Abstract
N2-fixing symbiotic root nodules of the actinorhizal host Datisca glomerata express Dgrca (D. glomerata Rubisco activase) mRNA, a transcript usually associated with photosynthetic organs or tissues. In northern blots a mature, 1700-nucleotide Dgrca mRNA was detected in green plant organs (leaves, flowers, and developing fruits) and in nodules but was not detected in roots. A second message of 3000 nucleotides was observed only in nodules. Both size classes of transcripts were polyadenylated. The larger transcript was 2- to 5-fold more abundant than the mature mRNA; it was hybridized to an intronic probe, indicating that a stable, incompletely spliced transcript was accumulating. Treatment with light on excised nodules did not alter the relative abundance of the two species. In in situ hybridizations the Dgrca message was expressed intensely in the nuclei of infected cells. The Dgrca transcripts also accumulated at lower levels in uninfected cortical cells adjacent to the periderm and the vascular cylinder. mRNA encoding the large subunit of Rubisco (DgrbcL) was abundant in mature infected cells and in the amyloplast-rich sheath of uninfected cortical cells lying between the infected cells and nodule periderm. The proteins Rubisco activase, Rubisco, and the 33-kD O2-evolving complex subunit did not accumulate to detectable levels, indicating that a functional photosynthetic apparatus was not prevalent in nodule tissue. Signals or factors required for the transcription of Dgrca appeared to be present in nodules, but efficient splicing and translation of the message were not observed in Frankia-infected tissue where transcript accumulation was highest.
Organogenesis of N2-fixing symbiotic root nodules represents an intricate interplay between two separate organisms: a plant and a microbial endosymbiont. Development of a functionally specialized organ, such as the symbiotic root nodule, depends on environmentally and developmentally induced cellular, physiological, and molecular events. Actinorhizal root nodules are induced by N2-fixing actinomycetes of the genus Frankia on woody dicotyledonous plants belonging to eight angiosperm families (Benson and Silvester, 1993). Actinorhizal symbioses contribute substantially to the global nitrogen economy, with particular impact on land reclamation, forestry, and management of sustainable ecosystems worldwide.
Functional aspects of the establishment of the endosymbiont, e.g. the infection process, N2 assimilation, O2 regulation, and C reduction, are shared between actinorhizal and legume nodules (for review, see Pawlowski and Bisseling, 1996; Pawlowski et al., 1996). However, there is considerable diversity in the morphological and organizational characteristics of these two nodule types. The actinorhizal nodule is a modified lateral root, having an organized meristem that arises from the pericycle and a central vascular cylinder surrounded by cortical cells, some of which contain Frankia. Legume nodules arise from interior cortical cell layers of the parent root and have peripheral vasculature (Hirsch and LaRue, 1997).
Molecular and physiological studies on a number of actinorhizal systems are gradually providing information on patterns of gene expression and biochemical processes relevant to nodule organogenesis. Homologs of genes coding for enzymes in primary N2 and C fixation, including Gln synthetase, Orn carbamoyl transferase, Suc synthase, and enolase, have been cloned from Alnus glutinosa by differential screening of cDNA libraries (Pawlowski et al., 1993). The characterization of expression of genes for leghemoglobin from Casuarina glauca (Jacobsen-Lyon et al., 1995; Gherbi et al., 1997), for a subtilisin-like protease (Ribeiro et al., 1995), for an enzyme involved in thiazole biosynthesis (Ribeiro et al., 1996), both from A. glutinosa, and for an A. glutinosa Cys protease of the papain superfamily (Goetting-Minesky and Mullin, 1994) is helping to identify metabolic processes in young and mature actinorhizal nodules. Recent reports of glutamate-and-Pro-rich, putative cell wall protein cDNA cell wall protein cDNA (Guan et al., 1997) and two Gly- and His-rich mRNAs expressed in the early infection zone (Pawlowski et al., 1997) contribute to an emerging picture of cell- and tissue-specific gene expression and their corresponding biochemical processes in nodule organogenesis.
To better understand actinorhizal nodule development, the relationship between nodule organization and function, and the cytological and biochemical diversity underlying different host-microbe N2-fixing symbioses, we initiated studies with Datisca glomerata. D. glomerata is an herbaceous perennial that grows in sandy soils of riparian ecosystems in California and northern Mexico (Davidson, 1973; Liston et al., 1989). It is closely related to the Indo-European species Datisca cannabina (Davidson, 1973); it is also related to begonias and cucurbits based on molecular systematics of rbcL (Rubisco large subunit genes (Swensen et al., 1994). The herbaceous nature of the foliage and the low tissue levels of phenolics render D. glomerata amenable to molecular biological studies. D. glomerata, with Coriaria, is distinguished from other actinorhizal hosts in that the symbiotic N2-fixing cells form a distinct, dense sector within the nodule cortex, to one side of the central vascular cylinder (Hafeez et al., 1984).
Because of our interest in gene expression in developing nodules of D. glomerata, we isolated a nodule cDNA clone, designated Dgrca (D. glomerata Rubisco activase), encoding a putative Rubisco activase. Rubisco activase normally accumulates in greening or photosynthetic tissues expressing Rubisco. The molecular mode of action of Rubisco activase has not been completely elucidated; however, it is postulated to cause conformational changes in Rubisco that promote the ordered binding of substrates required for optimal enzymatic activity and stability (Portis, 1990; Lan and Mott, 1991). Hence, it has a regulatory role in photosynthetic C reduction via the action of Rubisco. The significance of an mRNA encoding Rubisco activase in symbiotic root nodules of D. glomerata remains unknown, but might be attributed to: (a) a role in photosynthesis as a minor process that occurs in some nodules; (b) a role in O2 partitioning, whereby it activates the oxygenase function of Rubisco; and (c) part of a general induction of cellular processes during nodule organogenesis. In this paper we characterize the pattern of expression of Dgrca and other components of the photosynthetic apparatus to examine the possible role of Dgrca in nodules.
MATERIALS AND METHODS
Plant Material
Datisca glomerata (Presl) Baill seeds were obtained from plants in Vaca Hills, California. Plants were grown either in liquid culture medium consisting of one-quarter-strength Hoagland solution or in a 2:1 (v/v) mixture of Perlite and sand:peat:fir bark (1:1:1, v/v). Root nodules of D. glomerata were induced on greenhouse-grown seedlings by inoculation with crushed nodules of Ceanothus griseus var. horizontalis (Liu and Berry, 1991). Inoculated plants were fertilized with one-quarter-strength Hoagland medium. Nodules and root tips were excised from intact root systems of plants grown in soil or in liquid culture, and immediately transferred to liquid N2. Nodules were harvested 4, 5, 7, and 11 weeks after inoculation. Enhanced greening and vigorous growth of plants at 4 to 5 weeks after inoculation correlated with nodule lobe expansion and suggested that the onset of N2 fixation occurred at this time. Young leaves, flowers (sepals, anthers, stamens, and styles), and fruits containing immature seed (developing fruits) were collected from 5-month-old plants. To examine the effect of light on nodule greening and Dgrca mRNA levels, washed root systems on intact plants inoculated 17 weeks earlier were either wrapped in clear plastic and placed under continuous white light at 20°C for 16 to 20 h or placed in darkness for 20 h before excision of nodules. Nodules were also harvested from light-treated roots excised from plants 10 weeks after inoculation. Plant materials for RNA and DNA preparations were stored at −80°C.
RNA and DNA Isolation and Blot Analysis
Total RNA was isolated from D. glomerata nodules as previously described (Pawlowski et al., 1994). For blot analysis, poly(A−) RNA was recovered after two passages over oligo(dT25) magnetic beads (Dynal, Lake Success, NY) to remove poly(A+) RNA. RNA was partitioned on 1% agarose containing 6% formaldehyde (Sambrook et al., 1989), transferred to a nylon membrane (Zeta Probe, Bio-Rad), and hybridized as recommended by the manufacturer. Loading of equal amounts of total RNA for northern blots was determined from A260 titers and by visualization of ethidium bromide-stained rRNA bands. For cDNA library construction and blot analysis, poly(A+) RNA was obtained by two passes through oligo(dT)-cellulose columns (Boehringer Mannheim). Total DNA was obtained from young leaves of D. glomerata, essentially as described by Ribeiro et al. (1995), and transferred to the Zeta Probe nylon membrane. Hybridization of DNA blots was performed according to instructions (Bio-Rad). Autoradiography was carried out using preflashed Kodak XAR 5 film at −80°C with an intensifying screen (Lightning II Plus, DuPont).
Hybridization probes for RNA and DNA blots consisted of the 400-bp partial Dgrca cDNA insert, the full-length 1.7-kb cDNA insert, or intron A (128 bp) and intron C (160 bp) amplified by PCR. DNA probes were purified from agarose gels (JETSORB Gel Extraction kit, Genomed, Research Triangle Park, NC) and radiolabeled using [α-32P]dCTP (Amersham) and the Multiprime DNA Labeling System (Amersham) to specific activities of 3.5 to 7.0 × 104 Bq μg−1 (2.0–4.0 × 106 cpm ng−1). The intron A probe used in Figure 3B was labeled with both [α-32P]dCTP and [α-32P]dATP (Amersham).
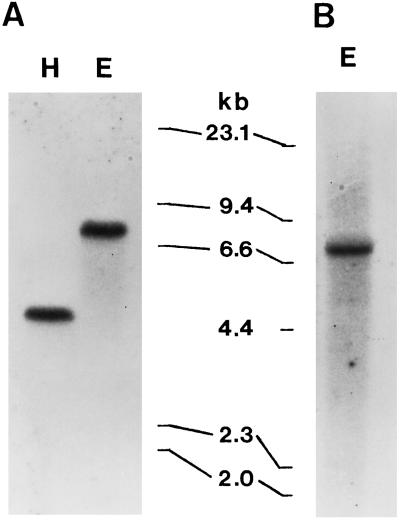
Figure 3.
Total DNA of D. glomerata hybridized to the 400-bp partial cDNA insert (A) and intron-A probe (B). DNA was treated with HindIII (lane H) or EcoRI (lanes E) before transfer to nylon membrane. Sizes (in kb) of phage Lambda HindIII fragments are shown in the middle. Autoradiography was carried out at −80°C for 3 (A) and 5 (B) d.
32P radiolabel was quantified from freshly hybridized nylon membranes using a phosphor imager (Storm, Molecular Dynamics, Sunnyvale, CA) and imaging plate (BAS IIIs, Fuji) and analyzed with ImageQuant software (version 4.0, Molecular Dynamics).
cDNA Library Construction and Screening
Two Lambda Zap cDNA libraries were made representing mRNAs from D. glomerata nodules harvested 4, 5, and 7 weeks after inoculation with Frankia. cDNA synthesis and ligation was carried out according to the protocol for Lambda Zap Express cDNA synthesis (Stratagene).
After mass excision, DNA was prepared from approximately 400 randomly selected phagemids and differentially screened with either radiolabeled-nodule or root cDNA. The average insert size of the first library was 500 to 600 bp. The second cDNA library was constructed to obtain a full-length clone. Double-stranded cDNA was fractionated on Sepharose CL-4B (Pharmacia); the 1- to 2-kb fraction was ligated to the Lambda Zap Express phage vector. The average size of cDNA inserts in this library was approximately 1.6 kb.
Primers
The oligonucleotide primers (Operon Technologies, Alameda, CA) for nucleotide sequencing of Dgrca cDNA and the Dgrca gene segment and for PCR of the Dgrca gene segment, intron A and intron C, are listed in Table I.
Table I.
Oligonucleotide primers used for sequencing or amplification of DNA fragments
| Purpose and Fragment | Name | Sequence (5′–3′) |
|---|---|---|
| Sequencing/primer walking | ||
| Dgrca cDNA | Dg195A | GTCCCCCAACGTTTGAT |
| rca3A | GGATTCTACATAGCTCCAG | |
| rca3B | CTGGAGCTATGTAGAATCC | |
| rca3C | CGTCGTCGTAAACCCGT | |
| Dgrca gene segment | rca3A | GGATTCTACATAGCTCCAG |
| rca31B | GCCTTTGACACATCTG | |
| rca31C | GCTTCTCCAGTGTCAT | |
| PCR amplification | ||
| Intron A | rcaInt3F | CATGACTCTCCCCAAC |
| rcaInt3R | CCTTGACCTTTACCTC | |
| Intron C | rcaInt1F | AGCAGTCAACAGATCC |
| rcaInt1R | TCCAGTTCCATTGTTG | |
| Dgrca gene segment | rca31C | GCTTCTCCAGTGTCAT |
| rca31D | CCAACCAATGTTCAGC | |
| rca31E | TCACGATACCGTTGCC |
In Situ Localization
Whole nodules were harvested 6 to 8 weeks after inoculation with Frankia and fixed in buffered 4% paraformaldehyde and 0.25% glutaraldehyde under a vacuum. Nodules were then dehydrated, embedded in paraffin, and sectioned. Nodule lobe sections were hybridized to 35S-radiolabeled RNA probes (see “RNA and DNA Isolation and Blot Analysis”), as described previously (van de Wiel et al., 1990; Ribeiro et al., 1995). Adjacent sections were used for hybridization to antisense and sense RNA probes. Sections were stained with ruthenium red and counterstained with toluidine blue.
A 500-bp HindIII fragment was obtained from the full-length clone of Dgrca and ligated to the Bluescript pBKS+ vector (Stratagene). The resulting subclone was either treated with SalI and transcribed with T7 RNA polymerase (antisense probe) or digested with EcoRI and transcribed with T3 RNA polymerase (sense probe). To obtain an antisense probe of DgrbcL, plasmid pDgrbcL, containing the rbcL coding sequence cloned in pBKS+, was linearized with SalI and transcribed with T7 RNA polymerase. The DgrbcL sense probe was generated with BamHI and T3 RNA polymerase. Probes for the nitrogenase subunit H gene (nifH) of Frankia were described by Ribeiro et al. (1995). Antisense and sense RNA probes were radiolabeled with [35S]UTP, as described by van de Wiel et al. (1990), without a cold chase. Maximum lengths of the probes were therefore 400 to 500 nucleotides.
Western Blots
Protein extracts were obtained from D. glomerata roots, root-free nodules and leaves, and leaves of soybean and tobacco by the following procedure: Tissue samples were frozen at −70°C, then homogenized with a mortar and pestle in 50 mm Tris-Cl, pH 7.4, 200 mm NaCl, and 10 mm MgCl2 (4 mL g−1 fresh weight tissue), and centrifuged for 15 min at 27,000g at 4°C. Equal amounts (by fresh weight) of clarified supernatants were used directly for SDS-PAGE (Laemmli, 1970). Protein determinations were performed according to the method of Bradford (1976). Immunodetection was performed according to the method of Sambrook et al. (1989) using rabbit polyclonal antibodies made to cotton Rubisco activase (provided by M. Salvucci, U.S. Department of Agriculture-Agricultural Research Service, Western Cotton Research Laboratory, Phoenix, AZ), to the soybean large subunit protein (Murphy, 1978), and to the OEC33 of pea (provided by S. Theg, University of California, Davis). Secondary anti-rabbit IgG conjugated to alkaline phosphatase was obtained from Sigma. Protein bands were visualized by staining with 5-bromo-4-chloro-3-indolyl phosphate (Sigma) and nitroblue tetrazolium (Eastman Kodak). At least two western-blot analyses using independent protein extractions were conducted with each primary antiserum.
RESULTS
We isolated an unusually expressed full-length (1634-bp) cDNA from symbiotic root nodules of D. glomerata. In BLASTX alignments (Altschul et al., 1990), the deduced amino acid sequence of the cDNA shared up to 89% similarity with Rubisco activases from many higher plants and cyanobacteria. We therefore designated our clone Dgrca. For clarity, only the three highest-scoring matches to sequences of apple, Arabidopsis, and tobacco are shown (Fig. 1).
Figure 1.
The deduced amino acid sequence of Dgrca (Dg) is conserved with respect to Rubisco activase coding sequences from other photosynthetic species, including apple (Md, P[N] = 1.8e−259; Watillon et al., 1993), Arabidopsis small peptide (At, P[N] = 5.8e−247; Orozco et al., 1993), and tobacco small peptide (Nt, P[N] = 4.8e−240; accession no. U35111).
The relative numbers of Dgrca clones in both libraries indicated that the corresponding mRNA was not a rare-class message in D. glomerata nodules. Initially, a 400-bp partial cDNA clone of Dgrca was isolated from approximately 400 clones picked at random. A screen of approximately 1.2 × 105 recombinant phage from a second library yielded over 20 separate clones that hybridized to a probe consisting of the partial Dgrca cDNA insert. The cDNAs ranged in size from 0.7 to 1.8 kb. The 3′-untranslated region varied in length; among five of the longest clones examined, two were found to contain 41 additional nucleotides immediately upstream of the poly(A+) tail, suggesting that multiple polyadenylation signals in Dgrca are utilized.
Two Size Classes of Dgrca RNA Are Present in Nodules
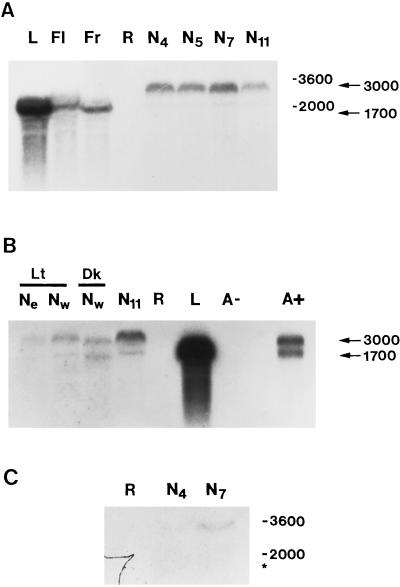
Dgrca mRNA was expressed in nodules and in photosynthetic organs of D. glomerata, including leaves, flowers, and immature fruits (Fig. 2A). Dgrca mRNA was not detected in the root samples. In the photosynthetic organs Dgrca mRNA was estimated to be 1700 nucleotides long. This corresponded to the size of the full-length cDNA clone (1634 bp), which was similar to Rubisco activase mRNAs from other plants (1650–1900 nucleotides). The data indicate that the 1700-nucleotide mRNA band represented the mature, fully spliced message.
Figure 2.
Expression of Dgrca mRNA in various organs of D. glomerata (A), in nodules with or without light treatment (B, lanes Ne, Nw, and Nw) and in oligo(dT)-cellulose fractions of nodule RNA (B, lanes A− and A+). Sizes in nucleotide of rRNA species are shown on the right of each panel. A, Total RNA from leaves (lane L), flowers (lane Fl), immature fruits (lane Fr), roots (lane R), and nodules harvested 4 to 11 weeks after Frankia inoculation (N4–N11) was hybridized to radiolabeled Dgrca cDNA insert. B, Total RNA from nodules harvested from excised roots (lane Ne) or from roots of whole plants (lanes Nw) after treatment with white light (Lt) or darkness (Dk) were hybridized to the full-length Dgrca cDNA probe. Total N11 RNA (lane N11), root RNA (lane R), and leaf RNA (lane L) are included for a comparison. The unbound fraction of total N11 RNA (lane A−) and the bound N11 RNA fraction (lane A+) after oligo(dT)-cellulose treatment were also hybridized to the full-length Dgrca cDNA probe. C, Total RNA from roots (lane R), N4 nodules, or N7 nodules was hybridized to a radiolabeled intron-A probe. The asterisk indicates the predicted position of the 1700-nucleotide mRNA species. For A and C and the first seven lanes of B, approximately 5 μg of total RNA was loaded into each lane; for lane A+ in B, 2.5 μg of poly(A+) RNA was used. Autoradiography was carried out with preflashed Kodak XAR 5 film at −80°C for 16.5 h (A), 6 d (B), and 5 d (C).
The two distinct size classes of Dgrca transcripts, 3000 and 1700 nucleotides, were observed in nodules harvested 4 to 11 weeks after Frankia inoculation (Fig. 2A). Only the 1700-nucleotide mRNA was detected in the photosynthetic organs, e.g. the flower, immature fruit, and leaves. Roots showed no detectable levels of either species. The 3000-nucleotide species was 2- to 5-fold more abundant than the 1700-nucleotide mRNA in total RNA from nodules harvested 11 weeks after inoculation (data not shown). The relative abundance of the 3000-nucleotide mRNA was reduced in the older (17-week) nodule samples, but was not altered significantly by exposure to white light (Fig. 2B, lanes 1–3). However, this light treatment was sufficient to cause faint greening in the mature nodules closest to the light source (data not shown).
To determine whether both size classes of mRNA were polyadenylated, poly(A+) and poly(A−) fractions were hybridized to the Dgrca cDNA insert in gel blots (Fig. 2B, lanes 7 and 9). Whereas no hybridization was detected in the poly(A−) fraction, both the 3000- and 1700-nucleotide species were observed in the poly(A+) fraction. The abundance ratio, quantified by phosphor imaging (data not shown), was nearly 1:1 in the nodule poly(A+) RNA fraction, as compared with 5:1 in total RNA. Attempts to obtain a 3-kb cDNA clone from several different poly(A+) RNA preparations, using random amplification of cDNA 3′ ends with two different primer pairs were unsuccessful.
Dgrca Is a Single-Copy Gene in D. glomerata
To determine the size of the rca gene family in D. glomerata, we hybridized total DNA to the 400-bp partial cDNA insert. A single 8.2-kb EcoRI fragment and a single 4.7-kb HindIII fragment (Fig. 3A) were radiolabeled; there was no evidence of two polymorphic Dgrca genes. We therefore conclude that Rubisco activase is encoded by a single gene or by a small, conserved gene family in D. glomerata, as in other plant species (Wernecke and Ogren, 1989; Wernecke et al., 1989; Rundle and Zielinski, 1991; Qian and Rodermel, 1993).
Occurrence of Intronic Sequences in the Dgrca Gene and 3000-Nucleotide mRNA
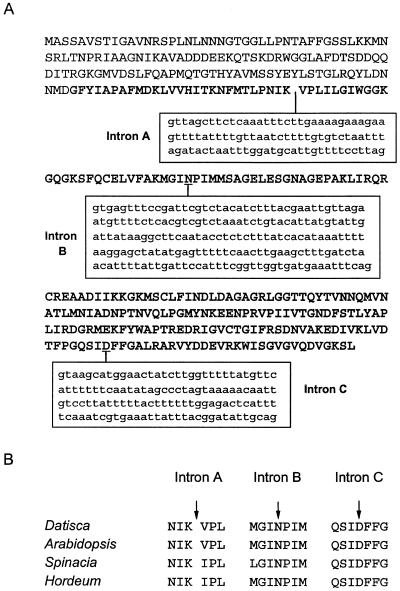
We were interested in whether the Dgrca gene was interrupted by introns that could account for the additional mass of the 3000-nucleotide mRNA. We examined Dgrca gene sequences from Arabidopsis (Orozco et al., 1993), spinach (Wernecke et al., 1989), and barley (Rundle and Zielinski, 1991). All three genes have four to six introns at conserved positions within the coding regions. A 1.2-kb fragment was amplified from the D. glomerata genome using PCR primers specific to the Dgrca coding region (Table I). This Dgrca gene segment contained three stretches of nucleotides, 96, 201, and 128 bp in length, that were not present in the Dgrca cDNA (Fig. 4A). The positions of these AT-rich stretches with respect to the coding region were identical to those of the introns in the rca genes from the other plants (Fig. 4B). Therefore, we refer to these genomic sequences as introns A, B, and C.
Figure 4.
A, The coding sequence (uppercase letters) of the Dgrca genomic DNA segment is interrupted by at least three intronic sequences (lowercase letters), designated Intron A, Intron B, and Intron C. B, The positions of these introns were highly conserved with respect to the Arabidopsis, spinach, and barley genes.
To generate hybridization probes, genomic DNA template was amplified with PCR primers annealing to coding sequences that immediately flanked intron A and intron C (Table I). In both RNA and DNA blots, the intron C PCR product hybridized to a wide number of species, possibly because it carried a repetitive sequence (data not shown). However, intron A hybridized to a single 8.2-kb EcoRI DNA fragment (Fig. 3B) and hybridized weakly to the 3000-nucleotide mRNA in blots of total nodule RNA (Fig. 2C).
Dgrca and DgrbcL mRNA Localization Patterns
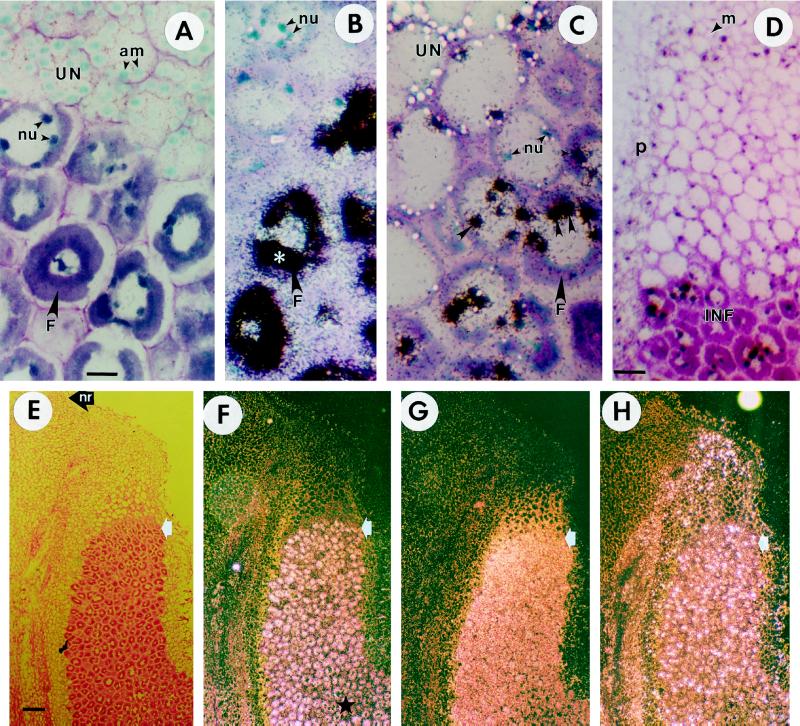
The mature Frankia-infected zone of the D. glomerata nodule consisted of a compact region of expanded host cells filled with Frankia vesicles arrayed peripherally around a central vacuole (Fig. 5, A and E). The mature tissue was distinguished by expression of nifH, seen as silver grains localized over the Frankia vesicles in the infected cells (Fig. 5, B and F). The youngest of these cells relative to the developmental gradient of the infection zone are indicated by white arrows in Figure 5, E through H. Beginning in the cells of the early infection zone, the host cytoplasm contained multiple nuclei (Fig. 5, A–C).
Figure 5.
In situ localization of Dgrca, DgrcbL, and nifH mRNA in D. glomerata nodule sections. A, Bright-field micrograph of the interface between Frankia-infected cells and mature, uninfected cells. Bar = 15 μm (for A–C). B, Bright-field micrograph of nifH expression in Frankia-infected cells. Silver grains indicating hybridization to antisense probes appear as black specks, particularly dense over the Frankia vesicles in the infected cells (white asterisk). C, Bright-field micrograph showing dense accumulations of silver grains indicating hybridization of the Dgrca antisense probe to nuclei of infected cells (arrowheads). Some Dgrca transcripts are detected in adjacent younger cells of the early infection stage. Mature, uninfected cells show scattered silver grains, with no significant accumulation of Dgrca mRNA. D, Bright-field micrograph of a longitudinal nodule section hybridized to the Dgrca antisense probe. The relatively large nuclei in the meristem are darkly stained with toluidine blue, but do not have silver grains. Infected cells accumulated Dgrca transcripts, whereas uninfected cells showed much less hybridization. Bar = 30 μm. E, Bright-field micrograph of a longitudinal section. The Frankia vesicles stained red are arrayed around central vacuoles in the mature, infected cells. Bar = 500 μm (for E–H). F, Dark-field micrograph showing nifH expression. Hybridization of transcripts to the nifH antisense probe, apparent as white specks, delineates the onset of N2 fixation in mature, infected cells (white arrows in E–H) within the developmental gradient of the infection zone. ★, A zone of senescence where nifH expression is reduced; the corresponding area in E has a vesiculated appearance because of degradation of the endosymbiont. G, Dark-field micrograph of DgrbcL expression in the zone before N2 fixation and in mature, infected cells. DgrbcL mRNA was less abundant in the zone of senescence. H, Dark-field micrograph of Dgrca expression in the infected cells and in surrounding uninfected cortical cells. nu, Nuclei; UN, uninfected cells; F, Frankia-infected cells; am, amyloplasts in uninfected cells; p, periderm; m, meristem; INF, infected cells; nr, base of a nodule root.
In contrast, the in situ expression pattern of Dgrca mRNA was complex. High levels of expression of Dgrca were observed in Frankia-infected cells, especially at early stages of vesicle differentiation before the onset of N2 fixation (Fig. 5, C, D, and G). The punctate appearance of the Dgrca signal (Fig. 5G) was attributed to the concentration of message in the nuclei of infected cells (Fig. 5C). Dgrca message was detected at lower levels in adjacent (younger) cells in the early stages of Frankia infection. Some Dgrca RNA was also detected in the nuclei of mature, infected cells expressing nifH and in the cortical cells immediately adjacent to infected cells (Fig. 5H). No Dgrca expression was observed in the nodule-lobe meristem (Fig. 5D) or in the nodule-root meristem (data not shown). The nuclei in the meristem and in the cells underlying the periderm in Figure 5D stained brightly with toluidine blue, but silver grains did not accumulate. Hybridization with the Dgrca sense probe was negative (data not shown).
DgrbcL mRNA was most abundant in the infected cells of the zone preceding N2 fixation (Fig. 5G). It was also detected in mature, infected cells and the uninfected cortical cells lying between the infected cells and nodule periderm. In both infected and uninfected cells, silver grains indicating DgrbcL expression were not densely clustered over a particular region of the cell, but appeared to be distributed throughout the cytoplasm. In carpels the DgrbcL antisense probe hybridized to photosynthetic parenchyma cells as expected, but not to immature, nonphotosynthetic anthers (data not shown). Hybridization of Dgrca, DgrbcL, and nifH was somewhat reduced in a region where Frankia vesicles were undergoing senescence (Fig. 5, E–H; see legend). Neither Dgrca nor DgrbcL antisense probes hybridized to periderm or vascular tissue.
Rubisco Activase and Other Proteins Associated with the Photosynthetic Apparatus Do Not Accumulate in D. glomerata Nodules
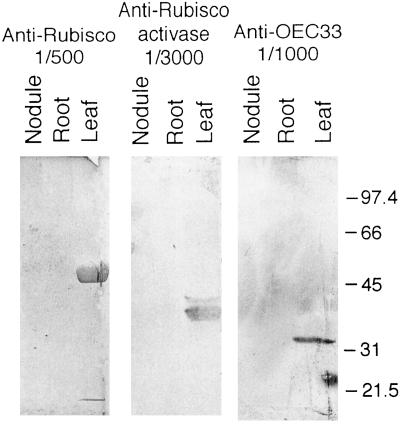
We performed western analyses to determine whether proteins of Rubisco activase, Rubisco large subunit, and the OEC33 were present in nodule extracts. All three proteins were detected in leaf extracts, but not in nodule or root extracts (Fig. 6). With polyclonal antibodies made to soybean Rubisco large subunit, a strong protein band was seen in leaf extracts at 50 to 60 kD (Fig. 6, left panel), representing the native form of the protein. We detected a protein of 40 kD in D. glomerata leaf extracts with antibodies made to tobacco Rubisco activase (Fig. 6, middle panel). This band was similar in size to Rubisco activase small polypeptides from other plant species (41–44 kD). Antibodies made to OEC33 from pea detected a 33-kD protein in the leaves of D. glomerata (Fig. 6, right panel).
Figure 6.
Western analyses for the Rubisco large subunit (left), Rubisco activase (middle), and OEC33 (right). Ten microliters of extracts of D. glomerata nodule, root, and leaf protein was partitioned on SDS-PAGE before reaction with antibodies (diluted as indicated). For Rubisco, 17, 2.0, and 59 μg of protein from nodule, root, and leaf were used, respectively; for Rubisco activase and OEC33, 32, 33, and 91 μg of protein from nodule, root, and leaf were used, respectively. Protein standards (in kilodaltons) are shown at the right.
DISCUSSION
We identified an mRNA encoding Rubisco activase in symbiotic root nodules of D. glomerata. Similarities between Dgrca and Rubisco activases from other organisms were evident in comparisons of their nucleotide and amino acid sequences, mRNA expression patterns in various organs, sizes of the gene families, and positions and sizes of three introns. Rubisco activase is conserved in photosynthetic organisms across a wide range of genera, including lower eukaryotes (Roesler and Ogren, 1990; Li et al., 1993), and has no reported alternative functions. A Rubisco activase-like GA-binding protein has been identified (Komatsu et al., 1996), but the deduced amino acid sequence of this gene shares limited identity with that of Dgrca, as well as with Rubisco activases from other species.
Rubisco activase functions as a heterodimer in most higher plants, with the exception of a monomeric form in maize (Salvucci et al., 1987). Large and small isoforms of Rubisco activase reported in Arabidopsis, spinach (Wernecke et al., 1989), and barley (Rundle and Zielinski, 1991) arise from alternative splicing of a single mRNA. In other species such as tobacco, the polypeptides are encoded by separate genes (Qian and Rodermel, 1993). In our RNA blots we detected two Dgrca transcripts expressed in D. glomerata nodules, a full-length, mature Dgrca mRNA of 1700 nucleotides, and an unusually large (3000 nucleotides), abundant transcript. The 1300-nucleotide size differential between these mRNAs was much greater than the difference of approximately 300 nucleotides that results from alternative splicing in other species. In addition, five separate Dgrca cDNA clones that were examined had identical 3′ sequences. Therefore, we found no evidence from either northern blots or from limited nucleotide sequence data to indicate that the alternative splicing of Rubisco activase mRNA observed in other species is occurring for Dgrca in D. glomerata nodules. Because Dgrca appears to be a single-copy gene, the 3000-nucleotide species is not likely to arise from transcription of a second gene.
Hybridization of the large transcript to intronic DNA suggested either that it was a form of heteronuclear RNA or that it represented an anomalous, incompletely spliced message. This possibility was supported by in situ localization of the Dgrca RNA primarily to nuclei of infected cells. Because the D. glomerata nodule is indeterminate, a developmental gradient from meristem to N2-fixing infected cells is present in the mature nodule, at least until 6 weeks after inoculation. Both mRNA species were observed in nodules up to 11 weeks postinoculation. The relatively low level of hybridization to the intron-A probe in RNA blots and the absence of intron-A hybridization in in situ experiments was attributed to its relatively short length (96 bp of intron A-specific sequence) and its AT-rich nature. Nucleotide sequence data of a 3-kb cDNA clone would reveal the additional sequences accounting for the larger transcript, but our attempts to obtain such a clone using random amplification of cDNA 3′ ends were unsuccessful. We do not know whether the 3000-nucleotide Dgrca transcript represents a novel form of alternatively spliced mRNA or whether it is translated. If it is translated, it must give rise to a protein that is not detected by the Rubisco activase antiserum used in our experiments.
The 3000-nucleotide RNA species was recovered from an oligo(dT)-cellulose column, indicating that at least a portion of this size population was polyadenylated. The ratio of the 3000-nucleotide species to the 1700-nucleotide species decreased from 5:1 in total nodule RNA preparations to about 1:1 in the nodule poly(A+) fraction, suggesting either that a significant portion of the 3000-nucleotide species did not have a poly(A+) tail and was labile, or that the 3000-nucleotide poly(A+) RNA was unstable during fractionation. Accumulation of unspliced or incompletely spliced heteronuclear RNA has been postulated to result from changes in the organization of the nucleus or from changes in splicing efficiency associated with promoter structure (Cramer et al., 1997), with heteronuclear RNA structure (for review, see Simpson and Filipowicz, 1996), or with the availability or action of components of the spliceosome complex (e.g. the SR proteins; Cáceres et al., 1997). Because of the complexity of the splicing apparatus and the splicing process, we have not pursued splicing as the biological basis for the presence of the 3000-nucleotide RNA.
Retention of transcripts within the nucleus has been reported for specific genes in a variety of organisms. Early expression of a male germ line-specific gene, Mst40, in spermatocytes of third larval instars of Drosophila melanogaster is associated with the nucleus; Mst40 mRNA was detected later in the cytoplasm (Russell and Kaiser, 1994). The D. melanogaster Hsr-omega gene, located in a heat-shock puff, undergoes alternative transcriptional termination in response to heat shock and other stimuli, generating a longer, nuclear-localized form of the transcript, which is polyadenylated and in which the introns are retained (Hogan et al., 1994). The authors suggest that the nuclear transcript may interact with proteins or have a regulatory role; for Dgrca, however, this remains to be determined. The O2 (Opaque-2) transcript, encoding a bZIP transcriptional regulator of zein storage protein synthesis in maize endosperm, is normally found in both the nucleus and cytoplasm (Dolfini et al., 1992). In one mutant an intron-containing O2 transcript accumulates in the nucleus. The efficiency of splicing of the waxy transcript in rice endosperm appears to vary naturally among cultivars, some of which accumulate a large pre-mRNA containing an intron (Wang et al., 1995). As expected, the titers of granule-bound starch synthase (Waxy protein) correlate directly with the level of mature waxy mRNA and inversely with that of the pre-mRNA. Genetic data indicate that splicing of the waxy intron is governed by cis-acting elements. It is curious that all of the intron-containing transcripts described above occur in organs or cells that are undergoing endoreduplication and, hence, share a common feature with the mature infected cells of D. glomerata nodules. Our findings suggest that the D. glomerata symbiotic nodule provides the signals or trans-acting factors necessary for transcription of Dgrca. However, other molecular components that are needed for efficient processing of the Dgrca transcript or for its transport from the nucleus do not appear to be functional or present in appropriate amounts.
The mature, 1700-nucleotide Dgrca mRNA is expected to be translatable. Our western-blot data indicate that Rubisco activase protein is present in leaves of D. glomerata but not in nodules or roots. The absence of detectable protein in nodules could result from a block in the export of mRNA to the cytoplasm, from a block in translation of the mRNA, and/or from rapid protein turnover. Low amounts of protein in tissues showing high levels of the corresponding transcript have been reported for a number of genes, including fbp1, a MADS-box transcription factor in stamens of wild-type petunia (Angenent et al., 1992); Sh1, which encodes an isoform of Suc synthase in maize embryos (Chourey and Taliercio, 1994); and an mRNA for S-adenosyl Met synthetase in poplar (Mijnsbrugge et al., 1996). In the case of an mRNA encoding the small subunit of ADP-Glc pyrophosphorylase, relatively low levels of the small subunit of ADP-Glc pyrophosphorylase in potato leaves are attributed to its instability in the absence of the large subunit (Nakata and Okita, 1996). Arrest of translation can also occur in response to environmental conditions, as observed for a mitochondrial mRNA-encoding adenine nucleotide translocator in young maize roots after O2 deprivation (Fennoy and Bailey-Serres, 1995).
In some species rbcL mRNA accumulates in etioplasts before greening (Tobin and Silvethorne, 1985; Berry et al., 1990); thus its presence in plastids of nonphotosynthetic organs is not without precedent. The localization of Dgrca transcripts coincided with that of the rbcL mRNA, but the former were most abundant in the nuclei of cells of the early infection zone, whereas the latter was abundant in the cytoplasm of mature, vesicle-containing cells, typically those expressing nitrogenase, leghemoglobin, Gln synthetase, and other proteins involved in N2 and C assimilation and O2 partitioning. Dgrca expression was also localized at low levels to the cytoplasm of uninfected cortical cells adjacent to the periderm and to cells surrounding the vascular cylinder. Differential expression patterns for Dgrca and DgrbcL can be expected, because these genes are transcribed in different subcellular compartments and are independently regulated in other developmental systems (Reski, 1994). The observed expression patterns may therefore reflect differences in the induction of nuclear- and plastid-coded genes or in plastid number and distribution within the nodule.
Nodule expression of Dgrca deviated from the organ-specific, light-regulated expression for rca genes observed in leaves of spinach (Orozco and Ogren, 1993) and in Arabidopsis (Liu et al., 1996). Dgrca nodule expression also deviated from the coordinate expression of other photosynthetic genes during photomorphogenesis and diurnal fluxes in photosynthetic capacity. White light treatment had no significant effect on the overall abundance of Dgrca mRNA or on the relative abundance of the two size classes of Dgrca transcripts in symbiotic root nodules. Circadian regulation of rca genes such as that observed in tomato (Martino-Catt and Ort, 1992) and Arabidopsis (Liu et al., 1996) has not yet been examined in Dgrca nodules. Because neither Rubisco activase nor Rubisco proteins accumulated to detectable levels in D. glomerata nodules, a role for these proteins in photosynthetic C reduction, oxygenase activity, or N2 accumulation in the nodule is unlikely. In stem nodules of Sesbania rostrata, photosynthetically competent chloroplasts were observed in cortical cells adjacent to N2-fixing infected cells, suggesting that they may have a role in C assimilation (James et al., 1996). In D. glomerata nodules, however, the absence of OEC33, a protein associated with the O2-evolving complex of the photosynthetic apparatus, suggests that Dgrca expression is not accompanied by the assembly of a functional photosynthetic apparatus or by additional chloroplast development.
The nodule greening and autofluorescence attributed to chlorophyll that we have observed in some mature D. glomerata nodules (P.A. Okubara and A.M. Berry, unpublished data) occurs much later after Frankia inoculation than does Dgrca mRNA expression. Although the relative abundance of 3000-nucleotide mRNA was not altered significantly by white light treatment, this treatment was sufficient to cause faint greening in the mature nodules closest to the light source (data not shown). We therefore postulate that the greening represents a distinct phenomenon not necessarily related to the induction of Dgrca transcription. The greening of organs derived from roots seems anomalous, yet development of functional chloroplasts in the pith cells of young poplar twigs (van Cleve et al., 1993) has been reported. Such cases of chloroplast differentiation may reflect the recruitment of stem-like traits during the development or evolution of certain organs, including the D. glomerata nodule. Greening has also been observed in some multinucleated giant cells derived from tomato roots after they have been infected with root knot nematodes from the genus Meloidogyne (V. Williamson, personal communication). The differentiation of certain root knots appears to be accompanied by chlorophyll accumulation, presumably through the ectopic expression of photosynthesis-associated mRNAs. The molecular or cellular basis for the unusual accumulation of Dgrca transcripts in nodules may well be revealed as progress is made in understanding the extensive cellular activity, strong induction of N2 metabolic pathways, and other phenomena that characterize nodule development.
The accession numbers for the sequences of Dgrca mRNA and the 1.2-kb Dgrca gene segment reported in this article are AF047352 and AF052424, respectively.
ACKNOWLEDGMENTS
The authors thank David Neale (U.S. Department of Agriculture Forest Service, Pacific Southwest, PWS, Davis, CA) for advice and use of facilities in construction of the first D. glomerata cDNA library; David Gilchrist, Chris Mau, and other members of the Center for Engineering Plants for Resistance Against Pathogens, Davis, CA, for use of the facilities for construction of the second cDNA library, for autoradiography, and for sequence alignments; Rich Jorgensen (University of California, Davis) for the use of his PCR facilities; Dean Lavelle (University of California, Davis) for diligent and expert sequencing of the Dgrca full-length cDNA and the Dgrca gene segment; Tony van Kampen (Wageningen Agricultural University) for sequencing of Dgrca clones; Michael E. Salvucci for the gift of Rubisco activase antibodies and for insightful discussions; Steve Theg for the gift of OEC33 antibodies; and Susan Swensen and Beth Mullin (University of Tennessee, Knoxville) for pDgRcbL.
Abbreviation:
- OEC33
oxygen-evolving complex 33-kD subunit
Footnotes
This work was supported by the California Agricultural Experiment Station (project no. 6258 to A.M.B.) and by a Katherine Esau Fellowship from the University of California, Davis, to K.P.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman JD. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Angenent GC, Busscher M, Franken J, Mol JNM, van Tunen AJ. Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell. 1992;4:983–993. doi: 10.1105/tpc.4.8.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DR, Silvester WB. Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol Rev. 1993;57:293–319. doi: 10.1128/mr.57.2.293-319.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JO, Breiding DE, Klessig DF. Light-mediated control of translational initiation of ribulose-1,5-bisphosphate carboxylase in amaranth cotyledons. Plant Cell. 1990;2:795–803. doi: 10.1105/tpc.2.8.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Misteli T, Screaton GR, Spector DL, Kariner AR . Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–237. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS, Taliercio EW. Epistatic interaction and functional compensation between two tissue- and cell-specific sucrose synthase genes in maize. Proc Natl Acad Sci USA. 1994;91:7917–7921. doi: 10.1073/pnas.91.17.7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson C. An anatomical and morphological study of Datiscaceae. ALISO. 1973;8:49–110. [Google Scholar]
- Dickey LF, Nguyen T-T, Allen GC, Thompson WF. Light modulation of ferredoxin mRNA abundance requires an open reading frame. Plant Cell. 1994;6:1171–1176. doi: 10.1105/tpc.6.8.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini SF, Landoni M, Tonelli C, Bernard L, Viotti A. Spatial regulation in the expression of structural and regulatory storage-protein genes in Zea mays endosperm. Dev Genet. 1992;13:264–276. [Google Scholar]
- Fennoy SL, Bailey-Serres J. Posttranscriptional regulation of gene expression in oxygen-deprived roots of maize. Plant J. 1995;7:287–295. doi: 10.1046/j.1365-313X.1998.00249.x. [DOI] [PubMed] [Google Scholar]
- Gherbi H, Duhoux E, Franche C, Pawlowski K, Nassar A, Berry AM, Bogusz D. Cloning of a full-length symbiotic hemoglobin cDNA and in situ localization of the corresponding mRNA in Casuarina glauca root nodule. Physiol Plant. 1997;99:608–616. [Google Scholar]
- Goetting-Minesky MP, Mullin BC. Differential gene expression in an actinorhizal symbiosis: evidence for a nodule-specific cysteine protease. Proc Natl Acad Sci USA. 1994;91:9891–9895. doi: 10.1073/pnas.91.21.9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C, Akkermans ADL, van Kammen A, Bissleing T, Pawlowski K. ag13 is expressed in Alnus glutinosa nodules in infected cells during endosymbiont degradation and in the nodule pericycle. Physiol Plant. 1997;99:601–607. [Google Scholar]
- Hafeez F, Akkermans ADL, Chaudhary AH. Observations on the ultrastructure of Frankia sp. in root nodules of Datisca cannabina L. Plant Soil. 1984;79:383–402. [Google Scholar]
- Hirsch AM, LaRue TA. Is the legume nodule a modified root or stem or an organ sui generis? Crit Rev Plant Sci. 1997;16:361–392. [Google Scholar]
- Hogan NC, Traverse KL, Sullivan DE, Pardue M-L. The nucleus-limited Hsr-omega-n transcript is a polyadenylated RNA with a regulated intranuclear turnover. J Cell Biol. 1994;125:21–30. doi: 10.1083/jcb.125.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen-Lyon K, Jensen EO, Joergensen J-E, Marcker KA, Peacock WJ, Dennis ES. Symbiotic and nonsymbiotic hemoglobin genes of Casuarina glauca. Plant Cell. 1995;7:213–223. doi: 10.1105/tpc.7.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James EK, Iannetta PPM, Nixon PJ, Whiston AJ, Peat L, Crawford RMM, Sprent JJ, Brewin NJ. Photosystem II and oxygen regulation in Sesbania rostrata stem nodules. Plant Cell Environ. 1996;19:895–910. [Google Scholar]
- Komatsu S, Masuda T, Hirano H. Rice gibberellin-binding phosphoprotein structurally related to ribulose-1,5-bisphosphate carboxylase/oxygenase activase. FEBS Lett. 1996;384:167–171. doi: 10.1016/0014-5793(96)00275-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lan Y, Mott KA. Determination of apparent Km values for ribulose 1,5-bisphosphate carboxylase oxygenase (Rubisco) activase using the spectrophotometric assay of Rubisco activity. Plant Physiol. 1991;95:604–609. doi: 10.1104/pp.95.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LA, Gibson JL, Tabita FR. The rubisco activase (rca) gene is located downstream from rbcS in Anabaena sp. strain CA and is detected in other Anabaena Nostoc strains. Plant Mol Biol. 1993;21:753–764. doi: 10.1007/BF00027109. [DOI] [PubMed] [Google Scholar]
- Liston A, Reiseberg AH, Elias TS. Morphological stasis and molecular divergence in the intercontinental disjunct genus Datisca (Datiscaceae) ALISO. 1989;12:525–542. [Google Scholar]
- Liu Q, Berry AM. The infection process and nodule initiation in the Frankia-Ceanothus root nodule symbiosis. Protoplasma. 1991;163:82–92. [Google Scholar]
- Liu Z, Taub CC, McClung CR. Identification of an Arabidopsis thaliana ribulose-1,5-bisphosphate carboxylase/oxygenase activase (RCA) minimal promoter regulated by light and the circadian clock. Plant Physiol. 1996;112:43–51. doi: 10.1104/pp.112.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino-Catt S, Ort DR. Low temperature interrupts circadian regulation of transcriptional activity in chilling-sensitive plants. Proc Natl Acad Sci USA. 1992;89:3731–3735. doi: 10.1073/pnas.89.9.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijnsbrugge KV, Van Montagu M, Inzé D, Boerjan W. Tissue-specific expression conferred by the S-adenosyl-l-methionine synthetase promoter of Arabidopsis thaliana in transgenic poplar. Plant Cell. 1996;37:1108–1115. doi: 10.1093/oxfordjournals.pcp.a029061. [DOI] [PubMed] [Google Scholar]
- Murphy TM. Immunochemical comparisons of ribulosebisphosphate carboxylases using antisera to tobacco and spinach enzymes. Phytochemistry. 1978;17:439–443. [Google Scholar]
- Nakata PA, Okita TW. Cis-elements important for the expression of the ADP-glucose pyrophosphorylase small-subunit are located both upstream and downstream from its structural gene. Mol Gen Genet. 1996;250:581–592. doi: 10.1007/BF02174446. [DOI] [PubMed] [Google Scholar]
- Orozco BM, McClung CR, Wernecke JM, Ogren WL. Molecular basis of the ribulose-1,5-bisphosphate carboxylase/ oxygenase activase mutation in Arabidopsis thaliana is a guanine-to-adenine transition at the 5′-splice junction of intron 3. Plant Physiol. 1993;102:227–232. doi: 10.1104/pp.102.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco BM, Ogren WL. Localization of light-inducible and tissue-specific regions of the spinach ribulose bisphosphate carboxylase oxygenase (Rubisco) activase promoter in transgenic tobacco plants. Plant Mol Biol. 1993;23:1129–1138. doi: 10.1007/BF00042347. [DOI] [PubMed] [Google Scholar]
- Pawlowski K, Bisseling T. Rhizobial and actinorhizal symbioses: what are the shared features? Plant Cell. 1996;8:1899–1913. doi: 10.1105/tpc.8.10.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski K, Kunze R, de Vries S, Bisseling T. Isolation of total, poly(A) and polysomal RNA from plant tissues. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual D5, Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–13. [Google Scholar]
- Pawlowski K, Ribeiro A, Bisseling T. Nitrogen fixing root nodule symbioses: legume nodules and actinorhizal nodules. In: El-Gewely MR, editor. Biotechnology Annual Review. Amsterdam, The Netherlands: Elsevier Science Publishers; 1996. pp. 151–184. [Google Scholar]
- Pawlowski K, Ribeiro A, Guan C-H, van Kammen AB, Akkermans A, Bisseling T (1993) Differential gene expression in root nodules of Alnus glutinosa. In NA Hegazi, M Fayez, M Monib, eds, Nitrogen Fixation with Non-Legumes. American University in Cairo Press, Cairo, Egypt, pp 185–190
- Pawlowski K, Twigg P, Dobritsa S, Guan CH, Mullin BC. A nodule-specific gene family from Alnus glutinosa encodes glycine- and histidine-rich proteins expressed in the early stages of actinorhizal nodule development. Mol Plant-Microbe Interact. 1997;10:656–664. doi: 10.1094/MPMI.1997.10.5.656. [DOI] [PubMed] [Google Scholar]
- Portis AR. Rubisco activase. Biochim Biophys Acta. 1990;1015:15–28. doi: 10.1016/0005-2728(90)90211-l. [DOI] [PubMed] [Google Scholar]
- Qian J, Rodermel S. Ribulose-1,5-bisphosphate carboxylase/oxygenase activase cDNAs from Nicotiana tabacum. Plant Physiol. 1993;102:683–684. doi: 10.1104/pp.102.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reski R. Plastid genes and chloroplast biogenesis. In: Mok DWS, Mok MC, editors. Cytokinins Chemistry, Activity, and Function. Boca Raton, FL: CRC Press; 1994. pp. 179–195. [Google Scholar]
- Ribeiro A, Akkermanns ADL, van Kammen A, Bisseling T, Pawlowski K. A nodule-specific gene encoding a subtilisin-like protease is expressed in early stages of actinorhizal nodule development. Plant Cell. 1995;7:785–794. doi: 10.1105/tpc.7.6.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A, Praekelt U, Akkermans ADL, Meacock PA, van Kammen A, Bisseling T, Pawlowski K. Identification of agthi1, whose product is involved in biosynthesis of the thiamine precursor thiazole, in actinorhizal nodules of Alnus glutinosa. Plant J. 1996;10:361–368. doi: 10.1046/j.1365-313x.1996.10020361.x. [DOI] [PubMed] [Google Scholar]
- Roesler KR, Ogren WL. Primary structure of Chlamydomonas reinhardtii ribulose 1,5-bisphosphate carboxylase/oxygenase activase and evidence for a single polypeptide. Plant Physiol. 1990;94:1837–1841. doi: 10.1104/pp.94.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle SJ, Zielinski RE. Organization and expression of two tandemly oriented genes encoding ribulosebisphosphate carboxylase/oxygenase activase in barley. J Biol Chem. 1991;266:4677–4685. [PubMed] [Google Scholar]
- Russell SRH, Kaiser K. A Drosophila melanogaster chromosome 2L repeat is expressed in the male germ line. Chromosoma. 1994;103:63–72. doi: 10.1007/BF00364727. [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Wernecke JM, Ogren WL, Portis AR. Purification and species distribution of Rubisco activase. Plant Physiol. 1987;84:930–936. doi: 10.1104/pp.84.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Simpson GG, Filipowicz W. Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organization of the spliceosomal machinery. Plant Mol Biol. 1996;32:1–41. doi: 10.1007/BF00039375. [DOI] [PubMed] [Google Scholar]
- Swensen SM, Mullin BC, Chase MW. Phylogenetic affinities of Datiscaceae based on an analysis of nucleotide sequences from the plastid rbcL gene. Syst Bot. 1994;19:157–168. [Google Scholar]
- Tobin EM, Silverthorne J. Light regulation of gene expression in higher plants. Annu Rev Plant Physiol. 1985;36:569–593. [Google Scholar]
- van Cleve B, Forreiter C, Sauter JJ, Apel K. Pith cells of poplar contain photosynthetically active chloroplasts. Planta. 1993;189:70–73. [Google Scholar]
- van de Wiel C, Scheres B, Franssen H, van Lierop M-J, Van Lammeren A, Van Kammen A, Bisseling T. The early nodulin transcript ENOD2 is located in the nodule parenchyma (inner cortex) of pea and soybean root nodules. EMBO J. 1990;9:1–7. doi: 10.1002/j.1460-2075.1990.tb08073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Zheng F-Q, Shen G-Z, Gao J-P, Snustad DP, Li M-G, Zhang J-L, Hong M-M. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995;7:613–622. doi: 10.1046/j.1365-313x.1995.7040613.x. [DOI] [PubMed] [Google Scholar]
- Watillon B, Kettmann R, Boxus P, Burny A. Developmental and circadian pattern of Rubisco activase messenger RNA accumulation in apple plants. Plant Mol Biol. 1993;23:501–509. doi: 10.1007/BF00019298. [DOI] [PubMed] [Google Scholar]
- Wernecke JM, Chatfield JM, Ogren WL. Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell. 1989;1:815–825. doi: 10.1105/tpc.1.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernecke JM, Ogren WL. Structure of an Arabidopsis thaliana cDNA encoding Rubisco activase. Nucleic Acids Res. 1989;17:2871. doi: 10.1093/nar/17.7.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]