Abstract
Taxonomy plays a central role in biological sciences. It provides a communication system for scientists as it aims to enable correct identification of the studied organisms. As a consequence, species descriptions should seek to include as much available information as possible at species level to follow an integrative concept of ‘taxonomics’. Here, we describe the cryptic species Epimeria frankei sp. nov. from the North Sea, and also redescribe its sister species, Epimeria cornigera. The morphological information obtained is substantiated by DNA barcodes and complete nuclear 18S rRNA gene sequences. In addition, we provide, for the first time, full mitochondrial genome data as part of a metazoan species description for a holotype, as well as the neotype. This study represents the first successful implementation of the recently proposed concept of taxonomics, using data from high-throughput technologies for integrative taxonomic studies, allowing the highest level of confidence for both biodiversity and ecological research.
Introduction
The biological discipline of taxonomy formally describes, classifies and names extant and extinct species1. It is therefore of central importance within the biological sciences, providing a communication system for scientists of all biological disciplines2. This is of pivotal importance as results and conclusions of studies that rely on incorrect identification and/or classification of the analysed organisms are highly questionable3. Despite all efforts in the description of organisms over the past centuries, many species remain to be discovered, classified, described, and named. It has been estimated that 3 to 100 million species exist on Earth4–7 with the most recent suggestions narrowing the extant diversity of life to approx. 8.7 million species8. These numbers clearly give the impression that still only a small fraction of all species on Earth (about 2 millions, with a rate of approx. 20,000 new species descriptions per year; see http://www.esf.edu/species/) have been scientifically described so far8.
Traditionally, morphological characteristics such as size, shape, colour, and anatomical structures are used to classify and describe species. With the rise of molecular biology in the middle of the 20th century, various new technologies have become available for taxonomic studies, such as the analysis of size, shape and the number of chromosomes9 or allozyme and isozyme electrophoretic analyses10,11. The development of the polymerase chain reaction (PCR) offered a broad variety of methods for sequence analyses of DNA12, including the study of random amplified polymorphic DNA fragments (RAPDs)13–15 and PCR-restriction fragment length polymorphisms (PCR-RFLPs)16,17. However, the continuous improvement of sequencing technologies and their decreasing costs promoted the application of nucleotide sequences in taxonomy. Various nuclear and particularly mitochondrial markers are commonly in use today. Due to their faster evolutionary rates (compared to most nuclear genes), mitochondrial protein-coding genes, and here in particular the cytochrome c oxidase subunit I (CO1), are regarded as powerful taxonomic markers for genetic diversity analysis at less inclusive levels, including families, genera and species18. Especially the 5′ end of CO1 has become extremely popular as it is flanked by two “universal” primer regions which allow successful amplification of a broad range of metazoans19–21. Therefore, this approximately 650 base-pair fragment was proposed as global standard for species identification - the so-called “barcode region” for animals22,23. Although different issues potentially affect the applicability of DNA barcodes and mitochondrial DNA in general for species identification (e.g., hybridizations, incomplete lineage sorting or the presence of mitochondrial pseudogenes)24, many studies successfully demonstrated its usefulness to delineate and identify species across a broad range of taxa25–30. Consequently, many recently published species descriptions include DNA barcode sequence data31–34. The application of high-throughput sequencing technologies allows even more detailed characterizations of species, particularly by incorporating complete genome sequence data35, as it has already been successfully implemented for a number of Bacteria and Archaea36–38.
Due to their high taxonomic diversity, ecological significance and many different life styles across aquatic and terrestrial environments, amphipod crustaceans represent excellent model organisms to study speciation and radiation of organisms. To date, more than 10,000 amphipod species have been described39, but various taxonomic uncertainties still remain, for example in the species-rich genus Gammarus40–42, the Tryphosa-group43, or in the subterranean genus Niphargus44. Additionally, recent studies indicate that species diversity of the Amphipoda is even larger than it had been expected based on the use of “traditional” morphological methods alone45–47.
Here we describe a cryptic species in the amphipod genus Epimeria from the North Sea, Epimeria frankei sp. nov. and also redescribe its sister species, Epimeria cornigera (Fabricius, 1779). Both descriptions include comprehensive morphological information, DNA barcodes, and complete nuclear 18S rRNA gene sequences. In addition, we provide full mitochondrial genome information for the first time for a holotype and a neotype within the Metazoa. This previously unmatched level of information for a taxonomic description represents the first implementation of the recently proposed concept of taxonomics, using comprehensive data based on modern molecular technologies for taxonomic studies35,48.
Results
DNA barcodes
A total number of 86 DNA barcodes of Epimeria cornigera (n = 23), Epimeria frankei (n = 57), and Gammarus locusta (n = 6) were analysed. For the analysed specimens of the genus Epimeria, the barcode fragment length ranged from a minimum of 616 to the full fragment length of 658 bp, including four barcode fragments with intermediate lengths (n = 4, 4.9%). The mean nucleotide frequencies were A = 0.29, C = 0.21, G = 0.16, and T = 0.34 for Epimeria cornigera, and A = 0.28, C = 0.22, G = 0.16, and T = 0.34 for Epimeria frankei. Intraspecific K2P distances ranged from 0% to 0.65% for Epimeria cornigera and 0% to 3.16% for Epimeria frankei. Interspecific distances between both species had values from 8.69% to 11.81% (Table 1). Epimeria cornigera had one unique BIN (AAU2062), whereas two BINs were identified for Epimeria frankei (AAU2061, ACS7228) (Table 1).
Table 1.
K2P distances between Epimeria cornigera, Epimeria frankei sp. nov., and Gammarus locusta.
| Species | Mean ISD | Max. ISD | n | BIN | Nearest Species | Distance to NN |
|---|---|---|---|---|---|---|
| Epimeria cornigera (Fabricius, 1779) | 0.14 | 0.65 | 23 | AAU2062 | Epimeria frankei | 8.69 |
| Epimeria frankei sp. nov. | 1.06 | 3.15 | 57 | AAU2061, ACS7228 | Epimeria cornigera | 8.69 |
| Gammarus locusta (Linnaeus, 1758) | 0.6 | 1.08 | 6 | AAC0402 | Epimeria frankei | 31.06 |
Divergence values were calculated for all sequences studied, based on the Nearest Neighbor Summary implemented in the Barcode Gap Analysis tool provided by the Barcode of Life Data System (BOLD). Align sequencing option: BOLD aligner (amino acid based HMM), ambiguous base/gap handling: pairwise deletion. ISD = intraspecific distance. BINs are based on the barcode analysis from 15-02-2017.
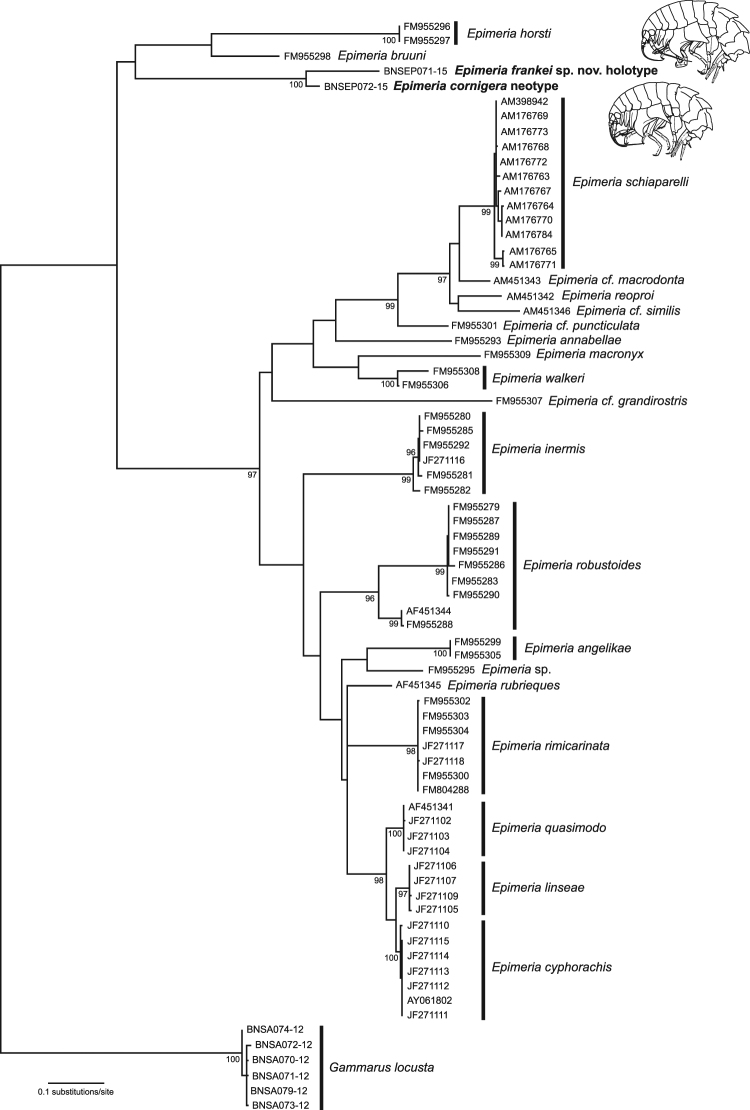
The NJ analyses based on K2P distances showed distinct clusters with bootstrap support values of 99% for all three species (Fig. 1). In the case of Epimeria frankei, our analysis revealed two distinct lineages with a minimum K2P distance of 2.17% and each corresponding to a distinct BIN.
Figure 1.
Neighbor-joining phylogenetic tree of the analysed COI sequences of Epimeria cornigera, Epimeria frankei sp. nov., and Gammarus locusta as outgroup. The optimal tree with the sum of branch length = 0.44256318 is shown. Evolutionary distances were computed using the Kimura 2-parameter model for substitution. Numbers next to internal branches are non-parametric bootstrap values (in %). Only values above 90% are shown. Asterisks indicate specimens with additional complete 18S rRNA gene sequence data. All codon positions were included. All ambiguous positions were removed for each sequence pair. There were a total of 658 positions in the final dataset.
Our phylogenetic analysis of all available CO1 sequences of Epimeria indicated Epimeria cornigera as sister taxon of Epimeria frankei with a bootstrap support of 100% (Fig. 2). Two species, Epimeria horsti Lörz 2008 and Epimeria bruuni Barnard 1969, were identified as their sister clade. However, this relationship only had low bootstrap support (40%, not shown). Furthermore, these four Epimeria species formed the sister clade of all other analysed Epimeria species.
Figure 2.
Maximum Likelihood phylogenetic tree of the partial COI sequence data set of 19 Epimeria species and Gammarus locusta as outgroup. Model of choice: HKY with a log likelihood score of −6124.0843, a discrete gamma distribution (+G, parameter = 1.0064) and a proportion of invariant sites (+I, 46% of sites). Numbers next to internal branches are non-parametric bootstrap values (in %). Only values above 90% are shown. Initial tree(s) for the heuristic search were obtained automatically by applying BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then by selecting the topology with superior log likelihood value. The tree is drawn to scale with branch lengths measured in substitutions per site. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 542 positions in the final dataset.
Small ribosomal subunit (18S rRNA gene)
Eight sequences of the complete 18S rRNA gene were successfully amplified and sequenced for the two species (Epimeria cornigera: n = 2, Epimeria frankei: n = 6). We found no insertions or deletions, resulting in a total length of 2,380 bp for both species. As a result of concerted evolution of rRNA gene clusters, we found no intragenomic or intraspecific variations. For all sequences of both species, average base frequencies were A = 0.22, C = 0.23, G = 0.28, and T = 0.27. In total, we observed 34 substitutions and a patristic distance of 2% between both species (Fig. 3; Table S1). Most substitutions (82%) were located in variable regions, with V1 = 1 substitution, V2 = 5 substitutions, V4 = 14, and V7 = 8 (Table S1).
Figure 3.
Unrooted Neighbour-joining phylogram of the complete 18S rRNA genes of Epimeria cornigera, Epimeria frankei sp. nov., and Gammarus locusta based on patristic distances.
Mitochondrial genome data: mitochondrial genome reconstruction
The lack of mitogenomic data from closely related species prevented a standard approach of mapping reads to a reference. We therefore implemented independent iterative mapping approaches for both Epimeria species. We selected three amphipod bait reference sequences: Gondogeneia antarctica (Chevreux, 1906) (Genbank: JN827386.1), Metacrangonyx goulmimensis Messouli, Boutin & Coineau, 1991 (Genbank: HE860501.1), and Onisimus nanseni (Sars, 1900) (Genbank: NC_013819.1). Regardless of the employed reference, default k-mer values of 31 produced no mappable reads. We therefore carried out the reconstruction using k-mer values of 19 and 25. Both of these values allowed reads to map to our chosen bait reference sequences. However, due to the high level of rearrangements in these distantly related amphipod mitochondria, the correct orientation and therefore continuous sequence of the Epimeria mitogenomes could not be recovered using a k-mer value of 19. This k-mer value resulted in multiple mapping initiation points along the reference, leading to a complete, yet fragmented sequence. In contrast, a k-mer value of 25 produced only a single initial mapping point on all three chosen bait reference sequences, allowing a single continuous sequence to be reconstructed and thereby overcoming the above mentioned problems caused by rearrangements.
Regardless of the bait reference sequence used, the final consensus was identical, providing additional reliability to the sequenced genomes. Automated annotations showed the presence of all expected tRNAs, rRNAs and coding genes in an invertebrate mitochondrial genome. Although iterative mapping and consensus sequence reconstruction were performed independently on each species’ genome, annotations showed very similar gene locations and lengths, providing additional support to our approach and final consensus sequences.
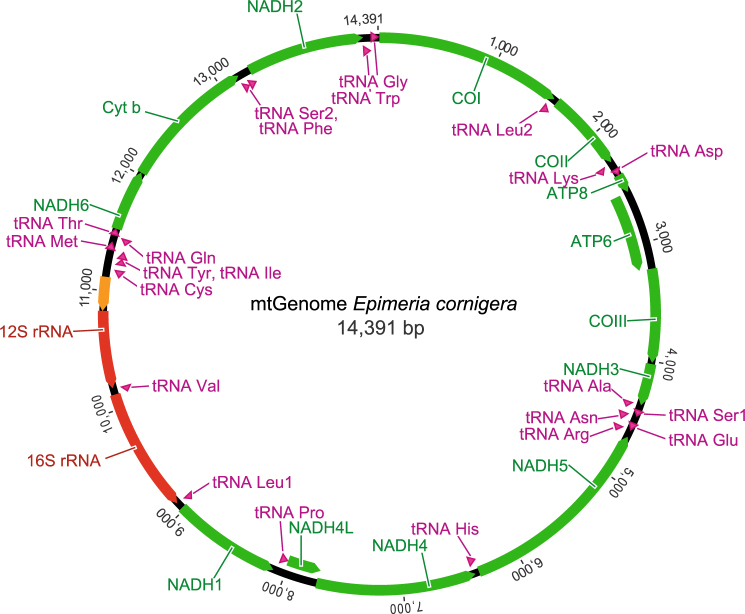
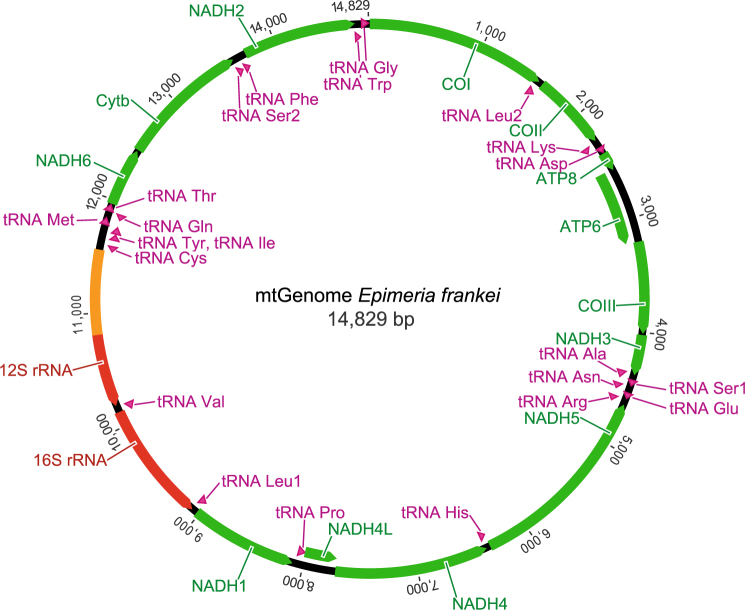
The complete mitochondrial genomes of Epimeria cornigera (14,391 bp) and Epimeria frankei (14,829 bp) are typical metazoan mitogenomes. Both consist of a single circular DNA molecule carrying 13 protein-coding genes, two subunits of the mitochondrial ribosomal RNA, 22 tRNAs, one for each amino acid except leucine and serine, which have two copies each, and a putative control region (Figs 4 and 5; Table 2). The heavy strands (H-strand) encode 25 genes (protein coding genes: 9, tRNAs: 16), whereas the remaining 12 genes (protein coding genes: 4, tRNAs: 6, rRNAs: 2) are on the light strands (L-strand). The gene organization of both genomes is similar to the supposed pancrustacean basic pattern49 except for the location of various tRNAs and a translocation of a fragment containing two protein-coding genes and one tRNA (NADH6, Cyt b, tRNASer2). There are no gene rearrangements between the two Epimeria species investigated in this study. Twenty three gene overlaps and 10 intergenic spacers were found for both mitogenomes. For Epimeria cornigera, the total length of overlaps is also 110 bp (ranging from 1 to 26 bp), whereas intergenetic spacers have values from 1 to 44 bp, resulting in a total length of 92 bp, respectively. For Epimeria frankei, the total lengths of overlaps are 110 bp, ranging from 1 to 26 bp, and 89 bp (1 to 44 bp) for intergenetic spacers. We found similar nucleotide compositions for both genomes, with frequencies of A = 0.36, C = 0.20, G = 0.10, and T = 0.34 for Epimeria cornigera and A = 0.35, C = 0.22, G = 0.10, and T = 0.33 for Epimeria frankei.
Figure 4.
Organization of the mitochondrial genome of Epimeria cornigera. Protein encoding genes are marked in green, ribosomal RNAs in red, tRNAs in violet, and the putative control region in orange. Abbreviations: ATP6/8: ATPase subunits 6/8; COI-III: Cytochrome c oxidase subunits I–III; NADH2-6/4 L: NADH dehydrogenase subunits 1–6/4 L, and Cyt b: Cytochrome b. All 22 transfer RNA genes are designated by tRNA-X and labeled according to the IUPAC-IUB single-letter amino acid codes. Arrowheads indicate the direction of transcription.
Figure 5.
Organization of the mitochondrial genome of Epimeria frankei sp. nov. Protein encoding genes are marked in green, ribosomal RNAs in red, tRNAs in violet, and the putative control region in orange. Abbreviations: ATP6/8: ATPase subunits 6/8; COI-III: Cytochrome c oxidase subunits I–III; NADH2-6/4 L: NADH dehydrogenase subunits 1–6/4 L, and Cyt b: Cytochrome b. All 22 transfer RNA genes are designated by tRNA-X and labeled according to the IUPAC-IUB single-letter amino acid codes. Arrowheads indicate the direction of transcription.
Table 2.
Gene structures of the mitochondrial genomes of Epimeria cornigera and Epimeria frankei sp. nov.
| Gene | Strand | Epimeria cornigera | Epimeria frankei sp. nov. | p-distance | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | Stop | Length (bp) | Amino acids | Intergenic§ | Start | Stop | Length (bp) | Amino acids | Intergenic§ | |||
| COI | H | 1 | 1563 | 1563 | 521 | −26 | 1 | 1563 | 1563 | 521 | −26 | 0.12 |
| tRNALeu2 | H | 1538 | 1596 | 59 | 0 | 1538 | 1596 | 59 | 0 | |||
| COII | H | 1597 | 2274 | 678 | 226 | 1 | 1597 | 2274 | 678 | 226 | 1 | 0.12 |
| tRNALys | H | 2276 | 2336 | 61 | −1 | 2276 | 2336 | 61 | −1 | |||
| tRNAAsp | H | 2336 | 2396 | 61 | 0 | 2336 | 2396 | 61 | 0 | |||
| ATP8 | H | 2397 | 2555 | 159 | 53 | −7 | 2397 | 2555 | 159 | 53 | −7 | 0.20 |
| ATP6 | H | 2549 | 3220 | 672 | 224 | −1 | 2549 | 3220 | 672 | 224 | −1 | 0.14 |
| COIII | H | 3220 | 4008 | 789 | 263 | 9 | 3220 | 4008 | 789 | 263 | 9 | 0.12 |
| NADH3 | H | 4018 | 4368 | 351 | 117 | −3 | 4018 | 4368 | 351 | 117 | −3 | 0.15 |
| tRNAAla | H | 4366 | 4426 | 61 | −3 | 4366 | 4426 | 61 | −3 | |||
| tRNASer1 | H | 4424 | 4476 | 53 | −2 | 4424 | 4476 | 53 | −2 | |||
| tRNAAsn | H | 4475 | 4535 | 61 | −4 | 4475 | 4535 | 61 | −4 | |||
| tRNAGlu | H | 4532 | 4594 | 63 | −3 | 4532 | 4593 | 62 | −3 | |||
| tRNAArg | H | 4592 | 4651 | 60 | −4 | 4591 | 4650 | 60 | −4 | |||
| NADH5 | L | 4648 | 6351 | 1704 | 568 | −3 | 4647 | 6350 | 1704 | 568 | −3 | 0.15 |
| tRNAHis | L | 6349 | 6408 | 60 | −20 | 6348 | 6407 | 60 | −20 | |||
| NADH4 | L | 6389 | 7723 | 1335 | 445 | −7 | 6388 | 7722 | 1335 | 445 | −7 | 0.13 |
| NADH4L | L | 7717 | 8013 | 297 | 99 | 2 | 7716 | 8012 | 297 | 99 | 2 | 0.18 |
| tRNAPro | L | 8016 | 8076 | 61 | 21 | 8015 | 8075 | 61 | 21 | |||
| NADH1 | L | 8098 | 9024 | 927 | 309 | −4 | 8097 | 9023 | 927 | 309 | −4 | 0.14 |
| tRNALeu1 | L | 9021 | 9080 | 60 | −6 | 9020 | 9080 | 61 | −6 | |||
| 16S rRNA | L | 9075 | 10123 | 1049 | −1 | 9075 | 10120 | 1046 | −1 | 0.08 | ||
| tRNAVal | L | 10123 | 10182 | 60 | −1 | 10120 | 10179 | 60 | −1 | |||
| 12S rRNA | L | 10182 | 10820 | 639 | 0 | 10179 | 10818 | 640 | 0 | 0.07 | ||
| ORF | L | 10821 | 11114 | 294 | 0 | 10819 | 11555 | 737 | 0 | |||
| tRNACys | L | 11115 | 11175 | 61 | 44 | 11556 | 11615 | 60 | 44 | |||
| tRNATyr | H | 11220 | 11279 | 60 | 2 | 11660 | 11719 | 60 | 2 | |||
| tRNAIle | H | 11282 | 11341 | 60 | −2 | 11722 | 11781 | 60 | −2 | |||
| tRNAMet | H | 11340 | 11402 | 63 | −3 | 11780 | 11842 | 63 | −3 | |||
| tRNAGln | L | 11400 | 11458 | 59 | −2 | 11840 | 11898 | 59 | −2 | |||
| tRNAThr | H | 11457 | 11517 | 61 | 1 | 11897 | 11958 | 62 | 1 | |||
| NADH6 | H | 11519 | 12022 | 504 | 168 | 9 | 11960 | 12466 | 507 | 169 | 6 | 0.15 |
| Cytb | H | 12032 | 13174 | 1143 | 381 | −5 | 12473 | 13615 | 1143 | 381 | −5 | 0.15 |
| tRNASer2 | H | 13170 | 13220 | 51 | 2 | 13611 | 13661 | 51 | 2 | |||
| tRNAPhe | H | 13223 | 13282 | 60 | −1 | 13664 | 13723 | 60 | −1 | |||
| NADH2 | H | 13282 | 14265 | 984 | 328 | 1 | 13723 | 14703 | 981 | 327 | 1 | 0.15 |
| tRNATrp | H | 14267 | 14328 | 62 | −1 | 14705 | 14766 | 62 | −1 | |||
| tRNAGly | H | 14328 | 14391 | 64 | 0 | 14766 | 14829 | 64 | 0 | |||
The letters H (heavy) or L (light) indicate that the gene is encoded either by the H- or L-strand. §Numbers correspond to the nucleotides separating adjacent genes. Negative numbers indicate overlapping nucleotides.
Mitochondrial genome data: protein-coding genes
We identified 13 protein-coding genes for both genomes, of which nine were encoded from the H-strand (ATP6, ATP8, COI, COII, COIII, Cyt b, NADH2, NADH4, NADH6) and four from the L-strand (NADH1, NADH4, NADH4L, NADH5) (Figs 4 and 5; Table 2). All proteins have typical invertebrate mitochondrial start codons (ATA, ATC, ATG, ATT, or TTG) and stop codons (TAA, TAG). Whereas 11 protein-coding genes have identical numbers of base pairs within both genomes, different lengths were found for two genes: NADH2 (Epimeria cornigera: 984 bp, 328 amino acids; Epimeria frankei: 981 bp, 327 amino acids) and NADH6 (Epimeria cornigera: 504 bp, 168 amino acids; Epimeria frankei: 507 bp, 169 amino acids). Nucleotide divergence (patristic distances) between the protein-coding genes of both mitogenomes ranged from a minimum of 0.12% for COI, COII and COIII to a maximum of 0.20% for ATP8 (Table 2).
Mitochondrial genome data: ribosomal and transfer RNA genes
A total of 22 tRNA genes with lengths from 51 bp (tRNASer2) to 64 bp (tRNAGly) were found within both mitochondrial genomes (Figs 4 and 5; Table 2). The data revealed differences in lengths (+/−1 bp) for four tRNAs: tRNAGlu, tRNALeu1, tRNACys, and tRNAThr. Sixteen tRNAs are encoded on the H-stand whereas six tRNAs are located on the L-strand. All tRNAs are capable of folding into a typically clover-leaf-like secondary structure except tRNASer1 and tRNASer2. As previously reported for other crustacean taxa, these two tRNAs cannot form a secondary structure due to their lack of the dihydrouracil arms50,51. Both rRNA genes were found on the L-strand. They are located between the tRNALeu1 and the putative control region and are separated by tRNAVal. Gene lengths of the 12S rRNA genes are 639 bp for Epimeria cornigera and 640 bp for Epimeria frankei, whereas the lengths of 16S rRNA genes are 1,049 bp (Epimeria cornigera) and 1,046 bp (Epimeria frankei). The nucleotide divergence (patristic distance) was 0.07% between the 12S rRNA genes and 0.08% between the 16S rRNA genes (Table 2).
Mitochondrial genome data: non-coding regions
The major non-coding regions (Epimeria cornigera: 294 bp, Epimeria frankei: 737 bp) were identified between the 12S rRNA and tRNACys genes, which are considered to represent the putative control regions (Table 2). It should be noted that the read depth in this region was relatively high in Epimeria cornigera compared to the rest of the genome. This could indicate the presence of a repetitive element that could not be be properly assembled, All other 10 non-coding regions are smaller, ranging from 1 to 44 bp for both genomes. The A+T contents of the control regions are higher than those of other mitogenome regions, with 0.78 for Epimeria cornigera and 0.81 for Epimeria frankei. Furthermore, both control regions consist of several repetitive motifs in tandem that are different among both genomes.
Morphological distinction
Epimeria frankei can be distinguished from E. cornigera by the shape of coxal plate 5. In E. frankei, the anteroventral margin of coxa 5 describes a nearly straight line or is only slightly concave, whereas this margin is strongly concave in E. cornigera (less pronounced in males). Coxa 5 is strongly extended posteroventrally in both species, coming along with an only slightly concave posterior margin in E. cornigera (distinctly concave in E. frankei). Additionally, urosomite 3 can be used as a diagnostic character. In E. frankei, urosomite 3 is pronounced in a distinctly upwards pointed projection (Fig. 8D), whereas urosomite 3 of E. cornigera is characterised by an inconspicuous blunt hump (Fig. 14D) or is not pronounced at all. The most conspicuous difference between the two species, however, can be observed on live or freshly caught individuals (observed on approx. 100 indiviuals of each species): E. frankei features an even, brownish or grey colouration with hints of purple or even pink (Fig. 6). In contrast, E. cornigera is characterised by an eye-catching red-white colouration (Fig. 6). Unfortunately, the colouration of individuals vanishes quickly when being exposed to ethanol, resulting in yellowish or pale white specimens in both species.
Figure 8.
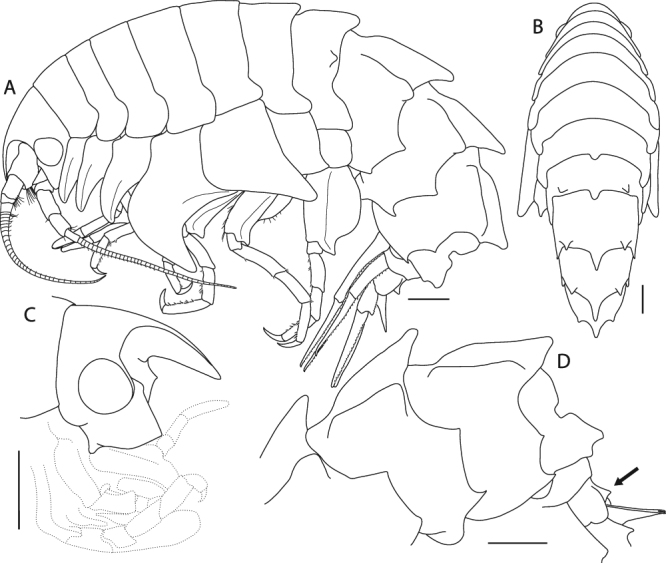
Epimeria frankei sp. nov., holotype, adult female, northern North Sea, NHMUK 2015.3264: (A) lateral habitus, (B) dorsal habitus, (C) head in lateral view, (D) pleon and urosome; all scales 1 mm.
Figure 14.
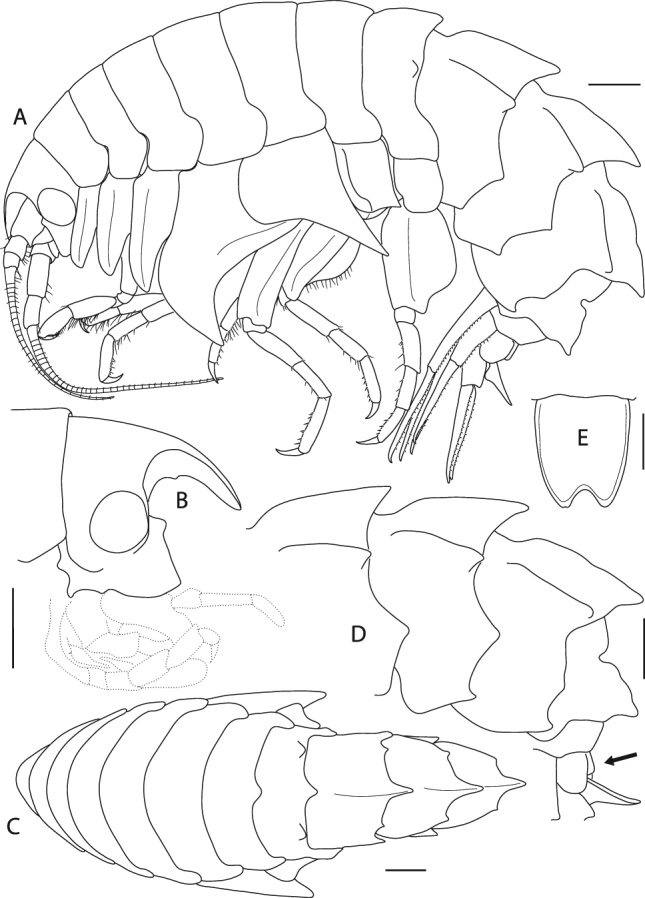
Epimeria cornigera, neotype, adult female, northern North Sea, NHMUK 2015.3268: (A) lateral habitus, (B) head in lateral view, (C) dorsal habitus, (D) pleon and urosome, (E) telson; scales: (A,B,C,D) 1 mm; (E) 0.5 mm.
Figure 6.

Photograph of freshly caught individuals of E. cornigera (top) and E. frankei sp. nov. (bottom); body colouration can clearly be used to discriminate the two species.
Discussion
The current study fully implements the concept of integrative taxonomy, proposed as a framework to bring together conceptual and methodological developments, combining as many different data types as possible to characterise species in a multifaceted and detailed way1,52,53. Here, two sibling amphipod species are described using comprehensive morphological, genetic, and mitogenomic data. This is the first use of genomic data (here: complete mitochondrial genomes) as part of a species description and a redescription within the Metazoa, setting a new desirable standard for future species descriptions35. We are convinced that modern species descriptions should seek for this integrative addition of comprehensive complementary information, ensuring the highest level of confidence for both biodiversity and ecological research.
Biology and geographic distributions
Species of the genus Epimeria are morphologically characterised by dorsal teeth and/or robustly elongated coxal plates 1–4. The coxal plates exhibit an ‘inter-locking’ mechanism which may have a function during the mating or as defence mechanism against predators54. A number of Epimeria species are frequently recorded from shallow to deep waters of the northern Atlantic and Arctic waters. Most Epimeria species are bottom-dwellers and were often assumed to be associated with other biota such as deep-sea corals, hydrozoans, sponges and holothurians in the North Atlantic55–57, but reliable information on their life styles is scarce. With the description of E. frankei, the number of species in the north-eastern Atlantic increases to a total of 5: E. cornigera (current study), E. frankei (sp. nov.; current study), E. loricata G.O. Sars 1879, E. parasitica M. Sars 1858 and E. tuberculata G.O. Sars 1893. The main morphological characteristics that can be used for their distinction are provided in the key below.
Former reports of E. cornigera s. lat. ranged from the NE Atlantic to the Mediterranean Sea. In fact, the current findings and the geographic information of the re-examined museum material suggest that E. cornigera s. str. may be restricted to the northern East Atlantic, whereas E. frankei seemingly exhibits a Mediterranean-Lusitanean distribution (Fig. 7). However, as also evidenced by the same type locality of the two species, both occur sympatrically on sandy bottoms of the North Sea and near the northern British coasts where water temperatures are probably suitable for both species.
Figure 7.
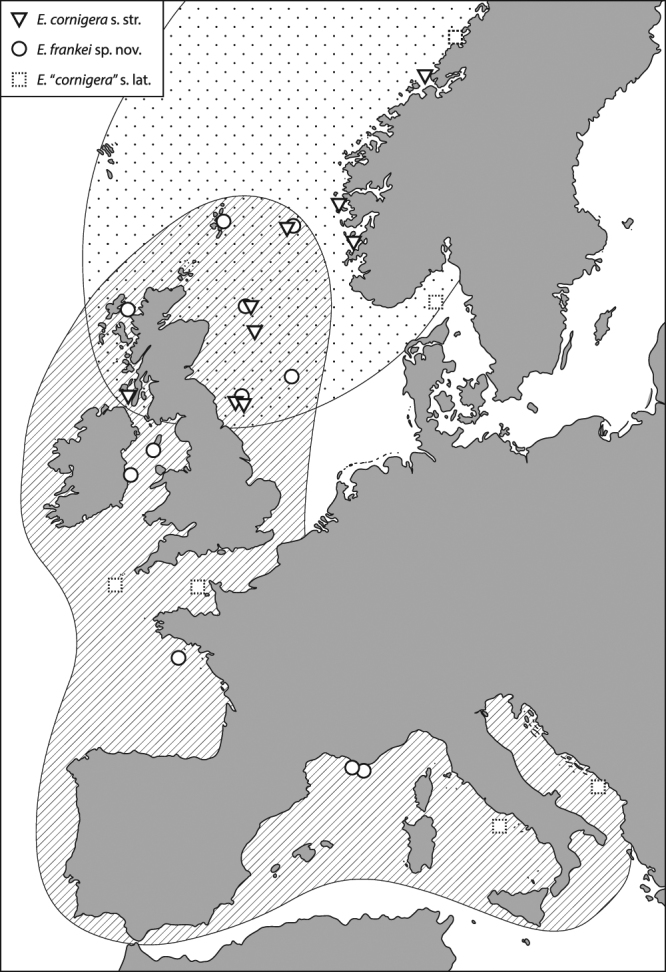
Suggested geographic distributions of E. cornigera and E. frankei sp. nov. considering the examined material of the current study and previous records of “Epimeria cornigera s. lat.” (i.e. published records of “E. cornigera” that cannot be assigned clearly) in the literature56,109,110,114,120–123. Initial map built with ArcGIS 10.3.1 (www.esri.com/arcgis) and modified with Adobe Illustrator (CS 6).
Key to the Epimeria species of the north-eastern Atlantic
- All pereonites dorsally carinate.... E. loricata
- Only pereonites (5) 6–7 dorsally carinate.... 2
- Apical tips of coxae 1–5 bluntly rounded; coxa 4 projecting ventrally with concave posteroapical margin; coxa 5 moderately extended posteroventrally (about as long as broad).... E. tuberculata
- Apical tips of coxae 1–5 rounded or subacute; coxa 4 strongly projecting ventrally and posteroapical margin strongly concave; coxa 5 strongly extended posteroventrally (at least twice as long as broad).... 3
- Epimeral plate 3 angular, produced to a large posterodistal projection, posterior margin straight.... E. parasitica
- Epimeral plate 3 biangular with subacute posterodistal tooth.... 4
- Coxa 5 anteroventral margin nearly straight or only slightly concave; urosomite 3 with pointed projection; body colouration of live animals single-coloured brown to grey with hints of purple.... E. frankei sp. nov.
- Coxa 5 anteroventral margin strongly concave; urosomite 3 not produced or only with blunt hump; body colouration of live animals white with red stripes.... E. cornigera
DNA barcodes
During the last few years it has become evident that DNA barcodes represent powerful diagnostic supplementary characters which accelerate and revive traditional morphological taxonomy by providing additional data58. Consequently, there is continuous progress of using DNA barcodes as part of species descriptions in the Crustacea24. The analysis of our obtained DNA barcode data revealed distinct clusters for both species with a minimum interspecific distance of 8.69% (K2P) (Table 1). This value is similar to interspecific distances known from other closely related Epimeria species, with minimum distances of 8.5%59. Furthermore, we found two lineages and two BINs (AAU2061, ACS7228) for Epimeria frankei (Fig. 1). However, as nuclear 18S rRNA gene sequences from specimens of both lineages were identical and no corresponding morphological differences were observed, these have to be considered as one species unless further data becomes available (Fig. 3). Therefore, the observed variability may be the result of phylogeographic effects in E. frankei60–62.
Phylogenetic analysis of the CO1 sequences
About 66% of the currently known species of Epimeria are found around Antarctica as part of a monophyletic lineage59,63–65, representing the best known example of a species flock within Antarctic amphipods66. This hypothesis is supported by our phylogenetic results: all studied species from Antarctica form a monophyletic clade with high bootstrap support (97%) (Fig. 2). Our analysis indicates a potential relationship of Epimeria cornigera and Epimeria frankei with two non-Antarctic species: Epimeria horsti and Epimeria bruuni. Both species are only known from waters around New Zealand so far67. However, this relationship is supported only by a low bootstrap value (40%). A more comprehensive analysis based on i) further, particularly non-Antarctic species, ii) more molecular data (both mitochondrial and nuclear) and iii) morphological characteristics is needed to reconstruct the phylogeny of this impressive amphipod genus in detail.
Mitochondrial genomes
Because modern high-throughput sequencing technologies allow for much faster and easier analyses of genomes, the analysis of mitogenomes will likely increase dramatically in the near future35. Compared to other crustacean groups, the number of available complete mitochondrial genomes for amphipods is quite high (n = 29, 2017-06-21) and these display various patterns of gene arrangements68. Mitogenomes of both Epimeria cornigera and Epimeria frankei represent typical circular metazoan mitogenomes with an identical gene arrangement (Table 2) and a patristic distance of 0.12% between both genomes. Patristic distances between all protein-coding genes range from 0.12% (COI, COII) to 0.20% (ATP6), and in the case of rRNA genes from 0.07 (12 rRNA) to 0.08 (16S rRNA). With a length of 737 bp the putative control region of Epimeria frankei is significantly longer than in Epimeria cornigera (294 bp). This region is the largest non-coding part of the mitochondrial DNA in general and includes the major regulatory elements for its replication and expression69. Furthermore, it is the most polymorphic part of the mitochondrial genome with hypervariable sites70. Nevertheless, we found two conserved parts shared between both genomes, covering 80 and 190 bp, respectively. Our results support the concept that the control region is divided into three polymorphic domains, being separated by two stretches with no interspecific variability71. However, more data have to be evaluated to analyse this phenomenon in more detail.
Material and Methods
Sample collection and DNA extraction
In total, 75 specimens were collected with beam trawls in the North Sea between 2012 and 2014 as part of the German Small-scale Bottom Trawl Survey (GSBTS72). The amphipods were preserved in ethanol (96%) directly after collection. All specimens were morphologically identified by JB and HN. For each specimen, total genomic DNA was extracted from one to three dissected pereopods using the QIAmp© Tissue Kit (Qiagen GmbH, Hilden, Germany) following the supplier’s extraction protocol.
Specimens were deposited in the collections of natural history museums (see below), whereas DNA extracts were listed and stored in the North Sea Fauna collection of the German Centre for Marine Biodiversity Research (DZMB) in Wilhelmshaven, Germany.
Amplification and sequencing: DNA barcodes and complete 18S rRNA gene sequences
Polymerase chain reaction (PCR) was used to amplify the DNA barcode fragment using the primer pair LCO1490 and HCO219819 or the newly designed degenerated amphipod-specific primer pair CO1-LCO-AMP1 (5′-THT CDA CHA ACC AYA AAG AYA TYG G-3′) and CO1-HCO-AMP2 (5′-ATD ACT TCW GGR TGV CCR AAR AAY C-3′). For the latter, we added derived M13 forward and reverse tails to both primers to provide known primer-binding sites for sequencing73. Therefore, the complete primer sequences were 5′-TGT AAA ACG ACG GCC AGT THT CDA CHA ACC AYA AAG AYA TYG G -3′ for CO1-LCO-AMP1 and 5′-CAG GAA ACA GCT ATG AC ATD ACT TCW GGR TGV CCR AAR AAY C-3′ for CO1-HCO-AMP2 (with underlined M13 tail sequences). Amplification reactions were carried out on a Mastercycler pro S system (Eppendorf, Hamburg, Germany), using illustraTM puReTaq Ready-To-Go PCR Beads (GE Healthcare, Buckinghamshire, UK) in a total volume of 20 µl, containing 17.5 µl sterile molecular grade H2O, 2 µl DNA template with an DNA amount between 2 and 150 ng/µl and 0.25 µl of each primer (20 pmol/µl) for amplification. For this gene fragment (CO1), PCR thermal conditions included an initial denaturation at 94 °C (5 min), followed by 38 cycles at 94 °C (denaturation, 45 s), 48 °C (annealing, 45 s), 72 °C (extension, 80 s), and a final extension step at 72 °C (7 min).
Furthermore, the complete nuclear 18S rRNA gene was amplified for selected individuals of both species (Epimeria cornigera: n = 2, Epimeria frankei: n = 6). Detailed information about used primer pairs, amplification reactions and temperature profiles are provided in previous studies74,75. For verification, negative and positive controls were included with each round of reactions. Three μl of the amplified products were inspected for size conformity by electrophoresis in a 1.5% agarose gel with GelRedTM using commercial DNA size standards. The remaining PCR product was purified with the QIAquick© PCR Purification Kit (Qiagen GmbH, Hilden, Germany). Purified PCR products were cycle sequenced and sequenced in both directions at a contract sequencing facility (GATC, Konstanz, Germany) using the M13 sequence tails for the DNA barcodes or the same primers as employed in the PCR in the case of the 18S rRNA gene. For this marker, some additional internal primers were used for sequencing: Forward: AF700F_mod, 1000 F, 1250FN_mod; Reverse: 700 R, 1155 R, 1500R_mod75,76. Complementary sequences were assembled with the Geneious version 9.0.4 program package77. BLAST searches were performed to confirm the identity of all new sequences78,79. Furthermore, Geneious was used to translate all COI sequences to amino acid sequences to check for presence of nuclear mitochondrial pseudogenes (numts).
All relevant voucher information, photos, DNA barcodes, used primer pairs and trace files are publicly accessible through the public data set “North Sea Epimera” (Dataset ID: DS-EPNS Epimeria North Sea; dx.doi.org/10.5883/DS-CRNS) on the Barcode of Life Data Systems (BOLD; www.boldsystems.org)80,81. All new sequence data were deposited in GenBank (CO1: MG191712-MG191786, 18S: MG191704-MG191711).
Mitochondrial genome: library building, sequencing and raw read processing
DNA extracts were built into double-barcoded Illumina sequencing libraries using a published protocol set82, with some minor modifications83. The optimal number of cycles for amplification was established using qPCR to avoid saturation. Sequencing was performed on the Illumina Nextseq. 500 at the University of Potsdam, Germany, using 150 base pair (bp) paired-end (PE) reads. 25,368,699 PE reads were sequenced for the holotype of Epimeria frankei and 20,329,986 for the neotype of Epimeria cornigera. Illumina adapter sequences were trimmed from read ends, and all reads less than 30 bp were discarded using Cutadapt 1.484. PCR duplicate sequences were removed using Fastuniq85. Forward and reverse sequences were interleaved into a single file using a perl script available in the MITObim package86.
Mitochondrial genome: sequence reconstruction
We carried out independent mitochondrial reconstructions for both species using MiTObim v1.886, an iterative mapping wrapper script for the Mira v4.0.2 assembler87. We implemented a direct reconstruction without prior mapping assembly using default parameters with alterations to mismatch values, k-mers and reference bait sequences. The utilised bait sequences consisted of: Gondogeneia antarctica (JN827386.1), Metacrangonyx goulmimensis (HE860501.1), and Onisimus nanseni (NC_013819.1). The interleaved fastq files were treated as single end data. Reconstructions were conducted using k-mer values of 19, 25 and 31. We implemented a reconstruction protocol based on that found in Westbury et al.88 by beginning with the most strict mismatch value (0%) and gradually increasing this value until the mitochondrial genome was complete. Mismatch values of 0–8% were implemented for Onisimus nanseni and Metacrangonyx goulmimensis whereas mismatch values of 0–6% were implemented for Gondogeneia antarctica. This resulted in 25 separate iterative mapping alignments.
Mira output maf files were converted to sam files and visualised using Geneious. For each independent iterative mapping alignment, a 20x minimum coverage was implemented for consensus building along with a strict 95% threshold for base calling. This resulted in 25 individual consensus sequences, nine constructed using Onisimus nanseni as reference, nine using Metacrangonyx goulmimensis as reference and seven from using Gondogeneia antarctica as reference. From this data, we constructed three reference-specific consensuses by aligning all consensus sequences produced via mapping to the same reference bait sequence, i.e. consensuses produced from different mismatch values but the same reference sequence, using Mafft89 and a 50% strict consensus base call. These three reference-specific consensus sequences were then aligned in Mafft and a final consensus was built using a 50% strict consensus base call. We performed automatic annotations with MITOS90 to check for the presence of genes and tRNAs in our final consensus sequence. The tRNAs were also annotated with ARWEN v1.291. Finally, gene limits were refined by comparison with orthologous mtDNA sequences of other amphipod genomes. Both new genomes were deposited in GenBank (accession numbers: Epimeria cornigera: MF361127, Epimeria frankei: MF361126).
Sequence analysis
DNA barcodes
In addition to the 75 CO1 sequences of both species obtained for this study, five already published sequences of Epimeria cornigera (accession numbers: KT208490, KT208918, KT209458, KT209498, KT209569) and six barcode sequences of the outgroup Gammarus locusta (Linnaeus, 1758) (KT208462, KT208572, KT208951, KT209189, KT209193, KT209211) were retrieved from GenBank28. Thus, the complete data set consisted of 86 DNA barcodes.
Intra- and interspecific nucleotide distances of the analysed amphipods were based on the Kimura 2-parameter model (K2P)92, using the analytical tools on the BOLD workbench (align sequences: BOLD aligner; ambiguous base/gap handling: pairwise deletion). We also used BOLD to calculate base frequencies of all barcodes. All barcode sequences were assigned to the Barcode Index Number (BIN) system which is implemented in BOLD81. All sequences were aligned using MUSCLE version 3.693 with default settings. In a further step, a neighbor-joining cluster analysis (NJ)94 was performed to reconstruct a phylogenetic tree based on K2P distances for all DNA barcode sequences using MEGA7.0.1895. Non-parametric bootstrap support values were obtained by resampling and analysing 1,000 replicates96.
In order to explore the phylogenetic position of Epimeria cornigera and Epimeria frankei, we reconstructed the phylogeny of the genus Epimeria in a maximum-likelihood framework using MEGA7.0.18. For the analysis, COI barcodes of the neotype of Epimeria cornigera and the holotype of Epimeria frankei were used as well as all available COI sequences of the DNA barcode fragment from NCBI GenBank with a length of at least 500 base pairs (n = 67, date of download: 01-02-2017), representing 16 of the 84 currently recognized species in the genus Epimeria (29%). A table of all included species and corresponding accession numbers is included in the supplementary information (Table S2). All sequences were aligned using MUSCLE version 3.693 with default settings and with the six sequences of Gammarus locusta as outgroup. The most appropriate model of sequence evolution was determined beforehand using the Decision Theory performance-based selection (DT) as implemented in jModeltest 2.1.197. The Hasegawa-Kishino-Yano model (HKY85)98 was shown to be the optimal nucleotide substitution model with the following parameters: nucleotide frequencies A: 0.32, C: 0.24, G: 0.15, T: 0.29; transition/transversion ratio = 3.4401; gamma distribution shape = 0.885; and proportion of invariable sites = 0.439. Node support for individual clades was estimated using 1,000 non-parametric bootstrap replicates96.
Small ribosomal subunit (18S rRNA)
All eight 18S rDNA sequences of the two Epimeria species and one outgroup sequence of Gammarus locusta (AF419222) were aligned with MUSCLE version 3.6 using default settings. The software package MEGA7.0.18 was used to calculate nucleotide frequencies, interspecific distances based on patristic distances and to perform a NJ cluster analysis for a graphical representation of nucleotide divergence. Secondary structure elements were identified using the CRW site99 and other published models100–102.
Mitochondrial genomes
An initial alignment of both genomes was obtained with Clustal W103 as implemented in BioEdit v7.2.6104. Start and end positions of all encoding genes were investigated using the Geneious software package. The nucleotide composition of both mitogenomes and patristic distances of all mitochondrial proteins were calculated with MEGA7.0.18. A graphical representation of both genomes was produced with the Geneious package.
Morphological analyses
Before examination, specimens were transferred to 70% ethanol and appendages were temporarily mounted in glycerin. Whole animals and appendages were then studied and drawn under a Leica M205c microscope equipped with a drawing tube. All material was later conserved in ethanol (95%). Pencil drawings were scanned and inked with the software Adobe Illustrator (CS6) on Wacom drawing tablets (Intuos 5, A4 and A3), following the methods described by Coleman105,106. Referring to the considerations of d’Udekem d’Acoz107 and Krapp-Schickel108, the following terminology was applied in the descriptions: seta = slender, flexible articulated structure; spine = robust, inflexible articulated structure (synonymous to ‘robust seta’); tooth = non-articulated, pointed ectodermal structure. Although officially listed in the collections of the Natural History Museum of Denmark, Copenhagen, Fabricius’ holotype is missing (pers. comm. Jørgen Olesen) and was thus presumed as lost in the current study. Furthermore, there is no exact information on the type locality, which can only be assumed to be located in the northern North Sea. Therefore, types were chosen considering the geographical area where both species were commonly found sympatrically. This was further validated by the descriptions and drawings of Sars55, which seem to refer to E. cornigera s. str. All type material was deposited in the Natural History Museum, London, UK (NHMUK). Further specimens from the current study were transferred to the natural history museums of Berlin and Frankfurt including their original accession numbers of the former collection of the Molecular Taxonomy (MT) group at DZMB. Additional material was examined in the collections of the NHMUK and the Museo Civico di Storia Naturale, Verona, Italy (MCSN).
Systematics
Epimeria frankei sp. nov. Beermann & Raupach (Figs 8, 9, 10, 11, 12 and 13).
Figure 9.
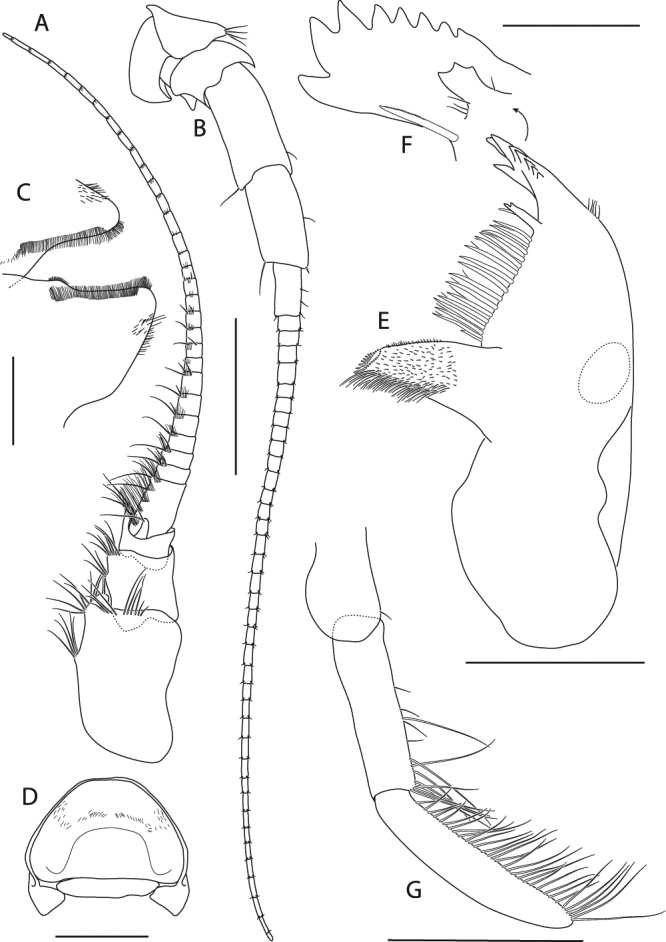
Epimeria frankei sp. nov., holotype, adult female, northern North Sea, NHMUK 2015.3264: (A) antenna 1, (B) antenna 2, (C) hypopharynx, (D) mandible, (E) mandible incisor and lacina mobilis, (F) mandibular palpus, (G) labrum; scales: (A,B) 1 mm; (C,D,E,G) 0.5 mm; (F) 0.25 mm.
Figure 10.
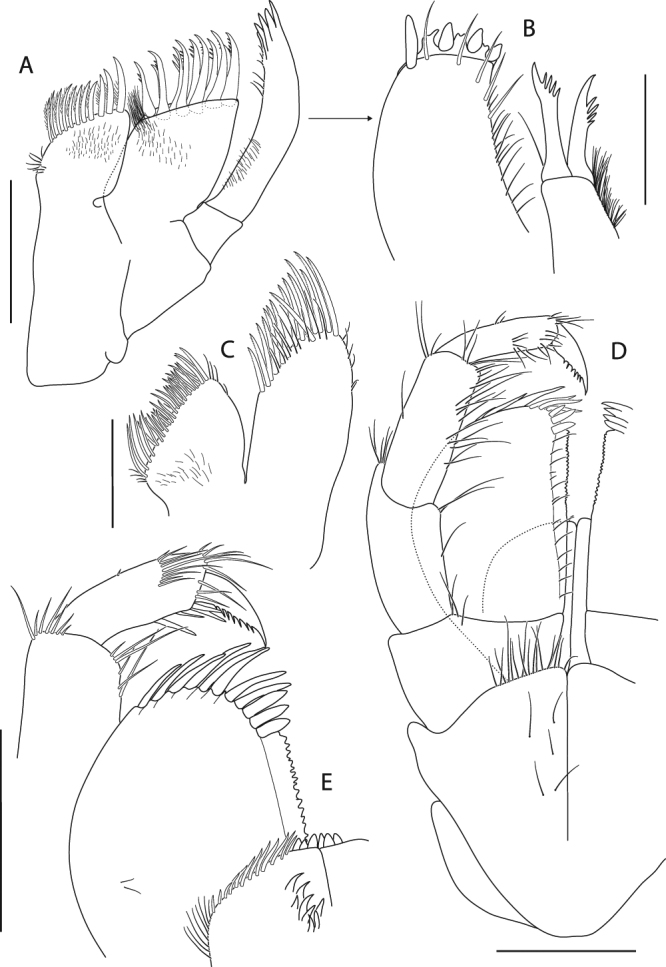
Epimeria frankei sp. nov., holotype, adult female, northern North Sea, NHMUK 2015.3264: (A) maxilla 1, (B) maxilla 1 palpus and outer plate distal ends lateral view, (C) maxilla 2, (D) maxilliped, (E) maxilliped palpus and outer plate; scales: (A,D,E) 0.5 mm; (B,C) 0.25 mm.
Figure 11.
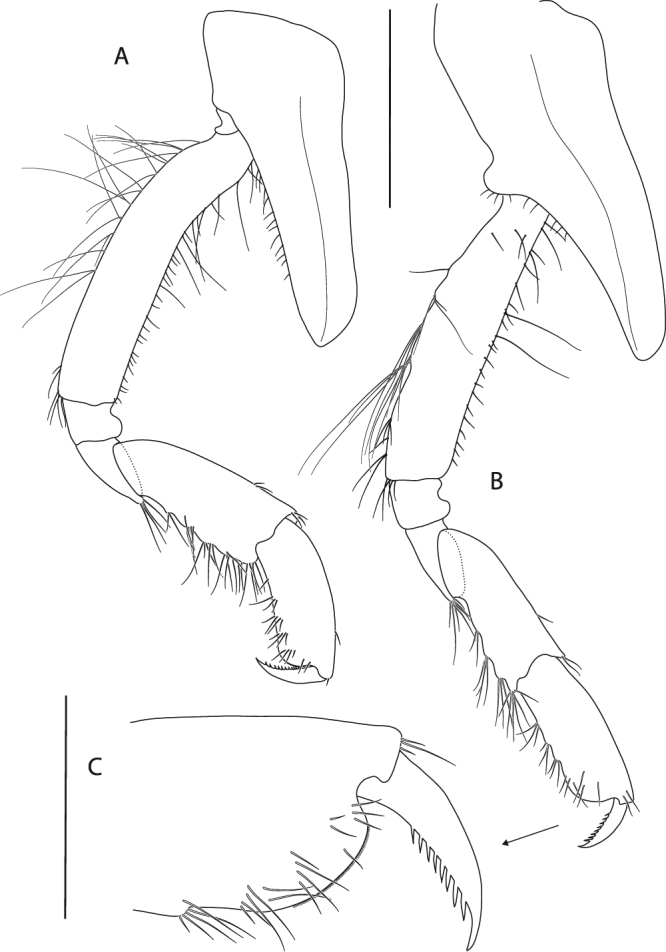
Epimeria frankei sp. nov., holotype, adult female, northern North Sea, NHMUK 2015.3264: (A) gnathopod 1, (B) gnathopod 2, (C) distal articles of gnathopod 2; scales: (A,B) 1 mm; (C) 0.3 mm.
Figure 12.

Epimeria frankei sp. nov., holotype, adult female, northern North Sea, NHMUK 2015.3264: (A) pereopod 3, (B) pereopod 4, (C) pereopod 5; all scales 1 mm.
Figure 13.
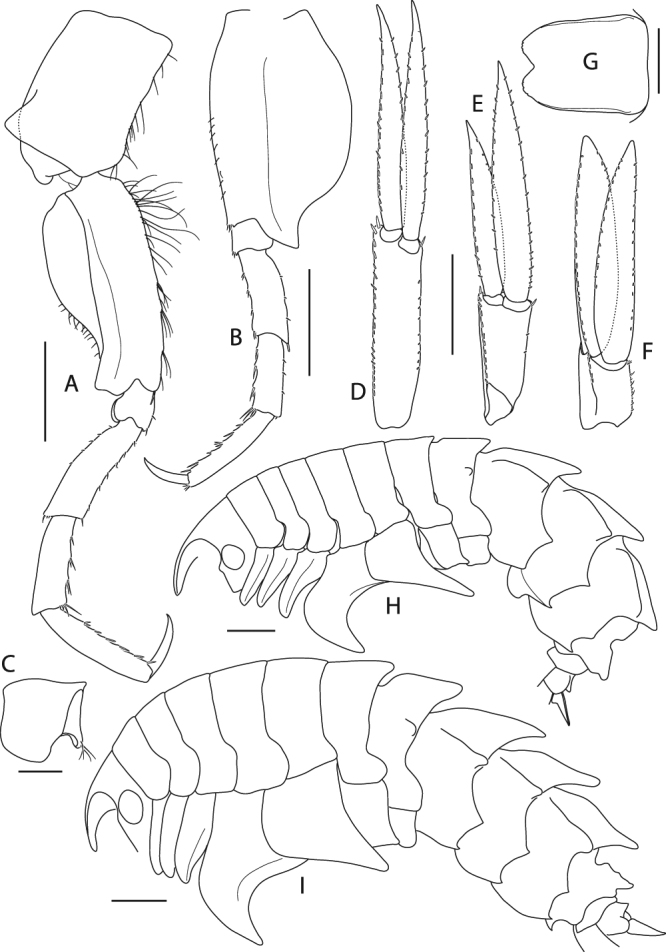
Epimeria frankei sp. nov., holotype, adult female, northern North Sea, NHMUK 2015.3264: (A) pereopod 6, (B) left pereopod 7, (C) right coxa 7, (D) uropod 1, (E) uropod 2, (F) uropod 3, (G) telson; Epimeria frankei sp. nov., paratype, adult male, northern North Sea, NHMUK 2015.3267: (H) lateral habitus; Epimeria frankei sp. nov., adult female, Mediterranean Sea, MCSN Ec1: (I) lateral habitus; scales: (A,B,D,E,F,H,I) 1 mm; (C,G) 0.5 mm.
Epimeria cornigera Chevreux & Fage109, 1925: 191, fig. 198–200. – Karaman110, 1972: 143. – Ledoyer111,112, 1977: 397; 1993: 616, fig. 423.
Material examined
Holotype (NHMUK 2015.3264), ovigerous female, 19 mm, northern North Sea – approx. 150 km west of Bergen (60.3104°, 2.4967°), in 97 m water depth, 05.viii.2013, coll. H. Neumann. Paratypes: two ovigerous females (NHMUK 2015.3265 - 3266), locality data identical to holotype; one adult male (Fig. 13; 13 mm; NHMUK 2015.3267), northern North Sea (60.3956°, 2.5788°), 86 m, 30.vii.2012, coll. H. Neumann.
Other material
One ovigerous female (MT07112), North Sea (57.9514°, −0.7692°), 101 m, 05.viii.2012, coll. H. Neumann. Fourteen males and 14 females (of which 7 ovigerous, 6 juvenile; MT06718, MT06742-6749, MT06755-6764, MT07102-7110), western North Sea (54.8732°, −0.9581°), 68 m, 07.iii.2012, coll. H. Neumann. Nine males (1 juvenile) and eight females (4 ovigerous, 4 juvenile; MT06787-7101, MT06712-6713), central North Sea (56.093°, 1.213°), 87 m, 06.iii.2012, coll. H. Neumann. One adult female (MT06708), northern North Sea (60.3659°, 2.4386°), 96 m, 31.vii.2012, coll. I. Rottgardt. Two adult females (1 ovigerous; MT06706-6707), northern North Sea (60.3583°, 2.5371°), 106 m, 31.vii. – 01.viii.2012, coll. I. Rottgardt. One juvenile female (MT06704), northern North Sea (60.3459°, 2.6711°), 101 m, 30.vii.2012, coll. H. Neumann. One juvenile female (MT06703), western North Sea (55.1097°, −0.5023°), 82 m, 07.ii.2012, coll. H. Neumann. One juvenile female (MT06701), western North Sea (55.4147°, −0.0218°), 75 m, 07.iii.2012, coll. H. Neumann. One male (?) and three ovigerous females (MT01164-1165, MT01167-1168), northern North Sea (60.3703°, 2.4784°), 100 m, 28.vii.2010, coll. H. Neumann. Two females (NHMUK unregistered), Shetland (North Sea), unknown water depth and collection date, coll. A.M. Norman. One adult female (NHMUK unregistered), Port Erin, Isle of Man (Irish Sea), unknown water depth, vii.1960, coll. I. Gordon. Pieces of three individuals (NHMUK 1952:5:7:152-155), Moray Firth (Scotland), unkown water depth and collection date, coll. Sp. Bate. Walker Collection (NHMUK 1925:9:8:877-887): One male, five females and four juveniles, Shiant and West Coast of Ireland, unkown water depth and collection date. Norman Collection (NHMUK 1911:11:8): One male and one female (16998–999), The Minch (Scotland), unknown water depth, 1866; diverse individuals (males, females, juveniles; 16934–943, 16914–933), Shetland (North Sea), unknown water depth, 1861; two females (16964–965), Clyde (Scotland), 1899; diverse individuals (males, females, juveniles; 16954–9563, 16975–976), unkown locality? (“Porcupine”), water depth and collection date. Approx. 100 specimens (males, females, juveniles; MCSN unregistered, “Rec Ledoyer sèr FVP 39”), south of Embiez (Toulon, France), 100–120 m, 26.vi.1975, coll. M. Ledoyer. Twenty individuals (males, females, juveniles; MCSN unregistered, “Rec Ledoyer Serie FVP, St. 33”), southeast of Canyon de Planir (south of Marseille, France), 90 m, 02.v.1975, coll. M. Ledoyer.
Etymology
Named in honour of Prof. Heinz-Dieter Franke for his continuous support and inspiration of the first author (JB) for many years. The name frankei is the Latin genitive derived from his name.
Diagnosis
Body dorsally smooth on pereonites 1–5; pereonite 6 roundly produced mid-dorsally; pereonite 7 with blunt mid-dorsal process; pleonites 1–3 strongly carinate, each with a blunt dorsolateral ridge; epimeral plates 1–3 biangular, with acute posterodistal teeth in epimera 2 and 3; urosomite 3 with distinctly upwards pointed projection; palp of maxilla 1 distally with 4 blunt spines and few subdistal setae; coxa 4 apical tip broadly rounded; coxa 5 anteroventral margin nearly straight or only slightly concave, apical tip of process broadly rounded; uropod 2 outer ramus distinctly shorter than inner ramus (about ¾).
Description
Based on female holotype, 19 mm.
Body (Fig. 8A,B) dorsally smooth on pereonites 1–5; pereonite 6 roundly produced mid-dorsally; pereonite 7 with blunt mid-dorsal process and a blunt dorsolateral process. Pleonites 1–3 with blunt carinae and each with a blunt dorsolateral ridge; epimeral plates 1–3 biangular, produced to blunt tooth in epimeron 1 and acute posterodistal teeth in epimera 2 and 3, angles on posterior margins subacute. Urosomite 1 with blunt carina, notched mid-dorsally; urosomite 2 shortest; urosomite 3 with distinctly upwards pointed projection (Fig. 8D).
Head (Fig. 8C) about as high as long; ventral lobus slightly rounded, anterior corner rectangular. Rostrum weakly curved, ventral margin almost straight, about as long as 1st peduncular article of antenna 1. Eyes large and rounded, bulging.
Antenna 1 (Fig. 9A) peduncular article 1 about 1.4 as long as articles 2–3 combined (length ratio 1: 0.5: 0.3); accessory flagellum consisting of a single tapering article, about 1/3 the length of the 1st flagellar article; 1st flagellar article about as long as peduncular article 2, primary flagellum with 30 articles.
Antenna 2 (Fig. 9B) slightly longer than antenna 1; peduncular article 1 shortest, article 2 partly overlapping articles 1 and 3; article 3 with distal cusp; article 4 slightly longer than article 5; flagellum with 38 articles.
Labrum (“upper lip”; Fig. 9D) ventrally rounded.
Hypopharynx (“lower lip”; Fig. 9C) with distally tapering lobes, densely coated with fields of setae on the medial margins.
Mandible (Fig. 9E) with 8-dentate incisor; lacinia mobilis triform (Fig. 9F); molar protruding, slender; mandibular palp (Fig. 9G) consisting of 3 articles, bearing long setae on the distal half of article 2 and the entire article 3 posteromarginally; setae length on palp article 3 increasing from proximal to distal.
Maxilla 1 (Fig. 10A) inner plate sub-triangular, with 11 stout plumose setae along the medial margin; outer plate truncate, with 11 mediodistally serrated spines (Fig. 10B); palp 2-articulate, article 2 about 5 times longer than the 1st article, curved inwards, distally serrate with 4 blunt spines and few subdistal setae (Fig. 10B).
Maxilla 2 (Fig. 10C) inner lobe broader and shorter than outer lobe, with numerous setae along the medial margin; outer lobe with a row of slender spines which are mediodistally serrate along with few short slender setae.
Maxilliped (Fig. 10D,E) inner plate about 2/3 the length of 1st palp article, apical ridge with blunt spines and stout setae; outer plate reaching about midpoint of palp article 2, ovoid, apical margin with progressively stouter, slender spines mediodistally, mediodistal region produced to a saw-like ridge; palp 4-articulate, medial margin of article 2 setose, article 3 with stout setae mediodistally, article 4 curved, with serrate medial margin.
Gnathopod 1 (Fig. 11A) coxa tapering distally and with broadly rounded apical tip, anterior margin nearly straight, posterior margin slightly concave and bearing a row of very fine setae; basis about as long as coxa, slightly curved anteriorly, posteromarginally with some long slender setae, setation of anterior margin sparse with few long slender setae proximally and a row of short slender setae mediodistally; ischium slightly wider than long; merus with longitudinal articulation of carpus, posterior margin oblique, with tuft of long setae posterodistally; carpus distally slightly expanded, with groups of long setae along the posterior margin and few short setae anterodistally; propodus about 0.8 of carpus length, posteriorly with tufts of setae; palm convex, with fine serration; dactylus curved, with serrate posterior margin.
Gnathopod 2 (Fig. 11B,C) coxa slightly longer than coxa 1, tapering distally with rounded apical tip, anterior margin sinuous, posterior margin distinctly convex medioapically with few very fine short setae mediomarginally; basis slightly shorter than coxa, nearly linear, distal half posteriomarginally with groups of long slender setae, sparse row of short setae along anterior margin with few long slender setae; ischium about as long as wide; merus and carpus of similar length and setation to gnathopod 1, but slightly narrower; propodus about as long as carpus, with tufts of setae along the posterior margin; palm obliquely convex, with fine serration and few setae on the lateral face (Fig. 11C); dactylus as for gnathopod 1.
Pereopod 3 (Fig. 12A) coxa subequal to coxa 2 but slightly longer and wider; basis about 0.7 as long as coxa, slightly sinuous but sublinear, posterior margin with groups of long slender setae on its distal half, anterior margin with sparse setation; ischium slightly longer than wide; merus length about 1.2 of carpus length, slightly curved posteriorly, with short setae along the posterior margin and tufts of setae antero- and posterodistally; carpus with tufts of stout setae posteromarginally; propodus about 1.2 the size of carpus, with 6 pairs of spine-like setae along the posterior margin; dactylus about half the size of propodus, curved.
Pereopod 4 (Fig. 12B) coxa strongly projecting ventrally and robust, anterior margin sinuous and strongly curved posteriorly, posterior margin excavate and with a centrally positioned rounded lobe, posteroapical margin strongly concave, apical tip broadly rounded; basis linear, posterior margin convex and with groups of long slender setae, anterior margin straight and with sparsely distributed slender setae; ischium about as long as wide, with a tuft of setae posterdistally; merus about 1.5 the size of carpus, sublinear but slightly curved posteriorly, with few short setae along the posterior margin and tufts of setae antero- and posterodistally; carpus with tufts of stout setae along the posterior margin; propodus about 1.2 the size of carpus, with 6 pairs of spine-like setae posteromarginally; dactylus about half the length of propodus, curved.
Pereopod 5 (Fig. 12C) coxa strongly extended posteroventrally, posterior margin evenly concave, anterior margin convex and describing a rounded right angle, anteroventral margin nearly straight, apical tip of process broadly rounded; basis rectangular, with posteriorly produced margin which expands to a bifid subrectangular lobe posterodistally, partly covering ischium when leg is retracted, anterior margin with long slender setae proximally and tufts of setae along the distal half; ischium longer than wide, lobate; merus about 1.2 the length of carpus, slightly curved anteriorly and slightly expanding distally, anterior margin with few very short setae, a single spine-like seta anterodistally and posterodistally; carpus slightly expanding distally, with 4 tufts of spine-like setae along anterior margin and posterodistally; propodus about 1.4 the size of carpus, with 5 pairs of spine-like setae anteromarginally; dactylus nearly half the size of propodus, curved.
Pereopod 6 (Fig. 13A) coxa subrectangular, longer than wide with an angular lateral lobe pointing posterodistally, anterior margin with fine setae; basis slightly curved posteriorly, with lobe roundly expanding proximally on the medial side of the posterior margin and posteriorly produced lateral margin which expands to a bifid subrectangular lobe distally, posteromedial lobe with row of short slender setae mediodistomarginally, anterior margin with long slender setae proximally and tufts of setae along the distal half; ischium longer than wide, lobate; merus about 1.2 the length of carpus, slightly curved anteriorly and slightly expanding distally, very short setae along the anterior margin and medioproximally on the posterior margin; carpus slightly expanding distally, with 4 tufts of stout setae anteromarginally; propodus about 1.3 the length of carpus, with 6 pairs of spine-like setae anteromarginally; dactylus large, more than half the length of propodus, curved.
Pereopod 7 (Fig. 13B,C) on right body side missing, except for the coxa; on left side all articles complete; coxa slightly longer than wide; basis much wider than pereopod 6, posteriorly lobate, posterodistal lobe subacute and partly covering ischium, with minute tooth anterodistally, anterior margin with few short setae; ischium about as long as wide, with minute tooth on the anterodistal margin; merus slightly longer than carpus, slightly curved anteriorly, anterior margin with pairs of short stout setae, posterodistal corner bearing a blunt spine; carpus with 4 tufts of stout setae anteromarginally; propodus 1.3 the length of carpus, with 6 groups of spine-like setae anteromarginally; dactylus about half the length of propodus, curved.
Uropod 1 (Fig. 13D) peduncle subrectangular, with small spines along medial and lateral margins, spines increasing in size from proximal to distal; rami subequal, about 1.2 the length of peduncle, margins bordered with small spines.
Uropod 2 (Fig. 13E) peduncle weakly expanding distally, lateral margin with row of small spines; outer ramus distinctly shorter than inner ramus (about ¾), inner ramus about double as long as peduncle, spination similar to rami of uropod 1.
Uropod 3 (Fig. 13F) peduncle shortest, with small spines along the sublateral margin, and short setae on the medial margin; rami subequal in length, spination similar to rami of uropod 1 and 2.
Telson (Fig. 13G) about 1.5 as long as wide, slightly notched to only 10% of its length, apices broadly rounded.
Variation
In general, males of E. frankei resemble their females except for smaller body sizes. They can, however, be distinguished from the females by some distinct morphological features of the trunk (Fig. 13H). In direct comparison, the female pereon segments are clearly broader than in males, which is particularly visible in dorsal view. Males, in return, tend to feature a proportionally more pronounced pleon. Furthermore, male coxal plates tend to be narrower and less tubby/rounded, resulting in a smaller coxal surface area. There is little intraspecific variation except for the sexual dimorphism described above. In the examined material, pereon segments 6–7 and pleon segments of specimens from the Mediterranean, however, exhibited higher mid-dorsal processes (Fig. 13I).
Epimeria cornigera (Fabricius, 1779) (Figs 14, 15, 16, 17, 18 and 19)
Figure 15.
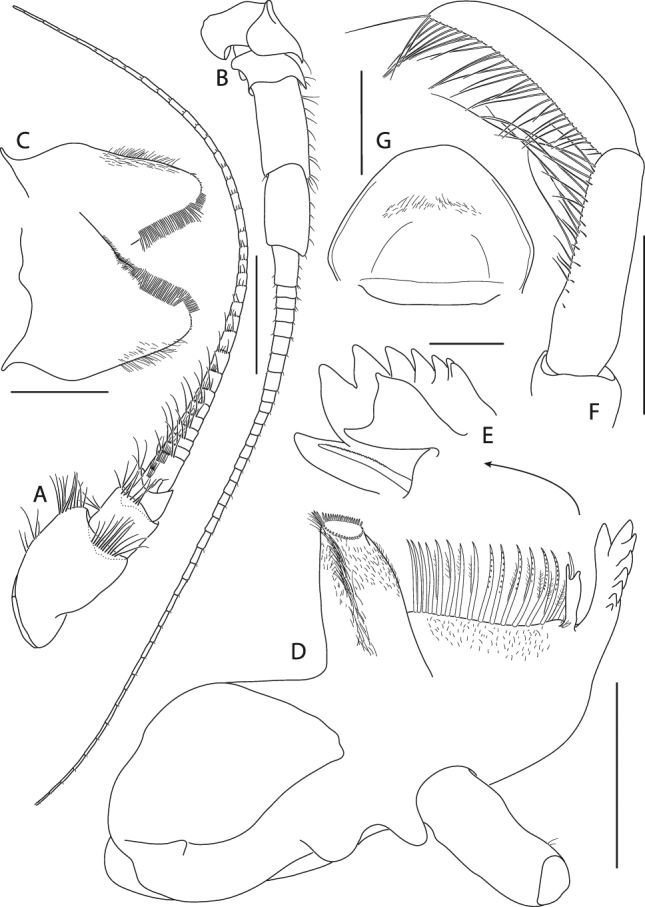
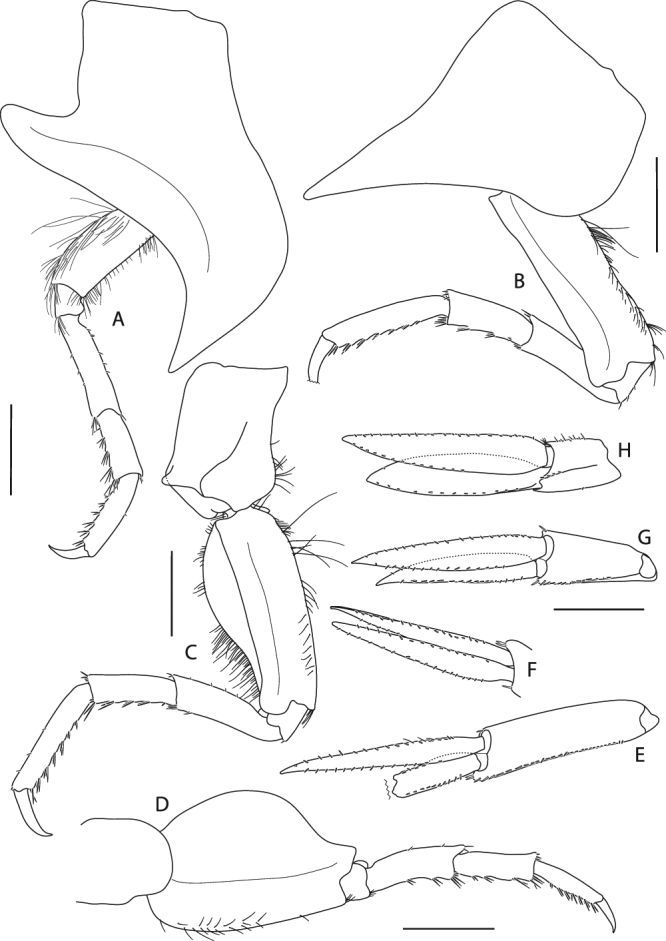
Epimeria cornigera, neotype, adult female, northern North Sea, NHMUK 2015.3268: (A) antenna 1, (B) antenna 2, (C) hypopharynx, (D) mandible, (E) mandible incisor and lacina mobilis, (F) mandibular palpus, (G) labrum; scales: (A,B) 1 mm; (C,D,F,G) 0.5 mm; (E) 0.1 mm.
Figure 16.
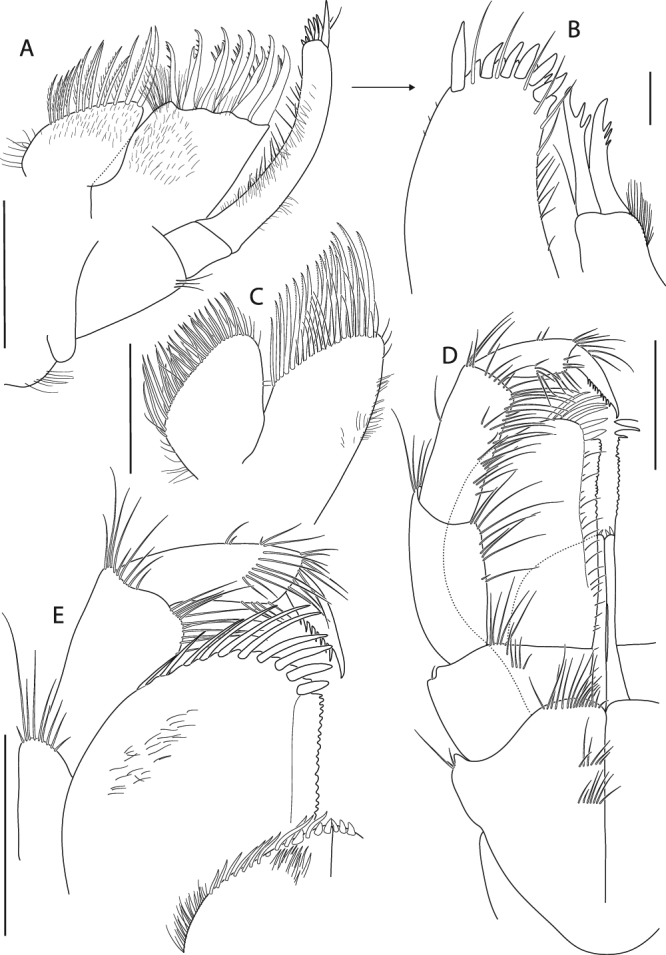
Epimeria cornigera, neotype, adult female, northern North Sea, NHMUK 2015.3268: (A) maxilla 1, (B) maxilla 1 palpus and outer plate distal ends lateral view, (C) maxilla 2, (D) maxilliped, (E) maxilliped palpus and outer plate; scales: (A,C,D,E) 0.5 mm; (B) 0.1 mm.
Figure 17.
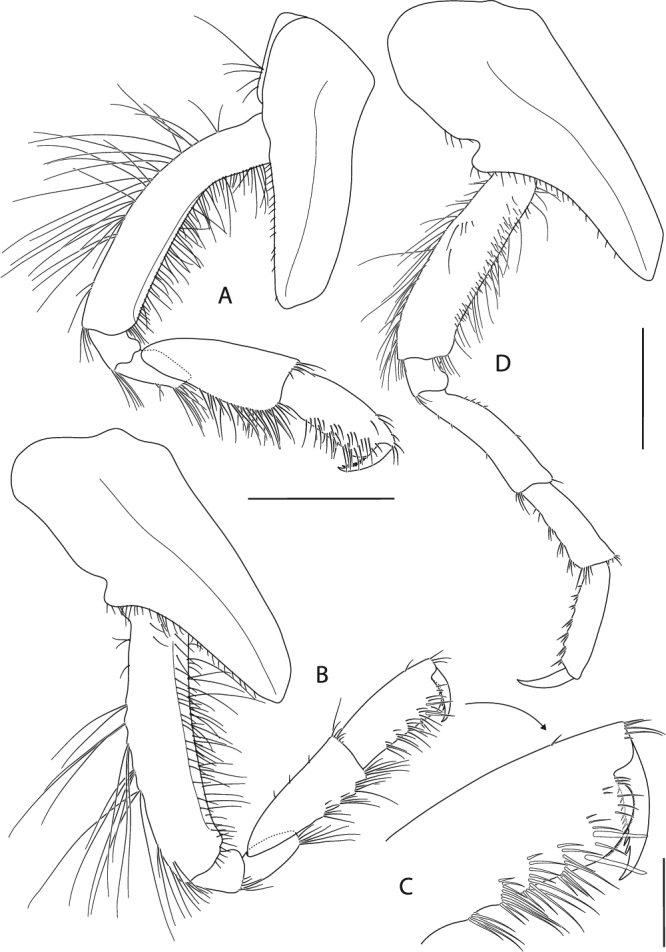
Epimeria cornigera, neotype, adult female, northern North Sea, NHMUK 2015.3268: (A) gnathopod 1, (B) gnathopod 2, (C) distal articles of gnathopod 2, (D) pereopod 3; scales: (A,B,D) 1 mm; (C) 0.25 mm.
Figure 18.
Epimeria cornigera, neotype, adult female, northern North Sea, NHMUK 2015.3268: (A) pereopod 4, (B) pereopod 5, (C) pereopod 6, (D) left pereopod 7 (drawn on habitus), (E) uropod 1 (outer ramus damaged), (F) rami of left uropod 1 (drawn on habitus), (G) uropod 2, (H) uropod 3; all scales 1 mm.
Figure 19.
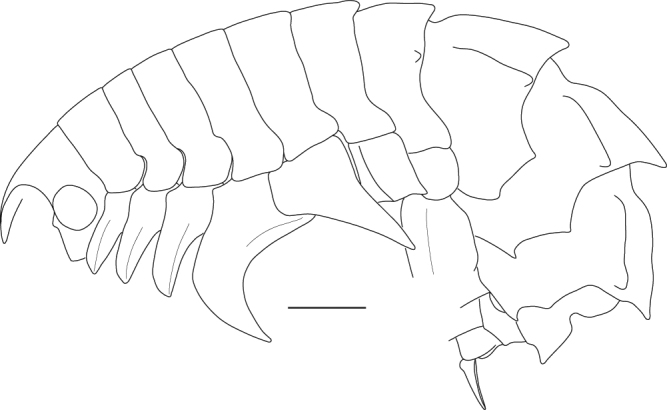
Epimeria cornigera, paratype, adult male, northern North Sea, NHMUK 2015.3269, lateral habitus; scale 1 mm.
Gammarus corniger Fabricius113, 1779: 383.
Epimeria tricristata Costa114, 1851: 46.
Acanthonotus owenii Bate115, 1857: 141. – Bate & Westwood116, 1863.
Acanthonotus testudo Bate117, 1862: 127.
Epimeria cornigera Boeck118, 1871: 185. – Sars55, 1893 364, pl 128. – Stebbing119, 1906: 323. – Stephensen56, 1938: 266. – Lincoln120, 1979: 436, fig. 207, 208a.
Type material examined
Holotype lost, type locality presumed as “coast of Norway”. Neotype (NHMUK 2015.3268), ovigerous female, 18 mm, northern North Sea – approx. 150 km west of Bergen (60.3104°, 2.4967°) in 97 m water depth, 05.viii.2013, coll. H. Neumann. Paratypes: two adult males, 14 mm (Fig. 19; NHMUK 2015.3269) and 14.5 mm (NHMUK 2015.3270) and seven adult females (6 ovigerous; NHMUK 2015.3271 − 3277), locality data identical to holotype.
Other material
Five males, three adult females (1 ovigerous) and one juvenile female (MT06700, MT06716-06717, MT06765-06769, MT06786), western central North Sea (57.394°, 0.2925°), 78 m, 10.iii.2012, coll. H. Neumann. One adult female (MT06718), western North Sea (54.8732°, −0.9581°), 68 m, 07.iii.2012, coll. H. Neumann. One male (MT06709), northern North Sea (60.3659°, 2.4386°), 96 m, 31.vii.2012, coll. I. Rottgardt. One adult female (MT06705), northern North Sea (60.3583°, 2.5371°), 106 m, 01.viii.2012, coll. I. Rottgardt. One ovigerous female (MT06702) western North Sea (55.1097°, −0.5023°), 82 m, 07.ii.2012, coll. H. Neumann. Two adult females (1 ovigerous; MT01169-01170), northern North Sea (60.3703°, 2.4784°), 100 m, 28.vii.2010, coll. H. Neumann. Two adult females (NHMUK 1985:233:2), North Sea (57.6000°, 0.5733°), 88–94 m, 1985, unknown collector. Five males, ten females (5 ovigerous) and 28 juveniles (NHMUK 1984:566:30), Irish Sea (Cumbrae Elbow.), 18 m (10 fathoms), 08.viii.1984, coll. R.J. Lincoln and G.A. Boxshall. One ovigerous female (NHMUK 1907:12:2:122), Irish Sea (Mull of Cantyre), unknown water depth and collection date, coll. University College of Dundee. One male and three ovigerous females (NHMUK 1887.21), Scottish coast? (“Loche Lyne”), unkown water depth and collection date, coll. J. Murray. Two ovigerous females (NHMUK 1986:606:2), North Sea (57.6050°, 0.5716°), 89 m, 24.viii.1985, coll. G. Cranmer. Norman Collection (NHMUK 1911:11:8): One male and four ovigerous females (16981-995), Trondheimsfjord (Norway), 365 m (200 fathoms), 1869; two adult females (16977-980), Hardangerfjord (Norway), 274–329 m, unknown collection date; ten adult females (16944-953), Durham Coast (western North Sea), unkown water depth and collection date. One adult female (MCSN 262), Irish Sea (53.7833°, −4.9666°), 80 m, 17.viii.1992, unknown collector. One ovigerous female (MCSN Z8-67), Bømlafjorden (Norway), water depth, sampling date and collector unknown. One male and one ovigerous female (MCSN 11177), Hjeltefjorden (60.4050°, 5.1169°), water depth and collection date unknown, coll. W. Vader.
Diagnosis
Body dorsally smooth on pereonites 1–5; pereonite 6 not produced or only slightly produced mid-dorsally; pereonite 7 with mid-dorsal process; pleonites 1–3 strongly carinate, each with a blunt dorsolateral ridge; epimeral plates 1–3 biangular, produced to subacute posterodistal teeth in epimera 2 and 3; urosomite 3 with only slightly produced rounded hump; palp of maxilla 1 distally with 6 blunt spines and few subdistal setae; coxa 4 apical tip subacute; coxa 5 anteroventral margin distinctly or strongly concave, apical tip of process subacute; uropod 2 outer ramus only slightly shorter than inner ramus (about 4/5).
Description
Based on female neotype, 18 mm.
Body (Fig. 14A,C) dorsally smooth on pereonites 1–5; pereonite 6 slightly produced mid-dorsally; pereonite 7 with mid-dorsal process and a rounded dorsolateral process posteriorly. Pleonites 1–3 strongly carinate, each with a blunt dorsolateral ridge; epimeral plates 1–3 biangular, produced to subacute posterodistal teeth on epimera 2–3 and broadly rounded on epimeron 1, angles on posterior margins blunt to broadly rounded. Urosomite 1 with apically rounded carina, notched mid-dorsally; urosomite 2 shortest; urosomite 3 with slightly produced rounded hump (Fig. 14D).
Head (Fig. 14B) about as high as long; ventral lobus slightly rounded, anterior corner rectangular. Rostrum curved, ventral margin rounded, about as long as 1st peduncular article of antenna 1. Eyes large and rounded, bulging.
Antenna 1 (Fig. 15A) peduncular article 1 about 1.5 the length of articles 2–3 combined (length ratio 1: 0.6: 0.3); accessory flagellum consisting of a single tapering article, about 1/3 the length of the 1st flagellar article; 1st flagellar article about as long as peduncular article 2, primary flagellum with 36 articles.
Antenna 2 (Fig. 15B) slightly longer than antenna 1; peduncular article 1 about as long as article 3, article 2 partly overlapping articles 1 and 3, with distal cusp; article 3 with distal cusp; article 4 slightly longer than article 5; flagellum with 39 articles.
Labrum (“upper lip”; Fig. 15G) ventrally rounded.
Hypopharynx (“lower lip”; Fig. 15C) with distally tapering lobes, densely coated with fields of setae on the medial margins.
Mandible (Fig. 15D) with 8-dentate incisor; lacinia mobilis bifid (Fig. 15E); molar protruding and slender; mandibular palp (Fig. 15F) consisting of 3 articles, bearing long setae on the distal half of article 2 and the entire article 3 posteromarginally, setae length on palp article 2 and 3 increasing from proximal to distal.
Maxilla 1 (Fig. 16A) inner plate sub-triangular, with 13 stout plumose setae along the medial margin; outer plate truncate, with 11 mediodistally serrated spines (Fig. 16B); palp 2-articulate, article 2 about 6 times longer than the 1st article, curved inwards, distally serrate with 6 blunt spines and few subdistal setae (Fig. 16B).
Maxilla 2 (Fig. 16C) inner lobe slightly broader and shorter than outer lobe, with numerous setae along the medial margin; outer lobe with a row of slender spines which are mediodistally serrate along with few short slender setae.
Maxilliped (Fig. 16D,E) inner plate about 2/3 the length of 1st palp article, apical ridge with blunt spines and stout setae; outer plate reaching about midpoint of palp article 2, ovoid, apical margin with slender spines which get stouter mediodistally, mediodistal region produced to a saw-like ridge; palp 4-articulate, mediodistal margin of article 1 with long setae, medial margin of article 2 strongly setose, article 3 with setae medio- and laterodistally, article 4 curved, with serrate medial margin.
Gnathopod 1 (Fig. 17A) coxa tapering distally and having a rounded subangular apical tip, anterior margin sinuous, posterior margin straight to slightly convex and bearing a row of short very fine setae along medioapically as well as longer setae subproximally; basis about as long as coxa, distinctly curved anteriorly, posteromarginally with tufts of long slender setae, anterior margin strongly setose with fine setae along its entire length; ischium slightly longer than wide, with tuft of setae posterodistally; merus with longitudinal articulation of carpus, posterior margin oblique, with group of setae posterodistally; carpus distally slightly expanded, with dense setation along the posterior margin and a tuft of shorter setae anterodistally; propodus about 0.8 of carpus length, posteriorly with setae along the margin and on the lateral face; palm convex, with fine serration; dactylus curved, with serrate posterior margin.
Gnathopod 2 (Fig. 17B,C) coxa slightly longer than coxa 1, tapering distally with a rounded subangular tip, anterior margin slightly sinuous, posterior margin distinctly convex medioapically with a row very fine short setae along the margin; basis slightly shorter than coxa, slightly curved anteriorly, posteromarginally with tufts of long slender setae, long slender setae along the entire anterior margin accompanied with few shorter slender setae; ischium slightly longer than wide, with group of setae posterodistally; merus similar in length and setation to gnathopod 1; carpus shape similar to gnathopod 1, with tufts of long setae on the posterior margin; propodus about 0.8 the length of carpus, with transverse rows of setae along the posterior margin; palm obliquely convex, with fine serration and 2 long blunt spines subdistally on the lateral face (Fig. 17C); dactylus as for gnathopod 1.
Pereopod 3 (Fig. 17D) coxa subequal to coxa 2 but slightly longer and wider; basis about 0.8 as long as coxa, sublinear but slightly curved anteriorly, posterior margin with tufts of long slender setae, anterior margin with long slender setae and a field of very short fine setae which extends to the lateral face; ischium about as long as wide; merus length about 1.3 of carpus length, slightly curved posteriorly and distally expanding, with few short setae at the posterior and anterior margins as well as groups of setae antero- and posterodistally; carpus slightly expanding distally, with tufts of stout setae posteromarginally; propodus about 1.1 the size of carpus, with 7 pairs of spine-like setae along the posterior margin; dactylus about half the size of propodus, curved.
Pereopod 4 (Fig. 18A) coxa strongly projecting ventrally and robust, anterior margin sinuous and strongly curved posteriorly, posterior margin excavate and with a centrally positioned rounded lobe, posteroapical margin strongly concave, apical tip subacute; basis linear, posterior margin convex and with long slender setae extending to the lateral face, anterior margin straight and with shorter slender setae; ischium slightly longer than wide, with a group of setae posterdistally; merus about 1.5 the size of carpus and sublinear, with few short setae along the margins and groups of setae antero- and posterodistally; carpus with tufts of stout setae along the posterior margin; propodus about 1.3 the size of carpus, with 6 pairs of spine-like setae posteromarginally; dactylus less than half the length of propodus, curved.
Pereopod 5 (Fig. 18B) coxa strongly extended posteroventrally, posterior margin slightly concave, anterior margin convex and strongly curved posteriorly, anteroventral margin distinctly concave, apical tip of process subacute; basis rectangular, with posteriorly produced margin which expands to a bifid subrectangular lobe posterodistally, partly covering ischium when leg is retracted, anterior margin with numerous setae along its entire length; ischium longer than wide, lobate; merus about 1.4 the length of carpus, slightly curved anteriorly, with short stout setae postero- and anterodistally; carpus slightly expanding distally, with tufts of spine-like setae along the anterior margin and posterodistally; propodus about 1.5 the size of carpus, with 7 tufts of spine-like setae anteromarginally; dactylus on right pereopod damaged, but clearly less than half the size of propodus, curved.
Pereopod 6 (Fig. 18C) coxa subrectangular, longer than wide with a laterally protruding ridge anteriorly and an angular lateral lobe pointing posterodistally, anterior margin with fine setae; basis slightly curved posteriorly, with lobe roundly expanding proximally on the medial side of the posterior margin and posteriorly produced lateral margin which expands to a bifid subrectangular lobe distally, posteromedial lobe with many slender setae mediodistomarginally, anterior margin with long slender setae proximally and few setae along the distal half; ischium longer than wide, lobate; merus about 1.3 the length of carpus, slightly curved anteriorly and slightly expanding distally, with very short setae along the posterior margin and tufts of setae postero- and anterodistally; carpus slightly expanding distally, with 5 groups of stout setae anteromarginally; propodus about 1.5 the length of carpus, with 7 groups of spine-like setae anteromarginally; dactylus about half the length of propodus, curved.
Pereopod 7 (Fig. 18D) on right body side missing; on left side all articles complete; coxa longer than wide; basis much wider than pereopod 6, posteriorly lobate, posterodistal lobe subacute and partly covering ischium, anterior margin with few short setae; ischium about as long as wide; merus slightly longer than carpus, slightly curved anteriorly, anterior margin with groups of short stout setae, posterodistal corner bearing two setae; carpus with 4 groups of stout setae anteromarginally; propodus about as long as carpus, with 7 groups of spine-like setae anteromarginally; dactylus about half the length of propodus, curved.
Uropod 1 (Fig. 18E,F) on right body side with damaged outer ramus, but undamaged on left body side; peduncle subrectangular, with small spines along medial margin, spine size slowly increasing from proximal to distal; rami subequal, about 1.2 the length of peduncle, margins bordered with small spines.
Uropod 2 (Fig. 18G) peduncle expanding distally, lateral margin with row of small spines; outer ramus slightly shorter than inner ramus (about 4/5), inner ramus about 1.7 the length of peduncle, spination similar to rami of uropod 1.
Uropod 3 (Fig. 18H) peduncle shortest, with small spines along the sublateral margin, and short setae on the medial margin; outer ramus slightly shorter than inner ramus, spination similar to rami of uropod 1 and 2.
Telson (Fig. 14E) about 1.5 as long as wide, slightly notched to about 15% of its length, apices pointedly rounded.
Variation
Similar to E. frankei, males of E. cornigera resemble their females, except for smaller body sizes. The described morphological differences between the sexes in E. frankei (see above) also apply to E. cornigera (Fig. 19).
Electronic supplementary material
Acknowledgements
We are grateful to Corinna Girschik and Rebekka Schüller for their help in the molecular laboratories of the DZMB. We also thank all crew members of the RV Walter Herwig III for their professional help and support as well as the BOLD team for managing the barcode data. Oliver Coleman (Museum für Naturkunde, Berlin) and Anne-Nina Lörz (University of Hamburg) kindly provided us with helpful comments on earlier parts of the manuscript. The first author further would like to thank Leonardo Latella, Roberta Salmaso (Museo di Storia Naturale, Verona) and particularly Miranda Lowe (Natural History Museum, London) for their hospitality and support. This study was part of the project “Molecular taxonomy of marine organisms (Metazoa) of the North Sea” supported by the German Federal Ministry of Education and Research (BMBF, grant no. 03F0664A) and the federal state of Niedersachsen. We would also like to thank Eva-Maria Priesterjahn for performing the Illumina library preparations and Alexandra Trinks and Johanna L.A. Paijmans for supervising her. The comments and suggestions of three anonymous reviewers greatly improved the quality of the manuscript.
Author Contributions
H.N. collected all specimens; J.B. conducted the morphological analysis and made the morphological species descriptions; M.V.W. and L.H. conducted the mitogenome analyses under supervision of M.H.; M.J.R. and F.D. performed all molecular sequence analyses; J.B. and M.J.R. wrote the manuscript with contributions of the other authors; J.B. and M.J.R. prepared all figures; original photograph in Fig. 6 was taken by H.N.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25225-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Padial JM, Miralles A, de la Riva I, Vences M. The integrative future of taxonomy. Front. Zool. 2010;7:16. doi: 10.1186/1742-9994-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuh R. The Linnean system and its 250-year persistence. Bot. Rev. 2003;69:59–78. [Google Scholar]
- 3.Bortulus A. Error cascades in the biological sciences: the unwanted consequences of using bad taxonomy in ecology. AMBIO. 2008;37:114–118. doi: 10.1579/0044-7447(2008)37[114:ecitbs]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Erwin TL. Tropical forests: their richness in Coleoptera and other arthropod species. Coleopts. Bull. 1982;36:74–75. [Google Scholar]
- 5.Stork N. How many species are there? Biodivers. Conserv. 1993;2:215–232. [Google Scholar]
- 6.May RR. Tropical arthropod species, more or less? Science. 2010;329:41–42. doi: 10.1126/science.1191058. [DOI] [PubMed] [Google Scholar]
- 7.May RR, Harvey PH. Species uncertainties. Science. 2009;323:687. doi: 10.1126/science.1170937. [DOI] [PubMed] [Google Scholar]
- 8.Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. How many species are there on earth and in the ocean? PloS Biol. 2011;9:e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quicke, D. L. J. Principles and Techniques of Contemporary Taxonomy. (Blackie Academic & Professional, 1993).
- 10.Mahamat H, et al. Isozyme analysis of Kenyan Phlebotomine sandflies (Diptera: Psychodidae) by isoleletric focusing (IEF) on Pharmacia Phast SystemTM. Biochem. Syst. Ecol. 1992;20:583–596. [Google Scholar]
- 11.Passamonti M, Mantovani B, Scali V. Karyotype and allozyme characterization of the Iberian Leptynia attenuata species complex (Insecta Phasmatodea) Zool. Sci. 1999;16:675–684. doi: 10.1016/s1055-7903(03)00156-8. [DOI] [PubMed] [Google Scholar]
- 12.Saiki R, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 13.Wilkerson RC, Parsons TJ, Albright DG, Klein TA, Braun MJ. Random amplified polymorphic DNA (RAPD) markers readily distinguish cryptic mosquito species (Diptera: Culicidae: Anopheles) Insect Mol. Biol. 1993;1:205–211. doi: 10.1111/j.1365-2583.1993.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 14.Frey JE, Frey B. Molecular identification of six species of scale insects (Quadraspidiotus sp.) by RAPD-PCR: assessing the field-specificity of pheromone traps. Mol. Ecol. 1995;4:777–780. [Google Scholar]
- 15.Costa FO, et al. Application of RAPD DNA fingerprinting in taxonomic identification of amphipods: a case-study with Gammarus species (Crustacea: Amphipoda) J. Mar. Biol. Assoc. UK. 2004;84:171–178. [Google Scholar]
- 16.Clark TL, Meinke LJ, Foster JE. PCR-RFLP of the mitochondrial cytochrome oxidase (subunit I) gene provides diagnostic markes for selected Diabrotica species (Coleoptera: Chrysomelidae) B. Entomol. Res. 2001;91:419–427. [PubMed] [Google Scholar]
- 17.Luchetti A, Mantovani B, Trentini M. Rapid identification of non-neosomic Tunga penetrans and Tunga trimamillata (Insecta Siphonaptera) specimens by PCR-RFLP methods. B. Insectol. 2005;58:15–18. [Google Scholar]
- 18.Arif IA, Khan HA. Molecular markers for biodiversity analysis of wildlife animals: a brief review. Anim. Biodiv. Conserv. 2009;32:9–17. [Google Scholar]
- 19.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994;3:294–299. [PubMed] [Google Scholar]
- 20.Geller J, Meyer C, Parker M, Hawk H. Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol. Ecol, Resour. 2013;13:851–861. doi: 10.1111/1755-0998.12138. [DOI] [PubMed] [Google Scholar]
- 21.Lobo J, et al. Enhanced primers for amplification of DNA barcodes from a broad range of marine metazoans. BMC Ecol. 2013;13:34. doi: 10.1186/1472-6785-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. P. Roy. Soc. Lond. B. Bio. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. P. Roy. Soc. Lond. B. Bio. 2003;270(Supplement):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raupach MJ, Radulovici AE. Looking back on a decade of barcoding crustaceans. Zookeys. 2015;539:53–81. doi: 10.3897/zookeys.539.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aliabadian M, et al. DNA barcoding of Dutch birds. Zookeys. 2013;365:25–48. doi: 10.3897/zookeys.365.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knebelsberger T, et al. A reliable DNA barcode reference library for the identification of the European shelf fish fauna. Mol. Ecol, Resour. 2014;14:1060–1071. doi: 10.1111/1755-0998.12238. [DOI] [PubMed] [Google Scholar]
- 27.Hendrich L, et al. A comprehensive DNA barcode database for Central European beetles with a focus on Germany: adding more than 3500 identified species to BOLD. Mol. Ecol, Resour. 2015;15:795–818. doi: 10.1111/1755-0998.12354. [DOI] [PubMed] [Google Scholar]
- 28.Raupach MJ, et al. The application of DNA barcodes for the identification of marine crustaceans from the North Sea and adjacent regions. PloS ONE. 2015;10:e0139421. doi: 10.1371/journal.pone.0139421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barco A, Raupach MJ, Laakmann S, Neumann H, Knebelsberger T. Identification of North Sea molluscs with DNA barcoding. Mol. Ecol, Resour. 2016;16:288–297. doi: 10.1111/1755-0998.12440. [DOI] [PubMed] [Google Scholar]
- 30.Hawlitschek O, et al. Comprehensive DNA barcoding of the herpetofauna of Germany. Mol. Ecol, Resour. 2016;16:242–253. doi: 10.1111/1755-0998.12416. [DOI] [PubMed] [Google Scholar]
- 31.Khalaji-Pirbalouty V, Raupach MJ. A new species of Cymodoce Leach, 1814 (Crustacea: Isopoda: Sphaeromatidae) based on morphological and molecular data, with a key to the Northern Indian Ocean species. Zootaxa. 2014;3826:230–254. doi: 10.11646/zootaxa.3826.1.7. [DOI] [PubMed] [Google Scholar]
- 32.Ng PKL, Meyer C. A new species of pea crab of the genus Serenotheres Ahyong & Ng, 2005 (Crustacea, Brachyura, Pinnotheridae) from the date mussel Leiosolenus Carpenter, 1857 (Mollusca, Bivalvia, Mytilidae, Lithophaginae) from the Solomon Islands. Zookeys. 2016;623:31–41. doi: 10.3897/zookeys.623.10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landschoff J, Lemaitre R. Differentiation of three common deep-water hermit crabs (Crustacea, Decapoda, Anomura, Parapaguridae) from the South African demersal abundance surveys, including the description of a new species of Paragiopagurus Lemaitre, 1996. Zookeys. 2017;676:21–45. doi: 10.3897/zookeys.676.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsukami S, Nakano T, Tomikawa K. A new species of the genus Nicippe from Japan (Crustacea, Amphipoda, Pardaliscidae) Zookeys. 2017;668:33–47. doi: 10.3897/zookeys.668.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raupach MJ, Amann R, Wheeler QD, Roos C. The application of “omics”-technologies for the classification and identification of animals. Org. Divers. Evol. 2016;16:1–12. [Google Scholar]
- 36.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson CC, et al. Microbial genomic taxonomy. BMC Genomics. 2014;14:913. doi: 10.1186/1471-2164-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun J, Rainey FA. Integrating genomics into the taxonomy and systematics of the Bacteria andArchaea. Int. J. Syst. Evol. Micr. 2014;64:316–324. doi: 10.1099/ijs.0.054171-0. [DOI] [PubMed] [Google Scholar]
- 39.Horton, T. et al. World AmphipodaDatabase. Accessed at, http://www.marinespecies.org/amphipoda on 2017-08-24 (2017).
- 40.Meyran JC, Monnerot M, Taberlet P. Taxonomic status and phylogenetic relationships of some species of Gammarus (Crustacea, Amphipoda) deduced from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 1997;8:1–10. doi: 10.1006/mpev.1996.0399. [DOI] [PubMed] [Google Scholar]
- 41.Costa FO, Henzler CM, Lunt DH, Whiteley NM, Rock J. Probing marine Gammarus (Amphipoda) taxonomy with DNA barcodes. Syst. Biodivers. 2009;7:365–379. [Google Scholar]
- 42.Hou Z, Li S. Intraspecific or interspecific variation: delimitation of species boundaries within the genus Gammarus (Crustacea, Amphipoda, Gammaridae), with description of four new species. Zool. J. Linn. Soc.-Lond. 2010;160:215–253. [Google Scholar]
- 43.Kilgallen NM, Lowry JK. The Tryphosa group (Crustacea: Amphipoda: Lysianassoidea: Lysianassidae: Tryphosinae) Zootaxa. 2014;3768:501–545. doi: 10.11646/zootaxa.3768.5.1. [DOI] [PubMed] [Google Scholar]
- 44.Delić T, Trontelji P, Rendŏs M, Fišer C. The importance of naming cryptic species and the conservation of endemic subterranean amphipods. Sci. Rep.-UK. 2017;7:3391. doi: 10.1038/s41598-017-02938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Havermans C, Nagy ZT, Sonet G, de Broyer C, Martin P. DNA barcoding reveals new insights into the diversity of Antarctic species of Orchomene sensu lato (Crustacea: Amphipoda: Lysianassoidea) Deep-Sea Res. Pt. II. 2011;58:230–241. [Google Scholar]
- 46.Verheye ML, Backeljau T, d’Udekem d’Acoz C. Looking beneath the tip of the iceberg: diversification of the genus Epimeria on the Antarctic shelf (Crustacea, Amphipoda) Polar Biol. 2016;39:925–945. [Google Scholar]
- 47.Havermans C, et al. Genetic and morphological divergences in the cosmopolitan deep-sea amphipod Eurythenes gryllus reveal a diverse abyss and a bipolar Species. PloS ONE. 2013;8:e74218. doi: 10.1371/journal.pone.0074218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reboleira ASPS, Enghoff H. Taxonomics – next-generation taxonomists. Org. Divers. Evol. 2016;16:679–680. [Google Scholar]
- 49.Kilpert F, Podsiadlowski L. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genomics. 2006;7:241. doi: 10.1186/1471-2164-7-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Cui Z. Complete mitochondrial genome of the Asian fiddler crab Charybdis japonica (Crustacea: Decapoda: Portunidae): gene arrangement of the marine brachyurans and phylogenetic considerations of the decapods. Mol. Biol. Rep. 2010;37:2559–2569. doi: 10.1007/s11033-009-9773-2. [DOI] [PubMed] [Google Scholar]
- 51.Ma H, et al. First mitochondrial genome for the red crab (Charybdis feriata) with implication of phylogenomics and genetics. Sci. Rep.-UK. 2015;5:11524. doi: 10.1038/srep11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dayrat B. Towards integrative taxonomy. Biol. J. Linn. Soc. 2005;85:407–415. [Google Scholar]
- 53.Schlick-Steiner BC, et al. Integrative taxonomy: a multisource approach to exploring biodiversity. Annu. Rev. Entomol. 2010;55:421–438. doi: 10.1146/annurev-ento-112408-085432. [DOI] [PubMed] [Google Scholar]
- 54.Moore PG. A functional interpretation of coxal morphology in Epimeria cornigera (Crustacea, Amphipoda, Paramphithoidae) J. Mar. Biol. Assoc. UK. 1981;61:749–757. [Google Scholar]
- 55.Sars, G. O. An account of the Crustacea of Norway with short descriptions and figures of all the species. Vol. 1: Amphipoda. Vol. 1. Amphipoda (Alb. Cammermeyers Forlag, 1893).
- 56.Stephensen K. The Amphipoda of North Norway and Spitsbergen with adjacent waters Part 2. Tromsø Museums Skrifter. 1938;3:141–278. [Google Scholar]
- 57.Stephensen K. Marine Amphipoda. The Zoology of Iceland. 1940;3:1–111. [Google Scholar]
- 58.DeSalle R, Egan MG, Siddall M. The unholy trinity: taxonomy, species delimitation and DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1905–1916. doi: 10.1098/rstb.2005.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lörz A-N, Maas EW, Linse K, Coleman CO. Do circum-Antarctic species exist in peracarid Amphipoda? A case study in the genus Epimeria Costa, 1851. Zookeys. 2009;18:91–128. [Google Scholar]
- 60.Lefébure T, et al. Phylogeography of a subterranean amphipod reveals cryptic diversity and dynamic evolution in extreme environments. Mol. Ecol. 2006;15:1797–1806. doi: 10.1111/j.1365-294X.2006.02888.x. [DOI] [PubMed] [Google Scholar]
- 61.Rock J, Ironside J, Potter T, Whiteley NM, Lunt DH. Phylogeography and environmental diversification of a highly adaptable marine amphipod. Gammarus duebeni. Heredity. 2007;99:102–111. doi: 10.1038/sj.hdy.6800971. [DOI] [PubMed] [Google Scholar]
- 62.Villacorta C, Jaume D, Oromi P, Juan C. Under the volcano: phylogeography and evolution of the cave-dwelling Palmorchestia hypogaea (Amphipoda, Crustacea) at La Palma (Canary Islands) BMC Biol. 2008;6:7. doi: 10.1186/1741-7007-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lörz A-N, Brandt A. Phylogeny of Antarctic Epimeria (Epimeriidae: Amphipoda) J. Mar. Biol. Assoc. UK. 2004;84:179–190. [Google Scholar]
- 64.d’Udekem d’Acoz C, Verheye ML. Epimeria of the Southern Ocean with notes on their relatives (Crustacea, Amphipoda, Eusiroidea) Euro. J. Taxon. 2017;359:1–553. [Google Scholar]
- 65.Verheye ML, Backeljau T, d’Udekem d’Acoz C. Locked in the icehouse: Evolution of endemic Epimeria (Amphipoda, Crustacea) species flock on the Antarctic shelf. Mol. Phylogenet. Evol. 2017;114:14–33. doi: 10.1016/j.ympev.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 66.Lecointre G, et al. Is the species flock concept operational? The Antarctic shelf case. PloS ONE. 2013;8:e68787. doi: 10.1371/journal.pone.0068787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lörz A-NE. Crustacea, Amphipoda) from New Zealand with a description of a new species. Zootaxa. 2008;1847:49–61. [Google Scholar]
- 68.Romanova EV, et al. Evolution of mitochondrial genomes in Baikalian amphipods. BMC Genomics. 2016;17:1016. doi: 10.1186/s12864-016-3357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pie MR, Oliveira-Neto JF, Boeger WA, Ostrensky A, Baggio RA. The organization of the mitochondrial control region in 2 brachyuran crustaceans: Ucides cordatus (Ocypodidae) and Cardisoma guanhum (Gecarcinidae) J. Hered. 2008;99:432–437. doi: 10.1093/jhered/esn024. [DOI] [PubMed] [Google Scholar]
- 70.Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. Am. J. Hum. Genet. 2000;67:1029–1032. doi: 10.1086/303092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grabowski, M. & Stuck, K. C. In Crustaceans and the biodiversity crisis, Vol. I (eds F. R. Schram & J. C. von Vaupel Klein) 333–344 (Brill Academic Publishers, 1999).
- 72.Ehrich S, et al. 20 years of the German Small-Scale Bottom Trawl Survey (GSBTS): A review. Senck. Marit. 2007;37:13–82. [Google Scholar]
- 73.Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes. 2007;7:544–548. [Google Scholar]
- 74.Raupach MJ, Held C, Wägele JW. Multiple colonization of the deep sea by the Asellota (Crustacea: Peracarida: Isopoda) Deep-Sea Res. Pt. II. 2004;51:1787–1795. [Google Scholar]
- 75.Raupach MJ, Mayer C, Malyutina M, Wägele JW. Multiple origins of deep-sea Asellota (Crustacea: Isopoda) from shallow waters revealed by molecular data. Proc. R. Soc. B. 2009;276:799–808. doi: 10.1098/rspb.2008.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dreyer H, Wägele JW. Parasites of crustaceans (Isopoda: Bopyridae) evolved from fish parasites: molecular and morphological evidence. Zoology. 2001;103:157–178. [Google Scholar]
- 77.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z, Schwatz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 79.Morgulis M, et al. Database indexing for production MEGABLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ratnasingham S, Hebert PDN. BOLD: The Barcode of Life data systems. Mol. Ecol. Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ratnasingham S, Hebert PDN. A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PloS ONE. 2013;8:e66213. doi: 10.1371/journal.pone.0066213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 6, pdb.prot5448 (2010). [DOI] [PubMed]
- 83.Fortes GG, Paijmans JL. Analysis of whole mitogenomes from ancient samples. Methods Mol. Biol. 2015;1347:179–195. doi: 10.1007/978-1-4939-2990-0_13. [DOI] [PubMed] [Google Scholar]
- 84.Martin W. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011;17:10–12. [Google Scholar]
- 85.Xu H, et al. FastUniq: A fast de novo duplicates removal tool for paired short reads. PloS ONE. 2012;7:e52249. doi: 10.1371/journal.pone.0052249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 2013;41:e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chevreux B, Wetter T, Suhai S. Genome sequence assembly using trace signals and additional sequence information. German Conference of Bioinformatics. 1999;99:45–56. [Google Scholar]
- 88.Westbury M, et al. A mitogenomic timetree for Darwin’s enigmatic South American Mammal Macrauchenia patachonica. Nat. Commun. 2017;8:15951. doi: 10.1038/ncomms15951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bernt M, et al. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 91.Laslett D, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 92.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 93.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 95.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 97.Darriba D, Taboada GL, Doallo R, Posada D. jModeltest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hasegawa M, Kishino H, Yano T-A. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 99.Cannone JJ, et al. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Crease TJ, Taylor D. The origin and evolution of variable-region helices in V4 and V7 of the small-subunit ribosomal RNA of branchiopod crustaceans. Mol. Biol. Evol. 1998;15:1430–1446. doi: 10.1093/oxfordjournals.molbev.a025871. [DOI] [PubMed] [Google Scholar]
- 101.Hwang UO, Ree HI, Kim W. Evolution of hypervariable regions, V4 and V7, of insect 18S rRNA and their phylogenetic implications. Zool. Sci. 2000;17:111–121. doi: 10.2108/zsj.17.111. [DOI] [PubMed] [Google Scholar]
- 102.Gillespie JJ, Johnston JS, Cannone JJ, Gutell RR. Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): structure, organization, and retrotransposable elements. Insect Mol. Biol. 2006;15:657–686. doi: 10.1111/j.1365-2583.2006.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids S. 1999;41:95–98. [Google Scholar]
- 105.Coleman CO. “Digital inking”: How to make perfect line drawings on computers. Org. Divers. Evol. 3, Electronic Supplement. 2003;14:1–14. [Google Scholar]
- 106.Coleman CO. Drawing setae the digital way. Zoosyst. Evol. 2009;85:305–310. [Google Scholar]
- 107.d’Udekem d’Acoz C. Contribution to the knowledge of European Liljeborgiidae (Crustacea, Amphipoda), with considerations on the family and its affinities. Bull. Inst. R. Sc. N. B.-S. 2010;80:127–259. [Google Scholar]
- 108.Krapp-Schickel T. New Antarctic stenothoids sensu lato (Amphipoda, Crustacea) Euro. J. Taxon. 2011;2:1–17. [Google Scholar]
- 109.Chevreux E, Fage L. Amphipodes. Faune de France. 1925;9:488. [Google Scholar]
- 110.Karaman GS. On some new or very interesting Amphipoda of the Adriatic Sea. Mem. Mus, Civ. S. N. Verona. 1972;20:99–147. [Google Scholar]
- 111.Ledoyer M. Contribution à l’etude de l’ecologie de la faune vagile profonde de la Méditerranée nord occidentale I. Les Gammariens. B. Mus. Civ. S. N. Verona. 1977;4:321–421. [Google Scholar]
- 112.Ledoyer, M. In The Amphipoda of the Mediterranean (ed S. Ruffo) 616-617 (Mémoirs de l’Institut Océanographique 13 (Part 3), 1993).
- 113.Fabricius, J. C. Reise nach Norwegen, mit Bemerkungen aus der Naturhistorie und Oekonomie. (Carl Ernst Bohn, 1779).
- 114.Costa, A. In Catalogo dei Crostacei italiani e di molti altri del Mediterraneo (ed Fr. Gugl. Hope) 48 (F. Azzolino, 1851).
- 115.Bate CS. A synopsis of the British edriophthalmous Crustacea - Part I. Amphipoda. Ann. Mag. Nat. Hist. 1857;19:135–152. [Google Scholar]
- 116.Bate, C. S. & Westwood, J. O. A history of the British sessile-eyed Crustacea. Vol. I., (John Van Voorst, 1863).
- 117.Bate, C. S. Catalogue of the specimens of amphipodous Crustacea in the collection of the British Museum. 399 (BritishMuseum, 1862).
- 118.Boeck ACA. Borealia et Arctica. Forhandlinger i Vidensk.-Selsk. i Christiana. 1871;1870:83–280. [Google Scholar]
- 119.Stebbing, T. R. R. In Das Tierreich, 21. Lieferung (ed. F. E. Schulze) 806 pp (R. Friedländer und Sohn, 1906).
- 120.Lincoln, R. J. British marine Amphipoda: Gammaridea. (British Museum (Natural History), 1979).
- 121.Enequist P. Studies on the soft-bottom amphipods of the Skagerak. Zool. Bidr. Upps. 1949;28:269–491. [Google Scholar]
- 122.Ellis JR, Martinez I, Burt GJ, Scott BE. Epibenthic assemblages in the Celtic Sea associated with the Jones Bank. Prog. Oceanogr. 2013;117:76–88. [Google Scholar]
- 123.Carpentieri P, Serpetti N, Colloca N, Criscoli A, Ardizzone G. Food preferences and rhythms of feeding activity of two co-existing demersal fish, the longspine snipefish, Macroramphosus scolopax (Linnaeus, 1758), and the boarfish Capros aper (Linnaeus, 1758), on the Mediterranean deep shelf. Mar. Ecol. 2016;37:106–118. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.