transformant Pa 1/14 was lowered to 30% of control activity and the wild-type-like transformant Pa 1/60 Mg-chelatase had nearly 100% of the control activity (100% was about 250 nkat kg−1 protein) (Fig. 3). The reduction in Mg-chelatase activity corresponds to the reduction of Chl H RNA contents and reflects the low-green pigmentation of the leaves.
Figure 3.
Analysis of Mg-chelatase activity in chloroplasts of selected Chl H antisense transformants and control plants. Mg-chelatase activity was determined in chloroplasts isolated from 5-week-old plants. Values represent means ± sd from three separate preparations.
Antisense Inactivation of Chl H Expression Leads to Lower Contents of Chlorophyll, Heme, and Carotenoids
The total amount of chlorophyll, non-covalently bound heme, and carotenoids was determined in leaf 4 of control and transgenic plants (Table I). The chlorophyll contents of the transformants declined in parallel with the deficiency in Mg-chelatase activity. Line Pa 1/14 contained only 5% of the control chlorophyll. The carotenoid levels were lowered to similar extent as the chlorophyll contents in the transgenic plants compared with the wild-type values, which is consistent with the parallel degradation of pigments and pigment binding proteins of the photosynthetic apparatus. In spite of the tremendous reduction of chlorophyll levels, the relative amounts of the carotenoid species analyzed did not significantly change either in low-light-grown or greenhouse-grown transformants relative to control plants. It could be expected that a block of the chlorophyll-synthesizing pathway would direct more protoporphyrin IX toward the heme pathway. However, the heme contents of the transgenic plants were also already lowered in young leaves (only 25% and 60% of the wild-type content in line Pa 1/14 and Pa 1/54, respectively) (Table I), which would be inconsistent with a feedback suppression in response to early transient heme accumulation.
Table I.
Influence of reduced Chl H mRNA levels on chlorophyll, heme, and carotenoid contents
| Plants | Chlorophyll | Heme | Carotenoids |
|---|---|---|---|
| nmol g−1 FW | |||
| SNN | 1,163.3 ± 72.3 | 35.6 ± 2.1 | 151.8 ± 13.9 |
| Pa 1/14 | 51.2 ± 7.9 | 8.4 ± 0.4 | 17.1 ± 0.2 |
| Pa 1/54 | 253.5 ± 16.8 | 21.1 ± 1.5 | 61.3 ± 9.2 |
| Pa 1/56 | 801.4 ± 66.3 | 30.1 ± 3.3 | 104.9 ± 12.7 |
| Pa 1/60 | 1,090.9 ± 9.3 | 35.2 ± 2.6 | 149.2 ± 2.3 |
Chlorophyll and heme were photometrically measured as described in “Materials and Methods.” Carotenoids were separated by HPLC but finally the total amount of carotenoids was calculated. Values represent means ± sd from three separate extractions.
Reduced Mg-Chelatase Activity Does Not Result in Accumulation of the Substrate Protoporphyrin IX
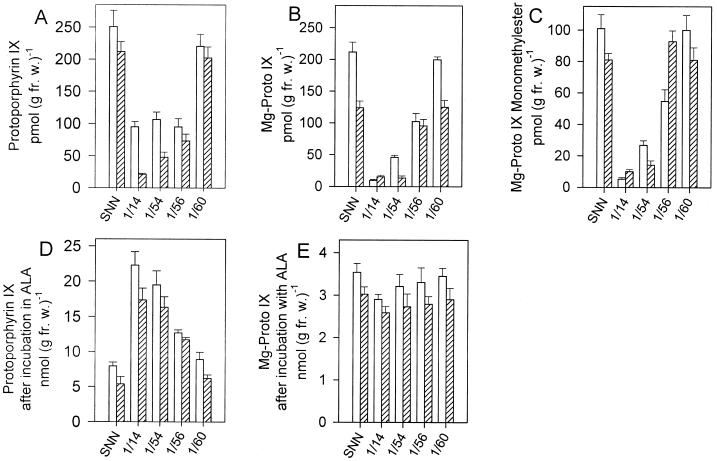
We previously characterized transgenic plants with deficiency in coproporphyrinogen oxidase and uroporphyrinogen decarboxylase, two preceding enzymes in the metabolic pathway. These plants accumulated their respective substrate, uroporphyrin(ogen) and coproporphyrin(ogen), in young leaves up to several hundred-fold compared with control plants and exhibited necrotic lesions (Kruse et al., 1995; Mock and Grimm, 1997). Accumulating protoporphyrin IX leads to light-dependent damage of plant cells, as is demonstrated upon application of diphenyl-ether-type herbicides, which inhibit protoporphyrinogen IX oxidase, the enzyme preceding Mg-chelatase (Matringe and Scalla, 1988; Witkowski and Halling, 1988). It was initially assumed that the transgenic plants with deficient Mg-chelatase activity accumulate protoporphyrin IX. However, less protoporphyrin IX was found in the extracts of transgenic lines than in control plants (Fig. 4A). The contents of Mg-protoporphyrin IX and Mg-protoporphyrin IX monomethylester were also reduced in the transgenic plants (Fig. 4, B and C). The uroporphyrin(ogen) and the coproporphyrin(ogen) contents were not increased (data not shown). The reduced steady-state level of protoporphyrin IX could be explained with a block at an earlier enzymatic step and a delayed supply of early precursors. ALA steady-state levels were below the detection limits.
Figure 4.
Analysis of porphyrins and Mg porphyrins in selected transgenic and wild-type plants. A through C, Contents of protoporphyrin IX, Mg-protoporphyrin (Mg-Proto) IX, and Mg-protoporphyrin IX monomethylester (Mg-Proto IX-MME) of leaf extracts from leaf 4 (white column) and 6 (striped column) were prepared and subjected to HPLC analysis with fluorescence detection as described in “Materials and Methods.” D and E, Leaf discs of leaf 4 (white columns) and 6 (striped columns) were incubated in 5 mm ALA in the dark for 4 h and analyzed for protoporphyrin IX and Mg-protoporphyrin IX accumulation. Values represent means ± sd from three separate extractions.
The site of inactivation in the pathway was identified by ALA incubation of discs from leaf 4 of Chl H antisense and control plants and subsequent determination of the content of accumulating protoporphyrin IX. ALA feeding led to higher accumulation of protoporphyrin IX in transgenic than in wild-type tissue (Fig. 4D), which can be explained by the corresponding Mg-chelatase deficiency in Chl H antisense plants. Mg-protoporphyrin IX contents were similar in leaf discs of all plants analyzed (Fig. 4E). Other metabolites between ALA and protoporphyrin IX did not accumulate, indicating that ALA synthesis is the limiting step for the formation of protoporphyrin IX and is apparently confined in response to Mg-chelatase deficiency.
Reduced Mg-Chelatase Activity Affects the Activity and Expression of Enzymes Involved in Early Steps of the Pathway
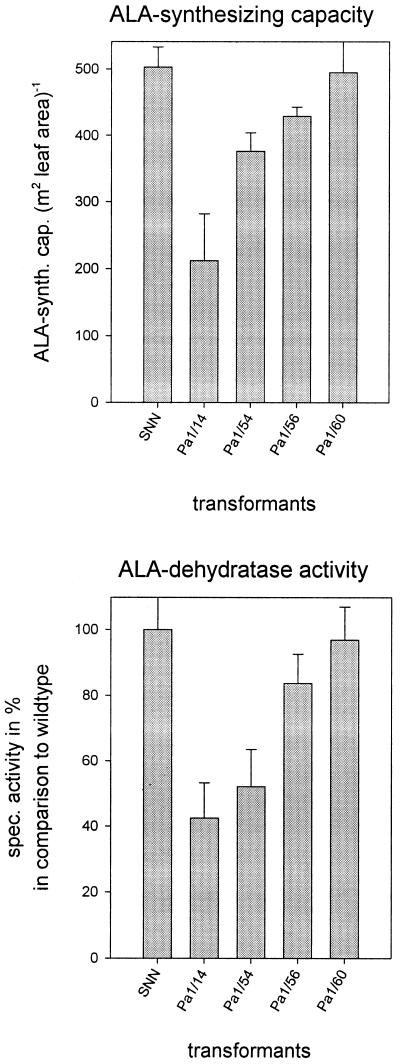
We examined activities and expression of enzymes of the early steps in the pathway. We assayed for ALA-synthesizing activity, which comprises mainly the glutamyl-tRNA reductase and the Glu 1-semialdehyde aminotransferase, and also for ALA-dehydratase. Discs of the fourth leaf of wild-type and transformant plants were incubated in 20 mm levulinic acid in the light (100 μmol photons m−2 s−1) to determine the accumulation of ALA (Fig. 5, top). The ALA-synthesizing capacity of Pa 1/14 yielded 40% of the control capacity. The rate of ALA synthesis of the other transformants was between the lowest and the control value (100%). The ALA-dehydratase assay of total leaf extracts revealed lower activities compared with control plants. The values were approximately reduced to the same extent as the ALA-synthesizing capacities in the same transgenic lines. ALA dehydratase activity of line Pa 1/14 gave rise to 40% of the wild-type activity. Both reduced activities of the early metabolic steps correlated with the reduced Mg-chelatase activities in the same transgenic line. Activities of uroporphyrinogen decarboxylase, coproporphyrinogen oxidase, and Fe-chelatase were not significantly altered in transgenic plants compared with wild-type plants (data not shown).
Figure 5.
Analysis of ALA-synthesizing capacity (top) and ALA- dehydratase activity (bottom) of control plants and selected transgenic plants expressing Chl H antisense RNA. Top panel, Leaf discs of the fourth leaf were incubated in the light for 6 h in levulinic acid, an inhibitor of ALA-dehydratase. Amounts of accumulated ALA were determined spectrophotometrically after reaction with Ehrlichs reagent. Values represent means ± sd from three separate experiments. Bottom panel, ALA-dehydratase activity was measured from total extracts of the fourth leaf. Values represent means ± sd from three separate preparations.
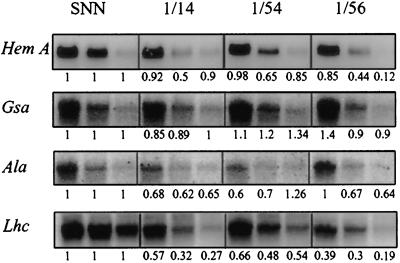
We also addressed the question of whether the loss of ALA synthesis and ALA-dehydratase activity could be explained by reduced transcriptional activities of the corresponding genes or by induced inactivation or degradation of the enzymes. Steady-state levels of RNA encoding glutamyl-tRNA reductase (Hem A), Glu 1-semialdehyde transferase (Gsa), and ALA-dehydratase (Ala) were measured from leaves 3, 5, and 7 of wild-type and transgenic plants (Fig. 6). As an example of a complex-regulated photosynthetic gene, transcript levels were also determined for light-harvesting chlorophyll-binding proteins of photosystem II (Lhc) (Fig. 6). As previously shown, chloroplast function affects Lhc gene expression (Oelmüller, 1989; Rapp and Mullet, 1991). Comparison of the transcript levels of each leaf analyzed from the selected transgenic lines and control plants revealed that Hem A and Ala RNA levels were reduced in the transgenic plants. Also, the Lhc transcript levels were highly reduced in response to antisense Chl H RNA expression. The Gsa RNA content was not reduced in transformants. The variation of its RNA levels did not affect the overall ALA-synthesizing capacity. Therefore, lower activities in ALA synthesis and of ALA-dehydratase can be attributed to the lower transcript levels for glutamyl-tRNA reductase and ALA-dehydratase, respectively. Steady-state levels of RNA encoding other enzymes in the pathway, such as uroporphyrinogen decarboxylase and coproporphyrinogen oxidase, were not influenced.
Figure 6.
Steady-state levels of RNA that encodes proteins of the early steps of the tetrapyrrole biosynthetic pathway and the antenna complex of photosystem II in antisense plants for CHL H (lines Pa 1/14, Pa 1/54, and Pa 1/56) and wild-type plants. Total RNA was isolated from leaves 3, 5, and 7 (counted from the top of each plant). Fifteen micrograms of RNA was loaded per lane, separated on a 1% (w/v) formaldehyde-agarose gel, and probed against cDNA encoding glutamyl-tRNA reductase (Hem A), Glu 1-semialdehyde aminotransferase (gsa), ALA-dehydratase (Ala), and the light-harvesting chlorophyll-binding proteins of photosystem II (Lhc). A cDNA probe for actin was subsequently hybridized to the RNA on the same filter to prove equal loading (data not shown). Numbers below each northern blot represent the relative levels of each transcript of the transgenic lines compared with those in the corresponding leaves of control plants.
DISCUSSION
The first visible symptoms of tobacco plants with reduced expression of CHL H were gradually slower growth rates and pale-green leaves. Chlorophyll deficiency (Table I) was correlated with the reduced steady-state level of Chl H RNA (Fig. 2) and lower Mg-chelatase activity (Fig. 3). In spite of diminished Mg-chelatase activity, the metabolic substrate protoporphyrin IX did not accumulate in transformants compared with control plants (Fig. 4). The gene expression for CHL I and CHL D was not affected. Inhibition of Chl H expression destabilized the Mg-chelatase complex and decreased the enzyme activity. The stoichiometric amounts of each subunit and the assembly of Mg-chelatase are still not entirely clear (Guo et al., 1998; Jensen et al., 1998; Gräfe et al., 1999), but it is sensible to assume that the reduced CHL H content reduces the number of functioning enzyme complexes. Because of low Mg-chelatase activity, non-metabolized protoporphyrin IX could preferentially be directed into the protoheme-synthesizing branch to give rise to excess heme. However, the measured steady-state heme contents of immature leaves were reduced to a similar extent as chlorophyll (Table I).
The leaf chlorosis of Chl H antisense plants resembles phenotypes of the xantha and chlorina barley mutants (Henningsen et al., 1993; Hansson et al., 1999) and the maize mutants blandy 4, l13, or oy-1040 (Mascia, 1978). Genetic and biochemical studies revealed that the barley mutants xantha f, g, and h have genetic lesions at three distinct mutant loci encoding the three Mg-chelatase subunits CHL H, CHL D, and CHL I, respectively (Jensen et al., 1996; Kannangara et al., 1997). The barley chlorina-125, -157, and -161 mutants contain point mutations in the Chl I gene (Hansson et al., 1999). The Arabidopsis mutant cs (Koncz et al., 1990) and virus-induced silencing of the sulfur allele in Nicotiana benthamiana (Kjemtrup et al., 1998) display a similar macroscopic phenotype like the Chl H antisense plants. The affected gene in both plant species encodes CHL I. Moreover, plants with Mg-chelatase deficiency phenotypically correspond to the yellow-green phenotype of transgenic plants expressing antisense RNA for Glu 1-semialdehyde aminotransferase (Höfgen et al., 1994; Härtel et al., 1997). These plants suffer from a lack of chlorophyll as result of deficient synthesis of ALA. The pale phenotype is attributed to the low ALA-synthesizing capacity.
It is important to emphasize that the ALA-synthesizing capacities and ALA-dehydratase activity are synchronously adjusted to the remaining chelating capacity of the Mg-chelatase in the Chl H antisense plants, implicating a mechanism that balanced activities of the early steps in response to the activities of late steps in the pathway. The transcript levels for glutamyl-tRNA reductase and ALA- dehydratase were gradually lower in parallel to their enzyme activities in the same analyzed transformants. Thus, it is suggested that the activities of the early steps of chlorophyll synthesis were determined by mechanisms controlling RNA stability or transcriptional activities.
Feedback-control mechanisms have previously been suggested to influence the activities in the early steps of tetrapyrrole biosynthesis in response to levels or synthesis of late metabolic intermediates and end products. In yeast and animals the heme pool controls ALA synthesis at various levels of ALA synthase expression, the equivalent enzyme to the plant ALA-forming C5 pathway (Ferreira and Gong, 1995). In plants, heme is also accepted as a negative effector of tetrapyrrole biosynthesis (Beale and Weinstein, 1990). Activity of recombinant glutamyl-tRNA reductase is reduced upon heme supplement (Pontoppidan and Kannangara, 1994). However, the in planta experimental evidence is still preliminary for a regulatory function of heme in the tetrapyrrolic pathway, especially on transcriptional control. We did not find any indication for heme accumulation in immature leaves of the Chl H-deficient transformants. The transformants already contained less heme in the fourth leaf than wild-type plants. Therefore, an initially increased heme pool is very unlikely to be involved in the feedback-controlled reduction of ALA synthesis.
In conclusion, lower ALA synthesis and the resulting protoporphyrin IX contents are directly correlated with the reduced Mg-chelatase activities. We favor a mechanism that coordinates synthesis of the tetrapyrrole precursor ALA at the level of gene expression and enzyme activities in response to the activities and the metabolic flux in the Mg porphyrin branch of the tetrapyrrolic pathway. This feedback control could involve the CHL H subunit, the assembled Mg-chelatase and its activity, or it could depend on a sensory mechanism for Mg-protoporphyrin levels, which were lower in the Chl H antisense plants due to reduced Mg-chelatase activity. Porphyrins have been previously proposed as mediating signals for the control of some nuclear genes (Johanningmeier and Howell, 1984; Susek and Chory, 1992). Feeding of Mg-protoporphyrin IX and Mg-protoporphyrin IX monomethylester to Chlamydomonas reinhardtii have been shown to function in triggering the modification of nuclear HSP70 gene expression in C. reinhardtii (Kropat et al., 1997).
The proposed feedback mechanism, which includes Mg-chelatase or its metabolic product, is consistent with the role of the GUN (genomes unregulated) genes (Susek et al., 1993). Their mutation affected coordination of nuclear genes with chloroplast function. The nature of the signal transduction chain that adjusts expression and activities for glutamyl-tRNA reductase and ALA-dehydratase with metabolic activities in the plastids remains unclear. It could include signaling via sensing the redox state of the quinone pool in the thylakoid membrane (Escoubas et al., 1995). The signal transduced from the Mg porphyrin-synthesizing pathway could trigger a regulator that either ties in gene expression with the metabolic pathway of tetrapyrroles or is part of an intracellular regulatory network for several physiological activities in plastids.
The Mg-chelatase activity requires the coordinated expression of all three subunits, including, most likely, post-translational modification to ensure the varying demands of newly synthesized chlorophyll. Maximal Mg-chelatase activity correlates with high ALA-synthesizing rates at the beginning of illumination during a 12-h light/12-h dark regime and allows the flow of most of the tetrapyrrole precursors into the chlorophyll-synthesizing branch (Papenbrock et al., 1999). Both activities are diminished in the dark period, at which time Fe-chelatase displays the highest activity.
Apart from the catalytic properties, two additional functions can be assigned to the active Mg-chelatase complex. The enzyme controls the metabolic flow rate by adjusting the activities of the limiting steps at the beginning of the pathway and by distributing protoporphyrin IX for chlorophyll and heme synthesis. Deregulated CHL H synthesis influenced these functions of Mg-chelatase. Antisense RNA inactivation of Mg-chelatase synthesis mimics constant low activities of Mg-chelatase, which consequently induce low ALA synthesis. Future experiments will substantiate regulatory roles of the CHL H subunit.
ACKNOWLEDGMENT
The technical assistance of Elis Fraust is acknowledged.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (Teilprojekt B15 of the SFB 363 to B.G.).
LITERATURE CITED
- Beale SI, Weinstein JD. Tetrapyrrole metabolism in photosynthetic organisms. In: Dailey HA, editor. Biosynthesis of Heme and Chlorophylls. New York: McGraw-Hill; 1990. pp. 287–391. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Castelfranco PA, Jones OTG. Protoheme turnover and chlorophyll synthesis in greening barley tissue. Plant Physiol. 1975;55:485–490. doi: 10.1104/pp.55.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelfranco PA, Walker CJ, Weinstein JD. Biosynthetic studies on chlorophylls: from protoporphyrin IX to protochlorophyllide. In: Chadwick DJ, Ackrill K, editors. The Biosynthesis of the Tetrapyrrole Pigments. Ciba Foundation Symposium 180. Chichester, UK: John Wiley; 1994. pp. 194–209. [DOI] [PubMed] [Google Scholar]
- Chomczinsky P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Escoubas JM, Lomas M, LaRoche J, Falkowski PG. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira GC, Gong J. 5-Aminolevulinate synthase and the first step of heme synthesis. J Bioenerg Biomembr. 1995;27:151–159. doi: 10.1007/BF02110030. [DOI] [PubMed] [Google Scholar]
- Gibson LCD, Willows RD, Kannangara CG, von Wettstein D, Hunter CN. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchein A. Magnesium protoporphyrin chelatase activity in Rhodopseudomonas spheroides studies with whole cells. Biochem J. 1972;127:97–106. doi: 10.1042/bj1270097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Lou M, Weinstein JD. Magnesium-chelatase from developing pea leaves. Plant Physiol. 1998;116:605–615. [Google Scholar]
- Gräfe S, Saluz HP, Grimm B, Hänel F. The role of the subunit CHLD in the chelation step of protoporphyrin IX. Proc Natl Acad Sci USA. 1999;96:1941–1946. doi: 10.1073/pnas.96.5.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm B. Novel insights into the control of tetrapyrrole metabolism of higher plants. Curr Opin Plant Biol. 1998;1:245–250. doi: 10.1016/s1369-5266(98)80112-x. [DOI] [PubMed] [Google Scholar]
- Hagège D, Werck-Reichhardt D, Schmitt P, Gaspar T. Deficiency in tetrapyrrole-containing compounds in a non-organogenic habituated sugarbeet cell line. Plant Physiol Biochem. 1992;39:649–654. [Google Scholar]
- Hansson A, Kannangara CG, von Wettstein D, Hansson M. Molecular basis for semidominance of missense mutations in the XANTHA-H (42 kDa) subunit of magnesium chelatase. Proc Natl Acad Sci USA. 1999;96:1744–1749. doi: 10.1073/pnas.96.4.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson M, Kannangara CG. ATPases and phosphate exchange activities in magnesium chelatase subunits of Rhodobacter sphaeroides. Proc Natl Acad Sci USA. 1997;94:13351–13356. doi: 10.1073/pnas.94.24.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Kruse E, Grimm B. Restriction of chlorophyll synthesis due to expression of glutamate-1-semialdehyde aminotransferase antisense RNA does not reduce the light-harvesting antenna size in tobacco. Plant Physiol. 1997;113:1113–1124. doi: 10.1104/pp.113.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningsen KW, Boynton JE, von Wettstein D. Mutants at xantha and albina loci in relation to chloroplast biogenesis in barley (Hordeum vulgare L.) Biologiske Skrifter Copenhagen Munksgaard. 1993;42:1–349. [Google Scholar]
- Hinchigeri SB, Hundle B, Richards WR. Demonstration that the BchH protein of Rhodobacter capsulatus activates S-adenosyl-l-methionine:magnesium protoporphyrin IX methyltransferase. FEBS Lett. 1997;407:337–342. doi: 10.1016/s0014-5793(97)00371-2. [DOI] [PubMed] [Google Scholar]
- Höfgen R, Axelsen KB, Kannangara CG, Schüttke I, Pohlenz HD, Willmitzer L, Grimm B, von Wettstein D. A visible marker for antisense mRNA expression in plants: Inhibition of chlorophyll synthesis with a glutamate 1-semialdehyde aminotransferase antisense gene. Proc Natl Acad Sci USA. 1994;91:1726–1730. doi: 10.1073/pnas.91.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L. Biochemical and genetic analysis of different patatin isoforms expressed in various cultivars of potato (Solanum tuberosum) Plant Sci. 1992;66:221–230. [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;228:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Hudson A, Carpenter R, Doyle R, Coen ES. Olive: a key gene required for chlorophyll biosynthesis in Antirrhinum majus. EMBO J. 1993;12:3711–3719. doi: 10.1002/j.1460-2075.1993.tb06048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Gibson LCD, Hunter CN. Determinants of the catalytic activity with the use of the purified I, D and H subunits of the magnesium protoporphyrin IX chelatase from Synechocystis PCC6803. Biochem J. 1998;334:335–344. doi: 10.1042/bj3340335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Willows RD, Petersen BL, Vothknecht UC, Stummann BM, Kannangara CG, von Wettstein D, Henningsen KW. Structural genes for Mg-chelatase subunits in barley: Xantha-f, -g and -h. Mol Gen Genet. 1996;250:383–394. doi: 10.1007/BF02174026. [DOI] [PubMed] [Google Scholar]
- Johanningmeier U, Howell S. Regulation of light-harvesting chlorophyll-binding proteins mRNA accumulation in Chlamydomonas reinhardtii. J Biol Chem. 1984;265:21820–21827. [PubMed] [Google Scholar]
- Kannangara CG, Vothknecht UC, Hansson M, von Wettstein D. Magnesium chelatase: association with ribosomes and mutant complementation studies identify barley subunit Xantha-G as a functional counterpart of Rhodobacter subunit BchD. Mol Gen Genet. 1997;254:85–92. doi: 10.1007/s004380050394. [DOI] [PubMed] [Google Scholar]
- Kjemtrup S, Sampson KS, Peele CG, Hguyen LV, Conkling MA, Thompson WF, Robertson D. Gene silencing from plant DANN carried by a Geminivirus. Plant J. 1998;14:91–100. doi: 10.1046/j.1365-313X.1998.00101.x. [DOI] [PubMed] [Google Scholar]
- Koncz C, Meyerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 1990;9:1337–1346. doi: 10.1002/j.1460-2075.1990.tb08248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Oster U, Rüdiger W, Beck CF. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc Natl Acad Sci USA. 1997;94:14168–14172. doi: 10.1073/pnas.94.25.14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse E, Mock HP, Grimm B. Reduction of coproporphyrinogen oxidase level by antisense RNA synthesis leads to deregulated gene expression of plastid proteins and affects the oxidative defense system. EMBO J. 1995;14:3712–3720. doi: 10.1002/j.1460-2075.1995.tb00041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse E, Mock HP, Grimm B. Isolation and characterisation of tobacco (Nicotiana tabacum) cDNA clones encoding proteins involved in magnesium chelation into protoporphyrin IX. Plant Mol Biol. 1997;35:1053–1056. doi: 10.1023/a:1005913315905. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Ball MD, Parham R, Rebeiz CA. Chloroplast biogenesis 65: enzymic conversion of protoporphyrin IX to Mg-protoporphyrin IX in a subplastidic membrane fraction of cucumber etiochloroplasts. Plant Physiol. 1992;99:1134–1140. doi: 10.1104/pp.99.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascia P. An analysis of precursors accumulated by several chlorophyll biosynthetic mutants of maize. Mol Gen Genet. 1978;161:237–244. [Google Scholar]
- Matringe M, Camadro JM, Joyard J, Douce R. Localization of ferrochelatase activity within mature pea chloroplasts. J Biol Chem. 1994;269:15010–15015. [PubMed] [Google Scholar]
- Matringe M, Scalla R. Studies on the mode of action of Acifluorfen-methyl in non-chlorophyllous soybean cells. Plant Physiol. 1988;86:619–625. doi: 10.1104/pp.86.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauzerall D, Granick S. The occurrence and determination of δ-aminolevulinic acid and porphobilinogen in urine. J Biol Chem. 1956;219:435–446. [PubMed] [Google Scholar]
- Mock HP, Grimm B. Reduction of uroporphyrin decarboxylase by antisense RNA expression affects activities of other enzymes involved in tetrapyrrole biosynthesis and leads to light-dependent necrosis. Plant Physiol. 1997;113:1101–1112. doi: 10.1104/pp.113.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmüller R. Photooxidative destruction of chloroplasts and its effects on nuclear gene expression and extraplastidic enzyme levels. Photochem Photobiol. 1989;49:229–239. [Google Scholar]
- Papenbrock J, Gräfe S, Kruse E, Hänel F, Grimm B. Mg-Chelatase of tobacco: identification of a Chl D cDNA sequence encoding a third subunit, analysis of the interaction of the three subunits with the yeast two-hybrid system and reconstitution of the enzyme activity by co-expression of recombinant CHL D, CHL H and CHL I. Plant J. 1997;12:981–990. doi: 10.1046/j.1365-313x.1997.12050981.x. [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Mock HP, Kruse E, Grimm B. Diurnal and circadian rhythms in tetrapyrrole biosynthesis: antagonistic maxima of magnesium chelatase and ferro chelatase. Planta. 1999;208:264–273. [Google Scholar]
- Pontoppidan B, Kannangara CG. Purification and partial characterisation of barley glutamyl tRNAGlu reductase. Eur J Biochem. 1994;225:529–537. doi: 10.1111/j.1432-1033.1994.00529.x. [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- Rapp J, Mullet J. Chloroplast transcription is required to express the nuclear genes RBSC and CAB:plastid DANN copy number is regulated independently. Plant Mol Biol. 1991;17:813–823. doi: 10.1007/BF00037063. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smith AG. Subcellular localization of two porphyrin-synthesis enzymes in Pisum sativum (pea) and Arum (cuckoo-pint) species. Biochem J. 1988;249:423–428. doi: 10.1042/bj2490423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- Susek RE, Chory J. A tale of two genomes: role of a chloroplast signal in coordinating nuclear and plastid gene expression. Aust J Plant Physiol. 1992;19:387–399. [Google Scholar]
- von Wettstein D, Gough S, Kannangara CG. Chlorophyll biosynthesis. Plant Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CJ, Willows RD. Mechanism and regulation of Mg-chelatase. Biochem J. 1997;327:321–333. doi: 10.1042/bj3270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willows RD, Gibson LCD, Kannangara CG, Hunter CN, von Wettstein D. Three separate proteins constitute the magnesium chelatase of Rhodobacter sphaeroides. Eur J Biochem. 1996;235:438–443. doi: 10.1111/j.1432-1033.1996.00438.x. [DOI] [PubMed] [Google Scholar]
- Witkowski DA, Halling BP. Accumulation of photodynamic tetrapyrroles induced by acifluorfen-methyl. Plant Physiol. 1988;87:632–638. doi: 10.1104/pp.87.3.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsebo KM, Hearst JE. Genetic-physical mapping of a photosynthetic gene cluster from R. capsulata. Cell. 1984;37:937–947. doi: 10.1016/0092-8674(84)90428-8. [DOI] [PubMed] [Google Scholar]