Abstract
Mycobacterium smegmatis FenA is a nucleic acid phosphodiesterase with flap endonuclease and 5′ exonuclease activities. The 1.8 Å crystal structure of FenA reported here highlights as its closest homologs bacterial FEN-family enzymes ExoIX, the Pol1 exonuclease domain and phage T5 Fen. Mycobacterial FenA assimilates three active site manganese ions (M1, M2, M3) that are coordinated, directly and via waters, to a constellation of eight carboxylate side chains. We find via mutagenesis that the carboxylate contacts to all three manganese ions are essential for FenA’s activities. Structures of nuclease-dead FenA mutants D125N, D148N and D208N reveal how they fail to bind one of the three active site Mn2+ ions, in a distinctive fashion for each Asn change. The structure of FenA D208N with a phosphate anion engaged by M1 and M2 in a state mimetic of a product complex suggests a mechanism for metal-catalyzed phosphodiester hydrolysis similar to that proposed for human Exo1. A distinctive feature of FenA is that it does not have the helical arch module found in many other FEN/FEN-like enzymes. Instead, this segment of FenA adopts a unique structure comprising a short 310 helix and surface β-loop that coordinates a fourth manganese ion (M4).
INTRODUCTION
Structure-specific DNA endonucleases of the FEN (flap endonuclease) and FEN-like family are widely distributed in Nature and play important roles in DNA replication, repair and recombination (1,2). FEN enzymes that incise 5′ flap junctions are exemplified by: (i) the N-terminal 5′ exonuclease domains of bacterial DNA polymerase I (3–7); (ii) free-standing bacterial FEN enzymes such as Escherichia coli ExoIX, Bacillus subtilis YpcP and Mycobacterium smegmatis FenA (7–9); (iii) bacteriophage enzymes T5 Fen (10,11) and T4 Fen/RNaseH (12,13); (iv) eukaryal FEN1 (14–16); and (v) archaeal FENs (17–19). Human ExoI is an exemplar of the FEN-like clade (20). Other members of the FEN/FEN-like family include the nucleotide excision repair nuclease Rad2 (21) and the Holliday junction resolvase GEN1 (22,23). The FEN/FEN-like family are metal-dependent phosphodiesterases. Crystal structures of family members highlight a conserved protein fold and an active site that includes either two or three divalent cations (8,10–13,16–24). Structures of T5 Fen, T4 RNaseH, human FEN1 and human ExoI in complexes with DNA (11,13,16,20,25,26) collectively indicate that the enzyme engages a 12-bp duplex DNA segment flanking the flap or nick via contacts to a helix-turn-helix motif, enforces a sharp bend in the template DNA strand at the flap/nick junction by insertion of a protein α-helix between the splayed-apart template strand nucleobases, and positions the scissile phosphodiester for metal-catalyzed hydrolysis. The 5′ flap of the scissile strand is threaded through a hole in the protein formed by a helical arch.
We recently identified M. smegmatis FenA (Figure 1B) as a stand-alone 5′ flap endonuclease on the basis of its homology to the N-terminal 5′ exonuclease domain of mycobacterial DNA polymerase I (Pol1) (7) (Figure 2B). A stand-alone FenA is also present in the human pathogen Mycobacterium tuberculosis (Figure 2B). The purified recombinant 319-aa FenA protein catalyzes manganese-dependent endonucleolytic incision of a nicked DNA with a 5′ flap, predominantly between the first and second nucleotides of the duplex segment of the flap strand. Initial FenA cleavage of the 5′-PO4 strand at a DNA nick (i.e. its exonuclease activity) also occurs between the first and second nucleotides of the duplex. In addition, FenA removes the 5′ App(dN) terminus of an aborted nick ligation reaction intermediate (7). Such lesions are formed under a variety of circumstances by bacterial NAD+-dependent DNA ligases (27) and especially by the bacterial non-homologous end-joining (NHEJ) ligases, DNA ligases D and C (28–30). The flap endonuclease and AppDNA removal activities of FenA point toward a role in gap repair during lagging strand DNA replication or NHEJ. The flap removal activity of FenA might also come into play in the mycobacterial single-strand annealing pathway of DNA double-strand break (DSB) repair that entails large DNA deletions between homologous regions flanking the DSB (31).
Figure 1.
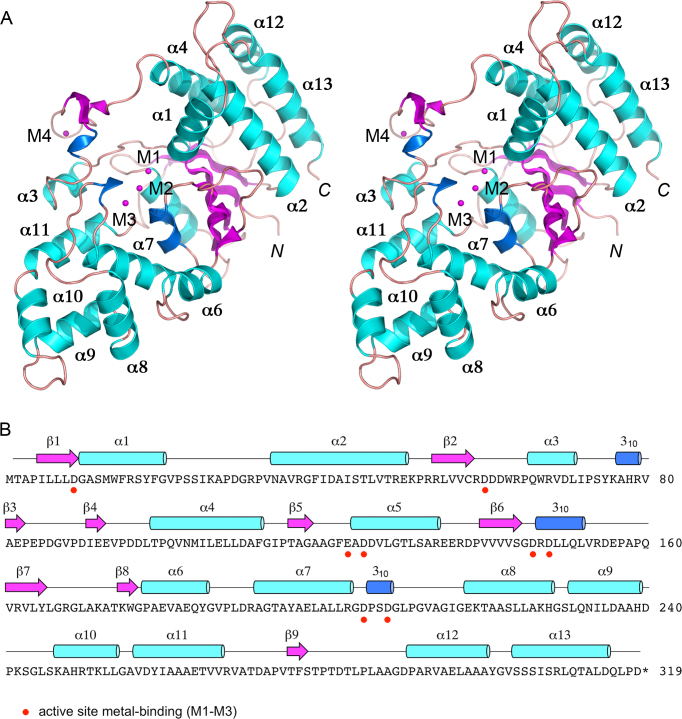
Overview of the FenA structure. (A) Stereo view of the tertiary structure of FenA, depicted as a cartoon model with magenta β strands, cyan α helices (numbered sequentially), and blue 310 helices. The N and C termini are indicated in italics. Three manganese ions in the active site (M1, M2, M3) and a fourth manganese (M4) engaged by the β3–β4 loop are shown as magenta spheres. (B) The secondary structure elements of FenA are shown above the FenA amino acid sequence, labeled sequentially and colored as in panel A. The eight acidic amino acid side chains that bind the active site metals are denoted by red dots below FenA the primary structure.
Figure 2.
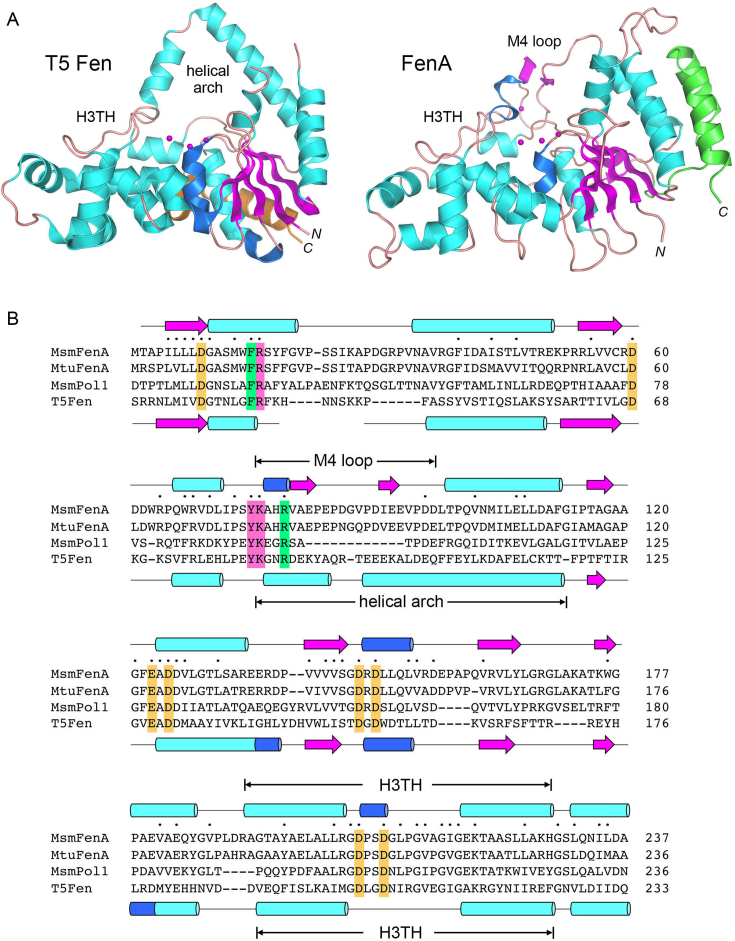
Comparison of mycobacterial FenA to T5 Fen. (A) Side-by-side superposition of the structures of bacteriophage T5 Fen with three Mg2+ ions in the active site (at left, from accession code: 5HMM) and FenA with three Mn2+ ions in the active site (at right). Cartoon models of the tertiary structures are shown colored according to secondary structure, except for the two C-terminal α helices that are colored gold in T5 Fen and green in FenA to highlight their different spatial locations. The N and C termini are labeled in italics. The conserved DNA-binding H3TH motifs are labeled. The helical arch of T5 Fen is not conserved in FenA, which has a distinctive M4 loop as shown. (B) The amino acid sequence of Mycobacterium smegmatis (Msm) FenA from aa 1 to 237 is aligned to the corresponding sequences of the homologous Mycobacterium tuberculosis FenA (aa 1 to 236), the N-terminal 5′ exonuclease domain of M. smegmatis Pol1 (aa 18–233) and bacteriophage T5 Fen (aa 3–233). Gaps in the alignment are denoted by dashes. Positions of side chain identity/similarity in all four polypeptides are denoted by dots above the MsmFenA sequence. Secondary structure elements of FenA and T5 Fen (colored as in Figure 1) are shown above and below their amino acid sequences. Amino acids subjected to mutational analysis in the present study are highlighted in shaded boxes: gold for amino acids coordinating metals M1, M2 and M3; magenta for nearby amino acids in the active site; green for amino acids that, in T5 Fen, make contacts to DNA.
Our goal in the present study was to determine the atomic structure of mycobacterial FenA in complex with its manganese cofactor, in order to gain insights into its mechanism and its relatedness to other FEN/FEN-like enzymes. We report that the FenA active site contains three manganese ions (M1, M2 and M3), similar to the three-magnesium structure determined for T5 Fen (11). A distinctive feature of FenA is that it does not have the helical arch module found in many other FEN/FEN-like enzymes. Rather, the analogous segment of FenA comprises a short 310 helix and surface β-loop that engages a fourth manganese ion (M4). We find that the direct and water-bridged side chain carboxylate contacts to all three active site manganese ions are essential for FenA’s flap endonuclease and 5′ exonuclease activities. Determining atomic structures of three nuclease-dead FenA Asp-to-Asn mutants revealed how these changes elicit a failure to bind one of the three active-site Mn2+ ions, in a distinctive fashion for each Asn change, without affecting overall protein structure. The structure of FenA with a phosphate anion coordinated by M1 and M2 in a state mimetic of a product complex suggests a mechanism for metal-catalyzed phosphodiester hydrolysis.
MATERIALS AND METHODS
Recombinant M. smegmatis FenA
We produced wild-type (WT) and mutant FenA proteins in E. coli as His10Smt3 fusions and isolated them from soluble extracts of 3-liter bacterial cultures by nickel-agarose chromatography as described previously (7). The His10Smt3 tags were removed with the Smt3-specific protease Ulp1, and native FenA proteins were separated from the tag by a second round of Ni-agarose chromatography. Proteins for crystallography were further purified by Superdex-200 gel filtration as described (7). Selenomethionine-(SeMet)-substituted WT FenA was produced as follows. Plasmid pET28-His10Smt3-FenA was transformed into E. coli B834(DE3), a methionine auxotroph. A single transformant was inoculated into 10 ml of LB medium containing 60 μg/ml kanamycin and incubated for 7 h at 37°C. The bacteria were harvested by centrifugation and then resuspended in 10 ml of complete LeMaster medium containing 60 μg/ml kanamycin. This step was repeated twice and the cells were finally resuspended in 400 ml of complete LeMaster medium (with kanamycin) containing 25 μg/ml SeMet (L-enantiomer). After overnight incubation, the culture volume was increased to 4 liters with fresh LeMaster medium containing kanamycin and SeMet, and growth was continued at 37°C with constant shaking until the A600 reached 0.4. The culture was adjusted to 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and 2% ethanol and incubated for 16 h at 17°C with continuous shaking. SeMet-FenA was then purified following the procedures described above for native FenA. The protein concentrations of the Superdex FenA preparations were determined by using the BioRad dye reagent with bovine serum albumin as the standard.
FenA-Ala and FenA-Asn missense mutations were introduced into the pET28b-His10Smt3-FenA expression plasmid by two-step overlap extension polymerase chain reaction (PCR) or by using the QuikChange method with NEB Phusion polymerase. The plasmid inserts were sequenced to verify that no unwanted coding changes were introduced during PCR amplification and cloning. WT and mutant FenA proteins used for nuclease assays were purified through the second Ni-agarose step described above, then concentrated by centrifugal ultrafiltration and stored at −80°C. Total protein concentrations of the concentrated FenA preparations were measured by using the Bio-Rad dye reagent. The concentrations of the FenA polypeptides were determined by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of 5 μg aliquots of each protein preparation in parallel with 1.25, 2.5, 5, 10 μg of bovine serum albumin (BSA) standards. The gels were stained with Coomassie blue, and the extents of dye binding to FenA and BSA polypeptides were quantified using Chemidoc XRS imager and ImageJ. The concentrations of the FenA proteins were calculated by interpolation to the BSA standard curve.
Crystallization, diffraction data collection and structure determination
FenA crystals were grown by sitting drop vapor diffusion at room temperature. A 1 μl aliquot of a solution of 4 mg/ml FenA and 10 mM MnCl2 in buffer containing 20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM DTT and 10% glycerol was mixed with 1 μl of precipitant solution and equilibrated in a 96-well plate against 150 μl of reservoir solution. The SeMet-WT FenA precipitant and reservoir solution solutions were 0.4 M NH4Cl, 28% PEG3350. The D125N precipitant solution was 0.2 M NaCl, 20% PEG3350 and the reservoir solution was 0.5 M LiCl. The D148N precipitant solution was 0.2 M LiCl, 20% PEG3350 and the reservoir solution was 0.5 M LiCl. The D208N precipitant solution was 0.2 M ammonium acetate, 20% PEG3350 and the reservoir solution was 0.5 M LiCl. The crystals were cryoprotected in a solution of 70% sucrose, 50 mM MnCl2 (for SeMet-WT, D125N and D148N) or 70% sucrose (D208N) before being flash-frozen in liquid nitrogen. X-ray diffraction data were collected from single crystals at APS beamline 24ID-C for SeMet-WT and D208N and at beamline 24ID-E for D148N and D125N. The SeMet-WT, D208N and D148N data were indexed with HKL2000 (32); D125N data were indexed with XDS (33). The cutoff criteria for X-ray diffraction data were CC(1/2) ≥0.6 and Rpim < 0.5 in the highest resolution shell. All FenA crystals were in space group P21. Phases were obtained using single-wavelength anomalous dispersion (SAD) data from a single crystal of SeMet-WT FenA, processed with SHELX (34) to locate two Se sites and one Mn site. After solvent flattening in Phenix (35), interactive model building was done using O (36). Refinement was accomplished with Phenix. Data collection and refinement statistics are compiled in Supplementary Table S1.
Nuclease assays
The nick and 5′ flap-nick substrates, prepared as described (7), were composed of an 18-mer DNAOH strand and a 5′ 32P-labeled 18-mer or 28-mer pDNA strand annealed to a 36-mer DNA template strand. Nuclease reaction mixtures (10 μl) containing 20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM MnCl2, 1 mM DTT, 1 pmol (0.1 μM) 32P-labeled DNA substrate and WT or mutant FenA (as specified in the figure legends) were incubated at 37°C for 30 min. The reactions were quenched by adding 10 μl of 90% formamide, 50 mM ethylenediaminetetraacetic acid (EDTA). The samples were heated at 95°C for 5 min and then analyzed by electrophoresis through a 40-cm 18% polyacrylamide gel containing 7.5 M urea in 44.5 mM Tris-borate, pH 8.3, 1 mM EDTA. The products were visualized and quantified by scanning the gel using a Typhoon FLA7000 imager. The extents of DNA cleavage were plotted as a function of input enzyme, with each datum being the average of at least three separate enzyme titration experiments (±SEM). The specific activities of the FenA preparations were calculated from the slopes of the titration curves in the linear range of FenA dependence, as implemented by linear regression in Prism. The standard errors of the linear regression fits for FenA proteins that had detectable activity varied from 2 to 16% of the calculated specific activity values. The specific activities of the mutant FenA enzymes were normalized to that of WT FenA (defined as 100%).
RESULTS
Structure of mycobacterial FenA in complex with manganese
The structure of FenA was determined using SAD phases obtained for a crystal of SeMet-substituted FenA in space group P21 grown from a solution of 0.1 mM SeMet-FenA and 10 mM MnCl2. The refined model at 1.8 Å resolution (Rwork/Rfree 0.174/0.205; Supplementary Table S1) comprised a continuous FenA polypeptide from amino acids 2–318 (Figure 1A). The secondary structure elements (12 β strands, 13 α helices and 3 310 helices) are displayed over the primary structure in Figure 1B. The tertiary structure includes a 7-strand β-sheet with topology β8↓•β7↑•β6↑•β1↑•β2↑•β5↑•β9↓, proceeding from front to back in the view in Figure 1A. The sheet is flanked on the upper right side by a five-helix bundle (α1, α2, α4, α12, α13) and on the lower left side by the other eight α helices, the 310 helices, and a surface β3–β4 loop (Figure 1A). The FenA model includes three Mn2+ ions in the active site (M1, M2 and M3) and a fourth Mn2+ (M4) engaged by the β3–β4 loop (Figure 1A). Eight acidic amino acids that coordinate the three manganese ions in the active site are denoted by red dots below the primary structure in Figure 1B.
Relatedness of FenA to other FEN family members
A DALI search (37) with the FenA structure identified other FEN/FEN-like proteins as homologs with Z-scores > 10 (see Supplementary Table S2). The top DALI hits, with Z scores greater than 20, were: E. coli ExoIX (Z score 25.1; 2.0 Å rmsd at 243 Cα positions with 25% amino acid identity); the N-terminal 5′ exonuclease domain of Taq DNA polymerase (Z score 23.1; 3.1 Å rmsd at 254 Cα positions with 26% amino acid identity); and bacteriophage T5 Fen (Z-score 22.5; 2.7 Å rmsd at 242 Cα positions with 21% amino acid identity). Figure 2A shows a side-by-side superposition of the structure of the 276-aa T5 Fen protein with three Mg2+ ions in the active site (11) on the present structure of FenA with three Mn2+ ions in the active site. The homology spans the N-terminal 240-aa segment of M. smegmatis FenA, the primary structure of which is aligned in Figure 2B to the corresponding segments of M. tuberculosis FenA, the 5′ exonuclease/Fen domain of mycobacterial Pol1 (6,7), and T5 Fen, revealing 64 positions of amino acid identity/similarity in all four proteins. The eight acidic amino acids that bind the three active site metal ions are strictly conserved and are highlighted in gold shading in Figure 2B. The secondary structure elements of FenA and T5 Fen are arrayed above and below their primary structures in Figure 2B and are mostly conserved, with exceptions as noted below.
FenA shares with T5 Fen and other FEN/FEN-like enzymes the helix–turn–helix motif (indicated by H3TH in Figure 2A and B) that engages the duplex DNA segment preceding the duplex-flap junction (11,13,16,20) as well as the ‘wedge’ helix (α1 in FenA) that enforces a sharp bend in the template DNA strand at the flap/nick junction (13,16,20). FenA deviates from T5 Fen in that it lacks the helical arch (Figure 2A) that creates an aperture through which the 5′ flap DNA is shown to thread, e.g. in T5 Fen, human FEN1 and human ExoI (11,16,20). Whereas the structures of some FEN family members that were crystallized without DNA reveal that segments corresponding to the helical arch are disordered (12,38), this is not the case in FenA. Rather, the FenA segment of primary structure between α3 and α4 adopts a distinctive secondary structure and fold (i.e. vis à vis T5 Fen), comprising a short 310 helix and a surface β-loop that coordinates the M4 manganese ion. The M4 β-loop is conserved in length and amino acid sequence in M. tuberculosis FenA, but is missing from the 5′ exonuclease domain of mycobacterial Pol1 (Figure 2B).
Another distinction between FenA and T5 Fen is that the two-helix bundles at their respective C-termini reside on different surfaces of the enzymes. These C-terminal α helices in T5 Fen (aa 262–290, colored gold in Figure 2A) pack against the α5 helix (aa 128–140) on the back surface of the protein as viewed in Figure 2A. By contrast, the C-terminal α12 and α13 helices of FenA (aa 291–318, colored green in Figure 2A) lie directly over the α2 and α4 helices on the rightward face of the enzyme as viewed in Figures 1A and 2A; the C-terminal helices are connected to the rest of the FenA polypeptide by a long loop (aa 268–290) that drapes across the back surface of the protein (Figure 1A and B). Equivalents of the FenA α12 and α13 helices are present in the same spatial position in human FEN1 and yeast Rad2 (16,21). However, the 251-aa E. coli ExoIX enzyme (the best homology hit to FenA per DALI) has no such C-terminal helices (8), nor does the 352-aa catalytic domain of human Exo1 (20).
Metal binding in the FenA active site
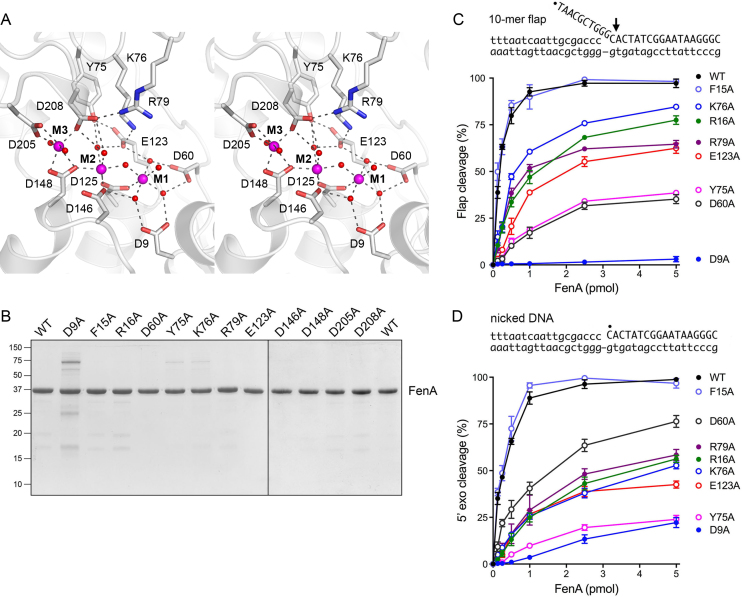
We observed three Mn2+ ions in the active site, affirmed by three discrete peaks of anomalous scattering in the SeMet-FenA dataset (Supplementary Figure S1) and in a native FenA dataset collected at the wavelength corresponding to Mn absorption peak (not shown). Each manganese ion is coordinated by a rich network of direct and water-mediated contacts to the carboxylate oxygens of conserved acidic side chains (shaded in gold in Figure 2B). The eight metal-binding residues in FenA are: Asp9, Asp60, Glu123 and Asp125 (site M1); Asp125, Asp146, Asp148 and Asp208 (site M2); Asp148, Asp205 and Asp208 (site M3) (Figure 3A). Sites M1 and M2 comprise what had been referred to previously as the ‘Cat1’ site. The M1 and M2 metal ions, spaced 3.6 Å apart, coordinate a bridging water. Asp125 makes direct bidentate contact to the M1 and M2 metals. The four remaining sites in the octahedral M1 complex are filled by waters bridged to side chains Asp9, Asp60 and Glu123. The four remaining positions in the octahedral M2 complex are occupied by the Asp146 and Asp148 Oδ2 atoms and by two waters—engaged to Asp208 and Glu123 Oϵ1, respectively (Figure 3A). The M3 site (previously named ‘Cat2’) is conserved among a subset of FEN/FEN-like family members, e.g. it is not present in E. coli ExoIX (8), notwithstanding the close overall homolog of ExoIX to FenA. The M3 manganese in FenA is located 4.1 Å away from M2. The M3 coordination complex is occupied by Asp148 Oδ1 and Oδ2, Asp205 Oδ2, Asp208 Oδ2 and two waters. Other conserved FenA side chains make noteworthy atomic contacts to the outer shell of the metal complexes. The Tyr75 OH makes bifurcated hydrogen bonds to an M2 water and Glu123 Oϵ2 and the Lys76 Nζ makes an ion pair with Glu123 Oϵ1 (Figure 3A). [The equivalent Tyr82 and Lys83 side chains in T5 Fen (Figure 2B; shaded in magenta) make the same contacts to the conserved metal-binding Glu128.] FenA Arg79 makes a salt bridge to Asp208 (Figure 3A). In the structure of T5 Fen bound to a flap duplex DNA, the equivalent arginine (Arg76; shaded in green in Figure 2B) makes a network of hydrogen bonds from its guanidinium nitrogens to the non-bridging phosphate oxygens of the first three nucleotides of the flap strand adjacent to the scissile phosphodiester (11).
Figure 3.
Active site architecture and structure-guided mutagenesis. (A) Stereo view of the FenA active site highlighting the octahedral coordination complexes of the manganese ions M1, M2 and M3 (depicted as magenta spheres). Selected amino acid side chains that coordinate the metals, the metal-bound waters (red spheres), and the metal-binding carboxylates are rendered as stick models against a cartoon trace of the FenA tertiary structure. Atomic contacts are denoted by dashed lines. (B) FenA mutants. Aliquots (5 μg) of the preparations of WT FenA and the indicated alanine mutants were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (in kilodaltons) of marker polypeptides are indicated on the left. (C and D) Mutational effects on flap endonuclease (C) and 5′ exonuclease activity (D). Reaction mixtures (10 μl) containing 20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM MnCl2, 1 mM DTT and 1 pmol (100 nM) of either 32P-labeled 10-mer flap-nicked duplex DNA (panel C) or 32P-labeled nicked duplex DNA (panel D), and increasing amounts of WT or mutant FenA were incubated for 30 min at 37°C. The DNA substrates are depicted above the graphs, with the 5′ 32P label denoted by •. The reaction products were analyzed by urea-PAGE and visualized and quantified by scanning the gel with a phosphorimager. (The predominant site of cleavage is indicated on the flap substrate by an arrow in C. The same phosphodiester is cleaved in the nicked DNA substrate in D.). The extents of cleavage of the substrates are plotted as a function of input FenA. Each datum is the average of at least three independent titration experiments ± SEM.
Effect of alanine mutations on FenA activity
To gauge the contribution of the metal-coordinating carboxylate residues to FenA activity, the Asp9, Asp60, Glu123, Asp146, Asp148, Asp205 and Asp208 side chains were mutated individually to alanine. (We showed previously that changing Asp125 to alanine abolishes FenA flap endonuclease and 5′ exonuclease activities (7)). We also introduced alanine in lieu of Tyr75, Lys76 and Arg79 in the active site (Figure 3A, discussed above), as well as two other conserved residues: Phe15 and Arg16 in the α1 helix (Figure 2B). The equivalent of Phe15 in T5 Fen (Phe32) demarcates the junction between the duplex and flap segments of the scissile strand, via a π-stack of the phenylalanine on the 5′-most paired nucleobase of the duplex (11). The counterpart of Arg16 in T5 Fen (Arg33) is located next to the 3′ end of the template strand of the flap duplex (11) and is a plausible candidate to interact with the 3′ template or duplex DNA segment of a flap-nick or Y-flap substrate.
Twelve FenA-Ala mutants were produced in E. coli in parallel with WT FenA. SDS-PAGE analysis of the recombinant protein preparations is shown in Figure 3B. To assay for flap endonuclease activity of FenA, a substrate was prepared by annealing a 5′ 32P-labeled 28-mer DNA strand and an unlabeled 18-mer DNAOH strand to a 36-mer template DNA strand, to form a singly nicked duplex with a 10-nt 5′ flap (Figure 3C). Reaction of WT FenA with 1 pmol (100 nM) nick flap substrate in the presence of 1 mM manganese resulted in FenA concentration-dependent conversion of the input 32P-labeled 28-mer DNA to a predominant 32P-labeled 11-nt product (via incision between the first and second nucleotides of the duplex segment) and a minor 12-nt species (cleaved at the vicinal phosphodiester) that were resolved from the substrate by urea-PAGE (7); the extent of flap cleavage as a function of FenA concentration is plotted in Figure 3C for WT FenA and 8 of the FenA-Ala mutants for which product formation was detectable at up to 5 pmol of input FenA. The D146A, D148A, D205A and D208A mutants, which formed no flap cleavage product at 5 pmol of input enzyme, were titrated up to 50 pmol of input FenA. The flap endonuclease-specific activities of the mutants, determined from the slope of the titration profile in the linear range as calculated by linear regression in Prism and then normalized to that of WT FenA, are shown in Table 1.
Table 1. Effects of FenA alanine mutations on flap endonuclease and 5′ exonuclease activities.
| FenA mutant | Specific Activity (% of WT FenA) | |
|---|---|---|
| Flap endonuclease | 5′ exonuclease | |
| D9A | 0.2 | 3 |
| F15A | 75 | 109 |
| R16A | 27 | 17 |
| D60A | 5 | 44 |
| Y75A | 6 | 5 |
| K76A | 35 | 18 |
| R79A | 29 | 20 |
| E123A | 15 | 18 |
| D146A | ≤0.1 | ≤0.1 |
| D148A | ≤0.1 | ≤0.1 |
| D205A | ≤0.1 | ≤0.1 |
| D208A | ≤0.1 | ≤0.1 |
| D60A-E123A | ≤0.1 | 0.1 |
In similar fashion, we assayed 5′ exonuclease activity of WT FenA and FenA-Ala mutants on a nicked DNA substrate, 5′ 32P-labeled at the nick (Figure 3D). FenA exonuclease action liberates 32P-dCMP as the predominant product, along with a minor dinucleotide species pCpA (7). The extents of 5′ exonuclease cleavage as a function of input FenA are plotted in Figure 3D. Again, the D146A, D148A, D205A and D208A mutants, which formed little or no exonuclease cleavage products at 5 pmol of input enzyme, were titrated up to 50 pmol of input FenA. The normalized 5′ exonuclease specific activities of the mutants are compiled in Table 1.
The salient findings were that loss of the carboxylate functional groups of Asp146 (M2 ligand), Asp148 (M2 and M3 ligand), Asp205 (M3 ligand) and Asp208 (M3 ligand and water-bridged M2 ligand) abolished flap endonuclease and 5′ exonuclease activities (i.e. the alanine mutants had ≤0.1% of the WT-specific activity), signifying that the M2 and M3 coordination complexes are essential for FenA function. Alanine mutation of Asp9, which coordinates two of the M1 waters, virtually eliminated the flap endonuclease (0.2% specific activity) and reduced 5′ exonuclease to 3% of WT activity. Yet, alanine substitutions of the other M1 water ligands—Asp60 and Glu123—elicited less severe reductions in flap endonuclease specific activity (to 5 and 15% of WT for D60A and E123A, respectively) and 5′ exonuclease-specific activity (to 44 and 18% of WT for D60A and E123A, respectively). Because Asp60 and Glu123 make atomic contacts to the same water molecule in the M1 coordination complex (Figure 3A), we considered the possibility that these two amino acids might be functionally redundant (at least in part). To address this point, we produced and purified a D60A-E123A double-mutant (Supplementary Figure S2A) and found that the simultaneous loss of both carboxylates abolished flap endonuclease and 5′ exonuclease activities (≤0.1% of WT) (Table 1). We conclude that all three active site manganese complexes are essential for FenA nuclease activity.
Alanine substitutions for conserved residues Tyr75, Lys76, and Arg79 that interact with the outer spheres of the metal complexes also resulted in lower flap endonuclease and 5′ exonuclease specific activity: 6 and 5% for Y75A; 35 and 18% for K76A and 29 and 20% for R79A (Table 1). The R16A mutation in α1 reduced endonuclease and exonuclease activity to 27 and 17% of WT, respectively. However, the F15A mutation was benign.
Effect of Asp to Asn substitutions
The six aspartates deemed essential for FenA activity based on alanine scanning (Asp9, Asp125, Asp146, Asp148, Asp205 and Asp208) were mutated conservatively to asparagine. The purified recombinant Asn mutants (Supplementary Figure S2A) were either inert or virtually inert when assayed for flap endonuclease and 5′ exonuclease activity (Table 2), i.e. the isosteric asparagine substitutions mimicked the effects of alanine substitution. The simplest explanation for the loss of function is that Asp-to-Asn mutation precludes binding of one or more of the active site manganese ions. Alternatively, Asn mutation might reduce affinity of manganese binding at sites M1, M2 or M3, such that the sites are not filled at the concentration of 1 mM Mn2+ established previously as optimal for WT FenA activity (7). To address the latter prospect, we tested whether increasing the manganese concentration in the flap endonuclease reaction mixtures to 5 and 10 mM might elicit a gain of activity for any of the ‘dead’ Asn mutants or for the D60A-E123A double-mutant. To achieve sensitivity in this experiment, the level of input mutant enzyme (10 pmol) was ∼10-fold greater than that sufficient for complete cleavage of the flap substrate by WT FenA. As shown in Supplementary Figure S2B, increasing manganese concentration to 5 and 10 mM had no significant rescuing effect on the D125N, D148N, D205N or D208N mutants. (Note that the four Mn2+ concentration-unresponsive mutations were at FenA aspartates that make direct atomic contact with M1, M2 or M3.) By contrast, the D9N and D60A-E123A proteins (mutated at acidic residues that make only water-mediated contacts to M1 or M2) were 8-fold and 11-fold more active in the presence of 5 and 10 mM Mn2+ than at 1 mM Mn2+, resulting in cleavage of 19% of the flap substrate (Supplementary Figure S2B). The activity of the D146N mutant (at an aspartate that contacts M2 directly) was increased 5-fold (to 32% flap cleavage) at 10 mM Mn2+ (Supplementary Figure S2B). That said, the apparent specific activities of the D9N, D60A-E123A and D146N proteins at higher manganese concentrations were still ≤1.2% of the flap endonuclease activity of WT FenA.
Table 2. Effects of Asp-to-Asn mutations on flap endonuclease and 5′ exonuclease activities.
| FenA mutant | Specific Activity (% of WT FenA) | |
|---|---|---|
| Flap endonuclease | 5′ exonuclease | |
| D9N | ≤0.1 | 0.2 |
| D125N | ≤0.1 | ≤0.1 |
| D146N | 0.4 | 0.3 |
| D148N | ≤0.1 | ≤0.1 |
| D205N | ≤0.1 | ≤0.1 |
| D208N | ≤0.1 | ≤0.1 |
Crystal structures of FenA Asp-to-Asn mutants
Structures of nuclease-dead FenA mutants D125N (at 2.2 Å resolution), D148N (1.8 Å resolution) and D208N (1.9 Å resolution) were determined by diffraction of crystals in space group P21 grown from FenA mutant protein solutions containing 10 mM MnCl2. Pairwise DALI comparison of the tertiary structure of WT FenA to D125N (rmsd 0.3 Å), D148N (rmsd 0.3 Å), and D208N (rmsd 0.1 Å) indicated they were virtually identical. Superposition of the active sites revealed that each of the Asn mutants lacked one of the three active site manganese ions. To wit, D125N was missing M2 whereas D148N and D208N were missing M3 (Figure 4). The presence of only two active site Mn2+ in the Asn mutants was affirmed by anomalous peaks coincident with the modeled metal ions (not shown). The effects of the Asn mutations on active site architecture are discussed in detail below.
Figure 4.
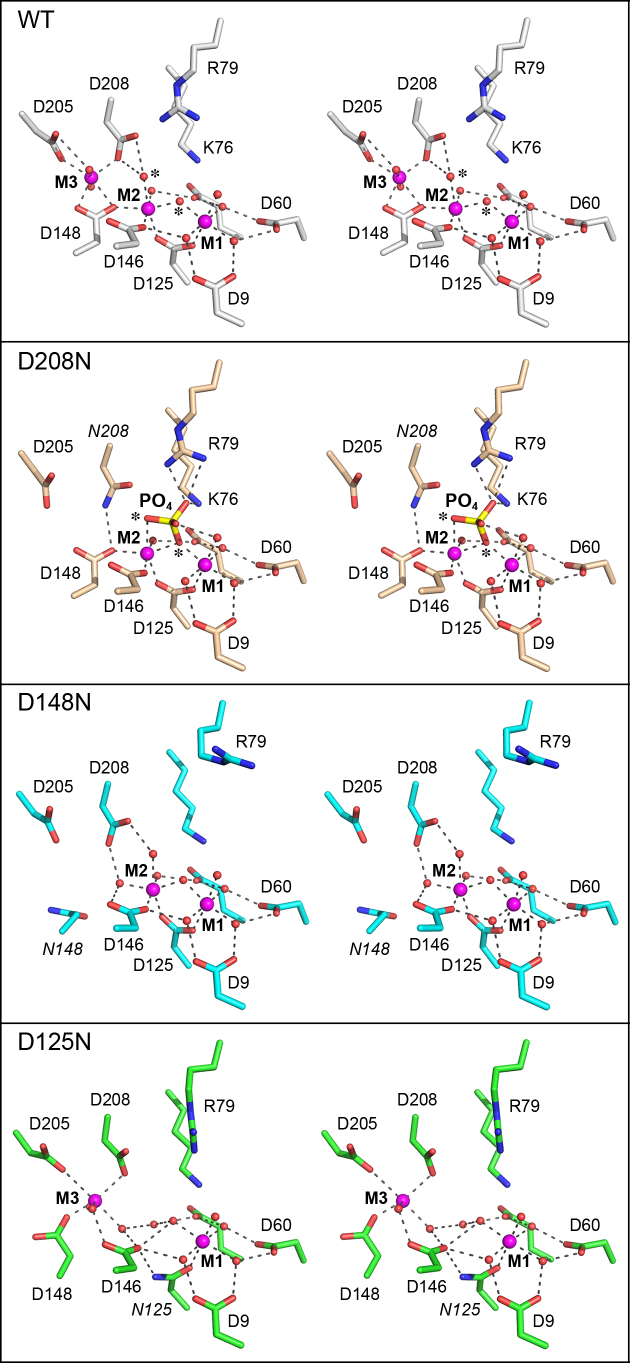
Active sites of FenA Asp-to-Asn mutants. The active sites of FenA WT (colored with gray carbons) and mutants D208N (beige carbons), D148N (cyan carbons) and D125N (green carbons) were superimposed and offset vertically. Stereo views are shown. The mutated Asn chains are labeled in italics. Manganese ions are magenta spheres and waters are red spheres. Atomic contacts are denoted by dashed lines.
The D125N change preserves the M1 site in which the Asn125 Oδ makes the same contact to M1 as the Asp125 Oδ2, but fails to fill the M2 site because the Asn Nδ does not accommodate direct M2 coordination. Rather, the Asn125 amide group rotates so that Nδ makes a hydrogen bond to Asp146 (Figure 4). The M3 site is also affected in D125N, whereby Asp148 rotates so that it makes only one direct contact to M3 (instead of two as in WT FenA) and the vacated position in the M3 coordination complex is taken by water bridged to Asp146 (in lieu of the Asp146 contact to the missing M2 manganese) (Figure 4). Asp208 maintains its M3 contact, but rotates absent the water-bridged interaction with M2, a consequence of which is that the Asp208-Arg79 salt bridge is lost and the Arg79 side chain rotates compared to the WT active site (Figure 4).
The D148N mutant fails to fill the M3 site and the Asn148 side chain rotates out of position so that it no longer makes the direct contact to the M2 manganese seen in WT FenA, in lieu of which the M2 coordination complex acquires a water bridged between Asp146 and Asp208 (Figure 4). The Arg79 side chain adopts a different rotamer in D148N whereby it points away from Asp208. The M1 site in D148N is unperturbed compared to WT FenA.
The D208N structure lacks M3 and instead the Asn208 Nδ makes a hydrogen bond to Asp148 (Figure 4). The salient feature of the D208N enzyme is the presence of a tetrahedron-shaped density in the active site that we modeled as a phosphate anion. [Note that phosphate was not included in the Fen-D208N plus manganese enzyme solution, nor was it supposed to be present in the commercial 0.2 M ammonium acetate, 20% PEG3350 precipitant solution used to grow the D208N crystals. However, by assaying all solutions for inorganic phosphate by the malachite green method, we determined that the commercial precipitant solution contained 0.4 mM inorganic phosphate.] The phosphate O1 oxygen is coordinated by the M2 manganese (at site in the M2 coordination complex occupied by a water in WT FenA) and its O2 oxygen is bridged between M2 and M1, in lieu of the bridging water seen in WT FenA. (The phosphate O1 and O2 atoms and the waters they replace are indicated by asterisks in the top two panels in Figure 4.) The phosphate O3 atom is engaged by Lys76 Nζ and the terminal guanidinium nitrogens of Arg79. The phosphate O4 atom is bridged by a water to the M1 manganese (Figure 4). The D208N structure with phosphate is instructive with respect to the FenA mechanism, insofar as we construe it to be a plausible mimetic of a product complex of the enzyme with the 5′-PO4 group of the incised flap strand, as discussed further below.
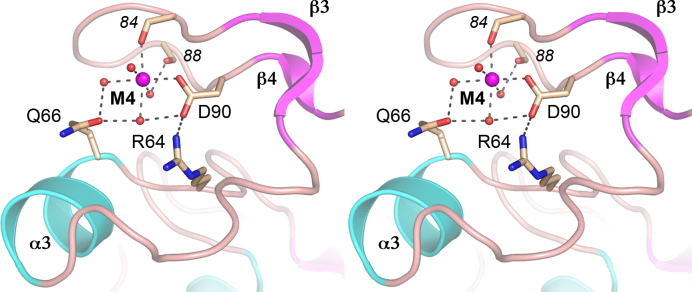
The M4 loop of FenA engages a fourth metal ion outside the active site
WT FenA and each of the Asn mutants that were crystallized contained a fourth Mn2+ engaged in the β3-β4 loop that packs against the FenA surface by virtue of metal-contacts and an Arg64-Asp90 salt bridge (Figure 5). The octahedral M4 complex is filled by Asp90 Oδ1, the Glu84 main-chain carbonyl oxygen, and four waters, one of which is bridged between Gln66 Oϵ and Asp90 Oδ2, while another is engaged to Gln66 Oϵ, and a third is coordinated to the Val88 main-chain carbonyl (Figure 5). The M4 side chain ligands are not conserved in ExoIX, Pol1, T5 Fen, T4 Fen/RNaseH or ExoI, consistent with this structural motif being a distinctive feature of FenA.
Figure 5.
Manganese engagement at the M4 loop. Stereo view of the M4 loop, depicted as a cartoon trace colored and labeled according to secondary structure. The M4 manganese is depicted as a magenta sphere and waters as red spheres. Selected amino acid side chains (labeled in plain font) and main-chain carbonyls (in italics) are shown as stick models with beige carbons. Atomic contacts are indicated by dashed lines.
In order to gauge whether the M4-coordinating amino acids are important for FenA activity, we simultaneously replaced Gln66 and Asp90 with alanine and purified the recombinant Q66A-D90A protein in parallel with WT FenA (Supplementary Figure S4A). Assays of the flap endonuclease activity (Supplementary Figure S4B) and 5′ exonuclease activity (Supplementary Figure S4C) as a function of input FenA showed that the titration profiles of the WT and Q66A-D90A enzymes were superimposable. The flap endonuclease specific activity of the Q66A-D90A mutant was 92% of WT; the 5′ exonuclease-specific activity of the Q66A-D90A mutant was 95% of WT. The benign effect of mutating the M4-coordinating amino acids stands in stark contrast to the severe defects accompanying mutations of the M1, M2 and M3 ligands. We infer that the distinctive M4 metal is not critical for FenA nuclease activity in vitro.
DISCUSSION
Mycobacterial FenA exemplifies a bacterial clade of stand-alone nucleic acid phosphodiesterases with flap endonuclease and 5′ exonuclease activities similar to those of the 5′ exonuclease domain of bacterial DNA Pol1, to which FenA is structurally homologous. The crystal structures of FenA reported here, together with the results of a structure-guided mutagenesis, firmly establish that FenA assimilates three manganese ions (its preferred metal cofactor) in the active site and that all three metals are essential for FenA nuclease activity. As a three-metal enzyme, FenA resembles bacteriophage T5 Fen, the structure of which has been solved with three magnesium ions in its active site (11), and which makes many of the same atomic contacts to the M1, M2 and M3 metals observed here for FenA. FenA differs from E. coli ExoIX, its nearest tertiary structural homolog among stand-alone FEN proteins, insofar as ExoIX lacks an M3 site and relies on two active site metals, at the positions analogous to M1 and M2 in FenA (8). Human FEN1 and ExoI also adhere to a two-metal mechanism with corresponding M1 and M2 sites (16,20).
The conservation of M1 and M2 in all FEN/FEN-like nucleases implies that they are direct catalysts of phosphodiester hydrolysis. In that vein, our structure of FenA mutant D208N with its active site populated by the M1 and M2 manganese ions and a phosphate bound between them provides mechanistic insights to FenA catalysis, as follows. Engagement of the phosphate displaces both the M1–M2 bridging water and an M2 water (denoted by asterisks in Figure 4, top panel) and replaces them with phosphate oxygens. We propose that the state of the FenA active site in the M1-M2•PO4 structure is an analog of the product state of the phosphodiesterase reaction, whereby the phosphate mimics the terminal 5′-PO4 of the cleaved DNA strand in the duplex segment of the flap-nick substrate. Because the water that functions as the nucleophile in the cleavage reaction becomes one of the non-bridging oxygens in the 5′-PO4 product, this means that either the M1-M2 bridging water or the M2 water is the attacking nucleophile.
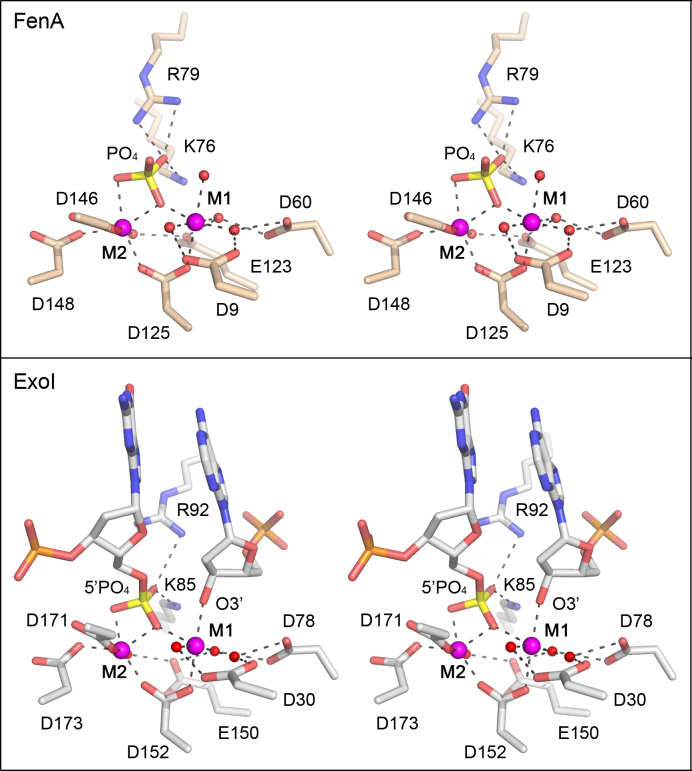
A comparison of the FenA M1-M2•PO4 structure to the structure of the product complex of the flap cleavage reaction of human ExoI, recently solved by the Beese lab (20), makes a strong case for the M2 water as the nucleophile for FenA. Figure 6 shows the active sites of the FenA product analog and the bona fide ExoI product complex superimposed and then offset vertically. The respective M1 and M2 metal coordination complexes (to Mn2+ in FenA and Mg2+ in Exo1) are virtually identical and the position and metal contacts of the FenA phosphate are the same as those of the DNA 5′-PO4 in the ExoI product complex. The 3′-OH of the cleaved flap strand in the Exo1 product complex (i.e. the leaving group in the phosphodiesterase reaction) is engaged directly by the M1 metal, and the O3′ atom occupies the same position as the M1 water in the FenA structure. The key point is that the O3′ leaving group is nearly apical to the 5′ phosphate oxygen atom that is liganded to M2 (O–P–O3′ angle of 165°), signifying that this apical 5′ phosphate oxygen derives from the water nucleophile. [By contrast, the 5′-phosphate oxygen atom that bridges M1 and M2 makes an O–P–O3′ angle of 57° in the ExoI product complex.] We surmise that the equivalent M2-bound phosphate oxygen atom in the FenA structure, which makes an O–P-wat angle of 167° to the M1 water (the putative product O3′), derives from the M2-bound water that is the likely nucleophile in the FenA reaction. This mechanism proposed for ExoI and invoked here for FenA differs from a model elaborated for T5 Fen, in which it is hypothesized that the water bridged between M1 and M2 in the T5 Fen active site is the nucleophile that attacks the scissile phosphodiester (11).
Figure 6.
Comparison of FenA product analog and ExoI product complex structures. Shown is a stereo view of the active sites of FenA D208N with Mn2+ at sites M1 and M2 and a phosphate anion mimetic of the product 5′ phosphate (top panel) superimposed on the active site of human ExoI in a DNA cleavage product complex with Mg2+ at sites M1 and M2 (bottom panel; from accession code: 5V0D). The deoxynucleotides at the 5′-PO4 and 3′-OH ends resulting from DNA cleavage are shown as stick models in the ExoI active site. M1 and M2 are depicted as magenta spheres and waters as red spheres. Atomic contacts are denoted by dashed lines.
The FenA M1 and M2 manganese complexes per se could potentially catalyze the chemical steps of phosphodiester hydrolysis: orientation and activation of the nucleophilic water (by M2 and Asp208), stabilization and charge neutralization of the phosphorane transition state (by joint M1–M2 coordination of a trigonal planar phosphate oxygen) and stabilization/expulsion of the O3′ leaving group (by contact to M1). An open question is the degree to which non-metal constituents of the FenA active site drive catalysis, e.g. by interactions of basic side chains with the scissile phosphodiester that might also stabilize the transition state. FenA Lys76 and Arg79 make electrostatic contacts to the same phosphate oxygen in the FenA product-like complex (Figure 6) and this phosphate oxygen corresponds to one of the trigonal planar oxygens in the proposed transition state of the ExoI reaction (20). ExoI has an arginine side chain, Arg92, at a position equivalent to Arg79 in FenA and, indeed, ExoI Arg92 contacts the 5′-PO4 oxygen atom that is not associated with one of the metals in the product complex (Figure 6). ExoI Lys85, though not equivalent to FenA Lys76 with respect to its position in the tertiary structure, also contacts the same 5′-PO4 oxygen as Arg92 (Figure 6). Whereas the ExoI Lys85 and FenA Lys76 Cα atoms to do not overlap, their Nζ atoms that contact the phosphates do coincide when the structures are aligned (Figure 6). Single-alanine mutations of Lys85 and Arg92 in human ExoI virtually abolished 5′ exonuclease activity (25). [The lysine and arginine equivalents in human FEN1, Lys93 and Arg100, also contact the scissile phosphate and mutating them singly to alanine reduced flap endonuclease activity by >400-fold (16,39). Alanine mutation of the lysine equivalent in T5 Fen (Lys83) reduced flap endonuclease activity by 1000-fold (40).] In the case of FenA, our mutational analysis excludes an essential role for Arg79 or Lys76 per se in transition state stabilization, insofar as changing Arg79 or Lys76 singly to alanine elicited only an ∼3-fold decrement in flap endonuclease activity and an ∼5-fold reduction in 5′ exonuclease activity (Table 1). To test whether Arg79 and Lys76 might be functionally redundant with respect to FenA activity, we purified a K76A-R79A double mutant of FenA (Supplementary Figure S4A) and assayed its flap endonuclease activity (Supplementary Figure S4B) and 5′ exonuclease activity (Supplementary Figure S4C) as a function of input enzyme. The flap endonuclease specific activity of K76A-R79A was 23% of WT, a value similar to the 29% activity seen for the R79A single mutant. The K76A-R79A 5′ exonuclease- specific activity was 9% of WT, which was half the specific activities of 18–20% of WT that we observed for the K76A and R79A single mutants. These results indicate only a modest additive effect on 5′ exonuclease function upon loss of both phosphate contacts and show that FenA retains significant flap endonuclease activity in the absence of both contacts. Thus, Arg79 and Lys76 are not redundantly essential à la the M1-coordinating residues Asp60 and Glu123.
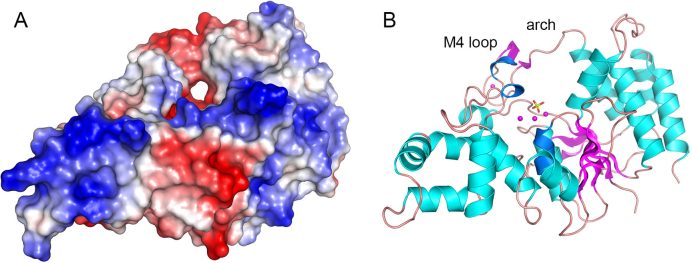
Although we do not have a structure of FenA bound to a flap DNA substrate, we can surmise from a comparison to the structure of a mutant T5 Fen bound to a flap DNA substrate, where T5 Fen contains M1 and M3 in the active site but lacks M2 (11), that the essential M3 manganese in FenA will contact the DNA phosphate group immediately 3′ of the scissile phosphodiester. An outstanding question anent FenA is how the enzyme engages a 5′ flap. The FenA protein structure does not have the helical arch module found in many other FEN/FEN-like enzymes. Instead it has a distinctive M4 loop in lieu of the first α helix of the helical arch. We envision two possible scenarios: (i) FenA accommodates the DNA 5′ flap in a unique manner involving the M4 loop; or (ii) the structure of the M4 loop is remodeled upon binding to DNA into a helical arch akin to that seen in T5 Fen, FEN1 and Exo1. The plausibility of the former model is suggested by a surface view of FenA, oriented in Figure 7A to highlight a through and through hole in the enzyme, the ‘roof’ of which is formed by the M4 loop as it reaches in an arch-like fashion to join the α4 helix (Figure 7B). This aperture is along the path predicted to be taken by the 5′ flap strand and could potentially flex to allow threading of the 5′ flap though the hole.
Figure 7.
Surface model of FenA highlights a hole that might accomodate a 5′ flap strand. Panel A shows a surface electrostatic model of WT FenA oriented so as to feature a through-and-through hole near the active site along the path predicted for the 5′ flap strand. Panel B shows a cartoon model of the tertiary structure in the same position used to generate the surface view. Manganese ions (magenta spheres) from the WT FenA structure, and a phosphate (stick model) imported from the D208N structure, are shown as active site landmarks in panel B.
DATA AVAILABILITY
Structural coordinates have been deposited in Protein Data Bank under accession codes 6C33, 6C34, 6C35 and 6C36.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by U.S. National Institutes of Health Grant AI64693. The MSKCC structural biology core laboratory is supported by National Cancer Institute grant P30-CA008748. X-ray diffraction data were collected at synchrotron facilities supported by grants and contracts from the National Institutes of Health (P41GM103403, HEI-S10RR029205) and the Department of Energy (DE-AC02-06CH11357).Funding for open access charge: U.S. National Institutes of Health [AI64693].
Conflict of interest statement. None declared.
REFERENCES
- 1. Finger L.D., Atack J.M., Tsutakawa S., Classen S., Tainer J., Grasby J., Shen B.. The wonders of flap endonucleases: structure, function, mechanism and regulation. Subcell. Biochem. 2012; 62:301–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tomlinson C.G., Atack J.M., Chapados B., Tainer J.A., Grasby J.A.. Substrate recognition and catalysis by flap endonucleases and related enzymes. Biochem. Soc. Trans. 2010; 38:433–437. [DOI] [PubMed] [Google Scholar]
- 3. Lyamichev V., Brow M.A., Dahlberg J.E.. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993; 260:778–783. [DOI] [PubMed] [Google Scholar]
- 4. Xu Y., Derbyshire V., Ng K., Sun X.C., Grindley N.D., Joyce C.M.. Biochemical and mutational studies of the 5′-3′ exonuclease of DNA polymerase I of Escherichia coli. J. Mol. Biol. 1997; 268:284–302. [DOI] [PubMed] [Google Scholar]
- 5. Lyamichev V., Brow M.A., Varvel V.E., Dahlberg J.E.. Comparison of the 5′ nuclease activities of Taq DNA polymerase and its isolated nuclease domain. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:6143–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mizrahi V., Huberts P.. Deoxy- and dideoxynucleotide discrimination and identification of critical 5′ nuclease domain residues of the DNA polymerase I from Mycobacterium tuberculosis. Nucleic Acids Res. 1996; 24:4845–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Uson M.L., Ghosh S., Shuman S.. The DNA repair repertoire of Mycobacterium smegmatis FenA includes the incision of DNA 5′ flaps and the removal of 5′ adenylylated products of aborted nick ligation. J. Bacteriol. 2017; 199:e00304–e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anstey-Gilbert C.S., Hemsworth G.R., Flemming C.S., Hodskinson M.R., Zhang J., Sedelnikova S.E., Stillman T.J., Sayers J.R., Artymiuk P.J.. The structure of Escherichia coli ExoIX – implications for DNA binding and catalysis in flap endonucleases. Nucleic Acids Res. 2013; 41:8357–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukushima S., Itaya M., Kato H., Ogasawara N., Yoshikawa H.. Reassessment of the in vivo function of DNA polymerase I and RNase H in bacterial cell growth. J. Bacteriol. 2007; 189:8575–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ceska T.A., Sayers J.R., Stier G., Suck D.. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature. 1996; 382:90–93. [DOI] [PubMed] [Google Scholar]
- 11. AlMalki F.A., Flemming C.S., Zhang J., Feng M., Sedelnikova S.E., Ceska T., Rafferty J.B., Sayers J.R., Artymiuk P.J.. Direct observation of DNA threading in flap endonuclease complexes. Nat. Struct. Mol. Biol. 2016; 23:640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mueser T.C., Nossal N.G., Hyde C.C.. Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell. 1996; 85:1101–1112. [DOI] [PubMed] [Google Scholar]
- 13. Devos J.M., Tomanicek S.J., Jones C.E., Nossal N.G., Mueser T.C.. Crystal structure of bacteriophage T4 5′ nuclease in complex with a branched DNA reveals how flap endonuclease-1 family nucleases bind their substrates. J. Biol. Chem. 2007; 282:31713–31724. [DOI] [PubMed] [Google Scholar]
- 14. Harrington J.J., Lieber M.R.. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994; 13:1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balakrishnan L., Barbara R.A.. Flap endonuclease 1. Annu. Rev. Biochem. 2013; 82:119–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsutakawa S.E., Thompson M.J., Arvai A.S., Neil A.J., Shaw S.J., Algasaier S.I., Kim J.C., Finger L.D., Jardine E., Gotham V.J.B. et al. Phosphate steering by Flap Endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability. Nat. Commun. 2017; 8:15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hwang K.Y., Baek K., Kim H.Y., Cho Y.. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat. Struct. Biol. 1998; 5:707–713. [DOI] [PubMed] [Google Scholar]
- 18. Hosfield D.J., Mol C.D., Shen B., Tainer J.A.. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998; 95:135–146. [DOI] [PubMed] [Google Scholar]
- 19. Chapados B.R., Hosfield D.J., Han S., Qiu J., Yelent B., Shen B., Tainer J.A.. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004; 116:39–50. [DOI] [PubMed] [Google Scholar]
- 20. Shi Y., Hellinga H.W., Beese L.S.. Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:6010–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mietus M., Nowak E., Jaciuk M., Kustosz P., Studnicka J., Nowotny M.. Crystal structure of the catalytic core of Rad2: insights into the mechanism of substrate binding. Nucleic Acids Res. 2014; 32:10762–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y., Freeman A.D.J., Declais A.C., Wilson T.J., Gartner A., Lilley D.M.J.. Crystal structure of a eukaryotic GEN1 resolving enzyme bound to DNA. Cell Rep. 2015; 13:2565–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S.H., Princz L.N., Klügel M.G., Habermann B., Pfander B., Biertümpfel C.. Human Holliday junction resolvase GEN1 uses as chromodomain for efficient DNA recognition and cleavage. Elife. 2015; 4:e12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim Y., Eom S.H., Wang J., Lee D.S., Suh S.W., Steitz T.A.. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995; 376:612–616. [DOI] [PubMed] [Google Scholar]
- 25. Orans J., McSweeney E.A., Iyer R.R., Hast M.A., Hellinga H.W., Modrich P., Beese L.. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011; 145:212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patel N., Atack J.M., Finger L.D., Exell J.C., Thompson P., Tsutakawa S., Tainer J.A., Williams D.M., Grasby J.A.. Flap endonucleases pass 5′-flaps through a flexible arch using a disorder-thread-order mechanism to confer specificity for free 5′-ends. Nucleic Acids Res. 2012; 40:4507–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chauleau M, Shuman S. Kinetic mechanism and fidelity of Escherichia coli NAD+-dependent DNA ligase (LigA). Nucleic Acids Res. 2016; 44:2298–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu H., Shuman S.. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3′-OH monoribonucleotide. J. Biol. Chem. 2008; 283:8331–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gong C., Martins A., Bongiorno P., Glickman M., Shuman S.. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 2004; 279:20594–20606. [DOI] [PubMed] [Google Scholar]
- 30. Zhu H., Shuman S.. Characterization of Agrobacterium tumefaciens DNA ligases C and D. Nucleic Acids Res. 2007; 35:3631–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta R., Barkan D., Redelman-Sidi G., Shuman S., Glickman M.S.. Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol. Microbiol. 2011; 79:316–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 33. Kabsch W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010; D66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sheldrick G.M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D. Biol. Crystallogr. 2010; D66:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M.. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991; A47:110–119. [DOI] [PubMed] [Google Scholar]
- 37. Holm L., Kaariainen S., Rosenstrom P., Schenkel A.. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008; 24:1780–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sakurai S., Kitano K., Yamguchi H., Hamada K., Okada K., Fukuda K., Uchida M., Ohtsuka E., Morioka H., Hakoshima T.. Structural basis for recruitment of human flap endonuclease I to PCNA. EMBO J. 2005; 24:683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsutakawa S.E., Classen S., Chapados B.R., Arvai A.S., Finger L.D., Guenther G., Tomlinson C.G., Thompson P., Sarker A.H., Shen B. et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011; 145:198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pickering T.J., Garforth S.J., Thorpe S.J., Sayers J.R., Grasby J.A.. A single cleavage assay fror T5 5′→3′ exonuclease: determination of the catalytic parameters for wild-type and mutant proteins. Nucleic Acids Res. 1999; 27:730–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural coordinates have been deposited in Protein Data Bank under accession codes 6C33, 6C34, 6C35 and 6C36.