Abstract
Mapping energy landscapes has proved to be a powerful tool for studying reaction mechanisms. Many complex biomolecular assembly processes, however, have remained challenging to access using this approach, including the aggregation of peptides and proteins into amyloid fibrils implicated in various disorders. Here we generalize the strategy used to probe energy landscapes in protein folding to determine the activation energies and entropies that characterise each of the molecular steps in the aggregation of the amyloid-β peptide (Aβ42), which is associated with Alzheimer’s disease. Our results reveal that interactions between monomeric Aβ and amyloid fibrils during fibril-dependent nucleation fundamentally reverse the thermodynamic signature of this process relative to primary nucleation, even though both processes generate aggregates from soluble peptides. By mapping the energetic and entropic contributions along the reactive trajectories, we show that the catalytic efficiency of Aβ42 fibril surfaces results from the enthalpic stabilisation of adsorbing peptides in conformations amenable to nucleation, driving a dramatic lowering of the activation energy barrier for nucleation.
The aberrant self-assembly of proteins and peptides into amyloid fibrils is implicated in the pathology of over 30 clinical disorders, including Alzheimer’s and Parkinson’s diseases1–3. Although it is a priority of both fundamental and translational research to characterise the molecular-level processes involved in amyloid formation4–7, this has proved challenging to achieve in practice7–9. Recently, however, advances in both experimental7, 10 and theoretical7, 11 methods have made it possible to acquire highly reproducible kinetic data in solution7, 10 under a wide range of experimental conditions, and to connect these measurements with the underlying microscopic processes through chemical kinetic rate laws5, 11. In the case of the 42-residue amyloid-β peptide (Aβ42), the aggregation of which is associated with Alzheimer’s disease, the key molecular steps in amyloid formation have been identified7 as the primary nucleation of new aggregates from monomers4, 6, 12, the addition of monomers to fibril ends resulting in their elongation13–15, and secondary nucleation of monomers involving the fibril surface7, 16.
This latter secondary nucleation process is a defining feature of Aβ42 aggregation7, 16, and several other amyloid systems8, 17, 18, and plays a key role in generating the high levels of toxicity associated with this peptide19–22, as well as potentially playing an important role in the spreading of the pathology in the brain23–25. The secondary nucleation process involves the surfaces of existing amyloid fibrils catalysing the formation of new pre-fibrillar aggregates from the soluble peptide26. Since the aggregates generated in this manner can grow into fibrils and subsequently catalyse additional nucleation events, the secondary nucleation step results in positive feedback and a chain reaction which underlies the autocatalytic nature of amyloid formation7, 9, 16. Furthermore, although the pre-fibrillar aggregates, also called oligomers, which are generated by both the primary and secondary nucleation processes are likely to be toxic to cells7, 19–22, once a small but critical concentration of Aβ42 fibrils is present, the concentration of toxic oligomers generated through the secondary nucleation process rapidly exceeds that generated by primary nucleation7.
Although the fundamental steps involved in Aβ amyloid formation have been identified, the molecular mechanisms underlying these nucleation and growth processes remain poorly understood. In the study of protein folding dynamics, the key to overcoming this same challenge has been to probe the energy landscape associated with the folding process, in practice by establishing the activation free energy, and the component enthalpic and entropic contributions from thermodynamic analysis of experimental data acquired across a range of temperatures. Here, we generalize this approach to describe complex protein self-assembly processes that involve multiple molecular events occurring simultaneously across heterogeneous size distributions of aggregates. We achieve this goal through advances in experimental and theoretical chemical kinetics that make it possible to decouple from measurements of the overall aggregation reaction the characteristic signatures of the individual steps in amyloid formation, each of which can then be analysed using the same approach used in protein folding dynamics.
Using this strategy, we determine the activation free energies of fibril elongation, primary nucleation and secondary nucleation in Aβ42 amyloid formation, and the enthalpic and entropic contributions to these barriers. The results uncover striking similarities between the interactions driving the elongation of fibrils and primary nucleation, but expose the secondary nucleation process as having an entirely different thermodyamic character from primary nucleation. This contrast in the thermodynamic signatures of the two nucleation processes occurs despite the fact that both processes generate new aggregates from monomers, indicating that the molecular nature of the fibril surface and its interactions with monomers plays a critical role in secondary nucleation. By probing directly the interactions between monomeric peptides and fibrils, we map the energy landscape that characterises secondary nucleation, revealing how the interactions between monomers and the surfaces of amyloid fibrils drive a dramatic reduction in the activation energy for nucleation.
Results
Global analysis of Aβ42 aggregation at different temperatures
We explored the kinetics of aggregation of the amyloid-β (M1–42) peptide, referred to here as Aβ42, across a range of concentrations and temperatures. In order to establish well-defined initial conditions for the kinetic experiments, and hence to obtain reproducible data, it is crucial to use highly purified sequence homogeneous recombinant Aβ42 together with repeated isolation of monomers through size-exclusion chromatography, and to control carefully the inertness of surfaces with which the peptide makes contact7, 10. Furthermore, to accurately decouple the temperature and concentration dependence of the individual reaction rates, the relative initial concentrations of monomeric peptide used at each temperature must be known to a very high degree of accuracy, since the reaction rates for this peptide are highly sensitive to the concentration of monomers in addition to the temperature7. To address this challenge, we arranged to carry out our experiments in parallel, using monomer solutions prepared simultaneously by dilution from a single master stock of Aβ42 and loaded in parallel into an array of fluorescence plate readers with the temperature of each device held constant at a different temperature.
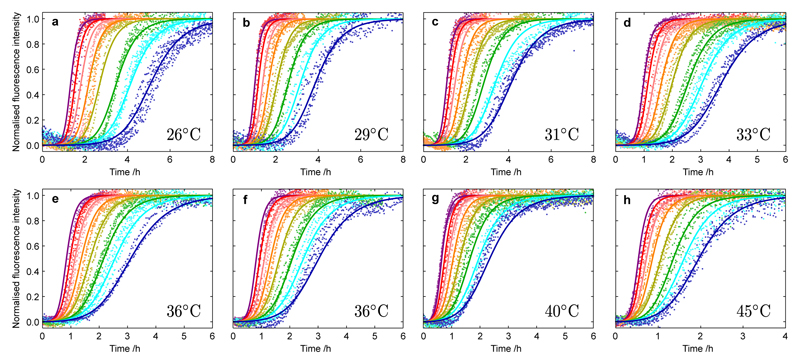
We first monitored the concentration-dependent kinetics of the aggregation of Aβ42 under quiescent condition for reactions beginning from purely monomeric peptide at seven different temperatures (Fig. 1). We carried out two sets of experiments, each involving four different temperatures, and included measurements at 36°C in both data sets to verify the high level of reproducibility across the two sets of data. At higher reaction temperatures, we observed an increase in the overall rate of formation of fibrils and characteristic changes in the time course of the reaction. At all temperatures and monomer concentrations we observe sigmoidal-like kinetic traces with a noticeable decrease in the sharpness of the transition as the temperature increases (Supplementary Fig. 2). To connect these macroscopic observations with the underlying microscopic processes, we applied an analytical approach, based on the integrated rate law that describes the aggregation of Aβ42, to determine the values of the rate constants controlling the reaction at each temperature5, 7, 27. Specifically, although there are distinct microscopic rate constants for fibril elongation (k+), primary nucleation (kn) and secondary nucleation (k2), the integrated rate law for Aβ aggregation7, 27 shows that the macroscopic reaction profiles, for reactions beginning from purely monomeric peptide, are controlled by just two combinations of these rate constants, k+kn and k+k2. Remarkably, at each temperature, we are able to fit essentially perfectly all of the data across multiple concentrations (Fig. 1a–g) using just these two kinetic parameters fixed globally across the entire data set, providing values for these combined rate parameters at each temperature (Fig. 3b,c).
Figure 1. Kinetics of Aβ42 aggregation from purely monomeric peptide at different temperatures and initial monomer concentrations.
Normalized experimental reaction profiles, monitored by ThT fluorescence, for Aβ42 aggregation from purely monomeric peptide for different initial concentrations of monomeric peptide and at different temperatures (a) 26°C, (b) 29°C, (c) 31°C, (d) 33°C, (e-f) 36°C, (g) 40°C and (h) 45°C in 20 mM sodium phosphate, 0.2 mM EDTA, 0.02% sodium azide, pH 8.0, with 6 µM ThT. The initial concentrations of monomers were 5.0 µM (purple), 4.5 µM (red), 4.0 µM (pink), 3.5 µM (orange), 3.0 µM (yellow), 2.5 µM (green), 2.2 µM (cyan) and 1.9 µM (blue). Note the different scales on the time axes; Supplementary Fig. 1 shows the data with the same scale for each panel. The data were recorded in two sets of four temperatures with measurements at 36°C included in both sets to act as a reference condition. The solid lines are global fits at each temperature using the analytical integrated rate law for Aβ42 aggregation. Two combinations of the microscopic rate constants, k+kn and k+k2, are used to globally fit the entire data set in each panel, in terms of rate constants for elongation (k+), primary nucleation (kn) and secondary nucleation (k2). The rate parameters determined at each temperature from the global fitting are plotted in Fig. 3a,b.
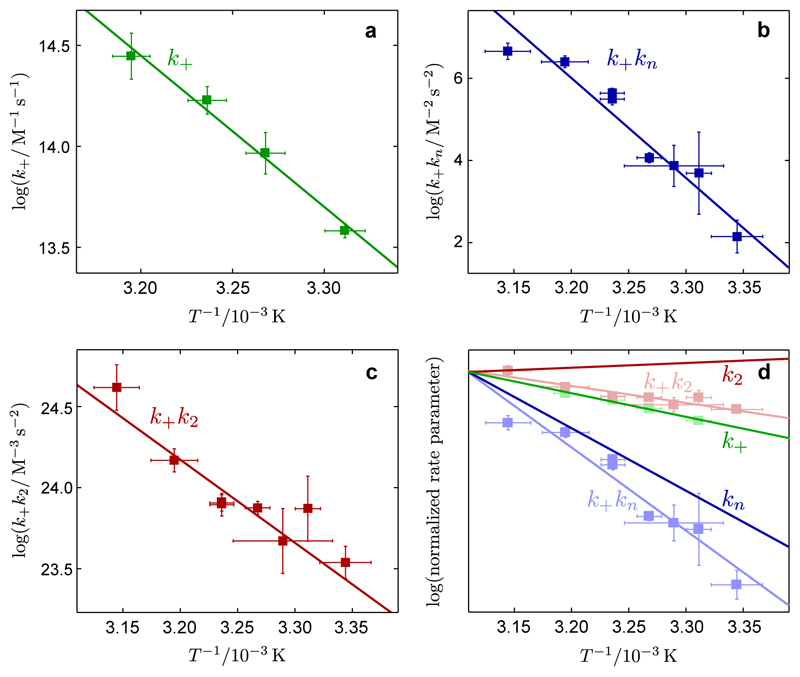
Figure 3. Arrhenius behavior of the microscopic rate constants for Aβ42 aggregation.
Arrhenius plots showing the temperature dependence of the rate parameters determined from the analytical fitting in Figs. 1, 2: (a) for k+, (b) for the combined rate parameter k+kn, (c) for the combined rate parameter k+k2, in terms of rate constants for elongation (k+), primary nucleation (kn) and secondary nucleation (k2). (d) shows how the data from (a-c), which is plotted as the faded data points and lines, are combined to give the temperature dependencies of the individual rate constants. Interestingly, while k+ and kn increase at higher temperatures, k2 has a weak temperature dependence with the opposite trend. The error bars for the rate parameters represent standard errors from the global fits shown in Figs. 1-2 and for the temperatures indicate the ranges of fluctuations recorded during each experiment.
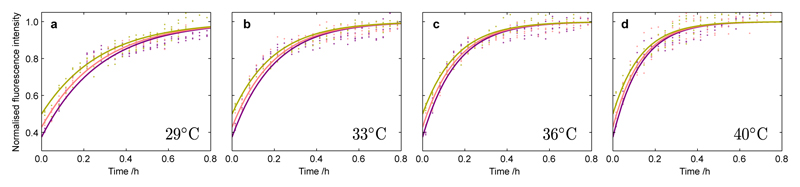
In order to decouple the values of the individual rate constants from the combined parameters k+kn and k+k2, it is necessary to determine one of the rate constants in isolation. To achieve this goal we measured reaction profiles from solutions containing not only monomeric peptide but also pre-formed fibrils (Fig. 2). At sufficiently high concentrations of pre-formed fibrils11 (here ca. 40% or greater of the total peptide concentration), the rate equations indicate that the elongation of fibrils is the dominant process and that the nucleation processes can be neglected, allowing determination of the elongation rate constant in isolation11. More precisely, the reaction profiles under these conditions depend primarily on the value k+/L(0)11, where L(0) is the mean length of the pre-formed fibrils present at the start of the reaction, which can be estimated independently using cryogenic transmission electron microscopy (Supplementary Fig. 3). In order to ensure that L(0) was consistent across all of our experiments, we used pre-formed fibrils generated at a single temperature (36°C). We also observed that the temperature at which pre-formed fibrils were generated did not significantly impact their ability to act as reactive fibril ends (Supplementary Fig. 3), and analyses by TEM and differential centrifugal sedimentation (Supplementary Fig. 4) were consistent with the prediction from the kinetic analysis7 that the mean length of fibrils, μ, generated in the aggregation reaction increases at higher temperatues, since Interestingly, by contrast to aggregation from purely monomeric peptide, the integrated rate law in the case of heavily pre-seeded reactions indicates that the normalized reaction profiles will be only weakly sensitive to the initial concentration of peptides11. The global fits to the data (Fig. 2a–d) in this case provide a direct estimate of the elongation rate constant k+ at each temperature. We find that k+ increases with temperature, which is reflected in our measurements (Fig. 2) by a steeper initial slope at higher temperatures.
Figure 2. Kinetics of pre-seeded Aβ42 aggregation at different temperatures and initial monomer concentrations.
Normalized experimental reaction profiles, monitored by ThT fluorescence, for Aβ42 aggregation in the presence of pre-formed fibrils for different initial concentrations of monomeric peptide and at different temperatures (a) 29°C, (b) 33°C, (c) 36°C and (d) 40°C. The initial concentrations of monomers were 5.0 µM (yellow), 4.0 µM (pink) and 3.0 µM (purple). The solid lines are global fits at each temperature using the analytical integrated rate law for Aβ42 aggregation. The data in each panel is fitted globally with a single parameter, k+/L(0), where L(0) is the average length of the pre-formed fibrils. The rate parameters determined at each temperature from the global fitting are plotted in Fig. 3c.
Arrhenius behaviour for combined rate constants
The global analyses in Figs. 1 and 2 provide values for the combined rate parameters k+kn and k+k2, and the elongation rate constant k+ at a variety of temperatures. These measurements together make it possible to decouple the values and temperature dependences of the individual rate constants k+, kn and k2, which describe transitions from well-defined initial to final states7, 11 on a free energy landscape. By analogy with protein folding28, 29, we extract the essential features of the free energy landscapes that are associated with the elongation of fibrils and the nucleation events by describing each of these processes as diffusive motions along one-dimensional free energy surfaces. Kramers theory28–31 provides the temperature dependence of the rate constants (k+, kn and k2) in the form of an Arrhenius equation which directly links a kinetic rate constant to the corresponding highest energy barrier along the potential landscape measured relative to the starting point: for a rate constant k, highest free energy barrier ΔG‡ϴ, and temperature T. It is important to note that, although the energy landscape may include multiple intermediate barriers, only the highest free energy barrier measured relative to the starting point enters the various rate constants and hence is directly probed by these experiments (see the Supplementary Note for a detailed discussion of the application of Kramers theory to biomolecular reactions). Where the rate laws contain products of the individual rate constants (i.e. k+kn or k+k2), the same temperature dependence is maintained with the component activation energies appearing summed in the exponent:
We first checked whether or not plots of the logarithms of the reaction rates against the inverse absolute temperatures were linear over the temperature range used (Fig. 3). Interestingly, we observed linearity of the Arrhenius plots in all three cases, indicating that well-defined activation energies characterize the distinct microscopic processes of fibril elongation, primary nucleation and secondary nucleation, and that any changes in heat capacity, ΔCp = ∂ΔH‡ϴ/∂T, between the starting states and transition states of the three processes were small over the temperature range studied. Since the free energy barrier ΔG‡ϴ associated with each process is related to the enthalpy ΔH‡ϴ and entropy ΔS‡ϴ of activation through the relationship ΔG‡ϴ = ΔH‡ϴ−TΔS‡ϴ, the enthalpies of the microscopic processes can be directly determined from the gradient of each plot according to ∂(log k)/∂(1/T) = −ΔH‡ϴ/R (Fig. 3a) or (Fig. 3b–c).
As a means to determine the free energy barrier ΔG‡ϴ, and hence also the entropy of activation using −TΔS‡ϴ = ΔG‡ϴ−ΔH‡ϴ, an independent estimate of the prefactor A is needed, although ΔG‡ϴ has only a logarithmic dependence on this value. The prefactor A can be thought of as the frequency of attempts at a given reaction, which is multiplied by to obtain the rate of successful (i.e. barrier-crossing) attempts at the reaction. We have in the past reported an estimate of the prefactor for amyloid fibril elongation, based on modeling the elongating monomer as a Gaussian chain that diffusively crosses the energy barrier and escapes from the metastable soluble state32. Given the fundamentally limited information about diffusion along the reaction coordinates for the nucleation processes, we partition all of the missing information into the free energy barrier in the rate equation. Other choices for the pre-factors are possible (for example, incorporating the process of monomer adsorption), and as such the free energy barrier is only meaningful if stated together with the corresponding pre-factor. The prefactor can be expressed as A ~ Dreff with a characteristic diffusion constant D ~ 10−9m2s−1 and the radius of the effective reaction volume reff ~ 3·10−11m in the case of the Aβ peptide32, 33. An analogous approach can be applied for the nucleation processes (see the Supplementary Note for the derivation), accounting for the different species involved in each process. Overall, the pre-factors for the nucleation processes are likely to involve characteric diffusion constants and effective length scales of the same order of magnitude as fibril elongation32, although the concentration dependence of the pre-factor must be scaled to account for the reaction order of each process.
Hence, the measurements of the molecular rate constants together with the Arrhenius plots provide values for the free energy, enthalpy and entropy of activation for fibril elongation (Fig. 3a), and the sums of these barriers for elongation and primary nucleation (Fig. 3b), and elongation and secondary nucleation (Fig. 3c). Moreover, using the temperature dependence of the elongation rate constant k+, we are able to resolve the temperature dependence of the nucleation rate constants kn and k2 from the combined rate parameters k+kn and k+k2 (Fig. 3d). We note that while the rates of both fibril elongation and primary nucleation increase strongly as temperature rises, the secondary nucleation rate is observed to have a much weaker dependence on temperature. These data explain the observed increase in the sharpness of the transition of the reaction profiles at lower temperatures (Supplementary Fig. 2), since the ratio of the rates of secondary to primary pathways increases7 at lower temperatures. We verified the continuation of this trend by monitoring aggregation reactions at temperatures as low as 10°C (Supplementary Fig. 2).
Activation energies of distinct molecular steps
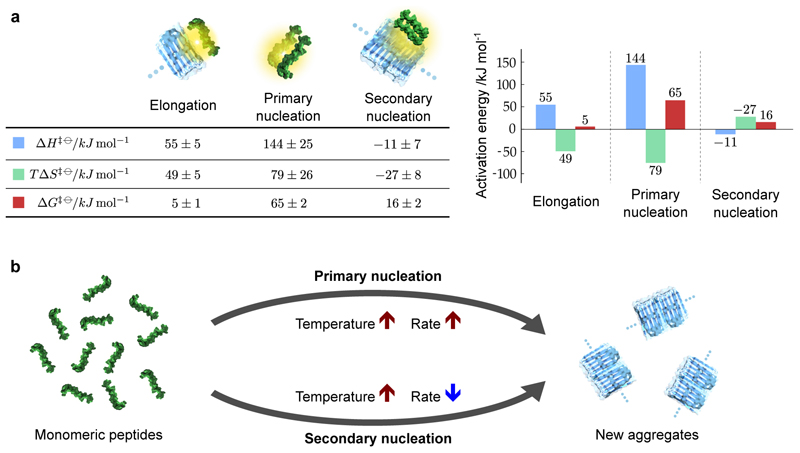
The values determined from Fig. 3 for the barriers in free energy, and the enthalpic and entropic contributions, reveal the individual energy barriers associated with the three key molecular steps in the aggregation of Aβ42 to form amyloid fibrils (Fig. 4). The results for elongation reveal a small free energy barrier with an enthalpic barrier and a favourable entropy of activation The enthalpic barrier for the addition of a monomeric peptide to a fibril, measured here for Aβ42 as is likely to represent the breakage of hydrogen bonds (and other interactions) associated with solvation and with the ensemble of internal structures of the Aβ peptide. By contrast, the favorable entropy of the transition state, (at 298K), can be attributed to the desolvation of hydrophobic segments of the incoming polypeptide and the fibril end. Remarkably, these values for the energy barriers associated with fibril elongation in solution are in agreement with previous two-state measurements of the elongation of surface-immobilized Aβ42 fibrils33, 35.
Figure 4. Activation energies of fibril elongation, primary nucleation and secondary nucleation in Aβ42 amyloid formation.
(a) The free energies of activation, and the enthalpic and entropic contributions, determined from our measurements. The activation energies for elongation and primary nucleation and primary nucleation consist of enthalpic barriers and favorable entropies of activation, whereas the enthalpic and entropic contributions to the free energy are reversed in sign for secondary nucleation. The values and standard errors shown were calculated by fitting the data shown in Fig. 3 with ΔH‡ϴ = −R ∂(log k)/∂(1/T) and ΔG‡ϴ = −RT log(k/A), which were combined to give TΔS‡ϴ = ΔH‡ϴ −ΔG‡ϴ. The entropic term is shown at T = 298K. The values are given per mole of reaction at a standard state of 1M. (b) Schematic showing the different temperature dependencies of the two nucleation processes that both generate aggregates from monomeric peptides. The structures used in the visual representations of each process are adapted from Refs. 45 and 46.
Moreover, the present strategy, which makes it possible to decouple and characterise the activation energies of complex processes in solution, also reveals the energy barriers for primary nucleation and secondary nucleation. The values determined for the barriers to primary nucleation (Fig. 4) follow the same overall trend to the elongation process indicating that fibril elongation and primary nucleation share fundamental qualitative similarities. The high activation energy for primary nucleation consists of an enthalpic barrier and a favorable entropy of activation These values have the same signs as the barriers for the elongation process, but have larger magnitudes, as would be expected for this slow nucleation process. Indeed, the free energy of activation determined here for primary nucleation is approximately 10 times larger than for fibril elongation. This increase occurs in spite of a more favourable entropy of activation and is driven by the much larger enthalpic barrier observed for primary nucleation relative to elongation.
The qualitative similarity in the data obtained for primary nucleation and fibril elongation implies that the hydrophobic effect also plays a dominant role in primary nucleation, and that the rate-limiting step in this process can be understood as an overall transition similar to fibril elongation, albeit requiring more extensive bond breaking. Specifically, the data show that the magnitude of the enthalpic barrier for primary nucleation is more than two times greater than the equivalent barrier for elongation, whereas the magnitude of the favourable entropic contributions are similar. The increase in the enthalpic barrier can be rationalised by noting that in forming a nucleus, the relevant hydrogen bonds associated with solvation must be broken in two monomers7, in contrast to the elongation process where the penalty corresponds to a single monomer being added to a partially desolvated existing fibril end 15, 32. Conversely, the rough similarity between the favourable entropies of activation measured for fibril elongation and primary nucleation is likely a consequence of the fact that in both processes an interface of similar size is desolvated. Interestingly, the data reveal a slightly less favourable entropy of activation for the elongation process, which may be explained by the fact that the terminal monomer at the fibril end in the elongation process is already partially desolvated relative to a free monomer. It is also interesting to note that the different magnitudes of the enthalpic barriers measured here for nucleation and elongation are in contrast to simple colloidal models of one-dimensional growth, where the energy barriers to nucleation and elongation are identical36. The increased enthalpic barrier for primary nucleation observed here thus reflects the large number of internal and solvent degrees of freedom associated with the peptide.
The results for secondary nucleation are in quantitative and qualitative contrast to those for fibril elongation and primary nucleation. The secondary nucleation process involves existing fibrils catalysing the nucleation of new aggregates from monomeric peptide. By definition, as a catalysed reaction, the energy barrier of secondary nucleation must be reduced relative to primary nucleation. This prediction is verified by the measurement of the activation energy of secondary nucleation as which represents a four-fold reduction in the energy barrier relative to primary nucleation, demonstrating the remarkably effective manner in which amyloid fibrils catalyse the nucleation of new aggregates from monomers. Strikingly, in addition to this dramatic lowering of the activation energy relative to primary nucleation, the enthalpic and entropic contributions to the free energy barrier are reversed in sign By contrast to the enthalpic barriers and favorable changes in entropy observed for primary nucleation and fibril elongation, the energy barrier for secondary nucleation is entirely entropic with a small negative enthalpic barrier In what follows we analyse in detail the energy landscape of secondary nucleation.
Mapping the energy landscape for secondary nucleation
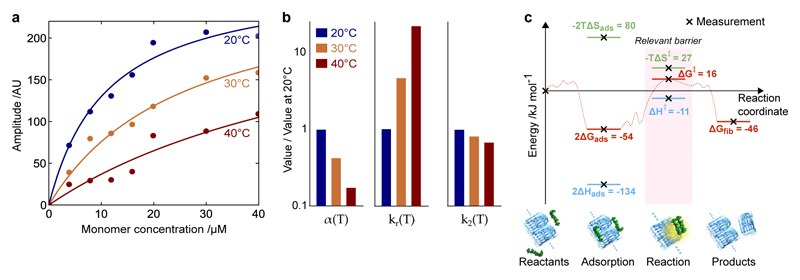
Elementary chemical reactions are typically associated with positive enthalpies of activation, while the data obtained here for secondary nucleation, which is a multimolecular reaction governed by diffusion dynamics, reveal a negative value. In order to gain additional mechanistic insight into the secondary nucleation process, we therefore mapped the reactive trajectory to construct a more complete picture of the energy landscape. To achieve this goal, we measured directly the interactions between monomeric peptides and the surfaces of fibrils as a function of concentration and temperature using surface plasmon resonance (SPR). The results (Fig. 5a) reveal strikingly that the surface coverage of monomers on the fibril surfaces decreases with increasing temperature.
Figure 5. Mapping the energy landscape for secondary nucleation.
(a) SPR measurements of the adsorption of monomers onto fibrils as a function of concentration and temperature. The data are fitted to the Langmuir isotherm to determine KD fitted at each temperature. (b) The variation in the surface coverage α(T) = 1/KD(T), the rate constant of the subsequent nucleation reaction kr(T), and the overall rate of secondary nucleation k2(T) = α(T)2kr(T). The corresponding values of KD are KD(20°C) = 11μM, KD(30°C) = 26μM, KD(40°C) = 64μM. (c) The energy landscape for secondary nucleation assuming a standard state of 1M, showing the trajectory from starting materials, through adsorption and reaction, to products. Since our experiments are typically at micromolar concentrations, the same landscape is shown assuming a standard state of 1μM in Supplementary Fig. 6, where ΔGads > 0. Note that, according to Kramers theory of diffusive reactions, the activation parameters determined from the temperature dependence of the rate constants correspond to the highest free energy barrier measured relative to the reactants.
In general, the rate of secondary nucleation can be written as r2(T, c) = kr(T)θ(T, c)n2, where θ(T, c) is the fibril surface coverage determined directly via SPR measurements39, c is the monomer concentration, n2 is the reaction order7, and kr(T) is the rate constant of the surface catalysed reaction. In the limit of low surface coverage, θ(T, c) = α(T)c, where α(T) is the inverse of the peptide-fibril equilibrium dissociation constant, α(T) = 1/KD(T) (see the Supplementary Note for a detailed derivation). In this case the rate of secondary nucleation reduces to r2(T, c) = k2(T)cn2, with k2(T) = kr(T)α(T)n2. The data reveal (Fig. 5b) that the surface occupancy of monomers on the fibril surface α(T) decreases with temperature, ∂α(T)/∂T < 0 (i.e. the adsorption is exothermic), while the rate of the subsequent surface catalysed reaction kr(T) increases with temperature, ∂kr/∂T > 0. At higher temperatures, the decrease in the concentration of monomers on the fibril surface α(T) is sufficient to outweigh the increase in the rate constant controlling the subsequent reaction kr(T), so that the overall rate constant of the secondary nucleation process k2(T) decreases as temperature increases (Fig. 5b).
Although the major role of the fibril, as the catalytic surface, is to provide a stabilisation of the transition state prior to nucleus formation, in this case driven by a favourable enthalpy of activation, it is interesting to note that the thermodynamic features uncovered here for surface-catalyzed secondary nucleation are reminiscent to those of classical heterogeneous catalysis of a reaction at a solid surface40.
This data for the adsorption of monomers onto the fibril surface also directly provides access to the change with adsorption in free energy through Furthermore, the corresponding change in enthalpy is given by the Clausius Clapeyron equation and the change in entropy may be calculated since (see the Supplementary Note for detailed calculations). By combining these direct measurements of the adsorption process with the results from bulk solution, we are able to map out the energy landscape for secondary nucleation (Fig. 5c) that characterises the transition from the initial reactants, through the adsorbed monomers on the fibril and the subsequent nucleation reaction, to the final products. Note that the free energy of the final products is known via equilibrium measurements of the critical concentration ccrit10, which gives at protein concentration cϴ = 1 M. Since our experiments are typically at micromolar concentrations, which are significantly closer to physiologically relevant conditions than a standard state of 1M, the energy landscape for secondary nucleation can also be calculated assuming a standard state of 1μM (Supplementary Fig. 6).
It is interesting to note from the energy landscape that the difference in enthalpy between the adsorbed state and the highest point on the energy landscape is given by which is similar to the enthalpic barrier determined for primary nucleation, We can speculate, therefore, that the secondary nucleation reaction that occurs after the adsoroption of monomers onto the fibril surface may share similar characteristics with primary nucleation in solution.
Discussion
The results presented here are the first determination of the thermodynamic signatures of the key microscopic steps in Aβ amyloid formation. The activation energies determined for primary nucleation and fibril elongation reveal that these processes are controlled by enthalpic barriers and favourable entropies of activation (ΔH‡ϴ > 0, ΔS‡ϴ > 0). By contrast, the results for secondary nucleation reveal remarkably that this process has a fundamentally different signature, with the relevant barrier being entropic rather than enthalpic in nature (ΔH‡ϴ < 0, ΔS‡ϴ < 0).
Although the primary and secondary nucleation processes both generate new aggregates from monomeric peptides, the results here show that these two processes have opposite temperature dependencies (Fig. 4b). These observations reveal that the interactions between monomeric Aβ and amyloid fibrils are not only able to change the rate constant characterising nucleation, but also to fundamentally reverse the thermodynamic signature of this process relative to primary nucleation. Moreover, our results show that Aβ42 monomers have an intermediate affinity for the surface of amyloid fibrils (KD ≈ 10μM), and indicate a mechanism involving the rapid adsorption (and desorption) of monomers prior to a rate-limiting reaction step on the fibril surface, a process analogous to classical heterogeneous catalysis. Indeed, the Sabatier principle states in qualitative terms that for efficient catalysis, the affinity between the catalyst and substrate should neither be too strong nor too weak, but rather intermediate to allow the substrate to bind and the products to dissociate. This classical concept was recently evaluated using DFT calculations leading to predictors of catalytic efficiency based on the interactions between surfaces and reactants41. In thermodynamic terms, the catalytic efficiency of Aβ42 fibril surfaces can therefore be explained through enthalpic stabilisation of conformations amenable to nucleation, resulting in a significant lowering of the activation energy barrier. Indeed, the surface-binding of monomers is likely to induce a significant structural change in the peptide, consistent with the very high specificity that has been observed for the secondary nucleation reaction, as demonstrated by the fact that fibrils constructed from other proteins and peptides, including Aβ40, are not able to efficiently catalyse nucleation of Aβ42 monomers7, 42.
Through mapping the energy landscapes that characterise the distinct molecular steps in the aggregation of Aβ42 into amyloid fibrils, the present study provides foundational insights into the molecular mechanism of secondary nucleation, which has been identified as a critical step in generating the high levels of toxicity associated with Aβ42 amyloid formation. Our results suggest that strategies that seek to interfere with the interactions between monomers and fibril surfaces, which drive the unique thermodynamic signature of secondary nucleation, may therefore be effective approaches to reducing the toxicity of Aβ aggregation. Indeed, we have recently shown that nature has designed molecular chaperones capable of binding to Aβ42 fibrils in competition with monomeric peptides, resulting in a lower population of surface bound Aβ42 monomers, inhibition of the overall rate of secondary nucleation, and the abolition of toxicity in living brain tissue9. Furthermore, the secondary nucleation of new aggregates on fibril surfaces is increasingly recognised as a central feature in several amyloid disorders beyond Alzheimer’s disease, ranging from Parkinson’s disease to type-II diabetes. It is likely that interactions between monomeric peptides and fibril surfaces will become a common target in the development of therapeutics which seek to ablate the toxicity associated with amyloid formation. Interestingly, the present work indicates that stabilisation of surface-bound monomers on the fibril surface could be a complementary approach.
More generally, the approach developed in this study extends the methods commonly used to probe the energy landscapes of two-state systems in protein folding to investigations of complex protein self-assembly, which involve a multitude of processes occurring simultaneously across heterogeneous populations. Probing energy landscapes has for decades proved a powerful tool to study reaction mechanisms. With our increasing ability to decouple and probe the rates of the individual processes driving self-assembly reactions, we expect this approach to have direct applicability across a diverse range of complex biochemical phenomena for which mechanistic information is currently challenging to achieve, including the more than thirty disease-related amyloid systems and a wide range of functional bio-molecular assembly processes involved in cellular structure and molecular trafficking.
Methods
Materials
We expressed the Aβ(M1-42) peptide (MDAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA) in Escherichia coli and purified the peptide as described previously44. Aliquots of purified Aβ42 were thawed and dissolved in 6 M GuHCl, and the monomer was isolated by two rounds of gel filtration on a Superdex 75 column in 20 mM sodium phosphate buffer, pH 8, with 200 μM EDTA and 0.02% NaN3. The centre of the monomer peak was collected on ice and lyophilized. The sample was again dissolved in 6 M GuHCl, and the monomer isolated by gel filtration on a Superdex 75 column in 20 mM sodium phosphate buffer, pH 8, with 200 μM EDTA and 0.02% NaN3 was typically found to have a concentration (determined by quantitative amino acid analysis purchased from BMC Uppsala) of 5-12 μM. The gel filtration step removes traces of pre-existing aggregates and exchanges the buffer to the one used in the fibril formation experiments.
Kinetic assays
The monomer was supplemented with 6 μM ThioflavinT (ThT) and was used to prepare a series of samples of Aβ42 with concentrations between 0.5 and 6 μM. All samples were prepared in low-bind Eppendorf tubes (Axygen, California, USA) on ice. Samples were pipetted into multiple wells of a 96 well half-area plate of black polystyrene with a clear bottom and PEG coating (Corning 3881, Massachusetts, USA), 100 μL per well. Assays were initiated by placing the 96-well plate at the designated temperature in a plate reader (Fluostar Omega, Fluostar Optima or Fluostar Galaxy, BMGLabtech, Offenburg, Germany). A series of control experiments7, 10 demonstrated that under the conditions used, the fluorescence from ThT is linearly related to the Aβ42 aggregate mass concentration.
Pre-formed fibrils
Kinetic experiments were set up as above for multiple samples of Aβ42 at 36°C. The fluorescence of added ThT was monitored for 1.5 h to verify the formation of fibrils. The samples were then collected from the wells into low-bind Eppendorf tubes (Axygen, California, USA) and sonicated for 2 min in a sonicator bath at room temperature to disrupt any fibril clusters. The previously established rate constants7 for elongation and secondary nucleation in Aβ42 aggregation show that at the concentrations of pre-formed fibrils applied here, the accelerating effect on the reaction is due primarily to elongation processes induced by the added reactive fibril ends.
Surface plasmon resonance studies
The SPR experiments were performed with a Biacore 3000 instrument (GE Healthcare), using C3 sensors, essentially as described previously39. The flow rate was 10 μl/min throughout using 20 mM phosphate, 0.2 mM EDTA, pH 8.0 with 0.005% Tween20 as the flow buffer. The central fraction of Aβ42 peptide monomer from gel filtration in 20 mM phosphate, 0.2 mM EDTA, pH 8.0, was collected and stored on ice for up to 8h until use. A solution of 10 μM of monomeric peptide was incubated for 2 h at 37°C to form fibrils, followed by tip sonication to produce short (ca. 50 nm) fibrils for immobilization. For the attachment of the fibrils to the surface of the sensor, the fibrils were diluted 10-fold into 10 mM sodium acetate buffer at pH 3.0. The carboxylic acid groups on the CM3 sensor surface were activated with a mixture of EDC and NHS to enable standard amine coupling chemistry. The injection of fibrils led to an increase of ca. 3000 RU. The subsequent incubation with monomer in 20 mM phosphate, 0.2 mM EDTA, pH 8.0 added ca. 9000 RU. Injection of monomers was followed by buffer flow. The linear parts of the dissociation curves were fitted to a linear function which was extrapolated to the beginning of the dissociation phase. The difference amplitude relative to baseline was taken to correspond to the monomer binding to the fibril surface. The amplitudes, A, were plotted as a function of monomer concentration, and fitted to A = A(∞)Kc/(1 + Kc), with K being the binding constant and c the monomer concentration.
Kinetic rate laws
Following our previous analysis7, 27, the generation of fibril mass, M, when both primary and secondary nucleation events occur is described by the integrated rate law:
| (1) |
where two particular combinations of the rate constants for primary nucleation (kn), elongation (k+) and fibril-catalysed secondary nucleation (k2) define much of the macroscopic behaviour; these parameters are related to the rate of formation of new aggregates through primary pathways and through secondary pathways where k2 = k− when n2 = 0. Indeed, Eq. 1 depends on the rate constants through these two parameters, λ and κ, alone since B± = (k∞ ± k̃∞)/(2κ), C± = ±λ2/(2κ2), and The initial concentration of soluble monomers is denoted m(0) and the exponents describing the dependencies of the primary and secondary pathways on the monomer concentration are given as nc and n2 respectively.
Kramers rate theory for multi-well potentials
Kramers rate theory for a double-well potential states that the rate at which the system diffuses from one minimum x1 to the other x2 is given by a prefactor A (which depends on the curvatures of the potential landscape at x1 and at the barrier x*) multiplied by the negative exponential of the highest free energy point measured relative to the starting point. When the potential landscape has greater than two local minima, the escape rate is still described by an equation of the same form provided that the local minima satisfy certain ordering conditions; in particular, for a multi-well potential landscape having local minima at x1, x2, … , xn there exists an ordering ≺ of these local minima (obtained by ordering the minima from deepest to shallowest) so that the expectation value for the average escape time from xk to the set ℳk = {xj|xj ≺ xk} satisfies Kramers formula. The Supplementary Note provides a detailed analysis of the temperature dependence of the individual microscopic steps involved in secondary nucleation.
Supplementary Material
Supplementary information accompanies this paper.
Acknowledgements
We thank Bengt Jönsson and Ingemar André for helpful discussions. We acknowledge financial support from the Schiff Foundation (SIAC), St John’s College, Cambridge (SIAC, TCTM), the Royal Physiographic Society (RC), the research school FLÄK of Lund University (SL, RC), the Swedish Research Council (SL) and its Linneaus Centre Organizing Molecular Matter (SL), the Crafoord Foundation (SL), Alzheimerfonden (SL), European Research Council (SL), nmc@lund (SL), Magdalene Collge, Cambridge (AKB), the Leverhulme Trust (AKB) and the Wellcome Trust (CMD, TPJK).
Footnotes
Author contributions
S. I. A. C., R. C., T. P. J. K. and S. L. designed the study. R. C., M. T. and S. L. performed the experiments. S. I. A. C., A. K. B., T. C. T. M. and T. P. J. K. analyzed the data. All authors discussed the results and contributed to writing the manuscript.
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information, and are available from the corresponding authors upon reasonable request.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Sipe JD, et al. Amyloid fibril protein nomenclature: 2012 recommendations from the nomenclature committee of the international society of amyloidosis. Amyloid. 2012;19:167–170. doi: 10.3109/13506129.2012.734345. [DOI] [PubMed] [Google Scholar]
- 4.Serio TR, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 5.Knowles TPJ, et al. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Culyba EK, Powers ET, Kelly JW. Amyloid-beta forms fibrils by nucleated conformational conversion of oligomers. Nat Chem Biol. 2011;7:602–609. doi: 10.1038/nchembio.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen SIA, et al. Proliferation of amyloid-beta42 aggregates occurs through a secondary nucleation mechanism. Proc Natl Acad Sci U S A. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meisl G, et al. Differences in nucleation behavior underlie the contrasting aggregation kinetics of the abeta40 and abeta42 peptides. Proc Natl Acad Sci U S A. 2014;111:9384–9389. doi: 10.1073/pnas.1401564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen SIA, et al. A molecular chaperone breaks the catalytic cycle that generates toxic abet oligomers. Nat Struct Mol Biol. 2015;22:207–213. doi: 10.1038/nsmb.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellstrand E, Boland B, Walsh DM, Linse S. Amyloid β-protein aggregation produces highly reproducible kinetic data and occurs by a two-phase process. ACS Chemical Neuroscience. 2010;1:13–18. doi: 10.1021/cn900015v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen SIA, et al. Nucleated polymerization with secondary pathways. i. time evolution of the principal moments. J Chem Phys. 2011;135:065105. doi: 10.1063/1.3608916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kar K, Jayaraman M, Sahoo B, Kodali R, Wetzel R. Critical nucleus size for disease-related polyglutamine aggregation is repeat-length dependent. Nat Struct Mol Biol. 2011;18:328–336. doi: 10.1038/nsmb.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oosawa F, Asakura S. Thermodynamics of the Polymerization of Protein. Academic Press; 1975: [Google Scholar]
- 14.Jarrett JT, Lansbury PT. Seeding "one-dimensional crystallization" of amyloid: a pathogenic mechanism in alzheimer’s disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 15.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong JS, Ansaloni A, Mezzenga R, Lashuel HA, Dietler G. Novel mechanistic insight into the molecular basis of amyloid polymorphism and secondary nucleation during amyloid formation. J Mol Biol. 2013;425:1765–1781. doi: 10.1016/j.jmb.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Ruschak AM, Miranker AD. Fiber-dependent amyloid formation as catalysis of an existing reaction pathway. Proc Natl Acad Sci U S A. 2007;104:12341–12346. doi: 10.1073/pnas.0703306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buell AK, et al. Solution conditions determine the relative importance of nucleation and growth processes in alpha-synuclein aggregation. Proc Natl Acad Sci U S A. 2014;111:7671–7676. doi: 10.1073/pnas.1315346111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 20.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 21.Walsh DM, et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 22.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–11. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 23.Knowles TPJ, et al. Observation of spatial propagation of amyloid assembly from single nuclei. Proc Natl Acad Sci U S A. 2011;108:14746–14751. doi: 10.1073/pnas.1105555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen SIA, et al. Spatial propagation of protein polymerization. Phys Rev Lett. 2014;112:098101. doi: 10.1103/PhysRevLett.112.098101. [DOI] [PubMed] [Google Scholar]
- 25.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501:45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrone F. Analysis of protein aggregation kinetics. Methods Enzymol. 1999;309:256–274. doi: 10.1016/s0076-6879(99)09019-9. [DOI] [PubMed] [Google Scholar]
- 27.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. Nucleated polymerization with secondary pathways. ii. determination of self-consistent solutions to growth processes described by non-linear master equations. J Chem Phys. 2011;135:065106. doi: 10.1063/1.3608917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: the energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 29.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 30.Kramers HA. Brownian motion in a field of forceand the diffusion model of chemical reactions. Physica. 1940;7:284. [Google Scholar]
- 31.Zwanzig R. Two-state models of protein folding kinetics. Proc Natl Acad Sci U S A. 1997;94:148–150. doi: 10.1073/pnas.94.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buell AK, et al. Frequency factors in a landscape model of filamentous protein aggregation. Phys Rev Lett. 2010;104:228101. doi: 10.1103/PhysRevLett.104.228101. [DOI] [PubMed] [Google Scholar]
- 33.Buell AK, et al. Detailed analysis of the energy barriers for amyloid fibril growth. Angew Chem Int Ed Engl. 2012;51:5247–5251. doi: 10.1002/anie.201108040. [DOI] [PubMed] [Google Scholar]
- 34.Zwanzig R. Diffusion in a rough potential. Proc Natl Acad Sci U S A. 1988;85:2029–2030. doi: 10.1073/pnas.85.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knowles TPJ, et al. Kinetics and thermodynamics of amyloid formation from direct measurements of fluctuations in fibril mass. Proc Natl Acad Sci U S A. 2007;104:10016–10021. doi: 10.1073/pnas.0610659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashchiev D, Auer S. Nucleation of amyloid fibrils. J Chem Phys. 2010;132:215101. doi: 10.1063/1.3447891. [DOI] [PubMed] [Google Scholar]
- 37.Mozurkewich M, Benson SW. Negative activation energies and curved arrhenius plots. 1. theory of reactions over potential wells. The Journal of Physical Chemistry. 1984;88:6429–6435. doi: 10.1021/j150669a073. [DOI] [Google Scholar]
- 38.Oliveberg M, Tan YJ, Fersht AR. Negative activation enthalpies in the kinetics of protein folding. Proc Natl Acad Sci U S A. 1995;92:8926–8929. doi: 10.1073/pnas.92.19.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saric A, et al. Physical determinants of the self-replication of protein fibrils. Nat Physics. 2016;12:874–880. doi: 10.1038/nphys3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roduner E. Understanding catalysis. Chem Soc Rev. 2014;43:8226–8239. doi: 10.1039/c4cs00210e. [DOI] [PubMed] [Google Scholar]
- 41.Medford A, et al. From the Sabatier principle to a predictive theory of transition-metal heterogeneous catalysis. Journal of Catalysis. 2015;328:36–42. [Google Scholar]
- 42.Cukalevski R, et al. The abeta40 and abeta42 peptides self-assemble into separate homomolecular fibrils in binary mixtures but cross-react during primary nucleation. Chem Sci. 2015;6:4215–4233. doi: 10.1039/c4sc02517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anwar J, Khan S, Lindfors L. Secondary crystal nucleation: Nuclei breeding factory uncovered. Angew Chem Int Ed Engl. 2015;54:14681–14684. doi: 10.1002/anie.201501216. [DOI] [PubMed] [Google Scholar]
- 44.Walsh DM, et al. A facile method for expression and purification of the Alzheimer’s disease-associated amyloid beta-peptide. FEBS J. 2009;276:1266–1281. doi: 10.1111/j.1742-4658.2008.06862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lührs T, et al. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crescenzi O, et al. Solution structure of the alzheimer amyloid beta-peptide (1-42) in an apolar microenvironment. similarity with a virus fusion domain. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.