Dear Editor,
In 2015, a Zika virus (ZIKV) outbreak began in South and Central America and in the Caribbean, and has since spread to both North America and Asia.1 It has been revealed that ZIKV is the primary cause of severe neurological pathologies, such as neonatal microcephaly and Guillain–Barré syndrome.2 ZIKV infection can also damage mouse testes, posing a potential threat to the mammalian reproductive system.3 Similar to the dengue virus (DENV) and the West Nile virus (WNV), ZIKV is a mosquito-borne flavivirus containing a single-stranded positive-sense RNA genome, which encodes three structural proteins (C, prM/M, and E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5).2,4,5 The mature form of the flavivirus C protein is ~12 kDa, and it plays a critical role in encapsulation of the RNA genome.2 In addition, the C protein interacts with intracellular lipid droplets for viral particle formation2 and inhibits host RNA silencing to suppress the immune response.6 The multiple functions of the flavivirus C protein in the viral life cycle make it an attractive target for drug development.
We set out to conduct structural studies of the ZIKV C protein in order to understand the role of the flavivirus C protein during encapsulation of the viral RNA genome. The flavivirus C protein is a highly basic macromolecule with a disordered N-terminus,7 which has caused problems in structural studies. Only an NMR structure of the DENV2 C protein (residues 21–100)8 and a 3.2-Å crystal structure of the WNV C protein (residues 24–98) have been previously reported.9 To overcome the poor yield and solubility problems of the ZIKV C protein, the core domain (residues 24–98) and a longer fragment (residues 2–104) (Supplementary information, Figure S1) from the ZIKV C protein were fused to the maltose-binding protein (MBP, residues 1–370), resulting in MBP-ZIKVCC (ZIKVCC) and MBP-ZIKVCL (ZIKVCL), respectively. Both fusion proteins are very soluble, and eluted as monodispersed peaks from a size exclusion column (Supplementary information, Figure S2) with the elution volumes matching those expected for the dimers.
ZIKVCC successfully produced two different crystal forms. Both crystals belong to the P21 space group, but the unit cell parameters are different. We were able to solve the two crystal structures of ZIKVCC at 2.0 and 2.9 Å, respectively. In the high-resolution structure, there are two ZIKVCC molecules in an asymmetric unit (ASU), forming a dimer with twofold non-crystallographic symmetry. These two fusion proteins dimerize solely via the capsid while the MBPs are on the outside of the dimer interface, participating in crystal packing (Supplementary information, Figure S3). In contrast, the low-resolution structure consists of two identical dimers in an ASU; however, superimposition of the dimers from these two structures individually shows that although the orientation of the MBP molecules is different, the central ZIKVCC molecules are identical (Supplementary information, Figure S4). These data indicate that ZIKVCC exists as a biological dimer in structure, and its oligomerization state is similar to that of the DENV C protein.7 For clarity, only the dimer of the ZIKVCC at high resolution is discussed in this study.
Within the ZIKVCC dimer, each protomer adopts the all-alpha helical conformation, from N- to C-termini, designated α1, α2, α3, and α4 (Fig. 1a and Supplementary information, Figure S5). These helices are connected by the loops between them. The root-mean-square deviations between the ZIKV C protomer and those of the WNV and DENV are 1.6 and 1.8 Å, respectively (for Cα; PDB IDs: 1SFK and 1R6R). The helical parts (α2, α3, and α4) aligned well, and the most distinct feature between these flavivirus C protomers lies in their N-termini (Supplementary information, Figure S6). The N-terminus of the ZIKVCC protomer is composed of a long, structured loop and a short helix (α1), whereas the N-termini of the WNV and DENV C protomers only consist of a much longer α1.8,9 An overlay of these structures shows that the N-termini of the ZIKV and WNV C protomers share the same orientation, but the N-terminus of the DENV C protomer adopts a completely different orientation (Supplementary information, Figure S6A and B). The diverse N-terminal structures of the flavivirus C proteins indicate the inherently flexible nature of this region, which might be able to adopt different conformations for various physiological processes.
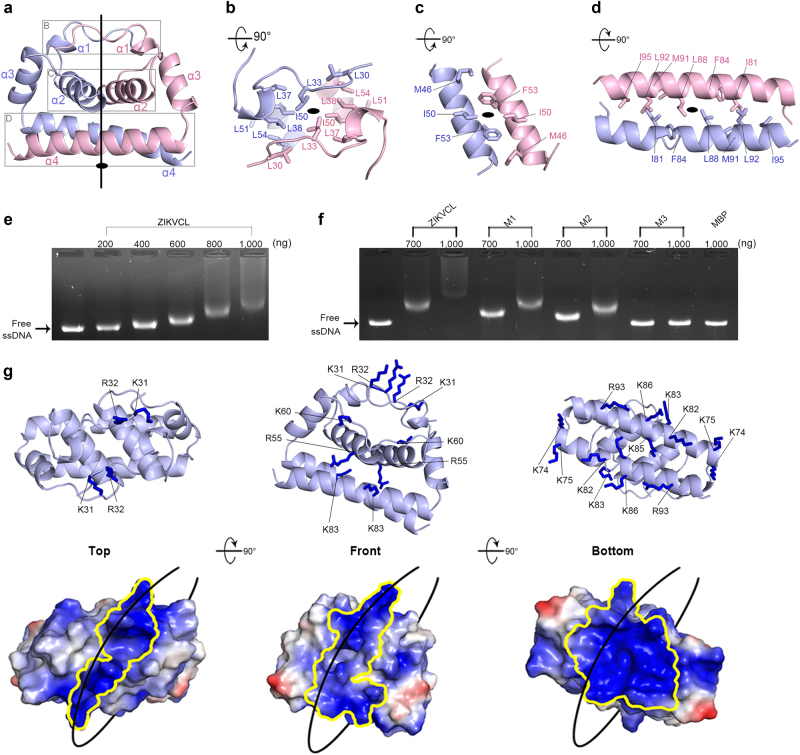
Fig. 1.
The overall structure of the ZIKV C protein shows a positively charged path on the dimeric ZIKV C protein surface that is essential for nucleic acid binding. a Ribbon diagram of the ZIKV C protein dimer colored with light blue (Protomer A) and light pink (Protomer B), respectively. b–d Close-up views of the dimer interface. Hydrophobic residues involved in dimerization include the N-terminal interface (b), the α2A–α2B interface (c), and the α4A–α4B interface (d). e EMSA assay of ZIKVCL binding to ssDNA in a dose-dependent manner. Lane 1: 100 ng free ssDNA; lanes 2–6: ZIKVCL protein of various concentrations was incubated with 100 ng free ssDNA. f EMSA assay of ZIKVCL mutants binding to ssDNA. Lane 1: 100 ng free ssDNA; lanes 2–3: WT ZIKVCL; lanes 4–5: M1; lanes 6–7: M2; lanes 8–9: M3; lane 10: MBP as a negative control. g A ribbon diagram labeled with positive residues on the surface of the ZIKV C protein dimer in different views and its corresponding electrostatic potential surface of the ZIKV C protein dimer. An uninterrupted positively charged path was identified as wrapping around the entire molecule
The two ZIKV C protomers associate into a dimer with a buried area of 2270 Å2, which is much larger than those for DENV C protein (1650 Å2)8 and WNV C protein (1530 Å2).9 The dimer contact interface can be roughly divided into three layers: N-terminusA-N-terminusB (layer I), α2A–α2B (layer II), and α4A–α4B (layer III). In layer I, it is notable that the structured N-terminal loop connected to α1 contributes to strong interactions between the two protomers, which are also stabilized by a core of hydrophobic residues (L30, L33, L37, L38, I50, L51, and L54 of both protomers; Fig. 1b). This feature explains why the ZIKV C dimer has a larger contact interface than the DENV and WNV C proteins (Fig. 1b and Supplementary information, Figure S6C). In layer II, α2A and α2B are aligned in an antiparallel style. Along the helix–helix interface, the hydrophobic residues M46, I50, and F53 (Fig. 1c), which are well conserved among flaviviruses, play a major role in the helix–helix interaction. In layer III, α4A and α4B also form an antiparallel helix pair. The conserved hydrophobic residues (I81, F84, L88, M91, L92, and I95) are responsible for the hydrophobic interactions (Fig. 1d). The conserved residues in the dimerization interface have been shown to be critical for its physiological role in flaviviruses.2 Mutation of these residues (F56K and F84K) could disrupt the protein folding, resulting in a soluble aggregate (Supplementary information, Figure S7A).
It has been reported that the flavivirus C protein has a high density of positive charge, and this property is critical for its affinity to both RNA and DNA.10,11 We used an electrophoretic mobility shift assay (EMSA) to test whether ZIKVCL could bind to M13mp18 single-stranded DNA (ssDNA). The EMSA assay showed that the ZIKV C protein could bind to the ssDNA in a dose-dependent manner (Fig. 1e). Since the flavivirus capsid core is still capable of encapsulating the RNA genome to produce the virions compared with the full-length protein,2 we can utilize our ZIKVCC structure as a model to study its function of packaging the RNA genome. In the crystal structure, we observe a continuous, positively charged area covering the major portion of the molecular surface. Starting from the bottom of the dimeric molecule, a cluster of basic residues (K74, K75, K82, K83, K85, K86, and R93) on α4A and α4B (Fig. 1g), which are facing the solvent, form a large positively charged surface. This positively charged area further extends to the front and the top (K31, R32, R55, and K60) of the homodimer, resulting in an uninterrupted path that wraps around the entire molecule (Fig. 1g). To find out whether these positively charged residues are involved in nucleic acid binding, we tested multiple mutants for ssDNA binding using the EMSA assay (Supplementary information, Figure S7B). As shown in Fig. 1f, M1 (mutant 1, K74E/K75E/K82E) mildly reduced the binding affinity to nucleic acids compared with the WT ZIKVCL, indicating that the positive residues at the bottom are involved in nucleic acid binding. M2 (mutant 2, K31E/R32E/K74E/K75E/K82E), which includes additional positive residues on the top of the molecule, lost much of its ability to bind to the nucleic acids, indicating that the positive residues on the top also participate in binding to the nucleic acids. Interestingly, M3 (mutant 3, K31E/R32E/R55E/K60E/K74E/K75E/K82E/K83E/K85E/K86E/R93E), which includes more positive residues on the front of the molecule, almost completely disrupted the interaction. These results indicate that the positively charged path that wraps around the ZIKV C molecule plays a pivotal role in nucleic acid binding. Moreover, K82, K83, K85, K86, and R93 on the bottom and K31 and R32 on the top of the C molecule have shown to be necessary for RNA binding in the Kunjin virus (a subtype of WNV).10
The ZIKV virion is essentially a condensed RNA-nucleocapsid kernel coated with an icosahedral shell composed of M and E proteins. The current model of the flavivirus virion8,9 suggests that the C protein interacts with the envelope proteins and the single-stranded RNA genome. However, mature flavivirus virions do not seem to have a defined shell composed of C proteins because the proteins could not be clearly identified by electron microscopy.12 Two hypotheses have been proposed: either internal symmetry of the nucleocapsid core is incompatible with the icosahedral shell, or their viral nucleocapsid cores might be disordered. The continuous distribution of the positive charge observed in our atomic model supports the latter. We propose that the single-stranded RNA genome of ZIKV likely wraps around individual C dimers to form a condensed but disordered “beads on a string” ribonucleocomplex structure as the virus kernel (Supplementary information, Figure S8). The viral RNA is typically in close contact with the C protein, and the interactions are predominantly between the sugar-phosphate backbone of the RNA and the positively charged side chains of the amino acids in the C protein, which is independent of the nucleotide sequence.
Electronic supplementary material
Acknowledgements
We thank Wen Cui and Hongliang Tian for data collection at beamline BL17U1 and BL18U1 of the Shanghai Synchrotron Radiation Facility (SSRF); Jun Li for data collection at beamline I04 of DIAMOND Synchrotron Radiation Facility (Britain); Xia Chen for technical support; Michael Rossmann, Lei Sun, and Erin Weber for discussion and advice. This work was supported by the National Key Basic Research Program of China (973 program; 2015CB859800), the National Natural Science Foundation of China (81772204 and 31528006), and the National Key Research Program of China (2016YFD0500300).
Competing interests
The authors declare no competing financial interests.
Footnotes
Ting Li, Qi Zhao, Xiaoyun Yang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0007-9.
References
- 1.Bogoch II, et al. Anticipating the international spread of Zika virus from Brazil. Lancet. 2016;387:335–336. doi: 10.1016/S0140-6736(16)00080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliveira ER, et al. The flavivirus capsid protein: Structure, function and perspectives towards drug design. Virus Res. 2017;227:115–123. doi: 10.1016/j.virusres.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Govero J, et al. Zika virus infection damages the testes in mice. Nature. 2016;540:438–442. doi: 10.1038/nature20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, et al. Mechanisms of activation and inhibition of Zika virus NS2B-NS3 protease. Cell Res. 2016;26:1260–1263. doi: 10.1038/cr.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian H, et al. The crystal structure of Zika virus helicase: basis for antiviral drug design. Protein Cell. 2016;7:450–454. doi: 10.1007/s13238-016-0275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuel GH, et al. Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proc. Natl Acad. Sci. USA. 2016;113:13863–13868. doi: 10.1073/pnas.1600544113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones CT, et al. Flavivirus Capsid Is a Dimeric Alpha-Helical Protein. J. Virol. 2003;77:7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma L, et al. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl Acad. Sci. USA. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dokland T, et al. West Nile virus core protein; tetramer structure and ribbon formation. Structure. 2004;12:1157–1163. doi: 10.1016/j.str.2004.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khromykh AA, Westaway EG. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 1996;141:685–699. doi: 10.1007/BF01718326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng HN, et al. DNA-binding property of recombinant capsid protein of Japanese encephalitis virus. Virus Genes. 2007;35:483–488. doi: 10.1007/s11262-006-0075-7. [DOI] [PubMed] [Google Scholar]
- 12.Sirohi D, et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.