ABSTRACT
Activation of CD4 T cells by dendritic cells leads to their differentiation into various effector lineages. The nature of the effector lineage is determined by the innate cues provided by dendritic cells to newly primed T cells. Although the cytokines necessary for several effector lineages have been identified, the innate cues that drive T follicular helper (Tfh) lineage cell development remain unclear. Here we found that following priming, CD4 T cells undergoing clonal expansion acquire a transient Tfh-like phenotype before differentiating into other effector lineages. In addition, we found that T cell-intrinsic myeloid differentiation antigen 88 (MyD88) signaling, which occurs downstream of interleukin-1 (IL-1) and IL-18 receptors, is critical for the primed CD4 T cells to transition out of the temporary Tfh lineage. Mice with T cell-specific deletion of MyD88 have a higher proportion of Tfh cells and germinal center (GC) B cells. These exaggerated Tfh cell and GC B cell responses, however, do not lead to protective immunity against infections. We demonstrate that T cell-intrinsic MyD88 is critical for effector lineage differentiation as well as production of the cytokines that are necessary for class switching. Overall, our study establishes that following priming and clonal expansion, CD4 T cells undergo a transitional Tfh-like phase and that further differentiation into effector lineages is dictated by T cell-intrinsic MyD88-dependent cues.
KEYWORDS: Borrelia burgdorferi, effector T cells, germinal centers, IL-1, IL-18, MyD88, Tfh cells, Th1/Th17 responses
INTRODUCTION
Among all the cell types of the adaptive immune system, CD4 T cells display the largest degree of plasticity, as they have the ability to differentiate into numerous sublineages in response to pathogen-specific and environmental cues (1, 2). The pathogen-specific signals are primarily derived from the innate immune system, while the environmental cues are defined by the tissue location and the global immune status of the host shaped by the microbiota and persistent infections (3). Differentiated CD4 T cell lineages orchestrate a broad range of effector activities, including providing help to CD8 T cells, activating B cells, and recruiting inflammatory cells, such as monocytes and neutrophils, to sites of infection and inflammation. Understanding CD4 T cell differentiation is therefore critical to gain insights into the function of the entire immune system. Naive CD4 T cells recognize cognate peptides presented in the context of major histocompatibility complex (MHC) class II molecules on dendritic cells (DCs), which results in clonal expansion of antigen-specific T cells. Costimulatory molecule engagement and the innate cytokine milieu present during the T cell priming dictate the fate of activated CD4 T cells (4, 5). T helper (Th) cell subsets (Th1, Th2, Th17, induced T regulatory [iTreg], and T follicular helper [Tfh] cells) are defined by the expression of a unique set of transcription factors as well as hallmark cytokines (4).
Tfh cells, defined by the expression of the Tfh cell master transcriptional factor B cell lymphoma 6 (BCL6), are important for the formation of productive germinal centers (GCs) and selection of high-affinity B cells (6–10). Tfh cells are characterized by their production of interleukin-21 (IL-21) and their selective expression of CXC chemokine receptor 5 (CXCR5), programmed death 1 (PD-1), and inducible costimulator (ICOS) (6). Like other CD4 T cell subsets, innate cytokines, namely, IL-6 and IL-21, have been proposed to be the drivers of Tfh cell differentiation (11). These cytokines induce the expression of BCL6, which promotes CXCR5 expression and GC formation, in addition to acting as a transcriptional repressor for alternative lineages (12–14). IL-6 is considered a key Tfh cell-priming cytokine, as it promotes the expression of BCL6 (15). However, an in vivo deficiency of IL-6 does not seem to impair Tfh cell differentiation (16). IL-12 has also been reported to be capable of inducing differentiation of IL-21-producing Tfh-like cells in humans; however, this finding could not be reproduced in murine models (17–19). A recent study has shown that during early Th1 cell differentiation, CD4 T cells pass through a Tfh-like phenotype and the local concentration of IL-2 dictates the fate of activated CD4 T cells to differentiate into Tfh cells versus non-Tfh lineage cells (20). Accumulating evidence also suggests that CD4 T cell lineages display a high degree of plasticity based on the cytokine milieu. Expression of BCL6 and IL-21 is not exclusive to Tfh cells, with other activated murine CD4 T cells also expressing these proteins (21–24). Human memory CD4 T cells with CXCR5 expression were reported to share functional properties with Tfh cells, but these cells also expressed canonical Th1, Th2, and Th17 cell transcription factors (25). These reports point to the existence of a cell-intrinsic regulator of Tfh cell fate determination. We therefore decided to investigate the early events in CD4 T cell differentiation in order to elucidate the role of innate cues in Tfh cell fate determination.
The importance of myeloid differentiation antigen 88 (MyD88) downstream of Toll-like receptors (TLRs) in DCs in driving T cell activation and differentiation is well established (26). Although MyD88 is a critical signaling adaptor downstream of TLRs, its function downstream of IL-1, IL-18, and IL-33 receptors in T cells is continuing to be unraveled (3). We have reported a critical role for T cell-intrinsic MyD88 in Th17 responses (27). Others have also shown that a lack of T cell-intrinsic MyD88 leads to compromised Th1 differentiation following protein immunization as a result of enhanced Treg suppression (28). In addition, T cell-intrinsic MyD88 has also been shown to be critical for priming of lymphocytic choriomeningitis virus (LCMV)-specific CD4 T cells (29). Pathogen recognition by DCs leads to the production of several inflammatory cytokines that shape the nature of adaptive immune responses. While priming cytokines like IL-6 and IL-12 have been prescribed functions in promoting specific CD4 T cell lineage commitment, the role of IL-1 family members in regulating early priming and lineage commitment of CD4 T cells is not entirely clear. In particular, whether T cell-intrinsic MyD88 regulates the early plasticity of T cell differentiation remains unknown.
In the present study, we examined the process of commitment by CD4 T cells with respect to lineage-specific markers and the role of innate cytokines in early CD4 T cell programming. Surprisingly, we found that the majority of activated CD4 T cells transition through a Tfh-like stage before differentiating into other effector lineages. Furthermore, we discovered that T cell-intrinsic MyD88, acting downstream of IL-1 and IL-18 receptors, is crucial for primed CD4 T cells to exit the transitional Tfh cell stage. T cell-specific deletion of MyD88 resulted in exaggerated Tfh lineage differentiation, which was accompanied by enhanced GC reactions. Our study provides novel insights into early CD4 T cell lineage commitment by identifying a previously unrecognized role for T cell-intrinsic MyD88 signaling in determining the fate of transitional Tfh lineage cells.
RESULTS
Activated CD4 T cells acquire a Tfh lineage phenotype before committing to other effector lineages.
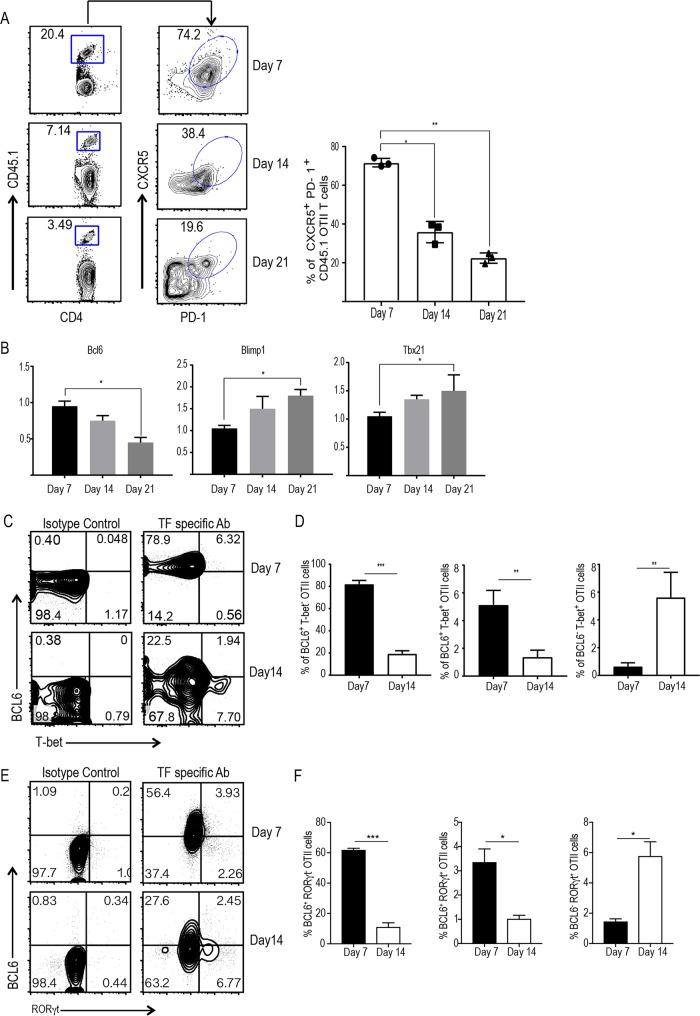
To investigate the early events of CD4 T cell differentiation in vivo, we used the OT-II T cell receptor (TCR) transgenic T cell transfer model. We closely mimicked the in vivo frequency and antigen-specific T cell response by transferring very low numbers (1 × 105) of purified OT-II T cells into wild-type (WT) mice. Following intravenous transfer of congenic OT-II cells, recipient mice were immunized with ovalbumin (OVA) mixed with lipopolysaccharide (LPS) emulsified in incomplete Freund's adjuvant. We tracked the expansion and differentiation of OT-II T cells on days 7, 14, and 21 in response to OVA immunization. Donor cells in the draining lymph nodes were collected and stained for congenic markers as well as T cell lineage markers. We found that the majority of transferred OT-II CD4+ T cells acquired a Tfh phenotype by day 7, as revealed by the high levels of expression of CXCR5 and PD-1 (Fig. 1A; see also Fig. S1A in the supplemental material). As expected, the transferred cell population (CD4+ CD45.1+) contracted after day 7. Notably, the proportion of Tfh-like cells among the remaining donor cells (CD4+ CD45.1+) gradually waned by day 14, and a majority of the donor T cells (CD4+ CD45.1+) lost this phenotype by day 21 (Fig. 1A). A small proportion of the donor cells retained Tfh lineage markers at day 21, suggesting that they potentially remained committed long-term Tfh cells rather than further differentiating into other lineages (Fig. 1A). We sorted the expanded OT-II T cells from the recipient mice and examined the transcriptional expression of Bcl6, B lymphocyte-induced maturation protein 1 (Blimp-1), and T-box transcription factor 21 (Tbx21; encoding T-bet) at various time points. Bcl6 expression was the highest at day 7 following immunization and gradually decreased over time, thereby mirroring the surface expression of Tfh lineage markers (Fig. 1B). Conversely, expression of transcription factors Blimp-1 and Tbx21 was the lowest at day 7 and gradually increased until day 21 (Fig. 1B). The kinetics of mRNA expression were reflected in the intracellular expression of these transcription factors. The majority of the cells expressed intracellular BCL6 at day 7 following immunization (Fig. S1B). Surprisingly, a small proportion of BCL6-expressing OT-II T cells also expressed the Th1-specific transcription factor T-bet (Fig. 1C and D) and the Th17-specific transcription factor retinoic acid-related orphan receptor γt (RORγt) (Fig. 1E and F). This suggests the existence of a partially differentiated state of CD4 T cells with the concomitant expression of Tfh and Th1/Th17 lineage transcription factors before terminal differentiation into effector T cells. Furthermore, the numbers of BCL6-expressing CD4 T cells were reduced by day 14 with a simultaneous increase in T-bet-expressing (T-bet+) BCL6-negative (BCL6−) cells (Fig. 1C and D) or RORγt+ BCL6− cells (Fig. 1E and F). Taken together, these results point to the possibility that activated CD4 T cells pass through a transient Tfh-like stage before effector differentiation into Th1 and Th17 lineage cells.
FIG 1.
The majority of primed CD4 T cells acquire Tfh lineage markers following clonal expansion. Sorted naive WT OT-II cells (CD45.1) were transferred into congenic (CD45.1) immunocompetent mice (1 × 105 cells/mouse). On the next day, the mice were immunized with OVA-LPS emulsified in IFA subcutaneously in the hind footpads. (A) The kinetics of OT-II T cell expansion and CXCR5+ PD-1+ OT-II cells at given time points (left) and the mean ± SEM percentage of expanded cells from three independent mice (right) are shown. (B) The relative expression of mRNA for transcription factors from sorted OT-II cells was evaluated by qPCR. (C and E) Flow plots of expanded OT-II T cells expressing lineage-specific transcription factors BCL6 and/or T-bet (C) or RORγt (E) on day 7 and day 14 postimmunization. (D and F) The mean ± SEM percentages of expanded OT-II cells expressing BCL6 and/or T-bet (D) and RORγt (F) on day 7 and day 14 postimmunization. Data are representative of those from three independent experiments with three mice for each group. (A, B, D, and F) P values were determined by an unpaired t test. *, P > 0.05; **, P > 0.01, ***, P > 0.001. TF, transcription factor; Ab, antibody.
Blocking the IL-1 family of cytokines leads to enhancement of Tfh lineage development.
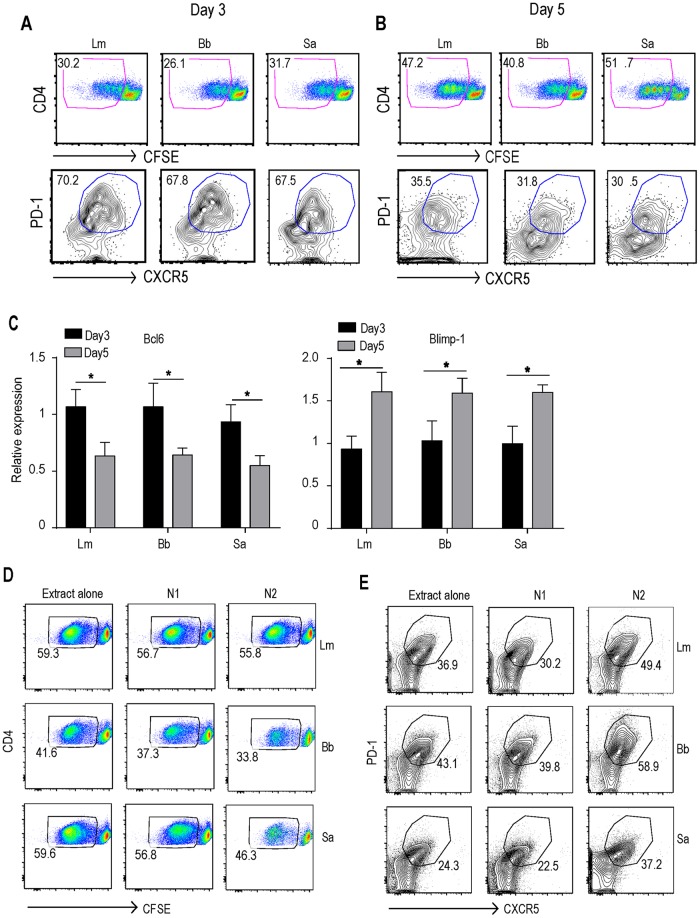
Use of the OT-II TCR transgenic T cells restricted our analysis to CD4 T cells of single epitope specificity, so we adopted a system that would allow us to study polyclonal antigen-specific T cell responses. We utilized an in vitro priming system where dendritic cells (DCs) are first fed with pathogen extract and then used to prime naive CD4 T cells. This approach has been successfully used previously to investigate pathogen-specific human CD4 T cell differentiation (30). We decided to use Staphylococcus aureus and Listeria monocytogenes, since these pathogens are known to induce an effective CD4 T cell response in vitro and in vivo (30, 31). We also used Borrelia burgdorferi, as it has recently been shown to be effective in inducing Tfh lineage commitment (32). Temporal analysis of the clonally expanded pathogen-specific CD4 T cells showed that the majority of activated CD4 T cells acquired a Tfh phenotype during the early stage (day 3) of priming (Fig. 2A). This was characterized by the surface expression of CXCR5 and PD-1 (Fig. 2A). Consistent with the in vivo data, this transient Tfh phenotype gradually diminished by day 5 (Fig. 2B). Analysis of Bcl6 and Blimp-1 expression in sorted carboxyfluorescein succinimidyl ester (CFSE)-negative cells corroborated the in vivo findings of higher levels of Bcl6 expression at early stages of differentiation, followed by the higher levels of Blimp-1 expression at later stages of differentiation (Fig. 2C). These in vitro results further support the hypothesis that, following priming, the majority of the clonally expanded CD4 T cells acquire Tfh-like characteristics before differentiating into other effector CD4 T cell lineages.
FIG 2.
Naive CD4 T cells acquire Tfh cell markers following in vitro pathogen priming. (A and B) CFSE-labeled CD4 T cells were cocultured with splenic CD11c+ DCs in the presence of 10 μg/ml of L. monocytogenes (Lm), B. burgdorferi (Bb), and S. aureus (Sa) bacterial extracts. Representative FACS plots show the proportion of primed and proliferating CFSElo, CXCR5+, and PD-1+ Tfh cells on day 3 (A) and day 5 (B) of coculture. (C) The relative expression of Bcl6 and Blimp-1 by sorted CFSElo CD4 T cells was quantified by qPCR. (D and E) CFSE-labeled CD4 T cells were cocultured with splenic DCs in the presence of 10 μg/ml of bacterial extract with different neutralizing antibodies (10 μg/ml). FACS plots of CFSElo cells (D) and CXCR5+ PD-1+ cells (gated on CFSElo) (E) on day 5. N1, neutralizing antibodies against IL-6, IL-12, IL-4, and IFN-γ; N2, conditions that used a recombinant antagonist for IL-1R (100 ng/ml) and anti-IL-18R (0.5 μg/ml) antibody. Data are representative of those from two independent experiments. (C) P values were determined by an unpaired t test. *, P > 0.05.
Our in vitro priming system allowed us to stimulate DCs with a complex mixture of microbial ligands and to test the role of specific innate cytokines via targeted neutralization. First, we individually neutralized innate cytokines, such as IL-12, IL-4, and IL-6, all of which are known to direct specific T effector lineage development. Surprisingly, we found that neutralization of IL-6, which has previously been implicated in Tfh cell differentiation, had no effect on early Tfh cell differentiation (Fig. S2). Additionally, neutralization of neither IL-12 nor IL-4 during priming impaired Tfh lineage development (data not shown). We then used a cocktail of antibodies that neutralized all three major priming cytokines (the N1 cocktail consisted ofanti-IL-6, anti-IL-12, and anti-IL-4) and observed only a marginal reduction in the proportion of T cells that acquired Tfh lineage markers (Fig. 2D and E). Next, we decided to abrogate IL-1β and IL-18 signaling since these cytokines are known to promote CD4 T cell responses but their role in Tfh cell fate determination remains elusive. Interestingly, simultaneous blocking of IL-1 receptor (IL-1R) and IL-1 receptor (IL-18R) signaling (with the N2 cocktail, which consists of an IL-1Ra antibody and anti-IL-18R) measurably enhanced the early proportion of Tfh lineage cells (Fig. 2D and E). There was also a decrease in the clonal expansion of primed CD4 T cells, confirming the findings of earlier studies on the role of IL-1 in CD4 T cell proliferation (Fig. 2D) (33). These data suggest that the IL-1 family of cytokines could be critical for the successful transition of Tfh-like cells into effector lineages.
T cell-intrinsic MyD88 plays a critical role in regulating the proportion of Tfh lineage cells.
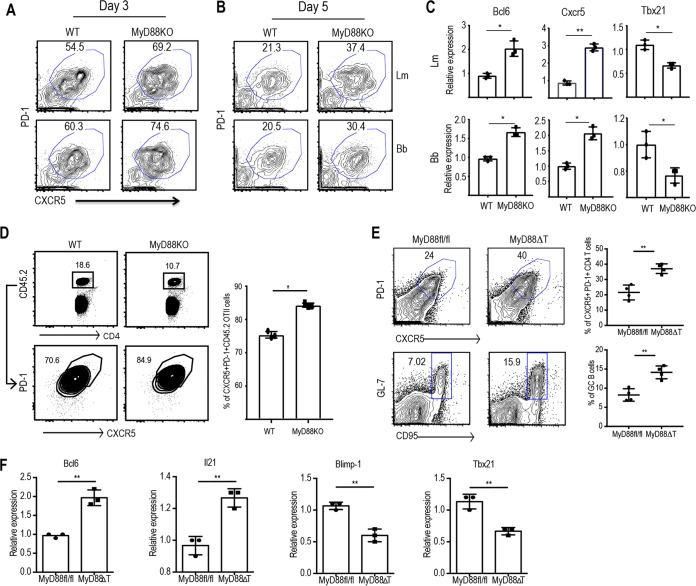
We have previously shown that IL-1R signaling in CD4 T cells is critical for the Th17 cell responses (27), and the role of IL-18 in Th1 cell biology is well-known (34). Since MyD88 is required for signal transduction downstream of both IL-1R and IL-18R (35), we explored the role of T cell-intrinsic MyD88 in early fate determination during CD4 T cell differentiation. We cocultured WT or MyD88-deficient naive CD4 T cells with WT splenic DCs in the presence of B. burgdorferi or L. monocytogenes extracts as described above. At 3 days after priming initiation, most CD4 T cells acquired the transient Tfh phenotype. However, we observed that a higher proportion of MyD88-deficient CD4 T cells acquired the Tfh phenotype (Fig. 3A), in agreement with the results of experiments where IL-1R and IL-18R signaling was simultaneously blocked. MyD88-deficient CD4 T cells continued to express enhanced Tfh lineage markers even on day 5 following priming (Fig. 3B). CFSE-negative CD4 T cells that lacked MyD88 expressed higher levels of Bcl6 and Cxcr5 than WT CD4 T cells (Fig. 3C). At the same time, Tbx21 was expressed at higher levels in WT T cells than MyD88-deficient T cells (Fig. 3C). While the levels of MyD88 expression did not change during the course of differentiation (Fig. S3), this set of results suggests that MyD88-mediated cues regulate the fate of Tfh cells and their transition into non-Tfh effector lineages. To test the role of T cell-intrinsic MyD88 in regulating Tfh lineage commitment in vivo, we transferred WT OT-II T cells or MyD88-deficient OT-II T cells into congenic mice. On the next day, recipient mice were immunized subcutaneously (in the footpad [fp]) with OVA and LPS emulsified in incomplete Freund adjuvant (IFA). The expansion and differentiation of donor cells were analyzed on day 7. MyD88-deficient OT-II T cells expanded less than the WT T cells at days 7 following priming, and their survival was affected at days 14 and 21, in agreement with the previous findings of the role of MyD88 in both clonal expansion and memory generation (Fig. 3D) (28). However, consistent with the in vitro findings, a higher proportion of MyD88-deficient OT-II T cells than WT OT-II T cells expressed Tfh cell markers at early stages following priming (Fig. 3D).
FIG 3.
T cell-intrinsic MyD88 plays a critical role in regulating the development of Tfh lineage cells. Naive CD4 T cells from WT and MyD88 KO mice were sorted and cocultured with CD11c+ splenic DCs in the presence of 10 μg/ml of L. monocytogenes or B. burgdorferi extract for 3 to 5 days. (A and B) FACS plots show the proportions of the CXCR5+ PD1+ Tfh cell populations on day 3 (A) and day 5 (B). (C) The relative expression of Bcl6, Cxcr5, and Tbx21 by sorted CFSElo CD4 T cells on day 5 upon stimulation with L. monocytogenes (top) and B. burgdorferi (bottom) extracts was quantified by qPCR. (D) Sorted naive WT OT-II cells (CD45.1) were transferred into congenic (CD45.1) immunocompetent mice (1 × 105 cells/mouse). On the next day, the mice were immunized with OVA-LPS emulsified in IFA subcutaneously in the hind footpads. A flow plot of the expanded OT-II cell population (left) and the proportion of CD4+ PD1+ CXCR5+ cells among the transferred cells (right) are shown. (E) Peyer's patches from MyD88fl/fl (control) and MyD88ΔT littermate mice were harvested, processed, and analyzed. (Left) Flow plots show the proportion of CXCR5+ PD-1+ Tfh cells (top) and CD19+ GL7+ CD95+ GC B cells (bottom) in control and MyD88ΔT mice. (Right) Quantification of flow data from five independent mice. (F) Relative expression of the respective genes in sorted CD4+ CD44hi CD62Llo T cells as quantified by qPCR. Data are representative of those from two independent experiments with three mice in each group. P values were determined by an unpaired t test. *, P > 0.05; **, P > 0.01.
T cell-intrinsic deletion of MyD88 results in enhanced polyclonal Tfh and GC responses in vivo.
One of the caveats of the OT-II T cell transfer model is the single TCR specificity, which may lead to confounding results. In order to examine a polyclonal CD4 T cell population, we generated mice that lacked MyD88 specifically in T cells. We crossed MyD88fl/fl mice (36) with transgenic mice expressing Cre under the control of the Lck promoter to generate MyD88fl/fl Lck-Cre Tg mice (here denoted MyD88ΔT mice). The mucosal immune system is constantly exposed to commensal microbes, and thus, there is continual antigen sampling by DCs and priming of CD4 T cells in Peyer's patches, leading to active Tfh cell and GC responses (37). We therefore analyzed the Tfh cell population in Peyer's patches and the mesenteric lymph nodes of MyD88ΔT mice and their control littermates. These experiments revealed that MyD88ΔT mice had a significantly higher proportion of Tfh lineage cells in Peyer's patches (Fig. 3E) than the MyD88fl/fl littermate controls. To test if the cells in Peyer's patches were bona fide Tfh cells that could support a robust GC reaction, we examined the status of the GCs in both MyD88ΔT mice and their littermate controls. The proportion of GC B cells correlated with exaggerated Tfh responses in MyD88ΔT mice (Fig. 3E). To quantify the proportions of different T cell lineages within the GCs, we sorted CD44hi CD62Llo CD4 T cells from the Peyer's patches and performed a quantitative PCR (qPCR) for Bcl6, Il21, Blimp1, and Tbx21. Consistent with the data obtained with OT-II cells, we found that MyD88-deficient CD4 T cells express more Bcl6 than WT CD4 T cells and that Blimp-1 and Tbx21 expression was inversely correlated with Bcl6 expression (Fig. 3F).
T cell-intrinsic MyD88 deletion leads to exaggerated Tfh and GC responses upon B. burgdorferi infection yet less efficient clearance of the pathogen.
B. burgdorferi-induced Lyme disease is a chronic inflammatory infection characterized by arthritis and myocarditis in humans (38). There is strong evidence for antibody-mediated control of B. burgdorferi infection and a role for both T cell-dependent and T cell-independent B cell responses in protective immunity (39–41). More recent work has demonstrated that the induction of robust Tfh cell and GC responses is necessary for the early clearance of B. burgdorferi (32). However, those Tfh responses were not sustained over time, unlike typical T cell-dependent antibody responses (32, 42, 43). Together, CD4 T cell help is critical for controlling the B. burgdorferi burden during the early phase of infection. This prompted us to characterize Tfh cell responses in vivo in response to B. burgdorferi infection. We infected WT mice with B. burgdorferi (strain 297, 105 CFU/mouse) intradermally and analyzed the Tfh cell and GC responses on days 14 and 21 postinfection in the spleen. Although naive mice did not show measurable Tfh cell and GC responses in the spleen (Fig. S4A and B), we found that intradermal infection with B. burgdorferi induced robust Tfh cell and GC responses by day 14 and that these responses persisted until day 21 of infection (Fig. S4C).
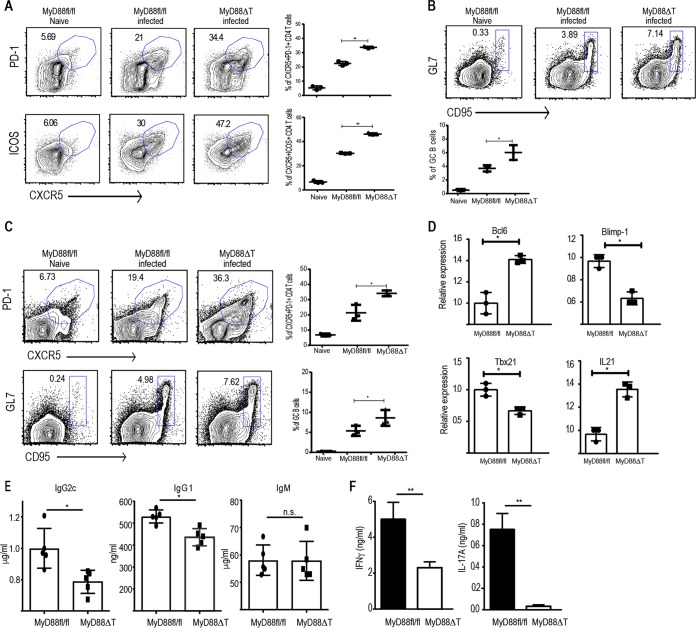
Since B. burgdorferi induces a robust Tfh cell and GC response in the spleens, we decided to test the role of T cell-intrinsic MyD88 in dictating the size of the Tfh cell and GC responses during an active infection. We infected MyD88fl/fl and MyD88ΔT mice with B. burgdorferi as described above. Of note, the proportion of Tfh cells and GC B cells in the spleen at steady state was comparable between MyD88fl/fl and MyD88ΔT mice (Fig. S4A and B). By day 14, MyD88ΔT mice had a significantly larger Tfh cell response in the spleen than the MyD88fl/fl littermate controls, as measured by the proportion of PD-1-expressing (PD-1+) CXCR5-expressing (CXCR5+) CD4 T cells (Fig. 4A). The proportion of ICOS-expressing CXCR5+ cells was also increased in MyD88ΔT mice, suggesting the greater localization of T cells in the B cell zone (Fig. 4A and S4D). Consequently, MyD88ΔT mice harbored a higher proportion of GC B cells than the controls, implying an exaggerated GC response (Fig. 4B). The elevated Tfh cell and GC responses were persistent and higher in MyD88ΔT mice, even at day 21 following B. burgdorferi infection (Fig. 4C). We sorted CD44hi CD62Llo CD4 T cells from the spleens of infected mice on day 14. Analysis of gene expression in the sorted cells revealed higher levels of Bcl6 and Il21 expression in the CD4 T cells from MyD88ΔT mice, while the expression of Blimp-1 and Tbx21 was higher in MyD88fl/fl mouse CD4 T cells (Fig. 4D).
FIG 4.
T cell-intrinsic MyD88 deletion leads to more robust Tfh cell and GC responses upon B. burgdorferi infection but defective Th1 and Th17 responses. MyD88fl/fl and MyD88ΔT mice were infected with B. burgdorferi strain 297 (105 CFU/mouse) intradermally. (A) Proportion of CD4+ T cells in the spleen that express Tfh cell markers on day 14 postinfection and quantification of CD4+ Tfh cells from three independent mice. (B) CD19+ GL7+ CD95+ GC B cells in the spleens of infected mice on day 14 postinfection and quantification of GC B cells from three independent mice. (C) Proportion of Tfh cells and GCs as well as their quantification in the spleens of infected mice on day 21 postinfection. (D) Relative expression of the indicated genes in sorted CD4+ CD44hi CD62Llo cells on day 14 postinfection as quantified by qPCR. (E) Enzyme-linked immunosorbent assay quantification of B. burgdorferi-specific immunoglobulins in the serum of B. burgdorferi-infected MyD88fl/fl and MyD88ΔT mice on day 21 postinfection. (F) CD4+ CD44hi cells from the spleens of B. burgdorferi-infected mice were sorted on day 14 and cocultured with splenic DCs in the presence of 50 μg/ml B. burgdorferi extract. After 48 h of coculture, the supernatants were collected to measure IL-17A and IFN-γ. Data are representative of those from two to three independent experiments with three mice in each group. P values were determined by an unpaired t test. *, P > 0.05; **, P > 0.01; n.s., not significant.
The ability of the mice to overcome B. burgdorferi infection is dependent on multiple innate and adaptive immunological responses (39). Previous studies have shown that B. burgdorferi-infected MyD88-deficient mice harbor a higher bacterial burden than WT mice, which was attributed to the defect in MyD88-dependent TLR signaling (44). However, IL-17-mediated antibody class switching has also been shown to be critical for the protection against B. burgdorferi (45). Gamma interferon (IFN-γ)-secreting CD4 T cells have been shown to be protective against B. burgdorferi infection. In light of our data pointing to a critical role of T cell-intrinsic MyD88 in early CD4 T cell fate determination, we decided to test the role of T cell-intrinsic MyD88 in the protection against B. burgdorferi infection. We quantified the bacterial burden in multiple tissues of MyD88fl/fl and MyD88ΔT mice at days 14 and 21 postinfection by qPCR using flaB-specific TaqMan probes. We observed a higher bacterial burden in the ears and skin of MyD88ΔT mice on days 14 and 21 than in those of the control mice (Fig. S4E). Analysis of the antibody responses from B. burgdorferi-infected mice showed that MyD88ΔT mice had significantly less IgG2c as well as IgG1 class-switched antibody than MyD88fl/fl mice (Fig. 4E). We did not observe defects in B. burgdorferi-specific IgM responses (Fig. 4E), presumably because B. burgdorferi-specific IgM has been reported to be largely T cell independent (32). Overall, these data argue that despite the higher initial Tfh cell and GC responses, the absence of MyD88 in T cells hampers CD4 T cell differentiation into non-Tfh lineage cells, resulting in reduced humoral responses. In particular, the defect in IgG2c could be due to the defect in suboptimal Th1 cell differentiation, as our previous results suggested.
We tested this idea by analyzing the functional status of CD4 T cells isolated from B. burgdorferi-infected mice on day 14. CD4 T cells were restimulated for 48 h with splenic DCs that had been fed B. burgdorferi extract. Supernatants were then analyzed for IFN-γ and IL-17A production. Consistent with our previous findings, MyD88-deficient CD4 T cells showed compromised IFN-γ and IL-17A production during B. burgdorferi-specific reactivation (Fig. 4F). These data establish that although T cell-intrinsic MyD88 deletion leads to higher Tfh responses, it also results in diminished effector differentiation and reduced titers of appropriate class-switched antibodies, thereby rendering mice inefficient in pathogen clearance.
Reexpression of MyD88 specifically in T cells is sufficient to reduce exaggerated Tfh responses.
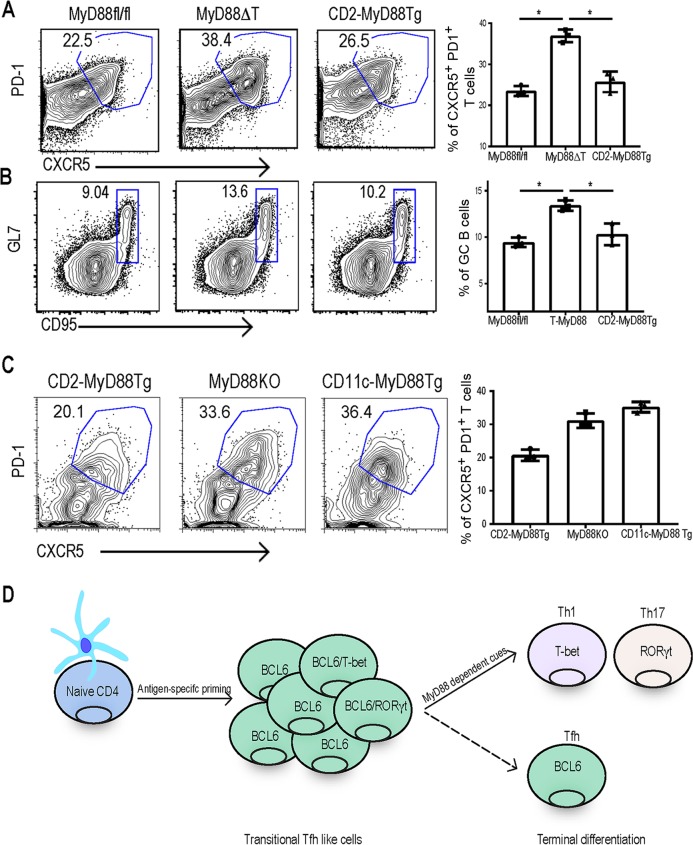
All our data presented above point to the fact that the absence of MyD88 in T cells leads to exaggerated Tfh responses. We therefore asked if reexpression of MyD88 specifically in T cells would reverse the Tfh lineage outcome. For this, we generated transgenic mice expressing MyD88 under the control of the human CD2 promoter and bred them to whole-body MyD88-deficient mice, here referred to as CD2-MyD88 Tg mice. We have successfully used this approach before to express MyD88 in DCs, in order to test the importance of DC-specific MyD88 in adaptive immunity (46). The CD2-MyD88 transgenic mice express MyD88 in T cells, but all other cellular compartments are still MyD88 deficient. Functional expression of the transgene was tested by analyzing T cell proliferation in response to IL-1β (Fig. S5A). Furthermore, B cells did not respond to TLR ligands that use MyD88-dependent signaling, confirming that MyD88 expression was limited to the T cell compartment (Fig. S5B). We cohoused these mice with MyD88fl/fl and MyD88ΔT mice for 3 weeks and examined the status of Tfh cells and GCs in the Peyer's patches. Isolated expression of MyD88 in T cells was sufficient to reverse the enhanced Tfh phenotype in CD4 T cells, and the proportion of Tfh cells in Peyer's patches of CD2-MyD88 Tg mice was comparable to that in MyD88fl/fl mice (Fig. 5A). Consequently, we also found a reduction in the steady-state GC responses in CD2-MyD88 Tg mice (Fig. 5B) compared to the responses in MyD88ΔT mice. Notably, expression of MyD88 only in dendritic cells did not reduce the proportion of Tfh lineage cells in Peyer's patches, suggesting a unique role for MyD88 in the T cell compartment in determining the transitional Tfh cell fate. Overall, the results from these genetic models show that T cell-intrinsic MyD88 is a critical regulator of effector T cell differentiation and the absence of MyD88-dependent signaling leads to exaggerated but dysfunctional Tfh lineage cells and germinal center reactions.
FIG 5.
Reexpression of MyD88 in T cells but not DCs reduces exaggerated Tfh responses. (A and B) MyD88fl/fl, MyD88ΔT, and CD2-MyD88 Tg mice were cohoused for 3 weeks before Peyer's patches were harvested, processed, and analyzed. (A) Flow plots show CD4+ T cells expressing surface Tfh cell markers CXCR5 and PD-1 (left) and their quantification for three independent mice (right). (B) Flow plots (left) and quantification (right) of CD19+ GL7+ CD95+ GC B cells. (C) CD2-MyD88 Tg, MyD88 KO, and CD11c-MyD88 Tg mice were cohoused for 3 weeks before Peyer's patches were harvested, processed, and analyzed for Tfh cell markers. Flow plots show the proportion of CD4+ T cells that express Tfh cell markers (left), and Tfh cells from three independent mice were quantified (right). Data are representative of those from three independent experiments with three mice in each group. P values were determined by an unpaired t test. *, P > 0.05. (D) Schematic representation of clonal expansion and differentiation of CD4 T cells into the transitional Tfh cell stage before terminal differentiation into effector lineages in a T cell-intrinsic MyD88 (IL-1 and IL-18)-dependent fashion.
DISCUSSION
Complex innate cytokine cues dictate the transition of naive T cells into lineage-committed effector T cells (3). Characterizing early stages of differentiation can provide clues into how T cells integrate diverse cytokines and generate a protective response. Here we have examined the early phenotypic and transcriptional kinetics of antigen-specific T cell differentiation in vivo and in vitro. Surprisingly, we found that early stages of clonally expanded antigen-specific CD4 T cells primarily acquired a Tfh-like phenotype (Fig. 5D). As the immune response progressed, Tfh cell characteristics waned with the emergence of effector T cell lineages (Fig. 5D). Our data point to an early plasticity during T cell differentiation, with Tfh-like cells acting as a transition stage but not the ultimate differentiation program. This is consistent with an earlier study reporting that T cells transiently go through the Tfh stage before differentiation into Th1 cells (20). The presence of Tfh cells during diverse immune challenges suggests that the Tfh stage is a central node of differentiation before the priming cytokines skew T cell differentiation into a particular lineage. This idea was further strengthened when we observed no defects in transitional Tfh cell differentiation when priming cytokines were neutralized.
Our finding that naive CD4 T cells transit through a Tfh-like state is consistent with several other reports showing that Tfh cell loci need to be actively suppressed in order to generate functional effector T cells (11). This implies that the Tfh-like transcriptional program is a default state adopted by CD4 T cells during clonal expansion. While several cell-intrinsic chromatin modulators have been identified to suppress the Tfh phenotype (47), no microenvironmental cue has been reported to restrain Tfh cell differentiation. Here, we found that T cell-intrinsic MyD88 signaling promotes the progression of Tfh-like CD4 T cells into effector T cell lineages. Specific deletion of T cell-intrinsic MyD88 led to exaggerated Tfh-like CD4 T cells and, consequently, enhanced GC responses in the Peyer's patches at steady state. Enhanced Tfh cell responses were observed in the spleens of MyD88ΔT mice following B. burgdorferi infection. This was directly correlated with inhibited expression of Tbx21, suggesting a fine balance between the Tfh and T effector cells regulated by MyD88-dependent signaling in T cells. It has been shown that specific members of the microRNA (miRNA) family, miR155 and miR17-92, enhance Tfh differentiation (48–50). miR155 expression directly correlated with the size of the Tfh cell population during inflammation (49). An independent study showed MyD88 to be a target of miR155 (51). The miR17-92 cluster also promotes Tfh cell and GC responses after protein immunization (46). Moreover, IL-1R expression was found to be downregulated by the miR17-92 cluster (46). We postulate that the aforementioned miRNAs might promote Tfh lineage commitment by inhibiting T cell-intrinsic MyD88/IL-1R signaling, thereby providing a novel link between the miRNA regulation of MyD88/IL-1R and Tfh cell differentiation.
It is well established that IL-6, IL-12, and IL-4 drive the effector T cell program by inducing or stabilizing lineage-specific transcription factors (4, 5). Here we show that the members of the IL-1 family of cytokines, especially IL-1 and IL-18, suppress the Tfh program for productive transcriptional conversion into appropriate effector lineages. The blocking of IL-1R or IL-18R signaling during naive T cell differentiation prevents the downregulation of Bcl6 and other Tfh cell-related factors, resulting in the concomitant expression of a transcription factor associated with effector T cell lineages, such as T-bet. It has been proposed earlier that there are distinct waves of Bcl6 expression during Tfh lineage development, and the first wave is induced in all activated T cells following their interaction with DCs (52). Our data suggest that it is important for these activated BCL6-expressing T cells to transition out of the Tfh lineage for effector differentiation, although a second wave of BCL6 is likely to be important for the development of long-lived Tfh cells. The unsuccessful transition into protective CD4 T cell lineages during infection resulted in defective protection against B. burgdorferi infection, as determined by a higher pathogen burden. It is important to note here that, even though Tfh-like cells were dominant in the absence of MyD88, the IgG1 and IgG2c levels were impaired. It has previously been reported that antibody responses against B. burgdorferi are not affected in the absence of MyD88 (53–55) but myeloid cell expression of MyD88 could still be critical for protection against B. burgdorferi (53, 54). Experiments performed using the whole-body MyD88-deficient mice do not account for the specific roles of MyD88 in myeloid cells, B cells, and T cells. Macrophages that lack MyD88 are defective in the phagocytosis and clearance of B. burgdorferi (53, 54), and consequently, a higher pathogen burden could result in higher IgM antibody responses in MyD88-deficient mice (55). Higher IgM antibody levels against B. burgdorferi in whole-body MyD88-deficient mice do not result in better protection (54), suggesting that the MyD88 in T cells and myeloid cells could be critical for protection against Borrelia. In our studies, MyD88 was intact in both myeloid cells and B cells, enabling us to specifically study its function in T cells. Here we show that despite an intact innate immune compartment, T cell-intrinsic deletion of MyD88 leads to defective Th1 and Th17 differentiation, thereby leading to compromised B. burgdorferi clearance. These data highlight the role of T cell-intrinsic MyD88 in generating optimal effector T cell response to mediate protection. Our findings are all the more relevant since loss-of-function mutations in MyD88 have been described in humans (56). Children with MyD88 deficiency succumb to several bacterial infections, which is often attributed to the impairment of the innate immune responses (56). Our data point toward the possibility that, in addition to defective innate immunity, these children might have suboptimal Th1 and Th17 lineage cells and protective antibodies that contribute to persistent invasive infections.
Collectively, our data identify a previously unknown role for T cell-intrinsic MyD88 as a critical sensor of microenvironmental cues during CD4 T cell priming which promotes effector T cell differentiation by suppressing Tfh cell programming.
MATERIALS AND METHODS
Mice.
MyD88fl/fl Lck-Cre Tg, MyD88fl/fl, MyD88 knockout (KO), CD11c-MyD88 Tg, and CD2-MyD88 Tg mice were bred and maintained at the animal facility of the University of Texas (UT) Southwestern Medical Center. Control C57BL/6 mice were obtained from the UT Southwestern Medical Center mouse breeding core facility. All mouse experiments were done per the protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the UT Southwestern Medical Center.
Generation of CD2-MyD88 Tg mice.
The Mus musculus MyD88-coding region was cloned into a modified human CD2 (hCD2) minigene-based vector, VA hCD2 (57), within EcoRI and SmaI sites. After removal of prokaryotic regions, the hCD2/MyD88 fragment was injected into C57BL/6 mouse blastocysts by the UT Southwestern Medical Center transgenic core facility. Seven founder lines were obtained and were then bred to MyD88 KO mice. F2 mice were tested for the expression of the transgene.
OT-II cell transfer and immunization.
CD45.2 WT OT-II or MyD88 KO OT-II T cells (1 × 105 cells/mouse) were transferred into congenic CD45.1 mice intravenously. On the next day, the mice were immunized with OVA (50 μg) and LPS (5 μg) emulsified in IFA in both footpads. The expansion of OT-II T cells was followed up to day 21.
Staining and flow cytometry.
Single-cell suspensions were prepared from Peyer's patches, lymph nodes, and spleen. Cells were stained with peridinin chlorophyll protein (PerCP)-conjugated anti-CD4, phycoerythrin (PE)-conjugated anti-CD44, PE-Cy7-conjugated anti-PD-1, allophycocyanin (APC)-conjugated anti-ICOS, PE-conjugated anti-CD95, APC-conjugated anti-GL7, and Pacific Blue-conjugated anti-CD19 (all from BioLegend) and with PerCP-conjugated anti-B220 and biotin-conjugated anti-CXCR5 (both from BD Biosciences) for 30 min on ice and were washed with fluorescence-activated cell sorting (FACS) buffer. For biotin-conjugated CXCR5, cells were incubated with streptavidin-conjugated BV421 for 30 min on ice, followed by 3 washes. The stained cells were then analyzed using a FACSCalibur or LSR-II (BD Biosciences) flow cytometer. Data were analyzed with FlowJo (version 10) software.
In vitro priming.
WT or MyD88 KO mouse naive CD4 T cells were isolated using a naive CD4 isolation kit (BioLegend) and were then labeled with CFSE (BioLegend). CFSE-labeled naive CD4 T cells were cocultured with splenic DCs fed with 10 μg/ml of bacterial extracts as described previously (30). Proliferating CFSElo CD4 T cells were analyzed for the Tfh phenotype on day 3 and day 5 of priming. For neutralization conditions, CFSE-labeled naive CD4 T cells and DCs were cocultured as mentioned above in the presence or absence of 10 μg/ml of neutralizing antibodies against specific cytokines.
B. burgdorferi infection.
B. burgdorferi infection experiments were conducted according to UT Southwestern Medical Center's IACUC-approved protocols. Low-passage-number cloned B. burgdorferi strain 297, which has proven infectivity and pathogenicity in mice, was used throughout the studies. Spirochetes were grown in Barbour-Stoenner-Kelly (BSK) complete medium (Sigma Chemical Co., St. Louis, MO) at 34°C to mid-log phase and then counted by dark-field microscopy using a Petroff-Hausser bacterial counting chamber. Spirochetes (105/mice) in BSK medium were inoculated intradermally at the middle posterior section of the neck. Control mice received BSK medium only. Mice were euthanized after 2 to 3 weeks of infection.
B. burgdorferi-specific antibody determination.
Ninety-six-well microtiter plates (Nunc) were coated with 20 μg/ml B. burgdorferi extract in bicarbonate buffer (pH 9.6) at 37°C overnight and blocked with phosphate-buffered saline (PBS) plus 10% fetal calf serum at room temperature for 3 h. After two washes with PBS–0.05% Tween 20, serially diluted sera (from 1:100 to 1:128,000) were applied and incubated at 4°C overnight. The wells were washed six times, biotinylated anti-IgM, anti-IgG2c, and anti-IgG1 were applied individually, and the plates were incubated at room temperature for 2 h. After six washes, the plates were incubated with streptavidin-peroxidase-conjugated anti-mouse immunoglobulin antibody at room temperature for 30 min. o-Phenylenediamine chromogenic substrate was added to all wells after six additional washes, and the absorbance of the plates was read at 490 nm.
Real-time PCR.
CD4+ CD44hi T cells from the Peyer's patches of cohoused mice or adoptively transferred OT-II T cells were sorted using a FACSAria cell sorter (BD Biosciences). CD4+ CD44hi T cells from the spleens of B. burgdorferi-infected mice were similarly sorted. Cells were lysed in the Trizol reagent, and RNA was extracted and reverse transcribed into cDNA. The relative expression of Bcl6, Blimp-1, Tbx21, CXCR5, and IL-21 was quantified using qPCR (AB QuantStudio 7 Flex). Values were normalized to those for β-actin, and relative expression changes were plotted.
Real-time PCR to determine B. burgdorferi burden in skin and ear.
The gene targeted for quantification of the B. burgdorferi burden was the flaB single-copy gene. The upstream primer was 5′-GTG CAT TTG GTT ATA TTG AG-3′, the downstream primer was 5′-CAG ACA GAG GTT CTA TAC A-3′, and the probe was 6-carboxyfluorescein (FAM)-5′-AAT AGA GCA ACT TAC AGA-3′-MGB. For quantitation of the host gene, the mouse β-actin gene was chosen. The upstream primer was 5′-AGA GGG AAA TCG TGC GTG AC-3′, the downstream primer was 5′-CAA TAG TGA TGA CCT GGC CGT-3′, and the TaqMan probe was FAM-5′CAC GGC CGC ATC CTC TTC TTC C-3′-black hole quencher 1. Plasmids containing the flaB gene of B. burgdorferi and the mouse β-actin gene served as standards. A plasmid containing the 1,100-bp flaB gene was obtained by PCR amplification using the flaB-specific upstream primer 5′-ATG ATT ATC AAT CAT AAT ACA TCA-3′ and downstream primer 5′-TTA TCT AAG CAA TGA CAA-3′. The PCR fragment was cloned into the pGEM-T vector (Promega, Madison, WI) and then propagated in competent Escherichia coli JM109 cells. The plasmid containing the 129-bp β-actin gene was obtained by PCR amplification using the upstream primer 5′-AGA GGG AAA TCG TGC GTG AC-3′ and the downstream primer 5′-CAA TAG TGA TGA CCT GGC CGT-3′ and cloned as described above. Plasmid DNA was quantified using a model 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). The plasmid control in the real-time PCR was used to ensure the efficacy of the assay and to allow compilation of standard curves for determination of the flaB copy number in mouse tissue. Tenfold serial plasmid dilutions ranging from 100 to 107 copies were prepared. The PCR mixture (total volume, 25 μl) consisted of 300 nM primer, 200 nM probe, 200 nM deoxynucleoside triphosphates, 3.5 mM MgCl2, 2 μl DNA, 1 U AmpliTaq Gold DNA polymerase, and 1× PCR buffer. Amplification and detection were performed using 40 cycles of 95°C for 10 s, 57°C for 30 s (for flaB), and 95°C for 10 s and 60°C for 30 s (for the β-actin gene). The β-actin gene and flaB PCRs used separate reaction mixtures. Individual samples were run in triplicate.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the C. Pasare lab for helpful discussion and critical reading of the manuscript.
This work was supported by grants from the National Institutes of Health (AI113125 and AI123176) to C.P.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00791-17.
REFERENCES
- 1.Yamane H, Paul WE. 2013. Early signaling events that underlie fate decisions of naive CD4(+) T cells toward distinct T-helper cell subsets. Immunol Rev 252:12–23. doi: 10.1111/imr.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitt N, Ueno H. 2015. Regulation of human helper T cell subset differentiation by cytokines. Curr Opin Immunol 34:130–136. doi: 10.1016/j.coi.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain A, Pasare C. 2017. Innate control of adaptive immunity: beyond the three-signal paradigm. J Immunol 198:3791–3800. doi: 10.4049/jimmunol.1602000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy KM, Reiner SL. 2002. The lineage decisions of helper T cells. Nat Rev Immunol 2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J, Yamane H, Paul WE. 2010. Differentiation of effector CD4 T cell populations. Annu Rev Immunol 28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 7.Fazilleau N, Mark L, McHeyzer-Williams LJ, McHeyzer-Williams MG. 2009. Follicular helper T cells: lineage and location. Immunity 30:324–335. doi: 10.1016/j.immuni.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King C. 2009. New insights into the differentiation and function of T follicular helper cells. Nat Rev Immunol 9:757–766. doi: 10.1038/nri2644. [DOI] [PubMed] [Google Scholar]
- 9.Nurieva RI, Chung Y. 2010. Understanding the development and function of T follicular helper cells. Cell Mol Immunol 7:190–197. doi: 10.1038/cmi.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu D, Vinuesa CG. 2010. The elusive identity of T follicular helper cells. Trends Immunol 31:377–383. doi: 10.1016/j.it.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity 41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science 325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu D, Batten M, Mackay CR, King C. 2009. Lineage specification and heterogeneity of T follicular helper cells. Curr Opin Immunol 21:619–625. doi: 10.1016/j.coi.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Choi YS, Eto D, Yang JA, Lao C, Crotty S. 2013. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol 190:3049–3053. doi: 10.4049/jimmunol.1203032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, Crotty S, Craft J. 2010. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol 185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner-Nanan B, Deenick EK, Tangye SG. 2009. Early commitment of naive human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol Cell Biol 87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt N, Morita R, Bourdery L, Bentebibel SE, Zurawski SM, Banchereau J, Ueno H. 2009. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity 31:158–169. doi: 10.1016/j.immuni.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler NS, Kulu DI. 2015. The regulation of T follicular helper responses during infection. Curr Opin Immunol 34:68–74. doi: 10.1016/j.coi.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, Schwartzberg PL, Hager GL, O'Shea JJ. 2011. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund R, Ahlfors H, Kainonen E, Lahesmaa AM, Dixon C, Lahesmaa R. 2005. Identification of genes involved in the initiation of human Th1 or Th2 cell commitment. Eur J Immunol 35:3307–3319. doi: 10.1002/eji.200526079. [DOI] [PubMed] [Google Scholar]
- 22.Mehta DS, Wurster AL, Weinmann AS, Grusby MJ. 2005. NFATc2 and T-bet contribute to T-helper-cell-subset-specific regulation of IL-21 expression. Proc Natl Acad Sci U S A 102:2016–2021. doi: 10.1073/pnas.0409512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suto A, Kashiwakuma D, Kagami S, Hirose K, Watanabe N, Yokote K, Saito Y, Nakayama T, Grusby MJ, Iwamoto I, Nakajima H. 2008. Development and characterization of IL-21-producing CD4+ T cells. J Exp Med 205:1369–1379. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei L, Laurence A, Elias KM, O'Shea JJ. 2007. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem 282:34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, Lavecchio EM, Punaro M, Pascual V, Banchereau J, Ueno H. 2011. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwasaki A, Medzhitov R. 2004. Toll-like receptor control of the adaptive immune responses. Nat Immunol 5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 27.Hu W, Troutman TD, Edukulla R, Pasare C. 2011. Priming microenvironments dictate cytokine requirements for T helper 17 cell lineage commitment. Immunity 35:1010–1022. doi: 10.1016/j.immuni.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schenten D, Nish SA, Yu S, Yan X, Lee HK, Brodsky I, Pasman L, Yordy B, Wunderlich FT, Bruning JC, Zhao H, Medzhitov R. 2014. Signaling through the adaptor molecule MyD88 in CD4+ T cells is required to overcome suppression by regulatory T cells. Immunity 40:78–90. doi: 10.1016/j.immuni.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou S, Kurt-Jones EA, Cerny AM, Chan M, Bronson RT, Finberg RW. 2009. MyD88 intrinsically regulates CD4 T-cell responses. J Virol 83:1625–1634. doi: 10.1128/JVI.01770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, Monticelli S, Lanzavecchia A, Sallusto F. 2012. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature 484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 31.Pepper M, Linehan JL, Pagan AJ, Zell T, Dileepan T, Cleary PP, Jenkins MK. 2010. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat Immunol 11:83–89. doi: 10.1038/ni.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elsner RA, Hastey CJ, Baumgarth N. 2015. CD4+ T cells promote antibody production but not sustained affinity maturation during Borrelia burgdorferi infection. Infect Immun 83:48–56. doi: 10.1128/IAI.02471-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Sasson SZ, Hu-Li J, Quiel J, Cauchetaux S, Ratner M, Shapira I, Dinarello CA, Paul WE. 2009. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc Natl Acad Sci U S A 106:7119–7124. doi: 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. 2001. Interleukin-18 regulates both Th1 and Th2 responses. Annu Rev Immunol 19:423–474. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi O, Akira S. 2002. MyD88 as a bottle neck in Toll/IL-1 signaling. Curr Top Microbiol Immunol 270:155–167. [DOI] [PubMed] [Google Scholar]
- 36.Hou B, Reizis B, DeFranco AL. 2008. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity 29:272–282. doi: 10.1016/j.immuni.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reboldi A, Cyster JG. 2016. Peyer's patches: organizing B-cell responses at the intestinal frontier. Immunol Rev 271:230–245. doi: 10.1111/imr.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petzke M, Schwartz I. 2015. Borrelia burgdorferi pathogenesis and the immune response. Clin Lab Med 35:745–764. doi: 10.1016/j.cll.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Tracy KE, Baumgarth N. 2017. Borrelia burgdorferi manipulates innate and adaptive immunity to establish persistence in rodent reservoir hosts. Front Immunol 8:116. doi: 10.3389/fimmu.2017.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skogman BH, Hellberg S, Ekerfelt C, Jenmalm MC, Forsberg P, Ludvigsson J, Bergstrom S, Ernerudh J. 2012. Adaptive and innate immune responsiveness to Borrelia burgdorferi sensu lato in exposed asymptomatic children and children with previous clinical Lyme borreliosis. Clin Dev Immunol 2012:294587. doi: 10.1155/2012/294587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golde WT, Piesman J, Dolan MC, Kramer M, Hauser P, Lobet Y, Capiau C, Desmons P, Voet P, Dearwester D, Frantz JC. 1997. Reactivity with a specific epitope of outer surface protein A predicts protection from infection with the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun 65:882–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elsner RA, Hastey CJ, Olsen KJ, Baumgarth N. 2015. Suppression of long-lived humoral immunity following Borrelia burgdorferi infection. PLoS Pathog 11:e1004976. doi: 10.1371/journal.ppat.1004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hastey CJ, Elsner RA, Barthold SW, Baumgarth N. 2012. Delays and diversions mark the development of B cell responses to Borrelia burgdorferi infection. J Immunol 188:5612–5622. doi: 10.4049/jimmunol.1103735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bockenstedt LK, Liu N, Schwartz I, Fish D. 2006. MyD88 deficiency enhances acquisition and transmission of Borrelia burgdorferi by Ixodes scapularis ticks. Infect Immun 74:2154–2160. doi: 10.1128/IAI.74.4.2154-2160.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oosting M, van de Veerdonk FL, Kanneganti TD, Sturm P, Verschueren I, Berende A, van der Meer JW, Kullberg BJ, Netea MG, Joosten LA. 2011. Borrelia species induce inflammasome activation and IL-17 production through a caspase-1-dependent mechanism. Eur J Immunol 41:172–181. doi: 10.1002/eji.201040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasare C, Medzhitov R. 2004. Toll-dependent control mechanisms of CD4 T cell activation. Immunity 21:733–741. doi: 10.1016/j.immuni.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Nurieva RI, Dong C. 2013. Transcriptional regulation of follicular T-helper (Tfh) cells. Immunol Rev 252:139–145. doi: 10.1111/imr.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM, Jeker LT. 2013. The microRNA cluster miR-17∼92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol 14:840–848. doi: 10.1038/ni.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu R, Kagele DA, Huffaker TB, Runtsch MC, Alexander M, Liu J, Bake E, Su W, Williams MA, Rao DS, Moller T, Garden GA, Round JL, O'Connell RM. 2014. miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation. Immunity 41:605–619. doi: 10.1016/j.immuni.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumjohann D, Ansel KM. 2013. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol 13:666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang B, Xiao B, Liu Z, Li N, Zhu ED, Li BS, Xie QH, Zhuang Y, Zou QM, Mao XH. 2010. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett 584:1481–1486. doi: 10.1016/j.febslet.2010.02.063. [DOI] [PubMed] [Google Scholar]
- 52.Baumjohann D, Okada T, Ansel KM. 2011. Cutting edge: distinct waves of BCL6 expression during T follicular helper cell development. J Immunol 187:2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- 53.Bolz DD, Sundsbak RS, Ma Y, Akira S, Kirschning CJ, Zachary JF, Weis JH, Weis JJ. 2004. MyD88 plays a unique role in host defense but not arthritis development in Lyme disease. J Immunol 173:2003–2010. doi: 10.4049/jimmunol.173.3.2003. [DOI] [PubMed] [Google Scholar]
- 54.Liu N, Montgomery RR, Barthold SW, Bockenstedt LK. 2004. Myeloid differentiation antigen 88 deficiency impairs pathogen clearance but does not alter inflammation in Borrelia burgdorferi-infected mice. Infect Immun 72:3195–3203. doi: 10.1128/IAI.72.6.3195-3203.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woods A, Soulas-Sprauel P, Jaulhac B, Arditi B, Knapp AM, Pasquali JL, Korganow AS, Martin T. 2008. MyD88 negatively controls hypergammaglobulinemia with autoantibody production during bacterial infection. Infect Immun 76:1657–1667. doi: 10.1128/IAI.00951-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku CL, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y, Vasconcelos J, Sirvent N, Guedes M, Vitor AB, Herrero-Mata MJ, Arostegui JI, Rodrigo C, Alsina L, Ruiz-Ortiz E, Juan M, Fortuny C, Yague J, Anton J, Pascal M, Chang HH, Janniere L, Rose Y, Garty BZ, Chapel H, Issekutz A, Marodi L, Rodriguez-Gallego C, Banchereau J, Abel L, Li X, Chaussabel D, Puel A, Casanova JL. 2008. Pyogenic bacterial infections in humans with MyD88 deficiency. Science 321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. 1995. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods 185:133–140. doi: 10.1016/0022-1759(95)00124-S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.