Summary
Mismatch Repair (MMR) increases the fidelity of DNA replication by identifying and correcting replication errors. Processivity clamps are vital components of DNA replication and mismatch repair, yet the mechanism and extent to which they participate in MMR remains unclear. We investigated the role of the Bacillus subtilis processivity clamp DnaN, and found that it serves as a platform for mismatch detection and coupling of repair to DNA replication. By visualizing functional MutS fluorescent fusions in vivo, we find that MutS forms foci independent of mismatch detection at sites of replication (i.e., the replisome). These MutS foci are directed to the replisome by DnaN clamp zones that aid mismatch detection by targeting the search to nascent DNA. Following mismatch detection, MutS disengages from the replisome, facilitating repair. We tested the functional importance of DnaN-mediated mismatch detection for MMR, and found that it accounts for 90% of repair. This high dependence on DnaN can be bypassed by increasing MutS concentration within the cell, indicating a secondary mode of detection in vivo whereby MutS directly finds mismatches without associating with the replisome. Overall, our results provide new insight into the mechanism by which DnaN couples mismatch recognition to DNA replication in living cells.
Keywords: Mismatch repair, MutS, DnaN, Bacillus subtilis
Introduction
Mismatch repair (MMR) increases the fidelity of DNA replication by identifying and correcting errors made by the replicative DNA polymerase [for review (Kunkel & Erie, 2005, Schofield & Hsieh, 2003)]. Upon detection of an error, MMR orchestrates its removal and accurate resynthesis of the surrounding DNA, ultimately increasing the fidelity of DNA replication by several hundred-fold [for review (Kunkel & Erie, 2005, Schofield & Hsieh, 2003, Iyer et al., 2006)]. Due to this important role in maintaining genome stability, MMR is found in all domains of life, with a high degree of conservation specifically among MutS and MutL proteins (Culligan et al., 2000). In bacteria, deletion of either mutS or mutL homologs leads to an increased mutation rate (Cox et al., 1972, Ginetti et al., 1996, Prudhomme et al., 1989, Cooper et al., 2012, Davies et al., 2011). This mutator phenotype is known to accelerate acquisition of multidrug resistant strains in hospital settings, while also enabling increased survival of bacterial pathogens in harsh environments, including growth inside the lungs of cystic fibrosis patients [(Prunier et al., 2003, Roman et al., 2004, Watson et al., 2004, Turrientes et al., 2010, Denamur et al., 2002, Oliver et al., 2000, Mena et al., 2008) and for review (Oliver & Mena, 2010, Chopra et al., 2003)]. MMR defects in eukaryotes are characterized by hypermutability and microsatellite instability; both of which have been linked to an increased predisposition for spontaneous tumorigenesis, as well as inherited conditions such as Lynch syndrome and Turcot syndrome (Hamilton et al., 1995, Nystrom-Lahti et al., 2002, Peltomaki, 2005, Fishel et al., 1993).
In bacteria, the MutS homodimer initiates MMR by detecting base-base mismatches or small insertion/deletion loops (IDLs) (Su & Modrich, 1986). In eukaryotes, base-base mismatches and small IDLs (1 or 2 extrahelical nucleotides) in DNA are primarily recognized by Msh2-Msh6 (MutSα), while larger IDLs (1–15 extrahelical nucleotides) are recognized by Msh2-Msh3 (MutSβ) heterodimers (Alani et al., 1995, Prolla et al., 1994, Habraken et al., 1996, Palombo et al., 1996). In all systems, following mismatch or IDL detection by a MutS homolog, MutL (MutLα or MutLβ in eukaryotes) is recruited to the site of the mismatch in a reaction that requires ATP (Schofield et al., 2001b). Following this step, MutL is hypothesized to facilitate removal of the mismatch by coordinating numerous DNA transactions including endonuclease nicking, helicase driven unwinding, and excision of the segment containing the misincorporated base(s) (Lahue et al., 1989).
Since replicative DNA polymerases have high fidelity, base pairing errors occur at a low frequency of one in 106–107 correctly paired bases [(Schaaper, 1993) and for review (Kunkel, 1992, Kunkel & Bebenek, 2000)]. In addition to the challenges posed by the low rate or error formation, base mispairs may also be obscured by DNA supercoiling, compaction and protein binding. MutS must also contend with other active processes on the DNA, including transcription, when searching for mismatches and IDLs in DNA [for review on chromosome organization (Jackson et al., 2012)]. Given these challenges, it has been proposed that MutS is coupled to DNA replication forks in order to facilitate efficient mismatch detection where mismatches are newly formed, and where the DNA is more likely to be free of protein impediments (Simmons et al., 2008, Smith et al., 2001). In support of this model, cytological studies conducted in B. subtilis, S. cerevisiae and human cells have shown that prokaryotic MutS-GFP and eukaryotic MutSα (Msh6-mCherry) form foci that are often coincident with DNA replication foci in vivo (Hombauer et al., 2011a, Simmons et al., 2008, Smith et al., 2001, Kleczkowska et al., 2001). Furthermore, in B. subtilis, MutS and mismatches were shown to alter localization of an essential DNA polymerase (Klocko et al., 2011). These results suggest that MutS is spatially coordinated with active replisomes in B. subtilis, S. cerevisiae, and human cells. In addition to possible spatial coupling between MMR and DNA replication, S. cerevisiae MMR was shown to be defective when Msh6 was unavailable during S phase (DNA replication), supporting the importance of temporal coupling of MutSα to DNA replication in eukaryotes (Hombauer et al., 2011b).
Studies in various model organisms indicate that DNA replication processivity clamps function in MMR (Flores-Rozas et al., 2000, Hombauer et al., 2011a, Kleczkowska et al., 2001, Lee & Alani, 2006, Lopez de Saro et al., 2006, Simmons et al., 2008, Shell et al., 2007). Processivity clamps exist as either a homodimer in bacteria (DnaN) or a homotrimer in archaea and eukaryotes (PCNA) (Kong et al., 1992, Krishna et al., 1994, Matsumiya et al., 2001). These clamps are loaded onto the 3′ termini of DNA by the clamp loader complex [e.g. (Georgescu et al., 2008, Jeruzalmi et al., 2001, Bowman et al., 2004)], and once loaded, DnaN and PCNA confer processive activity to replicative polymerases by tethering the polymerase to the DNA template (Stukenberg et al., 1991, Huang et al., 1981). MutS homologs contain a conserved DnaN clamp-binding motif, or PIP box [PCNA Interacting Protein] in eukaryotes, that mediates interactions between MutS proteins and their cognate processivity clamps (Flores-Rozas et al., 2000, Hombauer et al., 2011a, Kleczkowska et al., 2001, Lee & Alani, 2006, Lopez de Saro et al., 2006, Simmons et al., 2008, Shell et al., 2007, Monti et al., 2012). Studies in B. subtilis showed that deletion of the unstructured region of MutS (MutS800) containing a putative DnaN clamp-binding motif, reduced interaction with DnaN, yet MutS800 maintained the ability to preferentially bind mismatched DNA in vitro (Simmons et al., 2008). In vivo, the mutS800 allele eliminated functional MMR, and when translationally fused to gfp, failed to form foci demonstrating that although proficient in mismatch detection, MutS800 was defective for forming repair complexes in vivo (Simmons et al., 2008). Recent work in S. cerevisiae demonstrates that PCNA-associated MutSα accounts for 10–15% of MMR in vivo, and that Msh6-GFP (MutSα) foci are dependent upon interaction with PCNA through the Msh6 PIP box (Hombauer et al., 2011a). Processivity clamps are also proposed to function in downstream steps of MMR, such as facilitating activation of endonuclease activity in MutL homologs (Kadyrov et al., 2006, Pillon et al., 2010, Pillon et al., 2011, Pluciennik et al., 2010) and in re-synthesis of the gap in DNA following strand excision [(Gu et al., 1998, Umar et al., 1996) for review (Larrea et al., 2010, Lenhart et al., 2012)]. While it is clear that interactions between MutS and processivity clamps play a role in MMR [e.g. (Gu et al., 1998, Umar et al., 1996)], important questions remain about their significance (Clark et al., 2000, Flores-Rozas et al., 2000, Lopez de Saro et al., 2006), the mechanism(s) by which clamps influence MMR in vivo and the step during MMR that require processivity clamp interaction. Three main models have been used to explain the role of processivity clamps in MMR. These models include the hypothesis that clamps directly aid in mismatch binding (Flores-Rozas et al., 2000, Lau & Kolodner, 2003) or stabilize MutS at a mismatch (Simmons et al., 2008), clamps recruit MutS to sites of DNA replication (Hombauer et al., 2011a, Kleczkowska et al., 2001) or that clamps are required for DNA synthesis after mismatch removal and do not have an earlier role during MMR (Pluciennik et al., 2009). Otheret al. key questions are, what step of repair is affected by the clamp, as well as how MutS dynamics on DNA are effected by the presence of the clamp as MutS searches for and initiates repair of rare mismatches formed during replication.
In vivo studies of DNA replication in B. subtilis show that the DnaN processivity clamp exists in a “clamp zone” immediately following the progressing replication forks. DnaN clamps are retained on nascent DNA during Okazaki fragment maturation and accumulate until a steady state level is reached between actively loaded and unloaded clamps (Su’etsugu & Errington, 2011). Because DnaN clamp zones trail the replication fork, these zones have the potential to serve as platforms that maintain the spatial and temporal relationship between mismatch recognition and active replication forks. In this work, we used several separation-of-function MutS mutants that are defective in either mismatch detection or DnaN binding to determine when and where during repair the MutS-DnaN interaction is mechanistically significant in live cells. Using functional mutS-gfp fusions expressed from the mutS native locus, we report that DnaN clamp zones position MutS at newly replicated DNA prior to, and independent of, mismatch binding. After mismatch detection, MutS no longer remains coincident with the replication machinery, instead localizing to sites of repair. Importantly, ~90% of MMR in vivo is initiated through DnaN-clamp-zones, revealing a heavy reliance by MutS on the clamp during the initial steps of repair. We used the MutS800 mutant to uncouple MMR from DnaN and found that this mutant could account for only ~10% of in vivo repair. Remarkably, we were able to restore DnaN-independent MMR to wild-type levels by increasing the cellular levels of MutS800, illustrating that the functional significance of the DnaN-MutS interaction lies in maximizing the efficiency of mismatch detection in vivo. Interestingly, mismatch detection appeared to occur on replication fork proximal DNA. This observation contrasts with models where MMR is initiated through detection of a mismatch distal to the replisome and nascent DNA. Our findings indicate that B. subtilis MutS relies on mismatch detection on nascent DNA for efficient repair. Ultimately, by having MutS bind to DnaN clamp zones that closely trail replication forks, MMR and DNA replication become tightly coupled, allowing for efficient mismatch detection, MutL activation and subsequent repair in B. subtilis cells.
Results
B. subtilis MutSF30A is mismatch repair deficient due to loss of mismatch binding specificity
In order to determine if mismatch binding is necessary for MutS localization, we monitored MutSF30A, which has a mutation that should abolish mismatch recognition (Malkov et al., 1997). During high affinity interaction between MutS and mismatched DNA, the phenylalanine residue in the conserved GXFY(X)5DA motif stacks with the mismatched or unpaired base (Fig. 1A) (Lamers et al., 2000, Obmolova et al., 2000). Substitution of phenylalanine to alanine eliminates mismatch detection in vitro and functional MMR in vivo in several organisms (Bowers et al., 2000, Malkov et al., 1997, Schofield et al., 2001a). This mutation does not disrupt the ATPase mechanism, indicating that MutSF30A activities other than mismatch binding are unaffected (Jacobs-Palmer & Hingorani, 2007).
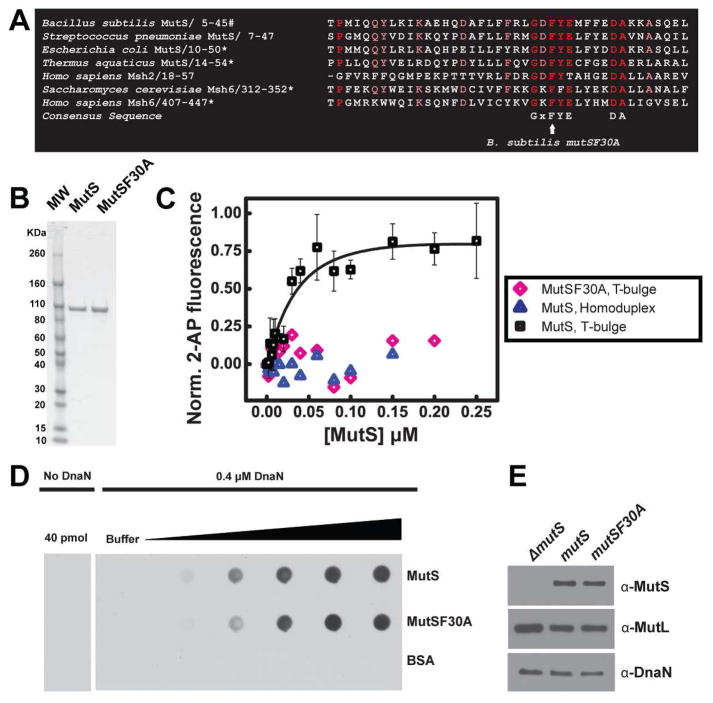
Figure 1. MutSF30A is unable to bind mismatches in vitro.
(A) The following conserved motif necessary for mismatch binding (GXFYXXXXXDA) is found in a wide range of eukaryotic and prokaryotic MutS homologs. Indicates (*) organisms previously demonstrated that substitution of the conserved phenylalanine residue (F) eliminates mismatch binding in vitro and prevents MMR in vivo. (B) 1 μg of purified MutS and MutSF30A protein electrophoresed on a 4–15% SDS-PAGE gradient gel. (C) In vitro binding of MutS and MutSF30A to T-bulge substrate. Legend: black squares and pink diamonds show MutS and MutSF30A interaction with T-bulge containing DNA, respectively, and blue triangles show MutS interaction with homoduplex DNA. (D) An immunodot blot (far western) analysis was performed to monitor interaction between MutS or MutSF30A with DnaN. Purified MutS and MutSF30A were blotted onto a nitrocellulose membrane over a range of 0.625 pmol to 40 pmols of dimer. Purified DnaN was incubated with the membrane and probed with affinity-purified antisera against DnaN in a 1:500 dilution. Shown (left most blot) is the purified antisera control against purified MutS and MutSF30A. Shown (right most blot) is the retention of DnaN by MutS and MutSF30A as described in “Experimental Procedures.” (E) In vivo steady state levels of MutS, MutL, and DnaN. A total of 5 μg of cell extract derived from the indicated strain was electrophoresed and immunoblotted in the indicated lanes.
We tested the corresponding mutSF30A mutation for the ability to support both mismatch binding in vitro and functional repair in B. subtilis. We purified B. subtilis MutSF30A using standard chromatography techniques without the use of an affinity tag (Fig. 1B). We found that purified MutS binds a T-bulge DNA substrate (containing an extrahelical thymidine) selectively with a Kd of 24 nM, while MutSF30A shows little binding to either a T-bulge or a homoduplex DNA substrate, precluding us from calculating a Kd (Fig 1C). Furthermore, we verified that the F30A mutation did not have an adverse affect on MutS binding to DnaN. An immunodot blot analysis shows comparable retention of DnaN by MutS and MutSF30A (Fig. 1D).
MutSF30A function was also tested in vivo by introducing an unmarked mutSF30A allele at the native mutS locus by allelic replacement (see “Experimental Procedures”). Immunoblot analysis confirmed that the mutant MutS protein, as well as the downstream gene product MutL, accumulated to wild-type levels in vivo (Fig. 1E). Using spontaneous rifampicin resistance as an indicator for mutation rate, we found that the mutSF30A allele conferred a mutation rate of 155.4 × 10−9 mutations/generation, significantly higher than the mutation rate of the wild type strain, which was 1.82 × 10−9 mutations/generation (Table 1). The mutSF30A mutation rate was indistinguishable from a strain with the mutSL::spec allele, which eliminates all in vivo MMR, showing an ~85 fold increase in mutagenesis to 154.5 × 10−9 mutations/generation. With these data we conclude that the B. subtilis mutSF30A allele is MMR defective in vivo due to a loss in mismatch binding specificity.
Table 1.
mutSF30A is defective for mismatch repair in vivo.
| Relevant genotype | No. of cultures | Mutation rate (10−9 mutations/generation)±[95%CI] | Relative mutation rate | Total MMR activity (%) |
|---|---|---|---|---|
| PY79 (wild type) | 51 | 1.82 [1.14–2.37] | 1 | 100% |
| mutSL::spc | 23 | 154.4 [146.6–162.2] | 84.9 | 0% |
| mutSF30A | 24 | 155.9 [147.5–163.3] | 85.5 | −0.7% |
| dnaN5 | 30 | 39.2 [33.7–44.7] | 21.6 | 75.5% |
Mismatch repair proficiency and analysis of the mutation rate of the mutSF30A strain compared to wild type cells and MMR deficient cells. The bracketed values represent the lower and upper bounds of the 95% confidence limit.
MutSF30A-GFP forms foci on DNA independent of mismatch binding
After demonstrating that B. subtilis MutSF30A is defective for mismatch binding, we sought to determine whether mismatch binding is a prerequisite for localization of MutS into discrete foci in vivo. We first determined that the mutS-gfp native locus allele exhibits ~90% of wild-type MMR activity, indicating that the gfp fusion has little impact on MutS function in vivo (Table S1). MutS-GFP foci were detected in only ~9% of untreated, exponentially growing cells, whereas treatment with the mismatch forming agent 2-aminopurine (2-AP) resulted in >45% of cells with MutS-GFP foci (Fig. 2A, C and S1). A similar increase in MutS-GFP foci occurred following introduction of a DNA polymerase mutant, polC mut-1 defective in proofreading (herein referred to as polCexo-), which substantially increases the frequency of errors during DNA replication (Sanjanwala & Ganesan, 1991). MutS-GFP focus formation was observed in ~25% of cells when polCexo- was the sole source of the replicative DNA polymerase in the cell (Fig. 2C). These results demonstrate that MutS-GFP focus formation responds to natural mismatches formed by normal bases during DNA replication.
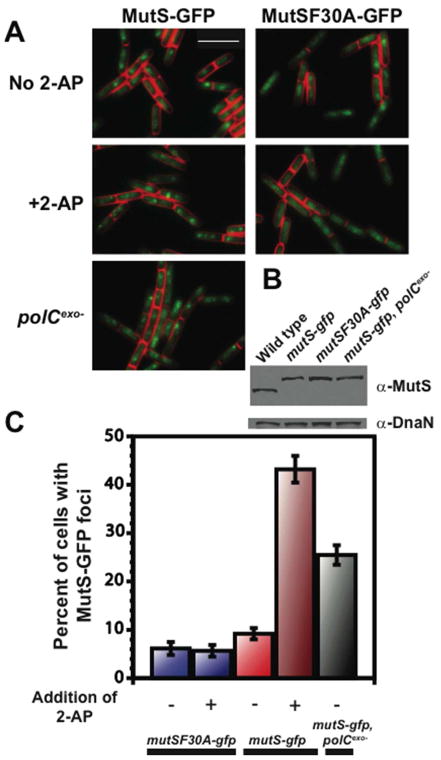
Figure 2. MutS-GFP forms foci independent of mismatch formation.
(A) Representative images of MutS-GFP or MutSF30A-GFP foci in cells with or without 600 μg/mL of 2-AP. The white bar is 4 μm. (B) An immunoblot of the indicated MutS derivative and DnaN as a loading control. (C) Bar graph of the groups represented in (A) showing the percentage of cells with MutS-GFP foci. Total number of cells scored for each condition was from left to right: n=1234, 1410, 2380, 1222, and 1797.
To test if mismatch binding was a prerequisite for MutS localization, we built a mutSF30A-gfp reporter allele at its native locus. Strikingly, MutSF30A-GFP formed foci during exponential growth in ~6% of cells (Fig. 2A and C). As a control, we determined via immunoblot that the cellular level of MutS and MutSF30A-GFP were indistinguishable (Fig. 2B). When MutSF30A-GFP cells were challenged with 2-AP, we did not observe an increase in foci above ~6% (Fig. 2C) supporting our data that MutSF30A is defective in mismatch recognition. Furthermore, MutL-GFP focus formation in mutSF30A cells was not stimulated by 2-AP treatment, indicating that MutS must be able to bind a mismatch in order to efficiently activate the downstream steps of repair in vivo, including recruitment of MutL (Supporting results and Fig. S2). Thus, MutSF30A fails to detect and respond to 2-AP formed mismatches in vivo, consistent with the above data showing loss of mismatch binding in vitro (Fig. 1C).
Interestingly, we did observe a small, though statistically significant difference in the percent of cells with foci between untreated mutSF30A-gfp and mutS-gfp cells (Fig. 2C and S1). This result is not explained by differences in binding of MutS or MutSF30A to DnaN since both proteins bind DnaN equally. Because we expect a small subset of cells to undergo MMR in the functional mutS-gfp background, we suggest that the slightly greater percentage of cells with MutS-GFP foci relative to the MutSF30A-GFP in untreated cells represents MutS-GFP foci engaged in repair. We conclude that MutS forms two types of foci: one licensed by mismatch detection and one that is mismatch-detection independent. Together our results demonstrate that MutS forms foci on DNA independent of, or prior to, mismatch binding in live cells.
MutS is staged at active replisomes prior to mismatch recognition
We investigated the localization dynamics of MutS foci before and after mismatch detection in order to better understand the spatial-temporal coupling of MutS to the replisome. We define the replisome as replication associated proteins (replicative polymerases, clamp loader components, processivity clamp, etc.) that localize as discrete foci at replication forks in vivo. In B. subtilis, the replisome occupies characteristic subcellular positions denoting the site of DNA synthesis (Berkmen & Grossman, 2006, Lemon & Grossman, 1998, Migocki et al., 2004). Immediately after replication initiation from oriC, origin regions and replicated DNA translocate away from the replisome toward the opposite cell poles (Webb et al., 1997, Teleman et al., 1998). Previously, it was shown that B. subtilis MutS-YFP colocalizes with the replisome in ~48% of cells (Smith et al., 2001). As shown above, the percentage of cells with MutS-GFP foci increase following 2-AP treatment (Fig. 2) (Klocko et al., 2011, Simmons et al., 2008, Smith et al., 2001), indicating that more repair complexes are formed. As DNA replication progresses, the newly replicated chromosomal DNA moves toward the cell poles (Webb et al., 1997), presumably taking replication errors away from the replisome. Thus, we hypothesized that the MutS-GFP foci would initially associate with mismatches in DNA at or near the replisome, and as repair and replication continued, the mismatch•MutS complex would move toward the cell poles, reducing colocalization with the replisome.
Initially, we tested our ability to spatially resolve replicated DNA from the replisome by monitoring the colocalization of CFP-Spo0J with DnaX-YFP. We grew cells slowly so that most cells would have one or two replisome foci during this analysis (Fig S3) (see “Experimental Procedures”). Spo0J, which localizes to and helps organize the origin of replication (oriC), should only colocalize with the clamp loader protein DnaX during replication of the origin region, and then translocate away from the replisome to the cell pole (Gruber & Errington, 2009, Sullivan et al., 2009). We found that in cells containing a single DnaX-mYFP focus, only 12.8% of replisome foci colocalized with CFP-Spo0J (n=297). Furthermore, inspection of these cells shows that most single replisome cells contain the expected origin-replisome-origin localization pattern along their longitudinal axis (Fig 3).
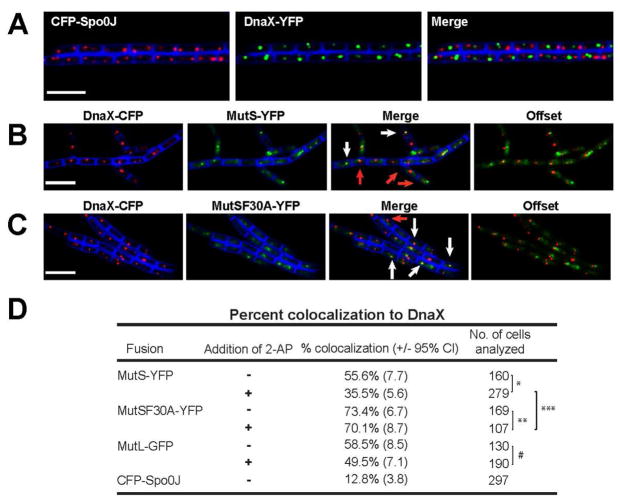
Figure 3. MutS-GFP colocalizes with the replisome prior to mismatch detection.
(A) Representative images of CFP-Spo0J and DnaX-YFP colocalization with the replisome. (B) Representative images of colocalization between MutS-YFP with DnaX-CFP following 2-AP treatment. White arrows denote MutS-YFP foci that colocalize with the DnaX-CFP, whereas red arrows denote MutS-YFP foci that do not colocalize with DnaX-CFP (C) Representative images of colocalization between MutSF30A-YFP with DnaX-CFP following 2-AP treatment. The vital membrane stain TMA-DPH is shown in blue, the white bar is 4 μm. (D) Scoring of colocalization of MutS-YFP and MutL-GFP at the replisome in the presence and absence of 2-AP, p-values are one- tailed; p= * 2.03×10−5; **2.77; ***4.77 × 10−10; # 0.0568
To test our hypothesis that MutS moves away from the replisome after mismatch binding, we introduced functional dnaX-cfp and mutS-yfp alleles (>90% MMR activity) (Table S1) into B. subtilis cells, with both fusions placed at their native locus and under control of their native promoters. During exponential growth, MutS-YFP foci colocalized with the replisome in ~56% of cells containing at least one DnaX-CFP focus and one MutS-YFP focus (Fig. 3B). When cells were treated with 2-AP to form mismatches, we observed a significant decrease in colocalization to ~35% (p =2.03 × 10−5) (Fig. 3B and D). These data support the hypothesis that mismatch recognition by MutS-YFP reduces colocalization with the replisome.
When the same experiment was performed with mutSF30A-yfp, we observed that MutSF30A-YFP foci colocalized with the replisome ~73% of the time in the absence of 2-AP challenge (Fig. 3C and D). When this strain was treated with 2-AP, there was no significant statistical difference in the position of MutSF30A-YFP foci compared with the untreated group (~70.1% colocalized: p=0.277). Thus, lacking the ability to detect mismatches in DNA, MutSF30A-YFP remains colocalized with the replisome. These results lead us to conclude that MutS-GFP foci, when not bound to mismatches, are staged near the active replisome, possibly due to physical coupling with a replication protein. Subsequently, upon encountering a mismatch, MutS disengages from the advancing replisome and remains behind on nascent DNA to direct the remaining steps in repair. This result provides insight into how mismatch recognition affects the dynamic association of MutS with the replisome in vivo.
Based on this model, we hypothesized that if MutS is positioned on newly replicated DNA through interaction with a replisome protein, then increasing expression of MutS or MutSF30A should increase the number of mismatch-independent foci by promoting this interaction in vivo. To this end, we constructed an in frame mutS deletion that maintains transcriptional control of mutL from its native promoter (Fig 1D & Fig. S4). We then expressed MutS-GFP or MutSF30A-GFP from an ectopic locus driven by an IPTG regulated promoter (Pspac). The ΔmutS, amyE::PspacmutS-gfp strain was 88.7% functional compared to ΔmutS, amyE::PspacmutS (Table S1). When either mutS-gfp or mutSF30A-gfp was ectopically expressed, we observed a 2–3 fold increase in the percent of untreated cells with foci (Fig. 4A, compare with Fig. 2C). This result was not affected by the presence or absence of mutL.
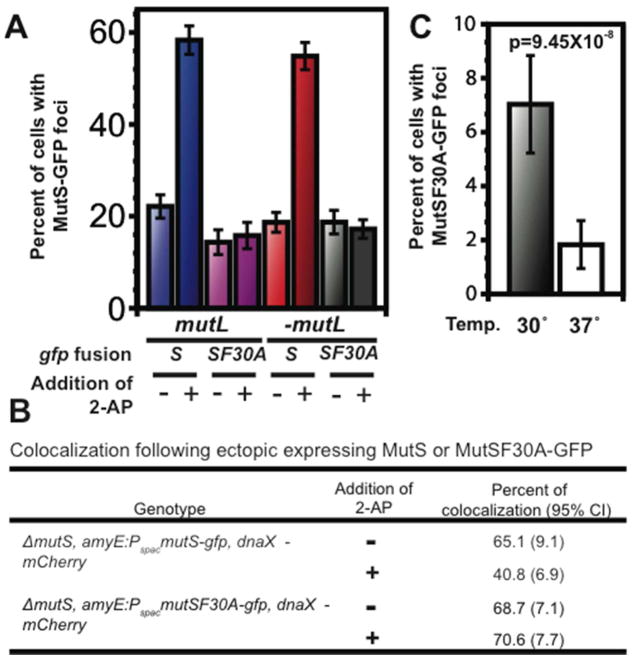
Figure 4. Elevated expression of MutS-GFP increases replisome-associated foci.
(A) Elevated expression of mutS-gfp and mutSF30A-gfp increases the percentage of cells with MutS foci. The first four groups contain a ΔmutS deletion and the expressed GFP fusion represents the sole copy of mutS within the cell. The second four groups represent a deletion of the entire mutSL locus with mutS- or mutSF30A-GFP expressed ectopically from a Pspac promoter (B) Elevated expression of mutS-gfp causes an increase in colocalization of MutS foci to DnaX-mCherry foci. Total number of cells scored is 106–196. (C) MutSF30A-GFP foci expressed from the mutS native locus in the dnaN5 background at 30 and 37 °C.
We then asked if increased expression of mutS-gfp and mutSF30A-gfp and the associated mismatch independent foci correlated with colocalization with the replisome marker dnaX-mcfp. We found that ectopic expression caused an increase in colocalization to ~65%, (Fig. 4B). When these cells were challenged with 2-AP, we expect a decrease in colocalization and indeed found ~41% were colocalized following 2-AP challenge (Fig. 4B). Ectopic expression of MutSF30A increased the percent of cells with foci, and colocalization of MutSF30A, which only forms mismatch-independent foci, remained at ~70% upon ectopic expression. These results show that increased expression of MutS increases the percentage of cells with foci colocalized to the replisome (Fig 4A), supporting the hypothesis that MutS is positioned at the replisome via a binding partner prior to mismatch identification.
It should be noted that in our colocalization experiments we used DnaX as a replisome marker instead of DnaN, which binds MutS in vitro, because the dnaN-mcfp fusion maintains an elevated mutation rate (25.3 × 10−9 mutations/generation) whereas dnaX-mcfp is wild type for mutation rate (data not shown). We determined that the smaller DnaX-mCherry foci colocalizes with DnaN-GFP foci in ~89% of cells, establishing DnaX as an appropriate substitute for DnaN in this analysis (Fig. S5).
MutSF30A-GFP foci are positioned at the replisome by DnaN prior to mismatch binding
The ability of MutSF30A-GFP to assemble into foci without mismatch identification suggests that MutS may be positioned at the replisome as a mechanism to spatially target newly formed mismatches in DNA. Furthermore, colocalization of MutSF30A with the replisome and MutSF30A binding to DnaN in vitro (Fig. 1E) suggest that MutSF30A localization is dependent on an interaction with a protein component of the replisome. In B. subtilis, DnaN forms large clamp assemblies termed “clamp zones” that form behind progressing replication forks (Su’etsugu & Errington, 2011). DnaN clamp zones contain ~200 accumulated clamps as clamp loading and unloading rates achieve equilibrium (Su’etsugu & Errington, 2011).
We hypothesized that a clamp zone facilitates the formation of mismatch-independent foci by recruiting MutS to the replisome via contact with DnaN. To test this hypothesis, we took advantage of the dnaN5 allele, which exhibits an increase in mutation frequency due to partial loss of MMR [(Simmons et al., 2008, Dupes et al., 2010) and Table S1]. The dnaN5 allele exhibits a temperature sensitive defect in MMR, which leads to a significant decrease in MutS-GFP focus formation at 37°C relative to 30°C. We determined that DnaN5 functions normally in DNA replication by measuring replication in vivo and found that dnaN5 is wild type for DNA synthesis and growth rate at both 30°C and 37°C [(Fig. S6) (Simmons et al., 2008, Dupes et al., 2010)]. We introduced dnaN5 into a strain bearing the mutSF30A-gfp allele at its native locus and scored the number of MutSF30A-GFP foci at 30°C and 37°C. At 30°C, we found that ~7% of cells contain a MutSF30A-GFP focus, results consistent with that of a dnaN+ strain (Fig. 4C and Fig. 1C). In contrast, at 37°C, the percentage of cells with MutSF30A-GFP foci decreased to <2% (p=9.45 × 10−8). Three prominent models have been used to explain the role of DnaN in mismatch repair. The first model suggests that MutS is stabilized or mismatch recognition affinity is increased through interaction with the replication clamp (Simmons et al., 2008, Flores-Rozas et al., 2000, Lau & Kolodner, 2003). The second model primarily supported from studies in human cell culture and S. cerevisiae predicts that MutS is recruited to sites of replication (Hombauer et al., 2011a, Kleczkowska et al., 2001). Finally, the third model suggests that the major role for DnaN clamp is during resynthesis of the DNA (Pluciennik et al., 2009). Our data show that interaction with DnaN is critical for formation of the mismatch-independent MutS foci in vivo. We further interpret these results to mean that DnaN clamp zones recruit and stage MutS immediately behind the advancing replication forks in vivo supporting the model that MutS is recruited to sites of replication before mismatch binding strongly supporting the second model.
DnaN clamp zones increase efficiency of mismatch detection by targeting MutS to nascent DNA
In defined in vitro MMR systems, purified MutS and MutSα can detect a mismatch in DNA without the need for a processivity clamp [e.g. (Tessmer et al., 2008, Zhai & Hingorani, 2010)]. We hypothesized that the association of MutS with DnaN might be necessary in vivo in order to restrict the search for mismatches to nascent DNA, making mismatch detection more efficient relative to MutS identifying a mismatch independent of DnaN binding. To test the key hypothesis that a DnaN clamp zone recruits MutS, we took advantage of the mutS800 allele, which lacks a C-terminal tether and is defective in DnaN binding, but is proficient for mismatch identification (Simmons et al., 2008). When the B. subtilis mutS800 allele was expressed from its native promoter, only ~9.5% of MMR activity is observed in vivo (Table 2). The mutS800 allele can support 97.4% MMR activity when this mutant protein is overexpressed from an IPTG driven Pspac promoter from an ectopic locus in a strain lacking the native mutS allele (Table 2). Immunoblotting shows that the PspacmutS800 protein level was 4-fold higher than the level produced from the mutS800 allele located at the mutS native locus (Figure S7). This result supports the hypothesis that increasing MutS800 concentration can compensate for the loss of interaction with DnaN and restore efficient MMR activity.
Table 2.
DnaN allows for efficient mismatch repair in vivo.
| Relevant genotype | No. of cultures | Mutation rate (10−9 mutations/generation)±[95%CI] | Relative mutation rate | Total MMR activity (%) |
|---|---|---|---|---|
| ΔmutS, amyE::PspacmutS | 25 | 3.09 [1.35–4.68] | 1 | 100 |
| ΔmutS | 28 | 159.3 [152.0–166.6] | 51.5 | 0 |
| mutS800 | 25 | 144.5 [134.9–154.2] | 46.7 | 9.5 |
| ΔmutS, amyE::PspacmutS800 | 23 | 7.10 [4.39–9.74] | 2.3 | 97.4 |
The ΔmutS designation indicates an in frame deletion of mutS, maintaining a functional mutL at its native locus (see “Materials and Methods”). The mutS800, allele was expressed from its native locus with mutL expressed ectopically from amyE using 1 mM IPTG. Brackets enclose the lower bounds and upper bounds of the 95% confidence limits. Percent MMR activity was determined using the following equation: [(R.M.R.null - R.M.R.strain)/(R.M.R.null-R.M.R.wild type)]•100. RMR=relative mutation rate. Mutations per culture (m) are as follows: ΔmutS, amyE::PspacmutS (1.78); ΔmutS (104.5); mutS800 (64.2); ΔmutS, amyE::PspacmutS800 (5.0).
When mutS800-gfp is expressed from the mutS native locus, the protein is defective in forming foci in response to mismatches in vivo (Simmons et al., 2008). Since mutS800 restores MMR activity when ectopically expressed, we asked if mutS800-gfp also forms foci following ectopic expression. Upon visualizing ectopically expressed MutS800-GFP in a ΔmutS background, we found that MutS800-GFP formed occasional foci in untreated cells (Fig. 5A); however, focus intensity was barely above the elevated background fluorescence and only formed in ~3.3% of cells. Upon 2-AP treatment, the percentage of cells with faint foci substantially increased to 14.1%, indicating that like MutS-GFP (Fig. 2), MutS800-GFP focus formation is responsive to an increase in mismatches in DNA. To verify this observation, we asked if mismatch binding by MutS800 was important for focus formation. To answer this question, mutSF30A800-gfp was placed under the control of an IPTG driven Pspac promoter and inserted at an ectopic locus in a ΔmutS background. This allele is defective in both DnaN clamp binding and mismatch detection. We predicted that overexpressed MutSF30A800 would fail to localize into foci if the observed localization of ectopically expressed mutS800 was dependent on mismatches and independent of DnaN. Indeed, we found that MutSF30A800 was blocked for focus formation (Fig. 5B) (<1% in both 2-AP treated and untreated samples). We conclude that when the DnaN tether on MutS is removed, mismatch binding becomes obligatory for focus formation in vivo (Fig. 5A, B, C). We further verified this observation by inserting the dnaN5 allele into the ΔmutS, amyE::PspacmutS-gfp background. At both 30°C and 37°C, MutS800-GFP formed foci in ~14% of cells, consistent with our results for the dnaN+ allele (Fig. 4C) and further confirming bypass of the DnaN role in MMR following overexpression (Fig. 5C, D).
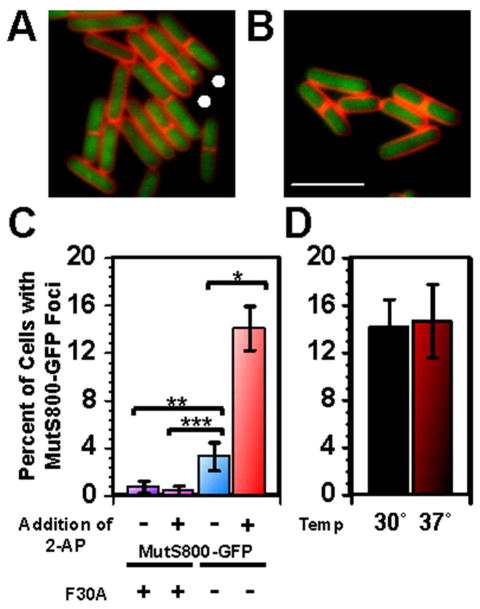
Figure 5. Mismatch detection by MutS800-GFP induces focus formation at nascent DNA when ectopically expressed.
(A) MutS800-GFP foci form in response to mismatches independent of DnaN (faint foci indicated by white circles) (B) MutSF30A800 fails to form foci. The vital membrane stain TMA-DPH is shown in red and the white scale bar is 4 μm. (C) Bar graph of ectopically expressed MutS800-GFP and MutSF30A800-GFP foci with and without 2AP. From left to right, total cells scored: 1114, 1154, 883, 1343. P-values are 1-tailed: *p=2.69×10−17, **p=1.20×10−5, ***p=3.22×10−7. (D) Bar graph of ectopically expressed MutS800-GFP foci within the dnaN5 background revealed no statistical difference at 30 °C and 37°C (p=0.38).
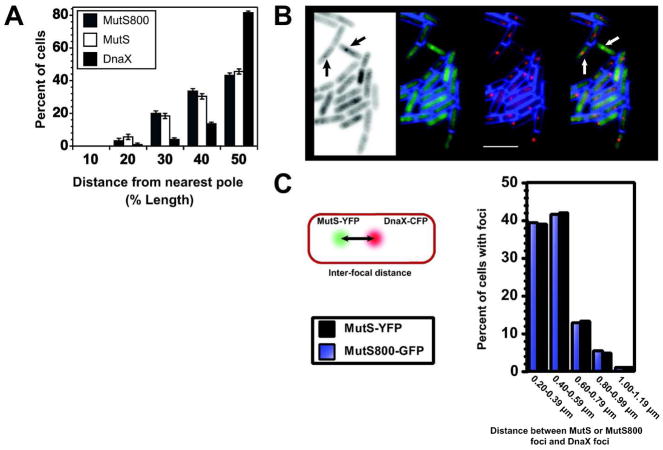
Because overexpression of mutS800 restores MMR to near wild type levels, bypassing the need for DnaN (Table 2), we asked where ectopically expressed MutS800 foci form in vivo. To address this question, we visualized and scored the subcellular location of ectopically expressed MutS800-GFP in comparison to natively expressed MutS-GFP. Upon scoring focus positions in the cell relative to the closest pole, we found that MutS800-GFP foci formed in the same subcellular positions as MutS-GFP foci (Fig. 6A). Moreover, following 2-AP challenge, MutS800-GFP colocalizes with DnaX-mCherry to almost the same extent as wild-type MutS-GFP (Fig 6B, 29.9±6.28% for MutS800-GFP and 35.5±5.6 for MutS-GFP: P=0.099). Finally, the vast majority of foci that do not colocalize with the replisome are replisome proximal (Fig. 6C). These results indicate that ectopically expressed MutS800-GFP also localizes to replisome proximal DNA for mismatch detection and initiation of repair; however, as shown previously, higher amounts of this mutant protein are required to achieve the same level of MMR as wild-type MutS (Table 2 and Fig. S7). Thus, MMR in B. subtilis is initiated and occurs predominantly in replisome proximal regions of DNA, and association of MutS with DnaN increases the efficiency of mismatch detection and repair by targeting MutS to nascent DNA. When DnaN-mediated mismatch detection is bypassed, as with MutS800 following overexpression, we found that MutS800 still forms foci in replisome proximal DNA in vivo (see discussion). Together, our results show that in order to initiate MMR, MutS must localize to the replisome or at least near the replisome in B. subtilis.
Figure 6. DnaN-independent focus formation of MutS800-GFP localizes to the same subcellular position as MutS-YFP.
(A) Position of MutS-GFP, ectopically expressed MutS800-GFP, and DnaX-GFP foci scored relative to cell length (n=125 cells for each group). (B) 29.9+6.3% (n=204) of the MutS800-GFP colocalizes with DnaX-mCherry. These results are not statistically different with p=0.099 when compared with MutS-YFP colocalization with DnaX-CFP 35.5+5.6 shown in Figure 3. The left most image is the negative image, MutS800-GFP, DnaX-mCherry, and a merge. The membrane is stained with TMA-DPH and is shown in blue, and the scale bar represents 4 μm. (C) We measured the inter-focal distance (IFD) between MutS-YFP and MutS800-GFP foci that failed to colocalize with DnaX-mCherry foci. No IFDs were measured less than 0.2 μm. We measured 94 MutS800-GFP, DnaX-mCherry and 105 MutS-YFP, DnaX-CFP focal pairs that failed to colocalize. Each bar represents the percentage of cells with IFD between the indicated distance.
Discussion
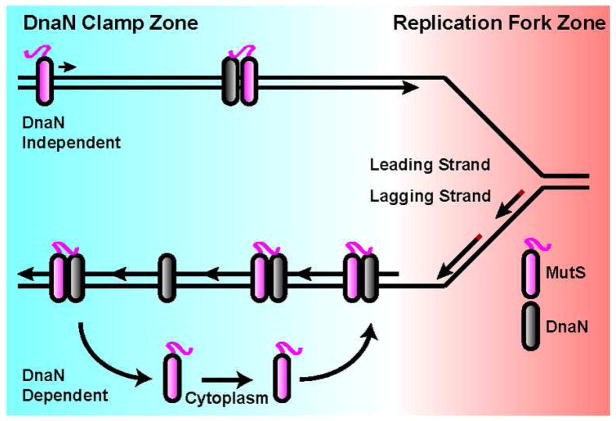
Three current models are used to explain the role of processivity clamps in mismatch repair: clamps stabilize MutS at a mismatch or increase mismatch binding affinity (Simmons et al., 2008, Flores-Rozas et al., 2000, Lau & Kolodner, 2003), clamps recruit MutS to sites of DNA replication (Hombauer et al., 2011a, Kleczkowska et al., 2001) or that clamps are required for DNA synthesis (Pluciennik et al., 2009). We have shown in this study that the B. subtilis processivity clamp, DnaN, facilitates ~90% of MMR and targets MutS to replisome proximal DNA prior to mismatch binding. A DnaN clamp zone forms in the wake of active replication forks (Su’etsugu & Errington), providing a platform for MutS to maintain a critical spatial and temporal relationship with the replisome (Fig. 7). We propose that DnaN-mediated targeting of MutS to nascent DNA allows for efficient mismatch detection by allowing for MutS to target newly formed errors.
Figure 7. Model for temporal coupling of MutS to DNA replication.
MutS relies on a DnaN clamp zone- to target MutS to nascent DNA. DnaN-dependent mode of mismatch detection represents 90% of repair. This figure is adapted from the clamp zone model (Su’etsugu & Errington).
MutS homologs spanning bacteria to humans exhibit the near-ubiquitous presence of a clamp-binding motif, suggesting that association with processivity clamps is important for MMR (Dalrymple et al., 2001). It has been known for decades that MutS is able to detect mismatches without accessory factors in vitro [e.g. (Prolla et al., 1994, Acharya et al., 1996, Su & Modrich, 1986)]. Nevertheless MutS800, which lacks the DnaN clamp-binding tether, is largely inactive for MMR while under control of its native promoter, retaining less than 10% activity. Interestingly, the same mutS800 allele restored MMR activity to 97% in vivo upon over expression. Our finding that MutS800 is capable of mismatch detection in the absence of DnaN in vivo led us to speculate that MutS800 could find mismatches and initiate repair at chromosomal locations distal to the replisome. To the contrary, we found that ectopically expressed MutS800-GFP formed foci at virtually identical subcellular positions as MutS-GFP, while MutSF30A800-GFP, which cannot bind mismatches or DnaN, was completely defective for focus formation. This result demonstrates that MutS800 binds mismatches in a replisome-proximal position like wild type MutS; however, a ~4-fold increase in concentration of the mutant protein is required to restore mismatch detection efficiency and compensate for the loss of interaction with DnaN. It was shown previously that B. subtilis MutL binds DnaN, and disruption of this contact causes complete loss of MMR in vivo (Pillon et al., 2010, Pillon et al., 2011). Overexpression of MutL mutants defective in binding DnaN fail to bypass the need for interaction with DnaN during MMR (Pillon et al., 2010, Pillon et al., 2011). We propose that MutS800 must identify mismatches in replisome proximal DNA to enable the downstream steps of repair, which include MutL recruitment and activation of its endonuclease activity. Current data suggests that these steps are dependent on interaction with DnaN, and may therefore require MutS to bind mismatches in the DnaN clamp zone in order to complete the downstream steps of MMR (Pillon et al., 2010, Pillon et al., 2011).
Consistent with our findings, in E. coli, when mutS800 is expressed from its native promoter, it confers an MMR defect, but is close to wild type for MMR when overexpressed from a plasmid [(Calmann et al., 2005, Lamers et al., 2000). Similarly, the equivalent mutant in P. aeruginosa, mutS798, complements a mutS deficient strain when overexpressed from a plasmid (Monti et al., 2012). Thus, in these systems, MutS is also capable of operating independent of DnaN, since both P. aeruginosa MutS798 and E. coli MutS800 fail to bind their cognate DnaN clamps (Calmann et al., 2005, Monti et al., 2012, Lopez de Saro et al., 2006). We propose that in these bacteria, when MutS is present at wild type levels, interaction with DnaN is important for targeting MMR to nascent DNA. When the protein is overexpressed, the requirement for DnaN is bypassed due to the increased likelihood of MutS directly binding mismatches in nascent DNA without DnaN association.
The model of a DnaN clamp zone that facilitates spatial and temporal coupling of mismatch detection with replication is an intriguing one, especially when considering the conservation of processivity clamp-binding motifs in MutS homologs (Dalrymple et al., 2001, Flores-Rozas et al., 2000). Consistent with the clamp zone model, fluorophore-labeled processivity clamps form foci in vivo in bacteria and eukaryotes [e.g. (Hombauer et al., 2011a, Kleczkowska et al., 2001)], suggesting that substantial local concentrations of clamps are present in organisms other than B. subtilis. The observation that a PCNA clamp zone may exist also agrees well with the higher proficiency of MMR on the lagging strand relative to the leading strand in S. cerevisiae (Pavlov et al., 2003). Other B. subtilis studies have shown that DnaN clamps are competent for protein recruitment to nascent DNA in vivo, as fluorophore-labeled peptides encoding a DnaN-binding motif are sufficient for forming replisome-localized foci (Simmons et al., 2008, Su’etsugu & Errington, 2011). A similar finding was reported for S. cerevisiae. When msh6 305-1242Δ, the unstructured region of msh6 that contains the PIP motif for binding PCNA (27-QSSLLSFF-34), was expressed from the native msh6 promoter this region was sufficient to form foci (Hombauer et al., 2011a). Furthermore, in human cell culture, overexpression of either MSH6 or MSH3 (which contain PIP motifs 4QSTLYSFF-11 and 21-QAVLSRFF-28, respectively) resulted in colocalization of MutSα and MutSβ with both PCNA and BrdU stain (Kleczkowska et al., 2001). Truncating the N-terminal 77 residues of MSH6 eliminated both MSH6 binding to PCNA in vitro and focus formation in vivo, indicating that localization of human MutS homologs to nascent DNA is dependent on interaction with PCNA (Kleczkowska et al., 2001). Collectively, these results support the hypothesis that a PCNA clamp zone present at nascent DNA facilitates mismatch detection in vivo. Our data further agrees well with the observation that MMR must occur concurrently with DNA replication in S. cerevisiae (Hombauer et al., 2011b). It was recently reported in S. cerevisiae, that PCNA-dependent MMR accounts for only 10–15% of MMR (Hombauer et al., 2011a), whereas DnaN-dependent MMR in B. subtilis accounts for ~90% of MMR by MutS. This is a notable difference in the orchestration of MMR between these organisms in vivo.
Another important finding from our study is that MutS localizes near the replisome independent of mismatch identification through the DnaN clamp zone. This conclusion is based on the observation that MutSF30A also forms foci that colocalize with the replisome, despite its inability to bind mismatches. Moreover, the MMR compromised dnaN5 mutant nearly abolishes MutSF30A-GFP localization in B. subtilis, indicating that MutSF30A foci are dependent on interaction with DnaN. Based on these data, we propose that MutS is coupled with the progressing replication fork prior to mismatch identification. An additional important finding is that after detecting a mismatch, MutS detaches from the replisome (DnaN) and remains at the mismatch site to conduct repair. This conclusion is based on the observation that MutS-YFP colocalizes less frequently with the replisome in 2-AP treated cells. Moreover, overexpression of MutS-YFP increases colocalization to the replisome in untreated cells, but there is little increase in colocalization following 2-AP treatment. Finally, the frequency of MutSF30A colocalization with the replisome is unchanged following 2-AP treatment, again, supporting the hypothesis that binding to mismatches causes release of MutS from the replisome. Overall, this study shows that MutS foci represent assemblies undergoing active repair of replication errors with two distinctive steps clearly identified: DnaN-coupled targeting of MutS to nascent DNA and release of MutS from the replisome following mismatch recognition.
Experimental Procedures
Bacteriological methods
All B. subtilis strains (Table S2) are isogenic derivatives of PY79 and grown according to standard procedures (Hardwood & Cutting, 1990). All oligos used in this study are listed in Table S3. To determine relative mutation rate, B. subtilis cells were grown at 37°C to OD600 of ~1.2, concentrated, and resuspended in 100 μL of 0.85% saline. A portion of the cells was serial diluted (10−6) and plated onto LB agar plates to determine total the viable count within the culture. The remaining resuspension was plated onto LB agar plates supplemented with 150 μg/mL rifampin. Mutation rate analysis was performed using MSS Maximum Likelihood Method with the 95% confidence interval, and statistical significance assessed using a one- or two-tailed T-test (Foster, 2006, Hall et al., 2009, Bolz et al., 2012).
Epifluorescence microscopy
Cells were prepared for live cell imaging essentially as described (Klocko et al., 2010, Simmons et al., 2009, Simmons et al., 2007). Briefly, strains were inoculated to a starting OD600 in S750 minimal media supplemented with 2% D-glucose. Cells were grown past 3 doublings to an OD600 of 0.4–0.5 and were split: one control culture and one culture challenged with 2-aminopurine to a final concentration of 600 μg/mL for one hour. Cell membranes were visualized using the fluorescent probe TMA-DPH at a working concentration of 10 μM. Replisome fusions were imaged with 0.5–1.0s exposures while MMR fusion proteins were imaged at 1.2–2.5s exposures. Scoring of cells as containing a MutS focus is outlined in Fig S7. The average cell focus encompasses ~4% of the cell area with an average intensity 2-fold greater than background. Colocalization and localization experiments were conducted as above except cells were grown in S750 minimal media supplemented with 1% L-arabinose. These conditions were used to produce cells with predominantly 1 DnaX-GFP focus per cell (avg. is 1.72 foci per cell with 39.1% of cells containing one focus) (Fig. S3). Image capture of both fusion proteins during colocalization experiments was performed in immediate succession and timed <2s of total capture to minimize any intracellular movement of either the MutS or replisomal fusions. For temperature release experiments, cells were grown and treated as above, but the prepared slide was incubated for 15 minutes at indicated temperature. Upon removal from the temperature-regulated chamber, slides were imaged for five minutes immediately upon removal.
Statistical analysis
Bar graphs are presented with error bars representing the 95% confidence interval, and statistical significance was determined using a one- or two-tailed T-test.
Immunoblotting
B. subtilis whole cell extracts were obtained basically as described (Rokop et al., 2004). Briefly, mid-exponential phase cultures were centrifuged and lysed by sonication (20 Hz), resuspended in lysis buffer [10 mM Tris-HCl (pH 7.0), 0.5 mM EDTA, 1 mM AEBSF, and 1X Protease Inhibitor Cocktail (Thermo Scientific)], and protein concentration was determined using Pierce BCA Protein Assay Kit (Thermo Scientific). Equal amounts of total protein were applied to each lane on a 4–15% gradient gel, and protein levels were probed with purified antisera: α-MutS (MI-1042), α-MutL (MI-1044), and α-DnaN (MI-1039). Immunoblots were developed as described previously (Simmons & Kaguni, 2003).
Immunodot blotting
Immunodot blotting was performed as previously described (Klocko et al., 2011, Walsh et al., 2012). Briefly, indicated proteins were immobilized onto a nitrocellulose membrane with the assistance of a Bio-dot microfiltration apparatus (Bio Rad). The membrane was incubated in blocking buffer (5% dry non-fat milk, 17.4 mM Na2HPO4, 2.6 mM NaH2PO4, 150 mM NaCl, 0.05% Tween-20) at 22°C for one hour. All subsequent washes and incubations took place in blocking buffer. After blocking, the membrane was incubated with 0.4 μM DnaN in blocking buffer for 3 hours at 22°C. The blot was subsequently washed three times and then incubated with affinity purified α-DnaN antisera overnight at 4°C. The blot was removed from primary antibody (MI 1038) and washed three times at 22°C and placed in secondary antisera (1:2000 α-Rabbit) for 2 hours at 22°C. The blot was washed 3 more times, followed by a wash in PBS (17.4 mM Na2HPO4, 2.6 mM NaH2PO4, 150 mM NaCl, 0.05% Tween-20) to remove excess milk proteins. The membrane was developed with chemiluminescence (Super-Signal Altra, Pierce) and expose to film as described (Klocko et al., 2011).
B. subtilis MutS–DNA interactions at equilibrium
Procedures used to purify untagged MutS and MutSF30A are detailed within the supplemental methods.
The mismatched DNA substrates for the MutS-DNA binding assay were prepared by annealing 37 nt 2-aminopurine (2-AP) labeled +T strand (5′ - TAA AGG AAG TCG TCT AT2-Ap TAT GGT ATG ACT AAG TGT A - 3′) with 36 nt (5′ - T ACA CTT AGT CAT ACC AT TAT AGA CGA CTT CCT TTA - 3′) or with 37 nt (5′ - T ACA CTT AGT CAT ACC ATG TAT AGA CGA CTT CCT TTA - 3′) strands to yield 2-AP(+T) and 2-AP(GT) duplexes, respectively. The matched substrate, 2-AP(GC) was prepared by annealing 37 nt 2-AP labeled strand (5′ - TAA AGG AAG TCG TCT AT2Ap CAT GGT ATG ACT AAG TGT A - 3′) with a 37 nt strand (5′ - T ACA CTT AGT CAT ACC ATG TAT AGA CGA CTT CCT TTA - 3′). The strands were heated to 95 °C, followed by slowly cooling to room temperature to obtain annealed duplex DNAs.
DNA binding was measured on a FluoroMax-3 fluorimeter (Jobin-Yvon Horiba Group; Edison, NJ). Titrations of 0.02 μM 2-AP(+T), 2-AP(GC) and 2-AP(GT) duplex DNAs with 0 – 0.4 μM MutS were performed in 3 ml quartz cuvettes in DNA binding buffer (20 mM Tris-HCl, pH 7.6, 50 mM NaCl, 5 mM MgCl2) at 25 °C. MutS was added incrementally to the sample, and fluorescence intensity was measured after mixing for 30 seconds (λEX = 315 nm and λEM = 375 nm). The data were corrected for intrinsic MutS fluorescence by subtracting data from parallel experiments with unlabeled DNA. Fluorescence intensity was plotted versus MutS concentration and the apparent dissociation constant (KD) for the interaction was obtained by fitting the data to a quadratic equation:
where D·M is the fraction of MutS•DNA, F0 is 2-AP(+T) fluorescence in the absence of protein and Fmax is maximal fluorescence, and Dt and Mt are total molar concentrations of DNA and MutS, respectively. The data were fit by non-linear regression using KaleidaGraph (Synergy Software)
Supplementary Material
Acknowledgments
We thank Dr. David Rudner (Harvard Medical School) and Dr. Heath Murray (Newcastle University) for strains. We would like to thank members of the Simmons lab for critical reading of this manuscript and the comments from two anonymous referees. This work was supported by grants from the Wendy Will Case Cancer Fund to L.A.S. and grants from the National Science Foundation to L.A.S. (MCB 1050948) and M.M.H. (MCB 1022203)
References
- Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Chi NW, Kolodner R. The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and insertions. Genes & development. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- Berkmen MB, Grossman AD. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol Microbiol. 2006;62:57–71. doi: 10.1111/j.1365-2958.2006.05356.x. [DOI] [PubMed] [Google Scholar]
- Bolz NJ, Lenhart JS, Weindorf SC, Simmons LA. Residues in the N-terminal domain of MutL required for mismatch repair in Bacillus subtilis. Journal of Bacteriology. 2012;194:5361–5367. doi: 10.1128/JB.01142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J, Tran PT, Liskay RM, Alani E. Analysis of yeast MSH2-MSH6 suggests that the initiation of mismatch repair can be separated into discrete steps. J Mol Biol. 2000;302:327–338. doi: 10.1006/jmbi.2000.4081. [DOI] [PubMed] [Google Scholar]
- Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- Calmann MA, Nowosielska A, Marinus MG. The MutS C terminus is essential for mismatch repair activity in vivo. J Bacteriol. 2005;187:6577–6579. doi: 10.1128/JB.187.18.6577-6579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I, O’Neill AJ, Miller K. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist Updat. 2003;6:137–145. doi: 10.1016/s1368-7646(03)00041-4. [DOI] [PubMed] [Google Scholar]
- Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. The Journal of Biological Chemistry. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- Cooper LA, Simmons LA, Mobley HL. Involvement of Mismatch Repair in the Reciprocal Control of Motility and Adherence of Uropathogenic Escherichia coli. Infect Immun. 2012;80:1969–1979. doi: 10.1128/IAI.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox EC, Degnen GE, Scheppe ML. Mutator gene studies in Escherichia coli: the mutS gene. Genetics. 1972;72:551–567. doi: 10.1093/genetics/72.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Meyer-Gauen G, Lyons-Weiler J, Hays JB. Evolutionary origin, diversification and specialization of eukaryotic MutS homolog mismatch repair proteins. Nucleic Acids Res. 2000;28:463–471. doi: 10.1093/nar/28.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci USA. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Dupes NM, Gerstenfeld TA, Simmons LA, Mekalanos JJ. DNA damage and reactive nitrogen species are barriers to Vibrio cholerae colonization of the infant mouse intestine. PLoS Pathog. 2011;7:e1001295. doi: 10.1371/journal.ppat.1001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E, Bonacorsi S, Giraud A, Duriez P, Hilali F, Amorin C, Bingen E, Andremont A, Picard B, Taddei F, Matic I. High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J Bacteriol. 2002;184:605–609. doi: 10.1128/JB.184.2.605-609.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupes NM, Walsh BW, Klocko AD, Lenhart JS, Peterson HL, Gessert DA, Pavlick CE, Simmons LA. Mutations in the Bacillus subtilis beta clamp that separate its roles in DNA replication from mismatch repair. J Bacteriol. 2010;192:3452–3463. doi: 10.1128/JB.01435-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MRS, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O’Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginetti F, Perego M, Albertini AM, Galizzi A. Bacillus subtilis mutS mutL operon: identification, nucleotide sequence and mutagenesis. Microbiology. 1996;142(Pt 8):2021–2029. doi: 10.1099/13500872-142-8-2021. [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Gu L, Hong Y, McCulloch S, Watanabe H, Li GM. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Research. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken Y, Sung P, Prakash L, Prakash S. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Current biology: CB. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SR, Liu B, Parsons RE, Papadopoulos N, Jen J, Powell SM, Krush AJ, Berk T, Cohen Z, Tetu B, et al. The molecular basis of Turcot’s syndrome. N Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- Hardwood CR, Cutting SM. Molecular Biological Methods for Bacillus. John Wiley & Sons; Chichester: 1990. [Google Scholar]
- Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011a;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 2011b;334:1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Hearst JE, Alberts BM. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. The Journal of Biological Chemistry. 1981;256:4087–4094. [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Jackson D, Wang X, Rudner DZ. Spatio-temporal organization of replication in bacteria and eukaryotes (nucleoids and nuclei) Cold Spring Harbor perspectives in biology. 2012:4. doi: 10.1101/cshperspect.a010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-Palmer E, Hingorani MM. The effects of nucleotides on MutS-DNA binding kinetics clarify the role of MutS ATPase activity in mismatch repair. J Mol Biol. 2007;366:1087–1098. doi: 10.1016/j.jmb.2006.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeruzalmi D, O’Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocko AD, Crafton KM, Walsh BW, Lenhart JS, Simmons LA. Imaging mismatch repair and cellular responses to DNA damage in Bacillus subtilis. J Vis Exp. 2010:1–4. doi: 10.3791/1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocko AD, Schroeder JW, Walsh BW, Lenhart JS, Evans ML, Simmons LA. Mismatch repair causes the dynamic release of an essential DNA polymerase from the replication fork. Mol Microbiol. 2011;82:648–663. doi: 10.1111/j.1365-2958.2011.07841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the b subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Krishna TSR, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. DNA replication fidelity. J Biol Chem. 1992;267:18251–18254. [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- Larrea AA, Lujan SA, Kunkel TA. SnapShot: DNA mismatch repair. Cell. 2010;141:730 e731. doi: 10.1016/j.cell.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Lau PJ, Kolodner RD. Transfer of the MSH2.MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. The Journal of Biological Chemistry. 2003;278:14–17. doi: 10.1074/jbc.C200627200. [DOI] [PubMed] [Google Scholar]
- Lee SD, Alani E. Analysis of interactions between mismatch repair initiation factors and the replication processivity factor PCNA. J Mol Biol. 2006;355:175–184. doi: 10.1016/j.jmb.2005.10.059. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- Lenhart JS, Schroeder JW, Walsh BW, Simmons LA. DNA Repair and Genome Maintenance in Bacillus subtilis. Microbiology and molecular biology reviews: MMBR. 2012;76:530–564. doi: 10.1128/MMBR.05020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Saro FJ, Marinus MG, Modrich P, O’Donnell M. The beta sliding clamp binds to multiple sites within MutL and MutS. J Biol Chem. 2006;281:14340–14349. doi: 10.1074/jbc.M601264200. [DOI] [PubMed] [Google Scholar]
- Malkov VA, Biswas I, Camerini-Otero RD, Hsieh P. Photocross-linking of the NH2-terminal region of Taq MutS protein to the major groove of a heteroduplex DNA. J Biol Chem. 1997;272:23811–23817. doi: 10.1074/jbc.272.38.23811. [DOI] [PubMed] [Google Scholar]
- Matsumiya S, Ishino Y, Morikawa K. Crystal structure of an archaeal DNA sliding clamp: proliferating cell nuclear antigen from Pyrococcus furiosus. Protein science: a publication of the Protein Society. 2001;10:17–23. doi: 10.1110/ps.36401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. Journal of Bacteriology. 2008;190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migocki MD, Lewis PJ, Wake RG, Harry EJ. The midcell replication factory in Bacillus subtilis is highly mobile: implications for coordinating chromosome replication with other cell cycle events. Mol Microbiol. 2004;54:452–463. doi: 10.1111/j.1365-2958.2004.04267.x. [DOI] [PubMed] [Google Scholar]
- Monti MR, Miguel V, Borgogno MV, Argarana CE. Functional analysis of the interaction between the mismatch repair protein MutS and the replication processivity factor beta clamp in Pseudomonas aeruginosa. DNA Repair (Amst) 2012;11:463–469. doi: 10.1016/j.dnarep.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Nystrom-Lahti M, Perrera C, Raschle M, Panyushkina-Seiler E, Marra G, Curci A, Quaresima B, Costanzo F, D’Urso M, Venuta S, Jiricny J. Functional analysis of MLH1 mutations linked to hereditary nonpolyposis colon cancer. Genes Chromosomes Cancer. 2002;33:160–167. [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- Oliver A, Mena A. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect. 2010;16:798–808. doi: 10.1111/j.1469-0691.2010.03250.x. [DOI] [PubMed] [Google Scholar]
- Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Current biology: CB. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, I, Mian M, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Lynch syndrome genes. Fam Cancer. 2005;4:227–232. doi: 10.1007/s10689-004-7993-0. [DOI] [PubMed] [Google Scholar]
- Pillon MC, Lorenowicz JJ, Uckelmann M, Klocko AD, Mitchell RR, Chung YS, Modrich P, Walker GC, Simmons LA, Friedhoff P, Guarne A. Structure of the endonuclease domain of MutL: unlicensed to cut. Mol Cell. 2010;39:145–151. doi: 10.1016/j.molcel.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon MC, Miller JH, Guarne A. The endonuclease domain of MutL interacts with the beta sliding clamp. DNA Repair (Amst) 2011;10:87–93. doi: 10.1016/j.dnarep.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluciennik A, Burdett V, Lukianova O, O’Donnell M, Modrich P. Involvement of the beta clamp in methyl-directed mismatch repair in vitro. J Biol Chem. 2009;284:32782–32791. doi: 10.1074/jbc.M109.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc Natl Acad Sci U S A. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla TA, Pang Q, Alani E, Kolodner RD, Liskay RM. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- Prudhomme M, Martin B, Mejean V, Claverys JP. Nucleotide sequence of the Streptococcus pneumoniae hexB mismatch repair gene: Homology of HexB to MutL of Salmonella typhimurium and to PMS1 of Saccharomyces cerevisiae. J Bacteriol. 1989;171:5332–5338. doi: 10.1128/jb.171.10.5332-5338.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier AL, Malbruny B, Laurans M, Brouard J, Duhamel JF, Leclercq R. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. The Journal of infectious diseases. 2003;187:1709–1716. doi: 10.1086/374937. [DOI] [PubMed] [Google Scholar]
- Rokop ME, Auchtung JM, Grossman AD. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol Microbiol. 2004;52:1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- Roman F, Canton R, Perez-Vazquez M, Baquero F, Campos J. Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. Journal of clinical microbiology. 2004;42:1450–1459. doi: 10.1128/JCM.42.4.1450-1459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjanwala B, Ganesan AT. Genetic structure and domains of DNA polymerase III of Bacillus subtilis. Mol Gen Genet. 1991;226:467–472. doi: 10.1007/BF00260660. [DOI] [PubMed] [Google Scholar]
- Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. The Journal of Biological Chemistry. 1993;268:23762–23765. [PubMed] [Google Scholar]
- Schofield MJ, Brownewell FE, Nayak S, Du C, Kool ET, Hsieh P. The Phe-X-Glu DNA binding motif of MutS. The role of hydrogen bonding in mismatch recognition. J Biol Chem. 2001a;276:45505–45508. doi: 10.1074/jbc.C100449200. [DOI] [PubMed] [Google Scholar]
- Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- Schofield MJ, Nayak S, Scott TH, Du C, Hsieh P. Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J Biol Chem. 2001b;276:28291–28299. doi: 10.1074/jbc.M103148200. [DOI] [PubMed] [Google Scholar]
- Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Molecular Cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Davies BW, Grossman AD, Walker GC. Beta clamp directs localization of mismatch repair in Bacillus subtilis. Mol Cell. 2008;29:291–301. doi: 10.1016/j.molcel.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Goranov AI, Kobayashi H, Davies BW, Yuan DS, Grossman AD, Walker GC. Comparison of responses to double-strand breaks between Escherichia coli and Bacillus subtilis reveals different requirements for SOS induction. J Bacteriol. 2009;191:1152–1161. doi: 10.1128/JB.01292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Grossman AD, Walker GC. Replication is required for the RecA localization response to DNA damage in Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:1360–1365. doi: 10.1073/pnas.0607123104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Kaguni JM. The DnaAcos allele of Escherichia coli: hyperactive initiation is caused by substitution of A184V and Y271H, resulting in defective ATP binding and aberrant DNA replication control. Mol Microbiol. 2003;47:755–765. doi: 10.1046/j.1365-2958.2003.03333.x. [DOI] [PubMed] [Google Scholar]
- Smith BT, Grossman AD, Walker GC. Visualization of mismatch repair in bacterial cells. Mol Cell. 2001;8:1197–1206. doi: 10.1016/s1097-2765(01)00402-6. [DOI] [PubMed] [Google Scholar]
- Stukenberg PT, Studwell-Vaughn PS, O’Donnell M. Mechanism of the sliding clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- Su SS, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su’etsugu M, Errington J. The replicase sliding clamp dynamically accumulates behind progressing replication forks in Bacillus subtilis cells. Mol Cell. 2011;41:720–732. doi: 10.1016/j.molcel.2011.02.024. [DOI] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Graumann PL, Lin DC, Grossman AD, Losick R. Chromosome arrangement within a bacterium. Current biology: CB. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- Tessmer I, Yang Y, Zhai J, Du C, Hsieh P, Hingorani MM, Erie DA. Mechanism of MutS searching for DNA mismatches and signaling repair. J Biol Chem. 2008;283:36646–36654. doi: 10.1074/jbc.M805712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrientes MC, Baquero MR, Sanchez MB, Valdezate S, Escudero E, Berg G, Canton R, Baquero F, Galan JC, Martinez JL. Polymorphic mutation frequencies of clinical and environmental Stenotrophomonas maltophilia populations. Applied and environmental microbiology. 2010;76:1746–1758. doi: 10.1128/AEM.02817-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- Walsh BW, Lenhart JS, Schroeder JW, Simmons LA. Far Western blotting as a rapid and efficient method for detecting interactions between DNA replication and DNA repair proteins. Methods in molecular biology. 2012;922:161–168. doi: 10.1007/978-1-62703-032-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ME, Jr, Burns JL, Smith AL. Hypermutable Haemophilus influenzae with mutations in mutS are found in cystic fibrosis sputum. Microbiology. 2004;150:2947–2958. doi: 10.1099/mic.0.27230-0. [DOI] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- Zhai J, Hingorani MM. Saccharomyces cerevisiae Msh2-Msh6 DNA binding kinetics reveal a mechanism of targeting sites for DNA mismatch repair. Proc Natl Acad Sci U S A. 2010;107:680–685. doi: 10.1073/pnas.0908302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.