Abstract
5′-Adenylylsulfate (APS) reductase was characterized in diverse marine algae. A cDNA encoding APS reductase from Enteromorpha intestinalis (EAPR) was cloned by functional complementation of an Escherichia coli cysH mutant. The deduced amino acid sequence shows high homology with APS reductase (APR) from flowering plants. Based on the probable transit peptide cleavage site the mature protein is 45.7 kD. EAPR expressed as a His-tagged recombinant protein catalyzes reduced glutathione-dependent reduction of APS to sulfite, exhibiting a specific activity of approximately 40 μmol min−1 mg protein−1 and Michealis-Menten kinetic constants of approximately 1.4 mm for reduced glutathione and approximately 6.5 μm for APS. APR activity and expression were studied in relation to the production of 3-dimethylsulfoniopropionate (DMSP), a sulfonium compound produced by many marine algae. A diverse group of DMSP-producing species showed extremely high enzyme activity (up to 400 times that found in flowering plants). Antibodies raised against a conserved peptide of APR strongly cross-reacted with a protein of 45 kD in several chlorophytes but insignificantly with chromophytes. In the chlorophyte Tetraselmis sp., APR activity varies significantly during the culture cycle and does not follow the changes in cellular DMSP content. However, a positive correlation was found between cell-based APR activity and specific growth rate.
S is an essential nutritional element for plant growth and development. Available in the environment primarily as sulfate it is imported into cells where it is incorporated into organic molecules as sulfate (e.g. extracellular polysaccharides like agar) or is reduced to sulfide before being incorporated as the thiol group of Cys. Cys serves as the central intermediate from which a diversity of primary and secondary metabolites and coenzymes are derived (Leustek and Saito, 1999). S-containing metabolites participate in a variety of cellular processes, including disease resistance (Alsher, 1989; Booth and Walker, 1997), tolerance to oxidation (Lappartient and Touraine, 1997), heavy metals (Ruegsegger and Brunold, 1992; Schafer et al., 1997), water stress (Storey et al., 1993), and developmental signaling (Bouarab et al., 1999).
Although S research in vascular plants has a 30-year history, only recently has the process been studied at the molecular level. Molecular and biochemical evidence has demonstrated significant differences in the first enzyme-catalyzing sulfate reduction among vascular plants, bacteria, and fungi. In all of the organisms, sulfate assimilation begins with the enzyme ATP sulfurylase (EC 2.7.7.4) that catalyzes the adenylation of sulfate to 5′-adenylylsulfate (APS). Adenylation is necessary because sulfate is relatively unreactive. APS is then reduced by APS reductase (APR; EC 1.8.99.-) in plants and some bacteria (Bick and Leustek, 1997; Abola et al., 1999; Bick et al., 2000), whereas in other bacteria and fungi APS is further phosphorylated at the 3′ position forming 3′-phospho-5′-adenylylsulfate (PAPS) before being reduced by PAPS reductase. Plant APR is unique in that it is able to use reduced glutathione (GSH) as a source of electrons. By contrast, bacterial APR and PAPS reductase require the redox factors thioredoxin (Trx) or glutaredoxin (Grx) as a source of electrons. The GSH-dependency of plant APR is probably mediated through a carboxyl terminal domain that functions as a Grx, which is lacking in the bacterial and fungal enzymes. In flowering plants, APR is thought to be a primary regulation point for the sulfate assimilation pathway (Leustek and Saito, 1999).
Most of what is known about sulfate assimilation in plants is from research on flowering plants. Little is known about the process in algae. Marine algae metabolize S in unique ways, thus they likely have special S requirements. For example, many produce large amounts of sulfated extracellular polysaccharides like agar. Others produce and accumulate in abundance a sulfonium compound known as 3-dimethylsulfoniopropionate (DMSP). DMSP synthesis is limited to very few species of flowering plants (Hanson and Gage, 1996) but is widespread among marine algae (Keller et al., 1989; Gröne, 1995). It probably serves as a compatible intracellular osmolyte and can accumulate to very high concentrations. Up to 0.5 m has been reported for some species (Keller et al., 1989). DMSP is synthesized from Met (Gage et al., 1997), therefore, it is dependent ultimately on the activity of the sulfate assimilation pathway. There is evidence that in some species DMSP synthesis is stimulated under N-limiting conditions (Gröne and Kirst, 1992). DMSP is the primary source for one of the most important biogenic gases in the atmosphere, dimethylsulfide (DMS), accounting for nearly 50% of the global biogenic S input into the atmosphere (Andreae and Raemdonck, 1983; Malin et al., 1992; Gröne, 1995).
As the first step toward understanding S metabolism in marine algae APR was studied in a variety of species, and a cDNA encoding the enzyme was cloned from the marine chlorophyte Enteromorpha intestinalis. The cDNA clone was used to express the enzyme for study and for production of a specific antibody. This antibody was used to study the regulation of APR in relation to DMSP production in another chlorophytic species Tetraselmis sp.
RESULTS
Cloning and Analysis of an APR cDNA from E. intestinalis
Functional complementation of an Escherichia coli cysH mutant with a cDNA library prepared from E. intestinalis resulted in the isolation of approximately 100 positive clones after screening 1 × 106 independent transformants. Twenty-five clones were selected for sequence analysis. All were found to be derived from the same gene but varied in the length of their 5′ and 3′ ends. The longest clone, referred to as EAPR, was 1,525 bp. It shows a continuous open reading frame beginning from nucleotide 1 to the termination codon at 1,336 to 1,338. Therefore, the translation initiator is not possible to predict, although a Met codon exists at position 68 to 70. An alignment of the EAPR open reading frame with Arabidopsis APR3 (Fig. 1) shows that the two have a high level of homology (56% identity, 63% similarity). The first 41 amino acids of EAPR show the properties of a transit peptide for plastid localization with a possible cleavage site at position 32 to 35 (LRAG) (Von Heijne et al., 1989). APR is known to be plastid localized in flowering plants (Rotte, 1998). If the EAPR protein were processed at the putative cleavage site (LRAG) the mature protein would be 411 amino acids with a predicted mass of 45.7 kD.
Figure 1.
Alignment of the deduced E. intestinalis EAPR amino acid sequence with Arabidopsis APR3 (AF01628). The sequence was aligned with PileUp program (Genetics Computer Group, Madison, WI). Putative reductase-domain and Grx-domain are positioned between amino acid numbers 36 to 116 and 117 to 446, respectively. Residues of important to the catalytic function and tertiary structures are indicated with dots, asterisks, or diamonds.
The alignment in Figure 1 shows that EAPR contains all of the hallmarks of plant APRs. The region from amino acids 83 to 310 is homologous with the sulfate assimilatory APR and PAPS reductase of bacteria and fungi (Berendt et al., 1995; Bick et al., 2000). This region has been defined as the reductase domain of plant APR (Bick et al., 1998) and includes a PP motif (indicated by dots), likely involved in nucleotide binding, and a region conserved in all of the assimilatory (P)APRs (indicated by asterisks) that is important for catalytic activity (Berendt et al., 1995). The region from amino acids 311 to 445 shows homology with Trx and Grx including the active site sequence (indicated with diamonds).
The genomic organization of the EAPR gene was studied in E. intestinalis by Southern blotting using the EAPR cDNA as a probe. A simple hybridization pattern was observed with all of the restriction enzymes tested (data not shown). This indicates that the sequence homologous to the EAPR cDNA likely exists as a single locus in the alga.
Characterization of the EAPR Product
E. coli JM96 strain (cysH mutant) is devoid of the enzyme responsible for the reduction of sulfate to sulfite (PAPS reductase). However, the strain expressing EAPR showed an APR activity of approximately 2 × 10−3 units mg−1 in the crude bacterial extract. Comparable APR activity was observed with a deletion subclone of the EAPR cDNA from which 249 bp was removed from the 5′ end (SstI site, indicated by an arrow in Fig. 1), indicating that the catalytic core of the enzyme lies beyond the 84th amino acid.
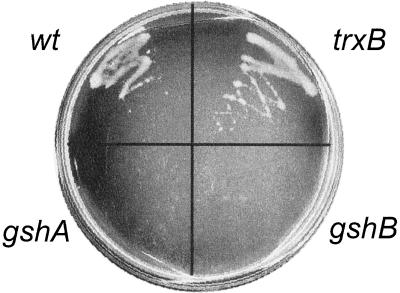
Plant APRs are unable to function with electrons donated from sources other than thiol compounds despite the existence of a C-terminal domain with sequence homology to Trx. To determine what electron donor EAPR protein uses during complementation, a test was performed using E. coli strains with mutations in gshA (γ-glutamyl-Cys synthetase) or gshB (GSH synthetase) required for GSH synthesis, or trxB (Trx reductase). The result indicates that EAPR cDNA clone was able to complement a cysH mutant of E. coli that also carries the trxB mutation (Fig. 2). However, it was unable to complement the strains lacking GSH (E. coli strains carrying cysH and gshA or gshB), although active EAPR protein was expressed in these bacterial strains as demonstrated by the in vitro APR activity measurements. Consequently, it is concluded that EAPR protein uses GSH, rather than Trx reductase, for catalysis.
Figure 2.
Complementation of E. coli cysH by E. intestinalis APR requires GSH, but not Trx. wt refers to the cysH strain carrying wild-type alleles for trxB, gshA, and gshB. The others are cysH strains carrying the designated mutation. The cultures were incubated for 40 h at 30°C on M9 plate without Cys.
The purified recombinant EAPR product has no PAPS reductase activity and exhibited a specific APR activity of approximately 40 units mg−1. This activity is significantly greater than the reported activities of APR products from Arabidopsis (0.2–4 units mg−1; Bick et al., 1998) but similar to APS reductase from Lemna (Suter et al., 2000). Moreover, the EAPR protein is not sensitive to inactivation by freeze thawing and storage in the thawed state as are Arabidopsis APRs, suggesting that the algal enzyme is a much more active and robust form of APR. The kinetics of the algal APR, however, is comparable with that reported for Arabidopsis APRs with apparent Kms of 1.39 mm for GSH and 6.5 μm for APS.
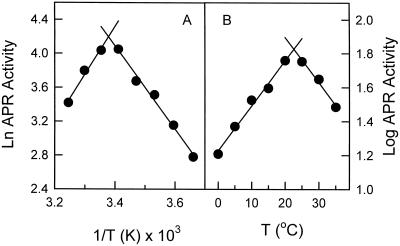
A strong temperature dependence of the activity was observed for the purified recombinant APR (Fig. 3). A sharp break was found between 20°C to 25°C with an apparent Ea, estimated from the slope of the Arrhenius plot, of 10.1 kcal mol−1 for the temperatures between 0°C to 20°C (slope = 5.098, r2 = 0.97, Fig. 3A). The Q10 below 20°C was estimated to be 1.9 (slope = 0.0276, r2 = 0.97, Fig. 3B), comparable with those found for many other biological processes (Berry and Raison, 1981). Similar responses were observed for APR activity in cell-free extracts of the phytoplankton Tetraselmis sp. and Emiliania huxleyi (data not shown).
Figure 3.
Temperature dependence of APR activity of purified EAPR recombinant protein. A, Arrhenius plot; B, log transformation. APR activity was determined as described in the “Materials and Methods” at the temperatures indicated in the graph.
Enzyme Activity and Antigenicity of APR from E. intestinalis and Several Marine Phytoplankton
APR and PAPS reductase activity were determined in several major groups of marine algae, including three chlorophytes (E. intestinalis, Tetraselmis sp., and Dunaliella salina), two diatoms (Thalassiosira weissflogii and Thalassiosira oceanica), two prymnesiophytes (I. galbana and E. huxleyi), and one dinoflagellate (Heterocapsa triquetra). These algae are from diverse phylogenetic origins and have different capacities for DMSP production (Table I). The results showed that most of the species tested have high APR activities ranging from approximately 100 up to 500 × 10−3 units mg−1, except for the dinoflagellate species (Table I). These activities are 100-fold greater than that from young leaves of two species of flowering plants Arabidopsis and Indian mustard (approximately 1 × 10−3 units mg−1). No significant PAPS reductase activity (< 1% of that for APR activity) was detected in any of the species examined, indicating that APR is likely the primary route for sulfate reduction in these organisms.
Table I.
Comparison of APS reductase activity among different plant species
| Algal Species | APS Reductase Activity in Crude Cell Lysates |
|---|---|
| × 10−3 units mg−1 | |
| Arabidopsis* (young leaves) | ≤1.3 |
| Indian mustard* (young leaves) | ≤1 |
| E. intestinalis | 4–20 |
| Tetraselmis sp. | 100–500 |
| D. salina* | 50–100 |
| T. weissflogii | 200 |
| T. oceanica | 80–130 |
| I. galbana | 100–250 |
| E. huxleyi | 100–400 |
| H. triquetra | 0.5–5 |
Arabidopsis and all phytoplankton species were laboratory-grown materials, whereas E. intestinalis were collected from the field (see “Materials and Methods”). Species with no capacity of DMSP synthesis were indicated with an asterisk. The range of activity was determined from samples collected at different growth stage, therefore reflecting the biological variations of the activity in different species.
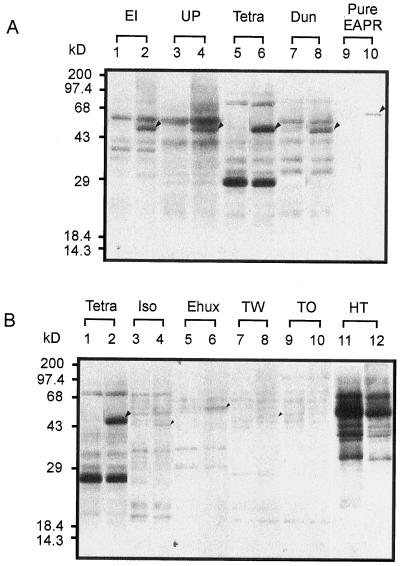
The cross-reactivity of an APR-specific peptide antiserum (Fig. 1) was tested against cell lysates from the algal species. The antibody cross-reacted significantly only with the chlorophytic species. The cross-reacting peptide in these species was approximately 45 kD (Fig. 4A), a size that is consistent with the mass of the mature protein predicted from the EAPR cDNA clone. The strongest cross reactivity was found with Tetraselmis sp., correlating with the extraordinarily high APR activity found in this species (Table I), suggesting that this high activity is achieved in part by high expression level of APR in this organism. Weak, specific cross-reactivity was observed with the prymnesiophyte, diatom, and dinoflagellate species examined (Fig. 4B), suggesting that APR from these species are divergent in the peptide sequence region (IAFSGAEDVA) conserved among APR from flowering plants and green algae. Similar species divergence in enzyme antigenicity has been observed for enzymes involved in other assimilatory pathways in marine algae, such as nitrate reductase and Gln synthetase (Gao et al., 1993; Robertson et al., 1996). The current result also supports the idea of a diverse evolutionary history for marine organisms.
Figure 4.
Comparison of ARS reductase protein antigenicity from marine algae. All of the lanes were loaded with equal amount of protein (6 μg) except for Ulva pertusa (30 μg) and the purified recombinant protein from pET-EAPR (2 ng). Odd-numbered lanes are controls blotted with preimmune antiserum, whereas even-numbered lanes are blotted with APR antiserum. A, EI (E. intestinalis): total activity was 120 pmol min−1; UP (U. pertusa): total activity was 16 pmol min−1; Tetra (Tetraselmis sp.): total activity was 1,300 pmol min−1; Dun (D. salina): total activity was 390 pmol min−1; Pure APR (purified recombinant protein from pET-EAPR): total activity was 90 pmol min−1. B, Tetra (Tetraselmis sp.): total activity was 1,300 pmol min−1; Iso (I. galbana): total activity was 1,480 pmol min−1; Ehux (E. huxleyi): total activity was 1,500 pmol min−1; TW (T. weissflogii): total activity was 1,400 pmol min−1; TO (T. oceanica): total activity was 800 pmol min−1; HT (H. triquetra): total activity was 30 pmol min−1.
Regulation of APR Activity and DMSP Production in Tetraselmis sp.
The high APR activity found in most marine algae examined in this study may reflect the special S needs of these organisms. Most of the species examined are known to produce large amounts of DMSP when N is limiting in the environment (Gröne, 1995). DMSP synthesis may ultimately be dependent on the activity of APR as it was demonstrated earlier that this is the first enzyme involved in sulfate reduction in these organisms and is most likely the regulation point in the sulfate assimilation pathway of flowering plants (Brunold, 1981; Brunold and Suter, 1984). As a first step toward understanding environmental regulation of APR and its role in the production of DMSP in marine organisms APR activity and intracellular DMSP contents were examined in Tetraselmis sp. cultures grown with limiting or sufficient N supplies. These conditions are known to alter the organism's ability to synthesize DMSP.
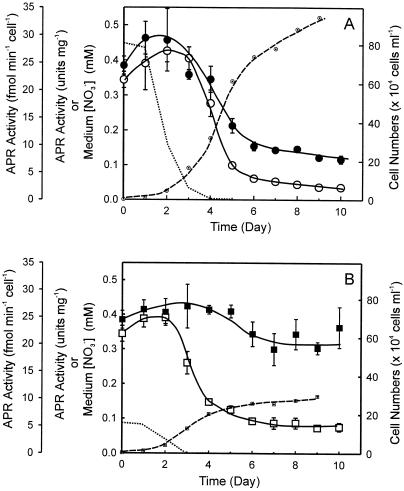
Shown in Figure 5 is APR activity in Tetraselmis sp. cells grown with abundant and limiting N supply during a culture cycle. For both of the treatments the highest activities were observed during the early log growth phase, the most rapid growth stage of batch cultures. As the cultures became depleted in N (d 3–5), APR activity decreased to <40% and reached a 10% to 25% level when the cultures reached stationary phase. For the abundant N treatment the pattern for cell-based APR activity was similar to that based on cellular protein content in these cultures (Fig. 5A, compare white circles with black circles). In contrast, APR activities in the cells grown with limited N supply, when expressed on cellular protein basis, did not change significantly during the culture cycle (Fig. 5B, compare black squares with white squares), although slightly higher activities were observed during the mid-log growth phase of these cells. The APR activities were comparable with those found in the early log growth-phased cells with abundant N supply. When the APR activity was expressed on a per-cell basis, the pattern of the activity change was similar to that found for cultures grown with abundant N supply with maximal activity during the rapid growth stage and low activity after N was depleted from the cultures. This result may reflect the organisms' ability to quickly mobilize cellular proteins after encountering N shortage. Since APR activity on protein basis was maintained through out the culture cycle, the results suggest that it may be spared from deactivation or proteolysis. Immunoblot analysis revealed that the changes in APR activity observed here are paralleled by changes in APR protein abundance (data not shown).
Figure 5.
APR activity from Tetraselmis sp. cultures grown with abundant N supply (A) and limited N supply (B). Dotted lines indicate the nitrate level in the cultures. Dash lines with small white circles or white squares are growth curves of the cultures indicated by the cell densities of the cultures. Black circles or squares represent APR activities normalized to cellular protein content, whereas white circles or squares represent APR activities based on cells. Each data point was the average of six measurements from four independent cultures.
It is interesting that under both of the growth conditions, APR activity on cell basis began declining when medium nitrate was reduced to approximately 35 μm and reached a plateau 1 d after the depletion of N from the media, suggesting a coordinated regulation between sulfate and nitrate assimilation. Moreover, the cell-based APR activity in cells grown with limited N supply plateaued, upon the depletion of nitrate from media, at a level twice that in cells grown with abundant N supply.
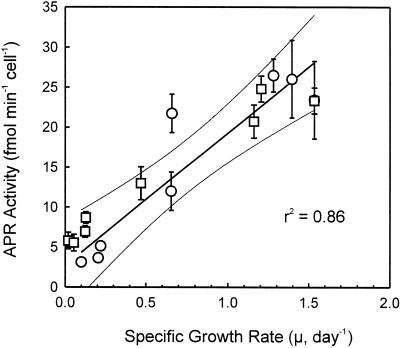
Given the observation that cell-based APR activity is inversely correlated with the growth curve and N availability under both of the growth conditions, a close coupling of grow rate and cell-based APR activity was hypothesized. Comparison of specific growth rate (μ) with cell-based APR activity (Fig. 6) further support the hypothesis. The result suggests that cell-based APR activity can be used as an index for estimating growth rate and thus productivity as well as the N-nutrition state of the cells.
Figure 6.
Correlation of APR activity and specific growth rate (μ). White circles were data points from cultures grown with abundant N supply, whereas white squares were data points from cultures grown with limited N supply. Each data point represents the average of six measurements from four independent cultures.
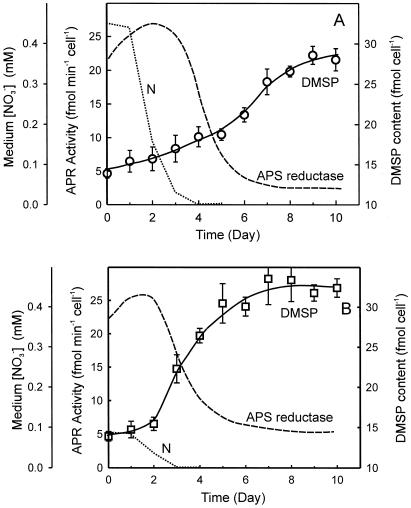
Intracellular DMSP content in Tetraselmis sp. cells grown under limited or abundant N supply conditions is illustrated in Figure 7. Both of the cultures exhibited similar intracellular DMSP content (approximately 15 fmol cell−1) when N was present in the medium. However, the increase in DMSP content after N was depleted was much more rapid in cells grown with limited N supply than those grown with abundant N supply. Further, after reaching stationary phase, the N-limited cultures attained a higher intracellular DMSP content compared with cells grown with abundant N. The changes in cellular DMSP content (Fig. 7, solid lines) did not correlate with the changes in APR activity, suggesting that APR does not directly limit or regulate DMSP synthesis and that the synthesis of DMSP may be partly or entirely supported by intracellular resources of assimilated S.
Figure 7.
APR activity and intracellular DMSP content during a growth cycle in batch cultures of Tetraselmis sp. grown under N-sufficient supply (A) and N-limited supply (B) conditions. Cell-based APR activity is indicated as the dashed lines, whereas intracellular DMSP content is indicated as the solid lines with white circles or squares. Dotted lines indicate the nitrate level in the cultures. Each data point was the average of six measurements from four independent cultures.
DISCUSSION
S metabolism is important to marine algae in that they produce and accumulate to high concentrations S compounds that most terrestrial plants do not produce. Unlike most terrestrial crop plants, many marine algae live in habitats that are characterized by limited N and abundant S supply. The concentration of sulfate in seawater is approximately 40 mm (Pickard and Emery, 1988). In response to this environment, many have evolved the ability to produce DMSP, a functional analog of the N-containing osmolyte Gly betaine. In those species that accumulate it, DMSP is likely a major sink for assimilated S in addition to protein, especially when N is not available. The ability to synthesize DMSP may play a critical role in the survival of these organisms in their environmental niches.
In the present study, as the first attempt to understand sulfate assimilation in marine organisms, a cDNA (EAPR) from E. intestinalis was cloned. The result indicates the structural and functional similarities between algal APR from marine chlorophyte algae and flowering plants. In addition, the result demonstrated the use of functional complementation approach as a powerful tool in investigating molecular and biochemical features of processes in marine algae.
Examination of temperature response of APR activity showed that the enzyme activity is strongly temperature dependent with a maximal activity at 20°C to 25°C. A sharp break at low temperatures (<30°C) for enzyme activity has only been observed for nitrate reductase (EC 1.6.6.1) previously, the first enzyme involved in nitrate reduction (Gao et al., 1993), but not for other enzymes that are also important in assimilatory processes in plants, e.g. RuBP carboxylase, PEPC Kinase, Gln sythetase, and Glu dehydrogenase (Ahmed et al., 1977; Descolas-Gros and De Billy, 1987; Cabello-Pasini, 1996; Robertson et al., 1996). However, unlike APR where a similar pattern was seen in both of the chlorophyte and chromophyte, a sharp break at low temperatures (15°C) for nitrate reductase was only observed in chromophytes. The chlorophytic nitrate reductase activity was linear up to 35°C (Gao et al., 2000). Such large differences in temperature dependence for important assimilatory enzymes may have significant impact on the geographic distribution and seasonal abundance of these organisms in the marine environment.
A significant correlation between the specific growth rate and APR activity on a per cell basis was found in Tetraselmis sp. cultures. Together with the observation that APR activity declines as N is depleted from the medium in cells grown with both of the limiting and the abundant N supply, the results suggest a coordinated regulation between sulfate and nitrate assimilation. Regulatory interactions between assimilatory S and N reduction have been observed in previous studies with various flowering plants (Reuveny et al., 1980; Neuenschwander et al., 1990; Brunold, 1993; Prosser et al., 1997; Lee, 1998; Koprivova et al., 2000). These interactions were thought to reflect a mechanism to coordinate and balance the flow of the two essential elements (S and N) into protein (Reuveny et al., 1980; Brunold and Suter, 1984). The previous studies showed that low nitrate concentration causes a drastic decrease in the specific APR activity after 24 h (Brunold and Suter, 1984) as well as the influx of sulfate into cells (Clarkson et al., 1989) in flowering plants. The reduction in APR is due to a decrease in the steady-state level of mRNA and protein (Lee, 1998). These results are consistent with our finding that high level of APR activity in the marine alga Tetraselmis sp. was observed when nitrate is plentiful. It is interesting that cell-based APS activity in cells grown with limited N supply plateaued at a level twice that found in cells grown with abundant N supply. One possible explanation for this observation is that APR is spared from proteolysis relative to other proteins in these cells.
The experiments did not reveal a direct correlation between APR activity and DMSP production as indicated by the intracellular DMSP content. This may be explained in part by the possibility that intracellular DMSP content does not represent accurately the rate of DMSP production. It also, in part, suggests an ability of the alga to mobilize intracellular S resources under the stress of N shortage. In this regard, the extraordinarily high APR activity, compared with flowering plants, may be of significance. Thus, whereas APR does not directly limit or regulate DMSP synthesis, its high activity may reflect a process for building an intracellular pool of reduced S, which could serve as a resource for support of DMSP biosynthesis when needed. To obtain a comprehensive understanding of the role of APR in DMSP production and its regulation in the marine organisms that have special S needs, further studies are required.
MATERIALS AND METHODS
Algal Materials and Growth Conditions
Enteromorpha intestinalis and Ulva pertusa were collected in early summer at the intertidal zone of the Gateway National Recreation Area at Sandy Hook, New Jersey (U.S. Department of the Interior collection permit no. SHU–98–03). The tissue was rinsed with ultrapure water and frozen immediately with dry ice. Frozen samples were stored at −80°C until use. Phytoplankton species (Tetraselmis species, CCMP897; Dunaliella salina, ATCC30861; Isochrysis galbana, CCMP1323; Emiliania huxleyi, CCMP1299, Thalassiosira weissflogii CCMP1336, Thalassiosira oceanica CCMP1005, and Heterocapsa triquetra CCMP449) were grown in f/2-enriched seawater medium (Guillard, 1975) at 19°C on a 12-h:12-h light:dark cycle at 200 μmol m−1 s−1, using NO3− as the sole N source. Artificial seawater medium (Sigma S-9883) with f/2-enrichments was used in the experiments assessing the role of APR in relation to DMSP production. The species with CCMP designations were obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (Bigelow Laboratory for Ocean Sciences, West Boothbay Harbor, ME). D. salina was obtained from the American Type Culture Collection (Fairfax, VA). All of the samples for analysis were harvested at noon by centrifugation, frozen in liquid N2, and stored at −80°C.
Construction of a cDNA Library from E. intestinalis and Cloning of EAPR
Total RNA was prepared as follows. Twenty-three grams (fresh weight) of frozen E. intestinalis was ground into powder with liquid N2 then mixed with 60 mL of extraction buffer containing 4 m guanidine thiocyanate, 25 mm sodium citrate (pH 7.0), 0.5% (w/v) sarcosyl, and 0.7% (v/v) β-mercaptoethanol. Then 0.1 volume of 2 m sodium acetate (pH 5.0) was added. The mixture was extracted with 40 mL of phenol (diethyl pyrocarbonate-water saturated) followed by an addition of 8 mL of chloroform:isoamyl alcohol (49:1). The mixture was centrifuged at 5,000g for 20 min at 4°C. The upper aqueous phase was transferred into a clean tube and re-extracted with 40 mL of chloroform:isoamyl alcohol (49:1). Total RNA was precipitated with an equal volume of ice-cold isopropanol. The RNA pellet was dissolved in 20 mL of extraction buffer and reprecipitated with 2 volumes of ethanol. The cDNA library was constructed in λTriplEx using the EcoRI (5′) and XbaI (3′) cloning sites. The library contained 1.2 × 106 independent clones.
The λ-phage cDNA library from E. intestinalis was converted to plasmid form by mass excission according to the manufacturer's instruction (CLONTECH Laboratories, Palo Alto, CA). The resulting plasmid library was then used to screen for clones that complement the Cys requirement of Escherichia coli strain JM96 (cysH), following the procedure described by Setya et al. (1996). JM96 was provided by the Coli Genetic Stock Center (Yale University, New Haven, CT).
DNA Sequencing and Nucleic Acid-Blot Analysis
DNA sequencing was carried out on both of the strands using plasmid primer sites with the original EAPR cDNA clone as well as deletion subclones on the parent vectors, using a DNA sequencer (model 373, Applied Biosystems, Foster City, CA).
For Southern blotting, E. intestinalis genomic DNA was isolated from materials obtained as described earlier (see “Algal Materials and Growth Conditions”). Four grams of tissue (frash weight) was first pulverized in liquid N2 and then mixed with 23 mL of preheated (65°C) extraction buffer containing 100 mm Tris [Tris(hydroxymethyl)-aminomethane]-HCl (pH 8.0), 500 mm NaCl, 8.3 mm NaOH, 1.25% (w/v) SDS, and 0.38% (w/v) sodium bisulfite. The mixture was then incubated at 65°C for 15 min, and 7 mL of 5 m potassium acetate was added. After standing on ice for 20 min, the mixture was centrifuged at 4,000g for 10 min. The DNA in the supernatant was precipitated by adding 0.7 volume of prechilled isopropanol and washed with 70% (v/v) ethanol. The DNA pellet was then resuspended in T5E buffer (50 mm Tris-HCl, pH 8.0, and 10 mm EDTA), containing DNase-free RNase A (final concentration of 20 μg/mL) and incubated at 37°C for approximately 1 h. The DNA was then subjected to buffer-saturated phenol/chloroform extraction twice and reprecipitated with ethanol. The final DNA pellet was resuspended in TE buffer (10 mm Tris HCl, pH 8.0, and 1 mm EDTA) and used for Southern blotting.
After digestion with the appropriate restriction enzymes, the genomic DNA were electrophoresed in 0.8% (v/v) agarose gels and transferred onto Zeta-Probe membrane (Bio-Rad Laboratories, Hercules, CA). The DNA probe, labeled by the random primer method (Life Technologies/Gibco-BRL, Cleveland) using [α-32P]dCTP, was made from the full-length cDNA of EAPR1. Hybridization was performed in 2× SSC containing 7% (w/v) SDS at 50°C.
Construction of Expression Plasmids and Purification of Recombinant EAPR
The EAPR cDNA, after removing 189 nucleotides from the 5′ end, was sucloned as a 1,360-bp NotI fragment into pET-30b (Novagen, Madison, WI). The plasmid, pET-EAPR was transformed into BL21(DE3) plysS. Transformants were grown at 37°C to an optical density of approximately 0.4 at 600 nm and then expression was induced with 1 mm isopropyl-β-d-thiogalactoside for 2 h before harvesting. Because of the association of a 55-amino acid-long fragment (which contains His-tag, thrombin, S-tag, and enterokinase cleavage site) with the recombinant EAPR enzyme, it is estimated that the resulting polypeptide is 466 amino acids with a predicted mass of approximately 52 kD.
The purification of recombinant EAPR enzyme was conducted using Ni-agarose (TALON, CLONTECH Laboratories). All of the procedures were carried out at room temperature (22°C–25°C). The bacterial cell pellet from a 100-mL culture was lysed by sonication in 10 mL of 20 mm Tris-HCl, pH 8.0, and 100 mm NaCl. The lysate was centrifuged at 12,000g for 10 min, and the supernatant was incubated with 1.5 mL of TALON resin for 30 min on a rotating shaker. The resin was washed with 5 volumes of lysis buffer and then packed into a disposable column. The column was further washed with 5 volumes of lysis buffer containing 10 mm imidazole. EAPR protein was eluted with lysis buffer containing 50 mm imidazole, then the buffer was replaced with 100 mm Tris-HCl, pH 8.5 containing 1 mm EDTA using microconcentrator unit (Microcon-30, Millipore, Bedford, MA). The pure protein was stored at −80°C.
The EAPR cDNA was also subcloned as a 1,310-bp (SstI and HindIII) fragment into pET-30a. The plasmid was transformed into BL21(DE3) plysS and induced only for APS activity determination.
(P)APS Reductase Activity and Protein Assays
Algal cells were harvested, frozen in liquid N2, and stored in −70°C before analysis. Algal cells were lysed in 100 mm Tris-HCl, pH 8.5, 1 mm EDTA, and the supernatant collected after centrifugation at 14,000g for 15 min. Microalgae were lysed by sonication. Macroalgae were ground to a fine powder in liquid N2. Enzyme activities were measured as described by Bick et al. (1998) using GSH (1 mm) as the electron donor and [35S]APS (approximately 500 Bq nmol−1) as subtrate. One unit of enzyme activity is defined as micromoles of product formed per minute. Apparent Km values were calculated by direct-fit to the Michaelis-Menten equation using Sigma-Plot (Jandel Scientific, version 5.0, SPSS, Inc., Chicago). Protein was estimated by Bradford assay (Bradford, 1979) using a bovine serum albumin standard.
Antibody Preparation and Immunoblotting
Antibodies were prepared in rabbits by Biosynthesis (Lewisville, TX) using the MAP peptide method. A peptide sequence conserved among APRs from flowering plants and EAPR was chosen for the procedure (NH2-IAFSGAEDVA).
Immunoblotting was carried out as described by Wang et al. (1993). The antibody was used at a dilution of 1:5,000. The secondary antibody was horseradish peroxidase-linked goat anti-rabbit diluted 1:10,000. The immune complexes were detected with the Renaissance Kit (DuPont NEN, Inc., Boston) and quantified with a densitometric video camera (model: Datavison 261 from Fernon Electronic Imaging, St. Michaels, MD) and the software NIH Image (version 1.0) provided by the National Institutes of Health (Bethesda, MD).
DMSP Measurements
DMSP content was measured indirectly as DMS produced after alkaline treatment of algal cells (White, 1982). Algal cells were collected by centrifugation and resuspended in 100 mm Tris-HCl, pH 8.0. Five to 10 μL of the cell suspensions was placed onto filter papers (8-mm diameter disc). The filter papers were placed into 2-mL glass vials containing 0.4 mL of 25% (w/v) NaOH and capped with Teflon-coated septum. The vials were incubated at room temperature for 4 h and then placed at 4°C overnight. DMS produced in the head space of the vials was removed with a gas-tight syringe and measured by gas chromatography on a Poraplot Q, fused silica column (10 × 0.53 mm, Chrompack, The Netherlands) using a Shimadzu GC-17A gas chromatograph outfitted with a flame ionization detector.
ACKNOWLEDGMENTS
We would like to thank Dr. Julie-Ann Bick for numerous helpful discussions and technical assistance and Dr. John Reinfielder (Rutgers Environmental Sciences Department) for generously sharing his culture collection.
Footnotes
This work was supported by the National Science Foundation (grant nos. IBN–9601146 and IBN–9817594), the Office of Naval Research (grant no. N00014–96–0212 to T.L.), and the Agricultural Research Service cooperative agreement (grant no. 58–6435–6–028 to O.M.E.S.).
LITERATURE CITED
- Abola AP, Willits MG, Wang RC, Long SR. Reduction of adenosine-5′-phosphosulfate instead of 3′-phosphoadenosine-5′phosphosulfate in Cys biosynthesis by Rhizobium meliloti and other members of the family Rhizobiaceae. J Bacteriol. 1999;181:5280–5287. doi: 10.1128/jb.181.17.5280-5287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed SI, Kenner RA, Packard TT. A comparative study of the glutamate dehydrogenase activity in several species of marine phytoplankton. Mar Biol. 1977;39:93–101. [Google Scholar]
- Alsher RG. Biosynthesis and antioxidant function of glutathione in plants. Physiol Plant. 1989;77:457–464. [Google Scholar]
- Andreae MO, Raemdonck H. Dimethylsulfide in the surface ocean and the marine atmosphere: a global view. Science. 1983;221:744–747. doi: 10.1126/science.221.4612.744. [DOI] [PubMed] [Google Scholar]
- Berendt U, Haverkamp T, Prior A, Schwenn JD. Reaction mechanism of thioredoxin: 3′-phospho-adenylylsulfate reductase investigated by site-directed mutagenesis. Eur J Biochem. 1995;233:347–356. doi: 10.1111/j.1432-1033.1995.347_1.x. [DOI] [PubMed] [Google Scholar]
- Berry JA, Raison JK. Responses of macrophytes to temperature. In: Lange OL, Aragno M, editors. Encyclopedia of Plant Physiology, New Series. 12A. Berlin: Springer-Verlag; 1981. pp. 277–338. [Google Scholar]
- Bick J, Aslund F, Chen Y, Leustek T. Glutaredoxin function for the carboxyl-terminal domain of the plant-type 5′-adenylylsulfate reductase. Proc Natl Acad Sci USA. 1998;95:8404–8409. doi: 10.1073/pnas.95.14.8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick J, Dennis JJ, Zylstra GJ, Nowack J, Leustek T. Identification of a new class of 5′-adenylylsulfate (APS) reductase from sulfate assimilating bacteria. J Bacteriol. 2000;182:135–142. doi: 10.1128/jb.182.1.135-142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick J, Leustek T. Plant sulfur metabolism: the reduction of sulfate to sulfite. Curr Opin Plant Biol. 1997;1:240–244. doi: 10.1016/s1369-5266(98)80111-8. [DOI] [PubMed] [Google Scholar]
- Booth E, Walker K. The effectiveness of foliar glucosinolate content raised by sulfur application on disease control in oilseed rape. In: Cram WJ, De Kok LJ, Stulen I, Brunold C, Rennenberg H, editors. Sulphur Metabolism in Higher Plants. Leiden, The Netherlands: Backhuys Publishers; 1997. pp. 327–329. [Google Scholar]
- Bouarab K, Potin P, Correa J, Kloreg B. Sulfated oligosaccharides mediate the interaction between a marine red alga and its green algal pathogenic endophyte. Plant Cell. 1999;11:1635–1650. doi: 10.1105/tpc.11.9.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1979;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brunold C. Regulation of adenosine 5′-phosphosulfate sulfotransferase in higher plants. In: Bothe H, Trebst A, editors. Biology of Inorganic Nitrogen and Sulfur. Heidelberg: Springer-Verlag; 1981. pp. 352–358. [Google Scholar]
- Brunold C. Regulatory interactions between sulfate and nitrate assimilation. In: De Kok LJ, Stulen I, Rennenberg H, Brunold C, Rauser WE, editors. Sulfur Nutrition and Assimilation in Higher Plants. The Hague, The Netherlands: SPB Academic Publishing; 1993. pp. 61–75. [Google Scholar]
- Brunold C, Suter M. Regulation of sulfate assimilation by nitrogen nutrition in the duckweek Lemna minor L. Plant Physiol. 1984;76:579–583. doi: 10.1104/pp.76.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Pasini A. Biochemical and environmental characterization of the light-independent carbon fixation in marine algae. PhD dissertation. Stony Brook: State University of New York; 1996. [Google Scholar]
- Clarkson OT, Saker LR, Purves JV. Depression of nitrate and ammonium transport in barley plants with diminished sulphate status of co-regulation of nitrogen and sulphate intake. J Exp Bot. 1989;40:953–963. [Google Scholar]
- Descolas-Gros C, De Billy G. Temperature adaptation of RuBP carboxylase: kinetic properties in marine Antarctic diatom. J Exp Mar Biol Ecol. 1987;108:147–158. [Google Scholar]
- Gage DA, Rhodes D, Nolte KD, Hicks WA, Leustek T, Cooper AJ, Hanson AD. A new route for synthesis of dimethylsulphoniopropionate in marine algae. Nature. 1997;387:891–894. doi: 10.1038/43160. [DOI] [PubMed] [Google Scholar]
- Gao Y, Smith GJ, Alberte RS. Nitrate reductase from the marine diatom Skeletonema costatum. Plant Physiol. 1993;103:1437–1445. doi: 10.1104/pp.103.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Smith GJ, Alberte RS. Temperature dependence of nitrate reductase activity and protein abundance in marine phytoplankton: ecological implications. J Phycol. 2000;36:304–313. [Google Scholar]
- Gröne T. Biogenic production of dimethylsulfide (DMS) and dimethylsulfoniopropionate (DMSP) in the marine epipelagic zone: a review. J Mar Syst. 1995;6:191–209. [Google Scholar]
- Gröne T, Kirst GO. The effect of nitrogen deficiency, methionine, and inhibitors of methionine metabolism on the DMSP contents of Tetraselmis subcordiformis (Stein) Mar Biol. 1992;112:497–503. [Google Scholar]
- Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of Marine Invertebrates Animals. New York: Plenum Press; 1975. pp. 29–60. [Google Scholar]
- Hanson AD, Gage DA. 3-Dimethylsulfoniopropionate biosynthesis and use by flowering plants. In: Keine RP, Visscher PT, Keller MD, Kirst GO, editors. Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds. New York: Plenum Press; 1996. pp. 75–86. [Google Scholar]
- Keller MD, Bellow WK, Guillard RRL. Dimethyl sulfide production in marine phytoplankton. In: Saltzmann EC, Cooper WJ, editors. Biogenic Sulfur in the Environment. Washington, DC: ACS Sympsium Series 393; 1989. pp. 167–182. [Google Scholar]
- Koprivova A, Suter M, den Camp RO, Brunold C, Kopriva S. Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol. 2000;122:737–746. doi: 10.1104/pp.122.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Touraine B. Comparison between demand-driven regulation of ATP sulfuphurylase activity and responses to oxidative stress. In: Cram WJ, De Kok LJ, Stulen I, Brunold C, Rennenberg H, editors. Sulphur Metabolism in Higher Plants. Leiden, The Netherlands: Backhuys Publishers; 1997. pp. 203–205. [Google Scholar]
- Lee S. Molecular analysis of sulfate assimilation in higher plants: effect of cysteine, sulfur and nitrogen nutrients; heavy metal stress; and genomic DNA cloning. PhD thesis. New Brunswick: Rutgers, The State University of New Jersey; 1998. [Google Scholar]
- Leustek T, Saito K. Sulfate transport and assimilation in plants. Plant Physiol. 1999;120:637–643. doi: 10.1104/pp.120.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin G, Turner SM, Liss PS. Sulfur: the plankton climate connection. J Phycol. 1992;28:590–597. [Google Scholar]
- Neuenschwander U, Suter M, Brunold C. Regulation of sulfate assimilation by light and O-acetyl-l-serine in Lemna minor L. Plant Physiol. 1990;97:253–258. doi: 10.1104/pp.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard GL, Emery WJ. Descriptive physical oceanography: an introduction. Oxford: Pergamon Press; 1988. pp. 12–18. [Google Scholar]
- Prosser IM, Schneider A, Hawkesford MJ, Clarkson DT. Changes in nutrient composition, metabolite, concentrations and enzyme activities in spinach in the early stage of S-deprivation. In: Cram WJ, De Kok LJ, Stulen I, Brunold C, Rennenberg H, editors. Sulphur Metabolism in Higher Plants. Leiden, The Netherlands: Backhuys Publishers; 1997. pp. 339–341. [Google Scholar]
- Reuveny Z, Dougall DK, Trinity PM. Regulatory coupling of nitrate and sulfate assimilation pathways in cultured tobacco cells. Proc Natl Acad Sci USA. 1980;77:6670–6672. doi: 10.1073/pnas.77.11.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DL, Smith GJ, Alberte RS. Isolation and characterization of glutamine synthetase from the marine diatom Skeletonema costatum. Plant Physiol. 1996;111:1169–1175. doi: 10.1104/pp.111.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotte C. Subcellular localization of sulfur assimilation enzymes in Arabidopsis thaliana (L.) Heynh. Diplomarbeit Thesis. Germany: Carl von Ossietzky Universität Oldenburg; 1998. [Google Scholar]
- Ruegsegger A, Brunold C. Effect of cadmium on γ-glutamylcysteine synthesis in maize seedlings. Plant Physiol. 1992;99:428–433. doi: 10.1104/pp.99.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer HJ, Greiner S, Rausch T, Haag-Derwer A. In seedlings of the heavy metal accumulator Brassica juncea Cu2+ differentially affects transcript amounts for γ-glutamylcysteine synthetase (γ-ECS) and metallothionein (MT2) FEBS Lett. 1997;404:216–220. doi: 10.1016/s0014-5793(97)00132-4. [DOI] [PubMed] [Google Scholar]
- Setya A, Murillo M, Leustek T. Sulfate reduction in higher plants: molecular evidence for a novel 5′-adenylylsulfate reductase. Proc Natl Acad Sci USA. 1996;93:13383–13388. doi: 10.1073/pnas.93.23.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey R, Gorham J, Pitman MG, Hanson AD, Gage D. Response of Melanthera biflora to salinity and water stress. J Exp Bot. 1993;44:1551–1560. [Google Scholar]
- Suter M, von Ballnoos P, Kopruva S, den Camp RO, Schaller J, Kuhlemeier C, Schurmann P, Brunold C. Adenosine 5′-phosphosulfate sulfotransferase and adenosine 5′-phosphosulfate reductase are identical enzymes. J Biol Chem. 2000;275:930–936. doi: 10.1074/jbc.275.2.930. [DOI] [PubMed] [Google Scholar]
- Von Heijne G, Steppuhn J, Herrmann RC. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Goffreda M, Leustek T. Characteristics of an Hsp70 homolog localized in higher plant chloroplasts similar to DnaK, the Hsp70 of prokaryotes. Plant Physiol. 1993;102:843–850. doi: 10.1104/pp.102.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RH. Analysis of dimethylsulfonium compounds in marine algae. J Mar Res. 1982;40:529–536. [Google Scholar]