Abstract
The stolbur phytoplasma vector Hyalesthes obsoletus is generally considered as a polyphagous species associated with numerous wild and cultivated plants. However, recent research in southeastern Europe, the distribution centre of H. obsoletus and the area of most stolbur-inflicted crop diseases, points toward specific host-plant associations of the vector, indicating specific vector-based transmission routes. Here, we study the specificity of populations associated with four host-plants using mitochondrial and nuclear genetic markers, and we evaluate the evolution of host-shifts in H. obsoletus. Host-plant use was confirmed for Convolvulus arvensis, Urtica dioica, Vitex agnus-castus and Crepis foetida. Mitochondrial genetic analysis showed sympatric occurrence of three phylogenetic lineages that were ecologically delineated by host-plant preference, but were morphologically inseparable. Nuclear data supported the existence of three genetic groups (Evanno’s ΔK(3) = 803.72) with average genetic membership probabilities > 90%. While populations associated with C. arvensis and U. dioica form a homogenous group, populations affiliated with V. agnus-castus and C. foetida constitute two independent plant-associated lineages. The geographical signal permeating the surveyed populations indicated complex diversification processes associated with host-plant selection and likely derived from post-glacial refugia in the eastern Mediterranean. This study provides evidence for cryptic species diversification within H. obsoletus sensu lato: i) consistent mitochondrial differentiation (1.1–1.5%) among host-associated populations in syntopy and in geographically distant areas, ii) nuclear genetic variance supporting mitochondrial data, and iii) average mitochondrial genetic distances among host-associated meta-populations are comparable to the most closely related, morphologically distinguishable species, i.e., Hyalesthes thracicus (2.1–3.3%).
Introduction
Knowledge of changes in insect genetic structure and the possibility of cryptic divergence are particularly important for pest populations because management strategies must be adapted to the ecological diversity of the pest [1–6]. Several planthopper species of the family Cixiidae (Hemiptera: Auchenorrhyncha: Fulgoromorpha) are vectors of plant-pathogenic phytoplasma bacteria whose host-plant choice determines the epidemiological pathways of the transmitted phytoplasma diseases [7–13]. The most promising method for revealing both the vector’s area of distribution and for predicting the disease impact on the targeted crop in the surrounding natural habitat is an assessment of the plants which simultaneously act as preferred host-plants of the vector and as the pathogen’s inoculum source, i.e., a dual host-plant [7, 14, 15]. Conversely, if host-plant preferences change and cause alterations in the vector’s feeding behavior (e.g. mono-, oligo- or polyphagous), such changes may initiate host-plant specialization and drive populations through successive stages of ambiguous, taxonomically indistinguishable but ecologically adapted populations. Such populations known as "host races", "ecological races" or "biotypes" ultimately lead toward true species status [16–18]. Of primary importance for elucidating the distribution, dispersal, impact and epidemiology of vector-transmitted plant diseases is knowledge of the evolutionary relationship between the vector and its host-plant(s). In a polyphagous vector, this is reflected through levels of host-plant association(s) and the extent of genetic segregation, i.e., specialization of the species in question.
Ever since Hyalesthes obsoletus Signoret, 1865 (Hemiptera: Cixiidae) rose to prominence as an important pest vector, inducing biomass losses exceeding agroeconomic thresholds, its role in the dissemination of stolbur phytoplasma ('Candidatus Phytoplasma solani', 16Sr XII-A subgroup) [19] has been thoroughly investigated [11, 13, 15, 20–25]. The frequent imprecise use of the term "host-plant"—referring to the multitude of wild and cultivated plants on which H. obsoletus feeds, developments, reproduces or even on which it is accidentally hosted—has led to a general acceptance of H. obsoletus being highly polyphagous (general overview given in S1 Table) but confuses which plants are utilized for larval development. The most common and widespread host plants are Convolvulus arvensis (field bindweed) and Urtica dioica (stinging nettle), both recognized as dual host-plants of primary importance in stolbur phytoplasma epidemiology [14, 25–27]. Each plant harbors a specific stolbur phytoplasma strain, tuf-a (type I) in U. dioica and tuf-b (type II) in C. arvensis, which lead to two independent plant-based epidemiologies [14]. H. obsoletus populations affiliated with these plants are well studied in central Europe [14, 25, 28], especially along the northernmost border of the species distribution range [7, 15, 29]. Recent research has also confirmed these associations and its epidemiological importance in southeastern Europe [13, 30].
Following the experimental verification of H. obsoletus as a vector of stolbur [23], research on its behavior and host-plant associations has been done mainly in central and western Europe. However, knowledge about (larval developmental) host-plants in the eastern Mediterranean, the species distribution center [31] remains scarce and neglected. For example, affiliation with Vitex agnus-castus as a common host-plant in the European Mediterranean has been neglected for a full century after being reported by Horváth [32], even after its confirmation as a true host-plant in Israel [33]. One reason for the neglect is founded on research focusing on agricultural weeds which act as vectors’ host-plant and inoculum source, rather than on the already known preferred plants from natural ecosystems [31]. Recent experimental verification of H. obsoletus associated with V. agnus-castus being a vector in the Montenegrin littoral, and the elucidation of stolbur epidemiological routes commencing from different dual host-plants, has encouraged further research to understand the genetic relationships among diverse host-associated populations [13, 34].
Sympatric co-occurrence of populations feeding on (a combination of) the three aforementioned host-plants is often observed, even in strict syntopy [7, 13, 30]. Unlike sympatry in the Montenegrin coastal zone where triple associations are recorded [13], Sharon et al. [33] documented a predominant use of V. agnus-castus in the eastern Mediterranean where adult presence on co-occurring U. dioica and C. arvensis was either lacking or very rare, respectively. Orenstein et al. [35] explained the absence as a phenological mismatch in relation to the insect’s life cycle. A peculiarity that further contributes to the difficulty of understanding H. obsoletus' host-plant associations across a wide distribution is the fact that it represents only one of the seven species constituting the Hyalesthes obsoletus species group: identical outer morphology, similarities of male genital structures and sympatric occurrences as well as the circum-Mediterranean distribution range of H. obsoletus that overlaps with H. lacotei (Dlabola, 1970) in south France, H. thracicus Hoch, 1986 in north Greece and Turkey, H. yozgaticus Hoch, 1986 in central Turkey (Anatolia), H. hani Hoch, 1986 in Lebanon, H. verticillatus Dlabola, 1994 in Israel and Syria, and H. flavovarius Kusnezov, 1935 in Uzbekistan [31, 36], has made research and exact delimitation of H. obsoletus and its developmental host plants difficult.
Studies of the genetic structure of plant-associated populations of H. obsoletus were initiated after severe outbreaks of stolbur-mediated Bois Noir disease of grapevine occurred in Germany, Austria and Switzerland. These outbreaks were linked to the recent northward colonization of new habitats by populations affiliated with U. dioica and the in situ formation of U. dioica and C. arvensis associated host-races among the peripheral populations on the species’ northernmost distribution range [7, 15, 25, 28, 37]. A study on the causes of H. obsoletus population expansion and genetic structure has identified an Israeli population comprised of individuals collected on Vitex sp. and Olea europaea (used as an outgroup for genetic comparison) as seemingly different from the central and west European populations associated with the traditional hosts C. arvensis and U. dioica [7, 15]. In addition, a recent study on the distribution, host association and stolbur phytoplasma vectoring ability of H. obsoletus populations in Serbia lead to the discovery of a new host association with Crepis foetida, stinking hawk’s-beard [38]. The first observation of H. obsoletus aggregation on C. foetida was made in 2006 on fallow meadows and recently abandoned arable land in east Serbia, near the Bulgarian border (Toševski I., unpublished data). The association was confirmed in the following years over a wider geographic range and preliminary data showed genetic differentiation of C. foetida populations relative to the C. arvensis- and U. dioica-associated populations [38]. Subsequent preliminary insights into genetic divergence relative to Mediterranean V. agnus-castus–affiliated populations raised questions regarding the cryptic differentiation potential of H. obsoletus [34].
On the Balkan Peninsula, the first reliable data regarding H. obsoletus plant associations [22] designated C. arvensis both as a suitable adult host-plant and as the source of the vectored pathogen (at that time known as the stolbur virus). During the following half century, according to faunistic records and summarized data from museum collections for this region, occurrence of H. obsoletus was indicated primarily on Vitex sp., less frequently Quercus sp. and only sporadically Urtica sp. and Convolvulus sp. [31]. The epidemiological role of the planthopper on the Balkan Peninsula has not been studied. However, an increasing pest potential channelled by global increases in temperature promoting population expansion and host-plant adaptation, has highlighted the importance of understanding the true relations between H. obsoletus and its developmental host-plants in southeastern Europe, the surmised origin of the recent colonization of western Europe [7, 15]. The ecological traits underlying the differentiation of H. obsoletus populations affiliated with C. arvensis and U. dioica at the northwestern edge of the range in western Europe, specific stolbur strains, different adult flight periods and the existence of two plant-based host races [7, 14, 39], represent natural selection processes driven by host-plant choice. This ostensibly polyphagous inhabitant of xerothermic grasslands, disturbed patches and any other available habitat with a preferred host-plant and an adequate soil for nymphal development connects natural and agricultural ecosystems by guiding semi-delineated stolbur transmission routes, which can be deeply affected by the population’s allegiance toward preferred dual host-plant [40].
The present study evaluates genetic differentiation and demographic history in four plant-associated populations of H. obsoletus from southeastern Europe with the aim of delineating population (host-plant) specialization and possible cryptic speciation. Using previously published genetic data analyzing the same mtDNA and microsatellite loci as the present study [7, 11, 15], we infer phylogenetic ancestry, population expansions and genetic uniqueness of these populations. We assess the evolution of host-race formation and specialization by separating plant- vs. geographic-based population structure. The insights about genetic distinctness can later be corroborated with other species traits such as acoustic signals, behavioral habits and/or subtle morphological structures and lead to proper species delimitation using an integrative approach [17, 41–44]. By testing the hypothesis that plant-affiliated populations in the species distribution center (sensu [31]) are highly specialized, we consider the potential for host race formation at its distribution edge [7].
Materials and methods
Sampling localities and sampling scheme
Surveys to collect Hyalesthes obsoletus populations associated with four target plants previously identified as preferred plants for this planthopper in southeastern Europe and eastern Mediterranean [13, 22, 31, 33, 38] were performed in 2011–2014 (Fig 1, Table 1). No specific permissions were required, as the study did not involve endangered or protected species. Collections were made from the end of June to the beginning of October, and were based on previous information on adult flight periods in populations associated with each of the four focal host-plants: Convolvulus arvensis (Ca), Urtica dioica (Ud), Vitex agnus-castus (Vac) and Crepis foetida (Cf) [31, 33, 38, 45]. To evaluate the planthopper’s natural occurrence (regardless of the crop species), we surveyed natural localities such as xerothermic meadows, fallow land, ruderal sites, degraded habitats along roads or rocky substrate of the Mediterranean littoral. At each location, insects were sampled with sweep nets and mouth aspirators from patches of a single host-plant, placed in 2 ml plastic tubes (Sarstedt) filled with 96% ethanol, transported in a portable filled cooler at 10°C to the laboratory and stored at 4°C. All field-collected individuals were examined under a stereomicroscope (Leica MZ7.5) and assigned to H. obsoletus by white collar and male genital morphology according to the taxonomic key provided by Hoch and Remane [31]. In addition to the sampling in the present study, we included previously reported H. obsoletus populations collected from crop plants in Romania and Russia (localities Radovanu and Mayak, designated with black outlined circles on Fig 1; [7]) that were not previously tested for mitochondrial diversity.
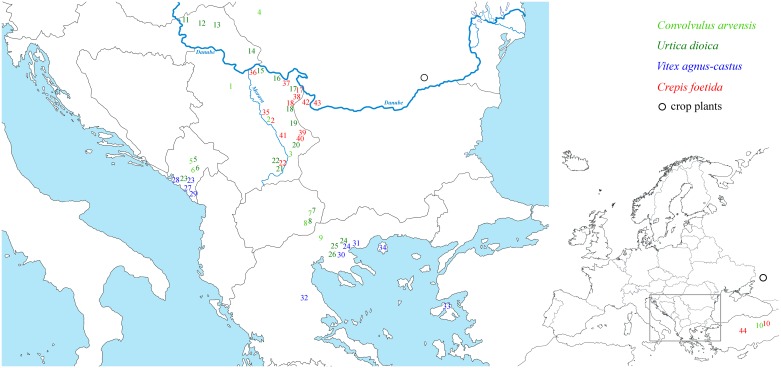
Fig 1. Sampling localities of Hyalesthes obsoletus populations and associated host-plants.
Numbers refer to localities listed in Table 1. The number’s color refers to the H. obsoletus population host-plant association as given on the map. Syntopic localities are designated with the same number in two host-plant corresponding colors. The sampling localities of two previously reported H. obsoletus populations collected on crop plants in Romania and Russia (Radovanu and Mayak) [7] are designated with black outlined circles. Reprinted from d-maps http://d-maps.com/carte.php?num_car=2068&lang=en and http://d-maps.com/carte.php?num_car=2232&lang=en under a CC BY license, with permission from Daniel Dalet, original copyright 2007–2018.
Table 1. Sampled locality data and summarized per population genetic diversity of Hyalesthes obsoletus sorted by corresponding host-plant and country of origin.
| Host-plant association | Country | Locality no. & name | GPS coordinates | Population no. | nDNA (microsatellite) analyzes | mtDNA analyzes | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | AR | HE | FIS | haplotype | frequency | |||||
| Convolvulus arvensis | Serbia | 1. Topola | N44 13.532 E20 40.224 | Pop1 | 20 | 4.774 | 0.791 | 0.054 | EC* | 6 |
| 2. Aleksinac | N43 36.010 E21 40.592 | Pop2 | 6 | 4.626 | 0.771 | -0.015 | EC* | 6 | ||
| 3. Predejane | N42 49.992 E22 07.912 | Pop3 | 9 | 4.956 | 0.837 | 0.164 | EC* | 6 | ||
| Romania | 4. Petrevo selo | N45 49.528 E21 31.548 | Pop4 | 9 | 4.763 | 0.816 | 0.151 | EC* | 6 | |
| Montenegro | 5. Martinići | N42 32.245 E19 10.763 | Pop5 | 17 | 4.412 | 0.777 | 0.264 | AB* | 6 | |
| 6. Podgorica | N42 26.919 E19 12.509 | Pop6 | 20 | 4.192 | 0.748 | 0.143 | AB* | 6 | ||
| Macedonia | 7. Hamzali | N41 29.860 E22 44.996 | Pop7 | 12 | 4.549 | 0.787 | 0.169 | EC* | 6 | |
| 8. Strumica | N41 26.505 E22 39.922 | Pop8 | 11 | 4.282 | 0.744 | 0.146 | EC* | 4 | ||
| πC | 1 | |||||||||
| ψC | 1 | |||||||||
| Greece | 9. Kilkis | N40 54.984 E22 49.101 | Pop9 | 6 | 4.550 | 0.721 | 0.216 | AB* | 1 | |
| XB | 1 | |||||||||
| WL | 2 | |||||||||
| WB | 2 | |||||||||
| Turkey | 10. Erzincan | N39 57.253 E38 38.063 | Pop10 | 3 | - | - | - | SK | 3 | |
| Urtica dioica | Serbia | 11. Gakovo | N45 55.880 E19 03.280 | Pop11 | 22 | 4.937 | 0.820 | 0.141 | EC* | 4 |
| FC* | 1 | |||||||||
| RC | 1 | |||||||||
| 12. Bačka Topola | N45 47.518 E19 35.604 | Pop12 | 20 | 4.934 | 0.826 | 0.157 | EC* | 4 | ||
| ωC | 2 | |||||||||
| 13. Bačko Petrovo selo | N45 43.693 E20 06.114 | Pop13 | 20 | 5.100 | 0.844 | 0.155 | EC* | 6 | ||
| 14. Vršac | N45 03.874 E21 11.208 | Pop14 | 12 | 4.590 | 0.804 | 0.148 | EC* | 6 | ||
| 15. Srednjevo | N44 39.685 E21 30.362 | Pop15 | 15 | 4.705 | 0.787 | 0.087 | EC* | 6 | ||
| 16. Boljetin | N44 31.740 E22 02.090 | Pop16 | 12 | 4.714 | 0.815 | 0.212 | EC* | 6 | ||
| 17. Negotin | N44 16.604 E22 30.484 | Pop17 | 6 | 4.644 | 0.803 | -0.003 | EC* | 6 | ||
| 18. Zaječar | N43 50.016 E22 17.334 | Pop18 | 20 | 4.807 | 0.816 | 0.072 | EC* | 4 | ||
| ξQ | 2 | |||||||||
| 19. Knjaževac | N43 30.610 E22 18.833 | Pop19 | 15 | 4.923 | 0.836 | 0.226 | EC* | 6 | ||
| 20. Grnčar | N43 01.270 E22 21.825 | Pop20 | 17 | 4.492 | 0.777 | 0.130 | EC* | 6 | ||
| 21. Vranje | N42 31.725 E21 54.319 | Pop21 | 12 | 4.380 | 0.76 | 0.120 | EC* | 6 | ||
| 22. Vranjska banja | N42 34.139 E21 58.367 | Pop22 | 11 | 4.425 | 0.753 | 0.060 | EC* | 6 | ||
| Montenegro | 23. Godinje | N42 13.421 E19 06.800 | Pop23 | 6 | 4.395 | 0.758 | 0.166 | EC* | 6 | |
| 5. Martinići | N42 32.245 E19 10.763 | Pop24 | 20 | 4.306 | 0.746 | 0.122 | EC* | 6 | ||
| 6. Podgorica | N42 26.919 E19 12.509 | Pop25 | 20 | 4.362 | 0.76 | 0.113 | EC* | 2 | ||
| αC | 2 | |||||||||
| βC | 2 | |||||||||
| Macedonia | 7. Hamzali | N41 29.860 E22 44.996 | Pop26 | 17 | 4.091 | 0.714 | 0.024 | EC* | 6 | |
| 8. Strumica | N41 26.505 E22 39.922 | Pop27 | 19 | 4.194 | 0.735 | 0.104 | EC* | 6 | ||
| Greece | 24. Arethousa | N40 45.767 E23 33.096 | Pop28 | 12 | 4.007 | 0.685 | -0.060 | EC* | 6 | |
| 25. Filadelfio | N40 45.246 E23 27.846 | Pop29 | 20 | 3.500 | 0.622 | -0.009 | EC* | 6 | ||
| 26. Profitis | N40 39.930 E23 17.331 | Pop30 | 6 | 3.602 | 0.643 | 0.193 | EC* | 3 | ||
| WB | 2 | |||||||||
| ρC | 1 | |||||||||
| Vitex agnus-castus | Montenegro | 27. Bar | N42 07.031 E19 04.581 | Pop31 | 18 | 4.198 | 0.747 | 0.038 | ZN | 5 |
| γN | 1 | |||||||||
| 23. Godinje | N42 13.421 E19 06.800 | Pop32 | 19 | 4.590 | 0.803 | 0.101 | ηN | 6 | ||
| 28. Kamenari | N42 28.591 E18 41.028 | Pop33 | 20 | 3.667 | 0.67 | 0.059 | ZN | 5 | ||
| ZO | 1 | |||||||||
| 29. Ulcinj | N41 56.515 E19 16.052 | Pop34 | 19 | 4.753 | 0.817 | 0.163 | ZN | 2 | ||
| ηN | 2 | |||||||||
| θN | 2 | |||||||||
| Greece | 30. Apollonia | N40 38.380 E23 29.960 | Pop35 | 10 | 4.732 | 0.819 | 0.168 | YM | 6 | |
| 31. Asprovalta | N40 45.124 E23 44.024 | Pop36 | 19 | 4.771 | 0.795 | 0.176 | YM | 6 | ||
| 24. Arethousa | N40 45.767 E23 33.096 | Pop37 | 20 | 5.154 | 0.855 | 0.204 | YM | 4 | ||
| ηM | 2 | |||||||||
| 32. Larisa | N39 38.544 E22 16.981 | Pop38 | 7 | 4.320 | 0.775 | 0.065 | YM | 6 | ||
| 33. Lesbos | N39 18.565 E26 8.379 | Pop39 | 5 | - | - | - | YM | 3 | ||
| σM | 2 | |||||||||
| 34. Thasos | N40 35.177 E24 37.525 | Pop40 | 3 | - | - | - | YM | 3 | ||
| Crepis foetida | Serbia | 2. Aleksinac | N43 36.010 E21 40.592 | Pop41 | 10 | 3.430 | 0.689 | 0.388 | JH | 4 |
| MH | 2 | |||||||||
| 35. Deligrad | N43 38.546 E21 33.444 | Pop42 | 20 | 3.552 | 0.688 | 0.193 | JH | 2 | ||
| MH | 2 | |||||||||
| μH | 2 | |||||||||
| 36. Požarevac | N44 39.310 E21 11.983 | Pop43 | 12 | 3.334 | 0.638 | 0.181 | JH | 4 | ||
| MH | 2 | |||||||||
| 37. Porečka reka | N44 24.267 E22 10.350 | Pop44 | 4 | - | - | - | JH | 1 | ||
| MH | 1 | |||||||||
| UH | 1 | |||||||||
| VH | 1 | |||||||||
| 17. Negotin | N44 16.604 E22 30.484 | Pop45 | 21 | 3.323 | 0.619 | 0.282 | JH | 3 | ||
| MH | 3 | |||||||||
| 38. Tamnič | N44 04.973 E22 32.048 | Pop46 | 7 | 3.365 | 0.613 | 0.165 | JH | 4 | ||
| MH | 2 | |||||||||
| 18. Zaječar | N43 50.016 E22 17.334 | Pop47 | 6 | 3.329 | 0.602 | 0.363 | JH | 4 | ||
| MH | 2 | |||||||||
| 39. Temska | N43 16.537 E22 31.840 | Pop48 | 12 | 3.370 | 0.656 | 0.200 | JH | 6 | ||
| 40. Pirot | N43 12.880 E22 31.585 | Pop49 | 19 | 3.689 | 0.687 | 0.257 | JH | 6 | ||
| 41. Jasenovik | N43 22.365 E22 02.441 | Pop50 | 20 | 3.743 | 0.715 | 0.147 | JH | 6 | ||
| 22. Vranjska banja | N42 34.139 E21 58.367 | Pop51 | 7 | 3.067 | 0.598 | 0.370 | JH | 2 | ||
| MH | 2 | |||||||||
| λH | 2 | |||||||||
| Bulgaria | 42. Vidin | N43 57.711 E22 51.258 | Pop52 | 6 | 2.800 | 0.563 | 0.198 | JH | 4 | |
| MH | 2 | |||||||||
| Romania | 43. Calafat | N43 59.926 E22 58.119 | Pop53 | 10 | 3.452 | 0.631 | 0.305 | JH | 4 | |
| MH | 2 | |||||||||
| Turkey | 10. Erzincan | N39 57.253 E38 38.063 | Pop54 | 8 | 4.379 | 0.768 | 0.175 | JH | 6 | |
| 44. Kırşehir | N39 26.021 E34 07.795 | Pop55 | 1 | - | - | - | JH | 1 | ||
Sample size (N); Allelic richness (AR); Expected heterozygosity (HE); Inbreeding coefficient (FIS).
Symbol "-" refers to genetic indices that are not calculated due to small sample size.
Mitochondrial (mtDNA) haplotypes designation according to Johannesen et al. [15]; First letter in haplotype name corresponds to COI-tRNA(Leu)-COII gene region haplotype designation and second letter to the 16S-tRNA(Leu)-ND1 gene region haplotype.
DNA extraction
Total DNA was extracted from each individual insect specimen using a non-destructive, partly modified sodium dodecyl sulfate (SDS) extraction method [46, 47]. Briefly, all specimens were punctured between hind legs and mesothorax and incubated overnight at 56°C in extraction buffer (SDS 0.5%, Tris 20 mM, EDTA 10 mM) with proteinase K (Fermentas) at a concentration of 187.5 μg/mL-1. After removing the insect, chloroform was added to the homogenate. The mixture was centrifuged at 4°C on 11000 rpm for 10 min. The chloroform step was repeated, after which the upper aqueous supernatant was precipitated with ice-cold isopropanol. This mixture was centrifuged at maximum speed for 15 min. The resulting DNA pellet was washed with 96% ethanol, air dried and re-suspended in 50 μl TE buffer (10 mM Tris, 1 mM EDTA, pH 7.6). To achieve a higher rate of DNA recovery for the dry museum specimen, and for specimens from localities in Turkey, Greece and Serbia with only few individuals, we used the Qiagen Dneasy Blood & Tissue Kit (Hilden, Germany) in accordance with the manufacturer’s instructions. Both extraction methods allowed us to preserve morphological features of specimens for later analyses. After DNA extraction, insect specimens were prepared as voucher dry specimens. They are housed at the Institute for Plant Protection and Environment collection (IPPE, Zemun, Serbia). Extracted DNA was kept at -20°C.
Mitochondrial genotyping
Mitochondrial DNA analyses of H. obsoletus were based on two concatenated gene regions (markers) previously characterized by Johannesen et al. [15, 37]: (i) cytochrome oxidase subunit I, tRNA for leucine and cytochrome oxidase subunit II region (COI-tRNA(Leu)-COII) and (ii) 16S ribosomal RNA, tRNA-Leu and reduced nicotinamide adenine dinucleotide (NADH) dehydrogenase subunit I region (16S-tRNA(Leu)-ND1). Amplifications were performed using primers S2792 and A3661 for the first gene region [48] and LR-N-12945 and N1-J-12261 for the second region [49] (Fig 2, S2 Table). Polymerase chain reactions (PCR) for both markers were performed in a 20-μL final reaction volume using Kapa Biosystems High Yield Reaction Buffer A (1.5 mM MgCl2, 1×), an additional 3.5 mM of MgCl2 for amplification of COI-tRNA(Leu)-COII and 1.5 mM for 16S-tRNA(Leu)-ND1, 0.5 mM of each dNTP, 0.4 μM of each primer, 1 U of KAPA Taq DNA polymerase (Kapa Biosystems, Inc., Woburn, MA, USA) and 1 μL of template DNA. Amplification was conducted in a Mastercycler ep gradient S (Eppendorf, Hamburg, Germany) using the following thermal profile for both markers: 95°C for 2 min, 35 cycles at 95°C for 30 s, 48°C for 1 min, 72°C for 1 min 30 s, and a final extension at 72°C for 10 min. Sequencing was performed on an ABI Prism 3700 automated sequencer (Macrogen Inc., Seoul, South Korea). Sequences were edited using FinchTV v.1.4.0 (http://www.geospiza.com) and aligned with ClustalW [50] within the MEGA v.5.2 software [51]. Whenever possible, six specimens per each H. obsoletus population were genotyped for mitochondrial markers, (Table 1). The nomenclature of newly identified haplotypes followed the designation system established by Johannesen et al. [15, 37]. Haplotypic nucleotide sequences were submitted to NCBI GenBank under the accession numbers KY368699-724 for COI-tRNA(Leu)-COII and KY368692-8 for 16S-tRNA(Leu)-ND1 (S3 Table).
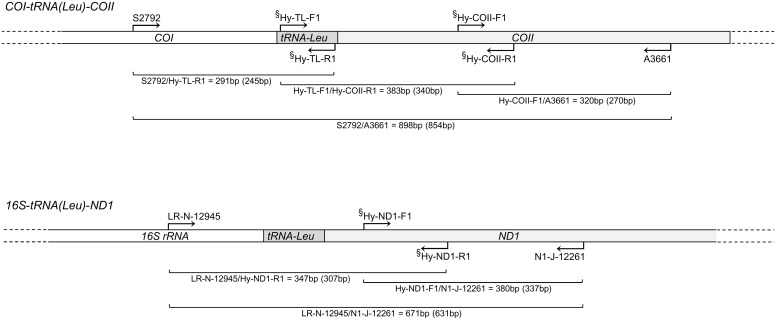
Fig 2. Schematic representation of COI-tRNA(Leu)-COII and 16S-tRNA(Leu)-ND1 mtDNA gene regions showing the binding site positions of the primers used for amplification of freshly collected Hyalesthes obsoletus specimens and dry museum H. thracicus specimen.
The amplicon length (bp) for each primer pair combination is given below the scheme (length excluding primers is given in parentheses). Primers marked with the symbol "§" were designed in this study and used for the amplification of short DNA fragments of the H. thracicus paratype specimen. Scheme not drawn to scale. Primer sequences are given in S2 Table.
Considering that orthology is of primary importance for phylogenetic studies, especially when cryptic speciation events are possibly involved [52], we performed analyses to confirm the orthologus status (and rule out nuclear pseudogenes of mitochondrial origin, Numts) of the amplified mitochondrial markers. To obtain reliable orthologous mitochondrial sequence of both marker genes, we made serial dilutions of genomic DNA for a subset of samples prior to performing the described PCR amplification protocols. We considered that in dilution of 1:6,250 any amplification reaction would contain less than one nuclear genome [53]. Sequences obtained from diluted samples were compared with the ones obtained using undiluted genomic DNA to confirm their identity and mitochondrial origin.
To achieve a better resolution for the evolutionary relations between and among H. obsoletus host-plant associated haplotype groups, we supplemented our study with Hyalesthes thracicus genetic data, morpho-phylogenetically the closest member of the same species group [31, 54]. We obtained an archival, museum, male paratype specimen collected in 1979 in the Lake Volvi surroundings (Greece), deposited in Prof. H. Hoch’s private collection (S1 Fig). The entomological tile and glue were carefully removed, where after the paratype was prepared for extraction as previously described. Considering that mtDNA in dry insect material is frequently highly degraded and fragmented, the archival specimen was analyzed using a set of four and two degenerate primers designed to amplify short fragments of the COI-tRNA(Leu)-COII and 16S-tRNA(Leu)-ND1 gene regions, respectively (Fig 2, S2 Table). Newly designed primers were degenerate due to an unknown sequence at the primer binding site. The primer design was based on a sequence alignment comparison among phylogenetically closely and more distantly related cixiid planthoppers: Hyalesthes philesakis Hoch, 1986, H. luteipes Fieber, 1876, H. ponticorum Hoch, 1986, H. aylanus Hoch, 1986, Reptalus panzeri (Löw, 1883), R. melanochaetus (Fieber, 1876), R. cuspidatus (Fieber, 1876) and Setapius apiculatus (Fieber, 1876). Six newly designed primers were combined with standard ones [48, 49], producing overlapping amplicons that allowed the recovery of the entire genetic region analyzed for fresh material (Fig 2; S2 Table). PCR conditions for short fragment amplification were the same as described for the full-length marker genes with the exception of an extension time that was reduced to 45 s. Nucleotide sequences of the H. thracicus COI-tRNA(Leu)-COII and 16S-tRNA(Leu)-ND1 gene regions were deposited in GenBank under the numbers KY368725 and KY368726, respectively.
Mitochondrial diversity and population differentiation
Genetic variations within and among host-plant associations were estimated with pairwise F-statistics [55] implemented in Arlequin v.3.5.1.2 [56]. Mitochondrial haplotype diversity was calculated for each of the 55 H. obsoletus populations (number and frequency of haplotypes). Standard molecular diversity indices (FST and θ) were calculated for the three metapopulations that were grouped according to the host-plant association and the cluster sharing between the populations associated with C. arvensis and U. dioica as host-plants: 1) C. arvensis + U. dioica, 177 individuals; 2) V. agnus-castus, 56 individuals; and 3) C. foetida, 83 individuals. To estimate the amount of genetic diversity within the selected groups (metapopulations) we calculated the population genetic parameter theta (θ) estimated from the number of alleles (θk), expected homozygosity (θH), segregating sites (θS) and pairwise differences (θπ). Theta (θ) represents the distribution of variation within or among populations when samples are considered to represent characteristics of the higher rank group from which they are sampled, in this case, the host-plant association [57, 58].
To study whether genetic diversity of each of the four host-plant associated groups depart from neutrality and to detect genetic signals of the demographic and/or spatial growth we calculated Tajima’s D [58] and Fu’s FS neutrality statistics [59]. Neutrality tests were performed in Arlequin v.3.5.1.2 [56]. Because U. dioica-associated populations are known to be undergoing demographic expansion in western Europe [8, 37], individuals associated with C. arvensis and U. dioica were treated both separately and as a meta-group to deduce the source of a departure from neutrality. Fu’s simulations are based on the infinite sites model of mutation. Negative values of Fu’s FS parameter indicate an excess in allele numbers as expected from a recent population expansion, while a positive value is evidence of an allele deficiency, as expected from a recent population bottleneck [60]. Tajima’s D test measures a difference in the number of pairwise differences and the number of segregating sites scaled to be the same in a neutrally evolving population. A negative Tajima’s D parameter signifies an excess of low frequency polymorphisms. A significant negative departure from these tests has been explained mainly as an excess of new mutations as a result of evolutionary forces, such as selective sweeps or population growth (signature of population expansion), while positive departure indicate a balancing selection.
The pairwise fixation index (FST) values and the Nei’s average number of pairwise differences [61] were calculated using Arlequin, to measure differentiation between all population pairs sorted according to host-plant association (as presented in Table 1). In total, calculations were performed for 51 H. obsoletus populations harboring at least five individuals. FST values range from 0 (including negative values) to 1, with 0 indicating no divergence between the populations, and 1 indicates that two populations are completely separate [55, 62]. Nei’s pairwise differences assume genetic differences that arise from mutations and changes in the frequency of haplotype in a population due to random sampling.
Phylogenetic analysis and haplotype network construction
Phylogenetic reconstruction and haplotype networks were based on newly identified haplotypes (this study) and all previously published H. obsoletus haplotypes [8, 11, 15, 37]. To obtain the best resolution of the phylogenetic relationships among the host-plant associated H. obsoletus populations, H. thracicus was used for tree rooting in all mtDNA phylogenetic analyses. Concatenated sequences of both mtDNA gene regions (1180 bp in length) were aligned and treated as a single locus. The maximum parsimony and neighbor-joining trees were generated in PAUP* v.4.0b10 [63] using the evolutionary model of nucleotide substitution that best fit the data, determined with jModelTest v.2.1.7 [64] under unconstrained prior distributions. Five hundred bootstrap replicates were performed to assess the branch support of the resulting tree topologies. The suggested substitution model was also used for the Bayesian-based phylogenetic analysis implemented with MrBayes v.3.1.2 [65, 66]. We used the following settings: two simultaneous runs executed for 1,000,000 generations, sampling every 100 generations and with a ‘burn-in’ of 25%. Posterior probabilities were assessed with Tracer v.1.5.0 [67] to ensure that sampling reached stationarity within the burn-in. The obtained trees were visualized in FigTree v.1.4 [68]. Average genetic distances among haplotypes were calculated within and between major phylogenetic clusters using PAUP* under the nucleotide substitution model as determined by jModelTest found for the phylogenetic tree reconstruction (see above). Additionally, genetic distances were calculated separately for haplotypes of the concatenated COI-COII genes, which was proven to be the most phylogenetically informative of the analyzed mtDNA markers for species-level identification and differentiation, e.g., [69–71].
Phylogenetic relationships between closely related species and such resulting from population-level processes (e.g., persistence of ancestral haplotypes, multifurcations, recombination and horizontal transfer) are often better visualized in reticulated graphs or networks [72]. MtDNA gene-region genealogies of H. obsoletus host-associated haplogroups were inferred employing two software packages with different network construction approaches. The software TCS v.1.21 [73] was used to construct a haplotype network based on statistical parsimony [74] with 95% confidence limits, while a median-joining network was calculated using Network v.4.612 (www.fluxus-engineering.com) keeping the parameter ε = 0. This method adds median vectors as uncollected or possibly extinct ancestral genotypes in order to reduce tree length to the minimum spanning trees combined within a single network [75].
Microsatellite genotyping and diversity
Microsatellites primers and PCR conditions follow Imo et al. [7, 76] and Maniyar et al. [28]. The microsatellite analysis was performed using a GA3130XL Genetic Analyzer (Applied Biosystems) (Mainz University). In total, 702 H. obsoletus individuals originating from 50 populations (Table 1) were genotyped at seven microsatellite loci previously used to delimit and to characterize population genetic structure and host races of H. obsoletus [7]. The loci were genotyped using GeneMapper v.4.0 (Applied Biosystems).
The presence of null alleles was evaluated using the EM algorithm (FreeNa) [77, 78]. Null allele frequencies were calculated per locus and population, as well as overall and per host-plant association. Deviations from Hardy-Weinberg (HW) were estimated using Micro-checker [79]. Linkage disequilibrium (LD) was checked with the web-based version of Genepop [80] using the settings 1,000 batches and 10,000 de-memorizations and iterations per batch. The sex-linked locus C147 was omitted in both analyses. Significance levels for multiple comparisons were adjusted by Bonferroni correction [81]. The allele number (A), the allelic richness (AR) and allele frequencies per locus were estimated in FSTAT v.2.9.3 [82] using all seven loci; whereas C147 was excluded in estimates of the population inbreeding coefficient (FIS). The mean expected heterozygosity per population (HE) was calculated in Arlequin v.3.5.1.2 [56]. Differences in the mean HE and AR per group of the host-plant associated populations were tested with t-tests using Statistica v.5.1 (StatSoft, Inc. 1997). Because sample sizes varied, we performed a linear regression analyses to test sample the dependency of sample size and genetic diversity measures.
Microsatellite population structure and differentiation
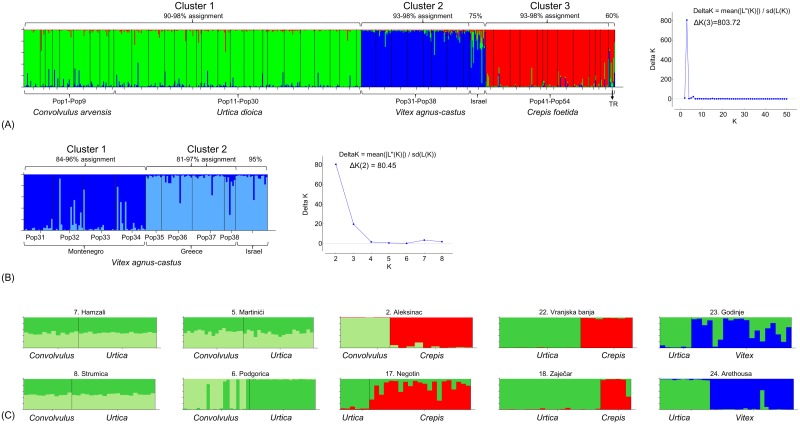
Analyses of the influence of host-plant affiliation and geographic separation on population structure were estimated using Bayesian clustering (Structure v.2.3.3) [83]. Following the procedure described in Imo et al. [7], we first estimated the highest level of genetic clustering, K, for all individuals based on their multi-locus genotypes. Alongside the 702 individuals collected in this study, we analyzed a single 20-member population associated with Vitex agnus-castus from Israel [7]. Based on the results of overall population clustering and knowing that the Bayesian analysis can be influenced by relative genotype frequencies, which may hide signals of a lower-level structure, we estimated association levels, i.e., K, within clusters for additional effects of geographic distance and host-plant. The Structure analysis for each test was repeated 20 times for each K with 50,000 burn-ins followed by 200,000 MCMC (Markov chain Monte Carlo) iterations using the admixture model and correlated allele frequencies. The web-based program Structure harvester [84] was used for visualizing the likelihood plateau of the distribution of LnP(D) and for inferring the most likely number of genetic clusters (ΔK) according to Evanno et al. [85]. Additionally, syntopic localities in which the geographical signal is excluded as a distortive factor were singled out in separate independent analyses. Genetic differentiation among clusters identified with Structure was estimated with the molecular variance analysis (Amova) in Arlequin v.3.5.1.2 [56].
Genetic clustering and identity among H. obsoletus populations associated with the four host-plants were further inferred with a maximum-likelihood phenogram using the Contml algorithm in Phylip v.3.69 [86]. Hyalesthes luteipes individuals sampled on Ulmus minor in Serbia were used as an outgroup root. This analysis was based on four loci (F56, F84, H120, G85) that amplified successfully in both species. The phenogram was visualized using FigTree v.1.4 [68]. It should be noted here that outgroups differed between the mtDNA and microsatellite analyses. MtDNA analysis was based on a single museum specimen of the nearest morphological relative H. thracicus. In contrast, microsatellite analysis (H. luteipes) relies on allele frequencies for which estimate several individual are required.
Results
Target host-plants were searched for associated H. obsoletus populations across the surveyed area (Fig 1) to obtain information on the geographic range of each insect-host association, distribution overlap (sympatry), population structure and genetic differentiation in syntopy. Surveys indicated that H. obsoletus populations in southeastern Europe are i) high density and very common in association with Ud; ii) rare and low density in association with Ca; iii) usually very common and in high density in association with Vac, however, it is restricted to the coastal distribution of the host-plant; and iv) high in number and very common in association with Cf in the eastern parts of the surveyed area but absent in the south (Fig 1). The number of collected individuals per population was always intended to be greater than or equal 6 and preferably 20. However, on some occasions this number was not achieved due to the end of the adult flight period or a low-density population (Pop10, 39, 40, 44 and 55; Table 1). In total, populations were collected in six countries with at least two identified H. obsoletus host-plant associations: Serbia (Ca, Ud and Cf), Romania (Ca and Cf), Montenegro (Ca, Ud and Vac), Macedonia (Ca and Ud), Greece (Ca, Ud and Vac) and Turkey (Ca and Cf). In addition to these locations, a single locality with Cf as a tentative host-plant was surveyed at a Danube locality in Bulgaria (Fig 1, locality 42) for the purpose of assessing colonization route and the distribution range of Cf-associated H. obsoletus populations. Locality 43 in Romania was surveyed to test whether the Danube acts as a natural barrier to the spread of Cf-associated H. obsoletus population into Central Europe (Fig 1). The collections made in Turkey were performed only on the two listed localities, without surveying other potential host-plants or their surroundings.
A total of 718 H. obsoletus adults were sampled from 55 populations at 44 localities in southeastern Europe and Turkey (Table 1, Fig 1). Eleven sites held syntopic host-plant populations of the planthopper: four Ca/Ud in Macedonia and Montenegro, two Ud/Vac in Greece and Montenegro, two Cf/Ca in Serbia and Turkey, and three Cf/Ud in Serbia. Ten H. obsoletus populations were sampled from C. arvensis, and populations from U. dioica were sampled at 20 localities. The V. agnus-castus-associated populations were sampled from four localities along the coastline area of Montenegro, from six localities in Thessaly (Greece), and from the Aegean islands Thasos and Lesbos 10 km from the nearest Greek or Turkish mainlands, respectively. The affiliation of H. obsoletus populations with C. foetida was observed mainly in eastern and southeastern Serbia, with a western limit along the river Morava (N = 11). The association with C. foetida was confirmed along the Danube in Bulgaria and Romania (Fig 1) but it was not observed in Montenegro, Macedonia or Greece. An additional nine adults were collected from C. foetida at two sites in Turkey, of which one, located in an eastern part of the Anatolian plateau (locality 10), held a syntopic C. arvensis population.
Population genetic diversity based on mitochondrial and microsatellite data
The mitochondrial genetic diversity analysis revealed highly significant differentiation among the three host-plant associated groups (Ca-Ud, Vac and Cf metapopulations), with 88% of genetic variance derived from host-plants (FST = 0.88, p < 0.001) and 12% attributed to within-group variations. Diversity estimates within and among host-associated metapopulations (Fig 3) were calculated for 316 individuals from the 55 H. obsoletus populations collected in our study (Table 1). Twenty-nine haplotypes were identified, three of which were previously found in the H. obsoletus populations associated with Ca and Ud from central Europe (AB, EC and FC; [15]), and 26 were newly described haplotypes from southeastern Europe and Turkey. Of the new haplotypes, three were found in association with Ca, six with Ud, eight with Vac and six with Cf (S3 Table). Overall, from 177 individuals collected from Ca-Ud, 56 from Vac and 83 sampled from Cf, 15, 8 and 6 haplotypes were recorded in each host-associated metapopulation, respectively.
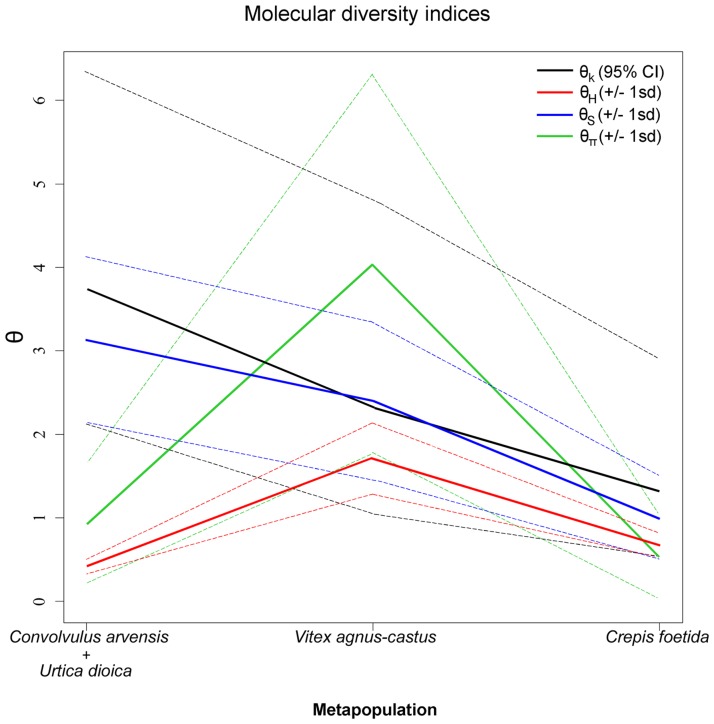
Fig 3. Mitochondrial diversity indices calculated 316 Hyalesthes obsoletus genotyped specimens of the three host-associated metapopulations.
Convolvulus arvensis and Urtica dioica (177 specimens), Vitex agnus-castus (56 specimens) and Crepis foetida (83 specimens). Indices are designated in colors: black = number of alleles (θk), red = expected homozygosity (θH), blue = segregating sites (θS) and green = pairwise differences (θπ).
Haplotypes identified in one of the three delineated metapopulations were never observed in the other two, i.e., haplotypes were host-specific at all localities (Table 1). For the two H. obsoletus populations collected on crop plants in Romania and Russia (localities Radovanu and Mayak; [7]), the Romanian locality exhibited haplotypes affiliated with both Ca-Ud (EC, including new haplotypes PC and ΦC) and Cf (JH and MH), while the Russian had a single haplotype (QB, S3 Table) derived from the AB haplotype of the Ca-Ud metapopulation. These two populations were not included in diversity estimates due to doubtful host-plant origin.
Comparison among the theta (θ) values of molecular diversity in these three host-plant associations revealed the highest mtDNA diversity estimates of number of alleles and segregating sites in the Ca-Ud H. obsoletus metapopulation (θk = 3.73 and θs = 3.13; Fig 3). In case of Vac- and Cf-associated metapopulations number of alleles θk was 2.32 and 1.31, and segregating sites θs was 2.39 and 1.00, respectively. Higher diversity estimates in the Ca-Ud metapopulation correlated with departures from the population equilibrium, indicating a recent population expansion, as suggested by the significantly negative values of Fu’s and Tajima’s neutrality indices (FS = -9.59, p < 0.001; D = -1.87, p < 0.01; S4 Table). On the contrary, neutrality indices were non-significant in Vac and Cf metapopulation (S4 Table). The observed lower level of expected homozygosity in the Ca-Ud metapopulation (θH = 0.42; Fig 3) compared to the other two associations (θH = 1.71 for Vac and 0.67 for Cf) could be a result of low-frequency alleles left over from geographical expansion [87]. Dividing this genetic cluster into two groups according to the host-plant (to deduce the source of the neutrality departure) revealed that the observed expansion signal derives from the U. dioica association with a highly significant negative value of neutrality indices (FS = -8.59, p < 0.001; D = -2.18, p < 0.001; S4 Table). Furthermore, genetic differentiation between the Ca- and Ud-affiliated H. obsoletus populations was low (FST = 0.25, p < 0.001), with 75% of the divergence deriving from the inner host-plant associated variance. A very low genetic polymorphism was detected in the genetic structure of the C. foetida metapopulation, while the V. agnus-castus association expressed the highest level of inner divergence (pairwise differences, θπ), corresponding to 90.5% (p < 0.001) of the genetic variation observed between the Montenegrin and Greek haplogroups.
Microsatellite analyses were performed on 702 individuals from 50 populations (S1 Appendix). Fifteen new alleles were observed compared to previous data [7] resulting in 162 detected alleles. All loci were polymorphic across all populations. The mean number alleles/locus across seven loci = 23, range: 16 (F84) − 30 (C147). Total of 28 (17%) detected alleles were private, occurring in one population only. Private alleles were recorded in each plant association: C. foetida = 2, C. arvensis = 6, V. agnus-castus = 9 and U. dioica = 11. The highest number of private alleles within a single sample population was 7, occurring within V. agnus-castus association (Pop36 and 37), while 11 (7%) were private relative to a host-plant association (V. agnus-castus = 5 and U. dioica = 6). No significant linkage disequilibrium was detected for any locus pair in any population after Bonferroni’s correction [81].
The mean null allele frequency over all loci and populations was 0.06 with the highest value detected on locus B82 (0.11), followed by E96 and G85 (0.10) and should not bias the following analyses. The frequencies of null alleles correlated with mtDNA divergence are as follows: C. arvensis and U. dioica had an approximate null allele frequency of 0.05 (microsatellites were developed from a U. dioica population; [76]), V. agnus-castus had 0.06, while C. foetida had one of 0.09.
Expected heterozygosity HE was independent of sample size (0.0005 < R2 < 0.16, p > 0.05). Hereafter we treated this parameter as an unweighted estimate. The mean genetic diversity estimates HE and AR did not differ significantly among populations affiliated with C. arvensis (HE = 0.777 and AR = 4.567), U. dioica (HE = 0.765 and AR = 4.455) and V. agnus-castus (HE = 0.785 and AR = 4.523), all p > 0.05 (Table 2). In contrast, the mean genetic diversity was significantly lower in the H. obsoletus populations associated with C. foetida (p < 0.05) than in the other host-associations (Table 2; HE = 0.651 and AR = 3.449). The observed signal is attributed to the Serbian, Romanian and Bulgarian populations, while the Turkish population had much higher values of diversity indices comparing to the European metapopulation (HE = 0.768 and AR = 4.379; Table 1).
Table 2. Comparison of microsatellite-based mean genetic diversity (AR—allelic richness and HE—expected heterozygosity) among four host-plant associated Hyalesthes obsoletus metapopulations.
| C. arvensis | U. dioica | C. foetida | V. agnus-castus | |
|---|---|---|---|---|
| Mean HE | 0.777 | 0.765 | 0.651* | 0.785 |
| Mean AR | 4.567 | 4.455 | 3.449* | 4.523 |
| No of populations | 9 | 20 | 13 | 8 |
* p < 0.05
C. foetida vs C. arvensis HE: t = 5.885, df = 20, p < 0.05; AR: t = 7.910, df = 20, p < 0.05
C. foetida vs U. dioica, HE: t = 5.355, df = 31, p < 0.05; AR: t = 6.945, df = 31, p < 0.05
C. foetida vs V. agnus-castus, HE: t = -5.291, df = 19, p < 0.05; AR: t = - 5.935, df = 19, p < 0.05
Evolutionary relatedness among Hyalesthes obsoletus host-associated lineages
A total of 51 host-associated H. obsoletus haplotypes were used for constructing a phylogenetic tree (Fig 4). Along with the previously noted 26 new haplotypes associated with the four host-plants detected in this study and the 22 previously described haplotypes associated with Ca, Ud or Vac [8, 11, 15, 37], phylogenetic analyses were supplemented with the data on the three new haplotypes (QB, ΦC, PC) collected from crop plants in Russia and Romania [7]. The Bayesian information criterion revealed the HKY+I (Hasegawa-Kishino-Yano, pinvar = 0.881) [88] to be the best substitution model for the ingroup sequences. This model was employed to estimate the pairwise genetic distances for the neighbor-joining tree and Bayesian analyses.
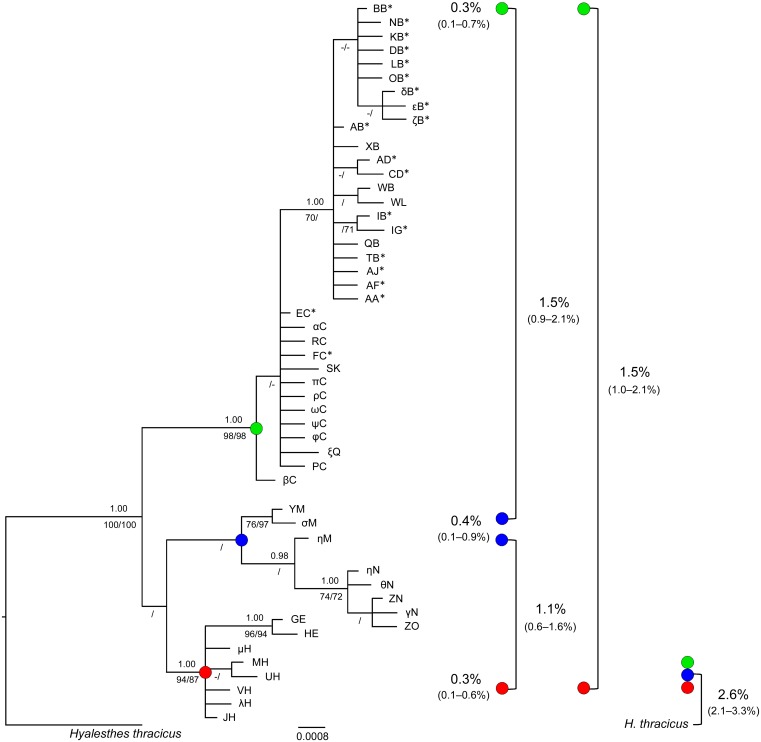
Fig 4. Bayesian phylogenetic tree inferred from 1180 bp of the COI-tRNA(Leu)-COII and 16S-tRNA(Leu)-ND1 concatenated mitochondrial gene regions sampled from the Hyalesthes obsoletus host-associated populations in this and previous studies.
The Bayesian posterior probabilities are noted above branches, and the maximum parsimony and neighbor-joining bootstrap support values appear below the branches in that order. Bootstrap values below 70% and posterior probabilities below 0.95 are omitted. The nonrecovered nodes are marked with an em dash "–". The main nodes are designated with colored circles corresponding to host-plant associated haplogroups as follows: green = Convolvulus arvensis and Urtica dioica, blue = Vitex agnus-castus, and red = Crepis foetida. Average within-group genetic distances for each of the three host-plant clusters are noted left of the corresponding colored circle, while between-group distances are given on the right. The range of within- and between-group distances is given in parentheses. Distances are calculated with correction by applying the HKY+I nucleotide substitution model (Hasegawa-Kishino-Yano, pinvar = 0.881) among three genotype groups for all 51 haplotypes according to the three major host-associated clusters; therefore, the Israeli GE and HE haplotypes are considered as Crepis foetida-cluster members, while the QB, ψC and PC collected on crop plant are members of the Convolvulus-Urtica group. Distances between each H. obsoletus host-associated haplogroup and H. thracicus used as the outgroup are presented in the figure’s bottom right corner. Haplotypes detected in previous studies [8, 11, 15, 37] are marked with an asterisk (*).
The obtained tree’s topology revealed a clear segregation of the H. obsoletus haplotypes into three phylogenetic clusters, each associated with host-plant preference: Ca-Ud, Vac and Cf (marked in green, blue and red in Fig 4, respectively). All three phylogenetic analyses resulted in the same general tree topology. Monophyly of the Ca-Ud and Cf mitochondrial lineages was supported by high Bayesian posterior probabilities (1.00) and bootstrap support values (87–98%), while values for the Vac lineage were lower (0.86, 53% and 56%) probably due to geographic diversity. In addition, the Vac and Cf lineages formed a joint branch, but with low support values (0.52, 53% and 58%) due to high inner-branch diversity. However, the bifurcations were supported using genealogical network analysis (see below).
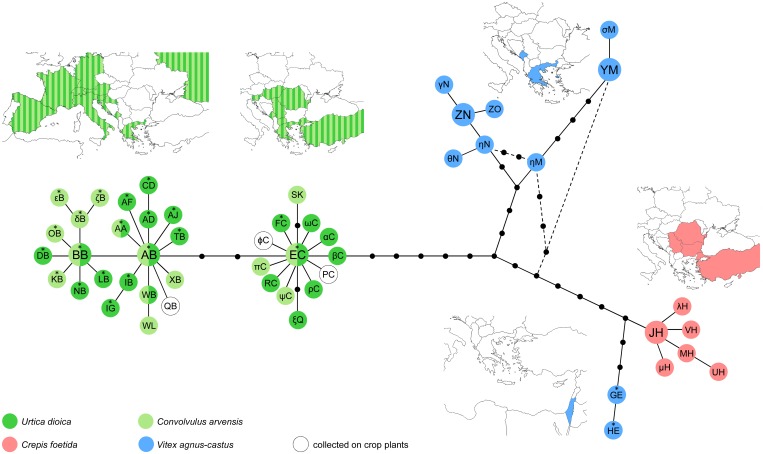
The mitochondrial genealogical network revealed a combination of clear and ambiguous host-plant affiliations, as well as geographic divisions within host-plant associated H. obsoletus haplotypes (Fig 5). Two alternative networks were obtained using both algorithms; however, these networks differed only in the bifurcation position of the major three branches (dashed lines in Fig 5). The populations affiliated with C. arvensis and U. dioica share five haplotypes and have ten and 17 private ones, respectively. They were separated into two phylogeographic sub-clades: a Western European haplogroup (BB- and AB-derived haplotypes) and an Eastern European haplogroup (EC-derived haplotypes). An exception to this geographic based distribution was the Russian haplotype QB, which belonged to the western haplogroup. It implicates a common host-plant association older than the phylogeographic division. V. agnus-castus haplotypes were paraphyletic, being divided into three subgroups consisting of the monophyletic Greek YM and Montenegrin ZN haplotype clades (three and five haplotypes, respectively) and a paraphyletic Israeli clade (two haplotypes). The two Israeli Vac haplotypes (GE and HE) were positioned closest to the C. foetida network branch as well as in the evolutionary tree (Fig 4). The six haplotypes affiliated with C. foetida were delimited as a monophyletic clade, all deriving from the JH haplotype. JH was detected in all Cf-associated H. obsoletus populations, which were collected over a wide geographic range, from east Turkey to east Serbia.
Fig 5. The phylogenetic haplotype network obtained using median-joining and statistical parsimony algorithms on concatenated COI-tRNA(Leu)-COII and 16S-tRNA(Leu)-ND1 mitochondrial gene regions of the 51 Hyalesthes obsoletus haplotypes identified in this and previous studies.
Haplotype colors correspond to the host-plant associations. Haplotypes detected in previous studies [8, 11, 15, 37] are marked with an asterisk (*). The most common haplotypes within each haplogroup are noted with enlarged circles. Dashed lines represent alternative variants of network formations obtained using both algorithms. Black dot vertices represent missing or unsampled haplotypes. Distribution maps are given above each host-associated H. obsoletus haplogroup. Each detected haplogroup’s country is designated on maps in the color corresponding to the associated host-plant. Because Convolvulus arvensis and Urtica dioica share a number of H. obsoletus haplotypes, BB-AB and EC haplogroup distribution is designated in two shades. Haplotypes detected in the two previously reported H. obsoletus populations collected on crop plants in Romania and Russia (Radovanu and Mayak) [7] are not colored.
The average pairwise genetic divergence between the three host-plant associated phylogenetic clusters varied between 1.1% and 1.5%, while the variance within clusters was 0.3–0.4% (Fig 4). Divergence estimates based only on the phylogenetically informative COI-COII gene region (711 bp) were higher, 2.5% (S5 Table). The genetic divergence between host-plant associated phylogenetic clusters and H. thracicus, the most closely related valid species in the species group, varied between 2.1% and 3.3% for the entire analyzed mtDNA segment and 2.9–3.8% for the COI-COII region (Fig 4, S5 Table). Thus, genetic distances among the three clusters of host-associated H. obsoletus haplotypes, on the one hand, and between H. thracicus as a morphologically delineated species on the other, suggested evolutionary relationships of three closely related, but separate, cryptic species delineated by host-plant choice.
Host-associated population differentiation: mitochondrial data
The measure of pairwise population differentiation due to genetic structure (FST) as well as Nei’s pairwise differences among and within populations clearly showed the existence of three genetically separate groups associated with different host-plants: 1) C. arvensis and U. dioica, 2) V. agnus-castus and 3) C. foetida (Fig 6). Additionally, the genetic structure analysis within associations revealed further population differentiation between some of the populations in the Ca-Ud group (Pop 5, 6 and 9) and within the Vac group.
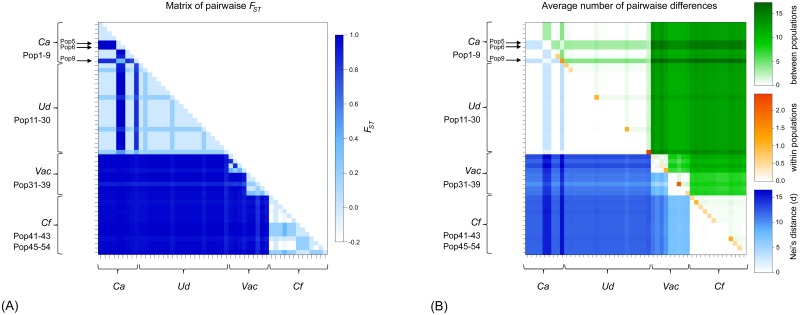
Fig 6. Plots representing parameters of mtDNA genetic differentiation between Hyalesthes obsoletus host-associated populations based on (A) pairwise FST values and (B) Nei’s average number of pairwise differences.
Associated host-plants: Convolvulus arvensis (Ca), Urtica dioica (Ud), Vitex agnus-castus (Vac), and Crepis foetida (Cf). Populations are listed in the same order as in Table 1. The three populations marked with arrows show genetic differentiation compared to all others in the same host-associated group (Ca-Ud). The blue elements below the diagonal of FST values range from 0 to 1, with 0 (including < 0) indicating no divergence between the populations and 1 indicating that two populations are completely separated. The green elements above the diagonal denote Nei’s average number of pairwise differences among populations, the orange diagonal elements denote Nei’s average number of pairwise differences within populations, and blue elements below the diagonal denote the net number of nucleotide differences among populations (Nei’s distance).
Genetic differentiation among the 51 host-associated H. obsoletus populations provided by the FST values were high (from 0.81 to 1.00) and significant (p < 0.01) when the populations associated with Ca-Ud, Vac and Cf were compared to each other (Fig 6; dark blue and green elements on plots). Conversely, the differentiation among populations associated with Cf, including the geographically distant population from Erzincan in east Turkey, was always low, FST = 0.00 to 0.20 (Fig 6). Within the population group associated with Vac populations originating from Montenegro and Greece were highly differentiated (FST = 0.73–1.00; p < 0.01), while differentiation among the Greek populations was low (FST = 0.00–0.30). Differentiation among most Ca and Ud population, regardless of geographic origin, was low, FST = 0.00 to 0.20. Nevertheless, three populations associated with Ca in Montenegro and Greece (Pop5, 6 and 9; marked with arrows in Fig 6) showed high and significant differentiation from the other populations within the Ca-Ud associations (FST = 0.70–1.00; p < 0.01). This finding was caused by the presence of the AB-group haplotypes in these populations (Table 1), while EC-group haplotypes were found in the other Ca-Ud populations. The genetic differentiation of these three populations was less prominent (FST = 0.40–0.50, p < 0.05) only in comparison with the Ud-associated Pop30 (Profitis, Greece) because of the mixed presence of the AB- and EC-group haplotypes in this population.
Estimates of Nei’s pairwise mean genetic distances among populations confirmed differentiation in relation to the host-plant (Fig 6B). The maximum pairwise difference was detected between the Ca-Ud and the Cf-associated populations (12–15) as well as between the Ca-Ud and Vac-associated populations (9–12). Within host-plants, pairwise genetic distances were 0.0–4.0 for the Ca-Ud associated populations, 0.0–1.2 for the Vac-associated populations in Montenegro and 0.0–2.1 in Greece (4–8 in between), and 0.0–1.0 for the Cf-associated populations. The within-population genetic distances were found for those associated with Cf, and were caused by the number and frequency of haplotypes (Fig 6B, orange elements).
In syntopy, i.e., co-occurrence of two host-plants harboring H. obsoletus (11 localities; Table 1), mtDNA haplotypes were always affiliated to the a priori defined host-plant association: AB or EC haplotype group with Ca or Ud, ZN or YM with Vac and JH associated with Cf. These findings unequivocally confirmed haplotype (lineage) specificity to host-plant.
Evidence of host-associated population differentiation: Microsatellite data
Bayesian clustering analysis performed with Structure based on the 50 H. obsoletus populations (702 individuals) from southeastern Europe and Turkey, and a single Vac-associated population from Israel (20 individuals) [7], supported the existence of three genetic groups (Evanno’s ΔK(3) = 803.72). The average genetic membership probabilities were > 90% and clustered as follows: the Ca- and Ud-associated individuals formed Cluster 1, Vac association formed Cluster 2, and the Cf-associated individuals formed Cluster 3 (Fig 7A), thus corroborating the mtDNA analysis results. The microsatellite analysis clustered the Israel Vac population with the Balkan Vac populations. This result contrasts the mtDNA analysis where these individuals cluster closer to the Cf metapopulation (Figs 4 and 5). However, the Israel population had a lower membership assignment of 75% to the Vac host-plant group in the overall analysis (Fig 7A). Estimates of molecular genetic variance (Amova) showed that 77% of the total genetic variance was host-plant affiliated (S6 Table). To elucidate the substructure of the three clusters, we analyzed each of them separately. Neither Cluster 1 (Ca and Ud, 29 population ΔK(2–28) = 0.06–11.14) nor Cluster 3 (Cf, 13 populations ΔK(2–12) = 0.18–5.32) showed signals of geographic differentiation, and for Ca and Ud there was no significant host-plant differentiation. In contrast, genetic variance in Cluster 2 (Vac) was divided between Montenegrin and the Greek metapopulations (approx. 400 km), geography explaining 60% of the total variance (S6 Table). The Structure analysis of all V. agnus-castus-related populations grouped Israel and Greek populations to the same subcluster (ΔK(2) = 80.45; Fig 7B) with 81–97% population membership assignment. An Amova of Montenegrin vs. Israel and Greek populations estimated 42% of the total genetic variance was caused by differentiation between the two clusters (S6 Table).
Fig 7. Bar plots of the Bayesian clustering analysis performed using Structure software on microsatellite data.
(A) 702 Hyalesthes obsoletus individuals from the 50 populations genotyped in this study and a single 20-member population associated with Vitex agnus-castus from Israel [7], suggesting a ΔK = 3 as the most likely number of genetic clusters; (B) 132 H. obsoletus individuals from the 8 populations associated with Vitex agnus-castus in Montenegro and Greece genotyped in this study and the aforementioned Israeli population [7], suggesting a ΔK = 2 as the most likely number of clusters; and (C) the ten syntopic localities of the H. obsoletus populations associated with two host-plants. Each column on the plots represents a single individual and the vertical black lines divide individuals by population. Colors represent proportional membership in each genetic cluster (green = Convolvulus arvensis and Urtica dioica, blue = Vitex agnus-castus, and red = Crepis foetida). Population membership assignments to the suggested clusters are designated above bar plots (A) and (B).
Among 11 syntopic sites harboring two host-associated H. obsoletus populations, ten were analyzed for genetic differentiation based on microsatellite data (Fig 7C). The population from C. arvensis in east Turkey (locality Erzincan, Pop10), syntopic with the C. foetida-associated population (Pop54), was removed from the analyses because the number of collected individuals was low (3 specimens). In three of the four analyzed syntopic Ca-Ud sites (Hamzali, Martinići and Strumica) no differentiation between host associations was found, agreeing with the general absence of the host-driven diversification signal for all Ca-Ud populations. However, in one Ca-Ud syntopic site (Podgorica), Evanno’s ΔK(2) = 76.94 suggested the existence of two clusters, hence showing a signal similar of host races as in southwest Germany (Fig 7C). At this site phylogeographic-related western AB-group haplotypes were found in the Ca-associated population (Pop 6) and eastern EC-group haplotypes were found in the Ud-association (Pop25). In all other syntopic localities (Ca/Cf, Ud/Cf or Ud/Vac), clear population-based host-plant affiliation were evident (population membership asignment 68–99%), but not for all individuals of the affiliated host-plant (Fig 7C). MtDNA analysis of these “wrongly assigned” individuals confirmed them as members of the source host-plant mtDNA haplogroup; hence, a genetic discrepancy was exhibited between mtDNA and microsatellite data (Figs 6 and 7).
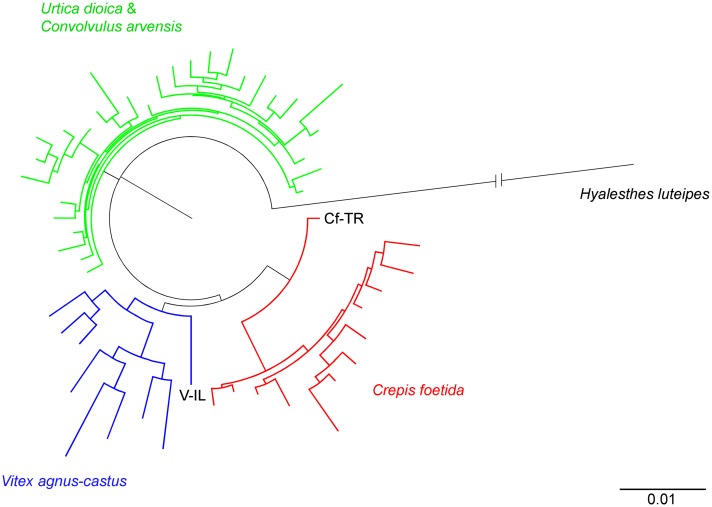
The maximum likelihood phenogram, based on 4 out of 7 microsatellite loci that amplified in the outgroup specimens of Hyalesthes luteipes, divided populations according to host-plant associated differentiation (Fig 8).
Fig 8. Maximum-likelihood phenogram based on allele frequencies calculated with four microsatellite loci for the 51 populations of H. obsoletus (722 individuals).
Individuals were associated with Convolvulus arvensis and Urtica dioica (green branches), Vitex agnus-castus (blue branches) and Crepis foetida (red branches) from Southeastern Europe and Turkey analyzed in this study and for the Israeli population associated with V. agnus-castus [7]. Hyalesthes luteipes was used as an outgroup to root the tree. The two most divergent populations of the Crepis foetida-associated genotype group from east Turkey (Cf-TR) and the Vitex agnus-castus-associated genotype group from Israel (V-IL) are designated.
Discussion
In recent years, several studies have documented specialized plant-associated life cycles of Hyalesthes obsoletus [7, 14, 15, 33, 38]. These observations lead Kosovac et al. [34] to hypothesize that H. obsoletus is a species complex undergoing cryptic speciation. In this paper, we analyzed genetic separation of four host-plant associated H. obsoletus populations in southeastern Europe, the ancestral distribution range of H. obsoletus. Our results clearly indicate three distinct genetic entities undergoing complex genetic diversification processes, and they question the taxonomic integrity of H. obsoletus sensu lato. Differentiation among three morphologically indistinguishable plant affiliated associations of H. obsoletus, 1) C. arvensis and U. dioica, 2) V. agnus-castus and 3) C. foetida show much higher segregation on an evolutionary time-scale compared with the previously identified C. arvensis and U. dioica host races in south Germany [7] or that suggested for Salvia sclarea- and Lavandula angustifolia-associated host-race formation in south France [11]. The following evidence for sibling species-level relatedness revealed in these host-plant relations are: i) clear and consistent mitochondrial genealogical divergence between host-associated populations in both syntopy and across the geographic distribution range (i.e. in allopatry); ii) nuclear genetic divergence supporting mitochondrial genealogy; and iii) the average genetic distance among mitochondrial haplotypes of host-associated populations (1.1–1.5%) is comparable to that of the most closely related morphologically separated species, i.e., Hyalesthes thracicus (2.1–3.3%).
Three cryptic genetic lineages deriving from four host-plant associations
This study was initially started as a survey to verify H. obsoletus’ affiliation with C. arvensis and U. dioica, the traditional host plants in southeastern Europe. Relations between these two associations were expected to be complex. Evidence for host races in the western European mtDNA sub-haplolineage (AB) was found in an area of recent immigration in Germany and northern France, but not in Western Europe south thereof [7, 15, 28]. The present study from southeastern Europe, where most Ca-Ud-associated individuals belong to the EC mtDNA sub-haplolineage also suggest a lack of host races associated with C. arvensis and U. dioica there. Combined data indicate an innate capability of H. obsoletus to utilize both plants in the ancestral distribution range. However, the finding of the core populations’ single syntopic locality with clear genetic differentiation on the host-plant level (Pop6 and 25, Podgorica, Montenegro), require further investigation. One reason for differentiation here may be phylogeographic bias rather than host-plant specialization. Individuals associated with the two plants belonged to the sub-lineages AB and EC, respectively. These sub-lineages are also genetically diverged for microsatellite loci [8].
The association with V. agnus-castus was described by Hoch and Remane [31] and need not have been a novelty for Europe. However, researchers have focused on preferences for crop weeds rather than its known wild-plant associations [31]. Prior to our study, the only recent data pointing to this plant association were from Israel, where it was experimentally confirmed as a true host [33]. In our study, we detected associations of H. obsoletus with V. agnus-castus in Montenegro and Greece (mainland and islands) and we believe that the association will be affirmed along the coast of the Adriatic Sea, as well as in other parts of the Mediterranean basin—such as the coastal regions of the Iberian, Apennine and Anatolian peninsula—as outlined in Hoch and Remane [31]. Recently, the V. agnus-castus association was confirmed in habitats surrounding vineyards along Bosnia and Herzegovina’s coastal area [89], while its occurrence in North Dalmatia (Croatia) was described in the late 19th century [32], but since forgotten. According to our data, H. obsoletus associated with V. agnus-castus on the Balkan Peninsula consists of two geographically separated genotype groups (Greek YM and Montenegrin ZN), which are linked genealogically by the haplotype ηM. The spatial separation between the groups is approximately 250 km along the coastline with suitable habitats for V. agnus-castus growth. Whether the two V. agnus-castus sub-lineages of H. obsoletus represent two subspecies separated by the Pindus mountain range as a barrier that prevents between-group interaction, or simply are phylogeographic variants, may be assessed by sampling in the intermediate range along the Albanian coastline.
One of the most interesting findings is a closer mtDNA genealogical relatedness of Israeli V. agnus-castus haplotypes to the C. foetida-associated haplo-group than to the Montenegrin and Greek haplotypes (Figs 4 and 5). Conversely, the Bayesian clustering analysis of nuclear data identified Greek and Israel V. agnus-castus specimens as monophyletic. Hence, a host shift and further diversification between populations associated with these two hosts has probably occurred in the area of Middle East; however the direction of the shift can only be speculated due to insufficient data. The host shift of a phytophagous insect to a new plant is a demanding, mainly because of the phytochemical barriers [90]. Populations that have undergone host shift could use ancestral and new host plants as a niche, but they might not be able to use them simultaneously, a phenomenon that can be detected by a host-plant associated fitness trade-offs on the ancestral host [2, 90, 91]. Therefore, it could be assumed the collection of specimens that are affiliated with one host-plant by preferred plant choice (e.g., V. agnus-castus), but that are linked to other association according to mtDNA genetic markers (i.e., C. foetida), as a relic from their host-shift past.
An unexpected result in this study was the genetic uniqueness and affiliation of H. obsoletus populations to C. foetida across a vast area ranging from east Turkey up to the Danube and Morava rivers on the Balkans (Fig 1). Although we have only few samples from Anatolia, some interesting and important findings have arisen because these specimens occur toward the eastern and southern borders of the H. obsoletus distribution. Güclü and Ozbek [92] reported the association with C. arvensis in Erzurum; we confirmed the association in 150-km distant Erzincan in eastern Turkey. The C. foetida association was encountered in Kırşehir and syntopically with C. arvensis in Erzincan. Much genetic specificity can be attributed to this host-plant association which indicates a detachment of this metapopulation from the rest of the H. obsoletus s.l. scope. A low number of private alleles indicates the tightness of this association and the low number of migrants [93] as well as the decrease in allelic richness of the European populations imply a drifting departure from the Middle Eastern ancestral area, contributing to the H. obsoletus hypothesized Levantine origin [15]. Meanwhile lower heterozygosity values, compared with other two associations, can be attributed to discrepancy forces such as inbreeding and genetic drift, probably due to westward range expansion. The eastward perimeter of the C. foetida-associated distribution is currently unknown, but according to available data, it expands to Erzincan in northeast Turkey, while westward we assume that Danube and Morava rivers are barriers toward central Europe, although a single population north of the Danube was recorded (Romania, locality 42). This association could have entered the European continent by crossing the Bosphorus and the Dardanelles or by following the Danube valley, like many other taxa that share this Balkan-Anatolian and Ponto-Panonian chorotype [94]. Haplotypes associated with C. foetida collected on crop plants in Radovanu (south Romania) and the presence of this association on both sides of the Danube in Bulgaria and Romania (Fig 1) support a Danube expansion rout.
Cryptic speciation in Hyalesthes obsoletus: A need for an integrative taxonomic approach
Due to its significance as a vector of plant pathogenic phytoplasma, H. obsoletus is the most well known representative of the south Palaearctic planthopper genus Hyalesthes. However, as the present study shows, its specific, plant-associated lineages moreover raise significant concern in the context of integrative taxonomy. This is further emphasized by fact that H. obsoletus is the type species, i.e., the name bearing type of the genus. Efforts made to locate H. obsoletus holotype specimen have failed [31], leading to the general conclusion that it is probably lost. However, according to the original description [95], the type specimen was collected in south France "France mérid. (Grenier), Chambéry (Cartereau)" as the type-locality, but a description of the host-plant is lacking. In addition, type material of a single H. obsoletus synonym, Liorhinus albolimbatus Kirchbaum, 1868 (synonymized by Fieber [96]) collected in Dalmatia is also probably lost [31]. Hence, attributing a type specimen to any plant association would be speculative, but knowing that all four wild host-plants are present in the vicinity of the type localities (http://ww2.bgbm.org/EuroPlusMed/query.asp) emphasizes a need for genetic analysis of this archival material (if located) and its mere importance for resolving H. obsoletus taxonomic and nomenclature status.
Hoch and Remane’s revision of genus Hyalesthes [31] was based on rich entomological collections. Cladistic analysis based on morphological characters revealed the existence of 33 species belonging to five monophyletic species groups [31, 36, 54, 97, 98]. The great morphological similarities among species within the same species group suggest a very shallow evolutionary history of these taxa. However, all within-group species can be reliably differentiated based on male genital armature [31], including H. obsoletus and its closest relative H. thracicus. These subtle phenotypic differences, and an average genetic distance of 2.6% (3.3% on COI-COII), suggest a recent common ancestor not older than the Quaternary glacial cycles [99–101]. The Mid-Atlantic Islands (Canaries and Madeira) and the east Mediterranean (Turkey-Anatolia) are two diversity centres of the genus Hyalesthes [54, 102]. Our genetic data indicate, in accordance with Hoch and Remane [31], an origin of H. obsoletus in the eastern Mediterranean. Micro-refugia as well as host-plant specialization likely expanded the distribution range of H. obsoletus westwards (Fig 5).
Haplotype divergence among the three host-plant associated H. obsoletus meta-populations was similar (ca. 2%) but morphological differences were not evident among them (A. Kosovac, J. Jović and I. Toševski, unpublished result). This lack of morphological difference may imply intense specialization to specific plant metabolites that rapidly separated the incipient species. Berlocher and Feder [17] argued for an incipient stage of sympatric speciation and host-associated species as the differentiation climax where populations are considered “operational species” when significant genetic, behavioral and morphological differences are present at two sympatric localities. Following the definition of sympatric speciation that implies the divergence of one evolutionary lineage into two in the absence of geographic isolation, we consider morphological delineation as the last step before true species status. In our study, we detected seven syntopic sites (two Ca/Cf, three Ud/Cf and two Ud/Vac) where plant-associated populations lacked genetic exchange. Hence, we view H. obsoletus as a species complex, for which cladogenesis is linked to morphological stasis [103]. The decoupling of genetic and morphological divergence is common for young cryptic species where morphological traits or other diagnosable features have not yet evolved [103, 104].
Following genetic and ecological (host-plant) differentiation of three entities within, what is up-to-date considered a single unique species under the name H. obsoletus, requires an integrative taxonomic approach. If the type material of H. obsoletus is considered lost, then the designation of a neotype, following the International code of Zoological nomenclature (ICZN), can lead to stabilization of the taxonomic and nomenclature ambiguity within the H. obsoletus species complex.
Influence of vectors’ divergence on stolbur phytoplasma epidemiology
Clarifying the true relationships of host associations of insect vectors is of particular epidemiological importance because these associations determine the breadth of pathogen transmission. For H. obsoletus, which acquires stolbur phytoplasma in the larval stage [24], knowledge of dual hosts is paramount. Disease cycles, which may or may not overlap, depend on the vectors’ plant preference [13, 14, 37]. Our results show that two opposing phenomena determine the epidemiology of stolbur phytoplasma vectored by H. obsoletus. First, the genetic homogeneity of H. obsoletus utilizing C. arvensis and U. dioica is opposed by two distinct plant associations of the pathogen, tuf-b and tuf-a, respectively [14]. Second, the V. agnus-castus and C. foetida cryptic species transmit only tuf-b [13, 38, 40]. Hence, epidemiological cycles are partly "un-coupled" relative to genetic diverged host-races or cryptic species and their host-plant associations. The multilocus genotypization of the pathogen strains in cross-transmission by naturally infected populations affiliated with C. arvensis, V. agnus-castus and C. foetida indicate the existence of intersections or meeting points in the epidemiological pathways of tuf-b strains associated with different host-plants and plant-specialized vector populations (A. Kosovac and J. Jović, unpublished data) [40]. However, considering the vector’s genetic structure it is more likely that when shared phytoplasma genotypes appear at meeting points in dual host-plants they are a consequence of feeding (i.e. transmission) by other vectors of minor or intermediate importance [9, 10, 13], than a result of H. obsoletus polyphagy. Although we do not exclude the possibility that H. obsoletus belonging to a specific association (e.g. C. arvensis) feeds on erroneous plants of another association (e.g. V. agnus-castus) within the same epidemiological cycle (tuf-b), this event is unlikely in many habitats of the species range due to differences in micro-geography, micro-climate and vegetation composition. Survival tests of genetic host-races on alternative plants [29] show that H. obsoletus is more dependent on the host-plant than was previously thought. Hence, host-plant preference, which is proven to be a crucially important trait for this insect, requires special attention in all further studies delimiting epidemiological cycles. Therefore, to verify that plants currently found in the literature are actual hosts for development, we encourage careful population sampling in the entire distribution range and molecular confirmation of larval presence on the plants’ roots to determine true host suitability and association. Only precise identification of dual hosts enables detecting pathogen transmission routes and epidemiological cycles for relevant management strategies, e.g., [13].
This thrilling evolutionary story of H. obsoletus occurs in natural ecosystems, unlike the majority of economically important insects, for which cryptic differentiation is usually attributed to survival and reproductive pressures in agro-ecosystems, e.g., [105–107]. The study’s insights into cryptic divergence of H. obsoletus support the notion that specialization and possibly cryptic species is more common than often appreciated in "generalist" insects [108]. It poses a challenge to taxonomic delimitation; erroneously delineating pest species has significant implications for success of short- or long-term control management.
Supporting information
(TXT)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors are grateful to Prof. Sanja Radonjić and Prof. Snježana Hrnčić (Biotechnical Faculty, University of Montenegro) for their help in collecting populations in Montenegro. We are indebted to Prof. Hannelore Hoch (Museum für Naturkunde, Berlin-Germany) for kindly providing us with paratype specimen of Hyalesthes thracicus. We owe special thanks to Dr. Tatyana Kastalyeva (All-Russian Research Institute of Phytopathology, Russian Agricultural Academy, ARRIP) for providing us hardly accessible publications in Russian.
Data Availability
DNA sequences are available in the GenBank database, accession numbers are listed in S3 Table. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by Ministry of Education, Science and Technological Development of the Republic of Serbia (III43001), Stiftung Rheinland-Pfalz für Innovation (15202-386261/861), and partly by the SCOPES program of the Swiss National Science Foundation (IZ73Z0_152414). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kabis MA, Khush GS. Genetic analysis of resistance to brown planthopper in rice (Oryza sativa L.). Plant Breeding. 1988; 100:54–58. [Google Scholar]
- 2.Via S. Ecological genetics and host adaptation in herbivorous insects: the experimental study of evolution in natural and agricultural systems. Annu Rev Entomol. 1990; 35:421–446. doi: 10.1146/annurev.en.35.010190.002225 [DOI] [PubMed] [Google Scholar]
- 3.McPheron BA. Population genetics and cryptic species In Tan K.-H., (Ed). Area wide control of fruit flies and other insect pests. Pernerbit Universiti sains Malaysia, Peneng; 2000; 483–490. [Google Scholar]
- 4.Hunter MD. Landscape structure, habitat fragmentation, and the ecology of insects. Agr Forest Entomol. 2002; 4:159–166. [Google Scholar]
- 5.Endersby NM, McKechnie SW, Ridland PM, Weeks AR. Microsatellites reveal a lack of structure in Australian populations of the diamondback moth, Plutella xylostella (L.). Mol Ecol. 2006; 15:107–118. doi: 10.1111/j.1365-294X.2005.02789.x [DOI] [PubMed] [Google Scholar]
- 6.Tsagkarakou A, Tsigenopoulos CS, Gorman K, Lagnel J, Bedford ID. Biotype status and genetic polymorphism of the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) in Greece: mitochondrial DNA and microsatellites. B Entomol Res. 2007; 97:29–40. [DOI] [PubMed] [Google Scholar]
- 7.Imo M, Maixner M, Johannesen J. Sympatric diversification vs. immigration: deciphering host-plant specialization in a polyphagous insect, the stolbur phytoplasma vector Hyalesthes obsoletus (Cixiidae). Mol Ecol. 2013; 22:2188–2203. doi: 10.1111/mec.12237 [DOI] [PubMed] [Google Scholar]
- 8.Johannesen J, Riedle-Bauer M. Origin of a sudden mass occurrence of the stolbur phytoplasma vector Hyalesthes obsoletus (Cixiidae) in Austria. Ann Appl Biol. 2014; 165(3):488–495. [Google Scholar]
- 9.Cvrković T, Jović J, Mitrović M, Krstić O, Toševski I. Experimental and molecular evidence of Reptalus panzeri as a natural vector of bois noir. Plant Pathol. 2014; 63(1):42–53. [Google Scholar]
- 10.Chuche J, Danet JL, Salar P, Foissac X, Thiéry D. Transmission of ‘Candidatus Phytoplasma solani’ by Reptalus quinquecostatus (Hemiptera: Cixiidae). Ann Appl Biol. 2016; 169(2):214–223. [Google Scholar]
- 11.Chuche J, Danet JL, Rivoal JB, Arricau-Bouvery N, Thiéry D. Minor cultures as hosts for vectors of extensive crop diseases: Does Salvia sclarea act as a pathogen and vector reservoir for lavender decline? J Pest Sci. 2018; 91(1):145–155. [Google Scholar]
- 12.Mitrović M, Jakovljević M, Jović J, Krstić O, Kosovac A, Trivellone V, et al. ‘Candidatus phytoplasma solani’ genotypes associated with potato stolbur in Serbia and the role of Hyalesthes obsoletus and Reptalus panzeri (hemiptera, cixiidae) as natural vectors. Eur J Plant Pathol. 2016; 144:619–630. [Google Scholar]
- 13.Kosovac A, Radonjić S, Hrnčić S, Krstić O, Toševski I, Jović J. Molecular tracing of the transmission routes of bois noir in Mediterranean vineyards of Montenegro and experimental evidence for the epidemiological role of Vitex agnus-castus (Lamiaceae) and associated Hyalesthes obsoletus (Cixiidae). Plant Pathol. 2016; 65:285–298. [Google Scholar]
- 14.Langer M, Maixner M. Molecular characterisation of grapevine yellows associated phytoplasmas of the stolbur group based on RFLP analysis of non-ribosomal DNA. Vitis. 2004; 43:191–199. [Google Scholar]
- 15.Johannesen J, Lux B, Michel K, Seitz A, Maixner M. Invasion biology and host specificity of the grapevine yellows disease vector Hyalesthes obsoletus in Europe. Entomol Exp Appl. 2008; 126:217–227. [Google Scholar]
- 16.Mullen SP, Shaw KL. Insect Speciation Rules: Unifying Concepts in Speciation Research. Annu Rev Entomol. 2014; 59:339–361. doi: 10.1146/annurev-ento-120710-100621 [DOI] [PubMed] [Google Scholar]
- 17.Berlocher SH, Feder JL. Sympatric speciation in phytophagous insects: Moving beyond controversy? Annu Rev Entomol. 2002; 47:773–815. doi: 10.1146/annurev.ento.47.091201.145312 [DOI] [PubMed] [Google Scholar]
- 18.Drès M, Mallet J. Host races in plant–feeding insects and their importance in sympatric speciation. Philos T Roy Soc B. 2002; 357(1420):471–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quaglino F, Zhao Y, Casati P, Bulgari D, Bianco PA, Wei W, et al. ‘Candidatus Phytoplasma solani’, a novel taxon associated with stolbur-and bois noir-related diseases of plants. Int J Syst Evol Micr. 2013; 63(8):2879–2894. [DOI] [PubMed] [Google Scholar]
- 20.Suhov KS, Vovk AM. Stolbur of solanaceous plants. USSR Academy of Sciences Publishers; 1949; 7–11. [Google Scholar]
- 21.Suhov KS, Razvyazkina MG. Biology of viruses and virus diseases of plants. Moscow: Soviet Science; 1955; 143–153. [Google Scholar]
- 22.Aleksić Ž, Šutić D, Aleksić D. Transmission intensity of stolbur virus by means of Hyalesthes obsoletus Sign. on some host plants. Plant Protection. 1967; 93–95:67–73. [Google Scholar]
- 23.Maixner M. Transmission of German grapevine yellows (Vergilbungskrankheit) by the planthopper Hyalesthes obsoletus (Auchenorrhyncha: Cixiidae). Vitis. 1994; 33:103–104. [Google Scholar]
- 24.Sforza R, Clair D, Daire X, Larrue J, Boudon-Padieu E. The role of Hyalesthes obsoletus (Hemiptera: Cixiidae) in the occurrence of bois noir of grapevines in France. J Phytopathol. 1998; 146:549–556. [Google Scholar]
- 25.Aryan A, Brader G, Mörtel J, Pastar M, Riedle-Bauer M. An abundant 'Candidatus Phytoplasma solani’ Stolbur tuf b phytoplasma strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus. Eur J Plant Pathol. 2014; 140:213–227. doi: 10.1007/s10658-014-0455-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kessler S, Schaerer S, Delabays N, Turlings TC, Trivellone V, Kehrli P. Host plant preferences of Hyalesthes obsoletus, the vector of the grapevine yellows disease ‘bois noir’, in Switzerland. Entomol Exp Appl. 2011; 139:60–67. [Google Scholar]
- 27.Landi L, Riolo P, Murolo S, Romanazzi G, Nardi S, Isidoro N. Genetic variability of stolbur phytoplasma in Hyalesthes obsoletus (Hemiptera: Cixiidae) and its main host plants in vineyard agroecosystems. J Econ Entomol. 2015; 108:1506–1515. doi: 10.1093/jee/tov103 [DOI] [PubMed] [Google Scholar]
- 28.Maniyar B, Kehrli P, Johannesen J. Population structure and incidence of the stolbur phytoplasma vector Hyalesthes obsoletus (Cixiidae) among geographic regions in Switzerland. J Appl Entomol. 2013; 137(8):589–600. [Google Scholar]
- 29.Maixner M, Albert A, Johannesen J. Survival relative to new and ancestral host plants, phytoplasma infection, and genetic constitution in host races of a polyphagous insect disease vector. Ecol Evol. 2014; 4(15):3082–3092. doi: 10.1002/ece3.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atanasova B, Jakovljević M, Spasov D, Jović J, Mitrović M, Toševski I, et al. The molecular epidemiology of bois noir grapevine yellows caused by ‘Candidatus Phytoplasma solani’ in the Republic of Macedonia. Eur J Plant Pathol. 2015; 142:759–770. [Google Scholar]
- 31.Hoch H, Remane R. Evolution und Speziation der Zikaden-Gattung Hyalesthes SIGNORET, 1865 (Homoptera Auchenorrhyncha Fulgoroidea Cixiidae). Marburger Entomologische Publikationen. 1985; 2:1–427. [Google Scholar]
- 32.Horváth G. Hemipterološki izlet u Primorje i Plitvička jezera. Glasnik Hrvatskoga Naravoslovnoga Društva (Societas historico-naturalis Croatica). 1891; VI(1–5):29–49. [Google Scholar]
- 33.Sharon R, Soroker V, Wesley SD, Zahavi T, Harari A, Weintraub PG. Vitex agnus-castus is a preferred host plant for Hyalesthes obsoletus. J Chem Ecol. 2005; 31:1051–1063. [DOI] [PubMed] [Google Scholar]
- 34.Kosovac A, Johannesen J, Krstić O, Mitrović M, Cvrković T, Toševski I, et al. Is Hyalesthes obsoletus a species complex undergoing cryptic speciation? More evidence of host-associated genetic differentiation in Southeast Europe. Mitt Klosterneuburg. 2016; 66:24–25. [Google Scholar]
- 35.Orenstein S, Zahavi T, Nestel D, Sharon R, Barkalifa M, Weintraub PG. Spatial dispersion patterns of potential leafhopper and planthopper (Homoptera) vectors of phytoplasma in wine vineyards. Ann Appl Biol. 2003; 142:341–348. [Google Scholar]
- 36.Dlabola J. Ergänzungen zur iranischen, israelischen und benachbarten Zikadenfaunen mit Beschreibungen 30 neuer Taxone (Homoptera, Auchenorrhyncha). Acta Mus Nat Pragae. 1994; 49B (1993) 1–4: 41–110. [Google Scholar]
- 37.Johannesen J, Foissac X, Kehrli P, Maixner M. Impact of vector dispersal and host-plant fidelity on the dissemination of an emerging plant pathogen. PLoS One. 2012; 7:e51809 doi: 10.1371/journal.pone.0051809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosovac A, Johannesen J, Krstić O, Mitrović M, Cvrković T, Maixner M, et al. Microsatellite and mtDNA evidence of genetic differentiation in Hyalesthes obsoletus populations associated with a new major host, stinking hawk’s-beard (Crepis foetida), in southeast Europe. Proceedings of the 3rd European Bois Noir Workshop, Barcelona. 2013; 18–19.
- 39.Maixner M, Langer M. Prediction of the flight of Hyalesthes obsoletus, vector of stolbur phytoplasma, using temperature sums. IOBC/WPRS Bulletin. 2006; 29:161.–. [Google Scholar]
- 40.Kosovac A, Krstić O, Jakovljević M, Cvrković T, Mitrović M, Toševski I, et al. Elucidation of 'Candidatus phytoplasma solani’ epidemiology through trac(k)ing transmission pathways using field, experimental and molecular data. Mitt Klosterneuburg. 2016; 66:9–11. [Google Scholar]
- 41.Scheffer SJ, Wiegmann BM. Molecular phylogenetics of the holly leafminers (Diptera: Agromyzidae: Phytomyza): species limits, speciation, and dietary specialization. Molecular Phylogenet Evol. 2000; 17:244–255. [DOI] [PubMed] [Google Scholar]
- 42.Toševski I, Caldara R, Jović J, Hernández-Vera G, Baviera C, Gassmann A, et al. Morphological, molecular and biological evidence reveal two cryptic species in Mecinus janthinus Germar (Coleoptera, Curculionidae), a successful biological control agent of Dalmatian toadflax, Linaria dalmatica (Lamiales, Plantaginaceae). Syst Entomol. 2011; 36(4):741–753. [Google Scholar]
- 43.Toševski I, Caldara R, Jović J, Hernández-Vera G, Baviera C, Gassmann A, et al. Host-associated genetic divergence and taxonomy in the Rhinusa pilosa Gyllenhal species complex: an integrative approach. Syst Entomol. 2015; 40:268–287. [Google Scholar]
- 44.Wilkins MR, Seddon N, Safran RJ. Evolutionary divergence in acoustic signals: causes and consequences. Trends Ecol Evol. 2013; 28:156–166. doi: 10.1016/j.tree.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 45.Cargnus E, Pavan F, Mori N, Martini M. Identification and phenology of Hyalesthes obsoletus (Hemiptera: Auchenorrhyncha: Cixiidae) nymphal instars. B Entomol Res. 2012; 102:504–514. [DOI] [PubMed] [Google Scholar]
- 46.Rees DJ, Emerson BC, Oromi P, Hewitt GM. Mitochondrial DNA, ecology and morphology: interpreting the phylogeography of the Nesotes (Coleoptera: Tenebrionidae) of Gran Canaria (Canary Islands). Mol Ecol. 2001; 10:427–434. [DOI] [PubMed] [Google Scholar]
- 47.Mahuku GS. A simple extraction method suitable for PCR-based analysis of plant, fungal, and bacterial DNA. Plant Mol Biol Rep. 2004; 22:71–81. [Google Scholar]
- 48.Brown JM, Abrahamson JG, Way PA. Mitochondrial DNA phylogeography of host-races of the goldenrod ball gallmaker, Eurosta solidaginis (Diptera: Tephritidae). Evolution. 1996; 50:777–786. doi: 10.1111/j.1558-5646.1996.tb03887.x [DOI] [PubMed] [Google Scholar]
- 49.Hedin MC. Molecular phylogenetics at the population/species interface in cave spiders of the southern Appalachians (Araneae: Nesticidae: Nesticus). Mol Biol Evol. 1997; 14:309–324. doi: 10.1093/oxfordjournals.molbev.a025766 [DOI] [PubMed] [Google Scholar]
- 50.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song H, Buhay JE, Whiting MF, Crandall KA. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA. 2008; 105:3486–13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calvignac S, Konecny L, Malard F, Douady CJ. Preventing the pollution of mitochondrial datasets with nuclear mitochondrial paralogs (numts). Mitochondrion. 2011; 11:246–254. doi: 10.1016/j.mito.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 54.Hoch H. Acht neue Arten der Gattung Hyalesthes Signoret, 1865 (Homoptera Fulgoroidea Cixiidae) aus dem östlichen Mittelmeergebiet. Marburger Entomologische Publicationen. 1986; 2:87–122. [Google Scholar]
- 55.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984; 38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- 56.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010; 10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 57.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983; 105:437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989; 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997; 147:915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holsinger K. Tajima’s D, Fu’s Fs, Fay and Wu’s H, and Zeng E et al.’s. In: Holsinger K (ed) Lecture notes in population genetics. 2012; 239–244.
- 61.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979; 76:5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992; 132:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and other methods). Sinauer Associates: Sunderland, MA, USA: 2002. [Google Scholar]
- 64.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest2: more models, new heuristics and parallel computing. Nat Methods. 2012; 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huelsenbeck JP, Ronquist F. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 2001; 17:754–755. [DOI] [PubMed] [Google Scholar]
- 66.Ronquist F, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003; 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- 67.Rambaut A, Drummond AJ. Tracer: MCMC Trace Analysis Tool Version v1.5.0. 2003–2009. http://beast.bio.ed.ac.uk/
- 68.Rambaut A. FigTree version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree/. 2012.
- 69.Hebert PD, Cywinska A, Ball SL. Biological identifications through DNA barcodes. P Roy Soc Lond B Bio. 2003; 270(1512):313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertin S, Picciau L, Ács Z, Alma A, Bosco D. Molecular identification of the Hyalesthes species (Hemiptera: Cixiidae) occurring in vineyard agroecosystems. Ann Appl Biol. 2010; 157:435–445. [Google Scholar]
- 71.Wang G, Li C, Zheng W, Song F, Guo X, Wu Z, et al. An evaluation of the suitability of COI and COII gene variation for reconstructing the phylogeny of, and identifying cryptic species in, anopheline mosquitoes (Diptera Culicidae). Mitochond DNA Part A. 2017; 28(5):769–777. [DOI] [PubMed] [Google Scholar]
- 72.Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Syst Biol. 2001; 50(4):580–601. [PubMed] [Google Scholar]
- 73.Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000; 9:1657–1660. [DOI] [PubMed] [Google Scholar]
- 74.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992; 132:619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999; 16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036 [DOI] [PubMed] [Google Scholar]
- 76.Imo M, Lüneburg J, Hankeln T, Seitz A, Johannesen J. Highly polymorphic di-and trinucleotide microsatellite markers for the grapevine yellows disease vector Hyalesthes obsoletus (Auchenorrhyncha: Cixiidae). Eur J Entomol. 2011; 108(1):161. [Google Scholar]
- 77.Chapuis MP, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2007; 24:621–631. doi: 10.1093/molbev/msl191 [DOI] [PubMed] [Google Scholar]
- 78.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J Roy Stat Soc B Met. 1977; 1–38. [Google Scholar]
- 79.Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004; 4:535–538. [Google Scholar]
- 80.Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995; 86:248–249. [Google Scholar]
- 81.Rice WR. Analyzing tables of statistical tests. Evolution. 1989; 43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x [DOI] [PubMed] [Google Scholar]
- 82.Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered. 1995; 86:485–486. [Google Scholar]
- 83.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003; 164:1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Earl DA, von Holdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012; 4:359–361. [Google Scholar]
- 85.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005; 14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 86.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.69. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle, WA. 2009.
- 87.Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol. 2008; 23:347–351. doi: 10.1016/j.tree.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 88.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985; 22:160–174. [DOI] [PubMed] [Google Scholar]
- 89.Đurić Z, Hrnčić S, Delić D. Morphological and molecular identification of Hyalesthes obsoletus Signoret (Auchenorrhyncha: Cixiidae) in Herzegovina vineyards. Mitt. Klosterneuburg. 2017; 67:177–181. [Google Scholar]
- 90.Bernays EA, Chapman RF. Host-plant selection by phytophagous insects (Vol. 2). Springer Science & Business Media; 2007. [Google Scholar]
- 91.Diegisser T, Tritsch C, Seitz A, Johannesen J. Infestation of a novel host plant by Tephritisconura (Diptera: Tephritidae) in northern Britain: host-range expansion or host shift? Genetica. 2009; 137:87–97. doi: 10.1007/s10709-009-9353-3 [DOI] [PubMed] [Google Scholar]
- 92.Güclü S, Ozbek H. Some studies on the biology of Hyalestes obsoletus Signoret (Homoptera: Cixidae) in the conditions of Erzurum. Türkiye Entomoloji Dergisi. 1988; 12:103–111. [Google Scholar]
- 93.Szpiech ZA, Rosenberg NA. On the size distribution of private microsatellite alleles. Theor Popul Biol. 2011; 80:100–113. doi: 10.1016/j.tpb.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taglianti AV, Audisio PA, Biondi M, Bologna MA, Carpaneto GM, De Biase A, et al. A proposal for a chorotype classification of the Near East fauna, in the framework of the Western Palearctic region. Biogeographia. 1999; 20:31–59. [Google Scholar]
- 95.Signoret V. Descriptions de quelques hémiptères nouveaux. Ann Soc Entomol Fr. 1865; 115–130. [Google Scholar]
- 96.Fieber FX. Berichtigungen zu Dr. Kirschbaum’s Cicaden der Gegend von Wiesbaden, Frankfurt a. M. und anderer Gegenden. Verhandlungen der Kaiserlich-Königlichen Zoologisch-botanischen Gesellschaft in Wien. 1872; 22:27–34. [Google Scholar]
- 97.Remane R, Hoch H. Sechs neue Arten der Gattung Hyalesthes Signoret, 1865 (Homoptera Fulgoroidea Cixiidae) von den Mittelatlantischen Inseln urid aus dem Irak. Marburger Entomologische Publicationen. 1986; 2:123–154. [Google Scholar]
- 98.Emeljanov AF. The new taxa of the tribe Pentastirini (Homoptera, Cixiidae) from Palaearctic. Zoologicheskii Zhurnal. 1995; 74(9):73–89. [Google Scholar]
- 99.Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 2004; 101:14812–14817. doi: 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hewitt GM. Some genetic consequences of ice ages and their role in divergence and speciation. Biol J Linn Soc. 1996; 58:247–276. [Google Scholar]
- 101.Hewitt GM. Post-glacial re-colonization of European biota. Biol J Linn Soc. 1999; 68:87–112. [Google Scholar]
- 102.Hoch H. Patterns of geographic distribution in the planthopper genus Hyalesthes Sign. (Homoptera: Fulgoroidea Cixiidae): a phylogenetic approach. Proceeding 2nd International congress concerning the Rhynchota fauna of Balkan and adjacent regions, Mikrolimni, Greece. 1986; 31–32.
- 103.Bickford D, Lohman DJ, Sodhi NS, Ng PK, Meier R, Winker K, et al. Cryptic species as a window on diversity and conservation. Trends in Ecol Evol. 2007; 22:148–55. [DOI] [PubMed] [Google Scholar]
- 104.Saez AG, Lozano E. Body doubles. Nature. 2005; 433:111 doi: 10.1038/433111a [DOI] [PubMed] [Google Scholar]
- 105.Brunner PC, Chatzivassiliou EK, Katis NI, Frey JE. Host-associated genetic differentiation in Thrips tabaci (Insecta; Thysanoptera), as determined from mtDNA sequence data. Heredity. 2004; 93(4):364–370. doi: 10.1038/sj.hdy.6800512 [DOI] [PubMed] [Google Scholar]
- 106.De Barro P, Ahmed MZ. Genetic networking of the Bemisia tabaci cryptic species complex reveals pattern of biological invasions. PLoS One. 2011; 6(10):e25579 doi: 10.1371/journal.pone.0025579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Horowitz AR, Ishaaya I. Dynamics of biotypes B and Q of the whitefly Bemisia tabaci and its impact on insecticide resistance. Pest Manag Sci. 2014; 70(10):1568–1572. doi: 10.1002/ps.3752 [DOI] [PubMed] [Google Scholar]
- 108.Loxdale HD, Lushai G, Harvey JA. The evolutionary improbability of ‘generalism’ in nature, with special reference to insects. Biol J Linn Soc. 2011; 103(1):1–18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TXT)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
DNA sequences are available in the GenBank database, accession numbers are listed in S3 Table. All other relevant data are within the paper and its Supporting Information files.