SUMMARY
Hibernating mammals survive hypothermia (<10°C) without injury, a remarkable feat of cellular preservation that bears significance for potential medical applications. However, mechanisms imparting cold-resistance, such as cytoskeleton stability, remain elusive. Using the first iPSC line from a hibernating mammal (13-lined ground squirrel), we uncovered cellular pathways critical for cold-tolerance. Comparison between human and ground squirrel iPSC-derived neurons revealed differential mitochondrial and protein quality control responses to cold. In human iPSC-neurons cold triggered mitochondrial stress, resulting in reactive oxygen species overproduction and lysosomal membrane permeabilization, contributing to microtubule destruction. Manipulations of these pathways endowed microtubule cold-stability upon human iPSC-neurons and rat (a non-hibernator) retina, preserving its light responsiveness after prolonged cold exposure. Furthermore, these treatments significantly improved microtubule integrity in cold-stored kidneys, demonstrating the potential for prolonging shelf-life of organ transplants. Thus, ground squirrel iPSCs offer a unique platform for bringing cold-adaptive strategies from hibernators to humans in clinical applications.
Keywords: Induced pluripotent stem cells, hibernation, microtubule cold stability, cold adaptation, ground squirrel, hypothermia, retina, mitochondria, lysosomal membrane permeabilization, organ storage
In Brief
iPSC-derived neurons from the hibernating 13-lined ground squirrel allow investigation of cellular cold resistance with implications for improved cold storage for organ transplants.
INTRODUCTION
Hibernation in mammals is a seasonal state of metabolic suppression and dormancy characterized by a decrease in body temperature (Andrews, 2007; Barnes, 1989; Carey et al., 2003; Geiser, 2004). Small hibernators such as 13-lined ground squirrels (GS; Ictidomys tridecemlineatus) use this remarkable strategy to survive extreme environmental stresses (Drew et al., 2007; Staples, 2016a; Storey and Storey, 2010). They routinely withstand a body temperature below 10°C that would otherwise cause severe injuries and organ failure in non-hibernating mammals (Quinones et al., 2014). For example, GS platelets display resistance to cold storage-induced damage (Cooper et al., 2012), and GS kidney tubular cells exhibit significantly less cell death when subjected to cold storage/rewarming compared to mice (Jain et al., 2016). If the mechanisms underlying such cold tolerance could be reproduced in non-hibernators, it would hold great promise for clinical applications.
Indeed, even with a mild temperature drop (2–3°C below normal body temperature), induced hypothermia has been shown to promote patient survival in critical care situations (Delhaye et al., 2012; Yenari and Han, 2012). At this temperature range, cold shock proteins (e.g. RBM3 and CIRP) are induced, which demonstrate promising neuroprotective effects (Bastide et al., 2017; Peretti et al., 2015; Zhu et al., 2016). However, hibernating GSs, whose body temperature drops from normal (~37°C) to near-freezing for days at a time, confront a much more strenuous challenge and yet avoid neurological damage (Arendt et al., 2003; von der Ohe et al., 2006). The mechanisms involved in cold adaptation at such hibernation-like temperatures remain unclear. One critical vulnerability is that the microtubule cytoskeleton quickly depolymerizes at near-freezing temperatures (Breton and Brown, 1998; Wallin and Strömberg, 1995; Weber et al., 1975). Microtubules have various cellular functions and are vital to many organs and the nervous system, as neurons require a stable microtubule cytoskeleton to maintain their elaborate dendritic and axonal processes as well as proper synaptic connections (Conde and Caceres, 2009; Kapitein and Hoogenraad, 2015; Penazzi et al., 2016). Hence, failure to protect the microtubules would result in catastrophic functional impairment. Although microtubule associated proteins (MAPs) can facilitate short-term (~1 h) microtubule stability in neurons at ~4°C (Bosc et al., 2003; Guillaud et al., 1998; Webb and Wilson, 1980), how GSs maintain their microtubules in days-long, hibernation bouts remains a mystery. Understanding the mechanisms of such remarkable cold adaptations during hibernation will be vital for harnessing the benefit of metabolic regulation and avoiding the harmful effects of such low temperatures (Dave et al., 2012; Forreider et al., 2016; Nathaniel, 2008).
In recent years, systematic profiling approaches have unveiled unique features of hibernators at gene and protein levels (Fedorov et al., 2014; Hampton et al., 2013; Hindle and Martin, 2013; Morin and Storey, 2009; Williams et al., 2005). However, hibernating species such as GSs are non-conventional animal models with annual reproductive cycles that hinder access to early developmental tissues. Consequently, hibernation research at molecular and cellular levels has been impeded by both a lack of transgenic models for in vivo genetic manipulations and a limited choice of cell types (Drew et al., 2011) for in vitro studies. To this end, induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006) are a valuable alternative that can establish various cell types and tissue models for in-depth mechanistic studies in vitro.
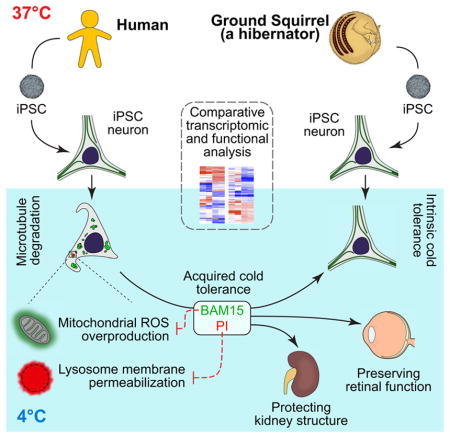
Here we report for the first time the establishment of iPSCs from a hibernating mammal (GS) to study their hibernation-specific features, inspiring novel pharmacological strategies to bestow cold adaptability to cells and organs from non-hibernating mammals. We found that neurons differentiated from GS-iPSCs retain intrinsic cold-resistant features such as microtubule stability. This enabled us to identify cellular pathways linking mitochondria-initiated oxidative stress and dysfunctional lysosomes with microtubule instability in the cold. Using drugs targeting these pathways, we demonstrate that human iPSC-derived neurons (iPSC-neurons) and rat retinal neurons can acquire cold-resistant features. Remarkably, these treatments functionally rescued cold-exposed rat retinal tissues whose structure and function would otherwise have been compromised. Furthermore, the same treatments prevented cold-induced microtubule and other damage in mouse kidneys undergoing conventional transplantation storage. Prospectively, GS-iPSCs can serve as a valuable platform for exploring the unique mechanisms of metabolic adaptation and stress responses in hibernators, and for facilitating the translation of hibernation research to medical applications.
RESULTS
GS iPSC-Neurons Recapitulate the Intrinsic Cold Resistance of Native GS Neurons
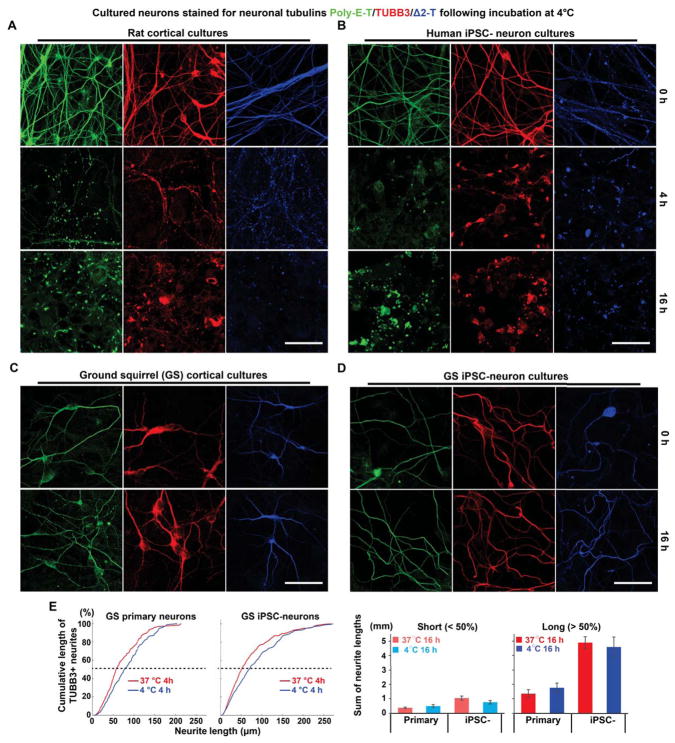
We first compared neuronal microtubule morphology from GSs and rats (a non-hibernator) at 4°C using antibodies against the neuron-specific β-tubulin isotype-III (TUBB3) and two α-tubulin isotypes that are enriched in neurons (Δ2-T, Poly-E-T). At physiological temperatures (37°C), rat neurons contained long, smooth microtubule processes; however, a 4-h incubation at 4°C induced microtubule fragmentation. Moreover, following an extended 16-h incubation at 4°C, most rat neuron microtubules disappeared, leaving abnormal tubulin aggregates (Figure 1A). Human iPSC-neurons exhibited similar cold-induced microtubule instability (Figure 1B). In stark contrast, cortical neurons cultured from neonatal GSs showed no signs of deterioration even after 16 h of cold exposure (Figure 1C).
Figure 1. Species-dependent Differences in Neuronal Microtubule Cold Stability.
(A) Immunofluorescence of TUBB3 (red), polyglutamylated tubulin (poly-E-T; green), or delta2-tubulin (Δ2-T; blue) in cultured rat primary cortical neurons incubated at 4°C for various durations. Scale bar: 40 μm.
(B) Immunofluorescence of TUBB3, poly-E-T, or Δ2-T in cultured human induced pluripotent stem cell-derived neurons (iPSC-neurons) incubated at 4°C for various durations. Scale bar: 40 μm.
(C) Immunofluorescence of TUBB3, poly-E-T, or Δ2-T in cultured 13-lined ground squirrel (GS) primary cortical neurons incubated at 4°C for various durations. Scale bar: 40 μm.
(D) Immunofluorescence of TUBB3, poly-E-T, or Δ2-T in GS iPSC-neurons incubated at 4°C for various durations. Scale bar: 40 μm. For other cell types differentiated from GS iPSCs, see also Figure S1.
(E) Cumulative plots and quantification of the lengths of manually traced TUBB3-positive microtubules (see STAR METHODS) from GS cortical primary neurons (n = 5 experiments each at 37°C and 4°C; cultures derived from 4 GS pups) and GS iPSC-neurons (n = 6 experiments each at 37°C and 4°C; cultures derived from 2 GS iPSC lines). Cumulative lengths of short (neurite lengths < 50% of the cumulative plots) and long microtubules (neurite lengths > 50% of the cumulative plots) were quantified separately. No significant difference in microtubule length attributed to cold temperature was found (p > 0.05; Student’s t-test for two-group comparison).
To overcome the limited accessibility of primary cultures of GS neurons and to facilitate mechanistic studies of microtubule cold resistance in vitro, we established GS iPSC lines from neonatal GS cortex using a modified reprogramming protocol and validated their pluripotency (Figures S1A, S1B and S1C). As with GS cortical primary neurons, microtubules of GS iPSC-neurons showed little difference between 37°C and 4°C incubations for 16 h (Figures 1D and 1E), suggesting that GS iPSC-neurons retain endogenous cold resistance.
Species-specific Gene Expression Profiles Highlight Cellular Pathways that Contribute to Microtubule Destruction in the Cold
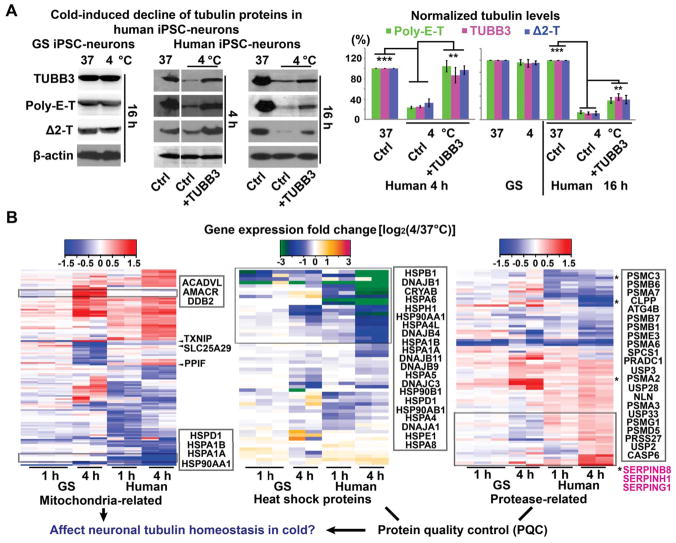
To elucidate how GS neurons preserve their microtubule integrity in the cold, we first examined tubulin protein expression. In agreement with microtubule morphology, protein levels of TUBB3, poly-E-T, and Δ2-T remained unchanged in GS iPSC-neurons following 16-h incubation at 4°C (Figure 2A). In contrast, in human iPSC-neurons following 4 h at 4°C, tubulin protein levels were significantly reduced (Figure 2A).
Figure 2. Species-dependent iPSC-neuronal Transcriptome Changes in Mitochondria and Protein Quality Control (PQC) in Response to Low Temperature.
(A) Western blots and quantification of TUBB3, poly-E-T and Δ2-T proteins in GS and human iPSC-neurons following 4- or 16-h cold exposure with or without TUBB3 overexpression (n = 5 experiments from 2 GS and 2 human cell lines; Student’s t-test between untreated controls with or without cold exposure, and between cold-exposed groups with or without the TUBB3 overexpression plasmid; ** p < 0.01; *** p < 0.001). See also Figure S2C.
(B) Heatmaps of transcriptomic alterations in GS and human iPSC-neuronal cultures induced by 4°C incubation for 1 h or 4 h (see STAR METHODS), highlighting two distinct categories of expression- genes related to mitochondrial functions, and genes encoding heat shock proteins (HSPs), proteases and proteinase inhibitors (SERPINs, magenta). Some genes exhibiting distinct alteration patterns between GS and human are emphasized. See also Figure S3, Data Files S1–S3.
These observed changes in microtubule homeostasis were not a secondary phenomenon attributable to cell death (Figure S2A). To test whether such microtubule destabilization resulted from a diminished pool of tubulin proteins, we overexpressed TUBB3 in human iPSC-neurons. TUBB3 overexpression temporarily preserved expression of all three proteins (TUBB3, poly-E-T, and Δ2-T) and long neurites for 4-h cold exposure (Figures 2A and S2D), likely by recruiting poly-E-T and Δ2-T to form more stable polymers. However, this rescue effect did not extend to the 16-h group (Figure S2D), perhaps due to cessation of TUBB3 production at 4°C and other cellular responses contributing to tubulin degradation.
We then used comparative RNA profiling in human and GS iPSC-neurons to systematically examine possible mechanisms of cold-induced tubulin degradation (Figure S3, Data Files S1, S2 and S3). Clustering analysis of differentially expressed genes (DEGs) and pathway analysis indicated that mitochondrial functions are among the most significantly changed pathways upon cold exposure (yellow cluster in Figure S3E and Data file S2, and Figure S3F). Among these most significantly changed pathways, two function-related gene clusters showed distinctly changing patterns in GS and human iPSC-neurons during cold exposure: 1) mitochondria-related genes and 2) genes participating in protein quality control (PQC), including heat shock proteins (HSPs), proteases, and proteinase inhibitor genes (Figures 2B and S3F). These differentially-affected pathways prompted us to investigate whether the mitochondrial and PQC responses to cold exposure contribute to microtubule destruction in human iPSC-neurons.
Cold Induces Mitochondrial Hyperpolarization and ROS Over-production in Human iPSC-Neurons
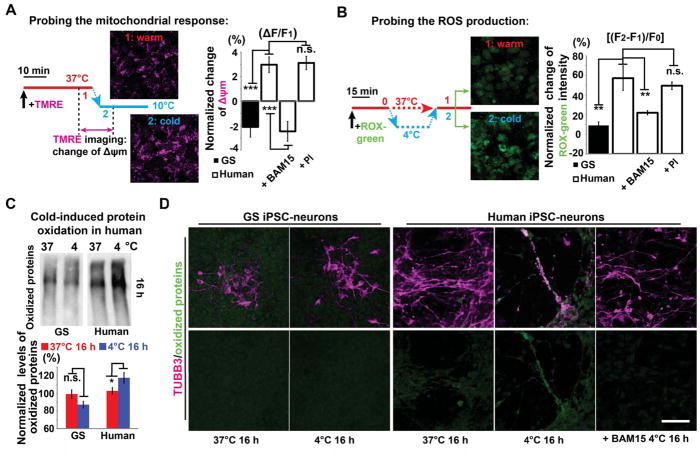
To examine whether mitochondria in human and GS iPSC-neurons respond differently to cold exposure, we monitored mitochondrial membrane potential (Δψm) changes. In human iPSC-neurons following a temperature decrease from 37°C to 10°C, we observed a hyperpolarizing Δψm (Figure 3A and Movie S1). In contrast, when GS iPSC-neurons were treated with the same experimental protocol, we instead observed a depolarizing Δψm (Figure 3A). Although this change is seemingly minute, it leads to a significant increase in reactive oxygen species (ROS) production (see DISCUSSION). Indeed, following 30-min cold exposure, we observed significantly higher cold-induced ROS production in human than in GS iPSC-neurons (Figure 3B), indicating that the immediate mitochondrial response results in prolonged oxidative stress. Importantly, when the human iPSC-neuron mitochondrial hyperpolarization was reversed by the addition of a low dose of mitochondrial uncoupler, BAM15 (0.1 μM, Figure 3A), we observed a significant decrease in ROS production, to a level comparable to GS iPSC-neurons in the cold (Figure 3B).
Figure 3. Cold-induced Mitochondrial Hyperpolarization and Oxidative Stress in Human iPSC-neurons.
(A) Live cell imaging protocol (left) of mitochondrial membrane potential (Δψm) with TMRE (see STAR METHODS), and quantification (right) of cold-induced Δψm changes in GS and human iPSC-neurons untreated controls, and treated with BAM15 or PI (n = 8 experiments; Student’s t-test between GS and human controls; ANOVA plus post hoc Tukey test for multiple human cold-exposed groups with or without drug treatment; *** p < 0.001; n.s. p > 0.05, not significant). BAM15: a mitochondrial uncoupling drug; PI: protease inhibitors. See also Movie S1.
(B) CellROX green imaging protocol (left) to assess cold-induced ROS production (see STAR METHODS), and quantification (right) of ROS production following 30-min cold exposure in GS and human iPSC-neurons (n = 8 experiments; Student’s t-test between GS and human comparisons; ANOVA plus post hoc Tukey test for multiple human cold-exposed groups with or without drug treatment; ** p < 0.01; n.s. p > 0.05, not significant).
(C) Western blots and quantification of oxidized proteins following 16-h incubation at 4°C (n = 5 experiments; Student’s t-test for two-group comparisons; * p < 0.05).
(D) Immunofluorescence of TUBB3 (magenta) and oxidized proteins (green). Note: after 16-h incubation at 4°C, minimal protein oxidation was detected on GS TUBB3+ processes even with digital enhancement, while in human neurons oxidized proteins were found on residual TUBB3+ microtubules, which could be reduced or completely mitigated by BAM15 treatment (n = 5 experiments). Scale bar: 50 μm. See also Figure S4. For all experiments in this figure, neurons were derived from 2 GS and 3 human iPSC lines for each group.
Because ROS may oxidize and disrupt microtubules (Wilson and Gonzalez-Billault, 2015), we hypothesized that the GS iPSC-neurons possess superior microtubule stability due to suppressed ROS generation as a consequence of muted hyperpolarizing Δψm in response to cold. To verify this, we directly probed cold-induced protein oxidation. We found that levels of oxidized proteins increased in human iPSC-neurons but remained unchanged in GS iPSC-neurons exposed to cold for 16 h (Figure 3C). Oxidized proteins are subject to protein-protein cross-linkages and aggregate formation (Gregersen and Bross, 2010; Grune et al., 1997). Indeed, immunolabeling of oxidized proteins revealed aggregates along residual microtubules after cold exposure in human but not in GS iPSC-neurons (Figures 3D and S4). Interestingly, the addition of BAM15 prevented accumulation of oxidized proteins along microtubules (Figure 3D). Thus, our findings suggest that overproduction of ROS via cold-induced mitochondrial hyperpolarization leads to microtubule damage and subsequent aggregation.
Cold Causes Lysosomal Membrane Permeabilization (LMP) in Human iPSC-neurons
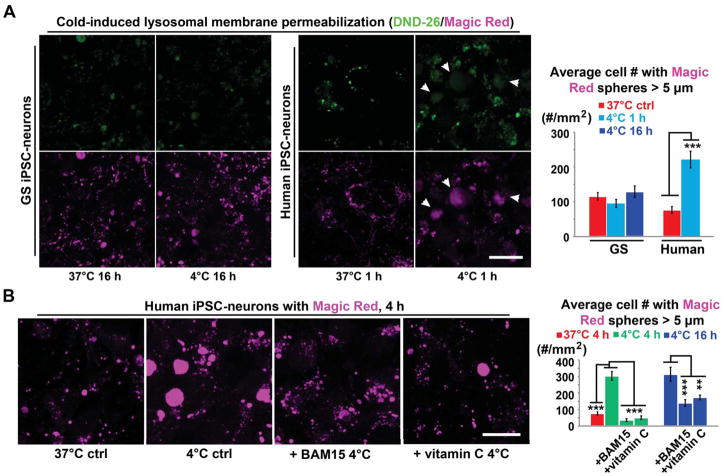
Damaged proteins would normally engage the PQC system for their removal (Goldberg, 2003; Zhang and Ye, 2014), with lysosomes providing an alternative pathway to the ubiquitin-proteasome machinery (Dunlop et al., 2009; Iwata et al., 2005; Lee et al., 2012). However, in cold-exposed human iPSC-neurons, we observed a PQC dysregulation (Figures 2B, S5B and S5C) and, surprisingly, evidence of compromised lysosomal integrity (i.e. lysosomal membrane permeability, LMP), which acidifies the cytosol and releases digestive enzymes. To assess the latter, we loaded GS and human iPSC-neurons with Magic Red and DND-26, which labeled discrete puncta of lysosomal vesicles. However, in cold-exposed human neurons, more diffuse cytosolic Magic Red and DND-26 signals were observed (Figure 4A; arrowheads) - typical signs of LMP and an acidified cytosolic environment. Thus, proteases leaked from lysosomes are poised to inadvertently digest cytosolic proteins such as adjacent microtubules. Notably, BAM15 and the antioxidant Vitamin C significantly reduced LMP (Figure 4B), demonstrating a link between cold-induced mitochondrial dysfunction and LMP.
Figure 4. Cold-induced Lysosomal Membrane Permeabilization (LMP) in Human iPSC-neurons.
(A) (Left panel) Live imaging of lysosomes in GS and human iPSC-neurons with DND-26 (green) and Magic Red (magenta; see STAR METHODS). Note: Following 1-h incubation at 4°C, accumulation of diffuse cytoplasmic fluorescence in marked human neurons (arrowheads) indicating LMP. Scale bar: 20 μm. (Right Panel) Quantification of cells containing diffused Magic Red signals (Diameter > 5μm) (n = 6 experiments; Student’s t-test for two-group comparison; *** p < 0.001).
(B) (Left panel) Live imaging of lysosomes in human iPSC-neurons with Magic Red under denoted conditions. Note: BAM15 (0.1 μM) or the antioxidant vitamin C (0.5 mM) alleviated LMP in human neurons treated following 4-h incubation at 4°C. Scale bar: 20 μm. (Right Panel) Quantification of cells containing diffused Magic Red signals (Diameter > 5μm) (n = 5 experiments; Student’s t-test between untreated controls with or without cold exposure; ANOVA plus post hoc Tukey test for multiple cold-exposed groups; ** p < 0.01; *** p < 0.001). See also Figure S5 for the involvement of other PQC components. For these experiments, neurons were derived from 2 GS and 3 human iPSC lines for each group.
Cold-exposed Human iPSC-neuronal Microtubules Are Protected by Uncoupling Mitochondria and Neutralizing the Adverse Effect of LMP
Motivated by our bioinformatics and experimental results, we sought to determine whether pharmacological intervention targeting the affected pathways could confer the GS’s microtubule cold resistance upon non-hibernating mammals. Indeed, a low dose of BAM15 preserved long neurites and tubulin proteins in human iPSC-neurons incubated at 4°C for 4 h (Figures 5A, 5B and 5C). Reinforcing the notion that this protection arises directly from the uncoupling effect of BAM15, transient overexpression of mitochondrial uncoupler proteins UCP1 or UCP2 (Laursen et al., 2015; Yamashita et al., 1999) in human iPSC-neurons likewise produced cold-stable neurites (Figures S6A, S6B and S6E).
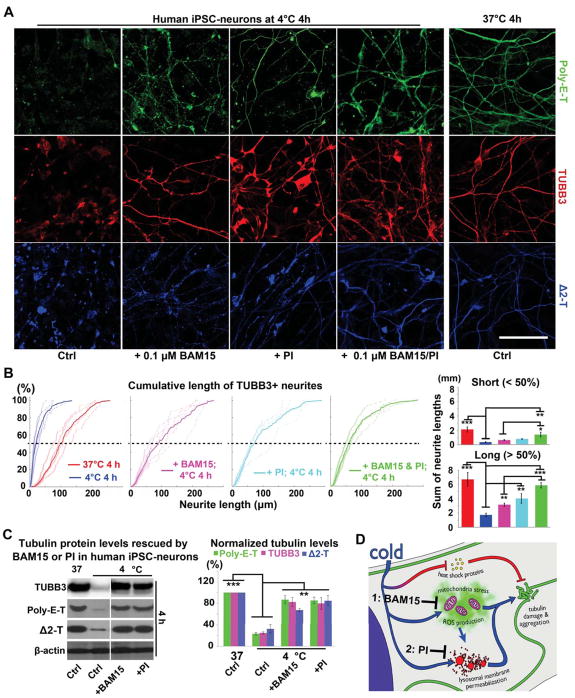
Figure 5. Morphological Protection of Human iPSC-neurons by BAM15/PI Pretreatments against Prolonged Cold Stress.
(A) Pre-treatment with BAM15 (0.1 μM; n = 17 experiments), PI (1:500; n = 24 experiments) or a combination of both (n = 20 experiments) preserved long neurites (poly-E-T: green; TUBB3: red; Δ2-T: blue) of human iPSC-neurons following 4-h incubation at 4°C. Scale bar: 40 μm. See also Figure S6.
(B) Cumulative plots and quantification of TUBB3+ neurite lengths (see STAR METHODS; BAM15: n = 6 experiments; PI: n = 5 experiments; BAM15 & PI: n = 6 experiments; Student’s t-test between untreated controls with or without cold exposure; ANOVA plus post hoc Tukey test for multiple cold-exposed groups treated for the same duration; * p < 0.05; ** p < 0.01; *** p < 0.001).
(C) Western blots and quantification confirming that BAM15 or PI pre-treatment maintained tubulin protein levels in human iPSC-neurons after 4-h incubation at 4°C (n = 5 experiments; Student’s t-test between untreated controls with or without cold exposure; ANOVA plus post hoc Tukey test for multiple cold-exposed groups; ** p < 0.01; *** p < 0.001).
(D) Proposed model depicting the mechanisms by which BAM15 and PI pre-treatments protect human iPSC-neurons from cold stress. For these experiments, human neurons were derived from 3 iPSC lines.
Additionally, we pretreated human iPSC-neurons with a protease inhibitor cocktail (PI) to alleviate the adverse effects of leaked proteases from LMP. PI effectively preserved long neurites in cold-exposed human iPSC-neurons (Figures 5A, 5B and 5C). Notably, PI did not alter Δψm hyperpolarization or ROS production (Figures 3A and 3B), suggesting that LMP induction lies downstream of mitochondrial activities. Remarkably, the combination of BAM15 and PI further preserved human iPSC-neuron microtubules to near-control levels (Figure 5B), suggesting that -- in addition to triggering LMP -- ROS may directly damage microtubules as demonstrated in Figures 3D and S4. A working model is demonstrated in Figure 5D.
BAM15 and Protease Inhibitors Preserve Morphology and Function in the Rat Retina during Cold Stress
Promising results from our in vitro experiments prompted us to test whether these treatments can also protect neural tissues of a non-hibernator when challenged with hibernation-like hypothermia. We used an ex vivo rat retina preparation, which retains intact neural structure and responds to light under healthy conditions.
Like human iPSC-neurons, cold-exposed retinal ganglion cells (RGCs) in rat retina exhibited considerable microtubule destruction, losing most of their dendritic microtubules and forming numerous tubulin aggregates along their axons (Figures 6A and 6B). In contrast, retinal tissues from hibernating GS showed no changes in microtubule morphology (data not shown). Strikingly, pre-incubation of rat retinas with BAM15 and PI greatly reduced the RGC microtubule defects caused by cold-exposure (Figures 6A and 6B).
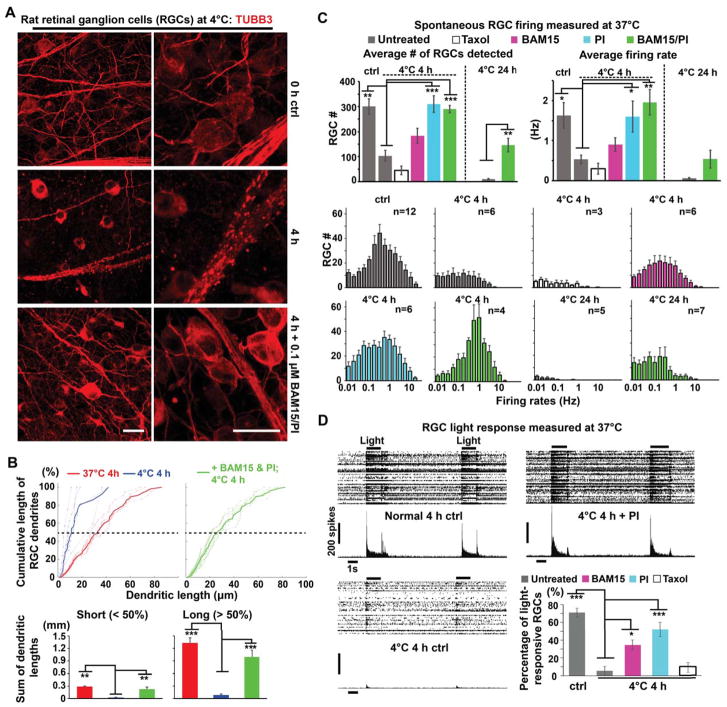
Figure 6. Morphological and Functional Protection of Rat Retinal Explants by BAM15/PI Pretreatments against Prolonged Cold Stress.
(A) Microtubule morphology in rat (n = 10 animals) retinal ganglion cells (RGCs) immunostained for TUBB3 (red). Scale bars: 20 μm.
(B) Cumulative plots and quantification of TUBB3+ RGC dendritic lengths in rat retina (see STAR METHODS; n = 6 animals; Student’s t-test for two-group comparisons; ** p < 0.01; *** p < 0.001).
(C) Quantification of multielectrode array (MEA) recordings of spontaneous RGC activity following exposure to 4°C for 0 h, 4 h, or 24 h. Treatments and their corresponding color legend are indicated. Upper-left panel: Numbers of active RGCs detected by MEA. Upper-right panel: Average firing rates for detected RGCs. Lower panel: Distributions of firing rates for detected RGCs. Student’s t-test between untreated controls with or without cold exposure, and the 24-h cold-exposed groups; ANOVA plus post hoc Tukey test for multiple cold-exposed groups treated for the same duration; * p < 0.05; ** p < 0.01; *** p < 0.001. See also Movie S2 for an animated example of MEA recordings of the RGC spontaneous firing. The number of animals used in each condition is provided in the histogram.
(D) Representative light responses of rat RGCs following 4-h cold exposure either untreated or treated with BAM15 (n = 5 animals), PI (n = 6 animals), or Taxol (n = 6 animals). Quantification is provided as the percentage of detected RGCs that were light-responsive (see STAR METHODS; Student’s t-test between untreated controls with or without cold exposure; ANOVA plus post hoc Tukey test for multiple groups cold-exposed for the same duration; * p < 0.05; *** p < 0.001). See also Movie S3 for an animated example of MEA recordings of the light response experiments.
We then used multi-electrode array (MEA) recordings to measure RGC activity. As shown in Figure 6C, incubation at 4°C for 4 h largely silenced rat RGCs and greatly reduced the average firing rates of remaining RGCs, whereas pre-treatment with BAM15 and/or PI significantly preserved rat RGC activity (Movie S2). In contrast, 10 μM Taxol (a classic microtubule stabilization drug) did not preserve RGC activity in cold-exposed rat retinas (Figure 6C), even though it preserved long neurites (Figure S2D). Further, we assessed the RGC light response, which is a strong indicator of the health and proper function of the retina (Figure 6D and Movie S3). In control rat retinas, approximately 70% of the RGCs sorted based on their spontaneous activities were light responsive, compared to only 5% in untreated rat retinas after 4°C for 4 h. However, RGCs in cold-incubated rat retinas pre-treated with BAM15 or PI, but not Taxol, displayed 7- to 11-fold increases in light-responsiveness. Taken together, these results indicate that uncoupling mitochondria and/or inhibiting lysosomal protease activity substantially enhanced cold-tolerance in cultured neurons and supported functional recovery of neural tissues from non-hibernating mammals.
BAM15 and Protease Inhibitors Improve Mouse Kidney Protection during Cold Storage and Rewarming
We further tested whether the microtubule protective effect of BAM15/PI treatment can be extended to non-neural tissues, specifically to organs awaiting transplantation, which are commonly cold-stored and re-warmed in conditions comparable to GS hibernation (McAnulty, 2010; Mitchell et al., 2011)(Figure S1E). As reported previously (Breton and Brown, 1998b; Mangino et al., 2008), we found that mouse kidneys stored in University of Wisconsin (UW) solution at 4°C for 24 h exhibited microtubule damage with drastically reduced tubulin labeling (especially in glomeruli), with limited recovery after re-warming (Figure 7). Remarkably, adding BAM15 and PI to the UW solution significantly improved tubulin signals in the cold, and bestowed prominent recovery of microtubule morphology after re-warming (Figure 7). In addition, BAM15 and PI also greatly reduced known adverse effects of extended cold storage to the kidney, such as protein/DNA damage caused by lipid peroxidation (Cotterill et al., 1989) and cell apoptosis (Jain et al., 2016)(Figures S7A and S7B). These results indicate that similar cellular mechanisms lead to cold-induced microtubule instability in various tissues and organs in non-hibernators. Thus BAM15/PI treatment may also confer cold protection to non-neural tissues and organs and prolong their shelf-life for transplantation.
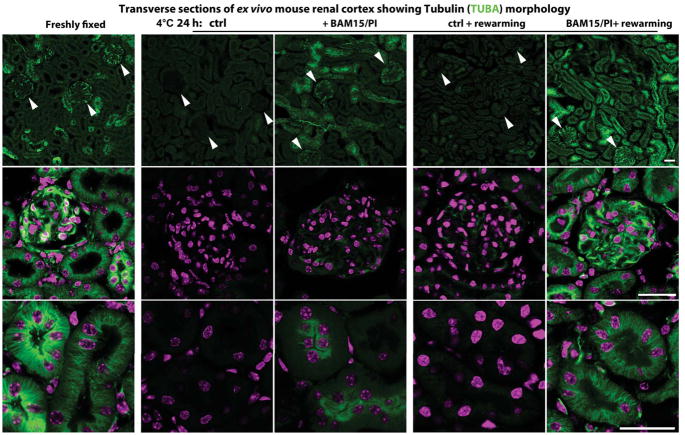
Figure 7. Protecting Mouse Kidneys with BAM15/PI during Cold Storage.
Immunofluorescence of α-tubulins (TUBA; green) and DAPI (magenta) in transverse sections of wild-type mouse kidneys under these conditions (n = 4–6 animals used for each condition): freshly fixed, after 24-h cold storage at 4°C (with or without BAM15/PI) or rewarming at 37°C (with or without BAM15/PI) for 30 min following 24-h cold storage. Note: Reduced tubulin signals in glomeruli (arrowheads in upper panel; middle panel) and renal tubular epithelium (lower panel) of mouse kidneys stored at 4°C for 24 h in standard University of Wisconsin (UW) Solution. Scale bars: 25 μm in all panels. See also Figure S7 for oxidative damages and cell apoptosis in mouse kidneys during cold storage.
DISCUSSION
GS iPSCs Are a Versatile Platform to Study Tissue/Cell type-specific Cold Adaptive Features of Hibernating Mammals
Hibernating animals display various tissue-specific, cold-adaptive features, each an incredible biological marvel waiting to be understood (Staples, 2016b). To empower effective in vitro study of these hibernation features in diverse cell and tissue types, we generated iPSCs from the GS. Most importantly, we demonstrated that cells derived from GS iPSCs retained native cold-resistant features and discovered a link between cold-induced mitochondrial stress, PQC dysfunction (e.g. LMP), and microtubule instability – problems that are apparently solved in hibernators. Inspired by this finding, we sought to alleviate these problems in non-hibernator cells and tissues by reducing ROS production, LMP, and protein degradation. These manipulations protected the microtubule integrity of human iPSC-neurons and preserved retinal functions in rat retina explants that were otherwise eradicated at near-freezing temperatures. Furthermore, the same treatments also protected mouse kidneys from microtubule and other cellular damage, in conditions mimicking cold storage for transplantation.
Cooling and therapeutic hypothermia (mild hypothermia) have been shown to be neuroprotective (Delhaye et al., 2012; Peretti et al., 2015; Yenari and Han, 2012), in which upregulation of cold shock proteins such as RBM3 appears to be essential (Bastide et al., 2017; Tong et al., 2013; Zhu et al., 2016). We observed a similar surge of RBM3 proteins in human and GS iPSC-neurons exposed to 33°C for 60 h, but not at hibernation-like temperatures (<10°C) (Figure S2B), consistent with previous reports (Tong et al., 2013; Zhu et al., 2016). These findings suggest that mild hypothermia and hibernation-like hypothermia may involve distinct sets of molecular mediators, or may reflect different translational regulations under these two conditions (Bastide et al., 2017; Qiang et al., 2013). While mice with various genetic manipulations can serve as powerful models for studying mild hypothermia (Peretti et al., 2015), the GS iPSC lines reported here provide a valuable alternative for investigating hibernation-like hypothermia.
Although iPSCs may not recapitulate all in vivo cellular features, due to possible epigenetic changes during the reprogramming process (Mertens et al., 2015), many intrinsic cellular features are preserved, facilitating in vitro disease modeling and drug screening (Avior et al., 2016). As an example, microtubule cold-stability is retained in GS iPSC-neurons.
Microtubules can be Protected by Multiple Levels of Regulation during Cold Exposure
Potential mechanisms for microtubule cold-stability in GS iPSC-neurons include: 1) Different isotypes or amino acid sequences of tubulins may confer cold-resistance as in Antarctic fish (Detrich and Parker, 1993). However, we found that the predicted protein sequences of the major tubulin types in GS are over 99% identical to those of human, mouse, or rat (data not shown). 2) Tubulins may undergo post-translational modifications (PTMs), which can alter microtubule stability (Hammond et al., 2008; Song et al., 2013). For example, a thorough proteomic study from GS revealed that low body temperature alters the PTMs of cytoskeletal regulatory proteins (Hindle and Martin, 2013), and PTMs have been implicated in metabolic and proteostatic regulation in hibernating bat brain (Zhang et al., 2014). However, we did not find significantly different expression patterns of common neuron-enriched PTMs, such as poly-E-T and Δ 2-T, in human and GS iPSC-neurons (Figures 1 and 2A); similar expression patterns were likewise found for tyrosinated or acetylated tubulins (data not shown). 3) MAPs have also been shown to stabilize microtubules (Billger et al., 1991). For example, in hibernating GS, hyper-phosphorylation of tau may play a role in stabilizing neuronal microtubules (Arendt and Bullmann, 2013). Neuronal microtubules are stabilized by MAP6 to withstand short-term cold exposure (~4°C for 30–60 mins) (Bosc et al., 2003; Guillaud et al., 1998). We found MAP6 was present but not differentially regulated by temperature in either human or GS iPSC-neurons (Data File S3). Nonetheless, these established mechanisms are apparently insufficient to bestow non-hibernating mammals with microtubule stability in hibernation-like hypothermia for prolonged periods of time (Figures 1 and 6; see also Taylor, 2006). Accordingly, here we have identified mitochondria-lysosome pathways as yet another level of regulation for microtubule stability.
Prevention of Cold-induced ROS Overproduction Stabilizes Microtubules
We observed that cold exposure induced Δψm hyperpolarization and ROS overproduction in human iPSC-neurons, whereas mitochondria in cold-exposed GS iPSC-neurons depolarized and produced significantly less ROS. Consistent with our findings, mild hypothermia has been reported to hyperpolarize isolated mouse mitochondria and increase ROS production (Ali et al., 2010). Owing to a steep nonlinear relationship between Δψm and ROS production for mitochondria near the normal ranges of Δψm (Starkov and Fiskum, 2003), the seemingly modest changes in Δψm observed here in fact correspond to substantial changes in ROS production, as demonstrated by direct ROS measurements (Figure 3B). Mitochondrial ROS generation is the major source of oxidative damage to proteins and membranes (Balaban et al., 2005) and likely contributes to the microtubule destruction observed in cold-exposed human iPSC-neurons and rat neural tissues. Hibernators, on the other hand, may have evolved strategies that evade oxidative stress by minimizing mitochondrial hyperpolarization. Consistent with this notion, in winter, the maximum Δψm of GS brain mitochondria during torpor (5°C) is slightly more depolarized compared to that during interbout arousals (37°C) (Ballinger et al., 2017). Intriguingly, in both states Δψm is more hyperpolarized than in spring, suggesting possible seasonal pre-conditioning. Mechanistically, either free fatty acids (Dedukhova et al., 1991; Skulachev, 1991), produced by augmented lipid metabolism during hibernation, or intrinsic uncoupling protein expression (Andrews et al., 2005; Boyer et al., 1998; Laursen et al., 2015; Liu et al., 2006) could result in higher proton leak, thus conferring protection against cold-induced ROS damage. Consistently, transient overexpression of UCP1/UCP2 in human iPSC-neurons enhanced microtubule cold-stability.
Cold-induced PQC Dysfunction Destabilizes Microtubules
In line with RNA-seq data, our experimental results indicate protein degradation pathways are compromised by cold exposure. Impairment of HSP/proteasome pathways may redirect protein degradation to the autophagosome/lysosome pathway (Dunlop et al., 2009; Iwata et al., 2005; Lee et al., 2012). However, protein levels of key autophagosome-lysosomal components did not change significantly in cold-exposed human iPSC-neurons (Figure S5D).
Instead, detrimental interactions may result from the juxtaposition of lysosomes and microtubules (Figure S5E) in cold-exposed human iPSC-neurons. Lysosomal dysfunction has been reported to impair microtubule structure (Fukuda et al., 2006). In turn, both destabilized microtubules and oxidative stress have been shown to lead to LMP (Boya and Kroemer, 2008; Roberg and Ollinger, 1998). Taken together, impaired PQC components coupled with ROS overproduction by stressed mitochondria may form a vicious cycle, resulting in irreversible protein damage and aggregation in the cold. Remarkably, we discovered that GS iPSC-neurons are free from all these adverse cellular responses to cold. A potential mechanism of PQC cold adaptation may be attributed to enhanced organelle membrane integrity in GS cells due to their unique lipid redistribution (Azzam et al., 2000) and may be explored further using the GS iPSC-model.
Pharmacological Interventions May Confer Cold Resistance upon Human Tissues for Clinical Applications
Our findings on microtubule cold-stability have compelling clinical applications, such as prolonging the survival of organs awaiting transplantation (Maathuis et al., 2007; Niemann et al., 2015). Prolonged cold storage has been shown to induce mild oxidative stress to organs (Fabre et al., 2002). Therefore, the standard cold storage medium, UW solution, is enriched in antioxidant components. Nonetheless, even with UW solution (Mangino et al., 2008) or other physiological solutions (Breton and Brown, 1998), cold-induced disruption and degradation of microtubules still occur. Here, we show that the addition of BAM15/PI to UW solution greatly enhanced tubulin preservation in the cold and promoted repolymerization upon rewarming - possibly by alleviating mitochondrial stress and hence reducing ROS production and tubulin damage caused by LMP.
This tissue-protective strategy is likely applicable to other transplantable organs. In addition, it demonstrates that, by understanding the biology of cold adaptation in hibernation, we can improve and broaden the applications of hypothermia, which has the benefit of metabolic suppression, but is often hampered by cold-induced cellular damage (Marion et al., 1997). With the pluripotency of GS iPSCs, unique cold-resistant features of different tissues can be explored more easily in vitro. Furthermore, extreme temperature is only one form of stressor to which the hibernator is adapted. We propose that GS iPSCs can be used to elucidate cellular mechanisms of adaptation to other insults such as metabolic stress, hypoxia/reperfusion injury, and inflammatory damage, potentially allowing us to harness protective effects for therapeutic uses.
STAR ★ METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests of reagents can be directed to and fulfilled by the Lead Contact, Dr. Wei Li (liwei2@nei.nih.gov). The following materials: GS-iPSC1 and GS-iPSC3 (13-lined Ground Squirrel iPSC lines) are properties of U.S. federal government and may be shared with research organizations under an MTA.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Thirteen-lined ground squirrels used in this study were maintained in the University of Wisconsin Oshkosh animal facility and housed in seasonally-varying light/dark cycles matching ambient for that location. All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Wisconsin Oshkosh and conform to the provisions of the Animal Welfare Act (NIH/DHHS) and the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Cortical tissues from 4 GS pups (1 female and 1 male at postnatal day 0; 1 female and 1 male at postnatal day 2) were provided courtesy of Prof. Dana Merriman at the University of Wisconsin Oshkosh. Male and female C57BL/6 mice and Sprague-Dawley rats were maintained in the National Institute of Neurological Disorders and Stroke (NINDS) animal facility and housed in a 12-h light/dark cycle. All animal procedures were approved by the Institutional Animal Care and Use Committee of the National Eye Institute (NEI)/NINDS and conform to the provisions of the Animal Welfare Act (NIH/DHHS). Post-mortem rat retinas were obtained from healthy adult rats of 2–6 months old and provided by the animal facility of NINDS. Post-mortem mouse kidneys were obtained from healthy adult mice of 2–12 months old and provided by the animal facility of NINDS.
Primary Cultures
Cortical tissues from postnatal day 0–2 (P0-2) GS pups (female) were dissected and dissociated using a neural tissue dissociation kit (Miltenyi Biotech, San Diego, CA). Neural precursor cells (NPCs) were then purified with magnet-activated cell sorting using magnetic anti-PSA-NCAM microbeads, myelin removal beads, and FcR blocking reagent (Miltenyi Biotech, San Diego, CA). Primary cultures of rat (embryonic day 18) and GS (P0-2) cortical neurons were differentiated from rat and GS NPCs following the protocol described in (Meberg and Miller, 2003), and subjected to the same temperature experiments as GS and human iPSC-neurons (described in a later section). Cells were maintained at 37°C with 5% CO 2 unless otherwise specified.
METHOD DETAILS
Reprogramming of neonatal GS NPCs into iPSC colonies
All cell culture media reagents were used as directed in the manufacturer’s instructions unless otherwise specified; the reagents are listed in Table S1. The purified P0-2 primary GS NPCs were expanded in 5% Matrigel-coated dishes at 37°C in NPC medium until 70–80% confluence was reached. The NPCs were then transfected with the Cytotune-iPS 2.0 Sendai Reprogramming kit (Thermo Fisher Scientific, Waltham, MA) for 24 hours. The transfected NPCs were dissociated by accutase digestion and passaged into new 5% matrigel-coated wells at a 1:3 ratio in iPSC maintenance medium. After 5–7 days, flat colonies emerged and were manually picked or lifted off with dispase digestion, mechanically dissociated with gentle pipetting, and passaged to a new Matrigel-coated culture well. Note: Due to the limited availability of newborn GS pups from which the GS NPCs were derived, the two GS iPSC lines (GS-iPSC1 and GS-iPSC3) that were successfully established were both female. Thus, an analysis of the influence or association of sex on the results of the study was not performed. Nonetheless, primary GS cortical cultures from both male and female pups did not reveal notable sex-dependent differences in our experiments.
Validation of GS iPSCs
GS iPSC samples were sent for G-banding karyotyping to assess genomic stability - chromosome counts and images provided by Cell Line Genetics (Madison, WI) (See Figure S1D). Pluripotency of GS iPSC colonies was characterized by alkaline phosphatase staining and OCT3/4/NANOG immunostaining (antibodies listed in the Key Resources Table). Using primers specific for the expression of 3 pluripotent marker genes (Pou5f1, Nanog, Sox2) and 10 differentiation markers (Dppa5, Pdx1, Nestin, Sox17, Otx2, Tp63, Foxa2, Gata4, Afp, Brachyury) we confirmed by qRT-PCR the pluripotency of the GS iPSC colonies (data not shown).
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Poly-E-T (1:1000) | AdipoGen | Cat# AG-20B-0020-C100 |
| Mouse monoclonal anti-TUBB3 (1:1000) | R&D Systems | Cat# MAB1195 |
| Mouse monoclonal anti-OCT3/4 (1:500) | Santa Cruz | Cat# sc-5279 |
| Rabbit polyclonal anti-NANOG (1:500) | Cosmo Bio | Cat# RCAB0004P-F |
| Mouse monoclonal anti-ALBUMIN (1:500) | Cedarlane | Cat# CL2513A |
| Rabbit monoclonal anti-HNF4α (1:500) | Cell Signaling | Cat# 3113S |
| Rabbit polyclonal anti-DESMIN (1:500) | Thermo Fisher | Cat# RB9014P1 |
| Mouse anti-TNNT (1:500) | Thermo Fisher | Cat# MS295P0 |
| Rabbit polyclonal anti- Δ2-T (1:1000) | Millipore | Cat# AB3203 |
| Rabbit polyclonal anti-Poly-E-T (1:2000) | AdipoGen | Cat# AG-25B-0030-C050 |
| Rabbit polyclonal anti-TUBB3 (1:2000) | BioLegend | Cat# 802001 |
| Mouse monoclonal anti-TUBA (1:200) | Sigma | Cat# F2168 |
| Rabbit monoclonal anti-LAMP1 (1:1000) | Cell Signaling | Cat# 9091 |
| Rabbit polyclonal anti-VCP (p97) (1:1000) | Cell Signaling | Cat# 2648 |
| Mouse monoclonal anti-VCP (p97) (1:5000) | Fitzgerald | Cat# 10R-P104a |
| Mouse monoclonal anti-PSMB2 (1:1000) | Santa Cruz | Cat# SC-515066 |
| Mouse monoclonal anti-PSMB5 (1:1000) | Santa Cruz | Cat# SC-393931 |
| Mouse monoclonal anti-HSP60 (1:1000) | Santa Cruz | Cat# 4868 |
| Mouse monoclonal anti-HSP70 (1:1000) | Santa Cruz | Cat# sc-7298 |
| Mouse monoclonal anti-HSP90 (1:1000) | Santa Cruz | Cat# sc-13119 |
| Rabbit monoclonal anti-β-actin (1:1000) | Cell Signaling | Cat# 4970 |
| Rabbit polyclonal anti-cleaved Caspase-3 (1:500) | Cell Signaling | Cat# 9661 |
| Chicken polyclonal anti-GFP (1:1000) | Aves Labs | Cat# GFP-1020 |
| Rabbit polyclonal anti-RBM3 (1:1000) | Proteintech | Cat# 14363-1-AP |
| Rabbit polyclonal anti-CIRP (1:1000) | Proteintech | Cat# 10209-2-AP |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cell Culture Media Formulations & Reagents | This paper | N/A |
| B27 supplement minus insulin | Thermo Fisher | Cat# A1895601 |
| CHIR99021 | Tocris | Cat# 4423 |
| IWP4 | Tocris | Cat# 5214 |
| B27 supplement (complete) | Thermo Fisher | Cat# 17504044 |
| RPMI | Thermo Fisher | Cat# 11875093 |
| b-FGF | R&D Systems | Cat# 233-FB-025 |
| HGF | R&D Systems | Cat# 294-HG-005 |
| Hepatocyte Maintenance Media w/ Supplement | Lonza | Cat# MM250 |
| Oncostatin M | R&D Systems | Cat# 295-OM-010 |
| MG-132 (5 μM) | Enzo Life Sci. | Cat# ENZ-51035 |
| BAM15 (0.05-5 μM) | Tocris | Cat# 5737 |
| Protease inhibitor cocktail set III (PI) (1:500) | Millipore | Cat# 539134 |
| PYR-41 (5-10 μM) | Tocris | Cat# 2978 |
| Taxol (0.1 or 10 μM) | Tocris | Cat# 1097 |
| CellROX Green (1 μM) | Thermo Fisher | Cat# C10444 |
| TMRE (50 nM) | Thermo Fisher | Cat# T669 |
| Ames’ medium | Sigma | Cat# A1420 |
| TRIzol | Thermo Fisher | Cat#15596018 |
| LysoTracker Green DND-26 | Thermo Fisher | Cat# L7526 |
| RIPA lysis and extraction buffer | Thermo Fisher | Cat# 89900 |
| SYBR Green PCR Master mix | Thermo Fisher | Cat# 4309155 |
| BrightGreen Express 2X qPCR MasterMix-ROX | ABM | Cat# MasterMix-ER |
| Critical Commercial Assays | ||
| CytoTune iPS 2.0 Sendai Reprogramming Kit | Thermo Fisher | Cat# A16517 |
| Protein Oxidation Detection Kit | Millipore | Cat# S7150 |
| Proteostat Aggresome Detection Kit | Enzo Life Sci. | Cat# ENZ-51035 |
| LipoJet in vitro transfection kit | SignaGen Laboratories | Cat# SL100468 |
| RNeasy kit | Qiagen | Cat# 74104 |
| TruSeq® Stranded mRNA LT - Set A | Illumina | Cat# RS-122-2101 |
| NextSeq® 500/550 High Output Kit v2 | Illumina | Cat# FC-404-2002 |
| Magic Red cathepsin B assay kit | Immuno-Chemistry Tech. | Cat# 938 |
| Click-iT lipid peroxidation imaging kit – Alexa Fluor 488 | Thermo Fisher | Cat# C10446 |
| Deposited Data | ||
| RNA-seq Data | This paper | GEO: GSE93935 |
| Experimental Models: Cell Lines | ||
| Ground Squirrel iPSC line (female) | This paper | GS-iPSC1 |
| Ground Squirrel iPSC line (female) | This paper | GS-iPSC3 |
| Ground Squirrel Cortical primary cultures (male/female) | This paper | N/A |
| Human iPSC line (female) | NIH NINDS Stem Cell Unit | SCU-i10 |
| Human iPSC line (male) | NIH NINDS Stem Cell Unit | SCU-i19 |
| Human iPSC line (male) | NIH NINDS Stem Cell Unit | SCU-i21 |
| Experimental Models: Organisms/Strains | ||
| Thirteen-lined ground squirrel | University of Wisconsin Oshkosh | N/A |
| Sprague Dawley Rats (males and females) | Charles River | RRID: RGD_737891 |
| Mouse: Wild-type C57BL/6 (males and females) | Jackson Labs | RRID: IMSR_JAX:000664 |
| Recombinant DNA | ||
| pCMV6-XL5-TUBB3 | Origene | Cat# SC116313 |
| pCMV6-Myc-DDK-Tubb3 | Origene | Cat# MR207181 |
| pCMV6-UCP1-IRES2-eGFP | GeneCopoeia | Cat# C0618-M61 |
| pCMV6-UCP2-IRES2-eGFP | GeneCopoeia | Cat# M0658-M61 |
| Software and Algorithms | ||
| Matlab rev.2015a | Mathworks | https://www.mathworks.com/ |
| JMP | SAS | https://www.jmp.com/en_us/home.html |
| MC Rack | MultiChannel Systems | https://www.multichannelsystems.com/software/mc-rack |
| Plexon Offline Sorter v.3.3.5 | Plexon Inc. | https://plexon.com/products/offline-sorter/ |
| ImageJ Gel Analysis tool | NIH | https://imagej.nih.gov/ij/ |
| Image J (Simple Neurite Tracer plugin) | (Longair et al., 2011) | https://imagej.net/Simple_Neurite_Tracer |
| Bcl2fastq Conversion Software v2.19.1 | Illumina | https://support.illumina.com/downloads/bcl2fastq-conversion-software-v2-19.html |
| TopHat v2.1.1 | (Trapnell et al., 2009) | https://ccb.jhu.edu/software/tophat/index.shtml |
| R Subread Package | (Liao et al., 2013) | http://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| DESeq2 | (Love et al., 2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Ensembl database (spetri2) | (Yates et al., 2016) | https://useast.ensembl.org/index.html |
| CLUSTAL Omega | (McWilliam et al., 2013) | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| NetAcet 1.0 Server | (Kiemer et al., 2005) | http://www.cbs.dtu.dk/services/NetAcet/ |
| ASEB | (Wang et al., 2012) | https://bioconductor.statistik.tu-dortmund.de/packages/2.12/bioc/html/ASEB.html |
| R Package piano | (Varemo et al., 2013) | https://bioconductor.org/packages/release/bioc/html/piano.html |
| R Package ggplot2 | (Wickham, 2009) | https://cran.r-project.org/web/packages/ggplot2/index.html |
The in vitro pluripotent status of GS iPSCs was also evaluated by their ability to differentiate into cell types from all 3 germ layers:
For Mesoderm differentiation: colonies were plated in a 6-well plate and the media was changed to RPMI containing 2% B27 without insulin and 12 μM CHIR99021. After another 24 h the media was replaced with RPMI containing 2% B27 supplement minus insulin and 5 μM IWP4. 48 h later the media was replaced with RPMI containing 2% B27 supplement (complete). Hereafter the media was changed every 3–4 days. Beating cardiomyocytes appeared around 6–10 days following initiation of the mesoderm differentiation protocol.
For Endoderm differentiation: colonies were plated in a 6-well plate and the medium was changed to RPMI and supplemented with 2% B27 and 100ng/ml Activin A. The medium was changed after 2–3 days. On day 5, the medium was changed to RPMI containing 2% B27 supplement and 10ng/ml b-FGF. The medium was changed after 2–3 days. On day 10, the medium was changed to RPMI containing 2% B27 supplement, and 20ng/mL HGF. On day 15, the medium was changed to Lonza Hepatocyte Culture Medium containing 20ng/ml oncostatin M. The medium was changed after 2–3 days. Cells were fixed and stained for HNF4a and Albumin.
For ectoderm differentiation: See Neuronal differentiation of human and GS iPSCs, and rat cortical primary cultures.
Pluripotency was also verified in vivo using a standard teratoma formation assay (Applied StemCell, CA, USA).
Neuronal differentiation of human and GS iPSCs, and rat cortical primary cultures
Three different human iPSC-derived NPC lines (2 male – SCU-i19, SCUi21, and 1 female – SCU-i10) were obtained from the Stem Cell Unit of the NINDS, courtesy of Dr. Barbara Mallon. Human and GS NPCs were first expanded in NPC medium for 3 days and then switched to neural differentiation medium (NDM) (Table S1) for at least 7 days resulting in a population of neurons and support cells. Experimental results were verified in all three human NPC lines. For GS neuronal differentiation, the first 2 days required GS NDM, afterwards the medium was changed to human NDM.
Evaluation of microtubule cold-stability
The temperature-dependent experiments in this study were performed at ambient CO2 concentration, which required the use of hibernate-medium (Table S1) to maintain the physiological pH. Experiments were conducted by first incubating cultured neurons (human iPSC-derived, GS iPSC-derived, GS primary, or rat primary cultures) or dissected retinas of adult rats in hibernate-medium for 60 min at room temperature. During this preparation time, drug treatments (see Chemicals, Peptides, and Recombinant Proteins, Key Resources Table) were introduced to test their effects on microtubule cold-stability. The cell culture dishes were then kept in an ambient 37°C incubator, or in a 4°C refrigerator. The cooling strategy in our experiments is comparable to conventional static tissue/organ cold storage (McAnulty, 2010; Mitchell et al., 2011) and another cell culture-based study (Rauen and de Groot, 1998). Cooling of the core body temperature of hibernating GSs transitioning from interbout arousal to torpor is provided in Figure S1E. The pH of hibernate-medium was stable and maintained the pH between 7.0 and 7.4 even after 16-h incubation at 4°C.
Multielectrode array (MEA) measurements
All experiments were carried out under dim red light. Rat retina explants were removed and cut into smaller pieces in hibernate-medium, and divided into different treatment groups for pre-incubation with drugs promoting microtubule cold-stability or a DMSO vehicle control at room temperature for 30 minutes. The cold-exposed groups were incubated in the refrigerator, while the fresh control samples were incubated on the bench in dark at room temperature. It has been shown that keeping enucleated eyes under hypothermic and ischemic conditions extends retinal viability in vitro (Reinhard et al., 2016; Schultheiss et al., 2016), as limited access to oxygen and nutrients might reduce mitochondrial metabolism and hence ROS production. Here we allowed the rat retinas to have full access to nutrients and oxygen in all experimental groups, hence low temperature was the only cause of stress in our experiments. After 4- or 24-h incubation, the cold-exposed retinal explants were placed on the bench for 10–15 min, and washed twice with fresh hibernate-medium. Retinal explants from all groups were then transferred to oxygenated (95% O2/5% CO2) bicarbonate-buffered AMES medium at room temperature. The sclera and RPE were removed prior to flat mounting the neural retina with the ganglion cell (RGC)-side facing down onto the MEA.
During MEA recording, the neural retinas were perfused with 95% O2/5% CO2-balanced AMES at 37°C to measure the spontaneous firing activity of the rat RGCs over the course of 15 min. To assess light responsiveness of the rat retina explants, the tissue was exposed to approximately 1-s flashes of light from either a bright white LED (Mightex, Pleasanton, CA) or a halogen lamp (OSRAM, Germany). For each retina, light-driven ganglion cell activity (action potential firing) was elicited in response to six flashes of light that were spaced at approximately either 10 or 15-s intervals. After the recording, the rat retinal explants were fixed with 4% paraformaldehyde for 2 h and then immunostained for neuronal tubulins. For MEA data processing and analyses, the firing spikes of RGCs were first filtered and recorded in MC Rack with a high-pass filter set at 200 Hz, a low-pass filter set at 3000 Hz, and the automatic standard deviation for spike detection threshold was set at −6. To remove false positives and to group spikes into clusters corresponding to individual cells, spike waveforms were sorted using Plexon Offline Sorter v.3.3.5 (Plexon Inc., Dallas, TX). Spikes were automatically clustered into a maximum of eight units (cells) per electrode using the built-in K-means clustering algorithm. The analysis and identification of light responsiveness was performed in MATLAB rev.2015a (The Mathworks, Natick, MA). Briefly, spike times were binned at 200 ms intervals, and the firing rates observed for a given cell during the first bin of each light flash were statistically compared to those during non-stimulated intervals. A cell was considered light-responsive if 1) the stimulated spike rate was larger than the mean baseline firing rate and 2) the outcome of a two-tailed, unequal variance t-test of these spike rates yielded a p-value < 0.01. This test underestimates the number of light-responsive cells because it detects ON, but not OFF light responses. OFF responses were clearly visible in most MEA recordings, but they constituted a minority of cell types and were not quantified here.
Western blotting and immunostaining
All antibodies are listed in the Key Resources Table. RIPA buffer was used to lyse the cells and extract proteins (Thermo Fisher, Waltham, MA). Total proteins or proteins in the soluble fraction after standard centrifugation removing precipitates and debris were run on SDS-PAGE. Standard protocols were used for western blotting and immunostaining. For immunostaining of oxidized proteins, fixed cells were first incubated with 2,4-dinitrophenylhydrazine (DNPH), then stained with an antibody recognizing the DNPH-protein complex (Protein oxidation detection kit, see Key Resources Table). All immunostaining and live cell confocal images were captured on a Zeiss LSM 510 system (Zeiss, Oberkochen, Germany). Western blots were quantified with the ImageJ Gel Analysis tool.
RNA-sequencing
GS and two different lines of human iPSC-neuronal cultures were maintained and prepared as described above. Experiments were conducted by first replacing NDM with fresh hibernate-medium and allowing the cultures to sit at room temperature for 30 min. GS neuronal cultures from 2 iPSC lines, and human neuronal cultures from 2 iPSC-NPC lines (SCU-i19 and SCU-i21) were used. The neuronal cultures from each cell line were divided into two groups: one was incubated in the refrigerator (4°C) and the other in an ambient 37°C incubator. Two incubation time points, 1 h and 4 h, were selected. At the end of cold-exposure, cells were collected and lysed in TRIzol (Thermo Fisher, Waltham, MA), total RNA was isolated and purified with miRNeasy kit (Qiagen, Hilden, Germany). TruSeq Stranded mRNA Library Prep Kit was used to generate the sequencing libraries (Illumina, San Diego, CA) following the manufacturer’s instructions. Briefly, 500 ng of total RNA was used for mRNA selection. After the reverse transcription to 1st strand cDNA, strand info was reserved during 2nd strand synthesis. The dsDNA fragments then had the addition of a single ‘A’ base and subsequent ligation of the adapter. The products were then purified and enriched with PCR to create the final cDNA library. The cluster generation and single indexed pair-end (2X75) sequencing was run on NextSeq 500 with the NextSeq 500 High Output Kit v2 (150 cycles). Up to 12 barcoded samples were pooled for one single run with Q30 > 85% and at least 30M passed filter reads per sample. The raw base call (.bcl) files were converted to demultiplexed fastq files with Bcl2fastq v2.19.1 for data analysis. The sequencing data are available at https://www.ncbi.nlm.nih.gov/geo/ under the accession number GSE93935.
Mapping, quantification and differential expression analysis of RNA-seq data
After adapter trimming, raw reads were assessed using the FastQC toolset (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads from different cell lines were aligned to the GS genome (Ensembl spetri2) (Yates et al., 2016) or to the human genome (UCSC hg19) (Speir et al., 2016) (http://genome.ucsc.edu/) separately with TopHat v2.1.1 (Trapnell et al., 2009) and allowed up to two mismatches per read for final read alignments. The complete parameter list for Tophat is “tophat2 -r 150 -g 20 --no-coverage-search --library-type fr-firststrand”. The statistics on the raw reads and alignments are shown in sheet ‘Mapping Statistics’, Data File S3. For quantification of gene expression, only the reads aligned to exons uniquely were counted using the R Subread package (Liao et al., 2013). A gene was included in further analysis if the maximum count-per-million > 3 within any included samples. Given the pair-wise design of the experiments, negative binomial GLM fitting and testing for the effect of treatment by control of the effect of different cell lines were performed with embedded functions in DESeq2 (Love et al., 2014). P values from differential expression tests were adjusted using the Benjamini-Hochberg procedure for multiple hypothesis testing.
Functional enrichment analysis of altered transcripts
Gene Ontology (GO) analysis was performed based on hypergeometric test using Piano package (Varemo et al., 2013), with the gene list of interest as foreground, and all identified genes as background. The annotation of gene to GO terms was retrieved and localized from Ensembl database.
Tubulin protein sequence homology analysis
Tubulin protein sequences for genes (TUBA1C, TUBB2A, TUBB2B, TUBB/TUBB5, TUBA4A, TUBB3, TUBB4A, and TUBB6) of human, mouse, rat and 13-lined ground squirrel were obtained from Uniprot database (2015) and aligned using CLUSTAL Omega program (McWilliam et al., 2013). Canonical protein sequences were used when multiple isoforms were available.
Acetylation prediction analysis of tubulin proteins
For all tubulin proteins mentioned above, we performed N-terminal and lysine acetylation events prediction using NetAcet (Kiemer et al., 2005) and ASEB (Wang et al., 2012) separately. During prediction, complete protein sequences were submitted directly. In ASEB analysis, all available lysine acetylation transferases (KAT) families were explored. K-sites with P < 0.05 were identified as acetylated.
Live cell imaging of mitochondrial membrane potential (TMRE)
GS and human iPSC-neuronal cultures were prepared as described above. Experiments were conducted by first removing the NDM and replacing it with hibernate-medium containing 50 nM TMRE (Thermo Fisher, Waltham, MA; see Key Resources Table). At this time, 1) 0.1 μM BAM15 or 2) 1:500 dilution of protease inhibitor cocktail III (Millipore-Sigma, Darmstadt, Germany) were delivered to the two drug treatment groups. Cells were returned to an ambient 37°C incubator for an additional 25 min. Additional hibernate-medium matching these treatment conditions was stored in 50 mL conical tubes in either a 45°C water bath or on ice to serve as a perfusion medium reservoir. Prior to imaging, TMRE-containing medium was discarded and replaced with fresh pre-warmed hibernate-medium. During the Z-stack time-lapse confocal imaging, the culture dish was continuously perfused; first with medium from the 45°C reservoir for 5 min, and then switched to the ice-cold reservoir. This manipulation reduced the temperature of the medium in the culture dish to 10°C (the lowest temperature the perfusion system could attain). Cells in the field of view were refocused, and the same Z-stack, time-lapse confocal images were taken for another 5 min. A maximum intensity projection of the Z-stack images was produced for data analysis. At least 5 different well-focused imaged cells per experiment were measured. We corrected for temperature-dependent TMRE fluorescence increases under the assumption that the total TMRE molecules absorbed into each cell should remain relatively constant within the 10-min imaging duration. Thus, we initially selected the whole cell area as the region of interest (ROI) to estimate the temperature dependent contribution of the TMRE dye’s fluorescence. To do this, we calculated the average TMRE fluorescence intensity from 10 frames of the 10°C recording and divided that by the average TMRE fluorescence intensity from 10 frames of the 37°C recording (F c-10/Fc-37). The mitochondrial areas from the same cells were then selected as ROIs to measure the mitochondrial TMRE intensity at 37°C (F m-37) and 10°C (F m-10). The corrected mitochondrial TMRE intensity at 10°C was calculated using the following equation : Fc.m-10 = Fm-10/(Fc-10/Fc-37). The percentage change in mitochondrial TMRE intensity change at 10°C was thus calculated as: (Fc.m-10 - Fm-37)/ Fm-37 x 100%.
Live cell imaging of ROS production (CellROX-green)
GS and human iPSC-neuronal cultures were prepared in two groups and pretreated with 1 μM CellROX-green (Thermo Fisher, Waltham, MA; see Key Resources Table) with the appropriate drug treatments or vehicle as described above. To explore the influence of cold stress on ROS production, cells were either maintained at 37°C (group 1; as normal controls) or exposed to 4°C (group 2). Cells in group 2 were maintained for 30-min at 37°C, acclimated to room temperature on the bench for 5 min before being placed in a 4°C refrigerator for 30 mi n; they were then gradually rewarmed at room temperature on the bench for 5 min prior to returning to 37°C. Z-stack confocal images were then obtained from the two groups at 37°C. Specifically, since CellROX-green only becomes fluorescent upon oxidation and DNA-binding, this method avoids temperature-dependent dye fluorescent artifacts that accompany imaging at different temperatures while recording the cumulative production of ROS. Maximum intensity projections of the Z-stack images were produced for data analysis. At least 5 different well-focused imaged areas from each experiment were measured. The percentage change in ROS production at 4°C was thus calculated as: (F2 – F1)/ F0 x 100%, in which F2 and F1 are the mean CellROX-green fluorescent intensity of the different areas from cultures in group 2 (F2) or group 1(F1) at the end of the temperature experiment, while F0 is the mean CellROX-green fluorescent intensity of the different areas from cultures in both group 1 and 2 at the end of the initial 30-min incubation at 37°C. Preliminary experiments indicated that prior to low temperature incubation (time 0), group 2 had similar CellROX-green fluorescent intensity to that of group 1 (data not shown).
Live cell imaging of lysosomes using Magic Red and DND-26
GS and Human iPSC-neuronal cultures were prepared in two groups and incubated with hibernate-medium containing Magic Red (1:1000; ImmunoChemistry Technologies, Bloomington, MN) at 37°C for 25 min, followed by the addition of LysoTracker Green DND-26 (50 nM; Thermo Fisher, Waltham, MA; see Key Resources Table) for 5 min. The dye-containing medium was then replaced with fresh hibernate-medium, and one group of cultures was incubated at 4°C for 1, 4 or 16 h, while the other group remained at 37°C serving as normal controls. The cold-exposed group was subjected to confocal imaging at various temperatures (10°C, room temperature, or 37°C), while the control group was imaged at either room temperature or 37°C. These imaging conditions did not influence significantly the occurrence of the lysosomal membrane permeability (LMP). Magic Red becomes fluorescent after being cleaved by the lysosomal protease cathepsin B, while DND-26 is an acidotropic probe that accumulates in the acidic lysosomal vesicles. When LMP occurs, Magic Red and DND-26 signals diffuse to the cytoplasm of the affected cells. In this work, we determined that cells have LMP if they contain a single Magic Red-stained particle of diameter > 5 μm, which is about the size of the neuronal cell nucleus.
Neurite tracing
Cultured neurons or rat retinal explants with or without treatments were fixed with 4% paraformaldehyde, permeabilized and washed with 0.1% Triton X-100 in phosphate buffered saline and stained by antibodies against TUBB3. TUBB3+ neurite or RGC dendrite paths in images of 188.94 x 188.94 μm2 from human iPSC-neuronal cultures and images of 67.48 x 67.48 μm2 from rat whole-mount retinas were traced with the ‘Simple Neurite Tracer’ plugin (Longair et al., 2011), ImageJ.
Detection of protein aggregates
After treatments, cultured neurons were fixed with 4% paraformaldehyde for 10 minutes and then permeabilized and washed with 0.1% Triton X-100 in phosphate buffered saline. Protein aggregates were visualized by a dye that intercalates into the denatured and aggregated proteins and becomes fluorescent (Proteostat aggresome detection kit, Enzo, Farmingdale, NY; see Key Resources Table) (Shen et al., 2011). For accumulation of protein aggregates on the neuronal microtubules, cells were stained with TUBB3 antibody and the protein aggregate dye, and images (188.94 μm x 188.94 μm) were taken. In each image, the number of protein aggregates residing on the microtubules was counted and divided by the total length of the TUBB3+ neurite paths.
Evaluation on mouse kidneys after cold storage
Healthy adult C57BL/6J mice under deep anesthetization were first gently perfused by warm 0.9% NaCl solution and hence sacrificed, then the mice were perfused a second time with standard University of Wisconsin Solution for human organ cold storage (UW solution; see Table S1) in control group, or UW solution supplemented with 0.2 μM BAM15/1:200 dilution of protease inhibitor cocktail III in treatment group. In experiments evaluating lipid peroxidation during cold storage, linoleamide alkyne (from the Click-iT lipid peroxidation imaging kit; Thermo Fisher, see Key Resources Table) was also included during perfusion. Mouse kidneys from the two groups were then removed and transferred to 12-well plates containing UW solution + linoleamide alkyne, or UW solution + BAM15/PI + linoleamide alkyne, and set at room temperature for 30 min. Fresh controls were fixed with 4% paraformaldehyde after this stage, while kidneys in cold-control group and treatment group were then incubated at 4°C. To evaluate tubulin re-polymerization following 24-h cold storage, the kidneys were placed at room temperature for 10 min, rinsed twice with fresh warm UW solution only, and then rewarmed in an ambient 37°C incubator for 30 min. The kidneys were then fixed with 4% paraformaldehyde for 2 h, processed for cryosections and immunostained for α-tubulin TUBA (see Key Resources Table). To evaluate lipid peroxidation following 24-h cold storage, kidneys were fixed and processed according to the manufacturer’s instruction (see Key Resources Table). To evaluate cell apoptosis following 72-h cold storage, the kidneys were fixed and immunostained for cleaved Caspase-3 (see Key Resources Table).
QUANTIFICATION AND STATISTICAL ANALYSIS
Matlab (Mathwork) and JMP Statistical Software (SAS) were used for data and statistical analysis. In this work, all experiments were repeated as indicated by ‘n’ provided in the Figure legends. The ‘n’ number represents experimental replications for each line of cultured cells, number of animals studied, or number of cells studied in karyotyping (Figure S1D). The total numbers of cell lines used in each experiment were also provided in the legends. Error bars in all figures indicate standard error of the mean. Neurons derived from 2 GS iPSC lines and 3 human iPSC-NPC lines (SCU-i10, SCU-i19 & SCU-i21; Barbara Mallon, Stem Cell Unit, NINDS) were used. Firstly, to determine if temperature had a significant effect, data from the normal control group and cold-exposed control group were analyzed by Student’s t-test. Secondly, to determine the significance of the effects of various treatments for the same duration, data from the treatment groups and cold-exposed control group were analyzed by ANOVA plus Tukey post hoc test. Statistical significance is defined as: * p < 0.05, ** p < 0.01 and *** p < 0.001. No inclusion criterion was used. The following exclusion criteria were used: For any given experiment, if the normal control (at 37°C) showed abnormal tubulin morphology, large numbers of dead cells, limited spontaneous RGC activity as evaluated by MEA or poor kidney histology, then the entire set of cultured cells or animal tissues/organs were removed from further analysis.
DATA AND SOFTWARE AVAILABILITY
RNA-seq data were at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93935 with access code GSE93935. Graphic presentation on RNA-seq data was done in R Package: gg plots (Wickham, 2009). No software was generated for this project.
Supplementary Material
List of gene clusters as shown in Figures S3D. To make the genes from GS and human comparable, gene ortholog mapping was performed according to Ensembl database. All DEGs (adjusted p < 0.05) from 4 conditions were mapped, 6823 orthologs were obtained. We clustered these orthologs by hierarchical clustering analyses of gene fold changes from all cell lines into 11 clusters. RPKM values of genes from each cluster are shown here. The DEGs within each cluster had more similar gene expression patterns than that from different clusters.
(A) Formation and characterization of GS iPSC colonies (see STAR METHODS). Colonies were positive for pluripotency assays such as alkaline phosphatase (AP) staining and immunostaining of the pluripotency markers OCT3/4 (green) and NANOG (red). Scale bar: 150 μm.
(B) Hematoxylin and eosin stained cross-sections of collected teratomas (see STAR METHODS) demonstrating differentiation of GS iPSCs into exogenous tissues (arrows) representing all three germ layers. Scale bar: 150 μm.
(C) (Left panel) GS iPSCs were differentiated into liver cells, a signature cell type of the endoderm, and stained by antibodies against liver cell markers ALBUMIN (green), HFN4 (red) and the cell nuclear dye DAPI (blue). (Right Panel) GS iPSCs were differentiated into muscle cells, a signature cell type of the mesoderm, and stained by antibodies against muscle cell markers Troponin T (TNNT, green), DESMIN (red) and DAPI (blue). Scale bars: 50 μm.
(D) Representative G-banded karyotype of GS iPSCs (n = 13 cells).
(E) Overlaid averaged temperatures during re-entrances into torpor from 7 GSs in one winter season (temperatures logged hourly using implanted iButton transponders). Each line represents a different animal. Records are aligned to the entrance into torpor, where 0 h is the timestamp before the first temperature drop > 3°C. The inset demonstrates the first month of hibernation from an example GS, highlighting torpor re-entrances used for this analysis in red. In 5 of 7 GSs, the body temperatures reached 10°C within 4–5 h, and all 7 GSs reached their stab le hibernation temperature in about 10 h, which is still within the time range of our cultured cell and organ cold storage experiments.
Representative experiments of human (top) and GS (bottom) TMRE intensity during temperature changes from normal (37°C) to near-hibernating (10°C) temperatures. Note the differential increases in intensity during cold exposure between species although neither the images nor their quantifications are corrected for temperature-dependent shifts in dye intensity. Quantifications after correction are presented in Figure 3A. ROIs for individual cells are overlaid with the raw images and color-coded to their intensity traces at the right. Thick black lines indicate the average normalized intensity in each experiment, and the green dashed line indicates the time point of the animation.
Recorded and spike-sorted action potentials were binned for each MEA electrode using 1-s bins. For each of the three conditions shown (left: fresh control; middle: 4-h cold exposure with vehicle control; right: 4-h cold exposure with BAM15 and PI pretreatment), a uniform intensity scaling was used for each electrode in which black = 0 spikes/s, white = 70 spikes/s. Note that each electrode may simultaneously record spikes from multiple RGCs. In this video, 5-min recordings from the total 15-min experiments were presented between 300 and 600 s.
Recorded and spike-sorted action potentials were binned for each MEA electrode using 1-s bins. For each of the four conditions shown, a uniform intensity scaling was used for each electrode in which black = 0 spikes/s, white = 190 spikes/s. Note that each electrode may simultaneously record spikes from multiple RGCs. For this experiment, six 1-s light flashes were presented beginning at 7 s and repeating every 15 s until the end of the experiment. The indicator in the center of the animation signals the timing of light flashes.
Table S1. Cell Culture Medium Formulations and Reagents, Related to STAR METHODS
To explore the differentially regulated pathways following cold stress in human and GS iPSC-neurons, the DEG clusters presented in Data File S1 were subjected to functional enrichment analysis using GO terms or KEGG pathways. The GO terms/KEGG pathways, statistical significant values, and the statistics of DEGs overlapping with the gene sets from these databases are listed in the table.
To explore the effect of cold-exposure on the transcriptome of human and GS iPSC-neurons, we performed differential expression analysis on all groups: GS 1h, GS 4h, Human 1h, and Human 4h. Neurons derived from 2 GS iPSC lines, and 2 human iPSC lines (SCU-i19 and SCU-i21) were used. The results of this analysis and related annotations are listed in the table which contains Ensemble gene ID, associated gene name, gene-counts normalized by library size for each gene in all samples, the mean expression level of each gene in all samples (base-Mean), log2 fold change, the significance p-value, and related descriptions for each gene. Genes with adjusted p < 0.05 were defined as DEGs.
(A) Cold-induced microtubule disruption is not a secondary effect of cell death (n = 5 experiments each from 2 GS and 3 human cell lines). (Upper left panel) GS iPSC-neurons maintained their long neurites (TUBB3, green) and viability (dying cells become permeable and stained by Propidium Iodide, magenta) during 16-h incubation at 4°C, but failed to survive when H2O2 (100 μM) was added. (Right panel) In contrast, 4-h incubation at 4°C disrupted most neurites in human iPSC-neurons without increasing cell death, while extended 24-h incubation caused a small increase in dead cells from 5% to 11% (Lower left panel; Student’s t-test for two-group comparisons; * p < 0.05; ** p < 0.01). Scale bars: 40 μm.
(B) Western blots and quantification of RBM3 and CIRBP proteins in awake and hibernating (torpor) GS retinas (n = 12 animals each), GS brains (n = 5 animals each), GS iPSC-neurons (n = 5 experiments from 2 cell lines for each group) and human iPSC-neurons (n = 5 experiments from 2 cell lines for each group). Note that RBM3 proteins were up-regulated in cultured neurons at mild hypothermia (33°C) with extended (60 h) incubation time, but not in other conditions tested (Student’s t-test for two-group comparisons; n.s., not significant, p > 0.05; *** p < 0.001). A, proteins extracted from active GS with a body temperature of 36°C; H, proteins extracted from hibernating GS with a body temperature of 2–4 °C.
(C) Western blots and quantification of total TUBB3, poly-E-T and Δ 2-T proteins in human iPSC-neurons following 4- or 16-h cold exposure (n = 5 experiments from 2 cell lines; Student’s t-test for two-group comparisons; * p < 0.05; ** p < 0.01; *** p < 0.001).
(D) Immunofluorescence of TUBB3 (red), poly-E-T (green), or Δ 2-T (blue) in human iPSC-neurons incubated at 4°C for 4 or 16 h (n = 5 experiments from 2 cell lines). Note: Overexpression of TUBB3 improved microtubule morphology only in the 4-h cold-exposed group; Taxol is a known drug that binds and stabilizes microtubules. Low dose Taxol (0.1 μM) did not protect human iPSC-neuronal microtubules following 16-h cold exposure. Scale bar: 40 μm.
(A) Factorial map of the principal-component analysis (PCA) of scaled mRNA RPKM values. Top variant mRNAs were used. The proportion of the variance explained by the principal components is indicated in parentheses.
(B) M-A plots showing changes in gene expression in GS and human iPSC-neurons. M: log2 mRNA fold change, A: log2 mean values of gene-counts normalized by library size. Red dots: significant up-regulated genes with adjusted p < 0.05; blue dots: significant downregulated genes with adjusted p < 0.05. Colored numbers in upper right correspond to the number of DEGs (upregulated: red; downregulated: blue).
(C) Venn diagrams indicating DEG overlap between the two time-points in human and squirrel.
(D) Venn diagrams indicating DEG overlap between human and squirrel orthologs at the two time-points.
(E) Clustering analysis of DEG orthologs with expression fold change. Sub-clusters are colored in red (785 genes), yellow (1150), grey (598), green (359), purple (167), pink (278), sky-blue (682), blue (499), black (549), orange (971), and claret (785) (top to bottom). Log2 of DEGs fold change was used.
(F) Cell condition-based functional enrichment of DEGs in human and GS. The significance of enrichment is colored ranging from magenta (significant) to green (insignificant).
(G) An example of TUBB3 predicted acetylation sites. TUBB3 protein sequences from human, mouse, rat and GS (ICTTR) were aligned. Identical amino acids in the same corresponding positions of multiple alignments are colored in red.
(A) In human iPSC-neurons, incubation at 4°C for 4 h was sufficient to cause accumulation of protein aggregates along the microtubule processes, while pre-incubation with MG-132, a proteasome-specific inhibitor known to induce cellular protein aggregate formation, did not show any further increase in aggregation (n = 8 experiments for each group from 3 cell lines; ANOVA plus post hoc Tukey test for multiple cold-exposed groups treated for the same duration; n.s. not significant; *** p < 0.001). This result is consistent with our finding that in human iPSC-neurons vasolin-containing proteins (VCPs) and chaperone proteins (HSPs) that regulate ubiquitin-mediated protein degradation were downregulated at 4°C (Figure S5), compromising the delivery of damaged proteins via VCP to proteasomes.
(B) Immunofluorescence of TUBB3 (blue), oxidized proteins (green) and a protein aggregate-binding dye (red; see STAR METHODS) in GS iPSC-neurons (n = 5 experiments from 2 cell lines). Scale bar: 20 μm.
(C) Immunofluorescence of TUBB3 (blue), oxidized proteins (green), and a protein aggregate-binding dye (red) in human iPSC-neurons (n = 6 experiments from 3 cell lines). Scale bar: 20 μm.
(A) Pre-treatment with PYR-41, an inhibitor of protein ubiquitination did not improve microtubule morphology (poly-E-T: green; TUBB3: red; Δ2-T: blue) during 4-h incubation at 4°C (n = 6 experiments from 3 cell lines). Scale bar: 30 μm.
(B) Western blots and quantification of valosin-containing protein (VCP) and molecular chaperones (HSP60 and HSP90) in human and GS iPSC-neurons after 16-h incubation at 4°C (n = 5 experiments for each group from 2 GS and 2 human iPSC lines; Student’s t-test for two-group comparisons; n.s. p > 0.05, not significant; ** p < 0.01).
(C) Western blots and quantification of proteasomal subunits PSMB2 and PSMB5, and lysosomal protein LAMP1 in human and GS iPSC-neurons after 4-h incubation at 4°C (n = 5 experiments for each group from 2 GS and 2 human iPSC lines; Student’s t-test for two-group comparisons; n.s. not significant; *** p < 0.001). Note that the LAMP1 antibody used in this study did not recognized GS LAMP1 proteins, probably because the epitope of LAMP1 proteins in GS is masked by GS-specific glycosylation.
(D) Representative western blots show that protein levels of key autophagosome components remained little changed in GS and human iPSC-neurons after 4-h cold exposure (n = 5 experiments for each group from 2 GS and 2 human iPSC lines).
(E) Immunofluorescence of TUBB3 (magenta) and LAMP1 (green; lysosomal marker) show evidence of lysosomal localization on microtubule processes of human iPSC-neurons. Scale bar: 20 μm.
(A) Immunofluorescence of TUBB3 (magenta) and GFP (green) in human iPSC-neurons transfected with plasmids overexpressing GFP only (CMV-GFP), UCP1 and GFP (CMV-UCP1-IRES2-GFP), or UCP2 and GFP (CMV-UCP2-IRES2-GFP) for 48 h and then incubated at 4°C for 4 h. Scale bar: 40 μm.
(B) Cumulative plots of TUBB3+ neurites of human iPSC-neurons following 4-h cold exposure (n = 5 experiments from 2 cell lines for each group).
(C) Immunofluorescence of TUBB3 (red), poly-E-T (green), or Δ2-T (blue) in human iPSC-neurons incubated at 4°C for 16 h. Note: High concentration of BAM15 (5 μM) produced poor microtubule morphology. Scale bar: 40 μm.
(D) Cumulative plots of TUBB3+ neurites of human iPSC-neurons (2 cell lines were tested; 0.1 μM BAM15: n = 6 experiments; 1:500 PI: n = 5 experiments; 0.1 μM BAM15 & 1:500 PI: n = 6 experiments) following 4- and 16-h cold exposure.
(E) Quantification on the lengths of TUBB3+ neurites of human iPSC-neurons under different experimental conditions (Student’s t-test between untreated controls with or without cold exposure; ANOVA plus post hoc Tukey test for multiple cold-exposed groups treated for the same duration; * p < 0.05; ** p < 0.01; *** p < 0.001).
(A) Kidneys from healthy wild-type mice were sectioned and visualized for proteins modified by lipid peroxidation (green; see STAR METHODS; n = 4 animals in each condition). Scale bars: 50 μm.
(B) Immunofluorescence of an apoptotic cell marker, cleaved Caspase-3 (red), and DAPI (blue) in transverse sections of wild-type mouse kidneys under these conditions (n = 4 animals for each condition): freshly fixed and following 4°C 72-h with or without BAM15/PI. Note that in the cold-exposed control group, high levels of cleaved Caspase-3 were found in the kidney cortex, including glomeruli (arrowhead). Scale bars: 50 μm.
Highlights.
iPSCs made from a hibernator (13-lined ground squirrel, Ictidomys tridecemlineatus)
Ground squirrel (GS) iPSC-differentiated neurons retain intrinsic cold-resistance
Human and GS neurons differ in their mitochondrial and lysosomal responses to cold
Drugs targeting these pathways improve cold tolerance in non-hibernator cells
Acknowledgments
We thank Drs. Robert Balaban, Jeffrey Diamond, Tiansen Li, Antonina Roll-Mecak, Rosa Puertollano, Zuhang Sheng, Yihong Ye and the members of the Li lab for critical discussions. Special thanks to Dr. Bing Zhou for his generous help and advice. Funding is provided by the Intramural Research Programs of the National Eye Institute (W.L.), Joint Research Fund for Overseas Natural Science of China #31528012 (Z.X. & W.L.), National Institute of Neurological Disorders and Stroke (B.S.M.), and Center for Human Immunology, Autoimmunity and Inflammation (J.C. & H.Z.).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, J.O. and W.L.; Methodology, J.O., Y.L., K.J.M., D.M., B.S.M., Z.X. and W.L.; Validation, J.O., J.M.B., T.Z., and Y.X.; Formal Analysis, J.O., J.M.B., Y.L. H.Z., J.C. and Z.X.; Investigation, J.O., J.M.B., T.Z., Y.X., H.Z. and J.C.; Resources, J.O, D.M., B.S.M. and W.L.; Data Curation, J.M.B., Y.L., J.C., Z.X. and W.L.; Writing, J.O., J.M.B., K.J.M. and W.L.; Visualization – J.O., J.M.B. and W.L.; Supervision, Z.X., B.S.M. and W.L.
DECLARATION OF INTERESTS
The authors declare no competing financial interests. A provisional patent for the composition and methods to protect mammalian tissue against cold and other metabolic stresses has been filed by the NEI under US Application Serial No. 62/547,945.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali SS, Marcondes MCG, Bajova H, Dugan LL, Conti B. Metabolic Depression and Increased Reactive Oxygen Species Production by Isolated Mitochondria at Moderately Lower Temperatures. Journal of Biological Chemistry. 2010;285:32522–32528. doi: 10.1074/jbc.M110.155432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews MT. Advances in molecular biology of hibernation in mammals. Bioessays. 2007;29:431–440. doi: 10.1002/bies.20560. [DOI] [PubMed] [Google Scholar]
- Andrews ZB, Diano S, Horvath TL. Mitochondrial uncoupling proteins in the CNS: in support of function and survival. Nat Rev Neurosci. 2005;6:829–840. doi: 10.1038/nrn1767. [DOI] [PubMed] [Google Scholar]
- Arendt T, Bullmann T. Neuronal plasticity in hibernation and the proposed role of the microtubule-associated protein tau as a “master switch” regulating synaptic gain in neuronal networks. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2013;305:R478–R489. doi: 10.1152/ajpregu.00117.2013. [DOI] [PubMed] [Google Scholar]
- Arendt T, Stieler J, Strijkstra AM, Hut RA, Rudiger J, Van der Zee EA, Harkany T, Holzer M, Hartig W. Reversible paired helical filament-like phosphorylation of tau is an adaptive process associated with neuronal plasticity in hibernating animals. J Neurosci. 2003;23:6972–6981. doi: 10.1523/JNEUROSCI.23-18-06972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avior Y, Sagi I, Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- Azzam NA, Hallenbeck JM, Kachar B. Membrane changes during hibernation. Nature. 2000;407:317–318. doi: 10.1038/35030294. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Ballinger MA, Schwartz C, Andrews MT. Enhanced oxidative capacity of ground squirrel brain mitochondria during hibernation. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2017;312:R301–R310. doi: 10.1152/ajpregu.00314.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes BM. Freeze avoidance in a mammal: body temperatures below 0 degree C in an Arctic hibernator. Science. 1989;244:1593–1595. doi: 10.1126/science.2740905. [DOI] [PubMed] [Google Scholar]
- Bastide A, Peretti D, Knight JR, Grosso S, Spriggs RV, Pichon X, Sbarrato T, Roobol A, Roobol J, Vito D, et al. RTN3 Is a Novel Cold-Induced Protein and Mediates Neuroprotective Effects of RBM3. Curr Biol. 2017;27:638–650. doi: 10.1016/j.cub.2017.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billger M, Strömberg E, Wallin M. Microtubule-associated proteins-dependent colchicine stability of acetylated cold-labile brain microtubules from the Atlantic cod, Gadus morhua. The Journal of Cell Biology. 1991;113:331–338. doi: 10.1083/jcb.113.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosc C, Andrieux A, Job D. STOP proteins. Biochemistry. 2003;42:12125–12132. doi: 10.1021/bi0352163. [DOI] [PubMed] [Google Scholar]
- Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- Boyer BB, Barnes BM, Lowell BB, Grujic D. Differential regulation of uncoupling protein gene homologues in multiple tissues of hibernating ground squirrels. Am J Physiol. 1998;275:R1232–1238. doi: 10.1152/ajpregu.1998.275.4.R1232. [DOI] [PubMed] [Google Scholar]
- Breton S, Brown D. Cold-induced microtubule disruption and relocalization of membrane proteins in kidney epithelial cells. J Am Soc Nephrol. 1998;9:155–166. doi: 10.1681/ASN.V92155. [DOI] [PubMed] [Google Scholar]
- Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83:1153–1181. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- Conde C, Caceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- Cooper ST, Richters KE, Melin TE, Liu ZJ, Hordyk PJ, Benrud RR, Geiser LR, Cash SE, Simon Shelley C, Howard DR, et al. The hibernating 13-lined ground squirrel as a model organism for potential cold storage of platelets. Am J Physiol Regul Integr Comp Physiol. 2012;302:R1202–1208. doi: 10.1152/ajpregu.00018.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotterill LA, Gower JD, Fuller BJ, Green CJ. Oxidative damage to kidney membranes during cold ischemia. Evidence of a role for calcium. Transplantation. 1989;48:745–751. doi: 10.1097/00007890-198911000-00004. [DOI] [PubMed] [Google Scholar]
- Dave KR, Christian SL, Perez-Pinzon MA, Drew KL. Neuroprotection: lessons from hibernators. Comp Biochem Physiol B Biochem Mol Biol. 2012;162:1–9. doi: 10.1016/j.cbpb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedukhova VI, Mokhova EN, Skulachev VP, Starkov AA, Arrigoni-Martelli E, Bobyleva VA. Uncoupling effect of fatty acids on heart muscle mitochondria and submitochondrial particles. FEBS Lett. 1991;295:51–54. doi: 10.1016/0014-5793(91)81382-i. [DOI] [PubMed] [Google Scholar]
- Delhaye C, Mahmoudi M, Waksman R. Hypothermia therapy: neurological and cardiac benefits. J Am Coll Cardiol. 2012;59:197–210. doi: 10.1016/j.jacc.2011.06.077. [DOI] [PubMed] [Google Scholar]
- Detrich HW, 3rd, Parker SK. Divergent neural beta tubulin from the Antarctic fish Notothenia coriiceps neglecta: potential sequence contributions to cold adaptation of microtubule assembly. Cell Motil Cytoskeleton. 1993;24:156–166. doi: 10.1002/cm.970240303. [DOI] [PubMed] [Google Scholar]
- Drew KL, Buck CL, Barnes BM, Christian SL, Rasley BT, Harris MB. Central nervous system regulation of mammalian hibernation: implications for metabolic suppression and ischemia tolerance. J Neurochem. 2007;102:1713–1726. doi: 10.1111/j.1471-4159.2007.04675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew KL, McGee RC, Wells MS, Kelleher-Andersson JA. Growth and differentiation of adult hippocampal arctic ground squirrel neural stem cells. J Vis Exp. 2011 doi: 10.3791/2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop RA, Brunk UT, Rodgers KJ. Oxidized proteins: mechanisms of removal and consequences of accumulation. IUBMB Life. 2009;61:522–527. doi: 10.1002/iub.189. [DOI] [PubMed] [Google Scholar]
- Fabre E, Conti M, Paradis V, Droupy S, Bedossa P, Legrand A, Benoit G, Eschwege P. Impact of different combined preservation modalities on warm ischemic kidneys: effect on oxidative stress, hydrostatic perfusion characteristics and tissue damage. Urol Res. 2002;30:89–96. doi: 10.1007/s00240-002-0237-6. [DOI] [PubMed] [Google Scholar]
- Fedorov VB, Goropashnaya AV, Stewart NC, Toien O, Chang C, Wang H, Yan J, Showe LC, Showe MK, Barnes BM. Comparative functional genomics of adaptation to muscular disuse in hibernating mammals. Mol Ecol. 2014;23:5524–5537. doi: 10.1111/mec.12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forreider B, Pozivilko D, Kawaji Q, Geng X, Ding Y. Hibernation-like neuroprotection in stroke by attenuating brain metabolic dysfunction. Prog Neurobiol. 2016 doi: 10.1016/j.pneurobio.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Ewan L, Bauer M, Mattaliano RJ, Zaal K, Ralston E, Plotz PH, Raben N. Dysfunction of endocytic and autophagic pathways in a lysosomal storage disease. Annals of Neurology. 2006;59:700–708. doi: 10.1002/ana.20807. [DOI] [PubMed] [Google Scholar]
- Geiser F. Metabolic rate and body temperature reduction during hibernation and daily torpor. Annu Rev Physiol. 2004;66:239–274. doi: 10.1146/annurev.physiol.66.032102.115105. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Gregersen N, Bross P. Protein misfolding and cellular stress: an overview. Methods Mol Biol. 2010;648:3–23. doi: 10.1007/978-1-60761-756-3_1. [DOI] [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Davies KJ. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997;11:526–534. [PubMed] [Google Scholar]
- Guillaud L, Bosc C, Fourest-Lieuvin A, Denarier E, Pirollet F, Lafanechere L, Job D. STOP proteins are responsible for the high degree of microtubule stabilization observed in neuronal cells. J Cell Biol. 1998;142:167–179. doi: 10.1083/jcb.142.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr Opin Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton M, Melvin RG, Andrews MT. Transcriptomic analysis of brown adipose tissue across the physiological extremes of natural hibernation. PLoS One. 2013;8:e85157. doi: 10.1371/journal.pone.0085157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindle AG, Martin SL. Cytoskeletal Regulation Dominates Temperature-Sensitive Proteomic Changes of Hibernation in Forebrain of 13-Lined Ground Squirrels. PLOS ONE. 2013;8:e71627. doi: 10.1371/journal.pone.0071627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Jain S, Keys D, Martin S, Edelstein CL, Jani A. Protection From Apoptotic Cell Death During Cold Storage Followed by Rewarming in 13-Lined Ground Squirrel Tubular Cells: The Role of Prosurvival Factors X-Linked Inhibitor of Apoptosis and PhosphoAkt. Transplantation. 2016;100:538–545. doi: 10.1097/TP.0000000000000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Hoogenraad CC. Building the Neuronal Microtubule Cytoskeleton. Neuron. 2015;87:492–506. doi: 10.1016/j.neuron.2015.05.046. [DOI] [PubMed] [Google Scholar]
- Kiemer L, Bendtsen JD, Blom N. NetAcet: prediction of N-terminal acetylation sites. Bioinformatics. 2005;21:1269–1270. doi: 10.1093/bioinformatics/bti130. [DOI] [PubMed] [Google Scholar]
- Laursen WJ, Mastrotto M, Pesta D, Funk OH, Goodman JB, Merriman DK, Ingolia N, Shulman GI, Bagriantsev SN, Gracheva EO. Neuronal UCP1 expression suggests a mechanism for local thermogenesis during hibernation. Proc Natl Acad Sci U S A. 2015;112:1607–1612. doi: 10.1073/pnas.1421419112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: crosstalk and redox signalling. Biochem J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Chan SL, de Souza-Pinto NC, Slevin JR, Wersto RP, Zhan M, Mustafa K, de Cabo R, Mattson MP. Mitochondrial UCP4 mediates an adaptive shift in energy metabolism and increases the resistance of neurons to metabolic and oxidative stress. Neuromolecular Med. 2006;8:389–414. doi: 10.1385/NMM:8:3:389. [DOI] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis MH, Leuvenink HG, Ploeg RJ. Perspectives in organ preservation. Transplantation. 2007;83:1289–1298. doi: 10.1097/01.tp.0000265586.66475.cc. [DOI] [PubMed] [Google Scholar]
- Mangino MJ, Tian T, Ametani M, Lindell S, Southard JH. Cytoskeletal involvement in hypothermic renal preservation injury. Transplantation. 2008;85:427–436. doi: 10.1097/TP.0b013e31815fed17. [DOI] [PubMed] [Google Scholar]
- Marion DW, Penrod LE, Kelsey SF, Obrist WD, Kochanek PM, Palmer AM, Wisniewski SR, DeKosky ST. Treatment of Traumatic Brain Injury with Moderate Hypothermia. New England Journal of Medicine. 1997;336:540–546. doi: 10.1056/NEJM199702203360803. [DOI] [PubMed] [Google Scholar]
- McAnulty JF. Hypothermic organ preservation by static storage methods: Current status and a view to the future. Cryobiology. 2010;60:S13–19. doi: 10.1016/j.cryobiol.2009.06.004. [DOI] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg PJ, Miller MW. Culturing hippocampal and cortical neurons. Methods Cell Biol. 2003;71:111–127. doi: 10.1016/s0091-679x(03)01007-0. [DOI] [PubMed] [Google Scholar]
- Mertens J, Paquola AC, Ku M, Hatch E, Bohnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell. 2015;17:705–718. doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T, Rotaru D, Saba H, Smith RA, Murphy MP, MacMillan-Crow LA. The mitochondria-targeted antioxidant mitoquinone protects against cold storage injury of renal tubular cells and rat kidneys. J Pharmacol Exp Ther. 2011;336:682–692. doi: 10.1124/jpet.110.176743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P, Jr, Storey KB. Mammalian hibernation: differential gene expression and novel application of epigenetic controls. Int J Dev Biol. 2009;53:433–442. doi: 10.1387/ijdb.082643pm. [DOI] [PubMed] [Google Scholar]
- Nathaniel TI. Brain-regulated metabolic suppression during hibernation: a neuroprotective mechanism for perinatal hypoxia-ischemia. Int J Stroke. 2008;3:98–104. doi: 10.1111/j.1747-4949.2008.00186.x. [DOI] [PubMed] [Google Scholar]
- Niemann CU, Feiner J, Swain S, Bunting S, Friedman M, Crutchfield M, Broglio K, Hirose R, Roberts JP, Malinoski D. Therapeutic Hypothermia in Deceased Organ Donors and Kidney-Graft Function. N Engl J Med. 2015;373:405–414. doi: 10.1056/NEJMoa1501969. [DOI] [PubMed] [Google Scholar]
- Penazzi L, Bakota L, Brandt R. Microtubule Dynamics in Neuronal Development, Plasticity, and Neurodegeneration. Int Rev Cell Mol Biol. 2016;321:89–169. doi: 10.1016/bs.ircmb.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Peretti D, Bastide A, Radford H, Verity N, Molloy C, Martin MG, Moreno JA, Steinert JR, Smith T, Dinsdale D, et al. RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature. 2015;518:236–239. doi: 10.1038/nature14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang X, Yang WL, Wu R, Zhou M, Jacob A, Dong W, Kuncewitch M, Ji Y, Yang H, Wang H, et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat Med. 2013;19:1489–1495. doi: 10.1038/nm.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones QJ, Ma Q, Zhang Z, Barnes BM, Podgoreanu MV. Organ protective mechanisms common to extremes of physiology: a window through hibernation biology. Integr Comp Biol. 2014;54:497–515. doi: 10.1093/icb/icu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen U, de Groot H. Cold-induced release of reactive oxygen species as a decisive mediator of hypothermia injury to cultured liver cells. Free Radic Biol Med. 1998;24:1316–1323. doi: 10.1016/s0891-5849(97)00456-5. [DOI] [PubMed] [Google Scholar]
- Reinhard K, Mutter M, Gustafsson E, Gustafsson L, Vaegler M, Schultheiss M, Muller S, Yoeruek E, Schrader M, Munch TA. Hypothermia Promotes Survival of Ischemic Retinal Ganglion Cells. Invest Ophthalmol Vis Sci. 2016;57:658–663. doi: 10.1167/iovs.15-17751. [DOI] [PubMed] [Google Scholar]
- Roberg K, Ollinger K. Oxidative stress causes relocation of the lysosomal enzyme cathepsin D with ensuing apoptosis in neonatal rat cardiomyocytes. Am J Pathol. 1998;152:1151–1156. [PMC free article] [PubMed] [Google Scholar]
- Schultheiss M, Schnichels S, Hermann T, Hurst J, Feldkaemper M, Arango-Gonzalez B, Ueffing M, Bartz-Schmidt KU, Zeck G, Spitzer MS. Hypothermia Protects and Prolongs the Tolerance Time of Retinal Ganglion Cells against Ischemia. PLoS One. 2016;11:e0148616. doi: 10.1371/journal.pone.0148616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Coleman J, Chan E, Nicholson TP, Dai L, Sheppard PW, Patton WF. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem Biophys. 2011;60:173–185. doi: 10.1007/s12013-010-9138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulachev VP. Fatty acid circuit as a physiological mechanism of uncoupling of oxidative phosphorylation. FEBS Lett. 1991;294:158–162. doi: 10.1016/0014-5793(91)80658-p. [DOI] [PubMed] [Google Scholar]
- Song Y, Kirkpatrick LL, Schilling AB, Helseth DL, Chabot N, Keillor JW, Johnson GV, Brady ST. Transglutaminase and polyamination of tubulin: posttranslational modification for stabilizing axonal microtubules. Neuron. 2013;78:109–123. doi: 10.1016/j.neuron.2013.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir ML, Zweig AS, Rosenbloom KR, Raney BJ, Paten B, Nejad P, Lee BT, Learned K, Karolchik D, Hinrichs AS, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples JF. Metabolic Flexibility: Hibernation, Torpor, and Estivation. Compr Physiol. 2016a;6:737–771. doi: 10.1002/cphy.c140064. [DOI] [PubMed] [Google Scholar]
- Staples JF. Comprehensive Physiology. John Wiley & Sons, Inc; 2016b. Metabolic Flexibility: Hibernation, Torpor, and Estivation. [DOI] [PubMed] [Google Scholar]
- Starkov AA, Fiskum G. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. Journal of Neurochemistry. 2003;86:1101–1107. doi: 10.1046/j.1471-4159.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. Metabolic rate depression: the biochemistry of mammalian hibernation. Adv Clin Chem. 2010;52:77–108. [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Taylor MJ. Advances in Biopreservation. CRC Press; 2006. Biology of Cell Survival in the Cold; pp. 15–62. [Google Scholar]
- Tong G, Endersfelder S, Rosenthal LM, Wollersheim S, Sauer IM, Buhrer C, Berger F, Schmitt KR. Effects of moderate and deep hypothermia on RNA-binding proteins RBM3 and CIRP expressions in murine hippocampal brain slices. Brain Res. 2013;1504:74–84. doi: 10.1016/j.brainres.2013.01.041. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Ohe CG, Darian-Smith C, Garner CC, Heller HC. Ubiquitous and temperature-dependent neural plasticity in hibernators. J Neurosci. 2006;26:10590–10598. doi: 10.1523/JNEUROSCI.2874-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin M, Strömberg E. Cold-stable and cold-adapted microtubules. Int Rev Cytol. 1995;157:1–31. doi: 10.1016/s0074-7696(08)62155-5. [DOI] [PubMed] [Google Scholar]
- Wang L, Du Y, Lu M, Li T. ASEB: a web server for KAT-specific acetylation site prediction. Nucleic Acids Res. 2012;40:W376–379. doi: 10.1093/nar/gks437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BC, Wilson L. Cold-stable microtubules from brain. Biochemistry. 1980;19:1993–2001. doi: 10.1021/bi00550a041. [DOI] [PubMed] [Google Scholar]
- Weber K, Pollack R, Bibring T. Antibody against tuberlin: the specific visualization of cytoplasmic microtubules in tissue culture cells. Proc Natl Acad Sci U S A. 1975;72:459–463. doi: 10.1073/pnas.72.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. [Google Scholar]
- Williams DR, Epperson LE, Li W, Hughes MA, Taylor R, Rogers J, Martin SL, Cossins AR, Gracey AY. Seasonally hibernating phenotype assessed through transcript screening. Physiol Genomics. 2005;24:13–22. doi: 10.1152/physiolgenomics.00301.2004. [DOI] [PubMed] [Google Scholar]
- Wilson C, Gonzalez-Billault C. Regulation of cytoskeletal dynamics by redox signaling and oxidative stress: implications for neuronal development and trafficking. Front Cell Neurosci. 2015;9:381. doi: 10.3389/fncel.2015.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Sato Y, Mori N. Difference in induction of uncoupling protein genes in adipose tissues between young and old rats during cold exposure. FEBS Lett. 1999;458:157–161. doi: 10.1016/s0014-5793(99)01143-6. [DOI] [PubMed] [Google Scholar]
- Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenari MA, Han HS. Neuroprotective mechanisms of hypothermia in brain ischaemia. Nat Rev Neurosci. 2012;13:267–278. doi: 10.1038/nrn3174. [DOI] [PubMed] [Google Scholar]
- Zhang T, Ye Y. The final moments of misfolded proteins en route to the proteasome. DNA Cell Biol. 2014;33:477–483. doi: 10.1089/dna.2014.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pan YH, Yin Q, Yang T, Dong D, Liao CC, Zhang S. Critical roles of mitochondria in brain activities of torpid Myotis ricketti bats revealed by a proteomic approach. Journal of Proteomics. 2014;105:266–284. doi: 10.1016/j.jprot.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Zhu X, Buhrer C, Wellmann S. Cold-inducible proteins CIRP and RBM3, a unique couple with activities far beyond the cold. Cell Mol Life Sci. 2016;73:3839–3859. doi: 10.1007/s00018-016-2253-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of gene clusters as shown in Figures S3D. To make the genes from GS and human comparable, gene ortholog mapping was performed according to Ensembl database. All DEGs (adjusted p < 0.05) from 4 conditions were mapped, 6823 orthologs were obtained. We clustered these orthologs by hierarchical clustering analyses of gene fold changes from all cell lines into 11 clusters. RPKM values of genes from each cluster are shown here. The DEGs within each cluster had more similar gene expression patterns than that from different clusters.
(A) Formation and characterization of GS iPSC colonies (see STAR METHODS). Colonies were positive for pluripotency assays such as alkaline phosphatase (AP) staining and immunostaining of the pluripotency markers OCT3/4 (green) and NANOG (red). Scale bar: 150 μm.
(B) Hematoxylin and eosin stained cross-sections of collected teratomas (see STAR METHODS) demonstrating differentiation of GS iPSCs into exogenous tissues (arrows) representing all three germ layers. Scale bar: 150 μm.
(C) (Left panel) GS iPSCs were differentiated into liver cells, a signature cell type of the endoderm, and stained by antibodies against liver cell markers ALBUMIN (green), HFN4 (red) and the cell nuclear dye DAPI (blue). (Right Panel) GS iPSCs were differentiated into muscle cells, a signature cell type of the mesoderm, and stained by antibodies against muscle cell markers Troponin T (TNNT, green), DESMIN (red) and DAPI (blue). Scale bars: 50 μm.
(D) Representative G-banded karyotype of GS iPSCs (n = 13 cells).
(E) Overlaid averaged temperatures during re-entrances into torpor from 7 GSs in one winter season (temperatures logged hourly using implanted iButton transponders). Each line represents a different animal. Records are aligned to the entrance into torpor, where 0 h is the timestamp before the first temperature drop > 3°C. The inset demonstrates the first month of hibernation from an example GS, highlighting torpor re-entrances used for this analysis in red. In 5 of 7 GSs, the body temperatures reached 10°C within 4–5 h, and all 7 GSs reached their stab le hibernation temperature in about 10 h, which is still within the time range of our cultured cell and organ cold storage experiments.
Representative experiments of human (top) and GS (bottom) TMRE intensity during temperature changes from normal (37°C) to near-hibernating (10°C) temperatures. Note the differential increases in intensity during cold exposure between species although neither the images nor their quantifications are corrected for temperature-dependent shifts in dye intensity. Quantifications after correction are presented in Figure 3A. ROIs for individual cells are overlaid with the raw images and color-coded to their intensity traces at the right. Thick black lines indicate the average normalized intensity in each experiment, and the green dashed line indicates the time point of the animation.
Recorded and spike-sorted action potentials were binned for each MEA electrode using 1-s bins. For each of the three conditions shown (left: fresh control; middle: 4-h cold exposure with vehicle control; right: 4-h cold exposure with BAM15 and PI pretreatment), a uniform intensity scaling was used for each electrode in which black = 0 spikes/s, white = 70 spikes/s. Note that each electrode may simultaneously record spikes from multiple RGCs. In this video, 5-min recordings from the total 15-min experiments were presented between 300 and 600 s.
Recorded and spike-sorted action potentials were binned for each MEA electrode using 1-s bins. For each of the four conditions shown, a uniform intensity scaling was used for each electrode in which black = 0 spikes/s, white = 190 spikes/s. Note that each electrode may simultaneously record spikes from multiple RGCs. For this experiment, six 1-s light flashes were presented beginning at 7 s and repeating every 15 s until the end of the experiment. The indicator in the center of the animation signals the timing of light flashes.
Table S1. Cell Culture Medium Formulations and Reagents, Related to STAR METHODS
To explore the differentially regulated pathways following cold stress in human and GS iPSC-neurons, the DEG clusters presented in Data File S1 were subjected to functional enrichment analysis using GO terms or KEGG pathways. The GO terms/KEGG pathways, statistical significant values, and the statistics of DEGs overlapping with the gene sets from these databases are listed in the table.
To explore the effect of cold-exposure on the transcriptome of human and GS iPSC-neurons, we performed differential expression analysis on all groups: GS 1h, GS 4h, Human 1h, and Human 4h. Neurons derived from 2 GS iPSC lines, and 2 human iPSC lines (SCU-i19 and SCU-i21) were used. The results of this analysis and related annotations are listed in the table which contains Ensemble gene ID, associated gene name, gene-counts normalized by library size for each gene in all samples, the mean expression level of each gene in all samples (base-Mean), log2 fold change, the significance p-value, and related descriptions for each gene. Genes with adjusted p < 0.05 were defined as DEGs.
(A) Cold-induced microtubule disruption is not a secondary effect of cell death (n = 5 experiments each from 2 GS and 3 human cell lines). (Upper left panel) GS iPSC-neurons maintained their long neurites (TUBB3, green) and viability (dying cells become permeable and stained by Propidium Iodide, magenta) during 16-h incubation at 4°C, but failed to survive when H2O2 (100 μM) was added. (Right panel) In contrast, 4-h incubation at 4°C disrupted most neurites in human iPSC-neurons without increasing cell death, while extended 24-h incubation caused a small increase in dead cells from 5% to 11% (Lower left panel; Student’s t-test for two-group comparisons; * p < 0.05; ** p < 0.01). Scale bars: 40 μm.
(B) Western blots and quantification of RBM3 and CIRBP proteins in awake and hibernating (torpor) GS retinas (n = 12 animals each), GS brains (n = 5 animals each), GS iPSC-neurons (n = 5 experiments from 2 cell lines for each group) and human iPSC-neurons (n = 5 experiments from 2 cell lines for each group). Note that RBM3 proteins were up-regulated in cultured neurons at mild hypothermia (33°C) with extended (60 h) incubation time, but not in other conditions tested (Student’s t-test for two-group comparisons; n.s., not significant, p > 0.05; *** p < 0.001). A, proteins extracted from active GS with a body temperature of 36°C; H, proteins extracted from hibernating GS with a body temperature of 2–4 °C.
(C) Western blots and quantification of total TUBB3, poly-E-T and Δ 2-T proteins in human iPSC-neurons following 4- or 16-h cold exposure (n = 5 experiments from 2 cell lines; Student’s t-test for two-group comparisons; * p < 0.05; ** p < 0.01; *** p < 0.001).
(D) Immunofluorescence of TUBB3 (red), poly-E-T (green), or Δ 2-T (blue) in human iPSC-neurons incubated at 4°C for 4 or 16 h (n = 5 experiments from 2 cell lines). Note: Overexpression of TUBB3 improved microtubule morphology only in the 4-h cold-exposed group; Taxol is a known drug that binds and stabilizes microtubules. Low dose Taxol (0.1 μM) did not protect human iPSC-neuronal microtubules following 16-h cold exposure. Scale bar: 40 μm.
(A) Factorial map of the principal-component analysis (PCA) of scaled mRNA RPKM values. Top variant mRNAs were used. The proportion of the variance explained by the principal components is indicated in parentheses.
(B) M-A plots showing changes in gene expression in GS and human iPSC-neurons. M: log2 mRNA fold change, A: log2 mean values of gene-counts normalized by library size. Red dots: significant up-regulated genes with adjusted p < 0.05; blue dots: significant downregulated genes with adjusted p < 0.05. Colored numbers in upper right correspond to the number of DEGs (upregulated: red; downregulated: blue).
(C) Venn diagrams indicating DEG overlap between the two time-points in human and squirrel.
(D) Venn diagrams indicating DEG overlap between human and squirrel orthologs at the two time-points.
(E) Clustering analysis of DEG orthologs with expression fold change. Sub-clusters are colored in red (785 genes), yellow (1150), grey (598), green (359), purple (167), pink (278), sky-blue (682), blue (499), black (549), orange (971), and claret (785) (top to bottom). Log2 of DEGs fold change was used.
(F) Cell condition-based functional enrichment of DEGs in human and GS. The significance of enrichment is colored ranging from magenta (significant) to green (insignificant).
(G) An example of TUBB3 predicted acetylation sites. TUBB3 protein sequences from human, mouse, rat and GS (ICTTR) were aligned. Identical amino acids in the same corresponding positions of multiple alignments are colored in red.
(A) In human iPSC-neurons, incubation at 4°C for 4 h was sufficient to cause accumulation of protein aggregates along the microtubule processes, while pre-incubation with MG-132, a proteasome-specific inhibitor known to induce cellular protein aggregate formation, did not show any further increase in aggregation (n = 8 experiments for each group from 3 cell lines; ANOVA plus post hoc Tukey test for multiple cold-exposed groups treated for the same duration; n.s. not significant; *** p < 0.001). This result is consistent with our finding that in human iPSC-neurons vasolin-containing proteins (VCPs) and chaperone proteins (HSPs) that regulate ubiquitin-mediated protein degradation were downregulated at 4°C (Figure S5), compromising the delivery of damaged proteins via VCP to proteasomes.
(B) Immunofluorescence of TUBB3 (blue), oxidized proteins (green) and a protein aggregate-binding dye (red; see STAR METHODS) in GS iPSC-neurons (n = 5 experiments from 2 cell lines). Scale bar: 20 μm.
(C) Immunofluorescence of TUBB3 (blue), oxidized proteins (green), and a protein aggregate-binding dye (red) in human iPSC-neurons (n = 6 experiments from 3 cell lines). Scale bar: 20 μm.
(A) Pre-treatment with PYR-41, an inhibitor of protein ubiquitination did not improve microtubule morphology (poly-E-T: green; TUBB3: red; Δ2-T: blue) during 4-h incubation at 4°C (n = 6 experiments from 3 cell lines). Scale bar: 30 μm.
(B) Western blots and quantification of valosin-containing protein (VCP) and molecular chaperones (HSP60 and HSP90) in human and GS iPSC-neurons after 16-h incubation at 4°C (n = 5 experiments for each group from 2 GS and 2 human iPSC lines; Student’s t-test for two-group comparisons; n.s. p > 0.05, not significant; ** p < 0.01).
(C) Western blots and quantification of proteasomal subunits PSMB2 and PSMB5, and lysosomal protein LAMP1 in human and GS iPSC-neurons after 4-h incubation at 4°C (n = 5 experiments for each group from 2 GS and 2 human iPSC lines; Student’s t-test for two-group comparisons; n.s. not significant; *** p < 0.001). Note that the LAMP1 antibody used in this study did not recognized GS LAMP1 proteins, probably because the epitope of LAMP1 proteins in GS is masked by GS-specific glycosylation.
(D) Representative western blots show that protein levels of key autophagosome components remained little changed in GS and human iPSC-neurons after 4-h cold exposure (n = 5 experiments for each group from 2 GS and 2 human iPSC lines).
(E) Immunofluorescence of TUBB3 (magenta) and LAMP1 (green; lysosomal marker) show evidence of lysosomal localization on microtubule processes of human iPSC-neurons. Scale bar: 20 μm.
(A) Immunofluorescence of TUBB3 (magenta) and GFP (green) in human iPSC-neurons transfected with plasmids overexpressing GFP only (CMV-GFP), UCP1 and GFP (CMV-UCP1-IRES2-GFP), or UCP2 and GFP (CMV-UCP2-IRES2-GFP) for 48 h and then incubated at 4°C for 4 h. Scale bar: 40 μm.
(B) Cumulative plots of TUBB3+ neurites of human iPSC-neurons following 4-h cold exposure (n = 5 experiments from 2 cell lines for each group).
(C) Immunofluorescence of TUBB3 (red), poly-E-T (green), or Δ2-T (blue) in human iPSC-neurons incubated at 4°C for 16 h. Note: High concentration of BAM15 (5 μM) produced poor microtubule morphology. Scale bar: 40 μm.
(D) Cumulative plots of TUBB3+ neurites of human iPSC-neurons (2 cell lines were tested; 0.1 μM BAM15: n = 6 experiments; 1:500 PI: n = 5 experiments; 0.1 μM BAM15 & 1:500 PI: n = 6 experiments) following 4- and 16-h cold exposure.
(E) Quantification on the lengths of TUBB3+ neurites of human iPSC-neurons under different experimental conditions (Student’s t-test between untreated controls with or without cold exposure; ANOVA plus post hoc Tukey test for multiple cold-exposed groups treated for the same duration; * p < 0.05; ** p < 0.01; *** p < 0.001).
(A) Kidneys from healthy wild-type mice were sectioned and visualized for proteins modified by lipid peroxidation (green; see STAR METHODS; n = 4 animals in each condition). Scale bars: 50 μm.
(B) Immunofluorescence of an apoptotic cell marker, cleaved Caspase-3 (red), and DAPI (blue) in transverse sections of wild-type mouse kidneys under these conditions (n = 4 animals for each condition): freshly fixed and following 4°C 72-h with or without BAM15/PI. Note that in the cold-exposed control group, high levels of cleaved Caspase-3 were found in the kidney cortex, including glomeruli (arrowhead). Scale bars: 50 μm.