Abstract
Postharvest aflatoxin contamination is a challenging issue that affects peanut quality. Aflatoxin is produced by fungi belonging to the Aspergilli group, and is known as an acutely toxic, carcinogenic, and immune-suppressing class of mycotoxins. Evidence for several host genetic factors that may impact aflatoxin contamination has been reported, e.g., genes for lipoxygenase (PnLOX1 and PnLOX2/PnLOX3 that showed either positive or negative regulation with Aspergillus infection), reactive oxygen species, and WRKY (highly associated with or differentially expressed upon infection of maize with Aspergillus flavus); however, their roles remain unclear. Therefore, we conducted an RNA-sequencing experiment to differentiate gene response to the infection by A. flavus between resistant (ICG 1471) and susceptible (Florida-07) cultivated peanut genotypes. The gene expression profiling analysis was designed to reveal differentially expressed genes in response to the infection (infected vs. mock-treated seeds). In addition, the differential expression of the fungal genes was profiled. The study revealed the complexity of the interaction between the fungus and peanut seeds as the expression of a large number of genes was altered, including some in the process of plant defense to aflatoxin accumulation. Analysis of the experimental data with “keggseq,” a novel designed tool for Kyoto Encyclopedia of Genes and Genomes enrichment analysis, showed the importance of α-linolenic acid metabolism, protein processing in the endoplasmic reticulum, spliceosome, and carbon fixation and metabolism pathways in conditioning resistance to aflatoxin accumulation. In addition, coexpression network analysis was carried out to reveal the correlation of gene expression among peanut and fungal genes. The results showed the importance of WRKY, toll/Interleukin1 receptor–nucleotide binding site leucine-rich repeat (TIR-NBS-LRR), ethylene, and heat shock proteins in the resistance mechanism.
Keywords: peanut, aflatoxin, Aspergilli, Aspergillus flavus, keggseq, KEGG enrichment analysis
Peanut (Arachis hypogaea L.), an oilseed crop, is a suitable substrate for fungal growth and mycotoxin production, and is the most susceptible species for aflatoxin production as compared with other oilseed crops such as soybean (Bean et al. 1972). Different mycotoxins are formed on peanuts, e.g., cyclopiazonic acid, zearalenone, trichothecene-toxins, and aflatoxin (Chang et al. 2013). The latter is the most common and destructive mycotoxin produced on peanut and other crops such as corn, cottonseed, rice, wheat, oat, and barley (Stubblefield et al. 1967; Cotty and Jaime-Garcia 2007; Mateo et al. 2011; Suárez-Bonnet et al. 2013; Dunham et al. 2017). Aflatoxin has received widespread attention since the discovery that it was the causative agent of “Turkey X disease,” a disease that killed 100,000 young turkeys on English poultry farms in 1960 (Spensley 1963). Aflatoxins are in an acutely toxic, carcinogenic, and immunosuppressive class of mycotoxins affecting animals including humans (Scheidegger and Payne 2003). In addition, aflatoxins are considered mutagenic agents as they cause oxidative damage to DNA (Verma 2004). Aflatoxins are classified in four major classes: B1, B2, G1, and G2 (Ehrlich et al. 2004); however, aflatoxin B1 is the most potent and carcinogenic naturally occurring substance known (Squire 1981).
Aflatoxin is produced in agricultural products mainly by contaminating Aspergillus flavus and A. parasiticus. Not only are the fungal products harmful, the fungus A. flavus is an ascomycetous fungus that can infect humans, plants, animals, and insects (Klich 2007). In humans, it is the second leading cause of invasive aspergillosis disease after A. fumigatus (Hedayati et al. 2007). A. flavus may infect peanut and lead to aflatoxin accumulation in the field (preharvest aflatoxin contamination) or during storage (postharvest aflatoxin contamination).
Abiotic stress is an important factor contributing to preharvest aflatoxin accumulation. Drought-tolerant genotypes, sufficient irrigation, and best management practices may reduce preharvest aflatoxin contamination since drought conditions and heat stress exacerbate aflatoxin contamination (Kisyombe et al. 1985; Holbrook et al. 2000a; Craufurd et al. 2006; Nigam et al. 2009). However, better understanding of the resistance mechanisms and the development of resistant genotypes for postharvest aflatoxin contamination is needed.
Different genetic factors that may affect Aspergillus spp. infection and/or aflatoxin accumulation have been proposed; however, the exact role of such factors remains unclear. Lipoxygenase (LOX) is a gene super family that encodes dioxygenases. It was found to have a critical role in many disease-response mechanisms of plants such as those against nematodes (Gao et al. 2008; Ozalvo et al. 2014), rust (Choi et al. 2008), downy mildew (Babitha et al. 2004, 2006), and insects (Wang et al. 2008; Tang et al. 2009; Yan et al. 2013). However, in peanut it has received most attention for its potential role in resistance to A. flavus. Burow et al. (2000) isolated the first peanut LOX, PnLOX1, from a seed cDNA library and found expression to be enhanced after infection by A. parasiticus. An opposite result was obtained by Tsitsigiannis et al. (2005) while studying two more LOXs, PnLOX2 and PnLOX3, where they observed reduced expression upon infection by A. flavus. Two additional LOXs were discovered later, which showed various responses to A. flavus inoculation (Müller et al. 2014). In addition, LOX expression differences have been observed upon interaction of Aspergillus spp. with plants other than peanut, e.g., soybean (Bean et al. 1972; Doehlert et al. 1993; Boué et al. 2005), maize (Gao et al. 2009; Huang et al. 2013), cottonseeds (Zeringue 1996), and almond (Mita et al. 2007).
Additionally, β-1,3-glucanases, chitinases, pathogenesis-related proteins 10 and 10.1, ribosome-inactivating proteins, and zeamatin may be related to A. flavus resistance (Fountain et al. 2014), along with WRKY transcription factors (Fountain et al. 2015b). Furthermore, the drought stress-responding compounds such as reactive oxygen species (ROS) are highly associated with aflatoxin production (Jayashree and Subramanyam 2000; Reverberi et al. 2012; Fountain et al. 2015a) and antioxidant enzymes are highly coexpressed with fungal growth under infection conditions (Fountain et al. 2016a).
Cultivated peanut, Ar. hypogaea, is an allotetraploid (2n = 4× = 40) that was formed by spontaneous doubling of a cross between two diploid progenitors, A. duranensis and A. ipaensis (Seijo et al. 2004). The whole-genome sequence of tetraploid peanut is not yet available. However, high-quality, well-annotated genomes of A. duranensis and A. ipaensis have been released (Bertioli et al. 2016; https://peautbase.org/). The two subgenomes together have a size of ∼2.7 Gb with 88,876 annotated proteins. The whole-genome sequence of A. flavus also has been released (https://www.aspergillusflavus.org/genomics/). The genome is 40 Mbp, containing 13,478 predicted genes on eight chromosomes. Aflatoxin biosynthesis is encoded by a 70-kbp gene cluster and has been extensively studied for A. flavus and A. parasiticus (Yu et al. 2004; Ehrlich et al. 2005; Georgianna and Payne 2009). Although only these two fungi are responsible for aflatoxin production in food products, the cluster region is conserved across other species such as A. bombycis and Emericella astellata (Amaike and Keller 2011). The aflatoxin biosynthetic pathway is responsive to environmental conditions such as temperature, stress, lipids, and salts (Bhatnagar et al. 2003), which makes breeding for resistance to aflatoxin production challenging.
In this study, we utilized the published peanut and A. flavus genomes to study the genes that respond to A. flavus infection and are differentially expressed during fungal interaction with resistant vs. susceptible peanut genotypes. Extended analysis comprising self-organizing maps, gene ontology (GO) term enrichment, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, and coexpression network analysis was conducted.
Materials and Methods
Plant material and infection
ICG 1471 and Florida-07 were planted on the Tifton Campus of the University of Georgia in June and harvested in October 2015. Thirty seeds from each genotype were inoculated with fungal spores, alongside 10 mock-treated seeds according to Korani et al. (2017). Briefly, seeds were surface sterilized for 15 min under UV light (LABCONCO purified class II biosafety cabinet, Kansas City, MO). The AF-70-GFP strain (Rajasekaran et al. 2008) was used for infection at a concentration of 1000 conidia/ml. The fungus was grown on potato dextrose agar (PDA) medium in petri dishes for 2 weeks at 30°, and conidia were suspended in 0.01% Tween-20 solution. The seeds were harvested at 16, 32, and 64 HAI (hours after inoculation) time points. The experiments were conducted using a randomized complete block design (10 seeds/block). Every individual seed was ground in liquid nitrogen and divided into three aliquots. The first portion was used for GFP quantification, the second for aflatoxin analysis, and the third for RNA-seq (RNA- sequencing) analysis. Sterilization, all infection procedures, and GFP and aflatoxin analyses were carried out according to the methods described previously (Korani et al. 2017). A Student’s t-test was used to test the differences in GFP expression and aflatoxin contamination between the two genotypes under infection conditions for every time point (R v3.2.2) (R Core Team 2014).
RNA extraction
For every time point and genotype, the third pulverized portion of six mock-treated seeds and six infected seeds was used for RNA extraction with QIAGEN RNeasy Plant Mini kit (QIAGEN, Valencia, CA) according to the manufacturer’s instructions. The quality of RNA was checked with an Agilent 2100 Bioanalyzer (Georgia Genomics Facility, University of Georgia, Athens, GA).
RNA sequencing
DNA was eliminated from the extracted RNA using DNase I, amplification grade (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Seventy-two RNA libraries were constructed using a KAPA stranded RNA-seq library preparation kit (KR0934-v1.13; Kapa Biosystems, Wilmington, MA) and the Illumina set B indexes (Illumina, San Diego, CA) according to the manufacturer’s instructions. The integrity analysis and quantification of the libraries were carried out using an Agilent 2100 Bioanalyzer and Qubit 2.0 Fluorometer (Georgia Genomics Facility, University of Georgia, Athens, GA). Sequencing was done on an Illumina HiSeq2500 in six lanes, with 12 samples pooled per lane (HudsonAlpha Institute for Biotechnology, Huntsville, AL).
Differential expression analysis
The sequence quality for all libraries was determined using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, v. 0.11.4 2015). Trimmomatic v0.36 (Bolger et al. 2014) was used to trim the low-quality bases and filter out low-quality sequences. The cleaned paired-end reads were aligned to a Bowtie-indexed (v1.1.0, Langmead et al. 2009) peanut synthetic tetraploid reference genome, containing the genomes of A. duranensis and A. ipaensis (Bertioli et al. 2016, https://peautbase.org/), using Tophat v2.0.14 (Trapnell et al. 2009). Only the cleaned paired-end reads of infected libraries were aligned to a Bowtie-indexed A. flavus NRRL3357 reference genome [National Center for Biotechnology Information (NCBI), txid5059] using Tophat v2.0.14. The raw counts were calculated using HTSeq v0.6.1p1 (Anders et al. 2015).
Differential expression analysis of counts was carried out using edgeR (Robinson et al. 2010). DESeq2 (Love et al. 2014) and Next MaSigPro (Nueda et al. 2014) were used to repeat the analysis for in silico validation. Supplemental Material, Table S1 shows different models used in the analysis. Two models were applied to test the differences between the genotypes (resistant and susceptible) due to the infection vs. control treatments; the second model was applied to test the differences between the responses of the fungal genes during fungal growth on the two genotypes for only high aflatoxin-contaminated treatments, since control treatments had no fungal growth and the low aflatoxin-contaminated treatment had limited fungal growth.
Cufflinks v2.2.1 (Trapnell et al. 2010) was used to calculate the fragments per kilobase of transcript per million mapped reads (FPKM), then the Z-score was calculated using R v3.2.2 (R Core Team 2014). The expression profile of differentially expressed genes was clustered using self-organization maps (SOMs) of the kohonen package (R Core Team 2014).
GO enrichment analysis
Libraries GenomicFeatures (Lawrence et al. 2013) and biomaRt (Durinck et al. 2005) were used to extract gene lengths and GO terms from annotation files, respectively. GO enrichment analysis of differentially expressed genes was implemented using GOseq v2.12 (Young et al. 2010) with a correction for gene-length bias.
KEGG enrichment analysis
KEGG enrichment analysis was carried out using the “keggseq” package for the three models described above. The KEGG enrichment analysis for a synthetic tetraploid genome requires merging the two subgenomes in one analysis. However, the available tools for KEGG analysis do not support combining two species. Therefore, we designed R packages to carry out this type of analysis designated keggseq. The P-value was calculated according to Yang et al. (2015) within the keggseq package, which is freely available to the public under Massachusetts Institute of Technology (MIT) license and can be downloaded from https://github.com/w-korani/keggseq.
The keggseq package provides some other advantages over the available tools. (1) It allows application of KEGG enrichment analysis for diploids or polyploids with any level of genome duplication; (2) it generates ready-to-publish plots and produces graphs of interested pathways that have differentially expressed enzymes marked; (3) it generates .csv files containing detailed information of enzymes included in pathways of interest; (4) it allows editing of gene identifiers (IDs) if the user wants to use an annotation different from KEGG annotation; (5) It is a run-time package since the data are downloaded directly from the KEGGs database, so it does not require an internal database for specific species; and (6) it is step-by-step and easily implemented.
De novo assembly of transcripts
The unmapped reads of ICG 1471 controls, remaining after alignment with A. duranensis, A. ipaensis, and A. flavus genomes, were converted back to paired-end fastq files using bamtools v2.25.0 (Barnett et al. 2011) and concatenated. Trinity v. 2.0.6 (Haas et al. 2013) was used to assemble the concatenated reads with normalization to maximum coverage of 50×. The transcripts were given IDs starting with “RC.”
The process was repeated for ICG 1471 treatments, Florida-07 controls, and treatments with given IDs, starting with RT, SC, and ST, respectively. The four assemblies were combined and the redundant transcripts were filtered out using EvidentialGene pipeline (http://arthropods.eugenes.org/). Since the assembly contained sequence from peanut and sequences from A. flavus, BLAST+ (basic local alignment search tool; Camacho et al. 2009) was used to cluster the assembly into peanut and fungal transcripts by applying BLASTn for the transcripts against the NCBI nucleotide database. Transcripts that matched plant sequences were identified as peanut transcripts and those that had fungal matches were defined as A. flavus transcripts. The peanut filtered assembly was merged with the peanut tetraploid assembly produced by Clevenger et al. (2016a). Blast2GO was used to annotate GO terms of the new transcripts (https://www.blast2go.com/). Differential expression analysis, GO, and KEGG enrichment analyses were carried out as described above in the first model (differences between resistant and susceptible genotypes due to the infection vs. control) (Table S1).
Coexpression network analysis
Differentially expressed gene analysis of the fungal response to the infection was carried out separately for each genotype using edgeR (Robinson et al. 2010). Since a fungal control treatment was lacking, the 16-hr treatment was used as control. In addition, differential expression analysis was carried out for both genotypes to test the treatment effect (controls vs. treatments) for time points 32 and 64 HAI for each genotype separately. The Z-scores for fungal and peanut genes were combined in one matrix/genotype with rows for gene ID and columns for time points. Pearson correlation analysis was done using the R v3.2.2 package (R Core Team 2014) as described by Musungu et al. (2016). Only pairs that showed correlation > 0.99 were loaded into cytoscape network v3.4.0 (Shannon et al. 2003). As the data set containing the correlated paired genes of the susceptible genotype was huge, the network was clustered only for the resistant genotype using the MCODE app (Bader and Hogue 2003), and then the genes that matched those of the susceptible genotype matrix were excluded from the clusters of the resistant genotype.
Data availability
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and supplemental figures and tables. Table S1: statistical models for differential expression analysis; Figure S1: sequence read integrity and mapping results; Figure S2: SOM clusters of differentially expressed genes between genotypes; Figure S3: KEGG pathway of α-linolenic acid metabolism; Figure S4: KEGG pathway of protein processing in the endoplasmic reticulum; Figure S5: KEGG pathway of the spliceosome; Figure S6: KEGG pathway of carbon fixation; Figure S7: KEGG pathway of carbon metabolism; Figure S8: expression profile of the novel transcripts; Figure S9: GO/KEGG enrichment analysis of differently expressed genes between peanut genotypes due to the infection vs. control of the newly assembled transcripts; Figure S10: SOM cluster groups of fungal differentially expressed genes; File S1.xlsx: Z-scores of differentially expressed genes; File S2.fasta: 2026 novel peanut transcripts; and File S3.txt: ICG 1471 coexpression network clusters. All raw data fastq sequences are deposited at the NCBI (http://www.ncbi.nlm.nih.gov/) under BioProject PRJNA417591. All raw sequences are deposited as BioSamples SAMN08000482: SAMN08000553. The keggseq package is freely available to the public under MIT license and can be downloaded from https://github.com/w-korani/keggseq. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5984860.
Results and Discussion
Fungal growth and aflatoxin accumulation
It was shown previously that the peanut genotype ICG 1471 is a strong candidate for resistance to aflatoxin accumulation upon in vitro inoculation of mature peanut seeds with A. flavus (Korani et al. 2017). In addition, ICG 1471 has been reported as a resource for resistance to pre- (Waliyar et al. 2003; Nigam et al. 2009) and postharvest (Waliyar et al. 2008) aflatoxin contamination. Therefore, it was used in this study along with Florida-07 (Gorbet and Tillman 2009), which was a susceptible genotype for both pre- and postharvest aflatoxin accumulation (Clevenger et al. 2016b; Korani et al. 2017).
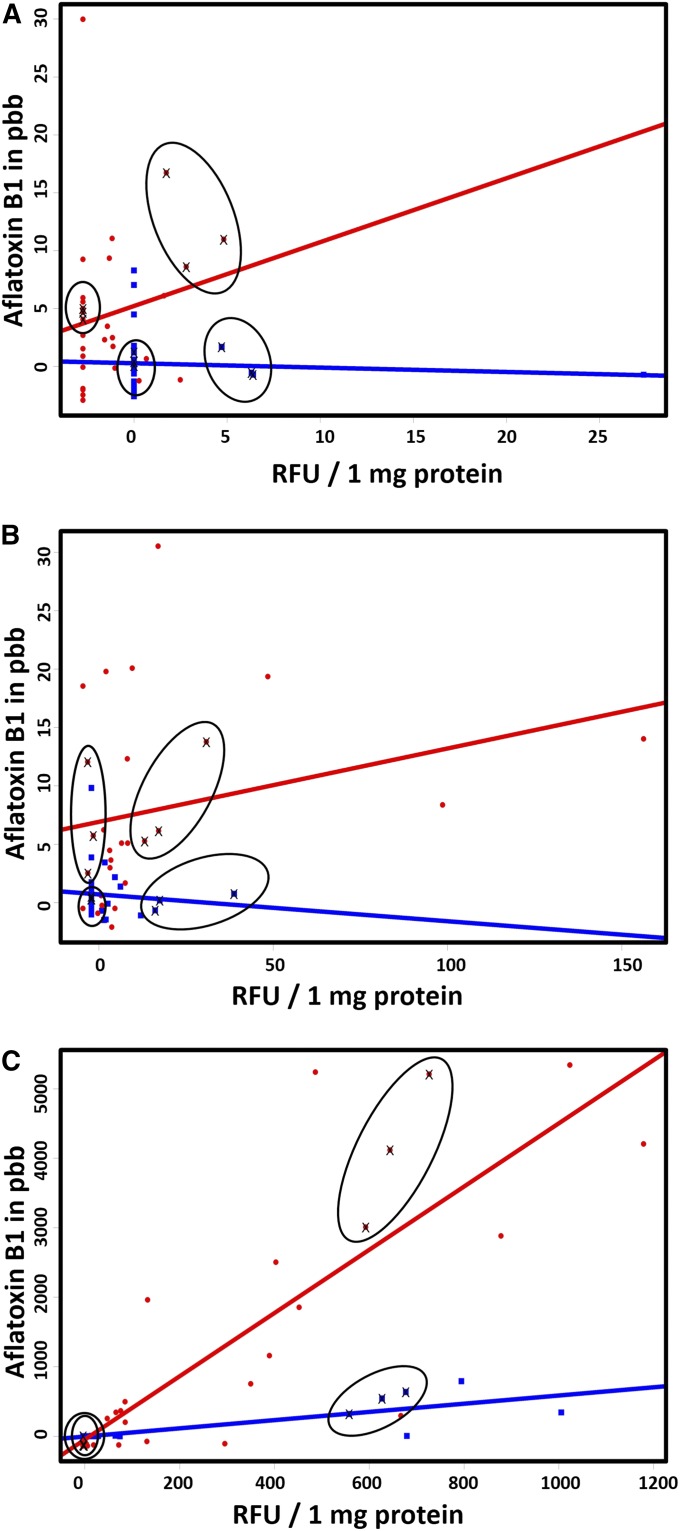
To estimate the dynamic change in gene expression, infected seeds and their controls were harvested at three different time points: 16, 32, and 64 HAI. Figure 1 shows the interaction between aflatoxin B produced by A. flavus and the progression of fungal growth estimated indirectly by relative fluorescence units of GFP protein signal. The data were consistent with our previous findings (Korani et al. 2017) as ICG 1471 showed less aflatoxin production as compared with Florida-07 (Table S1). A Student’s t-test revealed no significant differences between the two genotypes for GFP relative fluorescence for the three time points, yet aflatoxin levels between genotypes were significantly different for all three time points.
Figure 1.
Interaction between GFP signals and aflatoxin levels for 16 (A), 32 (B), and 64 (C) hours after inoculation. The red line and points represent Florida-07 data; the blue line and points represent ICG 1471 data; the cross marks show the samples that were chosen for RNA-sequencing analysis; and the ovals reveal the diverse range of aflatoxin contamination. RFU stands for GFP relative fluorescence unit.
The interaction plots revealed that not only do peanut genotypes interact differently with the fungus, but also every individual seed produced different amounts of aflatoxin within the same genotype/treatment/time point. This supports previous reports that aflatoxin accumulation is responsive to environmental influence (Blankenship et al. 1984; Kisyombe et al. 1985; Bhatnagar et al. 2003; Craufurd et al. 2006). The samples that were chosen for RNA-seq analysis were circled in the figure (Figure 1). Picking such samples with a diverse range of aflatoxin accumulation gave a realistic representation of the biological replication; however, it increases SD. Therefore, six biological replicates were used to study the differentially expressed genes due to genotypic effect.
Peanut genotypic differential expression analysis
The cleaned paired-end reads that were mapped to the synthetic tetraploid peanut and the A. flavus genomes are presented in Figure S1. Except for the highly fungal contaminated libraries (treatments of 64 HAI of Florida-07), 3.3–9.9 million paired-end reads were mapped to the peanut genome for every sample. This gave an average of 6 million paired-end reads/library and a total average of 5.6 million paired-end reads/library, including the highly contaminated libraries (which had 2.4–4.4 million mapped fragments/library). These results showed a reasonable coverage of the 2.7 Gb peanut genome (Bertioli et al. 2016).
In total, 4272 genes were differentially expressed between the two genotypes (resistant vs. susceptible) due to the infection by A. flavus (treatment vs. control). The Z-scores are provided in the supplemental materials (File S1.xlsx). The general pattern groups represented by SOM clusters are shown in Figure S2. Since the expression profile included the dynamic change across the three time points, some SOM clusters of these genes may have similar general trends and only differ slightly in the dynamic change from one time point to another. The clustering showed that some genes were downregulated due to the infection in the susceptible genotype and upregulated (Figure S2A) or unaffected (Figure S2B) in the resistant genotype. Conversely, some genes were upregulated due to the infection in the susceptible genotype and downregulated (Figure S2C) or unaffected (Figure S2) in the resistant genotype. A fifth group was upregulated in both genotypes but it was more highly expressed in the resistant genotype (Figure S2E).
To mitigate postharvest aflatoxin contamination of peanut, the resistance mechanisms have to be understood. Therefore, genes that are responsive to the infection (infection vs. mock-treatment) were studied. The large number of significant differentially expressed genes and GO terms generated by the analysis revealed the complexity of the interaction between A. flavus and peanut in terms of aflatoxin production and the significant effect of individual seed physiology on the process. In addition, it can be misleading, even after clustering/grouping, to assign a resistance function to a particular gene or small group of genes of interest; however, KEGG enrichment analysis gives clues to pathways that are associated with the resistance response.
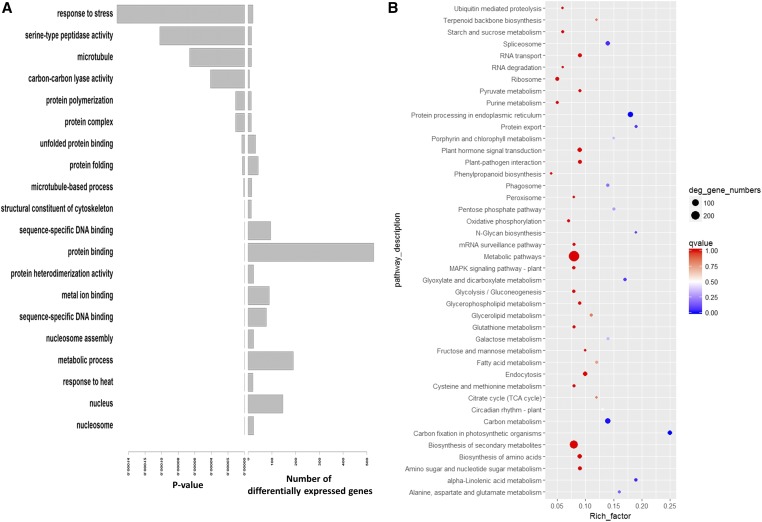
GO enrichment analysis of the differentially expressed genes generated 146 significant GO terms out of 1672 GO terms found in the annotation of the two subgenomes of peanut. The 20 most significant GO terms (Figure 2A) included several for protein processing, protein polymerization, protein complex, unfolded protein binding, protein folding, protein heterodimerization activity, and protein binding. The latter GO term represented 529 differentially expressed genes. On the other hand, KEGG enrichment analysis (Figure 2B) only generated five significant pathways: α-linolenic acid metabolism, protein processing in the endoplasmic reticulum, spliceosome, carbon fixation, and carbon metabolism (Figure S3, Figure S4, Figure S5, Figure S6, and Figure S7, respectively).
Figure 2.
Gene ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of peanut differently expressed genes between genotypes due to infection vs. control. (A) The 20 most significant GO terms extracted by GO enrichment analysis and (B) KEGG enrichment analysis carried out by the keggseq package. Rich_factor: the ratio of differentially expressed genes to the all genes that were annotated in the pathway.
As only five KEGG pathways were identified as significant for resistance (excluding individual seed effects), it can be assumed that they are the main keys controlling the defense mechanism in ICG 1471. The most interesting significant pathway is α-linolenic metabolism, which contains different components that have been reported as related to or responsive for biotic and abiotic stresses of plants. α-linolenic acid accounts for 0.37–1.11% of peanut total oil content (Ozcan 2010). Although it is a minor component of peanut oil, the pathway catabolizes α-linolenic acid into jasmonate and methyl-jasmonate and was significantly regulated in the resistant genotype ICG 1471. Jasmonates are synthesized though this pathway in two main cellular compartments: the chloroplast where α-linolenic acid is converted to 12-oxo-phytodienoic acid (OPDA) in a process initiated by chloroplast 13S-LOX (Bell et al. 1995), and the peroxisome where 12-OPDA is localized and converted to jasmonates (Stintzi 2000).
LOXs were documented to play a role in Aspergillus spp. infection and the subsequent aflatoxin contamination of different crops including peanut (Burow et al. 2000; Tsitsigiannis et al. 2005; Kumari et al. 2012; Müller et al. 2014), soybean (Bean et al. 1972; Doehlert et al. 1993; Boué et al. 2005), maize (Gao et al. 2009; Huang et al. 2013), cottonseeds (Zeringue 1996), and almond (Mita et al. 2007). Figure 3 shows nine LOXs that were found among the differentially expressed genes. BLASTp search against the NCBI nonredundant protein database was used to estimate their function; eight were predicted to generate 13-hydroxyperoxides and six had features of plastidial enzymes.
Figure 3.
Differentially expressed lipoxygenases. Left, middle, and right panels are 16, 32, and 64 hours after inoculation, respectively. The upper panel is plastidial genes and the lower panel is extraplastidial genes. All genes except Ad_25 are predicted to generate13-S-hydroxyperoxides. Ad_25 was not classified. FPKM: Z-scores of fragments per kilobase of transcript per million mapped reads.
Additionally, both 12-OPDA and jasmonates were documented to play independent roles in the wound response of Arabidopsis, and each influences the expression of an overlapping set of genes as well as different sets of responsive genes (Taki et al. 2005; Sham et al. 2015). In addition, numerous reports showed the importance of jasmonates in plant responses to biotic and abiotic stresses, e.g., insects (Thaler et al. 1996; McConn et al. 1997; Kessler et al. 2004), fungi (Vijayan et al. 1998; Thomma et al. 2000; Mei et al. 2006; Zeneli et al. 2006), and wounding, (Baldwin et al. 1997) and during development (Creelman and Mullet 1997). In particular, methyl-jasmonate was found to delay spore germination, and inhibit mycelial pigment formation and aflatoxin production of A. flavus (Goodrich-Tanrikulu et al. 1995). Interestingly, it was found to enhance aflatoxin production by A. parasiticus (Vergopoulou et al. 2001). However, Meimaroglou et al. (2009) showed that methyl-jasmonate might enhance or reduce aflatoxin production by A. parasiticus depending on its concentration. Moreover, fungal pathogens can manipulate, enhance, or suppress jasmonate signaling in plant hosts (Zhang et al. 2017).
Metabolic products such as 10-OPDA, which has a high phytotoxicity, are produced by the α-linolenic acid metabolism pathway enzyme 9S-LOX (Sherif et al. 2016). Another route through the α-linolenic acid metabolism pathway produces 8,11,14-heptadecatrienoic acid using α-dioxygenase 1 (DOX1) without LOX activity. Both the enzyme and its product were documented to increase in tobacco during interaction with Pseudomonas syringae pv syringae (Hamberg et al. 2003). In addition, DOX1 was upregulated in Arabidopsis after 12-OPDA treatment (Sham et al. 2015), which functioned to protect the plant from oxidative stress (De León et al. 2002).
Therefore, evidence suggests that, regardless of the direction of the effect, jasmonates and 12-OPDA produced by the α-linolenic pathway, in addition to other pathway components, have an important role in aflatoxin biosynthesis of Aspergillus spp. ICG 1471, as a resistant genotype, may regulate the synthesis of jasmonates to reduce aflatoxin production. On the other hand, Florida-07, as a susceptible genotype and a high-oleic variety, may not respond to infection with the same level of jasmonate production, possibly because of altered substrate amounts or membrane properties in a high-oleic fatty acid background, thereby resulting in elevated aflatoxin accumulation. Jasmonate levels will be tested in the future. Previously, high-oleic lines were shown to have double the aflatoxin contamination compared to normal oleic lines with similar genetic background under in vitro inoculation conditions (Xue et al. 2003). In the present study, the magnitude of aflatoxin contamination in Florida-07 is 15 times that of ICG 1471, which is a normal oleic variety. Therefore, the postharvest aflatoxin resistance conferred by ICG 1471 surpasses the effect of just having the normal oleic acid content.
Protein processing in the endoplasmic reticulum also was a significant pathway that may contribute to the resistance mechanism. However, its role may be integrated with the α-linolenic pathway since the endoplasmic reticulum contributes to the formation of peroxisomes (Hoepfner et al. 2005). Fountain et al. (2016b) showed that alternative carbon sources have different effects on aflatoxin and kojic acid production by the fungus; kojic acid has an important role in remediating damage resulting from ROS. These results reveal the importance of carbon fixation and metabolism pathways for aflatoxin production by Aspergillus spp. and protection of the fungus against oxidative damage.
As a synthetic reference genome of tetraploid peanut was used in our differential expression analysis, some genes/transcripts having roles in resistance to aflatoxin accumulation may not be represented within the two subgenomes. Therefore, de novo assemblies were constructed to capture such novel transcripts. Four assemblies were created for ICG 1471 control and treatments, and Florida-07 controls and treatments, which generated 61,176, 67,813, 90,543, and 109,068 total transcripts, respectively, and among them 413, 457, 551, and 505 were new transcripts, respectively.
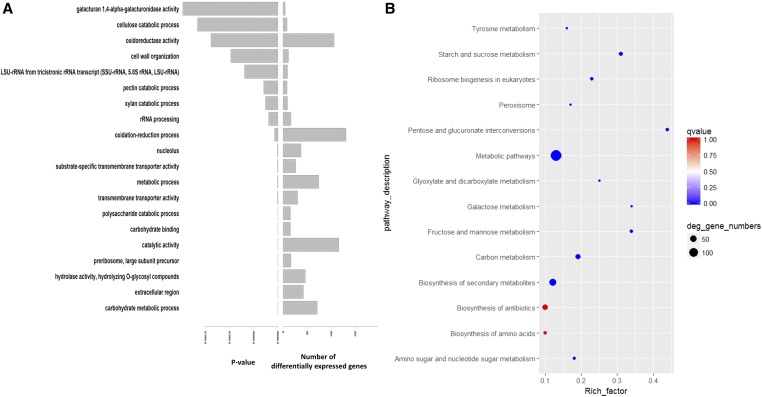
To validate the genes and pathways involved in resistance, analysis of differential expression between peanut genotypes was repeated using a combined reference transcriptome (88,626 transcripts) that included the 2026 novel transcripts (supplemental materials: File S2.fasta) and the previously published tetraploid peanut transcriptome (86,600 transcripts) (Clevenger et al. 2016a). The differential expression analysis generated 3879 significant genes for which Z-scores are given in the supplemental materials (File S1.xlsx). The expression profile of the novel transcripts also is given (Figure S8); out of the 2026 novel genes, 66 were differentially expressed. GO enrichment analysis identified 406 out of 8530 significant GO terms (Figure S9A). Most significant GO terms resulting from genomic analysis (using predicted transcripts) also were significant in transcriptomic analysis. However, interestingly, KEGG enrichment analysis generated four of the same significant pathways as with genomic analysis (Figure S9B), except for α-linolenic acid metabolism, which was near the significance threshold with a q-value of 0.06. These outputs confirmed the key roles of these five pathways and their respective genes in the resistance of peanut to aflatoxin produced by Aspergilli.
Differential expression of fungal genes and coexpression network analysis
The interaction between peanut seeds and Aspergilli encompasses responsive pathways inside the plant and those inside the fungi, and genes regulating the signaling between organisms. Furthermore, some fungal genes may be affected differentially by growth of the fungus on different peanut genotypes. To investigate host–pathogen interaction, differential expression analysis was carried out for fungal genes, which generated 1197 significant genes (Z-scores in supplemental materials, File S1.xlsx). SOM clusters of the expression patterns of these genes (Figure S10) and GO term enrichment analysis showed 97 significant GO terms, out of 4918 in total (Figure 4A). KEGG enrichment analysis identified eight significant pathways (Figure 4B): one interconversion pathway (pentose and glucuronate), one degradation pathway (valine, leucine, and isoleucine), and six metabolic pathways (fructose/mannose, galactose, starch/sucrose, glycerolipid, carbon, and metabolic pathways). Interestingly, seven of these pathways include carbohydrate processing. These results are in agreement with previous studies that showed changes in aflatoxin production by A. flavus or A. parasiticus using different sugar sources (Davis and Diener 1968; Abdollahi and Buchanan 1981). Growth of A. flavus on ICG 1471 may result in the production of different sugars than growth on Florida-07, leading to lower aflatoxin production by the fungus. A further consequence may be reduced kojic acid production and a subsequent increase in the sensitivity of the fungus to ROS. These two hypotheses need to be tested in future work.
Figure 4.
Gene ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed A. flavus genes due to growth of the fungus on resistant vs. susceptible genotypes. (A) The 20 most significant GO terms extracted by GO enrichment analysis and (B) KEGG enrichment analysis carried out by the keggseq package. Rich_factor: the ratio of the differentially expressed genes to all genes that were annotated in the pathway.
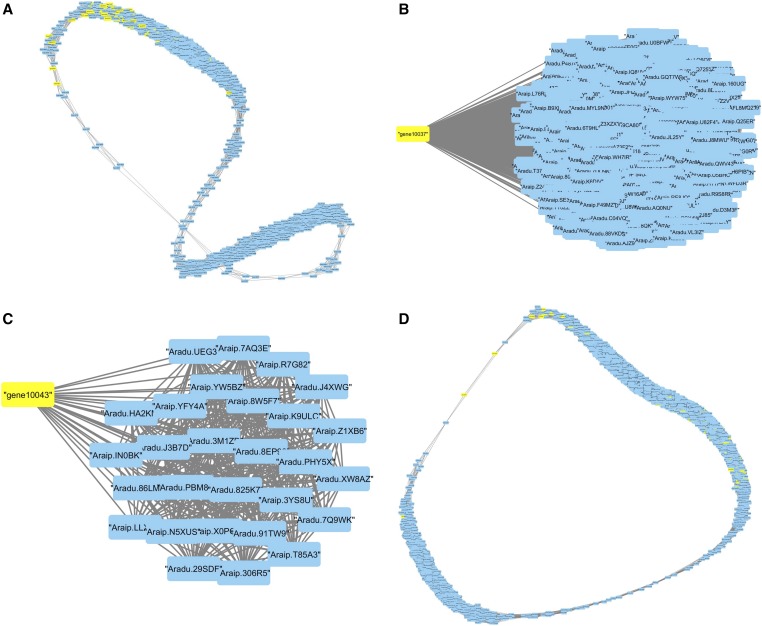
To further investigate the differential response of fungal genes due to host genotype, coexpression network analysis based on Pearson correlation was conducted (Figure 5). In total, 1265 and 1111 differentially expressed peanut and fungal genes, respectively, were found in A. flavus/ ICG 1471 interaction (for the time points of 64 and 32 HAI for the comparison of treatments vs. controls), which formed a matrix of 0.5 million correlated pairs (edges). More (6795 peanut and 1265 fungal genes) were differentially expressed in the A. flavus/Florida-07 interaction, which created a huge matrix of 14 million correlated pairs (edges). Figure 5 shows the interspecies peanut/A. flavus coexpression network for ICG 1471 (Figure 5A) and Florida-07 (Figure 5D). The MCODE cluster analysis of the ICG 1471 coexpression network generated 45 clusters (supplemental materials, File S3.txt). The most interesting clusters (subnetworks) were 1 and 15; subnetwork 1 had 1037 peanut genes and eight A. flavus genes (Figure 5B), including gene10037 (AflNA), and subnetwork 15 had 28 peanut genes and only one A. flavus gene, gene10043 (AflH) (Figure 5C). AflNA [averantin hydroxylase (EC 1.14.13.174)] and AflH [versiconal hemiacetal acetate reductase (EC 1.1.1.353)] encode two upstream enzymes regulating the aflatoxin biosynthetic pathway.
Figure 5.
Coexpression network analysis of peanut/A. flavus genes. (A) ICG 1471/A. flavus network. (B) subnetwork 1 of ICG 1471/A. flavus network. (C) subnetwork 15 of ICG 1471/A. flavus network. (D) Florida-07/A. flavus network; lines represent edges, blue rectangles are peanut nodes, and yellow rectangles are A. flavus nodes.
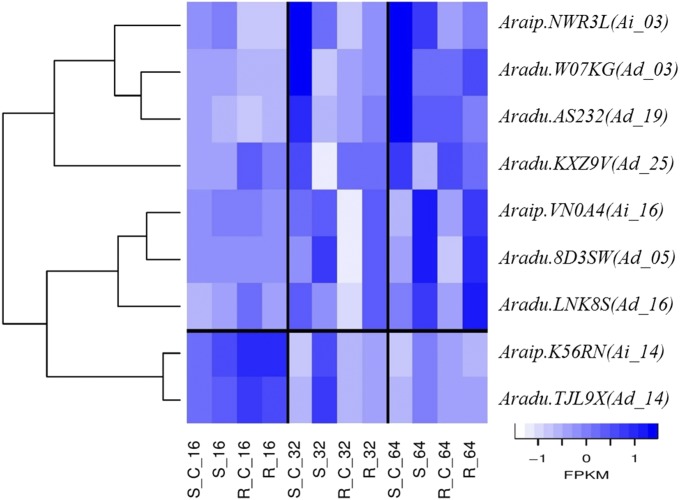
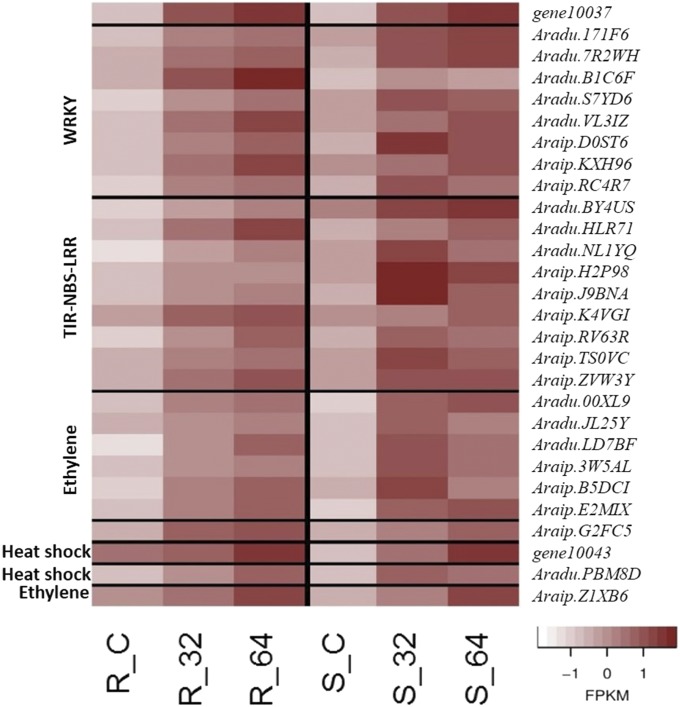
Out of the 1037 and 28 ICG 1471 peanut genes whose expression were highly correlated with gene10037 and gene10043 of A. flavus, respectively, 640 and 24 genes were not found in the Florida-07/A. flavus matrix. Among these genes, eight WRKY family transcription factors, nine toll/Interleukin1 receptor–nucleotide binding site leucine-rich repeat (TIRNBS-LRRs), six ethylene signaling proteins, and one heat shock protein were upregulated, and expression was correlated with gene10037. One heat shock and an ethylene signaling gene were upregulated, and expression was correlated with gene10043. Figure 6 represents the expression profile of these genes. Although gene expression was upregulated in both genotypes for all genes, ICG 1471 genes were coexpressed with gene10037 or gene10043 of A. flavus.
Figure 6.
Peanut coexpressed genes with gene10037 and gene10043 of A. flavus. FPKM: Z-scores of fragments per kilobase of transcript per million mapped reads.
Many plant disease-resistance genes encode NBS-LRR proteins (McHale et al. 2006; Sekhwal et al. 2015). Ethylene signaling genes were significantly upregulated in response to A. flavus infection of maize (Musungu et al. 2016). Heat shock proteins may play a role in plant defense by affecting R protein stability and their regulation (Lee et al. 2012). WRKY transcription factors were differentially expressed in the response of resistant and susceptible genotypes of maize to infection by A. flavus (Fountain et al. 2015b). In addition, they were found to have an effect on pathways for ethylene-jasmonate-mediated defense (Birkenbihl et al. 2012), plant response to heat stress (Li et al. 2010), and defense triggered by jasmonates, either negatively (Gao et al. 2011) or positively (Journot-Catalino et al. 2006). These eight WRKY genes may be important in controlling jasmonate defense mechanisms. In addition, the high correlations between expression of these genes in ICG 1471 and gene10037 of A. flavus reveals their importance in the defense mechanism and suggests that they may be involved in regulation of the α-linolenic acid metabolism pathway in ICG 1471.
In silico validation of differential expression analysis
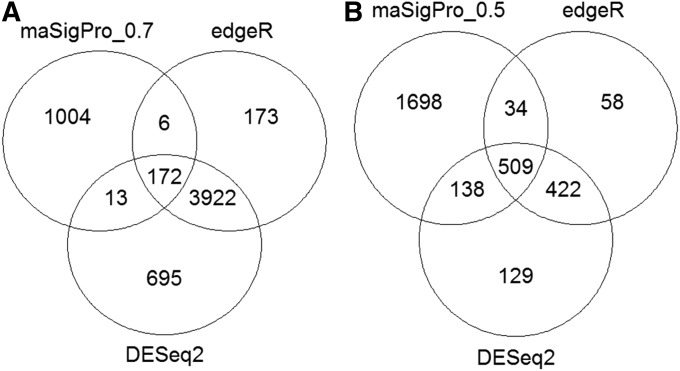
In this study, different complex factors were involved in the RNA-seq experiment, e.g., genotypic effect, A. flavus infection, and time-course dynamic change. Therefore, three analytical models were compared (Figure 7). Across all analyses, DESeq2 showed similar results to edgeR for identifying differentially expressed genes. On the other hand, Next maSigPro identified many genes that were not discovered by the other two methods and failed to extract many other genes that were determined to be differently expressed by the other two methods.
Figure 7.
Differential expression analysis with multiple programs for in silico validation of peanut genotypic differences due to A. flavus infection (A) and fungal/peanut-genotype effects (B).
EdgeR is one of the most common methods used for differential expression analysis of RNA-seq data. However, it is not a standard method to handle the time course experiments as it uses a negative binomial model, which deals with time points as independent factors (Robinson et al. 2010). Methods have been designed to account for time course experiments that used different models such as Next maSigPro (polynomial regression model) (Nueda et al. 2014), DyNB (nonparametric Gaussian processes regression negative binomial likelihood model) (Äijö et al. 2014), TRAP (β-negative binomial model) (Jo et al. 2014), SMARTS (input–output hidden Markov model) (Wise and Bar-Joseph 2015), EBSeq-HMM (empirical Bayes mixture model) (Leng et al. 2015), FunPat (different distribution models) (Sanavia et al. 2015), and timeSeq (negative binomial mixed-effect model) (Sun et al. 2016). All these methods had limitations and none was standardized to this type of analysis. Next, maSigPro was initially designed to analyze microarray data using polynomial regression and later was updated to handle RNA-seq data (Nueda et al. 2014). This method relies on R2 factor to extract the significant differentially expressed genes, which is considered a drawback since the threshold is user-defined (Spies and Ciaudo 2015). Although both edgeR and DESeq2 use a negative binomial model, DESeq2 has different implementation, tests, and normalization (Love et al. 2014). Both gave a reasonable level of analysis validation.
Conclusions
The objective of this study was to identify genetic factors and biochemical pathways that function to limit aflatoxin production in resistant peanut genotypes. Differential expression analysis revealed five important biochemical pathways regulating resistance. In addition, results captured the fungal pathways that are differentially affected by fungal infection and aflatoxin production on resistant vs. susceptible peanut genotypes. The study highlighted the critical role of the α-linolenic acid metabolism pathway and certain WRKY genes likely regulating the jasmonate-based defense pathways to mitigate aflatoxin production. To further estimate the effects of these components on aflatoxin production and/or identify effective QTL, we have created a population between ICG 1471 and Florida-07 that is being advanced to recombinant inbred lines.
Acknowledgments
We thank Jeff Cary, Unites States Department of Agriculture (USDA)-Agricultural Research Service-Southern Regional Research Center, New Orleans, for providing AF-70-GFP, the GFP-expressing A. flavus. This work was supported by the Peanut Foundation, the Agriculture and Food Research Initiative competitive grant 2012-85117-19435 of the USDA National Institute of Food and Agriculture, and the Feed the Future Innovation Lab for Collaborative Research on Peanut Productivity and Mycotoxin Control (Peanut and Mycotoxin Innovation Lab), supported by funding from the United States Agency for International Development. The authors declare no conflict of interest.
Author contributions: W.K. performed the experiments, designed keggseq, applied the data analysis, and wrote the manuscript. Y.C. assisted with experimental design and helped in laboratory work training. C.C.H. provided the peanut genotypes, field data, and assistance with aflatoxin analysis. P.O.-A. conceived and supervised the project, secured funding, and revised and submitted the manuscript.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5984860.
Communicating editor: J. Birchler
Literature Cited
- Abdollahi A., Buchanan R., 1981. Regulation of aflatoxin biosynthesis: induction of aflatoxin production by various carbohydrates. J. Food Sci. 46: 633–635. 10.1111/j.1365-2621.1981.tb04928.x [DOI] [Google Scholar]
- Äijö T., Butty V., Chen Z., Salo V., Tripathi S., et al. , 2014. Methods for time series analysis of RNA-seq data with application to human Th17 cell differentiation. Bioinformatics 30: i113–i120. 10.1093/bioinformatics/btu274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaike S., Keller N. P., 2011. Aspergillus flavus. Annu. Rev. Phytopathol. 49: 107–133. 10.1146/annurev-phyto-072910-095221 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitha M. P., Prakash H. S., Shetty H. S., 2004. Purification and properties of lipoxygenase induced in downy mildew resistant pearl millet seedlings due to infection with Sclerospora graminicola. Plant Sci. 166: 31–39. 10.1016/S0168-9452(03)00364-9 [DOI] [Google Scholar]
- Babitha M. P., Prakash H., Shetty H. S., 2006. Induction of lipoxygenase in downy mildew resistant seedlings of pearl millet in response to inoculation with Sclerospora graminicola. Int. J. Agric. Biol. 8: 560–564. [Google Scholar]
- Bader G. D., Hogue C. W., 2003. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4: 2 10.1186/1471-2105-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin I. T., Zhang Z.-P., Diab N., Ohnmeiss T. E., McCloud E. S., et al. , 1997. Quantification, correlations and manipulations of wound-induced changes in jasmonic acid and nicotine in Nicotiana sylvestris. Planta 201: 397–404. 10.1007/s004250050082 [DOI] [Google Scholar]
- Barnett D. W., Garrison E. K., Quinlan A. R., Stromberg M. P., Marth G. T., 2011. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27: 1691–1692. 10.1093/bioinformatics/btr174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean G. A., Schillinger J. A., Klarman W. L., 1972. Occurrence of aflatoxins and aflatoxin-producing strains of Aspergillus spp. in soybeans. Appl. Microbiol. 24: 437–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E., Creelman R. A., Mullet J. E., 1995. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 92: 8675–8679. 10.1073/pnas.92.19.8675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli D. J., Cannon S. B., Froenicke L., Huang G., Farmer A. D., et al. , 2016. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48: 438–446. 10.1038/ng.3517 [DOI] [PubMed] [Google Scholar]
- Bhatnagar D., Ehrlich K., Cleveland T., 2003. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 61: 83–93. 10.1007/s00253-002-1199-x [DOI] [PubMed] [Google Scholar]
- Birkenbihl R. P., Diezel C., Somssich I. E., 2012. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159: 266–285. 10.1104/pp.111.192641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship P. D., Cole R. J., Sanders T. H., Hill R. A., 1984. Effect of geocarposphere temperature on pre-harvest colonization of drought-stressed peanuts by Aspergillus flavus and subsequent aflatoxin contamination. Mycopathologia 85: 69–74. 10.1007/BF00436705 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boué S. M., Shih B. Y., Carter-Wientjes C. H., Cleveland T. E., 2005. Effect of soybean lipoxygenase on volatile generation and inhibition of Aspergillus flavus mycelial growth. J. Agric. Food Chem. 53: 4778–4783. 10.1021/jf058038o [DOI] [PubMed] [Google Scholar]
- Burow G. B., Gardner H. W., Keller N. P., 2000. A peanut seed lipoxygenase responsive to Aspergillus colonization. Plant Mol. Biol. 42: 689–701. 10.1023/A:1006361305703 [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. S., Sreedharan A., Schneider K. R., 2013. Peanut and peanut products: a food safety perspective. Food Control 32: 296–303. 10.1016/j.foodcont.2012.12.007 [DOI] [Google Scholar]
- Choi J., Alkharouf N., Schneider K., Matthews B., Frederick R., 2008. Expression patterns in soybean resistant to Phakopsora pachyrhizi reveal the importance of peroxidases and lipoxygenases. Funct. Integr. Genomics 8: 341–359. 10.1007/s10142-008-0080-0 [DOI] [PubMed] [Google Scholar]
- Clevenger J., Chu Y., Scheffler B., Ozias-Akins P., 2016a A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 7: 1446 10.3389/fpls.2016.01446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger J., Marasigan K., Liakos V., Sobolev V., Vellidis G., et al. , 2016b RNA sequencing of contaminated seeds reveals the state of the seed permissive for pre-harvest aflatoxin contamination and points to a potential susceptibility factor. Toxins (Basel) 8: 317 10.3390/toxins8110317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotty P. J., Jaime-Garcia R., 2007. Influences of climate on aflatoxin producing fungi and aflatoxin contamination. Int. J. Food Microbiol. 119: 109–115. 10.1016/j.ijfoodmicro.2007.07.060 [DOI] [PubMed] [Google Scholar]
- Craufurd P., Prasad P., Waliyar F., Taheri A., 2006. Drought, pod yield, pre-harvest Aspergillus infection and aflatoxin contamination on peanut in Niger. Field Crops Res. 98: 20–29. 10.1016/j.fcr.2005.12.001 [DOI] [Google Scholar]
- Creelman R. A., Mullet J. E., 1997. Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Biol. 48: 355–381. 10.1146/annurev.arplant.48.1.355 [DOI] [PubMed] [Google Scholar]
- Davis N. D., Diener U. L., 1968. Growth and aflatoxin production by Aspergillus parasiticus from various carbon sources. Appl. Microbiol. 16: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De León I. P., Sanz A., Hamberg M., Castresana C., 2002. Involvement of the Arabidopsis α‐DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J. 29: 61–72. 10.1046/j.1365-313x.2002.01195.x [DOI] [PubMed] [Google Scholar]
- Doehlert D. C., Wicklow D. T., Gardner H. W., 1993. Evidence implicating the lipoxygenase pathway in providing resistance to soybeans against Aspergillus flavus. Phytopathology 83: 1473–1477. 10.1094/Phyto-83-1473 [DOI] [Google Scholar]
- Dunham N. R., Peper S. T., Downing C. D., Kendall R. J., 2017. Aflatoxin contamination in corn sold for wildlife feed in Texas. Ecotoxicology 26: 516–520. 10.1007/s10646-017-1782-7 [DOI] [PubMed] [Google Scholar]
- Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., et al. , 2005. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21: 3439–3440. 10.1093/bioinformatics/bti525 [DOI] [PubMed] [Google Scholar]
- Ehrlich K. C., Chang P.-K., Yu J., Cotty P. J., 2004. Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl. Environ. Microbiol. 70: 6518–6524. 10.1128/AEM.70.11.6518-6524.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich K. C., Montalbano B. G., Cotty P. J., 2005. Divergent regulation of aflatoxin production at acidic pH by two Aspergillus strains. Mycopathologia 159: 579–581. 10.1007/s11046-005-1150-7 [DOI] [PubMed] [Google Scholar]
- Fountain J. C., Scully B. T., Ni X., Kemerait R. C., Lee R. D., et al. , 2014. Environmental influences on maize-Aspergillus flavus interactions and aflatoxin production. Front. Microbiol. 5: 40 10.3389/fmicb.2014.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J. C., Khera P., Yang L., Nayak S. N., Scully B. T., et al. , 2015a Resistance to Aspergillus flavus in maize and peanut: molecular biology, breeding, environmental stress, and future perspectives. Crop J. 3: 229–237. 10.1016/j.cj.2015.02.003 [DOI] [Google Scholar]
- Fountain J. C., Raruang Y., Luo M., Brown R. L., Guo B., et al. , 2015b Potential roles of WRKY transcription factors in regulating host defense responses during Aspergillus flavus infection of immature maize kernels. Physiol. Mol. Plant Pathol. 89: 31–40. 10.1016/j.pmpp.2014.11.005 [DOI] [Google Scholar]
- Fountain J. C., Bajaj P., Nayak S. N., Yang L., Pandey M. K., et al. , 2016a Responses of Aspergillus flavus to oxidative stress are related to fungal development regulator, antioxidant enzyme, and secondary metabolite biosynthetic gene expression. Front. Microbiol. 7: 2048 10.3389/fmicb.2016.02048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J. C., Bajaj P., Pandey M., Nayak S. N., Yang L., et al. , 2016b Oxidative stress and carbon metabolism influence Aspergillus flavus transcriptome composition and secondary metabolite production. Sci. Rep. 6: 38747 10.1038/srep38747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q.-M., Venugopal S., Navarre D., Kachroo A., 2011. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155: 464–476. 10.1104/pp.110.166876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Starr J., Göbel C., Engelberth J., Feussner I., et al. , 2008. Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol. Plant Microbe Interact. 21: 98–109. 10.1094/MPMI-21-1-0098 [DOI] [PubMed] [Google Scholar]
- Gao X., Brodhagen M., Isakeit T., Brown S. H., Göbel C., et al. , 2009. Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol. Plant Microbe Interact. 22: 222–231. 10.1094/MPMI-22-2-0222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgianna D. R., Payne G. A., 2009. Genetic regulation of aflatoxin biosynthesis: from gene to genome. Fungal Genet. Biol. 46: 113–125. 10.1016/j.fgb.2008.10.011 [DOI] [PubMed] [Google Scholar]
- Goodrich-Tanrikulu M., Mahoney N. E., Rodriguez S. B., 1995. The plant growth regulator methyl jasmonate inhibits aflatoxin production by Aspergillus flavus. Microbiology 141: 2831–2837. 10.1099/13500872-141-11-2831 [DOI] [PubMed] [Google Scholar]
- Gorbet D. W., Tillman B. L., 2009. Registration of ‘Florida-07’ peanut. J. Plant Regist. 3: 14–18. 10.3198/jpr2008.05.0276crc [DOI] [Google Scholar]
- Guo B., Xu G., Cao Y., Holbrook C., Lynch R., 2006. Identification and characterization of phospholipase D and its association with drought susceptibilities in peanut (Arachis hypogaea). Planta 223: 512–520. 10.1007/s00425-005-0112-0 [DOI] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., et al. , 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberg M., Sanz A., Rodriguez M. J., Calvo A. P., Castresana C., 2003. Activation of the fatty acid α-Dioxygenase pathway during bacterial infection of tobacco leaves formation of oxylipins protecting against cell. J. Biol. Chem. 278: 51796–51805. 10.1074/jbc.M310514200 [DOI] [PubMed] [Google Scholar]
- Hedayati M., Pasqualotto A., Warn P., Bowyer P., Denning D., 2007. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153: 1677–1692. 10.1099/mic.0.2007/007641-0 [DOI] [PubMed] [Google Scholar]
- Hoepfner D., Schildknegt D., Braakman I., Philippsen P., Tabak H. F., 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122: 85–95. 10.1016/j.cell.2005.04.025 [DOI] [PubMed] [Google Scholar]
- Holbrook C. C., Kvien C., Rucker K., Wilson D., Hook J., et al. , 2000a Preharvest aflatoxin contamination in drought-tolerant and drought-intolerant peanut genotypes. Peanut Sci. 27: 45–48. 10.3146/i0095-3679-27-2-1 [DOI] [Google Scholar]
- Huang P., Franco O., Yan Y., Keller N., Kolomiets M., 2013. Maize lipoxygenase LOX2 regulates pathogenesis of mycotoxin-producing Aspergillus flavus. Phytopathology 103: 62. [Google Scholar]
- Jaime-Garcia R., Cotty P. J., 2003. Aflatoxin contamination of commercial cottonseed in South Texas. Phytopathology 93: 1190–1200. 10.1094/PHYTO.2003.93.9.1190 [DOI] [PubMed] [Google Scholar]
- Jayashree T., Subramanyam C., 2000. Oxidative stress as a prerequisite for aflatoxin production by Aspergillus parasiticus. Free Radic. Biol. Med. 29: 981–985. 10.1016/S0891-5849(00)00398-1 [DOI] [PubMed] [Google Scholar]
- Jo K., Kwon H.-B., Kim S., 2014. Time-series RNA-seq analysis package (TRAP) and its application to the analysis of rice, Oryza sativa L. ssp. Japonica, upon drought stress. Methods 67: 364–372. 10.1016/j.ymeth.2014.02.001 [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N., Somssich I. E., Roby D., Kroj T., 2006. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302. 10.1105/tpc.106.044149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A., Halitschke R., Baldwin I. T., 2004. Silencing the jasmonate cascade: induced plant defenses and insect populations. Science 305: 665–668. 10.1126/science.1096931 [DOI] [PubMed] [Google Scholar]
- Kisyombe C. T., Beute M., Payne G., 1985. Field evaluation of peanut genotypes for resistance to Infection by Aspergillus parasiticus. Peanut Sci. 12: 12–17. 10.3146/pnut.12.1.0004 [DOI] [Google Scholar]
- Klich M. A., 2007. Aspergillus flavus: the major producer of aflatoxin. Mol. Plant Pathol. 8: 713–722. 10.1111/j.1364-3703.2007.00436.x [DOI] [PubMed] [Google Scholar]
- Korani W. A., Chu Y., Holbrook C., Clevenger J., Ozias-Akins P., 2017. Genotypic regulation of aflatoxin accumulation but not Aspergillus fungal growth upon post-harvest infection of peanut (Arachis hypogaea L.) seeds. Toxins (Basel) 9: 218 10.3390/toxins9070218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A., Sucharitha A., Devi P., 2012. Impact of 13-lipoxygenase on induction of resistance in peanut against aflatoxigenic Aspergillus continental. J. Biol. Sci. 5: 4–13. [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., et al. , 2013. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9: e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Yun H. S., Kwon C., 2012. Molecular communications between plant heat shock responses and disease resistance. Mol. Cells 34: 109–116. 10.1007/s10059-012-0121-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng N., Li Y., McIntosh B. E., Nguyen B. K., Duffin B., et al. , 2015. EBSeq-HMM: a Bayesian approach for identifying gene-expression changes in ordered RNA-seq experiments. Bioinformatics 31: 2614–2622. 10.1093/bioinformatics/btv193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhou X., Chen L., Huang W., Yu D., 2010. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 29: 475–483. 10.1007/s10059-010-0059-2 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo E. M., Gil-Serna J., Patino B., Jimenez M., 2011. Aflatoxins and ochratoxin A in stored barley grain in Spain and impact of PCR-based strategies to assess the occurrence of aflatoxigenic and ochratoxigenic Aspergillus spp. Int. J. Food Microbiol. 149: 118–126. 10.1016/j.ijfoodmicro.2011.06.006 [DOI] [PubMed] [Google Scholar]
- McConn M., Creelman R. A., Bell E., Mullet J. E., 1997. Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 5473–5477. 10.1073/pnas.94.10.5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale L., Tan X., Koehl P., Michelmore R. W., 2006. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 7: 212 10.1186/gb-2006-7-4-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C., Qi M., Sheng G., Yang Y., 2006. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 19: 1127–1137. 10.1094/MPMI-19-1127 [DOI] [PubMed] [Google Scholar]
- Meimaroglou D. M., Galanopoulou D., Markaki P., 2009. Study of the effect of methyl jasmonate concentration on aflatoxin B 1 biosynthesis by Aspergillus parasiticus in yeast extract sucrose medium. Int. J. Microbiol. 2009: 1–7. 10.1155/2009/842626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita G., Fasano P., De Domenico S., Perrone G., Epifani F., et al. , 2007. 9-Lipoxygenase metabolism is involved in the almond/Aspergillus carbonarius interaction. J. Exp. Bot. 58: 1803–1811. 10.1093/jxb/erm039 [DOI] [PubMed] [Google Scholar]
- Müller V., Amé M., Carrari V., Gieco J., Asis R., 2014. Lipoxygenase activation in peanut seed cultivars resistant and susceptible to Aspergillus parasiticus colonization. Phytopathology 104: 1340–1348. 10.1094/PHYTO-12-13-0338-R [DOI] [PubMed] [Google Scholar]
- Musungu B. M, Bhatnagar D., Brown R. L., Payne G. A., OBrian G., et al. , 2016. A network approach of gene co-expression in the Zea mays/Aspergillus flavus pathosystem to map host/pathogen interaction pathways. Front. Genet. 7: 206 10.3389/fgene.2016.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam S., Waliyar F., Aruna R., Reddy S., Kumar P. L., et al. , 2009. Breeding peanut for resistance to aflatoxin contamination at ICRISAT. Peanut Sci. 36: 42–49. 10.3146/AT07-008.1 [DOI] [Google Scholar]
- Nueda M. J., Tarazona S., Conesa A., 2014. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics 30: 2598–2602. 10.1093/bioinformatics/btu333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozalvo R., Cabrera J., Escobar C., Christensen S. A., Borrego E. J., et al. , 2014. Two closely related members of Arabidopsis 13‐lipoxygenases (13‐LOXs), LOX3 and LOX4, reveal distinct functions in response to plant‐parasitic nematode infection. Mol. Plant Pathol. 15: 319–332. 10.1111/mpp.12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan M. M., 2010. Some nutritional characteristics of kernel and oil of peanut (Arachis hypogaea L.). J. Oleo Sci. 59: 1–5. 10.5650/jos.59.1 [DOI] [PubMed] [Google Scholar]
- Rajasekaran K., Cary J. W., Cotty P. J., Cleveland T. E., 2008. Development of a GFP-expressing Aspergillus flavus strain to study fungal invasion, colonization, and resistance in cottonseed. Mycopathologia 165: 89–97. 10.1007/s11046-007-9085-9 [DOI] [PubMed] [Google Scholar]
- R Core Team , 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Reverberi M., Punelli M., Smith C. A., Zjalic S., Scarpari M., et al. , 2012. How peroxisomes affect aflatoxin biosynthesis in Aspergillus flavus. PLoS One 7: e48097 10.1371/journal.pone.0048097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanavia T., Finotello F., Di Camillo B., 2015. FunPat: function-based pattern analysis on RNA-seq time series data. BMC Genomics 16: S2 10.1186/1471-2164-16-S6-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidegger K. A., Payne G. A., 2003. Unlocking the secrets behind secondary metabolism: a review of Aspergillus flavus from pathogenicity to functional genomics. Toxin Rev. 22: 423–459. [Google Scholar]
- Seijo J. G., Lavia G. I., Fernández A., Krapovickas A., Ducasse D., et al. , 2004. Physical mapping of the 5S and 18S–25S rRNA genes by FISH as evidence that Arachis duranensis and A. ipaensis are the wild diploid progenitors of A. hypogaea (Leguminosae). Am. J. Bot. 91: 1294–1303. 10.3732/ajb.91.9.1294 [DOI] [PubMed] [Google Scholar]
- Sekhwal M. K., Li P., Lam I., Wang X., Cloutier S., et al. , 2015. Disease resistance gene analogs (RGAs) in plants. Int. J. Mol. Sci. 16: 19248–19290. 10.3390/ijms160819248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham A., Moustafa K., Al-Ameri S., Al-Azzawi A., Iratni R., et al. , 2015. Identification of Arabidopsis candidate genes in response to biotic and abiotic stresses using comparative microarrays. PLoS One 10: e0125666 10.1371/journal.pone.0125666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherif M., Becker E.-M., Herrfurth C., Feussner I., Karlovsky P., et al. , 2016. Volatiles emitted from maize ears simultaneously infected with two Fusarium species mirror the most competitive fungal pathogen. Front. Plant Sci. 7: 1460 10.3389/fpls.2016.01460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spensley P. C., 1963. Aflatoxin, the active principle in turkey ‘X’ disease. Endeavour 22: 75–79. 10.1016/0160-9327(63)90097-8 [DOI] [PubMed] [Google Scholar]
- Spies D., Ciaudo C., 2015. Dynamics in transcriptomics: advancements in RNA-seq time course and downstream analysis. Comput. Struct. Biotechnol. J. 13: 469–477. 10.1016/j.csbj.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire R. A., 1981. Ranking animal carcinogens: a proposed regulatory approach. Science 214: 877–880. 10.1126/science.7302565 [DOI] [PubMed] [Google Scholar]
- Stintzi A., 2000. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630. 10.1073/pnas.190264497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubblefield R. D., Shotwell O. L., Hesseltine C. W., Smith M. L., Hall H. H., 1967. Production of aflatoxin on wheat and oats: measurement with a recording densitometer. Appl. Microbiol. 15: 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Bonnet E., Carvajal M., Méndez-Ramirez I., Castillo-Urueta P., Cortés-Eslava J., et al. , 2013. Aflatoxin (B1, B2, G1, and G2) contamination in rice of Mexico and Spain, from local sources or imported. J. Food Sci. 78: T1822–T1829. 10.1111/1750-3841.12291 [DOI] [PubMed] [Google Scholar]
- Sun X., Dalpiaz D., Wu D., Liu J. S., Zhong W., et al. , 2016. Statistical inference for time course RNA-Seq data using a negative binomial mixed-effect model. BMC Bioinformatics 17: 324 10.1186/s12859-016-1180-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N., Sasaki-Sekimoto Y., Obayashi T., Kikuta A., Kobayashi K., et al. , 2005. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139: 1268–1283. 10.1104/pp.105.067058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Zhang Y., Liu B., Yu Z., Wu Y., 2009. Study on the relationship between lipoxygenase-3 and the characteristic of resisting storage insects of rice grain. J. Food Agric. Environ. 7: 334–338. [Google Scholar]
- Thaler J. S., Stout M. J., Karban R., Duffey S. S., 1996. Exogenous jasmonates simulate insect wounding in tomato plants (Lycopersicon esculentum) in the laboratory and field. J. Chem. Ecol. 22: 1767–1781. 10.1007/BF02028503 [DOI] [PubMed] [Google Scholar]
- Thomma B. P., Eggermont K., Broekaert W. F., Cammue B. P., 2000. Disease development of several fungi on Arabidopsis can be reduced by treatment with methyl jasmonate. Plant Physiol. Biochem. 38: 421–427. 10.1016/S0981-9428(00)00756-7 [DOI] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis D. I., Kunze S., Willis D. K., Feussner I., Keller N. P., 2005. Aspergillus infection inhibits the expression of peanut 13 S-HPODE-forming seed lipoxygenases. Mol. Plant Microbe Interact. 18: 1081–1089. 10.1094/MPMI-18-1081 [DOI] [PubMed] [Google Scholar]
- Vergopoulou S., Galanopoulou D., Markaki P., 2001. Methyl jasmonate stimulates aflatoxin B1 biosynthesis by Aspergillus parasiticus. J. Agric. Food Chem. 49: 3494–3498. 10.1021/jf010074+ [DOI] [PubMed] [Google Scholar]
- Verma R., 2004. Aflatoxin cause DNA damage. Int. J. Hum. Genet. 4: 231–236. 10.1080/09723757.2004.11885899 [DOI] [Google Scholar]
- Vijayan P., Shockey J., Lévesque C. A., Cook R. J., 1998. A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7209–7214. 10.1073/pnas.95.12.7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waliyar F., Traore A., Fatondji D., Ntare B., 2003. Effect of irrigation interval, planting date, and cultivar on Aspergillus flavus and aflatoxin contamination of peanut in a sandy soil of Niger. Peanut Sci. 30: 79–84. 10.3146/pnut.30.2.0002 [DOI] [Google Scholar]
- Waliyar F., Kumar P. L., Traoré A., Ntare B. R., Diarra B., et al. , 2008. Pre-and post-harvest management of aflatoxin contamination in peanuts, pp. 209–218 in Mycotoxins: Detection Methods, Management, Public Health and Agricultural Trade. CABI, Wallingford, UK. [Google Scholar]
- Wang R., Shen W., Liu L., Jiang L., Liu Y., et al. , 2008. A novel lipoxygenase gene from developing rice seeds confers dual position specificity and responds to wounding and insect attack. Plant Mol. Biol. 66: 401–414. 10.1007/s11103-007-9278-0 [DOI] [PubMed] [Google Scholar]
- Wise A., Bar-Joseph Z., 2015. SMARTS: reconstructing disease response networks from multiple individuals using time series gene expression data. Bioinformatics 31: 1250–1257. 10.1093/bioinformatics/btu800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H., Isleib T., Payne G., Wilson R. F., Novitzky W., et al. , 2003. Comparison of aflatoxin production in normal- and high-oleic backcross-derived peanut lines. Plant Dis. 87: 1360–1365. 10.1094/PDIS.2003.87.11.1360 [DOI] [PubMed] [Google Scholar]
- Yan L., Zhai Q., Wei J., Li S., Wang B., et al. , 2013. Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 9: e1003964 10.1371/journal.pgen.1003964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Chen X., Zhu C., Peng X., He X., et al. , 2015. RNA-seq reveals differentially expressed genes of rice (Oryza sativa) spikelet in response to temperature interacting with nitrogen at meiosis stage. BMC Genomics 16: 959 10.1186/s12864-015-2141-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A., 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11: R14 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Bhatnagar D., Cleveland T. E., 2004. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 564: 126–130. 10.1016/S0014-5793(04)00327-8 [DOI] [PubMed] [Google Scholar]
- Zeneli G., Krokene P., Christiansen E., Krekling T., Gershenzon J., 2006. Methyl jasmonate treatment of mature Norway spruce (Picea abies) trees increases the accumulation of terpenoid resin components and protects against infection by Ceratocystis polonica, a bark beetle-associated fungus. Tree Physiol. 26: 977–988. 10.1093/treephys/26.8.977 [DOI] [PubMed] [Google Scholar]
- Zeringue H., 1996. Possible involvement of lipoxygenase in a defense response in aflatoxigenic Aspergillus–cotton plant interactions. Can. J. Bot. 74: 98–102. 10.1139/b96-014 [DOI] [Google Scholar]
- Zhang L., Zhang F., Melotto M., Yao J., He S. Y., 2017. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 68: 1371–1385. 10.1093/jxb/erw478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and supplemental figures and tables. Table S1: statistical models for differential expression analysis; Figure S1: sequence read integrity and mapping results; Figure S2: SOM clusters of differentially expressed genes between genotypes; Figure S3: KEGG pathway of α-linolenic acid metabolism; Figure S4: KEGG pathway of protein processing in the endoplasmic reticulum; Figure S5: KEGG pathway of the spliceosome; Figure S6: KEGG pathway of carbon fixation; Figure S7: KEGG pathway of carbon metabolism; Figure S8: expression profile of the novel transcripts; Figure S9: GO/KEGG enrichment analysis of differently expressed genes between peanut genotypes due to the infection vs. control of the newly assembled transcripts; Figure S10: SOM cluster groups of fungal differentially expressed genes; File S1.xlsx: Z-scores of differentially expressed genes; File S2.fasta: 2026 novel peanut transcripts; and File S3.txt: ICG 1471 coexpression network clusters. All raw data fastq sequences are deposited at the NCBI (http://www.ncbi.nlm.nih.gov/) under BioProject PRJNA417591. All raw sequences are deposited as BioSamples SAMN08000482: SAMN08000553. The keggseq package is freely available to the public under MIT license and can be downloaded from https://github.com/w-korani/keggseq. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5984860.