Abstract
Multi-system metabolic disorders caused by defects in oxidative phosphorylation (OXPHOS) are progressive and severely crippling, often lethal, conditions. Inborn errors of OXPHOS function are termed Primary Mitochondrial Disorders (PMDs), and the use of nutritional interventions is routine in PMD supportive management. However, detailed mechanistic understanding and evidence for efficacy and safety is limited. Preclinical cellular and animal models systems are important tools to investigate PMD metabolic mechanisms and therapeutic strategies. This review assesses the mechanistic rationale and experimental evidence for nutritional interventions commonly used in PMDs, including micronutrients, metabolic agents, signaling modifiers, and dietary patterns, while highlighting important gaps in knowledge gaps and impediments for randomized controlled trials. The state-of-the-science for cellular and animal model systems that recapitulate mutations and clinical manifestations of specific PMDs are evaluated for their potential in determining pathological mechanisms, elucidating therapeutic health outcomes, and investigating the value of nutritional interventions for multiple mitochondrial disease conditions.
Keywords: primary mitochondrial OXPHOS disorders, nutritional interventions, electron transport chain, metabolism
Introduction
The classical view of mitochondria as simply cellular “powerhouses” has given way to an appreciation of a dynamic organelle network at the nexus of energy production, metabolic signaling, and cellular homeostasis. Multi-system metabolic disorders are caused by discrete genetic mutations that disrupt oxidative phosphorylation (OXPHOS). These Primary Mitochondrial Disorders (PMDs), sometimes referred to as mitochondrial encephalopathies or myopathies, are progressive, severely crippling, and often lethal diseases that affect 1 in 4,300 people (1). First identified in the late 20th century, research on PMD mechanisms and treatments has proven challenging due to their rarity, difficulties diagnoses, and genetic and phenotypic heterogeneity (2). For example, mutations in either nuclear genes or mitochondrial (mt)DNA encoding distinct proteins involved in OXPHOS can result in similar clinical manifestations. Conversely, the same genetic mutation can result in different clinical phenotypes, where the severity of PMDs may increase with the percentage of mutant mtDNA (heteroplasmy) in an age and cell type specific manner. Consequently, preclinical identification of therapeutic targets and clinical evaluation of interventions for PMD patients represent significant challenges, and no cures exist.
The use of nutritional interventions, including dietary supplements, in the supportive management of PMDs is an established approach intended to alleviate symptoms and/or delay disease progression (3, 4). However, the efficacy of supplements for specific biological endpoints or health outcomes is usually not known (4). Marketed dosages of nutritional supplements vary greatly, and the bioavailability and metabolism of various formulations are not always known. Nutritional interventions for PMD are empirically individualized to each patient, with the literature mostly comprising observational reports of non-standardized dosing regimens and limited evidence of broad efficacy (5). The ubiquity of supplements can present challenges for compliance in use of placebo controls and randomization in clinical trials for PMDs.
An understanding of the mechanistic rationale for using nutritional interventions in PMD patients is critical for investigating efficacy and safety. While some nutritional interventions for PMDs such as nicotinic acid and L-carnitine can be prescribed in FDA-approved drug formulations, most cannot, prompting patients and caregivers to seek out dietary supplement versions for reasons related to personal preference, cost, or convenience (4, 6). To this end, the development and use of robust preclinical model systems to investigate the pathogenic mechanisms of PMDs and strategically study the impact of nutritional interventions is tantamount (4). Herein, we survey nutritional interventions utilized in clinical settings for PMDs and describe key cellular and animal model systems of OXPHOS dysfunction that can be used to further elucidate the mechanisms and health outcomes of PMDs, and point to important areas in need of additional study.
Clinical Investigations of Nutritional Interventions
Nutritional interventions can be categorized as micronutrients, metabolic-modifying agents, or signaling-pathway modulators. Those currently used for PMDs, and their mechanistic rationale, are described (Figure 1; Supplemental Table S1).
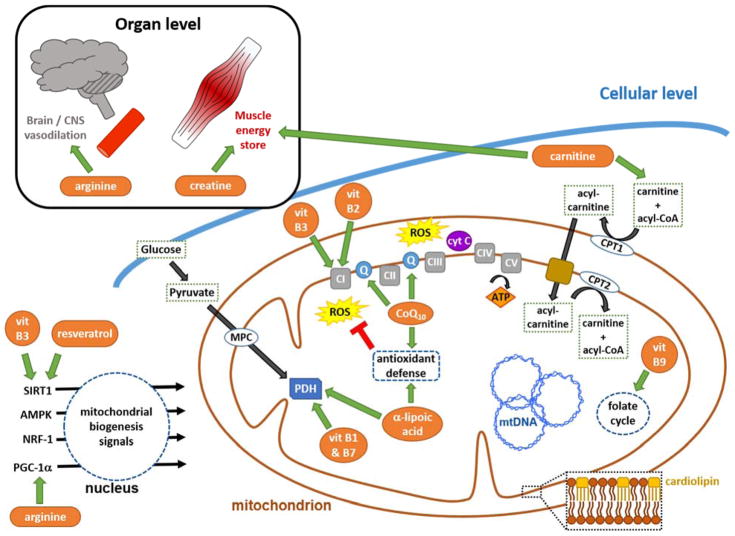
Figure 1. Mechanisms of nutritional interventions in PMDs.
At the organ level, arginine and creatine are used to improve vasodilation and energy storage. At the cellular level nutrients and metabolic modifiers and signaling activators are employed to support OXPHOS (vitamins B3 and B2, CoQ10) and metabolic pathways (vitamin B1, B7, and B9, L-carnitine, and α-lipoic acid), enhance antioxidant defenses (CoQ10 and α-lipoic acid), and promote mitochondrial biogenesis (vitamin B3, resveratrol, and arginine). Cardiolipin, dispersed throughout the mitochondrial inner membrane, is essential for optimal function of multiple metabolic enzymes. Abbreviations: CI-CV, OXPHOS complexes I-V; Q, ubiquinone; CoQ10, coenzyme Q10; CPT, carnitine palmitoyl transferase; cyt c, cytochrome c; MPC, mitochondrial pyruvate carrier; mtDNA, mitochondrial DNA; PDH, pyruvate dehydrogenase; ROS, reactive oxygen species, (superoxide, hydrogen peroxide and hydroxyl radical; vit, vitamin.
Micronutrients
Thiamine and biotin
Thiamine (vitamin B1) is a cofactor of α-ketoacid dehydrogenase complexes, which include pyruvate dehydrogenase complex (PDHc), α-ketoglutarate dehydrogenase, and branched-chain α-keto acid dehydrogenase; as well for transketolase, an enzyme of the pentose phosphate pathway, and 2-hydroxyacyl CoA lyase. While the thiamine requirement for healthy individuals is a daily intake of 1–2 mg, there is no evidence of adverse effects in patients receiving higher doses (7).
Thiamine is transported into cells by thiamine transporter 1 (THTR1) and THTR2. Mutations in the SLC19A3 gene (encoding THTR2) results in biotin-thiamine responsive basal ganglia disease and is recognized as a treatable cause of early-onset progressive neurodegenerative Leigh syndrome. A combination of biotin (2–10 mg/day) and thiamine (100–400 mg/day) has been found to be effective in THTR2 deficiency (8, 9). In current clinical practice, empiric supplementation of thiamine and biotin in the form of a B vitamin complex is commonly used (10), based on the premise it will enhance PDHc and/or OXPHOS flux, and thiamine-responsive PDHc deficiency has been reported (7). However, these thiamine and biotin combinations have not been studied in randomized control trials (RCTs), or in mitochondrial disease models.
Riboflavin
Riboflavin (vitamin B2) is the precursor of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN), which serve as essential cofactors for numerous mitochondrial dehydrogenases (Figure 1) (11). Riboflavin (50–400 mg daily) is a common intervention intended to enhance electron transport chain (ETC) efficiency in PMDs. However, these doses are not based on evidence from RCTs (12). Beneficial responses to riboflavin supplementation have been reported in patients with primary genetic defects of riboflavin transport (13, 14). Multiple acyl CoA dehydrogenase deficiency (MADD) is an autosomal recessive disorder of fatty acid and amino acid metabolism involving a defect of electron transport from FAD-containing CoA dehydrogenases to CoQ10 in the mitochondrial ETC, caused by mutations in flavoprotein dehydrogenase or flavoprotein genes ETFDH, ETFA or ETFB (15). Oral riboflavin supplementation (100–400 mg daily) often yields muscle strength improvement in patients with MADD. Mutations in ACAD9 (encoding acyl-CoA dehydrogenase 9) have been reported in some patients with mitochondrial ETC complex I deficiency and myopathy, and riboflavin supplementation (150–300 mg daily) yields improved exercise intolerance (16, 17). Neurological exam and muscle strength improvements, and muscle Complex I and skin fibroblast Complex II activity responses to riboflavin have been reported in some small, open label studies of patients with ETC complex I and II deficiencies (18–20). However, open-label studies in patients with a variety of PMDs (21, 22) did not show a significant response to riboflavin treatment in combination with other vitamins, and riboflavin supplementation in PMDs has not been reported in RCTs (23).
Nicotinic acid and nicotinamide riboside
Niacin (vitamin B3) refers to both nicotinic acid (NA) and its amide form nicotinamide (NAM), which are precursors to the essential cofactor nicotinamide adenine dinucleotide (NAD+) and NAD+ phosphate (NADP+) (24). In addition to crucial metabolic roles, these cofactors also play a central role in sirtuin activity and peroxisome proliferator activator receptor (PPAR) activation (25). NA is an FDA-approved drug for hypertriglyceridemia, while NAM is available as a dietary supplement in the United States.
Mitochondrial Complex I converts NADH to NAD+, and an increased cytosolic NADH:NAD+ ratio can shift the lactate dehydrogenase equilibrium and cause the lactic acidosis commonly seen in PMDs (26). Exercise and fasting are known to decrease the NADH/NAD+ ratio, resulting in activation of SIRT1 that induces a metabolic-fasting response and boosts mitochondrial biogenesis, fatty acid oxidation, and ATP production (27). However, disrupted NAD+ metabolism remains a relatively under-appreciated pathogenic factor in PMDs. NA and NR supplementation has not been reported in RCTs (28), though several studies in cellular and animal model systems indicate that niacin and its derivatives warrant further preclinical evaluation (see Sidebar).
Sidebar. Niacin and nicotinamide riboside studies in PMD model systems.
Model system studies have investigated the role of NADH/NAD+ ratio in PMDs. Leigh syndrome patient fibroblasts harboring mutations in ND4 and ND6 Complex I subunit genes revealed an increased relative NADH:NAD+ ratio but decreased absolute NADH and NAD+ concentrations. NA treatment normalized nutrient-sensing signaling network components (mTORC1 and AMPK), restored NADH/NAD+ redox balance, and improved cellular respiratory capacity (240). NR supplementation in patient fibroblasts harboring a NDUFS1 mutation increased cellular NAD+ content (241). C. elegans research suggests NAD+ deficiency contributors to global disruption of the nutrient-sensing network; NA treatment improved survival, decreased mitochondrial oxidant burden, normalized branched chain amino acid concentrations, and restored NADH (but not NAD+) levels (129). Complex I-deficient patients have elevated NADPH (242), and Ndufs4−/− mice revealed NADP+ levels also increase such that NADP+/NADPH ratio is unchanged (243). NR supplementation in Ndufs4−/− mice partially normalized the NADH/NAD+ ratio (244). NR induced mitochondrial biogenesis in Deletor mouse skeletal muscle and brown adipose tissue, preserved mitochondrial ultrastructure, and prevented mtDNA deletion accumulation (245). Collectively, these studies suggest that NADH deficiency can contribute to the phenotype of some PMDs, and that niacin supplementation might improve overall cellular function.
Folinic acid
Folinic acid (vitamin B9; 5-formyltetrahydrofolate) is a CNS-penetrant form of folic acid that functions as a B vitamin cofactor necessary for cellular one-carbon-transfer reactions (Figure 1). Cerebral folate deficiency (CFD) is characterized by 5-methyltetrahydrofolate (5-MTHF) deficiency in the cerebral spinal fluid (CSF) with normal peripheral total folate levels (29). Kearns–Sayre syndrome patients have CFD and beneficial responses to folinic acid have been reported (30, 31). However, CFD does not occur consistently in other PMDs. Empiric supplementation is relatively common in PMD patients with neurologic symptoms, especially in those documented to have CSF 5-MTHF deficiency. Folinic acid doses range from 0.5–2.5mg/kg daily in children or 2.5–25 mg daily in adults (12), and are typically well-tolerated. To date no results of RCTs for folinic acid for PMDs have been reported.
Metabolic-modifying agents
L-arginine
Although L-arginine is synthesized endogenously, metabolic demand may exceed biosynthetic capacity under certain stress or disease conditions. L-arginine is a precursor of proteins, polyamines, and creatine, can be converted to α-ketoglutarate (32), and can be decarboxylated to form agmatine, which possess antioxidant, anti-inflammatory, and neuromodulatory properties (33). By virtue of conversion to nitric oxide, L-arginine also has widespread impacts on gene expression and metabolic profiles via activation of PPAR-α and PGC-1α (34).
Beneficial effects of L-arginine supplementation have been reported in mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes (MELAS) (35), including improved serum arginine deficiency and prevention of stroke-like episodes (36). Consequently, L-arginine is considered a front-line treatment for stroke-like episodes in MELAS (10). While the initial rationale for testing L-arginine therapy in MELAS was to improve vasodilation (37), oral L-arginine (10 g/m2 body surface area/day) in MELAS patients increased de novo arginine synthesis rate, increased arginine and citrulline flux (36), and decreased plasma lactate levels (38). L-arginine therapy in three MELAS siblings yielded increased trends in mean percent maximum work at anaerobic threshold and a small increase in their VO2peak. L-arginine supplementation also slowed acidification, increased phosphorous to phosphocreatine (Pi/PCr) ratio, and decreased half-time of PCr recovery following moderate intensity exercise (39). While arginine supplementation has been investigated in open-label studies of MELAS patients, its efficacy in other PMDs has not been studied in model systems or RCTs.
L-carnitine
Carnitine is a hydrophilic quaternary amine with a major function in transporting long-chain fatty acids from the cytosol to the mitochondrial matrix for β-oxidation (Figure 1). Other functions include modulation of the acyl-CoA/CoA ratio, muscle storage of energy as acetyl-carnitine, and urinary excretion toxic acyl groups (40). L-carnitine contains a β-hydroxy moiety that can directly scavenge free radicals (41, 42).
Although carnitine is endogenously synthesized, the majority is obtained from the diet. PMD patients may have decreased free carnitine (43), which supplementation is aimed at restoring. However, circulating carnitine does not reflect tissue carnitine levels (44), and the evidence for clinical administration of L-carnitine is based only on case reports. Despite the lack of proven efficacy, L-carnitine supplementation (50–100 mg/kg/day) in PMD is common. Recently, concerns emerged of increased risks for cardiovascular disease and adverse cardiac events in subjects with high fasting plasma carnitine and its metabolite, trimethylamine-N-oxide (TMAO) (45). In addition, chronic dietary L-carnitine supplementation in mice markedly enhanced synthesis of trimethylamine and TMAO and increased atherosclerosis (45). These findings highlight the need to carefully consider the risk:benefit ratio with regard to nutritional supplements in PMD patients, especially when the evidence for efficacy and safety from rigorous studies is limited.
Creatine
Creatine transiently stores metabolic energy via the exchange of a high-energy phosphate bond with adenosine triphosphate (ATP) by creatine kinase (CK) (46). The bulk of creatine is stored in skeletal muscle, but it is also found in other high-energy demand tissues.
Creatine monohydrate supplementation can increase intracellular phosphocreatine levels in healthy individuals (47), and since PMD patients have low phosphocreatine levels (48, 49), supplementation may help buffer energy levels. In a Cochrane review of 12 mitochondrial disease patient RCTs, three trials studied creatine monohydrate alone, and one trial studied it in combination with coenzyme Q10 (CoQ10) and lipoic acid (23). Creatine monohydrate treatment significantly increased handgrip strength and decreased post-exercise lactate compared to placebo in a study of six MELAS and one mitochondrial myopathy patient (23). Creatine supplementation in combination with alpha-lipoic acid and CoQ10 in 16 patients with confirmed PMD had no clinically significant effect, despite a significant decrease in plasma lactate and urine 8-isoprostane levels (50).
Coenzyme Q10
CoQ10 is a lipid-soluble benzoquinone that transfers electrons in the ETC from Complexes I or II to Complex III and helps maintains mitochondrial inner membrane and OXPHOS complexes stability (51). In the ubiquinol form, CoQ10 inhibits lipid peroxidation and can protect mitochondrial inner membrane proteins and mtDNA from oxidative damage (52). Both endogenous synthesis and dietary sources contribute to CoQ10 levels in plasma, but leukocyte levels may better approximate tissue levels (53).
Mitochondrial OXPHOS dysfunction commonly causes increased reactive oxygen species (ROS) which can damage mitochondrial and cellular components. Therefore, a common empiric therapy is to use antioxidants in an attempt to protect from ROS-induced oxidative damage. Indeed, as a ROS scavenger with documented safety data, CoQ10 and its synthetic analogs are the most commonly recommended supplement option for PMDs (54). CoQ10 supplementation appears to be particularly effective in primary disorders of CoQ10 biosynthesis, which result in severe phenotypes including encephalomyopathy, ataxia, renal diseases, cerebellar atrophy, and hyperlactatemia (11).
Despite PMD clinical trials with CoQ10 supplementation, its therapeutic benefit has not been definitively established (23, 55, 56). Bioavailability of different CoQ10 formulations is an important consideration, and several animal studies investigating absorption and distribution of CoQ10 supplements have been performed in rodents (57, 58). CoQ10 enterally administered to rodents was shown to increase CoQ10 levels in blood and liver, but not in other tissues (56). While there have been few studies of CoQ10 distribution in human tissues following supplementation, studies in patients with neurodegenerative disorders provide evidence that uptake into brain occurs at high doses (up to 2400 mg/day) (56).
α-lipoic acid
Lipoic acid (α-LA) an essential redox cofactor of pyruvate dehydrogenase (59). It is synthesized de novo by mitochondrial lipoyl synthase (60), acts in both the cytosol and plasma membrane, readily crosses the blood–brain barrier (61), and is rapidly absorbed and reduced to dihydrolipoic acid (DHLA) in various tissues (62). The reduced form is an antioxidant (63) and also recycles other antioxidants, such as glutathione (GSH) and vitamins C and E (62). LA in combination with creatine and CoQ10 supplementation in a 16 patient RCT revealed no clinically significant effects (64). Given its reported potential benefits in disorders that involve mitochondrial dysfunction, such as diabetes (65) and dyslipidemia (66), further clinical and preclinical studies of α-LA in PMDs are warranted.
Signaling-pathway modulators
Resveratrol
Resveratrol is a bioactive polyphenol with reported antioxidant, anti-inflammatory, and anti-apoptotic activities (67), and the capacity to promote mitochondrial biogenesis in model organisms (68). This is thought to occur via SIRT1-dependent deacetylation of PGC-1α, (68, 69) a major regulator of mitochondrial biogenesis, although this mechanism remains controversial (70, 71). Resveratrol can correct ETC Complex I and IV defects in human fibroblasts via SIRT1 and AMPK-independent mechanisms that involve estrogen receptors (72). Additionally, the combination of resveratrol supplementation and metformin treatment stabilized OXPHOS super-complexes in HeLa cells (73). More studies into the mechanism by which resveratrol protects mitochondrial function, and might benefit PMD pathology, are clearly justified.
Ketogenic diet
A ketogenic diet (KD) modulates the amount of carbohydrates, protein, calories, and/or fats to shift metabolism towards β-oxidation and ketone body production, which in turn increase transcription of OXPHOS, TCA cycle, and glycolysis genes (74). Four KDs have been developed: the ‘classic’ KD (~90% of calories from fat); the medium-chain triglyceride diet; a low glycemic index (<50) diet; and a modified Atkins diet (≤20 g/day carbohydrates for adults) (75). A 20% fat KD was effective in reducing seizure frequency in PMD patients with OXPHOS defects (76), although no connection was found between the efficacy of the KD and changes in specific OXPHOS enzyme activities. An induction of brain mitochondrial uncoupling protein isoforms and a decrease in mitochondrial ROS was observed in mice following a classic KD, suggesting it might reduce mitochondria-mediated oxidative stress (77). However, KD remains controversial and exploratory in PMDs, with concerns of intolerance or disease exacerbation due to promoting secondary fatty acid oxidation defects and NAD+ deficiency.
Experimental models for PMDs
In this section, we describe salient examples of cellular and animal models used to investigate OXPHOS deficiencies, focusing on their use for testing nutritional interventions for PMDs. Key features of these model systems, along with some of their strengths and limitations are also highlighted (Figure 2; Supplemental Table S2).
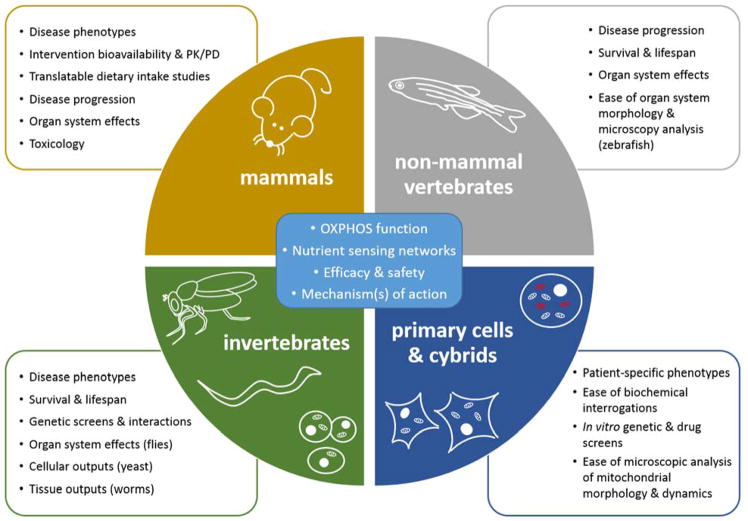
Figure 2. Key experimental strengths of model systems for PMD intervention studies.
Shared among the indicated model organisms and cellular systems is an ability to investigate genetic and biochemical characteristics of specific PMDs, nutrient sensing networks, and the efficacy, safety, and mechanism(s) of action for nutritional interventions. Individual model systems also have distinct experimental advantages, described in each quadrant. For example, primary cells allow for the study of patient-specific phenotypes and straightforward analysis of mitochondrial morphology and dynamics, invertebrates are well-suited for survival and lifespan investigations, and rodents permit assessments of nutritional intervention bioavailability and toxicology more directly relevant to human PMDs.
Cellular models
Cell culture models have been extensively used to investigate mitochondria functions and pathologic phenotypes due to OXPHOS defects. More recently, induced pluripotent stem cell (iPSC) technology has allowed the production of patient-derived replicating cells, with specific genetic OXPHOS defects, which can be differentiated into disease-relevant cell types.
Primary cells with OXPHOS defects
Primary cells from PMD patients (e.g. platelets, lymphocytes, fibroblasts, and myoblasts) provide a useful platform to characterize pathogenic mechanisms of OXPHOS deficiencies (78) and to test metabolic interventions (79). However, given that mitochondria are tailored for the specialized functions of different cell types, results from one cell type are not necessarily universal. Platelets and lymphocytes are easy to collect from peripheral blood and amenable to short-term in vitro experiments. Long-term propagation of lymphocytes in culture requires their transformation into lymphoblastoid cell lines (e.g. via Epstein-Barr virus). Lymphocytes and lymphoblasts are not ideal for certain biochemical OXPHOS studies due to their relatively low mitochondrial content. Skin fibroblasts are accessible by biopsy and can be propagated easily, and their abundant mitochondria with tubular morphology makes them well-suited for biochemical studies and investigations of mitochondrial network dynamics. Myoblasts are theoretically an optimal cell type to study mitochondrial defects because they are highly reliant on mitochondrial bioenergetics. However, myoblasts are typically procured via invasive muscle biopsy and pure cultures are not easily obtained. Myoblasts and myocytes can also be derived by transformation of primary skin fibroblasts (80).
Patient-derived skin fibroblasts have proven extremely useful to model biochemical deficiencies associated with PMDs and nutritional interventions (81). For example, patient fibroblasts with primary CoQ defects and Complex I deficiency have been used to study disease-related biochemical defects and to assess rescue by CoQ10 analogs (82, 83) and riboflavin (84). Other examples of successful use of primary skin fibroblasts for testing metabolic therapies for PMDs include using mitochondrial-targeted versions of vitamin E and CoQ10 to alleviate oxidative stress in cells from Friedreich Ataxia patients (85). Human fibroblasts from PMD patients have also been used as a screening platform for potential therapeutics. For example, mitochonic acid 5, a derivative of the plant hormone indole-3-acetic acid, was found to improve the survival of MELAS patient fibroblasts under metabolic stress (86). The utility of fibroblast and other primary cell lines is limited by their availability and replicative capacity in culture, which leads to senescence and metabolic remodeling. Genetic heterogeneity between patients can also affect analyses if the sample size of patient-derived cells with similar mutations is too small to achieve statistical significance.
Cybrid models
Cytoplasmic hybrids, or “cybrids,” are widely employed to study pathogenic mtDNA mutations in a relatively homogeneous nuclear genetic background. Virtually any cell type can be used as the source of cytoplasts, but fibroblasts, white blood cells, and platelets are most frequently employed due to their relative ease of access. Human cancer cell lines are most often used as the recipient cell lines (e.g. osteosarcoma). Cybrid technology has been used to generate immortalized human lines harboring specific pathogenic mtDNA mutations derived from PMD patients, including mt-tRNA mutations associated with MELAS (87), MERRF (myoclonic epilepsy with ragged red fibers) (88) and LHON (Leber’s hereditary optic neuropathy) (89). Since many mtDNA mutations are heteroplasmic, stochastic distribution or manipulation via short-term treatment with mtDNA-depleting agents (90) allows for the analysis of cybrids with a range of mtDNA mutation loads. Cybrids have been used to screen nutrients and metabolic intermediates that improve disease phenotypes under a variety of growth or stress conditions. One common paradigm is the substitution of glucose with galactose as the main carbon source, which forces cells to rely more heavily on ATP produced by OXPHOS than aerobic glycolysis (91, 92). In heteroplasmic cybrids with a high pathogenic mutation load, a ketogenic intervention induced a shift toward accumulation of more wild-type molecules (93), presumably by selective elimination of dysfunctional mitochondria containing high levels of mutant mtDNA (94). A similar heteroplasmy shift was also produced in MELAS neuroblastoma cybrids by a treatment with low-glucose medium supplemented with 1 mM L-arginine, which putatively functions by enhancing nitric oxide synthesis to induce mitochondrial biogenesis (95). Combined glutamate and aspartate supplementation improved the survival of NARP (neuropathy, ataxia and retinitis pigmentosa) cybrids in galactose medium (96), possibly by enhancing citrate synthesis (97). Riboflavin (0.06–0.08 μM) improved proliferation, ATP levels, and complex I+III activity, as well as decreased superoxide production in MELAS cybrids (98). Finally, antioxidants, including CoQ10, have also been successfully employed to improve survival and OXPHOS function in NARP and LHON cybrids (99).
Cybrid studies are useful for eliminating variation in nuclear genetic backgrounds between patient cells, but the interpretation of results can be limited if cancer cell backgrounds are employed that preferentially utilize anaerobic glycolysis over OXPHOS in culture. Thus, it is important to validate results from therapeutic intervention studies in cybrids and primary cells derived from PMD patients.
Induced pluripotent cells with OXPHOS defects
Stem cell technologies have allowed development of patient-derived iPSC lines of mitochondrial diseases (100–103) that allow for genetic manipulation and nutritional intervention studies. Importantly, iPSCs can be differentiated into disease-relevant cell types such as neurons and muscle cells. Generation and differentiation of iPSCs harboring pathogenic mtDNA mutations has allowed the study of mtDNA segregation and the effects of different mutation loads on relevant cells, such as cardiomyocytes (104). By exploiting natural mtDNA drift, combined with a contraction of mtDNA copy number upon induction of pluripotency and expansion upon differentiation, heteroplasmic iPSC lines with different proportions of mutant mtDNA can be obtained that model the variation observed in PMD patients. While these cells contain naturally occurring pathogenic mutations, they also allow for derivation of isogenic control lines via genetic correction of nuclear mutations, or by spontaneous segregation of heteroplasmic mtDNA in the parental proliferating fibroblasts. iPSCs derived from MELAS and Leigh syndrome patients have been characterized (105), and somatic cell nuclear transfer was used to replace mutant mtDNA from NARP fibroblasts to generate corrected isogenic control iPSCs, which showed normalization of the bioenergetic defects and associated gene expression profiles. Potential limitations of iPSCs include the high costs associated with generation and maintenance, complexity of the differentiation protocols, and paucity of stem cell metabolism knowledge.
Saccharomyces cerevisiae
Fundamental knowledge about mitochondria and their role in human disease has arisen from studies in the budding yeast Saccharomyces cerevisiae. In fact, evidence for cytoplasmic inheritance (mtDNA genes) and some of the first direct evidence of mtDNA itself was discovered in yeast (106). Many core nuclear-encoded mitochondrial proteins are homologous between yeast and mammals, including those involved in mitochondrial protein import, metabolite transport, fission and fusion, and mtDNA maintenance (107–110).
As a facultative anaerobe S. cerevisiae is can survive severe OXPHOS defects when grown on a fermentable fuel source, and thus provides an invaluable model to study mitochondrial defects (111). Mutant and regulated open reading frame collections are available that allow for systematic screening for genes involved with mitochondrial function, and many yeast mutants can be functionally corrected through complementation with human homologues (112, 113). It is also possible to generate mtDNA mutations by homologous recombination-based approaches in yeast (114), a capability which is limited or non-existent in other species. Studies in S. cerevisiae have been instrumental in uncovering pathologic mechanisms in mitochondrial diseases, including tafazzin interactions important in Barth Syndrome (115), cytochrome oxidase assembly disruption in Leigh syndrome (116, 117), and candidate genes responsible for a form of hereditary spastic paraplegia (118). Important limitations of the study of PMDs in yeast include the lack of Complex I and a full repertoire of cell death mechanisms, which can be a detriment to understanding all functional consequences of OXPHOS dysfunction (108, 119–122). Lastly, yeast mtDNA is larger (~80-kb) than mammalian mtDNA, contains introns, encodes only seven OXPHOS proteins and a non-conserved ribosomal protein (VAR1), and is unstable when mitochondrial translation is inhibited (123–125).
Invertebrate models
Caenorhabditis elegans
The nematode C. elegans is a powerful genetic model system that has been used to study mitochondrial biology (126–128). Its 3-day developmental cycle, 2-week lifespan, and isogenic offspring permit the rapid, economical study of large quantities of genetically identical animals (129). The expressed C. elegans proteome shares greater than 83% homology with human genes (130) and includes ~80% of genes for human inborn errors of metabolism (131, 132). Their mtDNA encodes 12 of the 13 proteins found in mammalian mtDNA, lacking only ATP8 (126). Limitations of C. elegans for PMD research include a simple nervous system that does not model the complex network of connections and cell interactions of mammals and lack of respiratory and vascular systems, eyes and liver that are often involved in PMD pathology. Finally, all somatic tissues in adults are post-mitotic.
Despite the aforementioned limitations, C. elegans has proven useful as a model organism to characterize global sequelae of primary mitochondrial ETC dysfunction (26, 133), as well as to evaluate the efficacy of nutritional interventions in PMD. In a C. elegans model of Complex I deficiency (NDUFV1), riboflavin supplementation yielded enhanced assembly and improved catalytic activities of complexes I and IV (134). In another model of mitochondrial Complex I deficiency (NDUFS2), resveratrol treatment significantly improved shortened gas-1(fc21) mutant lifespan regardless of the developmental stage at initiation. These worms display compensatory up-regulation of amino acid, fat, and other nutrient pathways in response to chronic Complex I dysfunction. Resveratrol reversed this metabolic response, and restore NADH and NAD+ levels (129) despite increasing the already elevated mitochondrial oxidant burden in this model.
Drosophila melanogaster
Drosophila melanogaster shares a high degree of genetic conservation with mammals, and has been effectively used to study mitochondrial disease states caused by mtDNA and nuclear gene mutations. Issues unique to multi-copy mtDNA have also been studied in Drosophila, including transmission, heteroplasmy, and mutant mtDNA threshold effects (135). Mitochondrial-targeted restriction enzymes that cleave wild-type mtDNA molecules have allowed selection of resistant molecules and generation of flies with mutations in cytochrome c oxidase I and ND2 (135, 136). However, the ability to generate mtDNA mutations in this manner is limited to naturally occurring restriction sites. Other mtDNA-based models of mitochondrial dysfunction in flies include a mutation in the ATP6 gene that models mitochondrial encephalomyopathies and tRNA mutations that characterized how the pathogenicity of mtDNA mutations can be altered by the nuclear genetic background (135, 137, 138). Finally, there are Drosophila models for Friedreich ataxia (139), mitochondrial deafness (135), Autosomal Spastic Ataxia with Leukoencephalophathy (140), and Leigh syndrome (141). These models have been informative in elucidating complexities of mitochondrial pathology (e.g. tissue-specificity, oxidative stress) (142–144), however their ability to model human pathological states varies (139, 142, 145). The unique lifecycle of Drosophila is well-suited for controlled feeding regimens to examine the effects of supplementation and specific diets on mitochondrial pathology over time in both adults and during development.
Vertebrate models
Danio rerio
The zebrafish, Danio rerio, is a powerful vertebrate model system of human diseases due to its rapid development, regular production of large numbers of offspring, and relatively inexpensive maintenance (146). Zebrafish have high genetic homology to mammals, and sequence conservation of mtDNA and nucleus-encoded mitochondrial proteins make it a valid model of human mitochondrial pathology (147). Zebrafish genetic investigations have traditionally been driven by forward genetic screens, however an increasing range of reverse-genetic techniques are becoming available. Transient knockdown of gene expression is possible with morpholino oligonucleotides, which allow for rapid and effective study of gene function (148). The use of transcription activator-like effector nucleases (149), and the CRISPR-Cas system (150) can be utilized to precisely modify the sequence at a particular locus in order to generate specific models of human disease.
The zebrafish possesses key organ systems including the brain, eyes, and hematologic system (151), which makes it a good model in which to study organ-specific mitochondrial pathology. Many characteristics of the zebrafish central nervous system are also conserved with humans and they develop tactile responses throughout the first few days of life (152), allowing the modeling of motor neuron diseases, myopathy, neuropathy and behavioral and cognitive endpoints. Due to their translucency, direct in vivo visualization of organ systems and mitochondria is possible, making zebrafish an attractive model for microscopy and testing of interventions. Limitations of zebrafish for PMD research include aquatic dietary formulations and feeding strategies that differ from other model organisms and less developed techniques to assess food intake (153) and energy expenditure (154).
Mouse models of OXPHOS deficiencies
Many mouse models of OXPHOS deficiencies (155, 156) or mtDNA dysfunction (157) have been developed (Supplemental Table S2), and insights into PMD mechanisms and nutritional interventions are discussed below. Although the pathogenesis of these mouse models is well-characterized in several cases, the number of studies conducted to investigate potential therapies are limited, opening a door for future intervention studies.
Mouse models of defects in mtDNA
Like humans, mice contain a double-stranded, circular mtDNA of ~16-kb that encodes thirteen core OXPHOS components, as well as two rRNAs and twenty-two tRNAs needed for mitochondrial ribosome biogenesis and translation (158). Since all other mitochondrial proteins are encoded in the nucleus (159), a common strategy in mouse models of PMD is to alter mtDNA by knock-out or overexpression of nuclear genes encoding mtDNA regulatory factors. However, transfer of specific mutant mtDNA species from tissues or cell lines into the mouse germline has also been accomplished.
Tfam mice
Mitochondrial transcription factor A, encoded by the Tfam gene, is an abundant mtDNA-binding protein, originally discovered as a transcriptional activator of mtDNA promoters (160). Tfam has additional roles in mtDNA metabolism, including packaging into nucleoids, mtDNA copy number regulation, segregation, and transcriptional priming of mtDNA replication (159). Global knock-out of the TFAM gene in mice results in severe mtDNA depletion, loss of mitochondrial respiration, and embryonic lethality (161). Surprisingly, heterozygous knock-outs (Tfam+/−) are viable despite having 30–70% mtDNA depletion in tissues. Tfam+/− mice are primed for antiviral innate immune activation due to enhanced level of cytoplasmic mtDNA (162). Thus, the Tfam+/− model may serve as a “sensitized” background for studying mtDNA instability in disease and the role of mtDNA in immune system activation.
To avoid embryonic lethality of global Tfam gene deletion, multiple tissue-specific Tfam KO models have been generated, and these have provided significant insight into the requirements of OXPHOS in tissues and model mitochondrial disease pathology to varying degrees (163, 164). For example, muscle-specific deletion of Tfam results in features that are common in human PMD including cytochrome c oxidase-negative and ragged-red fibers, myopathy, and hypertrophy of the heart (165, 166). The heart also displays a common metabolic switch towards glycolysis when OXPHOS is attenuated, as well as biochemical and physiological features of dilated cardiomyopathy present in Kearns-Sayre syndrome (167). Tfam-knock-out models have proven valuable in mechanistic studies that show the differential requirements for mtDNA and OXPHOS, variability in mitochondrial function between different tissues, and potential therapies (163, 164, 168).
TFB1M mice
Mitochondrial transcription factor B1 (TFB1M or mtTFB1) was originally identified as a homolog of the yeast mitochondrial transcription factor Mtf1p (or sc-mtTFB) (169). The two mammalian orthologs (TFB1M and TFB2M) are homologous to a class of site-specific, rRNA N6-adenine dimethyltransferases (170, 171). Knock-out of TFB1M in mice results in severely reduced 12S rRNA methylation, destabilization of the small mitochondrial ribosome subunit, and impaired mitochondrial translation (172). Transgenic mice that overexpress TFB1M have enhanced progressive hearing loss due to tissue-specific pathology in the inner ear, which can be rescued by reducing AMP kinase activity, suggesting a potential therapeutic route for deafness involving the A1555G mtDNA mutation (173, 174).
mtDNA mutator mice
DNA polymerase gamma (POLG) functions exclusively in mitochondria to replicate and repair mtDNA. The consequences of enhanced mtDNA mutagenesis can be modeled in mice that express a proofreading-deficient allele of its catalytic subunit (175, 176). These POLG mutants (a.k.a. mtDNA mutator mice) display systemic mitochondrial dysfunction, increased apoptosis, disrupted stem cell function, and progeroid phenotypes (175, 177). Mutator mice undergo a metabolic shift toward reliance on glycolysis to compensate for mitochondrial dysfunction in some tissues (178). There is some disagreement regarding whether point mutations or deletions (and other rearrangements) in mtDNA drive the aging-like phenotypes observed in these mice (179) and the precise cause of the progeroid phenotypes remains under investigation. Interventions that improve cell and tissue phenotypes in mtDNA mutator mice include targeting catalase to mitochondria, antioxidants, and endurance exercise (180–183).
Mouse model of MNGIE
Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) is caused by a deficiency in thymidine phosphorylase (TP) resulting in cellular imbalance of dNTPs and accumulation of thymidine and deoxyuridine. This imbalance in turn results in secondary reduction of mitochondrial deoxycytidine and mtDNA depletion, deletions, and point mutations (184). A mouse model has been generated by knocking out both thymidine phosphorylase (TP) and uridine phosphorylase (UP) (185). These TP−/−UP−/− mice display the same nucleoside accumulation as human patients and develop leukoencephalopathy, brain lesions, and in some cases OXPHOS deficiencies and brain mtDNA alterations (185, 186). Loss of motor control and gastrointestinal pathology observed in patients were not present in the disease-model mice.
Mouse models of pathogenic mtDNA mutations
Using cytoplasmic transfer of mtDNA into embryonic stem (ES) cells, chimeric mice have been generated that contain point mutations in the 16S rRNA (187, 188), OXPHOS genes (e.g., cytochrome c oxidase and ND6) (189, 190), and tRNAs (191), as well as, large-scale deletions (192, 193). These models have allowed mtDNA heteroplasmy, inheritance and pathology to be studied in vivo. Variable percentages of mutant mtDNA are found in mice generated in this fashion (194), and the severity of pathology observed correlates with higher loads of the mutant mtDNA (192, 193). Additionally, severe mutations appear to be selected against in the germline, while milder mutations persist for several generations (189). Because the genetic background is homogeneous, these mouse models allow the pathogenic consequences of specific mtDNA mutations to be studied without confounding effects of nuclear polymorphisms. These mice recapitulate many pathologies observed in human mitochondrial diseases including OXPHOS defects, lactic acidosis, anemia, muscle weakness, and renal failure (195, 196). Some of the mutations generated contribute to characterized diseases including MERRF (191), chronic progressive external ophthalmoplegia (CPEO) (192), LHON (190), and early-onset Pearson syndrome caused by heteroplasmic mtDNA deletions (197). Some of these models have been used to test gene therapy (198), and the antioxidant N-acetylcysteine was tested in ND6 mutant mice, resulting in increased longevity and decreased incidence of lymphoma (199). The development of new approaches to generate mice with mtDNA mutations should expand the number of these invaluable models for specific mitochondrial disorders (200, 201).
The Deletor Mouse
Adult-onset autosomal progressive external ophthalmoplegia (PEO) can be caused by mutations in adenine nucleotide translocator 1 (202), POLG (203), Twinkle mtDNA helicase (204), or ribonucleotide reductase subunit p53R2 (205). Common mtDNA mutations in Twinkle (13-bp duplication and a point mutation) have been recapitulated in the mutant Twinkledup mouse (206), which accumulates mitochondrial DNA deletions in all tissues (most prominently in heart and muscle), prompting the name “Deletor” mouse.
Biochemical and histological analyses of Deletor mice demonstrated OXPHOS dysfunction, cytochrome c oxidase (COX)-negative muscle fibers, abnormalities in mitochondrial ultrastructure, and increased autophagy. The number of abnormal, COX-deficient muscle fibers is similar to that of human PEO patients, and the delayed development recapitulates the late-onset PEO phenotype (206). However, unlike PEO pathology in humans, Deletor mice have a normal lifespan and no significant compromise of physical performance. Gene expression profiling of Deletor mice revealed induction of folate metabolism (methylenetetrahydrofolate dehydrogenase 2, Mthfd2), fasting-related lipid metabolism (fibroblast growth factor 21, Fgf21) (207), and phosphorylation of AKT1, all reflective of modulations in nutrient-sensing signaling networks and suggesting OXPHOS dysfunction in muscle tissue results in starvation-like response. The induction of Fgf21 suggests it may be secreted by skeletal muscle cells with mitochondrial myopathy and act as a systemic mediator of metabolism. Elevated serum Fgf21 has been demonstrated in adults with mitochondrial disease (208), and studies in patients and mouse models have implicated mtDNA instability or translational defects as prerequisites for Fgf21 secretion (209, 210). Current investigations are studying the potential for Fgf21 serum level as a biomarker for muscle myopathies caused by mtDNA defects. While nucleotide pool measurements in Deletor mice revealed imbalanced purine metabolism and tissue-specific induction of the folate cycle, folinic acid supplementation did not reverse the pathology (211). Significant increases in mitochondrial biogenesis and improved mitochondrial ultrastructure resulted after a two month KD in Deletor mice, although no alteration in the amount of mtDNA deletions nor skeletal muscle Complex I activity was observed (212). These reports prompted a pilot KD study in five mitochondrial myopathy patients and ten healthy subjects (213). While the trial was small and follow-up analyses are being performed, all of the KD subjects unexpectedly manifested muscle fiber damage after about 1.5 weeks of intervention (A. Suomalainen-Wartiovaara, personal correspondence). Therefore, while the KD may hold promise for some PMD patients, more preclinical research is needed to delineate appropriate applications.
Ndusf4−/− mice
Complex I dysfunction is one of the most common OXPHOS defects in PMD patients, with point mutations in NDUFS4 (NADH dehydrogenase-ubiquinone-Fe-S Protein 4) observed in patients with LHON, Leigh syndrome, and MELAS (214–216). A global knock-out mouse model for Complex I dysfunction (Ndufs4−/−) (217) displays a phenotypic progression that resembles Leigh syndrome encephalopathy and dies prematurely by day 50, (218). Rapamycin treatment has beneficial effects in this model of Leigh syndrome (219), suggesting that nutritional interventions that inhibit the mTORC1 pathway, such as indicated for the KD (220), may be beneficial for PMDs. Ndufs4+/− mice are another Leigh syndrome model with a less severe reduction Complex I activity and milder phenotypes (221).
Pdss2kd/kd Mice
CoQ10 deficiency is associated with at least five heterogeneous PMDs, but for most patients the genetic basis of the deficiency is unknown (222). Primary deficiencies in CoQ10 levels result from mutations in various genes required for ubiquinone biosynthesis, including PDSS1 or PDSS2 genes, which encode prenyl diphosphate dynthase subunits (223), the COQ2 gene, or the CoQ8/CABC1 gene. Although utilization of a ubiquinone as an electron carrier in the ETC is conserved across species, utilization of CoQ10 is not universal. Mice, for example, predominantly produce CoQ9 with small amounts of CoQ10.
Pdss2kd/kd mice, first identified by a kidney disease phenotype, harbor a homozygous mutation in PDSS2. Young Pdsskd/kd mice have normal ubiquinone levels, but adults have ~15–20% of normal CoQ9 and CoQ10 levels in brain, kidney, and muscle (224). Kidney Complex I+III activity is significantly reduced, although individual OXHPOS activities vary by tissue and age. Increased ROS production and oxidative stress are variably reported in kidney, but not observed in brain (224, 225). Metabolomic analyses of conditional CoQ deficiency in Pdss2kd/kd mouse liver revealed upregulation of multiple pathways including OXPHOS, the TCA cycle, and pathways that provide substrates for fatty acid and amino acid metabolism (225, 226). Despite significant CoQ9 deficiency and impaired Complex I and Complex II capacity, compensatory increases in Complex IV respiration was observed, and no overt symptomology was noted in liver-specific Pdss2 knockout mice.
Pilot studies of liposomal CoQ10 supplementation (~200 mg/kg/d) in Pdsskd/kd mice demonstrated a delayed onset of renal disease when administered from birth or weaning, and a partial rescue of proteinuria and interstitial nephritis (225, 227). However, kidney CoQ9 and CoQ10 content was not significantly altered, suggesting that CoQ10 might exerted its effect via antioxidant activity rather than enhanced OXPHOS, although oxidative stress was not measured. A subsequent study found limited therapeutic response to lifelong CoQ10 supplementation in a Pdss2 mouse model of fatal renal disease (226). However, significant responses were seen when the endogenous synthesis of CoQ9 and hydroquinone were increased with Probucol drug treatment, suggesting that targeting de novo supply of CoQ10 may be more effective than exogenous supplementation. Overall, these results suggest that understanding the mechanisms of CoQ10 supplementation benefits, including tissue-specific effects and the basis of responders versus non-responders, is a PMD research priority.
Barth Syndrome Mouse Model
Barth Syndrome is an X-linked disorder caused by pathogenic variants in the TAZ gene encoding Tafazzin (228), a transacylase that participates in acyl chain remodeling of cardiolipin, a major phospholipid of the mitochondrial inner membrane (229). Individuals with Barth Syndrome have clinical features that include early onset cardiomyopathy, neutropenia, skeletal myopathy, and growth abnormalities (228).
An inducible short hairpin RNA-induced (shRNA) mouse knockdown model of tafazzin deficiency was developed in 2011 (230, 231) with pathogenic features similar to Barth Syndrome patients, including left ventricular dilation and abnormal mass, decreased ejection fraction, stereotypical mitochondrial ultrastructural abnormalities, and abnormal cardiolipin species (230–232). However, the mice exhibit a late onset (7–8 months of age) cardiac functional abnormality, as opposed to the early onset cardiac phenotype observed in humans (233). Modifications of the shRNA induction protocol produced mice with a cardiac phenotype in utero and proved particularly useful in pinpointing the developmental stage in which cardiac non-compaction occurs in tafazzin deficiency (234). The tafazzin mouse model has proven useful in investigating other clinical phenotypes of Barth Syndrome, including exercise intolerance, OXPHOS defects, and abnormal in amino acid, folate, and lipid metabolism (232, 235–237). However the model has important limitations for studying cardiac arrhythmias (238), a major cause of morbidity and mortality in Barth Syndrome (233), and has not been used to address other cardinal features of Barth Syndrome like immune deficiency. Lastly, since this model is induced with doxycycline, potential effects of this agent on mitochondrial translation could complicate some findings (239).
Enhancing translatability of PMD model system studies
Regardless of the model system employed, rigor and reproducibility of preclinical nutritional interventions studies must be optimized such that the knowledge gained best informs approaches in PMD patients. While model systems for PMDs cannot completely recapitulate disease states observed in humans, they have the advantage of permitting detailed mechanistic experimentation in a genetically controlled population with nutritional interventions that are difficult to control for in a human PMD cohort. Preclinical nutritional intervention approaches can be reductionist, with systematic testing of single agents or combinations over time. Orthogonal testing of hypotheses in multiple models can be very powerful, as demonstrated by studies of NA and NR in patient-derived fibroblasts, C. elegans, Deletor, and Ndufs4−/− mice which help to elucidate the importance of NADH:NAD+ balance in PMDs (Sidebar).
To best inform clinical approaches, model system investigations must carefully consider the intervention formulation, dosing, and bioavailability. Rigorous characterization of identity, purity, and composition is especially important for dietary approaches, where patients have ready access to a large variety of nutritional supplements that are often far different than formulations used in biomedical research.
Bioavailability is a major area in need of further experimental evidence to support clinical use of nutritional interventions in PMDs, in particular as it relates to understanding mechanisms of action and efficacy in humans (Table 1) and in cases where it is unclear if intake beyond recommended dietary amounts confers any added health benefit. Animal models can serve as a bridge between cellular and human investigations, and their ability to do so is strengthened when an understanding of pharmacodynamics and pharmacokinetics in the animal system inform dosing regimens in patients. Without careful consideration of the relevance of intervention concentrations and human bioavailability wherever possible, preclinical research in model systems is unlikely to inform clinical practice. Additionally, model system investigations of bioavailability should consider several experimental measurement parameters such as the specific analyte (e.g., parent compound, metabolite, biomarker), physiological compartment (e.g., plasma, serum, tissue), and quantitative analytical method used.
Table 1.
Considerations to advance mechanistic and clinically translatable research in model systems.
| Analytical and measurement considerations | Administration considerations and biological variables |
|---|---|
Research rigor and reproducibility is enhanced through analytical confirmation of intervention purity, composition, and stability
|
Dosing
|
Adsorption
| |
Research comparability is enhanced with thorough reporting of intervention identity
|
Distribution
|
Experimental bioavailability quantification is dependent upon analytical methodology
|
Metabolism
|
Excretion
| |
Genetic variations
|
Conclusions
The use of nutritional interventions by the PMD community is pervasive and is often based on empirical evidence. Although the potential for metabolic cofactors as therapies is gaining emphasis, increased preclinical research on nutritional interventions is needed for the PMD patients. Cellular and animal model systems can recapitulate specific aspects of mitochondrial dysfunction and provide the means to thoroughly examine mechanistic interactions between interventions and underlying PMD biology (Figure 1). Advances in genetically engineered probes for redox imaging and multiple-omics provide new toolkits for objective measures of PMD progression at discrete sites in a controlled experimental setting. Through rigorous preclinical mechanistic studies, the potential exists to systematically evaluate specific nutrients, test dosing ranges, and query intervention efficacy and safety with enhanced confidence before initiating observational studies in patients. In some instances intervention outcomes have been unexpectedly negative, and an opportunity exists for cross-validation of both positive and negative nutritional intervention results between orthogonal PMD model systems. Moreover, indications of positive nutritional intervention effects in ‘healthy’ animals, such as resveratrol’s promotion of mitochondrial biogenesis, need to be tested in specific models of PMD to truly support their use invulnerable patient populations. Focused research in these areas will ultimately help to better inform cohort selection and outcome measurement criteria which are crucial for larger-scale RCT design for PMD interventions. Furthermore, insights gained from an enhanced emphasis on basic and preclinical research in model systems for PMDs could help build a foundation for significant advancements in our understanding of the mitochondria role in homeostasis and preventive healthcare.
Supplementary Material
Acronyms/Terms
- AMPK
AMP-activated protein kinase
- α-LA
α-lipoic acid
- CoQ10
coenzyme Q10
- ETC
electron transport chain
- iPSCs
induced pluripotent stem cells
- LHON
Leber’s hereditary optic neuropathy
- MADD
multiple acyl CoA dehydrogenase deficiency
- MELAS
mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes
- MERRF
myoclonic epilepsy with ragged red fibers
- MNGIE
mitochondrial neurogastrointestinal encephalomyopathy
- mtDNA
mitochondrial DNA
- NA
nicotinic acid
- NR
nicotinamide riboside
- OXPHOS
oxidative phosphorylation
- PEO
progressive external ophthalmoplegia
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator 1-α
- PMD
primary mitochondrial disorder
- POLG
DNA polymerase gamma
- PPAR
peroxisome proliferator-activated receptor
- ROS
reactive oxygen species
Literature Cited
- 1.Gorman GS, Schaefer AM, Ng Y, Gomez N, Blakely EL, Alston CL, Feeney C, Horvath R, Yu-Wai-Man P, Chinnery PF, Taylor RW, Turnbull DM, McFarland R. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol. 2015;77:753–9. doi: 10.1002/ana.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinnery PF, Turnbull DM. Epidemiology and treatment of mitochondrial disorders. Am J Med Genet. 2001;106:94–101. doi: 10.1002/ajmg.1426. [DOI] [PubMed] [Google Scholar]
- 3.Parikh S, Goldstein A, Koenig MK, Scaglia F, Enns GM, Saneto R, Anselm I, Cohen BH, Falk MJ, Greene C, Gropman AL, Haas R, Hirano M, Morgan P, Sims K, Tarnopolsky M, Van Hove JL, Wolfe L, DiMauro S. Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med. 2014 doi: 10.1038/gim.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camp KM, Krotoski D, Parisi MA, Gwinn KA, Cohen BH, Cox CS, Enns GM, Falk MJ, Goldstein AC, Gopal-Srivastava R, Gorman GS, Hersh SP, Hirano M, Hoffman FA, Karaa A, MacLeod EL, McFarland R, Mohan C, Mulberg AE, Odenkirchen JC, Parikh S, Rutherford PJ, Suggs-Anderson SK, Tang WH, Vockley J, Wolfe LA, Yannicelli S, Yeske PE, Coates PM. Nutritional interventions in primary mitochondrial disorders: Developing an evidence base. Mol Genet Metab. 2016;119:187–206. doi: 10.1016/j.ymgme.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinnery P, Majamaa K, Turnbull D, Thorburn D. Treatment for mitochondrial disorders. Cochrane Database Syst Rev. 2006:CD004426. doi: 10.1002/14651858.CD004426.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karaa A, Kriger J, Grier J, Holbert A, Thompson JL, Parikh S, Hirano M. Mitochondrial disease patients’ perception of dietary supplements’ use. Mol Genet Metab. 2016;119:100–8. doi: 10.1016/j.ymgme.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown G. Defects of thiamine transport and metabolism. J Inherit Metab Dis. 2014;37:577–85. doi: 10.1007/s10545-014-9712-9. [DOI] [PubMed] [Google Scholar]
- 8.Alfadhel M, Almuntashri M, Jadah RH, Bashiri FA, Al Rifai MT, Al Shalaan H, Al Balwi M, Al Rumayan A, Eyaid W, Al-Twaijri W. Biotin-responsive basal ganglia disease should be renamed biotin-thiamine-responsive basal ganglia disease: a retrospective review of the clinical, radiological and molecular findings of 18 new cases. Orphanet J Rare Dis. 2013;8:83. doi: 10.1186/1750-1172-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabarki B, Al-Shafi S, Al-Shahwan S, Azmat Z, Al-Hashem A, Al-Adwani N, Biary N, Al-Zawahmah M, Khan S, Zuccoli G. Biotin-responsive basal ganglia disease revisited: clinical, radiologic, and genetic findings. Neurology. 2013;80:261–7. doi: 10.1212/WNL.0b013e31827deb4c. [DOI] [PubMed] [Google Scholar]
- 10.Parikh S, Goldstein A, Koenig MK, Scaglia F, Enns GM, Saneto R, Anselm I, Cohen BH, Falk MJ, Greene C, Gropman AL, Haas R, Hirano M, Morgan P, Sims K, Tarnopolsky M, Van Hove JL, Wolfe L, DiMauro S. Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med. 2015;17:689–701. doi: 10.1038/gim.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath R. Update on clinical aspects and treatment of selected vitamin-responsive disorders II (riboflavin and CoQ 10) J Inherit Metab Dis. 2012;35:679–87. doi: 10.1007/s10545-011-9434-1. [DOI] [PubMed] [Google Scholar]
- 12.Parikh S, Saneto R, Falk MJ, Anselm I, Cohen BH, Haas R Medicine Society TM. A modern approach to the treatment of mitochondrial disease. Curr Treat Options Neurol. 2009;11:414–30. doi: 10.1007/s11940-009-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosch AM, Abeling NG, Ijlst L, Knoester H, van der Pol WL, Stroomer AE, Wanders RJ, Visser G, Wijburg FA, Duran M, Waterham HR. Brown-Vialetto-Van Laere and Fazio Londe syndrome is associated with a riboflavin transporter defect mimicking mild MADD: a new inborn error of metabolism with potential treatment. J Inherit Metab Dis. 2011;34:159–64. doi: 10.1007/s10545-010-9242-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green P, Wiseman M, Crow YJ, Houlden H, Riphagen S, Lin JP, Raymond FL, Childs AM, Sheridan E, Edwards S, Josifova DJ. Brown-Vialetto-Van Laere syndrome, a ponto-bulbar palsy with deafness, is caused by mutations in c20orf54. Am J Hum Genet. 2010;86:485–9. doi: 10.1016/j.ajhg.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsen RK, Olpin SE, Andresen BS, Miedzybrodzka ZH, Pourfarzam M, Merinero B, Frerman FE, Beresford MW, Dean JC, Cornelius N, Andersen O, Oldfors A, Holme E, Gregersen N, Turnbull DM, Morris AA. ETFDH mutations as a major cause of riboflavin-responsive multiple acyl-CoA dehydrogenation deficiency. Brain. 2007;130:2045–54. doi: 10.1093/brain/awm135. [DOI] [PubMed] [Google Scholar]
- 16.Gerards M, van den Bosch BJ, Danhauser K, Serre V, van Weeghel M, Wanders RJ, Nicolaes GA, Sluiter W, Schoonderwoerd K, Scholte HR, Prokisch H, Rotig A, de Coo IF, Smeets HJ. Riboflavin-responsive oxidative phosphorylation complex I deficiency caused by defective ACAD9: new function for an old gene. Brain. 2011;134:210–9. doi: 10.1093/brain/awq273. [DOI] [PubMed] [Google Scholar]
- 17.Garone C, Donati MA, Sacchini M, Garcia-Diaz B, Bruno C, Calvo S, Mootha VK, Dimauro S. Mitochondrial encephalomyopathy due to a novel mutation in ACAD9. JAMA Neurol. 2013;70:1177–9. doi: 10.1001/jamaneurol.2013.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernsen PL, Gabreels FJ, Ruitenbeek W, Hamburger HL. Treatment of complex I deficiency with riboflavin. J Neurol Sci. 1993;118:181–7. doi: 10.1016/0022-510x(93)90108-b. [DOI] [PubMed] [Google Scholar]
- 19.Penn AM, Lee JW, Thuillier P, Wagner M, Maclure KM, Menard MR, Hall LD, Kennaway NG. MELAS syndrome with mitochondrial tRNA(Leu)(UUR) mutation: correlation of clinical state, nerve conduction, and muscle 31P magnetic resonance spectroscopy during treatment with nicotinamide and riboflavin. Neurology. 1992;42:2147–52. doi: 10.1212/wnl.42.11.2147. [DOI] [PubMed] [Google Scholar]
- 20.Bugiani M, Lamantea E, Invernizzi F, Moroni I, Bizzi A, Zeviani M, Uziel G. Effects of riboflavin in children with complex II deficiency. Brain Dev. 2006;28:576–81. doi: 10.1016/j.braindev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Artuch R, Vilaseca MA, Pineda M. Biochemical monitoring of the treatment in paediatric patients with mitochondrial disease. J Inherit Metab Dis. 1998;21:837–45. doi: 10.1023/a:1005470702369. [DOI] [PubMed] [Google Scholar]
- 22.Matthews PM, Ford B, Dandurand RJ, Eidelman DH, O’Connor D, Sherwin A, Karpati G, Andermann F, Arnold DL. Coenzyme Q10 with multiple vitamins is generally ineffective in treatment of mitochondrial disease. Neurology. 1993;43:884–90. doi: 10.1212/wnl.43.5.884. [DOI] [PubMed] [Google Scholar]
- 23.Pfeffer G, Majamaa K, Turnbull DM, Thorburn D, Chinnery PF. Treatment for mitochondrial disorders. Cochrane Database Syst Rev. 2012;4:CD004426. doi: 10.1002/14651858.CD004426.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu Rev Nutr. 2008;28:115–30. doi: 10.1146/annurev.nutr.28.061807.155443. [DOI] [PubMed] [Google Scholar]
- 25.Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci. 2007;32:12–9. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Falk MJ, Zhang Z, Rosenjack JR, Nissim I, Daikhin E, Nissim I, Sedensky MM, Yudkoff M, Morgan PG. Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Mol Genet Metab. 2008;93:388–97. doi: 10.1016/j.ymgme.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tischner C, Wenz T. Keep the fire burning: Current avenues in the quest of treating mitochondrial disorders. Mitochondrion. 2015;24:32–49. doi: 10.1016/j.mito.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Pfeffer GMK, Turnbull DM, Thorburn D, Chinnery PF. Treatment for mitochondrial disorders (Review) Cochrane Database of Systematic Reviews. 2012 doi: 10.1002/14651858.CD004426.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyland K, Shoffner J, Heales SJ. Cerebral folate deficiency. J Inherit Metab Dis. 2010;33:563–70. doi: 10.1007/s10545-010-9159-6. [DOI] [PubMed] [Google Scholar]
- 30.Serrano M, Perez-Duenas B, Montoya J, Ormazabal A, Artuch R. Genetic causes of cerebral folate deficiency: clinical, biochemical and therapeutic aspects. Drug Discov Today. 2012;17:1299–306. doi: 10.1016/j.drudis.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 31.Pineda M, Ormazabal A, Lopez-Gallardo E, Nascimento A, Solano A, Herrero MD, Vilaseca MA, Briones P, Ibanez L, Montoya J, Artuch R. Cerebral folate deficiency and leukoencephalopathy caused by a mitochondrial DNA deletion. Ann Neurol. 2006;59:394–8. doi: 10.1002/ana.20746. [DOI] [PubMed] [Google Scholar]
- 32.Morris SM., Jr Arginine metabolism: boundaries of our knowledge. J Nutr. 2007;137:1602S–09S. doi: 10.1093/jn/137.6.1602S. [DOI] [PubMed] [Google Scholar]
- 33.Chai J, Luo L, Hou F, Fan X, Yu J, Ma W, Tang W, Yang X, Zhu J, Kang W, Yan J, Liang H. Agmatine Reduces Lipopolysaccharide-Mediated Oxidant Response via Activating PI3K/Akt Pathway and Up-Regulating Nrf2 and HO-1 Expression in Macrophages. PLoS One. 2016;11:e0163634. doi: 10.1371/journal.pone.0163634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nissim I, Horyn O, Daikhin Y, Chen P, Li C, Wehrli SL, Nissim I, Yudkoff M. The molecular and metabolic influence of long term agmatine consumption. J Biol Chem. 2014;289:9710–29. doi: 10.1074/jbc.M113.544726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koga Y, Povalko N, Nishioka J, Katayama K, Kakimoto N, Matsuishi T. MELAS and L-arginine therapy: pathophysiology of stroke-like episodes. Ann N Y Acad Sci. 2010;1201:104–10. doi: 10.1111/j.1749-6632.2010.05624.x. [DOI] [PubMed] [Google Scholar]
- 36.El-Hattab AW, Hsu JW, Emrick LT, Wong LJ, Craigen WJ, Jahoor F, Scaglia F. Restoration of impaired nitric oxide production in MELAS syndrome with citrulline and arginine supplementation. Mol Genet Metab. 2012;105:607–14. doi: 10.1016/j.ymgme.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddiq I, Widjaja E, Tein I. Clinical and radiologic reversal of stroke-like episodes in MELAS with high-dose L-arginine. Neurology. 2015;85:197–8. doi: 10.1212/WNL.0000000000001726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El-Hattab AW, Emrick LT, Williamson KC, Craigen WJ, Scaglia F. The effect of citrulline and arginine supplementation on lactic acidemia in MELAS syndrome. Meta Gene. 2013;1:8–14. doi: 10.1016/j.mgene.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodan LH, Wells GD, Banks L, Thompson S, Schneiderman JE, Tein I. L-Arginine Affects Aerobic Capacity and Muscle Metabolism in MELAS (Mitochondrial Encephalomyopathy, Lactic Acidosis and Stroke-Like Episodes) Syndrome. PLoS One. 2015;10:e0127066. doi: 10.1371/journal.pone.0127066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Hattab AW, Scaglia F. Disorders of carnitine biosynthesis and transport. Mol Genet Metab. 2015;116:107–12. doi: 10.1016/j.ymgme.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Li JL, Wang QY, Luan HY, Kang ZC, Wang CB. Effects of L-carnitine against oxidative stress in human hepatocytes: involvement of peroxisome proliferator-activated receptor alpha. J Biomed Sci. 2012;19:32. doi: 10.1186/1423-0127-19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribas GS, Vargas CR, Wajner M. L-carnitine supplementation as a potential antioxidant therapy for inherited neurometabolic disorders. Gene. 2014;533:469–76. doi: 10.1016/j.gene.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 43.DiMauro S, Hirano M, Schon EA. Approaches to the treatment of mitochondrial diseases. Muscle Nerve. 2006;34:265–83. doi: 10.1002/mus.20598. [DOI] [PubMed] [Google Scholar]
- 44.Stanley CA. Carnitine deficiency disorders in children. Ann N Y Acad Sci. 2004;1033:42–51. doi: 10.1196/annals.1320.004. [DOI] [PubMed] [Google Scholar]
- 45.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joncquel-Chevalier Curt M, Voicu PM, Fontaine M, Dessein AF, Porchet N, Mention-Mulliez K, Dobbelaere D, Soto-Ares G, Cheillan D, Vamecq J. Creatine biosynthesis and transport in health and disease. Biochimie. 2015;119:146–65. doi: 10.1016/j.biochi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 47.Harris RC, Soderlund K, Hultman E. Elevation of creatine in resting and exercised muscle of normal subjects by creatine supplementation. Clin Sci (Lond) 1992;83:367–74. doi: 10.1042/cs0830367. [DOI] [PubMed] [Google Scholar]
- 48.Tarnopolsky MA, Parise G. Direct measurement of high-energy phosphate compounds in patients with neuromuscular disease. Muscle Nerve. 1999;22:1228–33. doi: 10.1002/(sici)1097-4598(199909)22:9<1228::aid-mus9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 49.DeBrosse C, Nanga RP, Wilson N, D’Aquilla K, Elliott M, Hariharan H, Yan F, Wade K, Nguyen S, Worsley D, Parris-Skeete C, McCormick E, Xiao R, Cunningham ZZ, Fishbein L, Nathanson KL, Lynch DR, Stallings VA, Yudkoff M, Falk MJ, Reddy R, McCormack SE. Muscle oxidative phosphorylation quantitation using creatine chemical exchange saturation transfer (CrCEST) MRI in mitochondrial disorders. JCI Insight. 2016;1:e88207. doi: 10.1172/jci.insight.88207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez MC, MacDonald JR, Mahoney DJ, Parise G, Beal MF, Tarnopolsky MA. Beneficial effects of creatine, CoQ10, and lipoic acid in mitochondrial disorders. Muscle Nerve. 2007;35:235–42. doi: 10.1002/mus.20688. [DOI] [PubMed] [Google Scholar]
- 51.Rauchova H, Drahota Z, Lenaz G. Function of coenzyme Q in the cell: some biochemical and physiological properties. Physiol Res. 1995;44:209–16. [PubMed] [Google Scholar]
- 52.Ernster L, Dallner G. Biochemical, physiological and medical aspects of ubiquinone function. Biochim Biophys Acta. 1995;1271:195–204. doi: 10.1016/0925-4439(95)00028-3. [DOI] [PubMed] [Google Scholar]
- 53.Duncan AJ, Heales SJ, Mills K, Eaton S, Land JM, Hargreaves IP. Determination of coenzyme Q10 status in blood mononuclear cells, skeletal muscle, and plasma by HPLC with di-propoxy-coenzyme Q10 as an internal standard. Clin Chem. 2005;51:2380–2. doi: 10.1373/clinchem.2005.054643. [DOI] [PubMed] [Google Scholar]
- 54.Tarnopolsky MA. The mitochondrial cocktail: rationale for combined nutraceutical therapy in mitochondrial cytopathies. Adv Drug Deliv Rev. 2008;60:1561–7. doi: 10.1016/j.addr.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Hargreaves IP. Coenzyme Q10 as a therapy for mitochondrial disease. Int J Biochem Cell Biol. 2014;49:105–11. doi: 10.1016/j.biocel.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 56.Haas RH. The evidence basis for coenzyme Q therapy in oxidative phosphorylation disease. Mitochondrion. 2007;(7 Suppl):S136–45. doi: 10.1016/j.mito.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 57.Scalori V, Alessandri MG, Giovannini L, Bertelli A. Plasma and tissue concentrations of coenzyme Q10 in the rat after intravenous, oral and topical administrations. Int J Tissue React. 1990;12:149–54. [PubMed] [Google Scholar]
- 58.Lass A, Forster MJ, Sohal RS. Effects of coenzyme Q10 and alpha-tocopherol administration on their tissue levels in the mouse: elevation of mitochondrial alpha-tocopherol by coenzyme Q10. Free Radic Biol Med. 1999;26:1375–82. doi: 10.1016/s0891-5849(98)00330-x. [DOI] [PubMed] [Google Scholar]
- 59.Dorsam B, Fahrer J. The disulfide compound alpha-lipoic acid and its derivatives: A novel class of anticancer agents targeting mitochondria. Cancer Lett. 2016;371:12–9. doi: 10.1016/j.canlet.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 60.Wada H, Shintani D, Ohlrogge J. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc Natl Acad Sci U S A. 1997;94:1591–6. doi: 10.1073/pnas.94.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rochette L, Ghibu S, Muresan A, Vergely C. Alpha-lipoic acid: molecular mechanisms and therapeutic potential in diabetes. Can J Physiol Pharmacol. 2015;93:1021–7. doi: 10.1139/cjpp-2014-0353. [DOI] [PubMed] [Google Scholar]
- 62.Rochette L, Ghibu S, Richard C, Zeller M, Cottin Y, Vergely C. Direct and indirect antioxidant properties of alpha-lipoic acid and therapeutic potential. Mol Nutr Food Res. 2013;57:114–25. doi: 10.1002/mnfr.201200608. [DOI] [PubMed] [Google Scholar]
- 63.Borowczyk K, Krawczyk M, Kubalczyk P, Chwatko G. Determination of lipoic acid in biological samples. Bioanalysis. 2015;7:1785–98. doi: 10.4155/bio.15.95. [DOI] [PubMed] [Google Scholar]
- 64.Rodriguez MC, MacDonald JR, Mahoney DJ, Parise G, Beal MF, Tarnopolsky MA. Beneficial effects of creatine, CoQ(10), and lipoic acid in mitochondrial disorders. Muscle & Nerve. 2007;35:235–42. doi: 10.1002/mus.20688. [DOI] [PubMed] [Google Scholar]
- 65.Gomes MB, Negrato CA. Alpha-lipoic acid as a pleiotropic compound with potential therapeutic use in diabetes and other chronic diseases. Diabetol Metab Syndr. 2014;6:80. doi: 10.1186/1758-5996-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pashaj A, Xia M, Moreau R. alpha-Lipoic acid as a triglyceride-lowering nutraceutical. Can J Physiol Pharmacol. 2015;93:1029–41. doi: 10.1139/cjpp-2014-0480. [DOI] [PubMed] [Google Scholar]
- 67.Jardim FR, de Rossi FT, Nascimento MX, da Silva Barros RG, Borges PA, Prescilio IC, de Oliveira MR. Resveratrol and Brain Mitochondria: a Review. Mol Neurobiol. 2017 doi: 10.1007/s12035-017-0448-z. [DOI] [PubMed] [Google Scholar]
- 68.Viscomi C, Bottani E, Zeviani M. Emerging concepts in the therapy of mitochondrial disease. Biochim Biophys Acta. 2015;1847:544–57. doi: 10.1016/j.bbabio.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 69.Sugden MC, Caton PW, Holness MJ. PPAR control: it’s SIRTainly as easy as PGC. J Endocrinol. 2010;204:93–104. doi: 10.1677/JOE-09-0359. [DOI] [PubMed] [Google Scholar]
- 70.Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, Kim MK, Beaven MA, Burgin AB, Manganiello V, Chung JH. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–33. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beher D, Wu J, Cumine S, Kim KW, Lu SC, Atangan L, Wang M. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem Biol Drug Des. 2009;74:619–24. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 72.Lopes Costa A, Le Bachelier C, Mathieu L, Rotig A, Boneh A, De Lonlay P, Tarnopolsky MA, Thorburn DR, Bastin J, Djouadi F. Beneficial effects of resveratrol on respiratory chain defects in patients’ fibroblasts involve estrogen receptor and estrogen-related receptor alpha signaling. Hum Mol Genet. 2014;23:2106–19. doi: 10.1093/hmg/ddt603. [DOI] [PubMed] [Google Scholar]
- 73.Hofer A, Noe N, Tischner C, Kladt N, Lellek V, Schauss A, Wenz T. Defining the action spectrum of potential PGC-1alpha activators on a mitochondrial and cellular level in vivo. Hum Mol Genet. 2014;23:2400–15. doi: 10.1093/hmg/ddt631. [DOI] [PubMed] [Google Scholar]
- 74.Bough KJ, Wetherington J, Hassel B, Pare JF, Gawryluk JW, Greene JG, Shaw R, Smith Y, Geiger JD, Dingledine RJ. Mitochondrial biogenesis in the anticonvulsant mechanism of the ketogenic diet. Ann Neurol. 2006;60:223–35. doi: 10.1002/ana.20899. [DOI] [PubMed] [Google Scholar]
- 75.Kossoff EH, Hartman AL. Ketogenic diets: new advances for metabolism-based therapies. Curr Opin Neurol. 2012;25:173–8. doi: 10.1097/WCO.0b013e3283515e4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang HC, Lee YM, Kim HD, Lee JS, Slama A. Safe and effective use of the ketogenic diet in children with epilepsy and mitochondrial respiratory chain complex defects. Epilepsia. 2007;48:82–8. doi: 10.1111/j.1528-1167.2006.00906.x. [DOI] [PubMed] [Google Scholar]
- 77.Sullivan PG, Rippy NA, Dorenbos K, Concepcion RC, Agarwal AK, Rho JM. The ketogenic diet increases mitochondrial uncoupling protein levels and activity. Ann Neurol. 2004;55:576–80. doi: 10.1002/ana.20062. [DOI] [PubMed] [Google Scholar]
- 78.Valentino ML, Barboni P, Ghelli A, Bucchi L, Rengo C, Achilli A, Torroni A, Lugaresi A, Lodi R, Barbiroli B, Dotti M, Federico A, Baruzzi A, Carelli V. The ND1 gene of complex I is a mutational hot spot for Leber’s hereditary optic neuropathy. Ann Neurol. 2004;56:631–41. doi: 10.1002/ana.20236. [DOI] [PubMed] [Google Scholar]
- 79.Nakano K, Tarashima M, Tachikawa E, Noda N, Nakayama T, Sasaki K, Mizoguchi E, Matsuzaki M, Osawa M. Platelet mitochondrial evaluation during cytochrome c and dichloroacetate treatments of MELAS. Mitochondrion. 2005;5:426–33. doi: 10.1016/j.mito.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 80.Lattanzi L, Salvatori G, Coletta M, Sonnino C, Cusella De Angelis MG, Gioglio L, Murry CE, Kelly R, Ferrari G, Molinaro M, Crescenzi M, Mavilio F, Cossu G. High efficiency myogenic conversion of human fibroblasts by adenoviral vector-mediated MyoD gene transfer. An alternative strategy for ex vivo gene therapy of primary myopathies. J Clin Invest. 1998;101:2119–28. doi: 10.1172/JCI1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saada A. Mitochondria: mitochondrial OXPHOS (dys) function ex vivo--the use of primary fibroblasts. Int J Biochem Cell Biol. 2014;48:60–5. doi: 10.1016/j.biocel.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 82.Lopez LC, Luna-Sanchez M, Garcia-Corzo L, Quinzii CM, Hirano M. Pathomechanisms in coenzyme q10-deficient human fibroblasts. Mol Syndromol. 2014;5:163–9. doi: 10.1159/000360494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lopez LC, Quinzii CM, Area E, Naini A, Rahman S, Schuelke M, Salviati L, Dimauro S, Hirano M. Treatment of CoQ(10) deficient fibroblasts with ubiquinone, CoQ analogs, and vitamin C: time- and compound-dependent effects. PLoS One. 2010;5:e11897. doi: 10.1371/journal.pone.0011897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bar-Meir M, Elpeleg ON, Saada A. Effect of various agents on adenosine triphosphate synthesis in mitochondrial complex I deficiency. J Pediatr. 2001;139:868–70. doi: 10.1067/mpd.2001.118885. [DOI] [PubMed] [Google Scholar]
- 85.Jauslin ML, Meier T, Smith RA, Murphy MP. Mitochondria-targeted antioxidants protect Friedreich Ataxia fibroblasts from endogenous oxidative stress more effectively than untargeted antioxidants. FASEB J. 2003;17:1972–4. doi: 10.1096/fj.03-0240fje. [DOI] [PubMed] [Google Scholar]
- 86.Suzuki T, Yamaguchi H, Kikusato M, Matsuhashi T, Matsuo A, Sato T, Oba Y, Watanabe S, Minaki D, Saigusa D, Shimbo H, Mori N, Mishima E, Shima H, Akiyama Y, Takeuchi Y, Yuri A, Kikuchi K, Toyohara T, Suzuki C, Kohzuki M, Anzai J, Mano N, Kure S, Yanagisawa T, Tomioka Y, Toyomizu M, Ito S, Osaka H, Hayashi K, Abe T. Mitochonic Acid 5 (MA-5), a Derivative of the Plant Hormone Indole-3-Acetic Acid, Improves Survival of Fibroblasts from Patients with Mitochondrial Diseases. Tohoku J Exp Med. 2015;236:225–32. doi: 10.1620/tjem.236.225. [DOI] [PubMed] [Google Scholar]
- 87.King MP, Koga Y, Davidson M, Schon EA. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol Cell Biol. 1992;12:480–90. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Masucci JP, Davidson M, Koga Y, Schon EA, King MP. In vitro analysis of mutations causing myoclonus epilepsy with ragged-red fibers in the mitochondrial tRNA(Lys)gene: two genotypes produce similar phenotypes. Mol Cell Biol. 1995;15:2872–81. doi: 10.1128/mcb.15.5.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jun AS, Trounce IA, Brown MD, Shoffner JM, Wallace DC. Use of transmitochondrial cybrids to assign a complex I defect to the mitochondrial DNA-encoded NADH dehydrogenase subunit 6 gene mutation at nucleotide pair 14459 that causes Leber hereditary optic neuropathy and dystonia. Mol Cell Biol. 1996;16:771–7. doi: 10.1128/mcb.16.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ishikawa K, Hayashi J. Generation of mtDNA-exchanged cybrids for determination of the effects of mtDNA mutations on tumor phenotypes. Methods Enzymol. 2009;457:335–46. doi: 10.1016/S0076-6879(09)05019-8. [DOI] [PubMed] [Google Scholar]
- 91.Petrova-Benedict R, Buncic JR, Wallace DC, Robinson BH. Selective killing of cells with oxidative defects in galactose medium: a screening test for affected patient fibroblasts. J Inherit Metab Dis. 1992;15:943–4. doi: 10.1007/BF01800243. [DOI] [PubMed] [Google Scholar]
- 92.D’Aurelio M, Pallotti F, Barrientos A, Gajewski CD, Kwong JQ, Bruno C, Beal MF, Manfredi G. In vivo regulation of oxidative phosphorylation in cells harboring a stop-codon mutation in mitochondrial DNA-encoded cytochrome c oxidase subunit I. J Biol Chem. 2001;276:46925–32. doi: 10.1074/jbc.M106429200. [DOI] [PubMed] [Google Scholar]
- 93.Santra S, Gilkerson RW, Davidson M, Schon EA. Ketogenic treatment reduces deleted mitochondrial DNAs in cultured human cells. Ann Neurol. 2004;56:662–9. doi: 10.1002/ana.20240. [DOI] [PubMed] [Google Scholar]
- 94.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–40. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Desquiret-Dumas V, Gueguen N, Barth M, Chevrollier A, Hancock S, Wallace DC, Amati-Bonneau P, Henrion D, Bonneau D, Reynier P, Procaccio V. Metabolically induced heteroplasmy shifting and l-arginine treatment reduce the energetic defect in a neuronal-like model of MELAS. Biochim Biophys Acta. 2012;1822:1019–29. doi: 10.1016/j.bbadis.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sgarbi G, Casalena GA, Baracca A, Lenaz G, DiMauro S, Solaini G. Human NARP mitochondrial mutation metabolism corrected with alpha-ketoglutarate/aspartate: a potential new therapy. Arch Neurol. 2009;66:951–7. doi: 10.1001/archneurol.2009.134. [DOI] [PubMed] [Google Scholar]
- 97.Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, Chandel NS, DeBerardinis RJ. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7:1679–90. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garrido-Maraver J, Cordero MD, Monino ID, Pereira-Arenas S, Lechuga-Vieco AV, Cotan D, De la Mata M, Oropesa-Avila M, De Miguel M, Bautista Lorite J, Rivas Infante E, Alvarez-Dolado M, Navas P, Jackson S, Francisci S, Sanchez-Alcazar JA. Screening of effective pharmacological treatments for MELAS syndrome using yeasts, fibroblasts and cybrid models of the disease. Br J Pharmacol. 2012;167:1311–28. doi: 10.1111/j.1476-5381.2012.02086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mattiazzi M, Vijayvergiya C, Gajewski CD, DeVivo DC, Lenaz G, Wiedmann M, Manfredi G. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum Mol Genet. 2004;13:869–79. doi: 10.1093/hmg/ddh103. [DOI] [PubMed] [Google Scholar]
- 100.Prigione A, Lichtner B, Kuhl H, Struys EA, Wamelink M, Lehrach H, Ralser M, Timmermann B, Adjaye J. Human induced pluripotent stem cells harbor homoplasmic and heteroplasmic mitochondrial DNA mutations while maintaining human embryonic stem cell-like metabolic reprogramming. Stem Cells. 2011;29:1338–48. doi: 10.1002/stem.683. [DOI] [PubMed] [Google Scholar]
- 101.Hamalainen RH, Suomalainen A. Generation and Characterization of Induced Pluripotent Stem Cells from Patients with mtDNA Mutations. Methods Mol Biol. 2016;1353:65–75. doi: 10.1007/7651_2015_258. [DOI] [PubMed] [Google Scholar]
- 102.Hatakeyama H, Katayama A, Komaki H, Nishino I, Goto Y. Molecular pathomechanisms and cell-type-specific disease phenotypes of MELAS caused by mutant mitochondrial tRNA(Trp) Acta Neuropathol Commun. 2015;3:52. doi: 10.1186/s40478-015-0227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hick A, Wattenhofer-Donze M, Chintawar S, Tropel P, Simard JP, Vaucamps N, Gall D, Lambot L, Andre C, Reutenauer L, Rai M, Teletin M, Messaddeq N, Schiffmann SN, Viville S, Pearson CE, Pandolfo M, Puccio H. Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich’s ataxia. Dis Model Mech. 2013;6:608–21. doi: 10.1242/dmm.010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Folmes CD, Martinez-Fernandez A, Perales-Clemente E, Li X, McDonald A, Oglesbee D, Hrstka SC, Perez-Terzic C, Terzic A, Nelson TJ. Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cells. 2013;31:1298–308. doi: 10.1002/stem.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma H, Folmes CD, Wu J, Morey R, Mora-Castilla S, Ocampo A, Ma L, Poulton J, Wang X, Ahmed R, Kang E, Lee Y, Hayama T, Li Y, Van Dyken C, Gutierrez NM, Tippner-Hedges R, Koski A, Mitalipov N, Amato P, Wolf DP, Huang T, Terzic A, Laurent LC, Izpisua Belmonte JC, Mitalipov S. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nature. 2015;524:234–8. doi: 10.1038/nature14546. [DOI] [PubMed] [Google Scholar]
- 106.Schatz G. The Isolation of Possible Mitochondrial Precursor Structures from Aerobically Grown Baker’s Yeast. Biochem Biophys Res Commun. 1963;12:448–51. doi: 10.1016/0006-291x(63)90313-9. [DOI] [PubMed] [Google Scholar]
- 107.Reid GA, Schatz G. Import of proteins into mitochondria. Extramitochondrial pools and post-translational import of mitochondrial protein precursors in vivo. J Biol Chem. 1982;257:13062–7. [PubMed] [Google Scholar]
- 108.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baruffini E, Ferrero I, Foury F. Mitochondrial DNA defects in Saccharomyces cerevisiae caused by functional interactions between DNA polymerase gamma mutations associated with disease in human. Biochim Biophys Acta. 2007;1772:1225–35. doi: 10.1016/j.bbadis.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 110.Lasserre JP, Dautant A, Aiyar RS, Kucharczyk R, Glatigny A, Tribouillard-Tanvier D, Rytka J, Blondel M, Skoczen N, Reynier P, Pitayu L, Rotig A, Delahodde A, Steinmetz LM, Dujardin G, Procaccio V, di Rago JP. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis Model Mech. 2015;8:509–26. doi: 10.1242/dmm.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bulder CJ. Lethality of the Petite Mutation in Petite Negative Yeasts. Antonie Van Leeuwenhoek. 1964;30:442–54. doi: 10.1007/BF02046758. [DOI] [PubMed] [Google Scholar]
- 112.Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, Westermann B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:847–53. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barrientos A. Yeast models of human mitochondrial diseases. IUBMB Life. 2003;55:83–95. doi: 10.1002/tbmb.718540876. [DOI] [PubMed] [Google Scholar]
- 114.Bonnefoy N, Fox TD. Genetic transformation of Saccharomyces cerevisiae mitochondria. Methods Cell Biol. 2001;65:381–96. doi: 10.1016/s0091-679x(01)65022-2. [DOI] [PubMed] [Google Scholar]
- 115.Claypool SM, Boontheung P, McCaffery JM, Loo JA, Koehler CM. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol Biol Cell. 2008;19:5143–55. doi: 10.1091/mbc.E08-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barrientos A, Korr D, Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh’s syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mashkevich G, Repetto B, Glerum DM, Jin C, Tzagoloff A. SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J Biol Chem. 1997;272:14356–64. doi: 10.1074/jbc.272.22.14356. [DOI] [PubMed] [Google Scholar]
- 118.Coppola M, Pizzigoni A, Banfi S, Bassi MT, Casari G, Incerti B. Identification and characterization of YME1L1, a novel paraplegin-related gene. Genomics. 2000;66:48–54. doi: 10.1006/geno.2000.6136. [DOI] [PubMed] [Google Scholar]
- 119.Kerscher SJ. Diversity and origin of alternative NADH:ubiquinone oxidoreductases. Biochim Biophys Acta. 2000;1459:274–83. doi: 10.1016/s0005-2728(00)00162-6. [DOI] [PubMed] [Google Scholar]
- 120.Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest. 2005;115:2784–92. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schwimmer C, Rak M, Lefebvre-Legendre L, Duvezin-Caubet S, Plane G, di Rago JP. Yeast models of human mitochondrial diseases: from molecular mechanisms to drug screening. Biotechnol J. 2006;1:270–81. doi: 10.1002/biot.200500053. [DOI] [PubMed] [Google Scholar]
- 122.Cheng WC, Leach KM, Hardwick JM. Mitochondrial death pathways in yeast and mammalian cells. Biochim Biophys Acta. 2008;1783:1272–9. doi: 10.1016/j.bbamcr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bassett DE, Jr, Boguski MS, Hieter P. Yeast genes and human disease. Nature. 1996;379:589–90. doi: 10.1038/379589a0. [DOI] [PubMed] [Google Scholar]
- 124.Shadel GS, Clayton DA. Mitochondrial transcription initiation. Variation and conservation. J Biol Chem. 1993;268:16083–6. [PubMed] [Google Scholar]
- 125.Shadel GS. Yeast as a model for human mtDNA replication. Am J Hum Genet. 1999;65:1230–7. doi: 10.1086/302630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rea SL, Graham BH, Nakamaru-Ogiso E, Kar A, Falk MJ. Bacteria, yeast, worms, and flies: exploiting simple model organisms to investigate human mitochondrial diseases. Dev Disabil Res Rev. 2010;16:200–18. doi: 10.1002/ddrr.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hartman PS, Ishii N, Kayser EB, Morgan PG, Sedensky MM. Mitochondrial mutations differentially affect aging, mutability and anesthetic sensitivity in Caenorhabditis elegans. Mech Ageing Dev. 2001;122:1187–201. doi: 10.1016/s0047-6374(01)00259-7. [DOI] [PubMed] [Google Scholar]
- 129.McCormack S, Polyak E, Ostrovsky J, Dingley SD, Rao M, Kwon YJ, Xiao R, Zhang Z, Nakamaru-Ogiso E, Falk MJ. Pharmacologic targeting of sirtuin and PPAR signaling improves longevity and mitochondrial physiology in respiratory chain complex I mutant Caenorhabditis elegans. Mitochondrion. 2015;22:45–59. doi: 10.1016/j.mito.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Consortium CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–8. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 131.Sonnhammer EL, Durbin R. Analysis of protein domain families in Caenorhabditis elegans. Genomics. 1997;46:200–16. doi: 10.1006/geno.1997.4989. [DOI] [PubMed] [Google Scholar]
- 132.Kuwabara PE, O’Neil N. The use of functional genomics in C. elegans for studying human development and disease. J Inherit Metab Dis. 2001;24:127–38. doi: 10.1023/a:1010306731764. [DOI] [PubMed] [Google Scholar]
- 133.Maglioni S, Ventura N. C. elegans as a model organism for human mitochondrial associated disorders. Mitochondrion. 2016;30:117–25. doi: 10.1016/j.mito.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 134.Grad LI, Lemire BD. Riboflavin enhances the assembly of mitochondrial cytochrome c oxidase in C. elegans NADH-ubiquinone oxidoreductase mutants. Biochim Biophys Acta. 2006;1757:115–22. doi: 10.1016/j.bbabio.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 135.Sen A, Cox RT. Fly Models of Human Diseases: Drosophila as a Model for Understanding Human Mitochondrial Mutations and Disease. Curr Top Dev Biol. 2017;121:1–27. doi: 10.1016/bs.ctdb.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 136.Chen Z, Qi Y, French S, Zhang G, Covian Garcia R, Balaban R, Xu H. Genetic mosaic analysis of a deleterious mitochondrial DNA mutation in Drosophila reveals novel aspects of mitochondrial regulation and function. Mol Biol Cell. 2015;26:674–84. doi: 10.1091/mbc.E14-11-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Meiklejohn CD, Holmbeck MA, Siddiq MA, Abt DN, Rand DM, Montooth KL. An Incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 2013;9:e1003238. doi: 10.1371/journal.pgen.1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Holmbeck MA, Donner JR, Villa-Cuesta E, Rand DM. A Drosophila model for mito-nuclear diseases generated by an incompatible interaction between tRNA and tRNA synthetase. Dis Model Mech. 2015;8:843–54. doi: 10.1242/dmm.019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Anderson PR, Kirby K, Hilliker AJ, Phillips JP. RNAi-mediated suppression of the mitochondrial iron chaperone, frataxin, in Drosophila. Hum Mol Genet. 2005;14:3397–405. doi: 10.1093/hmg/ddi367. [DOI] [PubMed] [Google Scholar]
- 140.Bayat V, Thiffault I, Jaiswal M, Tetreault M, Donti T, Sasarman F, Bernard G, Demers-Lamarche J, Dicaire MJ, Mathieu J, Vanasse M, Bouchard JP, Rioux MF, Lourenco CM, Li Z, Haueter C, Shoubridge EA, Graham BH, Brais B, Bellen HJ. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Da-Re C, von Stockum S, Biscontin A, Millino C, Cisotto P, Zordan MA, Zeviani M, Bernardi P, De Pitta C, Costa R. Leigh syndrome in Drosophila melanogaster: morphological and biochemical characterization of Surf1 post-transcriptional silencing. J Biol Chem. 2014;289:29235–46. doi: 10.1074/jbc.M114.602938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 143.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 144.Toivonen JM, O’Dell KM, Petit N, Irvine SC, Knight GK, Lehtonen M, Longmuir M, Luoto K, Touraille S, Wang Z, Alziari S, Shah ZH, Jacobs HT. Technical knockout, a Drosophila model of mitochondrial deafness. Genetics. 2001;159:241–54. doi: 10.1093/genetics/159.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pesah Y, Pham T, Burgess H, Middlebrooks B, Verstreken P, Zhou Y, Harding M, Bellen H, Mardon G. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131:2183–94. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- 146.Steele SL, Prykhozhij SV, Berman JN. Zebrafish as a model system for mitochondrial biology and diseases. Transl Res. 2014;163:79–98. doi: 10.1016/j.trsl.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 147.Broughton RE, Milam JE, Roe BA. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 2001;11:1958–67. doi: 10.1101/gr.156801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 149.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–8. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stewart AM, Braubach O, Spitsbergen J, Gerlai R, Kalueff AV. Zebrafish models for translational neuroscience research: from tank to bedside. Trends Neurosci. 2014;37:264–78. doi: 10.1016/j.tins.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sager JJ, Bai Q, Burton EA. Transgenic zebrafish models of neurodegenerative diseases. Brain Struct Funct. 2010;214:285–302. doi: 10.1007/s00429-009-0237-1. [DOI] [PubMed] [Google Scholar]
- 153.Volkoff H, Peter RE. Feeding behavior of fish and its control. Zebrafish. 2006;3:131–40. doi: 10.1089/zeb.2006.3.131. [DOI] [PubMed] [Google Scholar]
- 154.Makky K, Duvnjak P, Pramanik K, Ramchandran R, Mayer AN. A whole-animal microplate assay for metabolic rate using zebrafish. J Biomol Screen. 2008;13:960–7. doi: 10.1177/1087057108326080. [DOI] [PubMed] [Google Scholar]
- 155.Vempati UD, Torraco A, Moraes CT. Mouse models of oxidative phosphorylation dysfunction and disease. Methods. 2008;46:241–7. doi: 10.1016/j.ymeth.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Torraco A, Peralta S, Iommarini L, Diaz F. Mitochondrial Diseases Part I: mouse models of OXPHOS deficiencies caused by defects in respiratory complex subunits or assembly factors. Mitochondrion. 2015;21:76–91. doi: 10.1016/j.mito.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Iommarini L, Peralta S, Torraco A, Diaz F. Mitochondrial Diseases Part II: Mouse models of OXPHOS deficiencies caused by defects in regulatory factors and other components required for mitochondrial function. Mitochondrion. 2015;22:96–118. doi: 10.1016/j.mito.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–35. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 159.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–25. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 160.Fisher RP, Clayton DA. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J Biol Chem. 1985;260:11330–8. [PubMed] [Google Scholar]
- 161.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–6. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 162.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–7. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Larsson NG, Rustin P. Animal models for respiratory chain disease. Trends Mol Med. 2001;7:578–81. doi: 10.1016/s1471-4914(01)02167-0. [DOI] [PubMed] [Google Scholar]
- 164.Torraco A, Diaz F, Vempati UD, Moraes CT. Mouse models of oxidative phosphorylation defects: powerful tools to study the pathobiology of mitochondrial diseases. Biochim Biophys Acta. 2009;1793:171–80. doi: 10.1016/j.bbamcr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li H, Wang J, Wilhelmsson H, Hansson A, Thoren P, Duffy J, Rustin P, Larsson NG. Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc Natl Acad Sci U S A. 2000;97:3467–72. doi: 10.1073/pnas.97.7.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson NG. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21:133–7. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 167.Hansson A, Hance N, Dufour E, Rantanen A, Hultenby K, Clayton DA, Wibom R, Larsson NG. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc Natl Acad Sci U S A. 2004;101:3136–41. doi: 10.1073/pnas.0308710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gineste C, Hernandez A, Ivarsson N, Cheng AJ, Naess K, Wibom R, Lesko N, Bruhn H, Wedell A, Freyer C, Zhang SJ, Carlstrom M, Lanner JT, Andersson DC, Bruton JD, Wredenberg A, Westerblad H. Cyclophilin D, a target for counteracting skeletal muscle dysfunction in mitochondrial myopathy. Hum Mol Genet. 2015;24:6580–7. doi: 10.1093/hmg/ddv361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol Cell Biol. 2002;22:1116–25. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–94. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 171.Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum Mol Genet. 2009;18:2670–82. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–97. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 173.McKay SE, Yan W, Nouws J, Thormann MJ, Raimundo N, Khan A, Santos-Sacchi J, Song L, Shadel GS. Auditory Pathology in a Transgenic mtTFB1 Mouse Model of Mitochondrial Deafness. Am J Pathol. 2015;185:3132–40. doi: 10.1016/j.ajpath.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Raimundo N, Song L, Shutt TE, McKay SE, Cotney J, Guan MX, Gilliland TC, Hohuan D, Santos-Sacchi J, Shadel GS. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell. 2012;148:716–26. doi: 10.1016/j.cell.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–23. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 176.Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–4. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 177.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–4. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 178.Saleem A, Safdar A, Kitaoka Y, Ma X, Marquez OS, Akhtar M, Nazli A, Suri R, Turnbull J, Tarnopolsky MA. Polymerase gamma mutator mice rely on increased glycolytic flux for energy production. Mitochondrion. 2015;21:19–26. doi: 10.1016/j.mito.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 179.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39:540–3. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 180.Dai DF, Chen T, Wanagat J, Laflamme M, Marcinek DJ, Emond MJ, Ngo CP, Prolla TA, Rabinovitch PS. Age-dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging Cell. 2010;9:536–44. doi: 10.1111/j.1474-9726.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hamalainen RH, Ahlqvist KJ, Ellonen P, Lepisto M, Logan A, Otonkoski T, Murphy MP, Suomalainen A. mtDNA Mutagenesis Disrupts Pluripotent Stem Cell Function by Altering Redox Signaling. Cell Rep. 2015;11:1614–24. doi: 10.1016/j.celrep.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shabalina IG, Vyssokikh MY, Gibanova N, Csikasz RI, Edgar D, Hallden-Waldemarson A, Rozhdestvenskaya Z, Bakeeva LE, Vays VB, Pustovidko AV, Skulachev MV, Cannon B, Skulachev VP, Nedergaard J. Improved health-span and lifespan in mtDNA mutator mice treated with the mitochondrially targeted antioxidant SkQ1. Aging (Albany NY) 2017;9:315–39. doi: 10.18632/aging.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Safdar A, Bourgeois JM, Ogborn DI, Little JP, Hettinga BP, Akhtar M, Thompson JE, Melov S, Mocellin NJ, Kujoth GC, Prolla TA, Tarnopolsky MA. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc Natl Acad Sci U S A. 2011;108:4135–40. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yadak R, Sillevis Smitt P, van Gisbergen MW, van Til NP, de Coo IF. Mitochondrial Neurogastrointestinal Encephalomyopathy Caused by Thymidine Phosphorylase Enzyme Deficiency: From Pathogenesis to Emerging Therapeutic Options. Front Cell Neurosci. 2017;11:31. doi: 10.3389/fncel.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Haraguchi M, Tsujimoto H, Fukushima M, Higuchi I, Kuribayashi H, Utsumi H, Nakayama A, Hashizume Y, Hirato J, Yoshida H, Hara H, Hamano S, Kawaguchi H, Furukawa T, Miyazono K, Ishikawa F, Toyoshima H, Kaname T, Komatsu M, Chen ZS, Gotanda T, Tachiwada T, Sumizawa T, Miyadera K, Osame M, Yoshida H, Noda T, Yamada Y, Akiyama S. Targeted deletion of both thymidine phosphorylase and uridine phosphorylase and consequent disorders in mice. Mol Cell Biol. 2002;22:5212–21. doi: 10.1128/MCB.22.14.5212-5221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lopez LC, Akman HO, Garcia-Cazorla A, Dorado B, Marti R, Nishino I, Tadesse S, Pizzorno G, Shungu D, Bonilla E, Tanji K, Hirano M. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum Mol Genet. 2009;18:714–22. doi: 10.1093/hmg/ddn401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Marchington DR, Barlow D, Poulton J. Transmitochondrial mice carrying resistance to chloramphenicol on mitochondrial DNA: developing the first mouse model of mitochondrial DNA disease. Nat Med. 1999;5:957–60. doi: 10.1038/11403. [DOI] [PubMed] [Google Scholar]
- 188.Sligh JE, Levy SE, Waymire KG, Allard P, Dillehay DL, Nusinowitz S, Heckenlively JR, MacGregor GR, Wallace DC. Maternal germ-line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc Natl Acad Sci U S A. 2000;97:14461–6. doi: 10.1073/pnas.250491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–62. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yokota M, Shitara H, Hashizume O, Ishikawa K, Nakada K, Ishii R, Taya C, Takenaga K, Yonekawa H, Hayashi J. Generation of trans-mitochondrial mito-mice by the introduction of a pathogenic G13997A mtDNA from highly metastatic lung carcinoma cells. FEBS Lett. 2010;584:3943–8. doi: 10.1016/j.febslet.2010.07.048. [DOI] [PubMed] [Google Scholar]
- 191.Shimizu A, Mito T, Hayashi C, Ogasawara E, Koba R, Negishi I, Takenaga K, Nakada K, Hayashi J. Transmitochondrial mice as models for primary prevention of diseases caused by mutation in the tRNA(Lys) gene. Proc Natl Acad Sci U S A. 2014;111:3104–9. doi: 10.1073/pnas.1318109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Inoue K, Nakada K, Ogura A, Isobe K, Goto Y, Nonaka I, Hayashi JI. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat Genet. 2000;26:176–81. doi: 10.1038/82826. [DOI] [PubMed] [Google Scholar]
- 193.Nakada K, Sato A, Sone H, Kasahara A, Ikeda K, Kagawa Y, Yonekawa H, Hayashi J. Accumulation of pathogenic DeltamtDNA induced deafness but not diabetic phenotypes in mito-mice. Biochem Biophys Res Commun. 2004;323:175–84. doi: 10.1016/j.bbrc.2004.08.073. [DOI] [PubMed] [Google Scholar]
- 194.Levy SE, Waymire KG, Kim YL, MacGregor GR, Wallace DC. Transfer of chloramphenicol-resistant mitochondrial DNA into the chimeric mouse. Transgenic Res. 1999;8:137–45. doi: 10.1023/a:1008967412955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kasahara A, Ishikawa K, Yamaoka M, Ito M, Watanabe N, Akimoto M, Sato A, Nakada K, Endo H, Suda Y, Aizawa S, Hayashi J. Generation of trans-mitochondrial mice carrying homoplasmic mtDNAs with a missense mutation in a structural gene using ES cells. Hum Mol Genet. 2006;15:871–81. doi: 10.1093/hmg/ddl005. [DOI] [PubMed] [Google Scholar]
- 196.Shimizu A, Mito T, Hashizume O, Yonekawa H, Ishikawa K, Nakada K, Hayashi J. G7731A mutation in mouse mitochondrial tRNALys regulates late-onset disorders in transmitochondrial mice. Biochem Biophys Res Commun. 2015;459:66–70. doi: 10.1016/j.bbrc.2015.02.070. [DOI] [PubMed] [Google Scholar]
- 197.Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2010;1797:113–28. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 198.Sato A, Kono T, Nakada K, Ishikawa K, Inoue S, Yonekawa H, Hayashi J. Gene therapy for progeny of mito-mice carrying pathogenic mtDNA by nuclear transplantation. Proc Natl Acad Sci U S A. 2005;102:16765–70. doi: 10.1073/pnas.0506197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yamanashi H, Hashizume O, Yonekawa H, Nakada K, Hayashi J. Administration of an antioxidant prevents lymphoma development in transmitochondrial mice overproducing reactive oxygen species. Exp Anim. 2014;63:459–66. doi: 10.1538/expanim.63.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Pinkert CA, Trounce IA. Production of transmitochondrial mice. Methods. 2002;26:348–57. doi: 10.1016/S1046-2023(02)00041-5. [DOI] [PubMed] [Google Scholar]
- 201.Kauppila JH, Baines HL, Bratic A, Simard ML, Freyer C, Mourier A, Stamp C, Filograna R, Larsson NG, Greaves LC, Stewart JB. A Phenotype-Driven Approach to Generate Mouse Models with Pathogenic mtDNA Mutations Causing Mitochondrial Disease. Cell Rep. 2016;16:2980–90. doi: 10.1016/j.celrep.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kaukonen J, Juselius JK, Tiranti V, Kyttala A, Zeviani M, Comi GP, Keranen S, Peltonen L, Suomalainen A. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–5. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- 203.Longley MJ, Clark S, Yu Wai Man C, Hudson G, Durham SE, Taylor RW, Nightingale S, Turnbull DM, Copeland WC, Chinnery PF. Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am J Hum Genet. 2006;78:1026–34. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–31. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 205.Tyynismaa H, Suomalainen A. Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep. 2009;10:137–43. doi: 10.1038/embor.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci U S A. 2005;102:17687–92. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkila S, Wenz T, Ruhanen H, Guse K, Hemminki A, Peltola-Mjosund KE, Tulkki V, Oresic M, Moraes CT, Pietilainen K, Hovatta I, Suomalainen A. Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet. 2010;19:3948–58. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 208.Davis RL, Liang C, Edema-Hildebrand F, Riley C, Needham M, Sue CM. Fibroblast growth factor 21 is a sensitive biomarker of mitochondrial disease. Neurology. 2013;81:1819–26. doi: 10.1212/01.wnl.0000436068.43384.ef. [DOI] [PubMed] [Google Scholar]
- 209.Suomalainen A, Elo JM, Pietilainen KH, Hakonen AH, Sevastianova K, Korpela M, Isohanni P, Marjavaara SK, Tyni T, Kiuru-Enari S, Pihko H, Darin N, Ounap K, Kluijtmans LA, Paetau A, Buzkova J, Bindoff LA, Annunen-Rasila J, Uusimaa J, Rissanen A, Yki-Jarvinen H, Hirano M, Tulinius M, Smeitink J, Tyynismaa H. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 2011;10:806–18. doi: 10.1016/S1474-4422(11)70155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lehtonen J, Forsstrom S, Viscomi C, Zeviani M, Moraes CT, Nakada K, Smeitink J, Wiesner RJ, Baris O, Isoniemi H, Hockerstedt K, Osterlund P, Hurme M, Jylhava J, Leppa S, Markkula R, Helio T, Mombelli G, Suomalainen Wariovaara A. Mitochondrial myopathy biomarker Fibroblast growth factor 21 is induced by muscle mtDNA instability and translation defects. Mitochondrion. 2015;24:S45–S46. [Google Scholar]
- 211.Nikkanen J, Forsstrom S, Euro L, Paetau I, Kohnz RA, Wang L, Chilov D, Viinamaki J, Roivainen A, Marjamaki P, Liljenback H, Ahola S, Buzkova J, Terzioglu M, Khan NA, Pirnes-Karhu S, Paetau A, Lonnqvist T, Sajantila A, Isohanni P, Tyynismaa H, Nomura DK, Battersby BJ, Velagapudi V, Carroll CJ, Suomalainen A. Mitochondrial DNA Replication Defects Disturb Cellular dNTP Pools and Remodel One-Carbon Metabolism. Cell Metab. 2016;23:635–48. doi: 10.1016/j.cmet.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 212.Ahola-Erkkila S, Carroll CJ, Peltola-Mjosund K, Tulkki V, Mattila I, Seppanen-Laakso T, Oresic M, Tyynismaa H, Suomalainen A. Ketogenic diet slows down mitochondrial myopathy progression in mice. Hum Mol Genet. 2010;19:1974–84. doi: 10.1093/hmg/ddq076. [DOI] [PubMed] [Google Scholar]
- 213.Ahola-Erkkila S, Auranen M, Isohanni P, Lundbom N, Piirila P, Pietilainen K, Suomalainen A. Pilot study: Modified Aktins diet trial for adult-onset mitochondrial myopathy. Mitochondrion. 2013;13:911. [Google Scholar]
- 214.Budde SM, van den Heuvel LP, Smeets RJ, Skladal D, Mayr JA, Boelen C, Petruzzella V, Papa S, Smeitink JA. Clinical heterogeneity in patients with mutations in the NDUFS4 gene of mitochondrial complex I. J Inherit Metab Dis. 2003;26:813–5. doi: 10.1023/b:boli.0000010003.14113.af. [DOI] [PubMed] [Google Scholar]
- 215.Petruzzella V, Vergari R, Puzziferri I, Boffoli D, Lamantea E, Zeviani M, Papa S. A nonsense mutation in the NDUFS4 gene encoding the 18 kDa (AQDQ) subunit of complex I abolishes assembly and activity of the complex in a patient with Leigh-like syndrome. Hum Mol Genet. 2001;10:529–35. doi: 10.1093/hmg/10.5.529. [DOI] [PubMed] [Google Scholar]
- 216.Ugalde C, Janssen RJ, van den Heuvel LP, Smeitink JA, Nijtmans LG. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum Mol Genet. 2004;13:659–67. doi: 10.1093/hmg/ddh071. [DOI] [PubMed] [Google Scholar]
- 217.Kruse SE, Watt WC, Marcinek DJ, Kapur RP, Schenkman KA, Palmiter RD. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab. 2008;7:312–20. doi: 10.1016/j.cmet.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Quintana A, Kruse SE, Kapur RP, Sanz E, Palmiter RD. Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc Natl Acad Sci U S A. 2010;107:10996–1001. doi: 10.1073/pnas.1006214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A, Uhde L, Hui J, Wall VZ, Gagnidze A, Oh K, Wasko BM, Ramos FJ, Palmiter RD, Rabinovitch PS, Morgan PG, Sedensky MM, Kaeberlein M. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342:1524–8. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.McDaniel SS, Rensing NR, Thio LL, Yamada KA, Wong M. The ketogenic diet inhibits the mammalian target of rapamycin (mTOR) pathway. Epilepsia. 2011;52:e7–11. doi: 10.1111/j.1528-1167.2011.02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ingraham CA, Burwell LS, Skalska J, Brookes PS, Howell RL, Sheu SS, Pinkert CA. NDUFS4: creation of a mouse model mimicking a Complex I disorder. Mitochondrion. 2009;9:204–10. doi: 10.1016/j.mito.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Quinzii CM, Hirano M, DiMauro S. CoQ10 deficiency diseases in adults. Mitochondrion. 2007;(7 Suppl):S122–6. doi: 10.1016/j.mito.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lopez LC, Schuelke M, Quinzii CM, Kanki T, Rodenburg RJ, Naini A, Dimauro S, Hirano M. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet. 2006;79:1125–9. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Quinzii CM, Garone C, Emmanuele V, Tadesse S, Krishna S, Dorado B, Hirano M. Tissue-specific oxidative stress and loss of mitochondria in CoQ-deficient Pdss2 mutant mice. FASEB J. 2013;27:612–21. doi: 10.1096/fj.12-209361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Peng M, Falk MJ, Haase VH, King R, Polyak E, Selak M, Yudkoff M, Hancock WW, Meade R, Saiki R, Lunceford AL, Clarke CF, Gasser DL. Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS Genet. 2008;4:e1000061. doi: 10.1371/journal.pgen.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Falk MJ, Polyak E, Zhang Z, Peng M, King R, Maltzman JS, Okwuego E, Horyn O, Nakamaru-Ogiso E, Ostrovsky J, Xie LX, Chen JY, Marbois B, Nissim I, Clarke CF, Gasser DL. Probucol ameliorates renal and metabolic sequelae of primary CoQ deficiency in Pdss2 mutant mice. EMBO Mol Med. 2011;3:410–27. doi: 10.1002/emmm.201100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Saiki R, Lunceford AL, Shi Y, Marbois B, King R, Pachuski J, Kawamukai M, Gasser DL, Clarke CF. Coenzyme Q10 supplementation rescues renal disease in Pdss2kd/kd mice with mutations in prenyl diphosphate synthase subunit 2. Am J Physiol Renal Physiol. 2008;295:F1535–44. doi: 10.1152/ajprenal.90445.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ades LC, Gedeon AK, Wilson MJ, Latham M, Partington MW, Mulley JC, Nelson J, Lui K, Sillence DO. Barth syndrome: clinical features and confirmation of gene localisation to distal Xq28. Am J Med Genet. 1993;45:327–34. doi: 10.1002/ajmg.1320450309. [DOI] [PubMed] [Google Scholar]
- 229.Vreken P, Valianpour F, Nijtmans LG, Grivell LA, Plecko B, Wanders RJ, Barth PG. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Commun. 2000;279:378–82. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- 230.Soustek MS, Falk DJ, Mah CS, Toth MJ, Schlame M, Lewin AS, Byrne BJ. Characterization of a transgenic short hairpin RNA-induced murine model of Tafazzin deficiency. Hum Gene Ther. 2011;22:865–71. doi: 10.1089/hum.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Acehan D, Vaz F, Houtkooper RH, James J, Moore V, Tokunaga C, Kulik W, Wansapura J, Toth MJ, Strauss A, Khuchua Z. Cardiac and skeletal muscle defects in a mouse model of human Barth syndrome. J Biol Chem. 2011;286:899–908. doi: 10.1074/jbc.M110.171439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kiebish MA, Yang K, Liu X, Mancuso DJ, Guan S, Zhao Z, Sims HF, Cerqua R, Cade WT, Han X, Gross RW. Dysfunctional cardiac mitochondrial bioenergetic, lipidomic, and signaling in a murine model of Barth syndrome. J Lipid Res. 2013;54:1312–25. doi: 10.1194/jlr.M034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ferreira C, Thompson R, Vernon H. Barth Syndrome. Seattle: University of Washington; 2014. [Google Scholar]
- 234.Phoon CK, Acehan D, Schlame M, Stokes DL, Edelman-Novemsky I, Yu D, Xu Y, Viswanathan N, Ren M. Tafazzin knockdown in mice leads to a developmental cardiomyopathy with early diastolic dysfunction preceding myocardial noncompaction. J Am Heart Assoc. 2012:1. doi: 10.1161/JAHA.111.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Huang Y, Powers C, Madala SK, Greis KD, Haffey WD, Towbin JA, Purevjav E, Javadov S, Strauss AW, Khuchua Z. Cardiac metabolic pathways affected in the mouse model of barth syndrome. PLoS One. 2015;10:e0128561. doi: 10.1371/journal.pone.0128561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Powers C, Huang Y, Strauss A, Khuchua Z. Diminished Exercise Capacity and Mitochondrial bc1 Complex Deficiency in Tafazzin-Knockdown Mice. Front Physiol. 2013;4:74. doi: 10.3389/fphys.2013.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Soustek MS, Baligand C, Falk DJ, Walter GA, Lewin AS, Byrne BJ. Endurance training ameliorates complex 3 deficiency in a mouse model of Barth syndrome. J Inherit Metab Dis. 2015;38:915–22. doi: 10.1007/s10545-015-9834-8. [DOI] [PubMed] [Google Scholar]
- 238.Kaese S, Verheule S. Cardiac electrophysiology in mice: a matter of size. Front Physiol. 2012;3:345. doi: 10.3389/fphys.2012.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, Jovaisaite V, Frochaux MV, Quiros PM, Deplancke B, Houtkooper RH, Auwerx J. Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang Z, Tsukikawa M, Peng M, Polyak E, Nakamaru-Ogiso E, Ostrovsky J, McCormack S, Place E, Clarke C, Reiner G, McCormick E, Rappaport E, Haas R, Baur JA, Falk MJ. Primary Respiratory Chain Disease Causes Tissue-Specific Dysregulation of the Global Transcriptome and Nutrient-Sensing Signaling Network. Plos One. 2013:8. doi: 10.1371/journal.pone.0069282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Felici R, Lapucci A, Cavone L, Pratesi S, Berlinguer-Palmini R, Chiarugi A. Pharmacological NAD-Boosting Strategies Improve Mitochondrial Homeostasis in Human Complex I-Mutant Fibroblasts. Mol Pharmacol. 2015;87:965–71. doi: 10.1124/mol.114.097204. [DOI] [PubMed] [Google Scholar]
- 242.Verkaart S, Koopman WJ, Cheek J, van Emst-de Vries SE, van den Heuvel LW, Smeitink JA, Willems PH. Mitochondrial and cytosolic thiol redox state are not detectably altered in isolated human NADH:ubiquinone oxidoreductase deficiency. Biochim Biophys Acta. 2007;1772:1041–51. doi: 10.1016/j.bbadis.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 243.Valsecchi F, Monge C, Forkink M, de Groof AJ, Benard G, Rossignol R, Swarts HG, van Emst-de Vries SE, Rodenburg RJ, Calvaruso MA, Nijtmans LG, Heeman B, Roestenberg P, Wieringa B, Smeitink JA, Koopman WJ, Willems PH. Metabolic consequences of NDUFS4 gene deletion in immortalized mouse embryonic fibroblasts. Biochim Biophys Acta. 2012;1817:1925–36. doi: 10.1016/j.bbabio.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 244.Karamanlidis G, Lee CF, Garcia-Menendez L, Kolwicz SC, Jr, Suthammarak W, Gong G, Sedensky MM, Morgan PG, Wang W, Tian R. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab. 2013;18:239–50. doi: 10.1016/j.cmet.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Khan NA, Auranen M, Paetau I, Pirinen E, Euro L, Forsstrom S, Pasila L, Velagapudi V, Carroll CJ, Auwerx J, Suomalainen A. Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol Med. 2014;6:721–31. doi: 10.1002/emmm.201403943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.