Abstract
The discovery of the 2-C-methyl-d-erythritol-4-phosphate pathway for the biosynthesis of isoprenoids raises the important question of the nature and regulation of the enzymes involved in this pathway. CLA1, a gene previously isolated from Arabidopsis, encodes the first enzyme of the 2-C-methyl-d-erythritol-4-phosphate pathway, 1-deoxy-d-xylulose-5-phosphate synthase. We demonstrate this enzyme activity by complementation of the cla1-1 mutant phenotype and by direct enzymatic assays. Based on mRNA and protein expression patterns this enzyme is expressed mainly in developing photosynthetic and non-photosynthetic tissues. The β-glucuronidase expression pattern driven from the CLA1 gene regulatory region supports the northern and protein data while also showing that this gene has some level of expression in most tissues of the plant. A mutation in the CLA1 gene interferes with the normal development of chloroplasts and etioplasts, but does not seem to affect amyloplast structure. Microscopic analysis also shows a pleiotropic effect of the CLA1 gene mutation in mesophyll tissue formation.
In higher plants isoprenoids are derived from isopentenyl diphosphate (IPP) and synthesized in at least two different compartments, the cytoplasm and the chloroplast. For a long time it was assumed that IPP was synthesized exclusively by the mevalonate pathway in all organisms (Spurgeon and Porter, 1981; Goldstein and Brown, 1990). However, independent studies have demonstrated that in eubacteria, green algae, and plants, IPP is also synthesized by a non-mevalonate pathway designated as the 2-C-methyl-d-erythritol-4-P (MEP) pathway (for review, see Rohmer, 1998, 1999; Lichtenthaler, 1999). Thus in plants cytosolic IPP is synthesized by the mevalonate pathway and plastidic IPP is synthesized by the MEP pathway (Lichtenthaler, 1999). In the MEP pathway IPP is synthesized from pyruvate and glyceraldehyde-3-P via novel intermediates (Rohmer et al., 1993; Eisenreich et al., 1996; Schwender et al., 1996; Lichtenthaler et al., 1997). Labeling and nuclear magnetic resonance studies showed that 1-deoxyxylulose 5-P (DXP) is the first intermediate in this pathway (Roh-mer et al., 1996; Arigoni et al., 1997). Genes encoding for the first enzyme in this pathway, DXP synthase, were isolated from several organisms and the enzymatic activity of their encoded proteins has been corroborated (Sprenger et al., 1997; Bouvier et al., 1998; Lange et al., 1998; Lois et al., 1998; Lichtenthaler, 1999). In addition to its role in IPP synthesis the DXP synthase in plants, as in Escherichia coli, seems also to be required for the synthesis of thiamin and pyridoxol (Julliard and Douce, 1991; Hill et al., 1996). The next gene involved in the MEP pathway has been isolated from E. coli, peppermint, and Arabidopsis (Takahashi et al., 1998; Lange and Croteau, 1999; Schwender et al., 1999). It encodes an enzyme that converts DXP to MEP (Takahashi et al., 1998). Finally a third intermediate product has been recently postulated, as 4-(cytidine- 5′-diphospho)-2-C-methyl-d-erythritol. This product is synthesized from MEP by an enzyme encoded by the ygbP gene from E. coli (Rohdich et al., 1999). Independently of this study we have proved that the latter intermediate is essential for the formation of IPP (Kuzuyama et al., 2000a).
The production of specific chloroplastic isoprenoids such as carotenoids and phytol has now been demonstrated to depend on the MEP pathway (Eisenreich et al., 1996; Arigoni et al., 1997; Knöss et al., 1997; Lichtenthaler et al., 1997; Zeidler et al., 1997). Thus the analysis of the regulation of the enzymes in the MEP pathway is important in understanding the biosynthesis and possible manipulation of such terpenoids in plants. The isolation of albino plant mutants in Arabidopsis resulted in the identification of a gene required for the synthesis of both chlorophyll and carotenoids, named CLA1 (Mandel et al., 1996). In the cla1-1 mutant plastid development is impaired at an early stage resulting in almost no thylakoid membrane proliferation; the plastids resemble an early proplastid stage. CLA1 is a single gene in the Arabidopsis genome and its disruption affects the expression of both nuclear- and chloroplast-encoded photosynthetic genes (Mandel et al., 1996). The CLA1 protein sequence has extensive identity with other reported DXP synthases.
In this report we demonstrate that the CLA1 gene encodes a functional DXP synthase. To understand the regulation of this gene, we performed a detailed analysis of the CLA1 gene mRNA expression and protein patterns. We show that the CLA1 gene transcripts and protein preferentially accumulate in young developing tissues. The microscopic analysis of different plastids in the cla1-1 mutant demonstrates that the disruption of the CLA1 gene affects the morphology of chloroplasts and etioplasts and alters the final stages of cellular morphogenesis in mesophyll tissue formation.
RESULTS
The Albino Phenotype of the cla1-1 Plant Can Be Rescued by the Addition of 1-Deoxy-d-Xylulose (DX)
The extensive amino acid similarity of the CLA1 gene to the published DXP synthases (Sprenger et al., 1997; Bouvier et al., 1998; Lange et al., 1998; Lois et al., 1998; Lichtenthaler, 1999) suggested that the CLA1 gene could encode a DXP synthase. To test whether the CLA1 protein functions as a DXP synthase we took advantage of the albino phenotype in the cla1-1 mutant. Synthetic DX, a non-phosphorylated version of the product of the DXP synthase, was supplemented on the growth medium of cla1-1 plants. This product was used to ensure penetration into the plant cells, as it was demonstrated to be efficiently incorporated into plastidic isoprenoids (Arigoni et al., 1997; Zeidler et al., 1997). As the cla1-1 mutation is lethal on soil, seed stocks are maintained as heterozygotes. Upon selfing, one-quarter of the progeny are albino on medium. After germination, such albino homozygous mutant plants were selected and transferred to plates containing 0.02% (w/v) DX. The development of these plants was assessed by visual inspection and their pigment content was quantified.
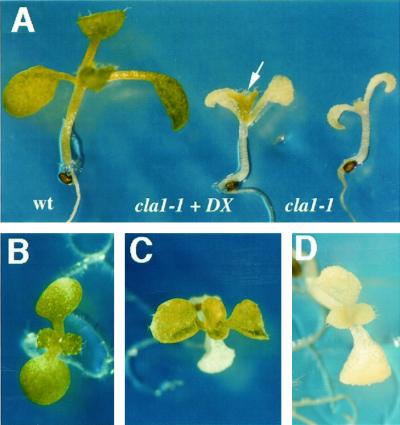
As shown in Figure 1, the first true leaves of the cla1-1 plants grown in germination media (GM) media developed the albino phenotype characteristic of this mutant (Fig. 1, A, right side and D). In contrast, cla1-1 plants grown on the same media supplemented with DX turned green (Fig. 1, A [middle plant] and C). For comparison, a Wassilewskija (WS) wild-type plant grown in GM media is shown in the left side of Figure 1, A and B. This green phenotype correlates with a substantial increase in chlorophyll and carotenoid content of the cla1-1 plants supplemented with DX compared with the ones grown in GM media (Table I). Greening observed in the leaves of the cla1-1 plants supplemented with DX is specific for this mutant, as other unrelated albino plants such as alb1-1 and alb2−1 (van der Veen and Blankenstijn de Vries, 1973; Relichova, 1976) remain albino (data not shown). The cla1-1 cotyledons have the capacity to respond to DX, but only upon direct exposure to this chemical during seed imbibition. We noticed however that DX at the concentration used in these experiments (0.02%) has a toxic effect in the early stages of development, as we detected yellowish seedlings in cla1-1 and wild-type plants when this compound was present during germination (data not shown).
Figure 1.
In vivo complementation of the albino phenotype in the cla1-1 seedlings by DX. A, Phenotypic analysis of 10-d-old seedlings of wild type grown on GM medium (left); cla1-1 grown on GM medium supplemented with 0.02% (w/v) of DX (center); and cla1-1 grown on GM medium (right). The arrow indicates the first pair of leaves on the DX supplemented mutant plant, where a green phenotype is clearly visible. An upper view is shown for a wild-type plant grown on GM medium (B), a 15-d-old cla1-1 seedling grown on GM medium supplemented with 0.02% (w/v) DX (C) in which the two pairs of true leaves can be seen, and a 15-d-old cla1-1 seedling grown on GM medium (D).
Table I.
Pigment quantification
| Sample | Chl a | Chl b | Chl Total | Carotenoids |
|---|---|---|---|---|
| mg pigment g−1 fresh wt | ||||
| Wild type | 590 | 242 | 832 | 130 |
| cla1-1 DX | 170 | 108 | 278 | 61 |
| cla1-1 | 11 | 19 | 30 | 10 |
Pigments were extracted from 15-d-old plants grown in GM or GM supplemented during 10 d with 0.02% DX.
The biochemical function of the recombinant CLA1 protein was also examined in vitro. The GST-CLA1 fusion protein expressed in E. coli was used to test for DXP synthase activity. The product obtained after incubation of pyruvate and glyceraldehyde-3-P with the CLA1 protein was treated with alkaline phosphatase and its identity was analyzed by gas chromatography-mass spectrometry (GC-MS) as the trimethylsilylated derivative. As shown in Table II the product obtained from this reaction were determined to be DX and are in agreement with the previously reported for peppermint DXP synthase (Lange et al., 1998).
Table II.
Identification of DX obtained by incubation of pyruvate/glyceraldehyde-3-P with CLA1 protein
| Sample | HPLC | GC | MS m/z (relative intensity) |
|---|---|---|---|
| min | % | ||
| Producta | 8.6 | 7.33 | 307 (21), 218 (100), 204 (27), 147 (91), 73 (57) |
| DXb | 8.5 | 7.30 | 307 (14), 218 (100), 204 (31), 147 (85), 73 (73) |
The data presented in this row correspond to those values obtained after the samples were treated with alkaline phosphatase in both crude enzyme extracts and an affinity-purified enzyme protein. The values are compared with those published for peppermint (Lange et al., 1998).
DX corresponds to the authentic DX molecule. Molecular ion peaks of trimethylsilyl ether products (calculated as m/z 350) were not observed.
Tissue- and Organ-Specific Expression of the CLA1 Gene in Arabidopsis
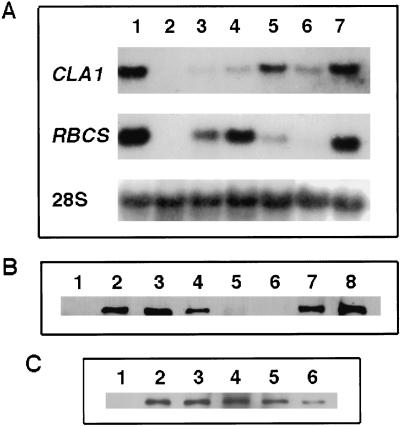
Although considerable information has been accumulated recently on the MEP pathway in plants, the expression pattern and regulation of the enzymes participating in this pathway are presently unknown. We decided to study the CLA1 gene expression pattern at the mRNA and protein levels. The RNA-blot hybridization data presented in Figure 2A demonstrate that the CLA1 mRNA is detected in all tissues examined, including non-photosynthetic tissues such as roots. CLA1 gene transcripts are especially abundant in seedlings (Fig. 2A, lanes 1 and 7) and in flower buds compared with the other organs analyzed (Fig. 2A, lane 5).
Figure 2.
Analysis of CLA1 gene transcript and protein accumulation in Arabidopsis plants. A, RNA-blot analysis of the CLA1 transcript. Five micrograms of total RNA was purified from 15-d-old wild-type seedlings (lane 1), cla1-1 plants (lane 2), and different tissues of wild-type plants including mature leaves (lane 3), cauline leaves (lane 4), buds (lane 5), roots (lane 6), and 5-d-old seedlings (lane 7). The probes used were CLA1 and RBCS, as well as rRNA as an RNA-loading control as indicated in the left side of the panel. B, CLA1 protein accumulation in different tissues. Western-blot analyses were performed using total protein extracts obtained from 15-d-old cla1-1 seedlings (lane 1), 15-d-old wild-type seedlings (lane 2), young rosette leaves (lane 3), mature rosette leaves (lane 4), roots (lane 5), 24-h-imbibed seeds (lane 6), flowers (lane 7), and immature siliques (lane 8). Fifteen micrograms of total protein extracts was loaded in each lane except for roots and seeds, where 30 and 45 μg was used, respectively. C, CLA1 protein developmental expression. Western-blot analysis shows CLA1 protein accumulation in 15-d-old cla1-1 seedlings (lane 1), 5-d-old (lane 2), 8-d-old (lane 3), 15-d-old (lane 4), 20-d-old (lane 5), and 25-d-old (lane 6) wild-type seedlings. In each lane, 15 μg of total protein was loaded.
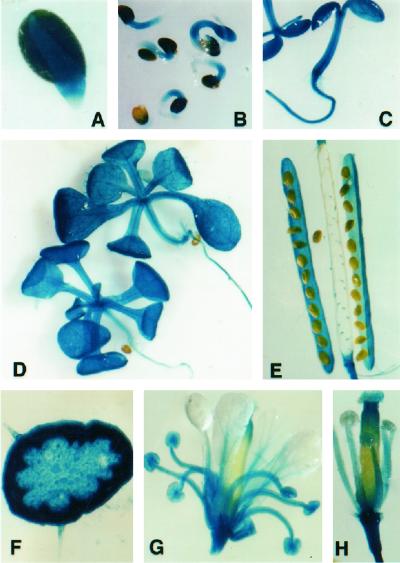
To obtain more insight into the participation of this gene in Arabidopsis development we investigated its expression pattern using a β-glucuronidase (GUS) reporter gene construct. A 1.4-kb upstream region from the CLA1 gene initiation codon was used to generate a translational fusion with the GUS reporter gene uidA. Eight independent transgenic plants from the T2 generation were analyzed for GUS expression. Although some variation in the intensity of staining was observed among the lines carrying this construct, all of them showed the same GUS staining pattern. According to the pattern observed the CLA1 gene is expressed very early in germinating seeds. As shown in Figure 3A, GUS activity was detected in the protruding root (with the exception of the root cap) 48 h after water imbibition. In 3-d-old seedlings (Fig. 3B), GUS is detected primarily in the hypocotyl and in the emerging cotyledons with faint staining in the root. In 5-d-old seedlings, GUS activity is detected in most of the plant, the cotyledons, the hypocotyl, and the root (Fig. 3C). In older seedlings, GUS staining is especially strong in the expanding leaves, including vascular tissue and trichomes (Fig. 3D), but a faint staining in the hypocotyl and root is also present. A transverse section of the inflorescence showed GUS activity in most of the cells: in the epidermis, including the trichomes, and in the cortex, vascular tissue, and pith (Fig. 3F). In the silique, GUS staining is observed in the locules and in the funiculus, but there is no expression detected in immature seeds (Fig. 3E). In the flower, GUS staining is intense in the sepals and the stamens (Fig. 3G), whereas in the petals, GUS activity is faint. Within carpels, staining is mostly restricted to the upper part of the stigma and in the stigmatic papillae (Fig. 3, G and H).
Figure 3.
Histochemical analyses of GUS activity in Arabidopsis plants expressing the GUS gene under the control of the CLA1 gene promoter. A, Water-imbibed (48-h) germinating seeds; B, 3-d-old seedlings; C, 5-d-old seedlings; D, 15-d-old seedlings; E, immature seeds and siliques from Arabidopsis; F, transverse section of the inflorescence; G, fully developed Arabidopsis flower; and H, individual stigma and anthers from a fully developed Arabidopsis flower.
Based on these two analyses we can conclude that the CLA1 gene is widely expressed throughout the Arabidopsis plant and that higher expression levels are found in the young tissues of the plant.
Characterization of the CLA1 Protein
To characterize the CLA1 gene product we performed western-blot analysis using polyclonal antibodies raised against a fusion protein between an E. coli glutathione S-transferase (GST) and most of the CLA1 protein. We detected a 70-kD protein in total extracts of 15-d-old wild-type seedlings (Fig. 2B, lane 2) that is not detected in extracts from cla1-1 plants (Fig. 2B, lane 1). The accumulation pattern of the CLA1 protein was analyzed in total protein extracts from different Arabidopsis tissues. As shown in Figure 2B, the CLA1 protein is detected in most plant tissues except in 24-h-water-imbibed seeds (Fig. 2B, lane 6). CLA1 is particularly abundant in young leaves, in buds, and in immature siliques, but barely detectable in roots. These results contrast with the northern and transgenic plant analyses in which the CLA1 gene is expressed at similar levels in both roots and mature leaves (Fig. 2A, lanes 3 and 6). The CLA1 protein accumulates most predominately in the young tissues of the plant (Fig. 2B, lanes 3 versus 4). When we compared the levels of CLA1 protein in extracts from seedlings of different ages we found that this protein increases as organs mature, reaching a maximum in 15-d-old plantlets as shown in Figure 2C, lane 4. After this stage the amount of CLA1 protein decreases in relation to the age of the plant (Fig. 2C).
Organelle and Tissue Morphology in the cla1-1 Plant
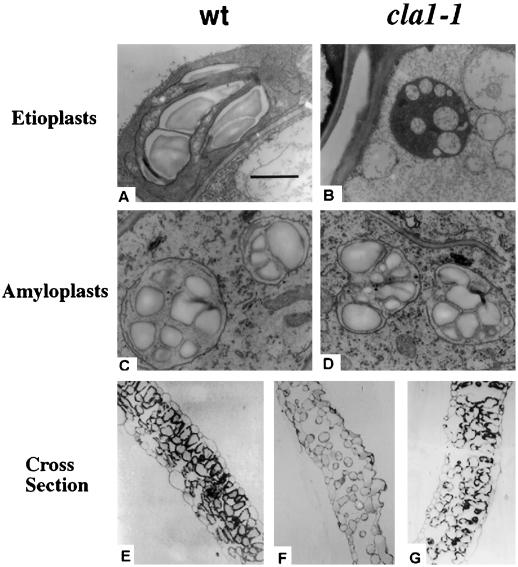
Our initial analysis demonstrated that the CLA1 protein in Arabidopsis is required for normal chloroplast differentiation (Mandel et al., 1996). Based on the CLA1 function and its mRNA and protein expression patterns we decided to re-analyze the structure of other plastid types in cla1-1 plants. Using transmission electron microscopy the morphology of etioplasts from the cotyledons of dark-adapted cla1-1 seedlings was analyzed. As cla1-1 seed stocks are heterozygous, seeds were initially germinated on Murashige and Skoog basal salt mixture media with light for 6 d and the albino homozygous mutant plants were transferred and kept in the dark during an additional 8 d. The same treatment was followed with wild-type plants to be used as controls. As shown in Figure 4B, the ultrastructure of the dark-adapted etioplasts in cla1-1 plant is altered in comparison with wild-type plastids (Fig. 4A). The prolamellar body in these organelles is absent and vesicles are present which seem to be associated with internal membranes. We also investigated the amyloplast structure in 10-d-old roots of the cla1-1 plant. It is interesting that as shown in Figure 4D, the morphology of this organelle does not seem to be altered compared with wild-type plants. Normal starch granules, characteristic of this plastid type, can be detected in the plastids of both wild-type and mutant plants.
Figure 4.
Microscopic analysis of the plastids and mesophyll tissue of the cla1-1 mutant. Transmission electron microscopic examination of plastids of wild-type (A and C) and cla1-1 (B and D) seedlings. Etioplasts were analyzed from cotyledons of seedlings that were dark-adapted for 4 d of wild type (A) and homozygous (B) cla1-1 mutants. Amyloplasts were analyzed from 10-d-old root seedlings of wild type (C) and cla1-1 (D) mutant. Transverse sections of the 10-d-old first leaf from plants of wild type (E), cla1-1 (F), and cla1-1 (G) mutant supplemented with 0.02% (w/v) DX.
A well-known event during leaf differentiation in dicot plants is the coordination with chloroplast development (Chory, 1992). Mutants have been isolated that partially uncouple such coordination, demonstrating that these processes can be separable (Mochizuki et al., 1996). We therefore asked if mutations in CLA1 have an effect on leaf cellular morphology. As shown in Figure 4F, the transverse leaf section of the cla1-1 plant shows an anomalous development of the mesophyll tissue compared with similar sectors from a wild-type plant (Fig. 4E). The proportion of air space compared with the mesophyll tissue is larger in the cla1-1 mutant than in the wild-type plant. Also for the cla1-1 mutant, the cells of the mesophyll tissue remain round and small; few palisade cells are present. This phenotype is unlikely to be the result of carbon or vitamin (thiamin or pyridoxol) deficiency as these are supplemented in the medium. The morphological abnormalities are reversible as soon as the plastid proceeds through its differentiation pathway. When cla1-1 plants are grown on Murashige and Skoog basal salt mixture media supplemented with DX they show a seminormal morphology of the mesophyll tissue including the presence of palisade cells (Fig. 4G).
DISCUSSION
In this report we demonstrate that the CLA1 gene encodes the previously reported DXP synthase (Sprenger et al., 1997; Bouvier et al., 1998; Disch et al., 1998; Lange et al., 1998; Lois et al., 1998; Lichtenthaler, 1999). Early work by Arigioni and coworkers (1997) showed that DX is an effective compound for phytol and carotenoid production in culture cells of Catharantus roseus. We also observed that DX has a striking capacity to restore pigment biosynthesis in the cla1-1 albino mutant. DX was efficiently absorbed by the root and transported into the leaves. It is still an open question whether DX is phosphorylated before it is converted to MEP.
The existence of two biosynthetic pathways for IPP production raises questions about the participation of each pathway in the synthesis of specific isoprenoid compounds and inter-pathway communication. Some exchange between the cytoplasmic and chloroplastic IPP pools has been suggested (Bach and Lichtenthaler, 1982; Arigoni et al., 1997; Nabeta et al., 1997). It is interesting that despite the albino phenotype of cla1-1 plants, low chlorophyll and carotenoid levels are detectable in this mutant (Table I). As this seems to be a null mutation, our interpretation is that cytosolic IPP probably moves into the plastids, resulting in limited pigment levels. However, this supply of cytosolic IPP is far too small to fulfill normal pigment biosynthesis requirements. Whether the supply of cytosolic IPP could be sufficient for the biosynthesis of other chloroplastic isoprenoids under specific physiological or developmental conditions needs to be defined.
This work is the first detailed characterization of DXP synthase expression patterns in plants. The CLA1 gene is widely expressed in photosynthetic and non-photosynthetic tissues and its expression is clearly modulated throughout plant development. The maximum mRNA levels of CLA1 correlate with the maturation stage of the leaves when there are massive requirements for chlorophylls and carotenoids. In non-photosynthetic tissues the CLA1 expression pattern supports the participation of the MEP pathway in the production of a variety of isoprenoids. It is interesting to note that even though substantial CLA1 mRNA levels were detected in roots by northern analysis and GUS staining of transgenic plants, the CLA1 protein levels in roots extracts were barely detectable. A potential post-transcriptional regulation mechanism for the CLA1 transcript might be operating in roots.
We demonstrated previously that CLA1 is required for proper chloroplast development (Mandel et al., 1996). The data presented here further substantiate the requirement for CLA1 to ensure development of etioplasts, but CLA1 does not seem to be required for amyloplast differentiation. It is apparent that expression of the CLA1 gene is not required for starch accumulation because the size and number of starch granules in the amyloplasts is similar in cla1-1 and wild-type plants. We have observed that the mesophyll tissue of cla1-1 is altered in comparison with wild-type plants. Similar phenotypes have been reported in other mutants that affect chloroplast development at an early stage such as Dcl-m and GHOST in tomato, Dag in Antirrhinum (Scolnik et al., 1987; Chatterjee et al., 1996; Keddie et al., 1996), and PAC and ATD in Arabidopsis (Reiter et al., 1994; van der Graaff, 1997). The common denominator in all is that the plastids are arrested early in development. One possibility is that early arrest interferes with production of a chloroplast signal to the cytoplasm and nucleus that directly influences the last stages of mesophyll differentiation (Susek and Chory, 1992; León et al., 1998).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis ecotypes Columbia or WS seeds were grown in Metromix 200 (Grace Sierra, Milpitas, CA) in controlled growth chambers at 24°C using a 16-h light/8-h dark photoperiod with cool-white illumination (20 μE m−2 s−1) for 3 to 4 weeks. Plants under sterile conditions were grown in Murashige and Skoog basal salt mixture supplemented with Gamborg's vitamins, 0.5% (w/v) MES [2-(N-morpholino)ethanesulfonic acid], 1% (w/v) Suc (GM media), and 0.7% (w/v) of phytoagar in the case of solid medium. Determination of total carotenoids and chlorophylls was conducted following the protocol reported by Lichtenthaler and Wellburn (1983).
In Vivo Complementation of the cla1-1 Albino Phenotype
The recessive albino cla1-1 mutant is lethal on soil (Mandel et al., 1996), thus seed stocks are maintained as heterozygotes. The cla1-1 heterozygous plants were germinated in GM media for 6 d. Homozygous albino cla1-1 plants were transferred to GM medium or GM supplemented with 0.02% (w/v) DX. As a control, WS wild-type plants were incubated in the same type of media.
Molecular Biology Techniques
Total RNA was isolated from different plant tissues using the procedure of Logemann et al. (1987) with minor modifications. For northern blots, RNA was fractionated by electrophoresis in 1.2% (w/v) agarose gels and transferred onto Hybond N+ nylon membranes (Amersham Corporation, Arlington Heights, IL). Hybridizations and washes were done at high stringency conditions according to standard procedures using 32P-radiolabeled probes (Church and Gilbert, 1984).
Production of the GST-CLA1 Fusion Protein and Antibody Preparation
To generate a fusion protein containing the CLA1 gene, a 2-Kb SspI-EcoRI fragment of the CLA1 cDNA was cloned downstream of the GST from the pGEX1 vector (Amersham Pharmacia Biotech, Buckinghamshire, UK). This fragment contains most of the CLA1 coding region with the exception of 197 bp of the putative chloroplast transit peptide. The generated plasmid, pGEX-CLA1, codes for the GST-CLA1 fusion protein without the putative chloroplast transit peptide. The integrity of the chimeric gene was verified by direct sequencing. The isopropylthio-β-d-galactoside-induced GST-CLA1 fusion protein was produced in Escherichia coli and purified by affinity chromatography using Glutathione Sepharose 4B resin (Amersham Pharmacia Biotech) according to the protocol published (Ausubel et al., 1989). For polyclonal antibody generation, purified GST-CLA1 protein (10 μg in 20 μL of phosphate-buffered saline) and complete Freund's adjuvant was injected intraperitoneal as 1:9 emulsion in a BALB/c mice (Harlow and Lane, 1988). Three additional injections (10 μg each), were administrated every 8 d starting 14 d after the initial injection. The ascites was collected 8 d later and titer was determined. In addition to recognizing the 70-kD CLA1 protein, this ascites fluid recognizes one abundant protein that is also present in the cla1-1 mutant plant.
Functional Assay of CLA1 Protein
DXP synthase activity was measured using 50 μg of the purified GST-CLA1 fusion protein. The reaction was done according to Kuzuyama et al. (2000b), in 100 mm Tris [tris(hydroxymethyl)aminomethane]-HCL (pH 8.0), 1 mm MgCl2, 2 mm dithiothreitol, 0.075 mm thiamine diphosphate, 20 mm glyceraldehyde-3-P, and 10 mm pyruvate at 37°C for 1 h and terminated by heating. Denatured proteins were removed by centrifugation at 15,000 rpm for 10 min. The supernatant was treated with 1 unit of bacterial alkaline phosphatase at 50°C for 1 h. The reaction mixture was treated with activated charcoal power and filtered. The filtrate was subject to HPLC-connected Shodex SUGAR KS-801 column (8 × 300 mm, SHOWA DENKO, Tokyo) heated at 80°C. The flow rate of water was 1 mL min−1. DX was detected at 8.6 min of refractive index detector. For comparison, authentic DX was detected at 8.5 min under the same condition consistent with that previously reported by Lange, et al. (1998).
To determine the product generated by the GST-CLA1 fusion protein, eluates after HPLC separation were collected, dried, and then derivatized with N,O bis(trimethylsilyl) acetamide:trimethylchlorosilane:N-trimethylsilyimidazole (2:3:2, v/v) in pyridine at 80°C for 20 min. GC-MS analysis was performed by using Finnigan MAT GCQ ion-trap GC-MS system (Thermoquest, San Jose, CA) equipped with a 30-m × 0.25-mm diameter fused silica capillary column coated with 0.25-μm film thickness of DB-5MS (J&W Scientific, Folson, CA). The oven temperature was programmed from 90°C (2-min hold) at 20°C min−1 to 150°C, at 10°C min−1 to 250°C, and then at 30°C min−1 to 300°C with a constant velocity at 40 cm min−1 He.
Western-Blot Analysis
Protein samples were quantified with Bradford reagent (Bio-Rad, Hercules, CA). Samples were separated by PAGE. To verify equal protein loading a parallel gel was run and stained with Coomassie Brilliant Blue R-250. The proteins were transferred onto nitrocellulose (Hybond C, Amersham Pharmacia Biotech) by electroblotting for 1 h at 200 mA in 25 mm Tris, 0.2 m Gly, and 20% (w/v) methanol. Immunodetection was done using a 1:1,000 dilution of the GST-CLA1 fusion protein polyclonal antibody. An antimouse immunoglobulin horseradish peroxidase-conjugate was used as a secondary antibody (Amersham Pharmacia Biotech), and detection was done with an enhanced chemiluminescence detection kit (Amersham Pharmacia Biotech).
Plasmid Construction
A CLA1-GUS translational fusion was constructed using a 1.4-kb fragment of the CLA1 5′-regulatory region (contained in the ESSA I FCA contig fragment no. 4, accession no. Z97339). Initially, a fragment of approximately 9 kb capable of complementing the cla1-1 mutant phenotype (Mandel et al., 1996) was subcloned into the pBluescript II vector and subjected to Exonuclease III/Mung Bean deletions. One of such deletions, containing approximately 1.4 kb upstream of the ATG codon from the CLA1 gene was PCR-amplified and used for expression analysis. The primers used for the PCR were: ATG-Nco (5′GCAGAAGAAGCCATGGGAGGTAC3′) that includes the CLA1 ATG codon, and BS-Hind (5′GGCCAAGCTTACGCCAAGCGCGCAAT3′) fromthe flanking vector sequence, plus a HindIII site at its end. The PCR fragment generated was first cloned as a translational fusion into a vector derived from pBluescript II KS(−) plasmids (Stratagene, La Jolla, CA) containing the uidA (GUS) gene followed by the nopaline synthase 3′ terminator from the pBin19 plasmid (Bevan, 1984), termed pBlueGUS. The entire fragment (CLA1 promoter:GUS and nopaline synthase-3′) was subcloned into the binary vector pBin19 (Bevan, 1984) generating the pBin/1458-G plasmid that was used for transformation into Arabidopsis.
Plant Transformation and Histochemical Analysis
Transgenic lines (Columbia) were constructed using Agrobacterium tumefaciens-mediated transformation by the vacuum infiltration method (Bechtold et al., 1993). Transgenic plants were identified by their capacity to develop roots and maintain green leaves in the presence of 50 μg mL−1 of kanamycin. They were then transferred to soil to get the transgenic seed and the following generations. GUS histochemical analysis was carried out according to a protocol previously described (Jefferson, 1987). The tissue was incubated at 37°C overnight (12 h). Destaining was accomplished by 30 min incubations with 3:1 (v/v) acetone:methanol solution. Whole tissues or sections were observed under bright-field microscopy (Type 104, Nikon, Tokyo).
Microscopy Techniques
For transmission electron microscopy, tissues were fixed with 6% (w/v) glutaraldehyde in phosphate-buffered saline (pH 7.2) for 10 h and post-fixed in 1% (w/v) osmium tetroxide in the same buffer for several hours. After dehydration in a graded series of ethanol and propylene oxide, samples were embedded in Epoxy resin. For electron microscopy, 60-nm thin sections were obtained and mounted on formvar-coated copper grids (Electron Microscopy Science, Fort Washington, PA). For contrast, 3% (w/v) uranyl acetate and 0.3% (w/v) lead citrate were used. Grids were observed with a transmission electron microscope (EM-10, Carl Zeiss, Jena, Germany) operating at 80 kV. For light microscopy, samples were treated as described above and 0.5-μm semi-thin sections were obtained. The sections were stained with 1% (w/v) toluidine blue and observed in bright field with a light microscope (Standard, Carl Zeiss).
ACKNOWLEDGMENTS
We want to thank Elizabeth Mata and Carlos González for their help in raising antibody and Paul Gaitan and Eugenio López for the synthesis of oligos. We thank Drs. Virginia Walbot, Analilia Arroyo, Helena Porta, Marcela Treviño, and Stuart Reichler for helpful comments on the manuscript.
Footnotes
This work was funded by Consejo Nacional de Ciencia y Tecnologia and Dirección General de Asuntos para el Personal Académico (grant nos. 110P–N9506 and IN205697) and by the Pew Charitable Trust.
LITERATURE CITED
- Arigoni D, Sagner S, Latzel C, Eisenreich W, Bacher A, Zenk MH. Terpenoid biosynthesis from 1-deoxy-d-xylulose in higher plants by intramolecular skeletal rearrangement. Proc Natl Acad Sci USA. 1997;94:10600–10605. doi: 10.1073/pnas.94.20.10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Siedman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1989. [Google Scholar]
- Bach TJ, Lichtenthaler HK. Inhibition of mevalonate biosynthesis and plant growth by the fungal metabolite mevinolin. In: Wintermanns JFGM, Kuiper PJC, editors. Biochemistry and Metabolism of Plant Lipids. Amsterdam: Elsevier; 1982. pp. 515–521. [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vie. 1993;316:1194–1199. [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acid Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, d'Harlingue A, Suire C, Backhaus RA, Camara B. Dedicated roles of plastid transketolases during the early onset of isoprenoid biogenesis in pepper fruits. Plant Physiol. 1998;117:1423–1431. doi: 10.1104/pp.117.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Sparvoli S, Edmunds C, Garosi P, Findlay K, Martin C. DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 1996;15:4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Chory J. A genetic model for light-regulated seedling development in Arabidopsis. Development. 1992;115:337–354. [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch A, Schwender J, Müller C, Lichtenthaler HK, Rohmer M. Distribution of the mevalonate and glyceraldehyde phosphate/pyruvate pathways for isoprenoid biosynthesis in unicellular algae and the cyanobacterium Synechocystis PCC 6714. Biochem J. 1998;333:381–388. doi: 10.1042/bj3330381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W, Menhard B, Hylands P, Zenk MH, Bacher A. Studies on the biosynthesis of taxol: the taxane carbon skeleton is not of mevalonoid origin. Proc Natl Acad Sci USA. 1996;93:6431–6436. doi: 10.1073/pnas.93.13.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 53–138. [Google Scholar]
- Hill RE, Himmeldrik K, Kennedy IA, Paulosky RM, Sayer BG, Wolf E, Spenser ID. The biogenetic anatomy of vitamin B6: a 13C NMR investigation of the biosynthesis of pyridoxol in Escherichia coli. J Biol Chem. 1996;271:30426–30435. doi: 10.1074/jbc.271.48.30426. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Report. 1987;5:387–405. [Google Scholar]
- Julliard JH, Douce R. Biosynthesis of the thiazole moitey of thiamin (vitamin B1) in higher plant chloroplasts. Proc Natl Acad Sci USA. 1991;88:2042–2045. doi: 10.1073/pnas.88.6.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddie JS, Carroll B, Jones JDG, Gruissem W. The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 1996;15:4208–4217. [PMC free article] [PubMed] [Google Scholar]
- Knöss W, Reuter B, Zapp J. Biosynthesis of the labdane diterpene marrubium in Marrubium vulgare via a non-mevalonate pathway. Biochem J. 1997;326:449–454. doi: 10.1042/bj3260449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuyama T, Takagi M, Kaneda K, Dairi T, Seto H. Formation of 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol from 2-C-methyl-d-erythritol 4-phosphate by 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase, a new enzyme in the nonmevalonate pathway. Tetrahedron Lett. 2000a;41:703–706. [Google Scholar]
- Kuzuyama T, Takagi M, Takahashi S, Seto H. Cloning and characterization of 1-deoxy-d-xylulose 5-phosphate synthase from Streptomyces sp. strain CL190, which uses both the mevalonate and nonmevalonate pathways for isopentenyl diphosphate biosynthesis. J Bacteriol. 2000b;182:891–897. doi: 10.1128/jb.182.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Croteau R. Isoprenoid biosynthesis via a mevalonate-independent pathway in plants: cloning and heterologous expression of 1-deoxy-d-xylulose-5-phos-phatereductoisomerase from peppermint. Arch Biochem Biophys. 1999;365:170–174. doi: 10.1006/abbi.1999.1168. [DOI] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, McCaskill D, Croteau R. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc Natl Acad Sci USA. 1998;95:2100–2104. doi: 10.1073/pnas.95.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León P, Arroyo A, Mackenzie S. Nuclear control of plastid and mitochondrial development in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:453–480. doi: 10.1146/annurev.arplant.49.1.453. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. The 1-deoxy-d-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Schwender J, Disch A, Rohmer M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 1997;400:271–274. doi: 10.1016/s0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans. 1983;11:591–592. [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Lois LM, Campos N, Rosa Putra S, Danielsen K, Rohmer M, Boronat A. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of d-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc Natl Acad Sci USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, León P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996;9:649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Susek R, Chory J. An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol. 1996;112:1465–1469. doi: 10.1104/pp.112.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeta K, Kawae T, Saitoh T, Kikuchi T. Synthesis of chlorophyll α and β-carotene from 2H and 13C-labeled mevalonates and 13C-labeled glycin in cultured cells of liverworts Heteroscyphus planus and Lophocolea heterophylla. J Chem Soc Perkin Trans. 1997;1:261–267. [Google Scholar]
- Reiter RS, Coomber SA, Bourett TM, Bartley GE, Scolnik PA. Control of leaf and chloroplast development by the Arabidopsis gene pale cress. Plant Cell. 1994;6:1253–1264. doi: 10.1105/tpc.6.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relichova J. Some new mutants. Arab Inf Serv. 1976;13:25–28. [Google Scholar]
- Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk M. Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc Natl Acad Sci USA. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M. Isoprenoid biosynthesis via the mevalonate-independent route, a novel target for antibacterial drugs? In: Jucker E, editor. Progress in Drug Research. Vol. 50. Basel: Birkhäuser Verlag; 1998. pp. 135–154. [DOI] [PubMed] [Google Scholar]
- Rohmer M. A mevalonate-independent route to isopentenyl diphosphate. In: Cane D, editor. Comprehensive Natural Product Chemistry, Isoprenoids including Steroids and Carotenoids. Vol. 2. Oxford: Pergamon Press; 1999. pp. 45–68. [Google Scholar]
- Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J. 1993;295:517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M, Seemann M, Horbach S, Bringer-Meyer S, Sahm H. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J Am Chem Soc. 1996;118:2564–2566. [Google Scholar]
- Schwender J, Müller C, Zeidler J, Lichtenthaler HK. Cloning and heterologous expression of a cDNA encoding 1-deoxy-d-xylulose-5-phosphate reductoisomerase of Arabidopsis thaliana. FEBS Lett. 1999;455:140–144. doi: 10.1016/s0014-5793(99)00849-2. [DOI] [PubMed] [Google Scholar]
- Schwender J, Seemann M, Lichtenthaler HK, Rohmer M. Biosynthesis of isoprenoids (carotenoids, sterols, prenyl side-chains of chlorophylls and plastoquinone) via a novel pyruvate/glyceraldehyde 3-phosphate non-mevalonate pathway in the green alga Scenedesmus obliquus. Biochem J. 1996;316:73–80. doi: 10.1042/bj3160073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik PA, Hinton P, Greenblatt IM, Giuliano G, Delanoy MR, Spector DL, Pollock D. Somatic instability of carotenoid biosynthesis in the tomato ghost mutant and its effect on plastid development. Planta. 1987;171:11–18. doi: 10.1007/BF00395063. [DOI] [PubMed] [Google Scholar]
- Sprenger GA, Schörken U, Wiegert T, Grolle S, de Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgeon SL, Porter JW. Introduction. In: Porter JW, Spurgeon SL, editors. Biosynthesis of Isoprenoid Compounds. New York: John Wiley & Sons; 1981. pp. 1–46. [Google Scholar]
- Susek R, Chory J. A tale of two genomes: role of a chloroplast signal in coordinating nuclear and plastid genome expression. Aust J Plant Physiol. 1992;19:387–399. [Google Scholar]
- Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phos-phatein an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E. Developmental mutants of Arabidopsis thaliana obtained after T-DNA transformation. PhD thesis. The Netherlands: Leiden University; 1997. [Google Scholar]
- van der Veen JH, Blankenstijn de Vries H. Double reduction in tetraploid Arabidopsis thaliana, studied by means of chlorophyll mutant with a distinct simplex phenotype. Arab Inf Serv. 1973;10:11–12. [Google Scholar]
- Zeidler JG, Lichtenthaler HK, May HU, Lichtenthaler FW. Is isoprene emitted by plants synthesized via novel isopentenyl pyrophosphate pathway? Z Naturforsch. 1997;52c:15–23. [Google Scholar]