Abstract
Background: Elucidating aging-related transcriptomic changes in human organs is necessary to understand the aging physiology and mechanisms, but little is known regarding the thyroid gland. We investigated aging-related transcriptomic alterations in the human thyroid gland and characterized the related molecular functions.
Methods: Publicly available RNA sequencing data of 322 thyroid tissue samples from the Genotype-Tissue Expression project were analyzed. In addition, our own 64 RNA sequencing data of normal thyroid tissue samples were used as a validation set. To comprehensively evaluate the associations between aging and transcriptomic changes, we performed a weighted gene coexpression network analysis and pathway enrichment analysis. The thyroid differentiation score was then used for further analysis, defining the correlations between thyroid differentiation and aging.
Results: The most significant aging-related transcriptomic change in thyroid was the downregulation of genes related to the mitochondrial and proteasomal functions (p = 3 × 10−6). Moreover, genes that are associated with immune processes were significantly upregulated with age (p = 3 × 10−4), and all of them overlapped with the upregulated genes in the thyroid glands affected by lymphocytic thyroiditis. Furthermore, these aging-related changes were not significantly different according to sex, but in terms of the thyroid differentiation, females were more susceptible to aging-related changes (p for trend = 0.03).
Conclusions: Aging-related transcriptomic changes in the thyroid gland were associated with mitochondrial and proteasomal dysfunction, loss of differentiation, and activation of autoimmune processes. Our results provide clues to better understanding the age-related decline in thyroid function and higher susceptibility to autoimmune thyroid disease.
Keywords: : thyroid gland, aging, RNA sequencing, Genotype-Tissue Expression project, mitochondria, autoimmunity
Introduction
In recent years, life expectancy has increased, resulting in more attention to the physiological changes and the related mechanisms of aging in order to find ways to enhance health in older age. Aging is a gradual and permanent, though not a pathological process. The hallmarks of aging include genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, stem cell exhaustion, altered intercellular communication, cellular senescence, and mitochondrial dysfunction (1).
Recently, Yang et al. catalogued age-related gene expression in nine human tissues using RNA sequencing (RNA-seq) data from the Genotype-Tissue Expression (GTEx) project including 105 thyroid tissue samples (2). In this study, multiple organs including lung and heart synchronously displayed mitochondrial dysfunction with aging, but the investigators did not find any functional alterations regarding aging in the thyroid. They analyzed 41,298 genes but only found three genes—PTCHD4, ZMAT3, and SP7—that had a relation with aging in the thyroid gland.
However, similar to other organs in the human body, the thyroid gland also displays morphological and physiological changes as an individual's age advances. A variety of morphological and physiological changes in the thyroid gland occur as part of the natural aging process. Histologic changes in the aging thyroid gland include increased interfollicular fibrosis, reduction in the size of follicles and their colloid content, flattening of the glandular epithelial cells, and decreased total weight of the thyroid gland (3). Aging-related physiological changes in thyroid function have been extensively investigated in a number of large studies (4–8). These studies demonstrated that the prevalence of subclinical thyroid dysfunction and levels of serum thyroid peroxidase and thyroglobulin antibodies rise with age. However, no study has evaluated changes in the gene expression profile of the human thyroid associated with aging.
The GTEx project is a well-established database for the scientific community to study the relationship between genetic variation and gene expression in various types of human tissues (9,10). The GTEx database (version [v] 6 release) has information on 8555 postmortem samples across 53 different types of human tissues from 544 individuals. It holds large-scale genomic and transcriptomic data obtained from using various types of next-generation sequencing methods. Among them, RNA-seq data is provided as ready-to-use file. When compared with the former version, GTEx v3 release, the v6 release database is greatly expanded and more informative. The total number of RNA-seq samples of various human tissues has been increased from 1641 to 8555. In particular, the number of thyroid tissue samples increased from 105 to 322 (age range 20–79 years). Therefore, these transcriptome data may be helpful for understanding the physiology of the aging thyroid gland, if they are analyzed adequately (11,12).
In this study, with weighted gene coexpression network analysis (WGCNA), we elucidated aging-related transcriptomic changes in human thyroid tissue using the public 322 RNA-seq data from the GTEx project and our own 64 RNA-seq data for the validation (13). We believe that the different analytic approach with a bigger dataset in our study compared with the study by Yang et al. (2) allows to expand the understanding on gene expression changes with aging in thyroid tissue, hence providing new insights into the mechanisms of aging-related thyroid diseases, which may have relevance for therapeutic strategies.
Materials and Methods
Subjects
The GTEx database (v6 release) provided 322 RNA-seq samples of normal thyroid glands from subjects in various age groups, which enabled us to study associations between changes in gene expression in thyroid tissue and aging (9,10). The age distribution and the frequency of lymphocytic thyroiditis (LT) are presented in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/thy). The frequency of LT was slightly higher in samples from female subjects, but there was no difference between age groups (9.82% vs. 5.24% in females and males, respectively; p = 0.82). Sixty-four LT-negative normal thyroid tissues from our previous research were also used in this study (13). These tissue samples consisted of normal tissue removed at surgery for well-differentiated thyroid carcinomas.
Weighted gene coexpression network analysis
WGCNA was used to identify associations of gene expression changes in thyroid gland tissue with aging or autoimmunity (14). In this analysis, pairwise correlations between the expression values of each gene were used to construct modules which denote the coexpression network. It is established by an undirected connection between each gene with significant relationships. The module eigengene is characterized as the first principal component of each module. It reflects the most illustrative gene expression of each sample in a module. This WGCNA approach has many advantages when compared with traditional differential expression analysis, including fine focus on coexpression gene patterns which allows precise identification of biologically meaningful modules containing related genes.
To identify the relationship between the modules and the clinical characteristics, the correlations between module eigengene and clinical traits, such as age, were determined. Moreover, WGCNA also computes correlation between gene and clinical traits. Aforementioned correlation tests were performed by determining Pearson correlation coefficients. The variance stabilizing transformed (VST) expression values from DESeq2 (v1.14.1) were used for the analysis according to the developer's instructions (15,16). DESeq2 is a type of R package for differentially expressed gene analysis using RNA-seq data. This method can be utilized not only for differentially expressed gene analysis, but also transforms raw expression level to log2 scale normalized values according to library size for subsequent analysis. Here, the VST values are used to eliminate the dependence of the variance on the mean, especially the high variance of the raw count data when the mean is low.
When we constructed the coexpression gene networks, according to the scale-free topology criterion, the optimal power was selected. The scale-free networks display highly heterogeneous features and their topologies are determined by a few greatly linked nodes (hubs) which connect the rest of the less linked nodes to the system (17).
Pathway enrichment analysis
We performed pathway enrichment analysis, which computes biologically meaningful pathways using overlaps of genes of interest with specific gene sets or pathways using the Molecular Signatures Database (MSigDB) version 6.0, a collection of annotated gene sets, subjecting genes involved in aging- and LT-related modules (18). Two databases from MSigDB were selected for the analysis: gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (19,20). The GO database includes three domains: cellular component, molecular function, and biological process. The GO cellular components describe an actual component of a cell, such as nucleus, or a group of gene products, such as ribosomes. The GO molecular function illustrates activities that take place at the molecular level, such as binding activity. The GO biological process defines sequential events achieved by one or more molecular functions, for example signal transduction. KEGG is a valuable source for understanding biological systems, organisms, and even the ecosystem. The database is constructed by information from genome sequencing and other high-throughput experiment results.
Mitochondrial copy number estimation
Eleven mitochondrial genes, which have average mapped reads higher than 100 (MT-RNR1, MT-RNR2, MT-ND1, MT-ND2, MT-CO1, MT-TS1, MT-CO2, MT-ND3, MT-ND5, MT-ND6, and MT-CYB) were subjected to the analysis. The average VST expression values across 11 mitochondrial RNAs (mtRNAs) were used to estimate mitochondrial copy number.
Thyroid differentiation score analysis
The Cancer Genome Atlas (TCGA) is a collaboration between the National Cancer Institute and the National Human Genome Research Institute to conduct comprehensive genomic research on 33 types of cancer. As described in TCGA study, we calculated the thyroid differentiation score (TDS) using 16 genes that have functions in thyroid metabolism: DIO1, DIO2, DUOX1, DUOX2, FOXE1, GLIS3, NKX2-1, PAX8, SLC26A4, SLC5A5, SLC5A8, TG, THRA, THRB, TSHR, and TPO (21). The VST expression values from DESeq2 were first centered at the median across all samples (15,16). Then, the TDS was determined as the average of the 16 genes in each sample:
TDS = Mean of median VST expression across 16 genes.
The association between aging and TDS were measured by one-way ANOVA and test for linearity using IBM SPSS Statistics Version 23.0.
Data visualization
We generated a chord diagram using NetworkAnalyst (22).
Results
WGCNA and the functional annotation of aging-related changes in the thyroid gland
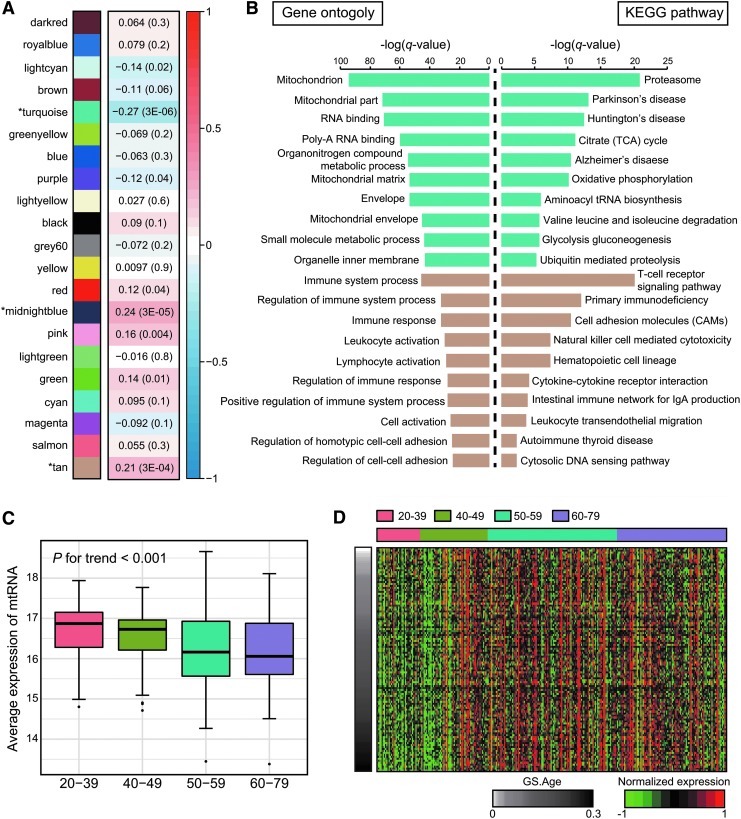
To identify significant gene expression changes in the thyroid gland associated with age, WGCNA was performed with 28,566 genes, excluding 22 LT-positive samples to eliminate the potential effects of autoimmune activity. We identified 21 modules with coexpressed genes (Fig. 1A), and the module with the maximal number of genes is shown in turquoise (n = 914) and the one with the minimal number of genes is shown in tan (n = 114). Each module from WGCNA analysis was subjected to pathway enrichment analysis. Within the 21 modules, eight of them exhibited significant correlations with age (p < 0.05) and we focused on the top three most significant modules for the main analysis, shown in turquoise (r = -0.27 and p = 3 × 10−6), midnight blue (r = 0.24 and p = 3 × 10−5), and tan (r = 0.21 and p = 3 × 10−4; Fig. 1A). The gene lists of the three aging-related modules are provided in Supplementary Table S2.
FIG. 1.
Aging-related weighted gene coexpression analysis of the thyroid gland. (A) Relationships between constructed modules and age. The Pearson correlation coefficient and the correlation test p-value (in brackets) between age and the module eigengene are presented. (B) The results of pathway enrichment analysis of two modules using gene ontology and KEGG pathway. (C) The significant association between average expression of mtRNA and age. (D) Heatmap representing the gene expression pattern of the tan module. KEGG, Kyoto Encyclopedia of Genes and Genomes; mtRNA, mitochondrial RNA.
Pathway enrichment analysis was then used to clarify the relationship between the modules and the pathways contained in GO and KEGG. Based on this analysis, the functional pathways that had the strongest associations with aging-related thyroid changes were identified (Fig. 1B). Among the top three most correlated modules, the GO or KEGG pathways were enriched in the turquoise or tan modules, while the second most significantly correlated module, midnightblue, did not exhibit any particular enrichment result. Moreover, we show all pathway enrichment results using every module, except for the top three most correlated modules, in Supplementary Figure S1.
Associations between mitochondrial and proteasomal dysfunction of the thyroid and aging
There were 662 downregulated and 254 upregulated genes discovered in the turquoise module, which was most significantly and negatively correlated with aging (r = −0.27 and p = 3 × 10−6) based on the WGCNA results. When the downregulated and the upregulated genes were subjected to pathway analysis separately, only downregulated genes showed enriched pathway results. They were enriched with mitochondrial functions, mitochondrial components, and metabolic processes by GO annotation, indicating that mitochondrial dysfunction is strongly associated with aging (Fig. 1B). The analysis performed using the KEGG pathway analysis further showed that the downregulated genes in the turquoise module were enriched in pathways of the proteasome and ubiquitin-mediated proteolysis, as well as those of mitochondrial function-related pathways including the tricarboxylic acid cycle and oxidative phosphorylation (OXPHOS). Table 1 shows individual genes involved in the tricarboxylic acid cycle or OXPHOS pathways associated with the turquoise module. Especially in case of OXPHOS, which comprises five complexes with many components, with the action taking place inside the mitochondria, all genes except for those involved in complex 4 were included in the turquoise module.
Table 1.
Genes Involved in Mitochondrial Function from the Aging-Related Turquoise Module
| KEGG pathway | Gene symbol | GSa.Age | pb.GS.Age | MMc.turquoise | p.MM.turquoise |
|---|---|---|---|---|---|
| TCA cycle | DLD | −0.26 | 3.69 × 10−6 | 0.90 | 9.36 × 10−110 |
| PDHB | −0.21 | 2.05 × 10−4 | 0.79 | 1.64 × 10−64 | |
| DLAT | −0.20 | 4.16 × 10−4 | 0.79 | 2.37 × 10−65 | |
| SDHC | −0.20 | 0.001 | 0.86 | 9.23 × 10−88 | |
| OGDH | −0.18 | 0.002 | 0.71 | 5.85 × 10−47 | |
| FH | −0.16 | 0.005 | 0.87 | 5.69 × 10−94 | |
| IDH2 | −0.15 | 0.008 | 0.52 | 5.38 × 10−22 | |
| SDHA | −0.14 | 0.019 | 0.60 | 3.03 × 10−30 | |
| MDH1 | −0.13 | 0.021 | 0.76 | 1.13 × 10−57 | |
| ACO2 | −0.11 | 0.066 | 0.69 | 2.27 × 10−44 | |
| PDHA1 | −0.08 | 0.150 | 0.57 | 1.88 × 10−27 | |
| OXPHOS | FADD | −0.30 | 8.13 × 10−8 | 0.67 | 9.84 × 10−41 |
| GSK3B | −0.28 | 6.55 × 10−7 | 0.69 | 3.64 × 10−43 | |
| NDUFS1 | −0.21 | 2.95 × 10−4 | 0.87 | 1.64 × 10−92 | |
| SDHC | −0.20 | 0.001 | 0.86 | 9.23 × 10−88 | |
| NDUFV3 | −0.19 | 0.001 | 0.64 | 1.56 × 10−36 | |
| ATP5A1 | −0.19 | 0.001 | 0.84 | 6.39 × 10−80 | |
| UQCRFS1 | −0.17 | 0.003 | 0.89 | 3.20 × 10−103 | |
| NDUFA9 | −0.14 | 0.017 | 0.79 | 1.05 × 10−65 | |
| SDHA | −0.14 | 0.019 | 0.60 | 3.03 × 10−30 | |
| ATP5F1 | −0.14 | 0.020 | 0.79 | 4.65 × 10−64 | |
| MAPK1 | −0.11 | 0.054 | 0.71 | 3.25 × 10−47 | |
| UQCRC2 | −0.11 | 0.060 | 0.69 | 1.63 × 10−43 | |
| NDUFB5 | −0.11 | 0.062 | 0.52 | 1.07 × 10−21 | |
| ATP5C1 | −0.11 | 0.071 | 0.76 | 3.48 × 10−57 | |
| ATP5B | −0.10 | 0.074 | 0.82 | 5.72 × 10−74 | |
| IDE | −0.10 | 0.082 | 0.68 | 1.35 × 10−41 | |
| EIF2AK3 | −0.08 | 0.155 | 0.61 | 2.03 × 10−31 | |
| NDUFV2 | −0.06 | 0.286 | 0.70 | 2.42 × 10−45 |
GS indicates gene significance, which denotes correlation between each gene and age.
p. denotes Pearson's correlation coefficient p-values.
MM indicates module membership, which denotes correlation between each gene and module eigengene.
KEGG, Kyoto Encyclopedia of Genes and Genomes; OXPHOS, oxidative phosphorylation; TCA, tricarboxylic acid.
We then closely analyzed the copy number of mitochondria according to the age group. Since the GTEx database does not provide genomic data regarding mitochondria, we first analyzed the correlation between mitochondrial copy number and average expression of mtRNA using TCGA whole-genome sequencing papillary thyroid carcinoma samples (Supplementary Fig. S2) (23,24). A strong positive correlation between two factors was observed, which can provide a valid reason for analyzing average expression of mtRNA for mitochondrial copy number estimation (Spearman's rank correlation coefficient; r2 = 0.63). As the age gets older, a gradual decrease of average mtRNA expression was indentified, which corresponds to declining mitochondrial function as shown in our aforementioned result (p for trend <0.001; Fig. 1C).
The genes showing associations between downregulated proteasome activity and aging are listed in Table 2. PSMA1, PSMA3, PSMA4, PSMA5, PSMA6, PSMB1, PSMB2, and PSMB5, which encode the 20S proteasome (core particle), showed decreased expression with aging. Furthermore, PSMC1, PSMC2, PSMC4, PSMD1, PSMD2, PSMD6, PSMD11, PSMD12, PSMD13, PSMD14, and PSME3, which encode the 19S proteasome (regulatory particle), were also identified as genes with reduced expression. Moreover, several ubiquitin-mediated proteolysis-related genes were found to be downregulated, including KEAP1 and CUL3. KEAP1 is a substrate adaptor protein for the CUL3-E3-ligase complex, which targets NRF2, a transcription factor that has a role in the cell's defense mechanism, for ubiquitination and degradation by the ubiquitin-proteasome system (UPS) (25,26).
Table 2.
Genes Involved in Proteasome Activity from the Aging-Related Turquoise Module
| KEGG pathway | Gene symbol | GSa.Age | pb.GS.Age | MMc.turquoise | p.MM.turquoise |
|---|---|---|---|---|---|
| Proteasome | PSMB2 | −0.22 | 1.68 × 10−4 | 0.83 | 1.71 × 10−77 |
| PSMD1 | −0.21 | 1.95 × 10−4 | 0.93 | 1.83 × 10−129 | |
| PSMC1 | −0.21 | 2.97 × 10−4 | 0.79 | 7.63 × 10−66 | |
| PSMA6 | −0.21 | 3.22 × 10−4 | 0.87 | 1.57 × 10−94 | |
| PSMA5 | −0.21 | 3.38 × 10−4 | 0.90 | 1.03 × 10−109 | |
| PSMD12 | −0.20 | 4.03 × 10−4 | 0.68 | 3.21 × 10−42 | |
| PSMD11 | −0.19 | 0.001 | 0.75 | 2.85 × 10−56 | |
| PSMA3 | −0.19 | 0.001 | 0.85 | 1.19 × 10−86 | |
| PSMA1 | −0.18 | 0.002 | 0.76 | 3.13 × 10−58 | |
| PSMC2 | −0.18 | 0.002 | 0.88 | 1.81 × 10−97 | |
| PSMA4 | −0.15 | 0.008 | 0.80 | 3.30 × 10−68 | |
| PSMB5 | −0.15 | 0.011 | 0.81 | 8.20 × 10−72 | |
| PSMD2 | −0.14 | 0.016 | 0.85 | 1.33 × 10−86 | |
| PSMD14 | −0.14 | 0.017 | 0.87 | 2.76 × 10−94 | |
| PSMB1 | −0.14 | 0.018 | 0.76 | 1.16 × 10−58 | |
| PSMD13 | −0.12 | 0.038 | 0.61 | 1.41 × 10−31 | |
| PSMC4 | −0.11 | 0.060 | 0.82 | 9.72 × 10−75 | |
| Ubiquitin mediated proteolysis | KEAP1 | −0.29 | 3.91 × 10−7 | 0.78 | 5.26 × 10−62 |
| TRIM32 | −0.24 | 2.93 × 10−5 | 0.60 | 1.63 × 10−30 | |
| CUL3 | −0.24 | 3.56 × 10−5 | 0.70 | 2.79 × 10−45 | |
| UBE3C | −0.23 | 5.34 × 10−5 | 0.79 | 1.06 × 10−65 | |
| UBE2J1 | −0.23 | 7.69 × 10−5 | 0.65 | 1.21 × 10−37 | |
| PRPF19 | −0.22 | 1.21 × 10−4 | 0.74 | 2.28 × 10−53 | |
| DDB1 | −0.20 | 0.001 | 0.77 | 7.73 × 10−61 | |
| RFWD2 | −0.17 | 0.003 | 0.72 | 1.65 × 10−48 | |
| UBE2K | −0.17 | 0.004 | 0.69 | 4.43 × 10−44 | |
| TRIP12 | −0.14 | 0.013 | 0.64 | 1.27 × 10−35 | |
| XIAP | −0.12 | 0.032 | 0.58 | 7.15 × 10−28 | |
| CDC20 | −0.09 | 0.111 | 0.66 | 2.84 × 10−38 |
GS indicates gene significance, which denotes correlation between each gene and age.
p. denotes Pearson's correlation coefficient p-values.
MM indicates module membership, which denotes correlation between each gene and module eigengene.
Lastly, we analyzed the key genes that have roles in mitochondrial function or proteasome activity including genes we mentioned above. We subjected 56 genes to the analysis, and most genes were expressed at a higher level than their median value in more than 50% of samples in age groups 20–39 and 40–49 years, whereas age groups 50–59 and 60–79 years exhibited a reduced proportion of samples with higher expression rate than the median value of each gene. This result again supports downregulated mitochondrial function and proteasome activity with aging (Supplementary Fig. S3A).
Associations between autoimmune activity of the thyroid and aging
The tan module, the third most relevant one associated with aging, is related to immune processes, immune responses, and immune cell activation in the enrichment analyses using the GO (Fig. 1B). In addition, the T-cell receptor signaling pathway and genes related to autoimmune thyroid disease were found in the KEGG pathway enrichment result.
Among the 114 genes in the tan module, 113 exhibited a positive correlation with aging. When subjects reached their early or late forties, the expression of genes in the tan module were found to be upregulated (Fig. 1D). As there is no further increase with aging, it does not imply a gradual increase of immune gene expression with aging but indicates that the time point of immune gene upregulation is occurring in the fifth decade. Moreover, to confirm whether these changes occurring with aging may relate to the phenotype or pathophysiological outcomes, we reviewed the preoperative thyroid function and antithyroid antibody status of 64 study subjects. We were able to demonstrate a significant positive correlation between age and serum thyrotropin levels (r = 0.262 and P = 0.038) and a significant negative correlation between age and serum free T4 levels (r = −0.267 and P = 0.034). In addition, thyroid peroxidase antibodies were positive in 12 of 61 subjects, and all of them were over age 40 (range 44–81 years old; Supplementary Table S3).
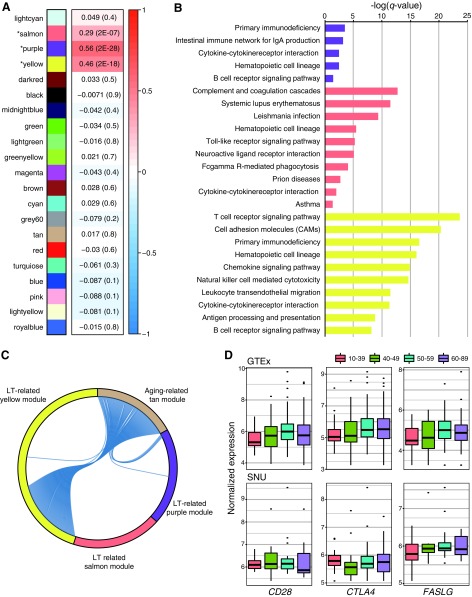
Next, we investigated whether aging-related changes in the expression of genes involved in immune processes were associated with alterations in autoimmune thyroid diseases, such as LT. We performed WGCNA to test the relevance of LT using all 322 GTEx samples, including 22 LT-positive samples (the diagnosis was based on GTEx clinical data) that were excluded from our primary analysis, and we identified 21 modules. Of those modules, three showed significant positive associations with autoimmunity (LT-related modules): purple (r = 0.56 and p = 2 × 10−28), yellow (r = 0.46 and p = 2 × 10−18), and salmon (r = 0.29 and p = 2 × 10−7; Fig. 2A). We also provide the gene list of the three LT-related modules (Supplementaty Table S4). Interestingly, all genes in the three LT-related modules were upregulated. When we further analyzed the three LT-related modules using the KEGG pathway enrichment analysis, the enrichment results indicated that they were also primarily related to immune response (Fig. 2B). In particular, the LT-related yellow module exhibited enrichment results highly similar to those of the aging-related tan module based on the KEGG pathway analysis (Fig. 1B).
FIG. 2.
LT-related weighted gene coexpression analysis of the thyroid gland. (A) Relationships between the constructed module and LT status. The Pearson correlation coefficient and correlation test p-value (in brackets) between LT status and the module eigengene are presented. (B) KEGG pathway enrichment results for three modules. (C) Chord diagram representing the relationship between the aging-related tan module and the three LT-related modules. (D) Box plots representing age-dependent changes in the expression of CD28, CTLA4, and FASLG. GTEx, Genotype-Tissue Expression; LT, lymphocytic thyroiditis; SNU, Seoul National University.
Because the aging-related tan module showed a relationship with the immune response, we investigated whether genes from the three LT-related modules overlapped with those from the aging-related tan module to clarify the associations between LT-related genes and aging-related genes (Fig. 2C). All 114 aging-related genes included in the tan module matched with genes from the LT-related modules; 108 genes from the tan module were included in the LT-related yellow module, and 6 genes were found in the LT-related purple module. Genes especially closely involved in the T-cell receptor signaling pathway that were upregulated in the LT-related yellow module and the aging-related tan module are listed in Table 3. In particular, CD28, CTLA4, and FASLG—which are well-known genes associated with various autoimmune diseases, including autoimmune thyroid disease—were identified in the aging-related tan module. Increasing trends with age were found in the expression of CD28 (r = 0.11 and P = 0.064), CTLA4 (r = 0.14 and P = 0.015), and FASLG (r = 0.12 and P = 0.042), although the trend for CD28 showed a lack of statistical significance due to its decrease in the age group of 60–89 years. (Fig. 2D). We validated this result with our 64 LT-negative normal thyroid samples and observed a similar expression pattern of FASLG (r = 0.26 and p = 0.045).
Table 3.
Genes Involved in the Immune Response from the LT-Related Tan Module
| KEGG pathway | Gene symbol | GSa.Age | pc.GS.Age | MMc.tan | p.MM.tan |
|---|---|---|---|---|---|
| T-cell receptor signaling pathway | ITK | 0.29 | 2.16 × 10−7 | 0.83 | 2.76 × 10−78 |
| ZAP70 | 0.26 | 6.83 × 10−6 | 0.9 | 1.88 × 10−106 | |
| CD247 | 0.24 | 3.14 × 10−5 | 0.9 | 1.88 × 10−107 | |
| PTPRC | 0.2 | 4.16 × 10−4 | 0.87 | 9.84 × 10−95 | |
| PDCD1 | 0.2 | 4.31 × 10−4 | 0.68 | 8.70 × 10−43 | |
| RASGRP1 | 0.2 | 0.001 | 0.9 | 1.43 × 10−110 | |
| GRAP2 | 0.18 | 0.002 | 0.86 | 8.31 × 10−89 | |
| CD8A | 0.17 | 0.003 | 0.72 | 6.64 × 10−49 | |
| CD8B | 0.17 | 0.004 | 0.73 | 3.11 × 10−50 | |
| CARD11 | 0.17 | 0.004 | 0.91 | 6.96 × 10−113 | |
| ICOS | 0.16 | 0.006 | 0.84 | 9.16 × 10−81 | |
| LCK | 0.16 | 0.005 | 0.9 | 1.34 × 10−107 | |
| CTLA4 | 0.14 | 0.015 | 0.78 | 2.16 × 10−63 | |
| CD28 | 0.11 | 0.064 | 0.83 | 9.08 × 10−76 | |
| CD3D | 0.03 | 0.573 | 0.84 | 6.08 × 10−81 | |
| Autoimmune thyroid disease | CTLA4 | 0.14 | 0.015 | 0.78 | 2.16 × 10−63 |
| FASLG | 0.12 | 0.042 | 0.67 | 1.01 × 10−40 | |
| CD28 | 0.11 | 0.064 | 0.83 | 9.08 × 10−76 |
GS indicates gene significance, which denotes correlation between each gene and age.
p. denotes Pearson's correlation coefficient p-values.
MM indicates module membership, which denotes correlation between each gene and module eigengene.
Using the same approach used for mitochondrial and proteasome genes, we then analyzed key genes that have roles in the T-cell receptor signaling pathway and autoimmune thyroid disease, and the results clearly show low expression levels of immune response genes in the 20–39 years age group compared with other age groups. This finding reflects the trend of an increased expression of immune response genes with aging (Supplementary Fig. S3B).
Associations between sex, thyroid differentiation, and aging-related changes in the thyroid gland
Since females are more susceptible to autoimmune thyroid disease and thyroid cancer than males, we analyzed aging-related changes in the thyroid gland by sex, excluding 22 LT-positive samples. In males (n = 199), aging-related changes in the gene expression pattern were almost the same as those analyzed in females (Supplementary Figs. S4A and S5A). Meanwhile, in the analysis of female subjects (n = 101), there were also blue (r = −0.17 and p = 0.09) and red (r = 0.16 and p = 0.1) modules associated with mitochondrial and immune-related genes, respectively, but the correlation with aging was not statistically significant (Supplementary Figs. S4B and S5B) possibly due to the relatively smaller number of samples compared with males. However, female subjects showed similar expression profiles of mitochondrial and immune-related genes as male subjects (Supplementary Fig. S6).
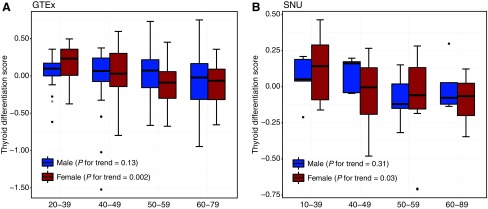
Next, we applied the TDS to observe changes in the gene expression patterns related to thyroid differentiation by age and sex. In particular, the TDS was significantly decreased with age in females (p for trend = 0.002; Fig. 3A), but it was not significant in males (p for trend = 0.13). Furthermore, we calculated the TDS using our RNA-seq data of 64 LT-negative samples and confirmed a similar declining trend in females (p for trend = 0.03; Fig. 3B) and no significant trend in males (p for trend = 0.31).
FIG. 3.
Aging-related changes of thyroid differentiation score according to sex using (A) GTEx and (B) our data.
Discussion
Aging is the most common risk factor for human diseases, and aging-related functional changes of organs usually act as contributory factors. Thus, studying the physiology or consequences of aging and its mechanisms is necessary to further extend life expectancy and maintain health. In this study, we identified aging-related transcriptomic changes in the thyroid that are associated with mitochondrial and proteasomal dysfunction, as well as the activation of autoimmune processes. Furthermore, no difference was found in aging-related changes according to sex, but in terms of thyroid differentiation, females were more susceptible to aging-related changes.
In aged tissues, genes involved in energy metabolism typically display downregulation, usually leading to mitochondrial malfunction (27). Recently, Yang et al. reported a study on age-dependent transcriptomic changes across multiple tissues, including the thyroid gland, using the GTEx database (2). In their study, numerous mitochondrial genes were downregulated in adipose tissue, artery, heart, lung, and blood with aging. However, only three genes with aging-related expression were found in the thyroid gland; thus, no further functional analyses were conducted. There are two major elements that distinguish our study from that of Yang et al., a different analytic method and the size of the dataset. The individual gene expression analysis approach and the smaller dataset (GTEx v3 release; n = 105) may have led to limitations in the results reported by Yang et al. (2). In addition, in the present study, a coexpressed gene network approach was applied, allowing us to identify abundant aging-related genes in the thyroid gland. Network analysis enables the construction of co-expressed networks based on similarities in gene expression and allows the functional annotation of unidentified genes. Generating coexpressed gene networks is straightforward and allowed us to explore the functions of clustered genes (28). In our coexpressed gene network analysis, we were able to identify decreased mitochondrial function as a major aging-related transcriptomic change in the thyroid gland, which is in consistent with the results of Yang et al. for other organs. In addition to changes in mitochondrial function, we found that decreased proteasomal activity was a biological process associated with aging. The proteasome is a major cellular component of proteolytic machinery that maintains proteostasis in collaboration with ubiquitins. Damaged proteins are degraded by the UPS, and dysfunction in this system with aging or aging-related diseases, such as neurodegenerative disorders and certain cancers, have been reported (1,29,30). The accumulation of misfolded proteins and oxidative stress have been portrayed as typical features of the aging process. Interestingly, the UPS and the mitochondria are highly interconnected; mitochondrial dysfunction can impair proteasome function via oxidative stress and low levels of ATP, while a decrease in UPS activity can impair mitochondrial function by affecting mitochondrial homeostasis, mitophagy, and the removal of damaged mitochondrial proteins (31). Collectively, we can conclude that mitochondrial dysfunction and impairment of the UPS are two hallmarks of aging across various types of organs, including the thyroid gland (Supplementary Table S5). Hence, metabolic processes such as proteostasis are reduced as a consequence.
Meanwhile, the thyroid gland is one of the most representative organs affected by autoimmune diseases. Improperly activated immune processes can induce an autoimmune response, and this autoimmune response to the thyroid plays a key role in causing thyroid dysfunction. Hashimoto's thyroiditis, also known as chronic LT, is a representative autoimmune disease of the thyroid. It affects approximately 5% of the population and typically begins between the ages of 30 and 50 (32,33). In the present study, gene expression patterns in the tan module, which was associated with the upregulation of immune activity with age, showed an increasing trend in the forties, even though the analysis was performed excluding LT-positive samples. For this reason, we identified LT-related modules and compared them with the age-associated tan module. We show that aging-associated gene expression profiles overlapped with gene expression patterns in tissues affected by autoimmune thyroid disease, indicating that there is a “link” between aging and upregulation of the immune response in the thyroid gland. Furthermore, we found that the preoperative serum thyrotropin and thyroid peroxidase antibody levels increased and free T4 levels decreased with aging, a finding that is consistent with previous studies (4–8). Although the causal relationship remains unclear, these results may reflect thyroid-specific changes associated with aging that were not identified in other tissues by Yang et al., and the data support that susceptibility to LT increases with age (Supplementary Table S5).
Thyroid-specific gene expression patterns have been used to evaluate the differentiation status of thyroid cancer, and the low expression of thyroid differentiation markers is a key feature of advanced thyroid cancer (34). The TDS, which was proposed by the TCGA study on papillary thyroid carcinomas (21), contains 16 genes involved in iodine metabolism. These markers might be affected by various conditions such as thyroid dysfunction, thyroiditis, thyroid hormone status, or iodine intake. We applied the TDS to normal thyroid tissue samples, and it significantly decreased with age in females. Thyroid disease, including autoimmune thyroiditis and thyroid cancer, is distinctively more frequent in females than in males, but the reason for this discrepancy has yet to be elucidated. The decrease of the TDS in females might reflect their susceptibility to thyroid disease. However, there was no significant difference between females and males in aging-related gene expression changes in mitochondrial biology or autoimmune activity. When the WGCNA was performed with female samples only, mitochondrial and immune-related modules showed no significant association with aging, which may have been due to the small number of subjects. Regardless of the WGCNA result, these transcriptomic changes in mitochondrial and autoimmune-related genes with aging could not explain the susceptibility of females to thyroid diseases, which suggests that the influence of extra-thyroidal factors might be the main reason for the predisposition of females to thyroid disease, despite the change in the TDS. In order to fully understand the susceptibility of females to thyroid disease, further studies are required.
As described, there were remarkable age-related changes in gene expression patterns observed in the thyroid. However, we also found that other important cellular signaling pathways are not dysregulated with aging. In particular, epigenetic alterations, one of the hallmarks of aging, did not show any relation with aging (1). Epigenetic alterations include histone demethylase activity, protein dealkylation, and demethylase activity, and those pathways were enriched in the royalblue module, however did not change with aging (r = 0.079 and p = 0.2; Supplementary Fig. S1). Also, in the present study, the T-cell-related immune response increased with aging, but humoral immunity did not exhibit a similar trend (salmon module; r = 0.055 and p = 0.3). Moreover, ribosome activity (black module; r = 0.09 and p = 0.1) and actin skeleton–related genes (lightgreen module; r = −0.016 and p = 0.8) did not change with aging.
The thyroid is an important organ that has essential roles in human physiology and is commonly affected by disease. Therefore, the characterization of changes in gene expression patterns during the aging process is important for the understanding of (patho)physiological changes and we believe that our findings provide new insights into the changes associated with aging in the thyroid.
In conclusion, the aging-related transcriptomic changes in the thyroid are consistent with well-known components of aging, such as mitochondrial dysfunction and the loss of proteostasis. Furthermore, we identified thyroid-specific changes associated with aging such as the upregulation of gene expression patterns related to the immune response, which overlap with the profile associated with autoimmune thyroid disease, and the susceptibility to aging-associated decreases in thyroid differentiation in females. Understanding changes in the gene expression of the “aging thyroid” may help in the search for targets to prevent and treat aging-related thyroid diseases.
Supplementary Material
Acknowledgments
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare, Republic of Korea (grant number HI13C1927).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153:1194–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J, Huang T, Petralia F, Long Q, Zhang B, Argmann C, Zhao Y, Mobbs CV, The GTEx Consortium, Schadt EE, Zhu J, Tu Z. 2015. Synchronized age-related gene expression changes across multiple tissues in human and the link to complex diseases. Sci Rep 5:15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mariotti S, Franceschi C, Cossarizza A, Pinchera A. 1995. The aging thyroid. Endocr Rev 16:686–715 [DOI] [PubMed] [Google Scholar]
- 4.Vanderpump MP, Tunbridge WM, French JM, Appleton D, Bates D, Clark F, Grimley Evans J, Hasan DM, Rodgers H, Tunbridge F, et al. . 1995. The incidence of thyroid disorders in the community: a twenty-year follow-up of the Whickham Survey. Clin Endocrinol (Oxf) 43:55–68 [DOI] [PubMed] [Google Scholar]
- 5.Hollowell JG, Staehling NW, Flanders WD, Hannon WH, Gunter EW, Spencer CA, Braverman LE. 2002. Serum TSH, T(4), and thyroid antibodies in the United States population (1988 to 1994): National Health and Nutrition Examination Survey (NHANES III). J Clin Endocrinol Metab 87:489–499 [DOI] [PubMed] [Google Scholar]
- 6.Surks MI, Hollowell JG. 2007. Age-specific distribution of serum thyrotropin and antithyroid antibodies in the US population: implications for the prevalence of subclinical hypothyroidism. J Clin Endocrinol Metab 92:4575–4582 [DOI] [PubMed] [Google Scholar]
- 7.Bremner AP, Feddema P, Leedman PJ, Brown SJ, Beilby JP, Lim EM, Wilson SG, O'Leary PC, Walsh JP. 2012. Age-related changes in thyroid function: a longitudinal study of a community-based cohort. J Clin Endocrinol Metab 97:1554–1562 [DOI] [PubMed] [Google Scholar]
- 8.Kim YA, Park YJ. 2014. Prevalence and risk factors of subclinical thyroid disease. Endocrinol Metab (Seoul) 29:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The GTEx Consortium 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The GTEx Consortium 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rung J, Brazma A. 2013. Reuse of public genome-wide gene expression data. Nat Rev Genet 14:89–99 [DOI] [PubMed] [Google Scholar]
- 12.Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG, Oezcimen A, Rocca-Serra P, Sansone SA. 2003. ArrayExpress–a public repository for microarray gene expression data at the EBI. Nucleic Acids Res 31:68–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo SK, Lee S, Kim SJ, Jee HG, Kim BA, Cho H, Song YS, Cho SW, Won JK, Shin JY, Park do J, Kim JI, Lee KE, Park YJ, Seo JS. 2016. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet 12:e1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin SM, Du P, Huber W, Kibbe WA. 2008. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res 36:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4:Article17 [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research N 2014. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159:676–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia J, Gill EE, Hancock RE. 2015. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 10:823–844 [DOI] [PubMed] [Google Scholar]
- 23.Reznik E, Miller ML, Senbabaoglu Y, Riaz N, Sarungbam J, Tickoo SK, Al-Ahmadie HA, Lee W, Seshan VE, Hakimi AA, Sander C. 2016. Mitochondrial DNA copy number variation across human cancers. Elife 5:DOI: 10.7554/eLife.10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reznik E, Wang Q, La K, Schultz N, Sander C. 2017. Mitochondrial respiratory gene expression is suppressed in many cancers. Elife 6: DOI: 10.7554/eLife.21592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villeneuve NF, Lau A, Zhang DD. 2010. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal 13:1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24:10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chistiakov DA, Sobenin IA, Revin VV, Orekhov AN, Bobryshev YV. 2014. Mitochondrial aging and age-related dysfunction of mitochondria. Biomed Res Int 2014:238463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serin EA, Nijveen H, Hilhorst HW, Ligterink W. 2016. Learning from Co-expression Networks: Possibilities and Challenges. Front Plant Sci 7:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morimoto RI. 2008. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev 22:1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller JN, Hanni KB, Markesbery WR. 2000. Impaired proteasome function in Alzheimer's disease. J Neurochem 75:436–439 [DOI] [PubMed] [Google Scholar]
- 31.Ross JM, Olson L, Coppotelli G. 2015. Mitochondrial and ubiquitin proteasome system dysfunction in ageing and disease: two sides of the same coin? Int J Mol Sci 16:19458–19476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pyzik A, Grywalska E, Matyjaszek-Matuszek B, Rolinski J. 2015. Immune disorders in Hashimoto's thyroiditis: what do we know so far? J Immunol Res 2015:979167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dayan CM, Daniels GH. 1996. Chronic autoimmune thyroiditis. N Engl J Med 335:99–107 [DOI] [PubMed] [Google Scholar]
- 34.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP, Xu B, Schultz N, Berger MF, Sander C, Taylor BS, Ghossein R, Ganly I, Fagin JA. 2016. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest 126:1052–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.