Abstract
Classical activation of M1 macrophages with lipopolysaccharide (LPS) is associated with a metabolic switch from oxidative phosphorylation to glycolysis. However, the generalizability of such metabolic remodeling to other modes of M1 macrophage stimulation, e.g. type II interferons (IFNs) such as IFNγ, has remained unknown as has the functional significance of aerobic glycolysis during macrophage activation. Here we demonstrate that IFNγ induces a rapid activation of aerobic glycolysis followed by a reduction in oxidative phosphorylation in M1 macrophages. Elevated glycolytic flux sustains cell viability and inflammatory activity, while limiting reliance on mitochondrial oxidative metabolism. Adenosine triphosphate (ATP) distributed by aerobic glycolysis is critical for sustaining IFN-γ triggered JAK (Janus tyrosine kinase)-STAT-1 (Signal Transducer and Activator of Transcription 1) signaling with phosphorylation of the transcription factor STAT-1 as its signature trait. Inhibition of aerobic glycolysis not only blocks the M1 phenotype and pro-inflammatory cytokine/chemokine production in murine macrophages and also human monocytes/macrophages. These findings extend on the potential functional role of immuno-metabolism from LPS- to IFNγ-linked diseases such as atherosclerosis and autoimmune disease.
Keywords: Immunometabolism, Inflammation, Interleukin-1 beta, Interferon gamma, Macrophage
Highlights
-
•
Interferon gamma (IFNγ) induces a rapid switch to aerobic glycolysis in M1 macrophages sustaining two signaling pathways.
-
•
Repurposing of the mitochondria, including reactive oxygen species production, allows for HIF-1α and IL-1β production.
-
•
ATP production by glycolysis allows for JAK activity, STAT-1 phosphorylation, iNOS and thereby nitric oxide production.
Inflammatory cells like macrophages, upon activation, switch their energy metabolism from the mitochondria to the processing of glucose. This switch feeds into the inflammatory action of macrophages and links it to the glucose concentration in the cell environment. Accordingly, under conditions of high glucose concentrations, as seen in patients with diabetes, a higher level of inflammatory activity might be seen. Performing these studies with the macrophage activator interferon gamma that plays a role in the development of narrowing of arteries (atherosclerosis) furthers the potential link of these findings to the more diffuse and aggressive nature of atherosclerosis in diabetic patients.
1. Introduction
The observation that cancer cells utilize aerobic glycolysis despite sufficient oxygen availability was first reported by Otto Warburg in 1923 (Warburg, 1923). Since then this phenotype has been observed in a number of highly proliferative cell types (Hsu and Sabatini, 2008; Vander Heiden et al., 2009). Moreover, it has recently been described that classically (lipopolysaccharide (LPS)) stimulated macrophages also display a Warburg metabolic phenotype (Tannahill et al., 2013; Palsson-McDermott et al., 2015; Yang et al., 2014; Mills et al., 2016). Macrophages are critical for both innate nonspecific host defense and the adaptive specific immune response (Akira et al., 2006), which broadly impacts diverse disease entities. Accordingly, glucose metabolism and aerobic glycolysis specifically may be of significance not only for LPS-linked infectious disease processes, but also interferon (IFN) γ-linked disease processes such as atherosclerosis and autoimmune diseases (McLaren and Ramji, 2009; Galkina and Ley, 2009, Shirai et al., 2016). This is of relevance as a “switch” from oxidative phosphorylation (OXPHOS) to aerobic glycolysis is not only a hallmark of T cell activation but required for T cell effector functions such as IFN-γ production (Chang et al., 2013; Peng et al., 2016). Of interest, while type II IFNs are known to lead to classical M1 macrophages, it is less well known if they induce a metabolic switch similar to LPS. Importantly, type I IFNs such as IFN-α have been shown induce alternative changes in the cellular metabolism of plasmacytoid dendritic cells consisting of increased fatty acid oxidation and OXPHOS (Wu et al., 2016).
Macrophages are not one homogenous cell population; in fact, various subtypes have been identified. The classical distinction is between the so-called classically activated M1 subtype and the so-called alternative activated M2 subtype, each with a seemingly different metabolic response to activation (Galván-Peña and O'Neill, 2015; Rodríguez-Prados et al., 2010). M1 macrophages rely on glycolysis (Tannahill et al., 2013; Palsson-McDermott et al., 2015; Yang et al., 2014; Mills et al., 2016), whereas M2 macrophages obtain energy from OXPHOS (Vats et al., 2006; Haschemi et al., 2012). Interestingly, the switch to aerobic glycolysis in M1 macrophages is accompanied by alterations in mitochondrial tricarboxylic acid (TCA) cycle activity leading to metabolite accumulation and increase in mitochondrial reactive oxygen species (ROS) production. This metabolic phenotype is associated with activation of hypoxia inducible factor-1α (HIF-1α)-responsive gene expression including those encoding for pro-inflammatory cytokines such as interleukin (IL)-1β (Liu et al., 2016; Tannahill et al., 2013; Mills et al., 2016). However, why macrophages switch to aerobic glycolysis, an energetically less efficient mode of metabolism, is likewise incompletely understood.
Here we report that IFN-γ increases aerobic glycolysis within minutes, followed by a decline in OXPHOS after several hours. This metabolic reprogramming is reversible, independent of nitric oxide (NO) production and alterations in mitochondrial TCA cycle metabolite accumulations. The metabolic switch allows for M1 macrophages to tolerate a reduction in mitochondrial adenosine triphosphate (ATP) production, and even more, it allows for mitochondrial ROS production, which stabilizes HIF-1α and contributes to the inflammatory response. Furthermore, intracellular glycolytic ATP is critical for the phosphorylation and pro-inflammatory activity of Signal Transducer and Activator of Transcription 1 (STAT-1) in the activation loop of the Janus tyrosine kinase (JAK)-STAT-1 pathway after IFN-γ stimulation. Finally, glycolysis inhibition not only blocked the M1 phenotype in murine cell lines but also in human monocytes and macrophages.
2. Materials and Methods
2.1. Key Resources Table
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Phospho-Stat1 (Tyr701) | Cell Signaling Technology | Cat# 7649 |
| Rabbit polyclonal anti-stat1 | Cell Signaling Technology | Cat# 9172 |
| Rabbit monoclonal anti-Phospho-IKKα (Ser176)/IKKβ (Ser177) | Cell Signaling Technology | Cat# 2078 |
| Rabbit monoclonal anti-Phospho-IκBα (Ser32) | Cell Signaling Technology | Cat# 2859 |
| Mouse monoclonal anti-IκBα (Amino-terminal Antigen) | Cell Signaling Technology | Cat# 4814 |
| Rabbit monoclonal anti-HIF-1α | Cell Signaling Technology | Cat# 14179 |
| Rabbit monoclonal Anti-Hexokinase II | Cell Signaling Technology | Cat# 2867 |
| Rabbit polyclonal anti-LDHa | Cell Signaling Technology | Cat# 2012 |
| Rabbit monoclonal anti-PKM2 | Cell Signaling Technology | Cat# 4053 |
| Rabbit monoclonal anti-Phospho-NF-κB p65 (Ser536) | Cell Signaling Technology | Cat# 3033 |
| Rabbit monoclonal anti-NF-κB p65 | Cell Signaling Technology | Cat# 8242 |
| Rabbit monoclonal anti- Phospho-Stat1 (Tyr701) | Thermo Fisher Scientific | Cat# 700349 |
| Rabbit polyclonal anti- iNOS (C-terminal region) | ECM Biosciences | Cat# NP2131 |
| Goat polyclonal anti-mouse IL-1 beta/IL-1F2 | R&D systems | Cat# AF-401 |
| Goat polyclonal anti-human IL-1 beta/IL-1F2 | R&D systems | Cat# AF-201 |
| Rabbit polyclonal anti-beta Actin | Abcam | Cat# ab8227 |
| Rabbit monoclonal APC/Cy7 anti-mouse CD86 | BioLegend | Cat# 105029 |
| GAPDH | Cell Signaling Technology | Cat# 5174 |
| Chemicals, peptides, and recombinant proteins | ||
| Lipopolysaccharides from Escherichia coli O127:B8 | Sigma-Aldrich | Cat# L4516 |
| Recombinant Murine IFN-γ | PeproTech | Cat# 315-05 |
| Recombinant Murine M-CSF | PeproTech | Cat# 315-02 |
| Recombinant Mouse IFN-beta | R&D systems | Cat# 8234-MB-010 |
| Recombinant Human M-CSF Protein | R&D systems | Cat# 216-MC |
| Recombinant Human IFN-γ | Peprotech | Cat# 300-02 |
| 2-Deoxy-d-glucose | Sigma-Aldrich | Cat# D8375 |
| Lactate Dehydrogenase A Inhibitor, FX11 | EMD Millipore | Cat# 427218 |
| d-(+)-Galactose | Sigma-Aldrich | Cat# G0750 |
| d-(+)-Glucose | Sigma-Aldrich | Cat# G8644 |
| Sodium oxamate | Sigma-Aldrich | Cat# O2751 |
| Nω-Nitro-l-arginine methyl ester hydrochloride | Sigma-Aldrich | Cat# N5751 |
| ATP Solution | Thermo Fisher Scientific | Cat# R0441 |
| Adenosine 5′-triphosphate, periodate oxidized sodium salt | Sigma-Aldrich | Cat# A6779 |
| Adenosine 5′-[γ-thio]triphosphate tetralithium salt | Sigma-Aldrich | Cat# A1388 |
| BzATP triethylammonium salt | Abcam | Cat# ab120444 |
| Perchloric acid | Sigma-Aldrich | Cat# 311421 |
| Potassium bicarbonate | Sigma-Aldrich | Cat# 60339 |
| Potassium phosphate monobasic | Sigma-Aldrich | Cat# P5655 |
| Potassium hydroxide | Sigma-Aldrich | Cat# 484016 |
| Tetrabutylammonium hydrogensulfate | Sigma-Aldrich | Cat# 155837 |
| Deuterium oxide | Sigma-Aldrich | Cat# 151882 |
| Oligomycin A | Sigma-Aldrich | Cat# 75351 |
| Oligomycin | Sigma-Aldrich | Cat# O4876 |
| FCCP | Sigma-Aldrich | Cat# C2920 |
| Antimycin A | Sigma-Aldrich | Cat# A8674 |
| Rotenone | Sigma-Aldrich | Cat# R8875 |
| Natural Streptolysin O (Hemolytic streptococcus) protein | Abcam | Cat# ab63978 |
| JAK inhibitor I | EMD Millipore | Cat# 420097 |
| 7-AAD Viability Staining Solution | BioLegend | Cat# 420403 |
| MitoSOX™ Red Mitochondrial Superoxide Indicator | Thermo Fisher Scientific | Cat# M36008 |
| Fluoroshield Mounting Medium With DAPI | Abcam | Cat# ab104139 |
| Ficoll-Paque PLUS | GE Healthcare Life Sciences | Cat# 17-1440-03 |
| Critical commercial assays | ||
| Proteome Profiler Mouse Cytokine Array Kit | R&D systems | Cat# ARY006 |
| Proteome Profiler Human Cytokine Array Kit | R&D systems | Cat# ARY005B |
| Avidin/Biotin Blocking Kit | Vector Laboratories | Cat# SP-2001 |
| ATP assay kit | MyBioSource | Cat# MBS841498 |
| Lactate Colorimetric/Fluorometric Assay Kit | BioVision | Cat# K607-100 |
| Deproteinizing Sample Preparation Kit | BioVision | Cat# K808-200 |
| ADP/ATP Ratio Assay Kit | Sigma-Aldrich | Cat# MAK135 |
| Griess Reagent Kit | Thermo Fisher Scientific | Cat# G-7921 |
| XTT Cell Viability Kit | Cell Signaling Technology | Cat# 9095 |
| Deposited data | ||
| NMR spectral data | ||
| Experimental models: cell lines | ||
| RAW 264.7 | ATCC | Cat# ATCC® TIB-71™ |
| Human monocytes | Mayo clinic | N/A |
| Experimental models: organisms/strains | ||
| C57BL/6J mice | The Jackson Laboratory | Cat# 000664 |
| Human carotid plaque | Mayo clinic | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software (version 7) | https://www.graphpad.com/scientific-software/prism/ |
| BD CellQuest™ Pro (Version 6.0) | BD Biosciences | https://www.bdbiosciences.com/documents/15_cellquest_prosoft_analysis.pdf |
3. Experimental Model and Subject Details
3.1. Mouse Strains
Female wild-type C57BL/6J mice, 8–12 weeks of age, were used in this study. The mice were bred and maintained under pathogen free conditions under a protocol approved by the Mayo Clinic Institutional Animal Use and Care Committee (IACUC).
3.2. Bone Marrow Derived Macrophages Isolation
Bone marrow-derived macrophages (BMDMs) were isolated from mice as previously described (Zhang et al., 2008). Briefly, bone marrow cells were flushed out from femur and tibia, resuspended and then grown in RPMI 160 (10% FBS and 1% penicillin and streptomycin) containing 10 ng/ml M-CSF. Fresh growth medium was added on day 3. Macrophages were harvested on day 7 and replated for further experimentation. All animal experiments were performed with Mayo Clinic Institutional Animal Use and Care Committee (IACUC) approval.
3.3. Raw264.7 Culture
Mouse leukemic monocyte/macrophage cell line (Raw264.7) was purchased from ATCC. Raw264.7 cells were cultured in high glucose DMEM containing 10% FBS and 1% penicillin and streptomycin.
Both BMDMs and RAW264.7 were used at 1 × 106/ml unless otherwise stated.
3.4. Human Monocytes and Macrophages
Human monocytes/macrophages were derived from patients undergoing carotid endarterectomy. Whole blood was collected at the time of the procedure and processed immediately. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation (Ficoll-Paque, GE) and subsequently incubated in RPMI 160 (10% FBS and 1% penicillin and streptomycin) for 2 h (Zhou et al., 2012). The floating cells were then washed out and remaining adherent cells were treated with or without 2-DG (10 mM) for 1 h, followed by IFN-γ stimulation for the indicated time periods. In order to derive human macrophages, monocytes were incubated in medium containing 20 ng/ml recombinant human M-CSF for 7 days. These macrophage were then similarly treated with or without 2-DG (10 mM) for 1 h, followed by IFN-γ stimulation for the indicated time periods. These human studies were approved by the Ethical Committee of Mayo Clinic and informed consent was obtained from all subjects.
4. Method Details
4.1. Macrophage Differentiation
BMDMs and Raw264.7 were plated for 24 h before the experiments. The cells were then stimulated with LPS or IFN-γ for 18–24 h to induce M1 macrophages. A concentration of 100 ng/ml was used for both, LPS and IFN-γ. For the recombinant IFN-γ used in this study, endotoxin level was of <0.10 EU per 1 μg of the protein, i.e. it was undetectable by the LAL method.
In order to derive human macrophages, monocytes from peripheral blood were incubated in medium containing 20 ng/ml recombinant human M-CSF for 7 days. Macrophages were then stimulated with 100 ng/ml human IFN-γ for the indicated time periods.
4.2. Metabolism Assay
ECAR and OCR of BMDMs were analyzed with an XF-24 Extracellular Flux Analyzer (Seahorse Bioscience) as described (Chang et al., 2013; Everts et al., 2014). For the real-time measurements, BMDMs were switched to seahorse media containing glucose, galactose or glucose plus different reagents (10 Mm 2-DG or 5 mM L-NAME) and incubated for 1 h. ECAR and OCR were measured 3–4 times at baseline and kinetics were recorded for the indicated times after injection with the different stimulators. For the endpoint measurements, BMDMs were stimulated in culture medium containing glucose or galactose, and other agents as indicated. After 24 h of stimulation with IFN-γ, BMDMs were switched to seahorse media containing either glucose or galactose and analyzed with the XF-24 Extracellular Flux Analyzer.
Metabolite levels were analyzed by NMR spectroscopy. 6 × 106 BMDMs in 10 ml culture medium were seeded in 100 mm petri dishes. These cells were stimulated with IFN-γ in the presence of glucose or galactose for 24 h. Cells were then rinsed with saline three times, quenched by adding 0.6 M HClO4 solution and finally frozen in liquid nitrogen. After neutralization with 2 M KHCO3 the extracts were spun down and the supernatants were collected.
In 500 μL of neutralized cell extract, 100 μL of phosphate buffer (pH 7.4) and 50 μL 0.1 M solution of TSP-d4 in D2O were added. Samples were vortexed for 20 s and transferred to 5 mm NMR tubes. NMR spectra were acquired on a Bruker 500 MHz Avance III HD spectrometer equipped with BBO cryoprobe and SampleCase auto sampler (Bruker Biospin, Rheinstetten, Germany). 1H NMR spectra were recorded using 1D noesy pulse sequence with presaturation (noesygppr1d), with 90° pulse (~13 μs), 4.68 s acquisition time, and 4 s relaxation delay. Spectra were referenced and phase and baseline corrected using the Topspin, version 3.5.
Metabolites were identified and quantified using the software program Chenomx NMR suite 8.2, by fitting the spectral lines of library compounds into the recorded NMR spectrum of the cell extracts. The quantification was based on peak area of TSP-d4 signal, and the metabolite concentrations were reported as μmol/mg protein.
4.3. Cytokines and Chemokines Measurement
The cytokine production from BMDMs after IFN-γ stimulation was measured using a mouse cytokine array (ARY006, R&D system) per manufacturer's instructions. Briefly, the cytokine array membranes were incubated with block buffer for 1 h at room temperature. In the meantime, the culture medium of BMDMs with different treatments was mixed with reconstituted cytokine detection antibody cocktail for 1 h. The culture medium was then added to the cytokine array membranes and incubated overnight at 4 °C. The next day, the membranes were washed three times and incubated with streptavidin-HRP for 30 min at room temperature. After washing three more times, the membranes were incubated with Chemi Reagent Mix for 1 min. The cytokines/chemokines expression density was developed by exposing membranes to a standard X-ray film.
4.4. Flow Cytometry
BMDMs were seeded in 24-well plate and stimulated with IFN-γ for 24 h in the presence of different reagents. Cells were collected by pipetting in PBS with 1 mM EDTA. Cells were then blocked by anti-mouse CD16/32 to reduce unspecific bindings. After incubation with different antibodies on ice for 15–20 min in the dark, cells were washed two times and resuspended in staining buffer. Cells were further stained with 7-AAD to exclude dead cells. Cells were analyzed by flow cytometry immediately. The final analysis only includes 7-AAD negative cells.
4.5. Mitochondrial ROS Measurements
Mitochondrial ROS was measured as previously described (West et al., 2011; Xu et al., 2015). Cells were plated in chamber slides (confocal microscope analysis) or 24-well plate (FACS analysis) and stimulated with IFN-γ in the presence of glycose or galactose for 24 h. After removal of the culture medium, cells were washed with warmed PBS and then incubated with MitoSOX red (Invitrogen) at 2.5uM for 30mins in phonel red- and serum-free DMEM (Invitrogen) at 37 °C. After washing with warm PBS (37 °C), cells were analyzed under the confocal microscope or by FACS immediately. Unstained cells were processed with same procedure but without stain with MitoSOX red.
4.6. P-STAT-1 Nuclear Translocation
BMDMs were seeded in chamber slides and pre-treatment with or without 2-DG for 1 h. Cells were then stimulated with IFN-γ for 30 min, washed with PBS and fixed with ½ methanol and ½ acetone for 15 mins. Blocking steps included Avidin/Biotin blocking kit (SP-2001, Vector) for 30 min followed by 5% normal horse serum in PBST for 30 min before staining. The primary antibody against p-STAT1 (Thermo Fisher 700,349) was prepared in PBST with 5% normal horse serum and incubated at 4 °C overnight; the fluorochrome-conjugated secondary antibody was incubated for 1 h. DAPI was used for nuclei staining. Images were analyzed using a laser scanning confocal microscope.
4.7. Lactate and ATP/ADP Ratio Measurements
Lactate production was measured by a commercial lactate assay kit (K607-100, BioVision) according to the manufacturer's instructions. Briefly, fresh supernatant of BMDMs with various treatments was collected and deproteinized by using a deproteinization sample preparation kit (K808-200, BioVision). After incubating with kit components, lactate level was quantified by measuring absorbance (OD 570 nm).
ATP/ADP ratio was analyzed by a commercial ATP/ADP ratio kit (MAK135, Sigma) according to the manufacturer's protocol. BMDMs were stimulated with IFN-γ in culture medium containing glucose or galactose for 24 h. After 30 min of treatment with DMSO or oligomycin (1 μM), cells were incubated in ATP reagent for 1 min and luminescence was read as the reflection of the ATP amount. After an additional 10 min of incubation, luminescence was read again to provide the background prior to ADP measurement. The final luminescence was tested as the reflection of the ADP amount. The ATP/ADP ration was calculated according to the manufacturer's protocol.
4.8. Nucleotide Measurements
Nucleotides were separated on a reversed-phase Discovery C18 columns (SIGMA, St. Louis, MO) with Hewlett-Packard series 1100 HPLC system (Agilent, Santa Clara, CA) as described previously (Dzeja et al., 2004). Briefly, a phosphate buffer with tetrabutylammonium sulfate and methanol mixture was used as mobile phase at 0.7 ml/min flow rate. Gradient elution was applied and the separation of nucleotides completed in 20 min. The nucleotide levels were normalized by protein amount.
4.9. Nitrite Determination
Nitrite was measured by the Greiss reagent kit (G-7921, Invitrogen) according to the manufacturer's instructions. Briefly, fresh supernatant of BMDMs with various treatments was collected and incubated with 20ul of a 1:1 mixture of reagent A (0.1% N-(1-naphthyl) ethylenediamine dihydrochloride) and reagent B (1% sulfanilic acid in 5% phosphoric acid). Absorbance at 548 was read to determine nitrite level.
4.10. Cell Viability
Cell viability was tested by using an XTT assay (Cell Signaling). Cells were plated in 96-well plate, after different intervention, 50ul XTT detection solution was added to each well and absorbance was read at 450 nm after 3 h incubation at 37 °C.
4.11. Western Blotting
Cells were lysed in RIPA buffer (9806, Cell signaling) supplemented with Complete Mini EDTA-Free protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Roche). Cell lysate was boiled in SDS sample buffer for 5 min, run on SDS gels, and transferred to PVDF membranes. The membranes were blocked for 1 h in TBS-T plus % nonfat dry milk and then incubated with primary antibodies overnight. After washing three to four times, the membranes were incubated with secondary antibodies for 1 h and washed three more times. ECL western blotting chemiluminescent substrates were added for 5 min. Bands of interest were developed by using an autoradiographic film.
4.12. Permeabilization
Permeabilization was performed as previously described (Walev et al., 2001). Cells were permeabilized by the pore-forming toxin Natural E. coli Streptolysin O (SLO, Abcam ab63978), which allows for delivery of molecules with up to 100 kDa mass to the cytosol. Briefly, SLO solution was added to HBSS without Ca2+ containing 30 mM HEPES to make permeabilization solution. SLO was used at 100 ng/ml. Raw264.7 cells were incubated in permeabilization solution at 37 °C in present with differentiation of ATP (0-10 mM). After 15 min of incubation, permeabliziation solution was taken out and fresh culture medium with/without 2-DG (10 mM) was added to the cells, followed by stimulation with IFN-γ for 1 h. Cells were subsequently lysed for Western blotting.
4.13. Statistical Analysis
Results were presented as mean ± standard error of the mean (sem). Unpaired Student's t-test was used to test the differences between two groups, based on the assessment of variance of the data. All data were analyzed by GraphPad Prism software (version 7). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Detailed statistical values were provided in the figure legends.
5. Results
5.1. Type II IFN Signaling Triggers Early, NO-independent, but Akt-Dependent Activation of Glycolysis
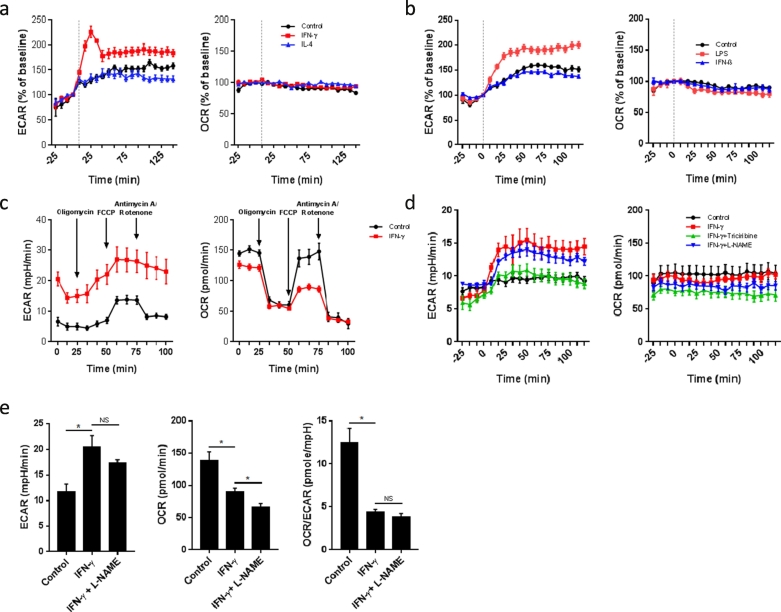
We first pursued a real-time analysis of the metabolic changes of bone marrow derived macrophages (BMDMs) activated with Type II IFN signaling. As shown in Fig. 1a, IFN-γ stimulation triggered an increase in real-time extracellular acidification rate (ECAR, an indicator of aerobic glycolysis) in BMDMs rather rapidly, i.e. in <15 min. On the contrary, oxygen consumption rate (OCR, an indicator of oxidative phosphorylation) remained stable early on (Fig. 1a). This metabolic switch was not observed in BMDMs stimulated with IL-4 (Fig. 1a), which induces alternative macrophage activation, or in response to type I IFNs like IFN-β (Fig. 1b). As outlined in Fig. 1c, IFN-γ led to a significant reduction in the maximum respiratory capacity (Fig. 1c), similar to LPS (Mills et al., 2016) but distinctly different from the metabolic effects of type I IFNs in dendritic cells (Wu et al., 2016).
Fig. 1.
IFNγ signaling triggers early, NO-independent, but Akt-dependent activation of glycolysis.
(a) Real-time measurement of ECAR and OCR of BMDMs, unstimulated (control) or stimulated with IFN-γ or IL-4, vertical line indicates time of cytokine injection. Data are representative of three independent experiments (n = 5, mean ± sem).
(b) Real-time measurement of ECAR and OCR of BMDMs, unstimulated (control) or stimulated with LPS or IFN-β, vertical line indicates time of cytokine injection. Data are representative of two independent experiments (n = 5, mean ± sem).
(c) ECAR and OCR profiles of BMDMs, measured 24 h after unstimulated conditions (control) or stimulation with IFN-γ (n = 5).
(d) Real-time measurement of ECAR and OCR of BMDMs, unstimulated (control) or stimulated with IFN-γ with or without 1-hour pretreatment with 5 mM L-NAME or 20 μM triciribine, vertical line indicates time of injection of IFN-γ. Data are representative of two independent experiments (n = 4, mean ± sem).
(e) ECAR, OCR and OCR/ECAR ratio of BMDMs, unstimulated (control) or stimulated with IFN-γ for 24 h with or without 1-hour pretreatment with 5 mM L-NAME. *p < .05, NS: no significant difference. Data are representative of two independent experiments (n = 3–5, mean ± sem).
NO has been shown to inhibit OXPHOS and redirect dendritic cells to use glycolysis for ATP production and survival, as measured by an increase in ECAR and a decrease in OCR (Everts et al., 2012). Pretreatment of classically stimulated macrophages with the cell-permeable NO synthesis inhibitor L-NAME decreased NO production (Fig. S1) but did not alter early metabolic reprogramming (Fig. 1d). We next examined the role of Akt, a known master regulator of metabolism. As showed in Fig. 1d, the Akt inhibitor triciribine completely inhibited the rapid switch to glycolysis. This is in keeping with the role of the Akt pathway for the Warburg Effect in cancer cells and the early glycolytic burst in dendritic cells (Everts et al., 2014). After 24 h of stimulation, ECAR remained elevated, OCR decreased in IFN-γ stimulated BMDMs, and the ECAR/OCR ratio remained unaffected by L-NAME (Fig. 1e). Thus, IFN-γ induces a rapid increase in aerobic glycolysis followed by a decrease in OXPHOS, which in distinction from dendritic cells was independent of NO synthesis (Everts et al., 2012) but in manner similar to dendritic cells was regulated by Akt (Everts et al., 2014).
5.2. Glycolysis Inhibition With 2-DG Blocks IFN-γ Induced Macrophages Differentiation and Activation
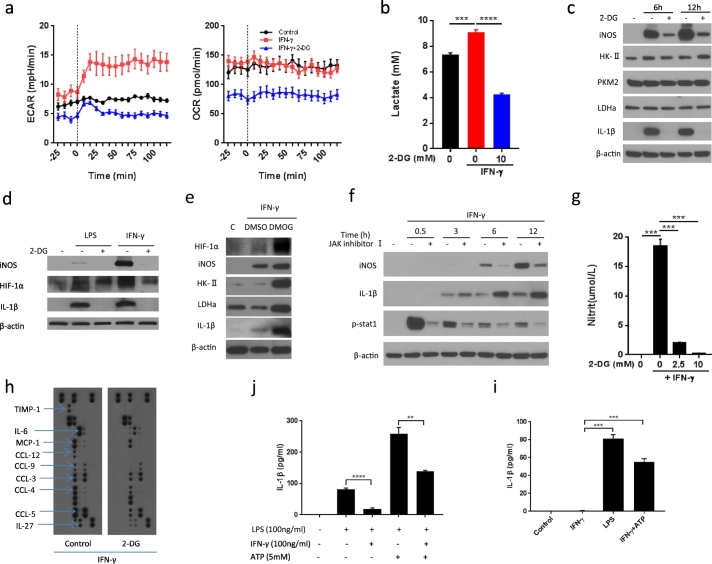
We next tested the functional implications of the metabolic switch in IFN-γ-activated BMDMs by using the glucose analog 2-deoxy-d-glucose (2-DG) as done extensively in previous studies to inhibit glycolysis (Tannahill et al., 2013; Yang et al., 2014; Everts et al., 2014; Mills et al., 2016). As outlined in Fig. 2a and b, 2-DG reduced not only ECAR and lactate production but also OCR. The reduction in OCR was noted even before stimulation and showed a plateau over the course of 120 min of real-time measurements. Importantly, 2-DG blocked the production of IL-1β in IFN-γ -activated BMDMs (Fig. 2c). The expression of IL-1β has been shown to be HIF-1α-dependent in LPS-stimulated macrophages (Tannahill et al., 2013). Likewise, we found that IFN-γ induced IL-1β production in BMDMs (Fig. 2d) and this response could be enhanced by the HIF-1α stabilizer DMOG (Fig. 2e). On the contrary, 2-DG decreased HIF-1α levels and IL-1β production in response to IFN-γ. IFN-γ firstly triggers the activation of the JAK-STAT-1 signaling pathway, but this was not the responsible pathway for IL-1beta expression. While potently reducing STAT-1 phosphorylation, JAK inhibition did not decrease IL-1β expression (Fig. 2f).
Fig. 2.
2-DG blocks IFN-γ induced macrophage activation.
(a) Real-time measurement of ECAR and OCR of BMDMs, unstimulated (control) or stimulated with IFN-γ with or without 1-hour pretreatment with 10 mM 2-DG, vertical line indicates initiation injection of IFN-γ. Data are representative of two independent experiments (n = 3–5, mean ± sem).
(b) Lactate production of Raw264.7 cells after stimulation with IFN-γ for 3 h ± 1 hour pre-treatment with 2-DG. ****p < .0001. Data are representative of two independent experiments (n = 4, mean ± sem).
(c) Immunoblot analysis of specified proteins in BMDMs after stimulation with IFN-γ for 6 and 12 h ± 1 hour pre-treatment with 2-DG (10 mM). Data are representative of three independent experiments.
(d) Immunoblot analysis of specified proteins in BMDMs after stimulation with LPS or IFN-γ for 12 h ± 1 hour pre-treatment with 2-DG (10 mM). Data are representative of three independent experiments.
(e) Immunoblot analysis of specified proteins in BMDMs after stimulation with IFN-γ for 48 h ± 3 hour pre-treatment with DMOG (200 μM).
(f) Immunoblot analysis of specified proteins in BMDMs after stimulation with IFN-γ for indicated time intervals ±1 hour pre-treatment with JAK inhibitor I.
(g) Nitric oxide (NO) production by BMDMs after stimulation with IFN-γ for 18 h ± 1 hour pre-treatment with 2-DG (10 mM). **p < 0.01, ***p < 0.001. Data are representative of two independent experiments (n = 4, mean ± sem).
(h) Cytokine production of BMDMs after stimulation with IFN-γ for 18 h ± 1 hour pre-treatment with 2-DG (10 mM).
(i) and (j) ELISA analysis of IL-1β production in BMDMs stimulated for 24 h with or without IFN-γ, LPS and ATP at indicated doses. **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are representative of two independent experiments (n = 4, mean ± sem).
Inducible NO synthase (iNOS) is another key marker of classically activated macrophages and the key controlling element of NO production of M1 macrophages. Intriguingly, a clear dose-response relationship was found between the concentration of glucose in the medium, IFN-γ-induced lactate production, and NO production (Fig. S2a and b). Moreover, iNOS expression time-dependently increased after IFN-γ as it does with LPS (Fig. 2c and d).
These responses to classical stimulation were blocked by 2-DG in a concentration-dependent manner (Fig. S2c and d and Fig. 2c, d, and g). Importantly, iNOS expression was not increased by the HIF-1α stabilizer DMOG (Fig. 2e) but potently blocked by JAK inhibition (Fig. 2f). Accordingly, IFN-γ mediates two key features of M1 activation, IL-1β and NO production, via two different transcription factor-signaling pathways, HIF-1α and STAT-1, respectively.
A comprehensive analysis of cytokine levels in the macrophage culture medium indicated that IFN-γ stimulated the release of several other key M1 macrophage-associated cytokines (e.g. IL-6 and MCP-1) and chemokines (CCL-3, CCL-4, CCL-5, CCL-9, and CCL-12) from murine BMDMs and this IFN-γ response was inhibited by 2-DG (Fig. 2h).
These data indicate that IL-1β expression aligns with HIF-1α- but not JAK-STAT-1-dependent gene expression whereas the opposite is true for iNOS. The expression of both of these as well as other cytokines/chemokines is blocked by 2-DG, indicating a possible link between metabolic reprogramming and both HIF-1α stabilization and JAK-STAT-1 activation.
Although IFN-γ can induce pro-IL-1β expression (Schindler et al., 1990; Collart et al., 1986), studies have showed that it can inhibit IL-1β production in macrophages stimulated by LPS and other cytokines (Mishra et al., 2013). Consistently, cleaved IL-1β production was reduced by IFN-γ in LPS-stimulated macrophages (Fig. 2j). Intriguingly, unlike LPS, IFN-γ stimulation did not induce cleaved IL-1β production in BMDMs unless combined with a second agent such as ATP (Fig. 2i), indicating that IFN-γ by itself cannot trigger inflammasome activation.
5.3. Glycolysis Inhibition With 2-DG Reduces IFN-γ-induced JAK-STAT1 Pathway Activation by Reducing ATP Production
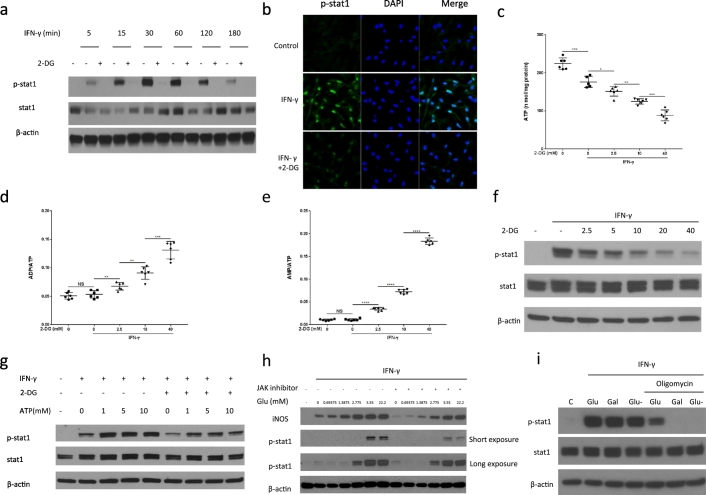
Based on the above findings, we next tested if glycolysis affected the activity of the JAK-STAT-1 pathway after IFN-γ stimulation. The JAK-STAT-1 pathway is unique in involving only one phosphorylation step of STAT-1 to directly activate gene transcription (Shuai and Liu, 2003). Intriguingly, 2-DG decreased STAT-1 phosphorylation in an immediate and sustained manner (Fig. 3a and Fig. S3a) and decreased the nuclear translocation of phosphorylated STAT-1 after IFN-γ stimulation (Fig. 3b). These findings were confirmed at different 2-DG and glucose concentrations (Fig. S3b–d).
Fig. 3.
2-DG reduces IFN-γ-induced JAK-STAT1 pathway activation by reducing ATP production.
(a) Immunoblot analysis of specified proteins in BMDMs as a function of time of IFN-γ stimulation ±1 hour pre-treatment with 2-DG (10 mM). Data are representative of two independent experiments.
(b) Immunofluorescence staining of p-STAT-1 in BMDMs after incubation with IFN-γ for 30 min ± 1 hour pre-treatment with 2-DG (10 mM). Data are representative of two independent experiments.
(c) to (e) ATP production, ADP/ATP, AMP/ATP ratio of Raw264.7 cells, measured by HPLC after stimulation with IFN-γ for 30 min ± 1 hour pre-treatment with 2-DG (10 mM). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, NS: no significant difference. Data are represented as mean ± sem (n = 6).
(f) Immunoblot analysis of specified proteins in Raw264.7 cells after stimulation with IFN-γ for 30 min ± 1 hour pre-treatment with indicated concentrations of 2-DG. Data are representative of two independent experiments,
(g) Immunoblot analysis of specified proteins in Raw264.7 cells after permeabilization with SLO (100 ng/ml), incubation with ATP (0–10 mM) for 15 min and 1-hour stimulation with IFN-γ (±10 mM 2-DG). Data are representative of two independent experiments.
(h) Immunoblot analysis of specified proteins in Raw264.7 cells after stimulation with IFN-γ for 9 h in medium containing different concentrations of glucose ±1 hour pre-treatment with JAK inhibitor I (100 nM). Data are representative of three independent experiments.
(i) Immunoblot analysis of specified proteins in Raw264.7 cells after stimulation with IFN-γ for 1 h in medium containing glucose or galactose ± oligomycin (1 μM). Data are representative of three independent experiments.
As JAK is an ATP-dependent kinase (Hammarén et al., 2015), we next tested the hypothesis that ATP production links aerobic glycolysis to the activation of the JAK-STAT-1 pathway by analyzing intercellular energy change and STAT-1 phosphorylation at different concentrations of 2-DG. As shown in Fig. 3c to e, 2-DG significantly decreased ATP and the ratios of ATP/ADP and ATP/AMP in a dose-dependent manner, correlating with a dose-dependent decrease in STAT-1 phosphorylation (Fig. 3f). Importantly, intracellular ATP delivery by transient membrane permeabilization was able to fully restore the decrease in STAT-1 phosphorylation by 2-DG (Fig. 3g). In contrast, extracellular ATP did not affect JAK-STAT-1 pathway activation and NO production (Fig. S3f–h). This observation supports the view that intracellular ATP generation is critical for downstream cell signaling in IFN-γ stimulated M1 cells. While 2-DG has been used as a glycolysis inhibitor, the current finding of the additional inhibitory effects of 2-DG on mitochondrial OXPHOS foils the conclusion that the rate of glycolysis is the main regulatory of the ATP pool for intracellular signaling in M1 macrophages. This point, however, is made by the observation that the inhibitory effect of an ATP-competitive JAK inhibitor on signature traits of M1 macrophages such as STAT-1 phosphorylation and iNOS expression could be overcome by increasing the extracellular glucose concentration, which translates into increased glycolytic throughput under conditions of IFN-γ stimulation (Fig. 3h). Moreover, we used oligomycin to block mitochondrial ATP production, which would commit the cell to intercellular ATP production from glycolysis. As a result, if there were to be a link, the dependency of STAT-1 phosphorylation on glycolysis should become much clearer. As shown in Fig. 3i, neither replacing glucose with galactose nor using oligomycin in isolation affected STAT-1 phosphorylation, indicating that both glycolytic ATP and mitochondrial ATP can support STAT-1 phosphorylation. STAT-1 phosphorylation was, however, blocked by galactose or glucose withdrawal after oligomycin treatment. Inhibition of STAT-1 phosphorylation in this setting could be overcome by increasing the extracellular glucose concentration (Fig. S3i). These data support ATP as the link between 2-DG treatment and STAT-1 phosphorylation.
5.4. Galactose Allows for Selective Glycolysis Interference and Confirms Its Significance in M1 Macrophages
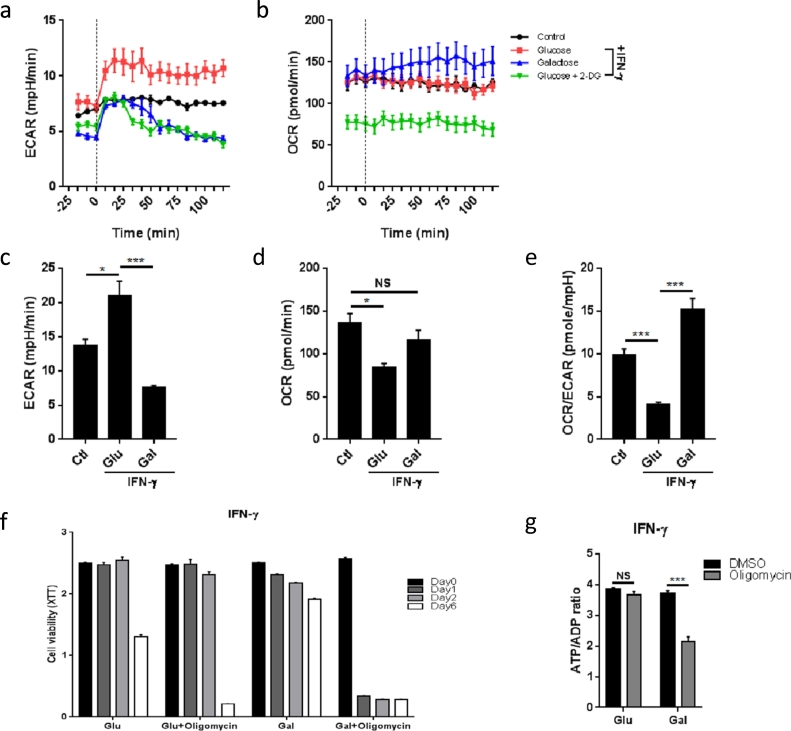
Although 2-DG is widely used in the field of immuno-metabolism and cancer as an inhibitor of hexokinase and glycolysis (Tannahill et al., 2013; Yang et al., 2014; Everts et al., 2014; Mills et al., 2016; Pelicano et al., 2006), it can exert effects beyond the metabolic block (Kurtoglu et al., 2007; Ralser et al., 2008; Wolf et al., 2016)). As described above, 2-DG also decreased oxygen consumption in IFN-γ activated macrophages, thereby confounding the ability to differentiate between the role of glycolysis and OXPHOS in M1 macrophages. As an alternative approach, glucose in the cell culture medium was replaced with galactose, which must be metabolized by the Leloir pathway in order to enter into glycolysis (Bustamante et al., 1977; Weinberg et al., 2010; Chang et al., 2013). The slower kinetics for galactose uptake versus direct entry of glucose into glycolysis potently reduces glycolytic flux. Furthermore, the Leloir pathway consumes ATP, and thus the net ATP yield from the conversion of galactose to pyruvate is zero. As shown by real-time metabolic monitoring (Fig. 4a and b), galactose rapidly reduced ECAR similar to 2-DG; but acute treatment with galactose did not reduce, in fact slightly increased OCR of IFN-γ-stimulated BMDMs (Fig. 4a and b). After 24 h of stimulation, BMDMs cultured in galactose maintained a significantly lower ECAR than cells activated in glucose or control BMDMs (Fig. 4c). Importantly, the OCR of BMDMs activated in galactose-containing medium was comparable to control BMDMs and significantly higher than the OCR of cells activated in glucose-containing medium (Fig. 4d and e). Under galactose conditions, IFN-γ stimulated macrophages had suppressed rates of glycolysis but unimpaired OXPHOS despite much higher than normal NO/nitrite levels. Aerobic glycolysis, however, does become critical for M1 macrophages when OXPHOS is impaired as noted with 2-DG. This is supported by the change of cell viability, which was reduced by 2-DG over time, but unaffected by galactose or even glucose depletion (Fig. S4a and b).
Fig. 4.
Galactose allows for selective interference with aerobic glycolysis and inhibits key aspects of activated M1 macrophages, which show metabolic plasticity.
(a) Real-time measurement of ECAR of BMDMs, unstimulated (control) or stimulated with IFN-γ after incubation in medium containing glucose, galactose or glucose +10 mM 2-DG for 1 h, vertical line indicates time of injection of IFN-γ. Data are representative of three independent experiments (n = 4–5, mean ± sem).
(b) Real-time measurement of OCR of BMDMs, unstimulated (control) or stimulated with IFN-γ after incubation in medium containing glucose, galactose or glucose +10 mM 2-DG for 1 h, vertical line indicates time of injection of IFN-γ. Data are representative of three independent experiments (n = 4–5, mean ± sem).
(c) ECAR of BMDMs, unstimulated (control) or stimulated with IFN-γ after incubation in medium containing glucose or galactose for 1 h. *p < 0.05, ***p < 0.001, NS: no significant difference. Data are representative of two independent experiments (n = 3–4, mean ± sem).
(d) OCR of BMDMs, unstimulated (control) or stimulated with IFN-γ after incubation in medium containing glucose or galactose for 1 h. *p < 0.05, ***p < 0.001, NS: no significant difference. Data are representative of two independent experiments (n = 3–4, mean ± sem).
(e) OCR/ECAR ratio of BMDMs, unstimulated (control) or stimulated with IFN-γ after incubation in medium containing glucose or galactose for 1 h. *p < 0.05, ***p < 0.001, NS: no significant difference. Data are representative of two independent experiments (n = 3–4, mean ± sem).
(f) Cell viability of BMDMs after stimulation with IFN-γ in medium containing glucose or galactose ± oligomycin (1 μM) at the indicated time intervals. Data are representative of two independent experiments (n = 6, mean ± sem).
(g) BMDMs were stimulated with IFN-γ for 24 h in medium containing glucose or galactose. ATP/ADP ratio was tested after treatment with DMSO or oligomycin for 30 min. ***p < 0.001, NS: no significant difference. Data are representative of two independent experiments (n = 4, mean ± sem).
5.5. Classically Activated Macrophages Display Metabolic Plasticity
As IFN-γ stimulated macrophages reprogram their metabolism by rapidly increasing aerobic glycolysis and decreasing OXPHOS over time, we next asked if these macrophages rely on glycolysis for their survival after IFN-γ stimulation. To then further test the metabolic flexibility of classically activated macrophages, we stimulated BMDMs in media with either glucose or galactose in the presence or absence of the OXPHOS inhibitor oligomycin. We found that BMDMs had similar viability when cultured in medium with glucose compared with galactose over 48 h (Fig. 4f). After 6 days of stimulation, cells cultured in galactose-containing medium displayed better viability than their counterparts grown in glucose-containing medium. Importantly, cells cultured in glucose-containing medium did not show reduced viability when treated with oligomycin (Fig. 4f), indicating that IFN-γ stimulated macrophages switched ATP production from OXPHOS to glycolysis as reported for LPS-activated macrophages (Mills et al., 2016). However, cells cultured in galactose died rapidly after oligomycin treatment (Fig. 4f), indicating that these cells relied on OXPHOS for their survival. In agreement, short exposure to oligomycin significantly reduced the ATP/ADP ratio in cells differentiated in galactose but not in those cells cultured in glucose (Fig. 4g). Thus, following classical activation with IFN-γ, M1 macrophages are not permanently committed to aerobic glycolysis and their metabolic reprogramming is reversible.
5.6. TCA Cycle Metabolites Accumulation Is Independent of Aerobic Glycolysis
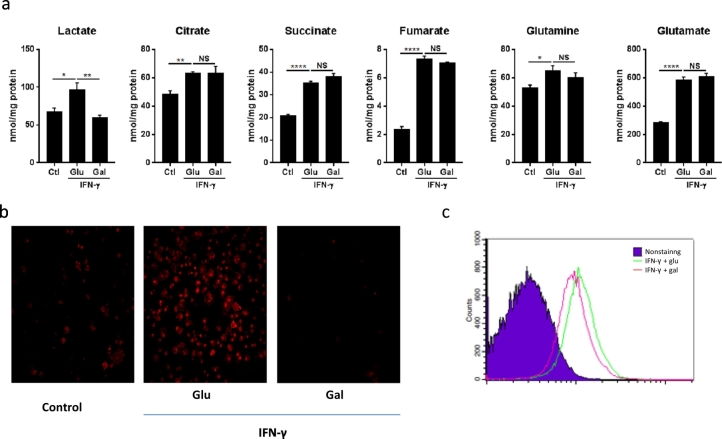
Increased rates of glycolysis and suppressed OXPHOS in LPS-stimulated macrophages leads to accumulation of TCA cycle metabolites (Tannahill et al., 2013; Jha et al., 2015; Mills et al., 2016), among which succinate has been deemed critical for HIF-α stabilization and its related IL-1β production. However whether IFN-γ stimulation also changes the levels of TCA cycle metabolites and whether this is by association or causal interaction has remained unknown. We therefore next performed targeted metabolic profiling of macrophages stimulated with IFN-γ in the presence of glucose or galactose. Cells stimulated in galactose displayed a reduction in lactate production associated with reduced rates of glycolysis (Fig. 5a). Importantly, IFN-γ stimulation resulted in significant accumulation of the TCA cycle metabolites citrate, succinate and fumarate (Fig. 5a). Intriguingly, this pattern of TCA cycle metabolites accumulation was fairly comparable under glucose and galactose conditions. Moreover, the levels of glutamine and glutamate, which feed into the TCA cycle in LPS-stimulated macrophages (Tannahill et al., 2013), were also significantly increased after FN-γ stimulation, and unaffected by galactose (Fig. 5a). Importantly, as shown in Fig. 5b and c, macrophages activated in galactose-containing medium produced significantly less mitochondrial ROS than cells activated in glucose-containing medium. Taken together, these findings indicate that galactose prevented the decrease in OXPHOS and increase in mitochondrial ROS production otherwise seen in M1 macrophages with classical stimulation without preventing an increase in succinate and fumarate levels while reducing citrate and glutamine levels.
Fig. 5.
TCA cycle metabolites accumulation is independent of aerobic glycolysis contrary to mitochondrial ROS production.
(a) BMDMs were incubated in in medium containing glucose or galactose for 1 h, followed by stimulation with IFN-γ for 24 h. Lysed cells were analyzed by NMR spectroscopy to determine metabolite levels. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. NS no significant difference. The data represent mean ± sem (n = 4).
(b) MitoSOX staining of Raw264.7 cells simulated with IFN-γ for 24 h in medium containing glucose or galactose. Data are representative of two independent experiments.
(c) MitoSOX FACS analysis of BMDMs simulated with IFN-γ for 24 h in medium containing glucose or galactose. Data are representative of two independent experiments.
5.7. Metabolic Reprogramming Allows for Repurposing of the Mitochondria to Sustain HIF-1α Signaling
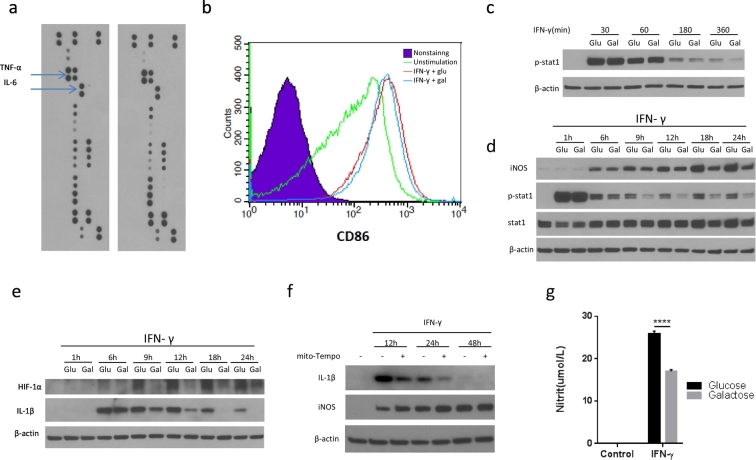
As outlined above, galactose reduced glycolysis but not OXPHOS, which allowed for a much better differentiation between the effects of aerobic glycolysis and those of OXPHOS in activated M1 macrophages. Importantly, we found that the effect size of galactose was much smaller than that of 2-DG. Specifically, there was no difference in the production of several cytokines/chemokines including TNF-α and IL-6 (Fig. 6a) or expression of M1 macrophage signature surface makers in BMDMs stimulated in galactose versus glucose containing medium (Fig. 6b). As illustrated in Fig. 6c and d, galactose led to a reduction in STAT-1 phosphorylation as well as iNOS expression and NO production over time (Fig. 6g). Likewise, HIF-1α and IL-1β levels were significantly reduced under galactose conditions over time (Figs. 6e). These observations are different from 2-DG, whose effects were of much faster onset (Figs. 2c, d, and 3a).
Fig. 6.
Galactose inhibits IFNγ-induced JAK-STAT-1 and HIF-1α pathway activation in M1 macrophages by modulating ATP and mitochondrial ROS production.
(a) Cytokine production of BMDMs after stimulation with IFN-γ for 24 h in medium containing glucose or galactose.
(b) FACS analysis of the M1 macrophage surface marker CD86 of BMDMs after stimulation with IFN-γ for 24 h in medium containing glucose or galactose.
(c, d, and e) Immunoblot analysis of specified proteins in BMDMs stimulated with IFN-γ for indicated time intervals in medium containing glucose or galactose. Data are representative of two independent experiments.
(f) Immunoblot analysis of specified proteins in BMDMs stimulated with IFN-γ for indicated time intervals with or without mito-tempo (1 mM) pretreatment for 2 h. Data are representative of two independent experiments.
(g) Nitric oxide (NO) production by BMDMs after stimulation with IFN-γ for 24 h in medium containing glucose or galactose. Data are representative of two independent experiments (n = 4, mean ± sem).
Similar to the differential dynamics of the activity of the JAK-STAT-1 pathway changes in intracellular ATP levels became evident within minutes with 2-DG and only within hours with galactose (Fig. S6a–h). These dynamics seemingly run in parallel with the activity of the JAK-STAT-1 pathway as outlined above. With regards to HIF-1α and IL-1β expression, prior work has pointed out the functional relevance of a switch from ATP to ROS production on mitochondrial level in classically stimulated macrophages, and ROS-induced stabilization of HIF-1α and related gene expression (Jha et al., 2015, Tannahill et al., 2013; Mills et al., 2016). In agreement and as mentioned above, galactose prevented the increase in mitochondrial ROS production otherwise so prominently noticed with classical stimulation under glucose conditions (Fig. 5b and c). Moreover, the mitochondrial ROS scavenger mito-tempo reduced IL-1β production in BMDMs stimulated with IFN-γ (Fig. 6f). Metabolic reprogramming to aerobic glycolysis in response to IFN-γ therefore seems to link to (and allow for) mitochondrial ROS production, HIF-1α stabilization and IL-1β expression as it does for ATP production and JAK-STAT-1 pathway activation with iNOS expression and NO production.
5.8. Conversion of Pyruvate to Lactate Is a Defining Step for Aerobic Glycolysis and IFN-γ M1 Activation
As the glycolytic cascade diverts at the level of pyruvate, we next performed experiments to test the significance of downstream pyruvate metabolism for IFN-γ activated macrophages. We first used the pyruvate analogue oxamate, a competitive inhibitor of lactate dehydrogenase A (LDHa), which rapidly blocked the switch to aerobic glycolysis (Fig. S7a and b) and significantly reduced HIF-1α and iNOS expression as well as NO production in IFN-γ-stimulated macrophages (Fig. S7c and d). However, oxamate has also been demonstrated to inhibit the carboxylation and entry of pyruvate into the mitochondria. We therefore utilized the more selective inhibitor FX-11 (O'Neill et al., 2016), which is a NADH competitive inhibitor of LDHa that does not affect the activities of LDHb and glyceraldehyde-3-phosphate dehydrogenase (GADPH) even at higher concentrations (Le et al., 2010). Using this inhibitor, we observed a reduction in ECAR (Fig. S7e and f) and in the production of several key M1 macrophage-associated cytokines including IL-1α, IL-1β, G-CSF, and IL-27, and chemokines including C5/C5a, CCL-12, CXCL-1, and CXCL-13 (Fig. S7g) as well as iNOS (Fig. S7h and i) and NO production (Fig. S7j and k). Furthermore, FX11 pretreatment reduced STAT-1 phosphorylation in IFN-γ stimulated BMDMs (Fig. S7l). We also examined the effect of Uk-5099, an inhibitor of mitochondrial pyruvate carrier activity, which had no effect on the IFN-γ-stimulated pathways (Fig. S7m). Collectively, these findings indicate that the last step in the aerobic glycolysis cascade is defining not only for this metabolic pathway but also for the signature traits of M1 macrophages.
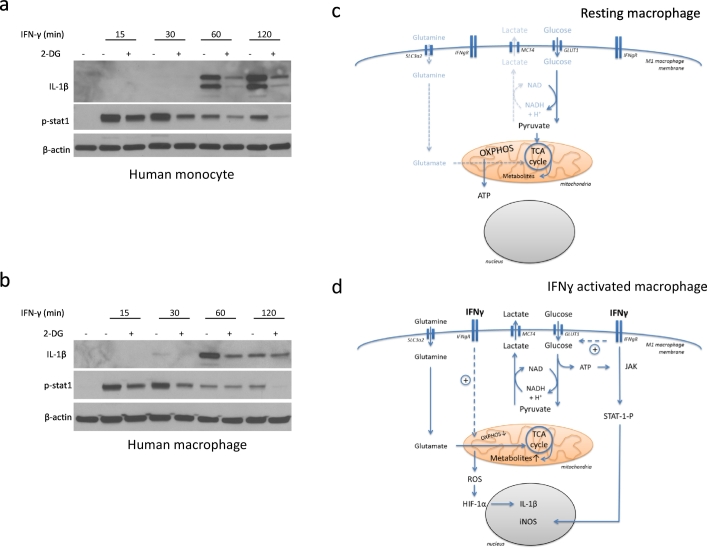
5.9. 2-DG Reduces Pro-Inflammatory Cytokines/Chemokines Production in Human Monocytes/Macrophages
Finally, we addressed the question how translational the outlined observations are. To further test this, we isolated peripheral blood monocytes and cultured macrophages from patients with atherosclerotic cardiovascular disease. As shown in Fig. 7a, 2-DG reduced IL-1β expression and STAT-1 phosphorylation in IFN-γ activated human peripheral blood monocytes. Moreover, 2-DG had the same effects on IFN-γ stimulated human macrophages (Fig. 7b). Thus, these data support the translation of the findings made in murine cell lines to human monocytes and macrophages. This may be of significance for disease processes influenced by IFN-γ and macrophages such atherosclerosis and might be of particular relevance for patients with diabetes, who are known to be prone to much more diffuse and complicated atherosclerosis.
Fig. 7.
2-DG reduces pro-inflammatory cytokines/chemokines production in human monocytes/macrophages.
(a) Immunoblot analysis of specified proteins in human monocytes after stimulation with IFN-γ for indicated time intervals, ±1 hour pre-treatment with 2-DG (10 mM).
(b) Immunoblot analysis of specified proteins in human macrophages after stimulation with IFN-γ for indicated time intervals, ±1 hour pre-treatment with 2-DG (10 mM).
(c) Conceptual outline. Under resting conditions, oxidative phosphorylation (OXPHOS) dominates macrophage cell metabolism even though a baseline level of aerobic glycolysis is present.
(d) Conceptual outline. Under conditions of M1 macrophage activation with IFN-γ, aerobic glycolysis is markedly increased and oxidative phosphorylation (OXPHOS) is markedly decreased. ATP generated from aerobic glycolysis feeds into the JAK-STAT1 signaling pathway and iNOS expression whereas the mitochondrial metabolic derangement feeds into reactive oxygen species (ROS) generation, HIF-1α stabilization, and IL-1β expression.
6. Discussion
Metabolic reprogramming of macrophages has been known for decades (Hard, 1970; Fukuzumi et al., 1996), but its implications for inflammatory cell function have emerged just in recent years (Tannahill et al., 2013; Palsson-McDermott et al., 2015; Yang et al., 2014; Gubser et al., 2013; Chang et al., 2013; Everts et al., 2014; Mills et al., 2016). In order to define the role of aerobic glycolysis in M1 macrophages, studies have relied on 2-DG as a competitive inhibitor to glucose in the first reaction step of glycolysis, which, may, however, not be as specific (Ralser et al., 2008). Indeed, here we found that 2-DG decreased both aerobic glycolysis and OXPHOS and had a profound dose-dependent effect on cellular ATP levels and cell viability. These findings challenge conclusions regarding the role of aerobic glycolysis reached by the use of 2-DG. As an alternative we used galactose, which is metabolized to glucose-6-phosphate at a very slow rate, thereby profoundly reducing glycolytic throughput. In agreement, we found that galactose reduced ECAR with little acute effect on OCR, thereby allowing for a much more specific evaluation of the role of aerobic glycolysis in M1 macrophages. Even under these conditions, IFN-γ differentiated macrophages into the M1 type phenotype based on the expression of signature surface markers and cytokines such as TNF-α and IL-6. However, HIF-1α and IL-1 β levels were significantly reduced by galactose, as was iNOS expression and NO production. Compared with 2-DG, the onset of these effects was more protracted and mirrored changes in cellular ATP levels, which declined much faster with 2-DG than with galactose. Taken together, these findings indicate that aerobic glycolysis is of particular significance for two gene transcription pathways in IFN-γ-stimulated macrophages: HIF-1α and STAT-1 (Tannahill et al., 2013; Mills et al., 2016).
Prior studies in dendritic cells indicated that the switch to aerobic glycolysis is vital to compensate for NO's inhibitory effect on mitochondrial ATP production (Everts et al., 2012 and Everts et al., 2014). Furthermore, in astrocytes NO increases glycolytic throughput via AMP-activated kinase (Almeida et al., 2004). However, no study has yet evaluated if glycolysis influences NO production and how NO production influences glycolysis in macrophages. This is of relevance though as NO is part of the cytotoxic activity of macrophages against invading microorganisms (MacMicking et al., 1997). Herein we found that NO production of activated M1 macrophages is dependent on aerobic glycolysis and increases as a function of extracellular glucose concentration. Furthermore, we found that NO production in M1 macrophages mirrored and followed an increase in iNOS expression (MacMicking et al., 1997). In keeping with the iNOS expression profile, there was no effect of iNOS inhibition with L-NAME on ECAR and OCR early after stimulation. Similarly, later on, contrary to what would be expected and what was shown for dendritic cells, iNOS inhibition with L-NAME did not increase OCR or changed the OCR/ECAR ratio. Thus, NO is not involved in the glycolytic reprogramming of M1 macrophages and is not as potent of a stimulus for aerobic glycolysis in these cells than it is in others (Albina and Mastrofrancesco, 1993; Mateo et al., 1995; Everts et al., 2014).
The JAK/STAT-1 pathway activated by IFN-γ entails the dimerization of the IFN receptor leading to the activation of JAK-1/JAK-2, which phosphorylates and thereby activates the transcription factor STAT-1. P-STAT-1 translocates into the nucleus and binds to IFN-response elements in the promoter region of different genes encoding for various cytokines, chemokines, and iNOS (Shuai and Liu, 2003). In agreement with previous studies, IFN-γ increased STAT-1 phosphorylation in M1 macrophages and this response was inhibited by 2-DG. Similarly, galactose had a profound inhibitory effect on STAT-1 phosphorylation, supporting the significance of aerobic glycolysis for the JAK/STAT-1 pathway. Glucose itself was unable to stimulate the activity of the JAK/STAT-1 pathway in the absence of IFN-γ. Accordingly, the activity of JAK needs to be primed and is upregulated (but not induced) by glucose and glycolytic throughput. While numerous molecular links are conceivable and were tested in an extensive series of experiments (Table S1), ATP was identified as the most likely pivotal link between glycolysis and JAK/STAT-1 pathway activity. Providing intracellular ATP overcame the inhibitory effect of 2-DG on STAT-1 phosphorylation. Furthermore, the inhibitory response of an ATP-competitive JAK inhibitor could be overcome by increasing the extracellular glucose concentration, which translates into increased glycolytic throughput under conditions of IFN-γ stimulation. Indeed, as an ATP-dependent kinase (Kesarwani et al., 2015), the activity of JAK increases as a function of intracellular ATP concentration until a plateau is reached. While aerobic glycolytic throughput can upregulate JAK activity by providing ATP, the initial activation of JAK is still required and relies on IFN-γ-induced receptor dimerization. Thus, aerobic glycolysis has a unique amplifying role for the signature signaling pathway triggered by IFN-γ in M1 macrophages: JAK/STAT-1, which translates into key M1 traits such as iNOS expression.
Previous studies identified HIF-1α as an important transcription factor for the classical phenotype of LPS-stimulated M1 macrophages. This was related to alterations in the TCA cycle with accumulation of succinate that then leads to the stabilization of HIF-1α and transcriptional activity (Tannahill et al., 2013). More recent studies indicate that it might not be succinate by itself that activates HIF-1α but rather succinate oxidation and mitochondrial ROS production (Lampropoulou et al., 2016; Mills et al., 2016). Indeed, one very unique observation of the current studies is the lack of differences in the TCA cycle metabolites succinate and fumarate between cells stimulated with glucose and galactose. The growth and stimulation of M1 macrophages in galactose-containing medium therefore, while inhibiting aerobic glycolysis, did seemingly not affect a TCA cycle point considered very critical for M1 macrophage activation and left OXPHOS unimpaired, allowing for ATP rather than ROS production. Future studies will have to define further the relationship between aerobic glycolysis, TCA cycle changes, OXPHOS and mitochondrial ROS production. The current results would suggest that M1 macrophage activation in response to IFN-γ is accomplished by different signaling pathways that include JAK/STAT-1 in a classical manner via the IFN-γ receptor as well as HIF-1α via so-called “repurposing” of the mitochondria.
As outlined, M1 macrophages just like other inflammatory cells and cancer cells are said to rely on aerobic glycolysis for their functional phenotype. Whether the switch to aerobic glycolysis is a terminal metabolic differentiation step in these cells has remained unknown. Herein we were able to show that a significant fraction of M1 macrophages is able to survive in glucose-containing conditions without a change of cell culture medium for 6 days. They remain inert to the effects of oligomycin early on, but become sensitive later on, indicating that these cells utilize aerobic glycolysis initially but revert to OXPHOS with glucose depletion. In marked distinction, M1 macrophages grown in galactose-containing medium survive longer but are sensitive to oligomycin from the beginning in keeping with the view that OXPHOS is their main mode of energy metabolism. Thus, there is plasticity in the metabolism of M1 macrophages, and these cells are able to conduct a metabolic switch not just once but multiple times and in either direction.
Previous studies in cancer cells indicated that the inhibition of the last step of aerobic glycolysis has a functional effect that is as profound as inhibition of the first, hexokinase-dependent step (Le et al., 2010). Herein we made similar observations using three different inhibitory approaches: oxamate, Uk-5099, and FX-11. The results with these three drugs were in agreement: pyruvate metabolism is of significance and it is the LDHa-catalyzed conversion step that defines aerobic glycolysis and its impact on M1 macrophages. These findings, in agreement with the classical Warburg effect, differ from studies on other immune cells such as dendritic cells, whose activation is not as sensitive to LDHa inhibition by oxamate (Everts et al., 2014). Accordingly, the functional significance of metabolic pathways differs among different inflammatory cell types. For the activation of M1 macrophages, the aerobic glycolytic cascade in its entity is seemingly of importance.
In summary (Fig. 7c and d), stimulation of resting macrophages with type II but not type I IFNs leads to a rapid increase in aerobic glycolysis and a protracted decrease in OXPHOS. This metabolic switch maintains M1 macrophage viability, ATP production to maximize JAK-STAT-1 pathway activity, and repurposing of the mitochondria for ROS production, stabilizing HIF-1a and its transcriptional activity. IFN-γ-activated M1 macrophages retain metabolic plasticity and can revert back to OXPHOS, and inhibition of aerobic glycolysis (at its first or last step) blocks M1 phenotype signature traits in murine and human monocyte/macrophage cell lines. These immuno-metabolic effects of IFN-γ independent of LPS may have important translational implications as they corroborate that IFN-γ can restore, at least partially, the immunometabolic paralysis to LPS that can evolve in monocytes from patients with sepsis and correlate with worse clinical outcomes (Cheng et al., Nature Immunology 2016; 17:406-413) In other clinical scenarios such as atherosclerosis, however, the impact may not be as beneficial (Chinetti-Gbaguidi et al., 2015; Shirai et al., 2016). Based on the current findings, high extracellular glucose concentrations, as seen in patients with (poorly controlled) diabetes mellitus, would be expected to translate into higher glycolytic and thus greater pro-inflammatory and pro-atherosclerotic activity of M1 macrophages, which may relate to the more aggressive nature of atherosclerosis in these patients. These redundancies and diversities underscore the complexity of the field and challenges its suitability as a target for therapeutic intervention. Indeed, the very recently published Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS) showed that IL-1β-directed therapy can reduce the risk of fatal and non-fatal myocardial infarction and stroke but at an increased risk of fatal sepsis (Ridker et al., 2017). Accordingly, any therapeutic approaches directed specifically at immunometabolism (and not only at one of its products as in the CANTOS trial) will require very careful evaluation.
Conflicts of Interest
The authors declared no conflict of interest.
Authorship Contributions
Feilong Wang, MD, designed and conducted experiments for this study, analyzed and interpreted the data, and drafted and edited the manuscript.
Song Zhang, PhD, designed and conducted experiments for this study, analyzed and interpreted the data, and provided critical input and review for the manuscript.
Ryounghoon Jeon, PhD, designed and conducted experiments for this study, analyzed and interpreted the data, and provided critical input and review for the manuscript.
Ivan Vuckovic, PhD, designed and conducted experiments for this study, analyzed and interpreted the data, and provided critical input and review for the manuscript.
Xintong Jiang, MD, conducted experiments for this study, analyzed and interpreted the data, and provided critical input and review for the manuscript.
Amir Lerman, MD, interpreted the data and provided critical input and review for the manuscript.
Clifford D. Folmes, PhD, conceptualized parts of the study, interpreted the data and provided critical input and review for the manuscript.
Petras, D. Dzeja, PhD, secured funding, conceptualized parts of the study, interpreted the data and provided critical input and review for the manuscript.
Joerg Herrmann, MD, secured funding, conceptualized the study and its experiments, interpreted the data and drafted and edited the manuscript.
Acknowledgments
This work was supported by the National Institute of Health/National Heart Lung Blood Institute (HL116952-03 and HL 85744-09, HL 121079-02), and U24DK100469 (Mayo Clinic Metabolomics Resource Core), a fellowship grant of the Department of Cardiovascular Diseases, Mayo Clinic, Rochester, MN, USA, and an endowment by the Sachs family.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.02.009.
Appendix A. Supplementary data
Supplementary material
References
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Albina J.E., Mastrofrancesco B.A.L.D.U.I.N.O. Modulation of glucose metabolism in macrophages by products of nitric oxide synthase. Am. J. Phys. Cell Phys. 1993;264:1594–1599. doi: 10.1152/ajpcell.1993.264.6.C1594. [DOI] [PubMed] [Google Scholar]
- Almeida A., Moncada S., Bolaños J.P. Nitric oxide switches on glycolysis through the AMP protein kinase and 6-phosphofructo-2-kinase pathway. Nat. Cell Biol. 2004;6:45–51. doi: 10.1038/ncb1080. [DOI] [PubMed] [Google Scholar]
- Bustamante E., Morris H.P., Pedersen P.L. Hexokinase: the direct link between mitochondrial and glycolytic reactions in rapidly growing cancer cells. Adv. Exp. Med. Biol. 1977;92:363–380. doi: 10.1007/978-1-4615-8852-8_15. [DOI] [PubMed] [Google Scholar]
- Chang C.H., Curtis J.D., Maggi L.B., Faubert B., Villarino A.V., O'Sullivan D., Huang S.C.C., van der Windt G.J., Blagih J., Qiu J. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti-Gbaguidi G., Colin S., Staels B. Macrophage subsets in atherosclerosis. Nat. Rev. Cardiol. 2015;12:10–17. doi: 10.1038/nrcardio.2014.173. [DOI] [PubMed] [Google Scholar]
- Collart M.A., Belin D., Vassalli J.D., De Kossodo S., Vassalli P. Gamma interferon enhances macrophage transcription of the tumor necrosis factor/cachectin, interleukin 1, and urokinase genes, which are controlled by short-lived repressors. J. Exp. Med. 1986;164:2113–2118. doi: 10.1084/jem.164.6.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzeja P.P., Terzic A., Wieringa B. Phosphotransfer dynamics in skeletal muscle from creatine kinase gene-deleted mice. Mol. Cell. Biochem. 2004;256:13–27. doi: 10.1023/b:mcbi.0000009856.23646.38. [DOI] [PubMed] [Google Scholar]
- Everts B. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B., Amiel E., Huang S.C.C., Smith A.M., Chang C.H., Lam W.Y., Redmann V., Freitas T.C., Blagih J., van der Windt G.J. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKK ɛ supports the anabolic demands of dendritic cell activation. Nat. Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzumi M., Shinomiya H., Shimizu Y., Ohishi K., Utsumi S. Endotoxin-induced enhancement of glucose influx into murine peritoneal macrophages via GLUT1. Infect. Immun. 1996;64:108–112. doi: 10.1128/iai.64.1.108-112.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkina E., Ley K. Immune and inflammatory mechanisms of atherosclerosis. Annu. Rev. Immunol. 2009;27:165–197. doi: 10.1146/annurev.immunol.021908.132620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván-Peña S., O'Neill L.A. Metabolic reprograming in macrophage polarization. M1/M2 macrophages: the arginine fork in the road to health and disease. Front. Immunol. 2015;5:275. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubser P.M., Bantug G.R., Razik L., Fischer M., Dimeloe S., Hoenger G., Durovic B., Jauch A., Hess C. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 2013;14:1064–1072. doi: 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- Hammarén H.M., Ungureanu D., Grisouard J., Skoda R.C., Hubbard S.R., Silvennoinen O. ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. 2015;112:4642–4647. doi: 10.1073/pnas.1423201112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hard G.C. Some biochemical aspects of the immune macrophage. Br. J. Exp. Pathol. 1970;51:97. [PMC free article] [PubMed] [Google Scholar]
- Haschemi A., Kosma P., Gille L., Evans C.R., Burant C.F., Starkl P., Knapp B., Haas R., Schmid J.A., Jandl C. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012;15:813–826. doi: 10.1016/j.cmet.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Jha A.K., Huang S.C.C., Sergushichev A., Lampropoulou V., Ivanova Y., Loginicheva E., Chmielewski K., Stewart K.M., Ashall J., Everts B. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Kesarwani M., Huber E., Kincaid Z., Evelyn C.R., Biesiada J., Rance M., Thapa M.B., Shah N.P., Meller J., Zheng Y. Targeting substrate-site in Jak2 kinase prevents emergence of genetic resistance. Sci. Rep. 2015;5 doi: 10.1038/srep14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtoglu M., Gao N., Shang J., Maher J.C., Lehrman M.A., Wangpaichitr M., Savaraj N., Lane A.N., Lampidis T.J. Under normoxia, 2-deoxy-d-glucose elicits cell death in select tumor types not by inhibition of glycolysis but by interfering with N-linked glycosylation. Mol. Cancer Ther. 2007;6:3049–3058. doi: 10.1158/1535-7163.MCT-07-0310. [DOI] [PubMed] [Google Scholar]
- Lampropoulou V., Sergushichev A., Bambouskova M., Nair S., Vincent E.E., Loginicheva E., Cervantes-Barragan L., Ma X., Huang S.C.C., Griss T. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 2016;24:158–166. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A., Cooper C.R., Gouw A.M., Dinavahi R., Maitra A., Deck L.M., Royer R.E., Vander Jagt D.L., Semenza G.L., Dang C.V. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Lu Y., Martinez J., Bi Y., Lian G., Wang T., Milasta S., Wang J., Yang M., Liu G. Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1α-dependent. Proc. Natl. Acad. Sci. 2016;113:1564–1569. doi: 10.1073/pnas.1518000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J., Xie Q.W., Nathan C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Mateo R.B., Reichner J.S., Mastrofrancesco B.A.L.D.U.I.N.O., Kraft-Stolar D.A.N.Y.A., Albina J.E. Impact of nitric oxide on macrophage glucose metabolism and glyceraldehyde-3-phosphate dehydrogenase activity. Am. J. Phys. Cell Phys. 1995;268:669–675. doi: 10.1152/ajpcell.1995.268.3.C669. [DOI] [PubMed] [Google Scholar]
- McLaren J.E., Ramji D.P. Interferon gamma: a master regulator of atherosclerosis. Cytokine Growth Factor Rev. 2009;20:125–135. doi: 10.1016/j.cytogfr.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Mills E.L., Kelly B., Logan A., Costa A.S., Varma M., Bryant C.E., Tourlomousis P., Däbritz J.H.M., Gottlieb E., Latorre I. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167:457–470. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B.B., Rathinam V.A., Martens G.W., Martinot A.J., Kornfeld H., Fitzgerald K.A., Sassetti C.M. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1 [beta] Nat. Immunol. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott E.M., Curtis A.M., Goel G., Lauterbach M.A., Sheedy F.J., Gleeson L.E., van den Bosch M.W., Quinn S.R., Domingo-Fernandez R., Johnston D.G. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H., Martin D.S., Xu R.A., Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- Peng M., Yin N., Chhangawala S., Xu K., Leslie C.S., Li M.O. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354:481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser M., Wamelink M.M., Struys E.A., Joppich C., Krobitsch S., Jakobs C., Lehrach H. A catabolic block does not sufficiently explain how 2-deoxy-d-glucose inhibits cell growth. Proc. Natl. Acad. Sci. 2008;105:17807–17811. doi: 10.1073/pnas.0803090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker P.M., Everett B.M., Thuren T., MacFadyen J.G., Chang W.H., Ballantyne C., Fonseca F., Nicolau J., Koenig W., Anker S.D., Kastelein J.J.P., Cornel J.H., Pais P., Pella D., Genest J., Cifkova R., Lorenzatti A., Forster T., Kobalava Z., Vida-Simiti L., Flather M., Shimokawa H., Ogawa H., Dellborg M., Rossi P.R.F., Troquay R.P.T., Libby P., Glynn R.J., CANTOS Trial Group Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 2017;377:1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Prados J.C., Través P.G., Cuenca J., Rico D., Aragonés J., Martín-Sanz P., Cascante M., Boscá L. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J. Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- Schindler R., Ghezzi P., Dinarello C.A. IL-1 induces IL-1. IV. IFN-gamma suppresses IL-1 but not lipopolysaccharide-induced transcription of IL-1. J. Immunol. 1990;144:2216–2222. [PubMed] [Google Scholar]
- Shirai T., Nazarewicz R.R., Wallis B.B., Yanes R.E., Watanabe R., Hilhorst M., Tian L., Harrison D.G., Giacomini J.C., Assimes T.L., Goronzy J.J., Weyand C.M. The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J. Exp. Med. 2016;213:337–354. doi: 10.1084/jem.20150900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K., Liu B. Regulation of JAK–STAT signalling in the immune system. Nat. Rev. Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats D., Mukundan L., Odegaard J.I., Zhang L., Smith K.L., Morel C.R., Wagner R.A., Greaves D.R., Murray P.J., Chawla A. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walev I., Bhakdi S.C., Hofmann F., Djonder N., Valeva A., Aktories K., Bhakdi S. Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-O. Proc. Natl. Acad. Sci. 2001;98:3185–3190. doi: 10.1073/pnas.051429498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. Metabolism of tumours. Biochem. Z. 1923;142:317–333. [Google Scholar]
- Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.S., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Brodsky I.E., Rahner C., Woo D.K., Erdjument-Bromage H., Tempst P., Walsh M.C., Choi Y., Shadel G.S., Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A.J., Reyes C.N., Liang W., Becker C., Shimada K., Wheeler M.L., Cho H.C., Popescu N.I., Coggeshall K.M., Arditi M. Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell. 2016;166:624–636. doi: 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Sanin D.E., Everts B., Chen Q., Qiu J., Buck M.D., Patterson A., Smith A.M., Chang C.H., Liu Z. Type 1 interferons induce changes in core metabolism that are critical for immune function. Immunity. 2016;44:1325–1336. doi: 10.1016/j.immuni.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Chi F., Guo T., Punj V., Lee W.P., French S.W., Tsukamoto H. NOTCH reprograms mitochondrial metabolism for proinflammatory macrophage activation. J. Clin. Invest. 2015;125:1579. doi: 10.1172/JCI76468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xie M., Yang M., Yu Y., Zhu S., Hou W., Kang R., Lotze M.T., Billiar T.R., Wang H. PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis. Nat. Commun. 2014;5:4436. doi: 10.1038/ncomms5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gonçalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2008;14:14. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Somasundaram R., Nederhof R.F., Dijkstra G., Faber K.N., Peppelenbosch M.P., Fuhler G.M. Impact of human granulocyte and monocyte isolation procedures on functional studies. Clin. Vaccine Immunol. 2012;19:1065–1074. doi: 10.1128/CVI.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material