ABSTRACT
Legionella pneumophila genes encoding LapA, LapB, and PlaC were identified as the most highly upregulated type II secretion (T2S) genes during infection of Acanthamoeba castellanii, although these genes had been considered dispensable on the basis of the behavior of mutants lacking either lapA and lapB or plaC. A plaC mutant showed even higher levels of lapA and lapB transcripts, and a lapA lapB mutant showed heightening of plaC mRNA levels, suggesting that the role of the LapA/B aminopeptidase is compensatory with respect to that of the PlaC acyltransferase. Hence, we made double mutants and found that lapA plaC mutants have an ~50-fold defect during infection of A. castellanii. These data revealed, for the first time, the importance of LapA in any sort of infection; thus, we purified LapA and defined its crystal structure, activation by another T2S-dependent protease (ProA), and broad substrate specificity. When the amoebal infection medium was supplemented with amino acids, the defect of the lapA plaC mutant was reversed, implying that LapA generates amino acids for nutrition. Since the LapA and PlaC data did not fully explain the role of T2S in infection, we identified, via proteomic analysis, a novel secreted protein (NttD) that promotes infection of A. castellanii. A lapA plaC nttD mutant displayed an even greater (100-fold) defect, demonstrating that the LapA, PlaC, and NttD data explain, to a significant degree, the importance of T2S. LapA-, PlaC-, and NttD-like proteins had distinct distribution patterns within and outside the Legionella genus. LapA was notable for having as its closest homologue an A. castellanii protein.
KEYWORDS: Acanthamoeba, acyltransferase, aminopeptidase, Legionella pneumophila, Legionnaires' disease, protease, type II secretion, intracellular infection
IMPORTANCE
Transmission of L. pneumophila to humans is facilitated by its ability to grow in Acanthamoeba species. We previously documented that type II secretion (T2S) promotes L. pneumophila infection of A. castellanii. Utilizing transcriptional analysis and proteomics, double and triple mutants, and crystal structures, we defined three secreted substrates/effectors that largely clarify the role of T2S during infection of A. castellanii. Particularly interesting are the unique functional overlap between an acyltransferase (PlaC) and aminopeptidase (LapA), the broad substrate specificity and eukaryotic-protein-like character of LapA, and the novelty of NttD. Linking LapA to amino acid acquisition, we defined, for the first time, the importance of secreted aminopeptidases in intracellular infection. Bioinformatic investigation, not previously applied to T2S, revealed that effectors originate from diverse sources and distribute within the Legionella genus in unique ways. The results of this study represent a major advance in understanding Legionella ecology and pathogenesis, bacterial secretion, and the evolution of intracellular parasitism.
INTRODUCTION
One of 62 species in Legionella (1), Legionella pneumophila is an inhabitant of water systems and the main agent of Legionnaires’ disease, a pneumonia of increasing prevalence (2, 3). Disease largely results from inhalation of contaminated droplets from aerosolizing devices (4). In aquatic habitats, the Gram-negative bacterium L. pneumophila flourishes as an intracellular parasite of amoebas, especially species of Acanthamoeba, Vermamoeba, and Naegleria (5, 6). Within lungs, L. pneumophila grows in macrophages, residing, as it does in amoebas, in a membrane-bound compartment, the Legionella-containing vacuole (LCV) (7, 8). Critical to infection is a type IV secretion system (T4SS), which translocates myriad effectors from the LCV into the host cytoplasm (9–11).
Type II secretion (T2S) is another major factor in L. pneumophila pathogenesis (12, 13). The T2S system (T2SS) promotes infection of the lungs, enhancing Legionella growth in macrophages, dampening the host cytokine response, and releasing damaging enzymes (14–16). While the T2SS does not impact entry into macrophages or evasion of lysosomes, it does promote replication in the LCV at 4 to 12 h postentry (17). T2S is also critical for L. pneumophila survival in water and for infection of Acanthamoeba castellanii, Naegleria lovaniensis, and Vermamoeba vermiformis (18–24). T2S also potentiates L. pneumophila biofilm formation and sliding motility (25, 26). During T2S, protein substrates first move across the inner membrane via the Sec or Tat pathway; once in the periplasm, they are recognized by the T2SS apparatus, which uses a pseudopilus to drive the proteins through an outer membrane channel (13, 27). On the basis of proteomic and enzymatic analysis of supernatants, we discerned that L. pneumophila T2S elaborates at least 27 proteins, including nearly 20 enzymes and novel proteins of unknown activity (13, 28, 29). T2S-dependent proteins known to promote infection of key amoebal hosts include the novel NttA protein, which facilitates infection of A. castellanii; the novel NttC protein, which fosters infection of V. vermiformis; and acyltransferase PlaC, metalloprotease ProA, and RNase SrnA, which enhance infection of V. vermiformis and N. lovaniensis (23, 24, 30, 31). Overall, L. pneumophila T2S is one of the best-characterized T2SSs, in terms of its output and functions (13). Yet there are gaps in knowledge; e.g., the exoproteins thus far linked to infection do not fully explain the impact of T2SS on L. pneumophila ecology. On the basis of a form of transcriptional analysis and proteomics, we have now used genetic, biochemical, bioinformatic, and structural biology methods to define a eukaryotic-protein-like aminopeptidase and a novel secreted protein that provide unique insight into the evolution of T2S and parasitism.
RESULTS
Transcriptional analysis identified T2SS-dependent LapA, LapB, and PlaC as hyperexpressed in a compensatory manner during infection of A. castellanii.
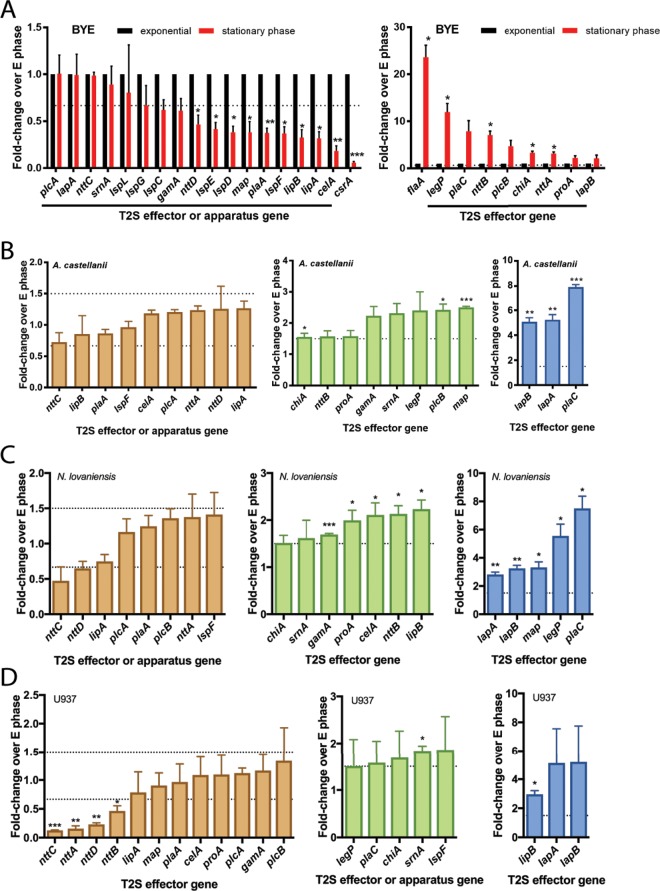
As a new means of uncovering important secreted proteins, we identified T2S substrates whose level of gene transcription is upregulated during infection of amoebas. We harvested wild-type strain 130b bacteria from both infected A. castellanii and exponential-phase buffered yeast extract (BYE) cultures, isolated RNA, and used quantitative reverse transcriptase PCR (qRT-PCR) to compare gene expression levels in intracellular versus extracellular bacteria. Since past studies identified some infectivity factors as being upregulated during the stationary phase (32), we also analyzed expression in stationary-phase cultures. Whereas five of the secreted-protein genes showed no change in expression in comparisons of log-phase versus stationary-phase bacteria, six others, along with the non-T2S csrA gene (33), were downregulated during the stationary phase (Fig. 1A, left). In contrast, eight other substrate genes, along with non-T2S flaA (32), had or trended toward increased expression in stationary phase (Fig. 1A, right). We also saw that the expression levels of genes encoding the T2SS apparatus were unchanged or downregulated in the stationary phase (Fig. 1A, left). Analyzing intracellularly grown bacteria, we found that 8/19 secreted-protein genes, along with lspF, which is representative of the T2SS apparatus, were expressed at a level that was equal to or slightly less than that seen in broth-grown bacteria (Fig. 1B, left). Eight other substrate genes were upregulated ~2-fold in intracellular bacteria (Fig. 1B, middle). Most notably, lapA, lapB, and plaC displayed 5- to 8-fold-higher levels in bacteria from within amoebas (Fig. 1B, right), suggesting that aminopeptidases LapA and LapB, which share 42% identity (30), and acyltransferase PlaC (34) are very important during intracellular growth.
FIG 1 .
Relative expression levels of genes encoding type II-secreted proteins (effectors) in wild-type L. pneumophila grown in broth or within amoebas and human macrophages. (A) Gene expression within wild-type 130b bacteria grown in BYE broth to the exponential phase (E phase) or stationary phase. qRT-PCR data are presented as means with standard errors of results from three independently grown cultures, with expression in the stationary phase normalized to expression in the E phase in broth. (B to D) Gene expression within 130b bacteria obtained from infected A. castellanii (B) or N. lovaniensis (C) at 44 h postinoculation or from infected human U937 macrophages (D) at 12 h postinoculation. qRT-PCR data are presented as means with standard errors of results from three independent infections (n = 3 each), with the intracellular expression normalized to the E phase in broth. Dashed lines indicate the customary 1.5-fold up- and/or downregulation threshold of biological interest. Asterisks indicate significant differences with >1.5-fold change relative to the E phase (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [one-sample t test]).
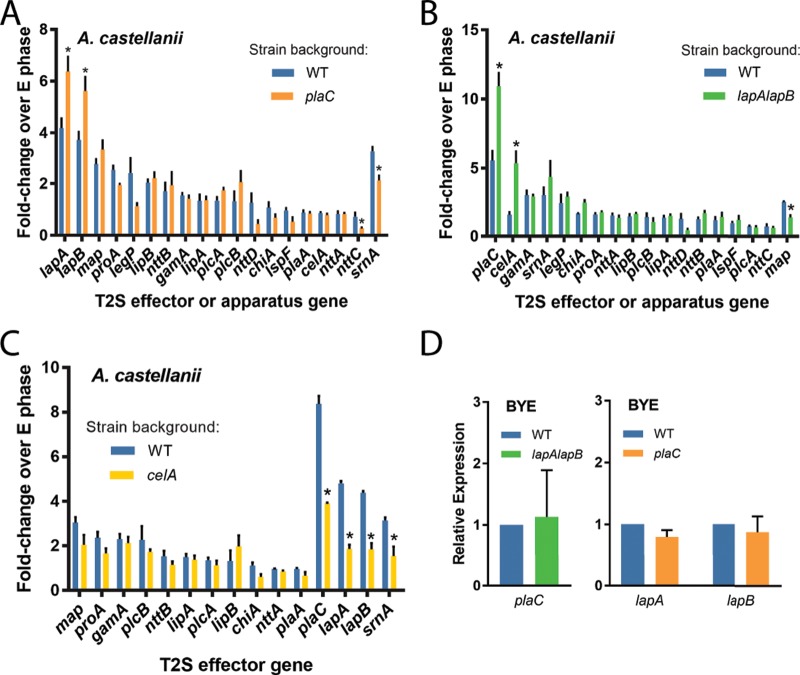
When we examined, in a similar way, gene expression in a plaC mutant (NU420) (23), we detected an even higher level of expression of lapA and lapB, while the expression levels of other T2S genes were unchanged or slightly decreased (Fig. 2A). In contrast, when we assayed transcript levels in a lapA lapB mutant (NU324) (30), there was a further heightening in levels of plaC mRNA (Fig. 2B). We also detected an increase in celA transcription in the lapA lapB mutant (Fig. 2B), but, when we examined expression in a celA mutant (NU353) (29), there was no compensatory increase in expression of lapA or lapB or of any other genes (Fig. 2C). These data suggested that the intracellular role(s) of LapA/LapB might be functionally compensatory with respect to that of PlaC, such that when the aminopeptidases are absent there is an increase in acyltransferase levels and vice versa. When the plaC mutant and lapA lapB mutant were grown in broth, no compensatory changes in expression were seen (Fig. 2D), implying that the functional redundancy between PlaC and LapA/LapB may be specific to intracellular growth.
FIG 2 .
Relative expression levels of genes encoding type II-secreted protein effectors in plaC mutant, lapA lapB mutant, and celA mutant bacteria grown in A. castellanii or in broth. (A to C) Gene expression within wild-type strain 130b (WT) versus plaC mutant NU420 (A), WT versus lapA lapB mutant NU324 (B), or WT versus celA mutant NU353 (C) obtained from infected A. castellanii at 44 h postinoculation. qRT-PCR data are represented as means with standard errors of results from three independent infections (n = 3 each), with expression normalized to the exponential (E) phase in broth. Asterisks indicate significant differences with >1.5-fold change in expression relative to the WT (*, P < 0.05 [Student’s t test]). (D) Gene expression in WT and lapA lapB mutant NU324 (left) or plaC mutant NU420 (right) during E phase in broth. Data are presented as means with standard errors of results from three independent cultures (n = 3), with expression normalized to the WT results.
Double mutants reveal key roles for LapA and PlaC in infection of A. castellanii.
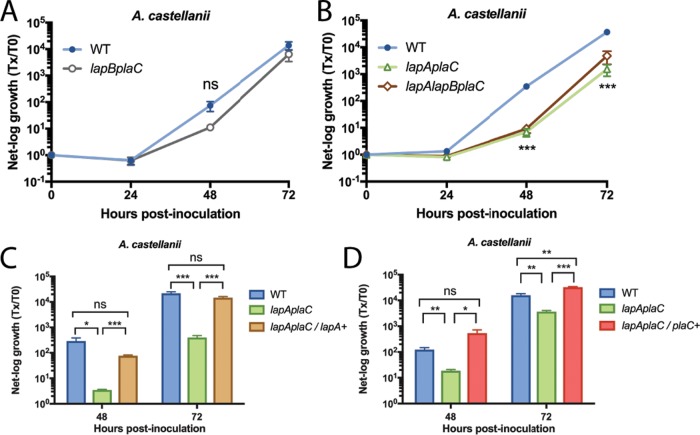
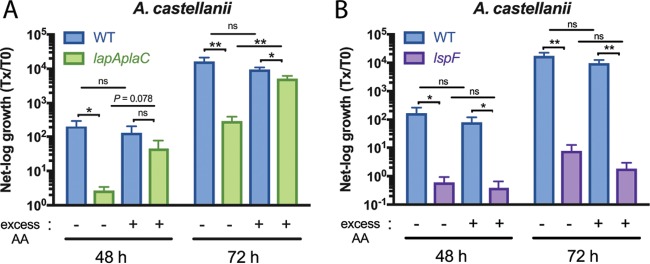
We previously observed that neither plaC mutant NU420 nor lapA lapB mutant NU324 is impaired for infection of A. castellanii (23, 30). Given our transcriptional data, we posited that a lapA plaC mutant, a lapB plaC mutant, and/or a lapA lapB plaC mutant might be impaired for infection due to the combined loss of two or more functionally redundant genes. Thus, we made double and triple mutants, and after observing their capacity to grow typically in BYE medium and survive normally in conditioned amoebal medium (see Fig. S1A and B [left panels] in the supplemental material), we tested them for infection of A. castellanii. Whereas the lapB plaC mutant (NU433) grew similarly to the wild type (Fig. 3A), the lapA plaC mutant (NU435) displayed an ~50-fold defect in infection (Fig. 3B). The lapA lapB plaC mutant (NU436) was no more defective than the lapA plaC mutant (Fig. 3B). Since lapA and plaC are monocistronic (Fig. S2A and B), the mutant defect was likely due to loss of lapA and plaC rather than polarity on downstream genes. When lapA was reintroduced into the lapA plaC mutant, there was restoration of infection (Fig. 3C), confirming the role of LapA. Similarly, the lapA plaC mutant was complemented by reintroduction of plaC (Fig. 3D), affirming the role of PlaC. Thus, we extended our transcriptional analysis of intracellular legionellae to include N. lovaniensis (Fig. 1C). Again, we saw that lapA, lapB, and plaC were upregulated (Fig. 1C, right). legP and map were also notably upregulated (Fig. 1C, right). Previously, we determined that the lapA lapB mutant is not impaired for infection of N. lovaniensis, whereas the plaC mutant has an ~10-fold defect (23). Given the transcription data, we compared the lapA lapB plaC mutant and plaC mutant for infection of N. lovaniensis, to possibly uncover another role for LapA (or LapB). However, the triple mutant was no more impaired than the single mutant (Fig. S3A). A like result was attained when we tested mutants in V. vermiformis (Fig. S3B). When analysis was expanded to include infected macrophages (Fig. 1D), lapA and lapB appeared to be among the most highly upregulated genes (right), whereas the change seen with plaC was modest (middle). Since the lapA lapB mutant and plaC mutant are not impaired in U937 cells (30, 34), we compared the lapA lapB plaC mutant to the wild type with respect to infection of macrophages. However, no new mutant phenotype was detected (Fig. S3C). Thus, though lapA and plaC expression was upregulated upon Legionella growth in two hosts, the compensatory link between LapA and PlaC was detected only in A. castellanii. Our mutant analysis, which was based on transcript data, uncovered a role for PlaC in infection of A. castellanii, expanding the idea of the protein’s importance beyond its role in infection of V. vermiformis and N. lovaniensis (23). A glycerophospholipid-cholesterol and -ergosterol acyl-transferase that has phospholipase A and lysophospholipase A activities, PlaC has been well studied biochemically (34, 35). More importantly, our transcriptional and mutational analyses revealed, for the first time, a role for the LapA aminopeptidase in an intracellular infection event; thus, we focused our next efforts on defining the biochemical, structural, and phylogenetic traits of LapA.
FIG 3 .
Intracellular infection of A. castellanii by L. pneumophila wild-type, lapB plaC mutant, lapA plaC mutant, and lapA lapB plaC mutant strains. (A and B) A. castellanii amoebas were infected with either wild-type strain 130b (WT) and lapB plaC mutant NU433 (A) or WT, lapA plaC mutant NU435, and lapA lapB plaC mutant NU436 (B). At the indicated times, net bacterial growth was determined by plating for CFU counts. Data are presented as means with standard errors of results from three independent experiments (n = 3 each). Tx/T0, determination time versus time 0. (C and D) Amoebas were infected with WT, lapA plaC mutant NU435, and complemented mutant NU435 containing plasmid-carried lapA (C) or plasmid-carried plaC (D). Bacterial growth data are presented as means with standard deviations of results from four infected wells and are representative of three independent experiments. Asterisks indicate significant differences in CFU recovery between a mutant and the WT or its complemented derivative (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [Student’s t test]). ns, not significant.
Extracellular replication and survival of the L. pneumophila wild-type strain, an nttD mutant, a lapA plaC mutant, and a lapA lapB plaC mutant in conditioned amoebal medium. (A) Wild-type strain 130b (WT), lapA plaC mutant NU435 and lapA lapB plaC mutant NU436 (left), and the WT strain and nttD mutant NU431 (right) were inoculated into BYE broth, and then, at various times postinoculation, bacterial growth was monitored spectrophotometrically. The data points represent means and standard deviations of results from duplicate cultures, and the results presented are representative of two independent experiments. (B) A. castellanii amoebas were seeded in-well, and then a 0.4-µm transwell insertion was added followed by bacterial suspensions to prevent contact between the WT strain, lapA plaC mutant NU435, and lapA lapB plaC mutant NU436 (left) or between the WT strain and nttD mutant NU431 (right) or with the host cells. At the indicated times, CFU counts were recorded from within the upper chamber. Data are presented as means and standard deviations of results from duplicate wells and are representative of two independent experiments. Download FIG S1, EPS file, 1.3 MB (1.3MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
L. pneumophila loci encoding type II-secreted proteins. The chromosomal regions of strain 130b containing lapA (A), plaC (B), and nttD (C) are depicted. The horizontal arrows denote the locations, orientations, and relative sizes of the genes. The “lpw” numbers indicated within the arrows are ORF designations used in the database. Gene names and annotations are indicated below the arrows. Sizes of the genes are given (in base pairs) above the arrows, and sizes of intergenic regions are noted between the arrows. (A) The genes adjacent but transcriptionally unlinked to lapA encode type IV secretion effectors (N. Shohdy, J. A. Efe, S. D. Emr, and H. A. Shuman, Proc Natl Acad Sci U S A 102:4866–4871, 2005; E. Portier, H. Zheng, T. Sahr, D. M. Burnside, C. Mallama, C. Buchrieser, N. P. Cianciotto, and Y. Héchard, Environ Microbiol 17:1338–1350, 2015; D. T. Isaac, R. K. Laguna, N. Valtz, and R. R. Isberg, Proc Natl Acad Sci U S A 112:E5208–E5217, 2015). (B) The gene upstream of plaC is predicted to encode part of an ABC transporter, whereas the downstream gene is likely involved in glucosamine-6-phosphate metabolism (A. Gaballa and J. D. Helmann, J Bacteriol 180:5815–5821, 1998; R. A. Tinsley, J. R. Furchak, and N. G. Walter, RNA 13:468–477, 2007). (C) The gene upstream of nttD is not annotated, although tertiary structure predictions suggest that its protein product is a lipase (L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass, and M. J. Sternberg, Nat Protoc 10:845–858, 2015). This protein may also be a substrate for the T2S based on the detection of the protein in wild-type supernatants and the presence of a signal peptide at the protein’s N terminus (28). The two genes downstream of nttD are predicted to be involved in DNA rearrangement and repair (M. M. Slupska, J. H. Chiang, W. M. Luther, J. L. Stewart, L. Amii, A. Conrad, and J. H. Miller, Genes Cells 5:425–437, 2000; C. Darrigo, E. Guillemet, R. Dervyn, and N. Ramarao, PLoS One 11:e0163321, 2016). Download FIG S2, EPS file, 2.3 MB (2.3MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Intracellular infection of N. lovaniensis, V. vermiformis, and human macrophages by the L. pneumophila wild-type, plaC mutant, or lapA lapB plaC mutant strain. N. lovaniensis amoebas (A), V. vermiformis amoebas (B), or U937 macrophages (C) were infected with wild-type strain 130b (WT) or plaC mutant NU420 (panels A and B only) or with lapA lapB plaC mutant NU436, and at 0, 24, 48, and 72 h postinoculation, CFU levels within the infected wells were determined. Bacterial growth resulting from intracellular infection is expressed as the ratio of the CFU count at t = 24, 48, or 72 h to the CFU count at t = 0. Data are presented as means and standard errors of results from three independent experiments (n = 3). Asterisks indicate significant differences in levels of CFU recovery between the WT strain and all mutants (*, P < 0.05 [Student’s t test]). Download FIG S3, EPS file, 0.7 MB (750.9KB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LapA is a protease-activated, broad-spectrum aminopeptidase.
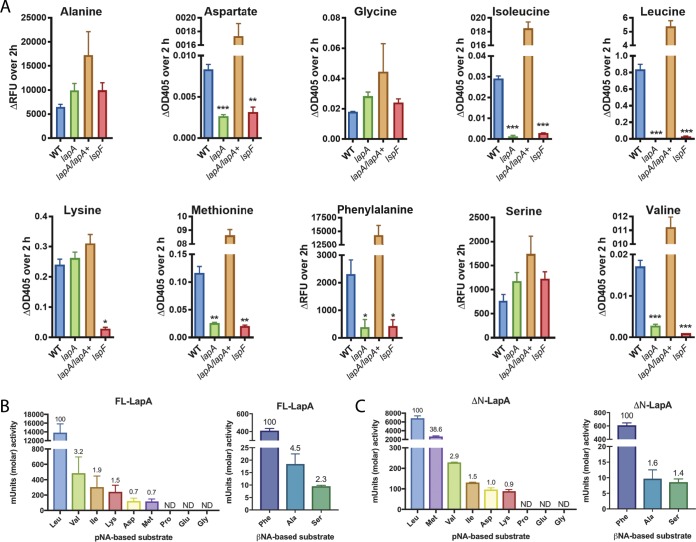
Past work found that LapA is active against leucine, phenylalanine, and tyrosine aminopeptides, based on the ability of wild-type supernatants but not lapA mutant supernatants to cleave those substrates (30). LapA is annotated as a leucine aminopeptidase, and the Merops database assigns LapA as a member of the M28 family of peptidases (subfamily E) (28, 30, 36). We more completely defined the specificity of LapA by testing wild-type, lspF mutant, and lapA mutant supernatants against additional aminopeptides. Wild-type samples had activity against alanine, aspartate, glycine, isoleucine, lysine, methionine, serine, and valine aminopeptides, in addition to leucine and phenylalanine aminopeptides (Fig. 4A). They had no activity against proline- or glutamate-containing substrates. The lspF mutant lost activity against aspartate, isoleucine, leucine, lysine, methionine, phenylalanine, and valine substrates but showed normal activity against alanine, glycine, and serine aminopeptides (Fig. 4A). Thus, 7/10 of the activities are mediated by T2S. The lapA mutant lacked activity against aspartate, isoleucine, methionine, and valine aminopeptides and cleaved alanine, glycine, lysine, and serine substrates (Fig. 4A). As described before (30), the mutant lacked activity against leucine and phenylalanine aminopeptides (Fig. 4A). When the complemented lapA mutant was examined, we saw restoration of the six activities that were lacking in the mutant (Fig. 4A). The complement trended toward elevated levels of activity against alanine and serine aminopeptides (Fig. 4A), suggesting that increased amounts of LapA might cleave additional substrates.
FIG 4 .
Aminopeptidase activities from L. pneumophila culture supernatants and purified recombinant LapA. Filter-sterilized supernatants from late-exponential-phase broth cultures of wild-type 130b (WT), lapA mutant NU320 (lapA), complemented lapA mutant NU320 containing plasmid-carried lapA (lapA/lapA+), and lspF mutant NU275 (lspF) were assayed for activity directed toward aminopeptidase substrates, with the N-terminal amino acid (AA) residue given. (A) Changes in background-adjusted absorbance at OD405 for the AA–p-nitroanilide substrates or fluorescence (expressed in relative fluorescence units [RFU]) for the AA–β-naphthylamide substrates were monitored kinetically over 2 h. Data are presented as means with standard errors of results from three independent experiments (n = 3 each). Asterisks indicate significant reductions in activity of a mutant relative to the WT (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [Student’s t test]). (B and C) Strains with purified, full-length LapA (FL-LapA) (B) or LapA missing its N-terminal PA domain (ΔN-LapA) (C) were incubated for 2 h at 37°C with various AA–p-nitroanilide substrates (left panels) or AA–β-naphthylamide substrates (right panels), and the levels of molar activity were determined (y axes). The relative activities against the various substrates are displayed above each bar, with the level of activity directed toward Leu-pNA (B and C, left) or toward Phe-βNA (B and C, right) arbitrarily set to 100. ND, not detected. Data are presented as means with standard errors of results from two independent experiments.
To confirm the activities of LapA, we purified recombinant LapA-His6-tagged protein containing 378 amino acids of the mature lapA product (minus the 23-residue signal sequence). As expected, purified LapA had activity against leucine, valine, isoleucine, aspartate, methionine, and phenylalanine aminopeptides, with its activity being greatest against the leucine substrate (Fig. 4B). LapA lacked activity against proline, glutamate, and glycine, compatible with supernatant tests. Although assaying of supernatants had not revealed LapA-dependent cleavage of alanine, lysine, and serine aminopeptides, due to the presence of LapB and a T2S-independent factor (Fig. 4A) (30), the testing of purified enzyme affirmed LapA’s ability to cleave these substrates (Fig. 4B). Combining these data with past results (30), we conclude that LapA is active against ≥10 substrates. Most substrates had nonpolar, amino acid targets, but, given its cleavage of aspartate, lysine, serine, and tyrosine aminopeptides, LapA has rather broad activity (37, 38). Typically of a member of the M28 family (39), LapA was inhibited by bestatin (Fig. S4A).
Dose-dependent inhibition and substrate binding pocket of LapA. (A) Purified full-length LapA was incubated at 37°C with a leucine aminopeptidase substrate and 2-fold-increasing concentrations of bestatin. The residual aminopeptidase activity compared to that seen with untreated LapA was graphed against the log concentration of the inhibitor, and the half-maximal inhibitory concentration (IC50) was determined via nonlinear regression and sigmoidal slope response (variable slope) curve fitting (GraphPad Prism, version 7.00 for Windows; Graphpad Software, Inc., La Jolla, CA). Data are presented as mean residual activity with standard errors of results from two independent experiments (n = 2). (B) The S. griseus aminopeptidase/l-leucine complex (PDB code: 1F2O) and the A. proteolytica aminopeptidase/bestatin complex (PDB code: 1XRY) were superimposed on the aminopeptidase domain of LapA. The LapA substrate binding pocket is shown as light orange sticks, with modeled l-leucine and bestatin shown as green sticks. Download FIG S4, EPS file, 1.1 MB (1.1MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LapA was predicted to contain an N-terminal, “protease-associated” (PA) domain, which in some M28 peptidases is necessarily excised in order to release a C-terminal peptidase domain (40, 41). That LapA might be processed is suggested from our prior two-dimensional (2-D) PAGE analysis of supernatants, which had detected truncated forms of LapA (28). To determine if processed LapA has activity, we purified a version of LapA that was missing the putative PA domain and repeated the enzyme assays. Truncated LapA had activity against the same substrates that had been cleaved by full-length LapA, with leucine aminopeptide remaining as the favored substrate (Fig. 4C). Interestingly, the activity against methionine aminopeptide and, to a lesser extent, the phenylalanine aminopeptide was elevated, whereas the activities against the other seven substrates were similar or slightly decreased (Fig. 4B and C). These data indicate that the removal of the PA domain of LapA, though not required, yields an enzymatically active protein. Also, full-length LapA and truncated LapA may have different substrate preferences.
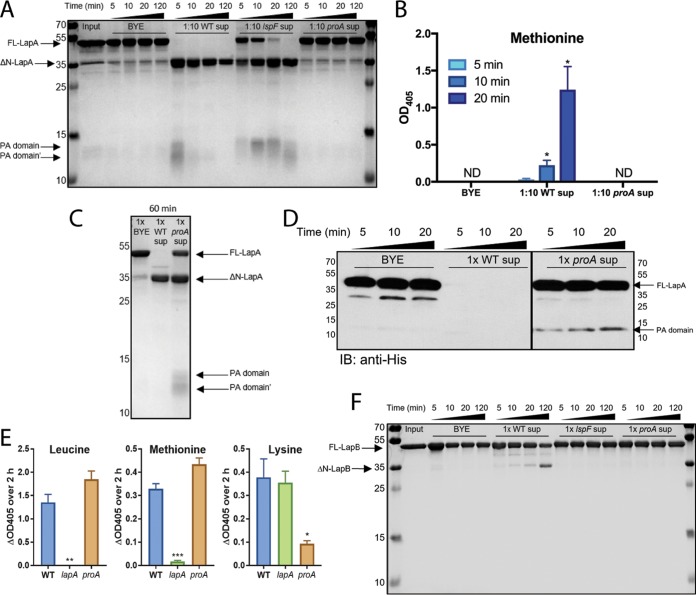
We posited that the T2SS-dependent ProA protease might be cleaving secreted LapA into its N-terminal PA domain and C-terminal peptidase domain. ProA is the major secreted protein of L. pneumophila and is known to cleave other exoproteins (19, 35, 42, 43). Hence, we incubated recombinant LapA, with its His6 tag N terminal to its PA domain, along with supernatants (diluted 1/10) obtained from the wild-type 130b strain, the lspF mutant, or the proA mutant, and assayed for the cleavage of LapA by PAGE and Coomassie staining (Fig. 5A). Wild-type supernatants rapidly cleaved the ~44-kDa LapA, resulting in the appearance of an ~35-kDa C-terminal portion, an ~13-kDa PA domain, and smaller cleavage/degradation products. However, proA mutant supernatants exhibited no evidence of cleavage over 2 h of incubation, appearing to be equivalent in that respect to the BYE medium control (Fig. 5A). The lspF mutant supernatants were also deficient for activity, although cleavage products did appear, most likely due to some release of ProA as a result of cell lysis. Compatibly with these data, purified LapA treated with (1/10-diluted) wild-type supernatants, but not proA mutant supernatants or BYE medium, cleaved methionine aminopeptide (Fig. 5B). Thus, we infer that ProA cleaves and activates LapA. Interestingly, when purified LapA was mixed for 1 h with undiluted rather than diluted supernatants, ProA-independent cleavage of LapA was seen (Fig. 5C). When immunoblots were used to examine the effect of undiluted proA mutant supernatants, the low-level cleavage event (the appearance of His-tagged PA) was detected after 5 min (Fig. 5D). However, undiluted proA mutant supernatants retained activity against leucine and methionine aminopeptides (Fig. 5E). Incidentally, the proA mutant supernatants still lacked activity against lysine aminopeptide, a LapB substrate (Fig. 5E), and ProA-dependent cleavage of LapB was observed on stained gels (Fig. 5F). Overall, these data showed that ProA is mainly responsible for activation of LapA but that LapA can also be cleaved, albeit to a lesser extent, by another secreted factor.
FIG 5 .
Effect of type II-secreted factors on LapA and LapB processing and activity. (A and B) Purified, full-length LapA (FL-LapA) (Input) was incubated at 37°C for the indicated times with either BYE medium or 1/10-diluted supernatants (sup) obtained from late-exponential-phase broth cultures of wild-type strain 130b (WT), lspF mutant NU275 (lspF), or proA mutant AA200 (proA) and then processed by SDS-PAGE followed by Coomassie staining for total protein detection (A) or incubated with methionine aminopeptidase substrate in order to assess LapA activation (B). Arrows to the left of the gel image in panel A denote the migration position of FL-LapA, LapA missing its N terminus (ΔN-LapA), the PA domain of LapA (PA domain), and smaller cleavage products (PA domain′). The identity of the minor bands that were present within the input material (including one that appears to nearly comigrate with ΔN-LapA) and were maintained during incubation with the BYE medium or culture supernatants is unknown, but they likely represented breakdown products of the recombinant LapA. The migration of molecular mass markers (in kilodaltons) is also denoted to the left of the gel in panel A. Enzyme activity data in panel B are presented as means with standard errors of results from three independent experiments (n = 3 each). Asterisks indicate significant increases in activity relative to t = 5 min (*, P < 0.05 [Student’s t test]). ND, not detected. (C and D) Purified FL-LapA was incubated at 37°C for the indicated times with either BYE medium or undiluted culture supernatants obtained from WT or proA mutant cultures and then processed for SDS-PAGE and Coomassie staining (C) or immunoblot analysis (IB) using anti-His antibodies directed against the hexahistidine tag embedded at the start of the PA domain (D). Arrows to the right side of the images in panels C and D denote the migration position of FL-LapA, ΔN-LapA, and the PA domain fragments, with the migration of the molecular mass markers denoted as well. Although the samples examined in panel D were on the same gel, they were not in adjacent lanes; therefore, we cropped out the intervening lanes that were not pertinent to the analysis. Data in panels C and D are representative of results of three and two experiments, respectively. (E) Filter-sterilized supernatants from late-exponential-phase broth cultures of wild-type 130b (WT), lapA mutant NU320 (lapA), and proA mutant AA200 (proA) were assayed for activity directed toward aminopeptidase substrates, with the N-terminal amino acid (AA) residue given. Changes in background-adjusted absorbance at OD405 for the AA–p-nitroanilide substrates were monitored kinetically over 2 h. Data are presented as means with standard errors of results from three independent experiments (n = 3 each). Asterisks indicate significant reduction in activity of a mutant relative to the WT (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [Student’s t test]). (F) Purified, full-length LapB (FL-LapB) (Input) was incubated at 37°C for the indicated times with either BYE medium or undiluted supernatants obtained from late-exponential-phase broth cultures of wild-type strain 130b (WT), lspF mutant NU275 (lspF), or proA mutant AA200 (proA) and then processed by SDS-PAGE followed by Coomassie staining for total protein detection. Arrows to the left of the gel image denote the migration position of FL-LapB and of LapB missing its N terminus (ΔN-LapB). The migration of molecular mass markers (in kilodaltons) is also denoted to the left of the gel. Data are representative of results from 3 independent experiments.
The structure of LapA.
To determine how LapA provides its broad specificity, we began structural studies of recombinant LapA. Crystals were readily obtained, and its structure was determined by molecular replacement, using the coordinates of LapB (PDB code: 5GNE) (41) as the search model, and electron density maps were refined to 1.9 Å. The structure of LapA contains two molecules in the asymmetrical unit, and all residues could be built except for the N-terminal His tags and residues within the interdomain linkers (residues 82 to 101) that connect the PA domain (residues 1 to 81) and aminopeptidase domains (residues 102 to 378). The PA domain is formed of four helices and three β-strands, and the aminopeptidase domain has nine helices and seven β-strands. Although LapA was purified as a monomer, in our structure, the PA domain of one chain was associated with the aminopeptidase domain of an adjacent chain, which formed a domain-swapped dimer (Fig. 6A). The domain-swapped monomers were essentially identical, with a root mean square deviation (RMSD) over Cα atoms of 0.083 Å (without linkers), although variations were seen within the interdomain sequence. As an M28 family metalloprotease, the peptidase domain of LapA binds two Zn ions in its active site, which are coordinated by the conserved residues His194, Asp207, Glu241, Glu242, Asp269, and His347 and one water molecule (Fig. 6B). Drawing on structures of aminopeptidases from Streptomyces griseus (PDB code: 1F20) (44) and Aeromonas proteolytica (PDB code: 1XRY) (45), the main determinant of specificity in this class of enzyme is a binding pocket directly adjacent to the active site, and in LapA, this is formed from the side chains of Asp269, Met270, Phe285, Cys314, Cys318, Phe333, Cys335, Phe339, and His342 and from the backbone of Ser319, Thr337, and Ser338 (Fig. S4B). This creates a relatively deep hydrophobic cavity, which can readily accommodate aromatic and aliphatic residues (Fig. 6C; see also Fig. S4B). In our structure of LapA, the PA domain covers the active site and substrate-binding pocket (Fig. S5) and thus represents the proenzyme form of LapA. A ring of predominantly hydrophobic residues on the surface of the enzymatic domain (Trp170, Leu198, Asp199, Tyr316, Phe339, His342, His347) provides a platform for the PA domain to dock, and this is mediated through its β-1 strand (Glu7), α2-helix (Ser39, Glu43, Thr46, Asp50), α2-α3 linker (His56, Phe57, Asn59), and α3-helix (His62).
FIG 6 .
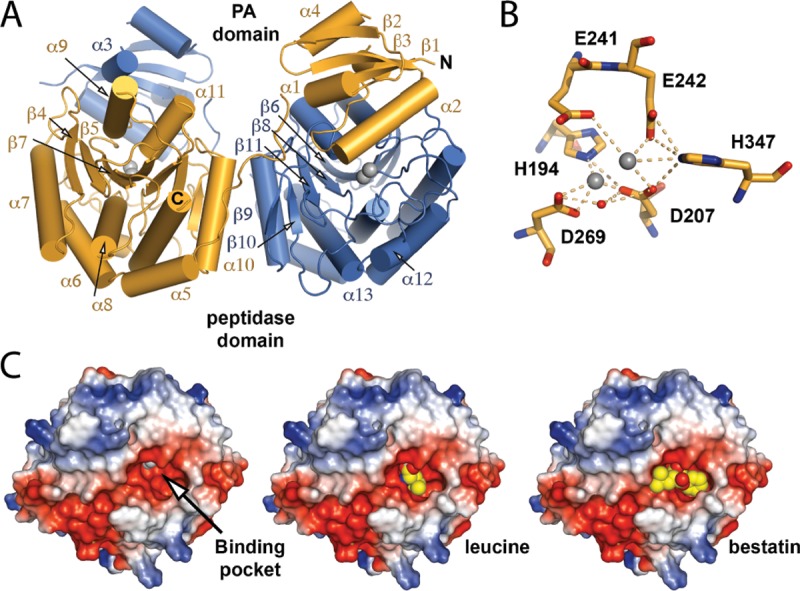
Overall structure of LapA. (A) The LapA domain-swapped dimer of the asymmetrical unit is presented as a cartoon with zinc ions drawn as gray spheres. (B) Active-site residues are shown as sticks, and zinc ions (gray) and water molecules (red) are drawn as spheres. (C) Electrostatic surface of the LapA aminopeptidase domain with active-site zinc ions as gray spheres (left). The electrostatic surface of LapA superimposed with (middle) the S. griseus aminopeptidase/l-leucine complex (PDB code: 1F2O) (48) or (right) the A. proteolytica aminopeptidase/bestatin complex (PDB code: 1XRY) (49), indicating potential binding of substrate amino acids, is shown. Leucine and bestatin are shown as spheres.
Interactions within the PA/aminopeptidase domain interface of LapA. (Left) The PA domain is shown as a cartoon, while the aminopeptidase domain is shown as a cartoon with a transparent surface. Interfacial residues are drawn as sticks. (Right) Magnification of the domain interface shown from three orientations. The secondary structure and the residues that make up the interface are annotated. Download FIG S5, EPS file, 3.1 MB (3.2MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LapA promotes nutrient acquisition during infection.
Given the enzymatic activity of LapA, which was revealed by structural analysis, we posited that LapA promotes L. pneumophila growth in A. castellanii by cleaving aminopeptides to release amino acids for uptake by the parasite. Thus, we asked if addition of a cocktail of amino acids to the Legionella–Acanthamoeba coculture could rescue growth of the lapA plaC mutant. To facilitate this experiment, we employed an amino acid cocktail (aspartate, alanine, glutamate, asparagine, glycine, serine, and proline) that was previously used to rescue the growth of a L. pneumophila mutant that was starved due to its lack of a T4SS effector (46). Thus, we used a reagent that was not inhibitory to L. pneumophila or its host. Moreover, the aspartate, alanine, and serine in the cocktail are predicted products of LapA activity (Fig. 4). Whereas amino acid supplementation had no effect on wild-type growth, it increased lapA plaC mutant numbers at 72 h postinoculation, and the results trended toward increases at 48 h (Fig. 7A). Thus, amino acid supplementation phenocopied the complementation results (Fig. 3C). The supplementation did not rescue the lspF mutant (Fig. 7B), which is compatible with this mutant lacking many exoproteins. Since neither the wild type nor the T2SS mutant was affected by addition of amino acids, the rescue of the lapA mutant was not a nonspecific effect on A. castellanii or L. pneumophila. Thus, the LapA mutant’s infection defect is tied to its loss of enzyme activity. Given that LapA, PlaC, and amino acid acquisition does not fully account for the ≥1,000-fold defect observed upon complete loss of T2S (23), we sought additional secreted proteins that promote infection of amoebas.
FIG 7 .
Intracellular infection of A. castellanii by the L. pneumophila wild-type strain, lapA plaC mutant, and lspF mutant in the presence of added amino acids. A. castellanii amoebas were infected with wild-type strain 130b (WT), lapA plaC mutant NU435, or lspF mutant NU275, and, at the indicated times, bacterial growth was determined. Where indicated, the medium in the infected wells was supplemented with 1 mM excess amino acids (AA) for the duration of the experiment. Growth data are presented as means with standard errors of results from three independent experiments (n = 3 each). Asterisks indicate significant differences in CFU recovery between strains under the given conditions (*, P < 0.05; **, P < 0.01 [Student’s t test]). ns, not significant.
Proteomic and mutational analyses identified NttD as a novel T2SS-dependent exoprotein that also promotes infection of A. castellanii.
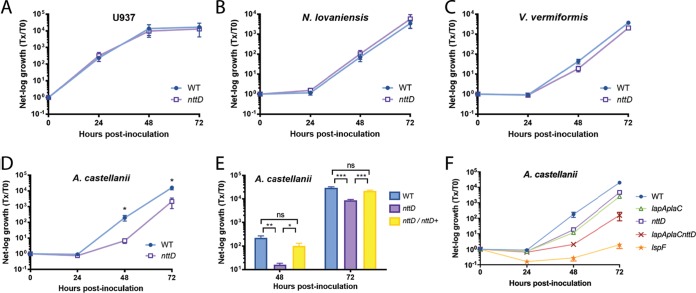
We next studied a 41-kDa protein that was previously detected in supernatants of L. pneumophila wild-type strain 130b grown in BYE broth but was absent in similarly derived supernatants of an lspF T2S mutant (28). Inspection of the 130b genome (47) revealed that the T2SS-dependent protein is encoded by open reading frame (ORF) L. pneumophila lpw10421, the second gene in a four-gene operon (Fig. S2C). Given our proteomics data and the bioinformatic analysis that is to follow, we named lpw10421 nttD, for novel type two secreted protein D, following the nomenclature used to denote nttA, nttB, and nttC (23, 24). NttD is not annotated but is noted to have domain-of-unknown-function 4785. An unpublished crystal structure for NttD from L. pneumophila strain Philadelphia-1 has been deposited by others in the Protein Data Bank (PDB code: 4KH9). NttD is a mainly β-strand protein, and analysis using the PISA server (48) indicated that it exists as a dimer (Fig. S6A), although we detected equilibrium between the monomer and dimer in solution (Fig. S6B). Each chain is made of three unique domains, with the N-terminal domain mediating dimerization. While tertiary-structure analysis using the Dali server (49) detected no homologies for the N-terminal domain (no Z score of >8), the C-terminal domain shared features with glycosidases. Yet when we tested purified protein and concentrated supernatants against glycosidase substrates, no activity was seen. In order to discern a role for NttD, we made a mutant of 130b inactivated for nttD (NU431). The mutant grew in BYE broth like the parental 130b strain (Fig. S1A, right), indicating that NttD is not required for extracellular survival, at least under standard conditions. From examination of BYE supernatants, the mutant had normal levels of phosphatase, protease, and lipase activities (not shown), indicating that loss of NttD does not cause a general defect in T2S (24, 50). Whereas the intracellular replication of the nttD mutant was not affected in the human U937 cell macrophages and Naegleria and Vermamoeba amoebas (Fig. 8A to C), it was impaired for infection of A. castellanii (Fig. 8D). At 48 and 72 h postinoculation, the NU431-infected acanthamoebae yielded ~30-fold-fewer legionellae. When infections were done in a transwell apparatus, such that bacteria were prevented from contacting host cells, the wild type and the mutant, though not growing, survived similarly (Fig. S1B, right), indicating that the reduced recovery of the mutant from the coculture was a result of defective infection rather than of impaired survival in the medium. When we tested a second nttD mutant (NU432) for infection of A. castellanii, the same defect was seen (not shown), implying that the loss of infectivity was due to the mutation in nttD rather than to a second-site mutation. Because a wild-type level of growth was achieved upon reintroduction of nttD into mutant NU431 (Fig. 8E), we concluded that NttD is required for infection of A. castellanii. Together, these data document NttD as a novel, T2SS-dependent exoprotein that promotes L. pneumophila infection of A. castellanii, thereby enhancing the range of Legionella in aquatic habitats. Discovering substantial roles for NttD, PlaC, and LapA in L. pneumophila infection of A. castellanii, we pondered to what degree these proteins account for the role of T2SS in infection. Thus, we tested a new mutant (NU441) that lacks lapA, plaC, and nttD. The triple mutant had a sizeable defect that went from ~90-fold at 48 h postinoculation to ~130-fold at 72 h (Fig. 8F). This defect was greater than that of the nttD mutant and lapA plaC mutant (Fig. 8F), indicating that NttD and LapA/PlaC operate in distinct pathways. The lapA plaC nttD mutant’s defect did not fully recapitulate the lspF mutant’s defect (Fig. 8F), indicating that there are additional T2S-dependent proteins that contribute to intracellular infection. However, the NttD, LapA, and PlaC data do explain, to a significant degree, the role of T2S in infection of A. castellanii.
FIG 8 .
Intracellular infection of macrophages and amoebas by the L. pneumophila wild-type strain, an nttD mutant, a complemented nttD mutant, and a lapA plaC nttD mutant. (A to E) U937 cells (A), N. lovaniensis (B), V. vermiformis (C), and A. castellanii (D and E) were infected with wild-type strain 130b (WT), nttD mutant NU431, or complemented nttD mutant NU431 containing plasmid-carried nttD, and, at 0, 24, 48, and 72 h postinoculation, CFU levels in the infected wells were assayed. Bacterial growth resulting from intracellular infection is expressed as the ratio of the CFU level at t = 24, 48, or 72 h to the CFU level at t = 0. Data in panels A to D are presented as means with standard errors of results from at least three independent experiments (n ≥ 3). Data in panel E are presented as means with standard deviations of results from four infected wells and are representative of three independent experiments. Asterisks indicate significant differences in the levels of CFU recovery between the mutant and the WT or the complemented mutant (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [Student’s t test]). (F) Amoebas were infected with the WT or lapA plaC mutant NU435, nttD mutant NU431, lapA plaC nttD mutant NU441, or lspF mutant NU275, and, at the indicated times, net bacterial growth was determined. Data are presented as means with standard errors of results from three independent experiments (n = 3 each). The lapA plaC nttD mutant displayed significantly reduced CFU recovery compared to the WT, the lapA plaC mutant, and the nttD mutant at 48 and 72 h postinoculation (P < 0.05 [Student’s t test]). The lspF mutant exhibited an even great reduction in growth compared to the WT, the lapA plaC mutant, and the nttD mutant at 24, 48, and 72 h (P < 0.01 [Student’s t test]).
NttD forms monomers and dimers in solution. (A) The crystal structure of the NttD dimer (PDB code: 4KH9) reveals three domains per monomer, which are numbered 1 (purple), 2 (yellow), and 3 (red) from the N terminus to the C terminus and are given for chain A. Chain B is depicted entirely in green. The NttD structure appears to form a stable homodimer, which is mediated through N-terminal domain 1. (B) Gel filtration profile of NttD showing that it forms both monomers and dimers in solution. Download FIG S6, EPS file, 1.4 MB (1.4MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bioinformatic analysis revealed distinctive distribution patterns for nttD-, plaC-, and lapA-related genes within and outside the Legionella genus.
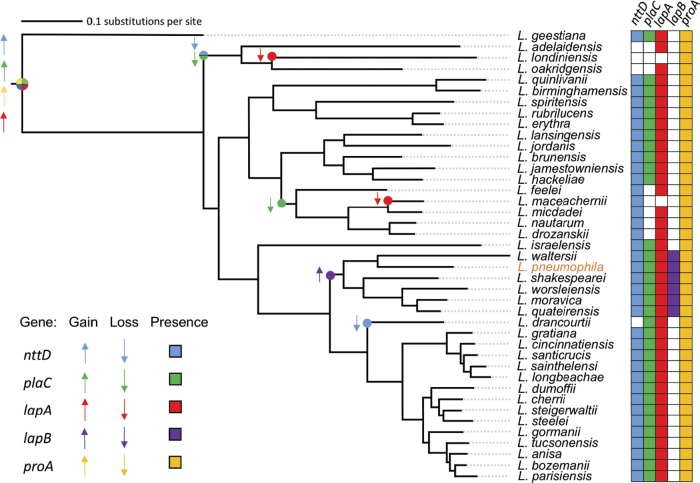
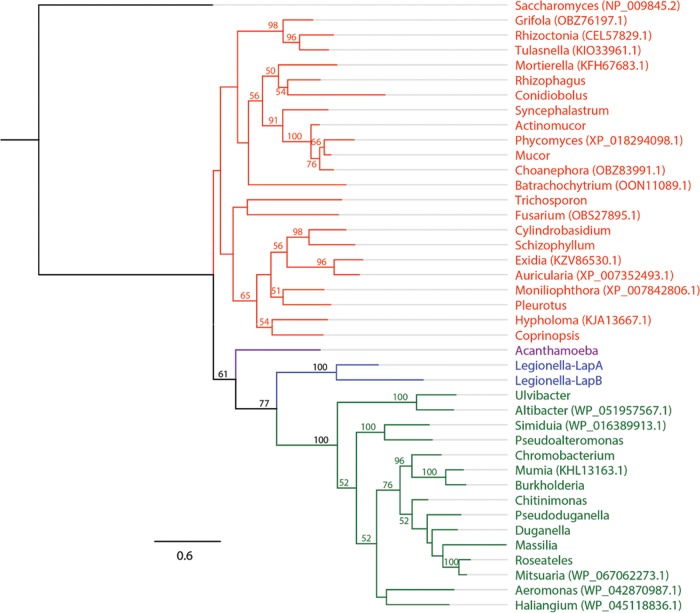
All sequenced L. pneumophila strains and all of the 40 other sequenced species of Legionella have complete T2SSs (see Table S1 in the supplemental material). Therefore, we performed BLAST analyses to discern the species- and genus-wide distribution of nttD, plaC, and lapA. The nttD gene was in all L. pneumophila strains, and an nttD-like gene was in 36/40 Legionella species (Fig. 9; see also Table S2A). On the basis of the distribution of nttD-like genes across the phylogenetic tree, nttD was found to be an ancestral gene that, though largely conserved, has been lost twice (Fig. 9). Further BLAST analysis found that NttD shares relatedness to hypothetical proteins in ≥9 genera of gammaproteobacteria and in one genus of deltaproteobacteria (Table 1). Similarly to nttD, plaC was conserved in the L. pneumophila species (Table S2B). A plaC-like gene was evident in 32 other Legionella species (Fig. 9; see also Table S2B). The 8 remaining species lacked a plaC-like gene, indicating that the gene occurs in ~80% of species. We surmise that plaC, like nttD, is an ancestral gene that, though reasonably conserved, has been lost twice (Fig. 9). Like nttD, plaC was lost prior to emergence of the adelaidensis-londiniensis-oakridgensis clade. Yet plaC appears to have also been lost prior to the advent of the feeleii-micdadei-drozanskii clade, resulting in a lower level of conservation than was determined for NttD (Fig. 9). Looking beyond Legionella, we found homologues of PlaC in genera of gammaproteobacteria, although these genera were different from those that had NttD homologues (Table 2). Interestingly, most non-Legionella homologues of PlaC, including the closest homologues, were in Cyanobacteria, a distant phylum that does not have a canonical T2SS (Table 2) (13). BLAST revealed that lapA occurs in all L. pneumophila strains (Table S2C). lapA-like genes were in 38 of the other species of Legionella (Fig. 9; see also Table S2C). Only L. maceachernii and L. londiniensis did not carry a lapA-like gene, due to two “recent” gene loss events. Thus, lapA is in ~95% of species, indicating that it is an ancestral gene that is even more prevalent than nttD or plaC (Fig. 9). LapB, though conserved within the L. pneumophila species (Table S2D), is present in only 12% of non-pneumophila species (Fig. 9; see also Table S2D). Seemingly, lapB is a recent gene acquisition, coincident with emergence of the clade containing L. pneumophila (Fig. 9). We also wondered about the prevalence of ProA, the T2SS-dependent protease that activates LapA and PlaC (Fig. 5) (35), and found that proA was 100% conserved (Fig. 9). Returning to the distribution of LapA-like proteins but looking beyond Legionella, BLAST analysis found that LapA’s closest prokaryotic homologues occur in betaproteobacteria (Table 3). This pattern of distribution of non-Legionella homologues was very different from that of NttD and PlaC, which included primarily gammaproteobacteria or cyanobacteria and no betaproteobacteria (Tables 1 and 2). Even more interestingly, the closest relative, which had 41% identity and 60% similarity with LapA, was an aminopeptidase of A. castellanii (Table 3). Other eukaryotic LapA-like proteins were in fungi, and in many cases, they were closer to LapA than the prokaryotic homologues (Table 3). Thus, LapA has strong eukaryotic-protein-like traits.
FIG 9 .
Distribution of genes encoding key type II-secreted proteins within the Legionella genus. Gain and loss events pertaining to genes encoding type II-secreted NttD, LapA, LapB, PlaC, and ProA were modeled on a maximum-likelihood tree of 41 Legionella species, which was constructed based on the protein sequence encoded by 78 nearly universal genes (9). Colored circles and arrows indicate a gene gain event (upward-pointing arrow) or loss event (downward-pointing arrow) based on maximum parsimony, and colored cells (right) indicate the presence of nttD, lapA, lapB, plaC, and proA within a given species.
TABLE 1 .
BLAST results for NttD outside the Legionella genusa
| Organismb | I, S, Cc | E value | Accession no. | Classification |
|---|---|---|---|---|
| Dyella japonica | 25, 45, 89 | 1E−22 | WP_035323882.1 | Gammaproteobacteria |
| Oleiagrimonas soli | 25, 44, 80 | 2E−21 | WP_043100684.1 | Gammaproteobacteria |
| Nannocystis exedens | 23, 43, 80 | 1E−19 | WP_096328563.1 | Deltaproteobacteria |
| Arenimonas oryziterrae | 24, 41, 85 | 3E−18 | WP_022968800.1 | Gammaproteobacteria |
| Kangiella aquimarina | 26, 44, 72 | 2E−16 | WP_018624440.1 | Gammaproteobacteria |
| Lysobacter capsici | 23, 40, 86 | 7E−13 | WP_082723375.1 | Gammaproteobacteria |
| Microbulbifer donghaiensis | 24, 46, 80 | 2E−12 | WP_073272160.1 | Gammaproteobacteria |
| Teredinibacter turnerae | 25, 45, 80 | 1E−11 | WP_018414733.1 | Gammaproteobacteria |
| Rhodanobacter spathiphylli | 22, 41, 83 | 1E−11 | WP_007808810.1 | Gammaproteobacteria |
| Stenotrophomonas maltophilia | 22, 38, 78 | 4E−10 | WP_042612098.1 | Gammaproteobacteria |
Analysis of non-Legionella homologues was done with the exclusion of hits from the Legionella genus only with homologues predicted using an E-value cutoff of <4 × 10−10 and an alignment length of at least 65% (80).
Only the top representative species per genus is given, based on the closest homology with NttD.
I, percent identity; S, percent similarity; C, percent coverage.
TABLE 2 .
BLAST results for PlaC outside the Legionella genus
| Organisma | I, S, Cb | E value | Accession no. | Classification |
|---|---|---|---|---|
| Anabaena cylindrical | 28, 47, 69 | 5E−32 | WP_015214616.1 | Cyanobacteria |
| Cylindrospermum stagnale | 27, 44, 70 | 4E−29 | WP_015210343.1 | Cyanobacteria |
| Endozoicomonas arenosclerae | 29, 43, 95 | 3E−27 | WP_062262828.1 | Gammaproteobacteria |
| Anabaenopsis circularis | 23, 41, 72 | 1E−26 | WP_096581537.1 | Cyanobacteria |
| Fischerella muscicola | 26, 42, 68 | 6E−26 | WP_051035591.1 | Cyanobacteria |
| Tolypothrix bouteillei | 25, 43, 69 | 7E−26 | WP_038073042.1 | Cyanobacteria |
| Chlorogloeopsis fritschii | 27, 42, 71 | 9E−26 | WP_016872501.1 | Cyanobacteria |
| Mastigocladopsis repens | 28, 43, 69 | 1E−25 | WP_033366504.1 | Cyanobacteria |
| Scytonema millei | 25, 42, 75 | 3E−25 | WP_069349443.1 | Cyanobacteria |
| Hassallia byssoidea | 26, 42, 68 | 2E−24 | KIF33255.1 | Cyanobacteria |
Only the top representative species per genus is given, based on the closest homology with PlaC. After the genera listed here, the next seven encoding a PlaC-like protein (E value, <1E−18) are five cyanobacteria followed by two gammaproteobacteria.
I, percent identity; S, percent similarity; C, percent coverage.
TABLE 3 .
BLAST results of LapA outside the Legionella genus
| Organisma | I, S, Cb | E value | Accession no. | Classification |
|---|---|---|---|---|
| Acanthamoeba castellanii | 41, 60, 70 | 7E−65 | XP_004341372.1 | Eukarya—protozoa |
| Chromobacterium sphagni | 40, 59, 70 | 2E−64 | WP_071111733.1 | Betaproteobacteria |
| Chitinimonas koreensis | 40, 59, 70 | 5E−64 | WP_028447211.1 | Betaproteobacteria |
| Burkholderia thailandensis | 39, 59, 69 | 5E−61 | WP_019254803.1 | Betaproteobacteria |
| Pseudogulbenkiania ferrooxidans | 38, 58, 70 | 8E−61 | WP_021478877.1 | Betaproteobacteria |
| Massilia yuzhufengensis | 39, 57, 70 | 2E−60 | WP_091877370.1 | Betaproteobacteria |
| Pleurotus ostreatus | 37, 53, 75 | 8E−60 | KDQ28511.1 | Eukarya—fungi |
| Duganella sacchari | 33, 53, 92 | 9E−60 | WP_072788963.1 | Betaproteobacteria |
| Janthinobacterium agaricidamnosum | 37, 57, 72 | 2E−59 | WP_038494963.1 | Betaproteobacteria |
| Syncephalastrum racemosum | 39, 58, 68 | 5E−59 | ORY97700.1 | Eukarya—fungi |
| Coprinopsis cinerea | 37, 57, 71 | 7E−59 | XP_001831818.1 | Eukarya—fungi |
| Actinomucor elegans | 36, 58, 68 | 9E−59 | AME15509.1 | Eukarya—fungi |
| Rhizophagus irregularis | 36, 57, 69 | 2E−58 | ESA22882.1 | Eukarya—fungi |
| Trichosporon asahii | 37, 56, 72 | 7E−58 | EKD01610.1 | Eukarya—fungi |
| Roseateles aquatilis | 38, 56, 70 | 1E−57 | WP_088383330.1 | Betaproteobacteria |
| Conidiobolus coronatus | 38, 57, 69 | 1E−57 | KXN70597.1 | Eukarya—fungi |
| Pseudoduganella violaceinigra | 34, 55, 81 | 3E−57 | WP_028103074.1 | Betaproteobacteria |
| Cylindrobasidium torrendii | 34, 53, 77 | 3E−57 | KIY62743.1 | Eukarya—fungi |
| Ulvibacter litoralis | 40, 55, 69 | 3E−57 | WP_093139368.1 | Bacteroidetes |
| Mucor circinelloides | 36, 58, 69 | 5E−57 | EPB89639.1 | Eukarya—fungi |
| Schizophyllum commune | 36, 55, 69 | 8E−57 | XP_003037075.1 | Eukarya—fungi |
| Pseudoalteromonas byunsanensis | 36, 56, 79 | 1E−56 | WP_070992923.1 | Gammaproteobacteria |
Only the top representative species per genus is given, based on the closest homology with LapA. After the genera listed here, the next 20 encoding a LapA-like protein (E value, <1E−53) are 15 genera of fungi and 1 genus each of betaproteobacteria, gammaproteobacteria, deltaproteobacteria, Actinobacteria, and Bacteroidetes.
I, percent identity; S, percent similarity; C, percent coverage.
Conservation of the T2SS apparatus genes. This table lists the T2SS apparatus genes present within L. pneumophila strains (A) and among different Legionella species (B). Download TABLE S1, PDF file, 0.03 MB (33.4KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of the nttD, plaC, lapA, and lapB genes. This table lists the nttD (A), plaC (B), lapA (C), and lapB (D) genes present within L. pneumophila strains and among different Legionella species. Download TABLE S2, PDF file, 0.1 MB (82.8KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The experimental data presented increase our understanding of T2S and host-parasite interactions in multiple ways. With the definition of NttD and LapA as major potentiators of infection of A. castellanii, the number of T2S effectors confirmed as being involved in infection of amoebas rises to seven (23, 24, 30, 31). Thus, approximately one-third of the known effectors are required for infection (13), implying that intracellular parasitism has played a large role in shaping the T2SS repertoire. That NttD, along with the previously described NttA and NttC, has a role in amoebal infection and yet lacks similarity to known enzymes suggests that many T2SS substrates may be specialized for the intra-amoebal lifestyle (23, 24). Although aminopeptidases in the M28 family have been linked to T2S and/or virulence (37, 51, 52), the results from this study of LapA mark the first documentation of a secreted aminopeptidase promoting an intracellular infection event. Thus, prokaryotic LapA-like proteins (Table 3) may be facilitating survival for bacteria such as Duganella and Burkholderia spp. that reside in amoebas (53). In establishing a link between LapA and PlaC, we have also revealed a unique example of functional redundancy operating during infection. While promoting growth in A. castellanii, NttD and the combination of LapA and PlaC were not needed for infection of other amoebas. This highlights how the importance of T2SS substrates varies in a host-specific fashion; presumably, each amoebal type represents a distinct environment and, hence, L. pneumophila invokes different exoproteins in order to efficiently establish its host range. Unlike NttD and LapA, PlaC was critical for infection of every amoebal host. There are probably commonalties across different amoebas, and so L. pneumophila repeatedly relies upon some of its effectors to flourish in aquatic habitats. The finding that ProA processes LapA and LapB adds to past work showing that ProA cleaves PlaA and PlaC and is required for strain 130b infection of V. vermiformis and N. lovaniensis (30, 35, 43). Another study found that a proA (mspA) mutant of strain JR-32 is impaired for infection of A. castellanii (19). Thus, ProA rivals PlaC in its broad significance. Although previous studies reported on the transcription of a few T2S genes (54–56), the current analysis represents the largest assessment of T2S effector expression by L. pneumophila. After examining 19 genes in two stages of extracellular growth and in three hosts, it is apparent that secreted-protein genes exhibit a variety of expression patterns rather than showing similar responses to conditions. From this, we intuit that the amounts of secreted protein made are controlled at the level of the individual or of subsets of effector-gene transcription more than at the level of apparatus gene transcription.
Bioinformatic analyses revealed further insights into T2SS and the origins of its effectors. Being found in the majority of Legionella species, the genes encoding NttD, PlaC, and LapA appear to be ancestral genes that underwent only a few loss events during the evolution of the genus. On the basis of the dispersal of homologues in other types of organisms, ancestral NttD was likely acquired from related gammaproteobacterial species, PlaC from cyanobacterial species, and LapA from protozoa or fungi. ProA is encoded by another ancestral T2SS gene, but in this case, no loss events were detected. Unlike NttD, PlaC, LapA, and ProA, LapB was only in the “recent” clade containing L. pneumophila. We posit that LapB arose from lapA duplication. This is supported by phylogenetic analysis of LapA and LapB, which form a monophyletic group exclusive of other bacteria or eukaryotes, and is based on the fact that LapB has acquired substitutions relative to the ancestral LapA (Fig. 10). Given that NttD, PlaC, LapA, and ProA promote infection but LapB does not, we speculate that ancestral, highly conserved T2SS effectors have the greater role in intracellular parasitism. This evolutionary pattern for T2SS effectors differs from that of most T4SS effectors, which are distributed in <25% of Legionella species (9).
FIG 10 .
Interkingdom phylogenetic tree of LapA-like proteins. A maximum-likelihood tree shows 41 aligned amino acid sequences of M28 family aminopeptidases. Bootstrap values (from 250 replicates) of >50 are given at the corresponding nodes. Aminopeptidase Y from Saccharomyces cerevisiae was used as the outgroup. The scale bar indicates the number of amino acid substitutions per site. Fungi are indicated in red, protozoa in purple, and bacteria in green, with the exception of L. pneumophila proteins, which are indicated in blue. NCBI accession numbers not listed in Table 3 are noted in parentheses after the respective genus designations.
The activities associated with PlaC and LapA provide hypotheses for how these T2S substrates promote infection. PlaC not only generates fatty acids but also can modify lipids within amoebal vacuoles (34, 35). Thus, PlaC may promote intracellular nutrient acquisition via direct lipid metabolism, or through modification of lipids in LCV membranes, which might in turn influence vesicular trafficking or anchoring of other effectors. LapA likely promotes replication through its ability to generate amino acids that are limiting during the intracellular lifecycle. Our analysis found a broad spectrum of activity for LapA, and we showed that amino acid supplementation rescues the growth of mutants lacking LapA. Interestingly, the leucine, methionine, isoleucine, and valine generated by LapA, coupled with arginine produced by LapB (30), account for all of the amino acids that cannot be synthesized by L. pneumophila or A. castellanii (46). It is intriguing to contemplate why LapA and PlaC, two very different exoenzymes, should overlap in terms of function. In one scenario, L. pneumophila utilizes both amino acids (generated by LapA) and lipids (generated by PlaC) for nutrition within the LCV. Thus, a growth defect is revealed only when both LapA and PlaC are absent, depriving the bacterium of two food sources. Alternatively, changes to LCV membranes mediated by PlaC might promote the import of nutrients beyond those targeted by LapA. Hence, without PlaC and LapA, intravacuolar bacteria again lose two food sources. To discern how PlaC and LapA operate, it would be helpful to learn their locations in the infected cell. Proteins secreted by T2S, including ProA, can be translocated out of the LCV (57), and thus, it is possible that LapA or PlaC or both have targets in the host cytoplasm. Overall, LapA and PlaC modulate different processes and yet accomplish a single task.
With the characterization of LapA, we have a striking example of a eukaryotic-protein-like, T2SS-dependent exoprotein, indicating that eukaryotic-protein-like effectors of L. pneumophila are not limited to the T4SS (58). LapA was likely acquired through horizontal gene transfer within A. castellanii or an ancestral protozoan. Indeed, the top-scoring LapA homologue outside Legionella belongs to A. castellanii and is a putative aminopeptidase. Also, LapA shares higher-than-average homology to its top-scoring eukaryotic homologue (41% identity, 60% similarity), compared to the average levels of identity and similarity of all other (primarily T4SS) eukaryotic-protein-like proteins of L. pneumophila (34% identity, 52% similarity) (59). Moreover, phylogenetic reconstruction of aligned amino acid sequences of LapA homologues revealed that LapA is not descended from other bacteria but shares a common ancestor with A. castellanii (Fig. 10). Interestingly, the A. castellanii aminopeptidase is secreted (60). Thus, legionellae benefit from secreting a host-like aminopeptidase as a means of scavenging limiting amino acids.
The structure of LapA is similar to those of several known aminopeptidases. The Dali server identified LapB (41) and A. proteolytica Apap (PDB code: 1AMP) (61) as having the highest tertiary homology (Z scores of 54.6 Å and 45.0 Å, respectively), although this is concentrated at the peptidase domain (see Fig. S7A in the supplemental material). For example, the positions of active-site residues in LapA and LapB structures are almost identical (Fig. S7B), while there is a significant reorganization of helices between PA domains. This accommodates differences in their substrate-binding pockets, with LapB’s cavity being wider and more negatively charged (Fig. S7C), compatible with a preference for positively charged lysine and arginine (30). Often, the role of the PA domain is to keep an enzyme in an inactive state prior to secretion (62), although for purified LapA, it did not prevent detection of activity. The effect of the PA domain is likely due to a transient association between the PA and peptidase domains in LapA, as we observed a monomeric and domain-swapped form in solution. Although domain swapping was not seen in the structure of LapB, it dimerizes in solution and its PA domain does not prevent detection of activity under some conditions (41). The LapA structure is the fifth structure for a Legionella T2SS effector, joining those of NttD, LapB, phosphatase Map, and VirK-like Lpg1832 (28, 41, 63–65).
Comparison of the L. pneumophila LapA and LapB tertiary structure, active-site residues, and substrate binding pocket. (A) Significant deviations between the LapA (orange) and LapB (PDB code: 5GNE) (purple) structures were observed within the PA domain helices, which are annotated. (B) Amino acid residues of the active sites of LapA (orange) and LapB (purple) are drawn as sticks, and zinc (Zn) and water (WAT) molecules are drawn as spheres. (C) LapA and LapB structures are shown as electrostatic surface potential data with active-site zinc ions drawn as gray spheres. Substrate binding sites are highlighted with a yellow dashed line. Download FIG S7, EPS file, 4.4 MB (4.5MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Although the PlaC, LapA, and NttD data explain, to a large extent, the impact of T2S on L. pneumophila infection of A. castellanii, future work should further define the Legionella T2SS output. Proteomic assessment of supernatants and subsequent (single) mutant analysis have heretofore been the means to identifying T2SS-dependent proteins that promote infection (13). These approaches helped reveal the nature of NttD as a potentiator of infection and will continue to offer new targets for analysis (28). Yet this strategy clearly failed to reveal the roles that LapA and PlaC have in A. castellanii infection (23, 30), undoubtedly due to the redundancy that exists between LapA and PlaC. The new-found importance of these two effectors was uncovered through a combination of transcriptional and mutational analyses. This approach has four steps: (i) identification of the genes that are upregulated during wild-type infection of the host; (ii) examination of the transcript profile of mutants that lack the upregulated genes, while being alert to compensatory increases in other transcripts; (iii) mutation of two or more upregulated genes that exhibit a compensatory link; and (iv) testing of mutants for loss of infection. Our data set predicts more substrates that may promote infection; e.g., two strongly upregulated genes during infection of A. castellanii and N. lovaniensis were legP encoding a putative protease and map encoding a phosphatase (Fig. 1B and C). Though neither a legP mutant nor a map mutant was impaired for infection (23), a legP map mutant might be defective. Interestingly, LegP and Map, like LapA, have eukaryotic-protein-like homologs (23, 28, 64).
MATERIALS AND METHODS
Strains, plasmids, and media.
L. pneumophila 130b (ATCC BAA-74) served as the wild type and parent for all mutants (25). proA mutant AA200, lspF mutant NU275, lapA mutant NU320, lapB mutant NU322, lapA lapB mutant NU324, celA mutant NU353, and plaC mutants NU367 and NU420 were described before (14, 15, 23, 29, 30, 66). Newly derived mutants lacking nttD, lapA, or plaC were made using methods of allelic exchange (23, 25). Complemented mutants were obtained by introducing intact copies of nttD, lapA, or plaC on a plasmid, analogously to past studies (24). Primers and plasmids used in mutant and complement constructions are listed in Table S3 in the supplemental material. Legionellae were routinely grown at 37°C on buffered charcoal yeast extract (BCYE) agar or in BYE broth (25). Escherichia coli DH5α (Life Technologies) was the host for the recombinant plasmids used in mutant construction and complementation. E. coli K-12 SHuffle (NEB) was used to get purified protein. E. coli cells were grown in LB medium. Unless otherwise noted, chemicals were from Sigma.
Primers and plasmids. This table lists the primers (A) and plasmids (B) that were used in this study. Download TABLE S3, PDF file, 0.1 MB (71.7KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Infection assays.
U937 cells (ATCC CRL-1593.2), A. castellanii (ATCC 30234), V. vermiformis (ATCC 50237), and N. lovaniensis (ATCC 30569) were infected with L. pneumophila as previously described (23). To assess survival of L. pneumophila in A. castellanii-conditioned media, we inoculated legionellae and amoebas on opposite sides of a 0.4-µm-pore-size transwell membrane (Corning; catalog no. 3470). To that end, 1 × 105 amoebas in 1 ml of protease-peptone-yeast extract (PY) medium (23) were seeded into 24-well plates, and then 1 × 104 wild-type or mutant bacteria were added in 100 µl PY medium to the transwell support. After periods of incubation, a 10-µl aliquot from the upper chamber was plated on BCYE agar to determine numbers of legionellae. To test the effect of amino acid supplementation on bacterial growth in A. castellanii, an amino acid cocktail (Lonza; catalog no. 13-114E) was added to the PY medium, giving 1 mM of each amino acid.
Quantitative reverse transcriptase PCR.
qRT-PCR was done as previously described (17). To monitor L. pneumophila transcription during extracellular growth, RNA was isolated from BYE cultures using a GeneJET RNA purification kit (Thermo Scientific). To examine bacterial RNA from infected host cells, U937 cells, A. castellanii, and N. lovaniensis were infected as described above, with the exception that the inoculation step was facilitated by centrifugation (500 × g, 5 min) of bacteria onto the monolayer, and the infected cells were lysed and processed using the GeneJET kit. Primers RW12 to RW69 (Table S3) used for qRT-PCR were designed using the Primer-BLAST tool at NCBI. lpw00031 (gyrB) and lpw16991 were used as references to normalize gene expression (67), and changes in expression were calculated using the threshold cycle (2−ΔΔCT) method (68).
Protein purifications.
Full-length LapA (FL-LapA; residues 1 to 378), N-terminal PA domain-truncated LapA (ΔN-LapA; residues 89 to 378), full-length LapB (FL-LapB; residues 1 to 373), and full-length NttD (FL-NttD; residues 1 to 373), minus N-terminal signal sequences, were amplified from 130b DNA and cloned into N-terminal His-tagged pET-46 Ek/LIC (EMD Millipore), using primers JG1 to JG8 (Table S3). Expression of the recombinant proteins in E. coli was induced with 1 mM IPTG (opyl-β-d-thiogalactopyranoside) at an optical density at 600 nm (OD600) of 0.6, and cells were harvested after overnight growth at 18°C with shaking. Cells were suspended in a mixture containing 20 mM Tris-HCl (pH 8), 200 mM NaCl, 5 mM MgCl2, 1 mg/ml DNase I, and 5 mg/ml lysozyme and were lysed by sonication. Recombinant proteins were purified using nickel-affinity chromatography (Qiagen) followed by gel filtration using a Superdex 75 column (GE Healthcare) equilibrated in 20 mM Tris–HCl (pH 8)–200 mM NaCl.
Enzyme assays.
Cell-free supernatants collected from late-log BYE cultures were assayed for protease, acid phosphatase, and lipase activities as previously described (50, 64). To assess secreted aminopeptidase activity, two previously described methods were used (30). Leucine, lysine, methionine, glycine, aspartate, isoleucine, proline, glutamate, and valine aminopeptidase activities were measured by the release of p-nitroanilide (p-NA) from l-leucine p-NA, l-lysine p-NA, l-methionine p-NA, glycine p-NA, l-aspartate p-NA (Chem-Impex), l-isoleucine p-NA (Chem-Impex), l-proline p-NA (MP Biomedicals), l-glutamate p-NA (Chem-Impex), and l-valine p-NA (Chem-Impex), respectively (30). Phenylalanine, alanine, and serine aminopeptidase activities were measured by release of β-naphthylamide (β-NA) from l-phenylalanine β-NA, l-alanine β-NA, and l-serine β-NA (Chem-Impex), respectively (30). Purified LapA-FL and LapA-ΔN were diluted to a concentration of 500 nM in the reaction buffers and assayed as described above. One unit of activity is equal to that which catalyzes the conversion of 1 µmol substrate/min, and molecular activity was expressed as the number of units per micromole of purified enzyme, based on standard curves corresponding to p-NA and β-NA (30, 69). Assays for α-mannosidase, β-mannosidase, β-galactosidase, and endo-1,4-β-mannanase were as previously described (70–72).
Assays for the cleavage, activation, and inhibition of aminopeptidases.
To assay for the ability of secreted proteins to cleave aminopeptidases, 10 µg of purified LapB-FL or LapA-FL was incubated at 37°C for up to 2 h with 50 µl of 1× or 1:10-diluted late-log-phase culture supernatants, respectively. Samples (10 µl) were collected on ice and were then subjected to SDS-PAGE. Protein was visualized with SimplyBlue SafeStain (Thermo Scientific). Immunoblots were done as previously described (17), with recombinant protein detected with mouse anti-His (Millipore; clone HIS.H8). For real-time monitoring of LapA-FL cleavage and resultant activation, LapA-FL (500 nM) was added to the assay buffer containing 3 mM p-NA-based substrate and the supernatant (1:10 final concentration). Absorbance was monitored at 405 nm. LapA inhibition assays were done using 500 nM LapA-ΔN incubated with serial dilutions of bestatin. Residual activity following treatment was determined based on release of p-NA from l-leucine–p-NA as described above.
Crystal structure determination.
Crystallization of LapA (10 mg/ml) was done using the sitting-drop vapor-diffusion method and growth at 293 K in 0.15 M ammonium sulfate–0.1 M 4-morpholineethanesulfonate (pH 5.5)–25% (wt/vol) polyethylene glycol 4000. Crystals were soaked for 30 to 60 s in cryoprotection solution (well solution supplemented with 20% glycerol) and flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K on Beamline I03 at the Diamond Light Source (DLS), United Kingdom. Data were processed using XDS (73) and scaled using AIMLESS (74). Initial phases were obtained using PHASER (75) with the structure of LapB (41) as the search model. Density modification was done with PARROT (76) followed by automated model building with ARPWARP (77). The remaining structure was manually built within COOT (78), and refinement was carried out with refmac (79) using noncrystallographic symmetry and translation-libration-screw groups. Ten percent of the reflections were omitted for cross-validation. Processing and refinement statistics for the final model are given in Table 4.
TABLE 4 .
Data collection and refinement statistics
| Parameter | LapA result(s)a |
|---|---|
| Crystal parameters | |
| Space group | P212121 |
| Cell dimensions (Å) | a = 76.60; b = 99.95; c = 104.02 |
| Data collection | |
| Beamline | DLS I03 |
| Wavelength (Å) | 0.97625 |
| Resolution (Å) | 99.55–1.87 (1.87–1.92) |
| No. of unique observations | 66,414 (4,808) |
| Rsymb | 0.063 (1.2) |
| <I>/σI | 12.5 (3.7) |
| Completeness (%) | 100 (100) |
| Redundancy | 6.5 (6.2) |
| Refinementc | |
| Rwork/Rfree (%) | 19.2/23.8 |
| No. of protein residues | 719 |
| No. of water molecules | 313 |
| No. of ions | 4 Zn ions |
| RMSD stereochemistryd | |
| Bond lengths (Å) | 0.022 |
| Bond angles (°) | 2.045 |
| Ramachandran analysise | |
| Residues in outlier regions (%) | 0.0 |
| Residues in favored regions (%) | 98.6 |
| Residues in allowed regions (%) | 100 |
Numbers in parentheses refer to the outermost resolution shell.
Rsym = ∑|I − <I>|/∑I, where I is the integrated intensity of a given reflection and <I> is the mean intensity of multiple corresponding symmetry-related reflections.
Rwork = ∑||Fo − |Fc||/∑Fo, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree = Rwork values calculated using 10% random data excluded from the refinement.
The RMSD stereochemistry value represents the deviation from ideal values.
Ramachandran analysis was carried out using Molprobity (81).
Accession number(s).
Coordinates and structure factors have been deposited in the wwPDB under PDB code 6ESL.
ACKNOWLEDGMENTS
We thank members of the Cianciotto laboratory past and present for helpful advice and assistance with some preliminary data. We also thank the beamline scientists at Beamline I03 of the Diamond Light Source, United Kingdom.
Footnotes
Citation White RC, Gunderson FF, Tyson JY, Richardson KH, Portlock TJ, Garnett JA, Cianciotto NP. 2018. Type II secretion-dependent aminopeptidase LapA and acyltransferase PlaC are redundant for nutrient acquisition during Legionella pneumophila intracellular infection of amoebas. mBio 9:e00528-18. https://doi.org/10.1128/mBio.00528-18.
REFERENCES
- 1.Bajrai LH, Azhar EI, Yasir M, Jardot P, Barrassi L, Raoult D, La Scola B, Pagnier I. 2016. Legionella saoudiensis sp. nov., isolated from a sewage water sample. Int J Syst Evol Microbiol 66:4367–4371. doi: 10.1099/ijsem.0.001357. [DOI] [PubMed] [Google Scholar]
- 2.Mercante JW, Winchell JM. 2015. Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin Microbiol Rev 28:95–133. doi: 10.1128/CMR.00029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkelman RL, Pruden A. 2017. Prevention of Legionnaires’ disease in the 21st century by advancing science and public health practice. Emerg Infect Dis 23:1905–1907. doi: 10.3201/eid2311.171429. [DOI] [Google Scholar]
- 4.van Heijnsbergen E, Schalk JA, Euser SM, Brandsema PS, den Boer JW, de Roda Husman AM. 2015. Confirmed and potential sources of Legionella reviewed. Environ Sci Technol 49:4797–4815. doi: 10.1021/acs.est.5b00142. [DOI] [PubMed] [Google Scholar]
- 5.Cianciotto NP, Hilbi H, Buchrieser C. 2013. Legionnaires' disease, p 147–217. In Rosenberg E, DeLong EF, Stackebrandt E, Thompson F, Lory S (ed), The prokaryotes—human microbiology, 4th ed. Springer, New York, NY. doi: 10.1007/978-3-642-30144-5. [DOI] [Google Scholar]
- 6.Hoffmann C, Harrison CF, Hilbi H. 2014. The natural alternative: protozoa as cellular models for Legionella infection. Cell Microbiol 16:15–26. doi: 10.1111/cmi.12235. [DOI] [PubMed] [Google Scholar]
- 7.Sherwood RK, Roy CR. 2016. Autophagy evasion and endoplasmic reticulum subversion: the yin and yang of Legionella intracellular infection. Annu Rev Microbiol 70:413–433. doi: 10.1146/annurev-micro-102215-095557. [DOI] [PubMed] [Google Scholar]
- 8.Steiner B, Weber S, Hilbi H. 2017. Formation of the Legionella-containing vacuole: phosphoinositide conversion, GTPase modulation and ER dynamics. Int J Med Microbiol. doi: 10.1016/j.ijmm.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Burstein D, Amaro F, Zusman T, Lifshitz Z, Cohen O, Gilbert JA, Pupko T, Shuman HA, Segal G. 2016. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet 48:167–175. doi: 10.1038/ng.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong KC, Ghosal D, Chang YW, Jensen GJ, Vogel JP. 2017. Polar delivery of Legionella type IV secretion system substrates is essential for virulence. Proc Natl Acad Sci U S A 114:8077–8082. doi: 10.1073/pnas.1621438114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escoll P, Song OR, Viana F, Steiner B, Lagache T, Olivo-Marin JC, Impens F, Brodin P, Hilbi H, Buchrieser C. 2017. Legionella pneumophila modulates mitochondrial dynamics to trigger metabolic repurposing of infected macrophages. Cell Host Microbe 22:302–316.e7. doi: 10.1016/j.chom.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Cianciotto NP. 2013. Type II secretion and Legionella virulence. Curr Top Microbiol Immunol 376:81–102. doi: 10.1007/82_2013_339. [DOI] [PubMed] [Google Scholar]
- 13.Cianciotto NP, White RC. 2017. Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect Immun 85:e00014-17. doi: 10.1128/IAI.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossier O, Starkenburg SR, Cianciotto NP. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires’ disease pneumonia. Infect Immun 72:310–321. doi: 10.1128/IAI.72.1.310-321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy-Simandle K, Stewart CR, Dao J, Debroy S, Rossier O, Bryce PJ, Cianciotto NP. 2011. Legionella pneumophila type II secretion dampens the cytokine response of infected macrophages and epithelia. Infect Immun 79:1984–1997. doi: 10.1128/IAI.01077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallama CA, McCoy-Simandle K, Cianciotto NP. 2017. The type II secretion system of Legionella pneumophila dampens the MyD88 and Toll-like receptor 2 signaling pathway in infected human macrophages. Infect Immun 85:e00897-16. doi: 10.1128/IAI.00897-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White RC, Cianciotto NP. 2016. Type II secretion is necessary for optimal association of the Legionella-containing vacuole with macrophage Rab1B but enhances intracellular replication mainly by Rab1B-independent mechanisms. Infect Immun 84:3313–3327. doi: 10.1128/IAI.00750-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liles MR, Edelstein PH, Cianciotto NP. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol Microbiol 31:959–970. doi: 10.1046/j.1365-2958.1999.01239.x. [DOI] [PubMed] [Google Scholar]
- 19.Hales LM, Shuman HA. 1999. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect Immun 67:3662–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossier O, Cianciotto NP. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect Immun 69:2092–2098. doi: 10.1128/IAI.69.4.2092-2098.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Söderberg MA, Rossier O, Cianciotto NP. 2004. The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures. J Bacteriol 186:3712–3720. doi: 10.1128/JB.186.12.3712-3720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Söderberg MA, Dao J, Starkenburg SR, Cianciotto NP. 2008. Importance of type II secretion for Legionella pneumophila survival in tap water and amoebae at low temperature. Appl Environ Microbiol 74:5583–5588. doi: 10.1128/AEM.00067-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyson JY, Pearce MM, Vargas P, Bagchi S, Mulhern BJ, Cianciotto NP. 2013. Multiple Legionella pneumophila type II secretion substrates, including a novel protein, contribute to differential infection of amoebae Acanthamoeba castellanii, Hartmannella vermiformis, and Naegleria lovaniensis. Infect Immun 81:1399–1410. doi: 10.1128/IAI.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyson JY, Vargas P, Cianciotto NP. 2014. The novel Legionella pneumophila type II secretion substrate NttC contributes to infection of amoebae Hartmannella vermiformis and Willaertia magna. Microbiology 160:2732–2744. doi: 10.1099/mic.0.082750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart CR, Burnside DM, Cianciotto NP. 2011. The surfactant of Legionella pneumophila is secreted in a TolC-dependent manner and is antagonistic toward other Legionella species. J Bacteriol 193:5971–5984. doi: 10.1128/JB.05405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duncan C, Prashar A, So J, Tang P, Low DE, Terebiznik M, Guyard C. 2011. Lcl of Legionella pneumophila is an immunogenic GAG binding adhesin that promotes interactions with lung epithelial cells and plays a crucial role in biofilm formation. Infect Immun 79:2168–2181. doi: 10.1128/IAI.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomassin JL, Santos Moreno J, Guilvout I, Tran Van Nhieu G, Francetic O. 2017. The trans-envelope architecture and function of the type 2 secretion system: new insights raising new questions. Mol Microbiol 105:211–226. doi: 10.1111/mmi.13704. [DOI] [PubMed] [Google Scholar]
- 28.DebRoy S, Dao J, Söderberg M, Rossier O, Cianciotto NP. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A 103:19146–19151. doi: 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce MM, Cianciotto NP. 2009. Legionella pneumophila secretes an endoglucanase that belongs to the family-5 of glycosyl hydrolases and is dependent upon type II secretion. FEMS Microbiol Lett 300:256–264. doi: 10.1111/j.1574-6968.2009.01801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossier O, Dao J, Cianciotto NP. 2008. The type II secretion system of Legionella pneumophila elaborates two aminopeptidases, as well as a metalloprotease that contributes to differential infection among protozoan hosts. Appl Environ Microbiol 74:753–761. doi: 10.1128/AEM.01944-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossier O, Dao J, Cianciotto NP. 2009. A type II secreted ribonuclease of Legionella pneumophila facilitates optimal intracellular infection of Hartmannella vermiformis. Microbiology 155:882–890. doi: 10.1099/mic.0.023218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byrne B, Swanson MS. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun 66:3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molofsky AB, Swanson MS. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol 50:445–461. doi: 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- 34.Banerji S, Bewersdorff M, Hermes B, Cianciotto NP, Flieger A. 2005. Characterization of the major secreted zinc metalloprotease-dependent glycerophospholipid:cholesterol acyltransferase, PlaC, of Legionella pneumophila. Infect Immun 73:2899–2909. doi: 10.1128/IAI.73.5.2899-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang C, Rastew E, Hermes B, Siegbrecht E, Ahrends R, Banerji S, Flieger A. 2012. Zinc metalloproteinase ProA directly activates Legionella pneumophila PlaC glycerophospholipid:cholesterol acyltransferase. J Biol Chem 287:23464–23478. doi: 10.1074/jbc.M112.346387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawlings ND, Barrett AJ, Bateman A. 2010. Merops: the peptidase database. Nucleic Acids Res 38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luckett JC, Darch O, Watters C, Abuoun M, Wright V, Paredes-Osses E, Ward J, Goto H, Heeb S, Pommier S, Rumbaugh KP, Cámara M, Hardie KR. 2012. A novel virulence strategy for Pseudomonas aeruginosa mediated by an autotransporter with arginine-specific aminopeptidase activity. PLoS Pathog 8:e1002854. doi: 10.1371/journal.ppat.1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang W, Li Z, Li C, Yu X, Wang F, Wan X, Wang Y, Ma L. 2016. High-level expression and characterization of the Bacillus subtilis subsp. subtilis str. BSP1 YwaD aminopeptidase in pichia pastoris. Protein Expr Purif 122:23–30. doi: 10.1016/j.pep.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Wilkes SH, Prescott JM. 1985. The slow, tight binding of bestatin and amastatin to aminopeptidases. J Biol Chem 260:13154–13162. [PubMed] [Google Scholar]
- 40.Gao X, Liu Z, Cui W, Zhou L, Tian Y, Zhou Z. 2014. Enhanced thermal stability and hydrolytic ability of Bacillus subtilis aminopeptidase by removing the thermal sensitive domain in the non-catalytic region. PLoS One 9:e92357. doi: 10.1371/journal.pone.0092357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang N, Yin S, Zhang W, Gong X, Zhang N, Fang K, Ge H. 2017. Crystal structure and biochemical characterization of an aminopeptidase LapB from Legionella pneumophila. J Agric Food Chem 65:7569–7578. doi: 10.1021/acs.jafc.7b02849. [DOI] [PubMed] [Google Scholar]
- 42.Flieger A, Neumeister B, Cianciotto NP. 2002. Characterization of the gene encoding the major secreted lysophospholipase A of Legionella pneumophila and its role in detoxification of lysophosphatidylcholine. Infect Immun 70:6094–6106. doi: 10.1128/IAI.70.11.6094-6106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang C, Hiller M, Flieger A. 2017. Disulfide loop cleavage of Legionella pneumophila PlaA boosts lysophospholipase A activity. Sci Rep 7:16313. doi: 10.1038/s41598-017-12796-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilboa R, Spungin-Bialik A, Wohlfahrt G, Schomburg D, Blumberg S, Shoham G. 2001. Interactions of Streptomyces griseus aminopeptidase with amino acid reaction products and their implications toward a catalytic mechanism. Proteins 44:490–504. doi: 10.1002/prot.1115. [DOI] [PubMed] [Google Scholar]
- 45.Chevrier B, D’Orchymont H, Schalk C, Tarnus C, Moras D. 1996. The structure of the Aeromonas proteolytica aminopeptidase complexed with a hydroxamate inhibitor. Involvement in catalysis of Glu151 and two zinc ions of the co-catalytic unit. Eur J Biochem 237:393–398. doi: 10.1111/j.1432-1033.1996.0393k.x. [DOI] [PubMed] [Google Scholar]
- 46.Price CT, Richards AM, Von Dwingelo JE, Samara HA, Abu Kwaik Y. 2014. Amoeba host-Legionella synchronization of amino acid auxotrophy and its role in bacterial adaptation and pathogenic evolution. Environ Microbiol 16:350–358. doi: 10.1111/1462-2920.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroeder GN, Petty NK, Mousnier A, Harding CR, Vogrin AJ, Wee B, Fry NK, Harrison TG, Newton HJ, Thomson NR, Beatson SA, Dougan G, Hartland EL, Frankel G. 2010. The genome of Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm secretion system effector proteins. J Bacteriol 192:6001–6016. doi: 10.1128/JB.00778-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 49.Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aragon V, Kurtz S, Flieger A, Neumeister B, Cianciotto NP. 2000. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect Immun 68:1855–1863. doi: 10.1128/IAI.68.4.1855-1863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sánchez-Porro C, Mellado E, Pugsley AP, Francetic O, Ventosa A. 2009. The haloprotease CPI produced by the moderately halophilic bacterium Pseudoalteromonas ruthenica is secreted by the type II secretion pathway. Appl Environ Microbiol 75:4197–4201. doi: 10.1128/AEM.00156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guenet C, Lepage P, Harris BA. 1992. Isolation of the leucine aminopeptidase gene from Aeromonas proteolytica. Evidence for an enzyme precursor. J Biol Chem 267:8390–8395. [PubMed] [Google Scholar]
- 53.Delafont V, Brouke A, Bouchon D, Moulin L, Héchard Y. 2013. Microbiome of free-living amoebae isolated from drinking water. Water Res 47:6958–6965. doi: 10.1016/j.watres.2013.07.047. [DOI] [PubMed] [Google Scholar]
- 54.Brüggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppée JY, Buchrieser C. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol 8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- 55.Broich M, Rydzewski K, McNealy TL, Marre R, Flieger A. 2006. The global regulatory proteins LetA and RpoS control phospholipase A, lysophospholipase A, acyltransferase, and other hydrolytic activities of Legionella pneumophila JR32. J Bacteriol 188:1218–1226. doi: 10.1128/JB.188.4.1218-1226.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jules M, Buchrieser C. 2007. Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics. FEBS Lett 581:2829–2838. doi: 10.1016/j.febslet.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 57.Truchan HK, Christman HD, White RC, Rutledge NS, Cianciotto NP. 2017. Type II secretion substrates of Legionella pneumophila translocate out of the pathogen-occupied vacuole via a semi-permeable membrane. MBio 8:e00870-17. doi: 10.1128/mBio.00870-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Al-Quadan T, Price CT, Abu Kwaik Y. 2012. Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol 20:299–306. doi: 10.1016/j.tim.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lurie-Weinberger MN, Gomez-Valero L, Merault N, Glöckner G, Buchrieser C, Gophna U. 2010. The origins of eukaryotic-like proteins in Legionella pneumophila. Int J Med Microbiol 300:470–481. doi: 10.1016/j.ijmm.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 60.Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, Bertelli C, Schilde C, Kianianmomeni A, Bürglin TR, Frech C, Turcotte B, Kopec KO, Synnott JM, Choo C, Paponov I, Finkler A, Heng Tan CS, Hutchins AP, Weinmeier T, Rattei T, Chu JS, Gimenez G, Irimia M, Rigden DJ, Fitzpatrick DA, Lorenzo-Morales J, Bateman A, Chiu CH, Tang P, Hegemann P, Fromm H, Raoult D, Greub G, Miranda-Saavedra D, Chen N, Nash P, Ginger ML, Horn M, Schaap P, Caler L, Loftus BJ. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol 14:R11. doi: 10.1186/gb-2013-14-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chevrier B, Schalk C, D’Orchymont H, Rondeau JM, Moras D, Tarnus C. 1994. Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family. Structure 2:283–291. doi: 10.1016/S0969-2126(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 62.Demidyuk IV, Shubin AV, Gasanov EV, Kostrov SV. 2010. Propeptides as modulators of functional activity of proteases. Biomol Concepts 1:305–322. doi: 10.1515/bmc.2010.025. [DOI] [PubMed] [Google Scholar]
- 63.Zhang N, Yin S, Liu S, Sun A, Zhou M, Gong X, Ge H. 2017. Crystal structure of lpg1832, a Virk family protein from Legionella pneumophila, reveals a novel fold for bacterial Virk proteins. FEBS Lett 591:2929–2935. doi: 10.1002/1873-3468.12773. [DOI] [PubMed] [Google Scholar]
- 64.Aragon V, Kurtz S, Cianciotto NP. 2001. Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect Immun 69:177–185. doi: 10.1128/IAI.69.1.177-185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dhatwalia R, Singh H, Reilly TJ, Tanner JJ. 2015. Crystal structure and tartrate inhibition of Legionella pneumophila histidine acid phosphatase. Arch Biochem Biophys 585:32–38. doi: 10.1016/j.abb.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 66.Moffat JF, Edelstein PH, Regula DP Jr., Cirillo JD, Tompkins LS. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea pig pneumonia model. Mol Microbiol 12:693–705. doi: 10.1111/j.1365-2958.1994.tb01057.x. [DOI] [PubMed] [Google Scholar]
- 67.Gunderson FF, Cianciotto NP. 2013. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio 4:e00074-13. doi: 10.1128/mBio.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Eisenthal R, Danson M. 2002. Enzyme assays: a practical approach, 2nd ed. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 70.Rangarajan M, Aduse-Opoku J, Hashim A, Paramonov N, Curtis MA. 2013. Characterization of the alpha- and beta-mannosidases of Porphyromonas gingivalis. J Bacteriol 195:5297–5307. doi: 10.1128/JB.00898-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uchiyama T, Miyazaki K, Yaoi K. 2013. Characterization of a novel beta-glucosidase from a compost microbial metagenome with strong transglycosylation activity. J Biol Chem 288:18325–18334. doi: 10.1074/jbc.M113.471342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agrawal P, Verma D, Daniell H. 2011. Expression of Trichoderma reesei beta-mannanase in tobacco chloroplasts and its utilization in lignocellulosic woody biomass hydrolysis. PLoS One 6:e29302. doi: 10.1371/journal.pone.0029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cowtan K. 2010. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr 66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamzin VS, Wilson KS. 1993. Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr 49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 78.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 79.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 80.Gomez-Valero L, Rusniok C, Rolando M, Neou M, Dervins-Ravault D, Demirtas J, Rouy Z, Moore RJ, Chen H, Petty NK, Jarraud S, Etienne J, Steinert M, Heuner K, Gribaldo S, Médigue C, Glöckner G, Hartland EL, Buchrieser C. 2014. Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires’ disease. Genome Biol 15:505. doi: 10.1186/PREACCEPT-1086350395137407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis IW, Murray LW, Richardson JS, Richardson DC. 2004. MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extracellular replication and survival of the L. pneumophila wild-type strain, an nttD mutant, a lapA plaC mutant, and a lapA lapB plaC mutant in conditioned amoebal medium. (A) Wild-type strain 130b (WT), lapA plaC mutant NU435 and lapA lapB plaC mutant NU436 (left), and the WT strain and nttD mutant NU431 (right) were inoculated into BYE broth, and then, at various times postinoculation, bacterial growth was monitored spectrophotometrically. The data points represent means and standard deviations of results from duplicate cultures, and the results presented are representative of two independent experiments. (B) A. castellanii amoebas were seeded in-well, and then a 0.4-µm transwell insertion was added followed by bacterial suspensions to prevent contact between the WT strain, lapA plaC mutant NU435, and lapA lapB plaC mutant NU436 (left) or between the WT strain and nttD mutant NU431 (right) or with the host cells. At the indicated times, CFU counts were recorded from within the upper chamber. Data are presented as means and standard deviations of results from duplicate wells and are representative of two independent experiments. Download FIG S1, EPS file, 1.3 MB (1.3MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
L. pneumophila loci encoding type II-secreted proteins. The chromosomal regions of strain 130b containing lapA (A), plaC (B), and nttD (C) are depicted. The horizontal arrows denote the locations, orientations, and relative sizes of the genes. The “lpw” numbers indicated within the arrows are ORF designations used in the database. Gene names and annotations are indicated below the arrows. Sizes of the genes are given (in base pairs) above the arrows, and sizes of intergenic regions are noted between the arrows. (A) The genes adjacent but transcriptionally unlinked to lapA encode type IV secretion effectors (N. Shohdy, J. A. Efe, S. D. Emr, and H. A. Shuman, Proc Natl Acad Sci U S A 102:4866–4871, 2005; E. Portier, H. Zheng, T. Sahr, D. M. Burnside, C. Mallama, C. Buchrieser, N. P. Cianciotto, and Y. Héchard, Environ Microbiol 17:1338–1350, 2015; D. T. Isaac, R. K. Laguna, N. Valtz, and R. R. Isberg, Proc Natl Acad Sci U S A 112:E5208–E5217, 2015). (B) The gene upstream of plaC is predicted to encode part of an ABC transporter, whereas the downstream gene is likely involved in glucosamine-6-phosphate metabolism (A. Gaballa and J. D. Helmann, J Bacteriol 180:5815–5821, 1998; R. A. Tinsley, J. R. Furchak, and N. G. Walter, RNA 13:468–477, 2007). (C) The gene upstream of nttD is not annotated, although tertiary structure predictions suggest that its protein product is a lipase (L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass, and M. J. Sternberg, Nat Protoc 10:845–858, 2015). This protein may also be a substrate for the T2S based on the detection of the protein in wild-type supernatants and the presence of a signal peptide at the protein’s N terminus (28). The two genes downstream of nttD are predicted to be involved in DNA rearrangement and repair (M. M. Slupska, J. H. Chiang, W. M. Luther, J. L. Stewart, L. Amii, A. Conrad, and J. H. Miller, Genes Cells 5:425–437, 2000; C. Darrigo, E. Guillemet, R. Dervyn, and N. Ramarao, PLoS One 11:e0163321, 2016). Download FIG S2, EPS file, 2.3 MB (2.3MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Intracellular infection of N. lovaniensis, V. vermiformis, and human macrophages by the L. pneumophila wild-type, plaC mutant, or lapA lapB plaC mutant strain. N. lovaniensis amoebas (A), V. vermiformis amoebas (B), or U937 macrophages (C) were infected with wild-type strain 130b (WT) or plaC mutant NU420 (panels A and B only) or with lapA lapB plaC mutant NU436, and at 0, 24, 48, and 72 h postinoculation, CFU levels within the infected wells were determined. Bacterial growth resulting from intracellular infection is expressed as the ratio of the CFU count at t = 24, 48, or 72 h to the CFU count at t = 0. Data are presented as means and standard errors of results from three independent experiments (n = 3). Asterisks indicate significant differences in levels of CFU recovery between the WT strain and all mutants (*, P < 0.05 [Student’s t test]). Download FIG S3, EPS file, 0.7 MB (750.9KB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Dose-dependent inhibition and substrate binding pocket of LapA. (A) Purified full-length LapA was incubated at 37°C with a leucine aminopeptidase substrate and 2-fold-increasing concentrations of bestatin. The residual aminopeptidase activity compared to that seen with untreated LapA was graphed against the log concentration of the inhibitor, and the half-maximal inhibitory concentration (IC50) was determined via nonlinear regression and sigmoidal slope response (variable slope) curve fitting (GraphPad Prism, version 7.00 for Windows; Graphpad Software, Inc., La Jolla, CA). Data are presented as mean residual activity with standard errors of results from two independent experiments (n = 2). (B) The S. griseus aminopeptidase/l-leucine complex (PDB code: 1F2O) and the A. proteolytica aminopeptidase/bestatin complex (PDB code: 1XRY) were superimposed on the aminopeptidase domain of LapA. The LapA substrate binding pocket is shown as light orange sticks, with modeled l-leucine and bestatin shown as green sticks. Download FIG S4, EPS file, 1.1 MB (1.1MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Interactions within the PA/aminopeptidase domain interface of LapA. (Left) The PA domain is shown as a cartoon, while the aminopeptidase domain is shown as a cartoon with a transparent surface. Interfacial residues are drawn as sticks. (Right) Magnification of the domain interface shown from three orientations. The secondary structure and the residues that make up the interface are annotated. Download FIG S5, EPS file, 3.1 MB (3.2MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NttD forms monomers and dimers in solution. (A) The crystal structure of the NttD dimer (PDB code: 4KH9) reveals three domains per monomer, which are numbered 1 (purple), 2 (yellow), and 3 (red) from the N terminus to the C terminus and are given for chain A. Chain B is depicted entirely in green. The NttD structure appears to form a stable homodimer, which is mediated through N-terminal domain 1. (B) Gel filtration profile of NttD showing that it forms both monomers and dimers in solution. Download FIG S6, EPS file, 1.4 MB (1.4MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conservation of the T2SS apparatus genes. This table lists the T2SS apparatus genes present within L. pneumophila strains (A) and among different Legionella species (B). Download TABLE S1, PDF file, 0.03 MB (33.4KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of the nttD, plaC, lapA, and lapB genes. This table lists the nttD (A), plaC (B), lapA (C), and lapB (D) genes present within L. pneumophila strains and among different Legionella species. Download TABLE S2, PDF file, 0.1 MB (82.8KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of the L. pneumophila LapA and LapB tertiary structure, active-site residues, and substrate binding pocket. (A) Significant deviations between the LapA (orange) and LapB (PDB code: 5GNE) (purple) structures were observed within the PA domain helices, which are annotated. (B) Amino acid residues of the active sites of LapA (orange) and LapB (purple) are drawn as sticks, and zinc (Zn) and water (WAT) molecules are drawn as spheres. (C) LapA and LapB structures are shown as electrostatic surface potential data with active-site zinc ions drawn as gray spheres. Substrate binding sites are highlighted with a yellow dashed line. Download FIG S7, EPS file, 4.4 MB (4.5MB, eps) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers and plasmids. This table lists the primers (A) and plasmids (B) that were used in this study. Download TABLE S3, PDF file, 0.1 MB (71.7KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.