ABSTRACT
A DNA microarray platform, based on the nucleotide sequences of the internal transcribed spacer regions (ITS1 and ITS2) of the rRNA gene, was developed to identify 32 fungal pathogens at the species level. The probe sequences were spotted onto polycarbonate slides with a mini-microarray printer, and after the hybridization, the results were visible with the naked eye. The performance of the microarray platform was evaluated against the commercial automated systems (Vitek 2 and BD Phoenix systems) and DNA sequencing (gold standard). A total of 461 blood culture bottles were tested: 127 positive for fungi, 302 positive for bacteria, and 32 that were negative. Once the microorganisms were identified by automated systems, fungal DNA was extracted directly from the blood culture bottles. The DNA products were tested using the microarray platform, and DNA sequencing was performed. The results of the microarray and DNA sequencing were concordant in 96.7% of cases, and the results from the automated systems and DNA sequencing were concordant in 98.4%. Of all the nucleotide sequences contained in the microarray platform, the microarray failed to identify four fungal isolates (one Candida parapsilosis, two Candida tropicalis, and one Cryptococcus neoformans). Of note, the microarray detected Candida krusei DNA in two blood cultures from the same patient, whereas the automated system was only positive for Enterococcus faecium. Our microarray system provided reliable and fast fungal identification compared to that from DNA sequencing and the automated systems. The simplicity of reading the results by the naked eye made this DNA platform a suitable method for fungal molecular diagnosis.
KEYWORDS: blood culture, fungal infection, microarrays
INTRODUCTION
The diagnosis of invasive fungal infections has been a challenge for clinicians and microbiologists, as an early diagnosis of the causative agent, together with a prompt institution of antifungal therapy, is associated with higher rates of patient survival (1–3). Despite medical and diagnostic advances in the field of fungal infections, there is a need for new diagnostic techniques and studies to leverage fast and precise identification of the pathogens (2).
The gold standard diagnosis of fungal infection occurs when fungi are recovered by culture or tissue biopsy. The modern automated blood culture systems are valuable methods for the identification of several invasive fungal infections, mainly for Candida spp., Fusarium spp., Cryptococcus spp., and Trichosporon spp. (4, 5). Non-culture-based diagnostic methods, such as the detection of fungal antigens or metabolites (6), and molecular methods using PCR have been introduced to ameliorate the identification of the fungal agent when conventional culture results are negative (7, 8). Galactomannan (GM), (1,3)-beta-d-glucan (BDG), and PCR methods have provided major improvements in the diagnoses of candidiasis and aspergillosis, and results from GM, BDG, and PCR methods were included among the mycological criteria for probable invasive fungal infections in immunocompromised patients (9–12). Promising and sophisticated laboratorial methodologies, such as matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (13, 14) and T2 magnetic resonance assays (15, 16), are now being used in the diagnoses of fungal infections. These new systems brought fast and accurate identification of a wide spectrum of fungal pathogens in the clinical laboratory, even though their results rely on commercial libraries. However, due to their high cost, they remain unaffordable for most hospitals in Brazil.
Recently, new commercial tests have been developed to improve the care of patients with invasive fungal infections. Luminex Molecular Diagnostics (Toronto, Canada) developed the fungal 23 analyte-specific reagent for the detection of several yeasts and molds of clinical importance (17). AdvanDx (Woburn, MA, USA) established a system using peptide nucleic acid probes for fluorescence in situ hybridization to establish fast species identification of Candida spp., including the identification from blood cultures (18). The PLEX-ID system (Abbott Molecular Inc., Des Plaines, IL) applies a broad-range PCR amplification coupled with electrospray ionization-mass spectrometry for the direct detection of pathogens without the need to wait for growth in culture (19). These methods can significantly reduce the turnaround time for the results; however, few studies compared the sensitivity and specificity of the methods with DNA sequencing identification.
Although DNA sequencing is time consuming and highly expensive and requires expertise to perform, it is an important complementary method of classical mycological identification of a fungal pathogen. Thus, there is a need for simpler, less expensive, and reliable molecular technologies, with an accuracy similar to that of DNA sequencing, for use in clinical practice. Several DNA microarray platforms have been developed to identify a diversity of microorganisms, such as viruses, bacteria, and fungi, simultaneously or individually, with good sensitivity and specificity (20–23). Due to the low stability of most fluorescent dyes and the expensive scanning equipment used in this technique, our research group developed a more affordable DNA microarray system for fungal identification at the genus and species levels. This platform was based on a panel of oligonucleotide probes that were designed on the basis of internal transcribed spacer (ITS) regions (24) of the rRNA gene to identify a variety of fungal species simultaneously in one polycarbonate slide, for which the results were visible to the naked eye (25). This array was previously studied by testing fungal DNA extracted from isolates grown in culture medium plates (25–27).
To improve the diagnosis of fungal infections, in our hospital, we carried out a study of the performance of our DNA microarray in the identification of fungi directly from blood culture bottles compared to the results from the automated systems and DNA sequencing.
MATERIALS AND METHODS
Study design.
The study was performed at the Clinical Hospital of the University of Campinas, Sao Paulo, Brazil, which is the referral hospital for tertiary care for more than 3 million inhabitants. This study was approved by the Institutional Ethical Committee (no. CAAE: 32955414.0.0000.5404).
From 2013 to 2016, we studied 461 blood cultures. Until August 2014, all blood cultures were performed using BacT/ALERT 3D (bioMérieux, Marcy-l'Etoile, France), and the identification of the microorganisms was done by the automated system Vitek2 (lab equipment, bioMérieux, Durham, NC, USA). Beginning in September 2014, the clinical microbiology laboratory changed its equipment and systems to BD Bactec FX and BD Phoenix (Becton Dickinson, Franklin Lakes, NJ, USA). The results of fungal identification obtained with the automated systems were compared with those from DNA microarray and DNA sequencing. As the product from the blood culture bottles used in this study came from hospitalized patients, the samples were analyzed by DNA microarray after the standard microbiological and biochemical tests were performed. The microarray results were not reported to the assistant physician, and they were not used for therapeutic purposes.
DNA extraction from blood culture bottles.
DNA extraction was performed according to the High Pure PCR kit (Roche Life Science, Indianapolis, IN, USA) specifications. Briefly, 200 μl of the contents of the blood culture bottles were added to 1 ml of alkaline solution (0.5 M NaOH, 0.05 M trisodium citrate dihydrate). After shaking, the samples were incubated at room temperature for 5 min and centrifuged, and the pellets were suspended in 1 ml of phosphate-buffered saline (PBS), followed by centrifugation at 16,000 × g for 5 min. The supernatants were discharged, and the pellets were suspended in 300 μl of PBS and 200 μl of tissue and lysis buffer (4 M urea, 200 mM Tris, 20 mM NaCl, and 200 mM EDTA [pH 7.4]). The mixtures were homogenized using a MagNA Lyser (Roche Life Science) at 5,000 × g for 10 min. The samples were then centrifuged at 10,000 × g for 2 min, and the supernatants were transferred to microtubes. Next, 200 μl of binding buffer (6 M guanidine-HCl, 10 mM urea, 10 mM Tris-HCl, 20% Triton X-100 [pH 4.4]) and 40 μl of proteinase K were added, and the mixtures were incubated at 70°C for 10 min and centrifuged at 16,000 × g for 2 min. The supernatants were transferred to new microtubes and 100 μl of isopropanol was added. The mixtures were centrifuged at 8,000 × g for 1 min, and the supernatants were discharged. Next, 500 μl of inhibitor removal buffer (5 M guanidine-HCl, 20 mM Tris-HCl [pH 6.6]) was added to the pellets followed by the addition of 20 μl of absolute ethanol. The samples were centrifuged at 8,000 × g for 1 min, and the supernatants were discharged. Then, 500 μl of wash buffer (20 mM NaCl, 2 mM Tris-HCl [pH 7.5]) was added to the samples along with 80 μl of absolute ethanol. The samples were centrifuged at 8,000 × g for 1 min, and this procedure was repeated twice. The supernatants were discharged, and the pellets were dissolved in 50 μl of elution buffer (10 mM Tris-HCl [pH 8.5]).
PCRs and DNA sequencing.
PCR was performed using the 5′-biotin-labeled fungus-specific universal primers ITS1-bio (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4-bio (5′-TCCTCCGCTTATTGATATGC-3′) (Sigma-Aldrich, St. Louis, MO, USA) (22) to amplify the entire ITS region and provide biotin labeling. The sizes of the amplified fragments varied from 426 to 930 bp depending on the fungal species. PCR was performed using PCR master mix (Promega, Fitchburg, WI, USA). The PCR mixtures were incubated in a Veriti 96-well thermal cycler (Applied Biosystems, Foster City, CA, USA) under the following conditions: 2 min for initial denaturation at 98°C, 40 cycles of DNA denaturation at 98°C for 30 s, primer annealing at 55°C for 30 s, and elongation at 72°C for 1 min, and a final elongation step at 72°C for 5 min. The PCR products were verified by electrophoresis in a 2% agarose gel at 100 V for 30 min. For DNA sequencing, the PCR products were used with the universal fungus-specific primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Sigma-Aldrich, St. Louis, MO), followed by purification with ExoSAP-IT for PCR product cleanup (Affymetrix USB, Cleveland, OH, USA), and the products were sequenced with the BigDye Terminator regent kit (Applied Biosystems) according to the manufacturer's protocols on an ABI Prism 3100 genetic analyzer (Applied Biosystems). The sequenced data were assembled and analyzed using BLAST ([Basic Local Alignment Search Tool] http://blast.ncbi.nlm.nih.gov), and the databases of the Centraalbureau voor Schimmelcultures (CBS) Fungal Biodiversity Centre, Institute of the Royal Netherlands Academy of Arts and Sciences (KNAW) (http://www.cbs.knaw.nl/index.php), and the Molecular Mycology Research Laboratory, University of Sidney (http://www.mycologylab.org), were used for genetic identification. For Fusarium, a partial portion of the translation elongation factor-1-alpha (TEF1α) gene was sequenced using HS392, HS393, EF11, and EF21 primers (28, 29). The DNA sequences were edited and assembled by Sequencher version 5.2.4 (Gene Codes Corp., Ann Harbor, MI, USA). For identification, a homology search for the sequences of the TEF1α gene was done using the BLAST tool of the NCBI database (GenBank), the database FUSARIUM-ID, and the Fusarium CBS database (http://www.cbs.knaw.nl/fusarium). To confirm the identity of our Fusarium species, we evaluated their positions with the maximum likelihood method. The TEF1α analysis tree was constructed. For these analyses, our sequences, together with the sequences retrieved from GenBank and the CBS database, were analyzed. The consensus sequences were computed with SeqMan from the Lasergene package (DNAStar Inc., Madison, WI, USA). The sequences were aligned with the program MAFFT (www.ebi.ac.uk/Tools/msa/mafft/), followed by manual adjustments with MEGA 6 (30) and BioEdit v7.0.5.2.
Visual DNA microarray slides.
The DNA microarray slide protocol was previously described by Sakai et al. (25) with modifications. Briefly, the oligonucleotide probes consisted of species/genus-specific nucleotide sequences with biotin-labeled poly(T) anchors at the ends of each nucleotide (Sigma-Aldrich), which were designed based on the ITS sequences (ITS1 and ITS2) of the typed strains (GenBank database, American Type Culture Collection, CBS) and the Medical Mycology Research Center, Chiba University, Japan. This microarray platform covered 13 genera and 32 species of pathogenic fungi (see Table S1 in the supplemental material). The probe sequences were spotted onto polycarbonate slides (NGK Insulators, Ltd., Aichi, Japan) with a KCS mini-microarray printer (Kubota Comps Corporation, Amagasaki, Japan). All slides had positive controls for fungi, where universal signals for fungi were enclosed in dotted lines framed at the right bottom of each slide. After the hybridization was completed, the results were visible to the naked eye. This microarray assay could be completed in ≤6 h.
Clinical data.
Demographic data were collected from the patients' records that had blood cultures collected for molecular analyses. The following was collected: underlying diseases, age, sex, outcome (death or survival), and use of antifungal therapy.
Accession number(s).
Sequences were deposited in GenBank under accession numbers MG599122 to MG599246 (see Table S2 in the supplemental material).
RESULTS
The inclusion of the blood culture bottles in our study occurred according to the routine procedures of the clinical microbiology laboratory, reflecting the population of our hospitalized patients during the study period. The 461 blood cultures were collected from 242 patients, and more than one blood culture from an individual patient was studied. One hundred twenty-seven blood culture bottles tested positive for fungi (77 patients), 302 bottles tested positive for bacteria, and 32 bottles tested negative after 7 days of incubation.
According to the automated systems, Vitek2 and BD Bactec FX, Candida spp. represented 77.2% (98 isolates) of all fungal isolates, of which Candida albicans (26 isolates), Candida tropicalis (29 isolates), Candida parapsilosis complex (12 isolates), Candida krusei (14 isolates), and Candida glabrata (11 isolates) were more frequently isolated than other Candida species (Table 1). For Candida species, the automated system identification was 100% concordant with the results of DNA sequencing for all the examined fungi except for the C. parapsilosis complex (83.3%), of which two isolates of Candida orthopsilosis were misidentified as C. parapsilosis; however, neither Vitek2 nor BD Phoenix has an identification database to differentiate the species of the C. parapsilosis complex. The GenBank accession numbers of the nucleotide sequences of fungal DNA are MG599122 to MG599246 in the Microarray_sequences_2017.sqn file. The full list of accession numbers is displayed in Table S2.
TABLE 1.
Identification of 127 blood cultures bottles positive for fungi according to automated systems Vitek2 and BD Phoenix, DNA sequencing, and DNA microarray
| Fungi | No. of isolates identified by: |
% agreement |
||||
|---|---|---|---|---|---|---|
| Microbiological identification | DNA sequencing | DNA microarray | Microbiology and DNA sequencing | DNA microarray and DNA sequencing | Microbiology and DNA microarray | |
| Candida albicans | 26 | 26 | 26 | 100 | 100 | 100 |
| Candida dubliniensis | 3 | 3 | 3 | 100 | 100 | 100 |
| Candida glabrata | 11 | 11 | 11 | 100 | 100 | 100 |
| Candida guilliermondii | 1 | 1 | 1 | 100 | 100 | 100 |
| Candida krusei | 14 | 14 | 14 | 100 | 100 | 100 |
| Candida lusitaniae | 1 | 1 | 1 | 100 | 100 | 100 |
| Candida parapsilosis | 12 | 10 | 9 | 83.3 | 90 | 75a |
| Candida orthopsilosisb | 0 | 2 | 0 | |||
| Candida pelliculosab | 1 | 1 | 100 | |||
| Candida tropicalis | 29 | 29 | 27 | 100 | 93.1 | 93.1 |
| Cryptococcus neoformans | 24 | 24 | 23 | 100 | 95.8 | 95.8 |
| Fusarium spp. | 2 | 2 | 2 | 100 | 100 | 100 |
| Histoplasma capsulatum | 1 | 1 | 1 | 100 | 100 | 100 |
| Saccharomyces cerevisiaeb | 2 | 2 | 100 | |||
| Total no. of isolates | 127 | 127 | 118 | 98.4c | 96.7d | 92.9e |
Percentage calculated by dividing the number of C. parapsilosis isolates identified by DNA microarray (9 isolates) by the total number of C. parapsilosis isolates identified by automated systems (12 isolates). Two C. orthopsilosis isolates were misidentified by the automated systems as C. parapsilosis. DNA microarray did not identify C. orthopsilosis as C. parapsilosis.
Absence of specific probes for identification by DNA microarray. Percentage agreement was calculated only for fungal species with oligonucleotide sequences contained in the platform.
Percentage calculated by dividing the number of isolates that were identically identified by automated systems and DNA sequencing (125 isolates) by the total number of isolates analyzed (127 isolates).
Percentage calculated by dividing the number of isolates identified in the DNA platform (118 isolates) by the number of isolates in the DNA sequencing whose oligonucleotide sequences were contained in the DNA platform (122 of the total of 127 isolates).
Percentage calculated by dividing the total number of isolates identified by DNA microarray (118 isolates) by the total number of isolates identified by microbiological identification (127 isolates).
The specific probes for the identification of the fungal species in the DNA microarray slant failed to identify one C. parapsilosis, two C. tropicalis, and one Cryptococcus neoformans. The microarray platform did not contain nucleotide sequences for C. orthopsilosis, resulting in the absence of identification, but the platform did not misidentify the two C. orthopsilosis isolates. For the same reason, the DNA microarray system also could not identify one Candida pelliculosa and two Saccharomyces cerevisiae. Cryptococcus spp. were the second most frequent yeast recovered in blood cultures, where 24 blood culture bottles yielded Cryptococcus, which was confirmed as C. neoformans by DNA sequencing. Two Fusarium isolates recovered from blood cultures were identified by DNA sequencing as the Fusarium solani species complex (FSSC) and as F. solani by DNA microarray. Our array has oligonucleotide sequences to identify FSSC and non-FSSC. The results of the identification of fungal isolates obtained with the automated systems and microarray agreed in 98.4% of cases, and the microarray results were concordant with DNA sequencing in 97.6% (Table 1). All 32 blood cultures that tested negative also tested negative by the microarray platform.
Positive blood cultures for bacteria were tested for DNA microarray at two different times: from 2014 to 2015, 93 bottles that tested positive for bacteria independent of the incubation time, and from 2015 to 2016, 209 bottles that tested positive for bacteria in less than 12 h. Among the 93 bottles, three blood cultures were collected on the same day from the same patient (patient RMPR) (Table 2). The automated system identified the fungal DNA of C. tropicalis in two blood culture bottles after 26 h 24 min and 39 h of incubation, and the third blood cultured was positive only for coagulase-negative Staphylococci after 28 h 19 min of incubation; however, DNA microarray identified fungal DNA in the three blood culture bottles. A second patient (patient PHD) (Table 2), who died of septic shock, had several blood cultures that tested positive for bacteria, including one that was positive for coagulase-negative Staphylococci and C. albicans, where C. albicans was also identified by DNA microarray. On the basis of these results, 209 bottles that were positive for bacteria in ≤12 h were examined. Among the 209 bacteria-positive blood cultures, DNA from C. krusei was identified in two blood cultures positive for Enterococcus faecium from a patient with Burkitt lymphoma (patient MM) (Table 2) after 10 h 43 min of incubation. These findings suggest that this platform can identify fungal DNA in mixed cultures of bacteria and fungi and in bottles where only bacteria were previously identified by the automated system (Table 2).
TABLE 2.
Description of the DNA microarray identification of fungi in blood culture bottles previously identified as positive for bacteria by the automated system
| Patient/age (yrs)/underlying condition/outcome | Blood culture bottle no. | Date (mo/day/yr) | Time to detection | Microorganism recovery in blood culture | DNA microarray result for fungi |
|---|---|---|---|---|---|
| RMPR/25/aplastic anemia plus BMT/death | 6262/12-1 | 08/30/2012 | 28 h 19 min | Staphylococcus coagulase negative | Candida tropicalis |
| 6977/12-1 | 08/30/2012 | 39 h | C. tropicalis | C. tropicalis | |
| 6977/12-2 | 08/30/2012 | 26 h 24 min | C. tropicalis | C. tropicalis | |
| PHD/13/septic shock/death | 6938/13-1 | 07/22/2013 | 9 h 36 min | Staphylococcus aureus | Negative |
| 6943/13-1 | 07/22/2013 | 74 h 52 min | S. aureus | Negative | |
| 7025/13-1 | 07/25/2013 | 21 h 36 min | P. aeruginosa | Negative | |
| 7025/13-2 | 07/25/2013 | 21 h 36 min | P. aeruginosa | Negative | |
| 7129/13-2 | 07/29/2013 | 35 h | Candida albicans | C. albicans | |
| 7181/13-2 | 07/31/2013 | 39 h 31 min | S. coagulase negative plus C. albicans | C. albicans | |
| MM/47/Burkitt lymphoma/death | 8392/15-1 | 10/09/2015 | 10 h 43 min | Enterococcus faecium | Candida krusei |
| 8392/15-2 | 10/09/2015 | 10 h 43 min | E. faecium | C. krusei | |
| 8392/15-3 | 10/09/2015 | 10 h 43 min | E. faecium | Negative | |
| 8262/15-1 | 10/14/2015 | 9 h 43 min | Pseudomonas aeruginosa plus Enterococcus faecalis | Negative | |
| 8262/15-2 | 10/14/2015 | 9 h 43 min | P. aeruginosa plus E. faecalis | Negative | |
| 8262/15-3 | 10/14/2015 | 9 h 43 min | P. aeruginosa plus E. faecalis | Negative |
The main clinical epidemiological data of the 242 patients who had blood cultures analyzed in this study were as follows: the mean age was 48.5 years, 54% were male, and 134 (54.3%) patients died. Oncohematologic (74 patients [18.6%]) and nonhematologic (39 patients [9.8%]) cancers were the most frequent underlying diseases, followed by cardiovascular (54 patients [13.6%]) and liver (38 patients [9.6%]) diseases. As our hospital is a tertiary referral hospital, the predominance of patients with malignancies reflected the population that was assisted in our hospital. Antifungal therapy was prescribed for 67 of the 77 patients with positive blood cultures for fungi. In this group of patients, the mortality rate was 55.7%. Among the 12 patients with fungal infections who did not receive antifungal therapy, nine (75%) died before the initiation of antifungal therapy.
DISCUSSION
An accurate identification of the etiologic agent is the central key for adequate antifungal treatment. Some studies have shown a direct relationship between early appropriate antifungal treatment and improved outcomes (31, 32). The appropriate prescription of an antifungal drug differs among fungal species; therefore, the delay in the detection of the infectious agent and its identification could lead to an increase in mortality rates (33, 34). Although the results from culture are the microbiologic criteria for diagnosing a fungal infection, the growth of a fungal agent may take days (Aspergillus spp.) or weeks (Histoplasma capsulatum), causing a delay in the initiation of antifungal therapy.
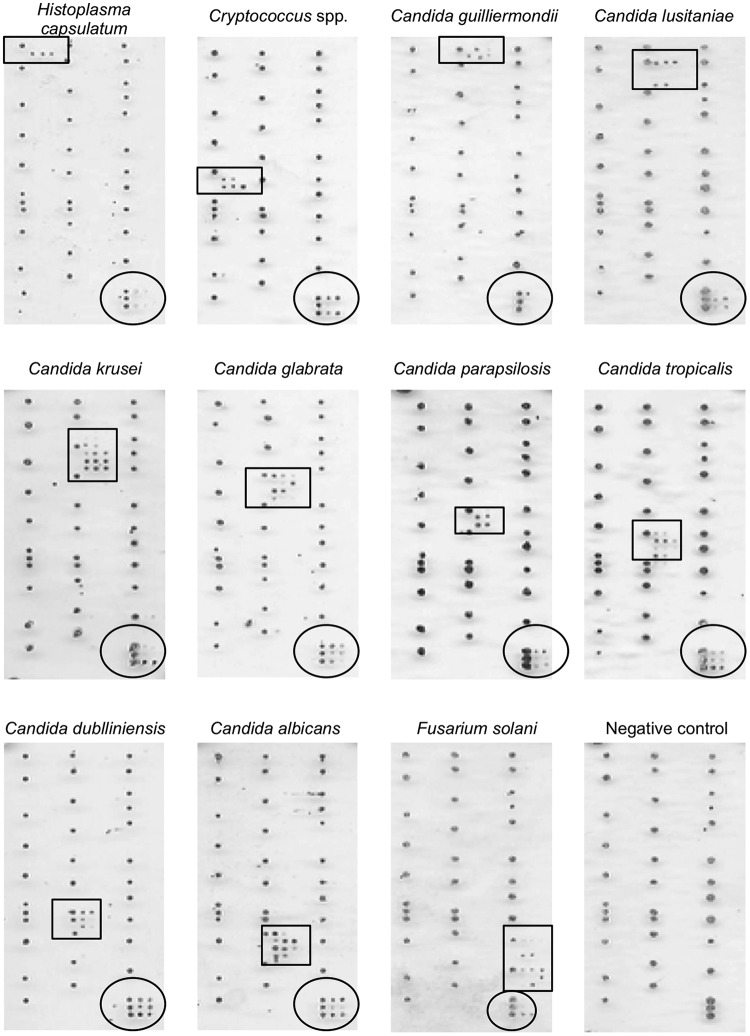
In the present study, a new molecular approach was evaluated for directly improving fungal diagnosis from the blood culture bottle. A DNA microarray platform was designed to cover 32 fungal pathogens in which the results were visible to the naked eye (Fig. 1). Conversely, microarray automated systems use fluorescence, nylon membranes, or DNA chips, which raise the cost of the tests. The use of our DNA microarray platform showed that the time required for performing the experiment and identifying the fungal agent from the blood culture bottle could be ≤6 h. The DNA microarray platform was demonstrated to be a potential tool to detect specific fungi and to identify fungal DNA in blood culture bottles that were also positive for bacteria. Considering the fungal species for which our platform contained oligonucleotides, our results showed a 96.7% concordance of the DNA microarray with DNA sequencing, whereas the DNA microarray missed the identification of four isolates (one C. parapsilosis, one C. neoformans, and two C. tropicalis). Previous studies on the identification of fungal isolates from blood culture bottles and DNA arrays designed on the basis of the ITS regions have shown similar results. Yoo et al. (35) tested a DNA platform for detecting fungi and bacteria in the same array using blood cultures from patients with sepsis and observed false-positive results with Candida spp., as well as one false negative in 17 blood culture bottles that were positive for fungi, resulting in 93% sensitivity for fungal detection in bloodstream infections. An Italian multicenter study evaluated a commercial microarray system for the identification of yeasts and described 95.5% agreement with traditional methods (36).
FIG 1.
Illustrative pictures of the results of the hybridization patterns of DNA microarray slants of different fungal species. Species-specific signals are enclosed in square frames, and universal signals for fungi are enclosed in round frames. The pictures show representative results of Histoplasma capsulatum, Cryptococcus spp., Candida guillermondii, Candida lusitaniae, Candida krusei, Candida glabrata, Candida parapsilosis, Candida tropicalis, Candida dubliniensis, Candida albicans, Fusarium solani, and the negative control.
DNA microarray platforms are shown to be highly sensitive and specific in studies that include fungal identification from fungi grown in culture media (24, 26, 27), from positive blood cultures (35–37), and from different clinical specimens, such as bronchial alveolar lavage fluid samples and blood samples (38, 39). Limitations that could impair the microarray sensitivity would be the presence of a low number of fungal cells in the clinical sample and inefficient DNA extraction. As with other molecular techniques (PCR, real-time PCR, loop-mediated isothermal amplification [LAMP], and sequencing), success in the detection of DNA depends on the number of detectable genome copies in the infected organisms per ml in clinical samples. Even though some studies have investigated different protocols and kits for extracting DNA from clinical samples to increase the efficiency of detection in samples with a low infection load, there is still a limit of detection that needs to be exceeded (23, 40).
Additionally, the DNA microarray demonstrated its capability for detecting fungal pathogens in bacteria-positive blood cultures. In our study, two blood cultures from the same patient (MM) (Table 2) were positive for E. faecium by automated methods, and when tested with the microarray platform, DNA from C. krusei was identified, suggesting that blood cultures could miss the identification of fungal pathogens in cases of mixed systemic infections. Faster-growing organisms can readily compete with the slower-growing ones in culture bottles and interfere with fungal growth; thus, we examined blood culture bottles that tested positive for bacteria in less than 12 h. Therefore, DNA microarray techniques are proposed to complement and increase the sensitivity of blood culture to promote the chance for an appropriate and early antifungal treatment.
One of the limitations of the technique was the absence of specific probes for C. orthopsilosis, C. pelliculosa, and Saccharomyces cerevisiae; however, the array supports the inclusion of other species with the design of new probes, including emerging fungi such as Candida auris. Moreover, this platform might be used as a screen for several Candida species, as approximately 90% of infections in hospitals are caused by Candida spp. (34).
Our study was not designed to include the clinical data of the patients for whom blood cultures were analyzed; however, by comparing the patients' clinical and epidemiologic findings, we observed a high mortality in patients with fungemia (55.7%). Previous studies performed in our hospital have also shown high mortality rates in patients with candidemia, ranging from 66% for non-intensive care unit (ICU) to 79% for ICU patients with candidemia (41). The high prevalence of patients with hematologic and nonhematologic cancers may reflect the referral population of our hospital, as described in previous studies (41, 42).
In conclusion, our DNA microarray platform demonstrated a high level of agreement with the results of the automated systems and DNA sequencing for the identification of Candida species and other fungi, which were detected directly from the blood culture bottles. This method provided reliable and fast fungal identification and could be a useful molecular tool in the identification of a wide range of pathogenic fungi. The construction of new platforms containing nucleotide sequences to cover emerging fungi, coupled with the simplicity of reading the results directly from plastic slides by the naked eye, make this DNA platform a suitable new approach for fungal molecular diagnosis.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by grants from the collaborative research project: Science and Technology Research Partnership for Sustainable Development (SATREPS), Japan, and University of Campinas, Brazil, no. 02P-29548-09, and Fundação de Amparo à Pesquisa do Estado de Sao Paulo (FAPESP) no. 2012/51158-0. L. L. Sturaro received a scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01908-17.
REFERENCES
- 1.Vena A, Bouza E, Valerio M, Padilla B, Pano-Pardo JR, Fernandez-Ruiz M, Diaz Martin A, Salavert M, Mularoni A, Puig-Asensio M, Munoz P, Project C. 2017. Candidemia in non-ICU surgical wards: comparison with medical wards. PLoS One 12:e0185339. doi: 10.1371/journal.pone.0185339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mercier T, Maertens J. 2017. Clinical considerations in the early treatment of invasive mould infections and disease. J Antimicrob Chemother 72:i29–i38. doi: 10.1093/jac/dkx031. [DOI] [PubMed] [Google Scholar]
- 3.Posch W, Heimdorfer D, Wilflingseder D, Lass-Florl C. 2017. Invasive candidiasis: future directions in non-culture based diagnosis. Expert Rev Anti Infect Ther 15:829–838. doi: 10.1080/14787210.2017.1370373. [DOI] [PubMed] [Google Scholar]
- 4.Fuller DD, Davis TE Jr, Denys GA, York MK. 2001. Evaluation of BACTEC MYCO/F Lytic medium for recovery of mycobacteria, fungi, and bacteria from blood. J Clin Microbiol 39:2933–2936. doi: 10.1128/JCM.39.8.2933-2936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albataineh MT, Sutton DA, Fothergill AW, Wiederhold NP. 2016. Update from the laboratory: clinical identification and susceptibility testing of fungi and trends in antifungal resistance. Infect Dis Clin North Am 30:13–35. doi: 10.1016/j.idc.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Mitsutake K, Miyazaki T, Tashiro T, Yamamoto Y, Kakeya H, Otsubo T, Kawamura S, Hossain MA, Noda T, Hirakata Y, Kohno S. 1996. Enolase antigen, mannan antigen, Cand-Tec antigen, and beta-glucan in patients with candidemia. J Clin Microbiol 34:1918–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau A, Chen S, Sleiman S, Sorrell T. 2009. Current status and future perspectives on molecular and serological methods in diagnostic mycology. Future Microbiol 4:1185–1222. doi: 10.2217/fmb.09.70. [DOI] [PubMed] [Google Scholar]
- 8.Baskova L, Buchta V. 2012. Laboratory diagnostics of invasive fungal infections: an overview with emphasis on molecular approach. Folia Microbiol (Praha) 57:421–430. doi: 10.1007/s12223-012-0152-3. [DOI] [PubMed] [Google Scholar]
- 9.Onishi A, Sugiyama D, Kogata Y, Saegusa J, Sugimoto T, Kawano S, Morinobu A, Nishimura K, Kumagai S. 2012. Diagnostic accuracy of serum 1,3-beta-d-glucan for Pneumocystis jirovecii pneumonia, invasive candidiasis, and invasive aspergillosis: systematic review and meta-analysis. J Clin Microbiol 50:7–15. doi: 10.1128/JCM.05267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE, European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy Infectious Diseases Mycoses Study Group Consensus Group. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heng SC, Morrissey O, Chen SC, Thursky K, Manser RL, Nation RL, Kong DC, Slavin M. 2015. Utility of bronchoalveolar lavage fluid galactomannan alone or in combination with PCR for the diagnosis of invasive aspergillosis in adult hematology patients: a systematic review and meta-analysis. Crit Rev Microbiol 41:124–134. doi: 10.3109/1040841X.2013.804033. [DOI] [PubMed] [Google Scholar]
- 12.Hou TY, Wang SH, Liang SX, Jiang WX, Luo DD, Huang DH. 2015. The screening performance of serum 1,3-beta-d-glucan in patients with invasive fungal diseases: a meta-analysis of prospective cohort studies. PLoS One 10:e0131602. doi: 10.1371/journal.pone.0131602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angeletti S. 2017. Matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology. J Microbiol Methods 138:20–29. doi: 10.1016/j.mimet.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Patel R. 2015. MALDI-TOF MS for the diagnosis of infectious diseases. Clin Chem 61:100–111. doi: 10.1373/clinchem.2014.221770. [DOI] [PubMed] [Google Scholar]
- 15.Neely LA, Audeh M, Phung NA, Min M, Suchocki A, Plourde D, Blanco M, Demas V, Skewis LR, Anagnostou T, Coleman JJ, Wellman P, Mylonakis E, Lowery TJ. 2013. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci Transl Med 5:182ra54. doi: 10.1126/scitranslmed.3005377. [DOI] [PubMed] [Google Scholar]
- 16.Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, Garey KW, Alangaden GJ, Vazquez JA, Groeger JS, Judson MA, Vinagre YM, Heard SO, Zervou FN, Zacharioudakis IM, Kontoyiannis DP, Pappas PG. 2015. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis 60:892–899. doi: 10.1093/cid/ciu959. [DOI] [PubMed] [Google Scholar]
- 17.Babady NE, Miranda E, Gilhuley KA. 2011. Evaluation of Luminex xTAG fungal analyte-specific reagents for rapid identification of clinically relevant fungi. J Clin Microbiol 49:3777–3782. doi: 10.1128/JCM.01135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shepard JR, Addison RM, Alexander BD, Della-Latta P, Gherna M, Haase G, Hall G, Johnson JK, Merz WG, Peltroche-Llacsahuanga H, Stender H, Venezia RA, Wilson D, Procop GW, Wu F, Fiandaca MJ. 2008. Multicenter evaluation of the Candida albicans/Candida glabrata peptide nucleic acid fluorescent in situ hybridization method for simultaneous dual-color identification of C. albicans and C. glabrata directly from blood culture bottles. J Clin Microbiol 46:50–55. doi: 10.1128/JCM.01385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simner PJ, Uhl JR, Hall L, Weber MM, Walchak RC, Buckwalter S, Wengenack NL. 2013. Broad-range direct detection and identification of fungi by use of the PLEX-ID PCR-electrospray ionization mass spectrometry (ESI-MS) system. J Clin Microbiol 51:1699–1706. doi: 10.1128/JCM.03282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou CC, Lee TT, Chen CH, Hsiao HY, Lin YL, Ho MS, Yang PC, Peck K. 2006. Design of microarray probes for virus identification and detection of emerging viruses at the genus level. BMC Bioinformatics 7:232. doi: 10.1186/1471-2105-7-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller MB, Tang YW. 2009. Basic concepts of microarrays and potential applications in clinical microbiology. Clin Microbiol Rev 22:611–633. doi: 10.1128/CMR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang A, Li JW, Shen ZQ, Wang XW, Jin M. 2006. High-throughput identification of clinical pathogenic fungi by hybridization to an oligonucleotide microarray. J Clin Microbiol 44:3299–3305. doi: 10.1128/JCM.00417-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leinberger DM, Schumacher U, Autenrieth IB, Bachmann TT. 2005. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. J Clin Microbiol 43:4943–4953. doi: 10.1128/JCM.43.10.4943-4953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiao CR, Huang L, Bouchara JP, Barton R, Li HC, Chang TC. 2005. Identification of medically important molds by an oligonucleotide array. J Clin Microbiol 43:3760–3768. doi: 10.1128/JCM.43.8.3760-3768.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai K, Trabasso P, Moretti ML, Mikami Y, Kamei K, Gonoi T. 2014. Identification of fungal pathogens by visible microarray system in combination with isothermal gene amplification. Mycopathologia 178:11–26. doi: 10.1007/s11046-014-9756-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Souza M, Matsuzawa T, Sakai K, Muraosa Y, Lyra L, Busso-Lopes AF, Levin ASS, Schreiber AZ, Mikami Y, Gonoi T, Kamei K, Moretti ML, Trabasso P. 2017. Comparison of DNA microarray, loop-mediated isothermal amplification (LAMP) and real-time PCR with DNA sequencing for identification of Fusarium spp. obtained from patients with hematologic malignancies. Mycopathologia 182:625–632. doi: 10.1007/s11046-017-0129-5. [DOI] [PubMed] [Google Scholar]
- 27.De Luca Ferrari M, Ribeiro Resende M, Sakai K, Muraosa Y, Lyra L, Gonoi T, Mikami Y, Tominaga K, Kamei K, Zaninelli Schreiber A, Trabasso P, Moretti ML. 2013. Visual analysis of DNA microarray data for accurate molecular identification of non-albicans Candida isolates from patients with candidemia episodes. J Clin Microbiol 51:3826–3829. doi: 10.1128/JCM.01050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muraosa Y, Schreiber AZ, Trabasso P, Matsuzawa T, Taguchi H, Moretti ML, Mikami Y, Kamei K. 2014. Development of cycling probe-based real-time PCR system to detect Fusarium species and Fusarium solani species complex (FSSC). Int J Med Microbiol 304:505–511. doi: 10.1016/j.ijmm.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 29.O'Donnell K, Kistler HC, Cigelnik E, Ploetz RC. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci U S A 95:2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farmakiotis D, Kyvernitakis A, Tarrand JJ, Kontoyiannis DP. 2015. Early initiation of appropriate treatment is associated with increased survival in cancer patients with Candida glabrata fungaemia: a potential benefit from infectious disease consultation. Clin Microbiol Infect 21:79–86. doi: 10.1016/j.cmi.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Garnacho-Montero J, Diaz-Martin A, Garcia-Cabrera E, Ruiz Perez de Pipaon M, Hernandez-Caballero C, Lepe-Jimenez JA. 2013. Impact on hospital mortality of catheter removal and adequate antifungal therapy in Candida spp. bloodstream infections. J Antimicrob Chemother 68:206–213. doi: 10.1093/jac/dks347. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Ruiz M, Guinea J, Lora-Pablos D, Zaragoza O, Puig-Asensio M, Almirante B, Cuenca-Estrella M, Aguado JM, CANDIPOP Project, GEIH-GEMICOMED (SEIMC), REIPI. 2017. Impact of fluconazole susceptibility on the outcome of patients with candidaemia: data from a population-based surveillance. Clin Microbiol Infect 23:672.e1–672.e11. doi: 10.1016/j.cmi.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Colombo AL, de Almeida Junior JN, Slavin MA, Chen SC, Sorrell TC. 2017. Candida and invasive mould diseases in non-neutropenic critically ill patients and patients with haematological cancer. Lancet Infect Dis 17:e344–e356. doi: 10.1016/S1473-3099(17)30304-3. [DOI] [PubMed] [Google Scholar]
- 35.Yoo SM, Choi JY, Yun JK, Choi JK, Shin SY, Lee K, Kim JM, Lee SY. 2010. DNA microarray-based identification of bacterial and fungal pathogens in bloodstream infections. Mol Cell Probes 24:44–52. doi: 10.1016/j.mcp.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Farina C, Russello G, Andreoni S, Bonetti C, Conte M, Fazi P, Lombardi G, Luzzaro F, Manso E, Marone P, Passera M, Rocchetti A, Sanna S, Vigano EF, Medical Mycology Committee (CoSM), Italian Society of Clinical Microbiology (AMCLI). 2012. Microarray technology for yeast identification directly from positive blood cultures. A multicenter Italian experience. Med Mycol 50:549–555. doi: 10.3109/13693786.2011.648216. [DOI] [PubMed] [Google Scholar]
- 37.Hsiue HC, Huang YT, Kuo YL, Liao CH, Chang TC, Hsueh PR. 2010. Rapid identification of fungal pathogens in positive blood cultures using oligonucleotide array hybridization. Clin Microbiol Infect 16:493–500. doi: 10.1111/j.1469-0691.2009.02828.x. [DOI] [PubMed] [Google Scholar]
- 38.Boch T, Reinwald M, Postina P, Cornely OA, Vehreschild JJ, Heussel CP, Heinz WJ, Hoenigl M, Eigl S, Lehrnbecher T, Hahn J, Claus B, Lauten M, Egerer G, Muller MC, Will S, Merker N, Hofmann WK, Buchheidt D, Spiess B. 2015. Identification of invasive fungal diseases in immunocompromised patients by combining an Aspergillus specific PCR with a multifungal DNA-microarray from primary clinical samples. Mycoses 58:735–745. doi: 10.1111/myc.12424. [DOI] [PubMed] [Google Scholar]
- 39.Boch T, Spiess B, Cornely OA, Vehreschild JJ, Rath PM, Steinmann J, Heinz WJ, Hahn J, Krause SW, Kiehl MG, Egerer G, Liebregts T, Koldehoff M, Klein M, Nolte F, Mueller MC, Merker N, Will S, Mossner M, Popp H, Hofmann WK, Reinwald M, Buchheidt D. 2016. Diagnosis of invasive fungal infections in haematological patients by combined use of galactomannan, 1,3-beta-d-glucan, Aspergillus PCR, multifungal DNA-microarray, and Aspergillus azole resistance PCRs in blood and bronchoalveolar lavage samples: results of a prospective multicentre study. Clin Microbiol Infect 22:862–868. doi: 10.1016/j.cmi.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 40.Ciardo DE, Lucke K, Imhof A, Bloemberg GV, Bottger EC. 2010. Systematic internal transcribed spacer sequence analysis for identification of clinical mold isolates in diagnostic mycology: a 5-year study. J Clin Microbiol 48:2809–2813. doi: 10.1128/JCM.00289-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagnani R, Resende MR, Trabasso P, Mikami Y, Schreiber AZ, Lopes AF, Muraosa Y, Kamei K, Moretti ML. 2015. Mortality related to candidemia and risk factors associated with non-Candida albicans. Infect Dis (Lond) 47:930–931. doi: 10.3109/23744235.2015.1075662. [DOI] [PubMed] [Google Scholar]
- 42.Moretti ML, Trabasso P, Lyra L, Fagnani R, Resende MR, de Oliveira Cardoso LG, Schreiber AZ. 2013. Is the incidence of candidemia caused by Candida glabrata increasing in Brazil? Five-year surveillance of Candida bloodstream infection in a university reference hospital in southeast Brazil. Med Mycol 51:225–230. doi: 10.3109/13693786.2012.708107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.