Abstract
HilD is an AraC-like transcriptional regulator that plays a central role in Salmonella virulence. HilD controls the expression of the genes within the Salmonella pathogenicity island 1 (SPI-1) and of several genes located outside SPI-1, which are mainly required for Salmonella invasion of host cells. The expression, amount, and activity of HilD are tightly controlled by the activities of several factors. The HilE protein represses the expression of the SPI-1 genes through its interaction with HilD; however, the mechanism by which HilE affects HilD is unknown. In this study, we used genetic and biochemical assays revealing how HilE controls the transcriptional activity of HilD. We found that HilD needs to assemble in homodimers to induce expression of its target genes. Our results further indicated that HilE individually interacts with each the central and the C-terminal HilD regions, mediating dimerization and DNA binding, respectively. We also observed that these interactions consistently inhibit HilD dimerization and DNA binding. Interestingly, a computational analysis revealed that HilE shares sequence and structural similarities with Hcp proteins, which act as structural components of type 6 secretion systems in Gram-negative bacteria. In conclusion, our results uncover the molecular mechanism by which the Hcp-like protein HilE controls dimerization and DNA binding of the virulence-promoting transcriptional regulator HilD. Our findings may indicate that HilE's activity represents a functional adaptation during the evolution of Salmonella pathogenicity.
Keywords: Salmonella enterica, post-transcriptional regulation, dimerization, virulence factor, pathogenesis, Hcp, HilD, HilE, protein–protein interaction, type 6 secretion system
Introduction
The genus Salmonella groups pathogenic bacteria for human and many animals; it comprises only two species, Salmonella enterica and Salmonella bongori, the former is further divided into six subspecies and more than 2500 serovars. Depending on the host, S. enterica serovar Typhimurium (S. Typhimurium) can cause diseases ranging from gastroenteritis to life-threatening systemic infection (1, 2). For instance, in humans, calves, and chickens, S. Typhimurium causes self-limiting gastroenteritis, whereas in laboratory mice, it causes a systemic infection resembling that produced by S. Typhi in humans; thus, S. Typhimurium is frequently used as a model to study the molecular mechanisms mediating the Salmonella virulence (1, 3, 4). Horizontal gene transfer events have greatly contributed to the evolution of the Salmonella pathogenicity (5, 6). Most of the genes gained by Salmonella are clustered in chromosomal regions called Salmonella pathogenicity islands (SPIs)3 (4, 5). SPI-1 is a 40-kb region conserved in the two Salmonella species, which includes 39 genes encoding a type 3 secretion system (T3SS-1), their chaperones and effector proteins, as well as some transcriptional regulators that control the expression of many virulence genes located within and outside SPI-1 (4, 7). The T3SSs are molecular syringes that extend from the membranes of several bacteria, composed of a basal body and a needle-like complex, through which effector proteins are injected from the bacterial cytoplasm into the cytoplasm of eukaryotic cells (8). Salmonella injects the SPI-1 effector proteins into the intestinal epithelial cells through the T3SS-1, which induces cytoskeletal rearrangements promoting the Salmonella invasion of these eukaryotic cells leading to enteritis (1, 4, 8).
The expression of the SPI-1 genes is controlled by several environmental clues, such as osmolarity, oxygen tension, pH, short- and long-fatty acid concentration, and bile (4, 9–11). In vitro, expression of the SPI-1 genes is induced at early stationary phase when Salmonella is grown in the nutrient-rich lysogenic broth (LB) (12, 13). Several regulators control the expression of the SPI-1 genes, including HilD, HilA, and InvF, all encoded in SPI-1, which act in a cascade fashion: HilD, a member of the AraC/XylS transcriptional regulators family, directly induces the expression of HilA, which in turn activates the expression of InvF; HilA and InvF activate the expression of all components of the T3SS-1, their chaperones, and effector proteins (4, 14). HilD induces the expression of HilA directly or through a positive feed-forward loop that it forms with HilC and RtsA (15, 16). HilC and RtsA are AraC-like transcriptional regulators that bind the same DNA sequence recognized by HilD; HilC is encoded within SPI-1, whereas RtsA is encoded in another island. HilD also controls the expression of many other virulence genes located outside SPI-1, including acquired and ancestral genes, directly, or indirectly through HilA, InvF, or other regulators (12, 17–25). In agreement with its role as a master transcriptional regulator for a high number of genes, the expression, concentration, and activity of HilD is tightly controlled. Transcription of hilD is positively autoregulated (15, 26) and is repressed by H-NS (27), whereas its translation is repressed by CsrA (28). In addition, Fur and FliZ control HilD at post-translational level (29, 30); the Lon protease degrades HilD, thus mediating its concentration (31, 32); and the CpxR/A two-component system decreases the stability of HilD through Lon-dependent and independent mechanisms (33). Furthermore, several compounds affect HilD: propionate and bile salts decrease its stability (9, 10); butyrate and oleate negatively affect its regulatory activity, whereas acetate and formate enhance this (11, 34–36); and l-arabinose affects HilD expression at post-transcriptional level (37).
The activity of HilD is also negatively controlled by HilE through protein–protein interaction (38). However, the specific effect of this interaction on HilD has remained unknown. The hilE gene resides in a region of the Salmonella chromosome similar to a pathogenicity island (38), supporting that it was acquired by horizontal transfer. The expression of hilE is positively regulated by the PhoPQ and PhoBR two-component systems, as well as by FimZ and LeuO (39, 40), whereas this is negatively regulated by Mlc and the small RNA IsmR (41, 42), which provides additional inputs controlling the activity of HilD.
Here we show that HilE negatively affects dimerization and DNA binding of HilD, by interacting with both the central and C-terminal regions of HilD, which mediate dimerization and DNA binding, respectively. Therefore, our results demonstrate how HilE regulates HilD activity. Additionally, our results revealed that HilE shares sequence and structural similarities with proteins called Hcp (hemolysin-coregulated protein), which are structural components of type 6 secretion systems in Gram-negative bacteria, supporting the hypothesis that HilE was adapted to act as an important regulatory protein during the Salmonella pathogenicity evolution.
Results
HilE interacts with the central region and with the C-terminal region of HilD
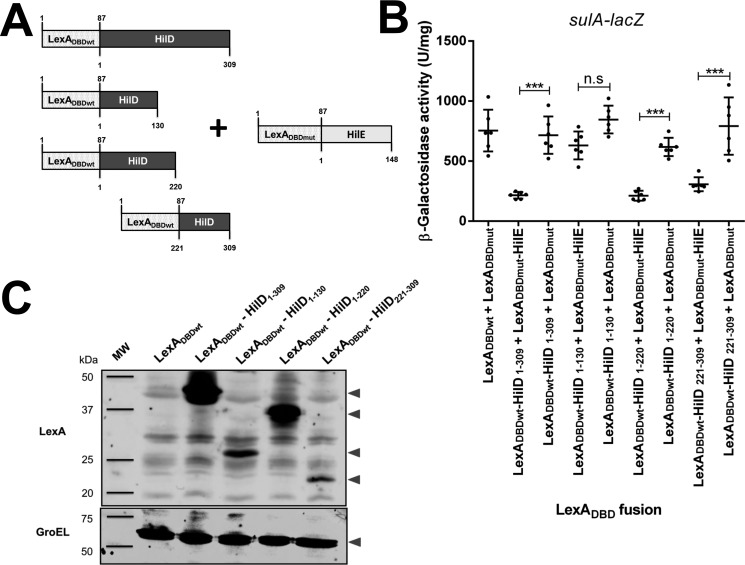
To investigate how HilE negatively affects the activity of HilD, we analyzed the interaction of HilE with different regions of HilD by using the LexA-based genetic system for heterodimerization, which is similar to a two-hybrid system to analyze protein–protein interactions (43, 44). It is important to remark that the interaction between HilE and HilD was previously demonstrated with this system (38). In the LexA-based genetic system for heterodimerization, the WT LexA DNA-binding domain (LexADBDwt) and a derivative mutant of this (LexADBDmut) are expressed from two different plasmids in the Escherichia coli SU202 reporter strain, which carries a chromosomal sulA–lacZ transcriptional fusion containing a LexA hybrid operator (43, 44). LexADBDwt and LexADBDmut cannot affect the expression of sulA–lacZ; however, when they are fused to proteins that interact between them, an active dimer of LexADBDwt and LexADBDmut is formed that is able to bind the hybrid LexA operator on sulA–lacZ and thus to repress the expression of this fusion. Fusion proteins of LexADBDwt with full-length or distinct regions of HilD and of LexADBDmut with full-length HilE were generated and assessed. The LexADBDwt–HilD fusions were named according to the amino acid position for the ends of the HilD fragment carried (Fig. 1A). As expected, the combination LexADBDwt–HilD1–309 (full-length HilD) + LexADBDmut–HilE (full-length HilE), but not LexADBDwt + LexADBDmut or LexADBDwt–HilD + LexADBDmut, mediated repression of sulA–lacZ (Fig. 1B), further confirming the interaction between HilD and HilE. Interestingly, the fusion proteins LexADBDwt–HilD1–220 or LexADBDwt–HilD221–309, in combination with LexADBDmut–HilE, also repressed the expression of sulA–lacZ (Fig. 1B), indicating that HilE interacts with the regions of HilD spanning amino acids 1–220 and 221–309. In contrast, the combination LexADBDwt–HilD1–130 + LexADBDmut–HilE did not repress the expression of sulA–lacZ, supporting that HilE does not interact with the N-terminal region of HilD from amino acids 1 to 130. However, the lack of interaction between LexADBDwt–HilD1–130 and LexADBDmut–HilE could be due to an incorrect folding of LexADBDwt–HilD1–130. As expected, the combination LexADBDwt–HilD1–130 + LexADBDmut, LexADBDwt–HilD1–220 + LexADBDmut, or LexADBDwt–HilD221–309 + LexADBDmut, used as a negative control, did not affect the expression of sulA–lacZ (Fig. 1B). Western blotting analysis showed expression signals for all the LexADBDwt–HilD fusion proteins tested (Fig. 1C). To note, the LexADBDwt–HilD221–309 fusion repressed the sulA–lacZ fusion, in combination with LexADBDmut–HilE (Fig. 1B), even when its expression level was lower than that of the LexADBDwt–HilD1–309 and LexADBDwt–HilD1–220 fusions (Fig. 1C), indicating that differences in the amount of the LexADBDwt–HilD fusion proteins assessed did not affect the results of these assays. In agreement with a previous report (45), expression of LexADBD was not detected in our Western blotting analysis (Fig. 1C). These results support that HilE interacts with regions spanning amino acids 130–220 and 221–309 of HilD.
Figure 1.
HilE interacts with two different regions of HilD. A, schematic representation of the LexADBDwt–HilD and LexADBDmut–HilE fusion proteins tested. The numbers indicate the residues of LexADBD, HilD, or HilE carried in the respective fusion protein. B, expression of the sulA–lacZ fusion was determined in the E. coli SU202 reporter strain containing the pair of plasmids pSR658 and pSR659 (LexADBDwt + LexADBDmut), pSR658–HilD1 and pSR659-HilE1 (LexADBDwt–HilD1–309 + LexADBDmut–HilE), pSR658–HilD1 and pSR659 (LexADBDwt–HilD1–309 + LexADBDmut), pSR658–HilD2 and pSR659-HilE1 (LexADBDwt–HilD1–130 + LexADBDmut–HilE), pSR658–HilD2 and pSR659 (LexADBDwt–HilD1–130 + LexADBDmut), pSR658–HilD4 and pSR659-HilE1 (LexADBDwt–HilD1–220 + LexADBDmut–HilE), pSR658–HilD4 and pSR659 (LexADBDwt–HilD1–220 + LexADBDmut), pSR658–HilD5 and pSR659-HilE1 (LexADBDwt–HilD221–309 + LexADBDmut–HilE), or pSR658–HilD5 and pSR659 (LexADBDwt–HilD221–309 + LexADBDmut). The β-gal activity was determined from samples collected of bacterial cultures grown in LB at 37 °C up to an A600 of 1.0. Expression of the LexADBD fusion proteins was induced by adding 1 mm IPTG to the medium. The data are the averages of three independent experiments performed in duplicate. The bars represent the standard deviations. ***, expression statistically significantly different compared with that reached in the absence of HilE (p < 0.001); n.s., no significant difference. C, expression of the LexADBDwt, LexADBDwt–HilD, LexADBDwt–HilD1–130, LexADBDwt–HilD1–220, and LexADBDwt–HilD221–309 proteins was analyzed by Western blotting using polyclonal anti-LexA antibodies. Whole cell lysates were prepared from samples of bacterial cultures grown in LB at 37 °C up to an A600 of 1.0. As a loading control, the expression of GroEL was also determined using polyclonal anti-GroEL antibodies. MW, protein molecular weight standards (Precision Plus ProteinTM; Bio-Rad). The arrowheads indicate the expected bands.
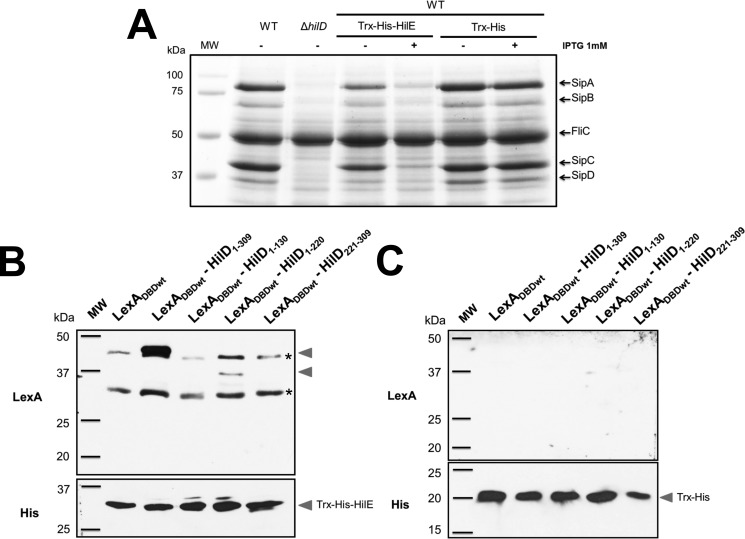
Pulldown assays were performed to confirm the HilE interactions with HilD. For this, we purified HilE fused to the Trx protein and a His6 tag (Trx–His–HilE) as described under “Experimental procedures.” The Trx–His–HilE fusion protein repressed the SPI-1–mediated protein secretion profile of the WT S. Typhimurium strain (Fig. 2A), which supports that it is able to negatively affect HilD. Trx–His–HilE was used as the bait protein in the pulldown assays; first, it was immobilized on Ni–NTA resin, and then whole-cell extracts containing either LexADBD or LexADBDwt–HilD1–309, LexADBDwt–HilD1–130, LexADBDwt–HilD1–220, LexADBDwt–HilD221–309 prey proteins were loaded to the Ni–NTA resin carrying Trx–His–HilE. As expected, Trx–His–HilE captured the LexADBDwt–HilD1–309 and LexADBDwt–HilD1–220 prey proteins, but not LexADBDwt–HilD1–130 or LexADBDwt (Fig. 2B), confirming the interaction of HilE with full-length HilD and the central region of HilD. LexADBDwt–HilD221–309 showed interaction with HilE in the LexA-based genetic system (Fig. 1B); however, in the pulldown assays, this interaction was no evident (Fig. 2B), which could be explained by the low level of expression showed by LexADBDwt–HilD221–309 (Fig. 1C). As a control, parallel pulldown assays were performed using Trx–His as the bait protein, which was unable to capture any of the LexADBDwt–HilD prey proteins tested (Fig. 2C). Taken together, these results indicate that HilE binds to both the central region and the C-terminal region of HilD.
Figure 2.
Pulldown assays showing the interaction between HilE and HilD. A, secretion of the SPI-1-encoded proteins SipA, SipB, SipC, and SipD was tested in the WT S. Typhimurium strain and its isogenic ΔhilD mutant, as well as in the WT S. Typhimurium strain carrying the pET32–HilE plasmid expressing Trx–His–HilE or the pET32b(+) vector expressing Trx–His. The bacterial cultures were grown for 9 h in LB at 37 °C, in the presence (+) or absence (−) of 1 mm IPTG to induce or not the expression of Trx–His–HilE or Trx–His. Supernatants of the cultures were analyzed in SDS-PAGE at 12%. FliC is a flagellar protein whose secretion is SPI-1–independent. B and C, bait proteins Trx–His–HilE (B) or Trx–His (C) immobilized on Ni–NTA resin were incubated with whole-cell extracts containing the LexADBDwt, LexADBDwt–HilD1–309, LexADBDwt–HilD1–130, LexADBDwt–HilD1–220, or LexADBDwt–HilD221–309 prey proteins. After washing, the proteins captured by the Trx–His–HilE or Trx–His bait proteins were analyzed by Western blotting using polyclonal anti-LexA antibodies. The Trx–His–HilE or Trx–His bait proteins were also detected with monoclonal anti-His6 antibodies. MW, protein molecular weight standards (Precision Plus ProteinTM; Bio-Rad). The arrowheads indicate the expected bands. Asterisks indicate bands showing cross-reaction with the polyclonal anti-LexA antibodies.
HilE does not affect the stability of HilD
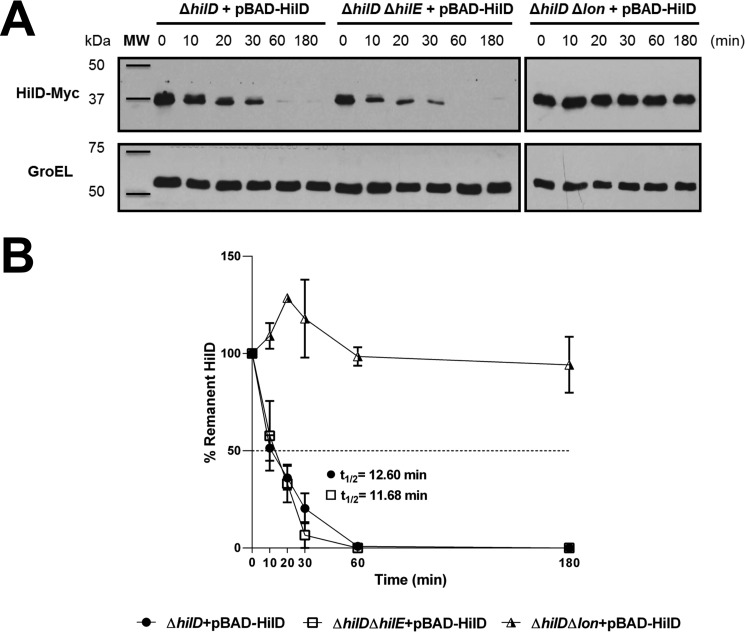
The interaction with HilE could affect the stability of HilD, to investigate this, we determined the in vivo half-life of HilD in presence or absence of HilE or the Lon protease, which degrades HilD. The cellular levels of Myc-tagged HilD (HilD–Myc) expressed from the pBAD-HilD plasmid, under an arabinose-inducible promoter, were monitored in the S. Typhimurium ΔhilD, ΔhilD ΔhilE, and ΔhilD Δlon mutants, at different times after adding a mix of transcription and translation inhibitors. The levels of HilD–Myc were similar over time in the ΔhilD and ΔhilD ΔhilE mutants, showing a half-life of HilD–Myc of 12.60 and 11.68 min, respectively; as expected, the stability of HilD–Myc was drastically increased in the ΔhilD Δlon mutant (Fig. 3, A and B). These results demonstrate that the interaction with HilE does not affect the stability of HilD.
Figure 3.
HilE does not affect the stability of HilD. A, stability of HilD–Myc was determined in the ΔhilD, ΔhilD ΔhilE, and ΔhilD Δlon mutants of S. Typhimurium carrying the pBAD-HilD1 plasmid, grown in LB at 37 °C. Expression of HilD–Myc, from the arabinose-inducible promoter of pBAD-HilD1, was induced with 0.05% l-arabinose for 45 min; then transcription and translation were halted by the addition of a mixture of antibiotics and glucose, and samples of bacterial cultures were taken at the indicated times. HilD–Myc was detected from whole-cell lysates of the samples by Western blotting using monoclonal anti-Myc antibodies. As a loading control, the expression of GroEL was also determined using polyclonal anti-GroEl antibodies. A representative Western blotting of three independent experiments is shown. The figure is composed by four different blots. B, densitometric analysis of the HilD–Myc bands from the Western blotting is indicated as the relative percentage of HilD–Myc at each time with respect to time 0. Intensity values of HilD–Myc bands were normalized with those respective of GroEL bands. The data are the averages of three independent experiments. The bars represent the standard deviation, and t½ indicates the half-life of HilD. MW, protein molecular weight standards (Precision Plus ProteinTM; Bio-Rad).
HilD acts as a dimer, and its central region mediates the interaction itself
Several AraC-like transcriptional regulators act as dimers (46–49). Therefore, we thought that the interaction of HilE with the central region of HilD could affect dimerization of HilD. To investigate this possibility, we first analyzed whether HilD indeed dimerizes by using the LexA-based genetic system for homodimerization (43, 44). In this case, LexADBDwt is expressed from a plasmid in the E. coli SU101 reporter strain, which carries a chromosomal sulA–lacZ transcriptional fusion containing the LexA WT operator (43, 44). LexADBDwt does not affect the expression of sulA–lacZ; however, when it is fused to a protein that itself interacts, an active dimer of LexADBDwt is formed, which is able to bind the WT LexA operator on sulA–lacZ and thus represses the expression of this fusion. The same LexADBDwt–HilD fusion proteins used before in the heterodimerization assay were now tested in the homodimerization assay. LexADBDwt alone and the fusion protein LexADBDwt–H-NS, whose dimerization capacity has been tested before (50), were used as negative and positive controls, respectively. Both LexADBDwt–H-NS and LexADBDwt–HilD1–309, but not LexADBDwt, repressed the expression of sulA–lacZ (Fig. 4A), indicating that HilD dimerizes. A Western blotting analysis showed expression signals for the LexADBDwt–HilD1–309 and LexADBDwt–H-NS fusion proteins (Fig. 4B). To confirm the dimerization of HilD, the size of the maltose-binding protein (MBP)–HilD fusion, purified in native conditions, was analyzed by gel-filtration chromatography. The MBP–HilD fusion protein eluted from the gel-filtration chromatography as a ∼178-kDa product (Fig. S1), which corresponds to a size similar to that expected for a dimer of this protein.
Figure 4.
HilD forms homodimers through its central region. A, expression of the sulA–lacZ fusion was determined in the E. coli SU101 reporter strain containing the plasmids pSR658 (LexADBDwt), pSR658–HilD1 (LexADBDwt–HilD1–309), or pSR658–HNS (LexADBDwt–H-NS). B, expression of LexADBDwt, LexADBDwt–HilD1–309, and LexADBDwt–H-NS was analyzed by Western blotting using polyclonal anti-LexA antibodies. As a loading control, the expression of GroEL was also determined using polyclonal anti-GroEL antibodies. MW, protein molecular weight standards (Precision Plus ProteinTM; Bio-Rad). The arrowheads indicate the expected bands. C, expression of the sulA–lacZ fusion was determined in the E. coli SU101 reporter strain containing the plasmids pSR658 (LexADBDwt), pSR658–HilD1 (LexADBDwt–HilD1–309), pSR658–HilD2 (LexADBDwt–HilD1–130), pSR658–HilD4 (LexADBDwt–HilD1–220), pSR658–HilD5 (LexADBDwt–HilD221–309), or pSR658–HilD3 (LexADBDwt–HilD130–309). The β-gal activity was determined from samples collected of bacterial cultures grown in LB at 37 °C up to an A600 of 1.0. Expression of LexADBDwt and the LexADBDwt fusion proteins was induced by adding 1 mm IPTG to the medium. The data are the averages of three independent experiments performed in duplicate. The bars represent the standard deviations. ***, expression statistically significantly different compared with that reached in the presence of LexADBDwt (p < 0.001).
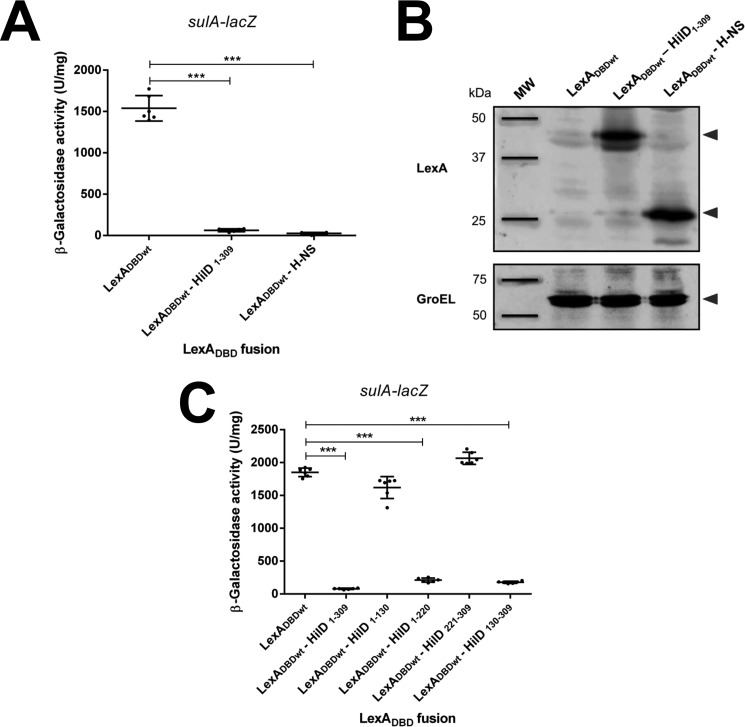
LexADBDwt–HilD1–220 also repressed the expression of sulA–lacZ (Fig. 4C), indicating that the region spanning amino acids 1–220 mediates the dimerization of HilD. In contrast, LexADBDwt–HilD1–130, LexADBDwt–HilD221–309, and the negative control LexADBDwt did not affect the expression of sulA–lacZ (Fig. 4C), supporting that the regions between amino acids 1 and 130 and amino acids 221 and 309 are not involved in the dimerization of HilD. These results suggest that the dimerization of HilD seems to be mediated by the region comprising amino acids 130–220. To further investigate this asseveration, we constructed and tested the LexADBDwt–HilD130–309 fusion protein, containing an extended N-terminal HilD region with respect to LexADBDwt–HilD221–309. As could be expected, LexADBDwt–HilD130–309 was able to represses the expression of sulA–lacZ (Fig. 4C), indicating that it interacts itself. Together, these results indicate that HilD dimerizes through its central region ranging from amino acids 130 to 220.
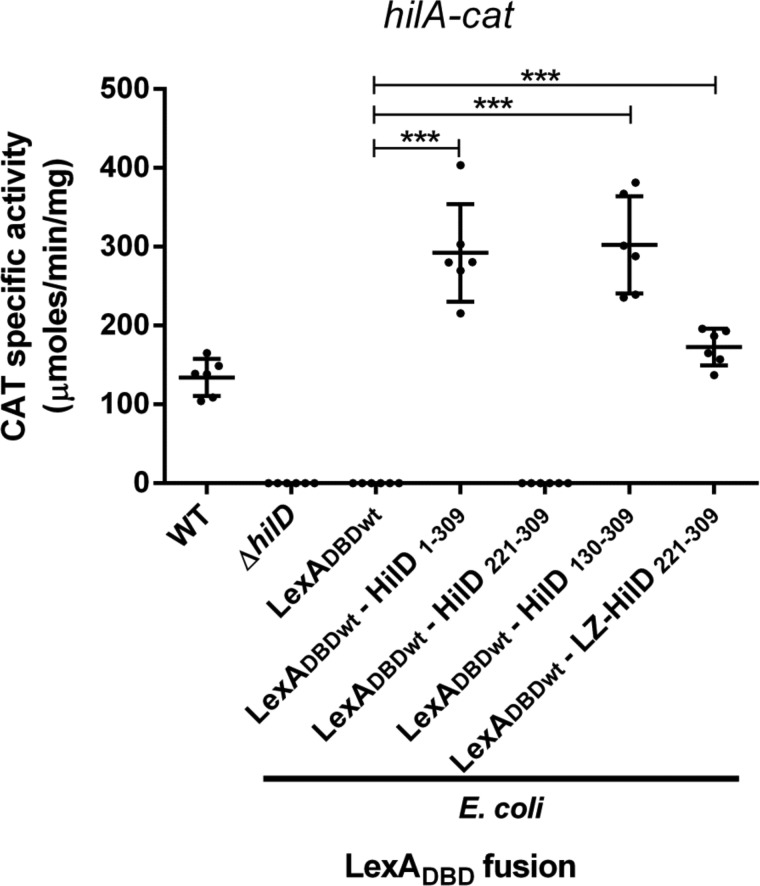
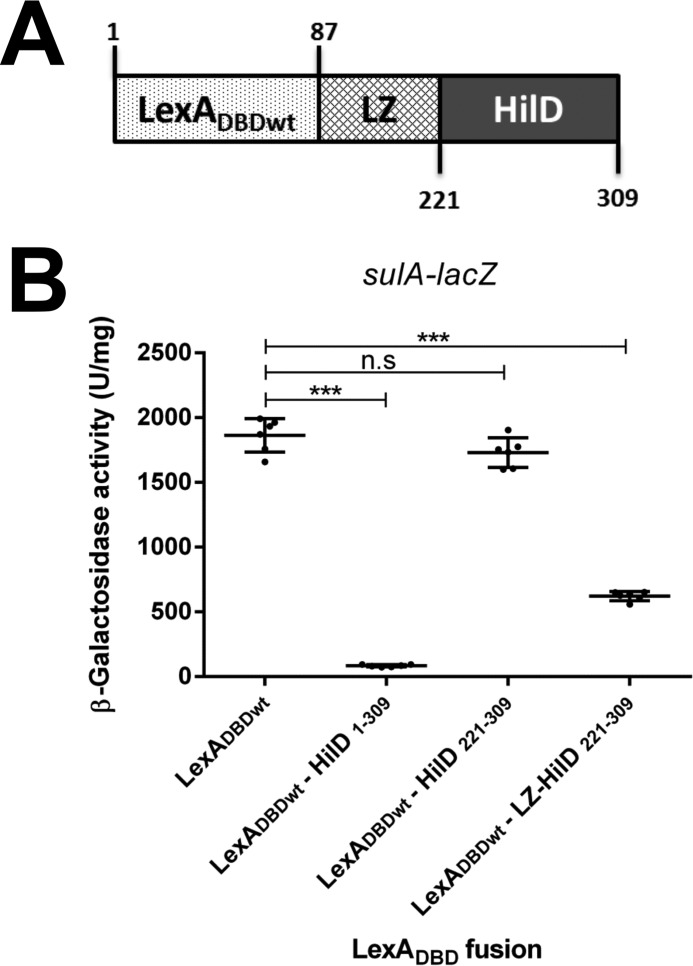
Next, we sought to determine whether dimerization is required for the regulatory activity of HilD. For this, the ability to induce the expression of a hilA–cat transcriptional fusion of LexADBDwt–HilD1–309, LexADBDwt–HilD221–309, and LexADBDwt–HilD130–309, which carry the DNA-binding domain of HilD, was tested in an E. coli K-12 strain. Because hilA is a gene directly and positively controlled by HilD (4, 51) and HilD is not present in E. coli K-12. As a negative control, LexADBDwt was also assessed; additionally, as a reference for the expression of hilA–cat in the presence or absence of HilD, the activity of this fusion was also determined in the WT S. Typhmurium strain and its isogenic ΔhilD mutant. The expression of hilA–cat was induced in the presence of LexADBDwt–HilD1–309 or LexADBDwt–HilD130–309, which show dimerization, but it was not induced by LexADBDwt–HilD221–309 that does not dimerize (Fig. 5). As expected, the expression of hilA–cat was also induced in the WT S. Typhimurium strain, but not in the ΔhilD mutant or in the presence of LexADBDwt (Fig. 5). These results support that HilD needs to form dimers to induce the expression of target genes. To confirm this, we generated and analyzed the chimeric protein LexADBDwt–LZ–HilD221–309, which is a fusion of LexADBDwt, the leucine zipper (LZ) motif of the GCN4 transcriptional factor of Saccharomyces cerevisiae, and the DNA-binding domain of HilD (amino acids 221–309) (Fig. 6A). The LZ motif has been used before as a dimerization module (52). The ability of the LexADBDwt–LZ–HilD221–309 fusion protein to undergo dimerization and to induce the expression of hilA was assessed and compared with that of the LexADBDwt–HilD221–309 and LexADBDwt–HilD1–309 fusion proteins. Both LexADBDwt–LZ–HilD221–309 and LexADBDwt–HilD1–309, but not LexADBDwt–HilD221–309, repressed the expression of sulA–lacZ in the E. coli SU101 reporter strain (Fig. 6B), indicating that LexADBDwt–LZ–HilD221–309 dimerizes through the heterologous LZ motif. Notably, LexADBDwt–LZ–HilD221–309 and LexADBDwt–HilD1–309, but not LexADBDwt–HilD221–309, induced the expression of the hilA–cat fusion in E. coli K-12 (Fig. 5), showing that the LZ motif generates dimers of the DNA-binding domain of HilD, which are able to induce expression of target genes. Overall, these results demonstrate that HilD dimerizes through its central region spanning amino acids 130–220, which is essential for the regulatory activity of HilD.
Figure 5.
Dimerization is required for the HilD regulatory activity. Expression of the hilA–cat fusion contained in the philA–cat1 plasmid was tested in the WT S. Typhimurium SL1344 and its isogenic ΔhilD mutant, as well as in the E. coli MC4100 strain carrying the plasmids pSR658 (LexADBDwt), pSR658–HilD1 (LexADBDwt–HilD1–309), pSR658–HilD5 (LexADBDwt–HilD221–309), pSR658–HilD3 (LexADBDwt–HilD130–309), or pSR658–HilD6 (LexADBDwt–LZ–HilD221–309). The CAT-specific activity was determined from samples collected of bacterial cultures grown in LB at 37 °C up to an A600 of 1.2. Expression of LexADBDwt, LexADBDwt–HilD1–309, LexADBDwt–HilD221–309, LexADBDwt–HilD130–309, and LexADBDwt–LZ–HilD221–309 was induced by adding 1 mm IPTG to the medium. The data are the averages of three independent experiments performed in duplicate. The bars represent the standard deviations. ***, expression statistically significantly different compared with that reached in the presence of LexADBDwt (p < 0.001).
Figure 6.
LexADBDwt–LZ–HilD221–309 dimerizes. A, schematic representation of LexADBDwt–LZ–HilD221–309. The numbers indicate the residues of LexADBD or HilD carried in this fusion protein. B, expression of the sulA–lacZ fusion was determined in the E. coli SU101 reporter strain containing the plasmids pSR658 (LexADBDwt), pSR658–HilD1 (LexADBDwt–HilD1–309), pSR658–HilD5 (LexADBDwt–HilD221–309), or pSR658–HilD6 (LexADBDwt–LZ–HilD221–309). The β-gal activity was determined from samples collected of bacterial cultures grown in LB at 37 °C up to an A600 of 1.0. Expression of LexADBDwt, LexADBDwt–HilD1–309, LexADBDwt–HilD221–309, and LexADBDwt–LZ–HilD221–309 was induced by adding 1 mm IPTG to the medium. The data are the averages of three independent experiments performed in duplicate. The bars represent the standard deviations. ***, expression statistically significantly different compared with that reached in the presence of LexADBDwt (p < 0.001); n.s., no significant difference.
HilE negatively affects the dimerization of HilD
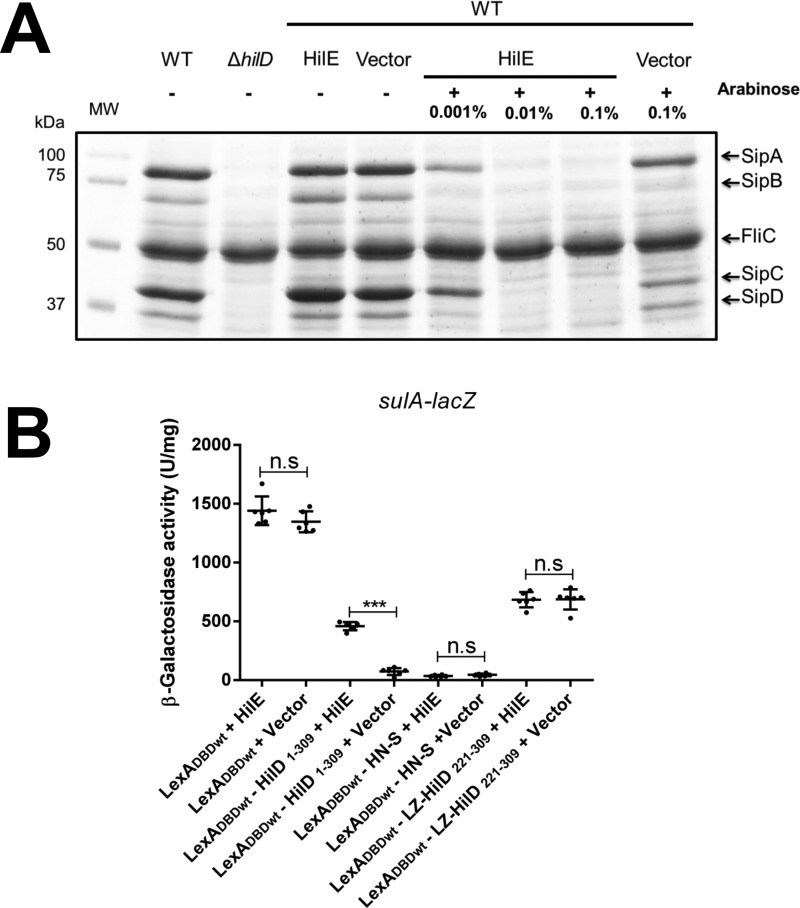
Based on the results described above, we now analyzed whether HilE affects the dimerization of HilD. For this, the LexADBDwt–HilD1–309 fusion protein, which carries the dimerization domain of HilD, was tested for homodimerization in the presence of the pA6–HilE1 plasmid expressing HilE or in the presence of the pMPM–A6Ω vector. Expression of HilE from pA6–HilE1 drastically reduced the SPI-1–mediated protein secretion profile of the WT S. Typhimurium strain (Fig. 7A), supporting that the amount of HilE reached from this plasmid can negatively affect HilD. The effect of HilE on LexADBDwt–LZ–HilD221–309 and LexADBDwt–H-NS, which dimerize through the LZ motif and the H-NS dimerization domain, respectively, was also assessed as negative controls. Interestingly, HilE affected the repression of sulA–lacZ mediated by the dimerization of LexADBDwt–HilD1–309; in contrast, it did not affect the repression of sulA–lacZ mediated by the dimerization of LexADBDwt–LZ–HilD221–309 or LexADBDwt–H-NS (Fig. 7B). These results indicate that HilE negatively affects the dimerization of HilD but not that of H-NS or the LZ motif tested.
Figure 7.
HilE inhibits the dimerization of HilD. A, secretion of the SPI-1-encoded proteins SipA, SipB, SipC, and SipD was tested in the WT S. Typhimurium strain and its isogenic ΔhilD mutant, as well as in the WT S. Typhimurium strain carrying the pA6–HilE1 plasmid expressing HilE from an arabinose-inducible promoter, or carrying the pMPM–A6Ω vector, in the presence (+) or absence (−) of increasing concentrations of l-arabinose. Bacterial cultures were grown for 9 h in LB at 37 °C, and supernatants were analyzed in SDS-PAGE at 12%. FliC is a flagellar protein whose secretion is SPI-1–independent. MW, protein molecular weight standards (Precision Plus ProteinTM; Bio-Rad). B, expression of the sulA–lacZ fusion was determined in the E. coli SU101 reporter strain containing the pair of plasmids pSR658 and pA6–HilE1 (LexADBDwt + HilE), pSR658 and pMPM–A6Ω (LexADBDwt + vector), pSR658–HilD1 and pA6–HilE1 (LexADBDwt–HilD1–309 + HilE), pSR658–HilD1 and pMPM–A6Ω (LexADBDwt–HilD1–309 + vector), pSR658–HNS and pA6–HilE1 (LexADBDwt–H-NS + HilE), pSR658–HNS and pMPM–A6Ω (LexADBDwt–H-NS + vector), pSR658–HilD6 and pA6–HilE1 (LexADBDwt–LZ–HilD221–309 + HilE), or pSR658-HilE6 and pMPM–A6Ω (LexADBDwt–LZ–HilD221–309 + vector). The β-gal activity was determined from samples collected of bacterial cultures grown in LB at 37 °C up to an A600 of 1.0. Expression of LexADBDwt, LexADBDwt–HilD1–309, LexADBDwt–HN-S, and LexADBDwt–LZ–HilD221–309 was induced by adding 1 mm IPTG to the medium. 0.1% l-arabinose was also added to induce the expression of HilE from pA6–HilE1. The data are the averages of three independent experiments performed in duplicate. The bars represent the standard deviations. ***, expression statistically significantly different compared with that reached in the absence of HilE (p < 0.001); n.s., no significant difference.
HilE directly affects the DNA-binding domain of HilD
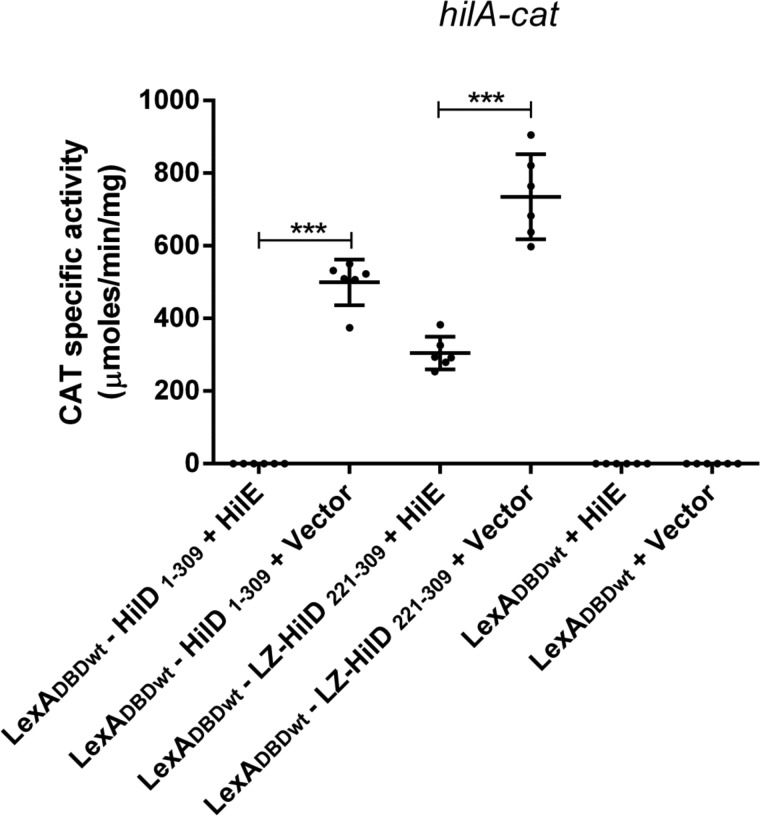
The results described above indicate that HilE affects the dimerization of HilD by interacting with the central region of this regulator, which in turn would indirectly inhibit its regulatory activity. However, our results revealed that HilE also interacts with the C-terminal region of HilD (Fig. 1B), carrying the DNA-binding domain, suggesting that HilE could also directly affect the DNA binding of HilD. To investigate this, we analyzed the effect of HilE on the ability of the LexADBDwt–HilD1–309 and LexADBDwt–LZ–HilD221–309 to induce the expression of the hilA–cat fusion in E. coli K-12. The presence of HilE completely blocked the activity of hilA–cat induced by LexADBDwt–HilD1–309, carrying the full-length HilD protein and partially inhibited the activity of this fusion mediated by LexADBDwt–LZ–HilD221–309, containing the DNA-binding domain of HilD fused to the heterologous LZ dimerization motif (Fig. 8). These results, together with the results indicating that HilE does not affect the dimerization mediated by the LZ motif (Fig. 7B), show that HilE can directly affect the DNA-binding domain of HilD, independently of its effect on the dimerization of this regulator. Therefore, the higher negative effect of HilE on the regulatory activity of LexADBDwt–HilD1–309 than that of LexADBDwt–LZ–HilD221–309 (Fig. 8) could be the result of a double effect of HilE on LexADBDwt–HilD1–309, on the dimerization and the DNA-binding domain, and just one effect of HilE on LexADBDwt–LZ–HilD221–309, on the DNA-binding domain.
Figure 8.
HilE directly affects the DNA-binding domain of HilD. Expression of the hilA–cat fusion contained in the philA–cat1 plasmid was tested in the E. coli MC4100 strain carrying the pair of plasmids pSR658–HilD1 and pK6–HilE1 (LexADBDwt–HilD1–309 + HilE), pSR658–HilD1 and pMPM–K6Ω (LexADBDwt–HilD1–309 + vector), pSR658–HilD6 and pK6–HilE1 (LexADBDwt–LZ–HilD221–309 + HilE), pSR658–HilD6 and pMPM–K6Ω (LexADBDwt–LZ–HilD221–309 + vector), pSR658 and pK6–HilE1 (LexADBDwt + HilE), or pSR658 and pMPM–K6Ω (LexADBDwt + vector). The CAT-specific activity was determined from samples collected of bacterial cultures grown in LB at 37 °C up to an A600 of 1.2. Expression of LexADBDwt, LexADBDwt–HilD1–309, and LexADBDwt–LZ–HilD221–309 was induced by adding 1 mm IPTG to the medium. 0.1% l-arabinose was also added to induce the expression of HilE from pK6–HilE1. The data are the averages of three independent experiments performed in duplicate. The bars represent the standard deviations. ***, expression statistically significantly different compared with that reached in the absence of HilE (p < 0.001).
HilE inhibits the DNA binding of HilD
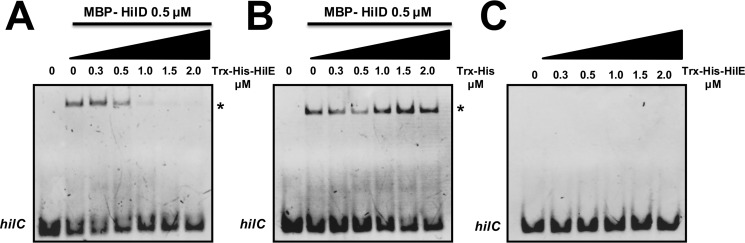
Our results strongly support that HilE inhibits the DNA-binding activity of HilD. To confirm this, we performed competitive electrophoretic mobility shift assays (EMSAs) with purified Trx–His–HilE and MBP–HilD proteins and a 50-bp fragment of the regulatory region of hilC carrying a HilD-binding site. As shown above, purified Trx–His–HilE interacts with HilD (Fig. 2B), and on the other hand, in a previous study we demonstrated that purified MBP–HilD binds to target genes in EMSAs (12). The hilC fragment was incubated with a constant concentration of MBP–HilD (0.5 μm) without or with increasing concentrations of Trx–His–HilE (0.3, 0.5, 1.0, 1.5, and 2 μm). Parallel binding reactions were performed with purified Trx–His instead Trx–His–HilE, as negative controls. From the concentration of 1.0 μm, Trx–His–HilE, but not Trx–His, drastically reduced the formation of the DNA/MBP–HilD complex (Fig. 9, A and B), indicating that Trx–His–HilE interferes with the DNA binding of MBP–HilD. Additional binding reactions ruled out a possible DNA-binding activity of Trx–His–HilE, even at the highest concentration tested (2.0 μm) (Fig. 9C). Thus, these results further confirm the interaction of HilE with HilD and demonstrate that this interaction inhibits the HilD binding to DNA.
Figure 9.
HilE inhibits the DNA-binding activity of HilD. Competitive nonradioactive EMSAs were performed to analyze the effect of HilE on the DNA-binding activity of HilD. A and B, a 50-pb DNA fragment of hilC, containing a HilD-binding site, was incubated with purified MBP–HilD (0.5 μm) and increasing concentrations (0, 0.3, 0.5, 1.0, 1.5, and 2.0 μm) of purified Trx–His–HilE (A) or Trx–His (B) proteins. C, EMSA was performed to evaluate DNA-binding activity of HilE. The 50-pb DNA fragment of hilC was incubated with increasing concentrations of purified Trx–His–HilE (0, 0.3, 0.5, 1.0, 1.5, and 2.0 μm). The DNA–protein complexes are indicated with asterisk and were resolved in a nondenaturing 6% polyacrylamide gel and stained with ethidium bromide.
HilE shares sequence and structure similarities with Hcp proteins from T6SSs
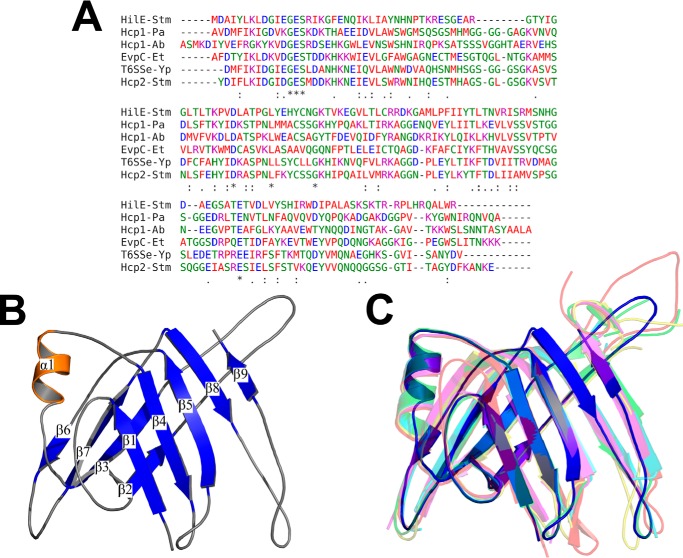
HilE was reported several years ago as a protein that does not present homology with any other protein in the databases (38). However, our recent BLASTp analysis revealed that the HilE sequence present an identity of ∼30% with Hcp proteins from type 6 secretion systems (T6SSs) of different Gram-negative bacteria (Fig. 10A and data not shown). To determine whether HilE also presents structural analogy with the Hcp proteins, it was modeled by I-TASSER server (53, 54). HilE was successfully modeled yielding a structure with a C-score of 1.07 and a TM-score of 0.86. The predicted structure is composed by a tight β-barrel domain with two β-sheets, with four and five β-strands each, flanked in one side by an α-helix (Fig. 10B). The overall modeled structure of HilE highly resembles that described for the monomeric subunits of Hcp proteins: Hcp1 of Pseudomonas aeruginosa (55), Hcp1 of Acinetobacter baumannii (56), EvpC of Edwardsiella tarda (57), a T6SS effector (T6SSe) of Yersinia pestis,4 and Hcp2 of S. Typhimurium (58) (Fig. 10C), showing TM values of 0.95, 0.89, 0.88, 0.86, and 0.78, respectively, with these proteins. TM values of >0.5 indicates structures sharing the same topology. These results are consistent with structural similarity shared between HilE and Hcp proteins.
Figure 10.
Sequence and structure comparison of HilE with Hcp proteins. A, sequence alignment of HilE and five Hcp proteins: Hcp1 of P. aeruginosa (Hcp1-Pa), Hcp1 of A. baumannii (Hcp1-Ab), EvpC of E. tarda (EvpC-Et), T6SSe of Y. pestis (T6SSe-Yp), and Hcp2 of S. Typhimurium (Hcp2-Stm). B, ribbon representation of the HilE predicted structure with I-TASSER server, colored by secondary structure: β-strands (blue), α-helix (orange) and loops (gray). C, overlapping of the HilE predicted structure (blue) with the crystallographic structure of the proteins Hcp1-Pa (PDB code 4KSR) (red), Hcp1-Ab (PDB code 4W64) (green), EvpC-Et (PDB code 3EAA) (yellow), T6SSe-Yp (PDB code 3V4H) (cyan), and Hcp2-Stm (PDB code 5XEU) (magenta).
Discussion
Negative regulation of HilE on the SPI-1 virulence genes is important for Salmonella fitness (59). Previously, it was shown that HilE negatively controls the expression of the SPI-1 genes by interacting with HilD, a central positive regulator for SPI-1 (38). In this study, we show that HilE specifically regulates the DNA-binding activity of HilD by inhibiting the dimerization and by directly acting on the DNA-binding domain of this regulator. In agreement with this, a recent study also found that HilE blocks the DNA binding of HilD (60). Our results indicate that HilD requires dimerization to induce the expression of target genes. Consistently, the HilD-binding sites on different genes have two direct repeat sequences (21, 27, 51). Several other AraC-like transcriptional regulators act as dimers, such as ToxT, ExsA, UreR, and AggR, which control expression of virulence genes in Vibrio cholerae, P. aeruginosa, Proteus mirabilis, and E. coli (45, 48, 49, 61, 62). HilE interacts with the central region of HilD and thus inhibits its dimerization, which indirectly would affect the DNA-binding activity of this regulator. Additionally, HilE interacts with the C-terminal region of HilD containing the DNA-binding domain, which negatively affects the transcriptional activity of HilD independently of its dimerization. This double interaction and effect supports a tight control of the HilD activity by HilE. In enteroagregative E. coli, the Aar protein interacts with the central region of AggR, containing the dimerization domain, inhibiting the dimerization and the DNA binding of this AraC-like regulator (62). Similarly, in P. aeruginosa, the ExsD protein interacts with the N-terminal region of ExsA, containing the dimerization domain, thus preventing the dimerization and DNA binding of this regulator (45, 61, 63). Neither Aar nor ExsD seem to directly compromise the DNA-binding domain of their target regulators. One example of a protein that directly acts on the DNA-binding domain of a transcriptional regulator is the control exerted by CarS on the CarA repressor in Myxococcus xanthus. CarS binds to the DNA-binding domain of CarA and thus inhibits its interaction with target genes; interestingly, the structure of CarS mimics the operator DNA recognized by CarA (64). Further studies, such as three-dimensional analysis, are required to determine the precise stoichiometry and the amino acids mediating the interaction between HilE and HilD, which is a matter of our current investigation.
Protein–protein interaction has been shown to regulate the stability of transcriptional regulators. For instance, FliT interacts with the FlhD4C2 complex, the central positive regulator of the flagellar genes, which enhances the degradation of FlhC subunit by the ClpXP protease (65). Our results discarded a negative effect of HilE on the stability of HilD in the growth conditions tested, further supporting that HilE only controls the regulatory activity of HilD. However, because HilD positively autoregulates (15, 26), HilE indirectly controls the expression and thus the amount of HilD.
Interestingly, we found that HilE shares sequence identity and structure analogy with Hcp proteins from different Gram-negative bacteria; thus, HilE could be postulated as a Hcp-like protein. Important to note, the Hcp-like proteins, including HilE, conserve high structure similarity but low sequence identity among them (55) (Fig. 10A). The Hcp proteins are structural components of the T6SSs, which are translocation machines that resemble an inverted phage tail, involved in different functions such as antibacterial activity and virulence (66, 67). Interestingly, the T6SSs components, including the Hcp proteins, are closely related to those of the phages tail, which work as channels for DNA delivery into bacteria (66, 67). Specifically, the Hcp proteins form a hexameric ring structure that stacks in a head-to-tail fashion, forming the tube through which T6SS effector proteins are either injected from the bacterial cytoplasm into the prey cell or released to the extracellular milieu (66–68). Furthermore, the Hcp proteins can also act as chaperones that help to the translocation of the T6SS effector proteins (69). S. Typhimurium possesses a functional T6SS, which is encoded in SPI-6 (70). Whether HilE has a structural/chaperone role in this T6SS remains to be determined.
The hilE gene is located in a genomic region that shows characteristics to be acquired by Salmonella (Fig. S2) (38); BLASTp analysis indicated that the other proteins also encoded in this Salmonella genomic island do not show homology with any protein related to the T6SSs. It is tempting to speculate that HilE diverged from an ancestral T6SS or phage Hcp protein. Thus, our results could illustrate the adaptation of a structural protein, during the evolution of Salmonella, to act as a regulator of virulence genes expression.
Experimental procedures
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Table S1. Bacterial cultures were grown in LB containing 1% tryptone, 0.5% yeast extract, and 1% NaCl at pH 7.5. When necessary, the media were supplemented with the following antibiotics: ampicillin (200 μg/ml), streptomycin (100 μg/ml), tetracycline (12 μg/ml), or kanamycin (20 μg/ml). Cultures for the determination of chloramphenicol acetyltransferase activity (CAT) and for the LexA-based genetic system assays were performed as we described previously (12, 28, 50).
Construction of plasmids
Plasmids and primers used in this study are listed in Table S1 and Table S2, respectively. To construct the plasmids pSR658–HilD1, pSR658–HilD2, pSR658–HilD3, pSR658–HilD4, and pSR658–HilD5, fragments of the hilD gene were amplified by PCR using the primers pairs HilD-SacI/HilDexR-PstI, HilD-SacI/HilD-130, HilD130–2/HilDexR-PstI, HilD-SacI/HilD-220, and HilD-221/HilDexR-PstI, respectively. The resulting PCR products were digested with SacI and PstI and cloned into the vector pSR658 digested with the same restriction enzymes. To construct the pSR659-HilE1 plasmid, the hilE gene was amplified by PCR using the primers HilE–SacI and HilE–HindIII-3′. The resulting PCR product was digested with SacI and HindIII and cloned into the vector pSR659 digested with the same restriction enzymes. To construct the plasmid pSR658–HilD6, the DNA fragment encoding 35 amino acids of the LZ motif from GCN4 of S. cerevisiae was amplified by PCR using the primers LZ-F and HilD221LZR and DNA of the pUT18C-zip plasmid as template. The fragment encoding the region of HilD from codons 221 to 309 (HilD221–309) was also amplified by PCR using the LZ-HilD221F and HilDexR-PstI primers. Then the LZ and HilD221–309 fragments were fused by overlap extension PCR using the LZ-F and HilDexR-PstI primers. The resulting PCR product was digested with XhoI and PstI and cloned into the pSR658 vector digested with the same restriction enzymes. To construct the pA6–HilE1 and pK6–HilE1 plasmids, the hilE gene was amplified by PCR using the HilE–NcoI-2 and HilE–His6 primers. The resulting PCR product was digested with NcoI and HindIII and cloned into the pMPM–A6Ω or pMPM–K6Ω vectors digested with the same restriction enzymes. To construct the plasmid pET32–HilE expressing the Trx–His–HilE fusion protein, the hilE gene was amplified by PCR using the HilE–NcoI-2 and HilE–PUT–BamHI primers. The resulting PCR product was digested with NcoI and BamHI and cloned into the pET32b(+) vector digested with the same restriction enzymes.
HilD dimerization assays
To test dimerization of HilD, the plasmids pSR658, pSR658–HilD1, or pSR658–HNS were transformed into the E. coli SU101 reporter strain for homodimerization assays, which carries the chromosomal sulA–lacZ transcriptional fusion (43, 44). Likewise, to determine the region of HilD containing the dimerization domain, the plasmids pSR658, pSR658–HilD1, pSR658–HilD2, pSR658–HilD3, pSR658–HilD4, or pSR658–HilD5 were transformed into the same reporter strain. Transformants were grown in LB with tetracycline and 1 mm IPTG to induce expression of LexADBD and LexADBD fusion proteins. The samples were collected at an A600 of 1.0 and used for the determination of β-gal activity.
To test whether HilE affects the dimerization of HilD, the E. coli SU101 reporter strain was first transformed with the plasmids pSR658, pSR658–HilD1, pSR658–HNS, or pSR658–HilD6 and then transformed with the pMPM–A6Ω vector or the pA6–HilE1 plasmid expressing HilE from the arabinose-inducible promoter. Transformants were grown in LB with tetracycline and ampicillin, as well as with 1 mm IPTG to induce the expression of LexADBD and LexADBD fusion proteins and with 0.1% l-arabinose to induce the expression of HilE. The samples were collected at an A600 of 1.0 and used for the determination of β-gal activity.
HilD–HilE heterodimerization assays
To test the interaction between HilD and HilE, the E. coli SU202 reporter strain for heterodimerization assays, which carries the sulA–lacZ transcriptional fusion with a hybrid LexA operator (43, 44) was first transformed with the plasmids pSR658, pSR658–HilD1, pSR658–HilD2, pSR658–HilD4, or pSR658–HilD5 and then transformed with the pSR659 vector or the pSR659-HilE1 plasmid. Transformants were grown in LB with tetracycline and ampicillin and with 1 mm IPTG to induce expression of LexADBD and LexADBD fusion proteins. The samples were collected at an A600 of 1.0 and used for the determination of β-gal activity.
β-gal assays
The β-gal assay and protein quantification to calculate specific activities were performed as previously described (71).
CAT assays
The CAT assays and protein quantification to calculate CAT-specific activities were performed as previously described (72).
Western blotting
Whole-cell extracts were prepared from bacterial samples collected at the indicated time points from LB cultures. 10 μg of each extract were subjected to electrophoresis in SDS-12% polyacrylamide gels and then transferred to 0.45-μm pore size nitrocellulose membranes (Merck), using a semidry transfer apparatus (Bio-Rad). The membranes containing the transferred proteins were blocked in 5% nonfat milk overnight and then incubated with anti-c-Myc (Sigma, catalog no. M4439) or anti-His6 (Roche, catalog no. 11922416001) monoclonal antibodies at 1:5000 and 1:1000 dilutions, respectively, or with anti-LexA (Abcam, catalog no. ab14553) or anti-GroEL (Sigma, catalog no. G6532) polyclonal antibodies at 1:10,000 and 1:100,000 dilutions, respectively. Horseradish peroxidase–conjugated anti-mouse (Sigma, catalog no. A9044) or anti-rabbit (Rockland, catalog no. 611-1302) at a dilution of 1:10,000 were used as the secondary antibodies. The bands on the blotted membranes were developed by incubation with the Western Lightning Chemiluminescence reagent plus (PerkinElmer) and exposed to Kodak X-Omat films or Amersham Imager 600 (GE Healthcare).
Expression and purification of MBP–HilD
Expression and purification of MBP–HilD were performed as we previously described (12).
Expression and purification of Trx–His and Trx–His–HilE
The Trx–His and Trx–His–HilE proteins were expressed in E. coli BL21/DE3 carrying the pET32b(+) or pET32–HilE plasmids, respectively, and purified from soluble bacterial extracts by using Ni–NTA–agarose columns (Qiagen). Bacterial cultures were grown in 250 ml of LB with ampicillin up to an A600 of 0.6, at 37 °C. Then the expression of Trx–His or Trx–His–HilE was induced by adding 1 mm IPTG, and the cultures were incubated at 30 °C for 4 h. Bacteria were collected by centrifugation at 4 °C, and the pellets were washed once with ice-cold binding buffer (5 mm imidazole, 500 mm NaCl, 20 mm Tris-HCl, pH 8.0) and resuspended in 30 ml of the same buffer. The cells were lysed with French press, and bacterial debris were removed by centrifugation at 4 °C. The soluble bacterial extracts were loaded into Ni–NTA–agarose columns previously equilibrated with 50 ml of binding buffer. The columns were washed with 100 ml of binding buffer and then with 100 ml of wash buffer (20 mm imidazole, 500 mm NaCl, 20 mm Tris-HCl, pH 8.0). The proteins were eluted with elution buffer (250 mm imidazole, 500 mm NaCl, 20 mm Tris-HCl, pH 8.0). The collected fractions were analyzed in a 12% SDS-PAGE. Those fractions containing the purified Trx–His and Trx–His–HilE proteins were loaded into a Slide-A-Lyzer 7K cassette (Thermo) and dialyzed at 4 °C in a buffer containing 20 mm Tris-HCl, pH 8.0, 40 mm KCl, 1 mm EDTA, and 20% (v/v) glycerol. Protein concentration was determined by the Bradford procedure. Aliquots of the purified Trx–His and Trx–His–HilE proteins were stored at −70 °C.
Electrophoretic mobility assays
EMSAs were performed using the purified MBP–HilD, Trx–His–HilE, or Trx–His proteins and a 50-bp DNA fragment of hilC containing a binding site of HilD. The 50-bp hilC fragment was generated by annealing the complementary primers HilCRR-F and HilCRR-R. For this, the primers, each at a final concentration of 15 μm, were boiled together at 95 °C for 10 min and then slowly cooling to room temperature. Competitive binding reactions were performed by mixing ∼100 ng of the hilC fragment with MBP–HilD (0.5 μm) and increasing amounts of Trx–His–HilE or Trx–His (0.3, 0.5, 1.0, 1.5, and 2.0 μm), in a total volume of 20 μl of binding buffer containing 100 μg/ml BSA, 30 mm HEPES, pH 7.5, 5 mm EDTA, 3 mm DTT, 200 mm KCl, 25 mm MgCl2, and 5% glycerol. For noncompetitive EMSAs, only increasing amounts of Trx–His–HilE purified protein (0.3, 0.5, 1.0, 1.5, and 2.0 μm) were used. Protein–DNA binding reactions were incubated at 37 °C for 20 min and then electrophoretically separated in 6% nondenaturing polyacrylamide gels in 0.5× Tris borate-EDTA buffer, at room temperature. The DNA fragments were stained with ethidium bromide and visualized with an Alpha-Imager UV Transilluminator (Alpha Innotech Corp.).
HilD stability assays
HilD stability assays were performed as we described previously (33). The values for HilD–Myc bands were normalized with respect to those for GroEL bands, and then the relative percentage of HilD–Myc at each indicated time, with respect to time 0, was calculated. The half-life time (t1/2) of HilD was calculated by one phase decay equation.
Protein secretion analysis
Protein secretion assays were performed as we described previously (28). The samples were analyzed in a 12% SDS-PAGE and stained with Coomassie Brilliant Blue R-250.
Gel-filtration assay
The purified protein MBP–HilD was subjected to gel filtration chromatography analysis by using AKTA-FPLC system (Superdex 200 HiLoadTM 26/60 column; GE Healthcare Life Sciences), with a flow rate of 0.5 ml/min and a pressure limit of 0.3 MPa, in a buffer containing 200 mm Tris-HCl, pH 8.0, 150 mm NaCl at 20 °C. The column was precalibrated by using a gel filtration molecular weight markers kit (Sigma–Aldrich) including cytochrome c from horse heart (12.4 kDa), carbonic anhydrase from bovine erythrocytes (29 kDa), albumin bovine serum (66 kDa), alcohol dehydrogenase from yeast (150 kDa), and β-amylase from sweet potato (200 kDa). The relative molecular mass of MBP–HilD was determined by comparison to the five-point molecular weight calibration curve.
Pulldown assays
Pulldown assays were performed with purified Trx–His–HilE or Trx–His fusion proteins and whole-cell extracts from E. coli SU101 expressing LexADBDwt, LexADBDwt–HilD1–309, LexADBDwt–HilD1–130, LexADBDwt–HilD1–220, or LexADBDwt–HilD221–309. To immobilize the bait protein, 15 μg of purified Trx–His–HilE or Trx–His were incubated for 1 h at 10 °C with 80 μl of Ni–NTA resin (Qiagen) previously equilibrated with lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 5 mm imidazole, pH 8.0) in a 1.5-ml microcentrifuge tube. The resin containing the immobilized bait protein was washed with 1 ml of wash buffer (70 mm NaH2PO4, 300 mm NaCl, 35 mm imidazole, pH 8.0) by centrifugation at 4000 × g for 1 min. The supernatant was carefully removed, and then the resin was incubated for 1 h at 10 °C with 80 μl of the whole-cell extract containing the respective prey protein. To remove the unbound proteins, the resin was washed five times with 1 ml of wash buffer by centrifugation at 4000 × g for 1 min. Finally, the resin was resuspended with 20 μl of SDS loading buffer containing 1.5% of β-mercaptoethanol and boiled for 5 min at 99 °C. After this, the samples were analyzed by Western blotting.
Sequence alignment and structure prediction of HilE
The sequence of HilE and that of some Hcp proteins were aligned using the Clustal Omega server (73). The sequence of HilE was submitted to I-TASSER server for structural modeling (53, 54), and a final model with a C score of 1.07, a TM score of 0.86, and a root mean square deviation of 2.7 Å was selected. All molecular graphics were done in PyMOL version 1.8 (PyMOL Molecular Graphics System, version 1.8).
Statistical analysis
Data from CAT and β-gal assays were analyzed using one-way analysis of variance with the Tukey's multiple comparison test. A p value of < 0.05 was considered significant. This statistical analysis was performed using Prism 6 program version 6.01 (GraphPad Software, San Diego, CA).
Author contributions
C. C. P.-A. and V. H. B. formal analysis; C. C. P.-A., G. V.-G., V. R. J.-G., E. R.-P., and V. H. B. investigation; C. C. P.-A. visualization; C. C. P.-A. methodology; C. C. P.-A. and V. H. B. writing-original draft; C. C. P.-A., G. V.-G., E. R.-P., and V. H. B. writing-review and editing; V. H. B. conceptualization; V. H. B. supervision; V. H. B. funding acquisition.
Supplementary Material
Acknowledgments
We thank F. J. Santana and M. Fernández-Mora for technical assistance, I. Gómez-Gómez for advice and assistance in the protein purification assays, D. Perez-Morales for standardization of the protein stability assays, and G. Soberón-Chavez and I. Martínez-Flores for critical reading of the manuscript.
This work was supported by Grant 254531 (to V. H. B.) and Predoctoral Fellowship 290934 (to C. C. P.-A.) from Consejo Nacional de Ciencia y Tecnología, Grant IN203415 (to V. H. B.) and a postdoctoral fellowship (to G. V.-G.) from Dirección General de Asuntos del Personal Académico de la Universidad Nacional Autónoma de México, and funds from the Posgrado en Ciencias Biológicas of the Universidad Nacional Autónoma de México (to C. C. P.-A.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1 and S2.
E. V. Filippova, A. Halavaty, G. Minasov, L. Shuvalova, I. Dubrovska, J. Winsor, L. Papazisi, and W. F. Anderson, unpublished observations.
- SPIs
- Salmonella pathogenicity islands
- SPI-1
- Salmonella pathogenicity island 1
- T3SS
- type 3 secretion system
- T6SS
- type 6 secretion system
- LexADBDwt
- LexA DNA-binding domain wildtype
- LexADBDmut
- LexA DNA-binding domain mutated
- MBP
- maltose-binding protein
- Trx
- thioredoxin
- LZ
- leucine zipper
- Hcp
- hemolysin-coregulated protein
- CAT
- chloramphenicol acetyltransferase
- LB
- lysogenic broth
- Ni–NTA
- nickel–nitrilotriacetic acid
- EMSA
- electrophoretic mobility assay
- IPTG
- isopropyl β-d-thiogalactopyranoside
- β-gal
- β-galactosidase
- PDB
- Protein Data Bank.
References
- 1. Haraga A., Ohlson M. B., and Miller S. I. (2008) Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6, 53–66 10.1038/nrmicro1788 [DOI] [PubMed] [Google Scholar]
- 2. Sánchez-Vargas F. M., Abu-El-Haija M. A., and Gómez-Duarte O. G. (2011) Salmonella infections: an update on epidemiology, management, and prevention. Travel. Med. Infect. Dis. 9, 263–277 10.1016/j.tmaid.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 3. Ohl M. E., and Miller S. I. (2001) Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52, 259–274 10.1146/annurev.med.52.1.259 [DOI] [PubMed] [Google Scholar]
- 4. Fàbrega A., and Vila J. (2013) Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26, 308–341 10.1128/CMR.00066-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Porwollik S., and McClelland M. (2003) Lateral gene transfer in Salmonella. Microbes Infect. 5, 977–989 10.1016/S1286-4579(03)00186-2 [DOI] [PubMed] [Google Scholar]
- 6. Ilyas B., Tsai C. N., and Coombes B. K. (2017) Evolution of Salmonella–host cell interactions through a dynamic bacterial genome. Front. Cell. Infect. Microbiol. 7, 428 10.3389/fcimb.2017.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hansen-Wester I., and Hensel M. (2001) Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3, 549–559 10.1016/S1286-4579(01)01411-3 [DOI] [PubMed] [Google Scholar]
- 8. Moest T. P., and Méresse S. (2013) Salmonella T3SSs: successful mission of the secret(ion) agents. Curr. Opin. Microbiol. 16, 38–44 10.1016/j.mib.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 9. Eade C. R., Hung C. C., Bullard B., Gonzalez-Escobedo G., Gunn J. S., and Altier C. (2016) Bile acids function synergistically to repress invasion gene expression in Salmonella by destabilizing the invasion regulator HilD. Infect. Immun. 84, 2198–2208 10.1128/IAI.00177-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hung C. C., Garner C. D., Slauch J. M., Dwyer Z. W., Lawhon S. D., Frye J. G., McClelland M., Ahmer B. M., and Altier C. (2013) The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol. Microbiol. 87, 1045–1060 10.1111/mmi.12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lawhon S. D., Maurer R., Suyemoto M., and Altier C. (2002) Intestinal short-chain fatty acids alter Salmonella Typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46, 1451–1464 10.1046/j.1365-2958.2002.03268.x [DOI] [PubMed] [Google Scholar]
- 12. Bustamante V. H., Martínez L. C., Santana F. J., Knodler L. A., Steele-Mortimer O., and Puente J. L. (2008) HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc. Natl. Acad. Sci. U.S.A. 105, 14591–14596 10.1073/pnas.0801205105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miao E. A., and Miller S. I. (2000) A conserved amino acid sequence directing intracellular type III secretion by Salmonella Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 97, 7539–7544 10.1073/pnas.97.13.7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Golubeva Y. A., Sadik A. Y., Ellermeier J. R., and Slauch J. M. (2012) Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics. 190, 79–90 10.1534/genetics.111.132779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ellermeier C. D., Ellermeier J. R., and Slauch J. M. (2005) HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57, 691–705 10.1111/j.1365-2958.2005.04737.x [DOI] [PubMed] [Google Scholar]
- 16. Saini S., Ellermeier J. R., Slauch J. M., and Rao C. V. (2010) The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog. 6, e1001025 10.1371/journal.ppat.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Petrone B. L., Stringer A. M., and Wade J. T. (2014) Identification of HilD-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 196, 1094–1101 10.1128/JB.01449-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martínez L. C., Banda M. M., Fernández-Mora M., Santana F. J., and Bustamante V. H. (2014) HilD induces expression of SPI-2 genes by displacing the global negative regulator H-NS from ssrAB. J. Bacteriol. 196, 3746–3755 10.1128/JB.01799-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Main-Hester K. L., Colpitts K. M., Thomas G. A., Fang F. C., and Libby S. J. (2008) Coordinate regulation of Salmonella pathogenicity island 1 (SPI1) and SPI4 in Salmonella enterica serovar Typhimurium. Infect. Immun. 76, 1024–1035 10.1128/IAI.01224-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martínez-Flores I., Pérez-Morales D., Sánchez-Pérez M., Paredes C. C., Collado-Vides J., Salgado H., and Bustamante V. H. (2016) In silico clustering of Salmonella global gene expression data reveals novel genes co-regulated with the SPI-1 virulence genes through HilD. Sci. Rep. 6, 37858 10.1038/srep37858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singer H. M., Kühne C., Deditius J. A., Hughes K. T., and Erhardt M. (2014) The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J. Bacteriol. 196, 1448–1457 10.1128/JB.01438-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cordero-Alba M., and Ramos-Morales F. (2014) Patterns of expression and translocation of the ubiquitin ligase SlrP in Salmonella enterica serovar Typhimurium. J. Bacteriol. 196, 3912–3922 10.1128/JB.02158-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thijs I. M., De Keersmaecker S. C., Fadda A., Engelen K., Zhao H., McClelland M., Marchal K., and Vanderleyden J. (2007) Delineation of the Salmonella enterica serovar Typhimurium HilA regulon through genome-wide location and transcript analysis. J. Bacteriol. 189, 4587–4596 10.1128/JB.00178-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Colgan A. M., Kröger C., Diard M., Hardt W. D., Puente J. L., Sivasankaran S. K., Hokamp K., and Hinton J. C. (2016) The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 12, e1006258 10.1371/journal.pgen.1006258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith C., Stringer A. M., Mao C., Palumbo M. J., and Wade J. T. (2016) Mapping the regulatory network for Salmonella enterica serovar Typhimurium invasion. MBio. 7, e01024–01016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olekhnovich I. N., and Kadner R. J. (2002) DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184, 4148–4160 10.1128/JB.184.15.4148-4160.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olekhnovich I. N., and Kadner R. J. (2007) Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J. Bacteriol. 189, 6882–6890 10.1128/JB.00905-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martínez L. C., Yakhnin H., Camacho M. I., Georgellis D., Babitzke P., Puente J. L., and Bustamante V. H. (2011) Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol. Microbiol. 80, 1637–1656 10.1111/j.1365-2958.2011.07674.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ellermeier J. R., and Slauch J. M. (2008) Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J. Bacteriol. 190, 476–486 10.1128/JB.00926-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chubiz J. E., Golubeva Y. A., Lin D., Miller L. D., and Slauch J. M. (2010) FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192, 6261–6270 10.1128/JB.00635-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takaya A., Kubota Y., Isogai E., and Yamamoto T. (2005) Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol. Microbiol. 55, 839–852 [DOI] [PubMed] [Google Scholar]
- 32. Boddicker J. D., and Jones B. D. (2004) Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect. Immun. 72, 2002–2013 10.1128/IAI.72.4.2002-2013.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De la Cruz M. A., Pérez-Morales D., Palacios I. J., Fernández-Mora M., Calva E., and Bustamante V. H. (2015) The two-component system CpxR/A represses the expression of Salmonella virulence genes by affecting the stability of the transcriptional regulator HilD. Front. Microbiol. 6, 807 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34. Gantois I., Ducatelle R., Pasmans F., Haesebrouck F., Hautefort I., Thompson A., Hinton J. C., and Van Immerseel F. (2006) Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 72, 946–949 10.1128/AEM.72.1.946-949.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang Y., Suyemoto M., Garner C. D., Cicconi K. M., and Altier C. (2008) Formate acts as a diffusible signal to induce Salmonella invasion. J. Bacteriol. 190, 4233–4241 10.1128/JB.00205-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Golubeva Y. A., Ellermeier J. R., Cott Chubiz J. E., and Slauch J. M. (2016) Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. MBio. 7, e02170–02115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. López-Garrido J., Puerta-Fernández E., Cota I., and Casadesús J. (2015) Virulence gene regulation by l-arabinose in Salmonella enterica. Genetics 200, 807–819 10.1534/genetics.115.178103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baxter M. A., Fahlen T. F., Wilson R. L., and Jones B. D. (2003) HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71, 1295–1305 10.1128/IAI.71.3.1295-1305.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baxter M. A., and Jones B. D. (2015) Two-component regulators control hilA expression by controlling fimZ and hilE expression within Salmonella enterica serovar Typhimurium. Infect. Immun. 83, 978–985 10.1128/IAI.02506-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Espinosa E., and Casadesús J. (2014) Regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by the LysR-type regulator LeuO. Mol. Microbiol. 91, 1057–1069 10.1111/mmi.12500 [DOI] [PubMed] [Google Scholar]
- 41. Gong H., Vu G. P., Bai Y., Chan E., Wu R., Yang E., Liu F., and Lu S. (2011) A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog. 7, e1002120 10.1371/journal.ppat.1002120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lim S., Yun J., Yoon H., Park C., Kim B., Jeon B., Kim D., and Ryu S. (2007) Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res. 35, 1822–1832 10.1093/nar/gkm060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dmitrova M., Younès-Cauet G., Oertel-Buchheit P., Porte D., Schnarr M., and Granger-Schnarr M. (1998) A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 257, 205–212 10.1007/s004380050640 [DOI] [PubMed] [Google Scholar]
- 44. Daines D. A., and Silver R. P. (2000) Evidence for multimerization of neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J. Bacteriol. 182, 5267–5270 10.1128/JB.182.18.5267-5270.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brutinel E. D., Vakulskas C. A., and Yahr T. L. (2010) ExsD inhibits expression of the Pseudomonas aeruginosa type III secretion system by disrupting ExsA self-association and DNA-binding activity. J. Bacteriol. 192, 1479–1486 10.1128/JB.01457-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gallegos M. T., Schleif R., Bairoch A., Hofmann K., and Ramos J. L. (1997) Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bustos S. A., and Schleif R. F. (1993) Functional domains of the AraC protein. Proc. Natl. Acad. Sci. U.S.A. 90, 5638–5642 10.1073/pnas.90.12.5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poore C. A., Coker C., Dattelbaum J. D., and Mobley H. L. (2001) Identification of the domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J. Bacteriol. 183, 4526–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prouty M. G., Osorio C. R., and Klose K. E. (2005) Characterization of functional domains of the Vibrio cholerae virulence regulator ToxT. Mol. Microbiol. 58, 1143–1156 10.1111/j.1365-2958.2005.04897.x [DOI] [PubMed] [Google Scholar]
- 50. Martínez L. C., Martínez-Flores I., Salgado H., Fernández-Mora M., Medina-Rivera A., Puente J. L., Collado-Vides J., and Bustamante V. H. (2014) In silico identification and experimental characterization of regulatory elements controlling the expression of the Salmonella csrB and csrC genes. J. Bacteriol. 196, 325–336 10.1128/JB.00806-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schechter L. M., and Lee C. A. (2001) AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella Typhimurium hilA promoter. Mol. Microbiol. 40, 1289–1299 10.1046/j.1365-2958.2001.02462.x [DOI] [PubMed] [Google Scholar]
- 52. Karimova G., Pidoux J., Ullmann A., and Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Roy A., Kucukural A., and Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., and Mekalanos J. J. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 312, 1526–1530 10.1126/science.1128393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruiz F. M., Santillana E., Spínola-Amilibia M., Torreira E., Culebras E., and Romero A. (2015) Crystal structure of Hcp from Acinetobacter baumannii: a component of the type VI secretion system. PLoS One 10, e0129691 10.1371/journal.pone.0129691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jobichen C., Chakraborty S., Li M., Zheng J., Joseph L., Mok Y. K., Leung K. Y., and Sivaraman J. (2010) Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS One 5, e12910 10.1371/journal.pone.0012910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin Q. P., Gao Z. Q., Geng Z., Zhang H., and Dong Y. H. (2017) Crystal structure of the putative cytoplasmic protein STM0279 (Hcp2) from Salmonella Typhimurium. Acta Crystallogr. F Struct. Biol. Commun. 73, 463–468 10.1107/S2053230X17010512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sturm A., Heinemann M., Arnoldini M., Benecke A., Ackermann M., Benz M., Dormann J., and Hardt W. D. (2011) The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7, e1002143 10.1371/journal.ppat.1002143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grenz J. R., Cott Chubiz J. E., Thaprawat P., and Slauch J. M. (2018) HilE regulates HilD by blocking DNA binding in Salmonella enterica serovar Typhimurium. J. Bacteriol. 10.1128/JB.00750-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brutinel E. D., Vakulskas C. A., and Yahr T. L. (2009) Functional domains of ExsA, the transcriptional activator of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 191, 3811–3821 10.1128/JB.00002-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Santiago A. E., Yan M. B., Tran M., Wright N., Luzader D. H., Kendall M. M., Ruiz-Perez F., and Nataro J. P. (2016) A large family of anti-activators accompanying XylS/AraC family regulatory proteins. Mol. Microbiol. 101, 314–332 10.1111/mmi.13392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thibault J., Faudry E., Ebel C., Attree I., and Elsen S. (2009) Anti-activator ExsD forms a 1:1 complex with ExsA to inhibit transcription of type III secretion operons. J. Biol. Chem. 284, 15762–15770 10.1074/jbc.M109.003533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. León E., Navarro-Avilés G., Santiveri C. M., Flores-Flores C., Rico M., González C., Murillo F. J., Elías-Arnanz M., Jiménez M. A., and Padmanabhan S. (2010) A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix. Nucleic Acids Res. 38, 5226–5241 10.1093/nar/gkq277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sato Y., Takaya A., Mouslim C., Hughes K. T., and Yamamoto T. (2014) FliT selectively enhances proteolysis of FlhC subunit in FlhD4C2 complex by an ATP-dependent protease, ClpXP. J. Biol. Chem. 289, 33001–33011 10.1074/jbc.M114.593749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Records A. R. (2011) The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol. Plant. Microbe. Interact. 24, 751–757 10.1094/MPMI-11-10-0262 [DOI] [PubMed] [Google Scholar]
- 67. Ho B. T., Dong T. G., and Mekalanos J. J. (2014) A view to a kill: the bacterial type VI secretion system. Cell. Host. Microbe. 15, 9–21 10.1016/j.chom.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gallique M., Bouteiller M., and Merieau A. (2017) The type VI secretion system: a dynamic system for bacterial communication? Front. Microbiol. 8, 1454 10.3389/fmicb.2017.01454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Silverman J. M., Agnello D. M., Zheng H., Andrews B. T., Li M., Catalano C. E., Gonen T., and Mougous J. D. (2013) Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol. Cell. 51, 584–593 10.1016/j.molcel.2013.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Folkesson A., Löfdahl S., and Normark S. (2002) The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res. Microbiol. 153, 537–545 10.1016/S0923-2508(02)01348-7 [DOI] [PubMed] [Google Scholar]
- 71. Oropeza R., Sampieri C. L., Puente J. L., and Calva E. (1999) Negative and positive regulation of the non-osmoregulated ompS1 porin gene in Salmonella Typhi: a novel regulatory mechanism that involves OmpR. Mol. Microbiol. 32, 243–252 10.1046/j.1365-2958.1999.01329.x [DOI] [PubMed] [Google Scholar]
- 72. Puente J. L., Bieber D., Ramer S. W., Murray W., and Schoolnik G. K. (1996) The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20, 87–100 10.1111/j.1365-2958.1996.tb02491.x [DOI] [PubMed] [Google Scholar]
- 73. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.